SUMMARY
Nucleotide deprivation and imbalance present detrimental conditions for animals and are thus expected to trigger cellular responses that direct protective changes in metabolic, developmental, and behavioral programs, albeit such mechanisms are vastly underexplored. Following our previous finding that Caenorhabditis elegans shut down germ cell proliferation in response to pyrimidine deprivation, we find in this study that endonuclease ENDU-2 regulates nucleotide metabolism and germ cell proliferation in response to nucleotide imbalance and other genotoxic stress, and that it affects mitotic chromosomal segregation in the intestine and lifespan. ENDU-2 expression is induced by nucleotide imbalance and genotoxic stress, and ENDU-2 exerts its function in the intestine, mostly by inhibiting the phosphorylation of CTPS-1 through repressing the PKA pathway and histone deacetylase HDA-1. Human EndoU also affects the response to genotoxic drugs. Our work reveals an unknown role of ENDU-2 in regulating nucleotide metabolism and animals’ response to genotoxic stress, which may link EndoU function to cancer treatment.
In Brief
Jia et al. show that an endonuclease plays a critical role in a nucleotide response system in nematodes and possibly mammalian cells. It affects nucleotide metabolism and reproductive development in response to nucleotide imbalance or other stresses, and it does so by inhibiting the phosphorylation of CTP synthase in the gut.
Graphical Abstract
INTRODUCTION
Nucleotides (NTs) are building blocks for DNAs and RNAs and indispensable for many important cellular processes, such as DNA damage response and energy production. Both the absolute levels and the balance between different NTs are critical for genome stability (Pai and Kearsey, 2017). Therefore, to survive the fluctuation in NT levels due to changes in diet or internal NT metabolic conditions, animals are expected to have evolved cellular mechanisms that sense and respond to NT level changes. Although there have been extensive studies regarding the impact of nucleotide imbalance on cell-cycle progression, DNA replication, and genome stability mainly using cultured cells or yeast (Pai and Kearsey, 2017), the mechanisms that regulate developmental and metabolic events in animal models in response to NT changes are underexplored.
We previously tested the hypothesis that animals likely have complex signaling systems that sense the level and/or balance of NTs to regulate germline proliferation, in which rapid DNA replication and RNA synthesis may demand healthy NT pools (Chi et al., 2016). We showed that the commonly used Caenorhabditis elegans food, Escherichia coli OP50 strain, provides less dietary pyrimidine than some other E. coli strains, such as the commonly used E. coli HT115 strain that has altered nucleotide metabolism (Chi et al., 2016). Cytidine deaminases CDD-1 and CDD-2 catalyze the conversion of cytidine NTs to uridine NTs. The combination of a CDD double-knockout mutant worm and a diet of E. coli OP50 leads to a significantly reduced thymidine pool, which causes a completely sterile phenotype due to the arrest of mitotic germline proliferation (Chi et al., 2016). Further analysis also implicates a likely nucleotide-sensing system in the intestine that regulates not only mitotic germline proliferation but also NT metabolism when animals encounter genotoxic stress due to NT deficiency or imbalance.
In this study, we isolated an endu-2 mutant from a NT-deficiency-related genetic screen and identified the ENDU-2 nuclease as a key factor in a system that regulates NT metabolism and germline proliferation in response to changes in NT levels and genotoxic stresses. ENDU-2 exerts its role in regulating nucleotide metabolism by promoting the activity of cytidine triphosphate (CTP) synthase CTPS-1 by controlling the phosphorylation of CTPS-1. We report the analysis of this regulatory pathway and evidence for the likely conservation of the function in mammals.
RESULTS
A Forward Genetic Screen Identified the Role of Intestinal ENDU-2 in Regulating Nucleotide Metabolism
To identify genes involved in the sensing and regulation of NT homeostasis, we performed an ethyl methane sulfonate (EMS)-induced mutagenesis screen for suppressors of the sterile phenotype of the cdd-1/2 double-knockout (cddko) mutant fed the E. coli OP50 strain. One of the suppressors we isolated, ku544, effectively suppressed the sterile phenotype (Figure 1A) and fully restored the germline morphology of the double mutant (100% wild type [WT], n = 200) (Figure S1A) (Chi et al., 2016). We were not able to test the brood size as both cddko worms fed with pyrimidine-rich cytR− bacteria and the suppressor strain showed severe egg-laying defects (Figures S1B and S1C). We thus indirectly examined fecundity by counting the eggs in adult animals. Our previous study showed feeding with high pyrimidine food (cytR− bacteria) strongly suppressed the germline proliferation defects of cddko animals (Chi et al., 2016). We found that adult cddko; ku544 animals contained a similar amount of eggs as adult cddko animals fed cytR− bacteria (4-day-old cddko; ku544 animals on OP50, 26 ± 2.7 eggs per worm; 4-day-old cddko on cytR−, 26.8 ± 4.6 eggs per worm; n = 9 each; p = 0.67, unpaired, two-tailed Student’s t test). These results indicate that ku544 is a strong suppressor of germline defects in cddko animals.
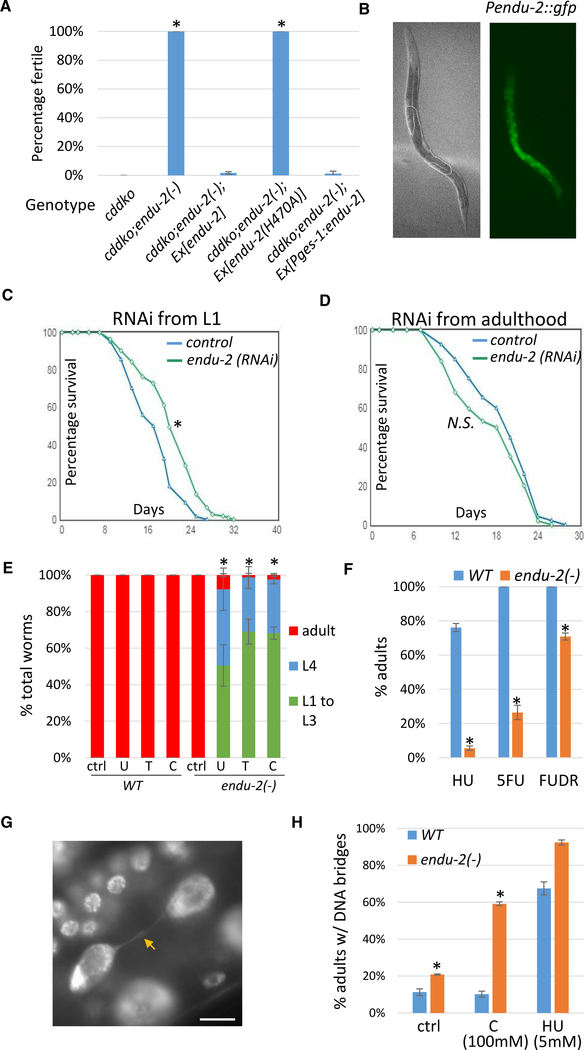
Figure 1. Mutating endu-2 in C. elegans Suppresses NT Deficiency-Induced Germline Proliferation Arrest and Renders Worms Hypersensitive to NT Supplementation and Genotoxic Drugs.
(A) Results of fertility test of worms fed low-pyrimidine food (E. coli OP50). Loss of endu-2 gene function suppressed sterility caused by cdd-1(−);cdd-2(−) double knockout (cddko). A transgene of wild-type (WT) endu-2, but not the endu-2(H470A) mutant gene, fully reversed the suppression. Expression of endu-2 specifically in the intestine using the Pges-1 promoter also fully reversed the suppression. The endu-2(−) allele is ku544. “Ex” indicates extrachromosomal transgenes. Error bars, standard deviation. Asterisks indicate significant differences. Tukey’s range test, family-wise error rate (FWER) = 0.01; see Method Details for details. The suppression of the germline morphology defect is described in the text and Figure S1A.
(B) Bright-field and fluorescence images showing the expression of a single-copy insertion of the Pendu-2::gfp reporter in the intestine. Dotted lines outline the distal region of the germline.
(C and D) Lifespan assays of worms with indicated RNAi treatment from L1 (C) or adult (D). *p < 0.01; N.S., not significant (p > 0.05), log-rank test. See text for details.
(E and F) High-level single-NT supplementation or genotoxic drugs caused severe developmental delay in endu-2(−) animals. WT or endu-2(−) worms (150–250 per replicate) were supplemented with 100 mM NTs uridine (U), thymidine (T), or cytidine (C) in (E), or hydroxyurea (HU) (10 mM), 5-FU (50 μM) or FUDR (50 μM) in (F).
(G) Fluorescence image showing the DNA bridge phenotype in a DAPI-stained worm intestine. The arrow points to the bridge connecting 2 daughter nuclei. Scale bar represents 10 μm.
(H) Percentage of adult worms having DNA bridges in the intestine.
(E, F, and H) Error bars, standard deviation. Asterisks indicate significant differences. Student’s t test between WT and endu-2(−) with the same treatment, unpaired, two-tailed, with Bonferroni correction, α = 0.0018 (E), 0.0033 (F and H); see Method Details for details.
Unless noted specifically, 50–100 worms were examined in each replicate, with at least 2 replicates for each experiment.
Genetic mapping and genome sequencing determined that ku544 is a C-to-T mutation in the endu-2 gene, which encodes a homolog of EndoU (endoribonuclease specific for uridylate) that has been linked to diverse physiological functions in multiple organisms (see Discussion). An alignment between ENDU-2 and its homolog in Xenopus and humans (Laneve et al., 2008; Renzi et al., 2006) demonstrated that all of the amino acids that are critical for its enzymatic activity are conserved in C. elegans (Figure S1D). Our ku544 mutation results in a T568I change in the ENDU-2 protein (Figure S1D). Mutating the corresponding threonine in XendoU (T278) caused an ~80% reduction in nuclease activity in an in vitro assay (Renzi et al., 2006), suggesting that the suppressor phenotype of endu-2(ku544/T568I) is likely due to a disruption of the nuclease activity of the enzyme. To provide further support for this notion, we created an endu-2(H470A) mutation, which was predicted to abolish the active site (Renzi et al., 2006) (Figure S1D). Consistently, while expressing a WT endu-2 transgene (Ex[endu-2]) in the cddko; endu-2(T568I) triple mutant reversed the suppressing effect of endu-2(T568I), a mutant endu-2(H470A) transgene (Ex[endu-2(H470A)]) failed to reverse the suppression (Figure 1A). Therefore, the loss of ENDU-2 nuclease activity recovers fertility that was shut down in C. elegans cddko worms fed low-pyrimidine food. In other words, WT ENDU-2 is critically involved in animals’ response to a deficiency in the pyrimidine pool. To simplify the presentation, we hereafter refer to the endu-2(ku544/T586I) allele as endu-2(−).
To examine the expression of endu-2, we fused its promoter region (8 kb upstream of the start codon) to gfp and made a single-copy integration line using the CRISPR-Cas9 technique, which permits the examination of potential germline expression that is frequently silenced in animals with multi-copy transgenes (Kelly et al., 1997). Strong expression was detected in the intestine (Figure 1B), which is consistent with a previous analysis using a multi-copy translational fusion GFP reporter (Ujisawa et al., 2018). However, no detectable fluorescence was seen in the germline. More important, expressing endu-2 specifically in the intestine of the cddko; endu-2(−) triple mutant using an intestine-specific promoter (Pges-1) reversed the rescue (Figure 1A), suggesting that ENDU-2 activity in the gut is sufficient to execute its role in germline proliferation arrest in the cddko mutant.
RNAi of endu-2 during Development Extends the Lifespan
We observed that endu-2 RNAi caused worms to lay eggs for an extended time, an indicator of increased lifespan (Huang et al., 2004). Further tests confirmed that endu-2 RNAi increased the lifespan of worms by ~20% (mean lifespan, control: 17.18 days, endu-2 RNAi: 20.48 days, p = 3.20E−06, log rank test) (Figure 1C). This effect is likely established during development, as the same RNAi treatment starting from adulthood did not enhance the lifespan (mean lifespan, control: 19.10 days, endu-2 RNAi: 17.45 days, p = 0.0677, log rank test) (Figure 1D). Both L1 and adult RNAi eliminated ENDU-2 protein expression (Figure S1E). These results are consistent with the observations that gene activities in innate immunity or short-term responses to pathogens or stress may compromise long-term survival or lifespan (Kudlow et al., 2012; Zhang et al., 2013).
endu-2(−) Worms Are Hypersensitive to NT Supplementation and Genotoxic Drugs
We next investigated whether endu-2(−) worms have altered responses to changes in pyrimidine levels. endu-2(−) worms showed no obvious defects when supplemented with low concentrations of uridine (U), thymidine (T), or cytidine (C) (Figure S1F). However, they showed significant growth delays compared to WT worms receiving high concentrations of pyrimidine supplements (Figure 1E), suggesting that endu-2(−) animals are sensitive to exogenous pyrimidines at high levels.
Next, we tested the sensitivity of endu-2(−) worms to hydroxyurea (HU). HU inhibits deoxyribonucleotide triphosphate (dNTP) biosynthesis, diminishes dNTP pools, and stalls DNA replication. We reasoned that since endu-2(−) affects pyrimidine metabolism (see below), it may be more sensitive to the dNTP pool reduction caused by HU. endu-2(−) worms were hypersensitive to HU treatment (Figure 1F). Fluorouracil (5-FU) and floxuridine (FUDR) are pyrimidine analogs that inhibit the biosynthesis of deoxythymidine triphosphate (dTTP). endu-2(−) worms also showed strong sensitivity to 5-FU and modest sensitivity to FUDR (Figure 1F). endu-2(−) worms, however, did not show increased sensitivity to UV treatment (Figure S1G), which supports the notion that endu-2(−) does not cause general sickness. Therefore, ENDU-2 is critically involved in animals’ response to genotoxicity.
ENDU-2 Is Necessary for the Integrity of Chromosome Segregation during Mitosis in the Intestine
To better understand the hypersensitivity observed in endu-2(−) worms treated with NTs or genotoxic drugs, we examined their nuclear morphology. DAPI staining of endu-2(−) worms revealed an increased incidence of DNA “bridges” (Bembenek et al., 2013) (Figure 1G) in the intestine, compared to WT (Figure 1H). This defect was significantly enhanced in NT-treated worms. When endu-2(−) worms were supplemented with 100 mM cytidine, nearly 60% of the worms showed the DNA bridge defect (Figure 1H). The DNA bridge defects were also slightly enhanced in endu-2(−) worms treated with HU (Figure 1H). The occurrence of DNA bridges is an indicator of chromosomal damage and has been shown to be caused by mutations in DNA damage response genes in C. elegans (Kalogeropoulos et al., 2004; Kniazeva and Ruvkun, 2019). Our results suggest that ENDU-2 is necessary for successful mitosis in many intestinal cells, which is consistent with a role for ENDU-2 in the DNA damage response.
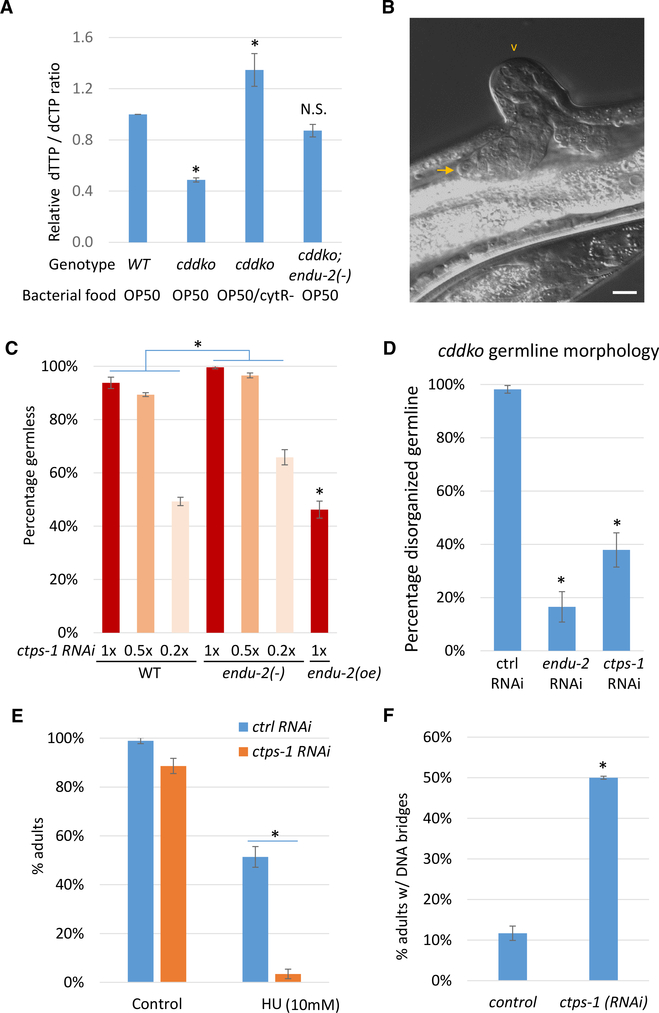
endu-2 Interacts with ctps-1 to Regulate NT Homeostasis
cddko worms had a significantly reduced dTTP:dCTP ratio in the NT pool that was recovered when fed pyrimidine-rich cytR− bacteria (Chi et al., 2016) (Figure 2A). The endu-2(−) mutation significantly restored the dTTP:dCTP ratio in the cddko background (Figure 2A), suggesting that ENDU-2 affects pyrimidine metabolism, which may be largely responsible for the suppression of sterility. In contrast, the endu-2(−) single mutant did not cause a significant change in the dTTP:dCTP ratio (Figure S2A), suggesting that ENDU-2 significantly affects relevant NT metabolic pathways only when NT metabolism is disturbed. To identify factors that act downstream of ENDU-2 to mediate its role in regulating pyrimidine metabolism, we carried out a candidate RNAi screen of pyrimidine metabolism genes (Figure S2B) and discovered a genetic interaction between endu-2 and ctps-1. ctps-1 encodes a homolog of CTPS that catalyzes the conversion of uridine triphosphate (UTP) to CTP (Figure S2B). CTPS has critical roles in NT metabolism (see Discussion). When WT worms were treated with ctps-1 RNAi, they showed a strong “germless” phenotype (>93%; Figures 2B and 2C). These “germless” worms were sterile, with very small germlines that contained a few undifferentiated germ cells (Figure 2B). Reducing the strength of RNAi by diluting ctps-1(RNAi) 5-fold with control (HT115) RNAi bacteria led to a reduction in the percentage of germless worms (~50%; Figure 2C). The endu-2(−) mutation enhanced the phenotype by diluted ctps-1(RNAi) as ctps-1(RNAi); endu-2(−) worms produced a higher percentage of germless worms than that caused by ctps-1(RNAi) alone. Conversely, endu-2 overexpression (oe) had a suppression effect on diluted ctps-1(RNAi), as ctps-1(RNAi); endu-2(oe) worms produced lower percentages of germless worms than that caused by ctps-1(RNAi) alone (Figure 2C). These data, including the similarity in loss-of-function phenotypes and phenotype enhancements, suggest that endu-2 and ctps-1 may act either in the same pathway or in parallel pathways to affect germline development and genotoxic responses.
Figure 2. endu-2 Acts with ctps-1 to Regulate NT Homeostasis.
(A) Bar graphs showing that the low dTTP:dCTP ratio in cddko mutants is significantly suppressed by endu-2(−). The cytR− mutation in E. coli alters NT metabolism and increases the pyrimidine level (Chi et al., 2016). Error bars, standard deviation. Asterisks indicate significant differences. N.S., not significant. Tukey’s range test, FWER = 0.01.
(B) A representative image of the “germless” phenotype of ctps-1(RNAi). The arrow points to the tip of the small germline. The arrowhead points to a commonly observed ventral protrusion of the vulva. The scale bar represents 10 μm.
(C) Percentage of worms showing the germless phenotype on varying dilutions of ctps-1 RNAi. RNAi dilution was done by mixing ctps-1(RNAi) bacteria with control RNAi bacteria. While endu-2(−) enhanced the defect of ctps-1(RNAi), endu-2 overexpression (oe) suppressed the defect. n = 2 (~200 worms each). Error bars, standard deviation. Asterisks indicate significant differences. Two-way ANOVA test was used to compare the response of WT and endu-2(−) to the serial dilution of ctps-1 RNAi. Two-tailed, unpaired Student’s t test was used to compare the response of WT and endu-2(oe) to undiluted ctps-1 RNAi. Bonferroni correction was applied, with α = 0.0024.
(D) Percentage of cddko worms showing disorganized germlines. RNAi of either endu-2 or ctps-1 suppressed the germline defect in cddko worms. Equal concentrations of RNAi bacteria (pyrimidine rich) and OP50 (pyrimidine poor) were mixed together and fed to worms to create a sensitized background.
(E) Percentage of worms reaching adulthood after 3 days with or without HU treatment, indicating that ctps-1(RNAi) worms are highly sensitive to HU.
(F) ctps-1(RNAi) adults had an increased number of DNA bridges in their intestines. Error bars in (D)–(F) represent standard deviation. Asterisks indicate significant differences.
In (D), Tukey’s range test, FWER = 0.01. In (E) and (F), Student’s t test, unpaired, two-tailed. Bonferroni correction was applied in €. α = 0.008 (E) and 0.01(F).
While E. coli OP50 is a pyrimidine-poor food that is causal to the sterility of cddko worms, E. coli HT115 (the common RNAi library parent strain) is a pyrimidine-rich food that fully suppresses the sterility of the cddko mutant (Chi et al., 2016). To create a more sensitive condition to observe the function of ctps-1, we diluted HT115 RNAi bacteria with E. coli OP50. cddko worms fed this OP50-diluted RNAi food had disorganized germlines due to the reduction in pyrimidine level (Figure S2C). Both OP50-diluted endu-2(RNAi) and OP50-diluted ctps-1(RNAi) partially rescued the germline defects in cddko worms (Figure 2D), suggesting that endu-2 and ctps-1 may function in the same pathway. Consistent with this notion, ctps-1(RNAi) also sensitized worms to HU treatment (Figure 2E) and the increased incidence of DNA bridges (Figure 2F). Moreover, the endu-2(−) mutation further enhances the sensitivity of ctps-1(RNAi) worms to HU treatment (Figure S2D), supporting the collaborative relation between the two proteins.
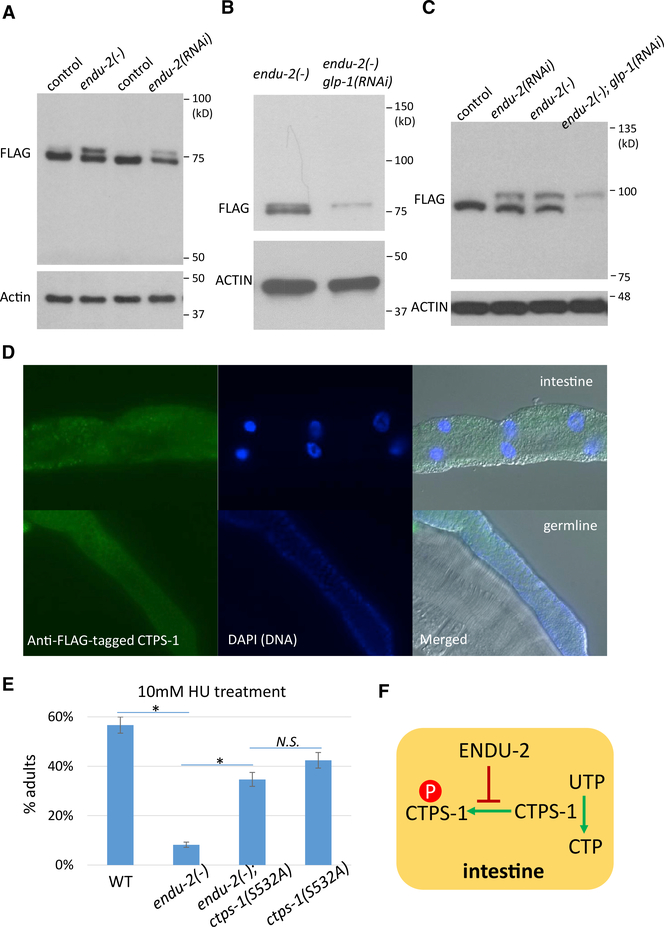
ENDU-2 Regulates CTPS-1 Phosphorylation in the Intestine
We then further investigated the functional relation between ENDU-2 and CTPS-1. To examine the potential regulation at the level of translation and/or post-translation, we inserted a FLAG tag to the C terminus of the endogenous ctps-1 locus. Western blot analysis showed that CTPS-1 produced a single strong band in WT worms. However, in endu-2(−) or endu-2(RNAi) worms, one extra band that migrated slightly slower was obvious (Figure 3A).
Figure 3. ENDU-2 Negatively Regulates CTPS-1 Phosphorylation in the Intestine.
(A) Western blot of FLAG::CTPS-1 in samples from indicated strains. Two bands were observed in both endu-2(−) and endu-2(RNAi) worms. The flag::ctps-1 background is included in all of the experiments for the detection of CTPS-1 (see Method Details). Samples were run on a 7.5% SDS-PAGE gel.
(B and C) Western blot of FLAG::CTPS-1 in endu-2(−) mutant worms treated with control or glp-1 RNAi that eliminates the germline. Samples were run on a 12% SDS-PAGE gel (B) or a 12.5% phos-tag gel (C) (see Method Details).
(D) Immunostaining of FLAG::CTPS-1 showed the expression of a FLAG-tagged CTPS-1 in the intestine (top panels) and the germline (bottom panels). See Figure S3A for negative controls.
(E) Disrupting the predicted phosphorylation site of CTPS-1 significantly suppressed the hypersensitivity of endu-2(−) animals to HU treatment. Error bars indicate standard deviation. Asterisks indicate significant differences, N.S., not significant. Tukey’s range test, FWER = 0.01.
(F) A model based on the genetic interaction and biochemical assays that show that ENDU-2 positively regulates CTPS-1 by blocking its phosphorylation.
The extra band could represent a splicing variant of ctps-1 or a modified CTPS-1 protein. Evaluation with a phos-tag gel, in which phosphorylated proteins migrate slower through the matrix, increased the separation between the two bands, compared to a regular PAGE gel of the same concentration (Figures 3B and 3C), suggesting that the higher-molecular-weight band that prominently appeared in the endu-2(−) lysate may represent a phosphorylated form of CTPS-1. The phosphorylation of CTPS-1 may be repressed by ENDU-2 in WT worms.
Whole-worm immunostaining demonstrated that CTPS-1 is expressed predominantly in the intestine and germline (Figures 3D and S3A). This observation raised an important question: in which of these two tissues is CTPS-1 modified? We treated endu-2(−) worms with glp-1(RNAi) to eliminate the germline and found that endu-2(−); glp-1(RNAi) worms only produced the larger-molecular-weight band (Figure 3C), indicating that the modification of CTPS-1 likely occurs in the intestine. Similar results were obtained when we tested endu-2(−); glp-1(e2141) double-mutant worms (Figure S3D). glp-1(e2141) is a temperature-sensitive mutant that is germless at 25°C (Kodoyianni et al., 1992). Given that endu-2 is strongly expressed in the intestine but not in the germline (Figure 1B), ENDU-2 likely represses the CTPS-1 modification locally in the intestine.
To characterize the endu-2-dependent modification of CTPS-1, we pulled down CTPS-1 proteins from WT endu-2(−); glp-1(RNAi) worms and endu-2(oe) worms and subjected the samples to mass spectrometry analysis. The search for post-translational modifications identified phosphorylations but not acetylation or ubiquitination of CTPS-1 (Figure S3B). Among the 3 identified phosphorylation sites, only phosphorylation of S532 was clearly repressed by the ENDU-2 activity (Figure S3B). Furthermore, we created the “phospho-dead” form of CTPS-1 in endu-2(−) mutants by introducing an S532A mutation. This mutation was found to suppress the hypersensitivity of endu-2(−) to HU treatment (Figure 3E) and the increased incidence of DNA bridges in endu-2(−) (Figure S3C), supporting the idea that the phosphorylation of CTPS-1 at S532 largely mediates the function of ENDU-2. Because endu-2(−) worms have phenotypes similar to those of ctps-1(RNAi) (suppression of cddko germline defects, hypersensitive to HU treatment, and increased number of intestine DNA bridges) (Figures 1 and 2), the above data suggest that ENDU-2 may regulate CTPS-1 by repressing CTPS-1 phosphorylation, which in turn represses the CTPS activity (Figure 3F).
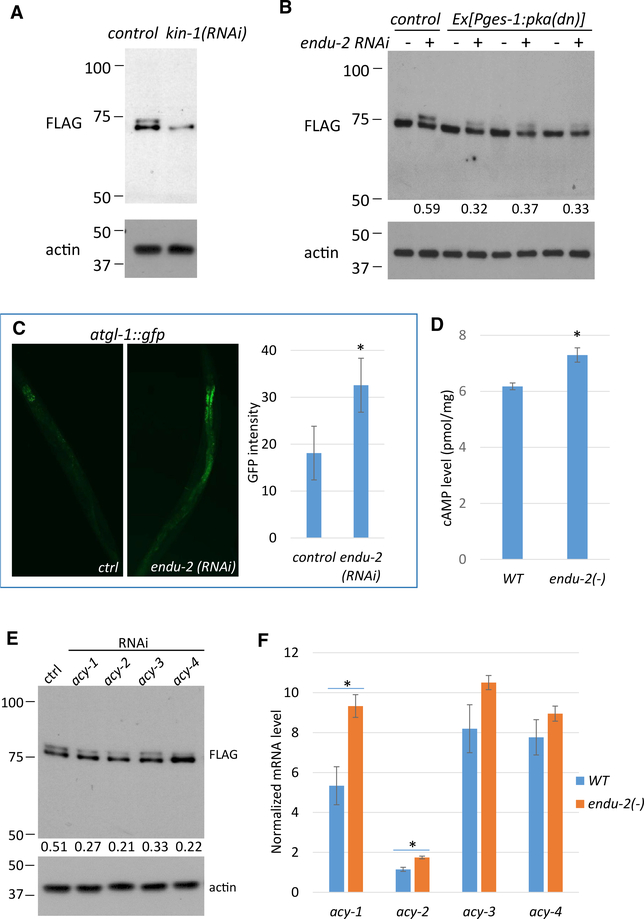
ENDU-2 Regulates CTPS-1 Phosphorylation through the PKA Pathway in the Intestine
To identify the kinase responsible for CTPS-1 phosphorylation, we applied the sequence surrounding S532 to search for potential kinases based on motif/kinase predictions (PhosphoMotif Finder; Amanchy et al., 2007) (Figure S4A) and then performed an RNAi screen of candidate kinase genes. When endu-2(−) worms were treated with kin-1 RNAi, the upper band of CTPS-1 disappeared (Figures 4A and S4B), indicating that kin-1 is required for CTPS-1 phosphorylation. kin-1 encodes the sole catalytic subunit of protein kinase A (PKA), and kin-1 RNAi eliminates PKA activity in C. elegans (Wang and Sieburth, 2013). In line with the local effect of ENDU-2, expressing dominant-negative PKA (Wang and Sieburth, 2013) specifically in the intestine, led to a significant reduction in CTPS-1 phosphorylation (Figure 4B).
Figure 4. ENDU-2 Regulates CTPS-1 Phosphorylation through the PKA Pathway in the Intestine.
(A) Western blot of the FLAG-tagged CTPS-1 protein. kin-1 RNAi eliminates CTPS-1 phosphorylation.
(B) Expressing dominant-negative PKA in the intestine reduced CTPS-1 phosphorylation. Numbers below the blot indicate the relative signal intensity between the upper and lower CTPS-1 band in each sample.
(C) GFP fluorescence image and quantitative data for the atgl-1::gfp reporter. Expression increased in endu-2(RNAi) animals.
(D) The whole-worm cAMP level was increased in endu-2(−) mutants.
(E) RNAi of acy genes reduced CTPS-1 phosphorylation. Numbers show the relative signal intensity between the upper and lower CTPS-1 bands in each sample.
(F) qPCR results for mRNA levels of the acy genes in WT or endu-2(−) worms. Error bars indicate standard deviation and asterisks indicate significant differences in all panels.
For (C) and (D), Student’s t test, unpaired, two-tailed, α = 0.01. For (F), Student’s t test, unpaired, 2-tailed with Bonferroni correction, α = 0.0018.
We then asked whether PKA mediates the regulation of ENDU-2 on CTPS-1 phosphorylation. ATGL-1 is a direct target of intestinal PKA, and an atgl-1::gfp reporter strain has been used previously as a reporter of intestinal PKA activity (Lee et al., 2014). We treated the atgl-1::gfp reporter strain with endu-2 RNAi and observed a significant increase in GFP fluorescence in the intestine (Figures 4C and S4C), supporting the fact that intestine PKA activity is increased in endu-2(−) mutants. The PKA pathway is known to be activated by an increased cytosolic cyclic AMP (cAMP) level (Sassone-Corsi, 2012). We found that the overall cAMP level was modestly but significantly increased in endu-2(−) worms (Figure 4D). It is likely that the increase in cAMP level in the intestine of endu-2(−) worms is much larger than the observed overall cAMP level change, since our data presented earlier (e.g., Figures 1A and 4A) suggest that ENDU-2 acts in the intestine for CTPS-1 phosphorylation and the roles on NT metabolism.
cAMP is synthesized by adenylyl cyclases (acy genes) (Sassone-Corsi, 2012). C. elegans has 4 acy genes, acy-1–acy4, and knocking down any acy gene in the endu-2(−) background leads to reduced CTPS-1 phosphorylation (Figure 4E). The expression of all 4 genes was increased in endu-2(−), but only changes in acy-1 and acy-2 were statistically significant (Figure 4F). It is important to note that all 4 genes are expressed in multiple tissues and that their expression levels in the intestine are weak (Govindan et al., 2009; Kim et al., 2012). Therefore, the effect of ENDU-2 on any of these acy genes specifically in the intestine could be significantly stronger than the difference shown in Figure 4F. Our data support the model in which ENDU-2 represses the phosphorylation of CTPS-1 at least in part by repressing PKA activity in the intestine, and the latter may be achieved at least partly through repressing the expression of acy genes and cAMP level change in the intestine.
ENDU-2 Regulates HDA-1 in Addition to PKA to Regulate CTPS-1 Phosphorylation
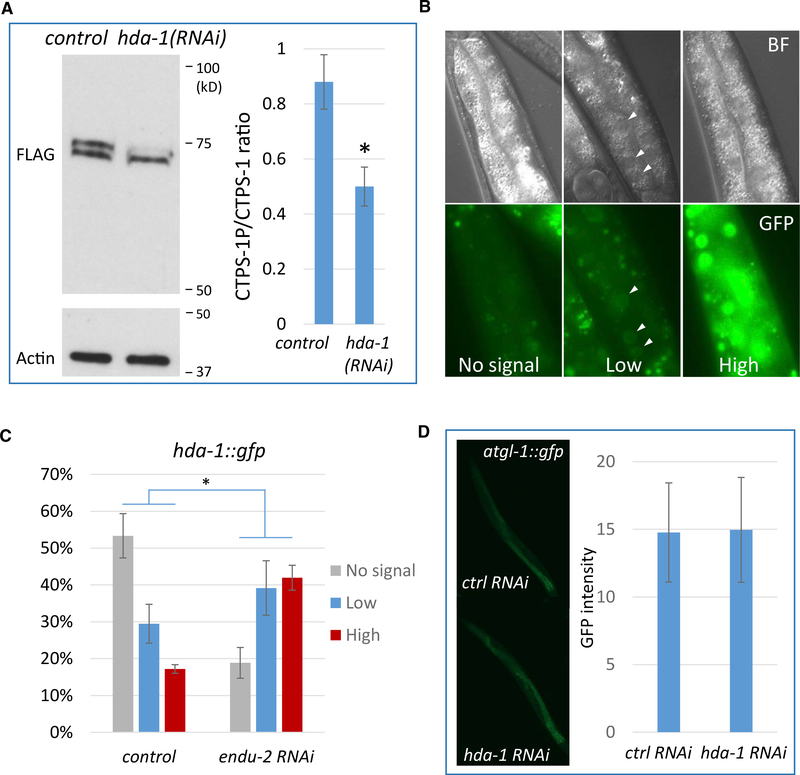
Although we demonstrated that PKA activity is necessary for CTPS-1 phosphorylation, our results also suggest that the reduction in PKA activity alone may not be sufficient to mediate the ENDU-2 role in CTPS-1 phosphorylation. To identify additional pathways, we carried out another RNAi screen of transcription factors that may regulate CTPS-1 phosphorylation and identified hda-1. hda-1 encodes an ortholog of human histone deacetylase 1 and 2 (Dufourcq et al., 2002). RNAi of hda-1 reduced CTPS-1 phosphorylation in endu-2(−) (Figure 5A).
Figure 5. ENDU-2 Regulates HDA-1 to Affect CTPS-1 Phosphorylation.
(A) Western blot of the FLAG-tagged CTPS-1 protein. hda-1 RNAi drastically reduced CTPS-1 phosphorylation. The CTPS-1P:CTPS-1 ratio is quantified on the right. n = 2. *p < 0.05 (Student’s t test, unpaired, 2-tailed).
(B and C) Fluorescence images (B) and quantitative (C) data for the hda-1::gfp reporter. endu-2(RNAi) increased the expression of the hda-1::gfp reporter in the intestine. *p < 0.01 (Pearson’s chi-square test).
(D) Fluorescence images and quantitative data for the atgl-1::gfp reporter. hda-1 RNAi did not affect the expression of an atgl-1::gfp reporter.
Error bars indicate standard deviation in all of the panels.
To test whether ENDU-2 also regulated HDA-1 expression, we used an hda-1::gfp reporter that is known to express in multiple tissues (Dufourcq et al., 2002; Matus et al., 2015). Given the intestinal role of ENDU-2, we focused on the intestinal expression of this reporter by dividing the intensity of the expression into 3 categories: “no signal,” “low signal” and “high signal” (Figure 5B). When the reporter strain was treated with endu-2 RNAi, a significant increase in GFP signal was observed, supporting the fact that ENDU-2 negatively regulates HDA-1 in the intestine (Figure 5C). Furthermore, when we treated atgl-1::gfp worms with hda-1 RNAi, we did not observe a change in GFP fluorescence (Figure 5D), indicating that HDA-1 does not affect intestinal PKA signaling. Therefore, ENDU-2 regulates HDA-1 in parallel to PKA to modulate the phosphorylation of CTPS-1.
endu-2 Expression Is Induced by Perturbation of NT Levels and Genotoxicity
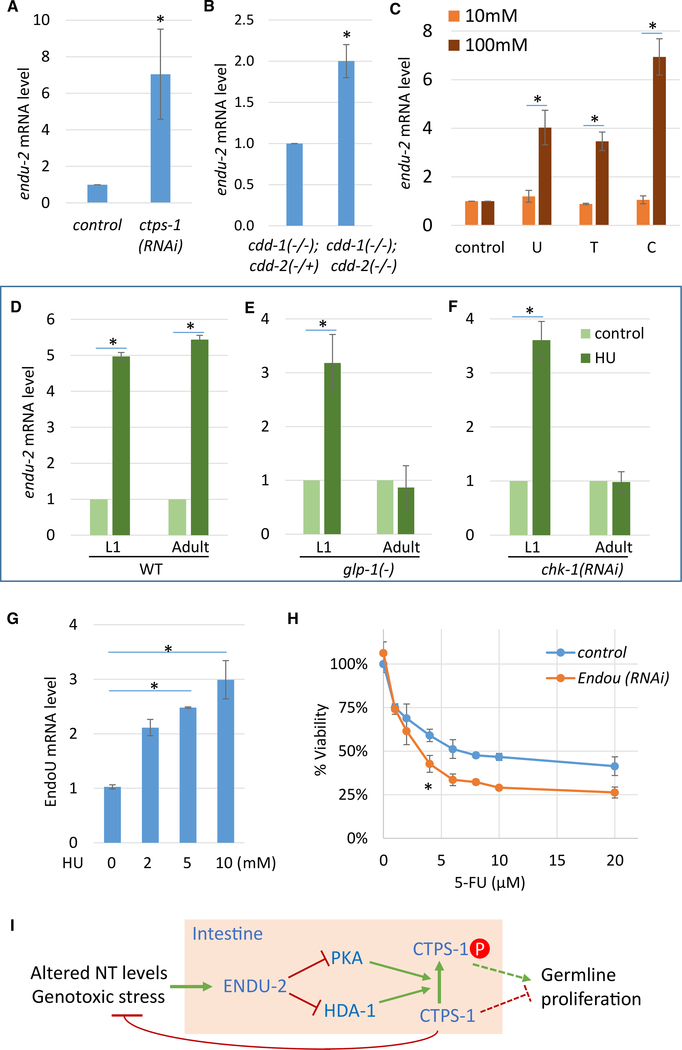
If ENDU-2 is critically involved in responding to NT deficiency or imbalance, its expression may be induced by these conditions. We found that the endu-2 mRNA level was upregulated in ctps-1(RNAi) and in cddko worms (Figures 6A and 6B). We also performed the NT supplementation experiment and found that endu-2 mRNA was induced by high concentrations of U, T, or C supplements (Figure 6C), demonstrating that endu-2 expression is induced by altered NT levels.
Figure 6. endu-2 mRNA Expression Is Induced by Perturbation of NT Levels and Genotoxicity.
(A–C) qPCR results for endu-2 mRNA level in young adults. An increase was seen with ctps-1(RNAi) (A), in cddko worms (B), or when WT worms were treated with 100 mM U, T, or C (C).
(D–F) qPCR results showing that endu-2 mRNA was up-regulated by HU treatment (D). glp-1(−) (E) or chk-1(RNAi) (F) blocked the induction of endu-2 by HU treatment in adults, but not in L1 larvae. L1 larvae were treated with 20 mM HU for 12 h. Adults were treated with 50 mM HU for 30 h. glp-1(e2141) worms were grown at 25°C to induce the germline loss and were then subjected to the same treatment as in (D). Error bars, SDs. Asterisks indicate significant differences in all of the panels.
(G) qPCR results for EndoU mRNA level. Expression was increased in HeLa cells treated with indicated concentrations of HU for 3 days. Tukey’s range test, FWER = 0.01, was used to calculate statistical significance.
(H) WST-8 cell proliferation assay results. EndoU RNAi-treated HeLa cells had reduced viability after 3 days of 5-FU treatment. *p < 0.0001 using 2-way ANOVA test.
(I) A model depicting the proposed role of ENDU-2 as a sensor and regulator of NT levels and genotoxic stresses. Arrows and T-bars indicate positive and negative regulation, respectively. The dashed arrow and the T-bar indicate that the mechanism by which germline proliferation is regulated by CTPS-1 is not known. It could be done through inhibitory regulation by non-phosphorylated CTPS-1, but a promotional regulation by phosphorylated CTPS-1 cannot be excluded (see Discussion).
(A–F) Student’s t test, unpaired, 2-tailed, and Bonferroni correction was carried out in (C)–(F). α = 0.01 (A and B), 0.0018 (C), and 0.008 (D)–(F).
HU treatment also strongly induced endu-2 expression in both adults and larvae (Figure 6D). HU treatment diminishes dNTP pools, stalls DNA replication, and causes a replication checkpoint-dependent cell-cycle arrest (Kalogeropoulos et al., 2004; Lee et al., 2010). Response to HU therefore could be attributed to NT level changes and/or the downstream checkpoint activation. In adults, the germline contains only dividing cells. The elimination of dividing cells using a glp-1 loss-of-function [glp-1(e2141)] mutant (Kodoyianni et al., 1992) abolished the induction of endu-2 by HU (Figure 6E), suggesting that the endu-2 response requires cell division in adults. chk-1 is required for replication checkpoint activation in early embryos and in the germline. chk-1 RNAi also induces DNA bridge formation (Kalogeropoulos et al., 2004; Lee et al., 2010). chk-1 RNAi abolished the endu-2 mRNA response to HU in adults but not in larvae (Figure 6F), suggesting that endu-2 is induced by HU through replication checkpoint activation in the germline in adults. It is important to note that endu-2 is not expressed in the germline. Therefore, the signal from checkpoint activation in the germline needs to be relayed to the intestine. endu-2 induction in larvae by HU was not affected by eliminating the germline or by chk-1 RNAi (Figures 6E and 6F), which is consistent with the transcriptional silencing of checkpoint signaling genes, including chk-1, in the majority of somatic cells (Vermezovic et al., 2012). In this case, endu-2 may be induced by HU through checkpoint-independent pathways or by NT level changes directly. To further examine the relations between endu-2 and ctps-1, we also tested the mRNA level of endu-2 in ctps-1(S532A) animals treated with HU. We found that abolishing CTPS-1 phosphorylation by the ctps-1(S532A) mutation did not affect the induction of endu-2 expression by HU (Figure S5A), which is consistent with the proposal that ctps-1 works downstream of endu-2.
Human EndoU Plays a Role in the Genotoxic Stress Response
Because ENDU-2 is structurally and biochemically conserved between human and C. elegans (Laneve et al., 2008; Ujisawa et al., 2018), its function in the genotoxic response observed in C. elegans may also be conserved in mammals. We examined the EndoU mRNA level and found that it was significantly increased after HU treatment in HeLa cells (Figure 6G), indicating that the expression of this enzyme is also induced by genotoxic stresses in mammalian cells. We then observed that EndoU RNAi treatment greatly enhanced the sensitivity of HeLa cells to 5-FU and FUDR (Figures 6H, S5B, and S5C), which is consistent with a likely role of EndoU in genotoxic stress response in mammalian cells.
DISCUSSION
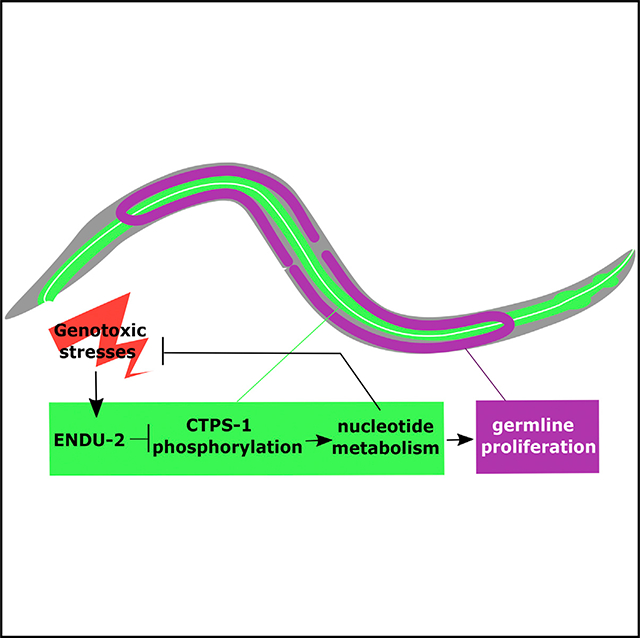
In this study, we identified C. elegans endu-2 as a key regulator in a pathway that responds to NT imbalance and genotoxic stresses. endu-2 may sense NT level change and/or genotoxic stresses through checkpoint-dependent and -independent pathways and positively regulates the CTPS-1 activity, likely by blocking its phosphorylation locally in the intestine through PKA signaling and HDA-1 (Figure 6I). Besides the suppression of the germline proliferation defect in cddko animals with low pyrimidine levels, the endu-2(−) mutation alone causes defects in chromosome segregation during mitosis in the intestine. In addition, endu-2(−) worms showed growth delay when fed high levels of NTs and have increased sensitivity to the genotoxic drugs HU, 5FU, and FUDR, but not to UV radiation. Like the C. elegans ortholog, human EndoU is induced by HU treatment, and knockdown of EndoU in HeLa cells renders them hypersensitive to 5FU and FUDR. These results suggest a prominent and conserved role of this endonuclease in the genotoxic stress response.
ENDU-2 Is a Conserved Nuclease with Functions in Diverse Cellular Processes
EndoU is a family of Ca2+-dependent endoribonucleases that is conserved across phyla. The RNA-binding and endoribonuclease activities of EndoU family members were first shown in Xenopus by in vitro assays (Laneve et al., 2003; Renzi et al., 2006) and have since been confirmed in diverse organisms ranging from bacteria to humans (Ivanov et al., 2004; Laneve et al., 2008, 2017; Michalska et al., 2018; Poe et al., 2014; Schwarz and Blower, 2014; Ujisawa et al., 2018). EndoU family members appear to be involved in many aspects of biology, including small nucleolar RNA biogenesis (Renzi et al., 2006), endoplasmic reticulum (ER) network formation (Schwarz and Blower, 2014), immune response (Poe et al., 2014), neurodegeneration (Laneve et al., 2017), and viral replication (Ivanov et al., 2004), and as antibacterial toxins (Michalska et al., 2018; Zhang et al., 2011). Human EndoU, also known as PP11 (placental protein 11), has been suggested to be a tumor marker because it is expressed in various tumors despite having no detectable expression in tissues other than the placenta (Laneve et al., 2008). In C. elegans, endu-2 has been shown to be one of many genes that alter expression in response to pathogen attack (Shapira et al., 2006) and play a role in the cold stress response (Ujisawa et al., 2018). Our results not only reveal a previously unknown role of EndoU in NT sensing and response but also provide mechanistic insights for such a specific function.
ENDU-2 Regulates Germline Proliferation in Response to NT Levels
NT balance is critical for genome integrity, and maintaining a healthy NT pool is crucial for reproductive success. We have shown previously that C. elegans actively shuts down germ cell proliferation by altering the GLP-1/Notch pathway in cddko mutant animals with a diminishing pyrimidine level (Chi et al., 2016). Here, we show that the loss of ENDU-2 function suppresses the germline shutdown by restoring the balance, indicating that the expression of ENDU-2 critically facilitates the shutdown of germ cell proliferation by preventing the changes that occurred in endu-2(−) mutants. Therefore, ENDU-2 appears to act as a safeguard for healthy germline development by sensing the NT pool and other genotoxic stresses that threaten reproductive success. Under these suboptimal conditions, endu-2 is induced, as we have shown, to shut down germline development to prevent damage and preserve resources for when the aversive condition improves (Figure 6I). Critically, as we have shown, the function of ENDU-2 is partially mediated by histone deacetylase-1 (HDA-1), which has been shown to affect germline development via the GLP-1/Notch pathway (Dufourcq et al., 2002), establishing a direct connection between NT sensing and germline development.
CTPS-1 Mediates at Least Part of the ENDU-1 Function in the NT Response Pathway
Among different NTs, cytidine NTs have unique functional properties during cell proliferation. CTP has the lowest cellular concentrations among all of the ribonucleotide triphosphates (Traut, 1994). CTP is also required for phospholipid biosynthesis (Chang and Carman, 2008; Gibellini and Smith, 2010), which is needed for cell membrane expansion during and after cell division. CTPS catalyzes the conversion of UTP to CTP, the final step in the de novo synthesis of cytidine NTs. CTPS activity is elevated in various cancers (Kizaki et al., 1980; van den Berg et al., 1993; Williams et al., 1978) and is an attractive target for the development of antineoplastic, antiviral, and antiparasitic drugs (Hofer et al., 2001; Jordheim et al., 2013; Schimmel et al., 2007) and immunosuppressants (Martin et al., 2014). CTPS activity is regulated through interactions with all 4 ribonucleotide triphosphates, including the inhibition by its product CTP. CTPS exists as inactive dimers and forms tetramers when activated (Endrizzi et al., 2004).There are 2 isoforms of CTPS in humans, CTPS1 and CTPS2. Both isoforms are regulated by phosphorylation (Chang et al., 2007; Han et al., 2005; Higgins et al., 2007; Kassel et al., 2010), but it is unknown whether and how the phosphorylation of CTPS is involved in the cellular stress response, especially in response to altered NT profiles. Previous studies have also found that CTPS can form large filamentous structures called cytoophidia in cells in various organisms, which is known to modulate CTPS activity and stability (Liu, 2016; Lynch et al., 2017). In addition, CTPS phosphorylation may affect the ability to form cytoophidia. The significance of forming cytoophidia regarding specific physiological functions remains to be investigated.
Our data indicate that ENDU-2 function is largely mediated through CTPS-1, based on the following observations: (1) genetic phenotypes and interaction between ctps-1 and endu-2 support their function in the same, or parallel, pathways; (2) ENDU-2 regulates the phosphorylation of CTPS-1 locally in the intestine; and (3) disrupting this CTPS-1 phosphorylation site suppresses the hypersensitivity of endu-2(−) to HU. We have also shown that ENDU-2 exerts its regulation on CTPS-1 through the PKA pathway and HDA-1. However, our results do not rule out additional pathways.
In our proposed model (Figure 6I), under NT deprivation or other genotoxic stress, elevated ENDU-2 activity represses CTPS-1 phosphorylation, which then contributes to the stoppage of germline proliferation, at least in part, by increased repressive activity of non-phosphorylated CTPS-1, although we have no evidence to exclude the contribution of reducing the activity of phosphorylated CTPS-1. While additional studies are needed to reach a firm conclusion regarding the role of CTPS-1 phosphorylation, the proposed model is supported by the data mentioned above. Several critical mechanistic questions remain to be addressed before creating a more comprehensive model for ENDU-2-mediated regulation of NT metabolism and germline development in response to NT deprivation or imbalance. First, identification of the direct targets of ENDU-2 is a critical step toward understanding how this endonuclease controls the activities of CTPS-1 and other downstream factors. Second, how CTPS-1 phosphorylation affects its enzymatic activity and how CTPS-1 activity in the intestine ultimately affects germline development remain attractive problems to tackle. Finally, how ENDU-2 expression and activity in the intestine is altered by changes in NT levels is potentially a critical question to investigate toward understanding the initial NT “sensing” mechanism.
The functional relation between ENDU-2 and CTPS-1 found in C. elegans is potentially also conserved in mammals. As mentioned above, there are 2 isoforms of CTPS in humans, CTPS1 and CTPS2, and both isoforms are regulated by phosphorylation. Specifically, CTPS1 is phosphorylated by glycogen synthase kinase 3 and CTPS2 by casein kinase 1, in which both phosphorylation events inhibit enzyme activity (Higgins et al., 2007; Kassel et al., 2010). There is evidence that PKA and protein kinase C (PKC) may also phosphorylate CTPS1, but the results are contradictory and the site of action of PKA is unknown (Chang et al., 2007; Han et al., 2005; Higgins et al., 2007). Our data indicate that C. elegans CTPS-1 undergoes PKA-mediated phosphorylation that inhibits the enzyme activity, suggesting that this regulation by PKA is likely conserved between worms and humans, although the predicted phosphorylation site S532 is not conserved in human CTPS genes.
Human EndoU Is a Potential Target for Combination Chemotherapy
C. elegans germ cells and human cancer cells share several key features, including robust proliferation, high nutrient demands, and possession of a subpopulation of stem cells (Hubbard, 2007). Studying the mechanism of the impact of NTs on germline proliferation may thus provide insight into the drug-resistance mechanism in cancer cells. Human EndoU is a tumor marker, as the protein expression is below the detection level outside the placenta but is present in several tumor tissues (Laneve et al., 2008). Consistent with its potential function in carcinogenesis, we show that C. elegans endu-2 is responsive to genotoxic stress and is important for successful mitosis in the intestine. At least some of the functions are likely conserved in humans, as we have shown that human EndoU is also induced by HU treatment, and knocking down EndoU rendered HeLa cells hypersensitive to 5-FU and FUDR, both of which are widely used chemotherapy drugs. It is important to note that EndoU RNAi alone does not affect cell viability, and a mouse strain containing a homozygous EndoU deletion is viable (Dickinson et al., 2016). EndoU is a potential target with low toxicity and known lead drugs to enhance the efficiency of chemotherapy drugs 5-FU and FUDR. A previous screen has already identified small molecules that inhibit EndoU activity in the micromolar range (Ragno et al., 2011).
STAR★METHODS
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Fan Jia (fan.jia@colorado.edu). All reagents generated in this study will be made available on request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
C. elegans culture and maintenance
Nematode stocks were maintained on nematode growth medium (NGM) plates seeded with bacteria (E. coli OP50) at 20°C, unless otherwise indicated. Hermaphrodites were used for all experiments. The following strains/alleles were obtained from the Caenorhabditis Genetics Center (CGC): N2 Bristol (termed wild-type), cdd-1(ok390), glp-1(e2141) and CB4856 Hawaiian strain. cdd-2(tm742) was obtained from the Mitani lab, (National BioResource Project, Tokyo, Japan). hjIs67[atgl-1p::atgl-1::gfp] was a gift from the Mak lab (Zhang et al., 2010) and qyIs92[hda-1 > HDA-1::GFP] was a gift from the Sherwood lab (Matus et al., 2015).
Cell culture
HeLa cells were maintained in 1x DMEM supplemented with 10% FBS and 1x Antibiotic Antimycotic solution (Sigma #A5955) at 37°C, 5% CO2 in a humidified incubator. For RNAi experiments, EndoU siRNA was purchased from Dharmacon (SMARTpool: siGE-NOME ENDOU siRNA for EndoU, M-012187–00-0005). Lipofectamine RNAiMAX Transfection Reagent (ThermoFisher, #13778075) was used for delivery of siRNA, following the manufacturer’s instructions. Knockdown efficiency was assessed by qPCR.
METHOD DETAILS
Isolation and genetic mapping of endu-2(ku544)
cddko worms are superficially WT when fed HT115 bacteria (Chi et al., 2016). Synchronized L4 cddko worms (P0) grown on HT115 were treated with EMS as described previously (Zhu et al., 2013). The treated P0 and F1 worms were maintained on HT115 food. Gravid F1 adults were bleached and the synchronized L1s (F2) were spotted onto OP50 bacteria. Fertile F3s found on OP50 plates were isolated as candidates carrying a suppressor mutation.
The ku544 allele was isolated from the screen above and was first backcrossed 4 times to cddko to remove background mutations. This initial backcross also confirmed that ku544 is a single recessive allele. The resulting ku544;cddko triple mutant was then crossed into the CB4856 Hawaiian strain and rough mapping with SNPs narrowed the suppressor mutation to a region with 9 potential candidates on chromosome X (Davis et al., 2005). Additional mapping using PCR Restriction Fragment Length Polymorphism identified a C-to-T mutation (T568I) in the coding region of endu-2.
Generation of transgenic lines
Point-mutations and small insertions were created using Crisper/Cas9 (Arribere et al., 2014). These strains include: endu-2(ku553) which contains the T568I mutation and an MfeI restriction site for genotyping [referred to as endu-2(−) in this report]; ctps-1(ku554) which contains a FLAG tag insertion into the 5′ end of the gene and is used as the background for all strains for western blot and immunostaining for the detection of CTPS-1 (Figures 3, 4A, 4E, and 5A); and ctps-1(ku555) which contains the S532A point mutation that abolishes the predicted phosphorylation site.
The primers used for creating these strains are:
ku553 sgRNA: aaattatcctggataggcag
ku553 repair template: gatggttctggaacttgcttggcaattgcctatccaggataatttatgacttgttcttg
ku554 sgRNA: ctcgtctattacagagatgc
ku554 repair template: ttatcaatttatctcgtctattacagagatggactacaaggacgacgacgacaagccaggcgaaaaacgcgtaaaatacattctggtg
ku555 sgRNA: taagaagcgaaccgacagct
ku555 repair template: atttgggatgttaagaagcgaaccgacgcctctgcggagcttatgcagatggctgatcag
Large single copy insertions were created using Crisper/Cas9 into Chromosome II (Dickinson et al., 2013) to generate MH5737 and MH5661. In MH5737 (kuIs114[Pendu-2:gfp:endu-2 3′UTR + unc-119]; unc-119), an 8kb promoter sequence upstream of the start codon of endu-2 drives the gfp expression. In MH5661 (kuIs112[Pendu-2(short):ha::endu-2:endu-2 3′UTR + unc-119]; unc-119), a 3kb promoter sequence upstream of endu-2 is fused to the rest of the endu-2 genomic sequence without the first intron [referred to as endu-2(oe) in this report]. A region of ~800bp immediately following the endu-2 stop codon was used as the 3′UTR sequence.
To create transgenic lines carrying endu-2 coding sequence, endu-2 genomic DNA including the 3′UTR was cloned into pPD95.77 (a gift from Andrew Fire, Addgene plasmid # 1495). The first intron of endu-2 was removed because it contains another protein coding gene kqt-2. This endu-2 genomic sequence without the first intron was fused with its own promoter (3kb upstream of endu-2) or the ges-1 promoter. H470A point-mutation was introduced into Pendu-2:endu-2 using PCR. These plasmids were injected into cddko using sur-5::dsRed as a co-injection marker to make cddko; Ex[Pendu-2:endu-2+sur-5::dsRed], cddko; Ex[Pges-1:endu-2] and cddko; Ex[Pendu-2:endu-2(H470A)+sur-5::dsRed] strains, respectively.
To construct intestine-specific PKA dominant-negative strains, the plasmid containing C. elegans dominant negative PKA, kin-2a(G310D) (Wang and Sieburth, 2013) (a gift from Derek Sieburth, USC) was fused to the ges-1 promoter as previously described (Jia et al., 2016). This new plasmid DNA was injected into ctps-1(ku554) worms together with sur-5::gfp as a co-injection marker. Three independent lines (ctps-1(ku554); Ex[Pges-1:pka(dn)+sur-5::gfp] #1~3) were used in the paper to account for variability between extrachromasomal arrays.
Fertility assay
Synchronized L1 worms were grown to L4 and individual worms were transferred to new plates. Worms were considered sterile if no viable progeny was observed one week after transfer. Fifty to a hundred worms were examined for each replicate and each experiment included at least two replicates. Two independent trials were carried out for each experiment.
Lifespan assay
Lifespan measurements were conducted at 20°C. The pre-fertile period of adulthood was defined as t = 0 for lifespan analysis and 80~100 worms were used per strain. Strains were grown for at least two generations ad libitum before lifespan analysis. For continuous RNAi, gravid adults (P0) were transferred to RNAi plates and F1 young adults were used for the assay. For adulthood-only RNAi, young adults grown on OP50 were transferred to RNAi plates and used for the assay. Results were analyzed by OASIS2 (Han et al., 2016). p values were calculated using the logrank (MantelCox) method. Two independent trials were carried out for each experiment.
Chemical supplementation of worm culture plates
Chemicals (Sigma) used in supplementation assays were: Uridine (U) (#U3003), thymidine (T) (#T1895), cytidine (C) (#C4654), 5-FU (#F6627), FUDR (#F0503) and hydroxyurea (HU) (#H8627). Supplements were directly added to cooled NGM medium before plates were poured. Overnight culture of OP50 bacteria were concentrated to OD600 = 3 and 100μL of the concentrate were added to each plate. Spotted plates were then treated with UV radiation for 30 minutes and were used within a week. For the supplementation assay, ~100 synchronized L1s were transferred to each plate and the plates were examined after three days. Each experiment contained at least two replicates and at least two independent experiments were carried out for each supplementation.
RNAi treatment
All RNAi by feeding used bacterial clones from the MRC RNAi library (Kamath et al., 2003) or the ORF-RNAi Library (Rual et al., 2004). The acy-4 RNAi construct was not found in the RNAi libraries and was made as previously described (Zhu et al., 2013). Feeding RNAi experiments were done according to (Chi et al., 2016). All RNAi started from synchronized L1 except for ctps-1 RNAi in Figures 2E and 2F. For these two experiments, synchronized L4 worms were fed control or ctps-1 RNAi plates until they reach adulthood. These adult worms were then bleached and the synchronized L1s were used for the experiments. RNAi for lifespan is described under “Lifespan Assay.” Gravid adults were bleached and synchronized L1s were used for the drug treatment or the DNA bridge assay.
dNTP measurement
dNTP extracts from worms were prepared as described by Pfister et al. (2015). Briefly, worms were washed in M9, re-suspended in 60% methanol and homogenized using an OMNI tissue homogenizer on ice. The homogenates were sonicated then incubated at −20°C for 2 hours. Samples were boiled for 3 minutes and centrifuged to remove cellular debris. The supernatant was dried and dissolved in sterile water and filtered through 0.2μM filters (Corning #431219). Determination of the dNTP pool size was based on DNA polymerase-catalyzed incorporation of radioactive dNTP into synthetic oligonucleotide templates as described by Sherman and Fyfe (1989) using α−32P-dATP as an incorporation label (Arnold et al., 2015). Each experiment contained 2–4 replicates and two independent experiments were carried out.
Immunocytochemistry
Whole-worm DAPI staining and antibody staining of dissected gonads was carried out as described by Chi et al. (2016). Primary antibody monoclonal ANTI-FLAG M2 (Sigma #F1804) was used at a dilution of 1:200. Secondary antibodies Alexa Fluor 488 goat anti-mouse (Invitrogen #A11001) were used at a dilution of 1:1000. Images were captured with Nomarski optics using a Zeiss Axioplan2 microscope and a Zeiss AxioCam MRm CCD camera. Image analysis and signal quantitation was performed with ImageJ. For fluorescent quantitation, at least 20 worms were measured for each sample.
cAMP measurement
cAMP concentrations were measured using a cyclic AMP ELISA kit (Cayman Chemical, #581001) as previously described (Lee et al., 2014). Briefly, young adult worms were washed in M9 and incubated in 0.1M HCl at RT for 20 minutes to inactivate phosphodiesterase. The worms were then washed and re-suspended in the ELISA buffer. Worms were homogenized using an OMNI tissue homogenizer on ice and the homogenates were sonicated. The worm extracts were collected by centrifugation and used for ELISA per the manufacturer’s instructions. Results were analyzed using 4 parametric logistic curve-fitting models. Each experiment contained 3–4 replicates and two independent experiments were carried out.
Mass spec analysis
Worm pellet was ground in liquid nitrogen and dissolved in lysis buffer (1x TBS, 0.5% NP40 [Sigma, #NP40S], 0.5mM EDTA, 1x Protease and Phosphatase Inhibitor Cocktail [ThermoFisher, #78440]) and were sonicated. Insoluble content was removed by centrifugation at 4°C. Protein concentration was determined using the Pierce Coomassie (Bradford) Protein Assay Kit (ThermoFisher, #1856209). Pull-down of FLAG::CTPS-1 was carried out using Anti-FLAG® M2 Magnetic Beads (Sigma, #M8823) per the manufacturer’s instructions. Mass-spec and data analysis was conducted at the Mass Spectrometry Facility at CU-Boulder.
Western blot with phos-tag gels
SuperSep Phos-tag, 12.5%, 17 well gels were purchased from Fisher (Wako Pure Chemical Corporation, #195–17991). Experiments were carried out using the same procedure for regular western blot experiments except that before transfer to PVDF membranes, gels were gently agitated in a transfer buffer containing 10mmol/L EDTA for 30 minutes. It is important to note that phos-tag gels cause ladders to migrate differently and is not indicative of the actual protein sizes.
Drug treatment for cells
For drug treatment, 5-FU (Sigma, #F6627) was dissolved in DMSO and FUDR (Sigma, #F0503) was dissolved in water as 100mM stock solutions. The stocks were added to the growth media to the desired final concentrations with or without RNAi and cell viability was tested after 3 days. Cell viability was determined using the WST-8 Cell Proliferation Assay Kit (Cayman Chemical, #10010199) following the manufacturer’s instructions. IC50 was calculated using Prism GraphPad. Each experiment contained 3 replicates and two independent experiments were carried out.
QUANTIFICATION AND STATISTICAL ANALYSIS
For Figures 1A, 2A, 2D, 3E, 6G and S3C, we first applied 1 way ANOVA to identify differences among the groups (p < 0.05 in all cases) and then used Tukey’s range test with a family-wise error rate (FWER) set to 0.01 to test differences between individual groups. For Figures 1E, 1F, 1H, 2C, 2E, 4F, 6C–6F, S2D, and S5, we kept the t test but used Bonferroni Correction for multiple-comparison correction based on the potential number of pairwise comparisons of means in the analysis (e.g., for an analysis with 6 group means, we divided our critical value, alpha = 0.05, by 6×5/2). For simplicity and consistency, we used this adjusted alpha both when comparing pairwise means and when comparing grouped means in ANOVA (e.g., Figure 2C); note that this approach is more conservative than alternative multiple comparison procedures often used in ANOVA, such as Tukey’s test.
Student’s t test was done in Excel. Pearson’s chi-square test (Figure 5C) and two-way ANOVA test (Figures 2C, 6H, S1C, and S5C) were performed in GraphPad Prism 5. One way ANOVA and Tukey’s range test was performed in Python 3.7.4 using scipy (1.2.0) and statsmodels (0.10.2) packages. For statistical analysis of lifespan results, see “Lifespan Assay” section above.
DATA AND CODE AVAILABILITY
This study did not generate any unique datasets or code.
Supplementary Material
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Monoclonal ANTI-FLAG M2 antibody produced in mouse | Sigma-Aldrich | Cat# F3165, RRID:AB_259529 |
| Anti-Actin antibody produced in rabbit | Sigma-Aldrich | Cat# A2066, RRID:AB_476693 |
| Rabbit Anti-HA-Tag Monoclonal Antibody, Unconjugated, Clone C29F4 | Cell Signaling Technology | Cat# 3724, RRID:AB_1549585 |
| Anti-mouse IgG, HRP-linked Antibody | Cell Signaling Technology | Cat# 7076, RRID:AB_330924 |
| Peroxidase-AffiniPure Goat Anti-Rabbit IgG (H+L) antibody | Jackson ImmunoResearch Labs | Cat# 111-035-003, RRID:AB_2313567 |
| Anti-FLAG® M2 Magnetic Beads | Sigma-Aldrich | Cat#M8823 |
| Bacterial and Virus Strains | ||
| Escherichia coli, cytR- | Caenorhabditis Genetics Center (CGC) | RRID:WB-STRAIN:WBStrain00041974 |
| Escherichia coli, OP50 | CGC | N/A |
| Escherichia coli, HT115(DE3) | CGC | N/A |
| RNAi feeding bacterial strain HT115(DE3) | GE Dharmacon (ORF RNAi library) | N/A |
| RNAi feeding bacterial strain HT115(DE3) | Source BioScience (Ahringer) | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Uridine | Sigma-Aldrich | Cat#U3003 |
| Thymidine | Sigma-Aldrich | Cat#T9250 |
| Cytidine | Sigma-Aldrich | Cat#C4654 |
| Hydroxyurea | Sigma-Aldrich | Cat#H8627 |
| 5-Fluorouracil (5-FU) | Sigma-Aldrich | Cat#F6627 |
| 5-Fluoro-2′-deoxyuridine (FUDR) | Sigma-Aldrich | Cat#F0503 |
| Critical Commercial Assays | ||
| WST-8 Cell Proliferation Assay Kit | Cayman Chemical | Cat#10010199 |
| Cyclic AMP ELISA Kit | Cayman Chemical | Cat# 581001 |
| Brilliant III Ultra-Fast SYBR Green QPCR Master Mix | Agilent Technologies | Cat# 600882 |
| SuperSep Phos-tag, 12.5%, 17 well gels | Wako Pure Chemical Corporation | Cat#195-17991 |
| Halt Protease and Phosphatase Inhibitor Cocktail | ThermoFisher | Cat#78440 |
| Pierce Coomassie (Bradford) Protein Assay Kit | ThermoFisher | Cat#1856209 |
| Lipofectamine RNAiMAX Transfection Reagent | ThermoFisher | Cat#13778075 |
| Pierce ECL Plus Western Blotting Substrate | ThermoFisher | Cat#32132 |
| Experimental Models: Cell Lines | ||
| HeLa cells | ATCC | Cat# CCL-2, RRID:CVCL_0030 |
| Experimental Models: Organisms/Strains | ||
| C. elegans: N2 | Caenorhabditis Genetics Center (CGC) | N/A |
| C. elegans: Strain VC208: cdd-1(ok390) | CGC | WB Strain: VC208 |
| C. elegans: Strain CB4037: glp-1(e2141) | CGC | WB Strain: CB4037 |
| C. elegans: Strain CB4856, Hawaiian strain | CGC | WB Strain: CB4856 |
| C. elegans: Strain cdd-2(tm742) | Mitani lab,National BioResource Project, Tokyo, Japan | N/A |
| C. elegans: Strain hjIs67[atgl-1p::atgl-1::gfp] | Zhang et al., 2010 | N/A |
| C. elegans: Strain qyIs92[hda-1 > HDA-1::GFP] | Matus et al., 2015 | N/A |
| C. elegans: Strain MH5737 (kuIs114[Pendu-2:gfp:endu-2 3′UTR + unc-119]; unc-119) | This paper | N/A |
| C. elegans: Strain MH5661 (kuIs112[Pendu-2(short):ha::endu-2:endu-2 3′UTR + unc-119]; unc-119) | This paper | N/A |
| C. elegans: Strain cddko: cdd-1(ok390); cdd-2(tm742) | Chi et al., 2016 | N/A |
| C. elegans: Strain cddko; Ex[Pendu-2:endu-2+sur-5::dsRed] | This paper | N/A |
| C. elegans: Strain cddko; Ex[Pges-1:endu-2] | This paper | N/A |
| C. elegans: Strain cddko; Ex[Pendu-2:endu-2(H470A)+sur-5::dsRed] | This paper | N/A |
| C. elegans: Strain endu-2(ku553) | This paper | N/A |
| C. elegans: Strain ctps-1(ku554) | This paper | N/A |
| C. elegans: Strain ctps-1(ku554, ku555) | This paper | N/A |
| C. elegans: Strain ctps-1(ku554); Ex[Pges-1:pka(dn)+sur-5::gfp] | This paper | N/A |
| Oligonucleotides | ||
| ku553 sgRNA: aaattatcctggataggcag | This paper | N/A |
| ku553 repair template: gatggttctggaacttgcttggcaattgcctatccaggataatttatgacttgttcttg | This paper | N/A |
| ku554 sgRNA: ctcgtctattacagagatgc | This paper | N/A |
| ku554 repair template: ttatcaatttatctcgtctattacagagatggactacaaggacgacgacgacaagccaggcgaaaaacgcgtaaaatacattctggtg | This paper | N/A |
| ku555 sgRNA: taagaagcgaaccgacagct | This paper | N/A |
| ku555 repair template: atttgggatgttaagaagcgaaccgacgcctctgcggagcttatgcagatggctgatcag | This paper | N/A |
| EndoU siRNA | Dharmacon | SMARTpool: M-012187-00-0005 |
| Recombinant DNA | ||
| Plasmid: pPD95.77 | pPD95.77 was a gift from Andrew Fire | Addgene Plasmid #1495 |
| Pladmid: pDD162 | Dickinson et al., 2013 | Addgene Plasmid #47549 |
| Plasmid: pHW154: Punc-47(FL)::kin-2a(G310D) | Wang and Sieburth, 2013 | N/A |
| Plasmid: L4440-acy-4RNAi | This paper | N/A |
| Software and Algorithms | ||
| OASIS2 | Han et al., 2016 | N/A |
| Fiji (ImageJ) | http://fiji.sc | Ver 1.50i; RRID:SCR_002285 |
| Prism 5 | GraphPad | N/A |
Highlights.
ENDU-2 nuclease regulates nucleotide metabolism and germ cell proliferation in worms
ENDU-2 expression is induced by nucleotide imbalance and other genotoxic stresses
ENDU-2 inhibits CTP synthase phosphorylation by repressing PKA and HDA-1 in the gut
ENDU-2 function may be conserved in mammalian cells
ACKNOWLEDGMENTS
We thank Derek Sieburth, Ho Yi Mak, David Sherwood, and Shohei Mitani for reagents; Thomas Lee of Mass Spectrometry Facility at the University of Colorado at Boulder for mass spectrometry analysis; Matt Keller for guidance on statistical analysis; and Emily Richard, Scott Ho, Aileen Sewell and other lab members for assistance and helpful discussions. Some strains were provided by the Caenorhabditis Genetics Center (supported by NIH P40 OD010440). This study was supported by the National Institutes of Health grant 5R01GM047869 (to M.H.) and the Howard Hughes Medical Institute Investigator fund.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2020.01.050.
REFERENCES
- Amanchy R, Periaswamy B, Mathivanan S, Reddy R, Tattikota SG, and Pandey A (2007). A curated compendium of phosphorylation motifs. Nat. Biotechnol 25, 285–286. [DOI] [PubMed] [Google Scholar]
- Arnold LH, Groom HC, Kunzelmann S, Schwefel D, Caswell SJ, Ordonez P, Mann MC, Rueschenbaum S, Goldstone DC, Pennell S, et al. (2015). Phospho-dependent Regulation of SAMHD1 Oligomerisation Couples Catalysis and Restriction. PLoS Pathog. 11, e1005194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arribere JA, Bell RT, Fu BX, Artiles KL, Hartman PS, and Fire AZ (2014). Efficient marker-free recovery of custom genetic modifications with CRISPR/Cas9 in Caenorhabditis elegans. Genetics 198, 837–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bembenek JN, Verbrugghe KJ, Khanikar J, Csankovszki G, and Chan RC (2013). Condensin and the spindle midzone prevent cytokinesis failure induced by chromatin bridges in C. elegans embryos. Curr. Biol 23, 937–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YF, and Carman GM (2008). CTP synthetase and its role in phospholipid synthesis in the yeast Saccharomyces cerevisiae. Prog. Lipid Res 47, 333–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YF, Martin SS, Baldwin EP, and Carman GM (2007). Phosphorylation of human CTP synthetase 1 by protein kinase C: identification of Ser(462) and Thr(455) as major sites of phosphorylation. J. Biol. Chem 282, 17613–17622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi C, Ronai D, Than MT, Walker CJ, Sewell AK, and Han M (2016). Nucleotide levels regulate germline proliferation through modulating GLP-1/ Notch signaling in C. elegans. Genes Dev. 30, 307–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis MW, Hammarlund M, Harrach T, Hullett P, Olsen S, and Jorgensen EM (2005). Rapid single nucleotide polymorphism mapping in C. elegans. BMC Genomics 6, 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson DJ, Ward JD, Reiner DJ, and Goldstein B (2013). Engineering the Caenorhabditis elegans genome using Cas9-triggered homologous recombination. Nat. Methods 10, 1028–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson ME, Flenniken AM, Ji X, Teboul L, Wong MD, White JK, Meehan TF, Weninger WJ, Westerberg H, Adissu H, et al. ; International Mouse Phenotyping Consortium; Jackson Laboratory; Infrastructure Nationale PHENOMIN, Institut Clinique de la Souris (ICS); Charles River Laboratories; MRC Harwell; Toronto Centre for Phenogenomics; Wellcome Trust Sanger Institute; RIKEN BioResource Center (2016). High-throughput discovery of novel developmental phenotypes. Nature 537, 508–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufourcq P, Victor M, Gay F, Calvo D, Hodgkin J, and Shi Y (2002). Functional requirement for histone deacetylase 1 in Caenorhabditis elegans gonadogenesis. Mol. Cell. Biol 22, 3024–3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endrizzi JA, Kim H, Anderson PM, and Baldwin EP (2004). Crystal structure of Escherichia coli cytidine triphosphate synthetase, a nucleotide-regulated glutamine amidotransferase/ATP-dependent amidoligase fusion protein and homologue of anticancer and antiparasitic drug targets. Biochemistry 43, 6447–6463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibellini F, and Smith TK (2010). The Kennedy pathway–De novo synthesis of phosphatidylethanolamine and phosphatidylcholine. IUBMB Life 62, 414–428. [DOI] [PubMed] [Google Scholar]
- Govindan JA, Nadarajan S, Kim S, Starich TA, and Greenstein D (2009). Somatic cAMP signaling regulates MSP-dependent oocyte growth and meiotic maturation in C. elegans. Development 136, 2211–2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han GS, Sreenivas A, Choi MG, Chang YF, Martin SS, Baldwin EP, and Carman GM (2005). Expression of Human CTP synthetase in Saccharomyces cerevisiae reveals phosphorylation by protein kinase A. J. Biol. Chem 280, 38328–38336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han SK, Lee D, Lee H, Kim D, Son HG, Yang JS, Lee SV, and Kim S (2016). OASIS 2: online application for survival analysis 2 with features for the analysis of maximal lifespan and healthspan in aging research. Oncotarget 7, 56147–56152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins MJ, Graves PR, and Graves LM (2007). Regulation of human cytidine triphosphate synthetase 1 by glycogen synthase kinase 3. J. Biol. Chem 282, 29493–29503. [DOI] [PubMed] [Google Scholar]
- Hofer A, Steverding D, Chabes A, Brun R, and Thelander L (2001). Trypanosoma brucei CTP synthetase: a target for the treatment of African sleeping sickness. Proc. Natl. Acad. Sci. USA 98, 6412–6416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, Xiong C, and Kornfeld K (2004). Measurements of age-related changes of physiological processes that predict lifespan of Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 101, 8084–8089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard EJ (2007). Caenorhabditis elegans germ line: a model for stem cell biology. Dev. Dyn 236, 3343–3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov KA, Hertzig T, Rozanov M, Bayer S, Thiel V, Gorbalenya AE, and Ziebuhr J (2004). Major genetic marker of nidoviruses encodes a replicative endoribonuclease. Proc. Natl. Acad. Sci. USA 101, 12694–12699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia F, Cui M, Than MT, and Han M (2016). Developmental Defects of Caenorhabditis elegans Lacking Branched-chain α-Ketoacid Dehydrogenase Are Mainly Caused by Monomethyl Branched-chain Fatty Acid Deficiency. J. Biol. Chem 291, 2967–2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordheim LP, Durantel D, Zoulim F, and Dumontet C (2013). Advances in the development of nucleoside and nucleotide analogues for cancer and viral diseases. Nat. Rev. Drug Discov 12, 447–464. [DOI] [PubMed] [Google Scholar]
- Kalogeropoulos N, Christoforou C, Green AJ, Gill S, and Ashcroft NR (2004). chk-1 is an essential gene and is required for an S-M checkpoint during early embryogenesis. Cell Cycle 3, 1196–1200. [PubMed] [Google Scholar]
- Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, Kanapin A, Le Bot N, Moreno S, Sohrmann M, et al. (2003). Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421, 231–237. [DOI] [PubMed] [Google Scholar]
- Kassel KM, Au R, Higgins MJ, Hines M, and Graves LM (2010). Regulation of human cytidine triphosphate synthetase 2 by phosphorylation. J. Biol. Chem 285, 33727–33736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly WG, Xu S, Montgomery MK, and Fire A (1997). Distinct requirements for somatic and germline expression of a generally expressed Caernorhabditis elegans gene. Genetics 146, 227–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Govindan JA, Tu ZJ, and Greenstein D (2012). SACY-1 DEADBox helicase links the somatic control of oocyte meiotic maturation to the sperm-to-oocyte switch and gamete maintenance in Caenorhabditis elegans. Genetics 192, 905–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kizaki H, Williams JC, Morris HP, and Weber G (1980). Increased cytidine 5′-triphosphate synthetase activity in rat and human tumors. Cancer Res. 40, 3921–3927. [PubMed] [Google Scholar]
- Kniazeva M, and Ruvkun G (2019). Rhizobium induces DNA damage in Caenorhabditis elegans intestinal cells. Proc. Natl. Acad. Sci. USA 116, 3784–3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodoyianni V, Maine EM, and Kimble J (1992). Molecular basis of loss-of-function mutations in the glp-1 gene of Caenorhabditis elegans. Mol. Biol. Cell 3, 1199–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudlow BA, Zhang L, and Han M (2012). Systematic analysis of tissue-restricted miRISCs reveals a broad role for microRNAs in suppressing basal activity of the C. elegans pathogen response. Mol. Cell 46, 530–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laneve P, Altieri F, Fiori ME, Scaloni A, Bozzoni I, and Caffarelli E (2003). Purification, cloning, and characterization of XendoU, a novel endoribonuclease involved in processing of intron-encoded small nucleolar RNAs in Xenopus laevis. J. Biol. Chem 278, 13026–13032. [DOI] [PubMed] [Google Scholar]
- Laneve P, Gioia U, Ragno R, Altieri F, Di Franco C, Santini T, Arceci M, Bozzoni I, and Caffarelli E (2008). The tumor marker human placental protein 11 is an endoribonuclease. J. Biol. Chem 283, 34712–34719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laneve P, Piacentini L, Casale AM, Capauto D, Gioia U, Cappucci U, Di Carlo V, Bozzoni I, Di Micco P, Morea V, et al. (2017). Drosophila CG3303 is an essential endoribonuclease linked to TDP-43-mediated neurodegeneration. Sci. Rep 7, 41559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SJ, Gartner A, Hyun M, Ahn B, and Koo HS (2010). The Caenorhabditis elegans Werner syndrome protein functions upstream of ATR and ATM in response to DNA replication inhibition and double-strand DNA breaks. PLoS Genet. 6, e1000801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Kong J, Jang JY, Han JS, Ji Y, Lee J, and Kim JB (2014). Lipid droplet protein LID-1 mediates ATGL-1-dependent lipolysis during fasting in Caenorhabditis elegans. Mol. Cell. Biol 34, 4165–4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JL (2016). The Cytoophidium and Its Kind: Filamentation and Compartmentation of Metabolic Enzymes. Annu. Rev. Cell Dev. Biol 32, 349–372. [DOI] [PubMed] [Google Scholar]
- Lynch EM, Hicks DR, Shepherd M, Endrizzi JA, Maker A, Hansen JM, Barry RM, Gitai Z, Baldwin EP, and Kollman JM (2017). Human CTP synthase filament structure reveals the active enzyme conformation. Nat. Struct. Mol. Biol 24, 507–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin E, Palmic N, Sanquer S, Lenoir C, Hauck F, Mongellaz C, Fabrega S, Nitschké P, Esposti MD, Schwartzentruber J, et al. (2014). CTP synthase 1 deficiency in humans reveals its central role in lymphocyte proliferation. Nature 510, 288–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matus DQ, Lohmer LL, Kelley LC, Schindler AJ, Kohrman AQ, Barkoulas M, Zhang W, Chi Q, and Sherwood DR (2015). Invasive Cell Fate Requires G1 Cell-Cycle Arrest and Histone Deacetylase-Mediated Changes in Gene Expression. Dev. Cell 35, 162–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalska K, Quan Nhan D, Willett JLE, Stols LM, Eschenfeldt WH, Jones AM, Nguyen JY, Koskiniemi S, Low DA, Goulding CW, et al. (2018). Functional plasticity of antibacterial EndoU toxins. Mol. Microbiol 109, 509–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pai CC, and Kearsey SE (2017). A Critical Balance: dNTPs and the Maintenance of Genome Stability. Genes (Basel) 8, 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfister SX, Markkanen E, Jiang Y, Sarkar S, Woodcock M, Orlando G, Mavrommati I, Pai CC, Zalmas LP, Drobnitzky N, et al. (2015). Inhibiting WEE1 Selectively Kills Histone H3K36me3-Deficient Cancers by dNTP Starvation. Cancer Cell 28, 557–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poe JC, Kountikov EI, Lykken JM, Natarajan A, Marchuk DA, and Tedder TF (2014). EndoU is a novel regulator of AICD during peripheral B cell selection. J. Exp. Med 211, 57–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragno R, Gioia U, Laneve P, Bozzoni I, Mai A, and Caffarelli E (2011). Identification of small-molecule inhibitors of the XendoU endoribonucleases family. ChemMedChem 6, 1797–1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renzi F, Caffarelli E, Laneve P, Bozzoni I, Brunori M, and Vallone B (2006). The structure of the endoribonuclease XendoU: from small nucleolar RNA processing to severe acute respiratory syndrome coronavirus replication. Proc. Natl. Acad. Sci. USA 103, 12365–12370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rual JF, Ceron J, Koreth J, Hao T, Nicot AS, Hirozane-Kishikawa T, Vandenhaute J, Orkin SH, Hill DE, van den Heuvel S, and Vidal M (2004). Toward improving Caenorhabditis elegans phenome mapping with an ORFeome-based RNAi library. Genome Res. 14 (10B), 2162–2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassone-Corsi P (2012). The cyclic AMP pathway. Cold Spring Harb. Perspect. Biol 10.1101/cshperspect.a011148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimmel KJ, Gelderblom H, and Guchelaar HJ (2007). Cyclopentenyl cytosine (CPEC): an overview of its in vitro and in vivo activity. Curr. Cancer Drug Targets 7, 504–509. [DOI] [PubMed] [Google Scholar]
- Schwarz DS, and Blower MD (2014). The calcium-dependent ribonuclease XendoU promotes ER network formation through local RNA degradation. J. Cell Biol 207, 41–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapira M, Hamlin BJ, Rong J, Chen K, Ronen M, and Tan MW (2006). A conserved role for a GATA transcription factor in regulating epithelial innate immune responses. Proc. Natl. Acad. Sci. USA 103, 14086–14091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman PA, and Fyfe JA (1989). Enzymatic assay for deoxyribonucleoside triphosphates using synthetic oligonucleotides as template primers. Anal. Biochem. 180, 222–226. [DOI] [PubMed] [Google Scholar]
- Traut TW (1994). Physiological concentrations of purines and pyrimidines. Mol. Cell. Biochem. 140, 1–22. [DOI] [PubMed] [Google Scholar]
- Ujisawa T, Ohta AT Ii, Minakuchi Y, Toyoda A, Ii M, and Kuhara A (2018). Endoribonuclease ENDU-2 regulates multiple traits including cold tolerance via cell autonomous and nonautonomous controls in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 115, 8823–8828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berg AA, van Lenthe H, Busch S, de Korte D, Roos D, van Kuilenburg AB, and van Gennip AH (1993). Evidence for transformation-related increase in CTP synthetase activity in situ in human lymphoblastic leukemia. Eur. J. Biochem. 216, 161–167. [DOI] [PubMed] [Google Scholar]
- Vermezovic J, Stergiou L, Hengartner MO, and d’Adda di Fagagna F (2012). Differential regulation of DNA damage response activation between somatic and germline cells in Caenorhabditis elegans. Cell Death Differ. 19, 1847–1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, and Sieburth D (2013). PKA controls calcium influx into motor neurons during a rhythmic behavior. PLoS Genet. 9, e1003831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JC, Kizaki H, Weber G, and Morris HP (1978). Increased CTP synthetase activity in cancer cells. Nature 271, 71–73. [DOI] [PubMed] [Google Scholar]
- Zhang SO, Box AC, Xu N, Le Men J, Yu J, Guo F, Trimble R, and Mak HY (2010). Genetic and dietary regulation of lipid droplet expansion in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 107, 4640–4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Iyer LM, and Aravind L (2011). A novel immunity system for bacterial nucleic acid degrading toxins and its recruitment in various eukaryotic and DNA viral systems. Nucleic Acids Res. 39, 4532–4552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Li J, Purkayastha S, Tang Y, Zhang H, Yin Y, Li B, Liu G, and Cai D (2013). Hypothalamic programming of systemic ageing involving IKK-β, NF-κB and GnRH. Nature 497, 211–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Shen H, Sewell AK, Kniazeva M, and Han M (2013). A novel sphingolipid-TORC1 pathway critically promotes postembryonic development in Caenorhabditis elegans. eLife 2, e00429. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not generate any unique datasets or code.