Abstract
Post-translational modifications play essential roles in finely modulating eukaryotic circadian clock systems. In plants, the effects of O-glycosylation on the circadian clock and the underlying mechanisms remain largely unknown. The O-fucosyltransferase SPINDLY (SPY) and the O-GlcNAc transferase SECRET AGENT (SEC) are two prominent O-glycosylation enzymes in higher plants, with both overlapped and unique functions in plant growth and development. Unlike the critical role of O-GlcNAc in regulating the animal circadian clock, here we report that nuclear-localized SPY, but not SEC, specifically modulates the pace of the Arabidopsis circadian clock. By identifying the interactome of SPY, we identified PSEUDO-RESPONSE REGULATOR 5 (PRR5), one of the core circadian clock components, as a new SPY-interacting protein. PRR5 can be O-fucosylated by SPY in planta, while point mutation in the catalytic domain of SPY abolishes the O-fucosylation of PRR5. The protein abundance of PRR5 is strongly increased in spy mutants, while the degradation rate of PRR5 is much reduced, suggesting that PRR5 proteolysis is promoted by SPY-mediated O-fucosylation. Moreover, multiple lines of genetic evidence indicate that PRR5 is a major downstream target of SPY to specifically mediate its modulation of the circadian clock. Collectively, our findings provide novel insights into the specific role of the O-fucosyltransferase activity of SPY in modulating the circadian clock and implicate that O-glycosylation might play an evolutionarily conserved role in modulating the circadian clock system, via O-GlcNAcylation in mammals, but via O-fucosylation in higher plants.
Keywords: circadian clock, O-fucosyltransferase, SPY, proteolysis, PRR5
INTRODUCTION
The circadian clock orchestrates numerous aspects of plant growth and development by synchronizing cellular events with external recurring light and dark cycles (Harmer, 2009; Nakamichi et al., 2012; Greenham and McClung, 2015; Fung-Uceda et al., 2018; Kim et al., 2018; Shalit-Kaneh et al., 2018; Zhang et al., 2018; Li et al., 2019). Maintenance of the proper circadian period is critical for providing higher plants with adaptive advantages in coordinating the internal daily physiological activities with metabolic homeostasis in changing environments (Greenham and McClung, 2015; Sanchez and Kay, 2016). The basic core circadian oscillator comprises interlocked transcription–translation feedback loops. A mutual transcription repression between TOC1 (TIMING OF CAB EXPRESSION 1, the founding member of the PSEUDO-RESPONSE REGULATOR [PRR] family) and CIRCADIAN CLOCK-ASSOCIATED 1 (CCA1) comprises the central loop of the core oscillator (Wang and Tobin, 1998; Gendron et al., 2012; Huang et al., 2012). In addition, CCA1 and LATE ELONGATED HYPOCOTYL (LHY) suppress transcription of (PRR7 and PRR9 (Kamioka et al., 2016). In turn, PRR9 and PRR7 together with PRR5 transcriptionally repress CCA1/LHY (Nakamichi et al., 2010; Wang et al., 2013; Greenham and McClung, 2015). Finally, the Evening complex, comprising EARLY FLOWERING 3 (ELF3), ELF4, and LUX ARRHYTHMO (LUX), inhibits the expression of PRRs and GI, hence relieving the inhibition of CCA1 and LHY expression indirectly (Nusinow et al., 2011; Herrero et al., 2012).
Post-translational modification also plays a vital role in ensuring the robustness of the core oscillator. For instance, many components of the core oscillator, including CCA1 and PRRs, are subjected to post-translational modifications such as phosphorylation (Sugano et al., 1998; Daniel et al., 2004; Fujiwara et al., 2008; Wang et al., 2010; Uehara et al., 2019), sumoylation (Hansen et al., 2017), and ubiquitination (Han et al., 2004; Baudry et al., 2010). Post-translational modifications are critical for maintaining protein activities and stability and subsequently play fundamental roles in an intricate control of the circadian clock. Although a number of post-translational modifications have been documented, whether and how O-glycosylation is involved in fine-tuning the circadian period in plants remains elusive. SPINDLY (SPY) and SECRET AGENT (SEC) are two prominent O-glycosyltransferases in higher plants (Olszewski et al., 2010). Both proteins contain a tetratricopeptide repeat (TPR) region at their N terminus that may act as a protein–protein interaction domain and an evolutionarily conserved C-terminal region related to animal O-linked N-acetylglucosamine transferases (OGTs) (Jacobsen et al., 1996). Very recently, SPY was biochemically characterized as a protein O-fucosyltransferase (POFUT) that competes with the O-GlcNAc transferase SEC to modulate the activity of DELLA proteins (Zentella et al., 2017). In addition, SPY also regulates the protein abundance of its interacting partners, such as DELLAs, TCP14, and TCP15 (Silverstone et al., 2007; Steiner et al., 2016), at the post-translational level. Nevertheless, it remains unclear whether O-glycosylation mediated by SPY is a prerequisite for the control of proteolysis of its interacting proteins.
SPY and SEC have multiple roles in regulating plant growth and development (Hartweck et al., 2006), such as cytokinin response, abiotic stress tolerance, and light signaling (Swain et al., 2001; Sothern et al., 2002; Tseng et al., 2004; Qin et al., 2011; Steiner et al., 2012). Strikingly, the double mutant of SEC and SPY is embryo lethal, indicating that they might be functionally redundant during reproductive development (Olszewski et al., 2010). In mice and flies, OGTs have been shown to control circadian period via O-GlcNAc modification of the circadian core components (Kim et al., 2012; Li et al., 2013). In Arabidopsis, whether and how O-glycosylation modulates the clock still remains unclear.
In this study, we report that nuclear-localized SPY, but not SEC, specifically modulates the Arabidopsis circadian clock. Biochemical and genetic evidences demonstrate PRR5 as a novel SPY nuclear interacting partner that mediates fine-tuning of the pace of the circadian clock. Notably, PRR5 can be O-fucosylated by SPY in planta, which promotes the proteolysis of PRR5 protein. Taken together, our findings provide novel insights into the unique role for SPY in modulating the circadian clock and suggest that O-glycosylation represents an evolutionarily conserved mechanism in fine-tuning the circadian clock system.
RESULTS
Nuclear-Localized SPY, but Not SEC, Specifically Modulates the Circadian Clock
To address if SPY and SEC have distinctive or overlapping roles in modulating the circadian clock, we crossed the respective mutants with circadian reporter lines, including CCA1:LUCIFER-ASE (CCA1:LUC), LNK2:LUC, and CAB2:LUC, to precisely assess their circadian phenotypes. Homozygous lines of F3 progeny harboring the circadian reporters were used to investigate their circadian phenotypes. As shown in Figure 1A and 1B, either a deletion in the TPR domain (M354 to Q376; spy-1) or a point mutation (G593S) in the catalytic domain (spy-3) (Supplemental Figure 1) significantly lengthens the circadian period relative to wild type (WT). This result indicates that both the TPR and the catalytic domains are essential for SPY function (Jacobsen et al., 1996). Similar results were obtained when LNK2:LUC and CAB2:LUC were used as reporters (Supplemental Figure 2).
Figure 1. Nuclear-Localized SPY, but Not SEC, Specifically Regulates the Circadian Clock.
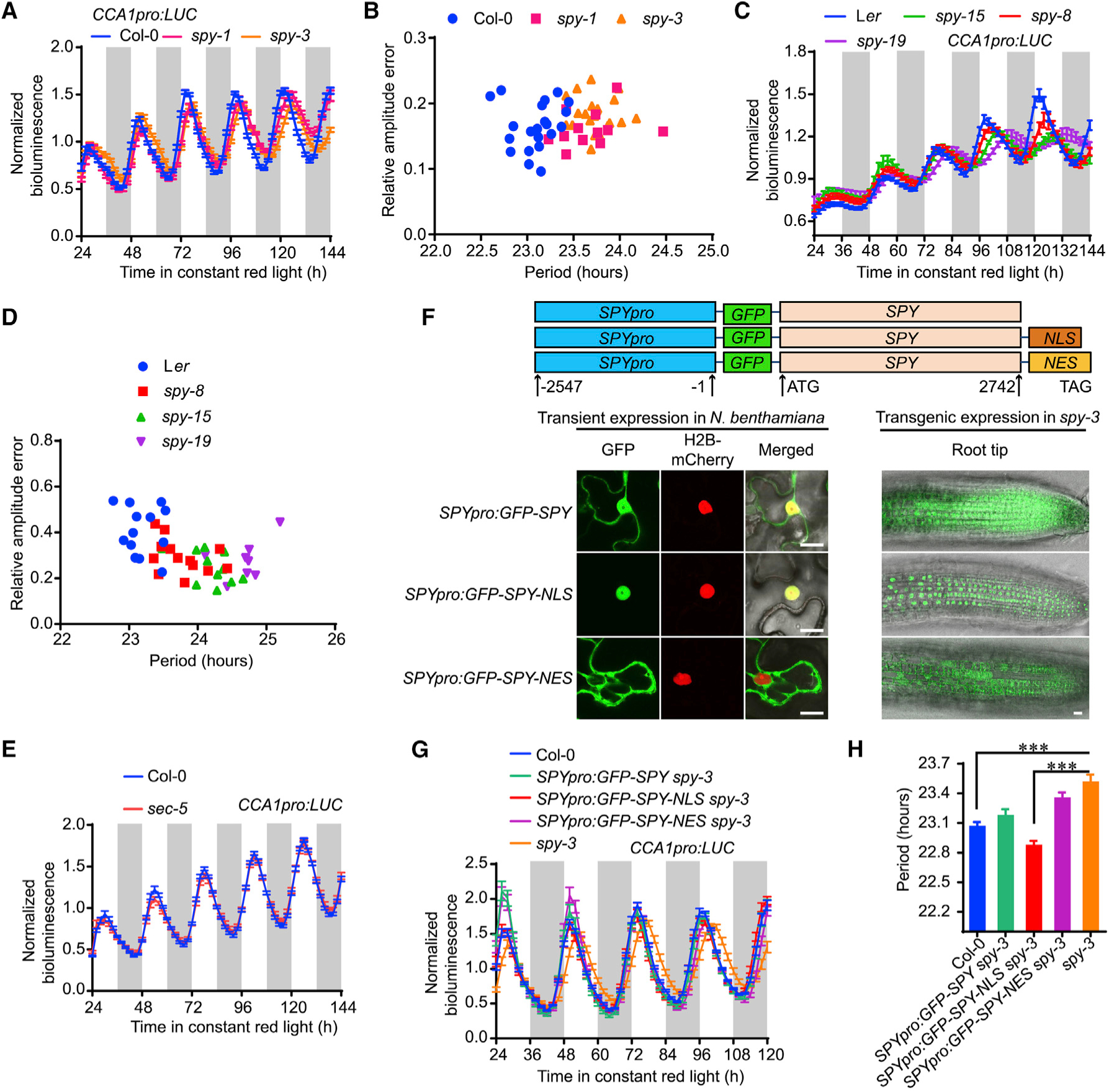
(A) Normalized bioluminescence activity of CCA1pro:LUC in spy-1, spy-3, and Col-0 plants in red light (20 μmol m−2 s−1). Data represent normalized means ± SE (n = 13–21). White and light-gray vertical bars indicate the subjective day and night, respectively.
(B) Scatterplot showing lengthened circadian periods in spy mutants with relatively normal amplitude errors, indicating strong circadian robustness in the estimation of circadian period.
(C) Normalized bioluminescence activity of CCA1:LUC in spy mutants with Ler background. Data represent means ± SE after normalization with the average bioluminescence intensity over 24 h to 144 h.
(D) Scatterplot showing the lengthening circadian period in spy mutants, with a relative amplitude error lower than 0.5.
(E) Null mutant of sec did not show aberrant circadian period phenotype, as indicated by bioluminescence trace of CCA1pro:LUC (n = 20).
(F) Subcellular localization of SPY with or without nuclear localization signal (NLS) and nuclear export signal (NES) in both the epidermal cells of transiently infiltrated N. benthamiana leaves and the root tips of stable transgenic Arabidopsis. Histone H2B-mCherry was used as a nuclear marker. Scale bars, 10 μm.
(G) Normalized bioluminescence activity of CCA1pro:LUC in SPY:GFP-SPY spy-3 and SPY:GFP-SPY-NLS spy-3. Data represent means ± SE (n = 18).
(H) The estimated circadian periods from Figure 1G showing that SPY:GFP-SPY-NLS could fully rescue the lengthened circadian period in spy-3 mutant. Data represent means ± SE (n = 18). *** indicates significant difference at p < 0.001 in t-test.
The circadian phenotypes of spy mutants in the Ler background were also investigated. Consistently, the circadian period of spy-8 was approximately 1 h longer than that of the WT (Figure 1C and 1D, Supplemental Figure 3). Importantly, the spy-15 and spy-19 mutants, caused by two separate point mutations in the C-terminal catalytic domain of SPY (Supplemental Figure 1A) that abolished protein -fucosyltransferase POFUT activity (Zentella et al., 2017), displayed even more lengthened circadian period than that of spy-8 (Figure 1D and Supplemental Figure 3). These results suggested that the POFUT activity of SPY was critical for its effect on circadian period. Since SEC is closely related to SPY in regulating plant growth and development, we also investigated whether SEC could regulate circadian period by using a previously characterized null mutant, sec-5 (Xing et al., 2018). However, no abnormal circadian phenotypes could be found in sec-5 (Figure 1E), indicating that SPY alone specifically modulates circadian period in Arabidopsis. To examine whether the expression of SPY is feedback regulated by the circadian clock, we examined the transcript and protein levels of SPY and found that they did not exhibit evident oscillation patterns under diurnal conditions (Supplemental Figure 1C–1E) at either the transcriptional or the translational level, suggesting that the expression of SPY is likely not feedback regulated by the circadian clock.
We next tested whether SPY nuclear presence is necessary for its circadian function. A native promoter-driven N-terminal GFP-tagged SPY coding sequence was fused with either the nuclear export signal (NES) or the nuclear localization signal (NLS) and transformed into spy-3 (Figure 1F). We first observed subcellular localization patterns of these proteins in both transiently transformed epidermal cells of Nicotiana benthamiana (Figure 1F, left) and stable transgenic plants in the spy-3 background (Figure 1F, right). As expected, GFP-SPY was distributed in both cytoplasm and nucleus, while NLS-tagged chimeric protein was found exclusively in the nucleus, and GFP-SPY-NES was found mainly in cytoplasm (Figure 1F). To evaluate if the subcellular localization of SPY was critical in modulating circadian period, we examined T3 transgenic lines with similar expression levels of exogenous chimeric SPY proteins (Supplemental Figure 4A). As shown in Figure 1G and 1H, SPY:GFP-SPY-NLS was more effective than SPY:GFP-SPY in complementing the longer circadian period phenotype of the spy-3 mutant, while SPY:GFP-SPY-NES was much less effective than SPY:GFP-SPY. However, SPY:GFP-SPY-NLS could not efficiently rescue the leaf serration and flowering time phenotypes of spy-3 (Supplemental Figure 4B and 4C), indicating that the mechanism for SPY-regulated flowering time is independent of its effect on the circadian clock. Taking these results together, we conclude that the nuclear-localized SPY protein is sufficient to rescue the long circadian period of the spy-3 mutant.
SPY Physically Interacts with and O-Fucosylates PRR5
To further investigate the underlying mechanism by which SPY modulates the circadian period, we aimed to identify SPY’s interacting partners through affinity purification followed by mass spectrometry (AP–MS). Since nuclear-localized SPY was sufficient for modulating circadian period, we employed both SPY:GFP-SPY and SPY:GFP-SPY-NLS transgenic plants for AP–MS with tissue harvested at ZT0 (Zeitgeber time) and ZT12, respectively, after entrainment under a 12/12 light/ dark cycle for 10 days. We obtained 15 and 19 overlapping nuclear-localized proteins as potential SPY interacting partners between SPY:GFP-SPY and SPY:GFP-SPY-NLS samples harvested at ZT0 and ZT12, respectively (Supplemental Figure 5A and 5C, Supplemental Data 1). Among the 15 identified common proteins from ZT0 samples, we identified TCP14 (Supplemental Figure 5B), a known SPY interacting protein (Steiner et al., 2012), but not any known circadian core components. In contrast, we identified PRR5 as a novel SPY interacting protein from samples harvested at ZT12 (Figure 2A, Supplemental Figures 5D and 6), the peak time of PRR5 protein accumulation.
Figure 2. SPY Physically Interacts with and O-Fucosylates PRR5.
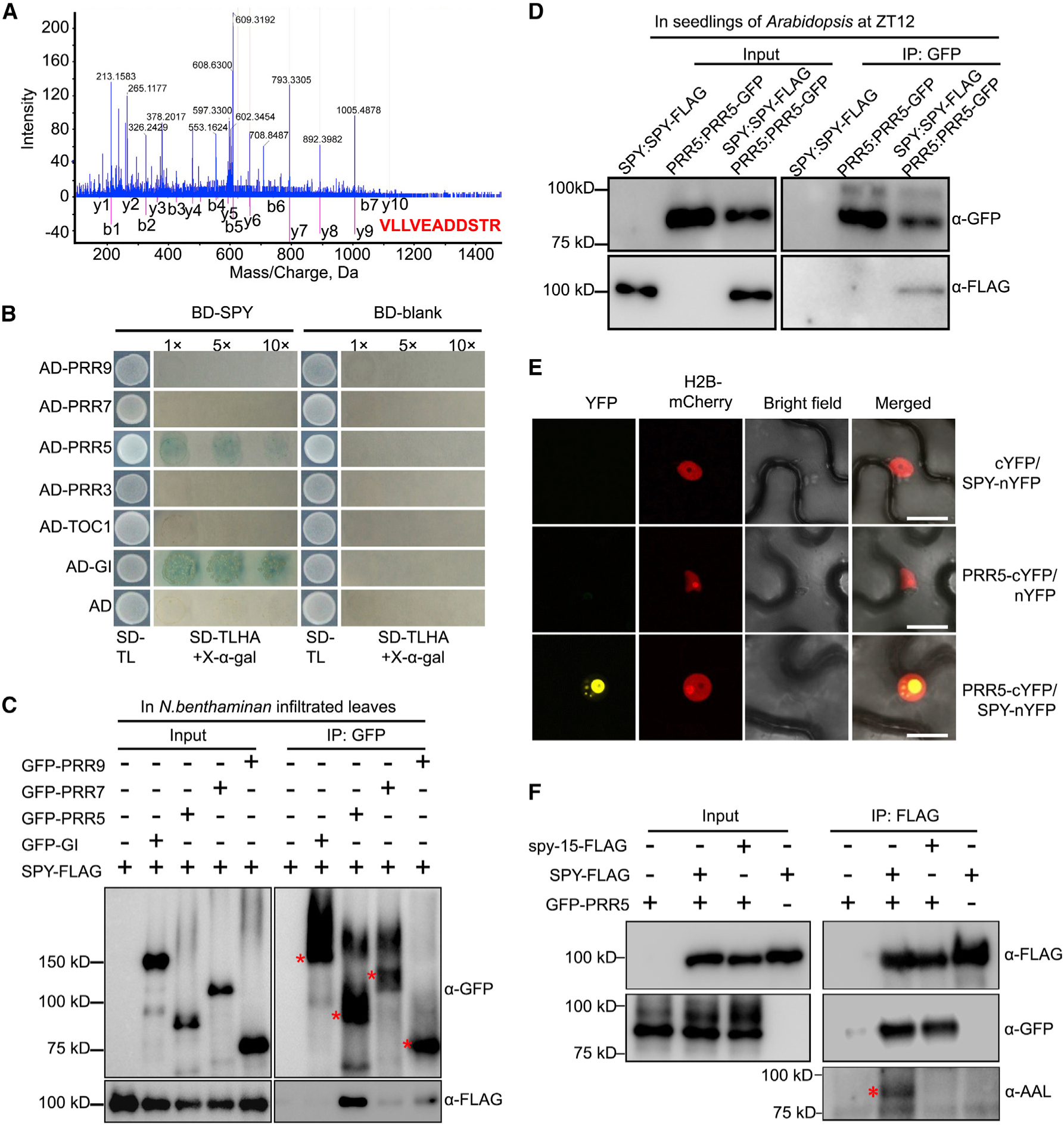
(A) Peptides of PRR5 protein were identified by affinity purification followed by mass spectrometry. Representative tandem mass spectrometry spectrum of the peptide corresponding to PRR5 protein (VLLVEADDSTR) is shown.
(B) Yeast two-hybrid assay showing that PRR5 interacts with SPY. Yeast clones were grown on synthetic dropout (SD) medium devoid of tryptophan and leucine (SD-TL) for initial screening (first column), before being transferred to SD medium devoid of tryptophan, leucine, histidine, and adenine, plus X-gal (SD-TLHA + 40 μg/ml X-gal) with dilution factor as indicated (second column).
(C) Co-immunoprecipitation analysis of SPY-FLAG with GFP-tagged circadian components. GFP-Trap beads were used to precipitate protein complexes extracted from co-infiltrated leaves of N. benthamiana. Red asterisks indicate the major bands of the correspondingly indicated proteins.
(D) Co-immunoprecipitation analysis of SPY with PRR5 in Arabidopsis. Both SPY-FLAG and PRR5-GFP were driven by their respective promoters. SPY-FLAG and PRR5-GFP alone were used as negative controls.
(E) SPY physically interacts with PRR5 in the nucleus in BiFC assay. H2B-mCherry was used as nuclear marker. Scale bars, 10 μm.
(F) PRR5 is O-fucosylated by SPY. Proteins were extracted from the co-infiltrated N. benthamiana leaves with indicated combinations for coimmunoprecipitation, with anti-FLAG antibody-conjugated beads. The antibodies used for detecting the respective protein are listed. The asterisk indicates the specific SPY-O-fucosylated PRR5 protein band. The representative data are from three individual biological replicates with similar results.
Yeast two-hybrid (Y-2-H) tests were conducted to determine whether other family members of PRR5 can interact with SPY. Consistent with previous findings, SPY interacted with GI in the Y-2-H assay (Figure 2B) (Tseng et al., 2004), but not any other PRR family members (Figure 2B). We further substantiated the SPY–PRR5 interaction in vivo by co-immunoprecipitation assay. Using transient co-expression of SPY-FLAG and GFP-PRR5 proteins in N. benthamiana leaves, we detected a strong interaction signal between SPY and PRR5, but not other PRR family members (Figure 2C, Supplemental Figure 7). Intriguingly, we failed to detect the physical interaction between SPY and GI in our co-immunoprecipitation assay, which could be due to their interaction being highly dynamic or weaker than the interaction between SPY and PRR5. Nonetheless, the lengthened circadian period phenotype cannot be explained by GI, as previous genetic evidence has shown (Tseng et al., 2004). Hence, we further pursued validation of the physical interaction between SPY and PRR5 in Arabidopsis. Notably, we found that PRR5 and SPY are able to interact in seedlings of Arabidopsis, driven by their own respective native promoters (Figure 2D), indicating that PRR5 interacts with SPY under endogenous conditions. We next utilized bimolecular fluorescence complementation (BiFC) analysis to examine the subcellular interaction pattern in planta between SPY and PRR5 proteins. We found strong fluorescence signals in N. benthamiana epidermal cells between SPY-nYFP and PRR5-cYFP in the nucleus (Figure 2E). Taken together, results of multiple approaches identify PRR5 as a novel nuclear interaction partner of SPY in vivo. To further investigate the consequence of SPY–PRR5 interaction, we examined whether PRR5 can be O-fucosylated by SPY through co-expression of GFP-PRR5 with FLAG-tagged WT SPY or FLAG-tagged spy-15, lacking POFUT activity (Zentella et al., 2017), in N. benthamiana. Furthermore, as Aleuria aurantia lectin (AAL) binds specifically to the terminally attached fucose, biotinylated AAL was used to determine whether PRR5 is O-fucosylated. We found that the PRR5 protein co-immunoprecipitated with WT SPY was specifically recognized by AAL (Figure 2F). In contrast, in the presence of spy-15 protein, PRR5 was not bound by AAL, indicating that PRR5 is O-fucosylated only by enzymatically active SPY.
Defining Domains Bridging Physical Interaction between SPY and PRR5
To further define the domains mediating the physical interaction between SPY and PRR5, we conducted Y-2-H in vitro and co-immunoprecipitation in vivo with various deletion combinations of both proteins. The PRR5 C terminus (173–558 aa) interacts with SPY in Y-2-H (Figure 3A) and when transiently co-expressed in N. benthamiana leaves. Further testing showed that both the PRR5 N terminus and the C-terminal 50 aa were dispensable for interacting with SPY (Figure 3B). For SPY, we found that the N-terminal fragment containing the TPR domain (1–431 aa), rather than the C-terminal fragment containing the OGT domain (432–914 aa), interacts with the PRR5 C terminus in Y-2-H (Figure 3C). We confirmed the interaction specificity between the PRR5 C terminus and the SPY TPR domain in co-immunoprecipitation assays (Figure 3D and 3E). The TPR domain truncated version of SPY (spy-8) abolishes the interaction with PRR5, while the POFUT null-activity version of SPY (spy-15) still interacts strongly with PRR5 (Figure 3F), further supporting that the TPR domain of SPY is required for mediating the interaction with PRR5.
Figure 3. Defining the Domain Requirement Bridging the Interaction between SPY and PRR5.
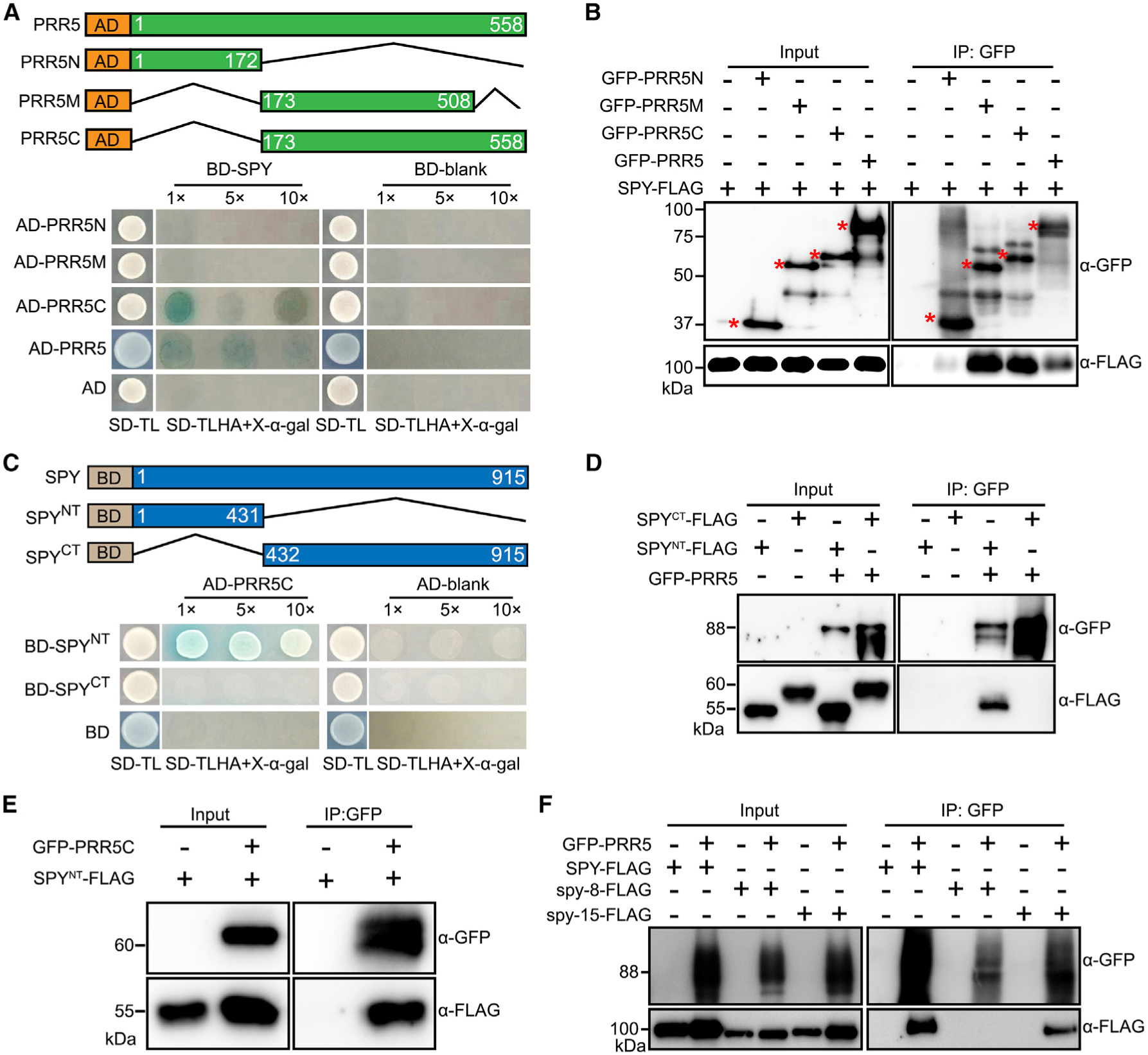
(A) Yeast two-hybrid assays showing that the C terminus of PRR5 is required for interacting with SPY. (Top) Schematic diagram of the constructs used for yeast two-hybrid assays. Numbers indicate the amino acid position. (Bottom) Yeast clones were grown on synthetic dropout (SD) medium devoid of tryptophan and leucine (SD-TL) for initial screening (first column), before being transferred to SD medium devoid of tryptophan, leucine, histidine, and adenine, plus X-α-gal (SD-TLHA + 40 μg/ml X-α-gal) with dilution factor as indicated (second column).
(B) Co-immunoprecipitation assay showing SPY interacts with PRR5 C terminus in vivo. SPY-FLAG was transiently expressed alone or co-expressed with PRR5N, PRR5M, and PRR5C. The input and IP samples were analyzed by immunoblotting using FLAG antibody and GFP antibodies, respectively.
(C) Yeast two-hybrid assays showing that SPY TPR region is essential for mediating the interaction between SPY and PRR5.
(D) Co-immunoprecipitation assay showing SPY TPR region is necessary and sufficient to interact with PRR5 in vivo. Assay performed as in B.
(E) Co-immunoprecipitation assay showing PRR5 C terminus is able to interact with SPY TPR region in vivo.
(F) Co-immunoprecipitation assay showing TPR domain mutation of SPY abolishes its interaction with PRR5, but catalytic domain mutation of SPY can interact with PRR5. The spy-8-FLAG contains a truncated TPR domain with from M354 to Q376, while spy-15-FLAG has a point mutation, E567K, which is considered as an essential amino acid residues for its catalytic activity.
SPY Promotes the Proteolysis of PRR5
To further investigate the biological significance of SPY-PRR5 physical interaction, we next examined the PRR5 protein subcellular localization pattern in WT and spy-3 using PRR5:PRR5-GFP. We found that the nuclear localization pattern of PRR5 was unchanged in the spy-3 mutant (Supplemental Figure 8), suggesting that SPY does not affect subcellular localization of PRR5 protein. Noticeably, an increase of fluorescence signal was observed in spy-3, indicating SPY may decrease PRR5 protein accumulation (Supplemental Figure 8). Hence, we further examined the PRR5 protein abundance in spy-3 over a 12 h light/12 h dark time course and constant red light condition. Consistent with the microscopy results, PRR5 protein abundance was appreciably increased in the spy-3 mutant in both light/dark and constant red light conditions, especially from ZT5 to ZT13 (Figure 4A and 4B, Supplemental Figure 9). Real-time qPCR demonstrated that PRR5 transcript levels are unchanged between Col-0 and spy-3 (Supplemental Figure 10), indicating that the SPY-mediated change in PRR5 accumulation is post-transcriptional. We tested if SPY activity alters PRR5 stability using cycloheximide (CHX) treatment and observed a slower loss of PRR5 abundance in spy-3, compared to that in the WT (Figure 4C and 4D). Taken together, our results suggest that SPY reduces PRR5 protein stability, likely by facilitating proteolysis.
Figure 4. SPY Facilitates the Proteolysis of PRR5 Protein.
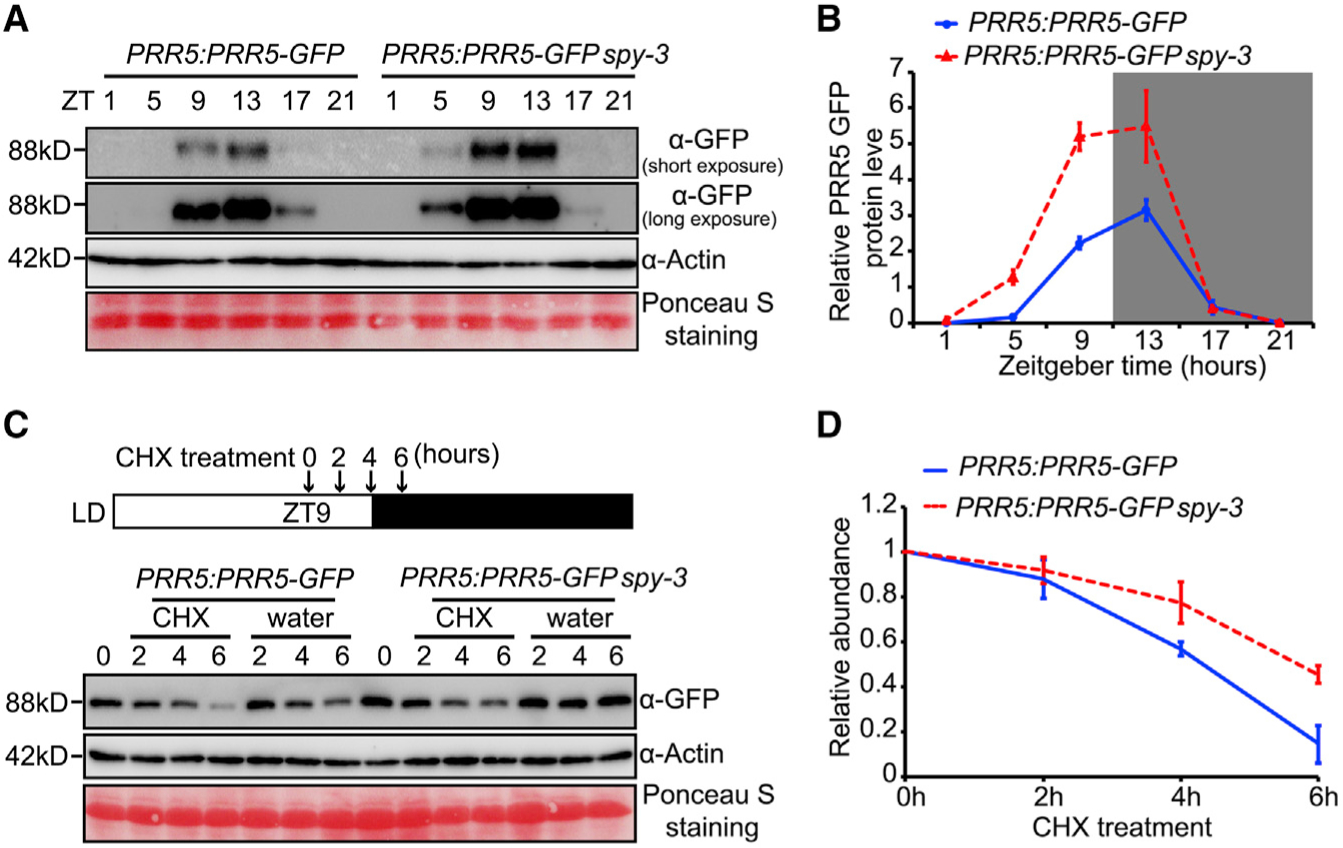
(A) Immunoblot analysis showing higher PRR5 protein accumulation in spy-3 mutant. Seedlings of PRR5:PRR5-GFP and PRR5:PRR5-GFP spy-3 were grown in 12 h L/12 h D photocycles. Samples were collected at indicated time points. PRR5-GFP protein abundance was detected with GFP antibody. Actin antibody and ponceau staining were used as loading controls.
(B) Quantitative analysis of PRR5 protein accumulation as shown in A with values normalized by corresponding Actin signals. Data are means ± SE from three biological replicates.
(C) PRR5 protein degradation is slower in spy-3 mutant. The cytoplasmic protein synthesis inhibitor, cycloheximide (CHX) was added at ZT9, and plants were harvested at 0, 2, 4 and 6 h after adding CHX, as indicated in the diagram. Control plants were harvested in parallel.
(D) Quantitative analysis of PRR5 protein abundance as shown in C. Data represent for means ± SE. from three individual biological replicates.
Transcriptional Repression of CCA1 by PRR5 Is Elevated in spy Mutants
PRR5 acts as a transcriptional repressor for CCA1 and LHY within the core feedback loops of circadian oscillator (Nakamichi et al., 2010; Wang et al., 2013). Since SPY facilitates the proteolysis of PRR5 protein, we reasoned that the expression of the direct targets of PRR5 might be reduced in the spy-3 mutant. We first checked the repression activity of PRR5 in the absence or presence of SPY in the infiltrated N. benthamiana leaves. As expected we found a significant transcriptional repression of CCA1 expression by PRR5. Importantly, SPY, but not SEC, diminished the transcription repression activity of PRR5 (Figure 5A and 5B). We further examined the promoter activity of CCA1 genetically by utilizing CCA1:LUC reporter crossed with spy-3 and spy-8 mutants, respectively. We monitored the bioluminescence signals generated by CCA1:LUC in both continuous light and continuous dark conditions. Lower bioluminescence signals were detected in spy-3 and spy-8 mutants starting from late subjective night to the early subjective daytime under continuous red light or continuous dark conditions (Figure 5C and 5D, Supplemental Figure 11A and 11B). In addition, we conducted RT-qPCR to determine the transcript levels of CCA1 and LHY in WT and spy-3, the two well-known target genes of PRR5. Consistently, we found that the transcript levels of CCA1 and LHY were much reduced in spy-3 in both light/dark (Figure 5E and 5F) and constant light conditions (Supplemental Figure 11C and 11D) at most of the tested time points. Taken together, we propose that the transcriptional repression of CCA1/LHY by PRR5 is modulated by SPY, likely at the post-translational level by affecting PRR5 protein abundance.
Figure 5. The Abundance of the Transcripts of CCA1 and LHY Are Regulated by SPY.
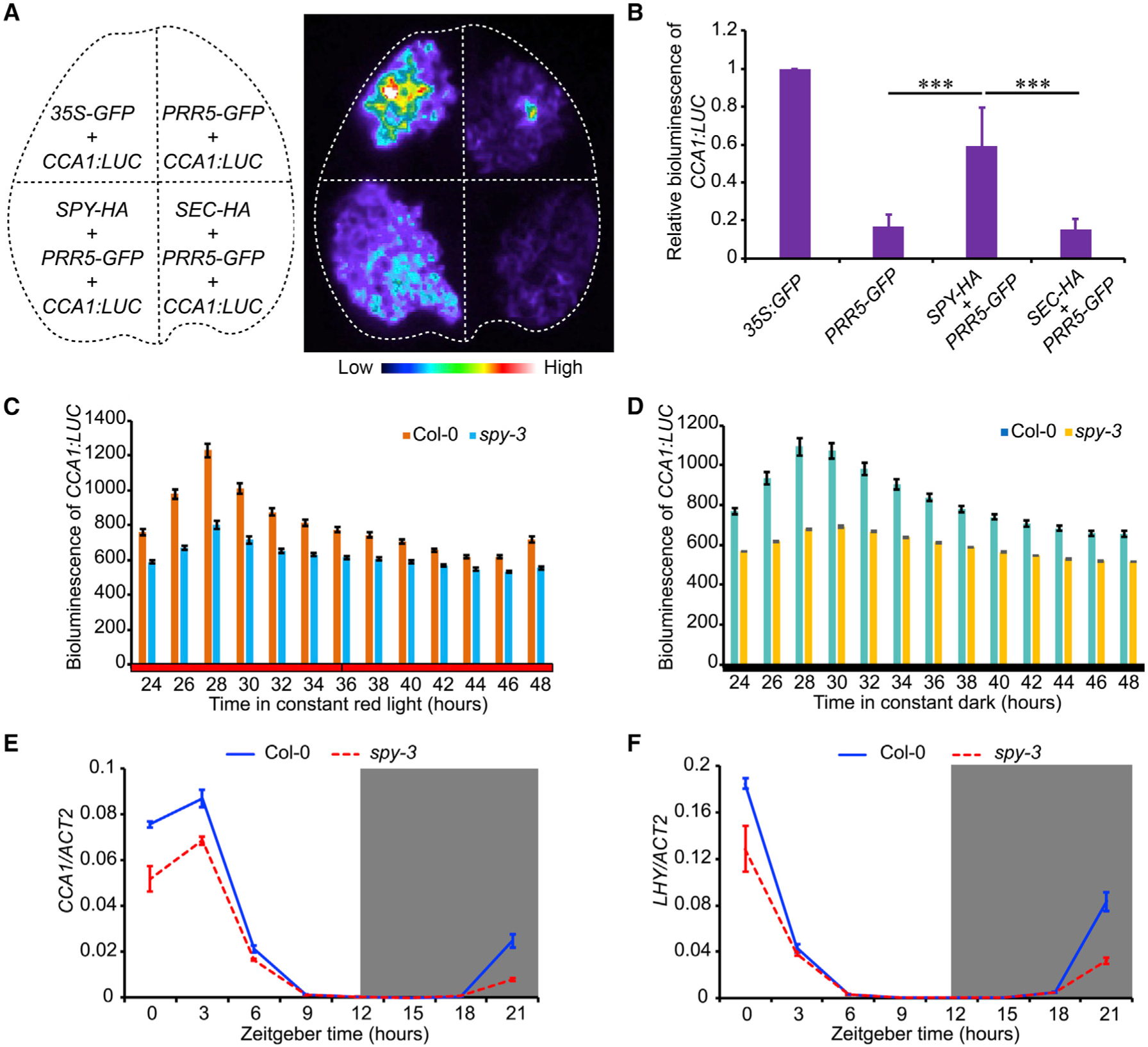
(A) Bioluminescence of CCA1:LUC in transiently transformed N. benthamiana leaves with indicated constructs shown in left panel. 35S:GFP was used as an effecter control.
(B) Quantitative analysis of bioluminescence intensity shown in A. Data represent mean ± SE. (n = 6). ***p < 0.001 (Student t-test).
(C and D) Bioluminescence of CCA1:LUC showing the lower transcription level of CCA1 in spy-3 in continuous red light or continuous darkness as denoted. Data represent mean ± SE (n = 15).
(E and F) Transcript levels of CCA1 (E) and LHY (F) in spy-3 mutant under LD condition. Data represent mean ± s.e. in three biological replicates. The gene expression level was normalized by ACT2 expression.
PRR5 Is a Major Genetic Downstream Target of SPY in Modulating the Circadian Clock
The finding that SPY facilitates the proteolysis of PRR5 is consistent with the lengthened circadian period phenotype in spy mutant, as elevated PRR5 protein level also lengthens circadian period while null mutant of PRR5 displays shorter circadian period phenotype (Nakamichi et al., 2005; Matsushika et al., 2007; Fujiwara et al., 2008; Baudry et al., 2010; Wang et al., 2010). To further investigate whether PRR5 genetically acts downstream of SPY, we examined the circadian phenotype of spy-3, prr5–1, and spy-3 prr5–1 mutants under free running conditions. Compared to wild type plants, spy-3 plants displayed a long period, while prr5–1 mutant displayed a short circadian period, consistent with previous studies (Nakamichi et al., 2005; Wang et al., 2010). Importantly, relative to the wild type controls, the double mutant of spy-3 prr5–1 displayed significantly shortened circadian period (Figure 6A–6C), but not to the extent of the shortened circadian period of prr5–1. Similarly, the null mutation of PRR5 is partially epistatic to spy-8 (Supplemental Figure 12A–12C). Reciprocally, we found that there was no additive or synergistic effect between PRR5 overexpression and spy-3 (Figure 6D and 6E), and PRR5 protein level is not dramatically different between 35S:PRR5-GFP and 35S:PRR5-GFP spy-3 (Supplemental Figure 13), further supporting the idea that PRR5 is a significant downstream target of SPY in the modulation of circadian period.
Figure 6. PRR5 Is a Genetic Downstream Target of SPY in Modulating Circadian Clock.
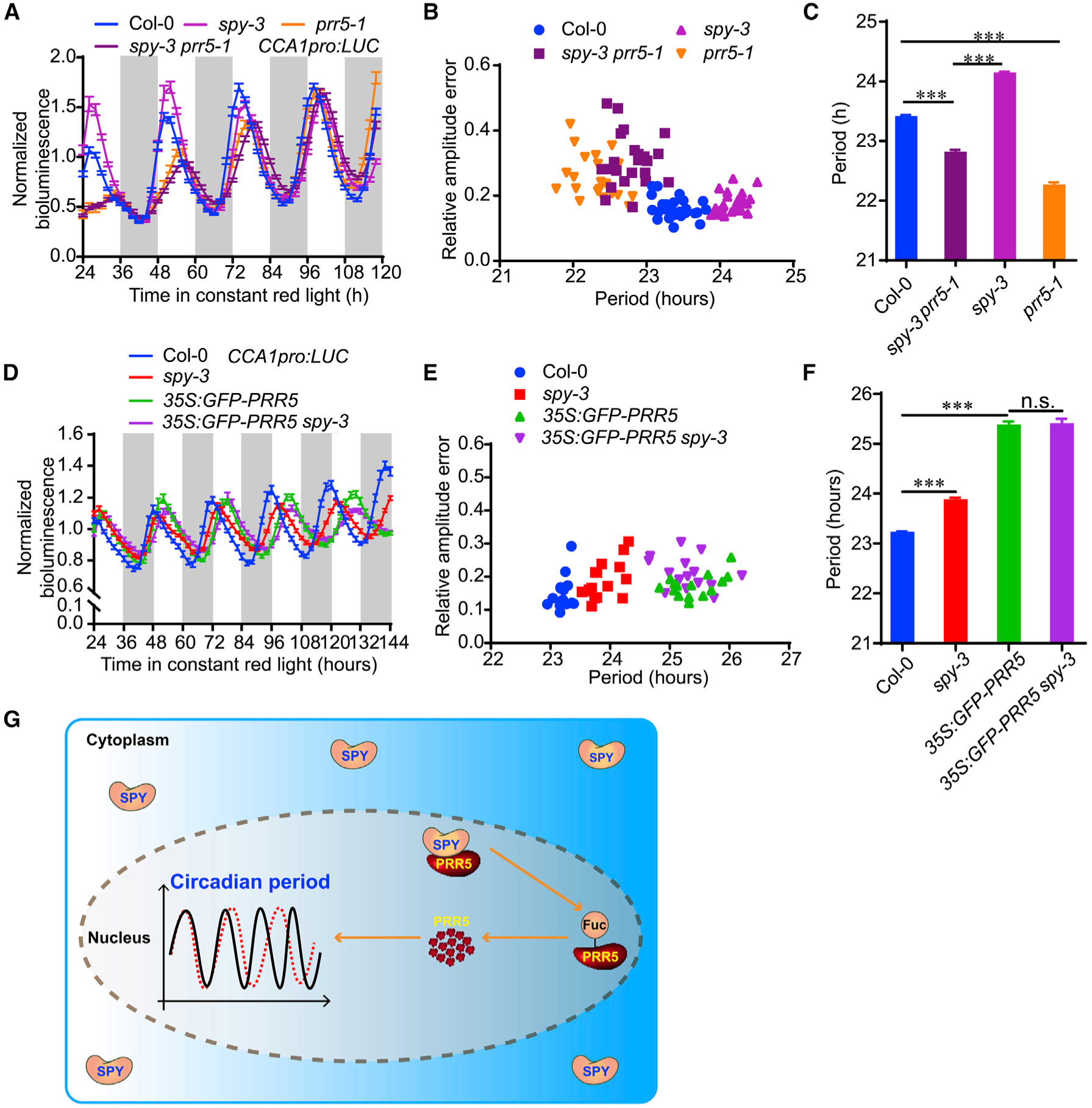
(A) Normalized bioluminescence activity of CCA1:LUC in spy-3, prr5–1, spy-3 prr5–1 and Col-0 plants in red light. Data represent mean ± SE (n = 15).
(B) Scatter plot showing that the lengthened circadian period phenotype of spy-3 could be partially rescued by prr5–1, the null mutation of PRR5.
(C) The estimated circadian period of spy-3, prr5–1, spy-3 prr5–1 and Col-0 plants. Data represent mean ± SE (n = 15 seedlings). ***p < 0.001 (Student t-test).
(D) Normalized bioluminescence activity of CCA1:LUC in spy-3, 35S:PRR5-GFP, 35S:PRR5-GFP spy-3 and Col-0 plants in continuous monochromatic red light. Data represent mean ± SE (n > 10).
(E) Scatter plot showing that the lengthened circadian period phenotype of 35S:PRR5-GFP is not increased by SPY mutation.
(F) The estimated circadian period of spy-3, 35S:PRR5-GFP, 35S:PRR5-GFP spy-3 and Col-0 plants. Data represent mean ± SE (n > 10). ***p < 0.001 (Student’s t-test).
(G) A proposed model of the molecular mechanism by which SPY fine-tunes circadian period in Arabidopsis. SPY fine-tunes circadian speed in nucleus, where it physically interacts with PRR5 and affects PRR5 protein stability.
Altogether, these findings support the notion that PRR5 is genetically downstream of SPY to specifically regulate circadian speed (Figure 6G). Other SPY related developmental phenotypes such as flowering time and leaf serration were not rescued by prr5–1 (Supplemental Figure 14A–14C), implicating a multifaceted role of SPY, probably mediated by distinct downstream targets.
DISCUSSION
Fine-tuning of the circadian period is critical for maintaining a proper daily recurring rhythm of cellular metabolism and energy homeostasis. A wide range of post-translational regulations have been demonstrated as key regulatory mechanisms in fine-tuning circadian period. OGT plays a pivotal role in mice and flies, respectively (Kim et al., 2012; Kaasik et al., 2013; Li et al., 2013), but less is known in higher plants. SPY and SEC were originally considered as two candidate OGTs in plants, each with a distinctive N-terminal TPR domain structure and C-terminal catalytic domain(Hartweck et al., 2006; Olszewski et al., 2010). However, recent studies reveal that SEC is an OGT, whereas SPY is a POFUT (Zentella et al., 2016, 2017). Here we show that SPY, but not SEC, specifically fine-tunes circadian period in Arabidopsis, indicating O-fucosylation, but not O-GlcNAcylation is a key determinant in plants. Consistently, spy mutant alleles with null-POFUT activities, such as spy-15 and spy-19, displayed even longer circadian period than the other alleles with TPR domain mutations, indicating that POFUT activity plays a pivotal role in SPY-mediated fine-tuning of circadian period.
It has been long known that SPY is a nucleocytoplasmic shuffling protein, and distinct subcellular-localized SPY appears to exert different physiological actions (Swain et al., 2002; Maymon et al., 2009). For instance, nuclear SPY modifies DELLA proteins to enhance its interaction with PIF4 and BZR1, which then act as negative regulators of GA (gibberellin) response, while cytoplasmic SPY specifically mediates cytokinin responses to control leaf serration phenotype (Swain et al., 2002; Maymon et al., 2009; Zentella et al., 2017). In the present study, we found that nuclear-localized SPY is necessary and sufficient for fine-tuning circadian period, and the null mutation of PRR5 dramatically reverts the lengthened circadian period in spy mutants, but not the leaf serration and flowering time phenotypes, consistent with SPY-PRR5 interaction occurring exclusively in the nucleus.
Our protein-protein interaction analysis indicated that SPY interacts with PRR5 in the nucleus, and that SPY TPR domain and PRR5 C-terminus are responsible in meditating SPY-PRR5 physical interaction in vitro and in vivo. Importantly, the spy-3 mutant (containing a missense mutation within the POFUT domain) displays significantly lengthened circadian period, suggesting that O-fucosyltransferase activity of SPY is required for its role in fine tuning the circadian clock. Moreover, the O-fucosyltransferase activity of SPY is critical for regulating PRR5 protein stability, as higher protein abundance of PRR5 is found in spy-3 mutant. In addition, coexpression of SPY and PRR5 in N. benthamiana resulted in O-fucosylation of PRR5, further supporting the idea that SPY O-fucosylates PRR5 to facilitate PRR5 proteolysis. Together, we propose that SPY TPR domain, mediates the protein-protein interaction with downstream targets, whereas its catalytic domain is essential for modulating target stability, likely through POFUT activity-mediated posttranslational modification. Consistently, overexpression of SPY TPR domain alone only phenocopied weak developmental phenotypes of spy mutant (Tseng et al., 2001), reinforcing the importance of SPY catalytic activity for function. Collectively, our findings and previous evidence (Silverstone et al., 2007; Steiner et al., 2016; Zentella et al., 2017) support the notion that SPY can regulate protein stabilities in plants. Further identification of the precise O-fucosylated amino acids in PRR5 protein by specialized mass spectrometry will be required to directly examine the consequence of O-fucosylation modification on PRR5 protein stability.
As SPY interacts with PRR5, and PRR5 protein accumulates in spy mutants, we reasoned that the lengthened circadian period in spy mutants could be due to the over-accumulation of PRR5 protein. SPY has been shown to interact with GI in Y-2-H and in vitro pull down assays (Tseng et al., 2004), confirmed in our tests (Figure 2B). However, we did not detect SPY-GI interaction in the AP-MS assay, which might be due to their weak interaction in plants. Alternatively, GI protein modified by SPY may be labile and subjected to rapid degradation. Nevertheless, the role of SPY in circadian period control is unlikely through its interaction with GI, as previous genetic evidence demonstrated that they likely function in parallel to control period (Tseng et al., 2004). By contrast, the null mutation of PRR5 largely reverts the lengthened circadian period phenotype of spy mutants, including mutations in its POFUT domain (spy-3) or TPR domain (spy-8). These results suggest that both types of spy mutants might over-accumulate PRR5 protein. Consistently, PRR5 overexpression caused long circadian period phenotype in Col-0, but not in spy-3 mutant, which already contained an elevated level of PRR5 protein (Figure 6D and 6E). The incomplete rescue of lengthened circadian period in spy mutants by prr5–1 could be due to the effects of SPY on other circadian clock regulators.
Interestingly, other developmental processes regulated by SPY, such as flowering time and leaf serration, is independent of PRR5, which could be mediated by other SPY downstream targets, such as GI or TCP14 (Tseng et al., 2004; Steiner et al., 2012). It is noteworthy that TCP14 was also identified as an interacting protein of SPY in our AP-MS analysis sampled at ZT0. Thus, the SPY interactome identified in this study could be a useful resource for the future investigation of the roles of SPY in physiology or development.
OGT-mediated protein O-GlcNAcylation has been extensively studied in mammalian circadian period regulation. In mice, BMAL1 and CLOCK undergo rhythmic O-GlcNAcylation, which stabilizes both proteins by counteracting ubiquitination-mediated proteolysis (Li et al., 2013). In Drosophila, PERIOD (dPER) is a direct target of OGT, controlling both the timing of dPER nuclear entry and dPER stability (Kim et al., 2012). In addition, transcriptional activity of dPER and dCLOCK can be modulated by O-GlcNAcylation, suggesting that OGT functions at multiple levels to regulate circadian period (Kaasik et al., 2013). In plants, however, we have found only O-fucosyltransferase activity (SPY) regulates clock function, and found no evidence for O-GlcNAcylation activity (SEC) in circadian processes. Intriguingly, SPY-like genes are absent in animals (Hartweck et al., 2006). Hence it appears that O-glycosylation plays an evolutionarily conserved role in regulating circadian period, via O-GlcNAcylation in animals, and O-fucosylation in plants. Whether and how O-fucosylation transduces nutrient signals or cellular energy homeostasis to core clock machinery awaits further exploration.
METHODS
Plant Materials and Constructs
The wild-types Arabidopsis thaliana used in this study were Columbia (Col-0) and Landsberg erecta (Ler) ecotype as indicated. The mutants of sec-5 (SALK_034290C) (Xing et al., 2018), spy-1, spy-3 (Qin et al., 2011) and prr5–1 (Wang et al., 2010) were from Col-0, while the mutants of spy-8, spy-15 and spy-19 were from Ler ecotype (Zentella et al., 2017) as previously reported. The spy-3 prr5–1 double mutant was generated in this study by genetically crossing spy-3 and prr5–1 mutants. The growth conditions were 12-h light/12-h dark, white light (300 μmol m−2 s−1), 22°C (LD); constant white light (300 μmol m−2 s−1), 22°C (LL) or constant darkness, 22°C (DD) as noted respectively. To examine circadian phenotypes, the surface sterilized seeds were grown on solidified Murashige and Skoog (MS) medium containing 3% sucrose under 12-h light/12-h dark conditions for 10 days, then transferred to constant red light (40 μmol m−2 s−1) or DD as indicated. For affinity purification assay followed by mass spectrometry, the two-week-old seedlings grown in LD condition were collected at the indicated time points. For making SPY:GFP-SPY spy-3, SPYpro:GFP-SPY-NLS spy-3, SPYpro:GFP-SPY-NES spy-3, SPYpro:LUC and SPYpro: LUC transgenic plants, the fragment of SPY promoter (−2547 to −1 bp, upstream of the start codon) was firstly amplified and inserted into PstI and KpnI sites of p1300 promoterless vector, followed by subcloning GFP-SPY, GFP-SPY-NLS, GFP-SPY-NES through KpnI and NcoI sites, and the corresponding constructs were transformed into Agrobacterium for floral dipping.
To generate 35S: SPY-HA and 35S: SEC-HA constructs, the respective fragments were amplified by PCR and then were subcloned into the PacI-PstI sites of the pCsVMV:HA-1300 vector. For making 35S: SPY-FLAG, 35S: SPYNT-FLAG and 35S: SPYCT-FLAG constructs, the fragments were amplified and then subcloned into the SacI-SpeI sites of the p35S:FLAG-1307 vector. To generate the 35S:GFP-SPY, 35S:GFP-SPYNLS, 35S:GFP-SPY-NES, 35S:GFP-GI, 35S:GFP-PRR5N, 35S:GFP-PRR5M and 35S:GFP-PRR5C constructs, the respective PCR fragments were subcloned into the KpnI and XhoI sites of the pENTR2B vector firstly, and then were subcloned into 35S:GFP vector via LR reaction. Transgenic line of PRR5:PRR5-GFP was generated previously (Fujiwara et al., 2008), and was used to cross with spy-3 to obtain PRR5:PRR5-GFP spy-3 plants. The 35S:GFP-PRR5, 35S:GFP-PRR7, 35S:GFPPRR9 and 35S:GFP-TOC1 constructs were made as previously described (Wang et al., 2013). For transient gene expression analysisN. benthamiana plants were grown in 25°C with 12 h Light/12 h Dark cycles for 4 to 6 weeks, and the leaves were infiltrated. All of the primers used for making the above constructs were listed in Supplemental Table 1.
Bioluminescence Assay and Circadian Period Estimation
The CCA1:LUC, CAB2:LUC, and LNK2:LUC reporter genes were reported previously (Wang et al., 2013; Xie et al., 2014). Bioluminescence assay was performed as previously described (Wang et al., 2013), and bioluminescence signals were obtained at constant red light or DD conditions as noted, after spraying luciferin on 7-day-old seedlings that were grown in 12 h light/12 h dark, by using a CCD camera (LN/1300-EB/1, Princeton Instruments). Raw bioluminescence data were imported into the Biological Rhythms Analysis software system (BRASS version 2.14; Southern and Millar, 2005; available from www.amillar.org) and analyzed with the Fourier transform–nonlinear least-squares suite of programs. Period lengths were estimated as variance-weighted periods±SE with a time window from 24 to 144 h, unless otherwise stated.
Yeast Two-Hybrid Assay
Yeast two-hybrid assay was performed according to the Match maker GAL4 Two-Hybrid System 3 manufacturer’s manual (Clontech). The prey plasmids including pGBKT7-SPY, pGBKT7-SPYNT and pGBKT7-SPCT were separately co-transformed with the bait plasmids, such as pGADT7-PRR5, pGADT7-PRR5N pGADT7-PRR5M, pGADT7-PRR5C into Saccharomyces cerevisiae strain AH109. The co-transformed yeast cells were cultured on synthetic dropout (SD) medium without tryptophan and leucine (SD-TL) for 3 days at 30°C. The transformants were then transferred onto SD/-Trp/-Leu/-His/-Ade (SD-TLHA) containing 40 μg/mL X-gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) for detecting the interacting activity. The primers used for yeast two hybrid assay were listed in Supplemental Table 1.
Affinity Purification Followed by Mass Spectrometry
Two-weeks-old Arabidopsis transgenetic seedlings harboring SPY-pro:GFP-SPY and SPYpro:GFP-SPY-NLS were harvested at ZT0 (Zeitgeber time, ZT0 represents the light on) and ZT12 respectively, and were quickly frozen in liquid nitrogen. 3 mL of liquid nitrogen ground tissue powders of each sample was used for protein extraction with 3 mL protein extraction buffer (10 mM Tris-HCl pH 7.5, 150 mM NaCl, 0.5 mM EDTA pH 8.0, 0.5% NP-40, 1 mM DTT, 1 mM PMSF, 2 μg/mL Chymostatin, 2 μg/mL Leupeptin, 2 μg/mL Pepstatin, 2 μg/mL Aprotinin, 50 mM MG132, 50 mM MG115, 50 mM ALLN, 2 mM NaF, 2 mM Na3VO4, and dd H2O complemented). After homogenization, the samples were sonicated for 5 min and centrifuged for 10 min at 8000 rpm at 4°C. The clear supernatant was then transferred to incubation with GFP-Trap beads (ChromoTek) for 1 h at 4°C with rotation. The beads were washed by using the ice-cold wash buffer I (10 mM Tris-HCl pH 7.5, 150 mM NaCl, 0.5 mM EDTA pH 8.0, 0.5% NP-40, 1 mM DTT, 1 mM PMSF) for four times, and then three times with the wash buffer II (10 mM Tris-HCl pH7.5, 150 mM NaCl, 0.5 mM EDTA pH 8.0, 1 mM DTT). For on-beads digestion, the beads were firstly resuspended with 45 μL elution buffer I (50 mM Tris-HCl pH 7.5, 2 M urea, 1 mM DTT) containing 5 μg/mL sequencing grade modified trypsin-promega, and were incubated at 30°C for 30 min. After centrifuging at 3000 g for 2 min at 4°C, the supernatant was transferred to a fresh vial. Then the beads were resuspended in 40 μL elution buffer II (50 mM Tris-HCl pH 7.5, 2 M urea, 5 mM iodoacetamide). After collecting the supernatant, repeat wash once with the buffer II, and combine all of the supernatant for incubation at 32°C overnight. After stopping the reaction, the samples were separated to 2 equal parts and individually used for spectral library building and quantitation analysis with SWATH method. For statistical analysis of t-test, quantitative data of the peptides were exported to MarkerView software. The putative SPY interacting proteins were identified based on the following criteria: proteins with at least two peptides that were present in both IP samples using GFP-SPY and GFP-SPY-NLS, and their abundance in comparison to that in the GFP negative control is over 1.5 fold.
Co-immunoprecipitation Assay
Agrobacteria containing 35S:GFP-PRRs and 35S:SPY-FLAG were coinfiltrated into 5 week-old N. benthamiana leaves. After 3 days, the infiltrated leaves were collected and ground in liquid nitrogen to fine powder. Protein extraction and immunoprecipitation were conducted as former description with GFP antibody (ab11120, Invitrogen) (Wang et al., 2013). GFP-tagged PRRs and FLAG tagged SPY and its deletions were detected by Western blotting using GFP antibody (ab6556, Abcam) FLAG antibody (M20008, Abmart), and Biotinylated-Aleuria aurantia Lectin (Vector labs, B-1395).
Bimolecular Fluorescence Complementation Assay
For Bimolecular Fluorescence Complementation (BiFC) assay, the full length coding region of SPY was subcloned into 2YN-pBI-nYFP, while the PRR5N (1–172aa), PRR5M (173–508aa) and PRR5C (173–558aa) were inserted into 2YC-pBI. All primers of these constructs for BiFC are in Supplemental Table 1. Agrobacteria containing the above respective plasmids were transformed into leaves of 5 weeks old N. benthamiana plants as indicated. Agrobacteria containing H2B-mCherry was used as a nuclear marker. The infiltrated leaf clipping were collected 3 to 4 days after infiltration, and the signals were examined with a Olympus FV1000MPE confocal microscope.
RNA Isolation and Real-Time qPCR
Tissues were harvested from two weeks old Arabidopsis seedlings grown on MS medium under LD or LL conditions, with a time course starting from ZT0 or LL24 as noted. Total RNA was extracted by using a TRIzol RNA extraction kit (Invitrogen) and treated with RNase-free DNase I (Thermo Fisher). The cDNA synthesis was performed with M-MLV reverse transcriptase (Promega) and oligo-dT. Quantitative real-time RT-PCR (qRTPCR) was performed by using SYBR Green Real-Time PCR Master Mix (TOYOBO, Osaka, Japan) according to the manufacturer’s instructions on a Applied Biosystems™ QuantStudio 3 instrument (Applied Bio-systems). Sequences and quantities of primers for the respective genes were listed in Supplemental Table 1. Gene expression was normalized by ACTIN2 expression. Mean values of 2–ΔCT were calculated from three biological replicates and three technical replicates as described previously (Wang et al., 2013).
Supplementary Material
ACKNOWLEDGMENTS
We thank Professors J.C. Jang and David E. Somers (Ohio State University) for their critical comments on the manuscript. We thank Dr. Zhuang Lu and Jingquan Li from the Plant Science Facility of the Institute of Botany, Chinese Academy of Sciences, for their excellent technical assistance on mass spectrometry and confocal microscopy, respectively. No conflict of interest declared.
FUNDING
This work was supported by the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB27030206), National Natural Science Foundation of China (No. 31770287) and National Key Research and Development Program of China (2016YFD0100600) to L.W., and the National Institutes of Health (R01 GM100051) to T.P.S.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information is available at Molecular Plant Online.
REFERENCES
- Baudry A, Ito S, Song YH, Strait AA, Kiba T, Lu S, Henriques R, Pruneda-Paz JL, Chua NH, Tobin EM, et al. (2010). F-box proteins FKF1 and LKP2 act in concert with ZEITLUPE to control Arabidopsis clock progression. Plant Cell 22:606–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel X, Sugano S, and Tobin EM (2004). CK2 phosphorylation of CCA1 is necessary for its circadian oscillator function in Arabidopsis. Proc. Natl. Acad. Sci. U S A 101:3292–3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara S, Wang L, Han L, Suh SS, Salome PA, McClung CR, and Somers DE (2008). Post-translational regulation of the Arabidopsis circadian clock through selective proteolysis and phosphorylation of pseudo-response regulator proteins. J. Biol. Chem 283:23073–23083. [DOI] [PubMed] [Google Scholar]
- Fung-Uceda J, Lee K, Seo PJ, Polyn S, De Veylder L, and Mas P (2018). The circadian clock sets the time of DNA replication licensing to regulate growth in Arabidopsis. Dev. Cell 45:101–113.e4. [DOI] [PubMed] [Google Scholar]
- Gendron JM, Pruneda-Paz JL, Doherty CJ, Gross AM, Kang SE, and Kay SA (2012). Arabidopsis circadian clock protein, TOC1, is a DNA-binding transcription factor. Proc. Natl. Acad. Sci. U S A 109:3167–3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenham K, and McClung CR (2015). Integrating circadian dynamics with physiological processes in plants. Nat. Rev. Genet 16:598–610. [DOI] [PubMed] [Google Scholar]
- Han L, Mason M, Risseeuw EP, Crosby WL, and Somers DE (2004). Formation of an SCF(ZTL) complex is required for proper regulation of circadian timing. Plant J. 40:291–301. [DOI] [PubMed] [Google Scholar]
- Hansen LL, Imrie L, Le Bihan T, van den Burg HA, and van Ooijen G (2017). Sumoylation of the plant clock transcription factor CCA1 suppresses DNA binding. J. Biol. Rhythms 32:570–582. [DOI] [PubMed] [Google Scholar]
- Harmer SL (2009). The circadian system in higher plants. Annu. Rev.Plant Biol 60:357–377. [DOI] [PubMed] [Google Scholar]
- Hartweck LM, Genger RK, Grey WM, and Olszewski NE (2006). SECRET AGENT and SPINDLY have overlapping roles in the development of Arabidopsis thaliana L. Heyn. J. Exp. Bot 57:865–875. [DOI] [PubMed] [Google Scholar]
- Herrero E, Kolmos E, Bujdoso N, Yuan Y, Wang M, Berns MC, Uhlworm H, Coupland G, Saini R, Jaskolski M, et al. (2012). EARLY FLOWERING4 recruitment of EARLY FLOWERING3 in the nucleus sustains the Arabidopsis circadian clock. Plant Cell 24:428–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Perez-Garcia P, Pokhilko A, Millar AJ, Antoshechkin I, Riechmann JL, and Mas P (2012). Mapping the core of the Arabidopsis circadian clock defines the network structure of the oscillator. Science 336:75–79. [DOI] [PubMed] [Google Scholar]
- Jacobsen SE, Binkowski KA, and Olszewski NE (1996). SPINDLY, a tetratricopeptide repeat protein involved in gibberellin signal transduction in Arabidopsis. Proc. Natl. Acad. Sci. U S A 93:9292–9296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaasik K, Kivimae S, Allen JJ, Chalkley RJ, Huang Y, Baer K, Kissel H, Burlingame AL, Shokat KM, Ptacek LJ, et al. (2013). Glucose sensor O-GlcNAcylation coordinates with phosphorylation to regulate circadian clock. Cell Metab. 17:291–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamioka M, Takao S, Suzuki T, Taki K, Higashiyama T, Kinoshita T, and Nakamichi N (2016). Direct repression of evening genes by CIRCADIAN CLOCK-ASSOCIATED1 in the Arabidopsis circadian clock. Plant Cell 28:696–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim EY, Jeong EH, Park S, Jeong HJ, Edery I, and Cho JW (2012). A role for O-GlcNAcylation in setting circadian clock speed. Genes Dev. 26:490–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Kim HJ, Vu QT, Jung S, McClung CR, Hong S, and Nam HG (2018). Circadian control of ORE1 by PRR9 positively regulates leaf senescence in Arabidopsis. Proc. Natl. Acad. Sci. U S A 115:8448–8453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MD, Ruan HB, Hughes ME, Lee JS, Singh JP, Jones SP, Nitabach MN, and Yang X (2013). O-GlcNAc signaling entrains the circadian clock by inhibiting BMAL1/CLOCK ubiquitination. Cell Metab. 17:303–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Wang Y, Zhang Y, Tian W, Chong K, Jang JC, and Wang L (2019). PRR5, 7 and 9 positively modulate TOR signaling-mediated root cell proliferation by repressing TANDEM ZINC FINGER 1 in Arabidopsis. Nucleic Acids Res. 47:5001–5015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushika A, Kawamura M, Nakamura Y, Kato T, Murakami M, Yamashino T, and Mizuno T (2007). Characterization of circadian-associated pseudo-response regulators: II. The function of PRR5 and its molecular dissection in Arabidopsis thaliana. Biosci. Biotechnol. Biochem 71:535–544. [DOI] [PubMed] [Google Scholar]
- Maymon I, Greenboim-Wainberg Y, Sagiv S, Kieber JJ, Moshelion M, Olszewski N, and Weiss D (2009). Cytosolic activity of SPINDLY implies the existence of a DELLA-independent gibberellin-response pathway. Plant J. 58:979–988. [DOI] [PubMed] [Google Scholar]
- Nakamichi N, Kiba T, Henriques R, Mizuno T, Chua NH, and Sakakibara H (2010). PSEUDO-RESPONSE REGULATORS 9, 7, and 5 are transcriptional repressors in the Arabidopsis circadian clock. Plant Cell 22:594–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamichi N, Kiba T, Kamioka M, Suzuki T, Yamashino T, Higashiyama T, Sakakibara H, and Mizuno T (2012). Transcriptional repressor PRR5 directly regulates clock-output pathways. Proc. Natl. Acad. Sci. U S A 109:17123–17128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamichi N, Kita M, Ito S, Sato E, Yamashino T, and Mizuno T (2005). The Arabidopsis pseudo-response regulators, PRR5 and PRR7, coordinately play essential roles for circadian clock function. Plant Cell Physiol. 46:609–619. [DOI] [PubMed] [Google Scholar]
- Nusinow DA, Helfer A, Hamilton EE, King JJ, Imaizumi T, Schultz TF, Farre EM, and Kay SA (2011). The ELF4-ELF3-LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature 475:398–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olszewski NE, West CM, Sassi SO, and Hartweck LM (2010). O-GlcNAc protein modification in plants: avolution and function. Biochim. Biophys. Acta 1800:49–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin F, Kodaira KS, Maruyama K, Mizoi J, Tran LS, Fujita Y, Morimoto K, Shinozaki K, and Yamaguchi-Shinozaki K (2011). SPINDLY, a negative regulator of gibberellic acid signaling, is involved in the plant abiotic stress response. Plant Physiol. 157:1900–1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez SE, and Kay SA (2016). The plant circadian clock: from a simple timekeeper to a complex developmental manager. Cold Spring Harb. Perspect. Biol 8:a027748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalit-Kaneh A, Kumimoto RW, Filkov V, and Harmer SL (2018). Multiple feedback loops of the Arabidopsis circadian clock provide rhythmic robustness across environmental conditions. Proc. Natl. Acad. Sci. U S A 115:7147–7152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstone AL, Tseng TS, Swain SM, Dill A, Jeong SY, Olszewski NE, and Sun TP (2007). Functional analysis of SPINDLY in gibberellin signaling in Arabidopsis. Plant Physiol. 143:987–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern MM, and Millar AJ (2005). Circadian genetics in the model higher plant, Arabidopsis thaliana. Methods Enzymol. 393:23–25. [DOI] [PubMed] [Google Scholar]
- Sothern RB, Tseng TS, Orcutt SL, Olszewski NE, and Koukkari WL (2002). GIGANTEA and SPINDLY genes linked to the clock pathway that controls circadian characteristics of transpiration in Arabidopsis. Chronobiol. Int 19:1005–1022. [DOI] [PubMed] [Google Scholar]
- Steiner E, Efroni I, Gopalraj M, Saathoff K, Tseng TS, Kieffer M, Eshed Y, Olszewski N, and Weiss D (2012). The Arabidopsis O-linked N-acetylglucosamine transferase SPINDLY interacts with class I TCPs to facilitate cytokinin responses in leaves and flowers. Plant Cell 24:96–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner E, Livne S, Kobinson-Katz T, Tal L, Pri-Tal O, Mosquna A, Tarkowska D, Mueller B, Tarkowski P, and Weiss D (2016). The putative O-linked N-acetylglucosamine transferase SPINDLY inhibits class I TCP proteolysis to promote sensitivity to cytokinin. Plant Physiol. 171:1485–1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugano S, Andronis C, Green RM, Wang ZY, and Tobin EM (1998). Protein kinase CK2 interacts with and phosphorylates the Arabidopsis circadian clock-associated 1 protein. Proc. Natl. Acad. Sci. U S A 95:11020–11025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swain SM, Tseng TS, and Olszewski NE (2001). Altered expression of SPINDLY affects gibberellin response and plant development. Plant Physiol. 126:1174–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swain SM, Tseng TS, Thornton TM, Gopalraj M, and Olszewski NE (2002). SPINDLY is a nuclear-localized repressor of gibberellin signal transduction expressed throughout the plant. Plant Physiol. 129:605–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng TS, Salome PA, McClung CR, and Olszewski NE (2004). SPINDLY and GIGANTEA interact and act in Arabidopsis thaliana pathways involved in light responses, flowering, and rhythms in cotyledon movements. Plant Cell 16:1550–1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng TS, Swain SM, and Olszewski NE (2001). Ectopic expression of the tetratricopeptide repeat domain of SPINDLY causes defects in gibberellin response. Plant Physiol. 126:1250–1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehara TN, Mizutani Y, Kuwata K, Hirota T, Sato A, Mizoi J, Takao S, Matsuo H, Suzuki T, Ito S, et al. (2019). Casein kinase 1 family regulates PRR5 and TOC1 in the Arabidopsis circadian clock. Proc. Natl. Acad. Sci. U S A 116:11528–11536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Fujiwara S, and Somers DE (2010). PRR5 regulates phosphorylation, nuclear import and subnuclear localization of TOC1 in the Arabidopsis circadian clock. EMBO J. 29:1903–1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Kim J, and Somers DE (2013). Transcriptional corepressor TOPLESS complexes with pseudoresponse regulator proteins and histone deacetylases to regulate circadian transcription. Proc. Natl. Acad. Sci. U S A 110:761–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZY, and Tobin EM (1998). Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell 93:1207–1217. [DOI] [PubMed] [Google Scholar]
- Xie Q, Wang P, Liu X, Yuan L, Wang L, Zhang C, Li Y, Xing H, Zhi L, Yue Z, et al. (2014). LNK1 and LNK2 are transcriptional coactivators in the Arabidopsis circadian oscillator. Plant Cell 26:2843–2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing L, Liu Y, Xu S, Xiao J, Wang B, Deng H, Lu Z, Xu Y, and Chong K (2018). Arabidopsis O-GlcNAc transferase SEC activates histone methyltransferase ATX1 to regulate flowering. EMBO J. 37:e98115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zentella R, Hu J, Hsieh WP, Matsumoto PA, Dawdy A, Barnhill B, Oldenhof H, Hartweck LM, Maitra S, Thomas SG, et al. (2016). O-GlcNAcylation of master growth repressor DELLA by SECRET AGENT modulates multiple signaling pathways in Arabidopsis. Genes Dev. 30:164–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zentella R, Sui N, Barnhill B, Hsieh WP, Hu J, Shabanowitz J, Boyce M, Olszewski NE, Zhou P, Hunt DF, et al. (2017). The Arabidopsis O-fucosyltransferase SPINDLY activates nuclear growth repressor DELLA. Nat. Chem. Biol 13:479–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wang Y, Wei H, Li N, Tian W, Chong K, and Wang L (2018). Circadian evening complex represses jasmonate-induced leaf senescence in Arabidopsis. Mol. Plant 11:326–337. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


