Introductory Paragraph:
Sequencing DNA derived from archaic bones has enabled genetic comparison of Neanderthals and anatomically modern humans (AMHs) and revealed that they interbred. However, interpreting what genetic differences imply about their phenotypic differences remains challenging. Here we introduce an approach for identifying divergent gene regulation between archaic hominins, like Neanderthals, and AMH sequences and find 766 genes likely to have been divergently regulated by Neanderthal haplotypes that do not remain in AMH. Divergently regulated genes include many involved in phenotypes known to differ between Neanderthals and AMHs, such as structure of the rib cage and supraorbital ridge development. They are also enriched for genes associated with spontaneous abortion, polycystic ovary syndrome, myocardial infarction, and melanoma. Phenotypes associated with modern human variation in these genes’ regulation in ~23,000 biobank patients further supports their involvement in immune and cardiovascular phenotypes. Comparing divergently regulated genes between two Neanderthals and a Denisovan revealed divergence in the immune system and in genes associated with skeletal and dental morphology that are consistent with the archaeological record. These results establish differences in gene regulatory architecture between AMHs and archaic hominins and provide an avenue for exploring phenotypic differences between archaic groups from genomic information alone.
Most aspects of archaic hominin biology cannot be directly studied due to their lack of preservation in fossils. The sequencing of DNA extracted from remains of extinct hominins has enabled the study of these groups’ origins and evolutionary histories on a scale not possible from fossils alone1-4. However, even with whole genome sequences available, the ability to infer traits of these hominins and how they differed from one another and anatomically modern humans (AMHs) is limited5. Greater morphological knowledge would be especially valuable for groups like the Denisovans that lack a substantial fossil record2,6. A key challenge in this task is the difficulty of mapping from genetic sequence differences to function.
Archaic hominins interbred with anatomically modern humans (AMHs)1,2,7, and as a result, more than one third of the Neanderthal genome remains in introgressed sequences in AMH genomes8,9. However, the factors that determined the patterns of Neanderthal ancestry in AMH genomes are not fully understood. The Neanderthal DNA that remains in modern Eurasian populations influences a range of traits, with a particular influence on immune, hair and skin, and neurological phenotypes10. This suggests differences between Neanderthals and AMHs that could have been selected for after interbreeding. There are only a small number of protein-coding differences between archaic hominins and modern humans11, but introgressed archaic sequences often exert their effects by modifying gene expression patterns10,12. One quarter of Neanderthal sequences remaining in AMHs have cis-regulatory effects, and Neanderthal alleles are particularly downregulated in the brain and testes13. Thus, divergent gene regulation between archaic and AMH sequences produces physiologically relevant effects.
While the functional effects of introgressed sequences have been studied in detail, much less is known about the functions of non-introgressed Neanderthal sequences. Understanding the functions of these regions would provide valuable insight into barriers to introgression, the role of selection in determining the landscape of archaic DNA in modern populations, and the phenotypic differences between archaic and modern humans. We addressed this challenge by quantifying divergence in gene regulation between archaic hominin and AMH sequences and associating divergently regulated genes with AMH phenotypes using existing annotations and a large biobank linked to electronic health records (EHRs)14. Our results demonstrate substantial divergence in gene regulation between hominins and have the promise to highlight previously inaccessible differences in archaic hominin biology.
Results
Quantifying gene regulatory divergence with PrediXcan
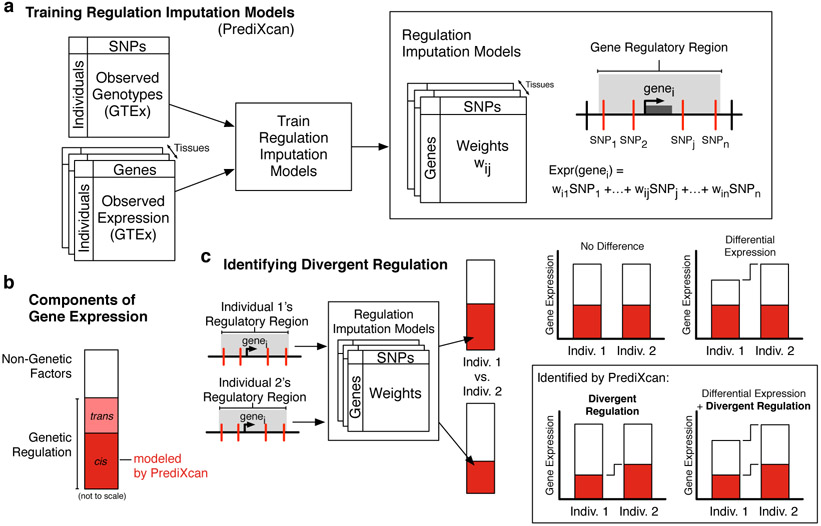
To identify archaic hominin sequences likely to have divergent gene regulatory effects compared to AMH sequences, we developed a statistic based on applying PrediXcan models to modern and archaic sequences. PrediXcan imputes the cis genetically regulated component of gene expression for genes in specific tissues using paired genotype and transcriptome data from human populations (Fig. 1A, B). Previous work has demonstrated that PrediXcan can impute the genetically regulated component of gene expression for thousands of genes, especially those whose regulatory architecture is dominated by common variants14. We considered accurate (FDR < 0.05) PrediXcan models of autosomal gene regulation from 44 tissues that were trained and evaluated on paired genotypes and normalized transcriptomes from the GTEx Consortium15, which consists of ~5% European ancestry and ~15% African ancestry individuals (Methods). The output of a PrediXcan model is not a direct proxy for gene expression in an individual. Instead, it is an estimate of the genetically regulated component of gene expression in reference to the distribution observed in the population used to train the model. Thus, differences in PrediXcan values between individuals reflect differences in variant genetic regulatory effects, not necessarily differences in overall gene expression (Fig. 1B, C). To emphasize this distinction, we refer to these differences as divergent regulation.
Fig. 1. Identifying divergent gene regulation between individuals using PrediXcan.
(a) Statistical models for imputing genetic regulation of gene expression (PrediXcan) were trained on genetic variants and normalized transcriptomes for 44 tissues from all individuals in the Genotype-Tissue Expression (GTEx) Project. Genetic variants within 1 Mb of each gene (Gene Regulatory Region indicated by gray box) were considered in the PrediXcan models; variants included in the models are illustrated by red vertical lines. (b) Gene expression levels are the result of genetic and non-genetic (e.g., environmental) factors. Our approach imputes the cis-genetic component of gene expression. (c) Our approach can identify divergent regulation between individuals, which reflects changes in the gene regulatory architecture, but does not necessarily imply differences in overall gene expression.
We consider the regulation of two classes of genes: 1) those that lack archaic ancestry in any variant in their PrediXcan model and 2) those with archaic ancestry in at least one modeled variant in at least one AMH (Fig. 2 A, Methods). We will first focus on the former group and refer to them as “genes without archaic regulatory regions” (GWARRs). For simplicity, we will refer to the latter as non-GWARRs.
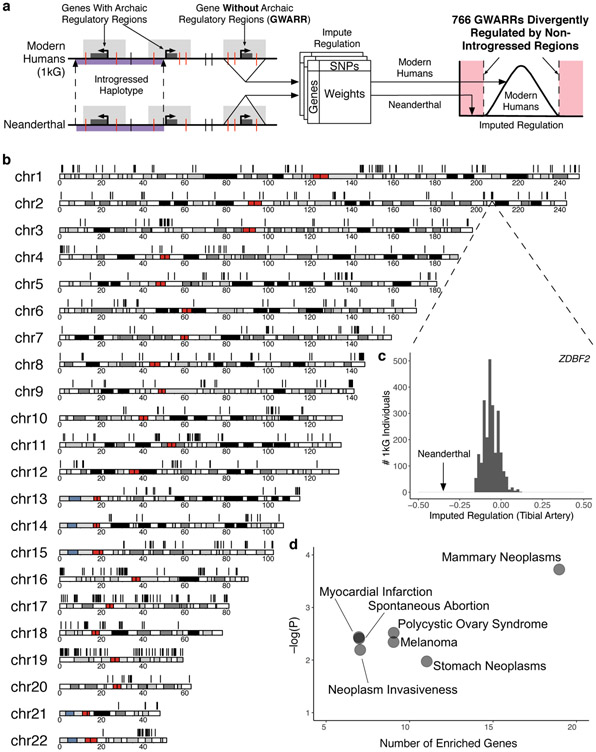
Fig. 2. Neanderthal sequences drive substantial divergent regulation compared to modern humans.
(a) Pipeline for comparing the effects of modern human and archaic hominin DNA on gene regulation in modern humans. We identified genes in modern humans without archaic introgression in their regulatory regions (GWARRs). We compared the imputed gene regulatory effects of Neanderthal sequences to the regulatory effects of the corresponding human sequences in individuals from the 1000 Genomes Project (1kG). These models do not directly model Neanderthal-specific variants (Supplementary Fig. 2), but most retain significant accuracy when applied to Neanderthal sequences (Supplementary Fig. 3). Genes for which the regulatory effect of the Neanderthal sequence was outside the range of all modern humans were labeled as divergently regulated (DR). (b) 766 GWARRs across the human genome (black lines) are divergently regulated by non-introgressed Neanderthal sequences in at least one tissue. (c) To illustrate the DR pattern, if the Altai Neanderthal sequence surrounding ZDBF2, a GWARR, were present in AMH genomes, it is predicted to drive regulation in tibial artery significantly lower than levels observed for all modern humans in 1kG (imputed regulation = −0.376, P = 0). Furthermore, the patterns of imputed regulation of ZDBF2 are similar across all populations (Supplementary Fig. 4), indicating little divergence has occurred in its regulation in more recent evolutionary history. (d) DR GWARRs are enriched for roles in several diseases, including spontaneous abortion, myocardial infarction, and melanoma, compared to all DR genes (FDR < 0.1, hypergeometric enrichment test on DisGeNET annotations).
Recent work has raised concerns about the accuracy of predictions based on genetic models when applied both across and within human populations due to demographic, environmental, and other confounding factors16-19. We address these concerns in detail in Supplementary Information, and summarize our results here. First, while the models we used decrease in performance when applied across human populations, they maintain substantial accuracy (Supplementary Fig. 1). However, as an additional challenge in our application, PrediXcan cannot directly model the effects of Neanderthal-specific variants, including both non-introgressed Neanderthal-derived variants and ancestral variants fixed on the AMH lineage (Supplementary Fig. 2). While such variants are important to regulation, they make up a small fraction (20%) of all variable sites between AMHs and Neanderthals, and as described in the next paragraph, we demonstrate that models trained on ancestral human sequences have significant accuracy when applied to Neanderthal sequences remaining in modern human genomes.
To estimate the performance of the approach applied to Neanderthal sequences, we used the fact that Eurasian populations contain both human and Neanderthal ancestry sequences in non-GWARRs’ regulatory regions. We first trained additional PrediXcan models for each non-GWARR using only the individuals with no Neanderthal ancestry in the gene’s regulatory region. We then applied these new models, which were trained only on human-ancestry sequences, to individuals with Neanderthal ancestry in the regulatory region and assessed their performance. We found only a 12% reduction in the number of models with significant accuracy compared to training without stratifying by ancestry in skeletal muscle, a representative tissue (Supplementary Fig. 1). Overall, the median decrease in accuracy (relative r2) in models ranged from 23% to 35% across tissues. Furthermore, the models are designed so that unobserved variants are treated as reference alleles, which pulls the prediction to the training population mean. Thus, missing variants may decrease accuracy, but they are unlikely to produce false positives. Altogether, these results indicate that our approach is informative about the regulatory effects of Neanderthal sequences, including thousands that no longer remain in AMHs.
Identifying Neanderthal divergently regulated (DR) genes
We applied the imputation models to each gene’s regulatory region from the high-quality genome sequence of the Altai Neanderthal7. This enabled us to estimate the effects of Neanderthal sequences on the regulation of 8587 GWARRs and 8854 non-GWARRs (Fig. 2A). We compared the gene regulatory effects of the Neanderthal sequence to the distributions observed when applying the same models to the corresponding regulatory regions of 2504 diverse AMH individuals from Phase 3 of the 1000 Genomes (1kG) Project20 and computed empirical P-values for the observed differences (Fig. 2A). Again, since our approach estimates the genetically controlled component of gene expression in AMHs, their output should not be seen as a direct proxy for gene expression (Fig. 1C, Supplementary Fig. 2). Thus, we use difference in the values for a gene between AMHs and Neanderthals as a proxy for differences in the regulatory architecture between the groups. We refer to genes for which the Neanderthal sequence’s value is outside the range observed over all 1kG individuals as Neanderthal divergently regulated (DR) genes (Fig. 2A, Supplementary Fig. 4).
Non-introgressed Neanderthal sequences divergently regulate 766 genes
Across all autosomes, non-introgressed Neanderthal sequences are predicted to divergently regulate 766 GWARRs in at least one tissue (Fig. 2B). We refer to these genes with predicted divergent regulation as DR GWARRs. DR GWARRs are found on all autosomes, with the greatest density on gene-rich chromosome 19 (Fig. 2 B). DR GWARRs are also observed across all tissues in GTEx (Supplementary Fig. 5), and are similarly likely to be upregulated or downregulated by the Neanderthal sequence (Supplementary Table 1).
Neanderthal sequences drive significantly more divergent regulation than observed when comparing sequences from an individual AMH to all others (12.4 times higher than maximum observed for an AMH, P < 0.02; Supplementary Fig. 6). Most genes exhibit similar regulatory effect distributions between human populations (Supplementary Fig. 7), and genes with large population differences are not enriched among DR GWARRs (P = 0.821, Fisher’s exact test). This suggests that the divergent regulation is specific to Neanderthals. Additionally, DR genes have a similar number of Neanderthal-specific alleles in their regulatory regions when compared to non-DR genes, indicating that the amount of unmodeled variation is not driving the differences (Supplementary Fig. 8).
To highlight bodily systems that were not receptive to Neanderthal sequences with divergent regulatory potential, we tested for enrichment of specific disease and phenotype associations among DR GWARRs compared to all DR genes. DR GWARRs were significantly enriched (FDR < 0.1, hypergeometric test on DisGeNET annotations with Benjamini-Hochberg (BH) multiple testing correction) for genes involved in spontaneous abortion, polycystic ovary syndrome, mammary neoplasms, myocardial infarction, melanoma, and stomach neoplasms (Fig. 2 D). Given their potential fitness effects, the DR GWARRs associated with spontaneous abortion (HSD17B1, IFI35, MUC4, IL20RA, TGFBI, TNFSF13, CD7) are of particular interest for further investigation.
We also tested for enrichment of Human Phenotype Ontology (HPO) annotations among DR GWARRs. While it did not pass multiple testing correction, the strongest enrichment was for genes involved in pectus carinatum, a deformity of the chest caused by overgrowth of the ribs and characterized by protrusion of the sternum (P = 4.3E-4; HP:0000768: GNPTG, HBA1, HBA2, MYH11, ORC4, SOS1, TNFRSF11B). The top associations also included other phenotypes that mirror physiological differences between humans and Neanderthals such as supraorbital ridge development (HP:0009891; HBA1, HBA2, PEX11A, and PEX13). Furthermore, many individual DR GWARRs function in human-specific phenotypes, including reproduction, neurotransmitter transport, circadian rhythm, and language (Supplementary Information). Overall, the large number of DR GWARRs suggests that there were substantial differences in gene regulation between modern humans and Neanderthals.
Divergent regulation of GWARRs is associated with clinical phenotypes in AMHs
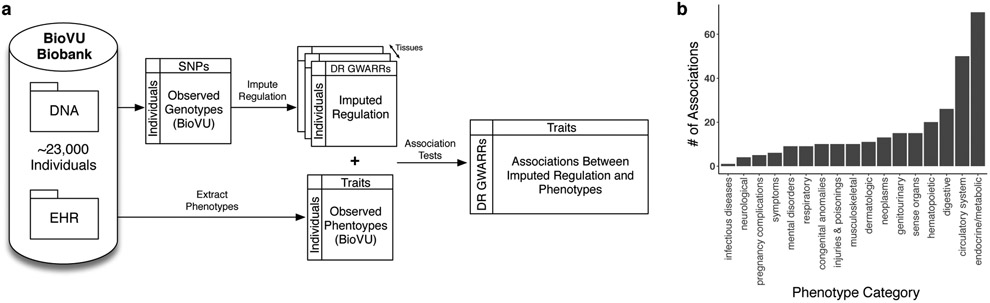
To gain further insight into organism-level effects of divergent regulation of GWARRs in modern humans, we quantified the association of their imputed regulation with clinical phenotypes using BioVU, Vanderbilt University’s biobank of patient DNA samples linked to de-identified EHRs. We used logistic regression to test for associations between the imputed regulatory profiles of Neanderthal DR GWARRs with phenotypes derived from the EHRs of ~23,000 individuals of European descent (Fig. 3A).
Fig. 3. Modern human variation in the regulation of GWARRs is associated with clinical phenotypes.
(a) Pipeline for associating variation in gene regulation with diverse clinical phenotypes. Using Vanderbilt’s BioVU biobank, human regulation of genes divergently regulated by non-introgressed Neanderthal sequences (DR GWARRs; Fig. 2) were imputed across ~23,000 European ancestry individuals. Combining these genes’ imputed regulation and phenotypes extracted from electronic health records (EHRs), we associated differences in imputed regulation to disease status using logistic regression controlling for standard covariates (Methods). (b) The number of associations between Neanderthal DR GWARRs and phenotypes in different phenotype categories at P < 1E–5. The endocrine and metabolic disorders phenotype category had the largest number of associations driven by many associations with T1D and other autoimmune diseases (Table 1). However, we caution against comparing across categories due to differences in sample size and power.
Variation in DR GWARR regulation in BioVU is associated with many phenotypes (22 at P < IE-7 and 284 at P < IE-5) across a broad range of phenotypic categories (Fig. 3B). The strongest associations include (Table 1): MSH5, PRSS16, VARS, and NCR3 with type 1 diabetes (T1D, Phecode: X250.1*; P = 1.3E-11, 5.2E-8, 7.1E-8, 8.0E-8, respectively), C11orf65 with transient mental disorders (Phecode: X291.1; P = 3.1E-9), SPINT1 with pulmonary embolism and infarction (Phecode: X452.1; P = 7.2E-8), and PSRC1 with hyperlipidemia (Phecode: X272.1; P = 3.1E-8). With the exception of C11orf65, each of these genes is known to function in pathways relevant to the associated diseases (Supplementary Information). Each of the genes associated with T1D is located in or proximal to the human major histocompatibility complex (MHC) locus on chromosome 6. Certain MHC alleles may have been acquired through adaptive introgression21; our results suggest that variation in other regions of the MHC that were not receptive to introgression is associated with disease. Driven by the large number of associations with T1D and other autoimmune diseases, the endocrine and metabolic disorders phenotype category had the largest number of associations (Fig. 3B), but the raw number of associations is difficult to compare across categories due to differences in sample size, power, and between-phenotype correlations. Furthermore, the directions of effect for these associations do not always suggest that regulation by the Neanderthal haplotype increases risk. Nonetheless, divergent regulation of genes for which Neanderthal sequences likely altered regulation is associated with risk for clinical phenotypes in modern human populations. This highlights genes and bodily systems for which the lack of Neanderthal ancestry near genes may be due to divergent gene regulatory function.
Table 1. Strongest associations between imputed regulation in BioVU and EHR-derived phenotypes for DR GWARRs.
Each gene associated with T1D is located in or near the MHC locus.
| Trait | Gene | Beta | P-value |
|---|---|---|---|
| Type 1 Diabetes | MSH5 | 3.81 | 1.28 × 10−11 |
| Transient Mental Disorders | C11orf65 | 11.6 | 3.14 × 10−09 |
| Hyperlipidemia | PSRC1 | −0.36 | 3.06 × 10−08 |
| Type 1 Diabetes with ophthalmic manifestations | PRSS16 | 1.33 | 5.20 × 10−08 |
| Type 1 Diabetes | VARS | 0.66 | 7.06 × 10−08 |
| Pulmonary Embolism | SPINT1 | 6.76 | 7.15 × 10−08 |
| Type 1 Diabetes with renal manifestations | NCR3 | 2.87 | 7.99 × 10 −08 |
Overall, the functions of DR GWARRs observed in the enrichment and biobank analyses suggest effects on a range of phenotypes, including reproductive, skeletal, cardiovascular, and immune traits. These systems are also influenced in AMHs by introgressed Neanderthal sequences10,22. This is consistent with a model in which these systems differed between Neanderthals and AMHs, and the genetic variants influencing these differences potentially had a range of fitness effects in the AMH context.
Genes in introgression deserts are not more likely to be divergently regulated
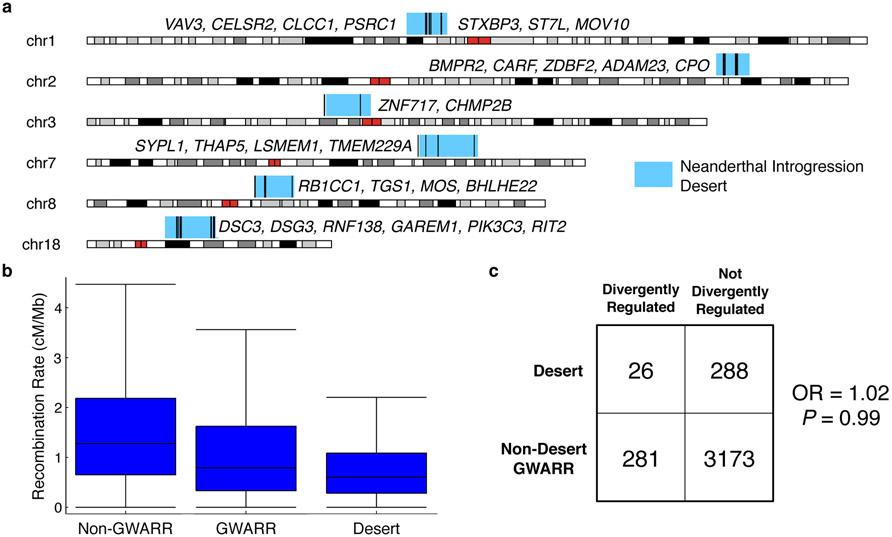
Given the potential importance of introgression deserts—long regions of the human genome significantly depleted of archaic ancestry—to human-specific biology, we examined the potential for divergent regulation by Neanderthal sequences among genes in six previously defined introgression deserts of greater than 8 Mb (Fig.4)8. Each desert contained at least one DR GWARR, and the deserts contained a total of 26 DR GWARRs. DR desert genes have been implicated—either in previous work or our biobank association tests—with a variety of traits important to humanness, including neural development (CELSR2, CHMP2B)23-25 and learning and spatial memory (CARF)26.
Fig 4. Genes in introgression deserts exhibit divergent regulation between modern humans and Neanderthals.
(a) Location of Neanderthal introgression deserts (blue boxes) and desert genes divergently regulated by Neanderthal sequences (black lines). Neanderthal DR genes are listed next to each desert. These genes have functions in a range of traits important to humanness, including neural development (CELSR2,CHMP2B) and spatial memory (CARF). (b) Recombination rate is significantly lower near genes (+/− 2 Mb) in introgression deserts than near other GWARRs or genes with archaic regulatory regions (Kruskal-Wallis test P ~ 0, Dunn’s post hoc analysis P ≈0.0). Box plots show the median, inner quartiles, and 95% confidence intervals, (c) Desert GWARRs are not significantly more likely to be DR compared to other GWARRs, even after controlling for recombination rate (OR = 1.02; Fisher’s Exact testP = 0.99).
Desert genes are not significantly more likely to be divergently regulated than other GWARRs (P = 0.60, permutation test). However, deserts have significantly lower recombination rates than other regions (Fig. 4 B), and the deserts also have significantly lower gene densities. Controlling for these factors, there was still no significant difference in the likelihood of desert genes being DR than other GWARRs (Fig. 4 C; matched recombination rate OR = 1.02; Fisher’s exact test P = 0.99; matched gene density OR = 1.04, P = 0.96). Recent work suggests that recombination rate influences the retention of introgressed sequences27, so it is possible that selection against a small number of diverged and deleterious regulatory Neanderthal haplotypes in these low recombination rate regions could have contributed to the formation of introgression deserts.
Imputing gene regulation in multiple archaic hominins
Due to the rapid degradation of most tissues and RNA, we are unlikely to ever be able to study gene expression levels directly from archaic samples. Archaic methylation status can be imputed for some regions of the genome28, but this approach is limited to the bone cells from which archaic DNA can be extracted. In the previous analyses, we focused on the gene regulatory effects of Neanderthal DNA in the AMH genomic context. However, comparing gene regulatory profiles from archaic hominins directly may also reveal attributes of tissues in archaic hominins and their differences from one another. This approach is particularly promising for groups, like the Denisovans, that lack a substantial fossil record.
We expanded our analysis and imputed the regulation of all genes in the high-quality genomes of the Altai Neanderthal, a Neanderthal from Vindija, Croatia3, and a Denisovan from the Altai cave29. To enable direct comparison, we reanalyzed the Altai Neanderthal using the smaller set of variants called in both Neanderthal genomes; the resulting imputed Altai values were concordant between the two variant sets (average Spearman ρ=0.81; Supplementary Fig. 9).
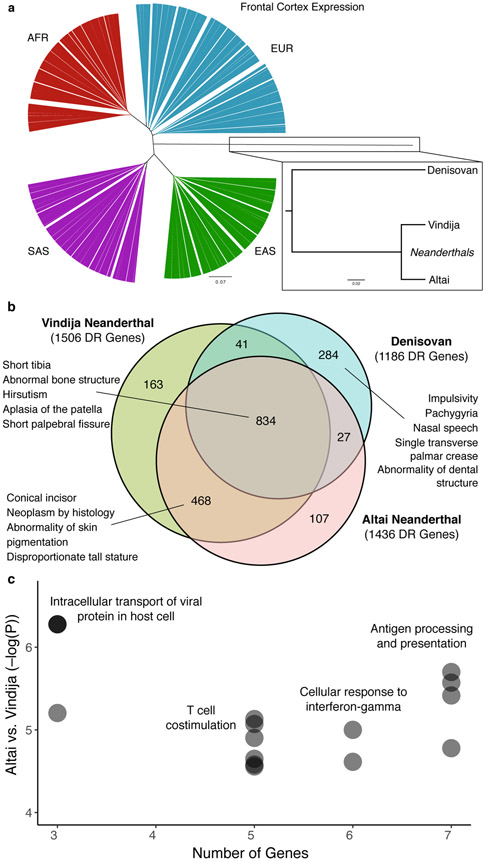
To obtain a global view of the similarity of regulatory patterns across tissues for each archaic individual compared to modern human populations from 1kG, we hierarchically clustered individuals based on the Pearson correlation of their regulatory profiles for all genes analyzed in each tissue. This revealed that, as expected, the three archaic individuals are closer to one another than to any AMH (Fig. 5A). Also, as expected, despite being separated by more than 50,000 years and nearly 5,000 kilometers, the two Neanderthals’ imputed regulatory profiles are more similar to one another than to the Denisovan (Fig. 5A, inset). Modern humans consistently group by continental population and all pairs of humans are more similar to one another than to any of the archaic individuals. Thus, the divergence of regulatory patterns in the archaic samples reflects our understanding of their evolutionary relationships with respect to one another and AMHs. These results held across all tissues analyzed and when we separated genes by the presence of archaic ancestry in their regulatory regions (Supplementary Figs. 10 and 11). We view these trees as a qualitative sanity check and caution against quantitative interpretation of the branch lengths as they are influenced by selective and demographic factors30, as well as unmodeled archaic alleles (Supplementary Fig. 2).
Fig. 5. Comparison of genome-wide regulatory profiles between two Neanderthals, a Denisovan, and modern humans.
(a) Hierarchical clustering of imputed gene regulation for all genes in the frontal cortex of archaic hominins and modern human populations from 1kG. Patterns are similar across all tissues (Supplementary Fig. 10) and when stratifying by presence of archaic ancestry in regulatory regions (Supplementary Fig. 11). (b) Venn diagram of divergently regulated genes identified in each archaic hominin vs. all AMHs. Examples of the top 10 enriched Human Phenotype Ontology annotations among genes divergently regulated in all archaic individuals, in both Neanderthals, and in the Denisovan are shown. All terms are given in Supplementary Tables 2-4. (c) Enrichment for GO Biological Process annotations among the 75 genes with the largest deviation (>1 standard deviation) in imputed regulation between the Altai and Vindija Neanderthals. Immune functions are significantly enriched for differences between the two Neanderthals. Only enrichments with FDR < 0.05 are plotted.
Differences in regulation between archaic hominins reflect potential phenotypic differences
To identify specific differences in gene regulation between AMHs and the archaic groups, we determined divergently regulated genes in each archaic hominin compared to AMHs and tested for enrichment of phenotype annotations from the HPO (Fig. 5B). Across all tissues, 97% of DR genes in the Altai Neanderthal were also DR in Vindija with the same direction of effect. Genes divergently regulated in all three archaic individuals compared to AMHs were nominally enriched for associations with short tibia (7.15x, P = 0.0017, hypergeometric test), abnormal bone structure (1.62x, P = 0.0034), hirsutism (2.61x, P = 0.0042), and many other traits (Supplementary Table 2). DR genes specific to Neanderthals and the Denisovan were both nominally enriched for phenotypes involving dental morphology (Supplementary Tables 3 and 4). The Neanderthal-specific DR set also included genes involved in skin pigmentation (1.96x, P = 0.0058) and stature (7.62x, P = 0.0063). The repeated enrichment for genes involved in skeletal and dental morphology is striking given the known differences between modern and archaic hominins in these traits. DR genes specific to the Denisovan were uniquely enriched for several phenotypes including impulsivity, cerebral cortex development (pachygyria and lissencephaly), hand morphology, and nasal speech (Supplementary Table 4). The potential Denisovan-specific differences in speech are further supported by recent results based on imputed DNA methylation changes31. However, we note that due to the large number of phenotype categories these associations did not pass FDR-based multiple testing correction. Collectively, these analyses highlight genes involved in known morphological differences between archaic hominins and AMHs and suggest additional phenotypic differences that cannot be directly studied from fossils.
To identify differences in regulation between the archaic individuals without comparison to AMHs, we analyzed genes with large magnitude (>1 SD of the GTEx distribution) differences in regulation between archaic individuals. As expected, the two Neanderthals have the fewest differences (75 genes vs. ~950 for each compared to the Denisovan). Immune response functions are significantly overrepresented among the genes different between the Neanderthals (Fig. 5C; FDR < 0.05), including transporting viral proteins (366.8x, P = 0.0015, hypergeometric test) and cellular response to interferon-gamma (12.03x, P = 0.0085). These 75 genes include 5 MHC class II genes. This suggests that gene regulatory differences between the two Neanderthals influenced immune function, possibly reflecting adaptations in these populations. The genes that differed in the Denisovan compared to both Neanderthals are associated with many more general terms. Altogether, these results identify thousands of candidate genes for which regulation has likely diverged between archaic hominins and modern humans.
Discussion
Our application of PrediXcan to archaic genomes is a powerful approach for studying the evolution of gene regulation and the biology of archaic groups. The molecular machinery and genetic architecture of gene regulation are largely conserved across humans, and most common human regulatory variants have similar effects across populations32,33. Our approach enabled us to study the regulation of many genes by archaic hominin sequences. However, accurate predictions cannot be made for all genes in all populations, especially for genes with regulatory architectures dominated by rare variants34,35 or trans effects36. Furthermore, since the imputation models are trained in modern humans, they do not incorporate the effects of archaic-specific alleles not present in human populations (Supplementary Fig. 2). Thus, it is likely for some genes that archaic-specific alleles could further modulate regulation. In these cases, the imputed effects are likely less accurate than in human populations, but any predicted deviations would still indicate divergence in regulatory architecture between archaic and AMH groups. As our understanding of the relationship between genotype and gene regulation improves and more tissues are characterized, our approach will enable testing of additional hypotheses about aspects of archaic hominin biology that are inaccessible to direct study.
In summary, there was substantial divergence in gene regulation between archaic hominins and modern humans. The affected genes influence a range of traits, including reproduction, skeletal development, language, and the immune system. Applying the regulation imputation models to a large, EHR-linked human biobank cohort further enabled the connection of divergent gene regulatory patterns with clinical phenotypes in modern human populations, in particular with autoimmune and cardiovascular disease. Our results suggest that divergent regulation may have been a barrier to Neanderthal introgression in some regions of the human genome; however, more work is needed to demonstrate this. We additionally show that imputing ancient gene regulatory profiles has promise for studying ancient phenotypes. This approach is also potentially applicable to more recent ancient human genomes, where there is less sequence divergence than among Neanderthals and AMHs, and could provide an opportunity to characterize gene regulation across diverse geographical and temporal ranges.
Methods
Modern and Archaic Genetic Data
We analyzed the high-coverage genome sequences of three archaic hominins. For most comparisons to modern humans, we used the high quality archaic genome from an ~122,000-year-old Neanderthal individual found in the Altai mountains (“Altai Neanderthal”)7, which was sequenced to 52x coverage and enabled PrediXcan analysis of the largest number of genes. For the comparisons that included multiple archaic individuals, we analyzed the 30x genome from a ~72,000-year-old Denisovan from the Altai mountains (“Denisovan”)29, and a 30x coverage genome of a ~52,000-year-old Neanderthal from Croatia (“Vindija Neanderthal”)3. For all three genomes, we considered only autosomal SNPs from the publicly available genomes.
To represent modern humans, we analyzed the genomes of 2504 individuals sequenced by the 1000 Genomes Project (1kG) and released in Phase 320. These include individuals from the European (EUR), African (AFR), East Asian (EAS), South Asian (SAS), and Admixed American (AMR) continental ancestry super-populations.
PrediXcan Gene Regulation Imputation Models
We considered PrediXcan models across 44 tissues from the PredictDB Data Repository (http://predictdb.org/; accessed Nov. 16, 2016). The models were trained on GTEx V6p using variants identified by 1kG (Phase 1) within 1 Mb of the gene. We considered only those models that explained a significant amount of variance in gene expression in each tissue (FDR < 0.05); this left us with 17,748 unique genes with an accurate model in at least one tissue (159,368 models total)14. We abbreviate the 44 tissues considered as follows: Adipose - Subcutaneous: ADPS, Adipose - Visceral Omentum: ABPV, Adrenal Gland: ADRNLG, Artery - Aorta: ARTA, Artery - Coronary: ARTC, Artery - Tibial: ARTT, Brain - Anterior Cingulate Cortex: BRNACC, Brain - Caudate: BRNCDT, Brain - Cerebellar Hemisphere: BRNCHB, Brain - Cerebellum: BRNCHA, Brain - Cortex: BRNCTX, Brain - Frontal Cortex: BRNFCTX, Brain - Hippocampus: BRNHPP, Brain - Hypothalamus: BRNHPT, Brain - Nucleus Accumbens basal ganglia: BRNNCC, Brain - putamen basal ganglia: BRNPTM, Breast: BREAST, Cells - Transformed Fibroblasts: FIBS, Colon - Sigmoid: CLNS, Colon - Transverse: CLNT, Esophagus - Gastroesophageal Junction: ESPGJ, Esophagus - Mucosa: ESPMC, Esophagus - Muscularis: ESPMS, Heart - Atrial Appendage: HRTAA, Heart - Left Ventricle: HRTLV, Liver: LIVER, Lung: LUNG, Cells- EBV-transformed Lymphocytes: LYMPH, Ovary: OVARY, Pancreas: PNCS, Pituitary: PTTY, Prostate: PRSTT, Skeletal Muscle: MSCSK, Skin - Not sun-exposed: SKINNS, Skin - Sun-exposed: SKINS, Small Intestine: SMINT, Spleen: SPLEEN, Stomach: STMCH, Testis: TESTIS, Thyroid: THYROID, Tibial Nerve: NERVET, Uterus: UTERUS, Vagina: VAGINA, Whole Blood: WHLBLD.
Imputation of Archaic Hominin and Modern Human Gene Regulation
Using the PrediXcan prediction program available from PredictDB, we applied the accurate prediction models to the relevant portions of the genome of the Altai Neanderthal to impute the effects of its sequence on gene regulation. The resulting predictions are normalized values in reference to the distribution observed in GTEx individuals used to train the original prediction models. To characterize regulatory patterns in modern human populations, we applied the same PrediXcan models to 2504 individuals from the 1kG20.
For all cross-archaic comparisons, we applied the same models to the sequenced Vindija Neanderthal, the Altai Neanderthal, and Denisovan, which were all recently processed with the same pipeline3. Imputed regulation based on the previous and new variant calls for the Altai Neanderthal were strongly correlated (0.78–0.85 across tissues; Supplementary Fig. 9).
Identification of Genes Divergently Regulated by Archaic Sequences
To identify genes divergently regulated by archaic compared to modern human sequences, we calculated an empirical P-value for the archaic predicted regulatory profile for each gene and tissue by calculating the proportion of modern humans who had a predicted value farther from the median of the full 1kG distribution for the tissue. Genes for which the archaic sequence is predicted to drive regulation completely outside the distribution observed in 1kG in at least one tissue were considered significantly divergently regulated (DR) genes (N = 2290), 766 of these were GWARRs (see next section for more on the GWARR definition). We plotted gene locations using karyoploteR37. We excluded all genes which were missing genotype calls at SNPs of at least one model.
Assessment of Imputation Accuracy on Neanderthal Sequences
It is not possible to directly assess the accuracy of gene regulation imputation models trained in AMH when applied to Neanderthal sequences. Therefore, we took several approaches to estimate prediction performance on diverged populations and its influence on downstream analyses.
First, we took advantage of the presence of many AMH individuals with Neanderthal-introgressed regulatory sequences for some genes. For each gene in which some GTEx individuals had Neanderthal ancestry within the region considered by the model (non-GWARRs), we trained a new “Introgressed Excluded” prediction model using only individuals without Neanderthal introgression in the regulatory region. (We considered variants in perfect LD (r2 = 1.0) with a Neanderthal tag variant as of Neanderthal ancestry.) These models mimic the situation for GWARRs; they are trained only on sequences without Neanderthal ancestry. We then applied each model to the individuals with Neanderthal ancestry and calculated the r2 for the PrediXcan prediction versus observed expression (Supplementary Fig. 2). A model was considered “imputable” if its output was significantly correlated with the observed expression (r > 0.1, P < 0.05). For each non-GWARR and tissue, we also calculated the relative performance of the model trained only on human-ancestry sequences (Introgressed Excluded) to that of the model trained on all sequences. In particular, we compared the human-only r2 to the r2 obtained by models trained on both human and Neanderthal ancestry sequences (r2Introgression-Excluded/r2All). Since the number of individuals without introgression in the regulatory region varies from gene to gene, we retrained the “All” models on a random set of individuals that matched the number without introgression. When testing prediction performance in the individuals with introgressed Neanderthal regulatory regions (the testing set), we required at least 50 individuals to ensure power to estimate performance. The r2 over all individuals with Neanderthal ancestry at the locus is shown as a large dot in Supplementary Fig. 2. We also resampled 50% of the testing set individuals 99 times and computed the r2 for each subsample to estimate the distribution of relative performance.
We also tested how well the PrediXcan models, which were trained on the primarily European ancestry GTEx population, generalize across populations. We applied PrediXcan models trained on GTEx LCLs to expression and genotype information from LCLs derived from 1kG European ancestry populations (CEU, GBR, FIN, TSI) and a sub-Saharan African population (YRI)38. We then compared the accuracy (r2 between observed and predicted values) between European and African ancestry individuals (Supplementary Fig. 3).
To estimate how much Neanderthal-specific variation the PrediXcan models trained on AMHs could be missing, we counted the number of Neanderthal-specific alleles present in the regulatory region of each gene (1 Mb up and downstream). For this analysis, Neanderthal-specific sites include any site where the Altai Neanderthal had at least one allele not observed in 1kG. To account for different overall evolutionary rates between genes, we computed the relative amount of Neanderthal-specific variation for each gene by dividing it by the total number of variants (Neanderthal-specific alleles plus all variable sites in 1kG). We then compared relative levels of Neanderthal-specific variation between DR and non-DR genes.
Divergent Regulation Between Humans
To aid interpretation of the number of divergently regulated genes observed with archaic sequences, we called DR genes in 50 random 1kG individuals, 10 from each continental population, using the same criteria as for archaic sequences: imputed regulation outside the range for all other 1kG individuals. For each population, we compared the distribution of the number of DR genes in each individual with the number identified in Neanderthal (Supplementary Fig. 6).
We also examined the stability of the imputed values across all 1kG populations. For all PrediXcan models in all tissues, we computed the median imputed regulation for each 1kG population. We then found the maximum difference between populations (Supplementary Fig. 7). Only 2.7% of all gene models have a maximum difference in population median regulation greater than 1 SD.
Classification and Comparison of Non-Introgressed and Desert Genes
We used the S*-based Neanderthal introgression map from Vernot et al.8 to identify the overlap between variants considered in gene regulation prediction models and introgressed sequences. After filtering out models that had no variants present in the Altai genome, we classified genes to be genes without archaic regulatory regions (GWARRs) if none of the variants considered in their prediction models were Neanderthal tag SNPs or in linkage disequilibrium (r2 > 0.8) with Neanderthal tag SNPs in Europeans (N = 8587). Genes with at least one introgressed Neanderthal SNP in their model were classified as “non-GWARRs.”
We also analyzed the effects of genes in introgression deserts that were recently identified using coalescent simulations based on demographic models8. By this definition, deserts are long regions where modern humans lack introgressed sequence. Desert regions >8 Mb long are significantly more common than expected from simulations, and they also exhibit higher levels of background selection. In our analyses, “desert” genes are the subset of GWARRs for which variants in their regulatory effect prediction models overlap the bounds of an introgression desert, excluding those that also include SNPs on introgressed haplotypes (N = 311).
We calculated the enrichment of DR GWARRs within and outside deserts by shuffling GWARR locations across the genome, constrained by chromosome. For each of 1000 permutations, we counted the number overlapping a desert to compute an empirical p-value. To evaluate DR enrichment accounting for recombination rate, we first calculated the recombination rate in 250 kb non-overlapping windows across the entire genome, using recombination maps calculated in African Americans27,39. We then intersected those windows with the regulatory region considered by PrediXcan for each gene. For each gene, we calculated the mean recombination rate across all windows overlapping the gene region, weighted by the number of base-pairs of overlap. We then binned genes by recombination rate (31 equal-width bins) and randomly selected 3454 GWARRs such that the overall distribution across bins was equal to the distribution of desert genes (the maximum without emptying a bin).We then performed a Fisher’s Exact test on DR status in desert genes vs. the recombination rate-matched GWARRs. To match by gene density, for each gene we counted the number of genes that overlapped the region considered by PrediXcan (1 Mb flanking on either side). We then repeated the binning and Fisher’s Exact test analyses as for recombination rate.
We identified gene regions overlapping human accelerated regions as those genes with at least one HAR within 1Mb 40. We then computed an odds ratio to assess the likelihood of certain classes of genes to be nearby a HAR compared to others.
Association and Enrichment Between Divergent Regulation and Phenotypes
To investigate potential phenotypic implications of DR GWARRs, we conducted two main analyses: gene set enrichment analysis and PrediXcan on Vanderbilt’s BioVU biobank.
To test for enrichment of genes known to be involved in particular human phenotypes or diseases, we performed gene set overrepresentation enrichment analysis on Disgenet disease annotations and human phenotype ontology terms between DR GWARRs and other DR genes using WebGestalt42. We used the hypergeometric test with BH multiple testing correction, a false discovery rate (FDR) threshold of 10%, and did not consider disease categories with fewer than 10 genes.
To explore the systems potentially impacted by divergent regulation of DR GWARRs in modern human populations, we used the PredixVU system at Vanderbilt University Medical Center to discover associations between predicted regulation and clinical phenotypes. The phenotypes were extracted from de-identified electronic medical records using ICD-9 codes that were organized into PheWAS codes in 17 groups (https://phewascatalog.org/phecodes) and were linked to genotypes from the BioVU biobank. In total, this involved ~23,000 subjects of European descent; the total number of cases and controls for each phenotype varied (on average 780 cases, 17176 controls). We considered only phenotypes with case counts greater than 30. The models used to impute regulation for these individuals were trained on HapMap SNPs, and there is a high correlation between the imputed values for the models trained on HapMap and 1kG14. For each phenotype, we used logistic regression to regress imputed regulation onto phenotype status, and included age, sex, and genetic principal components (3 for Europeans, 10 for African Americans) as covariates. N.B. associations between divergent regulation of a gene and a phenotype in this context are not necessarily in the same direction as the divergence in the Neanderthal.
Comparing Gene Regulation Among Archaic Hominins
To visualize global similarities between different groups for each tissue, we compared the imputed regulatory profiles of non-admixed 1kG populations (excluded: MXL, CLM, PUR, ACB, ASW, PJL, PEL) and the three archaic hominins. We hierarchically clustered each individual for each tissue using Pearson correlation on imputed regulation across all genes as the distance metric. We visualized the resulting trees using FigTree (Fig. 4A; Supplementary Fig. 10) (http://tree.bio.ed.ac.uk/software/figtree/). Results were similar when using Spearman correlation (Supplementary Fig. 12) and when stratifying by GWARR status (Supplementary Fig. 11).
To identify specific genes of interest to differences between the archaic groups, we generated lists of genes divergently regulated between the Altai Neanderthal, the Vindija Neanderthal, and the Denisovan. First, we called DR genes versus the 1kG individuals for each archaic individual, and then intersected the DR genes (Fig. 4B). We then conducted gene set ORA over the Human Phenotype Ontology using WebGestalt42, using only categories containing at least 10 genes.
To focus on genes with the largest differences in regulation, we computed the difference in predicted regulation between pairs of archaic individuals for each gene in each tissue for which it had an accurate model. We then picked genes that differed in imputed regulatory effect by greater than 1 (i.e., were >1 standard deviation apart with respect to the distribution of the GTEx training population). To identify general biological processes influenced by these genes that differed between the archaic hominins, we conducted gene set enrichment analyses on GO biological process terms versus the full GTEx project gene list using WebGestalt42.
Data Availability
All data reported in this paper are available in the project’s github repository (https://github.com/colbrall/neanderthal_predixcan_manuscript).
Code Availability
All code used in this paper is available from the project’s github repository (https://github.com/colbrall/neanderthal_predixcan_manuscript).
Supplementary Material
Acknowledgments
We thank Corinne Simonti, David Rinker, Mary Lauren Benton, Nicole Creanza, Emily Hodges, and Sharron Francis for helpful discussions and comments on the manuscript. L.L.C. was funded by NIH grant T32GM080178 to Vanderbilt University. J.A.C. was funded by NIH grants R01GM115836 and R35GM127087, the March of Dimes Prematurity Research Ohio Collaborative, and the Burroughs Wellcome Fund. E.R.G. acknowledges support from R01MH101820, R01MH090937, and R01MH113362, and benefited immensely from a Fellowship at Clare Hall, University of Cambridge.
This work was conducted in part using the resources of the Advanced Computing Center for Research and Education at Vanderbilt University, Nashville, TN, and was based in part on data from the PredixVU system of Vanderbilt University Medical Center. The research is solely the responsibility of the authors and does not necessarily represent the views of Vanderbilt University Medical Center.
Footnotes
Competing Interests
The authors report no conflicts interest.
References
- 1.Green RE et al. A draft sequence of the neandertal genome. Science 328, 710–722 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reich D et al. Genetic history of an archaic hominin group from Denisova cave in Siberia. Nature 468, 1053–1060 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Prüfer K et al. A high-coverage Neandertal genome from Vindija Cave in Croatia. Science 358, 655–658 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hajdinjak M et al. Reconstructing the genetic history of late Neanderthals. Nature 555, 652 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wolf AB & Akey JM Outstanding questions in the study of archaic hominin admixture. PLOS Genet. 14, e1007349 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sawyer S et al. Nuclear and mitochondrial DNA sequences from two Denisovan individuals. Proc. Natl. Acad. Sci 112, 15696 LP–15700 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prüfer K et al. The complete genome sequence of a Neanderthal from the Altai Mountains. Nature 505, 43–49 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vernot B et al. Excavating Neandertal and Denisovan DNA from the genomes of Melanesian individuals. Science 352, 235–239 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sankararaman S, Mallick S, Patterson N & Reich D The Combined Landscape of Denisovan and Neanderthal Ancestry in Present-Day Humans. Curr. Biol 26, 1241–1247 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Simonti CN et al. The phenotypic legacy of admixture between modern humans and Neandertals. Science 351, 737–741 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Castellano S et al. Patterns of coding variation in the complete exomes of three Neandertals. Proc. Natl. Acad. Sci 111, 6666–6671 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dannemann M, Prüfer K & Kelso J Functional implications of Neandertal introgression in modern humans. Genome Biol. 18, 61 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McCoy RC, Wakefield J & Akey JM Impacts of Neanderthal-Introgressed Sequences on the Landscape of Human Gene Expression. Cell 168, 916–927.e12 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gamazon ER et al. A gene-based association method for mapping traits using reference transcriptome data. Nat Genet 47, 1091–1098 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.GTEx Consortium. Genetic effects on gene expression across human tissues. Nature 550, 204–213 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martin AR et al. Human demographic history impacts genetic risk prediction across diverse populations. Am. J. Hum. Genet 100, 635–649 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martin AR et al. Clinical use of current polygenic risk scores may exacerbate health disparities. Nat. Genet 51, 584–591 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim MS, Patel KP, Teng AK, Berens AJ & Lachance J Genetic disease risks can be misestimated across global populations. Genome Biol. 19, 179 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mostafavi H, Harpak A, Conley D, Pritchard JK & Przeworski M Variable prediction accuracy of polygenic scores within an ancestry group. bioRxiv (2019). doi: 10.1101/629949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.The 1000 Genomes Project Consortium. A global reference for human genetic variation. Nature 526, 68–74 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abi-Rached L et al. The Shaping of Modern Human Immune Systems by Multiregional Admixture with Archaic Humans. Science 334, 89–94 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dannemann M & Kelso J The Contribution of Neanderthals to Phenotypic Variation in Modern Humans. Am. J. Hum. Genet 101, 578–589 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wada H, Tanaka H, Nakayama S, Iwasaki M & Okamoto H Frizzled3a and Celsr2 function in the neuroepithelium to regulate migration of facial motor neurons in the developing zebrafish hindbrain. Development 133, 4749 LP – 4759 (2006). [DOI] [PubMed] [Google Scholar]
- 24.Skibinski G et al. Mutations in the endosomal ESCRTIII-complex subunit CHMP2B in frontotemporal dementia. Nat. Genet 37, 806 (2005). [DOI] [PubMed] [Google Scholar]
- 25.Cox LE et al. Mutations in CHMP2B in Lower Motor Neuron Predominant Amyotrophic Lateral Sclerosis (ALS). PLoS One 5, e9872 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McDowell KA et al. Reduced cortical BDNF expression and aberrant memory in Carf knock-out mice. J. Neurosci 30, 7453–7465 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schumer M et al. Natural selection interacts with the local recombination rate to shape the evolution of hybrid genomes. Science 3684, 212407 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gokhman D et al. Reconstructing the DNA methylation maps of the Neandertal and the Denisovan. Science 1250368 (2014). [DOI] [PubMed] [Google Scholar]
- 29.Meyer M et al. A high-coverage genome sequence from an archaic Denisovan individual. Science 338, 222–226 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brawand D et al. The evolution of gene expression levels in mammalian organs. Nature 478, 343–348 (2011). [DOI] [PubMed] [Google Scholar]
- 31.Gokhman D et al. Extensive Regulatory Changes in Genes Affecting Vocal and Facial Anatomy Separate Modern from Archaic Humans. bioRxiv (2017). doi: 10.1101/106955 [DOI] [Google Scholar]
- 32.Martin AR et al. Transcriptome Sequencing from Diverse Human Populations Reveals Differentiated Regulatory Architecture. PLoS Genet. 10, 1004549 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kelly DE, Hansen MEB & Tishkoff SA Global variation in gene expression and the value of diverse sampling. Curr. Opin. Syst. Biol 1, 102–108 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hernandez RD et al. Singleton Variants Dominate the Genetic Architecture of Human Gene Expression. bioRxiv 219238 (2017). [Google Scholar]
- 35.Glassberg EC, Gao Z, Harpak A, Lan X & Pritchard JK Evidence for Weak Selective Constraint on Human Gene Expression. Genetics 211, 757–772 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu X, Li YI & Pritchard JK Trans Effects on Gene Expression Can Drive Omnigenic Inheritance. Cell 177, 1022–1034.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gel B & Serra E karyoploteR: an R/Bioconductor package to plot customizable genomes displaying arbitrary data. Bioinformatics 33, 3088–3090 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.The 1000 Genomes Project Consortium. An integrated map of genetic variation from 1,092 human genomes. Nature 491, 56–65 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hinch AG et al. The landscape of recombination in African Americans. Nature 476, 170–175 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Doan RN et al. Mutations in Human Accelerated Regions Disrupt Cognition and Social Behavior. Cell 167, 341–354.e12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lek M et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 536, 285–91 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang J, Vasaikar S, Shi Z, Greer M & Zhang B WebGestalt 2017: a more comprehensive, powerful, flexible and interactive gene set enrichment analysis toolkit. Nucleic Acids Res. 45, W130–W137 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data reported in this paper are available in the project’s github repository (https://github.com/colbrall/neanderthal_predixcan_manuscript).