Abstract
The design and creation of synthetic genomes provides a powerful approach to understanding and engineering biology. However, it is often limited by the paucity of methods for precise genome manipulation. Here, we demonstrate the programmed fission of the Escherichia coli genome into diverse pairs of synthetic chromosomes; and the programmed fusion of synthetic chromosomes to generate genomes with user-defined inversions and translocations. We further combine genome fission, chromosome transplant, and chromosome fusion to assemble genomic regions from different strains into a single genome. Thus, we program the scarless assembly of new genomes with nucleotide precision, a key step in the convergent synthesis of genomes from diverse progenitors. This work provides a set of precise, rapid, large-scale (megabase) genome-engineering operations for creating diverse synthetic genomes.
Efforts to minimize (1, 2), refactor (3), recode (4, 5), and reorganize chromosomes and genomes (2, 6) are providing new insights and opportunities. However, in Escherichia coli, the workhorse of synthetic biology, the methods necessary to realize a complete set of operations for synthetic genome design are missing. These operations include (i) the iterative replacement of genomic DNA with synthetic DNA, (ii) deletion of genomic DNA, (iii) translocation of large genomic sections, and (iv) inversion of large genomic sections as well as (v) methods for combining large genome sections from distinct strains for the convergent assembly of synthetic genomes. Each operation should be scarless and programmed with nucleotide precision so that genome designs can be precisely and rapidly realized.
Efficient, precise, and robust methods for iterative replacement (>100 kb per step) and deletion of genome sections have been reported (7); however, there has been less progress on creating methods for generating precisely programmed inversions or translocations in E. coli, with most current methods for inversions relying on sequence-specific recombinases. Moreover, methods for combining large (e.g., 0.5-Mb) sections from distinct genomes rely on classical conjugation (8) and its derivatives (5, 9). While these methods can be useful (5, 9), they are fundamentally limited because (i) they require large regions of homology [commonly at least 3 kb, and sometimes up to 400 kb, between the donor and recipient genomes (5)], (ii) undesired chimeras between the two genomes may result, and (iii) the site of crossover between the two genomes is not precisely specified. Indeed, in favorable cases, crossovers are only selected with kilobase resolution.
Chromosome fission and fusion have occurred in natural evolution (10, 11), and these processes may have accelerated evolution (10, 12, 13). The synthetic splitting and fusion of chromosomes has been explored to a limited extent, primarily in naturally recombinogenic organisms (13–18). One report excised up to 720 kb from a single region of the E. coli genome (19) by using natural homologous recombination in E. coli. Because the recombination frequency in E. coli is generally low (20), this approach is presumably very inefficient. A protelomerase of bacteriophage N15 and a Vibrio origin of replication were used to divide the circular E. coli chromosome into two linear subchromosomes. However, only one out of 22 arrangements tested was viable (21). Thus, the limited methods for splitting the E. coli genome are not general or efficient.
Here, we demonstrate that an E. coli genome, without any prior modification, can be efficiently split, by single-step programmed fission, into pairs of synthetic chromosomes. The resulting synthetic chromosomes enable precise, programmed fusions, genomic inversions and translocations; moreover, they provide a route to assemble new genomes through the precise, convergent assembly of large genomic fragments from distinct strains.
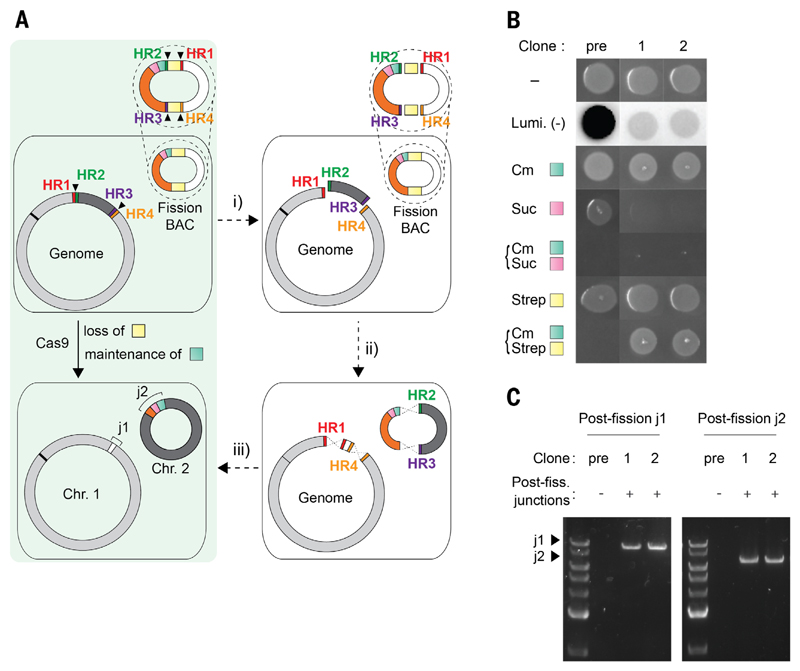
We designed and synthesized a system to precisely split the unmodified genome into two user-defined, circular chromosomes (Fig. 1A) and tested our approach by splitting the E. coli MDS42 (1) genome (data file S1) into a 3.43- and a 0.56-Mb chromosome. To achieve this, we first introduced Cas9 with appropriate spacers (table S1), the lambda-red recombination machinery, and a fission bacterial artificial chromosome (BAC) (data file S2) into cells. We implemented six Cas9-directed cuts in the DNA of these cells; two of these cuts target the genome, and four of these cuts target the fission BAC (data files S3 and S4). The two cuts in the genome create fragment 1 and fragment 2, and the four cuts in the fission BAC release linker sequence 1 and linker sequence 2. Chromosome 1 (3.43 Mb) containing the genomic origin of replication (oriC) was formed through lambda-red-mediated recombination between genomic fragment 1 and linker sequence 1, by virtue of their 50-base pair (bp) regions of homology (table S2). Similarly, chromosome 2 (0.56 Mb) was formed through lambda-red-mediated recombination between genomic fragment 2 and linker sequence 2 (Fig. 1A and fig. S1); this linker sequence contained its own replication and segregation machinery.
Fig. 1. Programmed genome fission splits the E. coli genome into two chromosomes.
(A) E. coli harbors a fission BAC, containing a double selection cassette (sacB-CmR shown as pink and green, respectively), rpsL (yellow), a luxABCDE operon (white), and the BAC replication machinery (orange). During fission, (i) Cas9 induces six cuts (black triangles), splitting the genome into fragment 1 (light gray, containing oriC indicated by black line) and fragment 2 (dark gray), and the fission BAC into four pieces (linker sequence 1, linker sequence 2, and two copies of rpsL). (ii) Homology regions (HRs) between fragments and their cognate linkers. (iii) Lambda-red recombination joins fragments and linkers to yield chromosomes 1 and 2 (Chr. 1 and Chr. 2). Junctions 1 and 2 (j1 and j2) are new junctions. (B) Growth and luminescence (Lumi.) of prefission (pre) and postfission (1 and 2) clones are consistent with the generation of two chromosomes (Chr. 1, ~3.43 Mb and Chr. 2, ~0.56 Mb). Cells were stamped in plain LB agar (-), 20 μg/ml chloramphenicol (Cm), 7.5% sucrose (Suc), 100 μg/mL streptomycin (Strep), or the indicated combination. (C) PCR of postfission (Post-Fiss.) clones across j1 and j2.
In the prefission strain, the fission BAC is non-essential and contains an SacB-CmR double selection cassette (this confers resistance to chloramphenicol and sensitivity to sucrose, but cells can grow on sucrose by losing the fission BAC), the luxABCDE operon (conferring luminescence), and rpsL (conferring sensitivity to streptomycin). After successful fission, the rpsL gene is lost, cells are resistant to streptomycin, the luxABCDE operon is removed from a strong promoter to chromosome 1 (leading to weaker luminescence), and the SacB-CmR double selection cassette becomes part of chromosome 2 and cannot be lost. Thus, correct postfission cells are selectively sensitized to sucrose.
After execution of the fission protocol, we enriched for cells that had undergone genome fission to generate two chromosomes, through growth on streptomycin and chloramphenicol (table S3). This selects for both loss of rpsL and maintenance of CmR in the SacB-CmR double selection cassette and therefore kills cells containing the fission BAC but allows growth of cells that have undergone programmed genome fission.
We characterized individual postfission clones by several independent methods. First, we examined the luminescence of cells and their growth on selective media (Fig. 1B). Successful clones had decreased luminescence with respect to prefission controls, gained sucrose sensitivity, and gained the ability to grow when challenged simultaneously with both chloramphenicol and streptomycin. Second, we performed polymerase chain reactions (PCRs) across the new junctions resulting from fission. Successful postfission clones exhibited bands of the expected size that were not present in prefission clones (Fig. 1C). This confirmed that both fission junctions were as expected. Third, we confirmed the expected restriction enzyme digestion pattern for the postfission genome by pulsed-field gel electrophoresis (fig. S2). Finally, we determined the replicon organization of the genome by de novo assembly; we achieved this by combining the results of short-read (300-bp paired end) and long-read (N50 of ~8.3 kb) sequencing in Unicycler (22) to generate one contig per replicon. The postfission assembly formed two circular contigs, which corresponded to the chromosomes expected from fission (fig. S3 and table S4). The copy number of each chromosome was as expected (table S5).
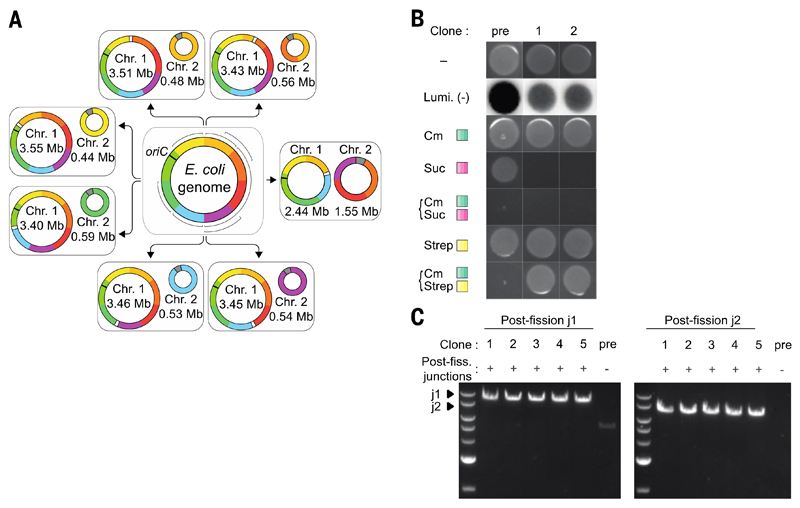
We demonstrated the scope and generality of fission by programming the splitting of the genome into five additional distinct and diverse pairs of chromosomes (Fig. 2 and (figs. S2 to S4). These included a pair in which chromosome 1 is 2.44 Mb and chromosome 2 is 1.55 Mb. Because chromosome 2 has BAC-derived replication and segregation machinery, our data are consistent with BACs being able to maintain megabases of DNA. The only constraints we imposed on the choice of fission sites were that they contained a protospacer adjacent motif (PAM) for Cas9 and lay greater than 30 bp outside any gene. Although a single 2-Mb fission test failed (fig. S5 and table S3), all other experiments we tried led to successful fission (figs. S1 to S4). Fission had only modest effects on the growth of cells (fig. S6). We observed that the genome fissions were present after approximately 105 generations of continuous growth (fig. S7).
Fig. 2. Fission can be performed throughout the E. coli genome.
(A) Successful Fissions performed. Each colour on the E. coli genome corresponds to ~0.5 Mb. We named the sections A to H. A is dark orange and the other sections are labeled alphabetically in a clockwise sequence. Linker sequence 1, white; oriC black bar; linker sequence 2, gray. Boundaries and homologies of each fission experiment are provided in table S2. Seven fissions are shown, including the 3.43, 0.56 Mb fission (Fig. 1). The 3.45, 0.54 Mb fission (purple Chr. 2) was performed using an E.coli genome in which a ~0.54 Mb section had been recoded (Fig. 3). (B) Growth and luminescence for the generation of the 2.44, 1.55 Mb fission; annotation as in Fig. 1B. Data for other fissions are shown in fig. S4. (C) PCR of clones across new junctions for 2.44 Mb, 1.55 Mb fission. Postfission clones (1 to 5) exhibit products of the expected size, whereas the prefission control does not. Junction PCR for other fissions in fig. S4.
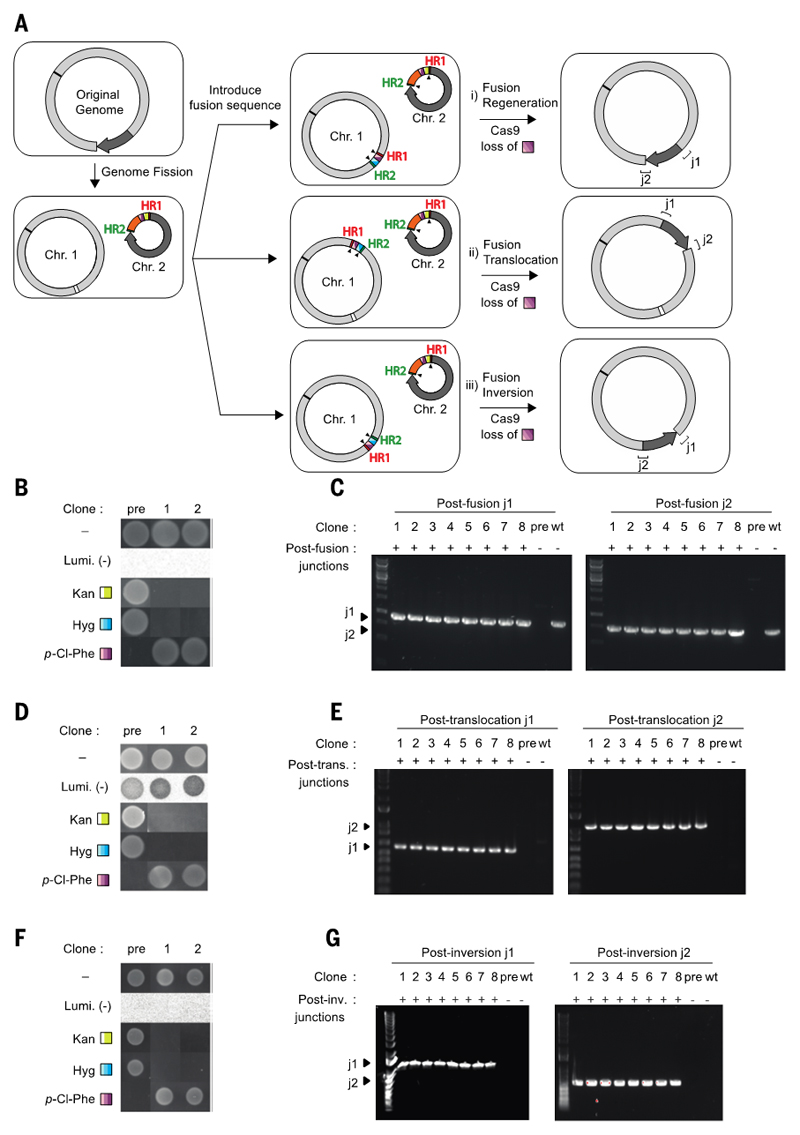
We demonstrated that the programmed fusion of synthetic chromosomes, generated by fission, enables the generation of precisely rearranged genomes (Fig. 3). We applied fission to a cell in which ~0.54 Mb, Section C (Fig. 2A and (figs. S1 and S3), of the genome is watermarked by 2521 synonymous codon changes (5) (data file S5); this brought the total number of successful fissions to 7 (Fig. 2A and fig. S1). The resulting cell contained chromosome 1 (3.45 Mb) and a watermarked chromosome 2 (0.54 Mb). After fission we replaced the SacB-CmR double selection cassette in chromosome 2 with an oriT-pheS*-KanR cassette (table S6). This cell provided a common intermediate for diverse fusions.
Fig. 3. Programmed chromosomal fusion enables translocations and inversions of large genomic segments from common fission intermediates.
(A) E. coli with two chromosomes (Chr. 1 ~3.45 Mb and Chr. 2 ~0.54 Mb) was generated by fission. The sequence of Chr. 2 is watermarked as described in the text. The colour coding is as in Fig. 1A, a pheS*-KanR double selection cassette (purple and yellow, respectively) is shown. A fusion sequence, consisting of a pheS*-HygR (purple and blue, respectively) double selection cassette flanked by HR1 and HR2, is introduced in the indicated positions and orientation in Chr. 1 by lambda red recombination. Protospacer directed cleavage (black arrows), lambda red recombination and selection for fusion products through the loss of pheS* on 4-chloro-phenylalanine yield the indicated products. (i) Regenerating the original genomic arrangement, (ii) translocation of the 0.54-Mb segment 700 kb away from its original position, and (iii) inversion of the 0.54-Mb segment. (B) Growth and luminescence (Lumi.) of pre- and post-fusion regeneration (1 and 2) clones. Hyg, hygromycin; Kan, kanamycin; p-Cl-Phe, 4-chloro-phenylalanine. (C) PCR of clones across new junctions for fusion regeneration. Post-fusion clones (1 to 8) exhibit products of the expected size, whereas the prefission control does not. (D, E) As in B, C, but for fusion translocation (trans.). (F, G) As in B, C, but for fusion inversion (inv.).
We first used fusion to regenerate the original genome. We prepared chromosome 1 for fusion by replacing its linker sequence 1 with a 'fusion sequence' for chromosome 2 (oligonucleotide sequences are provided in table S6). This contained a pheS*-HygR double selection cassette flanked by Cas9 cut sites and homology to fragment 2 in chromosome 2 (Fig. 3A). Fusion was initiated by Cas9-mediated cleavage at either side of the pheS*-HygR cassette in chromosome 1, and at the ends of the watermarked sequence in chromosome 2, and the resulting homologous ends were joined through lambda red-mediated recombination. We selected the fusion product on 4-chloro-phenylalanine.
We characterized postfusion clones by several independent methods. Successful clones were no longer sensitive to 4-chloro-phenylalanine or resistant to kanamycin or hygromycin (Fig. 3B). PCR across the new junctions generated by fusion led to bands of the correct size that were not present in the prefusion clones (Fig. 3C). We further demonstrated successful fusion by de novo assembly of short-read (300-bp paired-end) and long-read (N50 of ~20 kb) sequencing. The prefusion genome formed two circular contigs, whereas all postfusion assemblies formed a single circular contig, which corresponds to the expected fusion product (fig. S3).
We demonstrated that inserting the fusion sequence at different positions in chromosome 1 (500 or 700 kb away from linker sequence 1 (Fig. 3A and fig. S8), followed by initiation of fusion with chromosome 2, enables the selection of genomes bearing defined translocations (Fig. 3, D and E, and figs. S3 and S8). We also demonstrated that inserting the fusion sequence into chromosome 1 in an inverted orientation (Fig. 3A), followed by initiation of fusion with chromosome 2, enables the selection of genomes bearing defined inversions (Fig. 3, F and G, and fig. S3). An attempt at fusion 1.8 Mb away from the linker sequence did not lead to a stable translocation (fig. S8 and table S3).
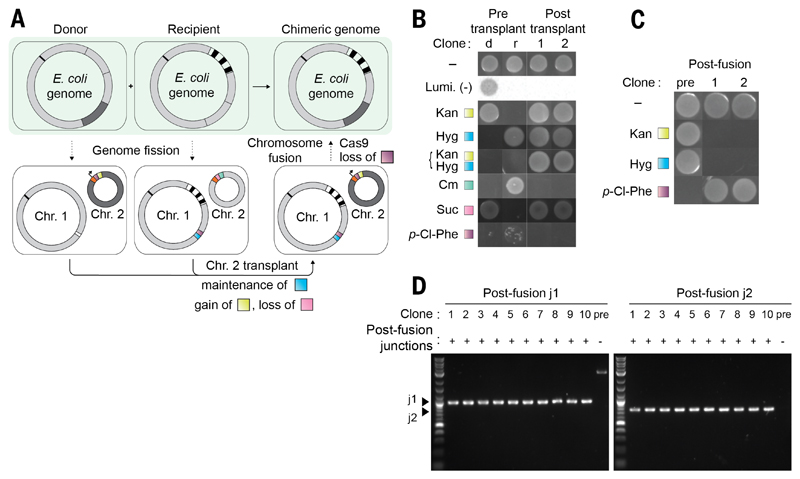
Next, we combined genome fission, bacterial conjugative transplant, and chromosome fusion to precisely combine defined sections of distinct genomes (Fig. 4A). This is a key step in the precise assembly of synthetic genomes from strains containing synthetic sections.
Fig. 4. Precise genome assembly from genomic segments of distinct strains.
(A) Precisely combining the watermarked region 1 (dark gray) from a donor strain and a watermarked region 2 (black striped) from a recipient strain into a single strain. Fission is performed in parallel in the donor and recipient strains. The resulting donor strain contains a watermarked Chr. 2 containing an oriT (black arrow) and a pheS*-KanR double selection cassette (purple and yellow); the remainder of Linker sequence 2 is orange. The resulting recipient strain contains an analogous non-watermarked Chr. 2, with a sacB-CmR cassette (pink and green). The linker sequence 1 (white) is replaced with a fusion sequence containing a pheS*-HygR cassette (purple and blue) in preparation for fusion. The donor cell is provided with a non-transferable F' plasmid. Mixing of donor and recipient cells facilitates conjugative transplant of Chr. 2 from the donor to the recipient; selection for KanR and against sacB-mediated sucrose sensitivity enables the isolation of cells that have gained a watermarked Chr. 2 and lost the non-watermarked Chr. 2. Subsequent genome fusion generates a strain in which the watermarked regions 1 and 2 have been precisely combined in a single chromosome. (B) Following chromosomal transplant by growth on selective media and luminescence. d, the pretransplant donor; r, pretransplant recipient. (C) After chromosomal fusion through growth on selective media. (D) PCR across the new junctions generated by chromosomal fusion yields products of the expected size in the post-fusion clones (1 to 10) but not in the prefusion control.
We began with two strains, each containing a different watermarked genomic section [section C or section A (Fig. 2 and fig. S1)], with the target of combining the watermarked sections in a single, chimeric genome. We defined one strain as the donor (data file S5) and the other strain as the recipient (data file S6). We performed fission on the genome of the donor to capture its watermarked sequence in chromosome 2. We then replaced the SacB-CmR double selection cassette in chromosome 2 with an oriT-pheS*-KanR cassette (table S6) and transformed a non-transferable F' plasmid (5) into the donor strain. These steps prepare the donor strain for transplant of chromosome 2 to the recipient.
In parallel, we performed fission on the genome of the recipient to split its genome, at the same position as the donor, into two synthetic chromosomes. This created a recipient containing a nonwatermarked chromosome 2 (the fission BAC used in the recipient, and therefore chromosome 2, contains a sacB-CmR cassette and does not contain oriT) and chromosome 1 that contains the second watermarked region. The linker sequence 1 in chromosome 1 of the recipient was then replaced with a fusion sequence containing a pheS*-HygR cassette flanked by regions of homology to the fragment of the original genome captured in chromosome 2.
To generate cells that contain both watermarked regions, we mixed donor and recipient cells. We selected for transfer of chromosome 2 from the donor to the recipient and recipient cells in which chromosome 2 from the donor had replaced the endogenous chromosome 2; we termed this overall process chromosome transplant. The resulting recipient cells contained chromosome 2 from the donor and chromosome 1 from the recipient. We generated a single, chimeric genome that contains both the watermarked sequences by fusion of the donor chromosome 2 and the recipient chromosome 1 (Fig. 4, B to D; fig. S3; and data file S7). All attempts at genome assembly were successful.
We demonstrated the efficient programmed, single-step fission of the unmodified E. coli genome into diverse megabase-scale chromosomes. These chromosomes provide a common intermediate for the facile creation of diverse genomes. The chromosomes in a single cell can be fused into a single genome to effect precise genomic translocations or precise and scarless inversions. This facilitates the realization of reorganized genome designs and the exploration of modular, synthetic syntenies that may be more amenable to engineering (2). Moreover, the transplant of chromosomes between cells, followed by fusion, enables the precise convergent assembly of new genomes. Our work provided the necessary set of precise, rapid, large-scale genome-engineering operations for creating diverse synthetic genomes.
Materials and Methods
Strains and plasmids used in this study
The positive-negative selection cassettes used in fission and fusion are -1/+1 (pheST251A_A294G -KanR), -2/+2 (sacB-CmR) and -1/+3 (pheST251A_A294G-HygR). -2/+2 and -1/+3 are described previously (7, 23). In -1/+1, pheST251A_A294G is dominant lethal in the presence of 4-chloro-phenylalanine, and KanR confers resistance to kanamycin. Both proteins are expressed polycistronically under control of the EM7 promoter. The -1/+3 cassette was synthesised de novo. The -1/+3 cassette is also referred to as pheS*-HygR. All cloning procedures described in this section were performed in E. coli DH10b, which carries an rpsLK43R mutation that confers resistance to streptomycin.
Fission BACs harboured the Linker 1 sequence (containing the luxABCDE operon), used to recircularize the genomic dsDNA breaks and complete Chromosome 1, as well as sacB-CmR and the Linker 2 sequence (containing the BAC partitioning elements) used to recircularize and complete the newly formed Chromosome 2. Fission BACs were constructed by Gibson assembly of two PCR products and two gBlocks (IDT): a PCR amplified luxABCDE tether construct, a PCR amplified sacB-CmR with BAC partitioning genes (repE, sopA-C), and two gBlocks containing rpsL constructs with flanking protospacers (see Data file S2 for an annotated Genbank file of a fission BAC; see Table S4 for homology sequences).
Fusion and conjugation experiments (see below) are performed with a BAC backbone in which the sacB-CmR cassette is replaced by an oriT-pheS*-KanR cassette. To generate this variant, plasmid pSC101_oriT-pheS*-Kan was first constructed by 4-piece Gibson assembly, where the PCR products were a pSC101 backbone, an oriT sequence, an EM7-pheS* sequence, and a kanamycin resistance sequence. This plasmid served as a template for amplification of the oriT-pheS*-KanR cassette for further recombination (see below).
The plasmid pKW20_CDFtet_pAraRedCas9_tracrRNA used throughout this study harbours the genes for Cas9 and the lambda-red recombination components alpha/beta/gamma under the control of an arabinose-inducible promoter, as well as a tracrRNA under its native promoter, as previously described (7).
The fission spacers are encoded in the plasmid pKW3_MB1Amp_TracrK_Spacer (Data file S3), which contains a pMB1 origin of replication, an ampicillin resistance marker, the spacer array under the control of its endogenous promoter as previously described (7), and a tracrRNA upstream of the spacer array (5).
For each fission procedure, the pKW3_MB1Amp_TracrK_Spacer plasmid was derivatized to contain a spacer/direct repeat array containing six spacers: four correspond to the target protospacer sequences for cutting the fission BAC, and two correspond to the target protospacer sequences for cutting the genome. The individual spacer arrays were assembled from overlapping oligos through multiple rounds of overlap extension PCR – the products were inserted by Gibson assembly between restriction sites AccI and EcoRI in the backbone of the pKW3_MB1Amp_TracrK_Spacer plasmid. All spacer array plasmids were verified by Sanger sequencing to be free of mutations; see Table S5 for the sequences of spacers used in this study. From this plasmid a derivative was constructed that swaps the ampicillin resistance gene for the kanamycin resistance gene; this plasmid was generated by Gibson assembly and named pKW5_MB1Kan_TracrK_Spacer (Data file S4). All fission experiments in this study were performed with the pKW3_MB1Amp_TracrK_Spacer plasmid, with the exception that the fission of a 1.54 Mb section (Section ABC) was performed with the pKW5_MB1Kan_TracrK_ Spacer plasmid.
The fusion spacers are encoded in an analogous plasmid as to that used for the fission spacers (pKW3_MB1Amp_TracrK_Spacer), except fusion uses only four spacers: two target Chromosome 1 (with the oriC origin of replication) and two target Chromosome 2 (with the BAC origin of replication). Fusion spacer plasmids were constructed in the same manner as described above.
Fission and fusion experiments were performed in the E. coli strain MDS42 (1), which has been utilized for recent genome engineering projects (5). The MDS42rpsLK43R strain (7) was used throughout our experiments with K43R mutation introduced into the rpsL gene to confer resistance to streptomycin in the absence of an additional wild-type copy of rpsL and sensitivity to streptomycin in the presence of any additional copy of wildtype rpsL.
The partially recoded MDS42rpsLk43R strain utilized for demonstration of Chromosome fusion, inversion and translocation contains a watermarked region 1 of approximately 0.5 Mb between 1,454,024 and 1,979,777 (Data file S5), with a defined serine codon and stop codon recoding scheme (TCG to AGC, TCA to AGT, and TAG to TAA). Similarly, the recipient strain of the chimeric genome generation experiment contains a watermarked region 2 with the same recoding scheme between coordinates 436,924-939,332 (Data file S6). Both of these strains were generated by GENESIS as described in detail previously (5). The resulting strain for watermarked region 1 contained an rpsL-KanR cassette at position 1,979,778-1,979,783, and the resulting strain for watermarked region 2 contained a sacB-CmR cassette at position 1,050,809-1,050,814 – these were removed from the genome by lambda-red mediated recombination prior to genome rearrangement experiments.
Fission procedure
To perform fission, the strains of interest were electroporated sequentially, first with pKW20_CDFtet_pAraRedCas9_tracrRNA, and then with the corresponding fission BAC. Cells harbouring both of these plasmids were made competent with 0.5% L-arabinose induction for 1 hour, as described previously (7), to express Cas9 and the lambda-red recombination machinery. 100 μL of induced electrocompetent cells were electroporated with ~8 μg of the corresponding pKW3_MB1Amp_TracrK_Spacer or pKW5_MB1Kan_TracrK_ Spacer plasmid, encoding spacer RNAs for the 6 necessary Cas9 cleavages. Cells were then recovered in 4 mL of SOB shaking at 37 °C for 1 hour. 80 μL of 25% L-arabinose were then added for a final concentration of 0.5%, and incubated for another hour. Cells were subsequently transferred to 100 mL of LB + 5 μg/mL tetracycline + 18 μg/mL chloramphenicol + 100 μg/mL ampicillin (for fissions performed with pKW3_MB1Amp_TracrK_Spacer) or 50 μg/mL kanamycin (for fissions performed with pKW5_MB1Kan_TracrK_Spacer), and incubated shaking at 37 °C for 4 hours.
Cells were then pelleted by centrifugation, resuspended in Milli-Q filtered water and spread in serial dilutions in LB agar plates supplemented with 5 μg/mL tetracycline + 100 μg/mL streptomycin + 18 μg/mL chloramphenicol + 100 μg/mL ampicillin (for fissions performed with pKW3_MB1Amp_TracrK_Spacer) or 50 μg/mL kanamycin (for fissions performed with pKW5_MB1Kan_TracrK_Spacer). From the resulting colonies, clones were identified that had some luminescent signal, but where this signal was lower than that of a pre-fission control. Candidate clones were assessed phenotypically by resuspending colonies in Milli-Q water and stamping them on the selection plates indicated in the main text. From overnight cultures of candidate clones, genomic DNA extractions were performed with the QIAgen Blood and Tissue kit, as per manufacturer’s instructions. From the genomic DNA template, PCR reactions were performed with primers targeting the hypothetical new junctions, i) flanking either side of the luxABCDE operon in the genome, and ii) flanking either side of the BAC backbone. PCR products were commonly analyzed alongside a GeneRuler 1 kb DNA Ladder (Thermo Scientific) on a 0.75% agarose gel.
Construction of watermarked Chr. 2 bearing oriT-pheS*-KanR
The partially recoded MDS42rpsLK43R strain (Data file S5) contains the watermarked region 1 (1,454,024 and 1,979,777). Fission was used to partition this watermarked region into Chr. 2. The sacB-CmR in the Linker sequence 2 of Chr. 2 was subsequently replaced by an oriT-pheS*-KanR using DOSER (7). Post-fission cells harbouring pKW20_CDFtet_pAraRedCas9_tracrRNA were made electrocompetent with of 0.5% L-arabinose induction for 1 hour. 100 μL of electrocompetent cells were electroporated with ~8 μg of oriT-pheS*-KanR PCR product, amplified from pSC101_oriT-pheS*-Kan with primers containing homology to the SacB-CmR cassette in Chr. 2 (see Table S6 for primer sequences). Cells were recovered for 4 hours (SOB, 200 rpm, 37 °C), pelleted by centrifugation, resuspended in 1 mL of Milli-Q water and plated on LB agar plates containing 7.5% sucrose and 50 μg/mL kanamycin. Clones that had undergone the replacement were identified by PCR flanking the oriT-pheS*-KanR integration site in Chr. 2, followed by Sanger sequencing. The phenotype of the correct clones was validated by stamping cell suspensions on 20 μg/mL chloramphenicol, 7.5% sucrose, 2.5 mM 4-chloro-phenylalanine, 50 μg/mL or a combination of these.
Insertion of pH with homologies for fusion
To recombineer pheS*-HygR selection cassettes into the positions targeted for fusion in the genome, the selection cassette was first PCR amplified with primers containing homology regions HR1 and HR2 (Table S2 for homology sequences, Table S6 for primer sequences). Briefly, post-fission MDS42rpsLK43R cells cured of the corresponding pKW3_MB1Amp_TracrK_Spacer fission plasmid, and harbouring pKW20_CDFtet_pAraRedCas9_tracrRNA were made electrocompetent with 1 hour of 0.5% L-arabinose induction as described previously (7), to express Cas9 and lambda-red machinery. 3 μg of column-purified HR1-pheS*-HygR-HR2 PCR product was electroporated into 100 μL of these competent cells. Transformed cells were recovered in 4 mL of super optimal broth (SOB) medium for 1 hour at 37 °C, diluted to 100 mL of LB with tetracycline (5 μg/mL), and incubated for 4 hours at 37 °C with shaking at 200 rpm. The culture was then spun down and re-suspended in 1 mL of Milli-Q water and spread in serial dilutions on selection plates of LB agar supplemented with 200 μg/mL hygromycin B.
Fusion procedure
After introduction of a pheS*-HygR at the desired locus for fusion, cells were made electrocompetent with L-arabinose induction, as described above, to drive expression of Cas9 and the lambda-red machinery from pKW20_CDFtet_pAraRedCas9_tracrRNA. 100 μL of electrocompetent cells were electroporated with ~8 μg of the corresponding pKW3_MB1Amp_TracrK_Spacer fusion plasmid, encoding spacer RNAs for 4 Cas9 cleavages that initiate fusion. Cells were recovered for 2 hours in 4 mL of SOB, with addition of 0.5% L-arabinose after 1 hour (as above). They were then transferred to 100 mL LB + 5 μg/mL tetracycline + 100 μg/mL ampicillin and recovered for 4 hours. Cells were then pelleted by centrifugation, resuspended in Milli-Q water and spread on LB agar plates containing 5 μg/mL tetracycline + 100 μg/mL ampicillin + 2.5 mM 4-chloro-phenylalanine. Resulting colonies were resuspended in Milli-Q water and stamped on selection plates (as above). Genomic DNA was prepared with the QIAgen Blood and Tissue kit as above, and PCR reactions were performed with primers flanking either side of the two new junctions generated by fusion.
Chromosomal transplant
The donor cell is post-fission MDS42rpsLk43R with a watermarked Chromosome 2 containing oriT-pheS*-KanR. The recipient cell is post-fission MDS42rpsLk43R with a non-watermarked Chromosome 2 containing SacB-CmR and a watermarked region in Chromosome 1 (see above). First, the luxABCDE operon in Chromosome 1 of the recipient cell was replaced with a pheS*-HygR cassette, as above. Then, the donor and recipient strains were respectively grown in LB + 50 μg/mL kanamycin or 18 μg/mL chloramphenicol, in the absence of tetracycline or ampicillin to isolate clones cured of both pKW3_fission_C and pKW20_CDFtet_pAraRedCas9_tracrRNA. Post-fission donor cells were subsequently electroporated with an immobilised F' plasmid - pJF146 – which carries apramycin resistance (5). This plasmid harbours the components required for conjugation but contains a truncated oriT sequence, which renders it incapable of self-transfer.
For conjugative transplant, the donor strain was grown overnight in 25 μg/mL apramycin, and the recipient in 200 μg/mL hygromycin B. 5 mL of cell suspension of OD600 = 2.4 were washed three times in LB + 2% glucose, and resuspended in 200 μL LB + 2% glucose. 100 μL of donor cell suspension and 100 μL of recipient suspension were gently mixed by pipetting, and the mixture spotted in 10-20 μL droplets on 37 °C pre-warmed TYE agar. The spots were left to dry and then incubated for 2 hours at 37 °C. Cells were washed off the plate with LB, diluted in 200 mL LB + 50 μg/mL kanamycin and 200 μg/mL hygromycin B, and incubated while shaking at 37 °C for 3 hours. The entire culture was pelleted by centrifugation, resuspended in Milli-Q water and plated in serial dilutions in LB agar supplemented with 50 μg/mL kanamycin + 200 μg/mL hygromycin B + 7.5% sucrose. Colonies were resuspended in Milli-Q and stamped on relevant selection plates.
Next-generation sequencing
For Illumina sequencing, E. coli genomic DNA was purified from overnight cultures using the DNEasy Blood and Tissue Kit (QIAgen) as per manufacturer’s instructions. Paired-end Illumina sequencing libraries were prepared using the Nextera XT Kit as per manufacturer’s instructions. Sequencing data was obtained in the Illumina MiSeq, running 2 x 300 or 2 x 75 cycles with MiSeq Reagent Kit v3.
For Oxford Nanopore sequencing, E. coli overnight cultures were purified with the PureGene Yeast/Bacteria Kit (QIAgen) as per manufacturer’s instructions, taking care to avoid vigorous shaking so as to minimise DNA shearing. Sequencing libraries were prepared with the Rapid Barcoding Kit (SQK-RBK004) with the following modifications. Purified DNA samples were diluted 1:10 in water and quantified using a Qubit Fluorometer with a dsDNA High Sensitivity assay. For a given sequencing run, equal masses of different undiluted DNA samples (at least 600 ng) were subject to fragmentation with 3.5 μL of barcoded fragmentation mix, adding water to a total volume of 20 μL, and incubated for 1min 30s at 30 °C followed by 1 minute at 80 °C. Ampure XP bead clean up was omitted and the barcoded libraries were pooled directly in equimass ratios, mixed with an equal volume of AMPure XP beads, washed twice with 500 μL of 70% EtOH and eluted in 11 μL of 10 mM Tris-HCl (pH 8.0) + 50 mM NaCl. 1 μL of pooled library was quantified in a Qubit Fluorometer with a dsDNA High Sensitivity assay, and 1 μL of RAP was added to the remaining 10 μL. The library loading mix was prepared with 11 μL of library, 34 μL of SQB, 25.5 μL of loading beads and 4.5 μL of nuclease-free water. The typical total input into the flowcell was ~1 μg of DNA. Sequencing was performed on a MinION Mk1B, using the MinKNOW software with standard 48 hour protocols.
Sequencing data analysis
Illumina reads were trimmed with Trimmomatic to remove adapter sequences (24). Oxford Nanopore (ONT) reads were basecalled using Guppy basecaller v2.3.1, with a mean quality cut-off filter of 6. Reads that passed the quality filter were subject to adapter trimming and demultiplexing with Porechop v0.2.4 (available at https://github.com/rrwick/Porechop), discarding the reads that contain internal adapters. Demultiplexed reads derived from the same physical DNA sample but obtained in different sequencing runs were in some instances pooled together to increase coverage of ONT reads. Hybrid de novo assemblies combining Illumina and ONT data were performed using Unicycler (22) with bridge application set to ‘normal’, except for Fission G which was performed in ‘bold’. Assemblies were visualised using Bandage (25). The assembly graphs shown in this work are a direct output of Unicycler and Bandage and were not subject to manual refinement.
Growth rate measurement and analysis
Clones resulting from fission and fusion experiments were streaked in LB agar + 100 μg/mL streptomycin plates. 5 colonies from each were picked and grown overnight in LB + 100 μg/mL streptomycin. Overnight cultures were diluted 1:100 in LB + 100 μg/mL streptomycin in a 96 well plate. OD600 measurements were taken every 5 minutes in a Tecan Infinite Microplate reader.
To determine doubling times, the growth curves were log2-transformed. At a linear phase of the curve during exponential growth, the first derivative was determined (d(log2(x))/dt) and ten consecutive time-points with the maximal log2-derivatives were used to calculate the doubling time for each replicate. A total of 5 independently grown biological replicates were measured for each strain. The mean doubling time and standard deviation from the mean were calculated for all n=5 replicates, except when indicated in the text.
Pulse-field gel electrophoresis analysis of fission strains
Clones resulting from fission experiments were grown overnight at 37 °C in 3mL of LB medium supplemented with appropriate antibiotics; 50 μg/mL of kanamycin for cells with Chr. 2 harbouring a pheST251A_A294G -KanR selection cassette, 20 μg/mL chloramphenicol for sacB-CmR, and 100 μg/mL streptomycin for the pre-fission MDS42 control. Genomic DNA agarose plugs were prepared from each strain using the BioRad CHEF Genomic DNA Kit, as per manufacturer’s instructions. Plugs were stored at 4 °C until used.
Plugs were cut in half (~50 μL per half plug), and subject to a restriction digest with AvrII. For this, half-plugs were rinsed in 0.1X BioRad CHEF Wash Buffer and equilibrated twice in 1X New England Biolabs Cutsmart buffer for 20 minutes at room temperature. The buffer was aspirated and each half-plug was then treated with 25U of AvrII in 100 μL of 1X NEB Cutsmart buffer, and incubated for 2 hours at 37 °C. Following restriction digestion the buffer was aspirated, 150 μL of BioRad CHEF Proteinase K solution was added to each half-plug, and plugs were incubated for another 30 minutes at 37 °C. Plugs were then rinsed with 800 μL of 0.5x TBE, and loaded into a 1% agarose pulse-field gel. Electrophoresis was conducted in a BioRad CHEF-DR-III pulse-field electrophoresis system, in 0.5x TBE, for 22 hours at 14 °C, with 20-120s switch times, 120° angle and 6 V/cm.
Analysis of genetic stability
Cells were grown at 37 °C in 3 mL volumes of LB medium supplemented with the appropriate antibiotic (e.g. 20 μg/mL of chloramphenicol for strains with Chr. 2 harbouring a sacB-CmR selection cassette or 50 μg/mL of kanamycin for strains with Chr. 2 harbouring a pheST251A_A294G -KanR selection cassette) while shaking at 200 rpm. The cultures were passaged by diluting 1/1500x once every twelve hours for a total of 5 days, which yielded a total estimated number of generations of 105 (26). To assess the genetic stability of post-passaged cells, the resulting cultures were used directly for the preparation of genomic DNA agarose plugs for pulse-field gel electrophoresis, as described above.
Supplementary Material
One sentence summary.
Technologies to precisely split, reorganize, and combine bacterial chromosomes to facilitate highly-programmable genome engineering.
Acknowledgements
We thank J. Houseley and J. Ajioka for providing equipment for pulse-field gel electrophoresis.
Funding
This work was supported by the Medical Research Council (MRC), UK (MC_U105181009 and MC_UP_A024_1008) and an ERC Advanced Grant SGCR, all to JWC.
Footnotes
Author contributions: K. W. designed and implemented the genome manipulation processes reported. K.W, D. d.l.T., and W. E. R. demonstrated scope. D. d.l.T implemented the de novo assembly approach. J.W.C defined the direction of research, supervised the project and wrote the paper with the other authors.
Competing interests: The authors declare no competing interests.
Data and materials availability: The sequences for de novo genome assemblies and DNA sequencing data have been deposited in NCBI’s GenBank and SRA databases, and their accession numbers are listed in table S4. All other data needed to evaluate the conclusions of the study are present in the paper or the supplementary materials.
References
- 1.Posfai G, et al. Emergent properties of reduced-genome Escherichia coli. Science. 2006;312:1044–1046. doi: 10.1126/science.1126439. [DOI] [PubMed] [Google Scholar]
- 2.Hutchison CA, 3rd, et al. Design and synthesis of a minimal bacterial genome. Science. 2016;351 doi: 10.1126/science.aad6253. aad6253. [DOI] [PubMed] [Google Scholar]
- 3.Chan LY, Kosuri S, Endy D. Refactoring bacteriophage T7. Mol Syst Biol. 2005;1 doi: 10.1038/msb4100025. 2005 0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lajoie MJ, et al. Genomically recoded organisms expand biological functions. Science. 2013;342:357–360. doi: 10.1126/science.1241459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fredens J, et al. Total synthesis of Escherichia coli with a recoded genome. Nature. 2019;569:514–518. doi: 10.1038/s41586-019-1192-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dymond JS, et al. Synthetic chromosome arms function in yeast and generate phenotypic diversity by design. Nature. 2011;477:471–476. doi: 10.1038/nature10403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang K, et al. Defining synonymous codon compression schemes by genome recoding. Nature. 2016;539:59–64. doi: 10.1038/nature20124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lederberg J, Tatum EL. Gene recombination in Escherichia coli. Nature. 1946;158:558. doi: 10.1038/158558a0. [DOI] [PubMed] [Google Scholar]
- 9.Isaacs FJ, et al. Precise manipulation of chromosomes in vivo enables genome-wide codon replacement. Science. 2011;333:348–353. doi: 10.1126/science.1205822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burt DW. Origin and evolution of avian microchromosomes. Cytogenet Genome Res. 2002;96:97–112. doi: 10.1159/000063018. [DOI] [PubMed] [Google Scholar]
- 11.Giannuzzi G, et al. Hominoid fission of chromosome 14/15 and the role of segmental duplications. Genome Res. 2013;23:1763–1773. doi: 10.1101/gr.156240.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cooper VS, Vohr SH, Wrocklage SC, Hatcher PJ. Why genes evolve faster on secondary chromosomes in bacteria. PLoS Comput Biol. 2010;6:e1000732. doi: 10.1371/journal.pcbi.1000732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Escudero JA, Mazel D. Genomic Plasticity of Vibrio cholerae. Int Microbiol. 2017;20:138–148. doi: 10.2436/20.1501.01.295. [DOI] [PubMed] [Google Scholar]
- 14.Ausiannikava D, et al. Evolution of Genome Architecture in Archaea: Spontaneous Generation of a New Chromosome in Haloferax volcanii. Mol Biol Evol. 2018;35:1855–1868. doi: 10.1093/molbev/msy075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Itaya M, Tanaka T. Experimental surgery to create subgenomes of Bacillus subtilis 168. Proc Natl Acad Sci U S A. 1997;94:5378–5382. doi: 10.1073/pnas.94.10.5378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luo J, Sun X, Cormack BP, Boeke JD. Karyotype engineering by chromosome fusion leads to reproductive isolation in yeast. Nature. 2018;560:392–396. doi: 10.1038/s41586-018-0374-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shao Y, et al. Creating a functional single-chromosome yeast. Nature. 2018;560:331–335. doi: 10.1038/s41586-018-0382-x. [DOI] [PubMed] [Google Scholar]
- 18.Ueda Y, et al. Large-scale genome reorganization in Saccharomyces cerevisiae through combinatorial loss of mini-chromosomes. J Biosci Bioeng. 2012;113:675–682. doi: 10.1016/j.jbiosc.2012.01.013. [DOI] [PubMed] [Google Scholar]
- 19.Yamaichi Y, Niki H. migS, a cis-acting site that affects bipolar positioning of oriC on the Escherichia coli chromosome. EMBO J. 2004;23:221–233. doi: 10.1038/sj.emboj.7600028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Raeside C, et al. Large chromosomal rearrangements during a long-term evolution experiment with Escherichia coli. MBio. 2014;5:e01377–01314. doi: 10.1128/mBio.01377-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liang X, Baek CH, Katzen F. Escherichia coli with two linear chromosomes. ACS Synth Biol. 2013;2:734–740. doi: 10.1021/sb400079u. [DOI] [PubMed] [Google Scholar]
- 22.Wick RR, Judd LM, Gorrie CL, Holt KE. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol. 2017;13:e1005595. doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schmied WH, et al. Controlling orthogonal ribosome subunit interactions enables evolution of new function. Nature. 2018;564:444–448. doi: 10.1038/s41586-018-0773-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wick RR, Schultz MB, Zobel J, Holt KE. Bandage: interactive visualization of de novo genome assemblies. Bioinformatics. 2015;31:3350–3352. doi: 10.1093/bioinformatics/btv383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Choe D, et al. Adaptive laboratory evolution of a genome-reduced Escherichia coli. Nat Commun. 2019;10 doi: 10.1038/s41467-019-08888-6. 935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Camacho C, et al. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.