Abstract
The actinobacterium Streptomyces sp. MC1 has previously shown the capacity to resist and remove Cr(VI) from liquid culture media. The aim of this work is to analyze the differential expression pattern of intracellular proteins when Streptomyces sp. MC1 is exposed to Cr(VI) in order to explain the molecular mechanisms of resistance that this microorganism possesses. For this purpose, 2D-PAGE and shotgun proteomic analyses (2D-nanoUPLC-ESI-MS/MS) were applied. The presence of Cr(VI) induced the expression of proteins involved in molecular biosynthesis and energy generation, chaperones with a key role in the repair of misfolded proteins and stress response, transcription proteins, proteins of importance in the DNA supercoiling, repair and replication, and dehydrogenases involved in oxidation–reduction processes. These dehydrogenases can be associated with the reduction of Cr(VI) to Cr(III). The results of this study show that proteins from the groups mentioned before are important to face the stress caused by the Cr(VI) presence and help the microorganism to counteract the toxicity of the metal. The use of two proteomic approaches resulted in a larger number of peptides identified, which is also transduced in a significant number of protein ID. This decreased the potential complexity of the sample because of the protein dynamic range, as well as increased the recovery of peptides from the gel after digestion.
Introduction
Chromium (Cr) is a toxic heavy metal that widely affects different cellular structures. Contamination with Cr(VI) caused by anthropogenic activities occurs mainly from industrial effluents that are released into the environment without proper treatments [1–4]. Despite the toxicity of heavy metals, some microorganisms isolated from contaminated environments have exhibited the capacity to resist the presence of these metals. Some of the mechanisms used by the Cr(VI)-resistant microorganisms are the biotransformation of Cr(VI) to Cr(III) and the chromium bioaccumulation inside the cell [4–6]. Within the heavy metal-resistant microorganisms, we can find Actinobacteria members, such as Streptomyces sp. MC1, which is the microorganism used in the present work [7–11].
One possible approach to understand the molecular mechanisms involved in the metal-microorganism interaction can be proteomics. The systematic study and comparison of the proteome of various metabolic and/or pathological conditions may throw light into those proteins whose presence, absence, or alteration correlates with certain physiological states. The techniques used in proteomics are grouped into two major categories: gel-based and gel-free technologies. The classical gel-based technique is two-dimensional electrophoresis and it is widely used in studies of heavy metal toxicity [12]. In recent years, some authors have focused their proteomic studies on alternative approaches such as the gel-free technique, an emerging tool for relative abundance analysis. Gel-free methods can eliminate the limitations of Molecular Weight (MW) and Isoelectric Point (pI) ranges, and they can increase the recovery rate and the dynamic range of proteins. Dynamic range is the ability of an analytical method to differentiate two proteins with large differences in concentration at the same point. Gel-free proteomics has a much higher capacity to detect ‘unique’ proteins, without differentiating all the different protein forms. The identification of protein expression induced or repressed in the presence of toxic concentrations of a heavy metal has the potential to help understand the molecular mechanisms involved in this condition of specific stress. However, little is known about the molecular mechanisms used by members of the genus Streptomyces against different heavy metals.
In our previous investigations, the addition of different concentrations of sulfate ions enhanced the removal of Cr(VI) by Streptomyces sp. MC1, removing 98% out of 20 μg ml−1 Cr(VI) from a liquid culture medium in the presence of 7.5mM sulfate ions [13]. Under these conditions, the differential expression of intracellular proteins was observed in one dimensional gels (1D-gels). When chromium was present, seven proteins were down-expressed and showed homology with the proteins involved in energy production, free radicals detoxification, and protein biosynthesis. In contrast, two proteins identified as Dihydrolipoamide dehydrogenase and S-Adenosyl-L-methionine synthase were overexpressed [13]. Even though a differential expression was identified, the 1D gel-based method has proved to be insufficient for a comprehensive study of protein expression because Cr(VI) toxicity is a complex mechanism that involves many biochemical and molecular processes.
In this work, we compare the differential expressions of intracellular proteins of Streptomyces sp. MC1 in the presence and absence of Cr(VI) using gel-based and gel-free methods. The main objective is to understand the homeostatic mechanisms used by the organism against the metal.
Materials and Methods
Microorganism and Culture Conditions
The microorganism used in this work was Streptomyces sp. MC1, provided by courtesy of Dra. María J. Amoroso. This microorganism was isolated from sugarcane in the province of Tucumán, Argentina (PROIMI collection, NCBI accession number AY741287) [14]. The microorganism maintenance was performed at 30 °C on solid minimal medium modified (MMm) by Villegas et al. [15], containing (g l−1): agar 15.0, glucose 10.0, L-asparagine 0.5, K2HPO4 0.5, MgCl2 0.17, and FeSO4·7H2O 0.01.
Liquid MMm was supplemented with 7.5 mM sulfate ions (as Na2SO4) in the presence or absence of 20 μg m l−1 Cr(VI) (as K2Cr2O7). The required volume of spore suspension to obtain 106 spores ml−1 was inoculated and incubated at 30°C and 180 rpm during 48 h. These conditions were determined based on our previous results [13]. All the assays were performed in triplicate.
Collection of Intracellular Proteins
Cells obtained from culture media in the presence and absence of Cr(VI) after 48 h of incubation were harvested by centrifugation at 8000×g during 10 min at 4°C (U-320R centrifuge) and washed twice with phosphate buffered saline (mM: NaCl 124; NaH2PO4 10; KH2PO4 3). Then, cells were frozen using liquid nitrogen and physically broken using a mortar and pestle. The powder obtained was recovered with Tris-EDTA buffer (Sucrose 11.29 g dl−1; Tris-HCl 1.5M pH 8.8 3.33 ml dl−1; EDTA 0.12g dl−1; DTT 1mM) and it was centrifuged at 6000×g during 15 min at 4 °C. Supernatants were used as samples of intracellular proteins.
Total protein content in supernatants was determined by the Bradford method (Bio-Rad) using bovine serum albumin as reference for protein concentrations (0–10 μg ml−1). Proteins were split into two aliquots and concentrated in two different ways. The first one was concentrated by lyophilization for shotgun proteomic analysis, while the second one was concentrated by supernatant filtration using disposable ultrafiltration devices with a 3kDa cut off (Vivaspin® 500 Centrifugal Concentrator) to perform the 2D electrophoresis analysis.
2D Electrophoresis
300 μl of rehydration solution (CHAPS 40 mg, DTT 16 mg, Immobiline pH Gradient 20 μl, Bromophenol blue 5 mg, and 1 ml of Denaturalization solution, containing Urea 7M, Tiourea 2M, and amberlite resin MB-150 SUPELCO) was added to 500 μg of intracellular proteins concentrated by ultrafiltration, as explained before, obtained from cultures in the presence and absence of Cr(VI). Then, 40 μl of solubilization solution (CHAPS 40 mg, DTT 16 mg, Tris-HCl −2M pH 8.8–20 μl, and 1000 μl of denaturalization solution) was also added. This mixture was centrifuged at 3500×g for 10 min.
Immobiline strips (GE, HealthCare) of 18 cm and non-lineal pH 4–7 were used for isoelectrofocus (IEF). Strips were passively rehydrated with protein samples at 20°C during 18 h. Focusing on IPGphor (GE, HealthCare) started at 500V for 1 h (lineal), then increased to 1000 V (6000 Vh) and 8000 V (27,000 Vh) in gradient, 10,000 V for 2 h (lineal), and finally was kept at 500 V until a total of 58,500 Vh was reached.
After IEF, strips were washed twice with 5 ml of base solution (Tris-1M pH 6.8–5 ml, Urea 36 g, SDS 1 g, H2OmQ 35 ml, Glycerol 30 ml). The strips were incubated with 10 ml of equilibrium solution No. 1 (DTT 167 mg and base solution 20 ml) and 10 ml of equilibrium solution No. 2 (Iodoacetamide 1.5 g and base solution 20 ml) during 15 min with soft shaking.
SDS-PAGE was performed on 12% of polyacrylamide gels using Bio-Rad Protean II xi cell, at 50 mA during 7 h. Finally, gels were stained with Coomassie R-250 0.1% and scanned and analyzed with Image Scanner III (GE Healthcare Life Sciences, LabScan 6.0 software). The differential spots were excised from gels for protein identification analysis. The 2D electrophoresis was carried out in triplicate.
Mass Spectrometry Analysis
The gels were rinsed with water and the spots of interest were excised from gels by cutting them with a clean scalpel. Spots excised were bleached with Water/Ethanol/Acetic Acid (50:40:10), and finally treated with DTT and Iodoacetamide, followed by in-gel digestion using Trypsin sequencing grade (Promega, Madison WI). The tryptic digested peptides were extracted from the gel using two cycles of Formic Acid/Acetonitrile/Water (1:2:97) and Water/Acetonitrile (50:50), respectively. The peptide clean-up and separation were carried out through nano-Ultra-Performance Liquid Chromatography (nanoAcquity UPLC) and analyzed by tandem mass spectrometry using Synapt G1 Q-TOF HDMS (Waters, Milford, MA) (Supporting data 1 and 2).
The aliquot with lyophilized proteins was resuspended in a solution of 100 mM ammonium bicarbonate (pH 8)/5% of acetonitrile, until a final protein concentration of 1 μg μl−1 was reached. Proteins were reduced and alkylated with DTT and Iodoacetamide, respectively, and digested with Trypsin sequencing grade (Promega, Madison WI). Peptides were concentrated in a vacuum centrifuge SpeedVac (Thermo, Savant). The analysis was performed using 2D-nano-Ultra-Performance Liquid Chromatography (2DnanoAcquity UPLC) coupled to tandem mass spectrometry (2D-nanoU-PLC-ESI-MS/MS) (Supporting data 2 and 3). The proteomic analyses were performed in triplicate of each biological replicate.
Bioinformatic Analysis
MASCOT server v2.5.1 (www.matrix-science.com, UK) in MS/MS ion search mode (local licenses) was applied to conduct peptide matches (peptide masses and sequence tags) and protein searches. This was carried out against a database customized by the combination of four Streptomyces spp. given the similarity or orthologous with Streptomyces sp. MC1: Streptomyces lividans TK 24, Streptomyces avermitilis MA-4680, Streptomyces scabiei 87.22, and Streptomyces griseus NBRC 13,350; from KEGG (Kyoto Encyclopedia of Genes and Genomes) v20160226 (30,918 sequences; 10,489,322 residues). We established the following parameters for the search: Carbamidomethyl (C) on cysteine was set as fixed, and variable modifications included asparagine and glutamine deamidation and methionine oxidation. Only one missed cleavage was allowed. Monoisotopic masses were counted. The precursor peptide mass tolerance was set at 20 ppm. Fragment mass tolerance was 0.3 Da and the ion score or expected cutoff was set at 5. The MS/MS spectra were searched with MASCOT using a 95% confidence interval (CI%) threshold (P < 0.05), while minimum score of 25 was used for peptide identification. Furthermore, the error tolerance mode was set up at MASCOT search to corroborate potential peptides unidentified during the first search. When the peptides identified match equally well to multiple protein ID, only those proteins that appeared in at least two or more replicates were considered to be included in the list.
The comparison of samples expression patterns was performed using ProteoIQ v2.8 (local license).
Results
2D Electrophoresis and Spot Identification
To identify the differential protein expressions induced in the presence of Cr(VI) in Streptomyces sp. MC1, 2D electrophoresis was carried out to separate the intracellular proteins obtained from cultures grown in the presence and absence of the metal.
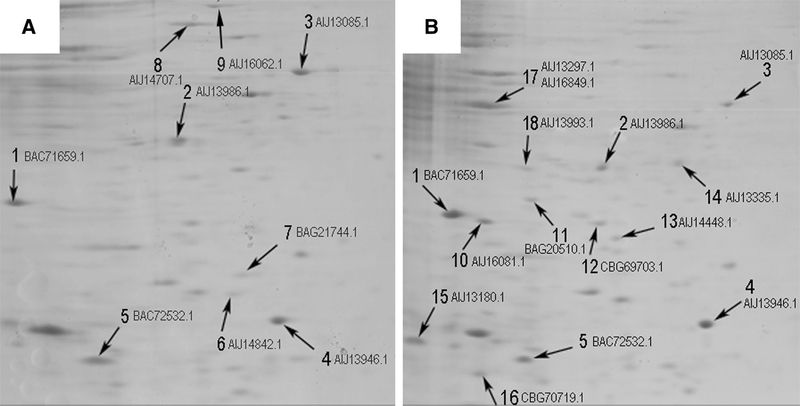
As shown in Fig. 1, a differential protein expression was observed and 18 differential spots were collected to identify the proteins of interest to understand the homeostatic mechanism used by Streptomyces sp. MC1 against the toxic heavy metal. Proteins identified are shown in Table 1.
Fig. 1.
2D gel electrophoresis obtained from intracellular proteins collected in the absence (a) and presence of Cr(VI) (b) in the culture media. Numbers and arrows indicate the selected spots analyzed for protein identification by 2D-nanoUPLC-ESI-MS/MS
Table 1.
Proteins identified in the selected spots from 2D gel obtained in the presence and absence of Cr(VI) in the culture medium
| Spota | Cr(VI) absence | Cr(VI) presence | Accessionb | Protein identificationb | Mascot scorec | Mass (kDa) |
|---|---|---|---|---|---|---|
| 1 | + | + | BAC71659.1 | Putative tellurium resistance protein | 2338 | 20.213 |
| 2 | + | + | AIJ13986.1 | Elongation factor Tu-1 | 2186 | 43.811 |
| 3 | + | + | AIJ13085.1 | Elongation factor Ts | 385 | 29.944 |
| 4 | + | + | AIJ13946.1 | 50S ribosomal protein L22 | 195 | 12.788 |
| 5 | + | + | BAC72532.1 | Putative cold shock protein | 740 | 7.115 |
| 6 | + | − | AIJ14842.1 | Hypothetical protein SLIV_19470 | 46 | 16.706 |
| 7 | + | − | BAG21744.1 | Putative ribose-5-phosphate isomerase | 47 | 17.753 |
| 8 | + | − | AIJ14707.1 | Hypothetical protein SLIV_18770 | 544 | 39.433 |
| 9 | + | − | AIJ16062.1 | 3-Oxoacyl-[acyl-carrier-protein] synthase II | 772 | 43.827 |
| 10 | − | + | AIJ16081.1 | Tellurium resistance protein TerE | 695 | 20.375 |
| 11 | − | + | BAG20510.1 | Putative 1L-myo-inositol-1-phosphate synthase | 642 | 39.380 |
| 12 | − | + | CBG69703.1 | Ribosome recycling factor | 122 | 20.728 |
| 13 | − | + | AIJ14448.1 | Hypothetical protein SLIV_17385 | 214 | 22.794 |
| 14 | − | + | AIJ13335.1 | Transcription termination factor Rho | 79 | 76.559 |
| 15 | − | + | AIJ13180.1 | d-3-phosphoglycerate dehydrogenase | 195 | 55.184 |
| 16 | − | + | CBG70719.1 | 10 kD chaperonin cpn10 | 197 | 11.039 |
| 17 | − | + | AIJ13297.1 | Cellulose-binding protein | 156 | 34.585 |
| AIJ16849.1 | Phenylalanine-tRNA ligase alpha subunit | 73 | 41.093 | |||
| 18 | − | + | AIJ13993.1 | DNA-directed RNA polymerase subunit beta’ | 462 | 145.199 |
The identification of spots is based on at least one peptide identified using tandem mass spectrometry analysis (MS/MS)
Function annotations were retrieved from NCBInr (https://www.ncbi.nlm.nih.gov)
The threshold was set up by the MASCOT server (local license) at a significance level of P ≤ 0.05 for random hit; scores greater than 25 were taken as a significant match for individual ion score. Significant scores indicate identity or extensive homology, based in Mowse algorithm (www.matrixscience.com)
Proteins overexpressed under the stress caused by the presence of the metal include those proteins involved in (i) protein biosynthesis, such as putative 1L-myo-inositol-1-phosphate synthase, ribosome recycling factor, transcription termination factor Rho, phenylalanine-tRNA ligase, and DNA-directed RNA polymerase; (ii) proteins involved in oxidation–reduction processes, such as D-3-phosphoglycerate dehydrogenase; (iii) proteins of response against stress, such as 10 kD chaperonin cpn10; and (iv) a protein of resistance to the metalloid tellurium, tellurium resistance protein TerE.
Spots observed in both groups that showed at least a two-fold intensity increase were selected as candidates for protein identification. Similar protein ID was identified in both conditions matching with putative tellurium resistance protein (spot 1), elongation factor Tu-1 (2), elongation factor Ts (3), 50S ribosomal protein L22 (4), and putative cold shock protein (5). The data obtained from the 2D gel analysis was not enough to determine the semi-quantitative differential expression among these proteins. The results from the gel analysis did not allow us to conclude whether the microorganism exposed to Cr(VI) could specifically respond to counteract the toxicity of this metal, even though the expression of proteins involved in different metabolic processes was significantly affected.
Shotgun Proteomic Analysis
As a complementary approach of 2D electrophoresis analysis to improve the relative abundance analysis, shotgun proteomics was performed. This particular method allows us to carry out a more comprehensive analysis of the differential expression pattern of intracellular proteins exposed to Cr(VI).
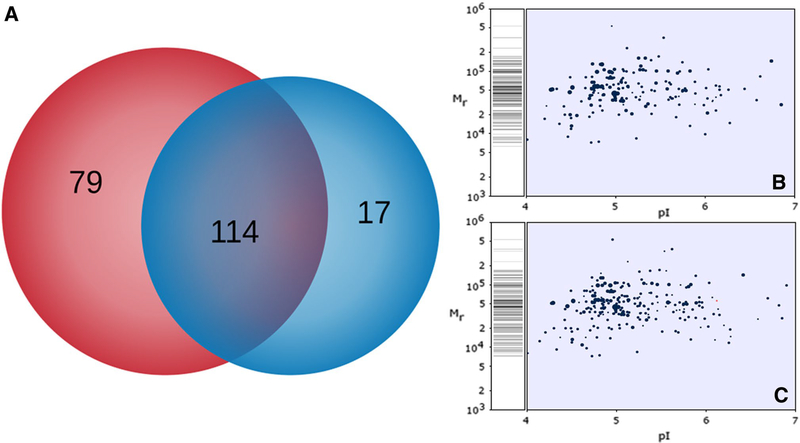
Figure 2 represents a Venn diagram of the discovery proteomic analysis using shotgun methodology through a gel-free technology. A number of 114 proteins were identified in common under both conditions; only 17 were found in the absence of Cr(VI) and 79 were observed exclusively when exposed to the metal. An in silico 2D-gel (theoretical MW/pI) representation was obtained from GPM tool with the aim to compare it with the experimental MW/pI.
Fig. 2.
Global proteomic analysis. Venn diagram of proteins identified in the presence (red) and absence of Cr(VI) in the culture media (blue); the protein group shared by both conditions is shown in violet (a). In silico 2D gel obtained from shotgun proteomic analysis of total intracellular proteins expressed in the absence (b) and presence of Cr(VI) (c) (Color figure online)
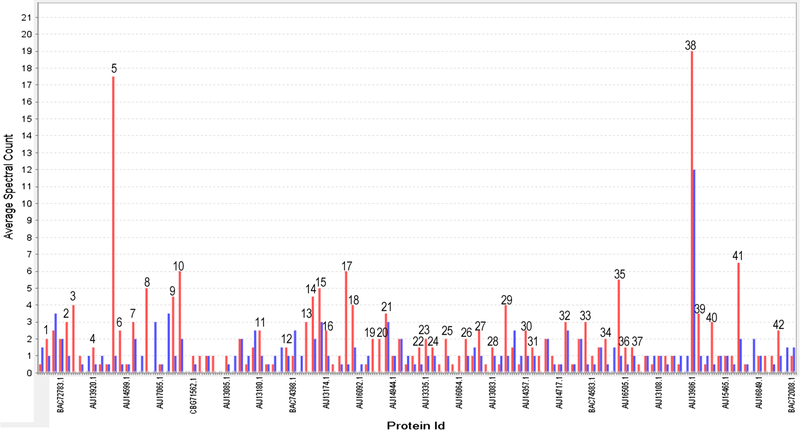
Figure 3 shows a comparative analysis of the relative abundance profiles for the 114 proteins observed under both conditions. Moreover, only 42 proteins showed a significant differential expression in the group exposed to Cr(VI). These proteins are specified in Supporting data 4 and can be mainly grouped into proteins involved in energy production (3 proteins), protein biosynthesis (17), oxidation–reduction processes (7), chaperones (3), and proteins involved in other metabolic processes (12).
Fig. 3.
Bar diagram of semi-quantitative expression of intracellular proteins obtained in the presence (red) and absence (blue) of Cr(VI) (Color figure online)
Furthermore, proteins detected only in the presence of Cr(VI) are shown in Supporting data 5. Similarly, they can also be mainly grouped into proteins involved in energy production (1), protein biosynthesis (33), oxidation–reduction processes (8), chaperones (1), and proteins involved in other metabolic processes (36).
The same types of proteins were found in both 2D electrophoresis and gel-free studies, including proteins involved in biosynthesis of macromolecules, chaperones of response against stress, and proteins involved in oxidation–reduction processes. These results indicate that these proteins are important to face the stress caused by the Cr(VI) presence and help the microorganism to counteract the toxicity of the metal. It is important to highlight that both techniques showed a major production of proteins by Streptomyces sp. MC1 in the presence of Cr(VI) than in the absence of this metal. However, there are considerable differences between both techniques. When we used gel-free techniques, the results were more exhaustive and comprehensive than those obtained using a gel-based technique.
Discussion
The study of the expression levels of proteins affected by the stress produced by the heavy metal is essential to understand the molecular mechanisms of resistance used by the microorganism. In this work, 2D-PAGE and shotgun proteomics (2D-nanoUPLC-ESI-MS/MS) were used to analyze the intracellular protein profile of Streptomyces sp. MC1 when exposed to Cr(VI) in the culture medium.
Much research has been devoted to proteome study using 2D-PAGE or shotgun proteomic analyses. Although gel electrophoresis is now a well-established and extensive technique, it has some shortcomings in terms of quantitative reproducibility, a limited range of MW and pI, difficulties in sample solubilization, as well as a low percentage of material recovery from the spot coming from the gel. Therefore, in recent years, some authors have focused their proteomic studies on alternative approaches, such as gel-free ones, and thus, a new tool for relative quantification analysis has emerged. Gel-free methods overcome the limitations in the range of MW and pI, increasing the percentage of material recovery and the dynamic range of proteins [12, 16, 17]. Although these new approaches initially were settled as substitutes for gel-based methods, in the view of our findings, they should probably be considered as complements.
In general, interactions between microorganisms and heavy metals are affected by numerous parameters, for example, organic or inorganic composition of media where microorganisms grow [8]. Previous studies related to heavy metal resistance mechanisms report a decrease of metal absorption and bioavailability when the microorganism grows in complex media because of the high concentration of organic components. In order to avoid the interferences mentioned before, MMm was used in the present work. Considering those limitations, Thompson et al. [18] studied the proteome of Pseudomonas putida F1 against 1mM Cr(VI) using two different media: LB (complex medium) and M9L(minimal medium). In LB-grown cells, they found up-regulation of proteins involved in inorganic ion transport, secondary metabolite biosynthesis and catabolism, and amino acid metabolism. In M9L-grown cells, the response was characterized by up-regulated proteins related to cell envelope biogenesis, inorganic ion transport, and motility. Likewise, DNA repair proteins and systems scavenging sulfur from alternative sources predominated under M9L-Cr(VI) conditions. The up-regulated proteins in both media belonged to different functional categories, such as transcription, inorganic ion transport/metabolism, and amino acid transport/metabolism. Thompson et al. [18] suggested that these proteins might serve as indicators of Cr(VI) stress in natural microbial communities. However, in the present work, we only found proteins of transcription processes, but not the other protein groups. These results indicate that looking for biomarkers of Cr(VI) resistance is not simple and more specific strategies have to be developed.
There are many studies related to proteome analyses of microorganisms resistant to heavy metals and other toxic compounds. However, no reports are observed about similar responses of the proteome of Streptomyces genus members against heavy metals. Several works have used 2D-PAGE or global proteomic analyses, which have been both applied in the present work. For example, Bar et al. [19] carried out a proteomic study using 2D-PAGE and MS in Klebsiella pneumoniae tolerant to Co(II) and Pb(II). They selected 13 differential spots, but only two proteins overexpressed in the presence of Co were identified. L-isoaspartate protein carboxymethyltransferase type II was proposed as an important protein for the repair and/or degradation of damaged proteins, while DNA gyrase A was described as a key protein for heavy metal tolerance. They concluded that the presence of heavy metals affected the expression of many genes, so transcription and supercoiling regulation could serve as a molecular mechanism of resistance. In the same line of research, Kılıç et al. [20] studied the proteomic profile of intracellular and membrane proteins of Pseudomonas aeruginosa when faced with 300 μg ml−1 Cr(VI). Making a comparison in protein expression levels, as done in this work, they found that many proteins were overexpressed in the presence of the metal. They found overexpression in the same type of proteins that we found in this work: chaperonins; proteins involved in biosynthesis and proteins responsible for energy production. Nevertheless, they reported another group of proteins involved in the detoxification of free radicals. Similar results were found in plants exposed to cadmium (Cd). Approximately 50% of the overexpressed proteins corresponded to proteins of response against stress. Considering the overexpressed proteins, antioxidant proteins, proteins related to carbohydrate metabolism, amino acid metabolism, among other, were reported [21].
Cherrad et al. [22] performed a proteomic analysis of secreted proteins when Botrytis cinerea was exposed to copper, zinc, nickel, and cadmium. For the analysis, these authors used 2D-PAGE and found that the production of oxidoreductases and cell wall degrading enzymes was modified in response to these metals. The amplitude of variations of the 55 unique proteins whose accumulation varied in the presence of at least one of the four metals was strongly correlated to the physicochemical properties of the metals. Cd exerted a more potent effect on the accumulation level of metal-sensitive secreted proteins compared to Cu, Zn, and Ni. By using Differential In-Gel Electrophoresis (DIGE), Dekker et al. [23] carried out a comparative proteomic analysis of Acidithiobacillus ferrooxidans in the presence and absence of 0.5 mM U(VI). They observed 17 up-regulated and one down-regulated proteins in the presence of the metal. Most of the up-regulated proteins were related to the general stress response or in reactive oxygen species detoxification.
In relation to gel-free proteomic analysis, we can mention the work carried out by Poirier et al. [24] with Pseudomonas fluorenscens BA3SM1 exposed to Zn, Cd, and Cu, and Yung et al. [25] who also carried out a label-free proteomic analysis of Caulobacter crescentus exposed to Cr, Cd, and U. These authors also found differential protein expressions in the presence of the metals.
There are several works describing proteomic analysis of actinobacteria resistant to toxic organic compounds. For example, Dávila Costa et al. [26] analyzed the proteome of Rhodococcus jostii RHA1 through LC–MS/MS and found differential expression of proteins involved in triacylglycerol accumulation after the addition of methyl viologen. On the other hand, Sineli et al. [27] found differential intracellular protein expression in Streptomyces sp. M7 when lindane was added to the culture media using quantitative proteomic analysis. However, no studies related to the proteome of Streptomyces genus members resistant to heavy metals have been found. Considering the previous statement, the aim of this study was to expand the knowledge in the response of Streptomyces sp. MC1 against the exposition to a toxic metal such as Cr(VI).
The results obtained in the present work yield data in accordance with the studies above. When faced to chromate ions, Streptomyces sp. MC1 induced the expression or overexpression of varied proteins involved in important metabolic processes. From these results, it can be stated that Streptomyces sp. MC1 induced the expression of proteins involved in biosynthesis and in energy generation, chaperones with a key role in the repair of misfolded proteins and stress response, transcription proteins, proteins of importance in the DNA supercoiling, repair and replication, and dehydrogenases involved in oxidation–reduction processes. The latter was found to be probably involved in the reduction of Cr(VI) to Cr(III).
The use of two proteomic approaches such as 2D-PAGE and shotgun (2D-nanoUPLC-ESI-MS/MS) improves the analysis of the resistance mechanism developed by Streptomyces sp. MC1 against Cr(VI). Moreover, the combination of the techniques results in a larger number of peptides identified transduced in a significant number of protein ID. Not only does this combination decrease the potential complexity of the sample because of the protein dynamic range, but it also increases the recovery of peptides from the gel after digestion.
Importantly, the lack of the sequenced genome as well as a specific protein database for Streptomyces sp. MC1 leads to the development of methodological strategies to reach preliminary proteomic results to understand the removal mechanisms used by this microorganism in the presence of Cr(VI) in the culture medium. One of the alternatives to overcome this difficulty is to use databases from orthologous microorganisms possessing a significant genetic similarity, as carried out in the present work. The importance of the integration and the comparison of strengths and limitations of proteomic databases is still debated in different fields of study [28]. The mass data obtained in this study can be re-analyzed when available the sequenced genome of the microorganism and/or the specific transcriptome databases in the presence and absence of Cr(VI). In the future, the availability of these bioinformatics data can contribute to deepen studies to understand the mechanisms related to the removal of heavy metals, as well as to determine whether those mechanisms are specific to particular metals. Moreover, these strategies can help to analyze their potential use as biomarkers of exposure to heavy metal contamination. Finally, one of the best options to validate proteins identified using mass spectrometry-based proteomics is western blot [29]. However, this approach is only possible with the specific antibodies for the strain used in the present work, which are not feasible at this time.
Supplementary Material
Acknowledgements
The authors thank the financial assistance of the National Agency for Scientific and Technological Promotion, Argentina (PICT 2013 No. 3170 to Dr. Villegas). The authors would also like to thank the English Scientific Writing Advice Group (GAECI) of the National University of San Luis for the revision of this article, and Mr. Doug Jennewein from USD-IT Research Computing for his help in the database installation and servers operation. Bonilla JO thanks CONICET for the awarded doctoral fellowship.
Footnotes
Compliance with Ethical Standards
Conflicts of interest The authors declare that they have no conflict of interest.
Electronic supplementary material The online version of this article (https://doi.org/10.1007/s00284-019-01790-w) contains supplementary material, which is available to authorized users.
Publisher’s Note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Cefalu WT, Hu FB (2004) Role of chromium in human health and in diabetes. Diabetes Care 27:2741–2751. 10.2337/diacare.27.11.2741 [DOI] [PubMed] [Google Scholar]
- 2.Cheung KH, Gu JD (2007) Mechanism of hexavalent chromium detoxification by microorganisms and bioremediation application potential: a review. Int Biodeterior Biodegrad 59(1):8–15. 10.1016/j.ibiod.2006.05.002 [DOI] [Google Scholar]
- 3.Viti C, Marchi E, Decorosi F, Giovannetti L (2014) Molecular mechanisms of Cr(VI) resistance in bacteria and fungi. FEMS Microbiol Rev 38:633–659. 10.1111/1574-6976.12051 [DOI] [PubMed] [Google Scholar]
- 4.Megharaj M, Avudainayagam S, Naidu R (2003) Toxicity of hexavalent chromium and its reduction by bacteria isolated from soil contaminated with tannery waste. Curr Microbiol 47:51–54. 10.1007/s00284-002-3889-0 [DOI] [PubMed] [Google Scholar]
- 5.Liu YG, Xu WH, Zeng GM, Li X, Gao H (2006) Cr(VI) reduction by Bacillus sp. isolated from chromium landfill. Process Biochem 41:1981–1986. 10.1016/j.procbio.2006.04.020 [DOI] [Google Scholar]
- 6.Elangovan R, Abhipsa S, Rohit B, Ligy P, Chandraraj K (2006) Reduction of Cr(VI) by a Bacillus sp. Biotechnol Lett 28(4):247–252. 10.1007/s10529-005-5526-z [DOI] [PubMed] [Google Scholar]
- 7.Alvarez A, Saez JM, Davila Costa JS, Colin VL et al. (2017) Actinobacteria: current research and perspectives for bioremediation of pesticides and heavy metals. Chemosphere 166:41–62. 10.1016/j.chemosphere.2016.09.070 [DOI] [PubMed] [Google Scholar]
- 8.Villegas LB, Rodriguez A, Pereira CE, Abate CM (2013) Cultural factors affecting heavy metals removal by actinobacteria. In: Actinobacteria, application in bioremediation and production of industrial enzymes. CRC Press, Boca Raton, pp 26–43 [Google Scholar]
- 9.Laxman RS, More S (2002) Reduction of hexavalent chromium by Streptomyces griseus. Miner Eng 15:831–837. 10.1016/S0892-6875(02)00128-0 [DOI] [Google Scholar]
- 10.Schmidt A, Haferburg G, Sineriz M, Merten D et al. (2005) Heavy metal resistance mechanisms in actinobacteria for survival in AMD contaminated soils. Chem Erde 65(S1):131–144. 10.1016/j.chemer.2005.06.006 [DOI] [Google Scholar]
- 11.Colin VL, Villegas LB, Abate CM (2012) Indigenous microorganisms as potential bioremediators for environments contaminated with heavy metals. Rev Int Biodeterior Biodegrad 69:28–37. 10.1016/j.ibiod.2011.12.001 [DOI] [Google Scholar]
- 12.Luque-García JL, Cabezas-Sanchez P, Camara C (2011) Proteomics as a tool for examining the toxicity of heavy metals. Trends Analyt Chem 30:703–716. 10.1016/j.trac.2011.01.014 [DOI] [Google Scholar]
- 13.Bonilla JO, Callegari EA, Delfini CD, Estevez MC, Villegas LB (2016) Simultaneous chromate and sulfate removal by Streptomyces sp. MC1. Changes in intracellular protein profile induced by Cr(VI). J Basic Microbiol 56(11):1212–1221. 10.1002/jobm.201600170 [DOI] [PubMed] [Google Scholar]
- 14.Polti MA, Amoroso MJ, Abate CM (2007) Chromium(VI) resistance and removal by actinomycete strains isolated from sediments. Chemosphere 67:660–667. 10.1016/j.chemosphere.2006.11.008 [DOI] [PubMed] [Google Scholar]
- 15.Villegas LB, Pereira C, Colin VL, Abate CM (2013) The effect of sulphate and phosphate ions on Cr(VI) reduction by Streptomyces sp. MC1, including studies of growth and pleomorphism. Int Biodeterior Biodegrad 82:149–156. 10.1016/j.ibiod.2013.01.017 [DOI] [Google Scholar]
- 16.Gevaert K, Van Damme P, Ghesquière B, Impens F et al. (2007) A la carte proteomics with an emphasis on gel-free techniques. Proteomics 16:2698–2718. 10.1002/pmic.200700114 [DOI] [PubMed] [Google Scholar]
- 17.Wöhlbrand L, Trautwein K, Rabus R (2013) Proteomic tools for environmental microbiology—a roadmap from sample preparation to protein identification and quantification. Proteomics 13:2700–2730. 10.1002/pmic.201300175 [DOI] [PubMed] [Google Scholar]
- 18.Thompson D, Chourey K, Wickham G, Thieman S et al. (2010) Proteomics reveals a core molecular response of Pseudomonas putida F1 to acute chromate challenge. BMC Genomics 11:311 10.1186/1471-2164-11-311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bar C, Patil R, Doshi J, Kulkarni MJ, Gade WN (2007) Characterization of the proteins of bacterial strain isolated from contaminated site involved in heavy metal resistance—a proteomic approach. J Biotechnol 128:444–451. 10.1016/j.jbiotec.2006.11.010 [DOI] [PubMed] [Google Scholar]
- 20.Kılıç NK, Stensballe A, Otzen DE, Dönmez G (2010) Proteomic changes in response to chromium(VI) toxicity in Pseudomonas aeruginosa. Bioresour Technol 101:2134–2140. 10.1016/j.biortech.2009.11.008 [DOI] [PubMed] [Google Scholar]
- 21.Lee K, Bae DW, Kim SH, Han HJ et al. (2010) Comparative proteomic analysis of the short-term responses of rice roots and leaves to cadmium. J Plant Physiol 167:161–168. 10.1016/j.jplph.2009.09.006 [DOI] [PubMed] [Google Scholar]
- 22.Cherrad S, Girard V, Dieryckx C, Gonçalves IR et al. (2012) Proteomic analysis of proteins secreted by Botrytis cinerea in response to heavy metal toxicity. Metallomics 4(8):835–846. 10.1039/c2mt20041d [DOI] [PubMed] [Google Scholar]
- 23.Dekker L, Arsène-Ploetze F, Santini JM (2016) Comparative proteomics of Acidithiobacillus ferrooxidans grown in the presence and absence of uranium. Res Microbiol 167(3):234–239. 10.1016/j.resmic.2016.01.007 [DOI] [PubMed] [Google Scholar]
- 24.Poirier I, Hammann P, Kuhn L, Bertrand M (2013) Strategies developed by the marine bacterium Pseudomona fluorescens BA3SM1 to resist metals: a proteome analysis. Aquat Toxicol 128–129:215–232. 10.1016/j.aquatox.2012.12.006 [DOI] [PubMed] [Google Scholar]
- 25.Yung MC, Ma J, Salemi MR, Phinney BS et al. (2014) Shotgun proteomic analysis unveils survival and detoxification strategies by Caulobacter crescentus during exposure to uranium, chromium, and cadmium. J Proteome Res 13:1833–1847. 10.1021/pr400880s [DOI] [PubMed] [Google Scholar]
- 26.Costa JSD, Silva RA, Leichert L, Alvarez HM (2017) Proteome analysis reveals differential expression of proteins involved in triacylglycerol accumulation by Rhodococcus jostii RHA1 after addition of methyl viologen. Microbiology 163:343–354. 10.1099/mic.0.000424 [DOI] [PubMed] [Google Scholar]
- 27.Sineli PE, Herrera HM, Cuozzo SA, Dávila Costa JS (2018) Quantitative proteomic and transcriptional analyses reveal degradation pathway of γ-hexachlorocyclohexane and the metabolic context in the actinobacterium Streptomyces sp. M7. Chemosphere 211:1025–1034. 10.1016/j.chemosphere.2018.08.035 [DOI] [PubMed] [Google Scholar]
- 28.Subba P, Narayana Kotimoole C, Prasad TSK (2019) Plant proteome databases and bioinformatic tools: an expert review and comparative insights. OMICS 23(4):190–206. 10.1089/omi.2019.0024 [DOI] [PubMed] [Google Scholar]
- 29.Ayaz A, Agarwal A, Sharma R, Kothandaraman N, Cakar Z, Sikka S (2018) Proteomic analysis of sperm proteins in infertile men with high levels of reactive oxygen species. Andrologia 50(6):e13015 10.1111/and.13015 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.