Abstract
DNA polymerases are vital for the synthesis of new DNA strands. Since the discovery of DNA polymerase I in Escherichia coli, a diverse library of mammalian DNA polymerases involved in DNA replication, DNA repair, antibody generation, and cell checkpoint signaling has emerged. While the unique functions of these DNA polymerases are differentiated by their association with accessory factors and/or the presence of distinctive catalytic domains, atomic resolution structures of DNA polymerases in complex with their DNA substrates have revealed mechanistic subtleties that contribute to their specialization. In this review, the structure and function of all 15 mammalian DNA polymerases from families B, Y, X, and A will be reviewed and discussed with special emphasis on the insights gleaned from recently published atomic resolution structures.
Keywords: DNA polymerase, Replication, DNA repair, Structural biology, DNA synthesis
Introduction
Without a way to replicate the genome, life as we know it would not be possible. DNA polymerases have evolved to fill this role, all of which perform a similar chemical reaction: the addition of a nucleotide monophosphate (dNMP) onto the end of a DNA primer sequence [1, 2]. As novel DNA polymerases are discovered and characterized, this nucleotidyl transferase reaction of DNA polymerization has been revealed to occur in diverse cellular situations aside from genome replication. These situations include the replication of mitochondrial DNA, synthesis from diverse substrates generated by DNA damage and during DNA repair, and error-rich DNA polymerization to provide antibody diversification [3, 4]. This cellular need for diverse types of DNA synthesis has led to the evolution of many specialized polymerases. Currently, 15 independent mammalian DNA polymerases have been characterized, each of which fulfills a specific biological role (Table 1) [3]. Based on sequence homology, mammalian DNA polymerases can be classified into four families: A, B, X, and Y [5, 6]. Generally, A-family DNA polymerases are involved in DNA lesion bypass and mitochondrial DNA replication and repair, B-family DNA polymerases are largely involved in genomic DNA replication, X-family DNA polymerases are involved in DNA repair and V(D)J recombination, and Y-family DNA polymerases are mainly involved in lesion bypass [7]. Each mammalian DNA polymerase will be discussed in detail in this review, with special emphasis given to the structural adaptations used to fill its biological niche.
Table 1.
Selected features of human DNA polymerases [4, 7, 40–42, 64, 67, 83, 84, 95, 114, 121, 135, 136, 259, 282, 288]
| Family | Pol | Catalytic subunit mass(kDa) | Structure without DNAa | DNA bound structurea | Fidelityb | Additional subunits | Biological pathwayc |
|---|---|---|---|---|---|---|---|
| B | Alpha (α) | 165 | 4B08 | 4Q5V, 5IUD | 10−4 | Subunit B, primase large, and primase small subunit | DNA replication |
| Delta (δ) | 100 | 3IAY | 10−5 | PolD2, PolD3, and PolD4 | DNA replication | ||
| Epsilon (ε) | 225 | 4M8O, 4PTF | 10−5 | p59, p12, and p17 | DNA replication | ||
| Zeta (ζ) | 353 | 3ABD | 10−3 f | Rev7, PolD2, and PolD3 | TLS | ||
| Y | Eta (η) | 78 | 3MFH | 10−2 | Monomer | TLS | |
| Iota (ι) | 80 | 1T3N, 2FLN, 4EYH | 10−1 | Monomer | TLS | ||
| Kappa (κ) | 76 | 1T94 | 2OH2, 4U6P, 6CST | 10−3 | Monomer | TLS | |
| REV1 | 138 | 2AQ4, 5WM8, 3BJY | N.A.d | Monomer | TLS | ||
| X | Beta (β) | 39 | 1BPB, 1RPL | 2BPG, 2FMS, 31SB | 10−4 | Monomer | BER |
| Lambda (λ) | 66 | 5CB1 | 1RZT, 2BCQ | 10−3 | Monomer | BER, NHEJ, TLS | |
| Mu (μ) | 55 | 4LZD | 2IHM, 4LZG | 10−3 | Monomer | NHEJ, V(D)J | |
| TdT | 56 | 1JMS | 4I27 | N.D.e | Monomer | V(D)J recombination | |
| A | Gamma (γ) | 140 | 3IKM | 4ZTZ, 5C53 | 10−5 | Two copies of subunit 2 | Mitochondrial DNA replication and repair |
| Theta (θ) | 290 | 4X0P | 10−3 | Monomer | TLS, MMEJ | ||
| Nu (v) | 100 | 4XVK | 10−3 | Monomer | ICL repair |
aRepresentative crystal structures discussed in this review
bApproximate error rate for single-base substitutions
cThis column focuses on the primary biological pathway(s) of each polymerase; others may also apply
dFidelity measurements are not applicable to REV1
eNo data could be found for the fidelity of TdT’s templated activity, although it is likely similar to other X family polymerases
fMeasurement from yeast homolog
The first detailed information regarding key amino acids necessary for DNA polymerase activity became available in the mid-1980s through structural studies of Escherichia coli DNA polymerase I [8]. Further advances, such as the crystallization of polymerases bound to oligonucleotides and time-lapse X-ray crystallography, have provided insight into DNA polymerase function in context of their DNA substrates [9]. As additional structures were published, including proteins from the Eukarya domain, it became increasingly clear that the catalytic subunits of these enzymes exhibit a common overall architecture [8, 10, 11]. This architecture has been likened to the structure of a human right hand, containing a palm, fingers, and thumb domain [12, 13]. When complexed with a DNA substrate, this hand can be envisioned to have DNA threaded through it, with the fingers binding the incoming nucleotide and coordinating the single-stranded template. The palm contains the catalytic core, which interacts with both the incoming nucleotide and dsDNA, while the thumb “grips” the dsDNA upstream of the polymerization site [13]. Many DNA polymerases exhibit global conformational changes upon the binding of a nucleotide, moving from an open to a closed state prior to catalysis. Continuing the hand analogy, this is accomplished by a shift in the fingers domain, in the same manner that a hand would transition from an open to closed position upon grabbing a rope [14–16].
Because the catalytic subunit of each DNA polymerase exhibits a similar structure, it should not be surprising that the chemical reaction that occurs is nearly identical in the case of each polymerase. In this reaction, DNA polymerases add a free dNMP to the end of a primer, creating a phosphodiester linkage [1, 2]. This nucleotide addition utilizes the 3ʹ hydroxyl group (3ʹ-OH) at the end of the primer as a nucleophile in an SN2 reaction, attacking the α phosphorous of the phosphate group of the incoming nucleotide triphosphate (dNTP) to form an incorporated nucleotide monophosphate and free pyrophosphate [2]. The conservation of this reaction between the mammalian DNA polymerases manifests itself in a generally universal mechanism. First, the DNA polymerase binds its DNA substrate, which usually consists of either gapped double-stranded DNA (dsDNA), or single-stranded DNA (ssDNA) with an annealed primer; in either case, the polymerase must access an exposed 3ʹ-OH. Then, a dNTP binds the complex (which is canonically complementary to the templating base along its Watson–Crick face), where it is poised for catalysis. Finally, catalysis occurs, during which the 3ʹ-OH is activated by a divalent cation (e.g., Mg2+) to form a phosphodiester bond between the primer terminus and the incoming dNMP, allowing the resultant pyrophosphate product to be released. Catalysis involves at least two divalent metals. A third metal has been observed in numerous crystallographic studies and the exact mechanistic role of the third metal ion remains an active area of research [17, 18]. DNA polymerase catalysis was visualized with pol β, η, and µ to study DNA synthesis via time-lapse X-ray crystallography, traversing from the ground state to the product state to observe phosphodiester bond formation [17, 19, 20]. As the number of published atomic resolution structures of DNA polymerase complexes increases, subtle, but still vitally important, structural specializations have been observed. Currently, all 15 mammalian DNA polymerases have published atomic-level structural information; the way these structures dictate their specialized functions will serve as a focal point of this review. See Table 1 for a summary of these DNA polymerases, their functions, and the families to which they belong.
B-family
During genomic replication, the six billion nucleotides that compose the human genome must be copied in a short amount of time, while minimizing the introduction of damaged and mismatched nucleotides [21]. Depending on their locations, erroneous base insertions may cause devastating cellular outcomes, including cell death, senescence, or carcinogenesis [22]. The monumental task of replicating the genome is performed by the B-family polymerases: DNA polymerases alpha (pol α), delta (pol δ), and epsilon (pol ε) (Fig. 1a). DNA polymerase zeta (pol ζ) is also a member of this family based on sequence homology, but is not responsible for bulk replication of the genome. Pol α acts as the initiator of DNA synthesis, using two separate active sites for dual activities, with its primase activity creating RNA primers on ssDNA and its polymerase activity extending them [23]. Pol δ and pol ε further extend those DNA primers, performing the bulk of DNA synthesis, and while controversial, many lines of evidence support pol ε replicating the leading strand and pol δ on the lagging strand [24]. Pol ζ, although homologous to the other members in the family, is not part of the traditional replisome, but assists when replication is stalled by DNA damage, such as during translesion synthesis, crosslink repair, and some forms of homologous recombination [25]. To ensure accurate replication of the genome, B-family polymerases have evolved to exhibit some of the highest levels of both processivity (the number of nucleotides inserted before the polymerase disassociates) and fidelity (the selection of the correct nucleotide to insert) [26]. The presence of a 3ʹ to 5ʹ exonuclease activity for proofreading improves fidelity by a factor of up to 100, shifting the error rate from around one misinsertion per 105 insertions to one misinsertion per 107 insertion events in pol δ and pol ε [27, 28]. The high processivity of B-family polymerases can be facilitated by interactions with proliferating cell nuclear antigen (PCNA), which acts like a clamp to keep the polymerases on the DNA [29, 30]. Working in concert, B-family polymerases compose a vital part of the DNA replication complex.
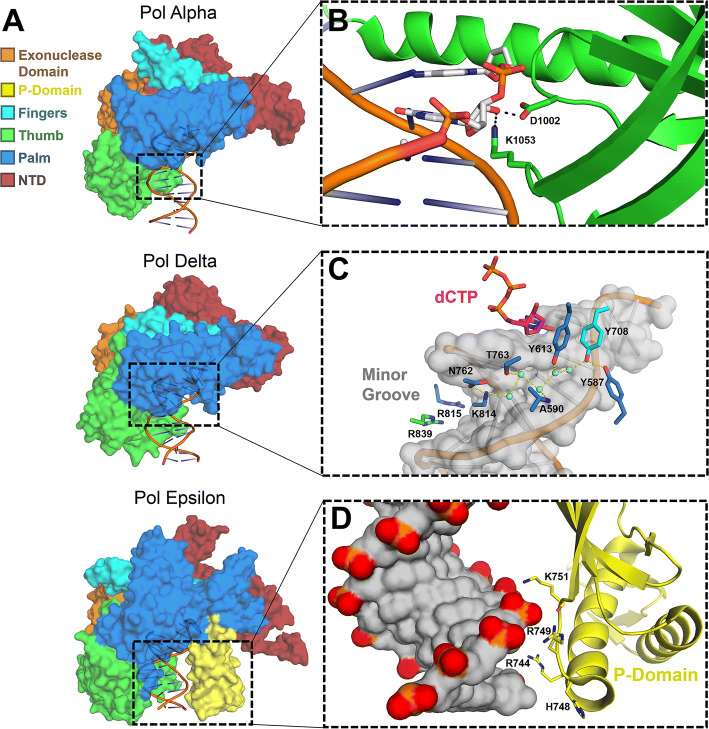
Fig. 1.
Overall structure and specific features of replicative B-family polymerases. a Surface representations of human pol α (5IUD), yeast pol δ (3IAY) and yeast pol ɛ (4M8O) are shown with their exonuclease (orange), P-domain (yellow), fingers (cyan), thumb (green), palm (blue), and N-terminal (red) domains highlighted. b A close-up view of the pol α thumb domain (green) making contact with the 2′-OH groups of the RNA of its DNA/RNA hybrid substrate (white sticks), the product of its primase activity (4Q5V). c The minor groove of nascent DNA probed by Pol δ through direct contacts (blue sticks), as well as through coordinated water-bridges (cyan spheres; 3IAY). d The P-domain (yellow), found only in Pol ɛ, provides additional contact to the nascent DNA (gray surface; 4M8O)
DNA polymerase alpha (pol α)
The B-family member DNA polymerase alpha (pol α) is involved in both DNA replication and cell-cycle regulation [31]. As the Greek letter α implies, pol α was one of the first DNA polymerases discovered and characterized [32]. It plays the crucial role of creating and extending primers at the beginning of genomic replication; therefore, pol α is tightly synchronized with the progression of the cell-cycle [33]. Pol α is made up of four subunits, the catalytic subunit, subunit B, DNA primase small subunit, and DNA primase large subunit (also named p180, p70, p49, and p58, respectively) [34]. Subunit B is a regulatory subunit, and the primase subunits together coordinate primase activity [35, 36]. Furthermore, solution studies of the primase subunits showed that both the large and small primase subunits are required for the primase activity [37]. Compared to other DNA polymerases, the catalytic subunit of pol α exhibits a moderate fidelity of nucleotide insertion, with an error rate of one base misinsertion per 1000 insertion events [7]. Although pol α contains an exonuclease domain, no proofreading activity has been observed; errors must, therefore, be corrected by other repair systems (Fig. 1a) [26, 38, 39].
The atomic resolution structure of the pol α catalytic subunit was initially obtained from a yeast homolog, which provided some of the first mechanistic insight [40]. Structures are now available of the human pol α catalytic subunit in multiple physiologically relevant ligand-bound states, including bound to a DNA:RNA hybrid and dsDNA helix substrates, which would be involved in DNA priming and DNA synthesis, respectively. Interestingly, the DNA in contact with pol α in the pol α:DNA complex structure is not in B form, but rather a hybrid A-B form, suggesting DNA sculpting by the enzyme during its polymerase activity [41]. Additionally, a structure of the catalytic subunit bound to its DNA:RNA substrate demonstrated that pol α directly coordinates certain 2ʹ-OH groups of the RNA primer, a unique substrate for pol α (Fig. 1b) [42]. Furthermore, this structure revealed the molecular contacts mediating inhibition by aphidicolin, a B-family polymerase inhibitor, which was shown to occlude the binding of the incoming nucleotide [42]. Altogether, these structures have acted to reveal the polymerase mechanism of pol α in both its native state and under conditions of perturbation via small molecule inhibitors.
Atomic resolution structures of the other three pol α subunits have also been obtained. The primase subunit structures were solved in complex with portions of both the B subunit and a majority of the catalytic subunit. This work provided particular insight on the conformational changes required for pol α to consistently create short RNA primers with its primase domain before transitioning to its polymerase domain. Specifically, these structures revealed an interdomain steric occlusion of the primase domains which limits primer elongation to ~ 10 ribonucleotides, as well as a global shift that allows the channeling of the primed DNA from the primase to the polymerase domain without dissociation [43]. Additionally, crystal structures of the large primase subunit revealed that an evolutionarily conserved iron-sulfur cluster is present in the protein’s core, an unusual finding as iron-sulfur clusters are almost exclusively found near the exterior of proteins [44]. Further investigation of the iron-sulfur cluster showed that the initiation of DNA synthesis may be triggered by the charge state of the iron-sulfur cluster in the primase domain, in which an electron is transferred from the polymerase domain to the primase domain via the DNA substrate. This altered charge state modifies the binding affinity and triggers the interdomain transport of DNA [45]. These structural advances in understanding all four subunits of pol α have been instrumental for gaining understanding of how DNA replication is initiated.
DNA polymerase delta (pol δ)
DNA polymerase delta (pol δ) is one of the major replicative DNA polymerases in eukaryotes and, in addition to participating in cell cycle checkpoints and multiple DNA repair pathways, is responsible for replication of the genome along with pol α and ε [46–49]. While the catalytic subunit of pol δ (POLD1) contains both the polymerase active site and exonuclease active site, the fully assembled pol δ is a heterotetramer comprised of POLD1 and three accessory subunits POLD2, POLD3, and POLD4 (also known as p125, p50, p66/p68, and p12, respectively) [50, 51]. These accessory subunits mediate many of the protein–protein interactions between pol δ and accessory proteins such as the clamp loader replication factor C (RFC) and PCNA [52–54]. When these binding regions are disrupted, or if PCNA is absent, processivity declines drastically, highlighting the critical role of PCNA in genomic replication [54]. Genomic replication is an exceedingly intricate process, which has made pol δ’s specific role in the replisome challenging to identify, and controversial to date [55–59]. Currently, one widely accepted model designates pol δ as responsible for the replication of the lagging strand of the replication fork, while pol ε concurrently replicates the leading strand [60, 61]. Conversely, some groups suggest pol δ may be the major polymerase on both strands with pol ε playing a less active role [56, 62]. In both models, however, pol δ replicates the lagging strand of the replication fork. Therefore, in either case, pol δ is an essential protein in eukaryotes via its vital role in genomic replication [63].
Because pol δ replicates billions of bases with each cell division, the mutational impact of misinsertion is amplified greater than other DNA polymerases that replicate on smaller scales. Accordingly, the fidelity of pol δ has evolved to be one of the highest among DNA polymerases, at a rate of ~ 1 base misinsertion per 105 nucleotides replicated [64, 65]. Without a functional proofreading domain, the fidelity of pol δ decreases 60-fold, but still retains a moderately high fidelity from its tight active site pocket [65, 66]. This active site pocket was structurally characterized in 2009 when a ternary structure was reported of Pol3, the yeast homolog to the human pol δ catalytic subunit, in complex with a DNA template-primer and an incoming dCTP [67]. The structure revealed that upon dNTP binding, a rotation of the fingers domain promoted correct insertion by shaping a tight pocket around the incoming nucleotide [67, 68]. The understanding of pol δ’s fidelity also grew from the aforementioned structure, because it includes the exonuclease domain of Pol3. In other polymerases with exonuclease domains, extensive interactions on the minor groove side of the DNA were shown to probe the base pairing properties of nucleosides up to two bases anteceding the next incoming base [69]. Disruption of these interactions by a mismatch is thought to trigger a conformational change to transfer the substrate to the exonuclease domain [70]. In the case of Pol3, these extensive minor groove interactions continued as far as 5 base pairs down the DNA, with many interactions mediated by coordinated waters (Fig. 1c). Therefore, pol δ would have up to five insertion events to “catch” the mistake, leading to a more sensitive proofreading potential than many other DNA polymerases [67].
DNA polymerase epsilon (pol ε)
When DNA polymerase epsilon (pol ε) was first discovered in 1986, it was initially thought to be a high molecular weight form of pol δ [71]. However, in 1990 it was determined to be a distinct polymerase that functions in concert with pols α and δ to replicate the genome [72]. Even now, after over 20 years of research into this enzyme, the exact role of pol ε in the replicative process is still debated [73]. Studies utilizing the specific error signatures of mutant yeast pol δ and pol ε, as well as the observation of pol ε specific incorporation of ribonucleotides, describe a model where pol ε and pol δ synthesize the leading and lagging strand, respectively. Still, alternative models have been put forward and have been the subject of multiple reviews [56, 58, 60, 74, 75]. DNA pol ε has similar features to the other B-family polymerases, yet unique domains and features which confer the greatest fidelity and processivity of any known mammalian DNA polymerase [76–78]. As a B-family polymerase, human pol ε is a heterotetramer composed of a large catalytic subunit encoded by POLE1 and three accessory subunits: POLE2, POLE3, and POLE4 (which encode proteins p261, p59, p12, and p17/CHRAC-17, respectively) [79]. A 20 Å cryo-EM structure of the yeast pol ε heterotetramer revealed that these accessory subunits extend from the catalytic subunit like a tail, interacting with the DNA helix, and are thought to increase processivity through direct interactions with DNA processivity factors. Additionally, structures of the human homologs of some of these subunits are also available [80–82].
Although a high-resolution structure of the human catalytic subunit has yet to be solved, ternary structures (with DNA substrate and incoming nucleotide) of the yeast homolog were determined [83, 84]. Similar to other high fidelity polymerases, the fingers domain closes upon nucleotide binding to form a tight pocket around the inserted nucleotide to both position the dNTP for catalysis and reduce the frequency of mismatch insertions. Interestingly, this pol ε pocket includes a unique residue in the major groove (Tyr481), not conserved in pol δ, possibly contributing to its superior fidelity and potentially reducing the insertion efficiency of modified nucleotides that are handled efficiently by other DNA polymerases [85, 86]. This structure also gave the first glimpse of an additional domain on pol ε not present in other members of the B-family, the P-domain (Fig. 1a, d). This domain was observed to wrap around the DNA and is named for its impact on processivity [83]. Further assaying this processivity, in the absence of PCNA, pol ε was observed to insert up to 27 nucleotides prior to dissociation, compared to pol δ’s three. Additionally, both the processivity and activity of pol ε were stunted with the removal of the P-domain, highlighting its essential role within pol ε’s function [83].
Similar to other DNA polymerases with exonuclease domains, residues along the minor groove survey the newly synthesized primer strand through extensive interactions. Upon disruption by a mismatched base pair, the exonuclease process is triggered [67, 69, 83]. A nearly identical system is seen in pol δ (Fig. 1c); however, pol ε contains an additional hydrogen bond from a coordinated water molecule to the nucleoside at the primer terminus (not observed in pol δ) which may aid in mismatch recognition [83]. When a mismatch is recognized, a conformational shift in the thumb domain guides the primer strand ~ 41 Å from the polymerase active site to the exonuclease domain where the exonuclease reaction occurs, similar to pol δ. Thus, structures of the yeast homolog of pol ε have revealed how the stringent polymerase active site and exquisitely sensitive exonuclease domain give rise to its nearly unmatched fidelity.
DNA polymerase zeta (pol ζ)
In the 1970s, REV1, REV3, and REV7 were identified as genes associated with a deficiency in mutagenesis [87–90]. Homology of REV3 to other genes encoding B-family DNA polymerases, as well as its role in mutagenesis, led to the hypothesis that REV3 encodes a lesion-associated DNA polymerase [91]. This was confirmed in 1996 when the yeast Rev3 and Rev7 proteins were purified as a heterodimeric complex and characterized as the sixth eukaryotic polymerase, DNA polymerase zeta (pol ζ) [92]. As the only error-prone member of the B-family, pol ζ has a significantly lower fidelity compared to other replicative polymerases [93]. However, pol ζ is an important part of DNA damage tolerance, in coordination with the Y-family polymerases which specialize in translesion DNA synthesis (TLS). As the first DNA polymerase found to specialize in TLS, its discovery and characterization led to a new era in the polymerase field as focus shifted to enzymes specializing in translesion DNA synthesis [94].
In 2010, the first published crystal structure of the multi-subunit pol ζ complex revealed a portion of the human Rev3 catalytic subunit in complex with Rev7 (the smaller, non-catalytic subunit), mapping their interaction to a region N-terminal of the polymerase domain [95]. Rev7 stimulates the activity of the catalytic Rev3 subunit by 20–30 fold in vitro [92] and mediates several protein–protein interactions with additional proteins, suggesting many potential roles of pol ζ in diverse cellular processes [95–99]. Although Rev3 and Rev7 are sufficient for pol ζ activity in vitro [92], a stable four-subunit heterotetramer complex can be purified with accessory subunits PolD2 and PolD3 from pol δ. This pol ζ holoenzyme (composed of Rev3, Rev7, PolD2, and PolD3) exhibits increased activity and enhanced processivity compared to the heterodimeric Rev3–Rev7 complex [100, 101]. Organization of this entire pol ζ heterotetramer was observed via electron microscopy (EM), generating a 23 Å resolution bilobal 3D model of yeast pol ζ [102]. The large lobe can be fit with the Rev3 catalytic subunit, and is connected to the smaller lobe which consists of the Rev3 C-terminal domain, as well as the regulatory subunits Rev7, pol31, and pol32 (yeast homologs of PolD2 and PolD3). Two stems of density connecting the lobes suggest dual linkers that may result in a more rigid association than seen in the more flexible assemblies of pols α and δ, which have a similar elongated bilobal structure [103, 104].
Pol ζ is specialized for the extension step of lesion bypass and can readily extend from matched, mismatched, and damaged primer-termini [105, 106]. Therefore, two polymerases act sequentially, with pol ζ facilitating extension after another TLS polymerase bypasses the lesion. Pol ζ is also recruited to sites of replication fork stalling caused by defective replicative polymerases [107]. Such compromised replisomes stall more frequently at DNA lesions, activating error-prone bypass and extension of mismatched primers by pol ζ. The activity of pol ζ is important for sustaining chromosome stability, but like most TLS mechanisms, occurs at the cost of increased mutagenesis. Because of this, pol ζ has been implicated in a large portion of cellular mutagenesis [108, 109]. While partial crystal structures and EM models provide some insight into its role in DNA damage bypass and extension, minimal structural information is available for pol ζ in the context of damage; therefore, the structural basis for lesion bypass and extension is unclear at this time.
Y-family
Despite the numerous pathways that work to repair DNA, some lesions can persist and block subsequent rounds of replication, as replicative polymerases are unable to accommodate most DNA lesions within their compact active sites [64, 66]. As a result, damage tolerance pathways have evolved to ensure that replication can be completed in the presence of DNA damage. Y-family DNA polymerases are specialized to ensure continuity at the replication fork via incorporation of a nucleotide opposite the lesion by TLS. When a replicative polymerase encounters a lesion, a polymerase switching event occurs, replacing the arrested polymerase with a TLS polymerase specialized for lesion bypass. The first genes encoding translesion polymerases were identified in 1971 by Jeffery Lemontt via a damage-induced mutagenesis study [87]. Due to their almost complete lack of primary sequence homology with replicative DNA polymerases, it took the discovery and characterization of several more TLS polymerases before they were connected as a family in 2001 [110].
The four main TLS polymerases (which include DNA polymerases eta (pol η), iota (pol ι), kappa (pol κ), and Rev1) belong to the Y-family of DNA polymerases and are uniquely adapted to bypass different DNA damage lesions. Crystal structures have provided insight into structurally distinct features that convey their diverse strategies for replicating through lesions. In addition to the canonical polymerase ‘right-hand’ domains described in the introduction, TLS polymerases contain an extra polymerase-associated domain (PAD) (sometimes called little finger [111] or wrist domain [112]) that extends from the C-terminus and makes extra DNA stabilizing contacts [113]. Another feature that sets apart these damage-associated polymerases is the absence of a 3ʹ–5ʹ exonuclease domain that proofreads newly synthesized DNA. While most high-fidelity replicative polymerases have this domain, most of the specialized polymerases do not and thus exhibit lower fidelity [114]. Here, we will further structurally analyze the unique mechanistic strategies used by each Y-family polymerase to allow the bypass of specific lesions.
DNA polymerase eta (pol η)
Originally identified in 1996 by sequence homology with prokaryotic TLS polymerases [115, 116], the yeast RAD30 gene was shown to encode DNA polymerase eta (pol η) in 1999 [117]. It was not long until the human homolog (POLH) was identified and characterized, making pol η one of the founding members of the Y-family of DNA polymerases [110]. The predominant biological role of pol η is replication through cyclobutane pyrimidine dimers (CPDs) [117] and 8-oxoguanine [118]. CPDs are a type of DNA lesion resulting from the UV-radiation catalyzed covalent linkage of two adjacent pyrimidines. While CPDs block most DNA polymerases, pol η is specialized to efficiently and correctly insert adenines (A) opposite each thymine (T) in the dimer, resulting in error-free bypass of the lesion (Fig. 2a). In humans, defects in pol η activity due to POLH mutations lead to the variant form of Xeroderma Pigmentosum (XP-V), a genetic disease associated with increased risk of sunlight-induced skin carcinomas [119, 120].
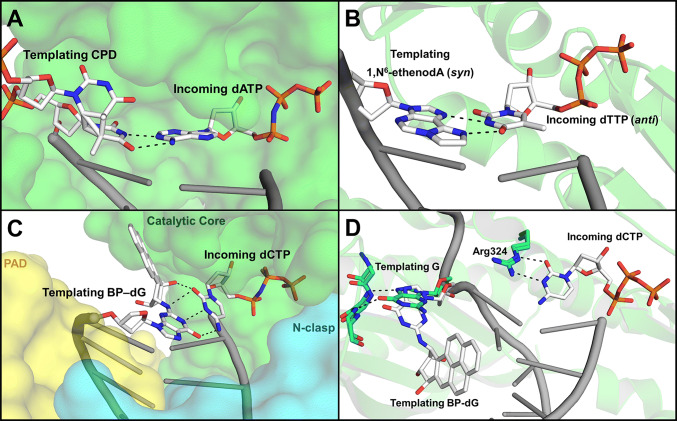
Fig. 2.
Y-family polymerases. Unique lesion bypass mechanisms of a templating lesion and incoming nucleotide are indicated (white sticks). a human Pol η shown in surface (green; 3MR3) b human Pol ι shown in cartoon (green; 2DPJ) c human Pol κ shown in surface with indicated catalytic core (green), PAD (yellow), and N-clasp (cyan) (4U7C) d yeast Rev1 shown in cartoon with undamaged templating guanine (green; 2AQ4), overlayed with damaged guanine template (white) and residues (cyan sticks; 5WM1)
Crystal structures have shown that, compared to other polymerases, the pol η fingers domain is positioned further from the templating base, enlarging the active site [121]. Importantly, this particularly wide active site is spacious enough to accommodate two bases of undamaged DNA or both bases of a CPD lesion (Fig. 2a) [121, 122]. It was hypothesized that since the structural constraints of the replicative polymerases are absent, pol η relies on the stability of Watson–Crick hydrogen bonding to determine which bases are correct for subsequent incorporation. Supporting this hypothesis, studies with yeast pol η used nucleotide base analogs to demonstrate that pol η is more dependent on Watson–Crick hydrogen bonding between the templating and incoming nucleotide than other polymerases [123]. This explains why pol η can replicate through some lesions (such as CPDs, 6–4 TC or CC photoproducts [124], and 8-oxoguanine) but is inhibited by lesions that largely disrupt Watson–Crick base pairing [125].
Since CPDs are known to confer bending of the double helix, it is surprising that crystal structures have shown that CPD-pol η complexes retain a straight B-form DNA molecule. This phenomenon is attributed to the close interaction between the pol η catalytic core and PAD domain, which creates a positively charged pocket for DNA binding. Described as a “molecular splint”, this pocket holds the DNA in a conformation that allows the bypass of CPD lesions [122]. After the incorporation of adenines across from the CPD, pol η processively inserts at least two additional bases past the lesion before steric hindrance and the loss of hydrogen bonding causes it to dissociate. The position-dependence of CPD-pol η interactions produce a structural basis for polymerase switching after lesion bypass [122]. While this discussion focused on the primary role of pol η in TLS, the polymerase has also been implicated in multiple other processes such as somatic hypermutation [126, 127] and homologous recombination [128, 129].
DNA polymerase iota (pol ι)
Human DNA polymerase iota (pol ι), a Y-family polymerase encoded by the POLI (RAD30B) gene, was shown to have TLS activity in 2000 [130]. While its existence has been suggested to be the result of POLH (pol η) gene duplication, pol ι has notably different biochemical properties that convey a unique mechanism for lesion bypass [131, 132]. Polymerases typically rely on the well-defined geometry of Watson–Crick base pair interactions, resulting in about the same catalytic efficiency for correct insertion for all four bases. Even though pol ι is often referred to as one of the most error-prone DNA polymerases, its activity is highly sequence context dependent, having higher efficiency and fidelity for nucleotide incorporation opposite template purines than opposite template pyrimidines [130, 133, 134].
Crystal structures of pol ι ternary complexes (with DNA substrate and incoming nucleotide), reveal features of the active site that provide insight into the insertion and base-pairing preferences elucidated from kinetic studies. Unlike replicative polymerases, a conformational change is seen in the DNA upon pol ι binding, while the enzyme and active site remains rigid [135]. Upon dNTP binding, the template base is forced to rotate about its glycosidic bond into syn-conformation, facilitating Hoogsteen base pairing with the Watson–Crick face of an incoming dNTP (anti). This mechanism relies on the narrow active site of pol ι to impose the required syn-conformation of the templating base. Several aliphatic active site residues (Leu62, Val64, and Gln59) impose constraints on the template, effectively preventing Watson–Crick base pairing. In addition, the syn-conformation is stabilized by other protein contacts and a small hydrophobic cavity which directly positions the sugar moiety of the template [136]. Holding the template securely in the syn-conformation effectively limits its hydrogen bonding opportunities to bases with Hoogsteen hydrogen bonding capability, resulting in low efficiency, low fidelity, and inaccurate lesion bypass [130, 134].
Structures of pol ι complexes with damaged templating DNA reveal that this Hoogsteen base pairing mechanism allows for the bypass of various lesions, specifically ones that impair Watson–Crick interactions [137, 138]. For example, N2-ethylguanine [139], O6-methylguanine [140], and 1,N6-ethenoadenine [141] are damaged bases that adopt the syn-conformation in the active site of pol ι to allow Hoogsteen base pairing (Fig. 2b). For minor groove adducts, rotation of the nucleotides into the syn-confirmation removes the damaged moiety from the active site by repositioning it into the major groove of the dsDNA, where it can be more readily accommodated. Once removed from the active site, these lesions no longer interfere with the interactions between the template base, polymerase, and incoming nucleotide, which allows for efficient lesion bypass [138]. While structures have shown pol ι using its Hoogsteen face via this mechanism, other studies suggest it can alternatively use its Watson–Crick face [142, 143], reinforcing its diverse TLS function in the bypass of many forms of lesions.
DNA polymerase kappa (pol κ)
Like other TLS polymerases, the POLK gene was discovered via homology searches for eukaryotic orthologs of the E. coli dinB gene [144]. The POLK gene was cloned in 1999 and its gene product, DNA polymerase kappa (pol κ), was demonstrated to have TLS activity in 2000 [145, 146]. The TLS function of pol κ remains somewhat unclear, with two main proposed cellular roles: (1) extension of primer termini, and (2) the incorporation of nucleotides to bypass bulky minor groove adducts [138]. While in vitro studies support both functions, many studies point to extension as its main cellular role. Although most polymerases are inefficient at extension from mismatched primer-termini, pol κ extends from mismatched [147] and lesion-containing termini [148–151] with comparable efficiency as non-damaged matched termini [146]. Mismatched termini could often result from lesion bypass by other TLS polymerases, suggesting pol κ may have an important cellular role as an extender of mismatched and lesion-containing termini resulting from TLS. Additionally, a recent publication identified pol κ as having a role in promoting DNA synthesis and replication stress recovery at sites of stalled forks [152]. Thus, the functional role of pol κ is an active area of interest in the field.
When a crystal structure of the pol κ catalytic core without DNA present was published in 2004, it was the first structure of any human Y-family polymerase [153]. Subsequently, in 2007, the structure of a pol κ-DNA ternary complex with an incoming nucleotide provided additional insight in the context of DNA. Specifically, it revealed the importance of a unique 100-residue N-terminal extension, called the N-clasp. The N-clasp extends from the thumb domain, causing DNA to be completely encircled near the primer terminus, likely stabilizing pol κ bound to not only matched, but also mismatched and lesion-containing termini (Fig. 2c) [154]. Mutational analysis has shown that the N-clasp facilitates extension from mismatched and likely damaged, bases [154]. It has been proposed that pol κ can extend a mismatch by locking the N-clasp around the primer-template, keeping it engaged on the distorted sugar phosphate backbones long enough for the misaligned 3ʹ OH to correctly re-position for nucleophilic attack [154].
Even though pol κ lacks proofreading activity, it has a moderate processivity and a low to moderate fidelity with error frequencies ranging for 10−2 to 10−4 [145, 146, 155]. A constrained active site is thought to mediate this, as well as extension, by accommodating only one traditional Watson–Crick base pair at a time [153, 154]. While pol κ may be better at extending from lesions than bypassing them, biochemical and kinetic studies have shown that purified pol κ is capable of inserting nucleotides opposite several templating lesions, albeit with varying efficiency. As one example, studies show efficient and accurate bypass for N2-adducted guanines via dCTP insertion, with larger adducts being physically accommodated by a gap in the pol κ structure between the catalytic core and PAD domain (Fig. 2c) [156]. A recent crystal structure of a pol κ ternary complex, with a DNA substrate containing a bulky BP-dG adduct and incoming dCTP in the active site, provided insight into a domain arrangement that opens the active site at the minor groove to accommodate the damage. Furthermore, this structure supported the previously reported indispensable role of the N-clasp during lesion bypass (Fig. 2c) [157]. Another recently published ternary structure reveals a more ordered N-clasp domain allowing higher resolution insight into the function of this flexible domain [158]. The flexibility and stability of these domains uniquely suit pol κ for the specific translesion functions of bypass and extension.
DNA polymerase Rev1
In 1996, the yeast DNA polymerase Rev1 was detected to have dCMP transferase activity [159]. Since this protein had previously been shown to be required for damage-induced mutagenesis, it was proposed that this newly identified function was important for mutagenic TLS. However, Rev1 was not officially recognized as a DNA polymerase until 1999, when deoxynucleotidyl transferase activity was detected in several enzymes homologous to Rev1 [94]. Interestingly, Rev1 differs from other polymerases in its selectivity of nucleotide incorporation. While replicative polymerases incorporate the correct nucleotide opposite all four template bases with nearly equivalent efficiencies, Rev1 displays a strong preference for the incorporation of cytosine (C) opposite a templating guanine (G) [159].
The yeast Rev1 ternary complex structure (with DNA substrate and incoming nucleotide) provided the first structural basis for its highly specific mechanism, revealing a lack of hydrogen bonding between a templating G and an incoming dCTP in the Rev1 active site [160]. Rather, the templating G is removed from the active site, while the incoming dCTP pairs with an arginine side chain (Arg324) (Fig. 2d). Publication of a human Rev1 crystal structure revealed that the key elements of this mechanism are conserved in the human enzyme [161]. In the human Rev1 structure, Leu358 is inserted into the DNA helix, displacing the templating G, which consequently swings out of the helix and into a hydrophobic pocket (His774, Lys681, and Phe525). This extrahelical position is stabilized via two hydrogen-bonding interactions between its Hoogsteen edge and main-chain amides of a protein loop (Fig. 2d, green). As in yeast, the Watson–Crick edge of the incoming dCTP pairs with protein residue Arg357, making this a protein-template-directed mechanism. The pattern of hydrogen bonding for both the templating G and incoming dCTP is such that it favors the identity of these two specific bases. In both positions, the substitution of another base would result in loss of hydrogen bonding, as well as electrostatic or steric repulsion [160]. Therefore, these highly specific hydrogen-bonding interactions reveal the mechanism of Rev1 specificity for dCTP incorporation opposite a templating G. Rev1 uses the same protein-template-directed mechanism of DNA synthesis to insert dCTP opposite damaged guanines. Recently, structures of yeast Rev1 in complex with substrates containing DNA lesions, such as BP-N2-dG [162], abasic sites [163], and γ-HOPdG adducts [164], have allowed for the visualization of this mechanism in the context of damage (Fig. 2d, white). The stabilization of the templating damaged G via Hoogsteen hydrogen bonding ensures its rotation into the extrahelical position where the bulky adduct can be accommodated in the major groove, removed from the active site. Therefore, hydrogen bonding of the templating G ensures accommodation of the adduct, while hydrogen bonding of the incoming dCTP ensures correct insertion [162]. Thus, these structural studies provide a basis for Rev1′s unique ability to dictate both the incoming and templating bases to ensure error-free bypass of damaged Gs.
While the mechanism is conserved, the yeast and human Rev1 structures differ in that the human Rev1 catalytic domain contains two large inserts in the palm and fingers domains, making it significantly larger [161]. The 40-residue palm domain insert, which extends away from the active site and is largely disordered, has been proposed to serve as an additional site for TLS co-factor binding and/or protein–protein interactions. Rev1 has been shown to have an important non-catalytic scaffolding role in TLS, simultaneously interacting with several polymerases and potentially facilitating polymerase switching at lesions and multi-polymerase TLS [165, 166]. The 54-residue fingers domain insert supports the selectivity of Rev1 for dCTP insertion, contributing to the hydrophobic pocket by acting as a “flap” to stabilize bulky N2-dG adducts in the template position. As both inserts are partially unstructured, it has been proposed they become ordered upon additional interactions with protein or bulky DNA lesions, respectively [161]. Overall, fitting with the theme of this review, these structural minutia collectively provide Rev1 with an incredibly unique TLS ability which is vital to cellular function.
X-family
The X-family is comprised of four small monomeric DNA polymerases involved in DNA repair: polymerase beta (pol β), polymerase lambda (pol λ), polymerase mu (pol μ), and terminal deoxynucleotidyl transferase (TdT). Pol σ (formerly pol κ) is considered a distant relative of the X-family; however, we have chosen not to include a discussion of pol σ due to the absence of significant structural characterization [167, 168]. Each X-family DNA polymerase plays a unique role in DNA repair. Pol β primarily functions as a gap-filling polymerase during the base excision repair (BER) of oxidative base damage [15]. The remaining X-family polymerases contribute specialized functions within the double-strand break (DSB) repair pathways of non-homologous end-joining (NHEJ) and V(D)J recombination [4, 169]. To provide specificity for this wide range of DNA substrates and biological functions, all X-family polymerases utilize an “8 kDa domain” (8kD, Fig. 3a, blue) to bind to the downstream ends of gapped DNA substrates. Additionally, pol λ, pol μ, and TdT contain an N-terminal “BRCA1 C-terminus” (BRCT, Fig. 3a, magenta) domain that facilitates their recruitment to the NHEJ and V(D)J pathways for DSB repair. Below, we further discuss these domains as well as other structural details which allow the X-family members to perform their distinct functional roles.
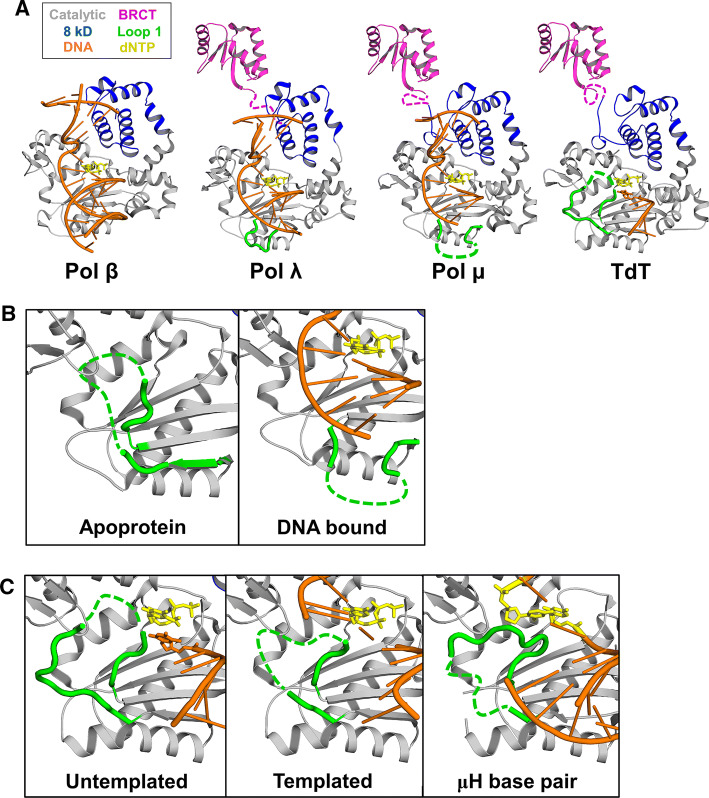
Fig. 3.
The different structural features of the polymerase X-family members and their substrates. a A representative pre-catalytic structure of each X-family member highlighting the similarities and differences between the DNA substrate (orange), the incoming dNTP (yellow sticks), and the structural domains: Catalytic (gray), BRCT (magenta), 8 kD (blue), and Loop 1 (green) (left to right) 2FMS, 2PFP, 5TXX, 4I27 b Structural comparison of Pol μ (gray), Loop 1 (green) and DNA (orange) between the apoprotein (4LZD) and DNA-bound (5TXX) structures. c Structural comparison of Loop 1 (green) in TdT (gray) with different DNA (orange) substrates (left to right) 4I27, 4QZ8 and 5D46
DNA polymerase beta (pol β)
In 1971, Chang and Bollum discovered the presence of a small molecular weight DNA polymerase in mammalian cells [170]. At only 39 kDa, this polymerase, named DNA polymerase beta (pol β), is still considered one of the simplest cellular DNA polymerases. Pol β, which exhibits moderate fidelity (Table 1), shares catalytic features with the high-fidelity replicative polymerases; for instance, both families’ exhibit rate-limiting product release and a conformational change of the catalytic domain upon nucleotide binding [171–173]. Biologically, pol β plays dual roles in the nuclear BER pathway, where it both cleaves out the 5′ dRP left by AP endonuclease cleaveage and fills the resulting gap [174]. Additionally, pol β has also been implicated in the mitochondrial BER and alternative-NHEJ pathways [175–177]. The biological implications and precise role of pol β in these scenarios remain to be fully elucidated.
Pol β is comprised of two domains, an N-terminal 8 kDa domain and a C-terminal 31 kDa catalytic domain, both of which contain key structural elements mediating its functional roles during BER (Fig. 3a) [11]. During BER, pol β binds its substrate via two helix-hairpin-helix (HhH) motifs, one a part of the 8 kDa domain and the other a part of the catalytic domain [15]. This places the lyase active site of the 8 kDa domain in position to excises the 5ʹ dRP [178–180]. The HhH motifs also stabilize a 90° bend of the DNA, giving the polymerase access to the 3′-primer terminus, and allowing the incoming nucleotide entry into the nascent base pair binding site [15]. The resultant pol β:DNA binary complex is in an “open” conformation. Selection of a correctly matched nucleotide occurs via an induced fit mechanism, requiring a transition to a “closed” conformation in which the N-helix (fingers) realigns the active site residues and metal ions with the DNA duplex and incoming nucleotide [172]. Once the active site is arranged into its “closed” catalytically competent conformation, the nucleotidyl transferase reaction is carried out, where ultimately the O3ʹ of the primer terminus performs a nucleophilic attack on the alpha-phosphate via catalytic metal coordination and proton abstraction by Asp256 [181–183].
Equipped with the structural simplicity of only two domains and a low molecular weight, pol β is an ideal candidate for X-ray crystallography. Consequentially, pol β was the first member of the X-family (and first eukaryotic polymerase overall) to be examined at atomic resolution, and has continued to serve as a model DNA polymerase for structural characterization of the near-universal nucleotidyl transferase mechanism used by DNA polymerases for DNA synthesis [11, 17, 181, 184, 185]. With the development of time-lapse X-ray crystallography with pol β, the entire catalytic cycle of the nucleotidyl transferase mechanism has been observed [17, 186]. Importantly, this system has been exploited to reveal how DNA polymerases process non-canonical DNA substrates such as mismatches, damaged nucleotides, anti-viral drugs, and therapeutic nucleotides [17, 86, 181, 185, 187–189]. This comprehensive use of pol β to study the polymerase mechanism has resulted in a large body of literature, including 339 X-ray structures deposited in the PDB, focused on pol β. With this in mind, we have kept this section brief and direct readers to Ref. [15] for a review dedicated to pol β.
DNA polymerase lambda (pol λ)
DNA polymerase lambda (pol λ) was first reported as a novel eukaryotic DNA polymerase in 2000 [190]. At the time, it was thought to play a role in meiosis; today, however, pol λ is primarily considered a gap filling polymerase and is associated with several DNA repair pathways [191–193]. Of the eukaryotic X-family members, pol λ is considered the most closely related to their common ancestor [194]. Consistent with this notion, pol λ functionally overlaps with both of its template-dependent X-family siblings (pol β and pol μ). Moreover, like pol β, pol λ participates in BER [192, 195, 196], and like pol μ, it participates in DSB repair though the NHEJ pathway [193] and has been implicated in TLS [191]. As the largest member of the X-family, pol λ has an 8 kDa domain, a catalytic domain, a Ser-Pro rich region, and a BRCT domain (Fig. 3a) [197, 198]. The 8 kDa domain contains its 5ʹ-dRP activity utilized during BER [195, 199], the Ser-Pro rich region serves as a target for post-translation modifications [200], and the BRCT domain is responsible for the recruitment of pol λ to NHEJ complexes [201]. Several catalytic features of pol λ activity, including its method of nucleotide discrimination and propensity to produce deletions, have been revealed by structural analysis and are discussed below.
The comparison of several X-ray crystallography structures representing different stages in the pol λ catalytic cycle indicate that the enzyme does not undergo a large-scale open to closed transition upon binding of the incoming nucleotide to the binary complex (pol λ:DNA), as has been observed with many other DNA polymerases [202]. In contrast, a more localized transition occurs involving a relocation of a key loop motif termed “loop 1″ and a corresponding shift of the template strand further into the active site to facilitate base pairing between it and the templating base. In addition, several active site residues reposition to form a binding pocket for the nascent base pair and establish interactions with the DNA minor groove that are important for base selectivity and catalysis. Consistent with the structural predictions, deletion of loop 1 results in significantly decreased fidelity, indicating that loop 1 plays a role in nucleotide selectivity [203].
Both NHEJ and long patch BER involve filling gaps longer than one nucleotide. Structural analysis of the pol λ in complex with a two-nucleotide gapped DNA substrate suggests that DNA binding by pol λ is directed largely by the 8 kDa domain, which anchors the polymerase at the 5′-end regardless of the conformation of the 3′-end [202]. Upon binding of the incoming dNTP, the 3′-primer-terminal nucleotide, as well as amino acid residues that form the active site and the nascent base pair binding pocket, assume an identical position as observed in the structure of the ternary complex with a one-nucleotide gap. The incoming dNTP binds opposite the 3′-template nucleotide of the gap, which is located in the active site. Though the gap is one nucleotide longer, the distance between the 3′-end of the gap and the 5′-end bound by the 8 kDa domain is the same as in the one-nucleotide gap structure. This is possible because the template strand is scrunched, placing the 5′-template nucleotide of the gap in an extra-helical conformation within an active site pocket created by three amino acid residues (Leu277, His511, and Arg514) [204]. Upon a translocation, pol λ subsequently inserts the second base in the 5ʹ templating position, filling the two-nucleotide gap. Of note, pol λ generates a high frequency of single-base deletions during DNA synthesis compared to other DNA polymerases [205]. The mechanism for producing single base deletions is thought to involve dNTP-induced misalignment of the template strand relative to the primer strand during catalytic cycling [206]. Therefore, the likelihood that pol λ will fill a two nucleotide gap with either one nucleotide (i.e., creating a single-base deletion) or two nucleotides, appears to be dependent on the stability of the misaligned intermediate [204].
DNA polymerase mu (pol µ)
DNA polymerase mu (pol μ) was first identified and biochemically characterized in 2000 [207]. This initial characterization showed that the enzyme has very high error rates on primer–template substrates and additionally possesses a template-independent activity, where it extends from a template lacking single-stranded DNA primer. Combined with the observation that it is expressed preferentially in secondary lymphoid tissues, this early characterization suggested that pol μ could act as a DNA mutator polymerase responsible for the somatic hypermutation of immunoglobulin genes. Today, in terms of metal utilization, template dependency, and indiscriminate use of both ribonucleotides and deoxyribonucleotides, pol μ is considered one of the most versatile DNA polymerases [208, 209]. As a result of this unique substrate specificity, pol μ specializes in the bridging of uncomplimentary DNA ends during NHEJ and V(D)J recombination, where this versatility allows for the impromptu selection of the most efficient nucleotide/metal ion combination.
Structural analysis has provided insight into the unique substrate specificity of pol µ, elucidating key features that facilitate its specialization in end-joining. The structure of pol μ is organized into three domains: a BRCT domain, an 8 kDa domain, and a catalytic domain. Relative to its X family siblings, pol β and pol λ, the pol μ 8 kDa domain contains fewer DNA contacts and lacks lyase activity [210]. However, there is an increase in the number of contacts between the upstream DNA and the helix-hairpin-helix (HhH) motif contained within the catalytic domain, which assists pol μ in binding non-complimentary DNA strands [210, 211]. Functionally, pol μ has very slow polymerization rates (0.006–0.076 s−1) for the correct insertion of deoxynucleotides [212]. In fact, the rates of correct insertion for pol µ are in the range of those obtained for the misincorporation of nucleotides by other DNA polymerases. Additionally, pol μ exhibits an increased fidelity for ribonucleotides [209, 213]. Importantly, this provides an advantage during NHEJ, during cell cycle timepoints when deoxyribonucleotide concentrations are very low. To this point, recent structures reveal that pol μ binds and incorporates ribonucleotides with canonical active site geometry and minimal distortion of the DNA substrate or nucleotide [213]. In addition, complementary incorporation kinetics utilizing wild type and rationally designed mutants of pol µ indicate that ribonucleotide accommodation involves synergistic interactions with multiple active site residues not found in polymerases with greater discrimination [213].
Structures representing various stages of the catalytic cycle indicate that pol µ is structurally rigid, where both the DNA and the enzyme lack large-scale conformational changes during dNTP binding and catalysis [214]. One exception is loop 1, which is oriented towards the DNA binding cleft of pol μ in the apoenzyme structure and is repositioned upon DNA binding to facilitate a catalytically competent position (Fig. 3b) [214]. Additionally, the inherent flexibility of this loop in pol μ appears to allow the accommodation of the different DNA substrates required for both its template-dependent and -independent activities. Supporting this hypothesis, deletion of the loop substantially reduces template-independent activity [215]. Biologically, this dual capacity could be advantageous to resolve microhomology-mediated NHEJ reactions. A ternary structure of Pol µ on a 2-nt gapped DNA substrate revealed that in contrast to pol λ, pol µ “skips” the first available template nucleotide, instead using the template base at the 5′ end of the gap to direct nucleotide binding and incorporation. This divergence from canonical 3′-end gap-filling provides additional insights into polymerase substrate choices during NHEJ [216]. Recently, time-lapse crystallography was employed to further examine the mechanism of pol µ during DSB repair [20]. These structures confirmed, using natural substrates, the lack of large protein or DNA conformational changes during catalysis, and identified several localized shifts of active site residues during metal binding and product dissociation.
TdT (terminal deoxynucleotidyl transferase)
Discovered in 1960, terminal deoxynucleotidyl transferase (TdT) is one of the first mammalian DNA polymerases to be identified, yet it remains one of the most poorly understood [169, 217]. TdT predominately incorporates nucleotides in a template-independent manner, using only single-stranded DNA as the nucleic acid substrate [218]. Despite its intriguing ability to create genomic material de novo, a specific biological role for TdT remained elusive for several decades. However, it is now known that TdT is responsible for the random addition of nucleotides to single-stranded DNA during V(D)J recombination [219, 220]. Importantly, this ability of TdT to intentionally create randomized genetic material during V(D)J recombination is vital for the mammalian immune system, as it promotes the immunoglobin and T cell antigen receptor diversity required for the neutralization of potential antigens [221–223].
Similar to pol μ, the structure of TdT is exceptionally rigid and consists of a BRCT domain, an 8 kDa domain, and a catalytic domain [224]. The first crystal structures of TdT, one with an oligonucleotide primer and another with an incoming ddATP-Co2+ complex, revealed steric clashes between the catalytic site and loop 1 (Fig. 3c). This long, “lariat-like” loop was proposed to explain the inability of TdT to accommodate duplex DNA [225]. In addition, these binary complexes revealed that the 8 kDa domain of TdT contacts the thumb domain to form a ring-like structure in which the single-stranded primer lies perpendicularly to the axis of the hole on the palm domain, and dNTPs diffuse through the hole into the enzyme’s active site. Later on, ternary complex structures of TdT bound to ssDNA with an incoming nucleotide were solved. Of note, there was no observed open to closed transition upon nucleotide binding, however, transient metal binding (occurring just before and leaving right after the chemistry step) in the active site was proposed to potentially drive efficient catalysis [226]. The combination of evidence from both structural and biochemical data deemed TdT incapable of interacting with duplex DNA substrates [224]. However, this widespread belief that TdT only possesses template-independent activity has recently been challenged.
TdT was recently shown to also have template-dependent activity in DSB repair, similar to pol μ, in the presence of higher concentrations dsDNA [227]. A series of crystal structures were recently solved with TdT bound to DSB substrates representing an annealed DNA synapsis containing one microhomology base pair and one nascent base pair (Fig. 3c). These structures revealed that the N-terminal 8 kDa domain and loop 1 cooperate to bridge the two DNA ends, providing a templating base in trans [228]. The structures from this and an additional study by the same group imply that TdT can direct DNA synapsis pre-assembly and alignment [227, 228], similar to pol µ [211]. Moreover, the structures indicate that TdT can halt its template-independent activity and switch into its template-dependent activity upon sensing the second DNA strand bridging the DSB. This is mediated through a reorganization of loop 1, which stabilizes the overlapping DSB strands (Fig. 3c). Consistent with this model, mutational analysis determined loop 1 to be essential for TdT to accommodate the opposing DNA strand during its template-dependent activity. Overall, this supports an intriguing model where TdT can single-handedly provide diversity via its template-independent activity and then accuracy via the template-dependent activity during V(D)J recombination. However, the precise biological role of these two activities attributed to TdT remains uncertain.
A-family
The members of the A-family of DNA polymerases share homology with E. coli DNA polymerase I, encoded by the polA gene. Mammalian cells contain at least three A-family DNA polymerases: gamma (pol γ), theta (pol θ), and nu (pol ν). DNA pol γ is responsible for the high fidelity replication and repair of mitochondrial DNA. Pols θ and ν, both low-fidelity nuclear enzymes, have been suggested to play roles in alternative nonhomologous end-joining and in the repair of interstrand DNA cross-links, respectively. Thus, the A-family polymerases have both similar and diverse biological functions. Structures of pol γ have shed new light on how the enzyme can uniquely adjust its processivity and fidelity to be efficient at both replication and repair [229]. Additionally, the crystal structures of pol ν and pol θ substrate complexes have elucidated unique polymerase structural diversity and identified several features of their respective fingers domains that confer their specialized functional attributes and provide insight into their poor DNA-synthesis fidelity [230–232].
DNA polymerase gamma (pol γ)
A novel RNA-dependent DNA polymerase activity in eukaryotic cells was reported in 1970 [233, 234]. By 1975, this polymerase was officially designated as DNA polymerase gamma (pol γ), although its cellular function as the main replicative and DNA repair polymerase in the midochondira was still undiscovered [235]. In 1977, pol γ was identified in mitochondria [236] and, not long after, evidence for its functional role was obtained by a study utilizing isolated brain synaptosomes [237]. Later, disruption of the yeast pol γ gene (MIP1) [238] and inhibition of mitochondrial DNA (mtDNA) replication by pol γ antibodies [239], provided further evidence for the mitochondrial role of pol γ. Accordingly, pol γ is critically important for mtDNA maintenance, and defects in pol γ-mediated mtDNA replication result in the loss of cellular respiration and ultimately induce mitochondrial genetic diseases, including mtDNA depletion disorders such as Alpers syndrome [240]. Additionally, human pol γ is known to be more susceptible than the nuclear DNA polymerases to inhibition by Nucleoside Reverse Transcriptase Inhibitors (NRTIs), which are designed to treat HIV; therefore, pol γ is likely responsible for a majority of the cellular toxicity caused by this key class of antiretroviral drugs [241, 242]. Of note, recent evidence has been mounting that pol γ is not acting alone in mitochondria, and this topic covered in Ref. [243].
The mtDNA replicated by pol γ is generally arranged in covalently closed circles and requires distinct molecular machinery from that used to replicate nuclear DNA. Two mechanisms of mtDNA synthesis have been proposed. The first is a conventional synchronous model, similar to that used to replicate nuclear DNA, where leading and lagging strand synthesis occur simultaneously [244]. The second mechanism is a strand-displacement (D-loop) model, where leading strand synthesis advances approximately two-thirds of the way around the circle before lagging strand synthesis is initiated [245–247]. Atomic level details, disclosed by a crystal structure of the human DNA pol γ heterotrimer, have helped elucidate the unique mechanism of mtDNA replication [229]. As was previously proposed based on a ~ 17 Å cryo-electron microscopy study [248], a single pol γA catalytic subunit interacts asymmetrically with a pol γB dimer (making contact primarily with only one pol γB monomer), as shown in Fig. 4a. Due to steric clashes, the authors were unable to model the enzyme as an A2B2 tetramer, which would potentially place two polymerases at a replication fork (a requirement for synchronous DNA synthesis). In agreement with the structural data, recent evidence, including ChIP-seq mapping of the mitochondrial ssDNA-binding protein (mtSSB) to the D-loop and the retention of primers at the two origins in RNaseH1 deficient cells, also points to the strand-displacement model as the more favored model of mtDNA replication [249].
Fig. 4.
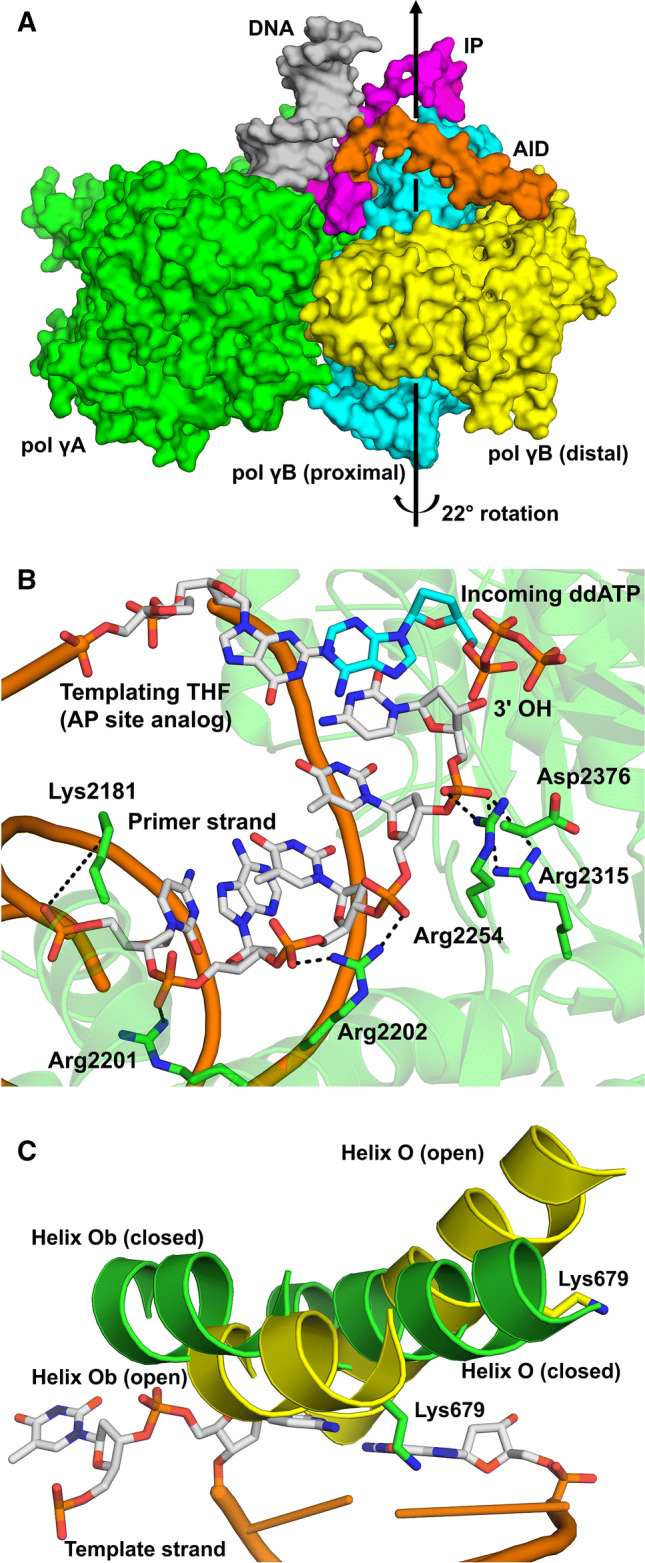
A-Family polymerases. a Domain and subdomain structure of pol γ (3IKM). b Ternary structure of pol θ (green) bypassing an AP site, highlighting the DNA contacts at the primer terminus (4X0P). c Overlay of pol ν open (yellow; 4XVL) and closed (green; 4XVK) binary complexes
The crystal structure revealed that pol γA adopts the canonical polymerase ‘right-hand’ configuration and undergoes a partial conformational change upon nucleotide binding. Although the overall fold of pol γA confirms its classification as a member of the A-family of DNA polymerases, pol γA uniquely possesses a large spacer domain (~ 400 residues) between its exonuclease and polymerase domains. This spacer contains a globular IP (Intrinsic Processivity) subdomain that provides the intrinsic processivity of pol γA and an extended AID (Accessory Interacting Determinant) subdomain that forms an interface with pol γB, further increasing the processivity of the holoenzyme (Fig. 4a). As a result, pol γA alone can synthesize ≥ 100-nucleotides prior to dissociation [250, 251], and its processivity is further enhanced by the presence of pol γB, which simultaneously accelerates the polymerization rate and suppresses the exonuclease activity [252]. Structurally, pol γB resembles class II aminoacyl tRNA synthetases and differs significantly from other processivity factors, such as sliding clamps and thioredoxin [229]. Structures of the ternary complex revealed that the dimeric Pol γB rotates 22° in ternary complexes, increasing the interaction between pol γA and the distal pol γB monomer (Fig. 4a) [253]. Importantly, this pol γA-distal pol γB interaction was shown to allosterically regulate the polymerase and exonuclease activities in the holoenzyme.
MtDNA is damaged by both endogenous and exogenous sources. Moreover, the leakage of electrons from the electron transport chain promotes the generation of reactive oxygen species, which are the major source of oxidative damage to mtDNA, and can result in deleterious mutations. Mitochondria are able to repair oxidative lesions due to their robust base excision repair (BER) system [254]. However, the lack of other efficient repair systems, such as mismatch repair and nucleotide excision repair (NER), results in the persistence of mutagenic lesions in mtDNA. Because unrepaired lesions can promote mtDNA mutations or even block mtDNA replication, the ability of pol γ to bypass such lesions by TLS (for their subsequent repair) is vital to maintaining the genetic integrity of mtDNA. Translesion DNA synthesis by the human DNA pol γ has been studied for a number of DNA lesions, including 7,8-dihydro-8-oxo-2′-deoxyguanosine, benzopyrene adducts, UV photoadducts, acrolein adducts, and several others [255].
DNA polymerase theta (pol θ)
Early biochemical characterization demonstrated the ability of DNA polymerase theta (pol θ) to bypass DNA lesions, hinting at a role in TLS [256, 257]. This was confirmed when it was shown to be involved in bypass of the oxidative lesion thymine glycol in vivo [256, 258]. Pol θ exhibits a very low fidelity of synthesis on normal DNA, similar to other TLS polymerases [259]. In addition to TLS, pol θ has recently been shown to play a central role in the alternative DNA double-strand break repair process termed microhomology-mediated end-joining [260–264]. Microhomology-mediated end-joining is a mutagenic and error-prone alternative to homologous recombination (HR) or NHEJ that utilizes short (2–6 bp) microhomologies to join two strands of dsDNA [265]. Furthermore, the ability of pol θ to extend ssDNA [266] and its 5′ deoxyribosephosphate lyase activity [267] point to roles in other various DNA repair pathways [256, 258, 266–269]. Additionally, depletion of pol θ leads to hypersensitivity to radiation [263, 270, 271] and decreased survival of cells deficient in homology-directed repair [260]. Fitting with its role in DNA repair and TLS, overexpression of POLQ, the gene encoding pol θ, is a predictive marker for multiple human cancers [260, 270, 272]. Underscoring this, inhibition of pol θ is a compelling chemotherapeutic strategy in multiple cancers [273].
Recently, crystal structures of the human pol θ polymerase domain were reported demonstrating how pol θ grasps the primer to bypass DNA lesions or extend poorly annealed DNA termini to mediate end-joining [231]. The structures revealed pol θ to have a core DNA polymerase fold with fingers, palm, and thumb domains. However, the structure identified unique, unresolved loops that likely confer its specialized functions. Because pol θ can bypass simple DNA lesions, it is noteworthy that the structure of the ternary complex (with DNA substrate and incoming nucleotide) exhibits extensive backbone contacts with the primer strand (Fig. 4b). Several of these contacts are unique to pol θ, including an essential contact for lesion bypass between Arg2254 and the primer terminus phosphate [231]. In addition, the closed pol θ substrate complex with an incoming ddGTP indicates that Asp2376 of the fingers domain interacts with Arg2254 of the thumb domain. This primer-grip strategy, coupled with the ability of pol θ to dimerize, could enable the enzyme to direct DNA synthesis from mismatched primer termini during repair of DNA double-strand breaks.
Pol θ is the only polymerase known to include a helicase domain. The pol θ helicase domain (pol θ-HLD) is a member of the superfamily II (SF2) helicases, most closely related to the Hel308 family of helicases, which are involved in the ATP-dependent 3′–5′ unwinding of lagging strands on replication forks [274–277]. Biochemical characterization of pol θ-HLD has demonstrated DNA-dependent ATPase activity but has not detected any helicase activity, although it is possible that the helicase activity employs an untested substrate or requires an accessory factor [260, 278, 279]. Recently, crystal structures of apo and nucleotide-bound forms of pol θ-HLD were reported. In common with the recently derived structure of the pol θ polymerase domain [231], the structure displays unexpected features corresponding to insertions in its amino acid sequence that appear to be responsible for its unique activities. Specifically, large hydrophobic regions containing polar contacts appear to enable the specific tetrameric formation of POLQ-HLD both in solution and in crystallo; suggesting that, at minimum, the enzyme acts as a dimer [230]. In light of this structural revelation, it has been proposed that two pol θ molecules could work together during the MMEJ annealing step, with one molecule operating on each side of a DNA break, thus preparing the annealed substrate for further processing by the polymerase domain [230].
DNA polymerase nu (pol ν)
DNA polymerase nu (pol ν) is a newly discovered A-family polymerase identified through its homology to the Drosophila Mus308 gene, which is thought to function in the repair of DNA interstrand cross-links [280]. The precise biological role of pol ν remains unknown, however, it has been shown to be uniquely prone to misincorporating dTTP opposite a template G [281, 282]. In addition, pol ν is capable of accurate bypass of major groove DNA lesions including thymine glycol and major groove DNA-peptide and DNA–DNA cross-links [283].
Pol ν is homologous to high-fidelity replicative polymerases, yet is error prone [282]. Instead of a simple open to closed movement of the O-helix (fingers domain) upon binding of a correct incoming nucleotide, recent structures of pol ν binary complexes (with DNA substrate) indicate it has an atypical open state and that closure requires the fingers domain to assume multiple conformations to contact the nascent base pair [232], see Fig. 4c. Importantly, in this unique open state, the O and Ob helices exclude the template from the active site while allowing the dNTP to bind, which may provide an explanation for the low fidelity of pol ν. In the closed conformation, residues of the fingers domain contact the nascent base pair. Of note, Lys679 is positioned over the nascent base on the major-groove side (Fig. 4c), and alanine substitution for this lysine results in a ~ tenfold increase in fidelity. This lysine may contribute to the stability of mismatches by forming hydrogen bonds to the non-Watson–Crick base pairs. Further analysis of these results points to a multifaceted role for Lys679 in substrate discrimination that is expected to be dependent on DNA sequence context [284].
Conclusion
DNA polymerases fulfill many vital roles in the cell ranging from the accurate replication of the genome, both nuclear and mitochondrial, to numerous functions associated with the repair and bypass of DNA lesions. To effectively execute these diverse roles, there are at least 15 mammalian polymerases, each specialized for different functions. While numerous biochemical experiments have aided in the characterization of the unique properties of these enzymes, it has largely been structural studies that have elucidated their specific mechanisms at the molecular level. Through the revelation of these individual mechanisms, insight into how DNA polymerases collectively work together to utilize and accommodate such a wide range of substrates has also been revealed. This review is an overview of all the mammalian polymerases, providing a brief introduction into each and discussing how their structure dictates their function. We aimed to provide insight from some of the hallmark structures and studies that have been done, as well as point to other studies and reviews where more specific information can be found.
As more information is learned about the diversity in DNA polymerase structure, function, and mechanism, it can be leveraged against human disease more effectively. Already, many existing cancer therapeutics work by modulating the DNA polymerase-mediated DNA damage response [285, 286]. For example, the perturbation of DNA polymerase activity can promote therapeutic resistance (via lesion bypass/TLS) or be utilized during combinatorial therapies with the design of specific inhibitors based on polymerase structure and mechanism. It is likely that our knowledge of DNA polymerase structure and function will continue to advance with the discovery of novel enzymes with a polymerase-like activity, novel interactions of accessory proteins, or crosstalk between known polymerases and pathways. This is evident from the recently discovered PrimPol, which in just the last few years has been shown to have both primase and polymerase activities [287]. Not currently classified into any of the mammalian DNA polymerase families, PrimPol is under active investigation in the field today.
Acknowledgements
Nicole M. Hoitsma and Dr. Amy M. Whitaker are co-authors and equally contributed to this paper. This work was supported by the National Institutes of Environmental Health Sciences of the National Institutes of Health under award number R35GM128562. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Nicole M. Hoitsma and Amy M. Whitaker contributed equally.
References
- 1.Wu S, Beard WA, Pedersen LG, Wilson SH. Structural comparison of DNA polymerase architecture suggest a nucleotide gateway to the polymerase active site. Chem Rev. 2014;114(5):2759–2774. doi: 10.1021/cr3005179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Delagoutte E. DNA polymerases: mechanistic insight from biochemical and biophysical studies. Front Biosci (Landmark Ed) 2012;17:509–544. doi: 10.2741/3941. [DOI] [PubMed] [Google Scholar]
- 3.Hubscher U, Nasheuer HP, Syvaoja JE. Eukaryotic DNA polymerases, a growing family. Trends Biochem Sci. 2000;25(3):143–147. doi: 10.1016/s0968-0004(99)01523-6. [DOI] [PubMed] [Google Scholar]
- 4.Yamtich J. Sweasy JB (2010) DNA polymerase family X: function, structure, and cellular roles. Biochim Biophys Acta. 1804;5:1136–1150. doi: 10.1016/j.bbapap.2009.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Braithwaite DK, Ito J. Compilation, alignment, and phylogenetic relationships of DNA polymerases. Nucleic Acids Res. 1993;21(4):787–802. doi: 10.1093/nar/21.4.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garcia-Diaz M, Bebenek K. Multiple functions of DNA polymerases. Crit Rev Plant Sci. 2007;26(2):105–122. doi: 10.1080/07352680701252817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hübscher U, Spadari S, Villani G, Maga G. DNA polymerases discovery, characterization and functions in cellular DNA transactions. Singapore: World Sci; 2010. [Google Scholar]
- 8.Ollis DL, Brick P, Hamlin R, Xuong NG, Steitz TA. Structure of large fragment of Escherichia coli DNA polymerase I complexed with dTMP. Nature. 1985;313:762. doi: 10.1038/313762a0. [DOI] [PubMed] [Google Scholar]
- 9.Beese LS, Derbyshire V, Steitz TA. Structure of DNA polymerase I Klenow fragment bound to duplex DNA. Science. 1993;260(5106):352. doi: 10.1126/science.8469987. [DOI] [PubMed] [Google Scholar]
- 10.Steitz TA. DNA polymerases: structural diversity and common mechanisms. J Biol Chem. 1999;274(25):17395–17398. doi: 10.1074/jbc.274.25.17395. [DOI] [PubMed] [Google Scholar]
- 11.Sawaya MR, Pelletier H, Kumar A, Wilson SH, Kraut J. Crystal structure of rat DNA polymerase beta: evidence for a common polymerase mechanism. Science. 1994;264(5167):1930–1935. doi: 10.1126/science.7516581. [DOI] [PubMed] [Google Scholar]
- 12.Kohlstaedt LA, Wang J, Friedman JM, Rice PA, Steitz TA. Crystal structure at 3.5 A resolution of HIV-1 reverse transcriptase complexed with an inhibitor. Science. 1992;256(5065):1783–1790. doi: 10.1126/science.1377403. [DOI] [PubMed] [Google Scholar]
- 13.Patel PH, Loeb LA. Getting a grip on how DNA polymerases function. Nat Struct Biol. 2001;8(8):656–659. doi: 10.1038/90344. [DOI] [PubMed] [Google Scholar]
- 14.Santoso Y, Joyce CM, Potapova O, Le Reste L, Hohlbein J, Torella JP, Grindley NDF, Kapanidis AN. Conformational transitions in DNA polymerase I revealed by single-molecule FRET. Proc Natl Acad Sci USA. 2010;107(2):715–720. doi: 10.1073/pnas.0910909107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beard WA, Wilson SH. Structure and mechanism of DNA polymerase β. Biochemistry. 2014;53(17):2768–2780. doi: 10.1021/bi500139h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doublié S, Sawaya MR, Ellenberger T. An open and closed case for all polymerases. Structure. 1999;7(2):R31–R35. doi: 10.1016/S0969-2126(99)80017-3. [DOI] [PubMed] [Google Scholar]
- 17.Freudenthal BD, Beard WA, Shock DD, Wilson SH. Observing a DNA polymerase choose right from wrong. Cell. 2013;154(1):157–168. doi: 10.1016/j.cell.2013.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gao Y, Yang W. Capture of a third Mg2+ is essential for catalyzing DNA synthesis. Science. 2016;352(6291):1334–1337. doi: 10.1126/science.aad9633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakamura T, Zhao Y, Yamagata Y, Hua YJ, Yang W. Watching DNA polymerase eta make a phosphodiester bond. Nature. 2012;487(7406):196–201. doi: 10.1038/nature11181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jamsen JA, Beard WA, Pedersen LC, Shock DD, Moon AF, Krahn JM, Bebenek K, Kunkel TA, Wilson SH. Time-lapse crystallography snapshots of a double-strand break repair polymerase in action. Nat Commun. 2017;8(1):253. doi: 10.1038/s41467-017-00271-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fragkos M, Ganier O, Coulombe P, Mechali M. DNA replication origin activation in space and time. Nat Rev Mol Cell Biol. 2015;16(6):360–374. doi: 10.1038/nrm4002. [DOI] [PubMed] [Google Scholar]
- 22.Campisi J. Aging, cellular senescence, and cancer. Annu Rev Physiol. 2013;75:685–705. doi: 10.1146/annurev-physiol-030212-183653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Muzi-Falconi M, Giannattasio M, Foiani M, Plevani P. The DNA polymerase alpha-primase complex: multiple functions and interactions. Sci World J. 2003;3:21–33. doi: 10.1100/tsw.2003.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kunkel TA, Erie DA. Eukaryotic mismatch repair in relation to DNA replication. Annu Rev Genet. 2015;49:291–313. doi: 10.1146/annurev-genet-112414-054722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gan GN, Wittschieben JP, Wittschieben BO, Wood RD. DNA polymerase zeta (pol [zeta]) in higher eukaryotes. Cell Res. 2008;18(1):174–183. doi: 10.1038/cr.2007.117. [DOI] [PubMed] [Google Scholar]
- 26.Johansson E, Dixon N. Replicative DNA polymerases. Cold Spring Harb Perspect Biol. 2013;5(6):a012799. doi: 10.1101/cshperspect.a012799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morrison A, Bell JB, Kunkel TA, Sugino A. Eukaryotic DNA polymerase amino acid sequence required for 3′—5′ exonuclease activity. Proc Natl Acad Sci USA. 1991;88(21):9473–9477. doi: 10.1073/pnas.88.21.9473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kunkel TA. Evolving views of DNA replication (In) fidelity. Cold Spring Harb Symp Quant Biol. 2009;74:91–101. doi: 10.1101/sqb.2009.74.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krishna TS, Kong XP, Gary S, Burgers PM, Kuriyan J. Crystal structure of the eukaryotic DNA polymerase processivity factor PCNA. Cell. 1994;79(7):1233–1243. doi: 10.1016/0092-8674(94)90014-0. [DOI] [PubMed] [Google Scholar]
- 30.Maga G, Frouin I, Spadari S, Hubscher U. Replication protein A as a “fidelity clamp” for DNA polymerase alpha. J Biol Chem. 2001;276(21):18235–18242. doi: 10.1074/jbc.M009599200. [DOI] [PubMed] [Google Scholar]
- 31.Foiani M, Lucchini G, Plevani P. The DNA polymerase alpha-primase complex couples DNA replication, cell-cycle progression and DNA-damage response. Trends Biochem Sci. 1997;22(11):424–427. doi: 10.1016/s0968-0004(97)01109-2. [DOI] [PubMed] [Google Scholar]
- 32.Yoneda M, Bollum FJ. Deoxynucleotide-polymerizing enzymes of calf thymus gland. I. large scale purification of terminal and replicative deoxynucleotidyl transferases. J Biol Chem. 1965;240:3385–3391. [PubMed] [Google Scholar]
- 33.Barnes R, Eckert K. Maintenance of genome integrity: how mammalian cells orchestrate genome duplication by coordinating replicative and specialized DNA polymerases. Genes. 2017;8(1):19. doi: 10.3390/genes8010019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schneider A, Smith RW, Kautz AR, Weisshart K, Grosse F, Nasheuer HP. Primase activity of human DNA polymerase alpha-primase. Divalent cations stabilize the enzyme activity of the p48 subunit. J Biol Chem. 1998;273(34):21608–21615. doi: 10.1074/jbc.273.34.21608. [DOI] [PubMed] [Google Scholar]
- 35.Starokadomskyy P, Gemelli T, Rios JJ, Xing C, Wang RC, Li H, Pokatayev V, Dozmorov I, Khan S, Miyata N, Fraile G, Raj P, Xu Z, Xu Z, Ma L, Lin Z, Wang H, Yang Y, Ben-Amitai D, Orenstein N, Mussaffi H, Baselga E, Tadini G, Grunebaum E, Sarajlija A, Krzewski K, Wakeland EK, Yan N, de la Morena MT, Zinn AR, Burstein E. DNA polymerase-alpha regulates the activation of type I interferons through cytosolic RNA:DNA synthesis. Nat Immunol. 2016;17(5):495–504. doi: 10.1038/ni.3409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Johnson LM, Snyder M, Chang LM, Davis RW, Campbell JL. Isolation of the gene encoding yeast DNA polymerase I. Cell. 1985;43(1):369–377. doi: 10.1016/0092-8674(85)90042-x. [DOI] [PubMed] [Google Scholar]
- 37.Copeland WC, Wang TS. Enzymatic characterization of the individual mammalian primase subunits reveals a biphasic mechanism for initiation of DNA replication. J Biol Chem. 1993;268(35):26179–26189. [PubMed] [Google Scholar]
- 38.Nick McElhinny SA, Kissling GE, Kunkel TA. Differential correction of lagging-strand replication errors made by DNA polymerases alpha and {delta} Proc Natl Acad Sci USA. 2010;107(49):21070–21075. doi: 10.1073/pnas.1013048107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pavlov YI, Frahm C, Nick McElhinny SA, Niimi A, Suzuki M, Kunkel TA. Evidence that errors made by DNA polymerase alpha are corrected by DNA polymerase delta. Curr Biol. 2006;16(2):202–207. doi: 10.1016/j.cub.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 40.Perera RL, Torella R, Klinge S, Kilkenny ML, Maman JD, Pellegrini L. Mechanism for priming DNA synthesis by yeast DNA polymerase alpha. Elife. 2013;2:e00482. doi: 10.7554/eLife.00482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Coloma J, Johnson RE, Prakash L, Prakash S, Aggarwal AK. Human DNA polymerase alpha in binary complex with a DNA:DNA template-primer. Sci Rep. 2016;6:23784. doi: 10.1038/srep23784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baranovskiy AG, Babayeva ND, Suwa Y, Gu J, Pavlov YI, Tahirov TH. Structural basis for inhibition of DNA replication by aphidicolin. Nucleic Acids Res. 2014;42(22):14013–14021. doi: 10.1093/nar/gku1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baranovskiy AG, Babayeva ND, Zhang Y, Gu J, Suwa Y, Pavlov YI, Tahirov TH. Mechanism of Concerted RNA-DNA Primer Synthesis by the Human Primosome. J Biol Chem. 2016;291(19):10006–10020. doi: 10.1074/jbc.M116.717405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vaithiyalingam S, Warren EM, Eichman BF, Chazin WJ. Insights into eukaryotic DNA priming from the structure and functional interactions of the 4Fe-4S cluster domain of human DNA primase. Proc Natl Acad Sci USA. 2010;107(31):13684–13689. doi: 10.1073/pnas.1002009107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.O’Brien E, Holt ME, Thompson MK, Salay LE, Ehlinger AC, Chazin WJ, Barton JK. The [4Fe4S] cluster of human DNA primase functions as a redox switch using DNA charge transport. Science. 2017;355(6327):1789. doi: 10.1126/science.aag1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Longley MJ, Pierce AJ, Modrich P. DNA polymerase delta is required for human mismatch repair in vitro. J Biol Chem. 1997;272(16):10917–10921. doi: 10.1074/jbc.272.16.10917. [DOI] [PubMed] [Google Scholar]
- 47.Kadyrov FA, Genschel J, Fang Y, Penland E, Edelmann W, Modrich P. A possible mechanism for exonuclease 1-independent eukaryotic mismatch repair. Proc Natl Acad Sci USA. 2009;106(21):8495–8500. doi: 10.1073/pnas.0903654106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nick McElhinny SA, Gordenin DA, Stith CM, Burgers PM, Kunkel TA. Division of labor at the eukaryotic replication fork. Mol Cell. 2008;30(2):137–144. doi: 10.1016/j.molcel.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zeng XR, Hao H, Jiang Y, Lee MY. Regulation of human DNA polymerase delta during the cell cycle. J Biol Chem. 1994;269(39):24027–24033. [PubMed] [Google Scholar]
- 50.MacNeill SA, Baldacci G, Burgers PM, Hubscher U. A unified nomenclature for the subunits of eukaryotic DNA polymerase delta. Trends Biochem Sci. 2001;26(1):16–17. doi: 10.1016/s0968-0004(00)01709-6. [DOI] [PubMed] [Google Scholar]
- 51.Chea J, Zhang S, Zhao H, Zhang Z, Lee EY, Darzynkiewicz Z, Lee MY. Spatiotemporal recruitment of human DNA polymerase delta to sites of UV damage. Cell Cycle. 2012;11(15):2885–2895. doi: 10.4161/cc.21280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu L, Rodriguez-Belmonte EM, Mazloum N, Xie B, Lee MY. Identification of a novel protein, PDIP38, that interacts with the p50 subunit of DNA polymerase delta and proliferating cell nuclear antigen. J Biol Chem. 2003;278(12):10041–10047. doi: 10.1074/jbc.M208694200. [DOI] [PubMed] [Google Scholar]
- 53.Ducoux M, Urbach S, Baldacci G, Hubscher U, Koundrioukoff S, Christensen J, Hughes P. Mediation of proliferating cell nuclear antigen (PCNA)-dependent DNA replication through a conserved p21(Cip1)-like PCNA-binding motif present in the third subunit of human DNA polymerase delta. J Biol Chem. 2001;276(52):49258–49266. doi: 10.1074/jbc.M106990200. [DOI] [PubMed] [Google Scholar]
- 54.Corrette-Bennett SE, Borgeson C, Sommer D, Burgers PM, Lahue RS. DNA polymerase delta, RFC and PCNA are required for repair synthesis of large looped heteroduplexes in Saccharomyces cerevisiae. Nucleic Acids Res. 2004;32(21):6268–6275. doi: 10.1093/nar/gkh965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Byrnes JJ, Downey KM, Black VL, So AG. A new mammalian DNA polymerase with 3′–5′ exonuclease activity: DNA polymerase delta. Biochemistry. 1976;15(13):2817–2823. doi: 10.1021/bi00658a018. [DOI] [PubMed] [Google Scholar]
- 56.Johnson RE, Klassen R, Prakash L, Prakash S. A major role of DNA polymerase delta in replication of both the leading and lagging DNA strands. Mol Cell. 2015;59(2):163–175. doi: 10.1016/j.molcel.2015.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Burgers PMJ, Gordenin D, Kunkel TA. Who is leading the replication fork, pol epsilon or pol delta? Mol Cell. 2016;61(4):492–493. doi: 10.1016/j.molcel.2016.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kunkel TA, Burgers PM. Dividing the workload at a eukaryotic replication fork. Trends Cell Biol. 2008;18(11):521–527. doi: 10.1016/j.tcb.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Burgers PM. Polymerase dynamics at the eukaryotic DNA replication fork. J Biol Chem. 2009;284(7):4041–4045. doi: 10.1074/jbc.R800062200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lujan SA, Williams JS, Kunkel TA. DNA polymerases divide the labor of genome replication. Trends Cell Biol. 2016;26(9):640–654. doi: 10.1016/j.tcb.2016.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Burgers PMJ, Kunkel TA. Eukaryotic DNA replication fork. Annu Rev Biochem. 2017;86:417–438. doi: 10.1146/annurev-biochem-061516-044709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kesti T, Flick K, Keranen S, Syvaoja JE, Wittenberg C. DNA polymerase epsilon catalytic domains are dispensable for DNA replication, DNA repair, and cell viability. Mol Cell. 1999;3(5):679–685. doi: 10.1016/s1097-2765(00)80361-5. [DOI] [PubMed] [Google Scholar]
- 63.Prindle MJ, Loeb LA. DNA polymerase delta in DNA replication and genome maintenance. Environ Mol Mutagen. 2012;53(9):666–682. doi: 10.1002/em.21745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schmitt MW, Matsumoto Y, Loeb LA. High fidelity and lesion bypass capability of human DNA polymerase delta. Biochimie. 2009;91(9):1163–1172. doi: 10.1016/j.biochi.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fortune JM, Pavlov YI, Welch CM, Johansson E, Burgers PM, Kunkel TA. Saccharomyces cerevisiae DNA polymerase delta: high fidelity for base substitutions but lower fidelity for single- and multi-base deletions. J Biol Chem. 2005;280(33):29980–29987. doi: 10.1074/jbc.M505236200. [DOI] [PubMed] [Google Scholar]
- 66.Kool ET. Active site tightness and substrate fit in DNA replication. Annu Rev Biochem. 2002;71:191–219. doi: 10.1146/annurev.biochem.71.110601.135453. [DOI] [PubMed] [Google Scholar]
- 67.Swan MK, Johnson RE, Prakash L, Prakash S, Aggarwal AK. Structural basis of high-fidelity DNA synthesis by yeast DNA polymerase delta. Nat Struct Mol Biol. 2009;16(9):979–986. doi: 10.1038/nsmb.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Prindle MJ, Schmitt MW, Parmeggiani F, Loeb LA. A substitution in the fingers domain of DNA polymerase delta reduces fidelity by altering nucleotide discrimination in the catalytic site. J Biol Chem. 2013;288(8):5572–5580. doi: 10.1074/jbc.M112.436410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Franklin MC, Wang J, Steitz TA. Structure of the replicating complex of a pol alpha family DNA polymerase. Cell. 2001;105(5):657–667. doi: 10.1016/s0092-8674(01)00367-1. [DOI] [PubMed] [Google Scholar]
- 70.Johnson SJ, Beese LS. Structures of mismatch replication errors observed in a DNA polymerase. Cell. 2004;116(6):803–816. doi: 10.1016/s0092-8674(04)00252-1. [DOI] [PubMed] [Google Scholar]
- 71.Crute JJ, Wahl AF, Bambara RA. Purification and characterization of two new high molecular weight forms of DNA polymerase delta. Biochemistry. 1986;25(1):26–36. doi: 10.1021/bi00349a005. [DOI] [PubMed] [Google Scholar]
- 72.Syvaoja JE. DNA polymerase epsilon: the latest member in the family of mammalian DNA polymerases. BioEssays. 1990;12(11):533–536. doi: 10.1002/bies.950121106. [DOI] [PubMed] [Google Scholar]
- 73.Stillman B. Reconsidering DNA polymerases at the replication fork in eukaryotes. Mol Cell. 2015;59(2):139–141. doi: 10.1016/j.molcel.2015.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nick McElhinny SA, Kumar D, Clark AB, Watt DL, Watts BE, Lundstrom EB, Johansson E, Chabes A, Kunkel TA. Genome instability due to ribonucleotide incorporation into DNA. Nat Chem Biol. 2010;6(10):774–781. doi: 10.1038/nchembio.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Miyabe I, Kunkel TA, Carr AM. The major roles of DNA polymerases epsilon and delta at the eukaryotic replication fork are evolutionarily conserved. PLoS Genet. 2011;7(12):e1002407. doi: 10.1371/journal.pgen.1002407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shcherbakova PV, Pavlov YI, Chilkova O, Rogozin IB, Johansson E, Kunkel TA. Unique error signature of the four-subunit yeast DNA polymerase epsilon. J Biol Chem. 2003;278(44):43770–43780. doi: 10.1074/jbc.M306893200. [DOI] [PubMed] [Google Scholar]
- 77.Shimizu K, Hashimoto K, Kirchner JM, Nakai W, Nishikawa H, Resnick MA, Sugino A. Fidelity of DNA polymerase epsilon holoenzyme from budding yeast Saccharomyces cerevisiae. J Biol Chem. 2002;277(40):37422–37429. doi: 10.1074/jbc.M204476200. [DOI] [PubMed] [Google Scholar]
- 78.Navas TA, Zhou Z, Elledge SJ. DNA polymerase epsilon links the DNA replication machinery to the S phase checkpoint. Cell. 1995;80(1):29–39. doi: 10.1016/0092-8674(95)90448-4. [DOI] [PubMed] [Google Scholar]
- 79.Pursell ZF, Kunkel TA. DNA polymerase ε: a polymerase of unusual size (and complexity) Prog Nucleic Acid Res Mol Biol. 2008;82:101–145. doi: 10.1016/S0079-6603(08)00004-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Asturias FJ, Cheung IK, Sabouri N, Chilkova O, Wepplo D, Johansson E. Structure of Saccharomyces cerevisiae DNA polymerase epsilon by cryo-electron microscopy. Nat Struct Mol Biol. 2006;13(1):35–43. doi: 10.1038/nsmb1040. [DOI] [PubMed] [Google Scholar]
- 81.Baranovskiy AG, Gu J, Babayeva ND, Kurinov I, Pavlov YI, Tahirov TH. Crystal structure of the human Polϵ B-subunit in complex with the C-terminal domain of the catalytic subunit. J Biol Chem. 2017;292(38):15717–15730. doi: 10.1074/jbc.m117.792705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Li Y, Pursell ZF, Linn S. Identification and cloning of two histone fold motif-containing subunits of HeLa DNA polymerase epsilon. J Biol Chem. 2000;275(30):23247–23252. doi: 10.1074/jbc.M002548200. [DOI] [PubMed] [Google Scholar]
- 83.Hogg M, Osterman P, Bylund GO, Ganai RA, Lundstrom EB, Sauer-Eriksson AE, Johansson E. Structural basis for processive DNA synthesis by yeast DNA polymerase varepsilon. Nat Struct Mol Biol. 2014;21(1):49–55. doi: 10.1038/nsmb.2712. [DOI] [PubMed] [Google Scholar]
- 84.Jain R, Rajashankar KR, Buku A, Johnson RE, Prakash L, Prakash S, Aggarwal AK. Crystal structure of yeast DNA polymerase epsilon catalytic domain. PLoS One. 2014;9(4):e94835. doi: 10.1371/journal.pone.0094835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Doublié S, Zahn KE. Structural insights into eukaryotic DNA replication. Front Microbiol. 2014;5:444. doi: 10.3389/fmicb.2014.00444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Schaich MA, Smith MR, Cloud AS, Holloran SM, Freudenthal BD. Structures of a DNA polymerase inserting therapeutic nucleotide analogues. Chem Res Toxicol. 2017;30(11):1993–2001. doi: 10.1021/acs.chemrestox.7b00173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lemontt JF. Mutants of yeast defective in mutation induced by ultraviolet light. Genetics. 1971;68(1):21–33. doi: 10.1093/genetics/68.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Prakash L. Effect of genes controlling radiation sensitivity on chemically induced mutations in Saccharomyces cerevisiae. Genetics. 1976;83(2):285–301. doi: 10.1093/genetics/83.2.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.McKee RH, Lawrence CW. Genetic analysis of gamma-ray mutagenesis in yeast. I. Reversion in radiation-sensitive strains. Genetics. 1979;93(2):361–373. doi: 10.1093/genetics/93.2.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lawrence CW, Das G, Christensen RB. REV7, a new gene concerned with UV mutagenesis in yeast. Mol Gen Genet. 1985;200(1):80–85. doi: 10.1007/BF00383316. [DOI] [PubMed] [Google Scholar]
- 91.Morrison A, Araki H, Clark AB, Hamatake RK, Sugino A. A third essential DNA polymerase in S. cerevisiae. Cell. 1990;62(6):1143–1151. doi: 10.1016/0092-8674(90)90391-q. [DOI] [PubMed] [Google Scholar]
- 92.Nelson JR, Lawrence CW, Hinkle DC. Thymine-thymine dimer bypass by yeast DNA polymerase zeta. Science. 1996;272(5268):1646–1649. doi: 10.1126/science.272.5268.1646. [DOI] [PubMed] [Google Scholar]
- 93.Zhong X, Garg P, Stith CM, Nick McElhinny SA, Kissling GE, Burgers PM, Kunkel TA. The fidelity of DNA synthesis by yeast DNA polymerase zeta alone and with accessory proteins. Nucleic Acids Res. 2006;34(17):4731–4742. doi: 10.1093/nar/gkl465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Vaisman A, Woodgate R. Translesion DNA polymerases in eukaryotes: what makes them tick? Crit Rev Biochem Mol Biol. 2017;52(3):274–303. doi: 10.1080/10409238.2017.1291576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hara K, Hashimoto H, Murakumo Y, Kobayashi S, Kogame T, Unzai S, Akashi S, Takeda S, Shimizu T, Sato M. Crystal structure of human REV7 in complex with a human REV3 fragment and structural implication of the interaction between DNA polymerase zeta and REV1. J Biol Chem. 2010;285(16):12299–12307. doi: 10.1074/jbc.M109.092403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kikuchi S, Hara K, Shimizu T, Sato M, Hashimoto H. Structural basis of recruitment of DNA polymerase zeta by interaction between REV1 and REV7 proteins. J Biol Chem. 2012;287(40):33847–33852. doi: 10.1074/jbc.M112.396838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Brandao LN, Ferguson R, Santoro I, Jinks-Robertson S, Sclafani RA. The role of Dbf4-dependent protein kinase in DNA polymerase zeta-dependent mutagenesis in Saccharomyces cerevisiae. Genetics. 2014;197(4):1111–1122. doi: 10.1534/genetics.114.165308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Fattah FJ, Hara K, Fattah KR, Yang C, Wu N, Warrington R, Chen DJ, Zhou P, Boothman DA, Yu H. The transcription factor TFII-I promotes DNA translesion synthesis and genomic stability. PLoS Genet. 2014;10(6):e1004419. doi: 10.1371/journal.pgen.1004419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Murakumo Y, Roth T, Ishii H, Rasio D, Numata S, Croce CM, Fishel R. A human REV7 homolog that interacts with the polymerase zeta catalytic subunit hREV3 and the spindle assembly checkpoint protein hMAD2. J Biol Chem. 2000;275(6):4391–4397. doi: 10.1074/jbc.275.6.4391. [DOI] [PubMed] [Google Scholar]
- 100.Makarova AV, Stodola JL, Burgers PM. A four-subunit DNA polymerase zeta complex containing Pol delta accessory subunits is essential for PCNA-mediated mutagenesis. Nucleic Acids Res. 2012;40(22):11618–11626. doi: 10.1093/nar/gks948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lee YS, Gregory MT, Yang W. Human Pol zeta purified with accessory subunits is active in translesion DNA synthesis and complements Pol eta in cisplatin bypass. Proc Natl Acad Sci USA. 2014;111(8):2954–2959. doi: 10.1073/pnas.1324001111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Gomez-Llorente Y, Malik R, Jain R, Choudhury JR, Johnson RE, Prakash L, Prakash S, Ubarretxena-Belandia I, Aggarwal AK. The architecture of yeast DNA polymerase zeta. Cell Rep. 2013;5(1):79–86. doi: 10.1016/j.celrep.2013.08.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Jain R, Hammel M, Johnson RE, Prakash L, Prakash S, Aggarwal AK. Structural insights into yeast DNA polymerase delta by small angle X-ray scattering. J Mol Biol. 2009;394(3):377–382. doi: 10.1016/j.jmb.2009.09.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Nunez-Ramirez R, Klinge S, Sauguet L, Melero R, Recuero-Checa MA, Kilkenny M, Perera RL, Garcia-Alvarez B, Hall RJ, Nogales E, Pellegrini L, Llorca O. Flexible tethering of primase and DNA Pol alpha in the eukaryotic primosome. Nucleic Acids Res. 2011;39(18):8187–8199. doi: 10.1093/nar/gkr534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Johnson RE, Washington MT, Haracska L, Prakash S, Prakash L. Eukaryotic polymerases iota and zeta act sequentially to bypass DNA lesions. Nature. 2000;406(6799):1015–1019. doi: 10.1038/35023030. [DOI] [PubMed] [Google Scholar]
- 106.Haracska L, Prakash S, Prakash L. Yeast DNA polymerase zeta is an efficient extender of primer ends opposite from 7,8-dihydro-8-Oxoguanine and O6-methylguanine. Mol Cell Biol. 2003;23(4):1453–1459. doi: 10.1128/MCB.23.4.1453-1459.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Northam MR, Robinson HA, Kochenova OV, Shcherbakova PV. Participation of DNA polymerase zeta in replication of undamaged DNA in Saccharomyces cerevisiae. Genetics. 2010;184(1):27–42. doi: 10.1534/genetics.109.107482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lemontt JF. Induction of forward mutations in mutationally defective yeast. Mol Gen Genet. 1972;119(1):27–42. doi: 10.1007/BF00270441. [DOI] [PubMed] [Google Scholar]
- 109.Quah SK, von Borstel RC, Hastings PJ. The origin of spontaneous mutation in Saccharomyces cerevisiae. Genetics. 1980;96(4):819–839. doi: 10.1093/genetics/96.4.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ohmori H, Friedberg EC, Fuchs RP, Goodman MF, Hanaoka F, Hinkle D, Kunkel TA, Lawrence CW, Livneh Z, Nohmi T, Prakash L, Prakash S, Todo T, Walker GC, Wang Z, Woodgate R. The Y-family of DNA polymerases. Mol Cell. 2001;8(1):7–8. doi: 10.1016/s1097-2765(01)00278-7. [DOI] [PubMed] [Google Scholar]
- 111.Ling H, Boudsocq F, Woodgate R, Yang W. Crystal structure of a Y-family DNA polymerase in action: a mechanism for error-prone and lesion-bypass replication. Cell. 2001;107(1):91–102. doi: 10.1016/s0092-8674(01)00515-3. [DOI] [PubMed] [Google Scholar]
- 112.Silvian LF, Toth EA, Pham P, Goodman MF, Ellenberger T. Crystal structure of a DinB family error-prone DNA polymerase from Sulfolobus solfataricus. Nat Struct Biol. 2001;8(11):984–989. doi: 10.1038/nsb1101-984. [DOI] [PubMed] [Google Scholar]
- 113.Trincao J, Johnson RE, Escalante CR, Prakash S, Prakash L, Aggarwal AK. Structure of the catalytic core of S. cerevisiae DNA polymerase eta: implications for translesion DNA synthesis. Mol Cell. 2001;8(2):417–426. doi: 10.1016/s1097-2765(01)00306-9. [DOI] [PubMed] [Google Scholar]
- 114.McCulloch SD, Kunkel TA. The fidelity of DNA synthesis by eukaryotic replicative and translesion synthesis polymerases. Cell Res. 2008;18(1):148–161. doi: 10.1038/cr.2008.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kulaeva OI, Koonin EV, McDonald JP, Randall SK, Rabinovich N, Connaughton JF, Levine AS, Woodgate R. Identification of a DinB/UmuC homolog in the archeon Sulfolobus solfataricus. Mutat Res. 1996;357(1–2):245–253. doi: 10.1016/0027-5107(96)00164-9. [DOI] [PubMed] [Google Scholar]
- 116.McDonald JP, Levine AS, Woodgate R. The Saccharomyces cerevisiae RAD30 gene, a homologue of Escherichia coli dinB and umuC, is DNA damage inducible and functions in a novel error-free postreplication repair mechanism. Genetics. 1997;147(4):1557–1568. doi: 10.1093/genetics/147.4.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Johnson RE, Prakash S, Prakash L. Requirement of DNA polymerase activity of yeast Rad30 protein for its biological function. J Biol Chem. 1999;274(23):15975–15977. doi: 10.1074/jbc.274.23.15975. [DOI] [PubMed] [Google Scholar]
- 118.Haracska L, Yu SL, Johnson RE, Prakash L, Prakash S. Efficient and accurate replication in the presence of 7,8-dihydro-8-oxoguanine by DNA polymerase eta. Nat Genet. 2000;25(4):458–461. doi: 10.1038/78169. [DOI] [PubMed] [Google Scholar]
- 119.Johnson RE, Kondratick CM, Prakash S, Prakash L. hRAD30 mutations in the variant form of xeroderma pigmentosum. Science. 1999;285(5425):263–265. doi: 10.1126/science.285.5425.263. [DOI] [PubMed] [Google Scholar]
- 120.Masutani C, Kusumoto R, Yamada A, Dohmae N, Yokoi M, Yuasa M, Araki M, Iwai S, Takio K, Hanaoka F. The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase eta. Nature. 1999;399(6737):700–704. doi: 10.1038/21447. [DOI] [PubMed] [Google Scholar]
- 121.Silverstein TD, Johnson RE, Jain R, Prakash L, Prakash S, Aggarwal AK. Structural basis for the suppression of skin cancers by DNA polymerase eta. Nature. 2010;465(7301):1039–1043. doi: 10.1038/nature09104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Biertumpfel C, Zhao Y, Kondo Y, Ramon-Maiques S, Gregory M, Lee JY, Masutani C, Lehmann AR, Hanaoka F, Yang W. Structure and mechanism of human DNA polymerase eta. Nature. 2010;465(7301):1044–1048. doi: 10.1038/nature09196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Washington MT, Helquist SA, Kool ET, Prakash L, Prakash S. Requirement of Watson-Crick hydrogen bonding for DNA synthesis by yeast DNA polymerase eta. Mol Cell Biol. 2003;23(14):5107–5112. doi: 10.1128/MCB.23.14.5107-5112.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Yu SL, Johnson RE, Prakash S, Prakash L. Requirement of DNA polymerase eta for error-free bypass of UV-induced CC and TC photoproducts. Mol Cell Biol. 2001;21(1):185–188. doi: 10.1128/MCB.21.1.185-188.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Haracska L, Washington MT, Prakash S, Prakash L. Inefficient bypass of an abasic site by DNA polymerase eta. J Biol Chem. 2001;276(9):6861–6866. doi: 10.1074/jbc.M008021200. [DOI] [PubMed] [Google Scholar]
- 126.Zhao Y, Gregory MT, Biertumpfel C, Hua YJ, Hanaoka F, Yang W. Mechanism of somatic hypermutation at the WA motif by human DNA polymerase eta. Proc Natl Acad Sci USA. 2013;110(20):8146–8151. doi: 10.1073/pnas.1303126110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Saribasak H, Rajagopal D, Maul RW, Gearhart PJ. Hijacked DNA repair proteins and unchained DNA polymerases. Philos Trans R Soc Lond B Biol Sci. 2009;364(1517):605–611. doi: 10.1098/rstb.2008.0188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kawamoto T, Araki K, Sonoda E, Yamashita YM, Harada K, Kikuchi K, Masutani C, Hanaoka F, Nozaki K, Hashimoto N, Takeda S. Dual roles for DNA polymerase eta in homologous DNA recombination and translesion DNA synthesis. Mol Cell. 2005;20(5):793–799. doi: 10.1016/j.molcel.2005.10.016. [DOI] [PubMed] [Google Scholar]
- 129.McIlwraith MJ, Vaisman A, Liu Y, Fanning E, Woodgate R, West SC. Human DNA polymerase eta promotes DNA synthesis from strand invasion intermediates of homologous recombination. Mol Cell. 2005;20(5):783–792. doi: 10.1016/j.molcel.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 130.Tissier A, McDonald JP, Frank EG, Woodgate R. poliota, a remarkably error-prone human DNA polymerase. Genes Dev. 2000;14(13):1642–1650. [PMC free article] [PubMed] [Google Scholar]
- 131.Vaisman A, Frank EG, McDonald JP, Tissier A, Woodgate R. poliota-dependent lesion bypass in vitro. Mutat Res. 2002;510(1–2):9–22. doi: 10.1016/s0027-5107(02)00248-8. [DOI] [PubMed] [Google Scholar]
- 132.Ishikawa T, Uematsu N, Mizukoshi T, Iwai S, Iwasaki H, Masutani C, Hanaoka F, Ueda R, Ohmori H, Todo T. Mutagenic and nonmutagenic bypass of DNA lesions by Drosophila DNA polymerases dpoleta and dpoliota. J Biol Chem. 2001;276(18):15155–15163. doi: 10.1074/jbc.M009822200. [DOI] [PubMed] [Google Scholar]
- 133.Washington MT, Johnson RE, Prakash L, Prakash S. Human DNA polymerase iota utilizes different nucleotide incorporation mechanisms dependent upon the template base. Mol Cell Biol. 2004;24(2):936–943. doi: 10.1128/MCB.24.2.936-943.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Zhang Y, Yuan F, Wu X, Wang Z. Preferential incorporation of G opposite template T by the low-fidelity human DNA polymerase iota. Mol Cell Biol. 2000;20(19):7099–7108. doi: 10.1128/mcb.20.19.7099-7108.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Nair DT, Johnson RE, Prakash L, Prakash S, Aggarwal AK. An incoming nucleotide imposes an anti to syn conformational change on the templating purine in the human DNA polymerase-iota active site. Structure. 2006;14(4):749–755. doi: 10.1016/j.str.2006.01.010. [DOI] [PubMed] [Google Scholar]
- 136.Nair DT, Johnson RE, Prakash S, Prakash L, Aggarwal AK. Replication by human DNA polymerase-iota occurs by Hoogsteen base-pairing. Nature. 2004;430(6997):377–380. doi: 10.1038/nature02692. [DOI] [PubMed] [Google Scholar]
- 137.Makarova AV, Kulbachinskiy AV. Structure of human DNA polymerase iota and the mechanism of DNA synthesis. Biochemistry (Mosc) 2012;77(6):547–561. doi: 10.1134/S0006297912060016. [DOI] [PubMed] [Google Scholar]
- 138.Washington MT, Carlson KD, Freudenthal BD, Pryor JM. Variations on a theme: eukaryotic Y-family DNA polymerases. Biochim Biophys Acta. 2010;1804:1113–1123. doi: 10.1016/j.bbapap.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Frank EG, Sayer JM, Kroth H, Ohashi E, Ohmori H, Jerina DM, Woodgate R. Translesion replication of benzo[a]pyrene and benzo[c]phenanthrene diol epoxide adducts of deoxyadenosine and deoxyguanosine by human DNA polymerase iota. Nucleic Acids Res. 2002;30(23):5284–5292. doi: 10.1093/nar/gkf643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Pence MG, Choi JY, Egli M, Guengerich FP. Structural basis for proficient incorporation of dTTP opposite O6-methylguanine by human DNA polymerase iota. J Biol Chem. 2010;285(52):40666–40672. doi: 10.1074/jbc.M110.183665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Nair DT, Johnson RE, Prakash L, Prakash S, Aggarwal AK. Hoogsteen base pair formation promotes synthesis opposite the 1, N6-ethenodeoxyadenosine lesion by human DNA polymerase iota. Nat Struct Mol Biol. 2006;13(7):619–625. doi: 10.1038/nsmb1118. [DOI] [PubMed] [Google Scholar]
- 142.Donny-Clark K, Shapiro R, Broyde S. Accommodation of an N-(deoxyguanosin-8-yl)-2-acetylaminofluorene adduct in the active site of human DNA polymerase iota: hoogsteen or Watson–Crick base pairing? Biochemistry. 2009;48(1):7–18. doi: 10.1021/bi801283d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Donny-Clark K, Broyde S. Influence of local sequence context on damaged base conformation in human DNA polymerase iota: molecular dynamics studies of nucleotide incorporation opposite a benzo[a]pyrene-derived adenine lesion. Nucleic Acids Res. 2009;37(21):7095–7109. doi: 10.1093/nar/gkp745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Gerlach VL, Aravind L, Gotway G, Schultz RA, Koonin EV, Friedberg EC. Human and mouse homologs of Escherichia coli DinB (DNA polymerase IV), members of the UmuC/DinB superfamily. Proc Natl Acad Sci USA. 1999;96(21):11922–11927. doi: 10.1073/pnas.96.21.11922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Ohashi E, Ogi T, Kusumoto R, Iwai S, Masutani C, Hanaoka F, Ohmori H. Error-prone bypass of certain DNA lesions by the human DNA polymerase kappa. Genes Dev. 2000;14(13):1589–1594. [PMC free article] [PubMed] [Google Scholar]
- 146.Johnson RE, Prakash S, Prakash L. The human DINB1 gene encodes the DNA polymerase Poltheta. Proc Natl Acad Sci USA. 2000;97(8):3838–3843. doi: 10.1073/pnas.97.8.3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Washington MT, Johnson RE, Prakash L, Prakash S. Human DINB1-encoded DNA polymerase kappa is a promiscuous extender of mispaired primer termini. Proc Natl Acad Sci USA. 2002;99(4):1910–1914. doi: 10.1073/pnas.032594399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Haracska L, Prakash L, Prakash S. Role of human DNA polymerase kappa as an extender in translesion synthesis. Proc Natl Acad Sci U S A. 2002;99(25):16000–16005. doi: 10.1073/pnas.252524999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Zhang Y, Wu X, Guo D, Rechkoblit O, Wang Z. Activities of human DNA polymerase kappa in response to the major benzo[a]pyrene DNA adduct: error-free lesion bypass and extension synthesis from opposite the lesion. DNA Repair. 2002;1(7):559–569. doi: 10.1016/s1568-7864(02)00055-1. [DOI] [PubMed] [Google Scholar]
- 150.Washington MT, Minko IG, Johnson RE, Wolfle WT, Harris TM, Lloyd RS, Prakash S, Prakash L. Efficient and error-free replication past a minor-groove DNA adduct by the sequential action of human DNA polymerases iota and kappa. Mol Cell Biol. 2004;24(13):5687–5693. doi: 10.1128/MCB.24.13.5687-5693.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Wolfle WT, Johnson RE, Minko IG, Lloyd RS, Prakash S, Prakash L. Replication past a trans-4-hydroxynonenal minor-groove adduct by the sequential action of human DNA polymerases iota and kappa. Mol Cell Biol. 2006;26(1):381–386. doi: 10.1128/MCB.26.1.381-386.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Tonzi P, Yin Y, Lee CWT, Rothenberg E, Huang TT. Translesion polymerase kappa-dependent DNA synthesis underlies replication fork recovery. Elife. 2018 doi: 10.7554/eLife.41426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Uljon SN, Johnson RE, Edwards TA, Prakash S, Prakash L, Aggarwal AK. Crystal structure of the catalytic core of human DNA polymerase kappa. Structure. 2004;12(8):1395–1404. doi: 10.1016/j.str.2004.05.011. [DOI] [PubMed] [Google Scholar]
- 154.Lone S, Townson SA, Uljon SN, Johnson RE, Brahma A, Nair DT, Prakash S, Prakash L, Aggarwal AK. Human DNA polymerase kappa encircles DNA: implications for mismatch extension and lesion bypass. Mol Cell. 2007;25(4):601–614. doi: 10.1016/j.molcel.2007.01.018. [DOI] [PubMed] [Google Scholar]
- 155.Zhang Y, Yuan F, Xin H, Wu X, Rajpal DK, Yang D, Wang Z. Human DNA polymerase kappa synthesizes DNA with extraordinarily low fidelity. Nucleic Acids Res. 2000;28(21):4147–4156. doi: 10.1093/nar/28.21.4147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Liu Y, Yang Y, Tang TS, Zhang H, Wang Z, Friedberg E, Yang W, Guo C. Variants of mouse DNA polymerase kappa reveal a mechanism of efficient and accurate translesion synthesis past a benzo[a]pyrene dG adduct. Proc Natl Acad Sci USA. 2014;111(5):1789–1794. doi: 10.1073/pnas.1324168111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Jha V, Bian C, Xing G, Ling H. Structure and mechanism of error-free replication past the major benzo[a]pyrene adduct by human DNA polymerase kappa. Nucleic Acids Res. 2016;44(10):4957–4967. doi: 10.1093/nar/gkw204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Jha V, Ling H. 2.0 A resolution crystal structure of human polkappa reveals a new catalytic function of N-clasp in DNA replication. Sci Rep. 2018;8(1):15125. doi: 10.1038/s41598-018-33371-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Nelson JR, Lawrence CW, Hinkle DC. Deoxycytidyl transferase activity of yeast REV1 protein. Nature. 1996;382(6593):729–731. doi: 10.1038/382729a0. [DOI] [PubMed] [Google Scholar]
- 160.Nair DT, Johnson RE, Prakash L, Prakash S, Aggarwal AK. Rev1 employs a novel mechanism of DNA synthesis using a protein template. Science. 2005;309(5744):2219–2222. doi: 10.1126/science.1116336. [DOI] [PubMed] [Google Scholar]
- 161.Swan MK, Johnson RE, Prakash L, Prakash S, Aggarwal AK. Structure of the human Rev1-DNA-dNTP ternary complex. J Mol Biol. 2009;390(4):699–709. doi: 10.1016/j.jmb.2009.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Rechkoblit O, Kolbanovskiy A, Landes H, Geacintov NE, Aggarwal AK. Mechanism of error-free replication across benzo[a]pyrene stereoisomers by Rev1 DNA polymerase. Nat Commun. 2017;8(1):965. doi: 10.1038/s41467-017-01013-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Nair DT, Johnson RE, Prakash L, Prakash S, Aggarwal AK. DNA synthesis across an abasic lesion by yeast REV1 DNA polymerase. J Mol Biol. 2011;406(1):18–28. doi: 10.1016/j.jmb.2010.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Nair DT, Johnson RE, Prakash L, Prakash S, Aggarwal AK. Protein-template-directed synthesis across an acrolein-derived DNA adduct by yeast Rev1 DNA polymerase. Structure. 2008;16(2):239–245. doi: 10.1016/j.str.2007.12.009. [DOI] [PubMed] [Google Scholar]
- 165.Pustovalova Y, Magalhaes MT, D’Souza S, Rizzo AA, Korza G, Walker GC, Korzhnev DM. Interaction between the Rev1 C-terminal domain and the PolD3 subunit of polzeta suggests a mechanism of polymerase exchange upon Rev1/Polzeta-dependent translesion synthesis. Biochemistry. 2016;55(13):2043–2053. doi: 10.1021/acs.biochem.5b01282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Wojtaszek J, Lee CJ, D’Souza S, Minesinger B, Kim H, D’Andrea AD, Walker GC, Zhou P. Structural basis of Rev1-mediated assembly of a quaternary vertebrate translesion polymerase complex consisting of Rev1, heterodimeric polymerase (Pol) zeta, and Pol kappa. J Biol Chem. 2012;287(40):33836–33846. doi: 10.1074/jbc.M112.394841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Wang Z, Castaño IB, Adams C, Vu C, Fitzhugh D, Christman MF. Structure/function analysis of the Saccharomyces cerevisiae Trf4/Pol sigma DNA polymerase. Genetics. 2002;160(2):381–391. doi: 10.1093/genetics/160.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Wang Z, Castano IB, De Las Penas A, Adams C, Christman MF. Pol kappa: a DNA polymerase required for sister chromatid cohesion. Science. 2000;289(5480):774–779. doi: 10.1126/science.289.5480.774. [DOI] [PubMed] [Google Scholar]
- 169.Motea EA. Berdis AJ (2010) terminal deoxynucleotidyl transferase: the story of a misguided DNA polymerase. Biochim Biophys Acta. 1804;5:1151–1166. doi: 10.1016/j.bbapap.2009.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Chang LM, Bollum FJ. Low molecular weight deoxyribonucleic acid polymerase in mammalian cells. J Biol Chem. 1971;246(18):5835–5837. [PubMed] [Google Scholar]
- 171.Bose-Basu B, DeRose EF, Kirby TW, Mueller GA, Beard WA, Wilson SH, London RE. Dynamic characterization of a DNA repair enzyme: NMR studies of [methyl-13C]methionine-labeled DNA polymerase beta. Biochemistry. 2004;43(28):8911–8922. doi: 10.1021/bi049641n. [DOI] [PubMed] [Google Scholar]
- 172.Freudenthal BD, Beard WA, Wilson SH. Structures of dNTP intermediate states during DNA polymerase active site assembly. Structure. 2012;20(11):1829–1837. doi: 10.1016/j.str.2012.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Werneburg BG, Ahn J, Zhong X, Hondal RJ, Kraynov VS, Tsai MD. DNA polymerase beta: pre-steady-state kinetic analysis and roles of arginine-283 in catalysis and fidelity. Biochemistry. 1996;35(22):7041–7050. doi: 10.1021/bi9527202. [DOI] [PubMed] [Google Scholar]
- 174.Podlutsky AJ, Dianova I, Wilson SH, Bohr VA, Dianov GL. DNA synthesis and dRPase activities of polymerase beta are both essential for single-nucleotide patch base excision repair in mammalian cell extracts. Biochemistry. 2001;40(3):809–813. doi: 10.1021/bi002064s. [DOI] [PubMed] [Google Scholar]
- 175.Kaufman BA, Van Houten B. POLB: a new role of DNA polymerase beta in mitochondrial base excision repair. DNA Repair. 2017;60:A1–a5. doi: 10.1016/j.dnarep.2017.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Prasad R, Caglayan M, Dai DP, Nadalutti CA, Zhao ML, Gassman NR, Janoshazi AK, Stefanick DF, Horton JK, Krasich R, Longley MJ, Copeland WC, Griffith JD, Wilson SH. DNA polymerase beta: a missing link of the base excision repair machinery in mammalian mitochondria. DNA Repair (Amst) 2017;60:77–88. doi: 10.1016/j.dnarep.2017.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Ray S, Breuer G, DeVeaux M, Zelterman D, Bindra R, Sweasy JB. DNA polymerase beta participates in DNA End-joining. Nucleic Acids Res. 2017 doi: 10.1093/nar/gkx1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Matsumoto Y, Kim K. Excision of deoxyribose phosphate residues by DNA polymerase beta during DNA repair. Science. 1995;269(5224):699–702. doi: 10.1126/science.7624801. [DOI] [PubMed] [Google Scholar]
- 179.Prasad R, Beard WA, Strauss PR, Wilson SH. Human DNA polymerase beta deoxyribose phosphate lyase. Substrate specificity and catalytic mechanism. J Biol Chem. 1998;273(24):15263–15270. doi: 10.1074/jbc.273.24.15263. [DOI] [PubMed] [Google Scholar]
- 180.Sobol RW, Prasad R, Evenski A, Baker A, Yang XP, Horton JK, Wilson SH. The lyase activity of the DNA repair protein beta-polymerase protects from DNA-damage-induced cytotoxicity. Nature. 2000;405(6788):807–810. doi: 10.1038/35015598. [DOI] [PubMed] [Google Scholar]
- 181.Whitaker AM, Smith MR, Schaich MA, Freudenthal BD. Capturing a mammalian DNA polymerase extending from an oxidized nucleotide. Nucleic Acids Res. 2017;45(11):6934–6944. doi: 10.1093/nar/gkx293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Lin P, Pedersen LC, Batra VK, Beard WA, Wilson SH, Pedersen LG. Energy analysis of chemistry for correct insertion by DNA polymerase beta. Proc Natl Acad Sci USA. 2006;103(36):13294–13299. doi: 10.1073/pnas.0606006103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Batra VK, Perera L, Lin P, Shock DD, Beard WA, Pedersen LC, Pedersen LG, Wilson SH. Amino acid substitution in the active site of DNA polymerase beta explains the energy barrier of the nucleotidyl transfer reaction. J Am Chem Soc. 2013;135(21):8078–8088. doi: 10.1021/ja403842j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Davies JF, Almassy RJ, Hostomska Z, Ferre RA, Hostomsky Z. 2.3 a crystal structure of the catalytic domain of DNA polymerase beta. Cell. 1994;76(6):1123–1133. doi: 10.1016/0092-8674(94)90388-3. [DOI] [PubMed] [Google Scholar]
- 185.Freudenthal BD, Beard WA, Perera L, Shock DD, Kim T, Schlick T, Wilson SH. Uncovering the polymerase-induced cytotoxicity of an oxidized nucleotide. Nature. 2015;517(7536):635–639. doi: 10.1038/nature13886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Freudenthal BD, Beard WA, Wilson SH. New structural snapshots provide molecular insights into the mechanism of high fidelity DNA synthesis. DNA Repair. 2015;32:3–9. doi: 10.1016/j.dnarep.2015.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Reed AJ, Suo Z. Time-dependent extension from an 8-oxoguanine lesion by human DNA polymerase beta. J Am Chem Soc. 2017;139(28):9684–9690. doi: 10.1021/jacs.7b05048. [DOI] [PubMed] [Google Scholar]
- 188.Vyas R, Reed AJ, Tokarsky EJ, Suo Z. Viewing human DNA polymerase beta faithfully and unfaithfully bypass an oxidative lesion by time-dependent crystallography. J Am Chem Soc. 2015;137(15):5225–5230. doi: 10.1021/jacs.5b02109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Reed AJ, Vyas R, Raper AT, Suo Z. Structural insights into the post-chemistry steps of nucleotide incorporation catalyzed by a DNA polymerase. J Am Chem Soc. 2017;139(1):465–471. doi: 10.1021/jacs.6b11258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Garcia-Diaz M, Dominguez O, Lopez-Fernandez LA, de Lera LT, Saniger ML, Ruiz JF, Parraga M, Garcia-Ortiz MJ, Kirchhoff T, del Mazo J, Bernad A, Blanco L. DNA polymerase lambda (Pol lambda), a novel eukaryotic DNA polymerase with a potential role in meiosis. J Mol Biol. 2000;301(4):851–867. doi: 10.1006/jmbi.2000.4005. [DOI] [PubMed] [Google Scholar]
- 191.Maga G, Villani G, Ramadan K, Shevelev I, Tanguy Le Gac N, Blanco L, Blanca G, Spadari S, Hubscher U. Human DNA polymerase lambda functionally and physically interacts with proliferating cell nuclear antigen in normal and translesion DNA synthesis. J Biol Chem. 2002;277(50):48434–48440. doi: 10.1074/jbc.M206889200. [DOI] [PubMed] [Google Scholar]
- 192.Braithwaite EK, Kedar PS, Stumpo DJ, Bertocci B, Freedman JH, Samson LD, Wilson SH. DNA polymerases beta and lambda mediate overlapping and independent roles in base excision repair in mouse embryonic fibroblasts. PLoS One. 2010;5(8):e12229. doi: 10.1371/journal.pone.0012229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Lee JW, Blanco L, Zhou T, Garcia-Diaz M, Bebenek K, Kunkel TA, Wang Z, Povirk LF. Implication of DNA polymerase lambda in alignment-based gap filling for nonhomologous DNA end joining in human nuclear extracts. J Biol Chem. 2004;279(1):805–811. doi: 10.1074/jbc.M307913200. [DOI] [PubMed] [Google Scholar]
- 194.Uchiyama Y, Takeuchi R, Kodera H, Sakaguchi K. Distribution and roles of X-family DNA polymerases in eukaryotes. Biochimie. 2009;91(2):165–170. doi: 10.1016/j.biochi.2008.07.005. [DOI] [PubMed] [Google Scholar]
- 195.Garcia-Diaz M, Bebenek K, Kunkel TA, Blanco L. Identification of an intrinsic 5′-deoxyribose-5-phosphate lyase activity in human DNA polymerase lambda: a possible role in base excision repair. J Biol Chem. 2001;276(37):34659–34663. doi: 10.1074/jbc.M106336200. [DOI] [PubMed] [Google Scholar]
- 196.Braithwaite EK, Prasad R, Shock DD, Hou EW, Beard WA, Wilson SH. DNA polymerase λ mediates a back-up base excision repair activity in extracts of mouse embryonic fibroblasts. J Biol Chem. 2005;280(18):18469–18475. doi: 10.1074/jbc.M411864200. [DOI] [PubMed] [Google Scholar]
- 197.Bebenek K, Pedersen LC, Kunkel TA. Structure-function studies of DNA polymerase λ. Biochemistry. 2014;53(17):2781–2792. doi: 10.1021/bi4017236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.van Loon B, Hubscher U, Maga G. Living on the edge: DNA polymerase lambda between genome stability and mutagenesis. Chem Res Toxicol. 2017;30(11):1936–1941. doi: 10.1021/acs.chemrestox.7b00152. [DOI] [PubMed] [Google Scholar]
- 199.DeRose EF, Kirby TW, Mueller GA, Bebenek K, Garcia-Diaz M, Blanco L, Kunkel TA, London RE. Solution structure of the lyase domain of human DNA polymerase lambda. Biochemistry. 2003;42(32):9564–9574. doi: 10.1021/bi034298s. [DOI] [PubMed] [Google Scholar]
- 200.Wimmer U, Ferrari E, Hunziker P, Hubscher U. Control of DNA polymerase lambda stability by phosphorylation and ubiquitination during the cell cycle. EMBO Rep. 2008;9(10):1027–1033. doi: 10.1038/embor.2008.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Mueller GA, Moon AF, DeRose EF, Havener JM, Ramsden DA, Pedersen LC, London RE. A comparison of BRCT domains involved in nonhomologous end-joining: introducing the solution structure of the BRCT domain of polymerase lambda. DNA Repair. 2008;7(8):1340–1351. doi: 10.1016/j.dnarep.2008.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Garcia-Diaz M, Bebenek K, Krahn JM, Blanco L, Kunkel TA, Pedersen LC. A structural solution for the DNA polymerase lambda-dependent repair of DNA gaps with minimal homology. Mol Cell. 2004;13(4):561–572. doi: 10.1016/s1097-2765(04)00061-9. [DOI] [PubMed] [Google Scholar]
- 203.Bebenek K, Garcia-Diaz M, Zhou R, Povirk LF, Kunkel TA. Loop 1 modulates the fidelity of DNA polymerase λ. Nucleic Acids Res. 2010;38(16):5419–5431. doi: 10.1093/nar/gkq261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Garcia-Diaz M, Bebenek K, Larrea AA, Havener JM, Perera L, Krahn JM, Pedersen LC, Ramsden DA, Kunkel TA. Template strand scrunching during DNA gap repair synthesis by human polymerase λ. Nat Struct Mol Biol. 2009;16:967. doi: 10.1038/nsmb.1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Bebenek K, Garcia-Diaz M, Blanco L, Kunkel TA. The frameshift infidelity of human DNA polymerase lambda. Implications for function. J Biol Chem. 2003;278(36):34685–34690. doi: 10.1074/jbc.M305705200. [DOI] [PubMed] [Google Scholar]
- 206.Bebenek K, Garcia-Diaz M, Foley MC, Pedersen LC, Schlick T, Kunkel TA. Substrate-induced DNA strand misalignment during catalytic cycling by DNA polymerase lambda. EMBO Rep. 2008;9(5):459–464. doi: 10.1038/embor.2008.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Dominguez O, Ruiz JF, Lain de Lera T, Garcia-Diaz M, Gonzalez MA, Kirchhoff T, Martinez AC, Bernad A, Blanco L. DNA polymerase mu (Pol mu), homologous to TdT, could act as a DNA mutator in eukaryotic cells. EMBO J. 2000;19(7):1731–1742. doi: 10.1093/emboj/19.7.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Nick McElhinny SA, Ramsden DA. Polymerase mu is a DNA-directed DNA/RNA polymerase. Mol Cell Biol. 2003;23(7):2309–2315. doi: 10.1128/MCB.23.7.2309-2315.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Martin MJ, Garcia-Ortiz MV, Esteban V, Blanco L. Ribonucleotides and manganese ions improve non-homologous end joining by human Polmu. Nucleic Acids Res. 2013;41(4):2428–2436. doi: 10.1093/nar/gks1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Moon AF, Garcia-Diaz M, Bebenek K, Davis BJ, Zhong X, Ramsden DA, Kunkel TA, Pedersen LC. Structural insight into the substrate specificity of DNA Polymerase μ. Nat Struct Mol Biol. 2006;14:45. doi: 10.1038/nsmb1180. [DOI] [PubMed] [Google Scholar]
- 211.Martin MJ, Juarez R, Blanco L. DNA-binding determinants promoting NHEJ by human Polµ. Nucleic Acids Res. 2012;40(22):11389–11403. doi: 10.1093/nar/gks896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Roettger MP, Fiala KA, Sompalli S, Dong Y, Suo Z. Pre-steady-state kinetic studies of the fidelity of human DNA polymerase mu. Biochemistry. 2004;43(43):13827–13838. doi: 10.1021/bi048782m. [DOI] [PubMed] [Google Scholar]
- 213.Moon AF, Pryor JM, Ramsden DA, Kunkel TA, Bebenek K, Pedersen LC. Structural accommodation of ribonucleotide incorporation by the DNA repair enzyme polymerase Mu. Nucleic Acids Res. 2017;45(15):9138–9148. doi: 10.1093/nar/gkx527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Moon AF, Pryor JM, Ramsden DA, Kunkel TA, Bebenek K, Pedersen LC. Sustained active site rigidity during synthesis by human DNA polymerase μ. Nat Struct Mol Biol. 2014;21:253. doi: 10.1038/nsmb.2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Juárez R, Ruiz JF, Nick McElhinny SA, Ramsden D, Blanco L. A specific loop in human DNA polymerase mu allows switching between creative and DNA-instructed synthesis. Nucleic Acids Res. 2006;34(16):4572–4582. doi: 10.1093/nar/gkl457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Moon AF, Gosavi RA, Kunkel TA, Pedersen LC, Bebenek K. Creative template-dependent synthesis by human polymerase mu. Proc Natl Acad Sci USA. 2015;112(33):E4530–E4536. doi: 10.1073/pnas.1505798112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Bollum FJ. Calf thymus polymerase. J Biol Chem. 1960;235:2399–2403. [PubMed] [Google Scholar]
- 218.Bollum FJ. Chemically defined templates and initiators for deoxypolynucleotide synthesis. Science. 1964;144(3618):560. doi: 10.1126/science.144.3618.560-b. [DOI] [PubMed] [Google Scholar]
- 219.Desiderio SV, Yancopoulos GD, Paskind M, Thomas E, Boss MA, Landau N, Alt FW, Baltimore D. Insertion of N regions into heavy-chain genes is correlated with expression of terminal deoxytransferase in B cells. Nature. 1984;311(5988):752–755. doi: 10.1038/311752a0. [DOI] [PubMed] [Google Scholar]
- 220.Bertocci B, De Smet A, Weill JC, Reynaud CA. Nonoverlapping functions of DNA polymerases mu, lambda, and terminal deoxynucleotidyl transferase during immunoglobulin V(D)J recombination in vivo. Immunity. 2006;25(1):31–41. doi: 10.1016/j.immuni.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 221.Kunkel TA, Gopinathan KP, Dube DK, Snow ET, Loeb LA. Rearrangements of DNA mediated by terminal transferase. Proc Natl Acad Sci USA. 1986;83(6):1867–1871. doi: 10.1073/pnas.83.6.1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Kepler TB, Borrero M, Rugerio B, McCray SK, Clarke SH. Interdependence of N nucleotide addition and recombination site choice in V(D)J rearrangement. J Immunol. 1996;157(10):4451–4457. [PubMed] [Google Scholar]
- 223.Komori T, Okada A, Stewart V, Alt FW. Lack of N regions in antigen receptor variable region genes of TdT-deficient lymphocytes. Science. 1993;261(5125):1171–1175. doi: 10.1126/science.8356451. [DOI] [PubMed] [Google Scholar]
- 224.Moon AF, Garcia-Diaz M, Batra VK, Beard WA, Bebenek K, Kunkel TA, Wilson SH, Pedersen LC. The X family portrait: structural insights into biological functions of X family polymerases. DNA Repair. 2007;6(12):1709–1725. doi: 10.1016/j.dnarep.2007.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Delarue M, Boule JB, Lescar J, Expert-Bezancon N, Jourdan N, Sukumar N, Rougeon F, Papanicolaou C. Crystal structures of a template-independent DNA polymerase: murine terminal deoxynucleotidyltransferase. EMBO J. 2002;21(3):427–439. doi: 10.1093/emboj/21.3.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Gouge J, Rosario S, Romain F, Beguin P, Delarue M. Structures of intermediates along the catalytic cycle of terminal deoxynucleotidyltransferase: dynamical aspects of the two-metal ion mechanism. J Mol Biol. 2013;425(22):4334–4352. doi: 10.1016/j.jmb.2013.07.009. [DOI] [PubMed] [Google Scholar]
- 227.Loc’h J, Rosario S, Delarue M. Structural basis for a new templated activity by terminal deoxynucleotidyl transferase: implications for V(D)J recombination. Structure. 2016 doi: 10.1016/j.str.2016.06.014. [DOI] [PubMed] [Google Scholar]
- 228.Gouge J, Rosario S, Romain F, Poitevin F, Beguin P, Delarue M. Structural basis for a novel mechanism of DNA bridging and alignment in eukaryotic DSB DNA repair. EMBO J. 2015;34(8):1126–1142. doi: 10.15252/embj.201489643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Lee YS, Kennedy WD, Yin YW. Structural insight into processive human mitochondrial DNA synthesis and disease-related polymerase mutations. Cell. 2009;139(2):312–324. doi: 10.1016/j.cell.2009.07.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Newman JA, Cooper CD, Aitkenhead H, Gileadi O. Structure of the helicase domain of DNA polymerase theta reveals a possible role in the microhomology-mediated end-joining pathway. Structure. 2015;23(12):2319–2330. doi: 10.1016/j.str.2015.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Zahn KE, Averill AM, Aller P, Wood RD, Doublie S. Human DNA polymerase theta grasps the primer terminus to mediate DNA repair. Nat Struct Mol Biol. 2015;22(4):304–311. doi: 10.1038/nsmb.2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Lee YS, Gao Y, Yang W. How a homolog of high-fidelity replicases conducts mutagenic DNA synthesis. Nat Struct Mol Biol. 2015;22(4):298–303. doi: 10.1038/nsmb.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Baltimore D. RNA-dependent DNA polymerase in virions of RNA tumour viruses. Nature. 1970;226(5252):1209–1211. doi: 10.1038/2261209a0. [DOI] [PubMed] [Google Scholar]
- 234.Temin HM, Mizutani S. RNA-dependent DNA polymerase in virions of Rous sarcoma virus. Nature. 1970;226(5252):1211–1213. doi: 10.1038/2261211a0. [DOI] [PubMed] [Google Scholar]
- 235.Weissbach A, Baltimore D, Bollum F, Gallo R, Korn D. Nomenclature of eukaryotic DNA polymerases. Science. 1975;190(4212):401–402. doi: 10.1126/science.1179222. [DOI] [PubMed] [Google Scholar]
- 236.Bolden A, Noy GP, Weissbach A. DNA polymerase of mitochondria is a gamma-polymerase. J Biol Chem. 1977;252(10):3351–3356. [PubMed] [Google Scholar]
- 237.Hubscher U, Kuenzle CC, Spadari S. Functional roles of DNA polymerases beta and gamma. Proc Natl Acad Sci USA. 1979;76(5):2316–2320. doi: 10.1073/pnas.76.5.2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Genga A, Bianchi L, Foury F. A nuclear mutant of Saccharomyces cerevisiae deficient in mitochondrial DNA replication and polymerase activity. J Biol Chem. 1986;261(20):9328–9332. [PubMed] [Google Scholar]
- 239.Lestienne P. Evidence for a direct role of the DNA polymerase gamma in the replication of the human mitochondrial DNA in vitro. Biochem Biophys Res Commun. 1987;146(3):1146–1153. doi: 10.1016/0006-291x(87)90767-4. [DOI] [PubMed] [Google Scholar]
- 240.Stumpf JD, Saneto RP, Copeland WC. Clinical and molecular features of POLG-related mitochondrial disease. Cold Spring Harb Perspect Biol. 2013;5(4):a011395. doi: 10.1101/cshperspect.a011395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Sohl CD, Szymanski MR, Mislak AC, Shumate CK, Amiralaei S, Schinazi RF, Anderson KS, Yin YW. Probing the structural and molecular basis of nucleotide selectivity by human mitochondrial DNA polymerase gamma. Proc Natl Acad Sci USA. 2015;112(28):8596–8601. doi: 10.1073/pnas.1421733112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Lewis W, Day BJ, Copeland WC. Mitochondrial toxicity of NRTI antiviral drugs: an integrated cellular perspective. Nat Rev Drug Discov. 2003;2(10):812–822. doi: 10.1038/nrd1201. [DOI] [PubMed] [Google Scholar]
- 243.Krasich R, Copeland WC. DNA polymerases in the mitochondria: a critical review of the evidence. Front Biosci (Landmark Ed) 2017;22:692–709. doi: 10.2741/4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Yang MY, Bowmaker M, Reyes A, Vergani L, Angeli P, Gringeri E, Jacobs HT, Holt IJ. Biased incorporation of ribonucleotides on the mitochondrial L-strand accounts for apparent strand-asymmetric DNA replication. Cell. 2002;111(4):495–505. doi: 10.1016/s0092-8674(02)01075-9. [DOI] [PubMed] [Google Scholar]
- 245.Clayton DA. Replication of animal mitochondrial DNA. Cell. 1982;28(4):693–705. doi: 10.1016/0092-8674(82)90049-6. [DOI] [PubMed] [Google Scholar]
- 246.Brown TA, Cecconi C, Tkachuk AN, Bustamante C, Clayton DA. Replication of mitochondrial DNA occurs by strand displacement with alternative light-strand origins, not via a strand-coupled mechanism. Genes Dev. 2005;19(20):2466–2476. doi: 10.1101/gad.1352105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Kasamatsu H, Vinograd J. Unidirectionality of replication in mouse mitochondrial DNA. Nat New Biol. 1973;241(108):103–105. doi: 10.1038/newbio241103a0. [DOI] [PubMed] [Google Scholar]
- 248.Yakubovskaya E, Lukin M, Chen Z, Berriman J, Wall JS, Kobayashi R, Kisker C, Bogenhagen DF. The EM structure of human DNA polymerase gamma reveals a localized contact between the catalytic and accessory subunits. EMBO J. 2007;26(19):4283–4291. doi: 10.1038/sj.emboj.7601843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Miralles Fuste J, Shi Y, Wanrooij S, Zhu X, Jemt E, Persson O, Sabouri N, Gustafsson CM, Falkenberg M. In vivo occupancy of mitochondrial single-stranded DNA binding protein supports the strand displacement mode of DNA replication. PLoS Genet. 2014;10(12):e1004832. doi: 10.1371/journal.pgen.1004832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Graves SW, Johnson AA, Johnson KA. Expression, purification, and initial kinetic characterization of the large subunit of the human mitochondrial DNA polymerase. Biochemistry. 1998;37(17):6050–6058. doi: 10.1021/bi972685u. [DOI] [PubMed] [Google Scholar]
- 251.Johnson AA, Tsai Y, Graves SW, Johnson KA. Human mitochondrial DNA polymerase holoenzyme: reconstitution and characterization. Biochemistry. 2000;39(7):1702–1708. doi: 10.1021/bi992104w. [DOI] [PubMed] [Google Scholar]
- 252.Johnson AA, Johnson KA. Exonuclease proofreading by human mitochondrial DNA polymerase. J Biol Chem. 2001;276(41):38097–38107. doi: 10.1074/jbc.M106046200. [DOI] [PubMed] [Google Scholar]
- 253.Szymanski MR, Kuznetsov VB, Shumate C, Meng Q, Lee YS, Patel G, Patel S, Yin YW. Structural basis for processivity and antiviral drug toxicity in human mitochondrial DNA replicase. EMBO J. 2015;34(14):1959–1970. doi: 10.15252/embj.201591520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Prakash A, Doublie S. Base excision repair in the mitochondria. J Cell Biochem. 2015;116(8):1490–1499. doi: 10.1002/jcb.25103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Copeland WC, Kasiviswanathan R, Longley MJ. Analysis of Translesion DNA Synthesis by the Mitochondrial DNA Polymerase gamma. Methods Mol Biol. 2016;1351:19–26. doi: 10.1007/978-1-4939-3040-1_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Seki M, Masutani C, Yang LW, Schuffert A, Iwai S, Bahar I, Wood RD. High-efficiency bypass of DNA damage by human DNA polymerase Q. EMBO J. 2004;23(22):4484–4494. doi: 10.1038/sj.emboj.7600424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Takata K, Shimizu T, Iwai S, Wood RD. Human DNA polymerase N (POLN) is a low fidelity enzyme capable of error-free bypass of 5S-thymine glycol. J Biol Chem. 2006;281(33):23445–23455. doi: 10.1074/jbc.M604317200. [DOI] [PubMed] [Google Scholar]
- 258.Yoon JH, Roy Choudhury J, Park J, Prakash S, Prakash L. A role for DNA polymerase theta in promoting replication through oxidative DNA lesion, thymine glycol, in human cells. J Biol Chem. 2014;289(19):13177–13185. doi: 10.1074/jbc.M114.556977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Arana ME, Seki M, Wood RD, Rogozin IB, Kunkel TA. Low-fidelity DNA synthesis by human DNA polymerase theta. Nucleic Acids Res. 2008;36(11):3847–3856. doi: 10.1093/nar/gkn310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Ceccaldi R, Liu JC, Amunugama R, Hajdu I, Primack B, Petalcorin MI, O’Connor KW, Konstantinopoulos PA, Elledge SJ, Boulton SJ, Yusufzai T, D’Andrea AD. Homologous-recombination-deficient tumours are dependent on Poltheta-mediated repair. Nature. 2015;518(7538):258–262. doi: 10.1038/nature14184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Kent T, Chandramouly G, McDevitt SM, Ozdemir AY, Pomerantz RT. Mechanism of microhomology-mediated end-joining promoted by human DNA polymerase theta. Nat Struct Mol Biol. 2015;22(3):230–237. doi: 10.1038/nsmb.2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Mateos-Gomez PA, Gong F, Nair N, Miller KM, Lazzerini-Denchi E, Sfeir A. Mammalian polymerase theta promotes alternative NHEJ and suppresses recombination. Nature. 2015;518(7538):254–257. doi: 10.1038/nature14157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Yousefzadeh MJ, Wyatt DW, Takata K, Mu Y, Hensley SC, Tomida J, Bylund GO, Doublie S, Johansson E, Ramsden DA, McBride KM, Wood RD. Mechanism of suppression of chromosomal instability by DNA polymerase POLQ. PLoS Genet. 2014;10(10):e1004654. doi: 10.1371/journal.pgen.1004654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Cancer Genome Atlas Research N Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474(7353):609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.McVey M, Lee SE. MMEJ repair of double-strand breaks (director’s cut): deleted sequences and alternative endings. Trends Genet. 2008;24(11):529–538. doi: 10.1016/j.tig.2008.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Hogg M, Sauer-Eriksson AE, Johansson E. Promiscuous DNA synthesis by human DNA polymerase theta. Nucleic Acids Res. 2012;40(6):2611–2622. doi: 10.1093/nar/gkr1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Prasad R, Longley MJ, Sharief FS, Hou EW, Copeland WC, Wilson SH. Human DNA polymerase theta possesses 5′-dRP lyase activity and functions in single-nucleotide base excision repair in vitro. Nucleic Acids Res. 2009;37(6):1868–1877. doi: 10.1093/nar/gkp035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Hogg M, Seki M, Wood RD, Doublie S, Wallace SS. Lesion bypass activity of DNA polymerase theta (POLQ) is an intrinsic property of the pol domain and depends on unique sequence inserts. J Mol Biol. 2011;405(3):642–652. doi: 10.1016/j.jmb.2010.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Seki M, Wood RD. DNA polymerase theta (POLQ) can extend from mismatches and from bases opposite a (6–4) photoproduct. DNA Repair. 2008;7(1):119–127. doi: 10.1016/j.dnarep.2007.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Higgins GS, Harris AL, Prevo R, Helleday T, McKenna WG, Buffa FM. Overexpression of POLQ confers a poor prognosis in early breast cancer patients. Oncotarget. 2010;1(3):175–184. doi: 10.18632/oncotarget.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Goff JP, Shields DS, Seki M, Choi S, Epperly MW, Dixon T, Wang H, Bakkenist CJ, Dertinger SD, Torous DK, Wittschieben J, Wood RD, Greenberger JS. Lack of DNA polymerase theta (POLQ) radiosensitizes bone marrow stromal cells in vitro and increases reticulocyte micronuclei after total-body irradiation. Radiat Res. 2009;172(2):165–174. doi: 10.1667/RR1598.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Kawamura K, Bahar R, Seimiya M, Chiyo M, Wada A, Okada S, Hatano M, Tokuhisa T, Kimura H, Watanabe S, Honda I, Sakiyama S, Tagawa M. DNA polymerase theta is preferentially expressed in lymphoid tissues and upregulated in human cancers. Int J Cancer. 2004;109(1):9–16. doi: 10.1002/ijc.11666. [DOI] [PubMed] [Google Scholar]
- 273.Wood RD, Doublie S. DNA polymerase theta (POLQ), double-strand break repair, and cancer. DNA Repair. 2016;44:22–32. doi: 10.1016/j.dnarep.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Singleton MR, Dillingham MS, Wigley DB. Structure and mechanism of helicases and nucleic acid translocases. Annu Rev Biochem. 2007;76:23–50. doi: 10.1146/annurev.biochem.76.052305.115300. [DOI] [PubMed] [Google Scholar]
- 275.Tafel AA, Wu L, McHugh PJ. Human HEL308 localizes to damaged replication forks and unwinds lagging strand structures. J Biol Chem. 2011;286(18):15832–15840. doi: 10.1074/jbc.M111.228189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Buttner K, Nehring S, Hopfner KP. Structural basis for DNA duplex separation by a superfamily-2 helicase. Nat Struct Mol Biol. 2007;14(7):647–652. doi: 10.1038/nsmb1246. [DOI] [PubMed] [Google Scholar]
- 277.Guy CP, Bolt EL. Archaeal Hel308 helicase targets replication forks in vivo and in vitro and unwinds lagging strands. Nucleic Acids Res. 2005;33(11):3678–3690. doi: 10.1093/nar/gki685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Maga G, Shevelev I, Ramadan K, Spadari S, Hubscher U. DNA polymerase theta purified from human cells is a high-fidelity enzyme. J Mol Biol. 2002;319(2):359–369. doi: 10.1016/S0022-2836(02)00325-X. [DOI] [PubMed] [Google Scholar]
- 279.Seki M, Marini F, Wood RD. POLQ (Pol theta), a DNA polymerase and DNA-dependent ATPase in human cells. Nucleic Acids Res. 2003;31(21):6117–6126. doi: 10.1093/nar/gkg814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Marini F, Kim N, Schuffert A, Wood RD. POLN, a nuclear PolA family DNA polymerase homologous to the DNA cross-link sensitivity protein Mus308. J Biol Chem. 2003;278(34):32014–32019. doi: 10.1074/jbc.M305646200. [DOI] [PubMed] [Google Scholar]
- 281.Arana ME, Potapova O, Kunkel TA, Joyce CM. Kinetic analysis of the unique error signature of human DNA polymerase nu. Biochemistry. 2011;50(46):10126–10135. doi: 10.1021/bi201197p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Arana ME, Takata K, Garcia-Diaz M, Wood RD, Kunkel TA. A unique error signature for human DNA polymerase nu. DNA Repair (Amst) 2007;6(2):213–223. doi: 10.1016/j.dnarep.2006.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Yamanaka K, Minko IG, Takata K, Kolbanovskiy A, Kozekov ID, Wood RD, Rizzo CJ, Lloyd RS. Novel enzymatic function of DNA polymerase nu in translesion DNA synthesis past major groove DNA-peptide and DNA-DNA cross-links. Chem Res Toxicol. 2010;23(3):689–695. doi: 10.1021/tx900449u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Beard WA, Wilson SH. Structures of human DNA polymerases nu and theta expose their end game. Nat Struct Mol Biol. 2015;22(4):273–275. doi: 10.1038/nsmb.3006. [DOI] [PubMed] [Google Scholar]
- 285.Jordheim LP, Durantel D, Zoulim F, Dumontet C. Advances in the development of nucleoside and nucleotide analogues for cancer and viral diseases. Nat Rev Drug Discov. 2013;12(6):447–464. doi: 10.1038/nrd4010. [DOI] [PubMed] [Google Scholar]
- 286.Korzhnev DM, Hadden MK. Targeting the translesion synthesis pathway for the development of anti-cancer chemotherapeutics. J Med Chem. 2016;59(20):9321–9336. doi: 10.1021/acs.jmedchem.6b00596. [DOI] [PubMed] [Google Scholar]
- 287.Rechkoblit O, Gupta YK, Malik R, Rajashankar KR, Johnson RE, Prakash L, Prakash S, Aggarwal AK. Structure and mechanism of human PrimPol, a DNA polymerase with primase activity. Sci Adv. 2016;2(10):e1601317. doi: 10.1126/sciadv.1601317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Korona DA, LeCompte KG, Pursell ZF. The high fidelity and unique error signature of human DNA polymerase ε. Nucleic Acids Res. 2011;39(5):1763–1773. doi: 10.1093/nar/gkq1034. [DOI] [PMC free article] [PubMed] [Google Scholar]