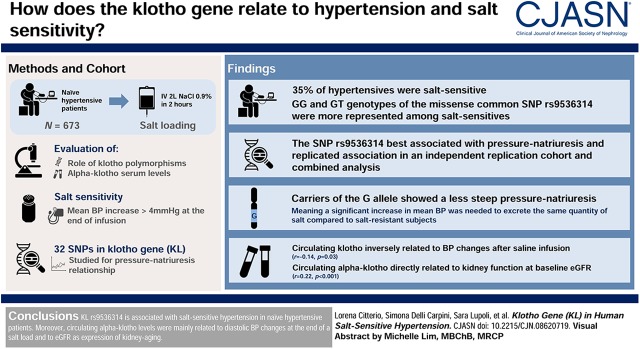
Visual Abstract
Keywords: hypertension, genetics and development, mice, humans, animals, dietary sodium chloride, blood pressure, alleles, genome-wide association study, transgenic mice, single nucleotide polymorphism, saline solution, natriuresis, sodium chloride, kidney, genotype, cohort studies
Abstract
Background and objectives
Hypertension is a common aging-related disorder. Salt intake is one of the main environmental factors contributing to the development of hypertension. Transgenic mice with one-half Klotho deficiency displayed a spontaneous BP increase and salt-sensitive hypertension in response to high sodium intake. Usually circulating levels of α-Klotho decrease with age, and this reduction may be stronger in patients with several aging-related diseases. This study aimed at exploring the association of Klotho with salt sensitivity in humans.
Design, setting, participants, & measurements
The role of Klotho polymorphisms and α-Klotho serum levels was evaluated in patients with hypertension who were treatment naive and underwent an acute salt-sensitivity test (discovery n=673, intravenous 2 L of 0.9% saline in 2 hours). Salt sensitivity was defined as a mean BP increase of >4 mm Hg at the end of the infusion. A total of 32 single nucleotide polymorphisms in the Klotho gene (KL), previously identified with a genome-wide association study, were used in the genetic analysis and studied for a pressure-natriuresis relationship.
Results
Of the patients with hypertension, 35% were classified as salt sensitive. The most relevant polymorphism associated with pressure natriuresis was the common missense single nucleotide polymorphism rs9536314, and the GG and GT genotypes were more represented among patients who were salt sensitive (P=0.001). Those carrying the G allele showed a less steep pressure-natriuresis relationship, meaning that a significant increase in mean BP was needed to excrete the same quantity of salt compared with patients who were salt resistant. KL rs9536314 also replicated the pressure-natriuresis association in an independent replication cohort (n=193) and in the combined analysis (n=866). There was an inverse relationship between circulating Klotho and mean BP changes after the saline infusion (r=−0.14, P=0.03). Moreover, circulating α-Klotho was directly related to kidney function at baseline eGFR (r=0.22, P<0.001).
Conclusions
KL rs9536314 is associated with salt-sensitive hypertension in patients with hypertension who are treatment naive. Moreover, circulating α-Klotho levels were mainly related to diastolic BP changes at the end of a salt load and to eGFR as an expression of kidney aging.
Introduction
An increase in salt sensitivity with advancing age has been noted in numerous studies (1–3). Nevertheless, the mechanism of salt sensitivity in individuals with both normotension and hypertension and its long-term significance remains unclear. Linkage analyses and association studies have suggested that genetic mechanisms might play a pivotal role in salt-sensitive hypertension (1,4).
An elegant paper, published in 2015, showed that transgenic mice with a one-half deficiency in the Klotho gene (KL[+/-]) demonstrated a spontaneous increase of BP and salt-sensitive hypertension in response to high sodium intake (5). Systolic BP in KL(+/-) mice began to increase at 15 weeks of age, reached a peak level at 17 weeks, and remained elevated thereafter, whereas it remained constant in wild-type (WT) mice. Interestingly, high sodium intake further increased BP in KL(+/-) mice but did not affect BP in WT mice.
The KL gene was originally identified as a novel aging suppressor gene in mice (6). Klotho protein is membrane bound and mainly expressed in the kidney (7). After cleavage, Klotho is secreted and released into the systemic circulation and has direct effects on tissues and cells that do not express Klotho (7). It has been observed that serum α-Klotho levels decreased with age after 40 years in humans (8). This decrease may also be observed in patients with several aging-related diseases such as cancer (9) and also in hypertensive nephrosclerosis, where both kidney and circulating Klotho are reduced (10).
The function of membrane-bound Klotho as the obligatory coreceptor of fibroblast growth factor 23 (11) was first described in inorganic phosphate metabolism. Subsequently, genetic studies reported single nucleotide polymorphisms (SNPs) in the KL gene that are associated with essential hypertension, hypertension phases, and BP regulation in different ethnic cohorts (12–14), as well as with kidney function (15), glucose metabolism, and cardiovascular risk factors (16,17).
Recently, a follow-up study showed that kidney damage associated with a reduction of α-Klotho levels may determine the upregulation of plasma aldosterone, supporting its role in salt-sensitive hypertension (18). Moreover, a study (10) in stroke-prone, spontaneously hypertensive rats in the early phase of hypertension (in which kidney damage involves the kidney medulla) showed that Klotho binds to the angiotensin II receptor type 1 to suppress angiotensin signal transduction, participating in the inactivation of the kidney renin-angiotensin-aldosterone system (RAAS) (18).
In this study, we examined the role of KL polymorphisms and circulating Klotho in salt-sensitive, salt-resistant hypertension and kidney sodium handling in a large cohort of white patients with hypertension who were treatment naive.
Materials and Methods
Study Participants
The participants enrolled in the study were newly discovered, unrelated, and never treated (naive) patients with mild hypertension who were referred for a hypertension assessment to the outpatient clinic at the San Raffaele Hospital between January 1997 and June 2019. Inclusion criteria for the study were as follows: age from 18 to 65 years; body mass index (BMI) <30 kg/m2; baseline sodium intake, evaluated as urinary sodium excretion <300 mEq/24 h, after at least 1 week of a normal sodium diet (target 5 g/d); and office systolic BP >140 mm Hg or diastolic BP >90 mm Hg as a mean of at least three consecutive measurements during the screening visit. Exclusion criteria included history of myocardial infarction, stroke, congestive heart failure, liver disease, secondary causes of hypertension excluded by routine methods, diabetes, women on oral contraceptives, severe hypertension (>180/110 mm Hg), drug or alcohol abuse, and creatinine clearance <70 ml/min.
This treatment-naive, hypertensive cohort is peculiar and unique because it was carefully phenotyped for BP, kidney function, kidney sodium handling, and the acute salt-sensitive test as continuous traits according to the American Heart Association statement on the salt-sensitive BP phenotype (19). In fact, this hypertensive cohort is characterized by deep hypertensive-related (intrinsic, exogenous, environmental, and molecular) phenotypes carefully selected to investigate hypertension at stage 1–2, minimizing confounding factors (20–23). All phenotypes were collected in the early hypertensive phase to avoid the effect of antihypertensive drugs and vascular or organ impairments (22). All clinical investigations have been conducted according to the Declaration of Helsinki principles. The Ethics Committee of the San Raffaele Scientific Institute approved the study and the written informed consent was signed by the participants before inclusion in the study.
Acute Salt-Sensitive Test
Patients with treatment-naive hypertension underwent an acute salt-loading test as previously reported (23–25) (Supplemental Figure 1). Briefly, after 2 hours of rest (equilibration), 2 L of 0.9% saline were infused intravenously over 2 hours (infusion), and patients were monitored for another 2 hours (recovery). Every 2 hours during the test, we collected venous blood samples to determine kidney function and electrolyte (creatinine, sodium, potassium) levels and urine samples to determine sodium and potassium excretion. The saline test was performed in two independent cohorts of patients with treatment-naive hypertension, the discovery (n=673) and the replication (n=193) cohorts, that were both selected at the outpatient clinic at the San Raffaele Hospital with the same inclusion and exclusion criteria during a different time frame. Hormonal parameters (plasma renin activity for the discovery cohort or renin for the replication cohort and aldosterone levels for both cohorts) were collected at the beginning and at the end of the test.
For the classification of salt-sensitive and salt-resistant hypertension, the mean BP was calculated as ([systolic BP − diastolic BP)/3] + diastolic BP. Patients were classified as salt resistant or salt sensitive according to mean BP changes after acute saline infusion: those who displayed a mean BP increase of >4 mm Hg (referring to the third tertile of distribution of BP variation at the end of infusion [23–26]) were considered salt sensitive. Conversely, patients with a smaller or negative BP change were classified as salt resistant. The slope of the relationship between BP and sodium excretion (pressure-natriuresis slope, mm Hg/μEq per minute) was calculated, per patient, by plotting sodium excretion (on the y axis) as a function of mean BP (on the x axis) which was observed under both basal conditions and after 2 hours of saline infusion, as discussed elsewhere (26).
Serum α-Klotho Measurement and Other Biochemistry
Human soluble α-Klotho was measured from serum using a specific assay kit (code number 27998; Immuno-Biological Laboratories, Takasaki, Japan). The Immuno-Biological Laboratories measurement range is 93.75–6000 pg/ml (intra-assay coefficient of variability, 4%; interassay coefficient of variability, 11%). Serum samples were diluted twofold in RIA buffer and processed as described in the protocol. Serum and urinary sodium and potassium were measured using flame photometry; serum and urinary creatinine were measured using an automated enzymatic method. Plasma renin activity and plasma aldosterone were measured by RIA in the discovery cohort, whereas in the replication cohort they were measured by chemiluminescence due to a methodologic upgrade. To assess kidney function, eGFR was calculated according to the CKD Epidemiology Collaboration equation (27).
Genotyping
DNA was extracted from peripheral blood collected at the beginning of the acute salt-loading test. The majority of patients in the discovery cohort (n=525) were genotyped using the Illumina 1M-Duo array (Illumina, San Diego, CA) within the HYPERGENES project (28), and the Illumina HumanOmniExpress array, within the InterOmics project (http://www.interomics.eu/).
The KL gene maps onto chromosome 13 (q13.1) and spans 50 kb, but for this study we extended our SNP selection to 40 (±20) kbp in the flanking regions to increase the length of the covered region. Next, we excluded all SNPs with a minor allele frequency <0.05, leaving a total of 32 genotyped SNPs that were considered for the analysis in treatment-naive hypertensive population (Supplemental Table 1 and Supplemental Figure 2); all SNPs passed the quality control threshold of the Hardy–Weinberg equilibrium (P>0.001). To exclude SNP redundancy in this area, we selected 15 tag SNPs (Table 1) that are in high linkage disequilibrium (LD; r2>0.80) with the remaining SNPs (Supplemental Table 1), covering the entire gene with extension to the 3′ and 5′ flanking regions. Tag SNPs were retrieved using Haploview 4.2 software (https://www.broadinstitute.org/haploview/haploview). HapMap phase 3 data were used for extracting the LD distribution across the KL region in white populations.
Table 1.
Tag single nucleotide polymorphisms spanning the KL gene region (±20 kb) significantly associated with pressure natriuresis at time 120 minutes
| SNP Rank | rs Code | Chr 13 Position (bp) | Alleles | Minor Allele | MAF | P Valuea | Homozygotes for the Major Alleleb | Heterozygotesb | Homozygotes for the Minor Alleleb |
|---|---|---|---|---|---|---|---|---|---|
| 1 | rs446647 | 33571078 | T/C | C | 0.22 | 0.69 | 0.007±0.036 | 0.006±0.026 | 0.002±0.016 |
| 2 | rs367243 | 33578412 | G/A | A | 0.20 | 0.74 | 0.007±0.036 | 0.005±0.026 | 0.005±0.014 |
| 3 | rs526906 | 33598987 | C/T | T | 0.14 | 0.37 | 0.006±0.027 | 0.003±0.045 | 0.008±0.019 |
| 4 | rs480830 | 33606723 | T/C | C | 0.46 | 0.49 | 0.007±0.029 | 0.008±0.037 | 0.003±0.026 |
| 5 | rs9526984 | 33609937 | A/G | G | 0.09 | 0.02 | 0.005±0.032 | 0.015±0.032 | 0.023±0.033 |
| 6 | rs1334928 | 33615948 | T/G | G | 0.38 | 0.09 | 0.010±0.032 | 0.003±0.035 | 0.006±0.023 |
| 7 | rs2320762 | 33617174 | T/G | G | 0.41 | 0.07 | 0.003±0.040 | 0.010±0.026 | 0.006±0.028 |
| 8 | rs657049 | 33622817 | A/G | G | 0.28 | <0.001c | 0.006±0.025 | 0.003±0.036 | 0.026±0.045 |
| 9 | rs9527026 | 33628239 | G/A | A | 0.15 | <0.000446c | 0.004±0.032 | 0.011±0.025 | 0.036±0.065 |
| 10 | rs9315202 | 33642016 | C/T | T | 0.30 | 0.06 | 0.006±0.039 | 0.010±0.021 | −0.001±0.032 |
| 11 | rs581971 | 33644398 | G/A | A | 0.47 | 0.61 | 0.008±0.029 | 0.005±0.038 | 0.007±0.022 |
| 12 | rs670657 | 33649054 | G/A | A | 0.17 | 0.59 | 0.007±0.027 | 0.004±0.043 | 0.008±0.015 |
| 13 | rs9536357 | 33651505 | G/A | A | 0.46 | 0.20 | 0.003±0.041 | 0.008±0.023 | 0.009±0.035 |
| 14 | rs9591501 | 33658270 | C/T | T | 0.09 | 0.08 | 0.005±0.032 | 0.013±0.033 | 0.023±0.029 |
| 15 | rs566996 | 33660785 | G/A | A | 0.25 | 0.58 | 0.008±0.037 | 0.004±0.024 | 0.008±0.029 |
SNPs are ranked by chromosome and position based on the GRCh37 (release 105) assembly. Chr13, chromosome 13; MAF, minor allele frequency.
Nominal P value of association.
Pressure natriuresis at time 120 minutes is expressed as mean±SD, adjusted for age, sex, and body mass index.
P<0.003, adjusted P value threshold after Bonferroni multiple comparison test.
Targeted genotyping for the rs9536314 SNP was performed using 5′ nuclease allelic discrimination assays, with allele-specific Minor Grove Binder (C_2983037_20 assay identifier, TaqMan SNP Genotyping Assay; Applied Biosystems, Foster City, CA).
Statistical Analyses
Continuous data were expressed as means±SD for normal variables, whereas dichotomous variables were presented as an absolute value. The general linear model was used to compare continuous variables among genotypic groups. We included into our models some covariates with known physiologic relevance for arterial BP and aging (sex, BMI, and age) and for eGFR for combined analysis only. Two-tailed association analysis was set at a nominal α=0.05 level, but after the use of a Bonferroni correction for multiple SNPs test, the threshold was then defined at α=0.003 for SNP association with pressure natriuresis (time 120 minutes). Regression analysis was used to estimate the potential relationship between serum α-Klotho and both BP variation during the acute salt-load test and eGFR at baseline. For database management and statistical analysis, we used the SPSS version 21 software package (SPSS, Chicago, IL).
Results
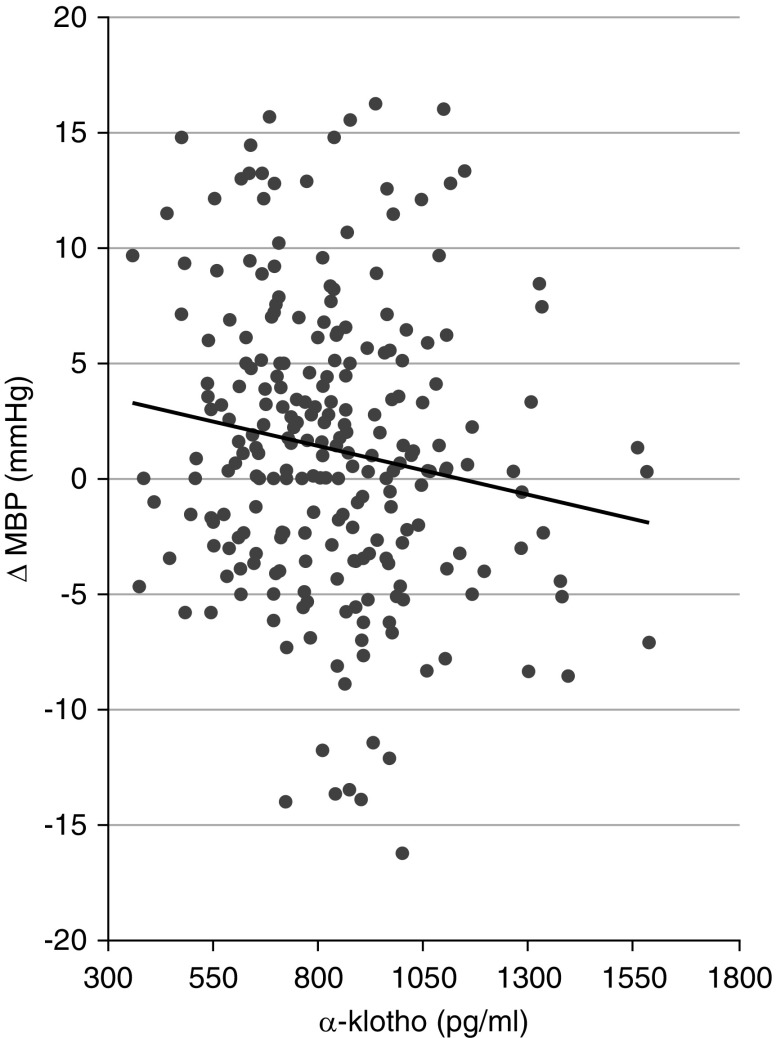
We enrolled 673 patients with treatment-naive hypertension from the outpatient clinic for hypertension of our institute into the discovery cohort. Clinical characteristics of the patients at baseline are reported in Table 2. According to the salt-sensitivity definition, 65% of patients with treatment-naive hypertension were classified as salt resistant, whereas 35% were classified as salt sensitive. Significantly lower plasma potassium at baseline was observed in patients who were salt resistant compared with those who were salt sensitive, whereas their urinary sodium excretion at baseline was similar (Table 2). Furthermore, kidney function expressed as creatinine clearance and eGFR (CKD Epidemiology Collaboration) at baseline was strictly related to circulating α-Klotho level (Supplemental Table 2). In a subgroup of 243 patients with treatment-naive hypertension (168 salt resistant and 75 salt sensitive), circulating α-Klotho was lower in salt-sensitive individuals without reaching statistical significance (Table 2). In the span of 24 hours, ambulatory BP monitoring for both systolic BP and diastolic BP during awake and sleeping periods resulted in values that were significantly higher in patients who were salt sensitive than in those who were salt resistant (Table 2). Furthermore, during the acute salt-sensitive test, the change in mean BP was found to be inversely correlated to serum α-Klotho levels (r=−0.15, P=0.02; data adjusted for sex, age, and BMI) as shown in Figure 1, similarly to the changes in systolic BP and mainly in diastolic BP (Supplemental Table 3).
Table 2.
Baseline clinical characteristics of the discovery treatment-naive hypertensive cohort that underwent the acute saline load
| Characteristic | Parameters | Discovery NHP Cohort (n=673) | Salt Resistant (n=436) | Salt Sensitive (n=237) |
|---|---|---|---|---|
| Anthropometric | Age, yr | 45±10 | 45±10 | 46±9 |
| Sex, female/male, n (%) | 126 (19%)/547 (81%) | 86 (20%)/350 (80%) | 40 (17%)/197 (83%) | |
| BMI, kg/m2 | 25±3 | 25±3 | 25±2 | |
| 24-h Ambulatory BP (mm Hg) | Systolic BP daytime | 142±11 | 141±10 | 144±13a |
| Diastolic BP daytime | 93±9 | 92±8 | 95±10a | |
| Systolic BP nighttime | 126±13 | 125±12 | 129±15a | |
| Diastolic BP nighttime | 79±11 | 77±10 | 81±12a | |
| Electrolytes and kidney function | Urinary sodium, mEq/24 h | 142±70 | 140±67 | 146±74 |
| Urinary potassium, mEq/24 h | 60±23 | 60±23 | 60±23 | |
| eGFR, ml/min per 1.73 m2 | 101±13 | 101±13 | 101±13 | |
| Plasma sodium, mEq/L | 142±2 | 142±2 | 142±3 | |
| Plasma potassium, mEq/L | 4.0±0.4 | 3.9±0.3 | 4.01±0.4a | |
| RAAS and water balance | Plasma renin activity, ng/ml per hour | 1.9±1.8 | 2.0±1.9 | 1.8±1.7 |
| Plasma aldosterone, ng/dl | 171±119 | 169±122 | 176±112 | |
| Klotho (n=243 NHPs) | ⍺-Klotho, pg/ml | 832±223 | 846±230 | 798±203 |
Normally distributed numeric data expressed as mean±SD unless otherwise indicated. NHP, patients with treatment-naive hypertension; BMI, body mass index; RAAS, renin-angiotensin-aldosterone system.
P<0.05 ANOVA, patients with salt resistance versus those with salt sensitivity.
Figure 1.
Inverse correlation between circulating α-Klotho and mean BP changes. ΔMBP, mean BP variation. r=−0.14; P=0.03, unadjusted; r=−0.15; P=0.02, adjusted for age, sex, and BMI.
KL Genetic Variants
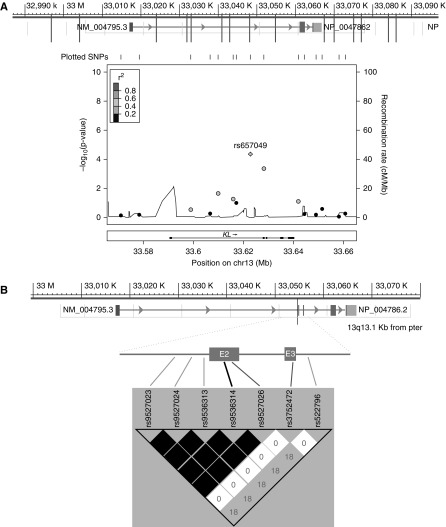
In the discovery cohort, most of our patients with treatment-naive hypertension (n=525) were genetically characterized in previous genome-wide studies, as described in Materials and Methods (Supplemental Figure 2). A total of 15 tag SNPs were selected for the genetic analysis of association. Three (rs9526984, rs657049, rs9527026) of the tag SNPs were significantly associated with BP variation and/or pressure natriuresis at time 120 minutes (Figure 2A), with nominal P values of association shown in Table 1. Only two SNPs (rs657049, rs9527026) passed the threshold of significance for the association with salt sensitivity after Bonferroni multiple comparison at P<0.003. Interestingly, rs9527026 is a synonymous lysine product (AAG→AAA), mapping in the first coding exon (exon 2) of the KL gene. To better characterize this exon region of association with further proxy SNPs, we performed an in silico analysis for the haplotype map of the region that includes exon 2 and the flanking introns, using HapMap phase 3 as reference. We found four additional SNPs in perfect LD with rs9527026 that were not present in our previous analysis (Figure 2B). Then, among the newly identified SNPs, we chose the common missense SNP rs9536314 as the potentially most interesting variant at the functional level because of its likely effect on Klotho structure or activity, and the remarkable WT phenylalanine conservation in homologous proteins in eukaryotic organisms (29) (T>G in F352V, minor allele frequency 0.17 for G allele in the white population) (Supplemental Figure 2). This single SNP was tested in the analysis of association with salt sensitivity in the discovery cohort (n=673).
Figure 2.
Association with pressure natriuresis of single nucleotide polymorphisms (SNPs) in KL region. (A) Tag SNPs covering the entire gene with extension to the 5′ and 3′ flanking regions and regional plot for association with pressure natriuresis at time 120 minutes. (B) Linkage disequilibrium (LD) map for the KL SNPs around exons 2–3. Numeric values represent pairwise LD as measured by r2. SNPs in high LD are represented by darker squares (r2>0.80). Chr13, chromosome 13; pter, p terminal; E2/3, exon 2/3.
We evaluated BP variation after the acute salt load across genotypes of rs9536314. The increase in systolic BP, diastolic BP, and consequently mean BP was significantly greater in the carriers of the G allele (Table 3). Moreover, the G minor allele was significantly more frequent in the salt-sensitive cohort than in the salt-resistant one (Supplemental Table 4). Next, we evaluated the effect of this polymorphism on the pressure-natriuresis relationship. Again, the carriers of the G allele showed a less-steep pressure-natriuresis curve (slope 0.013 μmol/min in GG+GT versus 0.004 μmol/min in TT genotypes, P=0.001; Table 4), meaning that G carriers, compared with homozygous T carriers, need to increase their BP to excrete the same amount of salt, similarly to how this occurs in subjects who are salt sensitive.
Table 3.
Effect of acute saline load in the discovery cohort of patients with treatment-naive hypertension according to KL rs9536314 genotypes
| Parameters | KL TG+GG (n=179) | KL TT (n=494) | P ANOVA |
|---|---|---|---|
| Baseline | |||
| Systolic BP, mm Hg | 138±14 | 140±15 | 0.12 |
| Diastolic BP, mm Hg | 89±10 | 91±10 | 0.11 |
| Mean BP, mm Hg | 106±10 | 107±11 | 0.08 |
| Urinary sodium, mEqa | 17±14 | 16±15 | 0.35 |
| Urinary potassium, mEqa | 7.5±7.4 | 7.0±8.2 | 0.51 |
| PRA, ng/ml per hour | 1.7±1.5 | 2.0±2.0 | 0.11 |
| Plasma aldosterone, ng/dl | 170±112 | 172±121 | 0.86 |
| End of infusion (time 120 minutes) | |||
| Systolic BP, mm Hg | 144±15 | 144±16 | 0.76 |
| Diastolic BP, mm Hg | 92±11 | 92±12 | 0.89 |
| Mean BP, mm Hg | 110±12 | 109±12 | 0.91 |
| Urinary sodium, mEqb | 55±33 | 53±31 | 0.51 |
| Urinary potassium, mEqb | 13±10 | 11±7 | 0.08 |
| End of recovery time (time 240 minutes) | |||
| PRA, ng/ml per hour | 0.6±0.5 | 0.6±0.5 | 0.92 |
| Plasma aldosterone, ng/dl | 51±43 | 55±49 | 0.40 |
| Change from baseline | |||
| Systolic BP, mm Hgd | 5.6±8.7 | 3.2±9.3 | 0.003c |
| Diastolic BP, mm Hgd | 2.3±6.1 | 0.9±6.2 | 0.01c |
| Mean BP, mm Hgd | 3.4±6.3 | 1.7±6.4 | 0.002c |
| Urinary sodium, mEqd | 37±33 | 37±30 | 0.82 |
| Urinary potassium, mEqd | 4.9±11 | 4.4±8.6 | 0.57 |
| PRA, ng/ml per houre | −1.1±0.1 | −1.4±0.09 | 0.11 |
| Plasma aldosterone, ng/dle | −121±8 | −123±5 | 0.81 |
Normally distributed numeric data are expressed as mean±SD. PRA, plasma renin activity.
Excretion during equilibration period (time −120/0 minutes).
Excretion during infusion period (time 0/120 minutes).
P<0.05.
Change from baseline to time 120 minutes (end of infusion period) for systolic BP, diastolic BP, mean BP, urinary sodium, and urinary potassium.
Change from baseline to time 240 minutes (end of recovery time) for PRA and aldosterone.
Table 4.
Salt-sensitive, pressure-natriuresis slopes by KL rs9536314 genotype in the discovery cohort of patients with treatment-naive hypertension, in the replication cohort, and in the combined cohort
| Cohort | Pressure Natriuresis Slope (mm Hg/μEq per minute) | P ANOVA | |
|---|---|---|---|
| KL Genotype TG+GG | KL Genotype TT | ||
| Discovery cohort (n=673) | 0.013±0.03 (n=179) | 0.004±0.03 (n=494) | 0.001 |
| Replication cohort (n=193) | 0.015±0.04 (n=47) | 0.005±0.03 (n=146) | 0.04 |
| Combined cohort (n=866)a | 0.013±0.03 (n=226) | 0.004±0.03 (n=640) | 0.000146 |
Normally distributed numeric data expressed as mean±SD. Data adjusted for age, sex, and body mass index.
The combined cohort was also adjusted for eGFR at baseline.
Serum α-Klotho levels at baseline were similar based on rs9536314 genotypes (TT 829±230.2 pg/ml versus GG+GT 838±202.6 pg/ml). We have not observed any difference in RAAS (evaluated as plasma renin activity and plasma aldosterone) at baseline (Supplemental Table 5), during the test (Table 3), or according to KL genotype classes.
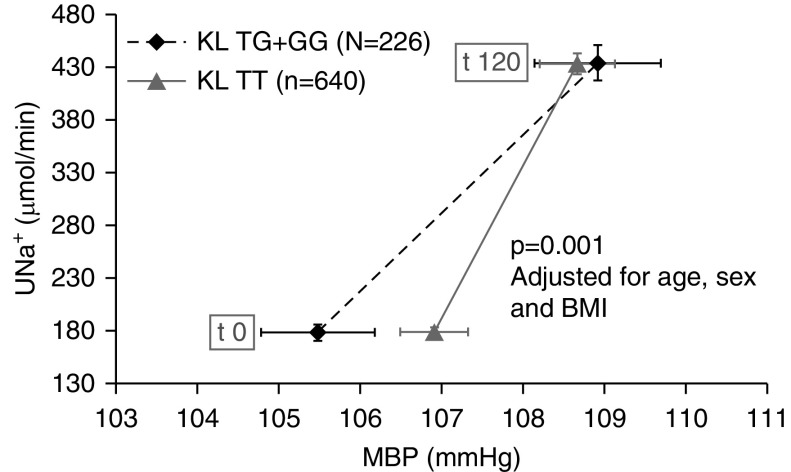
The rs9536314 association with the pressure-natriuresis curve (slope 0.015 μmol/min in GG+GT versus 0.005 μmol/min in TT genotypes, P=0.04; Table 4) was validated in an independent replication cohort (n=193 patients with treatment-naive hypertension; Supplemental Table 6). The discovery and replication cohorts were superimposable except for age, sex, BMI, and eGFR. Because the risk of the G allele effect was in the same direction in both discovery and replication analyses, we performed an adjusted combined analysis (n=866) to confirm the right-shifted pressure-natriuresis curve in patients carrying the G allele (P<0.001; Table 4). In the TT genotype, the pressure-natriuresis curve is steeper and shifted to the left, with a largely unchanged slope as for subjects who were salt resistant (Figure 3).
Figure 3.
Pressure-natriuresis relationship variation according to KL rs9536314 genotypes during acute salt load in the combined cohort. Urinary sodium excretion (UNa+) as a function of mean BP (MBP), at basal level (t 0) and after salt load (t 120). The pressure-natriuresis curve is drawn according to the KL rs9536314 polymorphism (carriers of the nonrisk genotype TT, gray curve; carriers of the risk allele G, black dashed curve). The significantly less-steep slope of the black dashed curve compared with the gray curve indicates the need for an increase in mean BP to excrete the salt load in carriers of the G allele. Horizontal whiskers refer to mean SEM for BP, vertical whiskers refer to SEM for urinary sodium excretion. BMI, body mass index.
Discussion
We have explored the association of KL polymorphisms and circulating Klotho levels in salt sensitivity in a large cohort of white patients with hypertension who were treatment naive. The genetic analysis showed that human salt-sensitive hypertension is associated with KL genotypes, whereas circulating α-Klotho is not associated with salt sensitivity but was mainly inversely correlated with diastolic BP changes after the acute sodium load. α-Klotho levels directly correlated with kidney function, even if in a normal range of eGFR, as represented in this cohort of patients with treatment-naive hypertension, further supporting the role of Klotho in kidney aging (7). Our findings are in line with recent experimental evidence suggesting the role of Klotho in kidney pathophysiology (10,15,30). Several studies have shown the association of KL gene polymorphisms with longevity and mortality in humans (8,14,29). Studies in Klotho-deficient mice have shown an association with extensive premature aging, reduced life span, skin and muscle atrophy, osteoporosis, and ectopic calcifications (31). Conversely, mice overexpressing Klotho showed a longer mean life span. Therefore, KL was defined an antiaging gene (6).
Klotho is abundantly expressed in the kidney, particularly in the distal convoluted tubule (6,32) but also in the kidney proximal tubule (33), parathyroid gland (34), ovary, testis, placenta, and in the choroid plexus in the brain (6). Recently, Klotho was found to be locally expressed in the adventitial area of the aorta, supporting the vascular protective effect of the Klotho protein (35). In humans, the secreted form of Klotho protein is predominant over the membrane form (7,36), and is released in systemic circulation after cleavage. Yamazaki et al. (8) reported that Klotho levels in serum decline with age in humans after 40 years of age. Indeed, during kidney aging, a decline in nephron size and number (due to increased glomerulosclerosis (37)) and tubulointerstitial changes (due to interstitial fibrosis leading to medulla hypotonicity) are observed, thus resulting in low GFR and a diminished ability for tubular salt and water reabsorption (38).
Klotho is also involved in kidney fibrosis progression. Klotho exhibits antifibrotic effects by inhibiting multiple growth factors including TGF-β1, Wnt/β-catenin, and IGF-1 (39–41). Administration of secreted Klotho or genetic manipulation to induce Klotho overexpression alleviate kidney fibrosis (15,40). However, kidney expression of Klotho is reduced in hypertension (10). Furthermore, Klotho restoration via epigenetic histone acetylation and DNA demethylation has been found as a critical mechanism of genistein’s antifibrosis function (42).
Previous studies in Klotho heterozygous-deficient mice demonstrated a causative link between Klotho and salt-sensitive hypertension (5). We have investigated the pressure-natriuresis relationship as a function of rs9536314 missense variant in the KL gene and observed a significantly less-steep and right-shifted curve in patients with treatment-naive hypertension carrying the GG and TG genotypes, compared with those with WT TT. Physiologic studies demonstrated that salt sensitivity is more prevalent in the old rather than the young population (43), likewise the association of age and salt sensitivity is more marked in individuals who are hypertensive compared with those who are normotensive (37,44). Although the Klotho level decreases and salt sensitivity increases with age, it is not clear whether Klotho deficiency by itself is associated with enhanced salt sensitivity in humans. Further studies in larger aged populations are needed.
Salt sensitivity may be considered as a result of alteration in glomerular filtration and tubular function, and/or as a consequence of increased kidney vascular tone and vascular stiffening with age along with the loss of efficacy of endogenous vasodilators (45). Accordingly, our data in patients with treatment-naive hypertension showed that circulating α-Klotho was directly related to eGFR at baseline and inversely related to BP increase after saline infusion before development of CKD. In this hypertensive cohort, we showed that salt-sensitive hypertension is associated with KL gene polymorphisms as well. In a previous paper by Arking et al. (29), the authors identified the so-called KL-VS haplotype of KL, which spans the exon 2 region and its flanking sequence, that includes six genetic variants in perfect LD, two of which were missense (F352V rs9536314 and C370S rs9527025). The KL-VS variants were associated with reduced human longevity and with increased cardiovascular risk (17,46). Among our patients with hypertension who were treatment naive, we characterized one of these variants, the synonymous rs9527026, that was found to be associated with pressure natriuresis. We then identified the proxy missense variant F352V (rs9536314) that was used to genotype a larger treatment-naive hypertensive cohort. In particular, we have demonstrated that subjects carrying the G allele of rs9536314 were salt sensitive. Indeed, in these patients, the pressure-natriuresis curve showed a significant shift to the right in both the discovery and replication cohorts, which represents an abnormal response to sodium load.
We also focused on the role of RAAS in relation to Klotho and salt sensitivity. Previous studies reported that angiotensin inhibited Klotho expression and that Klotho inhibited RAAS activation (47,48). In our patients, we have not observed any difference in RAAS at baseline and during the test according to both KL genotype classes.
A strength of this study is our cohort of patients with treatment-naive hypertension that was carefully phenotyped for BP, kidney function, kidney sodium handling, and acute salt sensitivity. All phenotypes were collected in the early hypertensive phase, thus avoiding the effects of antihypertensive drugs and vascular or organ impairments (22). Moreover, the reduction of environmental variability makes it possible to highlight the role of gene variants in salt sensitivity. Finally, the results of our study show the potential of the acute salt-load test to identify possible new pathways of salt sensitivity in humans.
This study has some important limitations. We have not found any significant relationship between serum α-Klotho at baseline and salt-sensitive status in neither of the KL genotypes. This may be due to the metabolic cascade of Klotho in that it is synthetized in the kidney but partially secreted (7,36). On the other hand, only a subset of our treatment-naive hypertensive cohort was analyzed for KL level, due to incomplete availability of biologic samples. Further studies with bigger sample sizes may be more able to detect this possible relationship. Several other aspects regarding Klotho function have not been investigated in this study. In particular, the reported acute salt load is not an appropriate stimulus to investigate the Klotho cascade in phosphate homeostasis, including fibroblast growth factor 23 (11). Other more-specific studies also need to be designed to explore the relationship among Klotho, salt, and inflammation.
In conclusion, for the first time we have shown the association between KL genotypes and salt-sensitive hypertension among patients with treatment-naive hypertension, implying a vascular effect after volume load. Moreover, circulating α-Klotho levels were inversely related to BP change during acute salt load and directly related to GFR as an expression of early and enhanced kidney aging.
Disclosures
Dr. Cusi is the Scientific Advisor of Bio4Dreams, an incubator of biotechnology startups unrelated with kidney research. All remaining authors have nothing to disclose.
Supplementary Material
Acknowledgments
We thank Mrs. Cinzia Scotti (Nephrology and Dialysis Unit, IRCCS San Raffaele Scientific Institute, Genomics of Renal Diseases and Hypertension Unit, Vita-Salute San Raffaele University, Milan, Italy) for technical assistance in preparing the manuscript.
Footnotes
Published online ahead of print. Publication date available at www.cjasn.org.
Supplemental Material
This article contains the following supplemental material online at http://cjasn.asnjournals.org/lookup/suppl/doi:10.2215/CJN.08620719/-/DCSupplemental.
Supplemental Table 1. All single nucleotide polymorphisms (SNPs) spanning KL gene region (±20 kb).
Supplemental Table 2. Correlation between α-Klotho and creatinine clearance or eGFR at baseline.
Supplemental Table 3. Correlation between α-Klotho and changes of systolic BP or diastolic BP at the end of salt infusion.
Supplemental Table 4. Salt-sensitive and salt-resistant patients’ distribution in the discovery naïve hypertensive cohort based on KL rs9536314, rs657049, and rs9527026 genotypes.
Supplemental Table 5. Baseline clinical characteristics of the discovery treatment-naive hypertensive cohort that underwent the acute saline load according to KL rs9536314 genotypes.
Supplemental Table 6. Baseline clinical characteristics of the replication treatment-naive hypertensive cohort that underwent the acute saline load and comparison with the discovery treatment-naive hypertensive cohort.
Supplemental Figure 1. Salt loading test scheme.
Supplemental Figure 2. Flow chart for the filtering of selected SNPs in KL region.
References
- 1.Kelly TN, He J: Genomic epidemiology of blood pressure salt sensitivity. J Hypertens 30: 861–873, 2012. [DOI] [PubMed] [Google Scholar]
- 2.AlGhatrif M, Wang M, Fedorova OV, Bagrov AY, Lakatta EG: The pressure of aging. Med Clin North Am 101: 81–101, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lin H, Lunetta KL, Zhao Q, Mandaviya PR, Rong J, Benjamin EJ, Joehanes R, Levy D, van Meurs JBJ, Larson MG, Murabito JM: Whole blood gene expression associated with clinical biological age. J Gerontol A Biol Sci Med Sci 74: 81–88, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arnett DK, Claas SA: Omics of blood pressure and hypertension. Circ Res 122: 1409–1419, 2018. [DOI] [PubMed] [Google Scholar]
- 5.Zhou X, Chen K, Lei H, Sun Z: Klotho gene deficiency causes salt-sensitive hypertension via monocyte chemotactic protein-1/CC chemokine receptor 2-mediated inflammation. J Am Soc Nephrol 26: 121–132, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kuro-o M, Matsumura Y, Aizawa H, Kawaguchi H, Suga T, Utsugi T, Ohyama Y, Kurabayashi M, Kaname T, Kume E, Iwasaki H, Iida A, Shiraki-Iida T, Nishikawa S, Nagai R, Nabeshima YI: Mutation of the mouse klotho gene leads to a syndrome resembling ageing. Nature 390: 45–51, 1997. [DOI] [PubMed] [Google Scholar]
- 7.Xu Y, Sun Z: Molecular basis of Klotho: From gene to function in aging. Endocr Rev 36: 174–193, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yamazaki Y, Imura A, Urakawa I, Shimada T, Murakami J, Aono Y, Hasegawa H, Yamashita T, Nakatani K, Saito Y, Okamoto N, Kurumatani N, Namba N, Kitaoka T, Ozono K, Sakai T, Hataya H, Ichikawa S, Imel EA, Econs MJ, Nabeshima Y: Establishment of sandwich ELISA for soluble alpha-Klotho measurement: Age-dependent change of soluble alpha-Klotho levels in healthy subjects. Biochem Biophys Res Commun 398: 513–518, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou X, Wang X: Klotho: A novel biomarker for cancer. J Cancer Res Clin Oncol 141: 961–969, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takenaka T, Inoue T, Miyazaki T, Kobori H, Nishiyama A, Ishii N, Hayashi M, Suzuki H: Klotho ameliorates medullary fibrosis and pressure natriuresis in hypertensive rat kidneys. Hypertension 72: 1151–1159, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Urakawa I, Yamazaki Y, Shimada T, Iijima K, Hasegawa H, Okawa K, Fujita T, Fukumoto S, Yamashita T: Klotho converts canonical FGF receptor into a specific receptor for FGF23. Nature 444: 770–774, 2006. [DOI] [PubMed] [Google Scholar]
- 12.Wang HL, Xu Q, Wang Z, Zhang YH, Si LY, Li XJ, Yang QH, Xiao H: A potential regulatory single nucleotide polymorphism in the promoter of the Klotho gene may be associated with essential hypertension in the Chinese Han population. Clin Chim Acta 411: 386–390, 2010. [DOI] [PubMed] [Google Scholar]
- 13.Nzietchueng R, El Shamieh S, Benachour H, Labat C, Herbeth B, Ndiaye NC, Masson C, Visvikis-Siest S, Benetos A: Klotho KL-VS genotype is involved in blood pressure regulation. Clin Chim Acta 412: 1773–1777, 2011. [DOI] [PubMed] [Google Scholar]
- 14.Gao LL, Ding X, Xie DM, Yang M, Dong BR: G-395A polymorphism in the promoter region of the KLOTHO gene and hypertension among elderly (90 years and older) Chinese individuals. Genet Mol Res 14: 15444–15452, 2015. [DOI] [PubMed] [Google Scholar]
- 15.Haruna Y, Kashihara N, Satoh M, Tomita N, Namikoshi T, Sasaki T, Fujimori T, Xie P, Kanwar YS: Amelioration of progressive renal injury by genetic manipulation of Klotho gene. Proc Natl Acad Sci U S A 104: 2331–2336, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rhee EJ, Oh KW, Yun EJ, Jung CH, Lee WY, Kim SW, Baek KH, Kang MI, Park SW: Relationship between polymorphisms G395A in promoter and C1818T in exon 4 of the KLOTHO gene with glucose metabolism and cardiovascular risk factors in Korean women. J Endocrinol Invest 29: 613–618, 2006. [DOI] [PubMed] [Google Scholar]
- 17.Arking DE, Becker DM, Yanek LR, Fallin D, Judge DP, Moy TF, Becker LC, Dietz HC: KLOTHO allele status and the risk of early-onset occult coronary artery disease. Am J Hum Genet 72: 1154–1161, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qian J, Zhong J, Yan M, Cheng P, Shi H, Hao C, Gu Y, Lai L: Circulating α-Klotho is related to plasma aldosterone and its follow-up change predicts CKD progression. Kidney Blood Press Res 43: 836–846, 2018. [DOI] [PubMed] [Google Scholar]
- 19.Elijovich F, Weinberger MH, Anderson CAM, Appel LJ, Bursztyn M, Cook NR, Dart RA, Newton-Cheh CH, Sacks FM, Laffer CL; American Heart Association Professional and Public Education Committee of the Council on Hypertension; Council on Functional Genomics and Translational Biology; Stroke Council : Salt sensitivity of blood pressure: A scientific statement from the American heart association [published correction appears in Hypertension 68: e62, 2016]. Hypertension 68: e7–e46, 2016. [DOI] [PubMed] [Google Scholar]
- 20.Wijmenga C, Zhernakova A: The importance of cohort studies in the post-GWAS era. Nat Genet 50: 322–328, 2018. [DOI] [PubMed] [Google Scholar]
- 21.Arcidiacono T, Simonini M, Lanzani C, Citterio L, Salvi E, Barlassina C, Spotti D, Cusi D, Manunta P, Vezzoli G: Claudin-14 gene polymorphisms and urine calcium excretion. Clin J Am Soc Nephrol 13: 1542–1549, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Citterio L, Lanzani C, Manunta P, Bianchi G: Genetics of primary hypertension: The clinical impact of adducin polymorphisms. Biochim Biophys Acta 1802: 1285–1298, 2010. [DOI] [PubMed] [Google Scholar]
- 23.Citterio L, Simonini M, Zagato L, Salvi E, Delli Carpini S, Lanzani C, Messaggio E, Casamassima N, Frau F, D’Avila F, Cusi D, Barlassina C, Manunta P: Genes involved in vasoconstriction and vasodilation system affect salt-sensitive hypertension. PLoS One 6: e19620, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Manunta P, Cusi D, Barlassina C, Righetti M, Lanzani C, D’Amico M, Buzzi L, Citterio L, Stella P, Rivera R, Bianchi G: Alpha-adducin polymorphisms and renal sodium handling in essential hypertensive patients. Kidney Int 53: 1471–1478, 1998. [DOI] [PubMed] [Google Scholar]
- 25.Manunta P, Burnier M, D’Amico M, Buzzi L, Maillard M, Barlassina C, Lanella G, Cusi D, Bianchi G: Adducin polymorphism affects renal proximal tubule reabsorption in hypertension. Hypertension 33: 694–697, 1999. [DOI] [PubMed] [Google Scholar]
- 26.Manunta P, Lavery G, Lanzani C, Braund PS, Simonini M, Bodycote C, Zagato L, Delli Carpini S, Tantardini C, Brioni E, Bianchi G, Samani NJ: Physiological interaction between alpha-adducin and WNK1-NEDD4L pathways on sodium-related blood pressure regulation. Hypertension 52: 366–372, 2008. [DOI] [PubMed] [Google Scholar]
- 27.Levey AS, Stevens LA, Schmid CH, Zhang YL, Castro AF 3rd, Feldman HI, Kusek JW, Eggers P, Van Lente F, Greene T, Coresh J; CKD-EPI (Chronic Kidney Disease Epidemiology Collaboration) : A new equation to estimate glomerular filtration rate [published correction appears in Ann Intern Med 155: 408, 2011]. Ann Intern Med 150: 604–612, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Salvi E, Kutalik Z, Glorioso N, Benaglio P, Frau F, Kuznetsova T, Arima H, Hoggart C, Tichet J, Nikitin YP, Conti C, Seidlerova J, Tikhonoff V, Stolarz-Skrzypek K, Johnson T, Devos N, Zagato L, Guarrera S, Zaninello R, Calabria A, Stancanelli B, Troffa C, Thijs L, Rizzi F, Simonova G, Lupoli S, Argiolas G, Braga D, D’Alessio MC, Ortu MF, Ricceri F, Mercurio M, Descombes P, Marconi M, Chalmers J, Harrap S, Filipovsky J, Bochud M, Iacoviello L, Ellis J, Stanton AV, Laan M, Padmanabhan S, Dominiczak AF, Samani NJ, Melander O, Jeunemaitre X, Manunta P, Shabo A, Vineis P, Cappuccio FP, Caulfield MJ, Matullo G, Rivolta C, Munroe PB, Barlassina C, Staessen JA, Beckmann JS, Cusi D: Genomewide association study using a high-density single nucleotide polymorphism array and case-control design identifies a novel essential hypertension susceptibility locus in the promoter region of endothelial NO synthase. Hypertension 59: 248–255, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arking DE, Krebsova A, Macek M Sr, Macek M Jr, Arking A, Mian IS, Fried L, Hamosh A, Dey S, McIntosh I, Dietz HC: Association of human aging with a functional variant of klotho. Proc Natl Acad Sci U S A 99: 856–861, 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aizawa H, Saito Y, Nakamura T, Inoue M, Imanari T, Ohyama Y, Matsumura Y, Masuda H, Oba S, Mise N, Kimura K, Hasegawa A, Kurabayashi M, Kuro-o M, Nabeshima Y, Nagai R: Downregulation of the Klotho gene in the kidney under sustained circulatory stress in rats. Biochem Biophys Res Commun 249: 865–871, 1998. [DOI] [PubMed] [Google Scholar]
- 31.Eren M, Boe AE, Murphy SB, Place AT, Nagpal V, Morales-Nebreda L, Urich D, Quaggin SE, Budinger GRS, Mutlu GM, Miyata T, Vaughan DE: PAI-1-regulated extracellular proteolysis governs senescence and survival in Klotho mice. Proc Natl Acad Sci U S A 111: 7090–7095, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee JW, Chou CL, Knepper MA: Deep sequencing in microdissected renal tubules identifies nephron segment-specific transcriptomes. J Am Soc Nephrol 26: 2669–2677, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hu MC, Shi M, Zhang J, Pastor J, Nakatani T, Lanske B, Razzaque MS, Rosenblatt KP, Baum MG, Kuro-o M, Moe OW: Klotho: A novel phosphaturic substance acting as an autocrine enzyme in the renal proximal tubule. FASEB J 24: 3438–3450, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ben-Dov IZ, Galitzer H, Lavi-Moshayoff V, Goetz R, Kuro-o M, Mohammadi M, Sirkis R, Naveh-Many T, Silver J: The parathyroid is a target organ for FGF23 in rats. J Clin Invest 117: 4003–4008, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ritter CS, Zhang S, Delmez J, Finch JL, Slatopolsky E: Differential expression and regulation of Klotho by paricalcitol in the kidney, parathyroid, and aorta of uremic rats. Kidney Int 87: 1141–1152, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matsumura Y, Aizawa H, Shiraki-Iida T, Nagai R, Kuro-o M, Nabeshima Y: Identification of the human klotho gene and its two transcripts encoding membrane and secreted klotho protein. Biochem Biophys Res Commun 242: 626–630, 1998. [DOI] [PubMed] [Google Scholar]
- 37.Kuro-O M: The Klotho proteins in health and disease. Nat Rev Nephrol 15: 27–44, 2019. [DOI] [PubMed] [Google Scholar]
- 38.Aucella F, Corsonello A, Leosco D, Brunori G, Gesualdo L, Antonelli-Incalzi R: Beyond chronic kidney disease: The diagnosis of Renal Disease in the Elderly as an unmet need. A position paper endorsed by Italian Society of Nephrology (SIN) and Italian Society of Geriatrics and Gerontology (SIGG). J Nephrol 32: 165–176, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sugiura H, Yoshida T, Shiohira S, Kohei J, Mitobe M, Kurosu H, Kuro-o M, Nitta K, Tsuchiya K: Reduced Klotho expression level in kidney aggravates renal interstitial fibrosis. Am J Physiol Renal Physiol 302: F1252–F1264, 2012. [DOI] [PubMed] [Google Scholar]
- 40.Doi S, Zou Y, Togao O, Pastor JV, John GB, Wang L, Shiizaki K, Gotschall R, Schiavi S, Yorioka N, Takahashi M, Boothman DA, Kuro-o M: Klotho inhibits transforming growth factor-beta1 (TGF-beta1) signaling and suppresses renal fibrosis and cancer metastasis in mice. J Biol Chem 286: 8655–8665, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou L, Li Y, Zhou D, Tan RJ, Liu Y: Loss of Klotho contributes to kidney injury by derepression of Wnt/β-catenin signaling. J Am Soc Nephrol 24: 771–785, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li Y, Chen F, Wei A, Bi F, Zhu X, Yin S, Lin W, Cao W: Klotho recovery by genistein via promoter histone acetylation and DNA demethylation mitigates renal fibrosis in mice. J Mol Med (Berl) 97: 541–552, 2019. [DOI] [PubMed] [Google Scholar]
- 43.Luft FC, Miller JZ, Grim CE, Fineberg NS, Christian JC, Daugherty SA, Weinberger MH: Salt sensitivity and resistance of blood pressure. Age and race as factors in physiological responses. Hypertension 17[Suppl 1]: I102–I108, 1991. [DOI] [PubMed] [Google Scholar]
- 44.Weinberger MH: Salt sensitivity of blood pressure in humans. Hypertension 27: 481–490, 1996. [DOI] [PubMed] [Google Scholar]
- 45.Sun Z: Aging, arterial stiffness, and hypertension. Hypertension 65: 252–256, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Arking DE, Atzmon G, Arking A, Barzilai N, Dietz HC: Association between a functional variant of the KLOTHO gene and high-density lipoprotein cholesterol, blood pressure, stroke, and longevity. Circ Res 96: 412–418, 2005. [DOI] [PubMed] [Google Scholar]
- 47.Zhou L, Mo H, Miao J, Zhou D, Tan RJ, Hou FF, Liu Y: Klotho ameliorates kidney injury and fibrosis and normalizes blood pressure by targeting the renin-angiotensin system. Am J Pathol 185: 3211–3223, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lim SC, Liu JJ, Subramaniam T, Sum CF: Elevated circulating alpha-klotho by angiotensin II receptor blocker losartan is associated with reduction of albuminuria in type 2 diabetic patients. J Renin Angiotensin Aldosterone Syst 15: 487–490, 2014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.