Abstract
Intervertebral disc degeneration (IDD) is a public health dilemma as it is associated with low back and neck pain, a frequent reason for patients to visit the physician. During IDD, nucleus pulposus (NP), the central compartment of intervertebral disc (IVD) undergo degeneration. Stem cells have been adopted as a promising biological source to regenerate the IVD and restore its function. Here, we describe a simple, two-step differentiation strategy using a cocktail of four factors (LDN, AGN, FGF and CHIR) for efficient derivation of notchordal cells from human embryonic stem cells (hESCs). We employed a CRISPR/Cas9 based genome editing approach to knock-in the mCherry reporter vector upstream of the 3’UTR of the Noto gene in H9-hESCs and monitored notochordal cell differentiation. Our data show that treatment of H9-hESCs with the above-mentioned four factors for six days successfully resulted in notochordal cells. These cells were characterized by morphology, immunostaining, and gene and protein expression analyses for established notochordal cell markers including FoxA2, SHH and Brachyury. Additionally, pan-genomic high-throughput single cell RNA-sequencing revealed an efficient and robust notochordal differentiation. We further identified a key regulatory network consisting of eight candidate genes encoding transcription factors including PAX6, GDF3, FOXD3, TDGF1 and SOX5, which are considered as potential drivers of notochordal differentiation. This is the first single cell transcriptomic analysis of notochordal cells derived from hESCs. The ability to efficiently obtain notochordal cells from pluripotent stem cells provides an additional tool to develop new cell-based therapies for the treatment of IDD.
Keywords: Notochord, Cell Differentiation, Single cell Transcriptomics, Stem cell, Gene-network, CRISPR/Cas9, RNA-Sequencing
Introduction:
Intervertebral disc degeneration (IDD) has been linked to low back pain, the third most common reason for doctor’s office visits, and has a significant direct and indirect economic costs to both the patient and society (Katz, 2006; St Sauver et al., 2013). Healthy intervertebral disc (IVD) is compartmentalized into well-defined regions of the outer annulus fibrosus (AF) and the inner nucleus pulposus (NP). During IDD, the discrete compartmental structure is disrupted, and dehydration of NP occurs due to a decrease in cellularity and proteoglycan content (Buckwalter, 1995; Urban & Roberts, 2003 ). First line therapies for IDD are symptomatic treatments that aim to relieve pain thus offering momentary benefits instead of a long-term cure (Mirza & Deyo, 2007). Although surgical therapies are relatively effective, they fail to regenerate the disc structure and are associated with additional complications and morbidities (Errico, 2005; Hanley et al., 2010). Therefore, it is necessary to identify an effective treatment modality that can effectively repair and restore the functional disc structure.
Cell based regenerative therapy constitutes a highly promising and attractive strategy for restoration of NP cellularity and reconstitution of the healthy disc structure (Chan & Gantenbein-Ritter, 2012; Leung, Chan, & Cheung, 2006; Sakai, 2008). The NP of adult human is derived from a rod-like midline structure of mesodermal origin known as notochord (T et al., 2017). Since adult NP do not possess self regeneration capabilities, the NP cell renewal depends upon availability of progenitor cells which commit to the NP lineage and undergo terminal differentiation and produce an appropriate proteoglycan-rich matrix which is essential for the functioning of IVD (DL, 2005; N et al., 2014). Among the progenitor cells which possess discogenic potential, the notochordal cells are considered ideal cells for regeneration of degenerated disc. However, due to limited availability of adult autologous or allogenic notochordal cells, there is constant quest to identify a viable cell source to facilitate this potentially effective regenerative strategy (Risbud, Schaer, & Shapiro, 2010).
Therefore, alternative cell sources such as multipotent or pluripotent stem cells including mesenchymal stem cells (MSCs), embryonic stem cells (ESCs), and induced pluripotent stem cells (iPSCs) have been investigated for potential differentiation into notochordal cells (Acosta, Lotz, & Ames, 2005; McCann, Bacher, & Seguin, 2011). Effective differentiation of these stem cells into functional notochordal cells is a critical step for successful cell-based regenerative therapy, as notochordal cells generate the terminally differentiated NP cells and maintain the disc homeostasis (Alini, Roughley, Antoniou, Stoll, & Aebi, 2002; Chan & Gantenbein-Ritter, 2012; McCann et al., 2011). In this regard, several studies attempted to differentiate MSCs, iPSCs or ESCs into notochord like cells using numerous strategies involving series of growth factors and cytokines (Chen et al., 2013; Chon, Lee, Jing, Setton, & Chen, 2013; Liu, Fu, Rahaman, Mao, & Bal, 2015; Liu, Rahaman, & Bal, 2014; Tang et al., 2018). Although these reports demonstrated successful differentiation into notochord-like cell, the employed strategies suffer from several limitations such as low yield of notochordal cells, inferior ability to produce native notochordal cells, longer duration of differentiation protocol, use of many expensive differentiation factors, lack of detailed characterization of notochordal cells and poor regenerative capacity of generated notochord cells (Chen et al., 2013; Chon et al., 2013; Liu et al., 2015; Liu et al., 2014; Tang et al., 2018).
The present study aimed to devise an effective strategy for robust differentiation of human embryonic stem cells (H9-hESCs) into notochordal cells. Thus, we developed a simple two-step protocol relying on four factors to direct hESC differentiation into notochordal cells. CRISPR/Cas9 genome editing was used to knock-in a red fluorescent reporter gene (mCherry) into the third exon of the endogenous NOTO gene in H9-hESCs. Due to its solitary expression in notochordal cells, the NOTO gene was selected for construction of the reporter vector which helps in effective screening of notochordal differentiation through spatial and temporal monitoring of NOTO-mCherry fluorescence. Our study further characterized the generated notochordal cells at both the transcript and protein levels and showed higher efficiency of our method to generate notochordal cells from hESCs. Additionally, single cell transcriptomic profiling during the course of differentiation events was employed to identify potential gene regulators, signaling mediators, and transcription factors for efficient differentiation of hESCs into notochordal cells.
Material and Methods:
Maintenance of pluripotent stem cell line:
H9-human embryonic stem cells (H9-hESc) were cultured in mTeSR™1 media (StemCell Technologies) on feeder free conditions in 6-well culture plates. To maintain the pluripotency, hESCs colonies after reaching 70% confluency, were washed with PBS and then treated with ReLeSR™ reagent (StemCell Technologies). Medium sized colonies were obtained and resuspended in mTeSR™1 media and plated on 0.1% Geltrex® (Peprotech) coated 6-well plates to expand H9-hESC colonies.
Construction of Noto-2A-mCherry reporter vector:
Human embryonic stem cells-H9 derived (hESCs) were genetically manipulated to knock-in the mCherry reporter gene into exon3 of the endogenous NOTO gene (Accession: NM_001134462.2). The targeting vector contains LoxP sites flanking both sides of a Neo cassette (LoxP-Neo-LoxP) for selection of positive clones. The mCherry sequence was inserted after exon 3 and before the 3’UTR sequence of the NOTO gene. A Diphtheria Toxin A (DTA) sequence was used to select clones with the correct orientation. CRISPR/Cas9 system and homologous recombination strategies were used to generate targeting vectors. All vectors were sequenced to verify the presence of NOTO-mCherry sequence using Sanger sequencing.
Noto-2A-mCherry reporter vector knock-in into H9-hESCs:
To knock-in the reporter vector in H9-hESCs, electroporation was utilized to reach the maximum transfection efficiency. H9-hESC colonies were treated with Accutase (Thermo Fisher Scientific) to create single cell suspensions, then mixed with targeting vectors and Cas9, followed by electroporation using the Gene Pulser Xcell™ electroporation system (Bio-Rad). Transfected cells were plated on DR4 MEF Feeder cells in complete hESC media supplemented with Y-27632 and small molecule L77507 to promote homology-directed repair (HDR) for 48hrs. Antibiotic selection was performed after 72 hrs of culture using 50 mg/ml G418 (Geneticin) and was continued until the colonies were manually picked. Afterwards, colonies were plated in 24-well plates and after the first passage, a portion of colonies were used to extract gDNA to perform PCR in order to identify positive clones. Individual clones of parental Noto-2A-mCherry H9-hESCs were nucleofected with Cre-puro plasmid for the excision of the Neo cassette and then plated on DR4 MEF feeder cells, and Puromycin selection was performed for 2 days. To examine the pluripotency of this genetically manipulated H9-hESCs harboring reporter vector, mRNA expression of pluripotent markers was analyzed using qPCR analysis for Sox2, Nanog Oct3/4 genes as reported previously (Drissi, Gibson, Guzzo, & Xu, 2015).
Establishment of two-step differentiation strategies to generate notochordal cells:
A simple, effective, two-step differentiation protocol was developed to generate notochordal cells from H9-hES cells. H9-hESC colonies were expanded and cultured in 0.1% Geltrex coated surface until it reached 80% confluency and were then split using ReLeSR reagent and seeded on 6-well plate in mTeSR™1 media. Two days later, mTeSR™1 medium was replaced with basal differentiation media containing IMDM, Ham’s F12, 150mM MTG, 0.5 mM AA;1X Pen/Strep; 0.5% B27; 0.5% N2; 0.05%BSA. Replacement of mTeSR-1 by basal differentiation media was considered as Day 0 of the differentiation protocol (Figure 1). For the first step, basal medium was supplemented with three growth factors for a period of four days. The three factors-LDN-193189 (0.25 μM) (Cayman) for inhibition of BMP signaling, human basic Fibroblast Growth Factor (FGF) (10ng/ml) (Peprotech), and AGN193109 (1μM) (Santa Cruz Biotechnology) as an inhibitor of pan retinoic acid signaling were used for early notochordal differentiation. These treatments were called LAF treatment. The second step of the differentiation protocol was commenced after four days of LAF treatment, which consisted of supplementation with CHIR (3 μM) (Cayman) for two additional days. This treatment condition was designated LAFC. During the course of the 6-day differentiation protocol, media was refreshed every 2 days with supplementation of growth factors. The parental H9-hESCs maintained without the addition of the four factors were considered the control group. To determine the efficiency of our established differentiation strategy, colonies were harvested at day 0 (control), day 4 (LAF treatment) and day 6 (LAFC treatment) and notochordal differentiation was monitored by evaluating the fluorescence of mCherry reporter using fluorescence microscopy (BioTek Lionheart LX Automated Microscope).
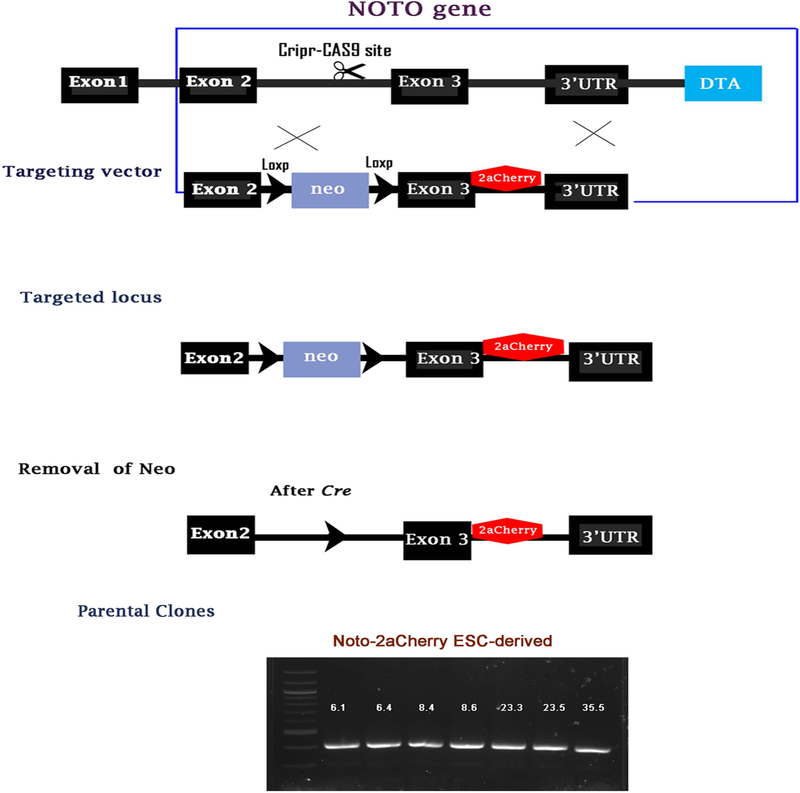
Figure 1: Generation of a Noto-2A-mCherry reporter line in H9-hESC:
(A-B) CRISPR mediated targeting of a Cherry reporter into the 3’UTR locus of the NOTO gene. H9-hESCs were genetically engineered to carry a cherry fluorescent protein reporter into the NOTO gene. In the targeting vector, the stop codon was removed and replaced with a viral 2A peptide followed by a cherry reporter coding sequence. This allows the gene product of NOTO to function properly and at the same time give accurate reporter expression to the endogenous gene. (C) After successful targeting and clonal selection, the Neo cassette is removed from different clonal lines by transient expression of Cre recombinase. (D) Characterization of NOTO-2A-mCherry knock-in in different clones of H9-hESCs. Precise mCherry integrated colonies present PCR products in selected clones.
Flow cytometry to determine the efficiency of notochordal differentiation:
To determine the efficacy of notochordal differentiation using our established protocol, we determined the frequency of cell populations expressing mCherry fluorescence by sorting Noto-2A-mCherry positive cells in all experimental groups (day 0, 4, and 6) using FACS Aria II cell sorter system (BD Biosciences, San Jose, CA). Briefly, colonies were digested using Accutase and single cells were suspended in a sorting buffer containing HBSS (Gibco), 1M HEPES, 0.02% fetal serum albumin supplemented with 10μM of ROCK inhibitor Y-27632. SYTOX Blue nucleic acid stain (Molecular Probes) was used to evaluate the percentage of cell death in each sample. Before sorting the cells, the flow cytometer was calibrated with mCherry calibration beads (Clontech).
We also examined the percentage of cells expressing the established notochordal markers SHH, FoxA2 and Brachyury (T) by intracellular antibody staining using a method reported previously (Khan & Poduval, 2011; 2012). Briefly, cells from the above three experimental groups (day 0, 4, 6) were dissociated into single cells using Accutase after washing with PBS, and then fixed with 4% paraformaldehyde for 10 minutes at room temperature and excess of paraformaldehyde was removed by washing once with wash buffer (PBS containing 1% BSA). Cell suspension were permeabilized with PBST (PBS containing 0.02% Tween 20) for 5 minutes at room temperature followed by washing with wash buffer. Permeabilized cells were then stained with APC-labeled anti-Brachyury polyclonal antibody (R&D System, # IC2085A), Alexa Fluor® 488-conjugated antibody against FoxA2 (R&D System, Catalog # IC2400G) and PE conjugated anti-SHH monoclonal antibody (R&D System, Catalog # IC4641P) for 30 minutes at room temperature. Appropriate isotype controls were used for staining. Efficiency of notochordal differentiation was analyzed by determining the percent positive cell population for the notochordal markers. Twenty thousand cells in each group were acquired using BD‐FACS Aria II (BD Biosciences, San Jose, CA) and data were analyzed using FlowJo software from Tree Star, Inc. (Ashland, OR) as described previously (Haseeb, Khan, Ashruf, & Haqqi, 2017; Khan, Ahmad, Ansari, & Haqqi, 2017).
RNA isolation, Reverse Transcription and qPCR:
RNA from different experimental conditions (day 0, 4 and 6) was isolated using TRIzol reagent (Invitrogen) as previously described (Akhtar, Khan, Ashruf, & Haqqi, 2017; Khan, Ahmad, & Haqqi, 2018; Khan, Haseeb, Ansari, Devarapalli, et al., 2017). RNA was treated with DNAse I (Invitrogen) and reverse transcribed to cDNA using random hexamers using qScript cDNA synthesis kit (Quantabio) following manufacturer’s instructions. Real‐time quantitative polymerase chain reaction (qPCR) was performed using PowerUp™ SYBR® Green master mix (Applied Biosystems) on the Analytik Jena RT-PCR detection system. All RT-qPCR experiments were performed with three biological replicates from each group and two technical replicates. Primer sequences for the all genes are listed in Supplementary Table 1. The mRNA expression of notochordal marker genes and pluripotency genes was normalized to GAPDH mRNA and relative expression levels were calculated using the 2−ΔΔCT method as described previously (Haseeb, Makki, Khan, Ahmad, & Haqqi, 2017; Khan, Ansari, & Haqqi, 2017; Khan, Haseeb, Ansari, & Haqqi, 2017).
High-density micromass culture for enhanced notochordal differentiation:
To enhance the effectiveness of our method for notochordal differentiation, we utilized a high-density micromass culture system to mimic the 3D in vivo avascular environment condition. After 6 days differentiation in monolayer, colonies were dissociated into single cells using Accutase treatment and re-suspended in basal differentiation medium supplemented with ROCK inhibitor (10μM) at cell density of 25×106 cells per ml. Each micromass was prepared by culturing high‐density cell pellets (25 ×104/10μl cells per pellet) in a dropwise fashion in 6-well culture plates without any surface coating. Immediately after seeding the micromass pellets, growth medium was carefully added dropwise from the edges of the plate to prevent dehydration of pellet. These pellets were incubated for 3 hrs at 37°C in 5% CO2, and then supplemented with 2 ml of growth medium. Each micromass was then grown for an additional 48 hrs before analysis of notochordal differentiation.
Single cell RNA-sequencing and transcriptomic analysis:
To determine the gene regulatory networks and signaling pathways involved in notochordal differentiation, unbiased pan transcriptomic analysis was performed at different stages of hESC differentiation (day 0, 4 and 6) using single cell RNA sequencing. Notochordal cells are precursors of terminally differentiated NP cells that play a pivotal role in the homeostasis of the NP (Alini et al., 2002; McCann et al., 2011; Risbud et al., 2010). Considering that both cell types exhibit parallel phenotypic and functional profiles, we used human nucleus pulposus cells (abbreviated as NPC) (ScienCell™ Research Laboratories; Catalog #4800) as a positive control to compare its global transcriptional profile with that of generated notochordal cells (day 4 and 6) to determine the efficacy of our differentiation strategy. Single-cell RNA-Seq libraries were prepared using Sure Cell WTA 3′ library prep kit (Illumina) along with the ddSEQ™ single cell Isolator according to the manufacturer’s instructions. The sequencing depth was 100,000 reads per cell and on average 300 cells per experimental group were analyzed using Seurat package in R as described previously (Satija, Farrell, Gennert, Schier, & Regev, 2015). Pseudo pools method employing a random pool of 20 cells of the same type within experimental group was used for the differential gene expression analysis. This ensures that a high proportion of genes are represented by an approximately normal count distribution and also allows for fitting of normalization strategies initially developed for bulk microarray or RNA-seq analysis. A total of 10 iterations of pseudo pool process were performed to eliminate cell-specific biases. The EdgeR Bioconductor package was used for differential gene expression analysis (Robinson, McCarthy, & Smyth, 2010). A mean fold change was calculated from all 10 permutations and significant differences in the expression levels were calculated by false discovery rate (FDR) corrected p-value. The genes with an FDR ≤0.05 were considered differentially expressed. Gene Ontology (GO) enrichment analyses for biological process and molecular function among different experimental groups were performed using GOrilla (Eden, Navon, Steinfeld, Lipson, & Yakhini, 2009), a web-based tool to identify enriched GO terms in ranked lists of genes. GO terms were visualized by REVIGO (Supek, Bosnjak, Skunca, & Smuc, 2011). Additionally, biological significance of enriched pathways and associated genes was determined by integrated network analysis using STRING (v: 11.0) (Szklarczyk et al., 2017) and networks were visualized using MCODE plugin in Cytoscape (v:3.7.1) (Otasek, Morris, Boucas, Pico, & Demchak, 2019; Shannon et al., 2003). To identify global transcriptional similarities among different experimental groups (day 0, 4, 6 and NPC), Heatmaps and Principal Component Analysis (PCA) were performed using ClustVis (Metsalu & Vilo, 2015).
Statistical analysis:
The data were presented as the Mean±SEM and statistically significant differences between the experimental groups and controls were analyzed using one-way ANOVA followed by post hoc analyses using the Tukey test. Unless otherwise noted, each experiment was repeated three times using three independent biological replicates. P<0.05 was considered to be statistically significant.
Results:
Generation of notochordal reporter (NOTO-2A-mCherry) pluripotent stem cells:
We utilized CRISPR/Cas9 based genome editing approach to construct NOTO-2A-mCherry red fluorescent reporter vector in H9-hESC lines. The experimental strategy used to generate the NOTO-2A-mCherry reporter ESC lines is shown in Figure 1A. The mCherry reporter sequence was inserted after exon 3 and before the 3’UTR sequence of the NOTO gene, which does not influence the endogenous expression or function of NOTO (Figure 1A). To construct the reporter vector, we used the 2A system to link exon 3 of the NOTO gene and mCherry reporter sequence to obtain co-expression of NOTO and mCherry at equimolar levels (Hsiao et al., 2008). The 2A self-cleaving peptide system links the reporter to the native open reading frame. Using CRISPR/Cas9 mediated gene editing followed by homologous recombination, we successfully inserted the NOTO-2A-mCherry reporter vector in H9-ESC lines (Figure 1B). Among the 36 generated colonies, four H9-hESC clones (#6, #8, #23, and #35) were selected for Neo removal using Cre mediated recombination (Figure 1C). These recombinants were analyzed to determine genotype and our results demonstrate that the zygosity of the clones does not affect the activation of NOTO-2A-mCherry. PCR characterization and Sanger-sequencing data showed that the mCherry sequence was precisely fused with the NOTO gene, with no mutation detected in engineered H9-hESC clones (Figure 1D, Data not shown). Two colonies from each selected clone were chosen to use as NOTO-2A-mCherry H9-hESC parental lines and used for differentiation into notochordal cells (Figure 1D).
Characterization of genetically engineered notochordal reporter pluripotent stem cells (Noto-2A-mCherry-H9-hESCs):
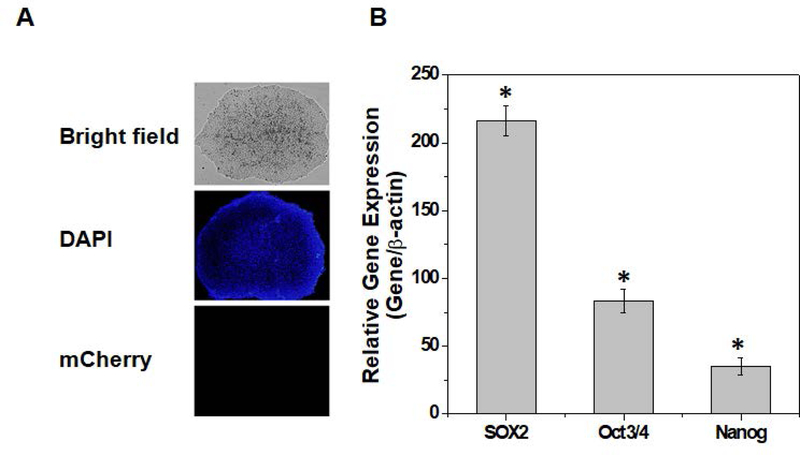
Parental clones of NOTO-2A-mCherry H9-hESC lines were maintained in feeder free culture conditions and colonies showed typical pluripotent cell morphology (Figure 2A). We next characterized the pluripotency of these parental clones by analyzing mRNA expression of stemness genes including NANOG, OCT4, and SOX2. Our results as shown in Figure 2B demonstrate significant expression of pluripotency marker genes in all of the selected clones (Figure 2B). Taken together these data establish that insertion of a red fluorescent reporter vector in H9-hESCs does not affect the pluripotent characteristics of engineered embryonic stem cells.
Figure 2: Characterization of a Noto-2A-mCherry reporter line in H9-hESC:
(A) Morphology of the Noto-2A-mCherry reporter colonies of H9-hESC in monolayer culture on a Geltrex coated plate. Undifferentiated colonies did not show mCherry fluorescence. (B) Pluripotency for derived colonies showing expression of stemness genes. RT-qPCR analyses showed induced expression of canonical stemness genes NANOG, OCT3/4, SOX2 in NOTO-2A-mCherry H9-hESC colonies. GAPDH served as the housekeeping gene and internal control. Represented gene expression data is relative to human H9-MSC derived from H9-hES cells.*P≤0.01, as compared to H9-MSCs.
Derivation of a two-step strategy for differentiation of H9-hESCs into notochordal cells:
Formation of the axial mesoderm from prechordal and notochordal progenitors requires a complex integration of signals for their induction and maintenance (Camus & Tam, 1999). Several reports demonstrated that notochordal differentiation from ESCs or iPSCs requires an array of growth factors (such as FGF),cytokines, and small molecules including BMP antagonists and retinoic acid inhibitors (Connolly, Patel, & Cooke, 1997; Streit & Stern, 1999). Here, we tested if a combination of the small molecules LDN93189, an inhibitor of BMP signaling, AGN193109, a pan receptor inhibitor of retinoic acid signaling and basic FGF2 treatment can induce NOTO-2A-mCherry activation in the parental H9-hESC lines (Figure 3A).
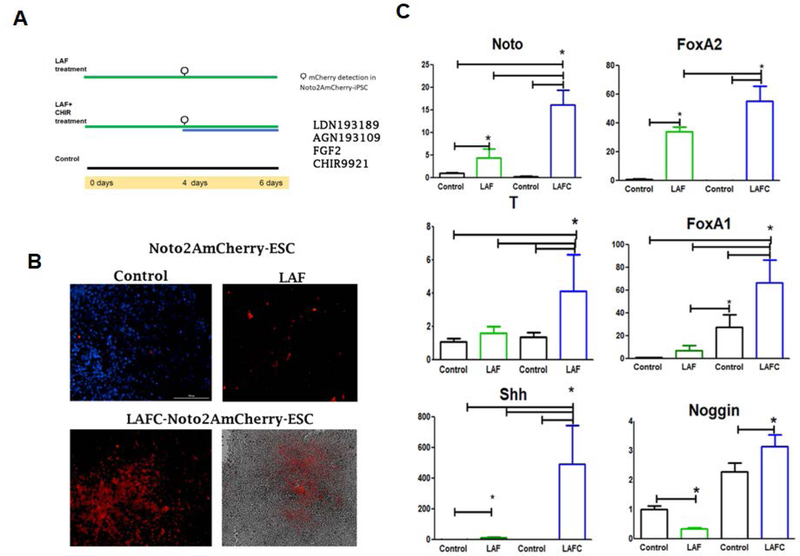
Figure 3: Derivation of two-step differentiation strategy for differentiation of H9-hESCs into notochordal cells:
(A) Schematic representation the two-step differentiation strategy of H9-hESCs differentiation into notochordal cells. (B) High magnification images of NOTO-mCherry positive cells during notochordal differentiation. NOTO reporter expressing cells observed in day 4 (LAF) and day 6 (LAFC treatments) of differentiation with a fluorescence microscope. (C) Expression of early notochord markers during differentiation. RT-qPCR analyses showed induced expression of NOTO, FOXA2, FOXA1, Brachyury (T),SHH, and Noggin (NOG) on 4 and 6 days of differentiation in H9-hESCs.GAPDH served as internal control and data is represented as expression relative to undifferentiated H9-hESCs (day 0). All data are represented as mean ± SEM from three independent experiments. *P≤0.01, as compared to control.
We considered the expression of NOTO-2A-mCherry as a key event during notochordal commitment based on robust documentation showing expression of NOTO as a crucial marker for notochordal formation. The fluorescence microscopic visualization of mCherry fluorescence demonstrated that treatment of H9-hESCs with LDN/AGN/FGF (LAF treatment) for four days showed slight activation of mCherry, indicating the initiation of notochordal differentiation (Figure 3B). Although mCherry fluorescence was observed only in small patches of cells (Figure 3B), the qPCR analyses showed upregulation of NOTO and FOXA2 mRNA expression compared to untreated parental group (day 0) (Figure 3C). At this stage of differentiation, no other notochordal marker expression was observed. Taken together, these data suggest that treatment with LDN/AGN/FGF (LAF) for four days is sufficient to initiate notochordal differentiation; however, differentiation is low indicating the requirement of additional signals for enhanced differentiation.
Previous studies have highlighted the crucial role of canonical Wnt and Nodal signaling for notochordal formation (Ukita et al., 2009; Wei & Wang, 2018). Therefore, we explored if activation of canonical Wnt signaling will be required for enhanced notochordal differentiation. Canonical Wnt signaling is dependent on β-catenin stabilization and its subsequent nuclear translocation mediates the activation of TCF/LEF family of transcription factors to regulate gene expression (Cadigan & Waterman, 2012). We used CHIR99021, a selective small molecule inhibitor of GSK3β, commonly used to activate canonical Wnt signaling. Due to upregulation of NOTO expression in response to LAF treatment, we decided to test whether initial induction of differentiation followed by addition of CHIR99021 for two days (LAFC treatment; LDN/AGN/FGF/CHIR, day 6) would increase differentiation (Figure 3A). Gross evaluation of LAFC treated cells (day 6) under fluorescence microcopy showed an increase in the fluorescence of NOTO-mCherry reporter expression. Interestingly, consistent with fluorescence observations in LAFC treated H9-hESCs (day 6), mRNA expression of the NOTO gene was dramatically increased when compared to both LAF (day 4) treated cells and the control group (day 0) (Figure 3C). Moreover, mRNA expression of key early notochordal markers such as Brachyury (T), SHH, FOXA1, FOXA2 and NOG (Noggin) were also significantly upregulated (Figure 3C). Additionally, our data show that there were no changes in expression of the late notochordal markers SOX9, SOX6, SOX5, YAP, Cytokeratins 8, 18 and 19. Taken together, these data suggest that cocktail of three factors (bFGF, LDN193189 and AGN193109) are critical for the generation of notochordal precursors and CHIR99021 treatment augments canonical Wnt signaling to induce NOTO expression. These data demonstrate that our two-step culture condition successfully differentiates pluripotent stem cells into early notochordal cells.
Characterization of H9-hESCs differentiation into notochordal cells:
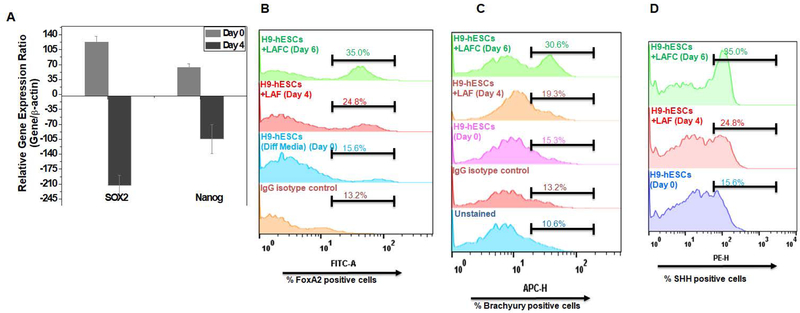
To further characterize the notochordal differentiation protocol, we examined whether our differentiation strategy favors commitment to mesodermal lineage through repression of prototypical pluripotency genes. To this end, we analyzed the expression of pluripotency genes throughout the differentiation protocol and our results demonstrate a significant repression of key stemness genes NANOG and Sox2 at day 6 of differentiation as compared to undifferentiated H9-hESC cells (Figure 4A). These results indicate that our two-step differentiation protocol using a cocktail of four factors significantly decreases the expression of pluripotency markers and effectively committed the ES cells towards a mesodermal phenotype.
Figure 4: Characterization of H9-hESCs differentiation into notochordal cells:
(A) RT-qPCR analyses showed decreased expression of NANOG and SOX2 on day 4 differentiation in H9-hESCs. GAPDH served as internal control and data is represented as expression relative to undifferentiated H9-hESCs (day 0). Flowcytometric histogram showing percent cells expressing (B) FOXA2, (C) Brachyury and (D) SHH during day 0, day 4 and day 6 of differentiation of H9-hESCs in monolayer. Cells were harvested and stained with anti-Barchyury-APC or anti-FOXA2-FITC or anti-SHH-PE labeled antibodies. All data are represented as mean±SEM from three independent experiments. *P≤0.01, as compared to undifferentiated control (day 0).
To further characterize the notochordal cells derived from H9-hESC differentiation, we analyzed the protein expression of early notochordal markers using intracellular antibody staining against SHH, FOXA2 and Brachyury (T) and determined the percentage of cells expressing these markers using flow cytometry. Flow cytometric histograms as shown in Figure 4 B–D demonstrate that treatment with three factors up to four days (LAF) showed a modest increase in cells expressing early notochordal markers (Figure 4B–D).When we treated cells grown in monolayer with our cocktail of four factors (LAFC, day 6) there was a significant increase in the percentage of cells expressing Brachyury (30.6%), FOXA2 (35%) and SHH (35%) (Figure 4B–D). These results recapitulate the finding of notochordal markers at the gene level indicating the robustness of our differentiation strategy.
Efficiency of differentiation strategy for notochordal differentiation in monolayer:
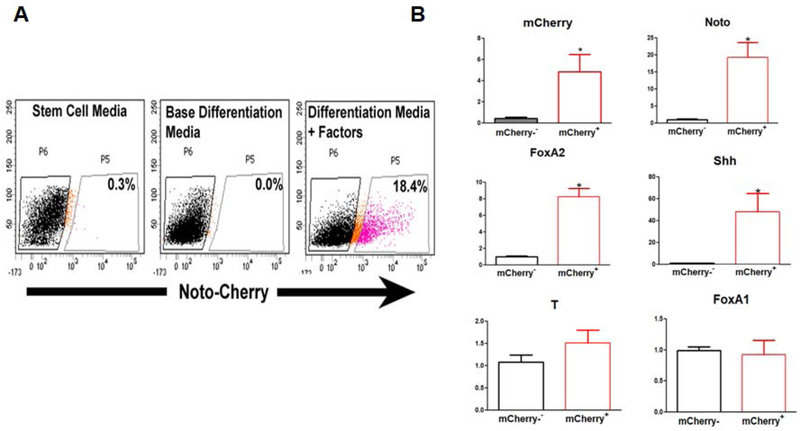
To determine the efficiency of hESC differentiation into notochordal cells, we first determined the percent population of Noto-mCherry+ cells (cells showing increased expression of NOTO protein) in H9-hESCs after 6 days of differentiation using FACS analyses. We obtained ~20% NOTO-mCherry+ cells from the live cell population of H9-hESCs on day 6 of monolayer culture (Figure 5A).
Figure 5: Efficiency of two step notochordal differentiation protocol in monolayer:
(A)FACS analyses and sorting of NOTO-RFP+ cells. FACs sorting and percent mCherry+positive cells during notochordal differentiation on day 0 and day 6. (B) Comparative gene expression profiles of notochordal phenotypic markers in FACS sorted NOTO-mCherry+and NOTO-mCherry− cells. RT-qPCR analyses showed the mRNA expression of NOTO, FOXA2, SHH, and Brachyury (T) on day 6 differentiation in sorted NOTO-mCherry+and NOTO-mCherry− cells. GAPDH served as the internal control and data is represented as expression relative to NOTO-mCherry− cells. All data are represented as mean±SD from two independent experiments. *P≤0.05, as compared to NOTO-mCherry− population.
We next characterized the pure population of notochordal cells (NOTO+) obtained from sorting of differentiated H9-hESCs (day 6). The expression of notochordal phenotypic marker genes NOTO, SHH, and FOXA2 were significantly upregulated in NOTO-mCherry positive (NOTO-mCherry+) compared to NOTO-mCherry negative cells (NOTO-mCherry−) (Figure 5 B). No significant differences were observed in the expression of Brachyury (T) and FOXA1 between NOTO-mCherry+ and NOTO-mCherry− cells, however, suggesting that both populations share the expression of some markers due to common mesodermal origin (Figure 5 B). It has been shown that bFGF, Wnt, and retinoic acid signaling pathways can also mediate the differentiation of others mesodermal lineages. Interestingly, NOTO-mCherry− population showed a slight increase in Sox9, Sox6 and Brachyury expression, which suggests a potential of differentiation of NOTO-mCherry− on other embryonic mesoderm types that could be needed to support the NOTO-mCherry+ population (data not shown). These data suggest that NOTO-mCherry− populations may differentiate into other mesodermal lineages but we cannot exclude the possibility that some of these cells still bear the capacity to further differentiate towards a notochordal cell phenotype.
High density micromass pellet culture improved the efficiency of notochordal differentiation:
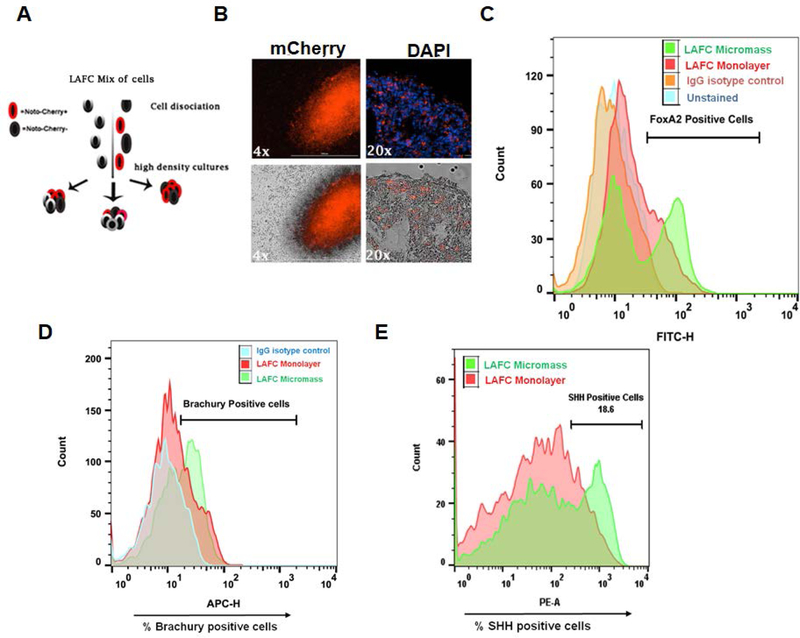
Our two-step differentiation protocol using cocktail of four factors in monolayer culture condition yielded about 20% of Noto+ cells. Thus, to determine if we could increase the number of NOTO+ cells we utilized a micromass system where early notochordal cells were cultured in 3D high density culture conditions. For this experiment, we did not sort Noto-mCherry+ from Noto-mCherry− cells, instead we used a mixed population (Noto+ and Noto−) (Figure 6A). The existing literature clearly demonstrates that high density culture mimics the mesenchymal cell condensation during disc development in embryonic stage. Moreover, 3D micromass culture offers a convenient in vitro model to study cell differentiation. Therefore, we used H9-hESC to generate high density condensations followed by culturing in basal media for two additional days. Results as shown in Figure 6B demonstrate that the proportion of cells expressing Noto-mCherry fluorescence was significantly higher when cultured in 3D conditions as compared to monolayer culture indicating that micromass condensation favors notochordal differentiation. To quantify the effectiveness of our differentiation strategy, in monolayer and micromass culture conditions, we examined the proportion of notochordal cells by analyzing percent cell populations expressing the notochordal markers by flow cytometry. Our differentiation protocol of H9-hESCs resulted in 43.6% FOXA2, 54% Brachyury (T) and 36.7% SHH positive cells in micromass condition as compared to 25% FoxA2, 34% Brachyury and 18.6% SHH positive cells in monolayer condition (Figure 6C-E). These results suggest that 3D high density micromass culture significantly improved the notochordal differentiation of H9-ESCs as compared to monolayer culture.
Figure 6: High density micromass pellet culture improved the efficiency of notochordal differentiation:
(A) Schematic representation of steps involved in high density micromass culture of H9-hESCs (B) High density cultures showed that proportion of notochordal cells expressing mCherry fluorescence at day 0 was significantly higher in 3D culture condition as compared to monolayer culture. Flowcytometric histogram showing percent cell population expressing (C) FoxA2 (D) Brachyury and (E) SHH in monolayer and 3D-micromass condition of H9-ESCs notochordal differentiation. Gating was done using isotype controls as reference. All data are represented as mean ± SEM from two independent experiments.
Single cell transcriptomic analysis reveals the role of a transcription factor regulatory network for notochordal differentiation:
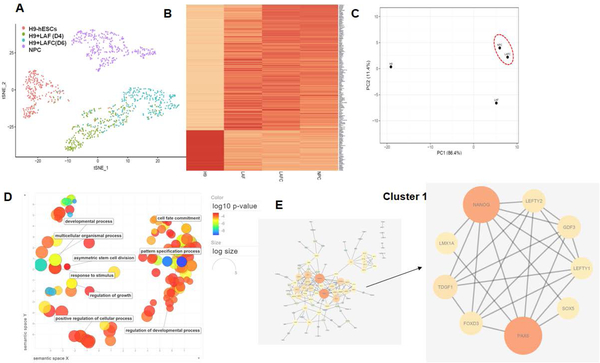
To determine the molecular signature and potential regulatory genes involved in notochordal differentiation, we performed pan-genomic high-throughput single cell RNA-sequencing over the course of H9-hESCs differentiation (day 0, 4 and 6) using a droplet based method. Human NP cells (NPC) were used as positive control for an efficient differentiation into notochordal cells. Transcriptomic profiling at the single cell level enabled us to decipher the molecular complexity and diversity during cell fate differentiation and provide a valuable resource to divulge the potential gene drivers of notochord differentiation. The cellular heterogeneity based on the transcriptomic profile at the single-cell level was performed by unsupervised analysis for cell clustering using a two dimensions t-distributed stochastic neighbor-embedding (t-SNE) method. Our t-SNE analysis of 250 cells on average, showed that day 0 and day 4hESCs were segregated into two distinct clusters and the day 6 differentiated hESCs cluster were close to the NPC cluster indicating the transcriptomic similarities between LAFC treated hESCs (day 6) and NPCs (Figure 7A). Further analyses of these data showed that 1,181 genes displayed significant differences in expression levels among four groups (FDR corrected P-value<0.05). Additionally, heatmap analysis showed the striking similarities between derived notochordal cells (LAFC, day 6) and human pulposus cells indicating that our two-step four factor based differentiation strategy resulted in successful differentiation of H9-hESCs in notochordal cells (Figure 7B).
Figure 7: Single cell transcriptomic analysis reveals the role of a transcription factor regulatory network during notochordal differentiation:
(A) tSNE plot of H9-hESCs on different days of notochordal differentiation on day 0 (control), day 4 (LAF), day 6 (LAFC). NP cells were also used for transcriptomic analysis, which showed that day 6 (LAFC treated) differentiation group clustered near NPC group indicating the global transcriptomic similarities between them. (B) Heatmap analysis using expression profile of 189 most differentially expressed genes between four groups showed similarity between differentiated H9-hESCs at day 6 and NPCs. (C) Principal component analysis (PCA) using expression profile of 189 genes among all experimental groups showed global transcriptional similarities between LAFC (day 6) and NPC group. (D) GO analysis (Biological process) in LAFC treated groups (day 6 of differentiation) as compared to undifferentiated H9-hESCs (control) showed enrichment of several biological processes involved in cell differentiation pathways. (E) Network analysis identified 2 different functional clusters among in PPI network constructed from differentially expressed genes in LAFC (day 6 of differentiation). MCODE analysis identified an enrichment of a functional cluster comprising of 8 genes including PAX6, GDF3, FOXD3, TDGF1, LMX1A, SOX5, LEFTY1 and LEFTY2 which is thought to be major driver of notochordal differentiation.
We next performed principal component analysis (PCA) using the top 189 most differentially expressed genes to further determine the transcriptomic similarities between the experimental groups. LAFC treated H9-hESC group (day 6 differentiation) clustered more closely to the NPC group, suggesting that the gene expression signature of derived notochordal cells were similar to NP cells (Figure 7C). These data further demonstrate the effective differentiation of hESCs into notochordal cells using our two-step differentiation strategy.
Further analysis showed the alterations in gene expression levels by treatment with the cocktail of four factors (LAFC) resulted in differential expression of 3,265 genes. Among these statistically significant genes, a total of 76 genes were upregulated (>10 fold, FDR P-value<0.01), whereas 69 genes were found to be significantly downregulated (>10 fold, FDR P-value<0.01). Table 1 depicts the list of the top 20 upregulated and downregulated genes in LAFC treated group as compared to untreated control group.
Table 1:
Top 20 most differentially expressed (up- and down-regulated) genes on day 6 (LAFC treated) of notochordal differentiation of H9-hESCs as compared to untreated parental control (day 0).
| SN | Gene | Mean Fold Change |
MeanPvalue | (Up/Downregulated |
|---|---|---|---|---|
| 1 | NR2F2 | 203.84 | 4.58E-77 | Upregulated in LAFC |
| 2 | DACH1 | 65.81 | 1.80E-22 | Upregulated in LAFC |
| 3 | DLK1 | 65.03 | 1.04E-230 | Upregulated in LAFC |
| 4 | LIX1 | 52.42 | 8.77E-59 | Upregulated in LAFC |
| 5 | IGFBP5 | 52.16 | 1.08E-49 | Upregulated in LAFC |
| 6 | LRP2 | 52.03 | 1.38E-49 | Upregulated in LAFC |
| 7 | DKK1 | 51.79 | 1.01E-43 | Upregulated in LAFC |
| 8 | PAX6 | 47.52 | 3.58E-65 | Upregulated in LAFC |
| 9 | WLS | 43.53 | 9.74E-71 | Upregulated in LAFC |
| 10 | SP5 | 43.07 | 7.41E-46 | Upregulated in LAFC |
| 11 | MYO3B | 42.74 | 5.31E-16 | Upregulated in LAFC |
| 12 | CNTN2 | 42.52 | 1.32E-91 | Upregulated in LAFC |
| 13 | ZNF503 | 41.44 | 5.98E-31 | Upregulated in LAFC |
| 14 | SOX1 | 38.76 | 1.76E-20 | Upregulated in LAFC |
| 15 | LMX1A | 37.67 | 1.53E-14 | Upregulated in LAFC |
| 16 | SCUBE2 | 27.90 | 8.79E-12 | Upregulated in LAFC |
| 17 | MEIS2 | 27.26 | 6.25E-11 | Upregulated in LAFC |
| 18 | PRTG | 27.12 | 2.65E-216 | Upregulated in LAFC |
| 19 | ILDR2 | 25.22 | 5.03E-11 | Upregulated in LAFC |
| 20 | POU3F2 | 25.04 | 4.77E-14 | Upregulated in LAFC |
| 21 | LINC00678 | 318.23 | 6.98E-107 | Downregulated in LAFC |
| 22 | MT2A | 147.65 | 7.99E-90 | Downregulated in LAFC |
| 23 | MT1G | 132.56 | 3.95E-48 | Downregulated in LAFC |
| 24 | MT1H | 94.17 | 6.23E-36 | Downregulated in LAFC |
| 25 | MT1E | 78.92 | 3.08E-43 | Downregulated in LAFC |
| 26 | NTS | 57.22 | 1.56E-31 | Downregulated in LAFC |
| 27 | LEFTY1 | 47.76 | 1.87E-21 | Downregulated in LAFC |
| 28 | TDGF1 | 44.30 | 1.22E-114 | Downregulated in LAFC |
| 29 | FOXD3.AS1 | 43.36 | 2.73E-66 | Downregulated in LAFC |
| 30 | LINC00458 | 41.53 | 1.53E-20 | Downregulated in LAFC |
| 31 | C9orf135 | 40.50 | 8.15E-33 | Downregulated in LAFC |
| 32 | CXCL5 | 34.55 | 4.98E-32 | Downregulated in LAFC |
| 33 | TMEM30B | 32.20 | 1.11E-16 | Downregulated in LAFC |
| 34 | NANOG | 30.64 | 4.16E-17 | Downregulated in LAFC |
| 35 | LOC101929194 | 29.66 | 2.63E-25 | Downregulated in LAFC |
| 36 | MT1F | 29.05 | 5.83E-16 | Downregulated in LAFC |
| 37 | D21S2088E | 28.16 | 6.74E-27 | Downregulated in LAFC |
| 38 | CHGA | 24.97 | 4.50E-60 | Downregulated in LAFC |
| 39 | LCK | 24.13 | 8.02E-55 | Downregulated in LAFC |
| 40 | NPFFR2 | 21.65 | 2.01E-13 | Downregulated in LAFC |
Gene ontology (GO) enrichment analysis was performed to relate these differentially expressed genes to biological processes and molecular function. Our analysis demonstrates the significant enrichment of several biological processes involved in cell differentiation, pattern specification processes, system development, regionalization, embryonic morphogenesis and cell fate commitment (Figure 7D). Additionally, GO analysis based on molecular function further identified molecular pathways related to Wnt-protein binding, signaling receptor binding, DNA-binding transcription repressor activity, and RNA polymerase II-specific transcriptional activity, suggesting the enrichment of pathways involving recruitment of transcription factors and further regulation of downstream genes involved in cell differentiation.
Further, we also performed functional cluster analysis using datasets of differentially expressed genes and identified several networks shown to possess relevant biological functions (Figure 7E). MCODE cluster analysis in protein-protein interaction (PPI) network identified an enrichment of a functional cluster comprising of 8 genes including PAX6, GDF3, FOXD3, TDGF1, LMX1A, SOX5, LEFTY1 and LEFTY2 (Figure 7E). These results provide candidate genes which might be closely associated with notochordal differentiation. Furthermore, these eight genes showed a high level of interaction among themselves (n=24), suggesting a possible physiological role during the differentiation process (Figure 7E). Interestingly, all of these genes code for transcription factors and thus form a regulatory network consisting of transcription factors that are known to be involved in cell fate and differentiation pathways. Our single cell transcriptomic data provide further support of our two-step differentiation strategy for efficient differentiation of hESC into notochordal cells. Transcriptomic profiling during various stages of cell differentiation revealed molecular diversity of notochord differentiation and identified potential genes and regulatory molecules controlling differentiation into notochordal cells.
Discussion:
Progressive loss of NP cellularity and inability of NP cells to maintain the normal homeostatic structure of the disc tissue is a characteristic of IDD (Aguiar, Johnson, & Oegema, 1999; Le Maitre, Pockert, Buttle, Freemont, & Hoyland, 2007). Cell-based regenerative therapies are an emerging, yet promising strategy for restoration of the degenerated disc. Generating NP cells or its precursor notochordal cells from pluripotent stem cells holds the potential to provide a constant cell source for future clinical applications to facilitate the long-term reconstitution of healthy disc tissue. However, the method to generate NP cells from pluripotent stem cells is in its infancy and available methods possess several limitations including reduced yield of differentiated cells, inferior ability to produce native-like NP cells and vulnerability to the challenging microenvironment of intervertebral discs (Acosta et al., 2005; Gorensek et al., 2004). These studies were undertaken to derive a simple, effective, and robust protocol to generate notochordal cells from human embryonic stem cells. In the present study, we established a two-step differentiation strategy using a cocktail of four factors consisting of small molecules and a growth factor to efficiently generate notochordal cells from hESCs over a differentiation period of six days.
Using state-of-the-art CRISPR/Cas9 based genome-editing, we constructed a fluorescent reporter vector for monitoring the notochordal differentiation. To construct this reporter, we selected the NOTO gene for genetic manipulation because of its selective expression during early stages of notochord development (Alten et al., 2012). An earlier study in human induced pluripotent stem cells (hiPSCs) designed a GFP-reporter construct in the promoter region of Brachyury gene (T) using lentivirus mediated transduction to monitor the cell differentiation into NP cell lineage (Tang et al., 2018). However, weak basal fluorescence of GFP, low transduction efficiency associated with lentiviral approach and ambiguity in considering the Brachyury as a specific notochordal and NP cell marker constitute technical limitations of this approach. Thus, in the present study, the red fluorescent reporter vector was generated by inserting mCherry into exon 3 of the NOTO gene in hESCs. The 2A self-cleaving peptide system was used to link the mCherry reporter with native open reading frame of the NOTO gene because it does not affect the endogenous function of NOTO gene and allows co-expression of NOTO and mCherry (Hsiao et al., 2008). Our well characterized Noto-2A-mCherry reporter in H9-hESC lines serve as a useful resource for notochordal differentiation in vitro which allow us to precisely determine the efficacy of our established differentiation strategy for successful differentiation of hESCs into notochordal/NP cells.
With NOTO-2A-mCherry as a selection marker and reporter for notochordal cells derivation, we have undertaken efforts to generate notochordal cells from parental hESCs. We established a simple method to generate Noto+ cells in a very reproducible and relatively quick fashion. A previous study demonstrated the combinatorial effect of FGF, AGN and BMP inhibition for the generation of NOTO positive cells, whereas a recent report by Tang et al showed that combination of BMP4, FGF2, WNT3a, Activin are needed to promote the induction of NOTO expression (Winzi, Hyttel, Dale, & Serup, 2011; Tang et al., 2018). Based on these published reports and our experience with stem cell differentiation (Gibson et al., 2017; Guzzo, Scanlon, Sanjay, Xu, & Drissi, 2014), our current method used bFGF in combination with two small molecules-LDN193189 and AGN193109 for a period of four days in parental hESC lines (denoted as LAF treatment). LDN193189 is a BMP signaling inhibitor that targets BMP type 1receptors ALK2 and ALK3. AGN193109 functions as a pan Retinoic Acid Receptor antagonist (RARα, β, and γ). Under these treatment conditions, patches of PSCs grow and a slight activation of mCherry was observed. After four days of differentiation, we added another small molecule called CHIR99021 to the previous cocktail of factors (denoted as LAFC treatment), which is a known GSK3β inhibitor and commonly used to enhance canonical Wnt signaling. We obtained an increase in the fluorescence of NOTO-mCherry reporter expression which is consistent with mRNA expression of NOTO, a direct target gene of the Wnt signaling pathway. Our detailed characterization studies suggest that bFGF and LDN193189 are critical for the generation of notochordal precursors and CHIR99021 treatment augments canonical Wnt signaling to induce NOTO expression. Additionally, our differentiation strategy further envisioned a method to temporally and spatially improve the frequency of notochordal cells by utilizing 3D high density culture conditions. High density micromass culture allow a convenient in vitro model to study cell differentiation which mimics the in vivo conditions of mesenchymal cells condensation observed in notochordal cell development. Our compelling data suggest that using differentiation strategy under micromass conditions instead of monolayer, greatly impact the efficiency of notochordal differentiation with 54% cell population showing positive expression for Brachyury.
Additionally, our study is the first to provide a pan transcriptomic landscape of notochortdal cells to understand the molecular signature and genetic determinants of notochordal differentiation. We utilized single cell RNA-sequencing instead of bulk RNA-sequencing as it reveals cellular heterogeneity which is masked by bulk RNA-sequencing. Our transcriptomic analysis revealed gene regulatory network which were significantly upregulated during the course of differentiation. Further, our comparative transcriptomic profiling using human NP cells, demonstrated that the genomic signature of derived notochordal cells using our established method (LAFC, day 6) share many similarities with authentic adult NP cells, indicating the success of our developed differentiation protocol. Moreover, our transcriptomic data reveal genome wide similarities between differentiated notochordal cells and human NP cells when cultured in monolayer. However, one would expect that these similarities would be even greater if cells are cultured in high density. This will be the object of our future studies focused on the influence of high density culture on driving terminal maturation of notochordal cellsindicating our two-step differentiation strategy under monolayer culture condition did not completely transform hESCs into notochordal cells. Furthermore, transcription factor (TF) regulatory network showed the enrichment of several TFs including PAX6, GDF3, FOXD3, TDGF1, LMX1A, SOX5, LEFTY1 and LEFTY2 which are considered potential drivers of notochordal differentiation. Earlier studies suggested that PAX6 controls progenitor cell identity and neuronal fate in response to graded SHH signaling (Ericson et al., 1997). Further Foxd3 has been shown to be an essential nodal-dependent regulator of mesoderm lineage in Zebrafish and GDF3 is considered as a BMP inhibitor and thus regulates cell fate in stem cells and early embryos (Chang & Kessler, 2010; Levine & Brivanlou, 2006). Finally, Sox5 is required for notochordal ECM formation, notochord cell survival, and development of the NP cell of intervertebral discs (Smits & Lefebvre, 2003). Further studies are needed to define the role these transcription factors in the differentiation of notochordal cells.
In summary, our study shows that notochordal cells can be efficiently derived from human embryonic stem cells through a simple, two-step differentiation protocol using a cocktail of four factors. The generated notochordal cells are a novel resource for the development of cell-based therapies for IDD and have the potential to have immense clinical utility.
Supplementary Material
Acknowledgements:
This work was supported by the National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS) grant (R21-AR-067903 to HD) and funds from Emory University School of Medicine.
Footnotes
Data Availability Statement:
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Conflict of Interest:
Authors do not have any conflict of interest.
References:
- Acosta FL Jr., Lotz J, & Ames CP (2005). The potential role of mesenchymal stem cell therapy for intervertebral disc degeneration: a critical overview. Neurosurg Focus, 19(3), E4. [DOI] [PubMed] [Google Scholar]
- Aguiar DJ, Johnson SL, & Oegema TR (1999). Notochordal cells interact with nucleus pulposus cells: regulation of proteoglycan synthesis. Exp Cell Res, 246(1), 129–137. [DOI] [PubMed] [Google Scholar]
- Akhtar N, Khan NM, Ashruf OS, & Haqqi TM (2017). Inhibition of cartilage degradation and suppression of PGE2 and MMPs expression by pomegranate fruit extract in a model of posttraumatic osteoarthritis. Nutrition, 33, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alini M, Roughley PJ, Antoniou J, Stoll T, & Aebi M. (2002). A biological approach to treating disc degeneration: not for today, but maybe for tomorrow. Eur Spine J, 11 Suppl 2(2), S215–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckwalter JA (1995). Aging and degeneration of the human intervertebral disc. Spine (Phila Pa 1976), 20(11), 1307–1314. [DOI] [PubMed] [Google Scholar]
- Cadigan KM, & Waterman ML (2012). TCF/LEFs and Wnt signaling in the nucleus. Cold Spring Harb Perspect Biol, 4(11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camus A, & Tam PP (1999). The organizer of the gastrulating mouse embryo. Curr Top Dev Biol, 45, 117–153. [DOI] [PubMed] [Google Scholar]
- Chan SC, & Gantenbein-Ritter B. (2012). Intervertebral disc regeneration or repair with biomaterials and stem cell therapy--feasible or fiction? Swiss Med Wkly, 142(142), w13598. [DOI] [PubMed] [Google Scholar]
- Chang LL, & Kessler DS (2010). Foxd3 is an essential Nodal-dependent regulator of zebrafish dorsal mesoderm development. Dev Biol, 342(1), 39–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Lee EJ, Jing L, Christoforou N, Leong KW, & Setton LA (2013). Differentiation of mouse induced pluripotent stem cells (iPSCs) into nucleus pulposus-like cells in vitro. PLoS One, 8(9), e75548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chon BH, Lee EJ, Jing L, Setton LA, & Chen J. (2013). Human umbilical cord mesenchymal stromal cells exhibit immature nucleus pulposus cell phenotype in a laminin-rich pseudo-three-dimensional culture system. Stem Cell Res Ther, 4(5), 120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly DJ, Patel K, & Cooke J. (1997). Chick noggin is expressed in the organizer and neural plate during axial development, but offers no evidence of involvement in primary axis formation. Int J Dev Biol, 41(2), 389–396. [PubMed] [Google Scholar]
- DL S. (2005). - Structure and function of the notochord: an essential organ for chordate. Development, 132(11), 2503–2512. [DOI] [PubMed] [Google Scholar]
- Drissi H, Gibson JD, Guzzo RM, & Xu RH (2015). Derivation and Chondrogenic Commitment of Human Embryonic Stem Cell-Derived Mesenchymal Progenitors. Methods Mol Biol, 1340, 65–78. [DOI] [PubMed] [Google Scholar]
- Eden E, Navon R, Steinfeld I, Lipson D, & Yakhini Z. (2009). GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics, 10(48), 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ericson J, Rashbass P, Schedl A, Brenner-Morton S, Kawakami A, van Heyningen V, … Briscoe J. (1997). Pax6 controls progenitor cell identity and neuronal fate in response to graded Shh signaling. Cell, 90(1), 169–180. [DOI] [PubMed] [Google Scholar]
- Errico TJ (2005). Lumbar disc arthroplasty. Clin Orthop Relat Res, 435(435), 106–117. [DOI] [PubMed] [Google Scholar]
- Gorensek M, Jaksimovic C, Kregar-Velikonja N, Knezevic M, Jeras M, Pavlovcic V, & Cor A. (2004). Nucleus pulposus repair with cultured autologous elastic cartilage derived chondrocytes. Cell Mol Biol Lett, 9(2), 363–373. [PubMed] [Google Scholar]
- Hanley EN Jr., Herkowitz HN, Kirkpatrick JS, Wang JC, Chen MN, & Kang JD (2010). Debating the value of spine surgery. J Bone Joint Surg Am, 92(5), 1293–1304. [DOI] [PubMed] [Google Scholar]
- Haseeb A, Khan NM, Ashruf OS, & Haqqi TM (2017). A Polyphenol-rich Pomegranate Fruit Extract Suppresses NF-kappaB and IL-6 Expression by Blocking the Activation of IKKbeta and NIK in Primary Human Chondrocytes. Phytother Res, 31(5), 778–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haseeb A, Makki MS, Khan NM, Ahmad I, & Haqqi TM (2017). Deep sequencing and analyses of miRNAs, isomiRs and miRNA induced silencing complex (miRISC)-associated miRNome in primary human chondrocytes. Sci Rep, 7(1), 15178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao EC, Yoshinaga Y, Nguyen TD, Musone SL, Kim JE, Swinton P, … Conklin BR (2008). Marking embryonic stem cells with a 2A self-cleaving peptide: a NKX2–5 emerald GFP BAC reporter. PLoS One, 3(7), e2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz JN (2006). Lumbar disc disorders and low-back pain: socioeconomic factors and consequences. J Bone Joint Surg Am, 88 Suppl 2, 21–24. [DOI] [PubMed] [Google Scholar]
- Khan NM, Ahmad I, Ansari MY, & Haqqi TM (2017). Wogonin, a natural flavonoid, intercalates with genomic DNA and exhibits protective effects in IL-1beta stimulated osteoarthritis chondrocytes. Chem Biol Interact, 274, 13–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan NM, Ahmad I, & Haqqi TM (2018). Nrf2/ARE pathway attenuates oxidative and apoptotic response in human osteoarthritis chondrocytes by activating ERK1/2/ELK1-P70S6K-P90RSK signaling axis. Free Radic Biol Med, 116, 159–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan NM, Ansari MY, & Haqqi TM (2017). Sucrose, But Not Glucose, Blocks IL1-beta-Induced Inflammatory Response in Human Chondrocytes by Inducing Autophagy via AKT/mTOR Pathway. J Cell Biochem, 118(3), 629–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan NM, Haseeb A, Ansari MY, Devarapalli P, Haynie S, & Haqqi TM (2017). Wogonin, a plant derived small molecule, exerts potent anti-inflammatory and chondroprotective effects through the activation of ROS/ERK/Nrf2 signaling pathways in human Osteoarthritis chondrocytes. Free Radic Biol Med, 106, 288–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan NM, Haseeb A, Ansari MY, & Haqqi TM (2017). A wogonin-rich-fraction of Scutellaria baicalensis root extract exerts chondroprotective effects by suppressing IL-1beta-induced activation of AP-1 in human OA chondrocytes. Sci Rep, 7(43789), 43789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan NM, & Poduval TB (2011). Immunomodulatory and immunotoxic effects of bilirubin: molecular mechanisms. J Leukoc Biol, 90(5), 997–1015. [DOI] [PubMed] [Google Scholar]
- Khan NM, & Poduval TB (2012). Bilirubin augments radiation injury and leads to increased infection and mortality in mice: molecular mechanisms. Free Radic Biol Med, 53(5), 1152–1169. [DOI] [PubMed] [Google Scholar]
- Khan NM, Sandur SK, Checker R, Sharma D, Poduval TB, & Sainis KB (2011). Pro-oxidants ameliorate radiation-induced apoptosis through activation of the calcium-ERK1/2-Nrf2 pathway. Free Radic Biol Med, 51(1), 115–128. [DOI] [PubMed] [Google Scholar]
- Le Maitre CL, Pockert A, Buttle DJ, Freemont AJ, & Hoyland JA (2007). Matrix synthesis and degradation in human intervertebral disc degeneration. Biochem Soc Trans, 35(Pt 4), 652–655. [DOI] [PubMed] [Google Scholar]
- Leung VY, Chan D, & Cheung KM (2006). Regeneration of intervertebral disc by mesenchymal stem cells: potentials, limitations, and future direction. Eur Spine J, 15 Suppl 3(3), S406–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine AJ, & Brivanlou AH (2006). GDF3, a BMP inhibitor, regulates cell fate in stem cells and early embryos. Development, 133(2), 209–216. [DOI] [PubMed] [Google Scholar]
- Liu Y, Fu S, Rahaman MN, Mao JJ, & Bal BS (2015). Native nucleus pulposus tissue matrix promotes notochordal differentiation of human induced pluripotent stem cells with potential for treating intervertebral disc degeneration. J Biomed Mater Res A, 103(3), 1053–1059. [DOI] [PubMed] [Google Scholar]
- Liu Y, Rahaman MN, & Bal BS (2014). Modulating notochordal differentiation of human induced pluripotent stem cells using natural nucleus pulposus tissue matrix. PLoS One, 9(7), e100885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCann MR, Bacher CA, & Seguin CA (2011). Exploiting notochord cells for stem cell-based regeneration of the intervertebral disc. J Cell Commun Signal, 5(1), 39–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metsalu T, & Vilo J. (2015). ClustVis: a web tool for visualizing clustering of multivariate data using Principal Component Analysis and heatmap. Nucleic Acids Res, 43(W1), W566–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirza SK, & Deyo RA (2007). Systematic review of randomized trials comparing lumbar fusion surgery to nonoperative care for treatment of chronic back pain. Spine (Phila Pa 1976), 32(7), 816–823. [DOI] [PubMed] [Google Scholar]
- N, K. V., J, U., M, F., C, N.-W., D, K., U, P., … S, R. (2014). - Cell sources for nucleus pulposus regeneration. Eur Spine J, 23(3), 013–3106. [DOI] [PubMed] [Google Scholar]
- Otasek D, Morris JH, Boucas J, Pico AR, & Demchak B. (2019). Cytoscape Automation: empowering workflow-based network analysis. Genome Biol, 20(1), 185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risbud MV, Schaer TP, & Shapiro IM (2010). Toward an understanding of the role of notochordal cells in the adult intervertebral disc: from discord to accord. Dev Dyn, 239(8), 2141–2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, & Smyth GK (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics, 26(1), 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai D. (2008). Future perspectives of cell-based therapy for intervertebral disc disease. Eur Spine J, 17 Suppl 4, 452–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satija R, Farrell JA, Gennert D, Schier AF, & Regev A. (2015). Spatial reconstruction of single-cell gene expression data. Nat Biotechnol, 33(5), 495–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, … Ideker T. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res, 13(11), 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits P, & Lefebvre V. (2003). Sox5 and Sox6 are required for notochord extracellular matrix sheath formation, notochord cell survival and development of the nucleus pulposus of intervertebral discs. Development, 130(6), 1135–1148. [DOI] [PubMed] [Google Scholar]
- St Sauver JL, Warner DO, Yawn BP, Jacobson DJ, McGree ME, Pankratz JJ, … Rocca WA (2013). Why patients visit their doctors: assessing the most prevalent conditions in a defined American population. Mayo Clin Proc, 88(1), 56–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streit A, & Stern CD (1999). Establishment and maintenance of the border of the neural plate in the chick: involvement of FGF and BMP activity. Mech Dev, 82(1–2), 51–66. [DOI] [PubMed] [Google Scholar]
- Supek F, Bosnjak M, Skunca N, & Smuc T. (2011). REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One, 6(7), e21800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, … von Mering C. (2017). The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res, 45(D1), D362–D368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- T, R., SV, N., GG, T., E, S., M, S., M, L., … RS, T. (2017). - Update on the Notochord Including its Embryology, Molecular Development, and. Cureus, 9(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang R, Jing L, Willard VP, Wu CL, Guilak F, Chen J, & Setton LA (2018). Differentiation of human induced pluripotent stem cells into nucleus pulposus-like cells. Stem Cell Res Ther, 9(1), 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ukita K, Hirahara S, Oshima N, Imuta Y, Yoshimoto A, Jang CW, … Sasaki H. (2009). Wnt signaling maintains the notochord fate for progenitor cells and supports the posterior extension of the notochord. Mech Dev, 126(10), 791–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban JP, & Roberts S. (2003). Degeneration of the intervertebral disc. Arthritis Res Ther, 5(3), 120–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei S, & Wang Q. (2018). Molecular regulation of Nodal signaling during mesendoderm formation. Acta Biochim Biophys Sin (Shanghai), 50(1), 74–81. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.