SUMMARY
Caloric restriction (CR) improves survival in nonhuman primates and delays the onset of agerelated morbidities including sarcopenia, which is characterized by the age-related loss of muscle mass and function. A shift in metabolism anticipates the onset of muscle-aging phenotypes in nonhuman primates, suggesting a potential role for metabolism in the protective effects of CR. Here, we show that CR induced profound changes in muscle composition and the cellular metabolic environment. Bioinformatic analysis linked these adaptations to proteostasis, RNA processing, and lipid synthetic pathways. At the tissue level, CR maintained contractile content and attenuated age-related metabolic shifts among individual fiber types with higher mitochondrial activity, altered redox metabolism, and smaller lipid droplet size. Biometric and metabolic rate data confirm preserved metabolic efficiency in CR animals that correlated with the attenuation of age-related muscle mass and physical activity. These data suggest that CR-induced reprogramming of metabolism plays a role in delayed aging of skeletal muscle in rhesus monkeys.
Graphical Abstract
In Brief
Sarcopenia is the age-related loss of muscle mass and function that is a major factor in frailty and loss of physical independence. In this rhesus monkey study, the authors investigate delayed aging by caloric restriction (CR) as a means to understand the biology of sarcopenia. They report beneficial changes with CR to reverse aging in muscle at the tissue level. Molecular and cellular assessments point to a role for growth and metabolic pathways in the effects of CR. The impact of CR on metabolism is linked to the retention of muscle mass and physical activity. The authors suggest that in addition to contributing to muscle aging, metabolism may also be an effective target for sarcopenia prevention.
INTRODUCTION
Sarcopenia is the age-associated decline in muscle mass and function. The physical basis for this condition is thought to be a combination of atrophy and loss of the constituent muscle fibers, although age-related changes in muscle composition may also be important (McGregor et al., 2014). The rhesus monkey (Macaca mulatta) is a highly translatable model for human aging with substantial genetic similarity to humans, a lifespan of several decades, and a spectrum of age-associated diseases that mirror those in humans (Balasubramanian et al., 2017). Caloric restriction (CR) significantly improves survival and delays the onset of age-related morbidities, such as cancer, cardiovascular disease, and diabetes (Colman et al., 2014; Mattison et al., 2017). This finding demonstrates that primate aging is malleable and adds weight to the concept that CR might be used as a research tool with which to uncover the biology of age-related disease. CR also protects against sarcopenia in rhesus (Colman et al., 2008; McKiernan et al., 2012), indicating that it impinges on factors that contribute to skeletal muscle aging.
The cellular composition of skeletal muscle is heterogeneous. In terms of contractile tissues, individual muscle fibers differ in the speed of contraction and maximal force generation, properties related to the isoform of the structural protein myosin that is expressed, and whether oxidative or non-oxidative metabolism is favored (Bassel-Duby and Olson, 2006). In addition to contractile content, muscle contains vasculature, satellite cells, adipocytes, and fibrotic material that plays an essential role in force transmission and repair (Mahdy, 2019). Mounting evidence indicates that changes in muscle quality drive sarcopenia. In humans, with disuse or age, fibrotic content accumulates and adipocytes and macrophages infiltrate the tissue (Brioche et al., 2016; Pagano et al., 2018). The same age-related outcomes are observed in rhesus monkeys (McKiernan et al., 2012; Pugh et al., 2013).
Our studies of aging in rhesus monkey and CR include a comprehensive investigation of age-related change including physical parameters. Similar to humans, muscle mass loss begins in middle age in monkeys at ~15 years of age (Colman et al., 2005; McKiernan et al., 2009), where age-related loss of quadricep bulk from ~15 to +25 years of age is ~23% for females and 27% for males. Vastus lateralis (VL) is one of the four constituent muscle groups within the quadriceps, and it is the one that is the most vulnerable to aging, with 40% lower tissue weight for old monkeys (~30 years of age) at necropsy compared with young adults of full stature (~8 years of age) (Pugh et al., 2013). VL comprises both oxidative and glycolytic fiber types (Pette and Staron, 2000) and succumbs to fiber atrophy and increased fibrosis with age in humans and monkeys (Lexell, 1995; McKiernan et al., 2012). Responses to aging are of a fiber-type-specific nature: slow twitch type I fibers are resistant to age-related atrophy, but fast twitch type II fibers exhibit a gradual decline in cross-sectional area beginning at middle age.
Defects in skeletal muscle energy metabolism with age have been documented in humans, rats, and mice (Figueiredo et al., 2009; Noland et al., 2009; Proctor et al., 1995), and these findings have been corroborated by meta-analysis of gene expression studies (de Magalhães et al., 2009; Mootha et al., 2003; Zahn et al., 2006). In humans, skeletal muscle mitochondrial activity declines with age (Proctor et al., 1995; Short et al., 2005). In rhesus monkeys, age-related changes in mitochondrial and redox metabolism occur in advance of the onset of muscle mass loss (Pugh et al., 2013) and before age-related declines in physical activity are detected (Yamada et al., 2013), suggesting that metabolism could play a causal role in skeletal muscle aging. A separate study has demonstrated that energy metabolism pathways are uniformly but modestly induced with CR in mice, and pathway level analysis confirmed the same in rhesus monkey skeletal muscle (Barger et al., 2015). To test this, we investigated the molecular and cellular phenotypes of delayed sarcopenia due to CR in rhesus monkeys and related these data to tissue, biometric, and functional outcomes.
RESULTS
Metabolic and Growth Networks Are Engaged by CR in Skeletal Muscle
The study cohort included adult (27.8 ± 1.7 years) male rhesus monkeys that had been on the University of Wisconsin (UW) Aging and Caloric Restriction study for 18 years (n = 5 control and n = 8 CR). At the outset of the study, adult monkeys were randomized into control and CR (30% restricted) diet groups. Body composition was assessed twice per year throughout the study by whole-body dual-energy X-ray absorptiometry (DEXA), and baseline measures showed equivalence among groups at the outset (Ramsey et al., 2000). Over the first few years, continued body weight increases were documented for the controls with an increase in lean mass and whole-body estimated skeletal muscle mass (ESM) up to middle age. CR animals were consistently smaller than controls, with a significantly lower body weight and lower absolute values of ESM (Colman et al., 2008, 2014). Fat mass was significantly lower for CR from 12 months of the study onward, and significant differences persisted beyond 25 years of age (Mattison et al., 2017). From middle age onward, a continuous decline in ESM occurred for controls (15% loss by ~19 years of age and 23% loss by 22 years of age), whereas a change in ESM for the CR monkeys over the same time frame was negligible. Monkeys in this study were just beyond the age of median lifespan (~26 years). By this time, unlike the CR animals, the control animals had experienced an age-related loss in body weight. As a result, differences in body weight and total lean mass between the groups were no longer statistically significant (Table 1). Focusing more specifically on the limbs, absolute values for upper leg lean mass (ULL) were consistently higher in controls than in CR animals, at least until the onset of age-related muscle mass loss; however, CR animals had proportionally higher ULL when values were adjusted for total body weight (Figure S1). Glucoregulatory parameters were assessed using blood taken around the time of DEXA. Fasting levels of glucose and insulin were numerically lower with CR, and the differences were statistically significant when age was used as a covariate (Table 1). The restricted monkeys were also significantly more insulin sensitive (insulin sensitivity index [Si]) based on the frequently sampled glucose tolerance test.
Table 1.
Body Mass and Glucoregulatory Measures
| Parametera | Control | Restricted |
|---|---|---|
| Age at biopsy | 27.7 ± 1.0 | 27.8 ± 0.8 |
| Body weight (g) | 11,362 ± 5,650 | 9,784 ± 968 |
| Total lean mass (g) | 8,047 ± 638 | 7,926 ± 541 |
| Adiposity | ||
| Fat (%) | 27.62 ± 14.45 | 18.32 ± 7.07 |
| Abdominal fat (%) | 37.81 ± 12.11 | 28.31 ± 12.11 |
| Circulating | ||
| Basal glucose (mg/dL) | 83 ± 11 | 59 ± 7b |
| Basal insulin (μU/mL) | 29 ± 22 | 15 ± 4b |
| Insulin sensitivity index; Si (310−4) | 1.97 ± 1.04 | 6.99 ± 2.77b |
Values are average ± interquartile range. Control n = 5 and restricted n = 8.
p < 0.05; p values adjusted with animal age at biopsy as a covariate.
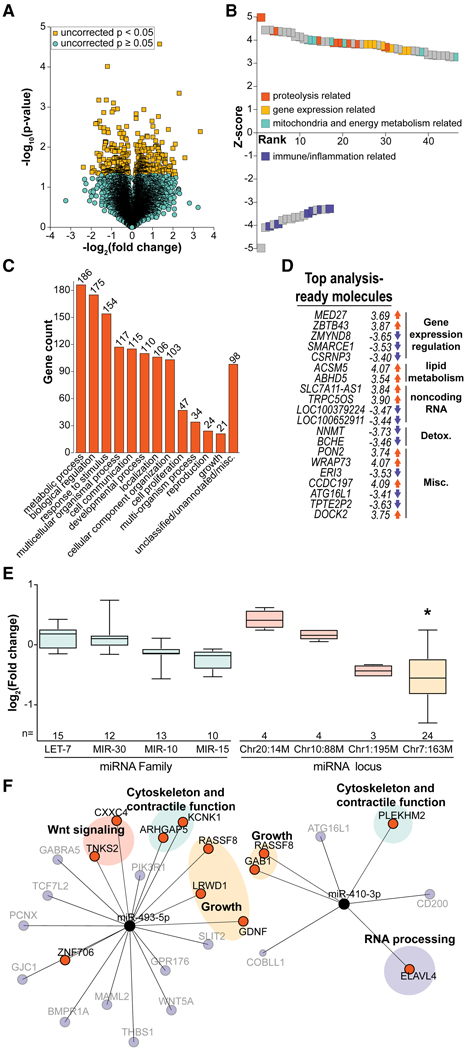
RNA was extracted and purified from VL biopsy samples that were taken at the same time that the DEXA was performed. Transcriptional data generated using the Affymetrix (Santa Clara, CA) M. mulatta microarray were used for pathway analysis of the CR response in monkey skeletal muscle (Barger et al., 2015), but an in-depth analysis of individual gene expression changes has not been previously reported. In this study, changes in gene expression were detected for 15,781 mRNA-hybridized non-redundant probe sets. Two-tailed Student’s t test revealed 404 transcripts with a fold change that passed an uncorrected p value threshold of0.05forCRcomparedwithcontrols(Figure 1A).Parametric analysis of gene set enrichment (PAGE) (Kim and Volsky, 2005) was used to derive Z scores for Gene Ontology (GO) term Biological Processes that were responsive to CR(TableS1).For the purposes of a robust pathway analysis, transcripts passing the more liberal uncorrected p of 0.05 were used, and 63 terms passing a false discovery rate (FDR) threshold of 0.05 were identified (Figure 1B). The majority of the GO terms had positive Z scores (46 terms) and were categorized as relating to proteolysis (8 terms), gene expression (7 terms), or mitochondria and energy metabolism (6 terms). The remaining 17 terms with negative Z scores principally consisted of immune and/or inflammatory processes (6 terms).
Figure 1. Gene Expression Analysis of mRNA and miRNA from Skeletal Muscle.
(A) Volcano plot displaying identified probes from the microarray (n = 5 or 8,control or CR, respectively). Probes passing an uncorrected p value threshold of p < 0.05 are highlighted in yellow</P>.</P>(B) PAGE analysis revealed GO terms that were rank ordered based on Z score and manually categorized as proteolysis related, gene expression related, immune and/or inflammation related, or mitochondria and/or energy metabolism related</P>. </P>(C) BAM analysis yielded 358 statistically significant genes between control and CR, which were non-exclusively categorized via GOSlim terms</P>.</P>(D) Ingenuity pathway analysis revealed the top 10 up- and downregulated analysis-ready molecules, which were categorized as related to gene expression regulation, lipid and fatty acid metabolism, noncoding RNA, detoxification, or miscellaneous</P>.</P>(E) Comparison of aggregate expression of microRNAs grouped together either due to genome location or miR family (n = 5 or 8, control or CR, respectively) (box, interquartile range; whiskers, minimum and maximum values; and bar, median)</P>. </P>(F) Network plot of selected miR members of chr7, miR-493–5p, and miR-4103p and putative targets present among the statistically significant transcripts changing as a result of CR. Edge length is an inverted, weighted score composite of transcript fold change and cumulative weighted context++ (TargetScan), describing the strength of the miR-target interaction. Orange targets are congruent in predicted direction with the direction of expression fold change of the miR (downregulated miRs = upregulated targets), whereas incongruent targets are blue and transparent. Targets are classified by function: growth, cytoskeletal and/or contractile function, Wnt signaling, or RNA processing. See also Figure S2 and Tables S1 and S2.
We next used Bayesian analysis of variance for microarrays (BAM), which uses a different criterion for assessing significance and can be more powerful for low n, high variance studies (Ishwaran and Rao, 2003). CR was associated with 501 differentially expressed probe sets identified, of which 358 were assigned to a non-redundant gene symbol (Table S1). We categorized these genes based on GOSlim biological process terms (Figure 1C) (Wang et al., 2017). Metabolic processes and biological regulation were the largest categories, with other relevant GO terms including development, reproduction, and growth. Taking a slightly different approach, we used ingenuity pathway analysis (IPA) to examine clinically relevant trends among the differentially expressed genes. Rhesus gene identifiers were first converted to corresponding human gene symbols for the IPA pipeline. IPA produces several categories of analysis from the input data. The top canonical pathways associated with CR included NANOG, a factor involved in mammalian stem cell pluripotency; UVA-induced MAPK signaling; and GDNF family ligand-receptor interactions (Table S1). The top diseases and disorders identified were all age-related disorders, including cancer, cardiovascular disease, organismal injury and abnormalities, neurological disease, and inflammatory response (Table S1). The molecular and cellular functions enriched in the data included cell death and survival, cellular assembly and organization, and cellular growth and proliferation, indicating changes to cell growth as a part of the CR response (Table S1). Finally, the 20 top analysis-ready molecules identified by IPA included several genes involved in gene expression regulation, lipid and fatty acid metabolism, and noncoding RNA (Figure 1D).
The presence of noncoding RNA in the IPA analysis led us to investigate whether resident microRNAs (miRNAs) might participate in or drive changes in skeletal muscle due to CR. Previous work implicated circulating miRNAs in the mechanisms of CR (Schneider et al., 2017). miRNAs are approximately 22-nucleotide-long RNA molecules that can direct repression of mRNA targets post-transcriptionally, thereby serving as regulators of gene expression (Bartel, 2018). Small RNA was extracted and sequenced identifying 1,703 miRNA species, of which 352 passed expression threshold cutoffs, including 272 known and 80 novel species (Table S2). No individual miRNA was statistically different between the diet groups; however, nearly all canonical and non-canonical miRNA loci are transcribed by RNA polymerase II (Bartel, 2018), leading to the idea that miRNA clusters might be co-regulated. Looking at high-abundance species only, we grouped miRNA either by family, based on sequence and target similarity, or by location, a shared locus on the genome. We then used an enrichment analysis strategy based upon parametric gene set analysis (Kim and Volsky, 2005) to examine whether these groups were, in aggregate, changing in response to CR. No miRNA families were up- or downregulated in CR skeletal muscle (Figure 1E); however, one miRNA locus appeared to be responsive CR. The miRNA locus on chromosome (chr) 7 comprises 24 miRNA species that collectively were significantly downregulated because of CR. miRNAs at the equivalent locus in humans (chr14q32) have been found to be associated with cancer and metabolic disease states (González-Vallinas et al., 2018; Nadal et al., 2014; Okamoto et al., 2016). In this study, among the statistically significant transcript changes identified via BAM analysis, 148 (41%) were targets of the 24 chr7 miRNAs. Two chr7 miRNA species, miR-493–5p and miR410–3p, showed the greatest overlap between putative target genes and CR-responsive genes detected by BAM analysis (4% and 5%, respectively). Both miR-493–5p and miR-410–3p were lower in abundance with CR, with opposing increases in transcript abundance of putative targets involved in growth, cytoskeleton and contractile function, WNT signaling, and RNA processing (Figures 1F and S2). These data are consistent with the concept that miRNAs may be acting locally in skeletal muscle to implement mechanisms of CR.
CR Induces a More Youthful Cellular Metabolic Environment
We next investigated the direct impact of CR on cellular metabolism and the overall metabolic environment. The structural properties of the reduced forms of NAD and NADP confer autofluorescence that can be stimulated and quantified at defined excitation and emission wavelengths, respectively (Figure 2A). Photons released following excitation at 780 nm are detected using spectral separation of the emission signals (Bird et al., 2004; Denk et al., 1990; Lakowicz et al., 1992). Fluorescence lifetime imaging microscopy (FLIM) quantifies the kinetics of photon emission from NAD(P)H, whereas multiphoton laser-scanning microscopy (MPLSM) counts photons released to generate an image of photon intensity per pixel. Fluorescence lifetime (τ) is the duration for which a population of fluorophores stay in an excited state; protein-bound and free NAD(P)H each have their own unique lifetimes that can collectively be described by a first-order decay curve comprising two exponential decay time constants (τ1 = free and τ2 = bound) and their fractional contribution (a1 and a2) to the decay curve. From these, a mean fluorescence lifetime (τm) for NAD(P)H can be calculated. Changes in any of these parameters reflect differences in the local cellular environment, for example, we previously reported a shift to longer τm from middle age to advanced age in rhesus VL (Pugh et al., 2013). Other skeletal muscle features detected by fluorescence microscopy include collagen cross-links that are distinguished by their hydroxylysyl pyridinoline emission signatures (red strings) (Provenzano et al., 2006) and lipofuscin deposits that are identified with fluorescence because of their very short τm signature (blue dots) (Di Guardo, 2015). As appropriate to the age of the animals of this cohort, both collagen cross-links and lipofuscin deposits were detected by FLIM, and although not quantified, these appeared to be less prominent in tissues from CR monkeys.
Figure 2. Small-Molecule Indices of Metabolic Changes.
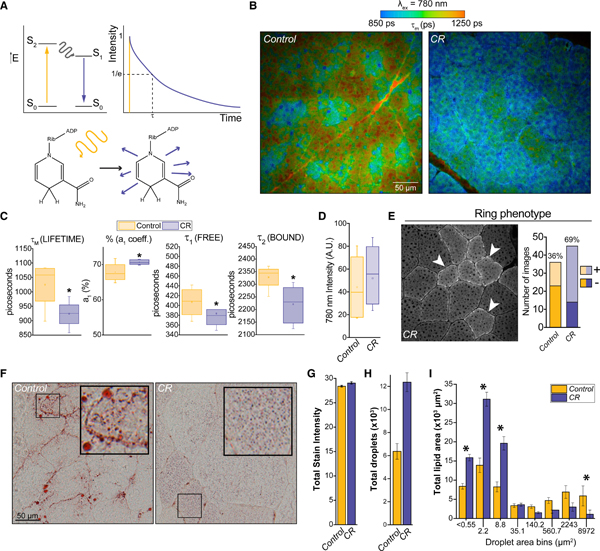
</P>(A) Schematic depicting the details of the fluorescence imaging of NAD(P)H; shown are a theoretical energy diagram (left) and fluorescence decay curve (right) that describe the excitation and emission of photons from the NAD(P)H molecule that forms the basis for the technique</P>.</P>(B) Sections of vastus lateralis prepared from control and CR rhesus monkeys were imaged using excitation wavelengths of 780 nm. Pulsed laser excitation and high-discrimination photon counting were used to generate aggregate decay curves from which fluorescent lifetime was calculated (n = 4 or 7, control or CR; 3 images per animal, representative images are shown). The weighted average of both short and long decay components (tm) in picoseconds were color mapped according to the scale shown. Scale bar, 50 μm</P>.</P>(C) Individual components of the exponential decay function for NAD(P)H in tissues from control (n = 4, 3 images per animal) and CR (n = 7, 3 images per animal) animals are shown in boxplots (box, interquartile range; whiskers, minimum and maximum values; bar, median; and small box, mean) (*p < 0.05)</P>.</P>(D) The total photon intensity at 780 nm (NAD(P)H). Boxplot is same as in (C)</P>.</P>(E) Representative intensity image highlighting the presence of the bright ring phenotype (left). Quantification (right)</P>.</P>(F) Representative images of control (left) and CR (right) muscle stained with oil red O. Scale bar, 50 μm</P>.</P>(G) Total stain intensity quantification</P>. </P>(H) Quantification of the total number of droplets (mean ± SEM; n = 5 for control or 8 for CR)</P>.</P>(I) Distribution of droplets into defined size bins. Asterisk indicates bins for which there is a statistically significant difference (p < 0.05) between diet groups(mean ± SEM). See also Figure S3 and Table S3</P>.
Decay curves for NAD(P)H in VL from control and CR monkeys were quantified and colored according to picoseconds of decay, with distinct τm signatures evident among distinct fiber types (Figure 2B). CR led to considerably shorter values of fluorescence lifetime as represented by the parameters τm, τ1, and τ2 (Figure 2C; Table S3), in each case opposing the previously reported age-related change in the NAD(P)H pool. These data demonstrate that the metabolic microenvironment in skeletal muscle is distinct for control and CR monkeys. Fluorescence intensity, while trending higher in CR, was not statistically different between diet groups (Figure 2D). At the individual fiber level, we observed a ring phenomenon where a bright ring of fluorescence was present on some of the fibers (Figure 2E). The proportion of fibers with this phenotype was substantially higher in VL from CR monkeys compared with controls, indicative of a change in spatial organization of NAD(P)H pools associated with the sarcolemma at the fiber periphery.
Prior studies in humans indicate that lipid utilization in skeletal muscle is attenuated with age, leading to larger and less numerous intracellular lipid droplets (Choi etal., 2016;Gueugneau et al., 2015). The same phenomenon has been observed with age in rhesus monkeys (Pugh et al., 2013). Using the neutral lipid stain oil red O (Figure 2F), we observed that although total intracellular stain intensity was not different between control and CR tissues, the total number of lipid droplets was substantially increased in CR animals, and the distribution of droplet areas was heavily skewed toward smaller droplets (Figures 2G–2I). The changes in lipid storage detected here are indicative of a more youthful cellular phenotype, indicating that CR-directed remodeling of lipid metabolism leads to increased accessibility of lipid fuels.
To determine whether the expression of lipid metabolism genes was changing, we used RT-PCR to quantify transcripts for genes involved in fatty acid biosynthesis, beta-oxidation, lipid biosynthesis, and lipid droplet trafficking (Figure S3). A trend toward lower expression of lipid synthesis genes was observed with CR, but none of the transcript levels were significantly different. Transcript levels of PLIN2 and PLIN3 were unaltered but PLIN5, which promotes association of lipid droplets with mitochondria (Kimmel and Sztalryd, 2014), and PLIN4, which is lowered in response to exercise (Pourteymour et al., 2015), show changes in the predicted direction, although neither reached statistical significance. Differences between control and CR in baseline transcript levels are not necessarily expected. Biopsy samples were collected from fasted animals so that lipid fuel utilization is predicted to be favored in skeletal muscle independent of diet group.
CR Preserves Fiber-Type-Specific Mitochondrial Activity and Contractile Content
Immunohistochemical detection of myosin isoforms in rhesus cryo-sectioned VL identifies three distinct fiber types: pure type I, pure type II, and mixed myosin fibers (MMFs) that express both myosin isoforms (Figure 3A). In our prior studies, we showed that in young monkeys, MMFs are the predominant fiber type but by the time of onset of sarcopenia (~15 years of age), the fiber type population shifts to equivalent proportions of MMF, pure type I, and pure type II. We reported a shift in VL fiber-type composition with CR toward a more youthful profile with MMF being the dominant fiber type, just as it is in young monkeys (Pugh et al., 2013). To investigate other cellular phenotypes of aging in this study, cross-sectional area, mitochondrial activity, and contractile content were measured in VL from control and CR monkeys.
Figure 3. Histological Assessments of Skeletal Muscle Composition and Mitochondrial Activity.
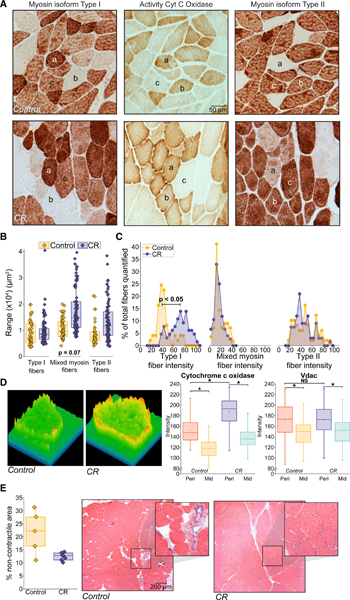
</P>(A) Representative images from control and CR muscle of histochemical stains for myosin isoform type I, cytochrome c oxidase, and myosin isoform type II. Equivalent fibers across adjacent stained sections are indicated as follows: a, pure type I; b, pure type II; and c, mixed myosin fibers (MMFs). Scale bar, 50 μm</P>.</P>(B) Quantification of fiber size distributions fortype I, II, and MMF (control n = 4 and CR n = 5; 6–16 fibers per type per animal) (box, interquartile range; whiskers, minimum and maximum values; bar, median, and small box, mean)</P>. </P>(C) Proportion of fibers at a given cytochrome c oxidase stain intensity for type I, II, and MMF (control n = 4 and CR n = 5; 6–16 fibers per type per animal)</P>. </P>(D) Assessment of mitochondrial activity and localization across fiber area. Left, 3D representation of cytochrome c oxidase staining across fiber area, highlighting heavier perimeter staining especially in CR animals. Right, quantification of cytochrome c oxidase intensity (control n = 4 and CR n = 5; 2 regions of each location type, 5 measurements per region, 1–5 fibers per image, 2–4 images per animal; total measurements n = 450 control and n = 710 CR) and VDAC (control n = 3, CR n = 4, 6 fibers per image, 2–4 images per animal; total measurements n = 108 control, n = 120 CR) in perimeter versus mid fiber. Boxplots are the same as in (B)</P>.</P>(E) Quantification (left) and representative images(right) of H&E staining of skeletal muscle highlighting the distribution of fiber contractile tissue and fibrotic non-contractile material (n = 5 control and n = 7 CR; 3–6 images per animal). Boxplots are the same as in (B). Scale bar, 200 μm See also Table S4.</P>.
Looking first at the fiber cross-sectional area, a trend toward greater fiber area for pure type II and MMF was detected for CR compared with controls, although neither reached statistical significance (Figure 3B; Table S4). The type I fibers were unaffected. Next, mitochondrial activity was assessed via enzymatic assay for cytochrome c oxidase (mitochondrial electron transport chain complex IV). Aging is associated with a decline in mitochondrial activity in type I and type II fibers in rhesus VL. In this study, type I fibers were found to have a significantly higher cytochrome c oxidase activity in tissues from CR monkeys than controls, where the distribution of fiber activity was skewed toward higher staining intensity (Figure 3C; Table S4). Type II fibers were unaffected. We previously reported greater activity in mitochondria localized to the perimeter than in the mid-fiber region and went on to show that age leads to a reduction in mitochondrial activity specifically in this sub-sarcolemmal region (Pugh et al., 2013). In this study, CR preserved the difference in mitochondrial activity stain intensity between the perimeter and the mid-fiber regions, demonstrating that compared with controls, higher mitochondrial activity of the sub-sarcolemmal population was maintained with CR (Figure 3D; Table S4). Immunodetection of the mitochondrial density marker voltage-dependent anion channel (VDAC) revealed a modest enrichment of mitochondria at the sarcolemmal periphery of oxidative fibers; however, there was no difference in mitochondrial distribution or density between control and CR, indicating that mitochondrial numbers do not explain how sub-sarcolemmal mitochondrial are more active in tissues from CR monkeys (Figure 3D). Notably, enzymatic detection of increased mitochondrial activity in the periphery in fibers from CR VL aligned with the ring phenomenon detected by FLIM.
Changes in muscle composition including a loss of contractile content is a hallmark of aging in humans (Lexell, 1995). Rhesus monkeys exhibit the same tissue-level phenotypes of muscle aging as humans and in a similar time frame, beginning in middle age. Levels of fibrosis were previously reported for the monkeys in this study and were ~14% for controls and ~6.5% for CR (McKiernan et al., 2012); however, these measures do not capture the extent of the loss of muscle functional content. In this study, histochemical analysis via hematoxylin and eosin staining was used to distinguish between the contractile (muscle fiber area) and non-contractile area (between fiber gaps and fibrotic material) (Figure 3E; Table S4). VL from CR monkeys included significantly more contractile content per total tissue area (~88%) than that of control monkeys, who, as expected for monkeys just past the age of median lifespan, had less contractile content (~78%).
CR Is Associated with Preserved Muscle Mass and Enhanced Metabolic Cost of Movement
As part of this longitudinal study of aging, measures of ESM were conducted each year via DEXA to document age-related changes. The average age of maximum or peak ESM can be calculated via breakpoint regression analysis of longitudinal DEXA body composition data (Colman et al., 2005). We previously reported that control monkeys reach peak ESM at ~16 years of age, but the CR animals do not reach peak ESM until later at ~18 years of age (McKiernan et al., 2012). In this study, consistent with the overall smaller size of the CR monkeys, peak appendicular ESM and peak ULL were numerically lower for CR than controls (Figure 4A). At the time of biopsy, measures of ESM and ULL expressed as a percent of peak were significantly higher for CR monkeys than controls (Table 2). Physical activity of the monkeys was measured daily using accelerometers as previously described (Yamada et al., 2013), and metabolic parameters were monitored via metabolic chambers on two occasions, one of which coincided with the timing of biopsy collection. Overall, measures of physical activity were numerically higher for the CR animals during the day, but the differences were not significant, likely due to the substantial variance in measures for the control group (Figure S4; Table 2). In our prior studies, we reported that the control monkeys had a significantly greater metabolic cost of movement than the CR monkeys (Yamada et al., 2013). The relationship between metabolic rate and activity is approximately linear, and the cost of activity can be calculated as the slope of the regression of metabolic rate (the dependent variable) on activity counts (Rezende et al., 2006). In this study, to understand how retention of muscle mass might relate to metabolic status, we conducted regressions of muscle mass as a percent of peak against insulin sensitivity, metabolic cost of movement, age, and activity levels. A negative correlation between ESM as a percent of peak and the metabolic cost of movement was observed for the CR animals (Figures 4B and S5). There was no relationship between muscle mass retained and metabolic cost of movement for the controls, suggesting that there may be a threshold below which muscle mass no longer has an influence on metabolic efficiency. The relationship between maintained muscle mass and circulating insulin levels was different between the two diet groups. A strong negative correlation (R2 = 0.74) between basal insulin and ESM as a percent of peak was detected for control monkeys, but in the CR monkeys where all the monkeys were insulin sensitive and muscle mass was largely retained, there was no relationship between parameters.
Figure 4. Whole-Body Measures of Muscle and Metabolism.
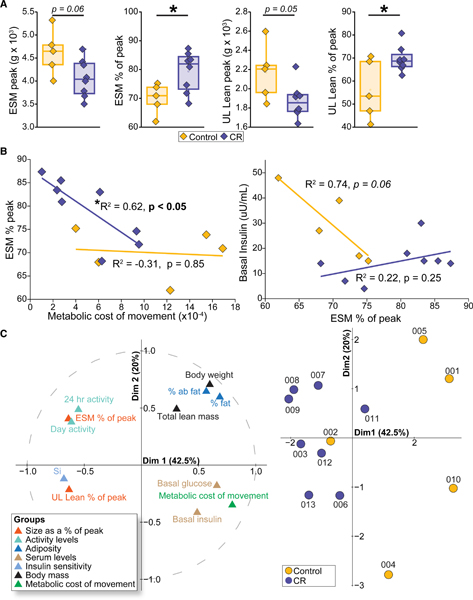
</P>(A) Indices of skeletal muscle mass (n = 5 control and n = 8 CR; *p < 0.05): ESM at peak, ESM as a % of peak, UL at peak, and UL lean mass as a % of peak</P>. </P>(B) Linear regressions of skeletal muscle mass measures with metabolic activity measures; ESM % of peak versus metabolic cost of movement (left) and basal insulin versus ESM % of peak (right)</P>.</P>(C) Multiple factor analysis of biometric indices: ESM % of peak; UL % of peak; accelerometer activity counts for morning, afternoon, day, night, and 24 h; ESM; UL Lean; ESM peak; UL lean peak; % abdominal fat; % fat; body weight; total body lean mass; insulin sensitivity index; basal serum glucose; and basal serum insulin. Left, variable correlation plot depicting the relationship between each variable and the first two principal components. Right, individuals map depicting principal component scores for control and CR animals (n = 5 control and n = 8 CR). See also Figures S4 and S5 and Table S5</P>.
Table 2.
Skeletal Muscle Mass and Activity Measures
| Parametera | Control | Restricted |
|---|---|---|
| Skeletal muscle mass | ||
| Estimated skeletal muscle mass (ESM) (g) | 3,230 ± 148 | 3,214 ± 185 |
| ESM peak (g) | 4,622 ± 426 | 4,062 ± 608 |
| ESM as % of peak | 70.0% ± 6.0% | 79.0% ± 10.0%b |
| UL lean (g) | 1,194 ± 124 | 1,303 ± 232 |
| UL lean peak (g) | 2,170 ± 282 | 1,873 ± 163 |
| UL lean as % of peak | 56.0% ± 21.0% | 70.0% ± 4.0%b |
| Activity | ||
| 24 h | 1,290.23 ± 1,020.48 | 1,918.90 ± 1,190.10 |
| Day | 1,992.58 ± 1,162.46 | 3,258.62 ± 2,482.52 |
| Night | 587.88 ± 125.42 | 579.18 ± 281.20 |
| Morning | 2,167.35 ± 630.25 | 4,322.17 ± 3,019.33 |
| Afternoon | 1,817.80 ± 1,471.75 | 2,195.07 ± 1,892.42 |
| Metabolic cost of movement (×10−4) | 10.93 ± 9.45 | 5.02 ± 4.44b |
Values are average ± interquartile range. Control n = 5 and restricted n = 8.
p < 0.05; p values adjusted with animal age at biopsy as a covariate.
A difference in variance was a common feature for the different measures conducted. Introducing age as a covariate clarified the differences between control and CR for some parameters, including fasting glucose and fasting insulin, which only reached significance when this approach was taken (Table 1). Analysis of the data from the perspective of variance revealed that body weight, metabolic cost of movement, and ULL individually all showed significantly greater variance for controls than CR (Table S5). Collectively across all biometrics, glucoregulatory function, physical activity, and metabolic assessment data, variance was significantly greater for control monkeys than CR. The compression of variance with CR is consistent with the concept that CR monkeys have a younger biological age. To understand how these data interact, we next conducted multiple factor analysis (MFA), which is essentially an extension of principal-component analysis (PCA) (Pagès, 2002). Multiple variable types are grouped into categories and weighted according to their contribution to the overall variance across the dataset. Looking at the contribution of each parameter to the variance across the first two dimensions, measures that could be viewed as somewhat related were clustered within the data space (Figure 4C). In particular, physical activity clustered with ESM expressed as a percent of peak, whereas somewhat unexpectedly, basal levels of glucose and insulin were found clustered with metabolic cost of movement, possibly connecting glucoregulatory function to metabolic efficiency. Notably, although many of the individual biometrics were not significantly different between groups, when considered together using MFA, the control and CR animals largely separate across the variable space in the first two dimensions, highlighting the advantage of considering multiple measures when distinguishing among heterogeneous populations.
DISCUSSION
One advantage of this study is the breadth of measures, including whole-body, cellular, and molecular indices quantified within a single cohort and conducted within a short time frame. There are multiple aspects to consider in skeletal muscle aging: the heterogeneity of muscle composition including contractile and non-contractile content, heterogeneity of fiber types that differ in metabolic and functional properties within the contractile content, and ultimately how these tissue and cellular-level changes translate to functional outcome (Egan and Zierath, 2013). Our prior studies have shown that changes in metabolism anticipate aging onset in skeletal muscle (Pugh et al., 2013), suggesting that metabolism might be a driver in skeletal muscle aging. If this were the case, then CR would need to impact metabolism and in particular mitochondrial function as part of its mechanisms in delaying sarcopenia. Data from this study are consistent with this concept. Aging causes a decline in the enzyme activity of complex IV of the electron transport chain, and this is opposed by CR. Aging leads to lower detected levels of NAD(P)H that are concomitant with changes in mitochondrial enzymatic activity, and CR opposes the impact of age in FLIM and MPLSM. Aging causes increases in lipid deposition within muscle fiber bundles and leads to larger, less numerous intracellular lipid droplets, suggesting altered fuel utilization with age. Here too, CR opposes this change. Cumulative data from this study demonstrate that CR abrogates tissue and cellular aging phenotypes of aging and that delayed sarcopenia is associated with attenuation of age-induced metabolic outcomes. These findings are consistent with a generalized model in which metabolism drives aging and age-related functional decline (Sun et al., 2016).
At the level of gene expression, the changes induced by CR in aging rhesus monkey skeletal muscle are consistent with evidence from human skeletal muscle aging and exercise studies. In humans, independent studies show that with exercise, the only currently effective treatment to counter sarcopenia, increased expression of genes involved in energy metabolism is predominant in the response to physical activity (Melov et al., 2007; Radom-Aizik et al., 2005). In frailty studies, diminished phosphocreatine recovery and a lower protein level of subunits of the electron transport system point to a loss in mitochondrial capacity as a contributing factor (Andreux et al., 2018). A recent transcriptional atlas of skeletal muscle aging in humans showed that mitochondrial pathways are among those most impacted by age and correlate strongest with oxygen consumption and physical capacity (Su et al., 2015). Single-fiber proteomic studies confirm fiber-type specificity in aging vulnerability and show that mitochondrial pathways are key in skeletal muscle aging (Murgia et al., 2017). In addition to mitochondrial protection with CR, transcriptional profiles point to enrichment of proteolysis pathways with CR. The ability to maintain the proteome and the integrity of protein turnover mechanisms is particularly important in skeletal muscle (Altun et al., 2010; Rom and Reznick, 2016; Sands et al., 2017). In contrast, the pathways that were downregulated by CR centered on inflammation and immune pathways. These outcomes relate to the idea of “inflammaging,” where local increases in sterile inflammation contribute to tissue-level dysfunction that may be especially important for skeletal muscle aging (Dalle et al., 2017; Franceschi et al., 2017). Overall, it appears that CR opposes the onset of age-related changes in skeletal muscle and shares mechanistic aspects with the beneficial effects of exercise.
In addition to these signature changes in skeletal muscle metabolism, molecular profiling has pointed to changes in growth signaling in the tissues from CR monkeys. This dual signature of metabolism and growth regulation was also observed in hepatic tissues from monkeys on short-term CR (Rhoads et al., 2018). Here, in skeletal muscle, at the individual gene, pathway, and network level, each analytic approach pointed to growth pathways as an intrinsic part of the CR response specifically. The recruitment of growth pathways may not be surprising given the smaller size of CR animals; however, it may have far-reaching consequences in terms of stem cell pluripotency, intracellular signaling cascades, and ligand-receptor interactions (Manning and Toker, 2017; Ward and Thompson, 2012). Growth pathways are integral in cell fate, functional dynamics including cytoskeleton assembly and organization, energetics, and growth. Studies in cells and in tissues of genetically diverse mice confirm the integration of metabolism and growth networks and indicate that mitochondria might be drivers rather than passengers in the aging process (Miller et al., 2019). An emerging theme from our work and that of others is the complex interplay between metabolic and growth regulation where changes in each define the range of response in the other, and both are coordinated through nutrient availability and nutrient-sensitive signaling (Bartke, 2017; Finkel, 2015; Ma and Gladyshev, 2017; Souder and Anderson, 2019). This may be of particular importance for immune function and inflammatory tone, where there is extensive overlap in signaling through these pathways and those responsive to growth signals (Brestoff and Artis, 2015; Hotamisligil, 2017).
The growing literature describes a role for nucleic acid signaling in the form of miRNA (Bartel, 2018), including their potential involvement in CR and genetic models of delayed aging (Makwana et al., 2017; Schneider et al., 2017; Victoria et al., 2015). Human and nonhuman primate miRNAs are highly homologous (Yue et al., 2008), indicating that their physiological frame of influence is likely to be conserved. Studies specifically investigating miRNA production in skeletal muscle have presented a complex picture with targets identified in growth, nutrient-sensing, and inflammatory pathways (Jung et al., 2019). In this study, miRNA induced by CR appear to target growth pathways including contractile processes that could arguably influence functional capacity independent of muscle bulk. We have previously published data showing that circulating miRNAs are altered with CR in monkeys (Schneider et al, 2017), and ongoing studies are investigating the impact of CR in adipose tissues. Together, these data raise the possibility that CR leads to recruitment of miRNAs as a means to regulate gene expression, both systemically and at the tissue-specific level.
The importance of age-related increases in variance among diverse measures is an emerging theme in the biology of aging research (Lowsky et al., 2014). The impact of age to increase variance in human gene expression within and among tissues was observed quite some time ago (Somel et al., 2006), with more recent studies in simple species such as the nematode showing divergence in transcription regulation even within individual cells (Mendenhall et al., 2017). Our data confirm this broadly observed phenomenon in which the control group showed consistently greater variance among a host of distinct measures. Human and nonhuman primate aging studies have had to deal with the dual challenge of genetic heterogeneity and increased variance as a function of age. Statistical approaches that interrogate relationships among parameters may be more revealing of the underlying biology than strategies seeking to identify individual parameters significantly altered with aging. MFA across datasets revealed unexpected patterns of connectivity, suggesting that disparate facets of muscle aging share underlying biology and that this is intimately connected to metabolic status. These approaches link the landscape of molecular and cellular change induced by CR to differences in functional outcomes including muscle mass retention, physical activity, and whole-body metabolic consequences of the CR diet. We show that the patterns of association among measured parameters are quite different for control and CR animals. Overall, this study reveals molecular and cellular underpinnings of delayed skeletal muscle aging with CR and provides compelling evidence for a role of metabolism not only in skeletal muscle aging but also in the mechanisms of CR.
Limitations
Apart from the size of the cohort, which is often an issue with NHP studies, the inclusion of only male monkeys is a limitation of this study. It will be important to discover if there is dimorphism in skeletal muscle aging and if the response to CR is equivalent for males and females. Given the importance of menopause in human aging including sarcopenia and frailty, the nonhuman primate model may hold some surprises as rhesus monkeys, unlike humans, retain menses until approximately the median lifespan (Walker and Herndon, 2008). The investigation of miRNAs in aging and in particular in skeletal muscle is a growing area of research (Jung et al., 2019) but one that is still in the early stages. It will be important to follow through on the exciting leads identified here to demonstrate functionality of the CR-sensitive skeletal muscle resident miRNAs. Another factor is the relatively compressed age range represented in this study; however, this strategy was engaged to maximize our ability to detect CR-induced differences in the tissue. The genetic heterogeneity of the rhesus monkey as a model can create problems in probing the underlying biology in any biomedical scenario; unlike mice, these monkeys are not clones. The challenges associated with this genetic backdrop are further compounded by the heterogeneity of variance as a function of age.
STAR★METHODS
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and request for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Rozalyn Anderson (rozalyn.anderson@wisc.edu). This study did not generate new unique reagents.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
All animal procedures were performed at the Wisconsin National Primate Research Center (WNPRC) under approved protocols from the Institutional Animal Care and Use Committee of the Office of the Vice Chancellor for Research and Graduate Education of the University of Wisconsin-Madison. This work is part of an ongoing longitudinal study of aging and CR in rhesus macaques (Macaca mulatta, taxon ID: 9544) at the WNPRC; as such, many of the data types analyzed were collected repeatedly throughout the longitudinal study. The study was initialized with adult animals (between 8 and 14 years of age) assigned to control (ad libitum fed) and CR (~30% restriction) diets. For the purposes of this investigation of advanced age males, analyses include biometric indices, blood draws, and biopsies collected for each animal within 6 months of the completion of 18 years on the longitudinal study.
METHOD DETAILS
Body weight of each animal was assessed throughout the study; the closest measurement to the muscle biopsy collection was used for the analyses herein. Appendicular lean mass and fat mass were assessed biannually using whole body dual energy x-ray absorptiometry (DEXA; Model DXP-L, GE/Lunar Corp., Madison, WI) scans as previously described (Yamada et al., 2013). ULL was determined by summing the lean mass from the thigh region of both limbs. Muscle mass loss for each individual animal was determined by dividing the lean mass (ULL in the case of UL Lean % of peak) at each time point by the maximum lean mass measured for that animal in the course of the study. Fat mass, determined by DEXA measurements, was used to calculate percent body fat. Percent abdominal fat was calculated using only DEXA measurement from the trunk region (excluding limbs).
Biopsy Collection
Vastus lateralis (VL) muscle biopsies were performed following standard surgical techniques immediately following DEXA. Biopsy tissue was bisected with one half of the sample flash frozen in liquid nitrogen and the other embedded in Optimal Cutting Temperature Medium (OCT, Sakura Inc., Torrance, CA) and frozen in liquid nitrogen. Samples were stored at −80°C until use. Serial cryostat sections 10 μm in thickness were cut at −14°C with a Leica Cryostat (Leica Biosystems, Buffalo Grove, IL). Sections were cut and used for staining on an as-needed basis.
Calorimetry
The details of the metabolic chambers have been described previously (Yamada et al., 2013). Briefly, twenty-four hour energy expenditure was measured in a standard cage enclosed within a transparent metabolic chamber; a portion of the exhaust air from the chamber was dried and analyzed for oxygen and CO2.
Accelerometer
Physical activity data collection has been previously described (Yamada et al., 2013). Briefly, commercial accelerometers attached to collars were used for a 3–4 week period, with the first five days being considered an adaptation period (data was discarded). The accelerometer samples activity counts every minute, and the resulting data were averaged for every hour, day (0600–1800 h), night (1800–0600 h), morning (0600–1200 h), afternoon (12001800 h), and 24 h periods.
Histochemistry
All specimens were processed simultaneously for each stain. Slide sections were stained with hematoxylin and eosin for muscle morphology and muscle fiber counts. Freshly cut cryostat sections were defrosted and air-dried for 10 min and stained for cytochrome c oxidase activity (Seligman et al., 1968). Briefly, sections were incubated in a solution of 0.1M phosphate buffer, pH 7.6, 0.5 mg/ml DAB (3,3’-diaminobenzidine), and 1 mg/ml cytochrome. Sections were incubated for 2 hr at room temperature and washed, dehydrated in a graded series of ethanol, cleared with three changes of xylene, and mounted under a glass coverslip with Permount (Fisher).
For fiber typing, slides were first processed for antigen retrieval. Slides were defrosted and air-dried for 10 min then fixed in 10% neutral buffered formalin for 10 min. Slides were washed thoroughly then placed in a polyethylene Coplin jar with 100mM sodium citrate buffer, pH 6.0, and microwaved until the solution reached 100°C. The Coplin jar was autoclaved at 121°C for 10 min, followed by slow pressure release. The slides were further cooled by successive dilutions with ddH2O until reaching room temperature then rinsed thoroughly with ddH2O. Sections were blocked with a solution of PBS, containing 1% BSA, 2% goat serum (Sigma), and 0.1% gelatin from cold-water fish skin for 30 min then blotted away. Primary antibodies diluted in Primary antibody diluent (Scytek, Provo, UT) were applied to the slides and incubated overnight at 4°C. Sections were rinsed thoroughly then incubated in biotinylated anti-mouse IgG or biotinylated anti-rabbit IgG (Vector Labs) for 1 h. Sections were rinsed, then incubated in peroxidase labeled avidin biotin complex (ABC) solution (Vector Labs) for 1 h and color developed with ImPACT NovaRED reagent (Vector Labs) for ~5–8 min and rinsed. Duration of incubation was optimized for each experiment to ensure adequate detection and to avoid over saturation. The primary antibodies used were anti-myosin Type I isoform (M8421, skeletal, slow myosin, monoclonal clone NOQ7.5.4D, Sigma-Aldrich, St. Louis, MO) and anti-myosin Type II isoform (M1570, skeletal, fast myosin, monoclonal clone MY32, Sigma-Aldrich), and anti-VDAC1/porin (ab15895, Abcam, Cambridge, MA).
For lipid staining, freshly cut cryostat sections were defrosted, air-dried for 10 min, fixed in 10% neutral buffered formalin for 1 h, and then rinsed thoroughly. Sections were incubated in a solution of 3 mg/ml Oil Red O in 36% triethyl-phosphate for 30 min (Koopman et al., 2001). Sections were rinsed with 3 changes of ddH2O and covered with a glass coverslip cemented with Clear-Mount (EMS, Hatfield, PA). Each experiment was conducted at least twice and equivalence of staining between batches confirmed.
Multiphoton Imaging
Autofluorescent detection and lifetime imaging was conducted using a multiphoton workstation at the University of Wisconsin Laboratory for Optical and Computational Instrumentation (LOCI) (Bird et al., 2004). The system is built around an inverted microscope (TE 2000, Nikon, Melville, NY) using illumination from a Ti:Sapphire mode-locking laser (tuning range of ~700–1000 nm, Coherent Mira, Coherent, Santa Clara, CA) pumped by an 8 W solid-state laser (Coherent Verdi) to generate pulse widths of approximately 120 fs at a repetition rate of 76 MHz. A H7422P GaAsP photon counting PMT (Hamamatsu) was used for intensity and lifetime imaging.
Cryosectioned tissue 10 μm thick was mounted on glass slides and dried. Fluorescence lifetimes were acquired with an electronic system for recording fast light signals by time correlated single photon counting (SPC-830, Becker & Hickl). The instrument response function of the optical system was calibrated using second harmonic generated signal from a ß-BaB2O4 crystal (Bird et al., 2004). A Nikon PlanApo VC 20X lens (N.A. = 0.75) was used for all imaging. For imaging of NAD(P)H the laser was tuned to 780 nm combined with 457/50 filtered emission. FLIM data were collected for 240 s using a pixel frame size for the multiphoton intensity images of 1024×1024, while lifetime images were 256 × 256. Acquisition was performed with WiscScan, a LOCI developed acquisition package software. Intensity data was analyzed in Fiji (Schindelin et al., 2012).
For analysis of fluorescence intensity and lifetime, data were imported into SPCImage (Becker & Hickl, v.2.9.1) where a Levenberg-Marquardt routine for nonlinear fitting was used to fit the fluorescence decay curve collected for each pixel in the 256 × 256 frame to a model multi-exponential decay function of the form:
where If (t) is the fluorescence intensity at time t after the excitation pulse, n is the total number of decay components in the exponential sum, and c is a constant pertaining to the level of background light noise. The variables ti and ai are the fluorescence lifetime and fractional contribution of the ith emitting species, respectively. Corrections were made for the noise using a calibration of the instrument response function (Bird et al., 2004). Data were assessed by the minimized Chi-square value generated during the fit so that analysis was unbiased. Data with a Chi-squared value of greater than 1.3 was deemed to be of a poor fit and was eliminated from the results. To eliminate background fluorescence a threshold for analysis was applied based on photon counts. Data were binned at >1000 photons/pixel to maintain high quality of the fit and was therefore collected using time correlated single photon counting techniques where the data was analyzed on a pixel-by-pixel analysis approach as described previously (Conklin et al., 2009).
Digital Image Capture and Analysis
Unless otherwise stated the stained slides were imaged with a 20x objective (10x for H&E staining used for non-contractile area analysis) in a Leica DM4000B microscope and photographed with a Retiga 4000R digital camera (QImaging Systems, Surrey, BC, Canada). Camera settings were optimized for each stain and for uniformity all images with were taken with identical settings, fixed light levels and fixed shutter speed. Images for Oil Red O lipid stained sections were captured within 24 h of tissue processing. Quantitative image analysis was performed using ImagePro Plus 6.3.1.542 Software (Media Cybernetics, Rockville, MD), FIJI (Schindelin et al., 2012), or Photoshop CS3 (Adobe Systems, San Jose, CA).
For cytochrome c activity stain, digital images were converted from color to monochrome and inverted, dark to light. For the analysis of the overall staining by fiber types, fibers were first identified as Type I, Type II, or mixed myosin from the presence or absence of staining for fiber type-specific myosin isoforms in adjacent sections. The cross-sections of fibers were outlined using a freehand drawing tool and the average intensity of the enclosed area was recorded using Photoshop. For perimeter and mid-fiber measures a pixel intensity line profile across individual fibers was created in FIJI for both cytochrome c oxidase and VDAC stains. Each pixel in the line profile was measured and data were smoothed over 10 pixels in single pixel increments. Intensity peaks (5 pixels) corresponding to cell perimeter and two points equidistant regions in the intervening space were identified.
For Oil red O lipid droplet detection, RGB digital images were color extracted to remove contributions from blue and green channels using Photoshop. The extracted images were imported into ImagePro Plus and a filter range of 60–255 was applied. Using the object detection tool, droplets were counted and size was measured and color-coded according to size categories. Unintentionally merged objects were segregated using the watershed split tool. The output of the automated droplet detection and size category classification were confirmed visually by comparison with the unprocessed original images. Data were exported as numbers of droplets within each category and area contained in size categories.
RNA Extraction
Roughly 50 mg of each rhesus muscle biopsy sample was homogenized in 1 mL TRIzol (ThermoFisher #15596018). Isolation of total RNA, including small RNA, was performed with the PureLink RNA Mini Kit (ThermoFisher #12183018A) according to the manufacturer’s protocol and RNA was eluted in 50 uL of RNase-free water.
RNA Expression Analysis
For mRNA gene expression analysis, 1 μg of total RNA from each sample was used to construct the microarray libraries with the GeneChip One-cycle Target Labeling and Control Reagents Kit (Affymetrix #900493) according to the manufacturer’s protocol. Fragmented and labeled DNA was hybridized to the Affymetrix M. mulatta microarray (Affymetrix #900655) according to the manufacturer’s instructions. Following hybridization, chips were read with a Hewlett-Packard GeneArray Scanner.
The MAS5 algorithm was used to generate signal intensity data from CEL files as previously described (Barger et al., 2015). Signal intensity data was then filtered to a set of unique transcripts with known Entrez Gene IDs. BAM uses a Bayesian hierarchical model to account for uncertainty, yielding gene effect estimates that are selectively shrunken relative to maximum likelihood estimates. This process generally shrinks the non-differentially expressed genes, enabling optimal balance against total false non-detections.
miRNA Expression Analysis
5 μg of total RNA from each sample was used to construct small-RNA libraries with the Illumina TruSeq Small RNA Sample Prep kit (Illumina, San Diego, CA, #RS-200–0012) according to the manufacturer’s instructions. Briefly, 3’ and 5’ adapters were sequentially ligated to small RNAs followed by cDNA synthesis, PCR amplification, extraction, and gel purification. Size, concentration, and purity of amplicons were confirmed on an Agilent Bioanalyzer High Sensitivity DNA chip (Agilent Genomics, Santa Clara, CA, #5067–4626). The libraries were pooled in equimolar amounts and sequenced on an Illumina HiSeq 2500 instrument to generate 1×100 base reads.
Bioinformatics for miRs
Genome build Mmul 8.0.1 for Macaca mulatta (Rhesus Monkey rheMac8) based on assembly accession GCA_000772875.3 was downloaded from NCBI and indexed with bowtie v1.0.0. The Perl script mapper.pl from the miRdeep2 package (Friedländer et al., 2012) patched with commit 791b689 (available on Github.com from the miRdeep2 authors) was used to trim the Illuimina TruSeq 3′ adaptor (-k TGGAATTCTCGGGTGCCAAGG) from input fastq reads (-e), discard reads shorter than 18 nucleotides (-l 18), and collapse duplicate reads (-m). After trimming operations, the filtered and collapsed reads were mapped with bowtie v1.0.0 to the Mmul 8.0.1 genome with no more than 0 mismatches allowed in the seed alignment (-n 0) and a total quality value of all mismatches in the aligned read not exceeding 80 (-e 80). The alignment mode was also performed with the options –best and –strata to report only the best alignment, based on quality, if two or more valid alignments were made.
Aligned reads were analyzed with miRDeep2.pl module, an algorithm based on a miRNA biogenesis model that uses NGS short-read data as input. It aligns reads to potential hairpin structures in a manner consistent with Dicer processing (Park et al., 2011), evaluates their minimum free energy, and assigns scores to measure the probability that the observed hairpins are true miRNA precursors. This information is coded into log-odd scores that predict how likely a given small RNA segment is a true miRNA. The miRDeep2 software performs this procedure for both known and putative (novel) miRNAs. Known mature and precursor noncoding miRNAs derived from rheMac8 were downloaded from miRbase 22 (miRbase.org; September 2018). Mature noncoding miRNAs identified in human (GRCh38; GCA_000001405.15) were designated as the phylogenetically related species for use filtering probable miRNA candidates.
In addition to identification of known and novel miRNAs, miRDeep2 also estimates expression levels for any detected miRNA. To test for differential expression among the control (C) and caloric-restricted (CR) sample groups, read counts of miRNAs were used as input into edgeR (Robinson et al., 2010). The significance of the negative-binomial test was adjusted with a Benjamini-Hochberg FDR correction at the 5% level (Reiner et al., 2003). Prior to analysis with edgeR, filtering of the miRNAs was performed, requiring a threshold of least 3 reads per million in 60% of individuals in control and caloric-restricted groups.
QUANTIFICATION AND STATISTICAL ANALYSIS
Rhesus monkeys are genetically heterogeneous, necessitating that the statistical approach was optimized for each experiment. For routine analysis Student’s t-test was applied with a cut off of p<0.05, FDR corrected or uncorrected as the analysis dictated. For biometric indices, ANCOVA models with age at biopsy as a covariate were used to test the mean differences between control and CR groups. Levene’s Test for Homogeneity Variance of Squared Deviations from Group Means (Levene, 1960) was used to test group differences in variability. The multivariate analog of the Levene Test suggested by Anderson (Anderson, 2006) was used as a multivariate test of differences in variance.
Percent Fiber per Functional Area
A linear mixed model (LMM) was performed to account for potential dependency among the measures. To correct for potential heterogeneity of variance across the age groups, adjustment of the degrees-of freedom were used (Kenward and Roger, 1997). Pairwise comparisons with Kenward-Roger dfs for heterogeneous variances and Tukey-Kramer type adjustments for multiple testing were made.
Cross-Sectional Area Analysis
A LMM was performed to account for potential dependency among the measures. Because of the relatively equal number of observations per animal and the expected low amount of dependency among observations residual degrees-of freedom were used (Bland and Altman, 1995).
Fiber Type Distribution
Pairwise comparisons were made using the mean ranks from nonparametric Kruskal-Wallis test, and a pairwise Fligner-Policello for unequal variances. For fiber type distribution in control and CR animals, a semi-parametric regression technique was applied. Generalized Estimating Equations (GEE) with exchangeable covariance structures were used to account for the dependency in the data and to determine significant differences in fiber numbers across groups.
Cytochrome c Oxidase Activity and VDAC Staining
a LMM was used to test differences in the groups. O’Brien’s Test for Homogeneity of Variance performed on the un-clustered data to determine variability. A heterogeneous compound-symmetric covariance structure for the LMM was specified including the containment method for degrees-of-freedom (df). Examination of the residuals from these models indicated no extreme departures from the normality assumption, but did confirm the heterogeneity of variance. Pairwise comparisons with Tukey-Kramer adjustments for multiple testing were applied to determine differences among groups.
Bright Periphery Fiber Images in 2-Photon Analysis
We assessed the proportion of intensity images that contained at least one fiber with the bright periphery phenotype by manual inspection.
Lipid Droplet Size and Area
GEE with exchangeable covariance structures were used to account for the dependency in the repeatedly measured data because of the typical non-normality associated with count and area data. For count data Poisson distributions were evaluated in the GEE. In preliminary analyses the variance of the number of droplets was often larger than the average number droplets, sometimes smaller, and when there were zero droplets there was zero variance. Since the number of droplets was modeled across multiple Bin Sizes and there were multiple fields for each subject, the data could be viewed as a mixture of Poisson distributions. Therefore, the Negative Binomial distribution was evaluated to model this over-dispersion. Due to zero droplets in some Bins, zero-inflated Poisson and Negative Binomial distribution were also evaluated. The natural logarithm of the sum for each plate was evaluated as an offset variable. For the pixel area of the droplets a GEE was modeled with a Gamma distribution.
BAM, PAGE, and Ingenuity Analysis
For gene expression data, individual Student’s t-tests were performed for each gene; those passing a threshold of p < 0.05 are used for broad strokes pathway and GO term analysis. For determination of the statistical significance of individual genes, the analysis of the microarray was performed via BAM, which uses a Bayesian hierarchical model to account for uncertainty, yielding gene effect estimates that are selectively shrunken relative to maximum likelihood estimates. This process generally shrinks the non-differentially expressed genes, enabling optimal balance against total false non-detections. For miRNA analysis, a Benjamini-Hochberg correction was applied to the p-values to account for multiple hypothesis testing. Assessment of group behavior of miRNA loci and families was performed via the method of Kim and Volsky as implemented in the R package PGSEA (Kim and Volsky, 2005).
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-Myosin Type I isoform, monoclonal NOQ37.5.4D | Sigma-Aldrich | Cat #: M8421; RRID: AB_2147479 |
| Biotinylated anti-mouse IgG | Vector Labs | Cat #: BA-2000; RRID: AB_2313581 |
| Biotinylated anti-rabbit IgG | Vector Labs | Cat #: BA-1100; RRID: AB_2336201 |
| Anti-Myosin Type II isoform, monoclonal MY-32 | Sigma-Aldrich | Cat #: M1570; RRID: AB_531175 |
| Anti-VDAC1 | Abcam | Cat #: ab15895; RRID: AB_2214787 |
| Biological Samples | ||
| Vastus lateralis biopsies | Macaca mulatta | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Cytochrome c | Sigma-Aldrich | C-2037 |
| 3,3’-diaminobenzidine tetrahydrochloride | Sigma-Aldrich | D-5905 |
| Peroxidase-labeled avidin biotin complex solution | Vector Labs | PK-6100 |
| Triethyl phosphate | Sigma-Aldrich | 538728–1L |
| Oil Red O | Alfa Aesar | A12989 |
| Sodium citrate | Fisher | BP327–1 |
| Phosphate buffered saline | Gibco | 14200–075 |
| Bovine serum albumin | Sigma-Aldrich | A2153–100G |
| Goat serum | Life Tech | 50062Z |
| Gelatin | Sigma-Aldrich | G7765–250ML |
| Primary antibody diluent | Scytek | ABB-500 |
| Formalin | Fisher | 245–684 |
| Clear-Mount | Electron Microscopy Sciences | 17985 |
| Hematoxylin | Sigma-Aldrich | MHS32 |
| Eosin | Sigma-Aldrich | HT110132 |
| Critical Commercial Assays | ||
| Illumina TruSeq RNA sample prep kit | Illumina | RS-122–2001 |
| Agilent DNA 1000 series chip assay | Agilent | 5067–1505 |
| TruSeq 100 bp SBS kit | Illumina | FC-104–5001 |
| Affymetrix GeneChip Rhesus microarray | Affymetrix | 900655 |
| GeneChip One-cycle target labeling and control reagents kit | Affymetrix | 900493 |
| Deposited Data | ||
| Raw and analyzed microarray (details) | This study | GEO: GSE64235 |
| Raw and processed miRNA-seq | This study | GEO: GSE139081 |
| Experimental Models: Organisms/Strains | ||
| Non-human primate: Macaca mulatta | Wisconsin National Primate Research Center | Colman et al., 2009 |
| Software and Algorithms | ||
| Networkx | https://networkx.github.io/ | N/A |
| SPCImage | Becker & Hickl | N/A |
| FactoMineR | http://factominer.free.fr/, CRAN | de Tayrac et al. (2009) |
| Factoextra | CRAN | N/A |
| FIJI | https://imagej.net/Welcome | Schindelin et al. (2012) |
| miRDeep2 | https://github.com/rajewsky-lab/mirdeep2 | Friedlander et al. (2012) |
Highlights.
Caloric restriction delays multiple features of skeletal muscle aging in monkeys
Pathways of growth and metabolism are recruited by caloric restriction
Mitochondrial metabolism is implicated in the mechanisms of delayed aging
Age-induced changes in physical measures correlate with
metabolic status
ACKNOWLEDGMENTS
We would like to acknowledge Scott Baum, Susan McKiernan, Derek Pavelec, and Yosuke Yamada, for consultation on sample procurement, data collection, and analysis. We thank Richard Weindruch for leadership of the study in earlier years. Funding for this study was provided by NIH grants AG037000, AG011915, AG047358, and AG040178 and the Department of Medicine, School of Medicine and Public Health, University of Wisconsin-Madison. The Wisconsin National Primate Research Center is supported by P51OD011106 and Research Facilities Improvement Programs RR15459–01 and RR020141–01. The study was conducted with the use of resources and facilities at the Geriatric Research Education and Clinical Center in the William S. Middleton Memorial Veterans Hospital, Madison, WI.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.cels.2019.12.002.
DATA AND CODE AVAILABILITY
The accession number for the microarray data reported in this paper is GEO: GSE64235 and the accession number for the miRNA data reported in this paper is GEO: GSE139081. All other data and analysis is available upon request.
Data processing and analysis utilized commercial and/or freely available software packages.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Altun M, Besche HC, Overkleeft HS, Piccirillo R, Edelmann MJ, Kessler BM, Goldberg AL, and Ulfhake B (2010). Muscle wasting in aged, sarcopenic rats is associated with enhanced activity of the ubiquitin proteasome pathway. J. Biol. Chem. 285, 39597–39608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson MJ (2006). Distance-based tests for homogeneity of multivariate dispersions. Biometrics 62, 245–253. [DOI] [PubMed] [Google Scholar]
- Andreux PA, van Diemen MPJ, Heezen MR, Auwerx J, Rinsch C, Groeneveld GJ, and Singh A (2018). Mitochondrial function is impaired in the skeletal muscle of pre-frail elderly. Sci. Rep. 8, 8548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian P, Mattison JA, and Anderson RM (2017). Nutrition, metabolism, and targeting aging in nonhuman primates. Ageing Res. Rev 39, 29–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barger JL, Anderson RM, Newton MA, da Silva C, Vann JA, Pugh TD, Someya S, Prolla TA, and Weindruch R (2015). A conserved transcriptional signature of delayed aging and reduced disease vulnerability is partially mediated by SIRT3. PLoS One 10, e0120738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP (2018). Metazoan microRNAs. Cell 173, 20–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartke A (2017). Somatic growth, aging, and longevity. NPJ Aging Mech. Dis. 3, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassel-Duby R, and Olson EN (2006). Signaling pathways in skeletal muscle remodeling. Annu. Rev. Biochem. 75, 19–37. [DOI] [PubMed] [Google Scholar]
- Bird DK, Eliceiri KW, Fan CH, and White JG (2004). Simultaneous twophoton spectral and lifetime fluorescence microscopy. Appl. Opt. 43, 5173–5182. [DOI] [PubMed] [Google Scholar]
- Bland JM, and Altman DG (1995). Calculating correlation coefficients with repeated observations: part 1–correlation within subjects. BMJ 310, 446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brestoff JR, and Artis D (2015). Immune regulation of metabolic homeostasis in health and disease. Cell 161, 146–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brioche T, Pagano AF, Py G, and Chopard A (2016). Muscle wasting and aging: experimental models, fatty infiltrations, and prevention. Mol. Aspects Med. 50, 56–87. [DOI] [PubMed] [Google Scholar]
- Choi SJ, Files DC, Zhang T, Wang ZM, Messi ML, Gregory H, Stone J, Lyles MF, Dhar S, Marsh AP, et al. (2016). Intramyocellular lipid and impaired myofiber contraction in normal weight and obese older adults. J. Gerontol. A Biol. Sci. Med. Sci. 71, 557–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colman RJ, Anderson RM, Johnson SC, Kastman EK, Kosmatka KJ, Beasley TM, Allison DB, Cruzen C, Simmons HA, Kemnitz JW, and Weindruch R (2009). Caloric restriction delays disease onset and mortality in rhesus monkeys. Scinece 325, 201–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colman RJ, Beasley TM, Allison DB, and Weindruch R (2008). Attenuation of sarcopenia by dietary restriction in rhesus monkeys. J. Gerontol. A Biol. Sci. Med. Sci. 63, 556–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colman RJ, Beasley TM, Kemnitz JW, Johnson SC, Weindruch R, and Anderson RM (2014). Caloric restriction reduces age-related and allcause mortality in rhesus monkeys. Nat. Commun. 5, 3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colman RJ, McKiernan SH, Aiken JM, and Weindruch R (2005). Muscle mass loss in rhesus monkeys: age of onset. Exp. Gerontol. 40, 573–581. [DOI] [PubMed] [Google Scholar]
- Conklin MW, Provenzano PP, Eliceiri KW, Sullivan R, and Keely PJ (2009). Fluorescence lifetime imaging of endogenous fluorophores in histopathology sections reveals differences between normal and tumor epithelium in carcinoma in situ of the breast. Cell Biochem. Biophys. 53, 145–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalle S, Rossmeislova L, and Koppo K (2017). The role of inflammation in age-related sarcopenia. Front. Physiol. 8, 1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Magalhaães JP, Curado J, and Church GM (2009). Meta-analysis of age-related gene expression profiles identifies common signatures of aging. Bioinformatics 25, 875–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Tayrac M, Lê S, Aubry M, Mosser J, and Husson F (2009). Simultaneous analysis of distinct Omics data sets with integration of biological knowledge: multiple factor analysis approach. BMC Genomics 10, 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denk W, Strickler JH, and Webb WW (1990). Two-photon laser scanning fluorescence microscopy. Science 248, 73–76. [DOI] [PubMed] [Google Scholar]
- Di Guardo G (2015). Lipofuscin, lipofuscin-like pigments and autofluorescence. Eur. J. Histochem. 59, 2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan B, and Zierath JR (2013). Exercise metabolism and the molecular regulation of skeletal muscle adaptation. Cell Metab. 17, 162–184. [DOI] [PubMed] [Google Scholar]
- Figueiredo PA, Powers SK, Ferreira RM, Appell HJ, and Duarte JA (2009). Aging impairs skeletal muscle mitochondrial bioenergetic function. J. Gerontol. A Biol. Sci. Med. Sci. 64, 21–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel T (2015). The metabolic regulation of aging. Nat. Med. 21, 1416–1423. [DOI] [PubMed] [Google Scholar]
- Franceschi C, Garagnani P, Vitale G, Capri M, and Salvioli S (2017). Inflammaging and ‘garb-aging’. Trends Endocrinol. Metab. 28, 199–212. [DOI] [PubMed] [Google Scholar]
- Friedlaänder MR, Mackowiak SD, Li N, Chen W, and Rajewsky N (2012). miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 40, 37–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Vallinas M, Rodríguez-Paredes M, Albrecht M, Sticht C, Stichel D, Gutekunst J, Pitea A, Sass S, Sánchez-Rivera FJ, Lorenzo-Bermejo J, et al. (2018). Epigenetically regulated chromosome 14q32 miRNA cluster induces metastasis and predicts poor prognosis in lung adenocarcinoma patients. Mol. Cancer Res. 16, 390–402. [DOI] [PubMed] [Google Scholar]
- Gueugneau M, Coudy-Gandilhon C, Théron L, Meunier B, Barboiron C, Combaret L, Taillandier D, Polge C, Attaix D, Picard B, et al. (2015). Skeletal muscle lipid content and oxidative activity in relation to muscle fiber type in aging and metabolic syndrome. J. Gerontol. A Biol. Sci. Med. Sci. 70, 566–576. [DOI] [PubMed] [Google Scholar]
- Hotamisligil GS (2017). Inflammation, metaflammation and immunometabolic disorders. Nature 542, 177–185. [DOI] [PubMed] [Google Scholar]
- Ishwaran H, and Rao JS (2003). Detecting differentially expressed genes in microarrays using Bayesian model selection. J. Am. Stat. Assoc. 98, 438–455. [Google Scholar]
- Jung HJ, Lee KP, Kwon KS, and Suh Y (2019). MicroRNAs in skeletal muscle aging: current issues and perspectives. J. Gerontol. A Biol. Sci. Med. Sci. 74, 1008–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenward MG, and Roger JH (1997). Small sample inference for fixed effects from restricted maximum likelihood. Biometrics 53, 983–997. [PubMed] [Google Scholar]
- Kim SY, and Volsky DJ (2005). PAGE: parametric analysis of gene set enrichment. BMC Bioinformatics 6, 144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel AR, and Sztalryd C (2014). Perilipin 5, a lipid droplet protein adapted to mitochondrial energy utilization. Curr. Opin. Lipidol. 25, 110–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koopman R, Schaart G, and Hesselink MK (2001). Optimisation of oil red O staining permits combination with immunofluorescence and automated quantification of lipids. Histochem. Cell Biol. 116, 63–68. [DOI] [PubMed] [Google Scholar]
- Lakowicz JR, Szmacinski H, Nowaczyk K, and Johnson ML (1992). Fluorescence lifetime imaging of free and protein-bound NADH. Proc. Natl. Acad. Sci. USA 89, 1271–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levene H (1960). Robust tests for equality of variances In Contributions to Probability and Statistics, Olkin I, ed. (Stanford University Press; ), pp. 278–292. [Google Scholar]
- Lexell J (1995). Human aging, muscle mass, and fiber type composition. J. Gerontol. A Biol. Sci. Med. Sci. 50, 11–16. [DOI] [PubMed] [Google Scholar]
- Lowsky DJ, Olshansky SJ, Bhattacharya J, and Goldman DP (2014). Heterogeneity in healthy aging. J. Gerontol. A Biol. Sci. Med. Sci. 69, 640–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma S, and Gladyshev VN (2017). Molecular signatures of longevity: insights from cross-species comparative studies. Semin. Cell Dev. Biol. 70, 190–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahdy MAA (2019). Skeletal muscle fibrosis: an overview. Cell Tissue Res. 375, 575–588. [DOI] [PubMed] [Google Scholar]
- Makwana K, Patel SA, Velingkaar N, Ebron JS, Shukla GC, and Kondratov RVKV (2017). Aging and calorie restriction regulate the expression of miR-125a-5p and its target genes Stat3, Casp2 and Stard13. Aging (Albany NY) 9, 1825–1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning BD, and Toker A (2017). AKT/PKB signaling: navigating the network. Cell 169, 381–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattison JA, Colman RJ, Beasley TM, Allison DB, Kemnitz JW, Roth GS, Ingram DK, Weindruch R, de Cabo R, and Anderson RM (2017). Caloric restriction improves health and survival of rhesus monkeys. Nat. Commun. 8, 14063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGregor RA, Cameron-Smith D, and Poppitt SD (2014). It is not just muscle mass: a review of muscle quality, composition and metabolism during ageing as determinants of muscle function and mobility in later life. Longev. Healthspan 3, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKiernan SH, Colman R, Lopez M, Beasley TM, Weindruch R, and Aiken JM (2009). Longitudinal analysis of early stage sarcopenia in aging rhesus monkeys. Exp. Gerontol. 44, 170–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKiernan SH, Colman RJ, Aiken E, Evans TD, Beasley TM, Aiken JM, Weindruch R, and Anderson RM (2012). Cellular adaptation contributes to calorie restriction-induced preservation of skeletal muscle in aged rhesus monkeys. Exp. Gerontol. 47, 229–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melov S, Tarnopolsky MA, Beckman K, Felkey K, and Hubbard A (2007). Resistance exercise reverses aging in human skeletal muscle. PLoS One 2, e465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendenhall A, Crane MM, Tedesco PM, Johnson TE, and Brent R (2017). Caenorhabditis elegans genes affecting interindividual variation in life-span biomarker gene expression. J. Gerontol. A Biol. Sci. Med. Sci. 72, 1305–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller KN, Clark JP, Martin SA, Howell PR, Burhans MS, Haws SA, Johnson NB, Rhoads TW, Pavelec DM, Eliceiri KW, et al. (2019). PGC1a integrates a metabolism and growth network linked to caloric restriction. Aging Cell 18, e12999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstråle M, Laurila E, et al. (2003). PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 34, 267–273. [DOI] [PubMed] [Google Scholar]
- Murgia M, Toniolo L, Nagaraj N, Ciciliot S, Vindigni V, Schiaffino S, Reggiani C, and Mann M (2017). Single muscle fiber proteomics reveals fiber-type-specific features of human muscle aging. Cell Rep. 19, 2396–2409. [DOI] [PubMed] [Google Scholar]
- Nadal E, Zhong J, Lin J, Reddy RM, Ramnath N, Orringer MB, Chang AC, Beer DG, and Chen G (2014). A microRNA cluster at 14q32 drives aggressive lung adenocarcinoma. Clin. Cancer Res. 20, 3107–3117. [DOI] [PubMed] [Google Scholar]
- Noland RC, Koves TR, Seiler SE, Lum H, Lust RM, Ilkayeva O, Stevens RD, Hegardt FG, and Muoio DM (2009). Carnitine insufficiency caused by aging and overnutrition compromises mitochondrial performance and metabolic control. J. Biol. Chem. 284, 22840–22852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto K, Koda M, Okamoto T, Onoyama T, Miyoshi K, Kishina M, Kato J, Tokunaga S, Sugihara TA, Hara Y, et al. (2016). A series of microRNA in the chromosome 14q32.2 maternally imprinted region related to progression of non-alcoholic fatty liver disease in a mouse model. PLoS One 11, e0154676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagano AF, Brioche T, Arc-Chagnaud C, Demangel R, Chopard A, and Py G (2018). Short-term disuse promotes fatty acid infiltration into skeletal muscle. J. Cachexia Sarcopenia Muscle 9, 335–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagès J (2002). Analyse factorielle multiple appliquée aux variables qualitatives et aux données mixtes. Rev. Stat. Appl. 50, 5–37. [Google Scholar]
- Park JE, Heo I, Tian Y, Simanshu DK, Chang H, Jee D, Patel DJ, and Kim VN (2011). Dicer recognizes the 50 end of RNA for efficient and accurate processing. Nature 475, 201–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pette D, and Staron RS (2000). Myosin isoforms, muscle fiber types, and transitions. Microsc. Res. Tech. 50, 500–509. [DOI] [PubMed] [Google Scholar]
- Pourteymour S, Lee S, Langleite TM, Eckardt K, Hjorth M, Bindesbøll C, Dalen KT, Birkeland KI, Drevon CA, Holen T, and Norheim F (2015). Perilipin 4 in human skeletal muscle: localization and effect of physical activity. Physiol. Rep. 3, e12481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proctor DN, Sinning WE, Walro JM, Sieck GC, and Lemon PW (1995). Oxidative capacity of human muscle fiber types: effects of age and training status. J. Appl. Physiol. (1985) 78, 2033–2038. [DOI] [PubMed] [Google Scholar]
- Provenzano PP, Eliceiri KW, Campbell JM, Inman DR, White JG, and Keely PJ (2006). Collagen reorganization at the tumor-stromal interface facilitates local invasion. BMC Med. 4, 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugh TD, Conklin MW, Evans TD, Polewski MA, Barbian HJ, Pass R, Anderson BD, Colman RJ, Eliceiri KW, Keely PJ, et al. (2013). A shift in energy metabolism anticipates the onset of sarcopenia in rhesus monkeys. Aging Cell 12, 672–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radom-Aizik S, Hayek S, Shahar I, Rechavi G, Kaminski N, and BenDov I (2005). Effects of aerobic training on gene expression in skeletal muscle of elderly men. Med. Sci. Sports Exerc. 37, 1680–1696. [DOI] [PubMed] [Google Scholar]
- Ramsey JJ, Colman RJ, Binkley NC, Christensen JD, Gresl TA, Kemnitz JW, and Weindruch R (2000). Dietary restriction and aging in rhesus monkeys: the University of Wisconsin study. Exp. Gerontol. 35, 1131–1149. [DOI] [PubMed] [Google Scholar]
- Reiner A, Yekutieli D, and Benjamini Y (2003). Identifying differentially expressed genes using false discovery rate controlling procedures. Bioinformatics 19, 368–375. [DOI] [PubMed] [Google Scholar]
- Rezende EL, Kelly SA, Gomes FR, Chappell MA, and Garland T (2006). Effects of size, sex, and voluntary running speeds on costs of locomotion in lines of laboratory mice selectively bred for high wheel-running activity. Physiological and Biochemical Zoology 79, 83–99. [DOI] [PubMed] [Google Scholar]
- Rhoads TW, Burhans MS, Chen VB, Hutchins PD, Rush MJP, Clark JP, Stark JL, McIlwain SJ, Eghbalnia HR, Pavelec DM, et al. (2018). Caloric restriction engages hepatic RNA processing mechanisms in rhesus monkeys. Cell Metab. 27, 677–688.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, and Smyth GK (2010). edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rom O, and Reznick AZ (2016). The role of E3 ubiquitin-ligases MuRF-1 and MAFbx in loss of skeletal muscle mass. Free Radic. Biol. Med. 98, 218–230. [DOI] [PubMed] [Google Scholar]
- Sands WA, Page MM, and Selman C (2017). Proteostasis and ageing: insights from long-lived mutant mice. J. Physiol. (Lond.) 595, 6383–6390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider A, Dhahbi JM, Atamna H, Clark JP, Colman RJ, and Anderson RM (2017). Caloric restriction impacts plasma microRNAs in rhesus monkeys. Aging Cell 16, 1200–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seligman AM, Karnovsky MJ, Wasserkrug HL, and Hanker JS (1968). Nondroplet ultrastructural demonstration of cytochrome oxidase activity with a polymerizing osmiophilic reagent, diaminobenzidine (DAB). J. Cell Biol. 38, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Short KR, Bigelow ML, Kahl J, Singh R, Coenen-Schimke J, Raghavakaimal S, and Nair KS (2005). Decline in skeletal muscle mitochondrial function with aging in humans. Proc. Natl. Acad. Sci. USA 102, 5618–5623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somel M, Khaitovich P, Bahn S, Pääbo S, and Lachmann M (2006)€. Gene expression becomes heterogeneous with age. Curr. Biol. 16, R359–R360. [DOI] [PubMed] [Google Scholar]
- Souder DC, and Anderson RM (2019). An expanding GSK3 network: implications for aging research. GeroScience 41, 369–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su J, Ekman C, Oskolkov N, Lahti L, Ström K, Brazma A, Groop L, Rung J, and Hansson O (2015). A novel atlas of gene expression in human skeletal muscle reveals molecular changes associated with aging. Skelet. Muscle 5, 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun N, Youle RJ, and Finkel T (2016). The mitochondrial basis of aging. Mol. Cell 61, 654–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Victoria B, Dhahbi JM, Nunez Lopez YO, Spinel L, Atamna H, Spindler SR, and Masternak MM (2015). Circulating microRNA signature of genotype-by-age interactions in the long-lived Ames dwarf mouse. Aging Cell 14, 1055–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker ML, and Herndon JG (2008). Menopause in nonhuman primates? Biol. Reprod. 79, 398–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Vasaikar S, Shi Z, Greer M, and Zhang B (2017). WebGestalt 2017: a more comprehensive, powerful, flexible and interactive gene set enrichment analysis toolkit. Nucleic Acids Res. 45, W130–W137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward PS, and Thompson CB (2012). Signaling in control of cell growth and metabolism. Cold Spring Harb. Perspect. Biol. 4, a006783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada Y, Colman RJ, Kemnitz JW, Baum ST, Anderson RM, Weindruch R, and Schoeller DA (2013). Long-term calorie restriction decreases metabolic cost of movement and prevents decrease of physical activity during aging in rhesus monkeys. Exp. Gerontol. 48, 1226–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue J, Sheng Y, and Orwig KE (2008). Identification of novel homologous microRNA genes in the rhesus macaque genome. BMC Genomics 9, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahn JM, Sonu R, Vogel H, Crane E, Mazan-Mamczarz K, Rabkin R, Davis RW, Becker KG, Owen AB, and Kim SK (2006). Transcriptional profiling of aging in human muscle reveals a common aging signature. PLoS Genet. 2, e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.