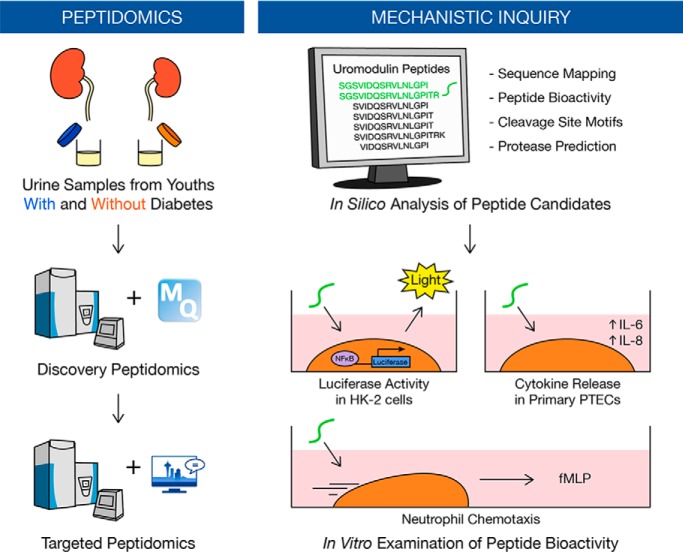
Dysregulated proteolysis is linked to early compensatory and maladaptive changes in the diabetic kidney. To elucidate the early kidney response to chronic hyperglycemia, we identified a urinary signature of uromodulin peptides associated with early type 1 diabetes before clinical manifestations of kidney disease and demonstrated novel bioactivity of two peptides in vitro using NFκB luciferase activity and neutrophil chemotaxis assays. These findings lay the groundwork for future validation studies.
Keywords: Peptidomics, diabetes, urine analysis, mass spectrometry, bioinformatics
Graphical Abstract
Highlights
Urinary peptide profiling of youths with type 1 diabetes before clinical injury.
Internal validation of uromodulin peptides by parallel reaction monitoring.
Discovery of novel bioactivity of uromodulin peptides in vitro.
In silico prediction of proteases involved in uromodulin processing.
Abstract
Chronic hyperglycemia is known to disrupt the proteolytic milieu, initiating compensatory and maladaptive pathways in the diabetic kidney. Such changes in intrarenal proteolysis are captured by the urinary peptidome. To elucidate the early kidney response to chronic hyperglycemia, we conducted a peptidomic investigation into urines from otherwise healthy youths with type 1 diabetes and their non-diabetic peers using unbiased and targeted mass spectrometry-based techniques. This cross-sectional study included two separate cohorts for the discovery (n = 30) and internal validation (n = 30) of differential peptide excretion. Peptide bioactivity was predicted using PeptideRanker and subsequently verified in vitro. Proteasix and the Nephroseq database were used to identify putative proteases responsible for peptide generation and examine their expression in diabetic nephropathy. A total of 6550 urinary peptides were identified in the discovery analysis. We further examined the subset of 162 peptides, which were quantified across all thirty samples. Of the 15 differentially excreted peptides (p < 0.05), seven derived from a C-terminal region (589SGSVIDQSRVLNLGPITRK607) of uromodulin, a kidney-specific protein. Increased excretion of five uromodulin peptides was replicated in the validation cohort using parallel reaction monitoring (p < 0.05). One of the validated peptides (SGSVIDQSRVLNLGPI) activated NFκB and AP-1 signaling, stimulated cytokine release, and enhanced neutrophil migration in vitro. In silico analyses highlighted several potential proteases such as hepsin, meprin A, and cathepsin B to be responsible for generating these peptides. In summary, we identified a urinary signature of uromodulin peptides associated with early type 1 diabetes before clinical manifestations of kidney disease and discovered novel bioactivity of uromodulin peptides in vitro. Our present findings lay the groundwork for future studies to validate peptide excretion in larger and broader populations, to investigate the role of bioactive uromodulin peptides in high glucose conditions, and to examine proteases that cleave uromodulin.
Diabetes mellitus is a metabolic disorder characterized by chronic hyperglycemia. It arises from either an insufficient production of insulin as a result of an autoimmune attack on beta cells of the pancreas (referred to as type 1 diabetes) or insulin resistance as a result of decreased sensitivity (referred to as type 2 diabetes) (1). Type 1 diabetes typically emerges in childhood or adolescence; whereas type 2 diabetes typically appears later in life and is often complicated by hypertension, obesity, or cardiovascular disease at time of diagnosis (2). Despite differences in etiology, both types predispose individuals to serious complications that threaten survival and quality of life (3).
A common microvascular complication is diabetic kidney disease. It is in fact the leading cause of end-stage renal disease, accounting for 30–50% of incident cases in Canada and worldwide (4, 5). The current standard of care is geared toward managing diabetes and delaying the onset of renal disease. Landmark clinical trials in diabetes, namely the Diabetes Control and Complications Trial (DCCT)1 and the Epidemiology of Diabetes Intervention and Complications (EDIC), have demonstrated that intensive glycemic control significantly reduces the risk of kidney disease (6); however, maintaining “normal” blood glucose levels may not be attainable for most individuals with diabetes. The search for better personalized and targeted therapies has been under way for decades; however, the greatest hurdle is our limited understanding of the early mechanisms responsible for initiating diabetic kidney disease.
Omics approaches have previously been employed to fill these gaps in knowledge. In a review of the existing literature on urinary peptidomics in diabetes, we noted that the primary focus had been placed on later stages of diabetic kidney disease, in which individuals with diabetes already present with some degree of impaired renal function such as microalbuminuria (7). Furthermore, studies rarely extended their interpretations of differentially excreted peptides beyond their potential roles as biomarkers. Applying a mechanistic approach to urinary peptidomics may provide new insights into the proteolytic milieu of the diabetic kidney. This approach combines the global study of naturally occurring peptides with the pursuit of elucidating mechanisms underlying normal and disease states (8, 9). Peptides represent an intriguing class of analytes because they are generated through the action of proteases on protein precursors. Akin to a chemical reaction, changes in peptide (product) levels reflect changes in protein (substrate) expression, proteolytic (enzyme) activity, or both. Furthermore, proteolysis of zymogens, prohormones, and other precursors is often required for protein maturation, activation, and regulation. Such is the case for angiotensinogen, which has no known bioactivity until it is cleaved by renin and subsequently angiotensin-converting enzyme to form the peptide hormone angiotensin (Ang) II: the principal regulator of blood pressure. As a result, peptides not only reflect proteolytic activity, but they may also act as potent bioactive mediators of disease.
Dysregulation of proteolytic activity has previously been linked to early compensatory and maladaptive changes in the diabetic kidney (10–12); although it is not fully understood. In this study, we aimed to elucidate the early kidney response to chronic hyperglycemia using urinary peptidomics. To do so, we carefully selected a population of otherwise healthy youths with type 1 diabetes and their non-diabetic peers as healthy controls, which enabled us to study the effect of chronic hyperglycemia in the diabetic kidney without potential confounding effects of comorbidities and medications. We identified a urinary signature of 15 differentially excreted peptides in the discovery cohort of thirty youths. Targeted methods using parallel reaction monitoring were developed and used to validate differential peptide excretion in a second cohort. We then employed in silico analyses to predict peptide bioactivity and to identify the potential proteases responsible for generating the peptides. Lastly, we demonstrated, for the first time to our knowledge, that a uromodulin peptide, whose excretion was elevated in those with diabetes, exhibited pro-inflammatory bioactivity.
EXPERIMENTAL PROCEDURES
Experimental Design and Statistical Rationale
This cross-sectional study included a discovery cohort and an internal validation cohort. Each cohort included 30 otherwise healthy youths with and without type 1 diabetes; each individual served as a biological replicate.
For the discovery phase, we performed power calculations to determine the appropriate total cohort size (n = 30) required to achieve a study power of 80% using the independent two-sample t test and Benjamini-Hochberg (BH) multiple testing correction with a false detection rate (FDR) of 0.0001 and an effect size d of 2. We thus obtained urine samples from 15 youths with type 1 diabetes and 15 healthy, non-diabetic, age- and sex-matched peers for the discovery phase.
For the validation phase, we performed power calculations based on the observed effect sizes from the discovery phase. To achieve a study power of 80% and an FDR of 0.05, we determined that the minimum number of samples to validate the top peptide candidates as determined by the BH adjustment was 9 per group (supplemental Table S1). To match the discovery analysis, we collected thirty urine samples for the validation cohort.
A total of 60 s-morning midstream urine samples were collected from 60 youths. All power calculations were performed with G*Power software.
Study Population
All sixty youths were under 19 years of age; had no history of hypertension, persistent microalbuminuria, renal disease, macrovascular disease, and chronic inflammatory disease; and had never used corticosteroid, anti-inflammatory, or anti-hypertensive drugs. In this study, youths with type 1 diabetes mellitus were in the earliest and uncomplicated stage of the natural history of diabetic kidney disease. Youths with type 1 diabetes were recruited and initially screened from multiple diabetes clinics in the Greater Toronto Area (e.g. Hospital for Sick Children, Markham-Stouffville Hospital, Credit Valley Hospital, and Charles H. Best Centre); whereas youths without diabetes were either family members of youths with type 1 diabetes or were healthy volunteers recruited at the Hospital for Sick Children and Toronto General Hospital.
Relevant clinical characteristics such as age, sex, glycated hemoglobin (HbA1c), blood pressure, body mass index (BMI), urinary albumin/creatinine ratios (ACR), and estimated glomerular filtration rate (eGFR) were compared between groups. We converted blood pressure and BMI into z-scores to account for age and sex differences in youths (13). eGFR was determined using the Larsson's formula (eGFR = 77.24 × CysC−1.2623). For the discovery cohort, clinical characteristics were measured at the time of study enrolment for both groups (Table I) and at time of urine collection for youths with diabetes only (supplemental Table S2). The mean times between the measurement of clinical characteristics and the urine collection was 2.7 ± 0.5 years and 2.5 ± 0.7 years for youths with diabetes and without diabetes, respectively. Groups were matched by age at time of urine collection and sex. For the validation cohort, we obtained clinical characteristics at or near the time of urine collection (Table II). Groups were matched based on age. The research ethics boards at the Hospital for Sick Children and Mount Sinai Hospital approved this study.
Table I. Clinical characteristics of the discovery cohort at time of study enrollment. Data are presented as mean ± standard deviation, except for sex (frequency). p values are shown between youths with and without type 1 diabetes (T1D).
| Clinical Characteristics | Youths without T1D (n = 15) | Youths with T1D (n = 15) | p |
|---|---|---|---|
| Age (years) | 13.3 ± 1.7 | 13.1 ± 1.5 | 0.7403 |
| Sex (females/males) | 6/9 | 6/9 | 1.0000 |
| Glycated hemoglobin, HbA1c (%) | 5.4 ± 0.3 | 8.5 ± 1.1 | 0.0000 |
| Diabetes duration (years) | 0.0 ± 0.0 | 7.1 ± 2.7 | 0.0000 |
| Body mass index, BMI (kg/m2) | 20.4 ± 2.1 | 21.4 ± 5.0 | 0.4804 |
| z-score BMI | 0.54 ± 0.89 | 0.65 ± 1.38 | 0.8047 |
| Systolic blood pressure, SBP (mmHg) | 109 ± 11 | 114 ± 10 | 0.1952 |
| z-score SBP | 0.03 ± 0.91 | 0.42 ± 0.70 | 0.2048 |
| Diastolic blood pressure, DBP (mmHg) | 63 ± 5 | 70 ± 7 | 0.0062 |
| z-score DBP | −0.01 ± 0.43 | 0.52 ± 0.66 | 0.0144 |
| Albumin/creatinine ratio, ACR (mg/mmol) | 0.5 ± 0.3 | 0.7 ± 0.6 | 0.1598 |
| Estimated glomerular filtration rate, eGFR (ml/min/1.73m2) | 118 ± 25 | 141 ± 27 | 0.0263 |
| Time between enrolment and urine collection (years) | 2.7 ± 0.5 | 2.5 ± 0.7 | 0.5899 |
Table II. Clinical characteristics of the validation cohort at time of urine collection. Data are presented as mean ± standard deviation, except for sex (frequency). p values are shown between youths with and without type 1 diabetes.
| Clinical characteristics | Youths without T1D (n = 15) | Youths with T1D (n = 15) | p |
|---|---|---|---|
| Age (years) | 16.0 ± 1.2 | 16.5 ± 1.1 | 0.2548 |
| Sex (females/males) | 5/10 | 6/9 | 0.7047 |
| Glycated hemoglobin, HbA1c (%) | 5.1 ± 0.3 | 9.1 ± 1.6 | 0.0000 |
| Diabetes duration (years) | 0.0 ± 0.0 | 10.4 ± 2.9 | 0.0000 |
| Body mass index, BMI (kg/m2) | 22.5 ± 4.8 | 23.3 ± 4.2 | 0.6781 |
| z-score BMI | 0.39 ± 1.67 | 0.64 ± 1.03 | 0.6395 |
| Systolic blood pressure, SBP (mmHg) | 120 ± 11 | 115 ± 11 | 0.2746 |
| z-score SBP | 0.56 ± 1.05 | 0.01 ± 1.05 | 0.1989 |
| Diastolic blood pressure, DBP (mmHg) | 64 ± 9 | 66 ± 8 | 0.5621 |
| z-score DBP | −0.20 ± 0.71 | −0.11 ± 0.67 | 0.7548 |
| Albumin/creatinine ratio, ACR (mg/mmol) | 0.6 ± 0.2 | 1.1 ± 1.1 | 0.1554 |
| Estimated glomerular filtration rate, eGFR (ml/min/1.73m2) | 102 ± 15 | 115 ± 24 | 0.0721 |
Collection, Handling, and Storage of Urines
We followed the Standard Protocol for Urine Collection and Storage, created by the Human Kidney and Urine Proteome Project (HKUPP) and the Human Proteome Organization (HUPO) (14). Following collection, all fresh urine samples were immediately kept at 4 °C until further processing. Urine samples were centrifuged at 1000 × g for 10 min to remove intact cells and debris. The supernatants were then aliquoted in either 15- or 50-ml Falcon tubes as well as 1.5-ml Eppendorf tubes and stored at −80 °C. This initial processing step was completed within 3 h of urine collection to obviate the need for urine preservatives and protease inhibitors. Samples were de-identified and randomized so that investigators were blinded to experimental groups during sample processing.
Discovery Peptidomics
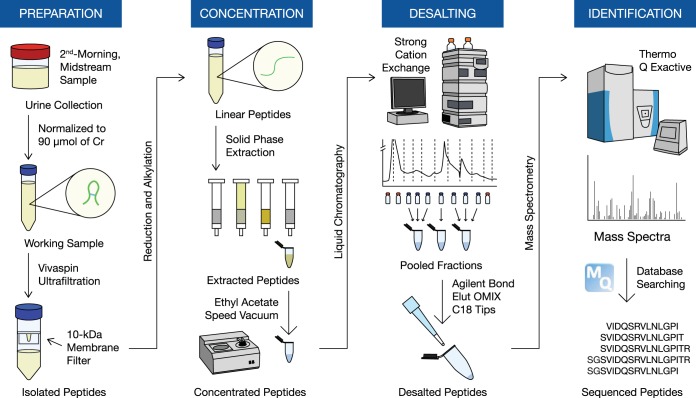
We employed previously published methods for the identification and quantification of the naturally occurring urine peptidome (15). Our workflow is summarized in Fig. 1. All 30 urine samples were processed and analyzed together in a single batch to avoid variability because of day-to-day sample preparation. Frozen samples were thawed, vortexed, and centrifuged at 1000 × g for 10 min to remove any remaining cells and debris. Sample volumes were normalized to 90 μmol of creatinine to account for degree of hydration. After adjusting the pH to 8 using ammonium bicarbonate, we used Vivaspin Centrifugal Concentrators (VivaProducts) with 10-kDa cut-off membranes to isolate urinary peptides. We added dithiothreitol to a final concentration of 2 mm and iodoacetamide to a final concentration of 4 mm to reduce and alkylate disulfide bonds. The peptides were then passed through solid-phase extraction using Oasis HLB cartridges (Waters Corporation, Milford, Massachusetts), after adjusting pH to 4 with formic acid. To remove urinary pigments, we added ethyl acetate, vortexed and centrifuged the sample, and then discarded the supernatant. The extracted peptides were speed-vacuumed until the final sample volume was less than 200 μl and subsequently stored at −20 °C until further processing.
Fig. 1.
Schematic outline of the urinary peptidomic protocol for the discovery phase.
Frozen peptides were thawed, topped off to 500 μl in strong cation exchange high-performance liquid chromatography (SCX-HPLC) running buffer (0.26 m formic acid in 5% acetonitrile), spun down at 17000 g for 1 min, and subsequently loaded onto a PolySULFOETHYL ATM column (The Nest Group, Inc., Southborough, Massachusetts) containing a hydrophilic, anionic polymer (poly-2-sulfoethyl aspartamide). We ran a 1-hour SCX-HPLC fractionation method on a linear gradient using the running buffer with incremental increases in 1 m ammonium formate as the elution buffer. We collected twelve fractions per sample at a rate of one fraction per 2 mins after the start of the elution gradient. All fractions were stored at −20 °C until the entire fractionation step for all thirty samples was completed. Overall, this fractionation step was carried out continuously for 2–3 weeks.
To reduce time per sample on the mass spectrometer, we selected, thawed, and pooled seven consecutive fractions, which covered the largest area of peptide abundance on the chromatogram. Pooled fractions were further desalted using Bond Elut OMIX C18 tips (Agilent Technologies, Santa Clara, California), eluted in 5 μl of 65% acetonitrile, and diluted with 60 μl of 0.1% formic acid in pure mass spectrometry-grade water. After desalting, we immediately loaded 40 μl of each of the three pooled fractions per sample onto a 96-well plate in individual wells, from which 18 μl from each well were injected onto a 3.3 cm C18 pre-analytical column (IntegraFrit capillary, New Objective; inner diameter of 150 μm; 5 μm bead size; Agilent Pursuit C18, Agilent Technologies) and then loaded onto a C18 resolving analytical column with dimensions 15 cm × 75 μm ID (PicoTip emitter, 8 μm tip, New Objective Agilent Pursuit C18, 3 μm bead size). These columns were operated on an EASY-nLC1000 system (Thermo Fisher Scientific, Waltham, Massachusetts), coupled to a Q Exactive Plus hybrid quadrupole-orbitrap mass spectrometer (Thermo Fisher Scientific) using a nano-electrospray ionization source. Each fraction ran on a 60-min gradient in data-dependent acquisition mode with full MS1 scan from 400 - 1500 m/z with a resolution of 70,000 and subsequent MS2 scan of the top 12 parent ions with a resolution of 17,500. Xcalibur software (v. 3.0.63; Thermo Fisher Scientific) was utilized to generate RAW files of each run. Mass spectrometry data have been deposited onto the ProteomeXchange Consortium via the PRIDE (16) partner repository with the dataset identifier PXD012210 (http://www.ebi.ac.uk/pride/archive/login).
All mass spectrometry raw data were analyzed by MaxQuant software (version 1.5.3.28) and were searched against the human Uniprot FASTA database (version July 2016 with 42,158 protein entries) using the built-in Andromeda search engine (17). To study endogenous peptides, we selected the “unspecific” digestion mode. Cysteine carbamidomethylation was the only fixed modification. Variable modifications included methionine oxidation, proline oxidation, and N-terminal acetylation with a maximum number of 5 modifications per peptide. The false discovery rate was set to 1% for both proteins and peptides with a minimum length of 6 amino acids and was determined by searching a randomized database. Potential contaminants were allowed in the search. The initial peptide tolerance was set to 20 ppm against a small “human-first-search” database. The main search peptide mass tolerance was 4.5 ppm, and the fragment mass MS/MS tolerance was set to 0.5 Da. Matching between runs was selected. Peptide intensities derived from extracted ion currents and were used as a measure of peptide quantification.
Peptidomic data analysis was done on Perseus software (version 1.5.5.3) (18). Non-human contaminants and reverse hits were manually checked and removed. We then filtered out peptides that were not quantified in all samples so that we could examine the most robust changes in peptide intensities between groups. Peptide intensities were log-transformed to approximate a normal distribution. Differential peptide intensities between groups were then determined using the independent t test (p < 0.05), followed by the BH adjustment (Q < 0.05).
Targeted Peptidomics
We initially developed targeted methods for selected reaction monitoring to validate the observed differences in urinary peptide excretion (using Tier 2 analyses). The top seven transitions were chosen for each precursor based on magnitude, order of intensity, co-elution, and peak shape. Linearity was determined for each of the detected crude heavy-labeled peptides (supplemental Fig. S1). We ultimately developed and performed a parallel reaction monitoring assay. Complete details about method development and optimization can be found in the Supplemental Methods. The master mix and list of transitions are summarized in supplemental Table S3.
Overall, sample preparation of urine was like what was described for discovery peptidomics with some notable exceptions. After thawing, urine volumes were normalized to 20 μmol of creatinine. Crude heavy-labeled peptide standards for each of the differentially excreted peptides were purchased from JPT Peptide Technologies (Berlin, Germany) and were spiked into the samples just prior to Vivaspin centrifugation. Steps involving SCX-HPLC fractionation and OMIX tip desalting were not performed. Instead, peptides were analyzed at the SickKids Proteomics, Analytics, Robotics, and Chemical Biology Centre (SPARC Biocenter in Toronto, Canada) facility, where they were loaded onto Evotips and injected in duplicate onto an Evosep One nLC system coupled to a Thermo Scientific Q Exactive HF-X hybrid quadrupole-orbitrap mass spectrometer on a 21-min unscheduled gradient. Raw files were imported into Skyline software (version 4.1). We then manually reviewed each precursor ion to ensure that all transitions were captured in the integration boundaries for relative quantification. Differential peptide excretion (deriving from peak area ratios of endogenous-to-heavy-labeled peptides) were determined using the Mann-Whitney test (p < 0.05) between groups in the validation phase.
Skyline files and raw data have been deposited in Panorama Public with a ProteomeXchange identifier PXD012389 (https://panoramaweb.org/lotso19.url).
Uromodulin Protein Excretion
Urine aliquots from the internal validation cohort were thawed to 4 °C, centrifuged at 2000 × g for 5 min, and analyzed in duplicate using enzyme-linked immunosorbent assay (ELISA). Human uromodulin ELISA (DY5144) and ancillary kits (DY008) were purchased from R&D Systems, Minneapolis, Minnesota. All plates were read at 450 and 540 nm using a Perkin Elmer 2103 EnVision Multilabel Plate Reader. Protein concentrations were extrapolated from optical densities using four-parameter logistic (4-PL) regression. Urinary measurements were normalized to urinary creatinine concentrations.
Cell Culture
Immortalized proximal tubular HK-2 cells were grown in 1:1 mix of Dulbecco's Modified Eagle Medium (DMEM) and Ham's F12 media supplemented with 10% fetal bovine serum, 10 ng/ml epidermal growth factor, 5 μg/ml transferrin, 5 μg/ml insulin, 0.05 μm hydrocortisone, 50 units/ml penicillin, and 50 μg/ml streptomycin at 37 °C with 5% CO2. Once the cells reached 50–60% confluency, cells were transfected for 16 h, serum-starved for 24 h, and then treated with peptide in fresh serum- and supplement-free DMEM/F12 for 24 h.
Human primary proximal tubular epithelial cells from a male donor were purchased from Lonza, Basel, Switzerland. The cells (at passage 6) were grown in DMEM media supplemented with 10% fetal bovine serum, 10 ng/ml epidermal growth factor, 5 μg/ml transferrin, 5 μg/ml insulin, 5 μg/ml selenium, 0.05 μm hydrocortisone, 50 units/ml penicillin, and 50 μg/ml streptomycin at 37 °C with 5% CO2. Once the cells reached 60–70% confluency, cells were serum-starved for 24 h and subsequently treated with peptide in fresh serum- and supplement-free DMEM for 24 h.
Luciferase Activity Assay
HK-2 and human primary proximal tubular epithelial cells were transfected with Renilla luciferase control reporter vector pRL-TK and a luciferase reporter for either NF-κB or AP-1. Purified peptides (70%) were purchased from JPT Peptide Technologies and were used in all in vitro experiments. Uromodulin protein (>95% purity) was purchased from Millipore Sigma, Burlington, Massachusetts (catalogue number AG733). Cells were serum-starved for 24 h and treated for 24 h with the uromodulin peptides (0.1 to 10 μm), uromodulin protein (0.2 to 20 μg/ml), angiotensin II (Ang II, 0.1 μm) or lipopolysaccharide (LPS, 0.1 μg/ml). In certain experiments, cells were pretreated with TAK-242 (0.3 μm) or gallein (10 μm) for 1 h. Reporter activities were measured using the Promega, Madison, Wisconsin dual-luciferase assay kit. The luciferase activity was normalized to the Renilla luciferase activity.
Neutrophil Chemotaxis Assay
Human primary neutrophils were isolated from peripheral blood from a healthy volunteer. Neutrophils were incubated with purified uromodulin peptide (1 μm) for 15 min at 37 °C and then placed on a 5% bovine serum albumin-coated microscope cover glass (22 × 40 mm) for 10 min at 37 °C. The cover glass was inverted onto Zigmond chambers: 100 μl of Hank's Balanced Salt Solution media in the right chamber, and 100 μl of media containing fMLP (1 μm) in the left chamber. We recorded neutrophil movement for 15 min at a rate of 0.05 frame per second using time-lapse video microscopy. Captured images were analyzed using cell-tracking Retrac software (version 2.1.01).
Cytokine Release from Kidney Tubular Cells
Human primary proximal tubular epithelial cells were serum-starved for 24 h and subsequently treated with purified uromodulin peptides (0.1 to 10 μm) or LPS (1 μg/ml) for 24 h. Conditioned media were collected, centrifuged at 2000 × g for 10 min, and kept at either 4 °C for immediate use or stored at −20 °C. Human Interleukin (IL)-6 (ab178013) and IL-8 (ab214030) SimpleStep ELISA kits were purchased from Abcam, Cambridge, UK. Conditioned media were analyzed in duplicate according to kit instructions. Plates were read as previously described.
Cytokine Measurements from Urine
Frozen urine aliquots were thawed and analyzed in duplicate by Eve Technologies (Calgary, Canada) using the Human Cytokine Array/Chemokine Array 42-Plex with IL-18 (HD42) (Millipore MILLIPLEX). The full list of analytes can be found in the supplemental Methods. The Luminex bead-based immunoassay was run on the Bio-Rad BioPlex 200 Systems. Analyte concentrations were normalized to urinary creatinine concentration as a measure of urinary excretion. We selected eight analytes a priori to characterize an inflammatory signal in urine based on the existing literature (19–22): IL-1β, IL-6, IL-8, IL-18, interferon gamma-induced protein 10 (IP-10/CXCL10), monocyte chemoattractant protein 1 (MCP-1/CCL2), macrophage inflammatory protein 1-beta (MIP-1β/CCL4), and tumor necrosis factor alpha (TNFα).
Bioinformatics
For the heatmap analysis, we converted log-transformed peptide intensities into z-scores and performed Euclidean hierarchical clustering using the R package pheatmap. PeptideRanker was used to predict bioactive peptides (23). Peptide sequences were aligned with precursor proteins using Peptide Extractor to visualize cleavage sites (9). We inferred protease activity using Proteasix (24). Structural information was extracted from the UniProtKB database. We used iceLogo to determine percent differences in amino acid proportion at each position and to visualize the consensus cleavage sites at the N- and C termini of urinary peptides (25). The MEROPS database (version 12.1, https://www.ebi.ac.uk/merops/index.shtml) was manually searched to identify proteases with specific cleavage sites and to collate cleavage site sequence logo motifs (26, 27).
Statistics
Normal distribution of each variable was examined using the Shapiro-Wilks normality test. We compared clinical characteristics between groups using the t test for continuous variables and the chi-squared test for categorical variables. Pearson correlations were performed on log-transformed urinary peptide excretion and uromodulin protein excretion. Differences in luciferase activities and protein concentrations were determined using the independent t test. Differential excretion of cytokines and chemokines was determined using the Mann-Whitney test on positive, non-zero data only. Imputation of zero-value intensities was performed with Perseus software from a normal distribution with a downshift of 1.8 and width of 0.3 standard deviations to determine fold changes for the cleavage site analysis. Plots were created with R software.
RESULTS
Urinary Peptidomic Analyses of Type 1 Diabetes
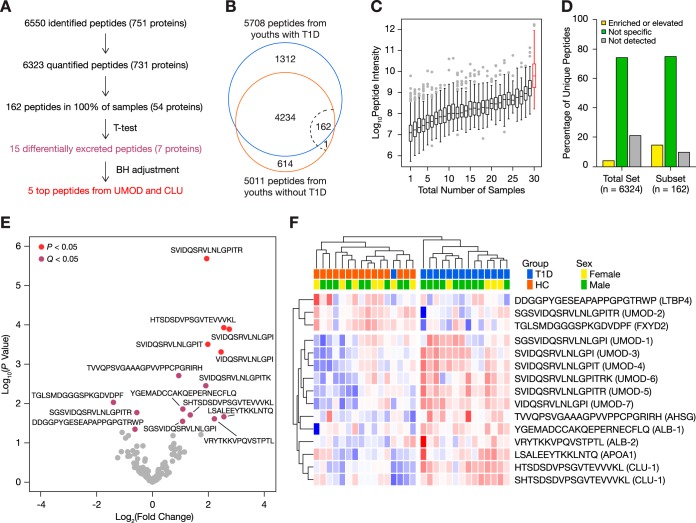
Overall, 6550 peptides deriving from 751 protein precursor groups were identified using our urinary peptidomics workflow (Fig. 2A, supplemental Tables S4 and S5). After the removal of false hits, unquantified peptides, and contaminants, our MaxQuant search provided label-free quantitation (LFQ) for 6323 peptides from 731 unique protein precursors (supplemental Tables S6 and S7): 5708 peptides were found in youths with diabetes, 5011 peptides originated from non-diabetic youths, and 4396 peptides were common to both groups (Fig. 2B). Of the 615 peptides found exclusively in the non-diabetic group, only ten peptides were found in at least ten samples—all deriving from the C-peptide region of insulin (supplemental Table S6). Notably, one C-peptide fragment (EAEDLQVGQVELGGGPGAGSLQP) was identified in all fifteen urine samples from youths without diabetes and in none from those with diabetes. On the other hand, no peptide was exclusively identified in urine from all youths with diabetes.
Fig. 2.
General characterization of the urinary peptidomes of 15 youths with type 1 diabetes and 15 healthy controls. A, Flow diagram describing the urinary peptidome. B, Venn diagram of urinary peptides. The dotted line represents the 100% cut-off, in which 162 peptides were quantified in every sample. C, Range of intensities of peptides found in one to thirty urines. Low-abundance peptides were found in relatively few samples, compared with high-abundance peptides. The range of intensities of the 162 peptides found in all 30 samples is highlighted in red. D, Kidney tissue origins of the total peptidome and the subset of 162 peptides using the Human Protein Atlas. E, Volcano plot of the 162 recurring peptides. Fifteen peptides were differentially excreted (independent two-sample Student's t test, p < 0.05, purple); however, only five survived the Benjamini-Hochberg adjustment (Q < 0.05, red). F, Heatmap representation of the 15 differentially excreted peptides with unsupervised clustering of samples. Log-transformed peptide intensities were converted into z-scores using means, wherein higher scores are indicated in red and lower scores in blue.
The most frequently observed protein precursors, associated with at least 100 urinary peptides, were serum albumin, collagen alpha-1(I) chain, hemoglobin subunit alpha and subunit beta, keratin type II cytoskeletal 6A, and apolipoprotein A1. Together, these six proteins accounted for 17% of the quantified peptidome (supplemental Table S7).
A subset of 162 peptides was found in all thirty samples (supplemental Table S8), representing 2.6% of the total peptidome. Although this subset mainly consisted of medium-to-high-abundance peptides, the range of intensities spanned four orders of magnitude (Fig. 2C). As expected, low-abundance peptides were not consistently detected across samples. We examined the tissue origins of peptides using the Human Protein Atlas database (28). Roughly 80% of unique peptides derived from proteins with known expression in the kidneys. Kidney-specific or -enriched proteins accounted for 15% of the 162 peptides, compared with 4% of the total peptidome (Fig. 2D). We thus decided to focus our further analyses on these 162 peptides as they reflected the most robust and consistent changes in the peptidome.
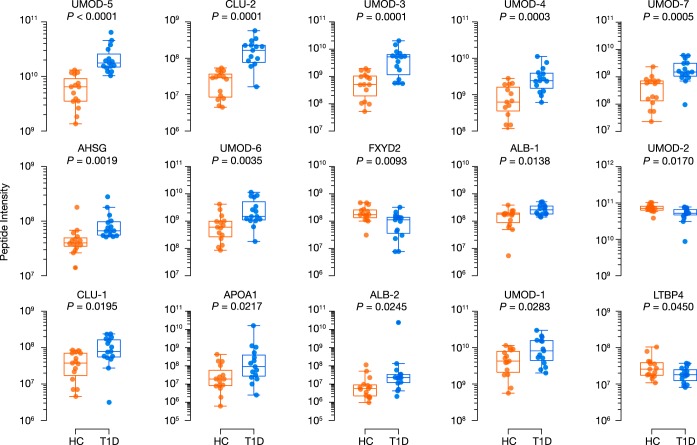
Fifteen peptides were differentially excreted between study groups (p < 0.05), deriving from alpha-2-HS-glycoprotein, albumin, apolipoprotein A1, clusterin, latent transforming growth factor beta binding protein 4, Na+/K+-transporting ATPase subunit gamma, and uromodulin (Fig. 2E, Table III). All but three peptides were more abundant in urines from youths with diabetes (Fig. 2F, Fig. 3). After BH adjustment, four uromodulin peptides and one clusterin peptide remained statistically significant (Q < 0.05). Unsupervised hierarchical clustering based on normalized peptide intensities segregated the two groups based on diabetes status, leaving one youth with diabetes among those without diabetes (Fig. 2F).
Table III. Summary of differentially excreted peptides from the discovery phase. p values were determined using the independent t-test and adjusted by Benjamini-Hochberg procedure (Q). Fold change represents the ratio of the mean log-transformed intensities of youths with type 1 diabetes (T1D) to healthy controls (HC). AHSG, alpha-2-HS-glycoprotein; ALB, albumin; APOA1, apolipoprotein A1; CLU, clusterin; LTBP4, latent transforming growth factor beta binding protein 4; FXYD2, Na+/K+-transporting ATPase subunit gamma; UMOD, uromodulin.
| Peptide | Name | Mass (Da) | Charge | Fold change (T1D–HC) | P | Q |
|---|---|---|---|---|---|---|
| SGSVIDQSRVLNLGPI | UMOD-1 | 1653.9050 | 2,3 | 2.11 | 0.0283 | 0.3298 |
| SGSVIDQSRVLNLGPITR | UMOD-2 | 1911.0538 | 2,3,4 | 0.68 | 0.0170 | 0.2768 |
| SVIDQSRVLNLGPI | UMOD-3 | 1509.8515 | 1,2 | 6.72 | 0.0001 | 0.0070 |
| SVIDQSRVLNLGPIT | UMOD-4 | 1610.8992 | 1,2,3 | 3.95 | 0.0003 | 0.0127 |
| SVIDQSRVLNLGPITR | UMOD-5 | 1767.0003 | 2,3,4 | 3.84 | <0.0001 | 0.0003 |
| SVIDQSRVLNLGPITRK | UMOD-6 | 1895.0952 | 2,3,4 | 3.75 | 0.0035 | 0.0820 |
| VIDQSRVLNLGPI | UMOD-7 | 1422.8195 | 1,2 | 5.48 | 0.0005 | 0.0159 |
| DDGGPYGESEAPAPPGPGTRWP | LTBP4 | 2208.9712 | 2,3 | 0.65 | 0.0450 | 0.4894 |
| TGLSMDGGGSPKGDVDPF | FXYD2 | 1735.7723 | 2 | 0.38 | 0.0093 | 0.1901 |
| SHTSDSDVPSGVTEVVVKL | CLU-1 | 1954.9848 | 2,3 | 2.55 | 0.0195 | 0.2891 |
| HTSDSDVPSGVTEVVVKL | CLU-2 | 1867.9527 | 2,3 | 5.87 | 0.0001 | 0.0096 |
| LSALEEYTKKLNTQ | APOA1 | 1636.8672 | 2,3 | 5.88 | 0.0217 | 0.2948 |
| YGEMADCCAKQEPERNECFLQ | ALB-1 | 2634.0607 | 2,3 | 2.12 | 0.0138 | 0.2503 |
| VRYTKKVPQVSTPTL | ALB-2 | 1715.9934 | 2,3,4 | 4.64 | 0.0245 | 0.3067 |
| TVVQPSVGAAAGPVVPPCPGRIRH | AHSG | 2421.3063 | 3,4 | 1.92 | 0.0019 | 0.0529 |
Fig. 3.
Boxplots for the fifteen differentially excreted peptides found in the discovery cohort. Differential excretion was determined using the independent two-sample t test after log-transforming peptide intensities (p < 0.05). HC, youths without T1D; T1D, youths with type 1 diabetes.
Validation of Differentially Excreted Peptides
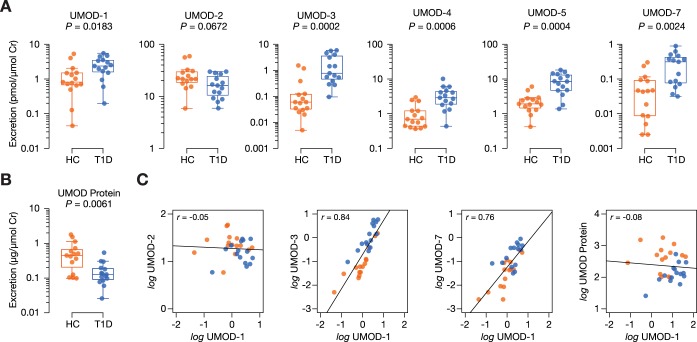
Parallel reaction monitoring takes advantage of mass spectrometry-based techniques to quantify multiple peptides simultaneously on quadrupole-orbitrap hybrid instrument using heavy-labeled internal standards (29). We successfully developed targeted methods for the relative quantification of six peptides: UMOD-1, UMOD-2, UMOD-3, UMOD-4, UMOD-5, UMOD-7. Each of the thirty urine samples were analyzed in duplicate injections onto the mass spectrometer with median technical coefficients of variation less than 12% for each peptide (Supplemental Table S9). All peptides except UMOD-2 were differentially excreted between groups in the validation cohort (p < 0.05) (Fig. 4A, Table IV). Fold changes were similar to those from the discovery phase. Notably, UMOD-2 was the lone peptide whose urinary excretion decreased in diabetes (fold change of 0.75, p = 0.0672). We also demonstrated that total uromodulin protein excretion was markedly reduced in diabetes (fold change of 0.28, p = 0.0061), similar to UMOD-2 (Fig. 4B), although there was no significant correlation between peptide and protein excretion (Fig. 4C, supplemental Table S10). We observed that the uromodulin peptides strongly correlated with one another, except for UMOD-2 (Fig. 4C, supplemental Table S10). These findings suggest UMOD-2 may derive separately from the other uromodulin peptides. To better understand this, we sought to examine whether the downward trend of UMOD-2 would be reflected when applying other urinary normalization methods (supplemental Table S11). In contrast to urinary excretion, urinary concentration of UMOD-2 was mildly increased in diabetes (fold change of 1.1, p = 0.7718). A similar trend was observed when UMOD-2 was normalized to total uromodulin protein (fold change of 2.8, p = 0.0561). The direction of change of the other uromodulin peptides remained consistent despite different normalization methods.
Fig. 4.
Evaluation of uromodulin peptides in validation cohort of thirty youths with and without type 1 diabetes. Differential excretion was determined using the Mann-Whitney test (p < 0.05). A, Urinary excretion of uromodulin peptides measured by parallel reaction monitoring. B, Urinary excretion of uromodulin protein measured using a commercial enzyme-linked immunosorbent assay kit. C, Representative correlation plots between uromodulin peptides and protein. HC, youths without T1D; T1D, youths with type 1 diabetes.
Table IV. Summary of differentially excreted uromodulin peptides from the validation phase. Differential excretion was determined using the Mann-Whitney test (p < 0.05). Fold change represents the ratio of the median peptide excretion of youths with type 1 diabetes (T1D) to the median peptide excretion of healthy controls (HC). AHSG, alpha-2-HS-glycoprotein; ALB, albumin; APOA1, apolipoprotein A1; CLU, clusterin; FXYD2, Na+/K+-transporting ATPase subunit gamma; LTBP4, latent transforming growth factor beta binding protein 4; UQ, unable to quantify; UMOD, uromodulin.
| Peptide | Name | Fold change (T1D:HC) | P |
|---|---|---|---|
| SGSVIDQSRVLNLGPI | UMOD-1 | 2.91 | 0.0183 |
| SGSVIDQSRVLNLGPITR | UMOD-2 | 0.75 | 0.0672 |
| SVIDQSRVLNLGPI | UMOD-3 | 12.83 | 0.0002 |
| SVIDQSRVLNLGPIT | UMOD-4 | 4.16 | 0.0006 |
| SVIDQSRVLNLGPITR | UMOD-5 | 4.41 | 0.0004 |
| SVIDQSRVLNLGPITRK | UMOD-6 | UQ | — |
| VIDQSRVLNLGPI | UMOD-7 | 6.94 | 0.0024 |
Novel Bioactivity of Uromodulin Peptides
We next investigated whether these uromodulin peptides might exert a functional or biological role in the kidney. PeptideRanker was used to predict bioactivity of the differentially excreted peptides (Table V). This server was trained using databases of known bioactive peptides (PeptidesDB, CAMP, and BIOPEP) (30–32), providing probabilities of bioactivity based on amino acid composition (23). Ang II, Ang (1–7), and vasopressin were also included as positive controls in our analysis. The top uromodulin peptides, UMOD-1 and UMOD-2, outscored Ang (1–7) with scores of 0.47 and 0.46, respectively.
Table V. Probability of bioactivity of differentially excreted peptides, as predicted by Peptide Ranker. The asterisk (*) denotes the peptides that were included as a positive control.
| Peptide | Protein/Name | Score |
|---|---|---|
| CYFQNCPRG/Vasopressin* | AVP | 0.90 |
| DDGGPYGESEAPAPPGPGTRWP | LTBP4 | 0.81 |
| DRVYIHPF/Angiotensin II* | AGT | 0.69 |
| TVVQPSVGAAAGPVVPPCPGRIRH | AHSG | 0.63 |
| YGEMADCCAKQEPERNECFLQ | ALB-1 | 0.59 |
| SGSVIDQSRVLNLGPI | UMOD-1 | 0.47 |
| SGSVIDQSRVLNLGPITR | UMOD-2 | 0.46 |
| SVIDQSRVLNLGPITRK | UMOD-6 | 0.35 |
| DRVYIHP/Angiotensin (1–7)* | AGT | 0.34 |
| TGLSMDGGGSPKGDVDPF | FXYD2 | 0.33 |
| SVIDQSRVLNLGPI | UMOD-3 | 0.29 |
| SVIDQSRVLNLGPIT | UMOD-4 | 0.29 |
| SVIDQSRVLNLGPITR | UMOD-5 | 0.29 |
| VIDQSRVLNLGPI | UMOD-7 | 0.24 |
| SHTSDSDVPSGVTEVVVKL | CLU-1 | 0.19 |
| HTSDSDVPSGVTEVVVKL | CLU-2 | 0.17 |
| LSALEEYTKKLNTQ | APOA1 | 0.12 |
| VRYTKKVPQVSTPTL | ALB-2 | 0.11 |
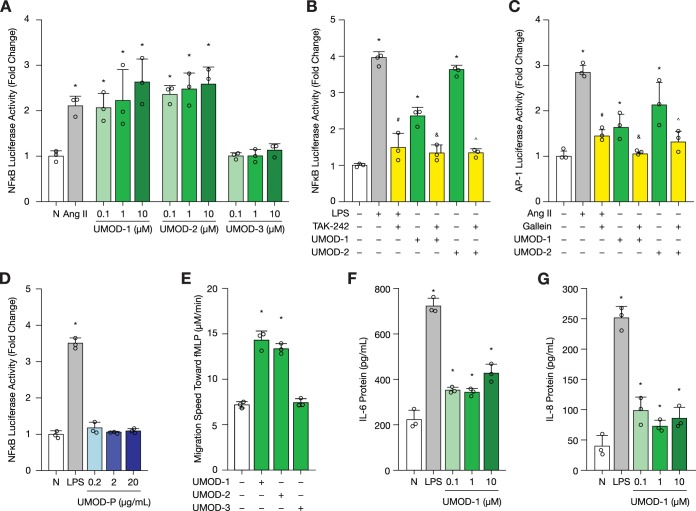
To verify the in silico prediction, we first performed NFκB and AP-1 luciferase activity assays in HK-2 cells. Both transcription factors are activated downstream of toll-like receptor 4 (TLR4) activation as part of the pro-inflammatory response (33). Chronic activation of inflammation is thought to contribute to diabetic kidney disease progression (34, 35). As an initial screen, cells were treated for 24 h with graded concentrations (0.1, 1, and 10 μm) of 70%-purified uromodulin peptides, UMOD-1, UMOD-2, and UMOD-3 (Fig. 5A). Ang II was used as a positive control based on previous findings that it promotes inflammation and oxidative stress in the kidney (36–38). At 0.1 μm, UMOD-1 and UMOD-2 significantly stimulated NFκB activity (p < 0.05). As predicted, UMOD-3 failed to elicit a response, even at the highest tested dose of 10 μm.
Fig. 5.
Uromodulin peptides are bioactive in vitro. Technical replicates are shown as open circles (n = 3). A, NFκB luciferase activity following uromodulin peptide treatment for 24 h in HK-2 cells at graded concentrations (0.1 to 10 μm). Angiotensin (Ang II, 0.1 μm) served as a positive control. B, NFκB luciferase activity following uromodulin peptide treatment (0.1 μm) for 24 h with or without TAK-242 pre-treatment (0.3 μm) for 1 h in HK-2 cells. Lipopolysaccharide (LPS, 0.1 μg/ml) served as a positive control. C, AP-1 luciferase activity following uromodulin peptide treatment (0.1 μm) for 24 h with or without gallein pre-treatment (10 μm) for 1 h in HK-2 cells. Ang II served as a positive control. D, NFκB luciferase activity following uromodulin protein treatment for 24 h in HK-2 cells at graded concentrations (0.2 to 20 μg/L). Lipopolysaccharide (LPS, 0.1 μg/ml) served as a positive control. E, Neutrophil chemotaxis following uromodulin peptide incubation toward the N-formyl-methionyl-leucyl-phenylalanine (fMLP) attractant. Migration was recorded for 15 min at a rate of 0.05 frames per second (fps) using time-lapse video microscopy. F–G, IL-6 and IL-8 protein levels in conditioned media of primary proximal tubular epithelial cells following UMOD-1 treatment for 24 h. LPS served as a positive control (1 μg/ml). *p < 0.05, compared with untreated cells (N). #p < 0.05, compared with cells treated with LPS alone. &p < 0.05, compared with cells treated with UMOD-1 alone. ∧p < 0.05, compared with cells treated with UMOD-2 alone.
Previous studies have shown that uromodulin protein activates NFκB in immune cells via TLR4 (39, 40). We thus inhibited TLR4 by pre-treating cells with TAK-242 (resatorvid) for 1 h, which significantly reduced LPS- and uromodulin peptide-induced NFκB activity (p < 0.05) (supplemental Fig. S2, Fig. 5B). LPS was used as a positive control for canonical TLR4-mediated NFκB activation (41, 42). Activation of NFκB signaling by uromodulin peptides, UMOD-1 and UMOD-2, may thus be mediated by TLR4 signaling. The same peptides also triggered an AP-1 response, which was attenuated with gallein, an inhibitor of G protein βγ subunit-dependent signaling (supplemental Fig. S3, Fig. 5C). Interestingly, uromodulin protein failed to stimulate NFκB luciferase activity in HK-2 cells at varying concentrations of 0.2, 2, and 20 μg/ml (Fig. 5D). These concentrations were selected to cover the reported range from 100 ng/ml to 25 μg/ml for urinary uromodulin concentration (5–8). In this study, the median urinary concentration for uromodulin protein was 2.5 μg/ml for youths with diabetes and 4.7 μg/ml for their non-diabetic peers. Similar results were observed in primary proximal tubular epithelial cells (supplemental Fig. S4).
Next, we conducted a neutrophil chemotaxis assay. Human blood neutrophils were first treated with one of the three previously tested uromodulin peptides, then placed in a Zigmond chamber, and finally subjected to a N-formyl-methionyl-leucyl-phenylalanine (fMLP) attractant gradient. Compared with untreated neutrophils or neutrophils treated with UMOD-3, UMOD-1 and UMOD-2 significantly enhanced neutrophil migration toward fMLP (p < 0.05) (Fig. 5E).
To bridge NFκB signaling and neutrophil chemotaxis, we examined whether UMOD-1 might induce cytokine release from primary proximal tubular epithelial cells. Interleukin (IL)-8 was specifically chosen as a neutrophil chemokine; whereas IL-6 was selected for its pro-inflammatory action. Both cytokines are downstream products of TLR-4 mediated NFκB signaling. We demonstrated that UMOD-1 promotes IL-6 and IL-8 secretion into serum-starved conditioned media of primary proximal tubular epithelial cells at 0.1 to 10 μm (p < 0.05) (Figs. 5F–5G).
Urinary Excretion of Cytokines and Chemokines
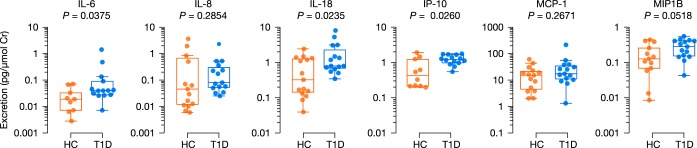
Our in vitro findings suggested a role for uromodulin peptides in inflammation. Accordingly, we speculated whether a pro-inflammatory signal based on increased urinary excretion of cytokines and chemokines could be detected in early diabetes. To do so, we used a Luminex bead-based immunoassay and adjusted urinary analyte concentrations to urinary creatinine concentrations. Eight analytes were selected a priori based on the literature on urinary markers of inflammation in diabetic kidney disease: IL-1β, IL-6, IL-8, IL-18, IP-10, MCP-1, MIP-1β, and TNFα (Fig. 6). IL-1β and TNFα, however, were omitted because more than two-thirds of the samples in each group could not be reliably quantified. Of the six remaining analytes, IL-6 (median fold change of 2.1), IL-18 (median fold change of 2.5), and IP-10 (median fold change of 2.8) were excreted to a greater extent by youths with diabetes, compared with their non-diabetic peers (p < 0.05).
Fig. 6.
Urinary cytokine and chemokine excretion of youths with and without diabetes. Differential excretion was determined using the Mann-Whitney test (p < 0.05). HC, youths without T1D; T1D, youths with type 1 diabetes.
Cleavage Site Analysis of Urinary Peptides
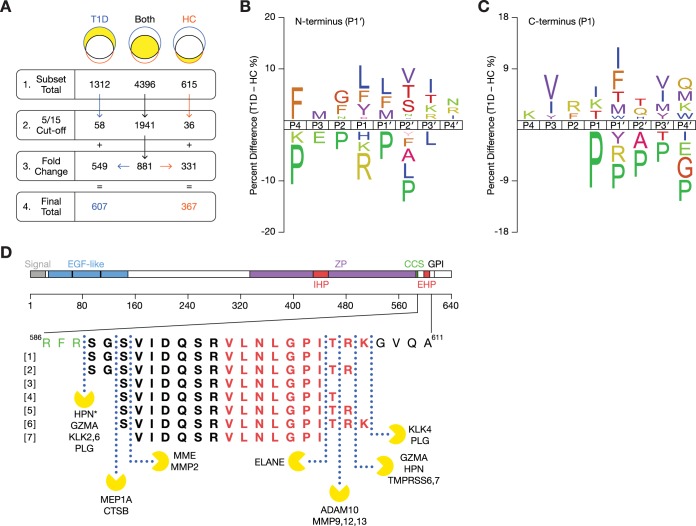
To investigate the proteolytic mechanisms altered in early diabetes, we first examined the cleavage sites associated with the N termini and C termini of urinary peptides. For this analysis, we initially included all 4396 peptides that were common to both groups and subsequently excluded peptides that were quantified in fewer than five samples per group. Missing LFQ data was imputed using the Perseus software. Of the 1941 eligible peptides, 549 peptides were increased (with a fold change of at least 1.5) and 331 peptides were decreased (with a fold change of 0.67 and below) in diabetes. We also added 94 peptides that were exclusively identified in one group if they were quantified in at least five samples: 58 from youths with diabetes and 36 from their non-diabetic peers. Thus, a total of 974 peptides was examined in our cleavage site analysis using iceLogo (25), in which the experimental set (n = 607 peptides that were elevated in diabetes) was compared with the reference set (n = 367 downregulated peptides) (Fig. 7A).
Fig. 7.
Understanding the proteolytic mechanisms responsible for generating the urinary peptidome. A, Flow diagram describing the selection of urinary peptides altered in diabetes for cleavage site analysis. From each section of the Venn diagram in Fig. 2B, peptides were eligible for inclusion if they were identified in at least five of the fifteen samples per group, where appropriate. Missing LFQ data was imputed using the Perseus software. Of the 1941 eligible peptides common to both groups, 549 peptides were increased (with a fold change of at least 1.5) in diabetes, and 331 peptides were decreased (with a fold change of 0.67 and below). A total of 974 peptides was examined in subsequent cleavage site analyses. B–C, Sequence logo of N- and C termini of urinary peptides using iceLogo. Significant differences in percentage of amino acids at each position in the cleavage are shown (p < 0.05). Positive values indicate increased percentages in type 1 diabetes. D, Proteolytic map of uromodulin and the predicted proteases. Differentially excreted peptides originate from a region near the C terminus of the precursor uromodulin protein, which is shown in bold at the bottom with predicted cleavage sites and proteases. Full-length uromodulin consists of 640 residues.
We noted a preponderance of proline residues at or near the N- and C termini of urinary peptides, especially peptides that were downregulated in diabetes (Fig. 7B–7C). In fact, proline residues accounted for 25% of all C termini of downregulated peptides, compared with 14% of C termini of peptides elevated in diabetes. Additionally, there was a higher percentage of trypsin-like cleavages (i.e. at the carboxyl side of an arginine or lysine residues) resulting in the N terminus of urinary peptides downregulated in diabetes; the opposite trend seems to be true at the C terminus of peptides elevated in diabetes (p < 0.05). Unlike the non-diabetic profile, the sequence motifs for diabetes were more diverse and highlighted a broader range of residues such as leucine, phenylalanine, and isoleucine. Notably, the top motif for the N terminus cleavage site (L*LV) corresponded to renin (26), which is secreted by juxtaglomerular cells of the kidney.
We next focused on the proteolytic mechanisms responsible for the uromodulin peptides. Using Peptide Extractor(9), we mapped all of the differentially excreted peptides to a specific region near the C terminus of uromodulin: 589SGSVIDQSRVLNLGPITRK607 (Fig. 7D). Adjacent regions include the hepsin consensus cleavage site at 586RFRS589 and a glycosylphosphatidylinositol (GPI) anchor site at S614, which anchors the uromodulin protein onto the cell membrane of the loop of Henle and distal tubular cells. This peptide region contains the external hydrophobic patch (EHP) motif, 598VLNLGPITRK607, which inhibits uromodulin polymerization and function (43). Given that urinary protein excretion of uromodulin was decreased in youths with diabetes (Fig. 4B), we hypothesize that changes in peptide excretion could reflect changes in proteolytic activity.
Proteasix (24) was used to predict the proteases responsible for generating the uromodulin peptides. A total of 156 combinations of predicted proteases and cleavage sites met the MEROPS threshold score. For each cleavage site, we selected the proteases according to the predicted probability as follows: (1) predicted probability fell above the 99th percentile of the population distribution of all sequences, (2) predicted probability was below, but was the closest to the 99th percentile, and (3) predicted probability was the furthest away from the MEROPS threshold score. After applying the criteria, nineteen protease/cleavage sites combinations remained (Table VI), and the predicted cleavage sites are illustrated in Fig. 7E. Of these predicted proteases, eight predominantly cleave at the carboxyl side of arginine or lysine (supplemental Fig. S5). According to our earlier sequence logo motif analyses, we speculate that the N-terminal cleavage of the peptide region (589SGSVIDQSRVLNLGPITRK607) occurs in normal, physiologic conditions (Fig. 7B), whereas the C-terminal cleavage events are increased in diabetes (Fig. 7C). This is supported by an increased percentage of cleavage events that are at the carboxyl side of isoleucine (UMOD-1, UMOD-3, and UMOD-7), lysine (UMOD-6), threonine (UMOD-4) of uromodulin peptides elevated in diabetes.
Table VI. Predicted proteases responsible for the generation of the differentially excreted uromodulin peptides. The asterisk (*) denotes a known cleavage site. The arrows (↑ and ↓) indicate the direction of the average fold change in Nephroseq gene expression datasets on kidney biopsies from adults with diabetic nephropathy, compared to living or cadaveric donors. The studies are referenced as follow: (1) Woroniecka Diabetes TubInt, (2) Schmid Diabetes TubInt, (3) Ju CKD Glom, (4) Ju CKD TubInt, (5) Woroniecka Diabetes Glom, and (6) ERCB Nephrotic Syndrome TubInt.
| Protease (gene symbol) | MEROPS ID | Predicted cleavage sites | Fold change (ref.) |
|---|---|---|---|
| A disintegrin and metalloproteinase domain-containing protein 10 (ADAM10) | M12.210 | GPIT RKGV | ↑ 1.7 (1,2) |
| Cathepsin B (CTSB) | C01.060 | FRSG SVID | ↑ 1.9 (1) |
| Granzyme A (GZMA) | S01.135 | TRFR SGSV | ↑ 4.6 (1,3–6) |
| PITR KGVQ | |||
| Hepsin (HPN) | S01.224 | TRFR SGSV* | ↓ 1.7 (2,3) |
| PITR KGVQ | |||
| Kallikrein-2 (KLK2) | S01.161 | TRFR SGSV | No change |
| Kallikrein-4 (KLK4) | S01.251 | ITRK GVQA | No change |
| Kallikrein-6 (KLK6) | S01.236 | TRFR SGSV | ↓ 4.1 (4,5) |
| Matrix metalloproteinase-2 (MMP2) | M10.003 | RSGS VIDQ | ↑ 2.7 (3,6) |
| Matrix metalloproteinase-9 (MMP9) | M10.004 | GPIT RKGV | ↑ 15.2 (6) |
| Matrix metalloproteinase-12 (MMP12) | M10.009 | GPIT RKGV | No change |
| Matrix metalloproteinase-13 (MMP13) | M10.013 | GPIT RKGV | No change |
| Meprin A (MEP1A) | M12.002 | FRSG SVID | No change |
| Neprilysin (MME) | M13.001 | RSGS VIDQ | ↓ 2.3 (3–5) |
| Neutrophil elastase (ELANE) | S01.131 | LGPI TRKG | No change |
| Plasminogen (PLG) | S01.233 | TRFR SGSV | ↓ 2.7 (3–4) |
| Matriptase-2 (TMPRSS6) | S01.308 | ITRK GVQA | No change |
| Matriptase-3 (TMPRSS7) | S01.072 | PITR KGVQ | Not found in database |
We also searched the Nephroseq v5 database (www.nephroseq.org, November 2018, University of Michigan, Ann Arbor, MI) for expression data in diabetic nephropathy and found nine proteases whose expression was significantly altered in cases of diabetic nephropathy (p < 0.05): a disintegrin and metallo-proteinase domain-containing protein 10, cathepsin B granzyme A, hepsin, kallikrein-6, matrix metalloproteinase-2, matrix metalloproteinase-9, neprilysin, and plasmin. Hepsin has previously been identified as a putative protease responsible for the N-terminal cleavage of UMOD-1 and UMOD-2. Meprin A and cathepsin B were predicted to cleave at carboxyl side of G590, resulting in the inert peptide UMOD-3. Neutrophil elastase was mapped to the C terminus of UMOD-1, UMOD-3, and UMOD-7.
DISCUSSION
It is generally accepted that chronic hyperglycemia disrupts the proteolytic milieu (10–12). The resulting peptide products could thus be used to examine the upstream interactions between proteolytic enzymes and protein substrates (9). In this study, we report that otherwise healthy youths with type 1 diabetes excrete a urinary peptide profile that is distinct from their non-diabetic peers and that these differences precede the onset of microalbuminuria or renal function decline. Seven of the fifteen differentially excreted peptides originated from a small region (589SGSVIDQSRVLNLGPITRK607) near the C terminus of uromodulin; and five were validated in a second cohort by parallel reaction monitoring. Furthermore, we discovered that UMOD-1, one of the validated peptides, displays novel bioactivity in vitro, by stimulating TLR4-dependent NFκB luciferase activity in HK-2 and primary proximal tubular epithelial cells, by inducing cytokine release from primary proximal tubular epithelial cells, and by enhancing neutrophil chemotaxis. A second peptide, UMOD-2, also exhibited similar bioactivity, but was not differentially excreted in the validation cohort. Interestingly, uromodulin protein failed to elicit a pro-inflammatory response in tubular epithelial cells. In silico analyses identified several proteases that could be responsible for generating these uromodulin peptides, such as hepsin, meprin A, cathepsin B, and neutrophil elastase. Our findings present new insights into the early kidney response to chronic hyperglycemia long before the clinical manifestation of renal dysfunction.
Uromodulin, also known as Tamm-Horsfall glycoprotein, is exclusively expressed in the loop of Henle and distal tubules of the kidney (44, 45). Naturally, its peptides have gained notoriety as potential kidney-specific biomarkers for diabetic nephropathy (46), hypertension (47, 48), chronic kidney disease (49), systematic lupus erythematosus (50), IgA nephropathy (51), and acute rejection following renal transplantation (52). Even though the exact peptide sequences differ between peptidomic studies, they typically originate from the same region as our peptides: 589SGSVIDQSRVLNLGPITRK607. These observations suggest that perturbations in specific proteolytic events associated with kidney injury may not be unique to diabetes after all. Still, our findings demonstrate that the urinary peptidome may reveal early signs of kidney injury in youths with diabetes, even when they present with no clinical symptoms of renal stress or disease. Additional validation studies are needed to examine how urinary excretion of uromodulin peptides changes over the course of diabetic kidney disease.
Beyond their potential utility as indicators of injury, very little is known about these uromodulin peptides. We thus sought to investigate whether they may play a direct mechanistic role in the kidney; and, for the first time to our knowledge, we demonstrated that two peptides, UMOD-1 and UMOD-2, were capable of stimulating pro-inflammatory responses in kidney tubular epithelial cells and enhanced neutrophil migration. These findings significantly enrich our current understanding of uromodulin biology because the glycoprotein is thought to be immunologically inert in the tubules, but active elsewhere (40, 53, 54). Namely, uromodulin was shown to induce the maturation of myeloid dendritic cells by activating TLR4-dependent NFκB signaling pathways (39). Uromodulin nanoparticles stimulated the NLRP3-inflammasome in antigen-presenting cells, triggering IL-1β secretion (40). In addition, uromodulin protein expression on membranes of cultured tubular epithelial cells enhanced neutrophil adherence and trans-epithelial migration (55). Neutrophils also preferentially attached to uromodulin protein immobilized on microtiters plates; however, no such interaction was observed with soluble uromodulin (56). A recent study reported that uromodulin abated renal and systemic oxidative stress by inhibiting the transient receptor potential cation channel, subfamily M, member 2 (TRPM2), as a protective mechanism against acute kidney injury (57). Altogether, uromodulin protein seems to rely on interactions with immune cells to trigger inflammation and may be protective in the kidney. In contrast, our in vitro data suggest that uromodulin peptides could mediate pro-inflammatory processes in the kidney directly.
We next examined how these peptides might be generated. Although basolateral expression has been observed, uromodulin is predominantly attached to the apical surface by a GPI anchor near the C terminus (58, 59). The bulk of the protein is subsequently shed into the urinary space following hepsin cleavage (60). More specifically, the type II transmembrane serine protease separates the active portion of the protein from our peptide region, which remains attached to the membrane and contains the inhibitory external hydrophobic patch (60–62). This cleavage event accounts for the N terminus of the two potentially bioactive peptides: UMOD-1 and UMOD-2. Highly expressed in the liver and renal tubules (63–65), hepsin is involved in cell growth and proliferation, blood coagulation, extracellular matrix remodeling (66–68). Studies have demonstrated that deficient hepsin expression produces longer, polymerization-incompetent isoforms of uromodulin containing the peptide region; results in intracellular accumulation of uromodulin; and predisposes tubules to stress (60, 62). Although renal expression of hepsin is downregulated in diabetic nephropathy (Nephroseq database), it is unclear whether hepsin expression or activity is reduced in the diabetic kidney before clinical injury.
Two other notable cleavage sites emerged from our in silico analyses: the N terminus of UMOD-3, UMOD-4, UMOD-5, and UMOD-6 by meprin A and cathepsin B; and the C terminus of UMOD-1, UMOD-3, and UMOD-7 by neutrophil elastase. Meprin A is a metalloproteinase found in the brush border of the proximal tubules, where it is constitutively shed into the urinary space by furin (69). In its active form, meprin A stimulates pro-inflammatory cytokine release and fibrosis in the diabetic kidney (70–73). Studies have also hinted at a protective role for meprin A, in which diminished expression and activity in the kidney were associated with later stages of diabetic kidney disease (74, 75). Urinary levels of meprin A were markedly elevated in microalbuminuric and late-stage disease but were negligible in normoalbuminuric individuals with diabetes and non-diabetic subjects (76). Cathepsin B, a lysosome-associated enzyme, is involved in autophagy, extracellular matrix remodeling, inflammasome activation, and sodium reabsorption in the kidney (77–80). Studies have shown that cathepsin B activity is decreased following in vitro stimulation of HK-2 cells with advanced glycation end-product (81) and in the kidneys of rats with streptozotocin-induced diabetes (82). Interestingly, lysosome-derived cathepsin B expression was elevated in glomeruli of a murine model of podocyte injury; and cathepsin B knockout mice were more resistant to injury and recovered more rapidly following injection of nephrotoxic serum (83). Urinary proteomic profiling in early diabetes has demonstrated increased urinary excretion of cathepsin B and other lysosome-associated enzymes (84, 85). Last, neutrophil elastase is secreted from neutrophil granules during acute inflammation and promotes reactive oxygen species generation (86, 87). Neutrophil infiltration into the kidney has been well-documented in later stages of diabetic kidney disease and contributes to disease progression and fibrosis (88–90).
Although it is possible that each peptide is generated de novo from uromodulin protein, we posit that they derive from peptide intermediates, similar to angiotensinogen and its peptide products. The 452-residue precursor is first shortened into the inactive Ang I decapeptide (1DRVYIHPFHL10) by renin. Subsequent angiotensin-converting enzyme activity produces the bioactive Ang II octapeptide (1DRVYIHPF8), which promotes sodium reabsorption via aldosterone secretion (91). Alternate processing of the Ang I peptide results in a myriad of bioactive and inert peptide sequences (92–95). Notably, Ang (1–7) and Ang II (1–8) have opposing biological effects (96, 97), underscoring the significance of a single residue. Likewise, the bioactive UMOD-1 (SGSVIDQSRVLNLGPI) and the inert UMOD-3 (SVIDQSRVLNLGPI) peptides differ in two residues at the N terminus, suggesting that the loss of the serine-glycine residues at the N terminus may render a peptide inactive and could act as a means to “turn off” peptide bioactivity. Interestingly, UMOD-2 excretion did not correlate with any of the other uromodulin peptides and may thus originate from a separate proteolytic mechanism. Although UMOD-2 was not differentially excreted in the validation cohort, it displayed a downward trend in diabetes. Its presence may be related to normal renal physiology or reflect decreased total uromodulin protein expression, rather than dysregulated processing in chronic hyperglycemia. Future studies will be necessary to define the proteolytic pathways and putative enzymes responsible for uromodulin processing.
Our study has several strengths. First, we carefully selected our study population to avoid possible confounding effects of concurrent medications and comorbidities. This allowed us to examine the early kidney response to chronic hyperglycemia in a homogenous cohort of youths with and without diabetes. Second, we conducted an internal validation of uromodulin peptides in a second, independent cohort. Finally, we provided three pieces of evidence of novel bioactivity of UMOD-1, whose urinary excretion was elevated in early type 1 diabetes, using three different cell systems. Our findings may open a new field of investigation into the mechanisms responsible for early kidney injury in type 1 diabetes and into therapies that target prevention, rather than management, of diabetic kidney disease.
Our study also has some important limitations. First, the cause-effect relationships between uromodulin peptides and chronic hyperglycemia cannot be fully discerned in a cross-sectional study. Second, the study population did not include individuals at later stages of diabetic kidney disease, and it is thus difficult to infer whether these peptides are associated with progressive disease. Finally, we were unable to experimentally verify the protease prediction because of the lack of a model system, which mimics the appropriate kidney cell type and naturally expresses uromodulin protein. Ultimately, an in-depth investigation into these proteolytic events will better define the pathophysiologic role of uromodulin peptides in the diabetic kidney.
In summary, we identified and validated a signature of uromodulin peptides associated with early, uncomplicated type 1 diabetes in two separate cohorts. Our discovery of novel bioactivity in uromodulin peptides suggests a potential role in mediating early changes in the diabetic kidney.
DATA AVAILABILITY
For discovery peptidomics, mass spectrometry data have been deposited onto the ProteomeXchange Consortium via the PRIDE (16) partner repository with the dataset identifier PXD012210 (http://www.ebi.ac.uk/pride/archive/login). For targeted peptidomics, Skyline files and raw data have been deposited in Panorama Public with a ProteomeXchange identifier PXD012389 (https://panoramaweb.org/lotso19.url).
Supplementary Material
Acknowledgments
We thank Christine Kerr, Daryl Baquillos, Laura Motran, Harriet Georgas, Denis Daneman, Veronica Talunay, Antoninus Soosaipillai, and the SPARC BioCentre facility members at SickKids Hospital in Toronto.
Footnotes
* J.A.D.V. is supported by the Banting & Best Diabetes Centre through the Yow Kam-Yuen Graduate Scholarship in Diabetes Research and Novo Nordisk Graduate Studentship and by an Ontario Graduate Scholarship. F.H.M. and E.S. have received funding through the Juvenile Diabetes Research Foundation Canadian Clinical Trials Network (JDRF-CCTN), Canadian Institutes of Health Research (CIHR), the Heart and Stroke Foundation of Canada, and Diabetes Canada. JWS is supported by operating grants from the Heart and Stroke Foundation of Canada and the CIHR CanSOLVE-CKD SPOR program. AK is supported by a Kidney Foundation of Canada operating grant, the Kidney Research Scientist Core Education and National Training (KRESCENT) program, Kidney Foundation of Canada Predictive Biomarker Grant, CIHR Catalyst Grant, and Canada Foundation for Innovation award. The authors declare that they have no conflicts of interest with the contents of this article.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- DCCT
- Diabetes Control and Complications Trial
- ACR
- Albumin/creatinine ratio
- Ang
- Angiotensin
- BH
- Benjamini-Hochberg
- BMI
- Body mass index
- DMEM
- Dulbecco's Modified Eagle Medium
- EDIC
- Epidemiology of Diabetes Intervention and Complication
- eGFR
- Estimated glomerular filtration rate
- ELISA
- Enzyme-linked immunosorbent assay
- FDR
- False discovery rate
- GPI
- Glycosylphosphatidylinositol
- HbA1c
- Glycated hemoglobin A
- HKUPP
- Human Kidney and Urine Proteome Project
- HUPO
- Human Proteome Organization
- IL
- Interleukin
- LFQ
- Label-free quantitation
- MS/MS
- Tandem mass spectrometry
- SCX-HPLC
- Strong cation exchange high-performance liquid chromatography
- TLR4
- Toll-like receptor 4.
REFERENCES
- 1. American Diabetes Association. (2014) Diagnosis and classification of diabetes mellitus. Diabetes Care 37, S81–S90 [DOI] [PubMed] [Google Scholar]
- 2. Harris M. I., Klein R., Welborn T. A., and Knuiman M. W. (1992) Onset of NIDDM occurs at least 4–7 yr before clinical diagnosis. Diabetes Care 15, 815–819 [DOI] [PubMed] [Google Scholar]
- 3. Foley R., Parfrey P., and Sarnak M. (1998) Clinical epidemiology of cardiovascular disease in chronic renal disease. Am. J. Kidney Dis. 32, S112–S119 [DOI] [PubMed] [Google Scholar]
- 4. Kim S. J., Gill J. S., Knoll G., Campbell P., Cantarovich M., Cole E., and Kiberd B. (2019) Referral for Kidney Transplantation in Canadian Provinces. J. Am. Soc. Nephrol., ASN.2019020127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Webster A. C., Nagler E. V., Morton R. L., and Masson P. (2017) Chronic kidney disease. Lancet 389, 1238–1252 [DOI] [PubMed] [Google Scholar]
- 6. The Diabetes Control and Complications Trial Research Group (1993) The effect of intensive treatment of diabetes on the development and progression of long-term complications in insulin-dependent diabetes mellitus. N. Engl. J. Med. 329, 977–986 [DOI] [PubMed] [Google Scholar]
- 7. Van J. A. D., Scholey J. W., and Konvalinka A. (2017) Insights into Diabetic Kidney Disease Using Urinary Proteomics and Bioinformatics. J. Am. Soc. Nephrol. 28, 1050–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Schulz-Knappe P., Schrader M., and Zucht H.-D. (2005) The peptidomics concept. Comb. Chem. High Throughput Screen. 8, 697–704 [DOI] [PubMed] [Google Scholar]
- 9. Guerrero A., Dallas D. C., Contreras S., Chee S., Parker E. A., Sun X., Dimapasoc L., Barile D., German J. B., and Lebrilla C. B. (2014) Mechanistic peptidomics: factors that dictate specificity in the formation of endogenous peptides in human milk. Mol. Cell. Proteomics 13, 3343–3351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Thrailkill K. M., Clay Bunn R., and Fowlkes J. L. (2009) Matrix metalloproteinases: their potential role in the pathogenesis of diabetic nephropathy. Endocrine 35, 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Madhusudhan T., Kerlin B. A., and Isermann B. (2016) The emerging role of coagulation proteases in kidney disease. Nat. Rev. Nephrol. 12, 94–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cocchiaro P., De Pasquale V., Della Morte R., Tafuri S., Avallone L., Pizard A., Moles A., and Pavone L. M. (2017) The Multifaceted Role of the Lysosomal Protease Cathepsins in Kidney Disease. Front. Cell Dev. Biol. 5, 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Must A., and Anderson S. E. (2006) Body mass index in children and adolescents: considerations for population-based applications. Int. J. Obes. 30, 590–594 [DOI] [PubMed] [Google Scholar]
- 14. Yamamoto T. (2010) The 4th Human Kidney and Urine Proteome Project (HKUPP) workshop. 26 September 2009, Toronto, Canada. Proteomics 10, 2069–2070 [DOI] [PubMed] [Google Scholar]
- 15. Smith C. R., Batruch I., Bauça J. M., Kosanam H., Ridley J., Bernardini M. Q., Leung F., Diamandis E. P., and Kulasingam V. (2014) Deciphering the peptidome of urine from ovarian cancer patients and healthy controls. Clin. Proteomics 11, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Vizcaíno J. A., Deutsch E. W., Wang R., Csordas A., Reisinger F., Ríos D., Dianes J. A., Sun Z., Farrah T., Bandeira N., Binz P. A., Xenarios I., Eisenacher M., Mayer G., Gatto L., Campos A., Chalkley R. J., Kraus H. J., Albar J. P., Martinez-Bartolomé S., Apweiler R., Omenn G. S., Martens L., Jones A. R., and Hermjakob H. (2014) ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat. Biotechnol. 32, 223–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cox J., Neuhauser N., Michalski A., Scheltema R. A., Olsen J. V., and Mann M. (2011) Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10, 1794–1805 [DOI] [PubMed] [Google Scholar]
- 18. Tyanova S., Temu T., Sinitcyn P., Carlson A., Hein M. Y., Geiger T., Mann M., and Cox J. (2016) The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 13, 731–740 [DOI] [PubMed] [Google Scholar]
- 19. Navarro-González J. F., and Mora-Fernández C. (2008) The role of inflammatory cytokines in diabetic nephropathy. J. Am. Soc. Nephrol. 19, 433–442 [DOI] [PubMed] [Google Scholar]
- 20. Ruster C., and Wolf G. (2008) The Role of chemokines and chemokine receptors in diabetic nephropathy. Front. Biosci. 13, 944. [DOI] [PubMed] [Google Scholar]
- 21. Navarro-González J. F., Mora-Fernández C., Muros de Fuentes M., and García-Pérez J. (2011) Inflammatory molecules and pathways in the pathogenesis of diabetic nephropathy. Nat. Rev. Nephrol. 7, 327–340 [DOI] [PubMed] [Google Scholar]
- 22. Wolkow P. P., Niewczas M. A., Perkins B., Ficociello L. H., Lipinski B., Warram J. H., and Krolewski A. S. (2008) Association of urinary inflammatory markers and renal decline in microalbuminuric type 1 diabetics. J. Am. Soc. Nephrol. 19, 789–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mooney C., Haslam N. J., Pollastri G., and Shields D. C. (2012) Towards the improved discovery and design of functional peptides: common features of diverse classes permit generalized prediction of bioactivity. PLoS ONE 7, e45012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Klein J., Eales J., Zürbig P., Vlahou A., Mischak H., and Stevens R. (2013) Proteasix: a tool for automated and large-scale prediction of proteases involved in naturally occurring peptide generation. Proteomics 13, 1077–1082 [DOI] [PubMed] [Google Scholar]
- 25. Colaert N., Helsens K., Martens L., Vandekerckhove J., and Gevaert K. (2009) Improved visualization of protein consensus sequences by iceLogo. Nat. Methods 6, 786–787 [DOI] [PubMed] [Google Scholar]
- 26. Rawlings N. D., Barrett A. J., Thomas P. D., Huang X., Bateman A., and Finn R. D. (2018) The MEROPS database of proteolytic enzymes, their substrates and inhibitors in 2017 and a comparison with peptidases in the PANTHER database. Nucleic Acids Res. 46, D624–D632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rawlings N. D., and Barrett A. J. (1999) MEROPS: the peptidase database. Nucleic Acids Res. 27, 325–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pontén F., Jirström K., and Uhlen M. (2008) The Human Protein Atlas–a tool for pathology. J. Pathol. 216, 387–393 [DOI] [PubMed] [Google Scholar]
- 29. Gallien S., Kim S. Y., and Domon B. (2015) Large-scale targeted proteomics using internal standard triggered-parallel reaction monitoring (IS-PRM). Mol. Cell. Proteomics 14, 1630–1644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Iwaniak A., Minkiewicz P., Darewicz M., Sieniawski K., and Starowicz P. (2016) BIOPEP database of sensory peptides and amino acids. Food Res. Int. 85, 155–161 [DOI] [PubMed] [Google Scholar]
- 31. Thomas S., Karnik S., Barai R. S., Jayaraman V. K., and Idicula-Thomas S. (2010) CAMP: a useful resource for research on antimicrobial peptides. Nucleic Acids Res. 38, D774–D780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Liu F., Baggerman G., Schoofs L., and Wets G. (2008) The construction of a bioactive peptide database in Metazoa. J. Proteome Res. 7, 4119–4131 [DOI] [PubMed] [Google Scholar]
- 33. Newton K., and Dixit V. M. (2012) Signaling in Innate Immunity and Inflammation. Cold Spring Harb. Perspect. Biol. 4, a006049–a006049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gurley S. B., Ghosh S., Johnson S. A., Azushima K., Sakban R. B., George S. E., Maeda M., Meyer T. W., and Coffman T. M. (2018) Inflammation and immunity pathways regulate genetic susceptibility to diabetic nephropathy. Diabetes 67, 2096–2106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lin M., Yiu W. H., Wu H. J., Chan L. Y. Y., Leung J. C. K., Au W. S., Chan K. W., Lai K. N., and Tang S. C. W. (2012) Toll-like receptor 4 promotes tubular inflammation in diabetic nephropathy. J. Am. Soc. Nephrol. 23, 86–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Konvalinka A., Zhou J., Dimitromanolakis A., Drabovich A. P., Fang F., Gurley S., Coffman T., John R., Zhang S.-L., Diamandis E. P., and Scholey J. W. (2013) Determination of an angiotensin II-regulated proteome in primary human kidney cells by stable isotope labeling of amino acids in cell culture (SILAC). J. Biol. Chem. 288, 24834–24847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lodha S., Dani D., Mehta R., Bhaskaran M., Reddy K., Ding G., and Singhal P. C. (2002) Angiotensin II-induced mesangial cell apoptosis: role of oxidative stress. Mol. Med. 8, 830–840 [PMC free article] [PubMed] [Google Scholar]
- 38. Wolf G., Bohlender J., Bondeva T., Roger T., Thaiss F., and Wenzel U. O. (2006) Angiotensin II upregulates toll-like receptor 4 on mesangial cells. J. Am. Soc. Nephrol. 17, 1585–1593 [DOI] [PubMed] [Google Scholar]
- 39. Säemann M. D., Weichhart T., Zeyda M., Staffler G., Schunn M., Stuhlmeier K. M., Sobanov Y., Stulnig T. M., Akira S., von Gabain A., von Ahsen U., Hörl W. H., and Zlabinger G. J. (2005) Tamm-Horsfall glycoprotein links innate immune cell activation with adaptive immunity via a Toll-like receptor-4-dependent mechanism. J. Clin. Invest. 115, 468–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Darisipudi M. N., Thomasova D., Mulay S. R., Brech D., Noessner E., Liapis H., and Anders H.-J. (2012) Uromodulin triggers IL-1 -dependent innate immunity via the NLRP3 inflammasome. J. Am. Soc. Nephrol. 23, 1783–1789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pugin J., Schürer-Maly C. C., Leturcq D., Moriarty A., Ulevitch R. J., and Tobias P. S. (1993) Lipopolysaccharide activation of human endothelial and epithelial cells is mediated by lipopolysaccharide-binding protein and soluble CD14. Proc. Natl. Acad. Sci. U.S.A. 90, 2744–2748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Akira S., and Takeda K. (2004) Toll-like receptor signalling. Nat. Rev. Immunol. 4, 499–511 [DOI] [PubMed] [Google Scholar]
- 43. Bokhove M., Nishimura K., Brunati M., Han L., de Sanctis D., Rampoldi L., and Jovine L. (2016) A structured interdomain linker directs self-polymerization of human uromodulin. Proc. Natl. Acad. Sci. 113, 1552–1557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tokonami N., Takata T., Beyeler J., Ehrbar I., Yoshifuji A., Christensen E. I., Loffing J., Devuyst O., and Olinger E. G. (2018) Uromodulin is expressed in the distal convoluted tubule, where it is critical for regulation of the sodium chloride cotransporter NCC. Kidney Int. 94, 701–715 [DOI] [PubMed] [Google Scholar]
- 45. Zimmerhackl L. B., Pfleiderer S., Kinne R., Manz F., Schuler G., and Brandis M. (1991) Tamm-Horsfall-Protein excretion as a marker of ascending limb transport indicates early renal tubular damage in diabetes mellitus type I. J. Diabet. Complications 5, 112–114 [DOI] [PubMed] [Google Scholar]
- 46. Krochmal M., Kontostathi G., Magalhães P., Makridakis M., Klein J., Husi H., Leierer J., Mayer G., Bascands J.-L., Denis C., Zoidakis J., Zürbig P., Delles C., Schanstra J. P., Mischak H., and Vlahou A. (2017) Urinary peptidomics analysis reveals proteases involved in diabetic nephropathy. Sci. Rep. 7, 15160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Mary S., Small H. Y., Siwy J., Mullen W., Giri A., and Delles C. (2017) Polymerization-incompetent uromodulin in the pregnant stroke-prone spontaneously hypertensive RatNovelty and significance. Hypertension 69, 910–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Carty D. M., Siwy J., Brennand J. E., Zürbig P., Mullen W., Franke J., McCulloch J. W., North R. A., Chappell L. C., Mischak H., Poston L., Dominiczak A. F., Delles C., and Delles C. (2011) Urinary proteomics for prediction of preeclampsia. Hypertension 57, 561–569 [DOI] [PubMed] [Google Scholar]
- 49. Good D. M., Zurbig P., Argiles A., Bauer H. W., Behrens G., Coon J. J., Dakna M., Decramer S., Delles C., Dominiczak A. F., Ehrich J. H. H., Eitner F., Fliser D., Frommberger M., Ganser A., Girolami M. A., Golovko I., Gwinner W., Haubitz M., Herget-Rosenthal S., Jankowski J., Jahn H., Jerums G., Julian B. A., Kellmann M., Kliem V., Kolch W., Krolewski A. S., Luppi M., Massy Z., Melter M., Neususs C., Novak J., Peter K., Rossing K., Rupprecht H., Schanstra J. P., Schiffer E., Stolzenburg J. U., Tarnow L., Theodorescu D., Thongboonkerd V., Vanholder R., Weissinger E. M., Mischak H., and Schmitt-Kopplin P. (2010) Naturally occurring human urinary peptides for use in diagnosis of chronic kidney disease. Mol. Cell. proteomics MCP 9, 2424–2437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Pejchinovski M., Siwy J., Mullen W., Mischak H., Petri M. A., Burkly L. C., and Wei R. (2018) Urine peptidomic biomarkers for diagnosis of patients with systematic lupus erythematosus. Lupus 27, 6–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wu J., Wang N., Wang J., Xie Y., Li Y., Liang T., Wang J., Yin Z., He K., and Chen X. (2010) Identification of a uromodulin fragment for diagnosis of IgA nephropathy. Rapid Commun. Mass Spectrom. 24, 1971–1978 [DOI] [PubMed] [Google Scholar]
- 52. Ling X. B., Sigdel T. K., Lau K., Ying L., Lau I., Schilling J., and Sarwal M. M. (2010) Integrative urinary peptidomics in renal transplantation identifies biomarkers for acute rejection. J. Am. Soc. Nephrol. 21, 646–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. El-Achkar T. M., and Wu X.-R. (2012) Uromodulin in kidney injury: an instigator, bystander, or protector? Am. J. Kidney Dis. 59, 452–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Chambers R., Groufsky A., Hunt J. S., Lynn K. L., and McGiven A. R. (1986) Relationship of abnormal Tamm-Horsfall glycoprotein localization to renal morphology and function. Clin. Nephrol. 26, 21–26 [PubMed] [Google Scholar]
- 55. Schmid M., Prajczer S., Gruber L. N., Bertocchi C., Gandini R., Pfaller W., Jennings P., and Joannidis M. (2010) Uromodulin facilitates neutrophil migration across renal epithelial monolayers. Cell. Physiol. Biochem. 26, 311–318 [DOI] [PubMed] [Google Scholar]
- 56. Toma G., Bates J. M., and Kumar S. (1994) Uromodulin (Tamm-Horsfall protein) is a leukocyte adhesion molecule. Biochem. Biophys. Res. Commun. 200, 275–282 [DOI] [PubMed] [Google Scholar]
- 57. LaFavers K. A., Macedo E., Garimella P. S., Lima C., Khan S., Myslinski J., McClintick J., Witzmann F. A., Winfree S., Phillips C. L., Hato T., Dagher P. C., Wu X.-R., El-Achkar T. M., and Micanovic R. (2019) Circulating uromodulin inhibits systemic oxidative stress by inactivating the TRPM2 channel. Sci. Transl. Med. 11, eaaw3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Jennings P., Aydin S., Kotanko P., Lechner J., Lhotta K., Williams S., Thakker R. V., and Pfaller W. (2007) Membrane targeting and secretion of mutant uromodulin in familial juvenile hyperuricemic nephropathy. J. Am. Soc. Nephrol. 18, 264–273 [DOI] [PubMed] [Google Scholar]
- 59. Rindler M. J., Naik S. S., Li N., Hoops T. C., and Peraldi M. N. (1990) Uromodulin (Tamm-Horsfall glycoprotein/uromucoid) is a phosphatidylinositol-linked membrane protein. J. Biol. Chem. 265, 20784–20789 [PubMed] [Google Scholar]
- 60. Brunati M., Perucca S., Han L., Cattaneo A., Consolato F., Andolfo A., Schaeffer C., Olinger E., Peng J., Santambrogio S., Perrier R., Li S., Bokhove M., Bachi A., Hummler E., Devuyst O., Wu Q., Jovine L., and Rampoldi L. (2015) The serine protease hepsin mediates urinary secretion and polymerisation of Zona Pellucida domain protein uromodulin. Elife 4, e08887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Tokonami N., Takata T., Beyeler J., Ehrbar I., Yoshifuji A., Christensen E. I., Loffing J., Devuyst O., and Olinger E. G. (2018) Uromodulin is expressed in the distal convoluted tubule, where it is critical for regulation of the sodium chloride cotransporter NCC. Kidney Int. 94, 701–715 [DOI] [PubMed] [Google Scholar]
- 62. Olinger E., Lake J., Sheehan S., Schiano G., Takata T., Tokonami N., Debaix H., Consolato F., Rampoldi L., Korstanje R., and Devuyst O. (2019) Hepsin-mediated processing of uromodulin is crucial for salt-sensitivity and thick ascending limb homeostasis. Sci. Rep. 9, 12287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Tsuji A., Torres-Rosado A., Arai T., Le Beau M. M., Lemons R. S., Chou S. H., and Kurachi K. (1991) Hepsin, a cell membrane-associated protease. Characterization, tissue distribution, and gene localization. J. Biol. Chem. 266, 16948–16953 [PubMed] [Google Scholar]
- 64. Wilson P. C., Wu H., Kirita Y., Uchimura K., Ledru N., Rennke H. G., Welling P. A., Waikar S. S., and Humphreys B. D. (2019) The single-cell transcriptomic landscape of early human diabetic nephropathy. Proc. Natl. Acad. Sci. 116, 19619–19625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Wu H., Malone A. F., Donnelly E. L., Kirita Y., Uchimura K., Ramakrishnan S. M., Gaut J. P., and Humphreys B. D. (2018) Single-cell transcriptomics of a human kidney allograft biopsy specimen defines a diverse inflammatory response. J. Am. Soc. Nephrol. 29, 2069–2080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Torres-Rosado A., O'Shea K. S., Tsuji A., Chou S. H., and Kurachi K. (1993) Hepsin, a putative cell-surface serine protease, is required for mammalian cell growth. Proc. Natl. Acad. Sci. U.S.A. 90, 7181–7185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Kazama Y., Hamamoto T., Foster D. C., and Kisiel W. (1995) Hepsin, a putative membrane-associated serine protease, activates human factor VII and initiates a pathway of blood coagulation on the cell surface leading to thrombin formation. J. Biol. Chem. 270, 66–72 [DOI] [PubMed] [Google Scholar]
- 68. Wilkinson D. J., Desilets A., Lin H., Charlton S., del Carmen Arques M., Falconer A., Bullock C., Hsu Y.-C., Birchall K., Hawkins A., Thompson P., Ferrell W. R., Lockhart J., Plevin R., Zhang Y., Blain E., Lin S.-W., Leduc R., Milner J. M., and Rowan A. D. (2017) The serine proteinase hepsin is an activator of pro-matrix metalloproteinases: molecular mechanisms and implications for extracellular matrix turnover. Sci. Rep. 7, 16693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Prox J., Arnold P., and Becker-Pauly C. (2015) Meprin α and meprin β: Procollagen proteinases in health and disease. Matrix Biol. 44-46, 7–13 [DOI] [PubMed] [Google Scholar]
- 70. Yura R. E., Bradley S. G., Ramesh G., Reeves W. B., and Bond J. S. (2009) Meprin A metalloproteases enhance renal damage and bladder inflammation after LPS challenge. Am. J. Physiol. Renal Physiol. 296, F135–F144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Herzog C., Haun R. S., Kaushal V., Mayeux P. R., Shah S. V., and Kaushal G. P. (2009) Meprin A and meprin α generate biologically functional IL-1β from pro-IL-1β. Biochem. Biophys. Res. Commun. 379, 904–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Herzog C., Haun R. S., Ludwig A., Shah S. V., and Kaushal G. P. (2014) ADAM10 is the major sheddase responsible for the release of membrane-associated meprin A. J. Biol. Chem. 289, 13308–13322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Mathew M. M., Han N. V., Murugesan A., Raj E. A., and Prasanth K. G. (2015) Evaluation of the protective effect of Pterocarpus marsupium on acetic acid-induced ulcerative colitis in rats. Inflammopharmacology 23, 195–201 [DOI] [PubMed] [Google Scholar]
- 74. Mathew R., Futterweit S., Valderrama E., Tarectecan A. A., Bylander J. E., Bond J. S., and Trachtman H. (2005) Meprin-alpha in chronic diabetic nephropathy: interaction with the renin-angiotensin axis. Am. J. Physiol. Renal Physiol. 289, F911–F921 [DOI] [PubMed] [Google Scholar]
- 75. Bylander J. E., Ahmed F., Conley S. M., Mwiza J.-M., and Ongeri E. M. (2017) Meprin metalloprotease deficiency associated with higher mortality rates and more severe diabetic kidney injury in mice with STZ-induced type 1 diabetes. J. Diabetes Res. 2017, 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Cao L., Sedighi R., Boston A., Premadasa L., Pinder J., Crawford G. E., Jegede O. E., Harrison S. H., Newman R. H., and Ongeri E. M. (2018) Undiagnosed kidney injury in uninsured and underinsured diabetic african american men and putative role of meprin metalloproteases in diabetic nephropathy. Int. J. Nephrol. 2018, 6753489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Conley S. M., Abais J. M., Boini K. M., and Li P.-L. (2017) Inflammasome activation in chronic glomerular diseases. Curr. Drug Targets 18, 1019–1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Anders H.-J., and Muruve D. A. (2011) The inflammasomes in kidney disease. J. Am. Soc. Nephrol. 22, 1007–1018 [DOI] [PubMed] [Google Scholar]
- 79. Liu D., Wen Y., Tang T.-T., Lv L.-L., Tang R.-N., Liu H., Ma K.-L., Crowley S. D., and Liu B.-C. (2015) Megalin/cubulin-lysosome-mediated albumin reabsorption is involved in the tubular cell activation of NLRP3 inflammasome and tubulointerstitial inflammation. J. Biol. Chem. 290, 18018–18028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Larionov A., Dahlke E., Kunke M., Zanon Rodriguez L., Schiessl I. M., Magnin J.-L., Kern U., Alli A. A., Mollet G., Schilling O., Castrop H., and Theilig F. (2019) Cathepsin B increases ENaC activity leading to hypertension early in nephrotic syndrome. J. Cell. Mol. Med., 23, 6543–6553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Liu W. J., Shen T. T., Chen R. H., Wu H.-L., Wang Y. J., Deng J. K., Chen Q. H., Pan Q., Huang Fu C., Tao J., Liang D., and Liu H. (2015) Autophagy-lysosome pathway in renal tubular epithelial cells is disrupted by advanced glycation end products in diabetic nephropathy. J. Biol. Chem. 290, 20499–20510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Peres G. B., Juliano M. A., Simões M. J., and Michelacci Y. M. (2013) Lysosomal enzymes are decreased in the kidney of diabetic rats. Biochim. Biophys. Acta 1832, 85–95 [DOI] [PubMed] [Google Scholar]
- 83. Höhne M., Frese C. K., Grahammer F., Dafinger C., Ciarimboli G., Butt L., Binz J., Hackl M. J., Rahmatollahi M., Kann M., Schneider S., Altintas M. M., Schermer B., Reinheckel T., Göbel H., Reiser J., Huber T. B., Kramann R., Seeger-Nukpezah T., Liebau M. C., Beck B. B., Benzing T., Beyer A., and Rinschen M. M. (2018) Single-nephron proteomes connect morphology and function in proteinuric kidney disease. Kidney Int. 93, 1308–1319 [DOI] [PubMed] [Google Scholar]
- 84. Suh M.-J., Tovchigrechko A., Thovarai V., Rolfe M. A., Torralba M. G., Wang J., Adkins J. N., Webb-Robertson B.-J. M., Osborne W., Cogen F. R., Kaplowitz P. B., Metz T. O., Nelson K. E., Madupu R., and Pieper R. (2015) Quantitative differences in the urinary proteome of siblings discordant for type 1 diabetes include lysosomal enzymes. J. Proteome Res. 14, 3123–3135 [DOI] [PubMed] [Google Scholar]
- 85. Magagnotti C., Zerbini G., Fermo I., Carletti R. M., Bonfanti R., Vallone F., and Andolfo A. (2019) Identification of nephropathy predictors in urine from children with a recent diagnosis of type 1 diabetes. J. Proteomics 193, 205–216 [DOI] [PubMed] [Google Scholar]
- 86. Brinkmann V., Reichard U., Goosmann C., Fauler B., Uhlemann Y., Weiss D. S., Weinrauch Y., and Zychlinsky A. (2004) Neutrophil extracellular traps kill bacteria. Science 303, 1532–1535 [DOI] [PubMed] [Google Scholar]
- 87. Wong S. L., Demers M., Martinod K., Gallant M., Wang Y., Goldfine A. B., Kahn C. R., and Wagner D. D. (2015) Diabetes primes neutrophils to undergo NETosis, which impairs wound healing. Nat. Med. 21, 815–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Takahashi T., Hato F., Yamane T., Inaba M., Okuno Y., Nishizawa Y., and Kitagawa S. (2000) Increased spontaneous adherence of neutrophils from type 2 diabetic patients with overt proteinuria: possible role of the progression of diabetic nephropathy. Diabetes Care 23, 417–418 [DOI] [PubMed] [Google Scholar]
- 89. Galkina E., and Ley K. (2006) Leukocyte recruitment and vascular injury in diabetic nephropathy. J. Am. Soc. Nephrol. 17, 368–377 [DOI] [PubMed] [Google Scholar]
- 90. Fardon N. J. M., Wilkinson R., and Thomas T. H. (2002) Abnormalities in primary granule exocytosis in neutrophils from Type I diabetic patients with nephropathy. Clin. Sci. 102, 69–75 [PubMed] [Google Scholar]
- 91. Davis J. O., Hartroft P. M., Titus E. O., Carpenter C. C. J., Ayers C. R., and Spiegel H. E. (1962) The role of the renin-angiotensin system in the control of aldosterone secretion. J. Clin. Invest. 41, 378–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Nehme A., Cerutti C., Dhaouadi N., Gustin M. P., Courand P.-Y., Zibara K., and Bricca G. (2015) Atlas of tissue renin-angiotensin-aldosterone system in human: A transcriptomic meta-analysis. Sci. Rep. 5, 10035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Velez J. C. Q., Ryan K. J., Harbeson C. E., Bland A. M., Budisavljevic M. N., Arthur J. M., Fitzgibbon W. R., Raymond J. R., and Janech M. G. (2009) Angiotensin I is largely converted to angiotensin (1–7) and angiotensin (2–10) by isolated rat glomeruli. Hypertension 53, 790–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Domenig O., Manzel A., Grobe N., Königshausen E., Kaltenecker C. C., Kovarik J. J., Stegbauer J., Gurley S. B., van Oyen D., Antlanger M., Bader M., Motta-Santos D., Santos R. A., Elased K. M., Säemann M. D., Linker R. A., and Poglitsch M. (2016) Neprilysin is a mediator of alternative renin-angiotensin-system activation in the murine and human kidney. Sci. Rep. 6, 33678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Brar G. S., Barrow B. M., Watson M., Griesbach R., Choung E., Welch A., Ruzsicska B., Raleigh D. P., and Zraika S. (2017) Neprilysin is required for angiotensin-(1–7)'s ability to enhance insulin secretion via its proteolytic activity to generate angiotensin-(1–2). Diabetes 66, 2201–2212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Pereira R. M., dos Santos R. A. S., Teixeira M. M., Leite V. H. R., Costa L. P., da Costa Dias F. L., Barcelos L. S., Collares G. B., and Simões e Silva A. C. (2007) The renin–angiotensin system in a rat model of hepatic fibrosis: Evidence for a protective role of Angiotensin-(1–7). J. Hepatol. 46, 674–681 [DOI] [PubMed] [Google Scholar]
- 97. Su Z., Zimpelmann J., and Burns K. D. (2006) Angiotensin-(1–7) inhibits angiotensin II-stimulated phosphorylation of MAP kinases in proximal tubular cells. Kidney Int. 69, 2212–2218 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
For discovery peptidomics, mass spectrometry data have been deposited onto the ProteomeXchange Consortium via the PRIDE (16) partner repository with the dataset identifier PXD012210 (http://www.ebi.ac.uk/pride/archive/login). For targeted peptidomics, Skyline files and raw data have been deposited in Panorama Public with a ProteomeXchange identifier PXD012389 (https://panoramaweb.org/lotso19.url).