ABSTRACT
Bile acid biotransformation is a collaborative effort by the host and the gut microbiome. Host hepatocytes synthesize primary bile acids from cholesterol. Once these host-derived primary bile acids enter the gastrointestinal tract, the gut microbiota chemically modify them into secondary bile acids. Interest into the gut-bile acid-host axis is expanding in diverse fields including gastroenterology, endocrinology, oncology, and infectious disease. This review aims to 1) describe the physiologic aspects of collaborative bile acid metabolism by the host and gut microbiota; 2) to evaluate how gut microbes influence bile acid pools, and in turn how bile acid pools modulate the gut microbial community structure; 3) to compare species differences in bile acid pools; and lastly, 4) discuss the effects of ursodeoxycholic acid (UDCA) administration, a common therapeutic bile acid, on the gut microbiota-bile acid-host axis.
KEYWORDS: C. difficile, bile acids, gut microbiota, ursodeoxycholic acid (UDCA), FXR
Introduction
Roughly a liter of bile is produced by human hepatocytes daily.1 Bile acids constitute about 50% of the organic component of bile.1 The primary bile acids produced in humans are cholic acid (CA) and chenodeoxycholic acid (CDCA). Once primary bile acids enter the gastrointestinal tract, over 50 chemically distinct secondary bile acids are produced.2 The chemical diversification of bile acids is a collaborative effort by the host (production of primary bile acids) and the gut microbiota (production of secondary bile acids). Some postulate that the gut microbiota act as an “endocrine organ” by altering host physiology via the production of metabolites, such as microbially derived secondary bile acids.3 Recently, interest in the gut microbiota-bile acid-host axis is expanding in diverse fields including gastroenterology, endocrinology, oncology, and infectious disease.1,3–12
Host bile acid metabolism
Bile acids are water-soluble, cholesterol derived amphipathic molecules of saturated hydroxylated C-24 sterols that are synthesized by hepatocytes.13 The liver is the only organ that contains all 14 enzymes that are required for de novo synthesis of bile acids.14,15 Cholic acid and CDCA are the main primary bile acids synthesized in humans and rodents (Figure 1).13,16,17 In rodents, a significant quantity of CDCA is converted by 6β-hydroxylation to muricholic acids, which are not detected in humans (Figure 1).3,18 These primary bile acids are further metabolized via N-acyl amidation to glycine or taurine producing conjugated bile acids, such as taurocholic acid (TCA).13 Conjugation is important for solubility of bile acids; at physiologic pH, unconjugated bile acids are only sparingly soluble.1,19 The ratio of glycine to taurine conjugated bile acids varies across mammals.16,20,21 Conjugated bile acids are actively secreted across the canalicular membranes into the bile ducts, which converge and empty into the gallbladder (except in species that lack a gallbladder such as horses, rats, and rabbits). Bile acids are just one component in bile, which also includes phospholipids, biliverdin/bilirubin, immunoglobulin A, mucus, various endogenous products (such as lipovitamins, corticosteroids, progesterone, testosterone), trace metals, and xenobiotics.1 Within the gallbladder, bile is concentrated 5–10 times, via removal of water and electrolytes, and acidified via Na+/H+ exchangers.1,22 Only about 50% of bile enters into the gallbladder, the remaining enters directly into the gastrointestinal tract.1 During a meal, cholecystokinin from the duodenum causes contraction of the gallbladder and concentrated bile is released into the proximal gastrointestinal tract.
Figure 1.
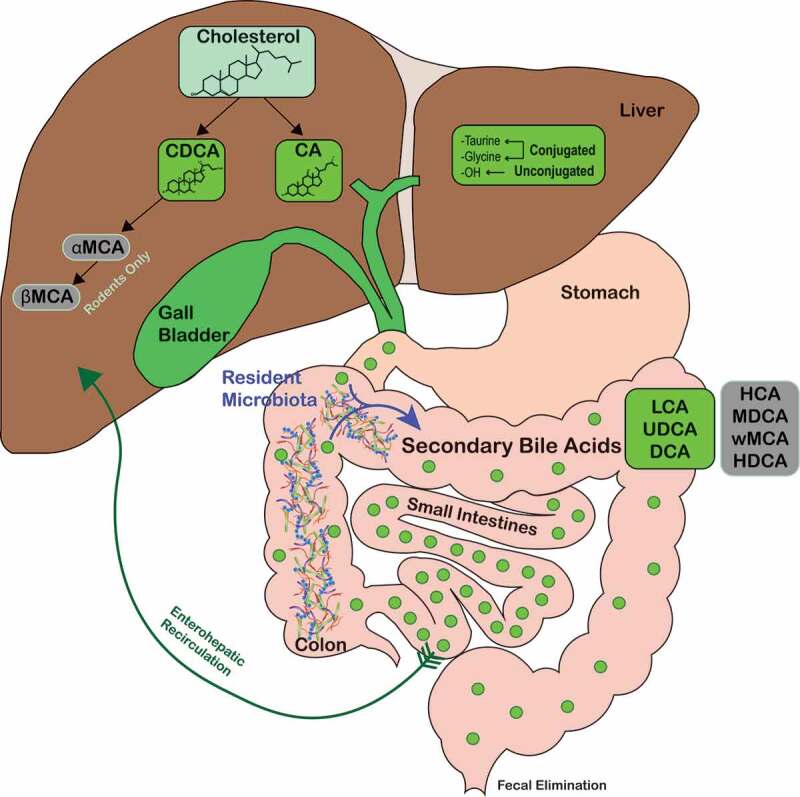
Overview of bile acid metabolism and enterohepatic recirculation.
Host-derived primary bile acids are synthesized by heptocytes (CA and CDCA in humans, and CA, CDCA, αMCA, and βMCA in rodents) and conjugated with either taurine or glycine. Primary bile acids are then secreted into the bile and stored in the gallbladder until secreted in the duodenum. Resident gut microbiota biotransform primary bile acids into secondary bile acids such as LCA, UDCA, and DCA (green shaded circles) and HCA, MDCA, ωMCA, and HDCA in rodents (gray-shaded circles). Abbreviations: CA, cholate; CDCA, chenodeoxycholate; DCA, deoxycholate; HCA, hyocholate; HDCA, hyodeoxycholate; LCA, lithocholate; MDCA, murideoxycholate; UDCA, ursodeoxycholate; αMCA, α-muricholate; βMCA, β-muricholate; ωMCA, ω-muricholate.
High concentrations of conjugated primary bile acids are noted within the duodenum, jejunum, and proximal ileum.13 The primary role of bile acids in the small intestine is to aid in fat emulsification and absorption. Bile acids undergo enterohepatic recirculation, a process which involves: (1). Passive absorption of conjugated and unconjugated bile acids in the small intestine and colon; (2) High-affinity active transport in the distal ileum.1,17,23 Absorbed bile acids enter into the portal bloodstream and are rapidly taken up by hepatocytes and resecreted into bile (Figure 1). A small fraction of bile acids escape enterohepatic recirculation and spill into systemic circulation, which allows bile signaling to occur in other organs and tissues.24,25 Enterohepatic recirculation is extremely efficient, with 95% of bile acids reabsorbed and only 5% lost into the feces.1 Hepatocytes maintain the bile acid pool by synthesizing bile acids to make up for fecal loss. In healthy humans, the total bile acid pool cycles about 10 times each day, which requires enterocytes and hepatocytes to transport about 20 g of bile acids every hour.5,26
Bile acids regulate their own synthesis and transport via the nuclear farnesoid X receptor (FXR; NR1H4), thus acting as hormones.27–30 Binding of bile acids to ileal FXR induces expression of fibroblast growth factor (FGF15/19).31 FGF15/19 travels via the portal bloodstream and binds to cell surface receptors on hepatocytes to repress bile acid synthesis (feedback inhibition) by inhibiting the rate-limiting enzyme cholesterol 7α- hydroxylase (CYP7A1) which allows the host to synthesize primary bile acids from cholesterol.28 Different bile acids have varying affinities to FXR.3,28,32 The bile acid-FXR pathway is also important for glucose homeostasis, lipid metabolism, protein synthesis, inflammation, and liver regeneration; however, this is beyond the scope of this review.6,27,28,33 Bile acids also interact with pregnane X receptors (PXR; NR1I2) and vitamin D receptors (VDR; NR1I1), which are reviewed elsewhere.4,28,34
Bile acids are biological detergents and can act as a host physicochemical defense system within the gut directly against both commensal resident microbes and enteric pathogens.1 Bile acids, via stimulation of FXR, can also induce expression of antimicrobial peptides.31 Therefore, bile acids are a major survival and colonization challenge to gut microbes. Regardless, gut commensals and some pathogens can elude the detrimental effects of bile acids and in some cases secure a fitness advantage.1
Diversification of host bile acids by gut microbes
The gastrointestinal microbiome is the most densely populated natural ecosystem and is comprised of over 1014 bacterial cells.14,35 Originally it was thought that microbial cells outnumbered human cells in the body by a ratio of 10:1;14,35 however, recent revised estimates suggest closer to 1.3–2.3:1.36 There are thousands of different bacterial species in the human gut.37 In healthy humans and animals, over 90–99% of the microbial community are dominated by two phyla, Firmicutes and Bacteroidetes, with fewer members in Proteobacteria, Actinobacteria, Verrucomicrobia, and Cyanobacteria.38,39 Diversity within this community is mainly at the genus, species, and strain level.14 It is estimated that 99% of functional genes in the human body are of microbial origin.40 By acting in an endocrine fashion, the gut microbiome can alter host physiology by producing metabolites, such as secondary bile acids.3,41
Secondary bile acids are produced by gut microbes via biotransformation of host-derived primary bile acids (Figure 2). When the small fraction (approximately 5%) of unabsorbed bile acids enters the distal ileum, cecum, and colon they undergo chemical diversification via three main microbial pathways: deconjugation, dehydrogenation, and dehydroxylation reactions.13
Figure 2.
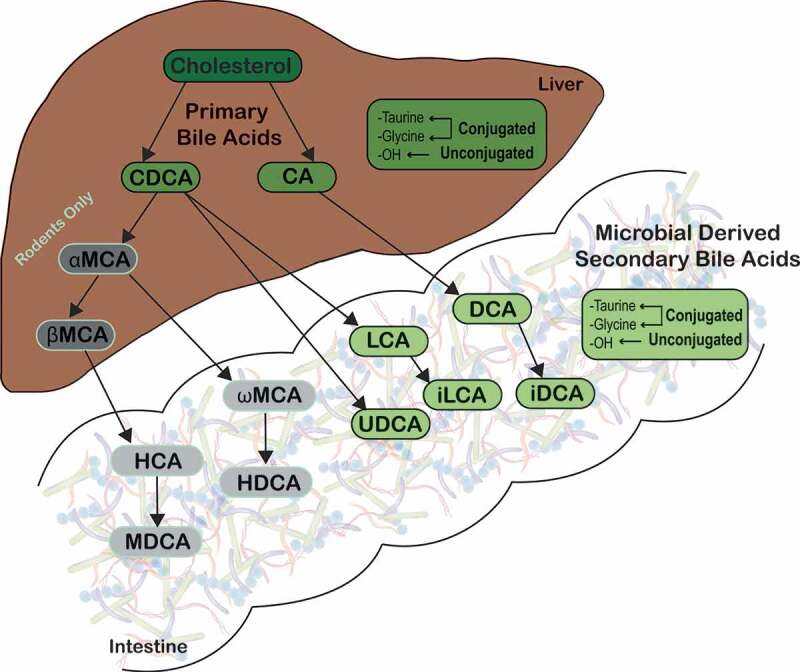
Microbial metabolism of host-derived primary bile acids.
Primary bile acids are synthesized from cholesterol by hepatocytes. Primary bile acids are then biotransformed by resident bacteria in the gastrointestinal tract to secondary bile acids, such as LCA and DCA, which predominate in humans. Abbreviations: CA, cholate; CDCA, chenodeoxycholate; DCA, deoxycholate; HCA, hyocholate; HDCA, hyodeoxycholate; LCA, lithocholate; MDCA, murideoxycholate; UDCA, ursodeoxycholate; αMCA, α-muricholate; βMCA, β-muricholate; ωMCA, ω-muricholate.
Deconjugation of host-derived primary bile acids occurs rapidly and via bile salt hydrolases (BSH), which are widespread in members of the gut microbiota.1,13,42 Based on metagenomic screening, three major phyla have BSHs: Firmicutes (30%), Bacteroidetes (14.4%), and Actinobacteria (8.9%).42 Within these phyla, the following genera are heavily studied: Clostridium, Bacteroides, Lactobacillus, Bifidobacterium, and Enterococcus (reviewed in Begley et al., 2005).1 The physiologic function of BSH is debated and the current three hypotheses are: (1) BSHs provide a nutritional advantage by liberating amino acids that can be used for carbon/nitrogen sources and energy generation via taurine as a terminal electron acceptor; (2) BSHs aid in incorporation of cholesterol and bile components into bacterial membranes; (3) BSHs provide a detoxification mechanism to diminish the inherent detergent properties of bile acids.1,43 Bile salt hydrolases appear to enhance bacterial colonization within the lower gastrointestinal tract, but appears to be strain specific.13 Some probiotic Lactobacillus strains have several BSH genes, thus highlighting the importance of deconjugation of bile acids for gut microbes.44–46 For the host, deconjugation of bile acids by gut microbes has several consequences. Unconjugated bile acids result in less effective emulsification of fat, less efficient enterohepatic recirculation of bile acids due to reduced distal ileal transporter affinity, and lowering of serum cholesterol levels.1,47,48 Despite the host impacts of microbial BSH activity, the bacterial physiologic advantage of BSHs remains elusive.13
Three distinct microbial hydroxysteroid dehydrogenases (HSDH), 3α-, 7α-, and 12α-, which result in oxidization and epimerization of specific hydroxyl groups on bile acids are present in gut microbes.13 These HSDHs can lead to the formation of over 20 diverse metabolites from the host-derived primary bile acids alone.13,14 Ridlon et al. speculated that these bile acid metabolites evolved as signaling molecules for microbes to communicate with other gut microbes (via microbe–microbe interactions) and/or alter host physiology (via microbe–host interactions).3
In the colon, nearly 100% of bile acids are microbially derived, and unconjugated bile acids undergo dehydrogenation carried out by a broad spectrum of bacteria.13,49 However, 7α-dehydroxylation is performed by only a few anaerobic species representing less than 0.025% of total gut microbiome and 0.0001% of total colonic microbiota.3,13,50 These are largely Clostridium spp. (C. hiranonis, C. hylemonae, C. sordelli, and C. scindens), which are members of the Firmicutes phyla.13,14,51–57 Removal of the 7α-hydroxyl group from host-derived primary bile acids requires multiple intracellular enzyme steps, which are encoded on the bai (bile acid inducible) operon.4,13,14,56,58,59 These reactions ultimately lead to formation of the secondary bile acids, deoxycholate (DCA) from CA and lithocholate (LCA) from CDCA (Figure 2).5,49,60 Additionally, DCA and LCA can be modified into other secondary bile acids by microbes, such as isoDCA (iDCA) and isoLCA (iLCA),49 however it is only partially understood which microbes conduct this conversion (Figures 2 and 3).5,60,62 Secondary bile acids can also undergo enterohepatic recirculation by passive colonic absorption and thus can be found in the liver and bile.3,23 In human feces, although secondary bile acids DCA and LCA predominate, there are over 50 different microbially derived secondary bile acids present.2
Figure 3.
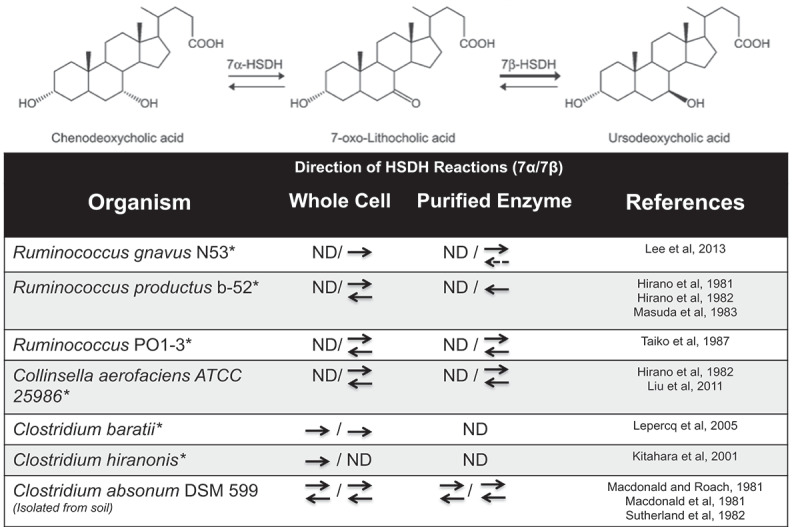
Directions of 7α/7β-HSDH reactions form UDCA-producing bacteria.
Epimerization reaction from CDCA to UDCA, which is catalyzed by the 7α-HSDH and 7β-HSDH enzymes. ND: Not determined; *: Found in the gastrointestinal tract. Dashed line denotes weak reaction. Figure modified from Lee et al. 2013.61
The microbial physiologic role of secondary bile acids remains elusive. It is suggested that microbially derived secondary bile acids are used in energy production as terminal electron acceptors, aid in formation of less membrane damaging bile acid pools, and alter enteric pathogen virulence, such as germination and outgrowth of Clostridioides difficile.1,9,13,63–65 In the host, secondary bile acids such as DCA and LCA can be cytotoxic molecules leading to oxidative stress, membrane damage, and colonic carcinogenesis (reviewed in Barrasa et al.).66 However, the secondary bile acid UDCA can protect colonic cells against apoptosis and oxidative damage.66 Additionally, in a colitis model, UDCA and LCA exerted anti-inflammatory properties.67 Thus, collectively highlighting the diverse and potentially divergent roles of microbially derived secondary bile acids. It is remarkable how little is known about secondary bile acids, including which gut microbes produce them, their microbial biologic function, and their impacts on host health and disease.
Gut microbial influence on composition of the host bile acid pools
When comparing gnotobiotic and conventional mice, bile acid pool size, composition, and concentrations are directly influenced by the gut microbiota and their metabolism of host-derived primary bile acids.68 The bile acid pools of conventional mice are more chemically diverse than gnotobiotic mice.68 The greatest diversification of bile acids was noted in biogeographic regions with dense and diverse microbial communities such as the cecum, colon, and feces.13,68
Disruptions in gut microbial communities result in derangements in bile acid metabolism.8,65,69 A classic example occurs after administration of antibiotics, which alter the gut microbiota community structure and function.18,65,68,70 Various antibiotics lead to rapid shifts in bile acid pools with host-derived primary bile acids predominating over microbially derived secondary bile acids in the large intestine and feces.18,65,70 This is not surprising since antibiotic treatment leads to depletion of 7α-dehydroxylation activity by the fecal microbiota.71 Subsequent bile acid dysmetabolism is also observed in liver cirrhosis and inflammatory bowel disease (IBD) patients with altered microbial ecosystems.8,72,73 Gut microbes are not only involved in formation of secondary bile acids but are also important for regulation of bile acid synthesis in hepatocytes.18 Therefore, it is not surprising that diseases, which alter the gut microbial ecosystem will also result in bile acid dysmetabolism.
Bile acid pool composition is modulated by interactions of microbially derived secondary bile acids with FXR.68 Recall, that bile acids differ in their activation of FXR (CDCA > DCA > LCA > CA), and thus, their ability to participate in feedback inhibition of host synthesis of primary bile acids.33 When comparing wild type and FXR-deficient mice raised conventionally versus rederived as gnotobiotic, the presence of the gut microbiota resulted in bile acid pools which were more potent activators of FXR and thus harbored more potential to inhibit host primary bile acid production.68 By altering host production of primary bile acids via FXR, the overall composition of the bile acid pool is modified.
The administration of probiotics also modifies the bile acid composition. Oral administration of Lactobacillus reuteri, a BSH-active microbe, resulted in increased intraluminal and circulating unconjugated bile acids, in addition to decreased inflammatory markers in hypercholesterolemic patients.74 Also, when mice were given VSL#3, comprised of eight different probiotics strains (Bifidobacterium breve, Bifidobacterium longum, Bifidobacterium infantis, Lactobacillus acidophilus, Lactobacillus plantarum, Lactobacillus paracasei, Lactobacillus bulgaricus, Streptococcus thermophiles), an increase in fecal bile acid deconjugation, fecal excretion of bile acids, and hepatic bile acid synthesis were noted.29 Using FXR- and FGF15-deficient mice, it was demonstrated that these changes in bile acid metabolism were dependent on the presence of the FXR-FGF system.29
These examples provide evidence that bile acid composition is influenced and dependent on the presence of the gut microbiota. Researchers propose that development of targeted microbial therapies could be used to manipulate host bile acid pools.75 For example, by coupling the bile acid metabolism of bacteria with the bai operon (found in C. scindens and C. hiranonis) to biotransform CA into DCA, one could then use other bacteria (such as Eggerthella lenta and Ruminococcus gnavus) to biotransform DCA into iso-DCA, which is considered to be less toxic to the host.49 Additional studies to orchestrate collaborative bile acid metabolism could provide novel therapeutic strategies in diseases associated with bile acid dysmetabolism, such as metabolic disease, obesity, IBD, and infectious enteric diseases such as C. difficile infection.3,4,7,8,50,65,73,76
Impact of bile acids on the gut microbiota community structure
As demonstrated, bile acid pools are a function of collaborative metabolism of the host and the gut microbiota. Resultant chemically diverse bile acids may function to directly shape the gut microbial community structure. Due to their lipophilic nature, bile acids display antimicrobial activity with bacterial membranes being their main targets.1 In addition to membrane damage, bile acids impose a fitness challenge to gut microbes through disruption of macromolecule stability by interfering with RNA secondary structures, causing DNA damage and promoting protein misfolding (reviewed in Begley et al. and more recently in Bustos et al.).1,43
Evidence of the direct antimicrobial effects of bile acids can be gleaned from murine models of biliary obstruction, which exhibit dramatic gut microbial community proliferation and increased bacterial translocation.31,77 These effects can be ameliorated with administration of bile acids resulting in inhibition of bacterial overgrowth.31 Bile acids also have indirect antimicrobial effects via FXR-induced antimicrobial peptide production and FXR-induced regulation of the host immune response.78,79
Significant changes in the microbiota community structure have been described in rats that were fed the primary bile acid, CA.80 CA supplemented in the feed resulted in expansion of the Firmicutes from 54% relative abundance in controls to 93–98% in CA-fed rats. This mainly consisted of an increase in Clostridium spp. from 39% relative abundance in controls to 70% in CA-fed rats.80 In general, a decrease in bile acid pools appears to favor Gram-negative outgrowth, which are capable of producing lipopolysaccharide (LPS) and some members harbor pathogenic potential.4,14,23,80 In contrast, outgrowth of Gram-positive Firmicutes, including some with 7α-dehydroxylation capabilities, is observed with increased bile acid pools and thus promotes secondary bile acid production.4,14,23,73,80
Another example of bile acids shaping the gut microbial ecosystem is in relation to colonization resistance against the enteric pathogen C. difficile. In humans with recurrent C. difficile infection (CDI), bile acid dysmetabolism, represented by an increase in primary bile acids and a decrease in secondary bile acids, is documented.81 Fecal microbiota transplantation (FMT), which leads to restoration of bile acid pools, particularly the microbially derived secondary bile acids, results in a 95% cure rate in recurrent CDI patients.81,82 Secondary bile acids directly inhibit spore germination and outgrowth of C. difficle.65,83,84 Administration of C. scindens, a 7α-dehydroxylating bacterium, partially restored colonization resistance against CDI in mice.56 Furthermore, administration of UDCA (Ursodiol) to a human patient with recurrent C. difficile ileal pouchitis prevented recurrence of CDI.85 Although the exact mechanisms of colonization resistance are unknown, it is apparent that secondary bile acids play an important role.7
It is evident that bile acids can alter the gut microbiota community structure through various mechanisms. In turn, the gut microbial communities can modulate bile acid pools, thus highlighting the interconnectedness of the gut microbiota-bile acid-host axis. Dysfunction in any of these components will have ramifications on the others. Numerous disease states are affected by the gut microbiota-bile acid-host axis and investigation into novel therapeutic strategies to modulate this axis are needed.1,3–12
Differences in bile acids profile between species
A major limitation in bile acid research is differences in bile acid profiles and metabolism among animals, therefore making it challenging to compare across species.5,16,60 A key example is the dramatic differences in UDCA in various species. This bile acid is considered to be the primary bile acid made by hepatocytes in bears, nutria, and beavers.16,60,86,87 However, UDCA has long been considered a secondary bile acid that is microbially synthesized in the gut of humans and rodents.15,86,88 Endogenous UDCA can be formed in two ways: (1) Epimerization of 7α-hydroxyl by 7α-hydroxysteroid dehydrogenase and 7β-hydroxysteroid dehydrogenase found in single species such as Ruminococcus gnavus, Clostridium absonum (only found in soil), or Clostridium baratii; or (2) By two separate bacterial species expressing one or the other HSDH enzyme (Figure 3).27,52,55,61,88–98 It remains unknown what additional gut microbes are also capable of converting CDCA into 7-oxo-LCA and then into UDCA.
Although the majority (>90%) of bile acid profiles in murine liver and bile are composed of muricholic acid (MCA) and CA, there is also evidence of UDCA and conjugated UDCA (with glycine and taurine).99 Recently, several murine studies reported that after antibiotic treatment there is an increase in total UDCA (combination of UDCA + taurine conjugated, TUDCA) in the large intestine and liver.18,68 Since antibiotic administration significantly reduces gut microbial populations, it was suggested that UDCA might be a primary bile acid synthesized by murine hepatocytes. This was further supported in gnotobiotic mice, which have TUDCA present in the absence of the microbiota.18,68 However, studies in conventional and gnotobiotic rats revealed hepatic UDCA only in conventional rats with intact gut microbiota and no TUDCA (which would be host conjugated).100 Unfortunately, this study did not evaluate bile acid profiles from intestinal luminal contents or feces.
Another key finding is that murine bile acid synthesis may be under potent positive feedback control via MCA and UDCA acting as antagonists of FXR/FGF15 system.101,102 Therefore, MCA and UDCA may counter the negative feedback inhibition effects of CDCA and CA on FXR/FGF15 in mice.101 It remains unclear if positive feedback on bile acid synthesis occurs in humans.
These studies highlight the inter-species differences in bile acid composition and abundance. Since there are differences in bile acid profiles and feedback responses between mice and humans, researchers have questioned if studies on the murine gut microbiota-bile acid-host axis can be utilized to gain insight into human diseases.6,103 Currently, this is a limitation of bile acid metabolism research, and the physiologic significance of inter-species variations remains unclear.
Ursodeoxycholic acid modulation of microbiota-bile acid-host axis
Bile from animals, including bear bile, have been extensively used therapeutically in China for over 2500 years.60 Globally, close to 1000 metric tons of UDCA are produced annually for pharmaceutical and nutraceutical use.86 Originally UDCA was obtained from bear bile, which is a primary bile acid in this species.60 Today, the most common formulation of UDCA is Ursodiol, which is chemically synthesized from CA.104 There are various other trade names globally for this product including Actigall, Deursil, and Urso. Currently, the Food and Drug Administration (FDA) approved formulation of UDCA, Ursodiol, is used to treat a variety of diseases including: cholesterol gallstones, primary biliary cirrhosis, primary sclerosing cholangitis, nonalcoholic fatty liver disease, chronic viral hepatitis C, recurrent colonic adenomas, cholestasis of pregnancy, and recurrent pancreatitis.3,102,105–112 UDCA has vast beneficial effects (anticholestatic, antifibrotic, antiproliferative, and anti-inflammatory) but the major effect on bile acid physiology is an increase in the hydrophilic bile acid pool by diluting the concentration of hydrophobic toxic secondary bile acids, DCA and LCA.3,113
In healthy humans administered UDCA (15 mg/kg/day) for 3 weeks, biliary and duodenal bile acid concentrations of UDCA and its conjugates (GUDCA and TUDCA) increased by 40% compared to baseline.114 A decrease in primary bile acids (CA and CDCA) and their conjugates as well as a decrease in the secondary bile acid DCA and its conjugates (GDCA and TDCA) was observed within biliary and duodenal bile.114 An increase in conjugates of the secondary bile acid LCA (GLCA and TLCA) was observed after UDCA treatment within biliary and duodenal bile samples.114 The gut microbiota was not evaluated in this study. As observed in the above study, LCA can increase during UDCA administration. This is thought to be from microbial conversion of UDCA to the hydrophobic toxic bile acid, LCA.88 Not only does this limit the UDCA available to provide beneficial therapeutic effects, it could be potentially harmful to the host, not to mention the gut microbiota. It has recently been suggested that pharmacological inhibition of microbial baiI gene, a component of the bai operon, could limit microbial conversion of UDCA into LCA and thus increase availability of UDCA.3 Impacts of higher concentrations of UDCA on the host and gut microbiota have not been investigated.
UDCA is incompletely absorbed in the small intestine, thus only a fraction reaches the large intestine where the greatest consortium of microbes are present.13,115,116 Several formulations of UDCA have been developed to try to increase colonic delivery, including conjugation with glutamate, nanosuspensions/nanoparticles, and microbial derivation.116–118 The UDCA-glutamate prevents absorption and biotransformation in the small intestine but takes advantage of the peptide bond cleavage within the colonic brush boarder enzymes to remove the glutamate thus allowing for colonic delivery of UDCA.116 UDCA nanosuspensions encapsulate this bile acid and allow for increased colonic transit time.117,118 UDCA could also be microbially derived from administration of UDCA-producing bacteria, such as Clostridium baratii or Ruminococcus gnavus N53 (Figure 3).61,90 Additional research is required to investigate the use of UDCA-producing bacteria in vivo. Lastly, 24-nor-UDCA is a shortened side chain of UDCA that undergoes chole-hepatic shunting, meaning that it passes through cholangiocytes into sinusoids via periductular capillary plexus.119,120 This formulation of UDCA is thought to enhance ductal targeting of UDCA administration. Effects of 24-nor-UDCA on bile acid profiles and the gut microbiota need to be characterized.
It remains unclear if and how exogenously administered UDCA shapes the gut microbial community structure and/or intestinal and fecal bile acid metabolome. It is evident that UDCA can alter liver and biliary bile acid pools, but intestinal contents and feces have not been fully evaluated. The host-derived primary bile acids within the small intestine are substrates for the microbially derived secondary bile acids. Therefore, characterization of this bile acid pool is imperative to accurately reflect physiologic effects of administering bile acids, such as UDCA. Further studies employing multi-omics approaches are required to investigate the impact of UDCA administration on the gut microbial community structure, bile acid metabolome, and the host.
Conclusions
In conclusion, there are two main sites of bile acid biosynthesis: hepatocytes within the host and microbes within the gastrointestinal tract. Bile acid pool diversity is attributed to a collaborative metabolism of the host and gut microbiota. As demonstrated, microbially derived secondary bile acids can not only affect the composition and function of the gut microbiota but also can modulate host physiology. Thus, secondary bile acids are hypothesized to be inter-kingdom signaling molecules.3 By understanding the dynamic intricacies in the gut microbiota-bile acid-host axis, insight into a variety of disease states (such as enteric pathogenesis, metabolic disease, obesity, IBD, and other chronic inflammatory conditions) may be elucidated. Expanding our knowledge may contribute to precision medicine through the development of novel targeted therapeutic strategies to manipulate the gut microbiota-bile acid-host axis during disease states with the ultimate goal of restoring health to the host and gastrointestinal ecosystem.
Funding Statement
JAW was funded by the Ruth L. Kirschstein National Research Service Award Research Training grant T32OD011130 by NIH. CMT is funded by the National Institute of General Medical Sciences of the National Institutes of Health under award number R35GM119438.
Disclosure of potential conflicts of interest
CMT is a scientific advisor to Locus Biosciences, a company engaged in the development of antimicrobial technologies. CMT is a consultant for Vedanta Biosciences.
References
- 1.Begley M, Gahan CG, Hill C.. The interaction between bacteria and bile. FEMS Microbiol Rev. 2005;29(4):625–651. doi: 10.1016/j.femsre.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 2.Setchell KD, Lawson AM, Tanida N, Sjovall J. General methods for the analysis of metabolic profiles of bile acids and related compounds in feces. J Lipid Res. 1983;24:1085–1100. [PubMed] [Google Scholar]
- 3.Ridlon JM, Bajaj JS. The human gut sterolbiome: bile acid-microbiome endocrine aspects and therapeutics. Acta Pharmaceutica Sinica B. 2015;5(2):99–105. doi: 10.1016/j.apsb.2015.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fiorucci S, Distrutti E. Bile acid-activated receptors, intestinal microbiota, and the treatment of metabolic disorders. Trends Mol Med. 2015;21:702–714. doi: 10.1016/j.molmed.2015.09.001. [DOI] [PubMed] [Google Scholar]
- 5.Kuipers F, Bloks VW, Groen AK. Beyond intestinal soap–bile acids in metabolic control. Nat Rev Endocrinol. 2014;10(8):488–498. doi: 10.1038/nrendo.2014.60. [DOI] [PubMed] [Google Scholar]
- 6.Nie YF, Hu J, Yan XH. Cross-talk between bile acids and intestinal microbiota in host metabolism and health. J Zhejiang Univ Sci B. 2015;16(6):436–446. doi: 10.1631/jzus.B1400327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Theriot CM, Young VB. Interactions between the gastrointestinal microbiome and clostridium difficile. Annu Rev Microbiol. 2015;69:445–461. doi: 10.1146/annurev-micro-091014-104115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duboc H, Rajca S, Rainteau D, Benarous D, Maubert M-A, Quervain E, Thomas G, Barbu V, Humbert L, Despras G, et al. Connecting dysbiosis, bile-acid dysmetabolism and gut inflammation in inflammatory bowel diseases. Gut. 2013;62(4):531–539. doi: 10.1136/gutjnl-2012-302578. [DOI] [PubMed] [Google Scholar]
- 9.Shen A. A gut odyssey: the impact of the microbiota on clostridium difficile spore formation and germination. PLoS Pathog. 2015;11:e1005157. doi: 10.1371/journal.ppat.1005157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gu Y, Wang X, Li J, Zhang Y, Zhong H, Liu R, Zhang D, Feng Q, Xie X, Hong J, et al. Analyses of gut microbiota and plasma bile acids enable stratification of patients for antidiabetic treatment. Nat Commun. 2017;8(1785). doi: 10.1038/s41467-017-01682-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wahlstrom A, Kovatcheva-Datchary P, Stahlman M, Backhed F, Marschall HU. Crosstalk between bile acids and gut microbiota and its impact on farnesoid X receptor signalling. Dig Dis. 2017;35(3):246–250. doi: 10.1159/000450982. [DOI] [PubMed] [Google Scholar]
- 12.Staley C, Weingarden AR, Khoruts A, Sadowsky MJ. Interaction of gut microbiota with bile acid metabolism and its influence on disease states. Appl Microbiol Biotechnol. 2017;101(1):47–64. doi: 10.1007/s00253-016-8006-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ridlon JM, Kang DJ, Hylemon PB. Bile salt biotransformations by human intestinal bacteria. J Lipid Res. 2006;47(241–259). doi: 10.1194/jlr.R500013-JLR200. [DOI] [PubMed] [Google Scholar]
- 14.Ridlon JM, Alves JM, Hylemon PB, Bajaj JS. Cirrhosis, bile acids and gut microbiota: unraveling a complex relationship. Gut Microbes. 2013;4(5):382–387. doi: 10.4161/gmic.25723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Russell DW. The enzymes, regulation, and genetics of bile acid synthesis. Annu Rev Biochem. 2003;72(1):137–174. doi: 10.1146/annurev.biochem.72.121801.161712. [DOI] [PubMed] [Google Scholar]
- 16.Hofmann AF, Hagey LR, Krasowski MD. Bile salts of vertebrates: structural variation and possible evolutionary significance. J Lipid Res. 2010;51(2):226–246. doi: 10.1194/jlr.R000042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hofmann AF. The enterohepatic circulation of bile acids in mammals: form and functions. Front Biosci. 2009;14:2584–2598. doi: 10.2741/3399. [DOI] [PubMed] [Google Scholar]
- 18.Zhang Y, Limaye PB, Renaud HJ, Klaassen CD. Effect of various antibiotics on modulation of intestinal microbiota and bile acid profile in mice. Toxicol Appl Pharmacol. 2014;277(2):138–145. doi: 10.1016/j.taap.2014.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hofmann AF, Mysels KJ. Bile acid solubility and precipitation in vitro and in vivo: the role of conjugation, pH, and Ca2+ ions. J Lipid Res. 1992;33:617–626. [PubMed] [Google Scholar]
- 20.Hofmann AF. Bile acids: trying to understand their chemistry and biology with the hope of helping patients. Hepatology. 2009;49(5):1403–1418. doi: 10.1002/hep.22789. [DOI] [PubMed] [Google Scholar]
- 21.Hofmann AF, Hagey LR. Bile acids: chemistry, pathochemistry, biology, pathobiology, and therapeutics. Cell Mol Life Sci. 2008;65(7–8):1220–1236. doi: 10.1007/s00018-008-7568-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hofmann AF. Bile acids: the good, the bad, and the ugly. News Physiol Sci. 1999;14:24–29. [DOI] [PubMed] [Google Scholar]
- 23.Chiang JY. Bile acids: regulation of synthesis. J Lipid Res. 2009;50(10):1955–1966. doi: 10.1194/jlr.R900010-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chiang JY. Bile acid metabolism and signaling. Compr Physiol. 2013;3(1191–1212). doi: 10.1002/cphy.c120023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wagner M, Trauner M. Recent advances in understanding and managing cholestasis. F1000Research. 2016;5:705. doi: 10.12688/f1000research.8012.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stellaard F, Sackmann M, Sauerbruch T, Paumgartner G. Simultaneous determination of cholic acid and chenodeoxycholic acid pool sizes and fractional turnover rates in human serum using 13C-labeled bile acids. J Lipid Res. 1984;25:1313–1319. [PubMed] [Google Scholar]
- 27.Kliewer SA, Mangelsdorf DJ. Bile acids as hormones: the FXR-FGF15/19 pathway. Dig Dis. 2015;33(3):327–331. doi: 10.1159/000371670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee FY, Lee H, Hubbert ML, Edwards PA, Zhang Y. FXR, a multipurpose nuclear receptor. Trends Biochem Sci. 2006;31(10):572–580. doi: 10.1016/j.tibs.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 29.Degirolamo C, Rainaldi S, Bovenga F, Murzilli S, Moschetta A. Microbiota modification with probiotics induces hepatic bile acid synthesis via downregulation of the Fxr-Fgf15 axis in mice. Cell Rep. 2014;7(12–18). doi: 10.1016/j.celrep.2014.02.032. [DOI] [PubMed] [Google Scholar]
- 30.Brestoff JR, Artis D. Commensal bacteria at the interface of host metabolism and the immune system. Nat Immunol. 2013;14(7):676–684. doi: 10.1038/ni.2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Inagaki T, Moschetta A, Lee Y-K, Peng L, Zhao G, Downes M, Yu RT, Shelton JM, Richardson JA, Repa JJ, et al. Regulation of antibacterial defense in the small intestine by the nuclear bile acid receptor. Proc Natl Acad Sci U S A. 2006;103:3920–3925. doi: 10.1073/pnas.0509592103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schaap FG, Trauner M, Jansen PL. Bile acid receptors as targets for drug development. Nat Rev Gastroenterol Hepatol. 2014;11(1):55–67. doi: 10.1038/nrgastro.2013.151. [DOI] [PubMed] [Google Scholar]
- 33.de Aguiar Vallim TQ, Tarling EJ, Edwards PA. Pleiotropic roles of bile acids in metabolism. Cell Metab. 2013;17(657–669). doi: 10.1016/j.cmet.2013.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Degirolamo C, Modica S, Palasciano G, Moschetta A. Bile acids and colon cancer: solving the puzzle with nuclear receptors. Trends Mol Med. 2011;17(10):564–572. doi: 10.1016/j.molmed.2011.05.010. [DOI] [PubMed] [Google Scholar]
- 35.Savage DC. Microbial ecology of the gastrointestinal tract. Annu Rev Microbiol. 1977;31(1):107–133. doi: 10.1146/annurev.mi.31.100177.000543. [DOI] [PubMed] [Google Scholar]
- 36.Sender R, Fuchs S, Milo R. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol. 2016;14(8):e1002533. doi: 10.1371/journal.pbio.1002533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci U S A. 2007;104:13780–13785. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA. Diversity of the human intestinal microbial flora. Science. 2005;308(5728):1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 2006;124(4):837–848. doi: 10.1016/j.cell.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 40.Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464(59–65). doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.O’Hara AM, Shanahan F. The gut flora as a forgotten organ. EMBO Rep. 2006;7(688–693). doi: 10.1038/sj.embor.7400731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jones BV, Begley M, Hill C, Gahan CG, Marchesi JR. Functional and comparative metagenomic analysis of bile salt hydrolase activity in the human gut microbiome. Proc Natl Acad Sci U S A. 2008;105:13580–13585. doi: 10.1073/pnas.0804437105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bustos AY, de Valdez F, Fadda G, Taranto MP. New insights into bacterial bile resistance mechanisms: the role of bile salt hydrolase and its impact on human health. Food Res Int. 2018;112(250–262):250–262. doi: 10.1016/j.foodres.2018.06.035. [DOI] [PubMed] [Google Scholar]
- 44.Kleerebezem M, Boekhorst J, van Kranenburg R, Molenaar D, Kuipers OP, Leer R, Tarchini R, Peters SA, Sandbrink HM, Fiers MWEJ, et al. Complete genome sequence of Lactobacillus plantarum WCFS1. Proc Natl Acad Sci U S A. 2003;100:1990–1995. doi: 10.1073/pnas.0337704100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pridmore RD, Berger B, Desiere F, Vilanova D, Barretto C, Pittet A-C, Zwahlen M-C, Rouvet M, Altermann E, Barrangou R, et al. The genome sequence of the probiotic intestinal bacterium Lactobacillus johnsonii NCC 533. Proc Natl Acad Sci U S A. 2004;101:2512–2517. doi: 10.1073/pnas.0307327101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.O’Flaherty S, Briner Crawley A, Theriot CM, Barrangou R. The lactobacillus bile salt hydrolase repertoire reveals niche-specific adaptation. mSphere. 2018:3. doi: 10.1128/mSphere.00140-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.De Smet I, Van Hoorde L, De Saeyer N, Vande Woestyne M, Verstraete W. In vitro study of Bile Salt Hydrolase (BSH) activity of BSH Isogenic Lactobacillus plantarum 80 strains and estimation of cholesterol lowering through enhanced BSH activity. 2009. doi: 10.3109/08910609409141371. [DOI] [Google Scholar]
- 48.Chikai T, Nakao H, Uchida K. Deconjugation of bile acids by human intestinal bacteria implanted in germ-free rats. Lipids. 1987;22:669–671. doi: 10.1007/bf02533948. [DOI] [PubMed] [Google Scholar]
- 49.Devlin AS, Fischbach MA. A biosynthetic pathway for a prominent class of microbiota-derived bile acids. Nat Chem Biol. 2015;11(9):685–690. doi: 10.1038/nchembio.1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ridlon JM, Kang D-J, Hylemon PB, Bajaj JS. Gut microbiota, cirrhosis, and alcohol regulate bile acid metabolism in the gut. Dig Dis. 2015;33(3):338–345. doi: 10.1159/000371678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wells JE, Williams KB, Whitehead TR, Heuman DM, Hylemon PB. Development and application of a polymerase chain reaction assay for the detection and enumeration of bile acid 7alpha-dehydroxylating bacteria in human feces. Clin Chim Acta. 2003;331:127–134. doi: 10.1016/s0009-8981(03)00115-3. [DOI] [PubMed] [Google Scholar]
- 52.Kitahara M, Takamine F, Imamura T, Benno Y. Clostridium hiranonis sp. nov., a human intestinal bacterium with bile acid 7alpha-dehydroxylating activity. Int J Syst Evol Microbiol. 2001;51:39–44. doi: 10.1099/00207713-51-1-39. [DOI] [PubMed] [Google Scholar]
- 53.Kitahara M, Takamine F, Imamura T, Benno Y. Assignment of Eubacterium sp. VPI 12708 and related strains with high bile acid 7alpha-dehydroxylating activity to Clostridium scindens and proposal of Clostridium hylemonae sp. nov., isolated from human faeces. Int J Syst Evol Microbiol. 2000;50 Pt 3:971–978. doi: 10.1099/00207713-50-3-971. [DOI] [PubMed] [Google Scholar]
- 54.Ridlon JM, Kang DJ, Hylemon PB. Isolation and characterization of a bile acid inducible 7alpha-dehydroxylating operon in Clostridium hylemonae TN271. Anaerobe. 2010;16:137–146. doi: 10.1016/j.anaerobe.2009.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Macdonald IA, White BA, Hylemon PB. Separation of 7 alpha- and 7 beta-hydroxysteroid dehydrogenase activities from Clostridium absonum ATCC# 27555 and cellular response of this organism to bile acid inducers. J Lipid Res. 1983;24:1119–1126. [PubMed] [Google Scholar]
- 56.Buffie CG, Bucci V, Stein RR, McKenney PT, Ling L, Gobourne A, No D, Liu H, Kinnebrew M, Viale A, et al. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature. 2015;517:205–208. doi: 10.1038/nature13828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dawson JA, Mallonee DH, Bjorkhem I, Hylemon PB. Expression and characterization of a C24 bile acid 7 alpha-dehydratase from Eubacterium sp. strain VPI 12708 in Escherichia coli. J Lipid Res. 1996;37:1258–1267. [PubMed] [Google Scholar]
- 58.Ridlon JM, Kang DJ, Hylemon PB, Bajaj JS. Bile acids and the gut microbiome. Curr Opin Gastroenterol. 2014;30(3):332–338. doi: 10.1097/MOG.0000000000000057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Doerner KC, Takamine F, LaVoie CP, Mallonee DH, Hylemon PB. Assessment of fecal bacteria with bile acid 7 alpha-dehydroxylating activity for the presence of bai-like genes. Appl Environ Microbiol. 1997;63:1185–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang DQ, Carey MC. Therapeutic uses of animal biles in traditional Chinese medicine: an ethnopharmacological, biophysical chemical and medicinal review. World J Gastroenterol. 2014;20(9952–9975). doi: 10.3748/wjg.v20.i29.9952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lee JY, Arai H, Nakamura Y, Fukiya S, Wada M, Yokota A. Contribution of the 7β-hydroxysteroid dehydrogenase from Ruminococcus gnavus N53 to ursodeoxycholic acid formation in the human colon. J Lipid Res. 2013;54:3062–3069. doi: 10.1194/jlr.M039834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zheng -M-M, Wang R-F, Li C-X, Xu J-H. Two-step enzymatic synthesis of ursodeoxycholic acid with a new 7β-hydroxysteroid dehydrogenase from Ruminococcus torques. Process Biochem. 2015;50:598–604. doi: 10.1016/j.procbio.2014.12.026. [DOI] [Google Scholar]
- 63.Mallonee DH, Hylemon PB. Sequencing and expression of a gene encoding a bile acid transporter from Eubacterium sp. strain VPI 12708. J Bacteriol. 1996;178:7053–7058. doi: 10.1128/jb.178.24.7053-7058.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sorg JA, Sonenshein AL. Bile salts and glycine as cogerminants for Clostridium difficile spores. J Bacteriol. 2008;190:2505–2512. doi: 10.1128/JB.01765-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Theriot CM, Koenigsknecht MJ, Carlson PE Jr, Hatton GE, Nelson AM, Li B, Huffnagle GB, Li J, Young VB. Antibiotic-induced shifts in the mouse gut microbiome and metabolome increase susceptibility to Clostridium difficile infection. Nat Commun. 2014;5:3114. doi: 10.1038/ncomms4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Barrasa JI, Olmo N, Lizarbe MA, Turnay J. Bile acids in the colon, from healthy to cytotoxic molecules. Toxicol In Vitro. 2013;27(2):964–977. doi: 10.1016/j.tiv.2012.12.020. [DOI] [PubMed] [Google Scholar]
- 67.Ward JBJ, Lajczak NK, Kelly OB, O'Dwyer AM, Giddam AK, Ní Gabhann J, Franco P, Tambuwala MM, Jefferies CA, Keely S, et al. Ursodeoxycholic acid and lithocholic acid exert anti-inflammatory actions in the colon. American journal of physiology. Gastrointestinal Liver Physiol. 2017;ajpgi.00256.02016. doi: 10.1152/ajpgi.00256.2016. [DOI] [PubMed] [Google Scholar]
- 68.Sayin SI, Wahlström A, Felin J, Jäntti S, Marschall H-U, Bamberg K, Angelin B, Hyötyläinen T, Orešič M, Bäckhed F. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab. 2013;17(2):225–235. doi: 10.1016/j.cmet.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 69.Jones ML, Tomaro-Duchesneau C, Prakash S. The gut microbiome, probiotics, bile acids axis, and human health. Trends Microbiol. 2014;22(6):306–308. doi: 10.1016/j.tim.2014.04.010. [DOI] [PubMed] [Google Scholar]
- 70.Zhao Y, Wu J, Li JV, Zhou N-Y, Tang H, Wang Y. Gut microbiota composition modifies fecal metabolic profiles in mice. J Proteome Res. 2013;12(6):2987–2999. doi: 10.1021/pr400263n. [DOI] [PubMed] [Google Scholar]
- 71.Samuel P, Holtzman CM, Meilman E, Sekowski I. Effect of neomycin and other antibiotics on serum cholesterol levels and on 7alpha-dehydroxylation of bile acids by the fecal bacterial flora in man. Circ Res. 1973;33:393–402. doi: 10.1161/01.res.33.4.393. [DOI] [PubMed] [Google Scholar]
- 72.Kakiyama G, Pandak WM, Gillevet PM, Hylemon PB, Heuman DM, Daita K, Takei H, Muto A, Nittono H, Ridlon JM, et al. Modulation of the fecal bile acid profile by gut microbiota in cirrhosis. J Hepatol. 2013;58(5):949–955. doi: 10.1016/j.jhep.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Devkota S, Chang EB. Interactions between diet, bile acid metabolism, gut microbiota, and inflammatory bowel diseases. Dig Dis. 2015;33(3):351–356. doi: 10.1159/000371687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jones ML, Martoni CJ, Prakash S. Letter to the editor regarding the report of Duboc et al: connecting dysbiosis, bile-acid dysmetabolism and gut inflammation in inflammatory bowel disease. Gut. 2013;62:654–655. doi: 10.1136/gutjnl-2012-303867. [DOI] [PubMed] [Google Scholar]
- 75.Foley MH, O’Flaherty S, Barrangou R, Theriot CM. Bile salt hydrolases: gatekeepers of bile acid metabolism and host-microbiome crosstalk in the gastrointestinal tract. PLoS Pathog. 2019;15(3):e1007581. doi: 10.1371/journal.ppat.1007581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Reeves AE, Theriot CM, Bergin IL, Huffnagle GB, Schloss PD, Young VB. The interplay between microbiome dynamics and pathogen dynamics in a murine model of Clostridium difficile Infection. Gut Microbes. 2014;2:145–158. doi: 10.4161/gmic.2.3.16333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Watanabe M, Fukiya S, Yokota A. Comprehensive evaluation of the bactericidal activities of free bile acids in the large intestine of humans and rodents. J Lipid Res. 2017;58(6):1143–1152. doi: 10.1194/jlr.M075143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Vavassori P, Mencarelli A, Renga B, Distrutti E, Fiorucci S. The bile acid receptor FXR is a modulator of intestinal innate immunity. J Immunol. 2009;183(6251–6261). doi: 10.4049/jimmunol.0803978. [DOI] [PubMed] [Google Scholar]
- 79.Cipriani S, Mencarelli A, Chini MG, Distrutti E, Renga B, Bifulco G, Baldelli F, Donini A, Fiorucci S, Ryffel B. The bile acid receptor GPBAR-1 (TGR5) modulates integrity of intestinal barrier and immune response to experimental colitis. PLoS One. 2011;6(10):e25637. doi: 10.1371/journal.pone.0025637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Islam KB, Fukiya S, Hagio M, Fujii N, Ishizuka S, Ooka T, Ogura Y, Hayashi T, Yokota A. Bile acid is a host factor that regulates the composition of the cecal microbiota in rats. Gastroenterology. 2011;141(5):1773–1781. doi: 10.1053/j.gastro.2011.07.046. [DOI] [PubMed] [Google Scholar]
- 81.Weingarden AR, Chen C, Bobr A, Yao D, Lu Y, Nelson VM, Sadowsky MJ, Khoruts A. Microbiota transplantation restores normal fecal bile acid composition in recurrent Clostridium difficile infection. Am J Physiol Gastrointest Liver Physiol. 2014;306:G310–319. doi: 10.1152/ajpgi.00282.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.van Nood E, Vrieze A, Nieuwdorp M, Fuentes S, Zoetendal EG, de Vos WM, Visser CE, Kuijper EJ, Bartelsman JFWM, Tijssen JGP, et al. Duodenal infusion of donor feces for recurrent Clostridium difficile. N Engl J Med. 2013;368:407–415. doi: 10.1056/NEJMoa1205037. [DOI] [PubMed] [Google Scholar]
- 83.Sorg JA, Sonenshein AL. Inhibiting the initiation of Clostridium difficile spore germination using analogs of chenodeoxycholic acid, a bile acid. J Bacteriol. 2010;192:4983–4990. doi: 10.1128/jb.00610-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sorg JA, Sonenshein AL. Chenodeoxycholate is an inhibitor of Clostridium difficile spore germination. J Bacteriol. 2009;191:1115–1117. doi: 10.1128/jb.01260-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Weingarden AR, Chen C, Zhang N, Graiziger CT, Dosa PI, Steer CJ, Shaughnessy MK, Johnson JR, Sadowsky MJ, Khoruts A.. Ursodeoxycholic acid inhibits clostridium difficile spore germination and vegetative growth, and prevents the recurrence of ileal pouchitis associated with the infection. J Clin Gastroenterol. 2015. doi: 10.1097/mcg.0000000000000427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hofmann AF, Hagey LR. Key discoveries in bile acid chemistry and biology and their clinical applications: history of the last eight decades. J Lipid Res. 2014;55(1553–1595). doi: 10.1194/jlr.R049437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hagey LR, Crombie DL, Espinosa E, Carey MC, Igimi H, Hofmann AF. Ursodeoxycholic acid in the Ursidae: biliary bile acids of bears, pandas, and related carnivores. J Lipid Res. 1993;34:1911–1917. [PubMed] [Google Scholar]
- 88.Takamine F, Imamura T. 7 beta-dehydroxylation of 3,7-dihydroxy bile acids by a Eubacterium species strain C-25 and stimulation of 7 beta-dehydroxylation by Bacteroides distasonis strain K-5. Microbiol Immunol. 1985;29:1247–1252. doi: 10.1111/j.1348-0421.1985.tb00915.x. [DOI] [PubMed] [Google Scholar]
- 89.White BA, Fricke RJ, Hylemon PB. 7 beta-Dehydroxylation of ursodeoxycholic acid by whole cells and cell extracts of the intestinal anaerobic bacterium, Eubacterium species V.P.I. 12708. J Lipid Res. 1982;23:145–153. [PubMed] [Google Scholar]
- 90.Lepercq P, Hermier D, David O, Michelin R, Gibard C, Beguet F, Relano P, Cayuela C, Juste C. Increasing ursodeoxycholic acid in the enterohepatic circulation of pigs through the administration of living bacteria. Br J Nutr. 2005;93:457–469. doi: 10.1079/bjn20041386. [DOI] [PubMed] [Google Scholar]
- 91.Hirano S, Masuda N. Epimerization of the 7-hydroxy group of bile acids by the combination of two kinds of microorganisms with 7 alpha-and 7 beta-hydroxysteroid dehydrogenase activity, respectively. J Lipid Res. 1981;22:1060–1068. [PubMed] [Google Scholar]
- 92.Hirano S, Masuda N. Characterization of NADP-dependent 7 beta-hydroxysteroid dehydrogenases from Peptostreptococcus productus and Eubacterium aerofaciens. Appl Environ Microbiol. 1982;43:1057–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Masuda N, Oda H, Tanaka H. Purification and characterization of NADP-dependent 7β-hydroxysteroid dehydrogenase from Peptostreptococcus productus strain b-52. Biochim Biophys Acta, Gen Subj. 1983;755:65–69. doi: 10.1016/0304-4165(83)90273-8. [DOI] [PubMed] [Google Scholar]
- 94.Taiko A, Teruaki A, KOBASHI K. Purification and characterization of 7β-hydroxysteroid dehydrogenase from ruminococus sp. of human intestine. J Biochem. 1987;102:613–619. doi: 10.1093/oxfordjournals.jbchem.a122095. [DOI] [PubMed] [Google Scholar]
- 95.Liu L, Aigner A, Schmid RD. Identification, cloning, heterologous expression, and characterization of a NADPH-dependent 7β-hydroxysteroid dehydrogenase from collinsella aerofaciens. Appl Microbiol Biotechnol. 2011;90:127–135. doi: 10.1007/s00253-010-3052-y. [DOI] [PubMed] [Google Scholar]
- 96.Macdonald IA, Hutchison DM, Forrest TP. Formation of urso-and ursodeoxy-cholic acids from primary bile acids by. Clostridium Absonum. J Lipid Res. 1981;22:458–466. [PubMed] [Google Scholar]
- 97.Macdonald IA, Roach PD. Bile salt induction of 7α-and 7β-hydroxysteroid dehydrogenases in Clostridium absonum. Biochim Biophys Acta, Lipids Lipid Metab. 1981;665:262–269. doi: 10.1016/0005-2760(81)90011-4. [DOI] [PubMed] [Google Scholar]
- 98.Sutherland JD, Macdonald IA. The metabolism of primary, 7-oxo, and 7 beta-hydroxy bile acids by Clostridium absonum. J Lipid Res. 1982;23:726–732. [PubMed] [Google Scholar]
- 99.Alnouti Y, Csanaky IL, Klaassen CD. Quantitative-profiling of bile acids and their conjugates in mouse liver, bile, plasma, and urine using LC-MS/MS. J Chromatogr B Analyt Technol Biomed Life Sci. 2008;873(2):209–217. doi: 10.1016/j.jchromb.2008.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Swann JR, Want EJ, Geier FM, Spagou K, Wilson ID, Sidaway JE, Nicholson JK, Holmes E. Systemic gut microbial modulation of bile acid metabolism in host tissue compartments. Proc Natl Acad Sci U S A. 2011;108 Suppl 1:4523–4530. doi: 10.1073/pnas.1006734107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Rudling M, Bonde Y. Stimulation of apical sodium-dependent bile acid transporter expands the bile acid pool and generates bile acids with positive feedback properties. Dig Dis. 2015;33(3):376–381. doi: 10.1159/000371690. [DOI] [PubMed] [Google Scholar]
- 102.Mueller M, Thorell A, Claudel T, Jha P, Koefeler H, Lackner C, Hoesel B, Fauler G, Stojakovic T, Einarsson C, et al. Ursodeoxycholic acid exerts farnesoid X receptor-antagonistic effects on bile acid and lipid metabolism in morbid obesity. J Hepatol. 2015;62(1398–1404). doi: 10.1016/j.jhep.2014.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Pavlidis P, Powell N, Vincent RP, Ehrlich D, Bjarnason I, Hayee B. Systematic review: bile acids and intestinal inflammation-luminal aggressors or regulators of mucosal defence? Aliment Pharmacol Ther. 2015;42:802–817. doi: 10.1111/apt.13333. [DOI] [PubMed] [Google Scholar]
- 104.Kanazawa T, Shimazaki A, Sato T, Hoshino T. Syntheses of Ursodesoxycholic Acid and Its Conjugated Bile Acid. Proc Jpn Acad. 1954;30:391–392. doi: 10.2183/pjab1945.30.391. [DOI] [Google Scholar]
- 105.Ikegami T, Matsuzaki Y. Ursodeoxycholic acid: mechanism of action and novel clinical applications. Hepatol Res. 2008;38(123–131). doi: 10.1111/j.1872-034X.2007.00297.x. [DOI] [PubMed] [Google Scholar]
- 106.Fischer S, Müller I, Zündt BZ, Jüngst C, Meyer G, Jüngst D. Ursodeoxycholic acid decreases viscosity and sedimentable fractions of gallbladder bile in patients with cholesterol gallstones. Eur J Gastroenterol Hepatol. 2004;16:305–311. doi: 10.1097/00042737-200403000-00010. [DOI] [PubMed] [Google Scholar]
- 107.Tsubakio K, Kiriyama K, Matsushima N, Taniguchi M, Shizusawa T, Katoh T, Manabe N, Yabu M, Kanayama Y, Himeno S. Autoimmune pancreatitis successfully treated with ursodeoxycholic acid. Intern Med. 2002;41:1142–1146. doi: 10.2169/internalmedicine.41.1142. [DOI] [PubMed] [Google Scholar]
- 108.Sinakos E, Wang J, Fu PP, Sharma S, Nagalingam A, Mells J, Handy J, Page AJ, Cohen C, Anania FA, et al. Bile acid changes after high-dose ursodeoxycholic acid treatment in primary sclerosing cholangitis: relation to disease progression. Hepatology. 2010;52(197–203). doi: 10.1002/hep.23631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Poupon RE, Bonnand AM, Queneau PE, Trépo C, JPí Z, Vetter D, Raabe JJ, Thieffin G, Larrey D, Grangé JD, et al. Randomized trial of interferon-alpha plus ursodeoxycholic acid versus interferon plus placebo in patients with chronic hepatitis C resistant to interferon. Scand J Gastroenterol. 2000;35:642–649. doi: 10.1080/003655200750023624. [DOI] [PubMed] [Google Scholar]
- 110.Serfaty L, De Leusse A, Rosmorduc O, Desaint B, Flejou JF, Chazouilleres O, Poupon RE, Poupon R. Ursodeoxycholic acid therapy and the risk of colorectal adenoma in patients with primary biliary cirrhosis: an observational study. Hepatology. 2003;38(1):203–209. doi: 10.1053/jhep.2003.50311. [DOI] [PubMed] [Google Scholar]
- 111.Carey EJ, Ali AH, Lindor KD. Primary biliary cirrhosis. Lancet. 2015;386(1565–1575). doi: 10.1016/s0140-6736(15)00154-3. [DOI] [PubMed] [Google Scholar]
- 112.Zhang L, Liu X-H, Qi H-B, Li Z, Fu X-D, Chen L, Shao Y. Ursodeoxycholic acid and S-adenosylmethionine in the treatment of intrahepatic cholestasis of pregnancy: a multi-centered randomized controlled trial. Eur Rev Med Pharmacol Sci. 2015;19:3770–3776. [PubMed] [Google Scholar]
- 113.Copaci I, Micu L, Iliescu L, Voiculescu M. New therapeutical indications of ursodeoxycholic acid. Rom J Gastroenterol. 2005;14:259–266. [PubMed] [Google Scholar]
- 114.Dilger K, Hohenester S, Winkler-Budenhofer U, Bastiaansen BAJ, Schaap FG, Rust C, Beuers U. Effect of ursodeoxycholic acid on bile acid profiles and intestinal detoxification machinery in primary biliary cirrhosis and health. J Hepatol. 2012;57(1):133–140. doi: 10.1016/j.jhep.2012.02.014. [DOI] [PubMed] [Google Scholar]
- 115.Crosignani A, Setchell KD, Invernizzi P, Larghi A, Rodrigues CM, Podda M. Clinical pharmacokinetics of therapeutic bile acids. Clin Pharmacokinet. 1996;30:333–358. doi: 10.2165/00003088-199630050-00002. [DOI] [PubMed] [Google Scholar]
- 116.Asciutti S, Castellani D, Nardi E, Morelli O, Clementi M, Chistolini F, Gentili G, Setchell KDR, O’Connell N, Pellicciari R, et al. A new amino acid derivative of ursodeoxycholate, (N-L-Glutamyl)-UDCA (UDCA-Glu), to selectively release UDCA in the colon. Anticancer Res. 2009;29:4971–4979. [PubMed] [Google Scholar]
- 117.Ma YQ, Zhang ZZ, Li G, Zhang J, Xiao HY, Li XF. Solidification drug nanosuspensions into nanocrystals by freeze-drying: a case study with ursodeoxycholic acid. Pharm Dev Technol. 2014;1–9. doi: 10.3109/10837450.2014.982822. [DOI] [PubMed] [Google Scholar]
- 118.De AK, Sana S, Datta S, Mukherjee A. Protective efficacy of ursodeoxycholic acid nanoparticles in animal model of inflammatory bowel disease. J Microencapsul. 2014;31(8):725–737. doi: 10.3109/02652048.2014.918666. [DOI] [PubMed] [Google Scholar]
- 119.Trauner M, Halilbasic E, Claudel T, Steinacher D, Fuchs C, Moustafa T, Pollheimer M, Krones E, Kienbacher C, Traussnigg S, et al. Potential of nor-Ursodeoxycholic Acid in Cholestatic and Metabolic Disorders. Dig Dis. 2015;33(3):433–439. doi: 10.1159/000371904. [DOI] [PubMed] [Google Scholar]
- 120.Fickert P, Pollheimer MJ, Silbert D, Moustafa T, Halilbasic E, Krones E, Durchschein F, Thüringer A, Zollner G, Denk H, et al. Differential effects of norUDCA and UDCA in obstructive cholestasis in mice. J Hepatol. 2013;58:1201–1208. doi: 10.1016/j.jhep.2013.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]


