Abstract
Genetic syndromes frequently present with overlapping clinical features and inconclusive or ambiguous genetic findings which can confound accurate diagnosis and clinical management. An expanding number of genetic syndromes have been shown to have unique genomic DNA methylation patterns (called “episignatures”). Peripheral blood episignatures can be used for diagnostic testing as well as for the interpretation of ambiguous genetic test results. We present here an approach to episignature mapping in 42 genetic syndromes, which has allowed the identification of 34 robust disease-specific episignatures. We examine emerging patterns of overlap, as well as similarities and hierarchical relationships across these episignatures, to highlight their key features as they are related to genetic heterogeneity, dosage effect, unaffected carrier status, and incomplete penetrance. We demonstrate the necessity of multiclass modeling for accurate genetic variant classification and show how disease classification using a single episignature at a time can sometimes lead to classification errors in closely related episignatures. We demonstrate the utility of this tool in resolving ambiguous clinical cases and identification of previously undiagnosed cases through mass screening of a large cohort of subjects with developmental delays and congenital anomalies. This study more than doubles the number of published syndromes with DNA methylation episignatures and, most significantly, opens new avenues for accurate diagnosis and clinical assessment in individuals affected by these disorders.
Keywords: DNA methylation, episignature, EpiSign, molecular diagnostics, uncertain clinical cases, VUS classification
Introduction
The past few years have seen the emergence of a critically important development in the molecular diagnosis of congenital disorders. DNA methylation episignatures, defined as the cumulative DNA methylation patterns occurring at multiple CpG dinucleotides across the genome, have been recognized to be intricately associated with many human traits, including age, sex, and disease status.1, 2, 3, 4, 5, 6 Specific patterns in the methylomes of individuals with defined congenital syndromes have recently received particular attention in clinical settings.7, 8, 9 The elucidation of DNA methylation patterns in a range of constitutional syndromes has led to the recognition that these episignatures represent an early event during embryo development, and thus are present in numerous tissues of the affected individuals, including peripheral blood, the most common source of DNA specimens in diagnostic laboratories.10,11 The stability of DNA methylation patterns provides ground for their use in clinical diagnosis. The conditions studied so far have demonstrated that the observed episignatures are specific to the syndromes in which they were discovered and that the observed patterns occur consistently across all of the individuals affected with the same syndrome;12 this promises that DNA methylation episignatures have a great potential to unlock the molecular diagnosis of congenital disorders, a feat which frequently cannot be achieved by conventional clinical and molecular assessments.13
We have previously been able to demonstrate that the episignatures of genetic syndromes can be used to reliably resolve ambiguous clinical cases associated with uncertain sequence variant or clinical findings and to detect disease through screening of cohorts of individuals with developmental delay and congenital anomalies but without a diagnosis.12, 13, 14, 15,16,17 In April of 2019, the first clinical genome-wide DNA methylation assay, “EpiSign,” which utilized genome-wide DNA methylation analysis for the screening of 14 syndromes known to harbor such episignatures, was launched. The computational assessment of DNA methylation data for these syndromes relies on the concurrent assessment of all of the conditions through the use of supervised and unsupervised classification algorithms; this results in acceptable performance in the moderate number of episignatures currently described.12,13 With an ongoing study of new syndromes, however, the number of conditions with episignatures to be included in the analysis will rise significantly, and this will introduce challenges to our current workflow. Specifically, the increased number of syndromes will increase the chance of overlap across different episignatures, and concurrent assessment of a large number of episignatures requires the implementation of novel computational approaches for disease classifications. To date, these questions have not been addressed, and the challenges of concurrent assessment for a very large number of DNA methylation episignatures are not known.
In the present study, we evaluate a large number of congenital syndromes for DNA methylation patterns, and we report 34 distinct and reliable episignatures. We demonstrate the implementation of a uniform approach for mapping DNA methylation signatures in numerous syndromes in order to enable their unbiased comparisons and assessments. We discuss the overlap, similarity, and hierarchical relationships across various episignatures, and we evaluate the extent to which these parameters cause challenges in episignature-based disease classification. Through the development of a supervised classification algorithm capable of simultaneous assessment of 34 episignatures, we demonstrate that the classification of closely related episignatures is feasible, and we show the power of this multiclass approach in resolving undiagnosed individuals with various forms of developmental delay and congenital anomalies.
Material and Methods
Subjects and Cohorts
The study cohort includes peripheral blood DNA samples from individuals who each have a confirmed diagnosis of one of 42 genetic syndromes (Table 1). These included samples collected from the Greenwood Genetic Center (Greenwood, South Carolina, USA), Amsterdam University Medical Center (Amsterdam, Netherlands), Radboud University Medical Center (Nijmegen, the Netherlands), Groupe DI France, Rouen University Hospital (Rouen, France), Université Paris Diderot (Paris, France), McGill University (Montreal, Canada), and Istituto di Ricovero e Cura a Carattere Scientifico (Rome, Italy), as well as specimens described in our previous publications.12,13,18, 19, 20, 21 The a priori motive for the selection of most of these syndromes was based on the involvement of their associated genes in transcriptional and epigenetic regulatory mechanisms and chromatin remodeling.22
Table 1.
Description of the Study Cohort
| Syndrome/Episignature | Abbreviation | Underlying Genes | Phenotype MIM Number | Training Cohort | Testing Cohort | Episignature Detected? |
|---|---|---|---|---|---|---|
| ADNP syndrome—5′ and 3′ terminal ends | ADNP_T | ADNP (outside c.2000-2340) | 615873 | 14 | 5 | yes |
| ADNP syndrome—central | ADNP_C | ADNP (c.2000-2340) | 615873 | 10 | 3 | yes |
| alpha-thalassemia mental retardation syndrome | ATRX | ATRX | 301040 | 13 | 5 | yes |
| autism, susceptibility to, 18 | AUTS18a | CHD8 | 615032 | 5 | 0 | yes |
| BAFopathies: Coffin-Siris 1–4 (CSS1–4) and Nicolaides-Baraitser (NCBRS) syndromes | BAFopathya | ARID1Aa, ARID1B, SMARCB1, SMARCA4, SMARCA2 | 614607, 135900, 614609, 614608, 601358 | 50 | 19 | yes |
| Börjeson-Forssman-Lehmann syndrome | BFLSa | PHF6 | 301900 | 4 | 0 | yes |
| cerebellar ataxia, deafness, and narcolepsy, autosomal dominant | ADCADN | DNMT1 | 604121 | 5 | 0 | yes |
| CHARGE syndrome | CHARGE | CHD7 | 214800 | 45 | 15 | yes |
| Chr7q11.23 duplication syndrome | Dup7 | Chr7q11.23 duplication | 609757 | 8 | 2 | yes |
| mental retardation, X-linked, syndromic, Claes-Jensen type (Claes-Jensen syndrome) | CJS | KDM5C | 300534 | 26 | 8 | yes |
| Cornelia de Lange syndrome 1–4 | CdLS | NIPBL, RAD21, SMC3, SMC1A | 122470, 614701, 610759, 300590 | 31 | 10 | yes |
| Down syndrome | Down | Chr21 trisomy | 190685 | 29 | 10 | yes |
| epileptic encephalopathy, childhood-onset | EEOCa | CHD2 | 615369 | 5 | 0 | yes |
| Floating-Harbor syndrome | FHS | SRCAP | 136140 | 15 | 5 | yes |
| genitopatellar syndrome | GTPTS | KAT6B | 606170 | 5 | 0 | yes |
| Hunter McAlpine syndrome | HMAa | 17q23.1-q24.2 duplication involving NSD1 | 601379 | 4 | 0 | yes |
| immunodeficiency-centromeric instability-facial anomalies syndrome 1 | ICF1 | DNMT3B | 242860 | 8 | 0 | yes |
| immunodeficiency-centromeric instability-facial anomalies syndrome 2–4 | ICF2_3_4 | CDCA7, ZBTB24, HELLS | 614069, 616910, 616911 | 7 | 0 | yes |
| Kabuki syndrome 1 and 2 | Kabukia | KMT2D, KDM6Aa | 147920, 300867 | 66 | 21 | yes |
| Kleefstra syndrome 1 | Kleefstra1a | EHMT1 | 610253 | 15 | 5 | yes |
| Koolen de Vreis syndrome | KDVSa | KANSL1 | 610443 | 6 | 0 | yes |
| mental retardation, autosomal dominant 51 | MRD51a | KMT5B | 617788 | 5 | 0 | yes |
| mental retardation, X-linked 93 | MRX93a | BRWD3 | 300659 | 5 | 0 | yes |
| mental retardation, X-linked 97 | MRX97a | ZNF711 | 300803 | 13 | 4 | yes |
| mental retardation, X-linked syndromic, Nascimento-type | MRXSNa | UBE2A | 300860 | 3 | 0 | yes |
| mental retardation, X-linked, Snyder-Robinson type | MRXSSRa | SMS | 309583 | 8 | 2 | yes |
| Rahman syndrome | RMNSa | HIST1H1E | 617537 | 6 | 0 | yes |
| Rubinstein-Taybi syndrome 1 and 2 | RSTSa | CREBBP, EP300 | 180849, 613684 | 30 | 9 | yes |
| SBBYSS syndrome | SBBYSSa | KAT6B | 603736 | 7 | 0 | yes |
| SETD1B-related syndrome | SETD1Ba | SETD1B | N/A | 8 | 0 | yes |
| Sotos syndrome | Sotos | NSD1 | 117550 | 47 | 15 | yes |
| Tatton-Brown-Rahman syndrome | TBRSa | DNMT3A | 615879 | 10 | 4 | yes |
| Wiedemann-Steiner syndrome | WDSTSa | KMT2A | 605130 | 12 | 4 | yes |
| Williams syndrome | Williams | Chr7q11.23 deletion | 194050 | 15 | 6 | yes |
| Cornelia de Lange syndrome 5 (females only) | CdLS5 | HDAC8 | 300882 | 8 | N/A | no |
| FG syndrome 1 | FG1a,b | MED12 | 305450 | 9 | N/A | no |
| Glass syndrome | Glassa,b | SATB2 | 612313 | 9 | N/A | no |
| KMT2C-related syndrome£ | KMT2Ca,b,c | KMT2C | 617768 | 4 | N/A | no |
| neurodevelopmental disorder with coarse facies and mild distal skeletal abnormalities | NEDCFSAa,b | KDM6B | 618505 | 5 | N/A | no |
| Rett syndrome | Rett | MECP2 | 312750 | 36 | N/A | no |
| Siderius-type X-linked syndromic mental retardation | MRXSSDa,b | PHF8 | 300263 | 9 | N/A | no |
| Smith-Magenis syndrome | SMSa,b | RAI1 | 309583 | 15 | N/A | no |
Indicates that these disorders (or some of their subtypes) were not evaluated in previous studies.
Indicates cohorts with no evidence of a reproducible episignature; this is potentially due to small sample size. A possibility of an episignature is not completely ruled out, and reanalysis using larger sample sizes is warranted.
The OMIM database, at the time of this study, has indicated that subjects with KMT2C mutations may be said to have “Kleefstra 2” syndrome. The DNA methylation signature found in Kleefstra 1 (caused by EHMT1), however, is completely absent in these subjects. It is acknowledged that these subjects have a distinct phenotype from Kleefstra syndrome and a name change is currently in process with OMIM. The numbers in the testing and training cohort columns indicate the sample counts available for each condition in each category. For cohorts with negative findings in the initial assessment, we did not further split the data into testing and training, and thus, the values in the testing column are indicated with N/A (not applicable).
Additional disease cohorts without established episignatures were used to assess the specificity of the classification models designed in this study. These cohorts included individuals diagnosed with Angelman syndrome (MIM: 105830), Prader-Willi syndrome (MIM: 176270), Beckwith-Wiedemann syndrome (MIM: 130650), Coffin-Lowry syndrome (MIM: 303600), Saethre-Chotzen syndrome (MIM: 101400), Fragile X syndrome (MIM: 300624), Silver-Russell syndrome (MIM: 180860), autism spectrum disorders, and RASopathies which have also been described previously.12,13,18
The underlying genetic variant from each subject used in the study was reviewed according to the American College of Medical Genetics (ACMG) guidelines for interpretation of genomic sequence variants,23 and only individuals confirmed to harbor pathogenic or likely pathogenic variants together with the clinical diagnosis were used to represent a syndrome.
Control specimens were healthy individuals without any developmental delay, intellectual disability, or congenital anomalies. The first set of controls used for mapping of the episignatures and training of the classification models included control specimens from the reference control cohort in the London Health Sciences Centre (LHSC) laboratory, along with additional control samples from the centers listed above. Controls that were used to measure the specificity of the developed classifier were compiled from five large databases of general population samples with various age and racial backgrounds.24, 25, 26, 27, 28
Unsolved cases that were screened in this study for the detection of potentially affected individuals were collected from all of the above sources over a period of four years. These samples were supplemented with a publicly available DNA methylation cohort of unresolved subjects that demonstrated various congenital anomalies and developmental delays.29
DNA Methylation Experiment
Peripheral whole-blood DNA was extracted using standard techniques. Following bisulfite conversion, DNA methylation analysis of the samples was performed using the Illumina Infinium methylation 450k or EPIC bead chip arrays according to the manufacturer’s protocol. These arrays cover between 450,000 and 860,000 human genomic methylation CpG sites, including 99% of RefSeq genes and 96% of CpG islands. The resulting methylated and unmethylated signal intensity data were imported into R 3.5.2 for analysis. Normalization was performed according to the Illumina normalization method with background correction done using the minfi package.30 Probes with detection p value > 0.01, those located on chromosomes X and Y, those known to contain a SNP at the CpG interrogation or single-nucleotide extension, and probes known to cross-react with chromosomal locations other than their target regions were removed. Arrays with more than 5% failure probe rates were excluded from the analysis. The methylation level for each probe was measured as a beta value, which was calculated from the ratio of the methylated signals versus the total sum of unmethylated and methylated signals, ranging between 0 (no methylation) and 1 (full methylation). All of the samples were examined for genome-wide methylation density, and those deviating from a bimodal distribution were excluded. Because samples were assayed using two different platforms (450k and EPIC), following normalization and quality controls, the downstream analyses were restricted to the probes shared across the two array types in order to maintain consistency in the computational workflow.
Selection of Cases and Matched Controls
We selected a random 75% subset of the affected subjects as a training cohort for the purpose of mapping of DNA methylation signatures and training of the classification models. The remaining 25% was used as a testing dataset for the assessment of the performance of the classification models developed later. All syndromes and their subtypes were equally represented in both of the training and testing cohorts. No division of the training and testing cohorts was performed for conditions with sample sizes less than 10 (Table 1). For every syndrome in the training cohort, a matched group of controls was selected through the use of the MatchIt package. Matching was performed based on age, sex, and the experimental batch. The sample size of the controls was increased until both the matching quality and the sample size were at their optimum and consistent across all diseases. This led to the determination of a control sample size four times larger than the case group in every comparison. Increasing the sample size beyond this value impaired the matching quality. After each matching trial, a principal component analysis (PCA) was performed to detect outliers and examine the data structures. Outlier samples and those with aberrant data structures were removed before a second matching trial was conducted. The iteration was repeated until no outlier sample was detected in the first two components of the PCA.
Mapping of DNA Methylation Episignatures
DNA methylation studies commonly consider two factors for the prioritization of CpG sites (probes, features, or predictors) that are important in various conditions. These factors are the level of methylation difference (effect size) and the probability that the observed difference is a false positive (p value). Because microarray technology is not sensitive enough to detect very small degrees of methylation change when measuring the methylation levels, and the number of tested CpGs is large, strict cut-offs are applied to both p value and methylation difference estimations during probe selection. In the literature, a range of cut-offs has been used for minimum methylation differences (5%–20%) and p values. The p value, specifically, can be varied based on the sample size and the confounding factors. In the current study, we have assessed 42 different syndromes which are expected to have varying levels and extents of methylation change. As examples, from our previous studies, we have observed that Sotos syndrome can be associated with robust changes in tens of thousands of probes, whereas this figure in Aref-Eshghi et al’s BAFopathies study hardly reaches 500.12,18 Therefore, the determination of a universal cutoff for methylation change and p value for all of the syndromes in the current study might not be a practical approach. In order to accommodate this level of heterogeneity across multiple conditions, instead, we determined a set of ~150 probes to be the most representative of the DNA methylation episignature for each condition, in line with what we had observed in our previous studies regarding the minimum number of probes needed for the classification of different syndromes.13
The following workflow was performed for each condition separately. We initially performed a multivariate linear regression modeling using the limma package.31 The methylation levels (beta value) were logit transformed into M-values (log2(beta/(1-beta))) in order to ensure homoscedasticity for linear modeling. The analysis was adjusted for blood cell type variations. The estimation of blood cell mixture was performed according to the algorithm developed by Houseman et al.32 The estimated values for each cell component were incorporated into the model matrix of the regression analysis as confounding variables. In situations where the samples were assayed in multiple batches or multiple arrays, we also adjusted the analysis for the top 10 principal components of the selected data. The p values obtained in linear modeling were moderated using the eBayes function. To prioritize the best set of probes for each analysis, we used the interaction between the effect size and p value by multiplying the absolute methylation difference between the affected subjects and controls by the negative value of the log-transformed p value (-log(p value)). The top 1,000 probes with the greatest obtained values were selected. Next, we performed a receiver’s operating curve characteristics analysis for every probe and measured the pairwise correlation coefficient between them. Selection of 100–150 probes from this list was conducted by first filtering out the half of the probes with the lowest area under the curve (AUC) and then removing another half from the remaining probes, which were highly correlated with each other. This was done by measuring the Pearson’s correlation coefficients and was carried out separately in cases and controls. The correlation coefficient cut-offs used for each condition were not constant because they yielded different levels of correlations across the selected probes, and thus we experimented with R-squared cutoffs <0.6–0.8 in order to reach the desired number of probes.
The final probes selected for each disorder contained those that were most differentiating, non-redundant, and not influenced by random data structures. To determine the robustness of the identified probes, before each analysis, 10%–20% of samples from the training cohort, depending on the sample size, were set aside and not used for feature selection. After each analysis, the patterns generated by the selected probes were compared between the samples used for the analysis with those that were not. Hierarchical clustering analysis with a heatmap and multidimensional scaling were used for this purpose. A robust episignature was expected to generate a similar pattern in both groups. In addition, we evaluated the methylation patterns of the other samples from the same experimental batch as the cases to rule out the possibility that the observed profile was related to the experimental batch structure. Furthermore, each condition was expected to present a unique profile significantly different from what was observed in controls. This entire process was repeated until all of the samples were used at least once during probe selection. Failure to adhere to any of these principles resulted in the conclusion that the identified probes were not reliable, and when that happened, that condition was excluded from further analysis. When a syndrome was caused by variation in multiple genes, each subtype was initially analyzed individually. If the probes specific to each subtype were not able to distinguish that subtype from the others, we concluded that they have indistinguishable profiles and thus treated them as one episignature.
Assessment of the Relationship between Episignatures
Probes co-occurring between every two episignatures were visualized using a circos plot.33 Further pairwise analysis for any two episignatures was performed using hierarchical clustering analysis with a heatmap as well as multidimensional scaling using the probes specific to each of the two pairs. We performed systematic analysis to determine the distance and similarities and the hierarchical order of the episignatures in order to visualize all episignatures in one dendrogram. For this analysis, we used all of the significant probes from all episignatures. For each syndrome, we aggregated the methylation levels of each probe by their median values across all of the samples with that condition in order to generate a reference methylome for that syndrome. The aggregated values were then used in a hierarchical clustering analysis to generate a dendrogram (Ward’s method on Euclidean distance). The episignatures clustering together in major branches of the dendrogram were further analyzed using a t-distributed stochastic neighbor embedding (t-SNE) analysis to visualize their degree of overlap and distinction. The analysis was performed using the Rtsne package according to the default parameters in order to reduce the dimensions of the data to two.34 The default perplexity parameter in the package (perplexity = 30) was used. For clusters with very small sample sizes, however, the perplexity parameter was reduced to the smallest value possible.
Construction of a Classification Algorithm for All of the Episignatures
Concurrent classification of individuals using multiple signatures can become a challenging task, potentially yielding inaccurate results as the data heterogeneity and the number of classes increase. We have previously demonstrated that support vector machines (SVMs), a class of supervised large margin classifiers, can provide enough power for differentiating disease groups from the healthy controls through the use of DNA methylation data, and that its performance remains acceptable given the small number of samples in rare syndromes (as few as five in some instances) and the relatively large number of predictors.12,13 Inherently, however, SVMs are binary classifiers, and their use for multiclass classification requires several modifications. The most common solutions for multiclass SVMs include one-against-one and one-against-all methods. In our previous studies, we have successfully implemented the one-against-one method for up to 16 classes13 in which every class is compared one by one with all other classes. Therefore, for n classes, this method will construct n × (n−1)/2 individual binary classifiers, and the final classification is made through a consensus reached by all of them. This approach can become challenging and impractical when the number of classes and predictors increases. For example, for 40 classes, 780 individual classifiers are needed, and this demands a great computational power. In addition, classes with a smaller number of samples or a milder DNA methylation change will yield less confident classifications. As the cumulative number of the predictors (probes) increases, the signal provided by such samples becomes diluted, and the classifications become less accurate. In these scenarios, the confidence scores generated for various disorders will be highly variable, making the one-against-one SVM less optimal for use in the clinical setting and diagnostic decision making. Therefore, in this study, we attempted to use the one-against-all SVM. For n classes, this method generates n−1 individual binary classifiers, each trained to distinguish the members of one class from the combined members of all of the remaining diseases and controls. This method significantly reduced the computational time and made it feasible to scale it up to a large number of classes.
The training of each SVM classifier was performed with a linear kernel using the e1071 R package. The training was only performed on the training data subset. To determine the best hyperparameter to be used in linear SVM (cost), and to measure the accuracy of the models, 10-fold cross-validation was performed during the training of each classifier. In this process, the training set was randomly divided into ten folds. Nine folds were used for training the model and one fold was used for testing. After we repeated this iteration for all of the ten folds, we calculated the mean accuracy and selected the hyperparameters with the most optimal performance. For every sample, the models were set to generate a score ranging between 0 and 1, representing the confidence of prediction for the specific class the SVM was trained to detect. Conversion of SVM decision values to these scores was carried out according to the Platt’s scaling method.35 A classification as one of the disorders was made when a sample received the greatest score for that class, a score that also needed to be greater than 0.5. The final models were applied to the training dataset in order to ensure the success of the training.
We ensured that the constructed models were not sensitive to the experimental batch structure of the methylation data by applying this structure to all of the samples assayed on the same batch that cases in the training dataset were drawn from. To confirm that the classifiers were not sensitive to the blood cell type compositions, we used methylation data from isolated blood cell populations of healthy individuals36 and supplied them to our models for prediction in order to examine the degree to which the resulting scores were varied across different blood cell types. Next, the models were applied to the testing cohort (25% subset of the affected cases not used for feature selection or training) in order to evaluate the predictive ability of the models on affected subjects. To determine the specificity of the models, we supplied a large number of DNA methylation arrays from healthy subjects. To understand whether the models were sensitive to other congenital disorders, we tested a large number of subjects with clinical and molecular diagnoses of such syndromes confirmed by the models.
Screening of Undiagnosed Subjects and Classification of Uncertain Cases
The final algorithm was used to classify subjects suspected of having any of the conditions used in the training, including those with no sequence variant information available, with inconclusive clinical assessment, or with DNA sequence variants of unknown significance (VUS). In addition, we used the algorithm to screen among a large group of individuals with various presentations of developmental delays and congenital anomalies but who had no established diagnosis despite routine clinical and molecular assessments including microarray copy-number variant (CNV) testing or exome sequencing. The subjects who were predicted to have the syndromes above were evaluated based on the available clinical and molecular information.
Data Availability
Some of the datasets used in this study are available publically and may be obtained from gene expression omnibus (GEO) using the following accession numbers. GEO: GSE116992, GSE66552, GSE74432, GSE97362, GSE116300, GSE95040, GSE104451, GSE125367, GSE55491, GSE108423, GSE116300, GSE89353, GSE52588, GSE42861, GSE85210, GSE87571, GSE87648, GSE99863, and GSE35069. These include DNA methylation data from patients with Kabuki syndrome, Sotos syndrome, CHARGE syndrome, immunodeficiency-centromeric instability-facial anomalies (ICF) syndrome, Williams syndrome, Chr7q11.23 duplication syndrome, Silver Russell syndrome, BAFopathies, Down syndrome, a large cohort of unresolved subjects with developmental delays and congenital abnormalities, and also several large cohorts of DNA methylation data from the general population. The rest of the data are not available due to the restrictions of the ethics approval.
Ethics Statement
The study protocol has been approved by the Western University Research Ethics Board (REB 106302) and the McMaster University Hamilton Integrated Research Ethics Boards (REB 13-653-T). Where applicable, participants provided informed consent prior to sample collection. All of the samples and records were de-identified before any experimental or analytical procedures were performed. The research was conducted in accordance with all relevant ethical regulations.
Results
Assessment of DNA Methylation Signatures in 42 Congenital Disorders
This study included peripheral blood DNA samples from a total of 787 subjects affected by 42 syndromes and their various subtypes. The syndrome names, their abbreviations, associated genes, OMIM identifiers, and the sample sizes are summarized in Table 1. Following genome-wide DNA methylation analysis using Infinium arrays and quality controls, ~400,000 probes passed detection quality filters in at least 95% of the samples, and these probes were used for subsequent analysis. Through the comparison of the training subset (Table 1 and Table S1) with age- and sex-matched samples selected from a pool of healthy controls (n = 749) for every condition, we prioritized between 100 and150 probes for each of their respective DNA methylation signatures. Of the conditions tested, eight did not have evidence of a reliable and replicable DNA methylation signature and were excluded from further assessment (Table 1), reducing the total training and testing cohort sample sizes to 540 and 152, respectively (Table 1 and Table S1), and limiting the total number of selected probes to 3,643 (Tables S2 and S3). The extent of DNA methylation changes varied across different conditions; Sotos syndrome; ICF syndrome; Tatton-Brown-Rahman syndrome (TBRS); mental retardation, X-linked syndromic, nascimento-type (MRXSN) syndrome; and autosomal dominant cerebellar ataxia, deafness, and narcolepsy (ADCADN) showed the most robust methylation changes (methylation differences of up to 60% between the cases and controls). BAFopathies, Cornelia de Lange syndrome (CdLS), Rubinstein-Taybi syndrome (RSTS), and mental retardation X-linked 97 (MRX97) presented some of the mildest DNA methylation patterns (with maximum DNA methylation difference between the cases and controls not greater than 20%).
As a general trend, we observed that different subtypes of the syndromes that result from multiple gene defects have highly similar DNA methylation profiles. This was found in Kabuki syndrome (Kabuki 1 and Kabuki 2), BAFopathies (CSS1, CSS2, CSS3, CSS4, and NCBRS), Cornelia de Lange syndrome (CdLS1, CdLS2, CdLS3, and CdLS4), and RSTS (RSTS1 and RSTS2), in which probes selected in each subtype generated a similar pattern in the other subtypes (Figure 1). Therefore, multiple subtypes of each of these syndromes were treated as a single entity in further analyses. The only exception to this rule was found for ICF syndrome. Despite a very robust shared DNA methylation pattern in the four ICF subtypes, it was observed that ICF1 could be fully distinguished from the other three ICF subtypes (ICF2, ICF 3, and ICF 4),19 and thus ICF syndrome type 1 and types 2–4 were treated as two separate episignature entities for the remainder of the study. One other exception was noted in CdLS5, resulting from mutations in HDAC8 for which no DNA methylation changes were observed, likely due to skewed inactivation of the mutated chromosome X in the peripheral blood of these individuals (all were females in our cohort).37
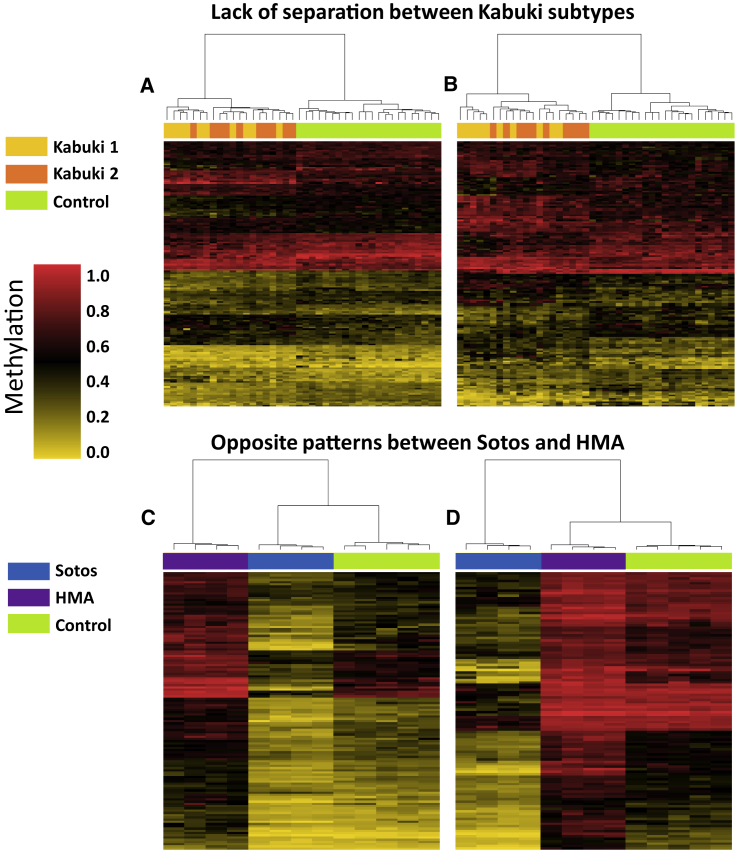
Figure 1.
Relationships across Various Syndromes and Their Subtypes
The plot shows clustering analysis with heatmap using probes specific to the DNA methylation of one syndrome (or its subtype) as compared with another. Rows indicate probes and columns indicate samples. The top pane colors indicate the classes. The heatmap color scale from gold to red represents the level of methylation from 0–1.
(A) Probes differentially methylated in Kabuki 1 (KMT2D) and controls do not provide distinction between subjects with Kabuki 1 and Kabuki 2 (KDM6A), although they differentiate both of them from the controls.
(B) The same pattern is observed when Kabuki-2-specific probes are used.
(C) Probes differentially methylated between individuals with Hunter McAlpine syndrome (HMA) (harboring duplication of NSD1) and controls generate a hypermethylation pattern in the HMA individuals. The same probes generate a mirror hypomethylation pattern in individuals with Sotos syndrome (loss of function of NSD1).
(D) The same mirror effect is observed when probes selected for Sotos syndrome are used.
Each of the genes studied here was found to be associated with a single DNA methylation signature with the exceptions of ADNP and KAT6B. KAT6B mutations result in two syndromes, genitopatellar syndrome (GTPTS) and Say-Barber-Biesecker-Young-Simpson syndrome (SBBYSS), each harboring a distinct DNA methylation signature. The patterns in GTPTS were found to be more robust than, and independent from, what was found in SBBYSS. ADNP was the only example of a syndrome caused by mutations in one gene but with two distinct DNA methylation signatures. The two signatures were distinguished by the mutation coordinates within ADNP: subjects who harbored variants within the central domain of c.2000–2340 (ADNP central–ADNP_C) showed a distinct pattern from those whose mutations resided in the regions outside c.2000–2340 (ADNP terminal–ADNP_T). These two groups were also treated as separate categories throughout the study, yielding a total of 34 episignatures in this manuscript.
In addition to the affected subjects, this study also includes apparently healthy individuals carrying pathogenic mutations in four genes: KDM5C (X-linked recessive, 14 obligate female carriers), KMT5B (autosomal dominant with incomplete penetrance, two healthy carriers), BRWD3 (X-linked recessive, one obligate female carrier), and UBE2A (X-linked recessive, six obligate female carriers). The key observation here was that healthy carriers may also present episignatures. The female KDM5C mutation carriers showed an intermediate pattern between the affected males and controls. Half of the KDM5C protein in the carrier females originates from the wild-type allele (KDM5C is not subject to X-linked inactivation). The single female carrier of a BRWD3 mutation also showed an intermediate methylation pattern between the affected males and controls. Despite an incomplete penetrance, the two healthy individuals with heterozygous KMT5B mutations demonstrated a methylation pattern similar to those of the affected cases (also heterozygous). The obligate female carriers of UBE2A, however, did not show any methylation changes, possibly due to skewed X chromosome inactivation.38
Relationship between Different DNA Methylation Signatures
The number of probes co-occurring at the episignatures of any two conditions was very small (<5%, Figure S1). However, pairwise analysis of the methylation patterns showed evidence of a relationship between some of them. We first evaluated syndromes arising from alternative dosage in shared genetic loci (i.e., loss of function versus gain of function). Two examples of such conditions in our cohort were Sotos syndrome versus Hunter McAlpine (HMA) syndrome (NSD1 loss of function versus NSD1 duplication, respectively) and Williams syndrome versus Chr7q11.23 duplication syndrome (Chr7q11.23 deletion versus duplication). In both sets of these pairs, symmetrical DNA methylation patterns were observed. This phenomenon was particularly striking in Sotos syndrome versus HMA syndrome; the former is drastically hypomethylated while the latter is distinctly hypermethylated (Figure 1). Another such example was noted in a single subject with duplication of ARID1A, which showed a mirrored DNA methylation pattern of all other BAFopathies resulting from loss-of-function mutations in the BAF complex genes including ARID1A. A common observation that was found in the pairwise comparisons of all syndromes was that in syndromes with extensive methylation changes, the patterns do not remain restricted to the probes selected for those conditions, and they also occur in probes specific to others. However, in any pairwise comparison, the probes from one syndrome alone may or may not fully distinguish the two syndromes from each other. Two examples of this phenomenon, one for a fully distinguishable pair (alpha-thalassemia mental retardation syndrome [ATRX] and Sotos syndrome) and one for a poorly distinguishable pair (Kabuki syndrome and BAFopathies) are illustrated in Figure 2. To systematically evaluate the relationship across all of the episignatures, we combined all of the identified probes and performed a clustering analysis to demonstrate the hierarchical order, as well as the similarities among various conditions, based on their DNA methylation profiles (Figure 3). The analysis generated two main clusters. The first was composed of syndromes with hypomethylation as the main pattern, including Sotos syndrome, ICF syndrome, Rahman syndrome (RMNS), Borjeson-Forssman-Lehmann syndrome (BFLS), and TBRS, which are also clinically related to each other in that growth abnormalities are major features they share. The other branch was subdivided into three subclusters. The smallest of these subclusters was composed of three syndromes: HMA syndrome, MRXSN syndrome, and SETD1B-related disorder, all with predominantly hypermethylated profiles, clustering together in the greatest distance from the hypomethylated episignatures. The other two clusters were composed of syndromes with mild-to-moderate DNA methylation patterns. Some of these, such as ATRX/ADNP_T or RSTS/CdLS generated pairs at the terminal branches of the dendrogram, indicating their high level of similarity. Of interest, the pair of Kabuki/BAFopathy, which was discussed earlier, clustered very close to each other. BAFopathies, specifically, had the most similar DNA methylation pattern to controls, clustering with them in a single branch. We projected the combined DNA methylation data of all of the probes from samples belonging to the major clusters identified here into three two-dimensional plots (Figure 4). This analysis indicated that despite similarities across some of these episignatures, they remain relatively distinct from each other when all of the selected probes are taken into account.
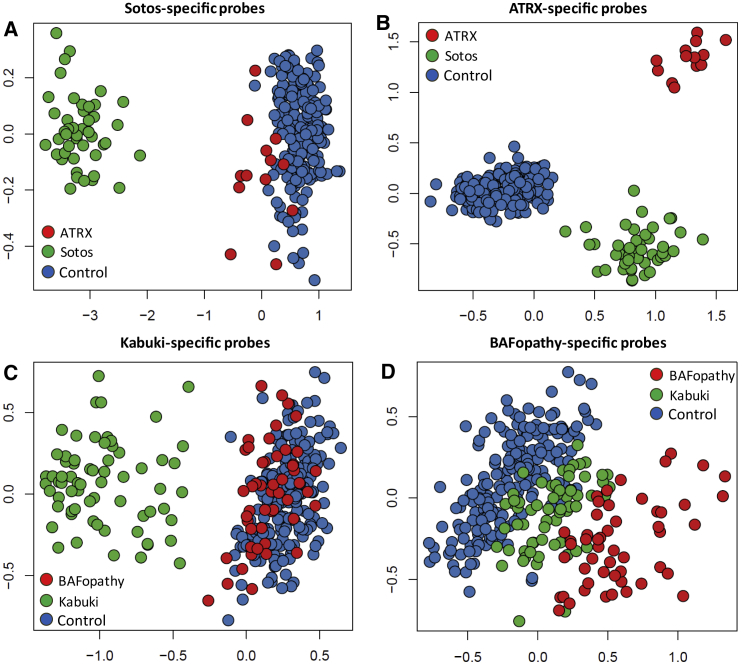
Figure 2.
DNA Methylation Episignature of One Syndrome in Others
The top two dimensions of multidimensional scaling plots (x axis = dim1, y axis = dim2) representing the pairwise distance across the samples with various episignatures:
(A) Sotos-syndrome-specific probes distinguish Sotos syndrome samples from controls, but they do not differentiate alpha-thalassemia mental retardation syndrome (ATRX) samples from the controls.
(B) ATRX-specific probes differentiate both Sotos syndrome and ATRX samples both from controls and from each other.
(C) Kabuki-syndrome-specific probes differentiate Kabuki syndrome samples from controls, but they do not distinguish the BAFopathy samples from controls.
(D) BAFopathy-specific probes generate an intermediate pattern for the Kabuki syndrome subjects between the BAFopathies and controls.
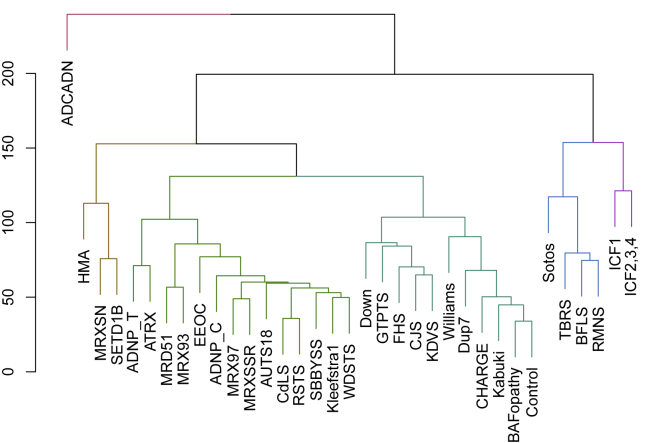
Figure 3.
Distance and Hierarchical Orders across 34 Episignatures
The dendrogram shows the distance and hierarchical orders of 34 episignatures. The y axis is the measure of distance or dissimilarity of either individual data points or clusters. The vertical position of the split in the dendrogram indicates the distance between every two points or clusters. The major splits are shown in different colors. Syndromes with very strong hypomethylation patterns are clustered together on the right, whereas those with hypermethylation episignatures are placed in a great distance to those in the left. As seen, BAFopathies are the most similar episignature to the controls, being consistent with their very mild DNA methylation changes.
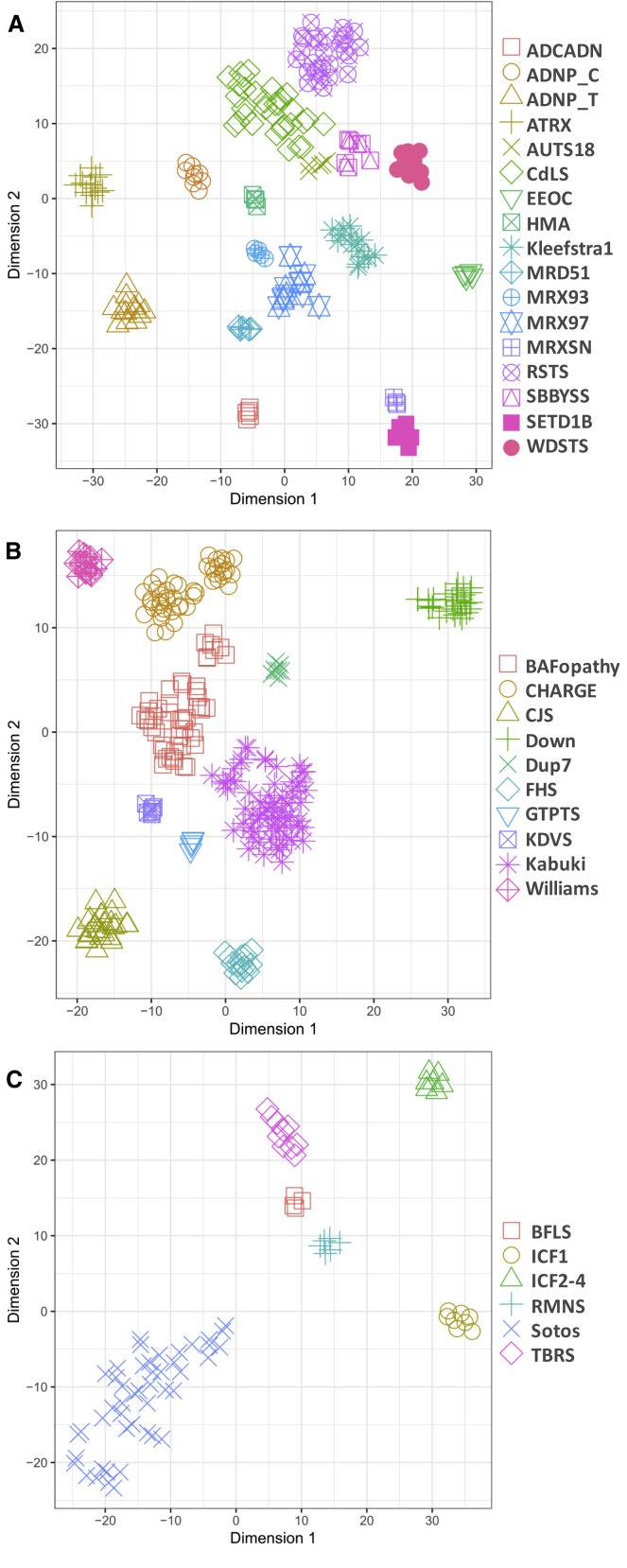
Figure 4.
Dimensionality Reduction of DNA Methylation Data from 34 Peripheral Blood DNA Methylation Episignatures
The members of the three major clusters identified in the dendrogram in Figure 3 were projected in three separate two-dimensional plots (A–C) using a t-distributed stochastic neighbor embedding (t-SNE). Despite similarities observed across some, the use of all of the probes from all episignatures together provides enough distinctions between them. A small subgrouping is observed for BAFopathies and CHARGE syndrome. This observation is not explained by the genes involved, mutation coordinates, mutation type, clinical presentations, age, or sex. It is also not replicated when probes specific to each of these conditions are used for this analysis.
Challenge of Disease Classification Using 34 Episignatures
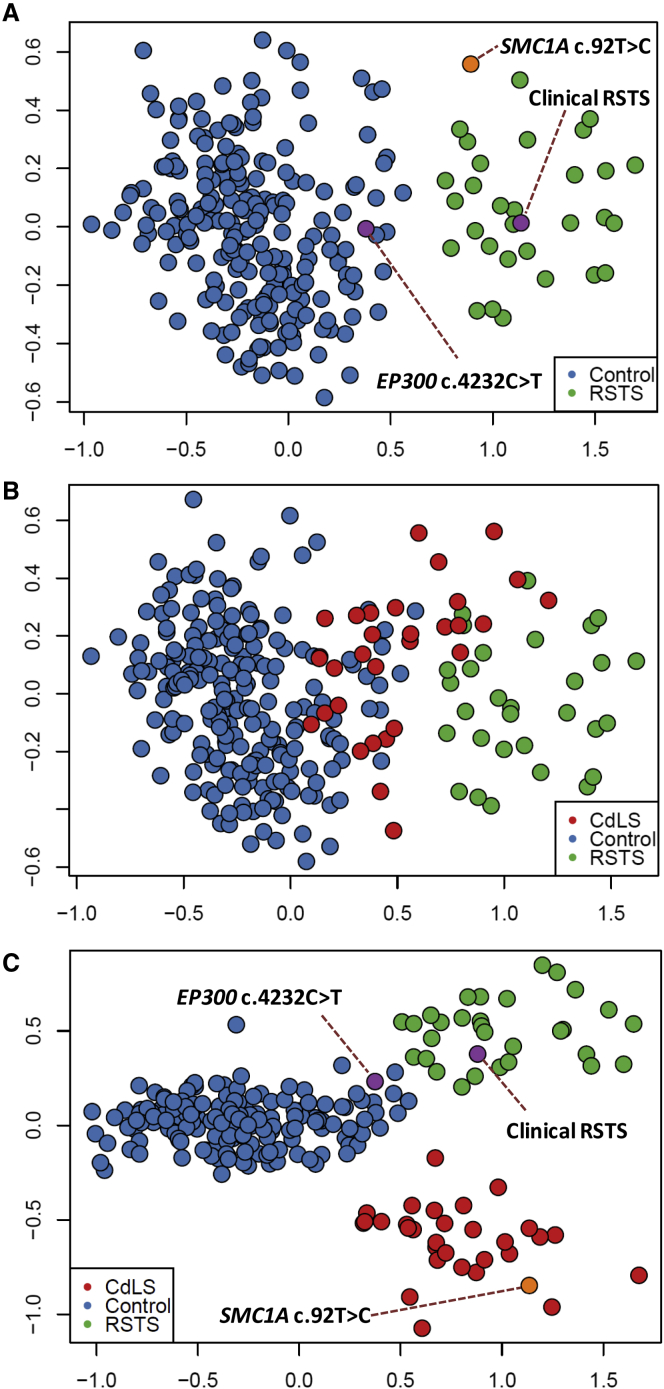
Binary classification of disease versus control using one episignature at a time is the most commonly used approach for determining if an individual is affected by a syndrome. Recognizing the considerable similarities among some of the 34 episignatures described in this study, we attempted to establish the accuracy of this approach. We examined the syndromes that are most closely related to each other as determined by using the dendrogram in Figure 3. Among these, we found several pairs, including RMNS/BFLS, MRXSN/SETD1B, Kabuki/BAFopathy, and RSTS/CdLS, for which an effort at classification using the episignature of only one pair was not always successful. An example of the workflow and the challenge is illustrated in Figure 5. The probes specific to RSTS generate a clear separation between the RSTS subjects and controls as demonstrated through the use of multidimensional scaling (Figure 5A). We added three subjects with uncertain diagnoses to this plot, one with a clinical diagnosis of RSTS but negative sequence finding in the RSTS-related genes, one with a de novo VUS in the RSTS2-related gene, EP300 (RefSeq accession number NM_001429.3, c.4232C>T; RefSeq NP_001420.2, p.Thr1411Ile), and the last subject with a rare variant in a CdLS-related gene, SMC1A (RefSeq NM_006306.2, c.92T>C; RefSeq NP_006297.2(LRG_773p2), p.Ile31Thr). Among the two RSTS-suspected subjects, the first one clustered with all confirmed RSTS cases, whereas the subject with EP300 VUS showed a pattern most similar to that of the controls (Figure 5A); this result indicates that the first individual was affected by RSTS, while the second was not. However, it was also noted that the subject with the SMC1A variant is situated closer to the RSTS subjects than to controls (Figure 5A). This raised the question of whether this latter subject was affected by RSTS or this classification was incorrect due to the overlapping nature of the RSTS and CdLS episignatures. To investigate this, we first added the known cases of CdLS to this analysis, which variably clustered with both controls and RSTS subjects (Figure 5B), confirming that the episignature of RSTS partially overlapped with that of CdLS. Repeating this analysis using probes from both episignatures, however, completely separated the two disorders from each other as well as from controls (Figure 5C). This analysis now clusters the subject with the SMC1A variant with other CdLS cases, indicating that the initial classification was not correct. This example indicates how attempts to classify disease by assessing one disorder at a time without the consideration of other episignatures can be error-prone.
Figure 5.
The Challenge of Disease Classification Using Closely Related Episignatures
The plot shows an attempt at disease classification of three subjects using DNA methylation data through unsupervised analysis.
(A) Multidimensional scaling of DNA methylation data from probes specific to RSTS episignature provides enough distinction between the Rubinstein-Taybi syndrome (RSTS) subjects and controls. The addition of two samples from individuals suspected to have RSTS (purple) clusters one of them with controls and the other with RSTS subjects. Another sample from an individual suspected to have Cornelia de Lange syndrome (CdLS; orange), however, is also situated closer to RSTS subjects than to controls.
(B) The addition of the CdLS samples to the analysis using the RSTS-specific probes demonstrates that these samples show an intermediate pattern between RSTS and controls.
(C) Incorporation of probes specific to CdLS in the analysis demonstrates that CdLS subjects are indeed distinct from RSTS cases. The uncertain sample from the individual suspected of having CdLS now clearly clusters with the other confirmed CdLS subjects.
Development of a Classification Algorithm for the Concurrent Detection of 34 Episignatures
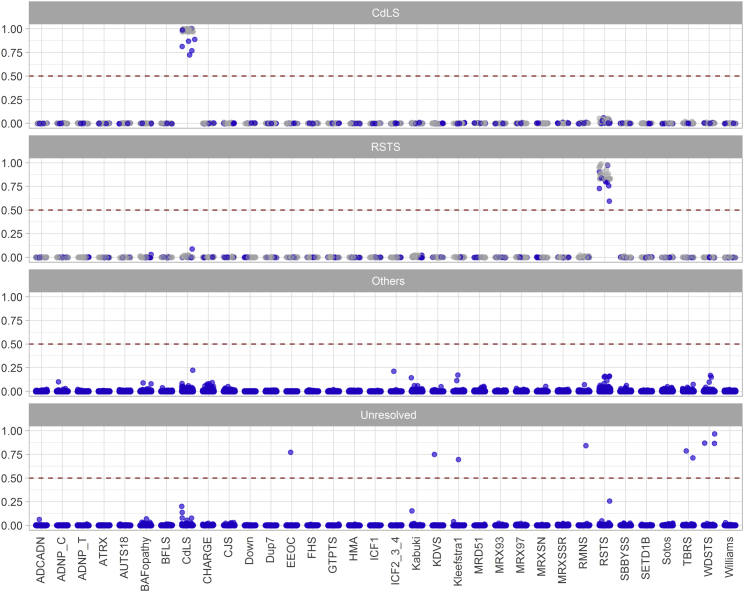
Concurrent assessment of multiple syndromes through the use of unsupervised analysis can become challenging and inaccurate when the number of classes increases to the scale presented in this manuscript. A supervised analysis may provide a more robust solution in these situations. We developed 34 individual SVM classifiers for the episignatures in this study, each trained to distinguish one disease class from the controls and also from the other 33 episignatures. The models were set to generate 34 scores ranging from 0–1, with higher scores representing a greater chance for any given subject of having a DNA methylation profile similar to each of the episignatures, respectively. The training was performed on the training cohort, during which 10-fold cross-validation was performed, resulting in an average accuracy of 99.9%. To control for the success of the procedure, the entire training cohort was supplied to the final models, which assigned correct classifications to all of the cases and controls used for training. Every sample was correctly classified into the category it belonged to, obtaining scores significantly greater from the other classes (Figure 6). We also confirmed that the classifiers were not sensitive to the batch structure of the data. To do this, we applied the classifiers to other samples processed in the same batch as the cases. All of these other samples received very low scores for all of the 34 classes. Additionally, we evaluated the extent to which the variation in blood cell type compositions influenced the scores. We did this by applying the classifiers to a total of 60 methylation array data files from six healthy individuals, each being assayed separately for whole blood, peripheral blood mononuclear cells, and granulocytes, as well as for seven isolated cell populations (CD4+ T, CD8+ T, CD56+ NK, CD19+ B, and CD14+ monocytes, neutrophils, and eosinophils). All of these samples received very low scores (<0.05) for all of the 34 classes and showed <5% average inter-cell-type variability in the scores.
Figure 6.
A Multiclass Classification Algorithm for Concurrent Classification of 34 Episignatures
Concurrent classification of the 34 episignatures is performed using 34 individual support vector machine (SVM) classifiers trained to distinguish each episignature from all others and from the methylation profile of the controls. For any given subject, each of which is represented with a point here, 34 models will generate 34 scores between 0 and 1 (y axis) representing the chance that the subject has a methylation profile similar to each of the 34 episignatures (x axis). The default cutoff of 0.5 is used for determining the class. However, most samples received scores close to 0 or 1, and thus for visualization, the points are jittered. Gray represents samples used in the training, and blue indicates those that were not used for training. The top two panels illustrate samples from the training and testing dataset with Cornelia de Lange syndrome (CdLS) and Rubinstein-Taybi syndrome (RSTS). These two categories were selected as examples among the 34 categories due to the challenge presented earlier in their unsupervised classification (Figure 5). As seen, each sample has received high scores only for the episignature it is supposed to have, and very low scores for all others. Samples with RSTS and CdLS have not been classified as one another. The third panel shows a trial performed for a large cohort of individuals from the general population (n = 2,315) as well as those with other developmental disorders not in the list of our episignatures (n = 442), all of which are scored close to zero. The final panel illustrates a cohort of unresolved subjects (n = 965) with various congenital anomalies among which a total of nine have been classified as potential cases of some of the syndromes in the study.
We next applied the model to the entire testing cohort, which was composed of 152 samples that were not used for feature selection or model training. All of these samples were assigned the expected class with scores similar to those of the training dataset; these results confirm that the models were robust in disease classification (Figure 6). To measure the specificity of our classifier, we tested whole blood methylation data from a total of 2,315 healthy subjects of various ethnic backgrounds (aged 0–94); all of this data received very low scores for all of the 34 episignatures (Figure 6). We also questioned whether the model could differentiate the above syndromes from other congenital or Mendelian disorders not included in the training cohort. The DNA methylation profiles of a total of 442 subjects, diagnosed with these types of syndromic conditions, were supplied to the algorithm for classification; and all of these profiles scored very low for all of the 34 categories (Figure 6), further confirming the specificity of our algorithm.
Screening of Unresolved Cohort and Classification of Uncertain Cases
We have previously demonstrated that individuals with neurodevelopmental syndromes lacking a diagnosis may be identified and diagnosed through screening of their DNA methylation profiles. Here, we tested two previously described cohorts of such individuals13 who have various developmental disorders and who have remained unresolved following the routine clinical assessments. This included 965 subjects, the majority of whom had undergone CNV microarray testing as part of the standard clinical workup, along with additional genetic testing in some cases, including targeted gene/panel or exome sequencing. These individuals had various forms of neurodevelopmental delays and congenital anomalies, including facial dysmorphism, developmental delay and/or intellectual disability, degenerative neural disease, autism, and various congenital organ defects, though none were suspected to have any of the syndromes described in this study. Applying our classifier to this cohort allowed the identification of nine subjects matching some of the newly described episignatures. This included the detection of three subjects with Wiedemann-Steiner syndrome (WDSTS), two subjects with TBRS, and four others with Kleefstra syndrome, RMNS, Koolen-de Vries syndrome (KDVS), and Epileptic encephalopathy, childhood-onset (EEOC), respectively (Figure 6 and Table S4). Most of these individuals were not available for further assessment; however, their reported clinical features were consistent with their predicted syndromes (Table S4). These features included macrosomia and macrocephaly in a TBRS-predicted individual, myoclonic seizures and behavioral problems in an EEOC-predicted individual, and speech problem with a bicuspid aortic valve in a KDVS-predicted individual. The subject presenting with the methylation profile of Kleefstra syndrome was initially reported to have an Angelman-like phenotype. An ultimate diagnosis for many such cases is Kleefstra syndrome.39 Of interest, the subsequent DNA sequencing identified a heterozygous splice site variant in EHMT1 (RefSeq NM_024757.4, c.3540+1G>C; RefSeq NP_079033.4, p.?), confirming our prediction. Another subject in this study who had a methylation profile similar to RMNS was the second case whose prediction was confirmed through sequencing. He was a two-year-old male presenting with developmental delay, hypotonia, abnormal brain MRI findings (ventriculomegaly), and cryptorchidism. The RMNS phenotype is highly variable and these findings can be observed in numerous syndromes. The genome sequencing, however, identified a de novo frameshift variant in HIST1H1E (RefSeq NM_005321.2, c.436_458del, RefSeq NP_005312.1, p.Thr146AspfsTer42), confirming the diagnosis of RMNS and the sensitivity of DNA methylation testing for screening of unresolved subjects.
In addition to these cases, we ascertained nine subjects with sequence variants of uncertain significance in six genes (CHD2, CREBBP, EHMT1, KDM6A, KMT2A, and PHF6). With the exception of one individual who had a missense variant in KDM6A (RefSeq NM_021140.2, c.871G>A, RefSeq NP_066963.2, p.Gly291Arg) and who represented the DNA methylation profile of Kabuki syndrome, all of the others were deemed to be negative for all of the episignatures described in this study (Table S5), providing strong functional evidence to rule out these provisional diagnoses (Table S5).
Discussion
Over the past decade, efforts have been made to improve the diagnostic yield of genetic disorders through means other than traditional sequence variant assessments. DNA methylation signatures have gained special interest through these endeavors, and their assessment in many syndromes has led to positive and reliable findings.11,40 Compared to the last year, the current study has nearly doubled the number of conditions that can effectively be diagnosed through DNA methylation testing.13 Meanwhile, besides improvements in screening and diagnosis, several repeating patterns are beginning to emerge with regards to DNA methylation signatures in these genetic syndromes.
After the analysis of 42 syndromes, it can be concluded that specific peripheral blood episignatures are to be found in the majority of individuals with congenital syndromes. The small portion of syndromes with negative findings may have very mild DNA methylation changes, or they may only represent episignatures in certain specific tissues, or in genetic loci not covered by the Infinium arrays. Some such syndromes with mild changes will likely eventually lead to positive findings through reassessment of larger cohorts, as has occurred for BAFopathies in which our primary analysis was negative.12 Yet which genetic syndromes are associated with DNA methylation changes remains unpredictable. We do not see a conclusive and consistent relationship between the gene function or clinical features and the presence of an episignature. There are several observations that might be worth consideration. Whenever the gene function involves a direct regulation of the methylation marks, an extensive level of changes in the methylome may be expected. Examples include DNMT1, DNMT3A, and DNMT3B, which encode various DNA methyltransferases41 and which are associated with very strong DNA methylation patterns. The observed patterns in these cases are also consistent with their immediate functionality, including a strong hypomethylation seen in our study in those with DNMT3A and DNMT3B defects. In other conditions, the changes most likely result from downstream pathways.10,13,42 The evidence for this comes from the general trend observed among multiple genetically heterogeneous conditions in which various gene defects result in similar episignatures. Most encoded genes of interest are part of a multi-protein complex or are key members of a single regulatory pathway. DNMT3B (ICF1), the only exception to this rule (distinguishable from ICF2–4), is involved in de novo methylation,43 a functionality that is absent in other ICF-related genes. Other interesting observations noted on several occasions throughout the analysis of these syndromes include a linear relationship between the defective protein dosage and the intensity of methylation changes, as well as the symmetrical patterns seen in protein loss versus gain. In all of these scenarios, the presence of one defective allele in the absence of clinical presentations was enough for the detection of DNA methylation changes. Similarly, it was noted that among X-linked disorders, a skewed X-inactivation may be the cause for concealing an episignature, as noted in CdLS5, which did not show any methylation profile. Of note, multiple reports have documented a skewed inactivation of the X chromosome harboring the mutated HDAC8 allele in the peripheral blood (but not some other tissues) of individuals with CdLS5.37 All of our CdLS5 subjects were females. Due to lack of X inactivation, male CdLS5 subjects might present a methylation pattern similar to those of subjects with other CdLS subtypes; this remains to be studied. These findings will undoubtedly pose more questions than answers regarding the underlying mechanisms of incomplete penetrance in Mendelian disorders. However, they do provide great potential for carrier screening and confirmation of DNA sequence variant pathogenicity in healthy carrier individuals with affected offspring.
While the biological interpretation of peripheral blood episgnatures in congenital disorders remains a daunting task requiring further experiments and study, their clinical diagnostic utility is obvious. We have previously demonstrated the use of episignatures for the classification of subjects with uncertain diagnoses, as well as for screening of unresolved cohorts using a smaller number of conditions.13 The current study demonstrates that these utilities can be accurately implemented using the newly mapped episignatures, although new challenges were introduced during the process which were not present in the analysis of a single syndrome or a smaller number of syndromes. As a general trend, consistent with our previous observations, we have found that the episignatures remain independent of each other. In cases where the patterns were mild, however, there is a chance of misclassification of other syndromes with stronger signatures as the first episignature. This challenge will not be resolved unless the episignatures of both syndromes are evaluated together, or a supervised algorithm is trained to distinguish the second episignature. This is an important observation in this study; it indicates that the overlap can be a basis for uncertain or incorrect classifications and that using DNA methylation for disease classifications should be performed with simultaneous consideration of all of the mapped episignatures. Through the development of a supervised algorithm that considers the methylation patterns of all of the syndromes during classification, we have shown here that one can avoid the chance of misclassification due to the closeness of some of the episignatures. This approach will ensure that the addition of new episignatures for disease classification will remain a practical and evolving topic.
Clinical episignature analysis could prove to be an efficient and effective diagnostic tool as part of a typical first-visit assessment for complex cases presenting with ambiguous phenotypes. Combined with CNV microarray and sequence analysis, clinical episignature analysis may provide higher diagnostic yield in a more efficient manner than do current standards of care.44 In the last year, our assessment of a cohort of 965 unresolved individuals with congenital anomalies and developmental delays identified 15 individuals with potential diagnoses of 14 syndromes along with more than a dozen individuals with other locus-specific methylation defects such as imprinting disorders and trinucleotide repeat expansions.13 Assessment of the same cohort through the use of the newly discovered episignatures in the current study has added another nine individuals to this list, representing an increased diagnostic yield. The success in applying epigenomics to screening and disease classification in congenital syndromes is highly contingent upon the mapping of DNA methylation episignatures from a large database of syndromes. This growing field will likely tackle many of the challenges being faced today in medical genetics practice with regards to the diagnosis of congenital disorders.
These findings demonstrate that the field of clinical and genetic diagnosis of hereditary disorders is rapidly entering a new era, i.e., clinical epigenomics. With the growing scientific knowledge and expanding clinical utility of DNA methylation episignatures, it becomes more necessary to engage expert groups, medical and laboratory regulatory bodies, and professional colleges in the development of clinical and laboratory guidelines and recommendations for an appropriate use of this new post-genomics clinical testing modality.
Declaration of Interests
The authors declare no competing interests.
Acknowledgments
We thank the staff, molecular geneticists, and clinical geneticists in centers across France, USA, Italy, Australia, the Netherlands, Japan, and Canada for the identification, evaluation, and diagnosis of the individuals with neurodevelopmental conditions presented in this study. Special thanks to Andrea Venema, Ab Chudley, Cindy Curry, Alasdair Hunter, Yanagi K., and Kaname T. for involvement in the collection and processing of samples for some of the cohorts in this study. We also thank the families of the affected subjects for providing consent and information. The study was partially financially supported by the Amsterdam Reproduction and Development Institute. Dedicated to the memory of Ethan Francis Schwartz, 1996–1998.
Published: February 27, 2020
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2020.01.019.
Web Resources
EpiSign, https://www.ggc.org/episign and https://genomediagnostics.amsterdamumc.nl/ngs-info/
French Rare disease network filière AnDDI-Rare, http://anddi-rares.org
Gene Expression Omnibus, https://www.ncbi.nlm.nih.gov/geo/
Online Mendelian Inheritance in Man, https://www.omim.org
Supplemental Information
References
- 1.Yousefi P., Huen K., Davé V., Barcellos L., Eskenazi B., Holland N. Sex differences in DNA methylation assessed by 450 K BeadChip in newborns. BMC Genomics. 2015;16:911. doi: 10.1186/s12864-015-2034-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Martin-Herranz D.E., Aref-Eshghi E., Bonder M.J., Stubbs T.M., Choufani S., Weksberg R., Stegle O., Sadikovic B., Reik W., Thornton J.M. Screening for genes that accelerate the epigenetic aging clock in humans reveals a role for the H3K36 methyltransferase NSD1. Genome Biol. 2019;20:146. doi: 10.1186/s13059-019-1753-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aref-Eshghi E., Zhang Y., Liu M., Harper P.E., Martin G., Furey A., Green R., Sun G., Rahman P., Zhai G. Genome-wide DNA methylation study of hip and knee cartilage reveals embryonic organ and skeletal system morphogenesis as major pathways involved in osteoarthritis. BMC Musculoskelet. Disord. 2015;16:287. doi: 10.1186/s12891-015-0745-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aref-Eshghi E., Schenkel L.C., Ainsworth P., Lin H., Rodenhiser D.I., Cutz J.C., Sadikovic B. Genomic DNA methylation-derived algorithm enables accurate detection of malignant prostate tissues. Front. Oncol. 2018;8:100. doi: 10.3389/fonc.2018.00100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jones M.J., Goodman S.J., Kobor M.S. DNA methylation and healthy human aging. Aging Cell. 2015;14:924–932. doi: 10.1111/acel.12349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Robertson K.D. DNA methylation and human disease. Nat. Rev. Genet. 2005;6:597–610. doi: 10.1038/nrg1655. [DOI] [PubMed] [Google Scholar]
- 7.Velasco G., Francastel C. Genetics meets DNA methylation in rare diseases. Clin. Genet. 2019;95:210–220. doi: 10.1111/cge.13480. [DOI] [PubMed] [Google Scholar]
- 8.Guerra J.V., Oliveira-Santos J., Oliveira D.F., Leal G.F., Oliveira J.R.M., Costa S.S., Krepischi A.C., Vianna-Morgante A.M., Maschietto M. DNA methylation fingerprint of monozygotic twins and their singleton sibling with intellectual disability carrying a novel KDM5C mutation. Eur. J. Med. Genet. 2019 doi: 10.1016/j.ejmg.2019.103737. [DOI] [PubMed] [Google Scholar]
- 9.Schulze K.V., Bhatt A., Azamian M.S., Sundgren N.C., Zapata G.E., Hernandez P., Fox K., Kaiser J.R., Belmont J.W., Hanchard N.A. Aberrant DNA methylation as a diagnostic biomarker of diabetic embryopathy. Genet. Med. 2019;21:2453–2461. doi: 10.1038/s41436-019-0516-z. [DOI] [PubMed] [Google Scholar]
- 10.Bend E.G., Aref-Eshghi E., Everman D.B., Rogers R.C., Cathey S.S., Prijoles E.J., Lyons M.J., Davis H., Clarkson K., Gripp K.W. Gene domain-specific DNA methylation episignatures highlight distinct molecular entities of ADNP syndrome. Clin. Epigenetics. 2019;11:64. doi: 10.1186/s13148-019-0658-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sadikovic B., Aref-Eshghi E., Levy M.A., Rodenhiser D. DNA methylation signatures in mendelian developmental disorders as a diagnostic bridge between genotype and phenotype. Epigenomics. 2019;11:563–575. doi: 10.2217/epi-2018-0192. [DOI] [PubMed] [Google Scholar]
- 12.Aref-Eshghi E., Rodenhiser D.I., Schenkel L.C., Lin H., Skinner C., Ainsworth P., Paré G., Hood R.L., Bulman D.E., Kernohan K.D. Genomic DNA methylation signatures enable concurrent diagnosis and clinical genetic variant classification in neurodevelopmental syndromes. Am. J. Hum. Genet. 2018;102:156–174. doi: 10.1016/j.ajhg.2017.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aref-Eshghi E., Bend E.G., Colaiacovo S., Caudle M., Chakrabarti R., Napier M., Brick L., Brady L., Carere D.A., Levy M.A. Diagnostic utility of genome-wide DNA methylation testing in genetically unsolved individuals with suspected hereditary conditions. Am. J. Hum. Genet. 2019;104:685–700. doi: 10.1016/j.ajhg.2019.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aref-Eshghi E., Schenkel L.C., Lin H., Skinner C., Ainsworth P., Paré G., Siu V., Rodenhiser D., Schwartz C., Sadikovic B. Clinical validation of a genome-wide DNA methylation assay for molecular diagnosis of imprinting disorders. J. Mol. Diagn. 2017;19:848–856. doi: 10.1016/j.jmoldx.2017.07.002. [DOI] [PubMed] [Google Scholar]
- 15.Schenkel L.C., Aref-Eshghi E., Skinner C., Ainsworth P., Lin H., Paré G., Rodenhiser D.I., Schwartz C., Sadikovic B. Peripheral blood epi-signature of Claes-Jensen syndrome enables sensitive and specific identification of patients and healthy carriers with pathogenic mutations in KDM5C. Clin. Epigenetics. 2018;10:21. doi: 10.1186/s13148-018-0453-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aref-Eshghi E., Bourque D.K., Kerkhof J., Carere D.A., Ainsworth P., Sadikovic B., Armour C.M., Lin H. Genome-wide DNA methylation and RNA analyses enable reclassification of two variants of uncertain significance in a patient with clinical Kabuki syndrome. Hum. Mutat. 2019;40:1684–1689. doi: 10.1002/humu.23833. [DOI] [PubMed] [Google Scholar]
- 17.Aref-Eshghi E., Schenkel L.C., Lin H., Skinner C., Ainsworth P., Paré G., Rodenhiser D., Schwartz C., Sadikovic B. The defining DNA methylation signature of Kabuki syndrome enables functional assessment of genetic variants of unknown clinical significance. Epigenetics. 2017;12:923–933. doi: 10.1080/15592294.2017.1381807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aref-Eshghi E., Bend E.G., Hood R.L., Schenkel L.C., Carere D.A., Chakrabarti R., Nagamani S.C.S., Cheung S.W., Campeau P.M., Prasad C. BAFopathies’ DNA methylation epi-signatures demonstrate diagnostic utility and functional continuum of Coffin–Siris and Nicolaides–Baraitser syndromes. Nat. Commun. 2018;9:4885. doi: 10.1038/s41467-018-07193-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krzyzewska I.M., Maas S.M., Henneman P., Lip K.V.D., Venema A., Baranano K., Chassevent A., Aref-Eshghi E., van Essen A.J., Fukuda T. A genome-wide DNA methylation signature for SETD1B-related syndrome. Clin. Epigenetics. 2019;11:156. doi: 10.1186/s13148-019-0749-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ciolfi A., Aref-Eshghi E. Frameshift mutations at the C-terminus of HIST1H1E result in a specific DNA hypomethylation signature. Clin. Epigenetics. 2020;12:7. doi: 10.1186/s13148-019-0804-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Velasco G., Grillo G., Touleimat N., Ferry L., Ivkovic I., Ribierre F., Deleuze J.F., Chantalat S., Picard C., Francastel C. Comparative methylome analysis of ICF patients identifies heterochromatin loci that require ZBTB24, CDCA7 and HELLS for their methylated state. Hum. Mol. Genet. 2018;27:2409–2424. doi: 10.1093/hmg/ddy130. [DOI] [PubMed] [Google Scholar]
- 22.Bjornsson H.T. The Mendelian disorders of the epigenetic machinery. Genome Res. 2015;25:1473–1481. doi: 10.1101/gr.190629.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., Grody W.W., Hegde M., Lyon E., Spector E., ACMG Laboratory Quality Assurance Committee Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kular L., Liu Y., Ruhrmann S., Zheleznyakova G., Marabita F., Gomez-Cabrero D., James T., Ewing E., Lindén M., Górnikiewicz B. DNA methylation as a mediator of HLA-DRB1∗15:01 and a protective variant in multiple sclerosis. Nat. Commun. 2018;9:2397. doi: 10.1038/s41467-018-04732-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Su D., Wang X., Campbell M.R., Porter D.K., Pittman G.S., Bennett B.D., Wan M., Englert N.A., Crowl C.L., Gimple R.N. Distinct epigenetic effects of tobacco smoking in whole blood and among leukocyte subtypes. PLoS ONE. 2016;11:e0166486. doi: 10.1371/journal.pone.0166486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johansson A., Enroth S., Gyllensten U. Continuous aging of the human DNA methylome throughout the human lifespan. PLoS ONE. 2013;8:e67378. doi: 10.1371/journal.pone.0067378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Van Baak T.E., Coarfa C., Dugué P.A., Fiorito G., Laritsky E., Baker M.S., Kessler N.J., Dong J., Duryea J.D., Silver M.J. Epigenetic supersimilarity of monozygotic twin pairs. Genome Biol. 2018;19:2. doi: 10.1186/s13059-017-1374-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ventham N.T., Kennedy N.A., Adams A.T., Kalla R., Heath S., O’Leary K.R., Drummond H., Wilson D.C., Gut I.G., Nimmo E.R., Satsangi J., IBD BIOM consortium. IBD CHARACTER consortium Integrative epigenome-wide analysis demonstrates that DNA methylation may mediate genetic risk in inflammatory bowel disease. Nat. Commun. 2016;7:13507. doi: 10.1038/ncomms13507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barbosa M., Joshi R.S., Garg P., Martin-Trujillo A., Patel N., Jadhav B., Watson C.T., Gibson W., Chetnik K., Tessereau C. Identification of rare de novo epigenetic variations in congenital disorders. Nat. Commun. 2018;9:2064. doi: 10.1038/s41467-018-04540-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aryee M.J., Jaffe A.E., Corrada-Bravo H., Ladd-Acosta C., Feinberg A.P., Hansen K.D., Irizarry R.A. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics. 2014;30:1363–1369. doi: 10.1093/bioinformatics/btu049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Houseman E.A., Accomando W.P., Koestler D.C., Christensen B.C., Marsit C.J., Nelson H.H., Wiencke J.K., Kelsey K.T. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics. 2012;13:86. doi: 10.1186/1471-2105-13-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krzywinski M., Schein J., Birol I., Connors J., Gascoyne R., Horsman D., Jones S.J., Marra M.A. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maaten L.V.D., Hinton G. Visualizing data using t-SNE. J. Mach. Learn. Res. 2008;9:2579–2605. [Google Scholar]
- 35.Platt J.C. Probabilities for support vector machines. In: Smola A., Bartlett P., Schölkopf B., Schuurmans D., editors. Advances in large margin classifiers. MIT Press; Cambridge: 2000. pp. 61–74. [Google Scholar]
- 36.Reinius L.E., Acevedo N., Joerink M., Pershagen G., Dahlén S.E., Greco D., Söderhäll C., Scheynius A., Kere J. Differential DNA methylation in purified human blood cells: implications for cell lineage and studies on disease susceptibility. PLoS ONE. 2012;7:e41361. doi: 10.1371/journal.pone.0041361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Deardorff M.A., Bando M., Nakato R., Watrin E., Itoh T., Minamino M., Saitoh K., Komata M., Katou Y., Clark D. HDAC8 mutations in Cornelia de Lange syndrome affect the cohesin acetylation cycle. Nature. 2012;489:313–317. doi: 10.1038/nature11316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nascimento R.M., Otto P.A., de Brouwer A.P., Vianna-Morgante A.M. UBE2A, which encodes a ubiquitin-conjugating enzyme, is mutated in a novel X-linked mental retardation syndrome. Am. J. Hum. Genet. 2006;79:549–555. doi: 10.1086/507047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tan W.H., Bird L.M., Thibert R.L., Williams C.A. If not Angelman, what is it? A review of Angelman-like syndromes. Am. J. Med. Genet. A. 2014;164A:975–992. doi: 10.1002/ajmg.a.36416. [DOI] [PubMed] [Google Scholar]
- 40.Aygun D., Bjornsson H.T. Clinical epigenetics: a primer for the practitioner. Dev. Med. Child Neurol. 2020;62:192–200. doi: 10.1111/dmcn.14398. [DOI] [PubMed] [Google Scholar]
- 41.Aref-Eshghi E., Schenkel L.C., Carere D.A., Rodenhiser D.I., Sadikovic B. Epigenetics in Human Disease. 2018. Epigenomic Mechanisms of Human Developmental Disorders; pp. 837–859. [Google Scholar]
- 42.Krzyzewska I.M., Maas S.M., Henneman P., Lip K.V.D., Venema A., Baranano K., Chassevent A., Aref-Eshghi E., van Essen A.J., Fukuda T. A genome-wide DNA methylation signature for SETD1B-related syndrome. Clin. Epigenetics. 2019;1:156. doi: 10.1186/s13148-019-0749-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baubec T., Colombo D.F., Wirbelauer C., Schmidt J., Burger L., Krebs A.R., Akalin A., Schübeler D. Genomic profiling of DNA methyltransferases reveals a role for DNMT3B in genic methylation. Nature. 2015;7546:243–247. doi: 10.1038/nature14176. [DOI] [PubMed] [Google Scholar]
- 44.Godler D.E., Amor D.J. DNA methylation analysis for screening and diagnostic testing in neurodevelopmental disorders. Essays Biochem. 2019;63:785–795. doi: 10.1042/EBC20190056. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Some of the datasets used in this study are available publically and may be obtained from gene expression omnibus (GEO) using the following accession numbers. GEO: GSE116992, GSE66552, GSE74432, GSE97362, GSE116300, GSE95040, GSE104451, GSE125367, GSE55491, GSE108423, GSE116300, GSE89353, GSE52588, GSE42861, GSE85210, GSE87571, GSE87648, GSE99863, and GSE35069. These include DNA methylation data from patients with Kabuki syndrome, Sotos syndrome, CHARGE syndrome, immunodeficiency-centromeric instability-facial anomalies (ICF) syndrome, Williams syndrome, Chr7q11.23 duplication syndrome, Silver Russell syndrome, BAFopathies, Down syndrome, a large cohort of unresolved subjects with developmental delays and congenital abnormalities, and also several large cohorts of DNA methylation data from the general population. The rest of the data are not available due to the restrictions of the ethics approval.