Abstract
The anterior segment of the vertebrate eye consists of highly organized and specialized ocular tissues critical for normal vision. The periocular mesenchyme, originating from the neural crest, contributes extensively to the anterior segment. During chick eye morphogenesis, the homeobox gene Six3 is expressed in a subset of periocular mesenchymal cells and in differentiating anterior segment tissues. Retrovirus-mediated misexpression of Six3 causes eye anterior segment malformation, including corneal protrusion and opacification, ciliary body and iris hypoplasia, and trabecular meshwork dysgenesis. Histological and molecular marker analyses demonstrate that Six3 misexpression disrupts the integrity of the corneal endothelium and the expression of extracellular matrix components critical for corneal transparency. Six3 misexpression also leads to a reduction of the periocular mesenchymal cell population expressing Lmx1b, Pitx2, and Pax6, transcription factors critical for eye anterior segment morphogenesis. Moreover, elevated levels of Six3 attenuate proliferation of periocular mesenchymal cells in vitro and differentiating anterior segment tissues in vivo. These results suggest that, in addition to its function in eye primordium determination, Six3 plays a role in regulating the development of the vertebrate eye anterior segment.
Keywords: eye, development, anterior segment, mesenchyme, Six3, misexpression, chick
INTRODUCTION
The anterior segment of the vertebrate eye consists of several ocular tissues, including the lens, the cornea, the iris, the ciliary body, and the trabecular meshwork. Proper spatial organization, differentiation, and maintenance of the anterior segment tissues are critical for normal visual function. The lack or loss of transparency of the lens and/or the cornea can cause visual impairment and lead to blindness. Defects and malformation of the ciliary body, which produces the aqueous humor, and of the trabecular meshwork, which allows passage of the aqueous humor through the Schlemm’s canal into the venous system, can result in abnormal intraocular pressure, which can then damage the optic nerve and retina, as has been found in certain types of glaucoma (Shields, 1998).
Formation of the anterior segment entails complex morphogenetic events involving cells derived from several distinct embryonic tissue origins. The lens and the corneal epithelium arise from embryonic ectoderm, whereas the endothelium and stroma of the cornea, the muscles of the ciliary body, the stroma of the iris, and the trabecular meshwork located at the junction between the cornea and iris contain cells derived from the periocular mesenchyme (Grainger, 1992; Graw, 1996). Fate-mapping studies in chick and mouse have shown that cranial neural crest cells contribute extensively to the periocular mesenchyme (Johnston et al., 1979; Trainor and Tam, 1995). During anterior segment morphogenesis, periocular mesenchymal cells migrate toward the margin of the optic cup and between the lens and the corneal epithelium to differentiate into various ocular tissues. Classical transplantation experiments show that differentiation of the cornea and development of the anterior chamber involve inductive signals from the lens (Coulombre and Coulombre, 1964; Genis-Galvez, 1966; Genis-Galvez et al., 1967). Recent studies using transgenic mice also demonstrate that formation of the corneal endothelium is required for normal development of the anterior segment (Reneker et al., 2000; Kidson et al., 1999). Thus, inductive tissue interactions play important roles in the proper formation of various anterior segment tissues.
Increasing evidence supports that transcription factors expressed in the eye primordium and/or in the periocular mesenchyme are critical for the patterning and differentiation of ocular tissues in the anterior segment. For example, the forkhead/winged-helix transcription factor Mf1, the bicoid class homeobox protein Pitx2, and the LIM homeodomain protein Lmx1b are all expressed in the periocular mesenchyme surrounding the developing optic cup, and loss-of-function mutations in each of these three genes result in anterior segment malformation in mouse (Kidson et al., 1999; Hong et al., 1999; Pressman et al., 2000; Lu et al., 1999). Mutations in the human FKHL7/FOXC1 gene (a homologue of the murine mf1 gene), the PITX2 gene, and the LMX1B gene cause the Axenfeld–Rieger anomaly, the Riegers and iridogoniodysgenesis syndrome, and the nail–patella syndrome, respectively (Semina et al., 1996; Gage and Camper, 1997; Kulak et al., 1998; Nishimura et al., 1998; Mears et al., 1998; Vollrath et al., 1998; McIntosh et al., 1998; Dreyer et al., 1998; Smith et al., 2000; Saleem et al., 2001; Alward, 2000). These disease conditions are characterized by anterior segment defects involving the cornea, the iris, and the trabecular meshwork, and all show a high probability of early onset glaucoma (Craig and Mackey, 1999). Moreover, these diseases are autosomal dominant, suggesting haploinsufficiency as a cause for the disorders (Craig and Mackey, 1999). The paired homeodomain transcription factor Pax6 is expressed in the neural tube-derived retina, as well as in the surface ectoderm destined to become the corneal epithelium and the lens (Grindley et al., 1995). Either an increase or decrease of Pax6 gene expression levels in the murine eye leads to anterior segment defects (Hill et al., 1991; Schedl et al., 1996). In humans, PAX6 mutations result in the autosomal dominant disorder aniridia, characterized by iris hypoplasia and progressive anterior segment dysgenesis leading to cataracts, corneal opacification, and glaucoma (reviewed by Glaser et al., 1995).
The Six family of genes encode homeobox transcription factors that were initially identified based on their homology with the Drosophila sine oculis gene, which when mutated causes malformation of the entire fly visual system (Cheyette et al., 1994; Oliver et al., 1995). The vertebrate Six3 and Six6 (Optx2) genes are closely related structurally and share similar but nonidentical expression domains in the anterior brain and the eye primordium (Oliver et al., 1995; Toy et al., 1998; Toy and Sundin, 1999; Jean et al., 1999). Misexpression of Six3 results in ectopic formation of the lens vesicle at the position of the otic vesicle (Oliver et al., 1996), ectopic formation of the retina at the mid/hindbrain junction (Loosli et al., 1999; Lagutin et al., 2001), or enlargement of the forebrain and defects in the optic stalk (Kobayashi et al., 1998). Similarly, misexpression of Six6 leads to an expanded retinal territory at the mid/hindbrain junction and increased eye size (Bernier et al., 2000; Zuber et al., 1999). In addition, overexpression of Six6 in the retinal pigmented epithelial cell cultures induces the expression of retinal-specific genes (Toy et al., 1998). In humans, mutations in the SIX3 gene have been linked to the holoprosencephaly syndrome (HPE2), which display a spectrum of anterior midline defects ranging from hypotelorism to cyclopia (Wallis et al., 1999; Pasquier et al., 2000; Wallis and Muenke, 2000), whereas mutations in the SIX6 gene have been associated with bilateral anophthalmia and pituitary anomalies (Gallardo et al., 1999). These data suggest that both Six3 and Six6 play important roles in vertebrate eye primordium determination as well as in neural retinal fate specification. In addition, other Six genes may also play roles in eye morphogenesis, as disruption of the Six5 gene is sufficient to cause cataract formation in the lens (Sarkar et al., 2000).
In this paper, we describe the expression patterns of Six3 in multiple ocular tissues during the critical period of anterior segment formation. We show that misexpression of Six3 during chick eye development causes morphological defects of the anterior segment and reduction of expression domains of several transcription factors critical for anterior segment development. Moreover, we provide evidence that elevated levels of Six3 influence cell proliferation of periocular mesenchyme and differentiating anterior segment tissues. These results support a regulatory role for Six3 in normal morphogenesis and differentiation of the eye anterior segment as well as in disease conditions involving congenital eye anomalies and glaucoma.
MATERIALS AND METHODS
Chick Embryos
White Leghorn chicken eggs were purchased from Spafas, Inc. Embryos were incubated at 38°C in a rotating humidified incubator. Developmental stages were determined according to Hamburger and Hamilton (1951).
Viral Stock Production and Injections
The replication-competent avian retroviral vector RCAS(A) (Hughes et al., 1987) was used to construct the different viruses expressing wild type and mutant Six3 cDNAs. The RCAS(A).Six3 viral construct was generated by cloning a 1179-bp cDNA fragment containing the coding region of the murine Six3 cDNA (from +179 NcoI site, to +1357 DraI site; Oliver et al., 1995) into the RCAS(A) vector. The RCAS(A).Six3V-E viral construct encoding the full-length murine Six3 cDNA with a missense mutation was generated by PCR using primers containing a single base change at the second position of codon 190 (GTG to GAG, i.e., V190E), based on sequence information of a Drosophila sine oculis mutation (V to D) at the same Valine residue (S. L. Zipursky and F. Pignoni, unpublished observations). The RCAS(A).ΔS viral construct was created by first deleting the sequence between the Bgl II (+199) and the XhoI (+692) of the murine Six3 cDNA and religating the filled-in ends, and then by replacing the starting codon with the Flag epitope tag. The sequences of the two mutant Six3 viral constructs (Six3.V-E and Six3.ΔS) were confirmed by DNA sequencing. Viral stocks with 1–2 × 108 cfu/ml titers were prepared by transfecting chick embryonic fibroblast cells (CEFs) with the viral DNA constructs. Culture media were collected and concentrated by centrifugation as described (Morgan and Fekete, 1996).
Concentrated viral stocks were mixed with 1/10 volume of 0.25% fast green dye (Morgan and Fekete, 1996) before injection. For stage 10 infection, the viral inoculum was injected into the anterior neural tube until an overflow of viral solution from the anterior neural pore occurred (0.2–0.4 μl). For stage 17 infection, the viral inoculum was injected into multiple sites (10–15) within the mesenchymal tissues surrounding the right optic cup. Eggs were sealed with tape and further incubated in a stationary position at 38°C for designated periods until embryos were harvested.
In Situ Hybridization
In situ hybridization was performed by using 14- to 20-μM-thick cryosections. Digoxigenin-labeled RNA probes were synthesized according to the manufacturer’s instructions (Boehringer Mann-heim). In situ hybridization was performed as previously described (Riddle et al., 1993; Yang and Cepko, 1996; Zhang and Yang, 2001). Chick Lmx1b cDNA (Riddle et al., 1995; Kania et al., 2000) was kindly provided by Dr. Randy L. Johnson (MD Anderson Cancer Center, Houston). Chick Pitx2 cDNA (Logan et al., 1998) was a gift from Dr. Cliff Tabin (Harvard Medical School, Boston). A partial chick Six3 cDNA clone (461 bp) was generated by RT-PCR using chick E4 eye cDNAs and degenerate oligonucleotide primers 5′-GARAGRYTIGGIMGITTYYTITGG and 5′-TGYCTTCTRTTYTTRAACCARTT, which corresponded to protein sequences ERLGRFLW and RRNKFWN, respectively. The identity of the chick Six3 clone was confirmed by DNA sequencing. A cDNA clone (572 bp) of the chick Kera gene encoding the keratocan protein core (Pellegata et al., 2000) was generated by RT-PCR using randomly primed chick E4 head cDNAs and primers 5′-ATGAGCTGGATCCTGAGC and 5′-ACAGCTGCAGTGTGTTAG, which corresponded to amino acid sequences NELDPEHW and ANTLQLF, respectively, and then sequenced to confirm its identity. For control or Six3 virus-infected embryos, a minimum of three embryos were sectioned and analyzed by in situ hybridization.
Histological Staining
Embryos were fixed in 4% paraformaldehyde in PBS overnight and embedded in paraffin following standard procedures. Then, 7-μm sections were dewaxed and stained with hematoxylin and eosin by using standard procedures.
Mesenchymal Cell Cultures
Embryonic day 4 (E4, stage 22–24) eyes with surrounding mesenchymal tissues were dissected and incubated with trypsin for 5 min at room temperature. The surface ectoderm and eyeballs were removed, and the remaining cells were collected and further dissociated by trypsin incubation (Altshuler and Cepko, 1992). Single-cell suspensions were plated on 10 μg/ml poly-d-lysine-coated six-well dishes at a density of 650 cells/mm2. Cells were infected with either 1 μl of control RCAS virus or different Six3 viruses at 1–2 × 108 cfu/ml and incubated at 37°C for 48 h in medium containing 42.8% DMEM, 50% F12, 5% fetal calf serum, 1% chick serum, 10 mM Hepes, pH 7.0, and penicillin/streptomycin. BrdU was added to culture media to a final concentration of 20 μM for 9 h before cells were fixed and processed for antibody staining.
Immunocytochemistry and Quantification
Cryosections of 16- to 20-μm thickness and monolayer cells used for antibody staining were fixed with 4% paraformaldelhyde in PBS. Sections or cells were incubated with primary antibodies and visualized by using either biotinylated secondary antibodies and the Vectastain ABC Elite Kit (Vector Laboratories) or Texas Red-conjugated (Jackson ImmunoResearch Laboratories) or Alexa 488-conjugated (Molecular Probes) secondary antibodies with costaining of cell nuclei by 4′−6′-Diamidino-2′-phenylindode (DAPI). Horseradish peroxidase staining using 3′, 3′-Diaminobenzidine (DAB) as chromogen was visualized by using Nomarski microscopy, whereas fluorescent signals were imaged by conventional fluorescent microscopy. For section immunostaining experiments, a minimum of three control or Six3 virus-infected eyes was analyzed.
The rabbit polyclonal antibody against the Six3 protein was raised by using the peptide antigen RLQHQAIGPSGMRSLAEPGC located near the C-terminal of the murine Six3 protein (Lagutin et al., 2001). To evaluate the specificity of the anti-Six3 antibody, the serum was diluted and preincubated three times with avian DF1 cells infected with either the Six3 virus or the RCAS virus, and the supernatants were then used to stain eye sections (Figs. 2E and 2F). The anti-Pax6 (Ericson et al., 1997), anti-type IX collagen (2C2; Irwin et al., 1985; Fitch et al., 1988), anti-keratan sulfate (I22; Funderbergh et al., 1982, 1986), anti-muscle cell specific antigen (13F4; Rong et al., 1987; Barrio-Asensio et al., 1999), and the anti-viral GAG protein (3C2; Stoker and Bissell, 1987) monoclonal antibodies were obtained from the Developmental Studies Hybrid-oma Bank (DSHB, Iowa City). The polyclonal antibodies p27 against the viral GAG protein were purchased from SPAFAS. The anti-BrdU antibody containing nuclease was obtained from Amer-sham.
FIG. 2.
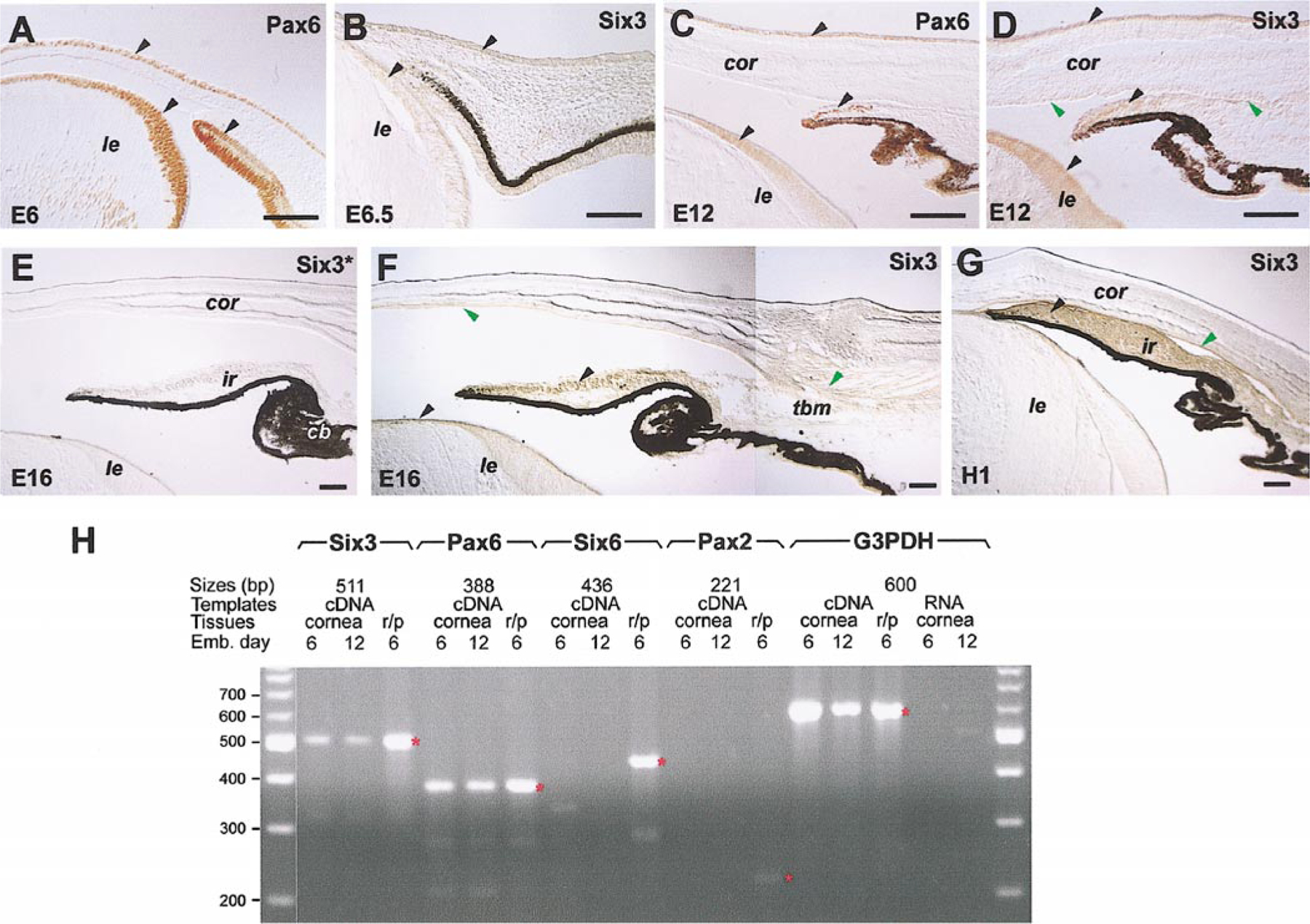
Expression of chick Six3 protein and RNA in the eye anterior segment. Immunocytochemical staining of Six3 protein (B, D–G) and Pax6 protein (A, C) is shown. Arrowheads point to tissues expressing either both proteins (black) or Six3 protein only (green). The anti-Six3 antibodies used in (E) and (F) have been preincubated with the Six3 virus-infected and the RCAS virus-infected DF1 cells, respectively. (H) Results of RT-PCR amplification of Six3, Six6, Pax6, Pax2, and G3PDH (control) using E6 and E12 corneal cDNAs, E6 retina, and pigmented epithelium cDNAs (r/p), or corneal RNAs without reverse transcription. Red asterisks (*) indicate the expected positions of amplified products. Scale bars, 100 μm. Abbreviations: cor, cornea; le, lens; ir, iris; tbm, trabecular meshwork.
For in vivo labeling with BrdU, embryos infected by viruses at stage 10 were windowed and 1 ml of PBS containing 100 μg BrdU was dripped on top of each embryo. The eggs were then returned to incubation for an additional 3 or 6 h. Embryos were fixed with 4% paraformaldehyde and cryosections were stained for BrdU incorporation by using a protocol previously described for paraffin embedded tissues (Belecky-Adams et al., 1996).
Percentages of BrdU marker-positive mesenchymal cells among total cells were determined by calculating the ratio of the fluorescent-labeled cells and the total DAPI-stained nuclei in a given dissociated cell sample. Cells were costained for the anti-viral P27 antibody to determine infection rates. For a given virus, 7–10 independent samples were quantified. The Student’s t test was used for statistical analyses. P values <0.02 were considered statistically significant.
RT-PCR Analyses
Total RNAs of E6 cornea, E12 cornea, as well as E6 retina and pigmented epithelium were isolated following the manufacturer’s instructions by using RNAzol B (Tel-Test, Inc., Friendswood, TX). Random hexamers and an MMLV reverse transcriptase (Life Technologies, Rockville, MD) were used to synthesize single-strand cDNAs. For RT-PCRs, products were amplified using 35 cycles at (95°C, 45 s; 52°C, 45 s; 72°C, 45 s). Equal volumes of PCR products were loaded and separated by agarose gel electrophoresis. The Six3 PCR product was sequenced to confirm the identity. The PCR primers used for each gene are: Six3, 5′-GCAAGCTGCAGGCCATGT and 5′-CATACATCACATTCCGAG; Six6 (Optx2), 5′-AACGAGTCGGTGCTGAGA and 5′-TGCTGCTGTAGCCTGTTC; Pax6, 5′-CAGCTTCACCATGGCCAA and 5′-TTACAGCGTAGGGCACAG; Pax2, 5′-ATAGTGGAGCTGGCTCACCA and 5′-AATCTCCCAAGCGAACATGGT; and G3PDH, 5′-CAGTGAGAAAGTCGGAGT and 5′-CAGCACCTCTGCCATCTC.
Western Blot Analysis
Chick embryonic fibroblast (CEF) cells infected with different Six3 viruses were cultured for 48 h. Cell extracts were prepared by incubating cells on ice in 50 mM Tris–HCl, pH 8.0, 250 mM NaCl, 0.5% NP40 for 15 min, followed by centrifuging cell lysates in a microfuge for 5 min at the highest speed and collecting the supernatants. E6 retinal and spinal cord extracts were similarly prepared by directly lysing tissues in the lysis buffer followed by centrifugation. Proteins were transferred to nitrocellulose membrane following SDS–PAGE. The anti-Six3 polyclonal antibodies (Lagutin et al., 2001) and HRP-conjugated secondary antibodies (Amersham) were used in ECL assays (Amersham) to detect Six3 protein signals.
TUNEL Assays
Cryosections of 20-μm thickness were used in TUNEL assays (Gavrieli et al., 1992) with fluorescein-conjugated nucleotides (Boehringer Mannheim) according to the manufacturer’s instructions. Apoptotic signals and cell nuclei stained by DAPI were visualized by using epifluorescent microscopy.
RESULTS
Expression of Six3 in Chick Ocular Tissues
As an initial step to elucidate the potential roles of Six3 in chick eye morphogenesis, a partial chick Six3 cDNA generated by RT-PCR was used in in situ hybridization analyses. Chick Six3 expression was first detected by whole-mount in situ hybridization at stage 5 in the anterior neural plate (data not shown; Bovolenta et al., 1998). At stage 14, in addition to the ventral forebrain and the prechordal plate, Six3 mRNA was detected in the neural tube-derived optic cup and in the ectoderm-derived nasal placode, lens vesicle, and presumptive corneal epithelium (Figs. 1A–1C). After the onset of neuronal differentiation, the stage 24 (embryonic day 4, E4) retina showed high levels of Six3 message, while the expression of Six3 in the pigmented epithelium, the lens, and the adjacent surface ectoderm declined (Fig. 1D). The expression of Six3 in the retina persisted throughout neurogenesis and remained in all three cellular layers at E16 when production of all retinal cell types was complete (Figs. 1D–1I). Interestingly, Six3-positive signals were also detected in mesenchymal cells located immediately adjacent to the pigmented epithelium (Figs. 1E and 1F).
FIG. 1.
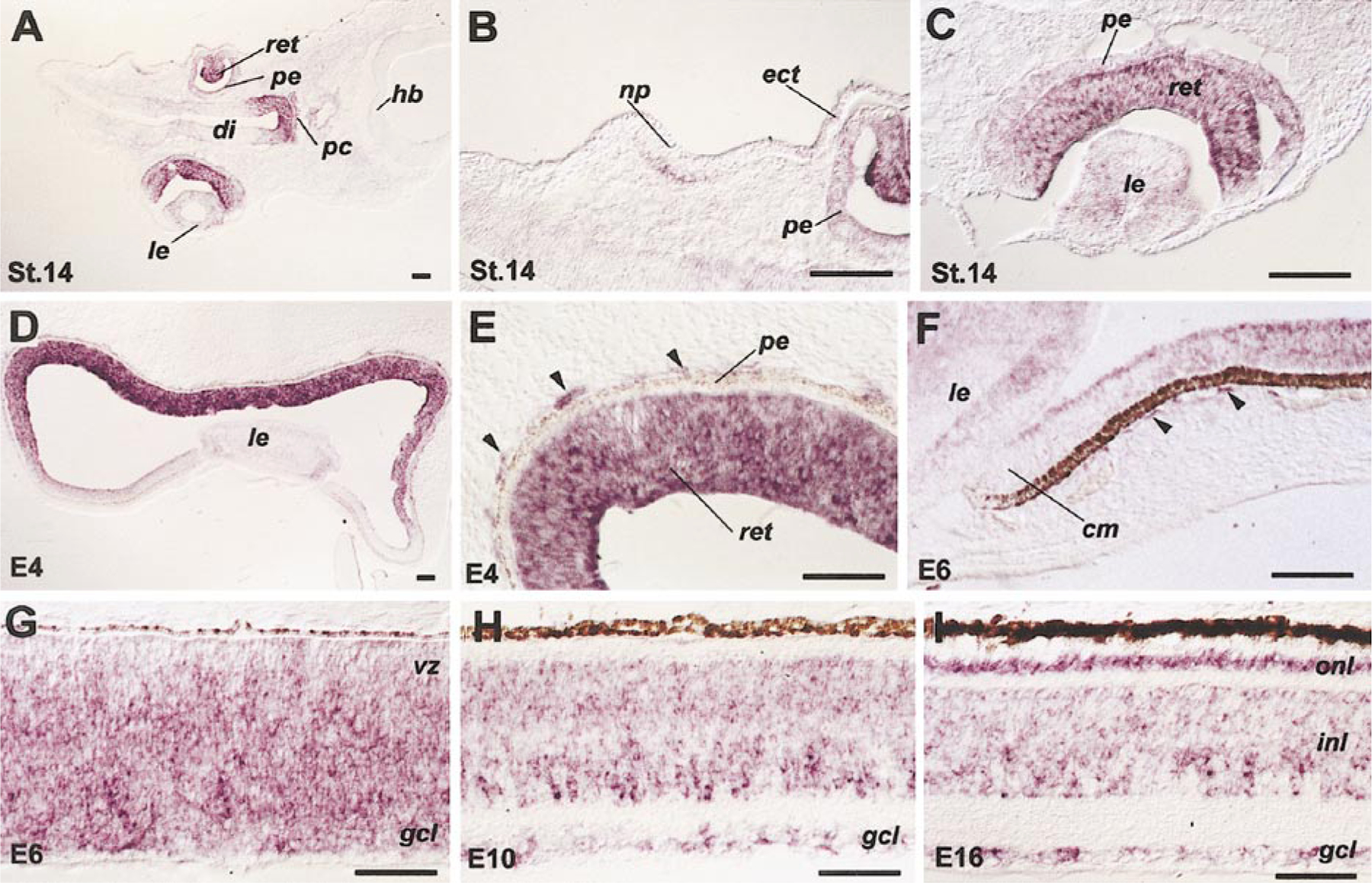
Expression patterns of chick Six3 mRNA as detected by in situ hybridization. Tissue sections of stage 14 head (A–C), stage 24 eye (D, E), E6 eye and retina (F, G), and E10 (H) and E16 (I) retinae hybridized with antisense probes are shown. Arrowheads (E, F) point to Six3-positive cells immediately adjacent to the pigmented epithelium. Scale bars, 50 μm. Abbreviations: cm, ciliary margin; di, diencephalon; ect, ectoderm; gcl, ganglion cell layer; hb, hindbrain; inl, inner nuclear layer; le, lens; np, nasal placode; onl, outer nuclear layer; pe, pigmented epithelium; pc, prechordal plate; ret, retina; vz, ventricular zone.
The expression patterns of Six3 protein were characterized by immunohistochemistry using Six3-specific antibodies. We first examined whether Six3 and Pax6 proteins were colocalized. In addition to its expression in the developing retina (data not shown), the chick Six3 protein was detected in the corneal epithelium and lens epithelium at E6.5 and E12 (Figs. 2B and 2D), coinciding with the expression of the Pax6 protein in these tissues (Figs. 2A and 2C). However, by E12, Six3 but not Pax6 protein is present in the corneal endothelium and the limbal region (Figs. 2C and 2D). After E16 and at hatching (H1), Six3 protein persisted in multiple anterior segment tissues, including the lens epithelium, the corneal endothelium, the iris, and the trabecular meshwork (Figs. 2F and 2G). Preincubation of the anti-Six3 antibodies with Six3-expressing cells abolished the Six3-staining signals (Figs. 2E and 2F), indicating that the antibodies specifically recognized the Six3 protein as an antigen. To further confirm the expression of chick Six3 in the developing cornea, RT-PCR using E6 and E12 corneal cDNAs was performed. The amplification results showed that Six3 transcripts were indeed expressed in the cornea at these stages, whereas Six6 (Optx2) was only detected in the retina and pigmented epithelium as expected but not in the cornea (Fig. 2H).
The RNA and protein expression patterns of Six3 together suggest that Six3 may be involved in the development of both neural tube- and mesenchyme-derived ocular tissues, especially during the morphogenesis and differentiation of the anterior segment of the eye.
Retrovirus-Mediated Misexpression of Wild Type and Mutant Six3 Proteins
To examine the potential role of Six3 during development of the anterior segment tissues, we constructed replication competent avian retroviruses expressing variants of the murine Six3 protein (Fig. 3A). In addition to a virus expressing the wild type Six3, two viruses producing mutant Six3 proteins were also constructed. Based on molecular genetic studies of Drosophila sine oculis mutations (F. Pignoni and S. L. Zipursky, unpublished observations), we constructed the Six3.V-E mutant cDNA encoding a single amino acid residue change (Valine190 to Glutamic acid, i.e., V190E) at a highly conserved position within the Six domain. The Six3.ΔS mutant contained partial deletions of the N-terminal portion and the Six domain as well as the Flag epitope tag. By Western blot analyses, the chick E6 retinal extract, but not the spinal cord extract, contained two major proteins recognized by the anti-Six3 antibodies, with the upper band corresponding to 34 kDa as predicted for the full-length endogenous chick Six3 protein (Fig. 3B). Chick embryonic fibroblast (CEF) cells infected by the wild type Six3 and Six3.V-E viruses produced murine Six3 variants with the expected molecular weight of 36 kDa, whereas CEF cells infected by the Six3.ΔS virus expressed a 26-kDa protein corresponding to the truncated Six3 protein (Fig. 3B). Furthermore, anti-Six3 immunocytochemical staining showed that all three virus-encoded murine Six3 variants were localized to the nuclei of infected CEF cells (Fig. 3C).
FIG. 3.
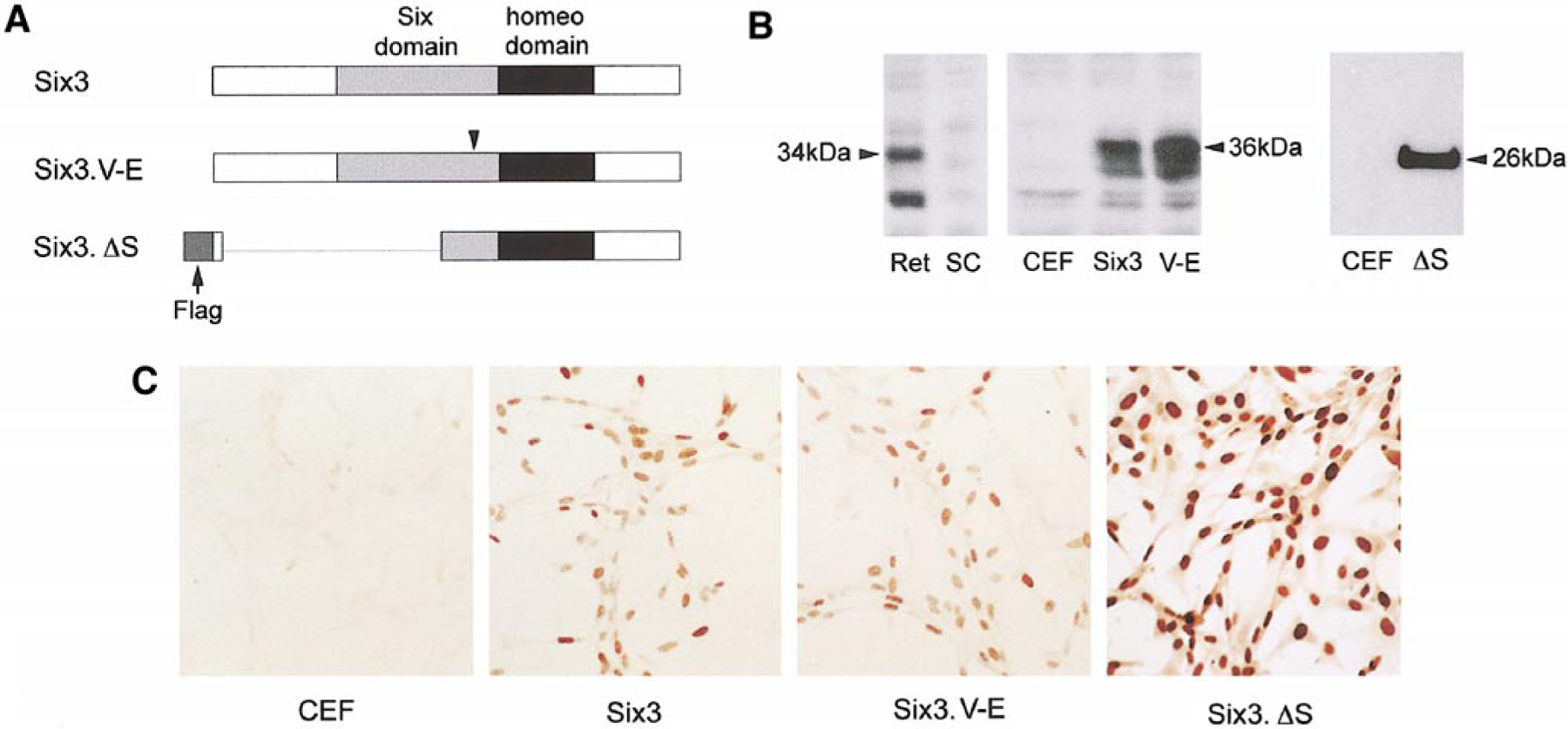
Viral-mediated expression of Six3 protein variants. (A) A schematic illustration shows the Six3 cDNA variants inserted into the RCAS(A) viral vector. The Six and the homeodomains are shown as gray and black boxes, respectively. The arrowhead indicates the position of the V190E mutation within the Six domain. The deleted region in Six3.ΔS is indicated with a line, and the arrow points to the N-terminal Flag epitope. (B) Western blot analyses using anti-Six3 antibodies show viral mediated expression of wild type and mutant Six3 proteins. The protein extracts used are derived from E6 chick retina (Ret), E6 chick spinal cord (SC), CEF cells (CEF), and RCAS.Six3 (Six3), RCAS.Six3.V-E (V-E), and RCAS.Six3.ΔS (ΔS) virus-infected CEF cells. (C) Immunocytochemical staining of CEF cells infected with different Six3 viruses demonstrates the expression and subcellular localization of the three Six3 variants.
Morphological Defects Caused by Six3 Misexpression
High-titer viruses were used to infect the developing optic primordium at stage 10 (E2) in order to misexpress wild type and mutant Six3 proteins. By E12, infection with wild type Six3 virus resulted in abnormal eye morphology, including corneal protrusion, eyelid hypoplasia, and, at low frequency, microphthalmia (Figs. 4A and 4B). By E16, Six3 virus-infected eyes displayed corneal opacification and iris hypoplasia (Figs. 4C–4F). In comparison to control RCAS virus-infected embryos, which rarely showed eye abnormality (1 of 60 embryos, or 1.6%), 78% of wild type Six3 virus-infected embryos displayed anterior segment anomalies represented by cornea protrusion (Table 1). Only 7.7% of the embryos infected with the Six3.V-E virus and 46% of the embryos infected with the Six3.ΔS virus showed anterior segment anomalies. Moreover, the eye phenotypes caused by Six3 mutant viruses were less severe compared with those caused by the wild type Six3 virus infection. These results indicate that viral-mediated misexpression of wild type Six3 affected anterior segment development. Furthermore, the deleted portions of the Six3 protein were involved in causing viral-induced morphogenetic defects, whereas the conserved Valine190 in the Six domain was critical for Six3 activity.
FIG. 4.
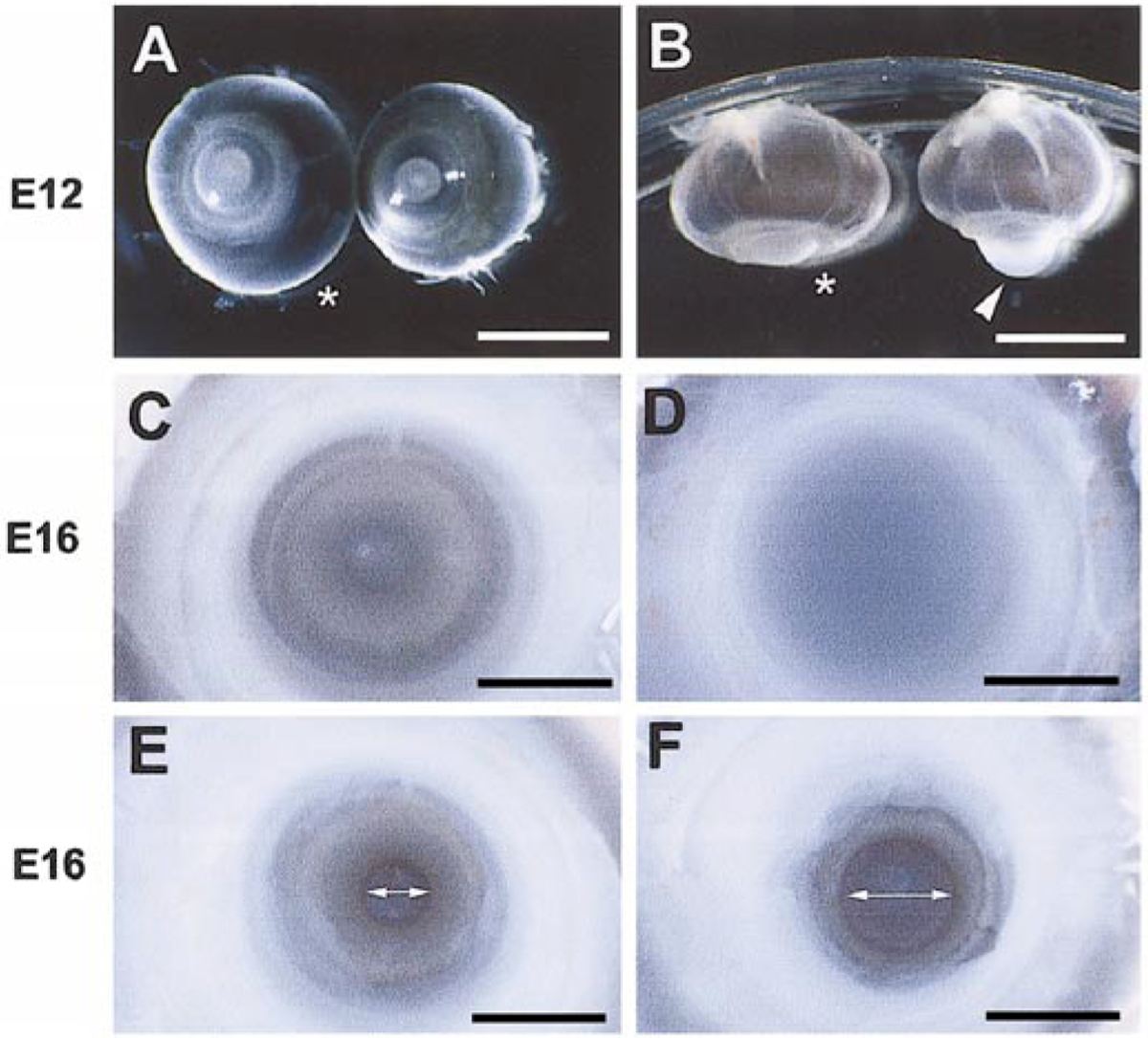
Anterior segment malformation caused by Six3 virus infection. (A, B) Frontal and ventral views, respectively, of E12 eyes infected by the Six3 virus and the control RCAS virus (asterisks) at stage 10. Arrowhead points to the protruding cornea of the Six3 virus-infected eye. (C, D) Frontal views of the corneas of the Six3 virus (D) and control RCAS virus (C)-infected eyes at E16. (E, F) Frontal views of the pupil in Six3 virus (F)- and control RCAS virus (E)-infected E16 eyes upon removal of the cornea. The two-headed arrows indicate pupil diameters. Scale bars, in (A, B), 5 mm; in (C–F), 250 μm.
TABLE 1.
Summary of Viral-Mediated Six3 Misexpression on Eye Morphogenesis
| Injection | Virus | Harvest | No. of survivors | No. with phenotypes | % with abnormalities |
|---|---|---|---|---|---|
| Stage 10 | RCAS(A) | E12 | 48 | 0 | |
| E16 | 4 | 1 | 1.6 | ||
| E18 | 8 | 0 | |||
| Six3 | E12 | 31 | 24 | ||
| E16 | 3 | 2 | 78.0 | ||
| E18 | 7 | 6 | |||
| Six3.V-E | E12 | 26 | 2 | 7.7 | |
| Six3.ΔS | E12 | 26 | 12 | 46.1 | |
| Stage 17 | RCAS(A) | E16 | 13 | 0 | 0 |
| Six3 | E16 | 8 | 6 | 75.0 |
Note. Viruses were injected at stage 10 into the neural tube or at stage 17 into the periocular mesenchyme and further incubated at 38°C until harvesting. Ocular abnormalities, primarily corneal protrusion, were scored by comparing the external appearance of Six3 viruses infected eyes with RCAS(A) control virus infected eyes at a given stage. Percentages of abnormality represent embryos harvested between E12 and E18
Histological staining of tissue sections revealed malformation of multiple anterior segment tissues due to misexpression of the wild type Six3 protein. At E12, some Six3 virus-infected embryos exhibited microphthalmic eyes compared with the control RCAS virus-infected or noninfected embryos (Figs. 5A and 5B). However, the laminar organization of the Six3 virus-infected neural retina appeared similar to that of the control RCAS virus-infected retina (data not shown). Six3 virus-infected eyes also showed retarded sclera and eyelid development in contrast to control eyes at E12 (Figs. 5C and 5D). By E16, the cornea of the Six3 virus-infected eyes showed increased thickness despite the presence of the three cellular layers, i.e., the corneal epithelium, the stroma, and the corneal endothelium (Figs. 5E and 5F). In Six3 virus-infected eyes, the corneal stromal cells (keratocytes) appeared to be distributed more sparsely. However, quantifications using thin sections showed no changes of stromal cell number per unit length of cornea (data not shown), suggesting that the corneal stroma was not properly condensed (Figs. 5G–5I). In addition, the structural integrity of the corneal endothelium appeared compromised, as indicated by the presence of vacuole-like gaps (Figs. 5G and 5H). The limbal region of the anterior segment, including the trabecular meshwork, also exhibited an abnormal morphology (Figs. 5J and 5K). Moreover, Six3 virus-infected eyes displayed underdeveloped ciliary bodies and shortened irises, i.e., ciliary body and iris hypoplasia (Figs. 5J and 5K). These histological results demonstrated that anterior segment tissue types containing mesenchyme components were affected by misexpression of Six3.
FIG. 5.
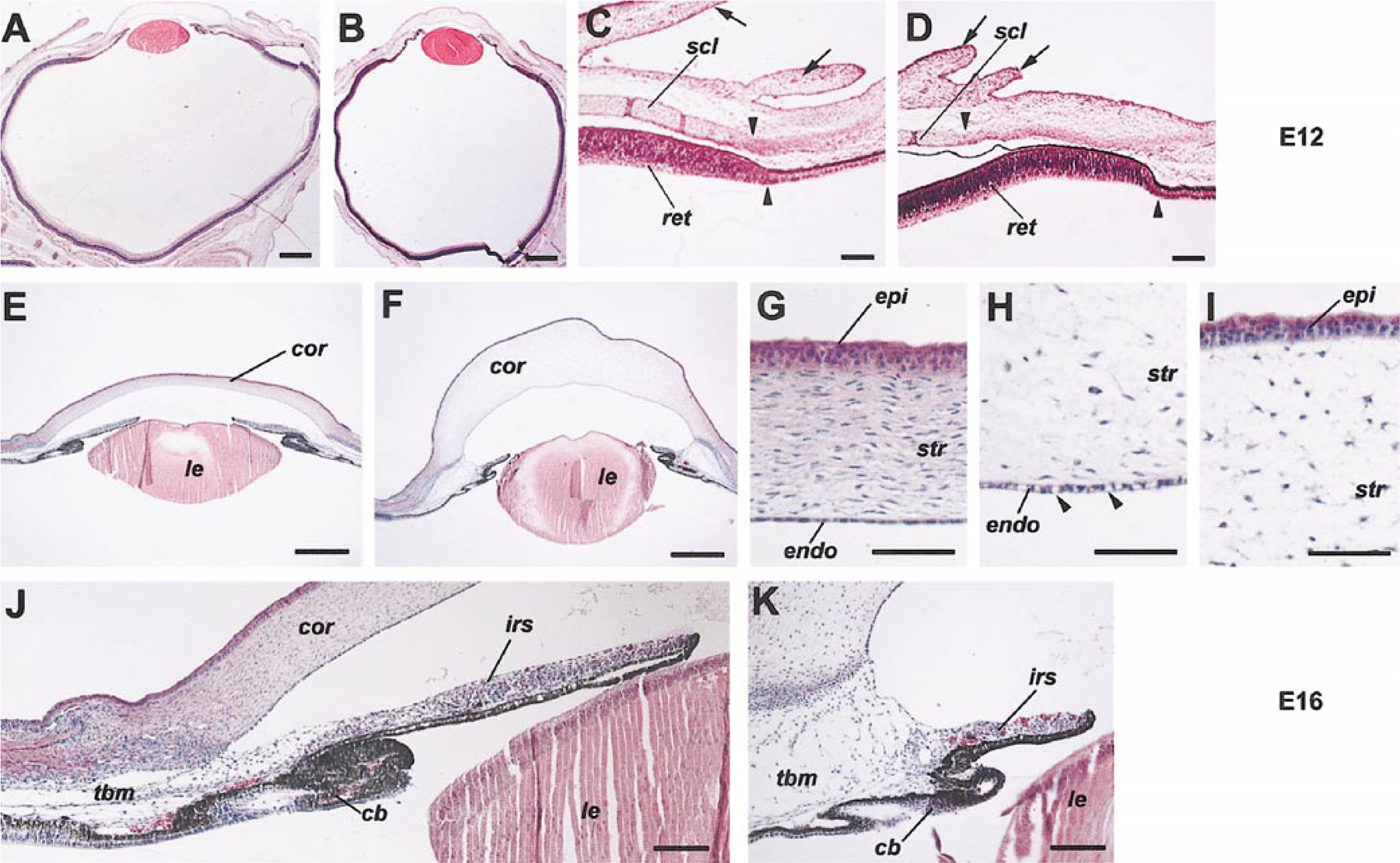
Disruption of anterior segment morphology by Six3 misexepression. H&E staining demonstrates anterior segment tissue defects caused by Six3 virus infection (B, D, F, H, I, K) compared with control RCAS virus infection (A, C, E, G, J) at stage 10. In the E12 sections (A–D) shown, arrows point to the eyelid; arrowheads indicate the termination points of the differentiated sclera and the peripheral retina. Among the E16 sections (E–K) displayed, (H) and (I) show partial views of Six3 virus-infected cornea, with arrowheads (H) pointing to the vacuoles in the corneal endothelium. Scale bars, in (A, B, E, F), 500 μm; in (C, D, J, K), 100 μm; in (G–I), 50 μm. Abbreviations: cb, ciliary body; cor, cornea; endo, corneal endothelium; epi, corneal epithelium; str, corneal stroma; ir, iris; le, lens; ret, retina; scl, sclera; tbm, trabecular meshwork.
Defective Corneal Differentiation Due to Six3 Misexpression
The transparency of the cornea is largely dependent on the composition and proper organization of extracellular matrix proteins in the corneal stroma. To determine possible effects of misexpressing Six3 on corneal differentiation, we examined the distribution of two corneal extracellular matrix components, type IX collagen and keratan sulfate proteoglycans (KSPGs). During normal corneal differentiation, collagen IX crosslinks other types of collagen into highly ordered fibrils, and the proteolytic processing of collagen IX may play a role in mesenchymal cell invasion of the corneal stroma (Fitch et al., 1998; Linsenmayer et al., 1998). Immunocytochemical analyses demonstrated that Six3 misexpression caused elevated collagen IX levels (2C2 monoclonal antibody; Fitch et al., 1998) in tissues lining the E12 anterior chamber, i.e., the corneal endothelium and the iris (Figs. 6A–6D). Keratocan is a major type of KSPG present in the developing and adult cornea and is implicated in corneal stroma differentiation and transparency (Rada et al., 1993; Dunlevy et al., 1998; Liu et al., 1998; Pellegata et al., 2000). In situ hybridization using a chick Keratocan (Kera) cDNA encoding the protein core of Keratocan detected an increased expression of Kera transcripts in Six3 virus-infected cornea and limbal regions (Figs. 6E and 6F). Consistently, immunocytochemical staining using the I22 monoclonal antibody, which recognized the keratan sulfate groups on different types of KSPGs, also revealed an increased presence of Keratan sulfate antigens in the Six3 virus-infected cornea (Figs. 6G and 6H) (Funderburgh et al., 1982, 1986). Taken together, these results suggest that overexpression of wild type Six3 disrupted corneal differentiation as reflected by altered expression of extracellular matrix molecules.
FIG. 6.
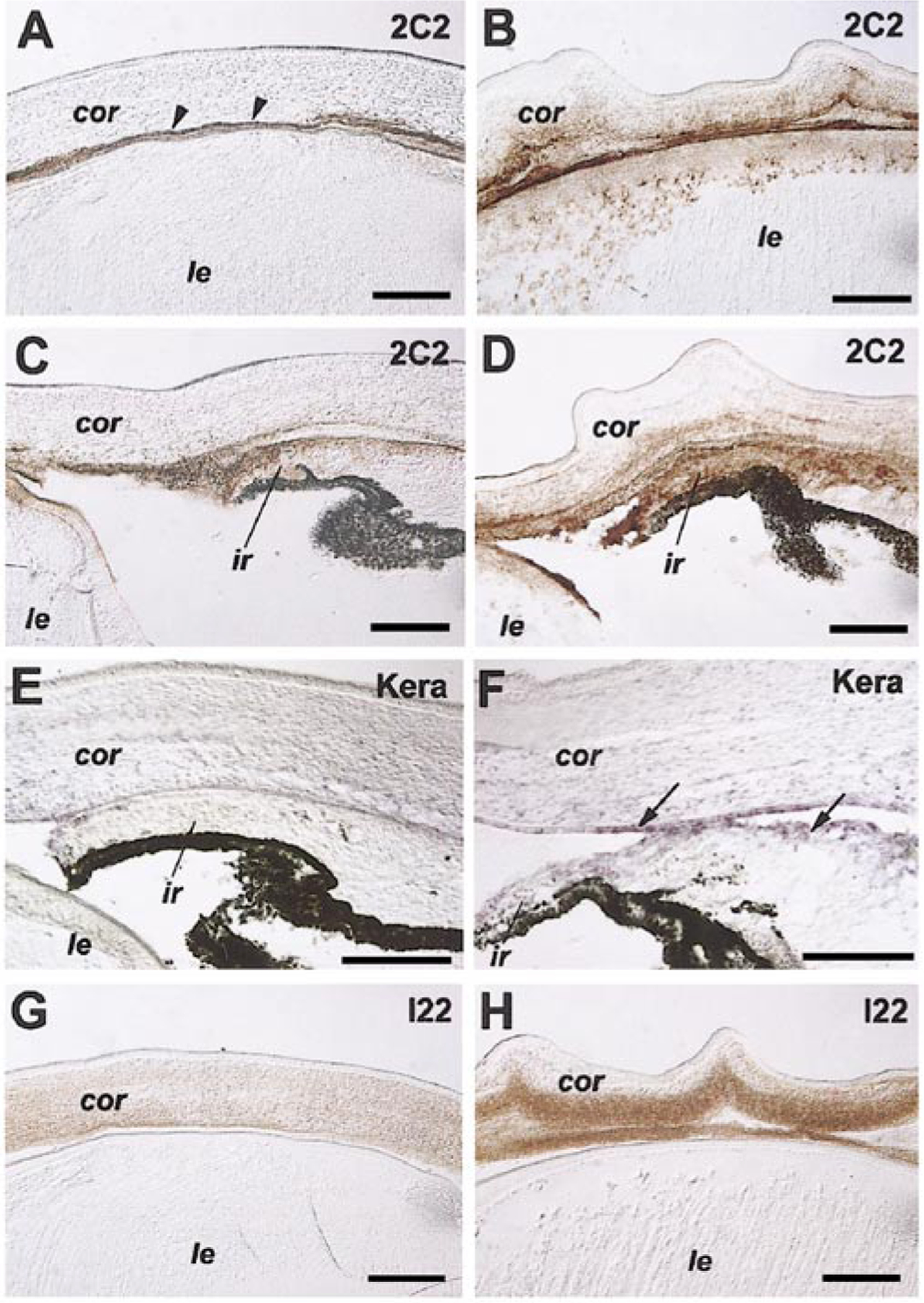
Effects of Six3 misexpression on corneal differentiation. Immunocytochemical staining (A–D, G, H) and in situ hybridization (E,F) of E12 eyes infected with the Six3 virus (B, D, F, H) and control RCAS virus (A, C, E, G) at stage 10 show altered expression patterns of type IX collagen (2C2 antibody; A–D), keratan sulfate (I22 antibody; G, H), and keratocan mRNA (kera antisense probe; E, F) in the cornea and iris. Arrowheads (A) point to corneal endothelium. Arrows (F) indicate in situ hybridization signals detected in corneal endothelium and in cells lining the anterior chamber. Scale bars, 100 μm. Abbreviations: cor, cornea; ir, iris; le, lens.
Effects of Six3 Misexpression on Molecular Markers of the Periocular Mesenchyme
Since misexpressing Six3 affected the morphogenesis of mesenchyme-derived anterior segment tissues, we next examined whether the expression of other transcription factors known to play roles in anterior segment development was influenced by Six3. In situ hybridization detected the expression of Lmx1b and Pitx2 mRNAs in regions surrounding the chick optic cup at stage 18 (E3; data not shown). Between E4 and E6, overlapping domains of the mesenchyme surrounding the eye expressed both Lmx1b and Pitx2 mRNAs (Figs. 7A and 7C). In Six3 virus-infected eyes, the size of expression territories of Lmx1b and Pitx2 near the ciliary margin was significantly reduced by E6.5 compared with controls; however, similar intensities of hybridization signals for Lmx1b and Pitx2 were observed (Figs. 7A–7D). To further evaluate whether Six3 virus infection decreased transcription of Lmx1b and/or Pitx2, RNAs from primary periocular mesenchymal cell cultures (see below) infected with either the RCAS control virus or the Six3 virus were assayed by semiquantitative RT-PCRs. No difference in the levels of Lmx1b and Pitx2 transcripts was detected when equal amounts of input RNAs were used (data not shown), suggesting that Six3 misexpression did not affect the levels of Pitx2 and Lmx1b transcription on a per cell basis, but more likely affected the numbers of periocular mesenchymal cells in the limbal region.
FIG. 7.
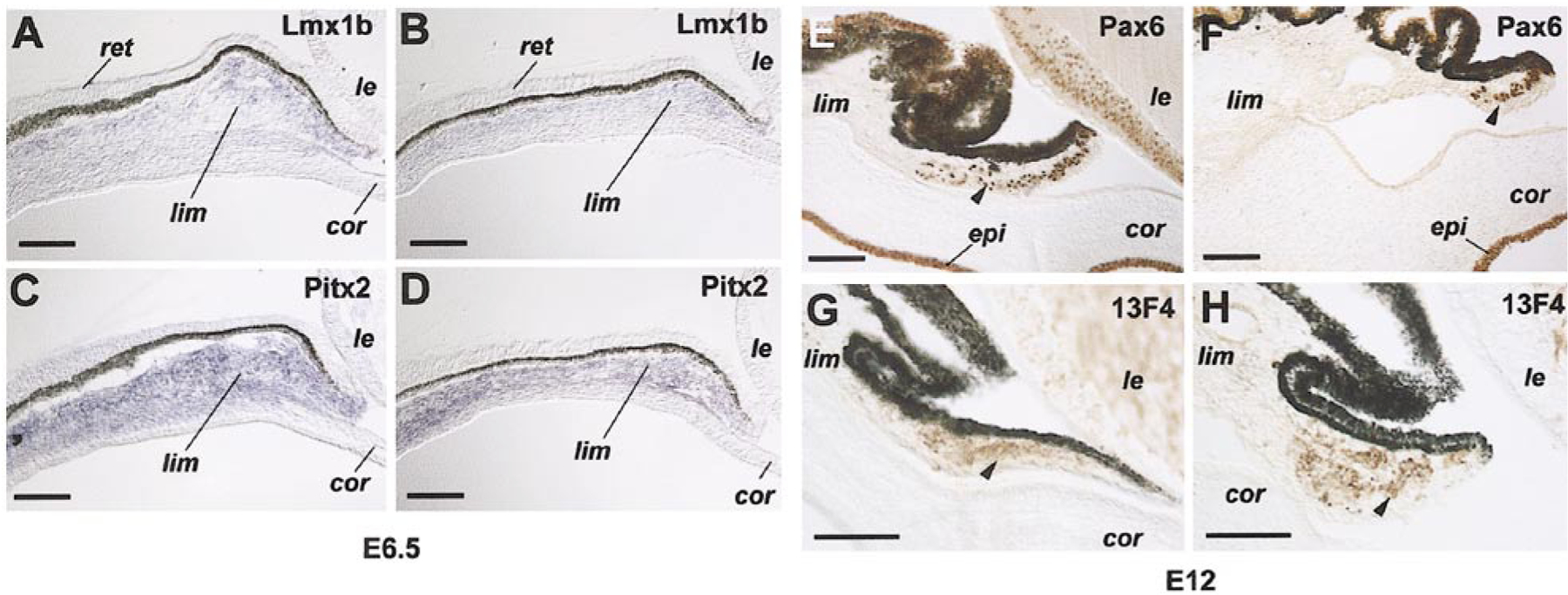
Altered expression patterns of periocular mesenchyme and anterior segment markers due to Six3 misexpression. (A–D) In situ hybridization signals of Lmx1b (A, B) and Pitx2 (C, D) in the limbal region of E 6.5 eyes infected by Six3 (B, D) and RCAS (A, C) viruses at stage10. (E–H) Immunocytochemical staining of Pax6 (E, F) and muscle marker 13F4 (G, H) in E12 anterior segments infected by Six3 (F, H) and RCAS (E, G) viruses at stage 10. Anti-viral protein immunostaining (3C2) of adjacent sections reveals extensive infection by the control RCAS virus and the Six3 virus of the regions analyzed (data not shown). Scale bars, 100 μm. Abbreviations: cor, cornea; epi, cornea epithelium; le, lens; lim, presumptive limbus; ret, retina.
During differentiation of the anterior segment, the corneal epithelium and a subset of iris stromal cells expressed the homeo and paired domain transcription factor Pax6 (Fig. 7E). Immunocytochemical staining showed that the expression of Pax6 protein in the corneal epithelium at E12 was not altered by Six3 virus infection (Figs. 7E and 7F). In contrast, the number of Pax6-positive cells within the iris stroma was markedly reduced in Six3 virus-infected eyes (Figs. 7E and 7F), even though the similar staining intensity remained in the residual Pax6-positive iris stromal cells. Furthermore, a muscle-specific marker (monoclonal antibody 13F4) (Rong et al., 1987; Barrio-Asensio et al., 1999) also revealed abnormal iris muscle organization in Six3 virus-infected eyes compared with the control virus-infected eye (Figs. 7G and 7H). Thus, Six3 misexpression altered the size of expression territories of key transcription factors in the periocular mesenchyme and their derivatives during the critical period of anterior segment formation.
Effects of Misexpressing Six3 on Mesenchymal and Anterior Segment Cell Proliferation
The ocular phenotypes caused by Six3 misexpression appeared to be associated with a reduction of the periocular mesenchymal cells that give rise to perspective anterior segment tissues. This effect potentially might be due to the influence of misexpressed Six3 on the specification or migration of neural crest cells originating from diencephalic and mesencephalic regions of the virally infected neural tube at stage 10. Alternatively, Six3 misexpression might affect the proliferation and/or apoptotic death of the periocular mesenchymal cells. To begin distinguishing these possibilities, we targeted viral infection directly to the periocular mesenchyme surrounding the optic cup at stage 17. Infection by the control RCAS virus did not give rise to any ocular abnormalities, whereas infection by the Six3 virus at stage 17 caused a similar percentage (75%) of anterior segment anomalies among injected embryos (Table 1). Since stage 17 periocular injection did not perturb the specification and initial migration of the head neural crest cells (data not shown), this result supported the notion that effects of Six3 misexpression were, at least in part, due to local perturbation of the periocular mesenchyme.
To determine whether Six3 misexpression caused anterior segment malformation by influencing cell proliferation, incorporation of the DNA synthesis precursor BrdU in vitro and in vivo was examined. At stage 23 (E4) when the first wave of mesenchymal cells begun migrating between the corneal epithelium and the lens, periocular mesenchymal cells near the ciliary margin of the eye expressed Six3 mRNA (Fig. 8A). By stage 25 (E5), both the newly formed corneal endothelium and the periocular mesenchymal cells were highly proliferative as indicated by extensive BrdU incorporation in vivo (Fig. 8B). To examine whether Six3 viral infection affected cell proliferation during this time window, periocular mesenchymal cells were cultured as a monolayer in vitro at E4 and infected with various viruses for 48 h. Quantification of BrdU incorporation showed that Six3 virus infection caused a statistically significant decrease in BrdU-positive cells from 47.6% found in the control RCAS virus-infected cultures to 40.0% (Fig. 8C). However, infection by the Six3.V-E virus did not result in a significant change of BrdU incorporation by this assay (44.5%) (Fig. 8C). Consistent with the in vitro proliferation assay, immunofluorescent microscopy revealed that at E6 Six3 virus-infected limbal region contained significantly less BrdU-labeled cells in vivo (Figs. 8D, 8E, 8H, and 8I). Furthermore, during the period of iris growth and differentiation (E12 to E16), the Six3 virus-infected iris showed a reduced number of BrdU-labeled cells in vivo compared with control-virus infected iris (Figs. 8F, 8G, 8J, and 8K). Together, these data showed that overexpression of the wild type Six3 protein reduced proliferation of periocular mesenchymal cells and their derivatives in vivo and in vitro.
FIG. 8.
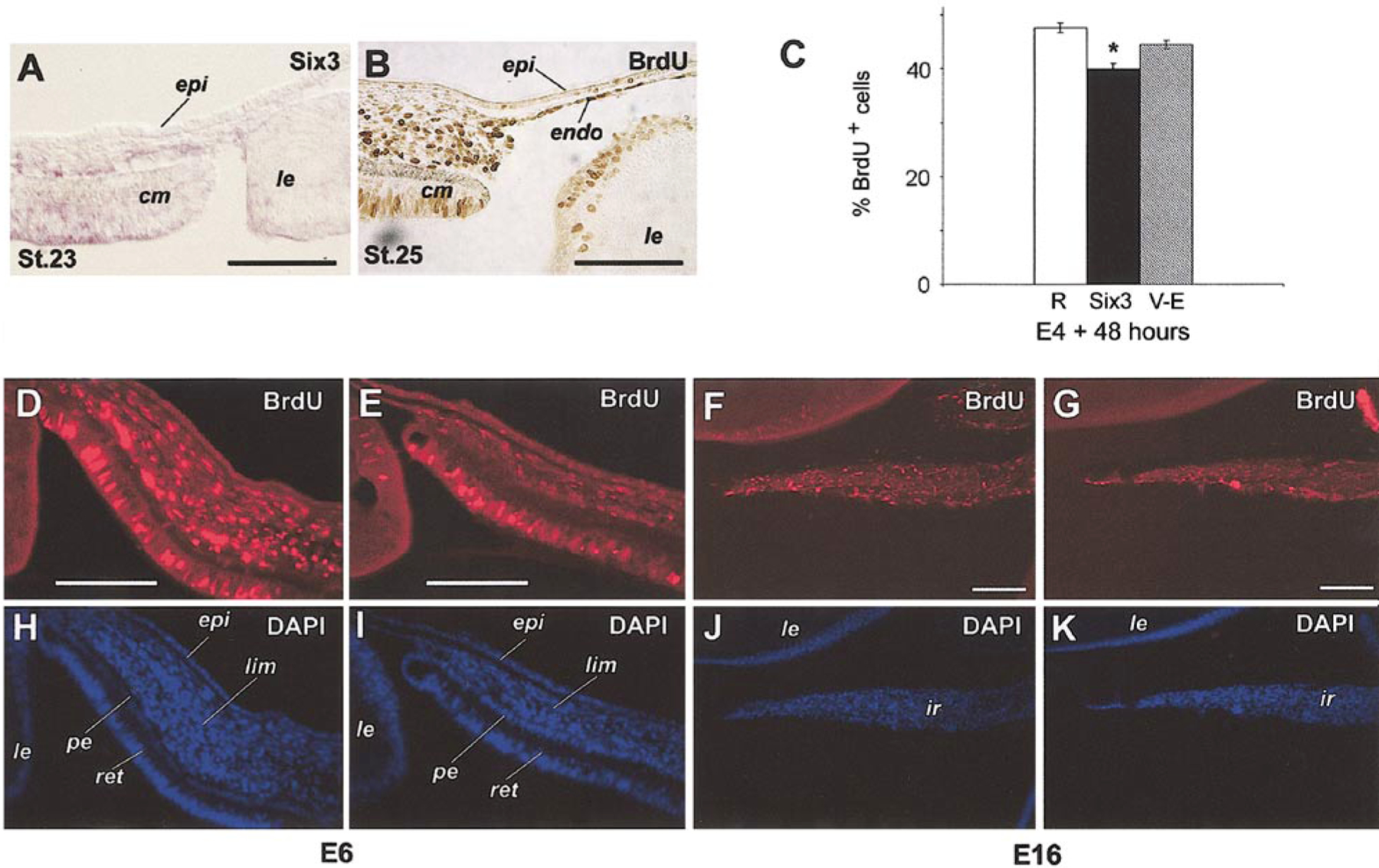
Effects of Six3 misexpression on cell proliferation. (A) In situ hybridization of Six3 in the anterior segment at stage 23 (E4). (B) Anti-BrdU immunostaining of eye anterior segment (3 h in vivo labeling) at stage 25 (E5). (C) Quantification of BrdU incorporation in periocular mesenchymal cells in vitro. Percentages of BrdU-positive cells among total cells are shown (average ± standard error). All cells were infected as indicated by coimmunostaining using anti-viral GAG antibody (P27) (data not shown); RCAS virus (R, n = 7), Six3 virus (Six3, n = 7; *, P = 0.015), Six3.V-E virus (V-E, n = 10). (D–G) Anti-BrdU immunostaining compares in vivo BrdU incorporation after infection by Six3 virus (E, G) and RCAS virus (D, F) at stage 10. (D, E) Infected limbal regions at E6 after 3 h of labeling. (F, G) Infected irises at E16 after 6 h of labeling. (H–K) DAPI staining of corresponding fields (D–G), respectively. Extensively viral infection of the same sections was detected by costaining with anti-viral GAG antibody (P27) (data not shown). Scale bars, 100 μm. Abbreviations: epi, corneal epithelium; ir, iris; le, lens; pe, pigmented epithelium; lim, presumptive limbus; ret, retina.
To rule out possible effects of Six3 misexpression on apoptotic cell death during anterior segment morphogenesis, we performed TUNEL assay. At E12, few TUNEL-positive cells were detected in anterior segment tissues infected by the RCAS virus. Similar extents of cell death were observed in Six3 virus-infected anterior segment, despite the presence of defective tissue morphology (Fig. 9). Therefore, the anterior segment defects caused by Six3 misexpression were unlikely due to altered patterns of apoptosis in the anterior portion of the eye at this stage.
FIG. 9.
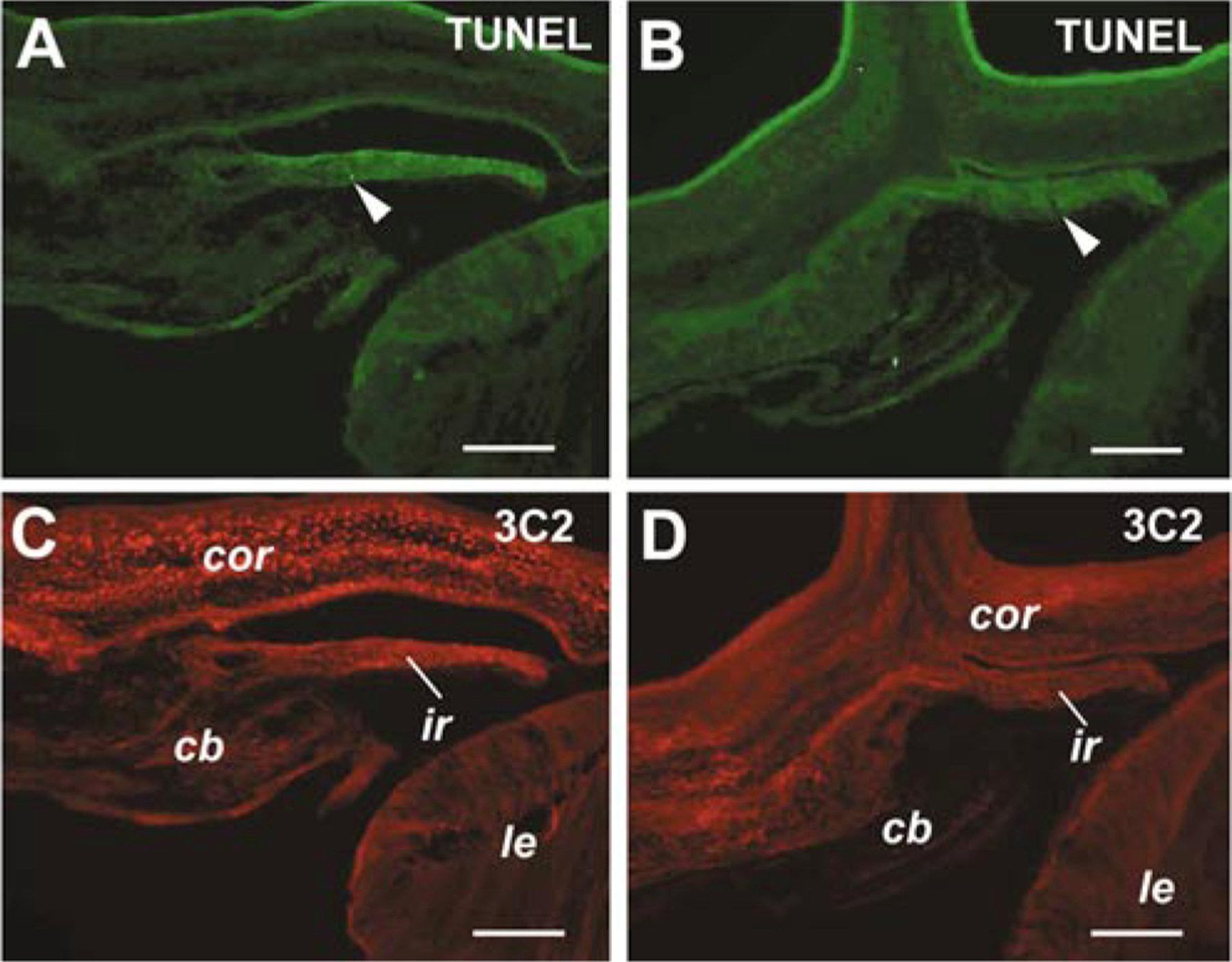
Effects of viral infection on cell death. TUNEL staining of E12 eyes infected at stage 10 by RCAS virus (A, C, same section) or Six3 virus (B, D, same section) are shown. Arrowheads point to detected apoptotic cells. Staining with the anti-viral GAG antibody 3C2 (C, D) demonstrates extensive infection of anterior segments by both RCAS and Six3 viruses. Scale bars, 100 μm. Abbreviations: cb, ciliary body; cor, cornea; ir, iris; le, lens.
DISCUSSION
In this paper, we provide evidence that the homeobox gene Six3 is expressed in the periocular mesenchyme and its derivatives, and that altering expression levels of Six3 leads to a range of defects in the anterior segment of the eye. Thus, our results reveal a potential regulatory role of Six3 in the morphogenesis of the vertebrate eye beyond the initial stage of eye primordium determination.
Expression of Six3 in the Periocular Mesenchyme and in Anterior Segment Tissues
Formation of the periocular mesenchyme becomes apparent at stage 18 (E3) in chick, as several genes, including Lmx1b and Pitx2, begin to express in overlapping zones surrounding the optic cup (data not shown; Pressman et al., 2000; Lu et al., 1999). Transcriptional activation of these genes within this region suggests that the optic cup provides inductive signals that positively regulate expression of these regulatory genes. In contrast to the broad periocular expression zone of Pitx2 and Lmx1b, Six3 transcripts are expressed in only a subset of periocular mesenchymal cells. This distinct pattern of Six3 expression in early periocular tissues suggests that regulatory mechanism(s) controlling Six3 expression in this region are distinct from mechanisms controlling the expression of other transcription factors also present in the periocular mesenchyme.
Despite the high homology between Six3 and Six6 as well as their similar expression domains in the anterior embryo (Oliver et al., 1995; Toy et al., 1998; Jean et al., 1999; Bovolenta et al., 1998), the expression patterns of these two genes in the periocular mesenchyme and the anterior segment are different. Our analyses show that Six3 is expressed in the presumptive trabecular meshwork, the cornea, and the iris, whereas Six6 is absent from these tissues. Therefore, although both Six3 and Six6 are likely to function in eye primordium determination and retinal neurogenesis, current data suggest that Six3 rather than Six6 may be required for anterior segment morphogenesis.
Requirements of Conserved Structural Motifs for Six3 Activity
The murine Six3 protein shares 86.4% over all amino acid sequence homology and identical Six and homeodomains with the chick Six3 protein (Kobayashi et al., 1998; Kawakami et al., 1996; Oliver et al., 1995; Bovolenta et al., 1996). The results of viral-mediated misexpression in vivo indicate that wild type murine Six3 protein effectively influences development of the chick anterior segment. The Six3.ΔS mutant shows reduced effectiveness in perturbing eye development, indicating that one or both of the deleted regions are required for the full activity of Six3 protein in causing anterior segment anomalies. The single missense mutation (Six3.V190E), which nearly abolishes the anterior eye phenotypes, is within the Six domain but outside of the deleted regions in Six3.ΔS, demonstrating that the replacement of the Valine190 residue by the charged Glutamic acid severely disrupted the activity of Six3. This is consistent with the fact that this Valine residue is absolutely conserved in all Six family proteins in vertebrate species (Kawakami et al., 2000). A Valine residue is also present in the same position in the Drosophila sine oculis gene product; a similar missense mutation (Valine to Aspartic acid) in So disrupts the activity of the endogenous wild type So protein when misexpressed during neurogenesis (F. Pignoni and S. L. Zipursky, personal communication). Our results are also consistent with a previous study in zebrafish demonstrating that deletion of the N-terminal half of the Six domain reduces the activity of the zebrafish Six3, whereas deletion of the C-terminal half of the Six domain completely abolishes the ability of Six3 to cause head defects in an overexpression assay (Kobayashi et al., 1998).
The molecular and genetic interactions between Six3 and other genes involved in anterior segment development are not well understood. In Drosophila, So protein forms complexes and synergizes with the eyes absent gene product via the Six domain to induce ectopic compound eye formation (Pignoni et al., 1997). The vertebrate Eya genes are expressed in the periocular mesenchyme and anterior segment tissues (Xu et al., 1997; Mishima and Tomarev, 1998), and mutations in human EYA1 lead to congenital cataracts and anterior segment anomalies (Azuma et al., 2000). However, a physical interaction of the vertebrate Six3 protein with Eya proteins has not been confirmed (Ohto et al., 1999). In chick, misexpression of Six3 does not appear to alter the expression pattern of Eya2 in the periocular mesenchyme (X.-M.Z. and X.-J.Y., unpublished data). Recent evidence indicates that Six3 and Six6 (Optx2) may act as transcription repressors during retina and brain formation (Zuber et al., 1999; Kobayashi et al., 2001). Like other homeobox transcription factors (Muhr et al., 2001), the Six domain of the Six3 protein contains two eh-1-related motifs responsible for interaction with the transcription corepressor Groucho (Kobayashi et al., 2001; Zhu et al., 2002). Interestingly, the single point mutation encoded by Six3.V-E is located outside of the two eh-1-related motifs. Thus, the Six domain may mediate molecular interactions with multiple proteins. Since the normal expression domains of Six3 only partially overlap with that of Pax6, Lmx1b, Pitx2, and Eya2 in periocular mesenchyme and in anterior segment, and since Six3 misexpression primarily affected the territories where these genes were expressed rather than their levels of expression, Six3 is unlikely to directly regulate transcription of these genes in the anterior segment.
Cellular Mechanisms Underlying Morphological Defects Caused by Six3 Misexpression
Increasing evidence indicates that anterior segment morphogenesis is sensitive to the expression levels of a number of transcription factors, including Lmx1b, Pitx2, Mf1/ FHKL7/FOXC1, and Pax6 (Pressman et al., 2000; Kidson et al., 1999; Lu et al., 1999; reviewed by Craig and Mackey, 1999; and Alward, 2000; Nishimura et al., 2001; Hill et al., 1991; Schedl et al., 1996). However, the molecular and cellular mechanisms by which these transcription factors regulate periocular mesenchyme patterning and differentiation remain largely unknown. Most of the anterior segment tissues disrupted by Six3 misexpression, including the presumptive trabecular meshwork, the corneal endothelium, the stroma of the ciliary body, and the stroma of the iris, express Six3 endogenously. Therefore, the abnormalities in these ocular tissues caused by Six3 virus infection are likely due to overexpression of Six3, which appears to have two effects. First, elevated Six3 expression attenuates cell proliferation in the mesenchyme. The observed reduction of proliferation in vitro and in vivo at E6 could explain the overall decrease in the mesenchymal cell population in the limbal region, which underlies the reduced territories of expression for Lmx1b and Pitx2. The iris hypoplasia caused by Six3 misexpression is reminiscent of Pax6 and Lmx1b loss-of-function phenotypes (Pressman et al., 2000; Glaser et al., 1995). However, the effects of these two transcription factors on mesenchymal and/or anterior segment tissue cell proliferation remain to be determined. Second, Six3 misexpression may also directly influence the differentiation of periocular mesenchymal derivatives. This is consistent with the altered expression of several corneal extracellular matrix markers and the corresponding corneal phenotypes. A similar effect on anterior segment tissue differentiation has been observed in the case of Lim1b loss-of-function mutation (Pressman et al., 2000). Further understanding of eye anterior segment morphogenesis will clearly require the identification of target genes of key transcription factors and cellular processes that they regulate.
Potential Roles for Six3 in Congenital Anterior Segment Anomalies and Glaucoma
The eye phenotypes caused by Six3 misexpression in chick overlap with anterior segment defects observed in animal models carrying mutations in Lmx1b, Pitx2, Mf1/ FHLK7, and Eya1 genes (Pressman et al., 2000; Lu et al., 1999; Kidson et al., 1999; Xu et al., 1999). Human mutations in these genes are associated with disease syndromes characterized by congenital anterior segment anomalies and early onset glaucoma. Consistent with our results demonstrating that Six3 is expressed in multiple developing ocular tissues and that anterior segment morphogenesis is sensitive to the expression levels of Six3, human patients carrying SIX3 mutations and showing mild holoprosencephaly also display microphthalmia and coloboma (Wallis et al., 1999; Wallis and Muenke, 2000). Therefore, the SIX3 gene is likely to play a role in normal eye development and SIX3 mutations may underlie human anterior segment anomalies, such as aniridia, corneal opacification, and congenital glaucoma.
ACKNOWLEDGMENTS
We thank Drs. Randy L. Johnson, Cliff Tabin, and Peter Gruss for providing cDNAs, and Francesca Pignoni and Larry Zipursky for communicating unpublished data and for discussion. We also thank Kristin Schmidt for her excellent technical support. This work was supported in part by grants from the Research to Prevent Blindness Foundation, the March of Dimes Birth Defect Foundation, the Karl Kirchgessner Foundation, and the NIH (EY12270) (to X.-J.Y.), and by grants from NIH (EY12162 and GM58462) and the American Lebanese Syrian Associated Charities (ALSAC) (to G.O.).
REFERENCES
- Altshuler D, and Cepko C (1992). A temporally regulated, diffusible activity is required for rod photoreceptor development in vitro. Development 114, 947–957. [DOI] [PubMed] [Google Scholar]
- Alward WL (2000). Axenfeld–Rieger syndrome in the age of molecular genetics. Am. J. Ophthalmol 130, 107–115. [DOI] [PubMed] [Google Scholar]
- Azuma N, Hirakiyama A, Inoue T, Asaka A, and Yamada M (2000). Mutations of a human homologue of the Drosophila eyes absent gene (EYA1) detected in patients with congenital cataracts and ocular anterior segment anomalies. Hum. Mol. Genet 9, 363–366. [DOI] [PubMed] [Google Scholar]
- Barrio-Asensio C, Murillo-Gonzalez J, Pena-Meliann A, and Puerta-Fonolla J (1999). Immunocytochemical study on the triple origin of the sphincter iris in the chick embryo. Dev. Genes Evol 209, 620–624. [DOI] [PubMed] [Google Scholar]
- Belecky-Adams T, Cook B, and Adler R (1996). Correlations between terminal mitosis and differentiated fate of retinal precursor cells in vivo and in vitro: Analysis with the “window-labeling” technique. Dev. Biol 178, 304–315. [DOI] [PubMed] [Google Scholar]
- Bernier G, Panitz F, Zhou X, Hollemann T, Gruss P, and Pieler T (2000). Expanded retina territory by midbrain transformation upon overexpression of Six6 (Optx2) in Xenopus embryos. Mech. Dev 93, 59–69. [DOI] [PubMed] [Google Scholar]
- Bovolenta P, Mallamaci A, and Boncinelli E (1996). Cloning and characterisation of two chick homeobox genes, members of the six/sine oculis family, expressed during eye development. Int. J. Dev. Biol (Suppl. 1), 73S–74S. [PubMed] [Google Scholar]
- Bovolenta P, Mallamaci A, Puelles L, and Boncinelli E (1998). Expression pattern of cSix3, a member of the Six/sine oculis family of transcription factors. Mech. Dev 70, 201–203. [DOI] [PubMed] [Google Scholar]
- Coulombre AJ, and Coulombre JL (1964). Lens development. The role of the lens in eye growth. J. Exp. Zool 156, 39–48. [DOI] [PubMed] [Google Scholar]
- Cheyette BN, Green PJ, Martin K, Garren H, Hartenstein V, and Zipursky SL (1994). The Drosophila sine oculis locus encodes a homeodomain-containing protein required for the development of the entire visual system. Neuron 12, 977–996. [DOI] [PubMed] [Google Scholar]
- Craig JE, and Mackey DA (1999). Glaucoma genetics: Where are we? Where will we go? Curr. Opin. Ophthalmol 10, 126–134. [DOI] [PubMed] [Google Scholar]
- Dreyer SD, Zhou G, Baldini A, Winterpacht A, Zabel B, Cole W, Johnson RL, and Lee B (1998). Mutations in LMX1B cause abnormal skeletal patterning and renal dysplasia in nail patella syndrome. Nat. Genet 19, 47–50. [DOI] [PubMed] [Google Scholar]
- Dunlevy JR, Neame PJ, Vergnes JP, and Hassell JR (1998). Identification of the N-linked oligosaccharide sites in chick corneal lumican and keratocan that receive keratan sulfate. J. Biol. Chem 273, 9615–9621. [DOI] [PubMed] [Google Scholar]
- Ericson J, Rashbass P, Schedl A, Brenner-Morton S, Kawakami A, van Heyningen V, Jessell TM, and Briscoe J (1997). Pax6 controls progenitor cell identity and neuronal fate in response to graded Shh signaling. Cell 90, 169–180. [DOI] [PubMed] [Google Scholar]
- Fitch J, Fini ME, Beebe DC, and Linsenmayer TF (1998). Collagen type IX and developmentally regulated swelling of the avian primary corneal stroma. Dev. Dyn 212, 27–37. [DOI] [PubMed] [Google Scholar]
- Fitch JM, Mentzer A, Mayne R, and Linsenmayer TF (1988). Acquisition of type IX collagen by the developing avian primary corneal stroma and vitreous. Dev. Biol 128, 396–405. [DOI] [PubMed] [Google Scholar]
- Funderburgh JL, Caterson B, and Conrad GW (1986). Keratan sulfate proteoglycan during embryonic development of the chicken cornea. Dev. Biol 116, 267–277. [DOI] [PubMed] [Google Scholar]
- Funderburgh JL, Stenzel-Johnson PR, and Chandler JW (1982). Monoclonal antibodies to rabbit corneal keratan sulfate proteoglycan. Curr. Eye Res 2, 769–776. [DOI] [PubMed] [Google Scholar]
- Gage PJ, and Camper SA (1997). Pituitary homeobox 2, a novel member of the bicoid-related family of homeobox genes, is a potential regulator of anterior structure formation. Hum. Mol. Genet 6, 457–464. [DOI] [PubMed] [Google Scholar]
- Gallardo ME, Lopez-Rios J, Fernaud-Espinosa I, Granadino B, Sanz R, Ramos C, Ayuso C, Seller MJ, Brunner HG, Bovolenta P, and Rodriguez de Cordoba S (1999). Genomic cloning and characterization of the human homeobox gene SIX6 reveals a cluster of SIX genes in chromosome 14 and associates SIX6 hemizygosity with bilateral anophthalmia and pituitary anomalies. Genomics 61, 82–91. [DOI] [PubMed] [Google Scholar]
- Gavrieli Y, Sherman Y, and Ben-Sasson SA (1992). Identification of programmed cell death in situ via specific labeling of nuclear DNA fragmentation. J. Cell Biol 119, 493–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genis-Galvez JM (1966). Role of the lens in the morphogenesis of the iris and cornea. Nature 210, 209–210. [DOI] [PubMed] [Google Scholar]
- Genis-Galvez JM, Santos-Gutierrez L, and Rios-Gonzalez A (1967). Causal factors in corneal development: an experimental analysis in the chick embryo. Exp. Eye Res 6, 48–56. [DOI] [PubMed] [Google Scholar]
- Glaser T, Walton DS, Cai J, Epstein JA, Jepeal L, and Maas RL (1995). PAX6 gene mutations in aniridia In “Molecular Genetics of Ocular Disease” (Wiggs JL, Ed.) pp. 51–82. Wiley-Liss, New York. [Google Scholar]
- Grainger RM (1992). Embryonic lens induction: Shedding light on vertebrate tissue determination. Trends Genet. 8, 349–355. [DOI] [PubMed] [Google Scholar]
- Graw J (1996). Genetic aspects of embryonic eye development in vertebrates. Dev. Genet 18, 181–197. [DOI] [PubMed] [Google Scholar]
- Grindley JC, Davidson DR, and Hill RE (1995). The role of Pax-6 in eye and nasal development. Development 121, 1433–1442. [DOI] [PubMed] [Google Scholar]
- Hamburger V, and Hamilton HL (1992). A series of normal stages in the development of the chick embryo. 1951. Dev. Dyn 195, 231–272. [DOI] [PubMed] [Google Scholar]
- Hill RE, Favor J, Hogan BL, Ton CC, Saunders GF, Hanson IM, Prosser J, Jordan T, Hastie ND, and van Heyningen V (1991). Mouse small eye results from mutations in a paired-like homeobox-containing gene. Nature 354, 522–525. [DOI] [PubMed] [Google Scholar]
- Hong HK, Lass JH, and Chakravarti A (1999). Pleiotropic skeletal and ocular phenotypes of the mouse mutation congenital hydrocephalus (ch/Mf1) arise from a winged helix/forkhead transcriptionfactor gene. Hum. Mol. Genet 8, 625–637. [DOI] [PubMed] [Google Scholar]
- Hughes SH, Greenhouse JJ, Petropoulos CJ, and Sutrave P (1987). Adaptor plasmids simplify the insertion of foreign DNA into helper-independent retroviral vectors. J. Virol 61, 3004–3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irwin MH, Silvers SH, and Mayne R (1985). Monoclonal antibody against chicken type IX collagen: Preparation, characterization, and recognition of the intact form of type IX collagen secreted by chondrocytes. J. Cell Biol 101, 814–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jean D, Bernier G, and Gruss P (1999). Six6 (Optx2) is a novel murine Six3-related homeobox gene that demarcates the presumptive pituitary/hypothalamic axis and the ventral optic stalk. Mech. Dev 84, 31–40. [DOI] [PubMed] [Google Scholar]
- Johnston MC, Noden DM, Hazelton RD, Coulombre JL, and Coulombre AJ (1979). Origins of avian ocular and periocular tissues. Exp. Eye Res 29, 27–43. [DOI] [PubMed] [Google Scholar]
- Kania A, Johnson RL, and Jessell TM (2000). Coordinate roles for LIM homeobox genes in directing the dorsoventral trajectory of motor axons in the vertebrate limb. Cell 102, 161–173. [DOI] [PubMed] [Google Scholar]
- Kawakami K, Ohto H, Takizawa T, and Saito T (1996). Identification and expression of six family genes in mouse retina. FEBS Lett. 393, 259–263. [DOI] [PubMed] [Google Scholar]
- Kawakami K, Sato S, Ozaki H, and Ikeda K (2000). Six family genes: Structure and function as transcription factors and their roles in development. Bioessays 22, 616–626. [DOI] [PubMed] [Google Scholar]
- Kidson SH, Kume T, Deng K, Winfrey V, and Hogan BL (1999). The forkhead/winged-helix gene, Mf1, is necessary for the normal development of the cornea and formation of the anterior chamber in the mouse eye. Dev. Biol 211, 306–322. [DOI] [PubMed] [Google Scholar]
- Kobayashi M, Nishikawa K, Suzuki T, and Yamamoto M (2001). The homeobox protein Six3 interacts with the Groucho corepressor and acts as a transcriptional repressor in eye and forebrain formation. Dev. Biol 232, 315–326. [DOI] [PubMed] [Google Scholar]
- Kobayashi M, Toyama R, Takeda H, Dawid IB, and Kawakami K (1998). Overexpression of the forebrain-specific homeobox gene six3 induces rostral forebrain enlargement in zebrafish. Development 125, 2973–2982. [DOI] [PubMed] [Google Scholar]
- Kulak SC, Kozlowski K, Semina EV, Pearce WG, and Walter MA (1998). Mutation in the RIEG1 gene in patients with iridogoniodysgenesis syndrome. Hum. Mol. Genet 7, 1113–1117. [DOI] [PubMed] [Google Scholar]
- Lagutin O, Zhu CC, Furuta Y, Rowitch DH, McMahon AP, and Oliver G (2001). Six3 promotes the formation of ectopic optic vesicle-like structures in mouse embryos. Dev. Dyn 221, 342–349. [DOI] [PubMed] [Google Scholar]
- Linsenmayer TF, Fitch JM, Gordon MK, Cai CX, Igoe F, Marchant JK, and Birk DE (1998). Development and roles of collagenous matrices in the embryonic avian cornea. Prog. Retin. Eye Res 17, 231–265. [DOI] [PubMed] [Google Scholar]
- Liu CY, Shiraishi A, Kao CW, Converse RL, Funderburgh JL, Corpuz LM, Conrad GW, and Kao WW (1998). The cloning of mouse keratocan cDNA and genomic DNA and the characterization of its expression during eye development. J. Biol. Chem 273, 22584–22588. [DOI] [PubMed] [Google Scholar]
- Logan M, Pagan-Westphal SM, Smith DM, Paganessi L, and Tabin CJ (1998). The transcription factor Pitx2 mediates situs-specific morphogenesis in response to left–right asymmetric signals. Cell 94, 307–317. [DOI] [PubMed] [Google Scholar]
- Loosli F, Winkler S, and Wittbrodt J (1999). Six3 overexpression initiates the formation of ectopic retina. Genes Dev. 13, 649–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu MF, Pressman C, Dyer R, Johnson RL, and Martin JF (1999). Function of Rieger syndrome gene in left–right asymmetry and craniofacial development. Nature 401, 276–278. [DOI] [PubMed] [Google Scholar]
- McIntosh I, Dreyer SD, Clough MV, Dunston JA, Eyaid W, Roig CM, Montgomery T, Ala-Mello S, Kaitila I, Winterpacht A, Zabel B, Frydman M, Cole WG, Franco-mano CA, and Lee B (1998). Mutation analysis of LMX1B gene in nail-patella syndrome patients. Am. J. Hum. Genet 63, 1651–1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mears AJ, Jordan T, Mirzayans F, Dubois S, Kume T, Parlee M, Ritch R, Koop B, Kuo WL, Collins C, Marshall J, Gould DB, Pearce W, Carlsson P, Enerback S, Morissette J, Bhattacharya S, Hogan B, Raymond V, and Walter MA (1998). Mutations of the forkhead/winged-helix gene, FKHL7, in patients with Axenfeld–Rieger anomaly. Am. J. Hum. Genet 63, 1316–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishima N, and Tomarev S (1998). Chicken Eyes absent 2 gene: Isolation and expression pattern during development. Int. J. Dev. Biol 42, 1109–1115. [PubMed] [Google Scholar]
- Morgan BA, and Fekete DM (1996). Manipulating gene expression with replication-competent retroviruses. Methods Cell Biol. 51, 185–218. [DOI] [PubMed] [Google Scholar]
- Muhr J, Andersson E, Persson M, Jessell TM, and Ericson J (2001). Groucho-mediated transcriptional repression establishes progenitor cell pattern and neuronal fate in the ventral neural tube. Cell 104, 861–873. [DOI] [PubMed] [Google Scholar]
- Nishimura DY, Searby CC, Alward WL, Walton D, Craig JE, Mackey DA, Kawase K, Kanis AB, Patil SR, Stone EM, and Sheffield VC (2001). A spectrum of FOXC1 mutations suggests gene dosage as a mechanism for developmental defects of the anterior chamber of the eye. Am. J. Hum. Genet 68, 364–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura DY, Swiderski RE, Alward WL, Searby CC, Patil SR, Bennet SR, Kanis AB, Gastier JM, Stone EM, and Sheffield VC (1998). The forkhead transcription factor gene FKHL7 is responsible for glaucoma phenotypes which map to 6p25. Nat. Genet 19, 140–147. [DOI] [PubMed] [Google Scholar]
- Ohto H, Kamada S, Tago K, Tominaga SI, Ozaki H, Sato S, and Kawakami K (1999). Cooperation of six and eya in activation of their target genes through nuclear translocation of Eya. Mol. Cell. Biol 19, 6815–6824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver G, Loosli F, Koster R, Wittbrodt J, and Gruss P (1996). Ectopic lens induction in fish in response to the murine homeobox gene Six3. Mech. Dev 60, 233–239. [DOI] [PubMed] [Google Scholar]
- Oliver G, Mailhos A, Wehr R, Copeland NG, Jenkins NA, and Gruss P (1995). Six3, a murine homologue of the sine oculis gene, demarcates the most anterior border of the developing neural plate and is expressed during eye development. Development 121, 4045–4055. [DOI] [PubMed] [Google Scholar]
- Pasquier L, Dubourg C, Blayau M, Lazaro L, Le Marec B, David V, and Odent S (2000). A new mutation in the six-domain of SIX3 gene causes holoprosencephaly. Eur. J. Hum. Genet 8, 797–800. [DOI] [PubMed] [Google Scholar]
- Pellegata NS, Dieguez-Lucena JL, Joensuu T, Lau S, Montgomery KT, Krahe R, Kivela T, Kucherlapati R, Forsius H, and de la Chapelle A (2000). Mutations in KERA, encoding keratocan, cause cornea plana. Nat. Genet 25, 91–95. [DOI] [PubMed] [Google Scholar]
- Pignoni F, Hu B, Zavitz KH, Xiao J, Garrity PA, and Zipursky SL (1997). The eye-specification proteins So and Eya form a complex and regulate multiple steps in Drosophila eye development. Cell 91, 881–891. [DOI] [PubMed] [Google Scholar]
- Pressman CL, Chen H, and Johnson RL (2000). LMX1B, a LIM homeodomain class transcription factor, is necessary for normal development of multiple tissues in the anterior segment of the murine eye. Genesis 26, 15–25. [PubMed] [Google Scholar]
- Rada JA, Cornuet PK, and Hassell JR (1993). Regulation of corneal collagen fibrillogenesis in vitro by corneal proteoglycan (lumican and decorin) core proteins. Exp. Eye Res 56, 635–648. [DOI] [PubMed] [Google Scholar]
- Reneker LW, Silversides DW, Xu L, and Overbeek PA (2000). Formation of corneal endothelium is essential for anterior segment development: A transgenic mouse model of anterior segment dysgenesis. Development 127, 533–542. [DOI] [PubMed] [Google Scholar]
- Riddle RD, Ensini M, Nelson C, Tsuchida T, Jessell TM, and Tabin C (1995). Induction of the LIM homeobox gene Lmx1 by WNT7a establishes dorsoventral pattern in the vertebrate limb. Cell 83, 631–640. [DOI] [PubMed] [Google Scholar]
- Riddle RD, Johnson RL, Laufer E, and Tabin C (1993). Sonic hedgehog mediates the polarizing activity of the ZPA. Cell 75, 1401–1416. [DOI] [PubMed] [Google Scholar]
- Rong PM, Ziller C, Pena-Melian A, and Le Douarin NM (1987). A monoclonal antibody specific for avian early myogenic cells and differentiated muscle. Dev. Biol 122, 338–353. [DOI] [PubMed] [Google Scholar]
- Saleem RA, Banerjee-Basu S, Berry FB, Baxevanis AD, and Walter MA (2001). Analyses of the effects that disease-causing missense mutations have on the structure and function of the winged-helix protein FOXC1. Am. J. Hum. Genet 68, 627–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar PS, Appukuttan B, Han J, Ito Y, Ai C, Tsai W, Chai Y, Stout JT, and Reddy S (2000). Heterozygous loss of Six5 in mice is sufficient to cause ocular cataracts. Nat. Genet 25, 110–114. [DOI] [PubMed] [Google Scholar]
- Schedl A, Ross A, Lee M, Engelkamp D, Rashbass P, van Heyningen V, and Hastie ND (1996). Influence of PAX6 gene dosage on development: Overexpression causes severe eye abnormalities. Cell 86, 71–82. [DOI] [PubMed] [Google Scholar]
- Semina EV, Reiter R, Leysens NJ, Alward WL, Small KW, Datson NA, Siegel-Bartelt J, Bierke-Nelson D, Bitoun P, Zabel BU, Carey JC, and Murray JC (1996). Cloning and characterization of a novel bicoid-related homeobox transcription factor gene, RIEG, involved in Rieger syndrome. Nat. Genet 14, 392–399. [DOI] [PubMed] [Google Scholar]
- Shields MB (1998). “Textbook of Glaucoma.” Williams &Wilkins, Baltimore. [Google Scholar]
- Smith RS, Zabaleta A, Kume T, Savinova OV, Kidson SH, Martin JE, Nishimura DY, Alward WL, Hogan BL, and John SW (2000). Haploinsufficiency of the transcription factors FOXC1 and FOXC2 results in aberrant ocular development. Hum. Mol. Genet 9, 1021–1032. [DOI] [PubMed] [Google Scholar]
- Stoker AW, and Bissell MJ (1987). Quantitative immunocytochemical assay for infectious avian retroviruses. J. Gen. Virol 68, 2481–2485. [DOI] [PubMed] [Google Scholar]
- Toy J, and Sundin OH (1999). Expression of the optx2 homeobox gene during mouse development. Mech. Dev 83, 183–186. [DOI] [PubMed] [Google Scholar]
- Toy J, Yang JM, Leppert GS, and Sundin OH (1998). The optx2 homeobox gene is expressed in early precursors of the eye and activates retina-specific genes. Proc. Natl. Acad. Sci. USA 95, 10643–10648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trainor PA, and Tam PP (1995). Cranial paraxial mesoderm and neural crest cells of the mouse embryo: Co-distribution in the craniofacial mesenchyme but distinct segregation in branchial arches. Development 121, 2569–2582. [DOI] [PubMed] [Google Scholar]
- Vollrath D, Jaramillo-Babb VL, Clough MV, McIntosh I, Scott KM, Lichter PR, and Richards JE (1998). Loss-of-function mutations in the LIM-homeodomain gene, LMX1B, in nail-patella syndrome. Hum. Mol. Genet 7, 1091–1098. [DOI] [PubMed] [Google Scholar]
- Wallis DE, Roessler E, Hehr U, Nanni L, Wiltshire T, Richieri-Costa A, Gillessen-Kaesbach G, Zackai EH, Rommens J, and Muenke M (1999). Mutations in the homeodomain of the human SIX3 gene cause holoprosencephaly. Nat. Genet 22, 196–198. [DOI] [PubMed] [Google Scholar]
- Wallis D, and Muenke M (2000). Mutations in holoprosencephaly. Hum. Mutat 16, 99–108. [DOI] [PubMed] [Google Scholar]
- Xu PX, Adams J, Peters H, Brown MC, Heaney S, and Maas R (1999). Eyal-deficient mice lack ears and kidneys and show abnormal apoptosis of organ primordia. Nat. Genet 23, 113–117. [DOI] [PubMed] [Google Scholar]
- Xu PX, Woo I, Her H, Beier DR, and Maas RL (1997). Mouse Eya homologues of the Drosophila eyes absent gene require Pax6 for expression in lens and nasal placode. Development 124, 219–231. [DOI] [PubMed] [Google Scholar]
- Yang XJ, and Cepko CL (1996). Flk-1, a receptor for vascular endothelial growth factor (VEGF), is expressed by retinal progenitor cells. J. Neurosci 16, 6089–6099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XM, and Yang XJ (2001). Temporal and spatial effects of Sonic hedgehog signaling in chick eye morphogenesis. Dev. Biol 233, 271–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu CC, Dyer MA, Uchikawa M, Kondoh H, Lagutin OV, and Oliver G (2002). Six3-mediated auto repression and eye development requires its interaction with members of the Groucho-related family of co-repressors. Development 129, 2835–2849. [DOI] [PubMed] [Google Scholar]
- Zuber ME, Perron M, Philpott A, Bang A, and Harris WA (1999). Giant eyes in Xenopus laevis by overexpression of XOptx2. Cell 98, 341–352. [DOI] [PubMed] [Google Scholar]


