Abstract
Pyruvate kinase M2 (PKM2) links metabolic and inflammatory dysfunction in atherosclerotic coronary artery disease; however, its role in oxidized LDL (Ox-LDL)-induced macrophage foam cell formation and inflammation is unknown and therefore was studied. In recombinant mouse granulocyte-macrophage colony-stimulating factor-differentiated murine bone marrow-derived macrophages, early (1–6 h) Ox-LDL treatment induced PKM2 tyrosine 105 phosphorylation and promotes its nuclear localization. PKM2 regulates aerobic glycolysis and inflammation because PKM2 shRNA or Shikonin abrogated Ox-LDL-induced hypoxia-inducible factor-1α target genes lactate dehydrogenase, glucose transporter member 1, interleukin 1β (IL-1β) mRNA expression, lactate, and secretory IL-1β production. PKM2 inhibition significantly increased Ox-LDL-induced ABCA1 and ABCG1 protein expression and NBD-cholesterol efflux to apoA1 and HDL. PKM2 shRNA significantly inhibited Ox-LDL-induced CD36, FASN protein expression, DiI-Ox-LDL binding and uptake, and cellular total cholesterol, free cholesterol, and cholesteryl ester content. Therefore, PKM2 regulates lipid uptake and efflux. DASA-58, a PKM2 activator, downregulated LXR-α, ABCA1, and ABCG1, and augmented FASN and CD36 protein expression. Peritoneal macrophages showed similar results. Ox-LDL induced PKM2- SREBP-1 interaction and FASN expression in a PKM2-dependent manner. Therefore, this study suggests a role for PKM2 in Ox-LDL-induced aerobic glycolysis, inflammation, and macrophage foam cell formation.
Keywords: lipid, cholesterol metabolism, oxidized low density lipoprotein
Oxidized LDL (Ox-LDL) promotes atherosclerosis by inducing macrophage foam cell formation and sterile inflammation (1–4). Macrophages treated with Ox-LDL accumulate modified lipids and generate an inflammatory response that involves interleukin 1β (IL-1β) production (5–7). Circulating monocytes derived from symptomatic atherosclerotic patients have a pro-inflammatory phenotype and elevated levels of glycolytic enzymes (8). Pyruvate kinase muscle isozyme M2 (PKM2) is a rate-limiting glycolytic enzyme that catalyzes the transfer of the phosphoryl group from phosphoenolpyruvate to ADP, thus generating ATP and pyruvate (9). A recent study suggests PKM2 as a link between metabolic and inflammatory dysfunction in atherosclerotic coronary artery disease (10). PKM2 is generally overexpressed in activated immune and cancer cells and participates in inflammation and the Warburg effect (also known as aerobic glycolysis) (11–13). Posttranslational modification of PKM2 by phosphorylation at tyrosine 105 promotes its nuclear translocation and inhibits PKM2 tetramer formation (14). In LPS-activated macrophages and cancerous cells, the monomer or dimer form of PKM2 translocates to the nucleus where it interacts with hypoxia-inducible factor-1α (Hif-1α) and induces expression of pro-inflammatory cytokine IL-1β and pro-glycolytic enzymes (15, 16). Ox-LDL-induced macrophage inflammation also involves Hif-1α (17).
Macrophage foam cell formation can be regulated at several levels, such as lipid uptake, biosynthesis, and efflux (18). PPARs and LXRs play an essential role in macrophage foam cell formation by affecting lipid uptake and efflux (1, 19). PPARγ regulates Ox-LDL-induced scavenger receptor CD36 (a class B scavenger receptor) expression and thereby affects macrophage lipid uptake and foam cell formation (20). PPARγ also regulates PKM2 expression in the fatty liver (21). LXRs regulate transcription of ABCA1 and ABCG1, which are involved in cholesterol efflux to apoA1 and HDL, respectively (22, 23). Endogenous lipid biosynthesis can also promote foam cell formation (24). SREBPs are transcription factors that regulate expression of lipogenic genes (25) and can modulate macrophage cholesterol biosynthesis (26). In hepatocellular carcinoma, PKM2 interaction with nuclear SREBP-1a leads to SREBP-1a stabilization and increased transcription of lipogenic genes and lipid biosynthesis (27). FASN, a target gene of SREBP-1a (28), can regulate macrophage foam cell formation by affecting lipid synthesis, uptake, and efflux (24). FASN-null macrophages exhibit decreased foam cell formation compared with the wild-type. The nuclear receptor, LXR-α, and its downstream target, ABCA1 expression, were increased, while the expression of CD36 was diminished in the FASN knockout macrophages (24).
The present study demonstrates the role of PKM2 in Ox-LDL-induced aerobic glycolysis, inflammation, and macrophage foam cell formation and deciphers the underlying mechanisms.
MATERIALS AND METHODS
Reagents
PKM2 activator DASA-58, 3-dodecanoyl-NBD cholesterol, an l-lactate assay kit, and GW9662 were procured from Cayman Chemical (Ann Arbor, MI). Shikonin, oligomycin, carbonyl cyanide-p-trifluoromethoxy-phenylhydrazone, rotenone, 2-deoxy-D-glucose, antimycin-A, disuccinimidyl suberate, PKM2 shRNA, apoA1 from human plasma, anti α-tubulin, anti β-actin, anti histone H3, and all other fine chemicals were purchased from Sigma (St. Louis, MO). PKM2, phospho-PKM2 (tyr105), and Hif-1α antibodies were from Cell Signaling Technologies (Danvers, MA). FASN and histone H1 antibodies were procured from Santa Cruz Biotechnology Inc. Antibody against ABCA1 was from Abcam (Cambridge, MA). ABCG1, CD36, and scavenger receptor class B type 1 (SRB1) antibodies were from Novus Biologicals (Littleton, CO). SREBP-1 antibody and the IL-1β murine ELISA kit were from BD Bioscience (San Jose, CA). TRIzol reagent and RevertAid cDNA synthesis kit were from Invitrogen. Recombinant mouse granulocyte-macrophage colony-stimulating factor was purchased from Peprotech. The 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanine perchlorate (DiI) dye and Amplex Red cholesterol assay kit were purchased from Thermo Fisher Scientific. nProtein A Sepharose™ 4 Fast Flow was from GE Healthcare Biosciences AB (Uppsala, Sweden). Seahorse XF assay media (102365-100) and Seahorse XF base media (102353-100) were from Agilent Technologies.
Cell culture and treatment
Bone marrow-derived macrophages (BMDMs) were isolated from the tibia and femur of 4- to 8-week-old C57BL/6J mice. All animal experiments were carried out with prior ethical approval by the Institutional Animal Ethical Committee, Council for Scientific and Industrial Research Central Drug Research Institute, as per the guidelines of the Committee for the Purpose of Control and Supervision of Experiments on Animals. Briefly, the bones were flushed with ice-cold PBS, followed by incubation with red blood cell lysis solution for 5 min to lyse the erythrocytes. A single-cell suspension was obtained by filtering through a 70 μm cell strainer. The cells were then plated in RPMI medium (Gibco) supplemented with 10% FBS (Gibco), 100 units/ml penicillin, 100 μg/ml streptomycin, 2 mM l-glutamine (Gibco), and 50 ng/ml recombinant mouse granulocyte-macrophage colony-stimulating factor at 37°C in a 5% CO2 incubator for 7 days as described earlier (29). BMDMs were incubated with PKM2 small molecule inhibitor Shikonin (1 μM) for 1 h followed by stimulation with Ox-LDL (40 μg/ml) for 24 h or PKM2 activator DASA-58 (50 μM) for 24 h. Peritoneal macrophages were isolated from 4- to 8-week-old C57BL/6J mice. One-half milliliter of 4% thioglycollate was injected into the mouse peritoneal cavity and peritoneal lavage was collected 3 days after the injection (30). Peritoneal fluid was collected in sterile PBS and centrifuged at 300 g for 6 min. Peritoneal cells were washed twice with PBS and then plated in 6-well plates. After 4 h incubation, all nonadherent cells were removed and the wells were washed with PBS. Adhered macrophages were cultured in DMEM medium supplemented with 10% FBS, 100 units/ml penicillin, 100 μg/ml streptomycin, and 2 mM l-glutamine. Adhered macrophages were used for future experiments (30).
Lipoprotein isolation, modification, and characterization
Human LDL (d = 1.019–1.063 g/ml) was isolated from the plasma of healthy volunteers by density gradient ultracentrifugation (31, 32). All procedures were approved by the Institutional Human Ethics Committee and were in agreement with Declaration of Helsinki principles. Informed consent was obtained from the human subjects or their representatives. The native LDL was dialyzed against PBS at 4°C for 12 h. The protein concentration of LDL was measured using a Pierce BCA protein assay kit (Thermo Fisher Scientific). Oxidation of native LDL (200 μg/ml) was performed in PBS by exposure to 5 μM of CuSO4 at 37°C for 24 h. The oxidation was terminated by the addition of sodium EDTA (0.2 mM) and butylated hydroxytoluene (50 μM). The extent of oxidation was monitored by calculating the thiobarbituric acid-reactive substances and differential electrophoretic mobility in 0.5% agarose gel. HDL (d = 1.063–1.210 g/ml) was also isolated after collecting the LDL fraction as described previously (33).
Cellular cholesterol measurement and Oil Red O staining
Total and esterified cholesterol content was measured by the Amplex Red cholesterol assay kit (Thermo Fisher Scientific) according to manufacturer’s instructions. Extraction of lipids was performed with the hexane and isopropanol method as described previously (34, 35). Briefly, macrophages were washed with ice-cold PBS, and cellular cholesterol was extracted with 800 μl of ice-cold hexane:isopropanol (3:2, v/v) with butylated hydroxytoluene (50 μg/ml) added in hexane and incubated for 30 min on ice with occasional gentle mixing. The mixture was taken out and centrifuged at 2,000 g for 5 min and the upper layer containing the organic phase was removed and kept. The lower aqueous phase was re-extracted with 300 μl of hexane:methanol (3:2, v/v) for 30 min on ice and again centrifuged at 2,000 g for 5 min. The organic phase was pooled and dried under a nitrogen stream. After cholesterol extraction, the cells were treated with NaOH (0.2 M) and SDS (0.1%, w/v), and subsequently protein concentrations were measured using a BCA assay kit. Total cholesterol (TC) and free cholesterol (FC) were measured by incubating 50 μl of extracted cholesterol diluted in reaction buffer with 50 μl of working reagent containing Amplex Red, cholesterol oxidase, and horseradish peroxidase without cholesterol esterase (TC) or with cholesterol esterase (FC) for 30 min at 37°C. Subsequently, fluorescence was measured on a fluorescence microplate reader (Omega, BMG Labtech, Germany) with excitation and emission wavelengths of 540 and 590 nm, respectively. Cholesteryl ester (CE) content was measured after subtracting FC from TC. A standard curve of cholesterol reference (0–8 μg/ml) was used to calculate TC, FC, and CE levels after normalization with protein.
Macrophages were fixed with 4% PFA followed by staining with 0.5% Oil Red O as described previously (36). Oil Red O-stained macrophages were visualized and images were acquired using a Leica QWin image system (Leica Microsystems, Germany).
Lipoprotein-deficient serum preparation and DiI labeling of Ox-LDL
Lipoprotein-deficient serum (LPDS) was prepared by density gradient centrifugation (37). The density of the human plasma was adjusted to 1.21 g/ml by the addition of solid KBr and NaCl. The subsequent solution was ultracentrifuged in an SW 41 Ti rotor (Beckman Coulter) at 220,000 g at 10°C for 48 h. The rich upper layer of lipoproteins was removed and the bottom portion was taken as the LPDS. LPDS was dialyzed extensively against 1× PBS (pH 7.4) for 48 h and then filtered, sterilized, and stored at −80°C. The lipid part of Ox-LDL was fluorescently labeled with a lipophilic dye, DiI (38). Briefly, Ox-LDL (1 mg/ml protein) was mixed with 1 ml of LPDS, filter sterilized through 0.45 μm filters, and incubated at 37°C for 8 h. The density of the solution was raised to 1.225 g/ml with the addition of solid KBr and loaded into an ultracentrifuge tube (Beckman Coulter); the top layer containing DiI-Ox-LDL was isolated after ultracentrifugation with a Beckman SW41 Ti rotor at 220,000 g at 4°C for 36 h, and re-isolation of DiI-Ox-LDL was performed. After re-isolation, DiI-Ox-LDL was extensively dialyzed against 1× PBS sterilized with a 0.22 μm filter and stored at 4°C.
Lentiviral vector production and transduction
Lentiviral particles were produced by cotransfection of packaging plasmid, envelop plasmid, and a plasmid encoding the gene of interest, PKM2 shRNA (TRCN0000366063;Sigma-Aldrich) or Scramble shRNA (SHC001;Sigma-Aldrich), in HEK293T cells using Lipofectamine 2000 (Invitrogen). The lentiviral particles were titrated and concentrated as described previously (39). On the sixth day, BMDMs were transduced with 1.5 × 106 PFU lentiviral particles. After 48 h of transduction, the effects of PKM2 knockdown in Ox-LDL-activated macrophages were studied.
DiI-labeled Ox-LDL uptake and binding assay
BMDMs were transfected with Scramble shRNA (1.5 × 106 PFU) or PKM2 shRNA (1.5 × 106 PFU) for 48 h followed by Ox-LDL treatment (40 μg/ml) for 24 h. Cells were washed with PBS and replenished with fresh medium. Subsequently, cells were incubated with DiI-Ox-LDL (10 μg/ml) either at 4°C for binding or 37°C for uptake assays. After 4 h incubation, cells were washed with 0.2% BSA containing PBS and harvested on ice. A minimum of 10,000 events were acquired by flow cytometry in FL-2 channel fluorescence and later analyzed using Cell Quest program FACS Calibur (Becton Dickinson) (40).
Western blotting
Whole cell extracts from BMDMs after various treatments were prepared in RIPA lysis buffer. The supernatants were collected after centrifugation at 16,000 g for 15 min and protein concentration was measured by using BCA reagent. Nuclear and cytosolic fractions were prepared as described previously (41). Briefly, after 24 h of Ox-LDL treatment, BMDMs were lysed in hypotonic buffer (20 mM HEPES, 10 mM KCl, 10% glycerol, 1 mM EDTA, 0.2% NP40, 1 mM PMSF, 5 μg/ml leupeptin, 1 mM Na3VO4, and 5 μg/ml aprotinin) and then centrifuged for 5 min at 800 g at 4°C. Supernatants containing cytosolic fractions were collected to a new tube, and pelleted nuclei were resuspended in hypertonic buffer (hypotonic buffer + 350 mM NaCl) for 30 min at 4°C and centrifuged at 16,000 g, and both nuclear and cytosolic lysates were probed for PKM2. Histone H1 and α-tubulin immunoblotting was done to assess the purity of the cytosolic and nuclear protein fractions. Subsequently, an equal concentration of proteins was boiled with Laemmli buffer, separated on 8–12% SDS-PAGE by Western blotting, and transferred to PVDF membrane. After blocking with 5% BSA in TBST, the membranes were probed with primary and secondary antibodies against several proteins of interest. Western blots were developed using a Gel Doc imaging system (Image Quant LS 500; GE Healthcare).
Co-immunoprecipitation and interaction studies
BMDMs treated with Ox-LDL were lysed as previously described (32). Pre-adsorption of PKM2 antibody on protein A/G-Sepharose beads was done in the presence of pre-adsorption buffer (16). The cell lysates were incubated with the immobilized PKM2 antibody and incubated at 4°C overnight. Immunoprecipitated proteins were subsequently washed, denatured, and separated on SDS-PAGE. Immunoblotting was performed with SREBP-1 and Hif-1α antibody for studying the interaction.
Analysis of PKM2 oligomers by cross-linking
For cross-linking of PKM2, Ox-LDL-treated cells (40 μg/ml) were washed with PBS (pH 8.0) and cross-linking (16) was performed using 1 mM disuccinimidyl suberate in PBS (pH 8.0) for 30 min at room temperature. The reaction was stopped by addition of Tris-HCl (pH 7.5) to a final concentration of 100 mM. Cell lysates consisting of an equal amount of proteins were analyzed by 8% SDS-PAGE and Western blot.
Cellular cholesterol efflux assay
BMDMs were transfected with Scramble shRNA (1.5 × 106 PFU) or PKM2 shRNA (1.5 × 106 PFU) for 48 h. To induce foam cell formation and equilibration with NBD-cholesterol, cells were pretreated with Ox-LDL (40 μg/ml) and NBD-cholesterol (1 μg/ml) for 12 h. Subsequently, cells were washed with PBS and incubated in phenol red-free RPMI medium containing 0.2% BSA with HDL (100 μg/ml) or apoAI (10 μg/ml) for another 6 h. The fluorescence-labeled NBD-cholesterol released from the cell into the medium was measured in a fluorimeter (Omega, BMG Labtech) at excitation and emission wavelengths of 485 and 540 nm, respectively. Cholesterol efflux was calculated from fluorescence in the medium relative to total fluorescence (cells and medium) and expressed as fold change (42).
RNA isolation and real-time RT-PCR
Total RNA was isolated from BMDMs with TRIzol reagent and reverse transcribed using a RevertAid cDNA synthesis kit (Thermo Fisher Scientific). Real-time quantitative PCR was carried out using LightCycler® 480 System II (Roche Applied Science, UK) using SYBR Green and gene-specific primers (31). The primers used are listed in Table 1. 18S was used as an internal standard to calculate the relative expression, and relative fold changes between the groups were calculated by using the comparative cycle threshold (2−ΔΔCT) method (43).
TABLE 1.
PCR primer sequences
| Gene | Primer | Sequence (5′→3′) |
| 18S | Forward | 5′GCAATTATTCCCCATGAACG3′ |
| Reverse | 5′GGCCTCACTAAACCATCCAA3′ | |
| IL-1β | Forward | 5′GCTTCAAATCTCGCAGCAGC3′ |
| Reverse | 5′TCACAGAGGATGGGCTCTTC3′ | |
| SLC2A1 | Forward | 5′ATGGATCCCAGCAGCAAG3′ |
| Reverse | 5′CCAGTGTTATAGCCGAACTGC3′ | |
| LDH | Forward | 5′GCTCCCCAGAACAAGATTACA3′ |
| Reverse | 5′TCGCCCTTGAGTTTGTCTTC3′ | |
| PKM2 | Forward | 5′AGGATGCCGTGCTGAATG3′ |
| Reverse | 5′TAGAAGAGGGGCTCCAGAGG3′ | |
| PPAR-α | Forward | 5′GTCCTCAGTGCTTCCAGAGG3′ |
| Reverse | 5′GGTCACCTACGAGTGGCATT3′ | |
| PPARγ | Forward | 5′AGTGGAGACCGCCCAGG3′ |
| Reverse | 5′GCAGCAGGTTGTCTTGGATGT3′ | |
| LXR-α | Forward | 5′GCTCTGCTCATTGCCATCAG3′ |
| Reverse | 5′TGTTGCAGCCTCTCTACTTGGA3′ |
Measurement of oxygen consumption rate and extracellular acidification rate
The oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) were measured using an XFp extracellular flux analyzer (Agilent Technologies) and as reported earlier (44). After Ox-LDL treatment (40 μg/ml), the cells were changed to unbuffered assay medium supplemented with 10 mM glucose, 1 mM pyruvate, 2 mM glutamine, (pH 7.4) for OCR and unbuffered base medium supplemented with 2 mM glutamine (pH 7.4) for ECAR measurement, respectively, and incubated for 45 min at 37°C. Baseline measurements were obtained before sequential injection of the mitochondrial inhibitors, oligomycin (1 μM), carbonyl cyanide-p-trifluoromethoxy-phenylhydrazone (1 μM), and antimycin (500 nM) plus rotenone (500 nM) for OCR and d-glucose (10 mM), oligomycin (1 μM), and 2-DG (50 mM) for ECAR. Extracellular l-lactate was measured in cell culture supernatant by l-lactate assay kit (Cayman Chemical) as described previously (45).
Statistical analysis
Data are presented as the mean ± SE. The difference between the two groups was determined by unpaired two-tailed Student’s t-test. A P-value <0.05 was considered statistically significant. All statistical analyses were performed with the GraphPad Prism 7.0 program (GraphPad Inc., La Jolla, CA).
RESULTS
Regulation of PKM2 by Ox-LDL in macrophages
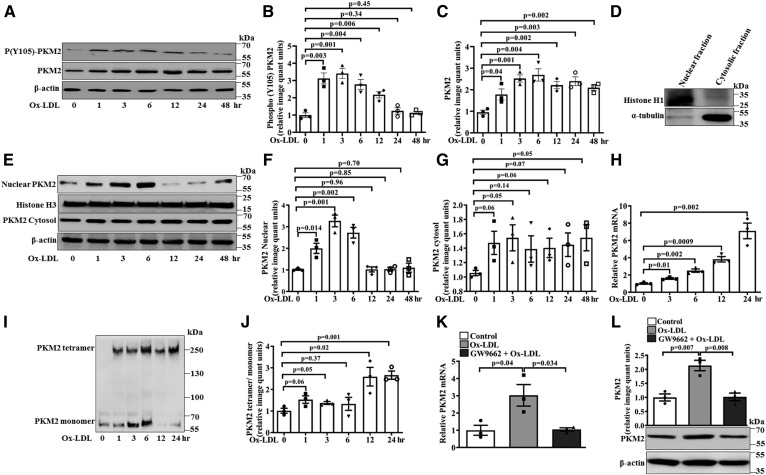
The monomeric or dimeric form of PKM2 regulates aerobic glycolysis and inflammation in activated immune cells (11). Therefore, the effect of Ox-LDL on PKM2 expression, tyrosine 105 phosphorylation, nuclear translocation, and oligomerization was evaluated. A significant increase in PKM2 tyrosine 105 phosphorylation was observed at 1 h (∼3.1-fold), 3 h (∼3.4-fold), 6 h (∼2.8-fold), and 12 h (∼2.2-fold) of Ox-LDL treatment when compared with the control (Fig. 1A, B). However, no significant change in phosphorylation was observed at 24 and 48 h (Fig. 1A, B) when compared with the control. Ox-LDL also induced a significant increase in PKM2 protein expression (1–48 h) when compared with the control (Fig. 1A, C). The effect of Ox-LDL on the PKM2 pathway was dose dependent because significant increases in PKM2 expression, PKM2 tyrosine 105 phosphorylation (supplemental Fig. S1A–C), and IL-1β mRNA and protein expression (supplemental Fig. S1D and E, respectively) were observed with increasing doses of Ox-LDL. The nuclear and cytosolic purity of Ox-LDL-treated samples was ascertained by histone H1 and α-tubulin immunoblotting (Fig. 1D). Ox-LDL induced a significant increase in the nuclear translocation of PKM2 at 1 h (∼2-fold), 3 h (∼3.3-fold), and 6 h (∼2.8-fold); although, at later time points, a decrease was observed when compared with the control (Fig. 1E, F). However, no significant increase in the cytosolic pool of PKM2 was observed (Fig. 1E, G). Concurrent with Ox-LDL-induced phosphorylation of PKM2 on tyrosine 105, we observed increased nuclear localization of PKM2. This effect was evident at 1 h and maximum at 3 h; however at later time points (12–48 h), a decrease was observed (Fig. 1E, F).
Fig. 1.
Ox-LDL regulates PKM2 tyrosine 105 phosphorylation, expression, and oligomerization in BMDMs. A: Western blot analysis of PKM2 expression and phosphorylation after Ox-LDL treatment (40 μg/ml). B: Bar diagram representing quantification of PKM2 phosphorylation as shown in A. C: Bar diagram representing quantification of PKM2 protein expression as shown in A. β-Actin was used as an internal control (n = 3). D: Purity assessment of nuclear and cytosolic preparations by immunoblotting with histone H1 and α-tubulin. E: Nuclear and cytosolic fraction subjected to Western blotting to detect PKM2 content at different time points of Ox-LDL treatment (n = 3). F: Relative quantification of PKM2 nuclear fraction as shown in E. G: Relative quantification of PKM2 cytosolic fraction as shown in E. H: Relative mRNA expression of PKM2 with respect to control gene 18S measured by quantitative PCR (n = 3). I: Western blot analysis of PKM2 after cross-linking with 1 mM disuccinimidyl suberate. J: Respective quantification of PKM2 monomer and tetramer form as shown in I (n = 3). K: Effect of GW9662 (10 μM, 24 h) pretreatment on Ox-LDL-induced (40 μg/ml, 6 h) PKM2 mRNA expression (n = 3). L: Effect of GW9662 on PKM2 protein expression (n = 3). Blots represent one of three similar experiments. Values represent mean ± SEM.
Similarly, Ox-LDL induced a time-dependent increase in PKM2 mRNA expression when compared with the control (Fig. 1H). At early time points of Ox-LDL treatment, an increase in the monomeric and tetrameric form of PKM2 was observed when compared with the control (Fig. 1I, J). However, at later time points (12–24 h), the tetrameric form of PKM2 dominated (Fig. 1I, J). Therefore, both nuclear and cytosolic functions of PKM2 were induced after Ox-LDL treatment. Because PPARγ mediates Ox-LDL-induced macrophage foam cell formation (20) and plays a role in PKM2 expression (21), its effect on PKM2 mRNA and protein expression was evaluated. Ox-LDL-induced PKM2 mRNA and protein expression was significantly attenuated in the presence of the PPARγ antagonist GW9662 when compared with cells treated with Ox-LDL alone (Fig. 1 K, L). Therefore, PPARγ regulates PKM2 expression in Ox-LDL-activated macrophages.
Ox-LDL induces aerobic glycolysis
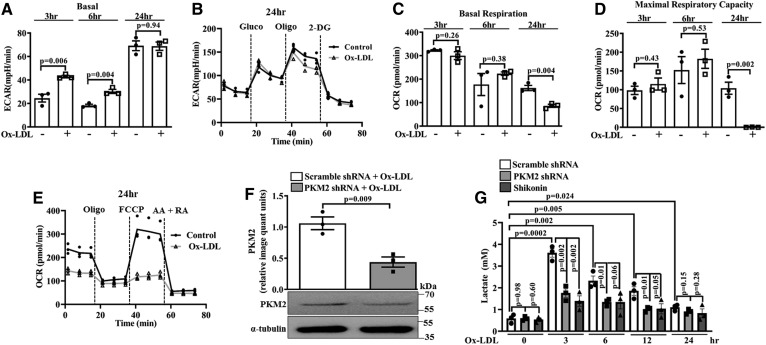
The Warburg effect or aerobic glycolysis is characterized by an increase in lactate production and ECAR (46). Ox-LDL induced a significant increase in the basal ECAR at 3 and 6 h when compared with the control (Fig. 2A). However, at 24 h, no significant change in the ECAR was observed when compared with the control (Fig. 2A, B). Ox-LDL also induced a significant decrease in the OCR and mitochondrial maximal respiratory capacity at 24 h (Fig. 2C–E); however, no significant change was observed at 3 and 6 h when compared with the control (Fig. 2C, D).
Fig. 2.
Ox-LDL induces aerobic glycolysis in BMDMs. Extracellular flux analysis and lactate measurement. A: Basal ECAR in control and Ox-LDL-treated (40 μg/ml) BMDMs (n = 3). B: Real-time changes in ECAR in control and Ox-LDL-stimulated (40 μg/ml) BMDMs (n = 3). C: Basal OCR in control and Ox-LDL-treated (40 μg/ml) BMDMs (n = 3). D: Maximal respiratory capacity in control and Ox-LDL-treated (40 μg/ml) BMDMs (n = 3). E: Real-time changes in OCR after pretreatment with Ox-LDL (40 μg/ml) (n = 3). F: Western blot analysis of PKM2 expression in Ox-LDL-activated (40 μg/ml) BMDMs after knockdown with PKM2 shRNA (1.5 × 106 PFU, 48 h) (n = 3). G: Time-dependent response of Ox-LDL (40 μg/ml) on extracellular l-lactate release in Shikonin- or PKM2 shRNA-pretreated BMDMs (n = 3). Values represent mean ± SEM.
To assess the role of PKM2 on Ox-LDL-induced lactate production, experiments were carried out in the presence of a PKM2 pharmacological inhibitor, Shikonin or PKM2 shRNA. As expected, PKM2 shRNA significantly reduced PKM2 protein expression when compared with Scramble shRNA (Fig. 2F). Ox-LDL induced a significant increase in the extracellular lactate levels at 3 h (∼3.6 mM), 6 h (∼2.3 mM), 12 h (∼1.8 mM), and 24 h (∼1.1 mM) when compared with the control (Fig. 2G). Cells pretreated with Shikonin or PKM2 shRNA and stimulated with Ox-LDL showed a significant decrease in lactate content when compared with Scramble shRNA- and Ox-LDL-treated cells (Fig. 2G). Therefore, the results suggest that PKM2 mediates Ox-LDL-induced aerobic glycolysis.
PKM2-Hif-1α axis fuels Ox-LDL-induced inflammation in BMDMs
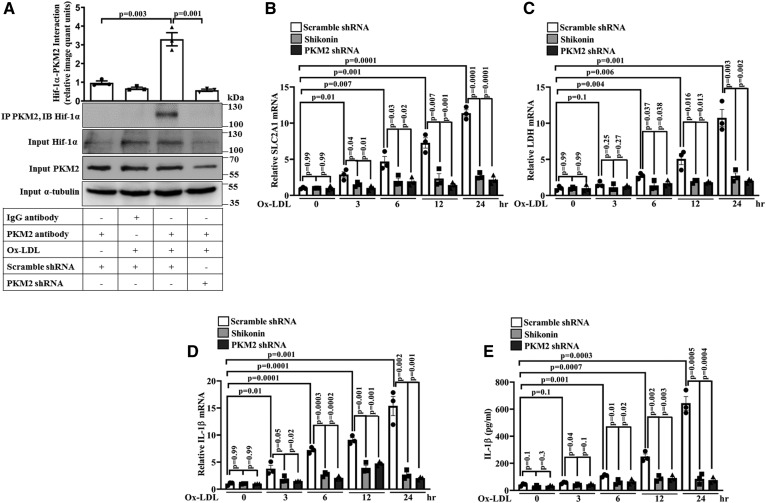
The PKM2-Hif-1α axis plays a vital role in macrophage immune response and aerobic glycolysis (16); therefore, its role in Ox-LDL-induced inflammation was assessed. Ox-LDL induced a significant increase in PKM2-Hif-1α interaction (Fig. 3A); although a significant increase in Hif-1α mRNA (supplemental Fig. S2) and protein expression was also observed (Fig. 3A). As expected, PKM2 shRNA significantly reduced PKM2 protein expression and therefore PKM2-Hif-1α interaction (Fig. 3A). Ox-LDL also induced a time-dependent increase in glucose transporter member 1 (SLC2A1), lactate dehydrogenase (LDH), and IL-1β mRNA expression and secretory IL-1β production when compared with the control (Fig. 3B–E). Shikonin or PKM2 shRNA significantly inhibited Ox-LDL-induced SLC2A1, LDH, and IL-1β mRNA expression and secretory IL-1β production when compared with cells treated with Ox-LDL alone (Fig. 3B–E). These results suggest the role of the PKM2-Hif-1α axis in Ox-LDL-induced inflammation and activation of Hif-1α target genes in BMDMs.
Fig. 3.
Ox-LDL induces PKM2-Hif-1α interaction and Hif-1α target genes SLC2A1, LDH, and IL-1β. A: Ox-LDL-treated (40 μg/ml, 6 h) BMDMs were subjected to immunoprecipitation with PKM2 antibody and immunoblotted with Hif-1α and PKM2 antibody (n = 3). A time-dependent RT-PCR mRNA analysis of SLC2A1 (B), LDH (C), and IL-1β (D). E: Secretory IL-1β measured by ELISA (n = 3) in Ox-LDL-treated (40 μg/ml) BMDMs in the presence or absence of Shikonin (1 μM, 1 h) or PKM2 shRNA (1.5 × 106 PFU, 48 h). In quantitative PCR, 18S was used as an internal control (n = 3). Values represent mean ± SEM.
Ox-LDL induces PKM2-Hif-1α axis, aerobic glycolysis, and inflammation in peritoneal macrophages
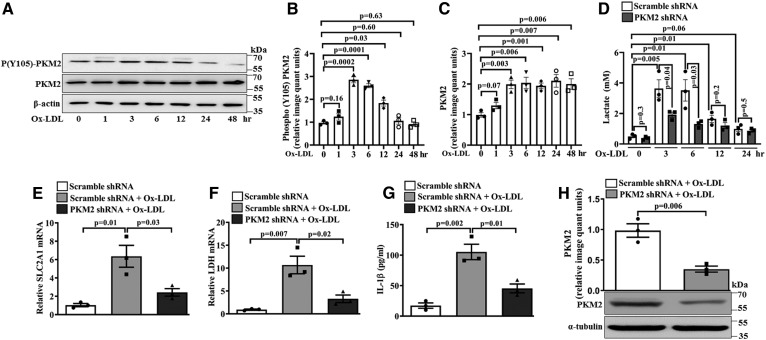
Experiments were carried out to assess whether Ox-LDL stimulates PKM2, aerobic glycolysis, and inflammation in peritoneal macrophages similarly to BMDMs. In peritoneal macrophages, Ox-LDL induced PKM2 tyrosine 105 phosphorylation and PKM2 protein expression in a time-dependent manner (Fig. 4A–C). A significant increase in PKM2 tyrosine 105 phosphorylation was observed at 3 h (∼2.8-fold), 6 h (∼2.6-fold), and 12 h (∼1.8-fold) of Ox-LDL treatment when compared with the control (Fig. 4A, B). However, no significant change in phosphorylation was observed at 24 and 48 h (Fig. 4A, B) when compared with the control. Furthermore, Ox-LDL induced a significant increase in PKM2 protein expression (3–48 h) compared with the control (Fig. 4A, C). Ox-LDL also induced a significant increase in extracellular l-lactate at 3 h (∼3.7 mM), 6 h (∼3.5 mM), and 12 h (∼1.7 mM) when compared with the control; however, at 24 h no significant change in l-lactate was observed (Fig. 4D). Cells pretreated with PKM2 shRNA and stimulated with Ox-LDL showed a significant decrease in lactate content when compared with Scramble shRNA- and Ox-LDL-treated cells (Fig. 4D). Similarly, Ox-LDL induced a significant increase in SLC2A1 and LDH mRNA expression and secretory IL-1β production when compared with the control (Fig. 4 E, F, and G, respectively). PKM2 shRNA significantly inhibited Ox-LDL-induced SLC2A1 and LDH mRNA expression and secretory IL-1β production when compared with Scramble shRNA- and Ox-LDL-treated cells (Fig. 4 E, F, and G, respectively). As expected, PKM2 knockdown with specific shRNA significantly abrogated Ox-LDL-induced PKM2 protein expression (Fig. 4H). Therefore, in peritoneal macrophages, Ox-LDL stimulates PKM2, aerobic glycolysis, and inflammation similarly to BMDMs.
Fig. 4.
Effect of Ox-LDL on PKM2 activation, Hif-1α target genes, l-lactate release, and inflammation in thioglycollate-induced murine peritoneal macrophages. A: Time-dependent PKM2 protein expression and tyrosine 105 phosphorylation in Ox-LDL-stimulated (40 μg/ml) cells (n = 3). B: Bar diagram representing quantification of PKM2 phosphorylation as shown in A. C: Bar diagram representing quantification of PKM2 protein expression as shown in A. β-Actin was used as an internal control (n = 3). D: Time-dependent response of Ox-LDL (40 μg/ml) on extracellular l-lactate release in Scramble shRNA-treated cells (1.5 × 106 PFU, 48 h) or PKM2 shRNA-treated cells (1.5 × 106 PFU, 48 h) (n = 3). Cells were treated with Ox-LDL (40 μg/ml, 24 h) in the presence of Scramble shRNA (1.5 × 106 PFU, 48 h) or PKM2 shRNA (1.5 × 106 PFU, 48 h), and samples were analyzed by quantitative PCR for the expression of SLC2A1 mRNA (E) and LDH mRNA (F). G: Secretory IL-1β measured by ELISA (n = 3). H: Relative PKM2 protein expression in Scramble shRNA-treated (1.5 × 106 PFU, 48 h) or PKM2 shRNA-treated (1.5 × 106 PFU, 48 h) and Ox-LDL-activated (40 μg/ml, 24 h) macrophages as assessed by Western blotting (n = 3). Blots represent one of three similar experiments. Values represent mean ± SEM.
PKM2 mediates Ox-LDL-induced foam cell formation and cholesterol efflux in BMDMs and peritoneal macrophages
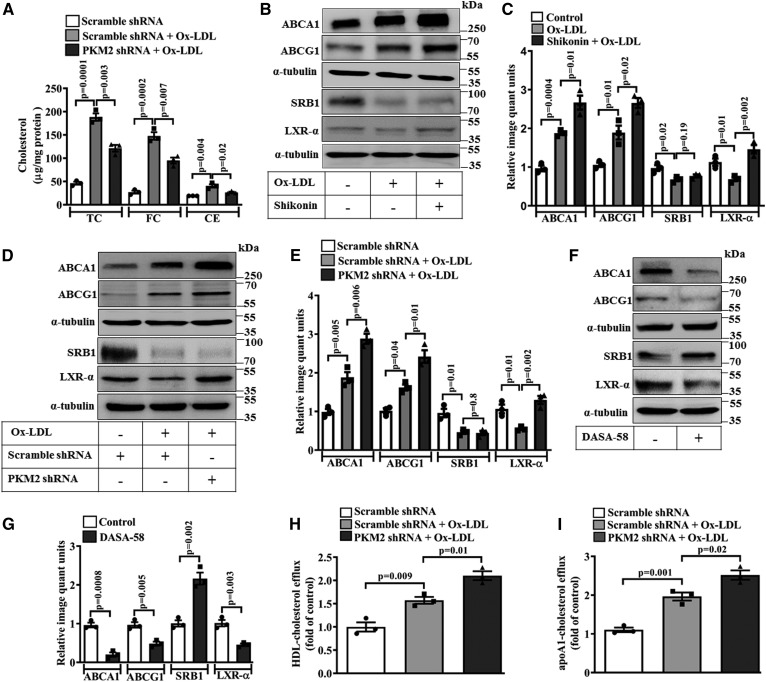
Ox-LDL treatment significantly increased cellular TC, FC, and CE (∼188.4, ∼147.9, and ∼40.5 μg/mg protein, respectively) in BMDMs when compared with the control (Fig. 5A). However, macrophages pretreated with PKM2 shRNA and stimulated with Ox-LDL showed a significant decrease in TC, FC, and CE when compared with Scramble shRNA- and Ox-LDL-treated cells (∼121.5, ∼95.2, and ∼26.3 μg/mg protein, respectively; Fig. 5A). PKM2 shRNA also decreased Ox-LDL-mediated intracellular lipid accumulation, as evident by Oil Red O staining (supplemental Fig. S3A). To delineate the underlying mechanisms by which PKM2 regulates foam cell formation, expression of cholesterol transporters (ABCA1, SRB1, and ABCG1) and transcription factors involved in cholesterol efflux (LXR-α, PPAR-α, and PPARγ) was assessed. Ox-LDL significantly induced the expression of ABCA1 (∼1.9-fold; Fig. 5B, C) and ABCG1 (∼1.8-fold; Fig. 5B, C) when compared with the control. Pretreatment with Shikonin or PKM2 shRNA significantly enhanced Ox-LDL-induced ABCA1 (P = 0.01 and P = 0.006, Fig. 5B, C and Fig. 5D, E; respectively) and ABCG1 (P = 0.02 and P = 0.01, Fig. 5B, C and Fig. 5D, E; respectively) expression when compared with cells treated with Ox-LDL alone. This suggests that PKM2 limits Ox-LDL-induced ABCA1 and ABCG1 expression.
Fig. 5.
PKM2 mediates Ox-LDL-induced foam cell formation and cholesterol efflux in BMDMs. A: Cellular TC, FC, and CE were measured in Ox-LDL-activated (40 μg/ml, 24 h) macrophage in the presence of Scramble shRNA (1.5 × 106 PFU, 48 h) or PKM2 shRNA (1.5 × 106 PFU, 48 h) (n = 3). B: Effect of Shikonin (1 μM, 1 h) pretreatment on ABCA1, ABCG1, SRB1, and LXR-α protein expression in Ox-LDL-stimulated (40 μg/ml, 24 h) macrophages as analyzed by Western blotting. C: Relative quantification of ABCA1, ABCG1, SRB1, and LXR-α protein expression as shown in B (n = 3). D: Effect of PKM2 shRNA (1.5 × 106 PFU, 48 h) on ABCA1, ABCG1, SRB1, and LXR-α protein expression in Ox-LDL-stimulated macrophages as analyzed by Western blotting. E: Bar diagram represents the relative densitometry quantification of ABCA1, ABCG1, SRB1, and LXR-α protein expression as shown in D (n = 3). F: Effect of DASA-58 treatment (50 μM, 24 h) on ABCA1, ABCG1, SRB1, and LXR-α protein expression as analyzed by Western blotting. G: Relative densitometry quantification of ABCA1, ABCG1, SRB1, and LXR-α protein expression as shown in F (n = 3). HDL-mediated cholesterol efflux (100 μg/ml, 6 h) (H) and apoA1-mediated cholesterol efflux (10 μg/ml, 6 h) (I) in BMDMs after PKM2 knockdown (1.5 × 106 PFU, 48 h) followed by incubation with Ox-LDL (40 μg/ml) and NBD cholesterol (1 μg/ml) for 12 h (n = 3). Blots represent one of three similar experiments. Values represent mean ± SEM.
mRNA expression of common transcription factors involved in cholesterol efflux was also assessed. Ox-LDL significantly induced PPAR-α and PPARγ mRNA expression; however, a significant reduction in LXR-α mRNA was observed (supplemental Fig. S3B–D). Pretreatment with PKM2 shRNA did not affect Ox-LDL-induced PPAR-α and PPARγ mRNA expression; however, significant recovery in LXR-α mRNA was observed (supplemental Fig. S3 B–D). Similarly, Ox-LDL induced a significant decrease in the LXR-α protein expression when compared with the control (P < 0.05, Fig. 5B–E). Pretreatment with Shikonin or PKM2 shRNA prevented the Ox-LDL-induced decrease in LXR-α protein expression (P < 0.01; Fig. 5B–E) when compared with cells treated with Ox-LDL alone. Although Ox-LDL reduced the expression of SRB1 when compared with the control (P < 0.05; Fig. 5B–E), pretreatment with Shikonin or PKM2 shRNA did not prevent the Ox-LDL-induced decrease in SRB1 expression (Fig. 5B–E). DASA-58, a small molecule activator of PKM2, significantly decreased the expression of ABCA1 (P = 0.008), ABCG1 (P = 0.005), and LXR-α (P = 0.003) when compared with the control (Fig. 5F, G). Surprisingly, a significant increase in SRB1 expression was observed with DASA-58 treatment when compared with the control (Fig. 5F, G).
Ox-LDL significantly increased HDL-dependent cholesterol efflux (P = 0.009, Fig. 5H) or apoA1-dependent cholesterol efflux (P = 0.001, Fig. 5I) in the BMDMs when compared with the control. However, pretreatment with PKM2 shRNA further augmented Ox-LDL-induced cholesterol efflux to HDL (P = 0.01, Fig. 5H) or apoA1 (P = 0.02, Fig. 5I) when compared with the Ox-LDL- and Scramble shRNA-treated groups. This correlates well with the significant increase in the ABCA1 and ABCG1 expression observed after PKM2 inhibition in Ox-LDL-treated macrophages. Moreover, treatment with the PKM2 activator, DASA-58, augmented Ox-LDL-induced intracellular TC, FC, and CE accumulation (supplemental Fig. S4A) and attenuated HDL- and apoA1-mediated cholesterol efflux (supplemental Fig. S4B, C) when compared with cells treated with Ox-LDL alone. Therefore, PKM2 seems to regulate macrophage foam cell formation by modulating ABCA1 and ABCG1 expression and cholesterol efflux.
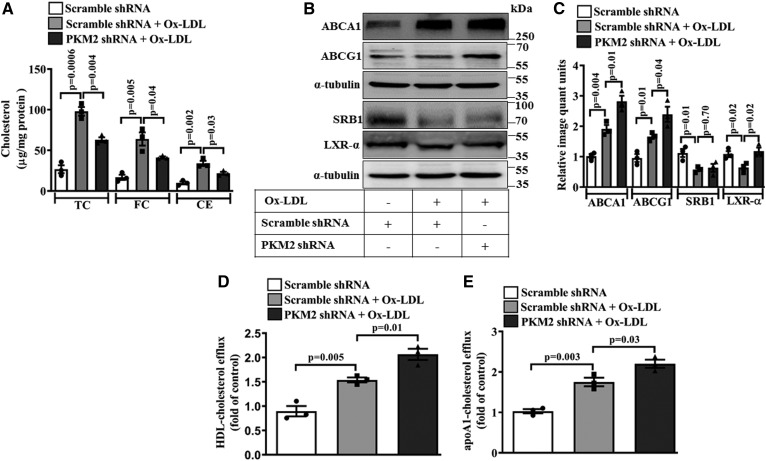
Similar to BMDMs, Ox-LDL treatment significantly increased cellular TC, FC, and CE (∼98, ∼64, and ∼34 μg/mg protein; respectively) in the thioglycollate-induced murine peritoneal macrophages when compared with the control (Fig. 6A). However, macrophages pretreated with PKM2 shRNA and stimulated with Ox-LDL showed a significant decrease in TC, FC, and CE when compared with Scramble shRNA- and Ox-LDL-treated cells (∼63.1, ∼41.2, and ∼21.9 μg/mg protein, respectively; Fig. 6A). Ox-LDL significantly induced the expression of ABCA1 (∼1.9-fold; Fig. 6B, C) and ABCG1 expression (∼1.7-fold; Fig. 6B, C) when compared with the control. Similar to BMDMs, pretreatment with PKM2 shRNA significantly enhanced the Ox-LDL-induced ABCA1 (P = 0.01; Fig. 6B, C) and ABCG1 expression (P = 0.04; Fig. 6B, C) when compared with Scramble shRNA- and Ox-LDL-treated cells. PKM2 inhibition did not affect the Ox-LDL-induced decrease in SRB1 expression (P = 0.01; Fig. 6B, C). Ox-LDL induced a significant decrease in the LXR-α protein expression (P = 0.02; Fig. 6B, C) when compared with the control. Pretreatment with PKM2 shRNA prevented Ox-LDL-induced downregulation of LXR-α expression (P = 0.02; Fig. 6B, C) when compared with Scramble shRNA- and Ox-LDL-treated cells. Ox-LDL significantly increased HDL-dependent cholesterol efflux (P = 0.005, Fig. 6D) or apoA1-dependent cholesterol efflux (P = 0.003, Fig. 6E) in peritoneal macrophages when compared with control cells. However, pretreatment with PKM2 shRNA further augmented Ox-LDL-induced cholesterol efflux to HDL (P = 0.01, Fig. 6D) or apoA1 (P = 0.03, Fig. 6E) when compared with Scramble shRNA- and Ox-LDL-treated cells. Therefore, similar to BMDMs, PKM2 regulates cholesterol efflux by modulating ABCA1 and ABCG1 expression in peritoneal macrophages also.
Fig. 6.
PKM2 supports foam cell formation and cholesterol efflux in thioglycollate-induced murine peritoneal macrophages. A: TC, FC, and CE in peritoneal macrophages after treatment with Ox-LDL (40 μg/ml, 24 h) in the presence of Scramble shRNA (1.5 × 106 PFU, 48 h) or PKM2 shRNA (1.5 × 106 PFU, 48 h) (n = 3). B: Effect of PKM2 (1.5 × 106 PFU, 48 h) or Scramble shRNA (1.5 × 106 PFU, 48 h) on Ox-LDL-induced (40 μg/ml, 24 h) ABCA1, ABCG1, SRB1, and LXR-α protein expression as assessed by Western blotting. C: Bar diagram represents the relative densitometry quantification of ABCA1, ABCG1, SRB1, and LXR-α protein expression as shown in B. HDL-mediated cholesterol efflux (100 μg/ml, 6 h) (D) and apoA1-mediated cholesterol efflux (10 μg/ml, 6 h) (E) in peritoneal macrophages pretreated with control or PKM2 shRNA followed by incubation with Ox-LDL (40 μg/ml) and NBD-cholesterol (1 μg/ml) for 12 h (n = 3). Blots represent one of three similar experiments. Values represent mean ± SEM.
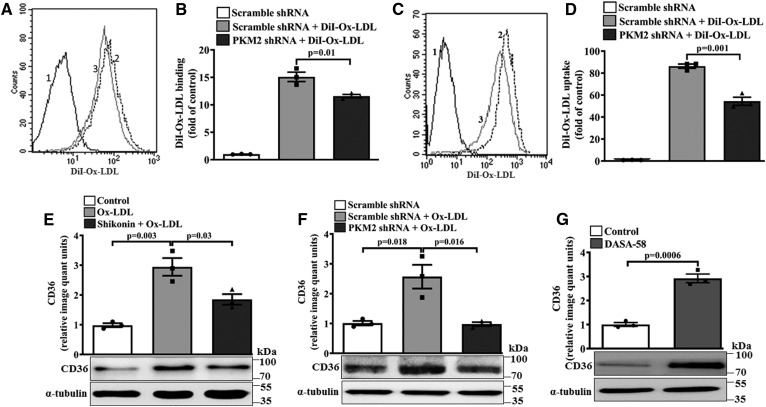
PKM2 regulates Ox-LDL binding and uptake and CD36 expression
The effect of PKM2 on scavenger receptor expression and cholesterol uptake was studied. PKM2 shRNA pretreatment significantly decreased the binding (P = 0.01; Fig. 7A, B) and uptake (P = 0.001; Fig. 7C, D) of DiI-Ox-LDL in macrophages when compared with Scramble shRNA- and DiI-Ox-LDL-treated cells. Ox-LDL induced a significant increase in scavenger receptor CD36 protein expression (∼2.9-fold, Fig. 7E) when compared with the control. However, pretreatment with Shikonin or PKM2 shRNA significantly reduced Ox-LDL-induced CD36 protein expression (P = 0.03 and P = 0.016; Fig. 7E and F, respectively) when compared with Ox-LDL-treated cells. Similarly, in peritoneal macrophages, PKM2 shRNA abrogated the Ox-LDL-induced CD36 protein expression when compared with Scramble shRNA- and Ox-LDL-treated cells (supplemental Fig. S5A). DASA-58 also significantly induced the expression of CD36 (∼3.2-fold, Fig. 7G). Therefore, PKM2 may regulate foam cell formation by regulating CD36 expression and Ox-LDL binding and uptake.
Fig. 7.
Effect of PKM2 on Ox-LDL binding, uptake, and CD36 expression. BMDMs were treated with Ox-LDL (40 μg/ml) in the presence of Scramble shRNA (1.5 × 106 PFU, 48 h) or PKM2 shRNA (1.5 × 106 PFU, 48 h) for 24 h. A: Histogram overlay showing DiI-Ox-LDL (10 μg/ml, 4 h) binding at 4°C in Ox-LDL-treated macrophages (40 μg/ml, 24 h) [1, autofluorescence; 2, Scramble shRNA + DiI-Ox-LDL; 3, PKM2 shRNA + DiI-Ox-LDL (n = 3)]. B: Fold change in DiI-Ox-LDL binding as shown in A (n = 3). C: Histogram overlay showing DiI-Ox-LDL (10 μg/ml, 4 h) uptake at 37°C in Ox-LDL-treated macrophages (40 μg/ml, 24 h) as assessed by flow cytometry (1, autofluorescence; 2, Scramble shRNA + DiI-Ox-LDL; 3, PKM2 shRNA + DiI-Ox-LDL). D: Fold change in DiI-Ox-LDL uptake as shown in C (n = 3). E: CD36 protein expression in Ox-LDL-activated (40 μg/ml, 24 h) macrophages in the presence or absence of Shikonin (1 μM, 1 h) as assessed by Western blotting. F: CD36 expression in Ox-LDL-activated (40 μg/ml, 24 h) macrophages in the presence of control shRNA (1.5 × 106 PFU, 48 h) or PKM2 shRNA (1.5 × 106 PFU, 48 h). G: DASA-58-stimulated (50 μM, 24 h) BMDMs analyzed for CD36 expression by Western blotting. α-tubulin was used as an internal control (n = 3). Values represent mean ± SEM.
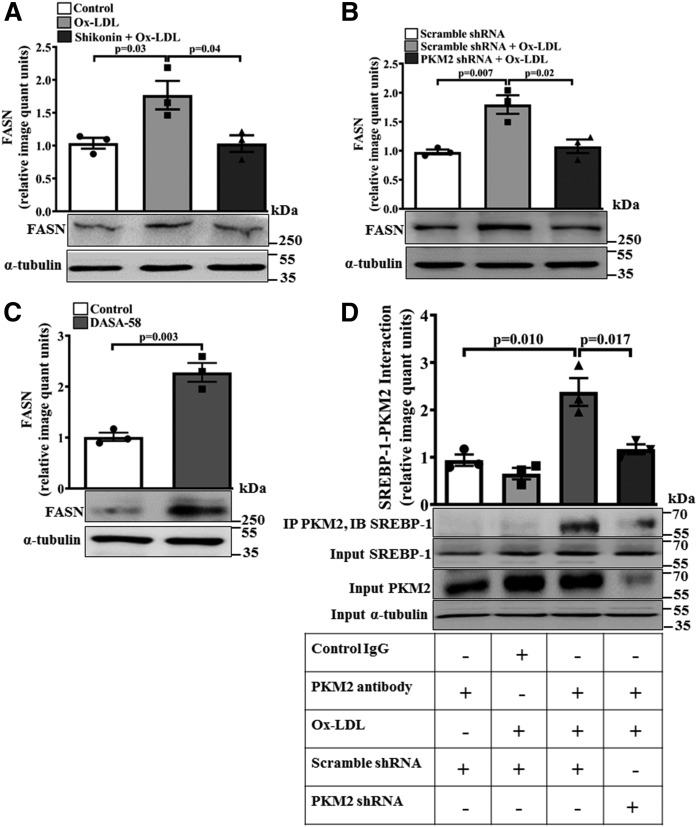
PKM2 regulates SREBP-1-dependent FASN expression
Because FASN plays a role in lipid biosynthesis (28), the effect of PKM2 on FASN protein expression was evaluated. Ox-LDL induced a significant increase in FASN protein expression when compared with the control (P = 0.03 and P = 0.007; Fig. 8A and B, respectively). However, pretreatment with Shikonin or PKM2 shRNA significantly attenuated Ox-LDL-induced FASN protein expression when compared with cells treated with Ox-LDL alone (P < 0.05; Fig. 8A, B). Similarly, in peritoneal macrophages, PKM2 knockdown inhibited Ox-LDL-induced FASN expression (supplemental Fig. S5B). DASA-58 significantly increased FASN protein expression (∼2.3-fold, Fig. 8C) when compared with the control. Therefore, PKM2 regulates Ox-LDL-induced FASN expression. Because SREBP-1 can drive FASN expression (28), its regulation by PKM2 was studied. Ox-LDL induced a significant increase in PKM2-SREBP-1 association in the macrophages (Fig. 8D). The inhibition of PKM2 did not affect the expression of SREBP-1 (Fig. 8D). Therefore, Ox-LDL induces PKM2-SREBP-1 interaction in a PKM2-dependent manner. While assessing any nonspecific effect of control shRNA on the pathways evaluated in the present study, it was observed that control shRNA had no significant effect on the expression of genes regulating cholesterol efflux, CD36, and FASN when compared with the control (supplemental Fig. S6). Therefore, the effects observed in the present study were specific to PKM2.
Fig. 8.
Ox-LDL induces PKM2-SREBP-1 interaction and SREBP-1 target gene FASN. A: FASN expression in the Ox-LDL-activated (40 μg/ml, 6 h) macrophages in the presence or absence of Shikonin (1 μM, 1 h) as assessed by Western blotting (n = 3). B: FASN protein expression in the Ox-LDL-activated (40μg/ml, 24 h) macrophages in the presence of control shRNA (1.5 × 106 PFU, 48 h) or PKM2 shRNA (1.5 × 106 PFU, 48 h) as assessed by Western blotting (n = 3) C: FASN expression in DASA-58-stimulated (50 μM, 24 h) BMDMs as analyzed by Western blot (n = 3). α-tubulin was used as an internal control in all the experiments (n = 3). D: Ox-LDL-treated (40 μg/ml, 6 h) BMDMs were subjected to immunoprecipitation with PKM2 antibody and immunoblotted with SREBP-1 and PKM2 antibody (n = 3). Blots represent one of three similar experiments. Values represent mean ± SEM.
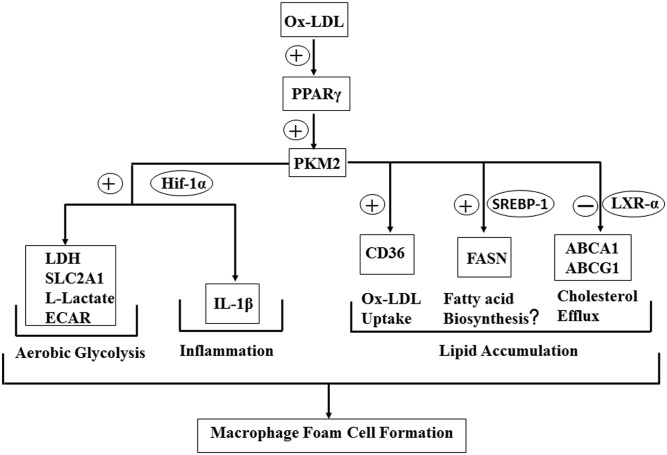
Therefore, in the present study, PKM2 regulates aerobic glycolysis, inflammation, and macrophage foam cell formation (Fig. 9).
Fig. 9.
Role of PKM2 in Ox-LDL-induced inflammation and foam cell formation.
DISCUSSION
PKM2 bridges metabolic and inflammatory dysfunction in atherosclerotic coronary artery disease (10). However, its role in macrophage foam cell formation is not clear. Here, we demonstrate the role of PKM2 in Ox-LDL-induced glycolytic switch, inflammation, and macrophage foam cell formation.
In atherosclerosis, macrophages play an important role in vascular cholesterol homeostasis by regulating Ox-LDL uptake and intracellular cholesterol trafficking (47). However, excessive accumulation of modified lipids leads to inflammation, foam cell formation, and disease progression (48). Therefore, targeting macrophage lipid accumulation and inflammation is an attractive strategy to prevent atherosclerosis.
During macrophage foam cell formation, there is transcriptional and posttranslational activation of PKM2 because an increase in PKM2 mRNA and protein expression and PKM2 phosphorylation is observed in Ox-LDL-stimulated macrophages. Usually the monomer or dimer form of PKM2 localizes to the nucleus (14), and nuclear translocation of PKM2 promotes a glycolytic and inflammatory phenotype (16). The tetrameric form of PKM2 is restricted to the cytosol and promotes glycolytic flux (49).
Phosphorylation of PKM2 on tyrosine 105 promotes its monomer or dimer conformation, nuclear translocation, and Warburg effect (16). In the present study, a shorter Ox-LDL treatment regimen induced similar modulation of PKM2. In a recent study, out of the several putative kinases that phosphorylate PKM2 at tyrosine 105 (50), Src kinase stands out because it plays an important role in Ox-LDL-induced foam cell formation (51). Therefore, it is quite possible that Src kinase activates PKM2 during Ox-LDL-induced macrophage foam cell formation. However, this needs detailed investigation. The mRNA for PKM2 continues to increase even up to 24 h while protein level stabilizes after 3h. This may be due to PKM2 posttranslational modifications like acetylation, which is known to limit PKM2 protein expression by promoting its lysosomal degradation (52). In the present study, the Ox-LDL-induced Warburg effect was evident due to a significant increase in lactate and ECAR. During the Warburg effect, an increase in glycolytic flux and a decrease in oxidative phosphorylation leads to lactate accumulation (46).
PKM2 supports the Ox-LDL-induced Warburg effect because a significant decrease in lactate was observed with PKM2 shRNA and Shikonin. It is well-known that the PKM2 tetramer conformation promotes glycolytic flux and limits the Warburg effect (16). Similarly, in the present study, longer treatment (12–24 h) of Ox-LDL promotes PKM2-dependent glycolytic flux, because there is an increase in PKM2 tetramer conformation. This is accompanied with decreased PKM2 tyrosine 105 phosphorylation, nuclear localization, ECAR, and lactate production. Therefore, Ox-LDL-induced time-dependent modulation of PKM2 phosphorylation correlates well with its nuclear cytosol localization and PKM2 tetramer formation. The functional effect of this modulation is evident from the ECAR and lactate production; although we observed a significant decrease in the OCR, which may be due to a broken Kreb’s cycle (53).
Because the PPARγ antagonist GW9662, significantly attenuated Ox-LDL-induced PKM2 expression, it can be speculated that PPARγ is upstream of PKM2 during foam cell formation. It is well reported that Ox-LDL-induced macrophage foam cell formation is mediated by PPARγ (20). Earlier reports also suggest that PPARγ supports the transcription of the PKM2 gene and promotes fatty liver disease (21). LPS induces the formation of a PKM2 and Hif-1α complex that can bind to the IL-1β promoter and regulate inflammation (16). PKM2 can also regulate Hif-1α activity and transcription of Hif-1α-dependent genes in hypoxia and cancer (15, 16). In the present study, Ox-LDL induced PKM2 and Hif-1α interaction, suggesting operation of similar mechanisms. PKM2 plays a role in Ox-LDL-induced Hif-1α turnover because Ox-LDL-induced Hif-1α mRNA and protein expression was significantly attenuated with PKM2 shRNA. Ox-LDL regulates PKM2-Hif-1α-induced transcription of the pro-glycolytic genes, SLC2A1 and LDH, and the pro-inflammatory gene, IL-1β, required for aerobic glycolysis in a PKM2-dependent manner because the effect was abrogated with PKM2 shRNA or Shikonin.
Deregulated lipid metabolism is common in foam cell formation, where SREBP-1a regulates lipid biosynthesis through transcriptional activation of lipogenic genes (27). Because PKM2 shRNA abrogated the Ox-LDL-induced significant increase in TC, FC, and CE, it can be concluded that PKM2 plays a role in Ox-LDL-induced macrophage foam cell formation. Because PKM2 inhibition augmented ABCA1 and ABCG1 protein expression and cholesterol efflux to apoA1 and HDL, it can be speculated that PKM2 regulates macrophage foam cell formation by promoting reverse cholesterol transport. LXR-α promotes HDL- and apoA1-mediated cholesterol efflux by directly increasing the transcription of ABCA1 and ABCG1 respectively in humans and mice (54–56). Although a significant decrease in LXR-α was observed with Ox-LDL, it is quite possible that PKM2 limits the effect of LXR-α on ABCA1 and ABCG1 expression because pharmacological inhibition of PKM2 further upregulates Ox-LDL-induced ABCA1 and ABCG1 expression and restores LXR-α expression. Moreover, the PKM2 activator, DASA-58, also downregulated LXR-α, ABCA1, and ABCG1 expression in macrophages. However, a role of other transcription factors cannot be ruled out. Although no change in SRB1 expression was observed with either PKM2 shRNA or Shikonin in Ox-LDL-treated cells, a significant increase was observed with DASA-58. Therefore, it can be speculated that the regulation of SRB1 by Ox-LDL is independent of PKM2.
An important step in foam cell formation is internalization of Ox-LDL by macrophage scavenger receptor CD36 (51). In the present study, PKM2 inhibition decreased Ox-LDL-induced CD36 expression and CD36-mediated binding and uptake of DiI-Ox-LDL in macrophages. Moreover, the PKM2 activator, DASA-58, increased the protein expression of CD36. Therefore, it can be speculated that PKM2 regulates foam cell formation by regulating CD36 expression and binding and uptake of Ox-LDL. Macrophages deficient in FASN showed decreased foam cell formation, and this involved upregulation of LXR-α and its downstream target ABCA1, increased apoA1-mediated cholesterol efflux, downregulation of scavenger receptor CD36, and decreased saturated fatty acid synthesis (24).
In the present study, Ox-LDL regulates FASN expression in a PKM2-dependent manner; however; detailed studies will be required to decipher the mechanistic role of FASN in the observed effects of PKM2.
Recently, it was reported that, in hepatocellular carcinoma, PKM2 interacts with nuclear SREBP-1a, and its enhanced phosphorylation at threonine 106 promotes SREBP-1a stabilization and increased transcription of lipogenic genes such as FASN (27). In the present study, because Ox-LDL induced a significant increase in PKM2-SREBP-1 interaction, it can be speculated that this interaction may regulate FASN expression.
The present study demonstrates the role of PKM2 in foam cell formation and inflammation. PKM2 interacts with Hif-1α and SREBP-1 promoting IL-1β and FASN expression, respectively. PKM2 regulates cholesterol uptake and efflux by affecting Ox-LDL binding, uptake, and expression of CD36, LXR-α, ABCA1, and ABCG1. Therefore, PKM2 can be an attractive target for preventing the foam cell formation and sterile inflammation observed during atherosclerosis.
Supplementary Material
Acknowledgments
The authors gratefully acknowledge the technical help provided by Mr. C. P. Pandey, Mrs. M. Chaturvedi, and Mr. A. L. Vishwakarma. CDRI communication no. 10036.
Footnotes
Abbreviations:
- BMDM
- bone marrow-derived macrophage
- CE
- cholesteryl ester
- DiI
- 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanine perchlorate
- ECAR
- extracellular acidification rate
- FC
- free cholesterol
- Hif-1α
- hypoxia-inducible factor-1α
- IL-1β
- interleukin 1β
- LDH
- lactate dehydrogenase
- LPDS
- lipoprotein-deficient serum
- OCR
- oxygen consumption rate
- Ox-LDL
- oxidized LDL
- PKM2
- pyruvate kinase M2
- SLC2A1
- glucose transporter member 1
- SRB1
- scavenger receptor class B type 1
- TC
- total cholesterol
This work was supported by the Council of Scientific and Industrial Research Project and Department of Biotechnology (DBT), India to M.K.B. M.K.B. is also supported by Ministry of Earth Sciences (MOES) and Department of Science and Technology (DST). A.K. and P.G. received fellowships from the University Grants Commission. The authors declare that they have no conflicts of interest with the contents of this article.
The online version of this article (available at https://www.jlr.org) contains a supplement.
REFERENCES
- 1.Tiwari R. L., Singh V., and Barthwal M. K.. 2008. Macrophages: an elusive yet emerging therapeutic target of atherosclerosis. Med. Res. Rev. 28: 483–544. [DOI] [PubMed] [Google Scholar]
- 2.Stewart C. R., Stuart L. M., Wilkinson K., Van Gils J. M., Deng J., Halle A., Rayner K. J., Boyer L., Zhong R., Frazier W. A., et al. 2010. CD36 ligands promote sterile inflammation through assembly of a Toll-like receptor 4 and 6 heterodimer. Nat. Immunol. 11: 155–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moore K. J., Sheedy F. J., and Fisher E. A.. 2013. Macrophages in atherosclerosis: a dynamic balance. Nat. Rev. Immunol. 13: 709–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Que X., Hung M. Y., Yeang C., Gonen A., Prohaska T. A., Sun X., Diehl C., Määttä A., Gaddis D. E., Bowden K., et al. 2018. Oxidized phospholipids are proinflammatory and proatherogenic in hypercholesterolaemic mice. Nature. 558: 301–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Janabi M., Yamashita S., Hirano K. I., Sakai N., Hiraoka H., Matsumoto K., Zhang Z., Nozaki S., and Matsuzawa Y.. 2000. Oxidized LDL-induced NF-κB activation and subsequent expression of proinflammatory genes are defective in monocyte-derived macrophages from CD36-deficient patients. Arterioscler. Thromb. Vasc. Biol. 20: 1953–1960. [DOI] [PubMed] [Google Scholar]
- 6.Hansson G. K., and Libby P.. 2006. The immune response in atherosclerosis: a double-edged sword. Nat. Rev. Immunol. 6: 508–519. [DOI] [PubMed] [Google Scholar]
- 7.Jiang Y., Wang M., Huang K., Zhang Z., Shao N., Zhang Y., Wang W., and Wang S.. 2012. Oxidized low-density lipoprotein induces secretion of interleukin-1β by macrophages via reactive oxygen species-dependent NLRP3 inflammasome activation. Biochem. Biophys. Res. Commun. 425: 121–126. [DOI] [PubMed] [Google Scholar]
- 8.Bekkering S., van den Munckhof I., Nielen T., Lamfers E., Dinarello C., Rutten J., de Graaf J., Joosten L. A. B., Netea M. G., Gomes M. E. R., et al. 2016. Innate immune cell activation and epigenetic remodeling in symptomatic and asymptomatic atherosclerosis in humans in vivo. Atherosclerosis. 254: 228–236. [DOI] [PubMed] [Google Scholar]
- 9.Semenova G., and Chernoff J.. 2012. PKM2 enters the morpheein academy. Mol. Cell. 45: 583–584. [DOI] [PubMed] [Google Scholar]
- 10.Shirai T., Nazarewicz R. R., Wallis B. B., Yanes R. E., Watanabe R., Hilhorst M., Tian L., Harrison D. G., Giacomini J. C., Assimes T. L., et al. 2016. The glycolytic enzyme PKM2 bridges metabolic and inflammatory dysfunction in coronary artery disease. J. Exp. Med. 213: 337–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Christofk H. R., Vander Heiden M. G., Harris M. H., Ramanathan A., Gerszten R. E., Wei R., Fleming M. D., Schreiber S. L., and Cantley L. C.. 2008. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. 452: 230–233. [DOI] [PubMed] [Google Scholar]
- 12.Palsson-McDermott E. M., Dyck L., Zasłona Z., Menon D., McGettrick A. F., Mills K. H. G., and O’Neill L. A.. 2017. Pyruvate kinase M2 is required for the expression of the immune checkpoint PD-L1 in immune cells and tumors. Front. Immunol. 8: 1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alquraishi M., Puckett D. L., Alani D. S., Humidat A. S., Frankel V. D., Donohoe D. R., Whelan J., and Bettaieb A.. 2019. Pyruvate kinase M2: a simple molecule with complex functions. Free Radic. Biol. Med. 143: 176–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hitosugi T., Kang S., Vander Heiden M. G., Chung T. W., Elf S., Lythgoe K., Dong S., Lonial S., Wang X., Chen G. Z., et al. 2009. Tyrosine phosphorylation inhibits PKM2 to promote the Warburg effect and tumor growth. Sci. Signal. 2: ra73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luo W., Hu H., Chang R., Zhong J., Knabel M., O’Meally R., Cole R. N., Pandey A., and Semenza G. L.. 2011. Pyruvate kinase M2 is a PHD3-stimulated coactivator for hypoxia-inducible factor 1. Cell. 145: 732–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Palsson-McDermott E. M., Curtis A. M., Goel G., Lauterbach M. A. R., Sheedy F. J., Gleeson L. E., van den Bosch M. W. M., Quinn S. R., Domingo-Fernandez R., Johnston D. G. W., et al. 2015. Pyruvate kinase M2 regulates Hif-1α activity and IL-1β induction and is a critical determinant of the Warburg effect in LPS-activated macrophages. Cell Metab. 21: 65–80. [Erratum. 2015. Cell Metab. 21: 347.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee S. J., Thien Quach C. H., Jung K-H., Paik J-Y., Lee J. H., Park J. W., and Lee K-H.. 2014. Oxidized low-density lipoprotein stimulates macrophage 18F-FDG uptake via hypoxia-inducible factor-1α activation through Nox2-dependent reactive oxygen species generation. J. Nucl. Med. 55: 1699–1705. [DOI] [PubMed] [Google Scholar]
- 18.Remmerie A., and Scott C. L.. 2018. Macrophages and lipid metabolism. Cell. Immunol. 330: 27–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ricote M., Valledor A. F., and Glass C. K.. 2004. Decoding transcriptional programs regulated by PPARs and LXRs in the macrophage: effects on lipid homeostasis, inflammation, and atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 24: 230–239. [DOI] [PubMed] [Google Scholar]
- 20.Nagy L., Tontonoz P., Alvarez J. G. A., Chen H., and Evans R. M.. 1998. Oxidized LDL regulates macrophage gene expression through ligand activation of PPARγ. Cell. 93: 229–240. [DOI] [PubMed] [Google Scholar]
- 21.Panasyuk G., Espeillac C., Chauvin C., Pradelli L. A., Horie Y., Suzuki A., Annicotte J. S., Fajas L., Foretz M., Verdeguer F., et al. 2012. PPARγ contributes to PKM2 and HK2 expression in fatty liver. Nat. Commun. 3: 672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rader D. J., Alexander E. T., Weibel G. L., Billheimer J., and Rothblat G. H.. 2009. The role of reverse cholesterol transport in animals and humans and relationship to atherosclerosis. J. Lipid Res. 50 (Suppl.): S189–S194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Phillips M. C. 2014. Molecular mechanisms of cellular cholesterol efflux. J. Biol. Chem. 289: 24020–24029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schneider J. G., Yang Z., Chakravarthy M. V., Lodhi I. J., Wei X., Turk J., and Semenkovich C. F.. 2010. Macrophage fatty-acid synthase deficiency decreases diet-induced atherosclerosis. J. Biol. Chem. 285: 23398–23409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jeon T. I., and Osborne T. F.. 2012. SREBPs: metabolic integrators in physiology and metabolism. Trends Endocrinol. Metab. 23: 65–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaplan M., Kerry R., Aviram M., and Hayek T.. 2008. High glucose concentration increases macrophage cholesterol biosynthesis in diabetes through activation of the sterol regulatory element binding protein 1 (SREBP1): inhibitory effect of insulin. J. Cardiovasc. Pharmacol. 52: 324–332. [DOI] [PubMed] [Google Scholar]
- 27.Zhao X., Zhao L., Yang H., Li J., Min X., Yang F., Liu J., and Huang G.. 2018. Pyruvate kinase M2 interacts with nuclear sterol regulatory element-binding protein 1a and thereby activates lipogenesis and cell proliferation in hepatocellular carcinoma. J. Biol. Chem. 293: 6623–6634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Amemiya-Kudo M., Shimano H., Hasty A. H., Yahagi N., Yoshikawa T., Matsuzaka T., Okazaki H., Tamura Y., Iizuka Y., Ohashi K., et al. 2002. Transcriptional activities of nuclear SREBP-1a, -1c, and -2 to different target promoters of lipogenic and cholesterogenic genes. J. Lipid Res. 43: 1220–1235. [PubMed] [Google Scholar]
- 29.Na Y. R., Gu G. J., Jung D., Kim Y. W., Na J., Woo J. S., Cho J. Y., Youn H., and Seok S. H.. 2016. GM-CSF induces inflammatory macrophages by regulating glycolysis and lipid metabolism. J. Immunol. 197: 4101–4109. [DOI] [PubMed] [Google Scholar]
- 30.Das R., Ganapathy S., Mahabeleshwar G. H., Drumm C., Febbraio M., Jain M. K., and Plow E. F.. 2013. Macrophage gene expression and foam cell formation are regulated by plasminogen. Circulation. 127: 1209–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tiwari R. L., Singh V., Singh A., Rana M., Verma A., Kothari N., Kohli M., Bogra J., Dikshit M., and Barthwal M. K.. 2014. PKCδ-IRAK1 axis regulates oxidized LDL-induced IL-1β production in monocytes. J. Lipid Res. 55: 1226–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rana M., Kumar A., Tiwari R. L., Singh V., Chandra T., Dikshit M., and Barthwal M. K.. 2016. IRAK regulates macrophage foam cell formation by modulating genes involved in cholesterol uptake and efflux. BioEssays. 38: 591–604. [DOI] [PubMed] [Google Scholar]
- 33.Havel R. J., Eder H. A., and Bragdon J. H.. 1955. The distribution and chemical composition of ultracentrifugally separated lipoproteins in human serum. J. Clin. Invest. 34: 1345–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Robinet P., Wang Z., Hazen S. L., and Smith J. D.. 2010. A simple and sensitive enzymatic method for cholesterol quantification in macrophages and foam cells. J. Lipid Res. 51: 3364–3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reis A., Rudnitskaya A., Blackburn G. J., Fauzi N. M., Pitt A. R., and Spickett C. M.. 2013. A comparison of five lipid extraction solvent systems for lipidomic studies of human LDL. J. Lipid Res. 54: 1812–1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yao S., Tian H., Miao C., Zhang D. W., Zhao L., Li Y., Yang N., Jiao P., Sang H., Guo S., et al. 2015. D4F alleviates macrophage-derived foam cell apoptosis by inhibiting CD36 expression and ER stress-CHOP pathway. J. Lipid Res. 56: 836–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Renaud J. F., Scanu A. M., Kazazoglou T., Lombet A., Romey G., and Lazdunski M.. 1982. Normal serum and lipoprotein-deficient serum give different expressions of excitability, corresponding to different stages of differentiation, in chicken cardiac cells in culture. Proc. Natl. Acad. Sci. USA. 79: 7768–7772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Al Gadban M. M., Smith K. J., Soodavar F., Piansay C., Chassereau C., Twal W. O., Klein R. L., Virella G., Lopes-Virella M. F., and Hammad S. M.. 2010. Differential trafficking of oxidized LDL and oxidized LDL immune complexes in macrophages: Impact on oxidative stress. PLoS One. 5: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nasri M., Karimi A., and Allahbakhshian Farsani M.. 2014. Production, purification and titration of a lentivirus-based vector for gene delivery purposes. Cytotechnology. 66: 1031–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu S., Huang Y., Xie Y., Lan T., Le K., Chen J., Chen S., Gao S., Xu X., Shen X., et al. 2010. Evaluation of foam cell formation in cultured macrophages: an improved method with Oil Red O staining and DiI-oxLDL uptake. Cytotechnology. 62: 473–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Harir N., Pecquet C., Kerenyi M., Sonneck K., Kovacic B., Nyga R., Brevet M., Dhennin I., Gouilleux-Gruart V., Beug H., et al. 2007. Constitutive activation of Stat5 promotes its cytoplasmic localization and association with PI3-kinase in myeloid leukemias. Blood. 109: 1678–1686. [DOI] [PubMed] [Google Scholar]
- 42.Zhang J., Cai S., Peterson B. R., Kris-Etherton P. M., and Vanden Heuvel J. P.. 2011. Development of a cell-based, high-throughput screening assay for cholesterol efflux using a fluorescent mimic of cholesterol. Assay Drug Dev. Technol. 9: 136–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rao X., Huang X., Zhou Z., and Lin X.. 2013. An improvement of the 2^(-delta delta CT) method for quantitative real-time polymerase chain reaction data analysis. Biostat. Bioinforma. Biomath. 3: 71–85. [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang J., Nuebel E., Wisidagama D. R. R., Setoguchi K., Hong J. S., Van Horn C. M., Imam S. S., Vergnes L., Malone C. S., Koehler C. M., et al. 2012. Measuring energy metabolism in cultured cells, including human pluripotent stem cells and differentiated cells. Nat. Protoc. 7: 1068–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vijayan V., Pradhan P., Braud L., Fuchs H. R., Gueler F., Motterlini R., Foresti R., and Immenschuh S.. 2019. Human and murine macrophages exhibit differential metabolic responses to lipopolysaccharide - a divergent role for glycolysis. Redox Biol. 22: 101147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Potter M., Newport E., and Morten K. J.. 2016. The Warburg effect: 80 years on. Biochem. Soc. Trans. 44: 1499–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yu X. H., Fu Y. C., Zhang D. W., Yin K., and Tang C. K.. 2013. Foam cells in atherosclerosis. Clin. Chim. Acta. 424: 245–252. [DOI] [PubMed] [Google Scholar]
- 48.van Tits L. J., Stienstra R., van Lent P. L., Netea M. G., Joosten L. A., and Stalenhoef A. F.. 2011. Oxidized LDL enhances pro-inflammatory responses of alternatively activated M2 macrophages: a crucial role for Krüppel-like factor 2. Atherosclerosis. 214: 345–349. [DOI] [PubMed] [Google Scholar]
- 49.Mazurek S., Boschek C. B., Hugo F., and Eigenbrodt E.. 2005. Pyruvate kinase type M2 and its role in tumor growth and spreading. Semin. Cancer Biol. 15: 300–308. [DOI] [PubMed] [Google Scholar]
- 50.Zhou Z., Li M., Zhang L., Zhao H., Sahin O., Chen J., Zhao J. J., Songyang Z., and Yu D.. 2018. Oncogenic kinase-induced PKM2 tyrosine 105 phosphorylation converts nononcogenic PKM2 to a tumor promoter and induces cancer stem-like cells. Cancer Res. 78: 2248–2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rahaman S. O., Lennon D. J., Febbraio M., Podrez E. A., Hazen S. L., and Silverstein R. L. L.. 2006. A CD36-dependent signaling cascade is necessary for macrophage foam cell formation. Cell Metab. 4: 211–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lv L., Li D., Zhao D., Lin R., Chu Y., Zhang H., Zha Z., Liu Y., Li Z., Xu Y., et al. 2011. Acetylation targets the M2 isoform of pyruvate kinase for degradation through chaperone-mediated autophagy and promotes tumor growth. Mol. Cell. 42: 719–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.O’Neill L. A. J., Kishton R. J., and Rathmell J.. 2016. A guide to immunometabolism for immunologists. Nat. Rev. Immunol. 16: 553–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Venkateswaran A., Laffitte B. A., Joseph S. B., Mak P. A., Wilpitz D. C., Edwards P. A., and Tontonoz P.. 2000. Control of cellular cholesterol efflux by the nuclear oxysterol receptor LXR alpha. Proc. Natl. Acad. Sci. USA. 97: 12097–12102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang N., Lan D., Chen W., Matsuura F., and Tall A. R.. 2004. ATP-binding cassette transporters G1 and G4 mediate cellular cholesterol efflux to high-density lipoproteins. Proc. Natl. Acad. Sci. USA. 101: 9774–9779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Teupser D., Kretzschmar D., Tennert C., Burkhardt R., Wilfert W., Fengler D., Naumann R., Sippel A. E., and Thiery J.. 2008. Effect of macrophage overexpression of murine liver X receptor-alpha (LXR-alpha) on atherosclerosis in LDL-receptor deficient mice. Arterioscler. Thromb. Vasc. Biol. 28: 2009–2015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.