Abstract
Accurate identification of compound–protein interactions (CPIs) in silico may deepen our understanding of the underlying mechanisms of drug action and thus remarkably facilitate drug discovery and development. Conventional similarity- or docking-based computational methods for predicting CPIs rarely exploit latent features from currently available large-scale unlabeled compound and protein data and often limit their usage to relatively small-scale datasets. In the present study, we propose DeepCPI, a novel general and scalable computational framework that combines effective feature embedding (a technique of representation learning) with powerful deep learning methods to accurately predict CPIs at a large scale. DeepCPI automatically learns the implicit yet expressive low-dimensional features of compounds and proteins from a massive amount of unlabeled data. Evaluations of the measured CPIs in large-scale databases, such as ChEMBL and BindingDB, as well as of the known drug–target interactions from DrugBank, demonstrated the superior predictive performance of DeepCPI. Furthermore, several interactions among small-molecule compounds and three G protein-coupled receptor targets (glucagon-like peptide-1 receptor, glucagon receptor, and vasoactive intestinal peptide receptor) predicted using DeepCPI were experimentally validated. The present study suggests that DeepCPI is a useful and powerful tool for drug discovery and repositioning. The source code of DeepCPI can be downloaded from https://github.com/FangpingWan/DeepCPI.
Keywords: Deep learning, Machine learning, Drug discovery, In silico drug screening, Compound–protein interaction prediction
Introduction
Identification of compound–protein interactions (CPIs; or drug–target interactions, DTIs) is crucial for drug discovery and development and provides valuable insights into the understanding of drug actions and off-target adverse events [1], [2]. Inspired by the concept of polypharmacology, i.e., a single drug may interact with multiple targets [3], drug developers are actively seeking novel ways to better characterize CPIs or identify novel uses of the existing drugs (i.e., drug repositioning or drug repurposing) [3], [4] to markedly reduce the time and cost required for drug development [5].
Numerous computational methods have been proposed to predict potential CPIs in silico to narrow the large search space of possible interacting compound–protein pairs and facilitate drug discovery and development [6], [7], [8], [9], [10], [11], [12]. Although successful results can be obtained using the existing prediction approaches, several challenges remain unaddressed. First, most of the conventional prediction methods only employ a simple and direct representation of features from the labeled data (e.g., established CPIs and available protein structure information) to assess similarities among compounds and proteins and infer unknown CPIs. For instance, a kernel describing the similarities among drug–protein interaction profiles [8] and the graph-based method SIMCOMP [13] were used to compare different drugs and compounds. In addition, the normalized Smith–Waterman score [9] is typically applied to assess the similarities among targets (proteins). Meanwhile, large amounts of available unlabeled data of compounds and proteins enable an implicit and useful representation of features that may effectively be used to define their similarities. Such an implicit representation of protein or compound features encoded by large-scale unlabeled data is not exploited well by most of the existing methods to predict new CPIs. Second, an increasing number of established DTIs or compound–protein-binding affinities (e.g., 1 million bioassays over 2 million compounds and 10,000 protein targets in PubChem [14]) raises serious scalability issues concerning the conventional prediction methods. For instance, many similarity-based methods [7], [9] require the computation of pairwise similarity scores between proteins, which is generally impractical in the setting of large-scale data. The aforementioned computational challenges highlight the need of more efficient schemes to accurately capture the hidden features of proteins and compounds from massive unlabeled data as well as the need of more advanced and scalable learning models that enable predictions from large-scale training datasets.
In machine learning communities, representation learning and deep learning (DL) are the two popular methods at present for efficiently extracting features and addressing the scalability issues in large-scale data analyses. Representation learning aims to automatically learn data representations (features) from relatively raw data that can be more effectively and easily exploited by the downstream machine learning models to improve the learning performance [15], [16]. Meanwhile, DL aims to extract high-level feature abstractions from input data, typically using several layers of non-linear transformations, and is a dominant method used in numerous complex learning tasks with large-scale samples in the data science field, such as computer vision, speech recognition, natural language processing (NLP), game playing, and bioinformatics [17], [18], [19]. Although several DL models have been used to address various learning problems in drug discovery [20], [21], [22], they rarely fully exploit the currently available large-scale protein and compound data to predict CPI. For example, the computational approaches proposed in the literature [20], [21] only use the hand-designed features of compounds and do not take into account the features of targets. Furthermore, these approaches generally fail to predict potential interacting compounds for a given novel target (i.e., without known interacting compounds in the training data); this type of prediction is generally more urgent than the prediction of novel compounds for targets with known interacting compounds. Although a new approach—AtomNet—has been developed [23] to overcome these limitations, it can only be used to predict interacting drug partners of targets with known structures, which is often not the case in the clinical practice. In addition, despite the promising predictive performance of conventional approaches reported on benchmark datasets [24], [25], [26], few efforts have been made to explore the extent to which these advanced learning techniques can promote the efficiency in the real drug discovery scenario.
In this article, we propose DeepCPI, a novel framework that combines unsupervised representation learning with powerful DL techniques for predicting structure-free CPIs. DeepCPI first uses the latent semantic analysis [27] and Word2vec [16], [28], [29] methods to learn the feature embeddings (i.e., low-dimensional feature representations) of compounds and proteins in an unsupervised manner from large compound and protein corpora, respectively. Subsequently, given a compound–protein pair, the feature embeddings of both compound and protein are fed into a multimodal deep neural network (DNN) classifier to predict their interaction probability. We tested DeepCPI on several benchmark datasets, including the large-scale compound–protein affinity databases (e.g., ChEMBL and BindingDB), as well as the known DTIs from DrugBank. Comparisons with several conventional methods demonstrated the superior performance of DeepCPI in numerous practical scenarios. Moreover, starting from the virtual screening initialized by DeepCPI, we identified several novel interactions of small-molecule compounds with various targets in the G protein-coupled receptor (GPCR) family, including glucagon-like peptide-1 receptor (GLP-1R), glucagon receptor (GCGR), and vasoactive intestinal peptide receptor (VIPR). Collectively, our computational test and laboratory experimentation results demonstrate that DeepCPI is a useful and powerful tool for the prediction of novel CPIs and can thus aid in drug discovery and repositioning endeavors.
Results
DeepCPI framework
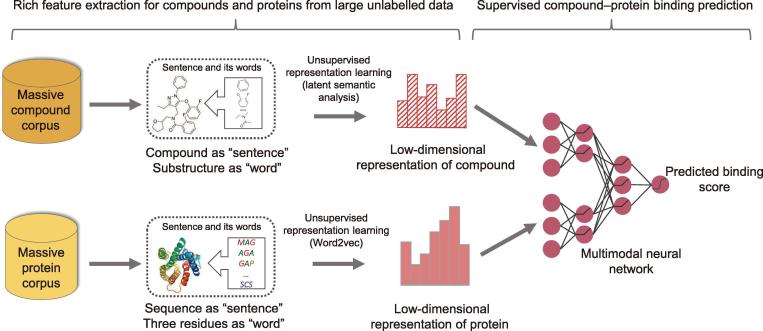
The DeepCPI framework comprises two main steps (Figure 1): (1) representation learning for both compounds and proteins and (2) predicting CPIs (or DTIs) through a multimodal DNN. More specifically, in the first step, we use several NLP techniques to extract the useful features of compounds and proteins from the corresponding large-scale unlabeled corpora (Figures S1 and S2; Materials and methods). Here, compounds and their basic structures are regarded as “documents” and “words”, respectively, whereas protein sequences and all possible three non-overlapping amino acid residues are regarded as “sentences” and “words,” respectively. Subsequently, the feature embedding techniques, including latent semantic analysis [27] and Word2vec [16], [28], are applied to automatically learn the implicit yet expressive low-dimensional representations (i.e., vectors) of compound and protein features from the corresponding large-scale unlabeled corpora. In the second step, the derived low-dimensional feature vectors of compounds and proteins are fed into a multimodal DNN classifier to make the predictions. Further details of the individual modules of DeepCPI, including the extraction of compound and protein features, DNN model, and implementation procedure, are described in Materials and methods.
Figure 1.
Schematic of the DeepCPI workflow First, motivated by the current NLP techniques, the unsupervised representation learning strategies (including latent semantic analysis and Word2vec) are used to obtain low-dimensional representations of compound and protein features from massive unlabeled data. Subsequently, these extracted low-dimensional feature representations of compounds and proteins are fed to a multimodal DNN to make the prediction. NLP, natural language processing; DNN, deep neural network.
Predictive performance evaluation
We mainly evaluated DeepCPI using compound–protein pairs extracted from the currently available databases, such as ChEMBL [30] and BindingDB [31]. We first used the bioactivity data retrieved from ChEMBL [30] to assess the predictive performance of DeepCPI. Specifically, the compound–protein pairs with half maximal inhibitory concentrations () or inhibition constants () were selected as positive examples, whereas pairs with or were used as negative examples. This data preprocessing step yielded 360,867 positive examples and 93,925 negative examples. To justify our criteria of selecting positive and negative examples, we mapped the known interacting drug–target pairs extracted from DrugBank [32] (released on November 11, 2015) to the corresponding compound–protein pairs in ChEMBL (Materials and methods). The binding affinities or potencies (measured by or ) of majority of the known interacting drug–target pairs were (>60% and 70% pairs for and , respectively) (Figure S3). Reportedly, is a widely-used and good indicator of strong binding affinities among compounds and proteins [33]. Therefore, we considered or as a reasonable criterion for selecting positive examples. There is no well-defined dichotomy between high and low binding affinities; thus, we used a threshold of (i.e., markedly higher than ) to select negative examples, which is consistent with the method reported elsewhere [23].
To evaluate the predictive performance of DeepCPI, we considered several challenging and realistic scenarios. A computational experiment was first conducted in which we randomly selected 20% pairs from ChEMBL as the training data and the remaining pairs as the test data. This scenario mimicked a practical situation in which CPIs are relatively sparsely labeled. ChEMBL may contain similar (redundant) proteins and compounds, which may lead to over-optimistic performance resulting from easy predictions. Therefore, we minimized this effect by only retaining proteins whose sequence identity scores were and compounds whose chemical structure similarity scores were (as computed based on the Jaccard similarity between their Morgan fingerprints). More specifically, for each group of proteins or compounds with sequence identity scores or chemical structure similarity scores , we only retained the protein or compound having the highest number of interactions and discarded the rest of the proteins or compounds in that group. The basic statistics of the ChEMBL and BindingDB datasets used in our performance evaluation are summarized in Tables S1 and S2, respectively.
Conventional cross validation, particularly leave-one-out cross validation (LOOCV), may not be an appropriate method to evaluate the performance of a CPI prediction algorithm, if the training data contain many compounds or proteins with only one interaction [34]. In such a case, training methods may learn to exploit the bias toward the proteins or compounds with a single interaction to boost the performance of LOOCV. Thus, separating the single interaction from other types of interactions during cross validation is essential [34]. Given a compound–protein interacting pair from a dataset, if the compound or protein only appeared in this interaction, we considered this pair as unique (Materials and methods). In this test scenario, we used non-unique examples as the training data and tested the predictive performance on unique pairs.
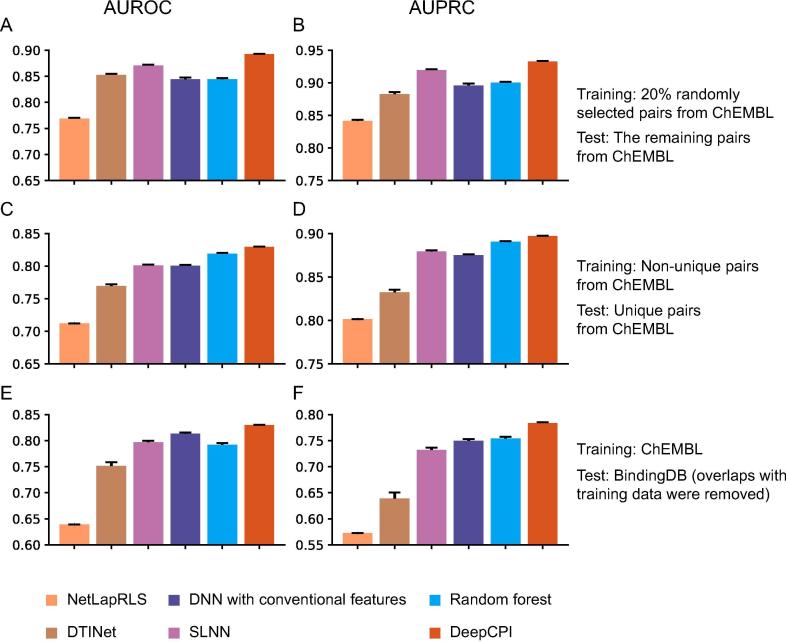
In all computational tests, three baseline methods were used for comparisons (Materials and methods). The first two were a random forest and a single-layer neural network (SLNN) with our feature extraction schemes. These were used to demonstrate the need for the DNN model. The third one was a DNN with conventional features (i.e., Morgan fingerprints [35] with a radius of three for compounds and pairwise Smith–Waterman scores for proteins in the training data), which was used to demonstrate the need for our feature embedding methods. Moreover, we compared DeepCPI with two state-of-the-art network-based DTI prediction methods—DTINet [12] and NetLapRLS [10] (Materials and methods)—in a setting where redundant proteins and compounds were removed; these two methods were not used in other scenarios (Figure S4) as the cubic time and quadratic space complexities concerning the large number of compounds exceeded the limit of our server. We observed that DeepCPI significantly outperformed the network-based methods (Figure 2A–D). Compared to these two network-based methods, DeepCPI achieved better time and space complexities (Materials and methods), demonstrating its superiority over network-based frameworks when handling large-scale data. In addition, DeepCPI outperformed the other three baseline methods (Figure 2A–D and Figure S4) and exhibited a better prediction accuracy and generalization ability of the combination of DL and our feature extraction schemes.
Figure 2.
Performance evaluation of DeepCPI
Performance of DeepCPI was evaluated in terms of AUROC and AUPRC using different training dataset and test dataset in comparison with other state-of-the-art methods. The methods for comparison include network-based NetLapRLS and DTINet, SLNN, DNN with conventional features, and random forest. Comparison of AUROC (A) and AUPRC (B) across different methods that were trained on 20% randomly selected compound–protein pairs from ChEMBL and tested on the remaining pairs from ChEMBL. Comparison of AUROC (C) and AUPRC (D) across different methods that were trained on non-unique pairs from ChEMBL and tested on unique pairs from ChEMBL. Comparison of AUROC (E) and AUPRC (F) across different methods that were trained on ChEMBL data and tested on BindingDB data (in which the overlapping compound–protein pairs were removed). In all cases, the redundant proteins and compounds were removed. The results are summarized over 10 trials and expressed as means ± SD. SLNN, single-layer neural network; DNN, deep neural network; AUROC, area under receiver operating characteristic curve; AUPRC, area under precision-recall curve.
Furthermore, we conducted two supplementary tests to assess the predictive performance of DeepCPI on BindingDB (Tables S1 and S2). BindingDB stores the binding affinities of proteins and drug-like small molecules [31] using the same criteria (i.e., or for positive examples and for negative examples) to label compound–protein pairs. The compound–protein pairs derived from ChEMBL and BindingDB were employed as the training and test data, respectively. Compound–protein pairs from BindingDB exhibiting a compound chemical structure similarity score of and a protein sequence identity score of compared with any compound–protein pair from ChEMBL were regarded as overlaps and removed from the test data. The evaluation results on the BindingDB dataset demonstrated that DeepCPI outperformed all of the baseline methods (Figure 2E and F; Figure S4). Collectively, these data support the strong generalization ability of DeepCPI.
We subsequently investigated the extraction of high-level feature abstractions from the input data using the DNN. We applied T-distributed stochastic neighbor embedding (t-SNE) [36] to visualize and compare the distributions of positive and negative examples with their original 300-dimensional input features and the latent features represented by the last hidden layer in DNN. In this study, DNN was trained on ChEMBL, and a combination of 5000 positive and 5000 negative examples randomly selected from BindingDB was used as the test data. Visualization (Figure S5) showed that the test data were better organized using DNN. Consequently, the final output layer (which was simply a logistic regression classifier) can more easily exploit hidden features to yield better classification results.
Finally, we compared the performance of DeepCPI with those of the following two DL-based models: AtomNet (a structure-based DL approach for predicting compound–protein binding potencies) [23] and DeepDTI [24] (a deep belief network-based model with molecule fingerprints and protein k-mer frequencies as input features) (Materials and methods). Specifically, we compared DeepCPI with AtomNet in terms of the directory of useful decoys from DUD-E [37]. DUD-E is a widely used benchmark dataset for evaluating molecular docking programs and contains active compounds against 102 targets (Table S3). Each active compound in DUD-E is also paired with several decoys that share similar physicochemical properties but have dissimilar two-dimensional topologies, under the assumption that such dissimilarity in the compound structure results in different pharmacological activities with high probability.
We adopted two test settings as reported previously [23] to evaluate the performance of different prediction approaches using DUD-E. In the first setting, cross validation was performed on 102 proteins, i.e., the data were separated according to proteins. In the second setting, cross validation was performed for all pairs, i.e., all compound–protein pairs were divided into three groups for validation. In addition to AtomNet, we also compared our method with a random forest model.
The tests on DUD-E showed that DeepCPI outperformed both the random forest model and AtomNet in the aforementioned settings (Table S4). In addition, in the second setting, our protein structure-free feature extraction schemes with the random forest model also greatly outperformed AtomNet, which requires protein structures and uses a convolutional neural network classifier. These observations further demonstrate the superiority of our feature extraction schemes. DeepCPI achieved a significantly larger area under receiver operating characteristic curve (AUROC) than the random forest model in the first test setting. The first setting was generally more stringent than the second one as protein information was not visible to classifiers during cross validation. Thus, this result indicates that DeepCPI has better generalization ability than the random forest model.
DeepDTI [24] requires high-dimensional features (14,564 features) as the input data; therefore, it can only be used for analyzing small-scale datasets. Thus, we mainly compared DeepCPI with DeepDTI using the 6262 DTIs provided by the original DeepDTI article [24]. We applied the same evaluation strategy as that applied in DeepDTI by randomly sampling the same number (6262) of unknown DTIs as negative examples and splitting the data into the training (60%), validation (20%), and test (20%) data. Our comparison showed that even on this small-scale dataset, DeepCPI continued to achieve a larger mean AUROC (0.9220) than DeepDTI (0.9158) (Table S5). Therefore, we believe that DeepCPI is superior to DeepDTI in terms of both predictive performance and scalability to large-scale compound affinity data.
Novel interaction prediction
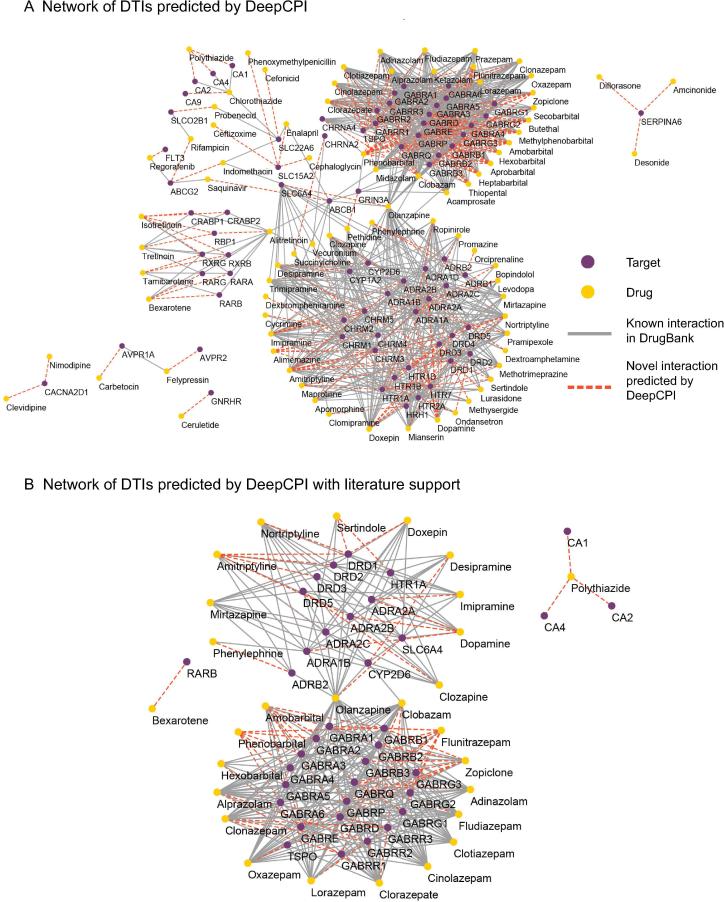
All known DTI data obtained from DrugBank [32] (released on November 11, 2015) were used to train DeepCPI. The novel prediction results on the missing interactions (i.e., the drug–target pairs that did not have an established interaction record in DrugBank) were then examined. Most of the top predictions with the highest scores could be supported by the evidence available in the literature. For example, among the list of the top 100 predictions, 71 novel DTIs were consistent with those reported in previous studies (Table S6). Figure 3 presents the visualization of a DTI network comprising the top 200 predictions using DeepCPI as well as the network of the 71 aforementioned novel DTIs.
Figure 3.
Network visualization of the novel DTIs predicted using DeepCPI
DeepCPI was trained on DrugBank data and used to predict novel DTIs (not recorded in DrugBank). A. A network comprising the top 200 novel DTI predictions. B. A network comprising 71 novel DTIs with literature support among the top 100 DTI predictions. Purple and yellow circles represent targets and drugs, respectively. Gray lines represent the known interactions derived from DrugBank, and red dashed lines represent the novel interactions predicted using DeepCPI. DTI, drug–target interaction.
We describe several examples of these novel predictions supported by the literature below (Table S6). Specifically, in addition to the known DTIs recorded in DrugBank, DeepCPI revealed several novel interactions in neural pharmacology. These interactions may provide new direction to further decipher the complex biological processes in the treatment of neural disorders. For instance, dopamine, a catecholamine neurotransmitter with a high binding affinity for dopamine receptors (DRs), was predicted by DeepCPI to also interact with the α2 adrenergic receptor (ADRA2A). Such a prediction representing a crosstalk within the evolutionally related catecholaminergic systems is supported by the known function of dopamine acting as a weak agonist of ADRA2A [38] as well as the evidence from multiple previous animal studies [39], [40]. Besides the intrinsic neurotransmitters, our prediction results involved various interesting interactions between other types of drugs and their novel binding partners. For instance, amitriptyline, a dual inhibitor of norepinephrine and serotonin reuptake, is commonly used to treat major depression and anxiety. Our predictions indicated that amitriptyline can also interact with three DR isoforms, including DRD1, DRD2, and DRD3. This result is supported by previous evidence, suggesting that amitriptyline displays binding to all three DR isoforms at sub-micromolar potencies [41].
While these antagonist potencies are relatively weak compared with those of other targets (e.g., solute carrier family 6 member 2 [14] and histamine H1 receptor [42]), this new predicted interaction may offer expansion in the chemical space of antipsychotics [41] and the treatment of autism [43]. Moreover, DeepCPI predicted that oxazepam, an intermediate-acting benzodiazepine widely used in the control of alcohol withdrawal symptoms, can also act on the translocator protein, an important factor involved in intromitochondrial cholesterol transfer. This prediction is also supported by previous data from radioligand binding assays [44] as well as by the observation that translocator protein is responsible for the oxazepam-induced reduction of methamphetamine in rats [45].
In addition to providing novel indications in neural pharmacology, our predictions showed that polythiazide, a commonly used diuretic, can act on carbonic anhydrases. This predicted interaction, which may be related to the antihypertensive function of polythiazide [38], is supported by the evidence that polythiazide serves as a carbonic anhydrase inhibitor in vivo [46]. Another important branch of novel interaction predictions exemplified by an enzyme–substrate interaction between desipramine and cytochrome CYP2D6 highlighted a potential novel indication predicted by DeepCPI from a pharmacokinetics perspective. Indeed, the predicted interaction between desipramine and CYP2D6 is supported by their established metabolic association [47], [48], thus offering important clinical implications in drug–drug interactions [49]. Overall, the novel DTIs predicted by DeepCPI and supported by experimental or clinical evidence in the literature further demonstrate the strong predictive performance of DeepCPI.
Validation by experimentation
As 30%–40% of the marketed drugs target GPCRs [50], [51], we applied DeepCPI to identify compounds acting on this class of drug targets. In this experiment, we used positive and negative examples from ChEMBL and BindingDB as well as the compound–protein pairs with affinities in ZINC15 [52], [53], [54] as the training data for DeepCPI. Briefly, we predicted potential interacting compounds using a dataset obtained from the Chinese National Compound Library (CNCL; http://www.cncl.org.cn/, containing 758,723 small molecules) with three class B GPCRs (GLP-1R, GCGR, and VIPR) involved in metabolic disorders and hypertension [55], [56]. These proteins are challenging drug targets, particularly for the development of small-molecule modulators. For each GPCR target, we ran the trained DeepCPI model on the CNCL dataset and selected the top 100 predictions with the highest confidence scores for experimental validation as detailed below.
Pilot screening
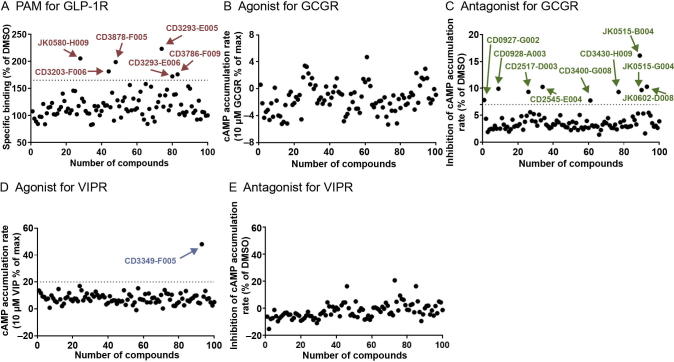
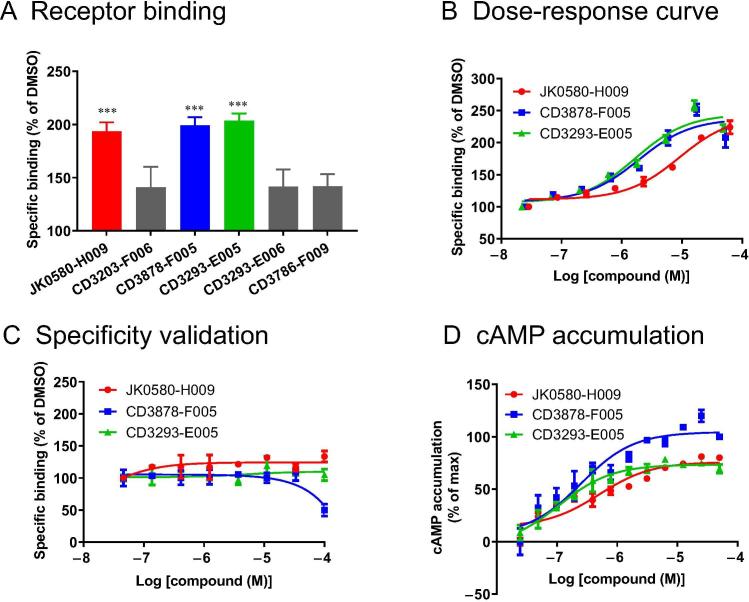
We first conducted several pilot screening assays as an initial experimental validation step to verify the top 100 compounds that were predicted by DeepCPI to act on the aforementioned three GPCRs. For GLP-1R, a whole-cell competitive binding assay was used to examine the effects of potential positive allosteric modulators (PAMs) (Figure 4A; Materials and methods). For GCGR and VIPR, a cAMP accumulation assay was conducted to evaluate the agonistic and antagonistic activities of the predicted compounds (Figure 4B–E). A total of six putative ligands showed a significant augmentation of radiolabeled GLP-1 binding compared with DMSO control, i.e., within the top 25% quantile of the maximum response (Figure 4A). Moreover, we discovered putative small-molecule ligands acting on GCGR and VIPR (Figure 4B–E). Among these, nine compounds exhibited significant antagonistic effects against GCGR (with 7% cAMP inhibition; Figure 4C), while one compound exhibited an obvious agonistic effect on VIPR (with 20% cAMP increase; Figure 4D). Thus, these hits were selected for further validation.
Figure 4.
Pilot screening of the predicted compounds acting on GLP-1R, GCGR, and VIPR
A. A whole-cell competitive ligand binding assay was used to assess the effects of potential PAMs on GLP-1R. GLP-1R was stably expressed in FlpIn-CHO cells, and the effects of the top 100 predicted compounds (10 µM) were studied using the binding assay. Compared to DMSO, six compounds (indicated by arrows) showed obvious enhancement of radiolabeled GLP-1 binding to GLP-1R (within the top 25% quantile of the maximum response). B. Agonist validation for GCGR. GCGR-expressing cells were exposed to 20 µM compounds, and data were normalized with 10 nM glucagon (100%). C. Antagonist validation for GCGR. GCGR-expressing cells were treated with 0.01 nM glucagon and 20 µM compounds, and data were normalized to the maximal response elicited by 0.01 nM glucagon. Nine compounds (indicated by arrows) showed antagonist effects on cAMP accumulation (>7% inhibition). D. Agonist validation for VIPR. CHO cells transfected with VIPR were incubated with 20 µM compounds for 40 min. Data were normalized to maximal response elicited by 10 µM VIP. CD3349-F005 showed a visible agonist effect on cAMP accumulation (>20% increase). E. Antagonist validation for VIPR. VIP and compounds were added at the concentration of 10 nM and 20 µM, respectively, and data were normalized to maximal response elicited by 10 nM VIP. For all panels, the compounds selected for further validation are labeled with CNCL identification numbers. GLP-1R, glucagon-like peptide-1 receptor; GCGR, glucagon receptor; VIP, vasoactive intestinal peptide receptor; VIPR, VIP receptor; PAM, positive allosteric modulator; CNCL, Chinese National Compound Library.
Confirmation of PAMs of GLP-1R
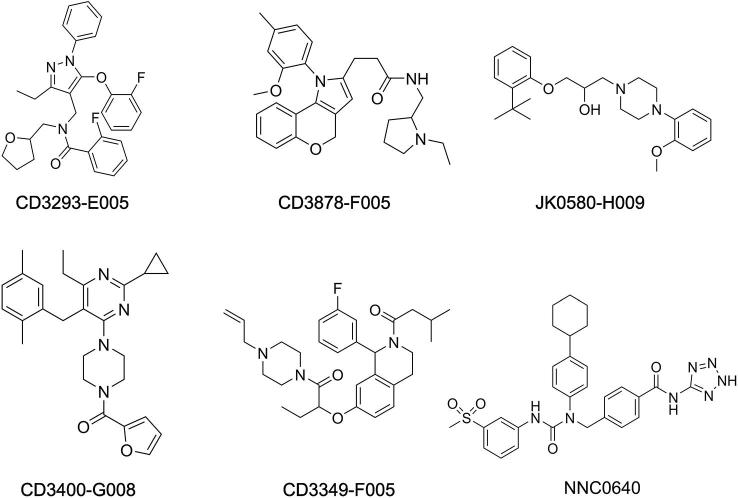
The six putative hits were examined for their binding to GLP-1R. Of these, three (JK0580-H009, CD3293-E005, and CD3848-F005; Figure S6) showed significant enhancement of GLP-1 binding to GLP-1R (Figure 5A). Their corresponding dose–response curves exhibited obvious positive allosteric effects, with half maximal effective concentration ( values within the low micromolar range (; Figure 5B). To test the specificity of the three compounds, we investigated their binding ability to GCGR, a homolog of GLP-1R. These compounds did not cross-react with GCGR (Figure 5C) but substantially promoted intracellular cAMP accumulation in the presence of GLP-1 (Figure 5D). Collectively, these results suggest that JK0580-H009, CD3293-E005, and CD3848-F005 are PAMs of GLP-1R.
Figure 5.
Experimental validation of the putative PAMs of GLP-1R
A. A receptor binding assay was used to validate the six hit compounds shown in Figure 4A. JK0580-H009, CD3878-F005, and CD3293-E005 displayed stable and significant enhancement of 125I-GLP-1 binding to GLP-1R (P < 0.0001). B. Dose–response characteristics of JK0580-H009, CD3878-F005, and CD3293-E005 in terms of the binding of 125I labelled GLP-1 (40 pM) to GLP-1R. C. Validation of specificity for compounds JK0580-H009, CD3878-F005, and CD3293-E005 using cells stably expressing GCGR. Compared to DMSO, there was no significant effect of the putative ligands on 125I-glucagon binding to GCGR. D. Effects of compounds JK0580-H009, CD3878-F005, and CD3293-E005 on cAMP accumulation. All compounds showed dose-dependent augmentation of intracellular cAMP levels in the presence of GLP-1. All measurements were performed with at least three independent experiments, and data are shown as mean ± SEM. ***, P < 0.0001 (one-way ANOVA followed by Dunnett’s post-test; in comparison to DMSO treatment).
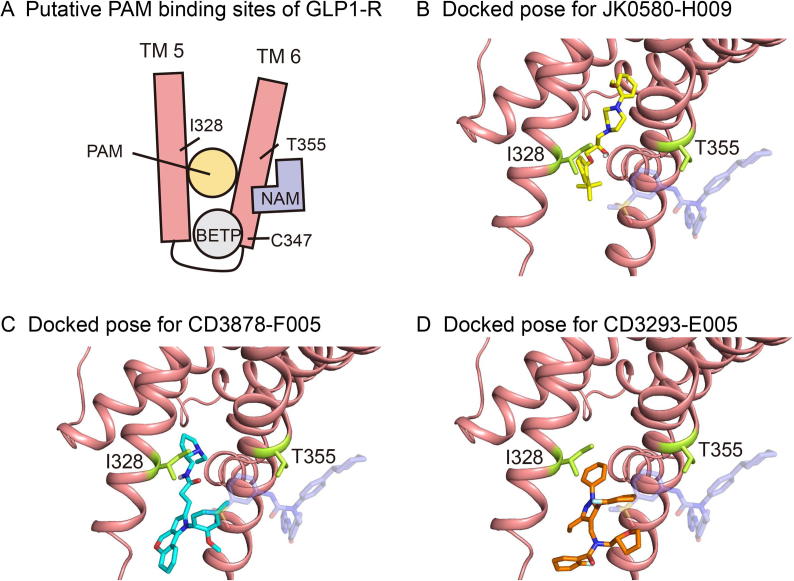
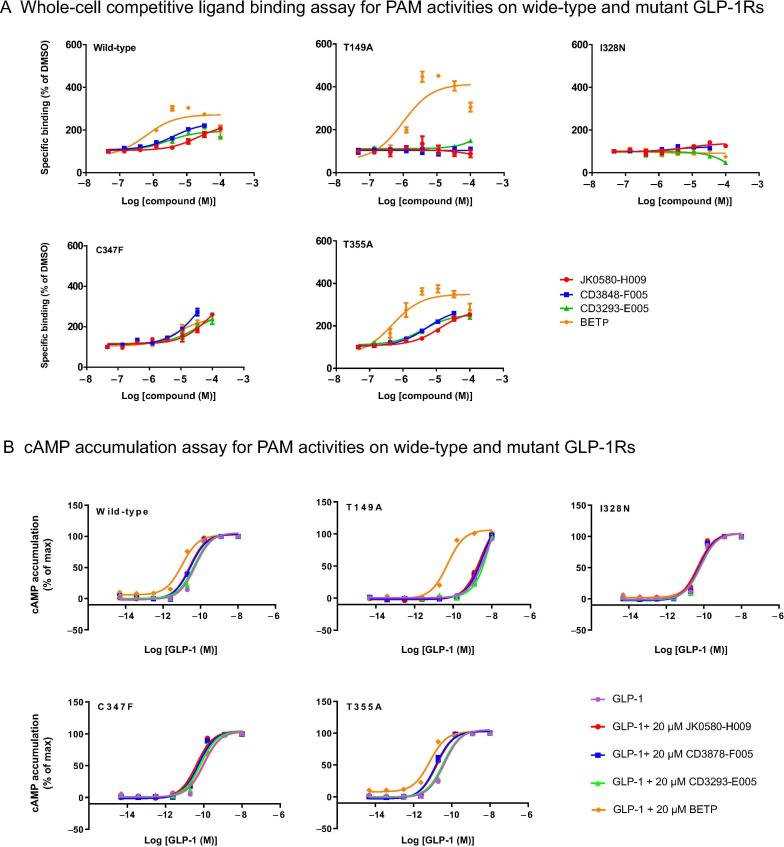
To explore possible binding modes of these new PAMs, we conducted molecular docking studies using AutoDock Vina [57] based on the high-resolution three-dimensional active structure of GLP-1R [58] (Figures 6 and Figure S6). We first used NNC0640 (Figure S6), a negative allosteric modulator of GLP-1R [59], as a control to demonstrate that our docking approach could recover the experimentally solved complex structure (Figure S7). Interestingly, our docking results indicated that the binding pocket for the predicted PAMs are located between transmembrane helix 5 (TM5) and TM6 of GLP-1R, which are distinct from that of NNC0640 (Figure 6) and consistent with the enlarged cavity in the active form of GLP-1R (Figure S8). Additionally, the docking results suggest that the binding sites of our predicted PAMs are located deeper inside the transmembrane domain of GLP-1R than that of the known covalently bound PAMs, including Compound 2 [59] and 4-(3-(benzyloxy)phenyl)-2-ethylsulfinyl-6-(trifluoromethyl)pyrimidine (BETP) [60], [61] (Figures 6A and S7). These findings reveal a novel route for discovering and designing new PAMs of GLP-1R. To further analyze these docking results, we produced four stable cell lines expressing mutant GLP-1Rs (C347F, T149A, T355A, and I328N). As a control, we first measured the activity of BETP, which is covalently bonded to C347 in GLP-1R. Consistent with the previous findings [59], T149A mutation diminished the binding between 125I-GLP-1 and GLP-1R, which could be restored by BETP treatment (Figure 7). Meanwhile, C347F mutation eliminated the covalent anchor of BETP and reduced its efficacy compared with that of wild-type GLP-1R (Figure 7).
Figure 6.
Molecular docking results of the predicted PAMs of GLP-1R
A. Schematic display of receptor binding sites of different PAMs including BETP and the compounds predicted by DeepCPI in the current study. T149, a key residue for GLP-1R allosteric modulation, is located on TM1 but not shown in this diagram. B. Docked poses for the predicted interaction between GLP-1R and JK0580-H009 (shown in yellow). C. Docked poses for the predicted interaction between GLP-1R and CD3878-F005 (shown in cyan) D. Docked poses for the predicted interaction between GLP-1R and CD3293-E005 (shown in orange). GLP-1R is shown in its active form (pink, PDB ID: 5NX2). The binding modes of the three PAMs are compared with that of NAM NNC0640 (light purple) adopted from PDB ID: 5VEX. Key residues for PAM and NAM binding, namely I328 and T355, are shown in stick and colored by lemon. TM, transmembrane; NAM, negative allosteric modulator; BETP, 4-(3-(benzyloxy)phenyl)-2-ethylsulfinyl-6-(trifluoromethyl)pyrimidine.
Figure 7.
Mutations in GLP-1R affect the activities of predicted PAMs
According to our docking results and previous reported binding sites for allosteric modulators, we established stable cell lines expressing T149A, I328N, C347F or T355A to identify the binding pocket for predicted PAMs. A. PAM activities of the predicted compounds on wild-type and mutant GLP-1 receptors, measured using a whole-cell competitive ligand binding assay at different concentrations. B. PAM activities of the predicted compounds on the wild-type and mutant GLP-1 receptors, measured by cAMP accumulation assay. All measurements were performed with at least three independent experiments. Data are shown as means ± SEM and fitted to a four-parameter logistic regression model.
However, none of the three predicted compounds exhibited binding to the T149A mutant, and their modulation behavior on C347F mutant generally aligned with that on wild-type, supporting its non-covalent binding nature (Figure 7, Table S7). These observations point to a divergent binding mode of the predicted PAMs different from that of BETP. Intriguingly, I328N mutation principally abolished the allosteric effects of the compounds (Figure 7), probably due to a large steric crash, as predicted by the docking study. In contrast, T355A mutation located at the other side of TM6 (Figure 6) showed a negligible impact on the PAM activities of the predicted compounds (Figure 7 and Table S7). Collectively, our mutagenesis results support the docking data, indicating that DeepCPI can discover potential PAMs of GLP-1R.
Validation of GCGR and VIPR modulators
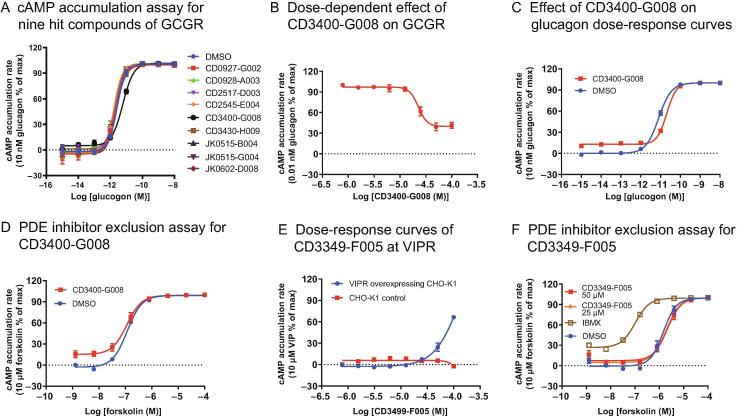
Nine hits with antagonistic effects against GCGR (Figure 4C) were identified in the pilot screening. Among these, CD3400-G008 (Figure S6) was confirmed to stably decrease glucagon-induced cAMP accumulation (Figure 8A). Subsequently, its dose dependency and estimated value of antagonism () were determined (Figure 8B). In addition, this compound led to a rightward shift of the glucagon dose–response curve, as measured by the cAMP accumulation assay (Figure 8C). This shift corresponded to an increase in the value of glucagon from to , although it did not affect forskolin-induced cAMP accumulation (Figure 8D), ruling out the possibility that CD3400-G008 decreases cAMP accumulation in a non-specific manner. Similarly, the agonistic effect of the putative VIPR agonist CD3349-F005 (Figures 4 and S6) was dose-dependent (Figure 8E), while its agonism specificity was confirmed using a phosphodiesterase (PDE) inhibitor exclusion assay (Figure 8F). The results showed that neither nor of CD3349-F005 affected forskolin-induced cAMP accumulation.
Figure 8.
Experimental validation of the hit compounds acting on GCGR and VIPR
A. Validation of the nine putative modulators identified in Figure 4C using cAMP accumulation assay. CD3400-G008 displayed a stable and remarkable antagonist effect on GCGR. B. Dose-dependent effect of CD3400-G008 on GCGR with respect to the inhibition of cAMP accumulation. GCGR-expressing cells were incubated with 0.01 nM glucagon and treated with different concentrations of CD3400-G008 to generate the dose–response curve (IC50 = 22.6 µM). C. Glucagon dose–response curves, in which its concentration was gradually increased from 0.01 pM to 10 nM, whereas that of CD3400-G008 was set to 20 µM; DMSO was used as negative control. D. Effect of CD3400-G008 on forskolin-induced cAMP accumulation. E. Dose–response curves of CD3349-F005 in CHO-K1 cells overexpressing VIPR and in the parental CHO-K1 cells. CD3349-F005 concentration was gradually decreased from 100 µM to 0.78 µM. F. PDE inhibitor exclusion assay was conducted using CHO-K1 cells to detect the agonist effects of CD3349-F005 on forskolin-induced cAMP accumulation; a potent PDE inhibitor IBMX was used as positive control. All measurements were performed with at least three independent experiments in quadruplicate. Data are shown as means ± SEM and fitted to a four-parameter logistic regression model. Max, maximum response; PDE, phosphodiesterase; IBMX, 3-isobutyl-1-methylxanthine.
Collectively, these data support the notion that DeepCPI prediction can offer a promising starting point for small-molecule drug discovery targeting GPCRs.
Discussion
In this article, we propose DeepCPI as a novel and scalable framework that combines data-driven representation learning with DL to predict novel CPIs (DTIs). By exploiting the large-scale unlabeled data of compounds and proteins, the employed representation learning schemes effectively extract low-dimensional expressive features from raw data without the requirement for information on protein structure or known interactions.
The combination of the effective feature embedding strategies and the powerful DL model is particularly useful for fully exploiting the massive amount of compound–protein binding data available from large-scale databases, such as PubChem and ChEMBL. The effectiveness of our method was fully validated using several large-scale compound–protein binding datasets as well as the known interactions between Food and Drug Administration (FDA)-approved drugs and targets. Moreover, we experimentally validated several compounds that were predicted to interact with GPCRs, which represent the largest transmembrane receptor family and probably the most important drug targets. This family constitutes >800 annotated and 150 “orphan” receptors. The latter are without known endogenous ligands and/or functions. Target-based drug discovery has been a focal point of research for decades. However, the inefficiency of mass random bioactivity screening promotes the application of in silico prediction and discovery of small-molecule ligands. Our DeepCPI model establishes a new framework that effectively combines feature embedding with DL for the prediction of CPIs at a large scale.
We conducted several functional assays to validate our prediction results regarding the identification of small-molecule modulators targeting several class B (i.e., the secretin-like family) GPCRs. GLP-1R is an established drug target for type 2 diabetes and obesity, and several peptidic therapeutic agents have been developed and marketed with combined annual sales of billions of dollars. As most therapeutic peptides require non-oral administration routes, discovery of orally available small-molecule surrogates is highly desirable. To the best of our knowledge, since the discovery of Boc5—the first non-peptidic GLP-1R agonist—more than a decade ago [62], [63], [64], little progress has been made in identifying “druggable” small-molecule mimetics for GLP-1. In this study, we identified three PAMs that were computationally predicted by DeepCPI and experimentally confirmed with bioassays to be specific to GLP-1R, thereby providing an alternative to discover non-peptidic modulators of GLP-1R.
The docking results of our predicted hits demonstrated that they could be fitted to similar sites corresponding to the binding pockets for previously known PAMs at GLP-1R in its active form. The experimental data generated by binding and cAMP accumulation assays confirmed the positive allosteric action of these hits. Overall, our modeling data, in conjunction with those obtained from mutagenesis studies, revealed the binding poses of the predicted interactions between these compounds and GLP-1R. These results offer new insights into the structural basis and underlying mechanisms of drug action.
Cross validation through different databases, supporting evidence from the known DTIs in the literature, and laboratory experimentation indicate that DeepCPI can serve as a useful tool for drug discovery and repositioning. In our follow-up studies, we intend to combine DeepCPI with additional validation experiments for the discovery of drug leads against a wide range of targets. Better prediction results may be achieved by incorporating other available data, such as gene expression and protein structures, into our DL model.
Materials and methods
DeepCPI
DeepCPI is an extension of our previously developed CPI prediction model [65]. We describe the building blocks of DeepCPI in the following three subsections.
Compound feature extraction
To learn good embeddings (i.e., low-dimensional feature representations) of compounds, we used the latent semantic analysis (also termed latent semantic indexing) technique [27], which is probably one of the most effective methods for document similarity analyses in the field of NLP. In latent semantic analysis, each document is represented by a vector storing the term frequency or term frequency-inverse document frequency information (tf-idf). This is a numerical statistic widely used in information retrieval to describe the importance of a word in a document. Subsequently, a corpus (i.e., a collection of documents) can be represented by a matrix, in which each column stores the tf-idf scores of individual terms in a document. Subsequently, singular value decomposition (SVD) is applied to obtain low-dimensional representations of features in documents.
In the context of compound feature extraction (Figure S1), a compound and its substructures can be viewed as a document and corresponding terms, respectively. Given a compound set , we use the Morgan fingerprints [35] with a radius of one to scan every atom of each compound in and then generate the corresponding substructures. Let denote the set of substructures generated from all compounds in . We then employ a matrix to store the word count information for these compounds, where represents the tf-idf value of the ith substructure in the th compound. More specifically, is defined as , where stands for the number of occurrences of the ith substructure in the jth compound, and . Here, represents the number of documents containing the th substructure. Basically, reweighs , resulting in lower weights for more common substructures and higher weights for less common substructures. This is consistent with an observation in the information theory that rarer events generally have higher entropy and are thus more informative.
After is constructed, it is then decomposed by SVD into three matrices, , such that . Here, is a diagonal matrix with the eigenvalues of on the diagonal matrix, and is a matrix in which each column is an eigenvector of corresponding to the th eigenvalue .
To embed the compounds into a low-dimensional space , where , we select the first columns of , which correspond to the largest eigenvalues in . Let denote the matrix with columns corresponding to these selected eigenvectors. Subsequently, a low-dimensional embedding of can be obtained by = , where is a matrix, in which the ith column corresponds to a -dimensional embedding (or embedded feature vector) of the ith compound. can be precomputed and fixed after being trained from a compound corpus. Given any new compound, its embedded low-dimensional feature vector can be obtained by left multiplying its tf-idf by (Figure S1).
Our compound feature embedding framework used the compounds retrieved from multiple sources, including all compounds labeled as active in bioassays on PubChem [14], all FDA-approved drugs in DrugBank [32], and all compounds stored in ChEMBL [30]. Duplicate compounds were removed according to their International Chemical Identifiers (InChIs). The total number of final compounds in our compound feature extraction module used to construct matrix was 1,795,801. The total number of different substructures generated from the Morgan fingerprints with a radius of one was 18,868. We set , which is a recommended value in latent semantic analysis [66].
Protein feature extraction
We applied Word2vec, a successful word-embedding technique widely used in various NLP tasks [16], [28], to learn the low-dimensional representations of protein features. In particular, we use the Skip-gram with a negative sampling method [28] to train the word-embedding model and learn the context relations between words in sentences. We first introduce some necessary notations. Suppose that we are given a set of sentences and a context window of size . Given a sentence that is represented by a sequence of words , where is the length of , the contexts of a word are defined as . That is, all the words appearing within the context window of size and centered at word in the sentence are regarded as the contexts of . We further use to denote the set of all words appearing in , and to denote the total number of occurrences of word appearing in the context window of the word in . Since each word can play two roles (i.e., center word and context word), the Skip-gram method equips every word with two -dimensional vector representations , where is used when is a center word and is used otherwise (both vectors are randomly initialized). Here, or basically represents the coordinates of in the lower dimensional (i.e., -dimensional) space after embedding. Subsequently, our goal is to maximize the following objective function:
| (1) |
where is the sigmoid function. Since the range of the sigmoid function is , can be interpreted as the probability of word being a context of word , and Equation 1 can be viewed as the log-likelihood of a given sentence set .
One problem in this objective function (i.e., Equation 1) is that it does not take into account any negative example. If we arbitrarily assign any large positive values to and , would invariably be predicted as 1. In this case, although Equation 1 is maximized, such embeddings are surely useless. To tackle this problem, a Skip-gram model with negative sampling [28] has been proposed, in which “negative examples” ∈ W and ) are drawn from a data distribution , where M represents the total number of words in and represents the total number of occurrences of word in Then, the new objective function can be written a follows:
| (2) |
where is the number of “negative examples” to be sampled for each observed word–context pair during training. Maximizing this objective function can be performed using the stochastic gradient descent technique [16].
For each observed word–context pair , Equation 2 aims to maximize its log-likelihood, while minimizing the log-likelihood of random pairs under the assumption that such random selections can well reflect the unobserved word–context pairs (i.e., negative examples) representing the background. In other words, the goal of this task is to distinguish the observed word–context pairs from the background distribution.
As in other existing schemes for encoding the features of genomic sequences [29], each protein sequence in our framework is regarded as a “sentence” reading from its N-terminus to C-terminus and every three non-overlapping amino acid residues are viewed as a “word” (Figure S2). For each protein sequence, we start from the first, second, and third amino acid residues from the N-terminus sequentially and then consider all possible “words” while discarding those residues that cannot form a “word”.
After converting protein sequences to “sentences” and all three non-overlapping amino acid residues to “words”, Skip-gram with negative sampling is employed to learn the low-dimensional embeddings of these “words”. Subsequently, the learnt embeddings of “words” are fixed, and an embedding of a new protein sequence is obtained by summing and averaging the embeddings of all “words” in all three possible encoded “sentences” (Figure S2). Of note, a similar approach has been successfully used to extract useful features for text classification using Word2vec [67].
In our study, the protein sequences used for learning the low-dimensional embeddings of protein features were retrieved from several databases, including PubChem [14], DrugBank [32], ChEMBL [30], Protein Data Bank [68] (www.rcsb.org), and UniProt [38]. All duplicate sequences were removed, and the final number of sequences for learning the protein features during the embedding process was 464,122. We followed the previously described principles [29] to select the hyper parameters of Skip-gram. More specifically, the embedding dimension was set to , the size of the context window was set to , and the number of negative examples was set to .
Multimodal DNN
Suppose that we are given a training dataset of compound–protein pairs and a corresponding label set , where stands for the total number of compound–protein pairs, indicates that compound and protein interact with each other, and otherwise. We first use the feature extraction schemes described earlier to derive the feature embeddings of individual compounds and proteins, and then feed these two embeddings to a multimodal DNN to determine whether the given compound–protein pair exhibits a true interaction.
We first introduce a vanilla DNN and then describe its multimodal variant. The basic DNN architecture comprises an input layer , an output layer , and hidden layers () between input and output layers. Each hidden layer contains a set of units that can be represented by a vector , where stands for the number of units in . Subsequently, each hidden layer can be parameterized by a weight matrix , a bias vector , and an activation function . More specifically, the units in can be calculated by , where , and the units in the input layer are the input features. We use the rectified linear unit function , which is a common choice of activation function in DL, perhaps due to its sparsity property, high computational efficiency, and absence of the gradient-vanishing effect during the back-propagation training process [69].
The multimodal DNN differs from the vanilla DNN in terms of the use of local hidden layers to distinguish different input modalities (Figure 1). In our case, the low-dimensional compound and protein embeddings are considered distinct input modalities and separately fed to two different local hidden layers. Subsequently, these two types of local hidden layers are concatenated and fed to joint hidden layers (Figure 1). Here, the explicit partition of local hidden layers for distinct input channels can better exploit the statistical properties of different modalities [70].
After we calculate the for the final (joint) hidden layer, the output layer is simply a logistic regression model that takes as its input and computes , where the output is the confidence score for classification, is the sigmoid function, , and are the parameters of the output layer . Since the sigmoid function has a range , can also be interpreted as the interacting probability of the given compound–protein pair.
To learn , , and all parameters , in the hidden layers from the training data set and the corresponding label set, we need to minimize the following cross-entropy loss:
The aforementioned minimization problem can be solved using the stochastic gradient descent and back-propagation techniques [19]. In addition, we apply two popular strategies in DL communities—dropout [71] and batch normalization [72]—to further enhance the classification performance of our DL model. In particular, dropout sets the hidden units to zero with a certain probability, which can effectively alleviate the potential overfitting problem in DL [71]. The batch normalization scheme normalizes the outputs of hidden units to zero mean and unit standard deviation, which can accelerate the training process and act as a regularizer [72].
Since positive and negative examples are possibly imbalanced, our classifier may learn a “lazy” solution. That is, in such a skewed data distribution case, the classifier can relatively easily predict the dominant class given any input. To alleviate this problem, we downsample the examples from the majority class in order for the numbers of positive and negative examples to be comparable during training. In our computational tests, we implement an ensemble version of the previously described DL model and use the average prediction over 20 models to obtain relatively more stable classification results.
Time and space complexities
Training
For compound feature extraction, given a compound, let denote the running time of generating its Morgan fingerprints with a radius of one. The time complexity for extracting the low-dimensional representations of compound features is , where stands for the number of compounds and stands for the total number of substructures in all compounds. Here, is required for generating the substructures of all compounds and is required for running SVD. The space complexity for the compound feature extraction module is . After training, we need space to store the selected eigenvectors of for future inference, where stands for the dimension of compound embedding.
For the protein feature extraction process, given a protein corpus , it takes time and space to scan and calculate the context information, where and stand for the set of all possible three non-overlapping amino acid residues and the context window size, respectively. Given a word–context pair , it takes time to compute the objective function in Equation 2, where stands for the dimension of a protein embedding and stands for the number of negative samples for each word–context pair. Suppose that we perform iterations of gradient descent, then, the time complexity of the protein feature extraction module is . After training, we need space to store the protein embeddings for future inference.
For a neural network, let and denote the numbers of units in layers and , respectively. Suppose that is the largest value among all layers, then, the time and space complexities for training our DL model are bounded by and , respectively, where stands for the total number of training samples, stands for the number of hidden layers in the neural network, and stands for the number of training iterations.
Prediction
During the prediction stage, given a compound–protein pair, we first compute the low-dimensional vector representations of their features. This operation takes time for the compound and time for the protein, where stands for the length of the protein sequence. Then, these two low-dimensional vector representations are fed to the deep multimodal neural network to make the prediction, which takes time. In our framework, we set = 1024 and = 256, which are small and can be considered constant (also see “Implementation of DeepCPI” below).
Mapping DrugBank data to ChEMBL
A known drug–target pair from DrugBank [32] is considered to be in ChEMBL [30] if compound–target pairs with identical InChIs for compounds or drugs and sequence identity scores for proteins are present in the latter dataset. Given two protein sequences and s', their sequence identity score is defined as = , where stands for the Smith–Waterman score [73].
Definition of unique compounds and proteins
Given a dataset, a compound is considered unique if its chemical structure similarity score with any other compound is <0.55, where the chemical structure similarity score is defined as the Jaccard similarity between two Morgan fingerprints with a radius of three of the corresponding compounds [35]. Similarly, a protein is considered unique if its sequence similarity score with any other protein is < 0.40.
DeepCPI implementation
The Morgan fingerprints of compounds were generated by RDKit (https://github.com/rdkit/rdkit). Latent semantic analysis and Word2vec (Skip-gram with negative sampling) were performed using Gensim [74], a Python library designed to automatically extract semantic topics from documents. Our DNN implementation was based on Keras (https://github.com/keras-team/keras)—a highly modular DL library. For all computational experiments, we used two local hidden layers with 1024 and 256 units, respectively, for both compound and protein input channels. The two local layers were then connected to three joint hidden layers with 512, 128, and 32 units, respectively. We set the dropout rate at 0.2. Batch normalization was added to all hidden layers. During training, we selected 20% of the training data as validation set to select the optimal training epoch.
Baseline methods
When testing on ChEMBL [30] and BindingDB [31], we compared our method with two network-based DTI prediction methods, including DTINet [12] and NetLapRLS [10], as well as with three constructed baseline methods as shown below.
For DTINet and NetLapRLS, Jaccard similarity between the Morgan fingerprints of the corresponding compounds with a radius of three and pairwise Smith–Waterman scores were used to construct compound and protein similarity matrices as required in both methods. The default hyperparameters of both methods were used.
For random forest with our feature extraction schemes, we set the tree number to 128 in all computational tests as previously recommended [75]. We randomly selected 20% of the training data as validation set to select the optimal tree depth from 1 to 30.
For SLNN with our feature extraction schemes, we used a local hidden layer with 1024 units for both compound and protein input channels. The two local layers were then connected and fed to a logistic layer to make the CPI prediction. We set the dropout rate to 0.2 and batch normalization was added to the hidden layer as in DeepCPI. We selected 20% of the training data as validation set to select the optimal training epoch.
For DNN with conventional features, instead of using our feature extraction schemes, Morgan fingerprints with a radius of three and pairwise Smith–Waterman scores were used as compound and protein features, respectively. These features were subsequently fed into the same multimodal neural network as in DeepCPI. We selected 20% of the training data as validation set to select the optimal training epoch.
We also compared our method DeepCPI with two other DL-based models, namely AtomNet [23] and DeepDTI [24].
When comparing with AtomNet, we experienced difficulty in reimplementing AtomNet. Therefore, we mainly compared the performance of DeepCPI with that of AtomNet on the same DUD-E dataset. For a fair comparison, we only used a single DeepCPI model instead of an ensemble version.
We also compared the performance of DeepCPI to that of DeepDTI on 6262 DTIs provided by the original DeepDTI article [24]. DeepDTI conducted a grid search to determine the hyper parameters of the model. Hence, for a fair comparison, we followed the same strategy to determine the hyper parameters of DeepCPI. Here, we reported the hyper parameter space that we searched. In particular, we selected a batch size from , dimensions of compound and protein features from , dropout rate from, and joint hidden layers with sizes from . We only used a single DeeCPI model instead of an ensemble version for a fair comparison.
Molecular docking
Compounds were docked using AutoDock Vina [57]. The GLP-1R model in its active form was extracted from a co-crystal structure of full-length GLP-1R and a truncated peptide agonist (PDB: 5NX2) [58]. The best docked poses were selected based on the Vina-predicted energy values.
Experimental validation
Cell culture
Stable cell lines were established using FlpIn Chinese hamster ovary (CHO) cells (Invitrogen, Carlsbad, CA) expressing either GLP-1R or GCGR and cultured in Ham’s F12 nutrient medium (F12) with 10% fetal bovine serum (FBS) and 800 μg/ml hygromycin-B at 37 °C and 5% carbon dioxide (CO2). Desired mutations were introduced to GLP-1R construct using the Muta-direct™ kit (Catalog No. SDM-15; Beijing SBS Genetech, Beijing, China) and integrated into FlpIn-CHO cells. VIPR overexpression was achieved through transient transfection using Lipofectamine 2000 (Invitrogen) in F12 medium with 10% FBS. Cells were cultured for 24 h before being seeded into microtiter plates.
Whole-cell competitive ligand binding assay
CHO cells stably expressing GLP-1R or GCGR were seeded into 96-well plates at a density of 3 × 104 cells/well and incubated overnight at 37 °C and 5% CO2. The radioligand binding assay was performed 24 h thereafter. For homogeneous binding, cells were incubated in binding buffer with a constant concentration of 125I-GLP-1 (40 pM, PerkinElmer, Boston, MA) or 125I-glucagon (40 pM, PerkinElmer) and unlabeled compounds at 4 °C overnight. Cells were washed three times with ice-cold PBS and lysed using 50 μl lysis buffer (PBS supplemented with 20 Tris–HCl and 1% Triton X-100, pH 7.4). Subsequently, the plates were counted for radioactivity (counts per minute, CPM) in a scintillation counter (MicroBeta2 Plate Counter, PerkinElmer) using a scintillation cocktail (OptiPhase SuperMix; PerkinElmer).
cAMP accumulation assay
All cells were seeded into 384-well culture plates (4000 cells/well) and incubated for 24 h at 37 °C and 5% CO2. For the agonist assay, after 24 h, the culture medium was discarded and 5 µl cAMP stimulation buffer [calcium- and magnesium-free Hanks’ balanced salt solution (HBSS) buffer with 5 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), 0.1% bovine serum albumin (BSA), and 0.5 mM 3-isobutyl-1-methylxanthine (IBMX)] was added to the cells. Subsequently, 5 µl compounds were introduced to simulate the cAMP reaction. For the PAM or antagonist assay, each well contained 2.5 µl cAMP stimulation buffer, 5 µl endogenous ligand (GLP-1, glucagon, or VIP) at various concentrations, and 2.5 µl testing compounds diluted in the cAMP assay buffer. After 40-min incubation at room temperature, cAMP levels were determined using the LANCE cAMP kit (Catalog No. TRF0264; PerkinElmer).
Specificity verification and PDE inhibitor exclusion assay
Two experiments were performed using the cAMP accumulation assay (antagonist mode) to study the specificity of the hit compounds acting on GCGR or VIPR. In the case of GCGR, glucagon was replaced by forskolin to investigate whether CD3400-G008 affects forskolin-induced cAMP accumulation. Forskolin concentration was gradually increased from 1.28 nM to 100 µM, whereas CD3400-G008 concentration was kept unchanged (20 µM). The PDE inhibitor exclusion assay was performed in CHO-K1 cells, in which IBMX-free stimulation buffer (calcium- and magnesium-free HBSS buffer with 5 mM HEPES and 0.1% BSA) was used. Concentrations of both IBMX (PDE inhibitor, positive control) and forskolin were gradually increased from 1.28 nM to 100 µM, and the agonistic effect of CD3349-F005 was examined at concentrations of 25 µM and 50 µM, respectively.
Availability
The source code of DeepCPI can be downloaded from https://github.com/FangpingWan/DeepCPI.
Authors’ contributions
FW, DY, MWW, and JZ conceived the project. JZ, DY, and MWW supervised the project. FW and JZ designed the computational pipeline. FW implemented DeepCPI, and performed the model training and prediction validation tasks. HH and JZ analyzed the novel prediction results. YZ, AD, XC, and DY performed the experimental validation. DY and MWW analyzed the validation results. HH and HG performed the computational docking and data analysis. TX and LC helped analyze the prediction results. FW, HH, MWW, and JZ wrote the manuscript with input from all co-authors. All authors read and approved the final manuscript.
Competing interests
The authors have declared no competing interests.
Acknowledgments
This work was supported in part by the National Natural Science Foundation of China (Grant Nos. 61872216 and 81630103 to JZ, 81872915 to MWW, 81573479 and 81773792 to DY), the National Science and Technology Major Project (Grant No. 2018ZX09711003-004-002 to LC), the National Science and Technology Major Project Key New Drug Creation and Manufacturing Program of China (Grant Nos. 2018ZX09735-001 to MWW, 2018ZX09711002-002-005 to DY), and Shanghai Science and Technology Development Fund (Grant Nos. 15DZ2291600 to MWW, 16ZR1407100 to AD). We acknowledge the support of the NVIDIA Corporation for the donation of the Titan X GPU used in this study.
Handled by Yi Xing
Footnotes
Peer review under responsibility of Beijing Institute of Genomics, Chinese Academy of Sciences and Genetics Society of China.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.gpb.2019.04.003.
Contributor Information
Dehua Yang, Email: dhyang@simm.ac.cn.
Ming-Wei Wang, Email: mwwang@simm.ac.cn.
Jianyang Zeng, Email: zengjy321@tsinghua.edu.cn.
Supplementary material
The following are the Supplementary data to this article:
Schematic of the compound feature embedding module (extraction of compound features) The substructures of each compound from a compound corpus are generated by Morgan fingerprints with a radius of one. The term (i.e., substructure) frequency-inverse document (i.e., compound) frequency information of all compounds in the corpus forms a matrix, where and are the total numbers of substructures and compounds in the corpus, respectively. Subsequently, SVD is used to decompose this matrix, and all eigenvectors corresponding to the largest eigenvalues,, are selected to construct a matrix , where is smaller than . This matrix can be precomputed and fixed after training. Then, for any new compound, its low-dimensional embedded feature vector can be easily obtained by simply left multiplying its tf-idf by. Details can be found in the Materials and methods section. tf-idf, term frequency-inverse document frequency; SVD, singular value decomposition.
Schematic of the protein feature embedding module (extraction of protein features) We first build a “word”-embedding set. In particular, we split each protein sequence from a corpus and represent it with three “sentences,” starting from the first, second, and third residue positions. Every three non-overlapping residues in a “sentence” are regarded as a “word.” For each “word” , all other “words” within a context window of size and centered at in any “sentence” are termed “the contexts” (or neighbors) of . The embeddings (i.e., the low-dimensional feature vectors) of “words” are learnt by maximizing the likelihood of their context words, while minimizing the likelihood of random “words” representing the background (i.e., maximizing Equation 2 in the main text). All “word”-embedding pairs are stored in an embedding set (implemented by a hash table). For a given protein sequence, its embedding is constructed by summing and averaging the embedding of all “words” in all three possible encoded “sentences.” The embedding of “words” can be precomputed and fixed after training. Details can be found in the Materials and methods section.
Statistics for mapped affinities of drug-target pairs from DrugBank Mapping the known interacting drug–target pairs obtained from DrugBank to the corresponding compound–protein pairs obtained from ChEMBL. Ranges of the measured values of IC50 and Ki in ChEMBL and their corresponding numbers of known interacting drug–target pairs mapped from DrugBank are plotted.
Additional performance evaluation of DeepCPI Supplementary results to Figure 2 on the performance comparison between different prediction methods. Unlike in Figure 2, the redundant proteins and compounds were not removed. All results are summarized over 10 trials and expressed as mean ± SD. SLNN, single-layer neural network; DNN, deep neural network; AUROC, area under receiver operating characteristic curve; AUPRC, area under precision-recall curve.
The t-SNE visualization of input features versus hidden features of the DNN Visualization of the distribution of positive and negative examples using t-SNE on the original 300-dimensional input features of the DNN (left) and the features represented by the last hidden layer of the DNN (right). DNN was trained on the ChEMBL dataset, and a combination of 5000 positive and 5000 negative examples randomly selected from BindingDB served as the test data and was also used for the visualization.
Supplementary Figure S6.
Chemical structures of the compounds used in this study
The docking result of the NNC0640-GLP-1R complex Recovery of the experimentally solved complex structure between NNC0640 and GLP-1R (PDB ID: 5VEX) using our docking method. The experimentally determined and docked positions of NNC0640 are shown in magenta and yellow, respectively. Blue and red balls represent nitrogen and oxygen atoms, respectively.
The docking results of three predicted PAMs binding to GLP-1R Docked poses of the predicted interactions between three PAMs and GLP-1R (PDB ID: 5NX2). Local views of the overall docked structure of JK0580-H009 (A), CD3878-F005 (B), and CD3293-E005 (C). For all panels, blue and red balls represent nitrogen and oxygen atoms, respectively, whereas blue and red threads represent nitrogen and carbon atoms from GLP-1R, respectively. Yellow (A), turquoise (B), and brown (C) balls represent carbon atoms, whereas the ball in white (B) represents a hydrogen atom and the balls in azure (C) represent fluorine atoms.
References
- 1.Keiser M.J., Setola V., Irwin J.J., Laggner C., Abbas A.I., Hufeisen S.J. Predicting new molecular targets for known drugs. Nature. 2009;462:175–181. doi: 10.1038/nature08506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lounkine E., Keiser M.J., Whitebread S., Mikhailov D., Hamon J., Jenkins J.L. Large-scale prediction and testing of drug activity on side-effect targets. Nature. 2012;486:361–367. doi: 10.1038/nature11159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Medina-Franco J.L., Giulianotti M.A., Welmaker G.S., Houghten R.A. Shifting from the single to the multitarget paradigm in drug discovery. Drug Discov Today. 2013;18:495–501. doi: 10.1016/j.drudis.2013.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walsh C.T., Fischbach M.A. Repurposing libraries of eukaryotic protein kinase inhibitors for antibiotic discovery. Proc Natl Acad Sci U S A. 2009;106:1689–1690. doi: 10.1073/pnas.0813405106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scannell J.W., Blanckley A., Boldon H., Warrington B. Diagnosing the decline in pharmaceutical R&D efficiency. Nat Rev Drug Discov. 2012;11:191–200. doi: 10.1038/nrd3681. [DOI] [PubMed] [Google Scholar]
- 6.Keiser M.J., Roth B.L., Armbruster B.N., Ernsberger P., Irwin J.J., Shoichet B.K. Relating protein pharmacology by ligand chemistry. Nat Biotechnol. 2007;25:197–206. doi: 10.1038/nbt1284. [DOI] [PubMed] [Google Scholar]
- 7.Martínez-Jiménez F., Marti-Renom M.A. Ligand-target prediction by structural network biology using nAnnoLyze. PLoS Comput Biol. 2015;11:e1004157. doi: 10.1371/journal.pcbi.1004157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Laarhoven T., Nabuurs S.B., Marchiori E. Gaussian interaction profile kernels for predicting drug–target interaction. Bioinformatics. 2011;27:3036–3043. doi: 10.1093/bioinformatics/btr500. [DOI] [PubMed] [Google Scholar]
- 9.Bleakley K., Yamanishi Y. Supervised prediction of drug–target interactions using bipartite local models. Bioinformatics. 2009;25:2397–2403. doi: 10.1093/bioinformatics/btp433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xia Z., Wu L.Y., Zhou X., Wong S.T. Semi-supervised drug-protein interaction prediction from heterogeneous biological spaces. BMC Syst Biol. 2010;4:S6. doi: 10.1186/1752-0509-4-S2-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Y., Zeng J. Predicting drug-target interactions using restricted Boltzmann machines. Bioinformatics. 2013;29:i126–i134. doi: 10.1093/bioinformatics/btt234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luo Y., Zhao X., Zhou J., Yang J., Zhang Y., Kuang W. A network integration approach for drug-target interaction prediction and computational drug repositioning from heterogeneous information. Nat Commun. 2017;8:573. doi: 10.1038/s41467-017-00680-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hattori M., Okuno Y., Goto S., Kanehisa M. Development of a chemical structure comparison method for integrated analysis of chemical and genomic information in the metabolic pathways. J Am Chem Soc. 2003;125:11853–11865. doi: 10.1021/ja036030u. [DOI] [PubMed] [Google Scholar]
- 14.Wang Y., Suzek T., Zhang J., Wang J., He S., Cheng T. PubChem BioAssay: 2014 update. Nucleic Acids Res. 2014;42:D1075–D1082. doi: 10.1093/nar/gkt978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bengio Y., Courville A., Vincent P. Representation learning: a review and new perspectives. IEEE Trans Pattern Anal Mach Intell. 2013;35:1798–1828. doi: 10.1109/TPAMI.2013.50. [DOI] [PubMed] [Google Scholar]
- 16.Mikolov T, Chen K, Corrado G, Dean J. Efficient estimation of word representations in vector space. arXiv 2013;1301.3781.
- 17.Zhang S., Liang M., Zhou Z., Zhang C., Chen N., Chen T. Elastic restricted Boltzmann machines for cancer data analysis. Quant Biol. 2017;5:159–172. [Google Scholar]
- 18.Hu H., Xiao A., Zhang S., Li Y., Shi X., Jiang T. DeepHINT: understanding HIV-1 integration via deep learning with attention. Bioinformatics. 2019;35:1660–1667. doi: 10.1093/bioinformatics/bty842. [DOI] [PubMed] [Google Scholar]
- 19.LeCun Y., Bengio Y., Hinton G. Deep learning. Nature. 2015;521:436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 20.Unterthiner T., Mayr A., Klambauer G., Steijaert M., Wegner J.K., Ceulemans H. Deep learning as an opportunity in virtual screening. Workshop Deep Learn Represent Learn. 2014;27:1–9. [Google Scholar]
- 21.Ramsundar B, Kearnes S, Riley P, Webster D, Konerding D, Pande V. Massively multitask networks for drug discovery. arXiv 2015;1502.02072.
- 22.Wan F., Hong L., Xiao A., Jiang T., Zeng J. NeoDTI: neural integration of neighbor information from a heterogeneous network for discovering new drug–target interactions. Bioinformatics. 2019;35:104–111. doi: 10.1093/bioinformatics/bty543. [DOI] [PubMed] [Google Scholar]
- 23.Wallach I, Dzamba M, Heifets A. AtomNet: a deep convolutional neural network for bioactivity prediction in structure-based drug discovery. arXiv 2015;1510.02855.
- 24.Wen M., Zhang Z., Niu S., Sha H., Yang R., Yun Y. Deep-learning-based drug–target interaction prediction. J Proteome Res. 2017;16:1401–1409. doi: 10.1021/acs.jproteome.6b00618. [DOI] [PubMed] [Google Scholar]
- 25.Öztürk H., Özgür A., Ozkirimli E. DeepDTA: deep drug–target binding affinity prediction. Bioinformatics. 2018;34:i821–i829. doi: 10.1093/bioinformatics/bty593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsubaki M., Tomii K., Sese J. Compound–protein interaction prediction with end-to-end learning of neural networks for graphs and sequences. Bioinformatics. 2019;35:309–318. doi: 10.1093/bioinformatics/bty535. [DOI] [PubMed] [Google Scholar]
- 27.Deerwester S., Dumais S.T., Furnas G.W., Landauer T.K., Harshman R. Indexing by latent semantic analysis. J Am Soc Inf Sci. 1990;41:391–407. [Google Scholar]
- 28.Mikolov T., Sutskever I., Chen K., Corrado G.S., Dean J. Distributed representations of words and phrases and their compositionality. Adv Neural Inf Process Syst. 2013;26:3111–3119. [Google Scholar]
- 29.Asgari E., Mofrad M.R. Continuous distributed representation of biological sequences for deep proteomics and genomics. PLoS One. 2015;10:e0141287. doi: 10.1371/journal.pone.0141287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bento A.P., Gaulton A., Hersey A., Bellis L.J., Chambers J., Davies M. The ChEMBL bioactivity database: an update. Nucleic Acids Res. 2014;42:D1083–D1090. doi: 10.1093/nar/gkt1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu T., Lin Y., Wen X., Jorissen R.N., Gilson M.K. BindingDB: a web-accessible database of experimentally determined protein-ligand binding affinities. Nucleic Acids Res. 2006;35:D198–D201. doi: 10.1093/nar/gkl999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wishart D.S., Knox C., Guo A.C., Shrivastava S., Hassanali M., Stothard P. DrugBank: a comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res. 2006;34:D668–D672. doi: 10.1093/nar/gkj067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Salvat R.S., Parker A.S., Choi Y., Bailey-Kellogg C., Griswold K.E. Mapping the Pareto optimal design space for a functionally deimmunized biotherapeutic candidate. PLoS Comput Biol. 2015;11:e1003988. doi: 10.1371/journal.pcbi.1003988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van Laarhoven T., Marchiori E. Biases of drug–target interaction network data. IAPR Inter Conf Pattern Recogn Bioinformatics. 2014:23–33. [Google Scholar]
- 35.Rogers D., Hahn M. Extended-connectivity fingerprints. J Chem Inf Model. 2010;50:742–754. doi: 10.1021/ci100050t. [DOI] [PubMed] [Google Scholar]
- 36.Lvd Maaten, Hinton G. Visualizing data using t-SNE. J Mach Learn Res. 2008;9:2579–2625. [Google Scholar]
- 37.Mysinger M.M., Carchia M., Irwin J.J., Shoichet B.K. Directory of useful decoys, enhanced (DUD-E): better ligands and decoys for better benchmarking. J Med Chem. 2012;55:6582–6594. doi: 10.1021/jm300687e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.UniProt Consortium UniProt: a hub for protein information. Nucleic Acids Res. 2015;43:D204–D212. doi: 10.1093/nar/gku989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cornil C.A., Ball G.F. Interplay among catecholamine systems: dopamine binds to α2-adrenergic receptors in birds and mammals. J Comp Neurol. 2008;511:610–627. doi: 10.1002/cne.21861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cornil C.A., Castelino C.B., Ball G.F. Dopamine binds to α2-adrenergic receptors in the song control system of zebra finches (Taeniopygia guttata) J Chem Neuroanat. 2008;35:202–215. doi: 10.1016/j.jchemneu.2007.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Von Coburg Y., Kottke T., Weizel L., Ligneau X., Stark H. Potential utility of histamine H3 receptor antagonist pharmacophore in antipsychotics. Bioorg Med Chem Lett. 2009;19:538–542. doi: 10.1016/j.bmcl.2008.09.012. [DOI] [PubMed] [Google Scholar]
- 42.Taylor J.E., Richelson E. High affinity binding of tricyclic antidepressants to histamine H1-receptors: fact and artifact. Eur J Pharmacol. 1980;67:41–46. doi: 10.1016/0014-2999(80)90006-0. [DOI] [PubMed] [Google Scholar]
- 43.Hellings J.A., Arnold L.E., Han J.C. Dopamine antagonists for treatment resistance in autism spectrum disorders: review and focus on BDNF stimulators loxapine and amitriptyline. Expert Opin Pharmacother. 2017;18:581–588. doi: 10.1080/14656566.2017.1308483. [DOI] [PubMed] [Google Scholar]
- 44.Schmoutz C.D., Guerin G.F., Goeders N.E. Role of GABA-active neurosteroids in the efficacy of metyrapone against cocaine addiction. Behav Brain Res. 2014;271:269–276. doi: 10.1016/j.bbr.2014.06.032. [DOI] [PubMed] [Google Scholar]
- 45.Spence A.L., Guerin G.F., Goeders N.E. The differential effects of alprazolam and oxazepam on methamphetamine self-administration in rats. Drug Alcohol Depend. 2016;166:209–217. doi: 10.1016/j.drugalcdep.2016.07.015. [DOI] [PubMed] [Google Scholar]
- 46.Scriabine A., Korol B., Kondratas B., Yu M., P'an S., Schneider J. Pharmacological studies with polythiazide, a new diuretic and antihypertensive agent. Proc Soc Exp Biol Med. 1961;107:864–872. doi: 10.3181/00379727-107-26780. [DOI] [PubMed] [Google Scholar]
- 47.Gueorguieva I., Jackson K., Wrighton S.A., Sinha V.P., Chien J.Y. Desipramine, substrate for CYP2D6 activity: population pharmacokinetic model and design elements of drug–drug interaction trials. Br J Clin Pharmacol. 2010;70:523–536. doi: 10.1111/j.1365-2125.2010.03731.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Spina E., Gitto C., Avenoso A., Campo G., Caputi A., Perucca E. Relationship between plasma desipramine levels, CYP2D6 phenotype and clinical response to desipramine: a prospective study. Eur J Clin Pharmacol. 1997;51:395–398. doi: 10.1007/s002280050220. [DOI] [PubMed] [Google Scholar]
- 49.Reese M.J., Wurm R.M., Muir K.T., Generaux G.T., John-Williams L.S., Mcconn D.J. An in vitro mechanistic study to elucidate the desipramine/bupropion clinical drug-drug interaction. Drug Metab Dispos. 2008;36:1198–1201. doi: 10.1124/dmd.107.020198. [DOI] [PubMed] [Google Scholar]
- 50.Stevens R.C., Cherezov V., Katritch V., Abagyan R., Kuhn P., Rosen H. The GPCR Network: a large-scale collaboration to determine human GPCR structure and function. Nat Rev Drug Discov. 2013;12:25–34. doi: 10.1038/nrd3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Filmore D. It's a GPCR world. Mod Drug Discovery. 2004;7:24–28. [Google Scholar]
- 52.Sterling T., Irwin J.J. ZINC 15–ligand discovery for everyone. J Chem Inf Model. 2015;55:2324–2337. doi: 10.1021/acs.jcim.5b00559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Irwin J.J., Sterling T., Mysinger M.M., Bolstad E.S., Coleman R.G. ZINC: a free tool to discover chemistry for biology. J Chem Inf Model. 2012;52:1757–1768. doi: 10.1021/ci3001277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Irwin J.J., Shoichet B.K. ZINC-a free database of commercially available compounds for virtual screening. J Chem Inf Model. 2005;45:177–182. doi: 10.1021/ci049714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Roth J.D., Erickson M.R., Chen S., Parkes D.G. GLP-1R and amylin agonism in metabolic disease: complementary mechanisms and future opportunities. Br J Pharmacol. 2012;166:121–136. doi: 10.1111/j.1476-5381.2011.01537.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Munro J., Skrobot O., Sanyoura M., Kay V., Susce M.T., Glaser P.E. Relaxin polymorphisms associated with metabolic disturbance in patients treated with antipsychotics. J Psychopharmacol. 2012;26:374–379. doi: 10.1177/0269881111408965. [DOI] [PubMed] [Google Scholar]
- 57.Trott O., Olson A.J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jazayeri A., Rappas M., Brown A.J., Kean J., Errey J.C., Robertson N.J. Crystal structure of the GLP-1 receptor bound to a peptide agonist. Nature. 2017;546:254–258. doi: 10.1038/nature22800. [DOI] [PubMed] [Google Scholar]
- 59.Song G., Yang D., Wang Y., de Graaf C., Zhou Q., Jiang S. Human GLP-1 receptor transmembrane domain structure in complex with allosteric modulators. Nature. 2017;546:312–315. doi: 10.1038/nature22378. [DOI] [PubMed] [Google Scholar]
- 60.Sloop K.W., Willard F.S., Brenner M.B., Ficorilli J., Valasek K., Showalter A.D. Novel small molecule glucagon-like peptide-1 receptor agonist stimulates insulin secretion in rodents and from human islets. Diabetes. 2010;59:3099–3107. doi: 10.2337/db10-0689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nolte W.M., Fortin J.-P., Stevens B.D., Aspnes G.E., Griffith D.A., Hoth L.R. A potentiator of orthosteric ligand activity at GLP-1R acts via covalent modification. Nat Chem Biol. 2014;10:629–631. doi: 10.1038/nchembio.1581. [DOI] [PubMed] [Google Scholar]
- 62.Su H., He M., Li H., Liu Q., Wang J., Wang Y. Boc5, a non-peptidic glucagon-like peptide-1 receptor agonist, invokes sustained glycemic control and weight loss in diabetic mice. PLoS One. 2008;3:e2892. doi: 10.1371/journal.pone.0002892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.He M., Su H., Gao W., Johansson S.M., Liu Q., Wu X. Reversal of obesity and insulin resistance by a non-peptidic glucagon-like peptide-1 receptor agonist in diet-induced obese mice. PLoS One. 2010;5:e14205. doi: 10.1371/journal.pone.0014205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.He M., Guan N., Gao W.W., Liu Q., Wu X.Y., Ma D.W. A continued saga of Boc5, the first non-peptidic glucagon-like peptide-1 receptor agonist with in vivo activities. Acta Pharmacol Sin. 2012;33:148–154. doi: 10.1038/aps.2011.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wan F, Zeng J. Deep learning with feature embedding for compound-protein interaction prediction. bioRxiv 2016;086033.
- 66.Bradford R.B. An empirical study of required dimensionality for large-scale latent semantic indexing applications. Proc ACM Int Conf Inf Knowl Manag. 2008:153–162. [Google Scholar]
- 67.Iyyer M., Manjunatha V., Boyd-Graber J., Daumé H., III Deep unordered composition rivals syntactic methods for text classification. Proc Conf Assoc Comput Linguist Meet. 2015;1:1681–1691. [Google Scholar]
- 68.Rose P.W., Prlić A., Bi C., Bluhm W.F., Christie C.H., Dutta S. The RCSB Protein Data Bank: views of structural biology for basic and applied research and education. Nucleic Acids Res. 2015;43:D345–D356. doi: 10.1093/nar/gku1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Glorot X., Bordes A., Bengio Y. Deep sparse rectifier neural networks. Proc 14th Int Conf Artif Intell Stat. 2011;15:315–323. [Google Scholar]
- 70.Srivastava N., Salakhutdinov R.R. Multimodal learning with deep boltzmann machines. Adv Neural Inf Process Syst. 2012;2:2222–2230. [Google Scholar]
- 71.Srivastava N., Hinton G., Krizhevsky A., Sutskever I., Salakhutdinov R. Dropout: a simple way to prevent neural networks from overfitting. J Mach Learn Res. 2014;15:1929–1958. [Google Scholar]
- 72.Ioffe S, Szegedy C. Batch normalization: accelerating deep network training by reducing internal covariate shift. arXiv 2015;1502.03167.
- 73.Smith T.F., Waterman M.S. Identification of common molecular subsequences. J Mol Biol. 1981;147:195–197. doi: 10.1016/0022-2836(81)90087-5. [DOI] [PubMed] [Google Scholar]
- 74.Rehurek R, Sojka P. Software framework for topic modelling with large corpora. Proc LREC 2010 Workshop New Challenges NLP Frameworks 2010.
- 75.Oshiro T.M., Perez P.S., Baranauskas J.A. How many trees in a random forest? Int Workshop Mach Learn Data Mining Pattern Recogn. 2012:154–168. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Schematic of the compound feature embedding module (extraction of compound features) The substructures of each compound from a compound corpus are generated by Morgan fingerprints with a radius of one. The term (i.e., substructure) frequency-inverse document (i.e., compound) frequency information of all compounds in the corpus forms a matrix, where and are the total numbers of substructures and compounds in the corpus, respectively. Subsequently, SVD is used to decompose this matrix, and all eigenvectors corresponding to the largest eigenvalues,, are selected to construct a matrix , where is smaller than . This matrix can be precomputed and fixed after training. Then, for any new compound, its low-dimensional embedded feature vector can be easily obtained by simply left multiplying its tf-idf by. Details can be found in the Materials and methods section. tf-idf, term frequency-inverse document frequency; SVD, singular value decomposition.
Schematic of the protein feature embedding module (extraction of protein features) We first build a “word”-embedding set. In particular, we split each protein sequence from a corpus and represent it with three “sentences,” starting from the first, second, and third residue positions. Every three non-overlapping residues in a “sentence” are regarded as a “word.” For each “word” , all other “words” within a context window of size and centered at in any “sentence” are termed “the contexts” (or neighbors) of . The embeddings (i.e., the low-dimensional feature vectors) of “words” are learnt by maximizing the likelihood of their context words, while minimizing the likelihood of random “words” representing the background (i.e., maximizing Equation 2 in the main text). All “word”-embedding pairs are stored in an embedding set (implemented by a hash table). For a given protein sequence, its embedding is constructed by summing and averaging the embedding of all “words” in all three possible encoded “sentences.” The embedding of “words” can be precomputed and fixed after training. Details can be found in the Materials and methods section.
Statistics for mapped affinities of drug-target pairs from DrugBank Mapping the known interacting drug–target pairs obtained from DrugBank to the corresponding compound–protein pairs obtained from ChEMBL. Ranges of the measured values of IC50 and Ki in ChEMBL and their corresponding numbers of known interacting drug–target pairs mapped from DrugBank are plotted.
Additional performance evaluation of DeepCPI Supplementary results to Figure 2 on the performance comparison between different prediction methods. Unlike in Figure 2, the redundant proteins and compounds were not removed. All results are summarized over 10 trials and expressed as mean ± SD. SLNN, single-layer neural network; DNN, deep neural network; AUROC, area under receiver operating characteristic curve; AUPRC, area under precision-recall curve.
The t-SNE visualization of input features versus hidden features of the DNN Visualization of the distribution of positive and negative examples using t-SNE on the original 300-dimensional input features of the DNN (left) and the features represented by the last hidden layer of the DNN (right). DNN was trained on the ChEMBL dataset, and a combination of 5000 positive and 5000 negative examples randomly selected from BindingDB served as the test data and was also used for the visualization.
The docking result of the NNC0640-GLP-1R complex Recovery of the experimentally solved complex structure between NNC0640 and GLP-1R (PDB ID: 5VEX) using our docking method. The experimentally determined and docked positions of NNC0640 are shown in magenta and yellow, respectively. Blue and red balls represent nitrogen and oxygen atoms, respectively.
The docking results of three predicted PAMs binding to GLP-1R Docked poses of the predicted interactions between three PAMs and GLP-1R (PDB ID: 5NX2). Local views of the overall docked structure of JK0580-H009 (A), CD3878-F005 (B), and CD3293-E005 (C). For all panels, blue and red balls represent nitrogen and oxygen atoms, respectively, whereas blue and red threads represent nitrogen and carbon atoms from GLP-1R, respectively. Yellow (A), turquoise (B), and brown (C) balls represent carbon atoms, whereas the ball in white (B) represents a hydrogen atom and the balls in azure (C) represent fluorine atoms.