Abstract
Induction of endogenous cardiomyocyte (CM) proliferation is one of the key strategies for heart regeneration. Increasing evidence points to the potential role of microRNAs (miRNAs) in the regulation of CM proliferation. Here, we used human embryonic stem cell (hESC)-derived CMs (hESC-CMs) as a tool to identify miRNAs that promote CM proliferation. We profiled miRNA expression at an early stage of CM differentiation and identified a list of highly expressed miRNAs. Among these miRNAs, miR-25 was enriched in early-stage hESC-CMs, but its expression decreased over time. Overexpression of miR-25 promoted CM proliferation. RNA sequencing (RNA-seq) analysis revealed that genes related to cell-cycle signal were strongly influenced by miR-25 overexpression. We further showed that miR-25 promoted CM proliferation by targeting FBXW7. Finally, the function of miR-25 in the regulation of CM proliferation was demonstrated in zebrafish. Our study suggested that miR-25 is a promising molecule for heart regeneration.
Keywords: miR-25, human embryonic stem cell, cardiomyocytes, proliferation
Introduction
Ischemic cardiomyopathy, one of the most common cardiac myopathies, originates because of inadequate oxygen and nutrient supply, causing myocardial infarction (MI).1 Adult mammalian cardiomyocytes (CMs) retain only limited endogenous renewal capacity,2,3 which is insufficient for the replacement of acute or chronic CM loss in ischemic cardiac injury.3 Several cell-based therapeutic approaches have been developed to treat ischemic heart diseases, including tissue engineering, stem cell transplantation, and mobilization of resident bone marrow cells, but ineffectiveness in clinical applications has fueled disenchantment with these approaches.4 The ability to use various combinations of cellular reprogramming transcription factors and other agents to directly transdifferentiate fibroblasts into CM-like cells offers a promising approach to treat MI,5,6 but the resistance of human cells to reprogramming is an important barrier to the clinical application of this strategy.7
An alternative, stimulation of endogenous CM proliferation, has emerged as an attractive option for promoting myocardial regeneration. Recent compelling evidence shows that pre-existing CMs in adult mammals have the potential to divide at a very low rate, and myocardial injury can increase the division rate.8 A more recent study has shown that mature adult CMs can reenter the cell cycle and form new CMs through dedifferentiation, proliferation, and redifferentiation.9 Increasing evidence points to the potential role of microRNAs (miRNAs) in the regulation of CM proliferation. miRNAs are a class of regulatory RNAs that measure ∼22 nt in length and post-transcriptionally regulate the expression of numerous genes via mRNA degradation and translational inhibition.10,11 Studies in rodents have identified several miRNAs that are involved in CM proliferation.9,12, 13, 14, 15, 16 For example, both miR-590 and miR-199a promote CM proliferation by targeting Homer1 and Hopx,12 miR-204 promotes CM proliferation by targeting Jarid2,15 and miR-210 promotes CM proliferation by targeting APC.16 These studies demonstrate that miRNAs play a crucial role in regulating CM proliferation.
Previous studies on CM proliferation have primarily used rodent models. However, the degree to which conclusions can be translated from rodents to human is unknown. Human pluripotent stem cells (hPSCs), including human induced pluripotent stem cells (hiPSCs) and human embryonic stem cells (hESCs),17,18 can be efficiently differentiated into CMs,8,19,20 offering a good opportunity to study CM proliferation in human cells. A recent large-scale screening using hPSC-derived CMs revealed that proliferative miRNAs in humans overlapped only minimally with those previously shown to stimulate rodent CM proliferation.21 In this study, we profiled miRNA expression at an early stage of CM differentiation and identified a list of highly expressed miRNAs. Further screening showed that miR-25 overexpression promotes CM proliferation. Importantly, we identified FBXW7 as a target of miR-25 for the promotion of CM proliferation. Our study suggests that miR-25 could be a potential molecule for cardiac regeneration.
Results
Profiling miRNA Expression in the Early Stage of CM Differentiation
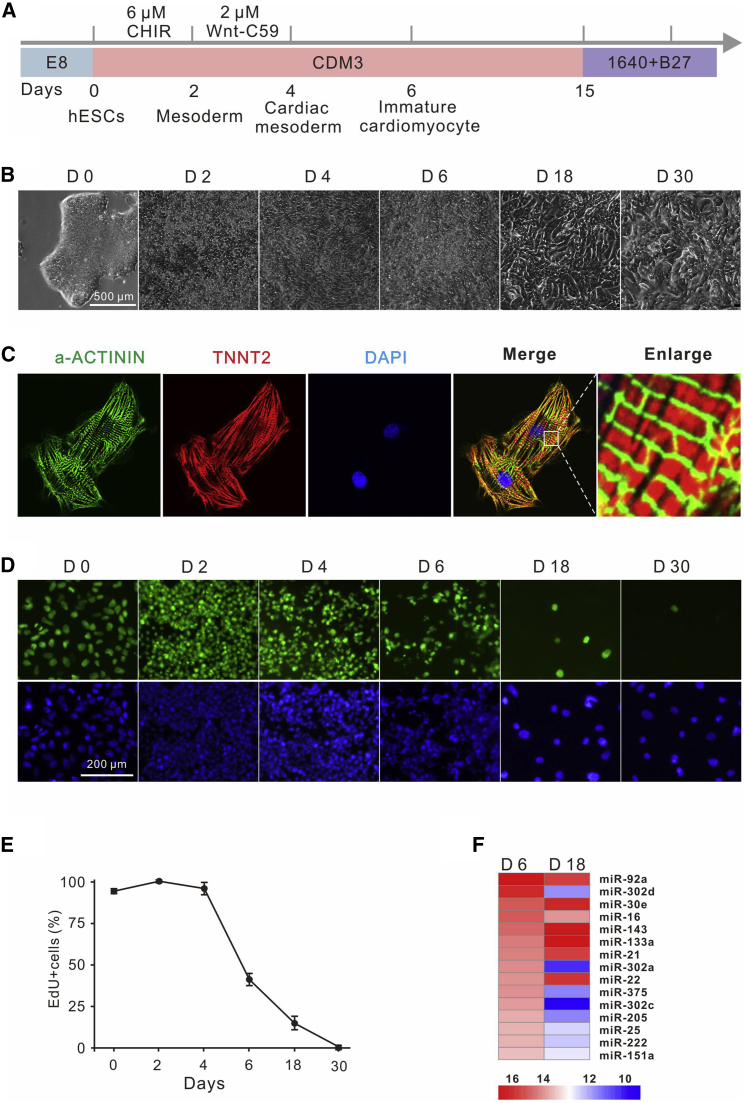
Previous studies have shown that early hPSC-CMs proliferate efficiently, similar to embryonic or fetal mammalian CMs, but their capacity for proliferation decreases over time,22, 23, 24 which offers us an opportunity to study which miRNAs regulate CM proliferation during this process. We used a monolayer-differentiation method to generate hPSC-CMs by temporally manipulating the canonical Wnt signaling pathway (Figure 1A). The CM transition requires several intermediate stages including mesoderm (day 2), cardiac mesoderm (day 4), and CM progenitor cells (day 6).19,25 The marker gene expression of each stage was confirmed by quantitative reverse transcription polymerase chain reaction (qRT-PCR) (Figure S1). The morphology of cells changed over time during differentiation (Figure 1B). Eight days after differentiation, the cells started contracting rhythmically (Video S1). Thirty days after differentiation, CMs showed regular sarcomeric structures, as illustrated by immunofluorescent (IF) α-actinin (α-ACTININ), cardiac troponin T (TNNT2), and 4′,6′-diamidino-phenylindole (DAPI) staining (Figure 1C). A 5-ethynyl-2′-deoxyuridine (EdU) cell proliferation assay revealed that cell proliferation decreased from 95.7% at day 4 to 1.9% at day 30 (Figures 1D and 1E).
Figure 1.
miR-25 Is Enriched in the Early Stage of CM Differentiation
(A) Schematic of chemically defined CM differentiation in vitro. (B) The morphology of cells during CM differentiation. (C) Thirty-day-old CMs showed regular sarcomeric structures, as illustrated by immunofluorescent staining of α-ACTININ (green) and TNNT2 (red). (D and E) Cell proliferation decreased from days 4 to 30 as revealed by EdU staining (D). EdU incorporation was quantified using ImageJ software (E). Approximately 2,000 cells were counted in each group. (F) The top 15 most abundant miRNAs at day 6 and their expression levels at day 18 were analyzed by miRNA-seq during hESC differentiation.
Cardiomyocytes with Regular Contraction
We hypothesized that miRNAs with high expression levels played a key role in regulating CM proliferation during CM differentiation. We profiled miRNA expression at the genome-wide level at two time points: days 6 and 18. We focused on miRNAs that highly expressed on day 6 but dramatically changed at day 18 (fold change >2 for upregulation, fold change <0.5 for downregulation) and identified 15 miRNAs. Among these miRNAs, expression of miR-30e, miR-143, miR-133a, miR-21, and miR-22 increased, whereas expression of miR-92a, miR-302/367 cluster, miR-16, miR-375, miR-205, miR-25, miR-222, and miR-151a decreased (Figure 1F). We hypothesized that the change of these miRNA expressions was associated with CM proliferation. Interestingly, miR-92a, miR-302/367 cluster, miR-16, miR-133, and miR-222 have been shown to regulate CM proliferation.13,26, 27, 28, 29
Overexpression of miR-25 Promotes hESC-CM Proliferation
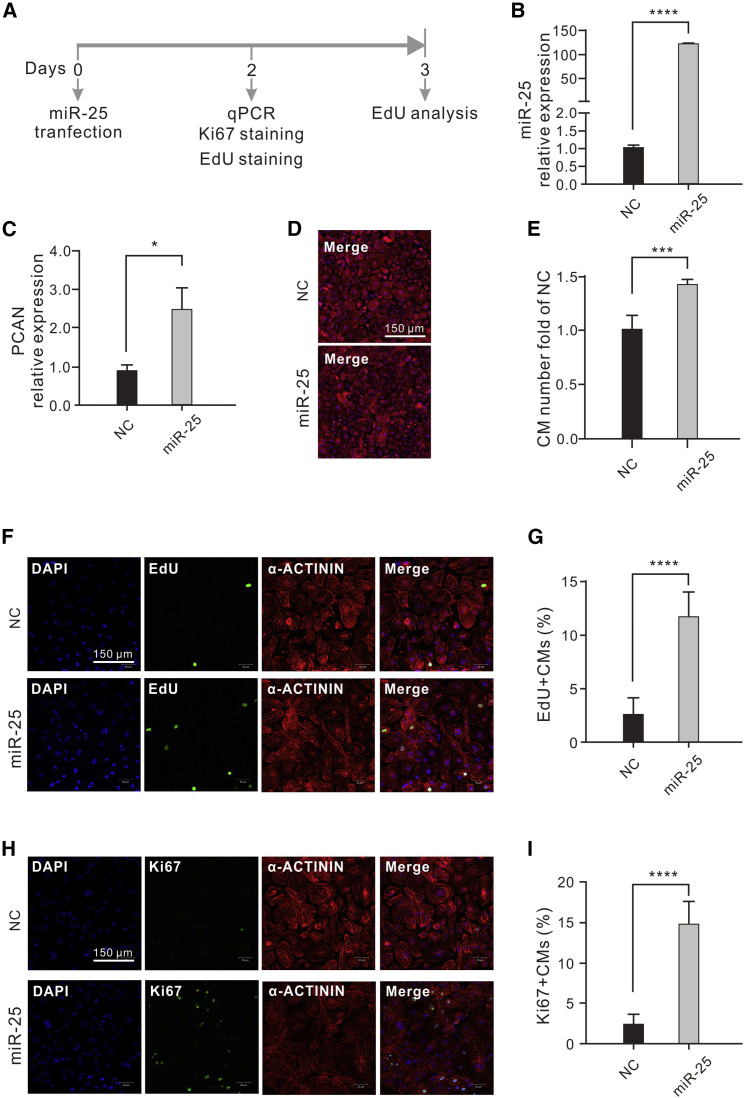
To identify additional miRNAs that promote CM proliferation, we knocked down miR-30e, miR-143, miR-21, and miR-22 in the 30-day-old hESC-CMs, but none of these miRNAs could promote CM proliferation. We overexpressed miR-375, miR-25, miR-205, and miR-151a in the 30-day-old hESC-CMs, and EdU cell proliferation assay revealed that miR-25 significantly promoted CM proliferation (Figure 2A; Figure S2). qRT-PCR showed that transfection of miR-25 mimics led to overexpression of miR-25 (Figure 2B) and proliferating cell nuclear antigen (PCNA) (Figure 2C). Transfection of miR-25 mimics led to CM proliferation as indicated by cell counts (Figures 2D and 2E). The proliferation of CMs was confirmed by an EdU cell proliferation assay. miR-25 transfection led to an increase in EdU-positive hESC-CMs from 2.5% to 11.6% (Figures 2F and 2G). We also used Ki-67 immunostaining to investigate the proliferation of CMs. The proportion of Ki-67-positive cells increased from 2.3% to 14.6% after miR-25 transfection (Figures 2H and 2I). In addition to its effect on hESC-CMs, miR-25 also promoted the proliferation of hiPSC-derived CMs (hiPSC-CMs) (Figures S3A–S3D). In summary, overexpression of miR-25 could promote proliferation of both hESC-CMs and hiPSC-CMs.
Figure 2.
miR-25 Promotes hESC-CM Proliferation
(A) Schematic of the experimental design. (B) qRT-PCR analysis showed that miR-25 expression was significantly increased in hESC-CMs transfected with miR-25 mimics (n = 3) NC = cells transfected with normal control mimics. (C) qRT-PCR analysis showed that the expression of proliferating cell nuclear antigen (PCNA) increased in hESC-CMs transfected with miR-25 mimics (n = 3). NC = cells transfected with normal control mimics. (D) hESC-CMs transfected with miR-25 mimics or NC were stained for sarcomeric α-ACTININ (red) and DAPI (blue). (E) Relative numbers of hESC-CMs treated with miRNA-25 or NC. (n = 3) (F) hESC-CMs transfected with miR-25 mimics or NC were stained with EdU (green), an antibody against α-ACTININ (red) and DAPI (blue). (F) Percentage of Edu+ hESC-CMs treated with miR-25 mimics or NC. Approximately 2,000 cells were counted in each group. (H) hESC-CMs transfected with miR-25 mimics or NC were stained with an antibody against Ki-67 (green), α-ACTININ (red) and DAPI (blue). (I) Percentage of Ki-67+ hESC-CMs treated with miR-25 mimics or NC. Approximately 2,000 cells were counted in each group. Statistical significance was calculated using Student's t test for paired samples. Data are shown as the mean ± SEM. *p < 0.05, ***p < 0.001, ****p < 0.0001.
We further examined whether overexpression of miR-25 influenced other properties of CMs. Overexpression of miR-25 did not influence sarcomeric structure (Figure S4A), cell size (Figures S4A and S4B), field potential duration (FPD; Fridericia corrected) (Figures S4C and S4D), or beat period (Figure S4E).
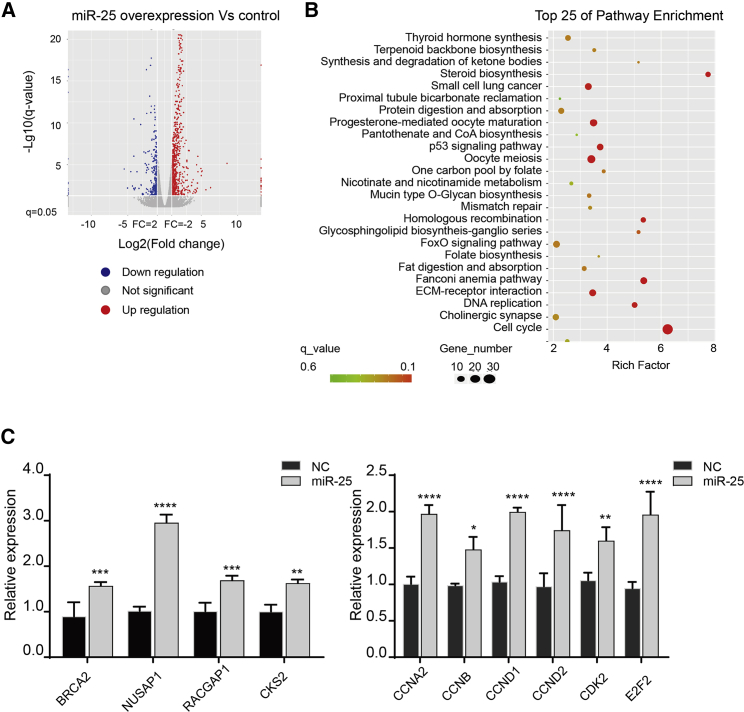
To better understand the molecular and cellular effects of miR-25 overexpression, we performed RNA sequencing (RNA-seq) analysis. A total of 967 differentially expressed genes (DEGs; q value < 0.05) were identified, including 696 upregulated genes and 271 downregulated genes (Figure 3A). Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis showed that 30 upregulated genes belonged to pathways related to the cell cycle, 16 genes were related to oocyte meiosis, and 10 genes were related to the p53 signaling pathway (Figure 3B). KEGG classification analysis showed that the most affected classes of genes were related to signal transduction, cancers, and cell growth and death (Figure S5A). Gene Ontology (GO) analysis revealed that the DEGs enriched in cellular components were related to proliferation, including condensed chromosome kinetochores and biological processes such as DNA replication (Figure S5B). qRT-PCR revealed that the expression of cell proliferation-associated genes (BRCA2, NUSAP1, RACGAP1, and CKS2) and cell-cycle-related genes (CCNA2, CCNB, CCND, CDK2, and E2F2) was increased (Figure 3C). These results indicated that miR-25 overexpression partly activated some cell-cycle-related genes that promote CM proliferation.
Figure 3.
miR-25 Overexpression Drives Cell-Cycle Progression in hESC-CMs
(A) Volcano map for the 967 differentially expressed genes in hESC-CMs overexpressing miR-25 compared with the normal control. (B) Scatterplots of the top 25 differentially regulated pathways identified in KEGG analyses. The vertical axis is the pathway term; the horizontal axis shows the rich factor for KEGG pathway enrichment. The q value denotes the significance of the pathway item. (C) qRT-PCR analysis revealed that miR-25 overexpression increased the expression of cell proliferation-associated genes and cell-cycle-related genes (n = 3). Statistical significance was calculated using Student's t test for paired samples. Data are shown as the mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
miR-25 Regulates hESC-CM Proliferation by Targeting FBXW7
To elucidate the mechanism of miR-25 in the regulation of CM proliferation, we used TargetScan and miRTarBase to predict miR-25 target genes. We focused on candidate targets that were predicted by both tools and obtained a list of 165 genes. Among these genes, BTG2, RECK, LATS2, CDKN1C, and FBXW7 have been reported to promote cancer cell proliferation.15,30, 31, 32, 33 A dual-luciferase reporter assay was performed to test whether these genes were direct targets of miR-25 in HEK293T cells. We constructed reporter constructs containing luciferase fused with the wild-type (WT) 3′ UTR of each gene. An initial screen revealed that fusion with the 3′ UTR of RECK, LATS2, and CDKN1C did not influence luciferase activity, whereas fusion with the 3′ UTR of BTG2 and FBXW7 decreased luciferase activity (Figures 4A and 4B; Figures S6A–S6H), suggesting that BTG2 and FBXW7 could be direct targets of miR-25. However, knockdown of BTG2 by small interfering RNA (siRNA) did not promote CM proliferation (Figure S7).
Figure 4.
FBXW7 Is a Direct Target Gene of miR-25
(A) A potential target site (highlighted in red) of miR-25 on the FRXW7 3′ UTR was predicted by TargetScan. The mutated target sequence is shown below. (B) A luciferase reporter assay showed that the predicted binding sequence was required for miR-25 inhibition (n = 3). (C) qRT-PCR showed that miR-25 overexpression decreased FBXW7 expression in hESC-CMs (n = 3). (D) Western blot analysis showed that miR-25 overexpression decreased FBXW7 expression in hESC-CMs. (E) Fold change expression of FBXW7 normalized by GAPDH as a internal control in hESC-CMs treated with miR-25 mimics or NC. (n = 3). (F) FBXW7 expression was knocked down by siFBXW7 in hESC-CMs (n = 3). (G) qRT-PCR showed that PCNA expression was significantly increased in hESC-CMs treated with siFBXW7 (n = 3). (H) EdU staining (green) revealed that FBXW7 knockdown increased CM proliferation. The number of EdU-positive cells is shown on the right. Nuclei were stained with DAPI (blue); CMs were stained with an antibody against α-ACTININ (red). Approximately 2,000 cells were quantified in each group. Scale bars, 150 μm. (I) Ki-67 staining (green) revealed that FBXW7 knockdown increased CM proliferation. The number of Ki-67-positive cells is shown on the right. Nuclei were stained with DAPI (blue); CMs were stained with an antibody against α-ACTININ (red). Approximately 2,000 cells were quantified in each group. Scale bars, 150 μm. Mut, mutant; siNC, siRNA negative control; WT, wild-type. Statistical significance was calculated using student's t test for paired samples. Data are shown as the mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
One miR-25 binding site was predicted on the FBXW7 3′ UTR (Figure 4A). The luciferase reporter assay revealed that miR-25 abolished the inhibitory effect when this site was mutated, indicating that it was an miR-25 binding site (Figure 4B). To investigate whether miR-25 inhibits FBXW7 in CMs, we transfected miR-25 mimics into CMs, which resulted in downregulation of FBXW7 expression as revealed by qRT-PCR (Figure 4C) and western blotting (Figures 4D and 4E). To test whether downregulation of FBXW7 expression led to CM proliferation, we transfected an siRNA against FBXW7 (siFBXW7) into hESC-CMs. Downregulation of FBXW7 expression was confirmed by qRT-PCR (Figure 4F). FBXW7 knockdown led to an increase in PCNA expression (Figure 4G), a marker gene of cell proliferation. Moreover, transfection of siFBXW7 led to hESC-CM proliferation as indicated by EdU and Ki-67 staining (Figures 4H and 4I). Taken together, these results demonstrated that miR-25 promoted hESC-CM proliferation at least partially via inhibition of FBXW7.
miR-25 Promotes CM Proliferation in Zebrafish
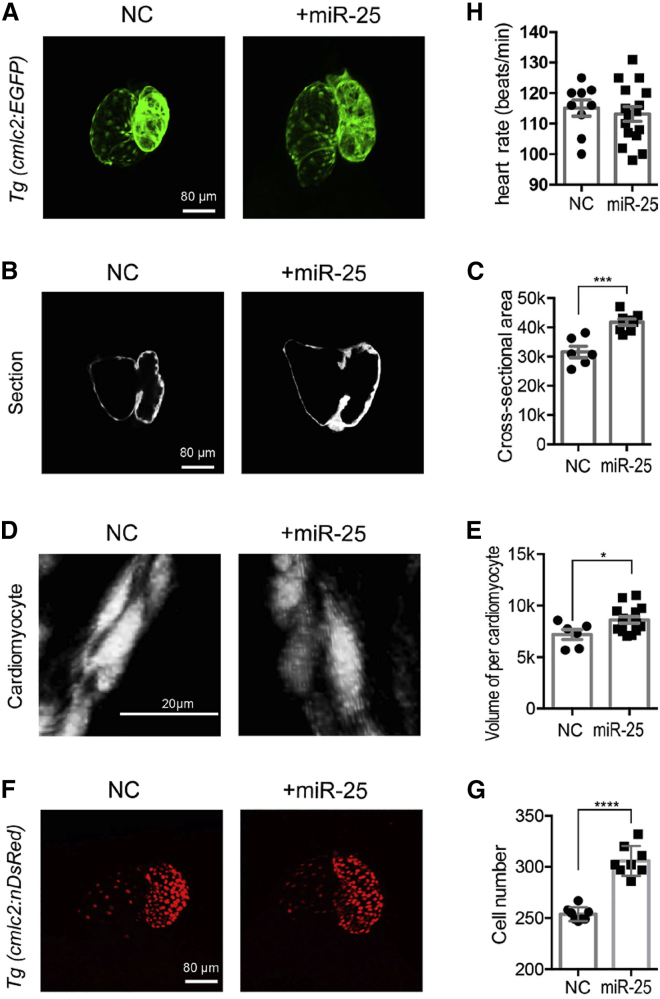
To investigate the regenerative potential of miR-25 at the organ level, we employed transgenic zebrafish with myocardium-specific GFP expression. miR-25 or normal control (NC) mimics were injected into the single-cell stage of zebrafish embryos. Heart morphology, size, and CM number were examined 72 h postfertilization (hpf). Confocal imaging revealed that miR-25 injection resulted in enlargement of the heart compared with the NC group (Figure 5A). Consistent with confocal imaging analysis, histological section analysis revealed that miR-25 injection significantly increased the cross-sectional area (Figures 5B and 5C). Furthermore, we analyzed the CM size by miR-25 injection, and the results revealed that miR-25 injection increased CM size (Figures 5D and 5E). To investigate whether CM number contributed to heart enlargement, we employed another transgenic zebrafish line with myocardium-specific RFP expression in the nuclei. Interestingly, miR-25 injection increased the number of CMs (Figures 5F and 5G), and it displayed a more significant effect on the proliferation of atrium CMs (Figure S8). Neither miR-25 nor NC injection had any influence on heart rate (Figure 5H). Collectively, these data demonstrated that miR-25 injection could increase both the size and the number of CMs in zebrafish.
Figure 5.
miR-25 Promotes CM Proliferation in Zebrafish
(A) Morphology of zebrafish hearts injected with miR-25 mimics or NC. Transgenic zebrafish with myocardium-specific GFP expression were used. (B) Histological section of the cross-sectional areas of miR-25 or NC injection. (C) The cross-sectional areas of miR-25 or NC injection. (n > 6) (D) The confocal picture of single CM injected with miR-25 or NC. (E) The volume of single CM treated with miR-25 or NC. (F) CM of zebrafish hearts injected with miR-25 or NC. (G) The number of CMs after miR-25 or NC injections. A transgenic zebrafish line with myocardium-specific RFP expression in the nuclei was employed. (H) miR-25 injection did not influence heart rate. Statistical significance was calculated using Student’s t test for paired samples. Data are shown as the mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
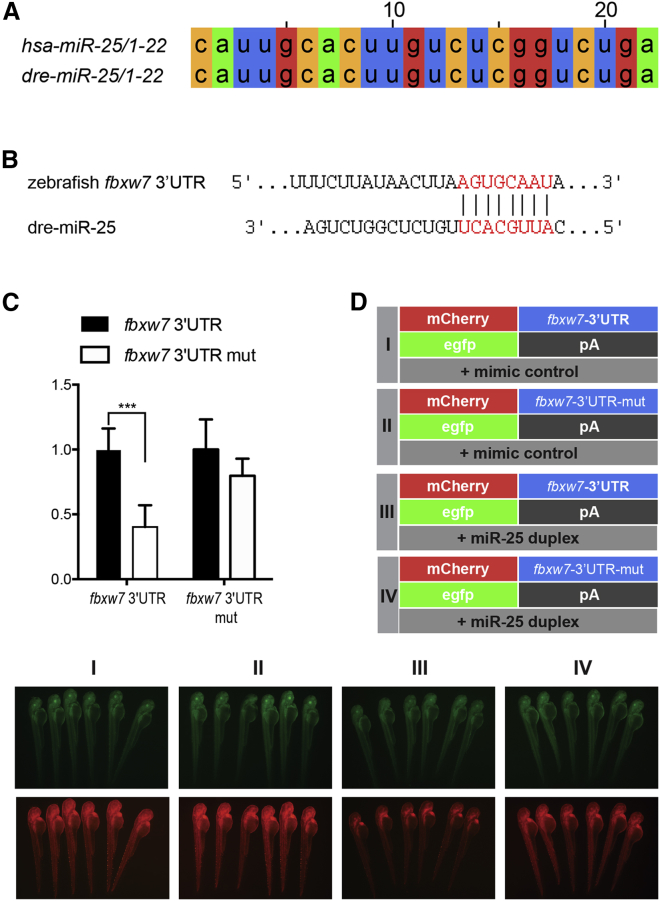
miR-25 Targets fbxw7 in Zebrafish
To test whether miR-25 targeted fbxw7 in zebrafish as well, we carried out a series of experiments. First, the sequence alignment analysis demonstrated that the zebrafish miR-25 (dre-miR-25) mature sequence was identical to the human miR-25 (hsa-miR-25) (Figure 6A). Moreover, we found that dre-miR-25 potentially targeted fbxw7 in zebrafish through TargetScanFish prediction (Figure 6B). In vitro luciferase assays revealed that miR-25 duplex significantly inhibited the expression of luciferase-fbxw7-3′ UTR compared with that of the control (Figure 6C). To test the functional interaction of miR-25 and fbxw7-3′ UTR in vivo, we performed the fluorescence sensor assay in zebrafish embryos. It was confirmed that miR-25 mimics repressed the expression of the mCherry-fbxw7-3′ UTR, but not mCherry-fbxw7-3′ UTR-mutant (mut), in which the miR-25 potential targeting site was mutated (Figure 6D). In summary, these results demonstrated that miR-25 also targeted fbxw7 in zebrafish.
Figure 6.
FBXW7 Is a Direct Target Gene of miR-25 in Zebrafish
(A) The sequence of miR-25 sequences is conserved in human and zebrafish. (B) A potential target site (highlighted in red) of miR-25 on the zebrafish fbxw7 3′ UTR was predicted by TargetScan. (C) Luciferase reporter assay showed that the predicted binding sequence was required for miR-25 inhibition. (D) mCherry sensors were co-injected with EGFP control as indicated. miR-25-duplex injection reduced the mCherry levels in mCherry-fbxw7-3′ UTR, whereas EGFP levels were unchanged. In the mutated sensor, no reduction in mCherry was noted. Experiments were repeated three times; for each group, six embryos were analyzed. Mut, mutant. Statistical significance was calculated using Student’s t test for paired samples. Data are shown as the mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
Discussion
Direct activation of endogenous CM proliferation to regenerate and repair the heart, both after injury and in chronic disease states, is emerging as one of the most promising strategies in cardiac regenerative medicine.34 Identification of genes that can be harnessed to promote CM proliferation would be helpful for cardiac regeneration. Previous studies in rodents revealed that forced overexpression of certain synthetic miRNAs can promote CM proliferation.12,34 In this study, we harnessed hESC-CMs to decipher miRNA functions in CM proliferation and demonstrated that miR-25 promoted CM proliferation. Importantly, we identified FBXW7 as a target of miR-25 to regulate human CM proliferation. FBXW7 is a cell-cycle regulatory factor that mediates the ubiquitin-dependent proteolysis of many positive cell-cycle regulators, including cyclin E1, c-Myc, c-Jun, and Notch.35,36 Low expression of FBXW7 has been observed in colorectal cancer.37 Inactivation of Fbxw7 in the T cell lineage of mice impairs cell-cycle exit during T cell differentiation.38 Therefore, FBXW7 plays an important role in the regulation of cell proliferation by modulating cell-cycle activities. In this study, we demonstrate that FBXW7 is a target of miR-25, and overexpression of miR-25 promotes CM proliferation by downregulating FBXW7. These results were confirmed in the zebrafish model. Therefore, miR-25 could serve as a novel molecule for cardiac regeneration.
miR-25 has multiple effects on heart failure. miR-25 expression in humans is initially decreased in the early stage of heart failure but is later increased in end-stage heart failure.39 Decreased miR-25 expression in diseased myocardium leads to re-expression of its target gene Hand2, a transcription factor that can cause cardiac dilation and dysfunction in the postnatal myocardium.40 Interestingly, another group has demonstrated that inhibition of miR-25 expression in diseased myocardium leads to overexpression of its target gene SERCA2a and improves cardiac function.20 Other effects of miR-25 include protection of CMs against oxidative damage41 and against apoptosis induced by sepsis.42 We first demonstrated that miR-25 promoted CM proliferation, extending the list of miR-25 functions.
Notably, multiple effects of miR-25 in the rodent heart have also been documented, some of which seem to contradict each other. Overexpression of miR-25 protects CMs against oxidative damage and sepsis-induced apoptosis.41,42 On the other hand, overexpression of miR-25 in the normal heart causes a significant loss of contractile function, CM fibrosis, and apoptosis.20,39 However, this study revealed that overexpression of miR-25 in zebrafish did not significantly alter the heart function. Inhibition of miR-25 improves cardiac contractility in the failing heart20 but can also induce atrial fibrillation,43 high blood pressure,39 mild heart dilation,39 and spontaneous cardiac dysfunction.40 Therefore, additional studies are required to investigate the therapeutic effects of the miR-25 expression during different stages of heart disease.
Materials and Methods
Cell Culture
The HEK293T (ATCC) cell line was grown in DMEM supplemented with 10% FBS, 1× penicillin-streptomycin. hESCs (Wicell) and hiPSCs (Kindly provided by Cellapy) were maintained in human PSCeasy Medium (Cellapy) on Matrigel-coated (Corning) plates and passaged using 0.5 mM EDTA every ∼3–4 days. All cells were incubated at 37°C with 95% air and 5% CO2.
In Vitro Differentiation
hESCs and hiPSCs were directly differentiated using small molecules following a previously described method.44 In brief, before differentiation, cells were dissociated using 0.5 mM EDTA at a ratio of approximately 1:10 and cultured in human PSCeasy Medium for ∼3–4 days. When the cell density reached ∼60%–70% confluence on day 0, the medium was changed to CDM3 basal medium containing 6 μM CHIR99021 (Selleck), an inhibitor of GSK3β that activates the Wnt signaling pathway. On day 2, the medium was changed to CDM3 containing 2 μM Wnt-C59 (Sigma), a Wnt inhibitor. From day 4 onward, the medium was replaced with fresh CDM3 every other day, and spontaneously contracting CMs were observed on day 8. From days 9 to 12, CMs were purified with Cardioeasy purification medium (Cellapy). CMs were maintained with medium composed of RPMI 1640 with 2% B27 serum-free supplement (Gibco) from day 15 onward, with the medium exchanged every 2 or 3 days.
Luciferase Reporter Assay
The 3′ UTR of the target gene containing putative binding sites for miR-25-3p was amplified by PCR from human genomic DNA and inserted into a psiCHECK-2 vector within XhoI and NotI restriction sites. Mutations were introduced in the seed region of the miR-25-3p binding site for comparison. The synthesized pre-miR-25 sequence was cloned into plko.1 to construct the miR-25 expression plasmid. For the luciferase reporter assay, HEK293T cells were preplated in 12-well plates. On the next day, the cells were cotransfected with 500 ng plko-miR-25 or control plasmid and 500 ng WT or mutated 3′ UTR of FBXW7 using Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer’s instructions. After transfection for 48 h, the cell lysates were harvested, and luciferase activity was measured using the Dual-Luciferase Reporter System (Promega). The activity of Renilla luciferase was normalized to that of firefly luciferase. The experiment was repeated three times independently.
Immunofluorescence
After 48 h of transfection, CMs were washed with PBS three times for 5 min each and fixed with 4% Paraformaldehyde Fix Solution (Sangon Biotech) for 15 min at room temperature and permeabilized with 0.3% Triton X-100 in PBS for 15 min, then blocked with 3% BSA for 1 h. Next, the cells were incubated with primary antibody in blocking solution overnight at 4°C. The following primary antibodies were used: mouse anti-α-ACTININ (1:500; Sigma) and rabbit anti-TNNT2 (1:500; Proteintech) were used for labeling the CMs. Rabbit anti-Ki-67 (1:300; Cell Signaling) was used to detect mitosis. After being washed with PBS three times, cells were incubated with secondary antibodies conjugated with Alexa Fluor 488 and 584 (Sigma) for 1 h at room temperature in the dark. For EdU staining, CMs were labeled with EdU for 24 h before immunofluorescence, and the Click-iT EdU Alexa Fluor 488 Imaging kit (Invitrogen) was used according to the manufacturer’s protocol. DAPI (1:1,000; Sigma) was used to stain nuclei. CM proliferation was calculated by counting Ki-67 (EdU)+, α-ACTININ+, and DAPT+ cells in every image. More than 2000 cells were counted. CM size was quantified using ImageJ.
Oligo Transfection
The synthetic miR-25 mimics, FBXW7 siRNA, and their NC were purchased from GenePharma (Shanghai). Differentiated CMs were isolated and replated at 80% confluency; after 3 days, when they resumed beating, they were transfected with 40 nM miRNA mimics or siRNA using Lipofectamine 3000 transfection reagent (Invitrogen). After 48 h of treatment, they were used for immunofluorescence, western blot, or qRT-PCR analysis.
RNA Isolation and qRT-PCR
Total RNA (including mRNA and miRNA) was extracted from treated CMs using TRIzol reagent (Ambion) following the manufacturer’s manual. For mRNA expression analysis, first-strand cDNA was synthesized using a 5× All-In-One RT MasterMix kit (abm), and mRNA expression was quantified with 2× SYBR Green qPCR Master Mix (bimake) according to the manufacturer’s protocol. GAPDH was used as an internal control. For miRNA, the cDNA was reverse transcribed using the miScript II RT kit (QIAGEN). The relative expression level of miR-25-3p was evaluated with the miScript SYBR Green PCR kit (QIAGEN) and normalized to hU6. qPCR was performed using the Bio-Rad Real-Time PCR Detection System, and the relative expression level was calculated using the 2−ΔΔCt method. The primer sequences used can be seen in Table S1.
Western Blot Analysis
Treated cells were harvested and lysed with Nonidet P-40 (NP-40) buffer (Beyotime) in the presence of 1 mM phenylmethanesulfonyl fluoride (Beyotime). After the cells were spun at 12,000 rpm for 10 min in a 4°C precooled centrifuge, the supernatant was collected for western blot analysis. Proteins were separated on an 8% SDS-PAGE gel and then transferred to a polyvinylidene fluoride (PVDF) membrane (Thermo). After being locked with 5% (w/v) BSA (Sigma) in TBS-T (0.1% Tween 20 in 1× TBS) buffer for 1 h at room temperature, the membrane was incubated with primary antibodies at 4°C overnight. The following antibodies were used: anti-FBXW7 (1:1,000; Abcam) and anti-GAPDH (1:2,000; Cell Signaling). After three washes in TBS-T for 5 min each, the membranes were incubated with secondary antibody at room temperature for 1 h, then washed three times and stained following the kit manufacturer’s recommendation. ImageJ was used to quantify the band intensities.
RNA-Seq and Bioinformatics Analysis
RNA-seq was performed by Shanghai Biotechnology Corporation (SBC). In brief, CMs transfected with miR-25-3p mimics or its NC were collected, and total RNA was extracted using an RNeasy Micro Kit (QIAGEN) following the manufacturer’s instructions. Total RNA integrity was checked with an Agilent Bioanalyzer 2100 (Agilent). The cDNA library was constructed using a TruSeq RNA Sample Prep Kit and sequenced on an Illumina HiSeq 2000 system (Illumina). Sequenced reads were mapped onto the human hg38 reference genome. The reads were converted into FPKM (fragments per kilobase of exon model per million mapped reads) for standardization of gene expression using StringTie (version 1.3.0). EdgeR was applied to identify DEGs between samples. KEGG and GO enrichment were analyzed for differentially expressed genes. The RNA-seq data has been deposited to SRA Database (accession number: PRJNA531900).
Electrical Activity Analysis
A multi-electrode array (MEA; Multi Channel Systems) was utilized to record the electrophysiological features of CMs, including beat period, field potential duration (FPD), and conduction velocity. FPD was subsequently corrected (FPDc) using Fridericia’s formula (FDP/3 √RR, where RR is the interspike interval). All experiments were performed in DMEM without FBS or antibiotics.
Ethics Statement
All zebrafish experimentation was carried out in accordance with the NIH Guidelines for the Care and Use of Laboratory Animals (https://oacu.od.nih.gov/regs/index.htm) and ethically approved by the Administration Committee of Experimental Animals, Jiangsu Province, China (approval ID: SYXK (SU) 2007-0021).
Zebrafish Strains and Breeding
Zebrafish were provided by the Zebrafish Center at Nantong University Jiangsu Key Laboratory of Neuroregeneration. Zebrafish embryos and adults were raised and maintained under the conditions described previously.45 Two transgenic zebrafish lines, Tg(cmlc2:nDsRed) and Tg(cmlc2:GFP), were used as described in a previous work.45
Injection of miRNA-25 Mimic and NC
miRNA-25 mimic 20 pmol and NC were injected into embryos in the one- to two-cell stages.
Target Prediction
Target prediction of miR-25 in zebrafish was carried out with TargetScanFish 6.2 (http://www.targetscan.org/fish_62/).
Whole-Embryo miRNA Sensor Assay
A whole-embryo miRNA sensor assay in zebrafish was carried out as described previously.45 egfp and mCherry coding sequences were cloned into the pCS2+ vector. The pCS2+-mCherry-fbxw7-3′ UTR control and the pCS2+-mCherry-fbxw7-3′ UTR-mut construct were generated. The pCS2+-EGFP vector was used as an injection control. The plasmids were linearized with NotI/KpnI and used as templates to synthesize the capped mRNAs using a mMESSAGE mMACHINE kit (Ambion). The RNAs were injected into the cytoplasm of one- to two-cell embryos (35 pg/embryo).
Microscopic Imaging
For microscopic imaging of zebrafish, embryos were embedded in 0.6% low-melting point agarose. Confocal imaging was performed with a Leica TCS-SP5 LSM. The anesthetic MS-222 (100% strength) was used to stop the heartbeat before confocal imaging. Analysis was performed using Imaris software.
Statistical Analysis
Statistical analysis was performed using GraphPad Prism 5. All data are presented as the mean ± SEM. An unpaired t test and one-way ANOVA were used to determine statistical significance between two and more than two groups, respectively. Significance levels are indicated as follows: *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
Author Contributions
B.W., M.X., M.L., and F.W. designed and performed the experiments; S.H. and X.C. analyzed the miRNA-seq data; F.L., L.Z., and Z.H. revised the manuscript; D.L. and Y.W. supervised the project. All authors read and approved the final manuscript.
Conflicts of Interest
X.C. works for Hangzhou Rongze Biotechnology Co., Ltd. The other authors declare no competing interests.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (grant 81870199); National Basic Research Program of China (grant 2015CB943300); Foundation for Innovative Research Group of the National Natural Science Foundation of China (grant 31521003); and Opening Program 2018 of the State Key Laboratory of Genetic Engineering (grant SKLGE1809).
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2020.01.013.
Contributor Information
Feng Lan, Email: fenglan@ccmu.edu.cn.
Dong Liu, Email: tom@ntu.edu.cn.
Yongming Wang, Email: ymw@fudan.edu.cn.
Supplemental Information
References
- 1.Anderson J.L., Morrow D.A. Acute Myocardial Infarction. N. Engl. J. Med. 2017;376:2053–2064. doi: 10.1056/NEJMra1606915. [DOI] [PubMed] [Google Scholar]
- 2.Senyo S.E., Lee R.T., Kühn B. Cardiac regeneration based on mechanisms of cardiomyocyte proliferation and differentiation. Stem Cell Res. (Amst.) 2014;13(3 Pt B):532–541. doi: 10.1016/j.scr.2014.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bergmann O., Bhardwaj R.D., Bernard S., Zdunek S., Barnabé-Heider F., Walsh S., Zupicich J., Alkass K., Buchholz B.A., Druid H. Evidence for cardiomyocyte renewal in humans. Science. 2009;324:98–102. doi: 10.1126/science.1164680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eschenhagen T., Bolli R., Braun T., Field L.J., Fleischmann B.K., Frisén J., Giacca M., Hare J.M., Houser S., Lee R.T. Cardiomyocyte Regeneration: A Consensus Statement. Circulation. 2017;136:680–686. doi: 10.1161/CIRCULATIONAHA.117.029343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ieda M., Fu J.D., Delgado-Olguin P., Vedantham V., Hayashi Y., Bruneau B.G., Srivastava D. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell. 2010;142:375–386. doi: 10.1016/j.cell.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qian L., Huang Y., Spencer C.I., Foley A., Vedantham V., Liu L., Conway S.J., Fu J.D., Srivastava D. In vivo reprogramming of murine cardiac fibroblasts into induced cardiomyocytes. Nature. 2012;485:593–598. doi: 10.1038/nature11044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Muraoka N., Yamakawa H., Miyamoto K., Sadahiro T., Umei T., Isomi M., Nakashima H., Akiyama M., Wada R., Inagawa K. MiR-133 promotes cardiac reprogramming by directly repressing Snai1 and silencing fibroblast signatures. EMBO J. 2014;33:1565–1581. doi: 10.15252/embj.201387605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Senyo S.E., Steinhauser M.L., Pizzimenti C.L., Yang V.K., Cai L., Wang M., Wu T.D., Guerquin-Kern J.L., Lechene C.P., Lee R.T. Mammalian heart renewal by pre-existing cardiomyocytes. Nature. 2013;493:433–436. doi: 10.1038/nature11682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deng S., Zhao Q., Zhen L., Zhang C., Liu C., Wang G., Zhang L., Bao L., Lu Y., Meng L. Neonatal Heart-Enriched miR-708 Promotes Proliferation and Stress Resistance of Cardiomyocytes in Rodents. Theranostics. 2017;7:1953–1965. doi: 10.7150/thno.16478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nair N., Gongora E. MicroRNAs as therapeutic targets in cardiomyopathies: myth or reality? Biomol. Concepts. 2014;5:439–448. doi: 10.1515/bmc-2014-0026. [DOI] [PubMed] [Google Scholar]
- 11.Palacín M., Reguero J.R., Martín M., Díaz Molina B., Morís C., Alvarez V., Coto E. Profile of microRNAs differentially produced in hearts from patients with hypertrophic cardiomyopathy and sarcomeric mutations. Clin. Chem. 2011;57:1614–1616. doi: 10.1373/clinchem.2011.168005. [DOI] [PubMed] [Google Scholar]
- 12.Eulalio A., Mano M., Dal Ferro M., Zentilin L., Sinagra G., Zacchigna S., Giacca M. Functional screening identifies miRNAs inducing cardiac regeneration. Nature. 2012;492:376–381. doi: 10.1038/nature11739. [DOI] [PubMed] [Google Scholar]
- 13.Tian Y., Liu Y., Wang T., Zhou N., Kong J., Chen L., Snitow M., Morley M., Li D., Petrenko N. A microRNA-Hippo pathway that promotes cardiomyocyte proliferation and cardiac regeneration in mice. Sci. Transl. Med. 2015;7:279ra38. doi: 10.1126/scitranslmed.3010841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pandey R., Ahmed R.P. MicroRNAs Inducing Proliferation of Quiescent Adult Cardiomyocytes. Cardiovasc. Regen. Med. 2015;2:e519. doi: 10.14800/crm.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liang D., Li J., Wu Y., Zhen L., Li C., Qi M., Wang L., Deng F., Huang J., Lv F. miRNA-204 drives cardiomyocyte proliferation via targeting Jarid2. Int. J. Cardiol. 2015;201:38–48. doi: 10.1016/j.ijcard.2015.06.163. [DOI] [PubMed] [Google Scholar]
- 16.Arif M., Pandey R., Alam P., Jiang S., Sadayappan S., Paul A., Ahmed R.P.H. MicroRNA-210-mediated proliferation, survival, and angiogenesis promote cardiac repair post myocardial infarction in rodents. J. Mol. Med. (Berl.) 2017;95:1369–1385. doi: 10.1007/s00109-017-1591-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thomson J.A., Itskovitz-Eldor J., Shapiro S.S., Waknitz M.A., Swiergiel J.J., Marshall V.S., Jones J.M. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 18.Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 19.Burridge P.W., Keller G., Gold J.D., Wu J.C. Production of de novo cardiomyocytes: human pluripotent stem cell differentiation and direct reprogramming. Cell Stem Cell. 2012;10:16–28. doi: 10.1016/j.stem.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wahlquist C., Jeong D., Rojas-Muñoz A., Kho C., Lee A., Mitsuyama S., van Mil A., Park W.J., Sluijter J.P., Doevendans P.A. Inhibition of miR-25 improves cardiac contractility in the failing heart. Nature. 2014;508:531–535. doi: 10.1038/nature13073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Diez-Cuñado M., Wei K., Bushway P.J., Maurya M.R., Perera R., Subramaniam S., Ruiz-Lozano P., Mercola M. miRNAs that induce human cardiomyocyte proliferation converge on the hippo pathway. Cell Rep. 2018;23:2168–2174. doi: 10.1016/j.celrep.2018.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Snir M., Kehat I., Gepstein A., Coleman R., Itskovitz-Eldor J., Livne E., Gepstein L. Assessment of the ultrastructural and proliferative properties of human embryonic stem cell-derived cardiomyocytes. Am. J. Physiol. Heart Circ. Physiol. 2003;285:H2355–H2363. doi: 10.1152/ajpheart.00020.2003. [DOI] [PubMed] [Google Scholar]
- 23.McDevitt T.C., Laflamme M.A., Murry C.E. Proliferation of cardiomyocytes derived from human embryonic stem cells is mediated via the IGF/PI 3-kinase/Akt signaling pathway. J. Mol. Cell. Cardiol. 2005;39:865–873. doi: 10.1016/j.yjmcc.2005.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cui L., Johkura K., Takei S., Ogiwara N., Sasaki K. Structural differentiation, proliferation, and association of human embryonic stem cell-derived cardiomyocytes in vitro and in their extracardiac tissues. J. Struct. Biol. 2007;158:307–317. doi: 10.1016/j.jsb.2006.11.009. [DOI] [PubMed] [Google Scholar]
- 25.Liu Q., Jiang C., Xu J., Zhao M.T., Van Bortle K., Cheng X., Wang G., Chang H.Y., Wu J.C., Snyder M.P. Genome-Wide Temporal Profiling of Transcriptome and Open Chromatin of Early Cardiomyocyte Differentiation Derived From hiPSCs and hESCs. Circ. Res. 2017;121:376–391. doi: 10.1161/CIRCRESAHA.116.310456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen J., Huang Z.P., Seok H.Y., Ding J., Kataoka M., Zhang Z., Hu X., Wang G., Lin Z., Wang S. mir-17-92 cluster is required for and sufficient to induce cardiomyocyte proliferation in postnatal and adult hearts. Circ. Res. 2013;112:1557–1566. doi: 10.1161/CIRCRESAHA.112.300658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Porrello E.R., Johnson B.A., Aurora A.B., Simpson E., Nam Y.J., Matkovich S.J., Dorn G.W., 2nd, van Rooij E., Olson E.N. MiR-15 family regulates postnatal mitotic arrest of cardiomyocytes. Circ. Res. 2011;109:670–679. doi: 10.1161/CIRCRESAHA.111.248880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu N., Bezprozvannaya S., Williams A.H., Qi X., Richardson J.A., Bassel-Duby R., Olson E.N. microRNA-133a regulates cardiomyocyte proliferation and suppresses smooth muscle gene expression in the heart. Genes Dev. 2008;22:3242–3254. doi: 10.1101/gad.1738708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu X., Xiao J., Zhu H., Wei X., Platt C., Damilano F., Xiao C., Bezzerides V., Boström P., Che L. miR-222 is necessary for exercise-induced cardiac growth and protects against pathological cardiac remodeling. Cell Metab. 2015;21:584–595. doi: 10.1016/j.cmet.2015.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.D’Uva G., Aharonov A., Lauriola M., Kain D., Yahalom-Ronen Y., Carvalho S., Weisinger K., Bassat E., Rajchman D., Yifa O. ERBB2 triggers mammalian heart regeneration by promoting cardiomyocyte dedifferentiation and proliferation. Nat. Cell Biol. 2015;17:627–638. doi: 10.1038/ncb3149. [DOI] [PubMed] [Google Scholar]
- 31.Chen H., Pan H., Qian Y., Zhou W., Liu X. MiR-25-3p promotes the proliferation of triple negative breast cancer by targeting BTG2. Mol. Cancer. 2018;17:4. doi: 10.1186/s12943-017-0754-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao H., Wang Y., Yang L., Jiang R., Li W. MiR-25 promotes gastric cancer cells growth and motility by targeting RECK. Mol. Cell. Biochem. 2014;385:207–213. doi: 10.1007/s11010-013-1829-x. [DOI] [PubMed] [Google Scholar]
- 33.Xiang J., Hang J.B., Che J.M., Li H.C. MiR-25 is up-regulated in non-small cell lung cancer and promotes cell proliferation and motility by targeting FBXW7. Int. J. Clin. Exp. Pathol. 2015;8:9147–9153. [PMC free article] [PubMed] [Google Scholar]
- 34.Chen J., Huang Z.-P., Seok H.Y., Ding J., Kataoka M., Zhang Z., Hu X., Wang G., Lin Z., Wang S. mir-17–92 cluster is required for and sufficient to induce cardiomyocyte proliferation in postnatal and adult hearts. Circ. Res. 2013;112:1557–1566. doi: 10.1161/CIRCRESAHA.112.300658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Minella A.C., Clurman B.E. Mechanisms of tumor suppression by the SCF(Fbw7) Cell Cycle. 2005;4:1356–1359. doi: 10.4161/cc.4.10.2058. [DOI] [PubMed] [Google Scholar]
- 36.Welcker M., Clurman B.E. FBW7 ubiquitin ligase: a tumour suppressor at the crossroads of cell division, growth and differentiation. Nat. Rev. Cancer. 2008;8:83–93. doi: 10.1038/nrc2290. [DOI] [PubMed] [Google Scholar]
- 37.Iwatsuki M., Mimori K., Ishii H., Yokobori T., Takatsuno Y., Sato T., Toh H., Onoyama I., Nakayama K.I., Baba H., Mori M. Loss of FBXW7, a cell cycle regulating gene, in colorectal cancer: clinical significance. Int. J. Cancer. 2010;126:1828–1837. doi: 10.1002/ijc.24879. [DOI] [PubMed] [Google Scholar]
- 38.Onoyama I., Tsunematsu R., Matsumoto A., Kimura T., de Alborán I.M., Nakayama K., Nakayama K.I. Conditional inactivation of Fbxw7 impairs cell-cycle exit during T cell differentiation and results in lymphomatogenesis. J. Exp. Med. 2007;204:2875–2888. doi: 10.1084/jem.20062299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li H., Xie Y., Liu Y., Qi Y., Tang C., Li X., Zuo K., Sun D., Shen Y., Pang D. Alteration in microRNA-25 expression regulate cardiac function via renin secretion. Exp. Cell Res. 2018;365:119–128. doi: 10.1016/j.yexcr.2018.02.029. [DOI] [PubMed] [Google Scholar]
- 40.Dirkx E., Gladka M.M., Philippen L.E., Armand A.S., Kinet V., Leptidis S., El Azzouzi H., Salic K., Bourajjaj M., da Silva G.J. Nfat and miR-25 cooperate to reactivate the transcription factor Hand2 in heart failure. Nat. Cell Biol. 2013;15:1282–1293. doi: 10.1038/ncb2866. [DOI] [PubMed] [Google Scholar]
- 41.Pan L., Huang B.J., Ma X.E., Wang S.Y., Feng J., Lv F., Liu Y., Liu Y., Li C.M., Liang D.D. MiR-25 protects cardiomyocytes against oxidative damage by targeting the mitochondrial calcium uniporter. Int. J. Mol. Sci. 2015;16:5420–5433. doi: 10.3390/ijms16035420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yao Y., Sun F., Lei M. miR-25 inhibits sepsis-induced cardiomyocyte apoptosis by targetting PTEN. Biosci. Rep. 2018;38 doi: 10.1042/BSR20171511. BSR20171511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chiang D.Y., Kongchan N., Beavers D.L., Alsina K.M., Voigt N., Neilson J.R., Jakob H., Martin J.F., Dobrev D., Wehrens X.H., Li N. Loss of microRNA-106b-25 cluster promotes atrial fibrillation by enhancing ryanodine receptor type-2 expression and calcium release. Circ. Arrhythm. Electrophysiol. 2014;7:1214–1222. doi: 10.1161/CIRCEP.114.001973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Burridge P.W., Holmström A., Wu J.C. Chemically defined culture and cardiomyocyte differentiation of human pluripotent stem cells. Curr. Protoc. Hum. Genet. 2015;87:21.3.1–21.3.15. doi: 10.1002/0471142905.hg2103s87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang X., Ling C.C., Li L., Qin Y., Qi J., Liu X., You B., Shi Y., Zhang J., Jiang Q. MicroRNA-10a/10b represses a novel target gene mib1 to regulate angiogenesis. Cardiovasc. Res. 2016;110:140–150. doi: 10.1093/cvr/cvw023. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Cardiomyocytes with Regular Contraction