Abstract
We use a hybrid fluorescence spectroscopic toolkit to monitor T4 Lysozyme (T4L) in action by unraveling the kinetic and dynamic interplay of the conformational states. In particular, by combining single-molecule and ensemble multiparameter fluorescence detection, EPR spectroscopy, mutagenesis, and FRET-positioning and screening, and other biochemical and biophysical tools, we characterize three short-lived conformational states over the ns-ms timescale. The use of 33 FRET-derived distance sets, to screen available T4L structures, reveal that T4L in solution mainly adopts the known open and closed states in exchange at 4 µs. A newly found minor state, undisclosed by, at present, more than 500 crystal structures of T4L and sampled at 230 µs, may be actively involved in the product release step in catalysis. The presented fluorescence spectroscopic toolkit will likely accelerate the development of dynamic structural biology by identifying transient conformational states that are highly abundant in biology and critical in enzymatic reactions.
Subject terms: Biophysics, Biological fluorescence, Kinetics, Single-molecule biophysics
T4 Lysozyme (T4L) is a model protein whose structure is extensively studied. Here the authors combine single-molecule and ensemble FRET measurements, FRET-positioning and screening and EPR spectroscopy to study the structural dynamics of T4L and describe its conformational landscape during the catalytic cycle by an extended Michaelis–Menten mechanism and identify an excited conformational state of the enzyme.
Introduction
Enzymes adopt distinct conformational states during catalysis1,2, where transiently populated (“excited”) states are often of critical importance in the enzymatic cycle. These states are short-lived and therefore “hidden” to many experimental techniques. Classical structural biology methods often struggle to fully capture enzymes during catalytic action because the conformational rearrangements often span several decades in time (ns-ms)3–8. Hence, there is an urgent need to develop experimental and analysis methods to overcome this challenge. Recently, we demonstrated by simulated experiments that a new analysis toolkit (“FRET on rails”) combined with molecular simulations can resolve short-lived conformational states of proteins9.
Here, we apply and extend the fluorescence analysis toolkit9,10, developed for dynamic structural biology, to interrogate the catalytic cycle of an enzyme11. In particular, the analysis (1) captures an excited, short-lived state and (2) identifies its potential relevance in the enzyme’s catalytic cycle. The presented approach may serve as a blueprint for future enzymologic studies with the well-established single-molecule multiparameter fluorescence detection (MFD) experiments in that it enables detecting hidden states by the unique time-resolution (picoseconds) and sensitivity (single-molecule) of fluorescence.
We use lysozyme (T4L) of the bacteriophage T4 as development platform and probe its conformational dynamics and structural features. Structurally, T4L12 consists of two interrelated subdomains, the N-terminal subdomain (NTsD) and the C-terminal subdomain (CTsD), differing in their folding behavior and stability13. A long α-helix (helix c) links the two subdomains (Fig. 1a). To date, more than 500 structural models of T4L are available within the Protein Data Bank (PDB). In this ensemble, T4L adopts several opening angles corresponding to a classic hinge-bending motion of the NTsD with respect to the CTsD. The enzymatic function of T4L is to cleave the glycosidic bond between N-acetylmuramic acid and N-acetylglucosamine of the saccharides of the bacterial cell wall14.
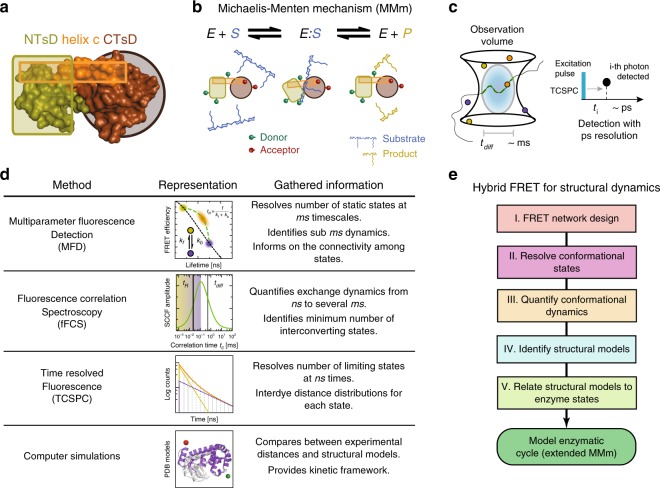
Fig. 1. Schematic of the high precision FRET and fluctuation analysis toolkit.
a Subdomain architecture of Phage T4 Lysozyme (T4L). b Upon cleavage of its substrate peptidoglycan (blue), the N-terminal lobe (green) and the C-terminal lobe (brown) of T4L exhibit a change of closure, which can be observed via the induced change in the FRET indicators14,15. c In MFD-experiments of freely diffusing single molecules, the emitted fluorescent photons (“bursts”) are detected with ps resolution (with respect to the exciting laser pulse) during the diffusion (on the ms time scale) of the molecule through the observation volume (diffusion time, tdiff). d The labeled T4L molecules are studied with different experimental methods. In single-molecule multiparameter fluorescence detection (MFD) experiments, the fluorescence bursts—averaged over ms—are analyzed e.g. with respect to their fluorescence lifetime 〈τD(A)〉F or FRET-efficiency E and displayed in multidimensional frequency histograms (2D MFD-histogram). Molecules that adopt a stable conformation during burst duration are located along the static FRET-line (black) (Supplementary Methods). Assuming that the two limiting states (yellow and blue) exchange on timescales faster than ms with exchange rate constants kf and kb, we find only a single population (orange) shifted towards a longer fluorescence lifetime that is located on the dynamic FRET-line (green) connecting these two limiting states. Thus, FRET-lines serve as a visual guide to interpret 2D MFD-histograms, with deviations from the static FRET-line being indicative for the dynamic averaging and dynamics at the sub-ms and ms timescales. Filtered fluorescence correlation spectroscopy (fFCS) computes the species-specific cross-correlation function (sCCF (green), Supplementary Methods). The observed anti-correlation reveals a characteristic relaxation time tR related to the inverse of the sum of the exchange rate constants, kf and kb. In eTCSPC, the distribution of the fluorescent photons (yellow/blue—individual states, orange—mixture) is detected with respect to an excitation pulse with ps resolution to reveal populations stable on the ns timescale (Supplementary Methods). Finally, in molecular simulations, the experimental results are compared to available structural models. e Flowchart for the hybrid FRET toolkit for determining structural dynamics. Based on a network of FRET variants, the conformational states and their exchange dynamics are determined, which are then used to identify the structural models. T4L variants with mutations altering their enzymatic activity relate the structural models to enzymatic states. Based on the gathered information, the enzymatic cycle can be modeled.
T4L in solution is thought to adopt conformations that are open to various degrees, and a covalent adduct of the protein and its processed enzymatic product can crystallize in a closed conformation14–16. Therefore, T4L is thought to follow a classical Michaelis–Menten mechanism (MMm) characterized as a two-state system (Fig. 1b). Here, an open and closed conformational state fulfils unique functions of substrate binding and substrate cleavage, respectively17. In a MMm, the product dissociates stochastically from the enzyme. For other enzymes, e.g. the Horseradish peroxidase18, an “active” product release state was identified. Recent experimental findings for T4L suggest the involvement of more than two states in catalysis19,20, where the turnover rate was estimated between 10 and 50 ms20–23, while the conformational dynamics fell within the ns to sub-ms range4,15,20–29. Such complex cases, with distinct interconverting conformational states, open additional reaction paths and yield disperse kinetics30.
For a full description of an enzymological cycle, the number of enzymatic states, their connectivity, the conformational structures of the states, and the states’ chemical function have to be unraveled. Technically, we achieve these objectives by a hybrid approach combining classic biochemical methods (mutagenesis & HPLC), probe-based spectroscopy, and molecular simulations. Förster resonance energy transfer (FRET) and electron paramagnetic resonance (EPR) spectroscopy probe distances between bioorthogonally introduced probes through dipolar coupling. In FRET spectroscopy, the coupling is measured between a donor (D) and acceptor (A) fluorophore.
In confocal MFD single-molecule FRET (smFRET) experiments, freely diffusing molecules are repeatedly excited by a pulsed light source, and the emitted fluorescence photon is detected with picosecond time-resolution by time-correlated single photon counting (TCSPC) for several milliseconds per molecule (diffusion time, tdiff)31 (Fig. 1c). smFRET experiments are ideal to study kinetics because no sophisticated strategies are necessary to synchronize molecules prior to the analysis. Consequently, it is possible to probe reliably protein kinetics over seven decades in time (sub ns-ms).
Distinct features of photon streams are highlighted by different representations (Fig. 1d). (1) A MFD-histogram is particularly valuable to reveal the number of states, identify dynamics, and to inform on state connectivities. A MFD-histogram is generated by analyzing two complementary FRET-indicators, the average intensity-based FRET-efficiency, E, and the fluorescence-averaged donor lifetime in the presence of acceptor, 〈τD(A)〉F, for individual single-molecule events31–33. (2) Filtered fluorescence correlation spectroscopy (fFCS) quantifies exchange dynamics among the states by determining relaxation times34,35. (3) The analysis of fluorescence decays reveals populations of states and equilibrium distance distributions. (4) Finally, these experimental distances can be translated to structural models by molecular simulations10,36,37.
Following the concepts that were established for simulated data to resolve the structure and dynamics of proteins by integrative studies with FRET experiments8,9 (Fig. 1e and more detailed in Supplementary Fig. 1), we start out with a systematic design of a FRET network for T4L to simultaneously monitor its dynamics and structural features. In step II, we use a combination of MFD to resolve the conformational states stable at the ns timescales. In step III, we quantify the conformational dynamics by employing fFCS and Monte Carlo simulations to resolve the connectivity of the conformational states. Following this, we perform a statistical analysis to further substantiate the existence of a hidden conformational state of T4L that was clearly identified above. In step IV, we identify structural models by using three distinct distance sets to screen an ensemble of structural models and to compare our identified states with models in the PDB. In the final step V, we derive an experimental energy landscape of T4L’s enzymatic cleavage cycle, based on shifting equilibrium, by mutating key active site residues that mimic functional enzyme states at various steps during substrate hydrolysis. Overall, our results demonstrate the potential of fluorescence spectroscopy to go beyond traditional experimental methods for obtaining a dynamic structural picture of enzymes in action. The existence of the identified hidden/excited conformational state is also corroborated by other analytical tools such as chromatography and EPR spectroscopy.
Results
Detecting T4L’s states by MFD
In our smFRET-experiments, we monitor the distance between a donor (D) and acceptor (A) attached to specific amino acids of a T4L variant (see Methods). We designed a network of 33 distinct T4L variants to probe hinge-bending motions of T4L from different spatial directions (Fig. 2a) that cover the whole protein.
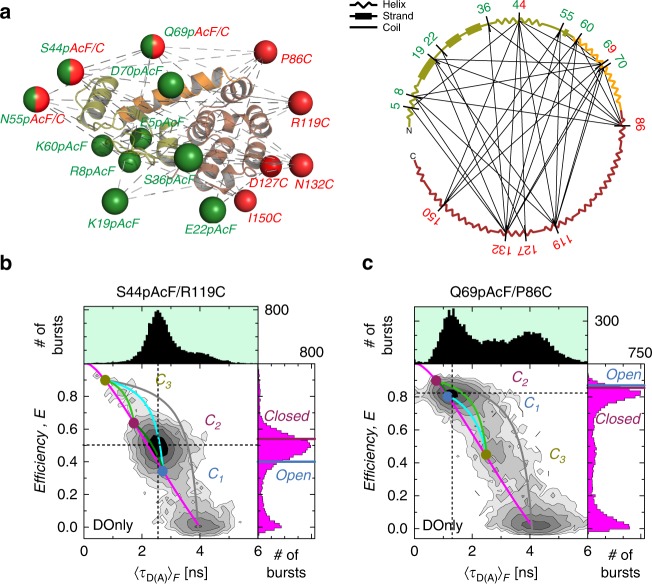
Fig. 2. FRET studies probe T4L structure and dynamics.
a Network representing 33 measured distinct FRET-variants of T4L. The donor (D) Alexa488-hydroxylamine and acceptor (A) Alexa647-maleimide are coupled to para-acetylphenylalanine (pAcF) and cysteine (C), respectively. The spheres in the left panel represent the average donor (green) and acceptor (red) position for a structure of T4L (PDB ID: 172L) that is determined by the FRET Positioning System (FPS)10. Positions 44, 55, and 69 are used for donor and acceptor labeling. Right panel shows the secondary structure elements (helix, strand or coil) of T4L. The labeling positions are indicated and network pairs are connected. The N-terminal lobe is shown in green, the connecting helix c in orange and the C-terminal lobe in brown. b, c The FRET-efficiency, E, and fluorescence lifetime of D in the presence of A, 〈τD(A)〉F, of the DA-labeled T4L variants (b) S44pAcF/R119C and (c) Q69pAcF/P86C are shown in two-dimensional MFD-histograms (center) with one-dimensional projections of E (right) and 〈τD(A)〉F (top). The magenta lines (static FRET-line) describe those molecules with a single conformational state. The limiting states (circles) of the dynamic FRET-lines (C1, blue; C2, purple; C3, yellow) are identified by eTCSPC (Supplementary Methods, Supplementary Note 4). The dynamic FRET-lines are shown in dark green (C1–C2), cyan (C1–C3), and light green (C2–C3) (Supplementary Methods). The gray lines trace molecules of high FRET-efficiency with a bleaching A. For comparison, the FRET-efficiencies of the X-ray structures for an open (blue, PDB ID: 172L) and closed (violet, 148L) state (determined by FPS) are shown as horizontal lines in the FRET-efficiency histograms (magenta) in the left 1D projection; for S44pAcF/R119C E(open) = 0.40 and E(closed) = 0.54 and for Q69pAcF/P86C E(open) = 0.87 and E(closed) = 0.86. Black dashed lines mark the center of the main peak.
In Fig. 2b, c, we present MFD-histograms with the two FRET-indicators for two exemplary variants of our FRET network. Three peaks are identified in the MFD-histogram. In both histograms, a major and a minor FRET peak are present. The peak located at a low FRET-efficiency E corresponds to molecules without, or with an inactive acceptor, fluorophore (DOnly).
For an open (PDB ID: 172L, blue) and a closed (PDB ID: 148L, magenta) conformation, FRET efficiencies E are predicted by experimentally calibrated dye models (see Methods, section 5.5)9,38. These E values are shown as horizontal lines in the marginal distributions of Fig. 2b, c. A comparison with the major peak (Fig. 2b: 0.3 < E < 0.7, Fig. 2c: E > 0.7) demonstrates that they are similar but not identical to known structural models. Next, we will show how dynamic exchange explains the observed peaks.
In MFD-histograms, FRET-lines (Supplementary Methods) serve as a unique guide to visualize conformational dynamics by peak shifts and splitting like in NMR relaxation dispersion measurements. A static FRET-line relates E and 〈τD(A)〉F for molecules in the absence of dynamics (magenta line, Fig. 2b, c). States that exchange on a time scale much slower than the observation time (quasi-static case) are separated in a MFD-histogram and follow the static FRET-line. However, a shift of a peak to the right with respect to the static FRET-line is a model-free indication for sub-ms dynamics31, because the FRET-efficiencies in a MFD-histogram are averaged over the observation time of the molecules (~ms). Thus, very fast exchanging states result in a single average peak that is shifted to the right of the static FRET-line. These peaks can be described by dynamic FRET-lines, which connect the exchanging states. For a visual representation of the possible transitions, the dynamic FRET-lines of the identified exchanging states are displayed in the MFD-diagrams (dark green, cyan, and light green). A dynamic FRET-line connecting high FRET states with the DOnly population (gray) demonstrates the lack of significant photobleaching or blinking of acceptor dyes.
In the presented data, the major populations are shifted to the right of the static FRET-line (Fig. 2b, c). This gives clear evidence for a dynamic exchange faster than ms. For molecules in very rapid (µs) exchange between an open and a closed conformation, we expected to detect a single averaged peak in MFD-histograms. Hence, taking fast exchange into account, the major peak of the smFRET data is in agreement with known X-ray structures14,39 and kinetic data4,13,20,21,23,24,29,40–42, most likely corresponding to the dynamic averaging of the hinge bending mechanism.
However, in 18 out of 33 MFD-histograms, we visually identify additional minor populations, which are in slow exchange with the major populations. Surprisingly, these minor populations (E > 0.8, Fig. 2b) and (0.2 < E < 0.6, Fig. 2c) can neither be assigned to the predicted average open and closed conformation (Supplementary Fig. 2). This is a first indicative for the existence of a third, conformationally excited, structurally distinct conformer.
In conclusion, MFD-histograms identify three conformers in T4L referred to as C1, C2, and C3. The conformers C1 and C2 are likely in fast exchange, while C3 is in slow exchange with C1 or C2. These conformers may represent limiting states in the exchange dynamics43,44.
Following the workflow presented in Fig. 1e, we next determine the kinetic signatures via fFCS and resolve remaining ambiguities by simulations of MFD-experiments.
Connectivity of states in a kinetic network
To construct a reaction scheme of T4L’s enzymatic cycle, the variant S44pAcF/I150C is used as pseudo-wildtype (“wt**”). At first, we carry out control experiments by comparing for this variant (DA)-labeled and reversely (AD)-labeled T4L variants. In this way, we could exclude potential dye artifacts (Supplementary Fig. 3a–c, Supplementary Note 1) because the kinetic behavior was independent of the labeling scheme. The MFD-histogram of S44pAcF/I150C (Fig. 3a) reveals a typical pattern: a major population C1/C2 (0.2 < E < 0.6) and a minor C3 population (E > 0.8) similar to the variants presented in Fig. 2b, c.
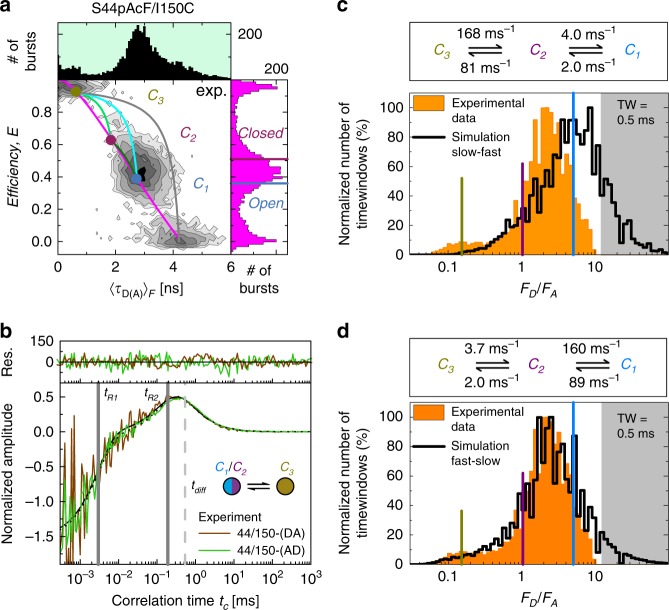
Fig. 3. Kinetic connectivity.
a The FRET-efficiency, E, and fluorescence lifetime of D in the presence of A, 〈τD(A)〉F, of the DA-labeled T4L variant S44pAcF/I150C (or wt**) is shown in the two-dimensional MFD-histograms (center) with the one-dimensional projections of E (right) and 〈τD(A)〉F (top). The magenta line (static FRET-line) represents the molecules with a single conformation. The dynamic FRET-lines represent the molecules changing their conformational state under monitoring. The limiting states (circles) of the dynamic FRET-lines (C1, blue; C2, purple; C3, yellow) are identified by eTCSPC (see Fig. 4). The dynamic FRET-lines are shown in dark green (C1–C2), cyan (C1–C3), and light green (C2–C3) (Supplementary Methods). The gray line represents the trace molecules of high FRET-efficiency with a bleaching A. The FRET-efficiencies of X-ray structures for an open (blue, PDB ID: 172L) and closed (violet, 148L) state (determined by FPS) are shown for comparison and represented as horizontal lines in the FRET-efficiency histograms. b Overlay of the normalized sCCFs (Supplementary Methods Equation (17)) of S44pAcF/I150C-(DA) and S44pAcF/I150C-(AD). The global fit with other variants shows two relaxation times (tR1 = 4.0 ± 2.3 µs, tR2 = 230 ± 28 µs) and a diffusion time tdiff = 0.54 ms. c, d Overlay of time-window analysis, slow-fast in c and fast-slow in d, respectively. The shaded area in gray indicates the region of donor only. See Supplementary Note 3 for data on the model and the simulation.
To unravel the kinetic behavior of an enzyme, one has to be aware that an enzymatic cycle with multiple states can be described by a transition rate matrix, which contains all exchange rate constants of the states. To recover T4L’s transition rate matrix, we determine a set of relaxation times by fFCS (see next paragraph) and the species fractions of the states by analysis of the fluorescence decays (for details see the section below). This analysis results in ambiguous solutions, which are resolved by simulating MFD experiments making use of the information contained in smFRET experiments.
Kinetic network of conformational states resolved by fFCS
By fFCS, we probe transitions in T4L on all relevant timescales34,35 to resolve the kinetic network of conformational states. fFCS uses species-specific information encoded as a characteristic pattern within the ns-regime of the polarization-resolved fluorescence decays34,45. This amplifies the contrast compared to conventional FCS for resolving relaxation times with high precision. We find very good agreement between the normalized species cross-correlation functions (sCCF) of the (AD)- and (DA)-labeled molecules. A global analysis of the sCCFs and the species auto-correlation functions (sACF) requires at least two relaxation times (tR1 = 4 μs and tR2 = 230 μs, Fig. 3b, Supplementary Note 2).
In summary, the two relaxation times obtained by sCCFs independently support the hypothesis of the interconversion between three states at sub-ms timescales. Moreover, in line with the MFD-histograms, we find a fast and a slow relaxation time.
Simulation of the kinetic network of T4 Lysozyme. The three identified conformers C1, C2, and C3 are assigned by their characteristic species fractions (see below) to the corresponding structural states open, closed, and excited, respectively.
Three distinct kinetic linear reaction schemes are possible: C1 ⇋ C2 ⇋ C3, C2 ⇋ C1 ⇋ C3, C1 ⇋ C3 ⇋ C2 while the cycle scheme C1 ⇋ C2 ⇋ C3 ⇋ C1 is unlikely due to the lack of burst across all FRET variants that connect C3 ⇋ C1 with an effective slower rate to satisfy equilibrium. These bursts would follow the dynamic FRET-lines as guides between states in the MFD-diagram (Fig. 3a and Supplementary Fig. 2). Nonetheless, the sequential closing, from the most open (lowest FRET-efficiency in the variant S44pAcF/150C) state to the most compact (highest FRET-efficiency in the variant S44pAcF/150C) is depicted by C1 ⇋ C2 ⇋ C3, so that we can discard the models C1 ⇋ C3 ⇋ C2 and C2 ⇋ C1 ⇋ C3. With the relaxation times determined by fFCS and the species fractions obtained by analysis of the fluorescence decays (see next section), we calculate the exchange rate constants and find two competing solutions (Supplementary Note 3, Equation (36)). The exchange between C1 and C2 can be either slow (Fig. 3c) or fast (Fig. 3d).
To solve this ambiguity, we simulate sm-experiments of the two possible solutions43 (Supplementary Note 3). The obtained simulations are compared with experimental histograms and fFCS (Fig. 3c, d, Supplementary Fig. 4). The corresponding p-values (C1–C2 fast vs. C1–C2 slow) of the respective 1D (p = 1 for 〈τD(A)〉F, p = 0.734 for E) and 2D MFD-histograms (p = 1) clearly demonstrate a better agreement of the experimental data with a fast exchange between C1 and C2 (Supplementary Note 3, Supplementary Table 1a, b).
To conclude, we experimentally determine all reaction rate constants that define the reaction network, and the resulting species fractions. This description covers µs – ms and captures the relevant global motions of T4L.
Characterization of the third conformer by eTCSPC
As demonstrated by fFCS analysis, T4L is highly dynamic. Hence, the FRET-efficiencies in smFRET-histograms only represent dynamic averages of states46. Therefore, for resolving the limiting states of the system, we record high-precision fluorescence decays by eTCSPC and analyze the distribution of the photon arrival times, t, with respect to the excitation pulse in fluorescence decays. This analysis benefits from polarization-free effects resulting from measuring at magic-angle detection, low background fluorescence, and the absence of photobleaching. Moreover, it can reveal DA-distance distributions and species populations47. To dissect the donor quenching by FRET (i.e., FRET-induced donor decay), we jointly analyze the DA- and DOnly-dataset, where the fluorescence lifetime distribution is shared with the DA-dataset. For physically meaningful analysis results, we explicitly consider the DA-distribution broadening due to the linkers by normal distributions47.
The analysis results of all 33 FRET-datasets are discussed using the variant S36pAcF/P86C shown in Fig. 4a (for other variants see Supplementary Note 4, Supplementary Fig. 5a). We display the experimental data by fluorescence decays of the DA- and the corresponding DOnly-sample (Fig. 4a). In agreement with the MFD-histograms and the fFCS data, 1-component models result in broad DA-distributions and/or are insufficient to describe the data (Fig. 4a, weighted residuals, violet). For S36pAcF/P86C, we obtain both, an unphysical distribution width and significant deviations in the weighted residuals, a strong indication that more than one conformer is found.
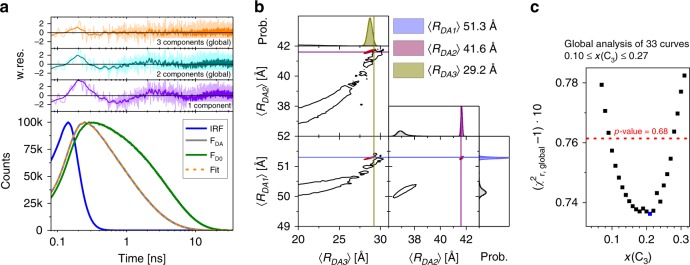
Fig. 4. Uncovering the third conformational state by eTCSPC.
The eTCSPC measurements of all FRET variants are analyzed by superposition of normal distributed DA-distances (due to the coupling of dyes with long and flexible linkers, details see Supplementary Methods). a Experimental fluorescence decay of the variant S36pAcF/P86C (FDA, gray, over laid in orange with best fit), the corresponding DOnly-reference sample (FD0, green), and instrumental response function (IRF, blue). At the top, the weighted residuals (w.res.) of different analyses by models with DA-distance distributions composed of one (violet), two (cyan), and three (orange) normal distributions are shown in corresponding colors. In the 2- and 3-component analysis, all FRET-measurements are jointly analyzed (global), and the species fractions of the states are shared among all 33 datasets. For the 1-component model of the variant S36pAcF/P86C, we find a mean DA-distance of 45.7 Å with a width of 17.6 Å. The analysis results of the 3-component model are summarized in Supplementary Table 2d–g. b Uncertainties, ΔRDA(kFRET), of the 3-component analysis for the variant S36pAcF/P86C. Other contributions to the total uncertainty, ΔRDA, i.e. κ2, are not shown for clarity. To the sides, the marginalized (projected) histograms of the sampled model parameters are shown (black: individual fit; red: global fractions). The lines highlight the most likely combination of distances. c Uncertainty estimation of the species fraction of the third (minor) state, x(C3), for the three-component analysis. The fraction x(C3) was varied from 0–0.32 followed by a subsequent minimization of all other model parameters. This yields the global, reduced χ2r,global of all 33 FRET eTCPSC measurements in dependence of x(C3). This curve has a minimum at x(C3) = 0.21 (χ2r,min =1.074). Points above the red line (χ2r =1.0761, p-value = 0.68) are significantly worse than the best analysis result as judged by an F-test (225 parameters, 100,000 channels).
The analysis of the fluorescence decays by a 2-component model yields an inconsistent assignment by the species fractions (Supplementary Fig. 5b, c). This is evident by significant differences among the species fractions (Supplementary Table 2b). Moreover, the DA-distances disagree with known structural models (compare Supplementary Table 2d, e).
In our effort to seek a consistent description of all measured fluorescence decays, we develop a joint/global model function. For such description, we treated all fluorescence decays as a single dataset sharing common species fractions for the states. This reduces the number of free parameters and dramatically stabilizes the optimization algorithm. Because the global 2-component model (Fig. 4, cyan, Supplementary Table 2c) shows no agreement with the data, we consequently used a 3-component model (Fig. 4a orange, Supplementary Table 2d–f) to describe the data.
To analyze the precision of this fit, the uncertainties ΔRDA of the obtained distances, 〈RDA〉, from the 3-component model need to be determined. ΔRDA depends on statistical uncertainties and systematic errors. We use the known shot noise of the fluorescence decays to estimate the statistical uncertainties, ΔRDA(kFRET), of the FRET-rate constant kFRET (Fig. 4b, Supplementary Table 2g). Moreover, we record polarization-resolved fluorescence decays of the donor and acceptor by eTCSPC to analyze the time-resolved anisotropy (Supplementary Table 3a, b) for estimating systematic errors, ΔRDA(κ2), due to the orientation factor κ2. In conclusion, we can demonstrate that ΔRDA(κ2) dominates the overall uncertainty of ΔRDA (Eq. (5), Supplementary Table 2d–g).
Moreover, we sample the model parameters of a 3-component model for individual datasets by a Markov chain Monte Carlo (MCMC) method. This demonstrates that, for given state populations, the mean distances 〈RDA,1〉, 〈RDA,2〉, and 〈RDA,3〉 are very well defined (compare red to black in Fig. 4b). This also shows that a global model, which interrelates the state populations among datasets, improves the capability to resolve interdye distances.
A global 3-component model has too many degrees of freedom (Supplementary Methods) to be exhaustive when sampling by MCMC. Hence, we vary the state population of the minor state, x(C3), while optimizing all other model parameters (support plane analysis). This way, we determine the dependency of x(C3) on the quality parameter χ2r of all measurements (Fig. 4c). This analysis (1) shows that the minor state population is in the range of 0.1–0.27 and best agrees with the data for 0.21 (Fig. 4c, p-value = 0.68), (2) provides an estimate for ΔRDA(kFRET) (Supplementary Table 2d–f), and (3) demonstrates that ΔRDA(κ2) dominates ΔRDA (Eq. (5)).
In summary, only a 3-component analysis describes all FRET samples and reference samples in a global model. This analysis recovers a set of physically meaningful average DA-distances that are grouped automatically and unbiased by their state populations. Additionally, the 3-component model is consistent with the fFCS data and with the dynamic FRET-lines displaying dynamically averaged sm-subpopulations in MFD (Fig. 2, Supplementary Fig. 2).
The integrated results are consistent with a view that T4L adopts three states (C1, C2, and C3), as opposed to the expected two conformational states based on structural pre-knowledge.
Structural features of conformational states
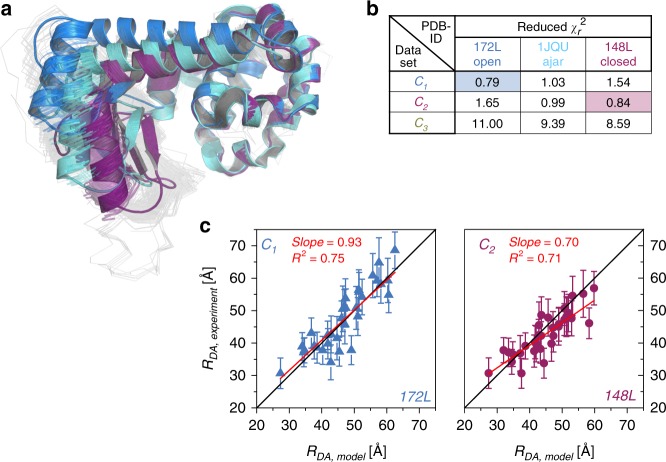
To compare the experimental distances 〈RDA,exp〉 obtained from the fluorescence decays—under consideration of their respective uncertainties ΔRDA—to the structural models deposited in the PDB, we cluster all available 578 structures of T4L and aligned them. We observed that the structural models of T4L group into open, ajar, and closed clusters (based on the proximity of the CTsD and NTsD, Supplementary Table 4) with an intra-cluster root mean-squared displacement of less than 1.8 Å. The representative structures of these clusters are given by PDB IDs 172L, 1JQU, and 148L for the open, ajar, and closed conformations, respectively (Fig. 5a).
Fig. 5. PDB screening.
a Overlay of the PDB structures used for screening. Blue, light blue and violet cartoons show the cluster representative of the open (172L), ajar (1JQU), and closed (148L) conformation of T4L. b Reduced χ2r for each distance set compared to the expected distances from the selected cluster representative. c The experimental distances RDA,experimental of the C1 and C2 dataset are plotted against the model distances RDA,Model from the best PDB structure representative and fitted linearly (red lines). The black lines show a 1:1-relationship. Error bars shown are determined from the support plane analysis (95% confidence interval) shown in Fig. 4 and described in the text.
Next, we apply the FRET positioning system (FPS)34 to compute an error function (χ2r,FPS) that compares the three sets of 33 distances 〈RDA,exp〉 to the modeled distances 〈RDA,model〉 by FPS. In χ2r,FPS, we consider explicitly the uncertainties, ΔRDA, of the distance 〈RDA,exp〉9. The overall agreement (minimum χ2r,FPS) for the distance sets for C1 and C2 is best for 172L and 148L, respectively (Fig. 5b). In Fig. 5c, 〈RDA,model〉 for 172L and 148L are compared to 〈RDA,exp,〉 of C1 and C2, respectively. A linear regression (red line) with a slope close to one demonstrates the absence of significant systematic deviations.
Structurally, the ajar state is more closed than the open state and more open than the closed state, most likely representing an intermediate conformation or it could arise from structural instabilities introduced by specific mutations such as W158L48. The deviation from the open and closed state is clearly reflected in the elevated χ2r,FPS. Consequently, within our precision we can safely assign C1 as open and C2 as closed state. Screening results of other structures in the PDB against the FRET data are very similar to the results for the discussed cluster representatives, as expected (Supplementary Fig. 6). However, none of the structures can be assigned to the C3 state as judged by the disagreement with the data (Fig. 5b, χ2r,FPS). Thus, we conclude that C3 is an excited conformational state of currently unknown structure.
Relevant functional states in the enzymatic cleavage cycle
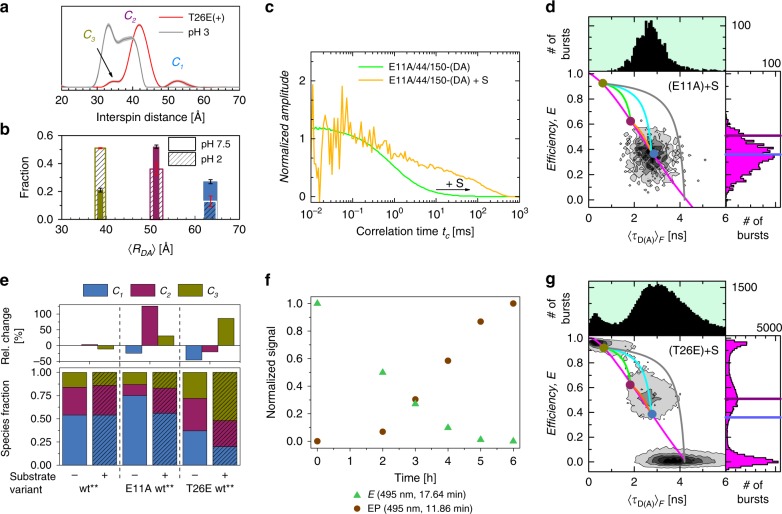
Detection of C3 by EPR. We use double electron-electron resonance (DEER) to provide additional support for the C3 state. Multiple DEER studies on T4L have shown interspin distributions for wt T4L49,50. Here, we show the distribution of interspin distances of the adduct form of the variant T26E/S44pAcF/I150C labeled with the appropriate spin label MTSSL to produce the variant T26E(+)- 44R1/150R1, which displays a satellite population with interspin distance of ~35 Å resembling the enzyme-product-complex EP within the catalytic cleavage cycle of T4L (Fig. 6a, Supplementary Fig. 5d). The most frequently observed distance falls at interspin distances of 42 Å with another less populated state at interspin distances of >50 Å. These may correspond to the various sub-states of the closed (C2) and open (C1) states, respectively. To ensure that this small population is not an artifact of the Tikhonov regularization algorithm51,52 or due to the rotamer populations of the spin label-carrying side chain, we lower the pH to influence the conformational equilibrium of the states53. The FRET-experiment with the variant S44pAcF/I150C shows an increase in the population of C3 at pH 2 (Fig. 6b), and the analogous DEER experiment at pH 3 shows a remarkably similar redistribution of interspin distances. Compared to physiological pH conditions (Fig. 6a, dashed trace), these distances exhibit a shortening that is consistent with the C3 state, thus validating our conclusion that EPR and FRET do show the excited state C3.
Fig. 6. Functional states of T4L.
a Interspin distribution of T26E(+)-44R1/150R1 and 44R1/150R1 at pH 3.0 from DEER experiments. Gray area surrounding the distribution corresponds to error estimates. b Population fraction of S44pAcF/I150C from eTCSPC at pH 7.0 (solid colors) and at pH 2.0 (dashed rectangles). The width of the bar represents the uncertainty in distance (filled bars: pH 7.5, hatched bars: pH 2). The error bars represent the statistical uncertainty in the amplitudes (black: pH 7.5, red: pH 2). C3 increases in population at low pH. Similar effect is shown in EPR DEER experiments (Supplementary Fig. 5d) error estimates are 95% confidence intervals as determined by support plane analysis (Supplementary Methods). c, d The effects of the substrate on E11A/S44C/I150C: c Overlay of normalized sCCF of E11A/S44C/I150C with and without substrate. Consistent with the larger rotational correlation, we observe a shift of tdiff towards longer times for the variant E11A/S44C/I150C when incubated with the substrate; d MFD-histograms for the variant E11A/S44C/I150C with substrate. Upon addition of the substrate, we observe a shift towards lower E values. e Species fractions of the variants S44pAcF/I150C (wt**), E11A/S44C/I150C (E11A wt**), and T26E/S44pAcF/I150C (T26E wt**) used to mimic free enzyme (E), enzyme-substrate complex (ES), and enzyme-product bound state (EP) without (−) and with (+) peptidoglycan. At the top, the relative change in fractions upon addition of peptidoglycan is shown. f, g Effects of the substrate on the variant T26E/S44pAcF/I150C: f Reverse-phase HPLC is used to monitor the adduct formation of the labeled T4L with peptidoglycan via plotting of the normalized signal at 17.64 min (E, green) and 11.86 min (EP, brown) after background subtraction; g MFD-histograms for the variant T26E/S44pAcF/I150C with substrate. An accumulation of the high FRET state is observed. The color code and FRET-lines for 2D and 1D MFD histograms (d) and g are as in Fig. 2.
Trapped reaction states of T4L
To mimic functional enzymatic states, we mutated the residues E11 and T26 at the active site using the backbone of the S44pAcF/I150C variant, also named wt**14,39,54. We use wt** because of the advantage in clearly resolving all three conformations of the free enzyme (E) by FRET. These mutations help identifying the role of C3 during enzyme catalysis: E11A, which inactivates T4L, causes the enzyme to bind its substrate S (peptidoglycan from Micrococcus luteus) while obviating the expected hydrolysis reaction54. Thus, in the presence of excess substrate, this mutation mimics the enzyme-substrate complex (ES). We monitor the effect of the substrate binding for the E11A mutation by FCS and compare the characteristic translational diffusion times, tdiff, in both the absence and presence of substrate. While tdiff is small (0.54 ms, Fig. 6c, green curve) without the substrate, it increases by several orders of magnitude when the large substrate is introduced (Fig. 6c, yellow curve). Moreover, the shift towards the larger donor anisotropy values upon incubation with substrate also provides additional evidence for substrate binding without cleavage (Supplementary Fig. 6e).
Sub-ensemble TCSPC analysis of the DA-subpopulation of the ES state (E11A/wt** in the presence of substrate, Fig. 6d, Supplementary Fig. 7a–d, Supplementary Note 5) reveals an increase of 125% in the population corresponding to C2 compared to the free enzyme state E, with a concomitant reduction of C1. In contrast, no effect of substrate binding for wt**-(DA) is observed because ES is not trapped (Fig. 6e).
Although the variant T26E cleaves the substrate, the formation of a covalent adduct (PDB ID 148L) prevents a release of the formed product14. Therefore, we use this intermediate adduct to mimic the product-bound enzyme state (EP). To confirm the adduct formation under our measurement conditions, we monitor the adduct formation of labeled T4L (T26E/wt** variant) by HPLC (Fig. 6f). T4L without substrate (E) elutes at ~18 min. After incubation with the substrate, the peak of E drops, and a new elution peak at ~12 min is detected with increasing incubation time (Fig. 6f, Supplementary Fig. 8), indicative of the adduct form of T4L (EP). Both ensemble (Fig. 6e) and sm MFD-measurements (Fig. 6g, Supplementary Fig. 7e–h) show a significant increase of the relative fraction of the C3 state, an effect also observed in the EPR measurements (Fig. 6a). In the T26E variant, the accumulation of the C3 state is connected to the inability of this variant to release a part of the product14. We conclude that the new excited conformational state must be involved in this step.
Discussion
In the following, we present the experimental evidence for the C3 state and its structural properties. To corroborate the existence of C3, we discuss our experiments in four aspects: (1) the kinetic behavior in sm-experiments, (2) the error statistics of data analysis, (3) the structural validation of the obtained FRET parameters, and (4) the effect of specific mutations.
Aspect 1: Kinetic behavior. Considering 18 out of 33 variants with FRET-pairs, the sm-experiments directly show the presence of an additional DA-subpopulation in the MFD-histograms, which differs significantly from C1 and C2 (Figs. 2, 3, Supplementary Fig. 2). This DA-subpopulation is either populated or depopulated with specific mutations that alter the overall catalytic activity of T4L. Moreover, our global fluctuation analysis recovers at least two relaxation times that are shared throughout all studied variants (Fig. 3, Supplementary Fig. 9). Applying kinetic theory, two relaxation times indicate at least three states in equilibrium, which are reproduced by Brownian dynamics simulations (Supplementary Fig. 4).
Aspect 2: Error statistics. Key to the analysis and determination of C3 by ensemble fluorescence decays is the use of global analysis of all 33 variants, which reduces the number of free parameters, increases fitting quality (Supplementary Methods), and gives a consistent description with sm-experiments. Moreover, assuming that CTsD and NTsD are rigid, there are six independent degrees of freedom in the system, which we significantly oversample by measuring 33 variants.
Aspect 3: Structural validation. In contrast to our 3-component model, the global analysis of eTCSPC data using a 2-component model yields two distance sets, which cannot describe the expected interdye distances of the known conformers (C1 (172L) and C2 (148L)). Furthermore, for the 2-component model, we do not observe the expected linear correlation between the modeled and experimental interdye distances, as shown in Supplementary Fig. 5b, c.
Aspect 4: Specific mutations. The final point to corroborate the existence of C3 are the results of a few specific mutations. The variant Q69pAcF/P86C is especially informative, as the donor is placed in the middle of helix c (Orange Fig. 1a), which connects both domains, while the acceptor is located in the middle of helix d, which is part of the CTsD (Brown Fig. 1a). According to FPS, the interdye distances for C1 (172L) and C2 (148L) states are hardly distinguishable by FRET, 〈RDA〉 of 34 and 35 Å, respectively. Assuming that both domains preserve their secondary structure, the compaction of T4L in C3 can only proceed by kinking the helix c. This conclusion is consistent with previous studies that identify V75 as the subdomain boundary and critical in protein stability of the pseudo-wild-type construct wt* with a boundary for the local stability to unfolding around residue N6813,55. Given the location of the dyes and the extension of the dye linkers, expected dye orientations will lead to an increase of the interdye distance for C3, i.e., a greater interdye distance is expected for C3 compared to the experimental distances for C1 (39 Å) and C2 (37 Å). The additional observed distance of 52.4 Å agrees with this hypothesis (Supplementary Table 2f, Fig. 2c).
An additional inactive variant (R137E)56,57, which disturbs the salt bridge between residues 22 and 137, reduces the population of C3 by ~50% (Supplementary Fig. 10d, Supplementary Table 2h), a phenotype also observed for the inactive variant E11A/S44C/I150C.
In conclusion, the existence of C3 demonstrates a greater level of complexity of the domain motions of T4L than a single hinge-bending motion, which is in agreement with recent indirect observations20,24,29. The complex exchange dynamics between the conformations with relaxation times of 4 and 230 µs and the small population of C3 may explain the difficulties of other experimental biophysical methods and MD simulations in identifying this exchange, and some heterogeneity in interspin distances observed in previous studies for similar conditions49.
Relating conformational states and enzyme function. A three-step process characterizes the T4L hydrolysis of peptidoglycans. First, the glycosidic bond of the substrate (S) is protonated by E11 followed by the simultaneous nucleophilic attack of water molecules, which are hydrogen-bonded to residues D20 and T26, on the C-1 carbon of S. As a result, the covalent adduct (ES) is observed in PDB ID 148L14. Second, the proton is presumably returned from D20 to E11 via solvent transfer. The third and final step is the product release from the active site to regenerate the enzyme to the original state.
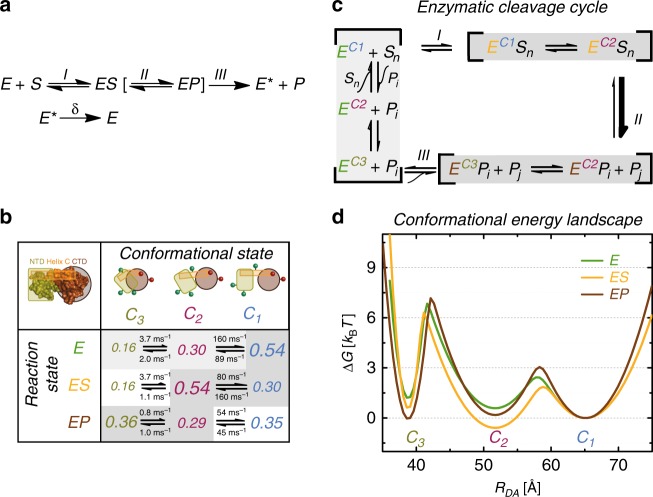
In view of the structural dynamics and to link T4L’s functional cycle to our three observed conformations, we use an extended MMm (eMMm) scheme as suggested by Kou et al.30 (Fig. 7a).
Fig. 7. Energy landscape of T4L.
a Extended Michaelis–Menten scheme. b T4L interconverts between three major C1, C2, and C3 conformational states. The population fractions of C1, C2, and C3 are normalized to the variant S44pAcF/I150C in the absence of substrate using the relative changes in Fig. 6e to correct for the influence of specific mutations in that absence. The different font sizes represent the species fractions xi for each conformer according to Supplementary Table 2h and satisfy x1 + x2 + x3 = 1. The three enzyme states are monitored via the following three enzyme variants: (i) the free enzyme state E via S44pAcF/I150C; (ii) the enzyme-substrate state ES via the inactive E11A/S44C/I150C with bound substrate; and (iii) the enzyme product state EP via the product adduct with T26E/S44pAcF/I150C after substrate cleavage. The reaction rate constants are calculated according to the process detailed in Supplementary Note 3, the confidence intervals of which are shown in Supplementary Table 4c. c The peptidoglycan chain with n subunits (Sn) is cleaved into two products (Pi and Pj with n = i + j) by T4L, both of which can be further processed by T4L until only the dimer of N-acetylglucosamine and N-acetylmuramic acid remains. The gray shaded steps indicate the conformational/reaction states observed. d Relative Gibbs free energy landscapes are calculated using , where kB is the Boltzmann constant and T is the temperature; kij are the reaction rate constants between states Ci and Cj for the data presented in (a). The activation energies are calculated according to assuming k0 = 103 ms−1 as an arbitrary constant. The distributions consider C1, C2, and C3 to follow a Gaussian distribution as a function of the interdye distance RDA. The Gaussian widths (σι) are adjusted to satisfy the energy differences and calculated activation energies. Each energy landscape is independently normalized to C1.
Here, this model considers a sequential succession of three steps to go from a free enzyme via a product bound state back to the relaxed free enzyme state. The substrate S binds reversibly to the enzyme E to form an enzyme-substrate complex ES and is converted to the product P, resulting in an EP complex with the product still bound to the enzyme. A transition of E to an excited state E* then releases P from the complex, followed by a relaxation of the free enzyme E* to E.
Next, we consider our results in the light of the eMMm framework. First, by using the S44/I150C backbone and two key functional T4L variants (E11A, T26E), we create the relationship between the conformational (C1, C2, and C3) and the above-described reaction states (E, ES, and EP) for purposes of elucidating the functional role of E* (Fig. 7). We observed a significant difference of the populations of the conformers (Fig. 6) between the three reaction states.
To connect conformational equilibria with catalysis, we monitor the relative changes in species fractions observed across the functional variants in both the absence and presence of substrate (upper panel of Fig. 6e) by generating a 3 × 3 state matrix (Fig. 7b). As indicated in the matrix, specific conformational states are favored in each enzyme reaction state (Fig. 7c,d). For this representation, we use the relative population changes as compared to the wt** to monitor the conformational populations of the different enzyme states.
In the free enzyme state E, the open conformation C1 is mostly populated to enable substrate binding, which initiates the catalytic cycle through the formation of ES. Through this cycle, the closed conformation C2 now becomes the most abundant conformation14. In this conformation, the glycosidic bond can be cleaved such that C2 connects both ES and EP. In our studies, we determine that the product release may occur in the compact conformation C3, a population that is greatly increased in EP. Thus, C3 links EP and E so that the original enzyme E is regenerated from EP, which closes the enzymatic cycle. Consequently, the compact state C3 now corresponds to the excited conformational state E* (Fig. 7a–d), the function of which is to disperse the product18,29 (Fig. 7b, d). These series of events show a sequential closing from the most open conformation C1 to the most compact form C3 along two coordinates: the reaction state and the conformational state.
Most strikingly, even under saturating conditions, which favor the ES and EP states, the enzyme was observed to remain in dynamic equilibrium between the conformational states, rather than transforming entirely into a single conformational state.
To visualize the relative energetic changes of the enzyme during the various steps of the catalytic cycle, we use the species fractions and reaction rate constants to compute the relative energy landscape based on the Arrhenius equation (Fig. 7d, Supplementary Fig. 11). We observe a sequential closing of the enzyme to populate C3. This is consistent with a ratchet model for providing directionality on the reaction58–60 beyond the directionality introduced by the excess of S. This also corroborates with our Monte Carlo simulations (Fig. 3) that are incompatible with the unlikely off pathway from C1 to C2 through C3 due to steric hindrances and with the fast hinge bending motion (4 μs) expected from structural models (PDB ID 172L and 148L).
All our evidence suggests that the conformational state C3, which appears to be more compact than any other structure known of T4L, is compulsory after rather than before catalytic cleavage. Thus, the compact nature of this structure suggests a functional role that is related to product release via an excited state E* (Fig. 7a). This mechanism can be an evolutionary advantage when directionality is required for function. On the contrary, considering a system with only two conformational states and without an active cleaning mechanism, the stochastic dissociation of the product can become rate-limiting given a high affinity of the product to the enzyme in the EP state. Indeed, a large surfeit of substrate is always characteristic of the in vivo conditions for T4L. Thus, the use of a three-state system to decouple the substrate access and product release can mitigate the occurrence of substrate inhibition in a two-state system when the route to the active site is clogged by excess substrate concentrations61.
In summary, we studied 33 distinct FRET-pairs to effectively oversample the anticipated simple hinge-bending motion of T4L. Due to the high precision, we identified three substrate-dependent fluorescence states that are in fast kinetic exchange. Inverting the positions of the dyes, e.g. S44pAcF/I150C-DA vs. S44pAcF/I150C-AD (Fig. 3b), rules out specific interactions of the dyes with T4L.
Functional variants change the relative populations of the fluorescence states that are determined in a substrate-dependent manner (Fig. 6). Given successive compaction via three conformational states (C1, C2, and C3), and three reaction states (E, ES, and EP), we considered an eMM reaction scheme (Fig. 7a, c) to provide a meaningful description of the data. Mutagenesis and stability studies indicate the stability of CTsD and a flexibility of the NTsD4,13,40–42,62, which may be a necessary principle for the construction of enzymes undergoing conformational changes during catalysis. The combination of known structural models and fluorescence data is used to create a proposed novel structural state in the catalytic cycle of T4L involving a rearrangement of the reactive NTsD with respect to the CTsD, which is deemed consistent with the eMMm for enzyme kinetics. For a complete structural insight, we are now using the obtained FRET-restraints to present a potential model of C3, which is left for a future report.
We anticipate that the presented integrative approach combining the fluorescence spectroscopic toolkit and computational information can accelerate the development of dynamic structural biology63 by resolving the behaviors of long- and short-lived excited states for purposes of characterizing their functional relevance. This approach is highly relevant as we move towards understanding biomolecular dynamics in situ, where “invisible” molecular effects (i.e., ionic, viscous, and crowding effects64) have the potential to modulate weak interactions with important repercussions in biological systems. The elucidation of excited conformational states is necessary for a thorough in-depth understanding of the mechanisms of enzymes. Thus, a comprehensive description of a dynamic molecular system contains intertwined kinetic and structural information, which is often difficult to obtain by traditional methods. Such information can be archived in the data bank PDB-dev65 so that excited conformational states gain the urgently needed visibility.
Methods
Sample preparation
T4L cysteine and amber (TAG) mutants are generated via site-directed mutagenesis in the pseudo-wild-type construct containing the mutations C54T and C97A (wt*), which was subsequently cloned into the pET11a vector (Life Technologies Corp.)66-68. All primer sequences are listed in Supplementary Table 6. The plasmid containing the gene with the desired mutant was co-transformed with pEVOL66 into BL21(DE3) E. coli (Life Technologies Corp.) and plated onto LB-agar plates supplemented with the respective antibiotics, ampicillin and chloramphenicol. A single colony was inoculated into 100 mL of LB medium containing the above-mentioned antibiotics and grown overnight at 37 °C in a shaking incubator. 50 mL of the overnight culture are used to inoculate 1 L of LB medium supplemented with the respective antibiotics and 0.4 g/L of pAcF (SynChem) and grown at 37 °C until an OD600 of 0.5 was reached. The protein production was induced for 6 h by addition of 1 mM IPTG and 4 g/L of arabinose.
Cells are harvested, lysed in 50 mM HEPES, 1 mM EDTA, and 5 mM DTT at pH 7.5 and purified using a monoS 5/5 column (GE Healthcare) with an eluting gradient from 0 to 1 M NaCl according to standard procedures. High-molecular weight impurities are removed by passing the eluted protein through a 30 kDa Amicon concentrator (Millipore), followed by subsequent concentration and buffer exchange to 50 mM PB, 150 mM NaCl pH 7.5 of the protein flow through with a 10 kDa Amicon concentrator. For the double cysteine mutant containing E11A, the temperature was reduced to 20 °C after induction and the cells are grown additional 20 h to increase the fraction of soluble protein. This mutant was produced and purified as described above, except that only ampicillin for selection and IPTG for induction are needed.
Site-specific labeling of T4L was accomplished using orthogonal chemistry. To probe T4L structure by FRET studies (Fig. 2a) we labeled the Keto group of the p-acetyl-L-phenylalanine (pAcF) amino acid at the N-terminal subdomain, hydroxylamine linker chemistry was used for the donor dye Alexa488 (Life Technologies Corp.). Cysteine mutants were labeled via a thiol reaction with maleimide linkers of the Alexa647 acceptor dye.
In one exceptional case of the sample S44pAcF/I150C-AD, the labeling was reversed in order to test the reproducibility of the filtered FCS (Fig. 3b) and FRET measurements with different dyes. Acceptor dye Alexa647 with hydroxylamine linker was used to label the pAcF and the Alexa488 donor dye with maleimide linker was used to label the cysteine residue of the mutant S44pAcF/I150C-AD.
For spin labeling, the S44C/I150C double mutant was diluted to a final concentration of ~10 µM in labeling buffer (50 mM MOPS, 25 mM NaCl, pH 6.8) and a 10-fold molar excess of a methanthiosulfonate nitroxide (MTSSL) was added overnight68. Next day, excess spin reagent was removed using a desalting column (HiPrep 26/10, GE Healthcare) according to the manufacturer’s instructions and concentrated with a 15 kDa Amicon concentrator (Millipore).
Binding of labeled T4L mutants to peptidoglycan from Micrococcus luteus (Sigma-Aldrich) was monitored by reverse phase chromatography using a C-18 column out of ODS-A material (4 × 150 mm, 300 Å) (YMC Europe GmbH, Dinslaken, Germany). The labeled protein (1 µM) was incubated with the substrate at 1 mg/mL in PBS. At various points of the reaction, 25 µL of mixed sample injected and further eluted with a gradient from 0 to 80% acetonitrile containing 0.01% trifluoroacetic acid for 25 min at a flow rate of 0.5 ml/min. The labeled complex elution was monitored by absorbance at 495 nm.
Single-molecule experiments
For single-molecule measurements with multiparameter fluorescence detection, we added 40 µM TROLOX to the measurement buffer to minimize the acceptor blinking and 1 µM unlabeled T4L to prevent any adsorption to the cover glass. A custom-built confocal microscope with a dead time-free detection scheme using 8 detectors (four green (τ-SPAD, PicoQuant, Germany) and four red channels (APD SPCM-AQR-14, Perkin Elmer, Germany)) was used for MFD and fFCS measurements. A time-correlated single photon counting (TCSPC) module with 8 synchronized input channels (HydraHarp 400, PicoQuant, Germany) was used to register the detected photon counts in the Time-Tagged Time-Resolved (TTTR) mode. For more details on TTTR please read69. The data was analyzed by established MFD procedures31,33,43 and software, a more detailed description is given in Supplementary Methods. Exemplary data analysis is shown in Supplementary Fig. 3 and Supplementary Note 1, MFD-histograms of all measurements are collected in Supplementary Fig. 2 and Supplementary Fig. 10.
Filtered FCS
Filtered FCS (fFCS) is a derivative of fluorescence correlation spectroscopy (FCS). In fFCS, the information on the fluorescent species contained in the time- and polarization- resolved fluorescence decays (exemplary shown in Supplementary Fig. 9a, c) was used to amplify the transitions between the species of interest32,34,45. For this, we arbitrarily constructed species-selective filters (exemplary shown in Supplementary Fig. 9b, d) based on the major and minor population in the smFRET experiment and calculated species-selective auto- (sACF) and cross-correlation functions (sCCF). The in total four curves (two sACF’s and two sCCF’s) are analyzed jointly using established fitting models (Supplementary Methods Equations (15–17))32,34,45. For more details see Supplementary Methods.
Fluorescence decay analysis
Fluorescence decays of all samples and single-labeled reference samples are collected on either an IBH-5000U (IBH, Scotland) or a Fluotime 200 (Picoquant, Germany) system. We collected high-precision fluorescence decays histograms with 30 million photons to precisely determine the FRET parameters of limiting states together with their corresponding structural properties. eTCSPC has the advantage of better photon statistics, polarization-free measurements due to magic-angle detection, a keenly evolved instrumental response function (IRF), low background fluorescence, and the absence of photobleaching at low excitation powers. As the fluorophores are coupled via long and flexible linkers, this resulted in a DA-distance distribution even for single protein conformations. For our data analysis, we assumed that the dyes rotate quickly (κ2 = 2/3) and diffuse slowly compared to the fluorescence lifetime (~ns)38. We validated the assumption of fast rotating dyes by time-resolved anisotropy measurements (Supplementary Table 3a–c). Moreover, we interpret the broadening of the DA-distance distributions, beyond what is expected from flexible linkers, as evidence for conformational heterogeneity of the host molecule. To dissect the donor-quenching by FRET (i.e., FRET-induced donor decay), we jointly analyzed the DA- and DOnly-dataset, where the donor fluorescence lifetime distribution was shared with the DA-dataset (Supplementary Methods, Supplementary Equations (20, 24-S27))47. We compared three different fit models (Supplementary Table 5). The results are summarized in Supplementary Table 2. We estimated the statistical uncertainty of the model parameters by making use of the known shot noise of the fluorescence decays. We randomly sampled the model parameters by a Markov chain Monte Carlo (MCMC) method to estimate their uncertainties for a single dataset (Supplementary Methods)70. The applied joint, global fit significantly reduced the overall dimensionality of the analysis but still left too many degrees of freedom (Supplementary Methods) for an exhaustive sampling by MCMC. Hence, we applied a support plane analysis to estimate the model parameter uncertainties, in which we systematically varied x3 while minimizing all other parameters.
Electron paramagnetic resonance
For double electron electron resonance (DEER) measurements of doubly spin labeled proteins, ~200 µM spin-labeled T4L containing 20% glycerol (v/v) was placed in a quartz capillary (1.5 mm i.d. × 1.8 mm o.d.; VitroCom) and then flash-frozen in liquid nitrogen. Sample temperature was maintained at 80 K. The four-pulse DEER experiment was conducted on a Bruker Elexys 580 spectrometer fitted with an MS-2 split ring resonator. Pulses of 8 ns (π/2) and 16 ns (π) are amplified with a TWT amplifier (Applied Engineering Systems). Pump frequency was set at the maximum of the central resonance, and the observe frequency was 70 MHz less than the pump frequency. Dipolar data are analyzed by using a custom program, LongDistances71, written in LabVIEW (National Instruments Co.). Distance distributions are acquired using Tikhonov regularization51.
Recovering the reaction network by Brownian dynamics simulations
To solve the ambiguity in the connectivity of states and kinetics of T4L, i.e. between the two possible analytical solutions of the transition rate matrix (Supplementary Methods Equation (31)), we used Brownian dynamics simulation of single-molecule and fFCS experiments. Simulations of single-molecule measurements are done via Brownian dynamics72-75. The spatial intensity distribution of the observation volume was assumed a 3D Gaussian. In contrast to other simulators, freely diffusing molecules in an “open” volume are used. Transition kinetics is modeled by allowing i → j transitions. The time that molecules spend in i and j states (ti and tj, respectively) are exponentially distributed with
| 1 |
Simulated photon counts are saved in SPC-132 data format (Becker & Hickel GmbH, Berlin, Germany) and treated as experimental data. To quantify the difference between the two possible, simulated models and the experimental data, we calculated the relative χ2 for the one-dimensional and two-dimensional MFD-histograms (Supplementary Note 3, Supplementary Table 1a, b).
Simulation of interdye distances and structural modelling
Accessible contact volume (ACV) simulations and interdye distances. The accessible volume (AV) considers dyes as hard sphere models connected to the protein via flexible linkers (modeled as a flexible cylindrical pipe)38. The overall dimension (width and length) of the linker is based on their chemical structures. For Alexa488 the five carbon linker length was set to 20 Å, the width of the linker is 4.5 Å and the dye radii R1 = 5 Å, R2 = 4.5 Å and R3 = 1.5 Å. For Alexa647 the dimensions used are: length = 22 Å, width = 4.5 Å and three dye radii R1 = 11 Å, R2 = 3 Å and R3 = 3.5 Å. Here, the dye distribution was modeled by the accessible contact volume approach (ACV)9, which is similar to the accessible volume (AV)38, but defines an area close to the surface as contact volume.
Similar approaches have been introduced before to predict possible positions for EPR and FRET labels10,36,37. The dye is assumed to diffuse freely within the AV and its diffusion is hindered close to the surface. The part of AV which is closer than 3 Å from the macromolecular surface (contact volume) is defined to have higher dye density ρDye,trapped. The spatial density ρDye along R is approximated by a step function: ρDye = [ρDye,free, R < 3 Å; ρDye,trapped, R ≥ 3 Å]. The ρDye,trapped/ρDye,free ratio is calculated from the fraction of the trapped dye xDye,trapped for each labeling position separately: ρDye,free/ρDye,trapped = VDye,trapped·(1 − xDye,trapped)/(xDye,trapped·VDye,free). For this, the fraction xDye,trapped was approximated by the ratio of the residual, r∞, and fundamental anisotropy, r0, determined by the time-resolved anisotropy decay of the directly excited dyes (Supplementary Table 3).
To account for dye linker mobility, we generated a series of ACV’s for donor and acceptor dyes attached to T4L placing the dyes at multiple separation distances. For each pair of ACV’s, we calculated the distance between dye mean positions (Rmp)
| 2 |
where and are all the possible positions that the donor fluorophore and the acceptor fluorophore can take. However, in ensemble TCSPC (eTCSPC) the mean donor–acceptor distance is observed:
| 3 |
which can be modeled with the accessible volume calculation.
The relationship between Rmp and 〈RDA〉 can be derived empirically following a third order polynomial from many different simulations. The 〈RDA〉 is not directly related to the distance between atoms on the backbone (Cα–Cα), except through the use of a structural model.
FRET positioning and Screening (FPS). FPS is done in four steps, and its flow is based on the recent work by Kalinin et al.10. In order to do our experimental design using the available PDB structures of T4L with respect to our FRET data, FPS calculates the donor and acceptor accessible volumes for each donor–acceptor labeling scheme. We then compute an error function for each conformational state C(i)
| 4 |
where N = 33 is the total number of FRET distances (〈RDA〉) and the overall theoretically computed absolute uncertainty (see next section).
In order to compare the structural models currently available in the PDB to our experimental results, we clustered all PDB models using the RMSD (Root Mean Squared Deviation) of Cα atoms as the similarity measure. Clustering allowed us to sort all PDB models into three distinct groups based on the similarity of their backbone shapes. We found that the structural models of T4L group into open, ajar, and closed clusters (based on the proximity of the CTsD and NTsD) with an intra-cluster RMSD of less than 1.8 Å. Representative structures of these clusters are given by PDB IDs 172L, 1JQU, and 148L for the open, ajar, and closed conformations, respectively (Fig. 5a). This was done using the agglomerative hierarchical complete-linkage clustering of the “fastcluster”76 software.
In Supplementary Table 4 we provide the complete breakdown of the three clusters. In Supplementary Fig. 6 we display the complete FRET-screening of the three clusters.
Calculation of uncertainties
The overall theoretically computed absolute uncertainty of the average inter-dye distance for the pair (i) is determined by the error-propagation rule:
| 5 |
In the following, we describe the computation of the four individual contributions expressed as absolute and relative distance uncertainties, ΔR and δR, respectively.
(1) Dye model. The relative distance error δRdye model usually considers the asymmetry of the AVs for random labeling of two equivalent labeling sites (in general two cysteines) with δRdye model ≈ 1.5 %47. However, in this work we labeled T4L specifically (one cysteine, one p-acetylphenylalanine) so that ΔRdye model = 0.
(2) Uncertainty of the Förster Radius R0. The relative distance error considers the uncertainty of the Förster Radius R0 that is usually dominated by κ2 errors related to the mutual orientation of donor and acceptor. At first, we use the experimental anisotropy decays r(t)dye recorded by eTCSPC and MFD and the wobbling in a cone (WIC) model to compute possible distribution of orientational factors, p(κ2)38,77. As input we determined the residual anisotropies of the donor fluorescence r3,D (Supplementary Table 3a), the directly excited acceptor fluorescence r3,A (Supplementary Table 3b) and the FRET-sensitized acceptor fluorescence r2,A(D) (Supplementary Table 3c). Based on these limits, we computed the distribution of the orientation factor p(κ2) (Supplementary Table 3d) for each FRET pair as described in Sindbert at al.38. Next, we compute how p(κ2) affects the experimentally observed interdye distance. Following the approach of Sindbert et al.38, we can assume that a DA pair is characterized by a single “true” DA distance RDA with κ2 ≠ 2/3. As we calculate the DA distance assuming κ2 = 2/3, we only recover an apparent DA distance, Rapp. Obviously, Rapp differs from RDA, . A distribution of κ2 relates for a single RDA to a distribution of apparent Rapp. For each FRET pair the distribution p(κ2) compiled in Supplementary Table 3d is transformed to a distribution of the relative distances Rapp / RDA = ξ. The standard deviation of the distribution p(ξ) is used as a relative approximate for the precision of the distance RDA.
| 6 |
The expectation value of p(ξ) can be used as an estimate of the accuracy:
| 7 |
For the WIC model with the given residual anisotropies, the precision dominates the relative uncertainty. Estimates for are very close to one. Hence, we estimate the overall uncertainty attributed to κ2 by
| 8 |
(3) Uncertainty of the D-only reference. The absolute uncertainty of the Donor-only reference ΔRreference considers the discrepancy between the photophysical properties of the donor in the FRET sample and the properties determined from the donor-only sample. This discrepancy is typically caused by unspecific labeling of the biomolecule and thus unknown fraction of donor–acceptor molecules with respect to acceptor-donor molecules (see also Peulen et al.47, Fig. 12). In this work, ΔRreference was set to zero, since specific labeling was used and the donor position is known exactly and accurate donor only sample was measured (Supplementary Table 2a).
(4) Statistical uncertainty (error of the fit). The state-specific asymmetric absolute statistical uncertainty is caused by the shot noise of the lifetime measurements (listed in Supplementary Table 2g). is calculated from the spread of obtained distances for the three states of the global fit using the shortest (〈Rnoise−〉) and longest distance (〈Rnoise+〉) below the 1σ-threshold (Supplementary Table 2g). As the distance 〈RDA〉 of the global fit with the lowest χr2 (x3 = 0.18) is not necessarily the average of 〈R noise-〉 and 〈R noise+〉, both and the resulting are not symmetric: .
For example, considering a 1σ-confidence interval, the fraction x3 of varies between 0.1 and 0.27 (see Fig. 4c). The corresponding and are the shortest and longest distance below the 1σ-threshold. The minimal χ2r,global for this distribution fit model is 1.0736 with the species fraction x3 = xmiddle = 0.18 with x1 = 0.44 and x2 = 0.38 and the state-specific mean distances are listed in the Supplementary Table 2d–f.
Controls for the FRET analysis
Since the problems inherent in the use of smFRET studies are connected with complexities related to the labels, we performed ten controls to check for any potential label artifacts. Please refer to the Supplementary Information for additional data and extended controls (Supplementary Note 6).
The labeling does not alter enzyme function with the labeled T26E variant indicating an expected adduct formation (Fig. 6, Supplementary Fig. 8).
Local quenching of the donor dye is considered when calculating distances and cross correlations (Supplementary Table 2).
The triplet state quenchers do not affect the observed relaxation times and amplitudes on the species cross-correlation (Supplementary Fig. 12).
The acceptor cis-trans isomerization does not contribute to the signal on the species correlation analysis (Supplementary Fig. 12).
The κ2 distributions indicated the validity of our assumption of κ 2 = 2/3. Supplementary Table 3 summarizes the residual anisotropies (r∞) of D—donor, A—acceptor and A(D) by the FRET-sensitized emission of acceptor used for calculating κ2 distributions31.
The existence and the population fraction of the new conformational state C3 with a confidence rate of 68% between 10 and 27% is consistent across our library of 33 variants with x3 = 21%. The variation of the experimental uncertainty is consistent with the determination in the literature that mutations slightly affect the conformational stability of T4L, which was measured in chemical denaturation experiments62. We thus attribute this variability of the species fractions to mutation effects.
All 33 variants provide a consistent view of the T4L conformational states, in which we determined after X-ray crystallography a consistency with the two limiting structures determined by T4L without outliers (Fig. 5, Supplementary Table 2d–f).
We oversample the FRET restraints to reduce the uncertainty introduced from each point mutation (Fig. 5, Supplementary Table 2d–f).
The thermodynamic stability and proper folding of our variants are verified by chemical denaturation with urea and by measuring CD spectra for both unlabeled and labeled T4L.
We fit time resolved fluorescence decays with various models to provide a consistent view of the conformational space (Supplementary Methods).
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Acknowledgements
We wish to acknowledge the support of Marina Rodnina, Philipp Neudecker, and M. Neal Waxham for their comments and suggestions on the paper, and Daniel Rohrbeck for support on the mutagenesis. We acknowledge the support of Christian A. Hanke for helping in organizing the data deposition at Zenodo. We also wish to acknowledge Evan Brooks for generating several of the T4L variants and in assisting with the DEER sample preparation. This research was supported by the European Research Council through the Advanced Grant 2014 hybridFRET (671208) (to C.A.M.S.), by the NIH (grant R01EY05216 to W.L.H. and grant 1P20GM121342 to HS), NSF CAREER MCB (grant 1749778 to H.S.) and the Jules Stein Professorship Endowment (to W.L.H.). H.S. wishes to acknowledge the support from the Alexander von Humboldt foundation and the Clemson Start-up funds. K.H. wishes to acknowledge the support of the NRW Research School BioStruct and the iGRASPseed International Graduate School of Protein Science & Technology.
Source Data
Author contributions
H.S. and K.H. purified and labeled the protein. H.S., T.P., K.H., and D.R. measured and analyzed the FRET experiments. T.P., M.D., and H.G. performed structural screening. M.R.F. performed and analyzed EPR experiments. W.H. performed study design and EPR analysis. S.F. developed fluorescence analysis tools. F.K. and R.K. developed fluorescence instrumentation and gave technical support. All authors discussed the results and commented on the paper. H.S., K.H., W.H., and C.A.M.S. wrote the paper. H.G. contributed to the writing of the paper. C.A.M.S. supervised the project.
Data availability
The source data for all variants and all used techniques and screening results for the PDB structures were uploaded to Zenodo under DOI 10.5281/zenodo.3376527 and are described in Supplementary Note 7. The Zenodo archive contains subfolders for all eTCSPC FRET data including reference measurements used for derivation of DA distances (eTCSPC_wildtype.zip; e.g. Figures 4, 5, and Supplementary Fig. 5), single-molecule raw data used for filtered FCS and MFD analysis (Single_molecule_wildtype.zip; e.g. Figures 2, 3, Supplementary Figs. 2, 4, and 9), single molecule data of functional variants used for derivation of the kinetic scheme (Single_molecule_functional_variants.zip; e.g. Figure 6, Supplementary Figs. 6, 7, 9, and 11), EPR data (EPR_wildtype.zip; e.g., Figure 6 and Supplementary Fig. 5), and an overview on the FRET screening of PDB structures (FRET_screening_of_PDB_structures.zip). The source data underlying Supplementary Fig. 8c is shown in the Source Data File. Further datasets of processed data and the analysis are available from the corresponding authors on reasonable request.
Code availability
Most general custom-made computer code is directly available from http://www.mpc.hhu.de/en/software. Additional computer code custom-made for this publication is available upon request from the corresponding authors. In-house programs are used (1) in the confocal multiparameter fluorescence detection experiments, (2) to elucidate the filtered fluorescence correlation spectroscopy curves, and (3) to analyze the fluorescence lifetime measurements. Software for analysis of single-molecule measurements and fluorescence correlation analysis and their simulation is available at http://www.mpc.uni-duesseldorf.de and software for analysis of fluorescence decays can be downloaded from http://www.fret.at/tutorial/chisurf/.
Competing interests
The authors declare no competing interests.
Footnotes
Peer review information Nature Communications thanks the anonymous reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Hugo Sanabria, Dmitro Rodnin, Katherina Hemmen.
Contributor Information
Hugo Sanabria, Email: hsanabr@clemson.edu.
Claus A. M. Seidel, Email: cseidel@hhu.de
Supplementary information
Supplementary information is available for this paper at 10.1038/s41467-020-14886-w.
References
- 1.Smock RG, Gierasch LM. Sending signals dynamically. Science. 2009;324:198–203. doi: 10.1126/science.1169377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hammes GG. Mechanism of enzyme catalysis. Nature. 1964;204:342–343. doi: 10.1038/204342a0. [DOI] [PubMed] [Google Scholar]
- 3.Kleckner IR, Foster MP. An introduction to NMR-based approaches for measuring protein dynamics. Biochim. Biophys. Acta. 2011;1814:942–968. doi: 10.1016/j.bbapap.2010.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McHaourab HS, Lietzow MA, Hideg K, Hubbell WL. Motion of spin-labeled side chains in T4 lysozyme. Correlation with protein structure and dynamics. Biochemistry. 1996;35:7692–7704. doi: 10.1021/bi960482k. [DOI] [PubMed] [Google Scholar]
- 5.Henzler-Wildman K, Kern D. Dynamic personalities of proteins. Nature. 2007;450:964–972. doi: 10.1038/nature06522. [DOI] [PubMed] [Google Scholar]
- 6.Mulder FA, Mittermaier A, Hon B, Dahlquist FW, Kay LE. Studying excited states of proteins by NMR spectroscopy. Nat. Struct. Biol. 2001;8:932–935. doi: 10.1038/nsb1101-932. [DOI] [PubMed] [Google Scholar]
- 7.Kilic, S. et al. Single-molecule FRET reveals multiscale chromatin dynamics modulated by HP1 alpha. Nat. Commun. 9, 235 (2018). [DOI] [PMC free article] [PubMed]
- 8.Hellenkamp B, Wortmann P, Kandzia F, Zacharias M, Hugel T. Multidomain structure and correlated dynamics determined by self-consistent FRET networks. Nat. Meth. 2017;14:174–180. doi: 10.1038/nmeth.4081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dimura M, et al. Quantitative FRET studies and integrative modeling unravel the structure and dynamics of biomolecular systems. Curr. Opin. Struct. Biol. 2016;40:163–185. doi: 10.1016/j.sbi.2016.11.012. [DOI] [PubMed] [Google Scholar]
- 10.Kalinin S, et al. A toolkit and benchmark study for FRET-restrained high-precision structural modeling. Nat. Meth. 2012;9:1218–1225. doi: 10.1038/nmeth.2222. [DOI] [PubMed] [Google Scholar]
- 11.van den Bedem H, Fraser JS. Integrative, dynamic structural biology at atomic resolution–it’s about time. Nat. Meth. 2015;12:307–318. doi: 10.1038/nmeth.3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matthews BW, Remington SJ. The three dimensional structure of the lysozyme from bacteriophage T4. Proc. Natl Acad. Sci. USA. 1974;71:4178–4182. doi: 10.1073/pnas.71.10.4178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Llinas M, Gillespie B, Dahlquist FW, Marqusee S. The energetics of T4 lysozyme reveal a hierarchy of conformations. Nat. Struct. Biol. 1999;6:1072–1078. doi: 10.1038/14956. [DOI] [PubMed] [Google Scholar]
- 14.Kuroki R, Weaver LH, Matthews BW. A covalent enzyme-substrate intermediate with saccharide distortion in a mutant T4 lysozyme. Science. 1993;262:2030–2033. doi: 10.1126/science.8266098. [DOI] [PubMed] [Google Scholar]
- 15.Goto NK, Skrynnikov NR, Dahlquist FW, Kay LE. What is the average conformation of bacteriophage T4 lysozyme in solution? A domain orientation study using dipolar couplings measured by solution NMR. J. Mol. Biol. 2001;308:745–764. doi: 10.1006/jmbi.2001.4614. [DOI] [PubMed] [Google Scholar]
- 16.Zhang XJ, Wozniak JA, Matthews BW. Protein flexibility and adaptability seen in 25 crystal forms of T4 lysozyme. J. Mol. Biol. 1995;250:527–552. doi: 10.1006/jmbi.1995.0396. [DOI] [PubMed] [Google Scholar]
- 17.Michaelis L, Menten ML, Johnson KA, Goody RS. The original Michaelis constant: translation of the 1913 Michaelis-Menten paper. Biochemistry. 2011;50:8264–8269. doi: 10.1021/bi201284u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zheng D, Lu HP. Single-molecule enzymatic conformational dynamics: spilling out the product molecules. J. Phys. Chem. B. 2014;118:9128–9140. doi: 10.1021/jp5014434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu M, Lu HP. Revealing multiple pathways in T4 Lysozyme substep conformational motions by single-molecule enzymology and modeling. J. Phys. Chem. B. 2017;121:5017–5024. doi: 10.1021/acs.jpcb.7b03039. [DOI] [PubMed] [Google Scholar]
- 20.Akhterov MV, et al. Observing lysozyme’s closing and opening motions by high-resolution single-molecule enzymology. ACS Chem. Biol. 2015;10:1495–1501. doi: 10.1021/cb500750v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu HP. Revealing time bunching effect in single-molecule enzyme conformational dynamics. Phys. Chem. Chem. Phys. 2011;13:6734–6749. doi: 10.1039/c0cp02860f. [DOI] [PubMed] [Google Scholar]
- 22.Tsugita A, Inouye M. Purification of bacteriophage T4 lysozyme. J. Biol. Chem. 1968;243:391–397. [PubMed] [Google Scholar]
- 23.Choi Y, et al. Single-molecule lysozyme dynamics monitored by an electronic circuit. Science. 2012;335:319–324. doi: 10.1126/science.1214824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yirdaw RB, McHaourab HS. Direct observation of T4 lysozyme hinge-bending motion by fluorescence correlation spectroscopy. Biophys. J. 2012;103:1525–1536. doi: 10.1016/j.bpj.2012.07.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Groot BL, Hayward S, van Aalten DM, Amadei A, Berendsen HJ. Domain motions in bacteriophage T4 lysozyme: a comparison between molecular dynamics and crystallographic data. Proteins. 1998;31:116–127. doi: 10.1002/(SICI)1097-0134(19980501)31:2<116::AID-PROT2>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 26.Lange OF, Grubmüller H. Full correlation analysis of conformational protein dynamics. Proteins. 2008;70:1294–1312. doi: 10.1002/prot.21618. [DOI] [PubMed] [Google Scholar]
- 27.Zacharias M. Combining elastic network analysis and molecular dynamics simulations by hamiltonian replica exchange. J. Chem. Theory Comput. 2008;4:477–487. doi: 10.1021/ct7002258. [DOI] [PubMed] [Google Scholar]
- 28.Bouvignies G, et al. Solution structure of a minor and transiently formed state of a T4 lysozyme mutant. Nature. 2011;477:111–114. doi: 10.1038/nature10349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu M, Lu HP. Probing protein multidimensional conformational fluctuations by single-molecule multiparameter photon stamping spectroscopy. J. Phys. Chem. B. 2014;118:11943–11955. doi: 10.1021/jp5081498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kou SC, Cherayil BJ, Min W, English BP, Xie XS. Single-molecule Michaelis-Menten equations. J. Phys. Chem. B. 2005;109:19068–19081. doi: 10.1021/jp051490q. [DOI] [PubMed] [Google Scholar]
- 31.Sisamakis E, Valeri A, Kalinin S, Rothwell PJ, Seidel CAM. Accurate single-molecule FRET studies using multiparameter fluorescence detection. Methods Enzymol. 2010;475:455–514. doi: 10.1016/S0076-6879(10)75018-7. [DOI] [PubMed] [Google Scholar]
- 32.Eggeling C, Fries JR, Brand L, Günther R, Seidel CAM. Monitoring conformational dynamics of a single molecule by selective fluorescence spectroscopy. Proc. Natl Acad. Sci. USA. 1998;95:1556–1561. doi: 10.1073/pnas.95.4.1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kühnemuth R, Seidel CAM. Principles of single molecule multiparameter fluorescence spectroscopy. Single Mol. 2001;2:251–254. doi: 10.1002/1438-5171(200112)2:4<251::AID-SIMO251>3.0.CO;2-T. [DOI] [Google Scholar]
- 34.Felekyan S, Kalinin S, Sanabria H, Valeri A, Seidel CAM. Filtered FCS: species auto- and cross-correlation functions highlight binding and dynamics in biomolecules. ChemPhysChem. 2012;13:1036–1053. doi: 10.1002/cphc.201100897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Böhmer M, Wahl M, Rahn HJ, Erdmann R, Enderlein J. Time-resolved fluorescence correlation spectroscopy. Chem. Phys. Lett. 2002;353:439–445. doi: 10.1016/S0009-2614(02)00044-1. [DOI] [Google Scholar]
- 36.Cai Q, et al. Nanometer distance measurements in RNA using site-directed spin labeling. Biophys. J. 2007;93:2110–2117. doi: 10.1529/biophysj.107.109439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Muschielok A, et al. A nano-positioning system for macromolecular structural analysis. Nat. Meth. 2008;5:965–971. doi: 10.1038/nmeth.1259. [DOI] [PubMed] [Google Scholar]
- 38.Sindbert S, et al. Accurate distance determination of nucleic acids via Förster resonance energy transfer: implications of dye linker length and rigidity. J. Am. Chem. Soc. 2011;133:2463–2480. doi: 10.1021/ja105725e. [DOI] [PubMed] [Google Scholar]
- 39.Shoichet BK, Baase WA, Kuroki R, Matthews BW. A relationship between protein stability and protein function. Proc. Natl Acad. Sci. USA. 1995;92:452–456. doi: 10.1073/pnas.92.2.452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McHaourab HS, Oh KJ, Fang CJ, Hubbell WL. Conformation of T4 lysozyme in solution. Hinge-bending motion and the substrate-induced conformational transition studied by site-directed spin labeling. Biochemistry. 1997;36:307–316. doi: 10.1021/bi962114m. [DOI] [PubMed] [Google Scholar]
- 41.Cellitti J, et al. Exploring subdomain cooperativity in T4 lysozyme I: structural and energetic studies of a circular permutant and protein fragment. Protein Sci. 2007;16:842–851. doi: 10.1110/ps.062628607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Llinas M, Marqusee S. Subdomain interactions as a determinant in the folding and stability of T4 lysozyme. Protein Sci. 1998;7:96–104. doi: 10.1002/pro.5560070110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kalinin S, Valeri A, Antonik M, Felekyan S, Seidel CA. Detection of structural dynamics by FRET: a photon distribution and fluorescence lifetime analysis of systems with multiple states. J. Phys. Chem. B. 2010;114:7983–7995. doi: 10.1021/jp102156t. [DOI] [PubMed] [Google Scholar]
- 44.Gopich IV, Szabo A. Theory of the energy transfer efficiency and fluorescence lifetime distribution in single-molecule FRET. Proc. Natl Acad. Sci. USA. 2012;109:7747–7752. doi: 10.1073/pnas.1205120109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Felekyan S, Sanabria H, Kalinin S, Kühnemuth R, Seidel CAM. Analyzing Förster resonance energy transfer with fluctuation algorithms. Methods Enzymol. 2013;519:39–85. doi: 10.1016/B978-0-12-405539-1.00002-6. [DOI] [PubMed] [Google Scholar]
- 46.Gopich IV, Szabo A. Single-macromolecule fluorescence resonance energy transfer and free-energy profiles. J. Phys. Chem. B. 2003;107:5058–5063. doi: 10.1021/jp027481o. [DOI] [Google Scholar]
- 47.Peulen TO, Opanasyuk O, Seidel CAM. Combining graphical and analytical methods with molecular simulations to analyze time-resolved FRET measurements of labeled macromolecules accurately. J. Phys. Chem. B. 2017;121:8211–8241. doi: 10.1021/acs.jpcb.7b03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sagermann M, Martensson LG, Baase WA, Matthews BW. A test of proposed rules for helix capping: implications for protein design. Protein Sci. 2002;11:516–521. doi: 10.1110/ps.39802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Islam SM, Stein RA, McHaourab HS, Roux B. Structural refinement from restrained-ensemble simulations based on EPR/DEER data: application to T4 lysozyme. J. Phys. Chem. B. 2013;117:4740–4754. doi: 10.1021/jp311723a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stein RA, Beth AH, Hustedt EJ. A straightforward approach to the analysis of double electron-electron resonance data. Methods Enzymol. 2015;563:531–567. doi: 10.1016/bs.mie.2015.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chiang YW, Borbat PP, Freed JH. The determination of pair distance distributions by pulsed ESR using Tikhonov regularization. J. Magn. Reson. 2005;172:279–295. doi: 10.1016/j.jmr.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 52.Chiang YW, Borbat PP, Freed JH. Maximum entropy: a complement to Tikhonov regularization for determination of pair distance distributions by pulsed ESR. J. Magn. Reson. 2005;177:184–196. doi: 10.1016/j.jmr.2005.07.021. [DOI] [PubMed] [Google Scholar]
- 53.Klein-Seetharaman J, et al. Long-range interactions within a nonnative protein. Science. 2002;295:1719–1722. doi: 10.1126/science.1067680. [DOI] [PubMed] [Google Scholar]
- 54.Hu D, Lu HP. Placing single-molecule T4 lysozyme enzymes on a bacterial cell surface: toward probing single-molecule enzymatic reaction in living cells. Biophys. J. 2004;87:656–661. doi: 10.1529/biophysj.104.042101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xue M, et al. How internal cavities destabilize a protein. Proc. Natl Acad. Sci. USA. 2019;116:21031–21036. doi: 10.1073/pnas.1911181116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rennell D, Bouvier SE, Hardy LW, Poteete AR. Systematic mutation of bacteriophage T4 lysozyme. J. Mol. Biol. 1991;222:67–88. doi: 10.1016/0022-2836(91)90738-R. [DOI] [PubMed] [Google Scholar]
- 57.Chen Y, Hu DH, Vorpagel ER, Lu HP. Probing single-molecule T4 lysozyme conformational dynamics by intramolecular fluorescence energy transfer. J. Phys. Chem. B. 2003;107:7947–7956. doi: 10.1021/jp022406z. [DOI] [Google Scholar]
- 58.Fersht, A. Structure and Mechanism in Protein Science: A Guide to Enzyme Catalysis and Protein Folding. First edn. (W. H. Freeman, 1998).
- 59.Ratzke C, Hellenkamp B, Hugel T. Four-colour FRET reveals directionality in the Hsp90 multicomponent machinery. Nat. Commun. 2014;5:4192. doi: 10.1038/ncomms5192. [DOI] [PubMed] [Google Scholar]
- 60.Vale RD, Oosawa F. Protein motors and Maxwell’s demons: does mechanochemical transduction involve a thermal ratchet? Adv. Biophys. 1990;26:97–134. doi: 10.1016/0065-227X(90)90009-I. [DOI] [PubMed] [Google Scholar]
- 61.Gohlke H, et al. Binding region of alanopine dehydrogenase predicted by unbiased molecular dynamics simulations of ligand diffusion. J. Chem. Inf. Model. 2013;53:2493–2498. doi: 10.1021/ci400370y. [DOI] [PubMed] [Google Scholar]
- 62.Cellitti J, Bernstein R, Marqusee S. Exploring subdomain cooperativity in T4 lysozyme II: uncovering the C-terminal subdomain as a hidden intermediate in the kinetic folding pathway. Protein Sci. 2007;16:852–862. doi: 10.1110/ps.062632807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lerner E, et al. Toward dynamic structural biology: Two decades of single-molecule Förster resonance energy transfer. Science. 2018;359:288. doi: 10.1126/science.aan1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ross JL. The dark matter of biology. Biophys. J. 2016;111:909–916. doi: 10.1016/j.bpj.2016.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vallat B, Webb B, Westbrook JD, Sali A, Berman HM. Development of a prototype system for archiving integrative/hybrid structure models of biological macromolecules. Structure. 2018;26:894–904. doi: 10.1016/j.str.2018.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lemke EA. Site-specific labeling of proteins for single-molecule FRET measurements using genetically encoded ketone functionalities. Methods Mol. Biol. 2011;751:3–15. doi: 10.1007/978-1-61779-151-2_1. [DOI] [PubMed] [Google Scholar]
- 67.Brustad EM, Lemke EA, Schultz PG, Deniz AA. A general and efficient method for the site-specific dual-labeling of proteins for single molecule fluorescence resonance energy transfer. J. Am. Chem. Soc. 2008;130:17664–17665. doi: 10.1021/ja807430h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fleissner MR, et al. Site-directed spin labeling of a genetically encoded unnatural amino acid. Proc. Natl Acad. Sci. USA. 2009;106:21637–21642. doi: 10.1073/pnas.0912009106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Picoquant. https://www.picoquant.com/images/uploads/page/files/14528/technote_tttr.pdf (2019).
- 70.Goodman J, Weare J. Ensemble samplers with affine invariance. Comm. Appl. Math. Comp. Sci. 2010;5:65–80. doi: 10.2140/camcos.2010.5.65. [DOI] [Google Scholar]
- 71.Altenbach, C. https://sites.google.com/site/altenbach/labview-programs/epr-programs/long-distances (2019).
- 72.Kask P, Palo K, Ullmann D, Gall K. Fluorescence-intensity distribution analysis and its application in biomolecular detection technology. Proc. Natl Acad. Sci. USA. 1999;96:13756–13761. doi: 10.1073/pnas.96.24.13756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dix JA, Hom EF, Verkman AS. Fluorescence correlation spectroscopy simulations of photophysical phenomena and molecular interactions: a molecular dynamics/monte carlo approach. J. Phys. Chem. B. 2006;110:1896–1906. doi: 10.1021/jp055840k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Enderlein J, Robbins DL, Ambrose WP, Goodwin PM, Keller RA. Statistics of single-molecule detection. J. Phys. Chem. B. 1997;101:3626–3632. doi: 10.1021/jp963261x. [DOI] [Google Scholar]
- 75.Laurence TA, Kapanidis AN, Kong XX, Chemla DS, Weiss S. Photon arrival-time interval distribution (PAID): a novel tool for analyzing molecular interactions. J. Phys. Chem. B. 2004;108:3051–3067. doi: 10.1021/jp036499b. [DOI] [Google Scholar]
- 76.Müllner D. fastcluster: Fast hierarchical, agglomerative clustering routines for R and Python. J. Stat. Softw. 2013;53:1–18. doi: 10.18637/jss.v053.i09. [DOI] [Google Scholar]
- 77.Dale RE, Eisinger J, Blumberg WE. The orientational freedom of molecular probes. The orientation factor in intramolecular energy transfer. Biophys. J. 1979;26:161–193. doi: 10.1016/S0006-3495(79)85243-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The source data for all variants and all used techniques and screening results for the PDB structures were uploaded to Zenodo under DOI 10.5281/zenodo.3376527 and are described in Supplementary Note 7. The Zenodo archive contains subfolders for all eTCSPC FRET data including reference measurements used for derivation of DA distances (eTCSPC_wildtype.zip; e.g. Figures 4, 5, and Supplementary Fig. 5), single-molecule raw data used for filtered FCS and MFD analysis (Single_molecule_wildtype.zip; e.g. Figures 2, 3, Supplementary Figs. 2, 4, and 9), single molecule data of functional variants used for derivation of the kinetic scheme (Single_molecule_functional_variants.zip; e.g. Figure 6, Supplementary Figs. 6, 7, 9, and 11), EPR data (EPR_wildtype.zip; e.g., Figure 6 and Supplementary Fig. 5), and an overview on the FRET screening of PDB structures (FRET_screening_of_PDB_structures.zip). The source data underlying Supplementary Fig. 8c is shown in the Source Data File. Further datasets of processed data and the analysis are available from the corresponding authors on reasonable request.
Most general custom-made computer code is directly available from http://www.mpc.hhu.de/en/software. Additional computer code custom-made for this publication is available upon request from the corresponding authors. In-house programs are used (1) in the confocal multiparameter fluorescence detection experiments, (2) to elucidate the filtered fluorescence correlation spectroscopy curves, and (3) to analyze the fluorescence lifetime measurements. Software for analysis of single-molecule measurements and fluorescence correlation analysis and their simulation is available at http://www.mpc.uni-duesseldorf.de and software for analysis of fluorescence decays can be downloaded from http://www.fret.at/tutorial/chisurf/.