Abstract
The human ovary orchestrates sex hormone production and undergoes monthly structural changes to release mature oocytes. The outer lining of the ovary (cortex) has a key role in defining fertility in women as it harbors the ovarian reserve. It has been postulated that putative oogonial stem cells exist in the ovarian cortex and that these can be captured by DDX4 antibody isolation. Here, we report single-cell transcriptomes and cell surface antigen profiles of over 24,000 cells from high quality ovarian cortex samples from 21 patients. Our data identify transcriptional profiles of six main cell types; oocytes, granulosa cells, immune cells, endothelial cells, perivascular cells, and stromal cells. Cells captured by DDX4 antibody are perivascular cells, not oogonial stem cells. Our data do not support the existence of germline stem cells in adult human ovaries, thereby reinforcing the dogma of a limited ovarian reserve.
Subject terms: Cell biology, Stem-cell niche
The outer lining or cortex of the human ovary determines fertility and has been postulated to contain oogonial stem cells. Here, the authors generate a single-cell map of the adult human ovarian cortex and show that DDX4 labels perivascular cells but no oogonial stem cells.
Introduction
The ovaries have two main functions: providing mature and developmentally competent oocytes, and producing hormones to support the female phenotype and pregnancy. In contrast to spermatozoa that are continuously produced throughout adulthood, oocytes are formed already during fetal life. In the female fetus, primordial germ cells (also called fetal germ cells, FGCs) proliferate and migrate to the fetal gonads where they become oogonia1. Oogonia proliferate and form cysts that are broken down by intersecting somatic cells into primordial follicles where meiosis is initiated. The oocytes within the primordial follicles remain dormant and arrested in the first meiotic division until the re-activation of follicles in the postnatal ovary2–4. The stock of dormant primordial follicles in the postnatal ovary, which resides within the ~1 mm thick ovarian cortex (Fig.1a), determines the fertile life span of the woman5,6. Upon activation, they mature in the process called folliculogenesis while migrating towards the inner part of the ovary, medulla (Fig. 1a)4,7. When the reserve of follicles is consumed, menopause commences and the woman cannot naturally conceive anymore.
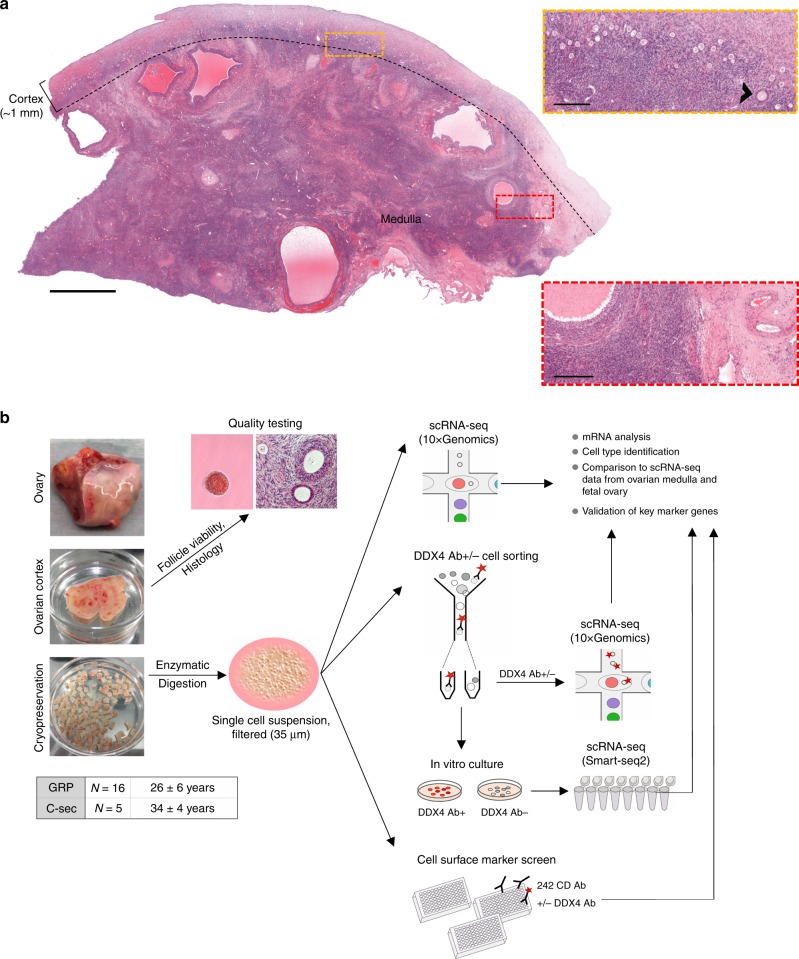
Fig. 1. Human ovary and study design.
a A representative ovary cross-section of a 26-year-old GRP showing the thin cortex layer (∼1 mm, used for fertility preservation) harboring clusters of small primordial follicles that form the ovarian reserve. Beneath the cortex is the inner part of the ovary, medulla, with looser connective tissue, denser vasculature, and larger growing follicles. Yellow rectangle is enlarged to highlight the ovarian reserve composed of primordial follicles resting in the cortex. One early growing (primary) follicle is indicated (arrowhead). Red rectangle is enlarged to highlight the more prominent vasculature in the medulla. Scale bars: 2 mm (overview), 250 µm (zoom-in). b Schematic representation of the study. In total, ovarian cortical tissue from 16 GRPs (mean age 26y) and five C-sec patients (mean age 34y) were used. Ovaries were trimmed upon tissue receival to separate the cortex. Cortex was cut into pieces, and the quality was assessed via general histology and viability of GRP follicles was further confirmed with neutral red staining. The samples were cryopreserved. Thawed ovarian cortex was enzymatically digested into single-cell suspension, filtered, and subjected either uncultured to single-cell transcriptome and surface proteome profiling using 10xGenomics technology and cell surface marker screening panel or cultured under oogonial stem cell (OSC) conditions and subjected to single-cell transcriptome analysis using Smart-seq2. In all assays, cells were stained with DDX4 Ab in order to relate the findings to DDX4 Ab+ (suggested OSCs) and Ab− ovarian cell populations. In the final data analysis, cortical cell populations were identified, related to cultured DDX4 Ab+ and Ab− cells, and compared with cells in the inner part of human ovaries21 and fetal ovarian cell populations22. Expression of key markers was validated with immunostainings, qPCR and RNA-FISH. Ab antibody, C-sec cesarean section, GRP gender reassignment patient, qPCR quantitative real-time PCR, RNA-FISH RNA-fluorescent in situ hybridisation, scRNA-seq single-cell RNA-sequencing.
Although it is well understood that the ovaries cease to function when women enter menopause at the age 50 on average6,8, there are reports suggesting that pre- and postmenopausal adult ovaries contain stem cells capable of differentiating into various cell lineages and even oocyte-like cells in vitro9–13. Oocyte forming stem cells, or oogonial stem cells (OSCs), are the most enigmatic and debated cell type in the ovarian cortex. They are hypothesized to stem from FGCs and be capable of giving rise to new oocytes13. These OSCs have been isolated from human ovarian tissue using fluorescence-activated cell sorting (FACS) and an antibody that targets the C-terminal domain of DDX4 protein14. DDX4 (a.k.a. VASA) is a cytoplasmic RNA helicase specifically expressed in the germline and commonly used as a marker for germ cells including oocytes in adult ovaries15–17. In contrast to all other types of germline cells, OSCs are suggested to express DDX4 on their cell membrane enabling the isolation via FACS13. Isolated OSCs were shown to express germline markers (e.g., DAZL, PRDM1, and IFITM3), proliferate in vitro and give rise to oocytes in a xeno-transplantation model13. The biological significance of these cells is poorly understood. In addition, many laboratories have not been able to repeat these findings and confirm the existence of OSCs18–20.
Recently, somatic cells from the inner part of human ovaries have been characterized by single-cell sequencing21. Here, we complete the map of cell types in human ovaries by providing an extensive characterization of germ cells and somatic cells in the outer layer of human ovaries, i.e., in the cortex. We use tissue samples provided by 21 patients with proven healthy follicles. Our results demonstrate six main cell types, but cannot provide support to the existence of OSCs. This data set will be a valuable tool for studying the role of specific cell populations in ovarian biology, dissecting causes of infertility, and developing assisted reproductive technologies or even contraceptives.
Results
Study setup
We performed single-cell RNA-sequencing (scRNA-seq) and surface marker screening to determine transcriptomes and cell surface proteomes of cells present in the ovarian cortex (Fig. 1b). Tissue from altogether 21 cesarean section (C-sec, N = 5) and gender reassignment patients (GRPs, N = 16) was used to have a comprehensive coverage of individuals and commonly used donor types (Fig. 1b). C-sec patients were proven fertile, and had no conditions or treatments known to affect ovarian function. GRPs were generally healthy and had no conditions known to affect ovarian function. All GRPs had androgen therapy prior to surgery for an average of 46 months (14–89 months).
In order to relate the findings to OSCs, we marked the cells with DDX4 antibodies (Ab) in all experiments; the single-cell mRNA sequencing (scRNA-seq) experiments were done on uncultured, unsorted cells, and sorted DDX4 Ab+ and Ab− cell fractions as well as on cultured, sorted DDX4 Ab+ and Ab− cells. The surface marker screen included DDX4 Ab as an additional marker. Data analysis included validation of markers by in situ hybridization (RNA-FISH), quantitative real-time PCR (qPCR) and immunostaining of ovarian samples, and merging the data with publicly available data sets of cells in the inner part of human ovaries21 as well as fetal ovaries containing FGCs22.
Six main cell populations in the human ovarian cortex
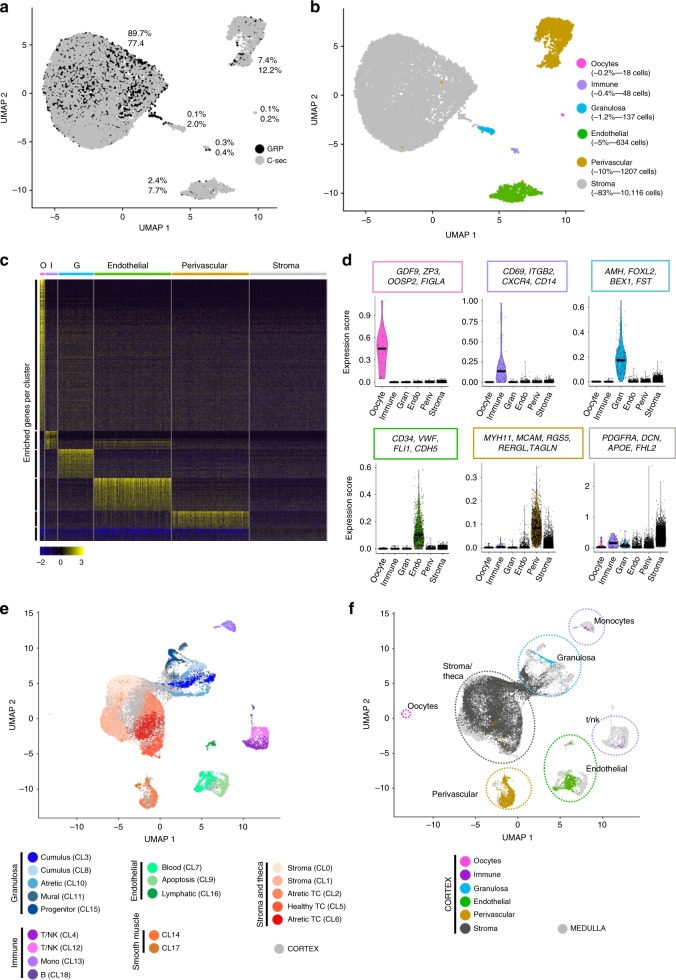
Ovarian tissue obtained from one GRP (20y) and three C-sec patients (28y, 32y, 36y) was used for unsorted single-cell profiling. The cortex containing the primordial follicles was carefully separated from medulla and processed to two independent libraries that were sequenced in parallel. After quality control and filtering (see Methods), a total of 12,160 cells were available for cell type characterization. Unbiased clustering and uniform manifold approximation and projection (UMAP) analysis revealed six main cell clusters and a similar contribution of cells from GRP and C-sec patients to all clusters (Fig. 2a and Supplementary Fig. 1a). Cell type identities were assigned using gene expression analysis (Fig. 2b, Supplementary Data 1). Distinct cell types were revealed based on enriched gene expression profiles (Fig. 2c). Signature genes were identified for each cell type and their expression score is shown as violin plots (Fig. 2d). The smallest population expressed the typical oocyte markers GDF9, ZP3, FIGLA, and OOSP2 (Fig. 2d, Supplementary Data 1). The immune cells expressed tissue-resident immune cell markers (e.g., CD69 and ITGB2) as well as mixed gene signatures of T-cells (CD2, CD3G, and CD8A) and antigen-presenting cells, such as CD14, HLA-DRA, B2M, and HLA-DQB1. Endothelial cells could be identified based on the expression of strong endothelial markers such as VE-cadherin (CDH5) and VWF. Granulosa cells were identified based on the expression of typical granulosa cell markers FOXL2 and anti-Müllerian hormone (AMH). Around 10% of ovarian cortex cells were annotated as perivascular cells expressing MYH11, MCAM, RGS5, RERGL, and TAGLN, with enriched gene signatures associated with both pericytes and smooth muscle cells23 (Supplementary Data 1). The majority of cells (83%) were broadly classified as stroma. There were no striking differences in gene expression between stromal cells but rather a shared expression of diverse mesodermal lineage markers (e.g., PDGFRA and DCN) and extracellular matrix proteins (e.g., COL1A1 and COL6A1) (Fig. 2d, Supplementary Data 1).
Fig. 2. Transcriptome analysis of unsorted ovarian cells.
Ovarian cortical cells from GRP and C-sec patients were sequenced on a single-cell level and integrated with ovarian medulla cells. a After quality control and filtration, 5715 GRP cells (black) and 6445 C-sec cells (grey) were further analyzed. Non-hierarchical cluster analysis revealed six main clusters, and both GRP and C-sec cells contributed to all clusters to a similar extent. b Main cell cluster identities were identified with the help of gene expression analysis. c Heatmap showing enriched genes per cluster. Clusters were downsampled to max. 300 cells per cluster for better visualization. d The color-coded violin plots show the expression scores of signature genes. e UMAP of merged scRNA-seq data of our unsorted ovarian cortex cells (grey) and the publicly available scRNA-seq data set of antral follicles and pieces of stroma picked from ovarian medulla (colored cells)21. Previous cluster annotations of both data sets are maintained21. f Same UMAP as in e, showing the contribution of our cortex cells (colored) to previously reported ovarian medulla clusters (grey). All identified cortical cell clusters joined the medullary cell clusters except from the oocytes that were unique to the cortex data set. The horizontal bars in violin plots indicate median gene expression per cluster. C-sec Cesarean section patient, CL cluster, G granulosa, GRP gender reassignment patient, I immune, TC theca cell, O oocytes, UMAP uniform manifold approximation and projection.
In order to obtain a complete map of cell types in adult human ovaries (Fig. 1a), we integrated our data with the recently published single-cell profiling of antral follicles and tissue fragments manually selected from the inner part of human ovary21. Although oocytes could not be detected in this data set21, most reported somatic cell types overlapped with the cortex cell types identified here including immune cells, perivascular (or smooth muscle) cells, and endothelial cells (Fig. 2e, f). Interestingly, the cortical granulosa cells clustered separately from the various granulosa cell types found in the antral follicles, whereas the theca/stroma cell cluster from the large antral follicle overlapped with our cortical stroma cluster (Fig. 2e, f). Collectively, this merged cell map demonstrates that similar vasculature and immune cell clusters can be found throughout ovaries; that there is a close relationship between theca cells and the general stromal cells; and that the granulosa cells are different in the cortical part of the ovary harboring the primordial follicles compared with the inner part containing growing antral follicles.
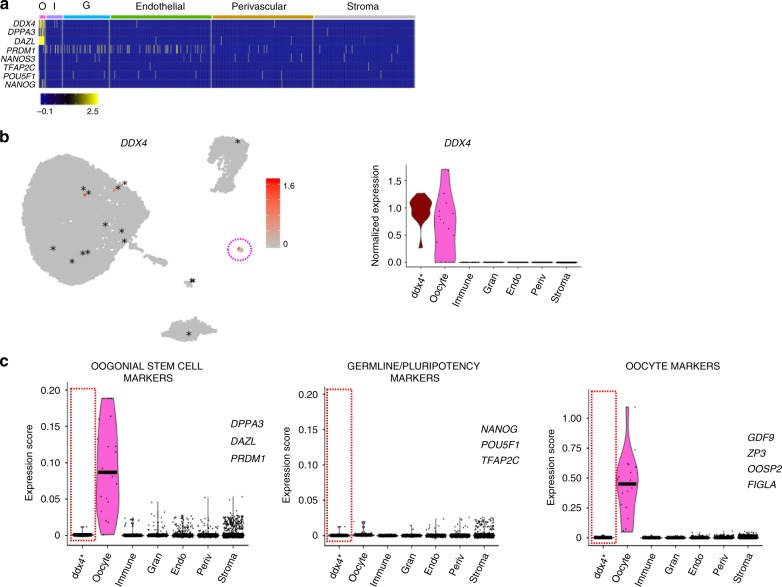
We next investigated the presence of OSCs in the cortex by studying the expression of reported OSC markers13 as well as germline stem cell and pluripotency markers. The OSC markers DDX4, DAZL, and DPPA3 were found in the oocytes only (Fig. 3a), whereas IFITM3 was detected to a varying extent in all cell types (Supplementary Fig. 1c). We noted that there were sporadic cells throughout the somatic types that expressed DDX4 hence possibly being OSCs (Fig. 3a, b). We manually pooled these 15 cells to their own cluster, marked as ddx4*, which showed a similar DDX4 expression level to oocytes (Fig. 3b) but no differential expression of any other reported OSC markers (DPPA3, DAZL, PRDM113), well-studied germline and pluripotency (POU5F1, NANOG, TFAP2C) or oocyte marker (GDF9, ZP3, OOSP2, FIGLA) when compared with somatic cells not expressing DDX4 (Fig. 3c and Supplementary Data 1). Highly expressed genes in the ddx4* cluster (Supplementary Data 1) showed no association with a distinct cell type but rather consisted of various abundantly expressed genes in the cortex. This might indicate that DDX4 expression in somatic cells is rather captured owing to intrinsic technical noise or transcriptional bursting reflecting stochastic gene activation24.
Fig. 3. Analysis of DDX4-expressing somatic cells of the ovarian cortex.
Somatic ovarian cortical cells expressing DDX4 were manually pooled for analysis of transcriptional profile. a Heatmap showing representative marker genes of OSCs and pluripotent cells by cell cluster. b Feature and violin plots highlighting all somatic cells expressing DDX4 transcript. Expression is mainly localized to the oocyte cluster (pink circle in feature plot) and dispersedly in 15 cells from other clusters (labeled with * for clarity). The 15 somatic cells expressing DDX4 were manually selected to form their own cluster (ddx4* in violin plot). c Violin plots displaying the expression scores of reported OSC marker genes (DPPA3, DAZL, PRDM1), germline and/or pluripotency-associated genes (POU5F1, NANOG, TFAP2C) and oocyte marker genes (GDF9, ZP3, OOSP2, FIGLA) across all cell clusters. Ddx4* cluster does not show differential expression of these markers. The horizontal bars in violin plots indicate median gene expression per cluster. G granulosa, I immune, O oocytes, UMAP uniform manifold approximation and projection.
DDX4 Ab+ ovarian cortical cells are perivascular cells
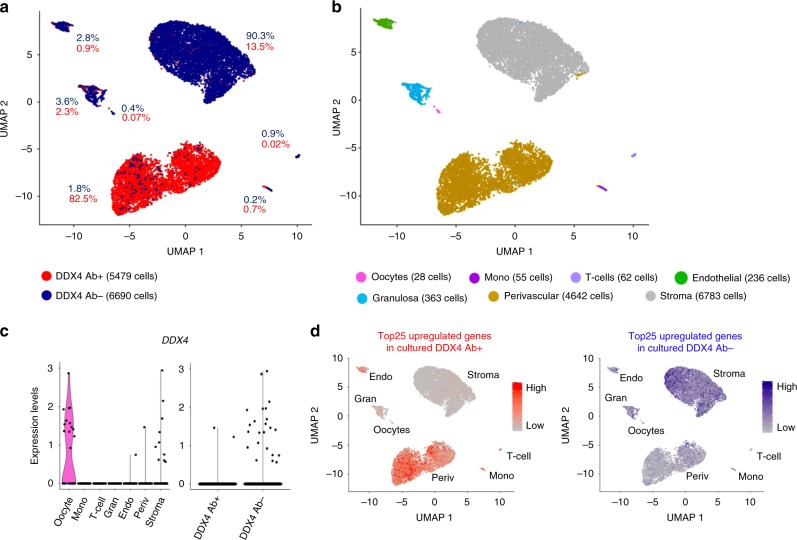
Although our data indicated that there are no DDX4-expressing germline stem cells in the human ovarian cortex, others have isolated such cells using polyclonal Ab (ab13840, Abcam) targeting the C-terminal domain of DDX4 protein11,13,14,25. We isolated DDX4 Ab-positive (DDX4 Ab+) and negative (DDX4 Ab−) cell populations using the same protocol and tissue source as in the previous reports13,14. We first confirmed that only the recommended DDX4 Ab is able to bind and isolate cells from the ovarian cortex. The Abcam Ab bound to a small fraction of ovarian cells (5–11.5%), whereas none of the other tested antibodies against the C-terminal domain enriched any cells above the negative controls (Supplementary Fig. 2a). Using the Abcam Ab, we sorted DDX4 Ab+ and DDX4 Ab− cells from three GRPs (20y, 22y, 26y) (Supplementary Fig. 2b). After quality control and filtration, there were 5479 Ab+ and 6690 Ab− cells available for analysis (Supplementary Fig. 2c). As the fraction of DDX4 Ab+ cells in this experiment was 7.7% (Supplementary Fig. 2b), the isolation enriched the population by sevenfold. Seven cell populations emerged from the two libraries (Fig. 4a, Supplementary Fig. 2c). Based on analysis of highly expressed genes (Supplementary Data 2), the same cell types were identified in this combined data set as in unsorted ovarian cortical cells with the exception of immune cells, which were now separated to T-cells and monocytes (Fig. 4b, Supplementary Fig. 2c, d). The majority (82.5%) of the DDX4 Ab+ cells contributed to a specific cell cluster, which was identified as perivascular cells (Fig. 4a, b and Supplementary Data 2). When considering the sevenfold enrichment of Ab+ cells, the DDX4 Ab− cells dominated all other clusters (Fig. 4a, b and Supplementary Fig. 2c). The top 25 highly expressed genes in Ab+ cells included several known perivascular marker genes such as MCAM, TAGLN, and ACTA2. The top 25 highly expressed genes in the Ab− cells included stromal cell markers, such as DCN and PDGFRA (Supplementary Data 2). Among the perivascular cell cluster, only one cell expressed DDX4 (Fig. 4c, left). In the whole data set, there were two cells in the DDX4 Ab+ and 23 cells in the DDX4 Ab− fractions that expressed DDX4 mRNA (Fig. 4c, right). Thirteen of these were oocytes. The somatic DDX4-expressing cells were pooled (ddx4*, 1 DDX4 Ab+ cell, 11 DDX4 Ab− cells, Supplementary Fig. 2e). Ddx4* cells did not express any germline marker except for IFITM3, which was in fact expressed in the majority of the ovarian cortex cells (Supplementary Fig. 2f, g). Ddx4* cells did not express any pluripotency or oocyte markers either (Supplementary Fig. 2g and Supplementary Data 2).
Fig. 4. Transcriptome analysis of sorted DDX4 Ab+ and Ab− ovarian cortex cells.
Ovarian cortex cells were sorted into DDX4 Ab+ or Ab− populations and sequenced on a single-cell level. a After quality control and filtration, 5479 of Ab+ and 6690 of Ab− cells were available for downstream analysis. Non-hierarchical cluster analysis revealed that 82.5% of the DDX4 Ab+ cells form a distinct cluster while all other clusters mainly consisted of DDX4 Ab− cells. b Differential gene expression analysis revealed seven main cell types. DDX4 Ab− cells mainly contributed to stroma, endothelial cells, granulosa cells, T-cells, monocytes, and oocytes. DDX4 Ab+ cells made up the cluster identified as perivascular cells. c Violin plot showing the expression of DDX4 among the different clusters (left) as well as among the sorted Ab+ and Ab− populations (right). d Comparison of sorted scRNA-seq data to single-cell transcriptome analysis of cultured DDX4 Ab+ and Ab− ovarian cells. Top 25 upregulated genes of cultured DDX4 Ab+ and Ab− cells were superimposed as expression score on to our uncultured sequenced DDX4 Ab+ and Ab− cells. The upregulated genes from cultured DDX4 Ab+ cells (scale bar depicting low expression in grey and high expression in red) are highly expressed in the perivascular and endothelial clusters, whereas the upregulated genes in cultured DDX4 Ab− cells (scale bar depicting low expression in grey and high expression in blue) are not found in these clusters. UMAP uniform manifold approximation and projection.
OSCs are usually cultured and expanded in vitro under specific conditions13,14, which could enrich a stem cell population that might go otherwise unnoticed. Therefore, we performed scRNA-seq on FACS-sorted DDX4 Ab+ and Ab− cells cultured under OSC conditions for several weeks. Even after culture, Ab+ and Ab− cells clearly clustered separately (Supplementary Fig. 2h). Highly expressed genes were compared with our uncultured, FACS-sorted ovarian cells. The top 25 upregulated genes in the cultured DDX4 Ab+ cells associated with the perivascular and endothelial cell clusters, whereas the top 25 upregulated genes in the cultured DDX4 Ab− cells did not (Fig. 4d and Supplementary Data 3). The same results were obtained when the top genes were overlaid with our uncultured, unsorted ovarian cells (Supplementary Fig. 2i). This suggested that in vitro culture does not change the identity of the DDX4 Ab+ cells.
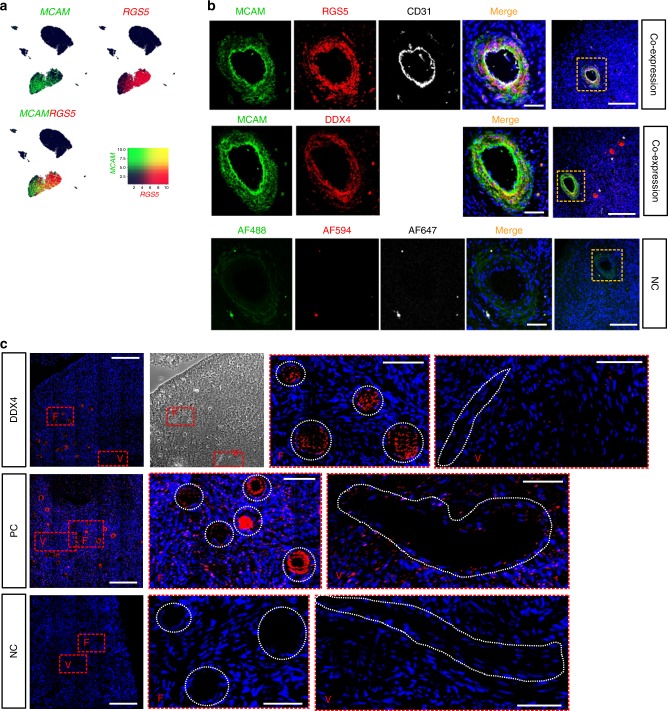
We next wanted to verify the localization of DDX4 Ab signal to perivascular cells in the human ovarian cortex. We chose two highly expressed markers from the perivascular cluster, MCAM and RGS5 (Fig. 5a), and carried out a co-staining with DDX4. DDX4 was found to be co-expressed with MCAM and RGS5 in the perivascular cells surrounding CD31-positive endothelial cells of blood vessels (Fig. 5b). In the same tissue section, oocytes within pre-antral follicles stained brightly for the DDX4 antibody but not for the perivascular markers (Fig. 5b last column, asterisks). Accordingly, DDX4 transcript was localized via RNA-FISH to the cytoplasm of oocytes only but not to somatic cells including perivascular cells of the ovarian cortex (Fig. 5c). These results suggested that the Abcam DDX4 Ab binds to an epitope on perivascular cells, which do not express DDX4 transcript.
Fig. 5. Validation of perivascular localization of DDX4 Ab signal.
a Feature plots displaying expression of MCAM (green) and RGS5 (red), both found among top highly expressed genes in DDX4 Ab+ cells, and their co-expression (yellow) on a transcriptional level. Both markers are highly expressed in cells of the perivascular cluster. Scale depicting low expression in dark blue and high expression in green, red, and yellow. b Immunostaining of human ovarian tissue sections with MCAM and RGS5, co-stained with the endothelial marker CD31, showed a distinct staining of cells surrounding endothelial cells of blood vessels. In a consecutive section, MCAM is co-expressed in DDX4 Ab+ cells. Oocytes staining brightly positive for DDX4 Ab+ are marked with white asterisks. As negative control, primary antibodies were omitted. DAPI (blue) was used as nuclear counterstain. Orange rectangles in last column (scale bars: 200 µm) demarcate the zoomed-in area (scale bars: 50 µm). Staining results were screened in three histological sections from two patients and a representative image is shown; all visible vessels or follicles were stained positive for MCAM/RGS5/DDX4 or DDX4 only, respectively. c RNA-FISH on ovarian cortex sections from a GRP donor showing the localization of DDX4 transcript in oocytes but not in blood vessel cells. Red rectangles are demarcating the zoomed-in areas containing follicles (F) or blood vessels (V) further highlighted with white dashed circles or lines, respectively. Nuclei were counterstained using Hoechst (blue). Positive control shows expression of Ubiquitin C. As negative control, probes targeting the bacterial DabB gene were used. Scale bars: 200 µm (zoomed-out), 100 µm (zoomed-in). RNA-FISH was performed on three histological sections of three donors, respectively, and representative images are shown. NC negative control, PC positive control.
Distinct surface marker profile on DDX4 Ab+ cells
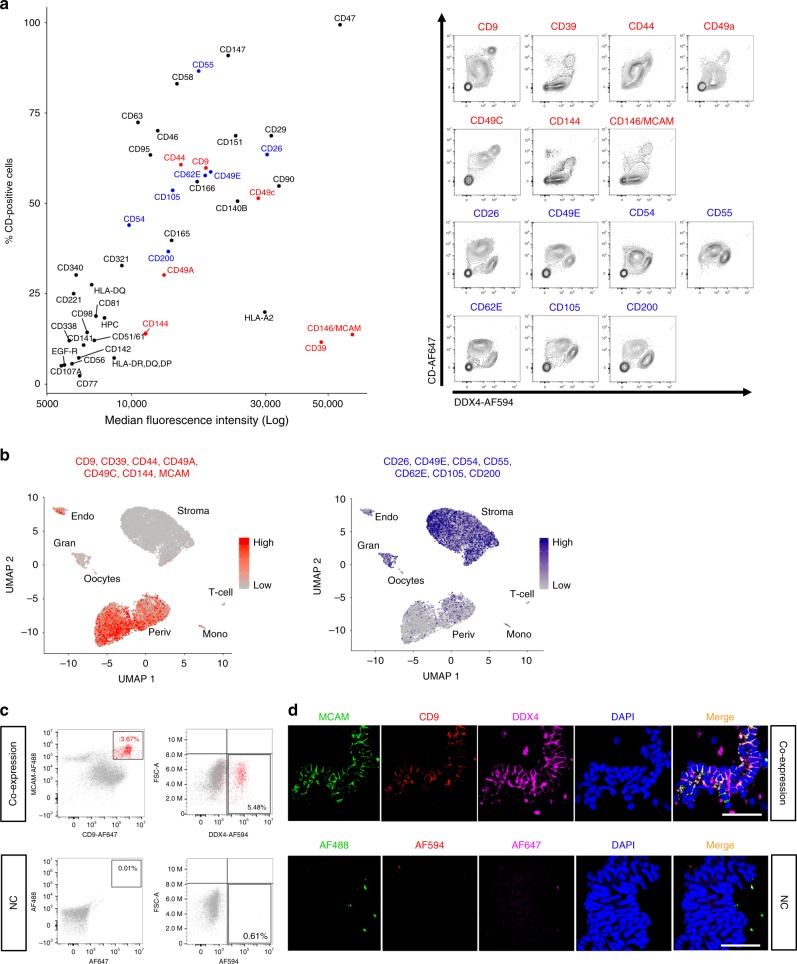
In order to study the identity of the DDX4 Ab+ cells using an independent approach, we carried out an extensive human cell surface marker profiling of ovarian cells co-stained with DDX4. Forty-three of the 242 surface markers present in the screen were found to be consistently expressed on ovarian cortex cells (Fig. 6a and Supplementary Tables 1–3). Of these, seven (CD9; CD39; CD44; CD49a; CD49c; CD144; and CD146, also known as MCAM) were brightly expressed on DDX4 Ab+ cells when compared with the DDX4 Ab− cell population, whereas seven (CD26; CD49e; CD54; CD55; CD62e; CD105; CD200) were weakly expressed on DDX4 Ab+ cells (Fig. 6a). Both positive and negative surface marker sets were used to generate expression scores and visualized in feature plots to associate them to cell clusters in our scRNA-seq data sets. Markers expressed on DDX4 Ab+ cells were again associated to the perivascular and endothelial cell clusters, whereas the surface markers absent from DDX4 Ab+ cells were not (Fig. 6b).
Fig. 6. Screening of ovarian cortex cells for surface proteins.
The surface of ovarian tissue cells was screened for the expression of 242 CD markers by flow cytometry. a Dot plot showing the log median fluorescence intensity (MFI, x axis) and the percentage of CD-positive cells (y axis) for those 43 CD markers that were repeatedly found to be present on ovarian tissue cells (left, CD marker expression values of repeat number three are shown as representatives). When co-stained with DDX4 Ab, seven of the markers were highly expressed (red) and seven were absent or lowly expressed (blue) on DDX4 Ab+ cells when compared with the DDX4 Ab− cell population. Co-expression of DDX4 and the 14 markers identified as high or low in DDX4 Ab+ cells is shown in representative contour plots (right). X axes display intensity of DDX4 Ab signal, y axes display intensity of CD marker signal. b Feature plots showing expression scores of the seven cell surface markers highly expressed in DDX4 Ab+ cells (scale bar depicting low expression in grey and high expression in red) and the seven markers lowly expressed in DDX4 Ab+ cells (scale bar depicting low expression in grey and high expression in blue) overlaid with our sorted scRNA-seq data. While markers that were found to be highly expressed in DDX4 Ab+ cells on a protein level are mainly expressed in perivascular cluster, CD markers lowly expressed in DDX4 Ab+ cells show a more disperse expression pattern. c FACS dot plots showing the co-expression of MCAM and CD9, two markers brightly expressed on DDX4 Ab+ cells, in regard to DDX4 Ab+ and Ab− cell populations. Around 3.67% of ovarian cortex cells were double positive for MCAM and CD9 (in red) and 5.48% are DDX4 Ab+. Around 80% of MCAM/CD9 double positive cells co-stained with DDX4 Ab (2.94% of MCAM+/CD9+/DDX4+ cells in total). Flow analysis was repeated in two independent experiments. d Unfiltered lysate of ovarian cortex cells cytospinned and stained for MCAM (green), CD9 (red), and DDX4 (magenta) showing co-expression of all three markers on blood vessels. As negative control, primary antibodies were omitted. DAPI (blue) was used as nuclear counterstain. Scale bars: 50 µm. Immunostaining was performed in duplicates in two independent experiments and a representative image is shown. All visible vessels stained positive for MCAM/CD9/DDX4. FACS fluorescence-activated cell sorting, MFI median fluorescence intensity, NC negative control, UMAP uniform manifold approximation and projection.
Markers of DDX4 Ab+ cells identified in the surface marker screen (MCAM and CD9, Fig. 6a) and scRNA-seq analysis (MCAM, CD9, and TAGLN, Supplementary Data 2), were validated via immunostaining to localize to small and large blood vessels in the cortex (Supplementary Fig. 3a, b). The gene expression of MCAM and TAGLN was further validated by qPCR to be enriched in the DDX4 Ab+ cell population when compared to the DDX4 Ab− fraction and bulk ovarian tissue cells (Supplementary Fig. 3c).
We next studied the extent to which DDX4 Ab stained perivascular cells. Altogether, 3.67% of the ovarian cells were double positive for the perivascular markers MCAM/CD9, and 80% of them were found to be also positive to DDX4 leading to 2.94% of live cells in the ovarian cortex being MCAM+/CD9+/DDX4+ (Fig. 6c). Immunostaining of unfiltered digested ovarian cortex cells likewise showed that all cells of a blood vessel were positive for DDX4 Ab with most of them also co-expressing MCAM and/or CD9 (Fig. 6d). These data collectively suggest that the majority of vascular cells are immunopositive for the DDX4 Ab while not expressing DDX4 transcript.
No detection of germline stem cells in the ovarian cortex
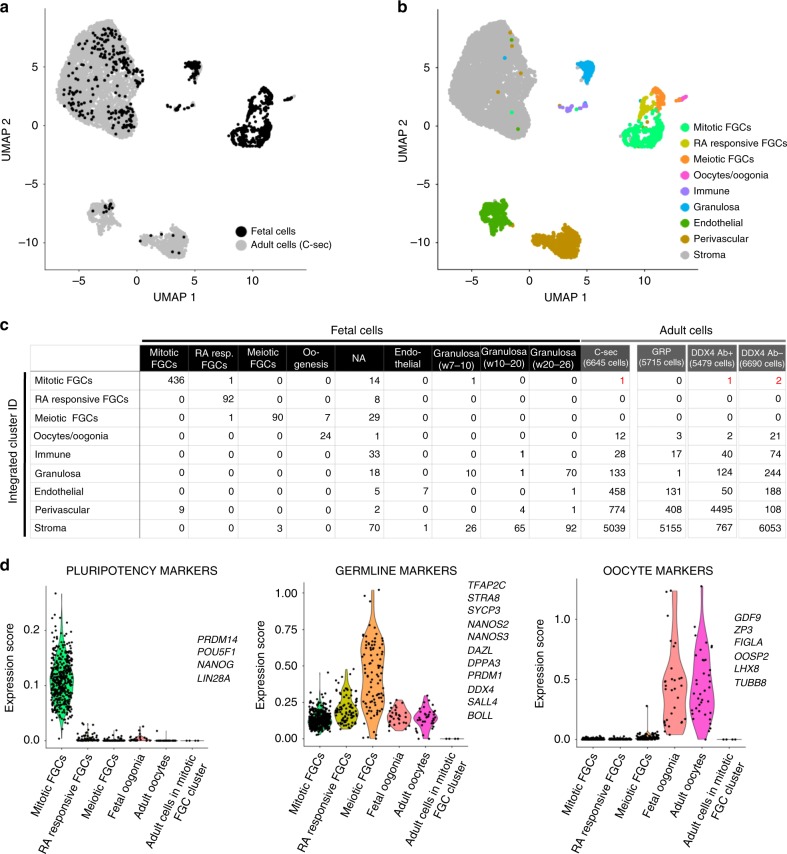
To further investigate the possible existence of germline stem cells in the ovarian cortex, regardless of immunopositivity for DDX4 Ab, we compared our data to a recently reported data set of human fetal ovaries consisting of single-cell transcriptomes of FGCs and somatic cells22. In these data, FGCs were classified into mitotic FGCs, retinoic acid (RA) responsive FGCs, meiotic FGCs, and oogonia, whereas granulosa cells were divided into week 7–10, week 10–20, and week 20–26 granulosa cells22. Owing to a bias caused by differences in cell numbers between the data sets (1123 fetal cells in22 vs. 24,329 adult cells in our data), fetal sequencing data were integrated with the individual adult scRNA-seq data sets, resulting in four separate analyses (fetal/C-Sec, fetal/GRP, fetal/DDX4 Ab+, and fetal/DDX4 Ab−) all yielding similar results. A representative data set (fetal/C-Sec) is shown in Fig. 7a, b. The integrated data set clustered to nine separate cell types with differing relative contributions by fetal and adult cells (Fig. 7a). Cell cluster identities were annotated based on gene expression analysis (Fig. 7b). FGCs of different stages formed separate clusters that were dominated by fetal cells, whereas fetal oogonia clustered together with adult oocytes. Similarly, fetal somatic cells annotated as endothelial cells clustered together with the adult endothelial cells (Fig. 7a–c). The week 20–26 granulosa cells fell into the same cluster with adult granulosa cells, whereas the majority of the earlier stages clustered with adult stroma cells (Fig. 7c and Supplementary Fig. 4). Although none of the sequenced 24,329 adult cells clustered with the meiotic and RA responsive FGCs, four cells clustered with the mitotic FGCs (one cell of C-sec sample, one cell of DDX4 Ab+ sample and two cells of DDX4 Ab− sample) (Fig. 7c and Supplementary Fig. 4), suggesting that they could potentially be germline stem cells that remain in adult ovaries. The transcription profile of these four cells was investigated further. Fetal mitotic FGCs showed a high expression of the pluripotency genes PRDM14, POU5F1, NANOG, and LIN28A, but none of these markers were found in the adult cells (Fig. 7d). Consistently, germline marker genes expressed in RA responsive and meiotic FGCs (TFAP2C, STAR8, SYCP3, NANOS2, NANOS3, DAZL, DPPA3, PRDM1, DDX4, SALL4, and BOLL) as well as oocyte marker genes expressed in fetal oogonia and adult oocytes (GDF9, ZP3, FIGLA, OOSP2, LIN28, TUBB8) were not detected in the four adult cells (Fig. 7d). Based on the gene expression profile of the four adult cells and their previous clustering behavior in adult data set analysis (Figs. 2b and 4b), these cells are considered to be granulosa, perivascular, and stroma cells.
Fig. 7. Investigation of potential germline stem cells in the adult human ovarian cortex.
Data sets of unsorted and sorted ovarian cortex cells were used individually for integration with scRNA-seq data set of human fetal tissue cells22. a UMAP plot showing sample origin of fetal (black) and adult cells (C-sec data, grey). b UMAP showing nine clusters representing different cell types annotated based on differential gene expression analysis. The same clusters as in adult tissue were found, as well as three new ones: mitotic fetal germ cells, retinoic acid responsive FGCs, and meiotic FGCs. c Table listing cell numbers from adult and fetal data sets per annotated cluster. One cell of C-sec sample, one cell of DDX4 Ab+ sample and two cells of DDX4 Ab− sample clustered with mitotic FGCs of the fetal sample (in red). These four cells were manually clustered together and studied further with violin plots. d Violin plots displaying expression scores of known pluripotency, germline and oocyte markers in the different FGC clusters in addition to adult oocyte cluster and the four adult ovarian cells clustering with mitotic FGCs. C-sec Cesarean section patient, FGCs fetal germ cells, GRP gender reassignemnt patient, NA non-annotated, UMAP uniform manifold approximation and projection.
Discussion
In this study, we provide a comprehensive map of cell populations in the human ovarian cortex, including both oocytes and their somatic cell niche. We identify oocytes, granulosa cells, immune cells, endothelial cells, perivascular cells, and stroma cells but do not find a germline stem cell population.
A concern in ovary research is the type of tissue available for research as healthy ovaries from young women are never surgically removed without severe indications. C-sec biopsies are small, GRP ovaries influenced by androgen treatment, and fertility preservation samples affected by the diagnosis and treatments. In addition, patient age has a major impact on follicle density and quality6,26. Typically, GRPs are young and have a high follicle density. Although all GRPs undergo androgen therapy prior to oophorectomy, their follicles do not differ in distribution, number and quality from untreated women27, and they can achieve pregnancy after discontinued hormone treatment28. In support of this, our data show that the ovarian cortex from GRPs has the same cell composition as tissue from C-sec patients. All tissue samples in our study were verified to have good quality and viable follicles, and the samples were handled according to protocols used in clinical fertility preservation29,30.
We succeeded in identifying oocytes from the cortical tissue samples. The oocytes likely stem from primordial follicles of the ovarian reserve that resides in the cortex31. The digested cell suspension underwent several filtering steps to exclude structures larger than 35 µm of size prior to analysis. This protocol likely excludes oocytes of growing follicle stages as oocytes from primordial follicles are ∼35 µm in size compared with primary follicle oocytes that already measure 40 µm32. In addition, the oocytes in our data clustered together with fetal oogonia, suggesting that they represent the earliest developmental stage of an oocyte, and stem from the primordial follicles established in the fetal ovaries. At last, the expression profile of the detected oocytes lacks markers of oocytes from antral follicles, such as NTF4, GPD1, and LCP217. The granulosa cells detected in our data set also appear to stem from very early follicles as they clustered together with fetal week 20–26 granulosa cells22 and lacked known markers of antral granulosa cells such as CYP19A1 and FSHR. In addition, our granulosa cells clustered separate from the ones isolated from antral growing follicles from the inner part of ovaries21. Primordial follicle granulosa cells control follicle activation33 and our data can help isolate these cells for further in vitro functional studies.
Ovaries undergo constant remodeling owing to folliculogenesis and ovulation, and immune cells have been reported to take part in both processes34–36. To further assure proper follicle maturation and restructuring of ovarian tissue, intense formation of new blood vessels composed of endothelial and perivascular cells is taking place in the adult human ovary37,38. Our data are consistent with the known cell types present in ovaries21. In our data set, macrophages and T-cells make up ∼0.5% of the ovarian cortex, endothelial cells ∼5% and perivascular cells ∼10%. These proportions do not necessarily reflect the true proportions in ovarian tissues, as the cells in our analyses were affected by tissue handling and dissociation methods, which could impact different cell types differently.
Steroidogenesis carried out by gonadotropin-sensitive theca and granulosa cells of growing follicles is an essential part of ovarian function. Theca cells are acquired during secondary follicle stage and they express STAR and CYP17A1, enabling them to synthesize androgens39. Androgens are further converted to estrogens by CYP19A1 in granulosa cells39. We do not detect theca cells in our data set or other cells with coherent signatures of gonadotropin responsiveness or steroidogenic activity, which is in agreement with known ovarian biology; the cortex contains predominantly primordial follicles that are independent of gonadotropins and steroidogenically inactive. However, some cells in the stroma cluster expressed STAR and CYP17A1, allowing to hypothesize about a possible presence of early theca cell precursors40. The close relationship between theca cells and stromal cells is further reflected by the difficulty to separate theca and stromal cell clusters even in large secondary follicles with visible theca cell layer growth21. Identification of factors that trigger theca cell recruitment and markers to identify the early theca will be needed to understand how this cell type relates to general stromal cells in cortex and medulla.
Some medical treatments such as chemotherapy lead to early exhaustion of ovarian reserve and premature ovarian insufficiency41. Some patients can benefit from fertility preservation via cryopreservation of ovarian cortical tissue that can be later transplanted for achieving pregnancy42. Reports of OSCs have given hope of new assisted reproductive technologies based on cells isolated from ovarian tissue with DDX4 Ab43–47, although some reports18–20 and clinical studies48 contradict the existence of OSCs and their clinical benefit. Our study cannot find OSCs in the ovarian cortex. Instead, our data imply that the DDX4 Ab+ cells that have been used to treat infertility in women45 have in fact been perivascular cells. Interestingly, we note that oocytes can be found among the FACS-sorted cells. The presence of oocytes in the DDX4 Ab+ cell fraction could help explain observations, suggesting that DDX4 Ab+ cells can give rise to oocytes13. Considering the clinical importance of OSCs, we also addressed their existence using an unbiased approach by merging our data with human FGCs, the suggested origin of OSCs in adult ovaries. None of the studied >24,000 cells expressed an FGC profile, indicating that no pre-meiotic FGCs remain in adult ovaries. Although this observation is in line with the known limited reproductive life span of women6, we cannot exclude that the OSCs are so rare that we missed their presence and the analysis of >24,000 cells might not be sufficient to capture very rare cell types. However, the original papers describing the isolation of DDX4 Ab+ cells propose that 1.7% of viable cells in sorting experiments are OSCs13,14. The smallest population we were able to capture with unbiased clustering approaches was the oocytes, corresponding to 0.15% of analyzed cells. Finally, it is possible that OSCs were sensitive to tissue digestion and hence lost during sample processing.
The FACS-based isolation protocol for OSCs that targets the C-terminal domain of DDX4 protein by a distinct antibody (ab13840, Abcam)14 has been criticized earlier. DDX4 is an RNA helicase found in the cytoplasm during germ cell development15 and its expression is maintained at a high level in oocytes16. Considering the function of DDX4 in RNA metabolism49,50, OSC isolation relying on membrane localization of DDX4 was questioned. Indeed, experimental51 and in silico data18 argue against an extracellular localization of the DDX4 C-terminus. Despite this, the expression of DDX4 C-terminus by DDX4 Ab on the surface of a cell line overexpressing DDX4 from a plasmid was recently reported52. In addition, several groups have been able to isolate a varying amount of cells from the ovarian cortex using this antibody, ranging from 1.7 to 42.7%9,13,19,20,25. In our experiments, the population size varies from 5 to 11.5%. The variation could be explained by the organization of the vasculature in the ovary. The initial manual trimming of the poorly vascularized ovarian cortex can lead to contamination with the highly vascularized medulla (see Fig. 1a), which could affect the amount of blood vessels and hence perivascular cells present in the sample. In addition, vascular density inversely correlates with follicle density in the cortex40, and this could lead to varying population size of DDX4 Ab+ cells. Our data also explain how DDX4 Ab+ “OSCs” have even been isolated from extragonadal tissues such as liver and kidney19. As all tissues contain blood vessels, perivascular cells are to be expected. We further show that no other Ab targeting DDX4 C-terminus can bind to cells in the ovarian cortex, and that the Abcam Ab stains blood vessels (and oocytes) in ovary tissue sections. We conclude that the DDX4 Ab (ab13840, Abcam) recognizes an epitope specifically expressed on perivascular cells.
Perivascular cells include pericytes and smooth muscle cells that originate from a common progenitor and express distinct sets of markers23. Depending on the type of blood vessel (fine capillary or big artery) one or both cell types can be found. Interestingly, tissue-resident pericytes have been suggested to contribute to the regeneration of various tissues including endometrium owing to their stem cell potential53. In this study, DDX4 Ab+ cells identified as perivascular cells express markers of pericytes and smooth muscle cells, suggesting that the epitope recognized by DDX4 Ab is expressed on both cell types. Whether this could explain the heterogeneity among isolated DDX4 Ab+ cells from the human ovary observed by others9 remains to be studied. In addition, the role of perivascular cells in monthly ovarian regeneration should be addressed in future studies.
Altogether, our data suggest that the adult human ovarian cortex consists of six main cell types and do not harbor germline stem cells. These data can be used (i) to isolate and study the specific populations further in vitro; (ii) to compare these normal cell profiles to those present in different disease conditions to understand causes of infertility; and (iii) to further develop artificial ovaries and other assisted reproductive technologies for infertility.
Methods
Ovarian tissue handling
In the described studies, tissue from 16 GRPs (20–38 yr) and five C-sec patients (28–37 yr) was used. Prior to gender reassignment surgery, all GRPs underwent androgen treatment for 14–89 months. All samples were collected at Karolinska University Hospital and transported in Dulbecco’s phosphate-buffered saline supplemented with calcium, magnesium, glucose, and pyruvate (DPBS++++, ThermoFisher Scientific, USA) to the research laboratory for processing. In GRP ovaries, cortex was separated from medulla and trimmed to a thickness of ∼1 mm using scalpels. C-sec biopsies were small surface cuts (max. 5 × 5 × 1 mm) and did not require trimming.
All tissue samples were cryopreserved and quality controlled using clinical fertility preservation protocols as outlined below29,30,54. For histological evaluation, fresh pieces of cortex were immediately fixed in 4% methanol-free formaldehyde (Thermo Fisher Scientific) and in Bouin’s solution (Sigma-Aldrich, USA). Upon dehydration and paraffin embedding, tissue was sectioned (4 µm) and stained with hematoxylin and eosin, and tissue quality was evaluated by counting and scoring follicles according to established criteria for follicle morphology evaluation55. Follicle densities in our C-sec cohort was 118.3 follicles mm−3 and within the normal range when compared with other studies56. GRPs were selected based on high follicle density determined prior to tissue freezing (see below) and showed a follicle density of 2962 follicles mm−3. Proportion of atretic follicles did not differ between C-sec and GRP tissues (13.3% ± 13.3 and 18.3% ± 2.4, respectively).
The density and viability of GRP follicles was studied with Neutral Red stain prior to tissue freezing and viability was found to be 80%. For assessment of follicle viability in GRP tissue, cortical tissue was chopped using McIIwain Tissue Chopper (Mickle Laboratory, UK) in digestion medium composed of McCoy’s (GIBCO, Thermo Fisher Scientific) medium containing 1 mg mL−1 human serum albumin (HSA, Vitrolife, Sweden), 1× GlutaMax (Thermo Fisher Scientific), 1× Insulin-Transferrin solution (GIBCO, Thermo Fisher Scientific), 40 µg mL−1 Liberase (Sigma-Aldrich), 0.4 mg mL−1 Collagenase IV (GIBCO, Thermo Fisher Scientific), and 0.2 mg mL−1 DNase I (BioRad, USA). The chopped pieces were then incubated in digestion medium containing Neutral Red (50 mg mL−1, Sigma-Aldrich) in +37 °C shaking for 30–50 min and the digestion was stopped using termination medium containing McCoy’s + 10% fetal bovine serum (FBS, Thermo Fisher Scientific). The proportion of viable (red) follicles of all follicles was evaluated under a stereomicroscope (Leica S9D) (Fig. 1b). For vitrification of ovarian tissue, our established vitrification protocol was used as follows57. Tissue was cut into pieces of 1 × 1 × 1 mm and transferred into vitrification solution containing 40% Ethylene Glycol (Sigma-Aldrich) and 10 mg mL−1 HSA for 2 min and 3 min. Then, pieces were transferred into cryo-tubes and stored in liquid nitrogen. For thawing, cryo-tubes were slightly opened and equilibrated in room temperature for 30 s. Closed tubes were then placed into a 37 °C water bath for 1.5 min, and the pieces were transferred through three different thawing solutions of decreasing sucrose and increasing HSA concentration for 2 min, 3 min, and 5 min. For slow-freezing of ovarian tissue, the standard protocol was followed30. In brief, cortical tissue was cut into pieces of a maximum size of 10 × 10 × 1 mm and pre-equilibrated in slow-freezing medium containing 7.5% of Ethylene Glycol and 10 mg mL−1 HSA in DPBS for 30 min shaking on ice. Pieces were transferred into cryo-tubes containing 1 mL of fresh slow-freezing solution and placed into controlled rate freezer (Kryo 360–1.7, Planer PLC, UK). For thawing of slow-frozen tissue, cryo-vials were placed in a 37 °C water bath for 1–2 min, transferred into thawing solutions with 10 mg mL−1 HSA and decreasing sucrose concentrations for 10 min, 10 min, and 10 min.
Dissociation of the ovarian cortex into single-cell suspension
Thawed human ovarian cortical tissue was chopped using scalpels into pieces of ∼0.3 mm3 and enzymatically digested in DMEM/F12 (Thermo Fisher Scientific) containing 5% FBS, 1 mg mL−1 collagenase IA (Sigma-Aldrich), 50 μg mL−1 Liberase and 1000 U DNase I (Roche, Sigma-Aldrich) in a shaking 37 °C water bath for max. 50 min. Digestion was stopped with medium containing 10% FBS and cell suspension was centrifuged for 7 min on 300 × g. Cells were resuspended in DPBS, 2% FBS and passed through a 40 μm cell strainer (VWR, USA), counted and used for subsequent experiments.
Flow cytometry and fluorescence-activated cell sorting
Dissociated ovarian cell suspension was prepared for flow cytometry (FC, surface marker screen using seven GRPs and validation of markers using three GRPs) and fluorescence-activated cell sorting (FACS, scRNA-seq using three GRPs) following the previously published oogonial stem cell isolation protocol14. All blocking and antibody incubation steps were carried out for 20 min on 4 °C. After blocking in DPBS containing 0.02% bovine serum albumin (BSA, Sigma-Aldrich and Normal Donkey Serum (1:50, EMD Millipore, Germany), samples were washed and stained using primary antibodies (DDX4, ab13840, Abcam; DDX4, SAB1300533, Sigma-Aldrich; DDX4, AP1403b, Abgent; MCAM (1:100), AF932, R&D Systems; CD9 (1:20), ab2215, Abcam) diluted in blocking solution. DDX4 antibody was titrated using concentrations ranging from 5 to 20 μg mL−1 in 100 μL of cell suspension (max. 1 × 106 cells) and recommended isotype control antibody (polyclonal rabbit IgG, ab171870, Abcam) was used as negative control. A distinct population of DDX4 Ab+ cells were found using a concentration of 10 μg mL−1 of DDX4 Ab (ab13840) in 100 μL, hence, all subsequent experiments were performed using this concentration (Supplementary Fig 2a). After 20 min, cells were washed and incubated with secondary antibodies diluted (1:250) in blocking solution: AF488 donkey anti-goat IgG (A11055), AF594 donkey anti-rabbit IgG (A21207), AF647 donkey anti-mouse IgG (A31571, all from Thermo Fisher Scientific). 4′,6-diamidino-2-phenylindole (DAPI; 0.2 μg mL−1, Thermo Fisher Scientific) was added to the final cell suspension as a live/dead marker. Before sorting, cells were transferred into round bottom tubes through a 35 µm nylon mesh (Falcon, Sigma-Aldrich). Negative and positive gates for analyzing and sorting cell populations were set using FMO (fluorescence minus one) controls, isotype controls as well as samples where primary antibody was omitted. The cells were either analyzed by FC using CytoFLEX S (Beckman Coulter, USA) equipped with 405, 638, 488, and 561 nm lasers or sorted using a BD FACSAria Fusion (BD Bioscience, USA) equipped with 405, 640, 488, 355, and 561 nm lasers. BD FACSDiva software or FlowJo v10 software (Tree Star) was used for downstream analysis and results are presented as representative FACS plots.
Single-cell mRNA sequencing of uncultured ovarian cells
Two runs of single-cell mRNA sequencing (scRNA-seq) of dissociated, uncultured ovarian tissue cells were performed. Each run consisted of two samples that were prepared and sequenced in parallel. In the first run, unsorted cortical cells from one GRP and three pooled C-sec patients were analyzed. The samples were digested into single cells in parallel and dead cells were removed (dead cell removal kit, MACS, Milteny Biotec, Bergisch-Gladbach, Germany). In the second run, cortex from three GRPs were pooled, digested into single cells, stained with DDX4 antibodies, and sorted into live (DAPI negative) DDX4 Ab+ and DDX4 Ab− cell populations. The cell suspensions were transported on 4 °C to the Eukaryotic Single Cell Genomics Facility (ESCG, SciLifeLab, Stockholm, Sweden) and prepared immediately for loading into the 10XGenomics Chromium controller for gem formation. The cell viability before loading was 91.1% (run 1, GRP), 92% (run 1, C-sec), 89% (run 2, DDX4 Ab+) and 95% (run 2, DDX4 Ab−). For scRNA-seq library preparation, 10XGenomics v2 (unsorted cells) or v3 (sorted populations) was used according to manufacturer’s instructions and sequenced on a NovaSeq 6000 using a S1 flow cell.
ScRNA-seq output files were converted using Cell Ranger 2.1.1 (unsorted cells) or Cell Ranger 3.0.1 pipeline (sorted cells) and aligned to the hg19 transcriptome using STAR aligner58. Analysis of filtered cells was performed in R version 3.5.159 using Seurat suite version 3.0.060,61. Details of the web summary of Cell Ranger statistics of unsorted and sorted data sets can be found in Supplementary Data 1 and 2, respectively. For downstream bioinformatics analysis of remaining cells after the initial Cell Ranger filtration based on correctly detected cellular barcodes, genes expressed in at least three cells were kept. In order to exclude potential doublets, cells expressing 200–7000 genes, and no more than 25% of mitochondrial genes were kept, resulting in 24,329 cells in total (12,160 unsorted and 12,169 sorted cells).
For integration of different data sets (unsorted GRP/C-sec), Seurat’s CCA integration tool was used. The sorted data sets were merged and regressed for batch effects.
All data sets were column-normalized and log-transformed prior to selection of highly variable genes, which were used for principal component analysis. Elbow plot function in Seurat was used to identify most significant PCs that were included when UMAP analysis was performed (dimensionality of 13 in unsorted and 12 in sorted data set). A resolution of 0.1 and a perplexity of 30 was chosen for analysis. In scRNA-seq analysis of the sorted data set, oocytes clustered with granulosa cells at a resolution of 0.1. In this case, oocytes were selected using CellSelector function in Seurat based on analysis of oocyte markers identified in the unsorted data set. Marker genes discriminating the different clusters were selected among highly expressed genes (p value < 0.01 and log(fold-change) > 0.25) using Wilcoxon Rank Sum test. In addition, the robustness of clustering was confirmed identifying clusters from the UMAP using the density based clustering method HDBSCAN62, which resulted in similar clusters and resolved the oocytes from the granulosa cells in both data sets (Supplementary Fig. 1b). Signature gene expression was calculated as percentage of sum counts of selected genes vs all expressed genes. The percentage ratio is added as feature value to each cell and multiplied by 100. Scaled gene expression values were used to generate heatmaps.
In vitro culture of DDX4 Ab+ and DDX4 Ab− ovarian tissue cells
Ovarian tissue of two C-sec patients (36y, 37y) was pooled before dissociated into single cells and sorted based on DDX4 Ab signal. Sorted DDX4 Ab+ and Ab− cell populations were cultured as follows19. Sorted cell populations were plated onto irradiated mouse embryonic fibroblasts (Applied StemCell, USA) in medium consisting of MEMα, 10% FBS, 1 mm sodium pyruvate, 1× Glutamax, 1% non-essential amino acids, 0.1 nm 2-mercaptoethanol, 1× N2-supplement (R&D Systems), 50 U mL−1 penicillin, 50 μg mL−1 streptomycin, 10 ng mL−1 recombinant human leukemia inhibitory factor (Merck Millipore), 1 ng mL−1 basic fibroblast growth factor (R&D Systems), 40 ng mL−1 glial cell-derived neurotropic factor (R&D Systems), and 10 ng mL−1 recombinant human epidermal growth factor (all from Life Technologies if not mentioned otherwise). After passage 3, cultured cells were plated onto mouse embryonic fibroblast-free 0.1% gelatin-coated plates (Sigma-Aldrich). DDX4 Ab+ cells and DDX4 Ab− cells were maintained for seven and six passages, respectively.
Single-cell mRNA sequencing of cultured ovarian cells
After culture, DDX4 Ab+ and Ab− cells were randomly picked by mouth pipetting into lysis buffer and cDNA libraries were prepared as follows19. Whole-cell lysates were reversely transcribed using SuperScript II reverse transcriptase (Invitrogen, USA) and resulting PCR products were preamplified using KAPA HiFi HotStart ReadyMix (KAPA Biosystems, USA). Spiked-in samples were sequenced on the Smart-seq2 platform using the TrueSeq dual-index sequencing primers (illumina, USA) according to manufacturer’s instructions on a HiSeq 2000.
STAR aligner58 was used to index the hg19 reference genome and align the resulting fastq files. Mapped reads were then counted in annotated genes using featureCounts. The annotations and reference genome were obtained from UCSC Genome Browser63. The count table from featureCounts was imported into R/Bioconductor and differential gene expression was performed using the EdgeR package and its general linear models pipeline. For gene expression analysis, genes with at least one count per million in three or more samples were used and normalized using TMM normalization. UMAP dimensionality reduction was performed in R with the uwot package using all genes with at least one count per million in three or more samples.
Integration of scRNA-seq data sets
The UMI count data of human fetal scRNA-seq were downloaded from http://github.com/zorrodong/germcell22. Female samples expressing 2000–9000 genes with no more than 1,100,000 transcripts were kept, resulting in 1123 cells in total. FindIntegrationAnchors and IntegrateData functions in Seurat61 were used with dimensionality of 15–40 for the integration of these fetal scRNA-seq data with the individual adult scRNA-seq data sets (C-sec, GRP, DDX4 Ab+, and DDX4 Ab−, respectively). The cell cycle scores based on G2/M and S phase markers were calculated by CellCycleScoring function, and these scores were regressed out using ScaleData function to remove the cell cycle effect. Identification of the clusters was determined using the FindClusters function with the resolution which classifies the FGCs into four clusters as reported in ref. 22 (resolution of 0.9–1.2).
Similarly, scRNA-seq data of the inner part of human ovaries, derived from 31 tissue samples of five ovaries, were obtained from gene expression omnibus using accession number GSE11812721. These data were processed with Seurat (v2.3.0) and scran (v1.12.0) to correct the patient bias by mutual nearest neighbors64, following the method described in ref. 21. Finally, 20,676 cells were clustered into 19 clusters as reported, which were then integrated with our 12,610 unsorted ovarian cortex cells, using FindIntegrationAnchors and IntegrateData functions in Seurat (v3.0.0) with dimensionality of 21. Cell cycle effect was removed as described above.
Quantitative real-time PCR (qPCR)
Total RNA was isolated using the RNeasy Mini Kit and treated with RNase-free Dnase (both from Qiagen, Germany). For cDNA synthesis, 10 ng of total RNA was reversely transcribed using random hexamers and Superscript III reverse transcriptase (Invitrogen, Thermo Fisher Scientific). Samples were subjected to qPCR using TaqMan probes for GAPDH (Hs02786624_g1), MCAM (Hs00174838_m1), and TAGLN (Hs01038777_g1) and TaqMan Fast Advanced Master Mix (all from Thermo Fisher Scientific) on the StepOne real-time PCR System (Applied Biosystems, USA). As negative control, the human embryonic stem cell line HS980 was included after culture in NutriStem hPSC XF medium (Biological Industries, Israel) on hrLN-521-coated plates (Biolamina, Sweden)65. The experiment was performed on three undissociated pieces of whole ovary tissue from two donors, respectively, three independently dissociated ovarian tissue samples, and three independently sorted DDX4 Ab+ and Ab− cell fractions. All samples were run in technical duplicates. Results are presented as mean ± SD (standard deviation of mean).
Fluorescent in situ hybridization (FISH)
RNA-FISH was performed using formaldehyde-fixed paraffin-embedded sections of the ovarian cortex from GRPs in biological triplicates. The RNAscope Multiplex Fluorescent Detection Kit v2 (323120, Advanced Cell Diagnostics, USA) was used according to manufacturer’s instructions. Signal from DDX4 (Hs-DDX4-O1-C3, 518571-C3), human-specific positive control probe targeting Ubiquitin C and negative control probe targeting the bacterial gene DabB (321831, all from Advanced Cell Diagnostics) was amplified using TSA with Cy3 (1:1500, Akoya Biosciences, USA). Nuclei were counterstained using Hoechst (Invitrogen, Thermo Fisher Scientific). Images were acquired on a Nikon spinning disk confocal microscope (Zeiss, Germany) and analyzed using Fiji/ImageJ software v2.0.
CD marker screen of human ovarian cortex cells
After dissociation of thawed ovarian cortex tissue into a single-cell suspension, cells were stained using the BD Lyoplate Screening Panels (BD Biosciences) of 242 monoclonal antibodies following manufacturer’s instructions. Altogether three runs were carried out. In the first run, pooled tissue cells from five GRP ovaries were used for the full plate of 242 CD markers. The subsequent two runs were performed with pooled tissue cells from four and six GRP ovaries, respectively, to verify the expression of the markers identified in the first run. DDX4 co-staining (10 μg mL−1, ab13840, Abcam) was carried out in all runs to identify markers on the surface of DDX4 Ab+ cells. All samples were analyzed in a 96-well format on a CytoFlex S flow cytometer (Beckman Coulter). Exclusion of dead cells was ensured by gating on the DAPI-negative cell population using FlowJo v10 software (Tree Star). Results are presented as percentage of positive live cells and median fluorescence intensity as well as representative contour plots.
Immunocytochemistry of dissociated ovarian cells
Ovarian tissue from three GRPs was dissociated separately into a bulk cell suspension passed through a 100 μm cell strainer (VWR, USA) to leave vessel structures intact and diluted to a concentration of 50,000 cells in 100 μL of DPBS (ThermoFisher Scientific) containing 2% FBS. Samples were cytospinned onto glass slides (VWR) using Cytocentrifuge (CytoSpin 4, Thermo Fisher Scientific) at 1200 rpm for 4 min. Slides were dried in room temperature for 2–4 h and fixed with 4% methanol-free formaldehyde for 10 min. Cells were permeabilized with 0.3% Triton X-100 (Sigma-Aldrich) in DPBS for 10 min, followed by blocking in DPBS/4% FBS/0.1% Tween-20 (P9416, Sigma-Aldrich) for 1 h. Primary antibodies (DDX4, 10 μg mL−1, ab13840, Abcam; CD9, 1:200, 312102, BioLegend; MCAM, 1:100, AF932, R&D systems) were diluted in blocking solution and cells were stained overnight at 4 °C, followed by incubation with secondary antibodies diluted 1:1000 in blocking solution for 2 h at room temperature (AF488 donkey anti-goat IgG, AF594 donkey anti-mouse IgG, AF647 donkey anti-rabbit IgG; all from Thermo Fisher Scientific, A11055, A21203, A31573, respectively). Cell nuclei were counterstained using Hoechst (1:1000, Invitrogen). Cells were mounted using fluorescent mounting medium (Dako Agilent, USA, California) and imaged with Olympus IX81 fluorescence microscope (Carl Zeiss Meditec, Germany) or Nikon spinning disk confocal microscope (Nikon). Acquired images were analyzed using Fiji/ImageJ software v2.0.
Immunohistochemistry of ovarian tissue sections
Immunohistochemistry was performed on freshly formaldehyde-fixed paraffin-embedded cortex sections (4 μm) of at least two GRPs in technical triplicates. After de-paraffinization and rehydration, antigen retrieval was performed in either 10 mm Sodium Citrate (pH 6) or 10 mm Tris/1 mm EDTA (pH 9) at 96 °C for 30 min. After blocking in 5% BSA and Normal Donkey Serum, tissue sections were incubated in primary antibody (DDX4, 10 μg mL−1, ab13840, Abcam; CD9, 1:200, 312102, BioLegend; MCAM, 1:100, AF932, R&D systems, RGS5, 1:50, MA5-25584, Invitrogen; CD31, 1:200, M0823, Dako; TAGLN, 1:100, MAB78861, R&D systems) overnight at 4 °C, followed by incubation with secondary antibody diluted 1:200 in blocking solution for 1 h at room temperature: AF488 donkey anti-goat IgG, AF594 donkey anti-mouse IgG, AF647 donkey anti-rabbit IgG (all from Thermo Fisher Scientific, A11055, A21203, A31573, respectively). Stained tissue sections were mounted in fluorescent mounting media (Dako Agilent) and imaged with Olympus IX81 fluorescence microscope (Carl Zeiss Meditec, Germany) or Nikon ultra fast widefiled microscope (Nikon, Germany). Acquired images were analyzed using Fiji/ImageJ software v2.0.
Ethics statement
Use of ovarian tissue in research was approved by Stockholm Region Ethical Review Board (Dnr. 2010/549–31/2, Dnr. 2015/798–31/2). Independent clinicians informed the patients about the study, and cortical tissue was biopsied from C-sec patients and whole ovaries collected from GRPs after written informed consent. Use of hESCs (HS980) was approved by the Swedish Ethical Review Authority (Dnr. 2011/745:31/3). HESCs were derived from whole inner cell mass of blastocysts after informed written consent from both parents65.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Acknowledgements
We thank all clinicians helping us with patient recruitment and tissue collection as well as all patients donating ovarian tissue to our studies. In particular, the irreplaceable help from Catarina Arnelo and Boel Niklasson is warmly acknowledged. Claus Yding Andersen is thanked for kindly teaching us controlled rate freezing and follicle viability assay. Richelle Björvang, Astrud Tuck, and Jasmin Hassan are thanked for the help with ovarian tissue handling. Sandra Petrus-Reurer is thanked for help with the experimental design. FC analysis and cell sorting were performed at the MedH Flow Cytometry core facility, Karolinska Institutet, Sweden. Sequencing was performed at the ESCG at Science for Life Laboratory, Stockholm, Sweden. We thank Tarja Schröder and Morphological Phenotype Analysis facility, Karolinska Institutet, Sweden, for H&E stains and tissue scans. Imaging of stained samples was performed at the Live Cell Imaging facility/Nikon Center of Excellence, Karolinska Institutet, Sweden. This work was supported by the Swedish Research Council (P.D, M.W., F.L., J.K.), the Swedish Research Council for Sustainable Development (P.D.), the Swedish Childhood Cancer Foundation (P.D.), Karolinska Institutet PhD student funding (M.W.), the Knut and Alice Wallenberg Foundation (J.K. and F.L.); Wallenberg Academy Fellow, Ragnar Söderberg Foundation, The Ming Wai Lau Center for Reparative Medicine, Center for Innovative Medicine/SLL (F.L.); the Scandinavia-Japan Sasakawa Foundation, the Japan Eye Bank Association, the Astellas Foundation for Research on Metabolic Disorders, and the Japan Society for the Promotion of Science Overseas Research Fellowships (M.Y.). This project has also received funding from the European Union’s Horizon 2020 research and innovation program under grant agreement No. 825100 (FREIA) (P.D., M.W.). Open access funding provided by Karolinska Institute.
Author contributions
M.W. carried out the experiments, analyzed data, performed sequencing analysis, and wrote the paper. M.Y., S.K., and A.D. carried out bioinformatic analyses. I.D. conceived the experiments and sorted cell populations. S. Panula and S. Petropoulos performed scRNA-seq experiment of cultured ovarian tissue cells. H.J. contributed to the surface marker screen verification. K. Palm and K. Pettersson coordinated patient recruitment, carried out surgeries, and evaluated patient health. O.H. conceived the experiments and supervised the project. J.K. contributed to essential resources and helped draft the manuscript. F.L. and P.D. conceived the experiments, analyzed data, supervised the project, and wrote the paper. All authors contributed to the preparation of the paper and approved the final version.
Data availability
The 10xGenomics and Smart-seq2 scRNA-seq data that support the finding of this study have been deposited in the ArrayExpress database at EMBL-EBI and are available under the accession codes ‘E-MTAb−8381’ and ‘E-MTAb−8403’, respectively.
Code availability
The scRNA-seq data analysis was performed using scripts in R, which are available in github, ‘SingleCellOvary [https://github.com/wagmag/SingleCellOvary]’.
Competing interests
The authors declare no competing interests.
Footnotes
Peer review information Nature Communications thanks Barbara Vanderhyden, Isabelle Stevant and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Fredrik Lanner, Pauliina Damdimopoulou.
Contributor Information
Fredrik Lanner, Email: Fredrik.Lanner@ki.se.
Pauliina Damdimopoulou, Email: Pauliina.Damdimopoulou@ki.se.
Supplementary information
Supplementary information is available for this paper at 10.1038/s41467-020-14936-3.
References
- 1.Sarraj MA, Drummond AE. Mammalian foetal ovarian development: consequences for health and disease. Reproduction. 2012;143:151–163. doi: 10.1530/REP-11-0247. [DOI] [PubMed] [Google Scholar]
- 2.Cohen PE, Holloway JK. Predicting gene networks in human oocyte meiosis. Biol. Reprod. 2010;82:469–472. doi: 10.1095/biolreprod.109.083014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.MacLennan M, et al. Oocyte development, meiosis and aneuploidy. Semin Cell Dev. Biol. 2015;45:68–76. doi: 10.1016/j.semcdb.2015.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hsueh AJ, et al. Intraovarian control of early folliculogenesis. Endocr. Rev. 2015;36:1–24. doi: 10.1210/er.2014-1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Findlay JK, et al. How is the number of primordial follicles in the ovarian reserve established? Biol. Reprod. 2015;93:111. doi: 10.1095/biolreprod.115.133652. [DOI] [PubMed] [Google Scholar]
- 6.Wallace WH, Kelsey TW. Human ovarian reserve from conception to the menopause. PLoS ONE. 2010;5:e8772. doi: 10.1371/journal.pone.0008772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McGee EA, Hsueh AJ. Initial and cyclic recruitment of ovarian follicles. Endocr. Rev. 2000;21:200–214. doi: 10.1210/edrv.21.2.0394. [DOI] [PubMed] [Google Scholar]
- 8.Gold EB. The timing of the age at which natural menopause occurs. Obstet. Gynecol. Clin. North Am. 2011;38:425–440. doi: 10.1016/j.ogc.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clarkson YL, et al. Initial characterisation of adult human ovarian cell populations isolated by DDX4 expression and aldehyde dehydrogenase activity. Sci. Rep. 2018;8:6953. doi: 10.1038/s41598-018-25116-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Parte S, et al. Detection, characterization, and spontaneous differentiation in vitro of very small embryonic-like putative stem cells in adult mammalian ovary. Stem Cells Dev. 2011;20:1451–1464. doi: 10.1089/scd.2010.0461. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 11.Silvestris E, et al. In vitro differentiation of human oocyte-like cells from oogonial stem cells: single-cell isolation and molecular characterization. Hum. Reprod. 2018;33:464–473. doi: 10.1093/humrep/dex377. [DOI] [PubMed] [Google Scholar]
- 12.Stimpfel M, et al. Isolation, characterization and differentiation of cells expressing pluripotent/multipotent markers from adult human ovaries. Cell Tissue Res. 2013;354:593–607. doi: 10.1007/s00441-013-1677-8. [DOI] [PubMed] [Google Scholar]
- 13.White YA, et al. Oocyte formation by mitotically active germ cells purified from ovaries of reproductive-age women. Nat. Med. 2012;18:413–421. doi: 10.1038/nm.2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Woods DC, Tilly JL. Isolation, characterization and propagation of mitotically active germ cells from adult mouse and human ovaries. Nat. Protoc. 2013;8:966–988. doi: 10.1038/nprot.2013.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smorag L, et al. The roles of DAZL in RNA biology and development. Wiley Interdiscip. Rev. RNA. 2014;5:527–535. doi: 10.1002/wrna.1228. [DOI] [PubMed] [Google Scholar]
- 16.Castrillon DH, et al. The human VASA gene is specifically expressed in the germ cell lineage. Proc. Natl Acad. Sci. USA. 2000;97:9585–9590. doi: 10.1073/pnas.160274797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Y, et al. Transcriptome landscape of human folliculogenesis reveals oocyte and granulosa cell interactions. Mol. Cell. 2018;72:1021–1034.e4. doi: 10.1016/j.molcel.2018.10.029. [DOI] [PubMed] [Google Scholar]
- 18.Zarate-Garcia L, et al. FACS-sorted putative oogonial stem cells from the ovary are neither DDX4-positive nor germ cells. Sci. Rep. 2016;6:27991. doi: 10.1038/srep27991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang H, et al. Adult human and mouse ovaries lack DDX4-expressing functional oogonial stem cells. Nat. Med. 2015;21:1116–1118. doi: 10.1038/nm.3775. [DOI] [PubMed] [Google Scholar]
- 20.Hernandez SF, et al. Characterization of extracellular DDX4- or Ddx4-positive ovarian cells. Nat. Med. 2015;21:1114–1116. doi: 10.1038/nm.3966. [DOI] [PubMed] [Google Scholar]
- 21.Fan X, et al. Single-cell reconstruction of follicular remodeling in the human adult ovary. Nat. Commun. 2019;10:3164. doi: 10.1038/s41467-019-11036-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li L, et al. Single-cell RNA-seq analysis maps development of human germline cells and gonadal niche interactions. Cell Stem Cell. 2017;20:858–873 e4. doi: 10.1016/j.stem.2017.03.007. [DOI] [PubMed] [Google Scholar]
- 23.Kumar A, et al. Specification and diversification of pericytes and smooth muscle cells from mesenchymoangioblasts. Cell Rep. 2017;19:1902–1916. doi: 10.1016/j.celrep.2017.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haque A, et al. A practical guide to single-cell RNA-sequencing for biomedical research and clinical applications. Genome Med. 2017;9:75. doi: 10.1186/s13073-017-0467-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ding X, et al. Human GV oocytes generated by mitotically active germ cells obtained from follicular aspirates. Sci. Rep. 2016;6:28218. doi: 10.1038/srep28218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ford JH. Reduced quality and accelerated follicle loss with female reproductive aging - does decline in theca dehydroepiandrosterone (DHEA) underlie the problem? J. Biomed. Sci. 2013;20:93. doi: 10.1186/1423-0127-20-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.De Roo C, et al. Ovarian tissue cryopreservation in female-to-male transgender people: insights into ovarian histology and physiology after prolonged androgen treatment. Reprod. Biomed. Online. 2017;34:557–566. doi: 10.1016/j.rbmo.2017.03.008. [DOI] [PubMed] [Google Scholar]
- 28.Light AD, et al. Transgender men who experienced pregnancy after female-to-male gender transitioning. Obstet. Gynecol. 2014;124:1120–1127. doi: 10.1097/AOG.0000000000000540. [DOI] [PubMed] [Google Scholar]
- 29.Kristensen SG, et al. A simple method to quantify follicle survival in cryopreserved human ovarian tissue. Hum. Reprod. 2018;33:2276–2284. doi: 10.1093/humrep/dey318. [DOI] [PubMed] [Google Scholar]
- 30.Rosendahl M, et al. Cryopreservation of ovarian tissue for a decade in Denmark: a view of the technique. Reprod. Biomed. Online. 2011;22:162–171. doi: 10.1016/j.rbmo.2010.10.015. [DOI] [PubMed] [Google Scholar]
- 31.Haino T, et al. Determination of follicular localization in human ovarian cortex for vitrification. J. Adolesc. Young. Adult Oncol. 2018;7:46–53. doi: 10.1089/jayao.2017.0028. [DOI] [PubMed] [Google Scholar]
- 32.Westergaard CG, Byskov AG, Andersen CY. Morphometric characteristics of the primordial to primary follicle transition in the human ovary in relation to age. Hum. Reprod. 2007;22:2225–2231. doi: 10.1093/humrep/dem135. [DOI] [PubMed] [Google Scholar]
- 33.Zhang H, et al. Somatic cells initiate primordial follicle activation and govern the development of dormant oocytes in mice. Curr. Biol. 2014;24:2501–2508. doi: 10.1016/j.cub.2014.09.023. [DOI] [PubMed] [Google Scholar]
- 34.Best CL, et al. Localization and characterization of white blood cell populations within the human ovary throughout the menstrual cycle and menopause. Hum. Reprod. 1996;11:790–797. doi: 10.1093/oxfordjournals.humrep.a019256. [DOI] [PubMed] [Google Scholar]
- 35.Wu R, et al. Macrophage contributions to ovarian function. Hum. Reprod. Update. 2004;10:119–133. doi: 10.1093/humupd/dmh011. [DOI] [PubMed] [Google Scholar]
- 36.Brannstrom M, et al. Localization of leukocyte subsets in the follicle wall and in the corpus luteum throughout the human menstrual cycle. Fertil. Steril. 1994;61:488–495. doi: 10.1016/S0015-0282(16)56581-9. [DOI] [PubMed] [Google Scholar]
- 37.Hazzard TM, Stouffer RL. Angiogenesis in ovarian follicular and luteal development. Baillieres Best Pract Res. Clin. Obstet. Gynaecol. 2000;14:883–900. doi: 10.1053/beog.2000.0133. [DOI] [PubMed] [Google Scholar]
- 38.Suzuki T, et al. Cyclic changes of vasculature and vascular phenotypes in normal human ovaries. Hum. Reprod. 1998;13:953–959. doi: 10.1093/humrep/13.4.953. [DOI] [PubMed] [Google Scholar]
- 39.Young JM, McNeilly AS. Theca: the forgotten cell of the ovarian follicle. Reproduction. 2010;140:489–504. doi: 10.1530/REP-10-0094. [DOI] [PubMed] [Google Scholar]
- 40.Delgado-Rosas F, et al. Superficial ovarian cortex vascularization is inversely related to the follicle reserve in normal cycling ovaries and is increased in polycystic ovary syndrome. Hum. Reprod. 2009;24:1142–1151. doi: 10.1093/humrep/dep008. [DOI] [PubMed] [Google Scholar]
- 41.Donnez J, et al. Ovarian tissue cryopreservation and transplantation: a review. Hum. Reprod. Update. 2006;12:519–535. doi: 10.1093/humupd/dml032. [DOI] [PubMed] [Google Scholar]
- 42.Donnez J, Dolmans MM. Fertility preservation in women. N. Engl. J. Med. 2017;377:1657–1665. doi: 10.1056/NEJMra1614676. [DOI] [PubMed] [Google Scholar]
- 43.Akahori T, Woods DC, Tilly JL. Female fertility preservation through stem cell-based ovarian tissue reconstitution in vitro and ovarian regeneration in vivo. Clin. Med. Insights Reprod. Health. 2019;13:1179558119848007. doi: 10.1177/1179558119848007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martin JJ, Woods DC, Tilly JL. Implications and current limitations of oogenesis from female germline or oogonial stem cells in adult mammalian ovaries. Cells. 2019;8:93. doi: 10.3390/cells8020093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Oktay K, et al. Oogonial precursor cell-derived autologous mitochondria injection to improve outcomes in women with multiple ivf failures due to low oocyte quality: a clinical translation. Reprod. Sci. 2015;22:1612–1617. doi: 10.1177/1933719115612137. [DOI] [PubMed] [Google Scholar]
- 46.Woods DC, Tilly JL. Autologous germline mitochondrial energy transfer (AUGMENT) in human assisted reproduction. Semin. Reprod. Med. 2015;33:410–421. doi: 10.1055/s-0035-1567826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fakih, M. H. et al. The AUGMENTSM treatment: physician reported outcomes of the initial global patient experience. JFIV Reprod. Med. Genet.3, 154 (2015).
- 48.Labarta E, et al. Autologous mitochondrial transfer as a complementary technique to intracytoplasmic sperm injection to improve embryo quality in patients undergoing in vitro fertilization-a randomized pilot study. Fertil. Steril. 2019;111:86–96. doi: 10.1016/j.fertnstert.2018.09.023. [DOI] [PubMed] [Google Scholar]
- 49.Fujiwara Y, et al. Isolation of a DEAD-family protein gene that encodes a murine homolog of Drosophila vasa and its specific expression in germ cell lineage. Proc. Natl Acad. Sci. USA. 1994;91:12258–12262. doi: 10.1073/pnas.91.25.12258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Noce T, Okamoto-Ito S, Tsunekawa N. Vasa homolog genes in mammalian germ cell development. Cell Struct. Funct. 2001;26:131–136. doi: 10.1247/csf.26.131. [DOI] [PubMed] [Google Scholar]
- 51.Nott TJ, et al. Phase transition of a disordered nuage protein generates environmentally responsive membraneless organelles. Mol. Cell. 2015;57:936–947. doi: 10.1016/j.molcel.2015.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Clarkson, Y. L. et al. Extracellular localisation of the C-terminus of DDX4 confirmed by immunocytochemistry and fluorescence-activated cell sorting. Cells8, pii: E578 (2019). [DOI] [PMC free article] [PubMed]
- 53.Spitzer TL, et al. Perivascular human endometrial mesenchymal stem cells express pathways relevant to self-renewal, lineage specification, and functional phenotype. Biol. Reprod. 2012;86:58. doi: 10.1095/biolreprod.111.095885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kristensen SG, et al. Isolation of pre-antral follicles from human ovarian medulla tissue. Hum. Reprod. 2011;26:157–166. doi: 10.1093/humrep/deq318. [DOI] [PubMed] [Google Scholar]
- 55.Gougeon A. Regulation of ovarian follicular development in primates: facts and hypotheses. Endocr. Rev. 1996;17:121–155. doi: 10.1210/edrv-17-2-121. [DOI] [PubMed] [Google Scholar]
- 56.Schmidt KL, et al. Density and distribution of primordial follicles in single pieces of cortex from 21 patients and in individual pieces of cortex from three entire human ovaries. Hum. Reprod. 2003;18:1158–1164. doi: 10.1093/humrep/deg246. [DOI] [PubMed] [Google Scholar]
- 57.Sheikhi M, et al. Preservation of human ovarian follicles within tissue frozen by vitrification in a xeno-free closed system using only ethylene glycol as a permeating cryoprotectant. Fertil. Steril. 2013;100:170–7.e1. doi: 10.1016/j.fertnstert.2013.03.018. [DOI] [PubMed] [Google Scholar]
- 58.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Team RC. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2013. [Google Scholar]
- 60.Butler A, et al. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 2018;36:411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stuart T, et al. Comprehensive integration of single-cell data. Cell. 2019;177:1888–1902.e21. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Campello RJGB, Moulavi D, Sander J. Density-Based Clustering Based on Hierarchical Density Estimates. Berlin, Heidelberg: Springer Berlin Heidelberg; 2013. [Google Scholar]
- 63.Haeussler M, et al. The UCSC Genome Browser database: 2019 update. Nucleic Acids Res. 2019;47(D1):p. D853–d858. doi: 10.1093/nar/gky1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Haghverdi L, et al. Batch effects in single-cell RNA-sequencing data are corrected by matching mutual nearest neighbors. Nat. Biotechnol. 2018;36:421–427. doi: 10.1038/nbt.4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rodin S, et al. Clonal culturing of human embryonic stem cells on laminin-521/E-cadherin matrix in defined and xeno-free environment. Nat. Commun. 2014;5:3195. doi: 10.1038/ncomms4195. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
The 10xGenomics and Smart-seq2 scRNA-seq data that support the finding of this study have been deposited in the ArrayExpress database at EMBL-EBI and are available under the accession codes ‘E-MTAb−8381’ and ‘E-MTAb−8403’, respectively.
The scRNA-seq data analysis was performed using scripts in R, which are available in github, ‘SingleCellOvary [https://github.com/wagmag/SingleCellOvary]’.