Abstract
Background
The grain number per panicle (GNP), which is one of three grain yield components, is an important trait for the genetic improvement of rice. Although the NAL1 and GNP1 genes regulating the rice GNP and grain yield have been cloned, their allelic diversity, functional differences in rice germplasms, and effects of their combination on GNP and grain yield remain unclear.
Results
Based on DNA sequences of these two genes in 198 cultivated rice (Oryza sativa) and 8–10 wild rice (Oryza rufipogon) germplasms, 16 and 14 haplotypes were identified for NAL1 and GNP1, respectively. The NAL1 gene had the strongest effects on GNP in indica (xian) and japonica (geng) subpopulations. In contrast, GNP1 had no significant effects in the geng subpopulation and was rare in the xian background, in which the superior GNP1 allele (GNP1–6) was detected in only 4.0% of the 198 germplasms. Compared with the transgenic lines with GNP1 or NAL1, the transgenic lines with both genes had a higher GNP (15.5%–25.4% and 11.6%–15.9% higher, respectively) and grain yield (5.7%–9.0% and 8.3%–12.3% higher, respectively) across 3 years. The two genes combined in the introgression lines in Lemont background resulted in especially favorable effects on the GNP.
Conclusions
Our results indicated that the GNP1 and NAL1 exhibited obvious differentiation and their combinations can significantly increase the grain yield in geng rice cultivars. These observations provide insights into the molecular basis of the GNP and may be useful for rice breeding of high yield potential by pyramiding GNP1 and NAL1.
Keywords: NAL1, GNP1, Pyramiding, Nucleotide diversity, Phenotypic variation
Background
Rice (Oryza sativa L.) is one of the most important staple food crops worldwide. Therefore, research aimed at increasing rice yield is critical for ensuring food production and satisfying the demands of a rapidly growing global population. Grain number per panicle (GNP), seed setting rate, effective panicle number per plant (EPN) and grain weight are the most important components of rice grain yield. Moreover, the GNP is also an important agronomic characteristic related to the ideal plant architecture (Virk et al. 2004). Increasing the rice grain yield by selective breeding involving grain yield components and the ideal plant architecture has been the key focus of rice breeding programs (Zhang 2007).
Map-based cloning methods have recently been applied to isolate several quantitative trait loci (QTLs) affecting yield-related traits, including Gn1a (Ashikari et al. 2005), DEP1 (Huang et al. 2009), OsSPL14 (Jiao et al. 2010), NAL1 (SPIKE, GPS, LSCHL4, and SS1) (Fujita et al. 2013; Takai et al. 2013; Zhang et al. 2014; Xu et al. 2015), and GNP1 (Wu et al. 2016). These genes control plant architecture and panicle type, ultimately influencing rice grain yield mainly through pleiotropic effects. Narrow leaf 1 (NAL1), which affects leaf morphogenesis by regulating polar auxin transport, was first cloned from a loss-of-function narrow-leaf rice mutant by Qi et al. (2008). Previous studies revealed that NAL1 is associated with pleiotropic effects that regulate the development of multiple traits related to the source (e.g., leaf width, leaf chlorophyll content, and photosynthetic efficiency), the sink (e.g., GNP), and the EPN in diverse genetic backgrounds (Fujita et al. 2013; Takai et al. 2013; Zhang et al., 2014; Xu et al. 2015; Yano et al. 2016). Another major QTL, GNP1, encodes the rice gibberellin biosynthesis gene GA20ox1. Increases in the GNP and rice grain yield involve enhanced cytokinin activity in panicle meristems. Additionally, rice plant height (PH) is regulated by OsGA20ox1 (Oikawa et al. 2004). In our previous studies, we found that introgression of the superior GNP1 allele from Teqing into Lemont lead to significantly increase in flag leaf length and area (Unpublished). So, GNP1 also has pleiotropic effects on the development of multiple traits related to the source (e.g. leaf size) and the sink (e.g., GNP). The superior NAL1 allele from the low-yielding Lemont variety can further increase 3.2%–3.8% grain yield of high-yielding Teqing variety in different environments (Xu et al. 2015), while the superior GNP1 allele from the high-yielding Teqing variety can increase 5.7%–9.6% grain yield of low-yielding Lemont variety in different environments (Wu et al. 2016). These results suggested that superior NAL1 and GNP1 alleles can potentially be used in high yield rice breeding. However, the allelic diversity and functional differences of GNP1 and NAL1 in rice germplasms remain unclear. Moreover, their relative importance and pyramiding effect on grain yield have not been characterized.
In this study, we re-sequenced GNP1 and NAL1 from 198 cultivated rice (Oryza sativa) and 8–10 wild rice (Oryza rufipogon) germplasms. The allelic frequencies and the associated differences in the GNP were compared between the xian and geng subpopulations. Furthermore, the potential utility of combining NAL1 and GNP1 to increase the rice grain yield was investigated by comparing the lines with either NAL1 or GNP1 with the lines carrying both NAL1 and GNP1. The results presented herein may help to clarify the comprehensive role of these two genes alone and in combination on the GNP and the potential rice grain yield. This study may also be useful for developing an efficient way to increase the GNP, thereby increasing the overall rice grain yield.
Results
Phenotypic and Genetic Variations in the Rice Germplasms
The 198 cultivated rice germplasms varied considerably regarding the GNP, flag leaf width (FLW), and PH (Additional file 1: Figure S1). The scree plot indicated that two principal components should be retained for the principal component analysis (Additional file 2: Figure S2a). The principal component score plot revealed the distribution of each accession in the rice diversity panel (Additional file 2: Figure S2b). Additionally, the highest log-likelihood scores of the population structure were obtained when the number of populations was set at 2 (K = 2; Additional file 2: Figure S2c), indicating the existence of two distinct subpopulations in the current panel (Additional file 2: Figure S2d, Additional file 3: Table S1). These findings along with the available accession information resulted in the identification of 146 xian and 43 geng varieties as well as three Aus and six admixed varieties. There were significant differences (P < 0.05) in the GNP and FLW, but not in the PH, between the xian and geng subpopulations over 3 years (Additional file 1: Figure S1).
Nucleotide Variations in GNP1 and NAL1
The 198 O. sativa germplasms were examined regarding the diversity in their GNP1 and NAL1 sequences. The 2.48 single nucleotide polymorphisms (SNPs) per kilobase (π = 2.48 × 10− 3) of the GNP1 locus was slightly fewer than 2.64 SNPs per kilobase (π = 2.64 × 10− 3) in O. rufipogon (Additional file 4: Table S2). A selection signal was not detected in the neutrality tests of the whole, xian, and geng populations (Additional file 4: Table S2). A sliding-window analysis indicated the π value was highest in the promoter region (Additional file 5: Figure S3a).
The nucleotide diversity at the NAL1 locus in the xian and geng germplasms and in O. rufipogon was 1.14 × 10− 3, 0.88 × 10− 3, and 1.0 × 10− 3, respectively (Additional file 4: Table S2). The π value in O. sativa (π = 1.65 × 10− 3) was nearly 2-fold higher than that in O. rufipogon. A neutrality test revealed a lack of selection signals in both subpopulations (Additional file 4: Table S2). The results of a sliding-window analysis revealed that in the geng subpopulation, the π value was highest in the third exon and in the 3′ untranslated region (UTR) (Additional file 5: Figure S3b).
Analysis of the GNP1 Haplotype
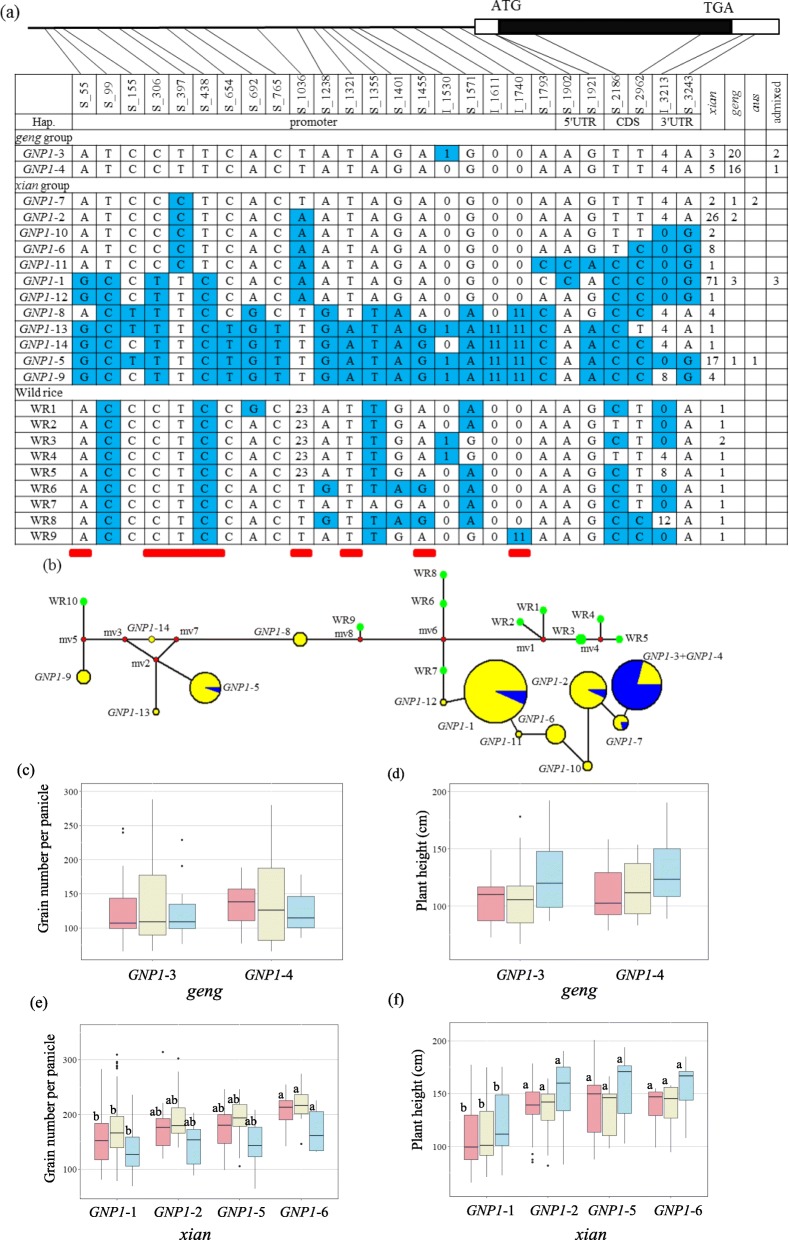
The complete GNP1 genomic sequence (3.7 kb) in the 198 analyzed accessions was re-sequenced. A total of 22 SNPs and four insertions/deletions (InDels) with a minimum allele frequency > 5% were considered for a haplotype analysis. Of these polymorphisms, 19 SNPs and three InDels were located in the promoter and 5′ UTR, including two 11-bp InDels in the promoter region. Additionally, one synonymous SNP and one nonsynonymous SNP leading to the replacement of an alanine by a valine were present in the coding region, with the remaining SNP and InDel located in the 3′ UTR (Fig. 1a). The amino acid variations detected in this study were the same as those identified in one of our previous studies (Wu et al. 2016).
Fig. 1.
GNP1 haplotype analysis. a The gene structure of GNP1. The coding region is indicated as black box, 3′ UTR and 5′ UTR are indicated as white boxes, and single nucleotide polymorphism (SNP) positions are connected to the haplotype table by lines (SNP frequency > 5%). Six potential intervals of promoters, marked with red stick, are found in the promoter region by promoter prediction software. Fourteen haplotypes, classified by SNP and insertion/deletion (InDel) variations, are detected in the collection (indicated as either blue or white). Numbers show the length of the InDel in bp. The number and subpopulation identity of varieties carrying each haplotype are indicated in columns to the right. The ten wild rice (O.rufipogon) varieties are indicated by WR1–9. b The phylogenetic network of GNP1. The phylogenetic network of haplotypes in Oryza sativa and O. rufipogon are constructed for GNP1. Each circle represents an allele. The size of the circles indicates the frequency of each allele. The green, yellow and blue circles indicate the allelic distribution in the wild rice, xian and geng species, respectively. The red median vectors (mv) represent a hypothesized sequence that is required to connect the existing sequences within the network with maximum parsimony. The solid lines represent one mutation step that interconnects adjacent alleles. c–d Grain number per panicle (c), plant height (d) are compared between the GNP1–3 and GNP1–4 haplotypes in geng rice (Oryza sativa). e–f Grain number per panicle (e), plant height (f) are compared among the GNP1–1,GNP1–2,GNP1–5 and GNP1–6 haplotypes in xian rice (Oryza sativa). Letters on histograms (a, and b) are ranked by Duncan’s test at P < 0.05. Pink, light yellow and light blue colors indicate in 2014, 2015 and 2016, respectively
Fourteen haplotypes (GNP1–1 to GNP1–14) were constructed based on the SNPs and InDels. A phylogenetic network for GNP1 alleles was constructed based on the 22 SNPs and four InDels in O. sativa and O. rufipogon (Fig. 1b). Accordingly, we determined that the most prevalent haplotypes were GNP1–1, GNP1–2, GNP1–3, GNP1–4, and GNP1–5, which were detected in 77, 28, 25, 22, and 19 varieties, respectively (Fig. 1a). The next most prevalent haplotype was GNP1–6, which occurred in eight varieties. The other haplotypes were relatively rare (i.e., only in one to five varieties). Haplotypes GNP1–3 (represented by Lemont) and GNP1–4 were mainly present in accessions belonging to the geng subpopulation (i.e., 76.6%), whereas the other four haplotypes (GNP1–1, GNP1–2, GNP1–5, and GNP1–6) were predominantly detected in xian varieties (i.e., 92.4%) (Fig. 1b). Moreover, there was relatively little diversity in the GNP1 sequence within the geng subpopulation, with only two haplotypes (GNP1–3 and GNP1–4) defined by one InDel at position I_1530 in the promoter region (Fig. 1a). In contrast, several GNP1 haplotypes were identified in the xian subpopulation, in which haplotype GNP1–9 had the most variability in the SNP and InDels (92.3% polymorphic sites) as well as a large genetic distance from GNP1–2 (Fig. 1b). Additionally, the GNP1 allele in the GNP1–9 haplotype (present in four xian accessions) is identical to that in Teqing with the superior GNP1 haplotype described by Wu et al. (2016). The differences in the GNP and PH were examined among varieties carrying haplotypes GNP1–1 to GNP1–6 because of the high frequency of these haplotypes in the rice germplasms. No differences in the GNP and PH were observed between the germplasms with the two major geng haplotypes (GNP1–3 and GNP1–4) in the geng subpopulation (Fig. 1c, d). However, in the xian subpopulation, the germplasms with the GNP1–6 haplotype had a higher GNP than those with GNP1–1. Additionally, the PH was lower for the germplasms with GNP1–1 than for the germplasms with the other three major xian haplotypes (GNP1–2, GNP1–5, and GNP1–6) (Fig. 1e, f). Thus, the germplasms carrying the GNP1–6 haplotype had a higher GNP and PH than those with GNP1–1.
In 198 panels, two protein types (GNP1-P1 and GNP1-P2) were identified based on one nonsynonymous SNP (Additional file 6: Figure S4a). There was no difference in GNP between the two protein types in xian subpopulation in 3 years except in 2014 (Additional file 6: Figure S4 b). Accessions carrying GNP1-P2 exhibited significantly higher PH than those carrying GNP1-P1 in xian subpopulation (Additional file 6: Figure S4 c).
To determine whether the SNPs or InDels in the promoter affected the GNP1 locus at the transcriptional level, we compared the relative expressions of GNP1–6 and GNP1–1. In young panicles, GNP1–6 was more highly expressed than GNP1–1 (Additional file 7: Figure S5a).
Analysis of the NAL1 Haplotype
We detected 19 SNPs and one InDel in the 4.4-kb NAL1 genomic region. Four SNPs were located in the promoter. One synonymous SNP was detected in the first exon, whereas one nonsynonymous SNP in the third exon resulted in the replacement of an arginine by a histidine. Two nonsynonymous SNPs were located in the fifth exon. Of the remaining 11 SNPs, seven and four were located in the intron and the 3′ UTR, respectively. A 5895-bp retrotransposon insertion was identified in the junction between the first intron and the second exon of NAL1 (Fig. 2a). However, the 10-amino acid deletion encoded in the fourth exon that was previously reported in the mutant Taichung 65 (Takai et al. 2013) was not detected in any of the 198 cultivated and eight wild rice germplasms, implying that this amino acid deletion is a rare mutation.
Fig. 2.
NAL1 haplotype analysis. a The gene structure of NAL1. The five exons are indicated as black boxes, 3′ UTR and 5′ UTR are indicated as white boxes, and single nucleotide polymorphism (SNP) positions are connected to the haplotype table by lines (SNP frequency > 5%). Sixteen haplotypes, classified by SNP and insertion/deletion (InDel) variations, are detected in the collection (indicated as either blue or white). Numbers show the length of the InDel in bp. The number and subpopulation identity of varieties carrying each haplotype are indicated in columns to the right. The eight wild rice varieties of O.rufipogon are indicated by WR1–4. b The phylogenetic network of NAL1. The phylogenetic network of haplotypes in Oryza sativa and O. rufipogon are constructed for NAL1. c–d Grain number per panicle (c), flag leaf width (d) are compared among the NAL1–2, NAL1–4, NAL1–5 and NAL1–6 haplotypes in geng rice (Oryza sativa). e–f Grain number per panicle (e), flag leaf width (f) are compared among the NAL1–1, NAL1–2 and NAL1–3 haplotypes in xian rice (Oryza sativa). Letters on histograms (a, and b) are ranked by Duncan’s test at P < 0.05.Light pink, light yellow and light blue colors indicate in 2014, 2015 and 2016, respectively
Sixteen haplotypes (NAL1–1 to NAL1–16) were identified (Fig. 2a). The NAL1 phylogenetic network revealed the most prevalent haplotypes were NAL1–1 to NAL1–5, which were present in 100, 20, 19, 18, and 13 germplasms, respectively (Fig. 2b). The next most prevalent haplotype was NAL1–6, which was detected in seven germplasms. The other haplotypes were present in fewer than five varieties each. The NAL1–1 and NAL1–3 haplotypes were prevalent in the xian subpopulation (i.e., 97% and 94.7%, respectively), whereas the other three haplotypes (NAL1–4, NAL1–6, and NAL1–5) were mainly present in the geng germplasms (i.e., 77.8%, 71.4%, and 69.2%, respectively) (Fig. 2b). One admixed pattern was identified for NAL1–2, which was included in 10 (50%) geng, 8 (40%) xian, and 2 (10%) Aus accessions.
In the geng subpopulation, the germplasms with the NAL1–2 and NAL1–4 haplotypes had a higher GNP and a greater FLW than the germplasms with the NAL1–6 haplotype (Fig. 2c, d). In the xian subpopulation, the germplasms containing the NAL1–1 and NAL1–3 haplotypes had a considerably higher GNP and greater FLW than the germplasms with the NAL1–2 haplotype (Fig. 2e, f).
In 198 panels, three protein types (NAL1-P1, NAL1-P2 and NAL1-P3) were observed based on three nonsynonymous SNPs (Additional file 8: Figure S6 a). Accessions carrying NAL1-P1 had significantly more GNP and larger FLW than those carrying NAL1-P2 in xian subpopulation (Additional file 8: Figure S6 b–c). Accessions carrying NAL1-P3 possessed significantly more GNP than those with NAL1-P2 in geng subpopulation in 2014 and 2016 (Additional file 8: Figure S6 d). And varieties with NAL1-P3 showed significantly enlarged FLW compared with those with NAL1-P2 in geng subpopulation (Additional file 8: Figure S6 e).
The real-time PCR results demonstrated that NAL1–2 was more highly expressed in young panicle than NAL1–4 or NAL1–6 in the geng germplasms (Additional file 7: Figure S5b). However, there were no expression-level differences in the rice young panicle among the NAL1–1, NAL1–2, and NAL1–3 haplotypes in the xian germplasms (Additional file 7: Figure S5c).
Association Analysis of SNPs and Traits
An association analysis of GNP1 identified a SNP (S_397) located at 1662 bp upstream of the ATG (chromosome 3: 36,149,264) that was significantly associated with the GNP and PH (Table 1).
Table 1.
Results of MLM association of SNPs extracted from the two genes (NAL1 and GNP1) with traits in 198 accessions
| Gene | Trait | GNP | FLW | PH | |||
|---|---|---|---|---|---|---|---|
| Site | P | R2 (%) | P | R2 (%) | P | R2 (%) | |
| NAL1 | S_1672 | 0.0019 | 6.70 | – | – | – | – |
| S_2452 | 0.0305 | 2.47 | 0.0071 | 4.21 | – | – | |
| GNP1 | S_397 | 0.0125 | 3.37 | – | – | 0.0073 | 4.18 |
R2 (%) Phenotypic variance explained, GNP grain number per panicle, FLW flag leaf width, PH plant height
For NAL1, a SNP (S_2452) located in the third exon (chromosome 4: 31,212,801) leading to the replacement of an arginine by a histidine was significantly associated with the GNP and FLW (Table 1). Another SNP (S_1672) located in the second intron was significantly associated with the GNP (Table 1). An allele with a 1-bp deletion was detected only in haplotype NAL1–6 and was the only variation between haplotypes NAL1–6 and NAL1–5 (Fig. 2a).
Effects of the Combination of Two Genes on the Grain Number per Panicle and the Grain Yield per Plant
On the basis of different major haplotypes for a single gene in 198 germplasms, the following four two-gene combinations were included in a comparative analysis of the xian subpopulation: GNP1–6/NAL1–1, GNP1–1/NAL1–1, GNP1–1/NAL1–2, and GNP1–1/NAL1–3 (Table 2). Six germplasms carrying GNP1–6/NAL1–1 had a higher GNP than the germplasms with the other three gene combinations, with average values of 201.7, 220.5, and 176.9 in 2014, 2015, and 2016, respectively (Table 2). Of the 71 xian germplasms with the GNP1–1 allele, 49, 6, and 6 had NAL1–1, NAL1–2, and NAL1–3, respectively. Six germplasms with the GNP1–1/NAL1–3 combination had a significantly higher GNP than the germplasms with GNP1–1/NAL1–2 (Table 2). However, the GNP did not significantly differ between the germplasms with GNP1–1/NAL1–1 or GNP1–1/NAL1–2 (Table 2), despite the observation that the germplasms with the NAL1–1 haplotype had a significantly higher GNP than the germplasms with NAL1–2 regardless of the genetic background (Fig. 2e). These observations implied that the GNP1–1 haplotype mainly affects the GNP.
Table 2.
Comparison of grain number among genotype combinations at NAL1 and GNP1 in xian subpopulation
| Haplotype | N | GNP | ||
|---|---|---|---|---|
| 2014 | 2015 | 2016 | ||
| GNP1–1/NAL1–3 | 6 | 197.9 ± 63.8a | 212.3 ± 71.6a | 161.3 ± 45.7a |
| GNP1–1/NAL1–1 | 49 | 158.4 ± 49.8ab | 169.1 ± 52.3ab | 130.7 ± 38.1ab |
| GNP1–1/NAL1–2 | 6 | 122.0 ± 26.2b | 125.4 ± 31.5b | 103.7 ± 24.0b |
| GNP1–1/NAL1–1 | 49 | 158.4 ± 49.8 | 169.1 ± 52.3 | 130.7 ± 38.1 |
| GNP1–6/NAL1–1 | 6 | 201.7 ± 40.2* | 220.5 ± 27.3* | 176.9 ± 40.4** |
N the number of xian varieties, GNP grain number per panicle
All data are present as mean ± SD; Letters are ranked by Duncan test at P < 0.05; The *, ** denotes significance of Student’s t test at P < 0.05, P < 0.01, respectively
Based on estimates of variance components for four traits, TGW and GNP were controlled mainly by VG, whereas VGEI was the main source for PN and GY in the introgression lines (ILs) with the Lemont background (LT-ILs) and the Teqing background (TQ-ILs) (Additional file 9: Table S3), indicating that PN and GY were highly environment-dependent, i.e., the two-gene combination had strong genotype by environment interaction on PN and GY (Wang et al. 2014). In LT-ILs and TQ-ILs, the GNP1 and NAL1 haplotypes were classified as strong or weak functional haplotypes based on their comparative performance in near-isogenic backgrounds (Xu et al. 2015; Wu et al. 2016). The Lemont carried the strong functional haplotype NAL1–4 (Xu et al. 2015), but a weak functional haplotype, GNP1–3, at GNP1. In contrast, the Teqing carried the weak functional haplotype NAL1–5, but a strong functional haplotype, GNP1–9, at GNP1 (Wu et al. 2016). The 194 LT-ILs were classified into the following four types according to their genotypes in the GNP1 (RM227–RM85) and NAL1 (RM317–RM255) intervals: GNP1WNAL1W (weak alleles at GNP1 and NAL1), GNP1SNAL1W (strong allele at GNP1, but a weak allele at NAL1), GNP1WNAL1S (weak allele at GNP1, but a strong allele at NAL1), and GNP1SNAL1S (strong alleles at GNP1 and NAL1). We observed that the GNP of the germplasms with GNP1SNAL1S, GNP1WNAL1S, or GNP1SNAL1W was higher than that of the germplasms with GNP1WNAL1W. Interestingly, the ILs carrying GNP1SNAL1S had a higher GNP than the lines with GNP1SNAL1W or GNP1WNAL1S (15.7%–16.4% and 26.3%–38.9% higher, respectively) (Table 3). Moreover, the germplasms with GNP1SNAL1S had a better average grain yield per plant (GYP) than the germplasms with GNP1WNAL1S in Beijing and Sanya (24.2% and 18.7% higher, respectively) because of the substantially higher GNP associated with GNP1SNAL1S. However, the germplasms with GNP1SNAL1S had a GYP that was similar to that of the germplasms with GNP1SNAL1W because of a slightly lower grain weight and EPN (Table 3). In the TQ-ILs, the GNP was higher with GNP1SNAL1S than with GNP1SNAL1W, which was similar to the results for the LT-ILs (Table 3). These findings suggested that in the xian and geng genetic backgrounds, the germplasms carrying both GNP1 and NAL1 had a considerably higher GNP than the germplasms carrying only GNP1 or NAL1. This was inconsistent with the GYP data because of a decrease in the grain weight and EPN. Specifically, GNP1SNAL1S and GNP1SNAL1W resulted in a similar GYP that was significantly higher than that of GNP1WNAL1S.
Table 3.
Grain yield and their related traits associated with the combination lines of NAL1 and GNP1
| GB | Regions | GNP1 | NAL1 | N | EPN | TGW | GNP | GYP |
|---|---|---|---|---|---|---|---|---|
| Lemont | Beijing | S | S | 17 | 9.0 ± 1.8a | 21.1 ± 2b | 196.1 ± 42.3a | 29.3 ± 9.4a |
| S | W | 14 | 9.6 ± 2.1a | 22.3 ± 3.1ab | 168.5 ± 37.9b | 29.3 ± 10.5a | ||
| W | S | 151 | 8.8 ± 1.5a | 22.2 ± 2ab | 141.2 ± 21.2c | 23.6 ± 5.9b | ||
| W | W | 12 | 9.9 ± 1.2a | 24.2 ± 2.1a | 114.6 ± 25.8d | 18.5 ± 2.6c | ||
| Sanya | S | S | 17 | 11.9 ± 2b | 22.1 ± 2.1b | 220.7 ± 45.7a | 47.6 ± 13a | |
| S | W | 14 | 14.1 ± 2.7a | 22.7 ± 2.8b | 190.7 ± 38.4b | 47.3 ± 12.4a | ||
| W | S | 151 | 12.6 ± 2.9ab | 22.1 ± 1.6b | 174.8 ± 28.4b | 40.1 ± 12.5b | ||
| W | W | 12 | 14.2 ± 2.8a | 24.0 ± 2.0a | 143.5 ± 34c | 41.1 ± 12.7b | ||
| Teqing | Beijing | S | S | 16 | 8.7 ± 1.3a | 22.4 ± 2.2a | 219.3 ± 35.7a | 28.8 ± 5.8ab |
| S | W | 217 | 9.4 ± 1.4a | 22.0 ± 2.0a | 195.8 ± 32.2b | 30.2 ± 6.8a | ||
| W | S | – | – | – | – | – | ||
| W | W | 19 | 8.9 ± 1.6a | 22.8 ± 1.8a | 164.6 ± 47.3c | 25.6 ± 7.5b | ||
| Sanya | S | S | 16 | 11.6 ± 2.2a | 23.8 ± 2.6a | 241.8 ± 30.0a | 47.0 ± 10.9a | |
| S | W | 217 | 12.2 ± 2.7a | 23.3 ± 2.0a | 219.0 ± 33.4b | 47.4 ± 14.0a | ||
| W | S | – | – | – | – | – | ||
| W | W | 19 | 12.5 ± 2.7a | 23.8 ± 2.2a | 188.5 ± 33.2c | 45.5 ± 11.5a |
GB genetic background, W weak functional lines (Teqing allele of NAL1, Lemont allele of GNP1), S strong functional lines (Lemont allele of NAL1, Teqing allele of GNP1) (Xu et al. 2015; Wu et al. 2016), N the number of lines, EPN effective panicle number per plant, TGW thousand grain weight (g), GNP grain number per panicle, GYP grain yield per plant (g), theoretical yield in Sanya, actual yield in Beijing; All data are present as mean ± SD; Letters are ranked by Duncan’s test at P < 0.05
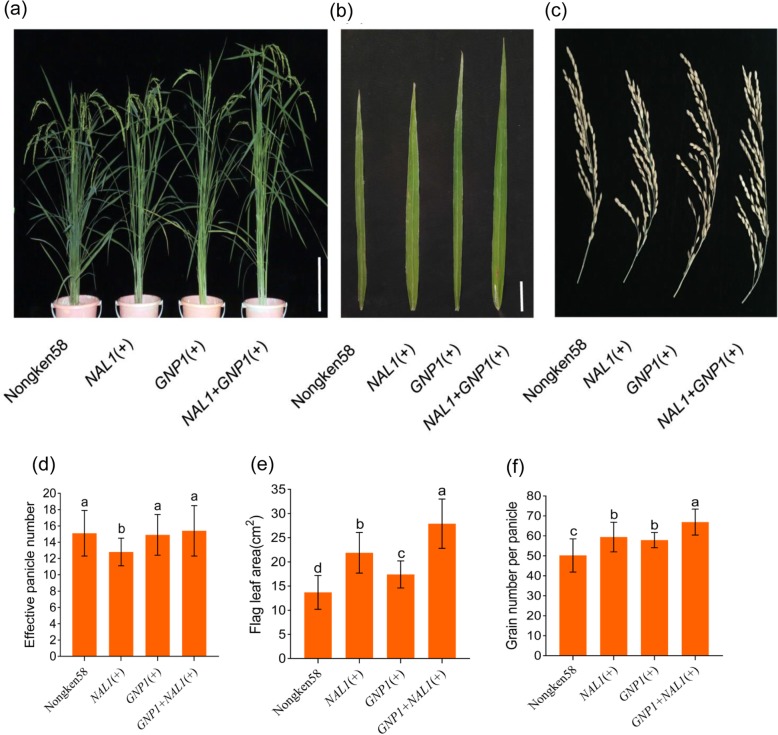
To further clarify the combined effects of GNP1 and NAL1 on the GYP in the same background, we compared the grain yield-related traits of Nongken58 (transgenic control, carrying haplotype NAL1–5 of NAL1 and GNP1–4 of GNP1) with those of homozygous transgenic lines with GNP1 derived from the high-yielding xian cultivar Teqing (with a strong functional GNP1 haplotype), NAL1 derived from the geng cultivar Lemont (with a strong functional NAL1 haplotype), or both GNP1 and NAL1. Relative to the corresponding values for Nongken58, the GNP1-carrying transgenic plants had a significantly higher GNP as well as a substantially higher PH and flag leaf length (FLL) (Fig. 3, Table 4). Additionally, the NAL1-positive transgenic plants had a significantly higher GNP, FLW, FLL, and PH, with a significantly lower EPN (Fig. 3, Table 4). Interestingly, compared with the GNP1- and NAL1-carrying transgenic plants, the transgenic plants with both GNP1 and NAL1 had a significantly higher GNP (15.5%–25.4% and 11.6%–15.9% higher, respectively) and PH (6.5%–21.0% and 8.6%–17.8% higher, respectively), but a significantly lower thousand grain weight (TGW) (3.1%–3.9% and 7.5%–7.6% lower, respectively) across 3 years (Table 4). The GYP of the transgenic plants with both GNP1 and NAL1 was 5.7%–9.0% higher than that of the GNP1-carrying transgenic plants because of a large increase in the GNP. Moreover, the combination of GNP1 and NAL1 resulted in a GYP that was 8.3%–12.3% higher than that of NAL1 alone because of a significant increase in the GNP and EPN (19.3%–21.7% higher) (Table 4).
Fig. 3.
Performance of three homozygous transgenic lines in the T3 generation under nature conditions. a–c Cross morphologies (a) flag leaf morphologies (b) panicle morphologies (c) of the lines with NAL1 or GNP1, the lines carrying both NAL1 and GNP1, and Nongken58 (transgenic control). (+) indicate transgene-positive. d–f Effective panicle number (d) flag leaf area (e) grain number per panicle (f) are compared the lines with either NAL1 or GNP1 with the lines carrying both NAL1. Error bars indicate SD. Letters on histograms (a, b, and c) are ranked by Duncan’s test at P < 0.05. Scale bars represent 10 cm (a) or 2 cm (b)
Table 4.
Performance of three homozygous transgenic lines under nature conditions
| Generation | Genotype | FLL | FLW | PH | EPN | GNP | TGW | GYP | +% CK | +% NAL1 | +% GNP1 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| T2 | Nongken58 | 21.3b | 1.0b | 69.4c | 19.0a | 58.2c | – | – | |||
| NAL1(+) | 24.5ab | 1.3a | 86.1b | 15.7b | 73.7b | – | – | ||||
| GNP1(+) | 27.1a | 1.0b | 83.2b | 18.3a | 68.1b | – | – | ||||
| GNP1 + NAL1(+) | 28.6a | 1.3a | 93.5a | 19.1a | 85.4a | – | – | ||||
| T3 | Nongken58 | 18.3b | 1.0b | 69.7c | 15.1a | 50.2c | 27.1a | 434.7 | |||
| NAL1(+) | 22.5a | 1.3a | 81.6b | 12.8b | 59.4b | 27.5a | 446.3 | 2.7 | |||
| GNP1(+) | 23.2a | 1.0b | 79.4b | 14.9a | 57.9b | 26.2ab | 460.1 | 5.8 | |||
| GNP1 + NAL1(+) | 26.6a | 1.4a | 96.1a | 15.4a | 66.9a | 25.4b | 501.4 | 15.3 | 12.3 | 9.0 | |
| T4 | Nongken58 | 23.3b | 1.2b | 96.0c | 13.5a | 70.7c | 26.2a | 530.3 | |||
| NAL1(+) | 30.6a | 1.5a | 107.6b | 10.9b | 90.6b | 26.8a | 553.9 | 4.5 | |||
| GNP1(+) | 32.4a | 1.2b | 110.6b | 12.6ab | 86.2b | 25.8ab | 567.2 | 7.0 | |||
| GNP1 + NAL1(+) | 34.6a | 1.5a | 117.8a | 13.0a | 101.1a | 24.8b | 599.7 | 13.1 | 8.3 | 5.7 |
(+) indicate transgene-positive, CK Nongken58, GNP grain number per panicle, EPN effective panicle number per plant, TGW thousand grain weight (g), FLL flag leaf length (cm), FLW flag leaf width (cm), PH plant height (cm), GYP grain yield per plant (g/m2)
Letters are ranked by Duncan’s test at P < 0.05
Discussion
GNP1 and NAL1 are Important for the Natural Variation in Rice Grain Production
In the rice germplasms investigated in this study, NAL1 was the most important locus for the GNP in both the xian and geng genetic backgrounds. These results were consistent with the findings of earlier studies involving near isogenic lines or transgenic rice lines carrying various alleles in diverse backgrounds (Fujita et al. 2013; Takai et al. 2013; Zhang et al. 2014; Xu et al. 2015; Yano et al. 2016). In previous investigations, sequence comparisons revealed that the NAL1 allele from xian and geng rice cultivars had relatively conserved differences in the coding region (mainly one single-base substitution), which may differentially affect the development of multiple traits related to leaf type, plant type, and panicle type (Fujita et al. 2013; Takai et al. 2013; Zhang et al. 2014; Xu et al. 2015). The single-base substitution located at amino acid position 233 in the trypsin-like serine and cysteine protease domain lead to the amino acid variation (arginine and histidine). The geng cultivars Nipponbare, Koshihikari, and Lemont, which have identical superior NAL1 alleles, carry the NAL1–4 haplotype. The introgression of the NAL1–4 alleles from geng rice germplasms into xian rice cultivars 9311, Takanari, and Teqing increased the GYP (Zhang et al. 2014), the leaf chlorophyll content (Zhang et al. 2014), the FLW (Xu et al. 2015), but decreased the photosynthetic rate per area (Takai et al. 2013). Taguchi-Shiobara et al. (2015) reported that the flag leaf width in transgenic plants of NAL1 varied depending on the ratio of an A allele (histidine-type, H-type) to G allele (arginine-type, R-type). In 198 panels, accessions carrying NAL1-P3 protein haplotype had more GNP in 2014 and 2016, and larger FLW in 3 years compared with those carrying NAL1-P2(Additional file 8: Figure S6d–e). The difference in the one amino acid variation (histidine/ arginine) in the third exon was also observed between the two protein types (Additional file 8: Figure S6a) The H-type variation decreases the EPN, but increases the GNP and the FLW relative to the corresponding levels due to the R-type variation in the geng genetic background (Yano et al. 2016), which is highly consistent with our results obtained with the 3 K RGP geng subpopulation (Wang et al. 2018) in the Rice SNP-Seek Database (Additional file 10: Figure S7). In the present study, a SNP (S_2452) in the third exon was also significantly associated with the GNP and FLW (Table 1). Thus, we presumed that the nucleotide variation (A/G) in the third exon is critical for determining the GNP and FLW. Interestingly, the GNP and FLW differed between the germplasms with NAL1–2 or NAL1–6 in the geng subpopulation, despite the lack of an amino acid variation (R/H) and the insertion of a retrotransposon. A sequence comparison indicated that NAL1–2 and NAL1–6 differed because of a single-base InDel (S_1672) in the second intron and four nucleotide variations in the 3′ UTR (Fig. 2a). Additionally, the panicle NAL1 expression levels significantly differed between the two haplotypes (Additional file 7: Figure S5b). Thus, we speculated the variability in the GNP between NAL1–2 and NAL1–6 might be due to the NAL1 expression level and/or the single-base InDel (S_1672) significantly associated with the GNP in the 198 analyzed germplasms (Table 1). Interestingly, even with a lack of an R/H variation encoded in the third exon, a retrotransposon insertion, and expression level differences (Additional file 7: Figure S5c), differences in the GNP were still observed between NAL1–2 and the other two major xian haplotypes (NAL1–1 and NAL1–3) in the xian genetic background. A sequence comparison revealed that NAL1–2 and the other two haplotypes (NAL1–1 and NAL1–3) differed regarding the presence of previously reported nucleotide variations in the fifth exon resulting in two amino acid substitutions (Takai et al. 2013; Fujita et al. 2013; Zhang et al. 2014). In 198 accessions, accessions with NAL1-P1 exhibited significantly more GNP and larger FLW than those with NAL1-P2 in xian subpopulation (Additional file 8: Figure S6b–c). The difference in the two nucleotide variations in the fifth exon was also observed between the two protein types (Additional file 8: Figure S6a). Thus, we speculated these nucleotide differences in the fifth exon also influence the GNP. Additionally, grain number is also likely affected by other factors, including gene interactions or the genetic background effects, with consequences for the plant characteristics and yield.
A previous study indicated that GNP1 increases the number of grains likely because of promoter sequence variations that alter GNP1 expression (Wu et al. 2016). In the present study, a sequence comparison confirmed that the GNP1 allele in the rare GNP1–9 haplotype, which is present in only four xian germplasms, is identical to that in Teqing with the superior GNP1 haplotype described by Wu et al. (2016). Therefore, the Teqing allele is likely relatively uncommon. Additionally, two previously reported multiple-base deletions (I_1611 and I_1740) in the promoter region were observed in the major haplotype GNP1–5 and in three rare haplotypes (GNP1–9, GNP1–13, and GNP1–14) (Fig. 1a). In contrast, there was no significant difference in the GNP between GNP1–5 and the other three major haplotypes (GNP1–1, GNP1–2, and GNP1–6), even though these haplotypes include the two multiple-base sequence variations in the promoter. Interestingly, there were significant differences in the GNP between GNP1–6 and GNP1–1 within the xian subpopulation, despite the absence of the two multiple-base sequence differences in the promoter region (Fig. 1e). In addition, no significant difference in GNP was observed between two protein types in 3 years except in 2014 (Additional file 6: Figure S4b). To determine whether the sequence variability between the GNP1–6 and GNP1–1 promoter regions influences GNP1 expression and contributes to the differences in the GNP, we analyzed the GNP1 expression levels in the young panicles of the germplasms carrying GNP1–6 or GNP1–1. The GNP1 transcript abundance in panicles was considerably greater in germplasms with GNP1–6 than in germplasms with GNP1–1 (Additional file 6: Figure S4a). Furthermore, we used PromPredict to predict the possible regulatory regions (Morey et al. 2011). Six potential promoter intervals, including one multiple-base deletion (I_1740) were identified in the promoter region (Fig. 1a). A single-base sequence variation (S_397) in the promoter contributed to the differences in the GNP and PH according to a gene-based association analysis (Table 1). However, the single-base alteration was not detected between Teqing and Lemont (Wu et al. 2016). These results implied that the nucleotide variation in the promoter region influences GNP1 expression, thereby affecting the GNP and PH.
Artificial Selection Patterns of NAL1 and GNP1
A comparison of the NAL1 sequences from the 198 O. sativa germplasms and the eight O. rufipogon germplasms revealed the NAL1–8 haplotype from the Xian haplogroup showed an allele that is identical to that of the wild variety WR4 (Fig. 2b), suggesting that it might be the original xian haplotype. Additionally, arginine was identified at amino acid position 233 in the NAL1 of all eight germplasms of O. rufipogon (Fig. 2a), from which O. sativa originated. Accordingly, the R-type allele appears to be the ancestral form, from which the H-type allele was derived. Moreover, our data indicated that the R-type allele has been retained in all wild rice germplasms as well as in 98.6% of the xian and 58.1% of the geng germplasms among the 198 analyzed germplasms (Additional file 11: Table S4) during the evolution across large geographical distances (Additional file 12: Figure S8a). These observations imply the R-type alleles have a competitive advantage over the H-type alleles under natural conditions. Furthermore, we clarified the NAL1 allele variation (R/H) based on the 3 K RGP database (Wang et al. 2018). We determined that 98.2% of the xian and 25.1% of the geng germplasms carry the R-type allele, whereas only 1% of the xian and 73.4% of the geng germplasms carry the H-type allele from 3 K RGP (Additional file 11: Table S4). These results suggest that the differences in the NAL1 allelic frequency might be related to the xian–geng differentiation. In the present study, the H-type allele appeared to be introgressed into a few xian landraces (2 of 146 among the 198 analyzed germplasms; 18 of 1785 in 3 K RGP). These landraces include a defined allele comprising the geng-like DNA flanking NAL1 in the xian background, which was likely the result of naturally occurring crosses and the subsequent introgression during domestication after the emergence of the H-type allele in the geng subpopulation. A 5895-bp retrotransposon insertion in the second exon of NAL1 was detected only in haplotype NAL1–4 of geng, suggesting that germplasms with haplotype NAL1–4 might be from the same ancestor. The NAL1–4 haplotype was present in 18 germplasms widely distributed in nine countries on five continents (Asia, Australia, Europe, Africa, and South America) (Additional file 12: Figure S8a). A phylogenetic network revealed that a retrotransposon insertion converted NAL1–10 to NAL1–4 (Fig. 2b).
The GNP1 haplotype of O. sativa ssp. geng, which is highly conserved (i.e., only one single-base variation), may have evolved from a unique ancient O. rufipogon germplasm. Additionally, the GNP1 nucleotide diversity in the geng landraces (π = 0.00085) was 68% lower than that of O. rufipogon (π = 0.00264). In contrast, the GNP1 nucleotide diversity in the xian landraces (π = 0.00255) decreased by only 3% (Additional file 4: Table S2). The nucleotide diversity of the geng GNP1 was similar to that of 111 randomly chosen gene fragments (π = 0.00111) (Caicedo et al. 2007), suggesting that the lower nucleotide diversity in geng than in xian or the wild progenitor was due to a genetic bottleneck effect rather than a selective sweep (Nagano et al. 2005).
Favorable Haplotypes/Alleles for Increasing the GNP in Rice
At the single-gene level, we identified favorable haplotypes for the two genes influencing the GNP. The NAL1–3 and NAL1–1 haplotypes as well as the GNP1–6 haplotype increased the GNP in the xian subpopulation. Therefore, these superior haplotypes may be ideal for increasing the GNP in xian rice cultivars. In the geng subpopulation, some cultivars with the superior haplotypes NAL1–2 and NAL1–4 (e.g., Lemont and Nipponbare) that resulted in increased grain number (sink-related trait) and wider flag leaves (source-related trait) may be the most appropriate germplasms for the molecular breeding of super high-yielding rice varieties with ideal plant characteristics and a balanced sink and source relationship. The findings described herein may be relevant for introducing superior haplotypes of a single gene into a single variety with the ideal genotype.
In a previous study, we determined that the rice grain yield of a near-isogenic line with the superior NAL1 allele from Lemont in a Teqing genetic background was 3.2%–3.8% higher than that of Teqing in three environments (Xu et al. 2015). Teqing is a well-known super-high-yielding and widely adaptable xian variety developed in China in 1984, with a grain yield that is 30% higher than that of Lemont under the growth conditions in the southern USA (Li et al. 1998). In other words, the superior NAL1 allele from the low-yielding Lemont germplasm can further increase the grain yield of high-yielding varieties such as Teqing. Previous studies on the molecular basis of complex traits proved that even though the germplasms themselves do not exhibit the target trait characteristics, they may still harbor favorable genes for increasing yield (He et al. 2006), salt tolerance (Zang et al. 2008), and drought tolerance (Wang et al. 2012). Therefore, to enhance quantitative traits, such as grain yield, the favorable alleles in some low-yielding varieties may be relevant for rice breeding programs.
Regarding GNP1, there were no significant differences in the GNP resulting from the GNP1–3 and GNP1–4 geng haplotypes. The superior GNP1–9 (Teqing) and GNP1–6 haplotypes were respectively detected in only 2% and 4% of the 198 germplasms (Fig. 1a). Therefore, we propose the superior GNP1 haplotype has no significant effects in the geng subpopulation and is rare in the xian background of the 198 analyzed cultivated rice germplasms. In a genome-wide association study of 3000 rice accessions, no significant SNP was identified in the GNP1 chromosomal regions affecting the GNP, implying the favorable GNP1 allele is relatively uncommon. The near-isogenic line NIL-GNP1TQ in the Lemont genetic background, which differs from Lemont only in an approximately 66.1-kb region containing GNP1 derived from Teqing, reportedly produces 5.7%–9.6% more grain yield than Lemont (Wu et al. 2016). These results suggest that the rare Teqing superior GNP1 allele may be useful for breeding high-yielding rice varieties. Many superior alleles mediating grain number, grain size, and grain weight, including OsSPL14 (Jiao et al. 2010), GS2 (Hu et al. 2015), OsglHAT1 (Song et al. 2015), are relatively rare at the natural population scale, but may be applicable for increasing rice yield in breeding programs (Jiao et al. 2010; Hu et al. 2015; Kim et al. 2018). Most parental inbred lines have only a few rare superior alleles, whereas high-yielding hybrid varieties possess several of these alleles (Huang et al. 2015). Increasing diversity via xian × geng crosses, which may enable the relatively rapid introduction of low-frequency superior alleles in specific lines, should be pursued in future studies involving hybrid rice breeding (Huang et al. 2015).
Two-Gene Combinations for Increasing the GNP and GYP in Rice
In this study, we proved that combining NAL1 and GNP1 increases the GNP in different genetic backgrounds. Specifically, the ILs carrying the strong NAL1 and GNP1 alleles (GNP1SNAL1S) in the Lemont genetic background had a higher GNP than the lines with only one strong allele (GNP1WNAL1S or GNP1SNAL1W) (Table 3). In the Teqing background, the GNP resulting from GNP1SNAL1S was significantly higher than that due to GNP1SNAL1W (Table 3). Moreover, the GNP of the transgenic lines carrying the favorable Teqing allele of GNP1 and Lemont allele of NAL1 was significantly higher than the GNP of transgenic lines carrying only GNP1 or NAL1 (Table 4). These results suggest these two genes may have a synergistic interactive effect on the regulation of grain number.
Compared with the NAL1 transgenic plants, the lines carrying NAL1 and GNP1 had a significantly higher flag leaf area (source-related trait), GNP (sink-related trait), and EPN, which increased the GYP by an average of 8.3%–12.3% (Fig. 3, Table 4). Similarly, the transgenic lines with both NAL1 and GNP1 had a higher flag leaf area and GNP than the transgenic plants with GNP1 alone, which resulted in an average increase in the GYP of 5.7%–9.0% (Fig. 3, Table 4). Our results imply that the combined effects of NAL1 and GNP1 may enhance the source–sink relationship, thus resulting in increased the yield in geng rice cultivar although they were also slight taller than single transgenic lines. Therefore, exploiting and pyramiding superior haplotypes (e.g., NAL1 from Lemont and GNP1 from Teqing) may enable the breeding of super-high-yielding rice varieties with optimized plant characteristics and source–sink relationships.
Conclusions
We identified 16 and 14 haplotypes for NAL1 and GNP1, respectively. The NAL1 gene had the strongest effects on GNP in xian and geng subpopulations while GNP1 had no significant effects in the geng subpopulation and was rare in the xian background. The transgenic lines with both genes exhibited higher GNP and grain yield than the transgenic lines with GNP1 or NAL1. The two genes combined in the introgression lines in Lemont background had favorable effects on the GNP. These results should contribute to rice breeding for high yield through pyramiding GNP1 and NAL1.
Methods
Plant Materials and Phenotyping
A total of 198 O. sativa germplasms varying regarding the GNP, FLW, and PH from 28 countries were obtained from the China National Crop Genebank of the Chinese Academy of Agricultural Science and the International Rice Research Institute. An additional 8–10 common wild rice (O. rufipogon) germplasms were obtained from the Guangxi Academy of Agricultural Sciences, Guangxi, China. Basic information for each germplasm is provided in Additional file 3: Table S1 and Additional file 13: Table S5. All germplasms were grown under normal field conditions at the following two locations over 3 years: Nanning (22.1°N, 107.5°E) in Guangxi province in 2014 and 2015 and Shenzhen (22.6°N, 114.1°E) in 2016. The field trial involved two rows of 12 plants each, with the rows separated by 25 cm and the plants in each row separated by 17 cm. The analysis was completed with three replicates. The fields were managed according to the local standard practices. At the heading stage, the FLW was measured for the main stem of each plant. At maturity, the GNP and PH of eight plants were measured.
Two sets of reciprocal ILs comprising 201 lines (32 BC2F8, 123 BC3F7, and 46 BC4F6) in the Lemont background (LT-ILs) and 252 lines (133 BC2F8, 96 BC3F7, and 23 BC4F6) in the Teqing background (TQ-ILs) were generated from the consecutive backcrossing and several generations of selfing between the xian cultivar Teqing and the geng cultivar Lemont (Mei et al. 2006). These lines were grown in Sanya (18.3°N, 109.3°E) in the winter of 2014 and in Beijing (40.2°N, 116.2°E) in the summer of 2015. At maturity, the EPN, TGW, GYP, and GNP were determined. After eliminating the lines with missing phenotypic data, the remaining lines were used for analyzing the effects of NAL1 and GNP1 on the EPN, GNP, TGW, and GYP in the two (LT and TQ) backgrounds.
DNA Extraction, PCR Amplification, Gene Cloning, and Sequencing
Fresh leaves were collected from the field-grown plants for a genomic DNA extraction with the cetyltrimethylammonium bromide method (Murray and Thompson 1980). The primers used to amplify the promoter, 5′ UTR, exon, intron, and 3′ UTR were designed based on the Nipponbare GNP1 and NAL1 alleles. The PCR was completed with KOD NEO DNA polymerase (Toyobo, Shiga, Japan) and a standard PCR protocol.
Because of the existence of heterozygous genotypes in O. rufipogon, the PCR product was ligated into the pGEM-T Easy Vector (Promega, USA) and then sequenced, after which one allele sequence was randomly selected. To ensure accuracy, the sequencing was performed twice with the ABI 3730 system at the Beijing Genomics Institute, China. Sequence contigs were assembled with the SEQUENCHER 4.1.2 program (Gene Codes Corporation, Ann Arbor, MI, USA). Details regarding the PCR primers and sequencing are listed in Additional file 14: Table S6.
Analyses of DNA Sequences and Population Structures
The DNA sequences were aligned with the MUSCLE program (Edgar 2004) and manually adjusted with BIOEDIT (Hall 1999). The number of segregating sites, nucleotide diversity (π), Watterson’s estimator (θ), and Tajima’s D and Fu and Li’s D of the neutrality test were calculated with the DNASP program (version 5.0) (Librado and Rozas 2009). The sliding-window method was employed to analyze the polymorphisms in the GNP1 and NAL1 genome sequences, with a window size of 100 bp and a step size of 20 bp, in which pairwise insertions and deletions were removed with DNASP (version 5.0). The evolutionary history was inferred based on the neighbor-joining method of the MEGA5 program (Tamura et al. 2011). The phylogenetic network was constructed with Network 4.611 (Hans-Jurgen et al. 1999) according to the corresponding user guide.
For population structure analyses, 24,460 evenly distributed SNPs were sampled to calculate the population structure (Q). We used a model-based Bayesian clustering analysis method implemented with the STRUCTURE program (version 2.3.4) (Pritchard et al. 2000). The Q matrix derived from the principal components was calculated with a Perl script and SNPs filtered with Plink (Purcell et al. 2007). The K matrix (kinship matrix) was obtained from the results of the relatedness analysis according to the EMMA method in GAPIT, which is an R software package (Lipka et al. 2012).
To minimize the effects of environmental variations, the best linear unbiased predictors (BLUPs) for each germplasm phenotype over 3 years were calculated with the R package rrBLUP (version 4.3) (Endelman 2011). The SNPs with a minor allele frequency less than 0.05 and/or a missing data rate exceeding 20% were removed and the remaining high-quality SNPs within NAL1 and GNP1 were used to perform gene-based association analyses via the mixed linear model with Q and K. The analyses were performed with the TASSEL program (Bradbury et al. 2007).
Gene Expression Analysis
Yong panicles were collected from five plants per germplasm at the panicle initiation stage for a subsequent gene expression analysis. Total RNA was extracted from the young panicles with the TRIzol reagent (Invitrogen), after which 2 μg total RNA was used as the template for a cDNA synthesis with SuperScript III reverse transcriptase (Invitrogen) in a final volume of 20 μl. A real-time PCR assay was performed with gene-specific primers (Additional file 14: Table S6) and a 7500 Real-Time PCR system (Applied Biosystems, Carlsbad, CA, USA) according to the manufacturer’s instructions. The rice Actin gene was used as the internal control. Each sample was analyzed with three technical replicates.
Complementation Test
For the NAL1 complementary test, a 13.8-kb DNA fragment from 3424 bp upstream of the transcription start site to 1450 bp downstream of the termination site was amplified from Lemont. The sequence was then cloned into pCAMBIA1300 to produce the pCAMBIA1300-NAL1 recombinant plasmid. For GNP1, the 5.5-kb full-length genomic DNA sequence from 3498 bp upstream of the transcription start site to 878 bp downstream of the termination site was amplified from Teqing. The PCR product was cloned into pCAMBIA1300 to generate the pCAMBIA1300-GNP1 recombinant plasmid. Additionally, a 19.3-kb fragment comprising the 13.8-kb NAL1 DNA fragment from Lemont and the 5.5-kb GNP1 genomic DNA from Teqing was inserted into the same pCAMBIA1300 vector. Specifically, the 5.5-kb GNP1 genomic sequence was amplified from Teqing with primers containing the SbfI and PstI sequences. The amplified fragment (5.5-kb) was digested with restriction enzymes and then inserted into pCAMBIA1300 at the SbfI and PstI sites to produce the pCAMBIA1300-GNP1–2 recombinant plasmid. The 13.8-kb NAL1 fragment was removed from pCAMBIA1300-NAL1 by a digestion with BamHI and SbfI. The resulting NAL1 fragment (13.8-kb) was inserted into the BamHI and SbfI sites of pCAMBIA1300-GNP1–2 to generate the pCAMBIA1300-GNP1 + NAL1 recombinant plasmid. The three recombinant plasmids were separately introduced into Agrobacterium tumefaciens strain EHA105 cells for the subsequent transformation of a geng variety, Nongken58. The primer sets used for the PCR amplifications are listed in Additional file 14: Table S6.
The three transgenic lines and Nongken58 were planted with a randomized block design (with three replicates) at a spacing of 25 × 17 cm in Sanya (18.3°N, 109.3°E), Hainan province, China during December 2015–April 2016, February–June, 2017, and December 2017–April 2018, respectively. At the heading stage, the FLL, FLW, and PH were measured for the main stem of each plant. At maturity, eight plants were sampled and dried in an oven at 70 °C for 5 days, after which the EPN, GNP, and TGW were calculated. Additionally, plants were harvested from a 2 m2 area in each plot and then air dried before determining the GYP.
Statistical Analysis
Differences in the phenotypic values and in the gene expression levels between the haplotypes (containing more than seven accessions) were examined by a one-way ANOVA or Student’s t-test. Duncan’s multiple range test was conducted to determine the significance of any differences (P < 0.05). The data analyses were performed with the SAS software. To dissect effect of genotype-by-environment interaction (GEI) on the tested traits, variance components were estimated using multiple-site analysis with all effects treated as random. Heritability across environments was computed using the estimated variance components as VG/(VG + VGEI/s + Ve/sr), where VG, VGEI, and Ve are the variances of genotype, GEI, and residual error, respectively, s is the number of environments, and r is the number of replicates. All analyses were conducted with the PBTools package (http://bbi.irri.org/products) developed by IRRI.
Supplementary information
Additional file 1: Figure S1. Comparison of grain number per panicle, flag leaf width and plant height between xian and geng rice (Oryza sativa L.) subpopulations in 198 accessions. (a–c) Cyan and orange colors indicate xian and geng, respectively. The *, **, *** denotes significance of Student’ s t test at P < 0.05, P < 0.01, and P < 0.001, respectively.
Additional file 2: Figure S2. Population structure analyses of 198 accessions. (a) Scree plot from GAPIT showing the selection of PCs for association study. (b) PCA plot based on the screen plot in the rice diversity panel. (c) Scree plot from STRUCTURE showing the selection of Q. (d) Bayesian clustering of 198 accessions using STRUCTURE program.
Additional file 3 : Table S1. Details of the accessions investigated and two grain number gene haplotypes.
Additional file 4 : Table S2. Nucleotide diversity of GNP1 and NAL1 genes. S, segregation sites; π, average number of nucleotide differences per site between random two sequences; θ, Watterson estimator; D1, Tajima’s D; D2, Fu and Li’D; D3, Fu and Li’F.
Additional file 5 : Figure S3. Sliding-windows analysis of GNP1 (a) and NAL1 (b) nucleotide diversity in O. sativa ssp. xian, O. sativa ssp. geng and O. rufipogon. The y-axis represents nucleotide diversity (π), and the genomic structure is shown at the bottom, where the black boxes indicate exons and the white boxes indicate introns and other noncoding regions.
Additional file 6 : Figure S4. Protein diversity of GNP1 (a). Comparison of grain number per panicle, and plant height between two protein types in xian subpopulations in 198 panel (b–c). Cyan and orange colors indicate GNP1-P1 and GNP1-P2, respectively. The *, **, *** denotes significance of Student’ s t test at P < 0.05, P < 0.01, and P < 0.001, respectively.
Additional file 7 : Figure S5. Expression level of NAL1 and GNP1 haplotypes in young panicle at the panicle initiation stage. (a) Expression level of GNP1–1 and GNP1–6 in xian subpopulation. (b) Expression level of NAL1–2, NAL1–4 and NAL1–6 in geng subpopulation. (c) Expression level of NAL1–1, NAL1–2 and NAL1–3 in xian subpopulation. Error bars indicate SD; Letters are ranked by Duncan’s test at P < 0.05. The * denotes significance of Student’ s t test at P < 0.05.
Additional file 8 : Figure S6. Protein diversity of NAL1 (a). Comparison of grain number per panicle, and flag leaf width among three protein types in xian and geng subpopulations, respectively (b–e). Cyan, orange, and lightpink colors indicate NAL1-P1, NAL1-P2 and NAL1-P3, respectively. The *, **, *** denotes significance of Student’ s t test at P < 0.05, P < 0.01, and P < 0.001, respectively.
Additional file 9 : Table S3. Variance components and heritabilty estimated by multiple-site analysis.
Additional file 10 : Figure S7. Comparison of grain number per panicle, flag leaf width and effective panicle number between R-type and H-type alleles located in the third exon of NAL1 gene in 3 K panel in xian and geng subpopulation, respectively. ***, P < 0.001 (Student’ s t test).
Additional file 11 : Table S4. Distribution of protein mutation sites diversity at amino acid position 233 of NAL1 gene. R, arginine; H, histidine.
Additional file 12 : Figure S8. Geographic distributions of different haplotypes of NAL1 and GNP1 among the 28 areas sampled.
Additional file 13 : Table S5. Details of the wild rice (Oryza rufipogon).
Additional file 14 : Table S6. Sequencing primers used in this study.
Acknowledgements
Not applicable.
Abbreviations
- EPN
Effective panicle number per plant
- FLL
Flag leaf length
- FLW
Flag leaf width
- GNP
Grain number per panicle
- H-type
Histidine-type
- ILs
Introgression lines
- LT-ILs
Introgression lines with the Lemont background
- PH
Plant height
- QTL
Quantitative trait loci
- R-type
Arginine-type
- TQ-ILs
Introgression lines with the Teqing background
Authors’ Contributions
YW and JLX designed and supervised the research. YW, LYZ, YTL, KC, and CCS performed the experiments. LYZ, CCW, XQZ, and SW analyzed data. YW and JLX wrote the paper. The author(s) read and approved the final manuscript.
Funding
This work was funded by the National Key Research and Development Program of China (2016YFD0100301), the National Natural Science Foundation of China (31671602), the Agricultural Science and Technology Innovation Program and the Cooperation and Innovation Mission (CAAS-ZDXT2018001), and the National Natural Science Foundation of China (31301285).
Availability of Data and Materials
All data generated or analyzed during this study are included in this published article and its supplementary information files.
Ethics Approval and Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12284-020-00374-8.
References
- Ashikari M, Sakakibara H, Lin S, Yamamoto T, Takashi T, Nishimura A, Angeles ER, Qian Q, Kitano H, Matsuoka M. Cytokinin oxidase regulates rice grain production. Science. 2005;309:741–745. doi: 10.1126/science.1113373. [DOI] [PubMed] [Google Scholar]
- Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES. TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics. 2007;23:2633–2635. doi: 10.1093/bioinformatics/btm308. [DOI] [PubMed] [Google Scholar]
- Caicedo AL, Williamson SH, Hernandez RD, Boyko A, Fledel-Alon A, York TL, Polato NR, Olsen KM, Nielsen R, McCouch SR, Bustamante CD, Purugganan MD. Genome-wide patterns of nucleotide polymorphism in domesticated rice. PLoS Genet. 2007;3:1745–1756. doi: 10.1371/journal.pgen.0030163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endelman JB. Ridge regression and other kernels for genomic selection with R package rrBLUP. Plant Genome J. 2011;4:250–255. doi: 10.3835/plantgenome2011.08.0024. [DOI] [Google Scholar]
- Fujita D, Trijatmiko KR, Tagle AG, Sapasap MV, Koide Y, Sasaki K, Tsakirpaloglou N, Gannaban RB, Nishimura T, Yanagihara S, Fukuta Y, Koshiba T, Slamet-Loedin IH, Ishimaru T, Kobayashi N. NAL1 allele from a rice landrace greatly increases yield in modern indica cultivars. Proc Natl Acad Sci U S A. 2013;110:20431–20436. doi: 10.1073/pnas.1310790110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall A. BioEdit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- Hans-Jurgen B, Peter F, Arne R. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- He GM, Luo XJ, Tian F, Li KG, Zhu ZF, Su W, Qian XY, Fu YC, Wang XK, Sun CQ, Yang JS. Haplotype variation in structure and expression of a gene cluster associated with a quantitative trait locus for improved yield in rice. Genome Res. 2006;16:618–626. doi: 10.1101/gr.4814006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Wang Y, Fang Y, Zeng L, Xu J, Yu H, Shi Z, Pan J, Zhang D, Kang S, Zhu L, Dong G, Guo L, Zeng D, Zhang G, Xie L, Xiong G, Li J, Qian Q. A rare allele of GS2 enhances grain size and grain yield in rice. Mol Plant. 2015;8:1455–1465. doi: 10.1016/j.molp.2015.07.002. [DOI] [PubMed] [Google Scholar]
- Huang X, Qian Q, Liu Z, Sun H, He S, Luo D, Xia G, Chu C, Li J, Fu X. Natural variation at the DEP1 locus enhances grain yield in rice. Nat Genet. 2009;41:494–497. doi: 10.1038/ng.352. [DOI] [PubMed] [Google Scholar]
- Huang X, Yang S, Gong J, Zhao Y, Feng Q, Gong H, Li W, Zhan Q, Cheng B, Xia J, Chen N, Hao Z, Liu K, Zhu C, Huang T, Zhao Q, Zhang L, Fan D, Zhou C, Lu Y, Weng Q, Wang ZX, Li J, Han B. Genomic analysis of hybrid rice varieties reveals numerous superior alleles than contribute to heterosis. Nat Commun. 2015;6:6258. doi: 10.1038/ncomms7258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, Wang Y, Xue D, Wang J, Yan M, Liu G, Dong G, Zeng D, Lu Z, Zhu X, Qian Q, Li J. Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat Genet. 2010;42:541–544. doi: 10.1038/ng.591. [DOI] [PubMed] [Google Scholar]
- Kim SR, Ramos JM, Hizon RJM, Ashikari M, Virk PS, Torres EA, Nissila E, Jena KK. Introgression of a functional epigenetic OsSPL14WFP allele into elite indica rice genomes greatly improved panicle traits and grain yield. Sci Rep. 2018;8:3833. doi: 10.1038/s41598-018-21355-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li ZK, Pinson SRM, Stansel JW, Paterson AH. Genetic dissection of the source-sink relationship affecting fecundity and yield in rice (Oryza sativa L.) Mol Breed. 1998;4:419–426. doi: 10.1023/A:1009608128785. [DOI] [Google Scholar]
- Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- Lipka AE, Tian F, Wang Q, Peiffer J, Li M, Bradbury PJ, Gore MA, Buckler ES, Zhang Z. GAPIT: genome association and prediction integrated tool. Bioinformatics. 2012;28:2397–2399. doi: 10.1093/bioinformatics/bts444. [DOI] [PubMed] [Google Scholar]
- Mei HW, Xu JL, Li ZK, Yu XQ, Guo LB, Wang YP, Ying CS, Luo LJ. QTLs influencing panicle size detected in two reciprocal introgressive line (IL) populations in rice (Oryza sativa L.) Theor Appl Genet. 2006;112:648–656. doi: 10.1007/s00122-005-0167-0. [DOI] [PubMed] [Google Scholar]
- Morey C, Mookherjee S, Rajasekaran G, Bansal M. DNA free energy-based promoter prediction and comparative analysis of Arabidopsis and rice genomes. Plant Physiol. 2011;156:1300–1315. doi: 10.1104/pp.110.167809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray MG, Thompson WF. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980;8:4321–4325. doi: 10.1093/nar/8.19.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano H, Onishi K, Ogasawara M, Horiuchi Y, Sano Y. Genealogy of the “green revolution” gene in rice. Genes Genet Syst. 2005;80:351–356. doi: 10.1266/ggs.80.351. [DOI] [PubMed] [Google Scholar]
- Oikawa T, Koshioka M, Kojima K, Yoshida H, Kawata M. A role of OsGA20ox1, encoding an isoform of gibberellin 20-oxidase, for regulation of plant stature in rice. Plant Mol Biol. 2004;55:687–700. doi: 10.1007/s11103-004-1692-y. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi J, Qian Q, Bu Q, Li S, Chen Q, Sun J, Liang W, Zhou Y, Chu C, Li X, Ren F, Palme K, Zhao B, Chen J, Chen M, Li C. Mutation of the rice Narrow leaf1 gene, which encodes a novel protein, affects vein patterning and polar auxin transport. Plant Physiol. 2008;147:1947–1959. doi: 10.1104/pp.108.118778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song XJ, Kuroha T, Ayano M, Furuta T, Nagai K, Komeda N, Segami S, Miura K, Ogawa D, Kamura T, Suzuki T, Higashiyama T, Yamasaki M, Mori H, Inukai Y, Wu J, Kitano H, Sakakibara H, Jacobsen SE, Ashikari M. Rare a previously unidentified histone H4 acetyltransferase enhances grain weight, yield, and plant biomass in rice. Proc Natl Acad Sci U S A. 2015;112:76–81. doi: 10.1073/pnas.1421127112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taguchi-Shiobara F, Ota T, Ebana K, Ookawa T, Yamasaki M, Tanabata T, Yamanouchi U, Wu J, Ono N, Nonoue Y, Nagata K, Fukuoka S, Hirabayashi H, Yamamoto T, Yano M. Natural variation in the flag leaf morphology of rice due to a mutation of the NARROW LEAF 1 gene in Oryza sativa L. Genetics. 2015;201:795–808. doi: 10.1534/genetics.115.181040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai T, Adachi S, Taguchi-Shiobara F, Sanoh-Arai Y, Iwasawa N, Yoshinaga S, Hirose S, Taniguchi Y, Yamanouchi U, Wu J, Matsumoto T, Sugimoto K, Kondo K, Ikka T, Ando T, Kono I, Ito S, Shomura A, Ookawa T, Hirasawa T, Yano M, Kondo M, Yamamoto T. A natural variant of NAL1, selected in high-yield rice breeding programs, pleiotropically increases photosynthesis rate. Sci Rep. 2013;23:2149. doi: 10.1038/srep02149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virk PS, Khush GS, Peng S. Breeding to enhance yield potential of rice at IRRI: the ideotype approach. Inst Rice Res Notes. 2004;29:S1–S9. [Google Scholar]
- Wang W, Mauleon R, Hu Z, Chebotarov D, Tai S, Wu Z, Li M, Zheng T, Fuentes RR, Zhang F, Mansueto L, Copetti D, Sanciangco M, Palis KC, Xu J, Sun C, Fu B, Zhang H, Gao Y, Zhao X, Shen F, Cui X, Yu H, Li Z, Chen M, Detras J, Zhou Y, Zhang X, Zhao Y, Kudrna D, Wang C, Li R, Jia B, Lu J, He X, Dong Z, Xu J, Li Y, Wang M, Shi J, Li J, Zhang D, Lee S, Hu W, Poliakov A, Dubchak I, Ulat VJ, Borja FN, Mendoza JR, Ali J, Li J, Gao Q, Niu Y, Yue Z, Naredo MEB, Talag J, Wang X, Li J, Fang X, Yin Y, Glaszmann JC, Zhang J, Li J, Hamilton RS, Wing RA, Ruan J, Zhang G, Wei C, Alexandrov N, McNally KL, Li Z, Leung H. Genomic variation in 3,010 diverse accessions of Asian cultivated rice. Nature. 2018;557:43–49. doi: 10.1038/s41586-018-0063-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XQ, Pang YL, Zhang J, Zhang Q, Tao YH, Feng B, Zheng TQ, Xu JL, Li ZK. Genetic background effects on QTL and QTL × environment interaction for yield and its component traits as revealed by reciprocal introgression lines in rice. Crop J. 2014;2:345–357. doi: 10.1016/j.cj.2014.06.004. [DOI] [Google Scholar]
- Wang Y, Zang JP, Sun Y, Ali J, Xu JL, Li ZK. Identification of genetic overlaps for salt and drought tolerance using simple sequence repeat markers on an advanced backcross population in rice. Crop Sci. 2012;52:1583–1592. doi: 10.2135/cropsci2011.12.0628. [DOI] [Google Scholar]
- Wu Y, Wang Y, Mi XF, Shan JX, Li XM, Xu JL, Lin HX. The QTL GNP1 encodes GA20ox1, which increases grain number and yield by increasing cytokinin activity in rice panicle meristems. PLoS Genet. 2016;12:e1006386. doi: 10.1371/journal.pgen.1006386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu JL, Wang Y, Zhang F, Wu Y, Zheng TQ, Wang YH, Zhao XQ, Cui YR, Chen K, Zhang Q, Lin HX, Li JY, Li ZK. SS1 (NAL1)- and SS2-mediated genetic networks underlying source-sink and yield traits in rice (Oryza sativa L.) PLoS One. 2015;10:e0132060. doi: 10.1371/journal.pone.0132060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano K, Yamamoto E, Aya K, Takeuchi H, Lo PC, Hu L, Yamasaki M, Yoshida S, Kitano H, Hirano K, Matsuoka M. Genome-wide association study using whole-genome sequencing rapidly identifies new genes influencing agronomic traits in rice. Nat Genet. 2016;48:927–934. doi: 10.1038/ng.3596. [DOI] [PubMed] [Google Scholar]
- Zang JP, Sun Y, Wang Y, Yang J, Li F, Zhou YL, Zhu LH, Xu JL, Li ZK. Dissection of genetic overlap of salt tolerance QTLs at the seeding and tillering stages using backcross introgressive lines in rice. Sci China Life Sci. 2008;51:583–591. doi: 10.1007/s11427-008-0081-1. [DOI] [PubMed] [Google Scholar]
- Zhang GH, Li SY, Wang L, Ye WJ, Zeng DL, Rao YC, Peng YL, Hu J, Yang YL, Xu J, Ren DY, Gao ZY, Zhu L, Dong GJ, Hu XM, Yan MX, Guo LB, Li CY, Qian Q. LSCHL4 from japonica cultivar, which is allelic to NAL1, increases yield of indica super rice 93-11. Mol Plant. 2014;7:1350–1364. doi: 10.1093/mp/ssu055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q. Strategies for developing green super rice. Proc Natl Acad Sci U S A. 2007;104:16402–16409. doi: 10.1073/pnas.0708013104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. Comparison of grain number per panicle, flag leaf width and plant height between xian and geng rice (Oryza sativa L.) subpopulations in 198 accessions. (a–c) Cyan and orange colors indicate xian and geng, respectively. The *, **, *** denotes significance of Student’ s t test at P < 0.05, P < 0.01, and P < 0.001, respectively.
Additional file 2: Figure S2. Population structure analyses of 198 accessions. (a) Scree plot from GAPIT showing the selection of PCs for association study. (b) PCA plot based on the screen plot in the rice diversity panel. (c) Scree plot from STRUCTURE showing the selection of Q. (d) Bayesian clustering of 198 accessions using STRUCTURE program.
Additional file 3 : Table S1. Details of the accessions investigated and two grain number gene haplotypes.
Additional file 4 : Table S2. Nucleotide diversity of GNP1 and NAL1 genes. S, segregation sites; π, average number of nucleotide differences per site between random two sequences; θ, Watterson estimator; D1, Tajima’s D; D2, Fu and Li’D; D3, Fu and Li’F.
Additional file 5 : Figure S3. Sliding-windows analysis of GNP1 (a) and NAL1 (b) nucleotide diversity in O. sativa ssp. xian, O. sativa ssp. geng and O. rufipogon. The y-axis represents nucleotide diversity (π), and the genomic structure is shown at the bottom, where the black boxes indicate exons and the white boxes indicate introns and other noncoding regions.
Additional file 6 : Figure S4. Protein diversity of GNP1 (a). Comparison of grain number per panicle, and plant height between two protein types in xian subpopulations in 198 panel (b–c). Cyan and orange colors indicate GNP1-P1 and GNP1-P2, respectively. The *, **, *** denotes significance of Student’ s t test at P < 0.05, P < 0.01, and P < 0.001, respectively.
Additional file 7 : Figure S5. Expression level of NAL1 and GNP1 haplotypes in young panicle at the panicle initiation stage. (a) Expression level of GNP1–1 and GNP1–6 in xian subpopulation. (b) Expression level of NAL1–2, NAL1–4 and NAL1–6 in geng subpopulation. (c) Expression level of NAL1–1, NAL1–2 and NAL1–3 in xian subpopulation. Error bars indicate SD; Letters are ranked by Duncan’s test at P < 0.05. The * denotes significance of Student’ s t test at P < 0.05.
Additional file 8 : Figure S6. Protein diversity of NAL1 (a). Comparison of grain number per panicle, and flag leaf width among three protein types in xian and geng subpopulations, respectively (b–e). Cyan, orange, and lightpink colors indicate NAL1-P1, NAL1-P2 and NAL1-P3, respectively. The *, **, *** denotes significance of Student’ s t test at P < 0.05, P < 0.01, and P < 0.001, respectively.
Additional file 9 : Table S3. Variance components and heritabilty estimated by multiple-site analysis.
Additional file 10 : Figure S7. Comparison of grain number per panicle, flag leaf width and effective panicle number between R-type and H-type alleles located in the third exon of NAL1 gene in 3 K panel in xian and geng subpopulation, respectively. ***, P < 0.001 (Student’ s t test).
Additional file 11 : Table S4. Distribution of protein mutation sites diversity at amino acid position 233 of NAL1 gene. R, arginine; H, histidine.
Additional file 12 : Figure S8. Geographic distributions of different haplotypes of NAL1 and GNP1 among the 28 areas sampled.
Additional file 13 : Table S5. Details of the wild rice (Oryza rufipogon).
Additional file 14 : Table S6. Sequencing primers used in this study.
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its supplementary information files.