Abstract
Liposomes are widely used as delivery systems in pharmaceutical, cosmetics and food industries, as well as a system for structural and functional study of membrane proteins. To accurately characterize liposomes, cryo-Electron Microscopy (cryo-EM) has been employed as it is the most precise and direct method to determine liposome lamellarity, size, shape and ultrastructure. However, its use is limited by the number of liposomes that can be trapped in the thin layer of ice that spans holes in the perforated carbon film on EM grids. We report a long-incubation method for increasing the density of liposomes in holes. By increasing the incubation time, high liposome density was achieved even with extremely dilute (in the nanomolar range) liposome solutions. This long-incubation method has been successfully employed to study the structure of an ion channel reconstituted into liposomes.
Keywords: liposomes, cryo-EM, sample preparation, membrane proteins
1. Introduction
In the past four years, about 1,000 protein structures have been determined to better than 4-Å resolution in EMDataBank using cryo-Electron Microscopy (cryo-EM). Among them, many are of detergent-solubilized membrane proteins [1–6]. However, the use of detergent to solubilize membrane proteins always raises the question whether the protein structure represents a biologically relevant state. As shown by both structural and functional studies, lipid membrane environment plays an essential role for the structural integrity and activity of membrane proteins [7–12]. One method to study membrane proteins in a lipid membrane environment is to use lipid nanodiscs, where the membrane protein resides in a small patch of lipid bilayer encircled by amphipathic scaffolding proteins [13]. This method has been employed to study the anthrax toxin pore at 22-Å resolution [14], TRPV1 ion channel in complex with ligands at 3–4 Å resolution [15], as well as other membrane proteins [13–23]. Another method, called “random spherically constrained” (RSC) single-particle cryo-EM, where the membrane protein is reconstituted into liposomes, was developed and employed to study the large conductance voltage- and calcium-activated potassium (BK) channels reconstituted into liposomes at 17-Å resolution [24]. Although both the nanodisc and RSC methods restore the lipid environment of membrane proteins, there is a major difference: the RSC method mimics the cell and provides an asymmetric environment (i.e. inside and outside conditions can be varied independently), whereas there is only one environment surrounding the membrane proteins in the nanodisc method. This is especially important for membrane proteins where an asymmetric environment is preferred (e.g. applying ligands to only one side of the membrane or applying transmembrane potential for voltage-gated ion channels or voltage-sensitive proteins or applying pressures to mechanosensitive channels).
Liposomes are widely used as carrier systems in pharmaceutics (e.g. encapsulate therapeutic agents for cancer therapy [25–27] and for neurological diseases [28]), food industries (e.g. encapsulate bioactive food compounds to improve flavoring and nutritional properties or protect the food from spoilage and degradation [29]) and cosmetics (e.g. as carriers of vitamins [30, 31]). Liposomes have been proven to be beneficial for stabilizing encapsulated agents, overcoming obstacles to cellular uptake, and improving delivery efficiency of compounds to targeted sites in vivo. It is critical to accurately characterize liposomes and drug-liposome interactions as biophysical properties of liposomes are known to influence biological activity, biodistribution, and toxicity. Among the available techniques (Dynamic Light Scattering (DLS), Size Exclusion Chromatography (SEC), Atomic Force Microscopy (AFM), and cryo-EM), cryo-EM is the most precise and direct method to determine liposome lamellarity, size, shape and ultrastructure, which may reveal clues to mechanism of action toward the clinical endpoints of efficacy and toxicity [32–41].
A major barrier for characterizing liposomes using cryo-EM is the difficulty of achieving high number of liposomes in the holes on holey TEM grids [42–44]. Previous methods used very high concentration of liposome [45] or continuous carbon film coated grids [46] or 2D crystal coated grids [44] or graphene coated grids [47]. These methods either require high concentration of liposomes which is not trivial to achieve when membrane proteins are reconstituted into liposomes (e.g. yield of the full length human BK protein is ~1μg / 150mm dish of cultured HEK293 cells and the recovery is about 25% after reconstitution and concentration), or result in higher background noise due to the usage of extra layer of substrate. Inspired by the work by Snijder et al [48] where multiple rounds of application and blotting (termed as multiple-application method) were used to reduce the required sample concentration, we present a long-incubation method (a single application followed with extended incubation) to increases liposome density in holes even with extremely dilute liposome solutions. As the incubation time increases, the density of liposomes in holes increases. After systematic tests, a hypothesis of the increased liposome densities in holes was proposed: liposomes are preferentially adsorbed on the surface of carbon; After the saturation of carbon regions by liposomes (i.e. monolayer coverage), liposomes are continuously accumulated (i.e. multilayer adsorption) in carbon regions; Eventually, the adsorbed liposomes are flushed (i.e. spill over) to holes by another application and blotting of either liposome solutions or liposome-free buffer solutions. The saturation of carbon regions by liposomes determines whether or not liposomes go into holes, while the accumulation of liposomes in carbon regions determines how many liposomes go into holes. This long-incubation method has been successfully employed to study the structure of an ion channel reconstituted into liposomes.
2. Methods
2.1. Liposome preparation
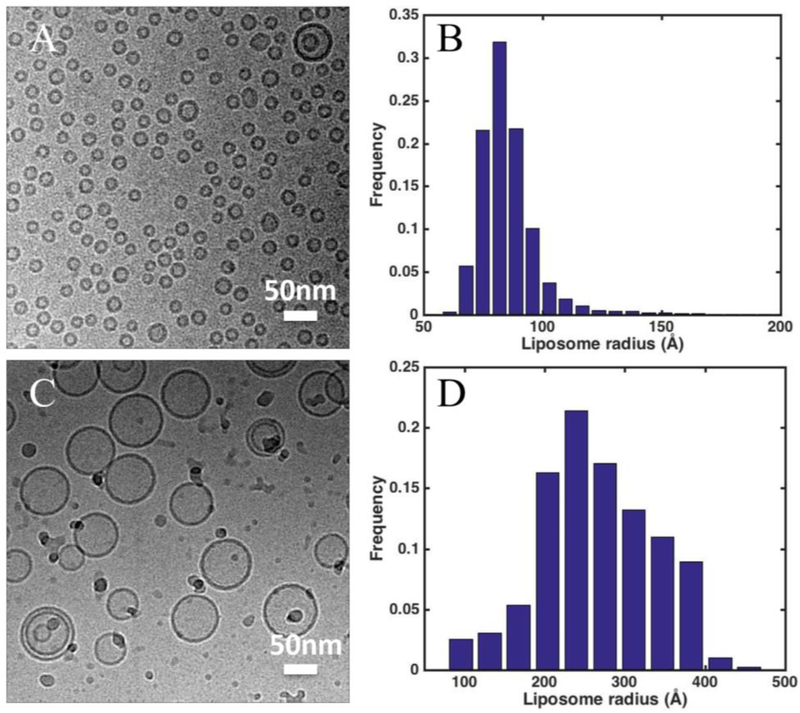
Liposomes were prepared in two ways: gel-filtration and extrusion (the resulting liposomes are termed as Liposome G and E, respectively). In gel-filtration method, 1-palmltoyl-2-oleoyl-sn-glycero-3-phophocoline (POPC) dissolved in chloroform from Avanti (Alabaster, AL) was dried under nitrogen. The thin film of lipids was then re-hydrated in buffer A (150 mM KCl, 20 mM HEPES, pH 7.3) in the presence of n-Decyl-β-D-maltopyranoside (DM) from Anatrace (Maumee, OH) with a lipid:detergent molar ratio of 1:3. Then appropriate 0.25 ml of the lipid-detergent mixture containing 0.4 mM lipid and 4 mM DM in buffer A was slowly loaded upon the top of a Sephadex G-50 (Sigma Aldrich, St. Louis, MO) column (hand packed in a 10 mm I.D x 300 mm height Econo column (Bio-Rad, Hercules, CA), 24 ml volume), which had been pre-equilibrated with running buffer A. The flow was driven by gravity at a speed of 0.2–0.3 ml/min, and the elution fractions were monitored by ÄKTA purifier system (GE Healthcare, Chicago, IL) and collected by Frac 920 (GE Healthcare, Chicago, IL). In extrusion method, POPC lipid dissolved in chloroform was dried under nitrogen, and then re-hydrated in buffer A. The lipid suspension was frozen and thawed 10 times, and extruded through a 50-nm polycarbonate membrane filter (Whatman, the United Kingdom) using a Lipex™ extruder (Northern Lipids Inc., Canada) [49]. Phospholipid concentration of liposomes from gel-filtration method and extrusion method were measured using Ascorbic Acid Colorimetric Assay [50]. Then the lipid concentration was adjusted to 2 mM. All operations were carried out at Room Temperature. Both methods produced unilamellar liposomes as shown in Fig. 1. The mean average sizes are 270 and 90 Å in radius for Liposome E and G, respectively.
Figure 1.
Cryo-EM images and size distribution of (A-B) gel-filtration liposomes (Liposome G) and (C-D) extruded liposomes (Liposome E) applied to TEM grids coated with continuous carbon film. The mean radius of Liposome G and E are 90 +/− 16 Å and 270 +/− 90 Å, respectively.
2.2. Expression, purification and reconstitution of human BK proteins
The BK proteins were expressed and purified as previously described [24]. Briefly, full-length human Slo1.1 protein (GI:507922) carrying an N-terminal FLAG tag was stably expressed in HEK293 cells. The BK protein was purified using an anti-FLAG affinity column (Sigma Aldrich, St. Louis, MO), where 4 mM of detergent dodecyl maltoside (DDM) (Anatrace, Maumee, OH) was exchanged for 4 mM of decyl maltoside (DM) (Anatrace, Maumee, OH) before elution with FLAG peptide (Sigma Aldrich, St. Louis, MO). Superose 6 (GE Healthcare, Chicago, IL) was used to further purify the tetrameric BK.
To reconstitute the BK protein for structural study, the purified BK protein was mixed with DM-solubilized POPC lipid (DM: POPC = 3:1) giving a final protein-to-lipid molar ratio at 1:1,000. The detergent was removed by Gel filtration on a hand packed Sephadex G-50 (Sigma Aldrich, St. Louis, MO) column (a 10 mm I.D x 300 mm height Econo column, 24 ml volume) with running buffer containing 20 mM HEPES, pH 7.3, 150 mM KCl, 2 mM EDTA.
2.3. Cryo-EM sample preparation, image collection and analysis
Three protocols were used to prepare cryo-EM samples: (A) on continuous carbon film coated grids: 2 μl of liposome solution was applied onto a glow-discharged continuous carbon coated TEM grid (EMCN, China), blotted immediately and fast frozen at 22 °C and 100% relative humidity in an FEI Vitrobot Mark IV (FEI, Hillsboro, OR) as for normal cryo-sample preparation. (B) multiple-application method: 2 μl of liposome solution was applied onto a glow-discharged holey carbon grid CFlat R2/2 (EMS, Hatfield, PA) and incubated for 10–20 s before blotted briefly from the edge. This procedure was repeated to the desired number of rounds of sample application [48]. After the last round of sample application, the sample was blotted and fast frozen at 22 °C and 100% relative humidity in an FEI Vitrobot Mark IV (FEI, Hillsboro, OR) as for normal cryo-sample preparation. (C) long-incubation method: 2 μl of liposome solution was applied onto a glow-discharged holey grid, either CFlat R2/2, CFlat R1.2/1.3 (EMS, Hatfield, PA), or Quantifoil R2/2 (Quantifoil, Germany), and incubated for 0.25–10 min at 22 °C and 100% relative humidity in an FEI Vitrobot Mark IV (FEI, Hillsboro, OR). After a brief blotting from the side of the grid, another 2 μl of solution with or without liposomes was applied to the grid. After 10–20 s, the grid was blotted and frozen as for normal cryo-sample preparation. Samples were imaged on an FEI G2 F20 (FEI, Hillsboro, OR) equipped with Eagle 4k camera (FEI, Hillsboro, OR), or an FEI T12 (FEI, Hillsboro, OR) equipped with a Gatan US4000 CCD camera (Gatan, Pleasanton, CA), or an FEI G2 F20 equipped with a K2 Summit Direct Detector (Gatan, Pleasanton, CA). The dose for each exposure was about 2,000 e−/nm2. Images were taken with a pixel size from 1.6 Å/pixel to 4.4 Å/pixel. Liposomes in each cryo-EM image were identified automatically using a home-made Matlab program (MathWorks, Natick, MA) as previously described [51]. Then the size distribution and mean size was analyzed using Matlab. For each condition, 3–5 grids were prepared and imaged. The collected images for each condition were similar, and only images from the same grid were used for statistical analysis.
The liposome concentration is estimated as outlined below. The average number of lipid molecules per liposome is:
where r is the radius of a liposome (from the center of the liposome to the center of the bilayer), APOPC is the area per lipid molecule (68 Å2) [52]. The liposome concentration is:
where Clipid is the lipid concentration.
3. Results
3.1. Multiple-application method
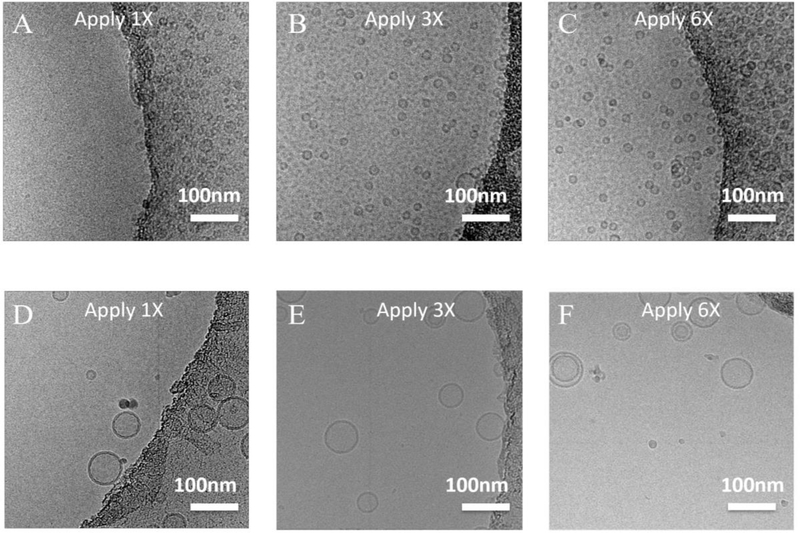
Liposomes were prepared using either gel filtration method (Liposome G) or extrusion method (Liposome E). Cryo-samples were prepared using the multiple-application method as previously described [48]. With one round of sample application (i.e. normal cryo-sample preparation method), there were few liposomes in holes (Fig. 2A&D) for both Liposome G and E. After 6 rounds of sample application, more liposomes went into holes for Liposome G (Fig. 2C). However, even after 6 rounds of sample application, only a few liposomes went into the holes for Liposome E (Fig. 2F). This is mainly due to the lower liposome concentration of Liposome E (although Liposome E and G have the same lipid concentration, the liposome concentration of Liposome E is about 9 times lower than that of Liposome G (58 nM vs. 520 nM)). The smaller diffusivity of Liposome E due to the large size (size ratio of Liposome E to Liposome G = 3 : 1, diffusivity ratio of Liposome E to Liposome G = 1 : 3 based on Stokes–Einstein equation) might also contribute to the lower liposome density. However, the effect is much smaller than that due to the liposome concentration difference: the ratio of characteristic diffusion distance of Liposome E to Liposome G is 1 : 1.7, which is much smaller than the liposome concentration ratio of Liposome E to Liposome G (1 : 9).
Figure 2.
Cryo-EM images of liposomes by the multiple-application method. (A-C) Liposome G was applied once, three times and six times. (D-F) Liposome E was applied once, three times and six times. Scale bar: 100 nm.
3.2. Long-incubation method
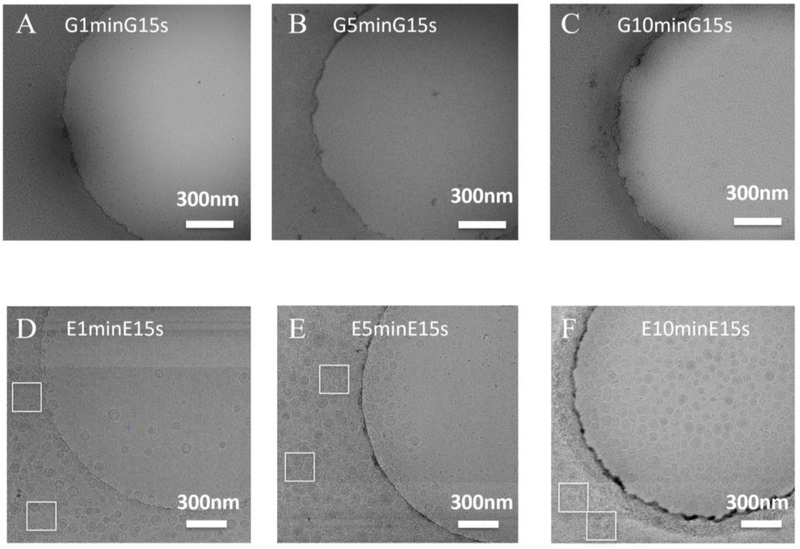
Although multiple-application of samples increased the liposome density in holes to some extent as seen in Fig. 2C for Liposome G, it did not work for Liposome E which has a lower liposome concentration. To reduce the required liposome concentration and the amount of sample to be used (i.e. one-time sample application vs six-time sample applications), a long-incubation method was developed. As shown in Fig. 3A&D, with a one-minute incubation, the liposome density in holes was comparable to that prepared by multiple-application method (six times of sample applications) (Fig. 2C&F). When the incubation time increased to 10 min (Fig. 3C), the liposome density of Liposome G in holes increased by a factor of three (Fig. 2C). Similar phenomena were observed for Liposome E (Fig. 3F & Fig. 2F). As shown in Fig. 3D (1-min incubation), the carbon region of the grid was not fully occupied by liposomes and few liposomes populated in holes. When the incubation time increased to 5 min, the carbon region was saturated by liposomes and liposomes over liposomes were observed, which showed multi-layer adsorption (Fig. 3E). More liposomes went into holes. With a 10-min incubation, the carbon region was occupied by too many liposomes which makes it impossible to identify individual liposomes (Fig. 3F). Many more liposomes went into holes. This suggests the long-incubation is the key step to increase the liposome density in holes.
Figure 3.
Cryo-EM images of liposomes prepared using the long-incubation method. (A-C) Liposome G was incubated for one minute (G1minG15s), 5 minutes (G5minG15s) and 10 minutes (G10minG15s) before blotting. Then Liposome G was applied again, incubated for 15 s, blotted and fast frozen. (D-F) Cryo-EM samples of Liposome E were prepared using the same method as in A-C. White boxes mark the carbon regions to show the adsorption of liposomes on carbon films. Scale bar: 300 nm.
3.3. Long-incubation followed by an application of different liposome solutions
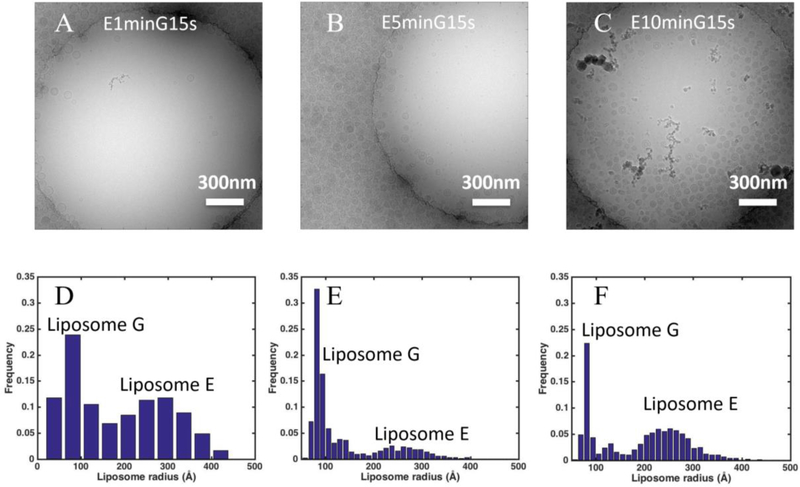
As shown in Fig. 3, long incubation increased liposome density in holes for both Liposome G and E. To track whether the increased liposome density came from the first round or the second round of sample application, Liposome E was added in the first round of sample application and Liposome G was added in the second round of sample application (Fig. 4A–C). As shown in Fig. 4D–F, two liposome populations were observed: Liposome E with a mean radius of 270 Å and Liposome G with a mean radius of 90 Å. The liposome coverage due to Liposome E increased from 2.5% to 6% and 18% when the incubation time increased from one minute to five and ten minutes (Fig. 5 A). However, the liposome coverage due to Liposome G was the same (~2%) with five-minute and 10-minute incubation of Liposome E as the incubation time for Liposome G was the same. The reason for a lower coverage of Liposome G in Fig. 4A lies in the fact that the carbon region was not saturated by Liposome E with the one-minute incubation (Fig. 4A and Fig. 3D). Newly added Liposome G preferably went to carbon regions. Therefore, the increase of liposome density in holes is due to the first round of sample application.
Figure 4.
Contributions of the first and second application of samples on liposome density in holes on a holey-carbon grid. (A-C) Representative cryo-EM images prepared with 1 minute, 5 minutes, and 10 minutes incubation of Liposome E followed by one application of Liposome G. (D-F) Size distributions of liposomes in holes (2 μm in diameter) from cryo-EM images prepared as shown in A-C. Scale bar: 300 nm. Analysis was based on 5, 10, and 5 images, 46, 321, and 482 liposomes for D, E and F, respectively.
Figure 5.
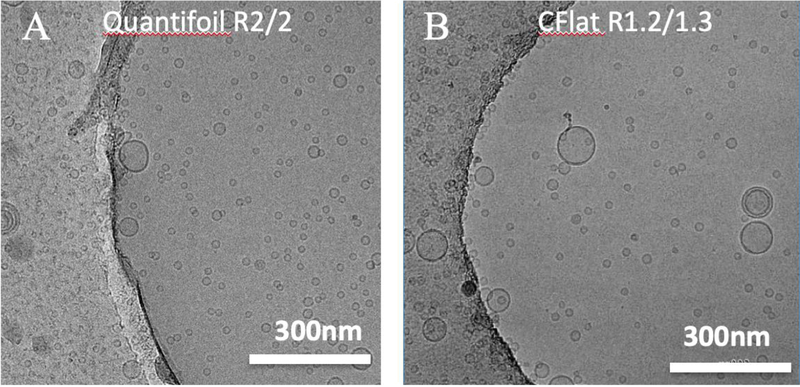
Cryo-EM images of Liposome G with 10-minute incubation followed by an application of Liposome G prepared on (A) Quantifoil R2/2 and (B) CFlat R1.2/1.3 grids. Scale bar: 300 nm.
3.4. Long-incubation method on different kinds of grids
In previous sections, only CFlat R2/2 grids were used. It would have been desirable to repeat the observations on other types of grids (e.g. different hole sizes, different manufactures). Thus, CFlat R1.2/1.3 grids were tested to see the effect of hole sizes, and Quantifoil R2/2 grids were tested to see the effect of grid manufacturing processes. With the sample hole size and distance between holes, a similar liposome density in holes was obtained (Fig. 5A). When the hole size becomes smaller, liposomes still enter holes and a slightly lower liposome density was observed (Fig. 5B). Thus the long-incubation methods works with common types of carbon grids used in the EM fields.
3.5. Long-incubation method used for studies of proteoliposomes
As shown in Sections 3.2–4, long-incubation method can be employed to increase liposome densities in holes for extremely low concentration liposome samples. Does it work for liposomes with reconstituted membrane proteins (proteoliposomes)? Here, the full length human BK proteins, whose yield is very low (about 1μg / 150mm dish of cultured HEK293 cells), were reconstituted into POPC liposomes using gel filtration method. As shown in Fig. 6, a good distribution of proteoliposomes in holes was achieved. The protein particles were easily identified in the cryo-EM images before and after liposome subtraction. In short, the long-incubation method works for both empty liposomes and proteoliposomes.
Figure 6.
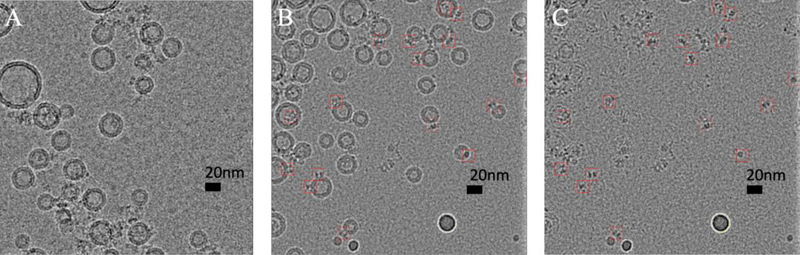
Liposome with reconstituted human BK channels prepared using long-incubation method. (A) A representative cryo-EM image of BK proteoliposomes. BK proteoliposomes were used in both the first and the second sample application. (B) A representative cryo-EM image of BK proteoliposomes. BK proteoliposomes was used in the first sample application, while proteoliposome-free buffer was used in the second sample application. (C) Liposomes are subtracted from the image shown in B. BK particles were marked with red boxes (15 nm). Scale bar: 20 nm.
4. Discussion
The liposome density observed in holes on the EM grid depends on the bulk liposome concentration, the interaction between liposomes and the carbon film, and the blotting process. Using the same sample and the same standard cryo-sample preparation method, there are at least 20 times more liposomes on carbon film (Fig. 1A&C) than in holes (Fig. 2A&D). However, the carbon film increases the background noise, thus decreases the signal-to-noise ratio (SNR) of the cryo-EM images, which is inferior for high-resolution studies (e.g. interaction between drugs and liposome, the structure of reconstituted membrane proteins). One remedy is to decrease the carbon film thickness: coating an ultra-thin carbon film over a holey carbon grid [46]. Another technique is to use multiple-application method [48]. The multiple-application method has been successfully employed to obtain high densities of biological macromolecules in holes with dilute samples (e.g. in micromolar range). However, this method did not work for Liposome E (e.g. in nanomolar range) as shown in Fig. 2D–F.
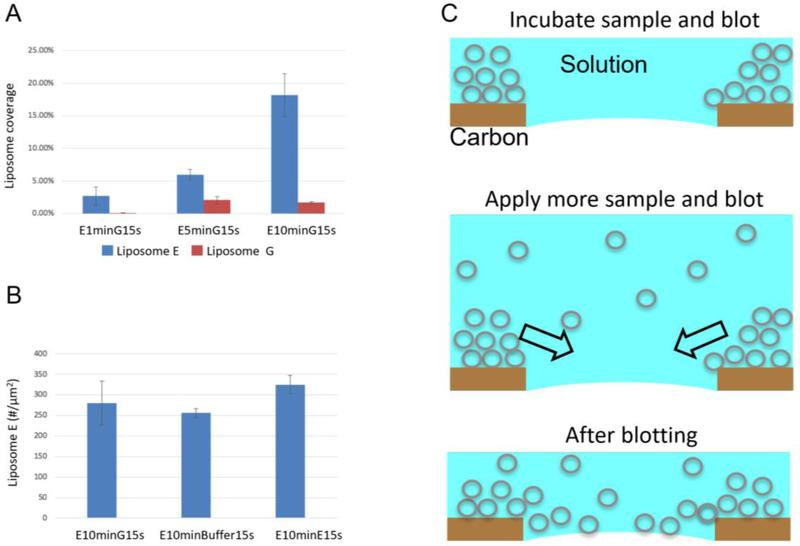
As shown by the number of liposomes in carbon region (Fig. 1A&C) and in holes (Fig. 2A&D), liposomes prefer carbon film over holes. As the incubation time increases from 15 s to 10 minutes, more and more liposomes accumulated/piled in carbon regions: there is still unoccupied area in carbon regions with one-minute incubation (white boxes in Fig. 3D), while liposomes overlapped with each other with five-min incubation (white boxes in Fig. 3E). As the majorities of liposomes are unilamellar (Fig. 1A&B), the overlapping of liposomes indicates that liposomes are piled up on carbon film as illustrated in Fig. 7C. After the first blotting, a thin layer of aqueous solution is left on the grid. With a second application of sample and blotting, the accumulated/piled liposomes in carbon regions are flushed into holes. The liposomes in holes is a combination of the accumulated liposomes from the first sample application and the liposomes from the second sample application. As shown in Fig. 7A, the liposome coverage due to the first application of sample (Liposome E) increased with increased incubation times. The liposome coverage due to the second application of sample (Liposome G) is constant as it is determined by the bulk liposome concentration of the applied liposome solution. However, if the incubation time is not long enough and the carbon region is not saturated (i.e. not fully covered), the liposomes from the second application of sample will preferably go to carbon regions to saturate the carbon film. Thus, the liposome coverage due to the second application of sample (Liposome G) is much lower with one-minute incubation than that with five-minute and 10-minute incubation of the first application of sample (Liposome E).
Figure 7.
Mechanism of the long-incubation method. (A) Liposome coverage with different incubation times. Analysis was based on 5, 10, and 5 images, 46, 321, and 482 liposomes for E1minG15s, E5minG15s and E10minG15s, respectively. (B) Liposome density when different solutions (Liposome G, liposome-free buffer A, and Liposome E) were used in the second sample application. Analysis was based on 3, 3, and 4 images, 975, 768 and 1,540 liposomes for E10minG15s, E10minBuffer15s, and E10minE15s, respectively. (C) A cartoon showing the long-incubation method.
In the long incubation method, the liposome density in holes is mainly determined by the incubation time after the first sample application. Therefore, it is possible to apply liposome-free buffers in the second sample application and obtain a similar liposome density in holes. To test this hypothesis, a liposome-free buffer A was used in the second application of sample. A slightly lower liposome density was observed (Fig. 7B) (256±11 vs. 325±22 liposomes / image). The accumulation of liposomes is the real step which increases the liposome density in holes. This is also the reason why the multiple-application method worked with dilute samples. The multiple application of dilute liposome solutions will saturate carbon regions (i.e. full monolayer coverage), but there is not enough time for liposomes to accumulate/pile in carbon regions. Thus, a long-incubation time is necessary to saturate carbon films and accumulate liposomes in carbon regions, which will be flushed into holes later. The flushing of accumulated liposomes into holes is a result of blotting from both side of the grid. To test this, the grid was blotted from both side after long-incubation, liposome density was similar to that using liposome-free solutions in the second sample application. However, when the grid was blotted from the same side of the liposomes, liposomes did not go into holes. Thus the blotting from both side is required to flush liposomes into holes.
The liposomes formed either by gel filtration or extrusion (9 or 27 nm in radius) were very stable. The size distribution was the same even after one month storage at 4 °C. No aggregation was observed. When liposomes were used to study the dipole potential, the liposome size was about 40 nm in radius [51], no fusion or aggregation was observed. It seems that the radius or curvature of liposomes has no effect on the tendency of liposomes to aggregate or fuse for liposomes up to 40 nm in radius. Study has shown when the ice thickness increases from 300 Å to 450 Å, the obviously detectable protein particles become undetectable in images recorded by a CCD camera. As liposomes were embedded in ice, small liposomes were preferred to have good contrast of liposomes and / or proteins reconstituted into liposomes.
5. Conclusion
Long-incubation method was developed to achieve desired liposome densities in cryo-EM images of liposomes even at extremely low liposome concentrations (e.g. in nanomolar range). It was employed to visualize liposomes produced by two commonly used methods (gel filtration and extrusion) and liposomes with reconstituted membrane proteins for high-resolution structural studies. We discovered that the saturation of carbon regions (i.e. monolayer coverage) determines whether liposomes go into holes, while the accumulation of liposomes in carbon regions (i.e. multilayer adsorption) determines how many liposomes go into holes.
Highlights.
A method was developed to achieve desired liposome densities in cryo-EM images of liposomes even at extremely low liposome concentrations (e.g. in nanomolar range)
The new method enables cryo-EM data collection for membrane proteins reconstituted into liposomes which could not be concentrated at a concentration high enough for standard cryo-EM sample preparation
The saturation of carbon regions (i.e. monolayer coverage) determines whether liposomes go into holes for cryo-EM images, while the accumulation of liposomes in carbon regions (i.e. multilayer adsorption) determines how many liposomes go into holes
The new method is also applicable to prepare cryo-samples of other biological samples
Acknowledgments
This work was supported by NIH Grant R01GM096458 to L. Wang.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference:
- [1].Liao M, Cao E, Julius D, Cheng Y, Structure of the TRPV1 ion channel determined by electron cryo-microscopy, Nature, 504 (2013) 107–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Fan G, Baker ML, Wang Z, Baker MR, Sinyagovskiy PA, Chiu W, Ludtke SJ, Serysheva II, Gating machinery of InsP3R channels revealed by electron cryomicroscopy, Nature, 527 (2015) 336–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Yan Z, Bai X.-c., Yan C, Wu J, Li Z, Xie T, Peng W, Yin C.-c., Li X, Scheres SHW, Shi Y, Yan N, Structure of the rabbit ryanodine receptor RyR1 at near-atomic resolution, Nature, 517 (2015) 50–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Hite RK, Yuan P, Li Z, Hsuing Y, Walz T, MacKinnon R, Cryo-electron microscopy structure of the Slo2.2 Na+ -activated K+ channel, Nature, 527 (2015) 198–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Hite RK, Tao X, MacKinnon R, Structural basis for gating the high-conductanceCa2+-activated K+ channel, Nature, 541 (2017) 52–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Tao X, Hite RK, MacKinnon R, Cryo-EM structure of the open high-conductanceCa2+-activated K+ channel, Nature, 541 (2017) 46–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Schmidt D, Jiang QX, MacKinnon R, Phospholipids and the origin of cationic gating charges in voltage sensors, Nature, 444 (2006) 775–779. [DOI] [PubMed] [Google Scholar]
- [8].Gonen T, Cheng Y, Sliz P, Hiroaki Y, Fujiyoshi Y, Harrison SC, Walz T, Lipid-protein interactions in double-layered two-dimensional AQP0 crystals, Nature, 438 (2005) 633–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Long SB, Tao X, Campbell EB, MacKinnon R, Atomic structure of a voltage-dependent K+ channel in a lipid membrane-like environment, Nature, 450 (2007) 376–383. [DOI] [PubMed] [Google Scholar]
- [10].Hilgemann DW, Getting ready for the decade of the lipids, Annu. Rev. Physiol, 65 (2003) 697–700. [DOI] [PubMed] [Google Scholar]
- [11].Hille B, Dickson EJ, Kruse M, Vivas O, Suh B-C, Phosphoinositides regulate ion channels, Biochim. Biophys. Acta, 1851 (2015) 844–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Lee AG, Biological membranes: the importance of molecular detail, Trends Biochem. Sci, 36 (2011) 493–500. [DOI] [PubMed] [Google Scholar]
- [13].Bayburt TH, Grinkova YV, Sligar SG, Self-Assembly of Discoidal Phospholipid Bilayer Nanoparticles with Membrane Scaffold Proteins, Nano Lett., 2 (2002) 853856. [Google Scholar]
- [14].Katayama H, Wang J, Tama F, Chollet L, Gogol EP, Collier RJ, Fisher MT, Three-dimensional structure of the anthrax toxin pore inserted into lipid nanodiscs and lipid vesicles, Proc. Natl. Acad. Sci. U. S. A, 107 (2010) 3453–3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Gao Y, Cao E, Julius D, Cheng Y, TRPV1 structures in nanodiscs reveal mechanisms of ligand and lipid action, Nature, 534 (2016) 347–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Jackson SM, Manolaridis I, Kowal J, Zechner M, Taylor NMI, Bause M, Bauer S, Bartholomaeus R, Bernhardt G, Koenig B, Buschauer A, Stahlberg H, Altmann K-H, Locher KP, Structural basis of small-molecule inhibition of human multidrug transporter ABCG2, Nat. Struct. Mol. Biol, 25 (2018) 333–340. [DOI] [PubMed] [Google Scholar]
- [17].Taylor NMI, Manolaridis I, Jackson SM, Kowal J, Stahlberg H, Locher KP, Structure of the human multidrug transporter ABCG2, Nature, 546 (2017) 504–509. [DOI] [PubMed] [Google Scholar]
- [18].Srivastava AP, Luo M, Zhou W, Symersky J, Bai D, Chambers MG, Faraldo-Gómez JD, Liao M, Mueller DM, High-resolution cryo-EM analysis of the yeast ATP synthase in a lipid membrane, Science (New York, N.Y.), 360 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Roh S-H, Stam NJ, Hryc CF, Couoh-Cardel S, Pintilie G, Chiu W, Wilkens S, The 3.5-Å CryoEM Structure of Nanodisc-Reconstituted Yeast Vacuolar ATPase Vo Proton Channel, Mol. Cell, 69 (2018) 993–1004.e1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Dang S, Feng S, Tien J, Peters CJ, Bulkley D, Lolicato M, Zhao J, Zuberbühler K, Ye W, Qi L, Chen T, Craik CS, Jan YN, Minor DL, Cheng Y, Jan LY, Cryo-EM structures of the TMEM16A calcium-activated chloride channel, Nature, 552 (2017) 426–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].McGoldrick LL, Singh AK, Saotome K, Yelshanskaya MV, Twomey EC, Grassucci RA, Sobolevsky AI, Opening of the human epithelial calcium channel TRPV6, Nature, 553 (2018) 233–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Autzen HE, Myasnikov AG, Campbell MG, Asarnow D, Julius D, Cheng Y, Structure of the human TRPM4 ion channel in a lipid nanodisc, Science (New York, N.Y.), 359 (2018) 228–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Chen Q, She J, Zeng W, Guo J, Xu H, Bai X-C, Jiang Y, Structure of mammalian endolysosomal TRPML1 channel in nanodiscs, Nature, 550 (2017) 415–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Wang L, Sigworth FJ, Structure of the BK potassium channel in a lipid membrane from electron cryomicroscopy, Nature, 461 (2009) 292–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Yuba E, Liposome-based immunity-inducing systems for cancer immunotherapy, Mol. Immunol, 98 (2018) 8–12. [DOI] [PubMed] [Google Scholar]
- [26].Børresen B, Hansen AE, Kjær A, Andresen TL, Kristensen AT, Liposome-encapsulated chemotherapy: Current evidence for its use in companion animals, Vet. Comp. Oncol, 16 (2018) E1–E15. [DOI] [PubMed] [Google Scholar]
- [27].Zununi Vahed S, Salehi R, Davaran S, Sharifi S, Liposome-based drug codelivery systems in cancer cells, Mater. Sci. Eng, C, 71 (2017) 1327–1341. [DOI] [PubMed] [Google Scholar]
- [28].Vieira DB, Gamarra LF, Getting into the brain: Liposome-based strategies for effective drug delivery across the blood–brain barrier, International Journal of Nanomedicine, 11 (2016) 5381–5414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Shukla S, Haldorai Y, Hwang SK, Bajpai VK, Huh YS, Han Y-K, Current Demands for Food-Approved Liposome Nanoparticles in Food and Safety Sector, Front. Microbiol, 8 (2017) 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Shashi K, Satinder K, Bharat P, A complete review on: Liposomes, International Research Journal of Pharmacy, 3 (2012) 10–16. [Google Scholar]
- [31].Mozafari MR, Liposomes: An overview of manufacturing techniques, Cellular and Molecular Biology Letters, 10 (2005) 711–719. [PubMed] [Google Scholar]
- [32].Lepault J, Pattus F, Martin N, Cryo-electron microscopy of artificial biological membranes, BBA - Biomembranes, 820 (1985) 315–318. [Google Scholar]
- [33].Aissaoui A, Chami M, Hussein M, Miller AD, Efficient topical delivery of plasmid DNA to lung in vivo mediated by putative triggered, PEGylated pDNA nanoparticles, J. Control. Release, 154 (2011) 275–284. [DOI] [PubMed] [Google Scholar]
- [34].Al-Ahmady Z, Lozano N, Mei KC, Al-Jamal WT, Kostarelos K, Engineering thermosensitive liposome-nanoparticle hybrids loaded with doxorubicin for heat-triggered drug release, Int. J. Pharm, 514 (2016) 133–141. [DOI] [PubMed] [Google Scholar]
- [35].Baxa U, Imaging of liposomes by transmission electron microscopy, in: Methods Mol. Biol, 2018, pp. 73–88. [DOI] [PubMed] [Google Scholar]
- [36].Bonnaud C, Vanhecke D, Demurtas D, Rothen-Rutishauser B, Petri-Fink A, Spatial SPION localization in liposome membranes, IEEE Trans. Magn, 49 (2013) 166–171. [Google Scholar]
- [37].Crawford R, Dogdas B, Keough E, Haas RM, Wepukhulu W, Krotzer S, Burke PA, Sepp-Lorenzino L, Bagchi A, Howell BJ, Analysis of lipid nanoparticles by Cryo-EM for characterizing siRNA delivery vehicles, Int. J. Pharm, 403 (2011) 237244. [DOI] [PubMed] [Google Scholar]
- [38].De Carlo S, Fiaux H, Marca-Martinet CA, Electron cryo-microscopy reveals mechanism of action of propranolol on artificial membranes, J. Liposome Res, 14 (2004) 61–76. [DOI] [PubMed] [Google Scholar]
- [39].Uhl P, Helm F, Hofhaus G, Brings S, Kaufman C, Leotta K, Urban S, Haberkorn U, Mier W, Fricker G, A liposomal formulation for the oral application of the investigational hepatitis B drug Myrcludex B, Eur. J. Pharm. Biopharm, 103 (2016) 159–166. [DOI] [PubMed] [Google Scholar]
- [40].Uhl P, Pantze S, Storck P, Parmentier J, Witzigmann D, Hofhaus G, Huwyler J, Mier W, Fricker G, Oral delivery of vancomycin by tetraether lipid liposomes, Eur. J. Pharm. Sci, 108 (2017) 111–118. [DOI] [PubMed] [Google Scholar]
- [41].Zhang P, Huang Y, Makhov AM, Gao X, Zhang P, Li S, Characterization of spherulites as a lipidic carrier for low and high molecular weight agents, Pharm. Res, 30 (2013) 1525–1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Haghiralsadat F, Amoabediny G, Sheikhha MH, Forouzanfar T, Arough JM, Helder MN, Zandieh-Doulabi B, A novel approach on drug delivery: Investigation of a new nano-formulation of liposomal doxorubicin and biological evaluation of entrapped doxorubicin on various osteosarcoma cell lines, Cell Journal, 19 (2017) 55–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Park SM, Cha JM, Nam J, Kim MS, Park SJ, Park ES, Lee H, Kim HR, Formulation optimization and in vivo proof-of-concept study of thermosensitive liposomes balanced by phospholipid, elastin-like polypeptide, and cholesterol, PLoS One, 9 (2014) 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Wang L, Ounjai P, Sigworth FJ, Streptavidin crystals as nanostructured supports and image-calibration references for cryo-EM data collection, J. Struct. Biol, 164 (2008) 190–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Frederik PM, Hubert DHW, Cryoelectron microscopy of liposomes, in: Methods Enzymol., 2005, pp. 431–448. [DOI] [PubMed] [Google Scholar]
- [46].Jensen KH, Brandt SS, Shigematsu H, Sigworth FJ, Statistical modeling and removal of lipid membrane projections for cryo-EM structure determination of reconstituted membrane proteins, J. Struct. Biol, 194 (2016) 49–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Palovcak E, Wang F, Zheng SQ, Yu Z, Li S, Betegon M, Bulkley D, Agard DA, Cheng Y, A simple and robust procedure for preparing graphene-oxide cryo-EM grids, J. Struct. Biol, 204 (2018) 80–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Snijder J, Borst AJ, Dosey A, Walls AC, Burrell A, Reddy VS, Kollman JM, Veesler D, Vitrification after multiple rounds of sample application and blotting improves particle density on cryo-electron microscopy grids, J. Struct. Biol, 198 (2017) 38–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Mayer LD, Hope MJ, Cullis PR, Vesicles Of Variable Sizes Produced By A Rapid Extrusion Procedure, Biochim. Biophys. Acta, 858 (1986) 161–168. [DOI] [PubMed] [Google Scholar]
- [50].Fiske CH, Subbarow Y, THE COLORIMETRIC DETERMINATION OF PHOSPHORUS, J. Biol. Chem, 66 (1925) 375–400. [Google Scholar]
- [51].Wang L, Bose PS, Sigworth FJ, Using cryo-EM to measure the dipole potential of a lipid membrane, Proc. Natl. Acad. Sci. U. S. A, 103 (2006) 18528–18533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Kučerka N, Tristram-Nagle S, Nagle JF, Structure of fully hydrated fluid phase lipid bilayers with monounsaturated chains, J. Membr. Biol, 208 (2006) 193–202. [DOI] [PubMed] [Google Scholar]
- [53].Stagg SM, Lander GC, Pulokas J, Fellmann D, Cheng A, Quispe JD, Mallick SP, Avila RM, Carragher B, Potter CS, Automated cryoEM data acquisition and analysis of 284 742 particles of GroEL, J. Struct. Biol, 155 (2006) 470–481. [DOI] [PubMed] [Google Scholar]