Abstract
Purpose
To assess testicular mRNA and protein expression levels of MRE11 and RAD50 in human azoospermia patients.
Methods
Patients diagnosed with maturation arrest at the spermatocyte stage (MA) and Sertoli cell-only syndrome (SCOS) were recruited through diagnostic testicular biopsy. Patients with normal spermatogenesis were studied as controls. In addition, knockdown of MRE11 and RAD50 was performed in GC-2spd(ts) cells to investigate their roles in cellular proliferation and apoptosis.
Results
mRNA and protein expression levels of MRE11 and RAD50 were measured using quantitative polymerase chain reaction, western blotting, and immunohistochemistry, respectively. Knockdown of both MRE11 and RAD50 utilized transfection with small interfering RNAs.
Conclusion
Our findings demonstrated altered expression levels of MRE11 and RAD50 in human testes with MA and SCOS, and showed that these alterations might be associated with impaired spermatogenesis. These results offer valuable new perspectives into the molecular mechanisms of male infertility.
Electronic supplementary material
The online version of this article (10.1007/s10815-019-01686-5) contains supplementary material, which is available to authorized users.
Keywords: Testes, Spermatogenic failure, MRE11, RAD50, GC-2spd(ts) cells
Introduction
Infertility affects approximately 15% of reproductive age couples and male factor infertility accounts for approximately half of all cases of infertility [1]. Azoospermia, defined as the complete absence of spermatozoa in the ejaculate, is identified in 15–20% of male factor infertility and can be classified as obstructive azoospermia (OA) and nonobstructive azoospermia (NOA) [2]. NOA includes maturation arrest (MA) at the spermatocyte stage and Sertoli cell-only syndrome (SCOS), two of the most severe forms of male factor infertility related to spermatogenic failure.
Previous work has identified hundreds of gene defects associated with abnormal spermatogenesis and these findings improved our understanding of the molecular mechanisms of spermatogenesis and possible pathologies of male infertility using multiple gene knockout mouse models [3]. However, the process of human male germ cell formation is somewhat different from that in mice. Thus, findings in mice are not substantially applicable to human [4]. Current research has demonstrated that genetic defects including numerical and structural chromosomal abnormalities (such as Y-chromosomal microdeletions and gene mutations) can result in human azoospermia [5]. However, the genetic causes and related molecular mechanisms in the majority of human cases remain unclear [6].
Precisely controlled by a highly organized cell machinery involving several thousand different genes, spermatogenesis encompasses three major stages: proliferation and differentiation of spermatogonia, meiotic division of spermatocytes, and formation of mature spermatozoa [7]. Meiotic division is a crucial stage in spermatogenesis during which diploid spermatocytes generate haploid spermatids. This process is initiated by the formation of DNA double-strand breaks (DSBs) at preferred sites [8–10]. Some DSBs are repaired as crossovers while the majority is repaired by homologous recombination [11]. The DSB repair process requires perfect coordination of certain proteins including SPO11, ATM, MLH1, RPA, RAD51, RAD18, MRE11, and RAD50 [12, 13]. Studies in animal models have demonstrated that defects in this process, such as deficient SPO11, ATM, MLH1, and RAD51, can cause aneuploidy or meiotic arrest of male germ cells and ultimately lead to male factor infertility [14–17].
The MRE11 complex, which is formed of two highly conserved subunit proteins (MRE11 and RAD50) and a third less conserved subunit protein (NBS1 in mammals and XRS2 in yeast), plays an essential role in break sensing, checkpoint activation, repair of DSBs [18, 19] and cellular proliferation, and apoptosis [20, 21]. The MRE11 protein has endonuclease and exonuclease functions to bind intrinsic DNA and the RAD50 protein has coiled-coil domains to hold the end of DNA breaks when binding with MRE11 [22, 23]. Germline mutations in MRE11 or RAD50 can cause ataxia telangiectasia-like disorder (ATLD) or Nijmegen breakage syndrome (NBS)-like disorder (NBSLD) [24, 25]. Studies have reported that mice carrying hypomorphic mutations in Mre11 and Rad50 (Mre11ATLD1/ATLD1 and Rad50S/S mice, respectively) exhibit impaired meiotic phenotypes of DSB repair, crossover formation, and male germ cell generation. This indicates that MRE11 and RAD50 play significant roles in spermatogenesis, especially in DSB repair during spermatocyte meiosis [26, 27]. However, little is known about the roles and functions of MRE11 and RAD50 in human spermatogenesis.
Interestingly, a previous study reported down-regulated mRNA expression levels of MRE11 and RAD50 in MA compared with OA patients using gene microarray technology, suggesting that these alterations may correlate with human spermatogenesis [28]. Therefore, the aim of this study was to evaluate the expression and localization of MRE11 and RAD50 in testicular tissue samples of azoospermia patients. Furthermore, GC-2spd(ts) cells were used to investigate the underlying molecular mechanisms of MRE11 and RAD50 in spermatogenesis.
Materials and methods
Patients and samples
This study was approved by the Medical Ethics Committee of Women’s Hospital, School of Medicine, Zhejiang University. All participants provided written informed consent prior to treatment. Azoospermia patients were recruited between May 2012 and April 2014. Patients with numerical chromosomal abnormality, Y-chromosomal microdeletion, hypogonadotropic hypogonadism, and those undergoing hormone treatments were excluded from the study. Testicular tissue samples were obtained from patients who underwent diagnostic testicular biopsy and stored at − 80 °C immediately. Histological examination was performed using standard procedures: fixed in Bouin’s solution (Sigma-Aldrich, St. Louis, MO, USA), embedded in paraffin, stained with hematoxylin (Sigma-Aldrich), and eosin (Sigma-Aldrich) and thereafter processed for histopathology by at least two pathologists. Diagnosis was carried out through a qualitative and quantitative evaluation of germinal epithelium in 25 seminiferous tubules and a calculation of the modified Johnsen score. According to the Johnsen score, testicular tissue samples were classified as OA with normal spermatogenesis, maturation arrest at the spermatocyte stage (MA), and Sertoli cell-only syndrome (SCOS). Patients diagnosed as OA with normal spermatogenesis (Johnsen score ≥ 9) were studied as normal controls.
Samples from patients recruited from May 2012 to April 2014 that contained both testicular tissue and paraffin-embedded sections were classified as first part samples. These were divided into three groups: OA (normal spermatogenesis, n = 20), MA (n = 20), and SCOS n = 20). In order to expand the total sample size to make clear the significance of the study, another 20 patients of each group recruited from 2017 January to 2018 January containing only paraffin-embedded sections obtained from hospital pathology department were used as second part samples.
GC-2spd(ts) cell culture
The mouse spermatocyte cell line, GC-2spd(ts), was purchased from the Conservation Genetics Chinese Academy of Sciences Kunming Cell Bank. This cell line was established from mouse pachytene spermatocytes and has been used as one of the most helpful models for studying regulatory mechanisms in male germ cells [29, 30]. GC-2spd(ts) cells were cultured in Dulbecco’s modified Eagle’s medium (Gibco, Gaithersburg, MD, USA) containing 10% fetal bovine serum (Gibco) at 37 °C with 5% CO2.
Quantitative real-time polymerase chain reaction
Total RNA of testicular tissue from the first part samples was extracted with the RNAiso Plus Kit (TaKaRa, Kusatsu, Japan) and reverse transcribed with the PrimeScript™ RT reagent Kit with gDNA Eraser (TaKaRa). RNA concentrations were measured at 260 nm, and purity was confirmed in A260/A280 ratio. qRT-PCR was then performed using the SYBR Premix Ex Taq (TaKaRa) on an ABI 7900 thermocycler according to the manufacturer’s protocol (Applied Biosystems, Foster City, CA, USA) as described previously [31]. Three replicate PCR experiments were conducted. The housekeeping gene, glyceraldehyde 3-phosphate dehydrogenase (GAPDH), served as the internal control for analyzing relative gene expression levels. The 2−ΔΔCt method was used to evaluate gene expression fold changes among the groups. All primers were synthesized by the TSINGKE Biological Technology (Beijing, China). The primer sequences are listed in Supplemental Table 1.
Western blot analysis
Total protein was extracted from testicular tissue of the first part samples and GC-2spd(ts) cells, according to the instructions of the Total Protein Extraction Kit for Animal Cultured Cells and Tissue (Invent Biotechnologies, Plymouth, MN, USA). For testicular tissue, 200 μL denaturing cell lysis buffer with 1% cocktail (Selleck, Houston, TX, USA) was added into the filter and grinded for 50–60 times, then incubated on ice for 5 min, centrifuged at 14,000 rpm for 2 min and the supernatant was collected. The protein concentration was measured using a bicinchoninic acid protein assay kit (Beyotime, Haimen, China) as described by the manufacturer. Equivalent amounts of proteins (20 μg) were separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and then transferred to polyvinylidene difluoride membrane (Millipore, Burlington, MA, USA). The membranes were blocked with 5% bovine serum albumin (Sigma-Aldrich) for 2 h at room temperature and incubated with primary antibodies against MRE11 (Atlas, Bromma, Sweden), RAD50 (Abcam, Cambridge, UK), and GAPDH (Abcam) at 4 °C overnight. The membranes were subsequently incubated with horseradish peroxidase (HRP)-conjugated secondary antibodies (Cell Signaling Technology, Danvers, MA, USA) for 1 h at room temperature. An enhanced chemiluminescence substrate (Thermo Fisher Scientific, Waltham, MA, USA) was used to visualize the results using an ImageQuant LAS 4000 mini (GE Healthcare, Pittsburgh, PA, USA).
Immunohistochemical analysis
Formaldehyde-fixed paraffin-embedded sections of testicular tissues of all the samples were cut into 5-mm sections and mounted onto slides. After quenching the endogenous peroxidase activity, the slides were incubated with blocking serum (Abcam) and then with primary antibodies against MRE11 and RAD50 at 4 °C overnight. The slides were then incubated with HRP-conjugated secondary antibodies and photographed with an Olympus BX20 microscope (Tokyo, Japan). To confirm the specificity of the primary antibodies, negative controls were performed in an identical way by replacing the primary antibodies with phosphate buffer saline (PBS) or normal IgG antibody (Abcam).
Small interfering RNA (siRNA) transfection of GC-2spd(ts) cells
GC-2spd(ts) cells were transfected with siRNA targeting mouse MRE11 or RAD50, or a non-targeting control siRNA, for 48–72 h using Lipofectamine™ RNAiMax transfection reagent (Thermo Fisher Scientific) according to the manufacturer’s protocol. Cells were divided into four groups: blank (untreated), control (transfected with non-targeting control siRNA), and MRE11 and RAD50 siRNAs. Knockdown of the target proteins was confirmed through western blot analysis. GAPDH was used as the loading control. siRNA was synthesized using RiboBio Co, Ltd. (Guangzhou, China). The siRNAs sequences were: MRE11 (sense: 5′-GCUUGGAGCUGCUUAGGAA dTdT-3′; antisense: 3′- dTdT CGAACCUCGACGAAUCCUU-5′) and RAD50 (sense: 5′-GGUAAUCACUCACGAUGAA dTdT-3′; antisense: 3′-dTdT CCAUUAGUGAGUGCUACUU-5′).
Cell proliferation analysis
GC-2spd(ts) cells were seeded and cultured in triplicate for each group at 3 × 103 cells/well in 96-well plates and reached 60–70% confluence after 24 h. Cell proliferation was determined using the Cell Counting Kit-8 (CCK-8) assay (Dojindo Molecular Technologies, Shanghai, China) according to the manufacturer’s protocol at 24, 48, and 72 h after siRNA transfection. In brief, 10 μL of the CCK-8 solution were added into each well and the absorbance was measured at 450 nm after 2 h of incubation at 37 °C using a spectrophotometer reader (Thermo Fisher Scientific). Cell proliferation was calculated using the optical density (OD) values.
Cell apoptosis analysis
Apoptosis was evaluated using an Annexin V-APC/7-AAD apoptosis kit (MultiSciences, Hangzhou, China) following the manufacturer’s protocol. Briefly, GC-2spd(ts) cells were seeded in triplicate for each group at 1 × 105 cells/well in 6-well plates, transfected as previously mentioned for 72 h and harvested by accutase (Thermo Fisher Scientific). Cells were collected and resuspended in 500 μL binding buffer. Then 5 μL of Annexin V-APC and 10 μL of 7-AAD were added and incubated at room temperature for 15 min in the dark. Apoptotic percentages were determined through flow cytometry using a FACSCalibur system (BD Biosciences, Franklin Lakes, NJ, USA).
Apoptosis was also analyzed using a one-step terminal deoxynucleotidyl transferase dUTP nick-end labeling (TUNEL) assay kit (Beyotime) according to the manufacturer’s protocol. After transfection for 72 h, cell slices were fixed with 4% paraformaldehyde at room temperature for 30 min. After permeabilization in 0.3% Triton X-100 for 5 min, the cell slices were incubated with TUNEL working solution at 37 °C for 1 h in the dark. Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI). The fluoresceine isothiocyanate labeled TUNEL-positive cells in five areas of the same size were selected randomly for each slice and examined using a laser confocal microscope (Olympus) by using 488 nm excitation. Cells with green fluorescence were considered as apoptotic.
Statistical analysis
Data are expressed as the means ± standard deviation (SD). Statistical comparisons were performed by ANOVA with Tukey’s HSD tests for post-hoc analysis using SPSS 19.0 software (IBM, Chicago, IL, USA). All in vitro experiments comprised three replicates and were performed at least twice independently. P < 0.05 was considered to indicate a statistically significant difference.
Results
Expression and localization of MRE11 and RAD50 in the testes of azoospermia patients
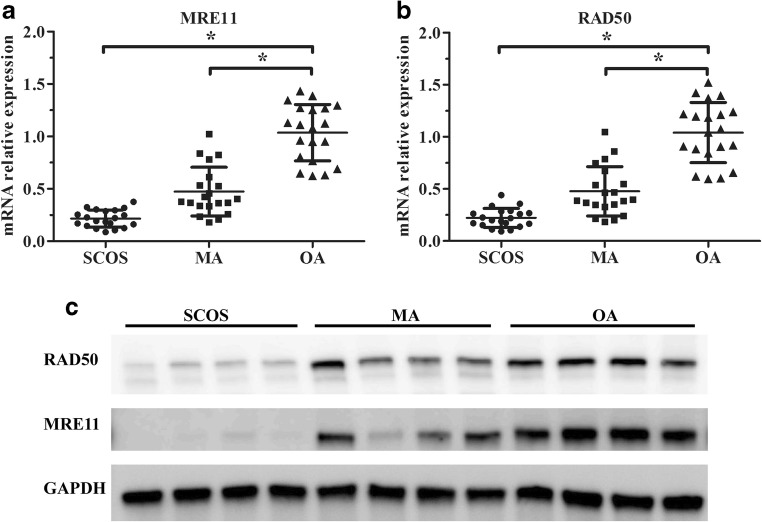
Expression levels of MRE11 and RAD50 mRNAs in testicular tissue samples from the three groups were quantified by qRT-PCR using the first part samples. The MA and SCOS groups demonstrated significantly decreased MRE11 and RAD50 mRNA expression compared with the OA group (Fig. 1a, b) using the first part samples. Western blot results of the first part samples showed that the expression level of MRE11 protein was significantly lower in the majority (12/20) of the MA and all of the SCOS samples compared with the OA samples. And in the SCOS and MA patient samples with low level of MRE11, interestingly, the expression level of RAD50 protein demonstrated decreased level corresponding to MRE11 at the same time when compared with the OA samples (Fig.1c).
Fig. 1.
mRNA and protein expression of MRE11 and RAD50 in human testicular tissue samples. Analysis of a MRE11 and b RAD50 mRNA expression by qRT-PCR in testicular tissue samples of SCOS, MA, and OA patients. Relative expression levels were determined by normalization to GAPDH. Each individual data point is shown along with horizontal and vertical bars representing the means ± SD. *P < 0.05. c Representative western blots of the MRE11 and RAD50 proteins in testes of SCOS, MA, and OA patients. GAPDH is the loading control.
We also examined the expression and localization of MRE11 and RAD50 proteins in testicular tissue sections of both the first and second part tissue samples using immunohistochemistry. In a representative OA testicular tissue sample, both MRE11 and RAD50 were stained predominantly in the nucleus, and were expressed at their highest levels in spermatogonia and spermatocytes, much weaker in round spermatids and Sertoli cells and absent in elongated spermatids and mature spermatozoa (Fig. 2a, b). Weak staining of MRE11 and RAD50 were found in Sertoli cells of SCOS samples (Fig. 2c, d). Simultaneous absent or weakly detectable staining of MRE11 and RAD50 were demonstrated within spermatogonia and spermatocytes in the majority of the MA samples (12 out of 20 in first part, and 13 out of 20 in second part samples) (Fig. 2g–j). The other MA testicular tissue samples showed normal staining of MRE11 and RAD50 (Fig. 2e, f). No signals were observed when the testicular tissue samples were incubated with PBS (Fig. 2k) or normal IgG antibody (Fig. 2l).
Fig. 2.
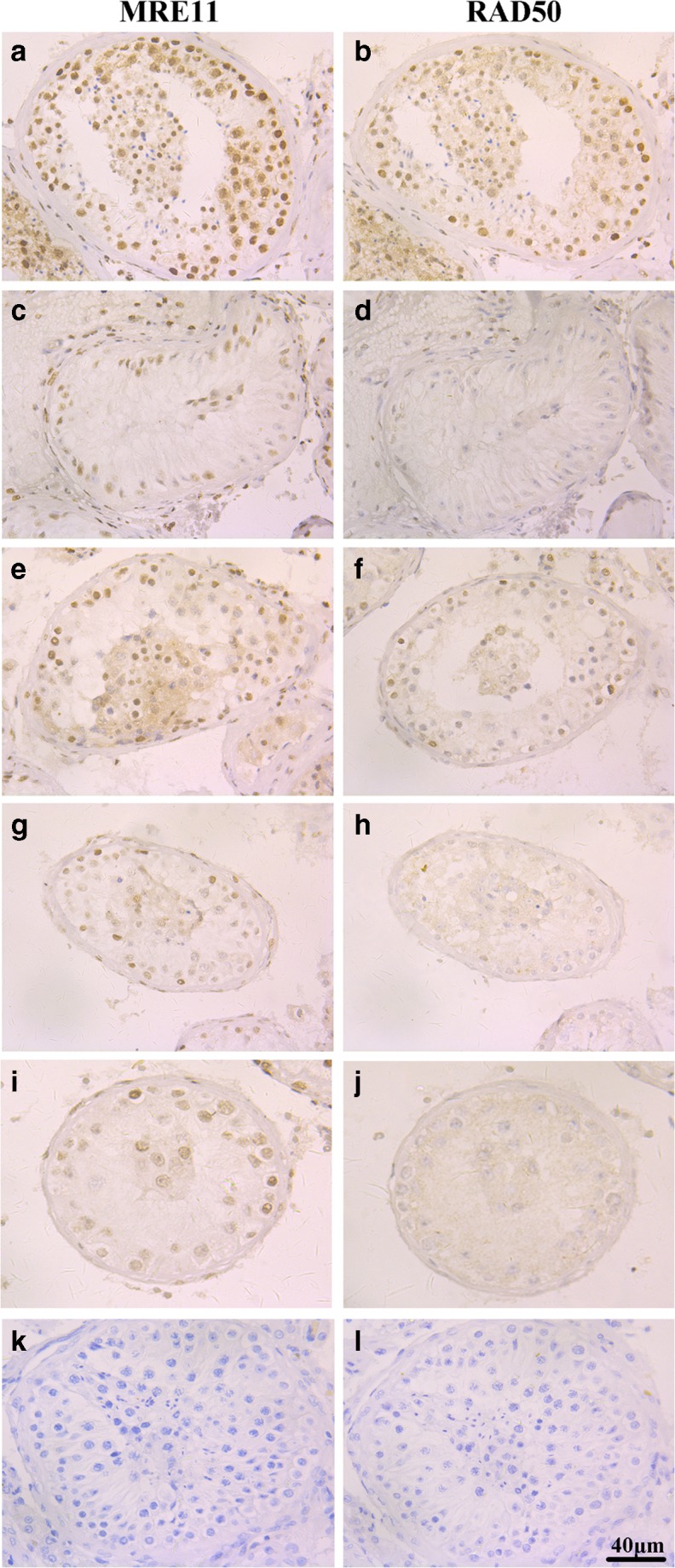
Immunohistochemistry analysis of MRE11 and RAD50 in human testicular tissue samples. MRE11 and RAD50 immunohistochemistry staining of representative testicular tissue sections from a, b a representative OA patient with normal spermatogenesis, c, d a representative patient with SCOS, e, f a representative patient with MA with normal levels of staining, and g–j two representative patients with MA with absent or weak staining of MRE11 and RAD50. Negative controls were incubated with PBS (K) and normal IgG antibody (L). The photos are at × 400 magnification and the scale bars represent 40 μm.
Down-regulation of MRE11 and RAD50 inhibited proliferation of GC-2spd(ts) cells
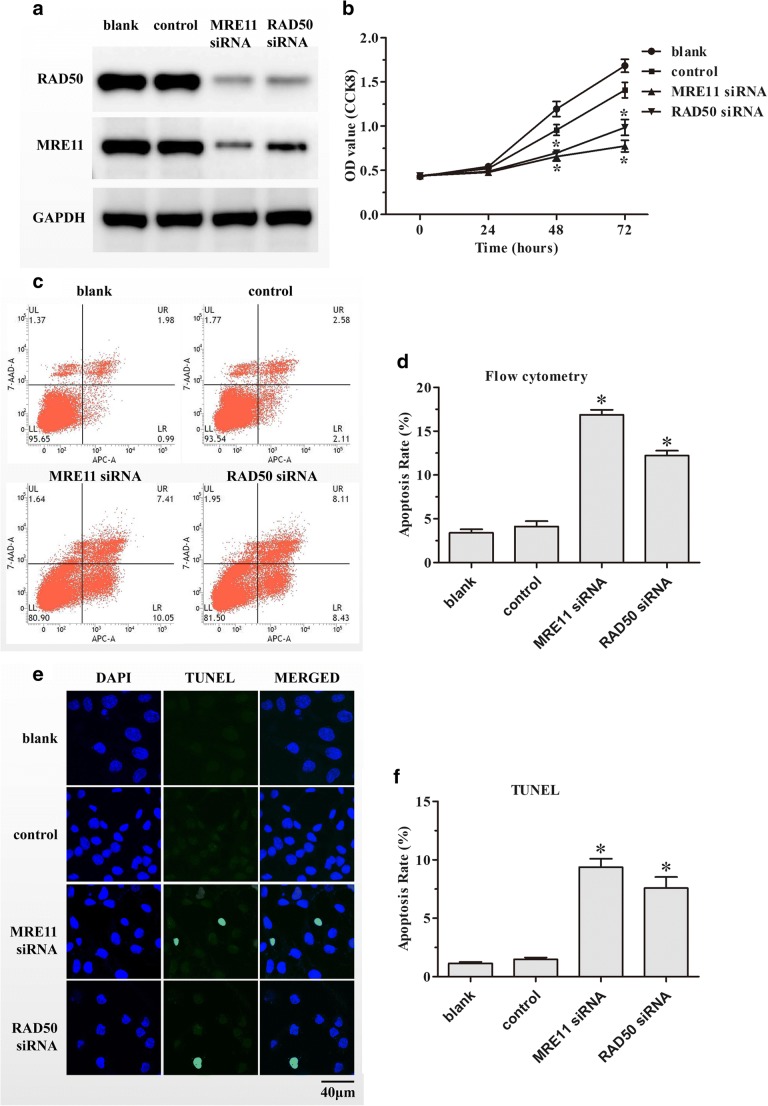
GC-2spd(ts) cell line is an immortalized mouse pachytene spermatocyte cell line and is one of the most useful models to study the regulatory mechanisms of male germ cells. To explore whether deficient expressions of MRE11 and RAD50 have functional effects on male germ cell proliferation, MRE11 and RAD50 protein levels were down-regulated in GC-2spd(ts) cells through transfection with siRNA. The efficiency of MRE11 and RAD50 interference was assessed using western blot analysis. As presented in Fig. 3a, the expression of MRE11 was significantly decreased in cells transfected with MRE11 siRNA and, simultaneously, showed a reduction of RAD50 compared with the NC or blank groups. Similarly, transfection with RAD50 siRNA significantly decreased the levels of both MRE11 and RAD50 in comparison with the NC or blank groups.
Fig. 3.
Effect of down-regulating MRE11 and RAD50 on cell proliferation and apoptosis of GC-2spd(ts) cells. a Down-regulation of MRE11 and RAD50 by transfecting GC-2spd(ts) cells with siRNA was analyzed through western blotting using GAPDH as the loading control. b Proliferation of GC-2spd(ts) cells after transfection with MRE11 or RAD50 siRNA was measured by the CCK8 assay. Apoptosis of GC-2spd(ts) cells after siRNA transfection for 72 h was analyzed by c flow cytometry and d TUNEL assays. The scale bars represent 40 μm. Each assay was performed in triplicate. Data are presented as means ± SD. *P < 0.05.
The CCK8 assay was conducted to evaluate the effects of MRE11 and RAD50 knockdown by siRNA on the proliferation of GC-2spd(ts) cells. Both the MRE11 and RAD50 siRNA groups exhibited significantly decreased proliferation 48 and 72 h after transfection compared with the NC or blank groups (Fig. 3b).
Down-regulation of MRE11 and RAD50 promoted apoptosis of GC-2spd(ts) cells
Flow cytometry was performed to examine whether knockdown of MRE11 and RAD50 induced apoptosis of GC-2spd(ts) cells. Both the MRE11 and RAD50 siRNA groups exhibited significantly increased percentages of apoptotic cells compared with the NC or blank groups (Fig. 3c, d). In addition, TUNEL staining was performed to confirm the apoptotic effect of knocking down MRE11 and RAD50 in GC-2spd(ts) cells. Our findings demonstrated that the numbers of TUNEL-positive cells increased significantly in both the MRE11 and RAD50 siRNA groups compared with the NC or blank groups (Fig. 3e, f).
Discussion
Mre11ATLD1/ATLD1 male mice exhibit a disturbed proportion of pachytene spermatocytes, increased synaptic defects, altered crossover formations, and impaired DSB repair [26]. Rad50S/S male mice display attrition of spermatogenic cells and increased germ cell apoptosis [27]. These findings indicated that both MRE11 and RAD50 perform essential roles in spermatogenesis. However, the pathophysiological functions of MRE11 and RAD50 in human testicular tissues with normal and abnormal spermatogenesis have never been explored. Combined with human testes microarray data [28], our results confirm deficient expression levels of MRE11 and RAD50 in patients with spermatogenic failure and suggest that these two proteins may play significant roles in human spermatogenesis.
In this study, we demonstrated that the expression levels of both MRE11 and RAD50 mRNAs and proteins were decreased in testicular tissue samples from patients with spermatogenic failure compared with the OA group. However, these results were acquired by using entire testicular tissue samples from patients with normal and abnormal spermatogenesis. Due to the highly complex mixture of various types of male germ cells, alterations of the expression of specific genes in azoospermia patients with spermatogenic failure may pinpoint the precise pathogenic molecular event responsible for the failure, or may show that it is simply a result of the absence of some specific germ cell types. IHC of testicular tissue sample was performed in order to compare the expression level of MRE11 and RAD50 in the same cell type to explore whether these alterations were resulted from the deletion of specific germ cell types in different groups. IHC results demonstrated that OA group samples presented strong staining while MA group samples with decreased expression level presented weak or negative staining of MRE11 and RAD50 in their common cell types, i.e., spermatogonia and spermatocytes. And late stage spermatogenic cells such as round spermatids, elongated spermatids, and mature spermatozoa presented weak or negative staining of MRE11 and RAD50. SCOS group samples presented weak staining in Sertoli cells. Considering that MA testicular tissues mainly consist of spermatogonia and spermatocytes, therefore, the decreased expression level of MRE11 and RAD50 in MA group compared with OA group mainly resulted from the lower expression of MRE11 and RAD50 in spermatogonia and spermatocytes of MA specimens, rather than a result of missing cell types. These findings demonstrated that deficient expression levels of MRE11 and RAD50 in spermatogenic failure may represent a true pathogenic molecular factor rather than merely a result of missing cells. And those patients with normal expression levels of MRE11 and RAD50 may have altered spermatogenesis triggered by other factors including dysfunction of other genes [32–39], epigenetic changes [40, 41], and environmental aspects [42].
A complex network of signaling pathways that affect the renewal, proliferation, apoptosis, and differentiation of male germ cells cooperate to determine cell fate during spermatogenesis [43]. Our in vitro experiments showed that knockdown of either MRE11 or RAD50 resulted in decreased expression levels of both proteins in GC-2spd(ts) cells, which was consistent with the simultaneous reduction of MRE11 and RAD50 in testes from humans with spermatogenic failure. This suggests that the intracellular stability of these two proteins is involved in specific protein–protein interactions. Previous studies have reported similar results of the effect of individual subunit on the stability of the MRE11/RAD50 complex [44, 45].
Our cell experimental data demonstrated that down-regulation of MRE11 and RAD50 inhibited proliferation and promoted apoptosis of GC-2spd(ts) cells. Meanwhile, increased apoptosis of germ cells was also observed in human MA and SCOS patients compared with OA individuals [35, 46]. These results are in accordance with the phenotypes of a disturbed proportion of pachytene spermatocytes and impaired DSB repair in Mre11ATLD1/ATLD1 male mice and increased germ cell apoptosis in Rad50S/S male mice. This indicates that MRE11 and RAD50 may play crucial roles in DSB repair, proliferation, differentiation, and apoptosis of male germ cells. Consequently, based on our findings and previous data from transgenic mice, deficient expression levels of MRE11 and RAD50 may be beneficial to predict sperm production or testicular histopathology, especially in nonobstructive azoospermia patients.
Abnormal expression of some genes, such as cell cycle regulatory genes (i.e., CDC25A and CCNA1 [32, 33]), apoptotic pathway genes (i.e., survivin, BAD, and BAX [34, 35]), RNA-binding genes (i.e., DAZ, DAZL,BOULE, and SAM68 [36, 37]), and hormone synthesis pathway genes (i.e., CYP1A1 and DAX-1A [38, 39]) correlate with spermatogenic failure in humans. Combined with our findings, this information contributes to knowledge regarding human male factor infertility involving spermatogenic failure and will be of great value to identify new potential targets to supplement testicular histological diagnosis and treat male factor infertility.
One limitation of this study is that testicular tissue samples from obstructive azoospermia patients having normal spermatogenesis were used as control group since normal testicular samples could be obtained for ethical reasons, which could lead to minor variations. In addition, specific DNA sequencing or whole genome sequencing of patients with spermatogenic failure will be investigated in subsequent study to explore the genetic causes of azoospermia. Moreover, findings in mouse GC-2spd(ts) cells might be different from human male germ cells and the functions of MRE11 and RAD50 in human testicular tissues still requires further researches.
In summary, the present study is the first to reveal decreased testicular expression levels of MRE11 and RAD50 in human patients with spermatogenic failure compared with those with normal spermatogenesis. In vitro GC-2spd(ts) cell experiments further demonstrated that decreased expression levels of MRE11 and RAD50 inhibited proliferation and promoted apoptosis of male germ cells. Altered testicular expression levels of MRE11 and RAD50 may lead to dysregulation of male germ cell proliferation and apoptosis and eventually impair spermatogenesis. We consider that deficient expression levels of MRE11 and RAD50 could serve as novel molecular markers of spermatogenic failure. Importantly, these results will facilitate our understanding of the mechanisms underlying male factor infertility and assist patients with their counseling, diagnosis, management, and treatment, especially for NOA patients.
Electronic supplementary material
(DOC 30 kb)
Acknowledgments
The authors thank all of the members of our laboratory for their support and valuable suggestions.
Funding information
This study was funded by the National Natural Science Foundation of China (No. 81601336, 81571500, 81771652, and 81801475), the National Key R&D Program of China (No. 2018YFC1004900), the Natural Science Foundation of Zhejiang Province of China (No. LQ15H040005, LQ17H040001, LGD19H040001, and LY17H040006), and the Medical Scientific Research Foundation of Zhejiang Province (No. 2015KYB200).
Compliance with ethical standards
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the Medical Ethics Committee of Women’s Hospital, School of Medicine, Zhejiang University and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Minhao Hu and Lejun Li contributed equally to this work.
References
- 1.Boivin J, Bunting L, Collins JA, Nygren KG. International estimates of infertility prevalence and treatment-seeking: potential need and demand for infertility medical care. Hum Reprod. 2007;22:1506–1512. doi: 10.1093/humrep/dem046. [DOI] [PubMed] [Google Scholar]
- 2.Massart A, Lissens W, Tournaye H, Stouffs K. Genetic causes of spermatogenic failure. Asian J Androl. 2012;14:40–48. doi: 10.1038/aja.2011.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Matzuk MM, Lamb DJ. The biology of infertility: research advances and clinical challenges. Nat Med. 2008;14:1197–1213. doi: 10.1038/nm.f.1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhu Z, Li C, Yang S, Tian R, Wang J, Yuan Q, Dong H, He Z, Wang S, Li Z. Dynamics of the transcriptome during human spermatogenesis: predicting the potential key genes regulating male gametes generation. Sci Rep. 2016;6:19069. doi: 10.1038/srep19069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wong WY, Thomas CM, Merkus JM, Zielhuis GA, Steegers-Theunissen RP. Male factor subfertility: possible causes and the impact of nutritional factors. Fertil Steril. 2000;73:435–442. doi: 10.1016/s0015-0282(99)00551-8. [DOI] [PubMed] [Google Scholar]
- 6.Cannarella R, Condorelli RA, Duca Y, La Vignera S, Calogero AE. New insights into the genetics of spermatogenic failure: a review of the literature. Hum Genet. 2019;138:125–140. doi: 10.1007/s00439-019-01974-1. [DOI] [PubMed] [Google Scholar]
- 7.Schultz N, Hamra FK, Garbers DL. A multitude of genes expressed solely in meiotic or postmeiotic spermatogenic cells offers a myriad of contraceptive targets. Proc Natl Acad Sci U S A. 2003;100:12201–12206. doi: 10.1073/pnas.1635054100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Massy B. Initiation of meiotic recombination: how and where? Conservation and specificities among eukaryotes. Annu Rev Genet. 2013;47:563–599. doi: 10.1146/annurev-genet-110711-155423. [DOI] [PubMed] [Google Scholar]
- 9.Keeney S, Lange J, Mohibullah N. Self-organization of meiotic recombination initiation: general principles and molecular pathways. Annu Rev Genet. 2014;48:187–214. doi: 10.1146/annurev-genet-120213-092304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lange J, Yamada S, Tischfield SE, Pan J, Kim S, Zhu X, Socci ND, Jasin M, Keeney S. The landscape of mouse meiotic double-strand break formation, processing, and repair. Cell. 2016;167:695–708. doi: 10.1016/j.cell.2016.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zakharyevich K, Tang S, Ma Y, Hunter N. Delineation of joint molecule resolution pathways in meiosis identifies a crossover-specific resolvase. Cell. 2012;149:334–347. doi: 10.1016/j.cell.2012.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Handel MA, Schimenti JC. Genetics of mammalian meiosis: regulation, dynamics and impact on fertility. Nat Rev Genet. 2010;11:124–136. doi: 10.1038/nrg2723. [DOI] [PubMed] [Google Scholar]
- 13.Gunes S, Al-Sadaan M, Agarwal A. Spermatogenesis, DNA damage and DNA repair mechanisms in male infertility. Reprod BioMed Online. 2015;31:309–319. doi: 10.1016/j.rbmo.2015.06.010. [DOI] [PubMed] [Google Scholar]
- 14.Baker SM, Plug AW, Prolla TA, Bronner CE, Harris AC, Yao X, Christie DM, Monell C, Arnheim N, Bradley A, Ashley T, Liskay RM. Involvement of mouse Mlh1 in DNA mismatch repair and meiotic crossing over. Nat Genet. 1996;13:336–342. doi: 10.1038/ng0796-336. [DOI] [PubMed] [Google Scholar]
- 15.Barlow C, Liyanage M, Moens PB, Tarsounas M, Nagashima K, Brown K, Rottinghaus S, Jackson SP, Tagle D, Ried T, Wynshaw-Boris A. Atm deficiency results in severe meiotic disruption as early as leptonema of prophase I. Development. 1998;125:4007–4017. doi: 10.1242/dev.125.20.4007. [DOI] [PubMed] [Google Scholar]
- 16.Romanienko PJ, Camerini-Otero RD. The mouse Spo11 gene is required for meiotic chromosome synapsis. Mol Cell. 2000;6:975–987. doi: 10.1016/s1097-2765(00)00097-6. [DOI] [PubMed] [Google Scholar]
- 17.Kuznetsov S, Pellegrini M, Shuda K, Fernandez-Capetillo O, Liu Y, Martin BK, Burkett S, Southon E, Pati D, Tessarollo L, West SC, Donovan PJ, Nussenzweig A, Sharan SK. RAD51C deficiency in mice results in early prophase I arrest in males and sister chromatid separation at metaphase II in females. J Cell Biol. 2007;176:581–592. doi: 10.1083/jcb.200608130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stracker TH, Theunissen JW, Morales M, Petrini JH. The Mre11 complex and the metabolism of chromosome breaks: the importance of communicating and holding things together. DNA Repair (Amst) 2004;3:845–854. doi: 10.1016/j.dnarep.2004.03.014. [DOI] [PubMed] [Google Scholar]
- 19.Borde V. The multiple roles of the Mre11 complex for meiotic recombination. Chromosom Res. 2007;15:551–563. doi: 10.1007/s10577-007-1147-9. [DOI] [PubMed] [Google Scholar]
- 20.Rupnik A, Lowndes NF, Grenon M. MRN and the race to the break. Chromosoma. 2010;119:115–135. doi: 10.1007/s00412-009-0242-4. [DOI] [PubMed] [Google Scholar]
- 21.Lavin MF, Kozlov S, Gatei M, Kijas AW. ATM-dependent phosphorylation of all three members of the MRN complex: from sensor to adaptor. Biomolecules. 2015;5:2877–2902. doi: 10.3390/biom5042877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de Jager M, Dronkert ML, Modesti M, Beerens CE, Kanaar R, van Gent DC. DNA-binding and strand-annealing activities of human Mre11: implications for its roles in DNA double-strand break repair pathways. Nucleic Acids Res. 2001;29:1317–1325. doi: 10.1093/nar/29.6.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hopfner KP, Karcher A, Craig L, Woo TT, Carney JP, Tainer JA. Structural biochemistry and interaction architecture of the DNA double-strand break repair Mre11 nuclease and Rad50-ATPase. Cell. 2001;105:473–485. doi: 10.1016/s0092-8674(01)00335-x. [DOI] [PubMed] [Google Scholar]
- 24.Stewart GS, Maser RS, Stankovic T, Bressan DA, Kaplan MI, Jaspers NG, Raams A, Byrd PJ, Petrini JH, Taylor AM. The DNA double-strand break repair gene hMRE11 is mutated in individuals with an ataxia-telangiectasia-like disorder. Cell. 1999;99:577–587. doi: 10.1016/s0092-8674(00)81547-0. [DOI] [PubMed] [Google Scholar]
- 25.Waltes R, Kalb R, Gatei M, Kijas AW, Stumm M, Sobeck A, Wieland B, Varon R, Lerenthal Y, Lavin MF, Schindler D, Dork T. Human RAD50 deficiency in a Nijmegen breakage syndrome-like disorder. Am J Hum Genet. 2009;84:605–616. doi: 10.1016/j.ajhg.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cherry SM, Adelman CA, Theunissen JW, Hassold TJ, Hunt PA, Petrini JH. The Mre11 complex influences DNA repair, synapsis, and crossing over in murine meiosis. Curr Biol. 2007;17:373–378. doi: 10.1016/j.cub.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bender CF, Sikes ML, Sullivan R, Huye LE, Le Beau MM, Roth DB, Mirzoeva OK, Oltz EM, Petrini JH. Cancer predisposition and hematopoietic failure in Rad50(S/S) mice. Genes Dev. 2002;16:2237–2251. doi: 10.1101/gad.1007902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bonache S, Algaba F, Franco E, Bassas L, Larriba S. Altered gene expression signature of early stages of the germ line supports the pre-meiotic origin of human spermatogenic failure. Andrology. 2014;2:596–606. doi: 10.1111/j.2047-2927.2014.00217.x. [DOI] [PubMed] [Google Scholar]
- 29.Hofmann MC, Hess RA, Goldberg E, Millan JL. Immortalized germ cells undergo meiosis in vitro. Proc Natl Acad Sci U S A. 1994;91:5533–5537. doi: 10.1073/pnas.91.12.5533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang H, Wen L, Yuan Q, Sun M, Niu M, He Z. Establishment and applications of male germ cell and Sertoli cell lines. Reproduction. 2016;152:R31–R40. doi: 10.1530/REP-15-0546. [DOI] [PubMed] [Google Scholar]
- 31.Le F, Wang LY, Wang N, Li L, Li LJ, Zheng YM, Lou HY, Liu XZ, Xu XR, Sheng JZ, Huang HF, Jin F. In vitro fertilization alters growth and expression of Igf2/H19 and their epigenetic mechanisms in the liver and skeletal muscle of newborn and elder mice. Biol Reprod. 2013;88:75. doi: 10.1095/biolreprod.112.106070. [DOI] [PubMed] [Google Scholar]
- 32.Cheng YS, Kuo PL, Teng YN, Kuo TY, Chung CL, Lin YH, Liao RW, Lin JS, Lin YM. Association of spermatogenic failure with decreased CDC25A expression in infertile men. Hum Reprod. 2006;21:2346–2352. doi: 10.1093/humrep/del163. [DOI] [PubMed] [Google Scholar]
- 33.Schrader M, Muller-Tidow C, Ravnik S, Muller M, Schulze W, Diederichs S, Serve H, Miller K. Cyclin A1 and gametogenesis in fertile and infertile patients: a potential new molecular diagnostic marker. Hum Reprod. 2002;17:2338–2343. doi: 10.1093/humrep/17.9.2338. [DOI] [PubMed] [Google Scholar]
- 34.Weikert S, Schrader M, Muller M, Schulze W, Krause H, Miller K. Expression levels of the inhibitor of apoptosis survivin in testes of patients with normal spermatogenesis and spermatogenic failure. Fertil Steril. 2005;83(Suppl 1):1100–1105. doi: 10.1016/j.fertnstert.2004.12.010. [DOI] [PubMed] [Google Scholar]
- 35.Jaiswal D, Trivedi S, Agrawal NK, Singh K. Dysregulation of apoptotic pathway candidate genes and proteins in infertile azoospermia patients. Fertil Steril. 2015;104:736–743. doi: 10.1016/j.fertnstert.2015.05.029. [DOI] [PubMed] [Google Scholar]
- 36.Kuo PL, Wang ST, Lin YM, Lin YH, Teng YN, Hsu CC. Expression profiles of the DAZ gene family in human testis with and without spermatogenic failure. Fertil Steril. 2004;81:1034–1040. doi: 10.1016/j.fertnstert.2003.08.047. [DOI] [PubMed] [Google Scholar]
- 37.Li LJ, Zhang FB, Liu SY, Tian YH, Le F, Lou HY, Huang HF, Jin F. Decreased expression of SAM68 in human testes with spermatogenic defects. Fertil Steril. 2014;102:61–67. doi: 10.1016/j.fertnstert.2014.03.036. [DOI] [PubMed] [Google Scholar]
- 38.Lardone MC, Parada-Bustamante A, Ebensperger M, Valdevenito R, Kakarieka E, Martinez D, Pommer R, Piottante A, Castro A. DAX-1 and DAX-1A expression in human testicular tissues with primary spermatogenic failure. Mol Hum Reprod. 2011;17:739–746. doi: 10.1093/molehr/gar051. [DOI] [PubMed] [Google Scholar]
- 39.Parada-Bustamante A, Molina C, Valencia C, Florez M, Lardone MC, Argandona F, Piottante A, Ebensperguer M, Orihuela PA, Castro A. Disturbed testicular expression of the estrogen-metabolizing enzymes CYP1A1 and COMT in infertile men with primary spermatogenic failure: possible negative implications on Sertoli cells. Andrology. 2017;5:486–494. doi: 10.1111/andr.12346. [DOI] [PubMed] [Google Scholar]
- 40.Ferfouri F, Boitrelle F, Ghout I, Albert M, Molina GD, Wainer R, Bailly M, Selva J, Vialard F. A genome-wide DNA methylation study in azoospermia. Andrology. 2013;1:815–821. doi: 10.1111/j.2047-2927.2013.00117.x. [DOI] [PubMed] [Google Scholar]
- 41.Ramasamy R, Ridgeway A, Lipshultz LI, Lamb DJ. Integrative DNA methylation and gene expression analysis identifies discoidin domain receptor 1 association with idiopathic nonobstructive azoospermia. Fertil Steril. 2014;102:968–973. doi: 10.1016/j.fertnstert.2014.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gabrielsen JS, Tanrikut C. Chronic exposures and male fertility: the impacts of environment, diet, and drug use on spermatogenesis. Andrology. 2016;4:648–661. doi: 10.1111/andr.12198. [DOI] [PubMed] [Google Scholar]
- 43.Neto FT, Bach PV, Najari BB, Li PS, Goldstein M. Spermatogenesis in humans and its affecting factors. Semin Cell Dev Biol. 2016;59:10–26. doi: 10.1016/j.semcdb.2016.04.009. [DOI] [PubMed] [Google Scholar]
- 44.Zhong ZH, Jiang WQ, Cesare AJ, Neumann AA, Wadhwa R, Reddel RR. Disruption of telomere maintenance by depletion of the MRE11/RAD50/NBS1 complex in cells that use alternative lengthening of telomeres. J Biol Chem. 2007;282:29314–29322. doi: 10.1074/jbc.M701413200. [DOI] [PubMed] [Google Scholar]
- 45.Kavitha CV, Choudhary B, Raghavan SC, Muniyappa K. Differential regulation of MRN (Mre11-Rad50-Nbs1) complex subunits and telomerase activity in cancer cells. Biochem Biophys Res Commun. 2010;399:575–580. doi: 10.1016/j.bbrc.2010.07.117. [DOI] [PubMed] [Google Scholar]
- 46.Kim SK, Yoon YD, Park YS, Seo JT, Kim JH. Involvement of the Fas-Fas ligand system and active caspase-3 in abnormal apoptosis in human testes with maturation arrest and Sertoli cell-only syndrome. Fertil Steril. 2007;87:547–553. doi: 10.1016/j.fertnstert.2006.07.1524. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOC 30 kb)