Abstract
Choby JE, Howard-Anderson J, Weiss DS (Emory University School of Medicine, Atlanta, GA, USA; Atlanta VA Medical Center, Decatur, GA, USA) Hypervirulent Klebsiella pneumoniae – clinical and molecular perspectives (Review).
Hypervirulent Klebsiella pneumoniae (hvKp) has emerged as a concerning global pathogen. hvKp is more virulent than classical K. pneumoniae (cKp) and capable of causing community-acquired infections, often in healthy individuals. hvKp is carried in the gastrointestinal tract, which contributes to its spread in the community and healthcare settings. First recognized in Asia, hvKp arose as a leading cause of pyogenic liver abscesses. In the decades since, hvKp has spread globally and causes a variety of infections. In addition to liver abscesses, hvKp is distinct from cKp in its ability to metastasize to distant sites, including most commonly the eye, lung and central nervous system (CNS). hvKp has also been implicated in primary extrahepatic infections including bacteremia, pneumonia and soft tissue infections. The genetic determinants of hypervirulence are often found on large virulence plasmids as well as chromosomal mobile genetic elements which can be used as biomarkers to distinguish hvKp from cKp clinical isolates. These distinct virulence determinants of hvKp include up to four siderophore systems for iron acquisition, increased capsule production, K1 and K2 capsule types, and the colibactin toxin. Additionally, hvKp strains demonstrate hypermucoviscosity, a phenotypic description of hvKp in laboratory conditions that has become a distinguishing feature of many hypervirulent isolates. Alarmingly, multidrug-resistant hypervirulent strains have emerged, creating a new challenge in combating this already dangerous pathogen.
Keywords: capsule, hypermucoviscous, hypervirulent, Klebsiella pneumonia, liver abscess, siderophores
Overview: Pathogenesis of Klebsiella pneumoniae
Klebsiella pneumoniae is an opportunistic pathogen capable of causing a variety of infections. Classically, K. pneumoniae is known to cause pneumonia, urinary tract infections and bacteremia in immunocompromised or frequently healthcare-exposed patients. In the Enterobacteriaceae family, K. pneumoniae isolates that have acquired resistance to carbapenems belong to the carbapenem-resistant Enterobacteriaceae (CRE) and represent a ‘critical concern’ of the World Health Organization [1]. K. pneumoniae is one of only a few Gram-negative rods capable of causing primary pneumonia [2] and is a leading cause of nosocomial pneumonia [3]. Additionally, K. pneumoniae is one of the most frequent causes of hospital-acquired urinary tract infections in the United States [3]. These types of nosocomial infections, most common amongst those at extremes of age or with underlying immunodeficiencies, are features of classical K. pneumonia (cKp).
In contrast, in the past few decades a distinct type of K. pneumoniae, hypervirulent K. pneumoniae (hvKp), has emerged as an important pathogen capable of causing community-acquired and, increasingly, hospital-acquired infections. Unlike cKp, hvKp often causes infection in otherwise healthy individuals [4-7]. hvKp was first recognized as a cause of pyogenic liver abscesses in Asia [5] and is beginning to be appreciated for the frequency at which it causes other types of diseases, outlined below. A defining feature of hvKp is a hypermucoid appearance on agar plates. Thus, hvKp has alternatively been referred to as hypermucoviscous (HMV) or hypermucoid K. pneumoniae to distinguish it from cKp. The ‘string test’ was developed as a phenotypic test for hypermucoviscosity; when a colony can be stretched at least five millimetres with an inoculation loop, the isolate is considered HMV and therefore hvKp [8].
The prevalence of hvKp amongst K. pneumoniae varies, but can be high in studies from hvKp-endemic areas, ranging from 12 to 45% [9-13]. Furthermore, these isolates have historically been largely susceptible to antibiotics. However, as we will explain, these strains are now becoming highly antibiotic-resistant, and thus, the clinical landscape of hypervirulent K. pneumoniae is changing drastically. In this review, we discuss potential sources of hvKp strains, the clinical features of hvKp infection, the molecular basis for its clinical identification and virulence, as well as the convergence of antibiotic-resistant and hypervirulent strains.
The source – hvKp in the gut
Klebsiella are ubiquitous Gram-negative, non-motile bacteria within the Enterobacteriaceae family, which includes well-studied human pathogens like Escherichia, Salmonella, Shigella and Yersinia. Klebsiella species are diverse, with relatively large genomes which enable metabolic flexibility, biochemical diversity and success in colonizing clinical and environmental niches [14]. K. pneumoniae is the most common human pathogen of this genus [2] and widely recognized as an opportunistic pathogen responsible for a large number of nosocomial infections. In mammals, K. pneumoniae is a common species in the gut [14, 15]. It can be readily isolated from human mucosal surfaces, faeces-contaminated hands [16], sewage-contaminated bodies of water, and even surfaces throughout hospitals [17, 18].
Despite its ubiquity in the environment, the reservoir leading to human infection with K. pneumoniae is often the patient’s own gut [19, 20]. Many different environmental sources may be responsible for initial gastrointestinal colonization by K. pneumoniae, including soil, vegetation or faeces-contaminated surfaces and water [14]. Hospital-acquired K. pneumoniae may be transmitted and acquired from hospital sources [21] or patient to patient [22, 23], and whilst this can happen with hypervirulent strains, the traditional antimicrobial susceptibility and community acquisition of hvKp do not suggest the hospital as the source of hvKp prior to intestinal carriage. The environmental source of hvKp that precedes gut colonization is unclear, but once in the gut, community person-to-person spread via the faecal–oral route in hvKp-endemic areas is a possible mechanism for carriage in the community.
hvKp can be a member of the gut microbiome, which likely contributes to its dissemination in communities and hospitals. As part of the recent Human Microbiome Project analyses in the United States, K. pneumoniae was identified in approximately 4% of stool and 10% of nares samples of volunteers [15]. Other studies have found K. pneumoniae in the stool of 10% [24] or 35% [17] of individuals. In hospitalized patients, however, rates of K. pneumoniae intestinal colonization are higher and have been reported between 19 and 38% [17, 19, 20, 24]. Studies of intestinal colonization in areas where hvKp is endemic provide more information about hvKp gastrointestinal carriage. Analysis of 43 patients with hvKp liver abscesses in Taiwan found a strong association between gastrointestinal colonization and liver abscess and identified carriage of hvKp isolates in healthy individuals as well [25]. Other studies identified carriage of hvKp in 5% of healthy Korean adults [26] and in 6% of nearly one thousand Asian adults surveyed [27]. These high rates of carriage of hvKp likely contribute to the hvKp infections in Asia.
There is also evidence suggesting that gut colonization by hvKp directly precedes infection in the same individual. These data are in line with other studies that generally conclude that K. pneumoniae infections stem from initial gut colonization [19, 20]. For example, about half of ICU K. pneumoniae infections were caused by the patient’s own gastrointestinal strain [19]. Additionally, using mouse models, hvKp genes required for gastrointestinal dissemination and colonization have been identified. This suggests that hvKp may use particular virulence determinants to access the liver [28]. In sum, hvKp proficiently colonizes the gastrointestinal tract which contributes to the person-to-person spread of hvKp and likely to the dissemination from the gut to other organs during disease.
Clinical presentation and risk factors
HvKp was first recognized as a unique clinical entity in the 1980s in Taiwan [4]. Community-dwelling patients, without hepatobiliary risk factors, were presenting with pyogenic liver abscesses caused by K. pneumoniae. Doctors and scientists soon discovered that these patients had an invasive type of K. pneumoniae with a tendency for metastatic spread to distant sites [7, 29]. Whilst originally observed predominantly in Southeastern Asia, there are now an increasing number of cases being reported worldwide, including in Europe and the United States [5, 7, 30-37]. Infections due to the hypervirulent form of K. pneumoniae are often diagnosed based on their clinical phenotype as there is no universally agreed upon marker for hypervirulence [7]. In particular, many of the early case reports (including those referenced below) are solely based on clinical presentation. Hypermucoviscous (HMV) and hypervirulent are often used synonymously in the literature; however, not all patients with a hypervirulent phenotype have hypermucoviscous bacteria and not all hypermucoviscous isolates lead to an invasive syndrome [38, 39].
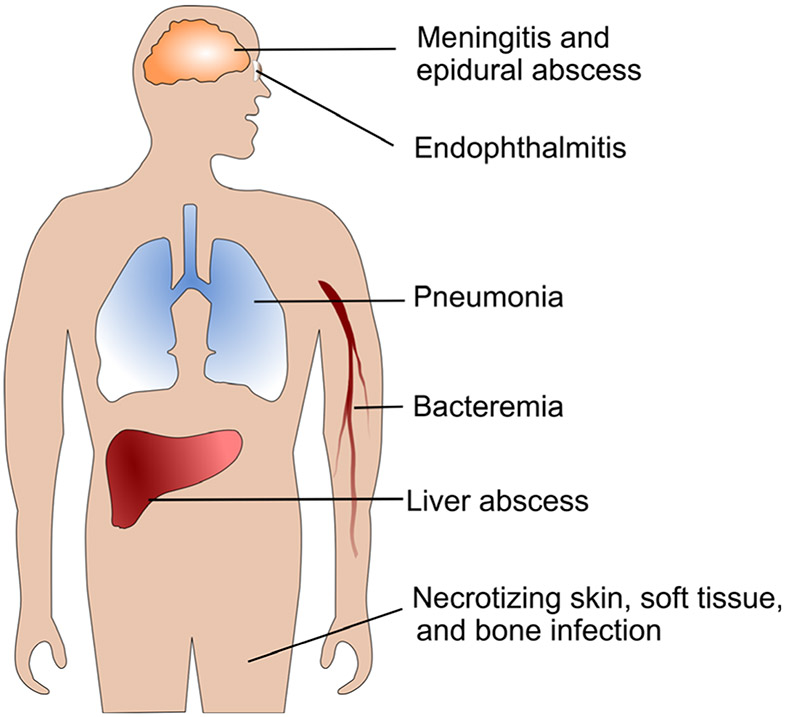
The symptoms of hvKp are nonspecific and can include fevers, chills, abdominal pain, nausea and vomiting, but may also be driven by the location of the metastatic infection [30]. In a large compilation of over 800 patients with K. pneumoniae liver abscesses in Taiwan, South Korea and the United States, 12% of patients had evidence of metastatic disease [30], although rates as high as 28% have been reported [5]. The most common metastatic sites include the eye, lung and CNS, each occurring in approximately one-third of patients who have metastatic disease [6, 30, 40, 41]. There have been numerous reports of patients with metastatic endophthalmitis or uveitis [4-6, 35, 40, 42-45], pulmonary infections including intraparenchymal disease or empyema [30, 35, 36, 40], and CNS disease including meningitis, brain abscess and epidural abscess [5, 6, 33, 40, 44, 46]. Additionally, patients can present with a liver abscess and severe skin, soft tissue and bone infections including necrotizing fasciitis [47, 48], neck and psoas abscesses [31, 33, 36, 40], and osteomyelitis [40, 49]. Less commonly, hvKp has been associated with seeding of other abdominal and pelvic organs including the prostate [37, 40], kidneys [31, 47] and spleen [7]. More recently, a high percentage of vascular thrombotic complications, including pulmonary embolus [31], has been reported, as well as at least one case of endocarditis [50]. Together, the presentations of hvKp (depicted in Fig. 1) are diverse and distinct from infections caused by cKp, underscoring the unique virulence of hvKp.
Fig. 1.
Hypervirulent K. pneumoniae infection sites. Common sites of primary and metastatic infections caused by hvKp
In addition to the pathognomonic syndrome of liver abscess with metastatic involvement, there are now many reported examples of hvKp causing primary extrahepatic disease. In a large observational study in Taiwan, 83 patients had community-acquired hypermucoviscous bacteremia, but only 37% of them presented with the characteristic invasive syndrome; 23% had no detected focal site of infection. Other patients had pneumonia, urinary tract infections, biliary tract infections, soft tissue disease and spontaneous bacterial peritonitis [51]. In another study of extrahepatic disease, 16 of 18 (89%) patients had HMV K. pneumoniae with primary sites of infection including renal abscess, soft tissue and muscle infection, CNS disease, mycotic aneurysm, lung abscess and septic arthritis [52]. Additional case reports have described isolated muscle abscesses, necrotizing fasciitis and osteomyelitis caused by HMV K. pneumoniae without concurrent liver abscess [34, 53, 54]. More research is needed to understand why some patients with HMV isolates present with the typical hypervirulent phenotype and some do not [39].
Whilst hvKp is more frequently associated with community-acquired disease [7, 29, 30], there have also been increasing reports of hypervirulent isolates causing healthcare-associated disease. In particular, pulmonary and ventilator-associated infections have been identified [10, 55-57] as well as healthcare-associated bacteremia [31]. In one hospital in China, there was an outbreak of carbapenem-resistant hvKp that caused fatal ventilator-associated pneumonias in five infected patients [55]. Increased circulation of hvKp strains in hospitals has potential to increase the overall burden of this pathogen.
Patients with hvKp tend to be younger, immunocompetent, present with severe disease compared to cKp [30, 51] and present from community settings [9, 38, 51, 58]. Diabetes is a significant risk factor for acquiring a hvKp infection as opposed to a cKp infection [58-62], and for developing metastatic complications in patients with a liver abscess [40, 63, 64]. However, not all studies have consistently found this association [6, 51], suggesting the connection between diabetes and hvKp could vary by region or clinical presentation. There has also been uncertainty as to whether Asians have an increased risk for hvKp and whether this represents a genetic or environmental association [29]. One study of patients of different ethnicities in Singapore demonstrated an association between capsular K1 liver abscess and Chinese patients (compared to Malay, Indian and Caucasian patients); however, this was not controlled for diet, socio-economic status or other important environmental exposures [65]. In a collection of 38 cases observed in the United States, almost half of the patients were non-Asian, and the ethnic diversity was greater than in Taiwan or South Korea [30]. Therefore, it is still unclear whether the emergence and prevalence of hvKp in Asia is a result of genetic susceptibility to hvKp in this population or other environmental or unknown factors.
Outcomes and treatment of hvKp
HvKp is associated with significant morbidity and mortality. Patients with the invasive syndrome have a mortality rate ranging from 3 to 31% [6, 30, 31, 41, 61, 64], and patients with HMV bacteremia have a mortality rate close to 35% or higher [51, 58]. In one case series of 12 French patients with the HMV phenotype, all 5 patients with bacteremia died [32]. Antimicrobial resistance also increases the risk for mortality [58, 66] which was 100% in one series of patients infected with carbapenemase-producing isolates [55]. Additional risk factors for mortality include gastrointestinal fistula, increased Acute Physiologic and Chronic Health Evaluation (APACHE) II or Pitt bacteremia scores, metastatic infection, septic shock, acute respiratory failure and gas formation on imaging [41, 58]. In addition to mortality, the morbidity from metastatic disease can be devastating. Approximately 70% of patients with ocular or CNS metastatic disease will develop long-term visual or neurologic disability [6, 45].
Management of hvKp infections requires both adequate source control and active antibiotic therapy, with usual treatment durations ranging from two to six weeks depending on the site and extent of infection [29, 30]. Drainage of liver abscesses has been associated with decreased risk of metastatic infection and mortality [41]. All K. pneumoniae are inherently resistant to ampicillin [67]. There have been no trials assessing which antibiotics are best suited for treating hvKp infections, but empiric antibiotic therapy should take into account both the site of infection and local antimicrobial resistance patterns (Table 1) [68]. Ultimately, antibiotic therapy should be tailored once in vitro susceptibilities of each isolate are available [30]. Intravitreal injections should also be used in patients with endophthalmitis [45]. Historically, antimicrobial resistance was rare with only 2% of isolates exhibiting resistance to tested first generation cephalosporins in Taiwan from 1997 to 2005 [6]. However, the prevalence of multidrug-resistant hvKp is increasing, possibly related to the increase in healthcare-associated infections, making this already morbid infection more difficult to treat [9, 10, 12, 55, 56, 58, 60, 69]. Carbapenem-resistant infections are the most concerning of the multidrug-resistant hvKp isolates, and the optimal treatment is still unknown. Therapeutic options to consider include ceftazidime/avibactam, meropenem/vaborbactam, imipenem/relebactam, eravacycline, plazomicin, cefiderocol and colistin, although these should be used only in consultation with expert opinion as many of these agents are not active against metallo-β-lactamases which can confer carbapenem resistance, and most have not been systematically evaluated for the treatment of carbapenem-resistant infections [68, 70].
Table 1.
| Clinical syndrome or site of infection |
Empiric antibiotic suggestionsa |
|---|---|
| Liver (or other intra-abdominal) abscess or pneumonia | β-lactam/β-lactamase inhibitors, third-generation cephalosporins, fluoroquinolones, carbapenemsb or aminoglycosidesc |
| Central nervous system infection | Third-generation cephalosporins or carbapenemsb |
| Endophthalmitis | Intravitreal antibiotics (cefazolin, ceftazidime, aminoglycosides or imipenem) plus intravenous antibiotics (variable, but usually cephalosporins) |
| Prostate abscess | Fluoroquinolones or trimethoprim-sulfamethoxazole |
This should be modified after in vitro susceptibility results are obtained.
This should be reserved for infections in which there is a strong suspicion of extended-spectrum-β-lactamase (ESBL) production.
This should only be considered in combination therapy.
Defining hvKp genotype and capsule type
When invasive K. pneumoniae infections first became a recognized clinical problem in Asia, many groups began tracking isolates in order to understand the differences between cKp and hvKp. Strategies to genetically and phenotypically classify hypervirulent isolates have elucidated trends amongst hvKp lineages that aid in their identification and tracking, but there is no unifying categorization that captures all hvKp strains. Clinical manifestations, sequence and capsule typing, the HMV phenotype, and presence of virulence-associated genes can all be used to differentiate hvKp from cKp strains. These details are discussed in this and the following section and are presented in Fig. 2.
Fig. 2.
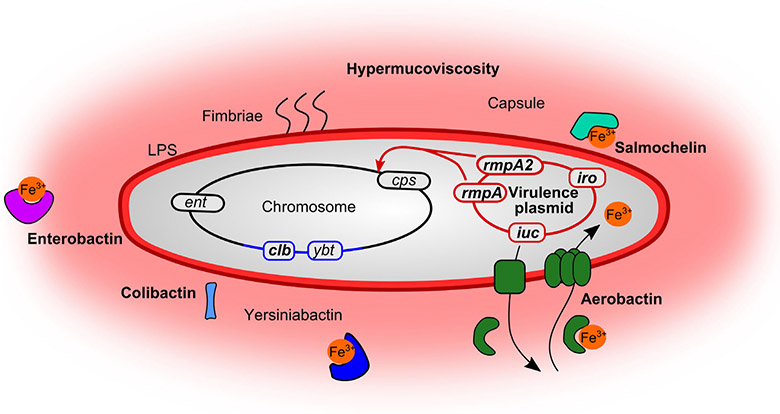
Defining features of hvKp. Defining features (in bold) of hvKp include both chromosomal and plasmid-encoded features that result in the enhanced virulence of hvKp. These are in addition to factors that contribute to the pathogenesis of all K. pneumoniae (not in bold): the siderophore systems enterobactin (ent) and yersiniabactin (ybt), as well as the fimbriae and LPS on the surface. Many hvKp encode yersiniabactin, as well as the secreted toxin colibactin (clb) within an integrative and conjugative element in the chromosome, shown in blue. Plasmid-encoded transcriptional regulators rmpA/rmpA2 (regulator of mucoid phenotype) regulate the chromosomal capsule (cps) loci causing hypercapsule associated with the hypermucoviscous phenotype of hvKp. hvKp strains are frequently of the K1 and K2 capsule types. Encoded on virulence plasmids are also the salmochelin (iro) and aerobactin (iuc) siderophore systems. The details of aerobactin export, binding to iron, and import for iron acquisition are representative of all the siderophores, as each system is typically complete for biosynthesis, export and import of the respective siderophore
The genomic structure of the K. pneumoniae species is diverse, with many distinct lineages. To categorize isolates within the species, multi-locus sequence typing (MLST) was used [71] and the resulting sequence type (ST) classifications remain clinically important today. Whole genome sequencing has enabled more in-depth descriptions of clonal groups (CG), of which there are hundreds, if not thousands [72]. The ST and CG phylogenetic nomenclature describes the genetic relatedness of isolates and therefore easily distinguishes K. pneumoniae (sometimes referred to as KpI) from related species K. quasipneumoniae (KpII) and K. variicola (KpIII).
Another identifier of hvKp is capsule type (K). Capsule is a polysaccharide synthesized by all K. pneumoniae that acts as a protective layer on the exterior of the bacterium, inhibiting phagocytosis, antimicrobial peptides, complement, and induction of the host inflammatory response [73]. Capsule is required for virulence in mouse models of cKp and hvKp infection [8, 74, 75]. The capsule genes are chromosomal and largely conserved across the species, but variation exists in the polysaccharide components of capsule and is the basis for capsule typing. The different polysaccharide variants that result from genetic differences are called K antigens and have been categorized serologically [76]. Presently, sequencing of the capsule synthesis wzi genes has become mainstream, and at least 134 distinct K loci have been identified [77]. The most common capsule locus associated with hvKp is K1, followed by K2, K5 and K57 [42, 65, 74, 78-82].
Despite the high frequency of K1 and K2 loci in hvKp isolates, capsule type does not fully account for hypervirulence; K1 and K2 capsule types co-occur with other virulence genes [78]. Attempts to genetically swap capsule loci between isolates to test the contribution of the capsule have been mixed; switching the capsule of a less virulent strain with that of a hvKp strain does not fully establish the virulent phenotype in mice, suggesting capsule is not the sole driver of virulence [83-85]. Capsule is however thought to prevent phagocytosis and increase hvKp survival and dissemination. Macrophage lectin receptors recognize repeated mannose or rhamnose sugars to induce phagocytosis. However, K1, K2, K5 and K57 capsule types lack these repeated sugars, therefore facilitating the evasion of lectinophagocytosis and macrophage killing [85-91]. K1 isolates were found to be more resistant than cKp isolates to extracellular and intracellular killing by neutrophils [92], which could aid in systemic dissemination [93] as well as limiting exposure to extracellular antibiotics [94]. It is likely that the evolution of hvKp with particular capsule types is the result of advantages conferred by these distinct structures.
The majority of hvKp strains belong to a small collection of clonal groups, indicating that only a few lineages of K. pneumoniae have developed hypervirulence. CG23 is the most dominant lineage and includes many sequence types: ST23, 26, 57 and 1633 [95]. Amongst these, ST23 strains are a leading cause of liver abscesses, and the most frequently observed hvKp across different surveys and collections [65, 82, 96-99]. ST23 is strongly associated with the K1 capsule type, and K1 ST23 strains are associated with four siderophores iron acquisition systems, the toxin colibactin (described in the following section) and invasive infections [100]. hvKp isolates exhibiting the K2 capsule come from a variety of sequence types [101]. An analysis of hvKp strains isolated from livers found that K2 isolates lacked the kfu iron uptake gene and allS, a gene for allantoin metabolism [82]. Allantoin is a degradation product from nucleic acids that some bacteria can use as a source of nitrogen [102]. K1 strains have both genes, and non K1/K2 isolates lack allS but some have kfu [82]. In an experimental model of infection, the absence of both allS and kfu did not reduce virulence, consistent with K2 isolates being fully virulent in humans [79]. Whilst ST23 is the dominant ST amongst hvKp isolates, there has been recent appreciation for other sequence types causing infections in Asia, the United States and Europe [26, 82, 98, 103]. It is likely that the dominance of K1 ST23 hvKp strains in surveys of Asian isolates might be skewed and that globally many different capsule and sequence types can be hvKp.
A defining feature of hvKp strains, which is used clinically to detect hypervirulence, is the presence of a virulence plasmid. Early work identified plasmid-encoded siderophore systems and the mucoid phenotype as key determinants of hvKp strains [104-106]. pK2044 [107] and pLVPK [108] are the most well-studied hvKp virulence plasmids and are highly similar [109, 110]; CG23 strains carry these plasmids whilst K2 hvKp strains often have smaller but similar virulence plasmids [82]. The most well-characterized version of pLVLK is 219 kb and has 13 insertion sequences, probably the result of sequential acquisition of horizontally transferred genes [108]. The CG23 associated plasmids contain virulence genes including the mucoid regulators rmpA and rmpA2, iron acquisition genes (described in detail below), and heavy metal resistance loci. Importantly, only about a third of the genes encoded by these virulence plasmids have known function [108]. The plasmid-encoded rmpA and rmpA2 regulate with capsule genes on the chromosome. Other plasmid-encoded genes, however, contribute to virulence in a capsule-independent manner.
In addition to virulence plasmids, hvKp strains have acquired other virulence genes through mobile genetic elements in the chromosome. More than a dozen distinct integrative and conjugative element (ICEs) exist in K. pneumoniae [111], across cKp and hvKp lineages. There are 14 different types of ICEs across hvKp, and genes encoding the siderophore yersiniabactin are a significant feature of most ICEs in cKp and hvKp strains [111]. ICEs are extremely prevalent in hvKp lineages, including in nearly 90% of CG23 [111, 112] and close to three quarters of all hvKp strains [113]. In addition to yersiniabactin genes, some ICEs carry other virulence genes; ICEKp1 encodes rmpA and the siderophore salmochelin [114], and ICEKp10 encodes colibactin synthetic genes [111]. The most common appears to be ICEKp10, found in 77% of ST23, 40% of ST258 (the dominant carbapenem-resistant cKp ST), and in 25 other STs [111].
Defining hvKp – hypermucoviscosity
The most obvious phenotypic feature of hvKp, albeit flawed, is HMV. HMV is observed in culture conditions, normally attributed to excess capsule synthesis. The ‘string test’ has been used to define HMV, in which a colony of bacteria can be stretched to a viscous string at least 5 millimetres from the surface of the agar plate [8]. As such an obvious identifier, HMV and hypervirulent were used interchangeably to describe hvKp isolates; however for reasons described below, hypervirulence more accurately describes this pathotype. RmpA was first described in 1989 as a regulator of mucoid phenotype and encoded by the virulence plasmid pKP100 [105], along with RmpB, another virulence plasmid-encoded regulator [115]. Further analyses showed that K1 hvKp isolates from Asia frequently encode a chromosomal rmpA [116], whilst many hvKp strains encode only plasmid copies of rmpA and rmpA2 [116-118], making rmpA a frequently used diagnostic marker of hypervirulence. Another gene required for HMV and virulence is magA [8], but this gene appears restricted to K1 isolates [74] and is involved in K1 capsule synthesis [119]. Newly described transcriptional regulators KvrA and KvrB also activate transcription of the chromosomal capsule synthesis genes and therefore are required for both full capsule production and HMV in hvKp strains. Accordingly, kvrA and kvrB deletion strains are less virulent than wild type [120].
Despite the importance of HMV to hvKp, the connection between capsule and HMV is still being elucidated. Genetic dissection of the capsule regulatory pathway in an hvKp strain recently uncovered RmpC, a novel transcriptional regulator. Whilst RmpA appears to coordinate capsule and HMV, RmpC only regulates capsule; an rmpC mutant synthesizes less capsule, but retains hypermucoviscosity. This suggests that capsule synthesis is not required for the HMV phenotype [121]. Indeed, early reports of RmpA and RmpB proposed that capsule overexpression was not the basis for HMV [105, 115]. These new data open the door to further understand the differences between excess capsule and HMV in hvKp.
Whilst the well-recognized definition of hvKp, especially for the common K1 ST23 isolates, is rmpA+/rmpA2+, magA + with HMV phenotype, there are many exceptions to this rule. magA seems to be a marker of the K1 capsule type only. [119]. Highlighting the diversity amongst isolates, there are examples of rmpA−/HMV+ [9, 122-125], rmpA−/rmpA2+/HMV+ [10], and rmpA+/HMV− [9, 118]. Further, in an attempt to identify new biomarkers for clinical testing of hvKp, the string test performed only moderately well as a predictor of cKp versus hvKp [117]. Studies evaluating the correlation between clinical definitions of hvKp and the string test found varying results, ranging from 51% [126] to 98% [8] between studies. The HMV phenotype likely contributes to the majority of hvKp strains, but the exceptions here further point to the diversity of K. pneumoniae capable of hypervirulence.
Iron acquisition sets hvKp up for success
Iron is a critical molecule for bacteria and humans, required for many cellular processes. In the human host, there is very little free iron in tissue or serum as the result of the insolubility of Fe3+ under physiological conditions and host restriction of Fe2+ and Fe3+. Iron availability is generally restricted by a coordinated group of proteins that even more severely limits bioavailable iron during infection in a process called nutritional immunity [127]. Therefore, invading bacterial pathogens encode high affinity iron acquisition systems to counteract host nutritional immunity processes. hvKp expresses four siderophores for iron acquisition, and the accumulation of siderophore systems in hvKp suggests iron acquisition is a critical component of the emergence of this pathogen.
Siderophores are small molecules synthesized inside the bacterium and then exported. Outside the cell, siderophores bind iron in the environment with extremely high affinity and are transported back into the cell, providing the bacterium with iron required for growth. Both hvKp and cKP encode the enterobactin (Ent) system, and often yersiniabactin (Ybt), on the chromosome. On the other hand, hvKp isolates also often synthesize aerobactin (Iuc), and salmochelin (Iro) systems [78], and an hvKp isolate produces a greater mass of siderophores relative to cKp isolates [128]. There is a strong trend of co-occurence of iro and iuc loci in hvKp strains in a recent analysis of ~ 2500 Kp genomes, suggesting salmochelin is present in these genomes because it is important to the success of hvKp as well [129]. An additional analysis of 97 genomes of the CG23 hvKp clonal group detected ybt, iuc and iro loci in nearly all genomes [112]. The host immune protein lipocalin-2 can bind iron-bound Ent and prevent its return to the cell [130] in an attempt to thwart iron acquisition by bacteria. Ybt, Iuc and Iro are lipocalin-2 resistant, suggesting these accessory siderophores in hvKp may help overcome restriction by lipocalin-2 nutritional immunity, as observed for other Gram-negative pathogens [127]. In addition to the Ybt, Iuc and Iro siderophores, other iron acquisition systems likely support the pathogenesis of hvKp. For example, the commonly studied NTUH-K2044 K1 ST23 isolate also encodes ferrichrome, ferric, ferrous and haem-iron uptake systems [131], underscoring the need of hvKp for iron.
Aerobactin (luc) has long been appreciated as a critical siderophore system of hvKp [104], and as such, it has been proposed as an anti-virulence target [132]. Encoded in just four genes with a relatively simple synthetic pathway [133], Iuc appears to be the only siderophore system required in hvKp isolates in laboratory experiments, and in mouse models of disease, inactivation of other siderophore systems had minimal effect [134, 135]. However, the occurrence of multiple siderophore systems in hvKp strains suggests siderophore systems in addition to Iuc play important roles in the human host, either in colonization or invasive disease that largely remains to be uncovered.
Additional hvKp virulence factors
Another interesting molecular feature of hvKp is the ability to synthesize the genotoxin colibactin. Colibactin is synthesized as part of secondary metabolism by nonribosomal peptide synthetases, polyketide synthases and other enzymes (pks genes) first described in E. coli [136]. The pks locus is over-represented in hvKp relative to cKp isolates, in the majority (up to 100%) of K1 isolates tested, usually encoded in a chromosomal integrative and conjugative element (ICE) [11, 82, 100, 137]. Colibactin damages DNA and disrupts the host cell cycle, but the exact mechanisms by which colibactin contributes to pathogenesis of hvKp are largely unknown. Genetic inactivation of colibactin synthesis in a pks + K1 ST23 isolate reduced the ability of this hvKP strain to colonize the gut and disseminate to other organs in a murine model of systemic infection as well as to induce cell death in the brain during meningitis [138]. Taken together, colibactin appears to support the colonization and pathogenesis of hvKp and may have contributed to the global spread of CG23 [112], of which K1 ST23 strains are prominent.
The ability of hvKp isolates to form biofilms may also contribute to hypervirulence. Biofilms are complex surface-associated communities with extracellular matrix that helps provide protection from the immune system and environmental insults. K. pneumoniae biofilm formation is well appreciated, but the genetic basis for biofilm formation is unclear. Varying reports attribute the biofilm phenotype to capsule [139, 140] and/or fimbriae [140-142], whilst others have shown lack of capsule enhances biofilm formation [143]. hvKp strains seem to be more robust biofilm producers in the laboratory relative to non-tissue-invasive isolates in a survey of more than 70 isolates [140]. Alternatively, other studies have found no difference in biofilm formation between invasive and non-invasive isolates [144]. Therefore, more research is needed to define the genetic basis for biofilms in hvKp strains as well as how capsule and HMV contribute to the biofilms observed in vitro.
Whilst iron acquisition systems, colibactin and possibly biofilms support the virulence of hvKp, a comprehensive understanding of virulence determinants is still lacking. The virulence plasmids encoded by hvKp isolates are generally 200 kb, yet only a handful of genes have been shown to be required for virulence. In addition to those discussed above, some virulence factors have been experimentally validated in mouse models of disease including: a metabolite transporter peg-344 [145], mrk fimbriae [28], the moaR and kva15 regulators [28], a kvgAS signalling system [28, 146], phospholipase D [147] and the allantoin metabolism gene cluster [148]. However, virulence factors identified across these various mouse models using different strains of hvKp surely only represent a fraction of the genes hvKp have acquired to cause disease in healthy hosts. An all-encompassing view of the hvKp virulence determinants is needed to improve diagnostics and identify new antibacterial targets.
Convergence of virulence and antibiotic resistance
In the first few decades after the emergence of hvKp, widespread antibiotic susceptibility of hvKp isolates allowed for uncomplicated treatment. Early reports investigating resistance amongst hvKp found very low levels of resistance, with less than 5% of hvKp bacteremia isolates producing extended-spectrum-β-lactamase (ESBL) genes [149], and 2% or less resistant to any single antibiotic tested [6]. However, almost four decades after the description of hvKp, it is now clear that antibiotic-resistant hvKp isolates have emerged and are of great clinical concern. Alarmingly, in a 2016 study from China, 20 of the 35 (57%) hvKp isolates causing bloodstream infections were carbapenemase producing [58]. There have also been reports of ESBL-producing hypervirulent isolates [9, 60, 62, 150], which will represent an additional clinical challenge. The spread of antibiotic-resistant isolates capable of invasive disease in healthy individuals has potential for great global impact. The relatively high prevalence of hvKp and carbapenem-resistant-cKP in China has been proposed as a major contributor to the convergence of resistance and virulence, and neighbouring regions should be aware of the potential for dissemination of such convergent clones [151]. It is clear, however, that isolates with both traits already exist in the United States [152, 153], Europe [150] and other areas outside China, either from dissemination or independent convergences.
Two types of convergence have been observed: cKp isolates acquiring virulence genes or entire virulence plasmids and hvKp isolates gaining chromosomal or plasmid-encoded antibiotic resistance genes. A sampling of recent examples is displayed in Table 2. Analyses of carbapenem-resistant K. pneumoniae clinical isolates have identified rates of hvKp near 10%, though this is usually assessed by the string test [10, 154-156]. Because genes that encode virulence or antibiotic resistance are often on mobile genetic elements like plasmids and ICE, it is not surprising that we are observing a convergence of virulence and antibiotic resistance.
Table 2.
A non-exhaustive listing of recent clinical isolates demonstrating multidrug resistance and hypervirulence
| Location | Sequence Type (ST) |
Capsule (K) | Clinical context | Notesa | Ref |
|---|---|---|---|---|---|
| Classical strains that acquired virulence genes | |||||
| China | ST11 | K47 | Ventilator-associated pneumonia | 5 isolates; widespread resistance; HMV+, 170 kb pLVPK-like plasmid of ST43. Retrospective analysis suggested ST11 isolates acquired virulence multiple times | [55] |
| ST11 | K47 | Retrospective study | pLVPK-like plasmid and two AR plasmids; unique feature of five tandem copies of blaKPC-2 | [174] | |
| India | ST147 | K54 | Sepsis and kidney injury | HMV+, limited acquisition of virulence genes, colistin resistant | [175] |
| United States | ST258 | K107 | Retrospective study | HMV+/rmpA−, a few virulence genes on hybrid AR plasmid; chromosomal siderophore receptor genes | [152] |
| Norway | ST15 | Two bacteremia isolates | 300 or 346 kb mosaic plasmids of pK2044-like virulence plasmid and conjugative AR plasmid | [176] | |
| China | ST15 | Elderly patients with pneumonia and other lung trauma | Clonal expansion of blaOXA-232 encoding isolate; virulence genes detected but not hypervirulent in laboratory models | [177] | |
| hvKp isolates acquiring resistance | |||||
| China | ST23 | K1 | Two bloodstream isolates | blaSHV-36 with colistin resistance in addition to pLVPK-like plasmid and canonical virulence genes | [178] |
| China | ST23 | K1 | Bloodstream, kidney abscess | Hybrid of pLVPK-like virulence plasmid with transposon-mediated integration of blaCTX-M-24 | [160] |
| United States | ST23 | K1 | Urine sample | blaSHV-36, fosA, oqxAB on chromosome; AR plasmid with blaKPC-2, blaTEM-1A and truncated blaOXA-9 | [153] |
| China | ST23 | K1 | Sepsis | pLVPK-like virulence plasmid with insertion of blaKPC-2 and dfrA14 genes | [158] [12] |
| China | ST36 | K62 | Bloodstream; burn wounds | pLVPK-like plasmid and AR plasmid encoding blaKPC-2, fosA, oqxAB, along with resistance to aminoglycosides, macrolides, sulfonamides and others | [179] |
| China | ST65 | K2 | Meningitis | Most canonical virulence genes on chromosome and pLVPK-like plasmid; encodes blaSHV-11, blaTEM-1, blaCTx-M-3, blaCTX-M-4 | [180] |
| China | ST65 | K2 | Infant bloodstream | Encodes ent and iuc but not ybt or kfu; hypervirulent in mouse model; blaSHV-11, blaTEM-53, decreased expression of ompK35/36 | [155] |
| France | ST86 | K2 | Carriage | Encodes blaCTX-M-3 | [150] |
| China | ST86 | K2 | Burn wound | blaKPC-2 and blaNDM-1; 215 kb virulence plasmid | [181] |
| Canada | ST86 | KL2 | UTI | pLVPK-like plasmid; plasmid with blaKPC-2 as well as blaSHV-1 and fosA | [182] |
AR, antimicrobial resistance; bla, beta-lactamase provides resistance to penicillins, first- and second-generation cephalosporins; blaCTX-M-24, blaCTX-M-3 and blaCTX-M-4, ESBLs with particular activity against cefotaxime; blaKPC-2, bla (K. pneumoniae carbapenem resistance) with carbapenemase activity; blaNDM-1, bla (New Delhi metallo-bla) with carbapenemase activity; blaOXA-32 and blaOXA-9, ESBLs with particular activity against oxacillin; blaSHV-36 and blaSHV-11, ESBLs; blaTEM-1A, blaTEM-1, and blaTEM-53, ESBLs; dfrA14, trimethoprim resistance gene; ESBL, extended-spectrum beta-lactamase provides additional resistance to monobactams and third-generation cephalosporins; fosA, fosfomycin resistance gene; kb, kilobase, length of DNA molecule; ompK35/36 outer membrane porins that allow entry carbapenems and cephalosporins, reduced expression increases resistance; oqxAB, resistance to olaquindox and some quinolones.
cKp strains that acquire virulence genes have potential to cause extremely problematic infections. The identity and quantity of virulence genes that a cKp strain has acquired likely will dictate its virulence potential, and a spectrum of virulence potential likely exists based on different gene acquisitions. The fatal outbreak in five patients caused by an ST11 cKp strain that became hvKp by acquiring a ~ 170 kb pLVPK-like plasmid demonstrates how devastating acquisition of entire virulence plasmids can be [55]. Retrospective analyses of carbapenem-resistant isolates have identified strains exhibiting HMV and other markers of virulence, which includes diverse sequence types. We recently identified a multidrug-resistant ST158 cKp strain demonstrating HMV yet lacking a pLVPK virulence plasmid; in a mouse model of infection, it is more virulent than cKp strains but less so than well-studied ST23 hvKp [152]. Another analysis of a collection of K. pneumoniae causing skin and soft tissue infections, which are typically caused by hvKp, identified four isolates of carbapenem-resistant strains, including one of which was a string test positive K2 ST14 type [157]. This isolate lacked the rmpA/rmpA2 genes and the yersiniabactin system, but encoded receptors (although not biosynthetic genes) for aerobactin and salmochelin. Accordingly, the isolate was not as virulent as classical hvKp NTUH-K2044 in mouse models of infection but appeared adapted to cause skin and soft tissue infections in a murine model of skin abscess formation [157]. The acquisition of siderophore receptors without biosynthetic genes suggests that some hvKp isolates might be capable of ‘stealing’ siderophores from other species, presumably in the gut environment or in polymicrobial skin infections. This isolate is consistent with ST14 having some virulence genes but also high rates of antibiotic resistance and causing nosocomial infections [78] and highlights the diversity of strains capable of causing invasive infections.
HvKp isolates have also acquired antimicrobial resistance genes. A survey of global K. pneumoniae isolates detected antimicrobial resistance genes in multiple K2 ST43 isolates [78]. A recent example includes a K1 ST23 hvKp isolated in the United States, which appeared to be asymptomatically colonizing a patient [153]. This strain encoded two plasmids, the first being a large hybrid virulence and resistance plasmid that encoded carbapenem resistance via blaKPC-2 similar to other hybrid plasmids reported in China [158, 159]. The second plasmid, which encodes blaKPC-2 andblaTEM-1A, is homologous to plasmids reported to be responsible for the dissemination of carbapenem resistance amongst Enterobacteriaceae [153]. Another example of an hvKp strain becoming multidrug resistant is strain 11492, isolated in China. 11492 is a K1 ST23 isolate, found to be as HMV and virulent as the model hvKP strain NTUH-K2044 but with widespread antibiotic resistance [160]. 11492 encodes most hvKp virulence genes on the chromosome but also has a hybrid virulence/antibiotic resistance plasmid that encodes salmochelin, rmpA/rmpA2, and blaCTX-M-24. Additional antibiotic resistance genes blaSHV-36, oqxA/oqxB (olaquindox and some quinolones [161]) and fosA (fosfomycin [162]) are encoded in the chromosome [160]. This strain represents the emergence of antibiotic resistance in an hvKp strain via acquisition of chromosomal and plasmid genes. Widespread, highly virulent hvKp strains armed with multidrug-resistant determinants are a great cause for concern. These examples, along with others presented in Table 2, show that the co-occurrence of virulence and antimicrobial resistance is becoming a global and widespread problem that must inform surveillance and treatment.
Outstanding questions and outlook
Global dissemination of hvKp.
Following early reports of hvKp in Asia, it is now apparent that hvKp has spread globally, including India [163, 164], Europe [32, 34, 35, 37, 165-168], Australia [169, 170] and the United States [5, 30, 31, 36, 53, 171, 172] and healthcare providers worldwide should recognize the threat of community-acquired hvKp in otherwise healthy individuals. The rise of hospital-acquired hvKp which may manifest in different presentations, and the convergence of multidrug resistance (MDR) and virulence either in cKp or hvKp strains, is particularly concerning. Methods to more rapidly and easily identify both hypervirulence and antibiotic resistance could improve treatment and patient outcomes.
The HMV phenotype
There are many outstanding questions regarding the basis and impact of HMV. As mentioned above, the connection between capsule synthesis and HMV is murky, and the biochemical nature of HMV is unclear. One might also speculate that HMV has important features distinct from capsule. Further analysis of the rmpC or similar mutants with reduced capsule but with HMV [121] could test the contribution of HMV to gastrointestinal colonization, organ invasion, immune evasion and desiccation tolerance. The HMV phenotype might also affect antimicrobial susceptibility testing, which could become a more frequent issue with increasing MDR.
Advanced diagnostics are needed for the rapid identification of hvKp in the clinic
Based on the diversity of STs, capsule (K) types and the presence of HMV in hvKp isolates, considerations must be made to efficiently diagnose an isolate as hvKp to inform treatment and patient care. Reliance on HMV has caused some hvKp isolates to be undetected, as demonstrated by the accounts of hvKp without a positive string test. Instead, a broader and more comprehensive diagnostic test is needed. A study of potential biomarkers for use in identifying hvKp isolates found that the presence of peg-344, iroB, iucA, plasmid-encoded rmpA/rmpA2 and quantification of siderophore production were accurate predictors of hvKp [117]. Some combination of these biomarkers should be considered as a diagnostic tool for rapid clinical identification of hvKp in the hospital.
The rise of MDR hvKp
One major question regarding the convergence of hypervirulence and MDR is restricted acquisition of antibiotic resistance genes by hvKP relative to cKp strains. Large genomic analysis of hvKp has identified relatively low diversity of capsule types and overall genome diversity, despite the emergence of CG23 in the late 1800s [112]. Others have speculated that there may be a barrier to horizontal gene transfer amongst hvKp clones which has largely limited acquisition of AR genes [173]. This barrier may include the upregulation of capsule which obfuscates the conjugation machinery and lowers the efficiency of transformation leading to DNA uptake [173]. In order to predict the spread of MDR in hvKp strains, it is important to understand the ecological or molecular barriers to acquisition of MDR plasmids by hvKp stains.
Surveillance and detection of MDR and hvKp
There are networks in place for detection and surveillance of carbapenem resistance in K. pneumoniae, and these networks should consider adding mechanisms to detect hvKp. Recent retrospective analyses of these surveillance collections have identified hypervirulent traits amongst carbapenem-resistant isolates in the United States, [152, 153], suggesting prospective surveillance could help uncover strains exhibiting both features.
Acknowledgements
We thank Victor Band, David Hufnagel, Hannah Ratner and Jessie Wozniak for critical evaluation of this manuscript. J.E.C. is supported by NIH Grant T32 DK108735. D.S.W. is supported by a Burroughs Wellcome Fund Investigator in the Pathogenesis of Infectious Disease award, NIH grant AI141883, and VA Merit Award I01 BX002788. J.H.A is in part supported by the National Center for Advancing Translational Sciences of the National Institutes of Health [UL1TR002378 and TL1TR002382]. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Conflict of Interest Statement
The authors declare no conflict of interest related to this review.
References
- 1.WHO. Global Priority List of Antibiotic-resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics. Geneva: World Health Organization; 2017 [Google Scholar]
- 2.Restuccia PA, Cunha BA. Klebsiella. Infect Control 1984; 5: 343–7. [DOI] [PubMed] [Google Scholar]
- 3.Magill SS, O’Leary E, Janelle SJ et al. Changes in Prevalence of Health Care-Associated Infections in U.S. Hospitals. N Engl J Med 2018; 379: 1732–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu YC, Cheng DL, Lin CL. Klebsiella pneumoniae liver abscess associated with septic endophthalmitis. Arch Intern Med 1986; 146: 1913–6. [PubMed] [Google Scholar]
- 5.Lederman ER, Crum NF. Pyogenic liver abscess with a focus on Klebsiella pneumoniae as a primary pathogen: an emerging disease with unique clinical characteristics. Am J Gastroenterol 2005; 100: 322–31. [DOI] [PubMed] [Google Scholar]
- 6.Fang CT, Lai SY, Yi WC, Hsueh PR, Liu KL, Chang SC. Klebsiella pneumoniae genotype K1: an emerging pathogen that causes septic ocular or central nervous system complications from pyogenic liver abscess. Clin Infect Dis 2007; 45: 284–93. [DOI] [PubMed] [Google Scholar]
- 7.Pomakova DK, Hsiao CB, Beanan JM et al. Clinical and phenotypic differences between classic and hypervirulent Klebsiella pneumonia: an emerging and under-recognized pathogenic variant. Eur J Clin Microbiol Infect Dis 2012; 31: 981–9. [DOI] [PubMed] [Google Scholar]
- 8.Fang CT, Chuang YP, Shun CT, Chang SC, Wang JT. A novel virulence gene in Klebsiella pneumoniae strains causing primary liver abscess and septic metastatic complications. J Exp Med 2004; 199: 697–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li W, Sun G, Yu Y et al. Increasing occurrence of antimicrobial-resistant hypervirulent (hypermucoviscous) Klebsiella pneumoniae isolates in China. Clin Infect Dis 2014; 58: 225–32. [DOI] [PubMed] [Google Scholar]
- 10.Liu Z, Gu Y, Li X et al. Identification and Characterization of NDM-1-producing Hypervirulent (Hypermucoviscous) Klebsiella pneumoniae in China. Ann Lab Med 2019; 39: 167–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lan Y, Zhou M, Jian Z, Yan Q, Wang S, Liu W. Prevalence of pks gene cluster and characteristics of Klebsiella pneumoniae-induced bloodstream infections. J Clin Lab Anal 2019: 33; e22838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang R, Lin D, Chan EW, Gu D, Chen GX, Chen S. Emergence of carbapenem-resistant serotype K1 Hypervirulent Klebsiella pneumoniae strains in China. Antimicrob Agents Chemother 2016; 60: 709–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu C, Shi J, Guo J. High prevalence of hypervirulent Klebsiella pneumoniae infection in the genetic background of elderly patients in two teaching hospitals in China. Infect Drug Resist 2018; 11: 1031–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bagley ST. Habitat association of Klebsiella species. Infect Control 1985; 6: 52–8. [DOI] [PubMed] [Google Scholar]
- 15.Conlan S, Kong HH, Segre JA. Species-level analysis of DNA sequence data from the NIH Human Microbiome Project. PLoS ONE 2012; 7: e47075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Casewell M, Phillips I. Hands as route of transmission for Klebsiella species. Br Med J 1977; 2: 1315–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davis TJ, Matsen JM. Prevalence and characteristics of Klebsiella species: relation to association with a hospital environment. J Infect Dis 1974; 130: 402–5. [DOI] [PubMed] [Google Scholar]
- 18.Rock C, Thom KA, Masnick M, Johnson JK, Harris AD, Morgan DJ. Frequency of Klebsiella pneumoniae carbapenemase (KPC)-producing and non-KPC-producing Klebsiella species contamination of healthcare workers and the environment. Infect Control Hosp Epidemiol 2014; 35: 426–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gorrie CL, Mirceta M, Wick RR et al. Gastrointestinal carriage is a major reservoir of Klebsiella pneumoniae infection in intensive care patients. Clin Infect Dis 2017; 65: 208–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martin RM, Cao J, Brisse S et al. Molecular epidemiology of colonizing and infecting isolates of Klebsiella pneumoniae. mSphere 2016; 1: e00261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jarvis WR, Munn VP, Highsmith AK, Culver DH, Hughes JM. The epidemiology of nosocomial infections caused by Klebsiella pneumoniae. Infect Control 1985; 6: 68–74. [DOI] [PubMed] [Google Scholar]
- 22.Snitkin ES, Zelazny AM, Thomas PJ et al. Tracking a hospital outbreak of carbapenem-resistant Klebsiella pneumoniae with whole-genome sequencing. Sci Transl Med 2012; 4: 148ra16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marchaim D, Chopra T, Pogue JM et al. Outbreak of colistin-resistant, carbapenem-resistant Klebsiella pneumoniae in metropolitan Detroit. Michigan. Antimicrob Agents Chemother 2011; 55: 593–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thom BT. Klebsiella in faeces. Lancet 1970; 2: 1033. [DOI] [PubMed] [Google Scholar]
- 25.Fung CP, Lin YT, Lin JC et al. Klebsiella pneumoniae in gastrointestinal tract and pyogenic liver abscess. Emerg Infect Dis 2012; 18: 1322–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chung DR, Lee H, Park MH et al. Fecal carriage of serotype K1 Klebsiella pneumoniae ST23 strains closely related to liver abscess isolates in Koreans living in Korea. Eur J Clin Microbiol Infect Dis 2012; 31: 481–6. [DOI] [PubMed] [Google Scholar]
- 27.Lin YT, Siu LK, Lin JC et al. Seroepidemiology of Klebsiella pneumoniae colonizing the intestinal tract of healthy Chinese and overseas Chinese adults in Asian countries. BMC Microbiol 2012; 12: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tu YC, Lu MC, Chiang MK et al. Genetic requirements for Klebsiella pneumoniae-induced liver abscess in an oral infection model. Infect Immun 2009; 77: 2657–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shon AS, Bajwa RP, Russo TA. Hypervirulent (hypermucoviscous) Klebsiella pneumoniae: a new and dangerous breed. Virulence 2013; 4: 107–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Siu LK, Yeh KM, Lin JC, Fung CP, Chang FY. Klebsiella pneumoniae liver abscess: a new invasive syndrome. Lancet Infect Dis 2012; 12: 881–7. [DOI] [PubMed] [Google Scholar]
- 31.McCabe R, Lambert L, Frazee B. Invasive Klebsiella pneumoniae infections, California, USA. Emerg Infect Dis 2010; 16: 1490–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Decre D, Verdet C, Emirian A et al. Emerging severe and fatal infections due to Klebsiella pneumoniae in two university hospitals in France. J Clin Microbiol 2011; 49: 3012–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Patel PK, Russo TA, Karchmer AW. Hypervirulent Klebsiella pneumoniae. Open Forum Infect Dis 2014; 1: ofu028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Monie M, Drieux L, Nzili B et al. Klebsiella pneumoniae necrotizing fasciitis of the leg in an elderly French woman. Clin Interv Aging 2014; 9: 1171–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sobirk SK, Struve C, Jacobsson SG. Primary Klebsiella pneumoniae Liver abscess with metastatic spread to lung and eye, a north-European case report of an emerging syndrome. Open Microbiol J 2010; 4: 5–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nadasy KA, Domiati-Saad R, Tribble MA. Invasive Klebsiella pneumoniae syndrome in North America. Clin Infect Dis 2007; 45: e25–8. [DOI] [PubMed] [Google Scholar]
- 37.Moore R, O’Shea D, Geoghegan T, Mallon PW, Sheehan G. Community-acquired Klebsiella pneumoniae liver abscess: an emerging infection in Ireland and Europe. Infection 2013; 41: 681–6. [DOI] [PubMed] [Google Scholar]
- 38.Togawa A, Toh H, Onozawa K et al. Influence of the bacterial phenotypes on the clinical manifestations in Klebsiella pneumoniae bacteremia patients: A retrospective cohort study. J Infect Chemother 2015; 21: 531–7. [DOI] [PubMed] [Google Scholar]
- 39.Catalan-Najera JC, Garza-Ramos U, Barrios-Camacho H. Hypervirulence and hypermucoviscosity: Two different but complementary Klebsiella spp. phenotypes? Virulence 2017; 8: 1111–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cheng DL, Liu YC, Yen MY, Liu CY, Wang RS. Septic metastatic lesions of pyogenic liver abscess. Their association with Klebsiella pneumoniae bacteremia in diabetic patients. Arch Intern Med 1991; 151: 1557–9. [PubMed] [Google Scholar]
- 41.Lee SS, Chen YS, Tsai HC et al. Predictors of septic metastatic infection and mortality among patients with Klebsiella pneumoniae liver abscess. Clin Infect Dis 2008; 47: 642–50. [DOI] [PubMed] [Google Scholar]
- 42.Fung CP, Chang FY, Lee SC et al. A global emerging disease of Klebsiella pneumoniae liver abscess: is serotype K1 an important factor for complicated endophthalmitis? Gut 2002; 50: 420–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chiu CT, Lin DY, Liaw YF. Metastatic septic endophthalmitis in pyogenic liver abscess. J Clin Gastroenterol 1988; 10: 524–7. [DOI] [PubMed] [Google Scholar]
- 44.Saccente M Klebsiella pneumoniae liver abscess, endophthalmitis, and meningitis in a man with newly recognized diabetes mellitus. Clin Infect Dis 1999; 29: 1570–1. [DOI] [PubMed] [Google Scholar]
- 45.Chou FF, Kou HK. Endogenous endophthalmitis associated with pyogenic hepatic abscess. J Am Coll Surg 1996; 182: 33–6. [PubMed] [Google Scholar]
- 46.Kuramochi G, Takei SI, Sato M, Isokawa O, Takemae T, Takahashi A. Klebsiella pneumoniae liver abscess associated with septic spinal epidural abscess. Hepatol Res 2005; 31: 48–52. [DOI] [PubMed] [Google Scholar]
- 47.Hu BS, Lau YJ, Shi ZY, Lin YH. Necrotizing fasciitis associated with Klebsiella pneumoniae liver abscess. Clin Infect Dis 1999; 29: 1360–1. [DOI] [PubMed] [Google Scholar]
- 48.Dylewski JS, Dylewski I. Necrotizing fasciitis with Klebsiella liver abscess. Clin Infect Dis 1998; 27: 1561–2. [DOI] [PubMed] [Google Scholar]
- 49.Goldman JM, Kowalec JK. Hepatic abscess and osteomyelitis from Klebsiella pneumoniae. JAMA 1978; 240: 2660. [PubMed] [Google Scholar]
- 50.Rivero A, Gomez E, Alland D, Huang DB, Chiang T. K2 serotype Klebsiella pneumoniae causing a liver abscess associated with infective endocarditis. J Clin Microbiol 2010; 48: 639–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee HC, Chuang YC, Yu WL et al. Clinical implications of hypermucoviscosity phenotype in Klebsiella pneumoniae isolates: association with invasive syndrome in patients with community-acquired bacteraemia. J Intern Med 2006; 259: 606–14. [DOI] [PubMed] [Google Scholar]
- 52.Ku YH, Chuang YC, Yu WL. Clinical spectrum and molecular characteristics of Klebsiella pneumoniae causing community-acquired extrahepatic abscess. J Microbiol Immunol Infect 2008; 41: 311–7. [PubMed] [Google Scholar]
- 53.Prokesch BC, TeKippe M, Kim J, Raj P, TeKippe EM, Greenberg DE. Primary osteomyelitis caused by hypervirulent Klebsiella pneumoniae. Lancet Infect Dis 2016; 16: e190–e5. [DOI] [PubMed] [Google Scholar]
- 54.Chang CM, Ko WC, Lee HC, Chen YM, Chuang YC. Klebsiella pneumoniae psoas abscess: predominance in diabetic patients and grave prognosis in gas-forming cases. J Microbiol Immunol Infect 2001; 34: 201–6. [PubMed] [Google Scholar]
- 55.Gu D, Dong N, Zheng Z et al. A fatal outbreak of ST11 carbapenem-resistant hypervirulent Klebsiella pneumoniae in a Chinese hospital: a molecular epidemiological study. Lancet Infect Dis 2018; 18: 37–46. [DOI] [PubMed] [Google Scholar]
- 56.Hyun M, Lee JY, Ryu SY, Ryoo N, Kim HA. Antibiotic resistance and clinical presentation of health care-associated hypervirulent Klebsiella pneumoniae infection in Korea. Microb Drug Resist 2019; 25: 1204–1209. [DOI] [PubMed] [Google Scholar]
- 57.Yan Q, Zhou M, Zou M, Liu WE. Hypervirulent Klebsiella pneumoniae induced ventilator-associated pneumonia in mechanically ventilated patients in China. Eur J Clin Microbiol Infect Dis 2016; 35: 387–96. [DOI] [PubMed] [Google Scholar]
- 58.Li J, Ren J, Wang W et al. Risk factors and clinical outcomes of hypervirulent Klebsiella pneumoniae induced bloodstream infections. Eur J Clin Microbiol Infect Dis 2018; 37: 679–89. [DOI] [PubMed] [Google Scholar]
- 59.Kim JK, Chung DR, Wie SH, Yoo JH, Park SW, Korean Study Group for Liver A. Risk factor analysis of invasive liver abscess caused by the K1 serotype Klebsiella pneumoniae. Eur J Clin Microbiol Infect Dis 2009; 28: 109–11. [DOI] [PubMed] [Google Scholar]
- 60.Liu C, Guo J. Hypervirulent Klebsiella pneumoniae (hypermucoviscous and aerobactin positive) infection over 6 years in the elderly in China: antimicrobial resistance patterns, molecular epidemiology and risk factor. Ann Clin Microbiol Antimicrob 2019; 18: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang JH, Liu YC, Lee SS et al. Primary liver abscess due to Klebsiella pneumoniae in Taiwan. Clin Infect Dis 1998; 26: 1434–8. [DOI] [PubMed] [Google Scholar]
- 62.Zhang Y, Zhao C, Wang Q et al. High prevalence of hypervirulent Klebsiella pneumoniae infection in China: geographic distribution, clinical characteristics, and antimicrobial resistance. Antimicrob Agents Chemother 2016; 60: 6115–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yoon JH, Kim YJ, Jun YH et al. Liver abscess due to Klebsiella pneumoniae: risk factors for metastatic infection. Scand J Infect Dis 2014; 46: 21–6. [DOI] [PubMed] [Google Scholar]
- 64.Lin YT, Liu CJ, Chen TJ, Fung CP. Long-term mortality of patients with septic ocular or central nervous system complications from pyogenic liver abscess: a population-based study. PLoS ONE 2012; 7: e33978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lee IR, Molton JS, Wyres KL et al. Differential host susceptibility and bacterial virulence factors driving Klebsiella liver abscess in an ethnically diverse population. Sci Rep 2016; 6: 29316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lin YT, Cheng YH, Juan CH et al. High mortality among patients infected with hypervirulent antimicrobial-resistant capsular type K1 Klebsiella pneumoniae strains in Taiwan. Int J Antimicrob Agents 2018; 52: 251–7. [DOI] [PubMed] [Google Scholar]
- 67.Donnenberg M 220-Enterobacteriaceae In: Bennett J, Dolin R, Blaser M, eds. Mandell, Douglas, and Bennett’s Principles and Practice of Infectious Diseases (Eighth Edition). Philadelphia, PA: Saunders, an imprint of Elsevier, 2015; 2503–17. [Google Scholar]
- 68.Russo TA, Marr CM. Hypervirulent Klebsiella pneumoniae. Clin Microbiol Rev 2019; 32: e00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Marr CM, Russo TA. Hypervirulent Klebsiella pneumoniae: a new public health threat. Expert Rev Anti Infect Ther 2019; 17: 71–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pouch SM, Patel G, Practice ASTIDCo. Multidrug-resistant Gram-negative bacterial infections in solid organ transplant recipients-Guidelines from the American Society of Transplantation Infectious Diseases Community of Practice. Clin Transplant 2019: 33; e13594. [DOI] [PubMed] [Google Scholar]
- 71.Diancourt L, Passet V, Verhoef J, Grimont PA, Brisse S. Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J Clin Microbiol 2005; 43: 4178–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wyres KL, Holt KE. Klebsiella pneumoniae population genomics and antimicrobial-resistant clones. Trends Microbiol 2016; 24: 944–56. [DOI] [PubMed] [Google Scholar]
- 73.Paczosa MK, Mecsas J. Klebsiella pneumoniae: Going on the Offense with a Strong Defense. Microbiol Mol Biol Rev 2016; 80: 629–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yeh KM, Kurup A, Siu LK et al. Capsular serotype K1 or K2, rather than magA and rmpA, is a major virulence determinant for Klebsiella pneumoniae liver abscess in Singapore and Taiwan. J Clin Microbiol 2007; 45: 466–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lawlor MS, Handley SA, Miller VL. Comparison of the host responses to wild-type and cpsB mutant Klebsiella pneumoniae infections. Infect Immun 2006; 74: 5402–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kauffmann F On the serology of the Klebsiella Group. Acta Pathologica Microbiologica Scandinavica 1949; 26: 381–406. [Google Scholar]
- 77.Wyres KL, Wick RR, Gorrie C et al. Identification of Klebsiella capsule synthesis loci from whole genome data. Microb Genom 2016; 2: e000102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Holt KE, Wertheim H, Zadoks RN et al. Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiella pneumoniae, an urgent threat to public health. Proc Natl Acad Sci USA 2015; 112: E3574–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yu WL, Ko WC, Cheng KC, Lee CC, Lai CC, Chuang YC. Comparison of prevalence of virulence factors for Klebsiella pneumoniae liver abscesses between isolates with capsular K1/K2 and non-K1/K2 serotypes. Diagn Microbiol Infect Dis 2008; 62: 1–6. [DOI] [PubMed] [Google Scholar]
- 80.Pan YJ, Fang HC, Yang HC et al. Capsular polysaccharide synthesis regions in Klebsiella pneumoniae serotype K57 and a new capsular serotype. J Clin Microbiol 2008; 46: 2231–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Follador R, Heinz E, Wyres KL et al. The diversity of Klebsiella pneumoniae surface polysaccharides. Microb Genom 2016; 2: e000073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Struve C, Roe CC, Stegger M et al. Mapping the evolution of hypervirulent Klebsiella pneumoniae. MBio 2015; 6: e00630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lin CL, Chen FH, Huang LY et al. Effect in virulence of switching conserved homologous capsular polysaccharide genes from Klebsiella pneumoniae serotype K1 into K20. Virulence 2017; 8: 487–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ofek I, Kabha K, Athamna A et al. Genetic exchange of determinants for capsular polysaccharide biosynthesis between Klebsiella pneumoniae strains expressing serotypes K2 and K21a. Infect Immun 1993; 61: 4208–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kabha K, Nissimov L, Athamna A et al. Relationships among capsular structure, phagocytosis, and mouse virulence in Klebsiella pneumoniae. Infect Immun 1995; 63: 847–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Athamna A, Ofek I, Keisari Y, Markowitz S, Dutton GG, Sharon N. Lectinophagocytosis of encapsulated Klebsiella pneumoniae mediated by surface lectins of guinea pig alveolar macrophages and human monocyte-derived macrophages. Infect Immun 1991; 59: 1673–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Pan YJ, Lin TL, Chen CT et al. Genetic analysis of capsular polysaccharide synthesis gene clusters in 79 capsular types of Klebsiella spp. Sci Rep 2015; 5: 15573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ho JY, Lin TL, Li CY et al. Functions of some capsular polysaccharide biosynthetic genes in Klebsiella pneumoniae NTUH K-2044. PLoS ONE 2011; 6: e21664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Corsaro MM, De Castro C, Naldi T, Parrilli M, Tomas JM, Regue M. 1H and 13C NMR characterization and secondary structure of the K2 polysaccharide of Klebsiella pneumoniae strain 52145. Carbohydr Res 2005; 340: 2212–7. [DOI] [PubMed] [Google Scholar]
- 90.van Dam JE, van Halbeek H, Kamerling JP et al. A bacteriophage-associated lyase acting on Klebsiella serotype K5 capsular polysaccharide. Carbohydr Res 1985; 142: 338–43. [DOI] [PubMed] [Google Scholar]
- 91.Kamerling JP, Lindberg B, Lonngren J, Nimmich W. Structural studies of the Klebsiella type 57 capsular polysaccharide. Acta Chem Scand B 1975; 29: 593–8. [DOI] [PubMed] [Google Scholar]
- 92.Wang L, Shen D, Wu H, Ma Y. Resistance of hypervirulent Klebsiella pneumoniae to both intracellular and extracellular killing of neutrophils. PLoS ONE 2017; 12: e0173638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lin JC, Chang FY, Fung CP et al. Do neutrophils play a role in establishing liver abscesses and distant metastases caused by Klebsiella pneumoniae? PLoS ONE 2010; 5: e15005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yang ZQ, Huang YL, Zhou HW, Zhang R, Zhu K. Persistent carbapenem-resistant Klebsiella pneumoniae: a Trojan horse. Lancet Infect Dis 2018; 18: 22–3. [DOI] [PubMed] [Google Scholar]
- 95.Brisse S, Fevre C, Passet V et al. Virulent clones of Klebsiella pneumoniae: identification and evolutionary scenario based on genomic and phenotypic characterization. PLoS ONE 2009; 4: e4982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wu H, Li D, Zhou H, Sun Y, Guo L, Shen D. Bacteremia and other body site infection caused by hypervirulent and classic Klebsiella pneumoniae. Microb Pathog 2017; 104: 254–62. [DOI] [PubMed] [Google Scholar]
- 97.Liao CH, Huang YT, Chang CY, Hsu HS, Hsueh PR. Capsular serotypes and multilocus sequence types of bacteremic Klebsiella pneumoniae isolates associated with different types of infections. Eur J Clin Microbiol Infect Dis 2014; 33: 365–9. [DOI] [PubMed] [Google Scholar]
- 98.Luo Y, Wang Y, Ye L, Yang J. Molecular epidemiology and virulence factors of pyogenic liver abscess causing Klebsiella pneumoniae in China. Clin Microbiol Infect 2014; 20: O818–24. [DOI] [PubMed] [Google Scholar]
- 99.Qu TT, Zhou JC, Jiang Y et al. Clinical and microbiological characteristics of Klebsiella pneumoniae liver abscess in East China. BMC Infect Dis 2015; 15: 161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Chen YT, Lai YC, Tan MC et al. Prevalence and characteristics of pks genotoxin gene cluster-positive clinical Klebsiella pneumoniae isolates in Taiwan. Sci Rep 2017; 7: 43120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Harada S, Ishii Y, Saga T, Aoki K, Tateda K. Molecular epidemiology of Klebsiella pneumoniae K1 and K2 isolates in Japan. Diagn Microbiol Infect Dis 2018; 91: 354–9. [DOI] [PubMed] [Google Scholar]
- 102.French JB, Neau DB, Ealick SE. Characterization of the structure and function of Klebsiella pneumoniae allantoin racemase. J Mol Biol 2011; 410: 447–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Arena F, Spanu T, Henrici De Angelis L et al. First case of bacteremic liver abscess caused by an ST260-related (ST1861), hypervirulent Klebsiella pneumoniae. J Infect 2016; 73: 88–91. [DOI] [PubMed] [Google Scholar]
- 104.Nassif X, Sansonetti PJ. Correlation of the virulence of Klebsiella pneumoniae K1 and K2 with the presence of a plasmid encoding aerobactin. Infect Immun 1986; 54: 603–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Nassif X, Fournier JM, Arondel J, Sansonetti PJ. Mucoid phenotype of Klebsiella pneumoniae is a plasmid-encoded virulence factor. Infect Immun 1989; 57: 546–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Tang HL, Chiang MK, Liou WJ et al. Correlation between Klebsiella pneumoniae carrying pLVPK-derived loci and abscess formation. Eur J Clin Microbiol Infect Dis 2010; 29: 689–98. [DOI] [PubMed] [Google Scholar]
- 107.Wu KM, Li LH, Yan JJ et al. Genome sequencing and comparative analysis of Klebsiella pneumoniae NTUH-K2044, a strain causing liver abscess and meningitis. J Bacteriol 2009; 191: 4492–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Chen YT, Chang HY, Lai YC, Pan CC, Tsai SF, Peng HL. Sequencing and analysis of the large virulence plasmid pLVPK of Klebsiella pneumoniae CG43. Gene 2004; 337: 189–98. [DOI] [PubMed] [Google Scholar]
- 109.Ye M, Tu J, Jiang J et al. Clinical and genomic analysis of liver abscess-causing Klebsiella pneumoniae identifies new liver abscess-associated virulence genes. Front Cell Infect Microbiol 2016; 6: 165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ramirez MS, Traglia GM, Lin DL, Tran T, Tolmasky ME. Plasmid-mediated antibiotic resistance and virulence in gram-negatives: the Klebsiella pneumoniae paradigm. Microbiol Spectr 2014; 2: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lam MMC, Wick RR, Wyres KL et al. Genetic diversity, mobilisation and spread of the yersiniabactin-encoding mobile element ICEKp in Klebsiella pneumoniae populations. Microb Genom 2018; 4 10.1099/mgen.0.000196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lam MMC, Wyres KL, Duchene S et al. Population genomics of hypervirulent Klebsiella pneumoniae clonal-group 23 reveals early emergence and rapid global dissemination. Nat Commun 2018; 9: 2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Marcoleta AE, Berrios-Pasten C, Nunez G, Monasterio O, Lagos R. Klebsiella pneumoniae asparagine tDNAs are integration hotspots for different genomic islands encoding microcin E492 production determinants and other putative virulence factors present in hypervirulent strains. Front Microbiol 2016; 7: 849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lin TL, Lee CZ, Hsieh PF, Tsai SF, Wang JT. Characterization of integrative and conjugative element ICEKp1-associated genomic heterogeneity in a Klebsiella pneumoniae strain isolated from a primary liver abscess. J Bacteriol 2008; 190: 515–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Nassif X, Honore N, Vasselon T, Cole ST, Sansonetti PJ. Positive control of colanic acid synthesis in Escherichia coli by rmpA and rmpB, two virulence-plasmid genes of Klebsiella pneumoniae. Mol Microbiol 1989; 3: 1349–59. [DOI] [PubMed] [Google Scholar]
- 116.Hsu CR, Lin TL, Chen YC, Chou HC, Wang JT. The role of Klebsiella pneumoniae rmpA in capsular polysaccharide synthesis and virulence revisited. Microbiology 2011; 157: 3446–57. [DOI] [PubMed] [Google Scholar]
- 117.Russo TA, Olson R, Fang CT et al. Identification of biomarkers for differentiation of hypervirulent Klebsiella pneumoniae from classical K. pneumoniae. J Clin Microbiol 2018; 56: e00776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Tan TY, Ong M, Cheng Y, Ng LSY. Hypermucoviscosity, rmpA, and aerobactin are associated with community-acquired Klebsiella pneumoniae bacteremic isolates causing liver abscess in Singapore. J Microbiol Immunol Infect 2019; 52: 30–4. [DOI] [PubMed] [Google Scholar]
- 119.Yeh KM, Lin JC, Yin FY et al. Revisiting the importance of virulence determinant magA and its surrounding genes in Klebsiella pneumoniae causing pyogenic liver abscesses: exact role in serotype K1 capsule formation. J Infect Dis 2010; 201: 1259–67. [DOI] [PubMed] [Google Scholar]
- 120.Palacios M, Miner TA, Frederick DR et al. Identification of two regulators of virulence that are conserved in Klebsiella pneumoniae classical and hypervirulent strains. MBio 2018; 9:e01443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Walker KA, Miner TA, Palacios M et al. A Klebsiella pneumoniae regulatory mutant has reduced capsule expression but retains hypermucoviscosity. MBio 2019; 10: e00089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Cubero M, Grau I, Tubau F et al. Hypervirulent Klebsiella pneumoniae clones causing bacteraemia in adults in a teaching hospital in Barcelona, Spain (2007–2013). Clin Microbiol Infect 2016; 22: 154–60. [DOI] [PubMed] [Google Scholar]
- 123.Suzuki K, Nakamura A, Enokiya T et al. Septic arthritis subsequent to urosepsis caused by hypermucoviscous Klebsiella pneumoniae. Intern Med 2013; 52: 1641–5. [DOI] [PubMed] [Google Scholar]
- 124.Wang J, Yan Y, Xue X, Wang K, Shen D. Comparison of pyogenic liver abscesses caused by hypermucoviscous Klebsiella pneumoniae and non-Klebsiella pneumoniae pathogens in Beijing: a retrospective analysis. J Int Med Res 2013; 41: 1088–97. [DOI] [PubMed] [Google Scholar]
- 125.Rafiq Z, Sam N, Vaidyanathan R. Whole genome sequence of Klebsiella pneumoniae U25, a hypermucoviscous, multidrug resistant, biofilm producing isolate from India. Mem Inst Oswaldo Cruz 2016; 111: 144–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Lin YC, Lu MC, Tang HL et al. Assessment of hypermucoviscosity as a virulence factor for experimental Klebsiella pneumoniae infections: comparative virulence analysis with hypermucoviscosity-negative strain. BMC Microbiol 2011; 11: 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Palmer LD, Skaar EP. Transition Metals and Virulence in Bacteria. Annu Rev Genet 2016; 50: 67–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Russo TA, Shon AS, Beanan JM et al. Hypervirulent K. pneumoniae secretes more and more active iron-acquisition molecules than "classical" K. pneumoniae thereby enhancing its virulence. PLoS ONE 2011; 6: e26734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Lam MMC, Wyres KL, Judd LM et al. Tracking key virulence loci encoding aerobactin and salmochelin siderophore synthesis in Klebsiella pneumoniae. Genome Med 2018; 10: 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Goetz DH, Holmes MA, Borregaard N, Bluhm ME, Raymond KN, Strong RK. The neutrophil lipocalin NGAL is a bacteriostatic agent that interferes with siderophore-mediated iron acquisition. Mol Cell 2002; 10: 1033–43. [DOI] [PubMed] [Google Scholar]
- 131.Hsieh PF, Lin TL, Lee CZ, Tsai SF, Wang JT. Serum-induced iron-acquisition systems and TonB contribute to virulence in Klebsiella pneumoniae causing primary pyogenic liver abscess. J Infect Dis 2008; 197: 1717–27. [DOI] [PubMed] [Google Scholar]
- 132.Russo TA, Gulick AM. Aerobactin synthesis proteins as antivirulence targets in hypervirulent Klebsiella pneumoniae. ACS Infect Dis 2019; 5: 1052–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Bailey DC, Alexander E, Rice MR et al. Structural and functional delineation of aerobactin biosynthesis in hypervirulent Klebsiella pneumoniae. J Biol Chem 2018; 293: 7841–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Russo TA, Olson R, MacDonald U, Beanan J, Davidson BA. Aerobactin, but not yersiniabactin, salmochelin, or enterobactin, enables the growth/survival of hypervirulent (hypermucoviscous) Klebsiella pneumoniae ex vivo and in vivo. Infect Immun 2015; 83: 3325–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Russo TA, Olson R, Macdonald U et al. Aerobactin mediates virulence and accounts for increased siderophore production under iron-limiting conditions by hypervirulent (hypermucoviscous) Klebsiella pneumoniae. Infect Immun 2014; 82: 2356–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Fais T, Delmas J, Barnich N, Bonnet R, Dalmasso G. Colibactin: more than a new bacterial toxin. Toxins 2018; 10: 151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Lai YC, Lin AC, Chiang MK et al. Genotoxic Klebsiella pneumoniae in Taiwan. PLoS ONE 2014; 9: e96292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Lu MC, Chen YT, Chiang MK et al. Colibactin Contributes to the Hypervirulence of pks(+) K1 CC23 Klebsiella pneumoniae in Mouse Meningitis Infections. Front Cell Infect Microbiol 2017; 7: 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Dzul SP, Thornton MM, Hohne DN et al. Contribution of the Klebsiella pneumoniae capsule to bacterial aggregate and biofilm microstructures. Appl Environ Microbiol 2011; 77: 1777–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Wu MC, Lin TL, Hsieh PF, Yang HC, Wang JT. Isolation of genes involved in biofilm formation of a Klebsiella pneumoniae strain causing pyogenic liver abscess. PLoS ONE 2011; 6: e23500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Kong Q, Beanan JM, Olson R et al. Biofilm formed by a hypervirulent (hypermucoviscous) variant of Klebsiella pneumoniae does not enhance serum resistance or survival in an in vivo abscess model. Virulence 2012; 3: 309–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Schroll C, Barken KB, Krogfelt KA, Struve C. Role of type 1 and type 3 fimbriae in Klebsiella pneumoniae biofilm formation. BMC Microbiol 2010; 10: 179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Huang TW, Lam I, Chang HY, Tsai SF, Palsson BO, Charusanti P. Capsule deletion via a lambda-Red knockout system perturbs biofilm formation and fimbriae expression in Klebsiella pneumoniae MGH 78578. BMC Res Notes 2014; 7: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Soto E, Dennis MM, Beierschmitt A et al. Biofilm formation of hypermucoviscous and non-hypermucoviscous Klebsiella pneumoniae recovered from clinically affected African green monkey (Chlorocebus aethiops sabaeus). Microb Pathog 2017; 107: 198–201. [DOI] [PubMed] [Google Scholar]
- 145.Bulger J, MacDonald U, Olson R, Beanan J, Russo TA. Metabolite transporter PEG344 is required for full virulence of hypervirulent Klebsiella pneumoniae strain hvKP1 after pulmonary but not subcutaneous challenge. Infect Immun 2017; 85: e00093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Lai YC, Lin GT, Yang SL, Chang HY, Peng HL. Identification and characterization of KvgAS, a two-component system in Klebsiella pneumoniae CG43. FEMS Microbiol Lett 2003; 218: 121–6. [DOI] [PubMed] [Google Scholar]
- 147.Lery LM, Frangeul L, Tomas A et al. Comparative analysis of Klebsiella pneumoniae genomes identifies a phospholipase D family protein as a novel virulence factor. BMC Biol 2014; 12: 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Chou HC, Lee CZ, Ma LC, Fang CT, Chang SC, Wang JT. Isolation of a chromosomal region of Klebsiella pneumoniae associated with allantoin metabolism and liver infection. Infect Immun 2004; 72: 3783–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Ko WC, Paterson DL, Sagnimeni AJ et al. Community-acquired Klebsiella pneumoniae bacteremia: global differences in clinical patterns. Emerg Infect Dis 2002; 8: 160–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Surgers L, Boyd A, Girard PM, Arlet G, Decre D. ESBL-Producing Strain of Hypervirulent Klebsiella pneumoniae K2. France. Emerg Infect Dis 2016; 22: 1687–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Chen L, Kreiswirth BN. Convergence of carbapenem-resistance and hypervirulence in Klebsiella pneumoniae. Lancet Infect Dis 2018; 18: 2–3. [DOI] [PubMed] [Google Scholar]
- 152.Wozniak JE, Band VI, Conley A et al. A Nationwide Screen of carbapenem-resistant Klebsiella pneumoniae reveals an isolate with enhanced virulence and clinically undetected colistin-heteroresistance. Antimicrob Agents Chemother 2019; 63: e00107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Karlsson M, Stanton RA, Ansari U et al. Identification of a carbapenemase-producing hypervirulent Klebsiella pneumoniae isolate, United States. Antimicrob Agents Chemother 2019; 63: e00519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Zhan L, Wang S, Guo Y et al. Outbreak by hypermucoviscous Klebsiella pneumoniae ST11 isolates with carbapenem resistance in a Tertiary Hospital in China. Front Cell Infect Microbiol 2017; 7: 182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Zhang Y, Zeng J, Liu W et al. Emergence of a hypervirulent carbapenem-resistant Klebsiella pneumoniae isolate from clinical infections in China. J Infect 2015; 71: 553–60. [DOI] [PubMed] [Google Scholar]
- 156.Yao B, Xiao X, Wang F, Zhou L, Zhang X, Zhang J. Clinical and molecular characteristics of multi-clone carbapenem-resistant hypervirulent (hypermucoviscous) Klebsiella pneumoniae isolates in a tertiary hospital in Beijing, China. Int J Infect Dis 2015; 37: 107–12. [DOI] [PubMed] [Google Scholar]
- 157.Krapp F, Morris AR, Ozer EA, Hauser AR. Virulence characteristics of carbapenem-resistant Klebsiella pneumoniae strains from patients with necrotizing skin and soft tissue infections. Sci Rep 2017; 7: 13533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Dong N, Lin D, Zhang R, Chan EW, Chen S. Carriage of blaKPC-2 by a virulence plasmid in hypervirulent Klebsiella pneumoniae. J Antimicrob Chemother 2018; 73: 3317–21. [DOI] [PubMed] [Google Scholar]
- 159.Yao H, Qin S, Chen S, Shen J, Du XD. Emergence of carbapenem-resistant hypervirulent Klebsiella pneumoniae. Lancet Infect Dis 2018; 18: 25. [DOI] [PubMed] [Google Scholar]
- 160.Shen D, Ma G, Li C et al. Emergence of a multidrug-resistant hypervirulent Klebsiella pneumoniae sequence Type 23 strain with a Rare bla CTX-M-24-harboring virulence plasmid. Antimicrob Agents Chemother 2019; 63: e02273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Hansen LH, Johannesen E, Burmolle M, Sorensen AH, Sorensen SJ. Plasmid-encoded multidrug efflux pump conferring resistance to olaquindox in Escherichia coli. Antimicrob Agents Chemother 2004; 48: 3332–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Arca P, Hardisson C, Suarez JE. Purification of a glutathione S-transferase that mediates fosfomycin resistance in bacteria. Antimicrob Agents Chemother 1990; 34: 844–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Shankar C, Veeraraghavan B, Nabarro LEB, Ravi R, Ragupathi NKD, Rupali P. Whole genome analysis of hypervirulent Klebsiella pneumoniae isolates from community and hospital acquired bloodstream infection. BMC Microbiol 2018; 18: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Remya P, Shanthi M, Sekar U. Occurrence and characterization of hyperviscous K1 and K2 serotype in Klebsiella pneumoniae. J Lab Physicians 2018; 10: 283–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Rafat C, Messika J, Barnaud G et al. Hypervirulent Klebsiella pneumoniae, a 5-year study in a French ICU. J Med Microbiol 2018; 67: 1083–9. [DOI] [PubMed] [Google Scholar]
- 166.Turton JF, Payne Z, Coward A et al. Virulence genes in isolates of Klebsiella pneumoniae from the UK during 2016, including among carbapenemase gene-positive hypervirulent K1-ST23 and ’non-hypervirulent’ types ST147, ST15 and ST383. J Med Microbiol 2018; 67: 118–28. [DOI] [PubMed] [Google Scholar]
- 167.Turton JF, Payne Z, Micah K, Turton JA. Capsular type K54, clonal group 29 and virulence plasmids: an analysis of K54 and non-K54 closely related isolates of Klebsiella pneumoniae. Epidemiol Infect 2018; 146: 1813–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Rossi B, Gasperini ML, Leflon-Guibout V et al. Hypervirulent Klebsiella pneumoniae in Cryptogenic Liver Abscesses, Paris. France. Emerg Infect Dis 2018; 24: 221–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Odouard C, Ong D, Shah PR et al. Rising trends of endogenous Klebsiella pneumoniae endophthalmitis in Australia. Clin Exp Ophthalmol 2017; 45: 135–42. [DOI] [PubMed] [Google Scholar]
- 170.Sturm E, Tai A, Lin B et al. Bilateral osteomyelitis and liver abscess caused by hypervirulent Klebsiella pneumoniae- a rare clinical manifestation (case report). BMC Infect Dis 2018; 18: 380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Kashani AH, Eliott D. The emergence of Klebsiella pneumoniae endogenous endophthalmitis in the USA: basic and clinical advances. J Ophthalmic Inflamm Infect 2013; 3: 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Mgbemena O, Serota DP, Kumar S, Wozniak JE, Weiss DS, Kempker RR. Peculiar purulence: Hypervirulent Klebsiella pneumoniae causing pyomyositis. Int J Infect Dis 2017; 65: 90–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Wyres KL, Wick RR, Judd LM et al. Distinct evolutionary dynamics of horizontal gene transfer in drug resistant and virulent clones of Klebsiella pneumoniae. PLoS Genet 2019; 15: e1008114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Dong N, Liu L, Zhang R et al. An IncR plasmid harbored by a hypervirulent carbapenem-resistant Klebsiella pneumoniae strain possesses five tandem repeats of the bla KPC-2: NTEKPC-Id Fragment. Antimicrob Agents Chemother 2019; 63: e01775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Bean DC, Agarwal A, Cherian BP, Wareham DW. Hypermucoviscous polymyxin-resistant Klebsiella pneumoniae from Kolkata, India: genomic and phenotypic analysis. J Global Antimicrob Resist 2019; 17: 1–2. [DOI] [PubMed] [Google Scholar]
- 176.Lam MMC, Wyres KL, Wick RR et al. Convergence of virulence and MDR in a single plasmid vector in MDR Klebsiella pneumoniae ST15. J Antimicrob Chemother 2019; 74: 1218–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Shu L, Dong N, Lu J et al. Emergence of OXA-232 carbapenemase-producing klebsiella pneumoniae that carries a pLVPK-like virulence plasmid among elderly patients in China. Antimicrob Agents Chemother 2019; 63: e02246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Lu Y, Feng Y, McNally A, Zong Z. The occurence of colistin-resistant hypervirulent Klebsiella pneumoniae in China. Front Microbiol 2018; 9: 2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Feng Y, Lu Y, Yao Z, Zong Z. Carbapenem-resistant hypervirulent Klebsiella pneumoniae of sequence type 36. Antimicrob Agents Chemother 2018; 62: e02644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Fu Y, Xu M, Liu Y, Li A, Zhou J. Virulence and genomic features of a bla CTX-M-3 and bla CTX-M-14 coharboring hypermucoviscous Klebsiella pneumoniae of serotype K2 and ST65. Infect Drug Resist 2019; 12: 145–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Liu Y, Long D, Xiang TX et al. Whole genome assembly and functional portrait of hypervirulent extensively drug-resistant NDM-1 and KPC-2 co-producing Klebsiella pneumoniae of capsular serotype K2 and ST86. J Antimicrob Chemother 2019; 74: 1233–40. [DOI] [PubMed] [Google Scholar]
- 182.Mataseje LF, Boyd DA, Mulvey MR, Longtin Y. Report on two hypervirulent Klebsiella pneumoniae producing a bla KPC-2 carbapenemase from a Canadian patient. Antimicrob Agents Chemother 2019; 63: e00517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Ghafourian S, Sadeghifard N, Soheili S, Sekawi Z. Extended Spectrum Beta-lactamases: Definition, Classification and Epidemiology. Curr Issues Mol Biol 2015; 17: 11–21. [PubMed] [Google Scholar]
- 184.Jia B, Raphenya AR, Alcock B et al. CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res 2017; 45: D566–D73. [DOI] [PMC free article] [PubMed] [Google Scholar]