Abstract
Organismal fitness is partly determined by how well the nutritional intake matches sex-specific metabolic requirements. Metabolism itself is underpinned by complex genomic interactions involving products from both nuclear and mitochondrial genomes. Products from these two genomes must coordinate how nutrients are extracted, used and recycled, processes vital for fuelling reproduction. Given the complicated nature of metabolism, it is not well understood how the functioning of these two genomes is modulated by nutrients. Here we use nutritional geometry techniques on Drosophila lines that only differ in their mtDNA, with the aim to understand if there is nutrient-dependent mitochondrial genetic variance for male reproduction. We first find genetic variance for diet consumption, indicating that flies are consuming different amounts of food to meet new physiological requirements. We then find an interaction between mtDNA and diet for fitness, suggesting that the mtDNA plays a role in modulating diet-dependent fitness. Our results enhance our basic understanding of nutritional health and our chimeric genomes.
Keywords: reproduction, mitochondria, nutrition, nutritional geometry
1. Introduction
A large determinant of organismal fitness is the acquisition of nutrients that fuel reproductive efforts [1]. In nature, species have to tailor their behaviour and physiology to maximize the metabolic and energetic functions underlying fitness, while working within the constraints of the resources available [2]. Beyond this external constraint, there are internal (genetic) constraints that modulate how nutrients are extracted, used and recycled; all of which have downstream fitness consequences. There are two main steps that influence nutrient metabolism. The first is the behavioural regulation that determines how much food is consumed [3]. This is based on environmental cues relating external diet quality/quantity and feedback about the animal's internal state that is provided by nutrient sensing pathways [4–6]. The second and potentially more important aspect is how nutrient composition shapes metabolic flux, with downstream effects on cellular processing, ultimately affecting fitness [7]. Both steps rely on a large number of genetically encoded elements and accordingly, there can be genetic variation in behavioural and metabolic processes. For instance, previous work has documented genetic variation in sex-specific nutritional requirements [8].
Importantly, however, genetic variation in the above study was restricted to the nuclear genome. Nuclear genes (nuDNA) are not the only genetic determinant of metabolic function. Genes encoded within the mitochondria (mtDNA) also play a major role in metabolism, signalling and its regulation [9]. We would therefore expect fitness to depend on the interaction between both genomes, and this supposition has been validated across several studies [10–12]. Despite the importance of mitochondrial function for metabolic regulation and efficiency, the effects of mitochondrial genetic variation on diet-dependent fitness remain under explored. Previous work in Drosophila has shown mtDNA-specific effects on mitochondrial physiology, with these effects being contingent on the diet that flies had been reared on [13]. More recently, studies have shown that changes in the mtDNA genome can also have diet-dependent effects on longevity [14,15]. What remains to be established is how changes in diet-dependent fitness are modified by the mtDNA genome. We can predict from these studies that changing the composition of dietary macronutrients (by changing environments and/or nutritional availability) can have drastic consequences on mitochondrial function by altering the production of mitochondrial metabolites and signalling molecules [16]. This process will ultimately have serious impacts on metabolic flux and, in turn, feedback into the evolutionary processes shaping mito-nuclear genotypes [17].
Here we aim to understand the effects of mtDNA variation and diet on male reproductive fitness in D. melanogaster. We use seven fly lines with an isogenic nuclear genome but each carrying a different naturally occurring mtDNA haplotype. We then apply nutritional geometry techniques to identify diets that maximize male fitness for a given line (see electronic supplementary material, methods for a brief summary of nutritional geometry principles). We recover previously described male-specific nutritional optima on carbohydrate-rich food when averaging across all lines; however, we find significant mitochondrial genetic variance underpinning optimal male nutrition. These results allude to complex genetic and nutritional interactions influencing life-history trait evolution.
2. Material and methods
(a). Drosophila stocks and maintenance
All flies were reared at 25°C and 50% humidity, on a 12:12 h light:dark cycle, 10 ml glass vials, on a cornmeal–molasses–agar medium (see electronic supplementary material, table S1 for recipe), with ad libitum live yeast added to each vial to promote female fecundity. For each line, flies were propagated by adult 4-day-old parents, with eggs laid kept at maximum 100 eggs.
For the experiment, we used seven Drosophila strains, all of which had the same isogenic nuclear background (w1118 – Bloomington Stock Center no. 5905) coupled to seven different mtDNA haplotypes from around the world [18]. These were w1118 (coevolved -WE), Barcelona (BAR); Dahomey (now called Benin) (DAH); Madang, Papua New Guinea (MAD); Mysore, India (MYS); Oregon, USA (ORE); Zimbabwe (ZIM).
(b). Synthetic diet and nutritional geometry
We used a modified liquid version of the synthetic diet described in Piper et al. [7], which is prepared entirely from synthetic components to enable precise control over nutritional value (see electronic supplementary material, table S1–S3). Four different diets were synthesized, which varied in the ratio of protein (P, individual amino acids) and carbohydrate (C, sucrose), while all other nutritional components were provided in fixed concentrations. Nutrient ratios used were [P:C] – 1:1, 1:2, 1:4 and 1:16, with the final concentration of each diet being 32.5 g l−1.
Groups of three virgin males from each line were collected and placed in vials that contained 0.8% agar and kept at 80% RH for 12 h to acclimatize to the vial. Following this period, all flies were supplied with one of the four artificial liquid diets using a 5 µl capillary tube. Feeding vials were changed daily during the 4-day feeding trial, and daily diet consumption was recorded. Diet consumption was summed across all days, to give one data point. Each tray contained five evaporation control vials which contained no flies.
(c). Non-competitive fertility
Following 4 days of feeding on experimental diets, non-competitive fertility was measured for all male flies. Females of the w1118 coevolved genotype were placed individually in vials containing standard yeast–molasses–cornmeal medium and left for 1 h to acclimatize. Following this period, a focal male was transferred to the vial directly from the feeding vial and left to mate with the tester female for 24 h. This timing was chosen to maximize the chance for mating to occur (96%) between the fly pair. While there is a chance that a double mating could have occurred, previous pilot experiments show the tester females to have a long refractory period. Focal males were then removed and discarded, and females were left to oviposit over two vials (48 h in the first vial and 48 h in the second). Total number of eclosing adult offspring 14 days following mating was counted and summed over both vials per female. Coevolved tester females were used in this experiment, as previous work has found mitochondrial genetic variance for female fitness components [10]. We therefore chose to keep the tester female genotype consistent across all treatments to avoid the female genotype influencing the male fitness response.
(d). Statistical analyses
We used a sequential model building approach [19] to determine if there was mitochondrial genetic variance for (i) total consumption of diets and (ii) diet-mediated fitness (for a full description of models and the nutritional geometry framework, see electronic supplementary material, methods). Models were fitted with maximum-likelihood and compared in a pairwise manner using parametric bootstrap analysis using the PBmodcomp function implemented in the package pbkrtest [20]. We ran an analysis of variance (ANOVA) with type III sums of squares on the full model in order to assess the significance of fixed terms in the model. We visualized nutritional landscapes based on untransformed data using non-parametric thin-plate splines implemented in the fields [21] package.
We used a permutation approach to assess to what degree fitness variation between mitochondrial lines is due to differences in diet consumption responses rather than the metabolic differences independent of consumption. This approach has been previously described [8] and is detailed in the electronic supplementary material, methods. The rationale is that if lines differ in fitness because they alter their consumption in line with the diet available and their physiological requirements, then breaking the association between consumption and line by permutation should result in lower mean predicted fitness than in the observed dataset.
3. Results
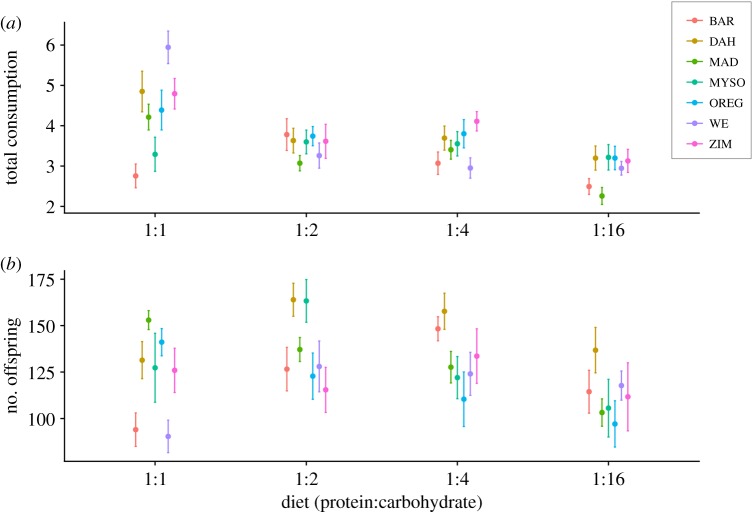
We find significant variation between the consumption of different diets, with protein-rich diets being consumed in larger quantities across all genotypes than carbohydrate-based diets (p < 0.05, figure 1). We also found significant mitochondrial genetic variation in consumption across diets (p = 0.048, 1.38% of variance in consumption, table 1A) and in diet-specific consumption (p < 0.001, 4.24% of variance, table 1A).
Figure 1.
(a) Total liquid consumption (µl) for the four different diets across all mitochondrial genotypes used in the experiment. (b) Total number of offspring sired for all mitochondrial genotypes used in this study across all diet treatments.
Table 1.
Full model of the nutritional effects on (A) diet consumption and (B) fitness. We include results from the parametric bootstrap model comparison, including the value of the test statistic (the log-likelihood ratio, LLR), degrees of freedom, p-value and percentage of the overall variance explained by each model, as well as the percentage of variance attributable to mitochondrial and diet-specific mitochondrial effects (Δ variance).
| A. diet consumption | |||||
|---|---|---|---|---|---|
| F | df | resid. df | p-value | ||
| (intercept) | 187.074 | 1 | 1.68 | 0.0102 | |
| diet | 18.482 | 3 | 356.16 | >0.001 | |
|
model comparison | |||||
| LLR | df | p-value | variance | Δ variance | |
| base model | 19.21% | – | |||
| mito | 2.461 | 1 | 0.04811 | 20.59% | 1.38% |
| diet-specific mito | 15.941 | 9 | 0.00101 | 24.83% | 4.24% |
| B. fitness | |||||
| F | df | Resid. df | p-value | ||
| (intercept) | 58.5258 | 1 | 4.27 | 0.0011 | |
| protein | 6.5259 | 1 | 357.46 | 0.01104 | |
| carbohydrate | 4.6687 | 1 | 354.85 | 0.0313 | |
| protein2 | 6.8662 | 1 | 355.42 | 0.0091 | |
| carbohydrate2 | 2.9447 | 1 | 352.75 | 0.0870 | |
| protein × carbohydrate | 0.0865 | 1 | 356.07 | 0.7688 | |
|
model comparison | |||||
| LLR | df | p-value | variance | Δ variance | |
| base model | 16.62% | – | |||
| mito | 9.8427 | 1 | 0.0018 | 20.29% | 3.67% |
| diet-specific mito | 9.7451 | 14 | 0.0495 | 26.40% | 6.11% |
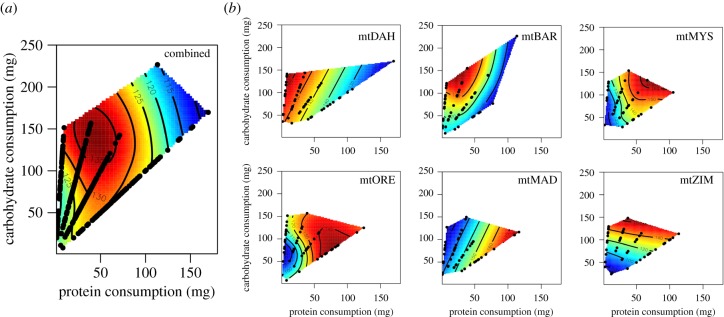
Analysing the relationship between diet and fitness, we recovered previous results whereby across all genotypes, male fitness is maximized by a moderate carbohydrate bias in the diet's macronutrient composition (P:C – 1:4, figure 2). However, we also found significant variation around this average (p = 0.0018, 3.67% of variance in fitness; table 1B, figure 2). Furthermore, there was evidence for genotypes showing differential fitness responses to diet variation (p = 0.0495, 6.11% of variance; table 1B, figure 2). For example, the fitness of male flies harbouring the DAH haplotype is maximized on a more carbohydrate-rich diet (P:C – 1:16, figure 2b), whereas ORE haplotype requires higher levels of protein to maximize fitness (P:C – 1:1, figure 2b). We further analyse these data using reaction norms (see electronic supplementary material) and find support for our nutritional geometry analysis.
Figure 2.
(a) Nutritional landscapes illustrating the effects of protein and carbohydrate intake on the expression of male traits. Black dots are individual data points. (b) Exemplary haplotype-specific nutritional fitness landscapes.
Using our permutation approach, we found that uncoupling behaviour (intake) and physiology tended to result in a reduction in fitness, but not statistically significantly so (p = 0.082). Thus, differences between genotypes in the behavioural responses to food composition might make some contribution to diet-dependent fitness (resulting in a tendency for reduced fitness when behaviour and physiology are dissociated), but genetic variation in fitness responses is dominated by the physiological and metabolic properties of the mitochondrial lines (resulting in a non-significant result).
4. Discussion
Nutrient acquisition and metabolism are important determinants of fitness components and phenotypic trait expression [22]. Genetic variation in fitness responses to nutrition are the result of two underlying processes. First, organisms with different genotypes can vary in how they change their behaviour and consume different amounts of food. Second, genotypes can differ in the functioning and efficiency of metabolism; a process critical for allocating resources to reproduction. Here we investigated this proposition in relation to the effects of mitochondrial genetic variation and nutrition on male fitness. By using the lines of D. melanogaster that couple diverse mtDNA haplotypes to the same isogenic nuclear background, we were able to isolate the effects of mitochondrial genetics on nutrient-dependent fitness. We applied nutritional geometry techniques across our mitochondrial panel and found evidence that different mtDNA lines require divergent nutrient compositions to maximize male fitness.
In line with previously described behavioural responses to holidic media [8], we found that flies consumed more of the protein than the carbohydrate diet. These results suggest that the diet-dependent modulation of consumption in our study was aimed at ensuring an adequate carbohydrate intake. In addition to these general responses, we found a small amount of genetic variation in total consumption (across diets) between the mtDNA haplotypes (table 1A, term ‘mito’), as well as more significant genetic variation for diet-specific feeding responses (term ‘diet-dependent mito’). We found a similar fitness responses across diets, with significant mitochondrial and diet-dependent mitochondrial genetic variance (table 1B). We also note that although our model explains only about a quarter of the variance in the measured responses—as expected for noisy traits like behaviour and male mating success—mitochondrial effects make a significant contribution to this figure (consumption: 5.62% of total variance, or 22.6% of the variance explained; fitness: 9.78% of total variance, 37% of variance explained).
Our results provide evidence that mitochondrial DNA influences male feeding behaviour and reproductive success, most likely due to their central role in metabolism and metabolic regulation. Consistent with previous work [8], the permutation analyses we performed suggest that genetic variation in fitness is more likely due to the physiological and metabolic properties of our mitochondrial genotypes than a consequence of altered feeding behaviour (non-significant permutation test). Nevertheless, the border-line p-value (p = 0.08) does not allow us to categorically completely rule out a contribution of mitochondria via the modulation of feeding.
The presence of diet-dependent effects of haplotypes on feeding and fitness reinforces the view that mitochondria are more than merely subordinate energy producers. They integrate metabolic flux and stress, signalling the physiological status of the cell to the nucleus [23,24]. Accordingly, changing the dietary composition (changing nutritional environments) will have a significant impact on the Krebs cycle intermediates and ultimately impact the metabolic flux balance of the cell [13,25]. Trying to pinpoint pathway(s) impacted by the complex interaction between diet and mito-nuclear genetics will require further experimentation. We can predict that Complex I is a very likely candidate to respond to both nutrition and mitochondrial effects as it is the starting point of OXPHOS and requires components encoded in both genomes. Moreover, many previous studies have linked this complex to many environmental responses [26–28].
Our finding of fitness variation among mitochondrial lines also supports the supposition that mtDNA variants may be a direct target of selection imposed by variation in dietary macronutrients [17]. Our study therefore contributes to a body of evidence suggesting that mtDNA is not just an ‘evolutionary bystander’ [29]. Indeed, empirical work by Aw et al. [27] has provided insights into mito-nuclear mechanisms that are affected by nutrition. Their study used similar Drosophila strains to our study; isogenic w1118 nuclear background coupled to haplotypes from Dahomey (DAH) and Australia (neither haplotype coevolved with the nuclear background). These authors performed cage experiments with populations composed from the two strains across several nutritional environments and found that the DAH haplotype increased in frequency on a carbohydrate-rich diet, but decreased on diets with higher protein content. Interestingly, in our study, we also found the DAH haplotype to perform best in a high carbohydrate environment. As frequency change in Aw et al.'s [27] experiment are due to performance differences in females (who transmit mitochondria), the consistent carbohydrate bias in the performance of DAH across their and our study suggests that the effects of the haplotype are similar in the two sexes.
While we use naturally occurring mtDNA haplotypes in our study, a caveat is that we only use a single nuclear background. As a consequence, we cannot differentiate between phenotypic effects that are due to mitochondrial haplotype alone, and those that arise from epistatic interaction between the haplotypes and the fixed nuclear background. Mossman et al. [30] have previously examined the role of interactions between mitochondrial genotype, nuclear genotype and diet (G × G × E) on development time in Drosophila. They used 12 nuclear backgrounds from the DGRP panel [31] coupled to a cross-species panel of six different mtDNA haplotypes (3 × D. melanogaster, 2 × D. simulans, 1 × D. mauritiana). They found significant G × G × E effects on development time; flies that developed on higher protein diets had shorter development times than those on higher carbohydrate foods, but the magnitude of this response depended on both the flies' mitochondrial and nuclear genotype. Nonetheless, authors did find mitochondrial genetic variance for development time. It remains to be seen whether mtDNA variation will alter the dietary response in natural populations which have high levels of both nuDNA and mtDNA genetic variance. Future work should aim to investigate the complex interaction between genomes and nutrition that drives life-history evolution in natural environments.
Supplementary Material
Supplementary Material
Acknowledgements
We thank Mark Hill and Filip Ruzicka for insightful discussions. Rebecca Finlay for help with running experiments.
Data accessibility
Data are available in the electronic supplementary material.
Authors' contributions
All authors contributed to experimental design and manuscript writing. M.F.C. and J.M. conducted the experiment; M.F.C. and M.R. performed statistical analyses. All authors agree to be held accountable for the content therein and approve the final version of the manuscript.
Competing interests
We declare we have no competing interests.
Funding
This study was funded by H2020 Marie Skłodowska-Curie Actions (grant no. 708362).
References
- 1.Stearns SC. 1992. The evolution of life histories. Oxford, UK: OUP. [Google Scholar]
- 2.Raubenheimer D, Simpson SJ. 2016. Nutritional ecology and human health. Annu. Rev. Nutr. 36, 603–626. ( 10.1146/annurev-nutr-071715-051118) [DOI] [PubMed] [Google Scholar]
- 3.Garlapow ME, Huang W, Yarboro MT, Peterson KR, Mackay TFC. 2015. Quantitative genetics of food Intake in Drosophila melanogaster. PLoS ONE 10, e0138129 ( 10.1371/journal.pone.0138129) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sun JH, et al. 2017. Drosophila FIT is a protein-specific satiety hormone essential for feeding control. Nat. Commun. 8, 14161 ( 10.1038/ncomms14161) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Corrales-Carvajal VM, Faisal AA, Ribeiro C. 2016. Internal states drive nutrient homeostasis by modulating exploration–exploitation trade-off. eLife 5, e19920 ( 10.7554/eLife.19920) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Itskov PM, Ribeiro C. 2013. The dilemmas of the gourmet fly: the molecular and neuronal mechanisms of feeding and nutrient decision making in Drosophila. Front. Neurobiol. 7, 12 ( 10.3389/fnins.2013.00012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Piper MD, et al. 2014. A holidic medium for Drosophila melanogaster. Nat. Methods 11, 100–105. ( 10.1038/nmeth.2731) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Camus MF, Fowler K, Piper MWD, Reuter M. 2017. Sex and genotype effects on nutrient-dependent fitness landscapes in Drosophila melanogaster. P. R. Soc. B-Biol. Sci. 284, 20172237 ( 10.1101/162107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wallace DC. 1999. Mitochondrial diseases in man and mouse. Science 283, 1482–1488. ( 10.1126/science.283.5407.1482) [DOI] [PubMed] [Google Scholar]
- 10.Camus MF, Dowling DK. 2018. Mitochondrial genetic effects on reproductive success: signatures of positive intrasexual, but negative intersexual pleiotropy. Proc. Biol. Sci. 285, 20180187 ( 10.1098/rspb.2018.0187) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Immonen E, Collet M, Goenaga J, Arnqvist G. 2016. Direct and indirect genetic effects of sex-specific mitonuclear epistasis on reproductive ageing. Heredity 116, 338–347. ( 10.1038/hdy.2015.112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jelic M, Arnqvist G, Kurbalija Novicic Z, Kenig B, Tanaskovic M, Andelkovic M, Stamenković-Radak M. 2015. Sex-specific effects of sympatric mitonuclear variation on fitness in Drosophila subobscura. BMC Evol. Biol. 15, 135 ( 10.1186/s12862-015-0421-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pichaud N, Messmer M, Correa CC, Ballard JWO. 2013. Diet influences the intake target and mitochondrial functions of Drosophila melanogaster males. Mitochondrion 13, 817–822. ( 10.1016/j.mito.2013.05.008) [DOI] [PubMed] [Google Scholar]
- 14.Nagarajan-Radha V, Rapkin J, Hunt J, Dowling DK. 2019. Interactions between mitochondrial haplotype and dietary macronutrient ratios confer sex-specific effects on longevity in Drosophila melanogaster. J. Gerontol. A 74, 1573–1581. ( 10.1093/gerona/glz104) [DOI] [PubMed] [Google Scholar]
- 15.Camus MF, O'Leary M, Reuter M, Lane N. 2020. Impact of mitonuclear interactions on life-history responses to diet. Phil. Trans. R. Soc. B 375, 20190416 ( 10.1098/rstb.2019.0416) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aw WC, Youngson NA, Ballard JW. 2016. Can we alter dietary macronutrient compositions and alleviate mitochondrial disease? J. Rare Dis. Res. Treatment 3, 31–37. [Google Scholar]
- 17.Ballard JW, Youngson NA. 2015. Review: can diet influence the selective advantage of mitochondrial DNA haplotypes? Biosci. Rep. 35, e00277 ( 10.1042/BSR20150232) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clancy DJ. 2008. Variation in mitochondrial genotype has substantial lifespan effects which may be modulated by nuclear background. Aging Cell 7, 795–804. ( 10.1111/j.1474-9726.2008.00428.x) [DOI] [PubMed] [Google Scholar]
- 19.Reddiex AJ, Gosden TP, Bonduriansky R, Chenoweth SF. 2013. Sex-specific fitness consequences of nutrient intake and the evolvability of diet preferences. Am. Nat. 182, 91–102. ( 10.1086/670649) [DOI] [PubMed] [Google Scholar]
- 20.Halekoh U, Højsgaard S. 2014. A Kenward-Roger approximation and parametric bootstrap methods for tests in linear mixed models: the R package pbkrtest. J. Stat. Softw. 59, 32 ( 10.18637/jss.v059.i09) [DOI] [Google Scholar]
- 21.Nychka D, Furrer R, Paige J, Sain S. 2015. fields: Tools for spatial data. See https://github.com/NCAR/Fields.
- 22.Lee KP, Simpson SJ, Clissold FJ, Brooks R, Ballard JWO, Taylor PW, Soran N, Raubenheimer D. 2008. Lifespan and reproduction in Drosophila: new insights from nutritional geometry. Proc. Natl Acad. Sci. USA 105, 2498–2503. ( 10.1073/pnas.0710787105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chandel NS. 2015. Evolution of mitochondria as signaling organelles. Cell Metab. 22, 204–206. ( 10.1016/j.cmet.2015.05.013) [DOI] [PubMed] [Google Scholar]
- 24.Chandel NS. 2014. Mitochondria as signaling organelles. BMC Biol. 12, 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Solon-Biet SM, et al. 2016. The ratio of macronutrients, not caloric intake, dictates cardiometabolic health, aging, and longevity in ad libitum-fed mice. Cell Metab. 19, 418–430. ( 10.3410/f.718306015.793513631) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Camus MF, Wolff JN, Sgrò CM, Dowling DK. 2017. Experimental support that natural selection has shaped the latitudinal distribution of mitochondrial haplotypes in Australian Drosophila melanogaster. Mol. Biol. Evol. 34, 2600–2612. ( 10.1093/molbev/msx184) [DOI] [PubMed] [Google Scholar]
- 27.Aw WC, et al. 2018. Genotype to phenotype: diet-by-mitochondrial DNA haplotype interactions drive metabolic flexibility and organismal fitness. PLoS Genet. 14, e1007735 ( 10.1371/journal.pgen.1007735) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morales HE, Pavlova A, Amos N, Major R, Kilian A, Greening C, Sunnucks P. 2018. Concordant divergence of mitogenomes and a mitonuclear gene cluster in bird lineages inhabiting different climates. Nat. Ecol. Evol. 2, 1258–1267. ( 10.1038/s41559-018-0606-3) [DOI] [PubMed] [Google Scholar]
- 29.Ballard JWO, Pichaud N. 2014. Mitochondrial DNA: more than an evolutionary bystander. Funct. Ecol. 28, 218–231. ( 10.1111/1365-2435.12177) [DOI] [Google Scholar]
- 30.Mossman JA, Biancani LM, Zhu CT, Rand DM. 2016. Mitonuclear epistasis for development time and its modification by diet in Drosophila. Genetics 203, 463–484. ( 10.1534/genetics.116.187286) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mackay TF, et al. 2012. The Drosophila melanogaster genetic reference panel. Nature 482, 173–178. ( 10.1038/nature10811) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are available in the electronic supplementary material.