Abstract
A Autosomal-dominant ELANE mutations are the most common cause of severe congenital neutropenia. Although the majority of congenital neutropenia patients respond to daily granulocyte colony stimulating factor, approximately 15 % do not respond to this cytokine at doses up to 50 μg/kg/day and approximately 15 % of patients will develop myelodysplasia or acute myeloid leukemia. “Maturation arrest,” the failure of the marrow myeloid progenitors to form mature neutrophils, is a consistent feature of ELANE associated congenital neutropenia. As mutant neutrophil elastase is the cause of this abnormality, we hypothesized that ELANE associated neutropenia could be treated and “maturation arrest” corrected by a CRISPR/Cas9-sgRNA ribonucleoprotein mediated ELANE knockout. To examine this hypothesis, we used induced pluripotent stem cells from two congenital neutropenia patients and primary hematopoietic stem and progenitor cells from four congenital neutropenia patients harboring ELANE mutations as well as HL60 cells expressing mutant ELANE. We observed that granulocytic differentiation of ELANE knockout induced pluripotent stem cells and primary hematopoietic stem and progenitor cells were comparable to healthy individuals. Phagocytic functions, ROS production, and chemotaxis of the ELANE KO (knockout) neutrophils were also normal. Knockdown of ELANE in the mutant ELANE expressing HL60 cells also allowed full maturation and formation of abundant neutrophils. These observations suggest that ex vivo CRISPR/Cas9 RNP based ELANE knockout of patients’ primary hematopoietic stem and progenitor cells followed by autologous transplantation may be an alternative therapy for congenital neutropenia.
Introduction
Autosomal dominant ELANE mutations encoding neutrophil elastase (NE) are the most common cause of severe congenital neutropenia (CN), an inherited bone marrow failure syndrome.1–3 Patients with CN suffer from severe life-threatening bacterial infections starting early after birth due to the absence or very low numbers of neutrophils in the peripheral blood (usually less than 500 cells per μL3). Hematopoietic stem and progenitor cells (HSPC) of CN patients fail to differentiate into mature neutrophils. This differentiation defect can be partially restored with daily or alternate-day subcutaneous injections of recombinant human granulocyte colony stimulating factor (rhG-CSF) in supra-physiological concentrations.4 Although rhG-CSF therapy improves the life expectancy and quality of life of CN patients, a subgroup does not respond to rhG-CSF. Additionally, about 15 % of CN patients developed myelodysplastic syndrome (MDS) or acute myeloid leukemia (AML) till now.3 There is a positive correlation between a rhG-CSF dose required to achieve acceptable neutrophil counts and a cumulative incidence to develop MDS or AML in CN patients.5 Therefore, CN patients, especially patients who either require high rhG-CSF dosages (above 50 μg/kg/day) and those who do not respond at all, need alternative therapeutic options. Hematopoietic stem cell transplantation (HSCT) would be a treatment of choice in CN patients, but it is associated with many adverse events, e.g. acute or chronic graft-versus-host-disease (GvHD), life-threatening infections, graft failure or graft rejection. Indeed, the overall survival of CN patients after HSCT is approximately 80 % only.
Recently established new technologies of CRISPR/Cas9-mediated gene editing in mammalian cells6,7 offer novel therapeutic options, especially for inherited monogenic disorders, including ELANE mutations associated CN. In this case, CRISPR/Cas9-mediated gene correction or knockout of the mutant gene in patient’s HSPC ex vivo followed by autologous transplantation of the corrected HSPC might be a better treatment than high dose rhG-CSF or allogeneic stem cell transplantation.
ELANE mutations induce unfolded protein response (UPR) and endoplasmic reticulum (ER) stress in HSPC of CN patients that leads to increased apoptosis and defective granulocytic differentiation.8–11 Therefore, inactivation of ELANE using CRISPR/Cas9-mediated knockout may abrogate UPR and ER stress caused by mutated ELANE with subsequent restoration of granulocytic differentiation. In support of this hypothesis, we recently identified a β-lactam-based inhibitor of human neutrophil elastase (NE), MK0339, which restored defective granulocytic differentiation of induced pluripotent stem cells (iPSC) and HL60 cells expressing mutated NE.12 In addition, a recent report by Nayak et al. demonstrated the restoration of the in vitro granulopoiesis of ELANE-CN patient-derived iPSC upon treatment with Sivelestat, another NE-specific smallmolecule inhibitor.12,13 Moreover, the fact that individuals showing mosaicism of inherited ELANE mutations have a higher proportion of ELANE mutated mature neutrophils hematopoietic cells in the bone marrow than in the blood14,15 supports the hypothesis that inactivation of ELANE mutations will improve neutrophil differentiation.
Another possibility to correct the disease phenotype is the direct correction of the specific gene mutation by the activation of homology-directed repair (HDR) of the mutated gene allele after cutting by CRISPR/Cas9 and cotransfection with a repair template. Most CN patients harbor inherited autosomal dominant missense or frameshift ELANE mutations that are distributed throughout all five exons and two introns.16 Therefore, CRISPR/Cas9-mediated correction of ELANE mutations would need to be patient/mutation specific. Since mutated ELANE may induce UPR and ER stress in edited cells, the introduction of new indels in the ELANE gene during the process of CRISPR/Cas9 based editing may be not beneficial for the integrity of the hematopoietic stem cell (HSC) pool.
The first pre-clinical CRISPR/Cas9-based gene therapy study of common inherited blood disorders, sickle cell disease, and β-thalassemia, was reported.17,18 In these settings, the β-globin gene locus was inactivated by the introduction of deletions in autologous HSPC by CRISPR/Cas9-mediated gene editing. This was done to mimic the hereditary persistence of fetal hemoglobin mutations in HSC.17,18
Here, we describe a CRISPR/Cas9 mediated ELANE KO by electroporation of HSPC and iPSC with ELANEspecific CRISPR/Cas9-sgRNA ribonucleoprotein (RNP) complexes. ELANE KO induces granulocytic differentiation of HSPC and iPSC of CN patients harboring ELANE mutations without affecting their phagocytic functions. These results suggest that it may be possible to use CRISPR/Cas9 based ELANE KO in autologous HSCT as a therapy for ELANE associated neutropenia.
Methods
Patients
Three healthy donors and five severe congenital neutropenia patients harboring ELANE mutations (ELANE-CN) were used in the study. Bone marrow and peripheral blood samples from patients were collected in association with an annual follow-up recommended by the Severe Chronic Neutropenia International Registry. Study approval was obtained from the Ethical Review Board of the Medical Faculty, University of Tübingen. Informed written consent was obtained from all participants of this study.
Cell culture
Human CD34+ HSPC were isolated from bone marrow mononuclear cell fraction using Ficoll gradient centrifugation followed by magnetic bead separation using Human CD34 Progenitor Cell Isolation Kit, (Miltenyi Biotech, #130-046-703). CD34+ cells were cultured in a density of 2 x 105 cells/mL in Stemline II Hematopoietic Stem Cell Expansion medium (Sigma Aldrich, #50192) supplemented with 10 % FBS, 1 % penicillin/streptomycin, 1 % L-Glutamine and a cytokine cocktail consisting of 20 ng/mL IL-3, 20 ng/mL IL-6, 20 ng/mL TPO, 50 ng/ml SCF and 50 ng/mL FLT-3L (all cytokines were purchased from R&D Systems). Human induced pluripotent stem cells (iPSC) were cultured on Geltrex LDEV-free reduced growth factor basement membrane matrix (Thermo Fisher Scientific, #A1413201) coated plates in a density of 2 x 105 cells/mL in StemFlex medium (Thermo Fisher Scientific, #A3349401) supplemented with 1 % penicillin/streptomycin. HL60 cells were maintained in RPMI-1640 supplemented with 10 % fetal bovine serum (FBS) (Gemini Bio Products, West Sacramento, CA, USA), 2 mM L-glutamine, and 1 % penicillin/streptomycin (Thermo Fisher Scientific) at 37°C and 5 % CO2.
Design of the ELANE-specific guide RNA (gRNA)
Specific CRISPR-RNA (crRNA) for the knockout of the ELANE gene (cut site: chr19 [CTGCGCGGAGGCCACTTCTG, +852,969 : -852,969], NM_001972.3 Exon 2, 161 bp; NP_001963.1 p.F54) was designed using the CCTop website.19
CRISPR/Cas9-gRNA RNP mediated ELANE KO in iPSC and HSPC
Electroporation was carried out using the Amaxa nucleofection system (P3 primary kit, #V4XP-3024) according to the manufacturer’s instructions. 1×106 human iPSC or CD34+ HSPC were electroporated with assembled gRNA (8 μg) and Cas9 (15 μg) protein (Integrated DNA Technologies).
Isolation of single cell iPSC clones
8 x 103 human iPSC were plated on Geltrex-coated 10cm dish in StemFlex medium (Thermo Fisher Scientific, #A3349401) and RevitaCell supplement (Thermo Fisher Scientific, #A2644501). The medium was changed every 24 hours without RevitaCell supplement. On day 7, single iPSC colonies were picked and transferred to the Geltrex-coated 96-well plates (one clone/well).
Colony Forming Unit (CFU) assay
CD34+ cells were resuspended in IMDM supplemented with 2 % FBS (Stemcell Technologies, #07700) and enriched Methocult (Stemcell Technologies, #H4435). The cell suspension was plated on 3.5 cm dishes (3×103 cells/dish) for 14 days.
In vitro phagocytosis assay
Cells were incubated with or without fluorescein-conjugated Staphylococcus aureus BioParticles (Invitrogen, #S2851) at a ratio of 100 particles per cell for two hours at 37°C, washed twice with PBS/ 2 % BSA, resuspended in 300 μL FACS buffer and analyzed by flow cytometry.
Statistical analysis
Differences in mean values between groups were analyzed using two-sided, unpaired Student’s t-tests using GraphPad Prism software.
Additional Material and Methods are available in the Online Supplementary Material and Methods.
Results
Inhibition of ELANE expression restored defective granulocytic differentiation of HL60 cell lines expressing endogenous ELANE mutations
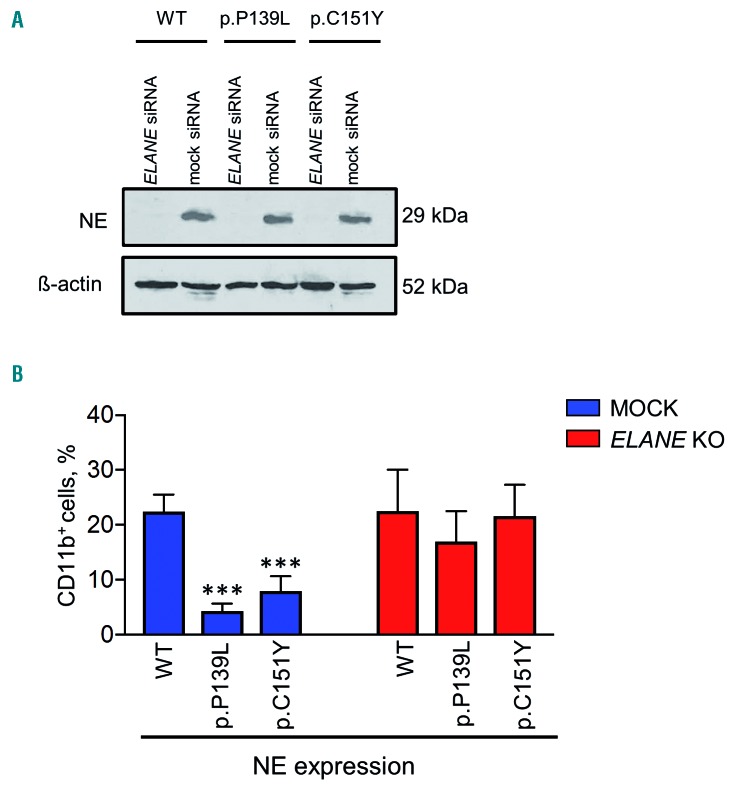
We created CRISPR/Cas9 edited mutant ELANE knockin HL60 human promyelocytic cell lines expressing either p.P139L or p.C151Y ELANE mutations. All-trans retinoic acid (ATRA) induced differentiation of wild-type and mutant HL60 clones revealed a typical impairment of granulocytic differentiation capacities in both mutant cell lines, as assessed by the significantly lower proportion of cells expressing CD11b granulocytic differentiation marker in p.P139L and p.C151Y mutant cell lines compared to the wild-type (P<0.0001 and P=0.00043, respectively) on day 5 of differentiation (Figure 1A-B and Online Supplementary Figure S1A-C). These findings are consistent with ELANE associated neutropenia patients phenotype.
Figure 1.
The effect of ELANE knock-down on the impaired myeloid differentiation of HL60 cells expressing mutant neutrophil elastase. (A) CRISPR/Cas9 edited human promyelocytic HL60 cells expressing p.P139L and p.C151Y mutant neutrophil elastase (NE) were electroporated with scrambled and anti-ELANE siRNA and maintained in the complete medium for five days. Western blot (WB) analysis at day 4 shows complete knock-down of NE detected with an anti-NE monoclonal antibody. For loading control, the membrane was stripped and re-probed with a β-actin-specific antibody. Representative WB membranes are depicted. (B) Myeloid differentiation was induced with 2 μM ATRA (all-trans retinoic acid). After five days, cells were labeled with CD11b myeloid differentiation surface marker and examined using Fluorescence-activated cell sorting (FACS) analysis. The proportion of CD11b-PE positive cells is indicated. Data represent means ± SD from four independent experiments. Two-sided, unpaired Student’s t-test P-values are shown, ***P<0.0001 and ***p=0.0043 for p.P139L and p.C151Y respectively compared to wild-type (WT).
As a proof-of-principle experiment, we have used RNA interference (RNAi) technology to knock down the expression of the ELANE gene in these cell lines. Thereby, we investigated the biological effects of inhibition of mutant NE on the granulocytic differentiation. Indeed, transfection of commercially available siRNA against the exon 4 of ELANE, completely knocked down the expression of NE in all cell lines. Production of CD11b positive cells was significantly restored in both mutant cell lines (P=0.00041 for p.P139L and P=0.00048 for p.C151Y), but not in wild-type cells (Figure 1A-B and Online Supplementary Figure S1A-C).
Design and validation of sgRNA targeting ELANE
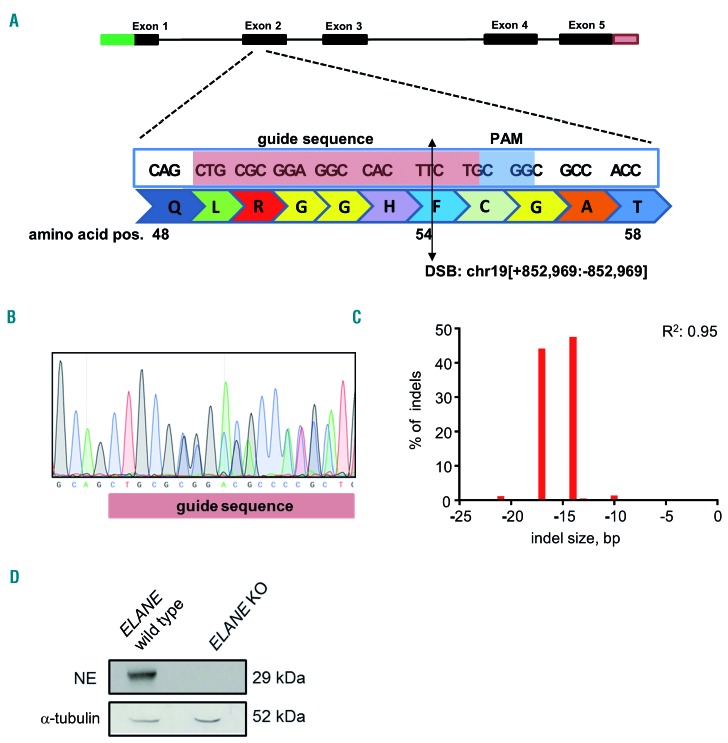
We further generated guide RNA (gRNA) specifically targeting exon 2 of ELANE by annealing CRISPR-RNA (crRNA) with trans-activating crRNA (tracrRNA). gRNA was incubated with recombinant Cas9 protein to generate CRISPR/Cas9-gRNA RNP complexes. The gRNA targeting exon 2 of ELANE (cut site: chr19[+852.969:-852.969], Figure 2A) was selected to introduce stop-codon mutations and to induce nonsense-mediated mRNA decay (NMD) of ELANE mRNA, which is caused by stop-codon or frameshift mutations at the beginning of ELANE mRNA. Based on our experimental analysis, the selected gRNA has high on-target activity with low off-target score (data not shown). To evaluate inhibition of NE expression by CRISPR/Cas9 RNP mediated targeting of exon 2 of ELANE, we generated an ELANE KO myeloid cell line THP-1, which has high basal expression levels of ELANE and NE. The efficiency of ELANE knockout in the total population of edited THP-1 cells was 77 %, as assessed by Sanger sequencing and tracking of indels by decomposition (TIDE) analysis (data not shown). The pure ELANE KO THP-1 cell clone has compound heterozygosity of 14 and 17 bp deletions on each allele (Figure 2B-C). The NE expression was completely absent in the pure ELANE KO THP-1 cell clone, as determined by Western blotting (WB) using anti-NE antibody against the C-terminus of NE protein (Figure 2D, Online Supplementary Figure S2A-B). These data suggest that sgRNA targeting ELANE that we designed led to a complete loss of NE protein.
Figure 2.
Establishment of CRISPR/Cas9 RNP-mediated ELANE knockout in THP-1 cells. (A) ELANE was targeted using single guide ribonucleoprotein (RNP)-mediated (highlighted in pink), which creates a double-strand break at NM_001972.2 exon 2, 161 bp after ATG; NP_001963.1, p.F54. Schematic presentation of the cut site by sgRNA. (B-D) Myeloid cell line THP-1 was electroporated with ELANE-specific CRISPR/Cas9 RNP. Single cell clones of ELANE KO THP-1 cells were generated, as described in the Material and Methods. Representative Sanger sequencing image (B), the bar chart of the TIDE assay (C) and representative Western blotting (WB) images of neutrophil elastase (NE) and α-tubulin protein expression (D) in single cell-derived ELANE KO THP-1 cells are depicted.
Restoration of the in vitro granulocytic differentiation in ELANE-CN iPSC clones after ELANE knockout
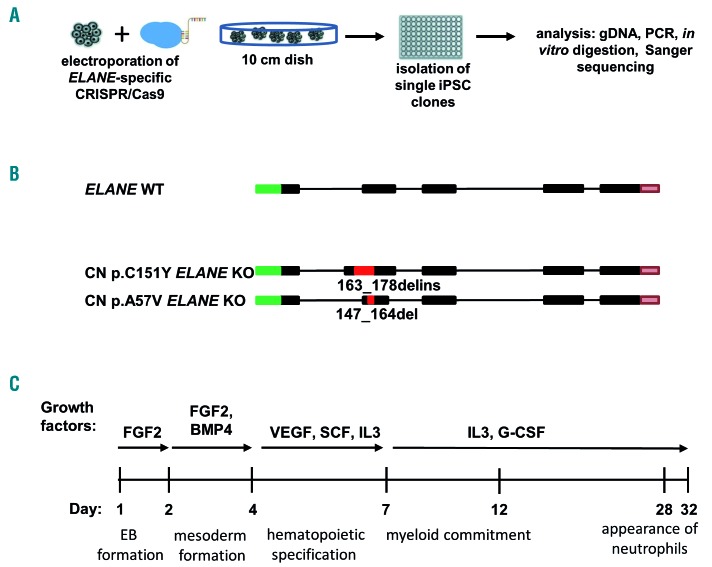
We generated iPSC from peripheral blood mononuclear cells (PB MNC) of two ELANE-CN patients, harboring ELANE mutations p.C151Y or p.A57V (CN p.C151Y iPSC and CN p.A57V iPSC, respectively). Additionally, iPSC of one healthy control (healthy ctrl iPSC) were evaluated. All three iPSC lines expressed elevated mRNA and protein levels of pluripotent stem cell-specific factors, displayed alkaline phosphatase activity and expression of pluripotent embryonic stem cell surface markers (Online Supplementary Figure 3A-C).
Figure 3.
Generation of ELANE KO CN iPSC clones. (A) Scheme of the ELANE-specific CRISPR/Cas9-gRNA ribonucleoprotein electroporation of induced pluripotent stem cells (iPSC) and generation of ELANE KO iPSC clones. Generation of single ELANE knockout iPSC clones was made by seeding single iPSC, subsequent picking of each clone and transferring them into 96 well plates. Screening of each iPSC clone was done by Cas9 in vitro digestion and Sanger sequencing. (B) Scheme of CRISPR/Cas9 introduced modifications in the ELANE gene in iPSC clones of congenital neutropenia (CN) patients. Red inserts show positions of indels in ELANE mRNA (NM_001972.2) and numbers refer to bp position after ATG. (C) Scheme of the EB-based hematopoietic/neutrophilic differentiation of iPSC.
Next, we used electroporation of iPSC clones with ELANE-specific CRISPR/Cas9-sgRNA RNP to generate pure ELANE KO CN iPSC clones. For this, electroporated iPSC were seeded on a geltrex coated culture dish and single-cell derived iPSC clones were isolated transferred to geltrex coated 96 well-plates for the subsequent selection of ELANE knockout clones (Figure 3A). Confirmed ELANE knockout iPSC clones have followed ELANE modifications: 274 bp del/ins in CN p.C151Y ELANE KO iPSC, and 17 bp del in CN p.A57V ELANE KO iPSC (Figure 3B). The editing efficiency of healthy ctrl iPSC was 97 % (Online Supplementary Figure S4A), therefore, we used the total population of gene-edited healthy control (ctrl) iPSC for further analysis. We did not detect any off-target activity of the gRNA for the selected cDNA sites in all studied iPSC, as assessed using Sanger sequencing (Online Supplementary Figure S4B and Tables S1, S2).
Applying a slightly modified in vitro embryoid body (EB)-based iPSC differentiation method that allows generation of hematopoietic cells and mature myeloid cells for approximately 30 days,22,23 we found an increase in the percentage of CD15+CD16+CD45+ granulocytes in ELANE KO CN-iPSC cell culture, as compared to CN-iPSC. The generation of granulocytes from ELANE KO CN-iPSC was comparable to iPSC generated from a healthy donor (Figure 3C, 4A, and Online Supplementary Figure S5A). Generation of immature hematopoietic cells (CD34+KDR+, CD34+CD43+, CD45−CD235+CD41a+ and CD45+CD34+ cells) and CD45+CD33+ myeloid progenitor cells in ELANE KO CN- and CN-iPSC lines were similar or increased, in comparison to corresponding MOCK treated iPSC lines (Online Supplementary Figure S6A).
Figure 4.
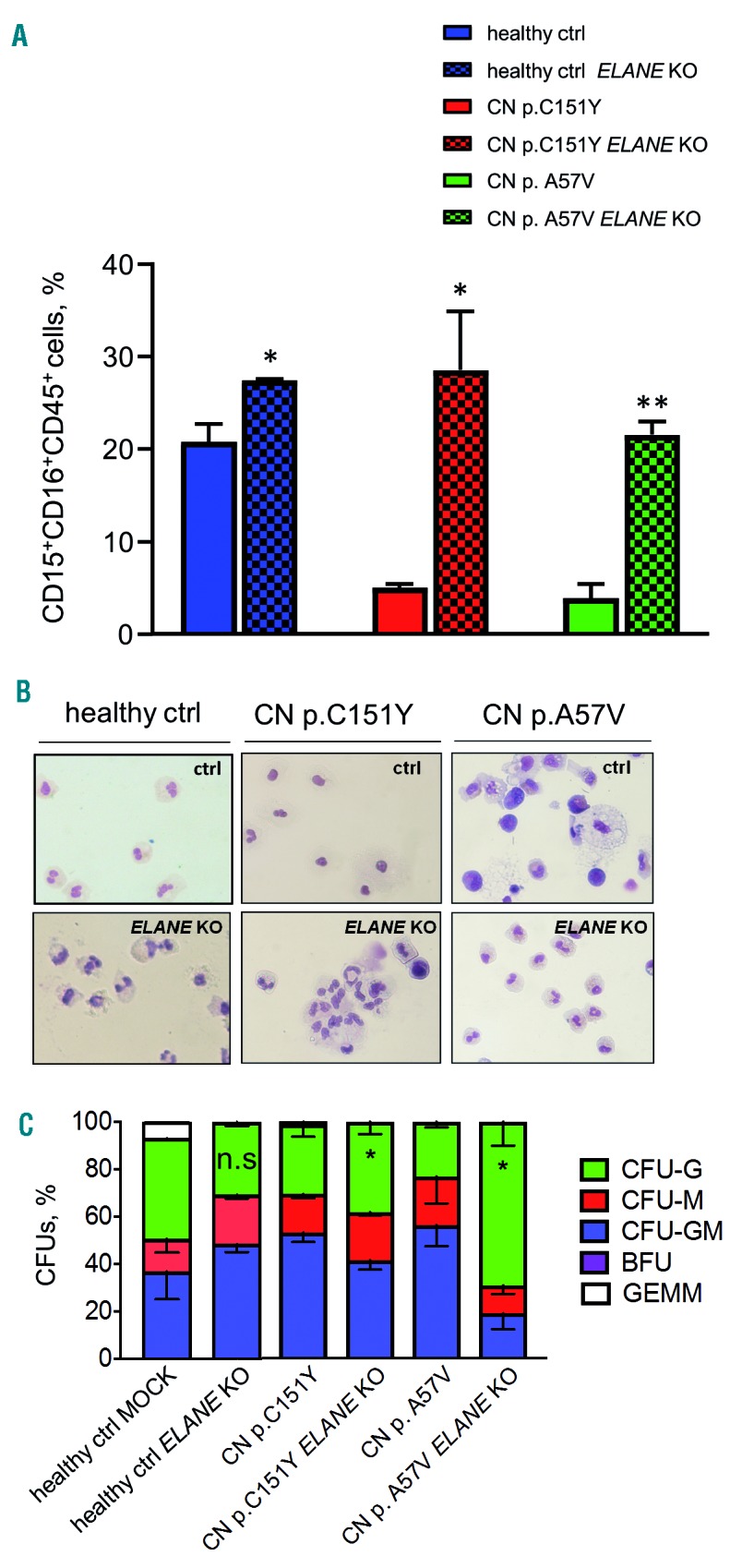
ELANE knockout restored granulocytic differentiation of ELANE-CN iPSC. (A) Flow cytometry analysis of suspension cells harvested from embryoid body (EB)-based granulocytic cell culture of respective iPSC clones on day 28 or 32 of differentiation. Data represent means ± standard deviation (SD) from two independent experiments. *P<0.05, **P<0.01. (B) Wright-Giemsa staining of cytospin preparations of suspension myeloid cells harvested from iPSC culture at day 28 or 32 of differentiation. Representative images are depicted. (C) Colony-forming unit (CFU) assay of CD34+ cells harvested from EB-based iPSC culture on day 14 of differentiation. Data represent means ± SD from two independent experiments. *P<0.05.
A CFU assay was performed with ELANE−/− iPSC-derived CD34+ cells from CN patients and showed elevated levels of CFU-G but reduced CFU-M colony numbers, as compared to CD34+ cells derived from MOCK treated CN iPSC clones (Figure 4C). These data suggest that ELANE knockout restores granulocytic differentiation in CN.
We did not observe any significant defects in in vitro granulocytic differentiation of ELANE KO iPSC generated from a healthy donor, as compared to MOCK treated cells (Figure 4A-C, Online Supplementary Figures S5 and S6).
ELANE knockout in HSPC of ELANE-CN patients restores diminished granulocytic differentiation
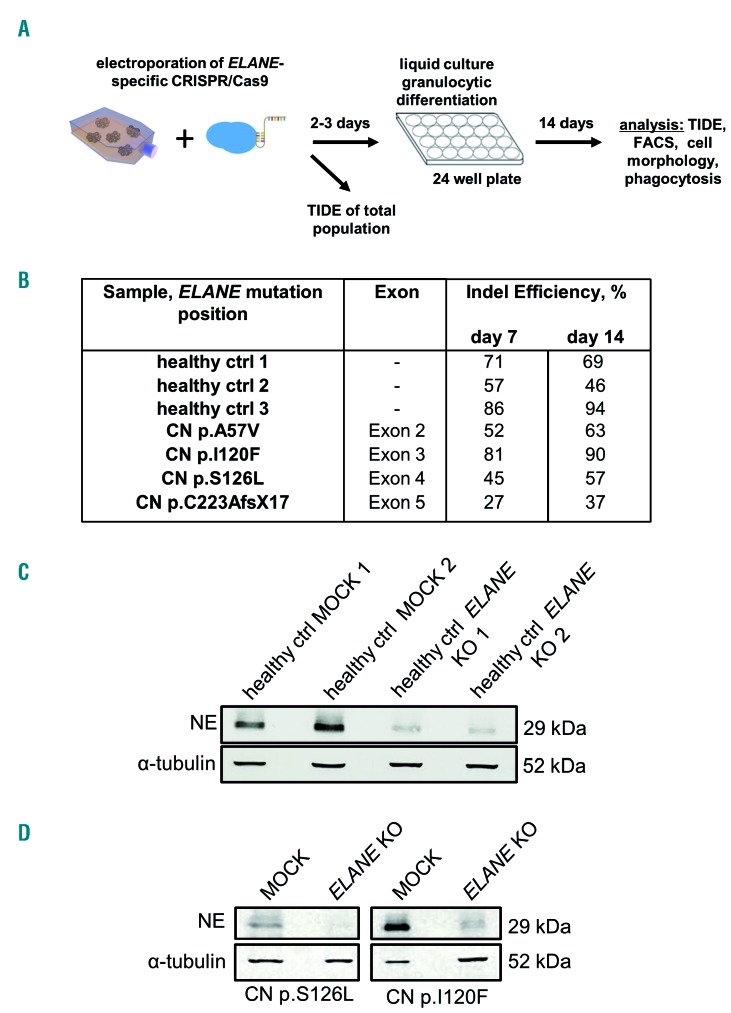
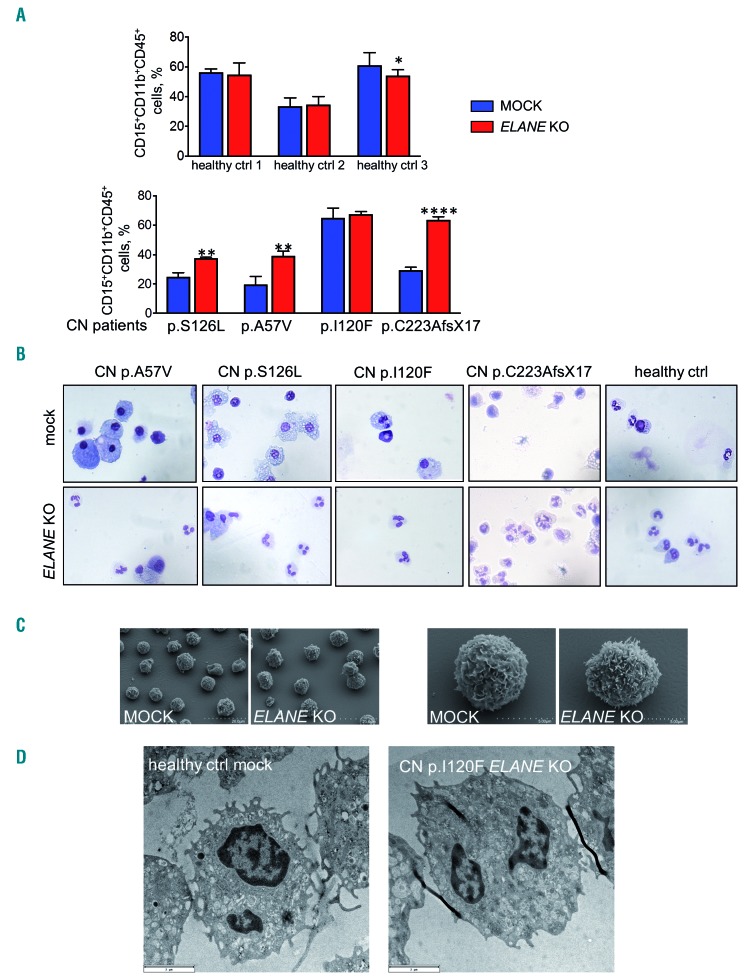
To further evaluate the clinical applicability of ELANE KO as a treatment option of ELANE-associated CN, we performed CRISPR/Cas9 RNP-mediated gene editing in primary bone marrow CD34+ HSPC of four ELANE-CN patients (Table 1) and three healthy donors and differentiated the cells towards neutrophils. ELANE knockout in CD34+ HSPC was performed by electroporation of human CD34+ HSPC with assembled ELANE specific sgRNA and Cas9 protein (Figure 5A). The editing efficiency varied between 27 % and 94 % (Figure 5B, Online Supplementary Figure S7). As expected, NE levels in neutrophils differentiated from the total population of edited cells were markedly reduced (Figure 5C-D, Online Supplementary Figure S8A-B). Moreover, ELANE KO leads to elevated granulocytic differentiation, as assessed by the percentage of CD15+CD11b+CD45+ cells (Figure 6A, Online Supplementary Figure S9A-B) and morphological examination of cytospin preparations of mature granulocytes generated on day 14 of the in vitro granulocytic differentiation using liquid culture (Figure 6B). At the same time, the ratio of ELANE KO cells increased from day 7 to day 14 of differentiation (Figure 5B). Simultaneously, the percentage of CD34+CD45+ cells was reduced in ELANE KO cells of CN patients, but not in healthy donor cells (Online Supplementary Figure S10A). In one patient (CN I120F), no difference in the percentage of CD15+CD11b+CD45+ cells between MOCK and ELANE KO samples was observed, but a clear improvement of granulocytic differentiation was detected in cytospin slides. This finding may be explained by relative mild neutropenia (Online Supplementary Table 3) and possible expression of CD15 in not fully mature myeloid cells in this patient.
Figure 5.
Efficient CRISPR/Cas9 RNP-based ELANE knockout in HSPC. (A) Scheme of the generation of ELANE KO HSPC using electroporation with ELANE-specific CRISPR/Cas9-gRNA ribonucleoprotein (RNP). (B) TIDE results of edited CD34+ HSPC at day 7 and 14 of liquid culture differentiation. (C and D) Hematopoietic stem and progenitor cells (HSPC) of healthy controls (C), or two CN patients (D) were electroporated with ELANE-specific CRISPR/Cas9 RNP, on day 14 of culture, cells were lysed in Laemmli buffer and Western blotting (WB) analysis using antineutrophil elastase (NE) antibody against C-terminus of NE was performed, staining with α-tubulin antibody was used as loading control. Representative WB images of cells from two independent experiments are depicted.
Figure 6.
ELANE KO restored granulocytic differentiation of ELANE-CN primary HSPC. (A) Differentiation capacity of ELANE knockout CD34+ cells was assessed by liquid culture differentiation after 14 days by investigating neutrophilic surface marker expression. Data represent means ± standard deviation (SD) from triplicates. *P<0.05, **P<0.01, ****P<0.0001. (B) Wright-Giemsa staining of differentiated cells was conducted on day 14 allowing morphologic discrimination of the cells. Representative images are depicted. (C-D) Electron micrographs of neutrophils generated on day 14 of liquid culture analyzed by scanning electron microscopy (SEM) (C) and transmission electron microscopy (TEM) (D). Representative SEM and TEM images are depicted. Typical neutrophil morphology is observed in all studied samples.
Scanning and transmission electron microscopy revealed that ELANE KO cells of both healthy control and one CN patient showed no significant differences in morphology or intracellular structures, compared with MOCK cells of a healthy donor (Figure 6C-D).
Altogether, these data suggest that ELANE KO cells have a differentiation advantage over the HSPC carrying mutated ELANE.
Neutrophils generated from ELANE KO HSPC exhibited unaffected ROS production, phagocytosis and chemotaxis upon activation in vitro
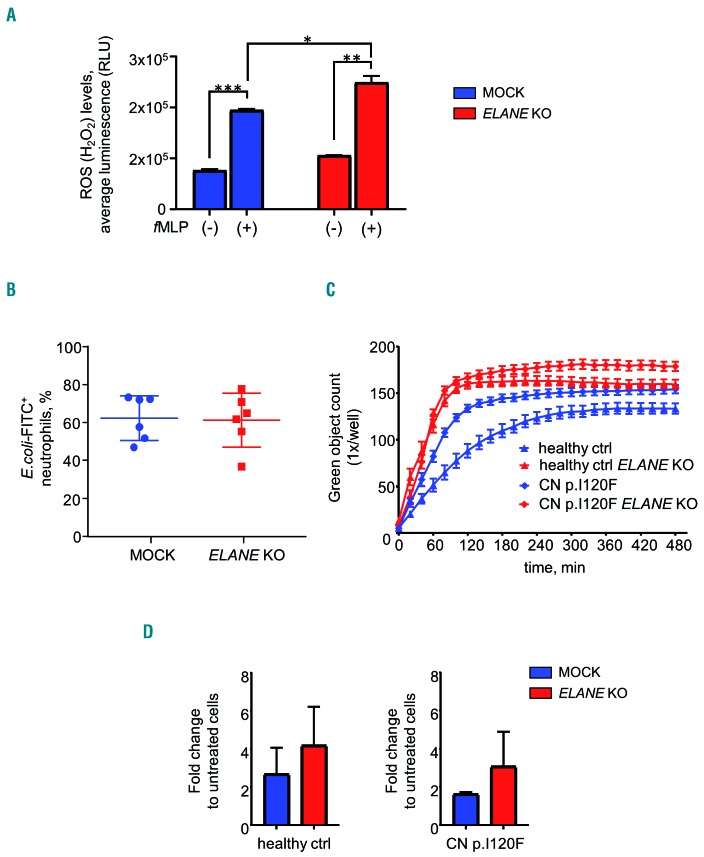
We further evaluated in vitro activation of neutrophils generated from ELANE KO HSPCs in liquid culture for 14 days. We first performed an assessment of H2O2 levels (ROS) in fMLP-activated ELANE KO neutrophils generated from a healthy donor. We detected no differences between ELANE WT and ELANE KO neutrophils (Figure 7A).
Figure 7.
Unaffected functions of ELANE KO neutrophils. (A, B, C) Reactive oxygen species (ROS) assay (A) and phagocytosis assay (B, C) of granulocytes generated on day 14 of liquid culture, as described in the Methods section. Data represent means ± SD from duplicates. *P<0.05, **P<0.01, ***P<0.0001. (C) Phagocytosis Kinetic using IncuCyte ZOOM System of granulocytes generated on day 14 of liquid culture as described in the Methods section. Data represent mean ± standard deviation (SD). (D) Chemotaxis depicted as fold change between fMLP-treated and untreated granulocytes generated on day 14 of liquid culture. Data represents mean ± SD, healthy control (ctrl) (n=2), CN patient (n=1).
Phagocytosis was evaluated by incubation of cells with fluorescein-conjugated Staphylococcus aureus BioParticles for two hours. Percentage of GFP+ granulocytes that engulfed bacteria were assessed by FACS using gating on granulocyte population in the dot plot of forward-scatter light (FSC) versus side-scatter light (SSC) channels. We did not detect any significant differences in phagocytosis of ELANE KO neutrophilic granulocytes, as compared to control MOCK cells (Figure 7B). As an independent evaluation of phagocytosis kinetics, we performed live cell imaging of neutrophils incubated with pHrodo Green E. coli Bioparticles Conjugate using IncuCyte ZOOM system and observed similar phagocytosis behavior of MOCK and ELANE KO neutrophils generated from a healthy donor or one CN patient (Figure 7C).
Chemotactic activity of fMLP-treated neutrophils was also comparable between MOCK and ELANE KO groups (Figure 7D).
Unaffected phagocytic activity of ELANE KO PMN in zebrafish embryos
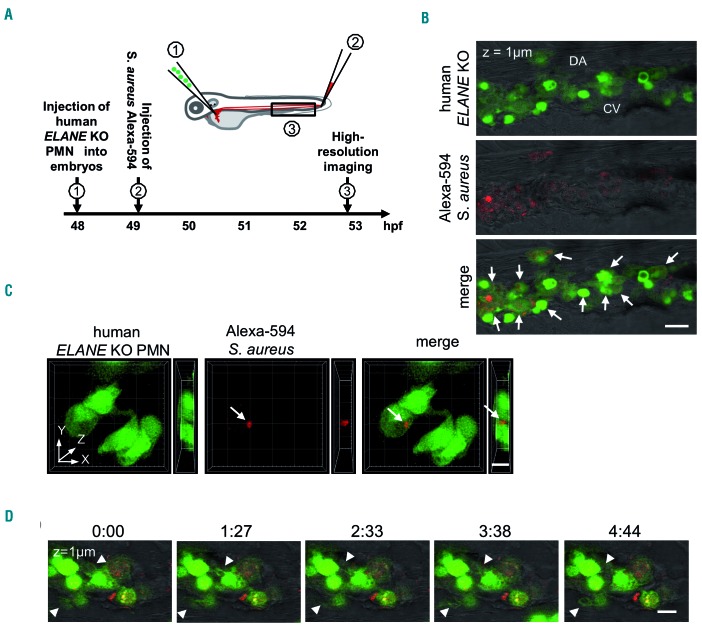
To evaluate the phagocytic activity of ELANE KO PMN in vivo, we transplanted fluorescently labeled polymorphonuclear leukocytes (PMN) generated from ELANE KO HD into zebrafish embryos (Figure 8A). PMN were injected into the duct of Cuvier, a wide circulation channel on the yolk sac connecting the heart to the trunk vasculature. Subsequently, Alexa-594-conjugated Staphylococcus aureus BioParticles were injected locally in the tail fin close to the caudal vein. Live imaging showed that human neutrophils migrated into the caudal hematopoietic tissue (CHT), which is equivalent to the fetal liver in mammals and provides a human-compatible environment.24,25 Confocal imaging of this region revealed that most of the ELANE KO neutrophils were found inside the perivascular pocket (Figure 8B) and many of them have engulfed bacteria (white arrows in Figure 8B and 8C, Online Supplementary Movie S1). Time-lapse in vivo imaging of the xenotrans-planted embryos also revealed that human ELANE KO PMN have the capability to form surface protrusion within the perivascular region (Figure 8D, Online Supplementary Movie S2). We could not detect a difference between transplanted human ELANE KO and control MOCK PMN in zebrafish embryos (data not shown). These observations indicate that human ELANE KO PMN are able to migrate and phagocyte in vivo.
Figure 8.
Human ELANE KO PMN are capable to migrate and to phagocyte S. aureus BioParticles in zebrafish embryos in vivo. (A) The scheme of in vivo phagocytosis assay in zebrafish embryos xenotransplanted with human fluorescently labeled polymorphonuclear leukocytes (PMN). (B) A representative confocal image highlighting the presence of transplanted human ELANE KO PMN in the caudal hematopoietic site of a zebrafish embryo at 53 hpf. White arrows indicate S. aureus BioParticles phagocytosed by human ELANE KO PMN. CV: caudal vein; DA: dorsal aorta. Scale bar: 20 mm. (C) Three-dimensional rendering of a z-stacks of 12 mm illustrating human ELANE KO PMN, one of them has engulfed S. aureus BioParticles (white arrow). Scale bar: 10 μm. (D) Still photographs from a time-lapse recording illustrating the phagocytic activity of transplanted human ELANE KO PMN in the zebrafish embryo. Arrowheads indicate the formation of neutrophil protrusions. Numbers indicate time in minutes. Scale bar: 10 μm.
ELANE KO restores deregulated expression of UPR gene BiP and anti-apoptotic factor Bcl-xl in ELANE KO iPSC derived cells of CN patients
We further evaluated the effects of ELANE KO on the expression of UPR gene BiP and anti-apoptotic factor Bcl-xl (Online Supplementary Figure S11). We analyzed pure ELANE KO HSPC (for Bcl-xl) or neutrophils (for BiP) generated from iPSC of two CN patients. We found that ELANE KO HSPC express elevated mRNA levels of Bcl-xl and BiP expression was markedly reduced in ELANE KO PMN, compared to cells carrying mutated ELANE. As expected, ELANE mRNA levels were severely diminished in ELANE KO cells (Online Supplementary Figure S11).
Discussion
The majority of patients suffering from congenital neutropenia respond well to daily treatment with rhG-CSF leading to a normal quality of life. However, in the last 25 years, we learned that CN is a preleukemic syndrome and that approximately 15 % of patients do not respond to even ultra-high dosages (>50 μg/kg/d) of rhG-CSF. Therefore, we are searching for other treatment modalities for CN patients that may prevent leukemic transformation and may be useful for those who are requiring high dosages of rhG-CSF or not responding at all to rhG-CSF. For these patients, the only available treatment is stem cell transplantation with the risk of transplant-associated adverse events such as acute or chronic GvHD.
In the present study, we described for the first time the establishment of an in vitro cellular model of CRISPR/Cas9 mediated gene therapy of CN associated with autosomal dominant ELANE mutations, the most frequent cause of CN. We tested ex vivo CRISPR/Cas9 RNP-based ELANE knockout in HSPC of CN patients that may be used for autologous transplantation as a therapeutic approach for ELANE-CN patients. Virus- and DNA-free application of CRISPR/Cas9 RNP markedly increases gene editing efficiency and simultaneously decreases the probability and frequency of off-target effects, because CRISPR/Cas9 RNP activity is preserved in cells for only approximately 48 hours. We recently reported the establishment of the fluorescent labeling of CRISPR/Cas9 RNP complexes for gene editing of primary hematopoietic stem cells and subsequent sorting of gene-modified cells for further applications.26 Implementation of this method will improve the efficiency of gene knockout or gene correction in HSPC, including ELANE knockout or correction of ELANE mutations.
In case of ELANE KO, different combinations of the ELANE gene editing are expected: we may generate unedited, monoallelic edited (of mutated or WT allele), or bi-allelic edited HSPC. Since we did not use any selection marker for edited HSPC, we were not able to estimate the proportion of HSPC with inactivation of the mutated ELANE allele. The fact that the proportion of ELANE KO cells was elevated upon granulocytic differentiation strongly argues for the differentiation advantage of the edited cells lacking ELANE (including loss of the mutated allele).
There are several potential unforeseen consequences of the ELANE gene knockout strategy. For example, Tidwell et al. reported the presence of two in-frame ATG codons in exon 2 and exon 4 of ELANE.27 They showed that the internal translation of NE can be initiated when the canonical translational start site and/or internal start sites in exon 2 are disrupted and that expression of internally-initiated ELANE is pathogenic. We found a marked reduction of ELANE mRNA, most probably due to the induction of nonsense-mediated mRNA decay (NMD) of ELANE mRNA after exposure of cells to ELANE-specific CRISPR/Cas9 sgRNA RNP. We also did not detect any additional NE protein bands on WB analysis of edited cells using antibody recognizing C-terminus of NE. Based on these observations, we concluded that our sgRNA is inducing loss of NE protein without activation of the pathogenic ELANE forms from the internal ATG.
We did not detect off-target activity in edited cells, but recent results from Alan Bradley have suggested that the introduction of CRISPR/Cas9 editing can cause multiple genomic changes far beyond the actual target.28 Therefore, for clinical applications, evaluation of the off-target activity of CRISPR/Cas9 on whole genome level using nextgeneration sequencing should be performed. In addition, it would be important to evaluate that the editing of ELANE occurred in the repopulating hematopoietic stem cell population and that these cells maintained their ability to engraft immunodeficient mice in vivo. Since most probably HSC are not expressing NE, we will not expect any damaging effects of the ELANE KO on the functions and integrity of HSC.
We demonstrated here, that CRISPR/Cas9 mediated ELANE KO in HSPC and iPSC of CN patients induces granulocytic differentiation and in vitro generated ELANE KO neutrophils have no defects in the phagocytic activity, ROS production, and chemotaxis. NE is a proteolytic enzyme of the neutrophil serine protease (NSP) family, including also cathepsin G (CG), proteinase 3 (PR3) and azurocidin (AZU1). NSP are stored in cytoplasmic granules, can be secreted into the extra- and peri-cellular space upon cellular activation and considered to be crucially involved in bacterial defense. NE and PR3 are very similar in their substrate specificity supporting a potentially redundant function for these two enzymes. Elane−/− mice have normal neutrophil counts, but there are conflicting results regarding the effect of NE-deficiency on neutrophil extravasation to sites of inflammation, phagocytosis, and neutrophil extracellular traps in mice. NE may or may not be essential for these processes.29–33 Papillon-Lefevre Syndrome (PLS) is the only human disorder known to cause NE deficiency. This rare autosomal recessive disease is due to loss-of-function mutations in the DPPI gene locus with the loss of the lysosomal cysteine protease cathepsin C/dipeptidyl peptidase I (DPPI). The activation of NSP, including NE, depends on the N-terminal processing activity of DPPI. Therefore, PLS patients exhibit a severe reduction in the activity and stability of all three NSP including NE. Intriguingly, patients with PLS have no defects in their ability to kill bacteria e.g. Staphylococcus aureus or E.coli, suggesting that redundancies in the neutrophil’s bactericidal mechanisms negate the necessity for serine proteases for killing common bacteria.34 Moreover, since the other serine proteases including CG, PR3 and neutrophil serine protease 4 remain intact, we do not expect for the resultant cells to develop any neutrophil-specific functional anti-bacterial or immunodeficiency phenotype in ELANE KO cells. Based on these observations, at this juncture, we believe that CRISPR/Cas9 based knockout of ELANE in HSPC of CN patients may restore defective granulopoiesis in CN patients without seriously impairing neutrophil functions. Further studies, including gene expression analysis to understand which pathways are affected by ELANE mutations and verifying that these pathways are indeed restored by ELANE KO, are essential to justify the therapeutic applications of ELANE KO technology in the future. It will also contribute to the understanding of the pathophysiology of the CN caused by ELANE mutations. Our first attempts to investigate intracellular signaling pathways affected by mutated ELANE revealed the restoration of mRNA expression of anti-apoptotic Bcl-xl factor that is downregulated in CN myeloid progenitor cells.35 Moreover, we found downregulation of mRNA levels of the key UPR player BiP, normally upregulated in HSPC of CN patients harboring ELANE mutations.8–10
CRISPR/Cas9 technology also allowed correction of the specific gene mutations. We selected the ELANE knockout approach since ELANE mutations are heterozygous gain- of-function gene defects that are distributed throughout all five exons and two introns of ELANE and specific ELANE mutations correction would require specific settings for each patient based on the mutation position. In addition, the requirement of the introduction of the donor repair template DNA in a gene correction approach requires the activation of HDR making it difficult to achieve efficient correction in primary HSPC. We and other investigators have reported that ELANE mutations induce UPR and ER stress.8–11 We also described deregulated signaling pathways in HSPC of CN patients downstream of ELANE mutations, such as diminished expression of transcription factors LEF-1, and C/EBPa,36–39 abrogated expression and phosphorylation of the adaptor protein HCLS1,40 elevated apoptosis35 and hyperactivated NAMPT/sirtuins.41 These intracellular defects may lead to the elevated fragility of HSPC during ex vivo gene manipulations and may affect gene correction efficiency. Moreover, gene editing strategy directed to the correction of ELANE mutations may lead to the creation of novel missense or frameshift mutations that may result in the novel mutant NE protein with damaging functions and potential generation of the pre-leukemic HSPC clones with proliferative advantage and possible leukemic transformation. Adeno-associated virus (AAV)-based vector may be used for the delivery of the donor repair template and is considered safer than retroviral constructs. Two groups recently published successful gene deletions as a gene therapy approach to cure sickle cell disease, a common inherited blood disorder.17,18 It should be noted that AAV-based expression constructs may induce anti-viral host immune responses and may non-specifically integrate into the host genome.
In summary, we report here for the first time a method of CRISPR/Cas9 mediated ELANE gene deletion in hematopoietic stem cells and iPSC from CN patients harboring ELANE mutations. The ELANE gene deletion resulted in the increase of granulocytic differentiation to functional normal mature neutrophils in these patients in vitro. Therefore, CRISPR/Cas9 based gene knockout of ELANE in CN patients harboring ELANE mutation might be a useful treatment option especially in patients requiring high G-CSF dosages or do not responding to G-CSF at all. In addition, it remains to be investigated in subsequent clinical studies whether, in CN patients harboring ELANE mutations, the ELANEgene knockout would also abrogate the leukemogenesis.
Acknowledgments
We would like to thank the FACS core facility of the UKT especially Stella Autenrieth for the assistance in flow cytometry; Michael Schindler and Esther Lehmann for their support in confocal microscopy. This work was supported by the Excellence Initiative of the Faculty of Medicine, University of Tübingen (JS), Jose Carreras Leukemia Foundation (JS, PM, MR), Madeleine Schickedanz Kinderkrebsstiftung (JS, MK, MN), DFG (JS, MK), intramural Fortüne program of the Medical Faculty of the Tübingen University (MK, DA, YX), German-Israeli Foundation for Scientific Research and Development (GIF) (KW, BD), German Cancer Consortium (PM), Else Kroner-Fresenius Stiftung (MK, BD), Fritz Thyssen Foundation (MR and BD), NIH R24 AI 049393 (VM, BF, CZ, DCD).
Footnotes
Check the online version for the most updated information on this article, online supplements, and information on authorship & disclosures: www.haematologica.org/content/105/3/598
References
- 1.Dale DC, Person RE, Bolyard AA, et al. Mutations in the gene encoding neutrophil elastase in congenital and cyclic neutropenia. Blood. 2000; 96 (7):2317–2322. [PubMed] [Google Scholar]
- 2.Skokowa J, Germeshausen M, Zeidler C, Welte K. Severe congenital neutropenia: inheritance and pathophysiology. Curr Opin Hematol. 2007; 14 (1):22–28. [DOI] [PubMed] [Google Scholar]
- 3.Skokowa J, Dale DC, Touw IP, Zeidler C, Welte K. Severe congenital neutropenias. Nat Rev Dis Primers. 2017;3:17032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Welte K, Gabrilove J, Bronchud MH, Platzer E, Morstyn G. Filgrastim (r-metHuG-CSF): the first 10 years. Blood. 1996;88 (6):1907–1929. [PubMed] [Google Scholar]
- 5.Rosenberg PS, Zeidler C, Bolyard AA, et al. Stable long-term risk of leukaemia in patients with severe congenital neutropenia maintained on G-CSF therapy. Br J Haematol. 2010;150 (2):196–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337 (6096):816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8 (11):2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grenda DS, Murakami M, Ghatak J, et al. Mutations of the ELA2 gene found in patients with severe congenital neutropenia induce the unfolded protein response and cellular apoptosis. Blood. 2007;110 (13): 4179–4187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nanua S, Murakami M, Xia J, et al. Activation of the unfolded protein response is associated with impaired granulopoiesis in transgenic mice expressing mutant Elane. Blood. 2011; 117(13):3539–3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nustede R, Klimiankou M, Klimenkova O, et al. ELANE mutant-specific activation of different UPR pathways in congenital neutropenia. Br J Haematol. 2016;172 (2):219–227. [DOI] [PubMed] [Google Scholar]
- 11.Kollner I, Sodeik B, Schreek S, et al. Mutations in neutrophil elastase causing congenital neutropenia lead to cytoplasmic protein accumulation and induction of the unfolded protein response. Blood. 2006;108 (2):493–500. [DOI] [PubMed] [Google Scholar]
- 12.Makaryan V, Kelley ML, Fletcher B, Bolyard AA, Aprikyan AA, Dale DC. Elastase inhibitors as potential therapies for ELANE-associated neutropenia. J Leukoc Biol. 2017;102 (4):1143–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nayak RC, Trump LR, Aronow BJ, et al. Pathogenesis of ELANE-mutant severe neutropenia revealed by induced pluripotent stem cells. J Clin Invest. 2015;125 (8):3103–3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ancliff PJ, Gale RE, Watts MJ, et al. Paternal mosaicism proves the pathogenic nature of mutations in neutrophil elastase in severe congenital neutropenia. Blood. 2002;100 (2): 707–709. [DOI] [PubMed] [Google Scholar]
- 15.Benson KF, Horwitz M. Possibility of somatic mosaicism of ELA2 mutation overlooked in an asymptomatic father transmitting severe congenital neutropenia to two offspring. Br J Haematol. 2002;118 (3):923; author reply -4. [DOI] [PubMed] [Google Scholar]
- 16.Makaryan V, Zeidler C, Bolyard AA, et al. The diversity of mutations and clinical outcomes for ELANE-associated neutropenia. Curr Opin Hematol. 2015;22 (1):3–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ye L, Wang J, Tan Y, et al. Genome editing using CRISPR-Cas9 to create the HPFH genotype in HSPCs: An approach for treating sickle cell disease and beta-thalassemia. Proc Natl Acad Sci U S A. 2016;113 (38): 10661–10665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Traxler EA, Yao Y, Wang YD, et al. A genome-editing strategy to treat beta-hemoglobinopathies that recapitulates a mutation associated with a benign genetic condition. Nat Med. 2016;22 (9):987–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stemmer M, Thumberger T, Del Sol Keyer M, Wittbrodt J, Mateo JL. CCTop: an intuitive, flexible and reliable CRISPR/Cas9 target prediction tool. PLoS One. 2015; 10 (4):e0124633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brinkman EK, Chen T, Amendola M, van Steensel B. Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res. 2014;42 (22):e168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bajoghli B, Kuri P, Inoue D, et al. Noninvasive in toto imaging of the thymus reveals heterogeneous migratory behavior of developing T cells. J Immunol. 2015; 195 (5):2177–2186. [DOI] [PubMed] [Google Scholar]
- 22.Lachmann N, Ackermann M, Frenzel E, et al. Large-scale hematopoietic differentiation of human induced pluripotent stem cells provides granulocytes or macrophages for cell replacement therapies. Stem Cell Reports. 2015;4 (2): 282–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dannenmann B, Zahabi A, Mir P, et al. Human iPSC-based model of severe congenital neutropenia reveals elevated UPR and DNA damage in CD34(+) cells preceding leukemic transformation. Exp Hematol. 2019;71-51-60. [DOI] [PubMed] [Google Scholar]
- 24.Hamilton N, Sabroe I, Renshaw SA. A method for transplantation of human HSCs into zebrafish, to replace humanised murine transplantation models. F1000Res. 2018; 7:594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Staal FJ, Spaink HP, Fibbe WE. Visualizing human hematopoietic stem cell trafficking in vivo using a zebrafish xenograft model. Stem Cells Dev. 2016;25 (4):360–365. [DOI] [PubMed] [Google Scholar]
- 26.Nasri M, Mir P, Dannenmann B, et al. Fluorescent labeling of CRISPR/Cas9 RNP for gene knockout in HSPCs and iPSCs reveals an essential role for GADD45b in stress response. Blood Adv. 2019;3 (1):63–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tidwell T, Wechsler J, Nayak RC, et al. Neutropenia-associated ELANE mutations disrupting translation initiation produce novel neutrophil elastase isoforms. Blood. 2014;123 (4):562–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kosicki M, Tomberg K, Bradley A. Repair of double-strand breaks induced by CRISPR-Cas9 leads to large deletions and complex rearrangements. Nat Biotechnol. 2018;36 (8):765–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Allport JR, Lim YC, Shipley JM, et al. Neutrophils from MMP-9- or neutrophil elastase-deficient mice show no defect in transendothelial migration under flow in vitro. J Leukoc Biol. 2002;71 (5):821–828. [PubMed] [Google Scholar]
- 30.Martinod K, Witsch T, Farley K, Gallant M, Remold-O’Donnell E, Wagner DD. Neutrophil elastase-deficient mice form neutrophil extracellular traps in an experimental model of deep vein thrombosis. J Thromb Haemost. 2016;14 (3):551–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hirche TO, Atkinson JJ, Bahr S, Belaaouaj A. Deficiency in neutrophil elastase does not impair neutrophil recruitment to inflamed sites. Am J Resp Cell Mol Biol. 2004;30 (4):576–584. [DOI] [PubMed] [Google Scholar]
- 32.Young RE, Thompson RD, Larbi KY, et al. Neutrophil elastase (NE)-deficient mice demonstrate a nonredundant role for NE in neutrophil migration, generation of proin-flammatory mediators, and phagocytosis in response to zymosan particles in vivo. J Immunol. 2004;172 (7):4493–4502. [DOI] [PubMed] [Google Scholar]
- 33.Belaaouaj A, McCarthy R, Baumann M, et al. Mice lacking neutrophil elastase reveal impaired host defense against gram negative bacterial sepsis. Nat Med. 1998;4 (5):615–618. [DOI] [PubMed] [Google Scholar]
- 34.Pham CT, Ivanovich JL, Raptis SZ, Zehnbauer B, Ley TJ. Papillon-Lefevre syndrome: correlating the molecular, cellular, and clinical consequences of cathepsin C/dipeptidyl peptidase I deficiency in humans. J Immunol. 2004;173 (12):7277–7281. [DOI] [PubMed] [Google Scholar]
- 35.Cario G, Skokowa J, Wang Z, et al. Heterogeneous expression pattern of pro- and anti-apoptotic factors in myeloid progenitor cells of patients with severe congenital neutropenia treated with granulocyte colony-stimulating factor. Br J Haematol. 2005;129 (2):275–278. [DOI] [PubMed] [Google Scholar]
- 36.Skokowa J, Cario G, Uenalan M, et al. LEF-1 is crucial for neutrophil granulocytopoiesis and its expression is severely reduced in congenital neutropenia. Nat Med. 2006;12 (10): 1191–1197. [DOI] [PubMed] [Google Scholar]
- 37.Skokowa J, Welte K. LEF-1 is a decisive transcription factor in neutrophil granulopoiesis. Ann N Y Acad Sci. 2007;1106:143–151. [DOI] [PubMed] [Google Scholar]
- 38.Skokowa J, Welte K. Dysregulation of myeloid-specific transcription factors in congenital neutropenia. Ann N Y Acad Sci. 2009;1176:94–100. [DOI] [PubMed] [Google Scholar]
- 39.Skokowa J, Welte K. Defective G-CSFR signaling pathways in congenital neutropenia. Hematol Oncol Clin North Am. 2013;27 (1): 75–88, viii. [DOI] [PubMed] [Google Scholar]
- 40.Skokowa J, Klimiankou M, Klimenkova O, et al. Interactions among HCLS1, HAX1 and LEF-1 proteins are essential for G-CSF-triggered granulopoiesis. Nat Med. 2012;18 (10):1550–1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Skokowa J, Lan D, Thakur BK, et al. NAMPT is essential for the G-CSF-induced myeloid differentiation via a NAD(+)-sirtuin-1-dependent pathway. Nat Med. 2009;15 (2):151–158. [DOI] [PubMed] [Google Scholar]