Significance
This study reveals that protein degradation plays a major role in the survival of the opportunistic bacterial pathogen Pseudomonas aeruginosa. Loss of multiple proteases, better known for their roles in proteostasis in response to stresses such as heat shock, accelerates cell death during growth arrest. This finding, coupled to the fact that the accumulation of misfolded and aggregated proteins in aging in eukaryotic cells is well appreciated to contribute to cellular damage and senescence, suggests that a general role for proteases in preserving bacterial proteostasis during aging has been overlooked. Our findings have implications for the study and treatment of infectious disease and highlight potentially conserved functions for proteases in combatting aging from bacteria to humans.
Keywords: Pseudomonas aeruginosa, growth arrest, survival, proteostasis, FtsH
Abstract
When nutrients in their environment are exhausted, bacterial cells become arrested for growth. During these periods, a primary challenge is maintaining cellular integrity with a reduced capacity for renewal or repair. Here, we show that the heat-shock protease FtsH is generally required for growth arrest survival of Pseudomonas aeruginosa, and that this requirement is independent of a role in regulating lipopolysaccharide synthesis, as has been suggested for Escherichia coli. We find that ftsH interacts with diverse genes during growth and overlaps functionally with the other heat-shock protease-encoding genes hslVU, lon, and clpXP to promote survival during growth arrest. Systematic deletion of the heat-shock protease-encoding genes reveals that the proteases function hierarchically during growth arrest, with FtsH and ClpXP having primary, nonredundant roles, and HslVU and Lon deploying a secondary response to aging stress. This hierarchy is partially conserved during growth at high temperature and alkaline pH, suggesting that heat, pH, and growth arrest effectively impose a similar type of proteostatic stress at the cellular level. In support of this inference, heat and growth arrest act synergistically to kill cells, and protein aggregation appears to occur more rapidly in protease mutants during growth arrest and correlates with the onset of cell death. Our findings suggest that protein aggregation is a major driver of aging and cell death during growth arrest, and that coordinated activity of the heat-shock response is required to ensure ongoing protein quality control in the absence of growth.
Most of our knowledge of bacterial physiology is derived from studying exponentially growing cells in nutrient-replete environments. While this study has provided a rich understanding of the complex and diverse processes at play during cellular growth and division, in many ways its scope is limited to what is likely just a fleeting period in the life of a bacterium. Awareness is mounting that most bacteria spend a majority of their lives not growing and dividing, as they quickly exhaust the nutrients in their environment and enter into a state of growth arrest (1–3). Yet comparatively few studies have investigated the molecular processes required for survival in this state, even though they are as important for evolutionary success as the ability to grow rapidly when nutrients once again become available (4). Moreover, successful strategies to survive extended periods of growth arrest can be important drivers in the emergence of antibiotic tolerance and resistance (5, 6), as well as in the establishment and persistence of biofilm communities (7–10). Thus, a better understanding of the underlying genetics and molecular mechanisms that promote survival during growth arrest is crucial for understanding how bacteria thrive in diverse ecological and clinical contexts.
Studies in Escherichia coli and other model species have provided valuable insight into the diverse strategies bacteria have evolved to survive growth arrest (3, 11, 12). These strategies are often dependent on the metabolic and biochemical capabilities of individual species, as well as on the nutrients available in the environment, highlighting the need to study growth arrest in evolutionarily and metabolically diverse bacteria under distinct growth-arrest regimes. For example, bacteria use varied alternative substrates as sources of electron donors or acceptors to generate ATP during growth arrest, and reroute their metabolic pathways in diverse ways to improve the efficiency of ATP generation (3). This energy is needed to protect the integrity of cellular components, such as nucleic acids and proteins, that cannot be easily replaced by nutrient-limited cells and are essential for survival (2, 3). However, how cells efficiently coordinate maintenance needs in the face of slim energy resources is poorly understood.
To understand the mechanisms underpinning the growth-arrested state, we have used the opportunistic pathogen Pseudomonas aeruginosa as a model organism (13–16). P. aeruginosa is well-adapted for survival in a variety of nutrient-limited or otherwise hostile environments, ranging from open ocean and freshwater sources, to the interior of surface-attached biofilms, to chronic wounds and the lungs of patients with cystic fibrosis (17, 18). Despite superficial differences, these environments often share a common physiological constraint: They can be limited for electron donors or acceptors, compelling organisms that survive within them to hone sophisticated strategies to cope with periods of energy-limited growth arrest (19–25).
Previously, we identified the ATP-dependent membrane protease FtsH as one of only a few genes that confer a general fitness advantage to P. aeruginosa during energy-limited growth arrest, regardless of the limitation that prompted entry into this state (15). FtsH has been well-studied during growth, and is involved in quality control of membrane proteins (26), regulation of the heat-shock response (27), and fine-tuning of lipopolysaccharide (LPS) levels by regulated degradation of the LPS biosynthetic enzymes LpxC and KdtA (28–34). This last activity is essential in E. coli, as loss of ftsH results in unbalanced lipid synthesis and lethal overproduction of LPS (28). Although many of the growth-related roles of FtsH are well characterized, its role during growth arrest in any organism has received less attention (35–37).
Recently, two studies (38, 39) attributed a role for FtsH in growth arrest survival through a novel cell death pathway in E. coli, in which dysregulation of an outer membrane (OM) signaling system putatively inhibits LpxC degradation by FtsH, resulting in overproduction of LPS and cell death upon nutrient depletion and growth arrest. Based on these results, we reasoned that one possibility FtsH is so important for survival of P. aeruginosa might be that FtsH plays a regulatory role during growth arrest by degrading LpxC, similar to its role in E. coli. However, given that LpxC levels are not regulated by FtsH in P. aeruginosa (37, 40), the plausibility of such a function was unclear.
Alternatively, several studies have pointed to a role for the heat-shock response during growth arrest in E. coli, suggesting that aggregation of misfolded or damaged proteins may become a critical problem for nongrowing cells (41–46). As a highly conserved component of the heat-shock response in many bacteria (32), we reasoned that FtsH might degrade these misfolded or damaged proteins during growth arrest in P. aeruginosa. Indeed, protein aggregation is a hallmark of aging across a wide range of cellular systems, from humans to bacteria (47–53), although whether protein aggregation represents a strictly pathological process has recently come into question (54–60). In this study, we sought to distinguish between these possibilities for the role of FtsH during growth arrest in P. aeruginosa using a genetic and cell biological approach.
Results
FtsH Maintains Cell Integrity during Growth Arrest.
We use the general term “growth arrest” when referring to two states: When cells enter stationary phase after growth in rich medium (lysogeny broth, LB), and when cells growing exponentially in LB are shifted to a buffered minimal medium devoid of organic carbon (carbon starvation medium, CSM). We refer to the former growth-arrested state as “stationary phase” and the latter as “carbon starvation.”
We constructed an isogenic markerless deletion of the ftsH ORF in P. aeruginosa strain UCBPP-PA14 and observed the morphology of the mutant strain during growth and growth arrest. Loss of ftsH did not affect cell length compared to WT during exponential growth in LB at 37 °C (SI Appendix, Fig. S1A, 3 h). Cell length decreased gradually as cells entered stationary phase after ∼8 h of growth, reaching a minimum of 2.8 µm for WT and 3.2 µm for ΔftsH, respectively (SI Appendix, Fig. S1A, 8 h). WT cell length did not change over the next 20 h in stationary phase while ΔftsH slowly increased in length to an average of 3.8 µm (SI Appendix, Fig. S1A, 28 h).
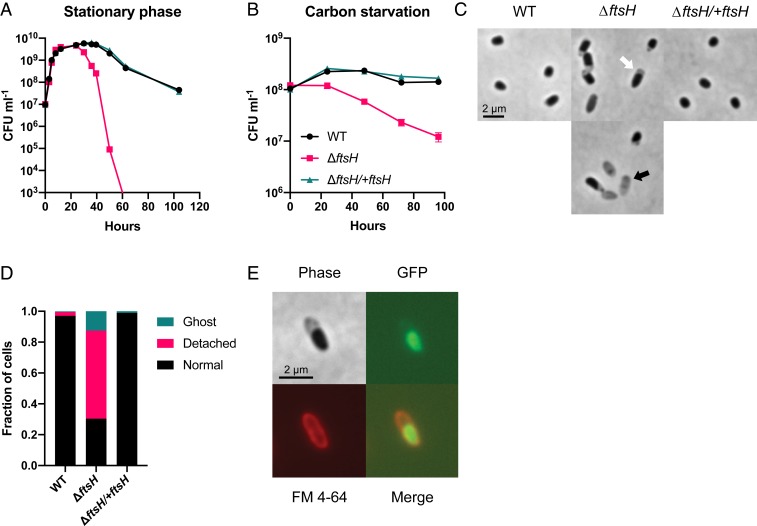
Death occurred after extended incubation in stationary phase for both WT and ΔftsH, although ΔftsH began to die earlier and to a greater extent than WT, becoming effectively unculturable after 60 h of total incubation time (Fig. 1A). Death of ΔftsH also occurred after >24 h of growth arrest in CSM, with ∼5- to 10-fold reduced viability after 96 h (Fig. 1B) and ∼100-fold reduced viability after 10 d (SI Appendix, Fig. S2A). In contrast, WT maintained full viability after 96 h of carbon starvation (Fig. 1B) and had ∼twofold reduced viability after 10 d (SI Appendix, Fig. S2A). Loss of ftsH also caused death when cells were starved for oxygen as a terminal electron acceptor or ammonium as a nitrogen source (SI Appendix, Fig. S2 B and C). Integration of the ftsH ORF with its native promoter at the glmS locus on the chromosome fully rescued the viability defects in growth arrest, confirming the role of FtsH in survival (the complemented strain ΔftsH/+ftsH) (Fig. 1 A and B).
Fig. 1.
FtsH maintains cell integrity during growth arrest. Loss of ftsH exacerbates cell death during stationary phase (A) and carbon starvation (B). Viability was below the limit of detection (∼3 × 102 CFU mL−1) for ΔftsH at 60 h in A. Representative data from at least three independent experiments are shown in A and the averages and SD of biological replicates are shown in B (n = 3). (C) Characteristic morphology of ΔftsH cells after 24 h of carbon starvation. The white arrow indicates a “detached” inner membrane and the black arrow indicates a “ghost cell.” (D) Quantification of the cellular morphologies described in C. A minimum of 300 cells were counted for each strain. (E) OM staining with FM 4-64 and cytoplasmic expression of GFP confirms that detachment occurs between the IM and OM.
Most ΔftsH cells lysed following death, as exhibited by a decrease in optical density and the appearance of only scattered “ghost” cells by microscopy (Fig. 1C and SI Appendix, Fig. S3). Before significant death and lysis of ΔftsH occurred, however, we observed many intact cells with what appeared to be a “detached” inner membrane (IM) (Fig. 1C). This cell morphology was largely absent in the WT and the complemented strain at the same time points (Fig. 1 C and D), and none of the strains displayed this morphology during growth. We stained carbon-starved ΔftsH cells expressing cytoplasmic GFP with the OM dye FM 4-64 to confirm separation between the OM and IM (Fig. 1E). The morphology of growth-arrested ΔftsH cells was strikingly similar to an LPS-overproducing strain of E. coli that also experienced rapid cell death during growth arrest (38). However, we ruled out the possibility of LPS overproduction as the cause of death of ΔftsH in our experiments, as LPS levels were similar between WT and ΔftsH during growth and growth arrest and cation supplementation did not prevent death (SI Appendix, Supplementary Text and Fig. S3). We conclude that FtsH is required to maintain cellular integrity during growth arrest, and that impaired cellular integrity in the absence of FtsH is not due to overproduction of LPS.
Identification of Genetic Interactions with ftsH.
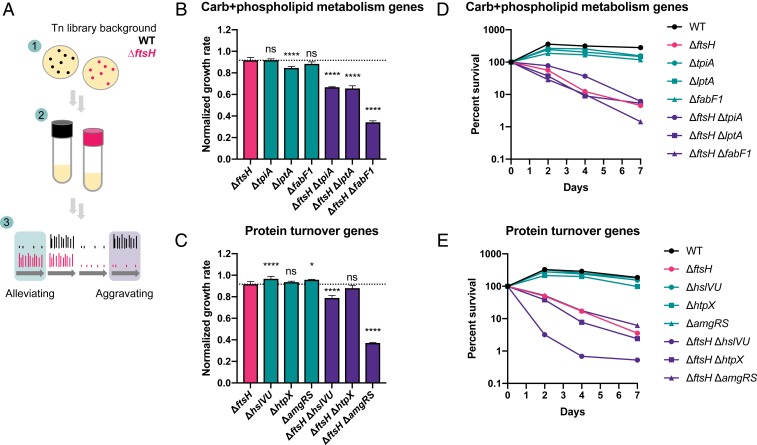
We performed transposon insertion sequencing (Tn-seq) in the ΔftsH background to identify genes that interact with ftsH on a genome-wide scale (61). In Tn-seq, a pooled transposon mutant library is grown under a condition of interest and then sequenced at the transposon–genome junction to identify where transposons are inserted and to quantify their abundance in the total population. Mutant abundance is then compared to a control population to determine the relative fitness of insertion at a particular locus under the condition of interest.
Initially, we attempted Tn-seq under growth-arrest conditions in the ΔftsH background using an experimental design similar to our previous study in the WT background (15). However, the reduced viability of the ΔftsH background during growth arrest resulted in contaminating growth of the trace amounts of E. coli donor carried over from the original conjugation (Materials and Methods). To avoid this problem, we instead compared insertions between the WT and ΔftsH transposon libraries after growth in liquid LB at 37 °C (Materials and Methods and Fig. 2A). We identified 36 genes that had a growth-aggravating interaction with ftsH: That is, mutations in these genes negatively affected fitness specifically in the ΔftsH background, which was defined as >10-fold reduced insertions in the ΔftsH background relative to the WT background (P < 0.05) (SI Appendix, Table S4 and Dataset S1). Aggravating interactions were detected among genes involved in cell envelope biogenesis and OM functions, as well as at the interface of carbohydrate and glycerophospholipid metabolism (SI Appendix, Fig. S4 and Table S4). Mutations in genes putatively involved in LPS biosynthesis (ssg, wapH), glycolysis (tpiA), fatty acid biosynthesis (fabF1), and phospholipid metabolism (lptA) were growth-aggravating in the ΔftsH background, suggesting that although FtsH does not regulate LpxC in P. aeruginosa, it could still play an integral role in glycerophospholipid metabolism and cell envelope biogenesis.
Fig. 2.
Mutations in metabolic and protein turnover genes aggravate growth and survival defects of ΔftsH. (A) Experimental design of Tn-seq screen. 1) Transposon libraries were generated in the WT (15) and ΔftsH backgrounds by selection on LB agar plates plus antibiotics at 37 °C. 2) An aliquot of each library was grown approximately seven to eight generations in liquid LB. 3) The read counts of the WT and ΔftsH libraries were compared to identify genes that are alleviating or aggravating for growth in the ΔftsH background. (B and C) Growth rate of carbohydrate and phospholipid metabolism and protein turnover mutants relative to ΔftsH in LB at 37 °C. All growth rates were normalized to the WT growth rate in LB at 37 °C and the dotted line indicates the normalized ΔftsH growth rate. Results of one-way ANOVA with Dunnett’s multiple comparisons test are denoted as follows: ns, not significant; *P < 0.05; ****P < 0.0001. Error bars show SD of biological replicates (n ≥ 3). (D and E) Survival of mutants during carbon starvation. Representative data from at least two independent experiments are shown.
Insertions in genes involved in posttranslational modification and protein turnover were also aggravating for growth, including the heat-shock proteases encoded by hslVU and htpX (62, 63) and the SsrA-binding protein encoded by smpB (64). We detected an aggravating interaction with the membrane-stress responsive two-component system encoded by amgRS, as observed previously in the P. aeruginosa strain PAO1 (65). AmgRS is a homolog of the CpxRA two-component system that regulates the envelope stress response in E. coli (66, 67), and loss of amgRS or ftsH in P. aeruginosa results in hypersensitivity to aminoglycoside-induced protein misfolding (65). Notably, htpX is regulated by AmgRS in P. aeruginosa and contributes to aminoglycoside resistance in conjunction with PA14_72930, another AmgRS-regulated gene of unknown function that was also aggravating for growth in our screen (SI Appendix, Table S4 and Dataset S1) (65).
We made isogenic markerless deletions of growth-aggravating genes from different functional categories to validate their interactions with ftsH. To this end, we made double mutants of ftsH with tpiA, lptA, and fabF1 (involved in “carbohydrate and phospholipid metabolism”) (SI Appendix, Fig. S4) and with hslVU, htpX, and amgRS (involved in “protein turnover”). The double mutants had significantly reduced growth rates in LB at 37 °C compared to the ΔftsH single mutant, with the exception of ΔftsH ΔhtpX (Fig. 2 B and C). The growth defects were greatest for ΔftsH ΔfabF1, with a 63% reduction in growth rate compared to ΔftsH, and for ΔftsH ΔamgRS, with a 60% reduction in growth rate (Fig. 2 B and C). Together, these data validate the results of our Tn-seq experiment and confirm a role for FtsH in promoting diverse cellular processes during growth.
Importantly, all double mutants, with the exception of ΔftsH ΔhslVU, showed similar survival profiles to the ΔftsH single mutant, indicating that while most of these genes are vital for growth in the ΔftsH background they do not affect survival during growth arrest (Fig. 2 D and E). In contrast, ΔftsH ΔhslVU had a survival defect ∼10-fold more severe than ΔftsH after 3 d of carbon starvation (Fig. 2E), indicating that HslVU plays a backup role to FtsH during growth and growth arrest. This result prompted us to systematically investigate the role of the ATP-dependent heat-shock proteases during growth arrest.
Heat-Shock Proteases Promote Survival during Growth Arrest.
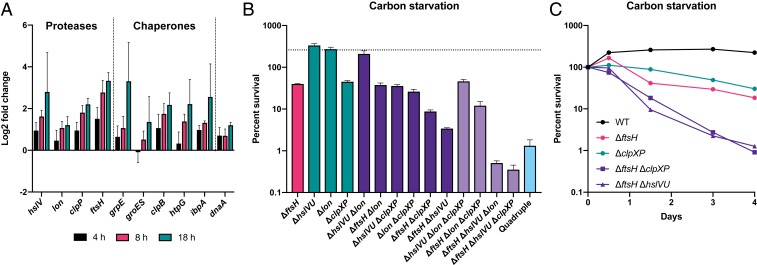
The heat-shock proteases classically act in conjunction with protein-folding chaperones to maintain proteostasis after exposure to high temperature, ethanol, or heavy metal stress (68, 69). To more broadly characterize the role of the heat-shock response during growth arrest, we measured expression of the major ATP-dependent proteases and chaperones in WT PA14 during carbon starvation by qRT-PCR (Fig. 3A). Following a substantial decrease in expression within the first hour of carbon starvation, most heat-shock regulated genes are induced over the next 18 h, whereas expression of the growth-related gene dnaA does not increase over time.
Fig. 3.
Heat-shock proteases overlap functionally to promote survival during carbon starvation. (A) The heat-shock response is induced during carbon starvation. Gene expression at the indicated time points is relative to 1 h after cells were shifted to CSM and is normalized to the expression of the housekeeping gene oprI. Expression of the DNA replication initiator dnaA was used as a negative control. Error bars show SD of biological replicates (n = 3). (B) Percent survival of protease mutants on day 3 of carbon starvation. The dotted line indicates WT survival. Error bars show SD of biological replicates (n = 3). (C) Survival time course of select mutants during carbon starvation. Representative data from at least two independent experiments are shown.
We predicted that further genetic perturbation of the protease network in P. aeruginosa would exacerbate survival defects during growth arrest. To test this prediction, we made all combinatorial deletions of the ATP-dependent protease-encoding genes ftsH, clpXP, lon, and hslVU, and assessed survival of the mutant strains during carbon starvation (Fig. 3B). Single deletion of lon, like hslVU, did not cause a survival defect after 3 d of carbon starvation, while deletion of clpXP caused a similar survival defect as ΔftsH (Fig. 3 B and C). We did not detect a fitness defect during carbon starvation for clpX or clpP transposon mutants in our previous study because there were few insertions in these genes in the WT transposon library (15) (Dataset S2). Although ΔclpXP had a similar growth rate as ΔftsH at 37 °C (see below), we noticed that ΔclpXP colonies took longer to appear upon plating after growth arrest. We reasoned that the reduced insertion rate at the clpXP locus is due to an extended lag time of mutant cells before beginning to grow (70), which leads to these mutants being outcompeted in the early stages of growth of the transposon libraries. These results emphasize that while Tn-seq is a powerful technique for studying single mutant fitness on a genome-wide scale, it frequently lacks resolving power for studying stress-related phenotypes of mutants with even a minor growth disadvantage under nonstressful conditions (71).
Unlike in the ΔftsH background, deletion of hslVU did not affect survival in the Δlon or ΔclpXP backgrounds, indicating that hslVU specifically backs up FtsH activity (Fig. 3B). Deletion of lon did not affect survival in the ΔftsH background, but exacerbated death in the ΔftsH ΔhslVU background, indicating that Lon plays a tertiary role in growth arrest survival (Fig. 3B). Deletion of clpXP also exacerbated death in the ΔftsH ΔhslVU background, as well as in the ΔftsH background (Fig. 3 B and C). Thus, ftsH and clpXP appear to contribute independently to survival during growth arrest. Finally, the ΔftsH ΔhslVU Δlon ΔclpXP quadruple mutant had a survival defect similar to the more sensitive triple mutants. However, suppressor mutations arise rapidly in this strain, making it difficult to determine an accurate growth rate and survival phenotype for the parent strain.
Heat and Alkaline pH Exacerbate Death during Growth Arrest.
The functional overlap of the heat-shock proteases during growth arrest suggests that they might act collectively to mitigate a common stress, which we hypothesized was caused by the accumulation of misfolded proteins. Thus, we tested the functional overlap between these proteases in two other conditions known to cause protein misfolding stress: Growth at high temperature or alkaline pH (72).
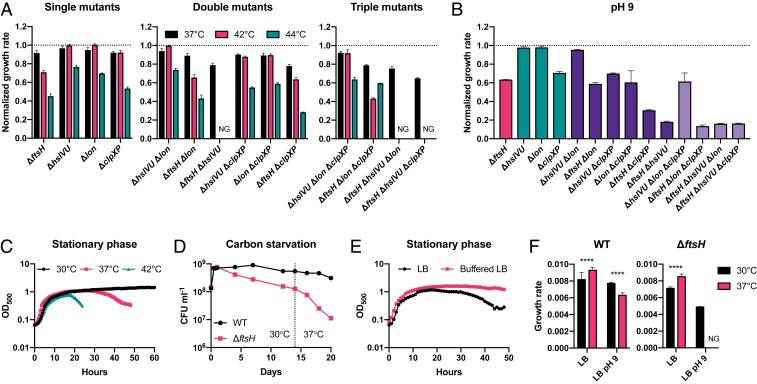
Functional overlap of the proteases during growth at high temperature was similar to that of growth arrest (Fig. 4A). While ΔftsH had only a minor growth defect compared to WT in LB at 37 °C (∼8% reduced growth rate), this defect became more pronounced at 42 °C and 44 °C (∼29% and ∼41% reduced growth rate, respectively). Once again, we observed functional redundancy between ftsH and hslVU, with loss of both genes resulting in no growth at 42 °C and 44 °C. Unlike in growth arrest, however, the temperature sensitivity of ΔftsH was greater than that of ΔclpXP, with the latter displaying temperature-sensitive growth only at 44 °C. The ΔhslVU and Δlon single mutants were also temperature-sensitive only at 44 °C. The pattern of overlap during growth in LB at pH 9 was strikingly similar to that of growth arrest (compare Fig. 4B and Fig. 3B), supporting the hypothesis that these two conditions invoke a common cellular crisis.
Fig. 4.
Heat and alkaline pH exacerbate growth and survival defects of protease mutants. (A) Growth rate of protease mutants normalized to the WT growth rate in LB at different temperatures. Error bars show SD of biological replicates (n ≥ 3). NG, no growth. (B) Normalized growth rate in LB pH 9 at 30 °C. Error bars show SD of biological replicates (n = 3). (C) Survival of ΔftsH during stationary phase is reduced at higher temperature. (D) Temperature upshift from 30 °C to 37 °C during carbon starvation accelerates the rate of death of ΔftsH. (E) Incubation in Mops buffered LB at 37 °C mitigates cell death of ΔftsH during stationary phase. (F) Higher temperature impedes growth of both WT and ΔftsH in alkaline pH. Growth is faster at 37 °C than at 30 °C for both WT and ΔftsH in neutral LB, but slower at 37 °C than at 30 °C in LB pH 9. Error bars show SD of biological replicates (n = 3). Unpaired t test: ****P < 0.0001; NG, no growth.
To determine whether the reduced growth rate of protease mutants at high temperature was caused by uniformly slower growth of individual cells or due to an increased rate of cell death, we used time-lapse microscopy to measure the growth rates of single cells of WT, ΔftsH, and ΔclpXP at high temperature and compared these rates to those derived from optical density measurements at the population level (SI Appendix, Fig. S5). The single-cell data for ΔftsH qualitatively matched the rates of biomass increase observed in bulk, indicating that a growth-rate defect, rather than an increased cell death rate, accounted for the apparent population growth-rate differences. In contrast, the single-cell data for ΔclpXP suggested that cell death may significantly contribute to the growth-rate defect observed in bulk at high temperature.
We reasoned that if growth arrest, high temperature, and alkaline pH indeed invoke a common stress, then these conditions should act synergistically to exacerbate growth and survival defects of protease mutants. Indeed, increased temperature correlated with earlier death of ΔftsH and ΔclpXP in stationary phase (Fig. 4C and SI Appendix, Fig. S6A), and a shift from 30 °C to 37 °C increased the death rate of ΔftsH during carbon starvation (Fig. 4D). Death of ΔftsH and ΔclpXP in stationary phase was delayed in buffered LB (Fig. 4E and SI Appendix, Fig. S6B), indicating that a combination of growth arrest and alkaline pH likely causes the dramatically greater loss of viability in stationary phase LB cultures compared to carbon starvation in buffered minimal medium (compare Fig. 1 A and B) (73). Higher temperature also exacerbated growth defects at alkaline pH, as WT grew slower at 37 °C than at 30 °C in LB pH 9, and ΔftsH failed to grow at 37 °C (Fig. 4F). In contrast, both strains grew faster at 37 °C than at 30 °C in neutral LB. Based on these data, we conclude that growth arrest, high temperature, and alkaline pH are synergistic stresses that require a coordinated response of the heat-shock proteases in order for cells to grow and survive.
Heat-Shock Proteases Delay Protein Aggregation during Growth Arrest.
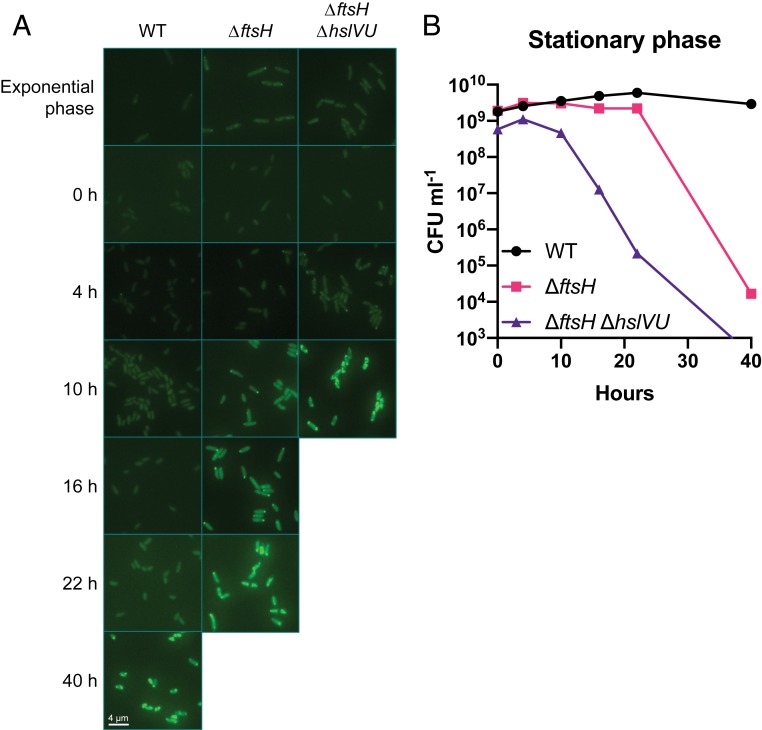
To indirectly assess the extent of protein misfolding during growth arrest, we visualized protein aggregates in single cells using fluorescently tagged IbpA, a small heat-shock protein that has been shown to bind to endogenous protein aggregates (74). IbpA fusion proteins constructed with monomeric fluorescent protein derivatives have been validated as a reliable reporter for monitoring protein aggregates in E. coli (58). A gene encoding IbpA-mVenus was cloned into the chromosome under the control of its native promoter in P. aeruginosa; when expressed, it rapidly formed fluorescent foci in both the WT and protease mutants upon heat shock (SI Appendix, Fig. S7). During exponential growth, and as cells entered stationary phase, IbpA-mVenus signal was predominantly low and diffusely distributed throughout the cytoplasm, indicating that protein aggregation was minimal under nutrient-replete growth conditions (Fig. 5A). Importantly, diffuse labeling during growth demonstrates that our reporter is not inherently aggregation-prone, a common artifact in these types of experiments (58, 75). Both ΔftsH and ΔftsH ΔhslVU developed one to two foci per cell during stationary phase that became brighter and more numerous over time, with an accelerated rate of foci formation in the double mutant. This pattern was also observed in WT, albeit with a delayed onset relative to the protease mutants. For all strains, the appearance of numerous foci correlated with cell death (Fig. 5B), and following death many cells lost fluorescence. Thus, aggregate formation is delayed, rather than completely prevented, by the activity of heat-shock proteases during growth arrest, and aggregate formation correlates with cell death in both the WT and protease-deficient strains.
Fig. 5.
Heat-shock proteases delay protein aggregation during growth arrest. (A) Fluorescence of cells expressing the IbpA-mVenus reporter during exponential growth and stationary phase at 37 °C; 0 h corresponds to the start of stationary phase (after 8 h of growth starting from an OD500 of 0.01). All images were taken using the same exposure time (400 ms) and are displayed with the same brightness and contrast. (B) Viability measurements corresponding to the time points in A. Viability was below the limit of detection (∼3 × 102 CFU mL−1) for ΔftsH ΔhslVU at 40 h. Representative data from two independent experiments are shown.
Discussion
Bacteria must adequately maintain their cellular integrity during extended periods of starvation in order to survive and to resume growth when nutrients become available. In this study, we describe a role for FtsH in promoting diverse metabolic processes during growth of P. aeruginosa and in maintaining cellular integrity during growth arrest. Unlike its posited role in E. coli (38, 39), our findings suggest that FtsH does not play a regulatory role in growth arrest survival of P. aeruginosa per se. Rather, FtsH appears to act in conjunction with the other major ATP-dependent heat-shock proteases to ensure ongoing protein quality control in the absence of growth. Protein aggregation appears to become a major problem during growth arrest, and could be caused by numerous factors, including misfolding of nascent peptides due to reduced abundance or activity of cotranslational folding chaperones, or misfolding of bulk proteins over time due to sustained and cumulative damage in the absence of growth and turnover.
We observed differing phenotypic interactions between ftsH and diverse functional classes of genes during growth and growth arrest of P. aeruginosa, raising the question of whether FtsH has divergent roles in these two states. Mutations in carbohydrate and lipid metabolic genes exacerbated the growth defect of ΔftsH but had no effect on ΔftsH survival during growth arrest (Fig. 2 B and D), while mutations in heat-shock proteases exacerbated both the growth and survival defects of ΔftsH (Fig. 2 C and E). What is the connection between FtsH and diverse metabolic pathways during growth? One possibility is that cells are more sensitive to protein misfolding and aggregation in the absence of FtsH-mediated protein quality control, which could interfere with the activity of growth-promoting metabolic enzymes. Evidence for this possibility is found in Mycobacteria, where disruption of proteostasis results in misfolding and reduced activity of multimodular lipid synthases (53). Another possibility is that, during growth, FtsH might still control flux through phospholipid and LPS metabolism in P. aeruginosa independent of a direct effect on the stability of LpxC. Recent studies have revealed complex cross-talk between phospholipid and LPS metabolism in E. coli, and point to a more nuanced role for FtsH in sensing and controlling flux through these pathways (76, 77). Perhaps an analogous role for FtsH exists in P. aeruginosa, with the regulatory architecture having diverged from that of E. coli.
Altered regulation of the heat-shock sigma factor σ32 in the absence of FtsH could also play a role during growth. During growth at moderate temperatures, σ32 levels are kept low by FtsH-mediated proteolysis in diverse species (31). Upon heat shock, σ32 becomes stabilized and induces expression of the heat-shock regulon. Overexpression of σ32 under noninducing conditions was shown to inhibit growth of Caulobacter crescentus by reprogramming gene expression away from a growth-promoting regime toward one favoring repair and maintenance (78). A similar problem could occur in P. aeruginosa, where loss of FtsH causes a global change in gene expression indirectly via elevated σ32. In support of this possibility, overexpression of σ32 down-regulates expression of multiple fatty acid biosynthetic genes in PA14, including fabF1 (79), whose deletion caused the most severe growth defect in the ΔftsH background in our experiments (Fig. 2B). If growth-promoting pathways are generally down-regulated in ΔftsH, then further perturbation to these pathways could severely compromise growth. This could explain why lipid metabolism genes are important for growth of ΔftsH yet dispensable for survival, as fatty acid and phospholipid synthesis would no longer be required once cells have stopped growing.
Like FtsH, ClpXP has also been implicated in adaptation to and survival of growth arrest in diverse organisms (80–85). Genetic perturbation of protein-folding chaperones and σ32 in E. coli also causes a survival defect during growth arrest (41, 42). Furthermore, oxidatively modified proteins that accumulate during aerobic growth arrest cause specific induction of heat-shock genes in E. coli (43–46). Together, these findings point to a generalized role for the heat-shock response in ensuring survival of nongrowing bacteria. Outside of these studies, however, the importance of heat-shock genes is overlooked in the literature on growth arrest. Most reviews on bacterial growth arrest do not discuss a role for the heat-shock response, and most references to heat shock refer mainly to its namesake role in temperature stress. Strikingly, of all of the deletion strains of P. aeruginosa we have tested, loss of FtsH or ClpXP caused by far the most severe survival defect during carbon starvation (ref. 15 and this study). Survival was considerably worse in protease mutant strains than for a mutant lacking the stress sigma factor RpoS, which is generally considered one of the principle molecular components required for adaptation to nutrient depletion (15, 86).
In contrast to the literature on growth arrest in bacteria, the importance of the heat-shock response and proteostasis in cellular aging is well described and appreciated in eukaryotic organisms (47, 48, 87–91). Defects in proteostasis are a hallmark of aging in eukaryotes, and our finding that growth-arrested bacteria sustain similar types of cellular damage as aging eukaryotic cells points to an equally important role for proteostasis in bacterial fitness. The role of proteostasis during bacterial growth arrest is in addition to its role during “replicative aging” of nutrient-replete, growing cells (50, 92–95). The latter phenomenon occurs when one daughter cell inherits the old cell pole while the other daughter is rejuvenated. Based on the literature and our data, we propose that cellular aging during growth arrest is at least equal in importance to replicative aging. Both replicative aging and aging in growth arrest are likely driven by the accumulation of protein aggregates (50, 52), and this process may be magnified in growth arrest due to the reduced capability of cells for growth and protein turnover.
The communal role of the heat-shock proteases during growth arrest is in contrast to the many varied and specific regulatory roles they play in the physiology of diverse bacteria. Perhaps growth arrest, along with high temperature and alkaline pH, was an important driver in the evolution of this ancestral proteolytic network to maintain protein quality control. Following its development, this redundant network could have been co-opted by different bacteria to regulate diverse cellular processes in addition to proteostasis, such as regulation of LPS and phospholipid metabolism by FtsH (32, 77), the DNA damage response by Lon and HslVU (96, 97), and RpoS stability, DNA replication, and cell division by ClpXP (98–100).
Our findings point to proteostasis as an important, yet underexplored, aspect of growth arrest physiology. If surviving growth arrest is as important for bacterial success in diverse ecological niches as we presume, this should compel us to investigate the role of proteostasis more thoroughly in these niches as a counterpart to the more commonly studied stresses that induce proteotoxicity, such as heat, peroxide, or antibiotic exposure during rapid growth. Indeed, opportunistic human pathogens like P. aeruginosa are unlikely to experience temperature stress much higher than 37 °C during chronic infection, a temperature at which P. aeruginosa grows optimally. On the other hand, cells can quickly become energy-limited for growth during chronic infection, and what was once an optimal growth temperature could now impede the survival of aging cells. Thus, proteases of the heat-shock response may hold potential as a therapeutic target for the treatment of P. aeruginosa and related bacterial infections.
Materials and Methods
Strains and Growth Conditions.
The strains, plasmids, and primers used in this study are listed in SI Appendix, Tables S1–S3. E. coli and P. aeruginosa were grown in LB (Difco) or on LB agar plates with appropriate antibiotics at 30 °C or 37 °C for all cloning and strain construction purposes. All gene deletions in P. aeruginosa strain UCBPP-PA14 were made as described previously (15). The LpxC inhibitor CHIR-090 was purchased from ApexBio and the OM dye FM 4-64 was purchased from Life Technologies.
Growth Arrest Survival Assays.
For studying growth arrest caused by carbon starvation, cells were grown in LB at 37 °C to an optical density at 500 nm (OD500) between 0.5 and 1, pelleted, and resuspended in CSM, which is derived from a minimal medium without an added carbon source (13). In CSM, magnesium and sulfate are supplied in the form of MgCl2 and NaSO4, respectively, instead of the standard MgSO4. This allows for magnesium concentrations to be titrated independently of sulfate when necessary. Starved cells were incubated aerobically at 37 °C with shaking at 250 rpm. For oxygen starvation, cells were resuspended in the same minimal medium plus 40 mM pyruvate and transferred into an anoxic glove chamber (Coy) containing an atmosphere of 15% CO2, 80% N2, and 5% H2 and incubated anaerobically at 33 °C without shaking. For nitrogen starvation, cells were resuspended in the same minimal medium plus 10 mM glucose and without NH4Cl and incubated aerobically at 37 °C with shaking at 250 rpm. CFU mL−1 were determined by viability plating over time in all conditions.
For studying growth arrest in stationary phase, overnight cultures grown in LB were back-diluted to an OD500 of 0.01 in LB or LB plus 50 mM Mops (buffered LB) and grown at the indicated temperature with shaking at 250 rpm. CFU mL−1 were determined by viability plating over time. Most strains reached stationary phase after ∼8 h of growth.
LPS Measurements.
Overnight cultures grown in 5 mL LB were back-diluted in triplicate to an OD500 of 0.01 in 5 mL LB and grown at 37 °C with shaking at 250 rpm. An OD 0.5 mL−1 equivalence of each culture was pelleted after 3 h of growth (corresponding to an OD500 between 0.2 and 0.3; exponential phase) and after 25 h of growth (corresponding to an OD500 between 4 and 5; stationary phase), and stored at −80 °C. LPS was extracted from frozen pellets as described previously (38). Gels were stained with Pro-Q Emerald 300 LPS Gel Stain Kit (Molecular Probes) according to the manufacturer’s protocol and the LPS bands were visualized by UV transillumination.
Generation of the Transposon Library in ΔftsH.
A transposon library was created in the ΔftsH background in the same manner as described for the WT strain (15). Briefly, the ΔftsH strain of P. aeruginosa and the E. coli strain SM10λpir carrying the transposon-bearing plasmid pIT2 were resuspended in 1 mL of LB from overnight streak plates on LB agar or LB agar plus carbenicillin (100 µg mL−1), respectively. The resuspended cells were adjusted to an OD500 of ∼50 for P. aeruginosa and ∼100 for E. coli. Next, 100 µL of each OD-adjusted strain was mixed together in an Eppendorf tube and two 50-µL aliquots of this mix were plated on sterile 0.2-µm filter discs placed on an LB agar plate. The conjugation spots were allowed to dry in a safety cabinet with laminar flow for 15 min and then the plate was incubated at 37 °C for 2.5 h. Following incubation, the two conjugation spots were resuspended in 3.5 mL of LB and 100-µL aliquots of this resuspension were plated on 33 LB agar plus tetracycline (60 µg mL−1) and chloramphenicol (10 µg mL−1) plates to select for transposon insertion mutants. The plates were incubated 27 h at 37 °C. Each plate yielded ∼4,500 mutants for a total of ∼150,000 mutants. Following incubation, colonies from all plates were pooled and resuspended in LB plus 15% glycerol. The density of the pooled library was adjusted to an OD500 of 10 and stored as 1-mL aliquots at −80 °C.
Tn-Seq Sample Preparation and Data Analysis.
Frozen aliquots of the WT and ΔftsH transposon libraries were thawed on ice for 15 min and diluted in 50 mL of LB to a concentration of ∼6 × 106 CFU mL−1. The cultures were grown at 37 °C with shaking at 250 rpm for 3.5 h, corresponding to approximately three to four cell doublings. An aliquot of each culture was then diluted in 50 mL of LB plus chloramphenicol (10 µg mL−1) to a concentration of ∼6 × 106 CFU mL−1 and grown at 37 °C with shaking at 250 rpm for 3 h, corresponding to approximately four doublings. Following growth, an OD 4 mL−1 equivalence of each LB plus chloramphenicol culture was pelleted and stored at −80 °C. Genomic DNA was extracted from the frozen samples for high-throughput sequencing, as described previously (15). Sequencing was performed at the Millard and Muriel Jacobs Genetics and Genomics Laboratory at the California Insitute of Technology. Sequences were mapped to the UCBPP-PA14 genome sequence using Bowtie (101) and analyzed using the ARTIST Tn-seq analysis pipeline in MATLAB, as described previously (15, 102).
Growth-Rate Measurements.
For batch growth-rate experiments, precultures of each strain were grown at 37 °C in LB then pelleted and resuspended at an OD500 of 0.05 in LB. Next, 150-µL aliquots of the resuspensions were dispensed into a 96-well microtiter plate and 50 µL of mineral oil was added to each well to prevent evaporation. Plates were incubated in a BioTek plate reader at the indicated temperature (37 °C, 42 °C, 41 °C, or 44 °C) with medium shaking and reading at OD500 every 15 min.
For microfluidic single-cell growth time-lapse experiments, precultures grown at 37 °C in LB were subcultured 1:200 in fresh LB and grown under the same conditions to OD500 ∼1.0. Cells were then diluted in fresh LB at room temperature to OD500 0.01 and loaded into a Millipore CellASIC microfluidic chamber bacteria plate (Cat: B04A-03-5PK). The plate was transferred to a prewarmed OKOLabs incubation cage (37 °C or 41 °C) around a Nikon Ti2 Eclipse microscope stage. Single cells of each strain were trapped in separate chambers on the plate using the CellASIC ONIX2 microfluidic system according to the manufacturer’s protocol. Fresh LB was flowed over the dispersed cells at a rate of ∼2.5 µL h−1. Phase-contrast images were taken with a 40×/0.95 NA objective every 5 min starting 30 min after the initial flow of fresh LB. Image analysis was performed in imageJ and data processing in Jupyter Lab. The imageJ plugin “StackReg” (http://bigwww.epfl.ch/thevenaz/stackreg/) was used to correct for drift in the time-lapse movies. Individual tracks of one or a few cells growing into isolated microcolonies were manually cropped out and inspected to ensure clonal growth. Binary masks of the cell area were generated for each frame and the total area per frame was quantified after excluding edge particles. Frames with zero area (due to the cell mass making transient contact with an edge) were removed and each track was divided by its first value to correct against initial cell area variability.
To calculate the growth rate, OD500 and cell area data were processed using Python 3.6 with help from the following Python libraries: pandas, numpy, scipy, matplotlib, and seaborn. OD500 data were smoothed using a savgol filter (scipy.signal.savgol_filter, window length: 9 time measurements, polyorder: 3). Outlier detection was performed by smoothing the averaged data for each genotype and identifying the data points >2 SDs away from the smoothed curve. Outliers were identified and replaced with the average smoothed value at their respective time points. Once outliers were mitigated, each individual sample was smoothed using the savgol filter. To estimate the growth rate for each sample, we searched for the line of best fit (fit via scipy.stats.linregress) with the largest slope using a sliding window of 250 min on the smoothed data. The sliding window was allowed to search intervals between 0 and 900 min.
qRT-PCR Measurements.
Triplicate overnight cultures grown in 5 mL LB were back diluted to an OD500 of 0.01 in 50 mL LB and grown at 37 °C with shaking at 250 rpm to an OD500 of 0.7, pelleted, and resuspended to an OD500 of 0.5 in CSM. Starved cells were incubated aerobically at 37 °C with shaking at 250 rpm for 1 h. Following this incubation, an OD 0.9 mL−1 equivalence of each culture (1.8 mL) was pelleted after 0, 4, 8, and 18 h of carbon starvation and stored at −80 °C until RNA extraction. RNA extraction and qRT-PCR were performed as described previously (103). Briefly, RNA was extracted using the RNeasy Mini Kit (Qiagen) and genomic DNA was depleted using the TURBO DNA-free Kit (Invitrogen). cDNA was synthesized from TURBO DNase-treated RNA using the iScript cDNA synthesis kit (Bio-Rad) and the reactions were run using a Fast 7500 Real-Time PCR System machine (Applied Biosystems). Expression of each gene was normalized to the expression of the housekeeping gene oprI.
Routine Microscopy.
Routine microscopy was performed using a Zeiss Axio Imager. An exposure time of 400 ms and 2,000 ms was used for cells expressing the IbpA-mVenus reporter in Fig. 5 and SI Appendix, Fig. S7, respectively. Within each experiment all images were adjusted for equal brightness and contrast using the image-processing software FIJI (104). Cell length in SI Appendix, Fig. S1 was measured using SuperSegger (105).
Data Availability Statement.
All strains used in the paper will be made available to readers.
Supplementary Material
Acknowledgments
We thank members of the D.K.N. laboratory for thoughtful discussions and critical feedback on the manuscript, and Lisa Racki (The Scripps Research Institute) for the gift of pLREX97. This manuscript derives from a chapter in D.W.B.’s doctoral thesis from the California Institute of Technology. This work was supported by the Millard and Muriel Jacobs Genetics and Genomics Laboratory at the California Institute of Technology and by the NIH (1R01AI127850-01A1 and 1R21AI146987-01).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1912082117/-/DCSupplemental.
References
- 1.Siegele D. A., Kolter R., Life after log. J. Bacteriol. 174, 345–348 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rittershaus E. S. C., Baek S.-H., Sassetti C. M., The normalcy of dormancy: Common themes in microbial quiescence. Cell Host Microbe 13, 643–651 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bergkessel M., Basta D. W., Newman D. K., The physiology of growth arrest: Uniting molecular and environmental microbiology. Nat. Rev. Microbiol. 14, 549–562 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kolter R., Growth in studying the cessation of growth. J. Bacteriol. 181, 697–699 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fridman O., Goldberg A., Ronin I., Shoresh N., Balaban N. Q., Optimization of lag time underlies antibiotic tolerance in evolved bacterial populations. Nature 513, 418–421 (2014). [DOI] [PubMed] [Google Scholar]
- 6.Levin-Reisman I., et al. , Antibiotic tolerance facilitates the evolution of resistance. Science 355, 826–830 (2017). [DOI] [PubMed] [Google Scholar]
- 7.Wentland E. J., Stewart P. S., Huang C. T., McFeters G. A., Spatial variations in growth rate within Klebsiella pneumoniae colonies and biofilm. Biotechnol. Prog. 12, 316–321 (1996). [DOI] [PubMed] [Google Scholar]
- 8.Werner E., et al. , Stratified growth in Pseudomonas aeruginosa biofilms. Appl. Environ. Microbiol. 70, 6188–6196 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Williamson K. S., et al. , Heterogeneity in Pseudomonas aeruginosa biofilms includes expression of ribosome hibernation factors in the antibiotic-tolerant subpopulation and hypoxia-induced stress response in the metabolically active population. J. Bacteriol. 194, 2062–2073 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fux C. A., Costerton J. W., Stewart P. S., Stoodley P., Survival strategies of infectious biofilms. Trends Microbiol. 13, 34–40 (2005). [DOI] [PubMed] [Google Scholar]
- 11.Kolter R., Siegele D. A., Tormo A., The stationary phase of the bacterial life cycle. Annu. Rev. Microbiol. 47, 855–874 (1993). [DOI] [PubMed] [Google Scholar]
- 12.Navarro Llorens J. M., Tormo A., Martínez-García E., Stationary phase in gram-negative bacteria. FEMS Microbiol. Rev. 34, 476–495 (2010). [DOI] [PubMed] [Google Scholar]
- 13.Glasser N. R., Kern S. E., Newman D. K., Phenazine redox cycling enhances anaerobic survival in Pseudomonas aeruginosa by facilitating generation of ATP and a proton-motive force. Mol. Microbiol. 92, 399–412 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Babin B. M., et al. , SutA is a bacterial transcription factor expressed during slow growth in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A. 113, E597–E605 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Basta D. W., Bergkessel M., Newman D. K., Identification of fitness determinants during energy-limited growth arrest in Pseudomonas aeruginosa. MBio 8, e01170-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bergkessel M., et al. , The dormancy-specific regulator, SutA, is intrinsically disordered and modulates transcription initiation in Pseudomonas aeruginosa. Mol. Microbiol. 112, 992–1009 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khan N. H., et al. , Isolation of Pseudomonas aeruginosa from open ocean and comparison with freshwater, clinical, and animal isolates. Microb. Ecol. 53, 173–186 (2007). [DOI] [PubMed] [Google Scholar]
- 18.Gellatly S. L., Hancock R. E. W., Pseudomonas aeruginosa: New insights into pathogenesis and host defenses. Pathog. Dis. 67, 159–173 (2013). [DOI] [PubMed] [Google Scholar]
- 19.Walters M. C. 3rd, Roe F., Bugnicourt A., Franklin M. J., Stewart P. S., Contributions of antibiotic penetration, oxygen limitation, and low metabolic activity to tolerance of Pseudomonas aeruginosa biofilms to ciprofloxacin and tobramycin. Antimicrob. Agents Chemother. 47, 317–323 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Borriello G., et al. , Oxygen limitation contributes to antibiotic tolerance of Pseudomonas aeruginosa in biofilms. Antimicrob. Agents Chemother. 48, 2659–2664 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dietrich L. E. P., et al. , Bacterial community morphogenesis is intimately linked to the intracellular redox state. J. Bacteriol. 195, 1371–1380 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Worlitzsch D., et al. , Effects of reduced mucus oxygen concentration in airway Pseudomonas infections of cystic fibrosis patients. J. Clin. Invest. 109, 317–325 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cowley E. S., Kopf S. H., LaRiviere A., Ziebis W., Newman D. K., Pediatric cystic fibrosis sputum can be chemically dynamic, anoxic, and extremely reduced due to hydrogen sulfide formation. MBio 6, e00767 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kopf S. H., et al. , Trace incorporation of heavy water reveals slow and heterogeneous pathogen growth rates in cystic fibrosis sputum. Proc. Natl. Acad. Sci. U.S.A. 113, E110–E116 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.DePas W. H., et al. , Exposing the three-dimensional biogeography and metabolic states of pathogens in cystic fibrosis sputum via hydrogel embedding, clearing, and rRNA labeling. MBio 7, e00796-16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Akiyama Y., Kihara A., Tokuda H., Ito K., FtsH (HflB) is an ATP-dependent protease selectively acting on SecY and some other membrane proteins. J. Biol. Chem. 271, 31196–31201 (1996). [DOI] [PubMed] [Google Scholar]
- 27.Tomoyasu T., et al. , Escherichia coli FtsH is a membrane-bound, ATP-dependent protease which degrades the heat-shock transcription factor sigma 32. EMBO J. 14, 2551–2560 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ogura T., et al. , Balanced biosynthesis of major membrane components through regulated degradation of the committed enzyme of lipid A biosynthesis by the AAA protease FtsH (HflB) in Escherichia coli. Mol. Microbiol. 31, 833–844 (1999). [DOI] [PubMed] [Google Scholar]
- 29.Katz C., Ron E. Z., Dual role of FtsH in regulating lipopolysaccharide biosynthesis in Escherichia coli. J. Bacteriol. 190, 7117–7122 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ito K., Akiyama Y., Cellular functions, mechanism of action, and regulation of FtsH protease. Annu. Rev. Microbiol. 59, 211–231 (2005). [DOI] [PubMed] [Google Scholar]
- 31.Narberhaus F., Obrist M., Führer F., Langklotz S., Degradation of cytoplasmic substrates by FtsH, a membrane-anchored protease with many talents. Res. Microbiol. 160, 652–659 (2009). [DOI] [PubMed] [Google Scholar]
- 32.Langklotz S., Baumann U., Narberhaus F., Structure and function of the bacterial AAA protease FtsH. Biochim. Biophys. Acta 1823, 40–48 (2012). [DOI] [PubMed] [Google Scholar]
- 33.Okuno T., Ogura T., FtsH protease-mediated regulation of various cellular functions. Subcell. Biochem. 66, 53–69 (2013). [DOI] [PubMed] [Google Scholar]
- 34.Bittner L.-M., Arends J., Narberhaus F., When, how and why? Regulated proteolysis by the essential FtsH protease in Escherichia coli. Biol. Chem. 398, 625–635 (2017). [DOI] [PubMed] [Google Scholar]
- 35.Lysenko E., Ogura T., Cutting S. M., Characterization of the ftsH gene of Bacillus subtilis. Microbiology 143, 971–978 (1997). [DOI] [PubMed] [Google Scholar]
- 36.Deuerling E., Mogk A., Richter C., Purucker M., Schumann W., The ftsH gene of Bacillus subtilis is involved in major cellular processes such as sporulation, stress adaptation and secretion. Mol. Microbiol. 23, 921–933 (1997). [DOI] [PubMed] [Google Scholar]
- 37.Fischer B., Rummel G., Aldridge P., Jenal U., The FtsH protease is involved in development, stress response and heat shock control in Caulobacter crescentus. Mol. Microbiol. 44, 461–478 (2002). [DOI] [PubMed] [Google Scholar]
- 38.Sutterlin H. A., et al. , Disruption of lipid homeostasis in the Gram-negative cell envelope activates a novel cell death pathway. Proc. Natl. Acad. Sci. U.S.A. 113, E1565–E1574 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.May K. L., Silhavy T. J., The Escherichia coli phospholipase PldA regulates outer membrane homeostasis via lipid signaling. MBio 9, e00379-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Langklotz S., Schäkermann M., Narberhaus F., Control of lipopolysaccharide biosynthesis by FtsH-mediated proteolysis of LpxC is conserved in enterobacteria but not in all gram-negative bacteria. J. Bacteriol. 193, 1090–1097 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Spence J., Cegielska A., Georgopoulos C., Role of Escherichia coli heat shock proteins DnaK and HtpG (C62.5) in response to nutritional deprivation. J. Bacteriol. 172, 7157–7166 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jenkins D. E., Auger E. A., Matin A., Role of RpoH, a heat shock regulator protein, in Escherichia coli carbon starvation protein synthesis and survival. J. Bacteriol. 173, 1992–1996 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dukan S., Nyström T., Bacterial senescence: Stasis results in increased and differential oxidation of cytoplasmic proteins leading to developmental induction of the heat shock regulon. Genes Dev. 12, 3431–3441 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ballesteros M., Fredriksson A., Henriksson J., Nyström T., Bacterial senescence: Protein oxidation in non-proliferating cells is dictated by the accuracy of the ribosomes. EMBO J. 20, 5280–5289 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fredriksson A., Ballesteros M., Dukan S., Nyström T., Defense against protein carbonylation by DnaK/DnaJ and proteases of the heat shock regulon. J. Bacteriol. 187, 4207–4213 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fredriksson A., Ballesteros M., Dukan S., Nyström T., Induction of the heat shock regulon in response to increased mistranslation requires oxidative modification of the malformed proteins. Mol. Microbiol. 59, 350–359 (2006). [DOI] [PubMed] [Google Scholar]
- 47.Hipp M. S., Park S.-H., Hartl F. U., Proteostasis impairment in protein-misfolding and -aggregation diseases. Trends Cell Biol. 24, 506–514 (2014). [DOI] [PubMed] [Google Scholar]
- 48.Kaushik S., Cuervo A. M., Proteostasis and aging. Nat. Med. 21, 1406–1415 (2015). [DOI] [PubMed] [Google Scholar]
- 49.David D. C., et al. , Widespread protein aggregation as an inherent part of aging in C. elegans. PLoS Biol. 8, e1000450 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lindner A. B., Madden R., Demarez A., Stewart E. J., Taddei F., Asymmetric segregation of protein aggregates is associated with cellular aging and rejuvenation. Proc. Natl. Acad. Sci. U.S.A. 105, 3076–3081 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kwiatkowska J., Matuszewska E., Kuczyńska-Wiśnik D., Laskowska E., Aggregation of Escherichia coli proteins during stationary phase depends on glucose and oxygen availability. Res. Microbiol. 159, 651–657 (2008). [DOI] [PubMed] [Google Scholar]
- 52.Maisonneuve E., Ezraty B., Dukan S., Protein aggregates: An aging factor involved in cell death. J. Bacteriol. 190, 6070–6075 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fay A., Glickman M. S., An essential nonredundant role for mycobacterial DnaK in native protein folding. PLoS Genet. 10, e1004516 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Narayanaswamy R., et al. , Widespread reorganization of metabolic enzymes into reversible assemblies upon nutrient starvation. Proc. Natl. Acad. Sci. U.S.A. 106, 10147–10152 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Petrovska I., et al. , Filament formation by metabolic enzymes is a specific adaptation to an advanced state of cellular starvation. eLife 3, e02409 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Munder M. C., et al. , A pH-driven transition of the cytoplasm from a fluid- to a solid-like state promotes entry into dormancy. eLife 5, e09347 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Leszczynska D., Matuszewska E., Kuczynska-Wisnik D., Furmanek-Blaszk B., Laskowska E., The formation of persister cells in stationary-phase cultures of Escherichia coli is associated with the aggregation of endogenous proteins. PLoS One 8, e54737 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Govers S. K., Mortier J., Adam A., Aertsen A., Protein aggregates encode epigenetic memory of stressful encounters in individual Escherichia coli cells. PLoS Biol. 16, e2003853 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pu Y., et al. , ATP-dependent dynamic protein aggregation regulates bacterial dormancy depth critical for antibiotic tolerance. Mol. Cell 73, 143–156.e4 (2019). [DOI] [PubMed] [Google Scholar]
- 60.Yu J., Liu Y., Yin H., Chang Z., Regrowth-delay body as a bacterial subcellular structure marking multidrug-tolerant persisters. Cell Discov. 5, 8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.van Opijnen T., Camilli A., Transposon insertion sequencing: A new tool for systems-level analysis of microorganisms. Nat. Rev. Microbiol. 11, 435–442 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sauer R. T., Baker T. A., AAA+ proteases: ATP-fueled machines of protein destruction. Annu. Rev. Biochem. 80, 587–612 (2011). [DOI] [PubMed] [Google Scholar]
- 63.Shimohata N., Chiba S., Saikawa N., Ito K., Akiyama Y., The Cpx stress response system of Escherichia coli senses plasma membrane proteins and controls HtpX, a membrane protease with a cytosolic active site. Genes Cells 7, 653–662 (2002). [DOI] [PubMed] [Google Scholar]
- 64.Keiler K. C., Waller P. R., Sauer R. T., Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science 271, 990–993 (1996). [DOI] [PubMed] [Google Scholar]
- 65.Hinz A., Lee S., Jacoby K., Manoil C., Membrane proteases and aminoglycoside antibiotic resistance. J. Bacteriol. 193, 4790–4797 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pogliano J., Lynch A. S., Belin D., Lin E. C., Beckwith J., Regulation of Escherichia coli cell envelope proteins involved in protein folding and degradation by the Cpx two-component system. Genes Dev. 11, 1169–1182 (1997). [DOI] [PubMed] [Google Scholar]
- 67.Ruiz N., Silhavy T. J., Sensing external stress: Watchdogs of the Escherichia coli cell envelope. Curr. Opin. Microbiol. 8, 122–126 (2005). [DOI] [PubMed] [Google Scholar]
- 68.Richter K., Haslbeck M., Buchner J., The heat shock response: Life on the verge of death. Mol. Cell 40, 253–266 (2010). [DOI] [PubMed] [Google Scholar]
- 69.Parsell D. A., Lindquist S., The function of heat-shock proteins in stress tolerance: Degradation and reactivation of damaged proteins. Annu. Rev. Genet. 27, 437–496 (1993). [DOI] [PubMed] [Google Scholar]
- 70.Kuroda A., et al. , Role of inorganic polyphosphate in promoting ribosomal protein degradation by the Lon protease in E. coli. Science 293, 705–708 (2001). [DOI] [PubMed] [Google Scholar]
- 71.Lee W., et al. , Genome-wide requirements for resistance to functionally distinct DNA-damaging agents. PLoS Genet. 1, e24 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tomoyasu T., Mogk A., Langen H., Goloubinoff P., Bukau B., Genetic dissection of the roles of chaperones and proteases in protein folding and degradation in the Escherichia coli cytosol. Mol. Microbiol. 40, 397–413 (2001). [DOI] [PubMed] [Google Scholar]
- 73.Vulić M., Kolter R., Alcohol-induced delay of viability loss in stationary-phase cultures of Escherichia coli. J. Bacteriol. 184, 2898–2905 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Laskowska E., Wawrzynów A., Taylor A., IbpA and IbpB, the new heat-shock proteins, bind to endogenous Escherichia coli proteins aggregated intracellularly by heat shock. Biochimie 78, 117–122 (1996). [DOI] [PubMed] [Google Scholar]
- 75.Landgraf D., Okumus B., Chien P., Baker T. A., Paulsson J., Segregation of molecules at cell division reveals native protein localization. Nat. Methods 9, 480–482 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Emiola A., Andrews S. S., Heller C., George J., Crosstalk between the lipopolysaccharide and phospholipid pathways during outer membrane biogenesis in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 113, 3108–3113 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Thomanek N., et al. , Intricate crosstalk between lipopolysaccharide, phospholipid and fatty acid metabolism in Escherichia coli modulates proteolysis of LpxC. Front. Microbiol. 9, 3285 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Schramm F. D., Heinrich K., Thüring M., Bernhardt J., Jonas K., An essential regulatory function of the DnaK chaperone dictates the decision between proliferation and maintenance in Caulobacter crescentus. PLoS Genet. 13, e1007148 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Schulz S., et al. , Elucidation of sigma factor-associated networks in Pseudomonas aeruginosa reveals a modular architecture with limited and function-specific crosstalk. PLoS Pathog. 11, e1004744 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Damerau K., St John A. C., Role of Clp protease subunits in degradation of carbon starvation proteins in Escherichia coli. J. Bacteriol. 175, 53–63 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Msadek T., et al. , ClpP of Bacillus subtilis is required for competence development, motility, degradative enzyme synthesis, growth at high temperature and sporulation. Mol. Microbiol. 27, 899–914 (1998). [DOI] [PubMed] [Google Scholar]
- 82.Gerth U., Krüger E., Derré I., Msadek T., Hecker M., Stress induction of the Bacillus subtilis clpP gene encoding a homologue of the proteolytic component of the Clp protease and the involvement of ClpP and ClpX in stress tolerance. Mol. Microbiol. 28, 787–802 (1998). [DOI] [PubMed] [Google Scholar]
- 83.Weichart D., Querfurth N., Dreger M., Hengge-Aronis R., Global role for ClpP-containing proteases in stationary-phase adaptation of Escherichia coli. J. Bacteriol. 185, 115–125 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gerth U., et al. , Clp-dependent proteolysis down-regulates central metabolic pathways in glucose-starved Bacillus subtilis. J. Bacteriol. 190, 321–331 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Michalik S., et al. , Life and death of proteins: A case study of glucose-starved Staphylococcus aureus. Mol. Cell. Proteomics 11, 558–570 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Battesti A., Majdalani N., Gottesman S., The RpoS-mediated general stress response in Escherichia coli. Annu. Rev. Microbiol. 65, 189–213 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Taylor R. C., Dillin A., Aging as an event of proteostasis collapse. Cold Spring Harb. Perspect. Biol. 3, a004440 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.López-Otín C., Blasco M. A., Partridge L., Serrano M., Kroemer G., The hallmarks of aging. Cell 153, 1194–1217 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Saez I., Vilchez D., The mechanistic links between proteasome activity, aging and age-related diseases. Curr. Genomics 15, 38–51 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Labbadia J., Morimoto R. I., The biology of proteostasis in aging and disease. Annu. Rev. Biochem. 84, 435–464 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Balchin D., Hayer-Hartl M., Hartl F. U., In vivo aspects of protein folding and quality control. Science 353, aac4354 (2016). [DOI] [PubMed] [Google Scholar]
- 92.Ackermann M., Stearns S. C., Jenal U., Senescence in a bacterium with asymmetric division. Science 300, 1920 (2003). [DOI] [PubMed] [Google Scholar]
- 93.Stewart E. J., Madden R., Paul G., Taddei F., Aging and death in an organism that reproduces by morphologically symmetric division. PLoS Biol. 3, e45 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Winkler J., et al. , Quantitative and spatio-temporal features of protein aggregation in Escherichia coli and consequences on protein quality control and cellular ageing. EMBO J. 29, 910–923 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rang C. U., Peng A. Y., Chao L., Temporal dynamics of bacterial aging and rejuvenation. Curr. Biol. 21, 1813–1816 (2011). [DOI] [PubMed] [Google Scholar]
- 96.Wu W. F., Zhou Y., Gottesman S., Redundant in vivo proteolytic activities of Escherichia coli Lon and the ClpYQ (HslUV) protease. J. Bacteriol. 181, 3681–3687 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kanemori M., Yanagi H., Yura T., The ATP-dependent HslVU/ClpQY protease participates in turnover of cell division inhibitor SulA in Escherichia coli. J. Bacteriol. 181, 3674–3680 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Schweder T., Lee K. H., Lomovskaya O., Matin A., Regulation of Escherichia coli starvation sigma factor (sigma s) by ClpXP protease. J. Bacteriol. 178, 470–476 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Vass R. H., Chien P., Critical clamp loader processing by an essential AAA+ protease in Caulobacter crescentus. Proc. Natl. Acad. Sci. U.S.A. 110, 18138–18143 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hill N. S., Zuke J. D., Buske P. J., Chien A. C., Levin P. A., A nutrient-dependent division antagonist is regulated post-translationally by the Clp proteases in Bacillus subtilis. BMC Microbiol. 18, 29 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Langmead B., Trapnell C., Pop M., Salzberg S. L., Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Pritchard J. R., et al. , ARTIST: High-resolution genome-wide assessment of fitness using transposon-insertion sequencing. PLoS Genet. 10, e1004782 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Perry E. K., Newman D. K., The transcription factors ActR and SoxR differentially affect the phenazine tolerance of Agrobacterium tumefaciens. Mol. Microbiol. 112, 199–218 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Schindelin J., et al. , Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Stylianidou S., Brennan C., Nissen S. B., Kuwada N. J., Wiggins P. A., SuperSegger: Robust image segmentation, analysis and lineage tracking of bacterial cells. Mol. Microbiol. 102, 690–700 (2016). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All strains used in the paper will be made available to readers.