Abstract
Microsomal cytochrome b5 (cytb5) is a membrane-bound protein capable of donating the second electron to cytochrome P450s (cytP450s) in the cytP450s monooxygenase reactions. Recent studies have demonstrated the importance of the transmembrane domain of cytb5 in the interaction with cytP450 by stabilizing its monomeric structure. While recent NMR studies have provided high-resolution insights into the structural interactions between the soluble domains of ~16-kDa cytb5 and ~57-kDa cytP450 in a membrane environment, there is need for studies to probe the residues in the transmembrane region as well as to obtain intermolecular distance constraints to better understand the very large size cytb5-cytP450 complex structure in a near native membrane environment. In this study, we report the expression, purification, functional reconstitution of 19F-labeled full- length rabbit cytb5 in peptide based nanodiscs for structural studies using NMR spectroscopy. Size exclusion chromatography, dynamic light scattering, transmission electron microscopy, and NMR experiments show a stable reconstitution of cytb5 in 4F peptide-based lipid-nanodiscs. The reported results demonstrate the use of 19F NMR experiments to study 19F-labeled (with 5-fluorotryptophan (5FW)) cytb5 reconstituted in peptide-nanodiscs and the detection of residues from the transmembrane domain by solution 19F NMR experiments. 19F NMR results revealing the interaction of the transmembrane domain of cytb5 with the full- length rabbit cytochrome P450 2B4 (CYP2B4) are also presented. We expect the results presented in this study to be useful to devise approaches to probe the structure, dynamics and functional roles of transmembrane domains of a membrane protein, and also to measure intermolecular 19F-19F distance constraints to determine the structural interactions between the transmembrane domains.
INTRODUCTION
Despite the recent advances in high-resolution structural studies of membrane proteins, obtaining their high-resolution dynamics continues to be a major challenge. Although both solution and solid-state NMR spectroscopy can be used to characterize the dynamics of membrane proteins, low sensitivity and resolution are the key limiting factors of NMR techniques for studies on challenging membrane proteins. One of the strategies used to overcome these challenges is to selectively label the protein with 19F nuclei as 19F NMR experiments can probe the local structure of a protein as well as to measure 19F-19F distance constraints for high-resolution structural studies[1–5]. In this study, we extend this approach to study a membrane-bound cytochrome b5 (cytb5).
Microsomal cytb5 is a 134–residue electron transport hemeprotein primarily associated with the endoplasmic reticulum of eukaryotic cells, and plays a vital role in the enzymatic function of cytochrome P450s[6–8]. Cytb5 can donate the second of the two required electrons to cytochrome P450s (CYP P450s), which can catalyze the metabolism of endogenous and exogenous substrates including fatty acids, sterols and over 50% clinical drugs[8–14]. Therefore, it is very important to measure the structural and dynamical interactions between the full-length membrane-bound cytb5 and cytP450s to better understand the catalytic mechanism of cytP450.
Cytb5 is comprised of three distinct regions: a heme containing soluble N-terminus (residue 1–89), a hydrophobic transmembrane C-terminus (residue 105–134), and a 15-residue flexible linker (residue 90–104) connecting the soluble and the transmembrane domains[6, 15–18]. Several studies have shown that the soluble domain of cytb5, lacking the transmembrane domain, aggregates to form oligomers and they do not interact with cytP450 in a physiologically-relevant manner[6, 19–21]. Moreover, our previous studies have demonstrated the significance of the lipid membrane on the structural folding and dynamics of cytb5 and cytP450 and the structural interactions between them and with membrane [22–26]. Therefore, to better understand the role of the transmembrane domain of cytb5, it is important to investigate the structure and dynamics of the full-length cytb5 in a membrane environment. However, the membrane-bound full-length cytb5 poses tremendous challenges for atomic-level structural studies by X-ray crystallography or solution-state nuclear magnetic resonance (NMR) spectroscopy[22–25]. The transmembrane residues of cytb5 are invisible to solution NMR spectroscopy[24], and only solid-state NMR studies are capable of probing the structure and dynamics of the transmembrane helix [25, 27]. Static solid-state NMR experiments on magnetically-aligned bicelles[27, 28] and polymer-nanodiscs[29] were used to determine the topology of the transmembrane helix of cytb5 and aligned bicelles were used to determine the tilt of the transmembrane helix of cytP450[30]. Remarkably, the interaction between the transmembrane domains of cytb5 and cytP450 reconstituted in a lipid bilayer was probed using dynamic nuclear polarization (DNP) based magic angle spinning (MAS) solid-state NMR experiments[31].
Tryptophan is abundant in membrane proteins, mostly located at the lipid-water interface, where it is thought to play a significant role as a membrane anchor [32–35]. Computational and experimental studies report that tryptophan residues of a membrane protein contribute to the stability and orientation of transmembrane domains, and play a significant role in membrane deformations[32, 35–39]. Cytb5 has 4 tryptophan residues: W27 is located in the N-terminus while the other three Trp residues are located in the transmembrane domain. In this study, the full-length cytb5 was labeled with 5-fluorotryptophan (5FW), and we demonstrate the use of 19F NMR experiments to probe the transmembrane domain of 19F-labeled cytb5 by solution NMR experiments. Recently lipid bilayer nanodiscs have been successfully used in the structural studies of membrane proteins [24, 40–43]. Here we used peptide-based lipid nanodiscs to reconstitute cytb5 or cyb5-CYP2B4 complex that exhibited stability for more than a week at room temperature [24, 44]. These nanodiscs exhibited a size ~8 nm diameter, and their fast tumbling nature allowed the detection of transmembrane residues of membrane-bound cytb5 by solution NMR.
Materials and Methods
Materials
Potassium phosphate monobasic and dibasic, glycerol, chloroform and 3,5-di-tert-butyl-4-hydroxytouene (BHT), 5-fluoroindole (98%) were purchased from Sigma-Aldrich. 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC), 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-L-serine (POPS), 1,2-dimyristoyl-sn-glycerol-3-phosphocholine (DMPC), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE), L-α-phosphatidylinositol (sodium salt) (PI), sphingomyelin (SM), and cholesterol (Chol) were purchased from Avanti Polar Lipids Inc.
Protein expression and purification
Plasmids containing the genes of cytb5 were transformed into Escherichia coli strain C41. 5-fluorotryptophan (5FW) labeling was obtained by adding 5-fluoroindole (5FI). A single colony was picked from the LB plate and transferred into 100 ml of LB medium containing 100 μg/ml carbenicillin. It was incubated overnight at 30 °C under shaking at 200 rpm. 10 ml of the overnight culture was centrifuged, decanted the supernatant, and transferred the pellets into 100 ml TB medium supplemented with a final concentration of 100 μg/ml carbenicillin. The culture was incubated at 35 °C under shaking at 180 rpm. After 2 hours, the above culture was centrifuged, decanted the supernatant, and transferred the pellets into 1 L M9 medium supplemented with 0.1 g of each of the amino acids (except tryptophan). The 5-fluorotryptophan (5FW) labeling of cytb5 was obtained by adding 5-fluoroindole 10 minutes before adding IPTG[45]. The other expression conditions and purification procedures were similar as reported previously [15]. The sample purity is verified by running SDS-PAGE gel (Figure 1B).
Figure 1. Reconstitution of cytochrome b5 in peptide-based lipid-nanodiscs.
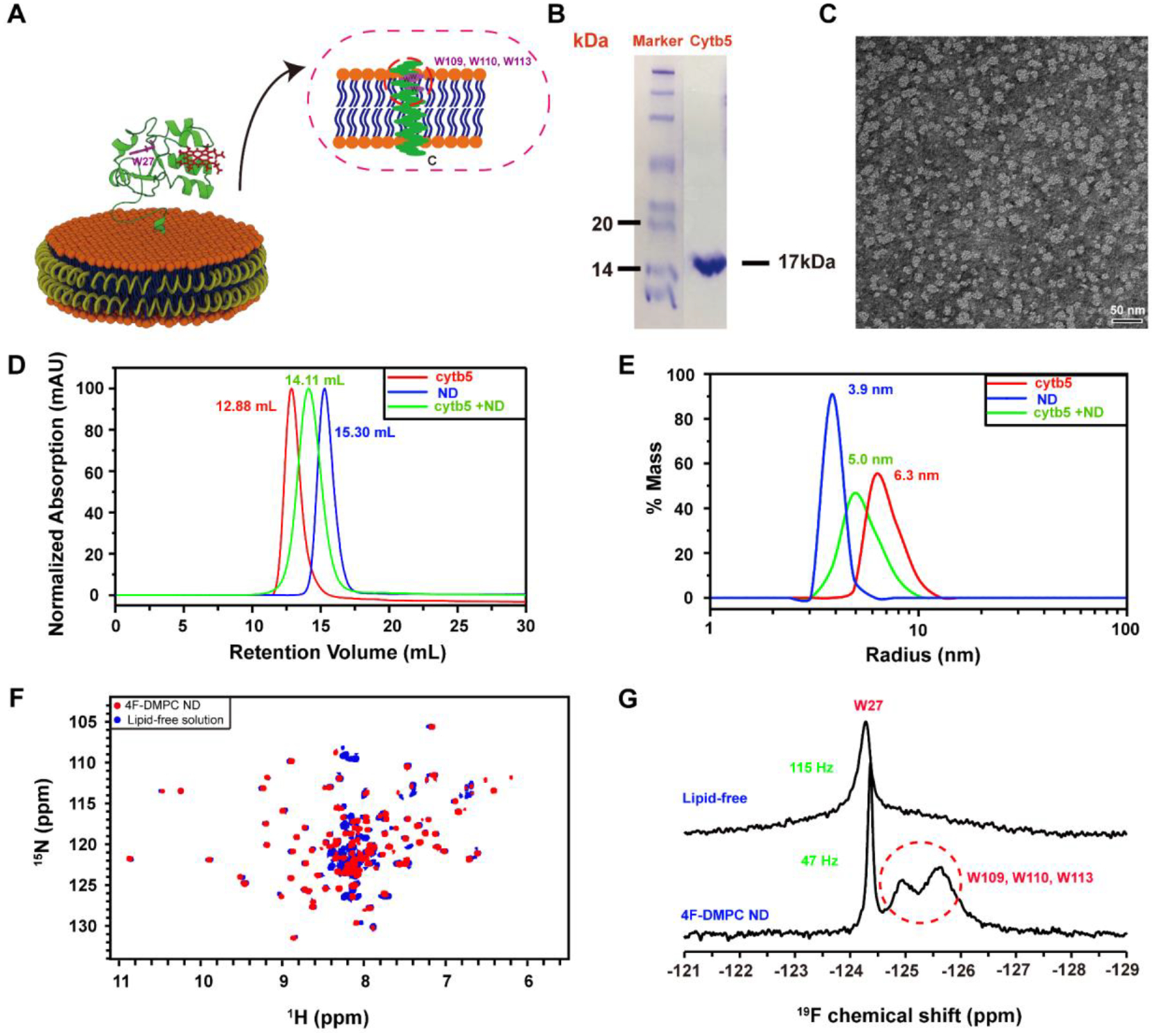
(A) Ribbon diagrams of cytochrome b5 in a nanodisc indicating 5F-tryptophan-labeled sites. (B) SDS-PAGE gel showing the purity 5FW-labeled cytb5. (C) TEM image of 4F-DMPC nanodiscs with a diameter of ~10 nm. (D) SEC elution profiles and (E) DLS profiles of cytb5 alone in buffer (red), empty 4F-DMPC nanodiscs (blue), and 4F-DMPC nanodiscs containing cytb5 (green). (F) 2D 1H-15N HSQC spectra of 15N-labeled cytb5 alone in buffer (blue) and reconstituted in the 4F-DMPC nanodiscs (red). (G) 1D 19F NMR spectra of 5F-tryptophan-labeled cytb5 alone in buffer (top trace) and reconstituted in the 4F-DMPC nanodiscs (bottom trace) at 298K.
Preparation of lipid-nanodiscs.
Lipid powders (except DMPC) were dissolved in HPLC-grade chloroform to make a stock solution at 20 mg/mL. DMPC powder was suspended in buffer A (40 mM potassium phosphate, pH 7.4) to make a stock solution at 20 mg/mL. A 18 residue helical amphipathic peptide scaffold (DWFKAFYDKVAEKFKEAF; referred to as 4F hereafter)[46, 47] was dissolved in buffer A to make a stock solution at 10 mg/mL. Nanodiscs were prepared as narrated below.
Preparation of 4F-DMPC nanodiscs:
DMPC and 4F stock solution were mixed together at a peptide:lipid ratio of 1:1.5 w/w, then incubated at 37 °C, 180 rpm overnight.
Preparation of 4F-POPC (or 4F-POPS, 4F-POPC/PS (8:2 molar ratio)) nanodiscs:
An aliquot of the dissolved POPC (or POPS, or POPC/PS (8:2 molar ratio) was transferred to an Eppendorf tube, and dried under a gentle flow of nitrogen gas, followed by a 5–10 minutes drying in vacuum. The resultant lipid film was hydrated with buffer A, and mixed with 4F peptide solution at 1:1 w/w peptide:lipid. The mixture underwent 3–5 freeze-thaw cycles between −80 °C and room temperature until it became clear, then it was placed in a shaker at room temperature overnight.
Preparation of 4F-Endoplasmic Reticulum (ER) lipids-nanodiscs:
Lipids for a biomimetic ER was mixed with the following molar ratios: 58% POPC, 7% POPS, 20% POPE, 7% POPI, 4% SM, 4% chol. The mixture was dried under nitrogen gas, followed by a 5–10 minutes drying in vacuum. The resultant lipid film was hydrated with buffer A, and then mixed with 4F peptide solution at a peptide:lipid ratio of 1:0.75 w/w. The mixture underwent three to five freeze-thaw cycles between −80 °C and room temperature, then was incubated at room temperature overnight. The nanodiscs were purified and characterized using size exclusion chromatography (SEC). A Superdex 200 Increase 300/10 GL column was used on an AKTA purifier (GE healthcare, Freiburg, Germany) by using 40 mM potassium phosphate buffer (pH 7.4) at a flow rate of 0.75 mL/min. The radius of nanodiscs was characterized by dynamic light scattering (DLS) experiments using Wyatt Technology® DynaPro® NanoStar®. The scattering curves were fitted by the isotropic sphere model.
Preparation of nanodiscs containing cytb5 or the cytb5-CYP2B4 complex.
To reconstitute cytb5 into nanodiscs, Cytb5 was mixed with the aforementioned purified nanodiscs at a molar ratio of 1:1.2 cytb5:nanodiscs. The mixture was then incubated at room temperature (or 37 °C) overnight. SEC was subsequently used for the purification of nanodiscs containing cytb5 from free cytb5 and empty nanodiscs. Fractions were collected and concentrated for NMR experiments. A previously published protocol was used to reconstitute the cytb5-CYP2B4 complex into 4F peptide based lipid-nanodiscs.[24] In brief, 4F peptide nanodiscs were first prepared by the mixing the desired amount of lipids and 4F peptide in 40 mM potassium phosphate buffer (pH 7.4) at room temperature. The solution was subjected to freeze-thaw cycles or overnight incubation to form nanodiscs via the self-assembly process. After that, the solution was purified using SEC to obtain nanodiscs by removing any free lipids and 4F peptide. These purified nanodiscs were characterized by DLS and TEM to determine the size and polydispersity. Then, cytb5 in buffer was added to the pre-formed 4F peptide lipid-nanodiscs. The mixture of nanodiscs and cytb5 was subjected to SEC purification to remove empty nanodiscs and any free (monomeric or aggregated) cytb5. The purified cytb5 reconstituted nanodiscs were then characterized by UV to confirm the presence of cytb5 (by monitoring the Soret band whose maximum absorption observed around 413 nm) and DLS to qualitatively determine the size of the nanodiscs. After the successful reconstitution of cytb5, a solution of unlabeled full-length CYP2B4 was added at a molar ratio of 1:1.2 cytb5:CYP2B4. The mixture was then incubated overnight at 37 °C. After the incubation, 4F peptide nanodiscs containing the cytb5-CYP2B4 complex were purified using SEC, and then characterized by UV and DLS.
NMR experiments
NMR spectra were acquired on a Bruker 500 MHz NMR spectrometer equipped with a 19F/1H/13C/15N probe. 40 mM potassium phosphate was used as the NMR buffer. Protein was dissolved in 90% NMR buffer and 10% D2O, or reconstituted in 4F-based nanodiscs as described above. 5FW-labeled cytb5 (0.3 mM) was used to record 19F NMR spectra. 19F NMR spectra were acquired with 18.8 kHz spectral width, 16 K complex points, and a recycle delay of 2 s. Proton decoupling was applied to obtain all 19F NMR spectra presented in this study.
Results and Discussion
19F solution NMR detects the resonances from the transmembrane domain
There are four tryptophans and three phenylalanines in cytb5. 19F-labeled cytb5 samples were obtained by using tryptophan with 5-fluoro-tryptophan (5FW) and phenylalanine with m-fluoro-phenylalanine. This procedure worked out well for the production of 19F-trptophan-labeled cytb5, but not for 19F-phenylalanine labeling. Hence, we only used 19F-tryptophan-labeled cytb5 in this study.
Cytb5 has four tryptophan (W27, W109, W110, W113), one located in the soluble domain and the other three are located in the transmembrane domain (Figure 1A). We labeled the tryptophans with 5-fluorotryptophan (5FW), and reconstituted the 5FW-labeled cytb5 in 4F-DMPC nanodiscs. The size of the nanodiscs was characterized by TEM (Figure 1C) and DLS (Figure 1E). The SEC profiles (Figure 1D) showed that cytb5 is successfully reconstituted in 4F-DMPC nanodiscs. 19F NMR spectra of 5FW-labeled cytb5 in the lipid-free solution (i.e., in buffer) and solution containing 4F-DMPC nanodiscs were acquired (Figure 1G). In the lipid-free solution, only a single narrow peak is observed, and a very low intensity broad peak can also be seen in the spectrum. On the other, in the solution containing 4F-DMPC nanodiscs, three resolved 19F peaks are observed. The narrowest peak is assigned to W27 residue located in the mobile soluble domain of the protein, whereas the two broad peaks marked with a red broken lined circle are assigned to W109, W110 and W113 (Figure 1G) which are located in the transmembrane domain of the protein. The W27 peak shows a line width of ~122 Hz in lipid-free solution, whereas it is ~49 Hz in 4F-DMPC nanodiscs, suggesting that cytb5 is aggregated in solution while it is a monomer in nanodiscs. On the other hand, the transmembrane Trp residues (W109, W110 and W113) are invisible in 2D 1H/15N HSQC NMR spectra of U-15N-cytb5 even when reconstituted in nanodiscs (Figure 1F), which is in agreement with our previous studies[24]. 19F NMR spectra of cytochrome b5 in lipid-free solution or reconstituted in nanodiscs were also acquired by varying the temperature of the sample (Figure 2). In the lipid-free solution, a single peak was observed regardless of the sample temperature (Figure 2A). However, in the 4F-DMPC nanodiscs, increasing sample temperature revealed four resolved 19F peaks due to the increasing tumbling rate of the cytb5-nanodiscs as well as due to the transition from the gel to liquid-crystalline phase (Figure 2B).
Figure 2. 19F NMR spectra of 5FW-labeled cytb5 reconstituted in nanodiscs composed with different types of lipids at different temperatures.
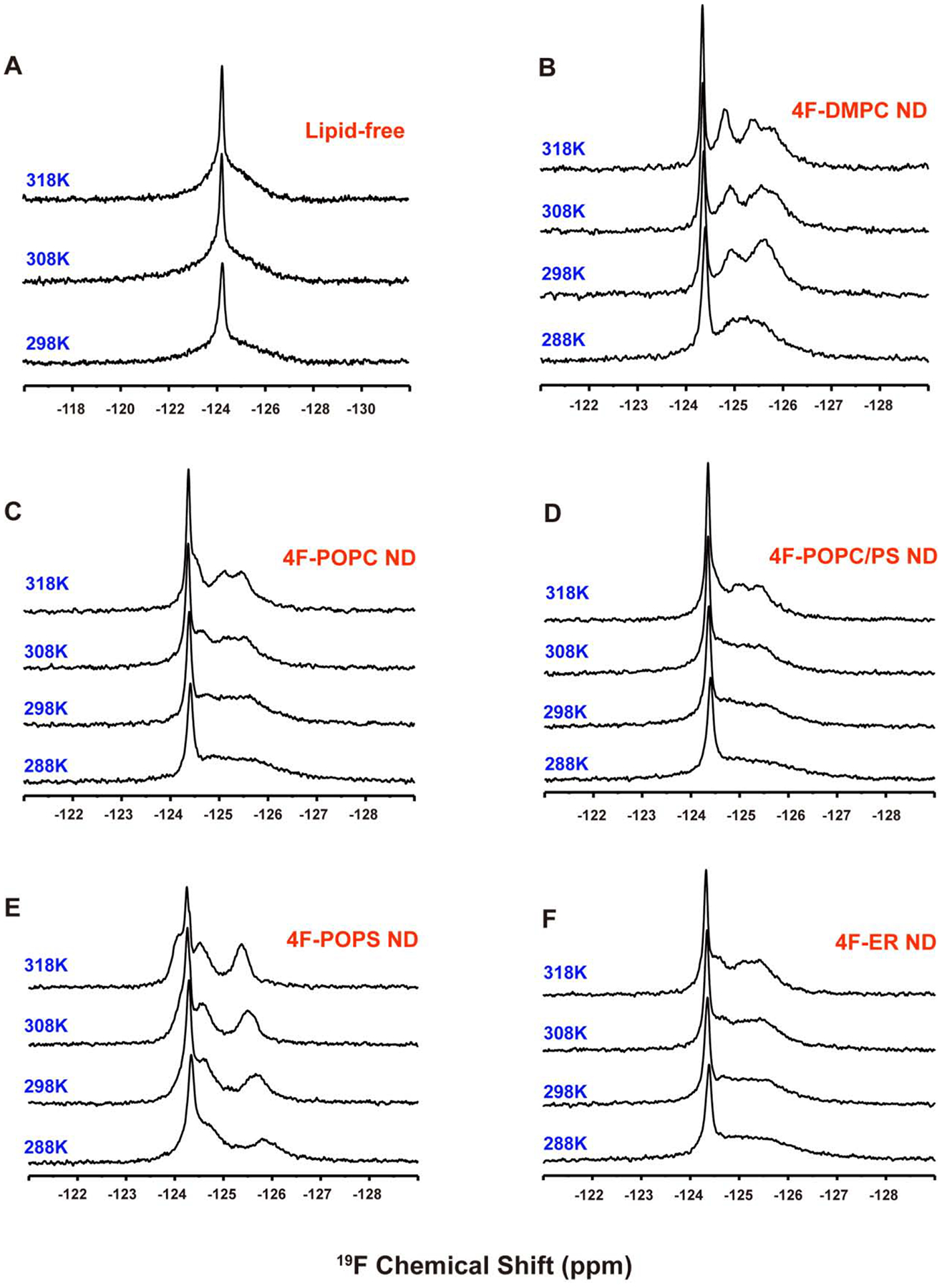
cytb5 in buffer (A) and reconstituted in the (B) 4F-DMPC, (C) 4F-POPC, (D) 4F-POPC/PS, (E) 4F-POPS, (F) 4F-ER nanodiscs at the indicated temperatures.
Solvent PRE
Solvent PRE measurements were used to detect the solvent accessible regions of the tryptophan residues. We used Gd-DTPA-BMA as the paramagnetic cosolvent in the 4F-nanodiscs containing 19F-labeled cytb5, and measured the solvent paramagnetic relaxation enhancement (sPRE); and the resultant 19F NMR spectra are shown in Figure 3. With the increasing concentration of Gd-DTPA-BMA, a small reduction in signal intensity is observed for W27, with a relative peak height of 84% at the 5 mM Gd-DTPA-BMA. However, for the other tryptophan residues, no observable reduction in signal intensity is observed even in presence of 10 mM Gd-DTPA-BMA, (Figure 3C). These observations suggest that W27 is exposed to the solution, whereas the other tryptophans are buried in the lipid bilayer.
Figure 3. 19F NMR experiments on nanodiscs containing 19F-cytb5.
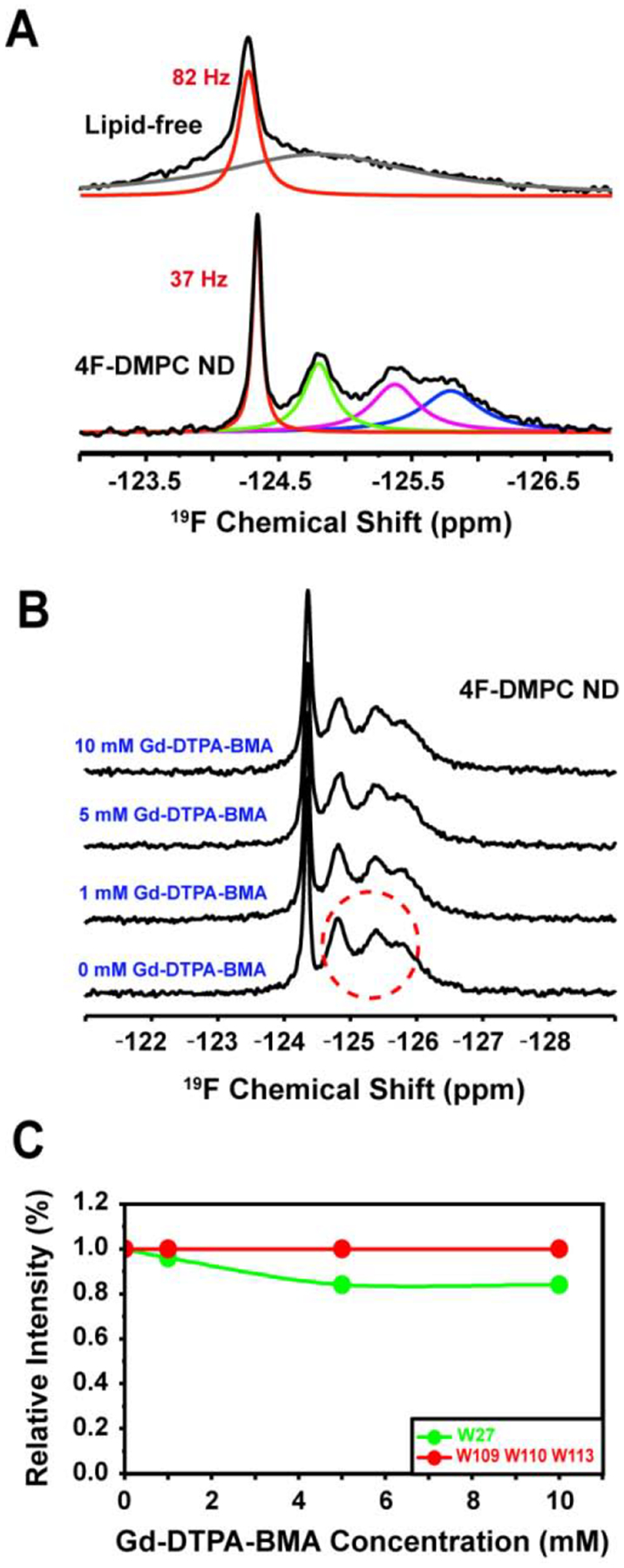
(A) 19F NMR spectra of 5F-tryptophan-labeled cytb5 alone in buffer and reconstituted in 4F-DMPC nanodiscs at 318 K. (B) 19F NMR spectra of cytb5 reconstituted in 4F-DMPC nanodiscs with varying concentration of Gd-DTPA-BMA. (C) NMR peak intensity observed for tryptophan residues of cytb5 in presence of Gd-DTPA-BMA.
Effects of different lipid composition on cytb5
Since the structure, folding and topology of a membrane protein could depend on the lipid composition of membrane mimetics used, we investigated the effects of varying the lipid composition on the reconstitution of cytb5. For this purpose, in addition to 4F-DMPC nanodiscs, 4F-POPC, 4F-POPC/PS (8:2 molar ratio) and 4F-POPS nanodiscs were prepared as described above. The diameters of these nanodiscs were measured to be ~8.4 nm for 4F-POPC, ~8.0 nm for 4F-POPC/PS (8:2 molar ratio), ~6.2 nm for 4F-POPS (Figure 4). We reconstituted 5FW-labeled cytb5 into these nanodiscs as described above. SEC and DLS profiles and 19F NMR spectra indicate that cytb5 is reconstituted into these nanodiscs that vary in the lipid composition (Figure 4).
Figure 4. Characterization of 4F-peptide nanodiscs containing 19F-cytb5.
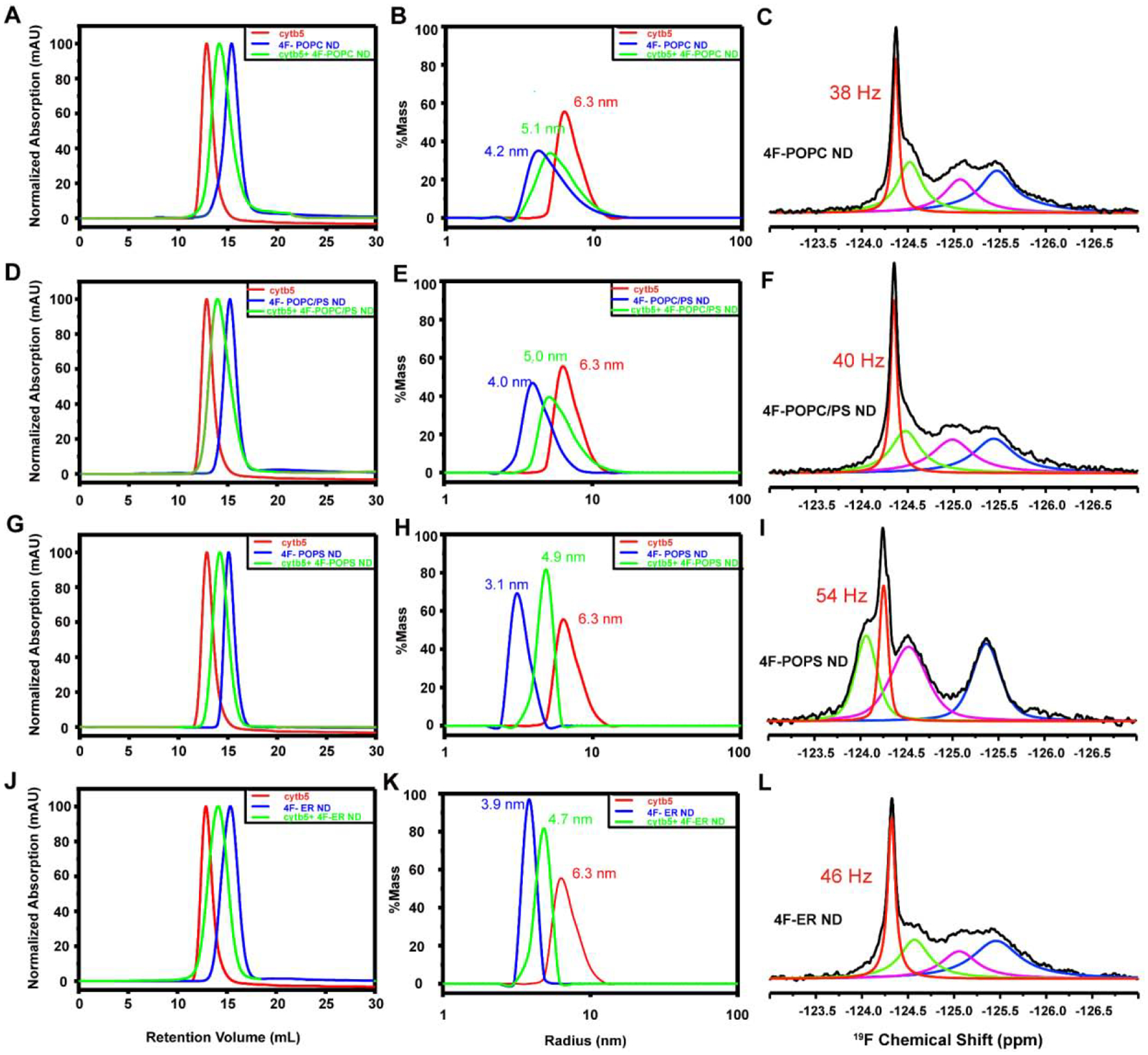
Reconstitution of cytb5 in 4F-POPC nanodiscs, 4F-POPC/PS nanodiscs, 4F-POPS nanodiscs and 4F-ER nanodiscs. SEC elution profiles (A, D, G, J) and DLS profiles (B, E, H, K) of cytb5 alone in buffer (red), empty nanodiscs (blue), and nanodiscs containing cytb5 (green). Fits of 19F NMR spectra of 5F-tryptophan-labeled cytb5 reconstituted in 4F-POPC (C), 4F-POPC/PS (F), 4F-POPS (I), and 4F-ER nanodiscs(L) at 310 K.
19F NMR spectra of cytb5 reconstituted in 4F-POPC, 4F-POPC/PS (8:2 molar ratio), or 4F-POPS nanodiscs were also acquired at various temperatures, and the spectra are shown in Figure 2. All of these spectra exhibit a narrow peak for W27 with a line width of 57 Hz in 4F-POPC, 55 Hz in 4F-POPC/PS, and 65 Hz in 4F-POPS. With the increasing temperature, besides the narrow peak from W27, three 19F NMR peaks from Trp residues in the transmembrane domain are also observed in all three types of nanodiscs at high temperatures (Figure 2). The presence of zwitterionic lipids like DMPC or POPC in nanodiscs resulted in the same 19F NMR peak positions indicating the uncharged lipids have a negligible effect on the soluble domains of cytb5. This is in complete agreement with previous NMR studies on membrane-bound cytb5 that reported a fast time scale of motion for residues from the soluble domain with negligible interaction with zwitterionic lipid membrane.[44] On the other hand, with a gradual increase in the concentration of negatively charged POPS lipids, a low-field shift (CSP <0.11 ppm) was observed for the 19F peak from W27 (Figures 2 and 4). However, for the residues from the transmembrane domain that exhibited broader lines as compared to that from W27, the increased chain length and the presence of an unsaturated bond in the neutral POPC lipids induced moderate chemical shifts as compared to that observed from DMPC nanodiscs. On the other hand, with the increasing concentration of negatively charged POPS, 19F spectra appeared to be significantly different from that observed from zwitterionic PC only lipids containing nanodiscs. The presence of 100 % POPS resulted in a maximum change in the chemical shift (Figure 2).
Cytb5 is primarily associated with the endoplasmic reticulum of eukaryotic cells. Therefore, biomimetic 4F-ER nanodiscs were developed to mimic the natural lipid membrane environment for cytb5. The diameter of the 4F-ER nanodiscs was determined to be ~8 nm (Figure 4). The SEC profiles and 19F NMR spectra (Figure 4) confirmed the successful reconstitution of cytb5 in 4F-ER nanodiscs.
Formation of cytb5-CYP2B4 complex in nanodiscs
Upon the addition of an unlabeled full-length CYP2B4 to nanodiscs containing cytb5, no obvious chemical shift perturbation was observed for the 19F peak from W27, but the intensity decreased significantly as compared to that observed for cytb5 alone (Figure 5), suggesting the formation of a productive cytb5-CYP2B4 complex in the nanodiscs. The complex formation was also confirmed by performing 2D TROSY-HSQC NMR experiments as reported previously.[23, 25] For the residues from the transmembrane domain, a small chemical shift perturbation was observed with a significant reduction in the signal intensity (Figure 5). The addition of BHT to the cytb5-CYP2B4 further broadened the 19F peak from W27 to beyond the detection limit, suggesting a tighter binding between cytb5 and CYP2B4 in presence of BHT. These results are consistent with our previous findings in DMPC/DHPC isotropic bicelles[23], and 22A-nanodiscs[24] by using 15N-cytb5 and CYP2B4.
Figure 5. 19F NMR revealing cytb5-cytP450 interactions in nanodiscs.
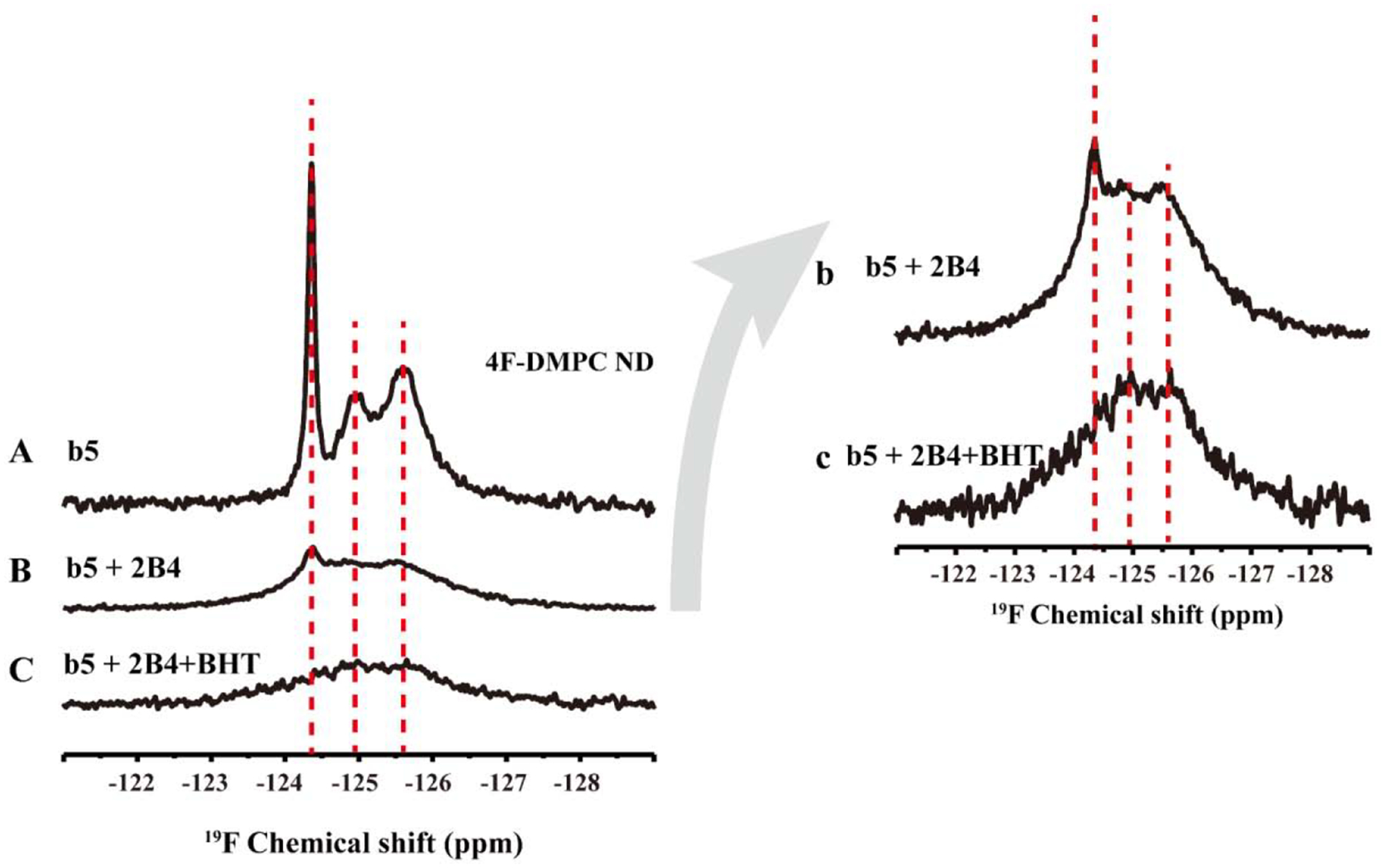
19F NMR spectra of 5F-tryptophan-labeled cytb5 alone in 4F-DMPC nanodiscs (A), cytb5 in complex with P450 2B4 (B), and cytb5-P450 in complex in presence of BHT at 298K. (b) and (c) are the magnification of (B) and (C), respectively.
Conclusions
In this study, wild-type cytb5 labeled with 5-fluorotryptophan (5FW) was successfully reconstituted in peptide-based nanodiscs with a varying lipid composition including 4F-DMPC, 4F-POPC, 4F-POPC/PS, 4F-POPS and 4F-ER nanodiscs. Simple 19F NMR spectra obtained from nanodiscs composed of DMPC, POPC, POPC/PS, or ER lipids and cytb5 at different temperatures demonstrated the feasibility of resolving the spectral lines from the transmembrane domains of the protein. A significant change in the chemical shift was observed in presence of a negative charged lipid POPS. Interaction of cytb5 with CYP2B4 resulted in broadening of all the four 19F spectral lines and further broadening was observed in presence of BHT indicating a tight complex formation. These results demonstrate the feasibility of 19F NMR experiments on nanodiscs containing single-pass transmembrane proteins.
Supplementary Material
Highlights.
19F-labeled cyt-b5 is expressed, purified and reconstituted in peptide nanodiscs.
19F NMR spectra of cytb5 reconstituted nanodiscs with varying lipid composition are reported.
19F solution NMR experiments reveal the interaction between cytb5 and cytP450.
ACKNOWLEDGEMENTS
This study was supported by NIH (GM084018 to A.R.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- [1].Liu JJ, Horst R, Katritch V, Stevens RC, Wuthrich K, Biased signaling pathways in beta2-adrenergic receptor characterized by 19F-NMR, Science, 335 (2012) 1106–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Kitevski-LeBlanc JL, Prosser RS, Current applications of 19F NMR to studies of protein structure and dynamics, Prog Nucl Magn Reson Spectrosc, 62 (2012) 1–33. [DOI] [PubMed] [Google Scholar]
- [3].Lindon JC, 19F NMR Spectroscopy, in: Worsfold P, Poole C, Townshend A, Miró M (Eds.) Reference Module in Chemistry, Molecular Sciences and Chemical Engineering, Academic Press, Oxford, 2016, pp. 141–151. [Google Scholar]
- [4].Danielson MA, Falke JJ, Use of 19F NMR to probe protein structure and conformational changes, Annu Rev Biophys Biomol Struct, 25 (1996) 163–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Boeszoermenyi A, Chhabra S, Dubey A, Radeva DL, Burdzhiev NT, Chanev CD, Petrov OI, Gelev VM, Zhang M, Anklin C, Kovacs H, Wagner G, Kuprov I, Takeuchi K, Arthanari H, Aromatic (19)F-(13)C TROSY: a background-free approach to probe biomolecular structure, function, and dynamics, Nat Methods, 16 (2019) 333–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Schenkman JB, Jansson I, The many roles of cytochrome b5, Pharmacol Ther, 97 (2003) 139–152. [DOI] [PubMed] [Google Scholar]
- [7].Porter TD, The roles of cytochrome b5 in cytochrome P450 reactions, J Biochem Mol Toxicol, 16 (2002) 311–316. [DOI] [PubMed] [Google Scholar]
- [8].Durr UH, Waskell L, Ramamoorthy A, The cytochromes P450 and b5 and their reductases--promising targets for structural studies by advanced solid-state NMR spectroscopy, Biochim Biophys Acta, 1768 (2007) 3235–3259. [DOI] [PubMed] [Google Scholar]
- [9].Guengerich FP, Wu ZL, Bartleson CJ, Function of human cytochrome P450s: characterization of the orphans, Biochem Biophys Res Commun, 338 (2005) 465–469. [DOI] [PubMed] [Google Scholar]
- [10].Guengerich FP, Cytochrome P450s and other enzymes in drug metabolism and toxicity, AAPS J, 8 (2006) E101–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Guengerich FP, Cytochromes P450, drugs, and diseases, Mol Interv, 3 (2003) 194–204. [DOI] [PubMed] [Google Scholar]
- [12].Peng HM, Im SC, Pearl NM, Turcu AF, Rege J, Waskell L, Auchus RJ, Cytochrome b5 Activates the 17,20-Lyase Activity of Human Cytochrome P450 17A1 by Increasing the Coupling of NADPH Consumption to Androgen Production, Biochemistry, 55 (2016) 4356–4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Sondhi V, Owen BM, Liu J, Chomic R, Kliewer SA, Hughes BA, Arlt W, Mangelsdorf DJ, Auchus RJ, Impaired 17,20-Lyase Activity in Male Mice Lacking Cytochrome b5 in Leydig Cells, Mol Endocrinol, 30 (2016) 469–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Simonov AN, Holien JK, Yeung JC, Nguyen AD, Corbin CJ, Zheng J, Kuznetsov VL, Auchus RJ, Conley AJ, Bond AM, Parker MW, Rodgers RJ, Martin LL, Mechanistic Scrutiny Identifies a Kinetic Role for Cytochrome b5 Regulation of Human Cytochrome P450c17 (CYP17A1, P450 17A1), PLoS One, 10 (2015) e0141252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Mulrooney SB, Waskell L, High-level expression in Escherichia coli and purification of the membrane-bound form of cytochrome b(5), Protein Expr Purif, 19 (2000) 173–178. [DOI] [PubMed] [Google Scholar]
- [16].Ozols J, Structure of cytochrome b5 and its topology in the microsomal membrane, Biochim Biophys Acta, 997 (1989) 121–130. [DOI] [PubMed] [Google Scholar]
- [17].Ozols J, Structure of cytochrome b5 and its topology in the microsomal membrane, Biochimica et Biophysica Acta (BBA) - Protein Structure and Molecular Enzymology, 997 (1989) 121–130. [DOI] [PubMed] [Google Scholar]
- [18].Balakrishnan S, Sarma SP, Engineering Aromatic-Aromatic Interactions To Nucleate Folding in Intrinsically Disordered Regions of Proteins, Biochemistry, 56 (2017) 4346–4359. [DOI] [PubMed] [Google Scholar]
- [19].Bendzko P, Usanov SA, Pfeil W, Ruckpaul K, Role of the hydrophobic tail of cytochrome b5 in the interaction with cytochrome P-450 LM2, Acta Biol Med Ger, 41 (1982) K1–K8. [PubMed] [Google Scholar]
- [20].Chiang JY, Interaction of purified microsomal cytochrome P-450 with cytochrome b5, Arch Biochem Biophys, 211 (1981) 662–673. [DOI] [PubMed] [Google Scholar]
- [21].Canova-Davis E, Waskell L, The identification of the heat-stable microsomal protein required for methoxyflurane metabolism as cytochrome b5, Journal of Biological Chemistry, 259 (1984) 2541–2546. [PubMed] [Google Scholar]
- [22].Zhang M, Huang R, Im SC, Waskell L, Ramamoorthy A, Effects of membrane mimetics on cytochrome P450-cytochrome b5 interactions characterized by NMR spectroscopy, J Biol Chem, 290 (2015) 12705–12718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Zhang M, Le Clair SV, Huang R, Ahuja S, Im SC, Waskell L, Ramamoorthy A, Insights into the role of substrates on the interaction between cytochrome b5 and cytochrome P450 2B4 by NMR, Sci Rep, 5 (2015) 8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Zhang M, Huang R, Ackermann R, Im SC, Waskell L, Schwendeman A, Ramamoorthy A, Reconstitution of the Cytb5-CytP450 Complex in Nanodiscs for Structural Studies using NMR Spectroscopy, Angew Chem Int Ed Engl, 55 (2016) 4497–4499. [DOI] [PubMed] [Google Scholar]
- [25].Ahuja S, Jahr N, Im SC, Vivekanandan S, Popovych N, Le Clair SV, Huang R, Soong R, Xu J, Yamamoto K, Nanga RP, Bridges A, Waskell L, Ramamoorthy A, A model of the membrane-bound cytochrome b5-cytochrome P450 complex from NMR and mutagenesis data, J Biol Chem, 288 (2013) 22080–22095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Mustafa G, Nandekar PP, Camp TJ, Bruce NJ, Gregory MC, Sligar SG, Wade RC, Influence of Transmembrane Helix Mutations on Cytochrome P450-Membrane Interactions and Function, Biophys J, 116 (2019) 419–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Durr UH, Yamamoto K, Im SC, Waskell L, Ramamoorthy A, Solid-state NMR reveals structural and dynamical properties of a membrane-anchored electron-carrier protein, cytochrome b5, J Am Chem Soc, 129 (2007) 6670–6671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Yamamoto K, Durr UH, Xu J, Im SC, Waskell L, Ramamoorthy A, Dynamic interaction between membrane-bound full-length cytochrome P450 and cytochrome b5 observed by solid-state NMR spectroscopy, Sci Rep, 3 (2013) 2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Ravula T, Ramadugu SK, Di Mauro G, Ramamoorthy A, Bioinspired, Size-Tunable Self-Assembly of Polymer-Lipid Bilayer Nanodiscs, Angew Chem Int Ed Engl, 56 (2017) 11466–11470. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- [30].Yamamoto K, Gildenberg M, Ahuja S, Im SC, Pearcy P, Waskell L, Ramamoorthy A, Probing the transmembrane structure and topology of microsomal cytochrome-p450 by solid-state NMR on temperature-resistant bicelles, Sci Rep, 3 (2013) 2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Yamamoto K, Caporini MA, Im SC, Waskell L, Ramamoorthy A, Transmembrane Interactions of Full-length Mammalian Bitopic Cytochrome-P450-Cytochrome-b5 Complex in Lipid Bilayers Revealed by Sensitivity-Enhanced Dynamic Nuclear Polarization Solid-state NMR Spectroscopy, Sci Rep, 7 (2017) 4116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Killian JA, von Heijne G, How proteins adapt to a membrane-water interface, Trends in Biochemical Sciences, 25 (2000) 429–434. [DOI] [PubMed] [Google Scholar]
- [33].Yau WM, Wimley WC, Gawrisch K, White SH, The preference of tryptophan for membrane interfaces, Biochemistry, 37 (1998) 14713–14718. [DOI] [PubMed] [Google Scholar]
- [34].de Planque MR, Kruijtzer JA, Liskamp RM, Marsh D, Greathouse DV, Koeppe RE 2nd, de Kruijff B, Killian JA, Different membrane anchoring positions of tryptophan and lysine in synthetic transmembrane alpha-helical peptides, J Biol Chem, 274 (1999) 20839–20846. [DOI] [PubMed] [Google Scholar]
- [35].de Jesus AJ, Allen TW, The role of tryptophan side chains in membrane protein anchoring and hydrophobic mismatch, Biochim Biophys Acta, 1828 (2013) 864–876. [DOI] [PubMed] [Google Scholar]
- [36].de Planque MR, Bonev BB, Demmers JA, Greathouse DV, Koeppe RE, Separovic F, Watts A, Killian JA, Interfacial anchor properties of tryptophan residues in transmembrane peptides can dominate over hydrophobic matching effects in peptide− lipid interactions, Biochemistry, 42 (2003) 5341–5348. [DOI] [PubMed] [Google Scholar]
- [37].de Planque MR, Killian JA, Protein-lipid interactions studied with designed transmembrane peptides: role of hydrophobic matching and interfacial anchoring, Mol Membr Biol, 20 (2003) 271–284. [DOI] [PubMed] [Google Scholar]
- [38].Bordag N, Keller S, α-Helical transmembrane peptides: a “divide and conquer” approach to membrane proteins, Chemistry and physics of lipids, 163 (2010) 1–26. [DOI] [PubMed] [Google Scholar]
- [39].Ridder AN, Morein S, Stam JG, Kuhn A, de Kruijff B, Killian JA, Analysis of the role of interfacial tryptophan residues in controlling the topology of membrane proteins, Biochemistry, 39 (2000) 6521–6528. [DOI] [PubMed] [Google Scholar]
- [40].Hagn F, Nasr ML, Wagner G, Assembly of phospholipid nanodiscs of controlled size for structural studies of membrane proteins by NMR, Nat Protoc, 13 (2018) 79–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Hagn F, Etzkorn M, Raschle T, Wagner G, Optimized phospholipid bilayer nanodiscs facilitate high-resolution structure determination of membrane proteins, J Am Chem Soc, 135 (2013) 1919–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Mak PJ, Denisov IG, Grinkova YV, Sligar SG, Kincaid JR, Defining CYP3A4 structural responses to substrate binding. Raman spectroscopic studies of a nanodisc-incorporated mammalian cytochrome P450, J Am Chem Soc, 133 (2011) 1357–1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Denisov IG, Sligar SG, Nanodiscs in Membrane Biochemistry and Biophysics, Chem Rev, 117 (2017) 4669–4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Ravula T, Barnaba C, Mahajan M, Anantharamaiah GM, Im SC, Waskell L, Ramamoorthy A, Membrane environment drives cytochrome P450’s spin transition and its interaction with cytochrome b5, Chem Commun (Camb), 53 (2017) 12798–12801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Crowley PB, Kyne C, Monteith WB, Simple and inexpensive incorporation of 19F-tryptophan for protein NMR spectroscopy, Chem Commun (Camb), 48 (2012) 10681–10683. [DOI] [PubMed] [Google Scholar]
- [46].Datta G, Chaddha M, Hama S, Navab M, Fogelman AM, Garber DW, Mishra VK, Epand RM, Epand RF, Lund-Katz S, Phillips MC, Segrest JP, Anantharamaiah GM, Effects of increasing hydrophobicity on the physical-chemical and biological properties of a class A amphipathic helical peptide, J Lipid Res, 42 (2001) 1096–1104. [PubMed] [Google Scholar]
- [47].Kariyazono H, Nadai R, Miyajima R, Takechi-Haraya Y, Baba T, Shigenaga A, Okuhira K, Otaka A, Saito H, Formation of stable nanodiscs by bihelical apolipoprotein A-I mimetic peptide, J Pept Sci, 22 (2016) 116–122. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


