Abstract
Gram-positive bacteria, including major clinical pathogens such as Staphylococcus aureus, are becoming increasingly drug-resistant. Their cell walls are composed of a thick layer of peptidoglycan (PG) modified by the attachment of wall teichoic acid (WTA), an anionic glycopolymer that is linked to pathogenicity and regulation of cell division and PG synthesis. The transfer of WTA from lipid carriers to PG, catalyzed by the LytR–CpsA–Psr (LCP) enzyme family, offers a unique extracellular target for the development of new anti-infective agents. Inhibitors of LCP enzymes have the potential to manage a wide range of bacterial infections because the target enzymes are implicated in the assembly of many other bacterial cell wall polymers, including capsular polysaccharide of streptococcal species and arabinogalactan of mycobacterial species. In this study, we present the first crystal structure of S. aureus LcpA with bound substrate at 1.9 Å resolution and those of Bacillus subtilis LCP enzymes, TagT, TagU, and TagV, in the apo form at 1.6–2.8 Å resolution. The structures of these WTA transferases provide new insight into the binding of lipid-linked WTA and enable assignment of the catalytic roles of conserved active-site residues. Furthermore, we identified potential subsites for binding the saccharide core of PG using computational docking experiments, and multiangle light-scattering experiments disclosed novel oligomeric states of the LCP enzymes. The crystal structures and modeled substrate-bound complexes of the LCP enzymes reported here provide insights into key features linked to substrate binding and catalysis and may aid the structure-guided design of specific LCP inhibitors.
Keywords: cell wall, teichoic acid, peptidoglycan, gram-positive bacteria, Staphylococcus aureus (S. aureus), Bacillus, X-ray crystallography, oligomerization, LytR–CpsA–Psr, size-exclusion chromatography with multiangle light scattering
Introduction
The discovery of penicillin nearly a century ago ushered in an era of targeting bacterial cell wall peptidoglycan (PG)2 biosynthesis as an effective approach to combat a wide variety of bacterial infections. To survive the onslaught of β-lactams and other classes of typically bactericidal antibiotics, bacteria have evolved a myriad of resistance countermeasures in parallel. As a result, new therapeutic agents are now urgently needed, and their development will rely on extensive research efforts on additional bacterial targets. Wall teichoic acid (WTA) is a Gram-positive bacterial cell wall polymer that is covalently attached to the N-acetylmuramic acid (MurNAc) C6-hydroxyl group of PG (Fig. S1). The inhibition of WTA biosynthesis is an attractive therapeutic approach because methicillin-resistant Staphylococcus aureus deficient in the production of WTAs are known to be resensitized to certain classes of β-lactam antibiotics (1, 2). This intriguing phenomenon is believed to stem from the role of WTA in guiding PG cross-linking through spatiotemporal localization of certain penicillin-binding proteins (PBPs) (1–3). The absence of WTA disables these select classes of PBPs through mislocalization and sensitizes the organism to β-lactams that target unaffected PBPs. Another physiological role of WTA is the regulation of cell division through localization of autolysins to the division septum for PG breakdown (4). In addition, this polymer is linked to pathogenicity because WTA-deficient mutants are defective in biofilm formation, host cell adherence, and colonization (5–7).
WTA is synthesized in the cytoplasm on undecaprenyl-phosphate (C55-P) for translocation across the lipid bilayer and subsequent attachment to PG (8). The ribitol-phosphate polymer of S. aureus WTA is anchored to PG through a sugar-based linkage unit comprised of phosphate–GlcNAc–N-acetylmannosamine–[glycerol-phosphate]2 (Fig. S1). The transfer of WTA to PG is catalyzed on the outer leaflet of the cytosolic membrane by members of the LytR–CpsA–Psr (LCP) protein family that are unique to predominantly Gram-positive bacteria (9). Deletion of Bacillus subtilis LCP enzymes (TagTBS, TagUBS, and TagVBS) has been shown to be lethal presumably because of the accumulation of nonfunctional lipid-bound WTA intermediates and depletion of the pool of lipid carriers required for PG synthesis (9, 10). In S. aureus, deletion of LCP enzymes (LcpASA, LcpBSA, and LcpCSA) resulted in WTA-deficient mutants with physiological and pathogenic defects (11, 12).
The LCP family represents an attractive class of drug targets in that the soluble catalytic region is on the extracellular face of the cytosolic membrane, and there are no mammalian orthologs (13). Furthermore, LCP enzymes are involved in the PG attachment of not only WTA but other secondary polymers of therapeutic interest, including Streptococcus pneumonia capsular polysaccharide (CP) and Mycobacterium tuberculosis arabinogalactan (14, 15).
Multiple copies of the lcp gene are often found in Gram-positive bacteria, and the gene products display catalytic and functional differences. B. subtilis TagU was found to have higher catalytic activity than TagT and TagV variants in that species, whereas M. tuberculosis Rv3484 was shown to be the only essential LCP enzyme in vivo (16, 17). In S. aureus, LcpA was discovered to be the primary WTA transferase, whereas LcpC acts as the primary CP transferase (18, 19). The natural PG acceptor substrate of LCP enzymes also appears variable. The disaccharide lipid-linked PG precursor, lipid II, was demonstrated to be a substrate of S. aureus LcpC but not for LcpA (18, 19). In addition, S. aureus LcpB was found to be incapable of utilizing cross-linked S. aureus PG as the acceptor substrate, suggesting that the attachment of WTA occurs prior to cross-linking of PG strands (20). In contrast, a study conducted on B. subtilis LCP enzymes demonstrated WTA ligation to mature cross-linked B. subtilis PG in vitro (16).
Given the observed functional variations among species, understanding the underlying molecular details becomes important. Prior crystallographic studies have been conducted on LCP enzymes with S. pneumoniae Cps2A and B. subtilis TagT characterized in complex with analogs of the polyprenyl substrate (Table S1). In the former, the bound lipids lack the saccharide headgroup that differentiates CP/WTA precursors from lipid II (9, 14). More recently, C30-PP–GlcNAc and C30-PP–GlcNAc–ManNAc were successfully co-crystallized with B. subtilis TagT; however, the saccharide headgroups were found in different orientations, and the expected specificity determining polar contacts between the saccharide substrate moieties and the enzyme active site were not observed (20). A notable structural variant of LCP enzymes is that of Actinomyces oris LcpA, which possesses unique structural features around the active site presumably associated with binding target proteins rather than PG for glycosylation (21).
Further structural characterization of LCP enzymes is required to investigate PG binding and to clarify the structural relationship between the donor lipid headgroup and the enzyme. In this study, we present four crystal structures of S. aureus and B. subtilis LCP enzymes for a comparative analysis that provides the molecular basis of residues assessed in published mutagenesis studies. Importantly, our structure of S. aureus LcpA complexed to C40-PP–GlcNAc provides clarity on the orientation of the saccharide headgroup and reveals an interface reliant on van der Waals and hydrophobic contacts rather than direct polar interactions. Furthermore, the structure of S. aureus LcpA provides a clinically relevant target for structure-guided design of inhibitors. We complement our crystallographic work by modeling PG-bound complexes, and we report hitherto unknown oligomeric states of various LCP enzymes in solution.
Results
Overall structure
Our crystallographic study yielded the first structure of the primary S. aureus WTA transferase, LcpASA (Fig. 1 and Table 1). The crystallized enzyme captured C40-PP–GlcNAc, an endogenous lipid with central features in keeping with the natural lipid donor substrate. The construct (residues 80–327; ΔTM) used for crystallization encompasses the extracellular catalytic region, known as the LCP domain, and lacks the single N-terminal transmembrane anchor (Fig. S2a). Our structure of LcpASA was solved by molecular replacement to 1.9 Å resolution, providing an excellent template for future structure-based drug design work.
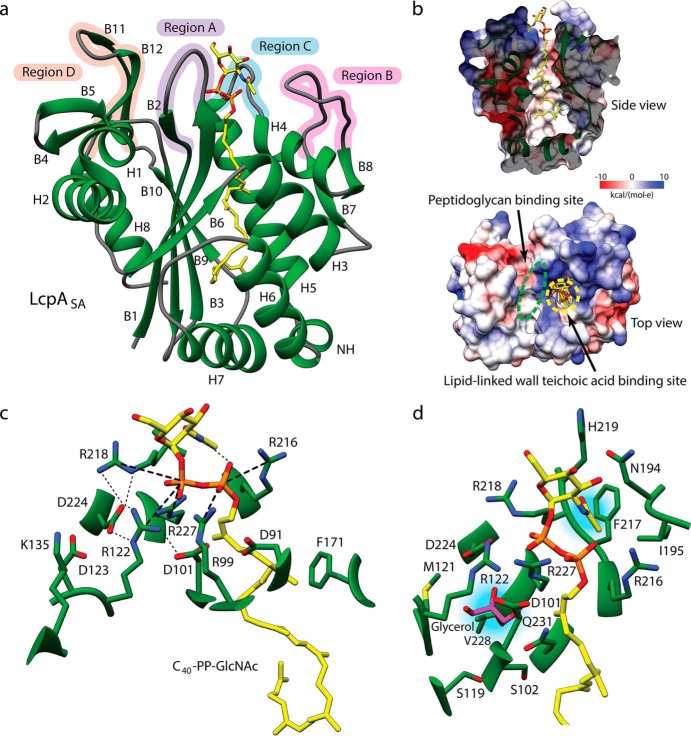
Figure 1.
Crystal structure of S. aureus LcpA. a, ribbon structure of LcpASA (green) bound to C40-PP–GlcNAc (yellow) with labeled structural features. H, helix; B, β-strand; NH, nonconserved helix. The active site is surrounded by four regions, designated here as A, B, C, and D. Heteroatoms are colored by type (oxygen, red; phosphorus, orange; nitrogen, blue). b, the lipid-binding pocket of LcpASA is shown in a cross-section of a side view with an electrostatic potential surface. The locations of the substrate-binding sites are outlined in the top view. c, conserved residues surrounding the pyrophosphate moiety of the bound lipid are depicted as sticks. Bonds are indicated by lines (salt bridges as thick dotted lines and hydrogen bonds as thin dotted lines). d, residues surrounding a buried glycerol molecule (purple) and the N-acetyl group of C40-PP–GlcNAc are depicted as sticks.
Table 1.
X-ray data collection and refinement statistics
| S. aureus LcpA | B. subtilis TagT | B. subtilis TagU (SeMet) | B. subtilis TagV | |
|---|---|---|---|---|
| Residues | 80–327 | 46–322 | 62–306 | 72–332 |
| Protein Data Bank code | 6UEX | 6UF5 | 6UF6 | 6UF3 |
| Data collection | ||||
| Wavelength (Å) | 0.97949 | 0.97964 | 0.97949 | 0.97949 |
| Spacegroup | C2221 | P41212 | P3221 | P212121 |
| Cell dimensions | ||||
| a, b, c (Å) | 89.25, 90.54, 94.75 | 65.66, 65.66, 143.07 | 48.79, 48.79, 234.27 | 40.99, 66.10, 81.72 |
| a, b, c (°) | 90, 90, 90 | 90, 90, 90 | 90, 90, 120 | 90, 90, 90 |
| Resolution (Å)a | 47.38–1.90 (1.97–1.90) | 48.37–2.80 (2.90–2.80) | 41.58–2.20 (2.28–2.20) | 36.64–1.6 (1.66–1.60) |
| CC1/2 | 0.998 (0.696) | 0.997 (0.655) | 0.999 (0.862) | 1 (0.709) |
| Rpim | 0.02126 (0.6351) | 0.05905 (0.5196) | 0.02982 (0.523) | 0.02434 (0.5561) |
| Rmeas | 0.05442 (1.643) | 0.2232 (1.989) | 0.09296 (1.665) | 0.0674 (1.561) |
| I/σI | 16.31 (1.36) | 13.10 (1.55) | 14.69 (1.46) | 19.00 (1.50) |
| Completeness (%) | 99.84 (99.80) | 99.84 (99.87) | 99.59 (99.11) | 99.82 (99.87) |
| Redundancy | 6.6 (6.6) | 13.9 (14.4) | 9.7 (10.0) | 7.6 (7.7) |
| Refinement | ||||
| Resolution (Å) | 47.38–1.90 | 48.37–2.80 | 41.58–2.20 | 36.64–1.60 |
| No. of reflections | 30,547 (3015) | 8230 (794) | 17,404 (1671) | 29,994 (2959) |
| Rwork/Rfreeb | 0.1922/0.2258 | 0.2307/0.2766 | 0.2445/0.2588 | 0.2004/0.2288 |
| No. of atoms | 2063 | 1881 | 1811 | 2133 |
| Protein | 1896 | 1879 | 1770 | 1966 |
| Ligand/ion | 110 | 0 | 36 | 0 |
| Water | 57 | 2 | 5 | 167 |
| B factors (Å2) | 67.32 | 63.12 | 93.25 | 36.23 |
| Protein | 65.98 | 63.15 | 92.57 | 35.69 |
| Ligand/ion | 91.91 | – | 129.37 | – |
| Water | 64.62 | 43.66 | 73.06 | 42.53 |
| RMSD | ||||
| Bond lengths (Å) | 0.008 | 0.005 | 0.006 | 0.006 |
| Bond angles (°) | 1.19 | 1.06 | 1.08 | 1.04 |
| Ramachandran (%) | ||||
| Favored | 97.96 | 94.98 | 96.41 | 98.79 |
| Allowed | 2.04 | 5.02 | 3.59 | 1.21 |
| Outliers | 0 | 0 | 0 | 0 |
| MolProbity | ||||
| Clashscore | 3.35 | 5.89 | 8.09 | 3.85 |
a The values in parentheses represent the highest-resolution shell.
b 5% of reflections were excluded from refinement and used to calculate Rfree.
The LCP domain of LcpASA is comprised of a six-stranded β-sheet sandwiched between multiple α-helices and several double-stranded β-sheets (Fig. 1a and Fig. S2b). A large hydrophobic lipid-binding pocket with a narrow opening and a wide base is formed by the central β-sheet and helices 3–7 (Fig. 1b). An electropositive region for binding the pyrophosphate moiety of the lipid donor substrate is found at the entrance of the hydrophobic pocket highlighting the location of the active site. LcpASA shows the highest overall structural similarity to Enterococcus faecalis EF0465 with an RMSD of 2.5 Å for 227 Cα pairs, indicating regions of significant difference (Table S1). However, structural comparison of just the highly conserved active-site residues shows that LcpASA is most similar to apo and C30-PP–GlcNAc–ManNAc–bound TagTBS with a closely matched RMSD of 0.85 Å for 10 Cα pairs in both instances and 1.3–1.6 Å for 44 common side chain heavy atoms therein. The active site of LcpASA is surrounded by four regions, designated here as regions A (residues 92–100), B (residues 188–201), C (residues 217–224), and D (residues 296–312), that display structural variability when compared with the LCP enzymes of other species (Fig. 1a). To this end, we have expanded our structural understanding of LCP enzymes by solving three additional structures from the prototypical Gram-positive bacterium, B. subtilis (Fig. 2 and Table 1). The first is the novel structure of TagUBS (residues 62–306; selenomethionine (SeMet)-substituted) phased by single isomorphous replacement and refined to 2.2 Å resolution. Additional insights into the architecture of the active site are provided by the structure of TagVBS (residues 72–332; 1.6 Å resolution) at a higher resolution than previously reported (2.6 Å resolution) and the structure of TagTBS (residues 46–322; 2.8 Å resolution) with additional electron density for a previously disordered and unmodeled region in the apo structure.
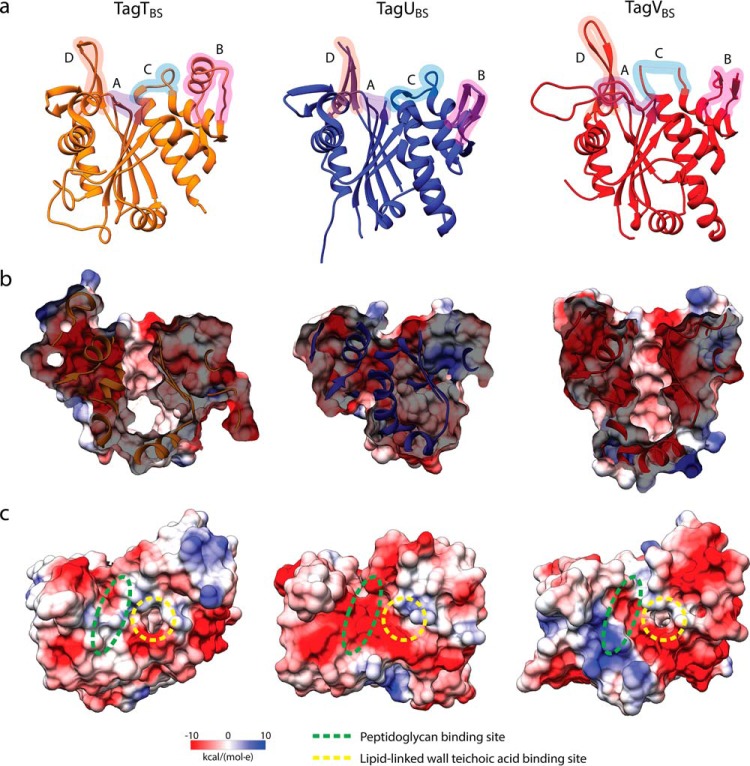
Figure 2.
Crystal structures of B. subtilis TagT, TagU, and TagV. a, ribbon structures of TagTBS (orange), TagUBS (blue), and TagVBS (red) with labeled regions surrounding the active sites (A, B, C, and D). b, the lipid-binding pockets are shown as cross-sections (side views) with electrostatic potential surfaces. c, the locations of the substrate-binding sites are outlined in the top views of the surfaces.
The electrostatic surfaces of the LCP enzymes are highly variable in addition to the electropositive region formed by conserved arginine residues at the active site. This electropositive region becomes more difficult to observe when the guanidinium side chains are not localized by a pyrophosphate group (Fig. 2c). Between our four structures, the most significant difference in secondary structure is found in region B, where LcpASA has a large loop, TagTBS has an α-helix, and both TagUBS and TagVBS have a double-stranded β-sheet (Figs. 1a and 2a). Regions A and C encompass flexible loops, and region D adopts a two-stranded β-sheet with an enrichment of aromatic residues that we predict bind to the carbohydrate groups of PG.
In the structure of TagUBS, the rearrangements of helices 3–7 on one side of the central β-sheet resulted in the collapse of the lipid-binding site (Fig. 2b). These structural differences are facilitated by the association of two protein molecules forming a modest interfacial surface area of 760 Å2 across a crystallographic 2-fold symmetry axis at one end of the lipid-binding site (helices 6 and 7 and β-strand 9) (Fig. S2c). The association is stabilized by hydrogen bonds and hydrophobic interactions mainly consisting of aromatic residues from the interior of the hydrophobic pocket. Notably, several crystal structures of LCP enzymes, including TagTBS here, display disorder of helix 6 (Fig. 2a). The exposure of the hydrophobic core to bulk solvent may be the driving force behind the dimerization of TagUBS. The hydrophobic and aromatic nature of this exposed region also suggests a possible surface for association with membrane.
Lipid-binding site
In the structure of S. aureus LcpA, we have fortuitously captured a lipid substrate produced by the overexpression strain BL21(DE3) Escherichia coli for the synthesis of O7-specific lipopolysaccharide (22). The clear density at the extended hydrophobic lipid-binding pocket (surface area ≈ 680 Å2; volume ≈ 550 Å3) allowed unambiguous modeling of a lipid tail and a monosaccharide-pyrophosphate headgroup (Fig. S2d). The electron density of the monosaccharide is in keeping with a GlcNAc moiety commonly appended to polyprenyl groups through the action of WecA in E. coli (23). The weak additional density extending from the C3-hydroxyl group of GlcNAc may indicate the presence of galactose presumably appended to GlcNAc by the enzyme WbbD in E. coli (24).
The interactions between LcpASA and C40-PP–GlcNAc are shown in Fig. 1c. Residues surrounding the lipid tail display low sequence conservation but are largely hydrophobic. Notably, the bulky aromatic side chain of Phe-171 introduces a kink at the third prenyl moiety, whereas the wide base of the pocket induces a twist in the lipid backbone to accommodate the remainder of the lipid tail. Invariant arginine residues form a positively charged entrance to the hydrophobic pocket, where their guanidinyl groups form salt bridges with the pyrophosphate moiety of the polyprenyl. Arg-99 and Arg-216 are in contact with the α-phosphate group and Arg-122, Arg-218, and Arg-227 are in contact with the β-phosphate group in keeping with earlier mutagenesis studies showing their importance in growth and activity in various species (summarized in Table S2) (9, 20, 25).
The orientation of the lipid-bound GlcNAc moiety is stabilized by an intramolecular hydrogen bond (3.0 Å) between the nitrogen atom of the N-acetyl group and a phosphoryl oxygen of the α-phosphate group (Fig. 1c). In addition, the position of the GlcNAc moiety is stabilized by hydrophobic and van der Waals interactions between the N-acetyl group and a shallow pocket outlined by residues Asn-194, Ile-195, Arg-216, Phe-217, Arg-218, and His-219 of regions B and C (Fig. 1d). These observations are also supported by the structure of TagTBS bound to C30-PP–GlcNAc–ManNAc (Fig. S3). The lack of specific interactions between the enzyme and the N-acetyl group likely allows for the significant observed differences in the secondary structure of region B as mentioned above. Region C possesses two highly conserved residues, a pyrophosphate-binding arginine (Arg-218) and an aspartate (Asp-224). The loop of region C is commonly held away from the active site by a resident acidic residue that forms a salt bridge with a nearby basic residue (Fig. S4). Region C is fully modeled in our structures of LcpASA, TagTBS, and TagUBS. A comparison between all available LCP enzyme structures reveals that ordering of the loop in region C depends heavily on the extension of helix 5 beyond the conserved aspartate (LcpASA Asp-224) to the adjacent residue (LcpASA Gly-223), commonly found to be glycine. We note that our structure of TagTBS differs from that of the previously reported apo and C40-PP bound structures in that region C is resolved. Interestingly, region C of our structure is conformationally distinct from that of the C30-PP–GlcNAc bound structure (RMSD of 2.7 Å for 7 Cα pairs) but more closely resembles that of the C30-PP–GlcNAc–ManNAc bound form (RMSD of 0.4 Å for 7 Cα pairs) with the loop directed away from the active site. The similarity indicates that this placement of region C is not induced by the presence of the second sugar as previously presumed.
The requisite catalytic metal is not observed in the electron density of LcpASA, despite the presence of the two conserved magnesium-binding aspartate residues (Asp-91 and Asp-101) at the opposite ends of region A and the appropriately positioned pyrophosphate moiety of the lipid substrate (Fig. 1c). We believe the metal ion may have been displaced during crystallization because region A is shifted slightly away from the active site through interactions with a neighboring molecule in the crystal; at the same time, this subtle motion likely inhibited the pyrophosphatase activity of the enzyme and fortuitously allowed capture of the intact lipid headgroup.
Our structure of TagUBS displayed no density for an endogenous lipid as the hydrophobic pocket is partially collapsed due to the rearrangements described above (significantly reduced surface area ≈ 130 Å2; volume ≈ 66 Å3). In contrast, small clusters of weak electron density, dimensions collectively in keeping with a potential bound lipid but of insufficient order for precise modeling, were observed within the lipid pocket of TagVBS (surface area ≈ 660 Å2; volume ≈ 500 Å3) and TagTBS (surface area ≈ 750 Å2; volume ≈ 630 Å3).
Interestingly, a cluster of ordered water molecules and a glycerol molecule from our cryoprotectant were found within the structure of LcpASA, highlighting the presence of an accessible hydrophilic pocket buried under the active site and adjacent to the lipid-binding pocket (Fig. 1d). The hydrophilic pocket is outlined by highly conserved residues including Ser-119, Arg-122, Asp-224, Arg-227, and Gln-231 (Fig. S5). We suggest that the conserved and accessible nature of this cryptic site could benefit the design of inhibitors with increased binding affinity.
Peptidoglycan-binding site
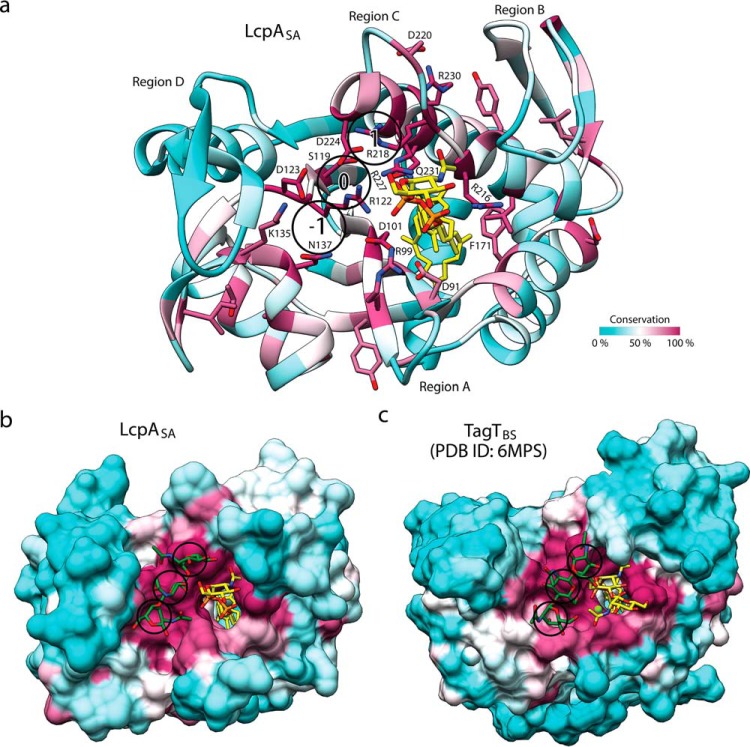
A groove outlined by regions A, C, and D (Fig. 3a and Fig. S4) has been proposed to be the general PG-binding site of LCP enzymes (9, 14, 20), but accurate assignment of binding features is hindered by structural variability surrounding the groove and supports the need to study the S. aureus variant described here for drug discovery efforts. Region A of LcpASA is fully modeled in our structure and exhibits low sequence conservation except for an invariant arginine residue, Arg-99. The side chain of Arg-99 is in contact with the α-phosphate group of the lipid; however, it is expected that Arg-99 will shift to avoid steric hindrance when a divalent cation is bound (Fig. 1c). The inability of Arg-99 to interact with the lipid in the catalytically active magnesium-bound state suggests that Arg-99 is involved in binding PG but not necessarily the lipid substrate. In the structure of LcpASA, the loop of region C is ordered and positioned to enlarge the putative PG-binding groove (Fig. 3). Region D, found across from region C on the opposite side of the PG groove, adopts a two-stranded β-sheet and often displays an enrichment of aromatic residues that could aid in the binding of PG through carbohydrate-aromatic interactions. In addition, LcpASA Asp-123, Lys-135, and Asn-137 are conserved residues with possible roles in PG binding because they are found away from the lipid substrate and within the putative PG-binding groove. Notably, TagTBS residues equivalent to Asp-123 and Lys-135 (TagTBS Asp-119 and Lys-131) were reported to be essential for activity (20).
Figure 3.
Peptidoglycan binding site of LCP enzymes. a, crystal structure of LcpASA colored by residue conservation. Residues that are over 80% conserved among the top 500 homologous sequences identified using ConSurf are depicted as sticks (27). The lipid is shown in yellow, and heteroatoms are colored by type (oxygen, red; phosphorus, orange; nitrogen, blue; magnesium, green). Saccharide-binding subsites (1, 0, and −1) identified by docking triGlcNAc are depicted within circles. b and c, the surfaces of LcpASA (b) and TagTBS (PDB code 6MPS) (c), colored by conservation, are depicted complexed to triGlcNAc (dark green) with the highest HADDOCK score.
To gain further insight into PG binding, we conducted docking studies using a data-driven biomolecular docking program, HADDOCK2.2 (26). A chitin oligosaccharide with three GlcNAc residues (triGlcNAc) was selected as the binding molecule to represent the glycan core of PG. triGlcNAc was docked onto two crystals structures: LcpASA in complex with C40-PP–GlcNAc and TagTBS in complex with C30-PP–GlcNAc–ManNAc (PDB code 6MPS) (Fig. 3 and Fig. S6). The C6-hydroxyl group belonging to the central GlcNAc of triGlcNAc was selected as the nucleophile, and distance constraints were imposed on its position relative to the CZ atom of the putative arginine general base (LcpASA Arg-122; TagTBS Arg-118; 3.5 Å) and the β-phosphorus group of the pyrophosphate moiety (electrophile; 2.5 Å). Conserved surface-exposed active-site residues in the vicinity of the electrophile were identified using ConSurf and selected as residues with potential to bind triGlcNAc (27). Regions A, B, C, and D adjacent to the active site were specified as fully flexible. 78 models of triGlcNAc-bound LcpASA clustered into 8 groups, and 98 models of triGlcNAc-bound TagTBS clustered into 12 groups (Tables S3 and S4). The top-scoring clusters of the two docking experiments produced structurally similar complexes that allowed mapping of saccharide-binding subsites in the predicted PG-binding groove. Subsite 0, adjacent to the β-phosphate group and above LcpASA Arg-122, houses the saccharide representing the MurNAc moiety of PG with the putative nucleophilic C6-hydroxyl group. Subsite 1 is adjacent to LcpASA Asp-224, and subsite −1 is between LcpASA Lys-135 and Arg-99. The variability in interactions with the glycan among top-scoring structures prevented accurate assignment of PG binding. Sequence conservation is low outside of the three subsites mapped in our docking study, and high variability was observed in docking experiments conducted with longer glycan strands. Importantly, there appears to be ample space in the groove to accommodate additional saccharides at either end of the identified subsites.
Catalytic mechanism
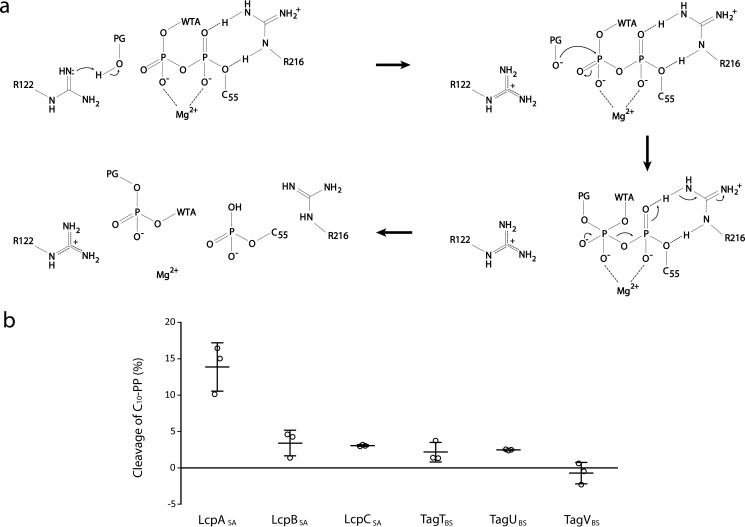
The reaction catalyzed by LCP enzymes is presumably similar to other biological phosphoryl-transfer reactions in which the nucleophile aligns with the electrophilic phosphorous (the β-phosphorous of C55-PP–WTA) and the leaving group (the α-phosphorous of C55-PP–WTA) for an in-line attack (Fig. 4a) (20, 28). It is unlikely that the reaction proceeds through the formation of a covalent enzyme intermediate because no residues are suitably positioned to play the role of the nucleophile in LcpASA and other variants (Fig. 3a). As such, we infer that the C6-hydroxyl group of PG MurNAc acts as the nucleophile and sits adjacent to the β-phosphorus group of the lipid substrate donor in the putative PG-binding groove. Furthermore, no conserved acidic residues that might typically act as the general base in deprotonation/activation of the C6-hydroxyl are appropriately localized. Based on the putative location of the nucleophile, we considered Arg-122 and Arg-218 of LcpASA as candidates to carry out the role of general base. Indeed, the equivalent TagTBS residues, Arg-118 and Arg-219, were reported to be essential for activity in vitro (20). There is precedent for enzymes to use arginine in a general base role, and commonalities among them are shared with LCP enzymes (29). Notably, the pKa of the arginine general base is believed to be depressed by adjacent positive charges, such as the cluster of arginine residues in the active site of LCP enzymes (29). Another common feature is the activation of the guanidinyl group by a nearby carboxylate group (29). In the structure of LcpASA, the two general base candidates are adjacent to the carboxylate groups of Asp-123 and Asp-224. Between the two aspartate residues, only the TagTBS residue (Asp-119) corresponding to LcpASA Asp-123 was found to be essential (20). It has been proposed that carboxylate groups may depress the pKa of a guanidinyl group by twisting the planar conformation, adopted by the protonated form, to eliminate resonance (29). Alternatively, the carboxylate group may facilitate proton exchange for rapid equilibrium between the deprotonated and protonated forms of guanidine (29). Exchange between the two forms is also expected to be rapid because of their relatively high solvent exposure (29). Of the two candidates, Arg-122 has a higher likelihood to be the general base because the adjacent carboxylate of Asp-123 is both conserved and essential. The only carboxylate group that the other general base candidate, Arg-218, can interact with belongs to Asp-224, a nonessential but invariant residue among LCP enzymes. The structure of LcpASA also shows that the α-phosphate group of the bound lipid forms a salt bridge with the guanidinyl group of Arg-216, a feature that is observed with other lipid-bound structures of LCP enzymes (9, 14, 20). Given its unique positioning, we propose that Arg-216 acts as the general acid with proton exchange of its guanidinium side chain promoted by access to bulk solvent. In support, the corresponding Corynebacterium glutamicum LcpA residue, Arg-257, was reported to be essential for growth in a complementation study (25).
Figure 4.
Enzymatic activity of LCP enzymes. a, proposed mechanism of WTA ligation to PG by LcpASA. b, enzymatic cleavage of C10-PP by S. aureus and B. subtilis ΔTM LCP enzymes. Pyrophosphatase activity was assessed by quantifying the amount of Pi released enzymatically after an 18-h incubation at 37 °C. The error bars represent the standard deviation of three independent experiments.
Collectively, we propose that catalysis is initiated when the transient deprotonated form of Arg-122 abstracts a proton from the PG MurNAc C6-hydroxyl group (Fig. 4a). In addition to coordinating the pyrophosphate moiety for catalysis, the magnesium ion and the invariant arginine residues are likely involved in stabilizing the transition state. This stabilization may occur through charge neutralization and the formation of stronger interactions with the trigonal bipyramidal transition state over the tetrahedral ground state. When the transition state collapses, a covalent bond between PG and WTA is formed, and the α-phosphate leaving group abstracts a proton from the guanidinium of Arg-216.
Pyrophosphatase activity and oligomerization
The catalytic release of Pi from pyrophosphate lipids by LCP enzymes has been reported (9, 14, 21, 25, 30). To compare the pyrophosphatase activity of S. aureus and B. subtilis LCP enzymes, we incubated 500 μm C10-PP with 50 μm purified LCP enzymes (Fig. 4b). A detergent (n-dodecyl β-d-maltoside) was included in the buffer to facilitate the release of endogenous lipids. LcpASA was found to exhibit the highest pyrophosphatase activity, with ∼14% of C10-PP enzymatically cleaved after an 18-h incubation at 37 °C. In contrast, TagVBS did not exhibit pyrophosphatase activity under these conditions, and the other enzymes were found to cleave 2–3% of C10-PP. These differences could stem from a number of factors including the affinity for the lipid substrate. Analysis of the transferase activity was prevented by difficulties in substrate production at sufficient yield.
LCP enzymes are generally regarded as monomeric proteins. A notable exception is C. glutamicum LcpA, which has been reported to dimerize under oxidizing conditions and tetramerize in reducing environments based on size-exclusion chromatography results (25). In our purification of S. aureus and B. subtilis LCP enzymes, small populations of oligomers were identified. To determine the oligomeric state of the complexes, size-exclusion chromatography with multiangle static light-scattering detection was performed (Fig. S7). LcpCSA, TagTBS, and TagVBS were found to assemble consistently into decamers with molecular masses of the oligomers (320.3 ± 8.4, 357.3 ± 2.9, and 330.8 ± 7.8 kDa) determined to be 10.4, 10.3, and 10.1 multiples of the corresponding monomers (30.7 ± 0.4, 34.8 ± 0.2, and 32.8 ± 0.8 kDa), respectively. Furthermore, TagTBS and TagVBS were also found to elute in a dimeric state (75.9 ± 0.9 and 65.9 ± 3.2 kDa, respectively), which suggests the decamers to be an assembly of dimers. Unfortunately, oligomers of the other LCP enzymes dissociated prior to analysis by size-exclusion chromatography with multiangle static light-scattering detection. Stable formation of the oligomers may require the missing transmembrane anchor and undiscovered binding partners.
Discussion
LCP-catalyzed secondary cell wall polymer attachment to peptidoglycan is an attractive therapeutic target because inhibition of this process is expected to disrupt peptidoglycan assembly, cell division, and pathogenicity. Furthermore, inhibitors of LCP enzymes would not be constrained by issues of membrane permeability. Here, we focused on the first atomic resolution analysis of the central LCP enzyme that mediates attachment of wall teichoic acid to peptidoglycan in S. aureus. Our crystal structures and modeled substrate-bound complexes provide a comprehensive understanding of key features linked to substrate binding and catalysis that should aid in the structure-guided design of inhibitors.
The structure of LcpASA in complex with C40-PP–GlcNAc provided clarification on the orientation of the saccharide headgroup and revealed a dependence on hydrophobic and van der Waals contacts rather than polar interactions with the enzyme (Fig. 1d). Another factor in the positioning of the GlcNAc moiety is an intramolecular hydrogen bond between the nitrogen atom of the N-acetyl group and a phosphoryl oxygen of the α-phosphate group (Fig. 1c).
The crystal contact at region A of the LcpASA structure inadvertently aided our structural investigation because it displaced the active-site magnesium ion and prevented the catalytic removal of the lipid headgroup. Despite the absence of the divalent cation, the pyrophosphate moiety of the lipid is appropriately positioned by salt-bridge interactions with multiple invariant arginine residues (Fig. 1c). One of these residues, Arg-218 of LcpASA, is found on the loop of region C, which we noted as being highly flexible in a comparison between all available LCP enzyme structures. We speculate that flexibility of the loop aids in product expulsion because displacement of the resident arginine residue would weaken interactions with the phosphate moieties of the products.
In our analysis of the lipid-binding pocket of LcpASA, we discovered the presence of a hydrophilic subpocket occupied by a glycerol molecule from the cryoprotectant (Fig. 1d). The binding of the glycerol molecule indicates that the buried pocket is not strictly enclosed and could accommodate exogenously added molecules. Importantly, the presence of this cryptic pocket is not restricted to LcpASA because the surrounding residues are highly conserved. In addition to the substrate-binding sites, the buried hydrophilic pocket and the shallow pocket occupied by the N-acetyl group should be considered in drug design strategies to increase binding specificity and affinity.
The results of our computational docking experiments led to the identification of three potential PG saccharide-binding subsites in proximity to conserved residues including Arg-99, Lys-135, Asn-137, and Asp-224 of LcpASA (Fig. 3). Additional information regarding the acceptor-binding site is acquired by comparing our LCP enzyme structures with that of A. oris LcpA, which catalyzes glycosylation with a protein acceptor (Fig. S4). Notably, the β-sheet of region B is extended by two antiparallel β-strands inserted into region C of A. oris LcpA. In addition, a loop with a 310 helix replaces β-strands 11 and 12 that make up region D in conventional LCP enzymes. This loop is shifted away from the active site, closing off one end of the putative acceptor substrate-binding groove while enlarging the opposite end, presumably for accommodating the protein substrate. These differences highlight the importance of the antiparallel β-strands of region D for PG binding in our structures. In addition to the differences in secondary structure, Asp-224 of LcpASA is replaced by an alanine in A. oris LcpA (A263), and conservation of this residue is low among enzymes closely related to A. oris LcpA. These findings support our proposed role of LcpASA Asp-224 in PG binding and not in the activation of the general base. The conservation of LcpASA Asp-123 among all LCP enzymes suggests that this aspartate is a more likely candidate for the execution of this role.
Lipid-bound structures of LCP enzymes have not provided clear insight into how the WTA intermediate is selected over the PG precursor, lipid II. Preference for the WTA intermediate may be facilitated by another enzyme that delivers C55-PP–WTA to LCP enzymes and possibly returns C55-P to the membrane for continuous PG synthesis. The paralogs, CapA1 and CapA2, are candidates for these proposed roles in S. aureus because CapA1 was reported to enhance the activity of LcpCSA and recognize C55-PP–WTA/CP preferentially over lipid II in catalyzing the cleavage of the pyrophosphate linkage (19). Hydrolysis of C55-PP–WTA/CP by CapA1 is perhaps a novel function because CapA2 is unable to catalyze this cleavage reaction (19). CapA enzymes are integral membrane proteins in the polysaccharide co-polymerase family known to facilitate export of bacterial cell wall polysaccharides and oligomerize into ring-like structures (31, 32). Our observation of decameric LCP enzymes raises the intriguing possibility of a direct interaction between LCP and CapA oligomers for the transport of lipid-linked WTA/CP from the membrane to the hydrophobic pocket of LCP enzymes.
Experimental procedures
Cloning, expression, and purification
Codon-optimized DNA sequences encoding for S. aureus LcpA (Uniprot Q99Q02), LcpB (Uniprot A0A0H3JTR6), and LcpC (Uniprot A0A0H3JNL8) and B. subtilis TagT (Uniprot Q7WY78), TagU (Uniprot Q02115), and TagV (Uniprot P96499) were used as templates for amplifying regions of interest. The PCR products were cloned into modified pET28a (Novagen) and pET41b (Novagen) vectors for expression with a thrombin-cleavable hexahistidine tag. The plasmids were transformed into E. coli BL21 (DE3) and grown in ZYP-5052 autoinduction medium at 37 °C for 4 h before lowering the temperature to 25 °C for 18 h. SeMet-substituted TagUBS was expressed using a metabolic inhibition protocol (33). pET28a encoding for an N-terminal His-tag was ideal for expression of LcpASA (residues 80–327), LcpBSA (residues 59–405), LcpCSA (residues 66–315) and TagTBS (residues 46–322). pET41b encoding for a C-terminal His tag was ideal for expression of TagUBS (residues 62–306) and TagVBS (residues 72–332). The cells were harvested by centrifugation and resuspended in 20 mm HEPES, pH 8, 150 mm NaCl, 25 mm imidazole for lysis using an Avestin EmulsiFlex-C5 homogenizer. Cell debris was pelleted by centrifugation at 125,000 × g for 45 min, and the supernatant was loaded on a 1-ml HisTrap FF column (GE Healthcare Life Sciences) equilibrated in the lysis buffer. The column was washed with 25 column volumes of the same buffer prior to elution with a linear imidazole gradient up to 500 mm in 25 ml. Purified enzymes were concentrated with an Amicon Ultra centrifugal filter (10-kDa molecular mass cutoff; EMD Millipore) for further purification by size-exclusion chromatography with a Superdex200 10/300 GL column (GE Healthcare Life Sciences) equilibrated with 20 mm HEPES, pH 8, 150 mm NaCl, 1 mm MgCl2. Peak fractions were pooled and concentrated to ∼10 mg/ml.
Crystallization and structure determination
The crystals were grown at room temperature using the sitting-drop vapor-diffusion method (1 μl of protein mixed with 1 μl of precipitant). 8.5 mg/ml LcpASA was crystallized in 2.4 m ammonium sulfate, 0.08 m citric acid, pH 5.2. 10 mg/ml TagTBS was crystallized in 0.2 m sodium malonate, 0.1 m Bis-tris propane, pH 6.5, 20% PEG 3350. 12 mg/ml TagUBS was crystallized in 1.6 m ammonium sulfate. 10 mg/ml TagVBS was crystallized in 0.3 m ammonium nitrate, 17% PEG 3350. The crystals were cryoprotected in mother liquor supplemented with 30% glycerol and flash-cooled in liquid nitrogen. The diffraction data were collected with the 08ID-1 Beamline of the Canadian Light Source and processed using AutoProcess. The structures of LcpASA, TagTBS, and TagVBS were solved by molecular replacement using Phaser-MR in Phenix with deposited structures (PDB code 3NXH or 4DE9) (14, 34). The structure of TagUBS was determined by single-wavelength anomalous diffraction phasing using a SeMet derivative. Model building and refinement were performed with Phenix and Coot (35, 36). Coordinates and structure factors were deposited in the Protein Data Bank with accession codes 6UEX (LcpASA), 6UF5 (TagTBS), 6UF6 (TagUBS), and 6UF3 (TagVBS).
Docking
Docking of triGlcNAc to crystal structures of LcpASA in complex with C40-PP–GlcNAc and TagTBS in complex with C30-PP–GlcNAc–ManNAc (PDB code 6MPS) was conducted with HADDOCK2.2 (26). The C6-hydroxyl group of the central GlcNAc was selected as the nucleophile, and distance constraints were imposed on its position relative to the CZ atom of the putative arginine general base (LcpASA Arg-122; TagTBS R118; 3.5 Å) and the β-phosphorus electrophile (2.5 Å). Conserved surface-exposed active-site residues in the vicinity of the electrophile were selected as passive residues with potential to bind triGlcNAc (LcpASA residues 91, 99, 101, 122, 123, 135, 137, 216, 218, 224, and 227; TagTBS residues 82, 95, 97, 118, 119, 131, 133, 217, 219, 224, and 227). These residues are at least 80% conserved among the top 500 homologous sequences complied using ConSurf (27). Regions A, B, C, and D surrounding the entrance of the lipid-binding pocket were specified as fully flexible regions. Each docking experiment was conducted using 4000 structures for rigid body docking, 1000 structures for semiflexible refinement, and 200 structures for explicit solvent refinement. Clustering of structures was performed using the root-mean-square deviation method with a 2 Å cutoff.
Pyrophosphatase assay
The pyrophosphatase activity of the different ΔTM LCP enzymes was assessed in triplicate with an end-point assay based on the release of Pi. Briefly, the reaction solutions were prepared in 20 mm HEPES, pH 8, 150 mm NaCl, 10 mm MgCl2, 1.4 mm n-dodecyl β-d-maltoside and contained 500 μm C10-PP (Sigma) and 50 μm of the particular LCP enzyme under scrutiny. These reaction mixtures were incubated at 37 °C for 18 h before diluting them ∼2.5-fold while simultaneously adding 1 unit/ml purine nucleoside phosphorylase and 200 μm 2-amino-6-mercapto-7-methylpurine ribonucleoside from an EnzChek phosphatase assay kit (Molecular Probes). Following a 10-min incubation period at room temperature, the absorbance of 20-μl aliquots was measured at 360 nm with a Synergy H4 multimode plate reader (BioTek). Pi released from the enzymatic cleavage of C10-PP was determined with control experiments lacking either or both C10-PP and an LCP enzyme.
Size-exclusion chromatography with multiangle static light scattering
Samples of freshly-purified ΔTM LCP enzymes (2 mg/ml in 100 μl) pooled from the oligomer peak after size-exclusion chromatography on a Superdex200 10/300 GL column (GE Healthcare Life Sciences) were subjected to size-exclusion chromatography analysis a second time (Superdex200 10/300 GL, 22 °C, and flow rate of 0.45 ml/min) but with in-line multiangle light-scattering analysis (miniDAWN TREOS coupled with an Optilab T-rEX differential diffractometer; Wyatt Technologies) to determine the oligomerization state. No reducing agent was included in the buffer because none of the enzymes contain cysteine residues that might form disulfide bonds. The data were analyzed using the ASTRA version 6.1 software package provided by the manufacturer, and the results cited herein represent the averages of three separate experiments.
Analysis software
Crystal structures were visualized and represented using UCSF Chimera and PyMOL (37, 38). The cavity size of the lipid-binding pockets was determined using CASTp with a default radius probe of 1.4 Å (39). Interactions were probed using LigPlot and PLIP (40, 41). Sequence alignments were produced using the ESPript 3.0 server (42). Sequence conservation was analyzed using ConSurf (27). The domain structures were visualized using Domain Graph (43). Scatterplots were generated using Interactive Dotplot (44).
Author contributions
F. K. K. L. and N. C. J. S. conceptualization; F. K. K. L. data curation; F. K. K. L. formal analysis; F. K. K. L. validation; F. K. K. L. and F. I. R. investigation; F. K. K. L. visualization; F. K. K. L. and F. I. R. methodology; F. K. K. L. writing-original draft; F. K. K. L., F. I. R., R. T. G., J.-P. S., E. D. B., and N. C. J. S. writing-review and editing; R. T. G., J.-P. S., E. D. B., and N. C. J. S. resources; N. C. J. S. supervision; N. C. J. S. funding acquisition; N. C. J. S. project administration.
Supplementary Material
Acknowledgments
We thank beamline staff for their assistance with X-ray data collection at the Canadian Light Source synchrotron facility (Beamline 08ID-1).
This work was supported by the Natural Sciences and Engineering Research Council (to F. K. K. L.), the Canadian Institutes of Health Research (to R. T. G., N. C. J. S., and E. D. B.), the Howard Hughes Medical Institute International Senior Scholar program (to N. C. J. S.), the Canada Foundation for Innovation (to N. C. J. S. and E. D. B.), the British Columbia Knowledge Development Fund (to N. C. J. S.), the Canada Research Chairs program (tier 1; to N. C. J. S. and E. D. B.), and the Joint Programming Initiative on Antimicrobial Resistance (to J.-P. S). The authors declare that they have no conflicts of interest with the contents of this article.
The atomic coordinates and structure factors (codes 6UEX, 6UF5, 6UF6, and 6UF3) have been deposited in the Protein Data Bank (http://wwpdb.org/).
This article contains Tables S1–S4 and Figs. S1–S7.
- PG
- peptidoglycan
- LCP
- LytR-CpsA-Psr
- WTA
- wall teichoic acid
- CP
- capsular polysaccharide
- PBP
- penicillin-binding proteins
- C10-PP
- geranyl-pyrophosphate
- C55-P
- undecaprenyl phosphate
- MurNAc
- N-acetylmuramic acid
- ManNAc
- N-acetylmannosamine
- triGlcNAc
- N,N′,N″-triacetylchitotriose
- PDB
- Protein Data Bank
- SeMet
- selenomethionine
- TM
- transmembrane
- RMSD
- root-mean-square deviation.
References
- 1. Campbell J., Singh A. K., Santa Maria J. P. Jr., Kim Y., Brown S., Swoboda J. G., Mylonakis E., Wilkinson B. J., and Walker S. (2011) Synthetic lethal compound combinations reveal a fundamental connection between wall teichoic acid and peptidoglycan biosyntheses in Staphylococcus aureus. ACS Chem. Biol. 6, 106–116 10.1021/cb100269f [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Farha M. A., Leung A., Sewell E. W., D'Elia M. A., Allison S. E., Ejim L., Pereira P. M., Pinho M. G., Wright G. D., and Brown E. D. (2013) Inhibition of WTA synthesis blocks the cooperative action of PBPs and sensitizes MRSA to β-lactams. ACS Chem. Biol. 8, 226–233 10.1021/cb300413m [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Atilano M. L., Pereira P. M., Yates J., Reed P., Veiga H., Pinho M. G., and Filipe S. R. (2010) Teichoic acids are temporal and spatial regulators of peptidoglycan cross-linking in Staphylococcus aureus. Proc. Natl. Acad. Sci. U.S.A. 107, 18991–18996 10.1073/pnas.1004304107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Schlag M., Biswas R., Krismer B., Kohler T., Zoll S., Yu W., Schwarz H., Peschel A., and Götz F. (2010) Role of staphylococcal wall teichoic acid in targeting the major autolysin Atl. Mol. Microbiol. 75, 864–873 10.1111/j.1365-2958.2009.07007.x [DOI] [PubMed] [Google Scholar]
- 5. Holland L. M., Conlon B., and O'Gara J. P. (2011) Mutation of tagO reveals an essential role for wall teichoic acids in Staphylococcus epidermidis biofilm development. Microbiology 157, 408–418 10.1099/mic.0.042234-0 [DOI] [PubMed] [Google Scholar]
- 6. Weidenmaier C., Peschel A., Xiong Y.-Q., Kristian S. A., Dietz K., Yeaman M. R., and Bayer A. S. (2005) Lack of wall teichoic acids in Staphylococcus aureus leads to reduced interactions with endothelial cells and to attenuated virulence in a rabbit model of endocarditis. J. Infect. Dis. 191, 1771–1777 10.1086/429692 [DOI] [PubMed] [Google Scholar]
- 7. Misawa Y., Kelley K. A., Wang X., Wang L., Park W. B., Birtel J., Saslowsky D., and Lee J. C. (2015) Staphylococcus aureus colonization of the mouse gastrointestinal tract is modulated by wall teichoic acid, capsule, and surface proteins. PLoS Pathog. 11, e1005061 10.1371/journal.ppat.1005061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Caveney N. A., Li F. K., and Strynadka N. C. (2018) Enzyme structures of the bacterial peptidoglycan and wall teichoic acid biogenesis pathways. Curr. Opin. Struct. Biol. 53, 45–58 10.1016/j.sbi.2018.05.002 [DOI] [PubMed] [Google Scholar]
- 9. Kawai Y., Marles-Wright J., Cleverley R. M., Emmins R., Ishikawa S., Kuwano M., Heinz N., Bui N. K., Hoyland C. N., Ogasawara N., Lewis R. J., Vollmer W., and Daniel R. A. Errington J. (2011) A widespread family of bacterial cell wall assembly proteins. EMBO J. 30, 4931–4941 10.1038/emboj.2011.358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. D'Elia M. A., Millar K. E., Bhavsar A. P., Tomljenovic A. M., Hutter B., Schaab C., Moreno-Hagelsieb G., and Brown E. D. (2009) Probing teichoic acid genetics with bioactive molecules reveals new interactions among diverse processes in bacterial cell wall biogenesis. Chem. Biol. 16, 548–556 10.1016/j.chembiol.2009.04.009 [DOI] [PubMed] [Google Scholar]
- 11. Chan Y. G., Frankel M. B., Dengler V., Schneewind O., and Missiakas D. (2013) Staphylococcus aureus mutants lacking the LytR–CpsA–Psr family of enzymes release cell wall teichoic acids into the extracellular medium. J. Bacteriol. 195, 4650–4659 10.1128/JB.00544-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Over B., Heusser R., McCallum N., Schulthess B., Kupferschmied P., Gaiani J. M., Sifri C. D., Berger-Bächi B., and Stutzmann Meier P. (2011) LytR–CpsA–Psr proteins in Staphylococcus aureus display partial functional redundancy and the deletion of all three severely impairs septum placement and cell separation. FEMS Microbiol. Lett. 320, 142–151 10.1111/j.1574-6968.2011.02303.x [DOI] [PubMed] [Google Scholar]
- 13. Hübscher J., Lüthy L., Berger-Bächi B., and Stutzmann Meier P. (2008) Phylogenetic distribution and membrane topology of the LytR–CpsA–Psr protein family. BMC Genomics 9, 617 10.1186/1471-2164-9-617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Eberhardt A., Hoyland C. N., Vollmer D., Bisle S., Cleverley R. M., Johnsborg O., Håvarstein L. S., Lewis R. J., and Vollmer W. (2012) Attachment of capsular polysaccharide to the cell wall in Streptococcus pneumoniae. Microb. Drug Resist. 18, 240–255 10.1089/mdr.2011.0232 [DOI] [PubMed] [Google Scholar]
- 15. Harrison J., Lloyd G., Joe M., Lowary T. L., Reynolds E., Walters-Morgan H., Bhatt A., Lovering A., Besra G. S., and Alderwick L. J. (2016) Lcp1 is a phosphotransferase responsible for ligating arabinogalactan to peptidoglycan in Mycobacterium tuberculosis. MBio. 7, e00972–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gale R. T., Li F. K. K., Sun T., Strynadka N. C. J., and Brown E. D. (2017) B. subtilis LytR–CpsA–Psr enzymes transfer wall teichoic acids from authentic lipid-linked substrates to mature peptidoglycan in vitro. Cell Chem. Biol. 24, 1537–1546.e4 10.1016/j.chembiol.2017.09.006 [DOI] [PubMed] [Google Scholar]
- 17. Malm S., Maaß S., Schaible U. E., Ehlers S., and Niemann S. (2018) In vivo virulence of Mycobacterium tuberculosis depends on a single homologue of the LytR–CpsA–Psr proteins. Sci. Rep. 8, 3936 10.1038/s41598-018-22012-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Schaefer K., Matano L. M., Qiao Y., Kahne D., and Walker S. (2017) In vitro reconstitution demonstrates the cell wall ligase activity of LCP proteins. Nat. Chem. Biol. 13, 396–401 10.1038/nchembio.2302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rausch M., Deisinger J. P., Ulm H., Müller A., Li W., Hardt P., Wang X., Li X., Sylvester M., Engeser M., Vollmer W., Müller C. E., Sahl H. G., Lee J. C., and Schneider T. (2019) Coordination of capsule assembly and cell wall biosynthesis in Staphylococcus aureus. Nat. Commun. 10, 1404 10.1038/s41467-019-09356-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Schaefer K., Owens T. W., Kahne D., and Walker S. (2018) Substrate preferences establish the order of cell wall assembly in Staphylococcus aureus. J. Am. Chem. Soc. 140, 2442–2445 10.1021/jacs.7b13551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Siegel S. D., Amer B. R., Wu C., Sawaya M. R., Gosschalk J. E., Clubb R. T., and Ton-That H. (2019) Structure and mechanism of LcpA, a phosphotransferase that mediates glycosylation of a Gram-positive bacterial cell wall–anchored protein. MBio. 10, e01580–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kim H., Kim S., and Yoon S. H. (2018) Metabolic network reconstruction and phenome analysis of the industrial microbe, Escherichia coli BL21(DE3). PLoS One 13, e0204375 10.1371/journal.pone.0204375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Alexander D. C., and Valvano M. A. (1994) Role of the rfe gene in the biosynthesis of the Escherichia coli O7-specific lipopolysaccharide and other O-specific polysaccharides containing N-acetylglucosamine. J. Bacteriol. 176, 7079–7084 10.1128/JB.176.22.7079-7084.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Riley J. G., Menggad M., Montoya-Peleaz P. J., Szarek W. A., Marolda C. L., Valvano M. A., Schutzbach J. S., and Brockhausen I. (2005) The wbbD gene of E. coli strain VW187 (O7:K1) encodes a UDP-Gal:GlcNAcα-pyrophosphate-Rβ1,3-galactosyltransferase involved in the biosynthesis of O7-specific lipopolysaccharide. Glycobiology 15, 605–613 10.1093/glycob/cwi038 [DOI] [PubMed] [Google Scholar]
- 25. Baumgart M., Schubert K., Bramkamp M., and Frunzke J. (2016) Impact of LytR–CpsA–Psr proteins on cell wall biosynthesis in Corynebacterium glutamicum. J. Bacteriol. 198, 3045–3059 10.1128/JB.00406-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. van Zundert G. C. P., Rodrigues JPGLM, Trellet M., Schmitz C., Kastritis P. L., Karaca E., Melquiond A. S. J., van Dijk M., de Vries S. J., Bonvin A. M. J. J. (2016) The HADDOCK2.2 web server: user-friendly integrative modeling of biomolecular complexes. J. Mol. Biol. 428, 720–725 10.1016/j.jmb.2015.09.014 [DOI] [PubMed] [Google Scholar]
- 27. Celniker G., Nimrod G., Ashkenazy H., Glaser F., Martz E., Mayrose I., Pupko T., and Ben-Tal N. (2013) ConSurf: using evolutionary data to raise testable hypotheses about protein function. Isr. J. Chem. 53, 199–206 10.1002/ijch.201200096 [DOI] [Google Scholar]
- 28. Lassila J. K., Zalatan J. G., and Herschlag D. (2011) Biological phosphoryl-transfer reactions: understanding mechanism and catalysis. Annu. Rev. Biochem. 80, 669–702 10.1146/annurev-biochem-060409-092741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Guillén Schlippe Y. V., and Hedstrom L. (2005) A twisted base?: The role of arginine in enzyme-catalyzed proton abstractions. Arch. Biochem. Biophys. 433, 266–278 10.1016/j.abb.2004.09.018 [DOI] [PubMed] [Google Scholar]
- 30. Grzegorzewicz A. E., de Sousa-d'Auria C., McNeil M. R., Huc-Claustre E., Jones V., Petit C., Angala S. K., Zemanová J., Wang Q., Belardinelli J. M., Gao Q., Ishizaki Y., Mikušová K., Brennan P. J., Ronning D. R., et al. (2016) Assembling of the Mycobacterium tuberculosis cell wall core. J. Biol. Chem. 291, 18867–18879 10.1074/jbc.M116.739227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Morona R., Van Den Bosch L., and Daniels C. (2000) Evaluation of Wzz/MPA1/MPA2 proteins based on the presence of coiled-coil regions. Microbiology 146, 1–4 10.1099/00221287-146-1-1 [DOI] [PubMed] [Google Scholar]
- 32. Tocilj A., Munger C., Proteau A., Morona R., Purins L., Ajamian E., Wagner J., Papadopoulos M., Van Den Bosch L., Rubinstein J. L., Féthière J., Matte A., and Cygler M. (2008) Bacterial polysaccharide co-polymerases share a common framework for control of polymer length. Nat. Struct. Mol. Biol. 15, 130–138 10.1038/nsmb.1374 [DOI] [PubMed] [Google Scholar]
- 33. Doublié S. (1997) Preparation of selenomethionyl proteins for phase determination. Methods Enzymol. 276, 523–530 10.1016/S0076-6879(97)76075-0 [DOI] [PubMed] [Google Scholar]
- 34. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., and Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 10.1107/S0021889807021206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Afonine P. V., Grosse-Kunstleve R. W., Echols N., Headd J. J., Moriarty N. W., Mustyakimov M., Terwilliger T. C., Urzhumtsev A., Zwart P. H., and Adams P. D. (2012) Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 68, 352–367 10.1107/S0907444912001308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Emsley P., Lohkamp B., Scott W. G., and Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D. Biol. Crystallogr. 66, 486–501 10.1107/S0907444910007493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., and Ferrin T. E. (2004) UCSF Chimera: a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
- 38. DeLano W. L. (2012) The PyMOL Molecular Graphics System, version 2.3, Schroedinger, LLC, New York [Google Scholar]
- 39. Tian W., Chen C., Lei X., Zhao J., and Liang J. (2018) CASTp 3.0: computed atlas of surface topography of proteins. Nucleic Acids Res. 46, W363–W367 10.1093/nar/gky473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Laskowski R. A., and Swindells M. B. (2011) LigPlot+: multiple ligand–protein interaction diagrams for drug discovery. J. Chem. Inf. Model. 51, 2778–2786 10.1021/ci200227u [DOI] [PubMed] [Google Scholar]
- 41. Salentin S., Schreiber S., Haupt V. J., Adasme M. F., and Schroeder M. (2015) PLIP: fully automated protein-ligand interaction profiler. Nucleic Acids Res. 43, W443–W447 10.1093/nar/gkv315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Robert X., and Gouet P. (2014) Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42, W320–W324 10.1093/nar/gku316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ren J., Wen L., Gao X., Jin C., Xue Y., and Yao X. (2009) DOG 1.0: illustrator of protein domain structures. Cell Res. 19, 271–273 10.1038/cr.2009.6 [DOI] [PubMed] [Google Scholar]
- 44. Weissgerber T. L., Savic M., Winham S. J., Stanisavljevic D., Garovic V. D., and Milic N. M. (2017) Data visualization, bar naked: A free tool for creating interactive graphics. J. Biol. Chem. 292, 20592–20598 10.1074/jbc.RA117.000147 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.