Abstract
Frequent injections of anti-vascular endothelial growth factor (anti-VEGF) agents are a clinical burden for patients with neovascular age-related macular degeneration (AMD). Genomic disruption of VEGF-A using adeno-associated viral (AAV) delivery of clustered regularly interspaced short palindromic repeats (CRISPR)-Cas9 has the potential to permanently suppress aberrant angiogenesis, but the factors that determine the optimal efficacy are unknown. Here, we investigate two widely used Cas9 endonucleases, SpCas9 and SaCas9, and evaluate the relative contribution of AAV-delivery efficiency and genome-editing rates in vivo to determine the mechanisms that drive successful CRISPR-based suppression of VEGF-A, using a mouse model of laser-induced choroidal neovascularization (CNV). We found that SpCas9 demonstrated higher genome-editing rates, greater VEGF reduction, and more effective CNV suppression than SaCas9, despite similar AAV transduction efficiency between a dual-vector approach for SpCas9 and single-vector system for SaCas9 to deliver the Cas9 orthologs and single guide RNAs (gRNAs). Our results suggest that successful VEGF knockdown using AAV-mediated CRISPR systems may be determined more by the efficiency of genome editing rather than viral transduction and that SpCas9 may be more effective than SaCas9 as a potential therapeutic strategy for CRISPR-based treatment of CNV in neovascular AMD.
Keywords: CRISPR, Cas9, genome editing, AMD, age-related macular degeneration, macular degeneration, CNV, VEGF, VEGFa, angiogenesis
Introduction
Anti-vascular endothelial growth factor (VEGF) therapy has revolutionized the management of neovascular age-related macular degeneration (AMD), but frequent intravitreal injections of anti-angiogenesis agents are costly, invasive, and a clinical burden.1, 2, 3, 4, 5 Gene therapy holds the promise for more durable VEGF suppression, but clinical trials have shown limited success. Subretinal injection of adeno-associated virus 2 (AAV2) expressing the soluble VEGF receptor sFLT1 showed no significant visual improvement in phase 2 clinical studies.6 Additional strategies being developed include lentiviral delivery of endostatin and angiostatin, subretinal AAV8 delivery of anti-VEGF antibody fragments, and intravitreal AAV2-7m8 expression of a soluble decoy VEGF receptor, with several clinical trials already underway.
Genome editing using clustered regularly interspaced short palindromic repeats (CRISPR)-associated Cas9 endonucleases has the potential to permanently reduce angiogenesis by disrupting VEGF-A at the genomic level. Cas9 endonucleases are derived from bacteria-adaptive immune systems and can be programmed using single guide RNAs (gRNAs) to introduce double-strand breaks (DSBs) at specific genomic loci, based on the presence of protospacer adjacent motif (PAM) sequences in the target gene. The DSBs trigger error-prone nonhomologous end joining (NHEJ) repair, which causes insertion or deletion mutations (indels) that result in nonsense or frameshift mutations that permanently disrupt the target gene.7 Streptococcus pyogenes-derived Cas9 (SpCas9) is the most widely used ortholog, as it provides the least restrictive PAM site (5′-NGG-3′) and previously showed its great potential in genome editing in vivo.8 However, the gene is large (4.1 kb) for the packaging capacity of AAV and must be deployed in a dual-vector approach to express the corresponding gRNAs.9 Other Cas9 orthologs from Staphylococcus aureus and Campylobacter jejuni (SaCas9 and CjCas9) are smaller in size and allow combined gRNA expression in a single AAV vector but have more restrictive PAM sequences.10,11
Our group first reported the use of lentiviral vectors expressing SpCas9 to suppress VEGF-A and angiogenesis in vitro using human retinal pigment epithelial (RPE) cells. Subsequent studies have demonstrated similar strategies targeting VEGF-A in mouse RPE12 and VEGF-receptor 2 in human endothelial cells,13 with in vivo applications using subretinal injections of preassembled Cas9 ribnonucleoproteins14 or recombinant AAV serotype 115 to reduce laser-induced choroidal neovascularization (CNV) in mouse eyes. However, the relative efficacies of different Cas9 orthologs for suppressing VEGF-A and CNV have not been compared, and the relative contribution of AAV transduction efficiency and in vivo genome-editing rates have not been explored.
In this study, we evaluate factors that dictate the effectiveness of AAV-mediated genome editing of VEGF-A by directly comparing two different Cas9 orthologs, SpCas9 and SaCas9, using a dual-vector approach for SpCas9 and an “all-in-one” single-vector system for SaCas9. We compared the AAV transduction efficiency and genome-editing rates in vivo to determine their relative contributions to effective suppression of VEGF and laser-induced CNV. Our findings provide a critical framework for clinical translation of CRISPR-based therapeutic approaches for neovascular AMD.
Results
Selection and Testing of gRNAs to Target VEGF-A
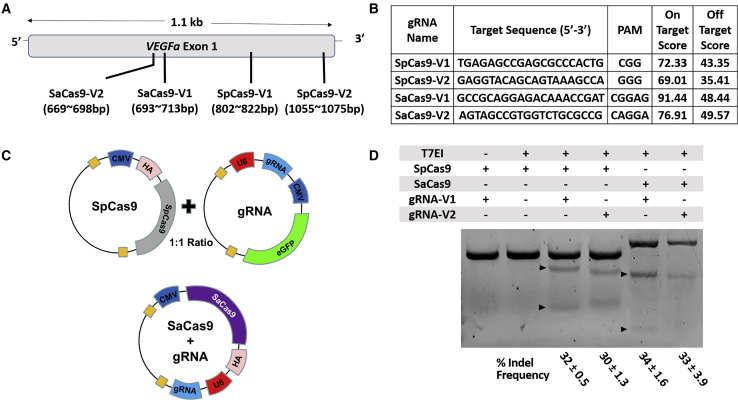
To identify the optimal gRNAs for SpCas9 or SaCas9, we performed in silico analysis (Benchling) as previously described.16 We chose two gRNAs with the highest predicted genome-editing rates (on-target score) targeting exon 1 of the mouse VEGF-A gene, in which an indel mutation is most likely to result in nonfunctional VEGF-A protein (Figures 1A and 1B). Due to the limited packaging capacity of AAV vectors (∼4.7 kb), SpCas9 and corresponding gRNAs were subcloned into two separate AAV cis constructs with a human influenza hemagglutinin (HA) tag for SpCas9 and enhanced green fluorescent protein (EGFP) reporter in the gRNA construct, while HA-tagged SaCas9 and gRNAs were subcloned into single AAV constructs (Figure 1C). Both Cas9 orthologs were driven by ubiquitous cytomegalovirus (CMV) promoters while the gRNAs were driven by U6 promoters. The gRNAs were tested in vitro by transfecting the constructs into NIH 3T3 cells and performing fluorescence-activated cell sorting (FACS) of GFP+ and HA-tag+ cells for SpCas9 and SaCas9, respectively. These studies showed successful indel formation in the mouse VEGF-A gene on T7E1 mismatch assays using both Cas9 orthologs and their corresponding gRNAs, with SpCas9 and SaCas9 demonstrating comparable efficiencies in generating indel mutations in vitro (Figure 1D; n = 3).
Figure 1.
Guide RNAs to Target Mouse VEGF-A Gene
(A) A schematic diagram of CRISPR target sequences in exon 1 of mouse VEGF-A on chromosome 17. (B) CRISPR target sequences and protospacer adjacent motif (PAM) sequences with on-target and off-target scores indicating predicted Cas9 efficiency and off-target probability, respectively. (C) Schematic diagrams illustrating dual AAV vector system to express SpCas9 and gRNA with EGFP reporter or single AAV vector system to express both SaCas9 and corresponding gRNA. Both HA-tagged Cas9 orthologs and EGFP are driven by CMV promoters, while gRNAs are driven by U6 promoters. (D) T7EI mismatch assays demonstrating frequency of indel formation in mouse VEGF-A gene using NIH 3T3 cells transfected with constructs expressing SpCas9 or SaCas9 with different corresponding gRNAs. % indel frequencies are expressed in mean ± SEM (n = 3).
AAV8 Transduction Efficiency in Mouse Retina
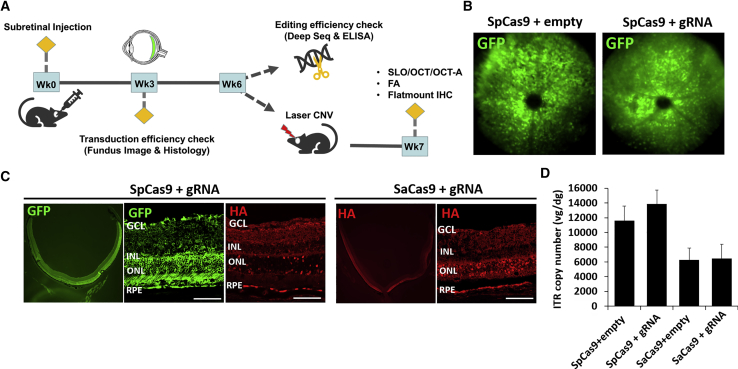
We chose the gRNAs with the highest cutting efficiency for each Cas9 endonuclease (V1), packaged the constructs into AAV serotype 8 (AAV8) vectors, then performed subretinal injections into mouse eyes to evaluate transduction efficiency using in vivo fundus imaging and ex vivo histological analyses (Figure 2A). We injected equivalent amounts of viral vectors carrying the two Cas9 orthologs, with a 1:1 ratio of SpCas9 and corresponding gRNA vectors (4 × 1011 vg/eye each) or SaCas9-gRNA only (4 × 1011 vg/eye total). AAV8 vectors expressing SpCas9 or SaCas9 with only the gRNA scaffold region and no targeting sequences were employed as controls (SpCas9-empty or SaCas9-empty). Three weeks after AAV injections, eyes that received the SpCas9 with control or VEGF-A-targeting gRNA vectors showed diffused GFP expression in vivo (Figure 2B). Histological analysis confirmed GFP and HA-tag expression mostly in the retinal pigment epithelium (RPE) and outer nuclear layer (ONL), consistent with the expected expression pattern using subretinal AAV8 delivery (Figure 2C).17,18 We also measured AAV vector genomes per mouse diploid genome (vg/dg) using the AAV8 inverted terminal repeats (ITRs) to quantify the efficiency of AAV transduction and found that eyes that received dual SpCas9 and gRNA vectors exhibited two times more AAV vector genomes than those that received SaCas9 as a single vector as expected (Figure 2D; n = 5). These results suggest that AAV8 transduction using the dual-vector system for SpCas9 and single-vector approach for SaCas9 were similar and both highly efficient.
Figure 2.
AAV8-Mediated Cas9 Expression in Mouse Eyes
(A) Schematic diagram of in vivo experiments, including subretinal AAV injection, evaluation for viral transduction efficiency at 3 weeks, measurements of gene-editing rates and VEGF-A suppression at 6 weeks, and creation and measurement of laser CNV at 6 and 7 weeks. (B) Representative fluorescence fundus image of mouse eyes that received SpCas9 with and without gRNA showing GFP fluorescence at 3 weeks after AAV injection. (C) Representative histological sections of mouse eyes that received SpCas9 or SaCas9 with corresponding gRNAs, with immunofluorescence labeling of GFP and HA-tag in photoreceptors and RPEs at 3 weeks after AAV injection (scale bars, 100 μm). (D) Bar graph comparing AAV transduction efficiency using qPCR of AAV ITR copy to measure viral genomes per diploid mouse genome (vg/dg) in eyes that received SpCas9 or SaCas9 with and without corresponding gRNAs (n = 5). Error bars, SEM.
SpCas9 Disrupts Vegf-a Gene and Suppresses VEGF-A Expression In Vivo
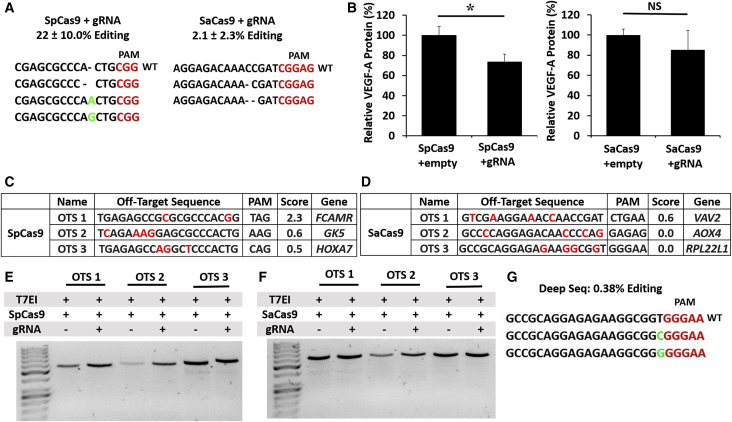
Six weeks after subretinal AAV injections, we performed deep sequencing of genomic DNA from isolated RPE cells and found that SpCas9 triggered greater indel formation (22.5% ± 10.0%, n = 4, average reads = 10402.25) than SaCas9 in vivo (2.1% ± 2.3%, n = 7, average reads = 641) (Figure 3A). AAV-mediated delivery of SpCas9 and gRNA resulted in 26.6% ± 4.1% reduction in VEGF-A protein in RPE-choroid tissues compared with SpCas9-empty control (p = 0.05, n = 9), while SaCas9-gRNA showed no statistically significant difference in VEGF-A suppression compared with SaCas9-empty (p = 0.9, n = 9) (Figure 3B).
Figure 3.
InVivoVEGF-A Genome Editing Efficiency, VEGF-A Protein Suppression, and Off-target Effects
(A) Representative deep-sequencing results showing indel mutations in vivo 6 weeks after injection of AAV expressing SpCas9 or SaCas9 and corresponding gRNAs. (B) Bar graphs comparing VEGF-A protein levels measured by ELISA in RPE-choroid tissues after injection of AAV-expressing SpCas9 or SaCas9 with and without gRNAs (n = 9). *p < 0.05, Student’s t test. NS, not significant. Error bars, SEM. (C and D) Predicted off-target sites for SpCas9 (C) or SaCas9 (D) targeting mouse VEGF-a, including off-target sequences (OTSs), PAM, off-target probability (higher score denotes higher probability of off-target mutation), and affected gene. (E and F) T7EI mismatch assays and deep-sequencing result comparing indel formation at off-target sites for SpCas9 (E)- or SaCas9 (F)-targeting mouse VEGF-a (n = 3). (G) Deep-sequencing results showing possible substitution mutation in SaCas9 OTS3.
We also evaluated the top 3 predicted off-target loci located within functional genes from 49 candidate loci for SpCas9-gRNA and 25 candidate loci for SaCas9-gRNA, using deep sequencing and T7 endonuclease I (T7EI) assays (Figures 3C–3G). We found no significant off-target indel mutation, except for a low rate of possible substitution mutations found at the RPL22L1 locus (0.13% ± 0.22%, n = 3, average total reads = 53601.33, indel reads = 241) for SaCas9 that was only detected in 1 of 3 samples evaluated by deep sequencing (Figure 3G). Since these substitution mutations are located within a G-C-rich region, which is more error-prone in next-generation sequencing,19,20 the possibility of a sequencing error cannot be excluded. By contrast, none of the eyes transduced with SpCas9 and gRNAs showed any detectable mutations at predicted off-target sites.
AAV Delivery of SpCas9 Reduces CNV in Mouse Eyes
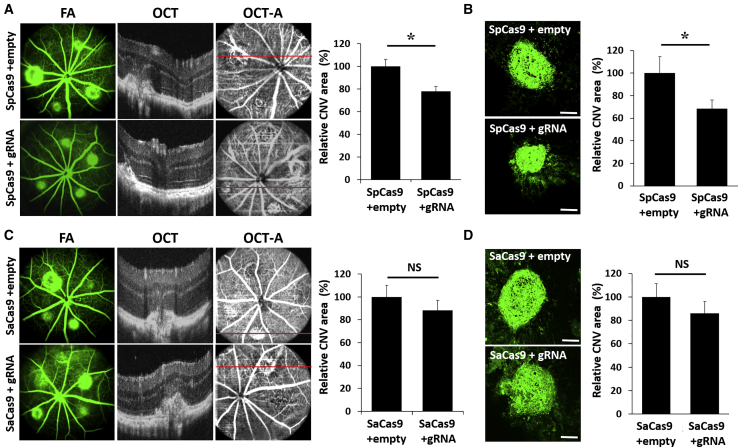
To compare the functional consequences of CRISPR-based VEGF suppression, we performed laser-induced CNV 6 weeks after AAV injections, then measured in vivo CNV size 1 week later using fundus fluorescein angiography (FA), optical coherence tomography (OCT), and OCT angiography (OCT-A) (Figures 4A and 4C), and by ex vivo staining of CNV with isolectin-B4 (IB4) (Figures 4B and 4D). Consistent with the degree of VEGF suppression, eyes that received SpCas9 with gRNA showed a 22% reduction in CNV area based on OCT-A in vivo (p = 0.006, n = 15) and 31.54% reduction based on IB4-staining ex vivo (p = 0.05, n = 20), compared with SpCas9 alone (Figures 4A and 4B). By contrast, SaCas9 showed no significant CNV reduction as seen on OCT-A (p = 0.39) or histology (p = 0.37, n = 16; Figures 4C and 4D). Our results indicate that SpCas9 and gRNA delivered using two AAV vectors is more effective at suppressing laser-induced CNV in mouse eyes than a single-vector platform for SaCas9-gRNA.
Figure 4.
CRISPR-Mediated Suppression of Laser-Induced CNV in Mouse Retina
(A and C) Representative fluorescein angiography (FA), optical coherent tomography (OCT), and OCT angiography (OCT-A) images of mouse eyes injected with AAV8 expressing SpCas9 (A) or SaCas9 (C) with or without corresponding gRNAs, 1 week after laser-induced CNV, with bar graphs showing CNV size measured from OCT-A images (n = 15). *p < 0.05, Student’s t test. NS, not significant. Error bars, SEM. (B and D) IB4-stained flat-mount immunohistochemistry of CNV lesions from eyes injected with AAV8-expressing SpCas9 (B) or SaCas9 (D) with and without corresponding gRNAs (scale bars, 100 μm), with bar graphs comparing CNV size measured from IB4 staining of flat-mount tissues (n = 20). *p < 0.05, Student’s t test. NS, not significant. Error bars, SEM.
Discussion
The management of neovascular AMD has been encumbered by the frequency and cost of intravitreal anti-VEGF injections. Although the eyes are well suited for gene therapy due to their immune privilege and ease of access, clinical studies employing AAV vectors to deliver anti-angiogenic factors have demonstrated limited success.6,21 Genome editing using CRISPR/Cas9 can permanently disrupt VEGF expression and provide a potential cure for neovascular AMD, but factors that determine the effectiveness of AAV-mediated genome editing in the eye have not been explored. Here, we evaluated two commonly used Cas9 orthologs using a dual-AAV system for SpCas9 and a single-AAV approach for SaCas9 to suppress VEGF-A expression and laser-induced CNV in mouse eyes. We found that despite similar viral transduction efficiency, SpCas9 demonstrated greater genome editing, VEGF reduction, and CNV suppression than SaCas9.
In our study, AAV delivery of SpCas9 and gRNAs only achieved up to 26% reduction of VEGF-A levels and 31% suppression of CNV. Although the clinical relevance of this partial VEGF suppression is unclear, the efficacy is somewhat comparable to the suppression of laser-induced CNV in mice using aflibercept, a soluble VEGF-binding protein that has been clinically approved to treat neovascular AMD, which shows a 43% reduction using our methods (Figures S1A and S1B) and is similar to published studies.22 Importantly, while most current pharmacotherapies require repeated, pulsatile treatments that result in fluctuations in VEGF suppression, evidence suggests that continuous, stable VEGF suppression using gene therapy23 or sustained delivery systems24 may be clinically effective even at lower therapeutic doses. For example, in a phase 2 clinical trial evaluating a refillable port delivery system for continuous release of the anti-VEGF antibody fragment ranibizumab, median time to refill was 15 months, as compared to the monthly or semi-monthly dosing frequencies of intravitreal injections.24 These results suggest that even partial, chronic suppression of VEGF using a CRISPR-based approach may be sufficient to control the disease process in neovascular AMD. In fact, complete VEGF suppression may not be preferred, as normal, homeostatic levels of VEGF are required to support the health of the choriocapillaris,25,26 and chronic over-suppression of VEGF may be associated with RPE or choroidal atrophy in AMD patients.27, 28, 29
Nevertheless, future studies may be necessary to further optimize genome-editing strategies for treating neovascular eye diseases. CRISPR-based approaches rely on the formation of indel mutations to disrupt the target gene. Because not all indel mutations result in null mutations and both alleles must be ablated to functionally disrupt gene expression, the rate of gene disruption may be stochastic and incomplete. Future studies using multiple gRNAs directed against the target gene in the future could trigger both indel mutations and gene truncation to enhance the efficiency of gene ablation.30
In addition to improving genome-editing efficacy, the cell-type specificity of targeting VEGF-A must also be optimized. VEGF is expressed in different retinal cell types including RPE,31 Muller cells,32 endothelial cells,33 astrocytes,34 and ganglion cells,35 which are not all effectively transduced by AAV8.17 We focused on VEGF expression in the RPE by employing subretinal AAV8, which has known tropism for RPE and photoreceptors in mice and nonhuman primates.36, 37, 38 However, the exact VEGF source involved in CNV pathogenesis has not been fully elucidated, and other VEGF sources such as Muller glia have been shown to play important roles in at least some other retinal neovascular conditions and models.32,39,40 Hence, additional studies using cell-type-specific approaches could help probe the pathologic sources of VEGF involved in CNV formation and potentially improve both the efficacy and specificity of CRISPR-based strategies for neovascular eye conditions.
Determining the optimal Cas9 endonuclease to target VEGF is a critical step for clinical translation of CRISPR-based therapies for CNV. Although viral-mediated genome editing has been used with some success to suppress VEGF and CNV,11,12,15,41 direct comparisons of different Cas9 orthologs are lacking. Here, we found SpCas9 demonstrated higher genome-editing rates, greater VEGF suppression, and more effective CNV reduction in vivo than SaCas9. Interestingly, we observed no clear difference between the two Cas9 orthologs using the T7E1 assay, suggesting that in vitro assays may not reliably predict genome-editing efficiency in vivo. Although genome-editing rates in vivo are difficult to quantify due to the small proportion of RPE cells within the RPE-choroid tissues evaluated in our study, our findings are consistent with reports showing superior genomic ablation using SpCas9 compared to SaCas9 and CjCas9 after AAV delivery to eyes of yellow fluorescence protein (YFP)-expressing transgenic mice (F. Li et al., 2019, ARVO, conference).
Though our data suggest SpCas9 exhibit superior genome-editing efficiency and VEGF-A suppression, SaCas9 still has significant translational potential due to its smaller size for AAV packaging (∼1 kb shorter than SpCas9). Previous animal studies have demonstrated effective in vivo editing of other target genes with SaCas9,42, 43, 44, 45 including CEP290 in murine and nonhuman primate retinae.43 Our studies using SaCas9 showed a trend toward VEGF-A suppression in vivo, although it did not reach statistical significance (p = 0.9). Further modifications such as increasing AAV dose or performing injections in multiple areas in the retina may help improve the efficacy of genome editing. Since genome-editing rates vary not only with the Cas9 enzyme but also target gene, gRNA location, and mode of delivery, further optimization for clinical translation is needed.
Although CRISPR-based strategies have been largely studied as a potential therapy for inherited retinal conditions, genome editing is well suited for treating multifactorial conditions such as AMD and CNV, as multiple gRNAs can be paired with a single Cas9 endonuclease to target multiple pro-angiogenic pathways simultaneously. Compared with intravitreal pharmacotherapies, CRISPR components can be genetically encoded to target specific cell types and minimize adverse systemic effects. Despite the potential advantage of providing an effective cure, however, genome manipulations are susceptible to off-target effects, and adverse effects are permanent. Future studies using unbiased, high-throughput off-target detection methods such as GUIDE-seq or DISCOVER-seq may be necessary to fully elucidate the potential risks before translation to human clinical studies.46, 47, 48 Together, our findings provide an important framework for further evaluation of CRISPR technology as a potential therapeutic approach for the management of CNV and other neovascular retinal conditions.
Materials and Methods
gRNA Design and AAV Constructs
gRNAs targeting exon 1 of the mouse VEGF-A gene were selected using gRNA design software (Benchling, San Francisco, CA) based on the predicted gene cleavage efficiency (on-target score) and probability for off-target binding (off-target score) as described previously.16 Off-target loci were predicted using the same software. We selected the two gRNAs with the highest on-target scores for each Cas9 ortholog then subcloned them into the gRNA scaffold of the two respective AAV vector systems. For SpCas9, we replaced the pMecp2 promoter of px551 (Addgene, 60975) and the hSyn promoter of px552 (Addgene, 60958) with a CMV promoter to drive SpCas9 and enhanced GFP in the two AAV vectors, while the VEGF-A targeting sequences were subcloned upstream of the U6-driven gRNA scaffold of px552. For SaCas9, we subcloned the targeting sequences upstream of the gRNA scaffold of px601 (Addgene, 61591). For control AAV constructs, the gRNA scaffold sequence was included, but without the targeting sequences. All plasmids were donated by Feng Zhang lab (Addgene, 60957, 60958, 61591).
Cell Culture, Transfection, and FACS
NIH 3T3 cells maintained in Dulbecco’s Modified Eagle Medium (DMEM) containing 10% fetal calf serum were transfected with the AAV constructs using JetPRIME (Polyplus, 133-06) according to manufacturer’s protocol. In brief, 20 μg of DNA was mixed with 1 mL transfection reagent and added to 1 × 106 cells in a 150-mm dish. After 48 h, incubation and confirmation of GFP or HA-tag expression, FACS was performed at the Flow Cytometry Core Facility at the UC Davis Comprehensive Cancer Center using the Astrios cell sorter (BD Biosciences). We performed live cell sorting for GFP+ cells, whereas HA-tag-expressing cells were fixed, permeabilized, and labeled with fluorescein isothiocyanate (FITC)-conjugated HA-tag antibody (Abcam, ab1208, 1:100) for 30 min at 4°C prior to cell sorting.
T7E1 Mismatch Assay
Frequency of indel mutations in vitro was conducted using a T7 endonuclease I mismatch detection assay. In brief, 200 ng of PCR products amplified from genomic DNA of target regions of the mouse Vegf-a gene were denatured and annealed for heteroduplex formation (95°C to 85°C at 2°C/s and 85°C to 25°C at 0.1°C/s) then incubated with T7 endonuclease I (NEB, E3321) for 15 min at 37°C. After stopping the reaction with 0.25 M EDTA, PCR products were isolated on a 2% Tris-borate-EDTA (TBE) agarose gel for analysis. Indel formation was calculated with the following equation: % indel = 100 × (1 − (1 − Fcut)1/2, where Fcut indicates the fraction of total cleaved DNA.
RPE Isolation, Genomic DNA Extraction, and DNA Sequencing
Six weeks after subretinal injection, RPEs were isolated using an established protocol with minor modification.49 In brief, after enucleation of the eye, anterior chamber, lens, and retina were removed, and the posterior eye cup was incubated with 0.05% trypsin-EDTA for 45 min at 37°C. Followed by gentle shaking of the eye cup to isolate the RPEs from the eye cup, cells pallets were collected for genomic DNA extraction.
Genomic DNA was extracted using either a DNeasy blood and tissue kit (QIAGEN, 69504) or a QIAamp DNA FFPE tissue kit (QIAGEN, 56404) according to manufacturer’s instructions, and the quantity and quality of gDNA were measured using a NanoDrop 2000c device (Thermo Fisher). PCR products were amplified from 100 ng of gDNA with forward and reverse primers using Phusion high-fidelity polymerase (NEB, M0530S) for 35 cycles (98°C for 10 s, 60°C for 30 s, and 72°C for 15 s) then purified with a MiniElute PCR purification kit (QIAGEN, 28004). The primer pairs used in this study are listed in Table S1. Sanger sequencing was performed at the UC Davis DNA sequencing facility. Deep sequencing of 200-bp PCR products of VEGF-A target sites amplified from genomic DNA was conducted at the DNA-sequencing core facility at Massachusetts General Hospital (Boston, MA, USA).
AAV Virus Production
The AAV cis constructs were packaged into AAV8 capsids and purified by the UC Davis Center for Vision Sciences Molecular Construct and Packaging core facility according to the procedures described in Flannery and Visel.50 Viral titers were determined by TaqMan; quantitative PCR and purity was assessed by SDS-polyacrylamide gel electrophoresis.
Animals, Subretinal Injections, and Tissue Collection
All animal experiments were approved and conducted under the guidelines of the Institutional Animal Care and Use Committee (IACUC) at UC Davis. Six- to eight-week-old male C57BL/6J mice (Jackson Laboratory, USA) were kept in a 12-h light/dark cycle room at the animal facility at UC Davis. For subretinal injections, animals were anesthetized with isoflurane inhalation, and eyes were dilated with 1% tropicamide and 2.5% phenylephrine. The eyes were prepared with 5% povidone iodine, followed by creation of a small conjunctival peritomy and a paralimbal sclerotomy using a 30G needle and insertion of a 31G Hamilton syringe to subretinally inject 2 μL of each AAV8 vector (4 × 1011 vg/eye). For the SpCas9 and gRNA dual vector system, viruses were mixed at a 1:1 ratio prior to injection. Six weeks after injection, eyes were enucleated, and RPE-choroid tissues were carefully dissected and homogenized to collect lysates for protein or genomic DNA extraction.
Quantitative Polymerase Chain Reaction (qPCR)
TaqMan qPCR was conducted at the Real-Time PCR Research and Diagnostic Core Facility at UC Davis. Each 10-μL reaction consists of 10 ng genomic DNA extracted from RPE-choroidal tissues, TaqMan Universal PCR master mix (Life Technologies, 4318157), 0.25 μM of probe for AAV ITR (5′-CACTCCCTCTCTGCGCGCTCG-3′), and 0.9 μM forward and reverse primers. We also used a TaqMan copy number reference assay (Applied Biosystems, 4458368) labeled with 2′-chloro-7′phenyl-1,4-dichloro-6-carboxy-fluorescein (VIC) fluorescence. FAM and VIC fluorescence were read every cycle, and AAV genomic copy numbers per mouse diploid genome (vg/dg) were calculated from standard curves.
Enzyme-Linked Immunosorbent Assay (ELISA)
Mouse VEGF-A Quantikine ELISA kits (Millipore, MMV00) were used to quantify VEGF-A protein according to the manufacturer’s guidelines. In brief, proteins were incubated on a plate coated with polyclonal antibodies against mouse VEGF-A protein for 2 h followed by horseradish peroxidase conjugate incubation. After washing the plate, substrate was applied and the plate was read at 450 nm with wavelength correction to 570 nm. Quantification of VEGF-A was calculated against a standard curve and normalized with total protein. Because the dual-vector SpCas9 system employs twice the amount of viral particles as the single-vector SaCas9 system, comparisons were made with each Cas9 ortholog and empty corresponding gRNA scaffolds (SpCas9-empty or SaCas9-empty) as controls.
Laser-Induced CNV and FA
Animals were anesthetized with isoflurane inhalation, and the pupils were dilated with 1% tropicamide and 5% phenylephrine. Mice were imaged using the MicronIV system (Phoenix, Pleasanton, CA, USA), with topical artificial tear gel (Genteal) applied to prevent corneal damage. Image-guided laser photocoagulation was performed with a 532-nm wavelength laser at 250 mW for 70 ms. CNV was evaluated with FA, OCT, and OCT-A 1 week after laser photocoagulation, and eyes with severe hemorrhage or where CNV lesion size could not be accurately assessed were excluded from analyses.
Scanning Laser Ophthalmoscopy (SLO) and OCT Imaging
SLO and OCT imaging were performed using a custom-built multimodal OCT and SLO mouse retinal imaging system developed at the small animal ocular imaging facility (EyePod) at UC Davis as previously reported.51 One week after inducing CNV, animals were anesthetized with isoflurane inhalation, and the pupils were dilated with 1% tropicamide and 2.5% phenylephrine prior to imaging. SLO and OCT images were acquired simultaneously with a SLO excitation laser (ORBIS 488LX, Coherent, Santa Clara, CA, USA) running at 488 nm with 100 μW at the mouse pupil and OCT with superluminescent diodes (SLD) light source (Broadlighter 860; Superlum Diodes, Cork, Ireland) operating at 860 nm with full width at half maximum (FWHM) of 132 nm and 600 μW at the mouse pupil, respectively. For SLO, reflectance and fluorescence images were averaged from 50 consecutive images. For OCT and OCT-A, 1080 horizontal B-scans acquired with a series of three multiple B- scans (BM-scans) per one location (spanning 540 × 360 pixels) corresponding to the SLO images were captured for three-dimensional cross-sectional visualization and co-registration. En-face depth average projections of the phase variance signal were used to generate OCT-A images. CNV size was measured by Fiji image-processing software (NIH).
Fluorescence Immunohistochemistry
Frozen sections were collected for immunofluorescence labeling as described previously.52 In brief, eyes cups were fixed with 4% paraformaldehyde (PFA) for 1 h and incubated in 30% sucrose in PBS overnight. Tissues were embedded the next day and sectioned at 12 μm thickness. After blocking with normal donkey serum, tissues were incubated with anti-GFP (Novusbio; NB100-1770) or anti-HA-tag (Cell Signaling; 3724) primary antibodies for either 2 h at room temperature or overnight at 4°C, followed by staining with Alexa Fluor 488 or 568-conjugated secondary antibodies and 4′,6-diamidino-2-phenylindole (DAPI). For measuring CNV size on flat-mount immunohistochemistry, eye cups were fixed with 4% PFA for 1 h and washed with PBS. Retinae were carefully removed, and the remaining RPE-choroid and scleral tissues were radially cut and labeled with Alexa Fluor 488-conjugated isolectin B4 (Invitrogen, I21411) overnight at 4°C. Fluorescence immunohistochemical images were captured with confocal microscopy (Olympus FV1000). CNV size was measured using Fiji image processing software (NIH).
Statistical Analysis
All data and plots were presented as mean ± SEM unless otherwise stated. Comparisons between groups were made using a two-tailed Student’s t test. p < 0.05 was considered statistically significant.
Author Contributions
G.Y. conceived of the study, and S.H.C. and G.Y. designed the experiments. S.H.C. performed the subretinal injections, laser-induced CNV, and all PCR and sequencing studies. I.N.M., U.N., A.N., N.S., E.T., E.L.N., and J.F assisted with immunohistochemistry. R.K.M., P.Z, and R.Z. assisted with EyePod imaging. G.Y. and S.H.C. analyzed data and wrote the manuscript; S.H.C. and G.Y. critically edited the manuscript.
Conflicts of Interest
G.Y. received research support from Alcon, Clearside Biomedical, and Iridex and personal fees for consultancy from Alimera, Allergan, Carl Zeiss Meditec, Genentech, Iridex, and Verily, all of which are unrelated to the contents of this study. The other authors declare no competing interests.
Acknowledgments
This study was supported by NIH (Bethesda, MD) K08 EY026101, R21 EY031108, and R01 026556; the E. Matilda Ziegler Foundation for the Blind (Darien, CT); the Barr Foundation for Retinal Research (Sacramento, CA); the ARVO Foundation (Rockville, MD); the Alcon Research Institute (Geneva, Switzerland); and the Macula Society (Beachwood, OH). Histological and ultrastructural studies were conducted at the Center for Vision Sciences (CVS) Structure-Function core facility; AAV production was conducted at the CVS Molecular Packaging and Construct core facility, and in vivo mouse imaging was conducted at the CVS “EyePod” Small Animal Ocular Imaging Laboratory, which are supported by NIH P30 EY012576. No funding organizations had any role in the design or conduct of this research. The content is solely the responsibility of the authors and does not necessarily represent the official views of the funding agencies.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtm.2020.01.006.
Supplemental Information
References
- 1.Rosenfeld P.J., Moshfeghi A.A., Puliafito C.A. Optical coherence tomography findings after an intravitreal injection of bevacizumab (avastin) for neovascular age-related macular degeneration. Ophthalmic Surg. Lasers Imaging. 2005;36:331–335. [PubMed] [Google Scholar]
- 2.Dugel P.U. Ranibizumab treatment of patients with ocular diseases. Int. Ophthalmol. Clin. 2006;46:131–140. doi: 10.1097/01.iio.0000212142.15794.30. [DOI] [PubMed] [Google Scholar]
- 3.Rosenfeld P.J., Heier J.S., Hantsbarger G., Shams N. Tolerability and efficacy of multiple escalating doses of ranibizumab (Lucentis) for neovascular age-related macular degeneration. Ophthalmology. 2006;113:623.e1. doi: 10.1016/j.ophtha.2006.01.027. [DOI] [PubMed] [Google Scholar]
- 4.Todorich B., Yiu G., Hahn P. Current and investigational pharmacotherapeutic approaches for modulating retinal angiogenesis. Expert Rev. Clin. Pharmacol. 2014;7:375–391. doi: 10.1586/17512433.2014.890047. [DOI] [PubMed] [Google Scholar]
- 5.Ellis M.P., Lent-Schochet D., Lo T., Yiu G. Emerging Concepts in the Treatment of Diabetic Retinopathy. Curr. Diab. Rep. 2019;19:137. doi: 10.1007/s11892-019-1276-5. [DOI] [PubMed] [Google Scholar]
- 6.Constable I.J., Pierce C.M., Lai C.M., Magno A.L., Degli-Esposti M.A., French M.A., McAllister I.L., Butler S., Barone S.B., Schwartz S.D. Phase 2a Randomized Clinical Trial: Safety and Post Hoc Analysis of Subretinal rAAV.sFLT-1 for Wet Age-related Macular Degeneration. EBioMedicine. 2016;14:168–175. doi: 10.1016/j.ebiom.2016.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cai B., Sun S., Li Z., Zhang X., Ke Y., Yang J., Li X. Application of CRISPR/Cas9 technologies combined with iPSCs in the study and treatment of retinal degenerative diseases. Hum. Genet. 2018;137:679–688. doi: 10.1007/s00439-018-1933-9. [DOI] [PubMed] [Google Scholar]
- 8.Hung S.S., Chrysostomou V., Li F., Lim J.K., Wang J.H., Powell J.E., Tu L., Daniszewski M., Lo C., Wong R.C. AAV-Mediated CRISPR/Cas Gene Editing of Retinal Cells In Vivo. Invest. Ophthalmol. Vis. Sci. 2016;57:3470–3476. doi: 10.1167/iovs.16-19316. [DOI] [PubMed] [Google Scholar]
- 9.Chew W.L., Tabebordbar M., Cheng J.K., Mali P., Wu E.Y., Ng A.H., Zhu K., Wagers A.J., Church G.M. A multifunctional AAV-CRISPR-Cas9 and its host response. Nat. Methods. 2016;13:868–874. doi: 10.1038/nmeth.3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Friedland A.E., Baral R., Singhal P., Loveluck K., Shen S., Sanchez M., Marco E., Gotta G.M., Maeder M.L., Kennedy E.M. Characterization of Staphylococcus aureus Cas9: a smaller Cas9 for all-in-one adeno-associated virus delivery and paired nickase applications. Genome Biol. 2015;16:257. doi: 10.1186/s13059-015-0817-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim E., Koo T., Park S.W., Kim D., Kim K., Cho H.Y., Song D.W., Lee K.J., Jung M.H., Kim S. In vivo genome editing with a small Cas9 orthologue derived from Campylobacter jejuni. Nat. Commun. 2017;8:14500. doi: 10.1038/ncomms14500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holmgaard A., Askou A.L., Benckendorff J.N.E., Thomsen E.A., Cai Y., Bek T., Mikkelsen J.G., Corydon T.J. In Vivo Knockout of the Vegfa Gene by Lentiviral Delivery of CRISPR/Cas9 in Mouse Retinal Pigment Epithelium Cells. Mol. Ther. Nucleic Acids. 2017;9:89–99. doi: 10.1016/j.omtn.2017.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang X., Zhou G., Wu W., Ma G., D’Amore P.A., Mukai S., Lei H. Editing VEGFR2 Blocks VEGF-Induced Activation of Akt and Tube Formation. Invest. Ophthalmol. Vis. Sci. 2017;58:1228–1236. doi: 10.1167/iovs.16-20537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim K., Park S.W., Kim J.H., Lee S.H., Kim D., Koo T., Kim K.E., Kim J.H., Kim J.S. Genome surgery using Cas9 ribonucleoproteins for the treatment of age-related macular degeneration. Genome Res. 2017;27:419–426. doi: 10.1101/gr.219089.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang X., Zhou G., Wu W., Duan Y., Ma G., Song J., Xiao R., Vandenberghe L., Zhang F., D’Amore P.A., Lei H. Genome editing abrogates angiogenesis in vivo. Nat. Commun. 2017;8:112. doi: 10.1038/s41467-017-00140-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yiu G., Tieu E., Nguyen A.T., Wong B., Smit-McBride Z. Genomic Disruption of VEGF-A Expression in Human Retinal Pigment Epithelial Cells Using CRISPR-Cas9 Endonuclease. Invest. Ophthalmol. Vis. Sci. 2016;57:5490–5497. doi: 10.1167/iovs.16-20296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Igarashi T., Miyake K., Asakawa N., Miyake N., Shimada T., Takahashi H. Direct comparison of administration routes for AAV8-mediated ocular gene therapy. Curr. Eye Res. 2013;38:569–577. doi: 10.3109/02713683.2013.779720. [DOI] [PubMed] [Google Scholar]
- 18.Xiong W., Cepko C. Distinct Expression Patterns of AAV8 Vectors with Broadly Active Promoters from Subretinal Injections of Neonatal Mouse Eyes at Two Different Ages. Adv. Exp. Med. Biol. 2016;854:501–507. doi: 10.1007/978-3-319-17121-0_67. [DOI] [PubMed] [Google Scholar]
- 19.Nakamura K., Oshima T., Morimoto T., Ikeda S., Yoshikawa H., Shiwa Y., Ishikawa S., Linak M.C., Hirai A., Takahashi H. Sequence-specific error profile of Illumina sequencers. Nucleic Acids Res. 2011;39:e90. doi: 10.1093/nar/gkr344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen Y.C., Liu T., Yu C.H., Chiang T.Y., Hwang C.C. Effects of GC bias in next-generation-sequencing data on de novo genome assembly. PLoS ONE. 2013;8:e62856. doi: 10.1371/journal.pone.0062856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heier J.S., Kherani S., Desai S., Dugel P., Kaushal S., Cheng S.H., Delacono C., Purvis A., Richards S., Le-Halpere A. Intravitreous injection of AAV2-sFLT01 in patients with advanced neovascular age-related macular degeneration: a phase 1, open-label trial. Lancet. 2017;390:50–61. doi: 10.1016/S0140-6736(17)30979-0. [DOI] [PubMed] [Google Scholar]
- 22.Saishin Y., Saishin Y., Takahashi K., Lima e Silva R., Hylton D., Rudge J.S., Wiegand S.J., Campochiaro P.A. VEGF-TRAP(R1R2) suppresses choroidal neovascularization and VEGF-induced breakdown of the blood-retinal barrier. J. Cell. Physiol. 2003;195:241–248. doi: 10.1002/jcp.10246. [DOI] [PubMed] [Google Scholar]
- 23.Grishanin R., Vuillemenot B., Sharma P., Keravala A., Greengard J., Gelfman C., Blumenkrantz M., Lawrence M., Hu W., Kiss S., Gasmi M. Preclinical Evaluation of ADVM-022, a Novel Gene Therapy Approach to Treating Wet Age-Related Macular Degeneration. Mol. Ther. 2019;27:118–129. doi: 10.1016/j.ymthe.2018.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Campochiaro P.A., Marcus D.M., Awh C.C., Regillo C., Adamis A.P., Bantseev V., Chiang Y., Ehrlich J.S., Erickson S., Hanley W.D. The Port Delivery System with Ranibizumab for Neovascular Age-Related Macular Degeneration: Results from the Randomized Phase 2 Ladder Clinical Trial. Ophthalmology. 2019;126:1141–1154. doi: 10.1016/j.ophtha.2019.03.036. [DOI] [PubMed] [Google Scholar]
- 25.Kurihara T., Westenskow P.D., Bravo S., Aguilar E., Friedlander M. Targeted deletion of Vegfa in adult mice induces vision loss. J. Clin. Invest. 2012;122:4213–4217. doi: 10.1172/JCI65157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saint-Geniez M., Maldonado A.E., D’Amore P.A. VEGF expression and receptor activation in the choroid during development and in the adult. Invest. Ophthalmol. Vis. Sci. 2006;47:3135–3142. doi: 10.1167/iovs.05-1229. [DOI] [PubMed] [Google Scholar]
- 27.Grunwald J.E., Pistilli M., Ying G.S., Maguire M.G., Daniel E., Martin D.F., Comparison of Age-related Macular Degeneration Treatments Trials Research Group Growth of geographic atrophy in the comparison of age-related macular degeneration treatments trials. Ophthalmology. 2015;122:809–816. doi: 10.1016/j.ophtha.2014.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grunwald J.E., Daniel E., Huang J., Ying G.S., Maguire M.G., Toth C.A., Jaffe G.J., Fine S.L., Blodi B., Klein M.L., CATT Research Group Risk of geographic atrophy in the comparison of age-related macular degeneration treatments trials. Ophthalmology. 2014;121:150–161. doi: 10.1016/j.ophtha.2013.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yiu G., Manjunath V., Chiu S.J., Farsiu S., Mahmoud T.H. Effect of anti-vascular endothelial growth factor therapy on choroidal thickness in diabetic macular edema. Am. J. Ophthalmol. 2014;158:745–751.e2. doi: 10.1016/j.ajo.2014.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tsai Y.T., Wu W.H., Lee T.T., Wu W.P., Xu C.L., Park K.S., Cui X., Justus S., Lin C.S., Jauregui R. Clustered Regularly Interspaced Short Palindromic Repeats-Based Genome Surgery for the Treatment of Autosomal Dominant Retinitis Pigmentosa. Ophthalmology. 2018;125:1421–1430. doi: 10.1016/j.ophtha.2018.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller J.W., Adamis A.P., Aiello L.P. Vascular endothelial growth factor in ocular neovascularization and proliferative diabetic retinopathy. Diabetes Metab. Rev. 1997;13:37–50. doi: 10.1002/(sici)1099-0895(199703)13:1<37::aid-dmr174>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 32.Pierce E.A., Avery R.L., Foley E.D., Aiello L.P., Smith L.E. Vascular endothelial growth factor/vascular permeability factor expression in a mouse model of retinal neovascularization. Proc. Natl. Acad. Sci. USA. 1995;92:905–909. doi: 10.1073/pnas.92.3.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aiello L.P., Northrup J.M., Keyt B.A., Takagi H., Iwamoto M.A. Hypoxic regulation of vascular endothelial growth factor in retinal cells. Arch. Ophthalmol. 1995;113:1538–1544. doi: 10.1001/archopht.1995.01100120068012. [DOI] [PubMed] [Google Scholar]
- 34.Stone J., Itin A., Alon T., Pe’er J., Gnessin H., Chan-Ling T., Keshet E. Development of retinal vasculature is mediated by hypoxia-induced vascular endothelial growth factor (VEGF) expression by neuroglia. J. Neurosci. 1995;15:4738–4747. doi: 10.1523/JNEUROSCI.15-07-04738.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stone J., Chan-Ling T., Pe’er J., Itin A., Gnessin H., Keshet E. Roles of vascular endothelial growth factor and astrocyte degeneration in the genesis of retinopathy of prematurity. Invest. Ophthalmol. Vis. Sci. 1996;37:290–299. [PubMed] [Google Scholar]
- 36.Vandenberghe L.H., Bell P., Maguire A.M., Cearley C.N., Xiao R., Calcedo R., Wang L., Castle M.J., Maguire A.C., Grant R. Dosage thresholds for AAV2 and AAV8 photoreceptor gene therapy in monkey. Sci. Transl. Med. 2011;3:88ra54. doi: 10.1126/scitranslmed.3002103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seitz I.P., Michalakis S., Wilhelm B., Reichel F.F., Ochakovski G.A., Zrenner E., Ueffing M., Biel M., Wissinger B., Bartz-Schmidt K.U., RD-CURE Consortium Superior Retinal Gene Transfer and Biodistribution Profile of Subretinal Versus Intravitreal Delivery of AAV8 in Nonhuman Primates. Invest. Ophthalmol. Vis. Sci. 2017;58:5792–5801. doi: 10.1167/iovs.17-22473. [DOI] [PubMed] [Google Scholar]
- 38.Natkunarajah M., Trittibach P., McIntosh J., Duran Y., Barker S.E., Smith A.J., Nathwani A.C., Ali R.R. Assessment of ocular transduction using single-stranded and self-complementary recombinant adeno-associated virus serotype 2/8. Gene Ther. 2008;15:463–467. doi: 10.1038/sj.gt.3303074. [DOI] [PubMed] [Google Scholar]
- 39.Jiang Y., Wang H., Culp D., Yang Z., Fotheringham L., Flannery J., Hammond S., Kafri T., Hartnett M.E. Targeting Müller cell-derived VEGF164 to reduce intravitreal neovascularization in the rat model of retinopathy of prematurity. Invest. Ophthalmol. Vis. Sci. 2014;55:824–831. doi: 10.1167/iovs.13-13755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang J., Xu X., Elliott M.H., Zhu M., Le Y.Z. Müller cell-derived VEGF is essential for diabetes-induced retinal inflammation and vascular leakage. Diabetes. 2010;59:2297–2305. doi: 10.2337/db09-1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Koo T., Park S.W., Jo D.H., Kim D., Kim J.H., Cho H.Y., Kim J., Kim J.H., Kim J.S. CRISPR-LbCpf1 prevents choroidal neovascularization in a mouse model of age-related macular degeneration. Nat. Commun. 2018;9:1855. doi: 10.1038/s41467-018-04175-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gaj T., Ojala D.S., Ekman F.K., Byrne L.C., Limsirichai P., Schaffer D.V. In vivo genome editing improves motor function and extends survival in a mouse model of ALS. Sci. Adv. 2017;3 doi: 10.1126/sciadv.aar3952. eaar3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maeder M.L., Stefanidakis M., Wilson C.J., Baral R., Barrera L.A., Bounoutas G.S., Bumcrot D., Chao H., Ciulla D.M., DaSilva J.A. Development of a gene-editing approach to restore vision loss in Leber congenital amaurosis type 10. Nat. Med. 2019;25:229–233. doi: 10.1038/s41591-018-0327-9. [DOI] [PubMed] [Google Scholar]
- 44.Ran F.A., Cong L., Yan W.X., Scott D.A., Gootenberg J.S., Kriz A.J., Zetsche B., Shalem O., Wu X., Makarova K.S. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015;520:186–191. doi: 10.1038/nature14299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yin C., Zhang T., Qu X., Zhang Y., Putatunda R., Xiao X., Li F., Xiao W., Zhao H., Dai S. In Vivo Excision of HIV-1 Provirus by saCas9 and Multiplex Single-Guide RNAs in Animal Models. Mol. Ther. 2017;25:1168–1186. doi: 10.1016/j.ymthe.2017.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smith C., Gore A., Yan W., Abalde-Atristain L., Li Z., He C., Wang Y., Brodsky R.A., Zhang K., Cheng L., Ye Z. Whole-genome sequencing analysis reveals high specificity of CRISPR/Cas9 and TALEN-based genome editing in human iPSCs. Cell Stem Cell. 2014;15:12–13. doi: 10.1016/j.stem.2014.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tsai S.Q., Zheng Z., Nguyen N.T., Liebers M., Topkar V.V., Thapar V., Wyvekens N., Khayter C., Iafrate A.J., Le L.P. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat. Biotechnol. 2015;33:187–197. doi: 10.1038/nbt.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wienert B., Wyman S.K., Richardson C.D., Yeh C.D., Akcakaya P., Porritt M.J., Morlock M., Vu J.T., Kazane K.R., Watry H.L. Unbiased detection of CRISPR off-targets in vivo using DISCOVER-Seq. Science. 2019;364:286–289. doi: 10.1126/science.aav9023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fernandez-Godino R., Garland D.L., Pierce E.A. Isolation, culture and characterization of primary mouse RPE cells. Nat. Protoc. 2016;11:1206–1218. doi: 10.1038/nprot.2016.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Flannery J.G., Visel M. Adeno-associated viral vectors for gene therapy of inherited retinal degenerations. Methods Mol. Biol. 2013;935:351–369. doi: 10.1007/978-1-62703-080-9_25. [DOI] [PubMed] [Google Scholar]
- 51.Zhang P., Zam A., Jian Y., Wang X., Li Y., Lam K.S., Burns M.E., Sarunic M.V., Pugh E.N., Jr., Zawadzki R.J. In vivo wide-field multispectral scanning laser ophthalmoscopy-optical coherence tomography mouse retinal imager: longitudinal imaging of ganglion cells, microglia, and Müller glia, and mapping of the mouse retinal and choroidal vasculature. J. Biomed. Opt. 2015;20:126005. doi: 10.1117/1.JBO.20.12.126005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chung S.H., Gillies M., Yam M., Wang Y., Shen W. Differential expression of microRNAs in retinal vasculopathy caused by selective Müller cell disruption. Sci. Rep. 2016;6:28993. doi: 10.1038/srep28993. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.