Abstract
Opportunistic viruses are a major problem for immunosuppressed individuals, particularly following organ or stem cell transplantation. Current treatments are non-existent or suffer from problems such as high toxicity or development of resistant strains. We previously published that a trafficking inhibitor that targets a host protein greatly reduces the replication of human cytomegalovirus. This inhibitor was also shown to be moderately effective against polyomaviruses, another family of opportunistic viruses. We have developed a panel of analogues for this inhibitor and have shown that these analogues maintain their high efficacy against HCMV, while substantially lowering the concentration required to inhibit polyomavirus replication. By targeting a host protein these compounds are able to inhibit the replication of two very different viruses. These observations open up the possibility of pan-viral inhibitors for immunosuppressed individuals that are effective against multiple, diverse opportunistic viruses.
Keywords: Human cytomegalovirus, mouse polyomavirus, antivirals, syntaxin 5, retrograde trafficking
1. Introduction
Viruses that maintain a lifelong association with their host remain a major problem for immunosuppressed individuals. Human cytomegalovirus (HCMV) and the JC and BK polyomaviruses are widespread throughout the human population. However, in healthy individuals an infection is largely asymptomatic. Both viruses persist in the host after establishing an initial infection. In the event of immunosuppression these viruses can reactivate, which may lead to end-organ disease, cancers, neurological disorders and ultimately mortality. HCMV reactivations are observed after solid organ and hematopoietic stem cells transplants[1-3]. Likewise, BK polyomavirus reactivation can occur in patients following kidney or allogeneic hematopoietic stem cells transplants[4, 5]. JC polyomavirus persists asymptomatically in the reno-urinary tract in most of the adult population, but may spread to the central nervous system in an immunocompromised host and result in progressive multifocal leukoencephalopathy (PML), causing fatal demyelination of neurons[6]. Any perturbation of the immune system provides a risk to patients already harboring these common pathogens.
Viruses, including the opportunistic pathogens mentioned above, are highly diverse. Thus, therapies are often developed against a single virus or group of highly related viruses. While treatments have remained elusive against some opportunistic viruses such as polyomaviruses, other therapies such as those used against HCMV suffer from high toxicity and development of resistant virus strains. Targeting a host factor required by a wide range of diverse viruses may allow for the development of a single therapy against multiple opportunistic agents. Many bacterial toxins and viruses are dependent upon endocytosis and subsequent trafficking to the endoplasmic reticulum for entry. This transport is dependent upon retrograde trafficking machinery. While other viruses may not use this trafficking machinery for entry, they may require them at a different stage in the production of infectious viral progeny, as has been shown for HCMV. Thus, inhibitors that target these factors may function as antiviral agents effective against a wide range of diverse viruses.
Retro2 was originally identified as one of two compounds that block the retrograde trafficking and downstream toxicity of ricin and Shiga-like toxins[7]. In the presence of the inhibitor, Syntaxin5 (STX5) is displaced from its normal Golgi localization pattern, disrupting a subset of retrograde trafficking to the Golgi. Importantly, no toxicity was observed indicating that endogenous retrograde cargos trafficked via alternative pathways in the presence of Retro2. Subsequent rounds of optimization led to the identification of two more potent inhibitors termed Retro94 and Retro 2.1[8, 9]. The dihydroquinazolinone Retro 2.1 exhibited 500-fold improvement of the EC50 against Shiga toxin challenge, 23 nM, and a 1000-fold enhancement, 54 nM, against ricin challenge[8]. These compounds have since been shown to effectively block entry of numerous pathogens, including the human BK and JC polyomaviruses and mouse polyomavirus (MuPyV)[10, 11]. Other viruses shown to be sensitive to inhibition by Retro2 include vaccinia virus[12, 13], filoviruses including the Ebola and Marburg viruses[14] and Enterovirus 71, which causes hand, foot, and mouth disease[15]. We previously found that STX5 is important for the assembly compartment of HCMV and that production of infectious HCMV virions is highly sensitive to Retro2 and Retro2.1[16]. Subsequently another herpesvirus, herpes simplex virus 2, was found to be susceptible to Retro2.1, albeit inhibition required a much higher concentration than HCMV[17]. Since several viruses all utilize STX5 at some point of their replication cycle, irrespective of the fact that STX5 is utilized for distinct roles, inhibitors that displace STX5 have been shown to be effective against multiple highly diverse viruses.
In this study we describe the generation of analogues that all displace STX5 from its normal Golgi apparatus. We then assayed these compounds for their ability to inhibit infection of both HCMV and MuPyV while exhibiting low or no toxicity on cells. Our studies have led to the identification of compounds with low cell toxicity that exhibit increased potency against diverse, unrelated opportunistic viruses.
2. Results
2.1. Synthesis of novel inhibitors
A previous structural activity relationship (SAR) investigation by Carney et al. found the addition of a fluorine atom at the C-6 position of the benzo moiety increases the compound’s potency as an inhibitor of host retrograde trafficking[18]. We prepared new analogues by introducing cyano and nitro groups in the C-6 position of the benzo moiety. Selection of cyano and nitro derivatives are based on the following facts. Over 30 nitrile-containing pharmaceuticals are prescribed for a diverse variety of medicinal indications with more than 20-25 additional nitrile-containing agents in clinical development[19]. Several small drug-like molecules containing a nitro group are in the pharmacy market or are being investigated; for example: Nitrofural, Nitroglycerin, Nitrofurantoin, Tinidazole.
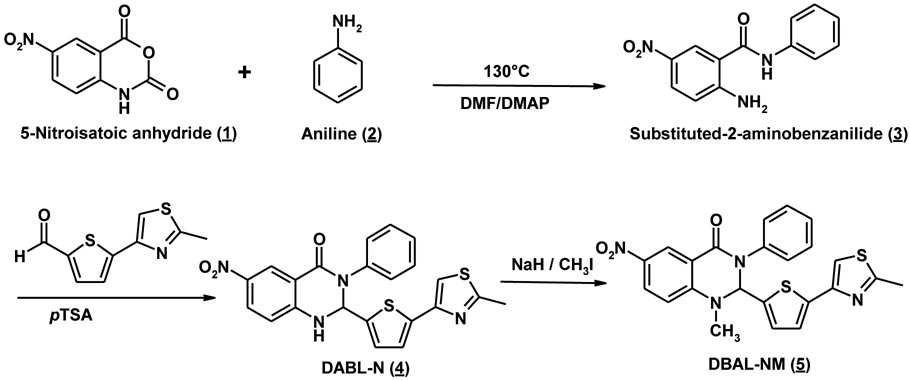
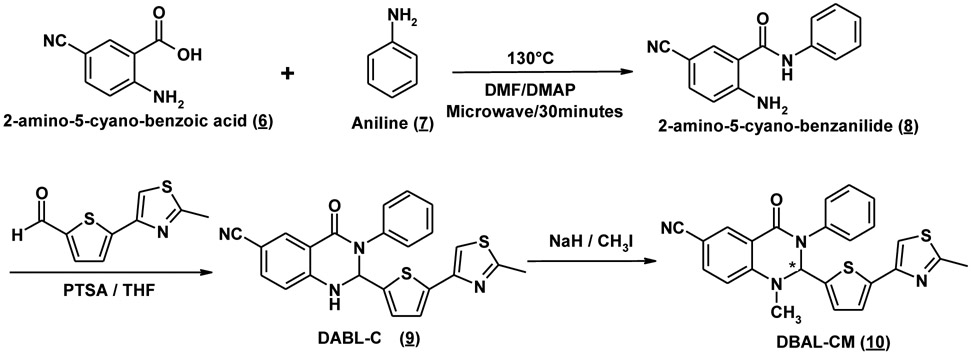
To synthesize these analogues, 1-Methyl-2-(5-(2-methylthiazol-4-yl)thiophen-2-yl)-3phenyl-2,3-dihydroquinazolin-4(1H)-one (Retro94) was synthesized as reported in the literature with minor modifications[20]. DABL-N (4), DABL-NM (5), DABL-C (9), and DABL-CM (10) were synthesized by following a similar procedure with minor modification as outlined in Schemes 1 & 2. The appropriate substituted 2-aminoanilides were prepared by using previously reported procedures or with modification using microwave-assisted chemical reaction. The reaction was carried out in a microwave safe glass vial that was irradiated in a microwave apparatus (LABMAT) at 130°C, 250 W to yield corresponding substituted 2-amino-5-cyanoanilides in quantitative yields in just 30 minutes compared to reported overnight heating. Substituted 2-aminobenzanilides were reacted with 5-(2-methyl-3-thiazol-4-yl)-2-thiophencarboxaldehyde at reflux temperature to yield the corresponding cyclic compound, obtained in moderate to good yields. The corresponding yellow colored cyclic compound was treated with NaH in dry THF to give a good yield of the desired compounds. Confirmation of the structure was obtained by 1H NMR, High resolution MS (HRMS), and purity was checked by reverse phase HPLC.
Scheme 1. General synthesis of DABL-N (4) and DABL-NM (5) analogues.
Scheme 2. General synthesis of DABL-C (9) and DABL-CM (10) analogues.
2.2. Effects on STX5 localization.
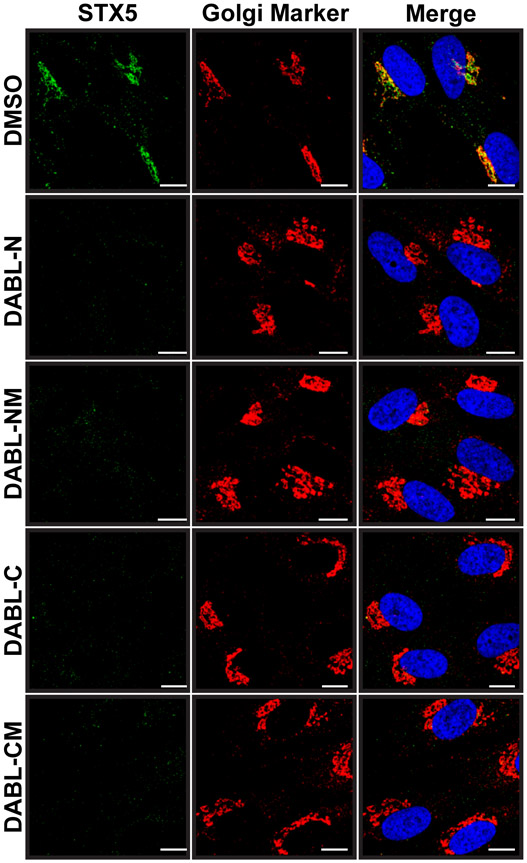
Although the exact target of Retro2 and subsequent optimized analogues has not yet been clearly defined, it is known that retrograde trafficking is blocked. A small imaging screen of factors involved in retrograde trafficking revealed that the cellular SNARE protein STX5, and to a lesser extent STX6, is dispersed from its normal Golgi localization and is instead found diffuse throughout the cell in the presence of Retro2 [7]. To show that the novel analogues are acting in a manner similar to Retro2, we stained for both STX5 and a Golgi apparatus marker (p115) in cells treated with the various compounds or with a DMSO vehicle control. As expected, STX5 retained its Golgi localization only in cells treated with the DMSO. In all cells treated with an analogue, STX5 was displaced from the Golgi and was barely detectable in the cell (Figure 1) as has been reported previously for Retro2 and Retro94[7, 16]. Thus, all compounds retain the ability to displace STX5 from the Golgi apparatus.
Figure 1. STX5 is displaced from Golgi.
Immunofluorescence of HDFs treated with 10 μM of the indicated compound. Cells were fixed and stained for STX5 (green) and the Golgi Marker p115 (red). Nuclei were stained with DAPI (blue).
2.3. Effects on HCMV replication
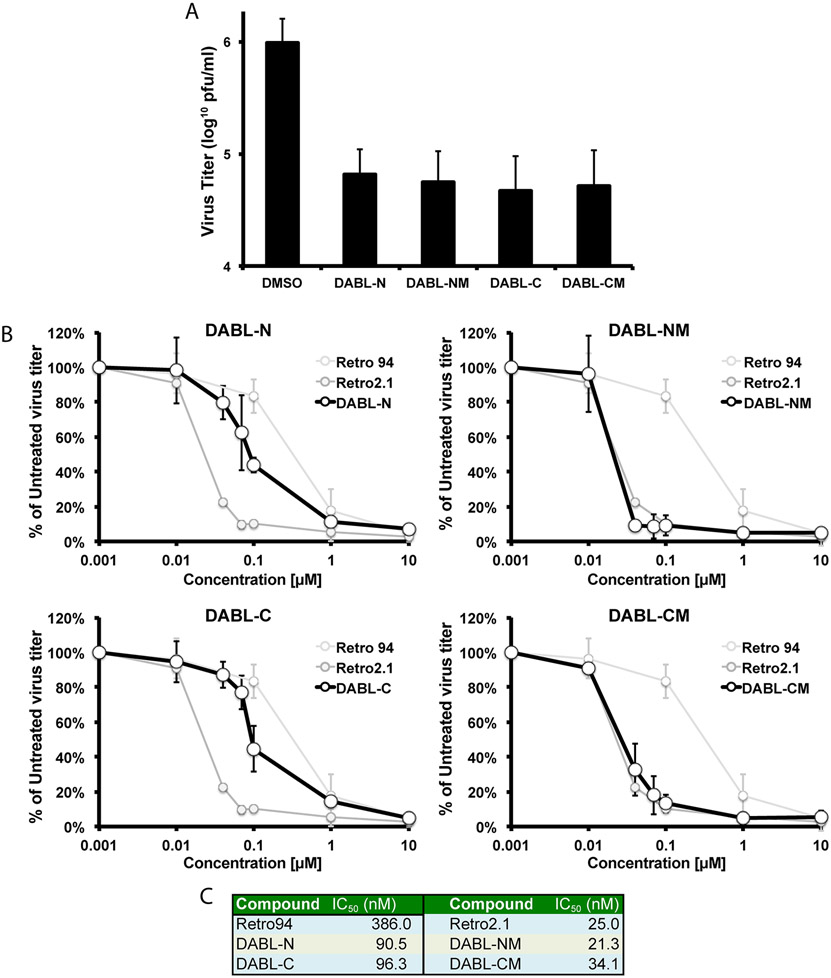
We next tested whether the analogues retained the ability of the compounds to inhibit the production of infectious HCMV virions. The compounds were added at a concentration of 10 μM at infection. Based on our previous work showing that the inhibition of HCMV is at a late stage[16], total virus, cell-free and cell-associated, were harvested 96 hours post-infection and titered for infectious virions. All analogues resulted in at least a one-log reduction in infectious virus titers (Figure 2A). Infectious virus titers were also analyzed over a 10-fold dilution series of the compounds. When percent inhibition was plotted relative to compound concentration, several analogues (DABL-NM, DABL-CM) exhibited a curve and IC50 values similar to the previously published Retro2.1 compound (Figures 2B&C). Two other analogues, DABL-N and DABL-C, although not as efficient, were still more effective than the previously published Retro94 compound. DABL-NM and DABL-CM were more potent compared to their corresponding analoguess DABL-N and DABL-C, which may be due to a better lipophilic nature of the compounds. Thus, the newly generated analogues were as effective or better than previously published compounds in inhibiting the production of infectious HCMV virions.
Figure 2. Production of infectious HCMV virions is inhibited.
(A) Following infection with HCMV (MOI: 3), 10 μM of indicated compounds or DMSO vehicle control were added at 2 hpi. Samples were harvested 96 hpi and analyzed for infectious virus titers. (B) Infectious virus titers following 10-fold dilutions of indicated compounds. Infections and sample harvest were as described in (A). Retro94 and Retro2.1 curves are shown for reference. (C) Calculated IC50 values for each compound. The graphed data are the average of three independent experiments.
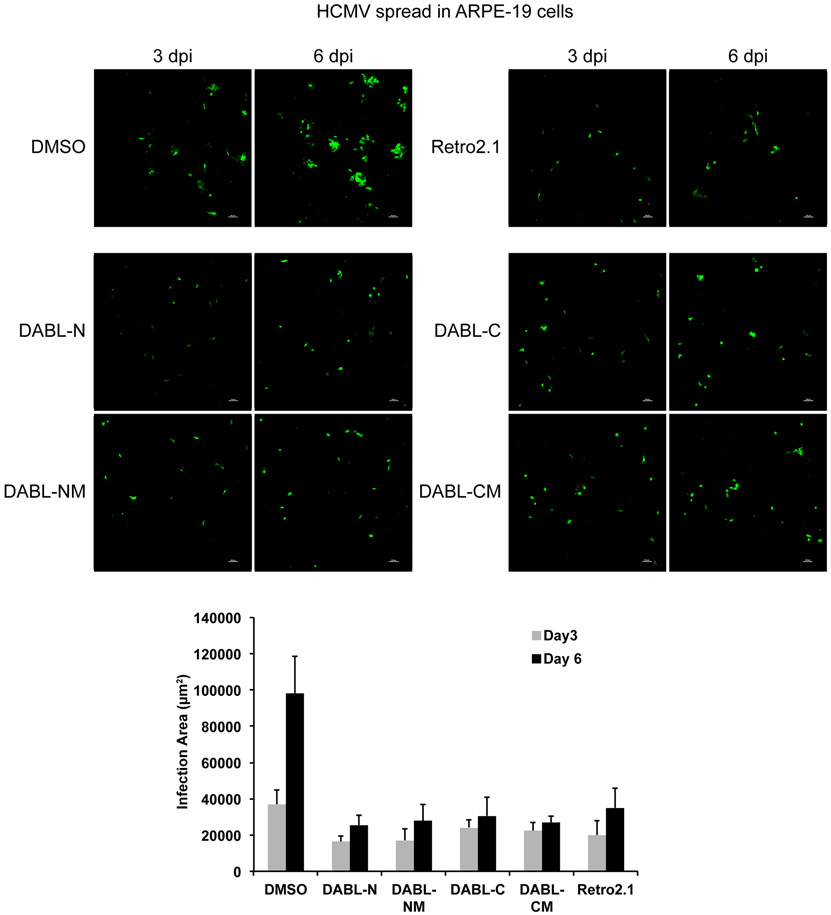
The previous titer data was obtained using human fibroblasts. However, a natural HCMV infection can occur in numerous cell types. We previously showed that inhibition of HCMV by Retro94 was observed for multiple cell-types and strains of HCMV[16]. Similarly, we analyzed the spread of HCMV through ARPE-19 cells and found spread to be reduced in the presence of all four DABL compounds (Figure 3). Thus, we observed inhibition of HCMV in both fibroblasts and epithelial cells.
Figure 3. DABL compounds inhibit HCMV spread in epithelial cells.
ARPE-19 cells infected with HCMV that expresses GFP and treated with 7.5 μM of the indicated compounds. Images were acquired 3 and 6 days post infection. Graph represents area of infection as calculated by GFP signal at 3 and 6 days post infection. Graph is average area from 3 distinct experiments.
2.4. Effects on cell viability
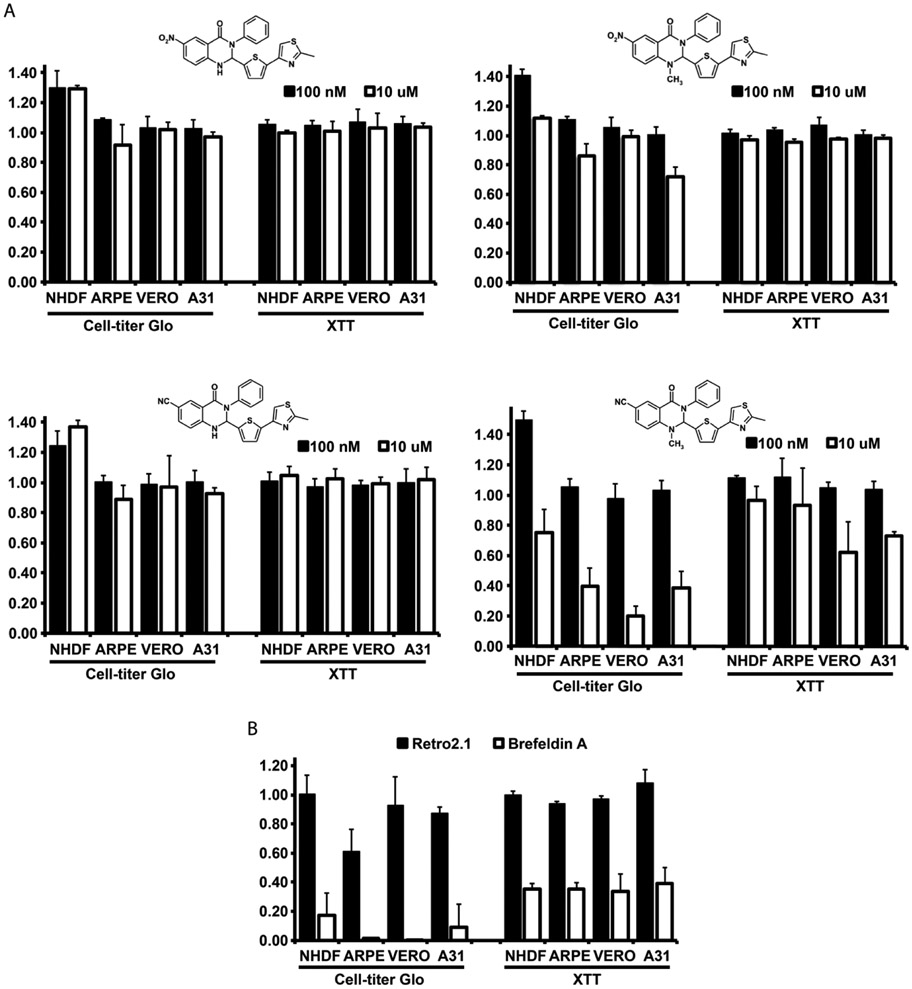
Adverse effects on cell fitness are a concern when displacing a cellular protein from its normal localization and function. To test the cytotoxicity and viability of several cell types in the presence of the compounds we used two distinct cell viability assays, as well as a visual inspection of the cell morphology. We assessed the cell viability after exposure to the drugs for 4 days using a CellTiter-Glo luminescent assay that quantifies ATP, and the XTT assay that quantifies NADPH. At a concentration of 100 nM, there was no difference in cell viability amongst any of the cell types as measured by either assay (Figure 4A). No visible effects were detected in cell morphology (data not shown). At the higher 10 μM concentration, we began to observe a cell-type dependent and drug dependent effect on cell viability. DABL-CM displayed the greatest degree of toxicity, particularly in the CellTiter-Glo assay. The other compounds, DABL-N, DABL-NM and DABL–C, exhibited much less toxicity and were tolerated better than the previously reported Retro2.1 compound (Figure 4B). The trafficking inhibitor Brefeldin A was used as a positive control, in which prolonged treatment is known to cause cell death. Generally, there was no difference amongst the cell types as the presence of toxicity in one cell type corresponded with toxicity in the others, albeit at varying levels. However, the primary human fibroblasts appeared to be the most resilient cell type tested, indicating that sensitivity may increase with prolonged culture times.
Figure 4. DABL compounds are well tolerated by cells.
A) Cell viability was measured using CellTiter-Glo or XTT assays after treatment of cells with 100 nM or 10 μM of the indicated compounds. Cell lines tested were primary human dermal fibroblasts (NHDF), pigmented retinal epithelial cells (ARPE), kidney epithelial cells from African green monkey (VERO) and mouse fibroblasts (A31). B) Viability assays were conducted as in A with 10 μM of Retro2.1 or Brefeldin A. The graphed data are the average of three independent experiments.
2.5. Effects on polyomavirus replication
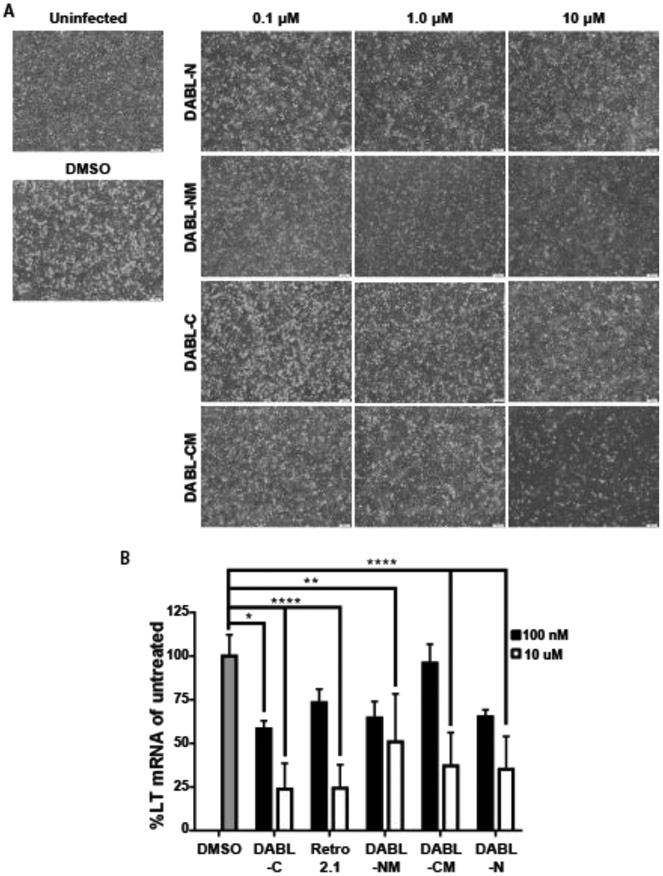
To assess the effect of the analogues on other opportunistic viruses, A31 monolayers were infected with mouse polyomavirus (MuPyV) at a low MOI on ice and subsequently treated with DMSO control, 100 nM, 1.0 μM or 10 μM of compound. The monolayers were imaged at six days post infection. Similar to what was observed for HCMV, virus spread was controlled at all concentrations of DABL-NM and DABL-CM (Figure 5A). While little to no protection was observed with 100 nM of DABL-C or DABL-N, the spread of polyomavirus was controlled by both compounds at 1.0 μM and 10 μM. Important to note, treatment with 10 μM DABL-CM resulted in clearing of the monolayer, confirming that this concentration was toxic to the cells, as was observed with the cell viability assays. Disruption of the monolayer was not observed for the other analogues at this higher concentration. To quantify MuPyV inhibition in the treated cells, levels of the viral large tumor antigen (LT) mRNA were measured by quantitative PCR (Figure 5B). Similar to the observations of viral disruption of the cell monolayer, at 10 μM all derivatives reduced viral mRNA levels 24 hours post infection. These results show that these analogues also block MuPyV, and do so at much lower concentrations than what was reported for the previously published parent compounds.
Figure 5. DABL compounds inhibit mouse polyoma virus.
(A) A31 cells uninfected or infected with mouse MuPyV and treated with increasing concentrations (100 nM, 1.0 μM or 10 μM) of the indicated compounds. (B) Total RNA was isolated 24 hpi from cells treated with the indicated concentration of compound. Levels of MuPyV LT mRNA were determined by quantitative PCR and normalized to untreated cells. Shown is mean (± SD), data is cumulative from two independent experiments (n=4 per treatment condition). Statistical significance was determined by 2-way ANOVA (*, **, **** p<0.05, 0.01, 0.0001).
3. Discussion
Through alterations of the parent Retro2.1 compound, we developed a series of analogues with nitro or cyano groups added to the quinazolinone alone or in combination with the addition of a methyl group. Similar to Retro2.1, these compounds induce cellular STX5 displacement from the Golgi upon treatment, leading to an inhibition of retrograde transport. The analogues demonstrate improved efficacy at inhibiting HCMV and MuPyV replication in infected cells, while exhibiting reduced toxicity in comparison with Retro2.1. With improved efficacy and reduced toxicity, these compounds are a promising target for in vivo studies examining the inhibition of cytomegalovirus and polyomavirus infections.
The use of a retrograde inhibitor for the treatment of opportunistic viral infections is attractive for multiple reasons. By disrupting a host cellular process utilized by the virus rather than targeting a viral protein directly, the action of these compounds is independent of structure or enzymatic activity of the viral proteins. This greatly reduces the risk of the emergence of viral mutants conferring resistance to these compounds. Additionally, immunosuppressed individuals are often at risk for the emergence of a variety of opportunistic pathogens. As these compounds inhibit multiple opportunistic viruses through a shared cellular pathway, they can be administered to treat or prevent multiple infections concurrently. Finally, the reduced cytotoxicity of these analogues is of particular importance in the context of BK virus-associated nephropathy in kidney transplant patients, where minimizing the potential nephrotoxic side effects of an antiviral to preserve kidney function is of the utmost importance[4, 5]. Together, these factors highlight the potential impact of a well-tolerated antiviral capable of targeting multiple opportunistic viruses through the modulation of a host process.
4. Experimental procedures
4.1. Chemical synthesis of DABL compounds
All reactions were carried out under an inert atmosphere with dry solvents under anhydrous conditions, unless otherwise stated. The materials obtained from Sigma-Aldrich and Thermo Fisher were used without further purification. All reactions were monitored by TLC on Silica Gel 60 F254 (Merck) using UV light detection. Silica gel (60-120 mesh size) for column chromatography was procured from SiliCycle (Quebec City, Canada) or compounds were purified using flash chromatography (Biotage). Microwave experiments were performed on CEM Discover focused microwave (250 MHz, 300W). 1H (500 MHz) NMR spectra were recorded on a Bruker Avance-500 spectrometer. Chemical shifts were recorded in δ (ppm) relative to the TMS signal, coupling constants (J) are given in Hz and multiplicities of the signals are reported as follows: s, singlet; d, doublet; dd, double doublet; t, triplet; m, multiplet; br, broad singlet. Purities of compounds were confirmed using 1H NMR and high-performance liquid chromatography (HPLC) analysis. Purity of final compounds were performed using a single Waters 610 pump on normal phase Phenomenex Lichorosorb Silica 60A column (5μ, 250 × 4.4mm) with 5%methanol in Methylene chloride at a flow rate of 1ml/min. The HPLC traces were monitored at 254 nm using Waters 2487 dual absorbance UV detector and Hitachi D-2500 Chromatography integrator. Reverse phased HPLC was run on a Vydac analytical column using two pumps and H2O: acetonitrile solvent system. Gradient program of 100% H2O to 100% acetonitrile in 60 minutes at a flow rate of 1 ml/min was used. The HPLC traces were monitored at 254 nm using Waters 2487 dual absorbance UV detectors. NMR and HPLC analysis showed that all tested compounds have >98% purity.
High Resolution Mass spectrometry (HRMS) was performed on Sciex 5800 MALDI TOF-TOF instrument, with the samples mixed 1:1 with MALDI matrix solution (5 mg/ml recrystallized CHCA (a-cyano-hydroxycinnamic acid), 2 mg/ml ammonium phosphate, 0.1% trifluoroacetic acid, 50% acetonitrile), with 1.0 ul of that 1:1 mixture spotted onto a stainless steel MALDI target plate and allowed to air dry. 500 laser shots at laser power 3200 were acquired and averaged from each sample/matrix spot, and the masses were automatically calculated from the observed times of flight by comparison to the times of flight from known standards in calibrant/matrix spots very close to the sample spots.
4.1.1. 5-Nitro-N-phenylbenzamide (3):
5-Nitroisatoic anhydride (208mg, 1 mmol) was mixed with aniline (1.1 mmol, 100mg), DMSO (0.5mL), and catalytic amount of DMAP (3mg) in a sealed tube and the resulting mixture was heated at 130°C overnight. The purple reaction mixture was cooled to room temperature and ethyl acetate was added to the mixture and sonicated to dissolve the entire residue. Flash chromatography [eluted with hexane : Ethyl acetate (8:2)] afforded compound 3 (160mg, 68%) as pale yellow powder. 1H-NMR (D4-MeOH) δ (ppm) 8.65 (d, 1H, J=2.5Hz), 8.12 and 8.10 (dd, 1H, J=2.5Hz, J=9.0Hz), 7.69-7.66 (m, 2H), 7.40-7.36 (m, 2H), 7.19-7.15 (m, 1H), 6.84 (d, 1H, J=9.5Hz).
4.1.2. 6-Nitro −2-(5-(2-methylthiazol-4-yl)thiophen-2-yl)-3-phenyl-2,3-dihydroquinazolin-4(1H)-one (DABL-N; 4):
5-Nitro-N-phenylbenzamide (3) (100mg, 0.39 mmol) and 5-(2-methylthiazol-4-yl)thiophene-2-carbaldehyde (90mg, 0.43 mmol) were dissolved in THF (8 mL) and a catalytic amount of PTSA (6 mg, 0.04 mmol) was added. The reaction mixture was refluxed overnight. After evaporation to dryness, the crude mixture was purified by flash chromatography on silica gel using hexane : Ethyl acetate (6:4) to yield compound 4 as pale yellow powder (101mg, 58%). 1H-NMR (CDCl3) δ (ppm) 8.88 (d, 1H, J=3.0Hz), 8.16 and 8.14 (dd, 1H, J=2.5Hz and J=9.0Hz), 7.38-7.34(m, 2H), 7.29-7.26 (m, 3H), 7.15 (s, 1H), 7.11 (d, 1H, J=3.5Hz), 6.80 (d, 1H, J=4.0Hz), 6.65 (d, 1H, 9.0Hz), 6.31 (d, 1H, J=2.5Hz), 2.70 (s, 3H, CH3); HRMS (M+H+) calculated for C22H16N4O3S2: 449.5196; found: 449.1263. Normal phase HPLC Rt: 3.20 min, Reverse phase HPLC Rt: 20.5 min
4.1.3. 6-Nitro-1-methyl-2-(5-(2-methylthiazol-4-yl)thiophen-2-yl)-3-phenyl-2,3-dihydroquinazolin-4(1H)-one (DABL-NM; 5):
6-Nitro-2-(5-(2-methylthiazol-4-yl) thiophen-2-yl)-3-phenyl-2,3-dihydroquinazolin-4(1H)-one (4) (50mg, 0.11 mmol) was carefully added to a suspension of NaH (6mg, 0.22 mmol, 10mg of 60 % in mineral oil, prewashed with hexanes) in anhydrous THF (10 mL). The resulting mixture was stirred at 0°C for 1 hr, followed by MeI (31mg, 0.22 mmol) dropwise added, and the reaction was allowed to proceed overnight at room temperature. The reaction flask was cooled down to 0°C, and the reaction mixture was quenched with saturated NaHCO3. The aqueous layer was extracted with CH2Cl2 (3 × 30ml), washed with 1N HCl (2 × 10mL), dried over MgSO4, filtered, and evaporated to dryness. The crude product was purified over silica gel column by eluting with hexane : Ethyl acetate (6:4) to give DABL-NM (5) in 71% yield. 1H NMR (CDCl3) δ (ppm) 9.0 (d, 1H, J=2.5Hz), 8.33 and 8.31 (dd, 1H, J=2.5Hz and J=9.0Hz), 7.43-7.39 (m, 2H), 7.35-7.28 (m, 3H), 7.19 (d, 1H, J=3.5Hz), 7.18 (ds, 1H), 6.85 (d, 1H, J=3.5Hz), 6.72 (d, 1H, J=9.0Hz), 6.09 (s, 1H), 3.16 (s, 3H, NCH3), 2.72 (s, 3H, CH3); HRMS (M+H+) calculated for C23H18N4O3S2: 463.5462; found: 463.1108. Normal phase HPLC Rt: 3.10 min, Reverse phase HPLC Rt: 20.76 min.
4.1.4. 5-Cyano-N-phenylbenzamide (8):
5-cyano-2-aminobenzoic acid (6) (100mg, 0.62 mmol) was mixed with aniline (7) (68mg, 0.75 mmol), DMSO (0.5mL), and a catalytic amount of DMAP (3mg) in a microwave glass vial that was irradiated in a microwave apparatus (LABMAT) at 130°C, 250 W for 30 min. After the reaction mixture was cooled to ambient temperature, the purple brown reaction mixture was diluted with ethyl acetate and sonicated to dissolve the entire residue. The organic fraction was washed with water (3 × 10 mL), dried over MgSO4, filtered, and evaporated to give the crude product. Flash chromatography [eluted with hexane : Ethyl acetate (8:2)] afforded 8 (130mg, 89%) as pale yellow powder. 1H-NMR (D4-MeOH) δ (ppm) 8.04 (d, 1H, J=1.8Hz), 7.66 (d, 2H, J=8.4Hz), 7.50 and 7.48 (dd, 1H, J=1.8Hz and J=9.0Hz), 7.37 (t, 2H, J=7.2Hz and J=7.8Hz), 7.17 (t, 1H, J=8.4Hz, J=7.2Hz), 6.86 (d, 1H, J=9.0Hz).
4.1.5. 6-Cyano-2-(5-(2-methylthiazol-4-yl)thiophen-2-yl)-3-phenyl-2,3-dihydroquinazolin-4(1H)-one (DABL-C, 9):
5-Cyano-N-phenylbenzamide (8) (200mg, 0.84 mmol) and 5-(2-methylthiazol-4-yl)thiophene-2-carbaldehyde (90mg, 0.43 mmol) were dissolved in THF (8 mL) and a catalytic amount of PTSA (6 mg, 0.04 mmol) was added. The reaction mixture was refluxed overnight. After evaporation to dryness, the crude mixture was purified by flash chromatography on silica gel [eluted with hexane : Ethyl acetate (6:4)] to yield 6-Cyano-2-(5-(2-methylthiazol-4-yl)thiophen-2-yl)-3-phenyl-2,3- dihydroquinazolin-4(1H)-one (8) as pale yellow powder (293mg, 81%). 1H-NMR (CDCl3) δ (ppm) 8.37 (d, 1H, J=1.8Hz), 7.60 and 7.59 (dd, J=1.8Hz and J=8.4Hz), 7.42-7.39(m, 2H), 7.33-7.29 (m, 3H), 7.19 (s, 1H), 7.17 (d, 1H, J=3.6Hz), 6.86 (d, 1H, J=3.6Hz), 6.77 (d, 1H, J=8.4Hz), 6.37 (d, 1H, J=3.0Hz), 5.37 (d, 1H, J=2.4Hz), 2.75 (s, 3H, CH3); HRMS (M+H+) calculated for C23H16N4OS2: 429.5315; found: 429.1397. Normal phase HPLC Rt: 3.44 min; Reverse phase HPLC Rt: 20.26 min
4.1.6. 6-Cyano-1-methyl-2-(5-(2-methylthiazol-4-yl)thiophen-2-yl)-3-phenyl-2,3-dihydroquinazolin-4(1H)-one (DABL-CM, 10):
6-Cyano-2-(5-(2-methylthiazol-4-yl)thiophen-2-yl)-3-phenyl-2,3- dihydroquinazolin-4(1H)-one (9) (120mg, 0.28 mmol) was carefully added to a suspension of NaH (14mg, 0.58 mmol, 23mg of 60 % in mineral oil, prewashed with hexanes) in anhydrous THF (10 mL). The resulting mixture was stirred at 0°C for 1 hr. MeI (63mg, 0.44 mmol) was added dropwise and the reaction was allowed to proceed overnight at room temperature. The reaction flask was cooled down to 0°C, and the reaction mixture was quenched with saturated NaHCO3. The aqueous layer was extracted with CH2Cl2 (3 × 30ml), washed with 1N HCl (2 × 10mL), dried over MgSO4, filtered, and evaporated to dryness. The crude product was purified over silica gel column by eluting with hexane : Ethyl acetate (6:4) to give DABL-CM (10) in 92% yield. 1H NMR (CDCl3) δ (ppm) 8.42 (d, IH, J=1.8Hz), 7.69 and 7.67 (dd, IH, J=2.4Hz and J=9.0Hz), 7.43-7.40 (m, 2H), 7.35-7.29 (m, 3H), 7.20 (d, IH, J=4.2Hz), 7.19 (s, IH), 6.84 (d, IH, J=3.6Hz), 6.73 (d, IH, J=9.0Hz), 6.07 (s, IH), 3.11 (s, 3H, N-CH3), 2.74 (s, 3H, CH3); HRMS(M+H+) calculated for C24H18N4OS2: 443.5581; found: 443.1668. Normal phase HPLC Rt: 3.21 min; Reverse phase HPLC Rt: 21 min.
4.2. Cell culture
Normal human dermal fibroblasts (HDFs) (Cell Applications Inc., 106-05N), normal human lung fibroblasts (MRC-5) (ATCC, CCL-171), BALB/3T3 clone A31 mouse fibroblasts (A31) (ATCC, CCL-163) and African green monkey kidney epithelial cells (VERO) were maintained in Dulbecco’s modified Eagle’s medium (DMEM) (Lonza). Human retinal pigmented epithelial cells (ARPE-19) were maintained in DMEM/Ham’s F12 (1:1 mix) (Lonza). All media was supplemented with 10% fetal bovine serum, 100 U/ml penicillin and 100 g/ml streptomycin, and 2 mM GlutaMAX (Gibco). Cells were maintained at 37°C with 5% CO2.
4.3. Cell viability assays
Cells used for viability assays in a 96 well plate were treated with DMSO or analogue compound (100 nm and 10 μM) for 96 hours. For the XTT assay, media was removed from all wells and replaced with fresh media. XTT and activating reagents (Biotium) were added to each well according to manufacturer’s instructions. Cells were incubated for 2 hours at 37°C, and absorbance was measured using a Synergy H1 Hybrid Reader (BioTek). For CellTiter-Glo Luminescent Cell Viability Assay (Promega), the media was replaced with fresh media and reagents were added to each well according to manufacturer’s instructions. Luminescence was measured using the Synergy H1 Hybrid Reader.
4.4. Immunofluorescence staining and confocal microscopy
Coverslips containing either HDFs were washed in phosphate-buffered saline (PBS) and fixed in 2% paraformaldehyde for 15 min at room temperature. Cells were blocked in PBS containing 10% human serum, 0.5% Tween-20, and 5% glycine. 0.1% Triton-X 100 was added for permeabilization. Primary antibodies used were against STX5 (Santa Cruz, sc-365124) and p115 (Proteintech, 13509-1-AP) as a Golgi marker. Secondary antibodies were Alexa Fluor 488 or 568 (ThermoFisher Scientific). Coverslips were mounted with ProLong Diamond Antifade Mountant containing DAPI (ThermoScientific). All images were acquired using a C2+ Confocal Microscope System (Nikon). Images were processed using NIS elements software. Images shown are volume renderings of Z-stacks.
4.5. HCMV production and infection assay
HCMV TB40/E was generated from BAC stocks. To produce virus stocks, BAC DNA was electroporated into MRC5 cells according to previously published protocols [21]. Virus-producing cells were saved in liquid nitrogen and used to infect cells in roller bottles to produce larger stocks of virus. Virus stocks were concentrated by high-speed centrifugation on a 20% sorbitol cushion at 20,000 rpm, for 1hr at 20°C in a Beckman SW32 rotor. Infections were done at a multiplicity of infection (MOI) 3 for single-step growth curves. Infections were done on NHDFs. Virus was incubated with cells for 2 hours before addition of fresh media, at which time DMSO or analogue compound (10 nM, 100 nM, 1 μM or 10 μM) was added. Virus was harvested at the indicated time-points for each experiment by scraping the cells into the medium, sonicating 10 times with 1 second pulses, vortexing for 15 seconds, and centrifuging at 13,000 rpm for 10 min. Supernatants were collected, aliquoted and flash-frozen in liquid nitrogen before storage at −80°C. HCMV was titrated by serial dilutions on MRC5 cells and quantified by the immunological detection of immediate-early proteins using antibodies that detect major immediate early antigens (EX2/3, gift from Jim Alwine[22] and monoclonal D4 antibody, generated by Neil Christensen). Images were acquired on a Nikon Eclipse Ti Inverted microscope and fluorescent nuclei were quantified using the NIS Elements Software.
4.6. Polyomavirus infection assay
The A2 strain of MuPyV was generated in baby mouse kidney cells as previously described[23]. A31 cells were seeded in a 6-well plate and infected the following day at an MOI of 0.1 for 1.5 hours on ice. The cells were washed twice with 10% FBS DMEM medium and treated with the indicated concentration of compound in 2 ml of 10% FBS DMEM. Images were taken at 6 days post infection on an Olympus IX73 using cellSens Dimension software. For MuPyV mRNA quantification, A31 cells were seeded in 12-well plates and infected the following day at an MOI of 1 for 1.5 hours at 37°C. The cells were washed twice with 10% FBS DMEM and treated with the indicated concentration of compound in 10% FBS DMEM for 24 hours. Total RNA was then isolated from the cells using TRI reagent (Sigma-Aldrich) and 2 μg of RNA was converted to cDNA. Levels of MuPyV LT mRNA were determined and normalized to TBP by quantitative PCR as previously described[11].
Supplementary Material
Acknowledgements
We would like to thank Kayla Keller for technical assistance and lab support and Neil Christensen for his generous help in generating a monoclonal antibody used in the assay for titering HCMV infectious virions. We would also like to thank Anne Stanley in the Mass Spectrometry and Proteomics Core Facility and Jyh-Ming Lin of NMR facility at Penn State College of Medicine for MS and NMR, respectively. This work was supported by grants from the H.G. Barsumian M.D. Memorial Fund (N.J.B), the W.W. Smith Charitable Trust (N.J.B, D.D.), the Penn State Cancer Institute (D.D., S.A.) and the National Institutes of Health (NS088367 and NS092662 (A.E.L)).
Abbreviations
- HCMV
human cytomegalovirus
- STX5
syntaxin 5
- MuPyV
mouse polyomavirus
- LT
large tumor antigen
References
- [1].Janeczko M, Mielcarek M, Rybka B, Ryczan-Krawczyk R, Noworolska-Sauren D, Kałwak K, Immune recovery and the risk of CMV/ EBV reactivation in children post allogeneic haematopoietic stem cell transplantation, Cent Eur J Immunol, 41 (2016) 287–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Fan J, Jing M, Yang M, Xu L, Liang H, Huang Y, Yang R, Gui G, Wang H, Gong S, Wang J, Zhang X, Zhao H, Gao H, Dong H, Ma W, Hu J, Herpesvirus infections in hematopoietic stem cell transplant recipients seropositive for human cytomegalovirus before transplantation, Int J Infect Dis, 46 (2016) 89–93. [DOI] [PubMed] [Google Scholar]
- [3].Ramanan P, Razonable RR, Cytomegalovirus infections in solid organ transplantation: a review, Infect Chemother, 45 (2013) 260–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Hirsch HH, Brennan DC, Drachenberg CB, Ginevri F, Gordon J, Limaye AP, Mihatsch MJ, Nickeleit V, Ramos E, Randhawa P, Shapiro R, Steiger J, Suthanthiran M, Trofe J, Polyomavirus-associated nephropathy in renal transplantation: interdisciplinary analyses and recommendations, Transplantation, 79 (2005) 1277–1286. [DOI] [PubMed] [Google Scholar]
- [5].Pahari A, Rees L, BK virus-associated renal problems--clinical implications, Pediatr Nephrol, 18 (2003) 743–748. [DOI] [PubMed] [Google Scholar]
- [6].Tan CS, Koralnik IJ, Progressive multifocal leukoencephalopathy and other disorders caused by JC virus: clinical features and pathogenesis, Lancet Neurol, 9 (2010) 425–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Stechmann B, Bai SK, Gobbo E, Lopez R, Merer G, Pinchard S, Panigai L, Tenza D, Raposo G, Beaumelle B, Sauvaire D, Gillet D, Johannes L, Barbier J, Inhibition of retrograde transport protects mice from lethal ricin challenge, Cell, 141 (2010) 231–242. [DOI] [PubMed] [Google Scholar]
- [8].Gupta N, Pons V, Noël R, Buisson DA, Michau A, Johannes L, Gillet D, Barbier J, Cintrat JC, (S)-N-Methyldihydroquinazolinones are the Active Enantiomers of Retro-2 Derived Compounds against Toxins, ACS Med Chem Lett, 5 (2014) 94–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Wessels E, Duijsings D, Niu TK, Neumann S, Oorschot VM, de Lange F, Lanke KH, Klumperman J, Henke A, Jackson CL, Melchers WJ, van Kuppeveld FJ, A viral protein that blocks Arf1-mediated COP-I assembly by inhibiting the guanine nucleotide exchange factor GBF1, Dev Cell, 11 (2006) 191–201. [DOI] [PubMed] [Google Scholar]
- [10].Nelson CD, Carney DW, Derdowski A, Lipovsky A, Gee GV, O’Hara B, Williard P, DiMaio D, Sello JK, Atwood WJ, A retrograde trafficking inhibitor of ricin and Shiga-like toxins inhibits infection of cells by human and monkey polyomaviruses, MBio, 4 (2013) e00729–00713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Maru S, Jin G, Desai D, Amin S, Shwetank, Lauver MD, Lukacher AE, Inhibition of Retrograde Transport Limits Polyomavirus Infection, mSphere, 2 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Sivan G, Weisberg AS, Americo JL, Moss B, Retrograde Transport from Early Endosomes to the trans-Golgi Network Enables Membrane Wrapping and Egress of Vaccinia Virus Virions, J Virol, 90 (2016) 8891–8905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Harrison K, Haga IR, Pechenick Jowers T, Jasim S, Cintrat JC, Gillet D, Schmitt-John T, Digard P, Beard PM, Vaccinia virus uses retromer-independent cellular retrograde transport pathways to facilitate the wrapping of intracellular mature virions during viral morphogenesis, J Virol, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Shtanko O, Sakurai Y, Reyes AN, Noël R, Cintrat JC, Gillet D, Barbier J, Davey RA, Retro-2 and its dihydroquinazolinone derivatives inhibit filovirus infection, Antiviral Res, 149 (2018) 154–163. [DOI] [PubMed] [Google Scholar]
- [15].Dai W, Wu Y, Bi J, Lu X, Hou A, Zhou Y, Sun B, Kong W, Barbier J, Cintrat JC, Gao F, Gillet D, Su W, Jiang C, Antiviral effects of Retro-2 and Retro-2.1 against Enterovirus 71 in vitro and in vivo., Antiviral Res, 144 (2017) 311–321. [DOI] [PubMed] [Google Scholar]
- [16].Cruz L, Streck NT, Ferguson K, Desai T, Desai DH, Amin SG, Buchkovich NJ, Potent Inhibition of Human Cytomegalovirus by Modulation of Cellular SNARE Syntaxin 5, J Virol, 91 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Dai W, Wu Y, Bi J, Wang J, Wang S, Kong W, Barbier J, Cintrat JC, Gao F, Jiang Z, Gillet D, Su W, Jiang C, Antiviral Effect of Retro-2.1 against Herpes Simplex Virus Type 2 In Vitro, J Microbiol Biotechnol, 28 (2018) 849–859. [DOI] [PubMed] [Google Scholar]
- [18].Carney DW, Nelson CD, Ferris BD, Stevens JP, Lipovsky A, Kazakov T, DiMaio D, Atwood WJ, Sello JK, Structural optimization of a retrograde trafficking inhibitor that protects cells from infections by human polyoma- and papillomaviruses, Bioorg Med Chem, 22 (2014) 4836–4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Fleming FF, Yao L, Ravikumar PC, Funk L, Shook BC, Nitrile-containing pharmaceuticals: efficacious roles of the nitrile pharmacophore, J Med Chem, 53 (2010) 7902–7917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Noel R, Gupta N, Pons V, Goudet A, Garcia-Castillo MD, Michau A, Martinez J, Buisson DA, Johannes L, Gillet D, Barbier J, Cintrat JC, N-methyldihydroquinazolinone derivatives of Retro-2 with enhanced efficacy against Shiga toxin, J Med Chem, 56 (2013) 3404–3413. [DOI] [PubMed] [Google Scholar]
- [21].Rabouille C, Kondo H, Newman R, Hui N, Freemont P, Warren G, Syntaxin 5 is a common component of the NSF- and p97-mediated reassembly pathways of Golgi cisternae from mitotic Golgi fragments in vitro, Cell, 92 (1998) 603–610. [DOI] [PubMed] [Google Scholar]
- [22].Harel NY, Alwine JC, Phosphorylation of the human cytomegalovirus 86-kilodalton immediate-early protein IE2, J Virol, 72 (1998) 5481–5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Lukacher AE, Wilson CS, Resistance to polyoma virus-induced tumors correlates with CTL recognition of an immunodominant H-2Dk-restricted epitope in the middle T protein, J Immunol, 160 (1998) 1724–1734. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.