Key Points
Question
What was the initial experience in Singapore with the outbreak of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)?
Findings
In this descriptive case series of the first 18 patients diagnosed with SARS-CoV-2 infection in Singapore between January 23 and February 3, 2020, clinical presentation was a respiratory tract infection with prolonged viral shedding from the nasopharynx of 7 days or longer in 15 patients (83%). Supplemental oxygen was required in 6 patients (33%), 5 of whom were treated with lopinavir-ritonavir, with variable clinical outcomes following treatment.
Meaning
These findings provide clinical features and course among patients diagnosed with SARS-CoV-2 infection in Singapore.
Abstract
Importance
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) emerged in Wuhan, China, in December 2019 and has spread globally with sustained human-to-human transmission outside China.
Objective
To report the initial experience in Singapore with the epidemiologic investigation of this outbreak, clinical features, and management.
Design, Setting, and Participants
Descriptive case series of the first 18 patients diagnosed with polymerase chain reaction (PCR)–confirmed SARS-CoV-2 infection at 4 hospitals in Singapore from January 23 to February 3, 2020; final follow-up date was February 25, 2020.
Exposures
Confirmed SARS-CoV-2 infection.
Main Outcomes and Measures
Clinical, laboratory, and radiologic data were collected, including PCR cycle threshold values from nasopharyngeal swabs and viral shedding in blood, urine, and stool. Clinical course was summarized, including requirement for supplemental oxygen and intensive care and use of empirical treatment with lopinavir-ritonavir.
Results
Among the 18 hospitalized patients with PCR-confirmed SARS-CoV-2 infection (median age, 47 years; 9 [50%] women), clinical presentation was an upper respiratory tract infection in 12 (67%), and viral shedding from the nasopharynx was prolonged for 7 days or longer among 15 (83%). Six individuals (33%) required supplemental oxygen; of these, 2 required intensive care. There were no deaths. Virus was detectable in the stool (4/8 [50%]) and blood (1/12 [8%]) by PCR but not in urine. Five individuals requiring supplemental oxygen were treated with lopinavir-ritonavir. For 3 of the 5 patients, fever resolved and supplemental oxygen requirement was reduced within 3 days, whereas 2 deteriorated with progressive respiratory failure. Four of the 5 patients treated with lopinavir-ritonavir developed nausea, vomiting, and/or diarrhea, and 3 developed abnormal liver function test results.
Conclusions and Relevance
Among the first 18 patients diagnosed with SARS-CoV-2 infection in Singapore, clinical presentation was frequently a mild respiratory tract infection. Some patients required supplemental oxygen and had variable clinical outcomes following treatment with an antiretroviral agent.
This case series describes the epidemiologic features, clinical presentation, treatment, and outcomes of the first 18 patients with confirmed coronavirus disease 2019 (COVID-19) in Singapore
Introduction
The third novel coronavirus in 17 years emerged in Wuhan, China, in December 2019.1 Phylogenetics has indicated that severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is closely related to bat-derived SARS-like coronaviruses.2 Early reports from Wuhan described the associated coronavirus disease 2019 (COVID-19) as a SARS-like atypical pneumonia in which 26% to 33% of patients required intensive care and 4% to 15% died.1,3,4 A large case series of 72 314 infected individuals has since refined these initial estimates in China to severe disease in 14% and a case-fatality rate of 2.3%.5
On January 23, 2020, the first imported SARS-CoV-2 infection in Singapore was detected in a visitor from Wuhan. Subsequently, COVID-19 has been diagnosed among other visitors and returning travelers, and from limited local transmission.6
While a proven effective antiviral treatment in COVID-19 is not available, the antiretroviral drug lopinavir-ritonavir has been proposed, as its potential effectiveness in the treatment of SARS was suggested in 2 case series.7,8 In Middle East respiratory syndrome, lopinavir-ritonavir also showed potential activity in marmosets,9 but in a mouse model it did not reduce virus titer or lung pathology, although it improved lung function.10
This case series describes the epidemiologic features, clinical presentation, treatment, and outcomes of the first 18 patients in Singapore with confirmed COVID-19.
Methods
Outbreak Response
On January 2, 2020, after first reports of an outbreak of atypical pneumonia in Wuhan, China, the Singapore Ministry of Health issued a health alert that patients with pneumonia and recent travel to Hubei Province should be screened for SARS-CoV-2 infection. All individuals with suspected SARS-CoV-2 infection were isolated with airborne and contact precautions, and attending staff wore personal protective equipment in accordance with the US Centers for Disease Control and Prevention guidelines.11 Extensive contact tracing followed by quarantine of asymptomatic contacts and hospital isolation and screening of symptomatic contacts was strictly enforced.
Data and Specimen Collection
Individuals confirmed to have COVID-19 by SARS-CoV-2 real-time reverse transcriptase–polymerase chain reaction (RT-PCR) were eligible for inclusion in this study (eMethods in the Supplement). Data were collected at the 4 hospitals that provided care for these patients.
Waiver of informed consent for collection of clinical data from infected individuals was granted by the Ministry of Health, Singapore, under the Infectious Diseases Act as part of the COVID-19 outbreak investigation. Written informed consent was obtained from study participants for collection of biological samples after review and approval of the study protocol by the institutional ethics committee.
Data from electronic health records were summarized using a standardized data collection form. Two researchers independently reviewed the data collection forms for accuracy.
Specimens (blood, stool, and urine samples; nasopharyngeal swabs) were collected at multiple time points in the first 2 weeks following study enrollment and tested by RT-PCR for the presence of SARS-CoV-2. RT-PCR cycle threshold values were collected. The cycle threshold value correlates with the number of copies of the virus in a biological sample, in an inversely proportional and exponential manner. Sequencing of PCR products of the RNA-dependent RNA polymerase (RdRp) gene were used to construct phylogenetic trees (eMethods in the Supplement).
Clinical Management
As part of standard of care, complete blood cell count, tests of kidney and liver function, and measurement of C-reactive protein and lactate dehydrogenase levels were performed. Respiratory samples were tested for influenza and other respiratory viruses with a multiplex PCR assay.
All patients received supportive therapy, including supplemental oxygen when saturations as measured by pulse oximeter dropped below 92%. Patients clinically suspected of having community-acquired pneumonia were administered empirical broad-spectrum antibiotics and oral oseltamivir. Coformulated lopinavir-ritonavir (400 mg/100 mg twice daily orally for up to 14 days) was prescribed to selected patients at the treating physicians’ discretion after shared decision-making and provision of oral informed consent. Corticosteroids were avoided, reflecting increased mortality with their use in severe influenza.12
Respiratory samples were sent daily for SARS-CoV-2 PCR on clinical recovery. Deisolation was contingent on at least 2 consecutive negative PCR assay results more than 24 hours apart.
Results
Epidemiologic Features
Between January 23 and February 3, 2020, 18 patients infected with SARS-CoV-2 were diagnosed in Singapore, with symptom onset from January 14 to January 30, 2020. All patients reported travel to Wuhan, China, in the 14 days prior to illness onset (eTable 1 in the Supplement). Four patients (22%) were identified through contact tracing, while 3 (17%) were identified through border screening. Of the 18 patients, 16 (89%) were Chinese nationals, while 2 (11%) were Singapore residents. There were 5 clusters comprising family, traveling companions, or other close contacts (eFigure 1 in the Supplement). Contact tracing of the 18 patients identified a total of 264 close contacts in Singapore (eFigure 2 in the Supplement). As of February 25, 2020, no infections had been detected among health care workers involved in the care of patients with COVID-19.
Clinical Features
Clinical features are summarized in the Table. Fever (13 [72%]), cough (15 [83%]), and sore throat (11 [61%]) were common symptoms. Rhinorrhea was infrequent (1 [6%]), while 6 patients (33%) had an abnormal chest radiograph finding or lung crepitations. No patients presented with a severe acute respiratory distress syndrome, and only 1 required immediate supplemental oxygen. Lymphopenia (<1.1 ×109/L) was present in 7 of 16 patients (39%) and an elevated C-reactive protein level (>20 mg/L) in 6 of 16 (38%), while kidney function remained normal.
Table. Clinical Features of Patients Infected With SARS-CoV-2.
| All patients (N = 18) | Did not require supplemental O2 (n = 12) | Required supplemental O2 (n = 6) | |
|---|---|---|---|
| Demographics | |||
| Age, median (range) y | 47 (31-73) | 37 (31-56) | 56 (47-73) |
| Male sex, No. (%) | 9 (50) | 7 (58) | 2 (33) |
| Any comorbidity, No. (%)a | 5 (28) | 1 (8) | 4 (67) |
| Signs and symptoms on presentation, No. (%) | |||
| Fever | 13 (72) | 7 (58) | 6 (100) |
| Cough | 15 (83) | 10 (83) | 5 (83) |
| Shortness of breath | 2 (11) | 1 (8) | 1 (17) |
| Rhinorrhea | 1 (6) | 1 (8) | 0 |
| Sore throat | 11 (61) | 8 (67) | 3 (50) |
| Diarrhea | 3 (17) | 3 (25) | 0 |
| Vital signs at presentation, median (range) | |||
| Temperature, °C | 37.7 (36.1-39.6) | 38.3 (36.6-39.6) | 37.7 (36.1-38.1) |
| Respiratory rate, breaths/min | 18 (16-21) | 18 (17-19) | 20 (16-21) |
| Pulse oximeter O2 saturation, % | 98 (95-100) | 98 (95-100) | 97 (95-98) |
| Systolic blood pressure, mm Hg | 131 (103-167) | 131 (104-167) | 136 (103-141) |
| Heart rate, /min | 97 (75-118) | 99 (75-118) | 91 (78-102) |
| Baseline investigations, median (range) | |||
| WBCs, ×109/L | 4.6 (1.7-6.3) | 4.6 (1.7-6.3) | 3.4 (2.6-5.8) |
| Hemoglobin, g/dL | 13.5 (11.7-17.2) | 13.9 (11.7-17.2) | 13.2 (11.7-14) |
| Platelets, ×109/L | 159 (116-217) | 159 (128-213) | 156 (116-217) |
| Neutrophils, ×109/L | 2.7 (0.7-4.5) | 2.8 (0.7-4.5) | 1.8 (1.2-3.7) |
| Lymphocytes, ×109/L | 1.2 (0.8-1.7) | 1.2 (0.8-1.6) | 1.1 (0.8-1.7) |
| C-reactive protein, mg/L (n = 16) | 16.3 (0.9-97.5) | 11.1 (0.9-19.1) | 65.6 (47.5-97.5) |
| LDH, U/L (n = 13) | 512 (285-796) | 424 (285-748) | 550 (512-796) |
| Abnormal chest radiograph, No. (%) | 6 (33) | 3 (25) | 3 (50) |
| Duration of symptoms, median (range) | |||
| Fever, d | 4 (0-15) | 1 (0-7) | 5 (4-15) |
| Any symptoms, d | 13 (5-24) | 12 (5-24) | 16 (10-20) |
Abbreviations: LDH, lactate dehydrogenase; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; WBC, white blood cell.
SI conversion factor: To convert LDH values to μkat/L, multiply by 0.0167.
Group not requiring supplemental oxygen: hypertension, diabetes. Group requiring supplemental oxygen: hypertension (n = 4), type 2 diabetes (n = 1), hyperlipidemia (n = 1).
The clinical course was uncomplicated for 12 patients (67%), but 6 patients (33%) desaturated and required supplemental oxygen (Figure 1). Chest radiographs showed no pulmonary opacities at presentation in 12 patients (67%) and remained clear throughout the acute illness in 9 (50%). Three patients with initially normal chest radiograph findings developed bilateral diffuse airspace opacities; of these, 2 had been persistently febrile for more than 1 week. Two individuals (11%) required admission to the intensive care unit because of increasing supplemental oxygen requirements, and 1 (6%) required mechanical ventilation. No concomitant bacterial or viral infections were detected, and there were no deaths as of February 25, 2020.
Figure 1. Time Course of Symptoms, Supplemental Oxygen Requirements, Hospital Admission, and Discharge of Patients Infected With SARS-CoV-2.
ICU indicates intensive care unit; PCR, polymerase chain reaction.
Clinical Outcomes
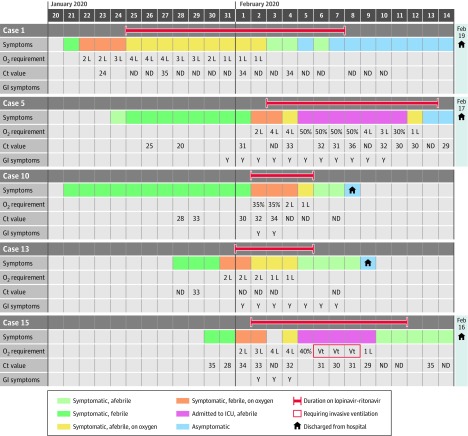
Of the 6 patients who required supplemental oxygen, 5 received lopinavir-ritonavir (Figure 2). For 3 of 5 patients (60%), initiation of lopinavir-ritonavir was followed by a reduction in supplemental oxygen requirements within 3 days, and viral shedding in nasopharyngeal swabs cleared within 2 days of treatment for 2 of 5 (40%).
Figure 2. Clinical Progress in Patients Treated With Lopinavir-Ritonavir.
Cycle threshold (Ct) value corresponds to the number of copies of the virus in a biological sample, in an inversely proportional and exponential manner. ICU indicates intensive care unit; GI, gastrointestinal; ND, not detected; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; Y, yes; Vt, mechanical ventilation.
Two patients, however, deteriorated and experienced progressive respiratory failure while receiving lopinavir-ritonavir, with 1 requiring invasive mechanical ventilation. Virus continued to be detected by nasopharyngeal swab or endotracheal tube aspirate for these 2 patients for the duration of their admission to intensive care.
Four of the 5 patients treated with lopinavir-ritonavir developed nausea, vomiting, and/or diarrhea, and 3 developed abnormal liver function test results. Only 1 individual completed the planned 14-day treatment course as a result of these adverse events.
Virologic Features
Median duration of viral shedding from first to last positive nasopharyngeal swab collected as part of clinical care was 12 days (range, 1-24), and 15 patients (83%) had viral shedding from the nasopharynx detected for 7 days or longer. Daily serial RT-PCR cycle threshold values for all 18 patients are shown in eFigure 3 in the Supplement. The time course of serial cycle threshold values by day of illness for 12 patients not receiving supplemental oxygen and 6 patients receiving supplemental oxygen (of whom 5 were treated with lopinavir-ritonavir) appeared similar (eFigure 4 in the Supplement).
Virus was detected by PCR in stool (4/8 patients [50%]) and in whole blood (1/12 [8%]); virus was not detected in urine (0/10 samples) (eTable 2 in the Supplement). Viral culture results were not available at the time of writing to determine the viability of virus detected outside the respiratory tract.
Sequences for phylogenetic analysis were available for 6 viruses (eFigure 5 in the Supplement). All clustered together with other SARS-CoV-2 sequences reported from China and other countries and are available in public databases.
Discussion
This descriptive case series reports the clinical features of the first 18 patients with laboratory-confirmed SARS-CoV-2 infection in Singapore, reporting epidemiologic features and clinical course in detail.
Perhaps reflecting the intensive efforts at contact tracing, 28% of patients were without fever vs 1.4% to 17% in 3 previously reported studies from Wuhan, China.1,4,13 In the current study, 6 of 18 patients (33%) experienced oxygen desaturation to 92% or less, in contrast to the 76% to 90% supplemental oxygen use from the other 3 studies.1,4,13
In 4 of 8 patients, virus was detected in stool, regardless of diarrhea, over 1 to 7 days. Viremia was detected in 6 patients1 and 1 of 5 patients14 in China but only in 1 of 12 in this study. In the family cluster in Shenzhen, the virus was not detected in urine or stool.14 In SARS, viremia was observed in the first week, with peak respiratory viral shedding in the second week and persistent stool viral shedding beyond the second week.15 In Middle East respiratory syndrome, viral shedding was greater in lower respiratory tract samples than in blood and stool.16 Viral load in nasopharyngeal samples and viremia was associated with disease severity in SARS.17
In this study, viral load in nasopharyngeal samples from patients with COVID-19 peaked within the first few days after symptom onset before declining. The duration of viral shedding from nasopharyngeal aspirates was prolonged up to at least 24 days after symptom onset. This was longer than reported from a comparable series from China.18 Toward the end of this period virus was only intermittently detected from nasopharyngeal swabs. It is unclear whether this is attributable to biological differences in the intensity of shedding or to sampling variability when low amounts of virus are present. Determining whether the virus remains transmissible throughout the period of detectability is critical to control efforts.
Five patients were treated with lopinavir-ritonavir within 1 to 3 days of desaturation, but evidence of clinical benefit was equivocal. While defervescence occurred within 1 to 3 days of lopinavir-ritonavir initiation, it was unable to prevent progressive disease in 2 patients. Decline in viral load as indicated by the cycle threshold value from nasopharyngeal swabs also appeared similar between those treated and not treated with lopinavir-ritonavir. The effectiveness of lopinavir-ritonavir treatment in COVID-19 needs to be examined in an outbreak randomized trial, given a lack of clear signal in this small case series.
Limitations
This study has several limitations. First, it was a case series of 18 patients who acquired their infection following travel to Wuhan, China. Findings from this study are valuable early data from a high-resource setting but may change as this outbreak continues to evolve and local transmission clusters emerge. Second, 9 individuals (50%) presented to the hospital more than 2 days after symptom onset. As a result, sample collection early during the course of illness was limited. Third, biological samples were collected systematically when possible, but not all patients consented to sample collection. Baseline laboratory data were also not available for all patients. Fourth, cycle threshold values are a quantitative measure of viral load, but correlation with clinical progress and transmissibility is not yet known.
Conclusions
Among the first 18 patients diagnosed with SARS-CoV-2 infection in Singapore, clinical presentation was frequently a mild respiratory tract infection. Some patients required supplemental oxygen and had variable clinical outcomes following treatment with an antiretroviral agent.
eMethods. Diagnostic Testing and RNA-Dependent RNA Polymerase (RdRp) Sequencing of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) From Nasopharyngeal Samples
eFigure 1. Case Map of Confirmed 2019-nCoV Cases in Singapore
eFigure 2. Workflow of Screening and Admission for Suspected COVID-19 (as of February 8, 2020).
eFigure 3A. Individual Plot of Serial Cycle Threshold (Ct) Values by Day of Illness for Each Patient
eFigure 3B. Serial Cycle Threshold Values for All Patients by Day of Illness
eFigure 4. Serial Cycle Threshold (Ct) Values by Day of Illness up to Day 14, Split by Patients Who (A) Required Supplemental Oxygen and (B) Did Not Require Supplemental Oxygen
eFigure 5. Phylogenetic Tree of Six Patients With Available Sequences
eTable 1. Epidemiologic Features of Patients Infected With 2019-nCoV
eTable 2. Cycle Threshold Values for Respiratory, Blood, Urine, and Stool Samples by Day of Illness
eTable 3. Singapore 2019 Novel Coronavirus Outbreak Research Team Members
eReferences
References
- 1.Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497-506. doi: 10.1016/S0140-6736(20)30183-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lu R, Zhao X, Li J, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565-574. doi: 10.1016/S0140-6736(20)30251-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhu N, Zhang D, Wang W, et al. ; China Novel Coronavirus Investigating and Research Team . A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382(8):727-733. doi: 10.1056/NEJMoa2001017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang D, Hu B, Hu C, et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA. Published online February 7, 2020. doi: 10.1001/jama.2020.1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu Z, McGoogan JM. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: summary of a report of 72 314 cases from the Chinese Center for Disease Control and Prevention. JAMA. Published online February 24, 2020. doi: 10.1001/jama.2020.2648 [DOI] [PubMed] [Google Scholar]
- 6.Wong JEL, Leo YS, Tan CC. COVID-19 in Singapore—current experience: critical global issues that require attention and action. JAMA. Published online February 20, 2020. doi: 10.1001/jama.2020.2467 [DOI] [PubMed] [Google Scholar]
- 7.Chan KS, Lai ST, Chu CM, et al. Treatment of severe acute respiratory syndrome with lopinavir/ritonavir: a multicentre retrospective matched cohort study. Hong Kong Med J. 2003;9(6):399-406. [PubMed] [Google Scholar]
- 8.Chu CM, Cheng VC, Hung IF, et al. ; HKU/UCH SARS Study Group . Role of lopinavir/ritonavir in the treatment of SARS: initial virological and clinical findings. Thorax. 2004;59(3):252-256. doi: 10.1136/thorax.2003.012658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chan JF, Yao Y, Yeung ML, et al. Treatment with lopinavir/ritonavir or interferon-β1b improves outcome of MERS-CoV infection in a nonhuman primate model of common marmoset. J Infect Dis. 2015;212(12):1904-1913. doi: 10.1093/infdis/jiv392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sheahan TP, Sims AC, Leist SR, et al. Comparative therapeutic efficacy of remdesivir and combination lopinavir, ritonavir, and interferon beta against MERS-CoV. Nat Commun. 2020;11(1):222. doi: 10.1038/s41467-019-13940-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Interim infection prevention and control recommendations for patients with known or patients under investigation for 2019 novel coronavirus (2019-nCoV) in a healthcare setting. Centers for Disease Control and Prevention. Updated February 21, 2020. Accessed February 27, 2020. https://www.cdc.gov/coronavirus/2019-nCoV/hcp/infection-control.html
- 12.Lansbury LE, Rodrigo C, Leonardi-Bee J, Nguyen-Van-Tam J, Shen Lim W. Corticosteroids as adjunctive therapy in the treatment of influenza: an updated Cochrane systematic review and meta-analysis. Crit Care Med. 2020;48(2):e98-e106. doi: 10.1097/CCM.0000000000004093 [DOI] [PubMed] [Google Scholar]
- 13.Chen N, Zhou M, Dong X, et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395(10223):507-513. doi: 10.1016/S0140-6736(20)30211-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chan JF, Yuan S, Kok KH, et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020;395(10223):514-523. doi: 10.1016/S0140-6736(20)30154-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peiris JS, Chu CM, Cheng VC, et al. ; HKU/UCH SARS Study Group . Clinical progression and viral load in a community outbreak of coronavirus-associated SARS pneumonia: a prospective study. Lancet. 2003;361(9371):1767-1772. doi: 10.1016/S0140-6736(03)13412-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oh MD, Park WB, Choe PG, et al. Viral load kinetics of MERS coronavirus infection. N Engl J Med. 2016;375(13):1303-1305. doi: 10.1056/NEJMc1511695 [DOI] [PubMed] [Google Scholar]
- 17.Hung IF, Cheng VC, Wu AK, et al. Viral loads in clinical specimens and SARS manifestations. Emerg Infect Dis. 2004;10(9):1550-1557. doi: 10.3201/eid1009.040058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zou L, Ruan F, Huang M, et al. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N Engl J Med. Published online February 19, 2020. doi: 10.1056/NEJMc2001737 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
eMethods. Diagnostic Testing and RNA-Dependent RNA Polymerase (RdRp) Sequencing of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) From Nasopharyngeal Samples
eFigure 1. Case Map of Confirmed 2019-nCoV Cases in Singapore
eFigure 2. Workflow of Screening and Admission for Suspected COVID-19 (as of February 8, 2020).
eFigure 3A. Individual Plot of Serial Cycle Threshold (Ct) Values by Day of Illness for Each Patient
eFigure 3B. Serial Cycle Threshold Values for All Patients by Day of Illness
eFigure 4. Serial Cycle Threshold (Ct) Values by Day of Illness up to Day 14, Split by Patients Who (A) Required Supplemental Oxygen and (B) Did Not Require Supplemental Oxygen
eFigure 5. Phylogenetic Tree of Six Patients With Available Sequences
eTable 1. Epidemiologic Features of Patients Infected With 2019-nCoV
eTable 2. Cycle Threshold Values for Respiratory, Blood, Urine, and Stool Samples by Day of Illness
eTable 3. Singapore 2019 Novel Coronavirus Outbreak Research Team Members
eReferences