Abstract
NADPH-oxidases (NOXs) purposefully produce reactive-oxygen-species (ROS) and are found in most kingdoms of life. The seven human NOXs are each characterized by a specific expression profile and a fine regulation to spatio-temporally tune ROS concentration in cells and tissues. One of the best known roles for NOXs is in host protection against pathogens but ROS themselves are important second messengers involved in tissue regeneration and the modulation of pathways that induce and sustain cell proliferation. As such, NOXs are attractive pharmacological targets in immunomodulation, fibrosis and cancer. We have studied an extensive number of available NOX inhibitors, with the specific aim to identify bona fide ligands versus ROS-scavenging molecules. Accordingly, we have established a comprehensive platform of biochemical and biophysical assays. Most of the investigated small molecules revealed ROS-scavenging and/or assay-interfering properties to various degrees. A few compounds, however, were also demonstrated to directly engage one or more NOX enzymes. Diphenylene iodonium was found to react with the NOXs’ flavin and heme prosthetic groups to form stable adducts. We also discovered that two compounds, VAS2870 and VAS3947, inhibit NOXs through the covalent alkylation of a cysteine residue. Importantly, the amino acid involved in covalent binding was found to reside in the dehydrogenase domain, where the nicotinamide ring of NADPH is bound. This work can serve as a springboard to guide further development of bona fide ligands with either agonistic or antagonistic properties toward NOXs.
Keywords: NADPH oxidase, NOX, Reactive oxygen species, ROS scavengers
Abbreviations: NOX, NADPH oxidase; DPI, diphenylene iodonium; ROS, reactive oxygen species; MCLA, 6-(4-methoxyphenyl)-2-methyl-imidazo[1,2-a]pyrazin-3(7H)-one; CBA, non-fluorescent coumarin boronic acid; PBS, Phosphate-Buffered Saline
Graphical abstract
Highlights
-
•
NOX inhibition by twenty-four compounds was tested using multiple assays.
-
•
Most of the evaluated compounds displayed significant ROS scavenging and/or assay-interfering effects.
-
•
DPI is demonstrated to engage the heme and the flavin cofactors of NOXs.
-
•
VAS2870 and VAS3947 are covalent bona-fide NOX inhibitors.
-
•
A conserved active-site cysteine can be targeted for inhibition.
1. Introduction
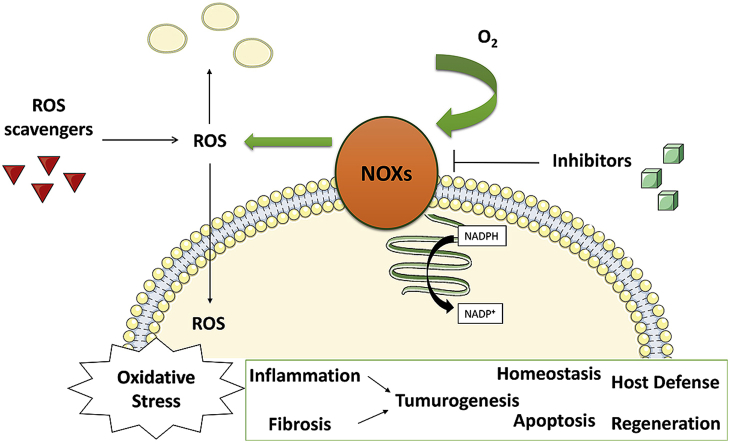
NADPH-oxidases (NOXs) are the only known enzymes whose specific function is reactive-oxygen-species (ROS) generation [[1], [2], [3], [4]]. The seven human NOXs share a conserved catalytic subunit, comprising an N-terminal membrane domain that embeds two hemes, and a C-terminal cytosolic domain that binds the FAD prosthetic group and the NADPH substrate [5]. The catalytic reaction starts with the reduction of the flavin by NADPH. This is followed by a step-wise electron transfer that leads to the reduction of the outer heme, located on the external side of the membrane, where ROS generation takes place. NOX1, NOX2, and NOX3 are regulated by various cellular proteins whereas NOX5 and DUOXs are activated by calcium through additional calcium-binding domains. NOX4 is the only constitutively active member of the family [6]. Generally, NOXs coordinate cell damage, stress responses and tissue regeneration. In particular, NOX2 is at the heart of the innate immunity by eliminating bacterial threats in the phagocytes. High levels of ROS derived from NOXs hyperactivity can result in genetic instability followed by excessive proliferation, activation of the DNA-damage response, proliferative senescence, and apoptosis [[7], [8], [9], [10], [11], [12]]. With their prototypical roles in ROS biology and redox signalling, NOXs are attractive drug targets in inflammation, fibrotic and degenerative processes, and cancer [3,[13], [14], [15]].
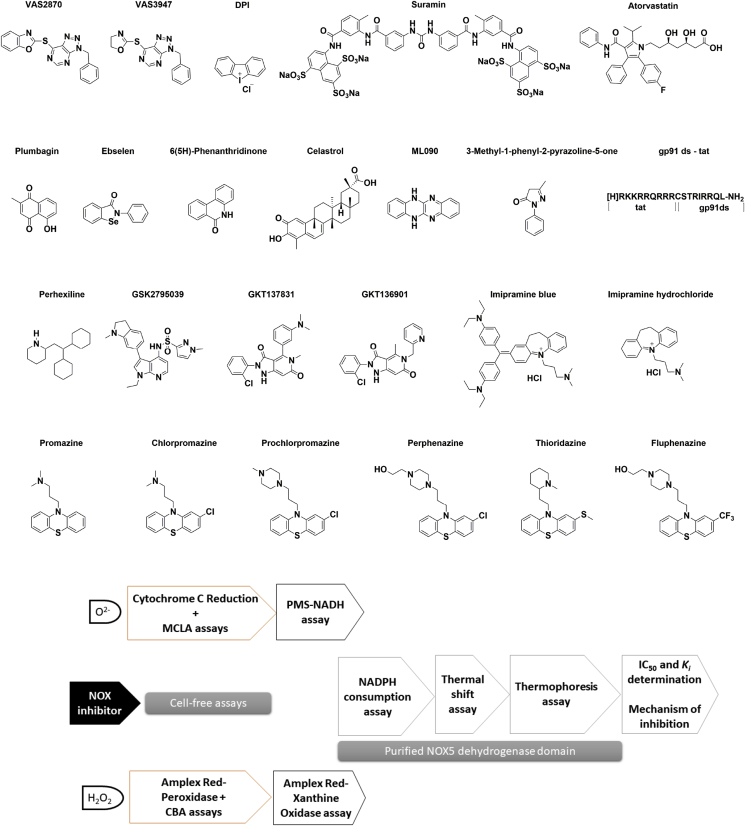
The functional complexity of NOXs suggests several potential mechanisms for a specific pharmacological response, such as the inhibition of the enzyme assembly or the competition with the NADPH and O2 substrates. Diphenylene iodonium (DPI), di-2-thienyliodonium, and apocynin have been the first molecules to be investigated as NOX inhibitors [[16], [17], [18], [19], [20], [21], [22]]. Indeed, DPI is widely employed in cell studies as negative control to assess NOX activity. Besides being reported as a flavoenzyme inhibitor [23], it has also been described as an inhibitor of cytochrome P-450 reductase [17], xanthine oxidase [24] and nitric-oxide synthase [25]. Suramin is a polysulfonated naphthylurea in use since the 1920s as an agent against human trypanosomiasis [26]. Suramin has also been reported to act as a NOX inhibitor in cell-free assays with no inhibitory activity in NOX over-expressing cells, probably due to lack of cell membrane permeability [27]. VAS2870 and VAS3947 are extensively used in current literature although there are contradictory observations regarding these compounds being bona fide NOX inhibitors [[27], [28], [29], [30], [31]]. Phenothiazine derivatives have been described as anti-NOX compounds and were studied with the aim of optimizing their chemical structure in terms of NOX selectivity and decrease of its assay-interference profile [32]. Additionally, perhexiline (used in angina pectoris and severe myocardial ischemia), plumbagin (a plant-derived naphthoquinone), ML090 (a quinoxaline derivative), 3-methyl-1-phenyl-2-pyrazoline, and imipramine have been identified as a possible inhibitors of NOXs [[33], [34], [35], [36], [37]]. Compounds such as GSK2795039 and GKT137831 (setanaxib) have been developed by pharmaceutical companies as NOX2 or NOX4/NOX1 selective inhibitors and reached clinical evaluation [28,31,[38], [39], [40]]. Lastly, the peptide tat-gp91ds specifically blocks the interaction between NOX2 and its activator p47phox [41]. Likewise, the inhibition of myocardial oxidative stress by statins, blockbuster cholesterol-lowering drugs, has been ascribed to their interference with Rac-induced NOX activation [[42], [43], [44]].
Scavenging of superoxide/hydrogen peroxide can represent an additional strategy to interfere with the functions of NOXs. For instance, phenanthridinones are antioxidant polyphenols with quinoid structures and were reported as NOX inhibitors [45]. Celastrol is a natural product used in the treatment of immunological diseases and has been described as a compound blocking the ROS production associated to NOXs [46,47]. Ebselen is a seleno-organic drug with antioxidant properties, mainly functioning as an effective scavenger of organic hydroperoxides [28,31,48]. However, there is also evidence of ebselen inhibiting ROS production by disturbing the assembly of NOX2-activating regulatory subunits [49,50].
Despite such a wealth of studies, a clear distinction between a ROS-scavenging molecule and a true NOX binder is often missing and disguised under the generic umbrella of the term “inhibitor”. The complexity of detection and quantification of ROS, short-lived entities in their biological milieu, has hindered the progress in the field. Therefore, our understanding of the inhibitors’ mechanism of action remains an open issue. The use of both orthogonal assays to validate the results from the primary screens and control assays to monitor the direct scavenging of ROS is of major importance. Additionally, direct binding assays may be necessary to discriminate a bona fide NOX inhibitor from assay-interfering compounds. Here, we describe a comprehensive experimental workflow for NOX inhibitor development and validation. We employed this strategy for the most widely used anti-NOX compounds to validate efficacy and selectivity using both cell-free and purified enzyme assays (Fig. 1). We found that most of the tested NOX inhibitors have very substantial off-target effects, which in many cases prohibit a convincing demonstration of their ability to directly engage the NOX targets. However, we also show that a few of the known NOX inhibitors are indeed proper NOX ligands with variable degrees of isoform selectivities. These data will help the interpretation of data arising from the usage of known NOX inhibitors and also lay down a much-needed strategy for future NOX drug discovery and optimisation.
Fig. 1.
The experimental workflow. (A) Chemical structure of reported NOX inhibitors and (B) experimental assays.
2. Materials and methods
2.1. Reagents
Roswell Park Memorial Institute (RPMI)1640 medium with l-glutamine and sodium bicarbonate, penicillin, streptomycin, Lithium Dodecyl sulfate (LiDS), MgCl2, flavin adenine dinucleotide disodium salt hydrate (FAD-Na2), β-nicotinamide adenine dinucleotide 2′-phosphate reduced tetrasodium salt (NADPH), β-Nicotinamide adenine dinucleotide, reduced disodium salt hydrate (NADH), 1-methoxy phenazine methosulfate, Nitroblue tetrazolium, Sodium Azide, Phosphate-Buffered Saline (PBS), Sodium Di-Hydrogen Phosphate (NaH2PO4·H2O), Isopropyl β-d-1-thiogalactopyranoside, Sodium chloride, Glycerol. Hepes, Leupeptin, Pepstatin, Phenylmethylsulfonyl fluoride. SigmaFastTM Protease Inhibitor Cocktail Tablets EDTA-Free, Hemin from Porcine, cytochrome c from equine heart, superoxide dismutase (SOD), Amplex Red, Sodium dithionite and horseradish peroxidase, Triton X-100 were purchased from Sigma-Aldrich. Potassium Phosphate Dibasic (K2HPO4) was purchased from Carlo Erba Reagents. Hydrochloric acid was purchased from Fluka. Magnesium Sulfate (MgSO4) and Acetonitrile were purchased from Merck. Fetal bovine serum was purchased from Invitrogen. 6-(4-methoxyphenyl)-2-methyl-imidazo [1,2-a]pyrazin-3(7H)-one (MCLA) was purchased from MedChemExpress. Non-fluorescent coumarin boronic acid (CBA) was synthesized in house. All tested inhibitors were purchased from Sigma-Aldrich except ML-090 (Cayman Chemical), GSK2795039 (MedChemExpress) and VAS3947 (Calbiochem). GKT136901 and GKT137831 were a kind gift of GenKyoTex SA (France). The detergent n-dodecyl-β-D-maltoside was purchased from Anatrace. Tat-gp91ds was purchased from Anaspec. The DYKDDDDK-FLAG peptide was purchased from China Peptides.
2.2. NOX expression and preparations
Bacterial NOX5 - The overexpression in E. coli and the preparation of cell membranes for the FLAG-(His)8-SUMO N-terminally tagged Cylindrospermum stagnale NOX5 was performed as reported [51]. The N-terminally strep-tagged dehydrogenase domain (residues 413–693; wild type and C-terminal mutant) and the FLAG-(His)8-SUMO N-terminally tagged transmembrane domain (residues 209–412; wild type and R256S mutant) were expressed and purified as described [5]. The C668S mutant of the dehydrogenase was prepared following the same protocols.
Human NOX2 membrane preparation and purification of cytosolic factors - X-CGD PLB-985 cells, transduced with the RD114-pseudotyped MFGS-gp91phox vector (PLB-985 cells from now on) were a kind gift from Henry Malech (NIH, Bethesda, USA) [52]. PLB-985 cells were cultured in suspension at 1 × 106 cells/mL in RPMI at 37 °C with 5% CO2. The medium was supplemented with 10% fetal bovine serum, 100 units/mL of penicillin and 100 μg/ml of streptomycin. Cells were centrifuged at 1000×g for 10 min, then resuspended in PBS and centrifuged again at 1000×g for 5 min and frozen at −80 °C. PLB-985 frozen pellets were resuspended at a concentration of 2 × 108 cells/ml in sonication buffer containing 10 mM Hepes (pH 7.4), 10 mM NaCl, 100 mM KCl, 12 mM EGTA, 3.5 mM MgCl2, 1 mM phenylmethylsulfonyl fluoride and supplemented with 2 μM leupeptin, 2 μM pepstatin, and protease inhibitors, just before sonication. The lysate was centrifuged at 2000 rpm for 5 min at 4 °C, and the supernatant was collected. The cell pellet was resuspended in sonication buffer and sonicated again on ice two times. The cell lysate was centrifuged at 2000 rpm for 5 min at 4 °C, and the supernatant was collected. Both supernatants were ultra-centrifuged (200,000×g for 30 min) at 4 °C (Optima MAX-XP Ultracentrifuge, Beckman Coulter). Protein concentration was assessed by Biuret Assay.
The full length human p67phox, p47phox and the constitutively active mutant Rac1 Q61L cloned into a pET-30a vector, with a N-terminal (His)6-tag, were a kind gift from Edgar Pick (Tel Aviv University, Israel). The recombinant proteins were expressed in E. coli Rosetta (DE3, pLysS) (Novagen), and bacteria were induced with 0.4 mM isopropyl β-d-thiogalactopyranoside at 18 °C for 16 h. The induced cells were suspended in lysis buffer (20 mM sodium phosphate buffer, 0.5 mM NaCl, and 20 mM imidazole, pH 7.4) supplemented with “Complete” EDTA-free protease inhibitor and 1% (v/v) Triton X-100. The bacteria were disrupted by sonication, the lysate was centrifuged at 34,000×g for 30 min at 4 °C, and the cleared cell-free extract was filtered through a 0.45-μm filter. The material was applied to Nickel-Sepharose 6 Fast Flow beads (GE Healthcare), and binding was allowed to proceed in the batch mode for 60 min on a rotary mixer at room temperature. The beads were washed with lysis buffer, followed by two washes with the washing buffer (20 mM sodium phosphate, 0.5 mM NaCl, and 40 mM imidazole, pH 7.4). The protein bound to the beads was eluted from the resin with the elution buffer (20 mM sodium phosphate, 0.5 mM NaCl, and 300 mM imidazole, pH 7.4) at a volume corresponding to ten times the volume of the beads. To improve the level of purity the proteins were subjected to further purification by gel filtration: p67phox on a Superdex 200 Hiload 16/60 fast protein liquid chromatography (FPLC) column, p47phox and Rac1 Q61L on a Superdex 75 Hiload 16/60 fast protein liquid chromatography (FPLC) column. 10-mg amounts of all the proteins were injected into the column, and chromatography was performed with Lysis buffer without imidazole, on a High-Pressure Liquid Chromatography (HPLC) system at a flow rate of 1 ml/min. The purified proteins were supplemented with 10% glycerol and stored at −80 °C.
Human NOX4 membrane preparation - The cDNAs encoding for the human NOX4 (isoform 1) and the human p22phox were originally obtained from Genescript. NOX4 was subcloned into a pEG BacMam vector (a kind gift from Eric Gouaux (Oregon Health and Science University, Portland)). The construct contained a Kozak sequence, followed by a (His)6 tag, an eGFP moiety and a TEV cleavage site at the N-terminus. P22phox was subcloned into the pDsRed-monomer-N1 vector with a DsRed moiety at the C-terminus. HEK293-EBNA1-6E cells were obtained from Yves Durocher (NRC-BRI, Montreal, Canada) [53]. Cells were grown in suspension using FreeStyle medium (Invitrogen). The day before transfection, cells were seeded at a density of 5 × 105 cells/ml. Transient NOX4-p22 co-transfection of HEK293 EBNA-1 was performed using polyethylenimine (linear MW 25000, Polyscience Europe Gmbh) in a ratio 1:3 (0.6 μg NOX4 vector: 0.4 μg p22 vector: 3 μg polyethylenimine, per ml of culture). After 24 h, co-transfected cells were harvested by centrifugation (1200×g, 5 min, 4 °C). Cell pellet was flash-frozen in liquid nitrogen and stored at −80 °C. Cell viability was evaluated by Trypan Blue exclusion assay (Trypan Blue solution w/v, Sigma). The cell pellet was thawed on ice and resuspended in sonication buffer (20% (v/v) glycerol, 120 mM NaCl, 1 mM EGTA in PBS pH 7.4) supplemented with protease inhibitors (SigmaFastTM Protease Inhibitor Cocktail Tablets, EDTA-Free, Sigma-Aldrich) at 4 °C. Cells were sonicated on ice. The cell lysate was then centrifuged at 500×g for 5 min at 4 °C, and the supernatant transferred in a fresh tube. The cell pellet was resuspended again in sonication buffer and the sample was sonicated again on ice. Cell lysate was then centrifuged at 500×g for 5 min at 4 °C, and the supernatant was collected. Both supernatants were pooled. The supernatant was ultra-centrifuged (160,000×g for 45 min) at 4 °C (Optima MAX-XP Ultracentrifuge, Beckman Coulter). Finally, the pellet was resuspended and diluted in sonication buffer to a final protein concentration of at least 30–40 mg/ml measured by the Biuret assay, aliquoted and stored in the −80 °C freezer. Protein quality was evaluated by measuring the fluorescence of the eGFP moiety and the DsRed moiety using fluorescence intensity-based detection on SDS-PAGE gel (BioRad; ChemiDoc MP imager; Alexa Fluor 488 for eGFP detection, Alexa Fluor 546 for DsRed detection).
2.3. Evaluation of NOX activity and NOX inhibitors in cell-free assays
Cytochrome c reduction assay on NOX2 and NOX5 membranes – This assay relies on the reduction of cytochrome c by the NOX-generated superoxide that can be monitored spectrophotometrically (absorbance at 550 nm). 156 μg of NOX2-p22 containing membranes were added to a reaction mixture containing 65 mM sodium phosphate buffer pH 7.0 with 1 mM EGTA, 1 mM MgCl2, 2 mM NaN3, 10 μM FAD, 100 μM LiDS, 160 nM recombinant cytosolic proteins p67phox, p47phox, Rac1 Q61L, and 200 μM cytochrome c. NOX5 containing membranes (45 μg) using PBS as assay buffer were added to a reaction mixture containing 2 mM NaN3, 200 μM FAD, 1 mM CaCl2 and 100 μM cytochrome c. Ligands were added to the reaction mixture at a concentration of 100 μM and incubated for 10 min at 25 °C. The reaction was initiated by the addition of 200 μM NADPH. Measurements were performed using a ClarioStar plate reader and Cary 100 UV–Vis spectrophotometer.
MCLA assay on NOX2 and NOX5 membranes - MCLA is a chemiluminescent (654 nm) reagent that is highly sensitive to superoxide. 20 μg of NOX2-p22 containing membranes were added to a reaction mixture containing 65 mM sodium phosphate buffer pH 7.0 with 1 mM EGTA, 1 mM MgCl2, 0.5 μM FAD, 100 μM LiDS, 160 nM recombinant cytosolic proteins p67phox, p47phox, Rac1 Q61L, and 1 μM MCLA. NOX5-containing membranes (12 μg) were added to a reaction mixture of PBS with 1 mM CaCl2, 0.5 μM FAD and 1 μM MCLA. Ligands were added to the reaction mixture at a concentration of 100 μM and incubated for 10 min at 25 °C. The reaction was initiated by the addition of 200 μM NADPH. Measurements were performed using a GloMax plate reader. The activities of NOX2 and NOX5 were evaluated also in the absence of the cytosolic proteins and CaCl2, respectively. SOD was used as an additional negative control. Control membranes not containing over-expressed NOX5 were further used to infer the noise-level signals.
Amplex red/peroxidase assay on NOX4 membranes – In this assay, hydrogen peroxide production is detected through the H2O2-dependent generation of the Resorufin by horseradish peroxidase. NOX4 containing membranes (100 μg) were added to a reaction mixture of PBS with 12.5 μM Amplex Red, 0.02 U/ml horseradish peroxidase and 40 μM FAD. The tested compounds (100 μM) were added to the mixture and incubated for 10 min at 4 °C. The reaction was started by adding 40 μM NADPH, a concentration chosen to minimise interference effects [54]. Measurements were performed using a ClarioStar plate reader (excitation 572 nm/emission 583 nm). Catalase as well as membranes transfected with an empty vector were used as negative control to estimate the background signal.
CBA assay on NOX4 membranes – The CBA assay is a fluorescent assay for the indirect detection of hydrogen peroxide relying on the enzymatic-independent reaction between hydrogen peroxide and the non-fluorescent coumarin boronic acid (CBA) with a 1:1 stoichiometry to form the fluorescent 7-hydroxy-coumarin. NOX4-p22 membranes (50 μg) were added to the PBS buffer containing 20 μM FAD. The tested compounds (100 μM) were added to the reaction mixture and incubated for 10 min at 4 °C. 200 μM CBA and, subsequently, 50 μM NADPH were added to start the reaction. Measurements were performed using a ClarioStar plate reader (excitation 320 nm/emission 430 nm). Because of the lower sensitivity, higher standard deviations were observed with this assay.
2.4. ROS-scavenging assays
Xanthine Oxidase assay - The inhibitory activity on xanthine oxidase was evaluated by measuring the effects of the tested inhibitors (100 μM) on the production of hydrogen peroxide (H2O2) from hypoxanthine, using the Amplex Red/Xanthine Oxidase Assay Kit (Thermofisher Scientific). The tested compounds and adequate amounts of xanthine oxidase were incubated for 15 min at 37 °C in a flat-black-bottom 96-well microplate (Thermofisher) with 100 μM Amplex Red reagent, 0.4 U/mL horseradish peroxidase, and 0.2 mM hypoxanthine. The production of H2O2 and consequently of resorufin was quantified at a ClarioStar plate reader based on the fluorescence generated (excitation 530 nm/emission 590 nm).
NADH-PMS assay - Superoxide radicals were generated in a phenazine methosulfate (PMS)-NADH system by oxidation of NADH and assayed by the reduction of nitroblue tetrazolium. In these experiments, the superoxide radicals were generated in Tris-HCl buffer (16 mM, pH 8.0) containing 80 μM nitroblue tetrazolium, 70 μM NADH and incubated with the tested compounds (100 μM). The reaction started by adding 5 μM phenazine methosulfate to the mixture. The reaction mixture was incubated at 25 °C for 5 min, and then the absorbance was read at 560 nm using a ClarioStar plate reader. SOD was used as a positive control. Decreased absorbance of the reaction mixture compared to the negative control indicated superoxide-scavenging activity from the tested compounds.
2.5. Kinetics evaluation of NOX inhibitors on the purified NOX5 dehydrogenase domain
The inhibitory activity of the tested compounds on the purified C. stagnale dehydrogenase domain was measured using 1 μM protein in a final volume of 130 μL of 50 mM Hepes pH 7.5 (and 300 μM FAD in the case of the wild-type protein) in a Cary 100 UV–visible spectrophotometer (Varian) equipped with a thermo-stated cell holder (T = 25 °C). Reaction rates were determined by following NADPH oxidation at 340 nm.
2.6. Microscale thermophoresis binding assay on purified NOX5 dehydrogenase domain
NOX5 dehydrogenase domain-suramin interactions were characterized by microscale thermophoresis. The ligand was titrated to a maximum concentration of 6.4 mM using a fixed concentration of the fluorescently labelled protein (~200 nM). Sixteen serially diluted titrations of suramin were prepared to generate one full binding isotherm. All binding reactions were carried out in a buffer containing Hepes 50 mM, NaCl 240 mM, 0.5% glycerol, 250 μM FAD and 0.1% pluronic acid at pH 7.5. Samples were loaded into NT.115 premium treated capillaries (Nanotemper Technologies) after the reaction was incubated at 25 °C for 10 min. Consequently, the samples were mounted in the Monolith NT.115 apparatus (Nanotemper Technologies) for binding measurements. The data for microscale thermophoresis analysis were recorded at 25 °C. All binding experiments were followed using a red fluorescence channel (red filter; excitation 605–645 nm, emission 680–685 nm). Data analysis was performed with software NTAffinityAnalysis (Nanotemper Technologies) where the binding isotherms were derived from the raw fluorescence data. The experiments were performed in triplicate and the resulting standard deviation calculated with NTAffinityAnalysis.
2.7. Thermal shift analysis on purified NOX5 dehydrogenase domain
Thermostability assays on wild-type dehydrogenase domain were performed on a TychoTMNT.6 system (NanoTemper Technologies GmbH, Munich, Germany) to assess the effect of the tested ligands on the stability of the purified protein. The experiments were conducted in the absence and presence of ligands. The samples were heated up from 35 °C to 95 °C for the duration of 3 min and their inflection point (Ti) was determined.
2.8. Characterization of DPI binding to NOX5
With regard to the flavin component, 20 μM of dehydrogenase proteins were reduced with 200 μM NADPH and incubated in the presence and absence of 200 μM DPI. UV–Vis spectra were recorded with Agilent Diode Array at 25 °C. With regard to the heme component, the experiments were carried out on the purified wild-type and R256S transmembrane domain of bacterial NOX5. An excess of sodium dithionite was mixed with 6 μM protein in the storage buffer. DPI (1 mM) was added to the solution in order to determine the ligand effect on the heme group of the protein. UV–Vis spectra were recorded with Agilent Diode Array at 25 °C. MS analysis was carried out upon heme extraction in an organic solvent. 30 μM of purified wild-type transmembrane domain of NOX5 was reduced with an excess of sodium dithionite, dissolved in the mixture acetonitrile:1.7 M HCl (8:2 vol/vol) and left stirring at room temperature for 20 min. The reaction was quenched by addition of MgSO4 and NaCl. The mixture was vortexed and then centrifuged for 5 min at 2500×g to separate the unfolded protein from the extracted heme. The same procedure was followed for the reduced transmembrane domain in the presence of 2 mM DPI. 10 μL of each extract sample were analysed by ESI-MS using a LCQFleet Thermo Scientific Ion Trap.
2.9. Characterization of VAS inhibitor binding to NOX5 by ESI-MS
1 μM wild-type or C668S mutant dehydrogenase domain (untreated or after incubation with VAS2870 or VAS3947) dissolved in a mixture of acetonitrile/water/formic acid 0.1% was analysed by ESI-MS using a LCQFleet Thermo Scientific Ion Trap.
2.10. Statistical analysis
All graphs were prepared using GraphPad Prism™ and all data are expressed as mean ± standard deviation. Sigmoidal curve-fitting of concentration-response curve, IC50-values and statistical tests were all performed with GraphPad Prism™. Concentration-response curves are presented as % of the control, with control being the untreated condition. For Ki determination, initial velocities obtained at multiple NADPH and inhibitor concentrations were fitted globally to competitive, uncompetitive and noncompetitive models. The best-fit model was identified based on the R2 parameter given by GraphPad Prism™ software (GraphPad Software, Inc., San Diego, CA).
3. Results and discussion
3.1. Characterization of NOXs and evaluation of their catalytic activity
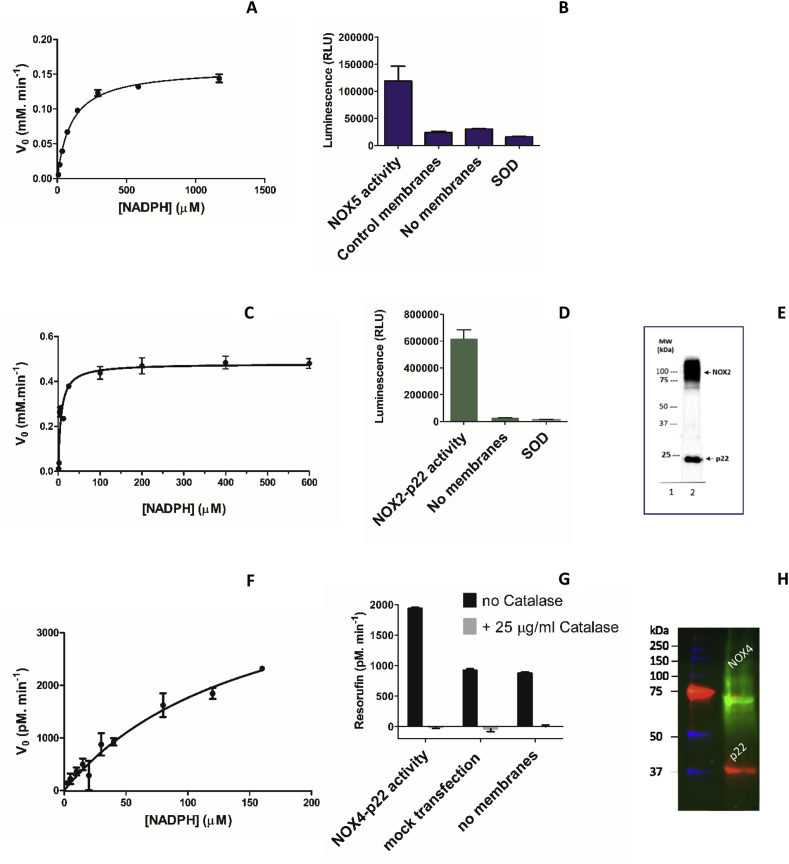
Our work relied on testing each compound on three NOX isoforms; human NOX2 membranes obtained from cultured PLB-985 cells, human NOX4 membranes from enzyme over-expressing HEK293 EBNA-1 cells, and Cylindrospermum stagnale NOX5 over-expressed in E. coli cell membranes (Fig. 2). We chose the C. stagnale NOX5 because it was successfully used for structural studies and is 40% identical in sequence to human NOX5 and two of its domains, dehydrogenase and transmembrane, are available in purified forms [5]. The enzymatic activities were evaluated using several methods: the cytochrome c reduction and MCLA assays were employed for the superoxide-producing NOX5 and NOX2 whereas the hydrogen peroxide-producing NOX4 was evaluated using a peroxidase-coupled and CBA assays. Under our experimental conditions, NOX5 displayed a KMNADPH of 101.4 ± 7.6 μM and a Vmax of 0.159 ± 0.003 mM min−1 (Fig. 2A) whereas NOX2 showed a KMNADPH of 6.6 ± 1.4 μM and a Vmax of 0.48 ± 0.02 mM min−1 (Fig. 2D). The analysis of NOX4 resulted in a KMNADPH of 156.5 ± 35.4 μM and a Vmax of 4524 ± 642 pM min−1, confirming the lower activity of this isoform (Fig. 2F) [[1], [2], [3]].
Fig. 2.
Characterization of NOXs. (A) Michaelis-Menten curve of NOX5 membranes using the cytochrome c reduction assay. (B) Superoxide generation by bacterial NOX5 using the MCLA assay. (C) Michaelis-Menten curve of human NOX2-p22 membranes using the cytochrome c reduction assay. (D) Superoxide generation by NOX2 using the MCLA assay. (E) Western-blot analysis of PLB-985 cell membranes by immunoblotting with NOX2 (54.1) and p22 monoclonal (44.1 antibodies). The arrows indicate the highly glycosylated 91 kDa NOX2, and the non-glycosylated 22 kDa p22 (lane 1: molecular weight marker; lane 2: NOX2-p22 containing membranes after separation on 12% polyacrylamide gel electrophoresis). (F) Michaelis-Menten curve of human NOX4-p22 HEK293 membranes using the Amplex Red-peroxidase assay. (G) Hydrogen peroxide generation by NOX4 and corresponding controls using the Amplex Red-peroxidase coupled assay. Full NADPH saturation could not be achieved because of assay interference. (H) In-gel fluorescence analysis of HEK cells heterologously expressing eGFP-NOX4 (predicted 96 kDa) and DsRed-p22 (predicted 50 kDa) expressing membranes. As often observed for membrane proteins, both fusion proteins display anomalous migration in a SDS PAGE due their hydrophobicity. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.2. Initial evaluation of NOX inhibitors
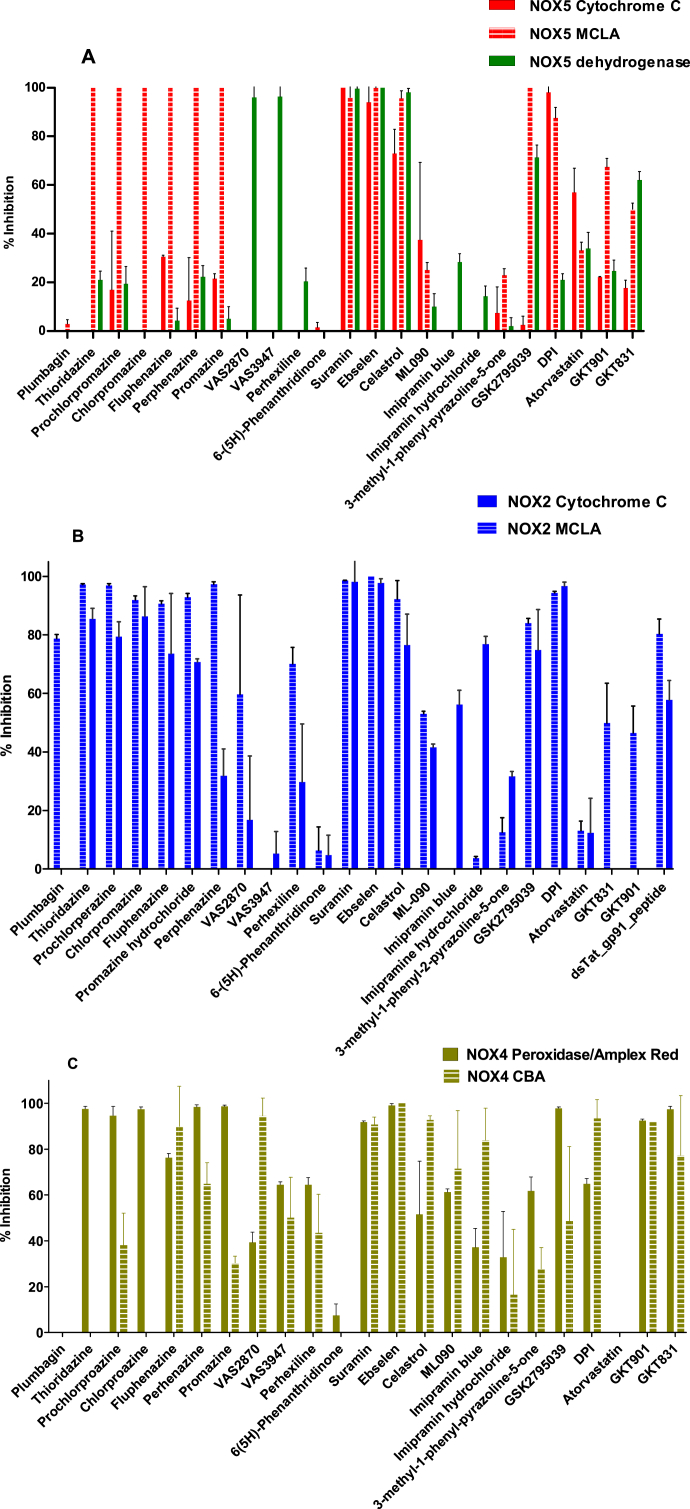
A total of 24 known NOX inhibitors were tested using a standardized protocol that would enable a close comparison of their anti-NOX activities. As many of these compounds were reported to be effective at single- or double-digit μM concentrations [28,31,38,39], we chose to probe them at 100 μM concentrations using 40–200 μM NADPH (Figs. 1A and Fig. 3A–C). A few molecules displayed full (suramin, ebselen, celastrol and DPI) or moderate (ML-090; 40–50%) inhibition on all NOXs. Differently from these pan-inhibitors, the other compounds featured some selectivity. The tat-gp91ds peptide strongly inhibited NOX2, as expected in light of its capacity to interfere with the enzyme activation [41]. The six tested phenothiazine derivatives and GSK2795039 displayed pronounced inhibition (>60%) of NOX2 and NOX4 whereas NOX5 was affected only when assayed with the MCLA-based protocol. VAS2870 and perhexiline displayed low-to-sustained (40–70%) inhibitions against NOX2 and NOX4 but not against NOX5. VAS3947 was active only against NOX4. Atorvastatin was instead active mostly on NOX5 (~50%) whereas the pyrazoline compound featured significant (>40% inhibition) activity only against NOX4. The other compounds exhibited more scattered patterns of inhibition with strong dependencies on the assays. Thus, GKT136901 and GKT137831 proved to be strong NOX4 inhibitors but moderately active against NOX2 and NOX5 only when assayed with MCLA. Likewise, imipramine blue, imipramine chloride, plumbagin, and phenanthridinone displayed assay-dependent inhibitory activities. Collectively, our initial survey demonstrated that most compounds do indeed feature the NOX inhibitory activities under the tested conditions. However, these activities often depend on the assay being used.
Fig. 3.
Activities of the NOX inhibitors (100 μM) evaluated on (A) bacterial NOX5, (B) human NOX2, and (C) human NOX4 membrane preparations.
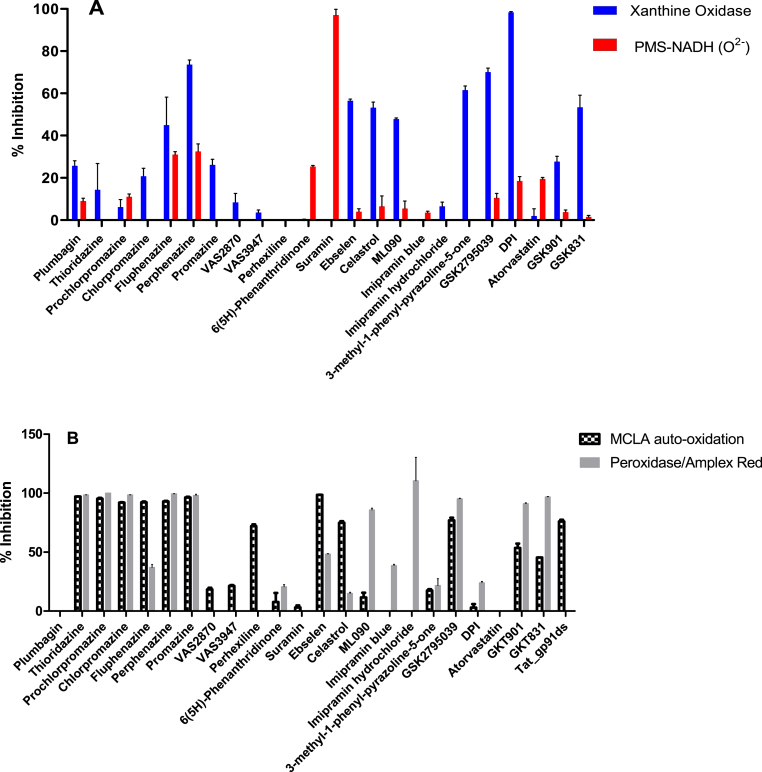
3.3. Evaluation of assay interferences and ROS-scavenging effects
NOX inhibition assays are prone to artefacts that may arise from redox and ROS-scavenging activities of the investigated compounds and/or interference with the reagents used in the enzymatic assays [[27], [28], [29], [30], [31]]. With regard to superoxide, scavenging was verified by a non-enzymatic assay whereby the superoxide generated by the reaction of phenazine methosulfate with NADH reacts with nitroblue tetrazolium to generate a colourful compound (absorption at 560 nm). As for hydrogen peroxide, we employed the H2O2-producing xanthine oxidase and its hypoxanthine substrate using the Amplex Red/peroxidase assay. Overall, most of the studied compounds displayed no significant interference with superoxide. The only exceptions were suramin, which proved to be a strong superoxide scavenger in agreement with previous data [55], and fluphenazine, perphenazine and 6(5H)-phenanthridinone, which showed some mild scavenging activities (Fig. 4A). The H2O2-interfering activities were instead more pervasive. Three phenothiazine derivatives (fluphenazine, perphenazine and promazine), ebselen, celastrol, DPI, 3-methyl-1-phenyl-2-pyrazoline, GKT137831 and ML090 showed powerful inhibitions of the hydrogen peroxide-producing activity of xanthine oxidase (Fig. 4A). These effects could arise from the direct inhibition of xanthine oxidase, the inhibition of the horseradish peroxidase used in the assay, interference with the Amplex Red probe, or a direct H2O2-scavenging effect. In order to clarify this issue and to further explore the redox reactivities of the tested compounds, we performed two additional assays. In the first one, the effect of the anti-NOX compounds on the spontaneous auto-oxidation of the MCLA probe was used as an indicator of some redox and/or off-target effect. In the second assay, the Amplex Red/peroxidase assay was executed by directly adding the inhibitors in the absence of a NOX enzyme (but in the presence of NADPH) to probe for interference with Amplex Red (Fig. 4B). The six phenothiazines, ebselen, GSK2795039, GKT136901 and GKT137831 showed a substantial interference in both assays. Moreover, celastrol and perhexiline interfered with the MCLA auto-oxidation whereas ML090 and the two imipramins displayed potent assay interference on the Amplex Red/peroxidase assay. Finally, we notice that the inhibition of MCLA auto-oxidation by tat-gp91ds is probably not surprising, because this peptidic inhibitor comprises a cysteine that can afford a mild anti-oxidant effect (Fig. 1A). These results were striking because all anti-NOX compounds showed some degree of assays interference, with the only exception of VA2870 and VAS3947.
Fig. 4.
Evaluation of ROS-scavenging and assay-interfering activities of NOX-inhibitors (100 μM) probed by (A) xanthine oxidase and phenazine methosulfate (PMS)-NADH assays and (B) MCLA auto-oxidation and peroxide-coupled assays.
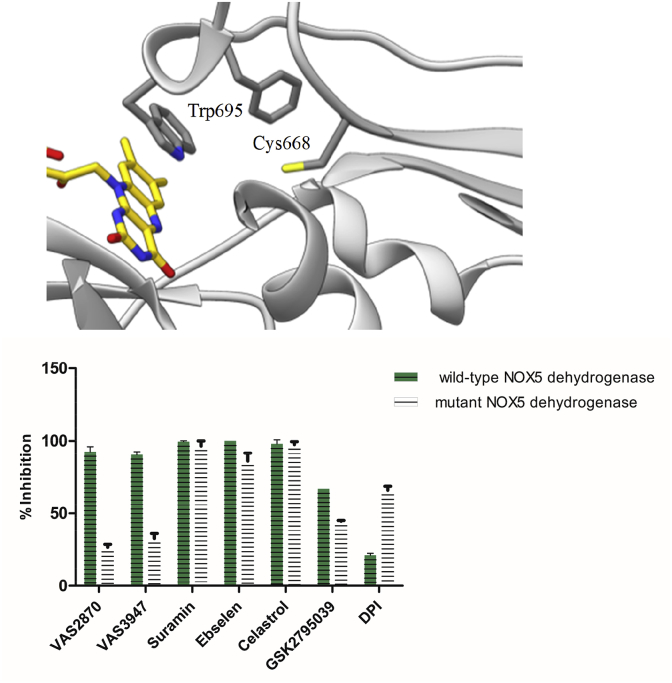
3.4. Inhibitor evaluation and validation using the purified dehydrogenase domain of NOX5
The outcome of the previous experiments indicated that the inhibitory activity of many of the tested compounds may well arise, in part or completely, from assay interference and/or ROS-scavenging activities. To further investigate the issue, we relied on the purified dehydrogenase domain of the bacterial NOX5, which can be expressed as stand-alone protein and is capable to oxidise NADPH using its FAD prosthetic group and oxygen as electron acceptor [5]. This enzymatic activity can be easily measured by a spectrophotometric NADPH-depletion assay to ascertain if certain compounds might function as competitive inhibitors by hindering access of the NADPH substrate. A low-activity domain mutant, where an engineered tryptophan side chain at the C-terminus occupies the binding site for the nicotinamide of NADPH, was used to corroborate the mechanism of inhibition (Fig. 5A) [5]. In fact, inhibitors selectively interfering with NADPH binding should be much more active on the wild-type dehydrogenase than on this mutant protein.
Fig. 5.
The dehydrogenase domain of NOX5 as model system for inhibition studies. (A) The dehydrogenase active site. Trp695 (green) is the stabilising amino acid belonging to the extension PWLEL added to the C-terminus (PDB 5O0X). (B) Inhibition studies (100 μM inhibitors) on the wild-type and C-terminal mutant dehydrogenase. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Only 7 compounds featured significant (>40%) inhibition of the isolated dehydrogenase domain (Fig. 3A). This finding was in sharp contrast with the widespread inhibitions observed against the full-length NOX5. Furthermore, 5 of the 7 hits (suramin, ebselen, celastrol, GSK2795039 and GKT831) displayed similar or identical degree of inhibition towards both wild-type and mutant dehydrogenase (Fig. 5B). Such a lack of selectivity, combined with the lack of any stabilising effect in thermal shift analysis (Supplementary Fig. 1), supported the notion that the inhibition exerted by these compounds is unlikely to arise from a specific interaction with the protein (e.g. with its NADPH-binding site) but rather from their previously-described non-specific effects (see Fig. 4A–B). This concept was validated by additional experiments on suramin. When assayed against the wild-type dehydrogenase domain, this compound showed uncompetitive inhibition with a sub-micromolar inhibition constant (Ki = 0.39 ± 0.05 μM). However, when tested with microscale thermophoresis, suramin featured a 1000-fold lower affinity, which likely reflects its propensity for protein-binding due to its heavily charged structure (Kd = 52.2 ± 1.1 μM; Supplementary Fig. 2). Beside these observations, a more encouraging pattern of data emerged from the characterization of VAS2870 and VAS3947, the other two compounds that proved to be strong dehydrogenase domain inhibitors. These anti-NOX and “non-interfering” (Fig. 3, Fig. 4) compounds displayed minimal inhibition (20–30%) against the mutant dehydrogenase as opposed to the 100% inhibition towards the wild-type dehydrogenase (Fig. 5B). This result was particularly informative as it was suggestive of a specific effect of VAS compounds to the dehydrogenase active site, as opposed to the non-specific effects featured by all other dehydrogenase inhibitors.
3.5. Specific NOX inhibitors and their mechanism of action
The extended analysis described in the previous paragraphs led to the conclusion that the DPI, VAS2870 and VAS3947, and tat-gp91ds are the most convincing bona fide antagonists of NOXs. As listed in Table 1, IC50 measurements proved them as effective low-micromolar or nanomolar NOX inhibitors while being mostly devoid of significant assay-interference or ROS-scavenging effects though DPI inhibits xanthine oxidase (a flavoenzyme) and the Cys-containing tat-gp91 interferes with MCLA (Fig. 4B). At this point, it is important to remark that we do not at all rule out that other compounds among our pool of investigated molecules may directly engage NOXs. However, the pronounced scavenging and interfering effects observed by us and others will inevitably confuse the evaluation of their bona fide binding and inhibitory activity (if any). For instance, we found that GSK2795039 displays a very low IC50 for NOX2 (0.0367 ± 0.0013 μM) but this value factors in the non-specific scavenging and interfering activities of this compound, as discussed above and in agreement with previous findings [[27], [28], [29], [30], [31]].
Table 1.
IC50 determination for selected NOX inhibitors.
| IC50 bacterial NOX5 (μM) | IC50 human NOX2 (μM) | IC50human NOX4c(μM) | |
|---|---|---|---|
| DPI | 3.28 ± 1.32a | 0.0562 ± 0.017a | 2.26 ± 1.18 |
| tat-gp91ds | Inactive at 100 μMb | 10.3 ± 1.2b | Inactive at 100 μMb |
| VAS3947 | Inactive at 100 μMa | Inactive at 100 μMa | 31.9 ± 24.3 |
IC50 determination using the MCLA assay.
IC50 determination using the cytochrome c reduction assay.
IC50 determination with the Amplex Red/HRP peroxidase assay.
Based on these data, we sought to clarify the inhibition mechanism of DPI and the VAS compounds by exploiting recombinant full-length NOX5 and its isolated domains as model enzymes for pharmacological and biophysical studies.
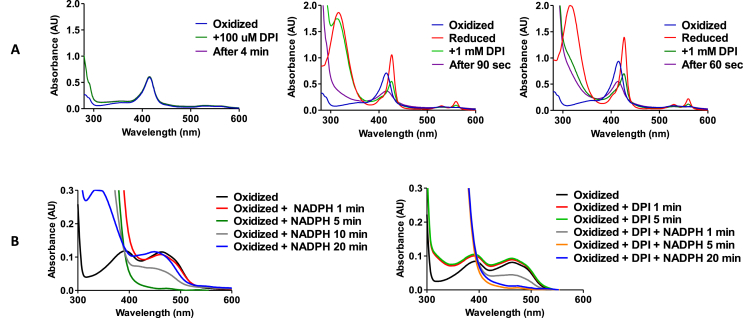
3.5.1. DPI targets the reduced state of the NOX prosthetic groups
The powerful inhibition of full-length NOX5 by DPI was confirmed by kinetic studies on the purified protein, which displayed an uncompetitive inhibition with a Ki = 0.98 ± 0.14 μM (Supplementary Fig. 3). Previous studies suggested that the main target of DPI are the heme groups of NOXs [19]. We sought to experimentally proof this hypothesis by using the recombinant purified transmembrane domain of NOX5 [5]. We first noticed that the oxidised heme spectrum is not perturbed by DPI (Fig. 6A). By contrast, when DPI was added to the dithionite-reduced protein, the rapid quenching of the heme absorbance at 427 nm and of the α-band at 558 nm was observed (Fig. 6A). After re-oxidation (upon consumption of dithionite following its reaction with DPI), the spectrum of the heme remained quenched. These experiments demonstrated that DPI reacts with the reduced transmembrane domain, possibly forming an adduct with heme. Next, this same experiment was performed with the low-activity Arg256Ser transmembrane mutant, which targets a conserved arginine side chain in the oxygen-binding site, adjacent to the extra-cellular heme of the protein [5]. The mutant yielded virtually the same result observed for the wild-type domain, showing that DPI binding does not depend on the fine structure of the heme-binding site (Fig. 6A). We further investigated this feature by HPLC/ESI-MS. Organic extraction carried out on the protein reduced with dithionite and treated with excess of the ligand, did not reveal any difference in the heme spectrum when compared to the spectrum measured on the protein not treated with DPI (Supplementary Fig. 4). Therefore, though stable in the protein environment, the DPI-heme adduct is reversible as it cannot withstand the extraction from the protein. We hypothesise that the adduct involves the formation of a σ-coordination complex between a phenyl group of DPI and the iron as proposed for other heme enzymes treated with heme-reacting compounds and featuring similar spectroscopic properties [56,57].
Fig. 6.
DPI targets the NOXs' prosthetic groups. (A) UV–visible absorption spectra of the oxidised (left), reduced (middle), and reduced R256S (right) NOX5's transmembrane domain after incubation with DPI. Protein reduction was achieved by adding dithionite that absorbs at 315 nm. (B) UV–visible absorption spectra of the mutant NOX5 dehydrogenase after incubation with NADPH (left) and NADPH plus DPI (right).
In addition to the established role as heme ligand, there is ample evidence in the literature that DPI can also bind to flavoproteins [17,58]. We indeed found that DPI strongly and uncompetitively inhibits the low-activity mutant of the NOX5 dehydrogenase domain (αKi = 0.42 ± 0.06 μM) though the inhibition of the wild-type domain is 200-fold weaker (αKi = 127 ± 13 μM; Supplementary Fig. 5). The pronounced inhibition by DPI of the mutant dehydrogenase is probably explained by the tightly bound FAD, that is retained by the protein even during purification, differently from the wild-type domain that instead requires the addition of exogenous FAD (300 μM) to allow activity measurements. The mutant dehydrogenase was therefore chosen for further mechanistic studies. We first noticed that the addition of NADPH leads to the fast reduction of the flavin. The prosthetic group is then slowly re-oxidised with concomitant consumption of NADPH (absorbance at 340 nm). However, upon re-oxidation, the wavelength of the absorbance peak characteristic of the oxidised flavin shifted from 462 nm to 450 nm (Fig. 6B). The latter wavelength peak corresponds to free FAD, demonstrating that the enzyme tends to release the FAD after its reduction. We then performed the same experiment in the presence of DPI. The incubation with the inhibitor did not affect the spectrum of the oxidised protein but, differently from the un-inhibited domain, no re-oxidation was observed after NADPH addition (Fig. 6B). These observations resemble previous findings on other flavoproteins treated with DPI: both flavin reduction by NADPH and retention of the flavin by the enzyme are necessary for DPI to react and afford inhibition through a stable reduced flavin-DPI adduct [17,58]. This mechanism is consistent with the observed pattern of uncompetitive inhibition and explains why the poorly FAD-retaining wild-type dehydrogenase is hardly affected by DPI. In summary, the NOX inhibition by DPI arises from its relatively non-specific reaction with the reduced states of both flavin and heme cofactors, possibly with a preponderant contribution of the two heme groups.
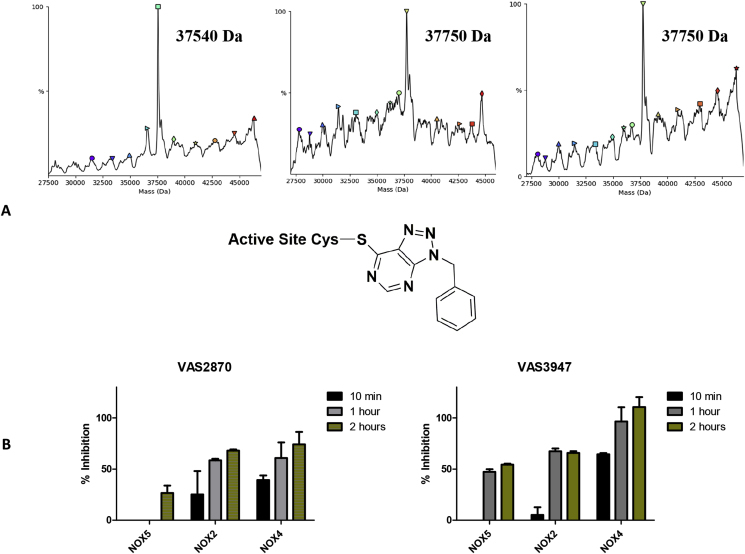
3.6. VAS2870 and VAS3947 alkylates a conserved active-site cysteine
VAS2870 and VAS3947 displayed very significant potency on the wild-type dehydrogenase domain of NOX5 with IC50 of 0.257 ± 0.106 μM and 6.09 ± 1.41 μM, respectively (Supplementary Fig. 6). In the course of the evaluation of these compounds, we considered their intense absorbance at 280 nm and noticed that it contributed to the absorbance of purified inhibited proteins that showed a higher absorbance after gel-filtration. This observation led us to hypothesise the formation of an adduct between the compounds and the enzyme, following previous literature indicating that VAS compounds can react with protein cysteines [59]. The gel-filtered proteins with or without VAS2870 were thereby analysed by ESI-MS. The results were unambiguous in that a 210 Da increase in the inhibitor-treated protein molecular weight was observed. This was fully consistent with the alkylation of a cysteine's thiol by the benzyl-triazolopyrimidine moiety (210 Da) of VAS2870, while the benzoxazoyl group acted as a leaving group (Fig. 7A). The same was observed for VAS3947, encompassing an oxazolyl ring as a leaving group, thus leaving a molecule of 210 Da covalently bound to the dehydrogenase domain (Fig. 7A).
Fig. 7.
VAS2870 and VAS3947 covalently inhibit NOX. (A) Deconvoluted mass spectra from NOX5 dehydrogenase without inhibitor (left), after incubation with VAS2870 (middle), and after incubation with VAS3947 (right) together with the structure of the Cys-inhibitor adduct. (B) The effect of incubation times on the NOXs inhibition.
Having showed that VAS2870 and VAS3947 act as covalent ligands of the dehydrogenase domain, we sought to identify which cysteine was involved in the covalent binding. The dehydrogenase domain of C. stagnale NOX5 contains four cysteines [5]. We focused on a conserved cysteine present in the active site as the one which could be targeted by the VAS derivatives (Cys668; Fig. 5A). Accordingly, we performed mutagenesis to replace Cys688 by a serine. The purified mutant exhibited Michaelis-Menten parameters (KMNADPH = 6.8 ± 1.5 and Kcat = 20 ± 2 min−1) similar to those of the wild-type protein (KMNADPH = 12 ± 3 μM, Kcat = 56 ± 5 min−1) [5]. Most interestingly, enzymatic assays performed with both VAS derivatives showed that the compounds could not inhibit the mutant domain. Consistently with this observation, we observed no increase in the mutated protein molecular weight upon incubation with the inhibitors, demonstrating that no adduct is formed in absence of Cys688 (Supplementary Fig. 7). Importantly, the targeted cysteine is absolutely conserved among NOXs. Indeed, VAS2870 and VAS3947 are powerful NOX4 inhibitors (Table 1 and Fig. 3). The potency of the inhibition increases with the time of incubation, as it can be expected from poorly soluble (logPVAS2870 = 3.6) covalent inhibitors. Consistently, with sufficiently long incubation times, also the inhibition of NOX2 and NOX5 became significant (Fig. 7B). The higher sensitivity to VAS compounds by the isolated dehydrogenase domain of NOX5 compared to the full-length protein (Table 1; Fig. 7; Supplementary Fig. 6) likely reflects a decreased reactivity and/or accessibility of the active-site cysteine when the domain is embedded in the membrane environment of the full-length protein. Interestingly, a role of this conserved residue in enzyme regulation has been suggested for the plant NOX5 orthologue RBOHD [60]. The key conclusion from these experiments is the demonstration of the VAS derivatives as direct active-site modifiers of NOXs.
4. Conclusions
Development of NOX inhibitors is especially challenging because many chemicals have redox and ROS-scavenging activities. These features have to be critically evaluated in order to deconvolute the inhibition arising from the specific binding of a compound to functional site(s) of the protein from non-specific effects. Considering the major role played in pathophysiological states by NOXs and, more in general by ROS, it seems paramount to assess the ROS-scavenging properties of any novel drug under development. Our survey of two dozen known anti-NOX compounds revealed that the vast majority of them feature assay-interfering and ROS-scavenging properties. We suspect that these non-specific effects greatly contribute to their (in many cases strong) NOX inhibition capacity. Our work also showed that orthogonal assays on different NOX isoforms and domains, together with recently developed technologies to probe protein-ligand binding, can effectively help to clarify the mode of inhibition by a target compound. DPI is firmly demonstrated to engage the heme and possibly the flavin cofactors. This compound suffers from many limitations because it acts on many targets and it is redox reactive. Nevertheless, it also features valuable properties. Its mode of inhibition (i.e. reacting only with the reduced prosthetic groups) implies that it will act only on activated NOXs. In addition, it may be the prototype of compounds that do not need to enter cell by targeting the ROS-producing heme on the external side of the cell-membrane. VAS2870 and VAS3947 demonstrate that a conserved active-site cysteine can be targeted for effective covalent inhibition. Importantly, this mode of inhibition affects the first step of the reaction (NADPH binding and FAD reduction), blocking NOXs at the beginning of their catalytic cycle. These findings open new avenues towards powerful and selective NOX inhibitors.
Author contributions
J.R., S.M., M.M. and M.C. prepared NOX2, NOX4, and NOX5, and performed all inhibition experiments. F.S.A. performed mutagenesis. F.C. supported the mass-spec experiments. S.V. and A.M. supervised chemical syntheses and interfering assays. J.R., F.M., and A.M. conceived the experiments and wrote the manuscript.
Funding
Research on NOX proteins was supported by the Associazione Italiana per la Ricerca sul Cancro (AIRC; IG19808) and the “Dipartimenti di Eccellenza (2018–2022)”. The project leading to these results has received funding from AIRC and from the European Union's Horizon 2020 research and innovation programme under the Marie Skłodowska-Curie grant agreement No 800924.
Declaration of competing interest
The authors declare that they have no conflict of interests.
Acknowledgements
We thank Edgar Pick (Tel Aviv University) for generously providing the plasmids for the expression of the NOX2-activating protein factors.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2020.101466.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Block K., Gorin Y. Aiding and abetting roles of NOX oxidases in cellular transformation. Nat. Rev. Canc. 2012;12:627–637. doi: 10.1038/nrc3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lambeth J.D., Neish A.S. Nox enzymes and new thinking on reactive oxygen: a double-edged sword revisited. Annu. Rev. Pathol. 2014;9:119–145. doi: 10.1146/annurev-pathol-012513-104651. [DOI] [PubMed] [Google Scholar]
- 3.Magnani F., Mattevi A. Structure and mechanisms of ROS generation by NADPH oxidases. Curr. Opin. Struct. Biol. 2019;59:91–97. doi: 10.1016/j.sbi.2019.03.001. [DOI] [PubMed] [Google Scholar]
- 4.Bedard K., Krause K.H. The NOX family of ROS-generating NADPH oxidases: physiology and pathophysiology. Physiol. Rev. 2007;87:245–313. doi: 10.1152/physrev.00044.2005. [DOI] [PubMed] [Google Scholar]
- 5.Magnani F., Nenci S., Millana Fananas E., Ceccon M., Romero E., Fraaije M.W. Crystal structures and atomic model of NADPH oxidase. Proc. Natl. Acad. Sci. 2017;114:6764–6769. doi: 10.1073/pnas.1702293114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nisimoto Y., Jackson H.M., Ogawa H., Kawahara T., Lambeth J.D. Constitutive NADPH-dependent electron transferase activity of the Nox4 dehydrogenase domain. Biochemistry. 2010;49:2433–2442. doi: 10.1021/bi9022285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weyemi U., Lagente-Chevallier O., Boufraqech M., Prenois F., Courtin F., Caillou B. ROS-generating NADPH oxidase NOX4 is a critical mediator in oncogenic H-Ras-induced DNA damage and subsequent senescence. Oncogene. 2012;31:1117–1129. doi: 10.1038/onc.2011.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ogrunc M., Di Micco R., Liontos M., Bombardelli L., Mione M., Fumagalli M. Oncogene-induced reactive oxygen species fuel hyperproliferation and DNA damage response activation. Cell Death Differ. 2014;21:998–1012. doi: 10.1038/cdd.2014.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ju H.-Q., Ying H., Tian T., Ling J., Fu J., Lu Y. Mutant Kras- and p16-regulated NOX4 activation overcomes metabolic checkpoints in development of pancreatic ductal adenocarcinoma. Nat. Commun. 2017;8:14437. doi: 10.1038/ncomms14437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crosas-Molist E., Bertran E., Rodriguez-Hernandez I., Herraiz C., Cantelli G., À Fabra. The NADPH oxidase NOX4 represses epithelial to amoeboid transition and efficient tumour dissemination. Oncogene. 2017;36:3002–3014. doi: 10.1038/onc.2016.454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grandvaux N., Soucy-Faulkner A., Fink K. Innate host defense: nox and Duox on phox's tail. Biochimie. 2007;89:1113–1122. doi: 10.1016/j.biochi.2007.04.008. [DOI] [PubMed] [Google Scholar]
- 12.O'Neill S., Brault J., Stasia M.-J., Knaus U.G. Genetic disorders coupled to ROS deficiency. Redox Biol. 2015;6:135–156. doi: 10.1016/j.redox.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pal R., Palmieri M., Loehr J.A., Li S., Abo-Zahrah R., Monroe T.O. Src-dependent impairment of autophagy by oxidative stress in a mouse model of Duchenne muscular dystrophy. Nat. Commun. 2014;5:4425. doi: 10.1038/ncomms5425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Loehr J.A., Wang S., Cully T.R., Pal R., Larina I.V., Larin K.V. NADPH oxidase mediates microtubule alterations and diaphragm dysfunction in dystrophic mice. Elife. 2018;7 doi: 10.7554/eLife.31732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Singel K.L., Segal B.H. NOX2-dependent regulation of inflammation. Clin. Sci. (Lond.) 2016;130:479–490. doi: 10.1042/CS20150660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu J., Risbood P., Kane C.T., Hossain M.T., Anderson L., Hill K. Characterization of potent and selective iodonium-class inhibitors of NADPH oxidases. Biochem. Pharmacol. 2017;143:25–38. doi: 10.1016/j.bcp.2017.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chakraborty S., Massey V. Reaction of reduced flavins and flavoproteins with diphenyliodonium chloride. J. Biol. Chem. 2002;277:41507–41516. doi: 10.1074/jbc.M205432200. [DOI] [PubMed] [Google Scholar]
- 18.O'Donnell B.V., Tew D.G., Jones O.T., England P.J. Studies on the inhibitory mechanism of iodonium compounds with special reference to neutrophil NADPH oxidase. Biochem. J. 1993;290:41–49. doi: 10.1042/bj2900041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Doussiere J., Gaillard J., Vignais P.V. The heme component of the neutrophil NADPH oxidase complex is a target for aryliodonium compounds. Biochemistry. 1999;38:3694–3703. doi: 10.1021/bi9823481. [DOI] [PubMed] [Google Scholar]
- 20.Doroshow J.H., Juhasz A., Ge Y., Holbeck S., Lu J., Antony S. Antiproliferative mechanisms of action of the flavin dehydrogenase inhibitors diphenylene iodonium and di-2-thienyliodonium based on molecular profiling of the NCI-60 human tumor cell panel. Biochem. Pharmacol. 2012;83:1195–1207. doi: 10.1016/j.bcp.2012.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Petrônio M.S., Zeraik M.L., Fonseca LMd, Ximenes V.F. Apocynin: chemical and biophysical properties of a NADPH oxidase inhibitor. Molecules. 2013;18:2821–2839. doi: 10.3390/molecules18032821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sun J., Ming L., Shang F., Shen L., Chen J., Jin Y. Apocynin suppression of NADPH oxidase reverses the aging process in mesenchymal stem cells to promote osteogenesis and increase bone mass. Sci. Rep. 2015;5:18572. doi: 10.1038/srep18572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Y., Trush M.A. Diphenyleneiodonium, an NAD(P)H oxidase inhibitor, also potently inhibits mitochondrial reactive oxygen species production. Biochem. Biophys. Res. Commun. 1998;253:295–299. doi: 10.1006/bbrc.1998.9729. [DOI] [PubMed] [Google Scholar]
- 24.Sanders S.A., Eisenthal R., Harrison R. NADH oxidase activity of human xanthine oxidoreductase. Eur. J. Biochem. 1997;245:541–548. doi: 10.1111/j.1432-1033.1997.00541.x. [DOI] [PubMed] [Google Scholar]
- 25.Stuehr D.J., Fasehun O.A., Kwon N.S., Gross S.S., Gonzalez J.A., Levi R. Inhibition of macrophage and endothelial cell nitric oxide synthase by diphenyleneiodonium and its analogs. Faseb. J. 1991;5:98–103. doi: 10.1096/fasebj.5.1.1703974. [DOI] [PubMed] [Google Scholar]
- 26.Voogd T.E., Vansterkenburg E.L., Wilting J., Janssen L.H. Recent research on the biological activity of suramin. Pharmacol. Rev. 1993;45:177. [PubMed] [Google Scholar]
- 27.Gatto G.J., Jr., Ao Z., Kearse M.G., Zhou M., Morales C.R., Daniels E. NADPH oxidase-dependent and -independent mechanisms of reported inhibitors of reactive oxygen generation. J. Enzym. Inhib. Med. Chem. 2013;28:95–104. doi: 10.3109/14756366.2011.636360. [DOI] [PubMed] [Google Scholar]
- 28.Augsburger F., Filippova A., Rasti D., Seredenina T., Lam M., Maghzal G. Pharmacological characterization of the seven human NOX isoforms and their inhibitors. Redox Biol. 2019;26:101272. doi: 10.1016/j.redox.2019.101272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dao V.T., Altenhöfer S., Elbatreek M.H., Casas A.I., Lijnen P., Meens M.J. Isoform-specific NADPH oxidase inhibition for pharmacological target validation. bioRxiv. 2018:382226. [Google Scholar]
- 30.Dao V.T.-V., Elbatreek M.H., Altenhöfer S., Casas A.I., Pachado M.P., Neullens C.T. Isoform-selective NADPH oxidase inhibitor panel for pharmacological target validation. Free Radic. Biol. Med. 2020;148:60–69. doi: 10.1016/j.freeradbiomed.2019.12.038. [DOI] [PubMed] [Google Scholar]
- 31.Altenhöfer S., Radermacher K.A., Kleikers P.W.M., Wingler K., Schmidt H.H.H.W. Evolution of NADPH oxidase inhibitors: selectivity and mechanisms for target engagement. Antioxidants Redox Signal. 2014;23:406–427. doi: 10.1089/ars.2013.5814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seredenina T., Chiriano G., Filippova A., Nayernia Z., Mahiout Z., Fioraso-Cartier L. A subset of N-substituted phenothiazines inhibits NADPH oxidases. Free Radic. Biol. Med. 2015;86:239–249. doi: 10.1016/j.freeradbiomed.2015.05.023. [DOI] [PubMed] [Google Scholar]
- 33.Van Heerebeek L., Meischl C., Stooker W., Meijer C.J.L.M., Niessen H.W.M., Roos D. NADPH oxidase(s): new source(s) of reactive oxygen species in the vascular system? J. Clin. Pathol. 2002;55:561–568. doi: 10.1136/jcp.55.8.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Killalea S.M., Krum H. Systematic review of the efficacy and safety of perhexiline in the treatment of ischemic heart disease. Am. J. Cardiovasc. Drugs. 2001;1:193–204. doi: 10.2165/00129784-200101030-00005. [DOI] [PubMed] [Google Scholar]
- 35.Ding Y., Chen Z.-J., Liu S., Che D., Vetter M., Chang C.-H. Inhibition of Nox-4 activity by plumbagin, a plant-derived bioactive naphthoquinone. J. Pharm. Pharmacol. 2005;57:111–116. doi: 10.1211/0022357055119. [DOI] [PubMed] [Google Scholar]
- 36.Munson J.M., Fried L., Rowson S.A., Bonner M.Y., Karumbaiah L., Diaz B. Anti-invasive adjuvant therapy with imipramine blue enhances chemotherapeutic efficacy against glioma. Sci. Transl. Med. 2012;4:127ra36. doi: 10.1126/scitranslmed.3003016. [DOI] [PubMed] [Google Scholar]
- 37.Klingenberg M., Becker J., Eberth S., Kube D., Wilting J. The NADPH oxidase inhibitor imipramine-blue in the treatment of burkitt lymphoma. Mol. Canc. Therapeut. 2014;13:833. doi: 10.1158/1535-7163.MCT-13-0688. [DOI] [PubMed] [Google Scholar]
- 38.Gorin Y., Cavaglieri R.C., Khazim K., Lee D.-Y., Bruno F., Thakur S. Targeting NADPH oxidase with a novel dual Nox1/Nox4 inhibitor attenuates renal pathology in type 1 diabetes. Am. J. Physiol. Ren. Physiol. 2015;308:F1276–F1287. doi: 10.1152/ajprenal.00396.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Teixeira G., Szyndralewiez C., Molango S., Carnesecchi S., Heitz F., Wiesel P. Therapeutic potential of NADPH oxidase 1/4 inhibitors. Br. J. Pharmacol. 2017;174:1647–1669. doi: 10.1111/bph.13532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hirano K., Chen W.S., Chueng A.L.W., Dunne A.A., Seredenina T., Filippova A. Discovery of GSK2795039, a novel small molecule NADPH oxidase 2 inhibitor. Antioxidants Redox Signal. 2015;23:358–374. doi: 10.1089/ars.2014.6202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rey F.E., Cifuentes M.E., Kiarash A., Quinn M.T., Pagano P.J. Novel competitive inhibitor of NAD(P)H oxidase assembly attenuates vascular O(2)(-) and systolic blood pressure in mice. Circ. Res. 2001;89:408–414. doi: 10.1161/hh1701.096037. [DOI] [PubMed] [Google Scholar]
- 42.Node K., Fujita M., Kitakaze M., Hori M., Liao J.K. Short-term statin therapy improves cardiac function and symptoms in patients with idiopathic dilated cardiomyopathy. Circulation. 2003;108:839–843. doi: 10.1161/01.CIR.0000084539.58092.DE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Violi F., Carnevale R., Pastori D., Pignatelli P. Antioxidant and antiplatelet effects of atorvastatin by Nox2 inhibition. Trends Cardiovasc. Med. 2014;24:142–148. doi: 10.1016/j.tcm.2013.09.006. [DOI] [PubMed] [Google Scholar]
- 44.Selemidis S., Sobey C.G., Wingler K., Schmidt H.H.H.W., Drummond G.R. NADPH oxidases in the vasculature: molecular features, roles in disease and pharmacological inhibition. Pharmacol. Ther. 2008;120:254–291. doi: 10.1016/j.pharmthera.2008.08.005. [DOI] [PubMed] [Google Scholar]
- 45.Gabor B., Monika H., Attila V., Krisztina F., Attila M., Laszlo O. Optimization of important early ADME(T) parameters of NADPH oxidase-4 inhibitor molecules. Med. Chem. 2012;8:174–181. doi: 10.2174/157340612800493674. [DOI] [PubMed] [Google Scholar]
- 46.Whitehouse S., Chen P.-L., Greenshields A.L., Nightingale M., Hoskin D.W., Bedard K. Resveratrol, piperine and apigenin differ in their NADPH-oxidase inhibitory and reactive oxygen species-scavenging properties. Phytomedicine. 2016;23:1494–1503. doi: 10.1016/j.phymed.2016.08.011. [DOI] [PubMed] [Google Scholar]
- 47.Jaquet V., Marcoux J., Forest E., Leidal K.G., McCormick S., Westermaier Y. NADPH oxidase (NOX) isoforms are inhibited by celastrol with a dual mode of action. Br. J. Pharmacol. 2011;164:507–520. doi: 10.1111/j.1476-5381.2011.01439.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nakamura Y., Feng Q., Kumagai T., Torikai K., Ohigashi H., Osawa T. Ebselen, a glutathione peroxidase mimetic seleno-organic compound, as a multifunctional antioxidant: implication for inflammation-associated carcinogenesis. J. Biol. Chem. 2002;277:2687–2694. doi: 10.1074/jbc.M109641200. [DOI] [PubMed] [Google Scholar]
- 49.Smith S.M.E., Min J., Ganesh T., Diebold B., Kawahara T., Zhu Y. Ebselen and congeners inhibit NADPH oxidase 2-dependent superoxide generation by interrupting the binding of regulatory subunits. Chem. Biol. 2012;19:752–763. doi: 10.1016/j.chembiol.2012.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Solbak S.M., Zang J., Narayanan D., Høj L.J., Bucciarelli S., Softley C. Developing inhibitors of the p47phox-p22phox protein-protein interaction by fragment-based drug discovery. J. Med. Chem. 2020;63(3):1156–1177. doi: 10.1021/acs.jmedchem.9b01492. [DOI] [PubMed] [Google Scholar]
- 51.Ceccon M., Millana Fananas E., Massari M., Mattevi A., Magnani F. Engineering stability in NADPH oxidases: a common strategy for enzyme production. Mol. Membr. Biol. 2017;34:67–76. doi: 10.1080/09687688.2018.1535141. [DOI] [PubMed] [Google Scholar]
- 52.Brenner S., Whiting-Theobald N.L., Linton G.F., Holmes K.L., Anderson-Cohen M., Kelly P.F. Concentrated RD114-pseudotyped MFGS-gp91phox vector achieves high levels of functional correction of the chronic granulomatous disease oxidase defect in NOD/SCID/beta -microglobulin-/- repopulating mobilized human peripheral blood CD34+ cells. Blood. 2003;102:2789–2797. doi: 10.1182/blood-2002-05-1482. [DOI] [PubMed] [Google Scholar]
- 53.Durocher Y., Perret S., Kamen A. High-level and high-throughput recombinant protein production by transient transfection of suspension-growing human 293-EBNA1 cells. Nucleic Acids Res. 2002;30:E9. doi: 10.1093/nar/30.2.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhao B., Summers F.A., Mason R.P. Photooxidation of Amplex red to resorufin: implications of exposing the Amplex red assay to light. Free Radic. Biol. Med. 2012;53:1080–1087. doi: 10.1016/j.freeradbiomed.2012.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sahu D., Sharma S., Singla R.K., Panda A.K. Antioxidant activity and protective effect of suramin against oxidative stress in collagen induced arthritis. Eur. J. Pharmaceut. Sci. 2017;101:125–139. doi: 10.1016/j.ejps.2017.02.013. [DOI] [PubMed] [Google Scholar]
- 56.Ortiz de Montellano P.R., Kerr D.E. Inactivation of catalase by phenylhydrazine. Formation of a stable aryl-iron heme complex. J. Biol. Chem. 1983;258:10558–10563. [PubMed] [Google Scholar]
- 57.Augusto O., Beilan H.S., Ortiz de Montellano P.R. The catalytic mechanism of cytochrome P-450. Spin-trapping evidence for one-electron substrate oxidation. J. Biol. Chem. 1982;257:11288–11295. [PubMed] [Google Scholar]
- 58.Covès J., Lebrun C., Gervasi G., Dalbon P., Fontecave M. Overexpression of the FAD-binding domain of the sulphite reductase flavoprotein component from Escherichia coli and its inhibition by iodonium diphenyl chloride. Biochem. J. 1999;342:465–472. [PMC free article] [PubMed] [Google Scholar]
- 59.Sun Q.-A., Hess D.T., Wang B., Miyagi M., Stamler J.S. Off-target thiol alkylation by the NADPH oxidase inhibitor 3-benzyl-7-(2-benzoxazolyl)thio-1,2,3-triazolo[4,5-d]pyrimidine (VAS2870) Free Radic. Biol. Med. 2012;52:1897–1902. doi: 10.1016/j.freeradbiomed.2012.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yun B.-W., Feechan A., Yin M., Saidi N.B.B., Le Bihan T., Yu M. S-nitrosylation of NADPH oxidase regulates cell death in plant immunity. Nature. 2011;478:264–268. doi: 10.1038/nature10427. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.