Abstract
Cells, the basic units of life, have striking differences at transcriptomic, proteomic and epigenomic levels across tissues, organs, organ systems and organisms. The coordination of individual immune cells is essential for the generation of effective immune responses to pathogens while immune tolerance is maintained to protect the host. In rheumatic diseases, when immune responses are dysregulated, pathologically important cells might represent only a small fraction of the immune system. Interrogation of the contributions of individual immune cells to pathogenesis and disease progression should therefore reveal important insights into the complicated aetiology of rheumatic diseases. Technological advances are enabling the high-dimensional dissection of single cells at multiple omics levels, which could facilitate the identification of dysregulated molecular mechanisms in patients with rheumatic diseases and the discovery of new therapeutic targets and biomarkers. The single-cell technologies that have been developed over the past decade and the experimental platforms that enable multi-omics integrative analyses have already made inroads into immunology-related fields of study and have potential for use in rheumatology. Layers of omics data derived from single cells are likely to fundamentally change our understanding of the molecular pathways that underpin the pathogenesis of rheumatic diseases.
Since the discovery of the cell, we have gained insights into everything from subcellular structures to genetic codes from this basic unit of life. However, the heterogeneity that exists between individual cells has become increasingly evident with the development of new single-cell technologies. For example, the introduction of next-generation sequencing (NGS) technology at the beginning of the 21st century marked a new chapter for genomic research1,2; billions of reads can now be routinely generated to help us to better understand the genome, transcriptome and epigenome at the single-cell level. The analysis of protein expression and post-translational modifications has been aided by the development of mass cytometry, which enables the simultaneous analysis of >100 protein markers in single cells3, and advances in single-cell technologies that enable the simultaneous analysis of multiple types of omics data are now providing researchers with opportunities to interrogate the heterogeneity of single cells at unprecedented depth.
Rheumatic diseases, which affect more than one-fifth of the population of the USA and millions of individuals worldwide4,5, have mostly unknown aetiologies. Small subsets of cells are thought to be important in the pathogenesis of a variety of rheumatic diseases, therefore studying the breakdown of immune tolerance and dysregulated pro-inflammatory pathways on a cell-by-cell basis presents a tremendous opportunity for rheumatology research.
In this Review, we look at the single-cell technologies currently available for researchers to use to better understand the heterogeneity of human cells and the pathogenic mechanisms of rheumatic diseases at different omics levels (FIG. 1). In particular, we discuss single-cell RNA sequencing (scRNA-seq), antigen receptor sequencing, mass cytometry, mass-spectrometry-based imaging and a variety of epigenomic platforms, as well as multi-omics technologies that enable simultaneous analyses of DNA, RNA and protein markers. We also summarize pioneering research that has used these powerful analytic platforms to elucidate complex immune cell networks in health and disease and discuss potential future applications of single-cell technologies in rheumatic disease research.
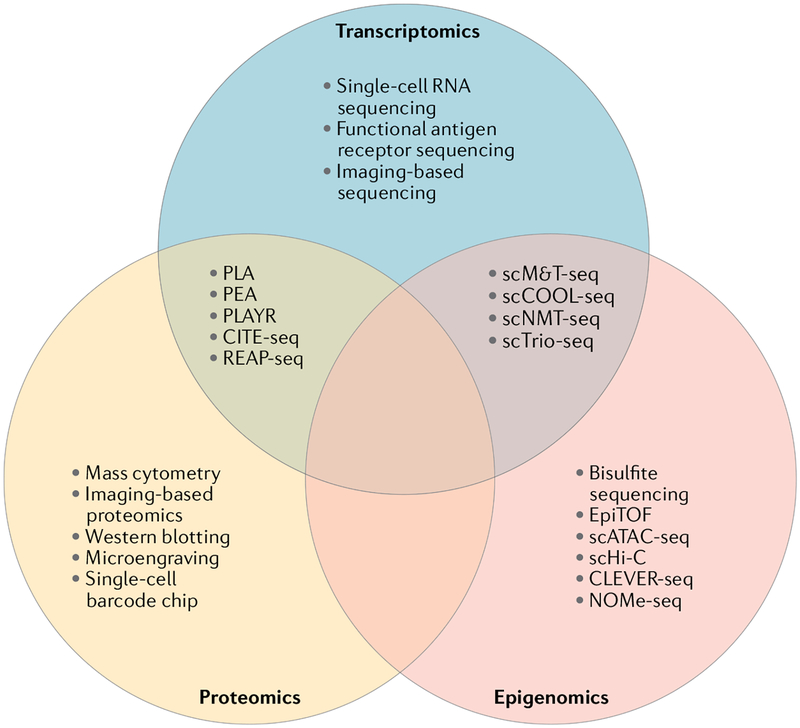
Fig. 1 |. Single-cell experimental platforms for omics analysis.
Venn diagram depicting single-cell technologies that can be used to interrogate the transcriptome, epigenome and proteome. Overlapping regions contain technologies that enable the integrative analysis of multiple omics in the same cells. CITE-seq, cellular indexing of transcriptomes and epitopes by sequencing; CLEVER-seq, chemical-labelling-enabled C-to-T conversion sequencing; EpiTOF, epigenetic landscape profiling using cytometry by time of flight; NOMe-seq, nucleosome occupancy and methylome sequencing; PEA, proximity extension assay; PLA, proximity ligation assay; PLAYR, proximity ligation assay for RNA; REAP-seq, RNA expression and protein sequencing; scATAC-seq, single-cell resolution in assay for transposase-accessible chromatin using sequencing; scCOOL-seq, single-cell chromatin overall omic-scale landscape sequencing; scHi-C, high-throughput variant of chromosome conforation capture performed on single cells; scM&T-seq, single-cell methylome and transcriptome sequencing; scNMT-seq, single-cell nucleosome, methylation and transcription sequencing; scTrio-seq; single-cell triple omics sequencing.
Conducting single-cell studies
Several collaborative projects have been launched that are devoted to advancing single-cell analyses for rheumatology research. For example, the Accelerating Medicines Partnership (AMP) rheumatoid arthritis (RA) and systemic lupus erythematosus (SLE) network aims to identify new therapeutic targets for RA and SLE and to understand disease mechanisms by leveraging the latest breakthroughs in single-cell technologies. Since its launch in 2014, the AMP RA and SLE network has made several important discoveries at the single-cell level and has uncovered molecular and cellular mechanisms that underlie the pathogenesis of rheumatic diseases6,7.
Collaborative programmes such as the AMP RA and SLE network highlight the fact that single-cell studies often require a team of investigators with expertise in different areas of biomedical research. To conduct a single-cell study, several important factors must be considered. First, high-quality clinical samples and meticulous medical records need to be collected by experienced physicians, as well as adequate control samples from healthy individuals. The detailed clinical information collected for individual samples ensures that disease-specific molecular signatures can be captured and that the effects of treatments or other unrelated medical events such as infections or vaccinations can be properly controlled. Notably, although samples from treatment-naive patients with new-onset disease present an excellent opportunity to identify dysregulated molecular mechanisms associated with rheumatic diseases, obtaining such samples before any medications have been used, especially those for symptom relief, is extremely challenging. Additionally, it is unlikely that the number of samples from patients with new-onset disease will be sufficient to fully represent the heterogeneity of clinical manifestations that occur in rheumatic diseases. Thus, single-cell studies are often conducted on samples from heterogeneous groups of patients, making the availability of detailed medical records essential for the success of a study.
Next, a power calculation is performed by biostatisticians to compute the number of samples required for downstream statistical analyses. Biostatisticians can also provide valuable input for experimental design. After a single-cell assay is performed, a joint effort is required by investigators with diverse backgrounds to accurately interpret the results, which normally takes much longer than the time taken to perform the actual assay. Because of the high cost and substantial resources required for most single-cell assays, it is often impractical for a single laboratory or institution to perform analyses on a large number of samples or on multiple cohorts of patients, making inter-institutional collaborations and free data-sharing partnerships appealing approaches for single-cell studies. Collaborative programmes are particularly important for rheumatology research, as a high degree of heterogeneity exists between patients and analyses of several patient populations with diverse demographic backgrounds and disease manifestations are often required. In the following sections, we outline some of the single-cell technologies that are currently available for use in rheumatic disease research.
Single-cell transcriptomics
scRNA-seq has become an essential tool for the study of biological systems in which cellular heterogeneity is prominent, such as the immune system. Each type of immune cell has specialized functions and transcription patterns, and cells of the adaptive immune system are further diversified owing to the expression of unique antigen receptors. In this section, we discuss single-cell transcriptomic analyses and paired antigen receptor sequencing. Single-cell genomic analyses are also important for our understanding of human diseases and have been comprehensively reviewed elsewhere8,9.
Gene expression analysis.
Since the first scRNA-seq study on mouse embryos was published in 2009 (REF10), this technology has rapidly improved in resolution, throughput and precision. It is now possible to analyse thousands of single cells simultaneously with great depth and accuracy. A variety of scRNA-seq methods have been established, most of which follow the same pattern of single-cell isolation, reverse transcription, cDNA amplification and library construction and all of which have different strengths11,12 (TABLE 1). Additional scRNA-seq technologies that preserve the tissue context or subcellular localization of transcripts have also been described13,14. These technologies, together with other powerful imaging-based methods, such as single-molecule RNA fluorescence in situ hybridization (FISH)15 and multiplexed error-robust FISH (MERFISH)16, enable spatially resolved transcriptomic analyses of single cells. The use of barcodes to label individual transcripts and mRNA from each cell further enhances the accuracy and throughput of scRNA-seq (BOX 1).
Table 1 |.
Single-cell RNA sequencing technologies
| Technique | Single-cell isolation method | Reverse transcription method | cDNA amplification method |
Throughput (number of cells analysed) | UMI | Refs |
|---|---|---|---|---|---|---|
| Tang et al. | Micropipetting | Poly(A)-based | PCR | 101 | No | 10 |
| inDrop | Droplet-based microfluidics system | Poly(A)-based | IVT | 104 | Yes | 20 |
| Drop-seq | Droplet-based microfluidics system | Poly(A)-based | PCR | 104 | Yes | 21 |
| Seq-Well | Microwells | Template-switching | PCR | 104 | Yes | 22 |
| CytoSeq | Microwells | Poly(A)-based | PCR | 105 | Yes | 23 |
| Div-seq | FACS | Template-switching | PCR | 103 (nuclei) | No | 24 |
| DroNc-seq | Droplet-based microfluidics system | Template-switching | PCR | 104 (nuclei) | Yes | 25 |
| STRT | Micropipetting | Template-switching | PCR | 102 | No | 159 |
| CEL-seq | Micropipetting | Poly(A)-based | IVT | 102 | No | 170 |
| SUPeR-seq | Micropipetting | Random primer-based | PCR | 102 | No | 171 |
| Quartz-seq | FACS | Poly(A)-based | PCR | 102 | No | 172 |
| MATQ-seq | Micropipetting | MALBAC primer-based | PCR | 102 | Yes | 162 |
| SMART-seq2 | Micropipetting | Template-switching | PCR | 102 | No | 173 |
| MARS-seq | FACS | Poly(A)-based | IVT | 103 | Yes | 161 |
| CEL-seq2 | Microfluidic devices | Poly(A)-based | IVT | 102 | Yes | 160 |
| Quartz-seq2 | FACS | Poly(A)-based | PCR | 103 | Yes | 174 |
| SMART-seq | Micropipetting | Template-switching | PCR | 102 | No | 175 |
| SC3-seq | Micropipetting | Poly(A)-based | PCR | 102 | Yes | 176 |
CEL-seq, cell expression by linear amplification and sequencing; CytoSeq, gene expression cytometry; Div-seq, RNA sequencing on divided cells; DroNc-seq, massively parallel single-nucleus sequencing with droplet technology; Drop-seq, droplet sequencing; FACS, fluorescence-activated cell sorting; IVT, in vitro transcription; MALBAC, multiple annealing and looping-based amplification cycles; MARS-seq, massively parallel single-cell RNA sequencing; MATQ-seq, multiple annealing and dC-tailing-based quantitative single-cell RNA sequencing; SC3-seq, single-cell mRNA 3′ end sequencing; seq2, second-generation platform; SMART-seq, switch mechanism at the 5′ end of RNA templates sequencing; STRT, single-cell tagged reverse transcription; SUPeR-seq, single-cell universal poly(A)-independent RNA sequencing; UMI, unique molecular identifier.
Box 1 |. Barcoding in single-cell RNA sequencing.
Unique molecular identifiers (UMIs) can be used to label individual RNA molecules with distinct barcodes156. Thus, after sequencing, it is possible to correct for the bias introduced by PCR amplification as reads with the same UMIs indicate that they were derived from the same starting templates. Specific barcodes can also be introduced to label cDNA from individual cells, thereby facilitating the simultaneous analysis of thousands of cells. For example, researchers used split-pool ligation-based transcriptome sequencing (SPLiT-seq) with a combinatorial barcoding strategy to analyse >150,000 single cells from the central nervous system of a developing mouse157. Another technique, single-cell combinatorial indexing RNA sequencing (sci-RNA-seq), introduces distinct barcodes to pools of cells during both first-strand synthesis and PCR amplification158; the two-barcode combinations label single cells without physically separating them. Strategies to label individual cells with unique barcodes have also been adopted for use with single-cell tagged reverse transcription (STRT)159, second-generation cell expression by linear amplification and sequencing (CEL-seq2)160, massively parallel single-cell RNA sequencing (MARS-seq)161, droplet sequencing21, the inDrop system20, gene expression cytometry23 and multiple annealing and dC-tailing-based quantitative single-cell RNA sequencing (MATQ-seq)162. Notably, gene expression cytometry23, the Seq-Well platform22 and a droplet-based approach163 have all been used successfully to barcode thousands of peripheral blood mononuclear cells for unbiased, highly multiplexed analyses of the immune system.
These methods all generate large and complex data sets that require specialized bioinformatic algorithms to facilitate accurate data debarcoding and interpretation. Advances in analytical tools and the challenges of using computational biology to analyse scRNA-seq data have been comprehensively reviewed elsewhere17,18. Notably, a powerful computational algorithm, termed demuxlet (demultiplexing and doublet identification from single-cell data), has been developed that utilizes natural genetic variations to identify the donors to whom individual cells belong in droplet-based scRNA-seq data sets in which cells from multiple donors are pooled and analysed together19. This method, which has the potential to enhance sample processing throughput and eliminate batch effects, was successfully used to study the transcriptomes of single peripheral blood mononuclear cells (PBMCs) from patients with SLE from pooled samples19.
The variety of scRNA-seq methods offered and the differences in throughput and sensitivity mean that selecting the most suitable platform for a study can be challenging. Usually, the choice of platform depends on the specific biological question being answered and the type of biological sample being analysed. For example, the high throughputs of the inDrop system20, droplet sequencing (Drop-seq)21, the Seq-Well platform22 and gene expression cytometry23 make them excellent options for profiling peripheral blood samples when coverage of a large number of immune cells is desired. For analysing solid tissue samples when isolating single cells would be technically challenging, a method for performing RNA-seq on divided cells (termed Div-seq)24 and massively parallel single-nucleus sequencing with droplet technology (DroNc-seq)25 are excellent alternatives that enable the sequencing of single nuclei. Importantly, owing to the small amounts of RNA that can be isolated from single cells, the quality of scRNA-seq results can be severely affected by technical or methodological problems. Furthermore, scRNA-seq is often unable to reliably detect transcripts expressed at medium-to-low concentrations, although assay sensitivity is continuously being improved. Rheumatic disease researchers are advised to consider all the advantages and limitations of scRNA-seq before performing this potentially resource-intensive technology. However, pioneering scRNA-seq studies have already been performed to investigate the heterogeneity of the human immune system and dysregulation in immune-mediated diseases at a single-cell level.
The results of scRNA-seq of cells from patients with SLE who have lupus nephritis, an important cause of morbidity and mortality in SLE, revealed the well-known interferon signature26,27 in renal tubular cells28. In these patients, the number of interferon-inducible transcripts correlated with histological evidence of chronicity, proteinuria and immunoglobulin deposition28. This study demonstrated that gentle tissue dissociation can be used on renal tissues to obtain a single-cell suspension for use in scRNA-seq and that transcriptomic data from a variety of renal cell subtypes can be analysed in conjunction with clinical data to dissect the heterogeneity of lupus nephritis. In RA, a portable microfluidic control system was assembled from 3D-printed parts to perform Drop-seq on synovial tissue samples29. scRNA-seq analysis of >20,000 cells revealed the identity of infiltrating haematopoietic cells and showed the presence of subtypes of fibroblasts characterized by distinct transcriptional profiles29. In accordance with the findings of this study29, an independent scRNA-seq study also found transcriptomically distinct subpopulations of fibroblasts in synovial tissue samples from patients with RA or osteoarthritis (OA)30. Three fibroblast subsets were defined by unique gene expression patterns and had different anatomical locations and cellular functions, including invasive migration, modulation of osteoclastogenesis and pro-inflammatory cytokine production. The relative proportions of the three subsets differed substantially between patients with RA and patients with OA, and the presence of large numbers of the CD34−THY1+ subset in RA synovium correlated with a high amount of immune cell infiltration, a high Krenn synovitis score and joint hypertrophy. Both studies29,30 provide excellent examples of how scRNA-seq can serve as a powerful platform to interrogate the heterogeneity of cells in tissue samples with complex cellular components and to characterize cell subsets on the basis of unique transcriptomic signatures. scRNA-seq has also been performed on fluorescence-activated cell sorting (FACS)-purified natural antibody-producing CD27+IgD+ B cells31, which are found in reduced numbers in patients with RA. The results of this study31 revealed an RA-specific pro-inflammatory transcription profile in this cell subset, highlighting that scRNA-seq can be used to interrogate the transcriptomic heterogeneity of cells that are important for pathogenesis when coupled with FACS purification using defined protein markers.
Notably, the clinical utility of scRNA-seq has been successfully demonstrated in other biomedical fields. For example, scRNA-seq analysis of cells from patients with melanoma receiving checkpoint inhibitor therapy revealed that the presence of a subset of CD8+ T cells was predictive of a positive clinical outcome and produced a combinatorial therapeutic strategy to enhance checkpoint inhibitor efficacy32. Such studies provide rheumatic disease researchers with valuable and helpful information in relation to experimental design and data analysis. Future studies in rheumatic diseases could be performed to identify immune cell subsets that correlate with patient responses to therapies that often have poor response rates. The results of such studies will have the potential to help researchers to understand the underlying pathological mechanisms of rheumatic diseases and to improve patient stratification and treatment outcomes.
Antigen receptor sequencing.
Several NGS-based platforms have been developed to profile the paired functional B cell receptor (BCR) sequences of individual B cells (BOX 2). Single-cell BCR sequencing of plasmablasts isolated from patients with RA identified BCRs specific for cyclic citrullinated peptides and other RA-associated autoantigens33. Sequencing of plasmablasts from anti-citrullinated protein autoantibody (ACPA)-positive individuals, who are at risk of developing RA, revealed both IgA-secreting and IgG-secreting clones responsive to common RA autoantigens, suggesting the potential involvement of mucosal immunity in the early stages of RA development34. Analysis of plasmablasts from patients with clinically evident RA that were collected longitudinally showed the presence of persistent IgA-producing cells that underwent continuous affinity maturation and produced ACPAs that formed pro-inflammatory immune complexes35. Single-cell functional BCR sequencing is likely to continue to provide new insights into the pathological functions of humoral immunity in rheumatic diseases.
Box 2 |. Methods for paired VH–VL sequence identification.
M icrowells can be used to capture single B cells, and a specialized PCR can then be performed in emulsion droplets to generate linkages between the variable regions of heavy-chain (VH) and light-chain (VL) transcripts164.
An axisymmetric flow-focusing device can be used to sequester single B cells into emulsion droplets with oligo(dT)-containing beads to capture mRNA. Beads are pooled and re-emulsified to generate linked VH–VL amplicons for sequencing165. This high-throughput technique enables the repertoire sequencing of millions of B cells in a single experiment.
VH and VL transcripts from single fluorescence-activated cell-sorting-purified B cell clones can be barcoded to facilitate the next-generation sequencing of paired full-length immunoglobulin VH and VL genes166.
Barcodes can be introduced to label mRNA from individual B cells during template switching and PCR am plification33.
An important utility of functional BCR analysis at the single-cell level is the in vitro expression of paired heavychain and light-chain sequences to construct clonespecific immunoglobulins. The resultant recombinant antibodies can be tested for antigen specificity using protein microarrays, and their importance in the pathogenesis of rheumatic diseases can be assessed using tissue culture or animal models. For example, BCR sequencing of plasmablasts from patients with idiopathic pulmonary arterial hypertension (IPAH) revealed the presence of monoclonal antibodies that recognize autoantigens frequently associated with connective tissue diseases36. Importantly, the treatment of cultured endothelial cells with recombinant antibodies that were generated on the basis of the sequencing results induced the secretion of pro-inflammatory cytokines and the expression of intercellular adhesion molecule 1, thereby implicating autoantibodies in the pathogenesis of IPAH36. Several autoimmune diseases are known to be mediated directly by pathogenic autoantibodies, such as anti-desmoglein 3 (DSG3) antibodies in pemphigus vulgaris37, but the clinical relevance of the autoantibodies detected in most rheumatic diseases is still unclear. Functional BCR sequencing of B cells from patients with rheumatic diseases has the potential to reveal new autoantibodies that could contribute to pathogenesis. Promising results from a study in which chimeric antigen receptor technology was used to target anti-DSG3-positive autoreactive B cells in a mouse model of pemphigus vulgaris38 suggest that the identification of pathogenic B cell clones could lead to the discovery of new antigen-specific therapeutic interventions for rheumatic diseases.
Similarly, established NGS-based single-cell methods exist to profile T cell receptor (TCR) sequences with paired α-chains and β-chains (BOX 3). In contrast to conventional bulk sequencing methods, single-cell analysis preserves information on functional TCRs, thereby enabling researchers to reconstruct clonespecific TCRs in vitro and to functionally characterize T cells with defined antigen specificities. Additionally, a yeast display-based platform can be used to determine the antigenic targets of TCRs with unknown specificities39. In this technique, yeast cells that express genetically encoded libraries of peptide-MHC complexes are selected through multiple rounds of enrichment using beads coated with recombinant TCRs. The antigen specificity of a TCR can subsequently be obtained by sequencing the enriched pool of yeast cells. Autoreactive T cells are thought to be involved in the pathogenesis of several rheumatic diseases; however, the cognate ligands for most rheumatic disease-related autoreactive T cells are unknown. The combination of single-cell TCR sequencing and yeast display-based antigen identification has been used successfully to discover the antigen specificities of tumour-infiltrating T cells from patients with colorectal adenocarcinoma and to identify common tumour antigens40. These complimentary techniques could therefore be adapted to sequence autoreactive T cells isolated from inflamed tissues relevant to rheumatic diseases such as synovial tissue, kidney and skin and to determine the antigen specificities of T cells that are highly enriched at these sites.
Box 3 |. Methods for paired α-chain and β-chain T cell receptor sequencing.
Reverse transcription of the lysate from single T cells can be perfored using primers tha t target T cell receptor (TCR) transcripts and T cell subset-specific markers, such as the transcription factors T-bet (for CD4+ T helper 1 (TH1) cells) and GATA3 (for CD4+Th2 cells)167. Nested PCR is then performed to am plify the cDNA of interest and to introduce barcodes for individual cells. This approach enables the accurate profiling of paired α-chains and β-chains to important phenotypic markers, thereby linking TCR specificity to the functional state of the same T cells.
A nother method, termed pairSEQ, enables paired TCR α-chains and β-chains to be decoded with out physically separating single cells168. mRNA from pools of T cells is reversed transcribed and TCR cDNA is am plified by PCR and barcoded before undergoing com bined high-throughput sequencing. Given the highly diverse nature of the TCR repertoire and the improbability of given α-chains and β-chains being present in identical pools of cells, the presence of the same α-chains and β-chains in multiple pools would infer that they are derived from the same T cell clone.
A flow-focusing device that facilitates the encapsulation of single cells in emulsion droplets for functional B cell receptor sequencing165 has also been used to profile functional TCRs169. This method enables the rapid analysis of millions of single T cells in an experiment and has the highest throughput reported to date.
The dysregulation of specific T cell subsets directly contributes to tissue damage and clinical symptoms in rheumatic diseases. For example, large numbers of T helper 17 (TH17) cells are found in the peripheral blood of patients with SLE and the kidneys of patients with lupus nephritis41,42, and the number of TH17 cells correlates with disease activity scores. Similarly, alterations in the numbers and immunomodulatory functions of regulatory T (Treg) cells in many rheumatic diseases and autoimmune diseases have led to the development of new therapies that aim to augment Treg cell functions43. The antigen specificities of these clinically important T cell subsets and the mechanisms by which they become activated in affected tissues or organs is a fascinating area of rheumatology research that, in our opinion, will be greatly accelerated by single-cell TCR sequencing technologies in the future.
Single-cell proteomics
The abundance of mRNA transcripts is not strictly reflective of the level of protein translation, both in the context of a population of cells and in single cells44. Additionally, the stability and function of a protein is modulated by post-translational modifications, which affect biological processes and cellular phenotypes. To begin to understand the heterogeneity of immune cells and proteome dysregulation associated with immune-mediated diseases, quantitative analyses of protein abundance in single cells, as well as patterns of post-translational modifications, provide a critical layer of biological information. In this section, we discuss mass cytometry, highly multiplexed imaging and other single-cell proteomic technologies.
Mass cytometry.
Mass cytometry uses lanthanide-labelled affinity reagents coupled with mass spectrometry to quantitatively measure proteins in individual cells with high precision and throughput. As many as45 isotopes are routinely used in current mass cytometry assays, although, in principle, >100 isotopes could be used for ultrahigh-content analyses of single cells45. Mass cytometry has been widely used to perform immune monitoring of phenotypic and functional protein markers in rheumatic diseases. Analyses of whole-blood samples from patients with SLE treated with Toll-like receptor ligands revealed differences in signalling protein activation and downstream cytokine production profiles compared with samples from healthy individuals46. Furthermore, a molecular signature defined by increased CC-chemokine ligand 2 (CCL2), CCL4 and IL-1 receptor antagonist in CD14hi classical monocytes was found by mass cytometry in patients with juvenile SLE47. This signature, which correlated with disease activity, could be abrogated by the use of the Janus kinase inhibitor ruxolitinib47.
Mass cytometry also facilitates the identification of unique immune cell subsets that are aberrantly expanded in patients with rheumatic diseases. Analyses of single-cell suspensions derived from inflamed solid tissues led to the discovery of previously uncharacterized immune cell populations, such as PD1hiCXCR5−CD4+ T cells that promote humoral immunity in inflamed RA synovial tissue48 and PD1+CD4+ T cells in the salivary glands of patients with primary Sjögren syndrome (pSS)49. Immunophenotyping of blood samples and paired labial salivary gland tissue samples from patients with pSS by mass cytometry revealed that changes in the frequencies of immune cell subtypes correlated with disease activity50. The identification of unique immune cell subsets can be accelerated by the use of mixed-effects modelling of associations of single cells, a statistical method in which the association between population clusters and disease status is tested while biological and technical confounding factors are controlled51. Using this statistical strategy to analyse mass cytometry data, a CD27-HLA-DR+ effector memory CD4+ T cell subset was discovered that correlated with RA disease severity51, demonstrating that this method has the potential to be applied broadly to single-cell rheumatic disease research. Importantly, the success of mass cytometry studies depends heavily on high-quality affinity reagents with high specificity and sensitivity; researchers should test reagents and protocols carefully before analysing precious clinical samples to ensure that quality data sets can be obtained. Together, the high content and throughput of mass cytometry make it an excellent experimental system for deep immunophenotyping and functional characterization of the immune system that can be applied to a wide range of cell types to identify differential protein markers.
Next-generation imaging.
Mass spectrometry imaging (MSI) is a powerful non-optical approach for the analysis of tissue specimens. Secondary ion mass spectrometry uses a finely focused primary ion beam to bombard the surface of a tissue sample, and the sputtered secondary ions from the tissue surface are captured and analysed by a mass spectrometer52. As a result, a high-resolution molecular image of the tissue surface can be generated. Two revolutionary MSI methods, multiplexed ion beam imaging (MIBI)53 and imaging mass cytometry54, achieve highly multiplexed images of fixed tissue samples. Both systems use lanthanide-labelled antibodies and follow conventional immunohistochemistry staining protocols. Through the use of MIBI or imaging mass cytometry at ultrafine resolution, the lanthanide-tagged tissue is converted to a stream of vaporized particles in a scanning process and is measured by a mass spectrometer. The spectra of lanthanides, which represent the abundance and distribution of the corresponding epitopes, are then assembled to construct a tissue image consisting of multiple biomarkers. Modern mass spectrometers offer excellent precision and restrict mass errors to a fraction of a Dalton. Different metal isotopes can be accurately resolved with minimal signal spill-over between tags. Signal amplification is not required in mass-spectrometry-based measurements; thus, the linear quantitative relationship across a tissue section is preserved. Both MIBI and imaging mass cytometry enable the quantitative measurement of an epitope at a dynamic range of more than five orders of magnitude and the simultaneous analysis of an unprecedented number of biomarkers55.
These next-generation imaging systems have been used to study tissue abnormalities in human diseases, mainly malignant tumours56. Their potential to study the pathological functions of tissue-resident immune cells and immune cell infiltrates in inflamed tissues in rheumatic diseases has not yet been explored. The majority of our understanding of the human immune system relies on circulating immune cells. However, given that subsets of immune cells can permanently reside in tissue niches, in situ analysis of these cells becomes essential to understand their function in immune surveillance and in disease. For example, tissue-resident memory T (TRM) cells have been found in nearly all non-lymphoid tissues57. In addition to providing local protection against frequently encountered pathogens, the dysregulation of TRM cells has been implicated in the pathology of several rheumatic diseases, including RA and juvenile idiopathic arthritis (JIA)58. Similarly to Trm cells, the localization of other immune cells to non-lymphoid tissues is indispensable for protecting tissues from invading pathogens59,60. Next-generation imaging technologies offer the opportunity to examine the functions of tissue-resident immune cells in immunosurveillance and in the pathophysiology of immune-mediated diseases. We envision that the use of such technologies will soon become widespread and will provide insights into the physiological and pathological functions of non-circulating immune cells.
Other types of single-cell proteomics.
Single-cell western blotting enables the analysis of individual cells seeded in microwells by thin-layer gel electrophoresis and photoimmobilization, followed by antibody hybridization61. This method separates denatured proteins by size in a similar manner to conventional western blotting, thereby offering additional precision and facilitating isoform-specific detection. Single-cell western blotting has been used to dissect the heterogeneity of neural stem cells and to track their lineage commitments during differentiation61. Alternatively, fluorescence-based imaging can be performed using microscopes commonly available in academic laboratories. By using multiple rounds of staining and de-staining with nanoparticles, up to 25 biomarkers can be reliably measured in single cells with quantum dot imaging62. An established protocol exists for quantum dot-based immunohistochemistry analysis63, which can be applied to analyse formalin-fixed and paraffin-embedded (FFPE) clinical samples for rheumatology research. Another method, co-detection by indexing (CODEX)64, provides high-parameter imaging with dimensionality comparable to mass-spectrometry-based methods and requires minimal modifications to conventional fluorescence microscopes. The invention of this analytic platform has enabled high-dimensional imaging of splenic tissues from lupus-prone MRL/lpr mice64.
Technologies to measure the signalling molecules that are secreted from single cells, such as cytokines, are powerful tools that facilitate the functional characterization of individual immune cells. Microengraving methods enable single cells to be trapped and cultured in microwells at sub-nanolitre volumes65. Secreted proteins can then be captured and detected by capping the microwells with substrate-coated glass slides. This technology, initially developed to accelerate the selection of hybridomas from polyclonal mixtures, has been used to interrogate single-cell variations in cytokine secretion by human PBMCs in response to immune stimulation66. Similarly, single-cell barcode chip (SCBC) uses a microfluidic system to simultaneously measure multiple molecules secreted from single cells and has been used to dissect the functional heterogeneity of cytotoxic T cells67.
Single-cell epigenomics
Single-cell epigenomic technologies provide researchers with exciting opportunities for the development of precision medicine. The epigenome is highly dynamic and is regulated by internal and external factors such as hormones, metabolites, microorganisms, environmental factors and ageing. The highly heterogeneous nature of rheumatic disease manifestations between patients suggests an important function for epigenomic variation in their pathogenesis. Advances in epigenomic technologies have helped researchers to interrogate multiple layers of the epigenome in individual cells68. These single-cell assays have yet to be used in rheumatic disease research, but it is hoped that they will provide opportunities to improve our understanding of the molecular mechanisms underlying rheumatic disease development. In this section, we discuss cutting-edge single-cell epigenomic technologies and describe areas of rheumatology research to which these exciting technologies could be applied in the future.
DNA methylation analysis.
The molecular links between DNA methylation and embryonic development, genomic stability, transcription regulation, gene imprinting, repetitive DNA silencing and X chromosome inactivation have been extensively studied69,70. Functionally, DNA methylation at promoters is mainly associated with gene silencing, whereas intragenic methylation at actively transcribed genes is essential for transcriptional fidelity. Enhancers and insulators are also regulated by DNA methylation, which affects transcription activities by altering their interactions with target promoters. Methylation at CpG sites is a classic epigenetic mechanism by which de novo cytosine modifications can be stably transmitted through mitosis and meiosis and between generations71. Several studies have also shown that DNA methylation patterns that are altered as a result of environmental perturbation remain evident for several generations after the external stimuli have been removed72. Importantly, both global disruption and locus-specific alterations to DNA methylation patterns are associated with human diseases, including rheumatic diseases.
The current gold standard for the detection of DNA methylation is bisulfite sequencing. The treatment of genomic DNA with sodium bisulfite converts unmethylated cytosines to uracils, whereas methylated cytosines are resistant to this chemical modification. This approach essentially transforms epigenetic marks into a genetic code that can be measured by microarray or NGS technology. The first human DNA methylome with single-base resolution was published in 2009 (REF73), and advances in NGS technologies facilitated single-cell DNA methylome profiling shortly thereafter. The first demonstration of single-cell DNA methylation analysis used reduced representation bisulfite sequencing (RRBS), in which restriction enzyme digestion is used to enrich genomic regions with high GC content before bisulfite sequencing is carried out74. Approximately 1.5 million CpG sites can be analysed in individual cells using this method. An alternative approach with improved CpG coverage, single-cell bisulfite sequencing (scBS-seq), used a modified version of post-bisulfite adaptor tagging (PBAT) to enable the retrieval of the methylation status of 1.8−7.7 million CpG sites75. A third method, single-cell whole-genome bisulfite sequencing (WGBS), enabled the analysis of large numbers of cells with a coverage of 1.4 million CpG sites76. Notably, WGBS has been used to profile the DNA methylomes of haematopoietic stem cells and progenitor cells77.
An important advantage of NGS-based methods relative to microarrays is the identification of new methylation sites that are not restricted to the content immobilized on the array. Although less prevalent than CpG dinucleotides, methylation at non-CpG residues occurs at higher rates in certain cell subtypes, such as embryonic stem cells78. The biological functions associated with non-CpG methylation have yet to be identified. Additionally, although microarrays measure CpG sites that represent nearly 99% of genes listed in the RefSeq database and 96% of CpG islands, only ~2% of the total CpG dinucleotides in the genome are covered79. DNA methylome sequencing can be used to identify differentially methylated regions (DMRs) located outside of the genetic regions that are associated with rheumatic diseases, thereby expanding the analysis to include regulatory elements such as enhancers and silencers.
Over the past decade, the model for the maintenance of DNA methylation dynamics has been revised to include active demethylation mediated by the TET enzyme family80, and the genome-wide distributions of two intermediates of the active demethylation process, 5-hydroxymethylcytosine (5hmC) and 5-formylcytosine (5fC), have been reported in single cells81. In vitro glucosylation of 5hmC followed by modification-dependent digestion with the restriction enzyme AbaSI and NGS (termed scAba-seq) enabled genome-wide 5hmC profiling in single cells81, whereas chemical labelling of 5fC with malononitrile resulted in a C-to-T conversion during PCR (termed CLEVER-seq), enabling researchers to map the genome-wide 5fC landscape82. Emerging evidence from similar techniques suggests an important function for DNA demethylation in the activation and development of B cells and T cells83. Together with bisulfite sequencing, analyses of distinct DNA modifications at single-base resolution and single-cell resolution is likely to be able to reveal aberrant DNA methylation patterns associated with rheumatic diseases.
DNA methylome studies in rheumatic diseases have mostly used microarray-based approaches, and single-cell analysis has yet to be reported. Microarray-based DNA methylome analyses have found DMRs in patients with SLE84, many of which were localized to type I interferon-inducible genes85–89. Altered DNA methylation patterns have also been associated with ethnicity90, clinical manifestations91–93 and disease activity94,95 in patients with SLE. Analyses of cells from monozygotic twins discordant for SLE revealed widespread alterations in the DNA methylation landscape in affected twins compared with unaffected twins96,97. Similarly, DNA methylome profiling of monozygotic twins discordant for ACPA-positive preclinical RA revealed DMRs between affected and unaffected twins98. Together with the presence of RA-associated DMRs in immune cells99,100 and synovial fibroblasts101–105, this evidence strongly suggests the involvement of DNA methylation in the pathogenesis of RA. Additionally, DMRs have been implicated in the pathogenesis of systemic sclerosis (SSc)106,107, pSS108 and JIA109.
Single-cell DNA methylome sequencing has been used in the field of cancer research to discover DMRs that have the potential to be used for diagnosis, to identify new therapeutic targets and to predict treatment responses110. These cancer research studies have laid a solid foundation for the application of single-cell DNA methylome analysis to clinical samples from patients with other diseases. We anticipate that the first single-cell DNA methylome analysis on clinical samples from patients with rheumatic diseases will soon be reported and will shed new light on the function of this important layer of epigenetic regulation in pathogenesis.
Histone modification analysis.
Histone post-translational modifications, often referred to as histone marks, are an important epigenetic mechanism for the regulation of chromatin dynamics111. Histone modifications are essential for successful haematopoietic cell development, effective immune responses against pathogens and the maintenance of immune tolerance112,113. The dynamics of histone modifications are maintained by several classes of chromatin-modifying enzymes, the dysregulation of which has been directly linked to human diseases114. Changes in the amount and distribution of histone modifications facilitate the establishment of pathological gene expression programmes in diseases such as cancer115 and the defective binding to histone post-translational modifications by reader proteins disrupts normal biological activities, which can also lead to the development of disease. For example, a genetic mutation in V(D)J recombination-activating protein 2 (RAG2) that abrogates its ability to bind histone H3 trimethylation at lysine 3 (H3K4me3) marks results in inefficient V(D)J recombination and immunodeficiency116. Together, the mis-writing, mis-erasing and mis-interpretation of histone post-translational modifications are signatures of a wide variety of human diseases117. Therapeutically targeting either chromatin-modifying enzymes or post-translational-modification-dependent protein-chromatin interactions therefore holds great promise for many diseases118.
To date, interrogating the dysregulated histone marks that are associated with rheumatic diseases has been challenging, mostly owing to technical limitations. Conventional assays such as chromatin immunoprecipitation sequencing (ChIP-seq) and immunoblotting require large numbers of cells and generate averaged overviews of heterogeneous populations of cells. However, ChIP-seq analysis of histone modifications at single-cell resolution has been reported119. Combining microfluidics-based barcoding of individual cells, immunoprecipitation of nucleosomes from a pool of cells and NGS enables the genome-wide locus-specific profiling of histone marks in single cells119. This technology, although powerful and able to demonstrate heterogeneity in histone marks at a single-cell level, has low sensitivity. A complementary technology, in which in situ hybridization and proximity ligation assays (PLAs) are integrated, also enables the locus-specific detection of histone marks in single cells120. However, this technology does not provide information about the genome-wide distributions of histone marks, and low throughput prevents it from being broadly applied.
New methods to facilitate genome-wide locus-specific analyses of histone marks in single cells are still required; however, one method has been developed to at least partially fill this technological gap. Epigenetic landscape profiling using cytometry by time-of-flight (EpiTOF) takes advantage of the high multiplexing power, single-cell resolution and the potential for quantitative measurement provided by mass cytometry to simultaneously detect the bulk concentrations of a variety of histone marks in individual cells121,122. Using this analytic platform, immune cell subtype-specific and haematopoietic lineage-specific histone modification profiles have been identified that predict immune cell identities at the single-cell level122. EpiTOF has the potential to serve as a powerful discovery platform to reveal dysregulated histone marks in patients with rheumatic diseases and to facilitate the identification of previously uncharacterized immune cell subtypes defined by unique histone modification patterns. An important use of EpiTOF data will be to provide guidance for ChIP-seq analyses, which can subsequently be used to discover locus-specific histone modification changes. The modulation of histone marks by manipulating chromatin-modifying enzymes can further inform about the reversibility of phenotypes of interest and provide a basis for the development of new therapies that target chromatin-modifying enzymes. Together with other epigenomic methods, such as chromatin structure sequencing and DNA methylome sequencing, single-cell histone modification analysis using techniques such as EpiTOF will help researchers to better understand the function of epigenetic regulations in the pathophysiology of rheumatic diseases.
Chromatin structure analysis.
The invention of assay for transposase-accessible chromatin using sequencing (ATAC-seq) has revolutionized the genome-wide analysis of chromatin structure123. This method makes use of the preferential insertion of sequencing adaptors into open chromatin by the hyperreactive Tn5 transposase. The sequencing reads therefore enable the inference of the chromatin accessibility landscape, which would have traditionally been mapped by DNase I hypersensitivity assays. Inaccessible sections of DNA of ~147 base pairs indicate the presence of nucleosomes, thereby providing a nucleosome positioning map similar to results obtained by micrococcal nuclease-based assays. Moreover, ATAC-seq enables the identification of transcription factor ‘footprints’, the chromatin occupancies of which are often identified by ChIP-seq.
Single-cell resolution in ATAC-seq (scATAC-seq) can be achieved using two independent approaches. One strategy uses combinatorial cellular indexing, in which individual cells are labelled with barcoded Tn5 transposase and then another indexed tag is introduced by PCR124. This method has been used to plot the single-cell chromatin accessibility maps of over 20,000 cells during Drosophila melanogaster embryonic development125. A second scATAC-seq approach involves microfluidic handling of individual cells followed by ATAC and PCR amplification using an integrated fluidics circuit126. Pooled chromatin accessibility landscapes from single cells resemble combined data derived from millions of cells; however, single-cell data sets preserve the individuality of single cells and showcase the heterogeneity within a population of cells.
Low-input ATAC-seq or scATAC-seq analyses of human immune cells have brought about many exciting discoveries. ATAC-seq has been used to map the epigenomic landscapes of haematopoietic stem cells and progenitor cells and has revealed a regulatory network that governs immune cell differentiation during haematopoiesis127,128. The lineage-specific chromatin dynamics of haematopoiesis were comprehensively characterized using data sets from ATAC-seq, RNA-seq and an advanced ChIP-seq method that requires as few as 500 cells129. The integration of scATAC-seq with TCR sequencing lead to the identification of clonespecific variability in chromatin accessibility within naive and memory CD4+ T cells and between TH1 cells, TH2 cells and TH17 cells, thereby uncovering dysregulated cis-elements and trans-elements associated with malignant clonal expansion in T cell leukaemia130. However, ATAC-seq data sets from patients with rheumatic diseases are scarce. CD4+CD28+KIR+CD11ahi cells and CD4+CD28+KIR−CD11alo cells purified from patients with SLE were characterized by genome-wide increases in chromatin accessibility, and CD4+CD28+ KIR+CD11ahi cells were enriched for differentially accessible regions in pro-inflammatory genes131. Similarly, using >1,000 naive B cells purified by FACS from patients with SLE, an SLE-specific chromatin accessibility landscape was identified that showed chromatin decondensation surrounding genes involved in B cell activation132. We envision that scATAC-seq will soon be used to analyse immune cells and clinically relevant tissues in rheumatic diseases.
In addition to ATAC-seq, several new technologies have facilitated the identification of the chromatin state in single cells. Single-cell DNase sequencing (scDNase-seq) can be used to identify sites that are hypersensitive to DNase I digestion, which mark open chromatin and active regulatory DNA elements133. Both fresh cells and FFPE tissues are compatible with scDNase-seq, making it a compelling option for rheumatology research. Another methodological advance has enabled Hi-C, a high-throughput variant of chromosome conformation capture, to be performed on single cells (scHi-C)134. Hi-C reveals the spatial organization of chromosomes by mapping contacts between DNA elements that are distantly located in the linear genomic sequence and megabase-sized topological domains where intradomain DNA segments frequently interact with each other. By performing Hi-C on primary human immune cells from patients with rheumatic diseases, genes that interact with non-coding disease-associated single-nucleotide polymorphisms have been identified135. Disruption of the architecture of topologically associated domains has also been linked to human diseases136. Hi-C and scHi-C are excellent experimental platforms for this mostly uncharted area of rheumatic disease research.
To better understand the molecular mechanisms by which differentially accessible chromatin states are created and maintained at specific loci, a systems-level analysis involving a combination of techniques such as EpiTOF, ChIP-seq and DNA methylome profiling is required to provide a comprehensive view of the epigenome. These technologies have complementary strengths that will enable researchers to link chromatin accessibility to histone and DNA modifications and have the potential to identify opportunities to alter chromatin states by manipulating chromatin-modifying enzymes.
Single-cell multi-omics technologies
Analysing DNA, RNA and proteins in individual cells can provide direct and powerful evidence for how layers of regulatory mechanisms work together to control cellular phenotypes. Several methods have been developed to conduct parallel analysis of either genomic, transcriptomic and epigenomic data or genomic, transcriptomic and proteomic data in single cells.
Parallel analyses of the genome, transcriptase and epigenome.
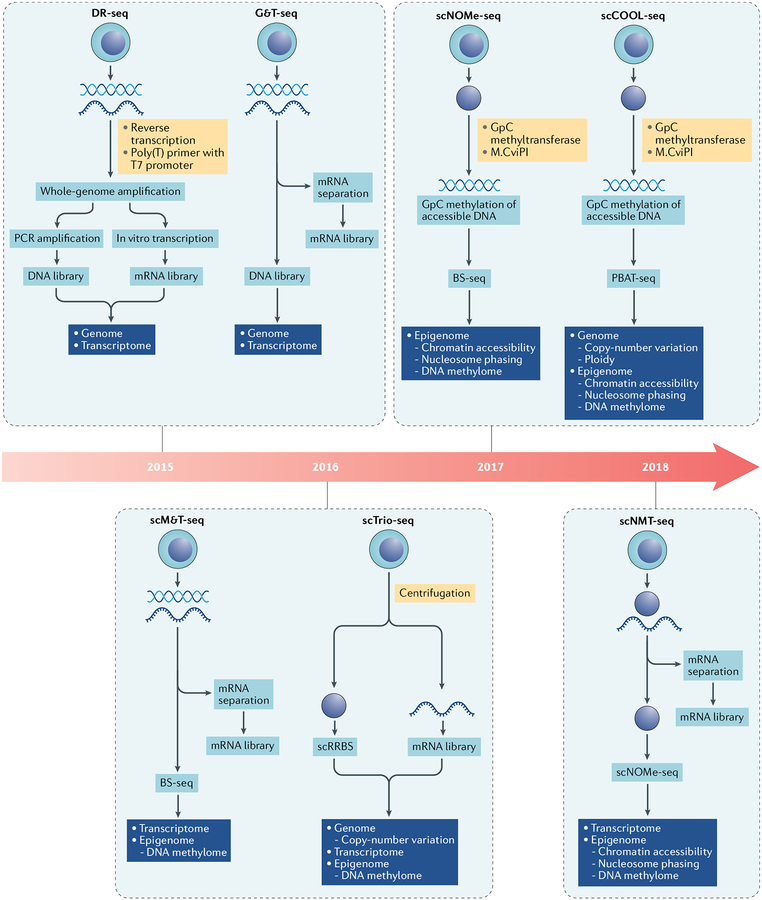
Following the introduction of the first single-cell integrated genomic DNA and mRNA sequencing (DR-seq) analysis137 in 2015, several NGS-based multi-omics methods have been described (FIG. 2). In DR-seq, a T7 promoter is added to mRNA transcripts, which can then be amplified using in vitro transcription to generate a transcriptomic library while, in parallel, a genomic library is constructed from the same sample using whole-genome amplification. An alternative strategy, genome and transcriptome sequencing (G&T-seq)138, involves the physical separation of mRNA from DNA using biotinylated poly(T) primers. The independently generated transcriptomic and genomic libraries are then analysed by NGS.
Fig. 2 |. Methods to simultaneously perform genomic, transcriptomic and epigenomic analysis.
Timeline of sequencing-based single-cell technologies for simultaneous analysis of combinations of the genome, transcriptome and epigenome, showing the basic methods for individual technologies and the biological info rmation that can be obtained using these platforms. BS-seq, bisulfite sequencing; DR-seq, DNA and mRNA sequencing; G&T-seq, genom e and transcriptome sequencing; PBAT-seq, post-bisulfite adaptor tagging sequencing; scCOOL-seq, single-cell chromatin overall omic-scale landscape sequencing; scM&T-seq, single-cell methylome and transcriptome sequencing; scNMT-seq, single-cell nucleosome, methylation and transcription sequencing; scNOMe-seq, single-cell nucleosome occupancy and methylome sequencing; scRRBS, single-cell reduced representation bisulfite sequencing; scTrio-seq; single-cell triple omics sequencing.
At the epigenomic level, several methods have been described to interrogate different layers of epigenetic regulation simultaneously, a few of which facilitate integrative analyses with genomics and/or transcriptomics. In single-cell methylome and transcriptome sequencing (scM&T-seq)139, mRNA is captured by poly(T)-coated beads for RNA-seq and genomic DNA is treated with bisulfite for DNA methylome analysis. Nucleosome occupancy and methylome sequencing (NOMe-seq)140,141 makes use of the GpC methyltransferase M.CviPI, which is isolated from chlorella virus, to methylate accessible GpC sites in vitro, leveraging the fact that GpC methylation does not occur naturally in the mammalian genome. After bisulfite conversion and high-throughput sequencing, both endogenous CpG methylation and M.CviPI-mediated GpC methylation can be evaluated bioinformatically to infer the methylome and chromatin accessibility, respectively. Single-cell chromatin overall omic-scale landscape sequencing (scCOOL-seq) integrates NOMe-seq and PBAT sequencing to enhance the sensitivity of methylome profiling and incorporates genomic analyses of copy-number variation and chromosome ploidy142. Another method that builds on NOMe-seq involves combining it with RNA-seq to create single-cell nucleosome, methylation and transcription sequencing (scNMT-seq)143. In this method, mRNA is purified from single-cell lysates using poly(T) oligonucleotides immobilized on magnetic beads for use in transcriptomic profiling and chromatin-containing genomic DNA is analysed by NOMe-seq. Yet another method, single-cell triple omics sequencing (scTrio-seq), enables researchers to interrogate copy-number variation, gene expression and DNA methylation simulateously144.
These powerful approaches have been used to investigate single-cell heterogeneity in biological contexts such as embryonic development and cancers137–139,142–144, but their utility for studying the human immune system and immune-mediated diseases has yet to be explored. Previous studies have shown differential DNA methylation in patients with rheumatic diseases108,145, and it is hoped that multi-omics analyses will reveal the functional effects of these methylation patterns on the transcriptome in the same cells. Multi-omics technologies can also be used to identify transcriptomic and epigenomic alterations in cells that carry chromosomal defects. The continuous maturation of these technologies in precision and throughput and improvements in computational algorithms to facilitate data interpretation will enable multi-omics analyses to maximize the data generated from precious clinical samples and yield one-of-a-kind data sets to facilitate integrative analyses.
Parallel analyses of the genome, transcriptase and proteome.
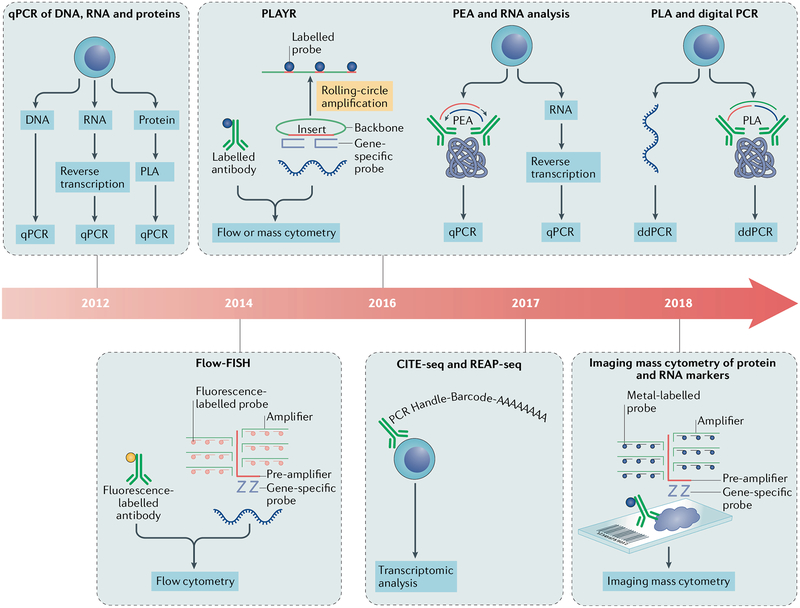
Flow cytometry and fluorescence microscopy are most frequently used to study protein expression; however, over the past few years, methods to reliably detect RNA using these technologies have been described (FIG. 3). One such technique modifies FISH to use branched DNA technology (flow-FISH)146, whereas other techniques such as PLA147,148 and proximity extension assay (PEA)149 simultaneously analyse protein and RNA at single-cell resolution. These technologies enable researchers to analyse both fresh frozen and FFPE samples from clinically important tissues, such as kidney tissue from patients with lupus nephritis or skin from patients with SSc. Following the advent of mass cytometry and mass spectrometry-based imaging systems, innovative methods have been developed to combine nucleic acid and protein marker measurements. In proximity ligation assay for RNA (PLAYR), the concept ofproximity ligation is applied to RNA quantification using flow cytometry or mass cytometry150. Sequence-specific probes target unique RNA transcripts to generate ligation products in a proximity-dependent fashion, which are then amplified by a rolling circle reaction to enhance assay sensitivity. This approach150 uses the full spectrum of the lanthanide series in mass cytometry, thereby enabling the parallel analyses of >40 RNA and/or protein targets. PLAYR has been used to monitor cell signalling cascades (protein phosphorylation) and cytokine production at a transcriptional level in specific immune cell subtypes (immunophenotypic markers) in stimulated PBMCs150. A method using RNAscope technology151 has also been described for detecting RNA in tissue samples in situ using imaging mass cytometry152. RNA signals from FFPE tissue samples are amplified by tree-like DNA scaffolds that have multiple binding sites for lanthanide-labelled probes. This highly sensitive assay152, which can detect fewer than ten copies of a specific mRNA molecule, enables multiplexed analyses of both RNA and protein markers at subcellular resolution. This method has been successfully used to measure the amount of CXC-chemokine ligand 10 expression and T cell infiltration in FFPE tissue samples from patients with breast cancer152. Rheumatic disease researchers could use a similar approach to analyse clinically relevant samples, such as synovial tissue from patients with RA.
Fig. 3 |. Methods to simultaneously perform nucleic acid and protein marker analysis.
Timeline of single-cell technologies that enable thesim ultaneous analysis of nucleic acid and protein markers, showing the basic methods for individual technologies and the readouts of these platforms. CITE-seq, cellular indexing of transcriptomes and epitopes by sequencing; ddPCR, droplet digital PCR; FISH, fluorescence in situ hybridization; PEA, proxim ity extension assay; PLA, proximity ligation assay; PLAYR, proximity ligation assay for RNA; qPCR, quantitative PCR; REAP-seq, RNA expression and protein sequencing.
Another class of technology converts protein concentrations into measurements of DNA abundance, which can be analysed by NGS-based methods along-side single-cell transcriptomic profiling (FIG. 3). Cellular indexing of transcriptomes and epitopes by sequencing (CITE-seq)153 and RNA expression and protein sequencing (REAP-seq)154 involve staining cells with epitope-specific affinity reagents conjugated with barcoded oligonucleotides that contain poly(A) tails. By taking advantage of the DNA polymerase activity of reverse transcriptase, the oligonucleotide labels can be amplified during sample processing for single-cell RNA-seq. The epitope concentrations inferred by the oligonucleotide reads can thus be obtained during transcriptomic analysis in single cells. CITE-seq and REAP-seq were initially benchmarked on cord blood and PBMCs, respectively. Both methods can be adapted with minimal modification to interrogate the immune systems in patients with rheumatic diseases. Importantly, performing CITE-seq or REAP-seq for rheumatology research can be facilitated by several commercially available DNA-barcoded antibodies and fully validated sample processing platforms. In principle, the DNA barcodes in these systems provide an almost unrestricted multiplexing capability, limited only by the availability of highly specific affinity reagents, and when coupled with a microfluidic device, CITE-seq and REAP-seq can be used to analyse hundreds of thousands of single cells simultaneously. Importantly, CITE-seq and REAP-seq have the potential to bridge decades of accumulated knowledge from cytometry research with the latest breakthroughs in single-cell RNA-seq. Cells important for pathogenesis that have been defined previously by immunophenotypic markers or autoreactive TCR binding to antigen-loaded MHC tetramers can now be identified computationally in scRNA-seq data sets using the same protein markers. Post-translational modifications to cell surface molecules, in particular glycosylation, have been implicated in the pathogenesis of many diseases155; CITE-seq and REAP-seq offer researchers the opportunity to interrogate the transcriptional programmes in cells that have distinct patterns of post-transcriptional modification on immunoregulatory cell surface molecules. The same concept applies to alternatively spliced isoforms of molecules such as CD45RA and CD45RO, which are used in flow cytometry to differentiate between naive and memory T cells and are difficult to quantitatively distinguish using transcript data. We anticipate that in the near future a protocol compatible with intracellular proteins or post-translational-modification-specific markers will be developed to enable the measurement of important markers alongside the transcriptome.
Conclusions
Over the past decade, several revolutionary single-cell technologies have been invented that offer the possibility to profile the genome, transcriptome, epigenome and proteome of individual cells to an unprecedented depth. Future efforts will require teams of basic researchers, clinicians, computational biologists and other researchers with multidisciplinary expertise in both academic institutions and in the biotechnology and pharmaceutical industries to work together. We anticipate that in the coming years, new single-cell technologies will be introduced that have improved resolution and throughput and that computational algorithms and methods will be developed that are specialized for the analysis of data from high-throughput single-cell technologies. Experiments that conceptually advance our understanding of the human immune system and rheumatic diseases using these new single-cell technologies are eagerly awaited. However, a fundamental question that should be considered is how these technologies can be combined to enrich our understanding of the human immune system, to identify causal pathogenic mechanisms in rheumatic diseases and, ultimately, to improve human health.
Key points.
Many analytical platforms are available for the quantitative analyses of the genome, epigenome, transcriptome and proteome of single cells, although these technologies have not been fully exploited in rheumatology research.
Single-cell RNA sequencing facilitates the simultaneous interrogation of the transcriptome of thousands of cells and transcript-based analyses of paired antigen receptor sequences.
Mass cytometry enables the deep immunophenotyping and functional characterization of protein markers that, when coupled with mass spectrometry imaging, provide information on the spatial relationships between molecules.
Many analytical platforms have been developed to investigate different layers of epigenomic regulation in single cells, including DNA methylation, histone modifications, chromatin accessibility and chromatin conformation.
High-dimensional multi-omics analyses enable the direct comparison of DNA, RNA and proteins and/or the epigenome in individual cells and offer great potential for understanding rheumatic diseases.
Acknowledgements
The authors thank the Autoimmunity Centers of Excellence (5U19AI110491–04 and 5UM1A110498–04), a research consortium supported by grants from the US National Institute of Allergy and Infectious Diseases (to P.J.U.), the Donald E. and Delia B. Baxter Foundation (to P.J.U.), E. F. Adler (to P.J.U.), the Henry Gustav Floren Trust (to P.J.U.) and the US NIH (5R01AI125197–02 to P.J.U. andP.K.).
Reviewer information
Nature Reviews Rheumatology thanks F. Mizoguchi, V. Malmstrom and K. Wei for their contribution to the peer review of this work.
Footnotes
Competing interests
The authors declare no competing interests.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Mardis ER The impact of next-generation sequencing technology on genetics. Trends Genet. 24, 133–141 (2008). [DOI] [PubMed] [Google Scholar]
- 2.Metzker ML Sequencing technologies — the next generation. Nat. Rev Genet 11, 31–46 (2010). [DOI] [PubMed] [Google Scholar]
- 3.Bendall SC et al. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science 332, 687–696 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Helmick CG et al. Estimates of the prevalence of arthritis and other rheumatic conditions in the United States, part I. Arthritis Rheum. 58, 15–25 (2008). [DOI] [PubMed] [Google Scholar]
- 5.Lawrence RC et al. Estimates of the prevalence of arthritis and other rheumatic conditions in the United States, part II. Arthritis Rheum. 58, 26–35 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang F et al. Defining inflammatory cell states in rheumatoid arthritis joint synovial tissues by integrating single-cell transcriptomics and mass cytometry. Preprint at bioRxiv 10.1101/351130 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Donlin LT et al. Methods for high-dimensonal analysis of cells dissociated from cyropreserved synovial tissue. Arthritis Res. Ther 20, 139 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gawad C, Koh W & Quake SR Single-cell genome sequencing: current state of the science. Nat. Rev Genet 17, 175–188 (2016). [DOI] [PubMed] [Google Scholar]
- 9.Shapiro E, Biezuner T & Linnarsson S Single-cell sequencing-based technologies will revolutionize whole-organism science. Nat. Rev Genet 14, 618–630 (2013). [DOI] [PubMed] [Google Scholar]
- 10.Tang F et al. mRNA-Seq whole-transcriptome analysis of a single cell. Nat. Methods 6, 377–382 (2009). [DOI] [PubMed] [Google Scholar]
- 11.Svensson V et al. Power analysis of single-cell RNA-sequencing experiments. Nat. Methods 14, 381–387 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ziegenhain C et al. Comparative analysis of single-cell RNA sequencing methods. Mol. Cell 65, 631–643 (2017). [DOI] [PubMed] [Google Scholar]
- 13.Crosetto N, Bienko M & van Oudenaarden A Spatially resolved transcriptomics and beyond. Nat. Rev. Genet 16, 57–66 (2015). [DOI] [PubMed] [Google Scholar]
- 14.Wang X et al. Three-dimensional intact-tissue sequencing of single-cell transcriptional states. Science 361, eaat5691 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Raj A, van den Bogaard P, Rifkin SA, van Oudenaarden A & Tyagi S Imaging individual mRNA molecules using multiple singly labeled probes. Nat. Methods 5, 877–879 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen KH, Boettiger AN, Moffitt JR, Wang S & Zhuang X RNA imaging. Spatially resolved, highly multiplexed RNA profiling in single cells. Science 348, aaa6090 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stegle O, Teichmann SA & Marioni JC Computational and analytical challenges in single-cell transcriptomics. 16, 133–145 Nat. Rev. Genet.( 2015). [DOI] [PubMed] [Google Scholar]
- 18.Grun D & van Oudenaarden A Design and analysis of single-cell sequencing experiments. Cell 163, 799–810 (2015). [DOI] [PubMed] [Google Scholar]
- 19.Kang HM et al. Multiplexed droplet single-cell RNA-sequencing using natural genetic variation. Nat. Biotechnol 36, 89–94 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klein AM et al. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell 161, 1187–1201 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Macosko EZ et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161, 1202–1214 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gierahn TM et al. Seq-Well: portable, low-cost RNA sequencing of single cells at high throughput. Nat. Methods 14, 395–398 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fan HC, Fu GK & Fodor SP Expression profiling. Combinatorial labeling of single cells for gene expression cytometry. Science 347, 1258367 (2015). [DOI] [PubMed] [Google Scholar]
- 24.Habib N et al. Div-Seq: Single-nucleus RNA-Seq reveals dynamics of rare adult newborn neurons.Science 353, 925–928 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Habib N et al. Massively parallel single-nucleus RNA-seq with DroNc-seq. Nat. Methods 14, 955–958 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bennett L et al. Interferon and granulopoiesis signatures in systemic lupus erythematosus blood. J. Exp. Med 197,711–723(2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baechler EC et al. Interferon-inducible gene expression signature in peripheral blood cells of patients with severe lupus. Proc. Natl Acad. Sci. USA 100, 2610–2615 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Der E et al. Single cell RNA sequencing to dissect the molecular heterogeneity in lupus nephritis. JCI Insight 2, 93009 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stephenson W et al. Single-cell RNA-seq of rheumatoid arthritis synovial tissue using low-cost microfluidic instrumentation. Nat. Commun 9, 791 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mizoguchi F et al. Functionally distinct disease-associated fibroblast subsets in rheumatoid arthritis. Nat. Commun 9, 789 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hu F et al. Impaired CD27+IgD+ B cells with altered gene signature in rheumatoid arthritis. Front. Immunol 9, 626(2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sade-Feldman M et al. Defining T cell states associated with response to checkpoint immunotherapy in melanoma. Cell 175, 998–1013 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tan YC et al. Barcode-enabled sequencing of plasmablast antibody repertoires in rheumatoid arthritis. Arthritis Rheumatol. 66, 2706–2715 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kinslow JD et al. Elevated IgA plasmablast levels in subjects at risk of developing rheumatoid arthritis. Arthritis Rheumatol 68, 237234.2383 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Elliott SE et al. Affinity maturation drives epitope spreading and generation of pro-inflammatory anti-citrullinated protein antibodies in rheumatoid arthritis. Arthritis Rheumatol. 70, 1946–1958 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blum LK et al. Circulating plasmablasts are elevated and produce pathogenic anti-endothelial cell autoantibodies in idiopathic pulmonary arterial hypertension. Eur. J. Immunol 48, 874–884 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Amagai M, Klaus-Kovtun V & Stanley JR Autoantibodies against a novel epithelial cadherin in pemphigus vulgaris, a disease of cell adhesion. Cell 67, 869–877 (1991). [DOI] [PubMed] [Google Scholar]
- 38.Ellebrecht CT et al. Reengineering chimeric antigen receptor T cells for targeted therapy of autoimmune disease. Science 353, 179–184 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Birnbaum ME et al. Deconstructing the peptide-MHC specificity of T cell recognition. Cell 157, 1073–1087 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gee MH et al. Antigen identification for orphan T cell receptors expressed on tumor-infiltrating lymphocytes. Cell 172, 549–563 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen DY et al. The potential role of Th 17 cells and Th 17-related cytokines in the pathogenesis of lupus nephritis. Lupus 21, 1385–1396 (2012). [DOI] [PubMed] [Google Scholar]
- 42.Shah K et al. Dysregulated balance of Th 17 and Th 1 cells in systemic lupus erythematosus. Arthritis Res. Ther 12, R53 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brusko TM, Putnam AL & Bluestone JA Human regulatory T cells: role in autoimmune disease and therapeutic opportunities. Immunol. Rev 223, 371–390 (2008). [DOI] [PubMed] [Google Scholar]
- 44.Vogel C & Marcotte EM Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat. Rev. Genet 13, 227–232 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Spitzer MH & Nolan GP Mass cytometry: single cells, many features. Cell 165, 780–791 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.O’Gorman WE et al. Single-cell systems-level analysis of human Toll-like receptor activation defines a chemokine signature in patients with systemic lupus erythematosus. J. Allergy Clin. Immunol 136, 1326–1336(2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.O’Gorman WE et al. Mass cytometry identifies a distinct monocyte cytokine signature shared by clinically heterogeneous pediatric SLE patients. J. Autoimmun 81, 74–89 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rao DA et al. Pathologically expanded peripheral T helper cell subset drives B cells in rheumatoid arthritis. Nature 542, 110–114 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Haskett S et al. Identification of novel CD4+ T cell subsets in the target tissue of Sjogren’s syndrome and their differential regulation by the lymphotoxin/LIGHT signaling axis. J. Immunol 197, 3806–3819 (2016). [DOI] [PubMed] [Google Scholar]
- 50.Mingueneau M et al. Cytometry by time-of-flight immunophenotyping identifies a blood Sjogren’s signature correlating with disease activity and glandular inflammation. J. Allergy Clin. Immunol 137, 1809–1821 (2016). [DOI] [PubMed] [Google Scholar]
- 51.Fonseka CY et al. Mixed-effects association of single cells identifies an expanded effector CD4+ T cell subset in rheumatoid arthritis. Sci. Transl. Med 10, eaaq0305 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McDonnell LA & Heeren RM Imaging mass spectrometry. Mass Spectrom. Rev 26, 606–643 (2007). [DOI] [PubMed] [Google Scholar]
- 53.Angelo M et al. Multiplexed ion beam imaging of human breast tumors. Nat. Med 20, 436–442 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Giesen C et al. Highly multiplexed imaging of tumor tissues with subcellular resolution by mass cytometry. Nat. Methods 11, 417–422 (2014). [DOI] [PubMed] [Google Scholar]
- 55.Bodenmiller B Multiplexed epitope-based tissue imaging for discovery and healthcare applications.CellSyst. 2, 225–238 (2016). [DOI] [PubMed] [Google Scholar]
- 56.Chevrier S et al. An immune atlas of clear cell renal cell carcinoma. Cell 169, 736–749 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mueller SN & Mackay LK Tissue-resident memory T cells: local specialists in immune defence. Nat. Rev. Immunol 16, 79–89 (2016). [DOI] [PubMed] [Google Scholar]
- 58.Clark RA Resident memory T cells in human health and disease. Sci. Transl. Med 7, 269rv261 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Davies LC, Jenkins SJ, Allen JE & Taylor R R. Tissue-resident macrophages. Nat. Immunol 14, 986–995 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fan X & Rudensky AY Hallmarks of tissue-resident lymphocytes. Cell 164, 1198–1211 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hughes AJ et al. Single-cell western blotting.Nat. Methods 11, 749–755 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zrazhevskiy P & Gao X Quantum dot imaging platform for single-cell molecular profiling. Nat. Commun 4, 1619 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xing Y et al. Bioconjugated quantum dots for multiplexed and quantitative immunohistochemistry. Nat. Protoc 2, 1152–1165(2007). [DOI] [PubMed] [Google Scholar]
- 64.Goltsev Y et al. Deep profiling of mouse splenic architecture with CODEX multiplexed imaging. Cell 174, 968–981 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Love JC, Ronan JL, Grotenbreg GM, van der Veen AG & Ploegh HL A microengraving method for rapid selection of single cells producing antigen-specific antibodies. Nat. Biotechnol 24, 703–707 (2006). [DOI] [PubMed] [Google Scholar]
- 66.Han Q, Bradshaw EM, Nilsson B, Hafler DA & Love JC Multidimensional analysis of the frequencies and rates of cytokine secretion from single cells by quantitative microengraving. Lab Chip 10, 1391–1400 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ma C et al. A clinical microchip for evaluation of single immune cells reveals high functional heterogeneity in phenotypically similar T cells. Nat. Med 17, 738–743(2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schwartzman O & Tanay A Single-cell epigenomics: techniques and emerging applications. Nat. Rev. Genet 16, 716–726 (2015). [DOI] [PubMed] [Google Scholar]
- 69.Bird A DNA methylation patterns and epigenetic memory. Genes Dev. 16, 6–21 (2002). [DOI] [PubMed] [Google Scholar]
- 70.Jones PA Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat. Rev. Genet 13, 484–492 (2012). [DOI] [PubMed] [Google Scholar]
- 71.Okano M, Bell DW, Haber DA & Li E DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99, 247–257 (1999). [DOI] [PubMed] [Google Scholar]
- 72.Reid MA, Dai Z & Locasale JW The impact of cellular metabolism on chromatin dynamics and epigenetics. Nat. Cell Biol 19, 1298–1306(2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lister R et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 462, 315–322 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Guo H et al. Single-cell methylome landscapes of mouse embryonic stem cells and early embryos analyzed using reduced representation bisulfite sequencing. Genome Res. 23, 2126–2135 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Smallwood SA et al. Single-cell genome-wide bisulfite sequencing for assessing epigenetic heterogeneity. Nat. Methods 11, 817–820 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Farlik M et al. Single-cell DNA methylome sequencing and bioinformatic inference of epigenomic cell-state dynamics. Cell Rep. 10, 1386–1397 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Farlik M et al. DNA methylation dynamics of human hematopoietic stem cell differentiation. Cell Stem Cell 19, 808–822 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ramsahoye BH et al. Non-CpG methylation is prevalent in embryonic stem cells and may be mediated by DNA methyltransferase 3a. Proc. Natl Acad. Sci. USA 97, 5237–5242 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bibikova M et al. High density DNA methylation array with single CpG site resolution. Genomics 98, 288–295(2011). [DOI] [PubMed] [Google Scholar]
- 80.Wu H & Zhang Y Reversing DNA methylation: mechanisms, genomics, and biological functions. Cell 156, 45–68 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mooijman D, Dey SS, Boisset JC, Crosetto N & van Oudenaarden A Single-cell 5hmC sequencing reveals chromosome-wide cell-to-cell variability and enables lineage reconstruction. Nat. Biotechnol 34, 852–856(2016). [DOI] [PubMed] [Google Scholar]
- 82.Zhu C et al. Single-cell 5-formylcytosine landscapes of mammalian early embryos and ESCs at single-base resolution. Cell Stem Cell 20, 720–731 (2017). [DOI] [PubMed] [Google Scholar]
- 83.Tsagaratou A, Lio CJ, Yue X & Rao A TET methylcytosine oxidases in T cell and B cell development and function. Front. Immunol 8, 220 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fernandez AF et al. A DNA methylation fingerprint of 1628 human samples. Genome Res. 22, 407–419 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Absher DM et al. Genome-wide DNA methylation analysis of systemic lupus erythematosus reveals persistent hypomethylation of interferon genes and compositional changes to CD4+ T cell populations. PLOS Genet. 9, e1003678 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Coit P et al. Genome-wide DNA methylation study suggests epigenetic accessibility and transcriptional poising of interferon-regulated genes in naive CD4+T cells from lupus patients. J. Autoimmun 43, 78–84 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Coit P et al. Epigenome profiling reveals significant DNA demethylation of interferon signature genes in lupus neutrophils. J. Autoimmun 58, 59–66 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mok A et al. Genome-wide profiling identifies associations between lupus nephritis and differential methylation of genes regulating tissue hypoxia and type 1 interferon responses. Lupus Sci. Med 3, e000183 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yeung KS et al. Genome-wide DNA methylation analysis of Chinese patients with systemic lupus erythematosus identified hypomethylation in genes related to the type I interferon pathway. PLOS ONE 12, e0169553 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Coit P et al. Ethnicity-specific epigenetic variation in naive CD4+ T cells and the susceptibility to autoimmunity. Epigenetics Chromatin 8, 49 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Coit P et al. Epigenetic reprogramming in naive CD4+ T cells favoring T cell activation and non-Th 1 effector T cell immune response as an early event in lupus flares. Arthritis Rheumatol. 68, 2200–2209 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Coit P et al. Renal involvement in lupus is characterized by unique DNA methylation changes in naive CD4+T cells. J. Autoimmun 61, 29–35 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Renauer P et al. DNA methylation patterns in naive CD4+ T cells identify epigenetic susceptibility loci for malar rash and discoid rash in systemic lupus erythematosus. Lupus Sci. Med 2, e000l0l (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hong KM et al. CD3Z hypermethylation is associated with severe clinical manifestations in systemic lupus erythematosus and reduces CD3Z-chain expression in T cells. Rheumatology 56, 467–476 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Jeffries MA et al. Genome-wide DNA methylation patterns in CD4+ T cells from patients with systemic lupus erythematosus. Epigenetics 6, 593–601 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Javierre BM et al. Changes in the pattern of DNA methylation associate with twin discordance in systemic lupus erythematosus. Genome Res. 20, 170–179 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ulff-Moller CJ et al. Twin DNA methylation profiling reveals flare-dependent interferon signature and B cell promoter hypermethylation in systemic lupus erythematosus. Arthritis Rheumatol. 70, 878–890 (2018). [DOI] [PubMed] [Google Scholar]
- 98.Gomez-Cabrero D et al. High-specificity bioinformatics framework for epigenomic profiling of discordant twins reveals specific and shared markers for ACPA and ACPA-positive rheumatoid arthritis. Genome Med. 8, 124 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Julia A et al. Epigenome-wide association study of rheumatoid arthritis identifies differentially methylated loci in B cells. Hum. Mol. Genet 26, 2803–2811 (2017). [DOI] [PubMed] [Google Scholar]
- 100.Liu Y et al. Epigenome-wide association data implicate DNA methylation as an intermediary of genetic risk in rheumatoid arthritis. Nat. Biotechnol 31, 142–147 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ai R et al. Joint-specific DNA methylation and transcriptome signatures in rheumatoid arthritis identify distinct pathogenic processes. Nat. Commun 7, 11849(2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ai R et al. DNA methylome signature in synoviocytes from patients with early rheumatoid arthritis compared to synoviocytes from patients with longstanding rheumatoid arthritis. Arthritis Rheumatol. 67, 1978–1980 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Nakano K, Whitaker JW, Boyle DL, Wang W & Firestein GS DNA methylome signature in rheumatoid arthritis. Ann. Rheum. Dis 72, 110–117 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Whitaker JW et al. An imprinted rheumatoid arthritis methylome signature reflects pathogenic phenotype. Genome Med. 5, 40 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.de la Rica L et al. Identification of novel markers in rheumatoid arthritis through integrated analysis of DNA methylation and microRNA expression. J. Autoimmun 41, 6–16 (2013). [DOI] [PubMed] [Google Scholar]
- 106.Altorok N, Tsou PS, Coit P, Khanna D & Sawalha AH Genome-wide DNA methylation analysis in dermal fibroblasts from patients with diffuse and limited systemic sclerosis reveals common and subset-specific DNA methylation aberrancies. Ann. Rheum. Dis 74, 1612–1620 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ding W et al. Genome-wide DNA methylation analysis in systemic sclerosis reveals hypomethylation of IFN-associated genes in CD4+ and CD8+ T cells. J. Invest. Dermatol 138, 1069–1077(2018). [DOI] [PubMed] [Google Scholar]
- 108.Altorok N et al. Genome-wide DNA methylation patterns in naive CD4+ T cells from patients with primary Sjogren’s syndrome. Arthritis Rheumatol. 66, 731–739 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Spreafico R et al. Epipolymorphisms associated with the clinical outcome of autoimmune arthritis affect CD4+T cell activation pathways. Proc. Natl Acad. Sci. USA 113, 13845–13850(2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Karemaker ID & Vermeulen M Single-cell DNA methylation profiling: technologies and biological applications. Trends Biotechnol. 36, 952–965 (2018). [DOI] [PubMed] [Google Scholar]
- 111.Kouzarides T Chromatin modifications and their function. Cell 128, 693–705 (2007). [DOI] [PubMed] [Google Scholar]
- 112.Smale SI, Tarakhovsky A & Natoli G Chromatin contributions to the regulation of innate immunity. Annu. Rev. Immunol 32, 489–511 (2014). [DOI] [PubMed] [Google Scholar]
- 113.Busslinger M & Tarakhovsky A Epigenetic control of immunity. Cold Spring Harb. Perspect. Biol 6, a019307 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Portela A & Esteller M Epigenetic modifications and human disease. Nat. Biotechnol 28, 1057–1068 (2010). [DOI] [PubMed] [Google Scholar]
- 115.Dawson MA & Kouzarides T Cancer epigenetics: from mechanism to therapy. Cell 150, 12–27 (2012). [DOI] [PubMed] [Google Scholar]
- 116.Matthews AG et al. RAG2 PHD finger couples histone H3 lysine 4 trimethylation with V(D)J recombination. Nature 450, 1106–1110 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Chi P, Allis CD & Wang GG Covalent histone modifications — miswritten, misinterpreted and mis-erased in human cancers. Nat. Rev. Cancer 10, 457–469 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Tough DF, Tak PP, Tarakhovsky A & Prinjha RK Epigenetic drug discovery: breaking through the immune barrier. Nat. Rev. Drug Discov 15, 835–853 (2016). [DOI] [PubMed] [Google Scholar]
- 119.Rotem A et al. Single-cell ChlP-seq reveals cell subpopulations defined by chromatin state. Nat. Biotechnol 33, 1165–1172(2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Gomez D, Shankman LS, Nguyen AT & Owens GK Detection of histone modifications at specific gene loci in single cells in histological sections. Nat. Methods 10, 171–177(2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Cheung P et al. Single-cell chromatin modification profiling reveals increased epigenetic variations with aging. Cell 173, 1385–1397(2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Cheung P et al. Single-cell epigenetics — chromatin modification atlas unveiled by mass cytometry. Clin. Immunol 196,40–48(2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Buenrostro JD, Giresi PG, Zaba LC, Chang HY & Greenleaf WJ Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213–1218(2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Cusanovich DA et al. Multiplex single cell profiling of chromatin accessibility by combinatorial cellular indexing. Science 348, 910–914 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Cusanovich DA et al. The ds-regulatory dynamics of embryonic development at single-cell resolution. Nature 555, 538–542 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Buenrostro JD et al. Single-cell chromatin accessibility reveals principles of regulatory variation. Nature 523, 486–490 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Buenrostro JD et al. Integrated single-cell analysis maps the continuous regulatory landscape of human hematopoietic differentiation. Cell 173, 1535–1548 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Corces MR et al. Lineage-specific and single-cell chromatin accessibility charts human hematopoiesis and leukemia evolution. Nat. Genet 48, 1193–1203 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Lara-Astiaso D et al. Immunogenetics. Chromatin state dynamics during blood formation. Science 345, 943–949 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Satpathy AT et al. Transcript-indexed ATAC-seq for precision immune profiling. Nat. Med 24, 580–590 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Gensterblum E et al. CD4+CD28+KIR+CD11ahi T cells correlate with disease activity and are characterized by a pro-inflammatory epigenetic and transcriptional profile in lupus patients. J. Autoimmun 86, 19–28(2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Scharer CD et al. ATAC-seq on biobanked specimens defines a unique chromatin accessibility structure in naive SLE B cells. Sci. Rep 6, 27030 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Jin W et al. Genome-wide detection of DNase I hypersensitive sites in single cells and FFPE tissue samples. Nature 528,142–146(2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Nagano T et al. Single-cell Hi-C reveals cell-to-cell variability in chromosome structure. Nature 502, 59–64 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Javierre BM et al. Lineage-specific genome architecture links enhancers and non-coding disease variants to target gene promoters. Cell 167, 1369–1384 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Lupianez DG, Spielmann M & Mundlos S Breaking TADs: how alterations of chromatin domains result in disease. Trends Genet. 32,225–237 (2016). [DOI] [PubMed] [Google Scholar]
- 137.Dey SS, Kester L, Spanjaard B, Bienko M & van Oudenaarden A Integrated genome and transcriptome sequencing of the same cell. Nat. Biotechnol 33, 285–289 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Macaulay IC et al. G&T-seq: parallel sequencing of single-cell genomes and transcriptomes. Nat. Methods 12, 519–522 (2015). [DOI] [PubMed] [Google Scholar]
- 139.Angermueller C et al. Parallel single-cell sequencing links transcriptional and epigenetic heterogeneity. Nat. Methods 13, 229–232 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Kelly TK et al. Genome-wide mapping of nucleosome positioning and DNA methylation within individual DNA molecules. Genome Res. 22, 2497–2506 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Pott S Simultaneous measurement of chromatin accessibility, DNA methylation, and nucleosome phasing in single cells. eLife 6, e23203 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Guo F et al. Single-cell multi-omics sequencing of mouse early embryos and embryonic stem cells.Cell Res. 27, 967–988 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Clark SJ et al. scNMT-seq enables joint profiling of chromatin accessibility DNA methylation and transcription in single cells. Nat. Commun 9, 781 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Hou Y et al. Single-cell triple omics sequencing reveals genetic, epigenetic, and transcriptomic heterogeneity in hepatocellular carcinomas. Cell Res. 26, 304–319 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Weeding E & Sawalha AH Deoxyribonucleic acid methylation in systemic lupus erythematosus: implications for future clinical practice. Front. Immunol 9, 875 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Porichis F et al. High-throughput detection of miRNAs and gene-specific mRNA at the single-cell level by flow cytometry. Nat. Commun 5, 5641 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Albayrak C et al. Digital quantification of proteins and mRNA in single mammalian cells. Mol. Cell 61, 914–924 (2016). [DOI] [PubMed] [Google Scholar]
- 148.Stahlberg A, Thomsen C, Ruff D & Aman P Quantitative PCR analysis of DNA, RNAs, and proteins in the same single cell. Clin. Chem 58, 1682–1691 (2012). [DOI] [PubMed] [Google Scholar]
- 149.Darmanis S et al. Simultaneous multiplexed measurement of RNA and proteins in single cells.Cell Rep. 14, 380–389 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Frei AP et al. Highly multiplexed simultaneous detection of RNAs and proteins in single cells. Nat. Methods 13,269–275(2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Wang F et al. RNAscope: a novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J. Mol. Diagn 14, 22–29 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Schulz D et al. Simultaneous multiplexed imaging of mRNA and proteins with subcellular resolution in breast cancer tissue samples by mass cytometry. CellSyst. 6, 531 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Stoeckius M et al. Simultaneous epitope and transcriptome measurement in single cells. Nat. Methods 14, 865–868 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Peterson VM et al. Multiplexed quantification of proteins and transcripts in single cells. Nat. Biotechnol 35, 936–939 (2017). [DOI] [PubMed] [Google Scholar]
- 155.Ohtsubo K & Marth JD Glycosylation in cellular mechanisms of health and disease. Cell 126, 855–867 (2006). [DOI] [PubMed] [Google Scholar]
- 156.Kivioja T et al. Counting absolute numbers of molecules using unique molecular identifiers. Nat. Methods 9, 72–74 (2011). [DOI] [PubMed] [Google Scholar]
- 157.Rosenberg AB et al. Single-cell profiling of the developing mouse brain and spinal cord with split-pool barcoding. Science 360, 176–182 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Cao J et al. Comprehensive single-cell transcriptional profiling of a multicellular organism. Science 357, 661–667 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Islam S et al. Characterization of the single-cell transcriptional landscape by highly multiplex RNA-seq. Genome Res. 21, 1160–1167 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Hashimshony T et al. CEL-Seq2: sensitive highly-multiplexed single-cell RNA-Seq. Genome Biol. 17, 77 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Jaitin DA et al. Massively parallel single-cell RNA-seq for marker-free decomposition of tissues into cell types. Science 343, 776–779 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Sheng K, Cao W, Niu Y, Deng Q & Zong C Effective detection of variation in single-cell transcriptomes using MATQ-seq. Nat. Methods 14, 267–270 (2017). [DOI] [PubMed] [Google Scholar]
- 163.Zheng GX et al. Massively parallel digital transcriptional profiling of single cells. Nat. Commun 8, 14049 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.DeKosky BJ et al. High-throughput sequencing of the paired human immunoglobulin heavy and light chain repertoire. Nat. Biotechnol 31, 166–169 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.DeKosky BJ et al. In-depth determination and analysis of the human paired heavy- and light-chain antibody repertoire. Nat. Med 21, 86–91 (2015). [DOI] [PubMed] [Google Scholar]
- 166.Busse CE, Czogiel I, Braun P, Arndt PF & Wardemann H Single-cell based high-throughput sequencing of full-length immunoglobulin heavy and light chain genes. Eur. J. Immunol 44, 597–603 (2014). [DOI] [PubMed] [Google Scholar]
- 167.Han A, Glanville J, Hansmann L & Davis MM Linking T cell receptor sequence to functional phenotype at the single-cell level. Nat. Biotechnol 32, 684–692 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Howie B et al. High-throughput pairing of T cell receptor alpha and beta sequences. Sci. Transl. Med 7, 301ra131 (2015). [DOI] [PubMed] [Google Scholar]
- 169.McDaniel JR, DeKosky BJ, Tanno H, Ellington AD & Georgiou G Ultra-high-throughput sequencing of the immune receptor repertoire from millions of lymphocytes. Nat. Protoc 11, 429–442 (2016). [DOI] [PubMed] [Google Scholar]
- 170.Hashimshony T, Wagner F, Sher N & Yanai I CEL-Seq: single-cell RNA-Seq by multiplexed linear amplification. Cell Rep. 2, 666–673 (2012). [DOI] [PubMed] [Google Scholar]
- 171.Fan X et al. Single-cell RNA-seq transcriptome analysis of linear and circular RNAs in mouse preimplantation embryos. Genome Biol. 16, 148 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Sasagawa Y et al. Quartz-Seq: a highly reproducible and sensitive single-cell RNA sequencing method, reveals non-genetic gene-expression heterogeneity. Genome Biol. 14, R31 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Picelli S et al. Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat. Methods 10, 1096–1098(2013). [DOI] [PubMed] [Google Scholar]
- 174.Sasagawa Y et al. Quartz-Seq2: a high-throughput single-cell RNA-sequencing method that effectively uses limited sequence reads. Genome Biol. 19, 29 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Ramskold D et al. Full-length mRNA-Seq from single-cell levels of RNA and individual circulating tumor cells. Nat. Biotechnol 30, 777–782 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Nakamura T et al. SC3-seq: a method for highly parallel and quantitative measurement of single-cell gene expression. Nucleic Acids Res. 43, e60 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]