Abstract
Diabetes mellitus is a metabolic disorder predominantly caused by the dysfunction of pancreatic β-cells. This dysfunction is partly caused by the dysregulation of pyruvate dehydrogenase (PDH), which acts as an important mediator of pyruvate oxidation after glycolysis and fuels the tricarboxylic acid cycle. Previous studies have reported decreased PDH expression in rodent models and humans with type 2 diabetes mellitus (T2DM), suggesting that PDH may play an important role in the development of T2DM. However, the mechanism by which PDH affects insulin secretion and β-cell development is poorly understood. Using immunofluorescence staining, the present study found that the expression of pyruvate dehydrogenase E1-α subunit (PDHA1; encoded by the PDHA1 gene) in the islets of type 2 diabetic mice (db/db mice) was lower than in wild-type mice, which indicated the possible association between PDHA1and diabetes. To further understand this mechanism, an inducible, islet-specific PDHA1 knockout mouse (βKO) model was established. The phenotype was authenticated, and the blood glucose levels and islet function between the βKO and control mice were compared. Though no changes were found in food intake, development status, fasting blood glucose or weight between the groups, the level of insulin secretion at 30 min after glucose injection in the βKO group was significantly lower compared with the control group. Furthermore, the performed of the βKO mice on the intraperitoneal glucose tolerance test was visibly impaired when compared with the control mice. Pancreatic tissues were collected for hematoxylin and eosin staining, immunohistochemical and confocal laser-scanning microscopy analysis. Examination of the islets from the βKO mouse model indicated that abolishing the expression of PDH caused a compensatory islet enlargement and impaired insulin secretion.
Keywords: pancreatic β-cell, pyruvate dehydrogenase, insulin, knockout mice, diabetes mellitus
Introduction
Diabetes mellitus is a metabolic disorder characterized by hyperglycemia as a result of β-cell dysfunction and insulin resistance (1,2). The pathogenesis of diabetes is not fully understood, but includes insulin resistance and a deficit of β-cell mass. Increasing attention has been paid to the significant role of mitochondrial energy metabolism in the development of diabetes mellitus (3,4). Mitochondria convert energy-storing molecules, such as glucose, free fatty acids and amino acids, into ATP via oxidative phosphorylation, and are involved in β-cell proliferation, and the synthesis and release of insulin (5).
Mitochondria are widely distributed in eukaryotic cells, providing energy for cellular activities, participating in important metabolic pathways, maintaining the cell ion balance and participating in signal transduction, all of which are important processes to maintain normal cellular activity (6).
Mitochondrial regulation controls various aspects of cellular homeostasis in the organism (7–9). Furthermore, the regulation of mitochondria contributes to the pathogenesis of metabolic disorders, including obesity, metabolic syndrome, type 2 diabetes mellitus (T2DM) and cancer (10,11). Previous studies have indicated that dysfunction in mitochondrial energy metabolism may be the fundamental cause of impaired insulin secretion by β-cells (11,12).
The primary role of β-cells is to release insulin in response to postprandial blood glucose levels (2). Glucose is transported into β-cells by diffusion and is subsequently metabolized during glycolysis and the tricarboxylic acid (TCA) cycle (5,7). Glycolysis leads to the production of pyruvate, which enters the mitochondria to be oxidized (8). Pyruvate is metabolized by the pyruvate dehydrogenase (PDH) complex (PDHc), a controller of glucose oxidation, which is present in the mitochondrial matrix, and is further converted to acetyl-coenzyme A (CoA) to obtain citrate that participates in the TCA cycle (12–14). This increases the ATP/ADP ratio and causes the plasma ATP-dependent K+ channel to close. The voltage-dependent Ca2+ channel opens and triggers the exocytosis of insulin (12,15). The metabolic coupling factor amplifies insulin secretion, meaning that glucose and products of glucose metabolism potentiate the exocytosis of insulin molecules without any further increase in cytosolic Ca2+ concentrations (11).
The most important enzyme to determine which path to take is PDHc, which catalyzes the initiating reaction in the TCA cycle and directly regulates insulin secretion. Impairment of the PDHc directly affects the growth and function of β-cells (7,9).
The PDHc is a composite of the mitochondrial enzyme system and a member of the 2-oxygen (generation) acid dehydrogenase complex family. PDHc is composed of thiamine-dependent tetrameric (α2β2) PDH (PDHA1; encoded by the PDHA1 gene), dihydrolipoamide acetyltransferase and flavin adenine dinucleotide-containing dihydrolipoamide dehydrogenase (E3), which is attached to the complex by the E3-binding protein (16,17).
PDH catalyzes the irreversible oxidative decarboxylation of pyruvate into acetyl-CoA and reduces NAD+ to NADH, which links the aerobic oxidation of glucose with the cyclic capacity of TCA, playing an important role in the energy metabolism of the mitochondrial respiratory chain and distinguishing between aerobic and anaerobic oxidation (18,19). When the levels of PDH are reduced, the proportion of energy supplied by glucose decreases, while the contribution of other energy-producing molecules, such as lipids and amino acids, increases (20). The activity of PDH is determined by the inhibitory effect of pyruvate dehydrogenase kinase (PDK) on the PDHc (17,21–23). PDK phosphorylates PDH-E1α, inactivating PDH. Loss of PDH activity leads to glucose metabolic disorders and tissue damage, which influence the growth, differentiation and functional expression of β-cells (4,18).
Previous studies have focused on the function of PDH (17,24,25). Mice with PDHA1 knocked out in the heart exhibited ventricular dysfunction, predominantly diastolic (26,27), while treatment with dichloroacetic acid has been reported to reverse ventricular dysfunction (28). The hyperinsulinemia-positive glucose clamping test in obese Wistar rats revealed higher plasma lactate levels (17,29). In the liver, under insulin resistance or obese conditions, PDH activity is abnormally reduced (30), glucose utilization is reduced and hepatic glycogen production is increased, leading to high levels of blood glucose (23,25,31).
Therefore, mitochondrial metabolism plays a significant role in the onset and development of diabetes. Previous studies have shown that the expression of PDHc is reduced in rodent models of T2DM and the human body (22,32), indicating the central role played by PDHc in the development of diabetes. However, the effect of PDHA1 on pancreatic β-cells has not been extensively explored. The present study aimed to clarify the association between PDHA1 and diabetes, assess the effect of PDHA1 on β-cell morphology and function, and elucidate the possible mechanism guiding PDHA1 action. The present study may provide a new theoretical framework to explain diabetes development and proposes a potential molecular target for the treatment of this disorder.
Materials and methods
Animals
B6.Cg-Tg (Ins1-cre/ERT) 1 lphi/J mice (cat. no. 024709; hereafter referred to as Ins-cre+/− mice) and B6.129P2-Pdha1tm1Ptl/J mice (cat. no. 017443; hereafter referred to as PDHA1flox/floxmice) were obtained from the Jackson Laboratory (n=4/group, 2 males and 2 females, ~20 g/each). Then, 5 db/db mice and 5 C57BL/6 mice were provided by the animal laboratory of the Southern Medical University (Guangzhou, China). All mice were given food and water ad libitum, housed in a clean laminar animal room at a controlled indoor temperature of 20–25°C and a humidity of 40–70% with a 12-h light/dark cycle.
Cre-loxP system
Based on the principle of the Cre-loxP system, PDHA1flox/flox mice were mated with Ins-cre+/− mice to obtain the F2 PDHA1flox/−Cre+/− mouse generation. These heterozygotes were further crossed with PDHA1flox/− Ins-cre+/− mice to produce PDHA1flox/flox Ins-cre+/− mice. Ins-cre recombinase effectively eliminates the sequence between two loxP sites. Moreover, the expression of Ins-cre recombinase can be triggered by the simultaneous action of insulin and exogenous tamoxifen (intraperitoneally injected tamoxifen for 7 consecutive days, 50 mg/kg) in mouse islets. Therefore, by controlling the time of exogenous tamoxifen injection, the PDHA1 gene could be specifically knocked out in the mouse islets (33,34). Mice with Ins-cre+/− genotypes were selected as the negative control (NC) group, while mice of the same age and gender with genotype PDHA1flox/flox Ins-cre+/− were the knockout experimental (βKO) group.
Genotypic identification
Then, ~3–4 mm length of tail was removed from the mouse with ophthalmic scissors and 100 µl each of rat tail lysate A (0.5% SDS, 0.1 M NaCl, 0.05 M EDTA, 0.01 M Tris-HCl pH 8.0 and protease K 100 g/ml) and B (NaOH 1 mmol/ml) added and then heated for 1 h. The supernatant contained mouse DNA (34). DNA was used for genotyping using PCR with mouse-specific primers (Table I) for the PDHA1 and Ins-cre genes. The reaction (20 µl total) contained 10 µl PCR mix (2X Taq Plus Master Mix, Invitrogen; Thermo Fisher Scientific, Inc.), 2 µl of each primer (primer concentration 10 µM) and 2 µl template DNA. For Ins-cre amplification, the PCR reaction conditions were as follows: 94°C for 2 min; 10 cycles of 94°C for 20 sec, 65°C for 15 sec and 68°Cf or 10 sec; 28 cycles of 94°C for 15 sec, 50°C for 15 sec and 72°C for 10 sec; and 72°C for 2 min. For PDHA1 amplification the PCR conditions were as follows: 94°C for 2 min; 10 cycles of 94°C for 20 sec, 65°C for 15 sec, 68°C for 10 sec; 28 cycles of 94°C for 15 sec, 60°C for 15 sec and 72°C for 10 sec; and 72°C for 2 min. The PCR products were separated using agarose gel electrophoresis (2%; 6 µl/well). Image analysis was performed using an Automatic Digital Gel Imaging System (Tanon 1600; Tanon Science & Technology, Co.). Each experiment was performed in triplicate.
Table I.
Primers used for genotyping.
| Primer name | Forward (5′-3′) | Reverse (3′-5′) |
|---|---|---|
| PDHA1 | AGCAGCCAGCACGGACTACT | GCAGCCAAACAGATTACACC |
| INS1-CRE | AGCAGCCAGCACGGACTACT | TGCGAACCTCATCACTCGT |
| INS1-CRE internal positive control | CTAGGCCACAGAATTGAAAGATCT | GTAGGTGGAAATTCTAGCATCATCC |
PDHA1, pyruvate dehydrogenase E1-α subunit.
Hematoxylin and eosin (H&E) staining
Mice were intraperitoneally injected with pentobarbital sodium solution at 150 mg/kg, with death determined after cessation of breathing for 2–3 min. When mice suffered pain and discomfort that affected their quality of life, they were euthanized with pentobarbital solution; this was set as the humane end point. The pancreas was removed from the mice, weighed, fixed for 24 h in 4% paraformaldehyde at 4°C and embedded in paraffin. The pancreas was sliced into 4-µm thick sections and stained with hematoxylin for 5 min at room temperature, alcohol hydrochloric acid for 1 sec and eosin for 1 sec. The tissue was dehydrated using graded ethanol, cleared in dimethylbenzene, sealed and observed under an inverted fluorescence microscope (IX53; Olympus Corporation, magnification, ×200; 3 fields analyzed per sample). Image-Pro Plus 5 (Media Cybernetics, Inc.) was used to count cells and measure cell size.
Western blot analysis
Pancreas tissue was collected, weighed and ground into a powder under liquid nitrogen. The samples were mixed with RIPA lysis buffer [50 mM Tris (pH 7.4), 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate]. The protein concentration was determined using the bicinchoninic acid method. Samples (20 µl) were separated using 10% SDS-PAGE, transferred to nitrocellulose membranes, then blocked with 5% skimmed milk at room temperature, incubated with the following primary antibodies for 14 h at 4°C: PDHA1 (1:1,000; cat. no. ab110334; Abcam) and GAPDH (1:1,000; cat. no. ab9484; Abcam). Membranes were washed three times with TBS with Tween 20 for 5 min each time and incubated with an horseradish peroxidase (HRP)-conjugated secondary antibody (1:500; cat. no. ab97023; Abcam) for 2 h at room temperature. Chemiluminescence was performed using Pierce™ Fast Western Blot ECL substrate (cat. no. 35055; Thermo Fisher Scientific, Inc.). Protein bands were visualized using the Odyssey Infrared Imaging System (LI-COR Biosciences). The results were quantitatively analyzed using Image-Pro Plus 6.0 software (Media Cybernetics, Inc.). The experiments were repeated three times for each sample.
Immunohistochemistry
The tissue samples were cut into 3–4-µm thick sections, dewaxing with xylene and hydration with alcohol (xylene1 10 min, xylene2 10 min, 95, 90, 80, 70 and 50% alcohol, each 5 min). Following permeabilization with 0.1% Triton X-100 and 3% H2O2, blocking with normal goat serum (cat. no. MP20008; Shanghai Yuanye Science & Technology) at room temperature for 30, 30 and 60 min, respectively. The slides were incubated with a primary antibody (PDHA1; 1:500; cat. no. ab110334; Abcam) at 4°C in a wet box for 14–16 h. Following this, the slides were washed with PBS for 3 min three times. A secondary HRP-conjugated antibody (1:300, cat. no. ab97023; Abcam) was added dropwise to each section, followed by incubation in a wet box at 37°C for 2 h. After washing for 3 min three times with PBS, the DAB color solution (OriGene Technologies, Inc.) was added, and tap water was used to stop development. The slides were counterstained with hematoxylin at room temperature for 3 min and washed with tap water until a blue color developed. The slides were observed and images were captured using a light microscope (Zeiss AG). As a negative control the primary antibody was substituted with PBS in parallel experiments.
Immunofluorescence
Tissue samples were cut into sections and processed as aforementioned. Slides were incubated with mouse anti-PDHA1 (1:200; cat. no. ab110334; Abcam) and rabbit anti-insulin (1:200; cat. no. ab63820; Abcam) overnight at 4°C in a wet box. The slides were washed for 3 min three times and incubated in the dark with the corresponding diluted secondary antibodies [1:500; cat. no. ab150077 (Alexa Fluor® 488-conjugated goat anti-rabbit IgG) and ab150116 (Alexa Fluor 594-conjugated goat anti-mouse IgG; Abcam) at 37°C for 2 h. The secondary antibody solution was discarded and the slides were washed three times. Slides were incubated with DAPI (1drop: 1 ml, ~1 min at room temperature. Images were captured under a laser confocal microscope and 5 visual fields containing pancreatic islet cells were selected under a the microscope (magnification, ×400); the number of PDHA1 positive cells and total number of cells in each visual field were counted by Image-Pro Plus 6.0 software (Media Cybernetics, Inc.). The rate of PDHA1 positive cells in islet cells was calculated.
Reverse transcription-quantitative PCR (RT-qPCR)
Total RNA from the islets, liver and adipose tissues were extracted using TRIzol (Vazyme). Absorbance at 260 and 280 nm (A260 and A280, respectively) was determined using an ultraviolet spectrophotometer, and the ratio of the optical density (OD) at these wavelengths (ODA260/A280) was 1.8–2.0. Complementary DNA was reverse transcribed using Hiscript II Q RT SuperMix for qPCR (Nanjing KeyGen Biotech Co., Ltd.), according to the manufacturer's instructions in a final volume of 20 µl. qPCR was performed using real-time PCR Master Mix (SYBR Green; KGA1339-1 200T, Nanjing KeyGen Biotech Co, Ltd.). The primers for PDHA1 were: Forward, 5′-GAGCTGAGCAGCTGTGTAAC-3′ and reverse, 5′-TGCCAATCGTTACAGGTATTACAG-3′. GAPDH forward, 5′-ACCACAGTCCATGCCATCAC-3′ and reverse, 5′-TCCACCACCCTGTTGCTGTA-3′. The thermocycling conditions were: 95°C for 30 sec, 95°C 10 sec; followed by 45 cycles of 60°C for 30 sec, 72°C for 30 sec; 60°C for 60 sec and 95°C for 15 sec. The gene expression was quantified with the 2−∆∆Cq method (35). Data were normalized to GAPDH mRNA levels. All reactions were performed in triplicate.
Intraperitoneal glucose tolerance test (IPGTT) and intraperitoneal insulin tolerance test (IPITT)
βKO and NC mice were selected (5/group) of the same age (8 weeks) and weight. Their body weight and fasting blood glucose levels were measured every 2 weeks. The food intake and nutritional status of the mice were also monitored. The mice were weighed and glucose was administered intraperitoneally (1.5 g/kg body weight) after 14 h fasting. Following this, blood samples (1–2 µl) were collected through the tail vein at 0, 15, 30, 60 and 120 min. The glucose levels were measured using a glucometer (Accu-Chek Active; Roche Diagnostics). At 1 week, the mice were fasted for 4 h, weighed and injected intraperitoneally with insulin (0.6 U/kg body weight). Blood samples (1–2 µl) were subsequently withdrawn from the tail vein at 0, 15, 30 and 60 min for blood glucose measurements.
Insulin concentration measurement
Glucose solution (5%; 1.5 g/kg) was injected into mice that had been fasting for 14 h. Blood samples (0.8–1 ml) were collected from the hearts of mice under anesthesia. Blood was collected from the heart at the moment of the injection or 30 min later, then euthanasia. The serum was blood was allowed to settle and separate for 30 min prior to centrifugation at 1,680 × g for 10 min, both at 4°C. An insulin ELISA kit (cat. no. EZRMI-13K; EMD Millipore) was used to measure the content of insulin in the mouse serum. Each sample was analyzed in triplicate. The samples and reaction solutions were added and mixed in the dark for 30 min. The OD value was determined using a UV spectrophotometer. A standard curve was constructed and the corresponding conversion of sample concentration was carried out.
Primary islet extraction
All experiments were repeated 3 times. The mouse epidermis was disinfected with ethanol, and the abdominal cavity was opened to allow isolation of the pancreatic tissue. The pancreases were rinsed with pre-cooled PBS solution, visible blood and nerves were removed and the pancreas was placed in a pre-cooled petri dish. Collagenase P (0.3 mg) was injected in each pancreas tissue. The tissue was then digested at 37°C for 15 min. The digestion was terminated using pre-cooled Hank's buffer (Gino Biomedical Technology Co., Ltd.), and after centrifugation (430 × g; 30 sec, 4°C), the digested tissue was filtered with a 40-mesh screen. Pancreatic islet cells (brown, cell mass) were selected with a 10 µl pipette under stereomicroscope. The selected islet cells used to extract RNA.
Statistical analysis
All experiments were repeated 3 times. Statistical analysis was performed using GraphPad 5 (GraphPad Software, Inc.), Image-Pro Plus 6.0 (Media Cybernetics, Inc.) and SPSS 22 (IBM Corp). Paired t-tests and ANOVA followed by a Bonferroni post-hoc analysis were used to determine differences between the βKO and NC mice. Data are presented as the mean ± SD. P<0.05 was considered to indicate a statistically significant difference.
Results
PDHA1 levels are lower in db/db mice
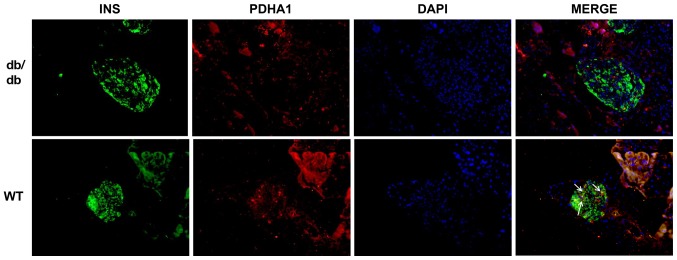
db/db mice are an animal model of T2DM that exhibit elevated blood glucose and insulin resistance in their adulthood (36). To investigate the expression of PDHA1 in diabetic mice, pancreas sections of db/db mice and C57BL/6 mice were analyzed using immunofluorescence. The cells that secreted insulin had green fluorescence, while cells expressing PDHA1 exhibited red fluorescence. The expression of PDHA1 in the islets of db/db mice was markedly downregulated compared with in C57BL/6 mice (Fig. 1), which was consistent with histology results showing that PDHA1 expression was decreased in T2DM mammals (36,37).
Figure 1.
PDHA1 expression is reduced in db/db mice. Decreased expression of the PDHA1 protein in db/db mice. The nucleus is stained blue, PDHA1 is red and INS is green. PDHA1, pyruvate dehydrogenase E1-α subunit; INS, insulin; WT, wild-type.
Generation and validation of the βKO mouse model
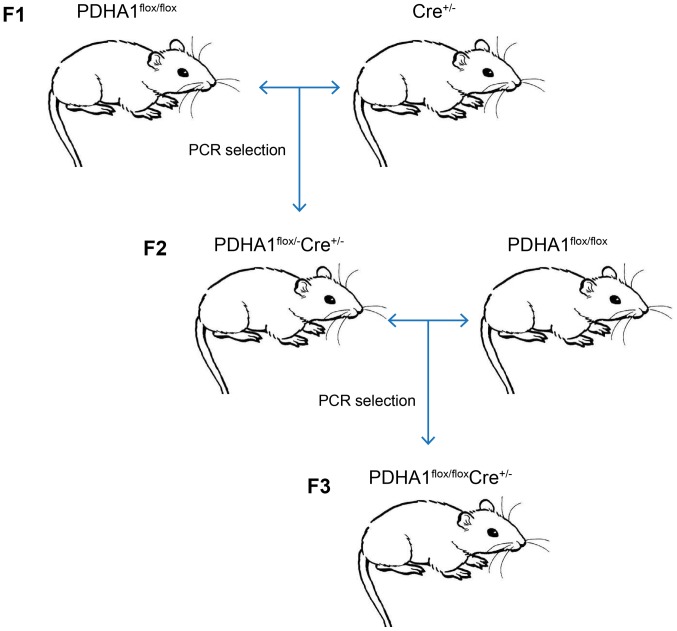
To further investigate the role of PDHA1 in the pancreas, a βKO mouse model was established. As the homozygous loss of PDHA1 may be embryonically lethal (37,38), the Cre-loxP method was used to generate mice with a pancreatic β-cell-specific knockout of PDHA1. Following the steps shown in Fig. 2, PDHA1flox/flox Ins-cre+/− mice were obtained by crossing PDHA1flox/− Ins-cre+/− mice and PDHA1flox/flox mice.
Figure 2.
Scheme showing breeding of PDHA1flox/flox Ins-cre+/− mice. PDHA1flox/flox mice were mated with Ins-cre+/− mice and then F2 PDHA1flox/− Ins-cre+/− mice generation was obtained. PDHA1flox/flox Ins-cre+/− mice were generated by crossing PDHA1flox/− Ins-cre+/− mice with PDHA1flox/flox mice. Genotyping was performed on 4-week-old mice using PCR. PDHA1, pyruvate dehydrogenase E1-alpha subunit.
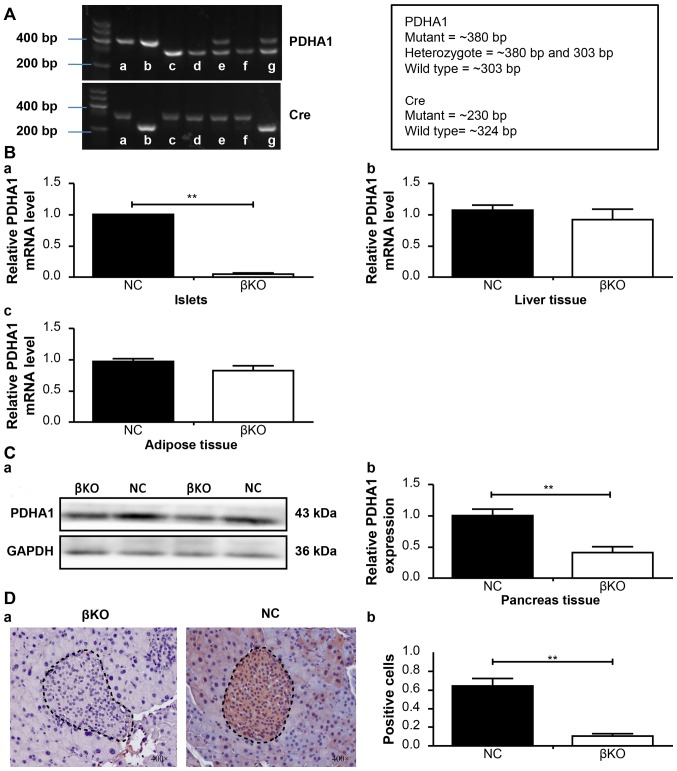
PCR analysis of DNA prepared from tail snips of the male knockout mice (4 weeks old) confirmed the recessiveness of the PDHA1 gene (303 bp fragment) and the dominance of PDHA1 (380 bp fragment), as well as that of the Ins-cre transgene (230 and 324 bp fragment; Fig. 3A). Subsequently, tamoxifen (5 mg/ml) was injected to induce the removal of the DNA between the loxP sites. PDHA1+/+ Ins-cre+/− mice were designated as βKO mice, while the PDHA1+/+ Ins-cre− mice from the same litter were referred to as NC mice. After 10 months of breeding, a total of 17 βKO mice were obtained, which were bright black and showed excellent growth and nutritional status.
Figure 3.
Generation and validation of βKO mouse model. (A) PCR genotyping results of PDHA1loxp/loxp mice and Ins-cre recombinase mice from DNA obtained from tail samples of 4-week-old mice; b is PDHA1flox/flox Ins-cre+/− and a is PDHA1flox/flox Ins-cre− mice; c, d and f wild type mice; e PDHA1flox/−, and g PDHA1+/− Ins-cre+/−. (B) Reverse transcription-quantitative PCR analysis of (Ba) islets, (Bb) liver and (Bc) adipose tissue from the βKO and NC groups. (C) PDHA1 protein was extracted from the pancreas of two groups of mice and then measured by (Ca) western blotting and (Cb) gray value analysis of the protein. (D) Immunohistochemical staining was performed to identify the effect of the PDHA1 gene in mouse pancreatic tissue. (Da) Immunohistochemical staining (magnification, ×400) was performed and (Db) the percentage of positive cells was calculated. The results show the mean ± SD of 5 mice. Data are expressed as the mean ± SD. **P<0.01. βKO, mouse PDHA1 knockout in β-cells; NC, negative control; PDHA1, pyruvate dehydrogenase E1-α subunit.
RT-qPCR and western blotting was used to verify the effect of PDHA1 knockout on the islets. RT-qPCR revealed that the relative PDHA1 mRNA levels in the pancreas islets were significantly reduced in the βKO mice (P<0.001) compared with the NC mice (Fig. 3B). Similar results were obtained in the western blot analysis; the expression of the PDHA1 protein was significantly lower in the βKO mice than in the NC mice (P<0.001; Fig. 3C), indicating that the expression of PDHA1 was almost completely abolished in the pancreatic islets of βKO mice.
To exclude the possible effect of loxP sites on target gene expression in other tissues or organs (14), the mRNA levels of PDHA1 in the liver and adipose tissue of βKO and NC mice were determined (Fig. 3B). The results showed no significant difference in PDHA1 expression in the liver and adipose tissues between the groups. To observe the distribution of PDHA1 in the pancreas of βKO and NC mice, immunohistochemistry was performed (Fig. 3D). It was found that the PDHA1 protein was predominantly distributed in the gland cells and the islets of the pancreas in the NC group; however, in the βKO group there was almost no brown-red PDHA1 staining in the islets (P<0.001), indicating that the expression of PDHA1 in the islets was significantly reduced. These results indicated that the mouse model had been successfully established.
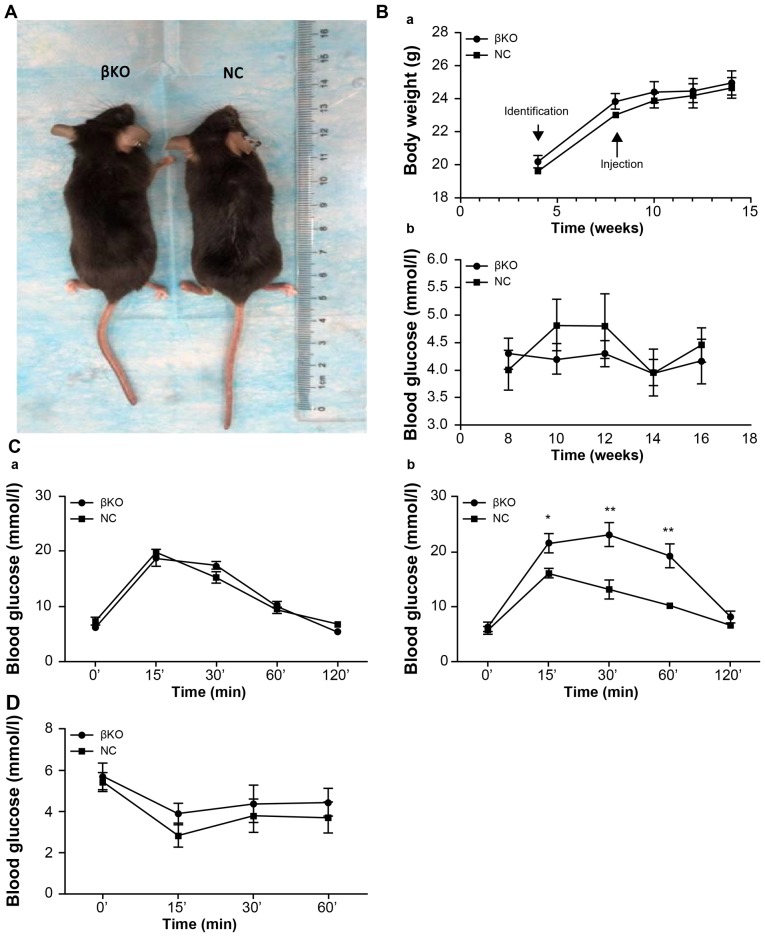
βKO mice exhibit impaired glucose, but not insulin, tolerance
Age- and weight-matched βKO and NC mice were compared. There were no differences in hair, morphology and nutritional status between the two groups (Fig. 4A). The weight of the mice increased with time; however, there were no significant differences between the two groups. Moreover, there were no significant alterations in fasting blood glucose levels (Fig. 4B). An IPGTT was performed in 8-week-old mice (Fig. 4Ca) and 12-week-old mice (Fig. 4Cb). βKO mice were less tolerant to glucose than the NC group (Fig. 4Cb). However, the IPITT values subsequently obtained showed no difference between the groups (Fig. 4D), indicating that the peripheral tissues of the βKO mice had similar levels of insulin sensitivity and showed no signs of insulin resistance compared with the NC mice. This indicated that the cause of hyperglycemia in the βKO mice was not related to insulin resistance in peripheral tissues, but instead was related to cellular dysfunction.
Figure 4.
βKO mice exhibit impaired glucose, but not insulin, tolerance. (A) The knockout mouse (left) and the control mouse (right). (Ba) Weight and (Bb) fasting blood glucose levels in male mice. (C) Intraperitoneal glucose tolerance test (Ca) before and (Cb) after injection with tamoxifen. (D) Intraperitoneal insulin tolerance test results from 12-week-old male mice. Results are expressed as the mean ± SD of 5 mice in all panels. *P<0.05, **P<0.01 vs. NC. βKO, mouse PDHA1 knockout in β-cells; NC, negative control; PDHA1, pyruvate dehydrogenase E1-α subunit.
Impaired glucose-stimulated insulin secretion in βKO mice
To investigate the variation in the function of the β-cells in knockout mice, serum insulin levels were measured at different times after glucose stimulation. It was found that serum insulin levels were significantly increased at 30 min (3.5±0.52 ng/ml) after glucose injection in NC mice. However, there was little change (0.78±0.26 ng/ml) in the levels of insulin in the βKO mice after glucose challenge (Fig. 5A).
Figure 5.
Glucose-stimulated insulin secretion is impaired in βKO mice. (A) Level of glucose-stimulated insulin secretion was determined in vivo in βKO and NC mice. The results show the mean ± SD from 3 mice. (B) Representative images of hematoxylin and eosin staining of islets from 16-week-old NC and βKO mice. (C) Quantification of the size of the islets in NC and βKO mice. **P<0.01. βKO, mouse PDHA1 knockout in β-cells; NC, negative control; PDHA1, pyruvate dehydrogenase.
This suggested that the impaired glucose tolerance in βKO mice may be related to impaired insulin secretion by β-cells. Considering that an abnormality in islet cells may impair secretion from islets, islet paraffin sections from βKO mice were analyzed. H&E staining (Fig. 5B) showed that the islets of the βKO mice were abnormally large compared with those of the NC mice. Most islets in the control group were tightly clustered and numerous, whereas those of the βKO mice were distributed loosely with an increased volume. The mean size of islets was significantly increased in βKO mice at 16 weeks compared with the NC mice (Fig. 5C). These results suggested that PDHA1 serves a significant role in the development of pancreas islets and that without PDHA1, the islet cells undergo significant changes in either morphology or function.
Discussion
Diabetes is a condition that affects individuals worldwide and can be caused by multiple factors, including hereditary, environmental and lifestyle factors, and can severely and negatively affect quality of life for patients (39). Interactions between various genes and environmental factors result in mutual causality, which can often aggravate symptoms (40,41). A number of previous medical studies have attempted to elucidate the effects of various factors on the growth and development of β-cells and insulin secretion (42) in order to explore the possible pathogenesis of diabetes.
The PDHA1 gene, which encodes PDHA1, is located on the short arm of the X chromosome (Xp22.12) and is ~17.5 kb in length (43,44), and contains a conservative thiamine diphosphate-binding region (2,44). PDH is a heterogeneous tetramer, whose components are encoded by the PDHA1 and PDHB genes (26). The expression level of PDHA1 is the primary determinant of PDH activity (45,46). When the activity of PDHA1 is impaired, pyruvate can no longer be metabolized into acetyl-CoA. This causes an increased flux in the production of acetyl-CoA from fats, amino acids and proteins, subsequently affecting metabolic states, such as fats and proteins, resulting in impaired cell growth and function (45). Another pathway by which mitochondria can be supplied with pyruvate is through pyruvate carboxylase (PC) (47), which catalyzes the conversion of pyruvate to oxaloacetic acid, which then either enters the TCA cycle or is used in gluconeogenesis (48). The Cre/loxP recombinase system is a conditional, inducible and spatiotemporal-specific gene targeting technology that is widely used in basic research into gene function (49). In the present study, this technique was used to construct a mouse model with an islet cell-specific knockout of the PDHA1 gene using adult male mice by regulating the injection time of tamoxifen. The expression levels of the PDHA1 protein were determined in pancreas sections of T2DM (db/db) and wild-type mice. The results of the present study suggested an association between the PDHA1 gene and T2DM, which is consistent with previous studies (5,22,24).
To verify the causal relationship between changes in PDHA1 protein levels and the development of T2DM, the Cre/loxP recombinase system was used (50,51) to construct transgenic mice with a specific knockout of the PDHA1 gene in β-cells. This model was then used to further investigate the role of PDHA1 in the development of T2DM by comparing the phenotypes of mice in the βKO and NC groups.
Homozygous Ins-cre mice exhibit embryonic lethality, and cannot survive and reproduce (34). Therefore, PDHA1flox/flox mice were bred with Cre+/− mice and tail tissue samples from the offspring were used to verify successful crossing. PDHA1flox/− Ins-cre+/− mice were then bred with PDHA1flox/flox mice; PDHA1flox/flox Ins-cre+/− mice were selected as the experimental group. Subsequently, tissues and cells were evaluated via RT-qPCR and western blot analysis. The experimental results indicated that β-cell-specific PDHA1 gene knockout mice had been successfully established.
Knocking out the PDHA1 gene in the β-cells of adult male mice did not significant alter the body weight, fasting blood glucose, body shape or nutritional status between the βKO and NC groups, which was consistent with the results of a study by Srinivasan et al (52). However, the authors of this previous study directly knocked out the PDHA1 gene in specific islet cells of embryos without tamoxifen induction, primarily studied the gene in immature mice, and did not discuss the morphology or phenotype of the βKO mice cells.
In the present study, the relationship between the PDHA1 gene and T2DM in adult mice (8 weeks old) was assessed. As the knockout affects only the β-cells of the islets, other tissues and organs may have compensated for the reduced expression of the PDHA1 gene. However, no differences in body weight and fasting blood glucose were observed between the two groups. It was found that the glucose tolerance of βKO mice was impaired compared with the control group, as determined by the higher blood glucose levels after injection of a glucose solution. The results of the present study suggested that the deletion of PDHA1 affected the homeostasis of glucose metabolism and the TCA cycle. Subsequent experiments should determine the levels of acetyl-CoA and the fatty acid content, to verify this conjecture.
To investigate whether the impaired homeostasis of glucose metabolism is due to the effects of insulin resistance in peripheral tissues (including the liver, fat and skeletal muscles) or the β-cells themselves, IPITT was performed. The results of the IPITT indicated that the elevated blood glucose level was associated with impaired islet function, not with peripheral resistance. The hypothesis that β-cell insulin secretion was functionally impaired was confirmed by ELISAs, contrary to the results of Nicholls et al (53). This discrepancy may be as a result of the models used; the study by Nicholls et al (53) was conducted at the cellular level and did not test insulin levels in animals. Furthermore, the previous study indirectly influenced the activity of PDH through PDK and did not directly interfere with the expression of PDH. Other studies have shown that PDH regulates insulin secretion in vitro (20,45,54).
The islets of the βKO group were irregularly enlarged with clear internal vacuoles, but no islet atrophy or necrosis was observed. These results, together with those from the glucose tolerance experiment, suggested that the morphological changes seen in the cells may be due to a compensatory increase in early diabetic islet cells, and an overall decrease in insulin secretion. The present study suggested that the PDHA1 gene plays an important role in the maintenance of β-cell status and insulin secretion in the islets, laying a foundation for further studies into mitochondrial energy metabolism.
The present study used knockout mice to verify the function of PDH in β-cells in adult mice, conducting for the first time, to the best of our knowledge, a comprehensive islet function test in knockout mice. Primary islet β-cells were extracted to allow further functional and morphological analyses. In conclusion, the present study successfully established the βKO model and explored the association between PDH and the morphology, and phenotype, of β-cells. The restoration of PDH expression in the pancreas of diabetic animals and patients may have clinically beneficial effects on β-cell function. The mechanisms responsible for the dysfunction of insulin secretion should be further explored and could aid in the development of a novel clinical therapy, providing new avenues for the diagnosis and treatment of diabetes.
Acknowledgements
Not applicable.
Glossary
Abbreviations
- IPGTT
intraperitoneal glucose tolerance test
- IPITT
intraperitoneal insulin tolerance test
- PC
pyruvate carboxylase
- PDH
pyruvate dehydrogenase
- PDHA1
pyruvate dehydrogenase E1-alpha subunit
- PDHc
PDH complex
- PDK
pyruvate dehydrogenase kinase
- T2DM
type 2 diabetes mellitus
- TCA
tricarboxylic acid cycle
- βKO
mouse PDHA1 knockout in β-cells
Funding
The present study was supported by a grant awarded by the National Natural Science Foundation of China (grant. no. 81770878); the Science and Technology Program of Guangzhou (grant no. 201604020007).
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
XW and JS conceived and designed the study. XW, SL, YY and YH performed the experiments. XB provided the mutants. XW drafted the manuscript. SL, DP, XB and JS analyzed the data. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The animal experiments were revised and approved by the Ethics Committee of the Southern Medical University (grant no. L2015069).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Aamodt KI, Powers AC. Signals in the pancreatic isletmicroenvironment influence β-cell proliferation. Diabetes Obes Metab. 2017;19(Suppl 1):S124–S136. doi: 10.1111/dom.13031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Seino S, Sugawara K, Yokoi N, Takahashi H. β-Cell signalling and insulin secretagogues: A path for improved diabetes therapy. Diabetes Obes Metab. 2017;19(Suppl 1):S22–S29. doi: 10.1111/dom.12995. [DOI] [PubMed] [Google Scholar]
- 3.Perry RJ, Zhang D, Zhang XM, Boyer JL, Shulman GI. Controlled-release mitochondrial protonophore reverses diabetes and steatohepatitis in rats. Science. 2015;347:1253–1256. doi: 10.1126/science.aaa0672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rovira-Llopis S, Bañuls C, Diaz-Morales N, Hernandez-Mijares A, Rocha M, Victor VM. Mitochondrial dynamics in type 2 diabetes: Pathophysiological implications. Redox Biol. 2017;11:637–645. doi: 10.1016/j.redox.2017.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaufman BA, Li C, Soleimanpour SA. Mitochondrial regulation of β-cell function: Maintaining the momentum for insulin release. Mol Aspects Med. 2015;42:91–104. doi: 10.1016/j.mam.2015.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morrow RM, Picard M, Derbeneva O, Leipzig J, McManus MJ, Gouspillou G, Barbat-Artigas S, Dos Santos C, Hepple RT, Murdock DG, Wallace DC. Mitochondrial energy deficiency leads to hyperproliferation of skeletal muscle mitochondria and enhanced insulin sensitivity. Proc Natl Acad Sci USA. 2017;114:2705–2710. doi: 10.1073/pnas.1700997114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang H, Lu J, Dolezal J, Kulkarni S, Zhang W, Chen A, Gorka J, Mandel JA, Prochownik EV. Inhibition of hepatocellular carcinoma by metabolic normalization. PLoS One. 2019;14:e0218186. doi: 10.1371/journal.pone.0218186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aleem S, Iqbal R, Shar T, Noreen S, Rafiq N, Javed I, Kosar S, Majeed HN, Sattar NA, Abid MK. Complications of Diabetes: An insight into genetic polymorphism and role of insulin. Recent Pat Inflamm Allergy Drug Discov. 2018;12:78–86. doi: 10.2174/1872213X12666180717165331. [DOI] [PubMed] [Google Scholar]
- 9.Bulthuis EP, Adjobo-Hermans MJW, Willems PHGM, Koopman WJH. Mitochondrial morphofunction in mammalian cells. Antioxid Redox Signal. 2019;30:2066–2109. doi: 10.1089/ars.2018.7534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu YQ, Jetton TL, Leahy JL. Beta-cell adaptation to insulin resistance. Increased pyruvate carboxylase and malate-pyruvate shuttle activity in islets of nondiabetic Zucker fatty rats. J Biol Chem. 2002;277:39163–39168. doi: 10.1074/jbc.M207157200. [DOI] [PubMed] [Google Scholar]
- 11.Srinivasan S, Guha M, Kashina A, Avadhani NG. Mitochondrial dysfunction and mitochondrial dynamics-The cancer connection. Biochim Biophys Acta Bioenerg. 2017;1858:602–614. doi: 10.1016/j.bbabio.2017.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mulder H. Transcribing β-cell mitochondria in health and disease. Mol Metab. 2017;6:1040–1051. doi: 10.1016/j.molmet.2017.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marchetti P, Bugliani M, De Tata V, Suleiman M, Marselli L. Pancreatic Beta cell identity in humans and the role of Type 2 diabetes. Front Cell Dev Biol. 2017;5:55. doi: 10.3389/fcell.2017.00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ciccarone F, Vegliante R, Di Leo L, Ciriolo MR. The TCA cycle as a bridge between oncometabolism and DNA transactions in cancer. Semin Cancer Biol. 2017;47:50–56. doi: 10.1016/j.semcancer.2017.06.008. [DOI] [PubMed] [Google Scholar]
- 15.Williams M, Caino MC. Mitochondrial dynamics in type 2 diabetes and cancer. Front Endocrinol (Lausanne) 2018;9:211. doi: 10.3389/fendo.2018.00211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Adeva M, González-Lucán M, Seco M, Donapetry C. Enzymes involved in l-lactate metabolism in humans. Mitochondrion. 2013;13:615–629. doi: 10.1016/j.mito.2013.08.011. [DOI] [PubMed] [Google Scholar]
- 17.Wijenayake S, Tessier SN, Storey KB. Regulation of pyruvate dehydrogenase (PDH) in the hibernating ground squirrel, (Ictidomys tridecemlineatus) J Therm Biol. 2017;69:199–205. doi: 10.1016/j.jtherbio.2017.07.010. [DOI] [PubMed] [Google Scholar]
- 18.Arumugam R, Horowitz E, Noland RC, Lu DH, Fleenor D, Freemark M. Regulation of islet β-cell pyruvate metabolism: interactions of prolactin, glucose, and dexamethasone. Endocrinology. 2010;151:3074–3083. doi: 10.1210/en.2010-0049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu J, Han J, Epstein PN, Liu YQ. Regulation of PDK mRNA by high fatty acid and glucose in pancreatic islets. Biochem Biophys Res Commun. 2006;344:827–833. doi: 10.1016/j.bbrc.2006.03.211. [DOI] [PubMed] [Google Scholar]
- 20.Kono M, Yoshida N, Maeda K, Skinner NE, Pan W, Kyttaris VC, Tsokos MG, Tsokos GC. Pyruvate dehydrogenase phosphatase catalytic subunit 2 limits Th17 differentiation. Proc Natl Acad Sci USA. 2018;115:9288–9293. doi: 10.1073/pnas.1805717115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jeoung NH. Pyruvate dehydrogenase kinases: Therapeutic targets for diabetes and cancers. Diabetes Metab J. 2015;39:188–197. doi: 10.4093/dmj.2015.39.3.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang S, Hulver MW, McMillan RP, Cline MA, Gilbert ER. The pivotal role of pyruvate dehydrogenase kinases in metabolic flexibility. Nutr Metab (Lond) 2014;11:10. doi: 10.1186/1743-7075-11-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krus U, Kotova O, Spégel P, Hallgard E, Sharoyko VV, Vedin A, Moritz T, Sugden MC, Koeck T, Mulder H. Pyruvate dehydrogenase kinase 1 controls mitochondrial metabolism and insulin secretion in INS-1 832/13 clonal beta-cells. Biochem J. 2010;429:205–213. doi: 10.1042/BJ20100142. [DOI] [PubMed] [Google Scholar]
- 24.Tso SC, Lou M, Wu CY, Gui WJ, Chuang JL, Morlock LK, Williams NS, Wynn RM, Qi X, Chuang DT. Development of dihydroxyphenyl sulfonylisoindoline derivatives as liver-targeting pyruvate dehydrogenase kinase inhibitors. J Med Chem. 2017;60:1142–1150. doi: 10.1021/acs.jmedchem.6b01540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hanson BJ, Capaldi RA, Marusich MF, Sherwood SW. An immunocytochemical approach to detection of mitochondrial disorders. J Histochem Cytochem. 2002;50:1281–1288. doi: 10.1177/002215540205001001. [DOI] [PubMed] [Google Scholar]
- 26.Gopal K, Almutairi M, Al Batran R, Eaton F, Gandhi M, Ussher JR. Cardiac-specific deletion of pyruvate dehydrogenase impairs glucose oxidation rates and induces diastolic dysfunction. Front Cardiovasc Med. 2018;5:17. doi: 10.3389/fcvm.2018.00017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sidhu S, Gangasani A, Korotchkina LG, Suzuki G, Fallavollita JA, Canty JM, Jr, Patel MS. Tissue-specific pyruvate dehydrogenase complex deficiency causes cardiac hypertrophy and sudden death of weaned male mice. Am J Physiol Heart Circ Physiol. 2008;295:H946–H952. doi: 10.1152/ajpheart.00363.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu CY, Tso SC, Chuang JL, Gui WJ, Lou M, Sharma G, Khemtong C, Qi X, Wynn RM, Chuang DT. Targeting hepatic pyruvate dehydrogenase kinases restores insulin signaling and mitigates ChREBP-mediated lipogenesis in diet-induced obese mice. Mol Metab. 2018;12:12–24. doi: 10.1016/j.molmet.2018.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Small L, Brandon AE, Quek LE, Krycer JR, James DE, Turner N, Cooney GJ. Acute activation of pyruvate dehydrogenase increases glucose oxidation in muscle without changing glucose uptake. Am J Physiol Endocrinol Metab. 2018;315:E258–E266. doi: 10.1152/ajpendo.00386.2017. [DOI] [PubMed] [Google Scholar]
- 30.Perry RJ, Samuel VT, Petersen KF, Shulman GI. The role of hepatic lipids in hepatic insulin resistance and type 2 diabetes. Nature. 2014;510:84–91. doi: 10.1038/nature13478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gudiksen A, Bertholdt L, Stankiewicz T, Tybirk J, Plomgaard P, Bangsbo J, Pilegaard H. Effects of training status on PDH regulation in human skeletal muscle during exercise. Pflugers Arch. 2017;469:1615–1630. doi: 10.1007/s00424-017-2019-6. [DOI] [PubMed] [Google Scholar]
- 32.Zhou YP, Priestman DA, Randle PJ, Grill VE. Fasting and decreased B cell sensitivity: Important role for fatty acid-induced inhibition of PDH activity. Am J Physiol. 1996;270:E988–E994. doi: 10.1152/ajpendo.1996.270.6.E988. [DOI] [PubMed] [Google Scholar]
- 33.Andersson KB, Winer LH, Mørk HK, Molkentin JD, Jaisser F. Tamoxifen administration routes and dosage for inducible Cre-mediated gene disruption in mouse hearts. Transgenic Res. 2010;19:715–725. doi: 10.1007/s11248-009-9342-4. [DOI] [PubMed] [Google Scholar]
- 34.Valny M, Honsa P, Kirdajova D, Kamenik Z, Anderova M. Tamoxifen in the mouse brain: Implications for Fate-Mapping studies using the Tamoxifen-Inducible Cre-loxP system. Front Cell Neurosci. 2016;10:243. doi: 10.3389/fncel.2016.00243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 36.Grosbellet E, Dumont S, Schuster-Klein C, Guardiola-Lemaitre B, Pevet P, Criscuolo F, Challet E. Circadian phenotyping of obese and diabetic db/db mice. Biochimie. 2016;124:198–206. doi: 10.1016/j.biochi.2015.06.029. [DOI] [PubMed] [Google Scholar]
- 37.Dugan LL, You YH, Ali SS, Diamond-Stanic M, Miyamoto S, DeCleves AE, Andreyev A, Quach T, Ly S, Shekhtman G, et al. AMPK dysregulation promotes diabetes-related reduction of superoxide and mitochondrial function. J Clin Invest. 2013;123:4888–4899. doi: 10.1172/JCI66218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miki Y, Tanji K, Mori F, Kakita A, Takahashi H, Wakabayashi K. Alteration of mitochondrial protein PDHA1 in Lewy body disease and PARK14. Biochem Biophys Res Commun. 2017;489:439–444. doi: 10.1016/j.bbrc.2017.05.162. [DOI] [PubMed] [Google Scholar]
- 39.American Diabetes Association. (2) Classification and diagnosis of diabetes. Diabetes Care. 2015;38(Suppl):S8–S16. doi: 10.2337/dc15-S005. [DOI] [PubMed] [Google Scholar]
- 40.Karaa A, Goldstein A. The spectrum of clinical presentation, diagnosis, and management of mitochondrial forms of diabetes. Pediatr Diabetes. 2015;16:1–9. doi: 10.1111/pedi.12223. [DOI] [PubMed] [Google Scholar]
- 41.Pastors JG, Warshaw H, Daly A, Franz M, Kulkarni K. The evidence for the effectiveness of medical nutrition therapy in diabetes management. Diabetes Care. 2002;25:608–613. doi: 10.2337/diacare.25.3.608. [DOI] [PubMed] [Google Scholar]
- 42.Bugger H, Abel ED. Molecular mechanisms of diabetic cardiomyopathy. Diabetologia. 2014;57:660–671. doi: 10.1007/s00125-014-3171-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mayers RM, Leighton B, Kilgour E. PDH kinase inhibitors: A novel therapy for Type II diabetes? Biochem Soc Trans. 2005;33:367–370. doi: 10.1042/BST0330367. [DOI] [PubMed] [Google Scholar]
- 44.Liu YQ, Moibi JA, Leahy JL. Chronic high glucose lowers pyruvate dehydrogenase activity in islets through enhanced production of long chain acyl-CoA: Prevention of impaired glucose oxidation by enhanced pyruvate recycling through the malate-pyruvate shuttle. J Biol Chem. 2004;279:7470–7475. doi: 10.1074/jbc.M307921200. [DOI] [PubMed] [Google Scholar]
- 45.Park S, Jeon JH, Min BK, Ha CM, Thoudam T, Park BY, Lee IK. Role of the pyruvate dehydrogenase complex in metabolic remodeling: Differential pyruvate dehydrogenase complex functions in metabolism. Diabetes Metab J. 2018;42:270–281. doi: 10.4093/dmj.2018.0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu Z, Yu M, Fei B, Fang X, Ma T, Wang D. miR-21-5p targets PDHA1 to regulate glycolysis and cancer progression in gastric cancer. Oncol Rep. 2018;40:2955–2963. doi: 10.3892/or.2018.6695. [DOI] [PubMed] [Google Scholar]
- 47.Gray LR, Tompkins SC, Taylor EB. Regulation of pyruvate metabolism and human disease. Cell Mol Life Sci. 2014;71:2577–2604. doi: 10.1007/s00018-013-1539-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jitrapakdee S, St Maurice M, Rayment I, Cleland WW, Wallace JC, Attwood PV. Structure, mechanism and regulation of pyruvate carboxylase. Biochem J. 2008;413:369–387. doi: 10.1042/BJ20080709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Branda CS, Dymecki SM. Talking about arevolution: The impact of site-specific recombinases on genetic analyses in mice. Dev Cell. 2004;6:7–28. doi: 10.1016/S1534-5807(03)00399-X. [DOI] [PubMed] [Google Scholar]
- 50.Yang F, Liu C, Chen D, Tu M, Xie H, Sun H, Ge X, Tang L, Li J, Zheng J, et al. CRISPR/Cas9-loxP-mediated gene editing as a novel site-specific genetic manipulation tool. Mol Ther Nucleic Acids. 2017;7:378–386. doi: 10.1016/j.omtn.2017.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Aguirre AJ, Meyers RM, Weir BA, Vazquez F, Zhang CZ, Ben-David U, Cook A, Ha G, Harrington WF, Doshi MB, et al. Genomic copy number dictates a gene-independent cell response to CRISPR/Cas9 targeting. Cancer Discov. 2016;6:914–929. doi: 10.1158/2159-8290.CD-16-0154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Srinivasan M, Choi CS, Ghoshal P, Pliss L, Pandya JD, Hill D, Cline G, Patel MS. ß-cell-specific pyruvate dehydrogenase deficiency impairs glucose-stimulated insulin secretion. Am J Physiol Endocrinol Metab. 2010;299:E910–E917. doi: 10.1152/ajpendo.00339.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nicholls LI, Ainscow EK, Rutter GA. Glucose-stimulated insulin secretion does not require activation of pyruvate dehydrogenase: Impact of adenovirus-mediated overexpression of PDH kinase and PDH phosphate phosphatase in pancreatic islets. Biochem Biophys Res Commun. 2002;291:1081–1088. doi: 10.1006/bbrc.2002.6567. [DOI] [PubMed] [Google Scholar]
- 54.Xu J, Han J, Long YS, Epstein PN, Liu YQ. The role of pyruvate carboxylase in insulin secretion and proliferation in rat pancreatic beta cells. Diabetologia. 2008;51:2022–2030. doi: 10.1007/s00125-008-1130-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.