Abstract
Background
The overuse of antimicrobials in food animals and the subsequent contamination of the environment have been associated with development and spread of antimicrobial resistance. This review presents information on antimicrobial use, resistance and status of surveillance systems in food animals and the environment in Africa.
Methods
Information was searched through PubMed, Google Scholar, Web of Science, and African Journal Online databases. Full-length original research and review articles on antimicrobial use, prevalence of AMR from Africa covering a period from 2005 to 2018 were examined. The articles were scrutinized to extract information on the antimicrobial use, resistance and surveillance systems.
Results
A total of 200 articles were recovered. Of these, 176 studies were included in the review while 24 articles were excluded because they were not relevant to antimicrobial use and/or resistance in food animals and the environment. The percentage of farms using antimicrobials in animal production ranged from 77.6% in Nigeria to 100% in Tanzania, Cameroon, Zambia, Ghana and Egypt. The most antibiotics used were tetracycline, aminoglycoside and penicillin groups. The percentage of multi drug resistant isolates ranged from 20% in Nigeria to 100% in South Africa, Zimbabwe and Tunisia. In the environment, percentage of multi drug resistant isolates ranged from 33.3% in South Africa to 100% in Algeria. None of the countries documented national antimicrobial use and resistance surveillance system in animals.
Conclusion
There is high level of antimicrobial use, especially tetracycline, aminoglycoside and penicillin in animal production systems in Africa. This is likely to escalate the already high prevalence of antimicrobial resistance and multi drug resistance in the continent. This, coupled with weak antimicrobial resistance surveillance systems in the region is a great concern to the animals, environment and humans as well.
Keywords: Antimicrobial use, Antimicrobial resistance, Surveillance, Food animals, Environment, Africa
Introduction
Food animals such as cattle, poultry and pigs have been extensively reared worldwide, not only as a source of food but also a source of income. The modes of productions are intensive [1], due to the rapidly increasing demand for livestock products driven by human population growth and urbanization [2, 3]. This has necessitated the uncontrolled use of antimicrobials [4], which has been associated with increase of antimicrobial resistance [5].
The subsequent contamination of soil, sediments, sludge, groundwater, wastewater, tap and surface water, and plants contribute to the emergence and spread of multi-drug (MDR) organisms in environment [6, 7]. Furthermore, unmonitored quantities of waste that contain antimicrobials generated by pharmaceutical manufacturers, hospitals, and livestock producers promote selection of resistomes in the environment, with potential spill over to animals and humans [8, 9]. Some of important organisms that have been found to circulate in different compartments include Campylobacter spp., Salmonella spp., Staphylococcus spp., Enterococcus spp. and ESBL- producing Enterobacteriaceae [10–12].
Although AMR is a global threat [13], the situation in Africa is compounded by a number of factors that include lack of access to appropriate antimicrobial therapy, weak of regulation in the use of antimicrobials for human and animal, weak surveillance systems, lack of updated antimicrobial use and treatment guidelines. Others are lack of continuing education on antimicrobial use (AMU) for prescribers, tendency for animal owners to stock drugs and engaging unskilled people to treat animals, high degree of drugs abuse by livestock keepers and unregulated disposal of waste in dumps [14–17].
Despite these facts, there is paucity of consolidated information on the antimicrobial use (AMU), antimicrobial resistance (AMR), surveillance and stewardship programmes in Africa. The few available reviews on antimicrobial use and resistance are mostly country specific [18, 19]. This study reviewed the use of antimicrobial agents, prevalence of antimicrobial resistance and status of surveillance systems in food-producing animals and the environment in Africa.
Methods
This review was carried out between October 2018 and April 2019. Pub-Med, Google Scholar, Web of Science, Africa Wide Information and African Journal Online databases were searched for information on AMU and AMR covering a period from 2005 to 2018. Full-length research articles and review papers written in English were considered. In addition, publications from Food and Agriculture Organization (FAO), World Health Organization (WHO), International Livestock Research Institute (ILRI), Office International des Epizooties (OIE) websites were also searched and reviewed.
Combinations of search terms used were ‘antimicrobial usage’, ‘antimicrobial use’, ‘antibiotic use’, ‘antimicrobial resistance’, ‘antimicrobial resistant’, ‘food-producing animals’, ‘food animals’, animal husbandry’, ‘animal farming’, ‘domestic animal farming’, ‘farmed animals’, ‘environment’, ‘environmental’, ‘waste water’, treated waste water’, ‘sea water’, ‘river water’, ‘effluent’, ‘irrigation water’, ‘surface water’, ‘soil’ and ‘vegetables’. Others were specific food animal descriptors such as ‘poultry’, ‘chickens’, ‘pigs’, ‘swine’, ‘cattle’, ‘beef cattle’, ‘dairy cattle’, ‘fish’, specific country by name, and the word ‘Africa’. The articles were scrutinized to extract information on the antimicrobial use, prevalence of AMR and availability of a surveillance system.
Results
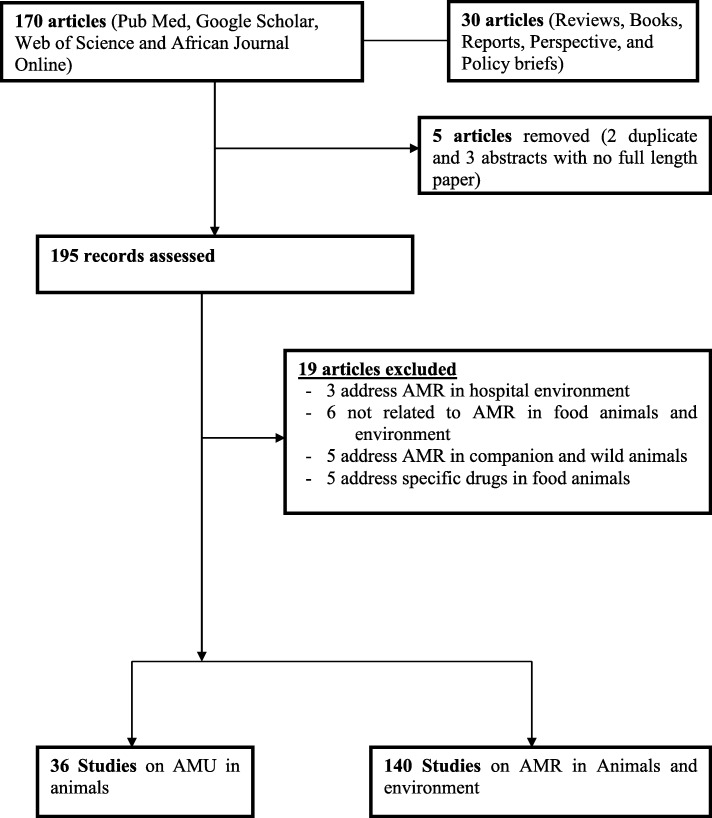
A total of 200 articles were recovered, of which 170 were original research articles and 30 were reviews, books, reports and perspectives or policy briefs. On further evaluation, five were removed, of which two reported similar data and three were abstracts whose full length papers could not be accessed. Of the 195 articles assessed, 19 were excluded from review because they were not relevant (Fig. 1), therefore only 176 studies were included in this review.
Fig. 1.
Flow diagram of number of articles obtained and those eliminated
Types of study and geographical distribution
The majority of the studies were cross-sectional in design, with only two being retrospective (one presenting data on AMU and the other on AMR patterns). Only three studies quantified the amount of antimicrobial using the defined daily dose per animal (DDDA). Some studies involved both surveillance of antimicrobial use and laboratory analysis of organisms’ (Table 1). Geographically, most (56%) of the studies were from the northern and southern Africa regions (Fig. 2).
Table 1.
Different study designs used in data collection on antimicrobial use and resistance
| Sample type | Study type | Approach | Number of articles |
|---|---|---|---|
| Food animals | Cross sectional | Surveillance (Questionnaire, observation and/or focus group discussion and in-depth interview) | 9 |
| Food animals | Cross sectional | Laboratory analysis of antimicrobial susceptibility and resistant organisms | 92 |
| Food animals | Cross sectional | Laboratory and surveillance | 14 |
| Fish | Cross sectional | Laboratory analysis of the resistant organism(s) | 7 |
| Environment | Cross sectional | Laboratory analysis of the resistant organism(s) | 26 |
| Food animals and environment | Cross sectional | Surveillance with laboratory analysis | 9 |
| Food animals and environment | Retrospective | Surveillance and laboratory analysis of the resistant organisms | 3 |
Food animals referred here includes all domestic animals farmed for food consumption (cattle, sheep, goats, poultry, camel, horse, rabbit and donkey); Fish involved those captured from natural water bodies (river, streams, dams and ocean) and the environment samples involved treated waste water, effluent, surface water, river and ocean water, sediments and soil
Fig. 2.
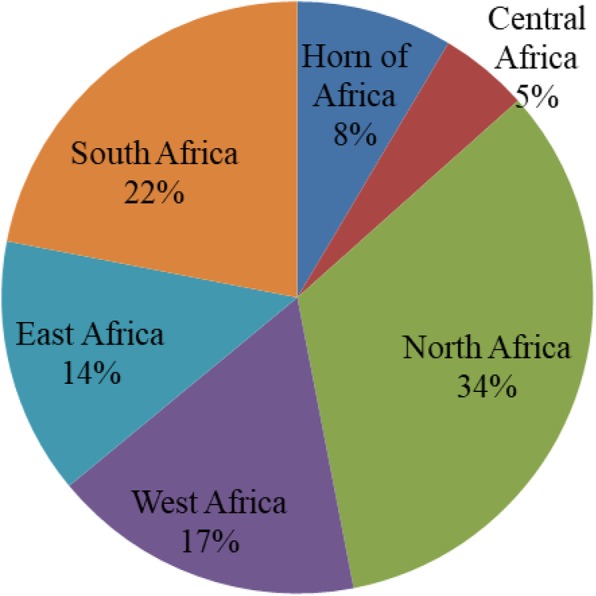
Geographical distribution of AMU and AMR studies in animal and environment
Antimicrobial use in animals
As shown in Table 2, antimicrobial agents were administered to different food animal species, mostly in poultry. The percentage of farms using antimicrobial agents ranged from 77.6% in Nigeria to 100% in Tanzania, Cameroon, Zambia, Ghana and Egypt. In total 14 different classes of antimicrobial agents were used, mainly tetracycline, aminoglycosides and penicillin. Antimicrobial agents such as macrolides, which are restricted for use in animal in some developed countries due to their impact in human health, were used in Africa.
Table 2.
Percentage of farms using antimicrobials by country, type of animal and class of antimicrobials
| Country | Food animal | % AMU | Class of antimicrobial | Reference |
|---|---|---|---|---|
| Ghana | Poultry | 98 | Tetracyclines, Aminoglycosides, Penicillins, Quinolones | [20] |
| Ghana | Cattle, goat, sheep, pig, poultry | 98 | Tetracyclines, Penicillins, Macrolides, Aminoglycosides, Sulphonamides, Benzimidazoles | [21] |
| Tanzania | Cattle, chickens, pigs | 100 | Tetracyclines, Sulphonamides, Penicillins, Aminoglycosides | [22] |
| Cameroon | Poultry | 100 | Aminoglycosides, Sulphonamides, Quinolones, Macrolides, Tetracyclines, Penicillins | [23] |
| Sudan | Poultry | 92 | Tetracyclines | [24] |
| Nigeria | Cattle, sheep, goats | 77.5 | Tetracyclines, Quinolones, Penicillins, Aminoglycosides | [25] |
| Zambia | Cattle | Aminoglycosides, Sulphonamides, Macrolides, Penicillins, Polypeptides, Tetracyclines | [17] | |
| Zambia | Cattle | 100 | Tetracyclines, Penicillins, | [26] |
| Tanzania | Cattle, goat, sheep, pigs, poultry | 74 | Tetracyclines, Penicillins, Macrolides, Aminoglycosides, Sulphonamides | [15] |
| Ghana | Pigs | 100 | Tetracyclines, Sulphonamides, Penicillins, Quinolones, Macrolides, Aminoglycosides | [27] |
| Tanzania | Cattle | 85 | Tetracyclines | [28] |
| Tanzania | Poultry | 90 | Tetracyclines, Sulphonamides, Dihydrofolate, Aminoglycosides, Quinolones | [14] |
| Sudan | Poultry, cattle, sheep, goats | 95 |
Tetracyclines, Penicillins, Macrolides, Sulphonamides, Aminoglycosides, Lincosamides, Streptogramins, Quinolones |
[29] |
| Ethiopia | Cattle, poultry | 80 | Tetracyclines, Penicillins, Sulphonamides | [30] |
| Nigeria | Poultry | 88.5 |
Tetracyclines, Aminoglycosides, Macrolides, Quinolones, Penicillins, Sulphonamides, Furanes, Polypeptides |
[31] |
| Uganda | Pigs | 40.6 | Dihydrofolate, Tetracyclines, Aminoglycosides, | [32] |
| Cameroon | Poultry | 80 | Tetracyclines, Macrolides, Phenocols, Aminoglycosides | [12] |
| Egypt | Poultry | 100 | Tetracyclines, Quinolones | [33] |
| Uganda | Poultry | 96.7 | Sulphonamides | [34] |
| Nigeria | Cattle | 77.6 |
Tetracyclines, Macrolides, Penicillins, Aminoglycosides, Sulphonamides, Quinolones |
[35] |
Antimicrobials use in the environment
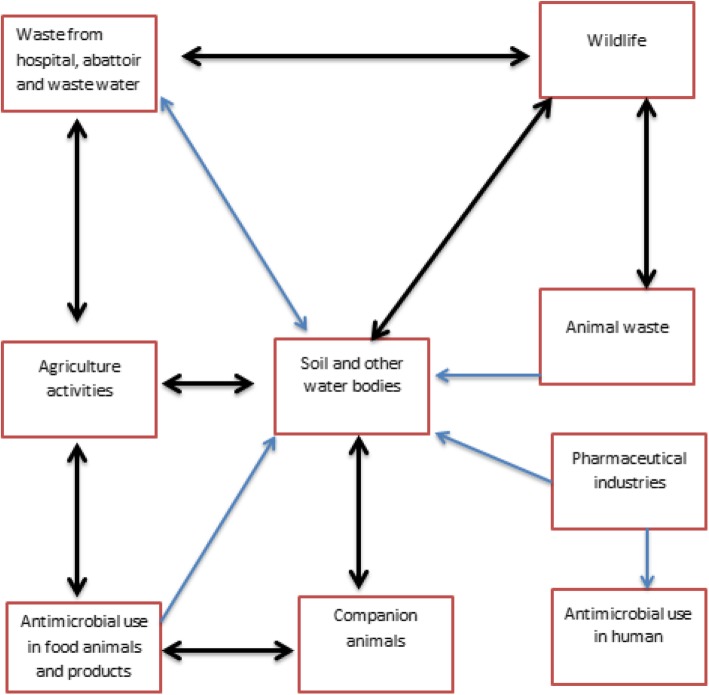
Figure 3 below summarizes the complex interactions between human, animal husbandry, veterinary medicine and the environment with potential for occurrence and spread of AMR resistomes. Such complexity complicates the interventions involved in curbing the burden of AMR.
Fig. 3.
Complex interactions involved in the spread antimicrobial resistance between sectors
Antimicrobial resistance in food animals
This review highlights different levels of antimicrobial resistance within and between countries. Proportion of MDR strains among E. coli, which is an indicator organism, is shown in Table 3. The multi-drug resistance is referred as non-susceptibility to more than three antimicrobial categories [54]. The prevalence of MDR E.coli ranged from 20% in Nigeria to 100% in South Africa, Zimbabwe and Tunisia, in most cases being above 80%. Of the 14 different classes of antimicrobial agents, higher prevalence of resistance was reported for tetracycline, sulphonamides and penicillin, which are the cheapest and therefore more widely used antibiotics.
Table 3.
Proportion of MDR strains among Escherichia coli isolated from food animals
| Sample type | % MDR | Antimicrobial class resisted | References |
|---|---|---|---|
| Poultry | 92.6 | Tetracycline, Penicillin, Quinolones, Phenocols, Sulphonamides, Cephalosporins | [36] |
| Cattle, goat, sheep, pig, poultry | 91.6 | Tetracycline, Penicillin, Phenocols, Aminoglycosides, Sulphonamides | [21] |
| Poultry | 42.9 | Tetracycline, Sulphonamides, Penicillin, Aminoglycosides, Quinolones, Phenocols, Cephalosporins | [37] |
| Poultry, pigs | 20 | Quinolones, Sulphonamides, Macrolides, Tetracycline, Phenocols, Penicillin, Aminoglycosides | [38] |
| Cattle | 100 | Phenocols, Penicillin, Tetracycline, Cephalosporins, Sulphonamides, Aminoglycosides, Quinolones | [39] |
| Poultry | 62 | Tetracycline, Quinolones, Sulphonamides | [40] |
| Cattle, pigs | 93.4 | Tetracycline, Sulphonamides, Macrolides | [41] |
| Cattle, pigs poultry | 45.5 | Tetracycline, Penicillin, Sulphonamides, Dihydrofolate, Penams, Macrolides, Cephalosporins, Clavam | [42] |
| Poultry | 83 | Tetracycline, Sulphonamides, Quinolones, Aminoglycosides, Dihydrofolate, Penicillin, Phenocols | [43] |
| Poultry | 80 | Quinolones, Tetracycline, Penicillin, Aminoglycosides, Sulphonamides, Phenocols | [44] |
| Poultry | 100 | Tetracycline, Penicillin, Quinolones, Aminoglycosides | [45] |
| Cattle, pigs, poultry | 65.5 | Tetracycline, Sulphonamides, Penicillin, Aminoglycosides, Quinolones, Clavam Glycopeptide, Cephalosporins, Dihydrofolate | [46] |
| Poultry | 40 | Tetracycline, Dihydrofolate, Phenocols Sulphonamides, Aminoglycosides | [47] |
| Fish | 54.5 | Cephalosporins, Aminoglycosides, Sulphonamides, Quinolones | [48] |
| Poultry | 90 | Penicillin, Clavam, Penams, Quinolones Cephalosporins, Aminoglycosides | [49] |
| Pigs | 80 | Penicillin, Penams, Tetracycline, Quinolones, Cephalosporins, Aminoglycosides | [50] |
| Poultry | 80 |
Penicillin, Macrolides, Aminoglycosides, Tetracycline, Sulphonamides, Phenocols, Dihydrofolate, Quinolones |
[33] |
| Poultry | 100 | Tetracycline, Quinolones, Dihydrofolate, Sulphonamides, Aminoglycosides | [51] |
| Poultry | 65 |
Tetracycline, Penicillin, Sulphonamides, Phenocols |
[52] |
| Fish | 100 | Aminoglycosides, Cephalosporins, Phenocols, Tetracycline, Sulphonamides, Dihydrofolate, Clavam, Penicillin | [53] |
Antimicrobial resistance in environment
In this review only a few studies reported AMR in the environment. These studies involved samples collected from domestic and biomedical waste, waste water, river sediments, surface and drinking water, treated waste water, river water and vegetables (Table 4). The prevalence of MDR E. coli in environmental samples ranged from 33.3% in South Africa to 100% in Algeria and South Africa. In total E. coli exhibited resistance to 16 different antimicrobial agents.
Table 4.
Percentage of MDR Escherichia coli from environmental samples
| Country | Sample type | %MDR | Antimicrobial class | References |
|---|---|---|---|---|
| South Africa | Treated Waste water | 75.9 | Lincosamides, Sulphonamides, Carbapenems, Quinolones, Penicillin, Tetracycline, Polypeptide, Dihydrofolate, Aminoglycosides, Cephalosporins Macrolides | [55] |
| South Africa | River water, Sediments | 84 |
Furans, Penicillin, Clavam, Quinolones, Phenocols, Dihydrofolate, Cephalosporins |
[56] |
| Ethiopia | Drinking water | 66.7 | Penicillin, Clavam, Quinolones, Cephalosporins, Tetracycline, Phenocols, Sulphonamides, Aminoglycosides | [57] |
| South Africa | River water | 100 | Penicillin, Tetracycline, Aminoglycosides, Cephalosporins, Quinolones, Dihydrofolate | [58] |
| South Africa | Treated waste water | 33.3 | Tetracycline, Penicillin, Furanes, Aminoglycosides, Cephalosporins, Phenocols, Quinolones, Polypeptide, Lipopeptides | [59] |
| Algeria | Treated waste water | 85 |
Cephalosporins, Quinolones, Sulphonamides, Aminoglycosides, Tetracycline, Phenocols |
[60] |
| Algeria | River water | 100 | Penicillin, Clavam, Monobactams | [61] |
| Egypt | River water | 82.5 |
Penicillin, Glycopeptides, Macrolides, Lincosamides, Dihydrofolate, Tetracycline, Sulphonamides |
[62] |
| Tunisia | Waste, Surface water | 76 | Aminoglycosides, Dihydrofolate, Quinolones, Sulphonamides, Phenocols, Tetracycline, Cephalosporins | [6] |
| Tanzania | Domestic, Biomedical waste, River sludge | 56 |
Aminoglycosides, Cephalosporins, Quinolones, Penicillin |
[63] |
Genetic relatedness of resistant genes in animals, humans and environment
We identified a few studies that had determined AMR in all three compartments, namely animals, humans and the environment. Although most of the studies involved phenotypic methods, few involved molecular characterizations as well. The most frequently used molecular methods were Polymerase Chain Reaction (PCR), Pulse Gel Field Electrophoresis (PGFE), and Multilocus Sequence Typing (MLST). A summary of AMR genes in the three compartments is shown in Table 5. Some genes such as blaCTX-M, blaTEM, blaSHV, blaOXA, aac(6′)- Ib-cr, tet(A), tet(B), sul1, sul2, and qnr were commonly detected in all of the three compartments, indicating a potential flow of AMR genes across humans, animals and environment [64–67].
Table 5.
A summary of resistant genes detected in humans, animals and environment
| Methodology | AMR genes | Reference | ||
|---|---|---|---|---|
| Human | Animals | Environment | ||
| Disc diffusion, CHROMagar™, Etest, MALDI-TOF-MS, MLST, PFGE, PCR, Sequencing |
blaCTX-M-15, blaTEM-1, blaOXA-1, aac(6′)- Ib-cr |
blaCTX-M-15, blaOXA-1, aac(6′)- Ib-cr, blaTEM-1 |
blaCTX-M-15, blaTEM-1, blaOXA-1, aac(6′)- Ib-cr |
[64] |
| Disk diffusion, PCR, sequencing |
blaTEM-1, blaOXA-10, blaCTX-M-15 |
blaTEM-1, blaTEM-57 |
[65] | |
| PRISMA guidelines | blaCTX-M-15 | blaCTX-M-15 | blaCTX-M-15 | [66] |
| MacConkey agar, Disk diffusion, PCR | blaKPC, blaOXA-48, blaNDM | blaKPC, blaOXA-48, blaNDM | blaKPC, blaOXA-48, blaNDM | [67] |
| PRISMA guidelines |
blaSHV, blaTEM, blaZ, tet(A), tet(B), tet(C), tet(K), vanA, vanB, vanC1, ermB, vanC2/3,erm, aac (6′)-aph (2″), aac(6′)-lb-cr, blaGES, blaGIM, blaIMP, blaKPC, blaNDM, blaNDM-1, blaPER, blaSIM, blaSPM, blaVIM, cfiA, cfxA, CjgyrA, CmeB, eis, embB, frxA, gidB, gyrA, gyrB, inhA, katG, mcr-1 plasmid, mtrR, mupA, nimA-J, parC, parE, penA, pilQ, pncA, ponA porB1b (penB), qepA, qnrA, qnrB, qnrC, qnrD, qnrS, rdxA, rpoB, Tet(O), tet(Q) |
blaSHV, blaTEM, blaZ, tet(A), tet(B), tet(C), tet(K), vanA, vanB, vanC1, ermB,vanC2/3, erm, ant (3″)-la, ermA, mphC, msrA |
blaSHV, blaTEM, blaZ, tet(A), tet(B), tet(C), tet(K), vanA, vanB, vanC1, ermB, vanC2/3, erm, aadA, aadA1, Bla, cat I, cat II, cmlA1, dfr18, dfrA1, dfrA1, floR, mefA, srC, strB, sul3, tet(D), tet(E), tet(G), tet(H), tet(J), tet(L), tet(Y) |
[68] |
|
MacConkey with 2 mg/L cefotaxime, Disc diffusion, VITEK®2 system and WGS |
blaCTX-M-15, blaTEM-1, blaOXA-1, aac(6′)- Ib-cr, strA, strB, aac(3)-IId, aadA1, qnrB1, qnrS1, sul1, sul2, dfrA14, dfrA17,dfrA1,dfrA18, tet(A), dfrA30, dfrA5, dfrA7, tet(D), blaSHV-11, blaSHV-1, blaACT-15, blaMIR-3, blaCMY-37, blaCMY-49, aadA2, aac(3)-IIa, qnrB29, qnrB48, oqxA, oqxB. |
blaCTX-M-15, blaTEM-1, blaOXA-1, aac(6′)- Ib-cr, strA, strB, aac(3)-IId, aadA1, qnrB1, qnrS1, sul1, sul2, dfrA14, dfrA17, dfrA1, dfrA18, dfrA30, dfrA5, dfrA7, tet(A), tet(D), aadA5 |
[69] | |
| MacConkey agar with 2 mg/mL cefotaxime, ChromAgar, Disk diffusion, WGS VITEK®2 | blaCTX-M-15, strA, strB, aac(6)-Ib-cr, qnrS1 | [70] | ||
| MacConkey agar with 2 mg/mL cefotaxime, Disk diffusion, VITEK®2, WGS CHROMagar, | blaCTX-M-15, aac(6′)Ib-cr, strA, strB, qnrS1 | [71] | ||
| MacConkey broth, Disc diffusion, API 20E, PCR, Sequencing | Tet(B) | Tet(B) | [41] | |
| Disk diffusion, MLST, WGS | strA, strB, aadA1, tet(B), blaTEM-1B, catA1, sul1, sul2, dfrA1 | strA, strB, aadA1, tet(B), blaTEM-1B, catA1, sul1, sul2, dfrA1 | [72] | |
| API 20E System, MALDI-TOF, PCR | blaCTX-M-1, blaCTX-M-15, blaTEM | blaCTX-M-1, blaTEM | [73] | |
| MacConkey and XLD agar, TSI, API 20 E, Disk diffusion, PCR | sul1 and tet(A) | sul1 and tet(A) | [74] | |
| MacConkey with cefotaxime (2 mg/l), MCA with ciprofloxacin, double-disc synergy test, PCR, MALDI, MLST, PFGE | blaTEM-1, blaCTX-M-15, sul1, sul2, tet(B), catA1, oqxA, dfrA17-aadA5, qepA, strA, tet(A), dfr12-orf-aadA2, qnrS1, aadA1, strA-B, tet(D), oqxB, sul3, dfrA7, aadA5, dfr1-sat-aadA1, qnrB1, aac(60)Ib-cr, blaOXA-1, blaSHV-2a, blaCTX-M variant 2 | blaTEM-1, blaCTX-M-15, sul1, sul2, tet(B), catA1, oqxA, dfrA17-aadA5, qepA, strA, tet(A), dfr12-orf-aadA2, qnrS1, aadA1, qnrB33, qnrB17, qnrB28, blaSHV-62 | [75] | |
| Slide agglutination, ETest, PCR |
blaTEM, blaSHV, blaCTX-M, tet(A), floR, sul2, dfrA1, dfr18, strA, strB, qnrVC3, gyrA, gyrB |
blaTEM, blaSHV, blaCTX-M, tet(A), floR, sul2, dfrA1, dfr18, strA, strB, qnrVC3, gyrA, gyrB |
[76] | |
Surveillance systems
Our review revealed that there were no country with a documented national AMU and AMR surveillance programme specific for either animals or environment. Most countries rely on point-prevalence rather than nation-wide surveillance programmes [19, 77]. Some countries have developed a national antimicrobial plan, as an initial stage towards developing AMU and AMR surveillance system in human, animal and environment. Fifteen countries have approved National action Plan for Antimicrobial resistance (NAP), while eight are waiting for the authority’s approval or they are heading towards finalization [78]. It was noted that there is support on capacity to develop surveillance and monitoring of AMU and AMR in food and agriculture provided by the World Health Organization in collaboration with Food and Agriculture Organization and World Organization for Animal Health (OIE) [79].
Other forms of supports included: (i) value chain analysis in animal health from farm to retail step, focusing on microbiology analysis [80]; (ii) training, technical support, strategic guidance and software tools for Web-based data entry and collaboration for the studies targeting animals, environment and other sectors [81]; (iii) training of laboratory technicians from human, agriculture and veterinary sectors from eight countries on laboratory surveillance and control of major food borne diseases [82]; and (iv) research projects on integrated surveillance of AMR in foodborne bacteria for Chad, Tanzania and Ethiopia [82]. Recently, the Africa Centre for Disease Control has launched a network called Antimicrobial Resistance and Surveillance Network comprising experts from animal, environmental and human health sectors. The network targets to mitigate harm from antimicrobial resistant organisms arising from animals, environment, agriculture and human [83]. According to WHO [82] other initiatives provided to countries in Africa included studies on ESBL-producing E.coli in animals and the environment organized in Egypt, Morocco and Sudan.
Discussion
This review involved studies with different designs in evaluation of AMU and AMR. Such variations, especially in sampling designs, complicate comparison of the reported data, and may impact on the reliability of data [32, 78]. There are also potential challenges such as: financial constraints, skilled human resource, lack of research facilities and lack of awareness on the role of animals and environment in the spread of antimicrobial resistances that differ within and between countries. As a consequence, most of the reported studies are from northern Africa, where the issues of AMU and AMR could be less critical than in other parts of Africa.
The review has revealed that more than 80% of farms use antimicrobial agents in animal production, driven by the increase in demand for food of animal origin and emerging trade opportunities [2, 84]. Some of the antimicrobial agents used such as macrolides that have been restricted for use elsewhere due health risk concerns [33, 35] were commonly used in Africa. We found higher AMR prevalence in tetracycline, aminoglycosides and penicillin, these are the cheapest antibiotics hence widely used. The challenge is that most farmers cannot afford alternative, relatively more expensive drugs. This coupled with low awareness on AMU and AMR is likely to further exacerbate the burden of AMR in the region. This review has revealed high prevalence of AMR, including MDR bacteria in the environment, probably associated with anthropogenic activities, application of manure in farming activities and agricultural wastes from community [61, 69, 85, 86]. There is potential spillage of resistant isolates from sewage, humans, companion and domestic animals and industries to the environment [69, 87].
A few studies compared AMR genes in humans, animals and environment and detected blaCTX-M, blaTEM, blaSHV, blaOXA, aac(6′)- Ib-cr, tet(A), tet(B), sul1, sul2, and qnr in all the three compartments [64–67] This finding seems to suggest that these MDR pathogens have high propensity to spread widely and cause infections that are difficult treat [18, 68, 88]. Due to limited laboratory capacity in most of the countries the identified AMR genes could just be a tip of iceberg, representing only a fraction of transmitted genes. This calls for more support to these countries to establish systematic AMR surveillance using more advanced techniques such as Whole Genome Sequencing for better understanding the magnitude, spread and evolution of MDR pathogens [67, 89]. Such information is critical in planning effective interventional measures.
The findings indicate that currently most countries in Africa do not have AMU and AMR surveillance systems and are at different stage of developing them. It may takes a long while before implementation. The different approaches used in different countries leads to inconsistency results which are difficult to compare at national, regional and international perspectives [86]. The results reported do not conform to the international reporting systems since there is no appropriate system for data collection, identifications, coordination and reporting [90]. It is important for countries in Africa to adopt internationally recommended surveillance systems that will take into account data collected from animal, agriculture, environment and human sectors. This review suggests that African countries should seek support from global AMR network to be assisted to develop and implement AMR surveillance under one health approach.
It is important to highlight a number of limitations that were uncovered during the review. First of all, information on AMU is based on percentage of farms using antimicrobial agents rather than the Defined Daily Doses Animals as recommended by the World Health Organization. Furthermore, most of the AMR studies were based on phenotypic rather than molecular techniques, thus limiting understanding of transmission dynamics. In addition, lack of standardized laboratory protocol may account for variations in the level of AMR reported within and between countries. More importantly, there is limited One Health approach surveillance of AMR; hence different sectors employ different approaches in monitoring AMU and AMR. Finally, weak enforcement of the available regulations leads to unmonitored production, distribution, handling, storage and sale of veterinary drugs. It is important that these limitations are taken into consideration as countries in the region continue to plan, develop, refine and implement their National One Health AMR plans.
Conclusion
There are very high levels of antimicrobial use and antimicrobial resistance, especially for tetracycline, aminoglycoside and penicillin in animal production systems in Africa. This is likely to escalate the already high prevalence of antimicrobial resistance and multi drug resistance in the continent. This, coupled with weak regulations and antimicrobial resistance surveillance systems in the region is a great concern to the animals, environment and humans as well. It is important that African countries strengthen their respective AMU and AMR regulations and surveillance systems to address the challenges identified in this review.
Supplementary information
Acknowledgements
Not applicable.
Availability of data and material
Data generated during this study is attached (supplementary file).
Abbreviations
- AGISAR
Advisory Group on Integrated Surveillance of Antimicrobial Resistance
- AMR
Antimicrobial resistance
- AMU
Antimicrobial use
- FAO
Food and Agriculture Organization
- ILRI
International Livestock Research Institute
- MDR
Multi drug resistance
- OIE
Office International des Epizooties
- WHO
World Health Organization
Authors’ contributions
Zuhura I. Kimera Did the literature search, generated the first draft and wrote the manuscript. Mecky I.N. Matee Designed the outline, reviewed the manuscript from the draft to the final work and provided relevant editing. Leonard E. G. Mboera Reviewed the manuscript from the draft to the final version, provided editing and assisted in reference formatting. Stephen E. Mshana and Mark M. Rweyemamu: Reviewed the first draft and coordinated the writing of the entire manuscript. The authors read and approved the final manuscript.
Funding
Not applicable.
Ethics approval and consent to participate
Not applicable in view of the nature of the study.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s13756-020-0697-x.
References
- 1.Ilea RC. Intensive livestock farming: global trends, increased environmental concerns, and ethical solutions. J Agric Environ Ethics. 2009;22:153–167. doi: 10.1007/s10806-008-9136-3. [DOI] [Google Scholar]
- 2.Schar D, Sommanustweechai A, Laxminarayan R, Tangcharoensathien V. Surveillance of antimicrobial consumption in animal production sectors of low- and middle- income countries : optimizing use and addressing antimicrobial resistance. PLoS One. 2018;15(3):1–9. doi: 10.1371/journal.pmed.1002521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ayukekbong JA, Ntemgwa M, Atabe AN. The threat of antimicrobial resistance in developing countries: causes and control strategies. Antimicrob Resist Infect Control. 2017;6(1):1–8. doi: 10.1186/s13756-017-0208-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thornton PK. Livestock production: recent trends, future prospects. Philos Trans R Soc B Biol Sci. 2010;365:2853–2867. doi: 10.1098/rstb.2010.0134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alonso CA, Zarazaga M, Ben Sallem R, Jouini A, Ben Slama K, Torres C. Antibiotic resistance in Escherichia coli in husbandry animals: the African perspective. Lett Appl Microbiol. 2017;64:318–334. doi: 10.1111/lam.12724. [DOI] [PubMed] [Google Scholar]
- 6.Ben Said L, Jouini A, Alonso CA, Klibi N, Dziri R, Boudabous A, et al. Characteristics of extended-spectrum beta-lactamase (ESBL) and pAmpC beta-lactamase-producing Enterobacteriaceae of water samples in Tunisia. Sci Total Environ. 2016;550:1103–1109. doi: 10.1016/j.scitotenv.2016.01.042. [DOI] [PubMed] [Google Scholar]
- 7.Gothwal R, Shashidhar T. Antibiotic pollution in the environment: a review. Clean Soil Air Water. 2015;43(4):479–489. doi: 10.1002/clen.201300989. [DOI] [Google Scholar]
- 8.World Bank . Drug-resistant infections: a threat to our economic future. 2017. [Google Scholar]
- 9.Oyekale AS, TO O. Healthcare waste management practices and safety indicators in Nigeria. BMC Public Health. 2017;17(1):1–13. doi: 10.1186/s12889-017-4794-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grace D. Review of evidence on antimicrobial resistance and animal agriculture in developing countries. International Livestock Research Institure. 2015. Available from: https://www.gov.uk/dfid-research-outputs/review-of-evidence-on-antimicrobial-resistance-and-animal-agriculture-in-developing-countries-201309
- 11.Bekele B, Ashenafi M. Distribution of drug resistance among enterococci and Salmonella from poultry and cattle in Ethiopia. Trop Anim Health Prod. 2010;42(5):857–864. doi: 10.1007/s11250-009-9499-0. [DOI] [PubMed] [Google Scholar]
- 12.Guetiya Wadoum RE, Zambou NF, Anyangwe FF, Njimou JR, Coman MM, Verdenelli MC, et al. Abusive use of antibiotics in poultry farming in Cameroon and the public health implications. Br Poult Sci. 2016;57(4):483–493. doi: 10.1080/00071668.2016.1180668. [DOI] [PubMed] [Google Scholar]
- 13.World Health Organization. Global action plan on antimicrobial resistance: WHO Press; 2015. p. 1–39. Available from: http://www.who.int/drugresistance/global_action_plan/en/. [cited 2018 Aug 13]
- 14.Nonga HE, Mariki M, MR KED. Antimicrobial usage and residue in Morogoro. Pakistan J Nutr. 2009;8(3):203–207. doi: 10.3923/pjn.2009.203.207. [DOI] [Google Scholar]
- 15.Caudell MA, Quinlan MB, Subbiah M, Call DR, Roulette CJ, Roulette JW, et al. Antimicrobial use and veterinary care among agro-pastoralists in northern Tanzania. PLoS One. 2017;12(1):1–18. doi: 10.1371/journal.pone.0170328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Afema JA, Byarugaba DK, Shah DH, Atukwase E, Nambi M, Sischo WM. Potential sources and transmission of salmonella and antimicrobial resistance in Kampala, Uganda. PLoS One. 2016;11(3):1–21. doi: 10.1371/journal.pone.0152130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mainda G, Bessell PB, Muma JB, McAteer SP, Chase-Topping ME, Gibbons J, et al. Prevalence and patterns of antimicrobial resistance among Escherichia coli isolated from Zambian dairy cattle across different production systems. Sci Rep. 2015;5:1–11. doi: 10.1038/srep12439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oloso NO, Fagbo S, Garbati M, Olonitola SO, Awosanya EJ, Aworh MK, et al. Antimicrobial resistance in food animals and the environment in Nigeria : a review. Int J Environ Res Public Health. 2018;15:1–23. doi: 10.3390/ijerph15061284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Founou LL, Founou RC, Essack SY. Antibiotic resistance in the food chain: a developing country-perspective. Front Microbiol. 2016;7(1881):1–19. doi: 10.3389/fmicb.2016.01881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boamah VE, Agyare C, Odoi H, Dalsgaard A. Antibiotic practices and factors influencing the use of antibiotics in selected poultry farms in Ghana. J Antimicrob Agents. 2016;2(2):1–8. [Google Scholar]
- 21.Donkor ES, Newman MJ, Yeboah-Manu D. Epidemiological aspects of non-human antibiotic usage and resistance: implications for the control of antibiotic resistance in Ghana. Trop Med Int Heal. 2012;17(4):462–468. doi: 10.1111/j.1365-3156.2012.02955.x. [DOI] [PubMed] [Google Scholar]
- 22.Katakweba AAS, Mtambo MMA, Olsen JE, Muhairwe AP. Awareness of human health risks associated with the use of antimicrobials among livestock keepers and factors that contribute to selection of antibiotic resistance bacteria within livestock in Tanzania. Livistock Rural Res Dev. 2012;24(10):1–14. [Google Scholar]
- 23.Kamini MG, Keutchatang FT, Huguette YM, Kansci Germain A, Gabriel MN. Antimicrobial usage in the chicken farming in Yaoundé, Cameroon: a cross-sectional study. Int J Food Contam. 2016;3(1):1–6. doi: 10.1186/s40550-015-0024-0. [DOI] [Google Scholar]
- 24.Sirdar MM, Picard J, Bisschop S, Gummow B. A questionnaire survey of poultry layer farmers in Khartoum state, Sudan, to study their antimicrobial awareness and usage patterns. Onderstepoort J Vet Res. 2012;79(1):1–8. doi: 10.4102/ojvr.v79i1.361. [DOI] [PubMed] [Google Scholar]
- 25.Adesokan HK, Akanbi IOIM, Akanbi IOIM, Obaweda RA, Vuuren V, Den Bogaard V, et al. Pattern of antimicrobial usage in livestock animals in South-Western Nigeria: the need for alternative plans. Onderstepoort J Vet Res. 2015;82(1):1–6. doi: 10.4102/ojvr.v82i1.816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mubita C, Syakalima M, Chisenga C, Munyeme M, Bwalya M, Chifumpa G, et al. Antibiograms of faecal Escherichia coli and enterococci species isolated from pastoralist cattle in the interface areas of the Kafue basin in Zambia - short communication. Vet Arh. 2008;78(2):179–185. [Google Scholar]
- 27.Osei SJ. Antibiotic types and handling practices in disease management among pig farms in Ashanti region, Ghana. J Vet Med. 2014;2014:1–8. doi: 10.1155/2014/531952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kimera ZI, Mdegela RH, Mhaiki CJN, Karimuribo ED, Mabiki F, Nonga HE, et al. Determination of oxytetracycline residues in cattle meat marketed in the Kilosa district, Tanzania. Onderstepoort J Vet Res. 2015;82(1):1–5. doi: 10.4102/ojvr.v82i1.911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eltayb A, Barakat S, Marrone G, Shaddad S, Sta C. Antibiotic use and resistance in animal farming : a quantitative and qualitative study on knowledge and practices among farmers in Khartoum, Sudan. Zoonoses Public Health. 2012;59:330–338. doi: 10.1111/j.1863-2378.2012.01458.x. [DOI] [PubMed] [Google Scholar]
- 30.Tufa TB, Gurmu F, Beyi AF, Hogeveen H, Beyene TJ, Ayana D, et al. Veterinary medicinal product usage among food animal producers and its health implications in Central Ethiopia. BMC Vet Res. 2018;14(409):1–7. doi: 10.1186/s12917-018-1737-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alhaji N.B., Haruna A.E., Muhammad B., Lawan M.K., Isola T.O. Antimicrobials usage assessments in commercial poultry and local birds in North-central Nigeria: Associated pathways and factors for resistance emergence and spread. Preventive Veterinary Medicine. 2018;154:139–147. doi: 10.1016/j.prevetmed.2018.04.001. [DOI] [PubMed] [Google Scholar]
- 32.Ikwap K, Erume J, Owiny DO, Nasinyama GW, Melin L, Bengtsson B, et al. Salmonella species in piglets and weaners from Uganda: prevalence, antimicrobial resistance and herd-level risk factors. Prev Vet Med. 2014;115(1–2):39–47. doi: 10.1016/j.prevetmed.2014.03.009. [DOI] [PubMed] [Google Scholar]
- 33.Dahshan H, Abd-Elall AMM, Megahed AM, Abd-El-Kader MA, Nabawy EE. Veterinary antibiotic resistance, residues, and ecological risks in environmental samples obtained from poultry farms, Egypt. Environ Monit Assess. 2015;187(2):1–10. doi: 10.1007/s10661-014-4218-3. [DOI] [PubMed] [Google Scholar]
- 34.Sasanya JJ, Ogawal Okeng JW, Ejobi F, Muganwa M. Use of sulfonamides in layers in Kampala district, Uganda and sulfonamide residues in commercial eggs. Afr Health Sci. 2005;5(1):33–39. [PMC free article] [PubMed] [Google Scholar]
- 35.Alhaji Nma Bida, Isola Tajudeen Opeyemi. Antimicrobial usage by pastoralists in food animals in North-central Nigeria: The associated socio-cultural drivers for antimicrobials misuse and public health implications. One Health. 2018;6:41–47. doi: 10.1016/j.onehlt.2018.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hamisi Z, Tuntufye H, Shahada F. Antimicrobial resistance phenotypes of Escherichia coli isolated from tropical free range chickens. Int J Sci Res. 2014;3(9):2012–2015. [Google Scholar]
- 37.Odwar JA, Kikuvi G, Kariuki JN, Kariuki S. A cross-sectional study on the microbiological quality and safety of raw chicken meats sold in Nairobi, Kenya. BMC Res Notes. 2014;7(627):1–8. doi: 10.1186/1756-0500-7-627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fortini D, Fashae K, García-Fernández A, Villa L, Carattoli A. Plasmid-mediated quinolone resistance and beta-lactamases in Escherichia coli from healthy animals from Nigeria. J Antimicrob Chemother. 2011;66(6):1269–1272. doi: 10.1093/jac/dkr085. [DOI] [PubMed] [Google Scholar]
- 39.Iweriebor BC, Iwu CJ, Obi LC, Nwodo UU, Okoh AI. Multiple antibiotic resistances among Shiga toxin producing Escherichia coli O157 in feces of dairy cattle farms in eastern cape of South Africa. BMC Microbiol. 2015;15(213):1–9. doi: 10.1186/s12866-015-0553-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fashae K, Ogunsola F, Aarestrup FM, Hendriksen RS. Antimicrobial susceptibility and serovars of Salmonella from chickens and humans in Ibadan, Nigeria. J Infect Dveloping Ctries. 2010;4(8):484–494. doi: 10.3855/jidc.909. [DOI] [PubMed] [Google Scholar]
- 41.Collins Njie Ateba CCB. Characterisation of Escherichia coli O157 strains from humans, cattle and pigs in the north-West Province, South Africa. Int J Food Microbiol. 2008;128(2):181–188. doi: 10.1016/j.ijfoodmicro.2008.08.011. [DOI] [PubMed] [Google Scholar]
- 42.Adenipekun EO, Jackson CR, Oluwadun A, Iwalokun BA, Frye JG, Barrett JB, et al. Prevalence and antimicrobial resistance in Escherichia coli from food animals in Lagos, Nigeria. Microb Drug Resist. 2015;21(3):358–365. doi: 10.1089/mdr.2014.0222. [DOI] [PubMed] [Google Scholar]
- 43.Adelowo OO, Fagade OE, Agersø Y. Antibiotic resistance and resistance genes in Escherichia coli from poultry farms, Southwest Nigeria. J Infect Dis Dev Ctries. 2014;8(9):1103–1112. doi: 10.3855/jidc.4222. [DOI] [PubMed] [Google Scholar]
- 44.Hussein AHM, Ghanem IAI, Eid AAM, Ali MA, Julie S, Li G, et al. Molecular and phenotypic characterization of Escherichia coli isolated from broiler chicken flocks in Egypt. Avian Dis. 2013;57(3):602–611. doi: 10.1637/10503-012513-Reg.1. [DOI] [PubMed] [Google Scholar]
- 45.Saidi B, Mafirakureva P, Mbanga J, Saidi B, Mafirakureva AP, Bc JM. Antimicrobial resistance of Escherichia coli isolated from chickens with Colibacillosis in and around Harare, Zimbabwe. Avian Dis. 2013;57(1):152–154. doi: 10.1637/10325-081512-Case.1. [DOI] [PubMed] [Google Scholar]
- 46.Katakweba AAS, Muhairwa AP, Lupindu AM, Damborg P, Rosenkrantz JT, Minga UM, et al. First report on a randomized investigation of antimicrobial resistance in fecal Indicator Bacteria from livestock, poultry, and humans in Tanzania. Microb Drug Resist. 2018;24(3):260–268. doi: 10.1089/mdr.2016.0297. [DOI] [PubMed] [Google Scholar]
- 47.Chah KF, Ugwu IC, Okpala A, Adamu KY, Andrea C, Ceballos S, et al. Detection and molecular characterisation of extended-spectrum Beta - lactamase-producing enteric bacteria from pigs and chickens in Nsukka, Nigeria. Glob Antimicrob Res. 2018;15:36–40. doi: 10.1016/j.jgar.2018.06.002. [DOI] [PubMed] [Google Scholar]
- 48.Brahmi S, Touati A, Dunyach-Remy C, Sotto A, Pantel A, J-PL High prevalence of extended-Spectrum Beta-lactamase producing Enterobacteriaceae in wild fish from the Mediterranean Sea in Algeria. Microb Drug Resist. 2017;00(00):1–9. doi: 10.1089/mdr.2017.0149. [DOI] [PubMed] [Google Scholar]
- 49.Belmahdi M, Bakour S, Al Bayssari C, Touati A, Rolain J-M. Molecular characterisation of extended-spectrum beta-lactamase- and plasmid AmpC-producing Escherichia coli strains isolated from broilers in Béjaïa, Algeria. J Glob Antimicrob Resist. 2016;6:108–112. doi: 10.1016/j.jgar.2016.04.006. [DOI] [PubMed] [Google Scholar]
- 50.Beshiru A, Igbinosa IH, Igbinosa EO. An investigation on Antibiogram characteristics of Escherichia coli isolated from piggery farms in Benin City, Nigeria. Ann Sci Technol. 2016;1(1):8–12. doi: 10.22366/ast.2016.01.002. [DOI] [Google Scholar]
- 51.Mnif B, Ktari S, Rhimi FM, Hammami A. Extensive dissemination of CTX-M-1- and CMY-2-producing Escherichia coli in poultry farms in Tunisia. Lett Appl Microbiol. 2012;55:407–413. doi: 10.1111/j.1472-765X.2012.03309.x. [DOI] [PubMed] [Google Scholar]
- 52.Tellevik MG, Blomberg B, Kommedal Ø, Maselle SY, Langeland N, Moyo SJ. High prevalence of faecal carriage of esbl-producing enterobacteriaceae among children in Dar Es Salaam, Tanzania. PLoS One. 2016;11(12):1–13. doi: 10.1371/journal.pone.0168024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Leila A, Agabou A, Chahed A, Kurekci C, Moreno E, Espigares M, et al. Isolation, molecular characterization and antimicrobial resistance of enterobacteriaceae isolated from fish and seafood. Food Control. 2018;88:54–60. doi: 10.1016/j.foodcont.2018.01.005. [DOI] [Google Scholar]
- 54.Magiorakos A, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an internatiojnal expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18(3):268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 55.Igwaran A. Molecular characterization and antimicrobial resistance pattern of Escherichia coli recovered from wastewater treatment plants in eastern Cape South Africa. Environ Res Public Heal. 2018;15(1237):1–10. doi: 10.3390/ijerph15061237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Abia ALK, Ubomba-Jaswa E, Momba MNB. High prevalence of multiple-antibiotic-resistant (MAR) Escherichia coli in river bed sediments of the Apies River, South Africa. Environ Monit Assess. 2015;187(10):1–13. doi: 10.1007/s10661-015-4879-6. [DOI] [PubMed] [Google Scholar]
- 57.Abera B, Kibret M, Goshu G, Yimer M. Bacterial quality of drinking water sources and antimicrobial resistance profile of Enterobacteriaceae in Bahir Dar city, Ethiopia. J Water, Sanit Hyg Dev. 2014;4(3):384–390. doi: 10.2166/washdev.2014.105. [DOI] [Google Scholar]
- 58.Ebomah KE, Adefisoye MA, Okoh AI. Pathogenic Escherichia coli strains recovered from selected aquatic resources in the eastern cape, South Africa, and its significance to public health. Int J Environ Res Public Health. 2018;15(7):1–10. doi: 10.3390/ijerph15071506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Adefisoye MA, Okoh AI. Identification and antimicrobial resistance prevalence of pathogenic Escherichia coli strains from treated wastewater effluents in eastern cape, South Africa. Microbiology. 2016;5(1):143–151. doi: 10.1002/mbo3.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Alouache S, Estepa V, Messai Y, Ruiz E, Torres C, Bakour R. Characterization of ESBLs and associated quinolone resistance in Escherichia coli and Klebsiella pneumoniae isolates from an urban wastewater treatment Plant in Algeria. Microb Drug Resist. 2013;20(1):30–38. doi: 10.1089/mdr.2012.0264. [DOI] [PubMed] [Google Scholar]
- 61.Tafoukt R, Touati A, Leangapichart T, Bakour S, Rolain JM. Characterization of OXA-48-like-producing Enterobacteriaceae isolated from river water in Algeria. Water Res. 2017;120:185–189. doi: 10.1016/j.watres.2017.04.073. [DOI] [PubMed] [Google Scholar]
- 62.Mervat A, Abo-State, Mahdy HM, Ezzat EHAES SM, MAE-B Antimicrobial Resistance Profiles of Enterobacteriaceae Isolated from Rosetta Branch of River Nile , Egypt Central Laboratory for Environmental Quality Monitoring ( CLEQM) World Appl Sci J. 2012;19(9):1234–1243. [Google Scholar]
- 63.Mwaikono KS, Maina S, Gwakisa P. Prevalence and antimicrobial resistance phenotype of enteric Bacteria from a municipal dumpsite. J Appl Environ Microbiol. 2015;3(3):82–94. [Google Scholar]
- 64.Ribeiro TG. Novais, Peixe L, Machado E. atypical epidemiology of CTX-M-15 among Enterobacteriaceae from a high diversity of non-clinical niches in Angola. J Antimicrob Chemother. 2016;71(5):1169–1173. doi: 10.1093/jac/dkv489. [DOI] [PubMed] [Google Scholar]
- 65.Eguale T, Birungi J, Asrat D, Njahira MN, Njuguna J, Gebreyes WA, et al. Genetic markers associated with resistance to beta-lactam and quinolone antimicrobials in non-typhoidal Salmonella isolates from humans and animals in Central Ethiopia. Antimicrob Resist Infect Control. 2017;6(1):13. [DOI] [PMC free article] [PubMed]
- 66.Seni J, Moremi N, Matee M, van der Meer F, DeVinney RMS, PJD Preliminary insights into the occurence of similar clones of extended-spectrum beta-lactamase-producing bacteria in humans, animals and the environment in Tanzania: a systematic review and meta-analysis between 2005 and 2006. Zoonoses Public Health. 2017;00:1–10. doi: 10.1111/zph.12387. [DOI] [PubMed] [Google Scholar]
- 67.Hamza E, Dorgham SM, Hamza DA. Carbapenemase-producing Klebsiella pneumoniae in broiler poultry farming in Egypt. Integr Med Res. 2016;7:8–10. doi: 10.1016/j.jgar.2016.06.004. [DOI] [PubMed] [Google Scholar]
- 68.Ekwanzala MD, Dewar JB, Kamika I, Momba MNB. Systematic review in South Africa reveals antibiotic resistance genes shared between clinical and environmental settings. Infect Drug Resist. 2018;11:1907–1920. doi: 10.2147/IDR.S170715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Moremi N, Manda EV, Falgenhauer L, Ghosh H, Imirzalioglu C, Matee M, et al. Predominance of CTX-M-15 among ESBL producers from environment and fish gut from the shores of Lake Victoria in Mwanza, Tanzania. Front Microbiol. 2016;7(1862):1–11. doi: 10.3389/fmicb.2016.01862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Seni J, Falgenhauer L, Simeo N, Mirambo MM, Imirzalioglu C, Matee M, et al. Multiple ESBL-producing Escherichia coli sequence types carrying quinolone and aminoglycoside resistance genes circulating in companion and domestic farm animals in Mwanza, Tanzania, harbor commonly occurring plasmids. Front Microbiol. 2016;7(142):1–9. doi: 10.3389/fmicb.2016.00142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mshana SE, Falgenhauer L, Mirambo MM, Mushi MF, Moremi N, Julius R, et al. Predictors of blaCTX-M-15 in varieties of Escherichia coli genotypes from humans in community settings in Mwanza, Tanzania. BMC Infect Dis. 2016;16(1):1–9. doi: 10.1186/s12879-016-1527-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kagambèga A, Lienemann T, Frye JG, Barro N, Haukka K. Whole genome sequencing of multidrug-resistant Salmonella enterica serovar Typhimurium isolated from humans and poultry in Burkina Faso. Trop Med Health. 2018;46(1):1–5. doi: 10.1186/s41182-018-0086-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Djeffal S, Bakour S, Mamache B, Elgroud R, Agabou A, Chabou S, et al. Prevalence and clonal relationship of ESBL- producing Salmonella strains from humans and poultry in northeastern Algeria. BMC Vet Res. 2017;13(132):1–9. doi: 10.1186/s12917-017-1050-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Christabel M, Budambula N, Kiiru J, Kariuki S. Characterization of antibiotic resistance in environmental enteric pathogens from Kibera slum in Nairobi-Kenya. J Bacteriol Res. 2012;4(4):46–54. doi: 10.5897/JBR12.008. [DOI] [Google Scholar]
- 75.Janatova M, Albrechtova K, Petrzelkova KJ, Dolejska M, Papousek I, Masarikova M, et al. Antimicrobial-resistant Enterobacteriaceae from humans and wildlife in Dzanga-Sangha protected area, Central African Republic Central African Republic. Vet Microbiol. 2014;171(3–4):422–431. doi: 10.1016/j.vetmic.2014.02.014. [DOI] [PubMed] [Google Scholar]
- 76.Ismail H, Smith AM, Tau NP, Sooka A, Keddy KH. Cholera outbreak in South Africa, 2008-2009: laboratory analysis of Vibrio cholerae O1 strains. J Infect Dis. 2013;208:2008–2009. doi: 10.1093/infdis/jit200. [DOI] [PubMed] [Google Scholar]
- 77.Perovic O, Schultsz C. Stepwise approach for implementing antimicrobial resistance surveillance in Africa. Afr J Lab Med. 2016;5(3):a482. doi: 10.4102/ajlm.v5i3.482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.WHO. Integrated Surveilance of Antimicrobial Resistance. Guidance from WHO Advisory Group. www.who.int. 2013. p. 1–100. Available from: http://apps.who.int/iris/bitstream/10665/91778/1/9789241506311_eng.pdf?ua=1. [cited 2018 Sep 3]
- 79.World Health Organisation for Animal Health . 2nd Conference on antimicrobial resistance and prudent use of antimicrobials in animals. 2018. [Google Scholar]
- 80.Ashley EA, Chua A, Dance D, Day NP, Dhorda M, Guerin P, Ranganathan N, Recht J, Rushton J, Smith C, Nigel Thomas PT, NJW . Antimicrobial Resistance in Low and Middle Income Countries: An Analysis of Surveillance Networks. 2017. [Google Scholar]
- 81.World Health Organisation . Intergrated Surveillance of Antimicrobial Resistance in Foodborne Bacteria: Application of a One Health Approach. Geneva: World Health Organization; 2017. pp. 1–88. [Google Scholar]
- 82.World Health Organization. Global Antimicrobial Resistance Surveillance System (GLASS) report: early implementation 2017–2018 Geneva: 2018.; 2018 p. 1–268. Available from: https://www.who.int/glass/resources/publications/early-implementation-report-2017-2018/en/. [cited 2018 Apr 22].
- 83.First Africa Centres for Disease Control and Prevention’s Southern Africa Regional Collaborating Centre Meeting. Lusaka, Zambia; 2017.
- 84.Ben L, Klibi N, Lozano C, Dziri R, Ben K, Boudabous A, et al. Science of the Total environment diversity of enterococcal species and characterization of high-level aminoglycoside resistant enterococci of samples of wastewater and surface water in Tunisia. Sci Total Environ. 2015;530–531:11–17. doi: 10.1016/j.scitotenv.2015.05.091. [DOI] [PubMed] [Google Scholar]
- 85.Majalija S, Francis O, Sarah W. G, Musisi-Lubowa VP and FM. Antibiotic Susceptibility Prifiles of Fecal Escherichia coli Isolated from Dip-Litter Broiler Chickens in Northern and Central Uganda. Vet Res 2010;3(4):75–80.
- 86.Manyi-Loh C, Sampson Mamphweli EM, AO Antibiotic use in agriculture and its consequential resistance in environmental sources : potential public health implications. Molecules. 2018;23(975):1–48. doi: 10.3390/molecules23040795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Moremi N, Claus H, Vogel U, Mshana SE. Faecal carriage of CTX-M extended-spectrum beta-lactamase-producing Enterobacteriaceae among street children dwelling in Mwanza city, Tanzania. PLoS One. 2017;12(9):1–11. doi: 10.1371/journal.pone.0184592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.De Boeck H, Lunguya O, Muyembe JJ, Glupczynski Y, Jacobs J. Presence of extended-spectrum beta-lactamase-producing Enterobacteriaceae in waste waters, Kinshasa, the Democratic Republic of the Congo. Eur J Clin Microbiol Infect Dis. 2012;31(11):3085–3088. doi: 10.1007/s10096-012-1669-8. [DOI] [PubMed] [Google Scholar]
- 89.Grundmann H and, Gelband H. Antimicrobial resistance surveillance with whole genome sequencing in Africa: It’s (about) time. Afr J Lab Med. 2018;7(2):1–3. [DOI] [PMC free article] [PubMed]
- 90.Dar OA, Hasan R, Schlundt J, Harbarth S, Caleo G, Dar FK, et al. Antimicrobials: access and sustainable effectiveness for exploring the evidence base for national and regional policy interventions to combat resistance. Lancet. 2016;387:285–295. doi: 10.1016/S0140-6736(15)00520-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data generated during this study is attached (supplementary file).