Abstract
Endeavors towards identifying key molecular markers for early diagnosis and treatment are driving the clinical study of papillary thyroid carcinoma (PTC). Recent studies have indicated that protein phosphatase, Mg2+/Mn2+ dependent, 1D (PPM1D) exerts an oncogenic function by increasing cell proliferation, migration and invasion in various cancer types. In addition, PPM1D has a high frequency of genetic alterations and has been proposed as a tumor driver in thyroid cancer, making PPM1D an attractive potential oncotarget for cancer treatment. The aims of the present study were to investigate the downstream targets of PPM1D and the potential molecular mechanisms of its oncogenic activities, as well as its clinical significance in PTC. As anticipated, PPM1D overexpression was confirmed in PTC clinical specimens. Furthermore, knockdown of PPM1D in thyroid cancer cell lines significantly suppressed the proliferation, migration and invasion but facilitated cell apoptosis. The protein levels of phosphorylated p38 mitogen-activated protein kinase (MAPK), p53 and Bax were increased in PPM1D-knockdown cells, while inhibition of p38 phosphorylation restored cell migration, proliferation and cell apoptosis. In addition, silencing of PPM1D expression induced nuclear translocation of p53 in K-1 and TPC-1 cells. The present results demonstrated that PPM1D regulated p38 MAPK and p53 signaling pathways to promote thyroid cancer progression. Collectively with the clinical results, these data qualified PPM1D as a potential diagnostic biomarker and therapeutic target in human thyroid cancer.
Keywords: papillary thyroid carcinoma, PPM1D/Wip1, p53, p38 MAPK, Bax, metastasis, apoptosis
Introduction
The incidence of thyroid cancer has increased worldwide due to the increased use of diagnostic imaging and surveillance (1), and creates a great burden on the health care system. Papillary thyroid cancer (PTC) is the most common endocrine malignancy and accounts for 80% of cases of differentiated thyroid cancer worldwide (2). Consistent with the majority of malignancies, thyroid carcinomas are usually associated with oncogenes that lead to aberrant cell proliferation, migration and invasion (3). However, a more detailed account of how oncogenes overcome the natural balance of tumor surveillance during the formation of PTC and contribute to tumor progression is required, and revealing these oncogenes and their mechanisms may provide approaches for cancer treatment and further improvement of clinical care.
Protein phosphatase, Mg2+/Mn2+ dependent, 1D (PPM1D), also referred to as wild-type p53 inducible protein 1 serine/threonine phosphatase, is a member of the protein phosphatase 2C family, and is recognized as an oncogene due to its roles in promoting tumorigenesis (4) and negative regulation of the DNA damage response system (5–7). A number of studies have demonstrated that PPM1D is involved in the development of a majority of human cancer types, including hepatocellular carcinoma, breast cancer, ovarian clear-cell carcinoma, bladder cancer and glioblastomas (8–10). Furthermore, PPM1D gene amplification and/or protein overexpression have been identified to contribute to tumorigenesis in in vivo (11) and in vitro studies (12–14). PPM1D protein overexpression was also identified to be significantly associated with poor clinical outcome in neuroblastoma and ovarian clear-cell carcinoma (15). Consecutive investigations have revealed that the oncogenic properties of PPM1D are mediated by inhibition of several tumor suppressor pathways, including p53, p38 mitogen-activated protein kinase (p38 MAPK), ataxia telangiectasia mutated and checkpoint kinase 1, therefore contributing to tumorigenesis, progression, invasion, distant metastasis and evasion of apoptosis (10,16).
Cellular homeostasis highly relies on fine-tuning signaling pathways that control the pace of cell proliferation and apoptosis, thereby preventing oncogenic cellular transformation through aberrant stress (17,18). The tumor suppressor p53 has a vital role in these pathways by transcriptionally upregulating target proteins, including WAF1, Bax and MDM2, which act to initiate cell cycle arrest or cell death under stresses. PPM1D was first identified as a target gene of p53 (19), but subsequent studies revealed that p53 may also be inactivated by PPM1D-induced dephosphorylation while cells switch from stress status to normal homeostasis (10,20). Previous studies indicated that the enhanced p53 pathway in PPM1D-knockout mice significantly impaired tumorigenesis in several tumor models (20,21), which draws attention to PPM1D as a potential anticancer target.
Furthermore, PPM1D also indirectly inactivates p53 through p38 MAPK (16). p38 MAPK is a component of the MAPK pathway, which is another protective signaling pathway in response to cellular stress (22). It was reported that PPM1D directly binds and inactivates p38 MAPK via dephosphorylation at Thr180 (23). In line with the aforementioned, p38 inactivation paralleled with p53 deactivation in vivo was also identified in a number of studies (24–26). However, the current knowledge on PPM1D is mostly based on studies on breast cancer or the subtypes of breast cancer, and whether PPM1D has any oncogenic properties via deactivation of p38 and p53 signaling pathways in PTC has so far remained elusive.
In the present study, PPM1D expression was examined in human PTC tissues as well as in paired adjacent non-cancerous tissues and a significant association between PPM1D overexpression and metastasis was revealed. The potential oncogenic properties of PPM1D were also confirmed in thyroid cell lines. A further mechanistic study indicated that the oncogenic activities of PPM1D in thyroid cancer cells are mediated by negative regulation of the p38 MAPK and p53 signaling pathways. These results contribute to the understanding of the effect of PPM1D overexpression in promoting PTC tumor progression, indicating that it may serve as a potential target for clinical treatment.
Materials and methods
Tissue specimens
A total of 89 thyroid cancer samples were obtained from patients who underwent surgery for thyroid cancer between August 2012 and February 2015 at Shanghai Cancer Center of Fudan University (Shanghai, China). Tissue specimens were frozen in liquid nitrogen immediately after surgical resection and stored at −80°C. All tissues were pathologically confirmed as PTC and final histological classification was obtained from paraffin-embedded sections. The study was performed in accordance with the Declaration of Helsinki and approved by the Institutional Research Ethics Committee of Shanghai Cancer Center, Fudan University (Shanghai, China). Written informed consent was obtained from all participants after reviewing the content and purpose of the study.
Cell culture and treatments
The human PTC original cell lines TPC-1 and K-1 were obtained from Dr Schweppe from the University of Colorado Cancer Center. STR profiling was performed to confirm cell authentication. All cells were grown in RPMI-1640 media (Sigma-Aldrich; Merck KGaA) supplemented with 10% heat-inactivated fetal bovine serum (FBS; Invitrogen; Thermo Fisher Scientific, Inc.), 100 IU/ml penicillin and 10 µg/ml streptomycin. Cell culture was performed at 37°C in a 90% humidified atmosphere with 5% CO2. A MAPK inhibitor (SB203580; Sigma-Aldrich; Merck KGaA) was dissolved in dimethyl sulfoxide (DMSO), then added into cell culture medium at a concentration of 100 nM for 24 h in order to inhibit p38 MAPK activity.
Small interfering (si)RNA and transfection
The specific siRNA targeting PPM1D and the scrambled siRNA used as a negative control were designed and purchased from GenePharma Co., Ltd. The sequences of the siRNA targeting PPM1D (siPPM1D) and scrambled siRNA were as follows: siPPM1D-1: 5′-CCGCACTCGTGCTTGCTTGAA-3′; siPPM1D-2: 5′-GTCACGTAACATGTCACAT-3′; negative control (NC): 5′-CCACCUCUGAUCGAUUUAUdTdT-3′. Cells were collected for further analysis of protein depletion after 48 h of transfection. All siRNAs were transfected into thyroid cancer cells using Lipofectamine™ 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) following the manufacturer's protocol.
Western blot analysis
Cell lysates were obtained from 1×106 cultured cells with a mixture of ProteoJET Mammalian Cell Lysis Reagent (Fermentas; Thermo Fisher Scientific, Inc.), phenylmethanesulfonyl fluoride and PhosSTOP (both from Roche Applied Science). Protein estimation was performed according to the Bradford method (Bio-Rad Laboratories, Inc.) with bovine serum albumin (BSA; Sigma-Aldrich; Merck KGaA) as a standard. Total protein (10 µg per lane) was resolved on 10–15% gradient pre-cast gels (Sigma-Aldrich; Merck KGaA) and transferred to a polyvinylidene difluoride membrane (EMD Millipore). After blocking in 5% non-fat milk for 2 h at room temperature, the membrane was probed with antibodies against human PPM1D (1:1,000 dilution; product code ab31270; Abcam), p38 (1:1,000 dilution; product no. 9212), phosphorylated p-p38 (1:1,000 dilution; product no. 4511), p53 (1:1,000 dilution; product no. 2527), Bax (1:1,000 dilution; product no. 5023), Histone H3 (1:1,000 dilution; product no. 4499; all from Cell Signaling Technology, Inc.) or GAPDH (1:5,000 dilution; product code ab9485; Abcam) overnight at 4°C. Subsequently, the membrane was incubated with HRP-conjugated goat anti-rabbit or HRP-conjugated goat anti-mouse IgG (1:10,000 dilution for both; product code ab205718 and product code ab19195, respectively; Abcam) for 2 h at room temperature. The indicated antibodies were detected with the SuperSignal West Pico ECL Chemiluminescent kit (Thermo Fisher Scientific, Inc.) and protein bands were quantified using ImageJ software (version 1.47v; National Institutes of Health, Bethesda).
Isolation of nuclei
Approximately 1×107 cells were collected and resuspended in hypotonic buffer (10 mM Tris, pH 7.9, 10 mM KCl, 1.5 mM MgCl2 and 0.05 mM DTT). Cell suspensions were homogenized with a pre-chilled Dounce homogenizer with 20 strokes and centrifuged at 100 × g for 15 min to retain the supernatant. 10X S100 buffer (0.3 M Tris, pH 7.9, 1.4 M KCl and 0.03 M MgCl2) was added to the supernatant, followed by centrifugation at 100,000 × g for 1 h and the supernatant was obtained for analysis of cytoplasmic proteins. The nuclear pellet was continuously lysed using high-salt extraction buffer (20 mM Tris, pH 7.9, 25% glycerol, 1.2 M KCl, 1.5 mM MgCl2 and 0.2 mM EDTA, pH 8.0) with two gentle stokes in the Dounce homogenizer. After centrifugation at 10,000 × g for 30 min, the supernatants containing nuclear extract were collected for analysis. All procedures were performed at 4°C.
Cell proliferation assay
Cell growth was determined using a Cell Counting Kit-8 (CCK-8) assay (Sigma-Aldrich; Merck KGaA) according to the manufacturer's protocol. In brief, cells were seeded in black wall/clear-bottom 96-well plates at 6×103 cells in 100 µl/well and allowed to attach overnight. At the indicated time-points, an aliquot of 10 µl CCK-8 solution was added to each well and the plate was incubated for 4 h at 37°C. The absorbance reading was performed at 450 nm using a spectrophotometer (Thermo Scientific™ NanoDrop™ 8000; Thermo Fisher Scientific, Inc.). Five replicates were used for each experimental condition. Cell-free medium was used as the blank group and cells treated with the solvent were used as a vehicle control. The viability of the cells was calculated as follows: Cell viability (%) = (each condition-blank group)/(vehicle control-blank group) ×100%.
Plate colony formation assay
Cells were trypsinized using 0.05% trypsin/EDTA (Thermo Fisher Scientific, Inc.) and 2×103 cells were seeded into 6-well plates and incubated at 37°C for 10 days. Colonized cells were washed with PBS followed by fixation with 10% methanol at room temperate (RT) for 5 min and staining with 5% Giemsa (Sigma-Aldrich; Merck KGaA) at RT for 10 min. The number of colonies >10 cells was counted under Leica DM4000B microscope (Leica Microsystems) using magnification of ×1.25, images were captured and scoring was performed.
Wound healing assay
Wound healing assays were performed to evaluate the migration of the transfected TPC-1 and K-1 cells. Cells (2×103) were seeded into 6-well plates. When cultured cells reached 90% confluence in the 6-well plate, one scratch was generated with a 200-µl pipette tip in each well to create a wound, followed by two washes with PBS. Migration of cells into the scraped area was recorded at 0 and 48 h after the scratches were made under a Leica DM4000B microscope (Leica Microsystems) using magnification of ×10.
Transwell invasion assays
Cell invasion was assessed using a modified Transwell chamber system (BD Biosciences) according to the manufacturer's protocols. In brief, cells were seeded onto Matrigel®-coated membrane inserts with a pore size of 8 µm. Medium containing 10% FBS, which served as the chemoattractant, was placed in the lower chamber. After 48 h of incubation, the cells were fixed with 75% methanol for 10 min at RT and stained with 0.1% crystal violet (Sigma-Aldrich; Merck KGaA) at RT for 10 min. The invaded cells on the lower surface of the filter that had penetrated through the Matrigel®-coated membrane were counted under an inverted microscope using magnification of ×10.
Apoptosis assay
Apoptotic cells were quantified by flow cytometry using an Annexin V-FITC/propidium iodide (PI) double-staining assay kit (BioVision, Inc.) according to the manufacturer's protocol. In brief, the cells were transfected with indicated siRNAs for 24 h, and were then harvested and washed 2 times with ice-cold PBS, re-suspended in 100 µl binding buffer and stained in the dark with 50 µl Annexin V-FITC and 50 µl propidium iodide at room temperature for 15 min. At least 10,000 events were recorded for each sample and the percentages of cells (viable, apoptotic and necrotic) were quantified by flow cytometry (Cytomics™ FC500 cytometer; Beckman Coulter, Inc.).
Immunofluorescence
Cells were washed twice with ice-cold PBS and fixed in 4% paraformaldehyde in PBS for 20 min at 4°C. After washing 3 times with PBS, cells were permeabilized with 0.1% Triton X-100 for 5 min at 4°C and incubated with 1% BSA in PBS for 30 min, followed by incubation with primary antibody to p53 (1:1,000 dilution; product no. 2527, Cell Signaling Technology, Inc.) at 4°C overnight. The cells were then washed in PBS three times, followed by incubation with the secondary antibodies, FITC-conjugated goat anti-rabbit antibody (1:500 dilution; cat. no. 65-6111 Invitrogen; Thermo Fisher Scientific, Inc.) for 1 h at room temperature in the dark. Cells were washed with PBS and nuclei were stained with 0.5 µg/ml DAPI in PBS containing Tween-20 for 5 min. Samples were mounted with immunofluorescence mounting medium (Dako Cytomation; Agilent Technologies, Inc.) and images were captured under a fluorescence microscope using magnification of ×20.
Immunohistochemical staining
Formalin-fixed and paraffin-embedded tissue sections were deparaffinized in xylene and hydrated through descending concentrations of ethanol prior to being placed in 3% hydrogen peroxide for 10 min at room temperature to inhibit endogenous peroxidase activity. The slides were incubated with blocking solution (10% BSA in 1X phosphate-buffered saline) for 1 h at room temperature, followed by incubation with primary antibody to PPM1D (5 µg/ml; cat. no. PA5-72839, Invitrogen; Thermo Fisher Scientific, Inc.) at 4°C overnight. A horseradish peroxidase-conjugated mouse secondary antibody (1:2,000 dilution; cat no. 65-6120, Invitrogen; Thermo Fisher Scientific, Inc.) was added, followed by incubation for 60 min at room temperature, followed by development with 3,3′-diaminobenzidine (DAB Substrate Chromogen System; Dako; Agilent Technologies, Inc.). Slides were fixed and images were captured by using the Olympus IX71 inverted microscope using the DP2-BSW Olympus image acquisition software system (Olympus, Corp.). The staining results were determined on the basis of the percentage of positive staining of tumor cell nuclei as follows: 0% (no staining); 1 (≤10%); 2 (10–50%) and 3 (>50%). The staining intensity was scored as follows: - (negative); + (moderately positive); and ++ (strongly positive), as previously reported (27). Two experienced pathologists who were blinded to the clinicopathological data of the patients confirmed the results.
Statistical analysis
Values are expressed as the mean ± standard error of the mean (SEM). Pearson's χ2 test was performed to compare differences in clinicopathological parameters across groups stratified by PPM1D protein expression. Univariate and multivariate logistic regression analyses (Cox proportional hazards model) were applied to assess the risk of lymph node metastasis and tumor size of ≥1 cm, and the odds ratio (OR) and 95% CI were reported. A dot plot displaying the distributions of the intensity of nuclear p53 across experimental groups was generated. Means between two groups were compared using Student's t-test. For a comparison of more than two group means, one-way analysis of variance (ANOVA) was applied followed by Tukey's post hoc test. Statistical analysis was performed using GraphPad Prism 5.1 (GraphPad Software, Inc.). P<0.05 was considered to indicate a statistically significant difference.
Results
Correlation of clinicopathological characteristics and expression of PPM1D in PTC tissue specimens
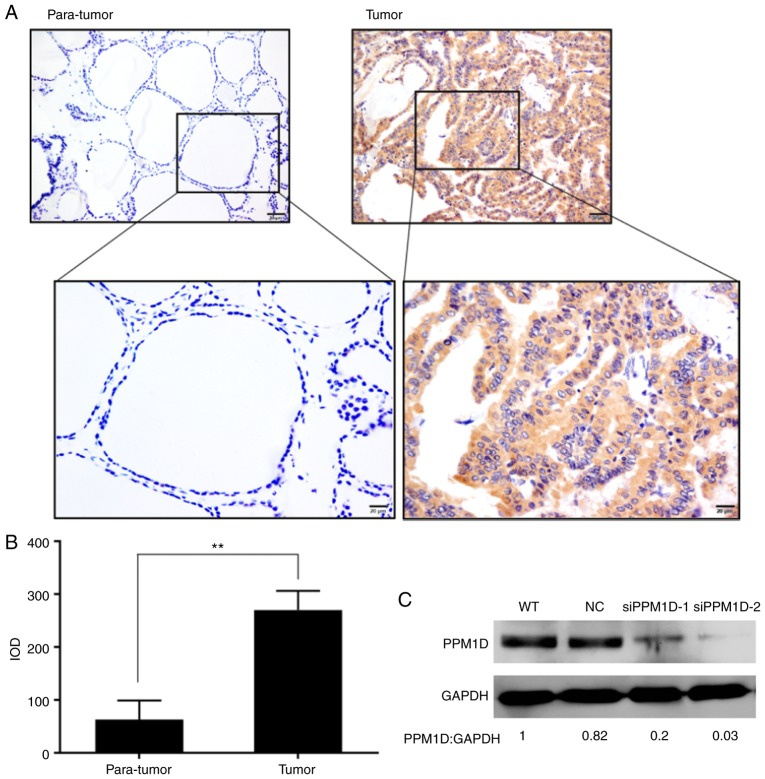
To understand the clinicopathologic significance of PPM1D expression in PTC, tissues from a total of 89 patients with PTC who had undergone tumor resection were analyzed by immunohistochemical staining. It was revealed that PPM1D expression was located within the cytoplasm of the PTC cells, whereas only a small number of scattered cells with positive staining were observed in adjacent non-tumor tissues (Fig. 1A). In addition, the intensity of PPM1D staining in cancer cells was significantly higher than that in non-tumor cells (Fig. 1B). The patients with PTC were classified into three groups including negative (−), moderately positive (+), and strongly positive (++) PPM1D expression; the associations between PPM1D expression and clinicopathological characteristics were assessed (Table I). Pearson's χ2-test indicated that PPM1D expression was significantly associated with tumor size (P=0.016) and lymph node metastasis (P=0.039), whereas no significant influence of PPM1D expression on other clinicopathological features, including age, sex and TNM stage, was observed (Table I). Next, in the univariate analysis, clinical features including tumor size >1 cm, TNM stage III–IV and PPM1D protein expression (+) were identified as risk factors of lymph node metastasis in PTC after adjustment for age, sex and multifocal lesions (Table IIA). A subsequent multivariate analysis revealed that a high level of PPM1D protein expression (++) was a significant risk factor for large tumor size (Table IIB). Based on the aberrantly high expression of PPM1D in PTC cells and its positive correlation with tumor size and lymph node metastasis, further exploration of the role of PPM1D in tumor formation, migration and invasion was performed.
Figure 1.
PPM1D protein expression is increased in thyroid cells. (A) The protein expression of PPM1D in PTC (tumor) and adjacent non-tumor tissues (para-tumor) tissues was examined by immunohistochemistry. Yellow-brown represents positive staining for PPM1D, localized in the cytosol (scale bars, 50 µm in the upper images and 20 µm in lower images). (B) PPM1D expression was quantified by determining the IOD scores. Values are expressed as the mean ± SEM from three independent experiments. P-values were determined using the independent-samples t-test, **P<0.01. (C) Cells transfected with the indicated siRNAs were harvested and subjected to western blot analysis. GAPDH served as a loading control. The relative quantification of protein bands as the ratio to their loading control were presented at the bottom of the western blotting. PPM1D, protein phosphatase, Mg2+/Mn2+ dependent, 1D; PTC, papillary thyroid carcinoma; IOD, integrated optical density; siRNA, small interfering RNA.
Table I.
Clinicopathological parameters by PPM1D protein expression in patients with PTC.
| PPM1D protein expression | ||||
|---|---|---|---|---|
| Clinicopathological parameters | − | + | ++ | P-value |
| Age (years) | 0.256 | |||
| <45 | 20 | 21 | 5 | |
| ≥45 | 13 | 27 | 3 | |
| Sex | 0.921 | |||
| Male | 7 | 12 | 2 | |
| Female | 26 | 36 | 6 | |
| Lymph node metastasis | 0.039a | |||
| Yes | 14 | 33 | 6 | |
| No | 19 | 15 | 2 | |
| Tumor size (cm) | 0.016a | |||
| ≤1 | 14 | 30 | 1 | |
| >1 | 19 | 18 | 7 | |
| Multifocal lesions | 0.738 | |||
| Yes | 5 | 10 | 1 | |
| No | 28 | 38 | 7 | |
| Bilateral | 0.635 | |||
| Yes | 12 | 22 | 4 | |
| No | 21 | 26 | 4 | |
| Extrathyroidal extension | 0.131 | |||
| Yes | 1 | 6 | 2 | |
| No | 32 | 42 | 6 | |
| TNM stage | 0.310 | |||
| I/II | 18 | 25 | 2 | |
| III/IV | 15 | 23 | 6 | |
-, Negative; +, moderate positive; ++, strong positive. The P-values were calculated using Pearson χ2-test.
P<0.05. PPM1D, protein phosphatase, Mg2+/Mn2+ dependent, 1D; PTC, papillary thyroid carcinoma.
Table II.
Clinicopathological and molecular factors in PTC.
| A, Clinicopathological and molecular factors associated with lymph node metastasis in PTC | ||||||
|---|---|---|---|---|---|---|
| Univariate analysis | Multivariate analysis | |||||
| Variables | OR | 95% CI | P-value | AOR | 95% CI | P-value |
| Sex, female vs. male | 1.469 | 0.548–3.937 | 0.445 | |||
| Age, <45 vs. ≥45 years | 2.214 | 0.933–5.257 | 0.072 | |||
| Multifocal lesions, yes vs. no | 1.163 | 0.382–3.544 | 0.791 | |||
| Tumor size, >1 vs. ≤1 cm | 6.394 | 2.457–16.393 | <0.001a | 6.394 | 2.208–18.182 | 0.001a |
| TNM stage, III–IV vs. I–II | 3.048 | 1.258–7.384 | 0.014a | 2.659 | 0.950–7.444 | 0.063 |
| PPM1D protein expression | ||||||
| − | Reference | Reference | ||||
| + | 2.986 | 1.188–7.503 | 0.020a | 2.512 | 0.852–7.406 | 0.095 |
| ++ | 4.071 | 0.713–23.623 | 0.114 | 5.446 | 0.809–36.661 | 0.081 |
| B, Clinicopathological and molecular factors associated with tumor size >1 cm in PTC | ||||||
| Univariate analysis | Multivariate analysis | |||||
| Variables | OR | 95% CI | P-value | AOR | 95% CI | P-value |
| Sex, female vs. male | 1.100 | 0.413–2.929 | 0.849 | |||
| Age, <45 vs. ≥45 years | 2.167 | 0.929–5.052 | 0.073 | |||
| Multifocal lesions, yes vs. no | 1.808 | 0.596–5.495 | 0.296 | |||
| Extrathyroidal extension, yes vs. no | 2.519 | 0.607–10.417 | 0.203 | |||
| Bilateral lesions, yes vs. no | 11.765 | 4.202–32.258 | <0.001a | 16.393 | 3.040–16.393 | 0.001a |
| PPM1D protein expression | ||||||
| − | Reference | Reference | ||||
| + | 0.442 | 0.179–1.092 | 0.077 | 0.455 | 0.137–1.512 | 0.199 |
| ++ | 5.158 | 0.568–46.834 | 0.145 | 17.218 | 1.393–212.831 | 0.027a |
P<0.05. PTC, papillary thyroid carcinoma; OR, odds ratio; AOR, adjusted odds ratio.
PPM1D regulates thyroid cancer cell growth
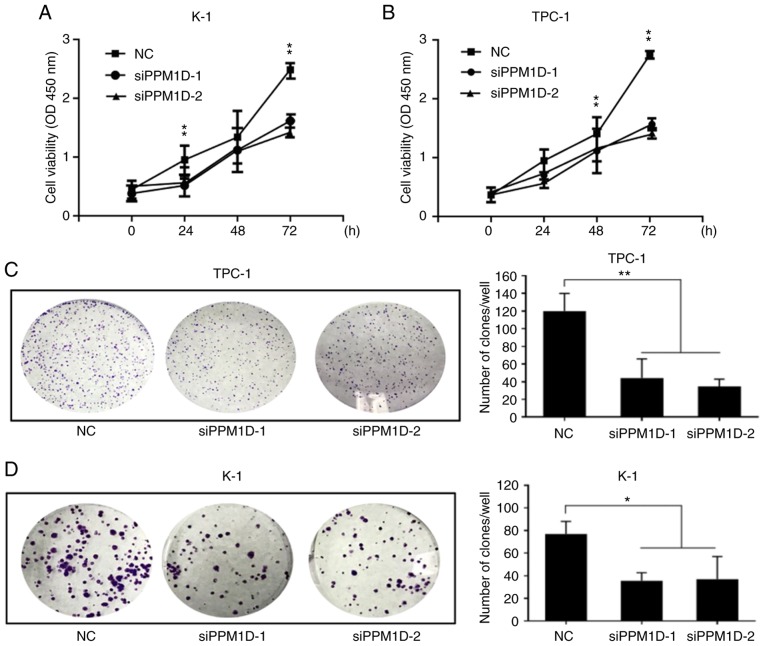
To investigate whether PPM1D is required for thyroid cancer cell growth, two siRNAs targeting PPM1D, siPPM1D-1 and siPPM1D-2, were used to knockdown PPM1D in TPC-1 and K-1 cells and one scrambled siRNA was used for the NC groups. Western blot analysis confirmed the efficient knockdown of PPM1D using the siPPM1Ds (Fig. 1C). The cell viability was then compared between the paired experimental groups. A CCK-8 assay revealed that PPM1D depletion led to a significant decrease in cell viability compared with that in the NC groups at 72 h for each of the two cell lines (Fig. 2A and B). Colony formation assays also indicated that PPM1D depletion reduced the colony number by 66.9 and 72.9% in TPC-1 cells and 56.6 and 52.8% in K-1 cells when using siPPM1D-1 and siPPM1D-2, respectively (Fig. 2C and D). In addition, statistical analysis revealed a significant decrease in the colony formation ability of siPPM1D-treated cells compared with that of NC cells (Fig. 2C and D). These results indicated that PPM1D promoted cell proliferation in TPC-1 and K-1 cell lines.
Figure 2.
PPM1D affects cell proliferation in vitro. (A and B) CCK-8 assay. TPC-1 and K-1 cells were treated with or without PPM1D siRNAs and the cell growth rate was determined at the indicated time-points by measuring the absorbance at 450 nm following incubation with a CCK-8 stain. Data for each condition were pooled from five wells and triplicate assays. (C and D) Plate colony formation assay of TPC-1 and K-1 cells with the same groups as described in (A and B). Representative images are provided on the left and statistical analysis of the cell clone formation rates is displayed on the right. Values are expressed as the mean ± SEM from three independent experiments. P-values were determined using ANOVA test followed by Tukey's post hoc test. *P<0.05, **P<0.01. PPM1D, protein phosphatase, Mg2+/Mn2+ dependent, 1D; CCK-8, Cell Counting Kit-8; siRNA, small interfering RNA.
Silencing of PPM1D inhibits the invasive ability of thyroid cancer cells
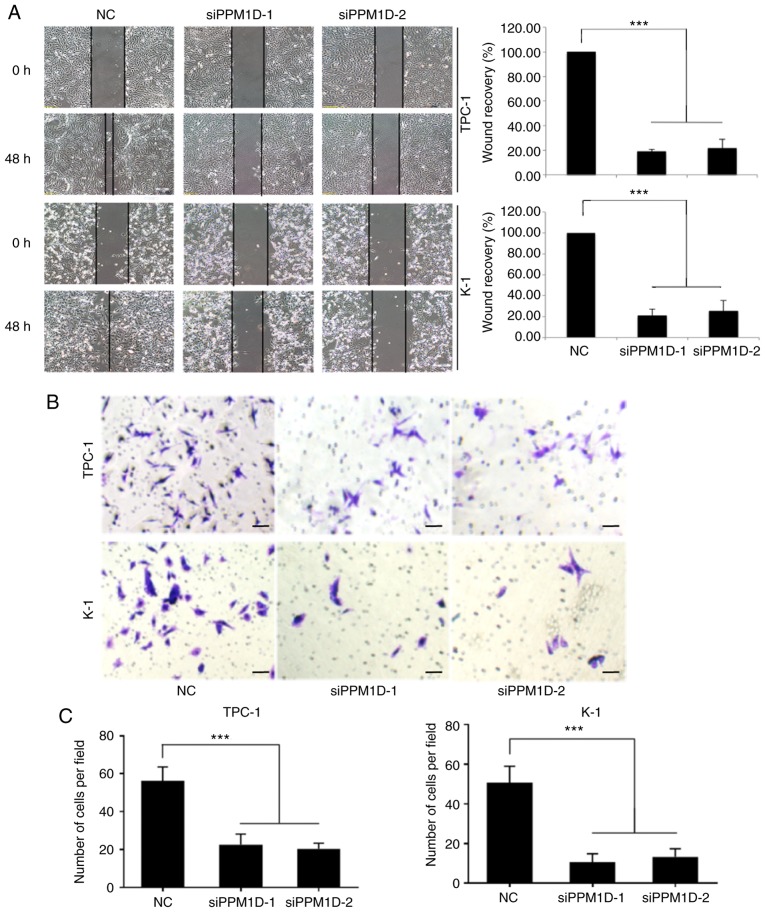
To investigate whether PPM1D is required for migration and invasion of TPC-1 and K-1 cells, a wound-healing assay and a Transwell invasion assay were performed. In the wound-healing assay, cells were cultured in serum-free medium and then allowed to migrate into a physical scratch. As presented in Fig. 3A, in the NC group, the cleared zone was almost filled with the cells at 48 h after the scratch wound was made. However, the siPPM1D-treated cells displayed a reduced ability to migrate and fill the cleared zone compared with that in the NC group. The statistical analysis (images on the right) indicated that the wound recovery rate was significantly reduced in siPPM1D-transfected cells compared with that in the NC group. The role of PPM1D in cell invasion was then evaluated using a Matrigel invasion assay, a modification of the Transwell assay. In brief, cells were placed onto Transwell with Matrigel pre-coated polycarbonate filters. The results indicated that cells in the control group and siRNA-treated cells were able to transgress through the Matrigel-coated membrane to the lower side of the filter, exhibiting invasive behavior. However, compared to the control group, a reduced number of cells transfected with siPPM1D passed though the Transwell insert (Fig. 3B). The quantitative data in Fig. 3C indicated a significant inhibition of cell invasion in PPM1D-silenced cells. Collectively, these results indicated that PPM1D is important for the migration and invasion of thyroid cancer cells in vitro.
Figure 3.
PPM1D promotes the cell migration and invasiveness of thyroid cancer cells in vitro. (A) Cell migration was quantified by wound healing assay. Cells were imaged immediately (0 h) and 48 h after scratches were created to assess the percentage of the wound-healed area. Representative images at different time-points are provided on the left (scale bar, 100 µm) and quantitative measurements are displayed on the right. (B and C) Cell invasion was assessed using a Transwell invasion assay. (B) Representative imagines (scale bar, 100 µm) and (C) Quantitative measurements. Values are expressed as the mean ± SEM of three independent experiments. P-values were determined using ANOVA test followed by Tukey's test. ***P<0.001. PPM1D, protein phosphatase, Mg2+/Mn2+ dependent, 1D.
PPM1D negatively regulates the p38 and p53 signaling pathways
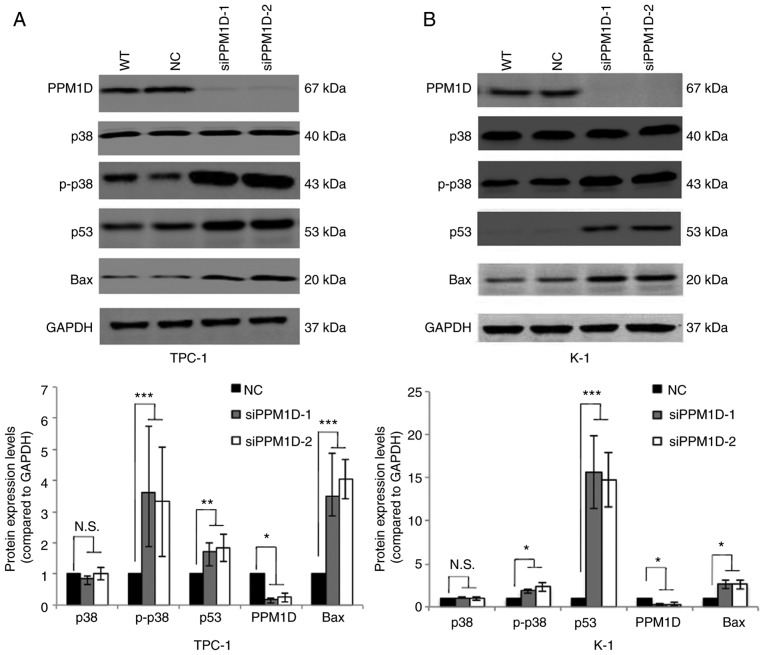
Next, it was investigated how PPM1D affects tumor cell behavior. Since PPM1D was first indicated to interact with p53 (19) and also reported to be involved in p38 MAPK activation, changes in the levels of key proteins in the p38 and p53 signaling pathways in thyroid cancer cells transfected with siPPM1D or scrambled siRNA were analyzed. The western blot results indicated that the protein levels of p-p38 MAPK, p53 and Bax were increased after treatment with siPPM1D (Fig. 4A and B, upper panel). Compared with the negative control group, knockdown of PPM1D increased the protein levels of p-p38 MAPK by >3-fold in TPC-1 cells and >1.8-fold in K-1 cells with concomitant upregulation of p53 by >1.7-fold in TPC-1 cells and >14-folds in K-1 cells and Bax by >3-fold in TPC-1 cells and >2-fold in K-1 cells (Fig. 4A and B, lower panel). These data indicated that PPM1D may exert its functions, at least partially, by inhibiting the p38 and p53 pathways.
Figure 4.
PPM1D suppresses p38 MAPK and p53 signaling pathways. p-p38 MAPK, p53 and Bax protein levels were upregulated in siPPM1D-treated thyroid cancer cells. (A and B) Western blot analysis of protein levels in TPC-1 and K-1 cells transfected with siPPM1Ds or scrambled RNA for 24 h (upper panel) and relative quantitative analysis of the protein levels in each group (lower panel). Values are expressed as the mean ± SEM of three independent experiments. P-values were determined using ANOVA test followed by Tukey's post hoc test. *P<0.05, **P<0.01, ***P<0.001. N.S., no significance; PPM1D, protein phosphatase, Mg2+/Mn2+ dependent, 1D; p-p38, phosphorylated p38; MAPK mitogen-activated protein kinase; WT, wild-type; NC, negative control; siPPM1D-1/siPPM1D-2: siRNAs against PPM1D; siRNA, small interfering RNA.
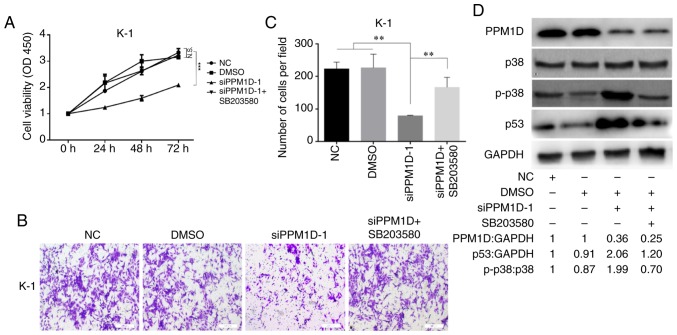
Inhibition of p38 activity reverses the inhibitory effect of PPM1D depletion on cell proliferation/invasion
Since overexpression of PPM1D was revealed to inactivate p38 (10), which may suppress the activity of p53, it was next examined whether PPM1D exerts its oncogenic role through regulation of p38 MAPK activity. After transfection with siPPM1D, K-1 cells were treated with or without SB203580, a chemical inhibitor of p38, and cell proliferation and invasion were compared among different groups at the indicated time-points. The results indicated decreased cell growth in response to PPM1D knockdown compared with that in the vehicle treatment DMSO or control group. However, the growth of siPPM1D-transfected cells was enhanced to a level similar to that of the control group when co-treated with the inhibitor across all time-points (Fig. 5A). Furthermore, the invasive capability was reduced in PPM1D-knockdown cells; however, concurrent inhibition of p38 restored the invasiveness of PPM1D-silenced K-1 cells (Fig. 5B). The number of invaded cells was significantly increased in the PPM1D-silencing group with p38 inhibition as compared with that in the group with PPM1D silencing only (Fig. 5C). Furthermore, the inhibitory effect of pharmacological inhibition of p38 on PPM1D silencing-induced expression of p53 was determined by western blot analysis. The results indicated that p-p38 MAPK and p53 were increased in PPM1D-depleted cells but remained suppressed in the presence of p38 inhibitor (Fig. 5D). Considering the crosstalk between the p53 and p38 signaling pathways during cell oncogenic transformation, these results indicated that the oncogenic properties of PPM1D were exerted via the p38 signaling pathway and that the p53 pathway has a direct and/or indirect contribution.
Figure 5.
p38 inhibitor restores cell growth and invasion in PPM1D-knockdown K-1 cells. (A) Cell growth was determined using a CCK-8 assay in cells with diminished PPM1D expression treated with or without SB203580. Each treatment was assayed in triplicate every 24 h for a total of 3 days. (B and C) The invasive ability of the cells was assessed using a Transwell cell invasion assay. (B) Representative images and (C) quantitative results. (D) Western blot analysis of PPM1D-silenced cells treated with or without p38 inhibitor for 24 h. The drug was used at a concentration of 100 nM. The relative quantification of protein bands as the ratio to their loading control are presented at the bottom of the western blotting. Values are expressed as the mean ± SEM of three independent experiments. P-values were determined using ANOVA test followed by Tukey's post hoc test. **P<0.01, ***P<0.001. N.S., no significance; PPM1D, protein phosphatase, Mg2+/Mn2+ dependent, 1D; CCK-8, Cell Counting Kit-8.
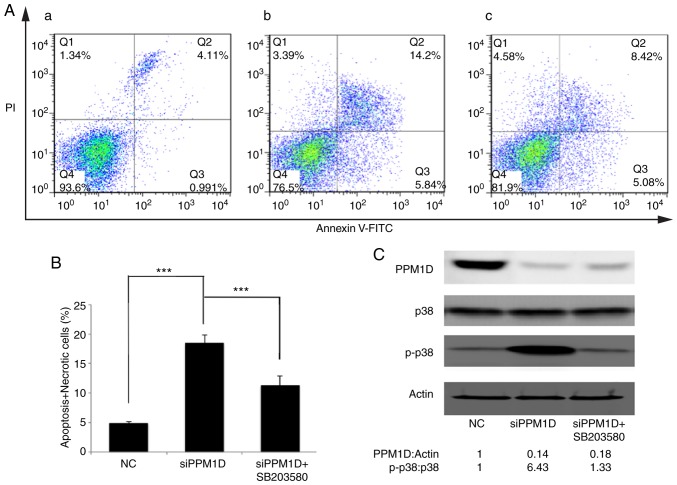
Inhibition of p38 activity counteracts PPM1D depletion-induced thyroid cancer cell apoptosis
The present study attempted to assess the effect of PPM1D in deregulating cell apoptosis and whether this was dependent on the p38 signaling pathway. PPM1D expression was silenced in K-1 cells. Concurrently, p38 activity was inhibited by SB203580. Cell apoptosis was detected by flow cytometry. As presented in Fig. 6A, viable cells were Annexin V-FITC- and PI-negative (Fig. 6A-Q4), dead cells were Annexin V-FITC-negative and PI-positive (Fig. 6A-Q1), cells in early apoptosis were Annexin V-FITC-positive and PI-negative (Fig. 6A-Q3), while cells in late apoptosis or already dead were Annexin V-FITC- and PI double-positive (Fig. 6A-Q2). The basal level of cell apoptosis was ~5%, as presented in Fig. 6A-a. Cell apoptosis was further evaluated in PPM1D-depleted K-1 cells with or without pharmacological inhibition of p38 MAPK activity and it was revealed that in K-1 cells, transfection with siPPM1D alone increased the number of apoptotic cells to 20.04% (Fig. 6A-b), but concurrent inhibition of p38 MAPK activity in PPM1D-depleted cells reduced cell apoptosis to 13.5% (Fig. 6A-c). These results indicated that PPM1D suppressed cell apoptosis by inhibiting the activity of p38 MAPK. All Annexin V-FITC-positive cells were combined to calculate the proportion of apoptotic cells. The statistical analysis based on the flow cytometric data indicated that cell apoptosis was significantly enhanced in siPPM1D-transfected K-1 cells compared with the NC group. By contrast, cell apoptosis was decreased when p38 activity was blocked in PPM1D-knockdown cells compared with the group with siPPM1D transfection alone (Fig. 6B). PPM1D knockdown and the reduced protein levels of p-p38 were confirmed by western blot analysis (Fig. 6C). Collectively, these results indicated that PPM1D was involved in the downregulation of p38 MAPK-mediated cell apoptosis.
Figure 6.
PPM1D silencing-induced K-1 cell apoptosis is suppressed by p38 inhibition. (A) K-1 cells treated with or without SB203580 were collected after transfection with siRNA specific to PPM1D. Apoptosis of K-1 cells was detected by flow cytometry. (a-c) Representative plots presenting Annexin V/PI staining. (a) NC, (b) PPM1D-1 siRNA-transfected, (c) PPM1D-1 siRNA-transfected + SB203580. (B) Analysis of apoptotic (early and late) cells from A. Values are expressed as the mean ± SEM of three independent experiments. P-values were determined using ANOVA test followed by Tukey's post hoc test. ***P<0.001. (C) Western blot analysis was performed to confirm the protein knockdown and pharmacological inhibition of p38. Actin was served as a loading control. The relative quantification of protein bands as the ratio to their loading control were presented at the bottom of the western blotting. PPM1D, protein phosphatase, Mg2+/Mn2+ dependent, 1D; NC, negative control (scrambled siRNA); siRNA, small interfering RNA; PI, propidium iodide.
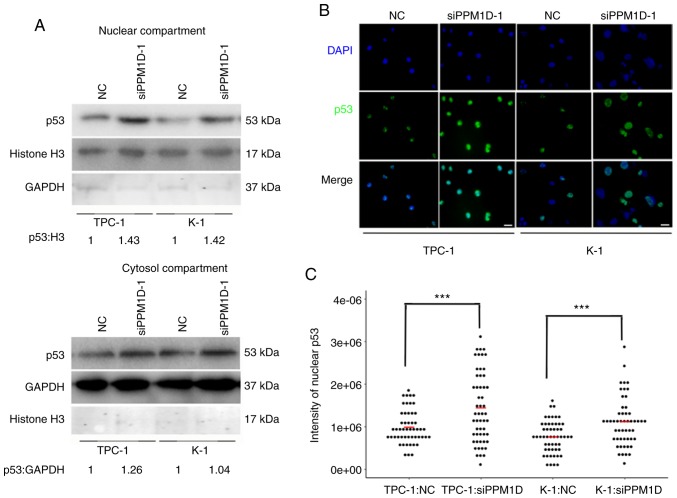
PPM1D regulates p53 nuclear translocation in thyroid cancer cells
Since p53 is able to transiently accumulate in the nucleus and act as a transcription factor of Bax in response to cell stress (19), it was further investigated whether downregulation of PPM1D increases p53 in the nucleus. The protein levels of p53 in the cytosolic and nuclear extract of TPC-1 and K-1 cells after PPM1D siRNA transfection were assessed by western blot analysis. In line with aforementioned findings, the two cell lines treated with siPPM1D exhibited increased p53 protein levels compared with those in the control groups (Fig. 7A). In addition, the increased accumulation of p53 in siPPM1D-transfected cells compared with that in the control groups was more evident in the nucleus compared with that in the cytosol, indicating that p53 translocates to the nucleus in response to PPM1D depletion (Fig. 7A, upper vs. lower panel). Next, to examine the nuclear distribution of p53, an indirect immunofluorescence assay was performed. In the imagines, p53 and nuclei were indicated by green and blue color, respectively (Fig. 7B). As indicated in a dot plot, the intensity of p53-positive nuclei was significantly increased after transfection with siPPM1D compared with that in the control groups (Fig. 7C), indicating more nuclear retention of p53 in PPM1D-knockdown cells. Collectively, the negative regulation of the p53 signaling pathway by PPM1D may lead to less nuclear accumulation of p53.
Figure 7.
PPM1D knockdown promotes nuclear retention of p53. (A) Protein lysates from cytoplasm and nuclei of TPC-1 and K-1 cells were subjected to western blot analysis. Histone H3 was measured as the control for the nuclear compartment, while GAPDH was used for the cytosol compartment. The relative quantification of protein bands as the ratio to their loading control are presented at the bottom of the western blotting. (B) Representative images of the indirect immunofluorescence assay suggesting nuclear translocation of p53 after transfection with siPPM1D in TPC-1 and K-1 cells (scale bar, 100 µm). (C) Quantification of nuclear staining intensity of p53 in B using ImageJ. n=50. P-values were determined using ANOVA test followed by Tukey's post hoc test. Values are expressed as the mean ± SEM of three independent experiments; the means of each group are indicated by the short red bar. ***P<0.001. PPM1D, protein phosphatase, Mg2+/Mn2+ dependent, 1D; siPPM1D, siRNA against PPM1D; siRNA, small interfering RNA.
Discussion
In the present study, PPM1D was identified as a molecular marker of metastasis and poor prognosis in patients with PTC and the oncogenic properties of PPM1D were also confirmed in vitro. PPM1D was indicated to promote cancer progression in PTC, as the analysis of 89 PTC patient samples indicated that high PPM1D protein expression was significantly associated with tumor size and lymph node metastasis. It was also demonstrated that PPM1D was overexpressed in PTC patient tissues compared with that in paired adjacent non-cancerous tissues. These results were in line with numerous previous studies reporting on the oncogenic properties of PPM1D in various types of cancer (5,28–30). These previous studies indicated that these features were largely associated with the ability of PPM1D to modulate the p38 MAPK and p53 signaling network.
In addition, the present study demonstrated that PPM1D exerted an essential influence on thyroid cancer cell proliferation and invasion in vitro. The growth rates of TPC-1 and K-1 cells transfected with siPPM1D were significantly decreased compared with those of control cells. Furthermore, significant decreases in colony formation and migration were observed in PPM1D-silenced cells. Of note, the cell proliferation of PPM1D-silenced K-1 cells was successfully restored by inhibiting the activity of p38 MAPK, indicating that PPM1D promotes cancer cell progression at least partially via regulation of p38 MAPK activity.
To confirm that PPM1D accelerates thyroid cancer cell progression through inhibition of the p38 MAPK and/or p53 signaling pathways in thyroid cancer cells, the association between PPM1D levels and the activities of p38 MAPK and p53 were delineated. The present study revealed that PPM1D knockdown in K-1 and TPC-1 cells led to increased protein levels of p53 and Bax, as well as enhanced p38 MAPK phosphorylation. As aforementioned, it is essential to consider the crosstalk between the p38 MAPK and p53 signaling pathways, and in this light, it was further demonstrated that knockdown of PPM1D induced apoptosis, which was partially reversed by additional inhibition of p38 MAPK activities. The present results demonstrated that silencing of PPM1D caused a significant upregulation of the p38 MAPK signaling pathway, which promoted the apoptosis of thyroid cancer cells. The mechanism underlying how PPM1D mediates p53 activity was further investigated, revealing that PPM1D may regulate nuclear translocation of p53. Since Bax is transcriptionally regulated by p53, it is possible that PPM1D deregulates Bax protein expression through the p53 signaling pathway. Since activation of p38 and expression of p53 are events that are commonly associated with apoptosis and that are situated downstream of the actual trigger of apoptosis, the direct target of PPM1D should be explored in the future.
In conclusion, the present results provide novel insight into the cancer biology of PTC; PPM1D was indicated to be a risk factor of metastasis and may serve as a potential therapeutic target. The downstream targets of PPM1D and the mechanisms underlying how these targets mediate PPM1D oncogenic activities remains to be further investigated.
Acknowledgements
We sincerely thank the University of Colorado Cancer Center Cell Bank for providing two thyroid cancer cell lines including K1 and TPC-1.
Glossary
Abbreviations
- PTC
papillary thyroid carcinoma
- PPM1D
protein phosphatase, Mg2+/Mn2+ dependent, 1D
Funding
The present study was supported by funds from the National Natural Science Foundation of China (grant nos. 81272934, 81572622, and 81772854 to QHJ; grant nos. 81472498 and 81772851 to YLW; grant no. 81502317 to WJW; grant no. 81702753 to TL); the Shanghai Municipal Planning Commission of Science and Research Fund for Young Scholars (grant no. 20154Y0050 to JX).
Availability of data and materials
Please contact the author for data requests.
Authors' contributions
QHJ conceived and designed the experiments. LTH selected the patients and collected the clinical samples. ZWL, DW and LTH performed the experiments. WJW analyzed the data and interpreted the results data along with TL and JX. TL, LTH and JX contributed the reagents/materials/analysis tools. ZWL and QHJ wrote the manuscript. YLW and YW critically revised manuscript for important intellectual content. All authors read and approved the final version of manuscript to be published and agree to be accountable for all aspects of the research in ensuring that questions in terms of the accuracy or integrity of any part of the research are appropriately investigated and resolved.
Ethics approval and consent to participate
For the use of the clinical materials for research purposes, the study was conducted in accordance with the Declaration of Helsinki. The Institutional Research Ethics Committee of Shanghai Cancer Center, Fudan University, approved the study, and informed consent was obtained from all participants and/or their legal guardians.
Patient consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
References
- 1.Cabanillas ME, McFadden DG, Durante C. Thyroid cancer. Lancet. 2016;388:2783–2795. doi: 10.1016/S0140-6736(16)30172-6. [DOI] [PubMed] [Google Scholar]
- 2.Lubitz CC, Sosa JA. The changing landscape of papillary thyroid cancer: Epidemiology, management, and the implications for patients. Cancer. 2016;122:3754–3759. doi: 10.1002/cncr.30201. [DOI] [PubMed] [Google Scholar]
- 3.Landa I, Ibrahimpasic T, Boucai L, Sinha R, Knauf JA, Shah RH, Dogan S, Ricarte-Filho C, Krishnamoorthy GP, Xu B, et al. Genomic and transcriptomic hallmarks of poorly differentiated and anaplastic thyroid cancers. J Clin Invest. 2016;126:1052–1066. doi: 10.1172/JCI85271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Le Guezennec X, Bulavin DV. WIP1 phosphatase at the crossroads of cancer and aging. Trends Biochem Sci. 2010;35:109–114. doi: 10.1016/j.tibs.2009.09.005. [DOI] [PubMed] [Google Scholar]
- 5.Li J, Yang Y, Peng Y, Austin RJ, van Eyndhoven WG, Nguyen KCQ, Gabriele T, McCurrachM E, Marks JR, Hoey T, et al. Oncogenic properties of PPM1D located within a breast cancer amplification epicenter at 17q23. Nat Genet. 2002;31:133–134. doi: 10.1038/ng888. [DOI] [PubMed] [Google Scholar]
- 6.Tamura S, Toriumi S, Saito JI, Awano K, Kudo TA, Kobayashi T. PP2C family members play key roles in regulation of cell survival and apoptosis. Cancer Sci. 2006;97:563–567. doi: 10.1111/j.1349-7006.2006.00219.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pullen KE, Ng HL, Sung PY, Good MC, Smith SM, Alber T. An alternate conformation and a third metal in PstP/Ppp, the M. tuberculosis PP2C-Family Ser/Thr protein phosphatase. Structure. 2004;12:1947–1954. doi: 10.1016/j.str.2004.09.008. [DOI] [PubMed] [Google Scholar]
- 8.Wagner EF, Nebreda AR. Signal integration by JNK and p38 MAPK pathways in cancer development. Nat Rev Cancer. 2009;9:537–549. doi: 10.1038/nrc2694. [DOI] [PubMed] [Google Scholar]
- 9.Guichard C, Amaddeo G, Imbeaud S, Ladeiro Y, Pelletier L, Maad IB, Calderaro J, Bioulac-Sage P, Letexier M, Degos F, et al. Integrated analysis of somatic mutations and focal copy-number changes identifies key genes and pathways in hepatocellular carcinoma. Nat Genet. 2012;44:694–698. doi: 10.1038/ng.2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang ZP, Tian Y, Lin J. Role of wild-type p53-induced phosphatase 1 in cancer. Oncol Lett. 2017;14:3893–3898. doi: 10.3892/ol.2017.6685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harrison M, Li J, Degenhardt Y, Hoey T, Powers S. Wip1-deficient mice are resistant to common cancer genes. Trends Mol Med. 2004;10:359–361. doi: 10.1016/j.molmed.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 12.Bulavin DV, Demidov ON, Saito S, Kauraniemi P, Phillips C, Amundson SA, Ambrosino C, Sauter G, Nebreda AR, Anderson CW, et al. Amplification of PPM1D in human tumors abrogates p53 tumor-suppressor activity. Nat Genet. 2002;31:210–215. doi: 10.1038/ng894. [DOI] [PubMed] [Google Scholar]
- 13.Cancer Genome Atlas Research N, corp-author. Integrated genomic characterization of papillary thyroid carcinoma. Cell. 2014;159:676–690. doi: 10.1016/j.cell.2014.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chew J, Biswas S, Shreeram S, Humaidi M, Wong HET, Dhillion MK, Teo H, Hazra A, Fang CC, López-Collazo D, et al. WIP1 phosphatase is a negative regulator of NF-kappaB signalling. Nat Cell Biol. 2009;11:659–666. doi: 10.1038/ncb1873. [DOI] [PubMed] [Google Scholar]
- 15.Tan DS, Maryou B, Lambros K, Rayter S, Natrajan R, Vatcheva R, Gao Q, Marchiò C, Geyer FC, Savage K, et al. PPM1D is a potential therapeutic target in ovarian clear cell carcinomas. Clin Cancer Res. 2009;15:2269–2280. doi: 10.1158/1078-0432.CCR-08-2403. [DOI] [PubMed] [Google Scholar]
- 16.Goloudina AR, Kochetkova EY, Pospelova TV, Demidov ON. Wip1 phosphatase: Between p53 and MAPK kinases pathways. Oncotarget. 2016;7:31563–31571. doi: 10.18632/oncotarget.7325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gorrini C, Harris IS, Mak TW. Modulation of oxidative stress as an anticancer strategy. Nat Rev Drug Discov. 2013;12:931–947. doi: 10.1038/nrd4002. [DOI] [PubMed] [Google Scholar]
- 18.Trachootham D, Alexandre J, Huang P. Targeting cancer cells by ROS-mediated mechanisms: A radical therapeutic approach? Nat Rev Drug Discov. 2009;8:579–591. doi: 10.1038/nrd2803. [DOI] [PubMed] [Google Scholar]
- 19.Fiscella M, Zhang H, Fan S, Sakaguchi K, Shen S, Mercer WE, Vande Woude GF, O'Connor PM, Appella E. Wip1, a novel human protein phosphatase that is induced in response to ionizing radiation in a p53-dependent manner. Proc Natl Acad Sci USA. 1997;94:6048–6053. doi: 10.1073/pnas.94.12.6048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu X, Nannenga B, Donehower LA. PPM1D dephosphorylates Chk1 and p53 and abrogates cell cycle checkpoints. Genes Dev. 2005;19:1162–1174. doi: 10.1101/gad.1291305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Demidov ON, Timofeev O, Lwin HNY, Kek C, Appella E, Bulavin DV. Wip1 phosphatase regulates p53-dependent apoptosis of stem cells and tumorigenesis in the mouse intestine. Cell Stem Cell. 2007;1:180–190. doi: 10.1016/j.stem.2007.05.020. [DOI] [PubMed] [Google Scholar]
- 22.Johnson GL, Lapadat R. Mitogen-activated protein kinase pathways mediated by ERK, JNK, and p38 protein kinases. Science. 2002;298:1911–1912. doi: 10.1126/science.1072682. [DOI] [PubMed] [Google Scholar]
- 23.Dudgeon C, Shreeram S, Tanoue K, Mazur SJ, Sayadi A, Robinson RC, Appella E, Bulavin DV. Genetic variants and mutations of PPM1D control the response to DNA damage. Cell Cycle. 2013;12:2656–2664. doi: 10.4161/cc.25694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu Y, Li N, Xiang R, Sun P. Emerging roles of the p38 MAPK and PI3K/AKT/mTOR pathways in oncogene-induced senescence. Trends Biochem Sci. 2014;39:268–276. doi: 10.1016/j.tibs.2014.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takekawa M, Adachi M, Nakahata A, Nakayama I, Itoh F, Tsukuda H, Taya Y, Imai K. p53-inducible wip1 phosphatase mediates a negative feedback regulation of p38 MAPK-p53 signaling in response to UV radiation. EMBO J. 2000;19:6517–6526. doi: 10.1093/emboj/19.23.6517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bulavin DV, Phillips C, Nannenga B, Timofeev O, Donehower LA, Anderson CW, Appella E, Fornace AJ., Jr Inactivation of the Wip1 phosphatase inhibits mammary tumorigenesis through p38 MAPK-mediated activation of the p16(Ink4a)-p19(Arf) pathway. Nat Genet. 2004;36:343–350. doi: 10.1038/ng1317. [DOI] [PubMed] [Google Scholar]
- 27.Wen D, Liao T, Ma B, Qu N, Shi RL, Lu ZW, Wang YL, Wei WJ, Ji QH. Downregulation of CSN6 attenuates papillary thyroid carcinoma progression by reducing Wnt/beta-catenin signaling and sensitizes cancer cells to FH535 therapy. Cancer Med. 2018;7:285–296. doi: 10.1002/cam4.1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fallahi P, Mazzi V, Vita R, Ferrari SM, Materazzi G, Galleri D, Benvenga S, Miccoli P, Antonelli A. New therapies for dedifferentiated papillary thyroid cancer. Int J Mol Sci. 2015;16:6153–6182. doi: 10.3390/ijms16036153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tang YL, Liu X, Gao SY, Feng H, Jiang YP, Wang SS, Yang J, Jiang J, Ma XR, Tang YJ, et al. WIP1 stimulates migration and invasion of salivary adenoid cystic carcinoma by inducing MMP-9 and VEGF-C. Oncotarget. 2015;6:9031–9044. doi: 10.18632/oncotarget.3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun GG, Wang YD, Liu Q, Hu WN. Expression of Wip1 in kidney carcinoma and its correlation with tumor metastasis and clinical significance. Pathol Oncol Res. 2015;21:219–224. doi: 10.1007/s12253-014-9811-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Please contact the author for data requests.