Abstract
Fifty four Trichoderma strains were isolated from soil samples collected from garlic and onion crops in eight different sites in Brazil and were identified using phylogenetic analysis based on combined ITS region, tef1-α, cal, act and rpb2 sequences. The genetic variability of the recovered Trichoderma species was analysed by AFLP and their phenotypic variability determined using MALDI-TOF. The strain clusters from both typing techniques coincided with the taxonomic determinations made from phylogenetic analysis. The phylogenetic analysis showed the occurrence of Trichoderma asperellum, Trichoderma asperelloides, Trichoderma afroharzianum, Trichoderma hamatum, Trichoderma lentiforme, Trichoderma koningiopsis, Trichoderma longibrachiatum and Trichoderma erinaceum, in the soil samples. We also identified and describe two new Trichoderma species, both in the harzianum clade of section Pachybasium, which we have named Trichoderma azevedoi sp. nov. and Trichoderma peberdyi sp. nov. The examined strains of both T. azevedoi (three strains) and T. peberdyi (12 strains) display significant genotypic and phenotypic variability, but form monophyletic clades with strong bootstrap and posterior probability support and are morphologically distinct from their respective most closely related species.
Introduction
One of the most important fungal diseases occurring in garlic (Allium sativum) and onion (Allium cepa) is white rot, caused by the sclerotium-forming fungus Sclerotium cepivorum, often causing severe losses in garlic and onion production worldwide [1]. In Brazil, the states of Paraná, Minas Gerais, São Paulo and Goiás produce 64% of the national Allium crop (mostly garlic and onion) [2]. Despite recent advances in Allium production in Brazil, production is not sufficient to fulfil internal demand, due to low productivity [3]. Despite the diversity of garlic and onion cultivars available to growers, the favorable humidity and temperature conditions for most of these cultivars are also conducive to white rot disease. In the absence of reliable conventional white rot control methods, biological control is being investigated as a viable option, particularly using species of the fungal antagonist, Trichoderma [4].
Trichoderma has been widely used in biological control due to its ecological plasticity, easy large-scale production and efficiency against many plant pathogens such as Fusarium, Pythium, Rhizoctonia, Sclerotinia, Botrytis and Verticillium [5–9]. Trichoderma species are common in rhizospheric and non-rhizospheric soils and in endophytic relationships with many plants, displaying antifungal properties as well as promoting growth and inducing plant resistance against pathogenic fungi [10–12]. Three Trichoderma asperellum strains, one Trichoderma harzianum strain and a fifth unidentified Trichoderma strain from the rhizosphere of garlic and onion crops in Costa Rica have been tested for their in vitro antagonism against S. cepivorum, following their identification using ITS sequences [13]. The combination of different biocontrol agents to obtain synergistic or additive effects has also been tested in the field to control S. cepivorum, where simultaneous application of four selected species of Trichoderma (Trichoderma hamatum, T. harzianum, Trichoderma oblongisporum and Trichoderma viride), in association with fungicides, was shown to be effective for the management of white rot disease [14].
The phylogenetic species concept, based on concordance of multiple gene genealogies, has revolutionized fungal taxonomy [15] and exposed weaknesses in traditional morphology-based identification. Taxonomic revisions and the recognition of previously cryptic speciation in Trichoderma has also made clear that the universal DNA barcode for fungi, the internal transcribed spacers 1 and 2 of the nuclear ribosomal RNA gene cluster (ITS), is no longer adequate to ensure accurate species determinations in many Trichoderma sections [16–18], where a multi-gene approach is now usually adopted. By 2015 [19], there were 256 accepted Trichoderma name combinations, a number that is regularly increasing.
Multi-gene phylogenetics provides a gold standard for fungal identification and species delimitation. However, its methodology is time-consuming, technically demanding and expensive. Phenotyping using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) provides an attractive alternative for rapid microbial identification and strain differentiation purposes and has been used in filamentous fungi such as species of Aspergillus, Fusarium, Penicillium, Trichoderma and Metarhizium, among others [20,21]. The major advantages of MALDI-TOF are its cost effectiveness, rapidity, low error rate and the possibility of distinguishing closely related species [22].
While correct species identification is important in the selection and validation of microbial biocontrol agents [23], assessment of infraspecific variation is also of importance to protect commercial strains and to understand the genetic resources available in natural populations. There are abundant reports of molecular genotyping techniques applied to fungal biocontrol agents available in the literature. One of the most attractive methods, however, due to its ability to efficiently generate large numbers of markers at low cost which are amenable to automated fluorescence-based scoring is AFLP (amplification fragment length polymorphism), which has been used to identify and differentiate closely related species of Trichoderma [24].
Due to the potential of Trichoderma species to control white rot disease, we aimed to collect strains from crop soils from multiple localities in some of the principal garlic and onion growing areas in Brazil. We also aimed to correctly identify the strains, under the current taxonomic framework, to the species level using multi-gene DNA sequence analysis and assess their genetic and phenotypic variation using AFLP and MALDI-TOF, respectively. Such data will be a valuable resource for ongoing biocontrol research in Brazil.
Materials and methods
Collection and isolation of Trichoderma strains
Trichoderma strains were isolated from soil samples collected from eight distinct garlic or onion crops in the Brazilian states of Santa Catarina (SC), Minas Gerais (MG), Rio Grande do Sul (RS) and São Paulo (SP) (Table 1; Fig 1 - Map). From each sample, 10 g of soil was placed in a 250 ml Erlenmeyer flask containing 90 ml of sterile distilled water. After stirring at 180 rpm for 40 min, serial dilutions were spread onto plates containing Martin's semi-selective medium (per litre: 18 g agar, 10 g dextrose, 0.5 g MgSO4, 0.5 g peptone, 0.5 g beef extract, 0.05 g bengal pink and 0.3 g chloramphenicol) and incubated at 28°C for 7 days. Isolated colonies with typical Trichoderma morphology were transferred to potato-dextrose agar (PDA; Difco) supplemented with 0.25 ml l-1 Triton X100 and 0.3 g l-1 chloramphenicol for the subsequent isolation of monosporic cultures. Fungal sample collection was carried out according to Brazilian legislation (IBAMA process 02001.006479/2010-93 and permit no 02/2008).
Table 1. Trichoderma species identified from garlic and onion crop soils in Brazil and Genbank accession numbers of their partial actin, calmodulin, rpb2, tef1-α and ITS sequences used in phylogenetic analysis.
| Species | Strain | Collection location | Crop | act | cal | rpb2 | tef1-α | ITS |
|---|---|---|---|---|---|---|---|---|
| T. koningiopsis | CEN1386 | Curitibanos, SC | Garlic | MK696725 | MK696671 | MK696779 | MK696617 | MK714859 |
| T. peberdyi | CEN1387 | Curitibanos, SC | Garlic | MK696727 | MK696673 | MK696781 | MK696619 | MK714861 |
| T. peberdyi | CEN1388 | Curitibanos, SC | Garlic | MK696728 | MK696674 | MK696782 | MK696620 | MK714862 |
| T. peberdyi | CEN1389 | Curitibanos, SC | Garlic | MK696729 | MK696675 | MK696783 | MK696621 | MK714863 |
| T. peberdyi | CEN1390 | Curitibanos, SC | Garlic | MK696730 | MK696676 | MK696784 | MK696622 | MK714864 |
| T. peberdyi | CEN1391 | Rio Paranaíba, MG | Garlic | MK696731 | MK696677 | MK696785 | MK696623 | MK714865 |
| T. peberdyi | CEN1392 | Rio Paranaíba, MG | Garlic | MK696732 | MK696678 | MK696786 | MK696624 | MK714866 |
| T. peberdyi | CEN1393 | Rio Paranaíba, MG | Garlic | MK696734 | MK696680 | MK696788 | MK696626 | MK714868 |
| T. hamatum | CEN1394 | Rio Paranaíba, MG | Garlic | MK696735 | MK696681 | MK696789 | MK696627 | MK714869 |
| T. hamatum | CEN1395 | Rio Paranaíba, MG | Garlic | MK696736 | MK696682 | MK696790 | MK696628 | MK714870 |
| T. asperelloides | CEN1396 | Rio Paranaíba, MG | Garlic | MK696737 | MK696683 | MK696791 | MK696629 | MK714871 |
| T. asperelloides | CEN1397 | Rio Paranaíba, MG | Garlic | MK696738 | MK696684 | MK696792 | MK696630 | MK714872 |
| T. peberdyi | CEN1398 | Bueno Brandão, MG | Garlic | MK696740 | MK696686 | MK696794 | MK696632 | MK714874 |
| T. longibrachiatum | CEN1399 | São Marcos, RS | Garlic | MK696741 | MK696687 | MK696795 | MK696633 | MK714875 |
| T. longibrachiatum | CEN1400 | São Marcos, RS | Garlic | MK696742 | MK696688 | MK696796 | MK696634 | MK714876 |
| T. longibrachiatum | CEN1401 | São Marcos, RS | Garlic | MK696744 | MK696690 | MK696798 | MK696636 | MK714878 |
| T. longibrachiatum | CEN1402 | São Marcos, RS | Garlic | MK696745 | MK696691 | MK696799 | MK696637 | MK714879 |
| T. azevedoi | CEN1403 | São Marcos, RS | Garlic | MK696746 | MK696692 | MK696800 | MK696638 | MK714880 |
| T. longibrachiatum | CEN1404 | São Marcos, RS | Garlic | MK696747 | MK696693 | MK696801 | MK696639 | MK714881 |
| T. koningiopsis | CEN1405 | São Marcos, RS | Garlic | MK696749 | MK696695 | MK696803 | MK696641 | MK714883 |
| T. koningiopsis | CEN1406 | São Marcos, RS | Garlic | MK696750 | MK696696 | MK696804 | MK696642 | MK714884 |
| T. koningiopsis | CEN1407 | São Marcos, RS | Garlic | MK696751 | MK696697 | MK696805 | MK696643 | MK714885 |
| T. asperelloides | CEN1408 | Monte Alto, SP | Onion | MK696753 | MK696699 | MK696807 | MK696645 | MK714887 |
| T. asperelloides | CEN1409 | Monte Alto, SP | Onion | MK696755 | MK696701 | MK696808 | MK696647 | MK714889 |
| T. afroharzianum | CEN1410 | Monte Alto, SP | Onion | MK696756 | MK696702 | MK696809 | MK696648 | MK714890 |
| T. asperelloides | CEN1411 | Monte Alto, SP | Onion | MK696757 | MK696703 | MK696810 | MK696649 | MK714891 |
| T. lentiforme | CEN1412 | Monte Alto, SP | Onion | MK696758 | MK696704 | MK696811 | MK696650 | MK714892 |
| T. asperelloides | CEN1413 | Monte Alto, SP | Onion | MK696759 | MK696705 | MK696812 | MK696651 | MK714893 |
| T. afroharzianum | CEN1414 | Monte Alto, SP | Onion | MK696760 | MK696706 | MK696813 | MK696652 | MK714894 |
| T. lentiforme | CEN1415 | São José do Rio Pardo, SP | Onion | MK696761 | MK696707 | MK696814 | MK696653 | MK714895 |
| T. lentiforme | CEN1416 | São José do Rio Pardo, SP | Onion | MK696762 | MK696708 | MK696815 | MK696654 | MK714896 |
| T. afroharzianum | CEN1417 | São José do Rio Pardo, SP | Onion | MK696763 | MK696709 | MK696816 | MK696655 | MK714897 |
| T. asperelloides | CEN1418 | São José do Rio Pardo, SP | Onion | MK696764 | MK696710 | MK696817 | MK696656 | MK714898 |
| T. asperelloides | CEN1419 | São José do Rio Pardo, SP | Onion | MK696765 | MK696711 | MK696818 | MK696657 | MK714899 |
| T. erinaceum | CEN1420 | São José do Rio Pardo, SP | Onion | MK696766 | MK696712 | MK696819 | MK696658 | MK714900 |
| T. erinaceum | CEN1421 | São José do Rio Pardo, SP | Onion | MK696767 | MK696713 | MK696820 | MK696659 | MK714901 |
| T. azevedoi | CEN1422 | Rio Paranaíba, MG | Onion | MK696768 | MK696714 | MK696821 | MK696660 | MK714902 |
| T. azevedoi | CEN1423 | Rio Paranaíba, MG | Onion | MK696769 | MK696715 | MK696822 | MK696661 | MK714903 |
| T. asperelloides | CEN1424 | Rio Paranaíba, MG | Onion | MK696770 | MK696716 | MK696823 | MK696662 | MK714904 |
| T. peberdyi | CEN1425 | Rio Paranaíba, MG | Onion | MK696771 | MK696717 | MK696824 | MK696663 | MK714905 |
| T. peberdyi | CEN1426 | Itobi, SP | Onion | MK696772 | MK696718 | MK696825 | MK696664 | MK714906 |
| T. asperelloides | CEN1427 | Itobi, SP | Onion | MK696773 | MK696719 | MK696826 | MK696665 | MK714907 |
| T. lentiforme | CEN1428 | Itobi, SP | Onion | MK696775 | MK696721 | MK696827 | MK696667 | MK714909 |
| T. lentiforme | CEN1429 | São José do Rio Pardo, SP | Onion | MK696776 | MK696722 | MK696828 | MK696668 | MK714910 |
| T. asperelloides | CEN1430 | São José do Rio Pardo, SP | Onion | MK696777 | MK696723 | MK696829 | MK696669 | MK714911 |
| T. asperelloides | CEN1431 | Sacramento, MG | Onion | MK696778 | MK696724 | MK696830 | MK696670 | MK714912 |
| T. peberdyi | CEN1457 | Curitibanos, SC | Garlic | MK696726 | MK696672 | MK696780 | MK696618 | MK714860 |
| T. peberdyi | CEN1458 | Curitibanos, SC | Garlic | MK696733 | MK696679 | MK696787 | MK696625 | MK714867 |
| T. asperelloides | CEN1459 | Bueno Brandão, MG | Garlic | MK696739 | MK696685 | MK696793 | MK696631 | MK714873 |
| T. longibrachiatum | CEN1460 | São Marcos, RS | Garlic | MK696743 | MK696689 | MK696797 | MK696635 | MK714877 |
| T. longibrachiatum | CEN1461 | São Marcos, RS | Garlic | MK696748 | MK696694 | MK696802 | MK696640 | MK714882 |
| T. longibrachiatum | CEN1462 | São Marcos, RS | Garlic | MK696752 | MK696698 | MK696806 | MK696644 | MK714886 |
| T. asperellum | CEN1463 | Monte Alto, SP | Onion | MK696754 | MK696700 | - | MK696646 | MK714888 |
| T. asperellum | CEN1464 | Itobi, SP | Onion | MK696774 | MK696720 | - | MK696666 | MK714908 |
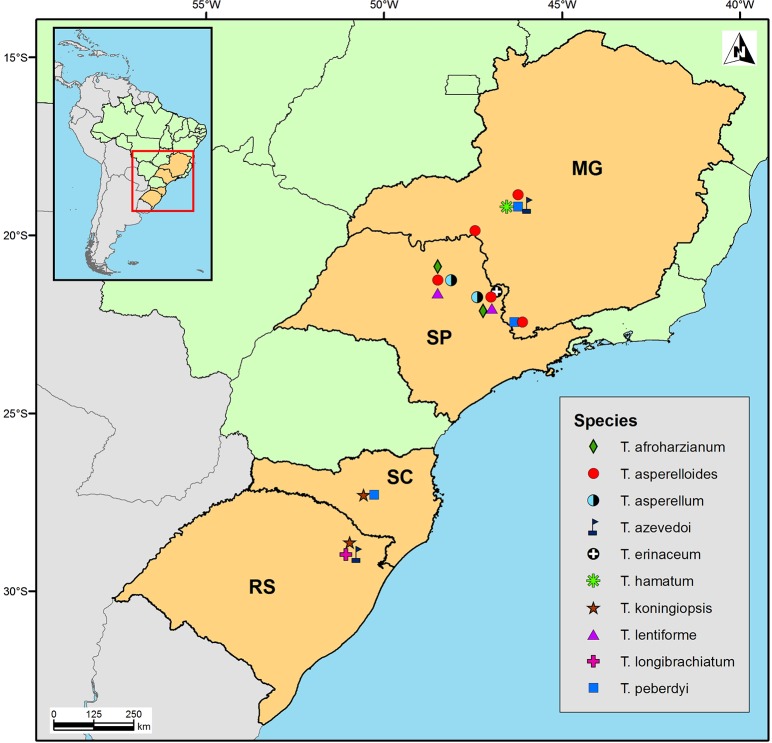
Fig 1. Map of Southeastern Brazil showing soil collection sites and recovered Trichoderma species.
Morphological characterization
For comparison of growth, colony appearance and morphological features, discs of fresh monosporic Trichoderma cultures were transferred to 9 cm Petri dishes containing 20 ml of either PDA, CMD (cornmeal dextrose agar) or SNA (synthetic low nutrient agar), which were cultured at 15, 20, 25, 30 and 35°C, with 12 hour photoperiod. Morphological characteristics, such as the aspects of phialides, conidia and chlamydospores were observed using a Nikon Eclipse Ci microscope fitted with a Nikon DS Ri2 camera. Microscopical measurements and analysis were carried out using NIS Elements (v. 4.30.01, Nikon) software, where means were based on 30 individual phialides and conidia from each specimen.
Phylogenetic analysis
Strains were cultivated on PDA for 72 h at 25°C prior to collection of mycelium, which was scraped from the agar surface, lyophilized and maintained at -80°C. Genomic DNA was purified from approximately 20 mg of the lyophilized mycelium, using a cetyl trimethyl ammonium bromide (CTAB) extraction method [25]. The nuclear ribosomal ITS1–5.8S rRNA–ITS2 region (ITS), actin (act), calmodulin (cal), translation elongation factor 1-α (tef1-α) and RNA Polymerase II subunit (rpb2) markers were amplified by PCR using a mix comprising approximately 2 ng genomic DNA, 1x PCR buffer with 2.0 mM MgCl2, 0.2 mM dNTPs, 1U Taq polymerase and 0.3 μM of each primer. Thermal cycling for all markers was standardized as 2 min at 95°C then 35 cycles of 20 sec at 95°C, 30 sec at the appropriate annealing temperature for the primers used and 90 sec at 72°C, followed by 7 min at 72°C. Primer sequences and annealing temperatures are given in Table 2. The internal act sequencing primer, Tact293F, was designed from a conserved region identified in a preliminary alignment of several Trichoderma act PCR products sequenced using the amplification primers, along with cognate reference sequences obtained from Genbank. The tef1-α reverse PCR primer, tef1080R, was designed from a conserved region identified in an alignment of the 3´ portion of the tef1-α gene, amplified from a selection of Trichoderma isolates using EF1–1018F and EF1–1620R primers. We obtained fewer artefact bands in PCRs using the tef1080R reverse primer compared with the more widely used tef997R primer. Since the 3´ portion of the tef1-α gene is much less variable than the 5´ portion, phylogenetic analysis was restricted to the portion amplified using tef71f and tef1080R primers. Furthermore, there is a richer representation of the 5´ portion of the tef1-α gene from related Trichoderma species in the databanks, further influencing our decision.
Table 2. Primers used for PCR and sequencing.
| Locus | Name | Primer sequence 5´-3´ | Tm | Reference |
|---|---|---|---|---|
| ITS | U1 | GGAAGKARAAGTCGTAACAAGG | 55 | [29] |
| U4 | RGTTTCTTTTCCTCCGCTTA | " | ||
| act | Tact1 | TGGCACCACACCTTCTACAATGA | 50 | [30] |
| Tact2 | TCTCCTTCTGCATACGGTCGGA | [31] | ||
| * | Tact511R | CTCAGGAGCACGGAAT | " | |
| * | Tact293F | GTGATCTTACCGACTACCTGATG | This study | |
| cal | CAL-228F | GAGTTCAAGGAGGCCTTCTCCC | 55 | [32] |
| CAL-737R | CATCTTTCTGGCCATCATGG | " | ||
| * | CAL-235F | TTCAAGGAGGCCTTCTCCCTCTT | " | |
| tef1-α | tef71f | CAAAATGGGTAAGGAGGASAAGAC | 50 | [33] |
| * | tef85f | AGGACAAGACTCACATC AACG | " | |
| * | tef954r | AGTACCAGTGATCATGTTCTTG | " | |
| * | tef997R | CAGTACCGGCRGCRATRATSAG | " | |
| tef1080R | GATACCAGCCTCGAACTCACC | This study | ||
| EF1–1018F | GAYTTCATCAAGAACATGAT | [34] | ||
| EF1–1620R | GACGTTGAADCCRACRTTGTC | " | ||
| rpb2 | RPB2_210up | TGGGGWGAYCARAARAAGG | 48 | Tom Gräfenhan; |
| RPB2_1450low | CATRATGACSGAATCTTCCTGGT | http://www.isth.info | ||
| * | RPB2 1150low | GGTTGTGATCRGGRAARGGAATG | " |
*Internal primers used for sequencing only.
PCR products were verified by agarose gel electrophoresis and were then prepared for sequencing using ExoSAP (Applied Biosystems, Foster City, CA, USA). Both DNA strands were sequenced using the Big Dye v.3.1 kit (Applied Biosystems), using appropriate primers (Table 2) and an ABI3730 DNA Analyzer (Applied Biosystems). Sequence reads were trimmed for quality, contigs assembled and any base calling mismatches resolved using Chromas Pro (v. 1.5, Technelysium Pty Ltd). Sequences, including references obtained from Genbank, were organized into matrices in Bioedit (v. 7.2.6) [26] and aligned using MAFFT v. 7 E-INS-i [27]. A concatenated matrix was assembled using Sequence Matrix (v. 1.8) [28].
An optimal partitioning scheme for each marker was determined in PartitionFinder 2 [35]. Maximum likelihood trees based on data from each marker as well as the concatenated matrix were constructed using IQ-TREE (v. 1.6.5) [36], where optimal nucleotide substitution models for each partition were selected using ModelFinder [37]. Branch support was estimated using the Ultrafast bootstrap (UFBoot) [38] with 1000 replicates. Branches with UFboot support of > = 95% were considered credible. Representative sequences of each Trichoderma isolate cluster for each marker were used in BLAST [39] searches of the Genbank database in order to make provisional Trichoderma species identifications. This information was used to select reference sequences for further analyses, also incorporating strains used in recent taxonomic and molecular phylogenetic treatments of appropriate Trichoderma sections [23,40,41], S1 Table.
The partitioned concatenated matrix was also analysed using the Bayesian Metropolis-coupled Markov Chain Monte Carlo method as implemented in MrBayes 3.2.6 [42], running on the CIPRES Science Gateway [43] and utilizing the Beagle library [44]. Model selection for each partition was made in PartitionFinder2 [35]. Two runs of eight MCMCMC chains with a heating temperature of 0.075 were conducted for ten million generations, sampling every 1000 generations. This runtime was sufficient for the convergence diagnostic, the standard deviation of split frequencies, to fall to a minimum of 0.005217. The first 25% of the trees were discarded (burn-in) prior to calculation of the 50% majority rule consensus tree.
AFLP genotyping
Genetic variability among a representative selection of 46 of the 54 Trichoderma isolates was evaluated using the amplified fragment length polymorphism method (AFLP; [45]; adapter and primer sequences given in Table 3) adapted for fluorescent detection. A one-step digestion and adapter-ligation protocol was adopted, which were performed in 20 μl volumes. A single reaction comprised 1X ligase buffer (Promega), 50 mM NaCl, 0.05 μg/μl bovine serum albumin, one unit T4 DNA ligase (Promega), five pmol EcoRI adapter, 50 pmol MseI adapter, five units EcoRI (EcoRI-HF high fidelity, NEB), five units of MseI and 100 ng genomic DNA. The reactions were incubated on a PCR machine at 37°C for two hours, then held at 17°C for one hour and then held at 4°C for two hours. Samples were then diluted five times by the addition of 80 μl H2O and stored at -80°C. The primers EcoRI+A and MseI+C were used for preselective PCR. A 20 μl PCR in 1X PCR Buffer with 2 mM Mg2+ contained 1 M Betaine, 0.25 mM dNTPs, 0.5 μM each primer, 1 U Taq polymerase and 2 μl of the diluted adapter-ligated DNA. Cycling conditions comprised an initial 72°C for 2 min to allow fill-in of the adapter ends, then 20 cycles of 94°C for 30 sec, 56°C for 1 min and 72°C for 2 min. Ramp rate was limited to 1°C per second. Following cycling, reactions were held at 72°C for 2 min and then 60°C for 30 min. Five μl of PCR products were subsequently analysed on a 1.5% agarose gel, producing a faint smear if the reaction was successful. The preselected DNA was then diluted five times by the addition of 80 μl H2O and stored at -20°C.
Table 3. AFLP adapter and primer sequences.
| Primer name | Primer sequence 5´-3´ |
|---|---|
| EcoRI-Adapter1 | CTCGTAGACTGCGTACC |
| EcoRI-Adapter2 | AATTGGTACGCAGTCTAC |
| EcoRI+A | GACTGCGTACCAATTCA |
| MseI_Adapter1 | GACGATGAGTCCTGAG |
| MseI_Adapter2 | TACTCAGGACTCAT |
| MseI+C | GATGAGTCCTGAGTAAC |
| (11) EcoRI-AC FAM | FAM+GACTGCGTACCAATTCAC |
| (14) EcoRI-AA VIC | VIC+GACTGCGTACCAATTCAA |
| (2) MseI+CTT | GATGAGTCCTGAGTAACTT |
| (3) MseI+CAT | GATGAGTCCTGAGTAACAT |
| (5) MseI+CAG | GATGAGTCCTGAGTAACAG |
| (6) MseI+CAA | GATGAGTCCTGAGTAACAA |
Genomic complexity was further reduced using selective PCR to produce resolvable AFLP profiles, using PCR primer pairs comprising one labelled EcoRI+2 and one unlabelled MseI+3 primer (Table 3). Selective primer combinations producing an adequate number of clear fluorescent peaks in preliminary screening were chosen from the range available in the Small Plant Genome Mapping Kit (Applied Biosystems). We found that whilst most MseI+3 primer variations gave satisfactory results, only the EcoRI+AC and EcoRI+AA primers were efficient in Trichoderma. A single 10 μl selective PCR reaction in 1X PCR buffer with 2 mM Mg2+ contained 0.15 μM each of MseI+3 primer and fluorochrome-labelled EcoRI+2 primer, 0.2 mM dNTPs, 0.5 U Taq polymerase and 2 μl of diluted preselected DNA. PCR cycling comprised an initial 94°C for 2 min, then 10 cycles of 94°C for 30 sec, 66°C for 30 sec and 72°C for 1 min. The annealing temperature was reduced by 1°C per cycle (touchdown). Then followed 25 cycles of 94°C for 30 sec, 56°C for 30 sec and 72°C for 1 min. Reactions were then held at 72°C for 3 min and at 60°C for 30 min. Six primer combinations (11x5, 11x6, 14x2, 14x3, 14x5, 14x6; Table 3) were selected for the full analysis. The fluorescent AFLP profiles were detected by mixing 1 μl PCR product with 9 μl HiDi formamide and 0.3 μl of the Genescan 600-LIZ v 2.0 molecular size ladder (Applied Biosystems). Samples were denatured at 95°C for 5 minutes and snap cooled on ice, prior to injection on an ABI 3730 DNA Analyzer (Applied Biosystems).
The raw AFLP data files were processed using PeakScanner (v. 2; Applied Biosystems). The table of peak area data was then imported into the R CRAN library program, RawGeno [46], for peak binning and filtering of low quality or partially overlapping peaks, thereby reducing the risk of size-homoplasy. The filtered AFLP profiles were then converted into a peak presence or absence binary matrix, totalling 364 characters. Data was analysed under the F81 (restriction; nst = 1 rates = invgamma) model in MrBayes 3.2.6 [42] using two runs of four MCMCMC chains, where two million generations were sampled every 1000 generations. This runtime was sufficient for the average standard deviation of split frequencies to fall to 0.007734. The first 25% of the trees were discarded (burn-in) prior to calculation of the 50% majority rule consensus tree. Intraspecific Nei-Li genetic distances (fragments, length = 4) were calculated in PAUP (v. 4.0a165) [47].
MALDI-TOF phenotyping and rapid identification
Samples of Trichoderma strains were collected from colonies cultivated on PDA plates, which were applied directly to a MSP96 plate (Bruker Daltonics GmbH, Bremen, Germany) and covered with 1 μl of MALDI matrix solution (‘Bruker HCCA’ or α-cyano-4-hydroxycinnamic acid, at a final concentration of 5 mg HCCA ml-1). After sample drying, analyses were performed on a MicroFlex MALDI-TOF mass spectrometer (Bruker Daltonics GmbH), fitted with a nitrogen laser (337 nm) of 20–65% offset intensity and spiral mode of acquisition, where an average of 400 shots (40 laser shots at 10 different regions of the target spot) at 60 Hz were conducted. Signals in the range 2000–20000 m/z were automatically collected with AutoConverter from the acquisition software (FlexControl 3.3; Bruker Daltonics GmbH). Data were exported to MALDI Biotyper software (3.0; Bruker Daltonics GmbH) and each consensus spectrum incorporated into a profile in the mean spectrum projection (MSP) database.
Based on the results of the phylogenetic analysis, the strains CEN1386, CEN1395, CEN1402, CEN1419, CEN1390, CEN1416 and CEN1420, were selected for the creation of a local Trichoderma spectrum database using the Biotyper MBT Explorer Software Module. The library was constructed by sampling colonies grown on individual PDA plates for five days, where material was collected from three distinct regions (colony edge, intermediate and central). The spectra obtained were then used to generate species profiles which were added to the database. Subsequent samples were analyzed in triplicate. Cluster analysis was conducted using the MSP Dendrogram Creation Standard Method (v.1.4) of MALDI Biotyper Software (v. 3.0).
Nomenclature
The electronic version of this article in Portable Document Format (PDF) in a work with an ISSN or ISBN will represent a published work according to the International Code of Nomenclature for algae, fungi, and plants, and hence the new names contained in the electronic publication of a PLOS ONE article are effectively published under that Code from the electronic edition alone, so there is no longer any need to provide printed copies.
In addition, new names contained in this work have been submitted to MycoBank from where they will be made available to the Global Names Index. The unique MycoBank number can be resolved and the associated information viewed through any standard web browser by appending the MycoBank numbers contained in this publication to the prefix http://www.mycobank.org/MB/. The online version of this work is archived and available from the following digital repositories: PubMed Central, LOCKSS.
Results and discussion
A total of 54 Trichoderma strains were isolated from crop soil samples from multiple sites representing the main growing areas of garlic and onion in Brazil. In quantitative terms, 11 strains were isolated from Rio Paranaíba, MG; one from Bueno Brandão, MG; one from Sacramento, MG; seven from Monte Alto, SP; nine from São José do Rio Pardo, SP; three from Itobi, SP; nine from São Marcos, RS; and five from Curitibanos, SC (Table 1; Fig 1).
All 54 presumptive strains were confirmed as Trichoderma species using the ITS oligonucleotide barcode identification program TrichOKEY2 [48] http://www.isth.info/. However, confident and unambiguous species identifications were not obtained in the searches, as expected, since previous studies have pointed out the limitations of ITS sequences to delimit Trichoderma species [23]. Furthermore, 12 isolates were returned as belonging to unidentified Trichoderma species. We therefore performed a full phylogenetic analysis on all 54 isolates with the addition of a further four phylogenetic markers: act, cal, tef1-α and rpb2. We first performed a ML phylogenetic analysis on the most highly substituted data set (tef1-α; Table 4). Clusters with high sequence similarity were identified and a representative sequence of each cluster used in BLAST [39] searches of the Genbank nucleotide database. Reference sequences for each marker were then selected from the Trichoderma (or its sexual morph, Hypocrea) species producing top hits, giving preference to type strains and those with most complete representation for our marker selection. We also selected reference sequences based on recent molecular taxonomic treatments of the Trichoderma sections and major clades identified in the initial ITS TrichOKEY screen (S1 Table).
Table 4. Partition statistics for each sequenced DNA locus generated in IQ-TREE.
| Partition | Sites | Invariable sites | Parsimony informative sites | Tree length | Best fit model (BIC) |
|---|---|---|---|---|---|
| act | 754 | 579 | 135 | 1.1454 | TIM3e+I+G4 |
| cal | 524 | 216 | 263 | 3.2113 | K2P+I+G4 |
| ITS | 685 | 405 | 144 | 1.8155 | TIM2+F+R3 |
| rpb2 | 800 | 471 | 295 | 2.7089 | TIMe+I+G4 |
| tef1-α | 752 | 272 | 405 | 9.7299 | TN+F+R4 |
Among the sequenced markers, the most informative, based on number of parsimony informative characters (PICS), was tef1-α, followed by rpb2, cal, ITS and act (Table 4). The tef1-α matrix, including reference sequences, was notably rich in indels, presenting a potential risk for alignment ambiguity, which we attempted to minimize by the use of MAFFT E-INS-i, which is among the most accurate of modern consistency-based programs [49]. Among the 54 isolates studied, sequence data were gathered for all five markers, except for two isolates, later identified as Trichoderma asperellum, for which the rpb2 marker could not be amplified, probably due to critical primer mismatch.
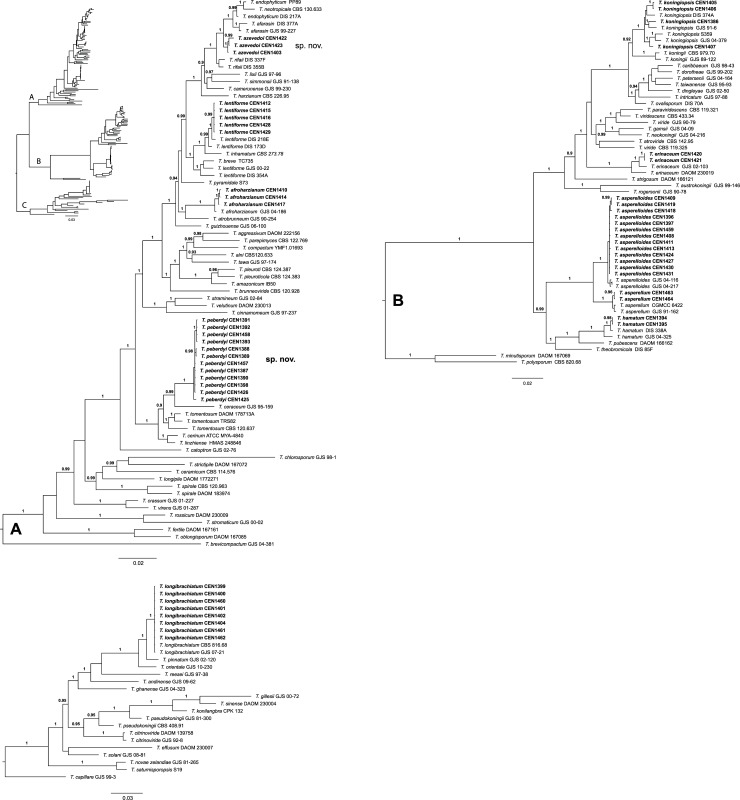
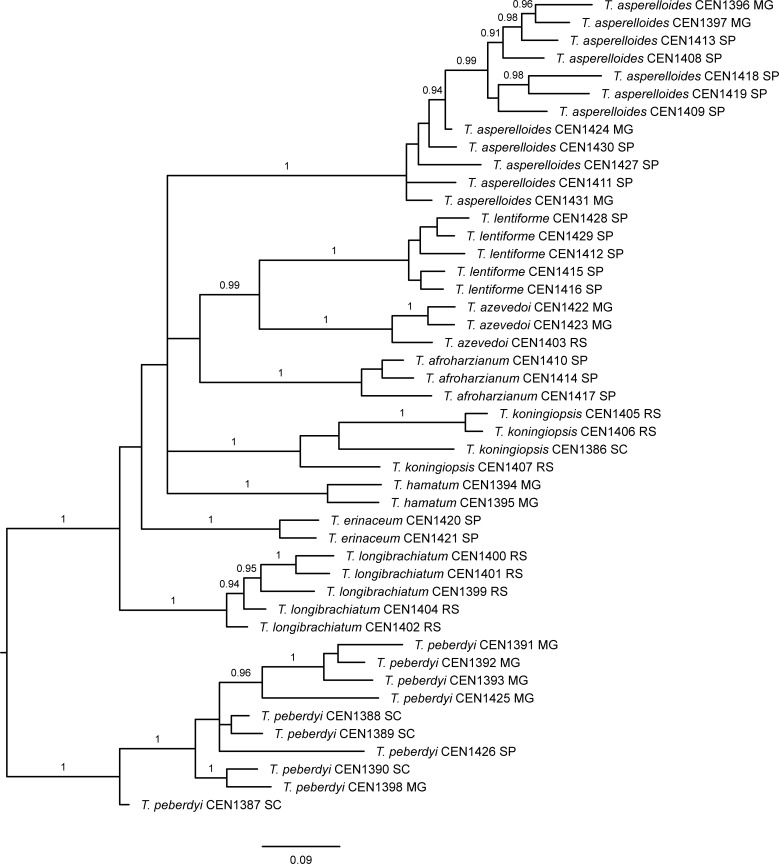
The Bayesian phylogram based on the concatenation of ITS, rpb2, act, cal and tef1-α sequences (Fig 2) permitted the unambiguous identification of eight species among the 54 isolates, based on their clustering with reference taxa. The isolates identified in the analysis included five Trichoderma lentiforme, two Trichoderma hamatum, three Trichoderma afroharzianum, four Trichoderma koningiopsis, two Trichoderma erinaceum, 13 Trichoderma asperelloides, two T. asperellum and eight Trichoderma longibrachiatum. The remaining 15 isolates formed two distinct clades: one comprising three isolates, most closely related to Trichoderma rifaii, Trichoderma afarasin, Trichoderma endophyticum and Trichoderma neotropicale and a second clade comprising 12 isolates, corresponding to the unidentified group in the TrichOKEY search, most closely related to Trichoderma ceraceum and Trichoderma tomentosum. Both clades are highly supported in the Bayesian analysis (PP = 1) (Fig 2) and possess 100% ultrafast bootstrap support in a maximum likelihood tree based on the same concatenated matrix (S1 Fig). We therefore suspected that the two unidentified clades represent new Trichoderma species, which we confirmed by examination of their distinctive growth and morphological characteristics.
Fig 2.
A-C. Midpoint rooted Bayesian phylogram (split into three parts as indicated in the overview), based on the concatenation of act, cal, ITS, rpb2 and tef1-α matrices. Posterior probabilities are given above branches (> 0.9) and the scale bar represents expected changes per site. Strains sequenced in the present study are in bold and are followed by CENxxx numbers. Two new Trichoderma species, T. azevedoi and T. peberdyi are indicated.
Taxonomy
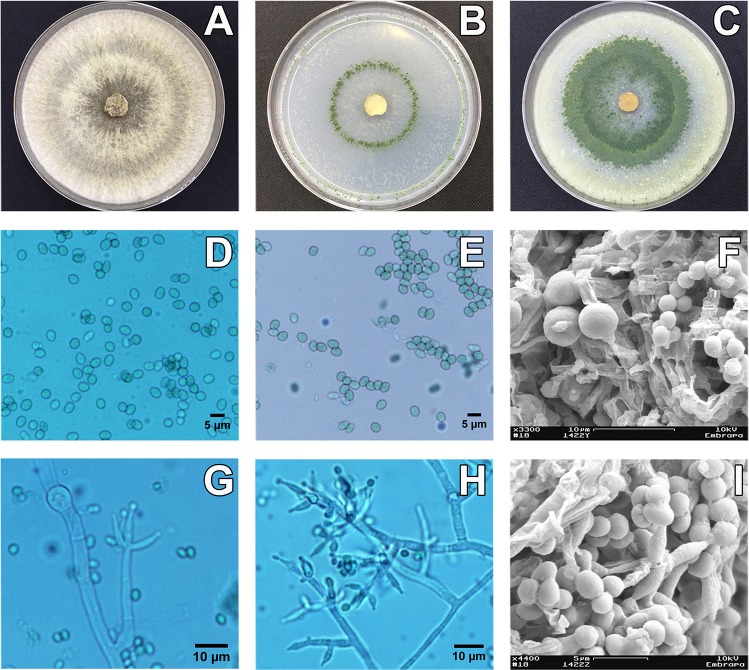
Trichoderma azevedoi Valadares-Inglis, M.C. & Inglis, P.W. sp. nov. Fig 3
Fig 3. Culture characteristics and morphology of T. azevedoi sp. nov. strain CEN1422 (holotype).
Panels A-I: Growth on three different media, PDA (A); SNA (B); CMD (C); and morphology of the conidia (D, E), phialides (G, H) and chlamydospores (G) using optical microscopy and conidia and chlamydospores using electron microscopy (F, I).
Mycobank MB830305. [urn:lsid:mycobank.org: 830305]
Etmology
Named in honour of João Lúcio Azevedo (São Paulo University—Brazil) for his contributions to mycology and microbial genetics in Brazil, including the mentoring of numerous professionals in Trichoderma studies.
Holotype
CEN1422, a freeze dried, metabolically inactive culture deposited in the Herbarium of Embrapa Recursos Genéticos e Biotecnologia (CEN). Collected in Rio Paranaíba—MG state, Brazil, 200 05’ 06” S, 510 00’ 02” E, from onion crop soil, 02/07/2015, by V. Lourenço Jr. & J.B.T. da Silva. An ex-holotype culture of CEN1422 has been deposited in the Embrapa Coleção de Microrganismos para o Controle de Fitopatógenos e Plantas Daninhas, with the accession number BRM46357.
Description
On CMD, colony radius 40 mm after 72h at 25 and 30°C and 12 h photoperiod. Mycelium hyaline with cottony pustules, sporulating heavily after 72 h at 30°C and 96 h at 25°C, turning green after 96 h, more abundantly in a broad ring about half-way to the plate center. At 20°C and 12 h photoperiod, colony radius 25 mm, mycelium hyaline with pustules of spores formed at 96h. No growth observed at 15 and 30°C. On SNA, colony radius 34 mm at 25°C and 40 mm at 30°C after 72 h with 12 h photoperiod. Mycelium hyaline with spores formed after 72 h in sparse clumps distributed throughout the plate. At 30°C mid-green spores are formed in a distinct thin concentric ring, approximately one third of the radius from the plate center to the edge. At 20°C spores are produced after 96 h similarly to 30°C. On PDA, colony radius 17 mm at 20°C, 62 mm at 25°C and 4 mm at 30°C, at 72 h with 12 h photoperiod. Mycelium cottony, with light green spores formed after 96 h, concentrated in the centre of the plate and in a broad concentric ring approximately half-way to the plate edge.
Conidiophores trichoderma-like, pyramidal with opposing branches or isolated, terminating in groups of three to five phialides. Phialides ampulliform to lageniform, constricted below the tip forming a narrow neck, measuring 7.71 ± 1.42 x 2.52 ± 0.32 μm (overall range 5.45–10.75 x 1.89–3.17 μm), base 1.46–2.55 μm (mean 1.99 μm). Conidia globose, subglobose to ovoid 3.90 ± 0.31 x 2.93 ± 0.22 μm (overall range: 3.54–4.65 x 2.55–3.33 μm). Chlamydospores common, terminal and intercalary, typically globose.
Sexual morph: Unknown. Known distribution: Brazil.
Other isolates examined
CEN1403, CEN1423. From garlic or onion crop soils.
Notes
Trichoderma azevedoi is closely related to Trichoderma rifaii (a member of the Trichoderma harzianum complex). However, T. rifaii is known only as an endophyte of Theobroma cacao and Theobroma gileri. T. azevedoi conidia (mean 3.90 x 2.93 μm) are much larger than T. rifaii (mean 2.6 x 2.4 μm) and T. azevedoi produces abundant clamydospores, which have not been reported in T. rifaii.
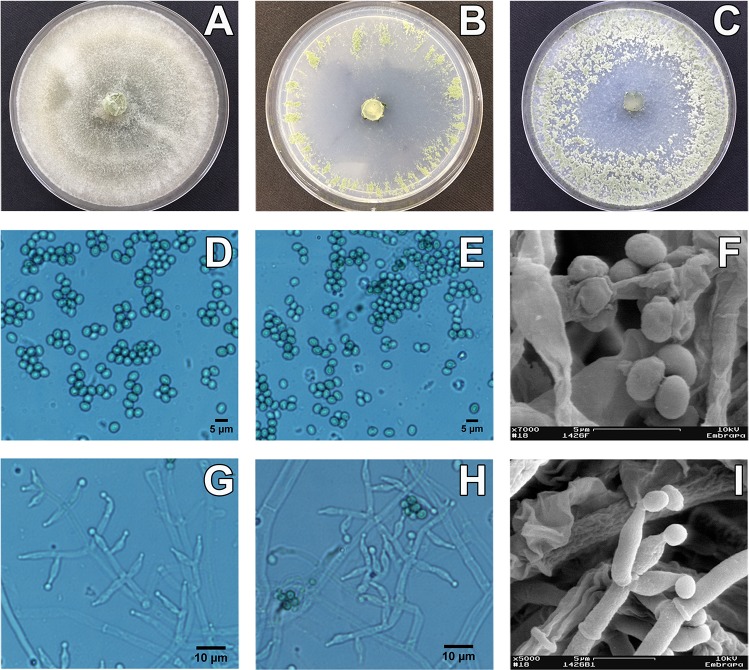
Trichoderma peberdyi Valadares-Inglis, M.C. & Inglis, P.W. sp. nov. Fig 4.
Fig 4. Culture characteristics and morphology of T. peberdyi sp. nov. strain CEN1426 (holotype).
Panels A-I: Growth on three different media, PDA (A); SNA (B); CMD (C); and morphology of the conidia (D, E) and phialides using optical microscopy (G, H) and conidia using electron microscopy (F, I).
Mycobank MB830304. [urn:lsid:mycobank.org: 830304]
Etmology
Named in honour of John F. Peberdy (Nottingham University, UK), for his important contributions to mycology and fungal biotechnology.
Holotype
CEN1426, a freeze dried, metabolically inactive culture deposited in the Herbarium of Embrapa Recursos Genéticos e Biotecnologia (CEN). It was isolated in Itobi—SP state, Brazil, 210 44’ 13” S, 460 58’ 30” E, from onion crop soil, on 02/09/2015, by V. Lourenço Jr & J.B.T. da Silva. An ex-holotype culture of CEN1426 has been deposited in the Embrapa Coleção de Microrganismos para o Controle de Fitopatógenos e Plantas Daninhas, with the accession number BRM46363.
Description
On CMD, colony radius 40 mm after 72h at 25 and 30°C with 12 h photoperiod. Colony hyaline in sterile zones with cottony aerial hyphae after 72 h at 25 and 30°C. Spores formed after 120 h at 25°C in pustules concentrated at the plate edge. Light green spores produced at 30°C after 120 h. At 20°C and 12h photoperiod, hyaline mycelium covering entire plate and no spores observed after 120h. No growth at 15 and 35°C. On SNA, colony radius 40 mm after 96 h at 25°C and 30°C, under 12h photoperiod. Colony hyaline with sparse cottony aerial hyphae. Light green spores produced in rays near plate edge after 120 h at 25°C. Sporulation less dense at 30°C. No spores formed at 20°C. No growth observed 15 and 35°C after 120h. On PDA, colony radius 40 mm at 25 and 30°C after 72h under 12h photoperiod. Mycelium cottony, with aerial hyphae covering the entire plate with conidia forming under cottony aerial hyphae after 120h at 25 and 30°C. No diffusible pigments or distinctive odours observed. Conidiophores trichoderma-like, pyramidal with opposing branches or isolated, terminating in groups of two to three phialides. Phialides ampulliform, 7.04 ± 1.01 x 2.67 ± 0.36 (range: 4.91–9.10 x 2.20–3.73 μm), base 1.46–2.55 (mean 1.99 μm). Conidia subglobose to ovoid 3.54–4.65 (3.90) x 2.55–3.33 (2.93) μm, thinning in the proximal region, produced in chains and aggregated in mucilaginous masses. Chlamydospores not observed.
Sexual morph: Unknown. Known distribution: Brazil.
Other isolates examined
CEN1387, CEN1388, CEN1389, CEN1390, CEN1391, CEN1392, CEN1393, CEN1398, CEN1457, CEN1458, CEN1425. All from garlic or onion crop soils.
Notes
Trichoderma peberdyi is closely related to Trichoderma tomentosum and Trichoderma ceraceum. In comparison with T. tomentosum, phialides of T. peberdyi are longer and possess a distinct neck, mostly curved towards the tip. T. peberdyi conidia are a much lighter green than T. tomentosum on SNA media and not produced in distinct concentric rings. T. peberdyi conidia are subglobose to ovoid, larger than T. tomentosum, a species that produces chlamydospores on CMD, unlike T. peberdyi. T. peberdyi is distinct from T. ceraceum by its lack of diffusible yellow pigment and absence of drops of clear green liquid into which conidia form. T. ceraceum is known only from the USA.
In trees based on individual markers (S2–S6 Figs), T. peberdyi sp. nov. was distinct from all other Trichoderma species and was well-supported in all but the act ML tree. T. azevedoi, however, appears to be closely related to other neotropical Harzianum clade species, but was clearly distinct and supported in the act and tef1-α ML trees (S2 and S6 Figs).
Geographical distribution of Trichoderma species in garlic and onion crop soils
The 54 isolates identified to species level, which were collected from eight different sites distributed in four southeastern Brazilian states, fell into three Trichoderma sections: Pachybasium, Trichoderma and Longibrachiatum (www.isth.info). The species diversity per collection site varied so that one site yielded a single Trichoderma species, two sites yielded two species, one site yielded three species and four sites yielded four species each (Fig 1; Table 1). In terms of crop, garlic (four sites) yielded six Trichoderma species and onion crops (six sites) yielded seven different species. Of the three most frequently isolated species in our analysis, T. asperelloides was isolated from six sites, T. longibrachiatum was isolated from a single site and one of the new species, T. peberdyi, was isolated from four sites (Fig 1).
Inferences on local species diversity are tentative at best, however, and would require a much larger quantitative study, possibly using a meta-barcoding approach (reviewed in Kredics et al., 2018). In a study on the diversity of Trichoderma species in the Colombian Amazon region, DNA barcoding of 107 strains using ITS and tef1 sequences showed that three common cosmopolitan species comprise 68% of the studied isolates, with T. harzianum sensu lato representing 38% of strains, followed by Trichoderma spirale at 17% and T. koningiopsis at 13%, whereas only four putative new taxa were suggested [50]. A larger study of 2078 Trichoderma strains collected from agricultural fields in Eastern China, representing four major agricultural provinces, identified 17 known species: T. harzianum (429 isolates), T. asperellum (425), T. hamatum (397), T. virens (340), T. koningiopsis (248), T. brevicompactum (73), T. atroviride (73), T. fertile (26), T. longibrachiatum (22), T. pleuroticola (16), T. erinaceum (16), T. oblongisporum (2), T. polysporum (2), T. spirale (2), T. capillare (2), T. velutinum (2), and T. saturnisporum (1) [51]. The authors showed that Trichoderma biodiversity in agricultural fields varied by region, crop, and season, where, for example, relative frequencies of T. hamatum and T. koningiopsis from rice crop soil were higher than those from wheat and maize soils, suggesting a crop preference of specific Trichoderma species. Although this study principally used ITS sequences to identify species and did not split the T. harzianum species complex along the lines of its currently accepted taxonomic framework [23], there is remarkable overlap with the species recovered in the present study of Brazilian garlic and onion crop soils, despite the large geographical separation. There is accumulating evidence that certain Trichoderma species have become highly adapted to agroecosystems. Sixty-five percent of Trichoderma species associated with the rhizosphere of maize were shared between samples collected from Austria, Tenerife, Madagascar and New Zealand, whereas Trichoderma species associated with endemic plants from the same regions were highly specific and diverse. All analysed rhizosphere samples, however, shared a global Trichoderma core community dominated by T. koningii and T. koningiopsis [52].
While a comprehensive worldwide survey of the distribution of Trichoderma species under the current rapidly evolving taxonomic framework does not yet exist, recent re-evaluations of existing international culture collections and new collecting efforts in under-sampled geographical locations have greatly expanded our knowledge. Pertinent to the new species recovered herein, those most closely related to T. azevedoi include T. T. rifaii, T. endophyticum and T. neotropicale, all of which have been reported to have neotropical distributions [23,33,53]. Species most closely related to T. peberdyi include T. ceraceum, first reported from the USA [54] and T. tomentosum, which is probably cosmopolitan (unpublished Genbank strain data). Among the other Trichoderma species recovered in the current study (Table 1), T. lentiforme has been reported to be neotropical [23], while the remaining species are of worldwide distribution [23,51,55,56].
Genotypic and phenotypic variability of Trichoderma strains
AFLP
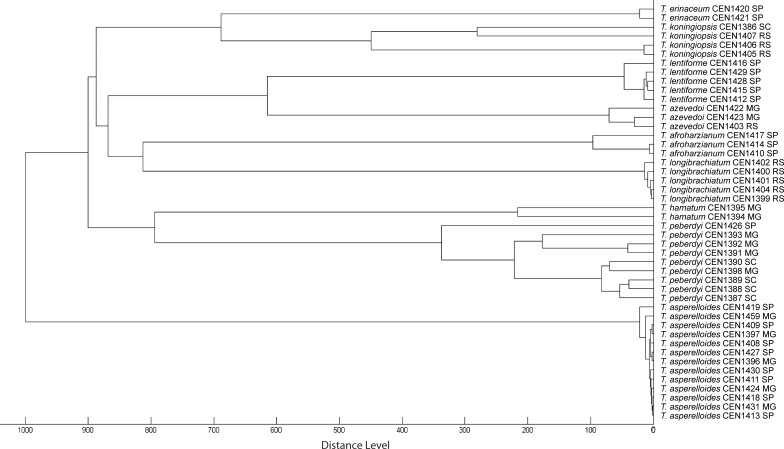
AFLP is a powerful and established molecular tool for the analysis of genetic variation in fungal populations [57]. The combination of six selective AFLP primer pairs (Table 3) yielded 364 binary characters in our selected sample of Trichoderma isolates, which were analysed using Bayesian phylogenetic inference (Fig 5). The AFLP clusters agreed closely with the species designations obtained by amplicon sequencing, where each species cluster possessed high posterior support. Closely related species in the Harzianum clade (T. lentiforme, T. azevedoi, T. afroharzianum and T. peberdyi), were clearly distinguished. However, posterior supports for some of the deeper branches of the tree were poor, as was topological congruence with the DNA sequence-based tree, suggesting that phylogenetic signals were probably saturated at this level or overcome by homoplasy. Modifications of the AFLP protocol, such as the use of EcoRI+3 primers for selective PCR, exclusion of smaller fragments or scoring of only major high rfu peaks, might improve phylogenetic resolution in deeper nodes of the resultant tree, but are likely to be at the expense of the ability to discriminate closely-related taxa. Fragment homology has been shown to decrease with greater time since divergence, so that AFLP data are probably best suited for examining phylogeographic patterns within species and among very recently diverged species [58].
Fig 5. AFLP midpoint rooted Bayesian phylogram.
Posterior probabilities are given above branches (>0.9) and the scale bar represents expected changes per site. Species names are followed by strain number and collection location by Brazilian state.
In T. asperelloides, which was one of the most commonly sampled taxa, there was no consistent, well-supported monophyletic grouping of strains according to geographical location (Fig 5). All isolates of this species, however, were collected in the south of MG and the north of SP states, which are contiguous regions (Fig 1) and where populations are possibly not clearly structured. In T. peberdyi sp. nov., another frequently sampled species, a well-supported clade of four isolates was apparent, all from the municipality of Rio Paranaíba in the mid-west of MG state (Table 1). The clade was further structured so that strain CEN1425, isolated from onion, was differentiated from the other three isolates, which were all isolated from garlic crop soil. The remaining T. peberdyi strains lacked clear and supported phylogeographic groupings, although most were isolated in the discontiguous SC state, where the outlier strain, CEN1387, was also isolated. Strain CEN1398 from Bueno Brandão-MG grouped with strain CEN1390 from SC. It is unclear if this pattern represents strain dispersal from SC or is the result of limited sampling of a contiguous population of the lineage, since the geographical range of T. peberdyi is currently unknown. T. peberdyi, along with our second newly described species, T. azevedoi sp. nov., possessed the widest geographical range observed in the current study. The two T. azevedoi strains collected from MG state formed a well-supported clade, distinct from the third strain collected from the distant RS state. The other Trichoderma species appeared to be common to only one or a few contiguous locations (Table 1; Fig 1), where mixing and dispersal of haplotypes over shorter distances is probably frequent. Elsewhere, in a study of Trichoderma spp. associated with the button mushroom, Agaricus bisporus, no clear trend was detected between AFLP clustering and geographic origin of isolated materials [59]. In terms of strain distinction, all of the Trichoderma species analysed by AFLP demonstrated significant genetic variability in the Bayesian phylogenetic analysis (Fig 5) and in calculated pairwise genetic distances (Table 5). T. peberdyi possessed the largest maximum intraspecific genetic distance, although the largest mean intraspecific distance was in T. koningiopsis.
Table 5. Intraspecific genetic distances based on ALFPs.
| Species | Mean Distance | Maximum Distance |
|---|---|---|
| T. afroharzianum | 0.0659 | 0.0797 |
| T. asperelloides | 0.0890 | 0.1209 |
| T. azevedoi | 0.0623 | 0.0742 |
| T. erinaceum | 0.0549 | 0.0549 |
| T. hamatum | 0.0769 | 0.0769 |
| T. koningiopsis | 0.1447 | 0.1786 |
| T. lentiforme | 0.0500 | 0.0659 |
| T. longibrachiatum | 0.0703 | 0.0906 |
| T. peberdyi | 0.1310 | 0.1978 |
MALDI-TOF
Matrix-assisted laser desorption/ionization mass spectrometry (MALDI-TOF MS) has become an attractive tool for the identification of microorganisms due to short processing time, reliable identification and low per-sample cost. Many filamentous fungi, such as Aspergillus, Fusarium, Penicillium and Trichoderma, have been identified by MALDI-TOF [20], where the technique can be used to complement DNA-based identification [60].
We selected 46 strains for analysis using MALDI-TOF, representing at least two of each Trichoderma species, previously identified by sequence analysis (Fig 2), with the exception of T. asperellum. Distance-based clustering of MALDI TOF spectra produced a dendrogram (Fig 6) with terminal clusters perfectly matching sequence-based identifications (Fig 2). Echoing the AFLP genotyping result (Fig 5), T. peberdyi was remarkable for the large phenotypic distance between strains in the MALDI-TOF dendrogram, second only to T. koningiopsis. In contrast, the T. asperelloides and T. longibrachiatum strains, which showed genetic variability in the AFLP analysis, were notably homogenous phenotypically. The most phenotypically diverse species was T. koningiopsis, which was sister to T. erinaceum, in agreement with the sequence-based phylogeny (Fig 2B). Otherwise, as was the case with AFLP genotyping, the topology of the MALDI-TOF dendrogram was not congruent with the sequence-based phylogeny, where both typing methodologies appear to be unsuitable for establishing deeper phylogenetic relationships in Trichoderma. No exclusive location-correlated groupings were observed in the MALDI-TOF species clusters, although some structure was evident in T. peberdyi, where a clade containing three strains from SC state was observed, which were joined by a fourth strain from MG (CEN1398). Three other T. peberdyi strains from MG state formed a sister group and a strain from SP was an outlier in the species. Genetically well-characterized Trichoderma species were previously examined using MALDI-TOF, where 129 strains representing 28 species in 8 phylogenetic clades were effectively identified to the species level, providing comparable resolution to ITS sequencing [61]. The authors claimed approximate agreement with the sequence-based phylogeny, which we did not reproduce herein, although this could be due to sampling differences between the two studies.
Fig 6. Dendrogram based on MALDI TOF analysis of Trichoderma strains isolated from garlic and onion crop soils.
Species names are followed by strain number and collection location by Brazilian state.
The major constraint on the use of MALDI-TOF for fungal identification, especially in environmental samples, is the lack of a comprehensive reference spectrum library [62]. Previously, MALDI-TOF was used to identify Metarhizium species, where accuracy was progressively improved with the addition of further correctly identified strains to the spectrum library until near perfect matches with DNA-based identifications were obtained [62]. Our analysis was principally directed towards clustering and detection of phenotypic diversity among the onion and garlic-associated strains. However, the technique would appear to be promising for the rapid identification of new Trichoderma isolates, since the MALDI-TOF MSP clusters we obtained agreed perfectly with sequence-based identifications. Similarly, MALDI-TOF could be exploited as a fast and economical means of large-scale pre-grouping and triage of anonymous isolates prior to selection of representative individuals for sequence-based phylogenetic identification.
Conclusions
The large variety of Trichoderma species and genotypes identified in a small sample (n = 54) of isolates from garlic and onion crops in South-eastern Brazil represents a considerable resource for the selection of antagonists for biocontrol programs. The biological diversity present is exemplified by the discovery of two new Trichoderma species in this sample. While Brazil is among the megadiverse countries, systematic studies on microbial diversity in the range of biomes in the country are currently few [63]. A much larger systematic survey of Trichoderma populations associated with both crop and natural soils would enable a clearer picture of the distribution of species in the region. Complimentary sampling of epiphytic and endophytic niches could also broaden the scope for discovery. Such programs also provide the opportunity to preserve distinctive and potentially valuable Trichoderma germplasm. Given the laborious nature of pure culture collection and amplicon sequencing for species identification, comprehensive geographical mapping of species could be more efficiently accomplished by metabarcoding, using a sufficiently discriminatory target sequence, such as tef1-α.
Supporting information
(PDF)
Ultrafast bootstrap values are given above branches (> = 90%) and the scale bar represents expected changes per site. Strains sequenced in the present study are in bold and are followed by CENxxx numbers. Two new Trichoderma species, T. azevedoi and T. peberdyi are indicated in bold type.
(PDF)
Ultrafast bootstrap values are given above branches (> = 90%) and the scale bar represents expected changes per site. Strains sequenced in the present study are followed by CENxxx numbers.
(PDF)
Ultrafast bootstrap values are given above branches (> = 90%) and the scale bar represents expected changes per site. Strains sequenced in the present study are followed by CENxxx numbers.
(PDF)
Ultrafast bootstrap values are given above branches (> = 90%) and the scale bar represents expected changes per site. Strains sequenced in the present study are followed by CENxxx numbers.
(PDF)
Ultrafast bootstrap values are given above branches (> = 90%) and the scale bar represents expected changes per site. Strains sequenced in the present study are followed by CENxxx numbers.
(PDF)
Ultrafast bootstrap values are given above branches (> = 90%) and the scale bar represents expected changes per site. Strains sequenced in the present study are followed by CENxxx numbers.
(PDF)
(NEX)
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
This work was funded by a grant to SCMM from Fundação de Apoio a Pesquisa do Distrito Federal (FAPDF competitive grant 0193.000.992/2015). http://www.fap.df.gov.br. The funders had no role in study design, data collection and analysis, decision to publish,or preparation of the manuscript.
References
- 1.Entwistle AR. Root diseases In: Rabinowitch HD editor. Onions and Allied Crops. CRC Press, Boca Raton, Florida; 1990. pp. 103–154. 10.1201/9781351075169 [DOI] [Google Scholar]
- 2.Oliveira ML, De Marchi BR, Mituti T, Pavan MA, Krause-Sakate R. Identification and sequence analysis of five allexiviruses species infecting garlic crops in Brazil. Trop Plant Pathol. 2014;39: 483–489. 10.1590/S1982-56762014000600011 [DOI] [Google Scholar]
- 3.Resende JTV de, Morales RGF, Zanin DS, Resende F V, Paula JT de, Dias DM, et al. Caracterização morfológica, produtividade e rendimento comercial de cultivares de alho. Hortic Bras. 2013;31: 157–162. 10.1590/S0102-05362013000100025 [DOI] [Google Scholar]
- 4.Clarkson JP, Payne T, Mead A, Whipps JM. Selection of fungal biological control agents of Sclerotium cepivorum for control of white rot by sclerotial degradation in a UK soil. Plant Pathol. 2002;51: 735–745. 10.1046/j.1365-3059.2002.00787.x [DOI] [Google Scholar]
- 5.Al-Sadi AM, Al-Oweisi FA, Edwards SG, Al-Nadabi H, Al-Fahdi AM. Genetic analysis reveals diversity and genetic relationship among Trichoderma isolates from potting media, cultivated soil and uncultivated soil. BMC Microbiol. 2015;15: 147 10.1186/s12866-015-0483-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Druzhinina IS, Seidl-Seiboth V, Herrera-Estrella A, Horwitz BA, Kenerley CM, Monte E, et al. Trichoderma: the genomics of opportunistic success. Nat Rev Microbiol. 2011;9: 749–759. 10.1038/nrmicro2637 [DOI] [PubMed] [Google Scholar]
- 7.Harman GE, Howell CR, Viterbo A, Chet I, Lorito M. Trichoderma species—opportunistic, avirulent plant symbionts. Nat Rev Microbiol. 2004;2: 43–56. 10.1038/nrmicro797 [DOI] [PubMed] [Google Scholar]
- 8.Quiroz Sarmiento VF, Ferrera Cerrato R, Alarcón A, Encarnación M, Hernández L. Antagonismo in vitro de cepas de Aspergillus y Trichoderma hacia hongos filamentosos que afectan el cultivo de ajo. Rev Mex Micol. 2008;26: 27–34. [Google Scholar]
- 9.Verma M, Brar SK, Tyagi RD, Surampalli RY, Valéro JR. Antagonistic fungi, Trichoderma spp.: Panoply of biological control. Biochem Eng J. 2007;37: 1–20. 10.1016/j.bej.2007.05.012 [DOI] [Google Scholar]
- 10.Bae S-J, Mohanta TK, Chung JY, Ryu M, Park G, Shim S, et al. Trichoderma metabolites as biological control agents against Phytophthora pathogens. Biol Control. 2016;92: 128–138. 10.1016/j.biocontrol.2015.10.005 [DOI] [Google Scholar]
- 11.Castillo FDH, Padilla AMB, Morales GG, Siller MC, Herrera RR, Gonzales CNA, et al. In vitro antagonist action of Trichoderma strains against Sclerotinia sclerotiorum and Sclerotium cepivorum. Am J Agric Biol Sci. 2011; 10.3844/ajabssp.2011.410.417 [DOI] [Google Scholar]
- 12.Garnica-Vergara A, Barrera-Ortiz S, Muñoz-Parra E, Raya-González J, Méndez-Bravo A, Macías-Rodríguez L, et al. The volatile 6-pentyl-2 H -pyran-2-one from Trichoderma atroviride regulates Arabidopsis thaliana root morphogenesis via auxin signaling and ETHYLENE INSENSITIVE 2 functioning. New Phytol. 2016;209: 1496–1512. 10.1111/nph.13725 [DOI] [PubMed] [Google Scholar]
- 13.Alvarado-Marchena L, Rivera-Méndez W. Molecular identification of Trichoderma spp. in garlic and onion fields and in vitro antagonism trials on Sclerotium cepivorum. Rev Bras Ciência do Solo. 2016;40: e0150454 10.1590/18069657rbcs20150454 [DOI] [Google Scholar]
- 14.Dilbo C. Integrated management of garlic white rot (Sclerotium cepivorum Berk) using some fungicides and antifungal Trichoderma species. J Plant Pathol Microbiol. 2015;06: 1000251 10.4172/2157-7471.1000251 [DOI] [Google Scholar]
- 15.Taylor JW, Jacobson DJ, Kroken S, Kasuga T, Geiser DM, Hibbett DS, et al. Phylogenetic species recognition and species concepts in fungi. Fungal Genet Biol. 2000;31: 21–32. 10.1006/fgbi.2000.1228 [DOI] [PubMed] [Google Scholar]
- 16.Kullnig-Gradinger CM, Szakacs G, Kubicek CP. Phylogeny and evolution of the genus Trichoderma: a multigene approach. Mycol Res. 2002;106: 757–767. 10.1017/S0953756202006172 [DOI] [Google Scholar]
- 17.Chaverri P, Castlebury LA, Samuels GJ, Geiser DM. Multilocus phylogenetic structure within the Trichoderma harzianum/Hypocrea lixii complex. Mol Phylogenet Evol. 2003;27: 302–313. 10.1016/s1055-7903(02)00400-1 [DOI] [PubMed] [Google Scholar]
- 18.Druzhinina I, Kubicek CP. Species concepts and biodiversity in Trichoderma and Hypocrea: from aggregate species to species clusters? J Zhejiang Univ Sci. 2005;6B: 100–112. 10.1631/jzus.2005.B0100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bissett J, Gams W, Jaklitsch W, Samuels GJ. Accepted Trichoderma names in the year 2015. IMA Fungus. 2015;6: 263–295. 10.5598/imafungus.2015.06.02.02 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chalupová J, Raus M, Sedlářová M, Šebela M. Identification of fungal microorganisms by MALDI-TOF mass spectrometry. Biotechnol Adv. 2014;32: 230–241. 10.1016/j.biotechadv.2013.11.002 [DOI] [PubMed] [Google Scholar]
- 21.Lopes RB, Faria M, Souza DA, Bloch C, Silva LP, Humber RA. MALDI-TOF mass spectrometry applied to identifying species of insect-pathogenic fungi from the Metarhizium anisopliae complex. Mycologia. 2014;106: 865–878. 10.3852/13-401 [DOI] [PubMed] [Google Scholar]
- 22.Hendrickx M. MALDI-TOF MS and filamentous fungal identification: a success story? Curr Fungal Infect Rep. 2017;11: 60–65. 10.1007/s12281-017-0277-6 [DOI] [Google Scholar]
- 23.Chaverri P, Branco-Rocha F, Jaklitsch W, Gazis R, Degenkolb T, Samuels GJ. Systematics of the Trichoderma harzianum species complex and the re-identification of commercial biocontrol strains. Mycologia. 2015;107: 558–590. 10.3852/14-147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Buhariwalla HK, Srilakshmi P, Kannan S, Kanchi RS, Chandra S, Satyaprasad K, et al. AFLP analysis of Trichoderma spp. from india compared with sequence and morphological-based diagnostics. J Phytopathol. 2005;153: 389–400. 10.1111/j.1439-0434.2005.00989.x [DOI] [Google Scholar]
- 25.Inglis PW, Pappas M de CR, Resende L V., Grattapaglia D. Fast and inexpensive protocols for consistent extraction of high quality DNA and RNA from challenging plant and fungal samples for high-throughput SNP genotyping and sequencing applications. Kalendar R, editor. PLoS One. 2018;13: e0206085 10.1371/journal.pone.0206085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hall TA. BIOEDIT: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/ NT. Nucleic Acids Symp Ser. 1999;41: 91–98. [Google Scholar]
- 27.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30: 772–780. 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vaidya G, Lohman DJ, Meier R. SequenceMatrix: concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics. 2011;27: 171–180. 10.1111/j.1096-0031.2010.00329.x [DOI] [PubMed] [Google Scholar]
- 29.Cheng T, Xu C, Lei L, Li C, Zhang Y, Zhou S. Barcoding the kingdom Plantae: new PCR primers for ITS regions of plants with improved universality and specificity. Mol Ecol Resour. 2016;16: 138–149. 10.1111/1755-0998.12438 [DOI] [PubMed] [Google Scholar]
- 30.Samuels GJ, Dodd SL, Lu B-S, Petrini O, Schroers H-J, Druzhinina IS. The Trichoderma koningii aggregate species. Stud Mycol. 2006;56: 67–133. 10.3114/sim.2006.56.03 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Samuels GJ, Ismaiel A. Trichoderma evansii and T. lieckfeldtiae: two new T. hamatum -like species. Mycologia. 2009;101: 142–156. 10.3852/08-161 [DOI] [PubMed] [Google Scholar]
- 32.Carbone I, Kohn LM. A method for designing primer sets for speciation studies in filamentous Ascomycetes. Mycologia. 1999;91: 553 10.2307/3761358 [DOI] [Google Scholar]
- 33.Hoyos-Carvajal L, Orduz S, Bissett J. Genetic and metabolic biodiversity of Trichoderma from Colombia and adjacent neotropic regions. Fungal Genet Biol. 2009;46: 615–631. 10.1016/j.fgb.2009.04.006 [DOI] [PubMed] [Google Scholar]
- 34.Rehner SA, Buckley E. A Beauveria phylogeny inferred from nuclear ITS and EF1-alpha sequences: evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia. 2005;97: 84–98. 10.3852/mycologia.97.1.84 [DOI] [PubMed] [Google Scholar]
- 35.Lanfear R, Frandsen PB, Wright AM, Senfeld T, Calcott B. PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol Biol Evol. 2016; msw260 10.1093/molbev/msw260 [DOI] [PubMed] [Google Scholar]
- 36.Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32: 268–274. 10.1093/molbev/msu300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 2017;14: 587–589. 10.1038/nmeth.4285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS. UFBoot2: improving the ultrafast bootstrap approximation. Mol Biol Evol. 2018;35: 518–522. 10.1093/molbev/msx281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215: 403–410. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- 40.Atanasova L, Druzhinina IS, Jaklitsch WM. Two hundred Trichoderma species recognized on the basis of molecular phylogeny In: Trichoderma: biology and applications. Wallingford: CABI; pp. 10–42. 10.1079/9781780642475.0010 [DOI] [Google Scholar]
- 41.Chen K, Zhuang W-Y. Discovery from a large-scaled survey of Trichoderma in soil of China. Sci Rep. 2017;7: 9090 10.1038/s41598-017-07807-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, et al. MrBayes 3.2: efficient bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61: 539–542. 10.1093/sysbio/sys029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miller MA, Pfeiffer W, Schwartz T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees 2010 Gateway Computing Environments Workshop (GCE). IEEE; 2010. pp. 1–8. 10.1109/GCE.2010.5676129 [DOI] [Google Scholar]
- 44.Ayres DL, Darling A, Zwickl DJ, Beerli P, Holder MT, Lewis PO, et al. BEAGLE: an application programming interface and high-performance computing library for statistical phylogenetics. Syst Biol. 2012;61: 170–173. 10.1093/sysbio/syr100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vos P, Hogers R, Bleeker M, Reijans M, Lee T van de, Hornes M, et al. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res. 1995;23: 4407–4414. 10.1093/nar/23.21.4407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Arrigo N, Holderegger R, Alvarez N. Automated scoring of AFLPs using RawGeno v 2.0, a free R CRAN library. 2012. pp. 155–175. 10.1007/978-1-61779-870-2_10 [DOI] [PubMed] [Google Scholar]
- 47.Swofford DL. PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods). Sinauer Associates, Sunderland, Massachusetts; 2012. pp. 166–187. 10.1159/000170955 [DOI] [Google Scholar]
- 48.Druzhinina IS, Kopchinskiy AG, Komoń M, Bissett J, Szakacs G, Kubicek CP. An oligonucleotide barcode for species identification in Trichoderma and Hypocrea. Fungal Genet Biol. 2005;42: 813–828. 10.1016/j.fgb.2005.06.007 [DOI] [PubMed] [Google Scholar]
- 49.Pais FS-M, Ruy P de, Oliveira G, Coimbra R. Assessing the efficiency of multiple sequence alignment programs. Algorithms Mol Biol. 2014;9: 4 10.1186/1748-7188-9-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.López-Quintero CA, Atanasova L, Franco-Molano AE, Gams W, Komon-Zelazowska M, Theelen B, et al. DNA barcoding survey of Trichoderma diversity in soil and litter of the Colombian lowland Amazonian rainforest reveals Trichoderma strigosellum sp. nov. and other species. Antonie Van Leeuwenhoek. 2013;104: 657–674. 10.1007/s10482-013-9975-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jiang Y, Wang J-L, Chen J, Mao L-J, Feng X-X, Zhang C-L, et al. Trichoderma biodiversity of agricultural fields in east china reveals a gradient distribution of species. PLoS One. 2016;11: e0160613 10.1371/journal.pone.0160613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zachow C, Berg C, Müller H, Monk J, Berg G. Endemic plants harbour specific Trichoderma communities with an exceptional potential for biocontrol of phytopathogens. J Biotechnol. 2016;235: 162–170. 10.1016/j.jbiotec.2016.03.049 [DOI] [PubMed] [Google Scholar]
- 53.Hoyos-Carvajal L, Bissett J. Biodiversity of Trichoderma in Neotropics In: The dynamical processes of biodiversity—case studies of evolution and spatial distribution. InTech; 2011. 10.5772/23378 [DOI] [Google Scholar]
- 54.Chaverri P, Samuels GJ. Hypocrea/Trichoderma (Ascomycota, Hypocreales, Hypocreaceae): Species with green ascospores. Stud Mycol. 2003; 1–113. [Google Scholar]
- 55.Druzhinina IS, Kubicek CP, Komon-Zelazowska M, Belayneh Mulaw T, Bissett J. The Trichoderma harzianum demon: complex speciation history resulting in coexistence of hypothetical biological species, recent agamospecies and numerous relict lineages. BMC Evol Biol. 2010;10: 94 10.1186/1471-2148-10-94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Druzhinina IS, Komoń-Zelazowska M, Ismaiel A, Jaklitsch W, Mullaw T, Samuels GJ, et al. Molecular phylogeny and species delimitation in the section Longibrachiatum of Trichoderma. Fungal Genet Biol. 2012;49: 358–368. 10.1016/j.fgb.2012.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Majer D, Mithen R, Lewis BG, Vos P, Oliver RP. The use of AFLP fingerprinting for the detection of genetic variation in fungi. Mycol Res. 1996;100: 1107–1111. 10.1016/S0953-7562(96)80222-X [DOI] [Google Scholar]
- 58.Althoff DM, Gitzendanner MA, Segraves KA. The utility of amplified fragment length polymorphisms in phylogenetics: a comparison of homology within and between genomes. Syst Biol. 2007;56: 477–484. 10.1080/10635150701427077 [DOI] [PubMed] [Google Scholar]
- 59.Vahabi K, Sharifnabi B, Zafari D. Genetic diversity of Trichoderma spp. associated with button mushroom, Agaricus bisporus, inferred from AFLP markers and ITS sequencing. Acta Phytopathol Entomol Hungarica. 2009;44: 239–253. 10.1556/APhyt.44.2009.2.3 [DOI] [Google Scholar]
- 60.Drissner D, Freimoser FM. MALDI-TOF mass spectroscopy of yeasts and filamentous fungi for research and diagnostics in the agricultural value chain. Chem Biol Technol Agric. 2017;4: 13 10.1186/s40538-017-0095-7 [DOI] [Google Scholar]
- 61.De Respinis S, Vogel G, Benagli C, Tonolla M, Petrini O, Samuels GJ. MALDI-TOF MS of Trichoderma: a model system for the identification of microfungi. Mycol Prog. 2010;9: 79–100. 10.1007/s11557-009-0621-5 [DOI] [Google Scholar]
- 62.Normand A-C, Cassagne C, Gautier M, Becker P, Ranque S, Hendrickx M, et al. Decision criteria for MALDI-TOF MS-based identification of filamentous fungi using commercial and in-house reference databases. BMC Microbiol. 2017;17: 25 10.1186/s12866-017-0937-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bruce T, de Castro A, Kruger R, Thompson CC, Thompson FL. Microbial diversity of brazilian biomes. 2012. pp. 217–247. 10.1007/978-1-4614-2182-5_13 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
Ultrafast bootstrap values are given above branches (> = 90%) and the scale bar represents expected changes per site. Strains sequenced in the present study are in bold and are followed by CENxxx numbers. Two new Trichoderma species, T. azevedoi and T. peberdyi are indicated in bold type.
(PDF)
Ultrafast bootstrap values are given above branches (> = 90%) and the scale bar represents expected changes per site. Strains sequenced in the present study are followed by CENxxx numbers.
(PDF)
Ultrafast bootstrap values are given above branches (> = 90%) and the scale bar represents expected changes per site. Strains sequenced in the present study are followed by CENxxx numbers.
(PDF)
Ultrafast bootstrap values are given above branches (> = 90%) and the scale bar represents expected changes per site. Strains sequenced in the present study are followed by CENxxx numbers.
(PDF)
Ultrafast bootstrap values are given above branches (> = 90%) and the scale bar represents expected changes per site. Strains sequenced in the present study are followed by CENxxx numbers.
(PDF)
Ultrafast bootstrap values are given above branches (> = 90%) and the scale bar represents expected changes per site. Strains sequenced in the present study are followed by CENxxx numbers.
(PDF)
(NEX)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.