Abstract
Malaria is caused by Plasmodium parasite. It is transmitted by female Anopheles bite. Thick and thin blood smears of the patient are manually examined by an expert pathologist with the help of a microscope to diagnose the disease. Such expert pathologists may not be available in many parts of the world due to poor health facilities. Moreover, manual inspection requires full concentration of the pathologist and it is a tedious and time consuming way to detect the malaria. Therefore, development of automated systems is momentous for a quick and reliable detection of malaria. It can reduce the false negative rate and it can help in detecting the disease at early stages where it can be cured effectively. In this paper, we present a computer aided design to automatically detect malarial parasite from microscopic blood images. The proposed method uses bilateral filtering to remove the noise and enhance the image quality. Adaptive thresholding and morphological image processing algorithms are used to detect the malaria parasites inside individual cell. To measure the efficiency of the proposed algorithm, we have tested our method on a NIH Malaria dataset and also compared the results with existing similar methods. Our method achieved the detection accuracy of more than 91% outperforming the competing methods. The results show that the proposed algorithm is reliable and can be of great assistance to the pathologists and hematologists for accurate malaria parasite detection.
Keywords: Malaria diagnosis, Microscopic images, Medical image processing, Parasite detection
Introduction
Malaria is one of the leading causes of death over the world with almost 400,000 deaths per year. More than 200 million cases of malaria are reported every year worldwide (WHO 2019). Malaria is caused by a protozoan parasite of the genus Plasmodium and it is usually diagnosed by an expert pathologist manually by examining the blood cells of the patient under a microscope. The manual examination is a tedious, time consuming, and error prone way to detect the malaria. Moreover, it is difficult to analyze each blood smear with full concentration as in many parts of the world the patient to doctor ratio is significantly below the World Health Organization (WHO) recommendations, one physician per 1000 population. The statistics show that over 45% of WHO member states do not meet this recommendation (WHO 2019). According to the malaria report of World Health Organization (WHO 2018) most of the deaths due to malaria occur in continental Africa and in 2017 among 435,000 deaths 93% were from Africa. This is because in that region malaria finds suitable environment to grow, and secondly, very less resources are available there to prevent this disease. In such situations, detection of malaria should be rapid and accurate to help in early identification and treatment. Therefore, development of automated systems is momentous for a quick, reliable, and timely detection of malaria.
Malaria is transferred into human body from the bites of female Anopheles mosquitoes. This parasite infects the red blood cells (RBC) and goes through a complex life cycle. During the life cycle, the malaria parasite grows and reproduces which damages the RBCs. At different stages of life, the parasite changes its shape which can be seen under microscope. There are five species of Plasmodium which cause malaria in humans: Plasmodium falciparum, Plasmodium vivax, Plasmodium malariae, Plasmodium ovale, Plasmodium knowlesi. Among these five species, P. falciparum and Plasmodium vivax are the most common (Warhurst et al. 1996; Mahmoud et al. 2019).
The manual method for malaria diagnosis is widely used because it is less expensive and can identify all species of malaria. This method is commonly used to find severity of malaria, testing of medicine against malaria, and also used to identify any parasites left after a treatment. For microscopic analysis of blood two kinds of blood smears are prepared: thick smear and thin smear. Thick smear can detect malaria more fast and accurately as compared to thin smear. On the other hand, the thin smear can detect species of malaria and can also identify severity of malaria (Warhurst et al. 1996; Das et al. 2015; Rosado et al. 2016; Jan et al. 2018). Besides having all these benefits, light microscopy has a huge disadvantage of extensive training, and correctness of result entirely depends on the skills of the microscopist. There exist some other techniques for detection of malaria, such as, microarrays (Patarakul 2008), polymerase chain reaction (PCR) (Johnston et al. 2006), rapid diagnostic test (RDT) (Moody 2002), quantitative buffy coat (QBC) (Clendennen et al. 1995), and immunofluorescent antibody testing (IFA) (She et al. 2007) etc. These techniques are either expensive or very complex to perform, hence the light microscopy still stays the commonly used technique for detection of malaria.
In recent years, the automatic detection of malaria has been an important research area. There are some key processing steps performed in almost every automatic malaria diagnostic system. The first step is the acquisition of digital microscopic blood images followed by a preprocessing performed in order to remove noise and artifacts from images. Third step is the separation of RBCs, white blood cells, platelets, and parasites, etc. In fourth step, some features are computed to differentiate between segmented objects and finally based on the computed features RBCs are classified in either infected or uninfected category.
In the malaria detection method presented by Yang et al. (2017), the thin blood smear images obtained through light microscopy are used and noise in images is removed by using mean filtering. Histogram thresholding is applied for segmentation and images are classified by using support vector machine (SVM). After preprocessing, the method of Linder et al. (2014) classifies the images using SVM on the basis of local binary pattern (LBP) and the algorithm of Mohammed and Abdelrahman (2017) does the same but uses normalized cross-correlation feasture. After segmentation, the classification of Kareem et al. (2012) is performed based on Hue Saturation Value (HSV), relative size, and geometry. The algorithm presented by Nasir et al. (2012) uses K-mean clustering to segment image followed by seeded region growing algorithm to remove any remaining unwanted region.
A number of malaria detection algorithms use the Otsu thresholding (Otsu 1979) to segment the objects of interests and then use different classifiers to separate the cells into affected and non-affected, e.g., Gatc et al. (2013), Malihi et al. (2013), Savkare and Narote (2015). Histogram based techniques to segment the blood cells from the image are also widely used in malaria detection algorithm e.g., Zou et al. (2010), Anggraini et al. (2011), Kaewkamnerd et al. (2012), Mushabe et al. (2013), Maiseli et al. (2014). Anggraini et al. (2011) removed the noise in thin blood smear through median filtering and segmentation was performed using Otsu thresholding (Otsu 1979). Naive bayes tree is used to classify cells based on the color intensity of cells. Le et al. (2008) proposed a technique based on supervised thresholding and tested it for both thin and thick blood smear.
Pan et al. (2018) presented an algorithm in which they classified cells by using deep convolutional neural networks. A deep learning based malaria detection system was presented by Hung et al. (2017). The detection and classification is performed using faster R-CNN followed by AlexNet for fine classification. The malaria detectors proposed by Liang (2016), Bibin et al. (2017), Poostchi et al. (2018), Vijayalakshmi and Kanna (2019) also use deep learning. Liang (2016) proposed a technique in which the images were first segmented and then to separate infected and uninfected cells a convolutional neural network was applied. Ross et al. (2006) used different morphological methods to classify thin blood smear microscopic images as malaria infected and uninfected. In malaria detection approach presented by Elter et al. (2011), the infected and uninfected cells were classified by using a SVM based on texture and morphological features. The method of Das et al. (2012) preprocessed images with illumination correction and noise reduction by geometric mean filter.
In this paper, we present a computer aided diagnosis system for malaria detection from the microscopic blood images. The algorithm is fully automatic and does not require any assistance from the user. There are two major advantages of the proposed method. First, instead of using the conventional preprocessing techniques, it exploits bilateral filtering to remove noise from the image. Second, we propose to use of object contours and 8-connected rule to identify the parasites in the cell. The performance of the proposed algorithm was evaluated on a publicly available malaria dataset and also compared with existing similar methods.
Materials and methods
The proposed algorithm works in two steps. First, the cell images were processed to remove the noise and enhance the quality of the image. Second, the image was processed to detect the parasite though adaptive thresholding and morphological operations. The Infected cells contain parasites which serve as discriminator between infected and uninfected cells.
Image preprocessing
The first step in the proposed algorithm is the image preprocessing to enhance the quality of the image and to remove any noise present in the image. This step is important as the performance of the later stages of the proposed algorithm depends on the quality of the image being fed to it. Due to poor acquisition, the images may be polluted with noise which must be removed before processing the image for parasite detection. Simple blurring filters e.g., average filter, are well-known for suppressing different noises in digital images, including, uniform noise, Gaussian noise, etc. However, in our case the images were low-resolution and content sensitive - the boundary of the parasite in the cell is very important and using simple blurring filters can deteriorate it resulting in the mis-detection of the parasite. Therefore, we need a filter that remove the noise but preserves the edges in the image. In our method, we used bilateral filter (Tomasi and Manduchi 1998) to remove the noise from the image.
In the conventional filtering methods, the filter weights depend on the spatial distance of the pixel from the filter center. Contrastingly, the filter weights in the bilateral filter not only depend on the spatial distance but also on the range differences i.e., color or intensity difference. The later characteristic of the filter helps it to achieve the edge preserving property. The bilateral filters are non-linear in nature as their weights depend on the contents of the underlying image. It is important to note that the computational complexity of bilateral filtering is no more than the conventional filtering. Let be the input original microscope color image of size . Each pixel consists of triplets - red, green, and blue components, represented as a vector . Here (x, y) are the spatial coordinates of the pixel in the image. The bilateral filtered image using window of size is obtained as:
| 1 |
where is the range kernel that smooths the intensity difference between the center pixel at (x, y) and its neighborhood pixel at (i, j), is the kernel that smooths based on the distance between the pixels (x, y) and (i, j). In our study, both of these kernels are based on Gaussian function,
| 2 |
| 3 |
where and are variance parameters. In (1), weight w normalizes the filtered value. It is computed as,
| 4 |
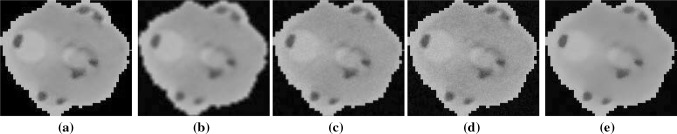
A sample input image from the test dataset is shown in Fig. 1a, and the results of applying different commonly used filters as preprocessing are shown in Fig. 1b–d. The results of applying ‘average filter’, ‘Gaussian lowpass filter’, and ‘median filter’ are shown in Fig. 1b–d, respectively. The result of applying the proposed preprocessing is shown in Fig. 1e. It can be observed that the result of average filtering (Fig. 1b) is very poor, though the noise is suppressed but it also deteriorated the boundaries of the objects which would result in poor parasite detection. The object boundaries in Gaussian lowpass filtered image (Fig. 1c) are better than average filtered image, however the noise is still presented in the filtered image. Similar observation can be made about the median filter (Fig. 1d). Compared to other preprocessing techniques, the noise present in the input image is greatly reduced by the proposed preprocessing technique without affecting the structural details present in the image.
Fig. 1.
a A sample input color image of a RBC, b result of preprocessing with an ‘averaging filter’, c result of preprocessing with an ‘Gaussian lowpass filter’, d result of preprocessing with an ‘median filter’, e result with proposed preprocessing
Parasite detection
The proposed algorithm detects the parasites from the image using its intensity values. Therefore, the preprocessed image is converted into grayscale I. This was achieved by processing each pixel by using the following formula:
| 5 |
Using (5), each pixel of the color image is converted into grayscale value. The resultant image is denoted as I. In the malaria dataset used in performance evaluation, each image contains one red blood cell (Fig. 5). To differentiate the infected and uninfected cells, different features have been used in literature e.g., edges and color. In this research we propose to use the object contours for this purpose instead of edges. We recall that an object contour is a closed curve compared to an edge which is a set of connected neighboring pixels. Therefore, contours are expected to be more helpful in detecting the objects inside a cell than the edges.
Fig. 5.
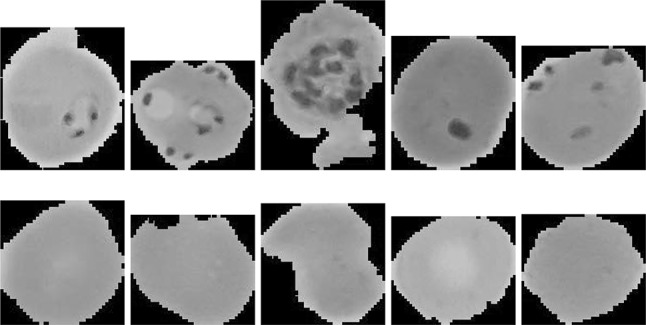
Sample microscope images of parasitized cells (top row) and uninfected cells (bottom row) taken from Malaria dataset
Numerous algorithms have been proposed to detect contours from grayscale and color images e.g., Catanzaro et al. (2009), Leordeanu et al. (2012), Yang et al. (2016), Li et al. (2018). Any of such approach can be used to detect the boundaries of the objects. However, we find that in the problem under discussion, any simple contour detection technique can be used as the images are colorless and textureless. To keep the proposed algorithm simple and efficient, we used Matlab built-in contour detection algorithm. It returns a contour matrix C that consists of two rows defining the contour levels.The very first column contains the contour levels A and number of vertices at each level while the remaining columns contains the coordinates (x, y) value of the vertices. In each column, the first row contains x value of the vertex and the second row corresponds to the y value of the vertex.
| 6 |
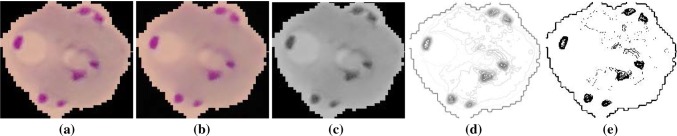
Figure 2d shows the contours detected from image in Fig. 2c.
Fig. 2.
a A sample input color image of an RBC, b image after preprocessing, c the grayscale image obtained from b by using (5), d contours detected in c, e image after adaptive thresholding
Adaptive thresholding is performed to segment parasites. In conventional thresholding techniques, a global threshold value is used for all pixels in the image whereas adaptive thresholding changes its threshold value for each pixel dynamically throughout the image based on its neighborhood. In our algorithm, the threshold value for each pixel was determined statistically which considered local intensity value of the neighbor pixels. This helps to remove the false positives, retaining only those pixel which are potential parasites. For a pixel in the contour image , in a window of size centered at that pixel (x, y) was processed to estimate its threshold value t.
| 7 |
If pixel value p(x, y) is greater than its respective threshold t(x, y), it is assigned foreground value otherwise it assumes background value. After performing the adaptive thresholding a binary image is obtained.
Figure 2e shows the result of adaptive thresholding the image in Fig. 2d. Figure 2e contains parasites, some artifacts, and boundaries of the cells. To get the parasites, we have to eliminate the outer edge and other little components. For this purpose, we will find the connectivity among pixel and its immediate neighbors in eight directions i.e., left, right, up, down, and diagonals. In the image , we used 8-connected components around the pixel (Fig. 3), the components that do not qualify this rule were discarded by leaving behind the islands which are the potential parasites (Fig. 4).
Fig. 3.
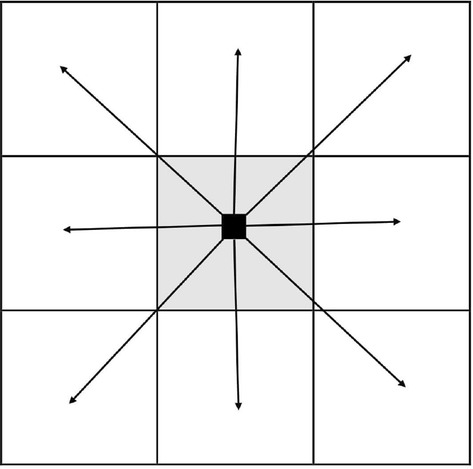
The 8-connectivity used to discard the edges and other small blobs. The objects, if any, obeying the connectivity were left which are considered parasites
Fig. 4.

Final image after removing the edges and other small blobs through 8-connected components
After removing edges and other artifacts from Fig. 2d, the results are shown in Fig. 4 which is our final image . The non-zero pixels in this image represent the presence of parasites. Finally, we computer number of connected components in and pass them through a threshold value to distinguish between the infected and uninfected cells. If number of components n are more than the threshold then that the cell is classified as infected, classified as uninfected otherwise. Different values of this threshold are experimented and results are discussed in next section.
Results and discussion
In this section, we report the performance of a proposed algorithm on a large publicly available dataset. We also compared the results of our algorithm with the other competing methods using different objective metrics to assess the effectiveness of the proposed method.
Dataset used for performance evaluation
The dataset we used to evaluate the performance of the proposed algorithm was taken from the Malaria Dataset provided by the National Library of Medicine (NLM), USA. The dataset is publicly available at https://ceb.nlm.nih.gov/repositories/malaria-datasets/ and was introduced by Rajaraman et al. (2018). The dataset was collected at Chittagong Medical College Hospital, Bangladesh with Giemsa-stained thin blood smear slides. The blood smears from 150 P. falciparum-infected and 50 healthy patients were taken. The resultant images were manually annotated by expert microscopists at the Mahidol-Oxford Tropical Medicine Research Unit in Bangkok, Thailand. To preserve the patients’ privacy, the images were de-identified before public release. Our test dataset comprises of 2000 images with equal instances of parasitized and uninfected cells. Few sample infected and uninfected images from the dataset are shown in Fig. 5.
Objective performance evaluation
In this section, we evaluated the performance of the proposed malaria parasite detection algorithm. The proposed algorithm was executed for each image in the dataset and on an image, there are four possible outcomes of the proposed algorithm (Table 1): true positives (TP), true negatives (TN), false positives (FP), and false negatives (FN). The true positives (TP) represent the infected cells detected without error and the true negatives (TN) denote the uninfected cells which are correctly detected. The false positives (FP) are the healthy cells detected as infected and the false negative (FN) are the infected cells detected as healthy.
Table 1.
Division of results in TP, TN, FN, and FP
| Infected | Uninfected | |
|---|---|---|
| Infected | True positive (TP) | False negative (FN) |
| Uninfected | False positive (FP) | True negative (TN) |
To objectively quantify the performance of the proposed algorithm five statistical metrics: Precision, Specificity, Recall, Accuracy, and F1-score (Fawcett 2006; Hajian-Tilaki 2013; Farid et al. 2018) were computed for whole dataset. Precision demonstrates how much the model is precise in terms of positive results. It computes out of all positive predicted values how many of them are actually positive.
| 8 |
Specificity measures the ratio of actual negatives that are correctly identified.
| 9 |
Recall computes how much of the actual positive results are captured correctly. It helps in false negative cases such that a patient was suffering from malaria but was diagnosed as healthy. It is computed as following:
| 10 |
Accuracy measures percentage of correctly classified cells from the overall assessments.
| 11 |
The F1 score is the weighted harmonic average of the precision and recall which ranges from 0 (worst) to 1 (best). Therefore, this score takes both false positives and false negatives into account and provides overall accuracy of the model.
| 12 |
In all experiments, the number of contours levels (6) was set to 20, and three values for window size (w) 19, 17, and 15 were experimented. The number of connected components n was also empirically evaluated with values 0, 1, and 2. The results of this evaluation are presented in Tables 2 and 3. These results show that the best values were produced by window size w of 19 and number of connected objects (n) in the infected image should be at least one while for uninfected cells n should be zero. For and , the algorithm achieves the best scores in accuracy, recall, and F1 score. The precision score for these settings is 0.9466 which is closest to the best results 0.9512, with a negligible difference, less than 0.005.
Table 2.
Resultant TP, TN, FN, and FP values for different values of ‘w’ and ‘n’ on the whole dataset
| w | n | TP | TN | FN | FP |
|---|---|---|---|---|---|
| 19 | > 0 | 886 | 950 | 114 | 50 |
| 19 | > 1 | 878 | 922 | 122 | 78 |
| 19 | > 2 | 840 | 943 | 160 | 57 |
| 17 | > 0 | 899 | 893 | 101 | 107 |
| 17 | > 1 | 848 | 930 | 152 | 70 |
| 17 | > 2 | 802 | 952 | 198 | 48 |
| 15 | > 0 | 869 | 897 | 131 | 103 |
| 15 | > 1 | 804 | 938 | 196 | 62 |
| 15 | > 2 | 760 | 961 | 240 | 39 |
Table 3.
Precision, recall, accuracy, and F1 score for different values of ‘w’ and ‘n’. The best results are marked in bold
| w | n | Precision | Recall | Accuracy | F1 score |
|---|---|---|---|---|---|
| 19 | > 0 | 0.9466 | 0.8860 | 0.9180 | 0.9153 |
| 19 | > 1 | 0.9184 | 0.8780 | 0.9000 | 0.8978 |
| 19 | > 2 | 0.9365 | 0.8400 | 0.8915 | 0.8856 |
| 17 | > 0 | 0.8936 | 0.8790 | 0.8960 | 0.8963 |
| 17 | > 1 | 0.9237 | 0.8480 | 0.8890 | 0.8843 |
| 17 | > 2 | 0.9435 | 0.8020 | 0.8770 | 0.8670 |
| 15 | > 0 | 0.8940 | 0.8690 | 0.8830 | 0.8813 |
| 15 | > 1 | 0.9284 | 0.8040 | 0.8710 | 0.8617 |
| 15 | > 2 | 0.9512 | 0.7600 | 0.8605 | 0.8449 |
We also compared the performance of the proposed algorithm with other existing similar methods, including those of Ross et al. (2006), Das et al. (2012), Hung et al. (2017) and Pan et al. (2018). Although this comparison would not be equitable or well earned as the datasets used in these methodologies were different, however it can provide a glance on the effectiveness of the proposed method. The results of this comparison are presented in Table 4. Our technique outperformed all competing methods with 0.9466 precision, 0.9180 accuracy, and 0.9500 specificity. The recall measure of Das method is better than ours, however its overall performance is significantly poor than our method. F1 score is considered to be more reliable as it considers both precision and recall. Our method achieved the best F1 score of 0.9153 outperforming the compared method. These statistics show that the proposed algorithm is reliable and effective in detection of the malarial parasites from microscope images of red blood cells.
Table 4.
Performance comparison of the proposed algorithm with other competing methods*. The best results are marked in bold
| Method | Precision | Specificity | Recall | Accuracy | F1 score |
|---|---|---|---|---|---|
| Ross | – | – | 0.8500 | 0.7300 | – |
| Das | – | 0.6890 | 0.9810 | 0.8400 | – |
| Hung | 0.7804 | 0.8519 | 0.7766 | 0.8215 | 0.7784 |
| Pan | 0.7439 | 0.8273 | 0.7402 | 0.7921 | 0.7420 |
| Proposed | 0.9466 | 0.9500 | 0.8860 | 0.9180 | 0.9153 |
Image preprocessing is the first step in most existing malaria detection algorithms. The objective of this step is to remove the noise from the image before applying the later steps. Usually, noise is removed though order statistics filters e.g., median filter or through the blur filters e.g., average filter and Gaussian lowpass filter. We showed in Sect. 2 that these filters may not be effective in many cases (Fig. 1). In this paper, we exploited the bilateral filtering and found that it produces better quality images that in turn improves the detection accuracy of the proposed method. Here we report the performance of the proposed algorithm with different filters as preprocessing step. We computed the performance measures without using any preprocessing, using average filter, Gaussian lowpass filter, median filter, and bilateral filter. Table 5 presents the results of this experiment. The results demonstrates that the best results in most metrics are achieved using bilateral filtering as preprocessing in the proposed method.
Table 5.
Performance comparison of different preprocessing algorithms. The best results are marked in bold
| Preprocessing method | Precision | Specificity | Recall | Accuracy | F1-score |
|---|---|---|---|---|---|
| No Preproprocessing | 0.8878 | 0.8840 | 0.9180 | 0.9010 | 0.9027 |
| Average filter | 0.9305 | 0.9420 | 0.6430 | 0.7975 | 0.7605 |
| Gaussian lowpass filter | 0.9315 | 0.9490 | 0.7740 | 0.8715 | 0.8576 |
| Median filter | 0.9076 | 0.9150 | 0.8350 | 0.8750 | 0.8698 |
| Bilateral filter | 0.9466 | 0.9500 | 0.8860 | 0.9180 | 0.9153 |
Few visual results of the proposed algorithm are presented in Figs. 6 and 7. The left-most column in each row shows the input image, the second, third, and fourth columns show the results of intermediate steps of the algorithm. The final results are shown in the right-most column. Figure 6 shows the five images from the test dataset that are infected and our method successfully detected them. Figure 7 shows the results of our algorithm on five uninfected images. We also report some failure cases of our method in Fig. 8. We observed that when the image is corrupted due to sever noise or its acquisition quality is poor, our algorithm is mistaken considering the noise bursts as parasites. Poor quality of the cell image can result in diluted parasite which gets removed during preprocessing or adaptive thresholding resulting in missed detection.
Fig. 6.
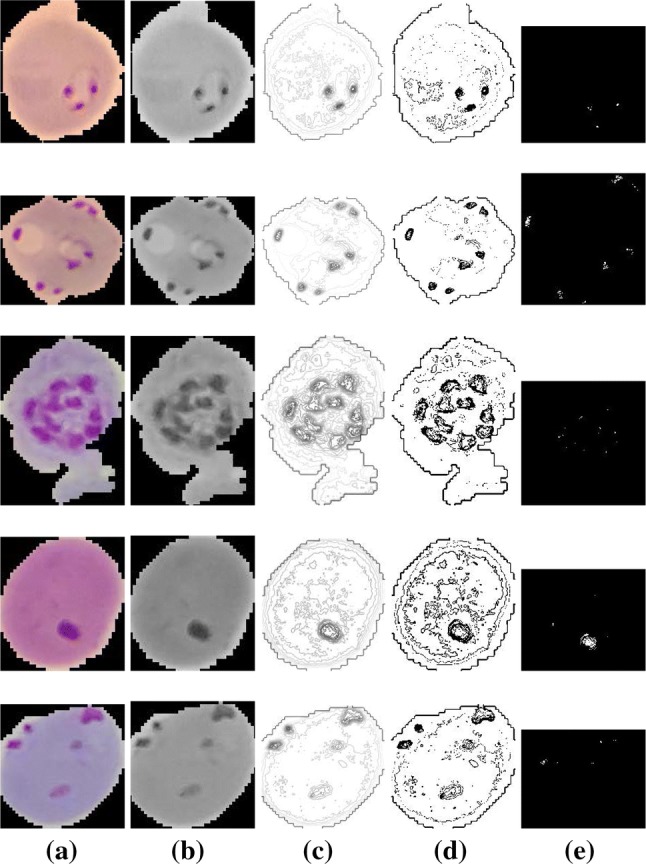
Successfully detected parasitized cells by our algorithm. a Input image, b preprocessed grayscale image, c contour image, d results of adaptive thresholding, e final results after refinement
Fig. 7.
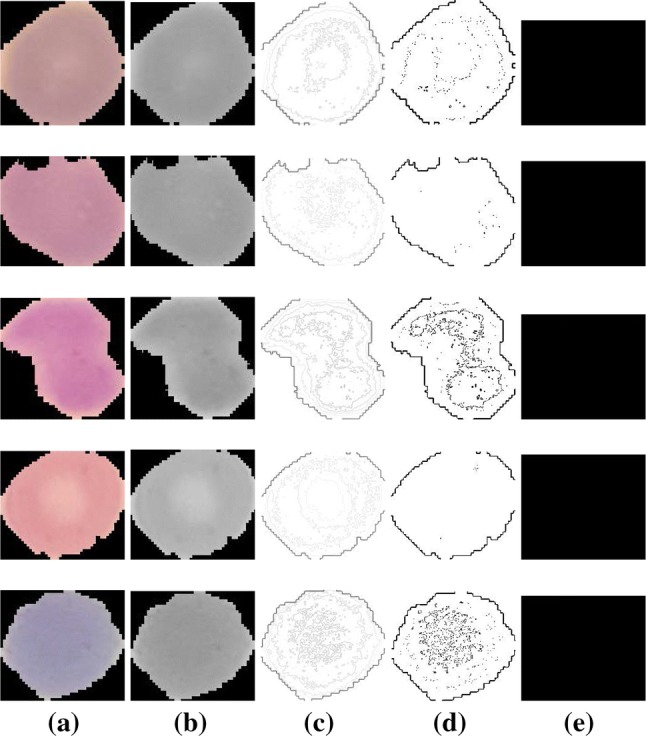
Successfully detected uninfected cells by our algorithm. a Input image, b preprocessed grayscale image, c contour image, d results of adaptive thresholding, e final results after refinement
Fig. 8.
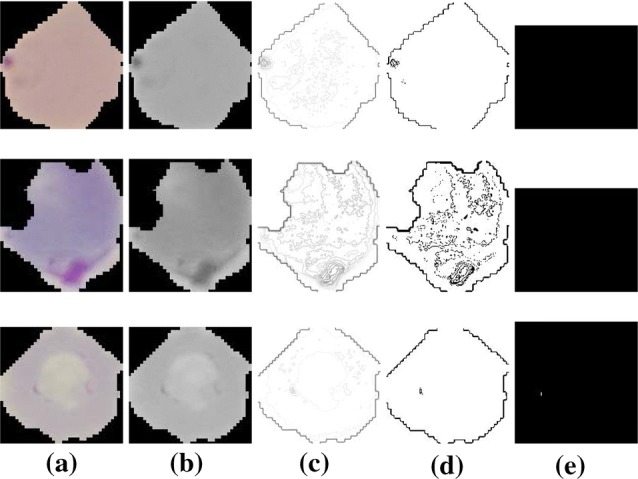
Failure cases. Top two rows show examples of parasitized cells which our method could not detect. The bottom row shows a case where cell was uninfected but detected as infected. a Input image, b preprocessed grayscale image, c contour image, d results of adaptive thresholding, e final results after refinement
The proposed algorithm is implemented in Matlab and is made available free of cost for peers on the project web-page (http://www.di.unito.it/~farid/Research/malaria.html). We also computed the time complexity of the proposed method. For this purpose, the proposed algorithm was executed over the whole dataset and time for each image was recorded and their average was computed. The experiment was executed on Intel® Core™ i5 processor with 8 GB RAM and 64-bit operating system. The results show that the proposed malaria detection algorithm is very fast and takes an average of 2.8 s to process one image. This time also includes the file input/output time. An efficient implementation of the algorithm can further improve the execution time.
Conclusions
In this paper, we presented a novel algorithm to automatically detect malaria from microscope blood smears. The algorithm preprocessed the cell images using bilateral filtering which has not been explored in previous techniques. Then by using adaptive thresholding and 8-connected rules, normal and infected cells were separated. The performance of the proposed algorithm was evaluated on standard malaria dataset consisting of large number of images with infected and healthy blood cells. The performance was computed using five statistical metrics and compared with existing similar techniques. The proposed method outperformed the compared method by achieving accuracy and F1 scores more than 0.91. The results reveal the efficacy of the proposed algorithm.
Author contributions
TF: Conceptualization; TF and MF: methodology; TF: software; TF and MF: validation; TF and MF: writing–original draft preparation.
Compliance with ethical standards
Conflict of interest
The authors declare they have no conflict of interests.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Informed consent
The research conducted in this paper utilized the retrospective and publicly available NIH Malaria database, so that no formal consent was necessary.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Anggraini D, Nugroho AS, Pratama C, Rozi IE, Pragesjvara V, Gunawan M (2011) Automated status identification of microscopic images obtained from malaria thin blood smears using bayes decision: a study case in Plasmodium falciparum. In: 2011 International conference on advanced computer science and information systems, IEEE, pp 347–352
- Bibin D, Nair MS, Punitha P. Malaria parasite detection from peripheral blood smear images using deep belief networks. IEEE Access. 2017;5:9099–9108. doi: 10.1109/ACCESS.2017.2705642. [DOI] [Google Scholar]
- Catanzaro B, Su B, Sundaram N, Lee Y, Murphy M, Keutzer K (2009) Efficient, high-quality image contour detection. In: Proceedings of the IEEE international conference on computer vision (ICCV), pp 2381–2388
- Clendennen TE, III, Long GW, Baird JK. Qbc® and giemsa-stained thick blood films: diagnostic performance of laboratory technologists. Trans R Soc Trop Med Hygiene. 1995;89(2):183–184. doi: 10.1016/0035-9203(95)90486-7. [DOI] [PubMed] [Google Scholar]
- Das DK, Ghosh M, Pal M, Maiti A, Chakraborty C. Machine learning approach for automated screening of malaria parasite using light microscopic images. Micron. 2012 doi: 10.1016/j.micron.2012.11.002. [DOI] [PubMed] [Google Scholar]
- Das DK, Mukherjee R, Chakraborty C. Computational microscopic imaging for malaria parasite detection: a systematic review. Malar J. 2015;260(1):1–19. doi: 10.1111/jmi.12270. [DOI] [PubMed] [Google Scholar]
- Elter M, Haßlmeyer E, Zerfaß T (2011) Detection of malaria parasites in thick blood films. In: IEEE engineering in medicine and biology society (EMBS), pp 5140–5144, 10.1109/IEMBS.2011.6091273 [DOI] [PubMed]
- Farid MS, Lucenteforte M, Grangetto M. DOST: a distributed object segmentation tool. Multimed Tools Appl. 2018;77(16):20839–20862. doi: 10.1007/s11042-017-5546-4. [DOI] [Google Scholar]
- Fawcett T. An introduction to ROC analysis. Pattern Recognit Lett. 2006;27(8):861–874. doi: 10.1016/j.patrec.2005.10.010. [DOI] [Google Scholar]
- Gatc J, Maspiyanti F, Sarwinda D, Arymurthy AM (2013) Plasmodium parasite detection on red blood cell image for the diagnosis of malaria using double thresholding. In: ICACSIS, pp 381–385, 10.1109/ICACSIS.2013.6761605
- Hajian-Tilaki K. Receiver operating characteristic (ROC) curve analysis for medical diagnostic test evaluation. Casp J Intern Med. 2013;4(2):627. [PMC free article] [PubMed] [Google Scholar]
- Hung J, Goodman A, Lopes S, Rangel G, Ravel D, Costa F, Duraisingh M, Marti M, Carpenter AE (2017) Applying faster R-CNN for object detection on malaria images. CoRR arXiv:1804.09548 [DOI] [PMC free article] [PubMed]
- Jan Z, Khan A, Sajjad M, Muhammad K, Rho S, Mehmood I. A review on automated diagnosis of malaria parasite in microscopic blood smears images. Multimed Tools Appl. 2018;77(8):9801–9826. doi: 10.1007/s11042-017-4495-2. [DOI] [Google Scholar]
- Johnston SP, Pieniazek NJ, Xayavong MV, Slemenda SB, Wilkins PP, da Silva AJ. Pcr as a confirmatory technique for laboratory diagnosis of malaria. Med Biol Eng Comput. 2006;44(3):1087–1089. doi: 10.1128/JCM.44.3.1087-1089.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaewkamnerd S, Uthaipibull C, Intarapanich A, Pannarut M, Chaotheing S, Tongsima S. An automatic device for detection and classification of malaria parasite species in thick blood film. BMC Bioinform. 2012;13(17):S18. doi: 10.1186/1471-2105-13-S17-S18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kareem S, Kale I, Morling RC (2012) Automated malaria parasite detection in thin blood films:-a hybrid illumination and color constancy insensitive, morphological approach. In: IEEE Asia Pacific conference on circuits and systems, IEEE, pp 240–243
- Le MT, Bretschneider TR, Kuss C, Preiser PR. A novel semi-automatic image processing approach to determine Plasmodium falciparum parasitemia in giemsa-stained thin blood smears. BMC Cell Biol. 2008;9(1):15. doi: 10.1186/1471-2121-9-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leordeanu Marius, Sukthankar Rahul, Sminchisescu Cristian. Computer Vision – ECCV 2012. Berlin, Heidelberg: Springer Berlin Heidelberg; 2012. Efficient Closed-Form Solution to Generalized Boundary Detection; pp. 516–529. [Google Scholar]
- Li S, Xu Y, Cong W, Ma S, Zhu M, Qi M. Biologically inspired hierarchical contour detection with surround modulation and neural connection. Sensors. 2018;18(8):2559. doi: 10.3390/s18082559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Z et al (2016) CNN-based image analysis for malaria diagnosis. In: IEEE international conference on bioinformatics and biomedicine (BIBM) pp 493–496. 10.1109/BIBM.2016.7822567
- Linder N, Turkki R, Walliander M, Mårtensson A, Diwan V, Rahtu E, Pietikäinen M, Lundin M, Lundin J. A malaria diagnostic tool based on computer vision screening and visualization of Plasmodium falciparum candidate areas in digitized blood smears. PLoS One. 2014;9(8):e104855. doi: 10.1371/journal.pone.0104855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmoud DM, Hussein HM, El Gozamy BMR, Thabet HS, Hassan MA, Meselhey RAA. Screening of Plasmodium parasite in vectors and humans in three villages in Aswan Governorate. Egypt J Parasit Dis. 2019;43(1):158–163. doi: 10.1007/s12639-018-1069-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiseli B, Mei J, Gao H, Yin S, Maiseli B (2014) An automatic and cost-effective parasitemia identification framework for low-end microscopy imaging devices. In: International conference on mechatronics and control (ICMC), pp 2048–2053, 10.1109/ICMC.2014.7231926
- Malihi L, Ansari-Asl K, Behbahani A (2013) Malaria parasite detection in giemsa-stained blood cell images. In: 8th Iranian conference on machine vision and image processing (MVIP), IEEE, pp 360–365
- Mohammed HA, Abdelrahman IAM (2017) Detection and classification of malaria in thin blood slide images. In: IEEE ICCCCEE, pp 1–5
- Moody A. Rapid diagnostic tests for malaria parasites. Clin Microbiol Rev. 2002;15(1):66–78. doi: 10.1128/CMR.15.1.66-78.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mushabe MC, Dendere R, Douglas TS (2013) Automated detection of malaria in giemsa-stained thin blood smears. In: IEEE engineering in medicine and biology society (EMBC), pp 3698–3701, 10.1109/EMBC.2013.6610346 [DOI] [PubMed]
- Nasir AA, Mashor M, Mohamed Z (2012) Segmentation based approach for detection of malaria parasites using moving k-means clustering. In: IEEE-EMBS conference on biomedical engineering and sciences, IEEE, pp 653–658
- Otsu N. A threshold selection method from gray-level histograms. IEEE Trans Syst Man Cybern. 1979;9(1):62–66. doi: 10.1109/TSMC.1979.4310076. [DOI] [Google Scholar]
- Pan W. David, Dong Yuhang, Wu Dongsheng. Machine Learning - Advanced Techniques and Emerging Applications. 2018. Classification of Malaria-Infected Cells Using Deep Convolutional Neural Networks. [Google Scholar]
- Patarakul K. Role of DNA microarray in infectious diseases. Chulalongkorn Med J. 2008;52:147–153. [Google Scholar]
- Poostchi M, Silamut K, Maude RJ, Jaeger S, Thoma G. Image analysis and machine learning for detecting malaria. Transl Res In-depth Rev Diagn Med Imaging. 2018;194:36–55. doi: 10.1016/j.trsl.2017.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajaraman S, Antani SK, Poostchi M, Silamut K, Hossain MA, Maude RJ, Jaeger S, Thoma GR. Pre-trained convolutional neural networks as feature extractors toward improved malaria parasite detection in thin blood smear images. PeerJ. 2018;6:e4568. doi: 10.7717/peerj.4568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosado L, Correia da Costa JM, Elias D, Cardoso SJ. A review of automatic malaria parasites detection and segmentation in microscopic images. Anti-Infect Agents. 2016;14(1):11–22. doi: 10.2174/221135251401160302121107. [DOI] [Google Scholar]
- Ross NE, Pritchard CJ, Rubin D, Duse A. Automated image processing method for the diagnosis and classification of malaria on thin blood smears. Med Biol Eng Comput. 2006;44:427–36. doi: 10.1007/s11517-006-0044-2. [DOI] [PubMed] [Google Scholar]
- Savkare SS, Narote SP (2015) Automated system for malaria parasite identification. In: International conference on communication, information computing technology (ICCICT), pp 1–4, 10.1109/ICCICT.2015.7045660
- She RC, Rawlins ML, Mohl R, Perkins SL, Hill HR, Litwin CM. Comparison of immunofluorescence antibody testing and two enzyme immunoassays in the serologic diagnosis of malaria. J Travel Med. 2007;14(2):105–111. doi: 10.1111/j.1708-8305.2006.00087.x. [DOI] [PubMed] [Google Scholar]
- Tomasi C, Manduchi R (1998) Bilateral filtering for gray and color images. In: Proceedings of the IEEE international conference on computer vision (ICCV), IEEE Computer Society, pp 839–846
- Vijayalakshmi A, Kanna BR. Deep learning approach to detect malaria from microscopic images. Multimed Tools Appl. 2019 doi: 10.1007/s11042-019-7162-y. [DOI] [Google Scholar]
- Warhurst D, Williams J. ACP broadsheet no 148. Laboratory diagnosis of malaria. J Clin Pathol. 1996;49(7):533. doi: 10.1136/jcp.49.7.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO (2018) World malaria report 2018. World Health Organization
- WHO (2019) Global Health Observatory (GHO) data . https://www.who.int/gho/health_workforce/physicians_density/en/. Accessed June 2019
- Yang D, Subramanian G, Duan J, Gao S, Bai L, Chandramohanadas R, Ai Y. A portable image-based cytometer for rapid malaria detection and quantification. PLoS One. 2017;12(6):e0179161. doi: 10.1371/journal.pone.0179161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Price B, Cohen S, Lee H, Yang M (2016) Object contour detection with a fully convolutional encoder-decoder network. In: Proceedings of the IEEE conference on computer vision and pattern Recognition (CVPR), IEEE computer society, Los Alamitos, CA, USA
- Zou L, Chen J, Zhang J, Garcia N (2010) Malaria cell counting diagnosis within large field of view. In: International conference on digital image computing: techniques and applications, pp 172–177, 10.1109/DICTA.2010.40