Abstract
Background
Microbiomes are extremely important for their host organisms, providing many vital functions and extending their hosts’ phenotypes. Natural studies of host-associated microbiomes can be difficult to interpret due to the high complexity of microbial communities, which hinders our ability to track and identify individual members along with the many factors that structure or perturb those communities. For this reason, researchers have turned to synthetic or constructed communities in which the identities of all members are known. However, due to the lack of tracking methods and the difficulty of creating a more diverse and identifiable community that can be distinguished through next-generation sequencing, most such in vivo studies have used only a few strains.
Results
To address this issue, we developed DISCo-microbe, a program for the design of an identifiable synthetic community of microbes for use in in vivo experimentation. The program is composed of two modules; (1) create, which allows the user to generate a highly diverse community list from an input DNA sequence alignment using a custom nucleotide distance algorithm, and (2) subsample, which subsamples the community list to either represent a number of grouping variables, including taxonomic proportions, or to reach a user-specified maximum number of community members. As an example, we demonstrate the generation of a synthetic microbial community that can be distinguished through amplicon sequencing. The synthetic microbial community in this example consisted of 2,122 members from a starting DNA sequence alignment of 10,000 16S rRNA sequences from the Ribosomal Database Project. We generated simulated Illumina sequencing data from the constructed community and demonstrate that DISCo-microbe is capable of designing diverse communities with members distinguishable by amplicon sequencing. Using the simulated data we were able to recover sequences from between 97–100% of community members using two different post-processing workflows. Furthermore, 97–99% of sequences were assigned to a community member with zero sequences being misidentified. We then subsampled the community list using taxonomic proportions to mimic a natural plant host–associated microbiome, ultimately yielding a diverse community of 784 members.
Conclusions
DISCo-microbe can create a highly diverse community list of microbes that can be distinguished through 16S rRNA gene sequencing, and has the ability to subsample (i.e., design) the community for the desired number of members and taxonomic proportions. Although developed for bacteria, the program allows for any alignment input from any taxonomic group, making it broadly applicable. The software and data are freely available from GitHub (https://github.com/dlcarper/DISCo-microbe) and Python Package Index (PYPI).
Keywords: Constructed community, Microbiome, 16S rRNA, Synthetic community, Taxonomic profiling, In vivo experimentation
Background
Multicellular eukaryotes live in association with complex communities of microorganisms (Zilber-Rosenberg & Rosenberg, 2008; Bordenstein & Theis, 2015; Rosenberg & Zilber-Rosenberg, 2016) that play important roles in host health and function (Huttenhower et al., 2012; Schlaeppi & Bulgarelli, 2015; Engel et al., 2016). Given the complexity of these systems and our inability to track and identify all members, it is often difficult to disentangle the factors influencing the structure and interactions among host-associated microbiomes. The development of synthetic model communities is a key strategy for addressing this issue (Busby et al., 2017). Next-generation sequencing of marker genes has demonstrated that both abiotic and biotic factors structure host-associated microbiomes (Spor, Koren & Ley, 2011; Huttenhower et al., 2012; Ofek-Lalzar et al., 2014; Adair & Douglas, 2017); however, the marker genes commonly used in these studies provide low taxonomic resolution, making it difficult to identify all microbes present in the community (Caporaso et al., 2011). Metagenomics studies provide insight into potential microbial function, but are not feasible for microbiomes within host tissues due to the presence of excess host DNA (Jiao et al., 2006; Feehery et al., 2013; Thoendel et al., 2016; Marotz et al., 2018). Accordingly, recent studies have utilized synthetic or simplified microbiome approaches to examine the drivers of host-associated microbiome assembly, interactions, and function (Bodenhausen et al., 2014; Lebeis et al., 2015; Timm et al., 2016; Niu et al., 2017). This approach involves adding previously characterized microbial strains to an axenic host organism, allowing for the investigation of colonization, shifts in community structure (Bodenhausen et al., 2014), microbe–microbe interactions, and host–microbe interactions. When such data are paired with genomic information, it becomes feasible to infer microbial strain metabolic potential. Despite the increased use and prioritization of synthetic systems by the research community (Busby et al., 2017), we currently lack adequate methods for systematically designing a microbial community that is identifiable by common sequencing techniques.
Until now, synthetic communities have been constructed from a functional perspective or with limited strains. For example, some researchers have focused on functional assets (characteristics) of microbes to create a specific metabolic output, often by combining a few bacterial (Shong, Jimenez Diaz & Collins, 2012; Mee et al., 2014; Shi et al., 2017) or fungal strains (Minty et al., 2013; Hu et al., 2017). Although useful for bio-engineering purposes, this approach is not as applicable to studies of microbiomes, in which diversity is much greater. Host-associated synthetic communities have also been restricted to a few strains, with confirmation through re-isolation, limiting researchers’ ability to extrapolate to more diverse communities (Bodenhausen et al., 2014; Niu et al., 2017; Herrera Paredes et al., 2018). Recent studies have linked host-associated microbiome function to microbial diversity (Turnbaugh et al., 2008; Laforest-Lapointe et al., 2017), requiring the incorporation of phylogenetic distance into synthetic community design. The design of phylogenetically diverse communities is associated with at least two major challenges: (1) creating a diverse community that can easily be distinguished through common high-throughput sequencing technologies, and (2) ensuring that community members possess the desired attributes (e.g., taxonomic composition and metabolic potential). Without advanced computational abilities, overcoming these challenges is formidable and time-consuming. Furthermore, manual bioinformatic workflows are difficult to document and error-prone, costing additional time and decreasing reproducibility.
In this paper, we describe an easy-to-use command-line program, Design of an Identifiable Synthetic Community of Microbes (DISCo-microbe), for creation of diverse communities of organisms that can be distinguished through next-generation sequencing technology for use in in vivo experiments. DISCo-microbe consists of two modules, create and subsample. The create module constructs a highly diverse community at a specified sequence difference from an input of aligned DNA/RNA sequences, e.g., 16S sequence. The module can either design a de novo community or design a community that includes targeted organisms. create solves problem (1) by easily generating a diverse community of members through an easily documentable method, ensuring reproducibility. The subsample module provides options for dividing the community into subsets, according to either the number of members or the proportions of a grouping variable, both of which can be specified by the user. subsample module solves problem (2) by allowing the user to subsample an already distinguishable community of members based on attributes of interest. Although this software was designed for construction of microbial communities, any DNA/RNA alignment can be used as input; consequently, users are not restricted to any particular organismal group or marker gene. DISCo-microbe is implemented in Python and is available through GitHub and PYPI.
Materials and Methods
DISCo-microbe is a command-line program written in Python and requires Biopython (Cock et al., 2009), which is automatically installed along with the program. We chose to implement DISCo-microbe in Python for easy portability to almost all systems. DISCo-microbe consists of two modules, create and subsample. We have written extensive documentation for DISCo-microbe following the principles outlined in (Seemann, 2013; Karimzadeh & Hoffman, 2018) including a quickstart tutorial that walks users through all commands, illustrating the ease of use and reproducibility of DISCo-microbe.
Workflow
Create module
The create module has two required arguments, an alignment of DNA or RNA sequences in FASTA format (–i-alignment) and a user-specified minimum sequence distance between community members (–p-editdistance). The module uses a greedy algorithm to construct a community maximizing the number of members at the user-specified sequence distance. The optional arguments for the create module include: (i) a community starter list (–p-include-strains), containing members the user would like to be included in the community; (ii) a seed number (–p-seed), for reproducibility; (iii) a metadata file (–i-metadata) for combination with the final community; iv) an option to output the FASTA file (–o-fasta) of the final community and; (v) an option to import a sequence distance database (–i-distance-database; described below).
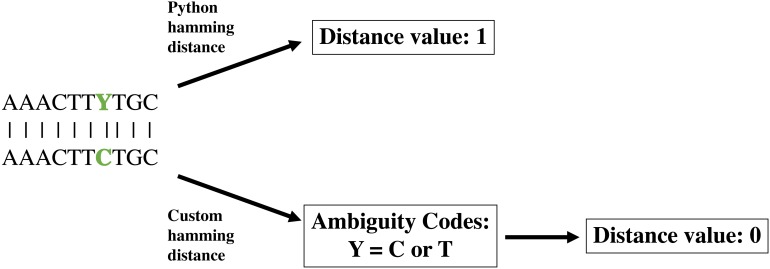
The create module operates in two distinct phases. The first phase creates a database of all pairwise sequence distances from the input alignment, calculated using a modified Hamming distance. The Hamming distance is a coding theory metric that measures the number of positions at which two sequences of equal length differ. Because the Hamming distance does not consider the nature of the differences, it can be problematic to determine the distance between molecular sequences, in which nucleotide ambiguities can be common; such ambiguities artificially inflate the number of differences between sequences, possibly causing the final community to be less distinguishable than expected (Fig. 1). To deal with IUPAC nucleotide ambiguities, we created a custom Hamming distance, termed the nucleotide Hamming distance, which accommodates nucleotide ambiguities and adjusts the distance value accordingly (Fig. 1). Furthermore, this metric can mitigate sequence errors introduced by PCR and sequencing technologies (Pfeiffer et al., 2018; Filges et al., 2019), allowing the identification of sequences containing up to d − 1 errors, where d is the user-specified minimum sequence distance. Lastly, we included an export of the distance database as a flat file for easy manipulation with command line utilities. This option also allows the user to load the database of previously calculated distances if a modification to the run parameters is wanted. Furthermore, the distance database is updated in real-time as distances are calculated, acting as a checkpoint to resume calculations with minimal lost time in the event that DISCo-microbe quits unexpectedly.
Figure 1. Demonstration of custom nucleotide Hamming distance.
Demonstration of Python Hamming distance and custom nucleotide Hamming distance, which takes into account nucleotide ambiguities.
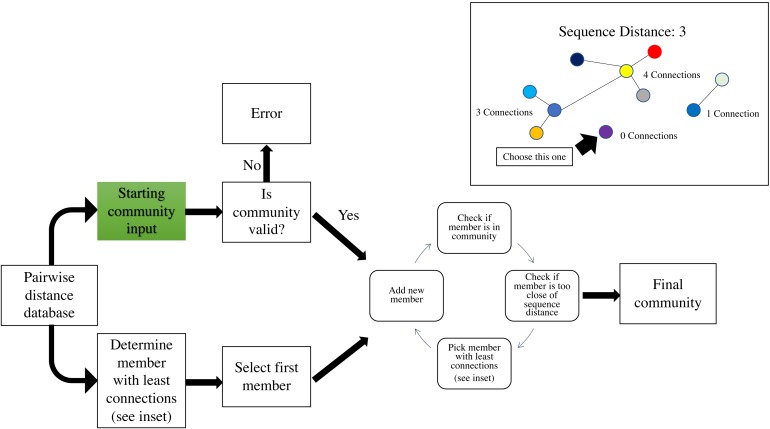
The second phase of the create module runs a greedy algorithm to construct a community. To initiate the community-building algorithm, the user can specify a starting community, which will be validated to determine that all pairwise distances meet the minimum requirement indicated by –p-editdistance. If the starting community is not valid at the indicated sequence distance, an error message with the conflicting sequence identifiers will be displayed. If a starting community is not specified, the individual with the fewest connections at the user-specified sequence distance (–p-editdistance) will be used to initiate the community (Fig. 2). If there is a tie for the fewest connections, one individual is selected at random. Once an initial community is established, the algorithm will iteratively add new members to the community by creating a list of possible members that meet a single requirement. The individual must meet the minimum sequence distance to any of the existing members; for example, if the user has specified a distance of 2, the module will check if the individual is at a distance of 0, 1 or 2 from any existing members. If this requirement is met, the individual is added to the list of potential community members. Next, the individual in the list with the fewest connections at the specified sequence distance (Fig. 2 inset) will be added to the community. Ties for the fewest connections are broken by randomly selecting an individual. The module will continue the process as described until there are no more individuals that meet the requirement for addition to the potential community member list. Current hierarchical clustering algorithms do not guarantee all sequences within a cluster are the specified distance from sequences within another cluster (Westcott & Schloss, 2015), which is essential to DISCo-microbe, motivating us to develop the currently implemented algorithm. Once the community list is complete, the program will output a tab-delimited text file of community members. The community list can be combined with metadata information (optional), such as taxonomic information, which is recommended if the user will be using the ‘subsample by proportions’ option later. A FASTA file of the community list can also be created if desired.
Figure 2. Workflow schematic of the loop that adds new members to the community, starting with the pairwise distance dictionary.
Inset: schematic of adding members with fewest connections at a specified DNA distance. Circles represent individuals, and lines indicate that the connected individuals are at a sequence distance of 3. Green indicates user input of file.
subsample module
The subsample module is designed to take the final output community from the create module and provide a subsample of the community. The module has multiple subsampling procedures. The first method is a random sampling (option: –p-num-taxa) of the indicated number of members, nfinal. The second method (option: –p-proportion) is for subsampling the specific proportions of a grouping variable. To illustrate the use of this option, we will refer to taxonomic information as the grouping variable; however, the user may provide any grouping variable for subsampling. For this option, the user will input two files: the community file from the create module with taxonomic information combined, and a file of the taxonomic groupings with desired proportions. DISCo-microbe will then generate a subsampling of the original community that is optimized to reflect the desired proportions. The optimization is accomplished through a greedy minimization of the sum of differences, , for the set TG of taxonomic groups specified in file 2 (taxonomic proportions file). Here, and are vectors of taxonomic group frequencies for the current and desired community, respectively, with and . The algorithm initializes fcurrent as the vector finput of taxonomic group frequencies of the community provided in file 1 (from create module) with members belonging to taxonomic groups in the set X, where groups not specified in file 2 are removed (X ≡ {x ∈ X|x⁄ ∈ TG}), and finput renormalized such that . Next, the algorithm will continuously iterate the following three steps:
(1) Determine the taxonomic group with largest difference in taxonomic group frequencies, .
(2) If the number of members in the taxonomic group identified in step 1 is less than 2 (ntmax < 2) break and output the current community; otherwise, randomly remove a member from tmax, resulting in fcurrent′.
(3) If , set otherwise stop the module and output the current community.
The user can modify the behavior of the algorithm by specifying both the number of members and the taxonomic proportions (–p-num-taxa and –p-proportion). Providing both options will force the algorithm to continue until the total number of members in the community, ntotal, is ≤nfinal (user-specified final number of members). Further, when both options are specified, step 2 of the greedy minimization is modified to not break iteration when ntmax < 2, and instead removes a member from the taxonomic group with the next-largest difference in frequencies, tnext, where ntnext ≥ 2. Additionally, if the force number option (option: –p-taxa-num-enforce) is used along with –p-num-taxa and –p-proportion, the algorithm will stop iteration when ntotal = nfinal regardless of whether the sum of frequency differences could be further minimized.
Test data set
The Ribosomal Database Project (Cole et al., 2014) file of 16S rRNA genes was downloaded (release 11.5, May 2019), and uncultured strains were using fasgrep (Lawrence et al., 2015). The alignment was trimmed to the V4 region, which is a commonly used region for next-generation sequencing of bacterial communities (Thompson et al., 2017). The initial file contained 239,244 sequences and was randomly subsampled to 10,000 sequences due to the computational intensity of building the community. A reference-based alignment against the SILVA database v. 132 (Pruesse et al., 2007) was created using the program SINA (Pruesse, Peplies & Glöckner, 2012). Alignment sites containing only gaps were removed using alncut (Lawrence et al., 2015). Additionally, 15 sequences aligned poorly and were removed, resulting in a final alignment of 9,985 sequences at a length of 502 bp. The 9,985-sequence alignment was used to create a highly diverse community at a minimum pairwise sequence distance of 3, with the seed set to 10 for reproducibility. Following construction, the subsample module was used to subsample the community to mimic the taxonomic composition a plant-associated microbiome. The final alignment, taxonomic proportion file, and commands used to create the community are available on GitHub for users to reproduce.
Benchmarking
We performed benchmarking on the distance database calculation and the full create command using hyperfine (https://github.com/sharkdp/hyperfine). Benchmarking was performed on a MacBook Air with 1.3 GHz Intel Core i5 with 10 replicate runs per benchmark. To perform the benchmarking, we subsampled the 16S ribosomal test dataset described above using the subsample command, to 50, 100, 250, 500, 1,000, 2,500, 5,000, 7,500, and the full 9,985 sequences for both the distance database calculation and the full create command.
Simulated Illumina data
We simulated 2 × 250 bp paired-end Illumina MiSeq sequencing data for the 16S rRNA RDP community described above using ART v2.5.8 (Huang et al., 2012) with the provided empirical error models for the Illumina MiSeq. We generated three different simulated sequencing data sets with 500 sequences per community member and two samples per simulation. The simulated data was analyzed using two post-processing workflows. The first workflow merges the forward and reverse reads using PEAR (Zhang et al., 2014) followed by dereplicating the sequences using FAST (Lawrence et al., 2015). The second workflow utilizes the dada2 pipeline (Callahan et al., 2016), a program commonly used in the analysis of microbial amplicon sequencing. The dada2 program models Illumina sequencing error and attempts to correct errors to recover the true sequence variants. The resulting sequences of both workflows were assigned to community members using the consensus BLAST (Altschul et al., 1990) method implemented in QIIME2 (Bolyen et al., 2019) with a 99% identity and 99% query length cutoff against the database of community member sequences. Using the community member assignment output, we determined the percent of sequences assigned to community members, percent of community members recovered, and for the dereplicated workflow, the accuracy of community member assignment. Unfortunately, the dada2 pipeline doesn’t provide a mapping of the predicted sequence variants to sequencing reads preventing us from determining the accuracy of community member assignment. Sequences that were unassigned by the consensus BLAST method were searched against the community member sequences using BLASTN keeping the top two hits.
Results
Workflow example
To demonstrate the applicability, usability, and ease of documenting workflows when using DISCo-microbe to construct identifiable diverse communities, we created and subsampled a community with a minimum sequence distance of 3 using 16S rRNA sequences from the RDP database. The initial sequence alignment contained the V4 region from 9,985 sequences with an average pairwise sequence distance of 10.6 ± 3.6%). Using the following create module
command:
| disco create –i-alignment RDP_aligned_sequences.fasta –p-editdistance 3 –p-seed 10 –i-metadata |
| RDP_Metadata_Taxonomy.txt –o-community-list RDP_Community_ED3_seed10.txt |
we constructed a community of 2,122 members that could be distinguished through next-generation sequencing. Using the following subsample module
command:
| disco subsample –i-input-community RDP_Community_ED3_seed10.txt –p-seed 10 –p-group-by Class –p-proportion RDP_Class_Proportions_file.txt |
the community was reduced to 784 community members with the approximate proportions of a plant–associated microbiome (Table 1; Cregger et al., 2018). The options for each module used above, along with the version of DISCo-microbe and Python, are the only documentation required to reproduce the design of this extremely complex community.
Table 1. Subsampled bacterial class proportions.
Bacterial class proportions used to subsample the community generated from the Ribosomal Database Project database and the actualized proportions of the resultant community.
| Bacterial class | Input proportions | Output proportions |
|---|---|---|
| Actinobacteria | 0.0885 | 0.0906 |
| Alphaproteobacteria | 0.1857 | 0.1875 |
| Anaerolineae | 0.004 | 0.0013 |
| Aquificae | 0.0003 | 0.0013 |
| Bacteroidia | 0.1 | 0.0982 |
| Betaproteobacteria | 0.1286 | 0.1301 |
| Chitinivibrionia | 0.004 | 0.0013 |
| Chloroflexia | 0.005 | 0.0051 |
| Deferribacteres | 0.0003 | 0.0013 |
| Deinococci | 0.0003 | 0.0026 |
| Deltaproteobacteria | 0.0418 | 0.0434 |
| Fibrobacteria | 0.0004 | 0.0026 |
| Fusobacteriia | 0.0003 | 0.0026 |
| Gammaproteobacteria | 0.4112 | 0.4133 |
| Gemmatimonadetes | 0.0073 | 0.0026 |
| Ktedonobacteria | 0.0097 | 0.0013 |
| Nitrospira | 0.0036 | 0.0051 |
| Planctomycetia | 0.009 | 0.0102 |
Benchmarking
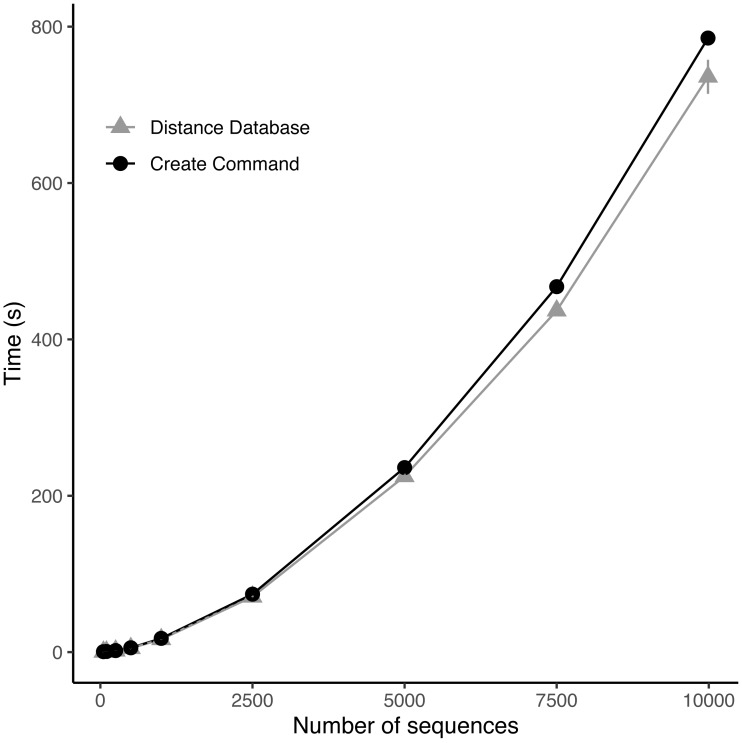
As the number of sequences increased the time to calculate the distance database and to create the full community increased exponentially (Fig. 3). Upon examination, the distance database was the most computationally expensive portion of the create module responsible for between 55 and 95% of the total time to create the community (Fig. 3). The full community construction with the alignment of 9,985 sequences using the create module took on average 13.09 min (±4.42 s) with 12.26 min (±4.01 s) being the distance database calculations.
Figure 3. Benchmarking of distance database and create module.
Benchmarking of custom nucleotide Hamming distance function for DNA and the create module at various numbers of 16S rRNA sequences subsampled from the Ribosomal Database Project dataset.
DISCo-microbe designs communities with members distinguishable by amplicon sequencing
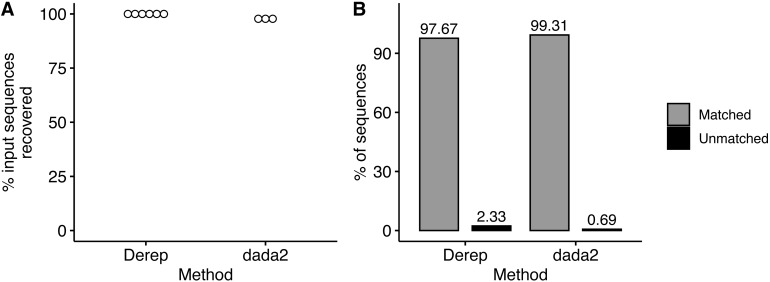
We simulated Illumina MiSeq sequencing data from the 2,120 member community constructed from the 9,985 16S rRNA sequences from the RDP database and described above. Unexpectedly, sequencing data was only generated from the first 2,065 community members due to an undocumented limit on the number of input sequences that ART (Huang et al., 2012) will process, however this does not change the overall results of the analysis. We noticed that ART simulated sequencing data consistent with empirically determined error rate of 0.24% errors per base (Pfeiffer et al., 2018). However, an average of 25% of the simulated sequences contained an error compared to an average of 6.4% of empirical sequences (Pfeiffer et al., 2018). Using the dereplication workflow, we were able to recover sequences from all 2,065 community members (Fig. 4A) and 97.7% (±0.0004%) of dereplicated sequences were assigned to a community member with the remaining 2.3% of sequences unassigned (Fig. 4B). Notably, none of the sequences were misclassified. Using the dada2 workflow, we recovered sequences from fewer of the community members (97.8% ± 0.0007%) compared to the dereplication workflow (Fig. 4A) but had a higher rate (99.3% ± 0.001) of sequence variants assigned to a community member (Fig. 4B). BLASTing unassigned sequences against the community member sequences mostly resulted in the top hit being the correct community member. Unexpectedly, one of the unassigned sequences from the dada2 workflow only had one nucleotide different from two community members. Upon further examination of these two community members we identified an alignment error in the alignment used to create the community that when corrected resulted in the two community members having a pairwise distance of 2 instead of the required 3.
Figure 4. Recovery and taxonomic assignment of sequences from Illumina simulator.
(A) The percent of sequences from the input community that were recovered from the Illumina simulator with the dereplication and the dada2 method. (B) The percent of sequences that had a taxonomic assignment or were not assigned.
Discussion
Microbial diversity is linked to function (Turnbaugh et al., 2008; Laforest-Lapointe et al., 2017), but understanding that diversity can be difficult due to the low resolution of taxonomic marker genes and the complexity of the microbial community, limiting our ability to identify and track individual community members. To tease apart the complex interactions within communities, there has been an increased demand for synthetic community systems (Busby et al., 2017). However, the generation of complex communities of organisms that can be easily distinguished through high-throughput methods can be difficult without strong computational skills. In general, two challenges are associated with the design of a synthetic community: (1) creation of a distinguishable community through common sequencing methods and (2) development of a community with the desired traits. Additionally, manual creation can lead to a lack of reproducibility due to the difficulty of documenting the workflow. In this paper, we describe an easy to use command-line program, Design of an Identifiable Synthetic Community of Microbes (DISCo-microbe), for the creation of diverse communities of organisms that can be distinguished through next-generation sequencing technology during in vivo experiments. DISCo-microbe solves the two previously mentioned problems using two modules, create and subsample.
The create module allows the user to construct a diverse community that is identifiable using common sequencing methods, thus solving the first problem. The ability to specify a minimum sequence distance allows flexibility in the construction of the community due to its robustness to sequencing errors introduced through PCR and sequencing (Pfeiffer et al., 2018). For example, if the user sets the minimum sequence distance to 5, sequences containing up to 2 sequencing errors ([d − 1]∕2) can be confidently assigned to the correct community member, sequences containing up to 4 errors (d − 1) can be identified, and it would take a minimum of 5 errors to assign a sequence to the incorrect community member. Usually, the smaller the minimum sequence distance, the more members will be included in the constructed community, potentially motivating users to set the minimum sequence distance to lowest setting of 1. However, at a minimum sequence distance of l, it only requires a single sequencing error to assign a sequence to the wrong community member. In order to implement the create module, we developed a custom nucleotide Hamming distance that accommodates nucleotide ambiguities. This is the first application of the Hamming distance algorithm incorporating IUPAC nucleotide ambiguity codes to measure distance between pairs of aligned sequences implemented in Python (see Šošić & Šikić, 2017 for an implementation in C). We determined that the most time-consuming step is the creation of the distance database due to the number of calculations required . Despite the large number of calculations required to create the distance database, the runtime for the create module on the largest community containing 9,985 sequences was only 13 min on a MacBook Air laptop.
The subsample module allows flexibility in the final constructed community. Specifically, it allows users to adapt the community to their experimental specifications, either by limiting the number of strains, specifying proportions of a grouping variable, or both. The subsample module eliminates major problem (2) by allowing users to tailor the already distinguishable community to include desired traits or proportions of members, examples of which are found in the detailed documentation.
Using simulated Illumina MiSeq data, we demonstrated the ability of DISCo-microbe to design diverse communities with members distinguishable by amplicon sequencing. We were able to identify sequences from 97.5% and 100% of community members when using the dada2 and dereplication workflows respectively. Notably, when using the dereplication workflow, we show that we do not have any misclassified sequences indicating that all members were distinguishable. Furthermore, the inability to assign 2.3% and 0.7% of sequences to community members in the dereplicate and dada2 workflows respectively were a result of multiple sequencing errors. The number of unassignable sequences in our simulated data is likely an overestimation compared to real data. Given that 25% of ART simulated Illumnia MiSeq reads had at least one error compared to the recently documented empirical rate of 6% (Pfeiffer et al., 2018). Despite the greater number of sequences being mutated than expected in a real sequencing run, we still show the ability to discriminate between community members with a high degree of accuracy and recall. Further investigation into the unassigned sequences using BLASTN demonstrated the ability to accurately assign all but one of these sequences based on their top BLAST hit against the community member sequences. Consequently, increasing the overall percent of sequences assigned to community members and percent of community members recovered without increasing our false positive rate. The only sequence unassignable by BLASTN was a dada2 sequence variant that only has a single nucleotide difference from two community members. Upon further investigation of these two community members we discovered errors in the alignment resulting in an overestimation of the distance between these two community members. This illustrates the dependence of DISCo-microbe on an accurate input alignment to determine the correct distance between individuals, and thus creating a community at the desired sequence distance. Notably, despite this alignment error the dereplication workflow along with BLASTN was able to accurately distinguish all community members making the community still identifiable.
Conclusions
DISCo-microbe is the first software designed for the construction of a diverse community of organisms that can be distinguished through low-cost, high-throughput amplicon sequencing for use in in vivo experiments. DISCo-microbe allows non-programmers to easily and reproducibly construct communities in which the members are identifiable through amplicon sequencing and the communities conform to user-specified attributes or numbers of members. DISCo-microbe is also the first software to implement a nucleotide specific Hamming distance in Python that takes into account nucleotide ambiguities in sequencing data. Although initially designed for bacterial community construction, the input of a nucleotide sequence alignment from any region allows the software to be used with any group of organisms. DISCo-microbe is designed for easy expansion of utilities; planned future versions will include new algorithms for community construction as well as new modules for creating a suite of tools for the design of constructed communities and processing of the resulting data.
Availability and requirements
Project name: DISCo-microbe
Project home page: https://github.com/dlcarper/DISCo-microbe
Operating system(s): platform-independent
Programming language: Python ≥ 3.4
Other requirements: BioPython
License: GNU General Public License v3.0
Abbreviations
- DNA
Deoxyribonucleic acid
- RNA
Ribonucleic acid
- rRNA
Ribosomal ribonucleic acid
- FASTA
Fast-all (file format)
- PYPI
Python Package Index
- PCR
polymerase chain reaction
Funding Statement
This research was sponsored by the Genomic Science Program, U.S. Department of Energy, Office of Science, Biological and Environmental Research as part of the Plant Microbe Interfaces Scientific Focus Area. Oak Ridge National Laboratory is managed by UT-Battelle, LLC, for the U.S. Department of Energy under contract DE-AC05-00OR22725. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Contributor Information
Dana L. Carper, Email: carperdl@ornl.gov.
David J. Weston, Email: westondj@ornl.gov.
Additional Information and Declarations
Competing Interests
The authors declare there are no competing interests.
Author Contributions
Dana L. Carper conceived and designed the experiments, performed the experiments, analyzed the data, prepared figures and/or tables, authored or reviewed drafts of the paper, wrote the majority of the source code, and approved the final draft.
Travis J. Lawrence performed the experiments, analyzed the data, authored or reviewed drafts of the paper, helped code the subsample module, and approved the final draft.
Alyssa A. Carrell performed the experiments, authored or reviewed drafts of the paper, wrote documentation for software, and approved the final draft.
Dale A. Pelletier and David J. Weston authored or reviewed drafts of the paper, and approved the final draft.
Data Availability
The following information was supplied regarding data availability:
Data and code are available at GitHub: https://github.com/dlcarper/DISCo-microbe.git.
References
- Adair & Douglas (2017).Adair KL, Douglas AE. Making a microbiome: the many determinants of host-associated microbial community composition. Current Opinion in Microbiology. 2017;35:23–29. doi: 10.1016/j.mib.2016.11.002. [DOI] [PubMed] [Google Scholar]
- Altschul et al. (1990).Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. Journal of Molecular Biology. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Bodenhausen et al. (2014).Bodenhausen N, Bortfeld-Miller M, Ackermann M, Vorholt JA. A synthetic community approach reveals plant genotypes affecting the phyllosphere microbiota. PLOS Genetics. 2014;10:e1004283. doi: 10.1371/journal.pgen.1004283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolyen et al. (2019).Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, Alexander H, Alm EJ, Arumugam M, Asnicar F, Bai Y, Bisanz JE, Bittinger K, Brejnrod A, Brislawn CJ, Brown CT, Callahan BJ, Caraballo-Rodríguez AM, Chase J, Cope EK, Da Silva R, Diener C, Dorrestein PC, Douglas GM, Durall DM, Duvallet C, Edwardson CF, Ernst M, Estaki M, Fouquier J, Gauglitz JM, Gibbons SM, Gibson DL, Gonzalez A, Gorlick K, Guo J, Hillmann B, Holmes S, Holste H, Huttenhower C, Huttley GA, Janssen S, Jarmusch AK, Jiang L, Kaehler BD, Bin KK, Keefe CR, Keim P, Kelley ST, Knights D, Koester I, Kosciolek T, Kreps J, Langille MGI, Lee J, Ley R, Liu Y-X, Loftfield E, Lozupone C, Maher M, Marotz C, Martin BD, McDonald D, McIver LJ, Melnik AV, Metcalf JL, Morgan SC, Morton JT, Naimey AT, Navas-Molina JA, Nothias LF, Orchanian SB, Pearson T, Peoples SL, Petras D, Preuss ML, Pruesse E, Rasmussen LB, Rivers A, Robeson MS, Rosenthal P, Segata N, Shaffer M, Shiffer A, Sinha R, Song SJ, Spear JR, Swafford AD, Thompson LR, Torres PJ, Trinh P, Tripathi A, Turnbaugh PJ, Ul-Hasan S, Van der Hooft JJJ, Vargas F, Vázquez-Baeza Y, Vogtmann E, Von Hippel M, Walters W, Wan Y, Wang M, Warren J, Weber KC, Williamson CHD, Willis AD, Xu ZZ, Zaneveld JR, Zhang Y, Zhu Q, Knight R, Caporaso JG. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nature Biotechnology. 2019;37:852–857. doi: 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordenstein & Theis (2015).Bordenstein SR, Theis KR. Host biology in light of the microbiome: ten principles of holobionts and hologenomes. PLOS Biology. 2015;13:e1002226. doi: 10.1371/journal.pbio.1002226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busby et al. (2017).Busby PE, Soman C, Wagner MR, Friesen ML, Kremer J, Bennett A, Morsy M, Eisen JA, Leach JE, Dangl JL. Research priorities for harnessing plant microbiomes in sustainable agriculture. PLOS Biology. 2017;15:e2001793. doi: 10.1371/journal.pbio.2001793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callahan et al. (2016).Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. DADA2: high-resolution sample inference from Illumina amplicon data. Nature Methods. 2016;13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso et al. (2011).Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, Fierer N, Knight R. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:4516–4522. doi: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cock et al. (2009).Cock PJA, Antao T, Chang JT, Chapman BA, Cox CJ, Dalke A, Friedberg I, Hamelryck T, Kauff F, Wilczynski B, De Hoon MJL. Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics. 2009;25:1422–1423. doi: 10.1093/bioinformatics/btp163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole et al. (2014).Cole JR, Wang Q, Fish JA, Chai B, McGarrell DM, Sun Y, Brown CT, Porras-Alfaro A, Kuske CR, Tiedje JM. Ribosomal database project: data and tools for high throughput rRNA analysis. Nucleic Acids Research. 2014;42:D633–D642. doi: 10.1093/nar/gkt1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cregger et al. (2018).Cregger MA, Veach AM, Yang ZK, Crouch MJ, Vilgalys R, Tuskan GA, Schadt CW. The Populus holobiont: dissecting the effects of plant niches and genotype on the microbiome. Microbiome. 2018;6:31. doi: 10.1186/s40168-018-0413-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel et al. (2016).Engel P, Kwong WK, McFrederick Q, Anderson KE, Barribeau SM, Chandler JA, Cornman RS, Dainat J, De Miranda JR, Doublet V, Emery O, Evans JD, Farinelli L, Flenniken ML, Granberg F, Grasis JA, Gauthier L, Hayer J, Koch H, Kocher S, Martinson VG, Moran N, Munoz-Torres M, Newton I, Paxton RJ, Powell E, Sadd BM, Schmid-Hempel P, Schmid-Hempel R, Song SJ, Schwarz RS, VanEngelsdorp D, Dainat B. The bee microbiome: impact on bee health and model for evolution and ecology of host-microbe interactions. mBio. 2016;7:1–9. doi: 10.1128/mBio.02164-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feehery et al. (2013).Feehery GR, Yigit E, Oyola SO, Langhorst BW, Schmidt VT, Stewart FJ, Dimalanta ET, Amaral-Zettler LA, Davis T, Quail MA, Pradhan S. A method for selectively enriching microbial DNA from contaminating vertebrate host DNA. PLOS ONE. 2013;8:e76096. doi: 10.1371/journal.pone.0076096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filges et al. (2019).Filges S, Yamada E, Ståhlberg A, Godfrey TE. Impact of polymerase fidelity on background error rates in next-generation sequencing with unique molecular identifiers/barcodes. Scientific Reports. 2019;9:3503. doi: 10.1038/s41598-019-39762-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera Paredes et al. (2018).Herrera Paredes S, Gao T, Law TF, Finkel OM, Mucyn T, Teixeira PJPL, Salas González I, Feltcher ME, Powers MJ, Shank EA, Jones CD, Jojic V, Dangl JL, Castrillo G. Design of synthetic bacterial communities for predictable plant phenotypes. PLOS Biology. 2018;16:e2003962. doi: 10.1371/journal.pbio.2003962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu et al. (2017).Hu J, Xue Y, Guo H, Gao M, Li J, Zhang S, Tsang YF. Design and composition of synthetic fungal-bacterial microbial consortia that improve lignocellulolytic enzyme activity. Bioresource Technology. 2017;227:247–255. doi: 10.1016/j.biortech.2016.12.058. [DOI] [PubMed] [Google Scholar]
- Huang et al. (2012).Huang W, Li L, Myers JR, Marth GT. ART: a next-generation sequencing read simulator. Bioinformatics. 2012;28:593–594. doi: 10.1093/bioinformatics/btr708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttenhower et al. (2012).Huttenhower C, Gevers D, Knight R, Abubucker S, Badger JH, Chinwalla AT, Creasy HH, Earl AM, Fitzgerald MG, Fulton RS, Giglio MG, Hallsworth-Pepin K, Lobos EA, Madupu R, Magrini V, Martin JC, Mitreva M, Muzny DM, Sodergren EJ, Versalovic J, Wollam AM, Worley KC, Wortman JR, Young SK, Zeng Q, Aagaard KM, Abolude OO, Allen-Vercoe E, Alm EJ, Alvarado L, Andersen GL, Anderson S, Appelbaum E, Arachchi HM, Armitage G, Arze CA, Ayvaz T, Baker CC, Begg L, Belachew T, Bhonagiri V, Bihan M, Blaser MJ, Bloom T, Bonazzi V, Paul Brooks J, Buck GA, Buhay CJ, Busam DA, Campbell JL, Canon SR, Cantarel BL, Chain PSG, Chen IMA, Chen L, Chhibba S, Chu K, Ciulla DM, Clemente JC, Clifton SW, Conlan S, Crabtree J, Cutting MA, Davidovics NJ, Davis CC, Desantis TZ, Deal C, Delehaunty KD, Dewhirst FE, Deych E, Ding Y, Dooling DJ, Dugan SP, Michael Dunne W, Scott Durkin A, Edgar RC, Erlich RL, Farmer CN, Farrell RM, Faust K, Feldgarden M, Felix VM, Fisher S, Fodor AA, Forney LJ, Foster L, Di Francesco V, Friedman J, Friedrich DC, Fronick CC, Fulton LL, Gao H, Garcia N, Giannoukos G, Giblin C, Giovanni MY, Goldberg JM, Goll J, Gonzalez A, Griggs A, Gujja S, Kinder Haake S, Haas BJ, Hamilton HA, Harris EL, Hepburn TA, Herter B, Hoffmann DE, Holder ME, Howarth C, Huang KH, Huse SM, Izard J, Jansson JK, Jiang H, Jordan C, Joshi V, Katancik JA, Keitel WA, Kelley ST, Kells C, King NB, Knights D, Kong HH, Koren O, Koren S, Kota KC, Kovar CL, Kyrpides NC, La Rosa PS, Lee SL, Lemon KP, Lennon N, Lewis CM, Lewis L, Ley RE, Li K, Liolios K, Liu B, Liu Y, Lo CC, Lozupone CA, Dwayne Lunsford R, Madden T, Mahurkar AA, Mannon PJ, Mardis ER, Markowitz VM, Mavromatis K, McCorrison JM, McDonald D, McEwen J, McGuire AL, McInnes P, Mehta T, Mihindukulasuriya KA, Miller JR, Minx PJ, Newsham I, Nusbaum C, Oglaughlin M, Orvis J, Pagani I, Palaniappan K, Patel SM, Pearson M, Peterson J, Podar M, Pohl C, Pollard KS, Pop M, Priest ME, Proctor LM, Qin X, Raes J, Ravel J, Reid JG, Rho M, Rhodes R, Riehle KP, Rivera MC, Rodriguez-Mueller B, Rogers YH, Ross MC, Russ C, Sanka RK, Sankar P, Fah Sathirapongsasuti J, Schloss JA, Schloss PD, Schmidt TM, Scholz M, Schriml L, Schubert AM, Segata N, Segre JA, Shannon WD, Sharp RR, Sharpton TJ, Shenoy N. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao et al. (2006).Jiao J-Y, Wang H-X, Zeng Y, Shen Y-M. Enrichment for microbes living in association with plant tissues. Journal of Applied Microbiology. 2006;100:830–837. doi: 10.1111/j.1365-2672.2006.02830.x. [DOI] [PubMed] [Google Scholar]
- Karimzadeh & Hoffman (2018).Karimzadeh M, Hoffman MM. Top considerations for creating bioinformatics software documentation. Briefings in Bioinformatics. 2018;19:693–699. doi: 10.1093/bib/bbw134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laforest-Lapointe et al. (2017).Laforest-Lapointe I, Paquette A, Messier C, Kembel SW. Leaf bacterial diversity mediates plant diversity and ecosystem function relationships. Nature. 2017;546:145–147. doi: 10.1038/nature22399. [DOI] [PubMed] [Google Scholar]
- Lawrence et al. (2015).Lawrence TJ, Kauffman KT, Amrine KCH, Carper DL, Lee RS, Becich PJ, Canales CJ, Ardell DH. FAST: FAST analysis of sequences toolbox. Frontiers in Genetics. 2015;6:172. doi: 10.3389/fgene.2015.00172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebeis et al. (2015).Lebeis SL, Paredes SH, Lundberg DS, Breakfield N, Gehring J, McDonald M, Malfatti S, Glavina del Rio T, Jones CD, Tringe SG, Dangl JL. Salicylic acid modulates colonization of the root microbiome by specific bacterial taxa. Science. 2015;349:860–864. doi: 10.1126/science.aaa8764. [DOI] [PubMed] [Google Scholar]
- Marotz et al. (2018).Marotz CA, Sanders JG, Zuniga C, Zaramela LS, Knight R, Zengler K. Improving saliva shotgun metagenomics by chemical host DNA depletion. Microbiome. 2018;6:42. doi: 10.1186/s40168-018-0426-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mee et al. (2014).Mee MT, Collins JJ, Church GM, Wang HH. Syntrophic exchange in synthetic microbial communities. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:E2149–E2156. doi: 10.1073/pnas.1405641111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minty et al. (2013).Minty JJ, Singer ME, Scholz SA, Bae C-H, Ahn J-H, Foster CE, Liao JC, Lin XN. Design and characterization of synthetic fungal-bacterial consortia for direct production of isobutanol from cellulosic biomass. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:14592–14597. doi: 10.1073/pnas.1218447110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu et al. (2017).Niu B, Paulson JN, Zheng X, Kolter R. Simplified and representative bacterial community of maize roots. Proceedings of the National Academy of Sciences of the United States of America. 2017;114:E2450–E2459. doi: 10.1073/pnas.1616148114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ofek-Lalzar et al. (2014).Ofek-Lalzar M, Sela N, Goldman-Voronov M, Green SJ, Hadar Y, Minz D. Niche and host-associated functional signatures of the root surface microbiome. Nature Communications. 2014;5:4950. doi: 10.1038/ncomms5950. [DOI] [PubMed] [Google Scholar]
- Pfeiffer et al. (2018).Pfeiffer F, Gröber C, Blank M, Händler K, Beyer M, Schultze JL, Mayer G. Systematic evaluation of error rates and causes in short samples in next-generation sequencing. Scientific Reports. 2018;8:10950. doi: 10.1038/s41598-018-29325-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruesse, Peplies & Glöckner (2012).Pruesse E, Peplies J, Glöckner FO. SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics. 2012;28:1823–1829. doi: 10.1093/bioinformatics/bts252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruesse et al. (2007).Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, Glockner FO. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Research. 2007;35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg & Zilber-Rosenberg (2016).Rosenberg E, Zilber-Rosenberg I. Microbes drive evolution of animals and plants: the hologenome concept. mBio. 2016;7:1–8. doi: 10.1128/mBio.01395-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlaeppi & Bulgarelli (2015).Schlaeppi K, Bulgarelli D. The plant microbiome at work. Molecular Plant-Microbe Interactions. 2015;28:212–217. doi: 10.1094/MPMI-10-14-0334-FI. [DOI] [PubMed] [Google Scholar]
- Seemann (2013).Seemann T. Ten recommendations for creating usable bioinformatics command line software. GigaScience. 2013;2(1):15. doi: 10.1186/2047-217X-2-15. 2047-217X-2-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi et al. (2017).Shi Y, Pan C, Wang K, Chen X, Wu X, Chen C-TA, Wu B. Synthetic multispecies microbial communities reveals shifts in secondary metabolism and facilitates cryptic natural product discovery. Environmental Microbiology. 2017;19:3606–3618. doi: 10.1111/1462-2920.13858. [DOI] [PubMed] [Google Scholar]
- Shong, Jimenez Diaz & Collins (2012).Shong J, Jimenez Diaz MR, Collins CH. Towards synthetic microbial consortia for bioprocessing. Current Opinion in Biotechnology. 2012;23:798–802. doi: 10.1016/j.copbio.2012.02.001. [DOI] [PubMed] [Google Scholar]
- Spor, Koren & Ley (2011).Spor A, Koren O, Ley R. Unravelling the effects of the environment and host genotype on the gut microbiome. Nature Reviews Microbiology. 2011;9:279–290. doi: 10.1038/nrmicro2540. [DOI] [PubMed] [Google Scholar]
- Thoendel et al. (2016).Thoendel M, Jeraldo PR, Greenwood-Quaintance KE, Yao JZ, Chia N, Hanssen AD, Abdel MP, Patel R. Comparison of microbial DNA enrichment tools for metagenomic whole genome sequencing. Journal of Microbiological Methods. 2016;127:141–145. doi: 10.1016/j.mimet.2016.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson et al. (2017).Thompson LR, Sanders JG, McDonald D, Amir A, Ladau J, Locey KJ, Prill RJ, Tripathi A, Gibbons SM, Ackermann G, Navas-Molina JA, Janssen S, Kopylova E, Vázquez-Baeza Y, González A, Morton JT, Mirarab S, Zech Xu Z, Jiang L, Haroon MF, Kanbar J, Zhu Q, Jin Song S, Kosciolek T, Bokulich NA, Lefler J, Brislawn CJ, Humphrey G, Owens SM, Hampton-Marcell J, Berg-Lyons D, McKenzie V, Fierer N, Fuhrman JA, Clauset A, Stevens RL, Shade A, Pollard KS, Goodwin KD, Jansson JK, Gilbert JA, Knight R. A communal catalogue reveals Earth’s multiscale microbial diversity. Nature. 2017;551:457–463. doi: 10.1038/nature24621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timm et al. (2016).Timm CM, Pelletier DA, Jawdy SS, Gunter LE, Henning JA, Engle N, Aufrecht J, Gee E, Nookaew I, Yang Z, Lu T-Y, Tschaplinski TJ, Doktycz MJ, Tuskan GA, Weston DJ. Two poplar-associated bacterial isolates induce additive favorable responses in a constructed plant-microbiome system. Frontiers in Plant Science. 2016;7:1–10. doi: 10.3389/fpls.2016.00497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh et al. (2008).Turnbaugh PJ, Bäckhed F, Fulton L, Gordon JI. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host & Microbe. 2008;3:213–223. doi: 10.1016/j.chom.2008.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Šošić & Šikić (2017).Šošić M, Šikić M. Edlib: a C/C ++ library for fast, exact sequence alignment using edit distance. Bioinformatics. 2017;33:1394–1395. doi: 10.1093/bioinformatics/btw753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westcott & Schloss (2015).Westcott SL, Schloss PD. De novo clustering methods outperform reference-based methods for assigning 16S rRNA gene sequences to operational taxonomic units. PeerJ. 2015;3:e1487. doi: 10.7717/peerj.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang et al. (2014).Zhang J, Kobert K, Flouri T, Stamatakis A. PEAR: a fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics. 2014;30:614–620. doi: 10.1093/bioinformatics/btt593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilber-Rosenberg & Rosenberg (2008).Zilber-Rosenberg I, Rosenberg E. Role of microorganisms in the evolution of animals and plants: the hologenome theory of evolution. FEMS Microbiology Reviews. 2008;32:723–735. doi: 10.1111/j.1574-6976.2008.00123.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The following information was supplied regarding data availability:
Data and code are available at GitHub: https://github.com/dlcarper/DISCo-microbe.git.