The Bacillus cereus group of bacteria includes species of high economic, clinical, biological warfare, and biotechnological interest, e.g., B. anthracis in bioterrorism, B. cereus in food intoxications, and B. thuringiensis in biocontrol. Knowledge about the ecology of these bacteria is hindered by our limited understanding of the regulatory circuits that control differentiation and specialization processes. Here, we uncover the participation of eight Rap quorum-sensing receptors in collective functions of B. thuringiensis. These proteins are highly multifunctional and redundant in their functions, linking ecologically relevant processes such as sporulation, biofilm formation, spreading, extracellular proteolytic activity, and probably other functions in species from the B. cereus group.
KEYWORDS: Bacillus cereus, Bacillus thuringiensis, biofilm formation, collective functions, quorum sensing, Rap-Phr, sporulation
ABSTRACT
Quorum sensing (QS) is a mechanism of synthesis and detection of signaling molecules to regulate gene expression and coordinate behaviors in bacterial populations. In Bacillus subtilis, multiple paralog Rap-Phr QS systems (receptor-signaling peptides) are highly redundant and multifunctional, interconnecting the regulation of differentiation processes such as sporulation and competence. However, their functions in the Bacillus cereus group are largely unknown. We evaluated the functions of Rap proteins in Bacillus thuringiensis Bt8741, which codes for eight Rap-Phr systems; these were individually overexpressed to study their participation in sporulation, biofilm formation, spreading, and extracellular proteolytic activity. Our results show that four Rap-Phr systems (RapC, RapK, RapF, and RapLike) inhibit sporulation, two of which (RapK and RapF) probably dephosphorylate Spo0F from the Spo0A phosphorelay; these two Rap proteins also inhibit biofilm formation. Four systems (RapC, RacF1, RacF2, and RapLike) participate in spreading inhibition; finally, six systems (RapC, -F, -F2, -I, and -I1 and RapLike) decrease extracellular proteolytic activity. We foresee that functions performed by Rap proteins of Bt8741 could also be carried out by Rap homologs in other strains within the B. cereus group. These results indicate that Rap-Phr systems constitute a highly multifunctional and redundant regulatory repertoire that enables B. thuringiensis and other species from the B. cereus group to efficiently regulate collective functions during their life cycle in the face of changing environments.
IMPORTANCE The Bacillus cereus group of bacteria includes species of high economic, clinical, biological warfare, and biotechnological interest, e.g., B. anthracis in bioterrorism, B. cereus in food intoxications, and B. thuringiensis in biocontrol. Knowledge about the ecology of these bacteria is hindered by our limited understanding of the regulatory circuits that control differentiation and specialization processes. Here, we uncover the participation of eight Rap quorum-sensing receptors in collective functions of B. thuringiensis. These proteins are highly multifunctional and redundant in their functions, linking ecologically relevant processes such as sporulation, biofilm formation, spreading, extracellular proteolytic activity, and probably other functions in species from the B. cereus group.
INTRODUCTION
Bacteria perform many functions that depend on multicellular-organism-like behaviors, such as cell differentiation and specialization. These collective functions allow the emergence of complex ecological interactions, including cooperation and division of labor in biofilms (1, 2). Collective functions are only evident and effective when performed by large groups in bacterial populations or communities (3–6). Some of the most-studied examples include bioluminescence by the squid symbiont Vibrio fischeri (7) and fruiting body formation during sporulation of Myxococcus xanthus (8).
In Gram-positive bacteria, collective functions and the molecular mechanisms for their control have been widely studied in Bacillus subtilis. In B. subtilis cultures, several mutually exclusive cell types (motile, competent, sporulating, cannibal, biofilm matrix producing, surfactant producing, and mining [9, 10]) mediate the emergence of ecological interactions such as cooperation, cheating, and cross-feeding (5, 6, 11). These phenomena, which ultimately affect the manifestation of collective traits such as sporulation efficiency, surface colonization, biofilm architecture complexity, etc. (2, 9, 12) depend on global modifications of transcriptional regulation; they are triggered by environmental cues, stress conditions, and cell-cell signaling and are tightly modulated by complex, overlapping regulatory circuits (13–15).
Bacteria detect cell density through quorum sensing (QS), which depends on self-produced signaling molecules that accumulate in the extracellular space as the population grows. Specific receptors in the cell membrane or in the cytoplasm recognize these signaling molecules and regulate downstream cellular processes (16–18). Collective traits such as virulence, competence, sporulation, and bioluminescence are regulated by QS. Gram-positive bacteria use small peptides as signaling molecules for QS (17).
The RRNPP protein family (Rgg, Rap, NprR, PlcR, and PrgX) are intracellular QS receptors that regulate several functions across Gram-positive bacteria (19–21). Genes coding for receptor proteins and their associated signaling peptides are encoded in transcriptional cassettes (22). To carry out signaling, peptides need to be secreted, matured by proteolysis and reinternalized at high cell density or quorum state (17, 20). Rgg, NprR, PlcR, and PrgX proteins are transcriptional activators that bind directly to DNA. Rap proteins, however, lack a DNA-binding domain, and they function by binding to and inhibiting response regulators or transcriptional activators (21, 23, 24). In high cell density, Phr signaling peptides bind to specific Rap proteins and release their inhibitory functions (25). Eleven Rap paralogs from B. subtilis strain 168 (RapA, -B, -C, -D, -E, -F, -G, -H, -I, -J, and -K), RapP from the B. subtilis 168 parental strain NCIB3610 (26), and Rap60 in strains carrying the plasmid pTA1060 (27) control diverse functions. The RapG-PhrG pair regulates the activation of DegU, a transcriptional regulator that controls aprE and comK genes encoding extracellular proteases and a transcription factor for competence in B. subtilis, respectively (15, 28); activity of ComA—the master regulator of competence genes—is inhibited by RapC, -D, -F, -G, -H, -K, and -60 (14, 27, 29–33); Spo0A—the transcriptional activator of many differentiation genes—is indirectly regulated by RapA, -B, -E, -H, -J, -P, and -60 (24, 27, 34–38). Hence, Rap protein paralogs from B. subtilis are highly multifunctional and redundant, and they connect several differentiation processes and coordinate collective traits.
Spo0A is activated by phosphorylation through a multicomponent phosphorelay system. Up to five kinases autophosphorylate in response to intracellular and environmental stress signals and transfer the phosphate group to Spo0F, which is then transferred to Spo0B and finally to Spo0A (39). Spo0A-P activates the transcription of multiple genes, including biofilm formation (at low concentrations) and early sporulation genes (at high concentrations [13]). Rap QS proteins prevent the phosphate transfer in the phosphorelay by binding to Spo0F (34, 40).
While the regulation of collective traits in B. subtilis is well known, these phenomena remain largely understudied in the Bacillus cereus group, which includes bacteria with clinical and biotechnological relevance (41). Although B. subtilis and the B. cereus group species share similar characteristics, such as the sporulation process and the Spo0A phosphorelay components, and have many protein families in common, they also present several genetic differences (42). In Bacillus thuringiensis (the most widely used biopesticide), the bifunctional QS receptor NprR, which is not present in B. subtilis (43–45), modulates the Spo0A phosphorelay through binding to Spo0F (similar to the activity of Rap proteins) and functions as a transcriptional activator through DNA binding. On the other hand, ComA and DegU response regulators are not encoded in B. thuringiensis. Additionally, Rap-Phr QS systems also differ in both groups. These QS systems have evolved by duplication and divergence mechanisms; even though multiple Rap protein paralogs are also found in the B. cereus group species, they have evolved independently, and no Rap homologs are shared between the two groups (46, 47). Therefore, it is not possible to predict the functions of Rap proteins in the B. cereus group based on what is known of Rap proteins from B. subtilis.
Some Rap-Phr systems from species of the B. cereus group have been studied. First, Rap BXA0205 and BA3790 from Bacillus anthracis strain A2012 were demonstrated to regulate sporulation initiation and to dephosphorylate Spo0F (48). Later, it was shown that Rap8 from B. thuringiensis HD73 regulates the sporulation process in vitro and in the insect cadaver as well as biofilm formation in vitro (49). A more recent study showed the participation of Rap6, -7, and -8—also known as RapC, RapK, and RapF, respectively (50)—in the modulation of the sporulation process in B. thuringiensis Bt407 (51). However, other Rap paralogs with unknown functions have been identified in the genomes of B. cereus group bacteria (47, 50), and these may be relevant to their ecology.
In this study, we evaluated the functions of Rap proteins using B. thuringiensis Bt8741 as a model. We generated eight Rap overexpression strains from Bt8741 to evaluate the role of each Rap paralog in sporulation efficiency, biofilm formation, spreading, and extracellular proteolytic activity. Once we assigned functions to Bt8741 Rap proteins, we identified homolog Rap proteins in the genomes of other strains and species from the B. cereus group; this allows the prediction of their functions based on Rap proteins from Bt8741 studied here.
(This article was submitted to an online preprint archive [52].)
RESULTS
Spo0F-binding residues from RapHBs are conserved in Rap proteins from Bt8741.
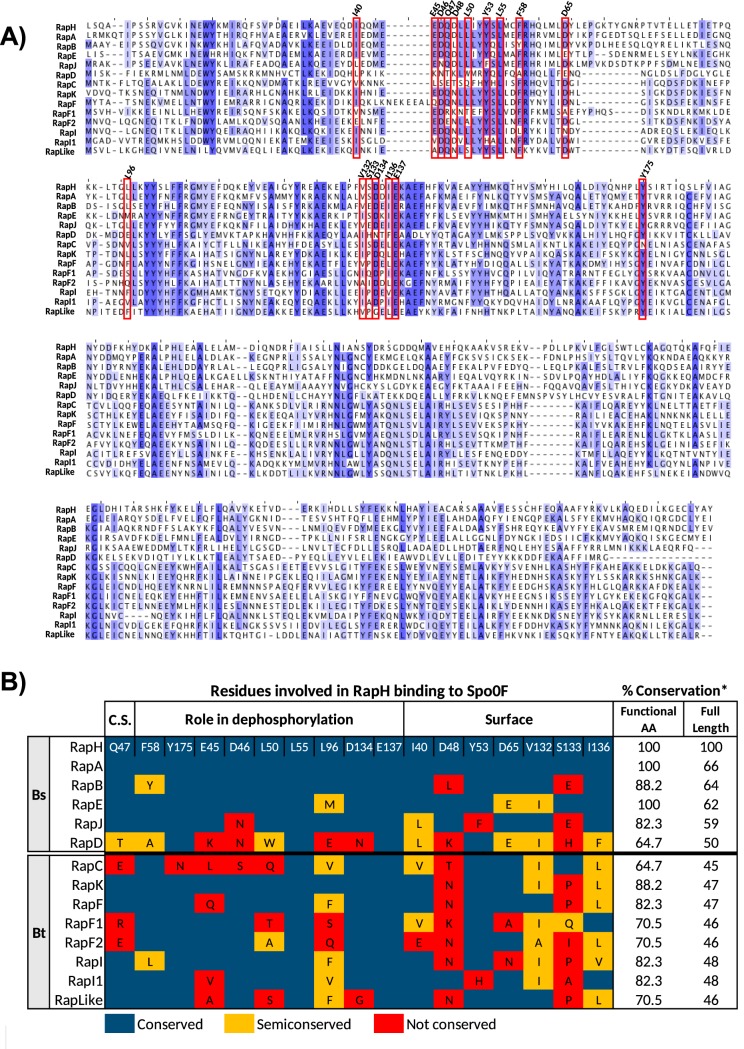
In order to predict the capacity of Rap proteins from Bt407 (a strain closely related to Bt8741) to bind to Spo0F, we analyzed the conservation of amino acids involved in Spo0F-binding by RapH from B. subtilis (RapHBs) (Fig. 1) (36). Rap proteins with high conservation of Spo0F-binding residues should retain the activity of inhibiting sporulation initiation, while Rap proteins with lower conservation could be specialized for other functions. We found more conservation of the functional amino acids of RapHBs in the sequences of both B. subtilis 168 and Bt407 compared to the corresponding full sequences (Fig. 1B). In B. subtilis 168, the full sequence conservation of the Rap proteins known to bind to Spo0F (RapA, -B, -E, and -J) compared to RapHBs ranged from 59% to 66%, and the functional amino acid conservation percentage, from 82% to 100%. In RapDBs, which does not bind to Spo0F, the full-length sequence is conserved at 50%, and the functional residues are only 64% conserved (Fig. 1B). In the case of Rap proteins from Bt407, the full sequence conservation in comparison to RapHBs ranged from 45% to 48%. On the other hand, conservation of the functional residues ranged from 64% to 88% (Fig. 1B). Since more conservation occurs in the Spo0F-binding functional residues, these amino acids could be important for the function of B. thuringiensis Rap proteins.
FIG 1.
Prediction of the capacity of Rap proteins from Bt8741 to bind and dephosphorylate Spo0F. (A) Multiple sequence alignment of the complete amino acid sequences of RapH, -A, -B, -E, -J, and -D from B. subtilis 168 and eight Rap proteins from Bt8741. Blue highlighting indicates highly conserved amino acids. Residues involved in the RapHBS-Spo0F binding are shown in red rectangles, and their position in RapHBs is shown on top of the alignment. (B) Conservation of residues involved in RapHBs binding to Spo0F. Residues were considered semiconserved when a functional amino acid of RapHBs was substituted with another amino acid with similar characteristics. Bs, Bacillus subtilis 168; Bt, Bacillus thuringiensis Bt407; C.S., catalytic site; *, percentage of conserved and semiconserved amino acids in pairwise alignment to RapHBs.
RapK exhibited the highest conservation percentage of Spo0F-binding residues (88%), followed by RapF, RapI, and RapI1 (82%), RapF1, RapF2, and RapLike (70%), and, finally, RapC (64%). We found that RapF1, RapF2, and RapC do not conserve the residue Q47, which is essential for the phosphatase activity of RapHBs (36). This analysis enables the prediction that some Rap protein paralogs from Bt8741 with a high conservation percentage of putative Spo0F-binding amino acids could dephosphorylate Spo0F. Indeed, RapK and RapF from Bt407, Rap8 from B. thuringiensis HD73 (ortholog to RapI from Bt407), and Rap BXA0205 and BA3790 from B. anthracis (homologs of RapK and RapF2, respectively) have been shown to participate in the modulation of sporulation (48, 49, 51). Previous to this work, RapF1, RapI1, and RapLike from Bt407 (or its homologs in other strains) had not been tested for their role in sporulation.
RapC, RapK, RapF, and RapLike control sporulation in Bt8741.
We constructed nine Rap overexpression strains in the Bt8741 wild-type (WT) background (see Table S1 in the supplemental material), one for each endogenous Rap protein identified in Bt407 (RapC, -K, -F, -F1, -F2, -I, and -I1 and RapLike) (50) and one for RapA from B. subtilis 168 (RapABs). We also generated a control strain of Bt8741 carrying the empty plasmid pHT315-PxylA (Table S1). Because some Rap proteins of B. subtilis are known to negatively regulate sporulation (24, 27, 34–37), the overexpression of Rap proteins involved in the regulation of sporulation in B. thuringiensis should result in the decrease of thermoresistant CFU (spores). A growth time course experiment confirmed that neither xylose addition nor Rap overexpression affected bacterial growth (Fig. S1).
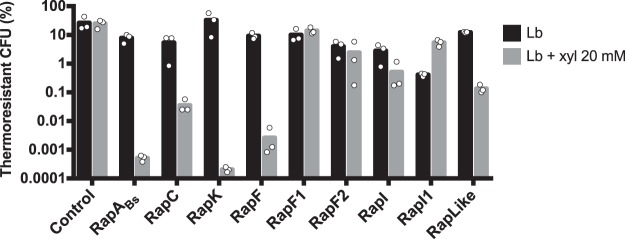
Sporulation efficiency of the control strain remained unchanged by the addition of xylose (Fig. 2). In contrast, the strain carrying PxylA′rapABs had decreased sporulation efficiency (from 7% to 0.0005%, ≈14,000-fold) caused by the induction with xylose. In fact, thermoresistant CFU were undetectable when RapABs was overexpressed (Fig. S2; CFU data of Fig. 2). We also found undetectable levels of spores in strains overexpressing RapK and RapF (Fig. S2); sporulation efficiency decreased ≈160,000-fold in the strain overexpressing RapK and ≈3,400-fold in the strain overexpressing RapF (Fig. 2). Strains carrying PxylA′rapC and PxylA′rapLike also exhibited reduced sporulation efficiency of ≈140-fold and ≈88-fold, respectively (Fig. 2). Finally, we found a slight ≈5-fold decrease in sporulation efficiency when RapI was overexpressed, and sporulation efficiency was not affected by the overexpression of RapF1, RapF2, or RapI1.
FIG 2.
Sporulation efficiency of Bt8741 carrying overexpression plasmids for Rap proteins with and without addition of inducer. In the cases where thermoresistant CFU were undetectable, we considered a value of 166 spores/ml, which is the detection limit for this assay. Columns represent the average of three individual measurements, shown as dots.
In this assay, we found that the addition of xylose to the medium and the presence of rap genes in plasmids had unspecific effects on growth and sporulation; e.g., in the control strain, the addition of xylose caused a decrease of ≈1 log10 in total and thermoresistant CFU (Fig. S2). For this reason, we used sporulation efficiency instead of CFU to identify Rap proteins that decrease sporulation. Additionally, our analysis was based on comparisons within each strain in induced versus not induced conditions, instead of comparing each overexpression strain against the control strain in induced conditions; this could be the source of discrepancies between our results and those of previous work (e.g., RapI versus the homolog Rap8 in B. thuringiensis HD73 [49]).
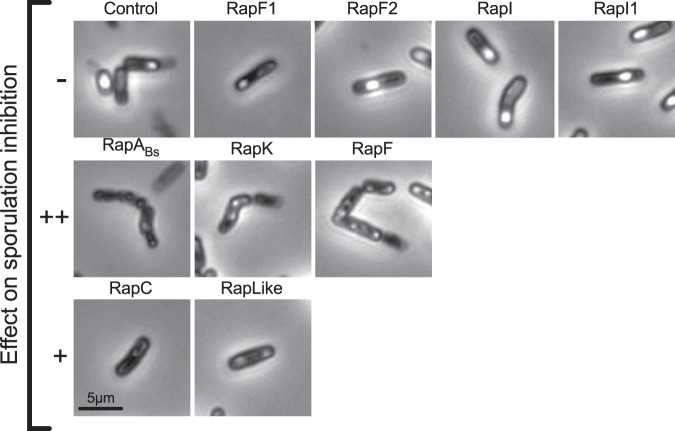
Samples of the Rap-overexpressing strain cultures at 72 h were observed with a microscope. We detected free spores and bacterial debris in all cultures when Rap proteins were not overexpressed (Fig. S3). Strains overexpressing Rap proteins that did not affect sporulation efficiency (RapF1, -F2, -I, and -I1) showed cell morphology similar to that of the control strain, i.e., bacilli with defined endospores. In samples from strains overexpressing RapABs, RapK, and RapF that acutely decreased sporulation efficiency, we observed chained, wrinkled cells with no spores (Fig. 3). Cells from strains overexpressing RapC and RapLike, were observed as rod shaped, and no spores were evident (Fig. 3).
FIG 3.
Cell morphology of strains with induced Rap protein overexpression at 72 h. Phase-contrast microscopy of 63× and 1.8× magnification. –, no effect; ++, undetectable thermoresistant CFU; +, decreased sporulation efficiency, detectable thermoresistant CFU.
The analysis of Spo0F-interacting residues (Fig. 1) was partially accurate at predicting the participation of Rap proteins in sporulation. RapK and RapF that highly conserve the Spo0F-binding residues had the strongest effect on sporulation inhibition (Table 1); however, RapI and RapI1 also had a high conservation of Spo0F-binding residues, and their overexpression had no effect on sporulation. Additionally, RapC and RapLike, which had lower conservation of Spo0F-binding residues, decreased sporulation efficiency.
TABLE 1.
Summarized results of the participation of Rap proteins in collective functions of Bt8741
| Phenotype for:c |
|||||||
|---|---|---|---|---|---|---|---|
| Namea | Nameb | Spo0F-binding prediction (%) | Sporulation | Biofilm formation | Spreading | Extracellular proteases | Multifunctional Rap? |
| RapABs | 100 | ++ | |||||
| RapC | Rap6 | 64.7 | + | ++ | + | Yes | |
| RapK | Rap8 | 88.2 | ++ | ++ | Yes | ||
| RapF | Rap7 | 82.3 | ++ | ++ | + | Yes | |
| RapF1 | Rap1 | 70.5 | + | ||||
| RapF2 | Rap4 | 70.5 | ++ | + | Yes | ||
| RapI | Rap5 | 82.3 | + | ||||
| RapI1 | Rap2 | 82.3 | + | ||||
| RapLike | Rap3 | 70.5 | + | ++ | + | Yes | |
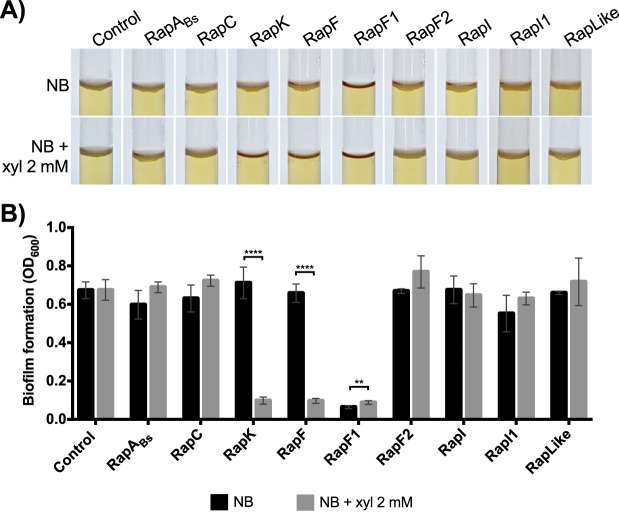
Overexpression of RapF and RapK prevents biofilm formation of Bt8741.
In nature, over 80% of bacteria live in biofilms (53); therefore, biofilm formation may be a relevant trait—albeit an understudied one—during the life cycle of B. thuringiensis. We quantified biofilm formation of the Rap overexpression strains in the air-liquid interphase at 48 h. Since 20 mM xylose in the medium caused a complete inhibition of biofilm formation in the Bt8741 control strain (not shown), we tested the effect of xylose concentration on this phenotype. We found that biofilm formation was not affected at 2 mM but was decreased at higher concentrations of 5, 10, and 15 mM (Fig. S4); therefore, overexpression of Rap proteins was performed with 2 mM xylose (54).
Overexpression of RapK and RapF caused the inhibition of biofilm formation of Bt8741 (Fig. 4A), evident by the significant decrease (P < 0.0001) in the optical density at 600 nm (OD600) measured in samples obtained from the surface of the cultures (Fig. 4B). On the other hand, biofilms were normally formed by strains overexpressing RapABs, -C, -F2, -I, and -I1 and RapLike (Fig. 4). The strain overexpressing RapF1 was unable to form biofilms even when overexpression was not induced (Fig. 4).
FIG 4.
Biofilm formation of Rap overexpression strains at 48 h. (A) Biofilms formed in the air-liquid interphase in glass tubes at 48 h. Biofilms are identified as a white layer on the surface of the medium. (B) Biofilm formation quantification of Rap overexpression strains in induced and not induced media after 48 h. Columns represent the average from 5 replicates ± standard deviation (SD). NB, nutrient broth; **, P < 0.005; ****, P < 0.0001.
In order to discard possible global growth defects in this assay when RapK and RapF were overexpressed, we measured planktonic growth through OD600 of the liquid media where biofilm formation was assessed. We found that planktonic growth was higher in conditions where a biofilm was not formed (Fig. S5). This suggests that RapK and RapF specifically inhibit biofilm formation (e.g., expression of genes related to synthesis of extracellular matrix components).
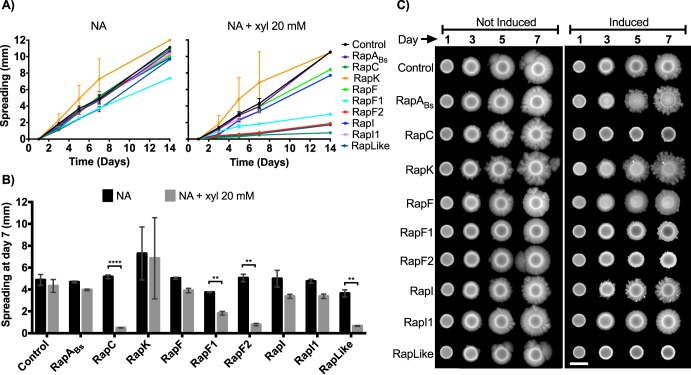
RapC, RapF1, RapF2, and RapLike regulate spreading of Bt8741 colonies.
Colonies of Bt8741 present a spreading phenotype that could be associated with its capacity to colonize hosts and habitats. Similar passive motility phenotypes have been described in other species of Bacillus, associated with the production of extracellular surfactant molecules (55–57). We observed that the overexpression of RapC, RapF1, RapF2, and RapLike caused a decrease in spreading (P < 0.05) of Bt8741 colonies at day 7 (Fig. 5A and B). The overexpression of RapC reduced 90% of the colony dispersion; RapF1 reduced 50%, RapF2 reduced 84%, and RapLike reduced 82% (Fig. 5B). We observed that the overexpression of RapC, RapF2, and RapLike completely eliminated this phenotype, while overexpression of RapF1 only decreased spreading (P < 0.05) (Fig. 5B and C).
FIG 5.
Spreading phenotype of Rap overexpression strains. (A) Spreading kinetics of colonies on agar. Each point represents the average from triplicates ± SD; only one data point is shown at day 14. (B) Spreading quantification of Rap overexpression colonies at day 7. Columns represent the average from triplicates ± SD. (C) Pictures of representative Rap overexpression strains spreading during 7 days. The scale bar indicates 5 mm. NA, nutrient agar; **, P < 0.005; ****, P < 0.0001.
The overexpression of RapABs, RapK, RapF, RapI, and RapI1 did not affect the spread of Bt8741 (P > 0.05) (Fig. 5B). In some cases, Rap overexpression affected colony morphology; i.e., colonies of strains overexpressing RapABs, RapK, and RapI showed an increased dendritic phenotype. However, the spreading phenotype, measured as colony radius, was still present (Fig. 5C).
Extracellular proteolytic activity is downregulated by RapC, -F, -F2, -I, and -I1 and RapLike in Bt8741.
In B. thuringiensis, the production of extracellular proteases is crucial during its necrotrophic phase, i.e., development in insect cadavers (43, 46). We tested the role of Rap proteins in extracellular proteolytic activity by measuring the effect of Rap overexpression on hydrolysis halos of colonies on milk agar (MA) plates. Overexpression of RapC, -F, -F2, -I, and -I1 and RapLike decreased the halo area (P < 0.05; Fig. 6B). In contrast, the proteolytic activity of the control strain and strains overexpressing RapABs, RapK, and RapF1 was not affected by the induction (P > 0.05; Fig. 6).
FIG 6.
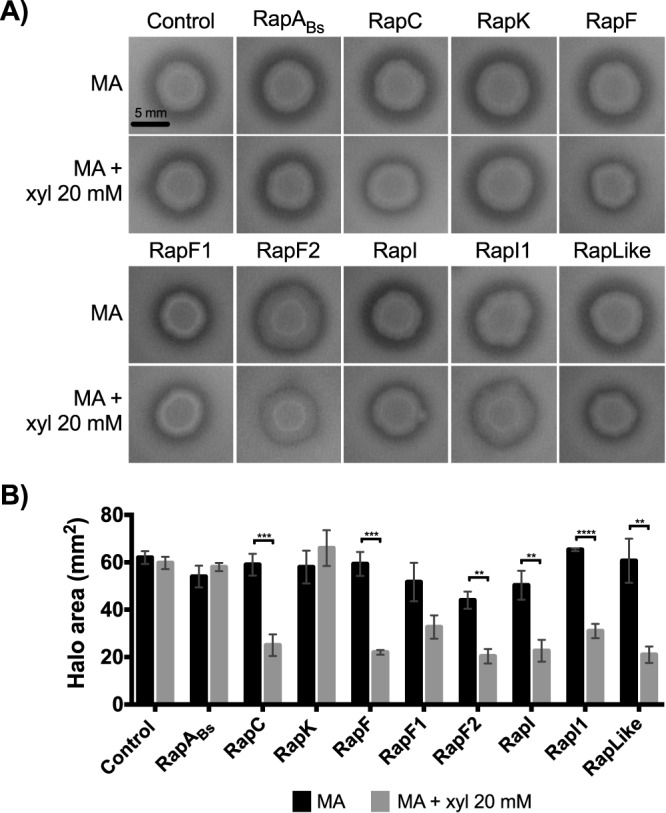
Extracellular proteolytic activity of Rap overexpression strains. (A) Effect of Rap protein overexpression in the hydrolysis halo of Rap overexpression strains colonies. (B) Hydrolysis halo area with and without Rap overexpression induction. Columns represent the average of 3 replicates ± SD. MA, milk agar; **, P < 0.005; ***, P < 0.0005; ****, P < 0.0001.
We noted a coincidence between Rap proteins that participate in inhibiting extracellular proteolytic activity and spreading (RapC, RapF2, and RapLike). However, we determined that the effects on spreading could not affect our measurements of extracellular proteases, because the spreading phenotype is not yet relevant at day 1 (Fig. 5), when proteolytic activity was assessed. Indeed, colony area was not affected by the overexpression of Rap proteins (except for RapF2), and the colony size of Rap-overexpressing strains was not reduced compared to that of the control strain (Fig. S6).
Are Rap paralogs a multifunctional and redundant regulatory repertoire across the B. cereus group?
Our phenotypic analyses showed that all Rap proteins participate in the regulation of at least one of the collective functions studied here (Table 1). We found that five out of eight Rap proteins from Bt8741 (RapC, RapK, RapF, RapF2, and RapLike) participate in more than one collective function, and all four collective functions were inhibited by more than one Rap protein.
In order to predict the functions of Rap paralogs across the B. cereus group, we analyzed Rap amino acid sequences from representative strains of B. thuringiensis, B. cereus, B. anthracis, B. weihenstephanensis, B. mycoides, B. pseudomycoides, and B. cytotoxicus (51) (Data Set S1). Since several Rap paralogs can be found in all species (47, 51), we hypothesized that the detection of Rap homologs to Bt8741 Raps could be useful in order to predict their functions. Seven out of eight Rap proteins from Bt8741 share homologs (identity, <90%) in other strains from the B. cereus group (Fig. 7). Homologs of RapK, RapF, and RapI were only found in other B. thuringiensis strains, while homologs of RapF1, RapF2, RapI1, and RapLike were found in strains from both B. thuringiensis and B. cereus. Notably, we did not detect RapC homologs in any strain (other than Bt407) or homologs to any Bt8741 Rap proteins in B. anthracis, B. weihenstephanensis, B. mycoides, B. pseudomycoides, or B. cytotoxicus; this was verified by querying amino acid sequences of Bt8741 Rap proteins against the NCBI GenBank database (as of November 2019) (Data Set S1). These results provide useful hypotheses for experimental testing of Rap protein functions in future studies.
FIG 7.
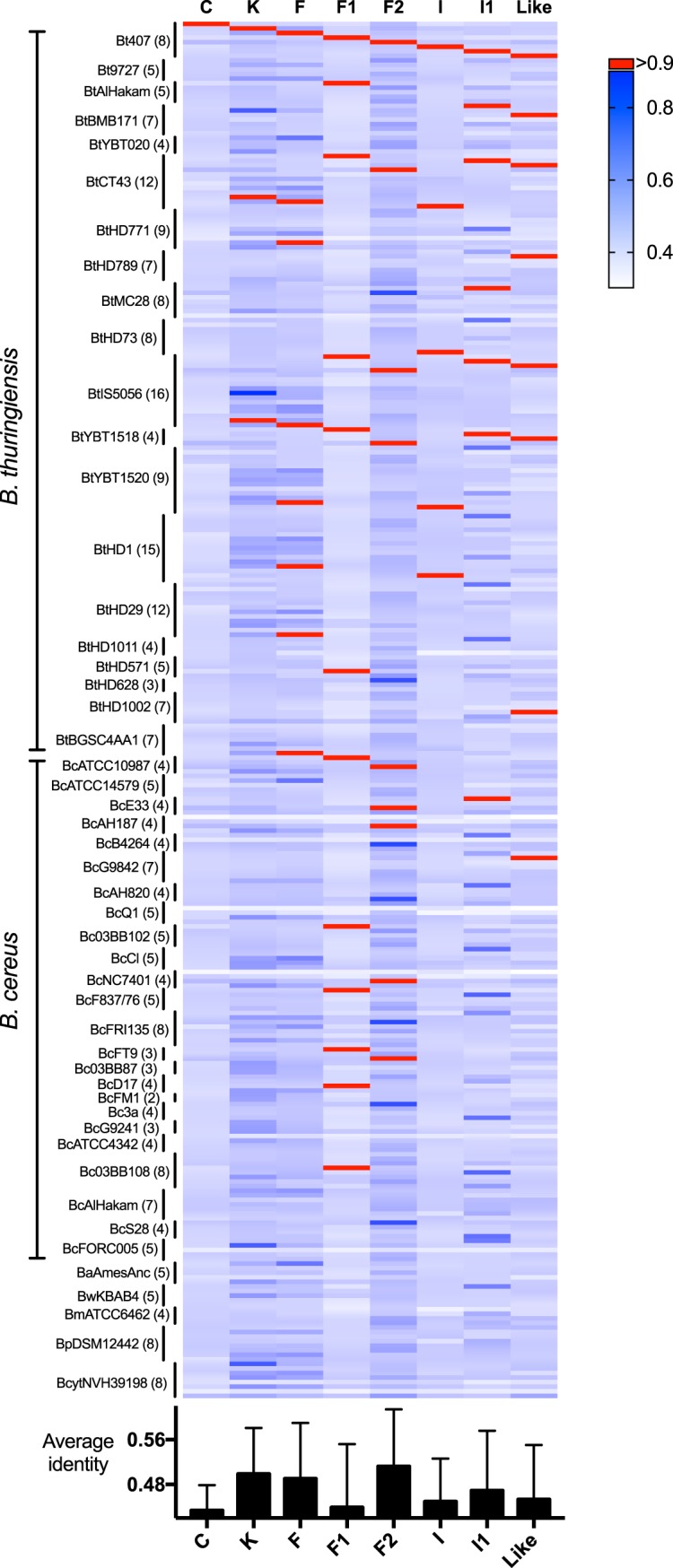
Homologs of Rap proteins from Bt8741 in strains from the B. cereus group. (Top) Heat map representing the identity of each Rap protein from Bt8741 against all Raps identified in strains from the B. cereus group (51). (Bottom) Average identity of each Rap protein from Bt8741 against all Raps from the B. cereus group. Bt, B. thuringiensis; Bc, B. cereus; Ba, B. anthracis; Bw, B. weihenstephanensis; Bm, B. mycoides; Bp, B. pseudomycoides; Bcyt, B. cytotoxicus. Numbers in parentheses indicate the number of Rap paralogs found in each strain. Values of identity for individual comparisons are available in Data Set S1.
DISCUSSION
Few studies have addressed multicellular behaviors such as differentiation, cell specialization, collective functions, and the resulting ecological interactions in species from the B. cereus group (11, 58). Similarly, molecular mechanisms for the control of differentiation processes in the B. cereus group bacteria remain understudied (48, 49, 51, 58, 59). In this work, we uncovered the functions of Rap protein paralogs from Bt8741. Rap proteins are highly redundant in B. subtilis, which could result in the compensation of phenotypes of rap deletion mutants. For this reason, we chose to generate overexpression strains of Bt8741. Through this clean, reductionist approach, we found that Rap-Phr paralogs in this strain regulate collective functions such as sporulation, biofilm formation, spreading motility, and production of extracellular proteases. Our results show that Rap paralogs constitute a regulatory repertoire that may allow B. thuringiensis populations to respond efficiently to environmental changes, contributing to the fitness of the population. Moreover, the functions uncovered here can be extrapolated to other B. thuringiensis and B. cereus strains that encode in their genomes Rap proteins highly conserved to Bt8741 Raps, but experimental confirmation is needed.
Recent studies have shown that bacteria benefit from keeping multiple Rap-Phr systems, as redundancy has been selected for due to the social advantages it provides (60). Because Rap proteins have a repressive function upon their target, the gain of a novel Rap-Phr system for the regulation of extracellular public good production enables a facultative cheating mechanism in which variants with an extra system exploit their ancestral strain. Here, we show that the production of extracellular public goods, such as biofilm matrix components (58, 61), extracellular proteases (62), and surfactants (such as kurstakins [43, 63, 64]), are likely controlled by Rap proteins in Bt8741. Therefore, the same facultative cheating mechanism could be expected during duplication of rap-phr genes in the B. cereus group, resulting in the presence of multiple Rap paralogs generated in an evolutionary process independent from that of B. subtilis (47). Multifunctionality may also provide evolutionary advantages. Because Rap-Phr systems are known to be parallel signaling pathways (47), they are not all activated simultaneously; instead, some of them may be active only under specific conditions, achieving the regulation of various differentiation processes and collective functions while optimizing energetic costs. Our phenotypic studies show that Raps from B. thuringiensis have specialized for a variety of functions, and diversification was probably facilitated by those mechanisms. The phylogeny of Raps from the B. cereus group (47, 51) indicates that this is the case for all species within the group, due to the presence of several Rap paralogs encoded in their genomes.
Sporulation in the Bacillus genus is essential for bacterial survival and dissemination in their habitats; it is also important for the biotechnological uses of Bacillus species. In B. subtilis, five Rap-Phr systems negatively regulate Spo0A phosphorelay by dephosphorylating Spo0F and therefore prevent the activation of Spo0A (34). We found that RapABs retained this function when it was overexpressed in Bt8741. Furthermore, four Rap-Phr systems from Bt8741 (RapK, RapF, RapC, and RapLike) also regulate sporulation in this species. We propose that RapK and RapF may function by dephosphorylating Spo0F; this suggestion is supported by the following findings: (i) both RapK and RapF retain a high conservation of Spo0F-binding residues from RapH, including the catalytic residue Q47; (ii) their overexpression resulted in an undetectable number of spores, similar to RapABs overexpression; (iii) the overexpression of RapABs, RapK, and RapF caused an identical cell morphology in the three overexpressing strains; and (iv) RapK and RapF also regulated biofilm formation, probably through the same activity on Spo0F.
Our results show that the participation of Rap proteins in sporulation cannot be fully predicted from the conservation of Spo0F-binding residues or the presence of the catalytic site residue Q47. This suggests that some Rap proteins could inhibit sporulation through other, unknown mechanisms. For this reason, experimental validation (e.g., direct measurement of Spo0F binding and bona fide phosphatase activity [21, 34, 44]) is essential for showing the participation of Rap proteins in the Spo0A phosphorelay. Further studies are needed in order to directly test the mechanisms by which RapC, RapK, RapF, and RapLike regulate sporulation in B. thuringiensis and other species from the B. cereus group.
We noted that the overexpression of RapABs, which completely prevented sporulation, did not affect biofilm formation in B. thuringiensis or any other phenotype studied here. This reflects the fact that Rap proteins are not regulators that establish promiscuous protein-protein interactions; instead, they coevolve with specific protein targets in each bacterial species. B. subtilis Rap proteins target DegU, ComA, and Spo0F (21), but Rap targets in the B. cereus group (besides Spo0F, which is highly conserved in B. subtilis and B. thuringiensis) have not been studied; however, the coincidence between Rap proteins that inhibit extracellular proteolytic activity and spreading suggests that one target regulator mediates both functions.
We propose that Rap proteins have diversified according to the ecological needs of each species. For example, B. subtilis is a soil-dwelling bacterium that can be found in root-associated biofilms (65); in B. subtilis, five Rap proteins modulate Spo0A-P levels (21, 66), affecting sporulation and biofilm formation. Here, we demonstrate that four Rap proteins modulated sporulation (RapC, RapK, RapF, and RapLike), while only two of these (RapK and RapF) affected biofilm formation. This highlights the importance of sporulation regulation in both species and shows that, probably, biofilm formation is not as essential in the life cycle of B. thuringiensis. In contrast, B. thuringiensis is a soil-inhabiting, insect-pathogenic, and necrotrophic bacterium (67). In this species, extracellular protease production is essential for nutrient scavenging, which is normally associated with the necrotrophic stage of bacterial development in the insect cadaver (43); it could also be relevant for adaptation against fluctuations in nutrient availability in the environment. While only 1 out of 11 Rap proteins from B. subtilis 168 modulates extracellular proteolytic activity (RapG) (28), B. thuringiensis has extended the modulation of extracellular protease production to 6 Rap-Phr systems (RapC, -F, -F2, -I, and -I1 and RapLike). The divergence of Raps within the B. cereus group suggests that Raps from B. anthracis, B. weihenstephanensis, B. mycoides, B. pseudomycoides, and B. cytotoxicus, which do not include any homologs to Bt8741 Raps, have specialized for functions that may contrast to those found here for Bt8741 Raps.
The B. cereus group comprises bacteria with clinical and biotechnological relevance, such as B. anthracis, B. cereus, and B. thuringiensis, as well as other environmental and facultative species (41). Understanding the regulatory processes of cell differentiation and specialization in these bacteria may enhance the use of biotechnologically relevant species, or the strategies to control human pathogens, through the intervention of their collective functions at the molecular level. For instance, B. anthracis and B. cereus are known for their pathogenic nature against mammals. Therefore, elucidating the role of Rap-Phr systems in the production of virulence factors in these species, such as anthrax toxin and capsule of B. anthracis or enterotoxins of B. cereus, could be of high relevance. Additionally, it is known that QS systems can be synthetically engineered (68, 69), and Rap-Phr systems could be manipulated in order to enhance B. thuringiensis survival, insect pathogenesis, or Cry protein production.
MATERIALS AND METHODS
Bacterial strains, media, and culture conditions.
Bacillus thuringiensis strain Bt8741 (46), a laboratory strain closely related to Bt407 (GenBank accession no. NC_018877.1), was used as the host for the overexpression of Rap proteins. Bacillus subtilis strain 168 was used for the amplification of rapA. Escherichia coli strain TOP10 was used for the construction and cloning of overexpression plasmids before transferring into Bt8741. Luria-Bertani (LB) broth (10 g liter−1 tryptone, 5 g liter−1 yeast extract, and 5 g liter−1 NaCl) and nutrient agar (8 g liter−1 nutrient broth and 15 g liter−1 agar) were used at 30°C for Bacillus cultures and at 37°C for E. coli and at 200 rpm for liquid cultures. Milk agar was prepared using nutrient agar supplemented with 5% skim milk (44). When needed, ampicillin (100 μg ml−1) or erythromycin (5 μg ml−1) was added to media. To induce expression from the xylA promoter in Bt8741, xylose was used to a final concentration of 20 mM (70) unless otherwise specified.
Analysis of putative Spo0F-binding amino acids in Raps from Bt407.
Based on the RapH residues involved in Spo0F binding in B. subtilis 168 (36), we determined the conservation of the corresponding residues in Raps from Bt407 in order to predict their capacity to bind to Spo0F. First, we analyzed the conservation of full-length Rap proteins from B. subtilis 168 and Bt8741 in comparison to RapH from B. subtilis 168 (RapHBs). For this, we performed pairwise alignments of the RapHBs amino acid sequence (GenBank accession no. NP_388565.2) with RapABs (GenBank accession no. (NP_389125.1), RapBBs (GenBank accession no. NP_391550.1), RapEBs (GenBank accession no. NP_390460.2), RapJBs (GenBank accession no. NP_388164.1), RapDBs (GenBank accession no. NP_391519.1) from B. subtilis 168, and each of the eight Raps from Bt407 (GenBank accession no. AFV21721.1, AFV22194.1, AFV22088.1, AFV16731.1, AFV19251.1, AFV22208.1, AFV16776.1, AFV17466.1), using the BLASTP tool (71). Then, all sequences were aligned together using the MAFFT version 7 online service (72) with the G-INS-i iterative refinement method (73). Finally, we identified in the alignment the amino acids of Rap protein sequences that correspond to the residues of RapHBs that participate in binding and dephosphorylation of Spo0F.
DNA manipulation.
All primers used in this study are listed in Table S2 in the supplemental material. DNA was isolated from B. subtilis 168 and Bt8741 using the PureLink genomic DNA minikit (Invitrogen, Carlsbad, CA). The QIAprep Spin miniprep kit (Qiagen, Germantown, MD) was used routinely for plasmid extraction and purification. Oligonucleotides were designed for amplifying each Rap gene from the Bt407 chromosome or plasmids (GenBank accession no. NC_018877.1, NC_018883.1, NC_018886.1, NC_018879.1, NC_018878.1) and the B. subtilis 168 genome (GenBank accession no. NC_000964.3) and synthesized by a commercial service (T4 Oligo, Irapuato, Mexico). PCR products and restriction reactions were purified using the PureLink quick PCR purification kit (Invitrogen). When needed, PCR products were isolated from 0.8% agarose gels using the Zymoclean gel DNA recovery kit (Zymo Research, Irvine, CA). The enzymes DreamTaq master mix, HindIII, SalI (Thermo Scientific, Waltham, MA), PstI, and T4 DNA ligase (New England Biolabs, Inc., Ipswich, MA) were used as recommended by the manufacturers.
Construction of Rap overexpression Bt8741 strains.
All strains and plasmids used in this study are listed in Table S1. rap genes from Bt407 were previously identified (50). We performed an independent search using the amino acid sequence of B. subtilis RapA (GenBank accession no. NP_389125.1) as a query in BLAST (71) and identified the same proteins. Additionally, to ensure the identity of the Rap proteins, the sequences were submitted to the NCBI conserved domain search tool (74) in order to determine the presence of a tetratricopeptide repeat-containing domain. For the construction of the overexpression plasmid pHT315-PxylA, the regulatory region of the xylose operon, including the xylA promoter (PxylA) and the repressor gene xylR, was amplified using PCR from the B. subtilis 168 genome using the primers GG1 and GG2 (Table S2). This PCR product was inserted into the HindIII and PstI sites of the pHT315 plasmid (75), and colonies were PCR checked using primers DS16 and DS17 (Table S2). The resulting plasmid, pHT315-PxylA, was transferred into E. coli TOP10 competent cells. Then, this plasmid was used for the inducible overexpression of Rap proteins with xylose in Bt8741. For this, rap genes encoded in the genome of Bt8741 (rapC, rapK, rapF, rapF1, rapF2, rapI, rapI1, and rapLike [50]) and rapA from B. subtilis 168 (RapABs [34]) were amplified using the corresponding primer pairs listed in Table S2 and inserted in-frame between the PstI and SalI sites of pHT315-PxylA. Nine overexpression plasmids, one for each Rap protein, were transferred into E. coli TOP10 competent cells. All plasmids were then transferred into Bt8741 electrocompetent cells using a protocol described in previous studies (44), generating nine Bt8741 strains for the overexpression of each Rap protein. Additionally, we transformed Bt8741 with pHT315-PxylA (without a rap gene), and the resulting strain was used as a control strain throughout the Rap induction experiments. We used a wild-type strain of Bt8741 for overexpression of Rap proteins; therefore, expression is expected from all Phrs present in the genome, which could antagonize Rap overexpression effects on the phenotypes studied. The complete sequence of pHT315-PxylA′rapI was verified using Illumina sequencing (MGH DNA Core, Cambridge, MA), and the rest of the PxylA′rap constructions were verified using Sanger sequencing (Unidad de Servicios Genómicos, Langebio-Cinvestav, Irapuato, Mexico) using primers GG26 and DS17 (Table S2).
Sporulation efficiency.
We assessed the effect of the overexpression of Rap proteins on sporulation efficiency in Bt8741. Preinoculums were prepared by picking a single colony of each strain into 5 ml of liquid medium and were grown overnight. Then, 1 ml of preinoculum was centrifuged, washed, and suspended in 1 ml of sterile phosphate-buffered saline (PBS). Glass culture tubes (25 mm diameter) with 5 ml of LB with erythromycin were inoculated with 50 μl (1% vol/vol) of preinoculum containing ≈107 CFU ml−1 and incubated for 72 h. All strains were cultured in triplicate in LB with and without the addition of xylose. To determine growth and sporulation, total and thermoresistant CFU were calculated by plating 10-fold serial dilutions in nutrient agar. For thermoresistant CFU, samples of 100 μl were incubated at 80°C for 20 min prior to diluting and plating. Sporulation efficiency was calculated as the percentage of thermoresistant CFU in total CFU.
Biofilm formation assay.
We evaluated the effect of the overexpression of Rap proteins on the capacity of Bt874 to form biofilms. For this assay, we used glass tubes (13 by 100 mm) with 3 ml nutrient broth plus erythromycin, with and without the addition of xylose, to a final concentration of 2 mM. Then, 3 μl of preinoculum was added in triplicate, and the inoculated tubes were incubated without agitation at 31°C ± 1°C for 48 hours. The culture medium was then removed with a syringe with a needle. The biofilm and ring attached to the wall of the tube, composed of cells from the biofilm, were suspended in 1.5 ml of sterile PBS, and the optical density (OD600) was measured. The OD600 was also measured from the removed liquid medium to address planktonic growth. At least 5 replicates of each treatment were performed.
Spreading assay.
The spreading phenotype of Rap overexpression Bt8741 variants was followed in colonies spotted on agar. For this assay, we used diluted nutrient agar (NA) (0.8 g liter−1 nutrient broth and 1.5 g liter−1 agar) with erythromycin and with or without the addition of xylose. Plates were air dried inside a biological hood for 60 min prior to inoculation. Then, 5 μl of preinoculum cultures was spotted in the center of the plate, dried for 5 min, and incubated at 30°C for 14 days. The inoculated agar plates were photographed at days 1, 3, 5, and 7 using a gel documentation system (Gel Doc XR+; Bio-Rad). The colony area was measured using Image Lab software (Bio-Rad), and radial growth was calculated. We subtracted from all observations the colony radius at day 1, which corresponds to the inoculated droplet area. Three replicates of each treatment were performed.
Extracellular proteolytic activity assay.
To evaluate the effect of Rap overexpression in extracellular proteolytic activity of Bt8741, 2 μl of preinoculum of each Rap overexpression strain was spotted in triplicate on milk agar with and without the addition of xylose. The hydrolysis halo area was measured after 24 h of incubation using Image Lab software (Bio-Rad). To correct for differences in colony growth, we subtracted the colony area.
Identification of Bt8741 Rap homologs in the B. cereus group.
We used a data set from previous work (51) consisting of Rap protein sequences from representative strains from the B. cereus group. The amino acid sequence of each Rap protein from Bt8741 was queried against each sequence from the B. cereus Rap protein data set using the tBlastn Blast2 online tool (71). We considered homologs the cases where identity was >90% between a Rap from Bt8741 and another Rap from the B. cereus group.
Statistics.
All the statistical analyses were performed using GraphPad Prism version 7.0a. Data obtained from the extracellular proteolytic activity assay, spreading (at day 7), and biofilm formation were analyzed with multiple t tests to search for differences between not induced and induced Rap protein overexpression conditions of each strain. Colony size data from the extracellular proteolytic activity assay were analyzed with a two-way analysis of variance (ANOVA), and the Tukey-Kramer test was used for multiple comparisons. A significance of P = 0.05 was used in all statistical tests.
Supplementary Material
ACKNOWLEDGMENTS
We thank all members of the Microbe-Plant Interactions group at CIDEA for their comments and suggestions. We thank the anonymous reviewers that helped to improve the manuscript during the review process. We gratefully acknowledge Gabriela Olmedo-Álvarez for her suggestions throughout the development of the experimental work and critical review of the manuscript. We thank Eneas Aguirre for critically reading the manuscript.
G.G. received a scholarship (no. 636324) from Conacyt. This work was partially funded by Conacyt (Fomix 267837 and Fordecyt 296368) to M.D.L.T.
J.R. and M.D.L.T. conceived the study. J.R. and G.G. designed the experimental work. G.G. conducted molecular protocols and lab experiments. G.G. conducted the computational analyses. G.G., M.D.L.T., and J.R. interpreted experimental data. G.G. wrote the first draft of the manuscript, and G.G., M.D.L.T., and J.R. added critical revisions.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Crespi BJ. 2001. The evolution of social behavior in microorganisms. Trends Ecol Evol 16:178–183. doi: 10.1016/s0169-5347(01)02115-2. [DOI] [PubMed] [Google Scholar]
- 2.Vlamakis H, Aguilar C, Losick R, Kolter R. 2008. Control of cell fate by the formation of an architecturally complex bacterial community. Genes Dev 22:945–953. doi: 10.1101/gad.1645008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shapiro JA. 1998. Thinking about bacterial populations as multicellular organisms. Annu Rev Microbiol 52:81–104. doi: 10.1146/annurev.micro.52.1.81. [DOI] [PubMed] [Google Scholar]
- 4.Jeckel H, Matthey N, Drescher K. 2019. Biophysics: common concepts for bacterial collectives. Elife 8:e47019. doi: 10.7554/eLife.47019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Vliet S, Ackermann M. 2015. Bacterial ventures into multicellularity: collectivism through individuality. PLoS Biol 13:e1002162. doi: 10.1371/journal.pbio.1002162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Claessen D, Rozen DE, Kuipers OP, Søgaard-Andersen L, Van Wezel GP. 2014. Bacterial solutions to multicellularity: a tale of biofilms, filaments and fruiting bodies. Nat Rev Microbiol 12:115–124. doi: 10.1038/nrmicro3178. [DOI] [PubMed] [Google Scholar]
- 7.Ruby EG. 1996. Lessons from a cooperative, bacterial-animal association: the Vibrio fischeri–Euprymna scolopes light organ symbiosis. Annu Rev Microbiol 50:591–624. doi: 10.1146/annurev.micro.50.1.591. [DOI] [PubMed] [Google Scholar]
- 8.Muñoz-Dorado J, Marcos-Torres FJ, García-Bravo E, Moraleda-Muñoz A, Pérez J. 2016. Myxobacteria: moving, killing, feeding, and surviving together. Front Microbiol 7:781. doi: 10.3389/fmicb.2016.00781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lopez D, Vlamakis H, Kolter R. 2009. Generation of multiple cell types in Bacillus subtilis. FEMS Microbiol Rev 33:152–163. doi: 10.1111/j.1574-6976.2008.00148.x. [DOI] [PubMed] [Google Scholar]
- 10.López D, Kolter R. 2010. Extracellular signals that define distinct and coexisting cell fates in Bacillus subtilis. FEMS Microbiol Rev 34:134–149. doi: 10.1111/j.1574-6976.2009.00199.x. [DOI] [PubMed] [Google Scholar]
- 11.Kovács ÁT, Dragoš A. 2019. Evolved biofilm: review on the experimental evolution studies of Bacillus subtilis pellicles. J Mol Biol 431:4749–4759. doi: 10.1016/j.jmb.2019.02.005. [DOI] [PubMed] [Google Scholar]
- 12.Kearns DB, Chu F, Rudner R, Losick R. 2004. Genes governing swarming in Bacillus subtilis and evidence for a phase variation mechanism controlling surface motility. Mol Microbiol 52:357–369. doi: 10.1111/j.1365-2958.2004.03996.x. [DOI] [PubMed] [Google Scholar]
- 13.Fujita M, González-Pastor JE, Losick R. 2005. High- and low-threshold genes in the Spo0A regulon of Bacillus subtilis. J Bacteriol 187:1357–1368. doi: 10.1128/JB.187.4.1357-1368.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Auchtung JM, Lee CA, Grossman AD. 2006. Modulation of the ComA-dependent quorum response in Bacillus subtilis by multiple Rap proteins and Phr peptides. J Bacteriol 188:5273–5285. doi: 10.1128/JB.00300-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mukai K, Kawata M, Tanaka T. 1990. Isolation and phosphorylation of the Bacillus subtilis degS and degU gene products. J Biol Chem 265:20000–20006. [PubMed] [Google Scholar]
- 16.Bassler BL, Losick R. 2006. Bacterially speaking. Cell 125:237–246. doi: 10.1016/j.cell.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 17.Waters CM, Bassler BL. 2005. Quorum sensing: cell-to-cell communication in bacteria. Annu Rev Cell Dev Biol 21:319–346. doi: 10.1146/annurev.cellbio.21.012704.131001. [DOI] [PubMed] [Google Scholar]
- 18.Williams P. 2007. Quorum sensing, communication and cross-kingdom signalling in the bacterial world. Microbiology 153:3923–3938. doi: 10.1099/mic.0.2007/012856-0. [DOI] [PubMed] [Google Scholar]
- 19.Declerck N, Bouillaut L, Chaix D, Rugani N, Slamti L, Hoh F, Lereclus D, Arold ST. 2007. Structure of PlcR: insights into virulence regulation and evolution of quorum sensing in Gram-positive bacteria. Proc Natl Acad Sci U S A 104:18490–18495. doi: 10.1073/pnas.0704501104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rocha-Estrada J, Aceves-Diez AE, Guarneros G, De La Torre M. 2010. The RNPP family of quorum-sensing proteins in Gram-positive bacteria. Appl Microbiol Biotechnol 87:913–923. doi: 10.1007/s00253-010-2651-y. [DOI] [PubMed] [Google Scholar]
- 21.Neiditch MB, Capodagli GC, Prehna G, Federle MJ. 2017. Genetic and structural analyses of RRNPP intercellular peptide signaling of Gram-positive bacteria. Annu Rev Genet 51:311–333. doi: 10.1146/annurev-genet-120116-023507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pottathil M, Lazazzera BA. 2003. The extracellular Phr peptide-Rap phosphatase signaling circuit of Bacillus subtilis. Front Biosci 8:32–45. doi: 10.2741/913. [DOI] [PubMed] [Google Scholar]
- 23.Core L, Perego M. 2003. TPR-mediated interaction of RapC with ComA inhibits response regulator-DNA binding for competence development in Bacillus subtilis. Mol Microbiol 49:1509–1522. doi: 10.1046/j.1365-2958.2003.03659.x. [DOI] [PubMed] [Google Scholar]
- 24.Perego M. 1997. A peptide export-import control circuit modulating bacterial development regulates protein phosphatases of the phosphorelay. Proc Natl Acad Sci U S A 94:8612–8617. doi: 10.1073/pnas.94.16.8612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Perego M. 2013. Forty years in the making: understanding the molecular mechanism of peptide regulation in bacterial development. PLoS Biol 11:e1001516-5. doi: 10.1371/journal.pbio.1001516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McLoon AL, Guttenplan SB, Kearns DB, Kolter R, Losick R. 2011. Tracing the domestication of a biofilm-forming bacterium. J Bacteriol 193:2027–2034. doi: 10.1128/JB.01542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boguslawski KM, Hill PA, Griffith KL. 2015. Novel mechanisms of controlling the activities of the transcription factors Spo0A and ComA by the plasmid-encoded quorum sensing regulators Rap60-Phr60 in Bacillus subtilis. Mol Microbiol 96:325–348. doi: 10.1111/mmi.12939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ogura M, Shimane K, Asai K, Ogasawara N, Tanaka T. 2003. Binding of response regulator DegU to the aprE promoter is inhibited by RapG, which is counteracted by extracellular PhrG in Bacillus subtilis. Mol Microbiol 49:1685–1697. doi: 10.1046/j.1365-2958.2003.03665.x. [DOI] [PubMed] [Google Scholar]
- 29.Solomon JM, Lazazzera BA, Grossman AD. 1996. Purification and characterization of an extracellular peptide factor that affects two different developmental pathways in Bacillus subtilis. Genes Dev 10:2014–2024. doi: 10.1101/gad.10.16.2014. [DOI] [PubMed] [Google Scholar]
- 30.Ogura M, Fujita Y. 2007. Bacillus subtilis rapD, a direct target of transcription repression by RghR, negatively regulates srfA expression. FEMS Microbiol Lett 268:73–80. doi: 10.1111/j.1574-6968.2006.00559.x. [DOI] [PubMed] [Google Scholar]
- 31.Bongiorni C, Ishikawa S, Stephenson S, Ogasawara N, Perego M. 2005. Synergistic regulation of competence development in Bacillus subtilis by two Rap-Phr systems. J Bacteriol 187:4353–4361. doi: 10.1128/JB.187.13.4353-4361.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hayashi K, Kensuke T, Kobayashi K, Ogasawara N, Ogura M. 2006. Bacillus subtilis RghR (YvaN) represses rapG and rapH, which encode inhibitors of expression of the srfA operon. Mol Microbiol 59:1714–1729. doi: 10.1111/j.1365-2958.2006.05059.x. [DOI] [PubMed] [Google Scholar]
- 33.Smits WK, Bongiorni C, Veening J-W, Hamoen LW, Kuipers OP, Perego M. 2007. Temporal separation of distinct differentiation pathways by a dual specificity Rap-Phr system in Bacillus subtilis. Mol Microbiol 65:103–120. doi: 10.1111/j.1365-2958.2007.05776.x. [DOI] [PubMed] [Google Scholar]
- 34.Perego M, Hanstein C, Welsh KM, Djavakhishvili T, Glaser P, Hoch JA. 1994. Multiple protein-aspartate phosphatases provide a mechanism for the integration of diverse signals in the control of development in B. subtilis. Cell 79:1047–1055. doi: 10.1016/0092-8674(94)90035-3. [DOI] [PubMed] [Google Scholar]
- 35.Jiang M, Grau R, Perego M. 2000. Differential processing of propeptide inhibitors of rap phosphatases in Bacillus subtilis. J Bacteriol 182:303–310. doi: 10.1128/jb.182.2.303-310.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parashar V, Mirouze N, Dubnau DA, Neiditch MB. 2011. Structural basis of response regulator dephosphorylation by Rap phosphatases. PLoS Biol 9:e1000589. doi: 10.1371/journal.pbio.1000589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parashar V, Jeffrey PD, Neiditch MB. 2013. Conformational change-induced repeat domain expansion regulates Rap phosphatase quorum-sensing signal receptors. PLoS Biol 11:e1001512. doi: 10.1371/journal.pbio.1001512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parashar V, Konkol MA, Kearns DB, Neiditch MB. 2013. A plasmid-encoded phosphatase regulates Bacillus subtilis biofilm architecture, sporulation, and genetic competence. J Bacteriol 195:2437–2448. doi: 10.1128/JB.02030-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Burbulys D, Trach KA, Hoch JA. 1991. Initiation of sporulation in B. subtilis is controlled by a multicomponent phosphorelay. Cell 64:545–552. doi: 10.1016/0092-8674(91)90238-t. [DOI] [PubMed] [Google Scholar]
- 40.Perego M. 2001. A new family of aspartyl phosphate phosphatases targeting the sporulation transcription factor Spo0A of Bacillus subtilis. Mol Microbiol 42:133–143. doi: 10.1046/j.1365-2958.2001.02611.x. [DOI] [PubMed] [Google Scholar]
- 41.Ehling-Schulz M, Koehler TM, Lereclus D. 2019. The Bacillus cereus group: Bacillus species with pathogenic potential. Microbiol Spectr 6:GPP3-0032–2018. doi: 10.1128/microbiolspec.GPP3-0032-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Anderson I, Sorokin A, Kapatral V, Reznik G, Bhattacharya A, Mikhailova N, Burd H, Joukov V, Kaznadzey D, Walunas T, Souza MÕ, Larsen N, Pusch G, Liolios K, Grechkin Y, Lapidus A, Goltsman E, Chu L, Fonstein M, Ehrlich SD. 2005. Comparative genome analysis of Bacillus cereus group genomes with Bacillus subtilis. FEMS Microbiol Lett 250:175–184. doi: 10.1016/j.femsle.2005.07.008. [DOI] [PubMed] [Google Scholar]
- 43.Dubois T, Faegri K, Perchat S, Lemy C, Buisson C, Nielsen-LeRoux C, Gohar M, Jacques P, Ramarao N, Kolstø A-B, Lereclus D. 2012. Necrotrophism is a quorum-sensing-regulated lifestyle in Bacillus thuringiensis. PLoS Pathog 8:e1002629. doi: 10.1371/journal.ppat.1002629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cabrera R, Rocha J, Flores V, Vázquez-Moreno L, Guarneros G, Olmedo G, Rodríguez-Romero A, de la Torre M. 2014. Regulation of sporulation initiation by NprR and its signaling peptide NprRB: molecular recognition and conformational changes. Appl Microbiol Biotechnol 98:9399–9412. doi: 10.1007/s00253-014-6094-8. [DOI] [PubMed] [Google Scholar]
- 45.Cabrera R, Rodríguez-Romero A, Guarneros G, de la Torre M. 2016. New insights into the interaction between the quorum-sensing receptor NprR and its DNA target, or the response regulator Spo0F. FEBS Lett 590:3243–3253. doi: 10.1002/1873-3468.12371. [DOI] [PubMed] [Google Scholar]
- 46.Rocha J, Flores V, Cabrera R, Soto-Guzmán A, Granados G, Juaristi E, Guarneros G, De La Torre M. 2012. Evolution and some functions of the NprR-NprRB quorum-sensing system in the Bacillus cereus group. Appl Microbiol Biotechnol 94:1069–1078. doi: 10.1007/s00253-011-3775-4. [DOI] [PubMed] [Google Scholar]
- 47.Even-Tov E, Omer Bendori S, Pollak S, Eldar A. 2016. Transient duplication-dependent divergence and horizontal transfer underlie the evolutionary dynamics of bacterial cell-cell signaling. PLoS Biol 14:e2000330-23. doi: 10.1371/journal.pbio.2000330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bongiorni C, Stoessel R, Shoemaker D, Perego M. 2006. Rap phosphatase of virulence plasmid pXO1 inhibits Bacillus anthracis sporulation. J Bacteriol 188:487–498. doi: 10.1128/JB.188.2.487-498.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fazion F, Perchat S, Buisson C, Vilas-Bôas G, Lereclus D. 2018. A plasmid-borne Rap-Phr system regulates sporulation of Bacillus thuringiensis in insect larvae. Environ Microbiol 20:145–155. doi: 10.1111/1462-2920.13946. [DOI] [PubMed] [Google Scholar]
- 50.Slamti L, Perchat S, Huillet E, Lereclus D. 2014. Quorum sensing in Bacillus thuringiensis is required for completion of a full infectious cycle in the insect. Toxins (Basel) 6:2239–2255. doi: 10.3390/toxins6082239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cardoso P de F, Perchat S, Vilas-Boas LA, Lereclus D, Vilas-Bôas GT. 2019. Diversity of the Rap-Phr quorum-sensing systems in the Bacillus cereus group. Curr Genet 65:1367–1381. doi: 10.1007/s00294-019-00993-9. [DOI] [PubMed] [Google Scholar]
- 52.Gastélum G, de la Torre M, Rocha J. 2019. Rap-protein paralogs of B. thuringiensis: a multifunctional and redundant regulatory repertoire for the control of collective functions. bioRxiv doi: 10.1101/784611. [DOI] [PMC free article] [PubMed]
- 53.Flemming HC, Wuertz S. 2019. Bacteria and archaea on Earth and their abundance in biofilms. Nat Rev Microbiol 17:247–260. doi: 10.1038/s41579-019-0158-9. [DOI] [PubMed] [Google Scholar]
- 54.Bhavsar AP, Zhao X, Brown ED. 2001. Development and characterization of a xylose-dependent system for expression of cloned genes in Bacillus subtilis: conditional complementation of a teichoic acid mutant. Appl Environ Microbiol 67:403–410. doi: 10.1128/AEM.67.1.403-410.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kinsinger RF, Shirk MC, Fall R. 2003. Rapid surface motility in Bacillus subtilis is dependent on extracellular surfactin and potassium ion. J Bacteriol 185:5627–5631. doi: 10.1128/jb.185.18.5627-5631.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kearns DB. 2010. A field guide to bacterial swarming motility. Nat Rev Microbiol 8:634–644. doi: 10.1038/nrmicro2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hölscher T, Kovács ÁT. 2017. Sliding on the surface: bacterial spreading without an active motor. Environ Microbiol 19:2537–2545. doi: 10.1111/1462-2920.13741. [DOI] [PubMed] [Google Scholar]
- 58.Fagerlund A, Dubois T, Økstad OA, Verplaetse E, Gilois N, Bennaceur I, Perchat S, Gominet M, Aymerich S, Kolstø AB, Lereclus D, Gohar M. 2014. SinR controls enterotoxin expression in Bacillus thuringiensis biofilms. PLoS One 9:e87532. doi: 10.1371/journal.pone.0087532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Raynor MJ, Roh JH, Widen SG, Wood TG, Koehler TM. 2018. Regulons and protein-protein interactions of PRD-containing Bacillus anthracis virulence regulators reveal overlapping but distinct functions. Mol Microbiol 109:1–22. doi: 10.1111/mmi.13961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Even-Tov E, Omer Bendori S, Valastyan J, Ke X, Pollak S, Bareia T, Ben-Zion I, Bassler BL, Eldar A. 2016. Social evolution selects for redundancy in bacterial quorum sensing. PLoS Biol 14:e1002386. doi: 10.1371/journal.pbio.1002386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vilain S, Pretorius J, Theron J, Brozel V. 2009. DNA as an adhesin: Bacillus cereus requires extracellular DNA to form biofilms. Appl Environ Microbiol 75:2861–2868. doi: 10.1128/AEM.01317-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gohar M, Gilois N, Graveline R, Garreau C, Sanchis V, Lereclus D. 2005. A comparative study of Bacillus cereus, Bacillus thuringiensis and Bacillus anthracis extracellular proteomes. Proteomics 5:3696–3711. doi: 10.1002/pmic.200401225. [DOI] [PubMed] [Google Scholar]
- 63.Gélis-Jeanvoine S, Canette A, Gohar M, Caradec T, Lemy C, Gominet M, Jacques P, Lereclus D, Slamti L. 2017. Genetic and functional analyses of krs, a locus encoding kurstakin, a lipopeptide produced by Bacillus thuringiensis. Res Microbiol 168:356–368. doi: 10.1016/j.resmic.2016.06.002. [DOI] [PubMed] [Google Scholar]
- 64.Béchet M, Caradec T, Hussein W, Abderrahmani A, Chollet M, Leclére V, Dubois T, Lereclus D, Pupin M, Jacques P. 2012. Structure, biosynthesis, and properties of kurstakins, nonribosomal lipopeptides from Bacillus spp. Appl Microbiol Biotechnol 95:593–600. doi: 10.1007/s00253-012-4181-2. [DOI] [PubMed] [Google Scholar]
- 65.Earl AM, Losick R, Kolter R. 2008. Ecology and genomics of Bacillus subtilis. Trends Microbiol 16:269–275. doi: 10.1016/j.tim.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mirouze N, Dubnau D. 2013. Chance and necessity in Bacillus subtilis development. Microbiol Spectr 1:TBS-0004-2012. doi: 10.1128/microbiolspectrum.TBS-0004-2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Argôlo-Filho RC, Loguercio LL. 2013. Bacillus thuringiensis is an environmental pathogen and host-specificity has developed as an adaptation to human-generated ecological niches. Insects 5:62–91. doi: 10.3390/insects5010062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang F, Kwan A, Xu A, Süel GM. 2015. A synthetic quorum sensing system reveals a potential private benefit for public good production in a biofilm. PLoS One 10:e0132948. doi: 10.1371/journal.pone.0132948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Geddes BA, Paramasivan P, Joffrin A, Thompson AL, Christensen K, Jorrin B, Brett P, Conway SJ, Oldroyd GED, Poole PS. 2019. Engineering transkingdom signalling in plants to control gene expression in rhizosphere bacteria. Nat Commun 10:3430. doi: 10.1038/s41467-019-10882-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Grandvalet C, Gominet M, Lereclus D. 2001. Identification of genes involved in the activation of the Bacillus thuringiensis inhA metalloprotease gene at the onset of sporulation. Microbiology 147:1805–1813. doi: 10.1099/00221287-147-7-1805. [DOI] [PubMed] [Google Scholar]
- 71.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 72.Katoh K, Rozewicki J, Yamada KD. 2017. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform 20:1160–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Katoh K, Kuma KI, Toh H, Miyata T. 2005. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res 33:511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Marchler-Bauer A, Bo Y, Han L, He J, Lanczycki CJ, Lu S, Chitsaz F, Derbyshire MK, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Lu F, Marchler GH, Song JS, Thanki N, Wang Z, Yamashita RA, Zhang D, Zheng C, Geer LY, Bryant SH. 2017. CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res 45:D200–D203. doi: 10.1093/nar/gkw1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Arantes O, Lereclus D. 1991. Construction of cloning vectors for Bacillus thuringiensis. Gene 108:115–119. doi: 10.1016/0378-1119(91)90495-w. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.