Abstract
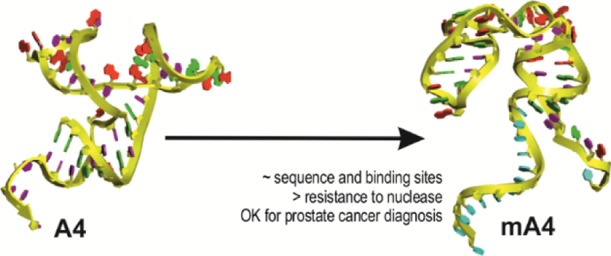
The RNA aptamer A4 binds specifically to tumor prostate cells. A4 was modified (mA4) by adding deoxyribonucleotides to its ends to remove the reactive 2′ hydroxyl groups of RNA’s sugar at the ends of the aptamer and to make it more stable to widespread RNase contamination in laboratories. Thus, mA4 would be more suitable to use in the clinical settings of prostate cancer (PCa). We aimed to characterize this optimized oligonucleotide to verify its potential as a diagnostic tool. The sequences and structures of A4 and mA4 were compared through in silico approaches to corroborate their similarity. Then, the degradation of mA4 was measured in appropriate media and human plasma for in vitro tests. In addition, the binding abilities of A4 to prostate cells were contrasted with those of mA4. The effects of mA4 were assessed on the viability, proliferation, and migration of human prostate cell lines RWPE-1 and PC-3 in three-dimensional (3D) cell cultures. mA4 showed configurational motifs similar to those of A4, displayed a half-life in plasma substantially higher than A4, and exhibited a comparable binding capacity to that of A4 and unaltered viability, proliferation, and migration of prostatic cells. Therefore, mA4 maintains the crucial 3D structures of A4 that would allow binding to its target, as suggested by in silico and binding analyses. mA4 may be a good PCa reporter as it does not change cellular parameters of prostate cells when incubated with it. Its additional deoxyribonucleotides make mA4 inherently more chemically stable than A4, avoiding its degradation and favoring its storage and handling for clinical applications. These characteristics support the potential of mA4 to be used in diagnostic systems for PCa.
1. Introduction
According to the Surveillance, Epidemiology, and End Results (SEER) Program from the National Cancer Institute in the United States, patients with metastatic stage of prostate cancer (PCa) diagnosis showed a five-year relative survival of only 30.5%, while patients with localized or regional lymph node stage at diagnosis had a five-year relative survival of 100% between 2009 and 2015.1 Consequently, the substantial gap between these two populations of patients suggests that the stage of PCa at diagnosis is crucial for patient survival rates and earlier diagnoses and treatments could improve them.2
PCa is screened by prostate-specific antigen (PSA) blood test, sometimes complemented by digital rectal examination. After a positive result, PCa is confirmed by histopathological examination of a biopsy.3 In the cases of negative ultrasound-guided biopsy in men with a high probability of sampling error, most urologists use prostate multiparametric magnetic resonance imaging (MRI) for PCa diagnosis.4 However, conflicting recommendations for the use of PSA in the clinical management of PCa5,6 evidenced that PSA is an unspecific biomarker with questionable utility in some cases.
On the one hand, high levels of PSA may not indicate PCa. In fact, PSA is produced by other glands, under other conditions and found even in females.7 Thus, there is a high chance of false-positive results with up to 67% of overdiagnosed and overtreated patients.8 On the other hand, about a quarter of men with low PSA levels have PCa and are underdiagnosed.9 In addition, PSA testing does not avoid the biopsy procedure for diagnosis, which is also susceptible of false-negatives because of sampling errors.10 These facts support the re-assessment of PSA and the search for more accurate and definitive biomarkers for the diagnosis and monitoring of PCa.
The approaches that analyze tumor components released in body fluids are called liquid biopsies.11 These liquid biopsies represent the current state of a cancer in an individual and their sampling is less invasive than surgical biopsies.12 Among other constituents, tumors release cells that may be detected in blood. These cells are known as circulating tumor cells (CTCs). The expression of surface markers on CTCs and their quantification have already been studied as factors for the clinical management of several cancers.13 For PCa, CTCs in blood samples are the most researched target in liquid biopsies.14
In this view, our research group used the prostate cell lines PC-3 and RWPE-1 as positive and negative targets, respectively, to select ligands of PCa CTCs by the systematic evolution of ligands by exponential enrichment in three-dimensional cell cultures (3D Cell-SELEX).15 This technology yielded several RNA aptamers that bind specifically to PC-3 cells. The A4 aptamer was one of the selected aptamers which showed a small size and a low free energy according to the minimum free energy (MFE) structure by the Sfold web server.16 Owing to these desirable characteristics in aptamers, A4 was further characterized, showing a high affinity for PC-3 cells.15 Therefore, we theorized that A4 may be used as a diagnostic tool which detects PCa CTCs in blood samples from patients.
However, factors and enzymes that degrade RNAs are highly efficient, as they show complementary and frequently reiterative mechanisms. In fact, RNAs may pose a great threat to the humans. For example, noncoding RNAs may interfere in the regulation of gene expression and viral RNAs may replicate to infect host cells.17 RNA aptamers are chemically more unstable than DNA aptamers because of their 2′ hydroxyl group that makes them more susceptible to hydrolysis and ribonuclease action.18,19
However, wobble base pairs in RNA aptamers, such as guanosine–uridine bonds, allow for more complex 3D structures, when compared to DNA structures.20 For these reasons, we maintained the unique sequence of the original RNA aptamer with its more intricate design and we added DNA ends with their higher chemical stability to a new version of A4 called mA4. The rest of the original RNA aptamer corresponds to sequences complementary to the primers used in the selection process that are identical in the other aptamers. At the 5′ end, we changed the sugar of nucleotides 1 to 14 to deoxyribose to improve RNase resistance and added a thymine spacer of 13 nucleotides. This spacer is important to avoid potential steric hindrance because of the interaction of the coupled biotin with labeled avidin molecules and allow target recognition.21 At the 3′ end, we changed the sugar of nucleotides 44 to 46 to deoxyribose and replaced the uracil base 46 with thymine, a canonical base in DNA, again to improve RNase resistance.
Thus, we combined the advantages of the two types of nucleotides in mA4, with the intention of reducing its susceptibility to degradation without perturbing its putative binding motifs. In the present work, we use in silico approaches to verify the similar composition and structure of A4 and mA4 and in vitro approaches to compare their resistance to nuclease in plasma and cell culture media. Enzyme-linked apta-sorbent assay (ELASA) is also used to prove whether mA4 maintains the binding profile of A4.
In addition to being specific and resistant to nuclease, nucleotide-based probes should not be prone to induce undesirable biological changes in order to be implemented in theranostic applications. Accordingly, we also characterize the biological effects of mA4 on the viability, proliferation, and migration of human prostate cell lines used to select it. The PC-3 cell line was isolated from bone marrow and it displays an androgen-independent growth, being considered a good model of advanced prostate adenocarcinoma22 and later of prostatic small cell carcinoma.23 Differently, RWPE-1 cell line was developed from the immortalization of nontumorigenic prostatic epithelial cells with human papilloma virus 18 representing a benign prostatic epithelium, which is androgen-sensitive.24
With these human cell lines, we can delimit the potential clinical uses of mA4 in the management of PCa patients. Thus, we hypothesize that mA4, the optimized version of A4, can also be used as a diagnostic tool for PCa.
2. Results and Discussion
2.1. Sequence of mA4 is Similar to That of A4
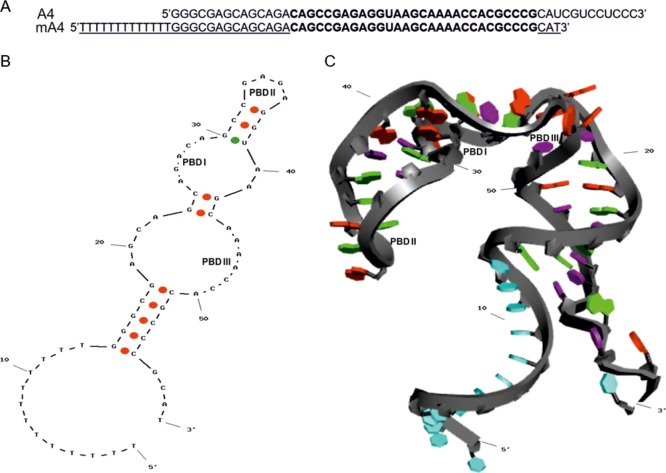
mA4 maintained the specific RNA sequence of A4 without the common flanking sequences for amplification and with additional random deoxyribonucleotides at both ends and a poly-T linker at the 5′ end for biotin coupling. The BLAST25 algorithm showed a good alignment between the all-DNA versions of the mA4 and A4 aptamers (Figure 1A). This alignment displayed an expect value of 3 × 10–23 and a score match of 86.1, with an 83% of cover and 100% of identity between the 46 common nucleotides. Consequently, mA4 has a similar sequence to that of A4, with a high-scoring segment. Most of the nucleotides of the two sequences were identical or homolog. As homolog nucleotides are able to maintain the function in aptamers with the same sequence, but different types of sugar in their nucleotides,26 mA4 may retain the binding abilities and affinities of A4.
Figure 1.
In silico similarity of mA4 to A4. (A) Alignment of mA4 and A4 aptamers by BLAST algorithm.25 Deoxyribonucleotides are represented underlined and identical ribonucleotides between the two aptamers are in bold. (B) Putative 2D structure of mA4 obtained using Mfold server. The green dot indicates a common base pair between the models for the prediction of RNA structures, MFE structure, and centroid structure. The red dots indicate base pairs only in the centroid structure. (C) 3D structure of mA4 based on its 2D structure, obtained using RNA Composer server and modified by Discovery Studio Visualizer software, v19.1.0.18287. Nucleobases in light blue indicate thymines, in red indicate adenines, in purple indicate cytosines and uridines, and in green indicate guanines. Potential binding domains are PBD I, PBD II, and PBD III.
2.2. In Silico Prediction of mA4 Folding Displays Potential Binding Structures
The most probable secondary structure of mA4 showed two single strands at the termini, three helical stems with two asymmetrical internal loops, and a hairpin tetraloop closed by a guanosine–uridine wobble base pair (Figure 1B). This structure displayed the lowest free energy (dG = −16.784 kcal/mol) with a GC content of 49.15%, which increased to 62.07% in the RNA region, and a melting temperature of 73.6 °C. Compared to A4,15 mA4 showed a free energy only 0.8 kcal/mol higher, one less weak guanosine–uridine bond, a high percentage of guanosine and cytosine bases, and a melting temperature 3.1 °C lower than that of A4. Therefore, mA4 is slightly more unstable than A4, but considering that its thymine spacer is the main difference between them and that it makes the mA4 sequence less stable, we ponder that mA4 still is a stable single-stranded oligonucleotide.
The longest strings of the two asymmetrical internal loops and the hairpin loop of mA4 share their RNA sequences and consensus motifs with conformations found in the secondary structure of A4.15 Because secondary structures of aptamers, such as hairpins, bulges, pseudoknots, G-quadruplexes, and so forth, provide information about relevant regions of binding because they establish noncovalent interactions with the target molecule,27,28 we defined three potential binding domains from the in silico prediction of mA4 folding. Potential binding domain I (PBD I) consists of nucleotides 25–29, PBD II consists of nucleotides 33–36, and PBD III consists of nucleotides 44–50 (Figure 1B). The mA4 tertiary structure also shows these three putative binding sites among the three helical stems (Figure 1C).These common architectural motifs are crucial, as 3D shapes are responsible for high-affinity noncovalent bindings between aptamers and their target molecules.29 Consequently, the 59-mer aptamer (mA4) would be able to bind to PC-3 cells in a similar way to that of the original 55-mer aptamer (A4).
2.3. mA4 Shows Resistance to Nucleases under the Tested Conditions
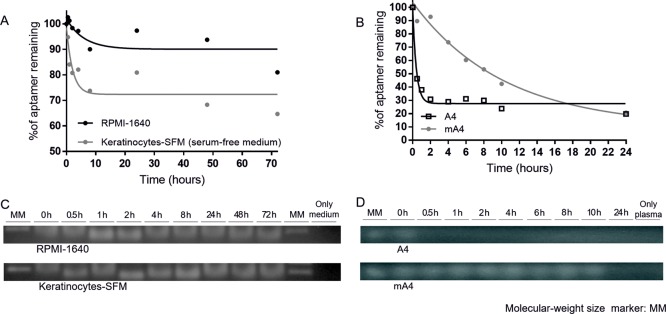
Prior to the biological assays, the in vitro stability of mA4 to nucleases was evaluated for 72 h in the RPMI-1640 medium and Keratinocyte serum-free medium (SFM) used to culture human tumor prostate PC-3 cells and human nontumor prostate RWPE-1 cells, respectively. Over this period, its half-life was 4.1 h in RPMI-1640 medium and 1.5 h in Keratinocyte-SFM, showing time-dependent degradation profiles (Figure 2A). Moreover, the fluorescence of ethidium bromide intercalated into the nucleotides of mA4 was conserved under the two conditions analyzed in the agarose gel (Figure 2C). This fact shows that a portion of mA4 remained undegraded 72 h after its incubation. The degradation of mA4 in Keratinocyte-SFM was faster than that in RPMI-1640. However, this apparent higher resistance to nuclease in RPMI-1640 may be put down to the differences in the nuclease profiles of the media30 rather than to conformation changes in the aptamer. In addition, mA4 was coupled with biotin, which would further protect it from nucleases.31
Figure 2.
Degradation profiles of mA4 and A4 in cell culture media and human plasma. (A) Nonlinear one-phase decay model to calculate the mA4 half-lives in media RPMI-1640 and Keratinocyte-SFM. (B) Nonlinear one-phase decay model to calculate the A4 and mA4 half-lives in human plasma. Values represent the mean of three independent experiments. (C) Representative images of the detection of mA4 incubated in the corresponding media for 72 h. (D) Representative images of the detection of A4 and mA4 incubated in human plasma for 24 h. Bands stained by ethidium bromide in 2% agarose gels (cropped gels). MM showing the band of 100 base pairs.
mA4 and A4 were also incubated in human blood plasma to test and compare their resistance to nucleases present in this biological fluid. mA4 demonstrated to be almost 25-fold more resistant to nuclease action than A4 (Figure 2B), with a half-life of 7.1 h, compared to the half-life of A4 which was only 0.3 h (18 min) in human plasma. It is remarkable that the staining profile of A4 fades after 0.5 h (Figure 2D).
Our results are similar to others comparing RNA and DNA aptamers, as they are in the same order of magnitude. For example, comparing anti-thrombin aptamers in human plasma, DNA aptamers were degraded after 7 h, while RNA aptamers remained only 10 min.32 Apart from the intrinsic higher resistance of DNA to blood plasma nucleases, the anticoagulant ethylenediamine tetraacetic acid (EDTA) used in the tubes for blood collection inhibits the action of endogenous DNases,33 favoring longer half-lives of present DNAs, including DNA aptamers incubated in blood plasma. The higher resistance to nuclease of mA4 makes it a better candidate for a future clinical diagnosis system with blood plasma as a sample used to detect PCa by liquid biopsy.
2.4. mA4 Binds Specifically to PC-3 Cells
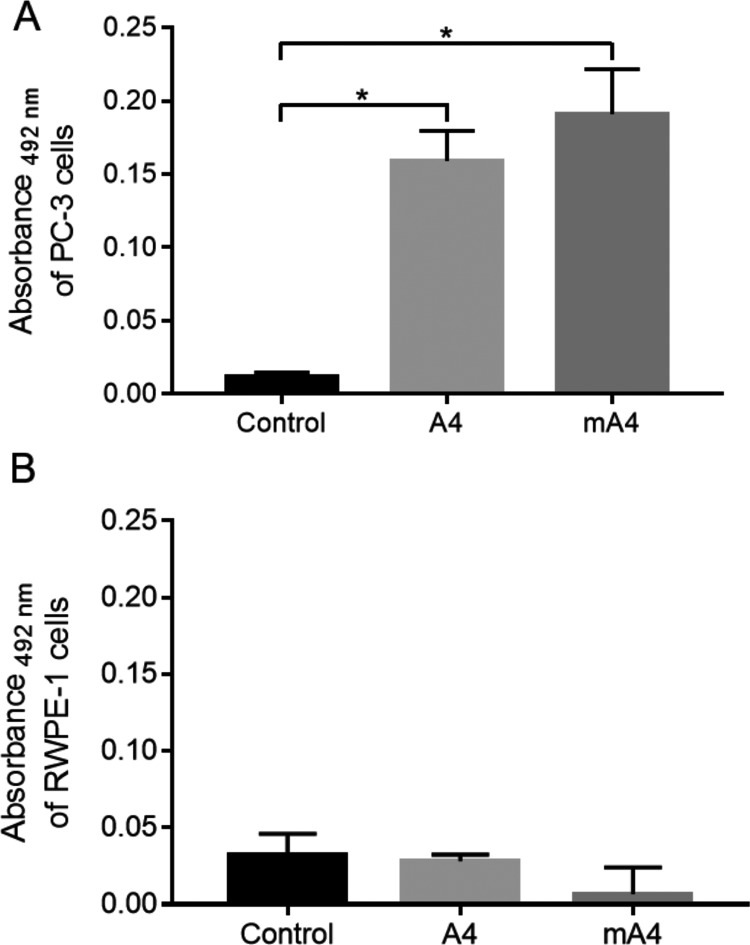
Because the ELASA is an easy, fast, and cost-effective technique to monitor the selection process of aptamers, comparable to the traditional flow cytometry approach,34 we used ELASA to compare the binding of mA4 and A4 to prostate cell lines used for A4 selection.
mA4 and A4 showed specificity in their binding, as both bind to PC-3 cells and do not bind to RWPE-1 cells, in comparison with the controls (Figure 3). In addition, the absence of the statistical difference between the binding of A4 and mA4 with PC-3 cells may indicate that they attach to these cells with comparable forces, in a similar way. Therefore, mA4 maintains the binding specificity of A4 to prostate cells.
Figure 3.
Binding of biotinylated A4 and mA4 to prostate cells shown by enzyme-linked apta-sorbent assay with streptavidin-conjugated horseradish peroxidase . (A) Absorbance at 492 nm of PC-3 cells. (B) Absorbance at 492 nm of RWPE-1 cells. Cells were incubated with no aptamer (control), aptamer A4, and aptamer mA4, both at a concentration of 2 μM. The data represent the means ± standard errors of the means of three independent experiments, run in triplicate. Data from cells incubated with aptamers were normalized subtracting the mean of the absorbance of the same cells incubated only with streptavidin-conjugated horseradish peroxidase. The data were statistically analyzed using the Kruskal–Wallis test with the Dunn post-test, *p < 0.05.
Although other studies, such as nuclear magnetic resonance spectroscopy, are needed to verify that a modified aptamer has mimetic structures of the native one,35 our results from the in silico studies and the ELASA suggest that mA4 may produce a scaffold on which nucleotides that interact with its molecular target are organized in structural motifs shared with the original sequence of A4.
2.5. Viability of Prostate Cells Grown under In Vivo-Like Conditions is Unaffected by mA4
Because different aptamers need different times to interact with their targets36 and produce a cellular response, we considered an interval of 48 h which is long enough to assess the potential effects of mA4 on cell lines. Consequently, these potential effects could be extrapolated to those of mA4 in a clinical sample tested for diagnosis, considering the necessary laboratory procedures and handling. In vitro assays were performed on the epithelial cell lines RWPE-1 and PC-3, as they were used in the negative and positive selections of the A4 aptamer, respectively. Although RWPE-1 cells are nonneoplastic human prostate cells and they represented the nontarget cells, PC-3 cells are neoplastic human prostate cells and they represented the target cells, which the aptamer should bind to in a potential diagnostic or treatment system.
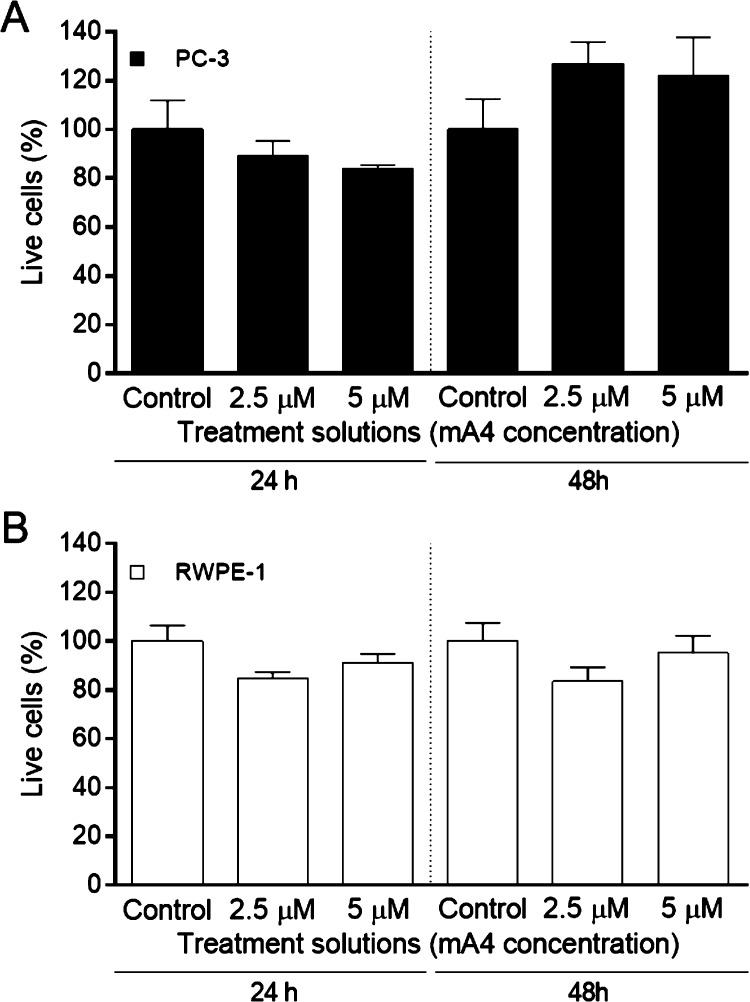
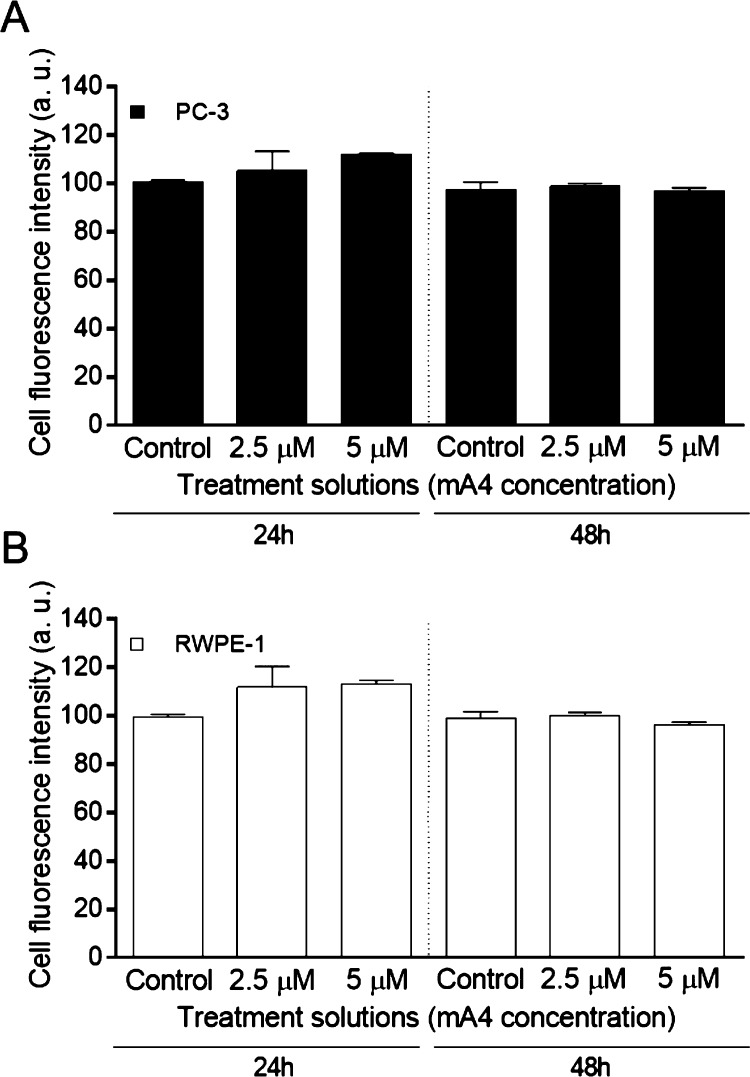
Viability of prostate cells cultured in magnetic 3D bioprinting was unaffected by the presence of mA4 at concentrations of 2.5 and 5 μM, after 24 and 48 h of incubation (Figure 4). A4 did not affect the viability of prostate cells under the tested conditions either (Figure S1).
Figure 4.
Effects on prostate cell viability induced by mA4 in 3D cultures. (A) Percentage of live PC-3 cells. (B) Percentage of live RWPE-1 cells. The data represent the means ± standard errors of the means of two independent experiments, run in triplicate, resulting from a spectrophotometric neutral red cell viability assay of the prostatic cell lines incubated with no aptamer (control) and with mA4 at 2.5 and 5 μM for 24 and 48 h. The data were statistically analyzed using the Kruskal–Wallis test with Dunn post-test for PC-3 cells and the one-way analysis of variance (ANOVA) test with Tukey post-test for RWPE-1 cells, *p < 0.05.
Despite cell viability remaining an issue for CTC isolation methods that use aptamers for diagnosis from human fluids, it is hard to find in the literature, cell viability tests with aptamers for diagnostic use. Contrary to our findings, most aptamers trigger cell death responses when they bind to their target cells, being used as therapeutic tools rather than diagnostic ones. For example, anti-EpCAM, anti-CD44, and bispecific anti-EpCAM-CD44 aptamers, at concentrations up to 4 μM, reduced the viability of ovarian tumor cell lines, which was correlated to the expression of these targets in the cells, after 72 h of incubation.37
The fact that mA4 did not kill its target cells PC-3 means that it could be used as a diagnostic tool reporting the presence of a molecular biomarker. If needed and tested, mA4 could report such a biomarker for a long period after its binding, as it does not kill the bearing cell. Besides being used as a diagnostic tool, this result does not exclude its use as a therapeutic tool. However, mA4 would need to be coupled with a drug to act as a targeted drug delivery system.
2.6. Prostatic Cells Proliferate at Their Normal Rate in the Presence of mA4 in an In Vivo-Like System
As cell viability is intimately related to cell proliferation, but not all viable cells are dividing, we tested cell proliferation of our prostate cell lines. PC-3 and RWPE-1 cells also maintained their proliferation ability (Figure 5), after incubation with mA4 for 48 h. Similar to our results, the incubation of the aptamer AS1411 with its target hepatocarcinoma cells failed to show significant differences in their proliferation profile at a concentration twice as high as the maximum concentration used by us, whereas when this aptamer was modified with the replacement of two internal thymines by two 5-(N-naphthylcarboxyamide)-2′-deoxyuridines to increase its affinity to its target nucleolin, it decreased the proliferation of these cells under the same conditions.38 Therefore, modifications that improve the interaction between aptamers and their targets could induce biological changes in target cells.
Figure 5.
Effects on prostate cell proliferation induced by mA4 in 3D cultures. (A) Fluorescence intensity of dividing PC-3 cells. (B) Fluorescence intensity of dividing RWPE-1 cells. The data represent the means ± standard errors of the means of two independent experiments, run in triplicate, resulting from a fluorimetric CytoSelect cell proliferation assay of the prostatic cell lines incubated with no aptamer (control) and with mA4 at 2.5 and 5 μM for 24 and 48 h. The data were statistically analyzed using the Kruskal–Wallis test with Dunn post-test for PC-3 cells and the one-way analysis of variance (ANOVA) test with Tukey post-test for RWPE-1 cells, *p < 0.05. Arbitrary units (a. u.).
In contrast to our results of PC-3 cells, most aptamers decrease cell proliferation when they bind to their target cells whose target molecules are involved in the signaling of growth factors.36 Thus, our results suggest that if the target molecule is involved in cell proliferation pathways, either the concentration of mA4 used in the proliferation assay, the expression of its molecular target in PC-3 cells, or the interactions between them are not enough to induce changes in the cell proliferation of PC-3 cells.
However, the concentrations used were higher to those previously reported by other researchers. For example, the RA16 aptamer, with its molecular target unknown, reduced the proliferation of its target cells NCI-H460 at a concentration of 300 nM,39 six times lower than the highest concentration used in our study. The U2 aptamer also decreased proliferation of glioblastoma cells at even lower concentrations, 25 and 50 nM, and after incubation for 24 and 48 h.40 Accordingly, the expression of the molecular target of mA4 may be low to induce observable changes in the proliferation of PC-3 cells. Another possibility is that the interaction between them is too weak and it needs to be enhanced.
2.7. mA4 Does Not Influence Migration of Prostatic Cell Lines
When these prostate cells were cultured in a monolayer, under the same conditions, the wound healing assay did not show significant differences in their migration profile, compared to the controls, after incubation with mA4 (Figure 6) or A4 (Figure S2) for 24 and 48 h. Although 3D cultures better mimic in vivo conditions and should be considered the first choice for in vitro tests,41 we did not find an alternative to replace the wound healing assay which is performed in two-dimensional (2D) cultures. However, the results of this assay were consistent with those of cell viability and proliferation that we performed in 3D cultures.
Figure 6.
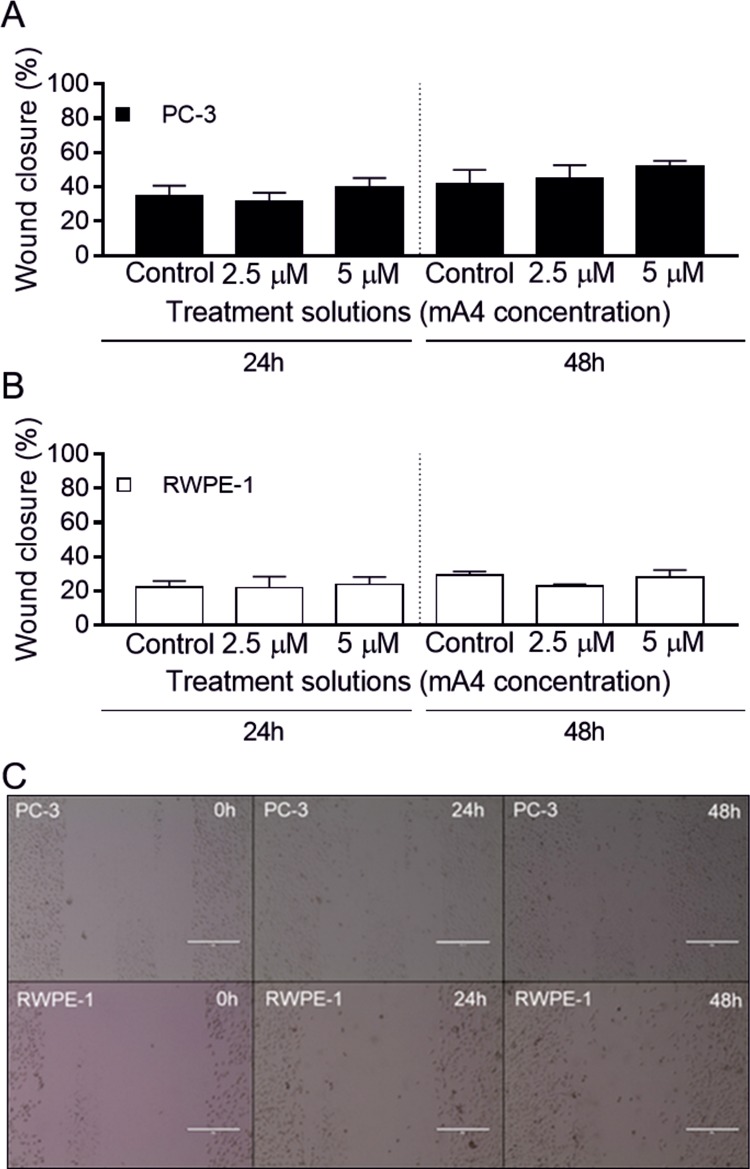
Effects on prostatic cell migration induced by mA4. (A) Wound closure percentage of PC-3 cells. (B) Wound closure percentage of RWPE-1 cells. The data represent the means ± standard errors of the means of two independent experiments, run in triplicate, resulting from a wound healing assay of the prostatic cell lines incubated with no aptamer (control) and with mA4 at 2.5 and 5 μM for 24 and 48 h. One-way analysis of variance with Tukey post-test was used to analyze the different conditions, *p < 0.05. C. Representative images of the wound healing assays of the cell lines treated with 5 μM of mA4 after 0, 24, and 48 h. Magnification × 10. Scale bar 400 μm.
Differently, the aptamer AXL reduced the migration of the ovary cancer cells SKOV3-IP1 after 24 and 48 h of incubation at a concentration of 100 nM.42 The aptamer U2 also decrease the migration of target glioblastoma multiforme cells, after 8 and 24 h of incubation at 50 nM,40 a concentration a hundredfold lower than our maximum concentration. Interestingly, the aptamer CL4, which shares the receptor EGFRvIII as its target with the aptamer U2, showed that the decrease in the migration of the target cells depends not only on the presence of the target but also on the expression of downstream proteins in the signaling pathways of its target.43 Therefore, we cannot rule out the chance that the target molecule of mA4 is involved in cell migration and there is a bypass that could constitutively activate the corresponding pathway in PC-3 cells. Cell proliferation and migration share the cellular pathways of microtubule polymerization that are involved in mitosis, intracellular trafficking, transcription, translation, and displacement of tumor cells.44 Therefore, our proliferation and migration results complement each other.
Considering that the binding of mA4 to its targets would inhibit their functions45 and that we used standard concentrations of the aptamer when compared to the other studies discussed, the absence of response in their target cells may indicate that its molecular target is either little expressed in PC-3 cells, involved in downstream signaling pathways constitutively activate, or simply not involved in viability, proliferation, or migration pathways.
3. Conclusions
In the present study, a practical model was applied to prove the feasibility of the optimized aptamer mA4 as a probe for in vitro diagnostic use. Our results indicated a high structural similarity between A4 and mA4, proposing the clinical application of mA4 as a theranostic tool with potentials in the field of PCa. As 3D cultures display cell proliferation rates similar to those found in tumor micro-environments,46 in vivo-like 3D magnetic cell cultures were used for characterization of mA4, which allowed us to increase our knowledge about mA4 and its interaction with prostatic cells. Because mA4 bound specifically to tumor prostate cells, is more resistant than A4 to nuclease present in plasma, and did not affect the behavior of prostate cells, we determine that mA4 can be directly used in in vitro diagnostic systems for PCa.
The potential use of mA4 in other theranostic applications, such as diagnostic imaging and treatment systems, still requires further investigation. mA4 did not showed therapeutic action itself against PCa cells. However, mA4 could be used as a targeted drug carrier, coupled with a drug, to avoid toxic effects in nontarget cells, which often limits the effect of treatments.47 As A4 and mA4 bind to PCa cells through molecular targets unknown to date,15 it would be interesting to identify the target molecule and its binding region to predict other potential biological actions of the aptamer in its target cells and to improve their interaction.48,49 Future findings will hopefully broaden the clinical applications of mA4.
4. Materials and Methods
4.1. Aptamers
Integrated DNA Technology (IDT, Coralville, IA, USA) synthesized mA4 and A4 and biotinylated them at their 5′ ends. The sequence of the RNA aptamer A4 was synthesized with its unique nucleotides without the sequences complementary to the primers used during its selection process (5′GGGCGAGCAGCAGACAGCCGAGAGGUAAGCAAAACCACGCCCGCAUCGUCCUCCC3′).15 This sequence was truncated and deoxyribonucleotides were added at its ends to design the aptamer mA4 yielding the following sequence, with the deoxyribonucleotides underlined: 5′TTTTTTTTTTTTTGGGCGAGCAGCAGACAGCCGAGAGGUAAGCAAAACCACGCCCGCAT3′. The sequences of both aptamers were aligned using BLAST25 to identify common nucleotides.
4.2. In Silico Elucidation of Secondary and Tertiary Structures
The secondary structure of mA4 was predicted using the Mfold web server (https://www.idtdna.com/Unafold/Home/Index).50 This secondary structure was translated into the dot-bracket notation (Vienna format) to be imported to the RNA Composer web server (http://rnacomposer.ibch.poznan.pl/Home),51 which predicted the tertiary structure of the RNA form of the aptamer.
The Discovery Studio Visualizer software (Dassault Systèmes BIOVIA, v19.1.0.18287, San Diego, CA, USA) was used to adapt the 3D RNA form of the aptamer to its 3D chimeric form, with its additional deoxyribonucleotides.
4.3. Aptamer Degradation Assay
Both RPMI-1640 (Cultilab, Campinas, SP, Brazil) and Keratinocyte-SFM (Thermo Fisher Scientific, Waltham, MA, USA) were supplemented with 100 μg/mL gentamicin (Thermo Fisher Scientific, Waltham, MA, USA), incubated with mA4 at a concentration of 33.34 μM at 37 °C for 0, 0.5, 1, 2, 4, 8, 24, 48, and 72 h, and then frozen at −80 °C.
Three blood samples were collected in tubes with EDTA (Vacuette K3EDTA, Greiner Bio-one, Kremsmünster, Austria) from PCa patients, together with the corresponding informed consent. This experiment was approved by the Research Ethics Committee of the Federal University of Uberlândia (CAAE 71108817.2.0000.5152) in accordance with all national and local guidelines and regulations.
After venipuncture, the tubes were centrifuged at 2000g for 15 min at 4 °C and the supernatants were mixed and used to incubate the aptamers A4 and mA4 separately, at a concentration of 33.34 μM at 37 °C for 0, 0.5, 1, 2, 4, 6, 8, 10, and 24 h. Then, the solutions were frozen at −80 °C.
The degradation of the aptamers at each time and in each diluent was analyzed after electrophoresis on a 2% agarose gel stained with ethidium bromide (Sigma-Aldrich, Saint Louis, MO, USA). The bands were imaged and their intensity were quantified using ImageJ program (v1.52o, NIH, Bethesda, MD, USA). A molecular-weight size marker (MM) of 100 pair bases (DNA Ladder 100 pb, Ludwig Biotecnologia, Alvorada, RS, Brazil) was used to assist in identifying the aptamers on the gel. Supplemented media or human blood plasma without aptamers were used as controls. Data were normalized considering fluorescence intensity at time 0 h as 100%. Exponential one-phase decay equation was used to get the half-life of mA4 and A4 under the tested in vitro conditions by PrismGraph program (GraphPad Software, v.7, La Jolla, CA, USA).
4.4. Cell Cultures
PC-3 (ATCC CRL-1435) and RWPE-1 (ATCC CRL-11609) human prostate cell lines (ATCC, Manassas, VA, USA) were cultured in a monolayer and 3D bioprinting was used for the in vitro assays. Both cell lines were successfully authenticated in 2018 by short tandem repeat analysis and monthly tested for mycoplasma contamination with negative results.
The PC-3 cell line was cultured in RPMI-1640 medium (Cultilab, Campinas, SP, Brazil), and the RWPE-1 cell line was cultured in Keratinocyte-SFM (Thermo Fisher Scientific, Waltham, MA, USA). Both cell lines were supplemented with 100 μg/mL gentamicin (Thermo Fisher Scientific, Waltham, MA, USA) and 10% fetal bovine serum (Gibco/Thermo Fisher Scientific, Waltham, MA, USA) and incubated at 37 °C in a 5% CO2 atmosphere. After trypsinization with 0.25% trypsin–EDTA solution (Corning, New York, NY, USA), cells were seeded on either plates for adherent monolayer cell culture or ultralow adhesion plates (Corning, New York, NY, USA) for 3D cell culture, according to each assay. Bio Assembler kits (Nano3D Biosciences, Houston, TX, USA) were used for 3D bioprinting, according to the manufacturer’s instructions.
4.5. Enzyme-Linked Apta-sorbent Assay
A day before the experiment, cells were plated in monolayers, in 96-well plates at a density of 5 × 104. Cells were blocked with phosphate buffer saline (PBS) containing 5% bovine serum albumin for 1 h at 37 °C followed by washing with PBS and incubation with 2 μM biotinylated aptamers A4 and mA4 for 1 h at 37 °C. After washing with PBS, cells were incubated with streptavidin-conjugated horseradish peroxidase (SAv-HRP) (BD Biosciences Pharmingen, San Diego, CA, USA) diluted (1:250) in PBS for 1 h at 37 °C. The wells were washed again with PBS, and the peroxidase reaction was initiated by orthophenylenediamine dihydrochloride (OPD) (Sigma-Aldrich, Saint Louis, MO, USA) in citrate-phosphate buffer with 3% H2O2 and terminated upon addition of sulphuric acid. The absorbance of each well was measured at 492 nm in a microplate reader to estimate the binding of the aptamers. In parallel, incubation of cells with PBS was used as a negative control and incubation of cells with SAv-HRP in PBS was used for data adjustment.
4.6. Cell Viability and Proliferation Assays
Cells were seeded in 96-well plates containing 2 × 103 cells per well and grown in 3D culture. After 24 h, cells were treated with the corresponding medium with mA4 at concentrations of 2.5 and 5 μM and without mA4 (control). Cellular viability and proliferation were analyzed, after 24 and 48 h of treatment. As an assay control, cellular viability was also evaluated after incubation of A4 at a concentration of 2 μM, with an additional death control with 1% Triton 100-X (Thermo Fisher Scientific, Waltham, MA, USA). Fetal bovine serum was not used for these treatments, as it could degrade the aptamers.
Cellular viability was assessed by an absorbance-based assay, performed with the neutral red reagent (Sigma-Aldrich, Saint Louis, MO, USA), according to the manufacturer’s instructions.
Cellular proliferation was assessed by a fluorescence-based assay, performed with the CytoSelect cell proliferation assay reagent (Cell Biolabs, San Diego, CA, USA), according to the manufacturer’s recommendations.
4.7. Wound Healing Assay
In 12-well plates, 2 × 105 cells per well were grown in monolayers to full confluence. A longitudinal scratch was made with a sterile pipette tip at the bottom of each well. Wells were washed with PBS and cells were treated with the corresponding medium with mA4 at concentrations of 2.5 and 5 μM and without mA4 (control). As an assay control, cellular migration was also evaluated after incubation of A4 at a concentration of 2 μM. Fetal bovine serum was not used for the treatments, as it could degrade the aptamers.
Images were acquired using an EVOS inverted microscope (Thermo Fisher Scientific, Waltham, MA, USA) at 0, 24, and 48 h after the treatments. The open (wounded) areas of the images were calculated using the ImageJ program (v1.52o, NIH, Bethesda, MD, USA) to assess cell migration.
4.8. Statistical Analyses
Statistical analyses were performed by PrismGraph program (GraphPad Software, v.7, La Jolla, CA, USA). The Kruskal–Wallis test with Dunn post-test or the one-way analysis of variance (ANOVA) test with Tukey post-test was used for multiple comparisons according to each set of data. The value of p < 0.05 was considered statistically significant for all tests.
Acknowledgments
This work was supported by the National Council for Scientific and Technological Development (CNPq), INCT-TeraNano grant number 465669/2014-0, and the Brazilian funding agency Coordination for the Improvement of Higher Education Personnel (CAPES).
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.9b03855.
Viability and migration of prostate cells incubated with A4 (PDF)
Author Present Address
§ Departamento de Imagens Médicas, Hematologia e Oncologia Clínica, Faculdade de Medicina de Ribeirão Preto, Universidade de São Paulo, Ribeirão Preto, SP, Brazil.
The authors declare no competing financial interest.
Supplementary Material
References
- Howlader N.; Noone A.; Krapcho M.; Miller D.; Brest A.; Yu M.; Ruhl J.; Tatalovich Z.; Mariotto A.; Lewis D.; et al. SEER Cancer Statistics Review, 1975–2016. https://seer.cancer.gov/csr/1975_2016/ (accessed Dec 22, 2019).
- Brawley O. W. Trends in Prostate Cancer in the United States. J. Natl. Cancer Inst. Monogr. 2012, 2012, 152–156. 10.1093/jncimonographs/lgs035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velonas V.; Woo H.; Remedios C.; Assinder S. Current Status of Biomarkers for Prostate Cancer. Int. J. Mol. Sci. 2013, 14, 11034–11060. 10.3390/ijms140611034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truong M.; Frye T. P. Magnetic Resonance Imaging Detection of Prostate Cancer in Men with Previous Negative Prostate Biopsy. Transl. Androl. Urol. 2017, 6, 424–431. 10.21037/tau.2017.03.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andriole G. L.; Crawford E. D.; Grubb R. L.; Buys S. S.; Chia D.; Church T. R.; Fouad M. N.; Isaacs C.; Kvale P. A.; Reding D. J.; et al. Prostate Cancer Screening in the Randomized Prostate, Lung, Colorectal, and Ovarian Cancer Screening Trial: Mortality Results after 13 Years of Follow-Up. J. Natl. Cancer Inst. 2012, 104, 125–132. 10.1093/jnci/djr500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröder F. H.; Hugosson J.; Carlsson S.; Tammela T.; Määttänen L.; Auvinen A.; Kwiatkowski M.; Recker F.; Roobol M. J. Screening for Prostate Cancer Decreases the Risk of Developing Metastatic Disease: Findings from the European Randomized Study of Screening for Prostate Cancer (ERSPC). Eur. Urol. 2012, 62, 745–752. 10.1016/j.eururo.2012.05.068. [DOI] [PubMed] [Google Scholar]
- Diamandis E. P.; Yu H. Nonprostatic Sources of Prostate-Specific Antigen. Urol. Clin. 1997, 24, 275–282. 10.1016/s0094-0143(05)70373-6. [DOI] [PubMed] [Google Scholar]
- Loeb S.; Bjurlin M. A.; Nicholson J.; Tammela T. L.; Penson D. F.; Carter H. B.; Carroll P.; Etzioni R. Overdiagnosis and Overtreatment of Prostate Cancer. Eur. Urol. 2014, 65, 1046–1055. 10.1016/j.eururo.2013.12.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tricoli J. V.; Schoenfeldt M.; Conley B. a. Detection of Prostate Cancer and Predicting Progression : Current and Future Diagnostic Markers Detection of Prostate Cancer and Predicting Progression : Current and Future Diagnostic Markers. Clin. Cancer Res. 2004, 10, 3943–3953. 10.1158/1078-0432.ccr-03-0200. [DOI] [PubMed] [Google Scholar]
- Murphy A. M.; McKiernan J. M.; Olsson C. A. Controversies in Cancer Screening. J. Urol. 2004, 172, 1822–1824. 10.1097/01.ju.0000140500.65341.9a. [DOI] [PubMed] [Google Scholar]
- Marrugo-Ramírez J.; Mir M.; Samitier J.; Marrugo-Ramírez J.; Mir M.; Samitier J. Blood-Based Cancer Biomarkers in Liquid Biopsy: A Promising Non-Invasive Alternative to Tissue Biopsy. Int. J. Mol. Sci. 2018, 19, 2877. 10.3390/ijms19102877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brock G.; Castellanos-Rizaldos E.; Hu L.; Coticchia C.; Skog J. Liquid Biopsy for Cancer Screening, Patient Stratification and Monitoring. Transl. Cancer Res. 2015, 4, 280–290. [Google Scholar]
- Strotman L. N.; Millner L. M.; Valdes R.; Linder M. W. Liquid Biopsies in Oncology and the Current Regulatory Landscape. Mol. Diagn. Ther. 2016, 20, 429–436. 10.1007/s40291-016-0220-5. [DOI] [PubMed] [Google Scholar]
- Campos-Fernández E.; Barcelos L. S.; de Souza A. G.; Goulart L. R.; Alonso-Goulart V. Research Landscape of Liquid Biopsies in Prostate Cancer. Am. J. Cancer Res. 2019, 9, 1309–1328. [PMC free article] [PubMed] [Google Scholar]
- Souza A. G.; Marangoni K.; Fujimura P. T.; Alves P. T.; Silva M. J.; Bastos V. A. F.; Goulart L. R.; Goulart V. A. 3D Cell-SELEX: Development of RNA Aptamers as Molecular Probes for PC-3 Tumor Cell Line. Exp. Cell Res. 2016, 341, 147–156. 10.1016/j.yexcr.2016.01.015. [DOI] [PubMed] [Google Scholar]
- Ding Y.; Chan C. Y.; Lawrence C. E. Sfold Web Server for Statistical Folding and Rational Design of Nucleic Acids. Nucleic Acids Res 2004, 32, W135. 10.1093/nar/gkh449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseley J.; Tollervey D. The Many Pathways of RNA Degradation. Cell 2009, 136, 763–776. 10.1016/j.cell.2009.01.019. [DOI] [PubMed] [Google Scholar]
- Ulrich Göringer H.; Adler A.; Forster N.; Homann M. Post-SELEX Chemical Optimization of a Trypanosome-Specific RNA Aptamer. Comb. Chem. High Throughput Screening 2008, 11, 16–23. [DOI] [PubMed] [Google Scholar]
- Wilson C.; Keefe A. Building Oligonucleotide Therapeutics Using Non-Natural Chemistries. Curr. Opin. Chem. Biol. 2006, 10, 607–614. 10.1016/j.cbpa.2006.10.001. [DOI] [PubMed] [Google Scholar]
- Zhu Q.; Liu G.; Kai M.; Zhu Q.; Liu G.; Kai M. DNA Aptamers in the Diagnosis and Treatment of Human Diseases. Molecules 2015, 20, 20979–20997. 10.3390/molecules201219739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C.; Liu J.; Zhang P.; Li J.; Ji H.; Yang X.; Wang K. A Recognition-before-Labeling Strategy for Sensitive Detection of Lung Cancer Cells with a Quantum Dot-Aptamer Complex. Analyst 2015, 140, 6100–6107. 10.1039/c5an01145k. [DOI] [PubMed] [Google Scholar]
- Kaighn M. E.; Narayan K. S.; Ohnuki Y.; Lechner J. F.; Jones L. W. Establishment and Characterization of a Human Prostatic Carcinoma Cell Line (PC-3). Invest. Urol. 1979, 17, 16–23. [PubMed] [Google Scholar]
- Tai S.; Sun Y.; Squires J. M.; Zhang H.; Oh W. K.; Liang C.-Z.; Huang J. PC3 Is a Cell Line Characteristic of Prostatic Small Cell Carcinoma. Prostate 2011, 71, 1668–1679. 10.1002/pros.21383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bello D.; Webber M. M.; Kleinman H. K.; Wartinger D. D.; Rhim J. S. Androgen Responsive Adult Human Prostatic Epithelial Cell Lines Immortalized by Human Papillomavirus 18. Carcinogenesis 1997, 18, 1215–1223. 10.1093/carcin/18.6.1215. [DOI] [PubMed] [Google Scholar]
- Zhang Z.; Schwartz S.; Wagner L.; Miller W. A Greedy Algorithm for Aligning DNA Sequences. J. Comput. Biol. 2000, 7, 203–214. 10.1089/10665270050081478. [DOI] [PubMed] [Google Scholar]
- Walsh R.; DeRosa M. C. Retention of Function in the DNA Homolog of the RNA Dopamine Aptamer. Biochem. Biophys. Res. Commun. 2009, 388, 732–735. 10.1016/j.bbrc.2009.08.084. [DOI] [PubMed] [Google Scholar]
- Stoltenburg R.; Reinemann C.; Strehlitz B. SELEX—A (r)Evolutionary Method to Generate High-Affinity Nucleic Acid Ligands. Biomol. Eng. 2007, 24, 381–403. 10.1016/j.bioeng.2007.06.001. [DOI] [PubMed] [Google Scholar]
- Cai S.; Yan J.; Xiong H.; Liu Y.; Peng D.; Liu Z. Investigations on the Interface of Nucleic Acid Aptamers and Binding Targets. Analyst 2018, 143, 5317. 10.1039/c8an01467a. [DOI] [PubMed] [Google Scholar]
- Ruscito A.; DeRosa M. C. Small-Molecule Binding Aptamers: Selection Strategies, Characterization, and Applications. Front. Chem. 2016, 4, 14. 10.3389/fchem.2016.00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahirwar R.; Nahar S.; Aggarwal S.; Ramachandran S.; Maiti S.; Nahar P. Silico Selection of an Aptamer to Estrogen Receptor Alpha Using Computational Docking Employing Estrogen Response Elements as Aptamer-Alike Molecules. Sci. Rep. 2016, 6, 21285. 10.1038/srep21285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni S.; Yao H.; Wang L.; Lu J.; Jiang F.; Lu A.; Zhang G. Chemical Modifications of Nucleic Acid Aptamers for Therapeutic Purposes. Int. J. Mol. Sci. 2017, 18, 1683. 10.3390/ijms18081683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krissanaprasit A.; Key C.; Fergione M.; Froehlich K.; Pontula S.; Hart M.; Carriel P.; Kjems J.; Andersen E. S.; LaBean T. H. Genetically Encoded, Functional Single-Strand RNA Origami: Anticoagulant. Adv. Mater. 2019, 31, 1808262. 10.1002/adma.201808262. [DOI] [PubMed] [Google Scholar]
- Barra G. B.; Santa Rita T. H.; Vasques J. d. A.; Chianca C. F.; Nery L. F. A.; Costa S. S. S. EDTA-Mediated Inhibition of DNases Protects Circulating Cell-Free DNA from Ex Vivo Degradation in Blood Samples. Clin. Biochem. 2015, 48, 976–981. 10.1016/j.clinbiochem.2015.02.014. [DOI] [PubMed] [Google Scholar]
- Nabavinia M. S.; Charbgoo F.; Alibolandi M.; Mosaffa F.; Gholoobi A.; Ramezani M.; Abnous K. Comparison of Flow Cytometry and ELASA for Screening of Proper Candidate Aptamer in Cell-SELEX Pool. Appl. Biochem. Biotechnol. 2018, 184, 444. 10.1007/s12010-017-2548-7. [DOI] [PubMed] [Google Scholar]
- Gelinas A. D.; Davies D. R.; Janjic N. Embracing Proteins: Structural Themes in Aptamer-Protein Complexes. Curr. Opin. Struct. Biol. 2016, 36, 122–132. 10.1016/j.sbi.2016.01.009. [DOI] [PubMed] [Google Scholar]
- Missailidis S.; Hardy A. Aptamers as Inhibitors of Target Proteins. Expert Opin. Ther. Pat. 2009, 19, 1073–1082. 10.1517/13543770903042337. [DOI] [PubMed] [Google Scholar]
- Zheng J.; Zhao S.; Yu X.; Huang S.; Liu H. Y. Simultaneous Targeting of CD44 and EpCAM with a Bispecific Aptamer Effectively Inhibits Intraperitoneal Ovarian Cancer Growth. Theranostics 2017, 7, 1373–1388. 10.7150/thno.17826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho Y.; Lee Y. B.; Lee J. H.; Lee D. H.; Cho E. J.; Yu S. J.; Kim Y. J.; Kim J. I.; Im J. H.; Lee J. H.; et al. Modified AS1411 Aptamer Suppresses Hepatocellular Carcinoma by Up-Regulating Galectin-14. PLoS One 2016, 11, e0160822 10.1371/journal.pone.0160822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H.; Zhang Y.; Yang H.; Qin M.; Ding X.; Liu R.; Jiang Y. In Vivo SELEX of an Inhibitory NSCLC-Specific RNA Aptamer from PEGylated RNA Library. Mol. Ther.--Nucleic Acids 2018, 10, 187–198. 10.1016/j.omtn.2017.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.; Peng L.; Liang Z.; Kou Z.; Chen Y.; Shi G.; Li X.; Liang Y.; Wang F.; Shi Y. Effects of Aptamer to U87-EGFRvIII Cells on the Proliferation, Radiosensitivity, and Radiotherapy of Glioblastoma Cells. Mol. Ther.--Nucleic Acids 2018, 10, 438–449. 10.1016/j.omtn.2018.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souza A. G.; Silva I. B. B.; Campos-Fernández E.; Barcelos L. S.; Souza J. B.; Marangoni K.; Goulart L. R.; Alonso-Goulart V. Comparative Assay of 2D and 3D Cell Culture Models: Proliferation, Gene Expression and Anticancer Drug Response. Curr. Pharm. Des. 2018, 24, 1689. 10.2174/1381612824666180404152304. [DOI] [PubMed] [Google Scholar]
- Kanlikilicer P.; Ozpolat B.; Aslan B.; Bayraktar R.; Gurbuz N.; Rodriguez-Aguayo C.; Bayraktar E.; Denizli M.; Gonzalez-Villasana V.; Ivan C.; et al. Therapeutic Targeting of AXL Receptor Tyrosine Kinase Inhibits Tumor Growth and Intraperitoneal Metastasis in Ovarian Cancer Models. Mol. Ther.--Nucleic Acids 2017, 9, 251–262. 10.1016/j.omtn.2017.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camorani S.; Crescenzi E.; Colecchia D.; Carpentieri A.; Amoresano A.; Fedele M.; Chiariello M.; Cerchia L. Aptamer Targeting EGFRvIII Mutant Hampers Its Constitutive Autophosphorylation and Affects Migration, Invasion and Proliferation of Glioblastoma Cells. Oncotarget 2015, 6, 37570–37587. 10.18632/oncotarget.6066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogden A.; Rida P. C. G.; Reid M. D.; Aneja R. Interphase Microtubules: Chief Casualties in the War on Cancer?. Drug Discovery Today 2014, 19, 824–829. 10.1016/j.drudis.2013.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel K. W.; Giangrande P. H. Therapeutic Applications of DNA and RNA Aptamers. Oligonucleotides 2009, 19, 209–222. 10.1089/oli.2009.0199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamura Y.; Mukohara T.; Shimono Y.; Funakoshi Y.; Chayahara N.; Toyoda M.; Kiyota N.; TAKAO S.; KONO S.; NAKATSURA T. Comparison of 2D- and 3D-Culture Models as Drug-Testing Platforms in Breast Cancer. Oncol. Rep. 2015, 33, 1837–1843. 10.3892/or.2015.3767. [DOI] [PubMed] [Google Scholar]
- Leong S.; McKay M. J.; Christopherson R. I.; Baxter R. C. Biomarkers of Breast Cancer Apoptosis Induced by Chemotherapy and TRAIL. J. Proteome Res. 2012, 11, 1240–1250. 10.1021/pr200935y. [DOI] [PubMed] [Google Scholar]
- Yoon S.; Rossi J. J. Emerging Cancer-Specific Therapeutic Aptamers. Curr. Opin. Oncol. 2017, 29, 366–374. 10.1097/cco.0000000000000389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K. Y.; Kang H.; Ryu S. H.; Lee D. S.; Lee J. H.; Kim S. Bioimaging of Nucleolin Aptamer-Containing 5-(N-Benzylcarboxyamide)-2’-Deoxyuridine More Capable of Specific Binding to Targets in Cancer Cells. J. Biomed. Biotechnol. 2010, 2010, 168306. 10.1155/2010/168306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. Mfold Web Server for Nucleic Acid Folding and Hybridization Prediction. Nucleic Acids Res. 2003, 31, 3406–3415. 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popenda M.; Szachniuk M.; Antczak M.; Purzycka K. J.; Lukasiak P.; Bartol N.; Blazewicz J.; Adamiak R. W. Automated 3D Structure Composition for Large RNAs. Nucleic Acids Res. 2012, 40, e112 10.1093/nar/gks339. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.