Abstract
Background
Clinical studies suggest obesity paradoxically increases survival during bacterial infection and sepsis but decreases it with influenza, but these studies are observational. By contrast, animal studies of obesity in infection can prospectively compare obese versus nonobese controls. We performed a systematic review and meta-analysis of animal investigations to further examine obesity's survival effect in infection and sepsis.
Methods
Databases were searched for studies comparing survival in obese versus nonobese controls. We performed a systematic review and meta-analysis of animal investigations to further examine obesity's survival effect in infection and sepsis. Methods. Databases were searched for studies comparing survival in obese versus nonobese animals following bacteria, lipopolysaccharide, or influenza virus challenges.
Results
Twenty-one studies (761 obese and 603 control animals) met the inclusion criteria. Obesity reduced survival in 19 studies (11 significantly) and the odds ratio (95% CI) of survival (0.21(0.13, 0.35); I2 = 64%, p < 0.01p < 0.01p < 0.01) but with high heterogeneity. Obesity reduced survival (1) consistently in both single-strain bacteria- and lipopolysaccharide-challenged studies (n = 6 studies, 0.21(0.13, 0.34); I2 = 64%, p < 0.01p < 0.01) but with high heterogeneity. Obesity reduced survival (1) consistently in both single-strain bacteria- and lipopolysaccharide-challenged studies (n = 6 studies, 0.21(0.13, 0.34); I2 = 64%, p < 0.01p < 0.01) but with high heterogeneity. Obesity reduced survival (1) consistently in both single-strain bacteria- and lipopolysaccharide-challenged studies (n = 6 studies, 0.21(0.13, 0.34); I2 = 64%, p < 0.01p < 0.01) but with high heterogeneity. Obesity reduced survival (1) consistently in both single-strain bacteria- and lipopolysaccharide-challenged studies (n = 6 studies, 0.21(0.13, 0.34); I2 = 64%, p < 0.01p < 0.01p < 0.01) but with high heterogeneity. Obesity reduced survival (1) consistently in both single-strain bacteria- and lipopolysaccharide-challenged studies (n = 6 studies, 0.21(0.13, 0.34); I2 = 31%, p=0.20 and n = 5, 0.22(0.13, 0.36); I2 = 0%, p=0.59, respectively), (2) not significantly with cecal ligation and puncture (n = 4, 0.72(0.08, 6.23); I2 = 75%, p < 0.01), and (3) significantly with influenza but with high heterogeneity (n = 6, 0.12(0.04, 0.34); I2 = 73%, p < 0.01). Obesity's survival effects did not differ significantly comparing the four challenge types (p=0.49). Animal models did not include antimicrobials or glycemic control and study quality was low.
Conclusions
Preclinical and clinical studies together emphasize the need for prospective studies in patients accurately assessing obesity's impact on survival during severe infection.
1. Introduction
Obesity is a growing problem in the developed world and underlies chronic comorbidities that reduce overall life expectancy (e.g., hypertension and diabetes) [1–5]. While 25% or more of adults admitted to intensive care units (ICUs) in the United States and other developed countries are overweight or obese, whether this negatively impacts ICU outcomes is unclear [6–10]. This point is highlighted by conflicting data regarding obesity's effects on mortality in patients with bacterial infection and sepsis or viral influenza, two common reasons for ICU admission. Although several individual studies suggest that obesity worsens or has no impact on survival in patients with sepsis, two recent systematic reviews and meta-analyses both reported that overweight or obese body mass indices (BMIs) were paradoxically associated with improved outcomes in sepsis [11–15]. By contrast, two other systematic reviews and meta-analyses and a retrospective analysis of a large patient database found that for patients with influenza pneumonia, obesity increased the risk of either a combined endpoint of ICU admission and death or death alone [16, 17]. Another pooled analysis found that obesity increased the risk of death with influenza and pneumonia [18].
Research into the mechanisms and effects of obesity has relied on both diet- and genetically induced animal obesity models (e.g., leptin or leptin-receptor-deficient mice) [19]. These models have also been used to examine the impact of obesity on the host response and outcome in conditions associated with critical illness, including bacterial and viral infection [20–23]. In contrast to clinical studies which have been observational, retrospective, and possibly confounded by differing baseline characteristics and methodology, animal studies employ prospective controlled designs and uniform subjects that differ primarily in body weight and obesity cause. We therefore performed a systematic review and meta-analysis of animal studies to further examine how obesity may impact survival with bacterial, lipopolysaccharide, or influenza infection.
2. Methods
This systematic review was prepared using the PRISMA (Preferred Reporting Items for Systematic Reviews and Meta-Analyses) guidance for literature review and extraction of data, and a completed PRISMA checklist is provided in the Supplementary Material. Complete methods are provided in the Supplementary Material.
2.1. Literature Search and Study Selection
Using published guidelines [24] and search strategies presented in the Supplementary Material, two authors (DJP and PQE) identified relevant studies in the following databases from inception through January 25, 2017, and without language restrictions: PubMed, EMBASE, Scopus, and Web of Science. Included studies were searched for additional references. Studies were included if they compared survival in a nonobese control versus obese group in an animal model with bacterial infection, bacterial lipopolysaccharide (LPS), or influenza viral infection challenge. Studies without reported animal weights were included if they compared animals with a diet or genotype known to produce obesity.
2.2. Data Extracted and Outcomes Examined
Data were extracted by two authors (WZ and PQE) for each survival experiment in a report as described in the supplemental methods. The primary outcome examined was the effect of obesity on the odds ratio of survival based on the number of animals reported living at the end of observation periods. Group sizes, animal weights, fat masses, and blood glucose levels were determined as described in the supplemental methods. Secondary outcomes, presented in the supplemental methods, included the effect of obesity on organ injury based on physiologic or histologic measures; bacterial or viral clearance assessed by reported bacteria or viral counts in blood or tissue; and inflammatory cytokine and leptin levels in serum or tissue. Study quality and risk of bias were assessed in studies based on the Systemic Review Center for Laboratory animal Experimentation (SYRCLE) grading system and as previously described [25, 26]. Criteria for this grading are further outlined in the supplemental methods.
2.3. Statistical Analysis
The odds ratio of survival with obesity versus a nonobese control was estimated using a random-effects model [27]. In retrieved studies in which more than one experiment was performed using the same type of obesity model (i.e., diet or genetic), if the survival results of these experiments were qualitatively similar and consistent, these results were pooled to provide a single survival effect for the study. Experiments comparing two obese groups to a common control group or with similar survival in obese and control groups were analyzed as described in the supplemental methods. The effects of obesity on survival were analyzed based on the type of obesity model employed, the animal species studied, and the type of infectious or septic challenge employed. Heterogeneity among studies was assessed using the Q statistic and I2 value and was considered moderate or greater for I2 ≥ 35% [28]. Publication bias was to be assessed by funnel plot and Egger's regression if sufficient data were available. All analyses were performed using R (version 3.4.0) package meta (version 4.9-1) [29, 30]. Two-sided p values ≤0.05 were considered significant.
3. Results
From 4,569 references identified in the literature search, 21 studies met inclusion criteria (Figure 1, Supplement References in the Supplementary Material). These 21 studies included 52 experiments employing 603 control and 761 obese animals. Table 1 provides details for each of the 52 experiments in the 21 studies regarding the animals and obesity and infection models employed. Table 2 summarizes for each experiment the weight, fat mass, and glucose level recorded in control and obese groups and the total number of animals and the number of survivors in control and obese groups. Of these 21 studies, 12 included one or more experiments studying diet-induced obesity models alone, 5 included one or more experiments studying genetic-induced obesity models alone, and 4 included experiments, some of which studied genetic-induced obesity and others which studied diet-induced obesity. Eighteen and three studies employed mouse and rat models, respectively. Six studies employed a single-strain bacterial challenge, five an LPS challenge, four a cecal ligation and puncture (CLP) challenge, and six an influenza virus challenge.
Figure 1.
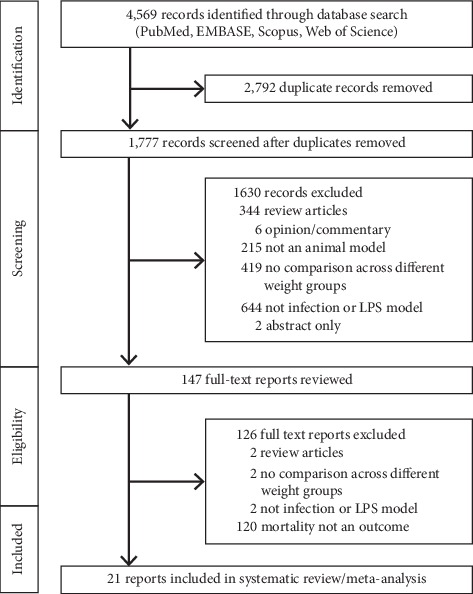
Flow diagram that summarizes the results of the literature search.
Table 1.
Study characteristics.
| Study (author, year) | Exp # | Species | Age (wk) | Sex | Obesity model | Challenge | Observation period | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Type | GT/DC | Type | Strain | Route | Dose | ||||||
| Single-strain bacteria models | |||||||||||
| Hsu, 2007 | 1 | Mouse | 8–12 | F | Gen | ob/ob | S. pneumoniae | n/a | IT | 105 CFU | 10 d |
| 2 | Mouse | 8–12 | M | Gen | ob/ob/lep | S. pneumoniae | n/a | IT | 105 CFU | 10 d | |
| Strandberg, 2009 | 3 | Mouse | 5–7 | M | Gen | ob/ob | S. aureus | n/a | IV | 5 × 107 CFU | 17 d |
| 4 | Mouse | 5–7 | M | DIO | HFD | S. aureus | n/a | IV | 5 × 107 CFU | 17 d | |
| Mancuso, 2014 | 5 | Mouse | 16–18 | F | Gen | CPEF/F | S. pneumoniae | n/a | IT | 5 × 104 CFU | 10 d |
| Svahn, 2015 | 6 | Mouse | 6 | M | DIO | HFDP | S. aureus | n/a | IV | 3.8–4.5 × 107 CFU | 17 d |
| 7 | Mouse | 6 | M | DIO | HFDS | S. aureus | n/a | IV | 3.8–4.5 × 107 CFU | 17 d | |
| 8 | Mouse | 6 | M | DIO | HFDS-HP | S. aureus | n/a | IV | 3.8–4.5 × 107 CFU | 17 d | |
| 9 | Mouse | 6 | M | DIO | HFDS-LP | S. aureus | n/a | IV | 3.8–4.5 × 107 CFU | 17 d | |
| Svahn, 2016 | 10 | Mouse | 6 | M | DIO | HFDS | S. aureus | n/a | IV | 3–5.4 × 107 CFU | 17 d |
| 11 | Mouse | 6 | M | DIO | HFDw6 | S. aureus | n/a | IV | 3–5.4 × 107 CFU | 17 d | |
| Wan, 2016 | 12 | Mouse | 3-4 | M | DIO | HFD | E. coli | n/a | IN | 109 CFU | 96 h |
| 13 | Mouse | 3-4 | M | DIO | HFD | E. coli | n/a | IN | 1010 CFU | 96 h | |
| Cecal ligation and puncture models | |||||||||||
| Tschop, 2010 | 14 | Mouse | 6–10 | M | Gen | ob/ob | Polymicrobial | n/a | IP | n/a | 240 h |
| 15 | Mouse | 6–10 | M | DIO | HFD | Polymicrobial | n/a | IP | n/a | 240 h | |
| 16 | Mouse | 6–10 | M | Gen | ob/ob | Polymicrobial | n/a | IP | n/a | 240 h | |
| Kaplan, 2012 | 17 | Mouse | 6 | M | DIO | HFD | Polymicrobial | n/a | IP | n/a | 30 h |
| Siegl, 2014 | 18 | Mouse | 19 | M | DIO | HFD | Polymicrobial | n/a | IP | n/a | 240 h |
| Kaplan, 2016 | 19 | Mouse | 6 | M | DIO | HFD | Polymicrobial | n/a | IP | n/a | 48 h |
| Lipopolysaccharide | |||||||||||
| Fagioni, 1998 | 20 | Mouse | 5 | F | Gen | ob/ob | E. coli 055:B5 | n/a | IP | 30 μg | 7 d |
| 21 | Mouse | 5 | F | Gen | ob/ob | E. coli 055:B5 | n/a | IP | 100 μg | 7 d | |
| 22 | Mouse | 5 | F | Gen | ob/ob | E. coli 055:B5 | n/a | IP | 300 μg | 7 d | |
| 23 | Mouse | 5 | F | Gen | db/db | E. coli 055:B5 | n/a | IP | 30 μg | 7 d | |
| 24 | Mouse | 5 | F | Gen | db/db | E. coli 055:B5 | n/a | IP | 100 μg | 7 d | |
| 25 | Mouse | 5 | F | Gen | db/db | E. coli 055:B5 | n/a | IP | 300 μg | 7 d | |
| Segersvard, 2003 | 26 | Rat | NR | M | DIO | HFD35 | E. coli | n/a | IP | 2 mg | 72 h |
| 27 | Rat | NR | M | DIO | HFD60 | E. coli | n/a | IP | 2 mg | 72 h | |
| Suto, 2007 | 28 | Mouse | NR | F | Gen | B6AY12w | E. coli 0111-B4 | n/a | IP | 50 μg | 7 d |
| 29 | Mouse | NR | F | Gen | B6Ay12w | E. coli 0111-B4 | n/a | IP | 100 μg | 7 d | |
| 30 | Mouse | NR | F | Gen | B6-ob/ob | E. coli 0111-B4 | n/a | IP | 100 μg | 7 d | |
| 31 | Mouse | NR | F | Gen | B6Ay12w | E. coli 0111-B4 | n/a | IP | 200 μg | 7 d | |
| 32 | Mouse | NR | F | Gen | B6Ay10m | E. coli 0111-B4 | n/a | IP | 50 μg | 7 d | |
| 33 | Mouse | NR | F | Gen | B6Ay10m | E. coli 0111-B4 | n/a | IP | 100 μg | 7 d | |
| Sakai, 2013 | 34 | Rat | 4 | M | DIO | HFD | E. coli 0111-B4 | n/a | IP | 10 mg/kg | 24 h |
| Fujiwara, 2014 | 35 | Rat | 4 | M | DIO | HFD | E. coli 0111-B4 | n/a | IP | 10 mg/kg | 12 h |
| 36 | Rat | 4 | M | DIO | HFD | E. coli 0111-B4 | n/a | IP | 10 mg/kg | 12 h | |
| Single-strain virus models | |||||||||||
| Smith, 2007 | 37 | Mouse | NR | NR | DIO | HFD | H1N1 influenza A | A/PR8 | IN | 2 HG units | 10 d |
| Easterbrook, 2011 | 38 | Mouse | 20 | M | DIO | HFD | H1N1 influenza A | CA/09 | IN | 2.5 × 105 pfu | 15 d |
| 39 | Mouse | 20 | M | DIO | HFD | H1N1 influenza A | NY312 | IN | 2.5 × 105 pfu | 15 d | |
| 40 | Mouse | 20 | M | DIO | HFD | H1N1 influenza A | Sw31 | IN | 50ul-SW31 | 15 d | |
| Milner, 2013 | 41 | Mouse | NR | M | DIO | HFD-UP | H1N1 influenza A | A/PR8 | PO | 5.3 × 105 TCID50 | 13 d |
| 42 | Mouse | NR | M | DIO | HFD-P | H1N1 influenza A | A/PR8 | PO | 5.3 × 105TCID50 | 13 d | |
| Radigan, 2014 | 43 | Mouse | 8–12 | NR | Gen | db/db | H1N1 influenza A | A/WSN/33 | IT | 500 pu | 14 d |
| 44 | Mouse | 8–12 | NR | Gen | db/db | H1N1 influenza A | A/WSN/33 | IT | 1500 pu | 14 d | |
| O'Brien, 2015∗ | 45 | Mouse | 11 | M | DIO | HFD | H1N1 influenza A | CA/09∗ | IN | 1 × 105 TCID50 | 10 d |
| 46 | Mouse | 8 | M | Gen | ob/ob | H1N1 influenza A | CA/09 | IN | 1 × 105 TCID50 | 10 d | |
| 47 | Mouse | 11 | M | DIO | HFD | H3N2 influenza A | HK68 | IN | 6.3 × 105 TCID50 | 10 d | |
| 48 | Mouse | 8 | M | Gen | ob/ob | H3N2 influenza A | HK68 | IN | 6.3 × 105 TCID50 | 10 d | |
| Milner, 2015 | 49 | Mouse | 14–16 | M | DIO | HFD | H1N1 influenza A | CA/09 | IN | 5.8 × 105 | 14 d |
| 50 | Mouse | 14–16 | M | DIO | HFD | H1N1 influenza A | CA/09 | IN | 1.3 × 103 | 14 d | |
| 51 | Mouse | 13–16 | M | Gen | LepRH-/- | H1N1 influenza A | CA/09 | IN | 5.8 × 105 | 14 d | |
| 52 | Mouse | 13–16 | F | Gen | LepRH-/- | H1N1 influenza A | CA/09 | IN | 5.8 × 105 | 14 d | |
B6-ob/ob: leptin-deficient mice; B6Ay10m and 12m: 10- and 12-week-old agouti peptide positive hyperphagic mice; CPE: lack functional carboxypeptidase enzyme; db/db: leptin receptor-deficient mice; DC: diet composition; DIO: diet-induced obesity; Exp: experiment; F: female; Gen: genetic-induced obesity; GT: genotype; HFD35: 35% of the energy from fat; HFD60: 60% of the energy from fat; HFD: high-fat diet; HFD-P: primed with virus; HFD-UP: unprimed; HFDP: polyunsaturated; HFDS-HP: high protein-to-carbohydrate ratio; HFDS-LP: low protein-to-carbohydrate ratio; HFDS: saturated; HFDw6: omega-6 fatty acid rich; IN: intranasal; IP: intraperitoneal; IT: intubation; IV: intravenous; LepRH-/-: lack leptin receptor signaling in hypothalamic neurons; M: male; n/a: not applicable; ob/ob: leptin-deficient mice; PO: oral administration; wk: week. ∗Oseltamivir treatment administered to animals.
Table 2.
Survival, weight, fat mass, and glucose level table.
| Study (author, year) | Exp # | Species | Obesity model | Weight (g) | Fat mass (g) | Glucose (mg/dl) | Control | Obese | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Type | GT/DC | Control | Obese | Control | Obese | Control | Obese | Tot | Surv | Rep | Tot | Surv | |||
| Single-strain bacteria models | |||||||||||||||
| Hsu, 2007 | 1 | Mo | Gen | ob/ob | NR | NR | NR | NR | NR | NR | 10 | 9 | 1 | 17 | 3 |
| 2 | Mo | Gen | ob/ob/lep | NR | NR | NR | NR | NR | NR | 10 | 9 | 1 | 18 | 8 | |
| Strandberg, 2009 | 3 | Mo | Gen | ob/ob | 28.8 ± 0.5 | 39.3 ± 1.1 | NR | NR | NR | NR | 15 | 12 | 0 | 18 | 5 |
| 4 | Mo | DIO | HFD | 28.8 ± 0.5 | 39.3 ± 1.1 | 5.5 ± 0.2 | 17.0 ± 0 | NR | NR | 21 | 18 | 0 | 18 | 8 | |
| Mancuso, 2014 | 5 | Mo | Gen | CPEF/F | 20 ± 0.5 | 47.5 ± 1 | NR | NR∗ | 120 ± 5 | 180 ± 5 | 6 | 2 | 0 | 6 | 3 |
| Svahn, 2015 | 6 | Mo | DIO | HFDP | 30 ± 1 | 35 ± 1 | 7.0 ± 0.5 | 14 ± 0.5 | NR | NR | 20 | 13 | 1 | 20 | 17 |
| 7 | Mo | DIO | HFDS | 30 ± 1 | 45 ± 1 | 7.0 ± 0.5 | 20.0 ± 0.5 | NR | NR | 20 | 13 | 1 | 20 | 4 | |
| 8 | Mo | DIO | HFDS-HP | 30 ± 1 | 45 ± 1 | 14.05 ± 0.5 | 20.0 ± 0.5 | NR | NR | 20 | 16 | 0 | 20 | 8 | |
| 9 | Mo | DIO | HFDS-LP | 30 ± 1 | 45 ± 1 | 14.05 ± 0.5 | 20.0 ± 0.5 | NR | NR | 20 | 17 | 0 | 20 | 8 | |
| Svahn, 2016 | 10 | Mo | DIO | HFDS | 39 ± 1 | 45 ± 1 | 15 ± 0.5 | 20.0 ± 0.5 | NR | NR | 20 | 14 | 1 | 20 | 3 |
| 11 | Mo | DIO | HFDw6 | 39 ± 1 | 42.5 ± 1 | 15 ± 0.5 | 20.0 ± 0.5 | NR | NR | 20 | 14 | 1 | 20 | 8 | |
| Wan, 2016 | 12 | Mo | DIO | HFD | 37.5 ± 1 | 47.5 ± 1 | NR | NR | 225 ± 40 | 350 ± 25 | 18 | 18 | 0 | 18 | 18 |
| 13 | Mo | DIO | HFD | 37.5 ± 1 | 47.5 ± 1 | NR | NR | 225 ± 40 | 350 ± 25 | 18 | 15 | 0 | 18 | 10 | |
| Cecal ligation and puncture models | |||||||||||||||
| Tschop, 2010 | 14 | Mo | Gen | ob/ob | NR | NR | NR | NR | NR | NR | 20 | 4 | 1 | 20 | 0 |
| 15 | Mo | DIO | HFD | NR | NR | NR | NR | NR | NR | 20 | 4 | 1 | 7 | 4 | |
| 16 | Mo | Gen | ob/ob | NR | NR | NR | NR | NR | NR | 11 | 3 | 0 | 11 | 0 | |
| Kaplan, 2012 | 17 | Mo | DIO | HFD | 23.4 ± 0.4 | 25.2 ± 0.4 | MRI∗ | MRI-inc∗ | NR | NR | 12 | 6 | 0 | 12 | 1 |
| Siegl, 2014 | 18 | Mo | DIO | HFD | 27.7 ± 0.2 | 34.4 ± 0.5 | NR | NR | NR | NR | 10 | 1 | 0 | 14 | 10 |
| Kaplan, 2016 | 19 | Mo | DIO | HFD | 27 ± 0.5 | 33 ± 1 | 2 ± 0.5 | 9.6 ± 2 | 165 ± 5 | 190 ± 5 | 12 | 4 | 0 | 12 | 2 |
| Lipopolysaccharide | |||||||||||||||
| Fagioni, 1998 | 20 | Mo | Gen | ob/ob | 20 | 40 | NR | NR | 100 ± 5 | 190 ± 5 | 5 | 5 | 0 | 5 | 3 |
| 21 | Mo | Gen | ob/ob | 20 | 40 | NR | NR | 100 ± 5 | 190 ± 5 | 5 | 5 | 0 | 5 | 2 | |
| 22 | Mo | Gen | ob/ob | 20 | 40 | NR | NR | 100 ± 5 | 190 ± 5 | 5 | 4 | 0 | 5 | 0 | |
| 23 | Mo | Gen | db/db | 20 | 40 | NR | NR | 110 ± 5 | 310 ± 50 | 5 | 4 | 0 | 5 | 4 | |
| 24 | Mo | Gen | db/db | 20 | 40 | NR | NR | 110 ± 5 | 310 ± 50 | 5 | 1 | 0 | 5 | 1 | |
| 25 | Mo | Gen | db/db | 20 | 40 | NR | NR | 110 ± 5 | 310 ± 50 | 5 | 0 | 0 | 5 | 0 | |
| Segersvard, 2003 | 26 | Ra | DIO | HFD35 | 415 ± 9 | 436 ± 8 | 3.4 ± 0.2∗∗ | 5.2 ± 0.2∗∗ | NR | NR | 12 | 10 | 1 | 16 | 9 |
| 27 | Ra | DIO | HFD60 | 415 ± 9 | 466 ± 5 | 3.4 ± 0.2∗∗ | 6.2 ± 0.2∗∗ | NR | NR | 12 | 10 | 1 | 16 | 10 | |
| Suto, 2007 | 28 | Mo | Gen | B6AY12- | 19.6 | 25.1 | NR | NR | NR | NR | 20 | 20 | 0 | 20 | 18 |
| 29 | Mo | Gen | B6Ay12- | 19.6 | 25.1 | NR | NR | NR | NR | 20 | 17 | 1 | 20 | 7 | |
| 30 | Mo | Gen | B6-ob/ob | 19.6 | 52.8 | NR | NR | NR | NR | 20 | 17 | 1 | 24 | 0 | |
| 31 | Mo | Gen | B6Ay12- | 19.6 | 25.1 | NR | NR | NR | NR | 15 | 6 | 0 | 15 | 0 | |
| 32 | Mo | Gen | B6Ay10m | 19.6 | 51 | NR | NR | NR | NR | 6 | 2 | 0 | 5 | 0 | |
| 33 | Mo | Gen | B6Ay10m | 19.6 | 51 | NR | NR | NR | NR | 11 | 2 | 0 | 10 | 0 | |
| Sakai, 2013 | 34 | Ra | DIO | HFD | 289 ± 3.5 | 310.6 ± 1.9 | 3.2 ± 0.3∗∗ | 6.8 ± 0.3∗∗ | 102.0 ± 2.2 | 116 ± 3.7 | 7 | 6 | 0 | 8 | 1 |
| Fujiwara, 2014 | 35 | Ra | DIO | HFD | 275 ± 1 | 294 ± 2.8 | NR | NR | NR | NR | 10 | 9 | 0 | 10 | 9 |
| 36 | Ra | DIO | HFD | 450 ± 1 | 504.4 ± 7.3 | NR | NR | NR | NR | 7 | 5 | 0 | 7 | 1 | |
| Single-strain virus models | |||||||||||||||
| Smith, 2007 | 37 | Mo | DIO | HFD | NR | NR | NR | NR | 86 ± 3 | 108 ± 9 | 18 | 17 | 0 | 18 | 10 |
| Easterbrook, 2011 | 38 | Mo | DIO | HFD | NR | NR | NR | NR | NR | NR | 5 | 5 | 0 | 5 | 1 |
| 39 | Mo | DIO | HFD | NR | NR | NR | NR | NR | NR | 5 | 5 | 0 | 5 | 5 | |
| 40 | Mo | DIO | HFD | NR | NR | NR | NR | NR | NR | 5 | 0 | 0 | 5 | 0 | |
| Milner, 2013 | 41 | Mo | DIO | HFD-UP | 30 ± 1 | 42.5 ± 1 | NR | NR | NR | NR | 4 | 0 | 0 | 4 | 0 |
| 42 | Mo | DIO | HFD-P | 30 ± 1 | 42.5 ± 1 | NR | NR | NR | NR | 46 | 46 | 0 | 46 | 44 | |
| Radigan, 2014 | 43 | Mo | Gen | db/db | 20–25 | 30–35 | NR | NR | NR | NR | 10 | 3 | 0 | 10 | 0 |
| 44 | Mo | Gen | db/db | 20–25 | 30–35 | NR | NR | NR | NR | 10 | 0 | 0 | 10 | 0 | |
| O'Brien, 2015 | 45 | Mo | DIO | HFD | 21 | 35 | NR | NR | NR | NR | 28 | 22 | 1 | 28 | 3 |
| 46 | Mo | Gen | ob/ob | 21 | 53 | NR | NR | NR | NR | 28 | 22 | 1 | 28 | 6 | |
| 47 | Mo | DIO | HFD | 21 | 35 | NR | NR | NR | NR | 28 | 21 | 1 | 28 | 0 | |
| 48 | Mo | Gen | ob/ob | 21 | 53 | NR | NR | NR | NR | 28 | 21 | 1 | 28 | 0 | |
| Milner, 2015 | 49 | Mo | DIO | HFD | 30 ± 1 | 42.5 ± 1 | NR | NR | 70 ± 5 | 105 ± 5 | 21 | 21 | 0 | 21 | 4 |
| 50 | Mo | DIO | HFD | 30 ± 1 | 42.5 ± 1 | NR | NR | 70 ± 5 | 105 ± 5 | 12 | 2 | 0 | 12 | 0 | |
| 51 | Mo | Gen | LepRH-/- | 30 ± 1 | 42.5 ± 1 | NR | NR | NR | NR | 20 | 18 | 0 | 10 | 5 | |
| 52 | Mo | Gen | LepRH-/- | 30 ± 1 | 42.5 ± 1 | NR | NR | NR | NR | 20 | 19 | 0 | 13 | 4 | |
Exp: experiment; db/db: leptin receptor-deficient mice; DC: diet composition; DIO: diet-induced obesity; Gen: genetic; GT: genotype; HFD: high-fat diet; Mo: mouse; NR: not recorded; ob/ob: leptin-deficient mice; Ra: Rat; Rep: repeating control animals; Surv: number of animals surviving; Tot: total number of animals studied; wk: week. ∗Noted increased fat mass on MRI but not quantitated; ∗∗total fat mass weights calculated based on animal total weights (see methods).
3.1. Animal Weights, Fat Masses, and Baseline Blood Glucose Levels
In all 43 experiments providing data, the weight of animals employed was greater in obese compared to control groups (Table 2). In the 9 experiments not providing weight data, either the genotype or diet of obese animals is recognized to produce increased weight. Fat mass and blood glucose were greater in obese animals in the 11 and 12 experiments, respectively, providing data.
3.2. Effect of Obesity on Survival
In each study including more than one experiment in the same obesity model type (diet or genetic), the effects of obesity on the odds ratio of survival (95% CI) (OR) in individual experiments were never qualitatively different and heterogeneity for the combined ORs for these experiments was never significant (p ≥ 0.13 in four studies and p ≥ 0.12 in five studies in diet and genetic models, respectively) (Figures and in the Supplementary Material). The survival results of these experiments were pooled in subsequent analysis. In the four studies that examined both diet and genetic models of obesity, the results of these experiments were also pooled except when the effects of obesity type on survival were compared.
Obesity was associated with reduced survival in 19 studies, and in 11 of these, the reductions were significant (Figure 2). Overall, obesity was associated with a reduced odds ratio (95% CI) of survival (0.21(0.13, 0.35); I2 = 64%, p < 0.01) but with moderate or more heterogeneity. The effect of obesity on the odds ratio of survival (95% CI) (OR) did not differ statistically significantly (p=0.19) comparing studies employing diet versus genetic obesity models (0.25 (0.11, 0.54); I2 = 71%, p < 0.01) versus (0.12 (0.06, 0.25); I2 = 54%, p=0.03) although there was moderate or more heterogeneity across each group of studies (Figure 3). This analysis examined 25 comparisons because 4 studies included experiments in both diet- and genetic-induced models. The effects of obesity on the OR also did not differ statistically (p=0.99) comparing studies including mice versus rats (0.22 (0.12, 0.39); I2 = 68%, p < 0.01 versus 0.22 (0.07, 0.63); I2 = 17%, p=0.30), but there was moderate or more heterogeneity across mouse but not the three rat studies (Figure 4). Finally, the effects of obesity on the OR did not differ significantly (p=0.49) comparing the four types of infectious or septic challenges (Figure 5). In studies with single-strain bacteria (n = 6) or LPS (n = 5) challenges, obesity reduced the ORs consistently (0.21(0.13, 0.34); I2 = 31%, p=0.20 and 0.22(0.13, 0.36); I2 = 0%, p=0.59). In CLP studies (n = 4), obesity reduced survival but not significantly (0.72(0.08, 6.23); I2 = 75%, p < 0.01). With influenza virus studies (n = 6), obesity reduced the OR in all studies (three significantly) but with moderate or more heterogeneity (0.12(0.04, 0.34); I2 = 73%, p < 0.01).
Figure 2.
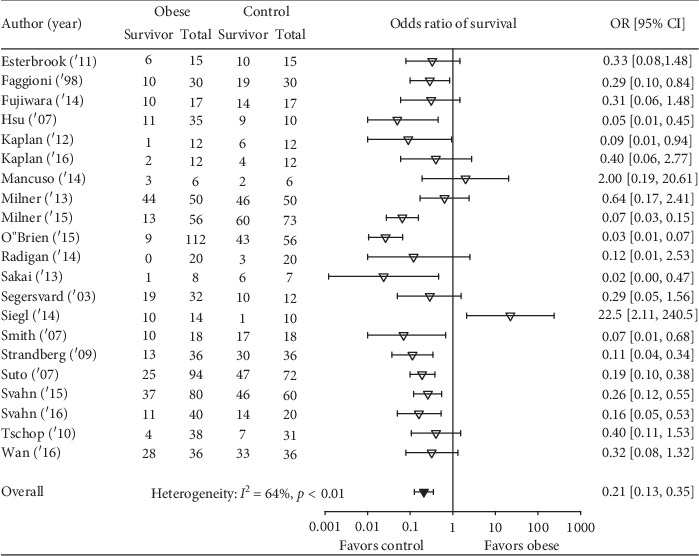
The number of total and surviving animals in obese and control groups for each of the 21 analyzed studies and the effects of obesity on the odds ratios (OR (95% CI)) of survival for each study. Also shown is the OR (95% CI) for the 21 studies and the associated I2 and its level of significance.
Figure 3.
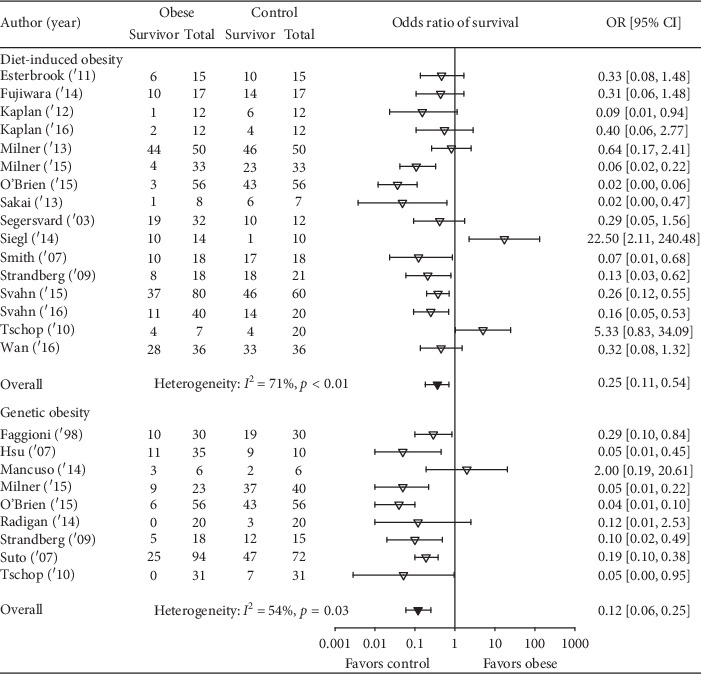
The number of total and surviving animals in obese and control groups for studies employing either a diet-induced obesity model or genetic-induced obesity model and the effects of obesity on the odds ratios (OR (95% CI)) of survival for each study and the overall OR (95% CI) for each type of obesity model and the associated I2 and its level of significance. As described in the results, because four studies examined both diet and genetic obesity models, this figure presents 25 comparisons, 16 with diet and 9 with genetic obesity models. The effects of obesity did not differ statistically significantly comparing the two types of models (p=0.19).
Figure 4.
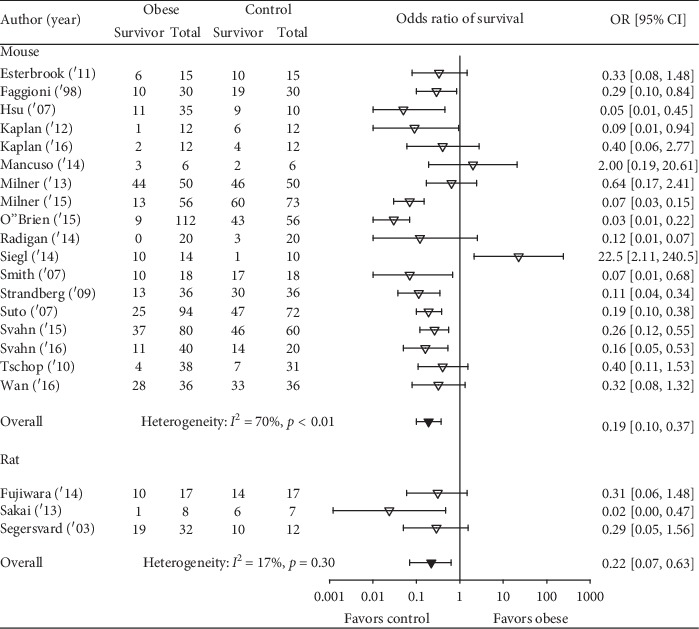
The number of total and surviving animals in obese and control groups for studies examining obesity in either mouse (18 studies) or rat (3 studies) and the effects of obesity on the odds ratios (OR (95% CI)) of survival for each study and the overall OR (95% CI) for each of the two species and the associated I2 and its level of significance. The effects of obesity did not differ statistically significantly comparing the two species (p=0.99).
Figure 5.
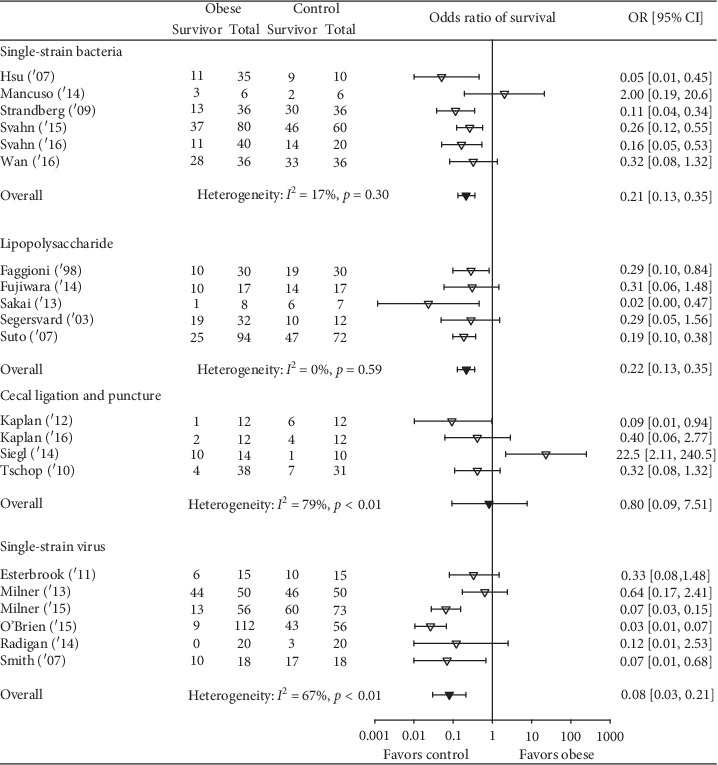
The number of total and surviving animals in obese and control groups for studies employing either a single strain bacterial infection model (n = 6 studies), a lipopolysaccharide model (n = 5 studies), a cecal ligation and puncture (polymicrobial infection) model (n = 4 studies), or a viral infection model (n = 6 studies) and the effects of obesity on the odds ratios (OR (95% CI)) of survival for each study and the overall OR (95% CI) for each type of infectious challenge model and the associated I2 and its level of significance. The effects of obesity did not differ significantly comparing the four model types (p=0.49).
The slope (±SE) for the relationship between the ratio of obese to control animal weights versus the ln(OR) with obesity in individual experiments was consistent, with an increasing detrimental effect of obesity on survival with increasing weight ratio, but this was not significant (−0.81 (0.52), p=0.20). A funnel plot and Egger's regression (p = 0.06) suggested potential publication bias ( in the Supplemental Material). Only one experiment reported treating animals with an antimicrobial agent (Table 1) and none employed cardiopulmonary monitoring and support or glucose control as would be done in patients.
3.3. Effect of Obesity on Measures of Organ Injury and on Microbe, Cytokine, and Leptin Levels for the Different Infectious Challenges
The effect of obesity on measures of organ injury and on microbe, host inflammatory cytokine, and leptin levels was then examined in experiments providing data based on the type of infectious or septic challenge that was employed. For organ injury, with a single bacterial strain challenge, lung wet-to-dry weight ratios (W/D) were significantly increased with obesity at 24 h in one experiment and at 24 h and 96 h in another (Table 3). With CLP challenge, blood urea nitrogen (BUN) and/or alanine aminotransferase (ALT) as measures of kidney and liver injury, respectively, were significantly increased with obesity in two experiments at 6 or 24 h and a histologic lung injury score was increased at 6 h in another. With LPS, aspartate aminotransferase (AST) and liver histology score were increased with obesity at 6 h in one, but lung septal thickness and lung W/D at 6 h were not significantly different in two others. With influenza virus challenge, in six experiments, lung histology scores and/or alveolar lavage protein concentrations were significantly increased with obesity at various days from 3 to 8 days following challenge, but in two experiments, there were no significant changes in these parameters. Across all infectious challenges, in the 12 experiments reporting that obesity significantly affected organ injury, these were all increased.
Table 3.
Effect of obesity compared to controls on parameters of organ injury.
| Author (year) | Exp # | Model | Site of infection | Significant changes in organ injury comparing obese and nonobese groups | Overall effect of obesity on measure of organ injury |
|---|---|---|---|---|---|
| Single-strain bacteria models | |||||
| Wan '16 | 12 | DIO | IN | Lung wet/dry ratio increased at 24 h with obesity | ↑ |
| 13 | DIO | IN | Lung wet/dry ratio increased at 24 and 96 h with obesity | ↑ | |
| Cecal ligation and puncture models | |||||
| Tschop '10 | 14 | Gen | IP | BUN as a marker of renal injury increased at 24 h with obesity ALT as a marker of liver injury increased at 24 h with obesity |
↑ |
| Kaplan '12 | 17 | DIO | IP | Histologic lung injury score increased at 6 h with obesity | ↑ |
| Kaplan '16 | 19 | DIO | IP | ALT as a marker of liver injury increased at 6 h with obesity | ↑ |
| Lipopolysaccharide | |||||
| Sakai '13 | 34 | DIO | IP | AST and liver histology score increased with obesity at 6 h | ↑ |
| Fujiwara ‘14 | 35 | DIO | IP | No significant differences in lung septal thickness or W/D at 6 h | NSD |
| 36 | DIO | IP | No significant differences in lung septal thickness or W/D at 6 h | NSD | |
| Single-strain virus models | |||||
| Smith ‘07 | 37 | DIO | IN | No significant difference in histologic lung injury score | NSD |
| Milner ‘13 | 42 | DIO | PO | Histologic lung injury increased at 5 d and BAL protein increased at 5 d and 6 d with obesity | ↑ |
| Radigan ‘14 | 43 | Gen | IT | BAL protein not significantly different at 4 d | NSD |
| 44 | Gen | IT | BAL protein significantly increased at 4 d with obesity | ↑ | |
| O'Brien | 45 | DIO | IN | Decreased lung epithelial regeneration and increased BAL albumin at 3 d and 6 d with obesity | ↑ |
| O'Brien | 46 | Gen | IN | Decreased lung epithelial regeneration and increased BAL albumin at 3 d and 6 d with obesity | ↑ |
| Milner ‘15 | 49 | DIO | IN | BAL protein and albumin increased at 4 d and BAL protein increased at 8 d with obesity | ↑ |
| 51 | Gen | IN | BAL protein increased at 8 d with obesity | ↑ | |
Exp: experiment; ALT: alanine aminotransferase; AST: aspartate aminotransferase; BAL: bronchoalveolar lavage; BUN: blood urea nitrogen; DIO: diet-induced obesity model; Gen: genetic model of obesity; NSD: no significant difference. ∗Remaining studies did not report organ injury data.
In experiments reporting microbial data, with single-strain bacteria, obesity significantly increased blood and/or tissue bacteria counts at ≥48 h in five experiments and had no significant effect in five others ( in the Supplementary Material). With CLP, obesity significantly increased bacteria counts in blood and tissue at 24 h in one experiment and had no significant effect in another. With influenza, obesity significantly increased lung virus titers in two experiments 4 or 5 days after challenge but had no significant effect in 9 others ( in the Supplemental Material). In summary, in all eight experiments reporting that obesity significantly affected microbial levels, these were all increased.
(for single bacteria strain, CLP, and LPS) and (for influenza virus) in the Supplemental Material also summarize data for cytokine and leptin levels. When the effect of obesity on significant changes in cytokines in experiments was examined across the four types of infectious challenges: for TNF, eight experiments reported significant increases and five decreases; for IL-1b, four reported increases and one a decrease in blood but an increase in lung tissue; for IL-6, five reported increases and one a decrease; for IL-10, two reported increases and one a decrease; and for MIP-2a, three reported increases and none a decrease. Overall, inflammatory cytokines were reported to be significantly increased in 23 cases and only decreased in 8. For leptin levels, five experiments reported significant increases and two decreases.
3.4. Risk of Bias and Study Quality
Randomization, blinding, sample size calculations, and/or numbers of animals withdrawn from the study were not possible or described in studies. Therefore, the risk of bias was unclear or high in all studies examined and study quality was judged to be low ( in the Supplemental Material).
4. Discussion
This systematic review retrieved 21 studies that assessed the effect of a diet or genetic animal obesity model on survival following a single bacterial strain, CLP, LPS, or viral influenza challenge. In 19 studies, obesity was associated with reduced odds ratios of survival, 11 statistically significantly. The negative effect of obesity did not differ significantly comparing diet versus genetic models, mouse versus rat models, or the four types of infectious challenges. However, while there was moderate or high heterogeneity for the effects of obesity across the 21 studies, obesity's adverse effects were consistent across the 6 and 5 studies employing either a single strain of bacteria or an LPS challenge. Across all studies, the negative effect of obesity on survival increased with increasing weight ratio (i.e., obese to control group) but not significantly. Consistent with these adverse survival effects, in the 16 individual experiments presenting quantifiable data, obesity significantly increased some measure of organ injury in 12 experiments and no experiment reported a significant decrease.
Microbial clearance data, while limited, provide one possible basis for the decreased survival and increased organ injury seen with obesity in these models. Of the 23 experiments reporting data, obesity significantly increased blood or tissue bacteria or virus counts in 8 experiments and no experiment reported a decrease. Obesity-related insulin resistance and hyperglycemia or other changes may have impaired microbial clearance and worsened survival and organ injury [31–39]. Blood and tissue inflammatory cytokine levels including either TNF-α, IL-1b, IL-6, IL-10, or MIP-2a were also increased with obesity in 22 of 30 cases and may have contributed to inflammatory organ injury and worsened survival. Adipose tissue (fat mass), a source of inflammatory cytokine production, was increased in all reports presenting data, and obesity has previously been associated with a proinflammatory state [40–46].
Different from these findings in animal obesity models, two recent systematic reviews of observational clinical studies and a retrospective analysis of a large patient database found that obese body mass indices (BMI) appeared to increase survival in septic patients [13–15]. Preclinical and clinical results may differ for several reasons. Patients with infection and sepsis receive antimicrobial agents and other measures to clear the infectious nidus. Those with cardiopulmonary instability are aggressively supported. Blood glucose levels are also controlled clinically to counter hyperglycemia's adverse effects on microbial clearance. Such supportive measures may negate obesity's detrimental effects on microbial clearance or inflammatory mediator release while unmasking potentially protective effects. In contrast, only one of the animal experiments analyzed here administered an antimicrobial agent and none employed glucose control or organ support. Also, patients typically receive chronic treatment, such as glucose, cholesterol, and triglyceride control that would negate the long-term effects of obesity on the vascular dysfunction potentially worsening sepsis outcomes.
However, improved survival with obesity in clinical sepsis studies may be confounded by several factors. For comparably severe infection, obese patients may be admitted to the ICU more frequently where treatment is more comprehensive than in the non-ICU setting [13, 14]. Obese patients may also present with infection more amenable to treatment than nonobese patients. Antibiotics or hemodynamic support may favor obese patients in the acute period. For example, weight-based antibiotic dosing produces higher antibiotic concentrations at infection sites in obese patients while non-weight-based fluid administration could reduce the risk of fluid overload. Finally, the timing and technique for measurement of BMI may be unreliable in some clinical sepsis studies.
Different from clinical sepsis studies, but more consistent with the present findings in animal models, two systematic reviews and meta-analyses of influenza virus infection, with 6 studies in one and 22 in the other, suggested that obese BMIs are associated with worsened combined outcomes including mortality and the need for ICU admission and organ system support [16, 17]. While these analyses' combined endpoints confound their interpretation, in one which reported survival alone across three studies, obesity increased mortality significantly.
Two important questions these preclinical studies do not address are the following. First, how does obesity impact the outcome of septic patients who survive their initial course of acute infection and inflammation but progress to later sepsis with more chronic inflammation? Inflammation in these patients is characterized by both pro- and anti-inflammatory host responses. These patients frequently require prolonged invasive intensive care unit support, and maladaptive anti-inflammatory responses are believed to predispose them to secondary infection. However, clinical studies have not yet attempted to differentiate the impact of obesity on outcomes in acute versus chronically ill septic patients. Furthermore, animal models needed to test this question, although necessary, would be complex, requiring both an initial and follow-up septic challenges and prolonged observation. The second question has to do with whether comorbidities impact the outcome of sepsis and presumably then, whether obesity's impact on outcome in septic patients is influenced by these comorbidities. Comorbidities such as heart, lung, and kidney disease are all known to worsen the outcome from sepsis. Interestingly, though, a recent retrospective analysis of a large patient database which noted a protective effect of obesity in septic patients did not find that such comorbidities influenced these effects [15].
The findings from this analysis in combination with data from clinical studies point to several questions that should be addressed in future preclinical studies. Would the use of antimicrobial agents blunt or reverse the harmful survival effects of obesity in preclinical models? Related to that question, does dosing antimicrobial therapy based on weight increase its effectiveness on either microbial clearance or survival? Would blood glucose control alter the effect of obesity on microbial clearance and outcome in preclinical obesity models? Similarly, would cardiopulmonary support with or without antimicrobial therapy and glucose control reverse the harmful effects of obesity in preclinical models?
The present study has limitations. Organ injury and microbe and cytokine data were not provided in many reports which prevent firm conclusions regarding the basis for obesity's adverse effects in these preclinical studies. Weights were not reported for 9 studies and only 11 studies reported animal fat masses, although the obesity models employed are recognized to produce increases in each. Most studies did not include baseline data prior to the start of obese diets or infectious challenge. The literature search was conducted through January 2017, but it retrieved a relatively large number of reports included in the analysis and the overall survival findings were very similar across studies. Finally, the risk of bias was unclear or high across studies and study quality was judged to be low.
Determining whether obesity improves or worsens survival in critically ill patients with infection or sepsis is important. While the animal studies examined here support an adverse effect, some clinical data suggest the opposite. However as noted, in almost all cases, animal models lacked the types of support (e.g., antimicrobial therapy or glucose control) patients receive. If obesity is indeed protective during sepsis, understanding these beneficial effects might lead to new therapeutic approaches. But if obesity is detrimental for the acutely infected patient, then developing therapeutic approaches to counteract those harmful effects are necessary. These preclinical and clinical experiences together emphasize the need for prospective clinical studies that can accurately assess obesity's impact on survival during severe infection.
5. Conclusions
The results of the preclinical studies examined here are not consistent with the reported protective effects obesity has in retrospective, observational studies of patients with bacterial infection and sepsis but are consistent with obesity's reported harmful effects during influenza. These preclinical and clinical studies together emphasize the need for prospective studies in patients accurately assessing obesity's impact on survival during severe infection whether from a bacterial or viral influenza source.
Acknowledgments
This work was supported by the Intramural Program of the NIH, Clinical Center, Critical Care Medicine Department. The authors thank Ms. Kelly Byrne for editorial assistance with this manuscript.
Abbreviations
- ALT:
Alanine aminotransferase
- AST:
Aspartate aminotransferase
- BMI:
Body mass indices
- BUN:
Blood urea nitrogen
- CLP:
Cecal ligation and puncture
- CI:
Confidence interval
- ICU:
Intensive care units
- IL:
Interleukin
- LPS:
Lipopolysaccharide
- MIP-2a:
Macrophage inflammatory protein 2-alpha
- OR:
Odds ratio of survival
- PRISMA:
Preferred Reporting Items for Systematic Reviews and Meta-Analyses
- SYRCLE:
Systemic Review Center for Laboratory animal Experimentation
- TNF:
Tumor necrosis factor
- W/D:
Wet-to-dry weight ratios.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
Authors' Contributions
WX extracted, analyzed, and interpreted data and helped write and edit the manuscript. DP helped design the study and conduct the literature search and edited the manuscript. JS performed statistical analysis and edited the manuscript. JW designed and conducted the literature search and edited the manuscript. XC performed data analysis and edited the manuscript. PQE helped design the study, extracted and analyzed data, and helped write and edit the paper.
Supplementary Materials
Supplementary Materials: search strategies and supplemental references showing the reports included in the analysis. Figure S1: this figure shows for each report including more than one experiment in the same diet-induced obesity model type the effects of obesity on the odds ratio of survival (95% CI) (OR) in individual experiments as well as the overall OR and its I2 and level of significance when these experiments were combined as described in the methods and results. The results of experiments in these comparisons were never qualitatively different and heterogeneity for overall ORs was not significant (p ≥ 0.13 in four reports with experiments which could be combined). Figure S2: this figure shows for each report including more than one experiment in the same genetic induced obesity model type the effects of obesity on the odds ratio of survival (95% CI) (OR) in individual experiments as well as the overall OR and its I2 and level of significance when these experiments were combined as described in the Methods and Results. The results of experiments in these comparisons were never qualitatively different and heterogeneity for overall ORs was not significant (p ≥ 0.12 in the five reports with experiments which could be combined). Figure S3: the funnel plot and Egger's statistic for the 25 studies described in the results. Table S1: effect of obesity compared to controls on bacteria counts and cytokine and leptin levels in bacteria-infected or LPS-challenged animals. Table S2: effect of obesity compared to controls on viral titer and cytokine and leptin levels in virus-infected animals. Table S3: summary of risk of bias scoring.
References
- 1.Ng M., Fleming T., Robinson M., et al. Global, regional, and national prevalence of overweight and obesity in children and adults during 1980–2013: a systematic analysis for the global burden of disease study 2013. Lancet (London, England) 2014;384(384):766–781. doi: 10.1016/S0140-6736(14)60460-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang Y., Beydoun M. A. The obesity epidemic in the United States gender, age, socioeconomic, racial/ethnic, and geographic characteristics: a systematic review and meta-regression analysis. Epidemiologic Reviews. 2007;29(1):6–28. doi: 10.1093/epirev/mxm007. [DOI] [PubMed] [Google Scholar]
- 3.Yang L., Colditz G. A. Prevalence of overweight and obesity in the United States, 2007–2012. JAMA Internal Medicine. 2015;175(8):1412–1413. doi: 10.1001/jamainternmed.2015.2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Flegal K. M., Kit B. K., Orpana H., Graubard B. I. Association of all-cause mortality with overweight and obesity using standard body mass index categories. JAMA. 2013;309(1):71–82. doi: 10.1001/jama.2012.113905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McGee D. L. Body mass index and mortality: a meta-analysis based on person-level data from twenty-six observational studies. Annals of Epidemiology. 2005;15(2):87–97. doi: 10.1016/j.annepidem.2004.05.012. [DOI] [PubMed] [Google Scholar]
- 6.Akinnusi M. E., Pineda L. A., El Solh A. A. Effect of obesity on intensive care morbidity and mortality: a meta-analysis. Critical Care Medicine. 2008;36(1):151–158. doi: 10.1097/01.ccm.0000297885.60037.6e. [DOI] [PubMed] [Google Scholar]
- 7.Dennis D. M., Bharat C., Paterson T. Prevalence of obesity and the effect on length of mechanical ventilation and length of stay in intensive care patients: a single site observational study. Australian Critical Care. 2017;30(3):145–150. doi: 10.1016/j.aucc.2016.07.003. [DOI] [PubMed] [Google Scholar]
- 8.Oliveros H., Villamor E. Obesity and mortality in critically ill adults: a systematic review and meta-analysis. Obesity. 2008;16(3):515–521. doi: 10.1038/oby.2007.102. [DOI] [PubMed] [Google Scholar]
- 9.Hogue C. W., Jr., Stearns J. D., Colantuoni E., et al. The impact of obesity on outcomes after critical illness: a meta-analysis. Intensive Care Medicine. 2009;35(7):1152–1170. doi: 10.1007/s00134-009-1424-5. [DOI] [PubMed] [Google Scholar]
- 10.Rahman A., Stapleton R. D., Heyland D. K. Not all critically ill obese patients are the same: the influence of prior comorbidities. ISRN Obesity. 2012;2012:7. doi: 10.5402/2012/743978.743978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Trivedi V., Bavishi C., Jean R. Impact of obesity on sepsis mortality: a systematic review. Journal of Critical Care. 2015;30(3):518–524. doi: 10.1016/j.jcrc.2014.12.007. [DOI] [PubMed] [Google Scholar]
- 12.Huttunen R., Laine J., Lumio J., Vuento R., Syrjanen J. Obesity and smoking are factors associated with poor prognosis in patients with bacteraemia. BMC Infectious Diseases. 2007;7(1):p. 13. doi: 10.1186/1471-2334-7-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pepper D. J., Sun J., Welsh J., Cui X., Suffredini A. F., Eichacker P. Q. Increased body mass index and adjusted mortality in ICU patients with sepsis or septic shock: a systematic review and meta-analysis. Critical Care. 2016;20(1):p. 181. doi: 10.1186/s13054-016-1360-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang S., Liu X., Chen Q., Liu C., Huang C., Fang X. The role of increased body mass index in outcomes of sepsis: a systematic review and meta-analysis. BMC Anesthesiology. 2017;17(1):p. 118. doi: 10.1186/s12871-017-0405-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pepper D. J., Demirkale C. Y., Sun J., et al. Does obesity protect against death in sepsis? A retrospective cohort study of 55,038 adult patients. Critical Care Medicine. 2019;47(5):643–650. doi: 10.1097/ccm.0000000000003692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun Y., Wang Q., Yang G., Lin C., Zhang Y., Yang P. Weight and prognosis for influenza A(H1N1)pdm09 infection during the pandemic period between 2009 and 2011: a systematic review of observational studies with meta-analysis. Infectious Diseases (London) 2016;48(11-12):813–822. doi: 10.1080/23744235.2016.1201721. [DOI] [PubMed] [Google Scholar]
- 17.Fezeu L., Julia C., Henegar A., et al. Obesity is associated with higher risk of intensive care unit admission and death in influenza A (H1N1) patients: a systematic review and meta-analysis. Obesity Reviews. 2011;12(8):653–659. doi: 10.1111/j.1467-789x.2011.00864.x. [DOI] [PubMed] [Google Scholar]
- 18.Kitahara C. M., Flint A. J., Berrington de Gonzalez A., et al. Association between class III obesity (BMI of 40–59 kg/m2) and mortality: a pooled analysis of 20 prospective studies. PLoS Medicine. 2014;11(7) doi: 10.1371/journal.pmed.1001673.e1001673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Speakman J., Hambly C., Mitchell S., Król E. The contribution of animal models to the study of obesity. Laboratory Animals. 2008;42(4):413–432. doi: 10.1258/la.2007.006067. [DOI] [PubMed] [Google Scholar]
- 20.Mittwede P. N., Clemmer J. S., Bergin P. F., Xiang L. Obesity and critical illness. Shock. 2016;45(4):349–358. doi: 10.1097/shk.0000000000000512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pini M., Castellanos K. J., Rhodes D. H., Fantuzzi G. Obesity and IL-6 interact in modulating the response to endotoxemia in mice. Cytokine. 2013;61(1):71–77. doi: 10.1016/j.cyto.2012.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khan M., Patrick A. L., Fox-Robichaud A. E. Development of a murine model of early sepsis in diet-induced obesity. BioMed Research International. 2014;2014:11. doi: 10.1155/2014/719853.719853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Petronilho F., Giustina A. D., Nascimento D. Z., et al. Obesity exacerbates sepsis-induced oxidative damage in organs. Inflammation. 2016;39(6):2062–2071. doi: 10.1007/s10753-016-0444-x. [DOI] [PubMed] [Google Scholar]
- 24.Stroup D. F., Berlin J. A., Morton S. C., et al. Meta-analysis of observational studies in epidemiology. JAMA. 2000;283(15):2008–2012. doi: 10.1001/jama.283.15.2008. [DOI] [PubMed] [Google Scholar]
- 25.Hooijmans C. R., Rovers M. M., de Vries R. B., Leenaars M., Ritskes-Hoitinga M., Langendam M. W. SYRCLE’s risk of bias tool for animal studies. BMC Medical Research Methodology. 2014;14:p. 43. doi: 10.1186/1471-2288-14-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wever K. E., Geessink F. J., Brouwer M. A. E., Tillema A., Ritskes-Hoitinga M. A systematic review of discomfort due to toe or ear clipping in laboratory rodents. Laboratory Animals. 2017;51(6):583–600. doi: 10.1177/0023677217705912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.DerSimonian R., Laird N. Meta-analysis in clinical trials. Controlled Clinical Trials. 1986;7(3):177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 28.Higgins J. P., Thompson S. G. Quantifying heterogeneity in a meta-analysis. Statistics in Medicine. 2002;21(11):1539–1558. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- 29. R Foundation for Statistical Computing, https://www.r-project.org/
- 30. Meta: General Package for Meta-Analysis, R Package Version, https://CRAN.R-project.org/package=meta.
- 31.Turina M., Fry D. E., Polk H. C., Jr. Acute hyperglycemia and the innate immune system: clinical, cellular, and molecular aspects. Critical Care Medicine. 2005;33(7):1624–1633. doi: 10.1097/01.ccm.0000170106.61978.d8. [DOI] [PubMed] [Google Scholar]
- 32.Jafar N., Edriss H., Nugent K. The effect of short-term hyperglycemia on the innate immune system. The American Journal of the Medical Sciences. 2016;351(2):201–211. doi: 10.1016/j.amjms.2015.11.011. [DOI] [PubMed] [Google Scholar]
- 33.Hair P. S., Echague C. G., Rohn R. D., Krishna N. K., Nyalwidhe J. O., Cunnion K. M. Hyperglycemic conditions inhibit C3-mediated immunologic control of Staphylococcus aureus. Journal of Translational Medicine. 2012;10:p. 35. doi: 10.1186/1479-5876-10-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Javid A., Zlotnikov N., Petrosova H., et al. Hyperglycemia impairs neutrophil-mediated bacterial clearance in mice infected with the lyme disease pathogen. PLoS One. 2016;11(6) doi: 10.1371/journal.pone.0158019.e0158019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rueda A. M., Ormond M., Gore M., Matloobi M., Giordano T. P., Musher D. M. Hyperglycemia in diabetics and non-diabetics: effect on the risk for and severity of pneumococcal pneumonia. Journal of Infection. 2010;60(2):99–105. doi: 10.1016/j.jinf.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 36.Alexiewicz J. M., Kumar D., Smogorzewski M., Klin M., Massry S. G. Polymorphonuclear leukocytes in non-insulin-dependent diabetes mellitus: abnormalities in metabolism and function. Annals of Internal Medicine. 1995;123(12):919–924. doi: 10.7326/0003-4819-123-12-199512150-00004. [DOI] [PubMed] [Google Scholar]
- 37.Kumar Nathella P., Babu S. Influence of diabetes mellitus on immunity to human tuberculosis. Immunology. 2017;152(1):13–24. doi: 10.1111/imm.12762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hulme K. D., Gallo L. A., Short K. R. Influenza virus and glycemic variability in diabetes: a killer combination? Frontiers in Microbiology. 2017;8:p. 861. doi: 10.3389/fmicb.2017.00861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kumar M., Roe K., Nerurkar P. V., et al. Impaired virus clearance, compromised immune response and increased mortality in type 2 diabetic mice infected with West Nile virus. PLoS One. 2012;7(8) doi: 10.1371/journal.pone.0044682.e44682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ghigliotti G., Barisione C., Garibaldi S., et al. Adipose tissue immune response: novel triggers and consequences for chronic inflammatory conditions. Inflammation. 2014;37(4):1337–1353. doi: 10.1007/s10753-014-9914-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shao L., Feng B., Zhang Y., Zhou H., Ji W., Min W. The role of adipose-derived inflammatory cytokines in type 1 diabetes. Adipocyte. 2016;5(3):270–274. doi: 10.1080/21623945.2016.1162358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Majdoubi A., Kishta O. A., Thibodeau J. Role of antigen presentation in the production of pro-inflammatory cytokines in obese adipose tissue. Cytokine. 2016;82:112–121. doi: 10.1016/j.cyto.2016.01.023. [DOI] [PubMed] [Google Scholar]
- 43.Frasca D., Blomberg B. B., Paganelli R. Aging, obesity, and inflammatory age-related diseases. Frontiers in Immunology. 2017;8:p. 1745. doi: 10.3389/fimmu.2017.01745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rogero M. M., Calder P. C. Obesity, inflammation, toll-like receptor 4 and fatty acids. Nutrients. 2018;10(4) doi: 10.3390/nu10040432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hube F., Birgel M., Lee Y. M., Hauner H. Expression pattern of tumour necrosis factor receptors in subcutaneous and omental human adipose tissue: role of obesity and non-insulin-dependent diabetes mellitus. European Journal of Clinical Investigation. 1999;29(8):672–678. doi: 10.1046/j.1365-2362.1999.00520.x. [DOI] [PubMed] [Google Scholar]
- 46.Winkler G., Kiss S., Keszthelyi L., et al. Expression of tumor necrosis factor (TNF)-alpha protein in the subcutaneous and visceral adipose tissue in correlation with adipocyte cell volume, serum TNF-alpha, soluble serum TNF-receptor-2 concentrations and C-peptide level. European Journal of Endocrinology. 2003;149(2):129–135. doi: 10.1530/eje.0.1490129. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Materials: search strategies and supplemental references showing the reports included in the analysis. Figure S1: this figure shows for each report including more than one experiment in the same diet-induced obesity model type the effects of obesity on the odds ratio of survival (95% CI) (OR) in individual experiments as well as the overall OR and its I2 and level of significance when these experiments were combined as described in the methods and results. The results of experiments in these comparisons were never qualitatively different and heterogeneity for overall ORs was not significant (p ≥ 0.13 in four reports with experiments which could be combined). Figure S2: this figure shows for each report including more than one experiment in the same genetic induced obesity model type the effects of obesity on the odds ratio of survival (95% CI) (OR) in individual experiments as well as the overall OR and its I2 and level of significance when these experiments were combined as described in the Methods and Results. The results of experiments in these comparisons were never qualitatively different and heterogeneity for overall ORs was not significant (p ≥ 0.12 in the five reports with experiments which could be combined). Figure S3: the funnel plot and Egger's statistic for the 25 studies described in the results. Table S1: effect of obesity compared to controls on bacteria counts and cytokine and leptin levels in bacteria-infected or LPS-challenged animals. Table S2: effect of obesity compared to controls on viral titer and cytokine and leptin levels in virus-infected animals. Table S3: summary of risk of bias scoring.


