In this review, Kim et al. discuss the importance of PARP family members and ADPRylation in gene regulation, mRNA processing, and protein abundance.
Keywords: ADP-ribosylation (ADPRylation), mono(ADP-ribose) (MAR), poly(ADP-ribose) (PAR), poly(ADP-ribose) polymerase (PARP), ribosome biogenesis, rRNA synthesis, PARP inhibitors (PARPi), DNA damage, RNA stability, mRNA translation, mRNA processing, mRNA splicing, stress responses
Abstract
ADP-ribosylation (ADPRylation) is a posttranslational modification of proteins discovered nearly six decades ago, but many important questions remain regarding its molecular functions and biological roles, as well as the activity of the ADP-ribose (ADPR) transferase enzymes (PARP family members) that catalyze it. Growing evidence indicates that PARP-mediated ADPRylation events are key regulators of the protein biosynthetic pathway, leading from rDNA transcription and ribosome biogenesis to mRNA synthesis, processing, and translation. In this review we describe the role of PARP proteins and ADPRylation in all facets of this pathway. PARP-1 and its enzymatic activity are key regulators of rDNA transcription, which is a critical step in ribosome biogenesis. An emerging role of PARPs in alternative splicing of mRNAs, as well as direct ADPRylation of mRNAs, highlight the role of PARP members in RNA processing. Furthermore, PARP activity, stimulated by cellular stresses, such as viral infections and ER stress, leads to the regulation of mRNA stability and protein synthesis through posttranscriptional mechanisms. Dysregulation of PARP activity in these processes can promote disease states. Collectively, these results highlight the importance of PARP family members and ADPRylation in gene regulation, mRNA processing, and protein abundance. Future studies in these areas will yield new insights into the fundamental mechanisms and a broader utility for PARP-targeted therapeutic agents.
ADP-ribosylation (ADPRylation) is a reversible posttranslational modification of proteins resulting in the covalent attachment of ADP-ribose (ADPR) units on substrate “acceptor” proteins. In this review, we discuss the broad role of ADPRylation and the poly(ADP-ribose) polymerase (PARP) enzymes that catalyze it in RNA-related processes, from rDNA transcription and ribosome biogenesis to mRNA processing and protein translation (Fig. 1). However, first we provide a brief introduction about PARPs and ADPRylation, as well as the NAD+ biosynthetic pathways that “feed” the PARP enzymes.
Figure 1.
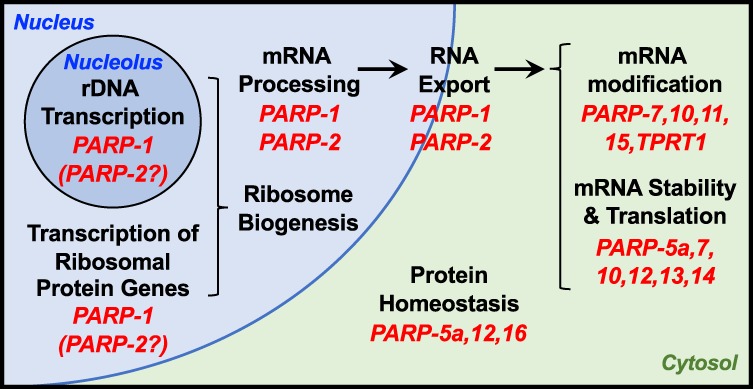
Role of PARPs and ADP-ribosylation in RNA biology. PARPs and ADPRylation play broad roles in RNA-related processes, from rDNA transcription and ribosome biogenesis to mRNA processing, protein translation, and proteostasis. See the text for descriptions.
PARPs and ADP-ribosylation
ADPRylation uses β-NAD+ as a donor of ADPR units, which are covalently linked to a variety of amino acid residues (e.g., Glu, Asp, and Ser) in substrate proteins (Schreiber et al. 2006; Gibson and Kraus 2012; Leung 2017). The modification may be in the form of a single ADP-ribose (ADPR) unit [i.e., mono(ADP-ribose), or MAR] or polymers of ADPR units [i.e; poly(ADP-ribose), or PAR] (Fig. 2). ADPRylation is catalyzed by the PARP family of enzymes (also known as the ARTD family) (Hottiger et al. 2010), consisting of 17 members that have distinct structural domains, activities, subcellular localizations, and functions (Amé et al. 2004; Schreiber et al. 2006; Vyas et al. 2013, 2014). PARPs function as ADPR “writers” that covalently attach ADP-ribose units on substrate proteins (Hottiger 2015; Gupte et al. 2017). PARP family members can be categorized according to their catalytic activities: (1) PARP “polyenzymes” (e.g., PARPs 1 and 2), which catalyze the polymerization of ADPR units in linear or branched chains through a process called PARylation (Gibson and Kraus 2012; Vyas et al. 2014; Gupte et al. 2017) and (2) PARP “monoenzymes” [i.e., mono(ADP-ribosyl) transferases or MARTs] (e.g., PARPs 7 and 16), which catalyze the addition of a single ADPR unit on target proteins through a process called MARylation (Gibson and Kraus 2012; Vyas et al. 2014; Gupte et al. 2017).
Figure 2.
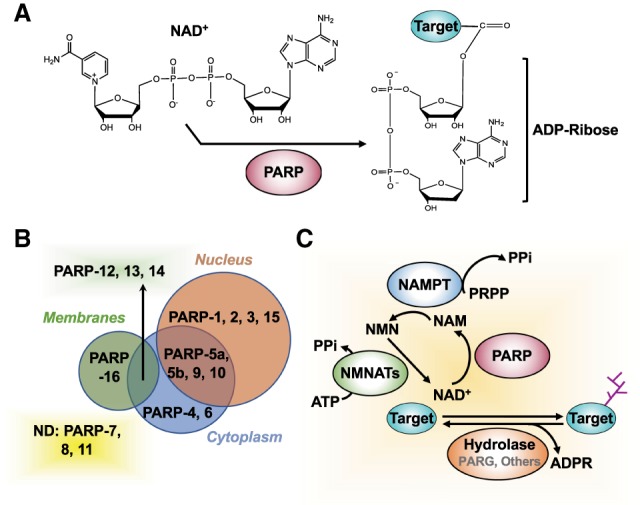
Overview of NAD+-dependent, PARP-mediated ADPRylation. (A) PARPs use NAD+ as a substrate to catalyze reversible ADPRylation of substrate proteins. (B) Subcellular localization of PARP family members. (C) NAD+ “feeds” PARP catalytic activity. This enzymatic activity generates nicotinamide (NAM) as a product, which is used by NAMPT in the NAD+ salvage pathway to regenerate NAD+. Several hydrolases that bind to specific structural features of MAR and PAR can hydrolyze the ADPR, returning the target proteins to their unmodified state.
Functional outcomes of site-specific ADPRylation
Historically, PARPs and ADPRylation have been studied in the context of DNA repair, with the primary focus on PARP-1 and PARylation in cancer (Morales et al. 2014; Ray Chaudhuri and Nussenzweig 2017). New findings on the diverse roles of PARPs in cellular processes beyond DNA repair (e.g., regulation of chromatin structure and gene expression, RNA processing, ribosome biogenesis, protein translation) have been complemented by recent advances that link ADPRylation to metabolism, inflammation, immunity, stress responses, hormonal signaling, and viral infections (Kim et al. 2005; Luo and Kraus 2012; Vyas and Chang 2014; Gupte et al. 2017). Although the functions of PARylation are well studied, much less is known about the functions of MARylation. Recent studies have begun to reveal novel and interesting functions for cytoplasmic PARP monoenzymes, such as PARP-7, PARP-12, PARP-14, and PARP-16, in molecular and cellular functions ranging from RNA processing and translation to stress granule formation and the unfolded protein response (Leung et al. 2011; Di Paola et al. 2012; Jwa and Chang 2012; Vyas et al. 2013, 2014; Roper et al. 2014; Ahmed et al. 2015; Bindesbøll et al. 2016; Iwata et al. 2016).
The growing understanding of the biological importance of ADPRylation has spurred interest in identifying the substrates of specific PARPs and the functional outcomes of the modification. The identification of specific sites of ADPRylation on a proteome-wide scale has lagged behind other PTMs until recently (Daniels et al. 2015). Although previous studies demonstrated effects of ADPRylation on target proteins, the sites were usually not mapped. Some examples where the sites have been mapped and functionally interrogated include DNA-binding transcription factors (e.g., NFAT [Olabisi et al. 2008], p53 [Kanai et al. 2007], STAT1α [Iwata et al. 2016], and C/EBPβ [Luo et al. 2017]), the chromatin insulator protein CTCF (Yu et al. 2004; Farrar et al. 2010), the RNA polymerase II (Pol II) transcription elongation factor NELF (Gibson et al. 2016), and the RNA helicase DDX21 (Kim et al. 2019). However, most of the aforementioned substrates are nuclear proteins modified by nuclear PARP polyenzymes. Further systemic identification and functional analysis of the substrates of cytoplasmic PARP monoenzymes and a demonstration of the biological importance of site-specific MARylation is needed.
Beyond protein substrates: ADPRylation of DNA and RNA
Recent studies have also identified nucleic acids as substrates for ADPRylation. ADPRylation of DNA was first identified as part of toxin-antitoxin systems in bacterial pathogens (e.g., Mycobacterium tuberculosis). DarT is a bacterial enzyme that ADPRylates thymidines on single-stranded DNA in a sequence-specific manner, a modification that can be removed by another enzyme, DarG (Jankevicius et al. 2016). In mammalian systems, nuclear PARPs 1 and 2 can PARylate and PARP-3 can MARylate phosphorylated DNA termini in vitro (Talhaoui et al. 2016; Munnur and Ahel 2017; Zarkovic et al. 2018), modifications that have been shown in some cases to be removed by ADPR hydrolases (Munnur and Ahel 2017). Likewise, ADPRylation of RNA by multiple PARP family members and a highly divergent microbial PARP homolog, TRPT1, as well as its erasure by cellular, viral, and bacterial macrodomain ADPR hydrolases, has been demonstrated in a variety of systems (Munir et al. 2018; Munnur et al. 2019). Thus, ADPRylation of both DNA and RNA may represent a widespread and physiologically relevant modification mediated by both PARP monoenzymes and polyenzymes.
Control of compartment-specific PARP functions by NAD+ synthases
The activity of the PARP enzymes is intimately tied to the synthesis of NAD+, which is consumed during ADPRylation reactions and thus, must be regenerated. While nutritional supplements such as Vitamin B3 act as dietary precursors for NAD+ biosynthesis, intracellular salvage pathways using nicotinamide (NAM) are the primary source of NAD+ in the cell (Verdin 2015). Nicotinamide mononucleotide adenylyltransferases (NMNATs) catalyze the final step in the NAD+ salvage pathway, combining nicotinamide mononucleotide (NMN) and ATP to make NAD+ (Ryu et al. 2015). Three different NMNATs, each with a distinct subcellular localization, control the subcellular levels of NAD+ in each compartment: NMNAT-1 is localized to the nucleus, NMNAT-2 is localized to the outer membrane of the golgi, and NMNAT-3 is localized to the mitochondria (Lau et al. 2009; Jayaram et al. 2011; Ryu et al. 2015). The specific subcellular localizations of these three NAD+ synthases leads to compartmentalized production of NAD+ (Zhang et al. 2012; Cambronne et al. 2016; Ryu et al. 2018), which has important functional consequences in the cell. For example, the control of nuclear and cytosolic levels of NAD+ by NMNAT-1 and NMNAT-2, respectively, regulate the PARP-1-dependent gene expression programs that drive adipocyte differentiation (Luo et al. 2017; Ryu et al. 2018). The control of subcellular PARP functions by compartmentalized NAD+ synthesis is just beginning to be understood and requires further study.
Role of PARP-1 and ADPRylation in the regulation of rDNA transcription and ribosome biogenesis
Ribosome biogenesis is a highly coordinated cellular process that involves the synthesis, processing, and modification of rRNAs, as well as the proper assembly of the rRNA with ribosomal proteins (Baßler and Hurt 2019). Emerging evidence highlights the role of ribosome biogenesis in various human cancers and its dysregulation promotes cellular growth and tumorigenesis (Ruggero and Pandolfi 2003; Aspesi and Ellis 2019). Enrichment of PARP-1 in the nucleolus, the site of ribosome biogenesis, was documented from the late 1980s to the early 2000s (Fakan et al. 1988; Desnoyers et al. 1996; Scherl et al. 2002). Subsequent studies have revealed an important role for PARP-1 in regulating multiple steps of ribosome biogenesis. A growing body of evidence supports the important role of PARP-1 in RNA polymerase I (Pol I)-dependent transcriptional regulation, preribosomal rRNA (pre-rRNA) processing, and rRNA modification (Boamah et al. 2012; Guetg et al. 2012). In addition, several studies have determined the physiological functions of PARP-1 in the regulation of ribosome biogenesis in response to DNA damage (Calkins et al. 2013; Bütepage et al. 2018). In this section, we describe in detail the growing awareness of the essential role of PARP-1 in the transcriptional regulation of rDNA and ribosome biogenesis under normal conditions and in various human diseases.
Brief overview of ribosome biogenesis
Ribosome biogenesis, which begins in the nucleolus, is the process by which cells generate new ribosomes to synthesize cellular proteins (Baßler and Hurt 2019). Ribosome biogenesis is a tightly controlled, energy-demanding process, comprising multiple steps including (1) synthesis of four different ribosomal RNA (rRNA) molecules (25S/28S, 18S, 5.8S, and 5S) and 79 ribosomal proteins by all three nuclear RNA polymerases (Pol I, II, and III) in eukaryotes, (2) rRNA processing and site-specific modifications including methylation and pseudouridylation by a small nucleolar ribonucleoprotein (snoRNP), and (3) assembly with rRNAs and ribosomal proteins to form preribosomal particles. After synthesis and assembly steps in the nucleolus and nucleus, the pre-40S and pre-60S ribosomal subunits exit to the cytoplasm to form mature ribosomal subunits (Fig. 3). The functional links among PARP-1, ADPRylation, and ribosome biogenesis are explored in more detail below.
Figure 3.
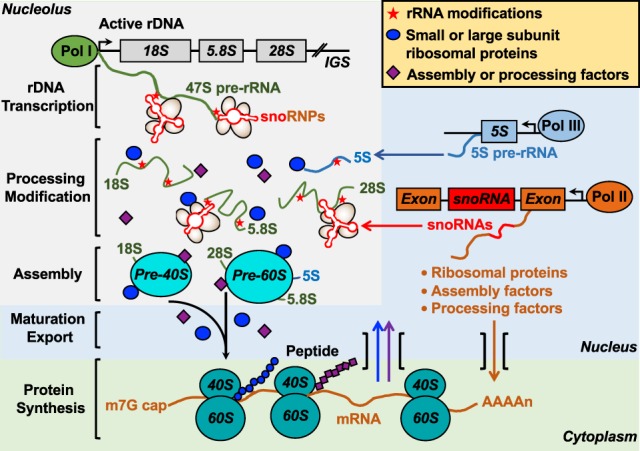
The steps in ribosome biogenesis. rRNA molecules (28S, 18S, 5.8S, and 5S), small and large subunit ribosomal proteins, and assembly or processing factors are synthesized by RNA polymerases (Pol I–III), and are subsequently modified and processed. rRNAs assemble with ribosomal proteins to form preribosome particles, which are exported to the cytoplasm to form mature ribosomal subunits and active ribosomes.
Role of PARP-1 in the formation of silent rDNA chromatin and transcriptional silencing
A newly discovered aspect of PARP-1 function is its role in modulating ribosome biogenesis in normal physiological conditions through PARP-1-mediated ADPRylation (Guetg et al. 2012). Tandemly repeated ribosomal RNA genes (rDNA) exist in two distinct epigenetic states: a permissive state allowing transcription and repressed state inhibiting transcription (Li et al. 2005). NoRC, a nucleolar chromatin remodeling complex comprising SNF2h and TIP5, plays an important role in establishing the silent state of rRNA genes by recruiting a DNA methyltransferase and a histone deacetylase to the rDNA promoter (Santoro et al. 2002). Interestingly, PARP-1 has been implicated in NoRC-mediated establishment of transcriptionally inactive rDNA chromatin during cell division (Guetg et al. 2012). PARP-1 interacts with NoRC-associated RNA (pRNA). This interaction is required for recruitment of TIP5, the large subunit of NoRC, to the promoter of silent rDNA after the passage of the replication fork. NoRC recruits and interacts with the histone deacetylase HDAC1 and DNA methyltransferase (DNMT), leading to epigenetic reprogramming, including histone modification and DNA methylation (Fig. 4A).
Figure 4.
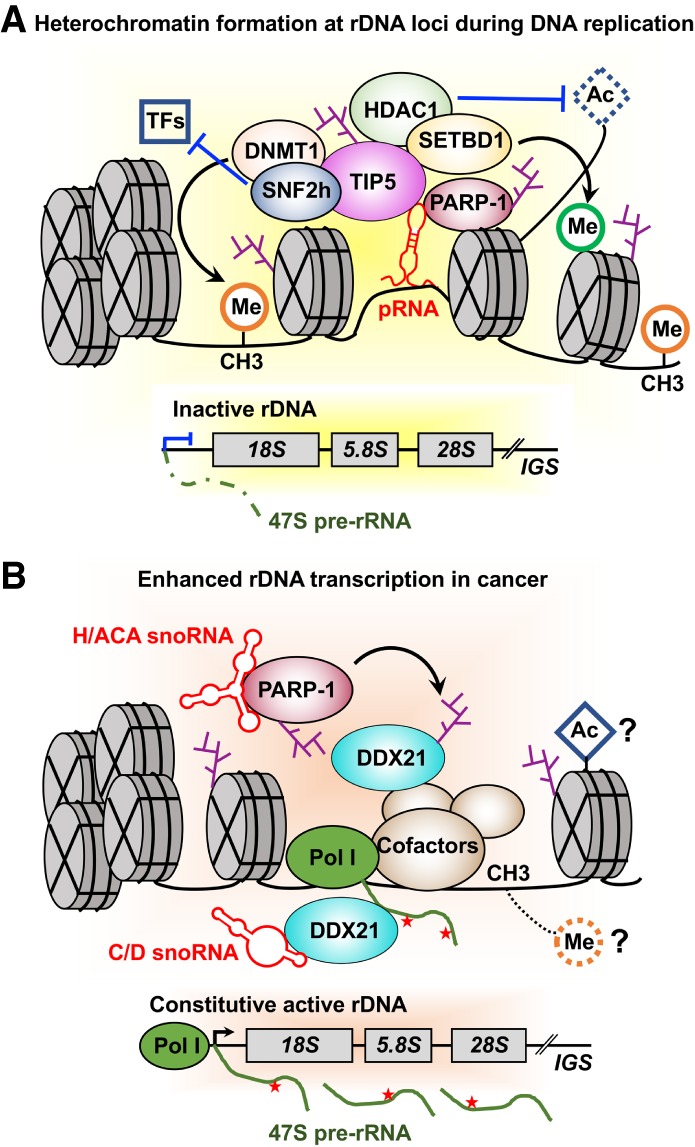
Dual roles of PARP-1 in the regulation of rDNA transcription. (A) pRNA-mediates PARP-1 and NoRC complex (TIP5 and SNF2h) interactions, leading to PARP-1 automodification and subsequent TIP5 or histone ADPRylation. TIP5 recruits the histone deacetylase HDAC1, DNA methyltransferase DNMT1, and histone lysine methyltransferase SETBD1 to the establish a silent state on rRNA genes. (B) snoRNAs interact with PARP-1 to activate PARP-1 catalytic activity. Subsequently, snoRNA-activated PARP-1 PARylates DDX21 to promote DDX21 association with the rDNA locus and retention in the nucleolus, promoting enhanced rDNA transcription in cancer.
This epigenetic reprogramming leads to impaired binding of various transcription factors to the rDNA promoter, with subsequent transcriptional silencing and heterochromatin formation. TIP5 is a key regulatory protein in the regulation of silent rDNA chromatin and heterochromatin formation. Importantly, pRNA-mediated TIP5-PARP-1 interactions lead to PARP-1 automodification and subsequent TIP5 and/or histone ADPRylation. Jointly, the studies described here indicate that PARP-1-mediated TIP5 and histone ADPRylation are essential for the formation of silent rDNA chromatin and transcriptional silencing by direct interaction with pRNA and the chromatin-remodeling complex NoRC.
Role of PARP-1 in Pol I-dependent transcription of rDNA
The first evidence of a functional link between rRNA processing and PARP-1 was proposed by Boamah et al. (2012) using Drosophila as a model system. PARP-1 plays an important role in the maintenance of nucleolar structure and function via the regulation of precursor rRNA processing, posttranscriptional modification, and preribosome assembly (Boamah et al. 2012). PARP-1 and its catalytic activity are required for (1) the maintenance of nucleolar integrity and (2) proper localization of nucleolar-specific proteins, such as fibrillarin, AJ1, nucleolin, and nucleophosmin in proximity to precursor rRNA in the nucleoli of Drosophila. Inhibition of PARP-1 enzymatic activity leads to nucleolar fragmentation and aberrant localization of nucleolar-specific proteins. PARP-1 deletion mutants exhibit a delay in rRNA processing and an increase in the levels of rRNA intermediates, such as 47S and 36S rRNA transcripts, which represses ribosome biogenesis (Boamah et al. 2012). The role of PARP-1 in regulating ribosome biogenesis will be discussed below.
Recent advances in chemical biology and protein engineering in the field have led to mass spectrometry-based identification of ADPRylation sites for PARP-1 protein substrates. This has led to the identification of a set of nucleolar proteins, which are key regulators in ribosome biogenesis, including DDX21, fibrillarin, nucleolar phosphoproteins numatrin/B23, and nucleolin/C23, as direct targets of PARP-1 enzymatic activity (Gagné et al. 2012; Chiou et al. 2013; Carter-O'Connell et al. 2014; Gibson et al. 2016; Kim et al. 2019). These PARP-1-ADPRylated proteins are known to be involved in rRNA transcription, pre-rRNA processing, and preribosome assembly, suggesting that PARP-1 and its enzymatic activity are required in nucleolar functions and, consequently, ribosome biogenesis.
In addition to the regulation of rRNA processing, PARP-1 can localize to the nucleolus, and PARP-1 accumulation in nucleoli is altered upon RNA polymerase I inhibition (Meder et al. 2005). Although this study suggests that PARP-1 (and PARP-2) does not affect the transcription of rDNA in murine fibroblasts, other studies have shown a role for PARP-1 in the regulation of rDNA transcription (Kurl and Jacob 1985; Guetg et al. 2012). Another study also reported that the nucleolar localization of PARP-1 is dependent upon active RNA synthesis (Desnoyers et al. 1996). Therefore, it is likely that PARP-1 nucleolar localization and its enzymatic activity are associated with the Pol I-dependent transcription of rDNA in nucleoli. These studies indicate that PARP-1 plays an important role in ribosome biogenesis, including rDNA transcription, processing, and ribosome assembly in the nucleolus. Emerging evidence has also implicated PARP-1 in the regulation of ribosome biogenesis in various human diseases, as discussed below.
Role of PARP-1 in the regulation of ribosomal biogenesis in pathological conditions
PARP inhibitors (PARPi), such as olaparib, rucaparib, niraparib, and talazoparib, are clinically important and have been approved by the FDA as monotherapies for treatment of recurrent, high-grade serous ovarian cancers with BRCA1/2 mutations (Bitler et al. 2017; McCann 2019). Olaparib has also been approved for the treatment of BRCA-mutated HER2-negative metastatic breast cancers (Robson et al. 2017), while niraparib has been shown to be efficacious in patients lacking BRCA mutations or HR deficiency (Mirza et al. 2016). These PARPi are currently being evaluated for their therapeutic potential in several other cancers. PARPi, acting through nuclear PARPs, are thought to control cancer cell growth primarily through inducing synthetic lethality in cancers that are deficient in homologous recombination (HR)-mediated DNA repair (e.g., in BRCA1/2 mutant cells) (Bryant et al. 2005). In the absence of functional BRCA1 or BRCA2 proteins, PARPi lead to the persistence of DNA lesions, resulting in chromosomal instability, subsequent cell cycle arrest, and apoptosis (Bryant et al. 2005; Farmer et al. 2005).
Growing evidence has shown that PARPi have therapeutic efficacy in BRCA1/2 wild-type cancers lacking other known HR or DNA repair defects. Interestingly, a recent study from our laboratory highlights the pathological significance of PARP-1-mediated site-specific ADPRylation events that are independent of PARP-1's role in DNA repair (Kim et al. 2019). Mechanistically, this study found that snoRNAs act as critical players in the activation of PARP-1 enzymatic activity in the nucleolus, which leads to DDX21 ADPRylation. DDX21 ADPRylation results in enhanced rDNA transcription, as well as breast cancer cell growth (Fig. 4B). Treatment with PARPi or mutation of the ADPRylation sites in DDX21 reduces DDX21 nucleolar localization, rDNA transcription, ribosome biogenesis, and cell growth (Kim et al. 2019). Thus, this study has uncovered an alternate molecular pathway for targeting breast cancer with PARPi irrespective of BRCA1/2 status by attenuating cancer-enhanced ribosome biogenesis. As such, this study strengthens the rationale for advancing the use of PARPi in clinical trials for the treatment of a broader array of cancers, including those with wild-type BRCA1/2.
Results from Guetg et al. (2012) using HEK-293T cells suggest an alternate mechanism for PARP-1's role in rDNA transcription, where PARP-1 and its enzymatic activity promote the formation of silent rDNA chromatin and transcriptional silencing of the rDNA locus. Increased numbers of ribosomes are required for the uncontrolled cellular proliferation and division of cancer cells compared with normal cells (Aspesi and Ellis 2019). PARP-1 and its enzymatic activity are significantly up-regulated in invasive cancer cells, as well as malignant tissues (Ossovskaya et al. 2010; Domagala et al. 2011). Thus, it is possible that the dual role of PARP-1 is based on the different amount of ribosomes present in cancer and normal cells to meet their need for protein synthesis and proliferation.
Interestingly, a recent study reported dispersed and less intense nucleolar PARP-1 staining in Alzheimer's disease (AD) compared with the distinct nucleolar localization in hippocampal pyramidal neurons in controls (Zeng et al. 2016). This study proposes that PARP-1 mislocalization from the nucleolus in AD (1) leads to hypermethylation of rDNA by DNA methyltransferase 1 (DNMT1), (2) subsequently reduces rDNA transcription and impairs ribosomal biogenesis, and (3) results in disruption of long-term memory formation (Fig. 5A,B). PARP-1-mediated DNMT1 ADPRylation inhibits the activity of DNMT1, subsequently preventing rDNA methylation and up-regulating rRNA expression. While these observations need to be confirmed, they suggest an interesting link between PARP-1 mislocalization and brain pathologies.
Figure 5.
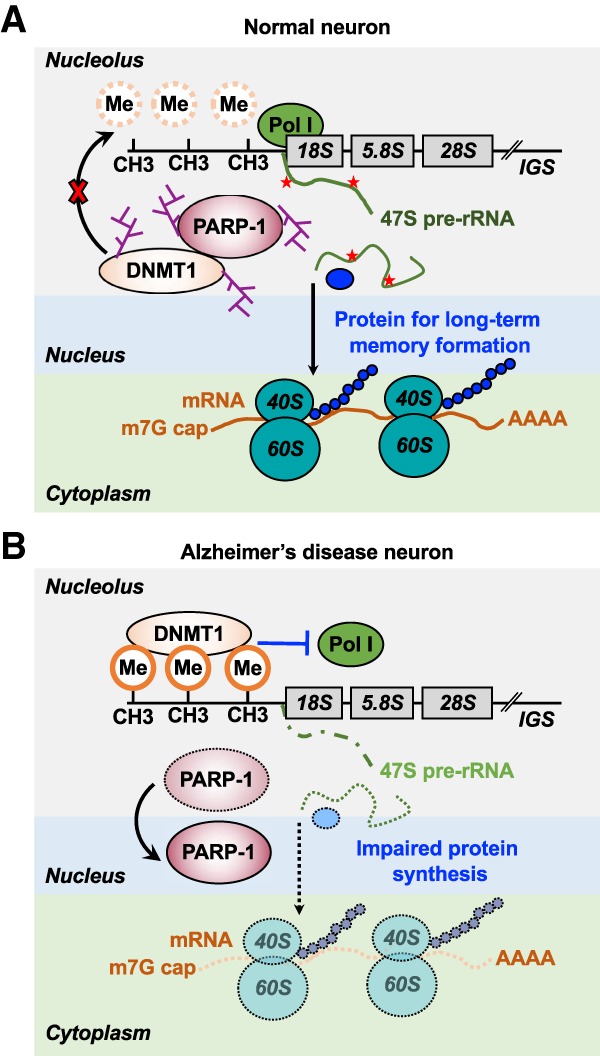
Role for PARP-1 in the regulation of rDNA transcription in neurons. (A) PARP-1-mediated DNMT1 ADPRylation prevents rDNA methylation by DNMT1, resulting in the production of rRNAs and subsequent ribosome biogenesis in normal neurons. (B) A substantial reduction in the nucleolar localization of PARP-1 leads to hypermethylation of rDNA by DNMT1, resulting in a reduction of rDNA transcription and ribosome biogenesis. Impaired ribosome biogenesis causes a disruption of long-term memory formation and the development of Alzheimer's disease.
Collectively, these studies indicate that PARP-1 and its enzymatic activity play an important role in the epigenetic regulation of rDNA in various aspects of biology. Based on the findings described above, PARP-1 regulates multiple areas of nucleolar function, including (1) establishment of transcriptionally inactive rDNA chromatin, (2) the maintenance of nucleolar integrity and structure, and (3) the regulation of Pol I dependent transcription of the rDNA.
Role of PARP-1 in the regulation of ribosomal biogenesis during DNA damage
Recent studies have shown that PARP-1 activation can regulate rDNA transcription in response to DNA damage (Calkins et al. 2013). DNA replication and rDNA transcription are inhibited following DNA damage mediated by UV light, γ radiation (IR), and cross-linking by cisplatin, resulting in the accumulation of cells in S phase. Inhibition of the DNA repair proteins, DNA-dependent protein kinase (DNA-PK), or PARP-1 prevents cisplatin-induced inhibition of rRNA synthesis (Calkins et al. 2013). This study showed that DNA-PK acts upstream of PARP-1 to recruit PARP-1 to chromatin at sites of DNA damage. Subsequent activation of PARP-1 leads to the inhibition of rRNA synthesis after DNA damage. Thus, DNA-PK-dependent PARP-1 activation may result in the maintenance of inherited silencing of rDNA genes and repression of rRNA synthesis (Fig. 6). However, PARP-1 translocates from the nucleolus to the nucleoplasm in the first 2 h after DNA damage and inhibition of rRNA synthesis has been observed between 12 and 24 h after DNA damage (Calkins et al. 2013). Thus, it is possible that nucleolar exit of PARP-1 after DNA damage, rather than direct silencing of rDNA by PARP-1 in nucleolus, affects the inhibition of rRNA synthesis in this process. Further studies of nucleolar-nucleoplasmic shuttling of PARP-1 after DNA damage are required to elucidate the role of PARP-1 in repression of rRNA synthesis following DNA damage.
Figure 6.
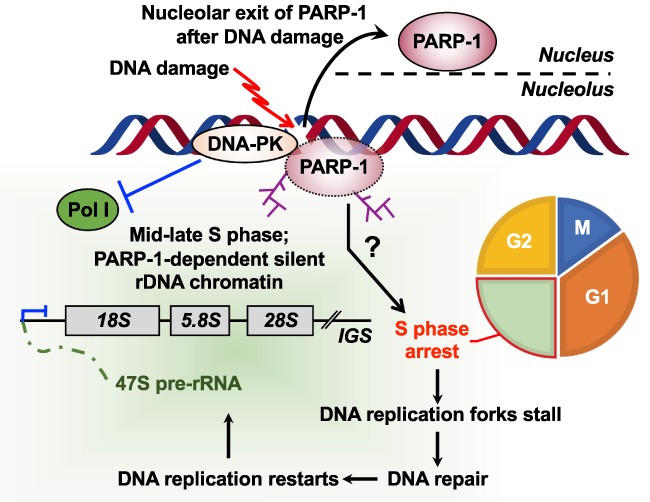
DNA damage-induced formation of silent rDNA chromatin. DNA damage leads to the accumulation of cells in S phase, with DNA replication forks stalled at sites of DNA damage. DNA-PK acts upstream of PARP-1 to recruit it to chromatin. Subsequent PARP-1 activation plays an essential role in the formation of silent rDNA chromatin at the time of replication.
Another possible mechanism by which PARP-1 could facilitate repression of rRNA synthesis is through direct or indirect interaction with target proteins with roles in rDNA transcription. For example, TARG1 localizes to transcriptionally active nucleoli through direct interaction with rRNA, as well as rRNA processing and ribosomal assembly factors, independent of ADPRylation (Bütepage et al. 2018). Interestingly, TARG1's nucleolar localization is abrogated by inhibition of rRNA transcription, indicating that TARG1 may function as a key regulator of rRNA synthesis. In addition, TARG1 relocalizes to the nucleoplasm upon DNA damage-induced PARP-1/PARP-2-dependent PARylation. TARG1 is mainly localized to the nucleolus in the absence of PAR, while it accumulates in the nucleoplasm in response to DNA damage-dependent PAR formation, indicating that TARG1 localization is strongly regulated by PARylation. These findings suggest that DNA damage-induced PARylation might serve to sequester TARG1 to the nucleoplasm, resulting in the loss of nucleolar function of TARG1 in rRNA synthesis (Bütepage et al. 2018).
Role of PARPs and ADPRylation in mRNA processing
Regulation of gene expression extends beyond transcription to include events that occur cotranscriptionally and posttranscriptionally. From the start of nascent mRNA transcription by RNA polymerase II until degradation in the cytoplasm, mRNAs are associated with RNA-binding proteins (RBPs). Many PARPs play key roles in these processes through various mechanisms, including (1) binding to RNAs, (2) interacting with, ADPRylating, and regulating RBPs and RNA processing factors, and (3) creating PAR, which can serve as a scaffold for noncovalent interactions with various RBPs and RNA processing factors. In this section, we explore the relationship among RNA processing events, PARP proteins, and ADPRylation.
Brief overview of mRNA processing
To reveal its coding potential, a pre-mRNA must undergo processing, including 5′ capping, splicing, and 3′ polyadenylation, to form mature mRNA before it can be exported to the cytoplasm for translation and degradation (Fig. 7). Nearly every step of this process has been shown to be regulated by PARP proteins. RNA processing events are tightly coordinated by RBPs (Castello et al. 2012), as the ribonucleoprotein (RNP) complex associated with RNA is dynamically remodeled sequentially by loss and gain of RBPs. Thus, RBPs serve a critical role in RNA metabolism. The most abundant cellular proteins contributing to RNP complex formation are members of the hnRNP and S/R protein families (Huang and Steitz 2005). The functions of RBPs are temporally and spatially regulated by phosphorylation (Matter et al. 2002), ubiquitylation (Bhandari et al. 2011), and ADPRylation (see below). RBPs have specificity for unique RNA targets, which regulates their splicing, stability, and localization.
Figure 7.
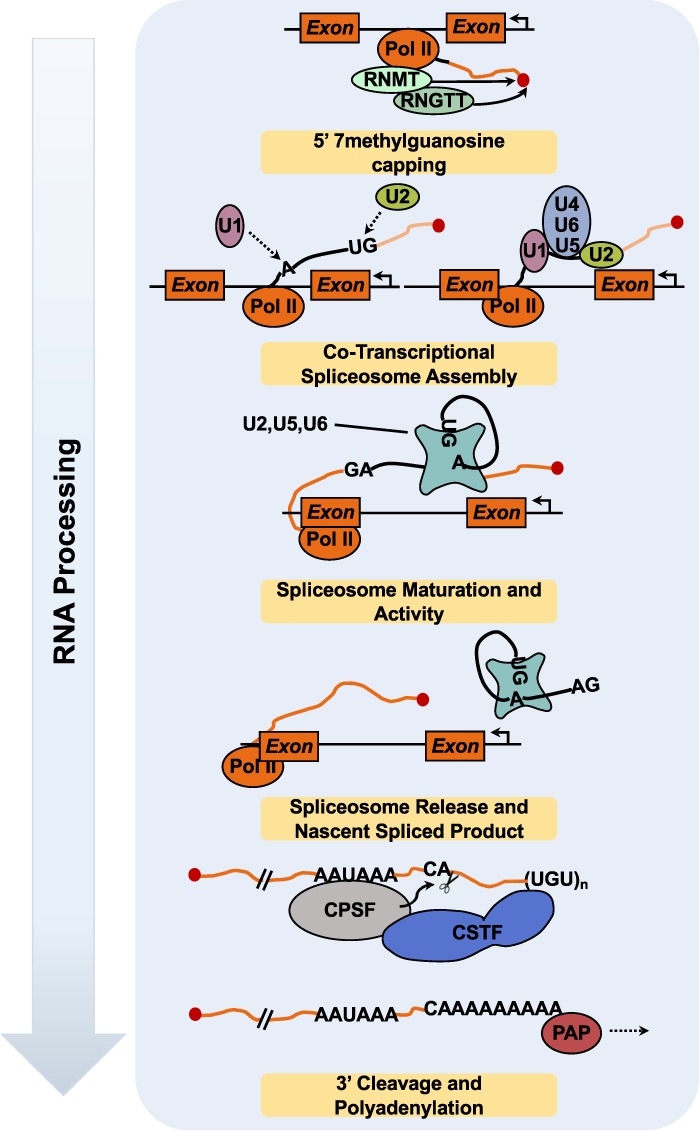
Overview of nuclear RNA processing events facilitating transformation from pre-mRNA to mRNA. Some RNA processing events occur cotranscriptionally, adding another level of regulation to the RNA processing. mRNAs are modified at their 5′ ends by the addition of a methylguanosine cap by RNA guanine-7 methyltransferase (RNMT) and RNA guanylyltransferase and 5′-phosphatase (RNGTT). RNMT recognizes and physically interacts with the CTD of RNA Pol II in close proximity to the 5′ end of the nascent RNA. RNMT (1) recruits RNGTT to add the guanosine to the 5′ end of the mRNA and (2) catalyzes the addition of a methyl group on the N7 position of the added guanosine. As RNA Pol II transcribes the length of the gene body, exon-intron boundaries are transcribed into the nascent pre-mRNA. The boundaries are identified via specific sequences that demarcate the exon–intron junctions. As the splice site motifs are transcribed, the spliceosome proteins snRNP U2 and U1 bind to their respective sequences, recruiting other spliceosome complex proteins (i.e., U5, U6, and U4). The spliceosome matures as U4 and U1 exit, followed by catalytic activation of the mature spliceosome. In a two-step process, the spliceosome forms a lariat structure with the exon and intron splice spites, cleaves the lariat, and ligates the exons together. Upon cleavage, the intron lariat spliceosome complex is released from the nascent spliced pre-mRNA. After RNA Pol II progresses into the 3′ UTR, the 3′ end is recognized by cleavage and polyadenylation specificity factor (CPSF) and cleavage stimulation factor (CSTF). These enzymes catalyze the cleavage of the pre-mRNA, leaving behind an exposed adenine, which is extended by polyadenylate polymerase (PAP).
PARP-1–chromatin complexes influence cotranscriptional mRNA processing
The nucleic acid-binding function of PARP-1 in its N-terminal zinc fingers has been implicated in regulation of the cotranscriptional aspect of mRNA processing. PARP-1 is a well-established chromatin architectural protein (Krishnakumar and Kraus 2010), and recently the effects of chromatin have been explored in relation to the regulation of splicing. Nucleosome occupancy regulates RNA polymerase (RNAP) kinetics, aiding in the recognition of weak splice sites (Schwartz and Ast 2010). Thus, factors impacting chromatin dynamics may regulate mRNA processing. A survey of the genomic distribution of dPARP-1 using nucleosome-ChIP-sequencing (nuc-ChIP-seq) in Drosophila S2 cells revealed that dPARP-1 preferentially binds to the +1 and +2 nucleosomes of active promoters and at internal exon/intron boundaries (Matveeva et al. 2016). These dPARP-1-bound nucleosomes have a high GC content; such nucleosomes are thought to mark weak splice sites and promote cotranscriptional splicing (Amit et al. 2012). Both depletion of dPARP-1 and inhibition of PARylation in S2 cells cause genome-wide changes in alternative splicing events (ASEs) without differential expression changes at constitutive exons. These changes in ASEs, however, occur at distinct loci in response to dPARP-1 depletion and dPARP-1 inhibition (Matveeva et al. 2016), highlighting the fact that the physical presence of PARP-1 is important for some molecular pathways, while its catalytic activity is necessary for others.
Changes in alternative splicing decisions correlate with PARP-1 nucleosome occupancy (Matveeva et al. 2016), suggesting that the PARP-1-mediated chromatin state affects RNAP II kinetics and, thus, cotranscriptional splicing (Fig. 8A). Deeper investigation of PARP-1 as a chromatin component revealed that PARP-1-mediated chromatin structure influences transcriptional elongation by RNAP II (Matveeva et al. 2019). Comparisons of PARP-1 nuc-ChIP-seq data with PRO-seq data, which delineates genomic sites of active transcription, showed that PARP-1 is enriched ∼25 bp downstream from RNAP II (Matveeva et al. 2019). Moreover, in locus-specific examples, PARP-1 depletion led to a corresponding decrease in RNAP II elongation, supporting previous results connecting PARP-1 and RNAP II elongation (Gibson et al. 2016). These results were supported at the genomic level by results from 3′NT-seq and NET-seq, which were used to measure changes in transcript length following PARP-1 depletion (Matveeva et al. 2019). Taken together, these results indicate PARP-1-bound nucleosomes decrease RNAP II elongation, facilitating increased cotranscriptional splicing.
Figure 8.
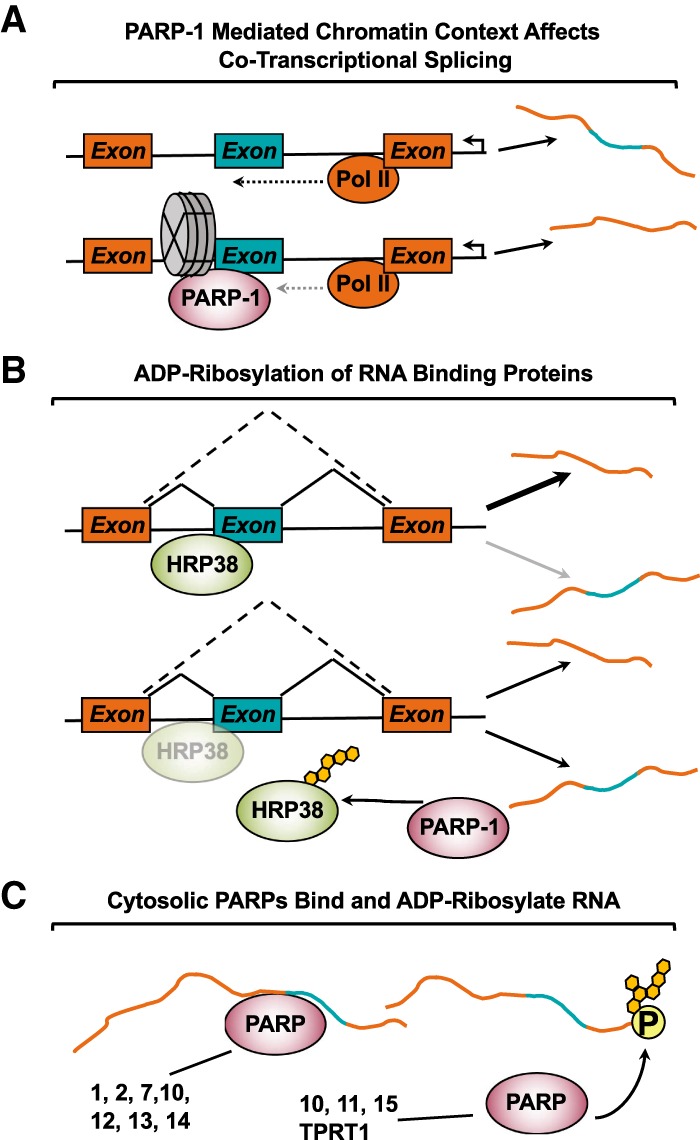
Examples of RNA processing events modulated by PARP activity. (A) PARP-1 binding to chromatin is enriched at nucleosomes +1/+2 within active promoters and at intron/exon boundaries. PARP-1 bound nucleosomes decrease RNA Pol II processivity, resulting in increased cotranscriptional splicing. (B) RNA-binding protein, HRP38, typically functions to inhibit splicing events. PARP-1 binds to and ADPRylates HRP38 in its RNA-binding domain, thereby attenuating HRP38's ability to bind RNA. The outcome is increased splicing of target transcripts. (C) Both PARP-1 and PARP-2 bind to RNA, resulting in the stimulation of their catalytic activities. Other cytosolic PARP family members (e.g., PARP-7, PARP-10, PARP-11, PARP-12, PARP-13, PARP-14, PARP-15) are known to be or are predicted to bind RNA. Furthermore, PARP-10, PARP-11, PARP-15, and TRPT1 were recently found to ADPRylate single-stranded RNAs at phosphorylated ends.
Interaction of mRNA processing factors with ADPR
In addition to the role of PARP-1-chromatin interactions, PARP family members have long been known to interact and regulate RNA binding and processing proteins through their catalytic activity. Kostka and Schweiger (1982) observed that the radioisotope tracer from [32P]-labeled NAD+ incubated with rat liver nuclei precipitated in particles carrying heterogeneous nuclear RNA (hnRNA), of which the majority is pre-mRNA. The radioactive signal was identified in ADPR moieties attached to RNPs with molecular weights of 36, 39, and 42 kDal. At the time, the biological function of ADPRylation was unknown. Since then, however, the relationships among PARPs, ADPRylation, and the regulation of mRNA processing have been met with growing appreciation.
Over a decade after the identification of ADPRylated RNPs, another group (Prasad et al. 1994) confirmed the interaction using similar methods to more specifically identify heterogeneous ribonucleoprotein particles (hnRNPs) A1 and A2/B1 as major targets of ADPRylation in HeLa cells. Proteomic approaches have further identified RBPs as ADPRylated substrates (Isabelle et al. 2010; Jungmichel et al. 2013; Zhang et al. 2013). Coimmunoprecipitation with PARP-1 and PARP-2 from human cells followed by mass spectrometry uncovered a host of hnRNPs (i.e., A1, A2/B1, C1/C2, G, H, K, E1, A3, L, M, U, and G) and other RNA processing proteins that interact with PARP-1 and PARP-2. The use of PAR-binding domains to directly pull-down ADPR interacting proteins reinforced the fact that a large number of hnRNPs and RBPs are in complex with ADPR in mammalian cells, which may occur in the absence of covalent modification of the proteins.
In addition to hnRNP members, serine-arginine-rich (S/R) family members interact with PAR chains as well. S/R factors function in both basal splicing and in the context of sequence-specific alternative splicing (Graveley 2000). Structurally S/R proteins are characterized by the presence of one to two RNA recognition motifs (RRMs) typically in their N-terminal region and an SR-rich domain (SR domain) in their C-terminal region. Malanga et al. (2008) observed that ASF/SF2, also known as serine- and arginine-rich splicing factor 1 (SRSF1), binds PAR via its RRM and RS domain. S/R proteins are phosphorylated by SR protein-specific kinases, regulating S/R protein function and localization (Huang and Steitz 2005). When SRSF1 was incubated with [32P]-labeled PAR, SRSF1-PAR high-affinity, salt-resistant complexes were observed (Malanga et al. 2008). The inhibition of SRSF1 phosphorylation corresponded to the presence of PAR in nuclear HeLa extracts. SRSF1 phosphorylation promotes splicing; therefore, the inhibitory action that PAR-bound SRSF1 has on its phosphorylation state could regulate alternative splicing or directly impact its RNA binding capacity. The function of PAR as a complex-forming scaffold or binding partner likely drives its ability to modulate the behavior of hnRNPs, S/R proteins, and other splicing factors.
Regulation of RNA-binding protein function by ADPRylation
The interaction between mRNA processing proteins and ADPR is not solely limited to binding of PAR, but includes direct ADPRylation of RBPs. PARylation of hnRNPs is conserved in Drosophila, with three hnRNPs (Hrp36, Hrp38, Hrp40) shown to be PARylated in vivo (Ji and Tulin 2009). hnRNPs bind to splice silencing sites and are widely accepted as canonical splicing repressors (Geuens et al. 2016). Immunofluorescent staining showed that PARylation of Hrp38 inhibited its localization to sites of active transcription. Furthermore, the RNA-binding ability of Hrp38 was negatively impacted by the presence of PAR (Ji and Tulin 2009), resulting in Hrp38 targets having increased production of the spliced isoform in a PARP-dependent manner. These results highlight the ability of hnRNP ADPRylation to modulate hnRNP activity (Fig. 8B).
RBP interactions with PARP family members or ADPR, as well as direct ADPRylation of RBPs, extends beyond the splicing factors and spliceosome components; RBPs that function in mRNA-processing events besides splicing are also regulated by PARPs and ADPRylation. Proteomic analysis identified PARP-1 within the 3′ end mRNA processing complex (Shi et al. 2009). 3′ end mRNA processing involves endonucleolytic cleavage of pre-mRNA by CPSF73, followed by synthesis of a poly(A) tail onto the 5′ cleaved products by poly(A) polymerase (PAP) (Fig. 7; Danckwardt et al. 2008). Key aspects of 3′ processing are associated with mRNA stability, export, and translation efficiency (Danckwardt et al. 2008). NAD+-dependent PARP-1 catalytic activity in an mRNA polyadenylation assay significantly decreased poly(A) tail length (Di Giammartino et al. 2013). PAP, the enzyme that catalyzes the creation of the poly(A) tail, directly interacts with PARP-1 and is ADPRylated in vitro. Activation of PARP-1 catalytic activity by heat shock decreased both PAP binding to mRNA and mRNA polyadenylation levels in 293T cells. Furthermore, heat shock caused dissociation of PAP from the 3′ end of actively transcribing non-heat-shock-dependent genes (Di Giammartino et al. 2013). These results indicate that, in contrast to prior studies highlighting the binding of PAR by RBPs, direct PARylation of core processing factors can modulate their activity and, thus, influence pre-mRNA processing.
Hu antigen R (HuR) is another example of an ADPRylated RBP. In lipopolysaccharide (LPS)-treated murine primary peritoneal macrophages, the stability of LPS-responsive pre-mRNAs decreases upon PARP-1 inhibition or depletion (Ke et al. 2017). These pre-mRNAs contain repeats of AUUUA motifs. HuR binds these adenylate–uridylate-rich elements (AU-rich elements; AREs), which are commonly enriched within 3′ UTRs and function as major mRNA destabilization elements (Zubiaga et al. 1995). In response to LPS, HuR interacts with PARP-1, resulting in the PARylation of HuR, which directs its proper subcellular localization and enhances its binding to LPS-responsive mRNA targets (Ke et al. 2017). PARP-1-mediated PARylation of HuR illustrates another aspect of RNA metabolism that is PARP regulated, namely mRNA stability. These studies demonstrate that PARylation of RBPs regulates the molecular functions of these proteins with regard to the formation of RNP complexes and facilitation of pre-mRNA processing.
As illustrated here, PARP family members have been shown to regulate mRNA processing by (1) catalytic activity directed toward RBPs, (2) direct protein–protein interaction with RBPs, and (3) changing the chromatin landscape to favor cotranscriptional splicing. The roles of interactions between PARP-1 and chromatin or regulatory proteins in these processes are well established; the role of direct interactions between PARP-1 and RNA is less well understood.
Function of PARP proteins binding directly to RNA
Recently, after speculation about the possibility, PARPs have been shown to bind directly to RNA. By using photoactivatable-ribonucleoside-enhanced cross-linking and immunoprecipitation (PAR-CLIP) in S2 Drosophila cells, dPARP-1 was identified as an RNA-binding protein (Matveeva et al. 2016). Furthermore, nucleosome-bound dPARP-1 was able to bind RNA concomitantly with splicing factors, forming a bridge that facilitates cotranscriptional splicing (Matveeva et al. 2016). Using the same assay in HeLa cells, a greater depth of sequencing allowed for higher resolution of the breadth and variety of mRNAs bound by PARP-1 (Melikishvili et al. 2017). PARP-1 binds target transcripts most frequently within the intron, indicating that PARP-1 preferentially binds to pre-mRNAs. While the majority of RNAs bound by PARP-1 in these assays were mRNAs, other types of RNAs bound by PARP-1 have been identified, including noncoding RNAs and small nuclear RNAs (Melikishvili et al. 2017; Kim et al. 2019), indicating that PARP-1 functions in the regulation of other RNA types as well. In contrast to intron-bound mRNAs, target mRNAs that were bound uniformly over the length of the transcript by PARP-1 exhibited a preference for exon boundaries. This favored binding pattern was enriched within a subset of transcripts that are alternatively spliced in a PARP-1-dependent manner (Melikishvili et al. 2017).
Interestingly, the binding of mRNAs stimulates PARP-1 catalytic activity (Melikishvili et al. 2017), as shown previously for PARP-2, which binds rRNA through its SAF/SAP domain (Léger et al. 2014). PARP-1 catalytic activity has also been shown to be stimulated in vitro and in cells by the binding of a selected set of snoRNAs (Kim et al. 2019). Thus, interactions between nucleic acid-binding PARPs (e.g., PARPs 1 and 2) and their target RNAs can activate PARP catalytic activity, promoting additional levels of regulation.
In the cytoplasm, further layers of mRNA processing modulate mRNA stability and decay. Cytoplasmic PARP family members (i.e., PARP-7, PARP-10, PARP-12, PARP-13, and PARP-14) have been predicted to bind RNA through RNA-binding CCCH-Zn fingers or RRMs (Bock et al. 2015). With the exception of PARP-10, these predicted RNA-binding PARPs also possess PAR-binding domains (i.e., WWE domains or macrodomains). PARP-13 has been shown to bind to viral RNAs, targeting them for decay (Guo et al. 2004; Zhu et al. 2011; Ficarelli et al. 2019; Meagher et al. 2019). PARP-13 also binds to cellular mRNAs as well (Todorova et al. 2014; Schwerk et al. 2019). PARP-13 recruits the exosome to mRNAs through protein–protein interactions with exosome component hRrp46p, targeting bound mRNAs for degradation (Guo et al. 2007). PARP-13 knockout results in major dysregulation of the transcriptome (Todorova et al. 2014). PARP-14, acting in the cytoplasm, also regulates mRNA stability. PARP-14 binds the RBP tristetraprolin to promote the selective posttranscriptional control of macrophage tissue factor expression by binding the ARE in the 3′ UTR of tissue factor mRNA, promoting its degradation (Iqbal et al. 2014). Tissue factor is required for the control of blood coagulation, and PARP-14-deficient mice exhibit thrombogenicity or an increased tendency of blood clotting (Iqbal et al. 2014). These examples highlight the role of RNA binding by PARP proteins, including cytosolic PARP monoenzymes, which allows for another layer of regulation in mRNA processing and metabolism. With these studies exploring RNA-mediated PARP activation, the field had yet to examine whether this catalytic activity could be directed toward RNA as a substrate.
ADPRylation of RNAs: a potential role in RNA processing?
For many years, proteins were assumed to be the only substrates for PARP enzymes. However, with recent evidence that DNA can be ADPRylated by the toxin Pierisin (Takamura-Enya et al. 2001), the question of whether RNA can be ADPRylated was revisited. Indeed, PARP-like proteins in bacteria and fungi can ADPRylate RNA (Munir et al. 2018). In biochemical assays with purified components, PARP-10, PARP-11, PARP-15, and PARP-TRPT1 ADPRylate the phosphorylated ends of RNA in a reversible manner (Munnur et al. 2019). Using [32P]-labeled NAD+ as an ADPR donor, PARP-10 robustly ADPRylates 5′ and 3′ phosphorylated single-stranded RNA (ssRNA). Unexpectedly, the remaining proposed RNA-binding PARPs, with the exception of PARP-7, were tested in the same manner and were unable to ADPRylate ssRNA, with or without phosphorylation (Fig. 8C). ADPRylation of RNA is a novel biochemical function of PARP family members, but these results need to be confirmed in vivo. In addition, further studies of RNA ADPRylation are necessary to understand its biological role and impact. Finally, the mechanism by which PARPs that lack RNA-binding domains direct their catalytic activity toward RNA requires further investigation.
Role of PARPs and ADPRylation in protein synthesis, protein degradation, and proteostasis
Altered protein homeostasis has been implicated in numerous pathological disorders, such as cancer and Alzheimer's disease. The maintenance of a homeostatic proteome (i.e., proteostasis) is intimately tied to RNA biology and is regulated at several stages, including ribosome biogenesis, ribosome function, mRNA translation, protein stability, protein folding, and clearance of misfolded proteins. Various PARPs and ADPRylation have been shown to regulate each of these processes to support protein homeostasis. In this section, we review current knowledge about the PARP-mediated regulation of protein synthesis and proteostasis under normal and pathological states.
Role of ADPRylation in the regulation of mRNA translation by bacterial toxins
As elaborated below, one of the initial indications of the role of PARPs as regulators of protein synthesis was from early studies demonstrating how toxins inhibit host mRNA translation. In addition, PARPs, especially cytosolic PARP enzymes, control mRNA translation in cells when they are subjected to stresses, such as viral infections. In the next two sections below, we describe the role of ADPRylation and PARPs in the regulation of mRNA translation during bacterial and viral infections.
Protein toxins are secreted by bacteria to disrupt host cellular pathways through covalent modifications of host proteins, which supports bacterial virulence. Over the past several decades, a number of bacterial toxins, such as those produced by Corynebacterium diphtheriae (i.e., diphtheria toxin) and Vibrio cholerae (i.e., cholera toxin), have been shown to MARylate host proteins (Simon et al. 2014). These toxins are A–B-type oligomeric molecules comprising a catalytic domain (A) mediating ADPRylation and a translocation domain (B) controlling entry of the toxin into the cell. During infections, the B domain is cleaved by proteases, such as furin, activating the catalytic A domain and releasing it into the cytosol (Fig. 9; Simon et al. 2014). The ADPR transferase activity of the A domain can modify a variety of host proteins, including the translation elongation factor EF2 (by diphtheria toxin and exotoxin A), G-protein subunits (by cholera toxin and pertussis toxin), and actin cytoskeleton by the C2 or C3-like exoenzymes (Deng and Barbieri 2008).
Figure 9.
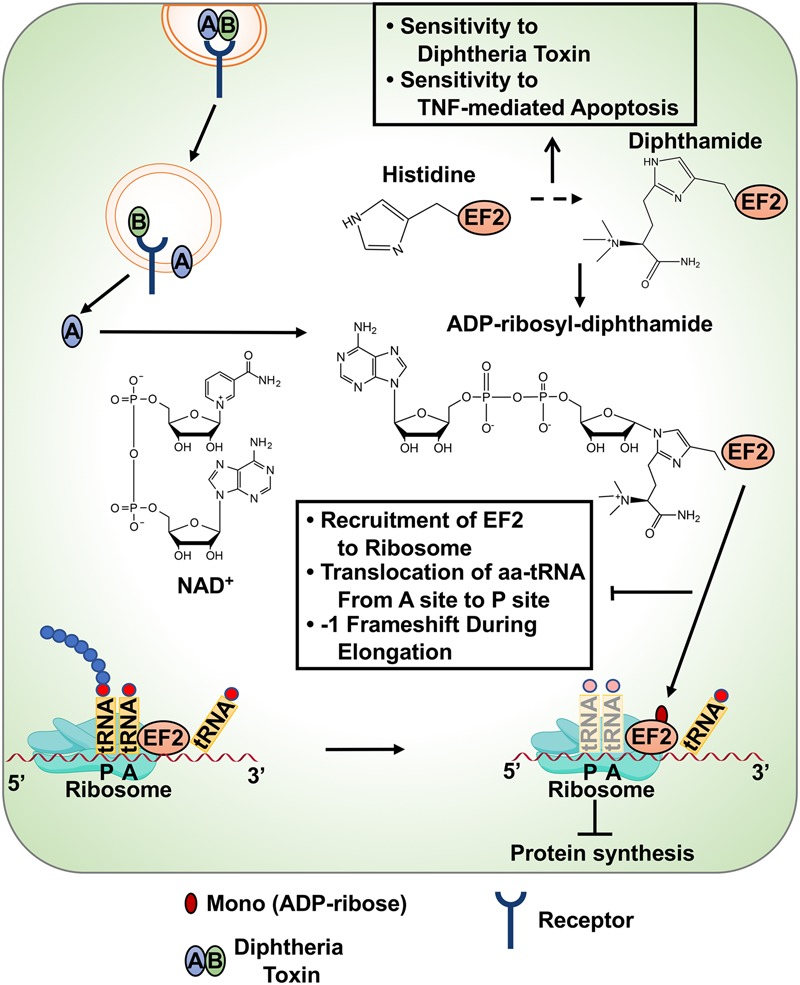
Diphtheria toxin-mediated regulation of mRNA translation. Diphtheria toxin is internalized in cells by binding to cell surface receptors and is subsequently activated by cleavage of the regulatory B domain from the catalytic A domain. The activated A domain MARylates the diphthamide residue in EF2 protein using NAD+ as a substrate. Histidine is modified to diphthamide by a multi-step biosynthetic pathway. ADPRylation of EF2 attenuates protein synthesis by inhibiting (1) EF2 incorporation into ribosomes, (2) transfer of aminoacyl tRNA from the A site to P site, and (3) reverse translocation and −1 frameshifting of ribosomes during translational elongation. Loss of the members of the diphthamide biosynthesis pathway results in a loss of sensitivity to diphtheria toxin and enhanced sensitivity to TNFα-mediated apoptosis.
A variety of ADPR acceptor residues in toxin substrates have been identified, including Arg, Thr, Asn, Gln, Cys, and His (Oppenheimer and Bodley 1981; Manning et al. 1984; West et al. 1985; Vandekerckhove et al. 1987; Lang et al. 2010, 2017; for detailed review, see Cohen and Chang 2018). Of note, diphtheria toxin, a highly potent toxin in humans, MARylates residue 715 in the eukaryotic elongation factor-2 (eEF2) (Foley et al. 1995), which is a variant histidine amino acid, termed as diphthamide (Van Ness et al. 1980). Thus far, diphthamide is found only in archaeal or eukaryotic EF2 proteins, with a single residue per molecule of EF2. Diphthamide is synthesized in a multi-step biosynthetic pathway. Knockout of genes encoding enzymes in this pathway in cancer cells results in a loss of EF2 ADPRylation and decreased sensitivity to diphtheria toxin, while enhancing sensitivity to TNFα-mediated apoptosis (Stahl et al. 2015).
How does EF2 ADPRylation affect mRNA translation? EF2 is a GTPase protein essential for mRNA translation, which catalyzes the transfer of peptidyl-tRNA from the ribosome A-site to P-site, moving the ribosome from 5′ to 3′ end of the mRNA template (Jørgensen et al. 2006). ADPRylation of EF2 has several effects on the function of ribosomes. Briefly, ADPRylation of EF2 results in (1) reduced ribosome association in pretranslocation state (Nygård and Nilsson 1985), (2) blocked translocation of aminoacyl tRNA from the A-site to P-site of ribosomes (Davydova and Ovchinnikov 1990), (3) errors in −1 frameshifting during translation elongation (Liu et al. 2012), and (4) blocked reverse translocation of ribosome (Fig. 9; Susorov et al. 2018). While these studies shed light on the mechanisms by which EF2 ADPRylation by toxins inhibits host mRNA translation during infections, mammalian PARPs also regulate the defense response to infections.
Role of ADPRylation and PARPs in the regulation of mRNA translation during viral infections
Cytokines, such as interferons, which are produced as a first line of defense against viral infections, induce the expression of PARP-7, PARP-10, PARP-12, and PARP-13. In addition, PARPs, such as PARP-12, PARP-9, and PARP-14, were identified as core interferon-stimulated genes in 10 different vertebrate species (Shaw et al. 2017). As mentioned in the previous section, with the exception of PARP-10, all of the interferon-stimulated PARPs belong to the CCCH-family, which contain RNA-binding CCCH-type zinc fingers. Thus, there is a potential role for these PARPs in mediating antiviral responses by altering the stability or translation of mRNAs (Wang et al. 2010; Atasheva et al. 2014). In the following section, we briefly discuss the various mechanisms by which PARPs regulate viral replication.
After Sindbis virus (SINV) infection, PARP-7 (also known as TIPARP) accumulates in the cytoplasm, where its binding to the SINV RNA leads to recruitment of the EXOSC5 complex to degrade the SINV mRNA, hence eliminating SINV viral particles from the cell (Fig. 10A). Consistent with this observation, Parp7 knockout mice are sensitive to SINV infections due to higher replication of the SINV virus (Kozaki et al. 2017). Similar to PARP-7, PARP-13, a CCCH PARP, binds to mRNAs or viral RNAs through its CCCH domain, resulting in the destabilization of the mRNAs by recruitment of mRNA degradation machinery (Fig. 10A; Bick et al. 2003; Guo et al. 2007; Zhu et al. 2011; Todorova et al. 2014). Although PARP-13 does not have catalytic activity, it is able to regulate important biological processes, such as miRNA silencing and stress granule assembly, by binding to and regulating the activity and function of other PARPs, such as PARPs 5a, 12, and 15 (Fig. 10B,C; Leung et al. 2011).
Figure 10.
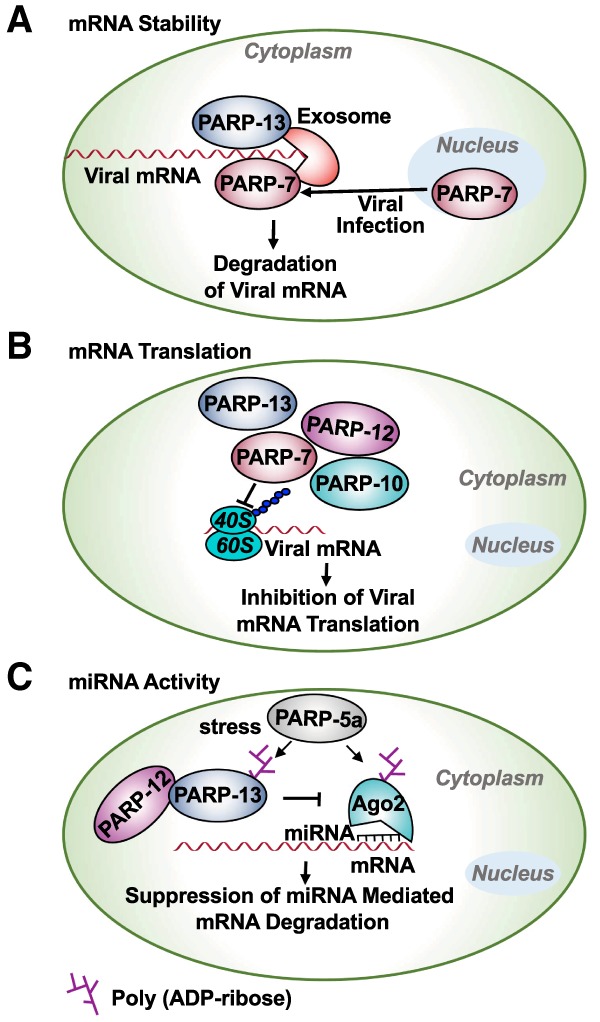
Role of PARPs in the regulation of mRNA functions. Several members of the PARP family control mRNA functions. These pathways play an important role in the regulation of response to viral infections and stress. (A) PARP-7 and PARP-13 bind to viral mRNAs and recruit the exosome complex, resulting in the degradation of the viral mRNAs, thus protecting against viral infections. (B) Interferon-responsive PARPs, such as PARP-7, PARP-10, PARP-12, and PARP-13, inhibit viral infections by inhibiting translation of the viral mRNAs. (C) In response to stress, PARP-13, PARP-12, and PARP-5a are recruited to stress granules, where PARP-5a PARylates PARP-13 and Ago2. These PARylation events inhibit miRNA silencing and increase expression of the genes required to elicit an antiviral response.
Macrophages are key immune cells that regulate inflammation by secreting cytokines and chemokines that regulate various pathways important for initiating inflammation. Multiple PARPs play key roles in this process. Notably, PARP-12 in macrophages localizes to stress granules, which are aggregates of ribonucleoproteins formed on stalled ribosomes under various stress conditions. PARP-12 is also associated with the translational machinery and suppresses mRNA translation (Welsby et al. 2014). Consistent with this, Atasheva et al. (2014) have shown that PARP monoenzymes, such as PARP-7, PARP-10, and PARP-12L (long isoform of PARP-12), inhibit viral protein synthesis by modulating the cellular translational machinery (Fig. 10B). Although the precise mechanism of this regulation is not completely known, a requirement for the RNA-binding domains and the catalytic activity of PARPs for this regulation suggests a complex regulation of translation and protein synthesis by these PARP enzymes (Atasheva et al. 2014).
Considerable evidence suggests that PARPs act as key players in antiviral response. Consistent with this observation, viruses have developed mechanisms to counteract PARP-mediated ADPRylation. RNA viruses, such as alphaviruses, encode highly conserved, nonstructural viral proteins containing macrodomains, which bind to ADPR and hydrolyze the ADPR from MARylated proteins. Recent studies have shown that mutations in the macrodomain of CHIKV virus that inhibit ADPR binding and hydrolase activity have decreased viral protein production and reduced virulence (McPherson et al. 2017; Abraham et al. 2018). Similar results have been observed for other virally encoded macrodomains, such as those from Coronavirus and Hepatitis E virus (Fehr et al. 2015, 2018; Li et al. 2016; Grunewald et al. 2019). In summary, the hydrolase activity of viral macrodomain proteins play a key role in virulence, but the substrates of the hydrolase activity of these macrodomains have yet to be identified. Moreover, PARPs may also regulate mRNA translation by modifying core ribosomal proteins. Whether the macrodomain hydrolases remove these modifications has yet to be studied.
ADPRylation of ribosomal proteins
Ribosomes are the basic machinery of mRNA translation. Eukaryotic ribosomes (80S ribosomes) are assembled from a large 60S subunit comprising 49 ribosomal proteins and a small 40S subunit comprising 33 ribosomal proteins, plus the rRNAs. As discussed above, ribosome biogenesis is a complex, multistep pathway that is regulated by PARP-1, but little is known about the impact of ADPRylation on ribosomal proteins and their functions within the ribosome. Recent studies have demonstrated that ribosomal proteins are indeed modified by PARPs. Zhen et al. (2017) found that ribosomal proteins are major targets of glutamate- and aspartate-directed ADPRylation in the breast cancer cell line MDA-MB-468 (Zhen et al. 2017). Although the precise effect of these modifications on ribosome function or mRNA translation is not yet understood, the sites of ADPRylation are located at the interfaces between the ribosomal subunits, suggesting that ADPRylation may disrupt protein–protein or protein–RNA interactions within the ribosome. Consistent with this observation, treatment with PARP inhibitors partially blocked DNA damage-induced translational suppression (Zhen et al. 2017). Another study profiling sites of ADPRylation in HeLa cells identified tyrosine residues as acceptors of ADPR in ribosomal proteins, with an enrichment of lysine at the +1 position (Hendriks et al. 2019). The precise impact of ribosomal protein ADPRylation on ribosome functions, including mRNA translation, has yet to be determined.
Regulation of protein ubiquitylation by PARPs and ADPRylation: ADPR-dependent ubiquitylation
Growing evidence has shown that PARPs regulate protein abundance in cells via posttranslational regulation under various stress conditions (Luo and Kraus 2012). Protein quality control is maintained by several pathways, including (1) ubiquitin-mediated protein degradation; (2) autophagy, which recycles proteins accumulated in autophagosomes; and (3) unfolded protein response pathways. Dysregulation of these pathways underlie numerous pathological conditions, such as cancer, inflammation, and neurodegenerative disorders. In this section, we review the role of PARPs in the regulation of protein quality control and its impact on pathologic conditions, such as neurodegenerative disorders.
Ubiquitylation is a major posttranslational modification that controls protein localization, stability, and activity (Mani and Gelmann 2005; Komander and Rape 2012). Protein ubiquitylation is a multistep process during which ubiquitin is attached to lysine residues in target proteins. This modification is mediated by three key types of enzymes: E1 ubiquitin-activating, E2 ubiquitin-conjugating, and E3 ubiquitin ligase enzymes. Improper function of these proteins is implicated in diseases, such as cancer (Mani and Gelmann 2005; Komander and Rape 2012). Early investigations of cross-talk between ADPRylation and ubiquitylation found that, in many cases, PARylation precedes ubiquitylation and promotes ubiquitin-dependent degradation through a “PAR-dependent ubiquitylation” pathway (Gibson and Kraus 2012). In this regard, RNF146, an E3 ubiquitin ligase, is recruited to PARylated target proteins by binding to PAR through its PAR-binding WWE domain (Zhang et al. 2011). This results in ubiquitylation of target proteins through the RING E3 ligase domain of RNF146. Substrates targeted by RNF146 for ubiquitylation include axin, BLZF1, and CASC3, which are substrates for tankyrases (PARP-5a and PARP-5b) (Fig. 11A; Zhang et al. 2011). Consistent with this, Bhardwaj et al. (2017) have identified several targets of tankyrases that are degraded by this pathway. This cross-talk between PARylation and ubiquitylation was also observed in CHFR-mediated ubiquitylation and degradation of PARP-1 under mitotic stress (Kashima et al. 2012).
Figure 11.
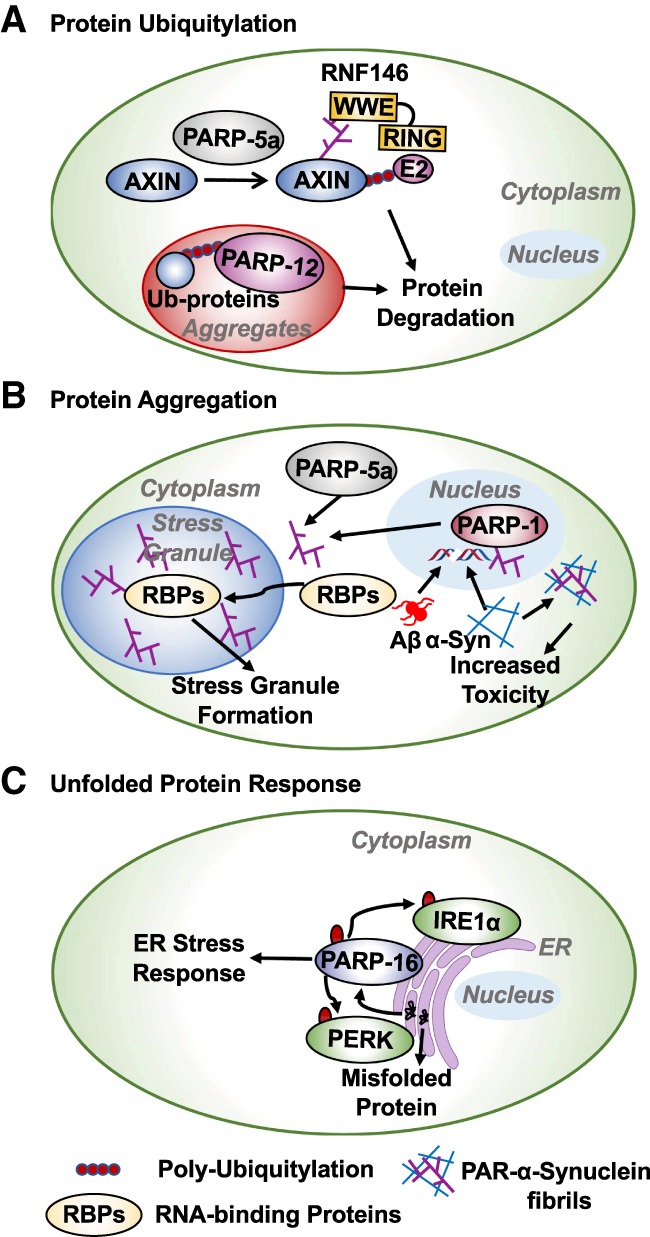
Role of PARPs in the regulation of protein homeostasis. The activity of both nuclear and cytosolic PARPs are required for protein homeostasis. (A) The WWE domain of RNF146 binds to PAR chains that are catalyzed by PARP-5a. RNF146 recruits an E2 ubiquitin-conjugating enzyme resulting in polyubiquitin-mediated degradation of PARP-5a substrates. PARP-12 binds to polyubiquitylated proteins in protein aggregates. (B) Toxic aggregates of β-amyloid and α-synuclein fibrils induce ROS-mediated DNA damage, causing the activation of PARP-1 and PAR synthesis. α-synuclein fibrils bound to PAR chains cause more cytotoxicity. The PAR chains generated by PARP-1 and PARP-5a recruit RNA-binding proteins (RBPs) to stress granules. PAR chains and modification of RBPs are required for stress granule assembly. (C) Unfolded proteins cause ER-stress and activate PARP-16. PARP-16 MARylates and activates PERK and IRE1α, which are required for ER stress response.
ADPR-dependent ubiquitylation has also been observed in bacteria and other mammalian systems. Recently, a putative MAR transferase motif was identified in members of the SidE effector family of the bacterial pathogen Legionella pneumophila, which supports ubiquitylation of targets without the need for E1 and E2 enzymes (Qiu et al. 2016). Interestingly, ubiquitin itself was found to be ADPRylated by SdeA, a SidE family member, using NAD+ as a substrate (Qiu et al. 2016). The cross-talk between MARylation and ubiquitylation is further exemplified by the finding that PARP-12 is enriched in aggregate structures containing ubiquitylated proteins in macrophages (Fig. 10A; Welsby et al. 2014). A recent study has also shown that PARP-12 regulates the stability of the four and a half LIM domain protein FHL-2, although MARylation of FHL-2 by PARP-12 was not observed (Shao et al. 2018).
Regulation of protein aggregation and proteostasis by PARPs and ADPRylation
Proteins containing glutamate repeats, low-complexity domains, or abnormal conformations tend to aggregate and cause cytotoxicity. Protein aggregate deposits can be found in samples from patients with neurodegenerative diseases, such as Parkinson's disease, amyotrophic lateral sclerosis, Alzheimer's disease (AD), and Huntington's disease, linking protein aggregation-mediated cytotoxicity to pathogenesis (Ross and Poirier 2005). The proteins that aggregate and the underlying mechanisms of their aggregation in these disorders are diverse, and PARPs (e.g., PARP-1) and PAR play a role in promoting these pathogenic effects.
Some of the first evidence of a role of PARP-1 in Alzheimer's disease was from a study by Love et al. (1999), in which they immunostained brain tissues for PAR and various cell type markers (e.g., MAP2 for neuronal cells, GFAP for astrocytes, and CD68 for microglia). They found that PAR is higher in brains from AD patients, specifically in neurons and astrocytes. They did not, however, observe a significant incorporation of PAR on tau or β-amyloid proteins, which commonly form aggregates in AD (Love et al. 1999). In contrast, Abeti et al. (2011) showed that PAR accumulates in the brains of aging TASTPM mice, which are a double-transgenic mouse model of Alzheimer's disease carrying two mutations associated with early-onset disease: the Swedish mutation in amyloid precursor protein and the M146V mutation in presenilin-1. They found that β-amyloid-mediated oxidative stress caused PARP-1 activation, depletion of NAD+ levels, and mitochondrial depolarization, which could be reversed by treating with inhibitors of PARP-1 or NADPH oxidase (Abeti et al. 2011).
Interestingly, in Parkinson's disease patients, PAR interacts with α-synuclein, accelerating α-synuclein fibrillarization, with the resulting α-synuclein fibrils exhibiting more toxicity (Kam et al. 2018). The α-synuclein aggregates accumulate in Lewy bodies in the neurons of Parkinson's disease patients causing neuronal dysfunction. Interestingly, recombinant α-synuclein fibrils activate PARP-1 through DNA damage, forming a feedback loop in which PAR plays a key role in α-synuclein fibrillarization that promotes cell toxicity through parthanatosis (Fig. 11B). This feedback loop can be disrupted by inhibition of PARP-1 activity or genetic depletion of PARP-1 (Kam et al. 2018).
Further elaborating these themes, mutations in RNA-binding proteins (RBPs), such as hnRNP A1, TDP-43, and FUS proteins, have been found to promote their aggregation in the cytoplasm and localization to stress granules (Kapeli et al. 2017). These mutations are causative for neurodegenerative disorders, including amyotrophic lateral sclerosis and frontotemporal dementia (Kapeli et al. 2017). Interestingly, PAR and members of the PARP-family regulate localization of these RBPs to stress granules and affect the formation of stress granules (Fig. 10B; Leung 2014; Bock et al. 2015). PAR chains produced by tankyrase recruit RBPs, such as TDP-43, to stress granules and drive the formation of stress-induced cytoplasmic TDP-43 foci (McGurk et al. 2018). Similar to TDP-43, hnRNP A1 binds to PAR chains through a PAR-binding motif and regulates the recruitment of hnRNP A1 to stress granules (Duan et al. 2019). Additionally, PARylation of hnRNP A1 regulates its nuclear-cytoplasmic shuttling (Duan et al. 2019). Given the important roles played by PARPs and PARylation in protein aggregation, inhibitors of PARP-1 or tankyrase could be effective in alleviating the neurodegenerative disease burden.
Protein folding is mediated by the chaperone network in the endoplasmic reticulum (ER). Accumulation of misfolded proteins in cells may result in ER stress and activation of the unfolded protein response (UPR) (Shacham et al. 2019). PARP-16, a tail-anchored ER protein, MARylates ER proteins involved in UPR, such as PERK and IRE1α, enhancing their kinase and endonuclease activities, respectively, during UPR (Fig. 11C; Jwa and Chang 2012). Similar to ER stress, amino acid starvation induces MARylation in a PARP-16-dependent manner, which triggers MARylation of Sec16, a key component of the Sec body (Aguilera-Gomez et al. 2016). This modification of Sec16 is required for Sec body formation during amino acid deprivation (Aguilera-Gomez et al. 2016). Thus, nuclear and cytosolic PARPs act together to control the function of various protein synthesis and protein quality control pathways.
Intriguingly, the levels of cellular NAD+, the substrate required for PARP activity, is associated with proteotoxicity and pathogenesis of neurodegenerative diseases (Ocampo et al. 2013; Sorrentino et al. 2017). Approaches to increase the levels of cellular NAD+ by treatment with NAD+ precursors are currently being evaluated (Hou et al. 2018; Rajman et al. 2018; Schöndorf et al. 2018). As shown previously in our laboratory, compartmentalization of NAD+ synthesis and function helps to balance nuclear and cytosolic PARP activity, and treatment with NAD+ precursors can disrupt this balance (Ryu et al. 2018). A greater understanding of the role of compartmentalized NAD+ synthesis and PARP-mediated ADPRylation is needed to fully understand its impact on mRNA translation and protein homeostasis.
Conclusions and perspectives
The regulation of RNA-dependent processes in cells by ADPRylation and PARP family members is required for general cell health and physiological processes. Growing evidence indicates that PARP-mediated ADPRylation events are key regulators of the protein biosynthetic pathway, leading from rDNA transcription and ribosome biogenesis to mRNA synthesis, processing, and translation. The impact of PARPs and ADPRylation is evident at nearly every step (Fig. 1). Ribosome biogenesis is an essential cellular process whose dysregulation is associated with various human diseases. In cancer, recent studies have begun to reveal novel mechanisms for PARP-1 and ADPRylation that govern the regulation of multiple steps in ribosome biogenesis, largely through regulation of rDNA transcription at rDNA loci. Furthermore, almost every step of mRNA processing—polyadenylation, splicing, and export—is impacted by PARP-1 function. Finally, additional studies have demonstrated a role for various PARP family members, including monoADPR transferases, in the regulation of mRNA translation and protein homeostasis (Fig. 1).
Most prior studies of PARPs and ADPRylation have focused on their roles in DNA damage repair and genome maintenance. Broadening our views of the molecular functions of PARPs and ADPRylation, especially with respect to previously unexplored areas, is an important step in exploiting PARPs to develop preventative and therapeutic approaches to human diseases. For example, the inhibitory effects of some current FDA-approved PARPi on ribosome biogenesis suggests a broader utility of PARPi in cancer treatment and possibly other diseases. Beyond the nuclear PARP polyenzymes, the therapeutic potential of the PARP monoenzymes is largely unexplored. In this regard, our knowledge of the regulation of mRNA translation by MARylation is largely restricted to bacterial toxins and infectious diseases. The regulation of mRNA translation by eukaryotic PARP enzymes and their role in diseases, such as cancer, requires further exploration.
Future studies should also address the molecular, biochemical, and cellular mechanisms of ADPRylation as a regulatory modification, including studies of site-specific ADPRylation of key players in mRNA processing, export, and mRNA translation. Likewise, more studies elucidating the distinct, but interrelated roles of the nuclear PARPs in the regulation of chromatin structure, transcription, and splicing are needed to better understand gene-specific regulation. While the pathogenic role of PARylation in these processes is very well understood in the context of DNA damage and stress, we do not yet fully understand their functions in normal physiological contexts. In addition, further studies are required to understand the role of NAD+-supported, PARP-mediated regulation of protein synthesis and protein quality control under normal and pathological conditions. We have no doubt that future studies of PARPs and ADPRylation will continue to yield exciting new fundamental knowledge, as well as therapeutically relevant discoveries.
Competing interest statement
W.L.K. is a founder, consultant, and member of the scientific advisory board for Ribon Therapeutics, Inc. He is also coholder of U.S. patent 9,599,606 covering a set of ADP-ribose detection reagents, which has been licensed to and is sold by EMD Millipore.
Acknowledgments
We thank members of the Kraus laboratory for critical comments and feedback on this work. The PARP-related research in the Kraus laboratory is supported by a grant from the National Institutes of Health/National Institute of Diabetes and Digestive and Kidney Diseases (R01 DK069710), grants from Cancer Prevention and Research Institute of Texas (RP160319 and RP190236), and funds from the Cecil H. and Ida Green Center for Reproductive Biology Sciences Endowment to W.L.K.
Footnotes
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.334433.119.
References
- Abeti R, Abramov AY, Duchen MR. 2011. β-Amyloid activates PARP causing astrocytic metabolic failure and neuronal death. Brain 134: 1658–1672. 10.1093/brain/awr104 [DOI] [PubMed] [Google Scholar]
- Abraham R, Hauer D, McPherson RL, Utt A, Kirby IT, Cohen MS, Merits A, Leung AKL, Griffin DE. 2018. ADP-ribosyl-binding and hydrolase activities of the alphavirus nsP3 macrodomain are critical for initiation of virus replication. Proc Natl Acad Sci 115: E10457–E10466. 10.1073/pnas.1812130115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilera-Gomez A, van Oorschot MM, Veenendaal T, Rabouille C. 2016. In vivo vizualisation of mono-ADP-ribosylation by dPARP16 upon amino-acid starvation. Elife 5: e21475 10.7554/eLife.21475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed S, Bott D, Gomez A, Tamblyn L, Rasheed A, Cho T, MacPherson L, Sugamori KS, Yang Y, Grant DM, et al. 2015. Loss of the mono-ADP-ribosyltransferase, Tiparp, increases sensitivity to dioxin-induced steatohepatitis and lethality. J Biol Chem 290: 16824–16840. 10.1074/jbc.M115.660100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amé JC, Spenlehauer C, de Murcia G. 2004. The PARP superfamily. BioEssays 26: 882–893. 10.1002/bies.20085 [DOI] [PubMed] [Google Scholar]
- Amit M, Donyo M, Hollander D, Goren A, Kim E, Gelfman S, Lev-Maor G, Burstein D, Schwartz S, Postolsky B, et al. 2012. Differential GC content between exons and introns establishes distinct strategies of splice-site recognition. Cell Rep 1: 543–556. 10.1016/j.celrep.2012.03.013 [DOI] [PubMed] [Google Scholar]
- Aspesi A, Ellis SR. 2019. Rare ribosomopathies: insights into mechanisms of cancer. Nat Rev Cancer 19: 228–238. 10.1038/s41568-019-0105-0 [DOI] [PubMed] [Google Scholar]
- Atasheva S, Frolova EI, Frolov I. 2014. Interferon-stimulated poly(ADP-Ribose) polymerases are potent inhibitors of cellular translation and virus replication. J Virol 88: 2116–2130. 10.1128/JVI.03443-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baßler J, Hurt E. 2019. Eukaryotic ribosome assembly. Annu Rev Biochem 88: 281–306. 10.1146/annurev-biochem-013118-110817 [DOI] [PubMed] [Google Scholar]
- Bhandari D, Guha K, Bhaduri N, Saha P. 2011. Ubiquitination of mRNA cycling sequence binding protein from Leishmania donovani (LdCSBP) modulates the RNA endonuclease activity of its Smr domain. FEBS Lett 585: 809–813. 10.1016/j.febslet.2011.02.007 [DOI] [PubMed] [Google Scholar]
- Bhardwaj A, Yang Y, Ueberheide B, Smith S. 2017. Whole proteome analysis of human tankyrase knockout cells reveals targets of tankyrase-mediated degradation. Nat Commun 8: 2214 10.1038/s41467-017-02363-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bick MJ, Carroll JW, Gao G, Goff SP, Rice CM, MacDonald MR. 2003. Expression of the zinc-finger antiviral protein inhibits alphavirus replication. J Virol 77: 11555–11562. 10.1128/JVI.77.21.11555-11562.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bindesbøll C, Tan S, Bott D, Cho T, Tamblyn L, MacPherson L, Grønning-Wang L, Nebb HI, Matthews J. 2016. TCDD-inducible poly-ADP-ribose polymerase (TIPARP/PARP7) mono-ADP-ribosylates and co-activates liver X receptors. Biochem J 473: 899–910. 10.1042/BJ20151077 [DOI] [PubMed] [Google Scholar]
- Bitler BG, Watson ZL, Wheeler LJ, Behbakht K. 2017. PARP inhibitors: clinical utility and possibilities of overcoming resistance. Gynecol Oncol 147: 695–704. 10.1016/j.ygyno.2017.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boamah EK, Kotova E, Garabedian M, Jarnik M, Tulin AV. 2012. Poly(ADP-Ribose) polymerase 1 (PARP-1) regulates ribosomal biogenesis in Drosophila nucleoli. PLoS Genet 8: e1002442 10.1371/journal.pgen.1002442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock FJ, Todorova TT, Chang P. 2015. RNA regulation by oly(ADP-Ribose) polymerases. Mol Cell 58: 959–969. 10.1016/j.molcel.2015.01.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, Kyle S, Meuth M, Curtin NJ, Helleday T. 2005. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature 434: 913–917. 10.1038/nature03443 [DOI] [PubMed] [Google Scholar]
- Bütepage M, Preisinger C, von Kriegsheim A, Scheufen A, Lausberg E, Li J, Kappes F, Feederle R, Ernst S, Eckei L, et al. 2018. Nucleolar-nucleoplasmic shuttling of TARG1 and its control by DNA damage-induced poly-ADP-ribosylation and by nucleolar transcription. Sci Rep 8: 6748 10.1038/s41598-018-25137-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calkins AS, Iglehart JD, Lazaro JB. 2013. DNA damage-induced inhibition of rRNA synthesis by DNA-PK and PARP-1. Nucleic Acids Res 41: 7378–7386. 10.1093/nar/gkt502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cambronne XA, Stewart ML, Kim D, Jones-Brunette AM, Morgan RK, Farrens DL, Cohen MS, Goodman RH. 2016. Biosensor reveals multiple sources for mitochondrial NAD+. Science 352: 1474–1477. 10.1126/science.aad5168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter-O'Connell I, Jin H, Morgan RK, David LL, Cohen MS. 2014. Engineering the substrate specificity of ADP-ribosyltransferases for identifying direct protein targets. J Am Chem Soc 136: 5201–5204. 10.1021/ja412897a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castello A, Fischer B, Eichelbaum K, Horos R, Beckmann BM, Strein C, Davey NE, Humphreys DT, Preiss T, Steinmetz LM, et al. 2012. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell 149: 1393–1406. 10.1016/j.cell.2012.04.031 [DOI] [PubMed] [Google Scholar]
- Chiou SH, Jiang BH, Yu YL, Chou SJ, Tsai PH, Chang WC, Chen LK, Chen LH, Chien Y, Chiou GY. 2013. Poly(ADP-ribose) polymerase 1 regulates nuclear reprogramming and promotes iPSC generation without c-Myc. J Exp Med 210: 85–98. 10.1084/jem.20121044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen MS, Chang P. 2018. Insights into the biogenesis, function, and regulation of ADP-ribosylation. Nat Chem Biol 14: 236–243. 10.1038/nchembio.2568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danckwardt S, Hentze MW, Kulozik AE. 2008. 3′ end mRNA processing: molecular mechanisms and implications for health and disease. EMBO J 27: 482–498. 10.1038/sj.emboj.7601932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels CM, Ong SE, Leung AK. 2015. The promise of proteomics for the study of ADP-ribosylation. Mol Cell 58: 911–924. 10.1016/j.molcel.2015.06.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davydova EK, Ovchinnikov LP. 1990. ADP-ribosylated elongation factor 2 (ADP-ribosyl-EF-2) is unable to promote translocation within the ribosome. FEBS Lett 261: 350–352. 10.1016/0014-5793(90)80589-B [DOI] [PubMed] [Google Scholar]
- Deng Q, Barbieri JT. 2008. Molecular mechanisms of the cytotoxicity of ADP-ribosylating toxins. Annu Rev Microbiol 62: 271–288. 10.1146/annurev.micro.62.081307.162848 [DOI] [PubMed] [Google Scholar]
- Desnoyers S, Kaufmann SH, Poirier GG. 1996. Alteration of the nucleolar localization of poly(ADP-ribose) polymerase upon treatment with transcription inhibitors. Exp Cell Res 227: 146–153. 10.1006/excr.1996.0259 [DOI] [PubMed] [Google Scholar]
- Di Giammartino DC, Shi Y, Manley JL. 2013. PARP1 represses PAP and inhibits polyadenylation during heat shock. Mol Cell 49: 7–17. 10.1016/j.molcel.2012.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Paola S, Micaroni M, Di Tullio G, Buccione R, Di Girolamo M. 2012. PARP16/ARTD15 is a novel endoplasmic-reticulum-associated mono-ADP-ribosyltransferase that interacts with, and modifies karyopherin-ß1. PLoS One 7: e37352 10.1371/journal.pone.0037352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domagala P, Huzarski T, Lubinski J, Gugala K, Domagala W. 2011. PARP-1 expression in breast cancer including BRCA1-associated, triple negative and basal-like tumors: possible implications for PARP-1 inhibitor therapy. Breast Cancer Res Treat 127: 861–869. 10.1007/s10549-011-1441-2 [DOI] [PubMed] [Google Scholar]
- Duan Y, Du A, Gu J, Duan G, Wang C, Gui X, Ma Z, Qian B, Deng X, Zhang K, et al. 2019. PARylation regulates stress granule dynamics, phase separation, and neurotoxicity of disease-related RNA-binding proteins. Cell Res 29: 233–247. 10.1038/s41422-019-0141-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fakan S, Leduc Y, Lamarre D, Brunet G, Poirier GG. 1988. Immunoelectron microscopical distribution of poly(ADP-ribose)polymerase in the mammalian cell nucleus. Exp Cell Res 179: 517–526. 10.1016/0014-4827(88)90289-3 [DOI] [PubMed] [Google Scholar]
- Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, Santarosa M, Dillon KJ, Hickson I, Knights C, et al. 2005. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 434: 917–921. 10.1038/nature03445 [DOI] [PubMed] [Google Scholar]
- Farrar D, Rai S, Chernukhin I, Jagodic M, Ito Y, Yammine S, Ohlsson R, Murrell A, Klenova E. 2010. Mutational analysis of the poly(ADP-ribosyl)ation sites of the transcription factor CTCF provides an insight into the mechanism of its regulation by poly(ADP-ribosyl)ation. Mol Cell Biol 30: 1199–1216. 10.1128/MCB.00827-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehr AR, Athmer J, Channappanavar R, Phillips JM, Meyerholz DK, Perlman S. 2015. The nsp3 macrodomain promotes virulence in mice with coronavirus-induced encephalitis. J Virol 89: 1523–1536. 10.1128/JVI.02596-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehr AR, Jankevicius G, Ahel I, Perlman S. 2018. Viral Macrodomains: unique mediators of viral replication and pathogenesis. Trends Microbiol 26: 598–610. 10.1016/j.tim.2017.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ficarelli M, Antzin-Anduetza I, Hugh-White R, Firth AE, Sertkaya H, Wilson H, Neil SJD, Schulz R, Swanson CM. 2019. CpG dinucleotides inhibit HIV-1 replication through zinc finger antiviral protein (ZAP)-dependent and -independent mechanisms. J Virol JVI.01337-19 10.1128/JVI.01337-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley BT, Moehring JM, Moehring TJ. 1995. Mutations in the elongation factor 2 gene which confer resistance to diphtheria toxin and Pseudomonas exotoxin A. Genetic and biochemical analyses. J Biol Chem 270: 23218–23225. 10.1074/jbc.270.39.23218 [DOI] [PubMed] [Google Scholar]
- Gagné JP, Pic E, Isabelle M, Krietsch J, Éthier C, Paquet E, Kelly I, Boutin M, Moon KM, Foster LJ, et al. 2012. Quantitative proteomics profiling of the poly(ADP-ribose)-related response to genotoxic stress. Nucleic Acids Res 40: 7788–7805. 10.1093/nar/gks486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geuens T, Bouhy D, Timmerman V. 2016. The hnRNP family: insights into their role in health and disease. Hum Genet 135: 851–867. 10.1007/s00439-016-1683-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson BA, Kraus WL. 2012. New insights into the molecular and cellular functions of poly(ADP-ribose) and PARPs. Nat Rev Mol Cell Biol 13: 411–424. 10.1038/nrm3376 [DOI] [PubMed] [Google Scholar]
- Gibson BA, Zhang Y, Jiang H, Hussey KM, Shrimp JH, Lin H, Schwede F, Yu Y, Kraus WL. 2016. Chemical genetic discovery of PARP targets reveals a role for PARP-1 in transcription elongation. Science 353: 45–50. 10.1126/science.aaf7865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graveley BR. 2000. Sorting out the complexity of SR protein functions. RNA 6: 1197–1211. 10.1017/S1355838200000960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunewald ME, Chen Y, Kuny C, Maejima T, Lease R, Ferraris D, Aikawa M, Sullivan CS, Perlman S, Fehr AR. 2019. The coronavirus macrodomain is required to prevent PARP-mediated inhibition of virus replication and enhancement of IFN expression. PLoS Pathog 15: e1007756 10.1371/journal.ppat.1007756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guetg C, Scheifele F, Rosenthal F, Hottiger MO, Santoro R. 2012. Inheritance of silent rDNA chromatin is mediated by PARP1 via noncoding RNA. Mol Cell 45: 790–800. 10.1016/j.molcel.2012.01.024 [DOI] [PubMed] [Google Scholar]
- Guo X, Carroll JW, Macdonald MR, Goff SP, Gao G. 2004. The zinc finger antiviral protein directly binds to specific viral mRNAs through the CCCH zinc finger motifs. J Virol 78: 12781–12787. 10.1128/JVI.78.23.12781-12787.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Ma J, Sun J, Gao G. 2007. The zinc-finger antiviral protein recruits the RNA processing exosome to degrade the target mRNA. Proc Natl Acad Sci 104: 151–156. 10.1073/pnas.0607063104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupte R, Liu Z, Kraus WL. 2017. PARPs and ADP-ribosylation: recent advances linking molecular functions to biological outcomes. Genes Dev 31: 101–126. 10.1101/gad.291518.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendriks IA, Larsen SC, Nielsen ML. 2019. An advanced strategy for comprehensive profiling of ADP-ribosylation sites using mass spectrometry-based proteomics. Mol Cell Proteomics 18: 1010–1026. 10.1074/mcp.TIR119.001315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hottiger MO. 2015. SnapShot: ADP-ribosylation signaling. Mol Cell 58: 1134–1134.e1. 10.1016/j.molcel.2015.06.001 [DOI] [PubMed] [Google Scholar]
- Hottiger MO, Hassa PO, Lüscher B, Schüler H, Koch-Nolte F. 2010. Toward a unified nomenclature for mammalian ADP-ribosyltransferases. Trends Biochem Sci 35: 208–219. 10.1016/j.tibs.2009.12.003 [DOI] [PubMed] [Google Scholar]
- Hou Y, Lautrup S, Cordonnier S, Wang Y, Croteau DL, Zavala E, Zhang Y, Moritoh K, O'Connell JF, Baptiste BA, et al. 2018. NAD+ supplementation normalizes key Alzheimer's features and DNA damage responses in a new AD mouse model with introduced DNA repair deficiency. Proc Natl Acad Sci 115: E1876–E1885. 10.1073/pnas.1718819115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Steitz JA. 2005. SRprises along a messenger's journey. Mol Cell 17: 613–615. 10.1016/j.molcel.2005.02.020 [DOI] [PubMed] [Google Scholar]
- Iqbal MB, Johns M, Cao J, Liu Y, Yu SC, Hyde GD, Laffan MA, Marchese FP, Cho SH, Clark AR, et al. 2014. PARP-14 combines with tristetraprolin in the selective posttranscriptional control of macrophage tissue factor expression. Blood 124: 3646–3655. 10.1182/blood-2014-07-588046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isabelle M, Moreel X, Gagné JP, Rouleau M, Ethier C, Gagné P, Hendzel MJ, Poirier GG. 2010. Investigation of PARP-1, PARP-2, and PARG interactomes by affinity-purification mass spectrometry. Proteome Sci 8: 22 10.1186/1477-5956-8-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata H, Goettsch C, Sharma A, Ricchiuto P, Goh WW, Halu A, Yamada I, Yoshida H, Hara T, Wei M, et al. 2016. PARP9 and PARP14 cross-regulate macrophage activation via STAT1 ADP-ribosylation. Nat Commun 7: 12849 10.1038/ncomms12849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankevicius G, Ariza A, Ahel M, Ahel I. 2016. The toxin-antitoxin system DarTG catalyzes reversible ADP-ribosylation of DNA. Mol Cell 64: 1109–1116. 10.1016/j.molcel.2016.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayaram HN, Kusumanchi P, Yalowitz JA. 2011. NMNAT expression and its relation to NAD metabolism. Curr Med Chem 18: 1962–1972. 10.2174/092986711795590138 [DOI] [PubMed] [Google Scholar]
- Ji Y, Tulin AV. 2009. Poly(ADP-ribosyl)ation of heterogeneous nuclear ribonucleoproteins modulates splicing. Nucleic Acids Res 37: 3501–3513. 10.1093/nar/gkp218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jørgensen R, Merrill AR, Andersen GR. 2006. The life and death of translation elongation factor 2. Biochem Soc Trans 34: 1–6. 10.1042/BST0340001 [DOI] [PubMed] [Google Scholar]
- Jungmichel S, Rosenthal F, Altmeyer M, Lukas J, Hottiger MO, Nielsen ML. 2013. Proteome-wide identification of poly(ADP-Ribosyl)ation targets in different genotoxic stress responses. Mol Cell 52: 272–285. 10.1016/j.molcel.2013.08.026 [DOI] [PubMed] [Google Scholar]
- Jwa M, Chang P. 2012. PARP16 is a tail-anchored endoplasmic reticulum protein required for the PERK- and IRE1α-mediated unfolded protein response. Nat Cell Biol 14: 1223–1230. 10.1038/ncb2593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kam TI, Mao X, Park H, Chou SC, Karuppagounder SS, Umanah GE, Yun SP, Brahmachari S, Panicker N, Chen R, et al. 2018. Poly(ADP-ribose) drives pathologic α-synuclein neurodegeneration in Parkinson's disease. Science 362: eaat8407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai M, Hanashiro K, Kim SH, Hanai S, Boulares AH, Miwa M, Fukasawa K. 2007. Inhibition of Crm1-p53 interaction and nuclear export of p53 by poly(ADP-ribosyl)ation. Nat Cell Biol 9: 1175–1183. 10.1038/ncb1638 [DOI] [PubMed] [Google Scholar]
- Kapeli K, Martinez FJ, Yeo GW. 2017. Genetic mutations in RNA-binding proteins and their roles in ALS. Hum Genet 136: 1193–1214. 10.1007/s00439-017-1830-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashima L, Idogawa M, Mita H, Shitashige M, Yamada T, Ogi K, Suzuki H, Toyota M, Ariga H, Sasaki Y, et al. 2012. CHFR protein regulates mitotic checkpoint by targeting PARP-1 protein for ubiquitination and degradation. J Biol Chem 287: 12975–12984. 10.1074/jbc.M111.321828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke Y, Han Y, Guo X, Wen J, Wang K, Jiang X, Tian X, Ba X, Boldogh I, Zeng X. 2017. PARP1 promotes gene expression at the post-transcriptional level by modulating the RNA-binding protein HuR. Nat Commun 8: 14632 10.1038/ncomms14632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MY, Zhang T, Kraus WL. 2005. Poly(ADP-ribosyl)ation by PARP-1: ‘PAR-laying’ NAD+ into a nuclear signal. Genes Dev 19: 1951–1967. 10.1101/gad.1331805 [DOI] [PubMed] [Google Scholar]
- Kim DS, Camacho CV, Nagari A, Malladi VS, Challa S, Kraus WL. 2019. Activation of PARP-1 by snoRNAs controls ribosome biogenesis and cell growth via the RNA helicase DDX21. Mol Cell 75: 1270–1285.e14. 10.1016/j.molcel.2019.06.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komander D, Rape M. 2012. The ubiquitin code. Annu Rev Biochem 81: 203–229. 10.1146/annurev-biochem-060310-170328 [DOI] [PubMed] [Google Scholar]
- Kostka G, Schweiger A. 1982. ADP-ribosylation of proteins associated with heterogeneous nuclear RNA in rat liver nuclei. Biochim Biophys Acta 696: 139–144. 10.1016/0167-4781(82)90021-5 [DOI] [PubMed] [Google Scholar]
- Kozaki T, Komano J, Kanbayashi D, Takahama M, Misawa T, Satoh T, Takeuchi O, Kawai T, Shimizu S, Matsuura Y, et al. 2017. Mitochondrial damage elicits a TCDD-inducible poly(ADP-ribose) polymerase-mediated antiviral response. Proc Natl Acad Sci 114: 2681–2686. 10.1073/pnas.1621508114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnakumar R, Kraus WL. 2010. PARP-1 regulates chromatin structure and transcription through a KDM5B-dependent pathway. Mol Cell 39: 736–749. 10.1016/j.molcel.2010.08.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurl RN, Jacob ST. 1985. Characterization of a factor that can prevent random transcription of cloned rDNA and its probable relationship to poly(ADP-ribose) polymerase. Nucleic Acids Res 13: 89–101. 10.1093/nar/13.1.89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang AE, Schmidt G, Schlosser A, Hey TD, Larrinua IM, Sheets JJ, Mannherz HG, Aktories K. 2010. Photorhabdus luminescens toxins ADP-ribosylate actin and RhoA to force actin clustering. Science 327: 1139–1142. 10.1126/science.1184557 [DOI] [PubMed] [Google Scholar]
- Lang AE, Qu Z, Schwan C, Silvan U, Unger A, Schoenenberger CA, Aktories K, Mannherz HG. 2017. Actin ADP-ribosylation at Threonine148 by Photorhabdus luminescens toxin TccC3 induces aggregation of intracellular F-actin. Cell Microbiol 19: e12636. [DOI] [PubMed] [Google Scholar]
- Lau C, Niere M, Ziegler M. 2009. The NMN/NaMN adenylyltransferase (NMNAT) protein family. Front Biosci (Landmark Ed) 14: 410–431. 10.2741/3252 [DOI] [PubMed] [Google Scholar]
- Léger K, Bär D, Savić N, Santoro R, Hottiger MO. 2014. ARTD2 activity is stimulated by RNA. Nucleic Acids Res 42: 5072–5082. 10.1093/nar/gku131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung AK. 2014. Poly(ADP-ribose): an organizer of cellular architecture. J Cell Biol 205: 613–619. 10.1083/jcb.201402114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung AKL. 2017. PARPs. Curr Biol 27: R1256–R1258. 10.1016/j.cub.2017.09.054 [DOI] [PubMed] [Google Scholar]
- Leung AK, Vyas S, Rood JE, Bhutkar A, Sharp PA, Chang P. 2011. Poly(ADP-ribose) regulates stress responses and microRNA activity in the cytoplasm. Mol Cell 42: 489–499. 10.1016/j.molcel.2011.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Santoro R, Koberna K, Grummt I. 2005. The chromatin remodeling complex NoRC controls replication timing of rRNA genes. EMBO J 24: 120–127. 10.1038/sj.emboj.7600492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Debing Y, Jankevicius G, Neyts J, Ahel I, Coutard B, Canard B. 2016. Viral macro domains reverse protein ADP-ribosylation. J Virol 90: 8478–8486. 10.1128/JVI.00705-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Bachran C, Gupta P, Miller-Randolph S, Wang H, Crown D, Zhang Y, Wein AN, Singh R, Fattah R, et al. 2012. Diphthamide modification on eukaryotic elongation factor 2 is needed to assure fidelity of mRNA translation and mouse development. Proc Natl Acad Sci 109: 13817–13822. 10.1073/pnas.1206933109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love S, Barber R, Wilcock GK. 1999. Increased poly(ADP-ribosyl)ation of nuclear proteins in Alzheimer's disease. Brain 122(Pt 2): 247–253. 10.1093/brain/122.2.247 [DOI] [PubMed] [Google Scholar]
- Luo X, Kraus WL. 2012. On PAR with PARP: cellular stress signaling through poly(ADP-ribose) and PARP-1. Genes Dev 26: 417–432. 10.1101/gad.183509.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X, Ryu KW, Kim DS, Nandu T, Medina CJ, Gupte R, Gibson BA, Soccio RE, Yu Y, Gupta RK, et al. 2017. PARP-1 controls the adipogenic transcriptional program by PARylating C/EBPβ and modulating its transcriptional activity. Mol Cell 65: 260–271. 10.1016/j.molcel.2016.11.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malanga M, Czubaty A, Girstun A, Staron K, Althaus FR. 2008. Poly(ADP-ribose) binds to the splicing factor ASF/SF2 and regulates its phosphorylation by DNA topoisomerase I. J Biol Chem 283: 19991–19998. 10.1074/jbc.M709495200 [DOI] [PubMed] [Google Scholar]
- Mani A, Gelmann EP. 2005. The ubiquitin-proteasome pathway and its role in cancer. J Clin Oncol 23: 4776–4789. 10.1200/JCO.2005.05.081 [DOI] [PubMed] [Google Scholar]
- Manning DR, Fraser BA, Kahn RA, Gilman AG. 1984. ADP-ribosylation of transducin by islet-activation protein. Identification of asparagine as the site of ADP-ribosylation. J Biol Chem 259: 749–756. [PubMed] [Google Scholar]
- Matter N, Herrlich P, König H. 2002. Signal-dependent regulation of splicing via phosphorylation of Sam68. Nature 420: 691–695. 10.1038/nature01153 [DOI] [PubMed] [Google Scholar]
- Matveeva E, Maiorano J, Zhang Q, Eteleeb AM, Convertini P, Chen J, Infantino V, Stamm S, Wang J, Rouchka EC, et al. 2016. Involvement of PARP1 in the regulation of alternative splicing. Cell Discov 2: 15046 10.1038/celldisc.2015.46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matveeva EA, Al-Tinawi QMH, Rouchka EC, Fondufe-Mittendorf YN. 2019. Coupling of PARP1-mediated chromatin structural changes to transcriptional RNA polymerase II elongation and cotranscriptional splicing. Epigenetics Chromatin 12: 15 10.1186/s13072-019-0261-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCann KE. 2019. Advances in the use of PARP inhibitors for BRCA1/2-associated breast cancer: talazoparib. Future Oncol 15: 1707–1715. 10.2217/fon-2018-0751 [DOI] [PubMed] [Google Scholar]
- McGurk L, Gomes E, Guo L, Mojsilovic-Petrovic J, Tran V, Kalb RG, Shorter J, Bonini NM. 2018. Poly(ADP-Ribose) prevents pathological phase separation of TDP-43 by promoting liquid demixing and stress granule localization. Mol Cell 71: 703–717.e9. 10.1016/j.molcel.2018.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPherson RL, Abraham R, Sreekumar E, Ong SE, Cheng SJ, Baxter VK, Kistemaker HA, Filippov DV, Griffin DE, Leung AK. 2017. ADP-ribosylhydrolase activity of chikungunya virus macrodomain is critical for virus replication and virulence. Proc Natl Acad Sci 114: 1666–1671. 10.1073/pnas.1621485114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meagher JL, Takata M, Gonçalves-Carneiro D, Keane SC, Rebendenne A, Ong H, Orr VK, MacDonald MR, Stuckey JA, Bieniasz PD, et al. 2019. Structure of the zinc-finger antiviral protein in complex with RNA reveals a mechanism for selective targeting of CG-rich viral sequences. Proc Natl Acad Sci 116: 24303–24309. 10.1073/pnas.1913232116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meder VS, Boeglin M, de Murcia G, Schreiber V. 2005. PARP-1 and PARP-2 interact with nucleophosmin/B23 and accumulate in transcriptionally active nucleoli. J Cell Sci 118: 211–222. 10.1242/jcs.01606 [DOI] [PubMed] [Google Scholar]
- Melikishvili M, Chariker JH, Rouchka EC, Fondufe-Mittendorf YN. 2017. Transcriptome-wide identification of the RNA-binding landscape of the chromatin-associated protein PARP1 reveals functions in RNA biogenesis. Cell Discov 3: 17043 10.1038/celldisc.2017.43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirza MR, Monk BJ, Herrstedt J, Oza AM, Mahner S, Redondo A, Fabbro M, Ledermann JA, Lorusso D, Vergote I, et al. 2016. Niraparib maintenance therapy in platinum-sensitive, recurrent ovarian cancer. N Engl J Med 375: 2154–2164. 10.1056/NEJMoa1611310 [DOI] [PubMed] [Google Scholar]
- Morales J, Li L, Fattah FJ, Dong Y, Bey EA, Patel M, Gao J, Boothman DA. 2014. Review of poly (ADP-ribose) polymerase (PARP) mechanisms of action and rationale for targeting in cancer and other diseases. Crit Rev Eukaryot Gene Expr 24: 15–28. 10.1615/CritRevEukaryotGeneExpr.2013006875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munir A, Banerjee A, Shuman S. 2018. NAD+-dependent synthesis of a 5′-phospho-ADP-ribosylated RNA/DNA cap by RNA 2′-phosphotransferase Tpt1. Nucleic Acids Res 46: 9617–9624. 10.1093/nar/gky792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munnur D, Ahel I. 2017. Reversible mono-ADP-ribosylation of DNA breaks. FEBS J 284: 4002–4016. 10.1111/febs.14297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munnur D, Bartlett E, Mikolčevič P, Kirby IT, Matthias Rack JG, Mikoč A, Cohen MS, Ahel I. 2019. Reversible ADP-ribosylation of RNA. Nucleic Acids Res 47: 5658–5669. 10.1093/nar/gkz305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nygård O, Nilsson L. 1985. Reduced ribosomal binding of eukaryotic elongation factor 2 following ADP-ribosylation. Difference in binding selectivity between polyribosomes and reconstituted monoribosomes. Biochim Biophys Acta 824: 152–162. 10.1016/0167-4781(85)90092-2 [DOI] [PubMed] [Google Scholar]
- Ocampo A, Liu J, Barrientos A. 2013. NAD+ salvage pathway proteins suppress proteotoxicity in yeast models of neurodegeneration by promoting the clearance of misfolded/oligomerized proteins. Hum Mol Genet 22: 1699–1708. 10.1093/hmg/ddt016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olabisi OA, Soto-Nieves N, Nieves E, Yang TT, Yang X, Yu RY, Suk HY, Macian F, Chow CW. 2008. Regulation of transcription factor NFAT by ADP-ribosylation. Mol Cell Biol 28: 2860–2871. 10.1128/MCB.01746-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oppenheimer NJ, Bodley JW. 1981. Diphtheria toxin. Site and configuration of ADP-ribosylation of diphthamide in elongation factor 2. J Biol Chem 256: 8579–8581. [PubMed] [Google Scholar]
- Ossovskaya V, Koo IC, Kaldjian EP, Alvares C, Sherman BM. 2010. Upregulation of poly (ADP-ribose) polymerase-1 (PARP1) in triple-negative breast cancer and other primary human tumor types. Genes Cancer 1: 812–821. 10.1177/1947601910383418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad S, Walent J, Dritschilo A. 1994. ADP-ribosylation of heterogeneous ribonucleoproteins in HeLa cells. Biochem Biophys Res Commun 204: 772–779. 10.1006/bbrc.1994.2526 [DOI] [PubMed] [Google Scholar]
- Qiu J, Sheedlo MJ, Yu K, Tan Y, Nakayasu ES, Das C, Liu X, Luo ZQ. 2016. Ubiquitination independent of E1 and E2 enzymes by bacterial effectors. Nature 533: 120–124. 10.1038/nature17657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajman L, Chwalek K, Sinclair DA. 2018. Therapeutic potential of NAD-boosting molecules: the in vivo evidence. Cell Metab 27: 529–547. 10.1016/j.cmet.2018.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray Chaudhuri A, Nussenzweig A. 2017. The multifaceted roles of PARP1 in DNA repair and chromatin remodelling. Nat Reviews Mol Cell Biol 18: 610–621. 10.1038/nrm.2017.53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robson M, Im SA, Senkus E, Xu B, Domchek SM, Masuda N, Delaloge S, Li W, Tung N, Armstrong A, et al. 2017. Olaparib for metastatic breast cancer in patients with a germline BRCA mutation. N Engl J Med 377: 523–533. 10.1056/NEJMoa1706450 [DOI] [PubMed] [Google Scholar]
- Roper SJ, Chrysanthou S, Senner CE, Sienerth A, Gnan S, Murray A, Masutani M, Latos P, Hemberger M. 2014. ADP-ribosyltransferases Parp1 and Parp7 safeguard pluripotency of ES cells. Nucleic Acids Res 42: 8914–8927. 10.1093/nar/gku591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross CA, Poirier MA. 2005. Opinion: what is the role of protein aggregation in neurodegeneration? Nat Rev Mol Cell Biol 6: 891–898. 10.1038/nrm1742 [DOI] [PubMed] [Google Scholar]
- Ruggero D, Pandolfi PP. 2003. Does the ribosome translate cancer? Nat Rev Cancer 3: 179–192. 10.1038/nrc1015 [DOI] [PubMed] [Google Scholar]
- Ryu KW, Kim DS, Kraus WL. 2015. New facets in the regulation of gene expression by ADP-ribosylation and poly(ADP-ribose) polymerases. Chem Rev 115: 2453–2481. 10.1021/cr5004248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu KW, Nandu T, Kim J, Challa S, DeBerardinis RJ, Kraus WL. 2018. Metabolic regulation of transcription through compartmentalized NAD+ biosynthesis. Science 360: eaan5780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro R, Li J, Grummt I. 2002. The nucleolar remodeling complex NoRC mediates heterochromatin formation and silencing of ribosomal gene transcription. Nat Genet 32: 393–396. 10.1038/ng1010 [DOI] [PubMed] [Google Scholar]
- Scherl A, Couté Y, Déon C, Callé A, Kindbeiter K, Sanchez JC, Greco A, Hochstrasser D, Diaz JJ. 2002. Functional proteomic analysis of human nucleolus. Mol Biol Cell 13: 4100–4109. 10.1091/mbc.e02-05-0271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schöndorf DC, Ivanyuk D, Baden P, Sanchez-Martinez A, De Cicco S, Yu C, Giunta I, Schwarz LK, Di Napoli G, Panagiotakopoulou V, et al. 2018. The NAD+ precursor nicotinamide riboside rescues mitochondrial defects and neuronal loss in iPSC and fly models of Parkinson's disease. Cell Rep 23: 2976–2988. 10.1016/j.celrep.2018.05.009 [DOI] [PubMed] [Google Scholar]
- Schreiber V, Dantzer F, Ame JC, de Murcia G. 2006. Poly(ADP-ribose): novel functions for an old molecule. Nat Rev Mol Cell Biol 7: 517–528. 10.1038/nrm1963 [DOI] [PubMed] [Google Scholar]
- Schwartz S, Ast G. 2010. Chromatin density and splicing destiny: on the cross-talk between chromatin structure and splicing. EMBO J 29: 1629–1636. 10.1038/emboj.2010.71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwerk J, Soveg FW, Ryan AP, Thomas KR, Hatfield LD, Ozarkar S, Forero A, Kell AM, Roby JA, So L, et al. 2019. RNA-binding protein isoforms ZAP-S and ZAP-L have distinct antiviral and immune resolution functions. Nat Immunol 20: 1610–1620. 10.1038/s41590-019-0527-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shacham T, Sharma N, Lederkremer GZ. 2019. Protein misfolding and ER stress in Huntington's disease. Front Mol Biosci 6: 20 10.3389/fmolb.2019.00020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao C, Qiu Y, Liu J, Feng H, Shen S, Saiyin H, Yu W, Wei Y, Yu L, Su W, et al. 2018. PARP12 (ARTD12) suppresses hepatocellular carcinoma metastasis through interacting with FHL2 and regulating its stability. Cell Death Dis 9: 856 10.1038/s41419-018-0906-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw AE, Hughes J, Gu Q, Behdenna A, Singer JB, Dennis T, Orton RJ, Varela M, Gifford RJ, Wilson SJ, et al. 2017. Fundamental properties of the mammalian innate immune system revealed by multispecies comparison of type I interferon responses. PLoS Biol 15: e2004086 10.1371/journal.pbio.2004086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Di Giammartino DC, Taylor D, Sarkeshik A, Rice WJ, Yates JR 3rd, Frank J, Manley JL. 2009. Molecular architecture of the human pre-mRNA 3′ processing complex. Mol Cell 33: 365–376. 10.1016/j.molcel.2008.12.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon NC, Aktories K, Barbieri JT. 2014. Novel bacterial ADP-ribosylating toxins: structure and function. Nat Rev Microbiol 12: 599–611. 10.1038/nrmicro3310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorrentino V, Romani M, Mouchiroud L, Beck JS, Zhang H, D'Amico D, Moullan N, Potenza F, Schmid AW, Rietsch S, et al. 2017. Enhancing mitochondrial proteostasis reduces amyloid-β proteotoxicity. Nature 552: 187–193. 10.1038/nature25143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl S, da Silva Mateus Seidl AR, Ducret A, Kux van Geijtenbeek S, Michel S, Racek T, Birzele F, Haas AK, Rueger R, Gerg M, et al. 2015. Loss of diphthamide pre-activates NF-κB and death receptor pathways and renders MCF7 cells hypersensitive to tumor necrosis factor. Proc Natl Acad Sci 112: 10732–10737. 10.1073/pnas.1512863112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susorov D, Zakharov N, Shuvalova E, Ivanov A, Egorova T, Shuvalov A, Shatsky IN, Alkalaeva E. 2018. Eukaryotic translation elongation factor 2 (eEF2) catalyzes reverse translocation of the eukaryotic ribosome. J Biol Chem 293: 5220–5229. 10.1074/jbc.RA117.000761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takamura-Enya T, Watanabe M, Totsuka Y, Kanazawa T, Matsushima-Hibiya Y, Koyama K, Sugimura T, Wakabayashi K. 2001. Mono(ADP-ribosyl)ation of 2′-deoxyguanosine residue in DNA by an apoptosis-inducing protein, pierisin-1, from cabbage butterfly. Proc Natl Acad Sci 98: 12414–12419. 10.1073/pnas.221444598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talhaoui I, Lebedeva NA, Zarkovic G, Saint-Pierre C, Kutuzov MM, Sukhanova MV, Matkarimov BT, Gasparutto D, Saparbaev MK, Lavrik OI, et al. 2016. Poly(ADP-ribose) polymerases covalently modify strand break termini in DNA fragments in vitro. Nucleic Acids Res 44: 9279–9295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todorova T, Bock FJ, Chang P. 2014. PARP13 regulates cellular mRNA post-transcriptionally and functions as a pro-apoptotic factor by destabilizing TRAILR4 transcript. Nat Commun 5: 5362 10.1038/ncomms6362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandekerckhove J, Schering B, Bärmann M, Aktories K. 1987. Clostridium perfringens iota toxin ADP-ribosylates skeletal muscle actin in Arg-177. FEBS Lett 225: 48–52. 10.1016/0014-5793(87)81129-8 [DOI] [PubMed] [Google Scholar]
- Van Ness BG, Howard JB, Bodley JW. 1980. ADP-ribosylation of elongation factor 2 by diphtheria toxin. Isolation and properties of the novel ribosyl-amino acid and its hydrolysis products. J Biol Chem 255: 10717–10720. [PubMed] [Google Scholar]
- Verdin E. 2015. NAD+ in aging, metabolism, and neurodegeneration. Science 350: 1208–1213. 10.1126/science.aac4854 [DOI] [PubMed] [Google Scholar]
- Vyas S, Chang P. 2014. New PARP targets for cancer therapy. Nat Rev Cancer 14: 502–509. 10.1038/nrc3748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vyas S, Chesarone-Cataldo M, Todorova T, Huang YH, Chang P. 2013. A systematic analysis of the PARP protein family identifies new functions critical for cell physiology. Nat Commun 4: 2240 10.1038/ncomms3240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vyas S, Matic I, Uchima L, Rood J, Zaja R, Hay RT, Ahel I, Chang P. 2014. Family-wide analysis of poly(ADP-ribose) polymerase activity. Nat Commun 5: 4426 10.1038/ncomms5426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N, Dong Q, Li J, Jangra RK, Fan M, Brasier AR, Lemon SM, Pfeffer LM, Li K. 2010. Viral induction of the zinc finger antiviral protein is IRF3-dependent but NF-κB-independent. J Biol Chem 285: 6080–6090. 10.1074/jbc.M109.054486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsby I, Hutin D, Gueydan C, Kruys V, Rongvaux A, Leo O. 2014. PARP12, an interferon-stimulated gene involved in the control of protein translation and inflammation. J Biol Chem 289: 26642–26657. 10.1074/jbc.M114.589515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West RE Jr, Moss J, Vaughan M, Liu T, Liu TY. 1985. Pertussis toxin-catalyzed ADP-ribosylation of transducin. Cysteine 347 is the ADP-ribose acceptor site. J Biol Chem 260: 14428–14430. [PubMed] [Google Scholar]
- Yu W, Ginjala V, Pant V, Chernukhin I, Whitehead J, Docquier F, Farrar D, Tavoosidana G, Mukhopadhyay R, Kanduri C, et al. 2004. Poly(ADP-ribosyl)ation regulates CTCF-dependent chromatin insulation. Nat Genet 36: 1105–1110. 10.1038/ng1426 [DOI] [PubMed] [Google Scholar]
- Zarkovic G, Belousova EA, Talhaoui I, Saint-Pierre C, Kutuzov MM, Matkarimov BT, Biard D, Gasparutto D, Lavrik OI, Ishchenko AA. 2018. Characterization of DNA ADP-ribosyltransferase activities of PARP2 and PARP3: new insights into DNA ADP-ribosylation. Nucleic Acids Res 46: 2417–2431. 10.1093/nar/gkx1318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng J, Libien J, Shaik F, Wolk J, Hernández AI. 2016. Nucleolar PARP-1 expression is decreased in Alzheimer's disease: consequences for epigenetic regulation of rDNA and cognition. Neural Plast 2016: 8987928 10.1155/2016/8987928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu S, Mickanin C, Feng Y, Charlat O, Michaud GA, Schirle M, Shi X, Hild M, Bauer A, et al. 2011. RNF146 is a poly(ADP-ribose)-directed E3 ligase that regulates axin degradation and Wnt signalling. Nat Cell Biol 13: 623–629. 10.1038/ncb2222 [DOI] [PubMed] [Google Scholar]
- Zhang T, Berrocal JG, Yao J, DuMond ME, Krishnakumar R, Ruhl DD, Ryu KW, Gamble MJ, Kraus WL. 2012. Regulation of poly(ADP-ribose) polymerase-1-dependent gene expression through promoter-directed recruitment of a nuclear NAD+ synthase. J Biol Chem 287: 12405–12416. 10.1074/jbc.M111.304469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wang J, Ding M, Yu Y. 2013. Site-specific characterization of the Asp- and Glu-ADP-ribosylated proteome. Nat Methods 10: 981–984. 10.1038/nmeth.2603 [DOI] [PubMed] [Google Scholar]
- Zhen Y, Zhang Y, Yu Y. 2017. A cell-line-specific atlas of PARP-mediated protein Asp/Glu-ADP-ribosylation in breast cancer. Cell Rep 21: 2326–2337. 10.1016/j.celrep.2017.10.106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Chen G, Lv F, Wang X, Ji X, Xu Y, Sun J, Wu L, Zheng YT, Gao G. 2011. Zinc-finger antiviral protein inhibits HIV-1 infection by selectively targeting multiply spliced viral mRNAs for degradation. Proc Natl Acad Sci 108: 15834–15839. 10.1073/pnas.1101676108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubiaga AM, Belasco JG, Greenberg ME. 1995. The nonamer UUAUUUAUU is the key AU-rich sequence motif that mediates mRNA degradation. Mol Cell Biol 15: 2219–2230. 10.1128/MCB.15.4.2219 [DOI] [PMC free article] [PubMed] [Google Scholar]


