Abstract
Phosphorus (P) is an essential nutrient for plant growth and productivity. Due to soil fixation, however, phosphorus availability in soil is rarely sufficient to sustain high crop yields. The overuse of fertilizers to circumvent the limited bioavailability of phosphate (Pi) has led to a scenario of excessive soil P in agricultural soils. Whereas adaptive responses to Pi deficiency have been deeply studied, less is known about how plants adapt to Pi excess and how Pi excess might affect disease resistance. We show that high Pi fertilization, and subsequent Pi accumulation, enhances susceptibility to infection by the fungal pathogen Magnaporthe oryzae in rice. This fungus is the causal agent of the blast disease, one of the most damaging diseases of cultivated rice worldwide. Equally, MIR399f overexpression causes an increase in Pi content in rice leaves, which results in enhanced susceptibility to M. oryzae. During pathogen infection, a weaker activation of defence‐related genes occurs in rice plants over‐accumulating Pi in leaves, which is in agreement with the phenotype of blast susceptibility observed in these plants. These data support that Pi, when in excess, compromises defence mechanisms in rice while demonstrating that miR399 functions as a negative regulator of rice immunity. The two signalling pathways, Pi signalling and defence signalling, must operate in a coordinated manner in controlling disease resistance. This information provides a basis to understand the molecular mechanisms involved in immunity in rice plants under high Pi fertilization, an aspect that should be considered in management of the rice blast disease.
Keywords: defence response, Magnaporthe oryzae, miR399, Oryza sativa, phosphate, rice, transcriptomics
High phosphate (Pi) fertilization and miR399 overexpression increases Pi content in rice leaves and enhances susceptibility to infection by the rice blast fungus Magnaporthe oryzae.

1. INTRODUCTION
Plants have evolved multiple mechanisms to perceive and respond to environmental stimuli, including biotic and abiotic stresses. Most research on plant responses to environmental stress has been conducted on plants subjected to an individual type of stress. In natural habitats, however, plants are simultaneously exposed to combinations of stress factors, and cross‐talk between responses induced by a particular type of stress may result in positive or negative impact over other stress.
Stress resulting from insufficient or excessive supply of nutrients can have an impact on disease resistance. Nutrient stress‐ and pathogen‐induced signalling pathways might interact with one another in a synergistic or antagonistic way and this interaction determines resistance or susceptibility to pathogen infection. Along with this, different results are found in the literature on the effect of addition of nutrients on disease severity, in which both increased and decreased disease severity have been reported. For instance, whereas nitrogen fertilization increases blast incidence in rice, it reduces disease severity caused by Verticillium spp. in Solanum species (Ballini et al., 2013; Veresoglou et al., 2013).
A nutrient of great importance for plant growth is phosphorus, which is absorbed through the roots almost exclusively in the form of inorganic phosphate (Pi). Though the overall content of phosphorus in soil is generally high, the low bioavailability of phosphate represents a limiting factor for plant growth in many natural and agricultural ecosystems (Hinsinger et al., 2011). As a consequence, Pi fertilizers are commonly used in modern farming to maximize crop yields, leading to a scenario of Pi excess in agricultural ecosystems. Excessive use of fertilizers and pesticides not only has a negative economic impact, but also causes a number of environmental problems due to soil pollution and water eutrophication, and raises serious concerns about food safety and animal health. However, the effect of Pi excess on disease resistance in crop species is poorly explored.
During pathogen infection, massive transcriptional reprogramming of gene expression occurs for the activation of plant defence responses (Jones and Dangl, 2006; Boller and He, 2009; Cook et al., 2015). This process is initiated by the recognition of pathogen‐associated molecular patterns (PAMPs) by host immune receptors at the plasma membrane, which triggers the induction of defence responses via multiple signalling cascades, so‐called PAMP‐triggered immunity (PTI) (Bigeard et al, 2015; Couto and Zipfel, 2016). Among others, PTI components include the production of reactive oxygen species (ROS), reinforcement of the cell wall, activation of protein phosphorylation processes, production of antimicrobial compounds, and accumulation of pathogenesis‐related (PR) proteins. Certain microbial pathogens are able to deliver effector proteins into the host cell that suppress PTI responses, resulting in plant susceptibility. In turn, many plants have evolved another layer of immunity through recognition of microbial effectors (or host proteins modified by effectors) by proteins encoded by resistance (R) genes. This recognition triggers a rapid and effective host defence response, so‐called effector‐triggered immunity (ETI) (Jones and Dangl, 2006). Whether nutrient stress can modulate host defence responses remains elusive, most probably because nutrient stress and disease resistance have been traditionally investigated separately from each other.
MicroRNAs (miRNAs) are small noncoding RNAs that mediate post‐transcriptional gene silencing through degradation or translational repression of target transcripts (Llave et al., 2002; Brodersen et al., 2008). They are essential regulators of gene expression in plant developmental processes and adaptation to biotic and abiotic stress (Navarro et al., 2006; Chen, 2009; Li et al., 2010; 2016; Shivaprasad et al., 2012; Campo et al., 2013; Jeong and Green, 2013; Baldrich et al., 2015; Baldrich and San Segundo, 2016; Song et al., 2019). Evidence also supports a regulation of nutrient homeostasis by certain miRNAs in plants (Liang et al., 2015; Paul et al., 2015; Chien et al., 2017). The best‐known examples of miRNAs involved in nutrient homeostasis are miR399 and miR827 (phosphate homeostasis), miR395 (sulphur homeostasis), and miR826 and miR5090 (nitrogen homeostasis).
The implication of miR399 in the phosphate starvation response (PSR) of Arabidopsis plants is well documented (Chiou et al., 2006; Hsieh et al., 2009; Puga et al., 2017; Ham et al., 2018). In brief, Pi deprivation rapidly increases miR399 accumulation, which causes degradation of PHOSPHATE2 (PHO2) transcripts encoding a ubiquitin‐conjugating E2 enzyme responsible for the ubiquitination and proteasome degradation of Pi transporters (Aung et al., 2006; Chiou et al., 2006; Liu et al., 2012). In this way, accumulation of miR399 in response to Pi starvation, and down‐regulation of PHO2, relieves negative post‐transcriptional control of Pi transporters for an increase in Pi uptake and translocation in Arabidopsis plants. While numerous studies have focused on the molecular mechanisms involved in the plant response to Pi starvation, mostly in the model dicotyledonous plant Arabidopsis, little attention has been paid to understand how crop species cope with Pi excess.
Rice is the most widely consumed staple food for a large part of the world's human population. However, rice production is severely threatened by the blast disease caused by the fungal pathogen Magnaporthe oryzae (Wilson and Talbot, 2009). In rice, the Pi starvation signal transduction pathway appears to be similar to that of Arabidopsis. OsPHO2 (originally named leaf tip necrosis 1, LTN1) is the AtPHO2 ortholog in rice (Hu et al., 2011). Despite the agronomic importance of rice, little information is available regarding the effect of Pi stress, either deficiency or excess, on blast disease resistance.
The aim of this study was twofold. First, to investigate whether Pi supply has an effect on blast disease resistance in rice. Second, to assess whether cross‐talk between miR399‐mediated signalling pathways and immune signalling pathways exists in rice. We show that growing rice plants under high Pi supply, as well as miR399 overexpression, increases Pi level in rice leaves and enhances susceptibility to M. oryzae infection. Transcriptome analysis revealed that miR399 overexpression affects the expression of genes involved in protein phosphorylation and transcription factor genes, as well as several defence‐related genes. On pathogen challenge, a weaker induction of defence gene expression is observed in miR399 overexpressing plants, as well as in wild‐type plants that have been grown under high Pi fertilization, which correlates well with their enhanced disease susceptibility phenotype. These data further support a link between Pi signalling and immune signalling in rice plants. Therefore, Pi excess caused by the indiscriminate use of Pi fertilizers in rice fields might have unintended consequences in rice production by facilitating blast infection.
2. RESULTS
2.1. High Pi fertilization enhances susceptibility of rice plants to the blast fungus M. oryzae
To assess whether Pi supply has an effect on disease resistance in rice, we grew rice (Oryza sativa japonica ‘Tainung 67’) plants for 15 days under different Pi regimes: high Pi, sufficient Pi, and low Pi (hereinafter high‐Pi, sufficient‐Pi, and low‐Pi plants). Compared to plants that have been grown under sufficient Pi supply, high‐Pi and low‐Pi plants accumulated higher and lower levels of free Pi in leaves, respectively (Figure 1a).
Figure 1.
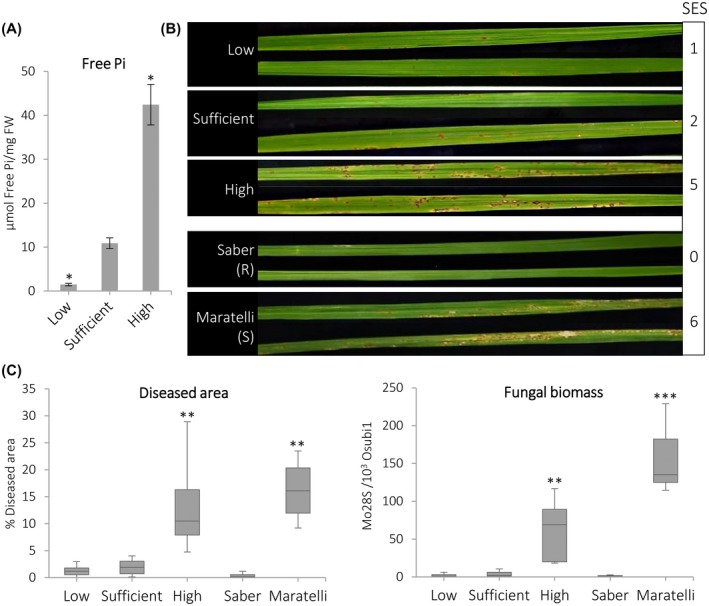
Susceptibility to infection by Magnaporthe oryzae in rice plants (Oryza sativa ‘Tainung 67’) grown under high‐Pi supply. (a) Free Pi content in leaves of rice plants grown under low, sufficient, or high Pi for 15 days (three‐ to four‐leaf stage). Bars represent mean ± SEM of four biological replicates with three plants per replicate. Comparisons have been made relative to the sufficient Pi condition (t test, *p < .05). (b) Disease symptoms at 7 days post‐infection (dpi) of low‐, sufficient‐, or high‐Pi treated plants. Representative results from one of three independent experiments that produced similar results are shown. Reference cultivars used were Saber (resistant) and Maratelli (susceptible). Standard evaluation system (SES) values are indicated on the right. (c) Percentage of diseased leaf area at 7 dpi (15 leaves per Pi treatment) (left panel). All the treatments were compared to the sufficient Pi supply (Wilcox test, **p ≤ .001). Fungal biomass was quantified at 7 dpi by quantitative PCR analysis using specific primers of M. oryzae Mo28S (right panel) and normalized to rice Ubiquitin 1 gene (eight biological replicates per Pi treatment). Comparisons have been made relative to the sufficient Pi supply (t test, **p ≤ .005, ***p ≤ .0001)
Rice plants at the three‐ to four‐leaf stage (high‐, sufficient‐, and low‐Pi plants) were challenged with M. oryzae. At the time of inoculation, no major differences were observed in growth in plants grown under different Pi treatments (only some reduction in shoot fresh weight in both low‐ and high‐Pi plants, compared to sufficient‐Pi plants) (Figure S1a,b). On pathogen inoculation, high‐Pi plants consistently displayed enhanced susceptibility to M. oryzae infection compared with sufficient‐ or low‐Pi plants (Figure 1b). The rice cultivars Maratelli and Saber were used as reference cultivars in infection experiments (susceptible and resistant to M. oryzae, respectively). Scoring of symptom severity was performed using the standard evaluation system (SES) ranging from 0 to 9 (0 = no symptom and 9 = highly susceptible). Whereas low‐ or sufficient‐Pi plants had SES values of 1 and 2, respectively, the high‐Pi plants showed SES values of 5 (Figure 1b). Quantification of diseased leaf area and fungal biomass in the infected leaves further confirmed enhanced susceptibility to M. oryzae in high‐Pi plants (Figure 1c). These results demonstrate that, when growing Tainung 67 rice plants under high‐Pi supply, these plants over‐accumulate Pi in leaves and exhibit enhanced susceptibility to M. oryzae infection.
Next, we investigated whether Pi treatment influences the development of blast disease not only in the rice cultivar Tainung 67, but also in other rice cultivars. To this end, rice cultivars with varying degrees of blast susceptibility were grown under different Pi regimes and then challenged with M. oryzae. The cultivars used were Nipponbare, Maratelli, Bomba, and Vialone Nano. For a comparison, the resistant cultivar Saber, characterized for the presence of the Pib resistance gene (Campos‐Soriano et al., 2013), was included in these experiments. Under high‐Pi conditions, all the susceptible rice genotypes assayed in this study over‐accumulated Pi in their leaves and showed increased susceptibility to blast infection (Figure 2). In contrast, high‐Pi treatment had no effect on the disease phenotype of the resistant cultivar Saber (Pib gene), suggesting that the Pib‐mediated resistance mechanisms to blast are not broken by high‐Pi treatment. Depending on the rice genotype, a small reduction in growth could be observed in both low‐Pi and high‐Pi plants compared with sufficient‐Pi plants, as revealed by fresh weight and dry weight measurements (Figure S2).
Figure 2.
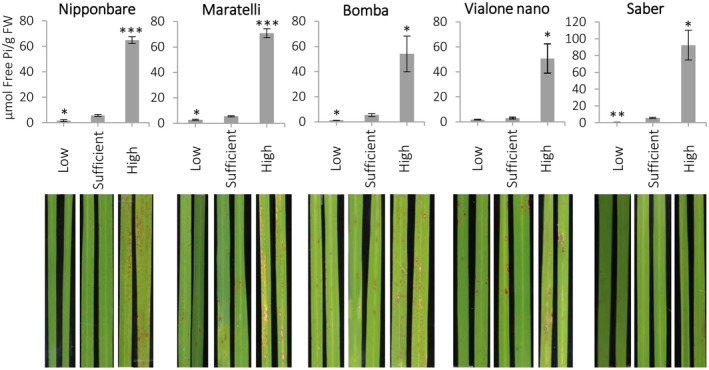
Susceptibility to infection by Magnaporthe oryzae in different rice genotypes grown under high‐Pi supply. Free Pi content in leaves of rice plants grown under low, sufficient, or high Pi for 15 days (three‐ to four‐leaf stage, upper panel) of the indicated rice genotypes. Bars represent mean ± SEM of four biological replicates with three plants per replicate. Comparisons have been made relative to the sufficient Pi condition (t test, *p < .05, **p < .005, ***p < .0001) Representative images of disease symptoms at 7 days post‐inoculation with M. oryzae spores (lower panel)
Under the experimental conditions assayed, low‐Pi treatment had a minimal impact on blast disease in all the rice genotypes (Figures 1 and 2). That low‐Pi plants perceive and react to the low‐Pi treatment used in this study was confirmed by induction of MIR399 expression and down‐regulation of its target gene OsPHO2, a typical response of plants to Pi starvation (Figure S3).
2.2. MIR399 overexpression increases susceptibility to M. oryzae infection
The rice miR399 family consists of 11 members (miR399a to miR399k; miRBase v22.1). Of them, miR399f has identical mature sequences in rice and Arabidopsis. Overexpression of miR399f results in over‐accumulation of Pi in leaves of Arabidopsis plants (Chiou et al., 2006). To further investigate the mechanisms by which over‐accumulation of Pi in rice plants enhances blast susceptibility we generated transgenic rice (Tainung 67) lines overexpressing miR399f (hereinafter referred to as miR399 OE plants; Methods S1) which were then challenged with the rice blast fungus.
The accumulation of miR399 precursor transcripts, and concomitant down‐regulation of OsPHO2, was confirmed through successive generations of the miR399 OE lines supporting that the MIR399f transgene was functional and stably inherited (Figure S4a). At the adult stage, miR399 OE plants exhibited leaf tip necrosis in older leaves, a typical trait of plants accumulating a high Pi level (Figure S4b). Five independent miR399 OE lines harbouring a single copy of the transgene inserted in their genome (Table S1) were selected for further studies.
The transgenic miR399 OE lines accumulated mature miR399 sequences at different levels, whereas OsPHO2 expression decreased in all the transgenic lines (Figure 3a, left and middle panels). Compared to wild‐type plants, miR399f OE plants accumulated a higher level of Pi in their leaves (Figure 3a, right panel).
Figure 3.
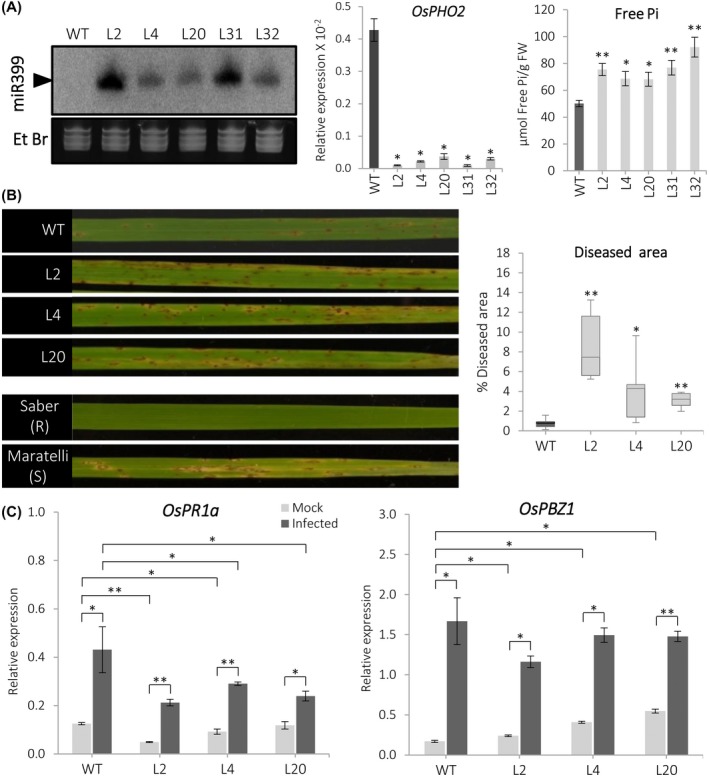
Susceptibility to infection by the rice blast fungus Magnaporthe oryzae in miR399 overexpressing (OE) rice plants. (a) Accumulation of mature miR399 sequences (left panel) and OsPHO2 transcripts (middle panel) in leaves of wild‐type (Tainung 67; WT) and five independent miR399 OE lines (independent homozygous lines, L2, L4, L20, L31, and L32) determined by small RNA northern blot and quantitative reverse transcription PCR analysis, respectively. Plants were grown under sufficient‐Pi conditions. Free Pi accumulation in miR399 OE lines and WT plants (right panel). Bars represent mean ± SEM of four biological replicates with three plants per replicate (t test, *p ≤ .05, **p < .005). (b) Susceptibility of miR399 OE plants to M. oryzae infection. Five independent homozygous lines (six plants each line) were assayed in each of four independent experiments with similar results (results for three miR399 OE lines are shown, L2, L4, and L20, similar results were obtained for the other lines). Reference cultivars: Saber (resistant) and Maratelli (susceptible). Right panel, diseased leaf area in infected leaves at 7 days post‐inoculation with M. oryzae spores (six leaves per line, t test, *p ≤ .05, **p ≤ .005; miR399 OE lines versus WT). (c) Accumulation of OsPR1a and OsPBZ1 transcripts in leaves of mock‐inoculated and M. oryzae‐inoculated wild‐type and miR399 OE plants (L2, L4, L20) at 48 hours post‐inoculation. Bars represent mean of three biological replicates (three pooled plants in each replicate) ± SEM (t test, *p ≤ .05, **p < .005, infected miR399 OE lines versus infected WT)
Three‐week‐old miR399 OE plants at the three‐ to four‐leaf stage were grown under Pi‐sufficient conditions and then challenged with M. oryzae. At 7 days post‐inoculation (dpi), few blast symptoms were visible in leaves of wild‐type plants, while leaves of miR399 OE lines were severely affected (Figure 3b, left panel). Quantification of diseased leaf area confirmed susceptibility to M. oryzae infection in miR399 OE plants (Figure 3b, right panel). These findings indicate that miR399 accumulation caused by miR399 overexpression has a negative effect on blast resistance in rice.
The OsPR1a and OsPBZ1 genes are recognized as molecular marker genes for the induction of the rice defence response to M. oryzae infection (Midoh and Iwata, 1996; Agrawal et al., 2001). Accordingly, we examined the expression of these defence‐related genes in leaves of miR399 OE lines and wild‐type plants during M. oryzae infection. As expected, OsPR1a and OsPBZ1 expression was induced in M. oryzae‐inoculated wild‐type plants compared to mock‐inoculated plants (Figure 3c). Although OsPR1a and OsPBZ1 expression was also activated by M. oryzae infection in miR399 OE plants, they were induced to a lesser extent in transgenic plants than wild‐type plants during M. oryzae infection (this different induction was more evident for OsPR1a) (Figure 3c). Intriguingly, in the absence of pathogen infection, the miR399 OE lines also exhibited a slight up‐regulation of OsPBZ1 compared with wild‐type plants (Figure 3c, right panel). A weaker induction of defence responses during infection is consistent with the observed phenotype of disease susceptibility in miR399 OE plants.
Knowing that miR399 OE plants over‐accumulate Pi in their leaves, and that OsPR1a and OsPBZ1 expression showed a weaker induction in miR399 OE plants during M. oryzae infection, it was of interest to assess whether Pi treatment has an effect on OsPR1a and OsPBZ1 expression in wild‐type plants. For this, we grew plants (Tainung 67) for 15 days under low‐, sufficient‐, and high‐Pi conditions. The expression of OsPBZ1 and OsPR1a was determined at different times after inoculation (24, 48, and 72 hours post‐inoculation [hpi]). OsPR1a and OsPBZ1 expression was induced in M. oryzae‐inoculated plants, but the level of induction of these genes differed among the Pi treatments (differences were clearly observed at 48 and 72 hpi; Figure 4). Thus, an increase in Pi supply is accompanied by a lower induction of OsPBZ1 during infection of wild‐type plants (Figure 4). Compared to sufficient‐Pi plants, OsPR1a is also activated to a lower extent in high‐Pi plants (at 48 hpi, although similar levels of OsPR1a expression occur in high‐ and sufficient‐Pi plants at 72 hpi). A weaker induction of defence gene expression in high‐Pi wild‐type rice plants during M. oryzae infection is in agreement with previous observations on susceptibility to M. oryzae infection in these plants (see Figure 1b; Tainung 67). However, the higher induction of OsPBZ1 and OsPR1a that occurs in low‐Pi plants relative to sufficient‐Pi plants (Figure 4) led to only a slight increase in disease resistance (see Figure 1a).
Figure 4.
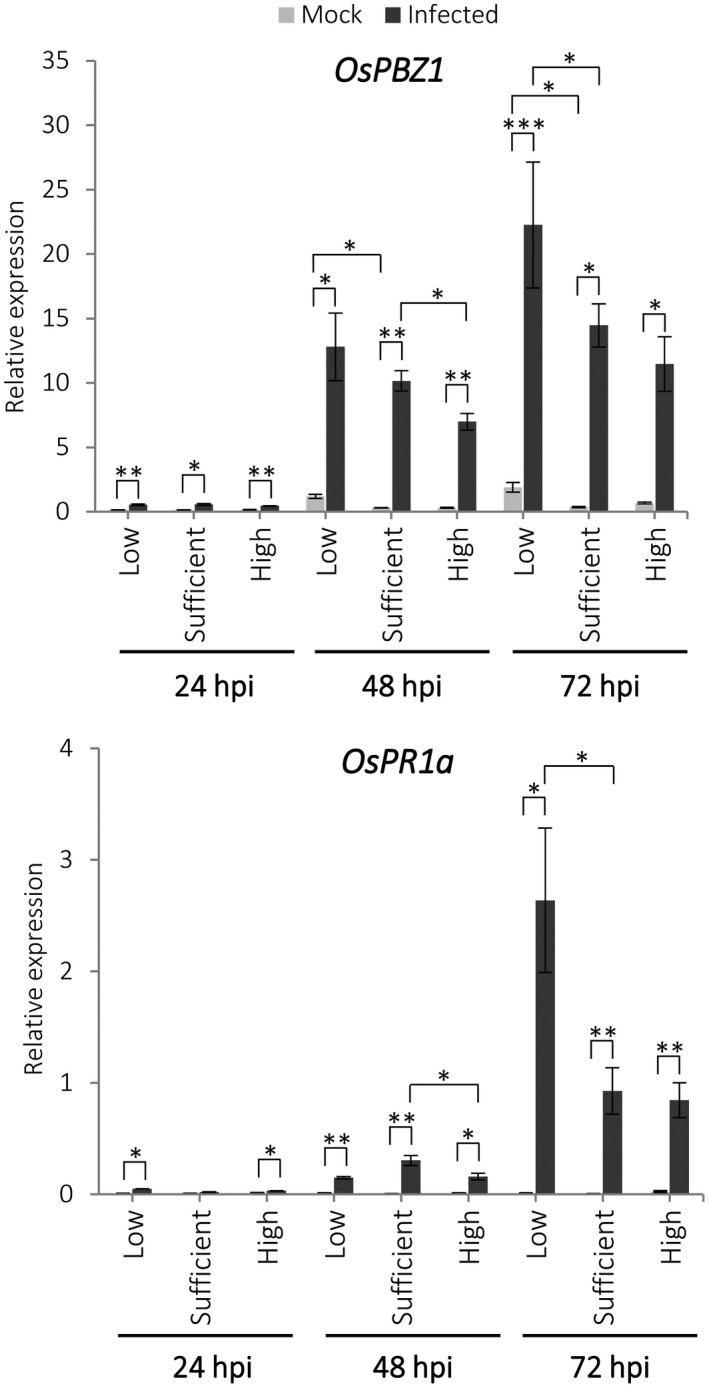
Expression of the defence marker genes OsPBZ1 and OsPR1a in mock‐ or Magnaporthe oryzae‐inoculated leaves of low‐, sufficient‐, or high‐Pi wild‐type (Tainung 67; WT) plants. Quantitative reverse transcription PCR was carried out at the indicated times after inoculation using the rice Ubiquitin 5 gene as the internal control. Bars represent means of three biological replicates, each one from a pool of three different plants ± SEM (t test, *p ≤ .05, **p < .005, ***p < .0001)
Collectively, the results obtained on blast disease assays in wild‐type plants grown under high Pi supply, together with those obtained in miR399 OE plants, support the fact that Pi accumulation negatively affects defence gene expression, hence blast resistance, in rice.
2.3. Transcript profiling of miR399 OE rice plants
To better understand the molecular mechanisms underlying blast susceptibility in miR399 OE rice plants, we performed transcriptome analysis of miR399 OE lines and wild‐type plants. Differentially expressed genes (DEGs) were identified based on significance level (p ≤ .05) and fold change (log2 fold change ≥0.5 or ≤0.5 as the threshold for up‐regulated and down‐regulated genes, respectively). Using these criteria, a total of 965 genes were found to be differentially expressed in miR399 OE plants relative to wild‐type plants, of which 690 genes were up‐regulated and 275 genes were down‐regulated (Table S2).
Gene ontology (GO) enrichment analysis of DEGs in miR399 OE plants was carried out by AgriGO (http://bioinfo.cau.edu.cn/agriGO; Du et al., 2010) and visualized by REVIGO (Supek et al., 2011). This analysis revealed statistically enriched terms in biological processes, cellular components, and molecular functions that were likely to be regulated by miR399 in rice. In the set of up‐regulated genes, the most abundant terms in the biological function category were “metabolism” (e.g., “phosphorus metabolism”), “response to stimulus” (e.g., “response to biotic stimulus”), and “post‐translational protein modification” (Figures 5a and S5). The GO terms over‐represented in down‐regulated genes clustered in the categories of “metabolism” (primary metabolism, phosphorus metabolism, protein metabolism), “response to stimulus”, “phosphorylation”, and “post‐translational protein modification” (Figures 5b and S6). The “cellular component” category from up‐regulated genes contained the terms “integral to membrane” and “nucleus”, whereas down‐regulated genes were enriched with the GO term “nucleus” (Figure S7).
Figure 5.
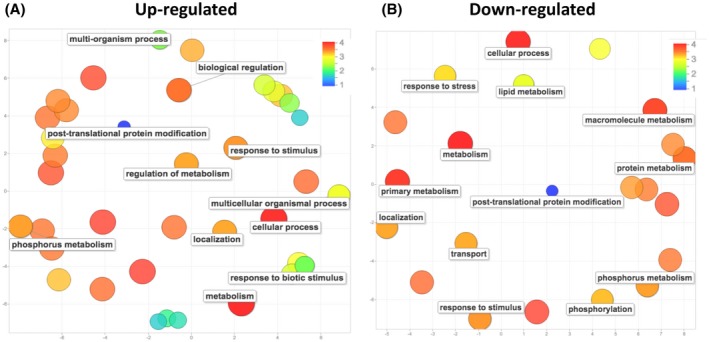
Gene ontology (GO) enrichment analysis of differentially expressed genes in miR399 overexpressing (OE) plants relative to wild‐type plants. The top GO terms enriched in miR399 OE plants were represented by using REVIGO after reducing redundancy (http://revigo.irb.hr/). (a) and (b) represent up‐regulated and down‐regulated genes, respectively. Circles represent GO terms and those clustered closer to each other represented similar GO terms. Circle colours (blue to yellow) represent the degree of GO enrichment (p value) and size is proportional to the frequency of the GO term in the GO database (larger and smaller discs represent more general and more specific terms, respectively)
AgriGO analysis also revealed that the most abundant subcategories in the molecular function category included “protein kinase activity” and “ATP binding”, in both up‐regulated and down‐regulated genes (Figures S8 and S9), whereas genes with “transcription factor activity” were highly represented among the up‐regulated genes in miR399 OE plants (Figure S8). The “zinc ion binding” and “calcium ion binding” subcategories were specifically enriched in up‐regulated genes (Figure S8), while “iron binding” genes were enriched in down‐regulated genes (Figure S9).
A heat map analysis of genes specifically annotated in the subcategories “protein phosphorylation and ATP binding” and “regulation of transcription” is presented in Figure S10 (left and middle panel; genes shown in the heat map are listed in Tables S3 and S4). Intriguingly, defence‐related genes were found among the set of both up‐regulated and down‐regulated genes in miR399 OE plants (Figure S10, right panel, and Table S5). These findings are further discussed below.
The expression of genes identified by RNA‐Seq analysis of miR399 OE plants was further investigated by quantitative reverse transcription PCR (RT‐qPCR). Genes that were classified into different functional categories were selected for this study, including genes involved in defence responses, diterpenoid phytoalexin biosynthesis, receptor protein kinase genes, protein kinase genes, and transcription factor (TF) genes (Figure 6). In agreement with results obtained by RNA‐Seq analysis, the expression of these genes was up‐regulated in miR399 OE plants compared to wild‐type plants.
Figure 6.
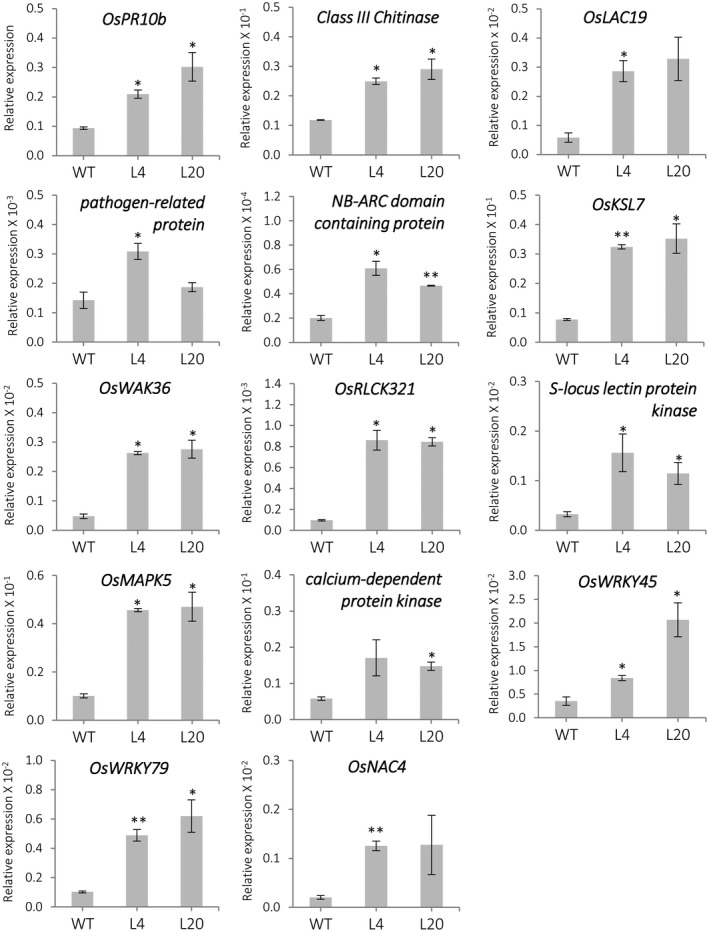
Expression of differentially expressed genes identified by RNA‐Seq analysis. Transcript levels were determined by quantitative reverse transcription PCR in leaves of wild‐type (WT) and miR399 overexpressing lines (lines L4 and L20, similar results were obtained for L2). Plants were grown under sufficient‐Pi conditions. The expression values were normalized to the rice Ubiquitin 1 gene. Gene‐specific primers sequences with its gene locus ID and heat maps code (from Figure S10) are presented in Table S6. Bars represents means of three biological replicates, each one from a pool of three different plants, ±SEM. Comparisons have been made relative to the WT (t test, *p ≤ .05, **p ≤ .005)
2.4. miR399 overexpression influences the expression of genes involved in protein phosphorylation and transcriptional regulation
The expression of a vast array of genes involved in protein phosphorylation was misregulated in miR399 OE plants (up‐regulation and down‐regulation) (Figure S10 and Table S3). Among the up‐regulated genes, we identified genes representing different families of receptor protein kinases and receptor‐like protein kinases, including wall‐associated kinases (WAKs; OsWAK36, OsWAK87, OsWAK41, OsWAK98), Ser/Thr receptor protein kinases (e.g., the PR5K receptor kinase) lectin‐like receptor protein kinases, and leucine‐rich repeat receptor protein kinases (Table S3). WAKs are known to play a key role in basal defence against the rice blast fungus (Delteil et al., 2016). Regarding the Ser/Thr protein kinase receptor PR5K, it contains an extracellular domain structurally related to pathogenesis‐related PR5 proteins involved in disease resistance (Wang et al., 1996). The receptor kinase LecRK‐I.9 has been reported to regulate jasmonic acid (JA) signalling and disease resistance in Arabidopsis (Balagué et al., 2017). The observed up‐regulation of receptor protein kinases for which a positive role in defence responses has been described is in apparent contradiction to the phenotype of blast susceptibility observed in miR399 OE plants.
In the set of down‐regulated genes in miR399 OE plants, there were three phytosulfokine receptor genes (members of the leucine‐rich repeat family of receptor kinases) (Figure S10, code PSKR‐1, PSKR‐2, PSKR‐3; Table S3). Binding of phytosulfokine peptides to phytosulfokine receptors differentially modulates Arabidopsis immune responses (Sauter, 2015). The expression of genes belonging to different families of protein kinases was also misregulated in miR399 OE plants (Figure S10 and Table S3). They included Ser/Thr protein kinases (code KIN), tyrosine protein kinases (code, PKDCC), and calcium‐dependent protein kinase (code CDPK) genes. Our transcriptome analysis also revealed that up to seven adenosine triphosphatase genes were up‐regulated in miR399 OE plants. Knowing that miR399 overexpression was associated with an increase in Pi content, the observed alterations in the expression of genes involved in protein phosphorylation and ATP binding might well be the consequence of Pi accumulation in leaves of miR399 OE plants.
Moreover, miR399 overexpression regulates the expression of a considerable number of TFs belonging to several TF families, most of them being up‐regulated in miR399 OE plants (Figure S10 and Table S4). They included WRKY, AP2 (APETALA2)‐related (e.g., ethylene‐responsive TFs), NAC (NAM, no apical meristem; ATAF1; and CUC2, cup‐shaped cotyledon), Myb (myeloblastosis), bZIP (basic leucine zipper), C2H2 (C2H2 zinc finger), and ZF1 (zinc finger) TF genes. It is generally assumed that WRKY TFs are important regulators of stress responses in plants, including rice blast resistance (Wei et al., 2013; Phukan et al., 2016). It should be noted that up to 22 WRKY TFs are up‐regulated in miR399 OE plants, including OsWRKY45 (Figure 6 and Table S4). OsWRKY45 has been reported to play a crucial role in resistance to blast infection, and its overexpression enhances resistance to infection (Shimono et al., 2007). As previously observed for receptor protein kinases involved in defence responses, an up‐regulation of OsWRKY45 in miR399 OE plants is in apparent contradiction to the phenotype of disease susceptibility in these plants. WRKY TFs have been also shown to regulate tolerance to Pi starvation in Arabidopsis and rice (Wang et al., 2014; Dai et al., 2016).
2.5. An increase in Pi content compromises the basal defence of rice plants during pathogen infection
As previously mentioned, RNA‐Seq analysis revealed misregulation of certain defence‐related genes in miR399 OE plants in the absence of pathogen infection, some of them being up‐regulated in these plants (e.g., members of the PR1, PR5, PR8, and PR10 families of PR genes) (Figures 6 and S10; Table S5). Genes involved in protection against oxidative stress and/or cell wall lignification were also misregulated in miR399 OE plants (e.g., peroxidase and laccase genes) (Figures 6 and S10; Table S5). Peroxidases and laccases contribute to plant cell‐wall lignification by catalysing the polymerization of monolignols (Miedes et al., 2014). Peroxidases also participate in maintenance of the cellular redox balance to protect against oxidative stress (Almagro et al., 2009). Unexpectedly, RNA‐Seq analysis of miR399 OE plants showed a basal expression level of several genes involved in diterpenoid phytoalexin biosynthesis, these genes being responsible for the production of major rice phytoalexins (momilactones, oryzalexin A, B, C, and D, oryzalexin E, and phytocassenes) (Yamane, 2013).
Because the miR399 OE rice plants exhibited enhanced susceptibility to M. oryzae infection, the observed expression of defence‐related genes (e.g., PR, phytoalexin biosynthesis genes) in these plants appears not to be effective enough to arrest pathogen infection. This observation posed intriguing questions related to regulation of defence gene expression in miR399 OE plants. We reasoned that miR399 overexpression, while causing a basal expression of some defence‐related genes under noninfection conditions, might interfere with the activation of defence responses during pathogen infection. To test this possibility, we monitored defence gene expression in miR399 OE and wild‐type plants that had been inoculated with M. oryzae or mock‐inoculated (48 hpi). Compared to wild‐type plants, a weaker induction of defence gene expression occurred in miR399 OE plants in response to M. oryzae infection, including OsPR10b, OsCHIT1, OsLAC19, OsWAK46, OsMAPK5, and OsWRKY45 (Figure 7). Similarly, a weaker response was observed in genes involved in diterpenoid phytoalexin biosynthesis (OsCPS2, OsKSL4, OsKSL7, Oryzalexin D synthase, Oryzalexin E synthase, OsMAS1). Whether miR399 OE plants fail to accumulate diterpenoid phytoalexins during M. oryzae infection remains to be investigated.
Figure 7.
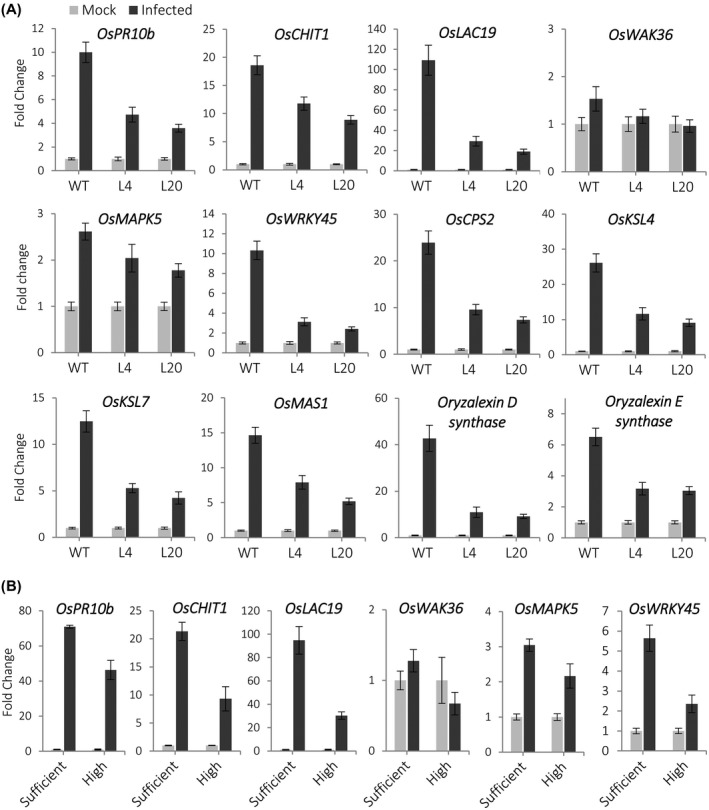
Induction of defence‐related genes in rice plants accumulating Pi in leaves on inoculation with Magnaporthe oryzae spores. (a) miR399 overexpressing (OE) plants grown under sufficient‐Pi conditions. Results obtained on lines L4 and L20 are shown (similar results were obtained for line L2). (b) Wild‐type (WT) plants that have been grown under high Pi supply. The expression values were normalized to the rice Ubiquitin 5 gene. Bars represent means of three biological replicates, each one from a pool of three different plants, ±SEM. In all the cases, the expression level in mock‐inoculated plants was set to 1.0
The observation that miR399 overexpression leads to an increase in Pi content and repression of the pathogen‐inducible host defence response prompted us to investigate whether genes for which a weaker induction occurred during infection in miR399 OE plants were also induced at a lower extent in rice plants that have been grown under high‐Pi conditions. In response to M. oryzae infection, all the defence‐related genes here examined (OsPR10b, OsCHIT1, OsLAC19, OsMAPK5, OsWRKY45, and OsWAK36) had a lower induction in high‐Pi plants compared to sufficient‐Pi wild‐type plants (Figure 7; fold‐induction is presented).
Collectively, the results presented in this study demonstrate that an increase in Pi level caused either by miR399 overexpression or treatment with high‐Pi represses the pathogen‐inducible host defence response. This, in turn, would negatively affect resistance to M. oryzae infection. These findings, together with the observation that defence gene expression decreases when increasing Pi supply, support the fact that Pi over‐accumulation (high‐Pi, miR399 OE plants) compromises the immune system in rice, hence fostering disease susceptibility.
3. DISCUSSION
Here we provide evidence that Pi, when in excess, is an important factor in determining susceptibility to infection by the foliar pathogen M. oryzae in rice. We show that an increase in Pi content caused by high Pi fertilization enhances blast susceptibility in different rice cultivars, (e.g., Tainung 67, Nipponbare, Maratelli, Bomba, and Vialone Nano). Equally, miR399f overexpression causes Pi accumulation and enhances blast susceptibility. Disease susceptibility in rice plants accumulating Pi in leaves was associated with a weaker induction of defence gene expression during pathogen infection. This finding indicates that these plants are not able to mount an effective defence response, which is in agreement with their phenotype of blast susceptibility. Contrary to what is observed in susceptible rice cultivars, Saber plants did not develop disease symptoms regardless of Pi supply, suggesting that resistance triggered by the Pib resistance gene is not affected by high‐Pi supply.
Pi excess might reduce the plant's ability to overcome pathogen infection in different ways. On the one hand, Pi excess may be responsible for metabolic changes in the host plant that jeopardize the plant defence response. Along with this, our transcriptome analysis showed that miR399 overexpression causes important perturbations in genes involved in metabolic processes, in particular lipid and carbohydrate metabolism. Another possibility, not excluding the previous one, is that fungal pathogenicity itself is affected by Pi availability and that the fungus modifies its pathogenicity programme in order to adapt to a situation of high Pi in the host tissue. An increase in Pi level might create a more favourable environment for pathogen growth that enables the pathogen to improve nutrient acquisition from the host plant, thereby promoting pathogenicity. Alternatively, high Pi content might also stimulate the production of pathogen virulence factors that suppress host defence mechanisms. In the case of nitrogen‐induced susceptibility to rice blast, it was reported that alterations in some metabolites (e.g., glutamine) are responsible for increased fungal pathogenicity (Huang et al., 2017). At present, however, it is not known whether an increase in Pi content in rice leaves has an effect on M. oryzae virulence. On the other hand, studies in Arabidopsis have shown that Pi content influences the structure of the root‐associated bacterial (Castrillo et al., 2017; Finkel et al., 2019) and fungal communities (Fabianska et al., 2019). It was proposed that changes in root fungal communities of Arabidopsis plants correlate with concurrent changes in the plant phosphate starvation response, suggesting that P‐deprived and P‐replete plants select different fungal taxa in their roots (Fabianska et al., 2019). Whether Pi content has an effect on potential rice pathogens needs to be further investigated. We also show that, even though the low‐Pi plants have activated the phosphate starvation response, low‐Pi treatment does not appear to affect blast severity in a substantial manner, at least under the experimental conditions here assayed. There is the possibility that activation of the phosphate starvation response allows the plant to cope with the low‐Pi stress condition imposed in this study, and more stressful conditions (e.g., lower Pi supply, longer periods of low Pi treatment) might be needed to observe a differential blast phenotype in low‐Pi rice plants.
Another finding of this study is that, under normal growth conditions, the miR399 OE plants show a basal expression of some defence‐related genes (e.g., OsWRKY45, diterpenoid phytoalexin biosynthesis genes) in the absence of pathogen infection. On pathogen challenge, however, the basal level of defence gene expression that is observed in Pi‐accumulating plants (both miR399 OE and wild‐type plants grown under high‐Pi supply) does not seem to be effective enough to arrest pathogen infection. These observations raise a number of interesting questions on how the plant perceives and responds to Pi excess by activating genes typically associated with the plant response to pathogen infection. A tempting hypothesis is that high Pi is perceived by the plant as a stressful situation and that this sensing triggers a basal expression of defence‐related genes by yet unknown signalling mechanisms in the absence of pathogen infection. Clearly, the observed weaker induction of M. oryzae‐inducible defence gene expression in Pi‐accumulating rice plants, and concomitant blast susceptibility, indicates that these plants respond in a less effective manner to pathogen infection, while supporting that there is an interplay between Pi‐ and pathogen‐induced signal transduction pathways in rice. Further investigation is needed to understand how signals coming from the two types of stresses, Pi excess and pathogen infection, are integrated during pathogen infection. Here, it is worth mentioning that whereas transcriptional reprogramming of gene expression in response to Pi starvation is widely described (Misson et al., 2005; Wasaki et al., 2006; Oono et al., 2013; Secco et al., 2013; Gho et al., 2018), the transcriptional response to Pi excess remains largely unknown. Very recently, a phosphate transporter from rice (OsPT8) has been reported to negatively regulate blast resistance, thus reinforcing the notion of cross‐talk between Pi homeostasis and immune signalling pathways in rice (Dong et al., 2019).
In addition, transgenic expression of a phytoplasma effector (SAP11AYWB) in Arabidopsis was found to increase miR399 accumulation, thereby increasing Pi level (Lu et al., 2014). During infection of SAP11AYWB Arabidopsis plants with the bacterial pathogen Pseudomonas syringae, the induction of defence responses was suppressed and the SAP11AYWB plants exhibited susceptibility to bacterial infection (Lu et al., 2014). In other studies, cross‐talk between Pi starvation and salt and drought stress signalling pathways, or hormone‐signalling pathways, has been described (Song and Liu, 2015; Abbas Khan et al., 2016; Baek et al., 2017; Castrillo et al., 2017). Knowing that the defence‐related hormones JA and ethylene (ET) function as signalling hormones in the rice defence against M. oryzae (Nasir et al., 2018), a major future challenge is to determine whether Pi content affects defence hormone signalling in rice for the control of immune responses. Collectively, these observations suggest that, although a function for miR399 in the two stress signalling systems, Pi signalling and immunity, can be postulated, we have yet to decipher the molecular mechanisms of this cross‐talk.
Phosphorus is a key component of many important biological molecules, including ATP, which exists not only in the cytoplasm, but also in the extracellular matrix. Of interest, plants are capable of sensing and responding to extracellular ATP during pathogen infection, a situation in which ATP is perceived as a damage‐associated molecular pattern (DAMP) for cellular damage caused during pathogen colonization (Tanaka et al., 2014). In this manner, extracellular ATP can be considered as a signalling molecule for the activation of Arabidopsis defence responses (Cao et al., 2014). More recently, extracellular ATP has been shown to play a role in the JA‐induced defence response of Arabidopsis through the direct activation of JA signalling (but not via JA biosynthesis) (Tripathi et al., 2018; Jewell et al., 2019). Evidence for a role of extracellular ATP as signalling molecule for the expression of defence responses in rice is, however, lacking.
Based on the results obtained in our transcriptome analysis of miR399 OE plants, a working model summarizing the impact of miR399 overexpression (and Pi accumulation) in regulating defence responses to pathogen infection in rice is presented in Figure 8, which includes processes related to pathogen recognition, signalling processes, and defence reactions. According to this model, miR399‐mediated perturbations in host gene expression would affect diverse physiological processes in rice plants under noninfection and infection conditions. Thus, miR399 overexpression (as well as growing rice plants under high Pi supply) would provoke a stressful situation in the plant that would have pleiotropic effects on metabolic processes and defence responses, and possibly also ATP signalling. The observation that miR399 overexpression alters the expression of diverse protein kinases and transcription factors potentially involved in pathogen recognition and/or defence signal transduction (e.g., receptor protein kinases, Ser/Thr kinases, among others) favours this possibility. These alterations would decrease the ability of the host plant to detect the invading pathogen and respond in a timely and appropriate manner, leading to enhanced susceptibility to M. oryzae infection.
Figure 8.
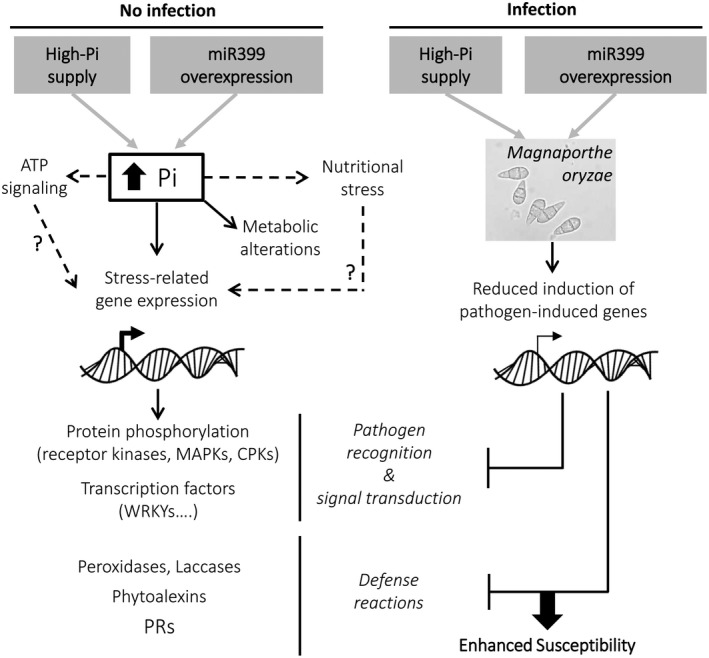
Proposed model for the role of miR399 and Pi homeostasis in the defence response to Magnaporthe oryzae. Under no infection conditions, high‐Pi supply in wild‐type plants and MIR399 overexpression (OE) entails Pi overaccumulation in rice leaves. This would trigger a state of nutritional stress with metabolic alterations and changes in ATP signalling, which ultimately would activate the expression of stress‐related genes, such as those involved in protein phosphorylation, transcription factors, peroxidases and laccases, and phytoalexin biosynthesis, as well as PR genes. On pathogen challenge, a weaker induction of these stress‐related genes would occur in high Pi‐treated plants and miR399 OE plants compared to wild‐type plants with a sufficient Pi supply. This, in turn, would lead to susceptibility to M. oryzae infection. Arrows and blunt ends, positive and negative regulation, respectively. Arrows with broken lines, unknown interlocked regulatory mechanisms
On the contrary, cross‐talk between Pi and other nutrients is well documented in plants and miRNAs have been shown to be involved in cross‐talk between nutrient deficiencies in Arabidopsis (Liang et al., 2015). In particular, miR399 has been proposed to participate in the regulation of multiple nutrient starvation responses in rice (Hu et al., 2015). Under this scenario, stress imposed by high Pi might also hamper the coordination of homeostatic pathways of Pi and other nutrients, and Pi excess caused by miR399 overexpression or high Pi supply might alter other nutrients’ accumulation. This nutritional imbalance would trigger an additional stress to the host plant that would impair the optimum defence response to pathogen infection (Figure 8). This piece of information will be useful in future studies to understand how plants integrate Pi signalling with immune responses.
The Arabidopsis miR399f has been reported to function as a positive regulator in tolerance to salt stress, while negatively regulating drought stress, through the control of two novel target genes in Arabidopsis, the ABF3 transcription factor and CSP41b (unknown function) genes (Baek et al., 2016). Although the results presented here indicate that miR399 overexpression is accompanied by down‐regulation of OsPHO2 in miR399 OE plants, the possibility that miR399f directs cleavage of still unknown target genes in rice should not be discarded.
Collectively, the results presented here support the finding that Pi accumulation in rice leaves negatively affects basal resistance to M. oryzae in rice by reducing the plant’s ability to cope with pathogen infection. Here, it is worth emphasizing that fungicides are widely used to protect rice plants against blast disease, which causes serious environmental problems. Indeed, rice blast is considered the most important fungus‐caused disease in plants in terms of scientific and economic relevance (Dean et al., 2012). Pi fertilization should then be considered on a cost–benefit basis in rice farming. The results presented here might lay a foundation for rationally optimizing fertilizer and pesticide use in rice production.
4. EXPERIMENTAL PROCEDURES
4.1. Plant material
Rice plants were grown at 28 ºC with a 14 hr:10 hr light/dark cycle. Transgenic rice (cultivar Tainung 67) plants were produced by Agrobacterium‐mediated transformation of embryogenic calli derived from mature embryos (Sallaud et al., 2003). Transgene copy number was assessed by PCR on genomic DNA as previously described (Yang et al., 2005). Primers are shown in Table S6. The susceptible rice cultivars Nipponbare, Maratelli, Vialone Nano, and Bomba and the resistant cultivar Saber harbouring the Pib resistance gene were used in this work.
4.2. Infection assays
Infection assays with the rice blast fungus M. oryzae (strain Guy 11) were carried out on soil‐grown rice plants at the three‐ to four‐leaf stage. Further experimental details can be found in Methods S1. The percentage of diseased leaf area was determined at 7 dpi with M. oryzae spores by using the APS Assess 2.0 program (Lamari, 2008). Fungal biomass on infected leaves was quantified by quantitative PCR (qPCR) using specific primers for the M. oryzae 28S DNA gene (Qi and Yang, 2002).
4.3. Pi treatment and measurement of Pi content
To assess the effect of Pi supply, rice plants were grown for 21 days in a mixture of 50% peat and vermiculite (>50 mg/L phosphorus) and 50% inorganic https://www.linguee.com/english-spanish/translation/quartz.html sand substrate. During the last 15 days, plants were fertilized with modified Hoagland half‐strength solution (2.5 mM Pi for high‐Pi, 0.25 mM Pi for sufficient‐Pi, and no Pi for low‐Pi supplies). The Pi content of rice leaves was determined as previously described (Ames, 1966; Versaw and Harrison, 2002).
4.4. Library preparation and RNA‐Seq analysis
Total RNA was extracted using the Maxwell 16 LEV Plant RNA Kit (Promega). Libraries were prepared from leaves of 3‐week‐old wild‐type and miR399 OE plants. Each biological replicate consisted of leaves from 15 plants. Two independent miR399 OE lines and three biological replicates for each genotype were examined (see Methods S1 for details on RNA‐Seq analysis). The RNA sequence data were deposited at the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO repository (accession number http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE137735).
4.5. Gene expression analyses
Total RNA was extracted from rice leaves using the TRIzol reagent (Invitrogen). For northern blot analysis, total RNAs were fractionated in a 17.5% denaturing polyacrylamide gel containing 8 M urea, transferred to nylon membranes, and hybridized with [γ32P]ATP‐labelled oligonucleotides (Table S6). Hybridization signals were detected using a phosphorimager (Bio‐Rad). Synthetic RNA oligonucleotides were used as size markers.
RT‐qPCR was performed in optical 96‐well plants using SYBR Green. Primers were designed using Primer‐BLAST (https://www.ncbi.nlm.nih.gov/tools/primer-blast/). Accumulation of mature miR399 sequences was determined by stem‐loop RT‐qPCR. Primers used for RT‐qPCR and stem‐loop RT‐qPCR are listed in Table S6. Further details on RT‐qPCR and stem‐loop RT‐qPCR can be found in Methods S1.
Supporting information
FIGURE S1 Treatment of rice plants (Oryza sativa “Tainung 67”) with different Pi concentrations. (a) Phenotype of plants that have been grown for 15 days under low‐, sufficient‐, or high‐Pi conditions. (b) Shoot fresh weight and dry weight of rice plants grown for 15 days under different Pi regimes. Data represent means of four biological replicates, each one from a pool of three different plants, ±SEM (one‐way analysis of variance and Tukey’s test, *p < .05, **p < .005)
FIGURE S2 Effect of treatment with low‐ and high‐Pi supply in the susceptible rice cultivars Nipponbare, Maratelli, Bomba, Vialone Nano, and the resistant cultivar Saber (harbouring the Pib resistance gene). Left panels, appearance of plants that have been grown for 15 days under different Pi regimes. Right panels, shoot fresh weight and dry weight of plants shown in left panels. Data represent means of four biological replicates, each one from a pool of three different plants, ±SEM (one‐way analysis of variance and Tukey’s test, *p < .05, **p < .005, ***p < .0001)
FIGURE S3 Effect of Pi treatment on the expression of Pi starvation‐responsive genes in rice (cv. Tainung 67). The accumulation of miR399f precursor transcripts and the miR399 target gene OsPHO2 was determined by quantitative reverse transcription PCR. The expression values were normalized to Ubiquitin 1 gene. Bars show the means of three biological replicates, with three different plants in each one, and SEM. Comparisons have been made relative to the sufficient Pi condition (t test, *p < .05)
FIGURE S4 Characterization of miR399f overexpressing (OE) rice plants. (a) Accumulation of miR399f precursor and OsPHO2 transcripts in miR399 OE lines (T0 generation). (b) Leaf tip necrosis in leaves of miR399 OE rice plants grown under controlled greenhouse conditions for 2 months
FIGURE S5 Gene ontology (GO) terms enriched in up‐regulated genes in miR399OE plants compared to wild‐type plants were generated by AgriGO according to biological process. Singular enrichment analysis (SEA), with the Rice Gramene gene model as a reference, by AgriGO analysis (hypergeometic test; Hochberg false discovery rate (FDR); p ≤ .05; “10” as minimum number of mapping entries). Boxes in the graph show GO terms, term definition, and statistical information. The significant terms are marked with colour, while nonsignificant terms are shown in white boxes. The colour intensity of a box positively correlates with the enrichment level of the term
FIGURE S6 GO terms enriched in down‐regulated genes in miR399 OE plants compared to wild‐type plants were generated by AgriGO according to biological process. Singular enrichment analysis (SEA), with the Rice Gramene gene model as a reference, by AgriGO analysis (Hypergeometic test; Hochberg FDR; p ≤ .05; “10” as minimum number of mapping entries). Boxes in the graph show GO terms, term definition, and statistical information. The significant terms are marked with colour, while nonsignificant terms are shown in white boxes. The colour intensity of a box positively correlates with the enrichment level of the term
FIGURE S7 GO terms enriched in miR399 OE plants compared to wild‐type plants were generated by AgriGO according to cellular component. (a) Functional categories in the group of up‐regulated genes. (b) Functional categories in the group of down‐regulated genes
FIGURE S8 GO terms enriched in up‐regulated genes in miR399 OE plants compared to wild‐type plants were generated by AgriGO according to molecular function. Singular enrichment analysis (SEA), with the Rice Gramene gene model as a reference, by AgriGO analysis (Hypergeometic test; Hochberg FDR; p ≤ .05; “10” as minimum number of mapping entries). Boxes in the graph show GO terms, term definition, and statistical information. The significant terms are marked with colour, while nonsignificant terms are shown in white boxes. The colour intensity of a box positively correlates with the enrichment level of the term
FIGURE S9 GO terms enriched in down‐regulated genes in miR399 OE plants compared to wild‐type plants were generated by AgriGO according to molecular function. Singular enrichment analysis (SEA), with the Rice Gramene gene model as a reference, by AgriGO analysis (Hypergeometric test; Hochberg FDR; p ≤ .05; “10” as minimum number of mapping entries). Boxes in the graph show GO terms, term definition, and statistical information. The significant terms are marked with colour, while nonsignificant terms are shown in white boxes. The colour intensity of a box positively correlates with the enrichment level of the term
FIGURE S10 Heat map visualization of differentially expressed genes in miR399 OE plants compared to wild‐type (WT) plants in the following subcategories: protein phosphorylation and ATP binding (left panel), transcriptional regulation (middle panel), and defence (right panel). Expression patterns are based on the expression levels obtained by RNA‐Seq (miR399 OE lines L4, L20 versus WT). Purple represents up‐regulated genes (log2 fold change ≥ 0.5) and green represents down‐regulated genes (log2 fold change ≤ −0.5). Data are means (n = 3). The full gene names and locus ID are listed in Tables S3, S4, and S5
TABLE S1 Determination of transgene copy number in miR399 OE plants by quantitative PCR using sucrose phosphate synthase gene (OsSPS) as endogenous monocopy reference gene (Ding et al., 2004). Primers sequences are shown in Table S6 (AtmiR399f and tNOS)
TABLE S2 List of up‐regulated and down‐regulated genes in miR399 OE plants compared to wild‐type plants. Significantly misregulated genes with p ≤ .05, FDR ≤ 0.05 (up‐regulated, log2 fold change ≥ 0.5; down‐regulated, log2 fold change ≤ −0.5)
TABLE S3 List of genes that are up‐regulated and down‐regulated in miR399 OE plants compared to wild‐type plants in the category of protein phosphorylation and ATP binding
TABLE S4 List of genes that are up‐regulated and down‐regulated in miR399 OE plants compared to wild‐type plants in the category of transcriptional regulation
TABLE S5 List of genes that are up‐regulated and down‐regulated in miR399 OE plants compared to wild‐type plants in the category of defence
TABLE S6 Sequences of oligonucleotides used for quantitative reverse transcription PCR analysis, northern blot, and transgene copy number determination
METHODS S1 Experimental procedures
ACKNOWLEDGEMENTS
M.B.P. was funded by a “la Caixa” scholarship for PhD studies in Spanish universities. We thank J. Freixanet for assistance in parts of this work. This research was supported by the Ministerio de Ciencia, Innovación y Universidades (MCIU)‐Agencia Estatal de Investigación (AEI) y el Fondo Europeo de Desarrollo Regional (FEDER) (BIO2015‐67212‐R and RTI2018‐101275‐B‐100) and the Academia Sinica, Taiwan (AS‐SS‐106‐03‐3). We also acknowledge support from the CERCA Programme (“Generalitat de Catalunya”) and the “Severo Ochoa Programme for Centres of Excellence in R&D” 2016‐2019 (SEV‐2015‐0533). The authors declare that there is no conflict of interest regarding the publication of this paper.
Campos‐Soriano L, Bundó M, Bach‐Pages M, Chiang S‐F, Chiou T‐J, San Segundo B. Phosphate excess increases susceptibility to pathogen infection in rice. Molecular Plant Pathology. 2020;21:555–570. 10.1111/mpp.12916
Lidia Campos‐Soriano and Mireia Bundó contributed equally to this work.
Funding information
M.B.P. was funded by a “la Caixa” scholarship for PhD studies in Spanish universities. This research was supported by the Ministerio de Ciencia, Innovación y Universidades (MCIU)‐Agencia Estatal de Investigación (AEI) y el Fondo Europeo de Desarrollo Regional (FEDER) (BIO2015‐67212‐R and RTI2018‐101275‐B‐100) and the Academia Sinica, Taiwan (AS‐SS‐106‐03‐3). We also acknowledge support from the CERCA Programme (“Generalitat de Catalunya”) and the “Severo Ochoa Programme for Centres of Excellence in R&D” 2016‐2019 (SEV‐2015‐0533).
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are openly available in the NCBI‐GEO repository at https://www.ncbi.nlm.nih.gov/geo/, reference number http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE137735.
REFERENCES
- Abbas, K.G. , Vogiatzaki, E. , Glauser, G. and Poirier, Y. (2016) Phosphate deficiency induces the jasmonate pathway and enhances resistance to insect herbivory. Plant Physiology, 171, 632–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal, G.K. , Rakwal, R. , Jwa, N.S. and Agrawal, V.P. (2001) Signalling molecules and blast pathogen attack activates rice OsPR1a and OsPR1b genes: a model illustrating components participating during defence/stress response. Plant Physiology and Biochemistry, 39, 1095–1103. [Google Scholar]
- Almagro, L. , Gómez Ros, L.V. , Belchi‐Navarro, S. , Bru, R. , Ros Barceló, A. and Pedreño, M.A. (2009) Class III peroxidases in plant defence reactions. Journal of Experimental Botany, 60, 377–390. [DOI] [PubMed] [Google Scholar]
- Ames, B.N. (1966) Assay of inorganic phosphate, total phosphate and phosphatases. Methods in Enzymology, 8, 115–118. [Google Scholar]
- Aung, K. , Lin, S.I. , Wu, C.C. , Huang, Y.T. , Su, C.L. and Chiou, T.J. (2006) pho2, a phosphate overaccumulator, is caused by a nonsense mutation in a microRNA399 target gene. Plant Physiology, 141, 1000–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek, D. , Chun, H.J. , Kang, S. , Shin, G. , Park, S.J. , Hong, H. et al (2016) A role for Arabidopsis miR399f in salt, drought, and ABA signaling. Molecules and Cells, 39, 111–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek, D. , Chun, H.J. , Yun, D. and Kim, M.C. (2017) Cross‐talk between phosphate starvation and other environmental stress signaling pathways in plants. Molecules and Cells, 40, 697–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balagué, C. , Gouget, A. , Bouchez, O. , Souriac, C. , Haget, N. , Boutet‐Mercey, S. et al (2017) The Arabidopsis thaliana lectin receptor kinase LecRK‐I.9 is required for full resistance to Pseudomonas syringae and affects jasmonate signaling. Molecular Plant Pathology, 8, 937–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldrich, P. and San Segundo B. (2016) MicroRNAs in rice innate immunity. Rice, 9, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldrich, P. , Campo, S. , Wu, M.T. , Liu, T.Z. , Hsing, Y.I.C. and San, S.B. (2015) MicroRNA‐mediated regulation of gene expression in the response of rice plants to fungal elicitors. RNA Biology, 12, 847–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballini, E. , Nguyen, T.T.T. and Morel, J.B. (2013) Diversity and genetics of nitrogen‐induced susceptibility to the blast fungus in rice and wheat. Rice, 6, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigeard, J. , Colcombet, J. and Hirt, H. (2015) Signaling mechanisms in pattern‐triggered immunity (PTI). Molecular Plant, 8, 521–539. [DOI] [PubMed] [Google Scholar]
- Boller, T. and He, S.Y. (2009) Innate immunity in plants: An arms race between pattern recognition receptors in plants and effectors in microbial pathogens. Science, 324, 742–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodersen, P. , Sakvarelidze‐Achard, L. , Bruun‐Rasmussen, M. , Dunoyer, P. , Yamamoto, Y.Y. , Sieburth, L. et al (2008) Widespread translational inhibition by plant miRNAs and siRNAs. Science, 320, 1185–1190. [DOI] [PubMed] [Google Scholar]
- Campo, S. , Peris‐Peris, C. , Siré, C. , Moreno, A.B. , Donaire, L. , Zytnicki, M. et al (2013) Identification of a novel microRNA (miRNA) from rice that targets an alternatively spliced transcript of the Nramp6 (Natural resistance‐associated macrophage protein 6) gene involved in pathogen resistance. New Phytologist, 199, 212–227. [DOI] [PubMed] [Google Scholar]
- Campos‐Soriano, L. , Valé, G. , Luppotto, E. and San Segundo, B. (2013) Investigation of rice blast development in susceptible and resistant rice cultivars using a gfp‐expressing Magnaporthe oryzae isolate. Plant Pathology, 62, 1030–1037. [Google Scholar]
- Cao, Y. , Tanaka, K. , Nguyen, C.T. and Stacey, G. (2014) Extracellular ATP is a central signaling molecule in plant stress responses. Current Opinion in Plant Biology, 20, 82–87. [DOI] [PubMed] [Google Scholar]
- Castrillo, G. , Teixeira, P.J.P.L. , Paredes, S.H. , Law, T.F. , De Lorenzo, L. , Feltcher, M.E. et al (2017) Root microbiota drive direct integration of phosphate stress and immunity. Nature, 543, 513–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook, D.E. , Mesarich, C.H. and Thomma, B.P. (2015) Understanding plant immunity as a surveillance system to detect invasion. Annual Review of Phytopathology, 53, 541–563. [DOI] [PubMed] [Google Scholar]
- Couto, D. and Zipfel, C. (2016) Regulation of pattern recognition receptor signalling in plants. Nature Reviews Immunology, 16, 537–552. [DOI] [PubMed] [Google Scholar]
- Chen, X. (2009) Small RNAs and their roles in plant development. Annual Review of Cell and Developmental Biology, 25, 21–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien, P.S. , Chiang, C.B. , Wang, Z. and Chiou, T.J. (2017) MicroRNA‐mediated signaling and regulation of nutrient transport and utilization. Current Opinion in Plant Biology, 39, 73–79. [DOI] [PubMed] [Google Scholar]
- Chiou, T.J. , Aung, K. , Lin, S.I. , Wu, C.C. , Chiang, S.F. and Su, C.L. (2006) Regulation of phosphate homeostasis by microRNA in Arabidopsis. The Plant Cell, 18, 412–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai, X. , Wang, Y. and Zhang, W.H. (2016) OsWRKY74, a WRKY transcription factor, modulates tolerance to phosphate starvation in rice. Journal of Experimental Botany, 67, 947–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean, R. , Van Kan, J.A.L. , Pretorius, Z.A. , Hammond‐Kosack, K.E. , Di Pietro, A. , Spanu, P.D. et al (2012) The top 10 fungal pathogens in molecular plant pathology. Molecular Plant Pathology, 13, 414–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delteil, A. , Gobbato, E. , Cayrol, B. , Estevan, J. , Michel‐Romiti, C. , Dievart, A. et al (2016) Several wall‐associated kinases participate positively and negatively in basal defence against rice blast fungus. BMC Plant Biology, 16, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, J. , Jia, J. , Yang, L. , Wen, H. , Zhang, C. , Liu, W. , et al (2004) Validation of a rice specific gene, Sucrose Phosphate Synthase, used as the endogenous reference gene for qualitative and real-time quantitative PCR detection of transgenes. Journal of Agricultural and Food Chemistry, 52, 3372–3377. [DOI] [PubMed] [Google Scholar]
- Dong, Z. , Li, W. , Liu, J. , Li, L. , Pan, S. , Liu, S. et al (2019) The rice phosphate transporter protein OsPT8 regulates disease resistance and plant growth. Scientific Reports, 9, 5408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du, Z. , Zhou, X. , Ling, Y. , Zhang, Z. and Su, Z. (2010) agriGO: a GO analysis toolkit for the agricultural community. Nucleic acids research, 38(suppl_2), W64–W70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabianska, I. , Gerlach, N. , Almario, J. and Bucher, M. (2019) Plant‐mediated effects of soil phosphorus on the fungal microbiota in Arabidopsis thaliana . New Phytologist, 221, 2123–2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel, O.M. , Salas‐González, I. , Castrillo, G. , Spaepen, S. , Law, T.F. , Pereira LimaTeixeira, P.J. et al (2019) The effects of soil phosphorus content on plant microbiota are driven by the plant phosphate starvation response. PLoS Biology, 17, e3000534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gho, Y.S. , An, G. , Park, H.M. and Jung, K.H. (2018) A systemic view of phosphate starvation‐responsive genes in rice roots to enhance phosphate use efficiency in rice. Plant Biotechnology Reports, 12, 249–264. [Google Scholar]
- Ham, B. , Chen, J. , Yan, Y. and Lucas, W.J. (2018) Insights into plant phosphate sensing and signalling. Current Opinion in Biotechnology, 49, 1–9. [DOI] [PubMed] [Google Scholar]
- Hinsinger, P. , Betencourt, E. , Bernard, L. , Brauman, A. , Plassard, C. , Shen, J. et al (2011) P for two, sharing a scarce resource: Soil phosphorus acquisition in the rhizosphere of intercropped species. Plant Physiology, 156, 1078–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh, L.C. , Lin, S.I. , Shih, A.C.C. , Chen, J.W. , Lin, W.Y. , Tseng, C.Y. et al (2009) Uncovering small RNA‐mediated responses to phosphate deficiency in Arabidopsis by deep sequencing. Plant Physiology, 151, 2120–2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, B. , Zhu, C. , Li, F. , Tang, J. , Wang, Y. , Lin, A. et al (2011) LEAF TIP NECROSIS1 plays a pivotal role in the regulation of multiple phosphate starvation responses in rice. Plant Physiology, 156, 1101–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, B. , Wang, W. , Deng, K. , Li, H. , Zhang, Z. , Zhang, L. et al (2015) MicroRNA399 is involved in multiple nutrient starvation responses in rice. Frontiers in Plant Science, 6, 188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, H. , Nguyen Thi Thu, T. , He, X. , Gravot, A. , Bernillon, S. , Ballini, E. et al (2017) Increase of fungal pathogenicity and role of plant glutamine in nitrogen-induced susceptibility (NIS) to rice blast. Frontiers in Plant Science, 8, 265 10.3389/fpls.2017.00265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jewell, J.B. , Sowders, J.M. , He, R. , Willis, M.A. , Gang, D.R. and Tanaka, K. (2019) Extracellular ATP shapes a defence‐related transcriptome both independently and along with other defence signaling pathways. Plant Physiology, 179, 1144–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong, D. and Green, P. (2013) The role of rice microRNAs in abiotic stress responses. Journal of Plant Biology, 56, 187–197. [Google Scholar]
- Jones, J. and Dangl, J. (2006) The plant immune system. Nature, 444, 323–329. [DOI] [PubMed] [Google Scholar]
- Lamari, L. (2008) Assess 2.0: Image Analysis Software for Plant Disease Quantification. St Paul, MN: APS Press. [Google Scholar]
- Li, Y. , Zhang, Q. , Zhang, J. , Wu, L. , Qi, Y. and Zhou, J.M. (2010) Identification of microRNAs involved in pathogen‐associated molecular pattern‐triggered plant innate immunity. Plant Physiology, 152, 2222–2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Z.Y. , Xia, J. , Chen, Z. , Yu, Y. , Li, Q.F. , Zhang, Y.C. et al (2016) Large scale rewiring of innate immunity circuitry and microRNA regulation during initial rice blast infection. Scientific Reports, 6, 25493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, G. , Ai, Q. and Yu, D. (2015) Uncovering miRNAs involved in cross‐talk between nutrient deficiencies in Arabidopsis. Scientific Reports, 5, 11813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, T.Y. , Huang, T.K. , Tseng, C.Y. , Lai, Y.S. , Lin, S.I. , Lin, W.Y. et al (2012) PHO2‐dependent degradation of PHO1 modulates phosphate homeostasis in Arabidopsis. The Plant Cell, 24, 2168–2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llave, C. , Xie, Z. , Kasschau, K.D. and Carrington, J.C. (2002) Cleavage of Scarecrow‐like mRNA targets directed by a class of Arabidopsis miRNA. Science, 297, 2053–2056. [DOI] [PubMed] [Google Scholar]
- Lu, Y.T. , Li, M.Y. , Cheng, K.T. , Tan, C.M. , Su, L.W. , Lin, W.Y. et al (2014) Transgenic plants that express the phytoplasma effector SAP11 show altered phosphate starvation and defence responses. Plant Physiology, 164, 1456–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Midoh, N. and Iwata, M. (1996) Cloning and characterization of a probenazole-inducible gene for an intracellular pathogenesis-related protein in rice. Plant and Cell Physiology, 37(1), 9–18. [DOI] [PubMed] [Google Scholar]
- Miedes, E. , Vanholme, R. , Boerjan, W. and Molina, A. (2014) The role of the secondary cell wall in plant resistance to pathogens. Frontiers in Plant Science, 5, 358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misson, J. , Raghothama, K.G. , Jain, A. , Jouhet, J. , Block, M.A. , Bligny, R. et al (2005) A genome‐wide transcriptional analysis using Arabidopsis thaliana Affymetrix gene chips determined plant responses to phosphate deprivation. Proceedings of the National Academy of Sciences of the United States of America, 102, 11934–11939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasir, F. , Tian, L. , Chang, C. , Li, X. , Gao, Y. , Tran, L.S.P. et al (2018) Current understanding of pattern‐triggered immunity and hormone‐mediated defence in rice in response to Magnaporthe oryzae infection. Seminars in Cell and Developmental Biology, 83, 95–105. [DOI] [PubMed] [Google Scholar]
- Navarro, L. , Dunoyer, P. , Jay, F. , Arnold, B. , Dharmasiri, N. , Estelle, M. et al (2006) A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science, 312, 436–439. [DOI] [PubMed] [Google Scholar]
- Oono, Y. , Kawahara, Y. , Yazawa, T. , Kanamori, H. , Kuramata, M. , Yamagata, H. et al (2013) Diversity in the complexity of phosphate starvation transcriptomes among rice cultivars based on RNA‐Seq profiles. Plant Molecular Biology, 83, 523–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul, S. , Datta, S. and Datta, K. (2015) MiRNA regulation of nutrient homeostasis in plants. Frontiers in Plant Science, 6, 232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phukan, U.J. , Jeena, G.S. and Shukla, R.K. (2016) WRKY Transcription factors: molecular regulation and stress responses in plants. Frontiers in Plant Science, 7, 760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puga, M.I. , Rojas‐Triana, M. , de Lorenzo, L. , Leyva, A. , Rubio, V. and Paz‐Ares, J. (2017) Novel insights in the regulation of Pi starvation responses in plants: facts and promises. Current Opinion in Plant Biology, 39, 40–49. [DOI] [PubMed] [Google Scholar]
- Qi, M. and Yang, Y. (2002) Quantification of Magnaporthe grisea during infection of rice plants using real‐time polymerase chain reaction and northern blot/phosphoimaging analyses. Phytopathology, 92, 870–876. [DOI] [PubMed] [Google Scholar]
- Sallaud, C. , Meynard, D. , van Boxtel, J. , Gay, C. , Bès, M. , Brizard, J.P. et al (2003) Highly efficient production and characterization of T‐DNA plants for rice (Oryza sativa L.) functional genomics. Theoretical and Applied Genetics, 106, 1396–1408. [DOI] [PubMed] [Google Scholar]
- Sauter, M. (2015) Phytosulfokine peptide signaling. Journal of Experimental Botany, 66, 5161–5169. [DOI] [PubMed] [Google Scholar]
- Secco, D. , Jabnoune, M. , Walker, H. , Shou, H. , Wu, P. , Poirier, Y. et al (2013) Spatio‐temporal transcript profiling of rice roots and shoots in response to phosphate starvation and recovery. The Plant Cell, 25, 4285–4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimono, M. , Sugano, S. , Nakayama, A. , Jiang, C.J. , Ono, K. , Toki, S. et al (2007) Rice WRKY45 plays a crucial role in benzothiadiazole‐inducible blast resistance. The Plant Cell, 19, 2064–2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivaprasad, P.V. , Chen, H.M. , Patel, K. , Bond, D.M. , Santos, B.A.C.M. and Baulcombe, D.C. (2012) A MicroRNA superfamily regulates nucleotide binding site‐leucine‐rich repeats and other mRNAs. The Plant Cell, 24, 859–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, L. and Liu, D. (2015) Ethylene and plant responses to phosphate deficiency. Frontiers in Plant Science, 6, 796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, X. , Li, Y. , Cao, X. and Qi, Y. (2019) MicroRNAs and their regulatory roles in plant–environment interactions. Annual Review of Plant Biology, 70, 489–525. [DOI] [PubMed] [Google Scholar]
- Supek, F. , Bošnjak, M. , Škunca, N. and Šmuc, T. (2011) REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS ONE, 6(7), e21800 10.1371/journal.pone.0021800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka, K. , Choi, J. , Cao, Y. and Stacey, G. (2014) Extracellular ATP acts as a damage‐associated molecular pattern (DAMP) signal in plants. Frontiers in Plant Science, 5, 446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi, D. , Zhang, T. , Koo, A.J. , Stacey, G. and Tanaka, K. (2018) Extracellular ATP acts on jasmonate signaling to reinforce plant defence. Plant Physiology, 176, 511–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veresoglou, S. , Barto, E. , Menexes, G. and Rillig, M.C. (2013) Fertilization affects severity of disease caused by fungal plant pathogens. Plant Pathology, 62, 961–969. [Google Scholar]
- Versaw, W. and Harrison, M.J. (2002) A chloroplast phosphate transporter, PHT2;1, influences allocation of phosphate within the plant and phosphate‐starvation responses. The Plant Cell, 14, 1751–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X. , Zafian, P. , Choudhary, M. and Lawton, M. (1996) The PR5K receptor protein kinase from Arabidopsis thaliana is structurally related to a family of plant defence proteins. Proceedings of the National Academy of Sciences of the United States of America, 93, 2598–2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H. , Xu, Q. , Kong, Y.H. , Chen, Y. , Duan, J.Y. , Wu, W.H. et al (2014) Arabidopsis WRKY45 transcription factor activates PHOSPHATE TRANSPORTER1;1 expression in response to phosphate starvation. Plant Physiology, 164, 2020–2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasaki, J. , Shinano, T. , Onishi, K. , Yonetani, R. , Yazaki, J. , Fujii, F. et al (2006) Transcriptomic analysis indicates putative metabolic changes caused by manipulation of phosphorus availability in rice leaves. Journal of Experimental Botany, 57, 2049–2059. [DOI] [PubMed] [Google Scholar]
- Wei, T. , Ou, B. , Li, J. , Zhao, Y. , Guo, D. , Zhu, Y. et al (2013) Transcriptional profiling of rice early response to Magnaporthe oryzae identified OsWRKYs as important regulators in rice blast resistance. PLoS ONE, 8, e59720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, R. and Talbot, N. (2009) Under pressure: investigating the biology of plant infection by Magnaporthe oryzae . Nature Reviews Microbiology, 7, 185–195. [DOI] [PubMed] [Google Scholar]
- Yamane, H. (2013) Biosynthesis of phytoalexins and regulatory mechanisms of it in rice. Bioscience, Biotechnology, and Biochemistry, 77, 1141–1148. [DOI] [PubMed] [Google Scholar]
- Yang, L. , Ding, J. , Zhang, C. , Jia, J. , Weng, H. , Liu, W. et al (2005) Estimating the copy number of transgenes in transformed rice by real‐time quantitative PCR. Plant Cell Reports, 23, 759–763. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
FIGURE S1 Treatment of rice plants (Oryza sativa “Tainung 67”) with different Pi concentrations. (a) Phenotype of plants that have been grown for 15 days under low‐, sufficient‐, or high‐Pi conditions. (b) Shoot fresh weight and dry weight of rice plants grown for 15 days under different Pi regimes. Data represent means of four biological replicates, each one from a pool of three different plants, ±SEM (one‐way analysis of variance and Tukey’s test, *p < .05, **p < .005)
FIGURE S2 Effect of treatment with low‐ and high‐Pi supply in the susceptible rice cultivars Nipponbare, Maratelli, Bomba, Vialone Nano, and the resistant cultivar Saber (harbouring the Pib resistance gene). Left panels, appearance of plants that have been grown for 15 days under different Pi regimes. Right panels, shoot fresh weight and dry weight of plants shown in left panels. Data represent means of four biological replicates, each one from a pool of three different plants, ±SEM (one‐way analysis of variance and Tukey’s test, *p < .05, **p < .005, ***p < .0001)
FIGURE S3 Effect of Pi treatment on the expression of Pi starvation‐responsive genes in rice (cv. Tainung 67). The accumulation of miR399f precursor transcripts and the miR399 target gene OsPHO2 was determined by quantitative reverse transcription PCR. The expression values were normalized to Ubiquitin 1 gene. Bars show the means of three biological replicates, with three different plants in each one, and SEM. Comparisons have been made relative to the sufficient Pi condition (t test, *p < .05)
FIGURE S4 Characterization of miR399f overexpressing (OE) rice plants. (a) Accumulation of miR399f precursor and OsPHO2 transcripts in miR399 OE lines (T0 generation). (b) Leaf tip necrosis in leaves of miR399 OE rice plants grown under controlled greenhouse conditions for 2 months
FIGURE S5 Gene ontology (GO) terms enriched in up‐regulated genes in miR399OE plants compared to wild‐type plants were generated by AgriGO according to biological process. Singular enrichment analysis (SEA), with the Rice Gramene gene model as a reference, by AgriGO analysis (hypergeometic test; Hochberg false discovery rate (FDR); p ≤ .05; “10” as minimum number of mapping entries). Boxes in the graph show GO terms, term definition, and statistical information. The significant terms are marked with colour, while nonsignificant terms are shown in white boxes. The colour intensity of a box positively correlates with the enrichment level of the term
FIGURE S6 GO terms enriched in down‐regulated genes in miR399 OE plants compared to wild‐type plants were generated by AgriGO according to biological process. Singular enrichment analysis (SEA), with the Rice Gramene gene model as a reference, by AgriGO analysis (Hypergeometic test; Hochberg FDR; p ≤ .05; “10” as minimum number of mapping entries). Boxes in the graph show GO terms, term definition, and statistical information. The significant terms are marked with colour, while nonsignificant terms are shown in white boxes. The colour intensity of a box positively correlates with the enrichment level of the term
FIGURE S7 GO terms enriched in miR399 OE plants compared to wild‐type plants were generated by AgriGO according to cellular component. (a) Functional categories in the group of up‐regulated genes. (b) Functional categories in the group of down‐regulated genes
FIGURE S8 GO terms enriched in up‐regulated genes in miR399 OE plants compared to wild‐type plants were generated by AgriGO according to molecular function. Singular enrichment analysis (SEA), with the Rice Gramene gene model as a reference, by AgriGO analysis (Hypergeometic test; Hochberg FDR; p ≤ .05; “10” as minimum number of mapping entries). Boxes in the graph show GO terms, term definition, and statistical information. The significant terms are marked with colour, while nonsignificant terms are shown in white boxes. The colour intensity of a box positively correlates with the enrichment level of the term
FIGURE S9 GO terms enriched in down‐regulated genes in miR399 OE plants compared to wild‐type plants were generated by AgriGO according to molecular function. Singular enrichment analysis (SEA), with the Rice Gramene gene model as a reference, by AgriGO analysis (Hypergeometric test; Hochberg FDR; p ≤ .05; “10” as minimum number of mapping entries). Boxes in the graph show GO terms, term definition, and statistical information. The significant terms are marked with colour, while nonsignificant terms are shown in white boxes. The colour intensity of a box positively correlates with the enrichment level of the term
FIGURE S10 Heat map visualization of differentially expressed genes in miR399 OE plants compared to wild‐type (WT) plants in the following subcategories: protein phosphorylation and ATP binding (left panel), transcriptional regulation (middle panel), and defence (right panel). Expression patterns are based on the expression levels obtained by RNA‐Seq (miR399 OE lines L4, L20 versus WT). Purple represents up‐regulated genes (log2 fold change ≥ 0.5) and green represents down‐regulated genes (log2 fold change ≤ −0.5). Data are means (n = 3). The full gene names and locus ID are listed in Tables S3, S4, and S5
TABLE S1 Determination of transgene copy number in miR399 OE plants by quantitative PCR using sucrose phosphate synthase gene (OsSPS) as endogenous monocopy reference gene (Ding et al., 2004). Primers sequences are shown in Table S6 (AtmiR399f and tNOS)
TABLE S2 List of up‐regulated and down‐regulated genes in miR399 OE plants compared to wild‐type plants. Significantly misregulated genes with p ≤ .05, FDR ≤ 0.05 (up‐regulated, log2 fold change ≥ 0.5; down‐regulated, log2 fold change ≤ −0.5)
TABLE S3 List of genes that are up‐regulated and down‐regulated in miR399 OE plants compared to wild‐type plants in the category of protein phosphorylation and ATP binding
TABLE S4 List of genes that are up‐regulated and down‐regulated in miR399 OE plants compared to wild‐type plants in the category of transcriptional regulation
TABLE S5 List of genes that are up‐regulated and down‐regulated in miR399 OE plants compared to wild‐type plants in the category of defence
TABLE S6 Sequences of oligonucleotides used for quantitative reverse transcription PCR analysis, northern blot, and transgene copy number determination
METHODS S1 Experimental procedures
Data Availability Statement
The data that support the findings of this study are openly available in the NCBI‐GEO repository at https://www.ncbi.nlm.nih.gov/geo/, reference number http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE137735.


