Abstract
PURPOSE:
To develop a motion-robust extension to the recently developed Echo-Planar Time-resolved Imaging (EPTI) approach, referred to as Propeller EPTI with Dynamic Encoding (PEPTIDE), by incorporating rotations into the rapid, multi-shot acquisition to enable shot-to-shot motion correction.
METHODS:
EPTI is a multi-shot EPI-based approach that allows extremely rapid acquisition of distortion- and blurring-free multi-contrast imaging and quantitative mapping. By combining k-space encoding rotations into the EPTI sampling strategy to repeatedly sample the low-resolution k-space center, PEPTIDE enables significant tolerance to shot-to-shot motion and B0 phase variations. Retrospective PEPTIDE datasets are created through combination of in vivo EPTI datasets with rotationally acquired protocols, to enable direct comparison of the two methods and their robustness to identical motion. PEPTIDE datasets are also prospectively acquired and again compared with EPTI, in the presence of true subject motion.
RESULTS:
PEPTIDE is shown to be motion robust to even severe subject motion (demonstrated >30° in-plane rotation, alongside translational and through-plane motion), while maintaining the rapid encoding benefits of the EPTI technique. The technique enables accurate quantitative maps to be calculated from even severe motion datasets. While the performance of the motion correction depends on the type and severity of motion encountered, in all cases PEPTIDE significantly increases image quality in the presence of motion comparative to conventional EPTI.
CONCLUSIONS:
PEPTIDE is a newly developed technique that combines a high degree of motion tolerance into the EPTI framework, enabling highly rapid acquisition of distortion- and blurring-free images at multiple TEs in the presence of motion.
Keywords: EPTI, PROPELLER, distortion-free, multi-contrast, EPI
Introduction:
Echo-planar Imaging (EPI) is a well-established technique for rapid MRI acquisition [1], with either single- or multi-shot varieties utilized in a wide range of applications, including diffusion [2, 3], perfusion [4,5] and fMRI [6,7]. Despite the fast acquisition enabled by EPI, the extended readout duration comes with well-known drawbacks: B0-inhomogeneity induced phase accrual along the phase encoding (PE)-direction, leading to geometric distortions of the image, and T2/T2* decay during the readout, resulting in spatial filtering (blurring). Additionally, while EPI allows rapid image acquisition, the extended readout duration places a limit on the timing between TEs, restricting its application to multi-echo techniques.
Techniques exist to compensate for issues such as geometric distortions in EPI, for example through field map-based corrections [8,9] or through repeat acquisitions with reversed phase-encoding directions [10]. These post-processing approaches perform well in general but do have limitations in areas with large susceptibility variation and do not correct for image blurring.
Echo-planar time-resolved Imaging (EPTI) [11] is a recently developed multi-shot EPI-based technique that is able to ‘time-resolve’ a time-series of multi-contrast images, free from image distortion and blurring, with a temporal spacing equal to the echo-spacing. When implemented in a gradient-echo and spin-echo (GESE) sequence, this enables the rapid acquisition of large time-series of multi-contrast T2 & T2*-weighted images, free from typical EPI-associated drawbacks, while achieving high spatiotemporal resolution. From this, the accurate calculation of a wide range of tissue information, including T2, T2*, proton-density, and tissue phase maps can be performed. Due to the rapid nature of the EPTI acquisition, these distortion-free maps can all be generated, with whole-brain coverage, from a single acquisition of <1 min (~1 s/slice for 1×1×3 mm3). Dynamic GE-EPTI has also been demonstrated for application to fMRI [11] and the EPTI technique has additionally recently been extended to 3D-EPI acquisitions [12,13].
In the multi-shot EPTI acquisition, each EPTI-shot is used to acquire a new segment of ky data in a sequential manner. This makes EPTI potentially susceptible to shot-to-shot physiological and bulk motion variations. A reconstruction method to mitigate image artifacts from shot-to-shot B0 phase variations has been successfully developed [11] but bulk motion still remains an issue. Radial and pseudo-radial trajectories are known for their inherently motion robust properties [14,15]. In addition, such trajectories also cope well with sub-Nyquist sampling and combine well with many advanced reconstruction techniques [16,17,18]. A hybrid radial-Cartesian acquisition scheme exists in which sets of parallel lines of data (resembling low-resolution standard Cartesian acquisitions) are acquired with varying rotations, often referred to as a PROPELLER acquisition [15]. This has some of the advantages of both Cartesian and radial acquisitions, while repeatedly sampling a region of central k-space. This continual resampling of the center in itself provides tolerance to inter-shot motion, as well as containing inherent “navigator”-like information that can be used for further motion- and phase-correction methods.
Proposed here is a motion robust extension to the EPTI technique, referred to as Propeller Echo-Planar Time-resolved Imaging with Dynamic Encoding (PEPTIDE). Combining a Propeller-style trajectory into the EPTI framework, along with dynamic-updating of the sensitivity-encoding information in the reconstruction, this work aims to substantially increase the robustness of the technique to motion.
Theory:
EPTI Review
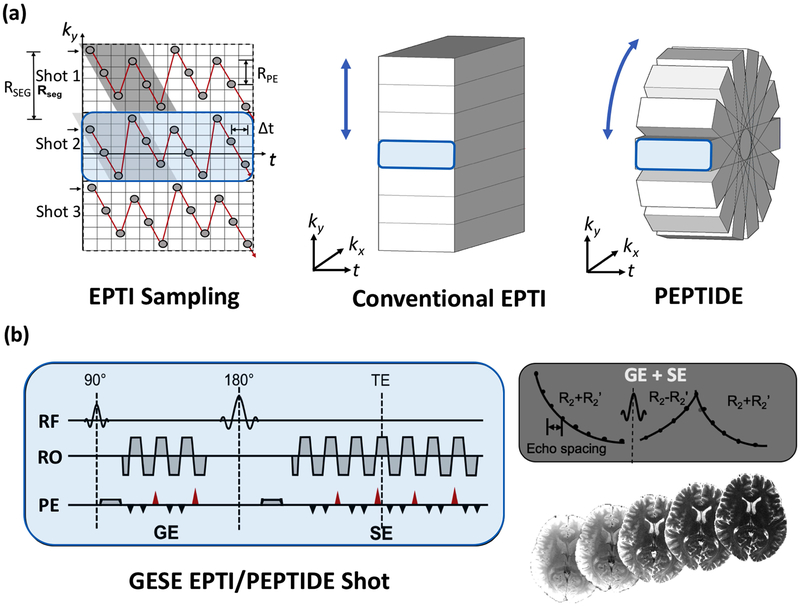
EPTI acquires k-t space through a multi-shot ky-segmented traversal, using a highly-undersampled zig-zag trajectory across each segment, as demonstrated in Figure 1a. The temporal dimension represents the echo-time of each phase-encode line during the echo-train readout. If ky-t space is fully sampled, a complete image with consistent phase-accumulation and signal decay can be generated for each echo time point from the corresponding kx-ky data. This gives a large time-series of contrast-varying, distortion-free images, with a temporal spacing (Δt) equal to that of the EPI echo-spacing.
Figure 1 –
PEPTIDE methodology. (a) Sampling of EPTI methods in ky-t space (left), demonstrating the segmented zig-zag pattern that allows neighboring data-points to be acquired with minimal time-spacing. This is shown in ky-kx-t space for a typical EPTI trajectory (middle), with complete sampling in kx. The proposed PEPTIDE trajectory (right) with rotation of the EPTI segments in the kx-ky plane, to enable motion-robust reconstruction. (b) Applying this trajectory with a combined gradient- and spin-echo sequence, complete reconstruction of k-t space provides time-resolved, distortion and blurring-free T2- and T2*-weighted images for each time point.
The segmented zig-zag traversal ensures that the phase changes between neighboring acquired k-space points are minimized while the jittered diagonal transversals of odd and even numbered ky-points mean that complementary neighboring ky-points are sampled in a spatiotemporal CAIPI-sampling pattern. This enables accurate reconstruction of the highly undersampled ky-t space through B0-inhomogeneity-informed parallel imaging, which employs GRAPPA-like compact kernels that utilize the small and spatially-smooth phase differences between the neighboring data points in ky-t space [19]. In typical brain imaging situations, ~ 25–40 ky encoding lines (RSEG) can be covered with each EPTI-shot when a ky sampling distance (RPE) of 4x Nyquist is used (Figure 1a). This results in a 7–9 EPTI-shot acquisition for 1 mm in-plane resolution imaging. This approach has previously been applied to the EPI readout in a gradient-echo (GE) sequence, as well as for a dual gradient- and spin-echo (GESE) EPTI sequence. The choice of sequence affects the signal curves that are being sampled in ky-t space, and hence the quantitative maps that can be derived from the acquisition.
PEPTIDE Sequence Design
PEPTIDE samples k-t space with a zig-zag segmental pattern, similar to EPTI. However, instead of sampling different segments with shifts along ky, PEPTIDE repeatedly acquires central segments but with varying rotations in the ky-kx plane (Figure 1a). As compared to EPTI, this is achieved in the sequence through replacement of the shot-to-shot ky shifts with shot-dependent rotation in the ky-kx plane.
Consistent with previous EPTI definitions, RPE is defined as the spacing between sequential phase-encode acquisitions, with each diagonal line of acquisitions covering a total distance in the phase-encode direction that is defined as RSEG. For EPTI the number of segments (Nseg) required for complete k-space coverage (-ky,max to +ky,max) is dependent on the value of RSEG. Similarly, for PEPTIDE, the number of acquired blades (Nb) required to achieve full k-space sampling is also depedent on RSEG and is related to the EPTI Nseg equivalent through Nb,full = (π/2) x Nseg,full , to account for the radial coverage. Hence, more shots are required to provide full Nyquist coverage in PEPTIDE. However, this comes with the benefit of motion robustness and the radial sampling with oversampled k-space center potentially provides the ability to still achieve reasonable reconstruction in the presence of certain levels of angular undersampling.
The PEPTIDE method is demonstrated here in a GESE sequence (Figure 1b).
PEPTIDE Reconstruction & Motion Correction
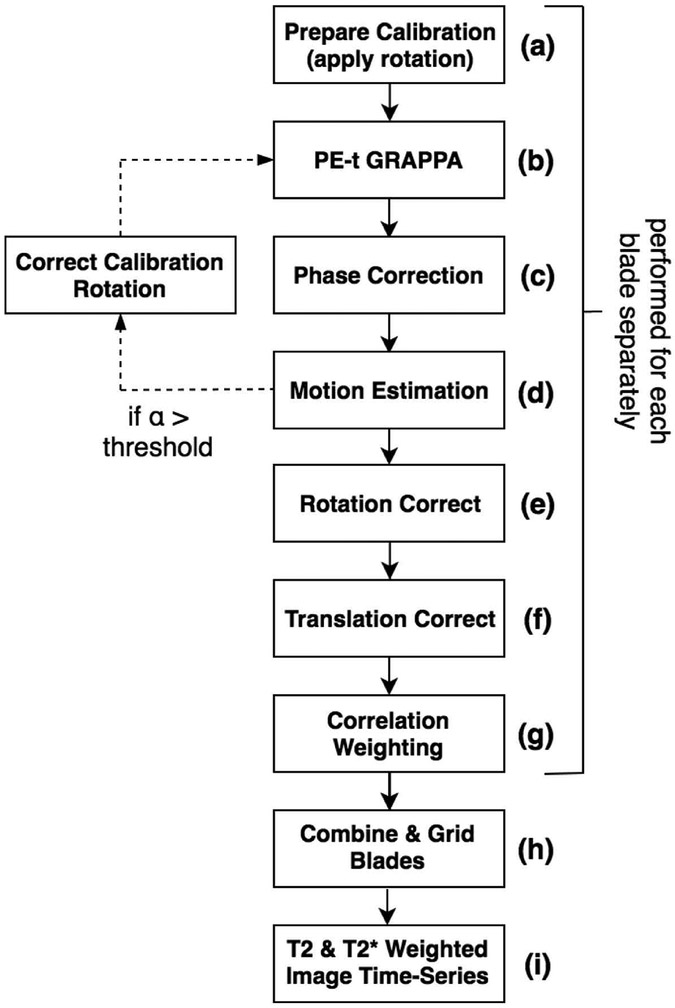
PEPTIDE reconstruction follows a pipeline as shown in Figure 2. (a) Adjustment is first made to the calibration data to match the blade being reconstructed (described later). (b) Each PEPTIDE blade can then be separately reconstructed using the B0-informed parallel imaging reconstruction described in [11]. This is equivalent to reconstruction of only the central segment of an EPTI dataset, which yields a time-series dataset with low resolution in one spatial dimension.
Figure 2 –
The implemented PEPTIDE reconstruction pipeline. Reconstructions are performed on a blade-by-blade basis, first applying a GRAPPA undersampling reconstruction, as for EPTI, prior to applying motion correction strategies, as for PROPELLER. Calibration data is matched to the orientation of each blade acquisition and can be further updated to compensate for calculated rotational subject motion. Finally, the reconstructed blades can be gridded and combined with appropriate weighting.
This then allows for a PROPELLER-style reconstruction/combination [15] across the blades and time-series, implementing various motion correction techniques for inter-blade motion/phase prior to combination through a gridding method, as follows. (c) Simple phase-correction is initially applied to each blade by subtracting a triangularly windowed phase of each blade from itself, to remove low-frequency spatially-varying components, as further described in [15]. This corrects for any offset in the center of the blade rotation, as well as helping remove B0 variation phase. (d) An affine (3 degrees-of-freedom) transformation is calculated between the blade data and a common reference, for each blade, to estimate the rotational and translational motion that has occurred. In this work the motion estimation followed the methods of [15], utilizing the centrally overlapping region of each blade to analyze both the real-component k-space correlations at various rotations as well as the peak of the complex data convolution, both against a fixed reference. Motion correction can be performed either for every timepoint within each PEPTIDE blade acquisition or, if the temporal footprint of each blade acquisition (<150 ms for the parameters demonstrated here) is deemed short enough, for a temporal average per blade. (e) An adjustment is made to the kx-ky trajectory to correct for the estimated rotational motion and (f) the appropriate phase-adjustment (linear phase slope) is applied to each blade k-space dataset to correct for the translational motion. (g) The translation and rotation corrected blades are then cross-correlated, so that a reduced weighting can be appropriately applied to blades that are poorly correlated in order to alleviate the artifacts due to through-plane or non-rigid motion [15,20].
The reconstructed and corrected blades can then be combined through a gridded process (h), across all time-points, to provide the multi-contrast image time-series after Fourier transform (i). In this implementation, the gridding was performed with an oversampling factor of 2 and a Kaiser-Bessel kernel width of 5 [21], with the application of a density compensation function (DCF) iteratively calculated using the methods described in [22]. Blade weighting was applied to the DCF with weighting coefficients calculated from the cross-correlation process.
Calibration Data
For the parallel imaging reconstruction, EPTI requires a short calibration scan of a small fully sampled ky-t region to train the GRAPPA-like kernels. It is proposed that PEPTIDE can use an identical single calibration scan for all blades by applying a rotation to the calibration data prior to calculation of the GRAPPA kernels for each blade. This avoids the necessity to collect additional reference data for each blade. This is similar to an approach used for slice-GRAPPA in SMS-PROPELLER [18], whereby the GRAPPA kernels are rotated prior to their application.
A further step is also proposed here whereby, for any reconstructed blade in which significant motion is detected, the estimated motion is used to update the rotation that needs to be applied to the calibration data for the GRAPPA kernel calculation of that blade. The GRAPPA kernel for that blade is then recalculated and the reconstruction reperformed to achieve improved results. In doing so, it is possible to dynamically update the parallel imaging reconstruction to ensure maximal accuracy across all blades and at all timepoints.
Methods:
All in vivo data were acquired with a consented institutionally approved protocol, on a Siemens Prisma 3T scanner with a 32-channel head coil (Siemens, Erlangen, Germany). Simulation experiments were performed, as well as two different methods of data acquisition, which will be referred to as i) retrospective PEPTIDE/EPTI and ii) prospective PEPTIDE/EPTI. One retrospective and three prospective subject acquisitions were performed (3 male, 1 female; mean age 34 years). Information and imaging parameters for these datasets are provided below. An overview of the datasets investigated can be found in Table 1. For all methods, reconstruction was performed offline using MATLAB (Naticks, MA, USA).
Table 1 –
Experimental Datasets
| EPTI | PEPTIDE | ||
|---|---|---|---|
| Simulation | No motion | RSEG=72,44,32,24,20 | RSEG=72,44,32,24,20 |
| Retrospective | No motion | 7 Segments | 3 Blades*, 5 Blades*, 7 Blades, 10 Blades* |
| Motion | 7 Segments: Motion1(non-center), Motion1(center), Motion2(non-center), Motion2(center) | 7 Blades: Motion1, Motion2, Motion1+2, Motion1+2+3 | |
| Prospective | No motion | 7 Segments, 3 Subjects | 10 Blades, 3 Subjects |
| Motion | 7 Segments, 3 Subjects | 10 Blades, 3 Subjects |
Further results presented in Supplementary
Simulated PEPTIDE/EPTI
Bloch simulations were performed to simulate EPTI and PEPTIDE acquired data. As described in [11], proton density, T2, T2*, B0 and coil sensitivity maps were acquired at 1.0×1.0×3.0mm2 resolution in a healthy volunteer using a 32-channel coil. This enabled simulation of fully sampled ky-t data with appropriately modelled T2/T2* decay and B0 inhomogeneity induced phase changes. From this the data was undersampled with both the EPTI and PEPTIDE sampling schemes and for varying segment sizes.
For all simulations, the echo-spacing was set as 1.05ms, matching the echo-spacing used to acquire PEPTIDE data on the scanner. Spatially uncorrelated Gaussian noise was added the k-space data to provide an SNR=40 at each timepoint. The calculated B0 map had a range of ±50Hz.
Undersampling was performed in-plane as RPE=4 for all cases, with RSEG values varied as 20,24,32,44 and 72 to provide closely matched spatial resolution for EPTI simulations with NSEG values of 11, 9, 7, 5 and 3, respectively. This undersampling was repeated with a PEPTIDE sampling pattern for the same range of RSEG values, using a corresponding number of blades for each, so that the same amount of simulated data was used for EPTI and PEPTIDE in each case.
The five EPTI and five PEPTIDE simulated datasets were reconstructed with their respective reconstruction algorithms, with no motion-correction applied to the PEPTIDE data (PE-t GRAPPA followed by gridding only). The reconstructions were compared against the fully-sampled reference and a normalized root mean square error (nRMSE) was calculated for each reconstructed image.
Retrospective PEPTIDE/EPTI
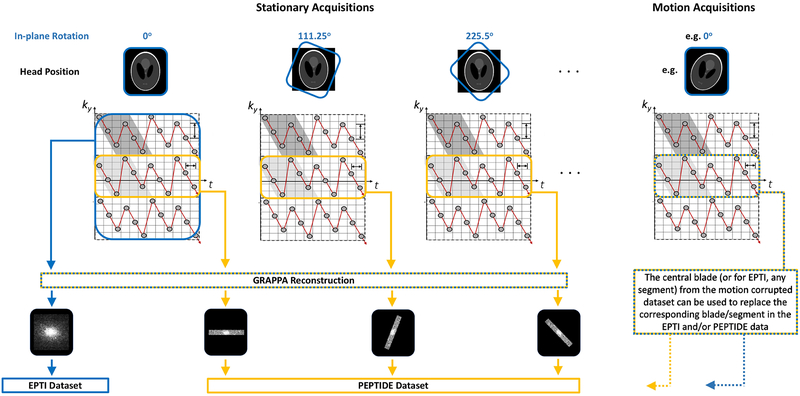
For initial evaluation of PEPTIDE feasibility and tolerance to precisely controlled motion, so-called retrospective PEPTIDE datasets were created. Each retrospective PEPTIDE dataset was formed through combination of different EPTI datasets that were acquired with varying in-plane rotations applied to the protocol prescription (Figure 3). The central segment from each of these EPTI acquisitions was then extracted, which provided data equivalent to rotated PEPTIDE blades. These ‘PEPTIDE’ blades were then processed and combined through the PEPTIDE reconstruction pipeline and compared against the equivalent EPTI reconstruction of the complete EPTI data from the acquisition with no rotation in the protocol prescription.
Figure 3 –
Retrospective dataset method overview. To compare the PEPTIDE and EPTI methods using identical in vivo data, complete EPTI acquisitions were performed with differing in-plane rotations. From this, the central ky-segments from each acquisition could be taken to form a PEPTIDE dataset, that could be compared with the reconstruction from a complete EPTI dataset. To investigate motion, additional acquisitions were also performed with shifts in the subject head position, from which a ky-segment could be taken to replace the corresponding segment in the stationary datasets for both PEPTIDE and EPTI.
EPTI data was acquired with the GESE sequence as described in [11]. The data was acquired using 7 ky-segments with RSEG=36 and RPE=4, and the following sequence parameters: reconstructed resolution=1.1×1.1×3.0 mm3, FOV=250×250×105 mm3, TRwhole-brain=4 s, echo time range of GE/SE=6.6–43.4 ms/60.7–135.3 ms, number of echo times (GE/SE) = 36/72, time per EPTI-segment/slice=150 ms and echo spacing=1.05 ms.
Ten repetitions were performed while the subject remained stationary, with in-plane rotation added to the protocol prescription of each acquisition incremented so as to provide a golden angle distribution and approximately full-sampling for PEPTIDE. The EPTI acquisition parameters resulted in additional lines of ky being sampled (i.e. increased spatial resolution in one direction) for ease of matching identical parameters between the EPTI and PEPTIDE sequences later – this additional data was not used in the comparative reconstructions.
Three additional scans were acquired, again with different in-plane rotations applied to the protocol prescription, prior to which the subject was asked to move their head into different head positions (varying directions and extents in each case). Calibration data was acquired before each EPTI acquisition, consisting of 36 full-FOV phase-encoding lines and 25 echo-times at the same echo-spacing as for the main acquisition, with the same in-plane rotation applied as the proceeding EPTI scan.
In total, 13 EPTI acquisitions were performed; 10 with varying in-plane rotations to the acquisition prescription and 3 which repeated a rotation prescription previously acquired but with a change in the head position of the subject. Retrospective PEPTIDE datasets were created using the central segments of these acquisitions as blades. Four “no-motion” PEPTIDE datasets were created, using a different number of blades, Nb, (10, 7, 5 and 3) from the acquisitions without subject head adjustment. As all the retrospective datasets had RSEG=36, this corresponded to varying levels of undersampling in the final reconstruction. Motion PEPTIDE datasets were created using 7 blades, matching the amount of data for EPTI, whereby one or more no-motion blades are replaced by the corresponding blade from an acquisition with subject head adjustment. The 7-ky segment “no motion” EPTI dataset used the unmodified, unrotated EPTI acquisition, while motion EPTI datasets replaced a segment with the corresponding segment from an acquisition with subject head adjustment. The impact of the different head position shifts was investigated for different segments in the EPTI data and for inclusion in a varying number of the PEPTIDE blades. An overview of these datasets can be seen in Table 1.
The EPTI datasets were reconstructed using the conventional EPTI reconstruction for comparison with the reconstruction of the retrospectively created PEPTIDE datasets. The PEPTIDE datasets were reconstructed with the single, non-rotation acquired, calibration data, as described in the PEPTIDE theory. In addition, PEPTIDE reconstructions were performed with the matching rotationally acquired calibration data for each blade, as a gold standard for validation of the shortened single calibration PEPTIDE method.
T2 and T2* maps were calculated by performing a 4-parameter fit to the signal equation [23]. The reconstructed time series were cropped of the edge timepoint images prior to processing to avoid artifacts from the PE-t GRAPPA reconstruction. Multiple averages would typically be preferred for more accurate fitting, however for simplification of comparison between cases with and without motion, single acquisition averages were used throughout the work.
Prospective PEPTIDE/EPTI
The PEPTIDE sequence was applied with sequence parameters otherwise identical to those used for the previous EPTI datasets. 10 blades were acquired, again with golden angle ordering in the acquisition. A 7 ky-segment EPTI dataset was also acquired for each subject for comparison. One calibration dataset was acquired per subject, for use with both the PEPTIDE and EPTI data.
Datasets were also acquired with deliberate subject motion occurring during the acquisition, with both the PEPTIDE and EPTI sequences, to investigate the effects of true intra-acquisition motion. Subjects were requested to make large deliberate motions, a few seconds after the scanning commenced, to mimic severe patient adjustment/non-compliance occurring after the start of the scan. The exact direction and type of motion was left at their discretion and they were informed that the motion could be in any direction, but they were requested to repeat the type and duration of motion as closely as possible in both scans (PEPTIDE and EPTI). Calibration data was reacquired prior to each motion dataset, so as to prevent any biases that could be introduced by the subject failing to return their head to an identical starting position.
T2 and T2* maps were calculated for the resulting reconstructions, applied as for the retrospective datasets.
Results:
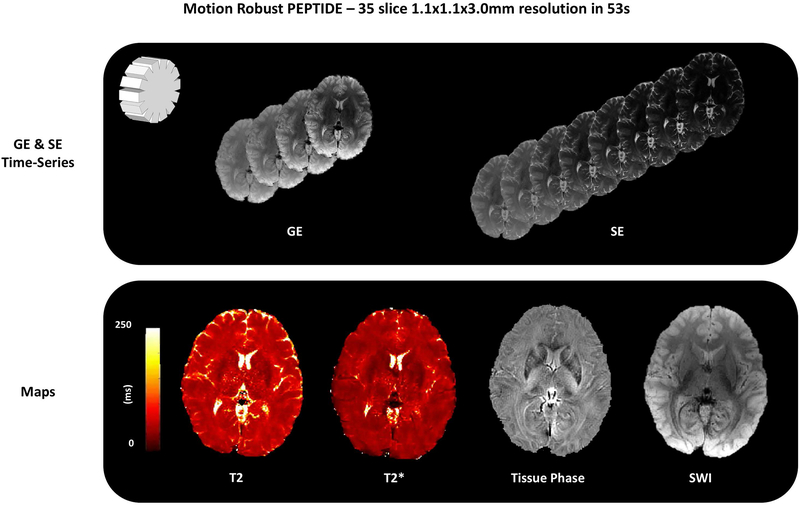
An example of PEPTIDE reconstructed images, and a selection of derived maps, for a representative imaging slice, is shown in Figure 4. The dataset was acquired using 10 PEPTIDE blades with a total acquisition time, Tacq, of 1.5 s/slice.
Figure 4 –
Example reconstructed images from a PEPTIDE acquisition. Through use of a gradient-echo and spin-echo sequence a time-series of both T2* and T2 images can be achieved through a single acquisition (a small sample of ~100 images shown). Results are demonstrated from a 35 slice, 1.1×1.1×3.0 mm3 resolution acquisition, taking 52s to acquire (10-blade PEPTIDE, total Tacq = 1.5 s/slice). Example T2, T2*, tissue phase and SWI maps calculated from these time-series are also demonstrated.
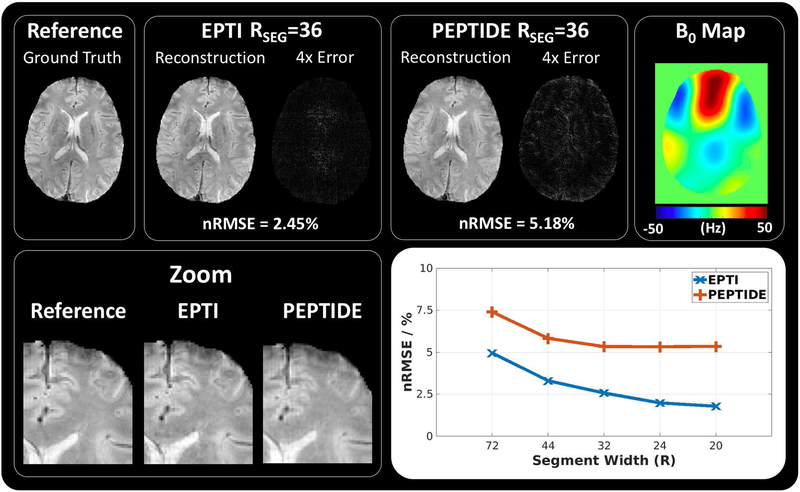
Results from the simulation experiments are shown in Figure 5. The simulated reference data, EPTI reconstruction and PEPTIDE reconstruction are shown for the RSEG=32 case (7 blades/segments), at 3 different reconstructed timepoints. The corresponding 4x intensity error maps are shown for each reconstruction, demonstrating a small increase in reconstruction errors for the PEPTIDE reconstruction, primarily focused at the edges. On the right, a zoomed in portion of these images demonstrates the overall high reconstruction accuracy in both cases, but with some edge errors in PEPTIDE. Below an overview of these results across different segment sizes is shown with a plot of averaged RMSEs for each simulated dataset reconstruction. Both EPTI and PEPTIDE demonstrate higher errors for larger segment sizes before reaching an error minimum for smaller segment sizes, with this minimum error appearing slightly earlier and lower for EPTI. For RSEG⩽32, PEPTIDE maintains a nRMSE of a little over 5% (mean: 5.34%) comparative to the simulated reference.
Figure 5 –
Simulations of EPTI and PEPTIDE. (top) Simulated ground truth data is compared to reconstructions of EPTI and PEPTIDE sampling, alongside the 4x error maps and calculated nRMSEs, demonstrated for the RSEG=36 (segment/blade width) case at TE=25ms. To the right the B0 map used in the simulation is shown. (bottom) On the left a zoomed-in region of each of the datasets is demonstrated. On the right an overview of the results is shown for RSEG values 72, 44, 32, 24 and 20, which use 3, 5, 7, 9 and 11 segments/blades respectively, for both EPTI and PEPTIDE.
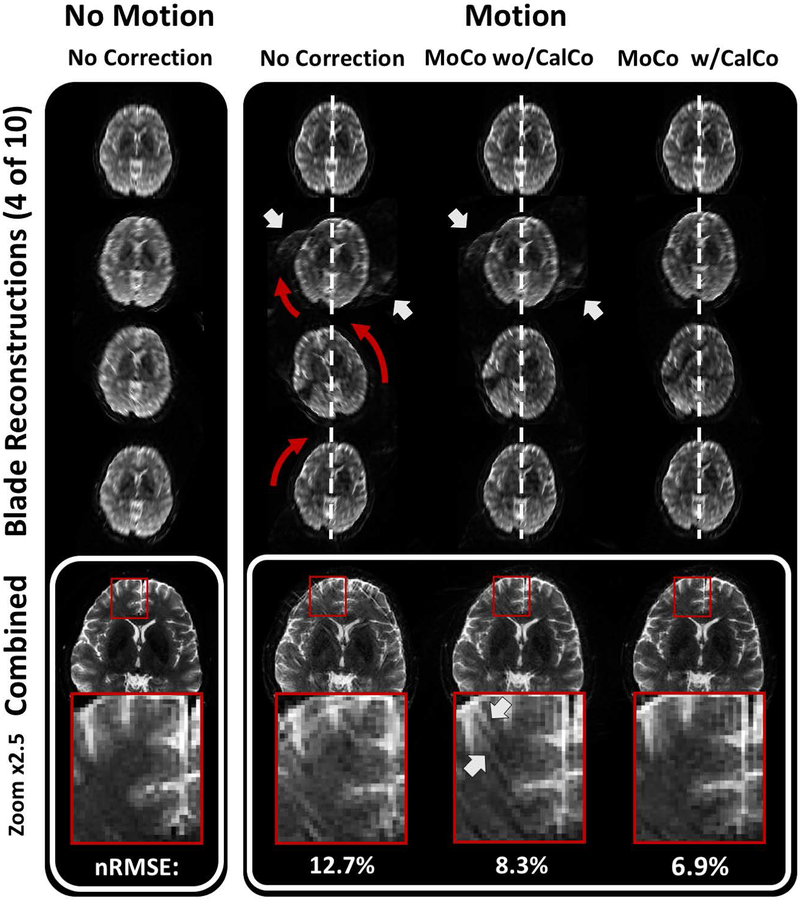
The reconstruction improvement from the motion correction process is demonstrated for the retrospective PEPTIDE dataset in Figure 6. The gridded individual blade reconstructions of the first four out of ten blades are shown on the top section of the figure, along with the combined image at the bottom section, for both a no motion (left) and motion case (right) through the various steps of motion correction. In the case of motion without correction, the individual blade reconstructions point to a clear right-left-right rotational motion during the data acquisition period of the second to the fourth PEPTIDE blade, as indicated by the orange arrows. While the image quality of the individual blade reconstructions is still relatively robust considering the amount of motion present, parallel imaging reconstruction errors along the undersampled PE direction of the motion corrupted blades are observed (white arrows), which is particularly strong for the second blade reconstruction. The motion and these reconstruction artifacts lead to severe degradation in the final combined reconstruction in this case. Application of the standard motion correction pipeline (without the additional step of correcting the calibration data) is shown in the middle motion case. The motion-correction clearly demonstrated an improvement, closer to the non-motion case, albeit with some parallel imaging artifacts still present in the individual blade images that manifest as additional streaks in the combined image. Including the motion correction of the calibration data is seen to remove these effects, leaving a final combined image very closely matching that of the non-motion case. Reconstruction of the blades performed with separate calibration datasets, acquired with the corresponding rotation to the protocol, showed similar performance to the results presented here, which used retrospective rotation of a single calibration dataset to match each blade.
Figure 6 –
PEPTIDE motion correction. Example of the PEPTIDE motion-correction process through various stages. Four out of ten reconstructed low-resolution blades are demonstrated above the final combined image and a zoomed region of interest for a dataset with no included motion (left) and a dataset with included motion (right) for a reconstruction without MoCo, with blade MoCo, and with MoCo that additionally includes the motion estimate into the GRAPPA calibration data. Red arrows highlight the rotations in head positions between the blades. White arrows indicate residual parallel imaging artifacts. The sequential improvement in image quality, towards that of the case with no motion, can be seen.
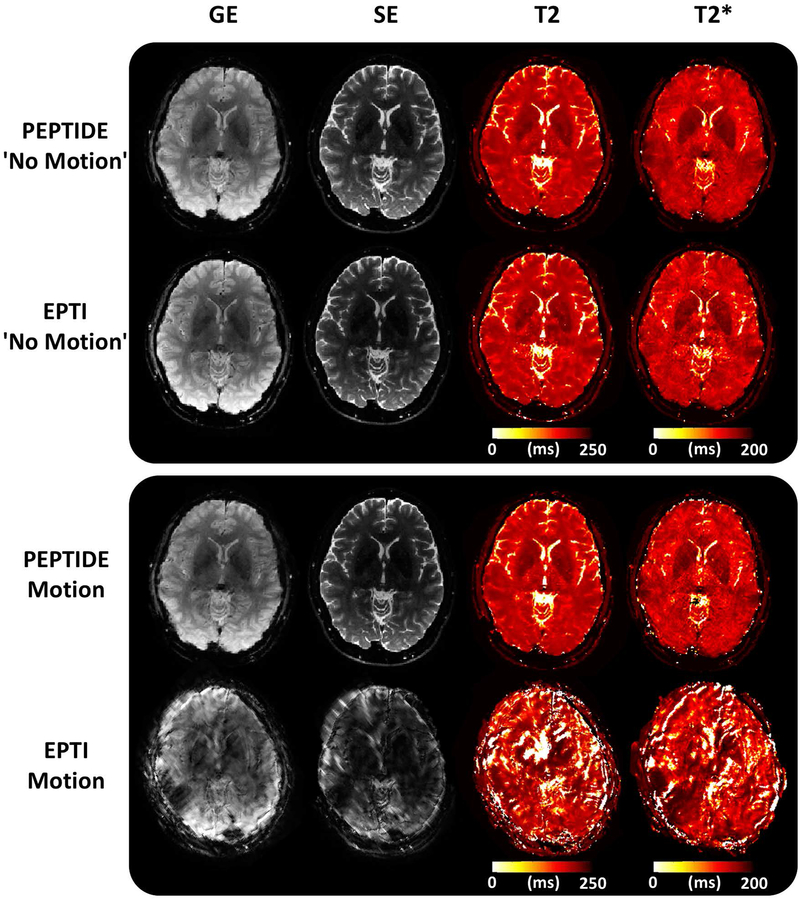
Figure 7 compares PEPTIDE and EPTI images, with an identical segment/blade number of 7, produced from the retrospective PEPTIDE/EPTI dataset. Demonstrated is a single time-point image from the GE and from the SE portion of the GESE acquisition, along with the corresponding T2 and T2* maps calculated from the full set of time-series images produced across the GESE readout, for cases without motion and with large motion in a single segment or blade. For the non-motion cases, PEPTIDE and EPTI can be seen to be similar in both the structural images and quantitative maps. With the inclusion of severe motion, the EPTI data becomes strongly corrupted, causing the corresponding reconstructions to be effectively unusable. In comparison, the PEPTIDE motion images remain of a high quality despite having identical motion (estimated to be ~20° in-plane rotation and ~1–2 pixels in-plane translation) and utilizing the same amount of acquisition time (same blade vs. segment number).
Figure 7 –
PEPTIDE and EPTI comparison. A single image from the gradient-echo (GE) and spin-echo (SE) acquired time series and calculated T2 and T2* maps are shown in a case without motion for PEPTIDE and EPTI, as well as for a case with identical severe (~20° rotation) motion for both - corresponding to the red color-coded motion from Figure 7. Both methods were reconstructed with the same number of blades/segments = 7.
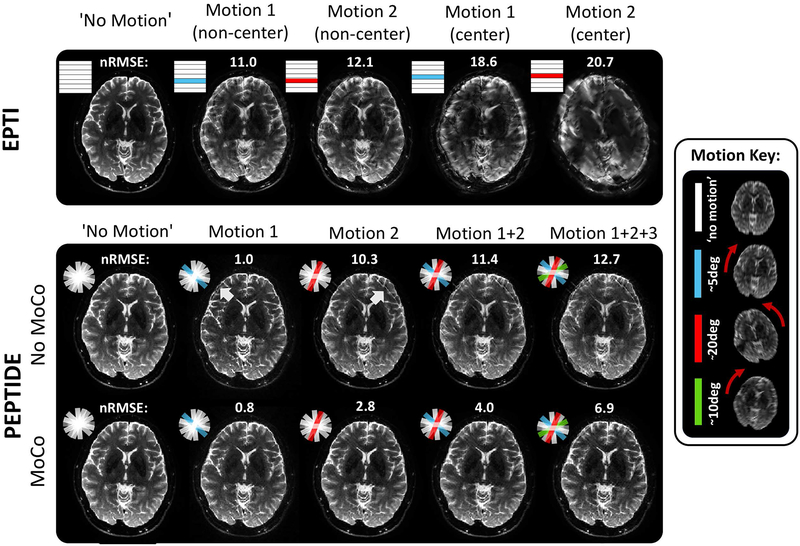
A further investigation into the effects of varying amounts of motion is shown in Figure 8. By again using the retrospective PEPTIDE/EPTI datasets, motion corrupted blades/segments are introduced into different segments of the EPTI dataset and into different blades of the PEPTIDE dataset as indicated by the color-coded segments/blades. For EPTI, a weaker (~5°)† and stronger (~20°)† extent of motion is demonstrated in both a non-central and central segment of the data. All cases produce visually reduced image quality comparative to the non-motion case, with predictably stronger degradation of image quality occurring when the motion is present in the central k-space segment. For PEPTIDE, the effect of including these different motion cases and in an increasing number of the PEPTIDE blades is demonstrated. Difference images of the reconstructions can be found in Supporting Information Figure S1. The error between motion and non-motion cases remains much lower for the PEPTIDE datasets (nRMSE: [2.8 – 6.9]% PEPTIDE, [11.0 – 20.7]% EPTI), and this is still somewhat the case even prior to motion-correction. With motion-correction, for the single blade of motion cases, most closely matches with the EPTI motion examples, relatively little image degradation can be seen. As more blades of large motion, in differing directions, are included in the PEPTIDE dataset (>40% of blades motion-corrupted) some artifacts begin to appear in the reconstructions. However, the majority of structural image features can still be clearly distinguished even in the extreme motion cases. The nRMSEs of the PEPTIDE reconstructions drop by 2–3 fold when motion correction (MoCo) reconstruction is utilized. The nRMSE of MoCo-PEPTIDE remains low even in the case of large motion corruption across 3 blades, with a resulting nRMSE of 6.9% that is significantly lower than that of the EPTI data containing milder motion for only one non-center segment (nRMSE of 11%).
Figure 8 –
Sensitivity of acquisitions to motion. The impact of motion on EPTI and PEPTIDE is demonstrated for varying degrees of motion. Different large head motions, of varying severity, are shown implemented into non-center and center segments of the EPTI acquisition (top). The PEPTIDE reconstruction is shown with these same blades of motion implemented along with increasing numbers of motion-corrupted blades, both with and without the motion correction steps of the reconstruction (bottom). A representation of the motion in each of the different blades is shown in the motion key (n.b. through-plane motion also occurred but is not depicted). Normalized root mean square error (nRMSE) is shown for each motion case reconstruction comparative to the non-motion case. Difference images for these reconstructions can be found in Supporting Information Figure S1.
Supporting Information Figure S2 examines the effect of the number of PEPTIDE blades (for a fixed RSEG) on the reconstruction quality of data acquisition without and with motion. With the parameters chosen, 10 blades are required for approximate full Nyquist sampling through the combination of blades, while 7 blades (matching the amount of data acquisition for the equivalent EPTI acquisition), 5 blades, and 3 blades all result in angular undersampling in the final reconstruction. The effect of the undersampling on the acquisition time is shown for each blade number. In the non-motion cases, a predictable loss of resolution occurred for the undersampled reconstructions in addition to some undersampling “streaking” artifacts, but otherwise the image quality degradation is not too severe for lower blade undersampling (e.g. between the 10 and 7 blade cases). With the inclusion of one motion corrupted blade (corresponding to the, “strong”, first motion corrupted blade in Figure 6 with ~20° rotation) image quality deteriorates to a greater extent with reducing number of blades, as the proportion of the blades that are motion corrupted increases.
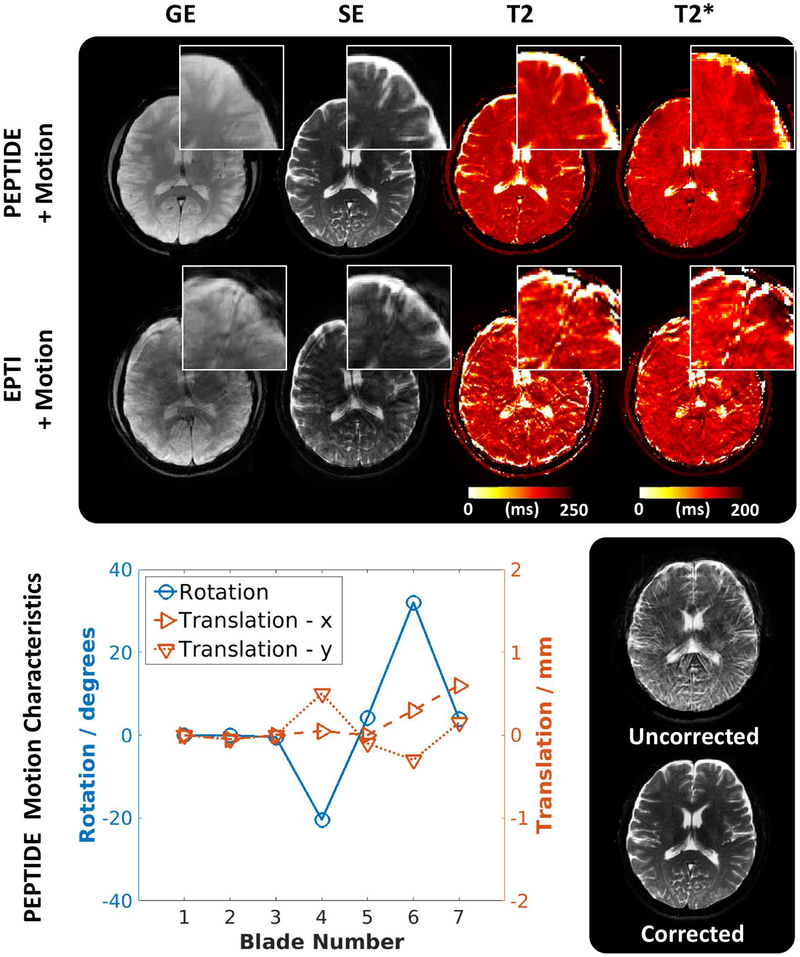
Prospective PEPTIDE acquired data also produced high-quality reconstructions, in good agreement with their EPTI equivalents, as shown in Figure 9. The PEPTIDE datasets with intra-acquisition motion still show a good ability at resolving anatomical details and no significant difference was found between the derived maps from datasets (PEPTIDE T2/T2* motion vs no-motion: P=0.39/0.82, paired two-tailed t-test). Figure 9 demonstrates a PEPTIDE acquisition with a calculated rotational range of −20° to 32°. For approximately the same motion, the degradation of the EPTI dataset is severe and the pixel-wise mapping values can no longer be considered to have equal means (EPTI T2/T2* motion vs no-motion: both P<<0.05, two-tailed t-test).
Figure 9 –
Prospectively acquired PEPTIDE/EPTI. (top) Demonstrative images from the GE and SE time-series and calculated maps for both PEPTIDE and EPTI acquired datasets in the presence of motion, showing zoomed in regions. (bottom) The calculated in-plane rotations and translations for the PEPTIDE dataset are shown next to an example of the improvement gained through application of motion correction.
Discussion:
We proposed PEPTIDE as a way of acquiring quantitative data, extending the EPTI methodology, and this has been shown in retrospective and prospective experiments to provide good motion robustness. Comparison of PEPTIDE and EPTI shows a significant improvement in motion tolerance, with a small trade-off in encoding efficiency in going from Cartesian to non-Cartesian sampling as is well known [24] and demonstrated by simulation tests in this work. This method should enable motion-robust quantitative imaging in a rapid time.
The PEPTIDE and EPTI simulations enabled examination of the accuracy of the two techniques against a realistically simulated ground truth as well as allowing an investigation into the effect of varying the segment width. The overall low nRMSE of both techniques is promising, although the increase in error with PEPTIDE relative to EPTI is worth noting. It is well-known that between non-Cartesian and Cartesian sampling there is a trade-off between sampling efficiency and motion robustness.
The retrospective PEPTIDE/EPTI investigation enabled comparison of in vivo PEPTIDE and EPTI data, including the addition of identical motion. It was demonstrated that in these cases of identical motion, PEPTIDE has the potential to considerably outperform EPTI, particularly if non-negligible motion were to occur during the central segment acquisition. The robustness of the PEPTIDE technique, even prior to motion correction, demonstrates the well-known motion-tolerance of pseudo-radial techniques. The PEPTIDE reconstruction pipeline, including the additional calibration data correction step, provides the potential to resolve time-series datasets from motion corrupted data that have very small errors in comparison to non-motion cases.
For the prospectively acquired datasets there is good agreement between the EPTI and PEPTIDE methods in stationary cases, with far greater tolerance to motion for the PEPTIDE method in the motion corrupted cases. The PEPTIDE method means that the motion-sensitivity is effectively reduced to the time period of a single blade acquisition (<150 ms). The potential for PEPTIDE to provide accurate quantitative information in the presence of motion is highlighted through good overall agreement between the PEPTIDE acquired maps and those of EPTI which has previously been well validated [11], as well as showing no statistical difference between PEPTIDE maps acquired with motion versus when acquired without.
In this work PROPELLER is investigated as a way to estimate and correct for motion in EPTI and appears to provide robust results. PROPELLER achieved this robustness to motion at a cost of having to oversample the center of k-space, which reduces acquisition efficiency and attainable sharpness when compared to an equivalent Cartesian acquisition. It should be noted that there are also a number of different approaches for estimating motion in Cartesian-based methods, including promising work with use of navigators [25][26] or tracking equipment [27][28], and these alternative methods could potentially be incorporated into EPTI. A comparison of how these methods perform comparative to the PEPTIDE approach could be a future direction of work.
There is also the potential for PEPTIDE to enable further acceleration, as the tolerance to motion does not appear to degrade substantially with moderate blade undersampling. However, the loss of spatial resolution concomitant with angular sampling requires care, with other techniques required to compensate for this effect. The total spatiotemporal undersampling rates demonstrated are also already very high, corresponding to approximate factors of R~32, R~44, and R~74 for the retrospective 10, 7, and 5 blade reconstructions respectively (total ky-t points reconstructed / acquired ky-t datapoints). Future work will explore the use of low-rank partial-separability reconstruction approaches [29] to alleviate these issues and potentially allow higher acceleration. Such a reconstruction is now being adapted to EPTI with promising results [13]. Modification of the PEPTIDE trajectory, for example through acquiring different undersampled sets of blades across the readout (e.g. GE vs SE) could further increase incoherent aliasing in its time-series data, which should work synergistically with such reconstruction.
With EPTI, sampling data can be acquired very rapidly. In the original work [11] it was shown that EPTI is typically SNR limited rather than encoding limited at 3T, and therefore it was proposed to acquire three averages to improve the calculated quantitative maps. In such a setup. the motion sensitivity will be increased in EPTI due to the longer scan. In comparison, with PEPTIDE the motion robustness should remain more consistent for the longer scan and an increased performance could be seen through more complete filling of k-space alongside the increased SNR benefit.
While PEPTIDE has been shown to have significantly improved tolerance to motion, there are still limits that are in-common with most PROPELLER type reconstructions. Through-plane motion is not corrected and is instead reweighted. As discussed, only a small number of blades were typically used in this work so as to keep the PEPTIDE technique as an ultra-fast acquisition, with a similar duration to EPTI. While the technique appears to still have some tolerance to motion with low blade numbers, low blade numbers could reduce the extent to which poorly correlated blades can be effectively discarded. In addition, although the calibration acquisition duration is kept short, the impact of motion occurring during the calibration data itself is feasible and has not been investigated, potentially requiring further work. Due to its rapid acquisition, obtaining calibration data both pre- and post-PEPTIDE could work as a safeguard.
The EPTI method aims to acquire data in a highly rapid and efficient manner and is therefore typically set so as to make full use of the scanner hardware within the approved safety limits. The rotational nature of PEPTIDE can force gradient waveforms (e.g. read and phase) that occur at the same time to “overlap” on a single physical axis for certain orientations/blades, potentially exceeding gradient amplitude or slew-rate limitations on that axis. For this reason, the sequence requires modification to check the feasibility of all blades prior to commencement of the scan, and it is possible that in some cases the PEPTIDE sequence will not be able to achieve the same echo-spacing as that of EPTI. In this work the PEPTIDE sequence was applied with conservative parameters so as to enable consistent application, resulting in an echo-spacing increase of approximately 7% comparative to that achievable in an equivalent EPTI scan.
The PEPTIDE reconstruction, being non-Cartesian, suffers from increased computational time comparative to EPTI. The undersampling reconstruction was applied as for EPTI but for each blade separately, causing increased overhead time, particularly from the necessity to rotate the calibration data and recalculate the GRAPPA weights for each blade. The motion correction and gridding steps are specific to PEPTIDE and their duration is dependent on the exact implementation. In this work, the non-optimized reconstruction is performed in MATLAB and in the most severe case can be up to an hour in duration per slice at 1.1mm resolution (Linux Server, Intel Xeon E5 @2.20GHz). More efficient implementation and use of GPUs are both possible and use of coil compression has shown promising early results for significant speed-up of this technique without noticeable loss in quality.
Other techniques and applications that have already been demonstrated with EPTI should also be applicable with PEPTIDE. Simultaneous multi-slice excitation has been demonstrated successfully with EPTI [11] as well as for PROPELLER [18] and, with appropriate application of the calibration data, could be applied to PEPTIDE. This would reduce the time required for full brain coverage, but there is potential that such a technique would still be sensitive to through-plane motion. Alternatively, extension of the technique to 3D, as has been initially demonstrated in EPTI [12], would lend itself very well to PEPTIDE; the increased dimensionality may allow increased undersampling of the “blades” while simultaneously providing full 3D navigator information for true motion correction in all three directions.
The EPTI and PEPTIDE methods are flexible in their application and could in theory be used in different sequence types. One potential extension is the inclusion of an inversion preparation pulse, with other necessary modifications, to additionally provide the capability of mapping T1. T1 mapping would enable a more complete quantitative description, calculated from a single scan. Another possibility is using the self-navigated nature of PEPTIDE, that makes it robust to strong shot-to-shot phase variations, in diffusion-weighted imaging [30]. The potential application of PEPTIDE in diffusion MRI could provide fast motion-robust multi-contrast diffusion-weighted images, and PEPTIDE has the potential to be combined with advanced diffusion acquisition methods [31,32] to achieve high-resolution high-SNR diffusion-weighted imaging without distortion and blurring.
Conclusion:
In conclusion, the PEPTIDE technique has been introduced as a motion tolerant extension to the EPTI technique, providing rapid acquisition of distortion- and blurring-free multi-contrast data. The technique has been shown to be able to provide significant improvement over EPTI in cases of strongly motion corrupted acquisitions, with quantitative maps from PEPTIDE acquired data that contain rotational head motions of >30° statistically similar to acquisitions without motion. There is potential application of the technique in a wide range of areas and many possible extensions to the technique have been described and will be explored in the near future.
Supplementary Material
Supporting Information Figure S1 – Difference Images for Figure 8. Error maps (3x intensity) are shown for the various retrospective motion cases, matching Figure 8, for EPTI, PEPTIDE without motion correction (moco) and PEPTIDE with moco. These are comparative to their non-motion cases (left).
Supporting Information Figure S2 – PEPTIDE blade undersampling. The PEPTIDE reconstruction is shown for decreasing numbers of blades, both with and without motion. N=10 is approximately equivalent to full sampling post-gridding, N=7 is equivalent to the number of blades required by the equivalent EPTI sequence, while the lower blade numbers correspond to increased acquisition acceleration.
Acknowledgements:
This work was supported in part by NIH research grants: R01MH116173, R01EB020613, R01EB019437, U01EB025162, P41EB015896, and the shared instrumentation grants: S10RR023401, S10RR019307, S10RR019254, S10RR023043.
Footnotes
Approximate in-plane rotational shift. In-plane translational shifts, as well as through-plane rotations and shifts, were also present.
References:
- 1.Mansfield P Multi‐planar image formation using NMR spin echoes. J Phys C: Solid State Phys. 1977;10:L55. [Google Scholar]
- 2.Warach S, Gaa J, Siewert B, Wielopolski P, Edelman RR. Acute human stroke studied by whole brain echo planar diffusion‐weighted magnetic resonance imaging. Ann Neurol. 1995;37:231–241. [DOI] [PubMed] [Google Scholar]
- 3.Butts K, de Crespigny A, Pauly JM, Moseley M. Diffusion‐weighted interleaved echo‐planar imaging with a pair of orthogonal navigator echoes. Magn Reson Med. 1996;35:763–770. [DOI] [PubMed] [Google Scholar]
- 4.Kucharczyk J, Vexler ZS, Roberts TP, et al. Echo‐planar perfusion‐sensitive MR imaging of acute cerebral ischemia. Radiology. 1993;188:711–717. [DOI] [PubMed] [Google Scholar]
- 5.Edelman RR, Siewert B, Darby DG, et al. Qualitative mapping of cerebral blood flow and functional localization with echo‐planar MR imaging and signal targeting with alternating radio frequency. Radiology. 1994;192:513–520. [DOI] [PubMed] [Google Scholar]
- 6.Biswal B, Zerrin Yetkin F, Haughton VM, Hyde JS. Functional connectivity in the motor cortex of resting human brain using echo‐planar MRI. Magn Reson Med. 1995;34:537–541. [DOI] [PubMed] [Google Scholar]
- 7.Kwong K Functional magnetic resonance imaging with echo planar imaging In: Pavone P, Rossi P, editors. Functional MRI. Syllabus (Bracco education in diagnostic imaging). Milano: Springer; 1996. p 73–90. [Google Scholar]
- 8.Jezzard P, Balaban RS. Correction for geometric distortion in echo planar images from B0 field variations. Magn Reson Med. 1995;34:65–73. [DOI] [PubMed] [Google Scholar]
- 9.Reber PJ, Wong EC, Buxton RB, Frank LR. Correction of off resonance‐related distortion in echo‐planar imaging using EPI‐based field maps. Magn Reson Med. 1998;39:328–330. [DOI] [PubMed] [Google Scholar]
- 10.Andersson JL, Skare S, Ashburner J. How to correct susceptibility distortions in spin‐echo echo‐planar images: application to diffusion tensor imaging. NeuroImage. 2003;20:870–888. [DOI] [PubMed] [Google Scholar]
- 11.Wang F, Dong Z, Reese TG, et al. Echo planar time‐resolved imaging (EPTI). Magn Reson Med. 2019; 81: 3599–3615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang F, Dong Z, Reese TG, Wald LL, Setsompop K. 3D-EPTI for Ultra-fast Multi-contrast and Quantitative Imaging. In Proceedings of the 27th Annual Meeting of ISMRM 2019. #0944. [Google Scholar]
- 13.Dong Z, Wang F, Reese TG, Bilgic B, Wald LL, Setsompop K. Echo Planar Time-Resolved Imaging (EPTI) with Subspace Constraint and optimized k-t trajectory. In Proceedings of the 27th Annual Meeting of ISMRM 2019. #0310. [Google Scholar]
- 14.Glover GH, Pauly JM. Projection Reconstruction Techniques for Reduction of Motion Effects in MRI. Magn Reson. Med 1992;28:275–289. [DOI] [PubMed] [Google Scholar]
- 15.Pipe JG. Motion Correction with PROPELLER MRI: Application to Head Motion and Free-Breathing Cardiac Imaging. Magn Reson Med. 1999;42:963–969. [DOI] [PubMed] [Google Scholar]
- 16.Ye JC, Tak S, Han Y, Park HW. Projection reconstruction MR imaging using FOCUSS. Magn Reson Med 2007;57:764–775. [DOI] [PubMed] [Google Scholar]
- 17.Block KT, Uecker M, Frahm J. Undersampled radial MRI with multiple coils. Iterative image reconstruction using a total variation constraint. Magn Reson Med. 2007;57:1086–1098. [DOI] [PubMed] [Google Scholar]
- 18.Norbeck O, Avventi E, Engström M, Rydén H, Skare S. Simultaneous Multi-Slice Combined With PROPELLER. Magn. Reson Med 2018;80:496–506. [DOI] [PubMed] [Google Scholar]
- 19.Dong Z, Wang F, Reese TG, et al. Tilted-CAIPI for Highly Accelerated Distortion-Free EPI with Point Spread Function (PSF) encoding. Magn Reson Med. 2019;81:377–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dong Z, Wang F, Ma X, Dai E, Zhang Z, Guo H. Motion-corrected k-space reconstruction for interleaved EPI diffusion imaging. Magn Reson Med. 2018;79:1992–2002. [DOI] [PubMed] [Google Scholar]
- 21.Beatty PJ, Nishimura DG, Pauly JM. Rapid gridding reconstruction with a minimal oversampling ratio. IEEE Trans Med Imaging. 2005;24:799–808. [DOI] [PubMed] [Google Scholar]
- 22.Pipe JG and Menon P. Sampling density compensation in MRI: Rationale and an iterative numerical solution. Magn Reson Med. 1999;41:179–186. [DOI] [PubMed] [Google Scholar]
- 23.Schmiedeskamp H, Straka M, Bammer R. Compensation of slice profile mismatch in combined spin‐and gradient‐echo echo‐ planar imaging pulse sequences. Magn Reson Med. 2012;67: 378–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bernstein MA, King KF, Zhou XJ. Handbook of MRI pulse sequences. Amsterdam: Elsevier Academic Press; 2004:1040. [Google Scholar]
- 25.Van Der Kouwe AJW, Benner T, Dale AM. Real‐time rigid body motion correction and shimming using cloverleaf navigators. Magn Reson Med. 2006;56:1019–1032. [DOI] [PubMed] [Google Scholar]
- 26.White N, Roddey C, Shankaranarayanan A, et al. PROMO: real-time prospective motion correction in MRI using image-based tracking. Magn Reson Med. 2010;63:91–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Slipsager JM, Ellegaard AH, Glimberg SL, et al. Markerless motion tracking and correction for PET, MRI, and simultaneous PET/MRI. PLoS ONE. 2019. 10.1371/journal.pone.0215524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eschelbach M, Aghaeifar A, Bause J, et al. Comparison of prospective head motion correction with NMR field probes and an optical tracking system. Magn Reson Med. 2019;81:719–729. [DOI] [PubMed] [Google Scholar]
- 29.Liang ZP. Spatiotemporal imaging with partially separable functions. In Proceedings of the IEEE Int. Symp Biomed Imag. 2007;pp.988–991. [Google Scholar]
- 30.Pipe JG, Farthing VG, Forbes KP. Multishot diffusion-weighted FSE using PROPELLER MRI. Magn Reson Med. 2002;47:42–52. [DOI] [PubMed] [Google Scholar]
- 31.Setsompop K, Fan Q, Stockmann J, et al. High-resolution in vivo diffusion imaging of the human brain with generalized slice dithered enhanced resolution: simultaneous multislice (gSlider-SMS). Magn Reson Med. 2018;79:141–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang F, Bilgic B, Dong Z, et al. Motion-robust sub-millimeter isotropic diffusion imaging through motion corrected generalized slice dithered enhanced resolution (MC-gSlider) acquisition. Magn Reson Med. 2018;80:1891–1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information Figure S1 – Difference Images for Figure 8. Error maps (3x intensity) are shown for the various retrospective motion cases, matching Figure 8, for EPTI, PEPTIDE without motion correction (moco) and PEPTIDE with moco. These are comparative to their non-motion cases (left).
Supporting Information Figure S2 – PEPTIDE blade undersampling. The PEPTIDE reconstruction is shown for decreasing numbers of blades, both with and without motion. N=10 is approximately equivalent to full sampling post-gridding, N=7 is equivalent to the number of blades required by the equivalent EPTI sequence, while the lower blade numbers correspond to increased acquisition acceleration.