Abstract
New strategies to characterize the effects of engineered nanomaterials (ENMs) based on omics technologies are emerging. However, given the intricate interplay of multiple regulatory layers, the study of a single molecular species in exposed biological systems might not allow the needed granularity to successfully identify the pathways of toxicity (PoT) and, hence, portraying adverse outcome pathways (AOPs). Moreover, the intrinsic diversity of different cell types composing the exposed organs and tissues in living organisms poses a problem when transferring in vivo experimentation into cell-based in vitro systems.
To overcome these limitations, we have profiled genome-wide DNA methylation, mRNA and microRNA expression in three human cell lines representative of relevant cell types of the respiratory system, A549, BEAS-2B and THP-1, exposed to a low dose of ten carbon nanomaterials (CNMs) for 48 h. We applied advanced data integration and modelling techniques in order to build comprehensive regulatory and functional maps of the CNM effects in each cell type.
We observed that different cell types respond differently to the same CNM exposure even at concentrations exerting similar phenotypic effects. Furthermore, we linked patterns of genomic and epigenomic regulation to intrinsic properties of CNM. Interestingly, DNA methylation and microRNA expression only partially explain the mechanism of action (MOA) of CNMs. Taken together, our results strongly support the implementation of approaches based on multi-omics screenings on multiple tissues/cell types, along with systems biology-based multi-variate data modelling, in order to build more accurate AOPs.
Keywords: Carbon nanomaterials, Systems toxicology, Mechanism of action, Toxicogenomics, Multi-omics, Adverse outcome pathway
Graphical abstract
Highlights
-
•
DNA methylation and microRNA expression partially explain the carbon nanomaterials (CNM) mechanism of action (MOA).
-
•
CNM geometrical properties impact several molecular districts of exposed cells.
-
•
Same CNM exposure exerts different MOA in different cell types even at concentrations causing similar phenotypic effects.
-
•
Multi-omics screenings on multiple cell types, along with multi-variate data modelling, allow building more accurate AOPs.
1. Introduction
Since the rapid expansion of the nanotechnology, nanotoxicology has emerged as an important discipline to ensure safe innovation. Nanomaterial toxicity is currently investigated by extensive animal studies, but more and more emphasis has been put into in vitro-based approaches as well as in understanding the relevant molecular mechanisms involved in engineered nanomaterials (ENMs) exposures. Several studies have already addressed the importance of profiling the transcriptome of exposed biological systems for understanding the molecular alterations caused in response to ENMs exposure (Nel, 2013; Nel et al., 2013; Marx-Stoelting et al., 2015; Jennen et al., 2011; Robinson et al., 2012; Tralau et al., 2015). However, the separate investigation of certain molecular domains does not allow to build a thorough landscape of the ENMs mechanism of action (MOA). Hence, to better model and predict the long-term adaptation of an exposed biological system, more comprehensive and integrative analyses, interrogating multiple molecular districts, need to be carried out. Still, to the best of our knowledge, there have not yet been to date comprehensive attempts to study ENMs MOA using a multi-omics approach.
In humans, exposure to ENMs mainly happens in production environments via the airways. We and others already showed that ENMs are able to exert toxic effects on the respiratory system, by using animal exposure models (Kinaret et al., 2017a; Rydman et al., 2015; Rydman et al., 2014). Apart from the pathological responses, ENMs MOA has been investigated in different tissues (Kinaret et al., 2017b; Fröhlich, 2017; Costa and Fadeel, 2016), by analyzing the molecular perturbations that these materials are able to induce on the normal transcriptional program of exposed cells and tissues.
However, responding to the constant pressure in developing reliable and efficient alternative screening methods, many studies tried to predict the potential ENMs effects in vivo by analyzing their effects in vitro (Drasler et al., 2017; Braakhuis, 2015; Törnqvist et al., 2014; Bachler et al., 2015). We recently demonstrated that carbon based nanomaterials have distinct transcriptional MOA between in vivo and in vitro experimental settings and that commonalities are to be found by using comprehensive gene network models (Kinaret et al., 2017b).
Changes in the levels of gene transcription can be usually appreciated at short term and acute responses, but they are less effective in explaining longer effects of ENMs exposure, which are of greater interest to model the real-life exposure scenarios. More recently, a number of studies have focused on the alterations caused by ENMs exposure at the level of DNA methylation (Chen et al., 2017; Sierra et al., 2017). Investigating epigenomic mechanisms, such as alteration of DNA methylation and microRNA expression, can indeed capture more persistent molecular changes underlying long-term transcriptional programs.
In this study, we exposed three human lung-derived cell lines, representative of major cell types of the respiratory system, to sublethal doses of ten different carbon nanomaterials for two days, assessed their genome-wide effects on three distinct molecular layers simultaneously, the DNA methylation, the microRNA and mRNA expression, and modelled their functional interactions.
By performing a comprehensive integrative analysis, we provide a broad picture of the cross-talk between regulatory factors (DNA methylation and miRNAs) and mRNA deregulation following the exposure to carbon nanomaterials. Further, we show how the MOA of the same carbon nanomaterials vary according to the cell type/tissue of origin, thus highlighting the importance of considering a heterogeneous and representative set of cell types for a target organ when testing the effects of engineered nanomaterials in vitro.
2. Results and discussion
2.1. Experimental setup
In this study, we investigated the effects of ten well characterized carbon nanomaterials (CNMs, Table 1) on three cell lines of human alveolar epithelium (A549), bronchial epithelium (BEAS-2B) and macrophages (PMA-differentiated THP-1). We focused on low dose exposure for 48 h and thoroughly characterized their effects by interrogating three different molecular districts genome-wide, the mRNA, microRNA, and DNA methylation.
Table 1.
Tested nanomaterials and their characteristics.
| Material name | Producer | Acronym | Type | Length (nm) | Diameter (nm) | Surface area (m2/g) | Aspect ratio | References |
|---|---|---|---|---|---|---|---|---|
| Carbon black (Evonik) | Evonik industries/Degussa | CBL | Particle | 14 | 14 | 265 | 1 | (Vippola et al., 2009) |
| Fullerene C60 (MTR) | MTR Ltd. | FUL | Sphere | 100 | 100 | 20 | 1 | (Lehto et al., 2014) |
| Graphite nanofibers (Sigma) | Sigma-Aldrich | GNF | Fiber | 10,000 | 140 | 32 | 71.4 | (Vippola et al., 2009) |
| Singlewalled carbon nanotube (Sigma) | Sigma-Aldrich | SIG_SW | Tube | 50,000 | 1.1 | 567 | 45,454 | (Vippola et al., 2009) |
| Singlewalled carbon nanotube (SES) | SES research | SES_SW | Tube | 1500 | 2 | 436 | 750 | (Vippola et al., 2009) |
| Multiwalled carbon nanotube (Bayer) | Bayer material science | BAY_MW | Tube | 1000 | 14.5 | 204 | 68.9 | (Vippola et al., 2009) |
| Multiwalled carbon nanotube (Mitsui) | Mitsui & Co. | MIT_MW | Tube | 13,000 | 50 | 22 | 260 | (Kinaret et al., 2017a) |
| Multiwalled carbon nanotube (SES) | SES research | SES_MW | Tube | 1500 | 20 | 60 | 75 | (Vippola et al., 2009) |
| Multiwalled carbon nanotube (cheaptubes) | Cheaptubes Inc. | CHT_MW | Tube | 30,000 | 11.5 | 180 | 2608 | (Rydman et al., 2015) |
| Multiwalled carbon nanotube (Sigma) | Sigma-Aldrich | SIG_MW | Tube | 100,000 | 15 | 119 | 6666 | (Vippola et al., 2009) |
2.2. Cell viability and cytotoxicity
Most of the in vitro exposure studies concentrate on acute effects (usually within 24 h), with relatively high ENM concentrations that do not correlate well with in vivo conditions or with the human long-term exposure scenarios (Landsiedel et al., 2014). Gangwal et al. estimated that a life-time (45 years) exposure duration of the alveolar mass retention for TiO2, Ag and CNT ranges from 10 to 50 μg/cm2. Based on the proposed model, the mass retention corresponds to in vitro studies with relatively high ENM concentrations ranging from 30 to 400 μg/ml (Gangwal et al., 2011).
Here, nanomaterial exposures to THP-1, BEAS-2B and A549 cell lines decreased the cell viability in dose-dependent manner, especially with the three highest tested doses of 50, 100 and 500 μg/ml (Fig. S1, Fig. S2, Fig. S3). Lactate Dehydrogenase (LDH) release was concordant with the cell viability measures in BEAS-2B and THP-1 showing a clear LDH increase after 48-h CNM exposure at the highest doses (Fig. S2, Fig. S3), while A549 seemed to be the least responsive cell line (Fig. S1). In particular, THP-1 was the most sensitive (Fig. S3), showing slightly more elevated LDH levels, when compared to other cell lines. This was especially observed with GNF and MIT_MW, that have a known toxic potential. The lower doses, ranging from 0.1 to 5 μg/ml, did not to cause major responses, with highly similar LDH levels and consistent cell viability. Given these results, the 10 μg/ml concentration was chosen for showing no significant cell death, but still indicating some responses as compared to the baseline, thus corresponding more to a subchronic rather than acute response.
2.3. Molecular effects
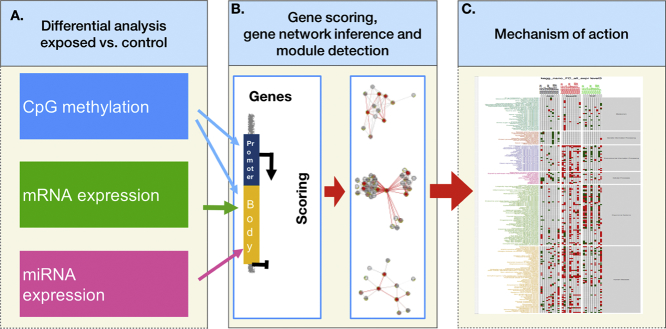
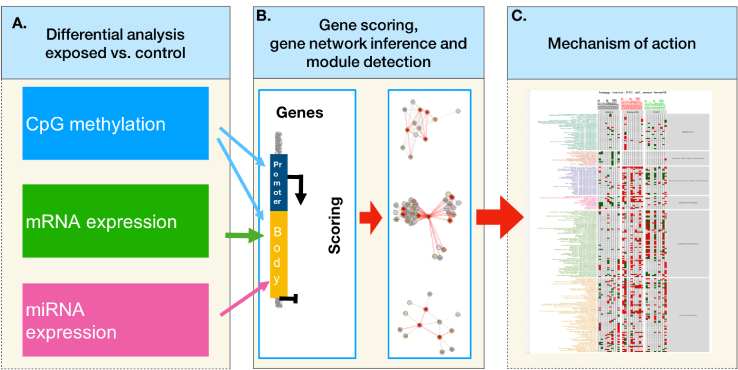
In addition to a variety of in vitro end point assessments, OMICS approaches allow for comparison of different in vitro systems as well as better correspondence to in vivo exposure scenarios (Kinaret et al., 2017b; Drasler et al., 2017). Combination of different OMICS methods can inform more thoroughly about ENMs MOA, which could be further used in ENM grouping approaches, as suggested by Riebeling et al. (2017). Gene expression profiling of co-cultures and 3D cultures, as well as systems taking into account exposure route such as Air Liquid Interface cultures (Latvala et al., 2016; Kletting, 2017), better resemble the in vivo exposure scenarios, but before clear conclusions about the ENM MOA can be drawn, cell type-specific responses need to be addressed by omics approaches (Drasler et al., 2017; Snyder-Talkington et al., 2015). The importance of multi-omics approaches in the study of MOA of toxicants has been demonstrated in several toxicogenomics studies (Jayapal, 2012; Gavin, 2016). On the other hand, focusing on one molecular layer alone might not be sufficient to fully describe the MOA of an external stimulus (Huang et al., 2017). The analysis of a single molecular layer can reveal information that is mainly related to reactive processes but not to the causative molecular processes, the integration of different layers, on the other hand, gives the possibility to describe the chain of causative changes involved in determining the observed phenotypes (Hasin et al., 2017). Here, we inferred the MOA of a panel of ten CNMs on three cell lines representative of major cell types of the respiratory system, A549, BEAS-2B and THP-1, by interrogating three molecular districts and systematically integrating their omics-derived data (Fig. 1).
Fig. 1.
Omics data integration and inference of mechanism of action procedure. (A) raw data from the three omics layers (DNA methylation, mRNA and miRNA expression) is preprocessed, and differential analysis for each layer is performed separately; (B) p-values and fold changes from each data layer are mapped to promoter and gene bodies of UCSC genes, combined in order to define ranks of genes, and used to extract high scoring gene modules; (C) enriched KEGG pathways are derived for the obtained modules in each exposure.
We defined the MOA of each CNM in each cell line as the ensemble of molecular and cellular pathways significantly altered after integration of genome wide DNA methylation, miRNA expression and gene expression.
By following a commonly accepted scheme of gene regulation (Consortium, 2012), we tested the hypothesis that hypomethylation in the gene promoter, paired with hypermethylation of the gene body and/or the under-expression of a targeting microRNA could explain gene upregulation. Likewise, hypermethylation of the promoter region and/or over-expression of a targeting microRNA could underlie gene down-regulation. For each exposure, we ranked the genes based on the concatenated regulatory effects of the three analyzed layers. Next, we inferred modules of high ranking genes that were enriched for protein-protein interactions and derived the enriched KEGG pathways. We then measured the effects of each nanomaterial in each cell line in terms of number of altered pathways as well as the direction of gene expression in these pathways.
2.4. Cell lines respond differently to CNM exposure
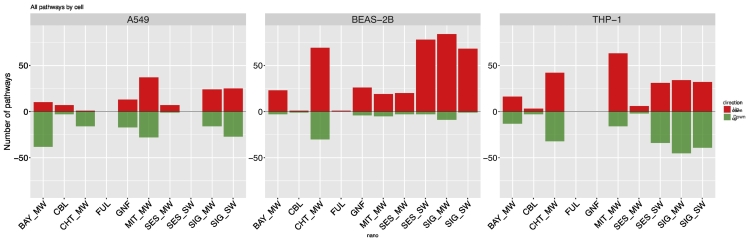
Based on the magnitude of the molecular events, overall, BEAS-2B cell line was the most sensitive to CNM exposure followed by THP-1, while A549 exhibited the smallest MOA (Fig. 2) (Scala et al., 2018). Interestingly, the MOA of BEAS-2B was largely unbalanced towards the induction of gene expression, while it was more balanced between the amount of up- and down-regulation in the other cell lines. BEAS-2B cell line, derived from normal human bronchial epithelium, might be more sensitive to stimuli than the other two cell lines considered in this study (A549 and THP-1), which are cancer-derived. Likewise, it is possible to hypothesize that the steady state transcriptome of non-cancerous BEAS-2B is generally kept at lower levels of activation. When analyzing the effects of the individual exposures in detail, considered at the level of the altered pathways, it appeared evident that fullerene (FUL) and carbon black (CBL), the spherical materials with the smallest aspect ratio, had a very marginal impact on all the three cell lines (Fig. 2). On the contrary, all the nanotubes investigated in this screening caused alteration of many molecular pathways in all the three cell lines, with the exception of SES_SW, to which A549 cells were essentially unresponsive. Interestingly, graphite fibers did not cause remarkable molecular alterations on the THP-1 cells, but had an impact to other two cell lines.
Fig. 2.
Bar plots reporting the number of up-regulated (red) and down-regulated (green) pathways in each exposure in the three cell lines. Pathway regulation is defined based on the median expression fold change of genes belonging to the pathway. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Our results overall suggest that same exposures may have significantly different MOA in different cell types. Specific steady-state transcriptional patterns of different cell types influence the cellular responses, signaling cascades and thus the CNM MOA. For this, it is expected that non-professional phagocytes, the epithelial cells BEAS-2B and A549, sense foreign substances differently than the professional phagocytes, such as macrophage-like cells derived from the THP-1 cell line. A549 cells originate from basal cells of the alveolar region (type II alveolar epithelial cells), whereas BEAS-2B cells are from bronchioles and trachea (Lieber et al., 1976; Schlinkert et al., 2015). The original anatomic location of the cells in the pulmonary tract reasonably also influences how environmental signals are processed. Epithelial cells from bronchioles are more likely to be directly exposed to foreign particles in the respiratory system in vivo, whereas basal cells, located under the first epithelial layer or in the alveolar regions, might not be affected by outer threats as rapidly and as severely.
Another difference between the two epithelial cell lines considered in this study, are organelles called lamellar bodies. Studying the uptake of different sized ultrafine particles of TiO2 in A549, Singh and collaborators noted that the uptake was limited to aggregated particles, found inside the lamellar bodies and membrane bound vacuoles (Singh et al., 2007). Similar results were regained in another study by Stearns et al., where the uptake of TiO2 particles was limited to aggregates found inside the lamellar bodies of the A549 cells after 24 h exposure (Stearns et al., 2001). They also postulated that single ultrafine particles are possibly not a target of phagocytosis by type II epithelium, but are internalized as aggregates, as also concluded by Churg et al. (Churg et al., 1998) Type II alveolar cells, including A549 cells, are the main producers of pulmonary surfactants, such as phospholipids that reduce the surface tension. The secreted surfactants cover the foreign particle surface by forming a biological corona (Sund et al., 2011). This protein-lipid coating can, in turn, change the modality of presentation of nanoparticles to the A549 cells, possibly explaining the lower level of molecular alterations observed in this study (Hohlfeld, 2002). Elsewhere, A549 cells were the least susceptible to different gold and silver nanomaterials as compared to BEAS-2B and primary lung epithelial cells, in line with our observations (Schlinkert et al., 2015). These results from several previous studies, do explain why also in our study set up, the A549 cell line was the least responsive. Bronchial epithelial cells such as BEAS-2B cell line on the other hand, have most likely evolved to have several response mechanisms, being among the first line of defense, constantly encountering a variety of inhaled particles. For example, it has been demonstrated by Zhang et al., that BEAS-2B cell line is more sensitive to cigarette smoke-induced cytotoxicity when compared to A549 (Zhang et al., 2017).
2.5. DNA methylation and microRNA expression partially explain CNM MOA
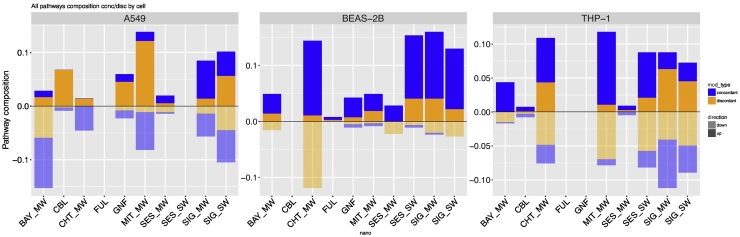
Next, we tested the hypothesis that the MOA of CNM would be explained by the regulatory model involving alteration of the DNA methylation at the level of the gene promoters and the regulation of microRNA expression (Fig. 3) (Scala et al., 2018).
Fig. 3.
Bar plots reporting the number of pathways derived from concordant (blue) and discordant (orange) genes in each exposure, for up-regulated (darker colors) and down-regulated (lighter colors) pathways in the three cell lines. Pathway regulation is defined based on the median expression fold change of genes belonging to the pathway. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Our results confirm that overall, the regulatory model exploited here explains a fair amount of the CNM MOA (62% in BEAS-2B, 53% in THP-1, and 50% in A549). When we focused on the proportion of the MOA resulting in gene induction, we observed a larger concordance in BEAS-2B (79%) and in THP-1 (65%) as compared to A549 (33%). On the contrary, the amount of the MOA proportion involving gene repression compatible with the methylation/miRNA regulatory model was highest in A549 (68%), moderate in THP-1 (39%), and minute in BEAS-2B (8%). Interestingly, the upregulated MOA of carbon black and Cheaptubes in A549 as well as SIG_MW in BEAS-2B are not explained at all by the regulatory model. Similarly, the downregulation of the Cheaptubes in BEAS-2B is entirely discordant with the tested regulatory model.
Omics studies are an emerging standard in the analysis of MOA of toxicants. However, focusing on a single molecular layer is not always sufficient to completely infer gene regulation patterns (Long et al., 2017) and the consequent alterations of the related molecular pathways. For example, in a similar experimental setting, where THP-1 cells were exposed to multi-walled carbon nanotubes, Oner et al. (Öner et al., 2016) inferred several pathways to be induced starting from genes showing promoter hypomethylation. However, our results indicate that the whole picture obtained through a multilayer approach is more complex and, especially in a “low-dose-short-time” setting, cannot be successfully portrayed by looking at a single molecular layer alone, as confirmed by the significant percentage of genes in our results, whose expression alteration does not follow the canonical regulation by DNA methylation.
2.6. Comparison of CNMs MOA
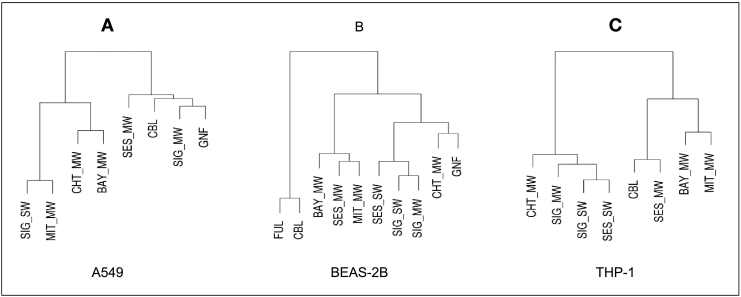
We used the motifs of pathway alteration to compute similarities of CNM MOA (Fig. 4). The patterns of MOA similarity in A549 were not easily attributable to CNM intrinsic properties. However, in this cell line, the two most similar MOA were exacerbated by SIG-SW, the material with the smallest diameter and highest surface area, and the known hazardous MIT_MW, which selectively induced pathways related to immune response (Fig. 4A).
Fig. 4.
Hierarchical clustering representing MOAs similarity of nanomaterials for each cell line. Distance between different exposures is computed using the complement of Jaccard index on shared positive and negative pathways. Clustering was performed using Ward's method.
Especially “environmental information processing” and “cellular processes” such as focal adhesion and cell adhesion were upregulated in BEAS-2B with several materials. It is known that induction of cell adhesion molecules, such as ICAM and VCAM are induced by pro-inflammatory cytokines, mediating leukocyte adhesion and extravasation (Atsuta et al., 1997; Veranth et al., 2004). In BEAS-2B we could appreciate a clear distinction between the particle and spherical materials, carbon black and fullerene, and the tubes and fibers (Fig. 4B). This difference was particularly evident at the level of the pathways related to regulation of actin cytoskeleton and focal adhesion. These pathways are altered by all the nanotubes and graphite nanofibers, but not by carbon black nanoparticles and fullerene nanospheres (Scala et al., 2018).
The uptake mechanism of BEAS-2B cells might involve an endocytic pathway in a size-dependent manner, as suggested by Kim et al. (Kim et al., 2011; Nymark et al., 2013) This might explain why most of the nanomaterials in our panel, being much longer than 40 to 50 nm, might not be internalized by BEAS-2B, but most likely trigger signaling cascades through surface contact. This is supported by high activation of ECM–receptor interaction, focal adhesion and cell adhesion molecules (CAMs), as well as regulation of actin cytoskeleton in BEAS-2B cells when exposed to tube-structured CNM. In contrast, all the three non-tube-structured nanomaterials, fullerene, graphite and carbon black, do not activate ECM receptor-interaction-pathways, regulation of actin cytoskeleton or focal adhesion (Scala et al., 2018). The small size of fullerene and carbon black particles might result in reduced interaction with the cell surfaces and overall signaling cascades. Interestingly, also the other non-tubular material, GNF, shows similar pattern in all three cell types. Moreover, poor ability of both type I and II epithelial cells to uptake carbon black particles has already been described in vivo (Corrin, 1970).
Interestingly cAMP signaling pathway is activated by SIG-SW, SIG-MW and SES-SW in BEAS-2B and THP-1 cells, but in opposite directions (Fig. 5) (Scala et al., 2018). Single-walled materials have the smallest diameter, whereas SIG-MW has the longest average length. Several signaling pathways are altered in opposite directions between the two cell lines, suggesting distinct sensing molecular mechanisms and receptor activation. For instance, the expression of genes belonging to the PI3K–Akt signaling pathway were found to be induced by all the nanotube materials in BEAS-2B. Instead, the same pathway was repressed by exposure with SIG-SW, CHT_MW, SIG-MW, SES-SW and MIT_MW nanotubes in THP-1 (Fig. 5) (Scala et al., 2018). These results suggest that, while in epithelial cells the regulation of PI3K-Akt signaling pathway genes is achieved by directly altering the DNA methylation levels, in THP-1 other epigenetic mechanisms may mediate a similar modulation of the same genes. Since alteration of the DNA methylation levels is thought to be more stable than other regulatory signals, it is reasonable to hypothesize that the induction of PI3K-Akt signaling pathway genes in epithelial cells could persist for longer time after exposure. These results are in concordance with the observations by Labib et al., who pinpointed transcriptomic alterations of PI3K-Akt signaling pathway in vivo as a driving event of long term MIT_MW exposure (Labib et al., 2015).
Fig. 5.
Selected pathways resulting from each exposure grouped by pathway category on rows and by exposed cell line on columns. Red and green cells are associated with significantly enriched KEGG pathways (FDR adjusted hypergeometric p-value <0.05), grey cells stand for no significant enrichment. Red cells are associated with enriched KEGG pathways whose genes have a positive median log fold-change in the corresponding comparison, green cells are associated with KEGG pathways whose genes have a negative median log fold-change in the corresponding comparison. Panel A reports mRNA expression median log fold-change directions, Panel B reports methylation median log fold-change directions and Panel C reports miRNA expression median log fold-change directions. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Similarly, we observed hypermethylation of the genes of the leukocyte transendothelial migration pathway in both epithelial cell lines when exposed to MIT_MW. Interestingly also Labib et al. concluded that the expression of the leukocyte extravasation pathway genes was still highly altered 56 days after lung exposure with the same material in mice in vivo (Labib et al., 2015). Our results further support the hypothesis that alteration of gene expression accompanied by DNA methylation changes could drive long term adaptation in exposed biological systems.
In THP-1, the MOA of CNM with high aspect ratio and higher diameter were grouped together, respectively (Fig. 4C). Exposure with higher diameter materials mainly altered pathways related to regulation of inflammatory and immune responses (TNF, cytokine-cytokine receptor interactions, IL17, RIG-I receptor and NOD-like receptor signaling pathways) (Fig. 5) (Scala et al., 2018). On the other hand, exposure to higher aspect ratio CNM produced changes in pathways of the intracellular signaling.
The endocrine system related pathways are altered in THP-1 and BEAS-2B, but not A549, by the CNMs with high surface area and aspect ratio, and small diameter, namely SIG_SW, CH, SIG_MW, SES_SW. In our screening, THP-1 is the most susceptible to endocrine system pathways alteration (Fig. 5) (Scala et al., 2018). Several ENMs have been shown to act as endocrine disrupting compounds (EDCs). Couleau et al. demonstrated that several EDCs reduced phagocytosis of differentiated THP-1 cells, and disturbed TNF-a, Il-1b and IL-8 cytokine secretions, as well as decreased ERK ½ phosphorylation was associated to EDC treatments (Couleau et al., 2015). Moreover, several studies have reported similar results, showing that EDCs directly affect the innate immune system (Bennasroune et al., 2012; Ohnishi et al., 2008; Roy et al., 2012; Watanabe et al., 2003).
Cancer pathways were mostly activated by the two epithelial cell lines. It has been shown that BEAS-2B cell line can be malignantly transformed in vitro via several compounds such as chromium, arsenite and cigarette smoke (Zhao and Klimecki, 2014). Proteoglycans in cancer-pathway was activated by most of the nanomaterials both, at the level of RNA expression as well as methylation, excluding only Cheaptubes and fullerene. The activation of proteoglycans, might also indicate a phenotypic change in the BEAS-2B cell line, towards cancer related signaling.
2.7. Conclusions
Here we show that the molecular alterations due to CNM exposure are highly dependent on the cell type as well as the geometrical properties of the nanoparticles. When information concerning changes in gene expression are systematically integrated with DNA methylation and miRNA expression, better understanding of ENM MOA can be drawn. Since epigenetic modulation is a molecular marker of long term stable adaptation, the interpretation of ENM MOA is highly enhanced by multi-omics approaches. This, in turn, helps in building more thorough landscape of the molecular effects caused by the exposure, thus supporting the definition of more accurate ENM adverse outcome pathways (AOPs).
3. Materials and methods
3.1. Nanomaterials and suspensions
Nanomaterials used in this study were previously characterized and reported in NANOATLAS (Vippola et al., 2009) as well as in our previous publications (Kinaret et al., 2017a; Rydman et al., 2015; Rydman et al., 2014; Palomäki et al., 2015). Providers and material characteristics are reported in Table 1.
Nanomaterial suspensions were prepared by weighing the materials into glass tubes and diluting them with 1% FBS-PBS to a stock concentration of 1 mg/ml. The stock was sonicated 2 × 15 min in bath sonicator (Elmasonic, USA) at room temperature, and additional 15 min to MIT_MW, GNF and FUL particles. Dilutions were prepared to corresponding cell culture media (see below) supplemented with 1% FCS and sonicated for 15 min and vortexed before the exposures.
3.2. Cell culturing and exposures
THP-1 cells (ATCC TIB-202) were grown in complete RPMI media, supplemented with 10% FBS and 1% Ultraglutamine. BEAS-2B cells (American Type Culture Collection through LGC Promochem AB (Borås, Sweden)) were grown in LHC-9 media and A549 (ATCC CCL-185) cells were grown in DMEM media including l-glutamine, supplemented with 10% FBS. Cells were grown in culturing flasks (75 cm2) at 37 °C in humidified atmosphere of 5% CO2. Exposures were performed in 12 -well plates, with concentration of 10 μg/ml in corresponding medias supplemented with 1% FCS, as follows: THP-1: 800,000 cells per well for RNA and DNA extractions, 900,000 cells/well to miRNA extractions; BEAS-2B: 100,000 cells/well to RNA, DNA and miRNA extractions; A549: 50,000 cells/well to RNA, DNA and miRNA extractions. THP-1 cells were differentiated with 50 nM PMA (phorbol-12-myristate-13-acetate) for 48 h before nanomaterial exposure. Exposures were conducted as triplicates with final concentration of 10 μg of ENM/ml in each well. The concentration of 10 μg/ml was chosen based on the cell viability (ATP) and cytotoxicity (LDH) screenings, using 8 different concentrations (Fig. S1, Fig. S2, Fig. S3). After 48-h incubation, the media was separated and the cells were washed with PBS and harvested according to the Qiagen AllPrep DNA/RNA extraction protocol (Qiagen, GmbH, Hilden, Germany).
3.3. Cell viability and LDH release
The viability (ATP) and cytotoxicity (LDH) screenings, using 8 different ENM concentrations 0,1; 0,5; 1; 5; 10; 50; 100 and 500 μg/ml (Fig. S1, Fig. S2, Fig. S3), were conducted in order to find an optimal exposure dose for 48-h nanomaterial exposures.
Cell viability, by quantitation of ATP was measured after 48 h exposures to eight different ENM concentrations, with CellTiter-Glo, Luminescent Cell Viability Assay (Promega Corporation, Madison, WI, USA). Cytotoxicity, based on the LDH release after 48 h exposures was measured from the supernatants with Cytotoxicity Detection Kit PLUS (LDH) (Roche Diagnostics GmbH, Mannheim, Germany, Fig. S1, Fig. S2, Fig. S3). Based on the results from both assays, 10 μg/ml concentration was chosen for inducing low levels of cytotoxicity and high cell viability.
3.4. Sample collection and extraction
DNA and mRNA were extracted and purified using Qiagen AllPrep 96 DNA/RNA extraction kit (Qiagen, Germany), following the protocol provided by the manufacturer.
Qiagen miRNeasy 96 extraction kit (Qiagen, Germany) was used to extract and purify miRNA from the exposed cells. RNA quality and quantity were confirmed by NanoDrop (ND-1000, Thermo Fisher Scientific Inc., Wilmington, NC, USA) and Bioanalyzer (Agilent Technologies, USA). Based on the RNA integrity values, the RNA samples with high RIN values (>9) were used in microarray analyses.
3.5. Microarray sample processing
mRNA transcriptome assay was performed as instructed by Agilent Quick Amp, two-color microarray-based gene expression analysis labeling protocol. 100 ng of dried total RNA was resuspended to 1,5 μl of purified water, and labeled with Cy3 or Cy5 dyes (Agilent, USA), and hybridized to Agilent SurePrint G3Human GE 8x60K DNA microarrays. Slides were scanned with Agilent scanner model G2505C, and data was extracted with Agilent feature extraction software (V11.5.1.1).
miRNA assays were performed according to the protocol provided by the manufacturer (miRNA Microarray System with miRNA Complete Labeling and Hyb Kit, Agilent). 100 ng of dried samples were resuspended in 2 μl of purified water and labeled with Cyanine 3-pCp dye and hybridized to Agilent SurePrint G3 Unrestricted Human miRNA_V21 8x60K microarrays. Slides were scanned with Agilent scanner model G2505C, and data was extracted with Agilent feature extraction software (V11.5.1.1).
Microarray data have been deposited in ArrayExpress database and are accessible through ArrayExpress accession numbers: E-MTAB-6396, E-MTAB-6396 and E-MTAB-6397.
3.6. Genome-wide DNA methylation
DNA was extracted together with RNA using Qiagen AllPrep 96 DNA/RNA extraction kit (Qiagen, Germany). 500 ng of DNA was bisulfite converted with the EZ-96 Methylation Kit (Zymo Research Corporation) according to the manufacturer's instructions. Bisulfite-treated DNA was amplified, fragmented and hybridized to the HumanMethylation450 BeadChip (Illumina) according to standard Illumina protocol and imaging was performed using Illumina iScan scanner.
3.7. Processing of each molecular layer (MRNA, MIRNA, DNA methylation)
For each cell line and each analyzed molecular level, data was imported and analyzed as described in the following.
3.8. mRNA
mRNA expression raw median values data from AgilentSurePrintG3_Human_GE_v3_8x60K were imported using limma read.maimages function. A quantile-based strategy was used to filter out low quality probes based on the value of negative control probes. In particular, probes having value higher than the 90% quantile of negative control probes in at least two samples were considered for subsequent analytical steps. Expression values were then log(2) transformed and normalized between samples using quantile normalization. Two rounds of batch correction using ComBat method from package SVA (Leek et al., 2012) were performed to eliminate the effect of labeling and slide. Multiple probes mapping to the same REFSEQ entry were summarized by the median value.
A limma based approach was employed in order to statistically estimate differences in average expression levels between each nanomaterial exposure and the corresponding controls.
3.9. miRNA
Raw median values data from Agilent microarrays were imported using limma read.maimages function. Probes were filtered in order to keep only probes detected for at least the 50% +1 samples in at least one treatment group. Expression values were log(2) transformed and then quantile normalized among samples. Probes were mapped to target miRNA by taking median value of probes for each target miRNA. Batch effect analysis revealed the presence in the dataset of a technical batch effect related to the array id. Batch effects correction was performed using ComBat method from the package SVA (Leek et al., 2012). Finally, a limma model was applied to statistically estimate difference in average miRNA expression.
3.10. CpG methylation
Raw methylation data files from Illumina Infinium 450 k array were imported with minfi package (Aryee et al., 2014). Data was then normalized using Subset-quantile Within Array Normalization (SWAN) (Makismovic et al., 2012) and represented as M-values. CpG probes were then filtered by a) removing all CpG probes with a detection p-value higher than 0,01 in at least on sample, b) removing all probes containing single nucleotide polymorphisms at the interrogation or the elongation site and c) removing all probes belonging to the set of known cross-hybridizing probes of the employed platform (Chen et al., 2013).
Batch effects evaluation was performed by detecting the presence of surrogate variables with sva function from SVA package. After estimating these variables, their value was discretized into n_samples^(1/3) bins using the discretize function from infotheo package (Meyer et al., 2008) and corrected using ComBat method from the package SVA (Leek et al., 2012). Finally a limma model was employed in order to statistically estimate differences in average methylation levels between each nanomaterial exposure and the corresponding controls (Smyth, 2005).
3.11. Multi-omics analysis
Integrative analysis of the three assayed datasets was performed for each exposure in each cell using SMITE toolkit (Wijetunga et al., 2017). This tool allows to rank genes by integrating results from differential analyses performed on one or more “modifier” layers.
UCSC genes were used as a reference point in this task. p-Values and fold changes were associated to each gene using the results of the mRNA differential analysis. In case of multiple transcript mapping to the same gene we took values from the transcript having the lowest p-value.
For each gene, two regulatory regions based on its genomic coordinates and the transcription orientation were defined: the promoter region, defined as 1 kb region flanking the transcription start site [TSS − 1 kb, TSS + 1 kb], and the gene body region, defined as the region spanning from TSS + 1 kb to the transcription termination site (TES) [TSS + 1 kb, TES]. A set of CpGs from the methylation layer and a set of miRNAs from the miRNA analysis were associated to each one of these regulatory regions. CpGs were associated to promoters or bodies by using their genomic position, while miRNA were assigned to all gene bodies in their set of target genes, computed as the top 10% scoring target genes, based on targetScan t-scores (Agarwal et al., 2015). For each gene region, the fold changes and weighted p-values from methylation differential analyses of all CpGs associated to the region were considered. These latter were respectively summarized by using the Stouffer method on the values, weighted by the CpG distance from the TSS. The same operation was performed on values from miRNA differential analysis but weighting values with the Sidak method. Next, miRNA and methylation p-values were logit transformed and rescaled using expression p-values as reference distribution.
Finally, genes were assigned with a score obtained by integrating the expression p-value and fold-change with the same values from the two modification layers using the weights provided in Table 2. The output of this integration step on each exposure is reported in (Scala et al., 2018). Finally, functional modules enriched for high scoring genes (p-value <0.01), were obtained by using a SpinGlass algorithm with 1000 randomizations on the Reactome (Croft et al., 2010) interaction network.
Table 2.
Weights and expected direction of interaction for each layer modelled in the SMITE integration model.
| Feature/relationship | mRNA expression | Gene promoter methylation | Gene body methylation | Targeting miRNA expression |
|---|---|---|---|---|
| Relationship with mRNA level | Dir Corr | Inv Corr | Dir Corr | Inv. Corr |
| Weight | 0.70 | 0.15 | 0.05 | 0.10 |
3.12. MOA definition
For each exposure in each cell-line, the list of the altered KEGG pathways was derived as follows. For each exposure, the set modG obtained by taking the union of genes constituting enriched functional modules was considered. The direction of modification layers in the associated regulatory regions was used to partition modG in two subsets, modG_ep and modG_not_ep. modG_ep was defined as the set of up-regulated genes with hypo-methylated promoter or targeted by down-regulated miRNAs along with the set of down-regulated genes with hyper-methylated promoter or up-regulated targeting miRNAs. modG_ep_not_ep was defined as the complement of modG_ep with respect to modG. Each of these three sets with enriched KEGG pathways was annotated using the enrichKEGG function from clusterProfiler package (Yu et al., 2012) with a p-value threshold of 0.05, thus deriving three sets of pathways modG_path, modG_ep_Path and modG_not_ep_Path. A positive or negative sign was associated to each computed pathway by deriving the sign of the median expression fold-change of the enriched genes. Distance between different exposures was computed in terms of shared activated pathways. In particular for a pair of exposures A and B, the distance between A and B was defined as dist(A,B) = 1 − [jacc_up(A,B) + jacc_down(A,B)]/2, where jacc_up(A,B) and jacc_down(A,B) are respectively the jaccard indices computed on common positive and common negative activated pathways between A and B.
The following are the supplementary data related to this article.
Viability (ATP) and cytotoxicity (LDH release) assays with eight exposure concentrations (0, 0.1, 0.5, 1, 5, 10, 100 and 500 μg/ml) to A549 cells, as well as lysed cells as positive controls. Titles of each graph (A–J) correspond to the material acronyms found from Table 1.
Viability (ATP) and cytotoxicity (LDH release) assays with eight exposure concentrations (0, 0.1, 0.5, 1, 5, 10, 100 and 500 μg/ml) to BEAS-2B cells, as well as lysed cells as positive controls. Titles of each graph (A–J) correspond to the material acronyms found from Table 1.
Viability (ATP) and cytotoxicity (LDH release) assays with eight exposure concentrations (0, 0.1, 0.5, 1, 5, 10, 100 and 500 μg/ml) to PMA-differentiated THP-1 cells, as well as lysed cells as positive controls. Titles of each graph (A–J) correspond to the material acronyms found from Table 1.
Excel spreadsheet, reporting in each sheet differential analysis summaries and SMITE gene scores for each exposure. For each gene are reported in the order the gene symbol, the SMITE score, the combined methylation log fold-change for promoter region, the combined methylation log fold-change for body region, the combined log fold-change of targeting miRNAs, mRNA expression log fold-change, combined methylation p-value for promoter region, combined p-values of targeting miRNAs, combined methylation p-value for body region, mRNA expression p-value.
Acknowledgements
This study was supported by the Academy of Finland (grant agreements 275151 and 292307), EU H2020 caLIBRAte project (grant agreement 686239), EU H2020 LIFEPATH (grant agreement 633666), and EU FP7 NANOSOLUTIONS project (grant agreement FP7-309329).
The authors would also wish to thank Hannu Norppa (FIOH) for providing the cell lines used in this study, Sirpa Hyttinen (FIOH) for her technical assistance, and Silvia Polidoro (HUGEF) for her valuable help with performing the genome-wide DNA methylation assays.
References
- Agarwal V., Bell G.W., Nam J.-W., Bartel D.P. elife. 2015;4:101. doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aryee M.J., Jaffe A.E., Corrada-Bravo H., Ladd-Acosta C., Feinberg A.P., Hansen K.D., Irizarry R.A. Bioinformatics. 2014;30:1363–1369. doi: 10.1093/bioinformatics/btu049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atsuta J., Sterbinsky S.A., Plitt J., Schwiebert L.M., Bochner B.S., Schleimer R.P. Am. J. Respir. Cell Mol. Biol. 1997;17:571–582. doi: 10.1165/ajrcmb.17.5.2685. [DOI] [PubMed] [Google Scholar]
- Bachler G., Losert S., Umehara Y., von Goetz N., Rodriguez-Lorenzo L., Petri-Fink A., Rothen-Rutishauser B., Hungerbuehler K. Part. Fibre Toxicol. 2015;12:3580. doi: 10.1186/s12989-015-0090-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennasroune A., Rojas L., Foucaud L., Goulaouic S., Laval-Gilly P., Fickova M., Couleau N., Durandet C., Henry S., Falla J. Int. J. Immunopathol. Pharmacol. 2012;25:365–376. doi: 10.1177/039463201202500206. [DOI] [PubMed] [Google Scholar]
- H.M.K.S.K.K.S.E.A. Braakhuis, 89 (2015) 1469–1495.
- Chen Y.-A., Lemire M., Choufani S., Butcher D.T., Grafodatskaya D., Zanke B.W., Gallinger S., Hudson T.J., Weksberg R. Epigenetics. 2013;8:203–209. doi: 10.4161/epi.23470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Xu M., Zhang J., Ma J., Gao M., Zhang Z., Xu Y., Liu S. Adv. Mater. Weinheim. 2017;29:1604580. doi: 10.1002/adma.201604580. [DOI] [PubMed] [Google Scholar]
- Churg A., Stevens B., Wright J.L. Am. J. Phys. Lung Cell. Mol. Phys. 1998;274:L81–L86. doi: 10.1152/ajplung.1998.274.1.L81. [DOI] [PubMed] [Google Scholar]
- Consortium T.E.P. Nature. 2012;489:57–74. [Google Scholar]
- Corrin B. Thorax. 1970;25:110–115. doi: 10.1136/thx.25.1.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa P.M., Fadeel B. Toxicol. Appl. Pharmacol. 2016;299:101–111. doi: 10.1016/j.taap.2015.12.014. [DOI] [PubMed] [Google Scholar]
- Couleau N., Falla J., Beillerot A., Battaglia E., D'Innocenzo M., Plançon S., Laval-Gilly P., Bennasroune A. PLoS One. 2015;10 doi: 10.1371/journal.pone.0131428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croft D., O'Kelly G., Wu G., Haw R., Gillespie M., Matthews L., Caudy M., Garapati P., Gopinath G., Jassal B.E.A. Nucleic Acids Res. 2010;39:D691–D697. doi: 10.1093/nar/gkq1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drasler B., Sayre P., Steinhäuser K.G., Petri-Fink A., Rothen-Rutishauser B. NanoImpact. 2017;8:99–116. [Google Scholar]
- Fröhlich E. J. Nanobiotechnol. 2017;15:84. doi: 10.1186/s12951-017-0320-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangwal S., Brown J.S., Wang A., Houck K.A., Dix D.J., Kavlock R.J., Hubal E.A.C. Environ. Health Perspect. 2011;119:1539–1546. doi: 10.1289/ehp.1103750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin A. University of Birmingham; 2016. Investigating the Mechanisms of Silver Nanoparticle Toxicity in Daphnia magna: A Multi-omics Approach. [Google Scholar]
- Hasin Y., Seldin M., Lusis A. Genome Biol. 2017;18:4181. doi: 10.1186/s13059-017-1215-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohlfeld J.M. Respir. Res. 2002;3(3):4. doi: 10.1186/rr176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S., Chaudhary K., Garmire L.X. Front. Genet. 2017;8:1005. doi: 10.3389/fgene.2017.00084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayapal M. 2012. Front Genet 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennen D., Ruiz-Aracama A., Magkoufopoulou C., Peijnenburg A., Lommen A., van Delft J., Kleinjans J. BMC Syst. Biol. 2011;5:139. doi: 10.1186/1752-0509-5-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H.R., Kim M.J., Lee S.Y., Oh S.M., Chung K.H. Mutat. Res. 2011;726:129–135. doi: 10.1016/j.mrgentox.2011.08.008. [DOI] [PubMed] [Google Scholar]
- Kinaret P., Ilves M., Fortino V., Rydman E., Karisola P., Lähde A., Koivisto J., Jokiniemi J., Wolff H., Savolainen K.E.A. ACS Nano. 2017;11:291–303. doi: 10.1021/acsnano.6b05652. [DOI] [PubMed] [Google Scholar]
- Kinaret P., Marwah V., Fortino V., Ilves M., Wolff H., Ruokolainen L., Auvinen P., Savolainen K., Alenius H., Greco D. ACS Nano. 2017;11 doi: 10.1021/acsnano.6b08650. (acsnano.6b08650–3796) [DOI] [PubMed] [Google Scholar]
- Kletting S. ALTEX. 2017;35:211–222. doi: 10.14573/altex.1607191. [DOI] [PubMed] [Google Scholar]
- Labib S., Williams A., Yauk C.L., Nikota J.K., Wallin H., Vogel U., Halappanavar S., Damoiseaux R., George S., Li M., Pokhrel S., Ji Z., France B., Xia T., Suarez E., Rallo R., Mädler L., Cohen Y., Hoek E., Nel A., Kroll A., Pillukat M.H., Hahn D., Schnekenburger J., Griffiths S.M., Singh N., Jenkins G.J., Williams P.M., Orbaek A.W., Barron A.R., Wright C.J., Doak S.H., Doak S.H., Griffiths S.M., Manshian B., Singh N., Williams P.M., Brown A.P., Jenkins G.J., Stone V., Johnston H., Schins R.P., Stone V., Pozzi-Mucelli S., Tran L., Aschberger K., Sabella S., Vogel U., Poland C., Balharry D., Fernandes T., Gottardo S., Hankin S., Hartl M.G., Hartmann N., Hristozov D., Hund-Rinke K., Johnston H., Marcomini A., Panzer O., Roncato D., Saber A.T., Wallin H., Scott-Fordsmand J.J., Guadagnini R., Kenzaoui B.H., Walker L., Pojana G., Magdolenova Z., Bilanicova D., Saunders M., Juillerat-Jeanneret L., Marcomini A., Huk A., Dusinska M., Fjellsbo L.M., Marano F., Boland S., Lai D.Y., Wilson V.S., Keshava N., Hester S., Segal D., Chiu W., Thompson C.M., Euling S.Y., Hamadeh H.K., Amin R.P., Paules R.S., Afshari C.A., Hamadeh H.K., Bushel P.R., Jayadev S., Martin K., DiSorbo O., Sieber S., Bennett L., Tennant R., Stoll R., Barrett J.C., Blanchard K., Paules R.S., Afshari C.A., Hamadeh H.K., Bushel P.R., Jayadev S., DiSorbo O., Bennett L., Li L., Tennant R., Stoll R., Barrett J.C., Paules R.S., Blanchard K., Afshari C.A., Bercu J.P., Jolly R.A., Flagella K.M., Baker T.K., Romero P., Stevens J.L., Waters M.D., Jackson M., Lea I., Halappanavar S., Saber A.T., Decan N., Jensen K.A., Guo C., Rogowski J., Koponen I.K., Levin M., Madsen A.M., Atluri R., Snitka V., Birkedal R.K., Rickerby D., Williams A., Wallin H., Yauk C.L., Vogel U., Husain M., Saber A.T., Guo C., Jensen K.A., Yauk C.L., Williams A., Vogel U., Wallin H., Halappanavar S., Halappanavar S., Jackson P., Williams A., Jensen K.A., Hougaard K.S., Vogel U., Yauk C.L., Wallin H., Poulsen S.S., Saber A.T., Williams A., Andersen O., Kobler C., Atluri R., Pozzebon M.E., Mucelli S.P., Simion M., Rickerby D., Mortensen A., Jackson P., Kyjovska Z.O., Molhave K., Jensen K.A., Yauk C.L., Wallin H., Halappanavar S., Vogel U., Poulsen S.S., Labib S., Wu D., Husain M., Williams A., Bogelund J.P., Andersen O., Kobler C., Molhave K., Kyjovska Z.O., Saber A.T., Wallin H., Yauk C.L., Vogel U., Halappanavar S., Bourdon J.A., Saber A.T., Halappanavar S., Jackson P.A., Hougaard K.S., Williams A., Vogel U., Wallin H., Yauk C.L., Bourdon J.A., Halappanavar S., Saber A.T., Williams A., Wallin H., Vogel U., Yauk C.L., Jackson P., Hougaard K.S., Vogel U., Casavant L., Williams A., Wade M., Yauk C.L., Wallin H., Halappanavar S., Moffat I., Chepelev N.L., Labib S., Bourdon-Lacombe J., Kuo B., Buick J.K., Lemieux F., Williams A., Halappanavar S., Malik A.I., Luijten M., Aubrecht J., Hyduke D.R., Fornace A.J., Swartz C.D., Yauk C.L., Webster A.F., Chepelev N., Gagne R., Kuo B., Williams A., Yauk C.L., Clewell H.J., Allen B.C., Wesselkamper S.C., Wang N.C., Lambert J.C., Hess-Wilson J.K., Zhao Q.J., Andersen M.E., Chepelev N.L., Moffat I.D., Labib S., Bourdon-Lacombe J., Kuo B., Buick J.K., Lemieux F., Malik A.I., Halappanavar S., Williams A., Yauk C.L., Jackson A.F., Williams A., Recio L., Waters M.D., Lambert I.B., Yauk C.L., Bourdon-Lacombe J.A., Moffat I.D., Deveau M., Husain M., Auerbach S., Krewski D., Bushel P.R., Williams A., Yauk C.L., Ma-Hock L., Treumann S., Strauss V., Brill S., Luizi F., Mertler M., Wiench K., Gamer A.O., Ravenzwaay B., Landsiedel R., Muller J., Huaux F., Moreau N., Misson P., Heilier J.F., Delos M., Arras M., Fonseca A., Nagy J.B., Lison D., Mercer R.R., Scabilloni J.F., Wang L., Battelli L.A., Friend S., Porter D.W., Pauluhn J., Dahm M.M., Evans D.E., Schubauer-Berigan M.K., Birch M.E., Deddens J.A., Birch M.E., Ku B.K., Evans D.E., Ruda-Eberenz T.A., Lee J.H., Lee S.B., Bae G.N., Jeon K.S., Yoon J.U., Ji J.H., Sung J.H., Lee B.G., Lee J.H., Yang J.S., Kim H.Y., Kang C.S., Yu I.J., Erdely A., Dahm M., Zeidler-Erdely P.C., Fernback J.E., Birch M.E., Evans D.E., Kashon M.L., Deddens J.A., Hulderman T., Bilgesu S.A., Battelli L., Schwegler-Berry D., Leonard H.D., McKinney W., Frazer D.G., Antonini J.M., Porter D.W., Schubauer-Berigan M.K., Han J.H., Lee E.J., Lee J.H., So K.P., Lee Y.H., Bae G.N., Lee S.B., Ji J.H., Cho M.H., Yu I.J., Maynard A.D., Baron P.A., Foley M., Kisin E.R., Methner M., Hodson L., Dames A., Geraci C., Guo N.L., Wan Y.W., Denvir J., Porter D.W., Pacurari M., Wolfarth M.G., Qian Y., Bourdon J.A., Saber A.T., Jensen K.A., Madsen A.M., Lamson J.S., Wallin H., Moller P., Loft S., Yauk C.L., Vogel U.B., Porter D.W., Hubbs A.F., Wu N., Wolfarth M.G., Sriram K., Leonard S., Battelli L., Schwegler-Berry D., Friend S., Andrew M., Chen B.T., Tsuruoka S., Endo M., Snyder-Talkington B.N., Dong C., Zhao X., Dymacek J., Porter D.W., Wolfarth M.G., Qian Y., Guo N.L., Snyder-Talkington B.N., Dymacek J., Porter D.W., Wolfarth M.G., Pacurari M., Denvir J., Qian Y., Guo N.L., Dymacek J., Snyder-Talkington B.N., Porter D.W., Wolfarth M.G., Castranova V., Qian Y., Guo N.L., Dymacek J., Guo N.L., Yang L., Allen B.C., Black M.B., Budinsky R.A., Dombkowski A., Cukovic D., LeCluyse E.L., Ferguson S.S., Rowlands J.C., Davis J.A., Gift J.S., Zhao Q.J., Clewell H.J., Bermudez E., Dodd D.E., Willson G.A., Campbell J.L., Thomas R.S., Philbert M.A., Auerbach S.S., Wetmore B.A., Devito M.J., Cote I., Rowlands J.C., Whelan M.P., Hays S.M., Meek M.E., Reiter L.W., Lambert J.C., Clewell H.J., Stephens M.L., Zhao Q.J., Wesselkamper S.C., Flowers L., Carney E.W., Pastoor T.P., Petersen D.D., Yauk C.L., Nong A., Wilson M.S., Wynn T.A., Liu J.Y., Brass D.M., Hoyle G.W., Brody A.R., Kerem B., Rommens J.M., Buchanan J.A., Markiewicz D., Cox T.K., Chakravarti A., Buchwald M., Tsui L.C., Raghu G., Collard H.R., Egan J.J., Martinez F.J., Behr J., Brown K.K., Colby T.V., Cordier J.F., Flaherty K.R., Lasky J.A., Lynch D.A., Ryu J.H., Swigris J.J., Wells A.U., Ancochea J., Bouros D., Carvalho C., Costabel U., Ebina M., Hansell D.M., Johkoh T., Kim D.S., King T.E., Kondoh Y., Myers J., Muller N.L., Nicholson A.G., Richeldi L., Selman M., Dudden R.F., Griss B.S., Protzko S.L., Schunemann H.J., Gasse P., Mary C., Guenon I., Noulin N., Charron S., Schnyder-Candrian S., Schnyder B., Akira S., Quesniaux V.F., Lagente V., Ryffel B., Couillin I., Piguet P.F., Vesin C., Grau G.E., Thompson R.C., Saito F., Tasaka S., Inoue K., Miyamoto K., Nakano Y., Ogawa Y., Yamada W., Shiraishi Y., Hasegawa N., Fujishima S., Takano H., Ishizaka A., Wilson M.S., Madala S.K., Ramalingam T.R., Gochuico B.R., Rosas I.O., Cheever A.W., Wynn T.A., Yoshizaki A., Yanaba K., Iwata Y., Komura K., Ogawa A., Akiyama Y., Muroi E., Hara T., Ogawa F., Takenaka M., Shimizu K., Hasegawa M., Fujimoto M., Tedder T.F., Sato S., Liu T., Jin H., Ullenbruch M., Hu B., Hashimoto N., Moore B., McKenzie A., Lukacs N.W., Phan S.H., Fichtner-Feigl S., Strober W., Kawakami K., Puri R.K., Kitani A., Jakubzick C., Choi E.S., Joshi B.H., Keane M.P., Kunkel S.L., Puri R.K., Hogaboam C.M., Gharaee-Kermani M., McGarry B., Lukacs N., Huffnagle G., Egan R.W., Phan S.H., Barron L., Wynn T.A., Bantsimba-Malanda C., Marchal-Somme J., Goven D., Freynet O., Michel L., Crestani B., Soler P., Leask A., Abraham D.J., Sato E., Koyama S., Robbins R.A., Donaldson K., Murphy F.A., DUFFIN R., Poland C.A., Ravichandran P., Rydman E.M., Ilves M., Vanhala E., Lehto M., Kinaret P.A., Pylkkänen L., Happo M., Hirvonen M.R., Greco D., Savolainen K., Wolff H., Alenius H., Sun B., Podila R., Shannahan J.H., Rao A.M., Brown J.M., Rydman E.M., Ilves M., Koivisto A.J., Kinaret P.A., Fortino V., Savinko T.S., Lehto M.T., Pulkkinen V., Vippola M., Hameri K.J., Matikainen S., Wolff H., Savolainen K.M., Wang P., Wang P., Ding L., Patlolla A., Kisin E.R., Azad N., Vietti G., Ibouraadaten S., Palmai-Pallag M., Yakoub Y., Bailly C., FENOGLIO I., Marbaix E., Lison D., Brule S., Wang X., Villeneuve D.L., Crump D., Garcia-Reyero N., Hecker M., Hutchinson T.H., LaLone C.A., Landesmann B., Lettieri T., Munn S., Nepelska M., Ottinger M.A., Vergauwen L., Whelan M., Villeneuve D.L., Crump D., Garcia-Reyero N., Hecker M., Hutchinson T.H., LaLone C.A., Landesmann B., Lettieri T., Munn S., Nepelska M., Ottinger M.A., Vergauwen L., Whelan M., Husain M., Kyjovska Z.O., Bourdon-Lacombe J., Saber A.T., Jensen K.A., Jacobsen N.R., Williams A., Wallin H., Halappanavar S., Vogel U., Yauk C.L., Shvedova A.A., Oberdörster G. Part. Fibre Toxicol. 2015;13:15. doi: 10.1186/s12989-016-0125-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landsiedel R., Sauer U.G., Ma-Hock L., Schnekenburger J., Wiemann M. Nanomedicine (London) 2014;9:2557–2585. doi: 10.2217/nnm.14.149. [DOI] [PubMed] [Google Scholar]
- Latvala S., Hedberg J., Möller L., Wallinder I.O., Karlsson H.L., Elihn K. J. Appl. Toxicol. 2016;36:1294–1301. doi: 10.1002/jat.3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leek J.T., Johnson W.E., Parker H.S., Jaffe A.E., Storey J.D. Bioinformatics. 2012;28:882–883. doi: 10.1093/bioinformatics/bts034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehto M., Karilainen T., Róg T., Cramariuc O., Vanhala E., Tornaeus J., Taberman H., Jänis J., Alenius H., Vattulainen I., Laine O. PLoS One. 2014;9 doi: 10.1371/journal.pone.0114490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieber M., Todaro G., Smith B., Szakal A., Nelson-Rees W. Int. J. Cancer. 1976;17:62–70. doi: 10.1002/ijc.2910170110. [DOI] [PubMed] [Google Scholar]
- Long M., Smiraglia D., Campbell M. Biomol. Ther. 2017;7:15. doi: 10.3390/biom7010015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makismovic J., Gordon L., Oshlack A. Genome Biol. 2012;13:R44. doi: 10.1186/gb-2012-13-6-r44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marx-Stoelting P., Braeuning A., Buhrke T., Lampen A., Niemann L., Oelgeschlaeger M., Rieke S., Schmidt F., Heise T., Pfeil R., Solecki R. Arch. Toxicol. 2015;89:2177–2184. doi: 10.1007/s00204-015-1602-x. [DOI] [PubMed] [Google Scholar]
- Meyer P.E., Schretter C., Bontempi G. Selected topics in signal processing. IEEE J. 2008;2:261–274. [Google Scholar]
- Nel A.E. J. Intern. Med. 2013;274:561–577. doi: 10.1111/joim.12109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nel A., Nel A., Xia T., Xia T., Meng H., Meng H., Wang X., Wang X., Lin S., Lin S., Ji Z., Ji Z., Zhang H., Zhang H. Acc. Chem. Res. 2013;46:607–621. doi: 10.1021/ar300022h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nymark P., Catalán J., Suhonen S., Järventaus H., Birkedal R., Clausen P.A., Jensen K.A., Vippola M., Savolainen K., Norppa H. Toxicology. 2013;313:38–48. doi: 10.1016/j.tox.2012.09.014. [DOI] [PubMed] [Google Scholar]
- Ohnishi T., Yoshida T., Igarashi A., Muroi M., Tanamoto K.-I. FEMS Immunol. Med. Microbiol. 2008;52:293–295. doi: 10.1111/j.1574-695X.2007.00355.x. [DOI] [PubMed] [Google Scholar]
- Öner D., Moisse M., Ghosh M., Duca R.C., Poels K., Luyts K., Putzeys E., Cokic S.M., Van Landuyt K., Vanoirbeek J.E.A. Mutagenesis. 2016;32:181–191. doi: 10.1093/mutage/gew053. [DOI] [PubMed] [Google Scholar]
- Palomäki J., Sund J., Vippola M., Kinaret P., Greco D., Savolainen K., Puustinen A., Alenius H. Nanotoxicology. 2015;9:719–728. doi: 10.3109/17435390.2014.969346. [DOI] [PubMed] [Google Scholar]
- Riebeling C., Jungnickel H., Luch A., Haase A. Adv. Exp. Med. Biol. 2017;947:143–171. doi: 10.1007/978-3-319-47754-1_6. [DOI] [PubMed] [Google Scholar]
- Robinson J.F., Pennings J.L.A., Piersma A.H. Methods Mol. Biol. 2012;889:347–371. doi: 10.1007/978-1-61779-867-2_22. [DOI] [PubMed] [Google Scholar]
- Roy A., Bauer S.M., Lawrence B.P. PLoS One. 2012;7 doi: 10.1371/journal.pone.0038448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rydman E.M., Ilves M., Koivisto A.J., Kinaret P.A.S., Fortino V., Savinko T.S., Lehto M.T., Pulkkinen V., Vippola M., Hämeri K.J.E.A. Part. Fibre Toxicol. 2014;11:838. doi: 10.1186/s12989-014-0048-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rydman E.M., Ilves M., Vanhala E., Vippola M., Lehto M., Kinaret P.A.S., Pylkkänen L., Happo M., Hirvonen M.-R., Greco D.E.A. Toxicol. Sci. 2015;147:140–155. doi: 10.1093/toxsci/kfv112. [DOI] [PubMed] [Google Scholar]
- G. Scala, V. Marwah, P. Kinaret, J. Sund, V. Fortino, D. Greco, Data in Brief, Submitted (2018). [DOI] [PMC free article] [PubMed]
- Schlinkert P., Casals E., Boyles M., Tischler U., Hornig E., Tran N., Zhao J., Himly M., Riediker M., Oostingh G.E.A. J. Nanobiotechnol. 2015;13:1. doi: 10.1186/s12951-014-0062-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sierra M.I., Rubio L., Bayón G.F., Cobo I., Menendez P., Morales P., Mangas C., Urdinguio R.G., Lopez V., Valdes A., Vales G., Marcos R., Torrecillas R., Fernández A.F., Fraga M.F. Nanotoxicology. 2017;11:857–870. doi: 10.1080/17435390.2017.1371350. [DOI] [PubMed] [Google Scholar]
- Singh S., Shi T., Duffin R., Albrecht C., Vanberlo D., Hohr D., Fubini B., Martra G., Fenoglio I., Borm P. Toxicol. Appl. Pharmacol. 2007;222:141–151. doi: 10.1016/j.taap.2007.05.001. [DOI] [PubMed] [Google Scholar]
- Smyth G.K. Springer-Verlag; New York: 2005. Limma: Linear Models for Microarray Data. [Google Scholar]
- Snyder-Talkington B.N., Dong C., Zhao X., Dymacek J., Porter D.W., Wolfarth M.G., Castranova V., Qian Y., Guo N.L. Toxicology. 2015;328:66–74. doi: 10.1016/j.tox.2014.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stearns R.C., Paulauskis J.D., Godleski J.J. Am. J. Respir. Cell Mol. Biol. 2001;24:108–115. doi: 10.1165/ajrcmb.24.2.4081. [DOI] [PubMed] [Google Scholar]
- Sund J., Alenius H., Vippola M., Savolainen K., Puustinen A. ACS Nano. 2011;5:4300–4309. doi: 10.1021/nn101492k. [DOI] [PubMed] [Google Scholar]
- Törnqvist E., Annas A., Granath B., Jalkesten E., Cotgreave I., Öberg M. PLoS One. 2014;9 doi: 10.1371/journal.pone.0101638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tralau T., Oelgeschläger M., Gürtler R., Heinemeyer G., Herzler M., Höfer T., Itter H., Kuhl T., Lange N., Lorenz N., Müller-Graf C., Pabel U., Pirow R., Ritz V., Schafft H., Schneider H., Schulz T., Schumacher D., Zellmer S., Fleur-Böl G., Greiner M., Lahrssen-Wiederholt M., Lampen A., Luch A., Schönfelder G., Solecki R., Wittkowski R., Hensel A. Arch. Toxicol. 2015;89:823–850. doi: 10.1007/s00204-015-1510-0. [DOI] [PubMed] [Google Scholar]
- Veranth J.M., Reilly C.A., Veranth M.M., Moss T.A., Langelier C.R., Lanza D.L., Yost G.S. Toxicol. Sci. 2004;82:88–96. doi: 10.1093/toxsci/kfh248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vippola M., Bard D., Sarlin E., Tuomi T., Tossavainen A. Finnish Institute of Occupational Health; 2009. Nanoatlas of Selected Engineered Nanoparticles. [Google Scholar]
- Watanabe H., Adachi R., Kusui K., Hirayama A., Kasahara T., Suzuki K. Int. Immunopharmacol. 2003;3:1601–1608. doi: 10.1016/S1567-5769(03)00182-6. [DOI] [PubMed] [Google Scholar]
- Wijetunga N.A., Johnston A.D., Maekawa R., Delahaye F., Ulahannan N., Kim K., Greally J.M. BMC Bioinf. 2017;18:534. doi: 10.1186/s12859-017-1477-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G., Wang L.-G., Han Y., He Q.-Y. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Li X., Xie F., Liu K., Liu H., Xie J. Environ. Toxicol. Pharmacol. 2017;54:40–47. doi: 10.1016/j.etap.2017.06.023. [DOI] [PubMed] [Google Scholar]
- Zhao F., Klimecki W.T. J. Appl. Toxicol. 2014;35:945–951. doi: 10.1002/jat.3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Viability (ATP) and cytotoxicity (LDH release) assays with eight exposure concentrations (0, 0.1, 0.5, 1, 5, 10, 100 and 500 μg/ml) to A549 cells, as well as lysed cells as positive controls. Titles of each graph (A–J) correspond to the material acronyms found from Table 1.
Viability (ATP) and cytotoxicity (LDH release) assays with eight exposure concentrations (0, 0.1, 0.5, 1, 5, 10, 100 and 500 μg/ml) to BEAS-2B cells, as well as lysed cells as positive controls. Titles of each graph (A–J) correspond to the material acronyms found from Table 1.
Viability (ATP) and cytotoxicity (LDH release) assays with eight exposure concentrations (0, 0.1, 0.5, 1, 5, 10, 100 and 500 μg/ml) to PMA-differentiated THP-1 cells, as well as lysed cells as positive controls. Titles of each graph (A–J) correspond to the material acronyms found from Table 1.
Excel spreadsheet, reporting in each sheet differential analysis summaries and SMITE gene scores for each exposure. For each gene are reported in the order the gene symbol, the SMITE score, the combined methylation log fold-change for promoter region, the combined methylation log fold-change for body region, the combined log fold-change of targeting miRNAs, mRNA expression log fold-change, combined methylation p-value for promoter region, combined p-values of targeting miRNAs, combined methylation p-value for body region, mRNA expression p-value.