Abstract
Family with sequence similarity (FAM46) proteins are newly identified metazoan-specific poly(A) polymerases (PAPs). Although predicted as Gld-2-like eukaryotic non-canonical PAPs, the detailed architecture of FAM46 proteins is still unclear. Exact biological functions for most of FAM46 proteins also remain largely unknown. Here, we report the first crystal structure of a FAM46 protein, FAM46B. FAM46B is composed of a prominently larger N-terminal catalytic domain as compared to known eukaryotic PAPs, and a C-terminal helical domain. FAM46B resembles prokaryotic PAP/CCA-adding enzymes in overall folding as well as certain inter-domain connections, which distinguishes FAM46B from other eukaryotic non-canonical PAPs. Biochemical analysis reveals that FAM46B is an active PAP, and prefers adenosine-rich substrate RNAs. FAM46B is uniquely and highly expressed in human pre-implantation embryos and pluripotent stem cells, but sharply down-regulated following differentiation. FAM46B is localized to both cell nucleus and cytosol, and is indispensable for the viability of human embryonic stem cells. Knock-out of FAM46B is lethal. Knock-down of FAM46B induces apoptosis and restricts protein synthesis. The identification of the bacterial-like FAM46B, as a pluripotent stem cell-specific PAP involved in the maintenance of translational efficiency, provides important clues for further functional studies of this PAP in the early embryonic development of high eukaryotes.
INTRODUCTION
PAPs are a branch of the nucleotidyltransferase (NTase) superfamily (1). Previous structural studies have established close relationship between PAPs and CCA-adding enzymes, another group of template-independent RNA polymerase (2). Eukaryotic PAPs share sequence similarity to the class-I archaeal CCA-adding enzymes, whereas bacterial PAPs are homologous to the class-II eukaryotic and bacterial CCA-adding enzymes (3). Eukaryotic PAPs can be classified into two subgroups. The ‘canonical’ PAPs, represented by nuclear PAPα, are responsible for adding long poly(A) tail during mRNA maturation. They are composed of three domains: an N-terminal catalytic domain containing the signature NTase motif, a central domain and a C-terminal RNA-binding domain (RBD). The ‘non-canonical’ PAPs, including but not limited to Gld-2, terminal uridylyltransferase (TUTs) and mitochondrial (mt-)PAP, add poly(A) tails or short terminal tails to a variety of RNA substrates including mRNA, snRNA, miRNA, aberrant rRNA and snoRNA (4). Given the diversity of their substrate preference, these non-canonical PAPs are recently renamed terminal nucleotididyltranferase (TENTs) (5). TENTs usually share a bipartite core PAP domain that lacks the RBD, and have different accessory domains to fulfil their diverse functions (4).
Most of the known eukaryotic PAPs are localized in the nucleus, and their activity and specificity relies on the association with other co-factors. For example, PAPα functions as a component of the cleavage and polyadenylation specificity factor (CPSF) complex (6). Caenorhabditiselegans (ce)Gld-2 separately forms complex with Gld-3 or RNP-8 to control gamete sex (7). In contrast, bacterial PAPs polyadenylate mRNAs in a non-discriminative manner, and usually require no partner (8,9). In terms of overall structure, bacterial PAPs are characterized by a seahorse-like shape, where the catalytic head domain is linearly aligned with the neck, body and tail domains involved in substrate RNA binding (9). This domain organization is distinct from that of eukaryotic PAPs.
Family with sequence similarity 46 (FAM46) is a group of predicted NTases found mainly in vertebrates (1). Human and mice both have four FAM46 proteins: namely FAM46A/B/C/D (also named TENT5A/B/C/D) (10). The amino acid sequences of these homologs share >40% overall sequence identity, but are not apparently associated with other protein families. Prior bioinformatics analysis suggested that the FAM46 proteins are non-classical PAPs (11), which was supported by a recent study on FAM46C, a putative suppressor for multiple myeloma (12). However, the detailed biological roles of other FAM46 proteins and the structural feature of this protein family still remain unclear.
Most eukaryotic mRNAs carry poly(A) tails, whose length is closely coupled with translational efficiency and mRNA stability (13). These tails are firstly added by nuclear PAPs during transcription termination, and usually shortened by deadenylases (14,15). The poly(A) tails can also be re-extended in the cytoplasm, which may promote translation and inhibit decay of certain mRNAs (16–18). This tail-length regulatory mechanism is considered to be an important regime of translation control in the early development of metazoan (19,20). A cytoplasmic PAP, Gld-2, was found to be responsible for this process during gametogenesis in C. elegans (21), Drosophila (22) and Xenopus (23). However, the cytoplasmic polyadenylation process is less understood in mammalian early embryos, partly due to the lack of information on corresponding PAPs.
In this study, we started from structure determination and revealed that FAM46B is a robust bacterial-like PAP. Being specifically expressed in human pluripotent stem cells, FAM46B is critical for cell viability, possibly by maintaining the stability of certain mRNAs. These results expand the knowledge of eukaryotic PAPs, and provide new insight into the translational control in early development of mammals.
MATERIALS AND METHODS
Protein expression and purification
The cDNAs of Xenopus tropicalis (xt), Homo sapiens (hs)FAM46B and Thermotoga maritima CCA adding enzyme (tmCCA) were purchased from YouBio. All constructs and mutants were expressed from a modified pET28 vector with an N-terminal His6-tag fusion followed by a PreScission protease (PSP) cleavage site. Recombinant plasmids were transformed into Escherichia coli Rosetta (DE3) cells. Transformed bacteria were cultured at 37°C in Terrific Broth (TB) medium until OD600 reached 0.6, and then induced with 0.1 mM isopropyl-1-thio-β-d-thiogalactopyranoside (IPTG). Induced bacteria were grown overnight at 18°C. Cells were lysed in 50 mM HEPES pH 7.5, 500 mM NaCl, 30 mM imidazole, 5 mM MgCl2, 5 mM KCl, 1 mM phenylmethanesulfonylfluoride (PMSF), 1 μM DNase I and 2 mM β-mercaptoethanol (β-ME) using a cell disruptor (JNBIO) and subjected to centrifugation at 40 000g for 1 h. The supernatant was filtered and applied to a Ni-NTA (first Ni-NTA) column (GE Healthcare) equilibrated with binding buffer A containing 20 mM HEPES pH 7.5, 500 mM NaCl, 30 mM imidazole, 2 mM MgCl2, 2 mM KCl and 2 mM β-ME. The column was extensively washed with buffer A and shortly with 20 mM HEPES pH 7.5, 500 mM NaCl, 70 mM imidazole, 2 mM MgCl2, 2 mM KCl and 2 mM β-ME. Proteins were eluted with 20 mM HEPES pH 7.5, 500 mM NaCl, 300 mM imidazole, 2 mM MgCl2, 2 mM KCl and 2 mM β-ME before incubated with 20 μg glutathione S-transferase (GST)-fused PSP to remove the His6-tag and dialyzed overnight against binding buffer B containing 20 mM HEPES pH 7.5, 300 mM NaCl, 2 mM MgCl2, 2 mM KCl and 2 mM β-ME. After dialysis, PSP was removed using a GST column. The protein was re-applied to a second Ni-NTA column equilibrated with binding buffer B, and eluted with binding buffer A. Eluted proteins were concentrated and loaded onto a HiLoad 16/60 Superdex 200 column (GE Healthcare) equilibrated with gel filtration buffer containing 20 mM HEPES pH 7.5, 150 mM NaCl, 2 mM MgCl2, 2 mM KCl and 1 mM dithiothreitol (DTT). Peak fractions corresponding to monomeric FAM46B were collected. tmCCA was purified as previously described (24), plus an extra ion-exchange step using a Resource Q column (GE Healthcare) to remove the nucleic acids bound to the protein. Cell lysis and protein purification were performed at 4°C.
Crystallization and structure determination
Crystals of xtFAM46B were obtained via hanging vapor diffusion by mixing 10 mg ml−1 protein with equal volume of reservoir solution containing 2% tacsimate pH 5.0, 0.15 M sodium citrate tribasic dehydrate pH 5.6 and 10% PEG 3350 at 10°C. During flash-cooling of the crystals in liquid N2, a cryo-solution containing 0.15 M sodium citrate tribasic dehydrate pH 5.6, 15% (w/v) PEG3350 and 10% glycerol was used. Diffraction datasets were collected at beamline BL19U1 of the Shanghai Synchrotron Radiation Facility (SSRF) and processed using the XDS suite (25). Phases were solved by molecular replacement using MolRep (26) with the atomic coordinate of mouse FAM46C (will be reported elsewhere) as search model. The structural model was built using COOT (27) and refined with Refmac5 (28) and Phenix (29). Structural validation was carried out using MolProbity (30). Structural illustrations were prepared using the PyMOL Molecular Graphics Systems (version 1.7.0.0, Schrödinger LLC; http://www.pymol.org/). Data collection and refinement statistics can be found in Table 1.
Table 1.
Crystallographic data collection and refinement
| xtFAM46B | |
|---|---|
| Data collection | |
| Space group | P 1211 |
| Cell dimensions | |
| a, b, c (Å) | 59.6, 76.1, 135.1 |
| α, β, γ (°) | 90, 96.5, 90 |
| Wavelength (Å) | 0.97915 |
| Resolution (Å)a | 47.1–2.69 (2.79–2.69) |
| No. reflections | 33266 (3214) |
| R sym | 0.083 (0.439) |
| I / σ(I) | 14.12 (3.65) |
| Completeness (%) | 99.2 (97.6) |
| Redundancy | 3.41 (3.36) |
| Refinement | |
| R work/Rfree | 0.204/0.252 |
| No. atoms | |
| Protein | 7988 |
| Ligand/ion | 39 |
| Water | 40 |
| B-factors | |
| Protein | 63.18 |
| Ligand/ion | 100.96 |
| Water | 44.02 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.002 |
| Bond angles (°) | 0.54 |
| Ramachandran | |
| Favored (%) | 96.5 |
| Outliers (%) | 0 |
aNumbers in parentheses represent values from the highest resolution shell.
In vitro CCA-adding assay
Truncated tRNAs (mini-tRNAs) with various three-termini were used to substitute full-length immature and mature tRNAs in the in vitro CCA-adding assay according to a previous study (31). The mini-tRNAs (mini-tRNA-D, mini-tRNA-DC and mini-tRNA-DCC), each conjugated with a 5′ biotin label, were purchased from Bioneer. For testing AMP/CMP incorporation, 10 μM FAM46B/C or tmCCA was incubated with 10 μM mini-tRNA and 200 μM ATP and/or CTP in a buffer containing 20 mM HEPES pH 7.5, 150 mM NaCl, 4 mM MgCl2 and 4 mM KCl at 37°C for 50 min before analyzed by 22% urea-PAGE. The biotinylated RNA substrates and products were incubated with HRP-conjugated streptavidin and visualized by chemiluminescence using the Chemiluminescent Nucleic Acid Detection Module Kit (Thermo Scientific) according to manufacturer's protocol. All experiments were repeated at least twice and the data showed satisfying consistency.
NTP consumption assay
16 μM protein was incubated with 80 μM A15 primer and 80 μM NTP in a buffer containing 20 mM HEPES pH 7.5, 150 mM NaCl, 4 mM MgCl2 and 4 mM KCl at 37°C for 20 min. The mixture was then analyzed by an HPLC system (Agilent) equipped with a reverse phase C18 ODS-2 Hypersil analytical column preceded by a C18 guard column (Thermo Scientific), with 100 mM potassium phosphate pH 6.5, 10 mM tetrabutyl ammonium bromide and 7.5% acetonitrile as running buffer. Nucleotides were detected by absorption at 254 nm and quantified by integration of the corresponding peaks.
NMP incorporation assay
10 μM protein was incubated with 10 μM 5′-biotinylated 15-mer RNA primer (Bioneer) and 200 μM NTP in a buffer containing 20 mM HEPES pH 7.5, 150 mM NaCl, 4 mM MgCl2 and 4 mM KCl at 37°C for 50 min. The mixtures were subjected to 20% urea–PAGE and the biotinylated RNA substrates and products were visualized by Chemiluminescence. All experiments were repeated at least twice and the data showed satisfying consistency.
Polyadenylation assay
10 μM protein was incubated with 5 μM 15-mer RNA primer, 500 μM ATP and 1 ul 1:1000 diluted [α-32P]-ATP (3000 Ci/mmol, PerkinElmer) at 37°C for 50 min. The mixtures were subjected to 20% urea–PAGE. The low range RNA marker (NEB) was pre-labeled with [γ-32P]-ATP (3000 Ci/mmol, PerkinElmer) using T4 polynucleotide kinase (NEB). The gels were dried, exposed to a phosphorimaging plate, and scanned using a Typhoon phosphorimager (GE Healthcare). All experiments were repeated at least twice and the data showed satisfying consistency.
Nucleotide binding assay
Binding of xtFAM46B to different adenosine nucleotides were analyzed by using isothermal titration calorimetry (ITC) at 25°C in the buffer containing 20 mM HEPES pH 7.5, 150 mM NaCl, 4 mM MgCl2 and 4 mM KCl. 100 μM protein was titrated by 1 mM adenosine nucleotides with 20 injections, 2 μl each. Resulting heat changes upon each injection was integrated using the PEAQ-ITC program provided by the manufacturer.
In vitro A-tailing experiment
Independent triplicate experiments were performed by incubating 5 μM FAM46B with 5 μM 15-mer random RNA (N15 RNA oligos, Integrated DNA Technologies) in a buffer containing 20 mM HEPES pH 7.5, 150 mM NaCl, 4 mM MgCl2 and 4 mM KCl at 37°C for 50 min. After incubation, samples were extracted with acid-phenol-chloroform pH 4.5 (Thermo Scientific), washed twice with 80% ethanol and eluted into ddH2O before subjected to 12% urea-PAGE. The poly(A)-added and poly(A) non-added RNA fragments were separated, individually pooled and extracted for preparing sequencing libraries with TruSeq small RNA library preparation kit (Illumina). Sequencing was performed on a Hiseq 2000 platform (Illumina) with 1 × 50 nt format.
Data analysis for in vitro A-tailing experiment
For all reads from the sequencing data, the first 15 bases were sub-stringed, and the index of the first 3 bases ‘TGG’ of the adapter was searched. The numbers of added A were calculated as ‘the index number—15’, if 90% of the bases were A between the first 15 bases and the adapter. If no adapter was found and 90% of the bases were A after the first 15 bases, then the number of A equals to 35. Reads without added A (0A) were classed as background, whereas reads with 5A or greater were classed as foreground. The frequencies of ATCG at each position were calculated both in background and foreground. At a particular position j, of a sequence in foreground, p = (pA, pC, pG, pT) denoted the probabilities of the four elements, and q = (qA, qC, qG, qT) denoted the corresponding background probabilities. Defining r = (rA, rC, rG, rT) by: ri = pi/qi (i = A, C, G, T), the sequence logos were drawn using this metric at each position at axis with ggseqlogo software (32).
Phylogenetic analysis
All protein sequences were downloaded from UniProt (https://www.uniprot.org/) and aligned by using MAFFT (https://www.ebi.ac.uk/Tools/msa/mafft/). Phylogenetic tree of whole protein sequence was constructed by using Bayesian Evolutionary Analysis Sampling Trees (BEAST) program, version 1.8.4. Each protein sequence data set was analyzed with the Blosum62 model, strict molecular clock model and coalescent constant size. Sampling and convergence of the runs were evaluated using the Tracer program, version 1.6 (http://beast.community/tracer) until effective sample size values exceeded 200 to ensure sufficient level of sampling. The length of Bayesian Monte Carlo Markov Chain (MCMC) was set 10 000 000, and sampled every 1000. States (the first 10%) were removed as burn-in. Maximum clade credibility trees were visualized with the FigTree program, version1.4.3 (http://beast.community/figtree) and iTOL (https://itol.embl.de/).
Bioinformatics analysis
The RNA-seq reads from published samples were mapped by the STAR mapper (33). The number of uniquely mapped reads was calculated on each annotated gene, and further normalized to fragment per kilobase per million (FPKM) by total number of uniquely mapped reads. For part of the ChIP-seq analysis, the raw sequencing reads of H3K4me3, H3K27me3 and H3K27ac in H9 hESCs (34) were mapped by bowtie 2 (35) with default parameter settings and MACS2 software was further used for the peak calling. The raw sequencing reads from OCT4, NANOG and SOX2 ChIP-seq data in HUES64 hESCs (36) were mapped to human genome (hg19) by bowtie 2 with default parameter settings, and MACS2 software (37) was further used for the peak calling. Graphics (for bioinformatics analyses) were done in R (http://www.r-project.org/), and ggplot2 (http://ggplot2.org/) was used for part of the graph preparation. The ChIP-seq peaks were visualized in IGV.
Microarray
Three replicates from differentiating hESCs during mesendodermal differentiation were used for RNA extraction. Quality-assessed RNA samples were hybridized to Affymetrix Gene Chip Human Gene 1.0 ST arrays (Affymetrix). Twelve raw data files generated from the Affymetrix scanner passed data quality control and were further performed with RNA normalization through the Affymetrix expression console. Further analysis was performed using SAM (significance analysis of microarrays) software (38).
hESC maintenance and mesendoderm induction
hESCs (H1 [Wi Cell Research Institute]) were maintained in mTeSR1 media (Stem Cell Technologies) on Matrigel (hESC qualified; Corning) coated dishes. For mesendoderm induction, media were changed from mTeSR1 to chemically defined medium (CDM) supplemented with Activin A (R&D Systems), BMP4 (bone morphogenetic protein 4, R&D Systems), bFGF (basic fibroblast growth factor, Invitrogen) and LY294002 (phosphoinositide-3-kinase/AKT serine/threonine kinase inhibitor, Sigma) at approximately 70% to 80% confluency (39,40).
In vitro differentiation assay
Embryoid body (EB) formation was performed as described previously (41). Briefly, hESCs were cultured on matrigel-coated six-well plates. Cells from one well were dissociated with collagenase IV (1 mg ml−1) for 5 min, and then split into small cell clumps. The small cell clumps were transferred into three wells of low-attachment six-well plate, and cultured in EB medium (knock-out DMEM, 20% knock-out serum replacement, 1 mM l-glutamine, 1% nonessential amino acids, 0.1 mM 2-mercaptoethanol and primocin). The medium was changed every 2 days. The embryoid bodies were cultured for 8 days followed by collection for qRT-PCR.
siRNA Transfection
Plates of 12-well (Corning) were coated with 400 μl of diluted Matrigel before addition of siRNA oligos. siRNAs (Qiagen) mixed with Opti-MEM I (GIBCO) were added into the wells. For higher transfection efficiency, a mixture of 2 μl DharmaFect1 (Dharmacon) transfection reagent with 98 μl Opti-MEM I was added into each well after siRNA addition. After incubation for ∼30 min at room temperature, single cell suspension of 2 × 106 hESCs in 800 μl of mTeSR1 with 10 μM Y-27632 (Stem Cell Technologies) was seeded into each well. 18 h later, Y-27632 was washed off. The working concentration of siRNA was 100 nM. Six hours after washing off Y-27632, RNA samples were collected for further assays unless specified. The siRNA sense sequences are: 5′-UAAGGCUAUGAAGAGAUAC (scramble), 5′-GGCGCCATTTGTCTTGGAATT (siRNA-1), 5′-CCGUGUGCCUCAUGAACCATT (siRNA-2).
Western blot
Experiments were performed as described (42). Briefly, cells were lysed in lysis buffer and cell lysate containing 30 μg protein was used for Western blots. Membranes were incubated with the primary antibody against EOMES (Cat No. ab23345, Abcam, 1:1000), FAM46B (Cat No. 23149-1-AP, Proteintech, 1:1000), GAPDH (Cat No. sc-47724, Santa Cruz Biotechnology, 1:5000), β-Tubulin (Cat No. 30303ES50, YEASEN, 1:1000) and Laimin B1 (Cat No. 16048, Abcam, 1:1000). The membranes were then incubated with IRDye 680LT Donkey anti-Rabbit IgG (Cat No. 926-68023, Li-COR Biosciences) or IRDye 800CW Donkey anti-Mouse IgG (Cat No. 926-32212, Li-COR Biosciences) as secondary antibodies and visualized on an Odyssey Infrared Imager (Li-COR Biosciences).
CRISPR knock-in
To generate hESCs with the 3 × Flag-tagged FAM46B, a two-step strategy was adopted, including (i) integration of PGK-Puro-IS-3 × Flag in the 5′ flanking region of the start codon, and (ii) excision of the sequence between TTAA by using an excision-only PiggyBac Transposase PBx, leaving behind 3 × Flag sequence linked to the start codon of FAM46B. Single clones were selected and one clone of the hESCs with Flag-tagged FAM46B protein was confirmed by sequencing and Western blot analysis.
Immunocytochemical staining
Immunofluorescence assay was performed according to the protocol described previously (43). Briefly, the attached cells were harvested at indicated stages, fixed with 4% paraformaldehyde, permeabilized in 0.4% Triton X-100 (Sigma-Aldrich) for intracellular antigens, blocked in 10% normal goat serum (Vector Laboratories), and then stained with antibodies to FAM46B (Cat No. 23149-1-AP, Proteintech, 1:200). Antibody labeling was visualized using DyLight 488-conjugated secondary antibodies (all from Jackson ImmunoResearch Laboratories, 1:1000). Nuclei were counterstained with Hoechst 33258 (Sigma-Aldrich, 1:2000). A Zeiss Observer microscope was used for slide observation and image capture.
qRT-PCR
Total RNA was prepared using the RNAprep pure Micro Kit (TIANGEN) following the manufacturer's instructions. cDNAs were generated by reverse-transcribed total RNA (1 μg) using ReverTra Ace reverse transcriptase (Toyobo). qRT-PCR was performed and analyzed by the ViiA™ 7 Real-Time PCR System (Life Technologies) with SYBR Green qPCR Master Mix (Roche). The qRT-PCR data were shown as fold changes normalized to internal control PBGD. The qRT-PCR primers are listed as follows:
| Gene | Primer sequence (5′-3′) | AT (°C) | PS (bp) |
|---|---|---|---|
| FAM46B | F-CCTGCCTACTAGACTTCCTGC | 60 | 149 |
| R-CTTGCCGCTCTTGTTGGACA | |||
| OCT4 | F-AGTGAGAGGCAACCTGGAGA | 60 | 81 |
| R-ACACTCGGACCACATCCTTC | |||
| NANOG | F-CATGAGTGTGGATCCAGCTTG | 60 | 191 |
| R-CCTGAATAAGCAGATCCATGG | |||
| SOX2 | F-GGGAAATGGGAGGGGTGCAAAAGAGG | 60 | 151 |
| R-TTGCGTGAGTGTGGATGGGATTGTG | |||
| PBGD | F-ATTACCCCGGGAGACTGAAC | 60 | 130 |
| R-GGCTGTTGCTTGGACTTCTC | |||
| SOX1 | F-ACCAGGCCATGGATGAAG | 60 | 67 |
| R-CTTAATTGCTGGGGAATTGG | |||
| GATA4 | F-GGAAGCCCAAGAACCTGAAT | 60 | 98 |
| R-GTTGCTGGAGTTGCTGGAA | |||
| T | F-ACCCAGTTCATAGCGGTGAC | 60 | 165 |
| R-CCATTGGGAGTACCCAGGTT | |||
| GAPDH | F-AGCCACATCGCTCAGACAC | 60 | 66 |
| R-GCCCAATACGACCAAATCC |
F, forward; R, reverse; AT, annealing temperature; PS, product size.
Flow cytometry
Cells were harvested at the indicated time points and stained with an Annexin V-FITC Apoptosis Detection Kit (BD Bioscience) to detect early apoptotic cells according to the manufacturer's instructions. Annexin V signals were detected by flow cytometry using a FACStar Plus Flow Cytometer (Becton-Dickinson).
MTT assay
A total of 1 × 105 hESCs per well were planted in 48-well plates in mTeSR1 and transfected with scramble or FAM46B siRNAs with the addition of Y-27632. Eighteen hours after transfection, medium was changed to mTeSR1. At 24, 30, 36 h after transfection, 10 μl of 5 mg ml−1 MTT [3-(4,5-dime-thylthiazolyl-2)-2,5-diphenyltetrazolium bromide, Sangon Bio-tech] solution was added and incubated for 4 h. DMSO (Sigma-Aldrich) was then added to solubilize the formazan product and the absorbance was measured at 570 nm.
RNA half-life analysis
H1 cells were treated with doxycycline for 12 h before incubation with 5 μg ml−1 actinomycin D (ActD, Sigma-Aldrich). Cells were collected at time points 0, 0.5, 1, 2, 3, 4 and 5 h after the addition of ActD. RNA was extracted using TRIzol (Sigma-Aldrich). Reverse transcription was performed with HiScript 1st Strand cDNA Synthesis Kit (Vazyme) following the manufacture's protocol. qRT-PCR was conducted with SYBR green (Bio-Rad) on an Bio-Rad CFX96 PCR machine. The qRT-PCR data were shown as fold changes normalized to internal control GAPDH. The qRT-PCR primers not included in the qRT-PCR method section are listed as follows:
| Gene | Primer sequence (5′-3′) | AT (°C) | PS (bp) |
|---|---|---|---|
| Wnt5A | F-CCCTTAGGCAGGTTGGCTTT | 60 | 90 |
| R-CACTGCAACTGAGGGGATGT | |||
| RICTOR | F-TGCAGAAGCACCCTTCACAA | 60 | 148 |
| R-AGCCAGTTATAGCACAACCCC | |||
| SKP2 | F-TGACATCGGATGCCCTCAAA | 60 | 249 |
| R-AAGCCAAATGGCATACCACAC | |||
| PAPBC4 | F-TGCAAGGTGGTGTGTGATGA | 60 | 70 |
| R- CAGCCTCTTGGGTCTCGAAG | |||
| RPS25 | F-GAAGATTCGAGGCTCCCTGG | 60 | 128 |
| R-CTGGAGCATCTCCACCCTTG |
F, forward; R, reverse; AT, annealing temperature; PS, product size.
FUNCAT assay and click chemistry
FUNCAT assay was performed as previously described (44). Briefly, 24 h after transfected with scramble or FAM46B siRNAs, 1 × 105 hESC cells of each sample were collected and lysed in ice-cold RIPA lysis buffer containing protease inhibitor cocktail (Sigma). HPG incorporated protein lysates were labeled selectively with Cy5-conjugated azide by copper-catalyzed azide-alkyne cycloaddition (CuAAC). Click reaction was performed following the protocol from Click Chemistry Protein Reaction Buffer Kit (Click Chemistry Tools). Finally, samples were heated at 70°C for 10 min before resolved by SDS-PAGE and imaged using a pharos FXTM Plus Molecular Imager (Bio-Rad). Loading consistency was confirmed by staining the gel with Coomassie Blue after fluorescent imaging. All experiments were repeated at least twice and the data showed satisfying consistency.
CRISPR knock-out
The inducible Cas9 FAM46B-knock-out hESC was generated by transfecting gRNA targeting the exons of FAM46B into the inducible Cas9 H1 cell line. Knock-out of FAM46B was induced by addition of 1 μg ml−1 doxycycline (Sigma-Aldrich), and cells were collected at different time points (0, 3, 6, 12, 24, 48 h) after doxycycline induction for analysis. The knock-out effect was of verified by qRT-PCR and Western blot using antibody against FAM46B. The gRNA sequences are 5′-AAACTATCGGCCTTCCCCGGACGGC-3′ (forward) and 5′-CACCGCCGTCCGGGGAAGGCCGATA-3′ (reverse).
RESULTS
Overall crystal structure of FAM46B
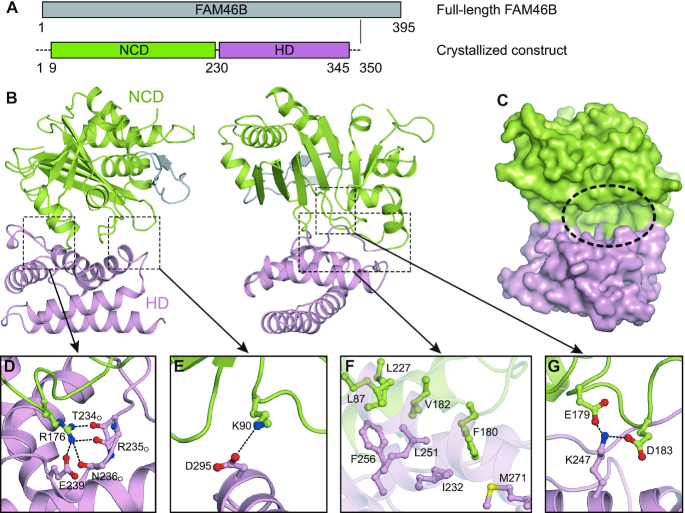
In order to obtain functional clues of FAM46 proteins, we solved a 2.7 Å crystal structure of nucleotide-free FAM46B protein from Xenopus tropicalis (xtFAM46B) with a Rfree of 0.252 (Table 1). In the structural model, residues 1−8, 198−200 and 347−350 are missing due to disordered configuration. The protein contains an N-terminal catalytic domain (NCD, residues 9−230 resolved in the structure) and a C-terminal helical domain (HD, residues 231−346), which together form a massive cleft (Figure 1A−C). Inside the cleft, the three NTase consensus carboxylates, Asp91, Asp93 and Glu167 are present at the putative catalytic site, but we did not observe the electron density for catalytic metal ion. A conserved Arg176 directs the loop linking the two domains via hydrogen bonds and a salt bridge with Thr234, Arg235O, Asn236O and Glu239 (Figure 1D). The other side of the cleft is shielded by a salt bridge between Lys90 from the NCD and Asp295 from the HD (Figure 1E). The bottom of the cleft is featured by an extensive hydrophobic network formed by residues from both NCD and HD (Figure 1F). Glu179 and Asp183 sit in the center of the cleft and interact with Lys247 protruding from the HD (Figure 1G).
Figure 1.
Overall structure of FAM46B. (A) Schematic representation showing the organization of crystallized construct based on full-length FAM46B. NCD, N-terminal catalytic domain; HD, helical domain. Borders of each domain are indicated by residue numbers. (B) Structure of FAM46B. Domains of FAM46B are indicated and colored as in A. The two featured β-hairpin is colored gray. (C) Surface representation of FAM46B with the inter-domain cleft indicated by dashed ellipse. (D–G) Interactions between NCD and HD. Side chains of involved residues are shown as ball-and-stick models in the same color as the domains they belong to.
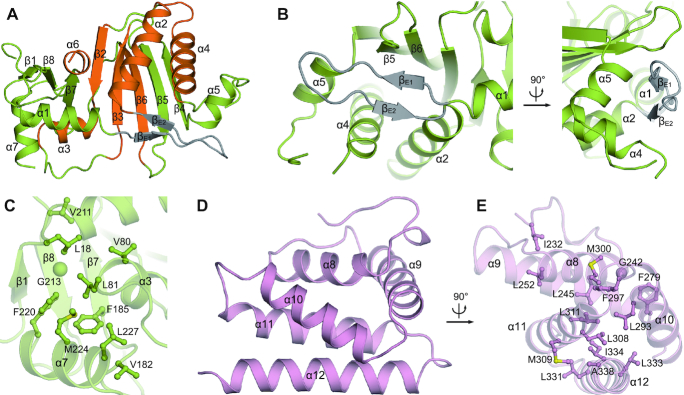
The NCD is built of a central eight-stranded β-sheet (β1−β8) surrounded by seven α-helices (α1−α7) involving a typical NTase core (Figure 2A). An extended β-hairpin, composed of βE1, βE2 and an elongated connecting loop, is situated between α1 and α2. This FAM46-specific β-hairpin stretches vertically to the central β-sheet, and covers the valley created by α4, α5, β5 and β6 (Figure 2B). Beside the NTase core, α7 is closely associated with β1, α3, β7 and β8 via hydrophobic interactions (Figure 2C). The HD is composed of 5 α-helices. α8, α9 and α10 are packed into a triangle platform, whereas α11 and α12 form an antiparallel pair underneath (Figure 2D). The five α-helices are tightly associated via hydrophobic interactions (Figure 2E).
Figure 2.
Structural details of FAM46B domains. (A) Overall structure of NCD with secondary elements assigned. The NTase core is specified in orange. (B) The ambient interactions of the featured β-hairpin in FAM46B NCD. (C) The local hydrophobic cluster next to the NTase core. Side chains of involved residues are shown as ball-and-stick models. (D) Overall structure of HD. (E) Hydrophobic interactions that stabilize HD.
FAM46B is a bacterial-like PAP
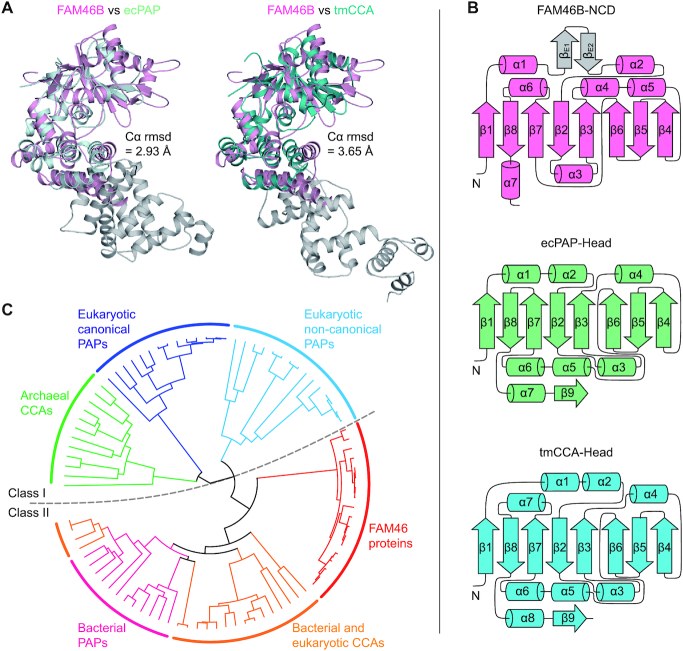
The overall architecture of FAM46B does not conform to the previously calculated FAM46C model (11). Compared to other known eukaryotic PAPs such as the classical PAPα and non-classical Gld-2 or mt-PAP (45–47), the NCD of FAM46B is double-sized in terms of amino acid residue numbers, and contains more secondary elements (Supplementary Figure S1A and B). To our knowledge, FAM46 proteins are likely to bear the largest catalytic domain among all known PAPs. Compared to Gld-2, FAM46B has a substantially wider NCD and deeper cleft (Supplementary Figure S1C and D). In addition, the relative position of FAM46B NCD and HD also differs from that of the catalytic domain and central domain of Gld-2 (Supplementary Figure S1E). This somewhat ‘complex’ architecture of FAM46B, which is obviously distinguished from other known eukaryotic PAPs, prompted us to look for its other potential relatives in the Dali server (48). Intriguingly, FAM46B shows high topological similarity with the class-II bacterial PAPs and CCA-adding enzymes. When FAM46B is individually superimposed with Escherichia coli PAP I (ecPAP) (9) and Thermotoga maritima CCA-adding enzyme (tmCCA) (24,49), the root mean standard deviation (r.m.s.d.) values of aligned Cα atoms are 2.93 Å and 3.65 Å, respectively, although the sequence identity between FAM46B and these two prokaryotic enzymes is only ∼15% (Figure 3A and Supplementary Figure S2). In particular, the central eight-stranded β-sheet in the NCD is highly consistent with the bacterial PAPs/CCA-adding enzymes, and the HD of FAM46B also resembles their neck domains (Figure 3A and B). The conserved inter-domain salt bridges in ecPAP and tmCCA, which are shown to be involved in the coordination of nucleotides (i.e. Asp151-Arg200 in ecPAP, and Asp130-Arg180 in tmCCA), are equivalent to the aforementioned Glu179-Lys247 pair in FAM46B (Figure 1G and Supplementary Figure S3A). Phylogenetic analysis also revealed that compared to eukaryotic PAPs, FAM46 proteins are more closely related to class-II bacterial PAPs and CCA-adding enzymes (Figure 3C and Supplementary Figure S3B).
Figure 3.
FAM46B is a bacterial-like PAP. (A) Structural comparison between FAM46B and ecPAP (left) or tmCCA (right). FAM46B (magenta) is separately superimposed with ecPAP (Protein Data Bank code 3AQM, pale cyan) and tmCCA (3H39, teal). The r.m.s.d. values of aligned Cα atoms are shown. The body and tail domains of ecPAP and tmCCA are colored grey. (B) The topology diagram of the FAM46B NCD (top), as compared with those of the head domains of ecPAP (middle) and tmCCA (bottom), colored as in A. (C) The phylogenetic analysis revealing a close relationship between FAM46 proteins and bacterial PAPs. Subtypes of PAPs and CCAs are color-specified. Class I (cool colors) and II (warm colors) PAPs/CCAs are separated by a dashed line.
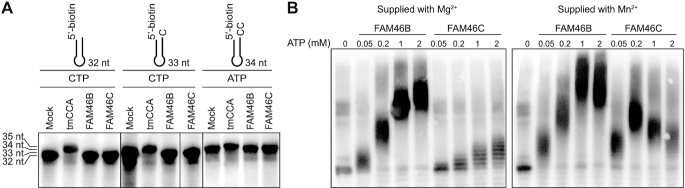
Following this clue, we tested whether FAM46B and FAM46C possess PAP and/or CCA-adding activities. CCA-adding enzymes catalyze the consecutive 3′-terminal CCA extension of precursor tRNAs with CTP and ATP as donors, while PAPs selectively add a string of nontemplated AMPs to the 3′-end of target mRNAs by consuming ATP molecules. 5′-biotinylated 15-mer poly(A) RNA (A15) and mini-tRNA substrates were individually incubated with FAM46B or FAM46C in the presence of corresponding nucleotides, and the products were visualized using streptavidin-conjugated horseradish peroxidase (HRP). Both proteins failed to add AMP or CMP to mini-tRNAs lacking 3′ A, CA and CCA (Figure 4A). On the other hand, FAM46B and FAM46C were able to elongate A15 in the presence of Mg2+, and the lengths of the products were apparently coupled to ATP concentration (Figure 4B). When Mg2+ was replaced by Mn2+, both proteins became more potent in catalyzing polyadenylation (Figure 4B). Notably, FAM46B showed much higher PAP activity than FAM46C, especially in Mg2+-present conditions. FAM46B is able to efficiently extend oligoA substrates ranging from A5 to A30, but shows no apparent binding to AMP, ATP or ATPγS (Supplementary Figure S3C and D).
Figure 4.
FAM46B has PAP activity. (A) Test for CCA-adding activity for human FAM46B and mouse FAM46C. 10 μM 5′-biotinylated mini-tRNA primers with different 3′ tails as indicated were individually incubated with 10 μM protein in the presence of 2 mM ATP or CTP for 50 min. tmCCA was used as a positive control. (B) Polyadenylation activity for human FAM46B and mouse FAM46C. 10 μM 5′-biotinylated A15 primer was incubated with 10 μM FAM46B or FAM46C in the presence of 4 mM Mg2+ or Mn2+ for 50 min. ATP was supplied with increasing concentrations as indicated.
tmCCA and ecPAP rely on two charged residues (Asp174 and Arg177 in tmCCA; Asp194 and Arg197 in ecPAP), which are conserved among class-II bacterial PAPs, in recognizing adenosine and cytosine nucleotides. However, structural comparison between FAM46B and tmCCA/ecPAP revealed that these two conserved residues are not present in FAM46B (Supplementary Figure S3E). Considering the lack of the nucleotide-binding affinity of FAM46B that also differs from tmCCA and ecPAP, we infer that FAM46B and FAM46C are bacterial-like poly(A) polymerases in terms of overall folding but may possess a different nucleotide binding manner from the class-II bacterial PAPs. In the rest part of the paper, human FAM46B has been used for all the experiments.
The catalytic properties of FAM46B
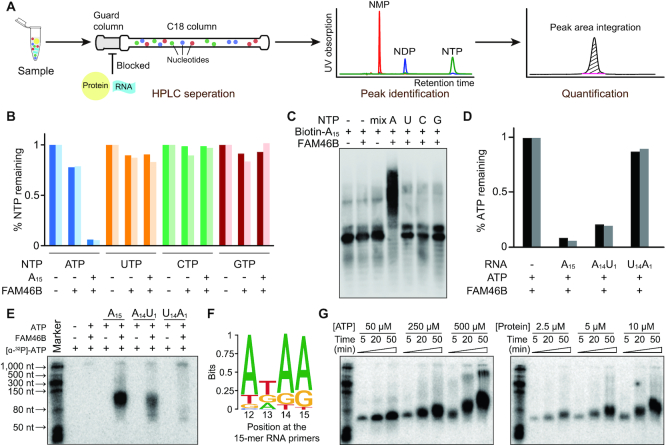
To investigate the catalytic properties of FAM46B, we firstly examined the nucleotide selection of FAM46B. An HPLC-based method was developed to monitor the incorporation of each monophosphate nucleotide (NMP). FAM46B and A15 primer were individually incubated with corresponding label-free triphosphate nucleotides (NTPs). After reaction, the mixture was applied to a C18 column with an upstream guard column to block the protein and RNA primer/product (Figure 5A). Thus, we were able to monitor the consumption of NTPs by quantifying the remaining nucleotides. When ATP was supplied, we observed decreased area of the ATP peak but no AMP peak after incubation, suggesting that ATP has been hydrolyzed and added to the A15 primer (Supplementary Figure S4A). In contrast, UTP, CTP or GTP was barely consumed under the same conditions (Figure 5B and Supplementary Figure S4B). Consistent with the HPLC assay, FAM46B only extends 5′-biotinylated A15 in the presence of ATP (Figure 5C).
Figure 5.
The substrate sequence preference of FAM46B. (A) Schematic drawing showing the experimental process of HPLC-based NTP consumption assay. (B) Consumption of different NTPs by human FAM46B. Results from two independent experiments are presented. (C) In vitro polyadenylation assay showing FAM46B extends 5′-biotinylated A15 primer only in the presence of ATP. (D) ATP consumption of FAM46B in the presence of different RNA primers. (E) Polyadenylation assay showing the AMP incorporation to different RNA primers by FAM46B. (F) Position-dependent nucleotide frequency of substrates extended by FAM46B. (G) Progressive polyadenylation activity of FAM46B, shown by ATP (left panel) or protein (right panel) concentration-dependent ATP incorporation in the presence of A15 primer.
In order to figure out whether FAM46B has preference on the oligonucleotide primers, we compared the ATP consumption of FAM46B in the presence of A15, A14U1 or U14A1. For three cases, the remaining ATP portions were approximately 10%, 30% and 90%, respectively (Figure 5D). Similar results were observed in AMP incorporation assay using α-32P-labeled ATP ([α-32P]-ATP), where A14U1 led to less AMP incorporation than A15, and U14A1 showed only little ATP incorporation (Figure 5E). To further explore the potential sequence preference of FAM46B over the substrates, we used a pool of random sequences of 15 nt as primers for an in vitro adenosine (A)-tailing assay (Supplementary Figure S5). It turned out that FAM46B indeed prefers an AxAA sequence at 3′-end of the substrates (Figure 5F). Further experiments indicated that FAM46B extended A15 primer in a ATP/protein concentration-dependent manner, and the majority of poly(A) products were below 150 nt in length (Figure 5G). These results suggest that FAM46B has strong selectivity on RNA molecules that are adenosine-rich close to 3′-end, and the base type at the very 3′-end of substrate RNA also impacts polyadenylation efficiency.
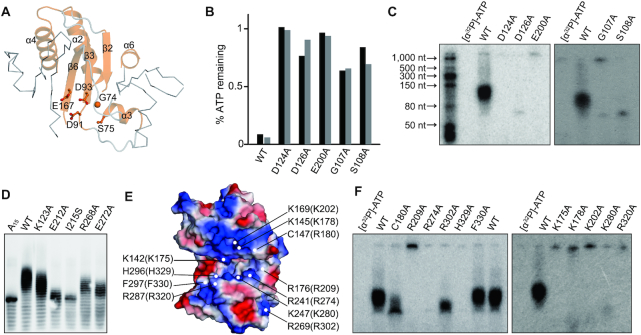
The catalytic center and potential RNA-binding area of FAM46B
All NTases contain three critical motifs in the catalytic center: hG[GS], [DE]h[DE]h and h[DE]h (h indicates a hydrophobic amino acid). To verify these functional motifs in FAM46B, we mutated corresponding key residues, namely Gly107, Ser108, Asp124, Asp126 and Glu200, individually into alanine and performed AMP incorporation and ATP consumption assays (Figure 6A). Unexceptionally, these mutants failed to hydrolyze ATP in the presence of A15, and lost polyadenylation activity (Figure 6B and C). We also did mutagenesis analysis for the residues involved in NCD-HD interaction (Figure 1D−G). Except for Lys123 (corresponding to Lys90 in xtFAM46B as shown in Figure 1E), mutation of other selected residues at NCD-HD interface substantially reduced PAP activity of FAM46B (Figure 6D).
Figure 6.
Critical sites of FAM46B for polyadenylation activity. (A) The NTase core of FAM46B. The consensus NTase motif is highlighted as cartoon representation in orange. The conserved key residues are shown as sticks-and-ball models. (B) ATP consumption of mutants regarding key residues shown in A. Results from two independent experiments are presented. (C) AMP incorporation of mutants regarding residues shown in A. (D) Polyadenylation activity of human FAM46B mutants regarding residues located in the interface between NCD and HD. (E) Surface electrostatic potential of xtFAM46B. Conserved negatively charged residues that may be involved in RNA binding, together with two hydrophobic residues close to the catalytic center, are specified. Corresponding residues of human FAM46B are indicated in the parentheses. (F) AMP incorporation of mutants regarding residues shown in D.
Surface electrostatic potential analysis reveals that positively-charged patches exist around the catalytic center of FAM46B (Figure 6E). The mutations of patch-involving residues Lys175, Lys202, Arg209, Arg274, Lys280 and His329 to alanine all deprived the PAP activity of FAM46B. In contrast, the mutation of Arg302, an HD residue located at the opposite side of the central cleft, still remained partially active (Figure 6F). We also tested two non-charged residues close to the positively charged patches, namely Cys180 and Phe330. C180A had reduced PAP activity, and F330A was as potent as the wild type (Figure 6F). These data suggest that these positively-charged patches may be involved in the RNA binding. Similarly, the neck domain of ecPAP also possesses a positively-charged area that has been proved critical for RNA binding (9).
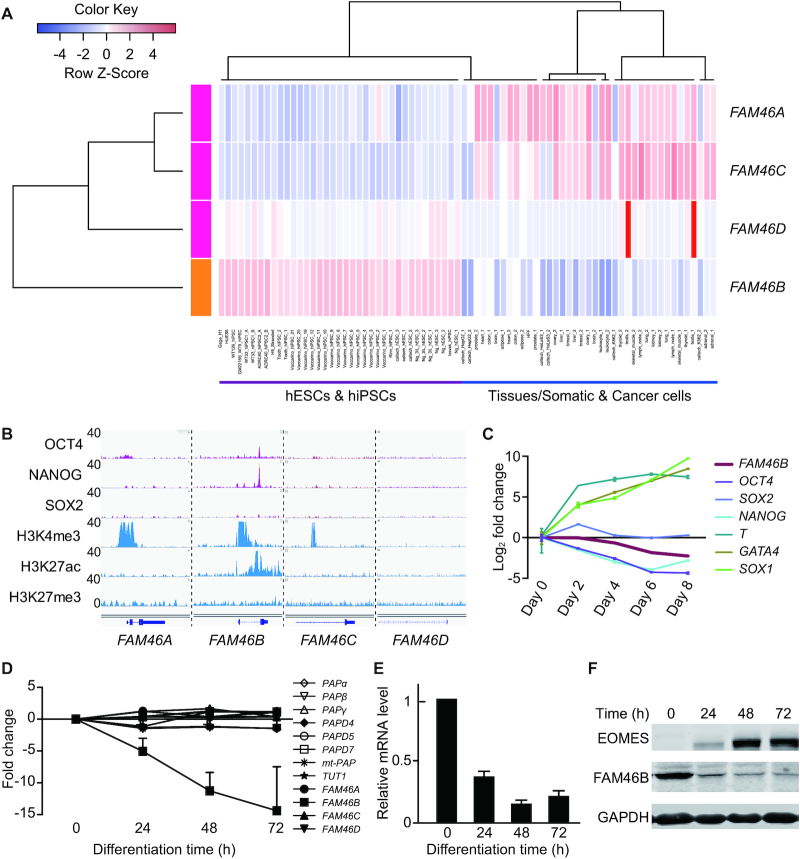
FAM46B is specifically expressed during human early development
In order to get an overview of the expression profile of FAM46 family members during mammalian early embryogenesis, we analyzed single-cell RNA-seq data of human and mouse pre-implantation embryos (50,51). FAM46B and FAM46C are maternal genes, and highly expressed in human embryos before zygotic genome activation (ZGA) (Supplementary Figure S6A). Similar transcriptional dynamics was observed also for mouse Fam46 family during early development (Supplementary Figure S6B). Following ZGA, the transcription level of FAM46B and FAM46C is sharply decreased. Interestingly, in contrast to Fam46b in mouse embryos, FAM46B is reactivated in human blastocyst, uniquely and highly expressed in human epiblast (Supplementary Figure S6A and B). We then investigated the transcription specificity of FAM46 members by comparing large RNA-seq datasets of multiple human embryonic stem cell (hESC) lines and human induced pluripotent stem cell (hiPSC) lines together with various cancer cell lines and somatic tissues (Figure 7A). The analysis showed that FAM46B is uniquely expressed in hESCs and hiPSCs, but not in cancer or somatic cells.
Figure 7.
FAM46B is specifically expressed in human pluripotent stem cells. (A) Expression profiles of FAM46 family genes in 12 hESC (H1, H6 and H9), 25 hiPSC samples, 32 normal somatic tissues and 6 cancer cell lines. The rows represent the transcription FAM46 family genes. The RNA-seq data from ENCODE was analyzed, and the RNA expression level was shown as FPKM (fragment per kilobase per million reads). (B) The comparison of chromatin status and pluripotent transcription factor binding sites around the promoters of FAM46 family genes. The promoter of FAM46B is bound by pluripotent transcription factors OCT4, NANOG and SOX2, and marked with transcriptionally active histone marks H3K4me3 and H3K27ac. (C) The mRNA level of FAM46B is gradually decreased during embryoid formation. Fold changes relative to day 0 are plotted for Day 2–8. Error bar indicate s.d. (D) Microarray data showing the relative expression alteration of FAM46 genes together with other canonical (PAPα, PAPβ and PAPγ) and noncanonical PAPs (PAPD4/Gld-2, PAPD5, PAPD7 and mt-PAP) at various time points after induced differentiation of hESCs. Note the declining tendency of FAM46B. Fold changes relative to day 0 are plotted for Day 1−3. Error bar indicate s.d. (E) Relative mRNA levels of FAM46B at various time points after induced differentiation determined by qPCR. mRNA level of FAM46B at day 0 is set to 1. Error bar indicates s.d. (n = 3). (F) FAM46B protein levels at various time points after induced differentiation determined by Western blot. EOMES is the marker for mesendoderm differentiation.
Consistent with FAM46B transcription in human pluripotent cells, ChIP-seq data shows that, in contrast to FAM46A/C/D, the promoter region of FAM46B is marked with transcriptionally active histone marks (H3K4me3, H3K27ac). Notably, the co-binding site of pluripotency transcription factors OCT4 and NANOG is embedded in the intron of FAM46B, indicating FAM46B transcription might directly regulated by OCT4 and NANOG (Figure 7B). These data imply a strong coupling between FAM46B expression and human pluripotency.
Expression of FAM46B is coupled to human pluripotency
To explore the pattern of FAM46B transcription during hESC differentiation, we first employed a typical spontaneous differential model: embryoid body formation. The quantitative real-time PCR (qRT-PCR) results showed that FAM46B expression level was gradually declined during embryoid formation (Figure 7C). Importantly, FAM46B is also down-regulated in neuroectodermal and mesendormal cells differentiated from hESCs (52), indicating that FAM46B inactivation might be required for lineage specification and germ layer formation during the embryonic development in humans (Supplementary Figure S6C).
As FAM46B is still expressed in the early time of mesendoderm differentiation (Supplementary Figure S6C), we analyzed its transcription dynamics in detail during the mesendoderm differentiation from hESCs. We noticed that the mRNA level of FAM46B showed approximately 5-, 11- and 15-fold decrease at 24, 48 and 72 h after induced differentiation, respectively (Figure 7D). In contrast, the mRNA levels of FAM46A/C/D, as well as other known canonical and non-canonical PAPs, remained steady. Subsequent qRT-PCR analysis confirmed that the mRNA level of FAM46B sharply declines following mesendodermal induction (Figure 7E). In order to monitor the endogenous FAM46B protein, we knocked-in a 3 × Flag-tag to the 5′ end of FAM46B reading frame (Supplementary Figure S6D). Western blot analysis indicated that the FAM46B protein level decreased in a time-dependent manner upon mesendodermal induction (Figure 7F). We also examined the transcriptional dynamics of FAM46 family during human somatic cell reprogramming (53). Intriguingly, FAM46B expression level is gradually increased following pluripotency acquisition, while FAM46A is gradually decreased, and FAM46C/D are not expressed during the whole reprogramming process (Supplementary Figure S6E). Altogether, these data suggest that FAM46B is specifically expressed in human pluripotent cells and may play crucial roles in human pluripotent cells.
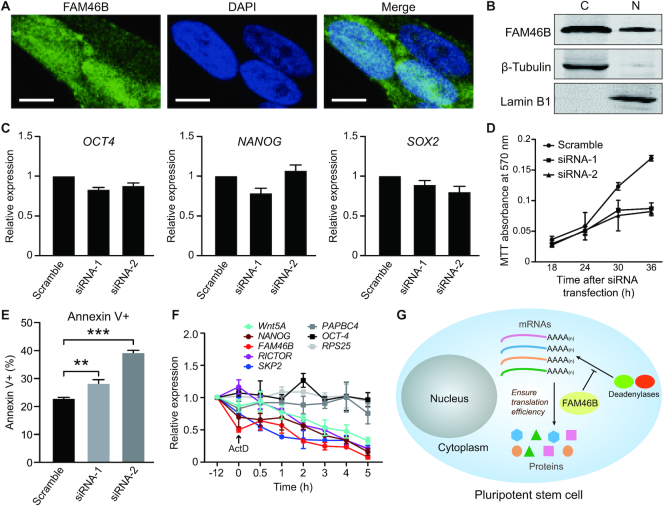
FAM46B is essential for the viability of hESCs
To explore the biological role of FAM46B, we tested its subcellular localization in hESCs. Immunofluorescent staining indicated a ubiquitous distribution of endogenous FAM46B (Figure 8A). Subsequent nuclear/cytosolic fractionation assay revealed a cytoplasmic preference of FAM46B residence, though it also appeared in the nuclear fraction (Figure 8B). Given the differentiation-coupled down-regulation feature of FAM46B, we investigated whether FAM46B is directly associated with pluripotency maintenance of hESCs. Knock-down of FAM46B by siRNAs did not significantly alter the mRNA levels of the pluripotency-related genes OCT4, NANOG and SOX2 (Figure 8C and Supplementary Figure S7A and B). On the other hand, instead of causing cell differentiation, FAM46B knock-down severely deteriorated viability of hESCs as soon as within 30 h, as indicated by MTT assays (Figure 8D), and the growth defect was clearly observable under microscope (Supplementary Figure S7C). According to the flow cytometry analysis, apoptotic hESCs (characterized by the annexin+ proportion) were increased after a 24-hour treatment of FAM46B siRNA (Figure 8E and Supplementary Figure S7D). We also tried to knock out FAM46B in human embryonic cells using CRISPR/Cas9 system. The surviving hESCs had at a least one chromosome copy intact, while perished hESCs had frame-shift insertions or deletions (data not shown) on FAM46B induced by Cas9 for both chromosome copies. These results suggest that FAM46B may not be directly involved in pluripotency maintenance, but is essential for the viability of hESCs.
Figure 8.
Characterization of FAM46B in hESCs. (A) Subcellular localization of endogenously expressed FAM46B protein in hESCs shown by immunofluorescent staining. Scale bar, 5 μM. (B) Cytosolic (C)/nuclear (N) fractionation experiment showing the subcellular localization of endogenous flag-tagged FAM46B in hESCs. (C) Knock-down of FAM46B did not significantly alter the expression level of OCT4, NANOG and SOX2. Error bar indicates s.d. (n = 3). (D) Knock-down of FAM46B inhibits cell viability in MTT assay. (E) Flow-cytometry assay showing knock-down of FAM46B leads to hESC apoptosis. Significance was calculated with two-sided Student's t-test (n = 3). **P < 0.01, ***P < 0.001. (F) mRNA half-life of selected genes in response to conditional knock-out of FAM46B in hESCs. Inhibition of cellular transcription by adding ActD was set to time point 0 and indicated by an arrow. (G) Model showing that FAM46B promotes translation in pluripotent cells.
To understand how FAM46B affects the viability of hESCs, we checked whether FAM46B plays a role in protein synthesis by using the fluorescent non-canonical amino acid labeling (FUNCAT) strategy (Supplementary Figure S8). In order to avoid severe cell death, hESCs had to be collected at 24 h after transfection of siRNAs. Newly synthesized proteins were labeled using homopropargylglycine (HPG) and detected via click chemistry. When FAM46B was knocked-down, the amount of newly synthesized proteins in hESCs was somewhat declined (Supplementary Figure S8B). We measured mRNA half-life of 8 selected genes, which are functionally linked with FAM46 proteins or mediate pluripotency, in FAM46B conditional knock-out hESCs. Within 5 h of treatment of actinomycin D, mRNA levels of Wnt5A, NANOG, FAM46B, RICTOR and SKP2 were substantially declined, whereas those of other genes remained relatively steady (Figure 8F, Supplementary Figure S8C). These data indicate that FAM46B may be involved in maintaining the translation efficiency of at least some genes in hESCs through preventing degradation of their mRNAs (Figure 8G).
DISCUSSION
In this study, we have identified FAM46B as a novel type of eukaryotic non-classical PAP. The striking structural similarity with bacterial PAP distinguishes FAM46B from other known eukaryotic PAPs. Consistently, FAM46B also shares similar functional properties with its bacterial relatives but not eukaryotic non-classical PAPs, as proven by our biochemical analysis. First, like bacterial PAPs, FAM46B actively catalyzes polyadenylation without extra partners in vitro. For eukaryotic ceGld-2, a partner (ceGld-3) is indispensable for its enzymatic efficiency, as ceGld-3 promotes the stability of ceGld-2 by sheltering a hydrophobic area on its surface, and provides extra interface for binding the RNA substrate (46). While FAM46B has no prominent hydrophobic area on the surface, and its large NCD contains plenty of potential RNA binding sites, a Gld-3-like partner thus seems unnecessary for FAM46B. Second, although favoring A-rich over U-rich sequence, FAM46B lacks strict discrimination on RNA substrates at least in vitro. This is also a feature inherited from bacterial PAP (8).
These bacterial-like features of FAM46B provide valuable clues for explaining the biological role of FAM46B. The early embryos of metazoans are transcriptionally inactive, which entails robust cytoplasmic polyadenylation to stabilize the mRNAs and increase translational efficiency (19,20). Except for FAM46B reported in this study, only a couple of eukaryotic cytoplasmic PAP have been identified and characterized, such as Gld-2 (4). While Gld-2 is essential in C. elegans, Drosophila and Xenopus, knock-out of this PAP shows no visible phenotype in mice (54). This evidence implies that, other PAPs functionally overlapping Gld-2 are likely to exist in mammals. Given the fact that FAM46B is an active PAP with endogenously cytoplasmic localization and unique and dynamic expression profile in human pre-implantation embryos and hESCs/hiPSCs, it is tempting to speculate that FAM46B is involved in cytoplasmic polyadenylation of mRNA in the early stage of human embryogenesis. Interestingly, FAM46B and FAM46C have been found to be essential for the meiosis in mammalian oocytes (55). Re-analysis of single-cell RNA-seq datasets of human germline and embryonic development covering stages from primordial germ cells (PGCs) to hatched blastocyst indicates that the mRNA levels of FAM46B and FAM46C dramatically increase in PGCs and the zygote stage of early development, till ZGA initiation (50,56). These studies imply that FAM46B and/or FAM46C may function as early as during gametogenesis. According to our results, FAM46B is most likely involved in the overall translational control of the hESCs without significantly altering the transcriptional profile. Given its potent PAP activity, a reasonable speculation is that FAM46B may counterbalance the deadenylation of mRNAs to maintain the poly(A) tail length of mRNAs at a steady level (14,15), so as to ensure the translational efficiency. Understanding the detailed mechanism on how FAM46B influence translation in hESCs required future studies based on transcriptome-wide characterization of mRNA poly(A) tail-lengths and quantitative proteomics.
The prominent negative correlation between expression level and cell differentiation suggests that FAM46B is not all-time needed during the embryonic development. The mRNA level of FAM46B in Xenopus shows high consistency: it grows steadily from two-cell stage until blastula stage and then sharply dives to almost zero (57). Thus, it is possible that FAM46B also functions in non-mammals. On the other hand, functions of Gld-2 are mostly described in gametogenesis (21–23), and its mRNA level remains unchanged after hESC differentiation. FAM46B and Gld-2 may account for cytoplasmic polyadenylation in different situations of early animal development. This arouses interesting questions: why the animal embryos require a bacterial-like PAP before they run into complexity? To answer this question, future studies are demanded to reveal the exact RNA substrates and putative regulatory factors of FAM46B, for which many possibilities need to be considered. For instance, some non-classical PAPs are found to catalyze the addition of single nucleotide to microRNAs (58), or require non-protein partners (59). In addition, it might be worthwhile to seek the connections between early animal embryos and bacteria in the control of poly(A) tail length. In contrary to eukaryotes, polyadenylation in bacteria usually leads to the degradation of regulatory RNAs and eventually causes translation inhibition (8). In this context, the observation that hESCs are extraordinarily sensitive to FAM46B depletion is somewhat analogous to the situation in bacteria: even moderate overexpression of PAP I is lethal to E. coli (60).
The crystal structure of FAM46B provides important clues in the future research of FAM46 proteins. Cytoplasmic polyadenylation is suggested to be active in early animal embryos and neuronal synapses (18). Coincidently, existing evidence of FAM46 proteins reveals its relationship with development and neuronal disorders. It has been reported that FAM46A is expressed in tooth germs and may be involved in the formation of tooth enamel (61). Mutations in FAM46A are found responsible for several autosomal recessive disorders in humans, including osteogenesis imperfecta and retinitis pigmentosa (62–64). Overexpression of FAM46D is closely associated with autism spectrum disorder-like behaviors in a mouse model (65). Human FAM46B shows 55%, 55% and 43% sequence identity to human FAM46A, FAM46C and FAM46D, respectively, which implies that these FAM46 proteins would share extensive structural and biochemical similarity to FAM46B. Thus, the feature as bacterial-like PAPs is the key to understand the functional mechanisms of FAM46 proteins in their various biological roles.
DATA AVAILABILITY
The X-ray crystallographic coordinates and structure factor files for xtFAM46B structure have been deposited in the Protein Data Bank (PDB) under the accession number 6JYJ.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the staff of SSRF for the help with the collection of crystal diffraction data, Prof. Xianghuo He and Prof. Qiurong Ding for the technical assistance.
Author contributions: S.G, and H.L. conceived project. J.L.H. performed most of the biochemical experiments. H.Z. crystallized xtFAM46B, solved the structure and did phylogenetic analysis of FAM46 proteins. H.L., P.Z. and H.B. did most of the experiments on hESCs, including microarray, CRISPR knock-in and examination on phenotypes of FAM46B knock-down under the supervision of H.T.Y.. M.Z.Y. performed in vitro differentiation of hESCs and ChIP-seq analysis. W.S. performed in vitro tailing assay. L.L., J.X.F. and B.Y. helped with cell culture, protein purification and urea PAGE experiments. T.Z. and Y.Z. handled autoradiography for the NMP incorporation assay. F.L., J.Y.Y. and C.N. contributed to FUNCAT assay. Q.G. and Y.L. performed additional bioinformatics analysis. Y.C. helped with X-ray diffraction data collection. Q.Z. and M.S.Z. tested subcellular localization of FAM46B. S.L. advised on protein purification and structure determination. Y.W., W.C. and J.W. supervised bioinformatics analysis. J.L.H., H.L., H.Z., J.W., H.T.Y. and S.G. wrote the paper.
Notes
Present address: Wei Sun, Department of Pharmaceutical Chemistry and Cardiovascular Research Institute, University of California San Francisco, CA 94158, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Key R&D Program of China [2018YFA0508300 to S.G., 2017YFA0103700 to H.T.Y., 2018YFA0107003 to J.W.]; National Natural Science Foundation of China [31470729, 31722016, 81772977 to S.G., 81520108004 to H.T.Y.]; Natural Science Foundation of Guangdong Province [2014TQ01R584, 2014A030312015]; Innovative Team Program of Guangzhou Regenerative Medicine and Health Guangdong Laboratory [2018GZR110103002 to S.G.]; The Shenzhen Science and Technology Program [KQTD20180411143432337 to W.C.]. Program for Guangdong Introducing Innovative and Entrepreneurial Teams [2016ZT06S029 to J.W.]. Funding for open access charge: National Key R&D Program of China [2018YFA0508300].
Conflict of interest statement. None declared.
REFERENCES
- 1. Kuchta K., Knizewski L., Wyrwicz L.S., Rychlewski L., Ginalski K.. Comprehensive classification of nucleotidyltransferase fold proteins: identification of novel families and their representatives in human. Nucleic Acids Res. 2009; 37:7701–7714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Tomita K., Yamashita S.. Molecular mechanisms of template-independent RNA polymerization by tRNA nucleotidyltransferases. Front. Genet. 2014; 5:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Yue D., Maizels N., Weiner A.M.. CCA-adding enzymes and poly(A) polymerases are all members of the same nucleotidyltransferase superfamily: characterization of the CCA-adding enzyme from the archaeal hyperthermophile Sulfolobus shibatae. RNA. 1996; 2:895–908. [PMC free article] [PubMed] [Google Scholar]
- 4. Laishram R.S. Poly(A) polymerase (PAP) diversity in gene expression–star-PAP vs canonical PAP. FEBS Lett. 2014; 588:2185–2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Warkocki Z., Liudkovska V., Gewartowska O., Mroczek S., Dziembowski A.. Terminal nucleotidyl transferases (TENTs) in mammalian RNA metabolism. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2018; 373:20180162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Colgan D.F., Manley J.L.. Mechanism and regulation of mRNA polyadenylation. Genes Dev. 1997; 11:2755–2766. [DOI] [PubMed] [Google Scholar]
- 7. Kim K.W., Nykamp K., Suh N., Bachorik J.L., Wang L., Kimble J.. Antagonism between GLD-2 binding partners controls gamete sex. Dev. Cell. 2009; 16:723–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sarkar N. Polyadenylation of mRNA in prokaryotes. Annu. Rev. Biochem. 1997; 66:173–197. [DOI] [PubMed] [Google Scholar]
- 9. Toh Y., Takeshita D., Nagaike T., Numata T., Tomita K.. Mechanism for the alteration of the substrate specificities of template-independent RNA polymerases. Structure. 2011; 19:232–243. [DOI] [PubMed] [Google Scholar]
- 10. Menezes M.R., Balzeau J., Hagan J.P.. 3′ RNA uridylation in epitranscriptomics, gene regulation, and disease. Front. Mol. Biosci. 2018; 5:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kuchta K., Muszewska A., Knizewski L., Steczkiewicz K., Wyrwicz L.S., Pawlowski K., Rychlewski L., Ginalski K.. FAM46 proteins are novel eukaryotic non-canonical poly(A) polymerases. Nucleic Acids Res. 2016; 44:3534–3548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mroczek S., Chlebowska J., Kulinski T.M., Gewartowska O., Gruchota J., Cysewski D., Liudkovska V., Borsuk E., Nowis D., Dziembowski A.. The non-canonical poly(A) polymerase FAM46C acts as an onco-suppressor in multiple myeloma. Nat. Commun. 2017; 8:619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Moore M.J., Proudfoot N.J.. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell. 2009; 136:688–700. [DOI] [PubMed] [Google Scholar]
- 14. Goldstrohm A.C., Wickens M.. Multifunctional deadenylase complexes diversify mRNA control. Nat. Rev. Mol. Cell Biol. 2008; 9:337–344. [DOI] [PubMed] [Google Scholar]
- 15. Chen C.Y., Shyu A.B.. Mechanisms of deadenylation-dependent decay. Wiley Interdiscip. Rev. RNA. 2011; 2:167–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Salles F.J., Lieberfarb M.E., Wreden C., Gergen J.P., Strickland S.. Coordinate initiation of Drosophila development by regulated polyadenylation of maternal messenger RNAs. Science. 1994; 266:1996–1999. [DOI] [PubMed] [Google Scholar]
- 17. Barkoff A., Ballantyne S., Wickens M.. Meiotic maturation in Xenopus requires polyadenylation of multiple mRNAs. EMBO J. 1998; 17:3168–3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Weill L., Belloc E., Bava F.A., Mendez R.. Translational control by changes in poly(A) tail length: recycling mRNAs. Nat. Struct. Mol. Biol. 2012; 19:577–585. [DOI] [PubMed] [Google Scholar]
- 19. Richter J.D. Cytoplasmic polyadenylation in development and beyond. Microbiol. Mol. Biol. Rev. 1999; 63:446–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Subtelny A.O., Eichhorn S.W., Chen G.R., Sive H., Bartel D.P.. Poly(A)-tail profiling reveals an embryonic switch in translational control. Nature. 2014; 508:66–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wang L., Eckmann C.R., Kadyk L.C., Wickens M., Kimble J.. A regulatory cytoplasmic poly(A) polymerase in Caenorhabditis elegans. Nature. 2002; 419:312–316. [DOI] [PubMed] [Google Scholar]
- 22. Cui J., Sackton K.L., Horner V.L., Kumar K.E., Wolfner M.F.. Wispy, the Drosophila homolog of GLD-2, is required during oogenesis and egg activation. Genetics. 2008; 178:2017–2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Barnard D.C., Ryan K., Manley J.L., Richter J.D.. Symplekin and xGLD-2 are required for CPEB-mediated cytoplasmic polyadenylation. Cell. 2004; 119:641–651. [DOI] [PubMed] [Google Scholar]
- 24. Toh Y., Takeshita D., Numata T., Fukai S., Nureki O., Tomita K.. Mechanism for the definition of elongation and termination by the class II CCA-adding enzyme. EMBO J. 2009; 28:3353–3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kabsch W. Xds. Acta Crystallogr. D. Biol. Crystallogr. 2010; 66:125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Vagin A., Teplyakov A.. MOLREP: an automated program for molecular replacement. J Appl Cryst. 1997; 30:1022–1025. [Google Scholar]
- 27. Emsley P., Cowtan K.. Coot: model-building tools for molecular graphics. Acta Crystallogr. D. Biol. Crystallogr. 2004; 60:2126–2132. [DOI] [PubMed] [Google Scholar]
- 28. Murshudov G.N., Vagin A.A., Dodson E.J.. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D. Biol. Crystallogr. 1997; 53:240–255. [DOI] [PubMed] [Google Scholar]
- 29. Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W. et al.. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D. Biol. Crystallogr. 2010; 66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chen V.B., Arendall W.B. 3rd, Headd J.J., Keedy D.A., Immormino R.M., Kapral G.J., Murray L.W., Richardson J.S., Richardson D.C.. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D. Biol. Crystallogr. 2010; 66:12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tomita K., Ishitani R., Fukai S., Nureki O.. Complete crystallographic analysis of the dynamics of CCA sequence addition. Nature. 2006; 443:956–960. [DOI] [PubMed] [Google Scholar]
- 32. Wagih O. ggseqlogo: a versatile R package for drawing sequence logos. Bioinformatics. 2017; 33:3645–3647. [DOI] [PubMed] [Google Scholar]
- 33. Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R.. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013; 29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ziller M.J., Edri R., Yaffe Y., Donaghey J., Pop R., Mallard W., Issner R., Gifford C.A., Goren A., Xing J. et al.. Dissecting neural differentiation regulatory networks through epigenetic footprinting. Nature. 2015; 518:355–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Langmead B., Salzberg S.L.. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012; 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gifford C.A., Ziller M.J., Gu H., Trapnell C., Donaghey J., Tsankov A., Shalek A.K., Kelley D.R., Shishkin A.A., Issner R. et al.. Transcriptional and epigenetic dynamics during specification of human embryonic stem cells. Cell. 2013; 153:1149–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W. et al.. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008; 9:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tusher V.G., Tibshirani R., Chu G.. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. U.S.A. 2001; 98:5116–5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Brons I.G., Smithers L.E., Trotter M.W., Rugg-Gunn P., Sun B., Chuva de Sousa Lopes S.M., Howlett S.K., Clarkson A., Ahrlund-Richter L., Pedersen R.A. et al.. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature. 2007; 448:191–195. [DOI] [PubMed] [Google Scholar]
- 40. Chng Z., Teo A., Pedersen R.A., Vallier L.. SIP1 mediates cell-fate decisions between neuroectoderm and mesendoderm in human pluripotent stem cells. Cell Stem Cell. 2010; 6:59–70. [DOI] [PubMed] [Google Scholar]
- 41. Wang J., Xie G., Singh M., Ghanbarian A.T., Rasko T., Szvetnik A., Cai H., Besser D., Prigione A., Fuchs N.V. et al.. Primate-specific endogenous retrovirus-driven transcription defines naive-like stem cells. Nature. 2014; 516:405–409. [DOI] [PubMed] [Google Scholar]
- 42. Liang J., Wang Y.J., Tang Y., Cao N., Wang J., Yang H.T.. Type 3 inositol 1,4,5-trisphosphate receptor negatively regulates apoptosis during mouse embryonic stem cell differentiation. Cell Death Differ. 2010; 17:1141–1154. [DOI] [PubMed] [Google Scholar]
- 43. Cao N., Liu Z., Chen Z., Wang J., Chen T., Zhao X., Ma Y., Qin L., Kang J., Wei B. et al.. Ascorbic acid enhances the cardiac differentiation of induced pluripotent stem cells through promoting the proliferation of cardiac progenitor cells. Cell Res. 2012; 22:219–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yang J.Y., Deng X.Y., Li Y.S., Ma X.C., Feng J.X., Yu B., Chen Y., Luo Y.L., Wang X., Chen M.L. et al.. Structure of Schlafen13 reveals a new class of tRNA/rRNA- targeting RNase engaged in translational control. Nat. Commun. 2018; 9:1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bard J., Zhelkovsky A.M., Helmling S., Earnest T.N., Moore C.L., Bohm A.. Structure of yeast poly(A) polymerase alone and in complex with 3′-dATP. Science. 2000; 289:1346–1349. [DOI] [PubMed] [Google Scholar]
- 46. Nakel K., Bonneau F., Eckmann C.R., Conti E.. Structural basis for the activation of the C. elegans noncanonical cytoplasmic poly(A)-polymerase GLD-2 by GLD-3. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:8614–8619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lapkouski M., Hallberg B.M.. Structure of mitochondrial poly(A) RNA polymerase reveals the structural basis for dimerization, ATP selectivity and the SPAX4 disease phenotype. Nucleic Acids Res. 2015; 43:9065–9075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Holm L., Rosenstrom P.. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010; 38:W545–W549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Li F., Xiong Y., Wang J., Cho H.D., Tomita K., Weiner A.M., Steitz T.A.. Crystal structures of the Bacillus stearothermophilus CCA-adding enzyme and its complexes with ATP or CTP. Cell. 2002; 111:815–824. [DOI] [PubMed] [Google Scholar]
- 50. Yan L., Yang M., Guo H., Yang L., Wu J., Li R., Liu P., Lian Y., Zheng X., Yan J. et al.. Single-cell RNA-Seq profiling of human preimplantation embryos and embryonic stem cells. Nat. Struct. Mol. Biol. 2013; 20:1131–1139. [DOI] [PubMed] [Google Scholar]
- 51. Deng Q., Ramskold D., Reinius B., Sandberg R.. Single-cell RNA-seq reveals dynamic, random monoallelic gene expression in mammalian cells. Science. 2014; 343:193–196. [DOI] [PubMed] [Google Scholar]
- 52. Jang J., Wang Y., Lalli M.A., Guzman E., Godshalk S.E., Zhou H., Kosik K.S.. Primary Cilium-Autophagy-Nrf2 (PAN) Axis Activation Commits Human Embryonic Stem Cells to a Neuroectoderm Fate. Cell. 2016; 165:410–420. [DOI] [PubMed] [Google Scholar]
- 53. Cacchiarelli D., Trapnell C., Ziller M.J., Soumillon M., Cesana M., Karnik R., Donaghey J., Smith Z.D., Ratanasirintrawoot S., Zhang X. et al.. Integrative Analyses of Human Reprogramming Reveal Dynamic Nature of Induced Pluripotency. Cell. 2015; 162:412–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Nakanishi T., Kumagai S., Kimura M., Watanabe H., Sakurai T., Kimura M., Kashiwabara S., Baba T.. Disruption of mouse poly(A) polymerase mGLD-2 does not alter polyadenylation status in oocytes and somatic cells. Biochem. Biophys. Res. Commun. 2007; 364:14–19. [DOI] [PubMed] [Google Scholar]
- 55. Pfender S., Kuznetsov V., Pasternak M., Tischer T., Santhanam B., Schuh M.. Live imaging RNAi screen reveals genes essential for meiosis in mammalian oocytes. Nature. 2015; 524:239–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Dang Y., Yan L., Hu B., Fan X., Ren Y., Li R., Lian Y., Yan J., Li Q., Zhang Y. et al.. Tracing the expression of circular RNAs in human pre-implantation embryos. Genome Biol. 2016; 17:130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Tan M.H., Au K.F., Yablonovitch A.L., Wills A.E., Chuang J., Baker J.C., Wong W.H., Li J.B.. RNA sequencing reveals a diverse and dynamic repertoire of the Xenopus tropicalis transcriptome over development. Genome Res. 2013; 23:201–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Burns D.M., D’Ambrogio A., Nottrott S., Richter J.D.. CPEB and two poly(A) polymerases control miR-122 stability and p53 mRNA translation. Nature. 2011; 473:105–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Mellman D.L., Gonzales M.L., Song C., Barlow C.A., Wang P., Kendziorski C., Anderson R.A.. A PtdIns4,5P2-regulated nuclear poly(A) polymerase controls expression of select mRNAs. Nature. 2008; 451:1013–1017. [DOI] [PubMed] [Google Scholar]
- 60. Cao G.J., Sarkar N.. Identification of the gene for an Escherichia coli poly(A) polymerase. Proc. Natl. Acad. Sci. U.S.A. 1992; 89:10380–10384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Etokebe G.E., Kuchler A.M., Haraldsen G., Landin M., Osmundsen H., Dembic Z.. Family-with-sequence-similarity-46, member A (Fam46a) gene is expressed in developing tooth buds. Arch. Oral. Biol. 2009; 54:1002–1007. [DOI] [PubMed] [Google Scholar]
- 62. Lagali P.S., Kakuk L.E., Griesinger I.B., Wong P.W., Ayyagari R.. Identification and characterization of C6orf37, a novel candidate human retinal disease gene on chromosome 6q14. Biochem. Biophys. Res. Commun. 2002; 293:356–365. [DOI] [PubMed] [Google Scholar]
- 63. Barragan I., Borrego S., Abd El-Aziz M.M., El-Ashry M.F., Abu-Safieh L., Bhattacharya S.S., Antinolo G.. Genetic analysis of FAM46A in Spanish families with autosomal recessive retinitis pigmentosa: characterisation of novel VNTRs. Ann. Hum. Genet. 2008; 72:26–34. [DOI] [PubMed] [Google Scholar]
- 64. Doyard M., Bacrot S., Huber C., Di Rocco M., Goldenberg A., Aglan M.S., Brunelle P., Temtamy S., Michot C., Otaify G.A. et al.. FAM46A mutations are responsible for autosomal recessive osteogenesis imperfecta. J. Med. Genet. 2018; 55:278–284. [DOI] [PubMed] [Google Scholar]
- 65. Hamilton S.M., Spencer C.M., Harrison W.R., Yuva-Paylor L.A., Graham D.F., Daza R.A., Hevner R.F., Overbeek P.A., Paylor R.. Multiple autism-like behaviors in a novel transgenic mouse model. Behav. Brain Res. 2011; 218:29–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The X-ray crystallographic coordinates and structure factor files for xtFAM46B structure have been deposited in the Protein Data Bank (PDB) under the accession number 6JYJ.