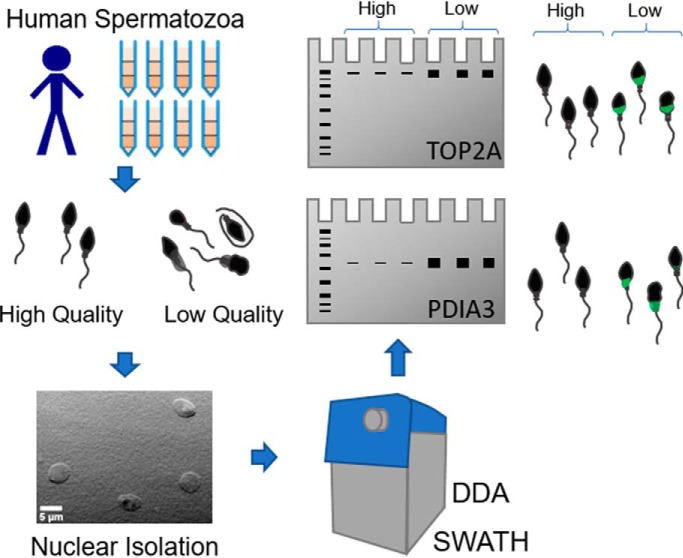
Human spermatogenesis is a far from perfect process and even fertile men produce high numbers of defective spermatozoa. These cells are often characterized by chromatin defects. Following the isolation of normal and abnormal sperm cells, a label free comparison (SWATH) of nuclei from these populations demonstrates a retention of nuclear proteins in abnormal cells. Validation of mass spectrometry results by immuno-analysis reveals Topoisomerase 2A as a promising biomarker of sperm nuclear quality.
Keywords: SWATH-MS, mass spectrometry, biomarker: prognostic, subcellular analysis, cell fractionation, male fertility, nucleus, spermatozooa
Graphical Abstract
Highlights
Human spermatozoa possess cells of poor morphology that lack nuclear integrity.
These cells can be isolated by density separation.
Mass spectrometry reveals their nuclei contain excess protein.
TOP2A is a promising marker of this poor nuclear development.
Abstract
Male infertility is widespread and estimated to affect 1 in 20 men. Although in some cases the etiology of the condition is well understood, for at least 50% of men, the underlying cause is yet to be classified. Male infertility, or subfertility, is often diagnosed by looking at total sperm produced, motility of the cells and overall morphology. Although counting spermatozoa and their associated motility is routine, morphology assessment is highly subjective, mainly because of the procedure being based on microscopic examination. A failure to diagnose male-infertility or sub-fertility has led to a situation where assisted conception is often used unnecessarily. As such, biomarkers of male infertility are needed to help establish a more consistent diagnosis. In the present study, we compared nuclear extracts from both high- and low-quality spermatozoa by LC-MS/MS based proteomic analysis. Our data shows that nuclear retention of specific proteins is a common facet among low-quality sperm cells. We demonstrate that the presence of Topoisomerase 2A in the sperm head is highly correlated to poor head morphology. Topoisomerase 2A is therefore a potential new biomarker for confirming male infertility in clinical practice.
A spermatozoon is a unique cell that is well equipped for its main purpose; to deliver the paternal DNA to the oocyte (1, 2). Spermatogenesis, a process that involves intense cell division and morphological changes, is far from being a perfect process (3, 4). In fact, so poor is human spermatogenesis, that all men, even fertile men, produce defective spermatozoa (5). However, when the number of defective cells reaches a critical level, a man can become either subfertile or infertile (5).
One of the major issues of male-factor infertility is the actual diagnosis. Typically, men are classified as infertile or sub-fertile after looking at the amount of sperm they produce, the motility of the spermatozoa and the morphology of the cells. Although measurements of total sperm counts and sperm motility are consistent worldwide (nowadays commonly measured by computer), analysis of sperm morphology is proving to be problematic (6). According to the World Health Organisation Manual (1992), a spermatozoon can have several morphological abnormalities including defects within the head, tail and/or midpiece, or cells may possess excess residual cytoplasm. Detailed analysis on how sperm morphology should be assessed has been set out (7). However, even though sperm morphology is guided by specific criteria, inter- and intra-laboratory differences are prevalent (8) with one report showing user variation in excess of 12% (9). To illustrate how variable the analysis of sperm morphology can be, the initial examination of 1745 sperm samples over time from 1980 to 1994 showed that sperm morphology had a significant decline (10); however, the independent re-assessment of the same samples revealed no significant morphology drop. Instead, the previously observed decline was because of inconsistent analysis of sperm morphology over this time period (10). The inability to assess sperm morphology and understand its impact on fertility leads to unnecessary treatment and associated costs to many couples. Indeed, of 1391 couples that were referred to assisted conception, including male and female infertility cases, 45.6% ultimately conceived spontaneously (11). This suggests that many couples are misdiagnosed and better prognostic markers of fertility, particularly regarding male fertility potential, are desperately needed to avoid overtreatment of patients (12, 13).
The most common morphological abnormality reported is that pertaining to the shape of the sperm head. Of concern, changes to head morphology may reflect the quality and/or packaging of the DNA (14). During the transition from a round spermatid to elongating spermatid to spermatozoon, a dramatic reduction in the size of the nucleus occurs (15), such that a sperm nuclear volume is about 1/7th the size of any somatic cell (16). This is accomplished as the family of histones are replaced by two smaller and highly charged, proteins known as protamines (PRM1, PRM2). Estimates suggest that 90% of genomic DNA is bound to PRM1 or PRM2, while the remaining 10% maintains canonical histone-DNA interaction (17). Following histone-protamine replacement, DNA compaction achieves an almost crystalline level when inter- and intra-molecular disulfide binds are formed between PRM1 and PRM2 (18–20). Such dense packaging affords critical protection of the genomic DNA from environmental factors and offers a hydrodynamic shape for motility. However, at times, it is evident that chromatin compaction does not occur correctly. Indeed, poor chromatin compaction is observed in cohorts of infertile men (21–23) and is also associated with lower in vitro fertilization (IVF)1 outcomes (24).
Armed with this background knowledge, the IVF industry has sought to increase success rates by passing sperm ejaculates through density gradients. During this process, higher dense cells typically pass through the gradient and pellet at the bottom of the tube, whereas lower dense cells (including those with increased nuclear volume) pellet within the gradient (25). In general, higher dense cells possess better levels of motility, morphology and chromatin compaction (26) and as such, would be the same ones responsible for natural fertility.
In previous studies, we have compared the proteomic composition of high- and low-dense spermatozoa and demonstrated protein changes occur between these two cell populations (27). Of interest, many proteins found to be in higher or lower abundance within poor quality cells were “common” across different donors. Although male-factor infertility is often suggested to have a very heterogeneous etiology, our dataset suggested the opposite and potentially that the mechanisms leading to defective sperm formation may be more common than we previously understood (27). In this analysis, which was performed on intact spermatozoa, we noted that very few nuclear proteins were identified. Therefore, to understand if the nuclear content of poor-quality spermatozoa is also impacted during spermatogenesis, we decided to perform subcellular fractionation of the spermatozoa. This approach allowed us to then compare the nuclear protein composition between high- and low-quality spermatozoa and investigate their association with sperm morphology (28). In our quest to identify protein biomarkers that would better predict the fertility status of men, we found that nuclear protein retention of proteins, including Topoisomerase 2A (TOP2A), may be a hallmark of low-quality sperm cells. Such markers could be added to a panel of proteins for better diagnosis of men's fertility status in the future, rather than relying on subjective measurements of sperm morphology.
EXPERIMENTAL PROCEDURES
All chemicals were purchased from Sigma-Aldrich at the highest research grade, with the exception of albumin and ammonium persulphate (Research Organics, Cleveland, OH), Percoll (GE Healthcare, Castle Hill, Australia), HEPES (Gibco, Invitrogen Australia, Melbourne, Australia) and 10 × Ham's F-10 (MP Biomedical, Seven Hills, NSW, Australia). d-glucose, sodium hydrogen carbonate, sodium chloride, potassium chloride, calcium chloride, potassium orthophosphate and magnesium sulfite were all AnalaR grade. Chloroform, methanol and formaldehyde were purchased from Fronine (Riverstone, Australia) at the highest purity available. Ultrapure water was from Fluka (Castle Hill, Australia), Micrococcal Nuclease (MNase) and 10× MNase buffer were from NEB biolabs (Arundel, Australia), Tris from ICN biochemicals (Castle Hill, Australia), precast-SDS gels from BioRad (Gladsville, NSW, Australia) and Complete mini protease inhibitor from Roche (Mannheim, Germany).
Preparation of Human Spermatozoa and Ethics
Institutional and State Government ethical approval was secured for the use of human semen samples in this research program. The work was carried out in accordance with The Code of Ethics of the World Medical Association (Declaration of Helsinki) for experiments involving humans. The study population was comprised of donors (n = 8) aged from 25 to 70 years and free of any detectable organic disease. None of the men reported any obvious infertility-impairment defects such as varicocele. The semen samples were produced by masturbation after a minimum of 3 days of abstinence. Samples were allowed to stand at 37 °C for at least 30 min for liquefaction to occur before being processed. Sperm numbers, morphology and total motility were assessed, according to the WHO criteria for fertility (29). Semen samples were then processed over a three-tier Percoll density gradient. Working solution was made by diluting Percoll with Biggers, Whitten & Whittingham (BBW) solution in a 9:1 ratio. A 2 ml aliquot of working solution (considered 100% Percoll) was placed in a 15 ml falcon tube, and overlaid with 2 ml of 60% Percoll, followed by 2 ml of 30% Percoll. The 60 and 30% Percoll solutions were obtained by diluting the working solution with BWW. Semen samples were gently layered over the gradients. Samples were then centrifuged at 500 × g for 30 min, and both the spermatozoa at the bottom of the 100% Percoll solution (high quality cells) and at the interface between the 100 and 60% layers (low quality cells), were collected into separate tubes. Cells were then washed with 5 ml BBW and centrifuged at 500 × g for 15 min. Pellets were resuspended in 1 ml BWW and cell counts were performed. 10 μl of anti-CD45-coated dynabeads (Thermofisher, 11153D) were washed 3 times in BWW and added to the sample to ensure that no contaminating leukocytes were present and placed on a rotator at room temperature for 30 mins (1000 × g, 30 °C). Dynabeads were removed using a magnet, and the cells were pelleted at 500 × g for 3 min and washed twice with BWW.
Sperm Swelling and DAPI Staining
Approximately 0.5 × 106 spermatozoa were resuspended in 150 μl of buffer 3 (50 mm sodium borate, 1% (w/v) SDS and 10 mm EDTA; pH 9.0) for 50 mins. The reaction was quenched by the addition of an equal volume of 4% paraformaldehyde. Cells were air dried onto glass slides and stained by overlaying ∼50 μl of 10 mm DAPI (4′,6-diamidino-2-phenylindole). Cells were imaged using fluorescence microscopy.
Image J Analysis
Fluorescent images were converted to 8-bit and the threshold set to create a binary image. The plugin 'Analyze particles' was used to measure the area of fluorescence, excluding areas <50 pixels and with the setting 'Exclude edges' selected. At least 100 cells were imaged and compared between groups using students t test (p < 0.05).
Sperm Nuclei Isolation
A total of 30 × 106 spermatozoa (determined using a hemocytometer) were resuspended in 750 μl of buffer 1 (1 mm phenylmethylsulfonyl fluoride (PMSF), 1% (w/v) cetyl-triethylammonium bromide (CTAB), 10 mm DTT and 50 mm Tris, pH 8.0) for 15 min on ice. Afterward, 750 μl of buffer 2 (buffer 1 without DTT) was added and then incubated for 45 min at 4 °C with gentle rotation. Cells were then centrifuged at 3000 × g for 5 min at room temperature and the cell pellet, which consisted of nuclei, was gently resuspended in 500 μl of buffer 2 and incubated for 30 min. Isolated nuclei were then gently washed twice in 500 μl and once in 300 μl of washing buffer (10 mm Tris and 1 mm PMSF; pH 8.0) using 5-min centrifugations at 3000 × g and 4 °C.
Nuclei Precipitation and Peptide Generation
Nuclei were resuspended in 450 μl of tris-buffered saline, supplemented with 10 mm DTT, and incubated for 30 min at 37 °C. An aliquot of 50 μl of 10x MNase buffer and 0.5 μl (1,000 U) of MNase were added, and samples were incubated for a further 30 min at 37 °C. Immediately following MNase digestion, one volume of chloroform and two volumes of methanol were added to two volumes of sample, and the mixture vortexed vigorously. This sample was centrifuged (10,000 × g, 2 min) and, after phase separation, the upper phase was removed. Three volumes of methanol were added to the remaining lower phase and the mixture gently inverted twice. The solution was then centrifuged (10,000 × g, 15 min) to pellet the precipitated protein. The supernatant was discarded, and the pellet allowed to air dry. Trypsin was added to the pellet and digested overnight at 37C. Samples were centrifuged at 17,000 × g for 20mins, transferred to sample vials and acidified.
Data Dependent Acquisition (DDA) and Library Generation
To generate the spectral library, 11 of the samples were randomly selected and fragmentation spectra acquired in DDA mode on a Sciex 6600 triple ToF mass spectrometer as previously described (26). Each file was loaded into Protein Pilot v5.0.1 (AB Sciex) and searched using the Paragon algorithm (5.0.1.0, 4895) in “thorough ID” mode, trypsin as the digestion enzyme, with no cys alkylation, acetylation emphasis as a special factor, and ID focus on biological modifications. The files were searched against the human SwissProt database (downloaded January 2017), which contained 42,324 proteins. Also include in the search database were proteins identified from the CRAPome (30). The final spectral library included 389 proteins, comprised of 1331 peptides and 8797 spectra with a global false discovery rate (FDR) of 1%. An in-depth explanation of how FDR is calculated by the paragon algorithm, the handling of multiple spectra for peptide ids and the assignment of variable modifications is explained elsewhere (31).
Sequential Window Acquisition of all Theoretical Masses (SWATH) Analysis
To obtain peptide quantitative data, the 16 samples (8 high- and 8 low-quality spermatozoa) prepared above, were again run through liquid-chromatography, coupled to a 6600 Triple ToF mass spectrometer, running a SWATH acquisition method. For the acquisition of SWATH data, the settings were as follows: a MS1 scan was performed followed by 100 m/z isolation windows with 1m/z overlap. The window sizes were determined using the Sciex Variable Window Calculator 1.0 using a DDA file generated above. The total cycle time was 2.8s. To quantify peptides, the spectral library generated above was imported into PeakView (2.2). Files acquired by SWATH-MS were imported using the SWATH-MS/MSAll Micro app (1.0). The generated library was imported, excluding modified and non-unique peptides. Retention times were aligned by manually selecting peptides present across all samples from proteins known to be in high abundance. Peptides were taken from an even distribution across the elution time. All samples were manually inspected to ensure the peptide was detected and selected correctly by the SWATH algorithm before elution time recalibration. After recalibration, it was ensured that linearity between samples was uniform. Following retention time alignment, the samples were processed to match to the spectral library within a 5 min RT window. The top six most abundant ions (excluding the precursor ion) were used for identifying, with a mass tolerance of 10 ppm for matching between the library and acquired SWATH data. The area under the curve for identified fragment ions was calculated automatically by the SWATH algorithm and summed for matched peptides (99% confidence).
Perseus
Following processing in PeakView, the raw data was exported into MarkerView (version 1.3.0.1). From here, the data was again exported as a raw text file, which allows for analysis using the freeware Perseus software (version 1.5.2.6) (www.perseus-framework.org). In Perseus, the data was log-transformed then normalized to the median of each individual sample. Sample groups were compared using a paired Welchs t test. Proteins with a fold change greater than 2.0 and p value > 0.05 were considered significant.
Immunofluorescence
Immunofluorescence was performed as described elsewhere, using a dilution of 1/50 for all primary antibodies (32). Cell morphology was scored using standard WHO guidelines. Fluorescent secondary antibodies used were: goat anti-rabbit 488 (Abcam, a1108), donkey anti-sheep- FITC (Sigma, F7634).
Immunoblotting
Whole cells were prepared as described above, or cell nuclei prepared as above (all steps prior to trypsin digestion). Proteins were solubilized in SDS extraction buffer (2% (w/v) SDS, 10% (w/v) sucrose, 114 mm Tris and 1 tablet of complete mini protease inhibitor: pH 7.5). Immunoblotting was performed as described elsewhere (27). For blots performed on whole cells, ∼5 μg of protein was used for analysis, and alpha tubulin used as a loading control. For protein extracts from nuclei, 10 × 106 nuclei were used for protein extraction, and a silver stain loaded with equal protein used for immunoblotting as the loading control. Antibodies used and their dilutions were as follows: mouse monoclonal antibody against TOP2A (Abcam, ab52934) diluted 1/1000, sheep polyclonal antibody against PDIA3 (Bio-techne, AF8219) diluted 1/1000, mouse monoclonal against alpha tubulin (Sigma, T5168) diluted 1/2000, rabbit polyclonal against histone H3 (Sigma, H0164) diluted 1/3000. Secondary anti bodies used for detection: goat anti-rabbit HRP (Sigma, DC03L), goat anti-mouse HRP (Invitrogen, A16072), rabbit anti-sheep HRP (Sigma, 402100).
Experimental Design and Statistical Rationale
For the mass spectrometric analysis of sperm nuclei, ejaculates were processed through a Percoll gradient to isolate high- and low-quality spermatozoa populations. Isolated cells were treated with a detergent to isolate nuclei and treated with MNase prior to trypsin digestion of whole nuclei to generate peptides for analysis. 11 samples were used for library generation (5 high quality/6 low quality, randomly selected), and 16 samples for SWATH data generation (high/low quality from 8 donors). 8 donors were selected as an appropriate sample size, as we reached the threshold power of detection at 6 donors, with minimal increases at 7 and none at 8. For sperm chromatin swelling, immunoblotting and immunocytochemistry experiments, high- and low-quality sperm/nuclei from 3 separate donors were used. For statistical analysis of SWATH data using Perseus software, data was log2 transformed, median normalized and a Welch's t test performed (Welch's t test was used as the data was assumed to be normally distributed, and as the welches t test perform as well at the students t test in situation of equal variance, and out performs in situations of unequal variance (33)). A p value >0.05 and >2-fold change was used to threshold changes. For chromatin swelling an unpaired 2 tailed Welch's t test was used, and for immunocytochemistry counts a paired 2 tailed student's t test was used.
RESULTS
Isolation, Purity and Biochemical Properties of Human Sperm Nuclei
We compared the composition of nuclear protein between high- and low-quality human spermatozoa, aiming to identify protein abundance changes which could be used to better classify the fertility potential of sperm samples. To achieve this, we first separated sperm samples into two populations; one with high-quality spermatozoa and the other with low-quality cells, using a discontinuous Percoll gradient protocol adapted from a previously described one (34). In brief, three different density gradients of Percoll are overlaid and the sperm samples placed on top, then centrifuged. Spermatozoa that pellet at the bottom of the tube are of higher density compared with those that pellet at the 100/60% Percoll interface (Fig. 1A). We have previously characterized these populations and shown that the majority (>90%) of the lower dense cells have distinctly poor morphology and motility (herein referred to as low quality) In contrast, cells of high density are enriched with morphologically normal, motile and highly fertile sperm (27) (herein referred to as high quality). Images of spermatozoa from each fraction are shown in Fig. 1B. Cells in the low-quality fraction often show morphological defects, including amorphous heads (Fig. 1B, upper image, black arrows), which contrast with cells from the high-quality fraction, where morphologically normal cells are enriched (Fig. 1B, lower image, white arrow) (Raw semen analysis of donors used for SWATH can be found in supplemental Table S1).
Fig. 1.
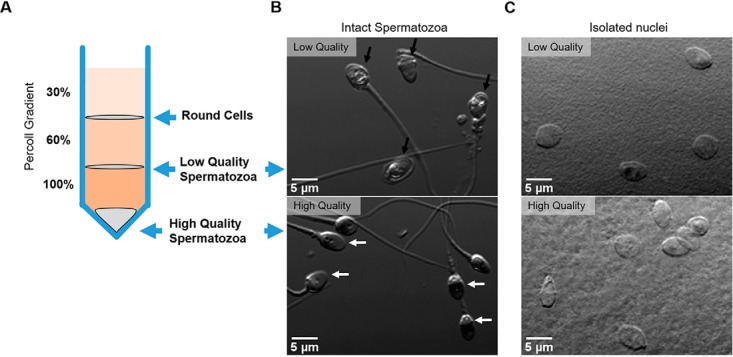
Isolation of high and low quality spermatozoa through Percoll density gradients. A, The ejaculate is placed over a discontinuous Percoll gradient as shown. High quality spermatozoa have a greater density than low quality cells. B, Phase contrast image of intact spermatozoa obtained from either the high- (lower) or low- (upper) quality Percoll fractions. Black arrows indicate amorphous heads, white arrow indicates normal morphology. C, Direct contrast image of nuclei obtained following CTAB detergent treatment from high- (lower) or low- (upper) quality human spermatozoa. Scale bar = 5 μm.
To isolate the nucleus, intact sperm cells were treated with the detergent CTAB as previously described (35). To characterize the purity of nuclei preparations, direct interference contrast (DIC) images were taken (Fig. 1C). Counting over 400 nuclei derived from either high (Fig 1B, lower), or low (Fig. 1B, upper) quality cells using direct interference contrast (DIC) demonstrated there was no evidence of fractured tails or midpiece. In addition, no round cell contamination was present suggesting the population of nuclei are highly pure, which has also been previously reported by others (35). In confirmation of this, after proteomic analysis of the isolated nucleus from both high- and low-quality sperm, the major non-nuclear sperm proteins (e.g. Tubulin, protein kinase A anchoring protein 3 and 4, and several heat shock proteins) were not apparent.
Differences in Chromatin Compaction Within the Low-quality Fractions
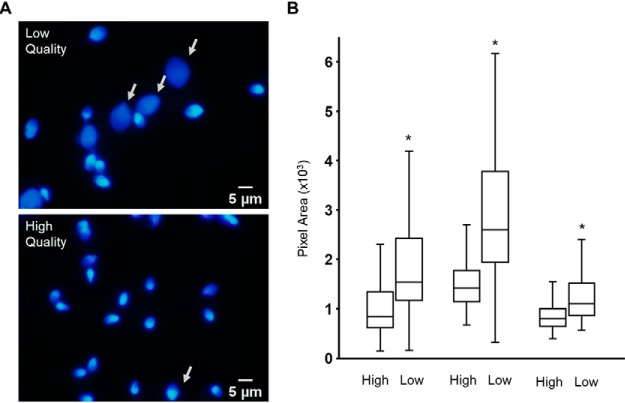
Although cells from low quality fraction showed a higher percentage of abnormal morphology than the high-quality fraction, this was not the case when we observed only the nucleus. Both populations produced very similar images with very little evidence of gross differences regarding nuclear morphology. As such, to determine if there were biochemical changes, we challenged spermatozoa with SDS/EDTA (36). Previous work has shown that when incubated for short periods of time, sperm with poorly compacted chromatin (the definition in this case, being that they look “granular” under electron microscope) decondense rapidly, showing the chromatin was not tightly compact (compact sperm nuclei are very electron dense) (36). As seen in Fig. 2A, the chromatin of some spermatozoa from the low quality population undergoes rapid decondensation, or enlargement, following SDS/EDTA treatment (Fig 2A, white arrow). To quantify this, the area of nuclei in both populations following treatment was measured using Image J. Examples from three donors are shown (Fig. 2B). In each case, there is a significant increase in the DAPI stained area from high- to low-quality cells. However, the total number of cells that showed this increase varies between donors. This data demonstrates that, despite no gross differences in nuclear morphology observed, chromatin derived from low-quality spermatozoa are more susceptible to SDS-EDTA treatment than those derived from high quality cells and are therefore likely to be less compact.
Fig. 2.
The nuclei of low quality spermatozoa are more susceptible to decondensation following treatment with SDS/EDTA. Following Percoll fractionation A, high- (upper) and low- (lower) quality spermatozoa were challenged with SDS/EDTA then stained with DAPI. Decondensed nuclei are shown (white arrow); scale bar = 5 μm. B, Box and whisker plot of DAPI fluorescent areas from >100 cells in high- and low-quality populations of sperm isolated from three donors and subjected to decondensation (*p < 0.00001).
Generation of SWATH Library
To determine quantitative differences between the nuclear composition from both high- and low-quality sperm population, we performed a proteomic analysis using Sequential Window Activation of all Theoretical spectra, or SWATH. Following trypsin digestion, 11 samples consisting of 5 high- and 6 low-quality fractions from 6 donors were randomly selected and used to generate the spectral library by running the mass spectrometer in data-dependent mode. The library generated consisted of 389 proteins, which comprised of 1331 peptides and 8797 spectra at a global false discovery rate (FDR) of 1%. In comparison, proteomic analysis of somatic nuclei preparations usually produce ∼2500 protein identifications (28). However, it needs to be recognized that to protect the DNA, spermatozoa possess a highly compact nuclei, which has been described as crystal-like with (almost) no nucleoplasm (18–20). This explains why we find reduced amounts of total protein identifications compared with somatic cell nuclei (28).
A comparison of our list of sperm nuclear proteins to the only previously published study (34) is shown in supplemental Fig. S1. We were initially concerned that only a ∼50% overlap of proteins was found between both studies. However, it appears that (1) sample purity and (2) reporting of redundant hits explain the discrepancies. Examining the list unique to De Mateo et al. (35), we could identify some proteins that are unlikely to be associated with the nucleus, suggesting that a minor contamination of the nuclear preparation may have occurred. Examples include albumin, protein kinase A anchoring protein 3 or 4, many of the mitochondrial proteins (37), fibronectin (38), ferritin (39), lactate dehydrogenase (40), prostatic acid phosphate (41) and tubulin. However, perhaps the greatest discrepancy is the way protein hits were reported. De Mateo et al. appear to have used redundancy in reporting protein hits (i.e. if one peptide matches to multiple proteins, then all proteins are reported), whereas our report uses non-redundant hits (that is, if one peptide is found to match to many proteins, but within the same protein group, and a second peptide is found that matches to only one protein isoform, then only this protein isoform is reported). There is no “correct” method given redundant reporting gives many false positives, but leaves few false negatives, whereas non-redundant reporting gives fewer false positives, but may fail to identify proteins that are present. However, the two approaches greatly exaggerate differences between the lists. For example, 15 isoforms of histone H2B are reported by De Mateo et al. (35), whereas our list only contains histone H2B2E, H2BFM and H2B1A. However, all 15 H2B isoforms reported by De Mateo et al. would not have a unique peptide (something our study requires), because they only differ by a few amino acids at the N terminus of the protein, which is rich in lysine and arginine (the same amino acid cut by trypsin). Thus, in this example alone, we only have 3 “overlapping proteins,” whereas in reality, if both methods used non-redundant reporting, the lists would be extremely close. Therefore, our criterion was more stringent and that likely explains the difference between the identifications reported by both studies. The protein identifications from both lists (De Mateo et al., and ours) are given in supplemental Table S2.
Quantitative SWATH Analysis
To quantitatively compare proteins from high- and low-quality sperm nuclei, eight men gave donations and the sperm nuclei were isolated and run on the Triple-ToF 6600 in SWATH mode. Relative quantification was performed using the freeware statistical analysis package, Perseus (www.perseus-framework.org). Data was log2 transformed to reduce sample spread and then normalized to the median value for each individual sample to account for variation. Using a paired Welchs t test, we identified significant protein changes only in proteins with the minimum criteria cut off value of p < 0.05 and a fold change no less than 2. In total, 26 proteins were significantly different between the groups, which represents 6.7% (26 of 389) of the total protein identified here. Proteins with significant regulation can be found in Table I.
Table I. Significantly regulated proteins that are more abundant in low-quality human spermatozoa following Percoll density gradient separation.
| Uniprot Accession # | Gene name | Protein Name | Log2 Fold Change | p value |
|---|---|---|---|---|
| P27487 | DPP4 | Dipeptidyl peptidase 4 | -5.20 | 6.01E-06 |
| O00468 | AGRN | Agrin | -4.82 | 5.30E-05 |
| P54107 | CRISP1 | Cysteine-rich secretory protein 1 | -4.44 | 0.004512 |
| P35052 | GPC1 | Glypican-1 | -3.78 | 0.000514 |
| P11388 | TOP2A | DNA topoisomerase 2-alpha | -3.61 | 0.000816 |
| Q7Z6Z7 | HUWE1 | E3 ubiquitin-protein ligase HUWE1 | -3.50 | 0.00303 |
| P26641 | EEF1G | Elongation factor 1-gamma | -3.26 | 0.000194 |
| P26640 | VARS | Valine–tRNA ligase | -3.25 | 0.000107 |
| P04083 | ANXA1 | Annexin A1 | -3.23 | 0.001318 |
| P02751 | FN1 | Fibronectin | -3.067 | 0.003391 |
| Q8IX12 | CCAR1 | Cell division cycle and apoptosis regulator protein 1 | -3.01 | 0.012372 |
| Q15046 | KARS | Lysine–tRNA ligase | -2.97 | 6.36E-05 |
| P15144 | ANPEP | Aminopeptidase N | -2.82 | 0.004911 |
| P29692–2 | EEF1D | Isoform 2 of Elongation factor 1-delta | -2.69 | 0.000117 |
| P26583 | HMGB2 | High mobility group protein B2 | -2.59 | 0.03805 |
| P30101 | PDIA3 | Protein disulfide-isomerase A3 | -2.58 | 0.000584 |
| P06748 | NPM1 | Nucleophosmin | -2.52 | 0.003851 |
| P24534 | EEF1B2 | Elongation factor 1-beta | -2.49 | 0.003658 |
| Q5BJF6–7 | ODF2 | Isoform 7 of Outer dense fiber protein 2 | -2.47 | 0.00268 |
| O95473 | SYNGR4 | Synaptogyrin-4 | -2.31 | 0.015401 |
| P49913 | CAMP | Cathelicidin antimicrobial peptide | -2.25 | 0.005459 |
| P30084 | ECHS1 | Enoyl-CoA hydratase, mitochondrial | -2.24 | 0.001973 |
| Q04837 | SSBP1 | Single-stranded DNA-binding protein, mitochondrial | -2.12 | 0.007349 |
| P07355–2 | ANXA2 | Isoform 2 of Annexin A2 | -2.09 | 0.035633 |
| P14923 | JUP | Junction plakoglobin | -2.08 | 0.015926 |
| P39023 | RPL3 | 60S ribosomal protein L3 | -2.01 | 0.000688 |
Protamine 2 and Histones Do Not Change Within the Low-quality Cells
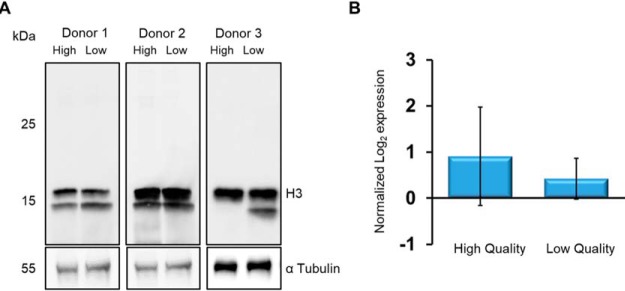
Several reports have suggested that infertile men lack protamine 2 specifically (42), or have a change in the ratio of protamine 1/protamine 2 (43, 44). Furthermore, others have suggested that within infertile spermatozoa, there are increased amounts of histone (21). However, our SWATH data indicated that neither protamine 2, nor any of the histone proteins were significantly different. To confirm this, we compared high- and low-quality sperm fractions using antibodies against histone H3. As shown (Fig. 3A), we found no change in the level of H3 when normalized to the loading control α-Tubulin. The quantitative SWATH data for comparison are plotted (Fig 3B).
Fig. 3.
Levels of histone H3 are consistent between high and low quality spermatozoa populations. A, Immunoblot of SDS extracted proteins from high- and low-quality spermatozoa isolated from three ejaculates, probed using anti-H3. Cross-reacting bands were visualized with chemiluminescence after addition of horse-radish peroxidase conjugated secondary antibody. Lower Blot: following recording of chemiluminescence, the nitrocellulose was stripped of primary antibody, re-blocked and probed with anti-α Tubulin. The chemiluminescent signal obtained was used a loading control. B, The abundance of histone H3 according to the SWATH analysis between high- and low-quality sperm cells demonstrates no significant difference between the populations (n = 8, p > 0.05).
SWATH Analysis Suggests That Nuclear Retention of Proteins Is a Hallmark of Low Dense Spermatozoa
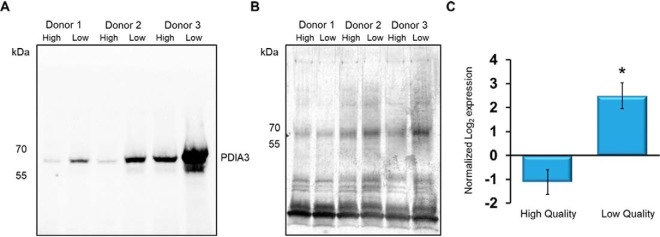
Inspection of Table I demonstrates that all proteins significantly different between groups, were exclusively more abundant within the low-quality fraction cells. From a germline developmental point of view, this data can be reasonably explained. During spermatogenesis, the sperm genome is packaged to near crystalline-state (45). During these events, there is a characteristic reduction in nuclear volume to around one-seventh the size of any somatic cell (16) that enables a more hydrodynamic head shape and protects the DNA from toxic insult (45). The processes that lead to a reduction in nuclear volume are unclear but may involve ubiquitin-dependent degradation (46) and/or manchette removal of protein and nuclear compaction (47–53). Less likely, but potentially still a factor, are tubulobulbular complexes (54) and ectoplasmic specialization removal of proteins (55). The finding that low-dense spermatozoa have more abundant proteins suggests a failure in one or more of these processes, by which nuclear volume is reduced. To validate the SWATH analysis, we performed immunoblotting. The proteins topoisomerase 2 alpha (TOP2A) and protein disulfide isomerase A3 (PDIA3) were selected because of their significant fold change and their potential importance within the nucleus of a developing spermatozoon. Nuclei from high- and low-quality human spermatozoa were taken, the proteins extracted and separated via SDS-PAGE. Following transfer, the nitrocellulose was probed with antibodies against these proteins. As shown, PDIA3 protein was more abundant within the low-quality sperm nuclei (Fig. 4A). Inter-donor variation of the changes was seen. For example, donor 1 (Fig 4A) appeared to have less PDIA3 than donor 3, despite near equal loading (Fig 4B). However, in all cases, PDIA3 was consistently higher in the nucleus of the low-quality sperm fractions. This was in accord with the proteomic data plotted as normalized expression (Fig. 4C) (note that the negative value associated with the high-quality sperm cells is because of the normalization against the median).
Fig. 4.
PDIA3 is detected in greater abundance in spermatozoa derived from the low quality fractions. A, Human sperm ejaculates were taken from individual donors and high- and low-quality fractions were isolated through Percoll density gradients. Nuclei isolated from spermatozoa were then lysed and separated via SDS-PAGE. Following transfer, the nitrocellulose was probed with primary antibodies against PDIA3. Cross-reacting bands were visualized with chemiluminescence after addition of horse-radish peroxidase conjugated secondary antibody. The approximate position of the molecular mass markers are shown. B, An equal sized aliquot of the same sample used for immunoblot was loaded into SDS page, run and total protein was detected via silver stain. C) PDIA3 abundance according to the SWATH analysis between high- and low-quality sperm cells demonstrates significantly more abundance within the latter (n = 8, *p < 0.01).
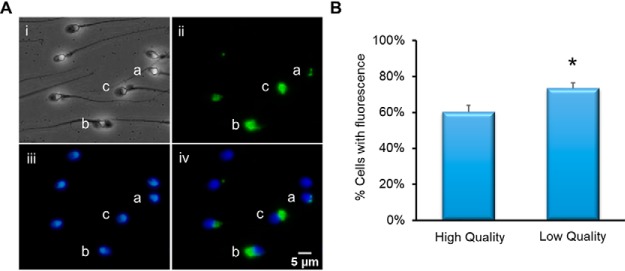
Immunocytochemical analysis of intact sperm against PDIA3 demonstrated great variation in staining (Fig. 5A). The protein was shown to be located to several different area of the cell including the redundant nuclear envelope (Fig. 5A, ii, a), perinuclear theca (Fig. 5A, ii, b) and residual cytoplasm (Fig. 5B, ii, c). Note, it is for this reason (particularly the residual cytoplasm staining), that the immunoblot in Fig. 4 was performed against isolated nuclei. We observed that a greater number of spermatozoa were positive with PDIA3 in the low-quality sperm fraction compared its high-quality counterpart. In most cases, staining was associated with poor sperm morphology, consistent with the notion that PDIA3 was higher in low quality cells. However, given the variation in staining patterns, we did not pursue this further.
Fig. 5.
Immunocytochemical analysis of PDIA3. Spermatozoa were fixed and settled onto poly-lysine slides. Following permeabilization with 0.2% Triton-X and blocking with 3% BSA, the cells were incubated with primary anti-PDIA3 antibody then secondary alexafluor-488-conjugated antibody and counterstained with DAPI. A, (i) Phase image of spermatozoa, (ii) Immunofluorescent image using anti-PDIA3 antibody. Staining can be observed in a- redundant nuclear envelope, b-perinuclear theca, c-residual cytoplasm. (iii) DAPI staining of spermatozoon DNA, (iv) composite image of DAPI and PDIA3 fluorescence. Scale bar = 5 μm. B, Comparison of cells with detected PDIA3 fluorescence between high- and low-quality cell populations (n = 3, *p = <0.01).
Top 2A Is Associated with Low-quality Sperm Heads
Immunoblotting demonstrated that TOP2A (Fig. 6A) was more abundant within the low-quality fractions, which is consistent with the quantitative SWATH data (Fig. 6B). Immunohistochemical staining demonstrated that TOP2A was localized to the redundant nuclear envelope (Fig. 7A). The redundant nuclear envelope consists of excess vestments of the nuclear envelope after compaction of the nucleus during sperm development. The proximity of TOP2A with DAPI-stained nuclei is shown (Fig. 7A, iv). The overlay demonstrates that TOP2A is part of the nuclear compartment of the sperm cell.
Fig. 6.
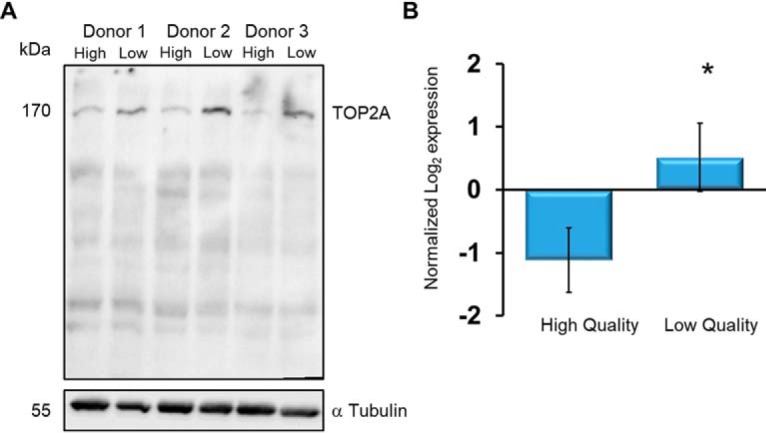
TOP2A is more abundant in spermatozoa derived from the low quality fractions. Human spermatozoa from individual donors were fractionated through Percoll density gradients. Intact cells were lysed and run into SDS-PAGE. Following transfer, the nitrocellulose was probed with primary antibodies against A, TOP2A (upper blot). Cross-reacting bands were visualized with chemiluminescence after addition of horse-radish peroxidase conjugated secondary antibody. Lower Blot: Following recording of chemiluminescent, the nitrocellulose was stripped of primary antibody, re-blocked and probed with anti-α Tubulin. B, The TOP2A abundance according to the SWATH analysis between high- and low-quality sperm cells demonstrates significantly more abundance within the latter (n = 8, *p = 0.0001).
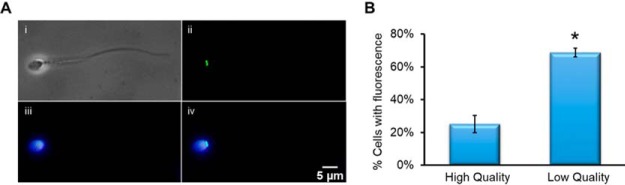
Fig. 7.
Immunofluorescent detection of TOP2A in spermatozoa demonstrates the protein is associated with amorphous heads. Spermatozoa were fixed and settled onto poly-lysine slides. Following permeabilization with 0.2% Triton-X and blocking with 3% BSA, the cells were incubated with primary anti-TOP2A antibody then secondary alexafluor-488-congucated antibody and counterstained with DAPI A, (i) Phase image of spermatozoa, (ii) Immunofluorescent image using anti-TOP2A antibody fluorescence within spermatozoa, (iii) DAPI staining of spermatozoon DNA, (iv) composite image of DAPI and TOP2A fluorescence. Scale bar = 10 μm. B, Comparison of cells with detected TOP2A fluorescence between high- and low-quality cell populations (n = 3, *p = <0.0001).
During our analysis, we noted that some sperm cells were stained with TOP2A, whereas others were not. To investigate this further, we counted the number of sperm within the high- and low-quality fractions that were positive for TOP2A and found a significant difference with more sperm positive for TOP2A in the low-quality fractions (Fig. 7B).
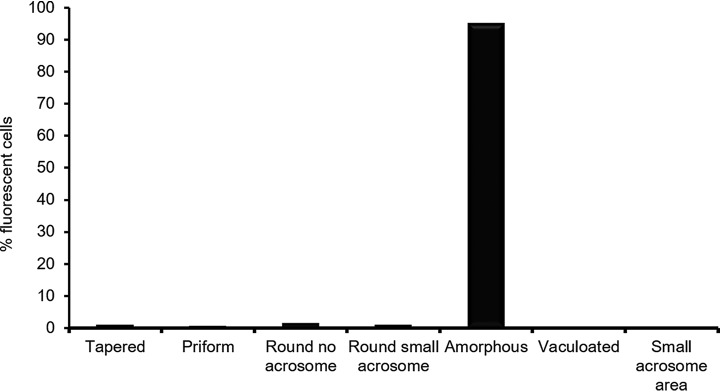
Finally, we determined the morphology of spermatozoa that were positive for TOP2A staining. To achieve this, we isolated both high- and low-quality spermatozoa and then proceeded to characterize the morphology of sperm cells positive for TOP2A in both populations. Regardless of their origin from the Percoll fraction, we found that the overwhelming majority of spermatozoa positive for TOP2A demonstrated amorphous heads (Fig. 8). As the high quality Percoll fraction is enriched for morphologically normal spermatozoa, this data explain why TOP2A is more abundant in the lower quality cells.
Fig. 8.
TOP2A is associated with amorphous sperm heads. Cells from either the high- or low-quality Percoll fractionation were stained immunostained for TOP2A. Those cells positive for the protein, were then assessed in terms of their morphology. The graph shows that combined cell counts for both high- and low-quality spermatozoa (n = 3 donors, 100 cells counted/fraction).
DISCUSSION
Male infertility is a growing medical condition. Although solutions are available through a variety of assisted reproductive technologies (ART), there are many drawbacks. First, the technology is expensive, with the average cost/cycle sitting around $12–18K. Second, there is no guarantee of success. In the United States (2015), 464 reporting clinics performed 231,936 cycles, of which only 60,778 resulted in live births (26% success rate). Third, it is highly medicalized, with the most common form of assisted conception, namely intra-cellular sperm injection (ICSI), involving hormone stimulation of the female followed by oocyte retrieval. Yet for many couples, it is evident that the referral for assisted conception was unnecessary (11). Despite these concerns, the number of couples resorting to ART increases by ∼10% each year and will continue to grow as the public becomes increasingly aware of ART options and availability (56).
The problem faced by health practitioners and in vitro fertilization clinics alike is the classification of men requiring help. Currently, diagnosis of male-factor infertility relies on 2 criteria. The first of these includes unprotected sexual intercourse without success for at least 1 year. However, ∼50% of couples that fail in the first year of trying, will go on to conceive in the second year without any medical intervention (57) and as such, further criteria is required. To this end, a semen analysis is performed that classifies the male patient as fertile or infertile based on the guidelines published by the World Health Organisation in 1980 (29). However, because of the inadequacies found in correctly predicting infertility, revised values were released in 1987, 1992, 1999 and again in 2010 (29). The semen analysis considers total sperm numbers and percentages of motile and morphologically normal cells. On the surface, a semen analysis would benefit the classification of fertile and infertile men. However, several reports shows that a semen analysis falls short of a true diagnosis (58–70).
An alternative method for the diagnosis of male-factor infertility is to build up a panel of protein biomarkers whose expression correlates with sperm fertilizing potential. The advantage of using a panel of markers is the ability to cover more aspects of “fertility” than a semen analysis alone would be capable of. For example, with men classified as “idiopathic infertile,” we have shown that Outer dense fiber 1 (ODF1) was virtually absent from their spermatozoa (29). ODF1 is a chaperone that regulates the joining of the sperm head to the tail. Despite a “normal” semen profile, sperm lacking ODF1 are quite fragile and easily decapitate over time, which may explain their infertility (32). Of interest, this current report found a second ODF family member to be more abundant in the sperm nucleus of low-quality cells, namely ODF2. Initially, this finding was surprising, as ODF2 is a well-recognized, major protein of the sperm flagella (71, 72) and not the nucleus. However, upon further inspection, we noted our proteomic platform had detected a unique peptide from the ODF2-isoform 9 transcript. This isoform (Q5BJF6–7, NP_702910) is a testis expressed transcript and arises because of an alternate start site, giving the protein a unique N-terminal. Inspection of the sequence demonstrated that the N terminus of ODF2-isoform 9 has a nuclear localization signal (73). Indeed, and in concert with this idea, ODF2 specific isoforms have been located in the nuclei of germ cells during spermatogenesis (74).
Although lack of ODF1 is a biomarker of infertility for some men, it certainly does not work for all men. As such, a panel of markers will be required to diagnose an array of male-factor infertility phenotypes. Previously, alterations to the PRM1/2 ratio in mature sperm have been often associated with poor reproductive outcomes (75, 76). In this study we find no change in either PRM2 or the detected histone peptides between our populations (Fig. 3, supplementary Table S3). Although our results are may initially appear to be at odds with previous findings, these studies are often comparisons of fertile and infertile men. When protamine ratios and levels have been Percol separated fractions from the same individual, little (77) to no change (78) is observed. Both PDIA3 and TOP2A hold some promise to identify samples with poor sperm head shape. PDIA3 has been previously localized to the rat sperm membrane and is a chaperone known to be involved in the reduction of disulfide bonds of several proteins (79). Included in this list is the major histocompatibility complex class 1 (MHC) (80). Although a somatic protein, the reductase activity of PDIA3 leads to unfolding of the MHC protein, causing it to be targeted for destruction (80). Such a mechanism of action may explain why PDIA3 was largely associated with poor quality spermatozoa. Although the very heterogeneous signals we obtained through PDIA3, staining of human spermatozoa made it difficult to investigate this phenomenon further.
TOP2A is a protein normally involved in the alteration of DNA topology. Specifically, the enzyme activity of TOP2A causes the induction of transient double strand breaks within DNA. These temporary breaks allow one DNA strand to pass through another, creating or releasing torsional stress (81). The enzyme is vastly important to spermatogenesis, as topoisomerase activity is required for successful meiosis (82). Additionally, long term treatment of mice with clinical doses of etoposide (a TOP2A inhibitor), causes chromosomal and sperm morphology aberrations (83). This is not surprising, as in addition to its role in spermatogenesis, TOP2A is involved in the histone to protamine transition (84). This is of interest, as with infertile men, either the level of protamine's or the ratio between protamine 1 and protamine 2 within the sperm population can be different compared with fertile men (85). Given this data, our initial hypothesis was that expression of TOP2A may be related to sperm chromatin compaction through aberrant histone or protamine levels. However, both SWATH and immunoblotting demonstrated this to be incorrect. Rather, we found that in virtually all cases, TOP2A was associated with poor head morphology.
The association between the etiology of poor sperm head morphology and TOP2A expression is yet to be explained. One idea is that during spermatogenesis, nuclear compaction occurs at the end of the cell elongation stage. At the same time, proteins that are not necessary for sperm function are removed. Several processes may affect this, including ubiquitin-dependent degradation and/or manchette removal of protein and nuclear compaction (47–53). With respect to the former, previous work in our lab shows no difference in proteins labeled with anti-ubiquitin antibody between high- and low-quality sperm cells (data not shown). As such, there is a lack of evidence to support this idea. However, many genetic knockout models suggest manchette formation is critical for nuclear compaction and by implication, nuclear protein removal. Examples include KIF3A (47), MNS1 (48), HOOK1 (49), RIM-BP3 (50), MEIG1 (51) SPEM1 (52) and katanin p80 (53). The manchette is a skirt-like structure that forms around the elongating sperm head (86). As the manchette moves laterally from the sperm head toward the flagella, it not only shapes the cells, but also helps compact the nucleus (86). Thus, as low-quality cells contain more abundant nuclear proteins, we speculate that this finding has largely something to do with poor manchette formation. Unfortunately, this model will be difficult to test using human testis sections as one cannot distinguish between high and low sperm quality at the stage of development in the testis.
In conclusion, we found that retention of nuclear proteins is a hallmark of poor-quality spermatozoa, as defined here. Furthermore, TOP2A is highly associated with amorphous sperm heads, and therefore a potential new biomarker for male-factor infertility in clinical practice.
DATA AVAILABILITY
All raw MS files (DDA and SWATH), and the generated libraries (containing annotated spectra, in both .group and .mzid) can be on the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) (PXD014210) via the MassIVE partner repository with the dataset identifier MSV000083954. Processed data is available in supplemental Table S3.
Supplementary Material
Footnotes
* The authors declare that they have no conflicts of interest with the contents of this article.
 This article contains supplemental Figures and Tables.
This article contains supplemental Figures and Tables.
1 The abbreviations used are:
- IVF
- in vitro fertilization
- LC-MS/MS
- liquid chromatography tandem-mass spectrometry
- MS
- mass spectrometry
- MW
- molecular weight
- ICSI
- intracellular sperm injection
- PMSF
- Phenylmethylsulfonyl fluoride
- CTAB
- 1% (w/v) cetyl-triethylammonium bromide
- DAPI
- 4′,6-diamidino-2-phenylindole
- BWW
- Biggers Whitten & Whittingham
- DDA
- data dependent acquisition
- SWATH
- sequential window acquisition of all theoretical masses.
REFERENCES
- 1. Salicioni A. M., Platt M. D., Wertheimer E. V., Arcelay E., Allaire A., Sosnik J., and Visconti P. E. (2007) Signalling pathways involved in sperm capacitation. Soc. Reprod. Fertil. Suppl. 65, 245–259 [PubMed] [Google Scholar]
- 2. Schultz N., Hamra F. K., and Garbers D. L. (2003) A multitude of genes expressed solely in meiotic or postmeiotic spermatogenic cells offers a myriad of contraceptive targets. Proc. Natl. Acad. Sci. U.S.A. 100, 12201–12206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Jung A., Eberl M., and Schill W. (2001) Improvement of semen quality by nocturnal scrotal cooling and moderate behavioural change to reduce genital heat stress in men with oligoasthenoteratozoospermia. Reproduction 121, 595–603 [DOI] [PubMed] [Google Scholar]
- 4. Selit I., Basha M., Maraee A., El-Naby S., Nazeef N., El-Mehrath R., and Mostafa T. (2013) Sperm DNA and RNA abnormalities in fertile and oligoasthenoteratozoospermic smokers. Andrologia 45, 35–39 [DOI] [PubMed] [Google Scholar]
- 5. McLachlan R. I., and de Kretser D. M. (2001) Male infertility: the case for continued research. Med. J. Aust. 174, 116–117 [DOI] [PubMed] [Google Scholar]
- 6. Cooper T. G., Noonan E., Von Eckardstein S., Auger J., Baker H., Behre H. M., Haugen T. B., Kruger T., Wang C., and Mbizvo M. T. (2010) World Health Organization reference values for human semen characteristics. Human Reproduction Update 16, 231–245 [DOI] [PubMed] [Google Scholar]
- 7. Menkveld R. (2010) Clinical significance of the low normal sperm morphology value as proposed in the fifth edition of the WHO Laboratory Manual for the Examination and Processing of Human Semen. Asian J. Androl. 12, 47–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Franken D. R., Menkveld R., Kruger T. F., Sekadde-Kigondu C., and Lombard C. (2003) Monitoring technologist reading skills in a sperm morphology quality control program. Fertility Sterility 79, 1637–1643 [DOI] [PubMed] [Google Scholar]
- 9. Cooper T. G., Neuwinger J., Bahrs S., and Nieschlag E. (1992) Internal quality control of semen analysis. Fertility Sterility 58, 172–178 [PubMed] [Google Scholar]
- 10. Horte A., Vierula M., Toppari J., and Suominen J. (2001) Reassessment of sperm morphology of archival semen smears from the period 1980–1994. Int. J. Androl. 24, 120–124 [DOI] [PubMed] [Google Scholar]
- 11. Brandes M., Hamilton C., De Bruin J., Nelen W., and Kremer J. (2009) The relative contribution of IVF to the total ongoing pregnancy rate in a subfertile cohort. Human Reproduction 25, 118–126 [DOI] [PubMed] [Google Scholar]
- 12. De Kretser D. (1997) Male infertility. Lancet 349, 787–790 [DOI] [PubMed] [Google Scholar]
- 13. Krausz C. (2011) Male infertility: pathogenesis and clinical diagnosis. Best Practice Res. Clin. Endocrinol. Metabolism 25, 271–285 [DOI] [PubMed] [Google Scholar]
- 14. Auger J., Jouannet P., and Eustache F. (2016) Another look at human sperm morphology. Hum Reprod 31, 10–23 [DOI] [PubMed] [Google Scholar]
- 15. Clermont Y. (1963) The cycle of the seminiferous epithelium in man. Am. J. Anat. 112, 35–51 [DOI] [PubMed] [Google Scholar]
- 16. Champroux A., Torres-Carreira J., Gharagozloo P., Drevet J., and Kocer A. (2016) Mammalian sperm nuclear organization: resiliencies and vulnerabilities. Basic Clin. Androl. 26, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ward W. S. (2009) Function of sperm chromatin structural elements in fertilization and development. MHR 16, 30–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Balhorn R., Corzett M., Mazrimas J., and Watkins B. (1991) Identification of bull protamine disulfides. Biochemistry 30, 175–181 [DOI] [PubMed] [Google Scholar]
- 19. Shalgi R., Seligman J., and Kosower N. S. (1989) Dynamics of the thiol status of rat spermatozoa during maturation: analysis with the fluorescent labeling agent monobromobimane1. Biol. Reproduction 40, 1037–1045 [DOI] [PubMed] [Google Scholar]
- 20. Saowaros W., and Panyim S. (1979) The formation of disulfide bonds in human protamines during sperm maturation. Experientia 35, 191–192 [DOI] [PubMed] [Google Scholar]
- 21. Foresta C., Zorzi M., Rossato M., and Varotto A. (1992) Sperm nuclear instability and staining with aniline blue: abnormal persistance of histones in spermatozoa in infertile men. Int. J. Androl. 15, 330–337 [DOI] [PubMed] [Google Scholar]
- 22. De Iuliis G. N., Thomson L. K., Mitchell L. A., Finnie J. M., Koppers A. J., Hedges A., Nixon B., and Aitken R. J. (2009) DNA damage in human spermatozoa is highly correlated with the efficiency of chromatin remodeling and the formation of 8-hydroxy-2′-deoxyguanosine, a marker of oxidative stress1. Biol. Reproduction 81, 517–524 [DOI] [PubMed] [Google Scholar]
- 23. Zandemami M., Qujeq D., Akhondi M. M., Kamali K., Raygani M., Lakpour N., Shiraz E. S., and Sadeghi M. R. (2012) Correlation of CMA3 staining with sperm quality and protamine deficiency. Lab. Med. 43, 262–267 [Google Scholar]
- 24. Franken D. R., Franken C. J., De La Guerre H., and De Villiers A. (2002) Normal sperm morphology and chromatin packaging: comparison between aniline blue and chromomycin A3 staining. Andrologia 31, 361–366 [DOI] [PubMed] [Google Scholar]
- 25. Jayaraman V., Upadhya D., Narayan P. K., and Adiga S. K. (2012) Sperm processing by swim-up and density gradient is effective in elimination of sperm with DNA damage. J. Assisted Reproduction Gen. 29, 557–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hammoud S., Liu L., and Carrell D. T. (2009) Protamine ratio and the level of histone retention in sperm selected from a density gradient preparation. Andrologia 41, 88–94 [DOI] [PubMed] [Google Scholar]
- 27. Netherton J., Hetherington L., Ogle R., Velkov T., and Baker M. (2017) Proteomics analysis of good and poor quality human sperm demonstrates several proteins are routinely aberrantly regulated. Biol. Reproduction 99, 95–08. [DOI] [PubMed] [Google Scholar]
- 28. Castillo J., Amaral A., and Oliva R. (2014) Sperm nuclear proteome and its epigenetic potential. Andrology 2, 326–338 [DOI] [PubMed] [Google Scholar]
- 29. WHO. (2010) WHO laboratory manual for the examination of human semen and sperm-cervical mucus interation., Cambridge University Press, United Kingdom [Google Scholar]
- 30. Mellacheruvu D., Wright Z., Couzens A. L., Lambert J.-P., St-Denis N. A., Li T., Miteva Y. V., Hauri S., Sardiu M. E., Low T. Y., Halim V. A., Bagshaw R. D., Hubner N. C., al-Hakim A., Bouchard A., Faubert D., Fermin D., Dunham W. H., Goudreault M., Lin Z.-Y., Badillo B. G., Pawson T., Durocher D., Coulombe B., Aebersold R., Superti-Furga G., Colinge J., Heck A. J. R., Choi H., Gstaiger M., Mohammed S., Cristea I. M., Bennett K. L., Washburn M. P., Raught B., Ewing R. M., Gingras A.-C., and Nesvizhskii A. I. (2013) The CRAPome: a contaminant repository for affinity purification–mass spectrometry data. Nat. Methods 10, 730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Shilov I. V., Seymour S. L., Patel A. A., Loboda A., Tang W. H., Keating S. P., Hunter C. L., Nuwaysir L. M., and Schaeffer D. A. (2007) The paragon algorithm, a next generation search engine that uses sequence temperature values and feature probabilities to identify peptides from tandem mass spectra. Mol. Cell. Proteomics 6, 1638–1655 [DOI] [PubMed] [Google Scholar]
- 32. Hetherington L., Schneider E. K., DeKretser D., Muller C. H., Hondermarck H., Velkov T., and Baker M. A. (2016) Deficiency in outer dense fiber 1 is a marker and potential driver of idiopathic male infertility. Mol. Cell. Proteomics 15, 3685–3693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Delacre M., Lakens D., and Leys C. (2017) Why psychologists should by default use Welch's t-test instead of Student's t-test. Int. Rev. Soc. Psychol. 30, 92–101 [Google Scholar]
- 34. de Mateo S., Castillo J., Estanyol J. M., Ballesca J. L., and Oliva R. (2011) Proteomic characterization of the human sperm nucleus. Proteomics 11, 2714–2726 [DOI] [PubMed] [Google Scholar]
- 35. de Mateo S., Castillo J., Estanyol J. M., Ballescà J. L., and Oliva R. (2011) Proteomic characterization of the human sperm nucleus. Proteomics 11, 2714–2726 [DOI] [PubMed] [Google Scholar]
- 36. Bedford J., Bent M., and Calvin H. (1973) Variations in the structural character and stability of the nuclear chromatin in morphologically normal human spermatozoa. Reproduction 33, 19–29 [DOI] [PubMed] [Google Scholar]
- 37. Aitken R. J., and Baker M. A. (2008) The role of proteomics in understanding sperm cell biology. Int. J. Androl. 31, 295–302 [DOI] [PubMed] [Google Scholar]
- 38. Fusi F., and Bronson R. (1992) Sperm surface fibronectin expression following capacitation. J. Androl. 13, 28–35 [PubMed] [Google Scholar]
- 39. Kwenang A., Kroos M., Koster J., and Van Eijk H. (1987) Iron, ferritin and copper in seminal plasma. Human Reproduction 2, 387–388 [DOI] [PubMed] [Google Scholar]
- 40. Goldberg E., and Hawtrey C. (1967) The ontogeny of sperm specific lactate dehydrogenase in mice. J. Exp. Zool. 164, 309–316 [DOI] [PubMed] [Google Scholar]
- 41. Yam L. T. (1974) Clinical significance of the human acid phosphatases: a review. Am. J. Med. 56, 604–616 [DOI] [PubMed] [Google Scholar]
- 42. Mohammad H. N.-E., Mohammad S., Shahnaz R., Maryam A., Shahla R., Fariba M., and Mohammad M. (2005) Effect of sperm DNA damage and sperm protamine deficiency on fertilization and embryo development post-ICSI. Reproductive Biomed. Online 11, 198–205 [DOI] [PubMed] [Google Scholar]
- 43. Balhorn R., Reed S., and Tanphaichitr N. (1988) Aberrant protamine 1/protamine 2 ratios in sperm of infertile human males. Experientia 44, 52–55 [DOI] [PubMed] [Google Scholar]
- 44. Aoki V. W., Moskovtsev S. I., Willis J., Liu L., Mullen J. B. M., and Carrell D. T. (2005) DNA integrity is compromised in protamine-deficient human sperm. J. Androl. 26, 741–748 [DOI] [PubMed] [Google Scholar]
- 45. Balhorn R. (2007) The protamine family of sperm nuclear proteins. Genome Biol. 8, 227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Nakamura N. (2013) Ubiquitination regulates the morphogenesis and function of sperm organelles. Cells 2, 732–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lehti M. S., Kotaja N., and Sironen A. (2013) KIF3A is essential for sperm tail formation and manchette function. Mol. Cell. Endocrinol. 377, 44–55 [DOI] [PubMed] [Google Scholar]
- 48. Zhou J., Yang F., Leu N. A., and Wang P. J. (2012) MNS1 is essential for spermiogenesis and motile ciliary functions in mice. PLoS Genetics 8, e1002516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Mendoza-Lujambio I., Burfeind P., Dixkens C., Meinhardt A., Hoyer-Fender S., Engel W., and Neesen J. (2002) The Hook1 gene is non-functional in the abnormal spermatozoon head shape (azh) mutant mouse. Human Mol. Gen. 11, 1647–1658 [DOI] [PubMed] [Google Scholar]
- 50. Zhou J., Du Y.-R., Qin W.-H., Hu Y.-G., Huang Y.-N., Bao L., Han D., Mansouri A., and Xu G.-L. (2009) RIM-BP3 is a manchette-associated protein essential for spermiogenesis. Development 136, 373–382 [DOI] [PubMed] [Google Scholar]
- 51. Zhang Z., Shen X., Gude D. R., Wilkinson B. M., Justice M. J., Flickinger C. J., Herr J. C., Eddy E. M., and Strauss J. F. (2009) MEIG1 is essential for spermiogenesis in mice. Proc. Natl. Acad. Sci. U.S.A. 106, 17055–17060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zheng H., Stratton C. J., Morozumi K., Jin J., Yanagimachi R., and Yan W. (2007) Lack of Spem1 causes aberrant cytoplasm removal, sperm deformation, and male infertility. Proc. Natl. Acad. Sci. U.S.A. 104, 6852–6857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. O'Donnell L., Rhodes D., Smith S. J., Merriner D. J., Clark B. J., Borg C., Whittle B., O'Connor A. E., Smith L. B., and McNally F. J. (2012) An essential role for katanin p80 and microtubule severing in male gamete production. PLoS Genetics 8, e1002698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Russell L. D. (1979) Spermatid-Sertoli tubulobulbar complexes as devices for elimination of cytoplasm from the head region of late spermatids of the rat. Anatomical Record 194, 233–246 [DOI] [PubMed] [Google Scholar]
- 55. Denison F. C., Smith L. B., Muckett P. J., O'Hara L., Carling D., and Woods A. (2011) LKB1 is an essential regulator of spermatozoa release during spermiation in the mammalian testis. PLoS ONE 6, e28306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Schultz R. M., and Williams C. J. (2002) The science of ART. Science 296, 2188–2190 [DOI] [PubMed] [Google Scholar]
- 57. Snick H., Snick T., Evers J., and Collins J. (1997) The spontaneous pregnancy prognosis in untreated subfertile couples: the Walcheren primary care study. Human Reproduction 12, 1582–1588 [DOI] [PubMed] [Google Scholar]
- 58. Aston K. I., Uren P. J., Jenkins T. G., Horsager A., Cairns B. R., Smith A. D., and Carrell D. T. (2015) Aberrant sperm DNA methylation predicts male fertility status and embryo quality. Fertil. Steril. 104, 1388–1397 e1385 [DOI] [PubMed] [Google Scholar]
- 59. Esteves S. C., Zini A., Aziz N., Alvarez J. G., Sabanegh E. S., and Agarwal A. (2012) Critical appraisal of World Health Organization's new reference values for human semen characteristics and effect on diagnosis and treatment of subfertile men. Urology 79, 16–22 [DOI] [PubMed] [Google Scholar]
- 60. David G., Jouannet P., Martin-Boyce A., Spira A., and Schwartz D. (1979) Sperm counts in fertile and infertile men. Fertil. Steril. 31, 453–455 [PubMed] [Google Scholar]
- 61. MacLeod J., and Wang Y. (1979) Male fertility potential in terms of semen quality: a review of the past, a study of the present. Fertil. Steril. 31, 103–116 [DOI] [PubMed] [Google Scholar]
- 62. van Zyl J. A., Menkveld R., van Kotze T. J., Retief A. E., and van Niekerk W. A. (1975) Oligozoospermia: a seven-year survey of the incidence, chromosomal aberrations, treatment and pregnancy rate. Int. J. Fertil. 20, 129–132 [PubMed] [Google Scholar]
- 63. Smith K. D., Rodriguez-Rigau L. J., and Steinberger E. (1977) Relation between indices of semen analysis and pregnancy rate in infertile couples. Fertil. Steril. 28, 1314–1319 [DOI] [PubMed] [Google Scholar]
- 64. Eliasson R. (2010) Semen analysis with regard to sperm number, sperm morphology and functional aspects. Asian J. Androl. 12, 26–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Papillon-Smith J., Baker S. E., Agbo C., and Dahan M. H. (2015) Pregnancy rates with intrauterine insemination: comparing 1999 and 2010 World Health Organization semen analysis norms. Reprod. Biomed. Online 30, 392–400 [DOI] [PubMed] [Google Scholar]
- 66. Sanchez V., Wistuba J., and Mallidis C. (2013) Semen analysis: update on clinical value, current needs and future perspectives. Reproduction 146, R249–258 [DOI] [PubMed] [Google Scholar]
- 67. Wang C., and Swerdloff R. S. (2014) Limitations of semen analysis as a test of male fertility and anticipated needs from newer tests. Fertil. Steril. 102, 1502–1507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Wang Y., Yang J., Jia Y., Xiong C., Meng T., Guan H., Xia W., Ding M., and Yuchi M. (2014) Variability in the morphologic assessment of human sperm: use of the strict criteria recommended by the World Health Organization in 2010. Fertil. Steril. 101, 945–949 [DOI] [PubMed] [Google Scholar]
- 69. Menkveld R., Wong W. Y., Lombard C. J., Wetzels A. M., Thomas C. M., Merkus H. M., and Steegers-Theunissen R. P. (2001) Semen parameters, including WHO and strict criteria morphology, in a fertile and subfertile population: an effort towards standardization of in-vivo thresholds. Hum. Reprod. 16, 1165–1171 [DOI] [PubMed] [Google Scholar]
- 70. Ombelet W., Wouters E., Boels L., Cox A., Janssen M., Spiessens C., Vereecken A., Bosmans E., and Steeno O. (1997) Sperm morphology assessment: diagnostic potential and comparative analysis of strict or WHO criteria in a fertile and a subfertile population. Int. J. Androl. 20, 367–372 [DOI] [PubMed] [Google Scholar]
- 71. Chen J. V., and Megraw T. L. (2013) Cenexin1 and Odf2: splice variants with diverged cilium functions. Cell Cycle 12, 869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Soung N. K., Kang Y. H., Kim K., Kamijo K., Yoon H., Seong Y. S., Kuo Y. L., Miki T., Kim S. R., Kuriyama R., Giam C. Z., Ahn C. H., and Lee K. S. (2006) Requirement of hCenexin for proper mitotic functions of polo-like kinase 1 at the centrosomes. Mol. Cell. Biol. 26, 8316–8335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Ba A. N. N., Pogoutse A., Provart N., and Moses A. M. (2009) NLStradamus: a simple Hidden Markov Model for nuclear localization signal prediction. BMC Bioinformatics 10, 202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Rivkin E., Tres L. L., and Kierszenbaum A. L. (2008) Genomic origin, processing and developmental expression of testicular outer dense fiber 2 (ODF2) transcripts and a novel nucleolar localization of ODF2 protein. Mol. Reprod. Dev. 75, 1591–1606 [DOI] [PubMed] [Google Scholar]
- 75. Oliva R. (2006) Protamines and male infertility. Hum. Reprod. Update 12, 417–435 [DOI] [PubMed] [Google Scholar]
- 76. Nanassy L., Liu L., Griffin J., and Carrell D. (2011) The clinical utility of the protamine 1/protamine 2 ratio in sperm. Protein Pept Lett. 18, 72–777 [DOI] [PubMed] [Google Scholar]
- 77. Castillo J., Simon L., de Mateo S., Lewis S., and Oliva R. (2011) Protamine/DNA ratios and DNA damage in native and density gradient centrifuged sperm from infertile patients. J. Androl. 32, 324–332 [DOI] [PubMed] [Google Scholar]
- 78. Mengual L., Ballescá J. L., Ascaso C., and Oliva R. (2003) Marked differences in protamine content and P1/P2 ratios in sperm cells from percoll fractions between patients and controls. J. Androl. 24, 438–447 [DOI] [PubMed] [Google Scholar]
- 79. Frickel E.-M., Frei P., Bouvier M., Stafford W. F., Helenius A., Glockshuber R., and Ellgaard L. (2004) ERp57 is a multifunctional thiol-disulfide oxidoreductase. J. Biol. Chem. 279, 18277–18287 [DOI] [PubMed] [Google Scholar]
- 80. Antoniou A. N., Ford S., Alphey M., Osborne A., Elliott T., and Powis S. J. (2002) The oxidoreductase ERp57 efficiently reduces partially folded in preference to fully folded MHC class I molecules. EMBO J. 21, 2655–2663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Champoux J. J. (2001) DNA topoisomerases: structure, function, and mechanism. Annu. Rev. Biochem. 70, 369–413 [DOI] [PubMed] [Google Scholar]
- 82. Cobb J., Reddy R. K., Park C., and Handel M. A. (1997) Analysis of expression and function of topoisomerase I and II during meiosis in male mice. Mol. Reproduction Develop. 46, 489–498 [DOI] [PubMed] [Google Scholar]
- 83. Marchetti F., Pearson F. S., Bishop J. B., and Wyrobek A. J. (2005) Etoposide induces chromosomal abnormalities in mouse spermatocytes and stem cell spermatogonia. Human Reproduction 21, 888–895 [DOI] [PubMed] [Google Scholar]
- 84. Cobb J., Miyaike M., Kikuchi A., and Handel M. A. (1999) Meiotic events at the centromeric heterochromatin: histone H3 phosphorylation, topoisomerase IIα localization and chromosome condensation. Chromosoma 108, 412–425 [DOI] [PubMed] [Google Scholar]
- 85. Bianchi P., Manicardi G., Bizzaro D., Bianchi U., and Sakkas D. (1993) Effect of deoxyribonucleic acid protamination on fluorochrome staining and in situ nick-translation of murine and human mature spermatozoa. Biol. Reproduction 49, 1083–1088 [DOI] [PubMed] [Google Scholar]
- 86. Kierszenbaum A. L., Rivkin E., and Tres L. L. (2007) Molecular biology of sperm head shaping. Soc. Reproduction Fertility Suppl. 65, 33. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw MS files (DDA and SWATH), and the generated libraries (containing annotated spectra, in both .group and .mzid) can be on the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) (PXD014210) via the MassIVE partner repository with the dataset identifier MSV000083954. Processed data is available in supplemental Table S3.