Abstract
Silencing of transposable elements (TEs) is established by small RNA-directed DNA methylation (RdDM). Maintenance of silencing is then based on a combination of RdDM and RNA-independent mechanisms involving DNA methyltransferase MET1 and chromodomain DNA methyltransferases (CMTs). Involvement of RdDM, according to this model should decrease with TE age but here we show a different pattern in tomato and Arabidopsis. In these species the CMTs silence long terminal repeat (LTR) transposons in the distal chromatin that are younger than those affected by RdDM. To account for these findings we propose that, after establishment of primary RdDM as in the original model, there is an RNA-independent maintenance phase involving CMTs followed by secondary RdDM. This progression of epigenetic silencing in the gene-rich distal chromatin is likely to influence the transcriptome either in cis or in trans depending on whether the mechanisms are RNA-dependent or -independent.
Subject terms: Gene silencing, Plant molecular biology
RNA-directed DNA methylation (RdDM) is thought to silence newly inserted transposable elements (TEs) with RNA-independent mechanisms becoming more prominent as TEs age. Here, the authors show that RdDM continues to silence the oldest intact distal TEs in tomato and Arabidopsis suggesting a second, later phase of RdDM.
Introduction
Epigenetic control of genomes is associated with chemical modifications of DNA and histone1, or both, that are often linked to silencing of transposable elements (TEs) as part of a genome defense system2. There are also effects of these modifications on gene expression, chromosome behavior and differentiation of pericentric heterochromatin and distal euchromatin. Full understanding of genomes, therefore, requires knowledge of factors affecting the establishment and maintenance of epigenetic marks. The simplest explanation of these factors invoke DNA sequence elements or structures that are associated with recruitment of DNA- or histone-methyltransferases to chromatin domains through the RNA-directed DNA methylation (RdDM) and other pathways2. Once established, DNA methylation can be maintained through cell division by different factors depending on the sequence context2,3.
To investigate factors influencing epigenetic marks in plants we focus here on tomato in which there are large and well-separated pericentric and distal chromatin domains4,5. In Arabidopsis, in contrast, the pericentric region is relatively small and largely restricted to narrow domains around the centromere and small knobs on chromosome 46. DNA methylation in tomato, as in other plants, occurs at CG, CHG and CHH sequence contexts (H indicates A, T, or C). Loss of CG methylation in the met1 mutant disrupts growth rate, flowering time, and gametogenesis in Arabidopsis7–9. In contrast the loss of CHG or CHH methylation does not show any obvious phenotype in Arabidopsis10, presumably because the levels of CHG/CHH methylation in WT is lower than CG methylation11,12.
To gain insights into the roles of CHG methylation in tomato, we utilized CRISPR to knock out tomato homologs of KYP and CMT3 which are required for maintenance of CHG methylation13,14. Genome-wide methylation profiles reveal that, as in Arabidopsis, SlKYP and SlCMT3 are required for CHG methylation maintenance, and CHH methylation to a lesser extent. However, unlike Arabidopsis, single slkyp or slcmt3 mutants have pleiotropic phenotypes affecting growth, development and reproduction. Our analysis of these mutants together with the slnrpd1 mutant15 identifies anti-correlation between CMT3-mediated and RdDM-mediated TE silencing in distal chromatin. In terms of intact long terminal repeats (LTR) transposons, CMT3 tends to target younger elements than RdDM.
Results
CRISPR-based mutation of SlKYP and SlCMT3
Tomato is a good epigenetic model plant because it has a clearly delineated heterochromatic region in the pericentromere. There are mutants and knock down lines of epigenetic pathway genes and a well characterized effect of DNA methylation in fruit ripening15–18. To extend our previous analysis of DNA methylation in tomato we used CRISPR small guide RNAs (sgRNAs) to target tomato orthologs of AtCMT3 and AtKYP—the major genes involved in CHG methylation in Arabidopsis13,14. AtKYP binds methylated CHG, mediates H3K9 di-methylation that, in turn, recruits CMT319,20. The SlMET1l (Solyc01g006100) and SlMET3l (Solyc12g100330)21,22 loci are two tomato orthologs with similarity to AtCMT3 (Supplementary Fig. 1a) and we refer to them as (Sl)CMT3a and (Sl)CMT3b. The tomato ortholog of AtKYP is SlSDG5 (Solyc02g094520) (Supplementary Fig. 1b,c)23 and we refer to it as (Sl)KYP.
Consistent with the functions of their Arabidopsis orthologs the CMT3a, CMT3b, and KYP mutations (Supplementary Fig. 2) had no effect on CG methylation (Supplementary Figs. 3a and 4a)24. However, also as in Arabidopsis, CHG methylation in cmt3a and kyp was lower than WT with kyp having a stronger effect than cmt3a (Supplementary Fig. 3b). There was also a genome-wide reduction of CHH methylation in kyp (Supplementary Fig. 3c). In cmt3b there was no effect in any context (Supplementary Fig. 4). From these results we conclude that SlCMT3a rather than SlCMT3b is the primary functional homolog of AtCMT3. Most (89%) of the differentially methylated regions (DMRs) (CHG hypomethylation) in kyp (Supplementary Fig. 5a,b) overlapped with cmt3a DMRs (Supplementary Fig. 5c). The CHH kyp and cmt3a hypoDMRs represent subsets of kyp and cmt3a CHG hypoDMRs (Supplementary Fig. 5c).
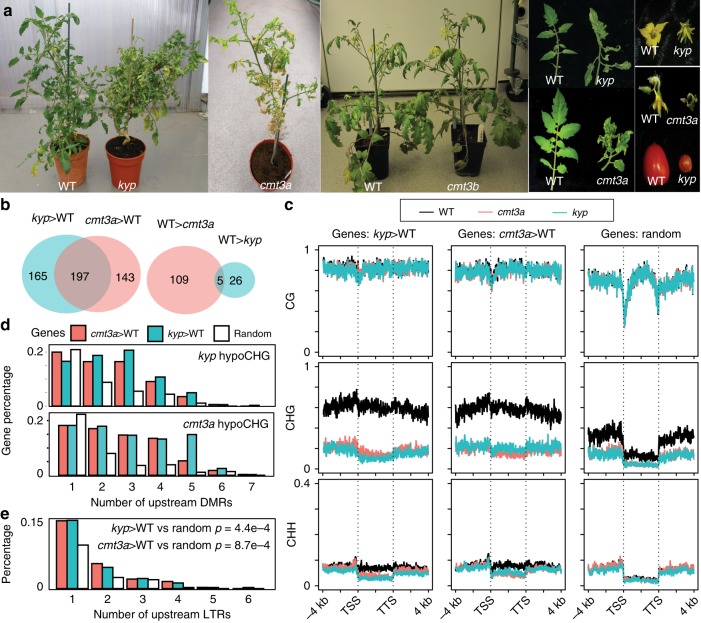
The kyp and cmt3a but not cmt3b mutants were stunted with wrinkled, asymmetric leaves with yellow spots, few and abnormal flowers, reduced fruit production and with no seeds (Fig. 1a). As DNA methylation may cause transcriptional silencing we predicted that the disrupted growth would be due to CHG hypomethylation in promoter regions leading to gene over-expression. Consistent with that idea, there were 362 upregulated genes and only 31 that were downregulated in kyp (Supplementary Data 1). Similarly, 340 genes were upregulated in cmt3a and 114 were downregulated (Supplementary Data 2). There is an overlap in half of the upregulated but very few of the downregulated genes in kyp and cmt3a (Fig. 1b).
Fig. 1. SlKYP and SlCMT3a are required for tomato growth, development and reproduction.
a Mutants phenotypes. One-month-old plants, leaves from a single branch, flowers and fruits of WT and mutants were shown. b Venn diagrams showing overlap analysis of differentially expressed (DE) genes; kyp > WT, upregulated genes in kyp; cmt3a > WT, upregulated genes in cmt3a; WT > cmt3a, downregulated genes in cmt3a; WT > kyp, downregulated genes in kyp. The upregulated genes are not mis-annotated TEs because 271 of 340 cmt3a-upregulated genes and 289 of 362 kyp-upregulated genes are well annotated protein-coding sequences with high confidence (Supplementary Data 1 and 2). c Average DNA methylation levels across DE genes or random genes and flanking regions in WT and mutants. TSS and TTS indicate transcription start site and termination site, respectively. d Number of kyp hypoCHG DMRs and cmt3a hypoCHG DMRs within upstream 2 kb regions of DE genes or random genes. Y-axis represents the percentage of genes that have different numbers of DMRs in their upstream 2 kb regions. e Number of LTR TEs within upstream 2 kb regions of DE genes or random genes. Y-axis represents the percentage of genes that have different numbers of LTRs in their upstream 2 kb regions. T-tests are performed between DE genes and random genes and p-values are shown.
Corresponding to the proposed link of DMRs with CMT- and KYP-mediated effects on gene expression, there was a dramatic reduction of CHG methylation and a slight reduction of CHH methylation in the gene body and flanking region of upregulated genes in kyp and cmt3a compared with random genes (Fig. 1c). Consistent with this pattern, there were more hypoCHG DMRs (both kyp DMRs and cmt3a DMRs) in upregulated genes in mutants compared with random genes (Fig. 1d).
It is likely that the CMT- and KYP-mediated DMRs and differentially expressed genes are related to TEs. The pericentric distribution of CMT-dependent CHG methylation and KYP-dependent CHG and CHH methylation (Supplementary Figs. 5d,e and S6), for example, coincides with the chromosomal distribution of Gypsy and Copia LTR elements identified by RepeatModeler25 and anti-correlates with the distribution of TIR elements and protein-coding genes (Supplementary Fig. 7). In addition, the overexpressed genes were more associated with LTR TEs than the random genes (Fig. 1e and Supplementary Fig. 8).
KYP/CMT3a and RdDM in pericentric and distal chromatin
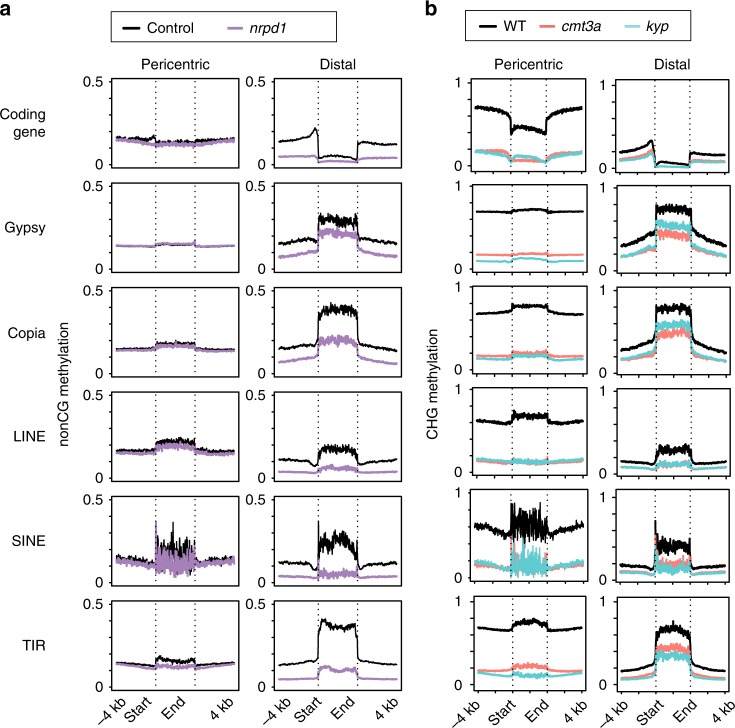
Therefore, to better understand CMT- and KYP-dependent CHG and CHH methylation, we analyzed the methylation pattern of TEs in the wild type and mutant plants. As the tomato chromosomes are predominantly pericentric heterochromatin we looked separately at the pericentric and distal chromosome regions (Fig. 2 and Supplementary Figs. 6 and 9). For comparison, we include data from an RdDM-defective nrpd1 mutant that has a greater effect on CHH methylation in the distal chromosome arms than in the pericentric region15.
Fig. 2. DNA methylation of TEs in pericentric and distal chromatin.
a Average non-CG methylation over coding genes and different TE families in control and nrpd1. b Average CHG methylation in WT, kyp and cmt3a. Distal and pericentric chromatins are plotted separately. Different genomic elements are aligned at the 5′ end or the 3′ end, and average methylation for all cytosines within each 100 bp interval is plotted. The dotted lines represent the points of alignments of coding-gene transcriptional start site or annotated repeat elements start site and coding-gene transcription termination site or annotated repeat elements end site, respectively. TIR, terminal inverted repeats; LINE, long interspersed nuclear elements; SINE, short interspersed nuclear elements.
As in Arabidopsis26, the CHH methylation in distal TEs is primarily due to NRPD1 and KYP in pericentric regions (Supplementary Fig. 9a,b). At CHG NRPD1 has little effect (Supplementary Fig. 9c) but both KYP and CMT3a are influential at both pericentric and distal TEs (Fig. 2b). In the pericentric regions, however, the patterns of CMT3a- and KYP-mediated DNA methylation were not specific for TEs (Fig. 2b). There was a high level of CHG methylation and strong CHG hypomethylation both within these elements and also in the flanking DNA (Fig. 2b). The lack of specificity of cmt3a and kyp CHG hypomethylation in the pericentric region is reinforced by the strong CHG hypomethylation pattern at the transcribed and flanking region of protein-coding genes (Fig. 2b). This pattern is also consistent with the preferential pericentric distribution of upregulated genes in cmt3a and kyp (Supplementary Fig. 10).
In the distal chromatin, however, the TEs were specific foci of CHG and CHH methylation (Fig. 2a, b). NRPD1 mediates CHG and CHH methylation (non-CG methylation) and CMT3a and KYP mediates CHG methylation in the distal chromosomal domains (Fig. 2 and Supplementary Fig. 9) and the cmt3a and kyp CHG DMRs were less dense than in the pericentric region (Supplementary Fig. 5d, e). There were higher levels of CHG and CHH DNA methylation in the distal TEs than their flanking DNA and, with Gypsy, Copia and TIR class 2 type elements, there was a proximity effect: the CHG methylation in the flanking DNA decreased with increasing distance from the transposon (Fig. 2b). In all TEs there was CHG (or non-CG for nrpd1) hypomethylation in the mutants (Fig. 2a, b). In contrast there was very little differential DNA methylation in protein-coding genes in the distal chromatin (Fig. 2a, b). From these findings we conclude that there is a high degree of specificity of CMT3a, KYP and NRPD1 for transposons in the distal but not in the pericentric regions of tomato chromosomes (Fig. 2a, b).
CMT3a effects anti-correlate RdDM at distal TEs
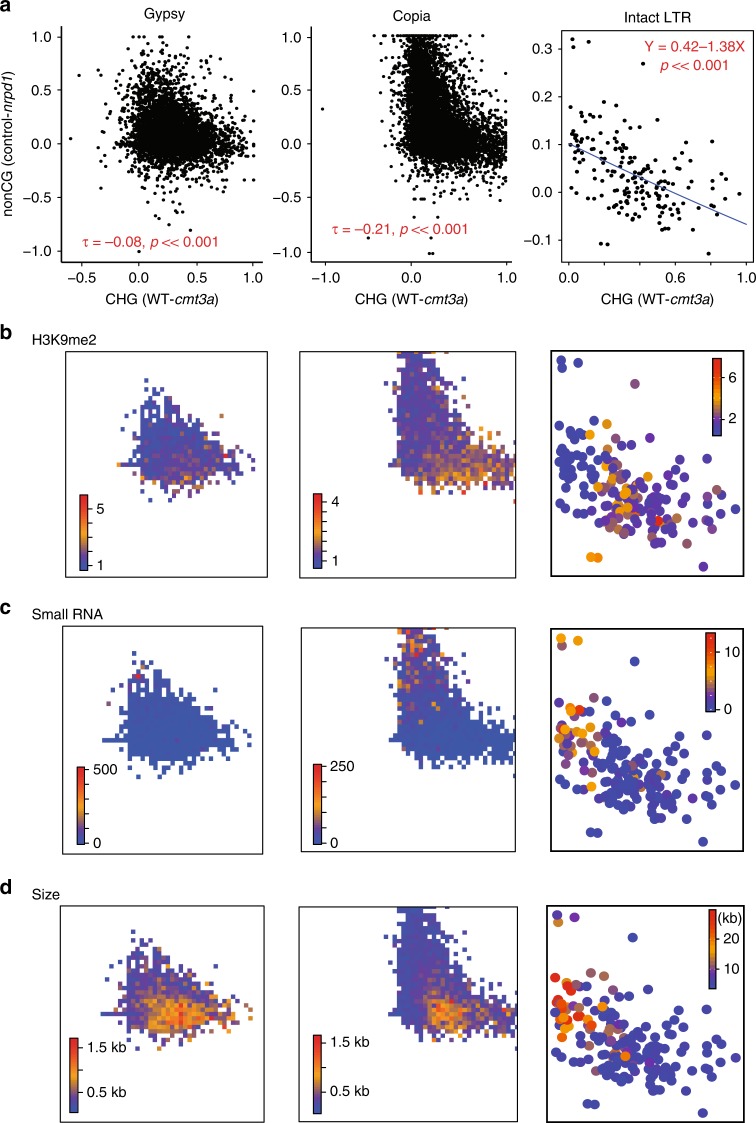
To investigate whether CMT3a-mediated CHG methylation coincides with NRPD1-mediated non-CG methylation in the distal chromatin (Fig. 2), we plotted the change in non-CG methylation due to nrpd1 vs the change in cmt3a-mediated CHG methylation of each TE in each TE family (Fig. 3a and Supplementary Fig. 11a). With most element types (except LINE) there was an inverse relationship of non-CG hypomethylation in nrpd1 with CHG hypomethylation in cmt3a (Fig. 3a and Supplementary Fig. 11a). This inverse relationship was most pronounced with the Gypsy, Copia and TIR elements (Fig. 3a and Supplementary Fig. 11a). The intact LTRs identified by LTRpred27,28 also show a similar negative correlation between non-CG hypomethylation in nrpd1 and CHG hypomethylation in cmt3a (Fig. 3a).
Fig. 3. CMT3a-mediated CHG methylation anti-correlates NRPD1-mediated non-CG methylation in distal LTR TEs.
a Scatter plots of hypomethylation of each TE. X-axis represents the reduction of CHG methylation in cmt3a, y-axis represents the reduction of non-CG methylation in nrpd1. Kendall’s tau test is performed for Gypsy and Copia. The Kendall rank correlation coefficients and p values relative to the null hypothesis of no correlation are indicated in red. Linear regression is performed for intact LTRs (the blue line). Regression model and the p-value relative to the null hypothesis that the line has zero gradient are indicated in red. b–d H3K9me2 (b), small RNA (c) and size (d) profiling of each TE. For Gypsy and Copia, color key represents average levels of H3K9me2 (b), small RNA (c) or size (d) of no less than three dots within each colored square. For intact LTR, color key represents levels of H3K9me2 (b), small RNA (c) or size (d) of each dot.
Not surprisingly, given the biochemistry of plant epigenetics2,3, the distal nrpd1-hypomethylated elements were less associated with H3K9me2 and more associated with small RNA than the targets of CMT3a (Fig. 3b, c and Supplementary Fig. 11b,c). These trends were similar for distal Gypsy, Copia, TIR and for intact LTRs. With element size, however, there were opposite trends in distal Gypsy, Copia and TIR vs intact LTRs. With Gypsy, Copia and TIR the nrpd1-affected elements tended to be smaller (Fig. 3d and Supplementary Fig. 11d), as reported previously24,26. However, with the intact LTRs, the CMT3a targets were smaller (Fig. 3d).
To explain these data with the intact LTRs we invoke TE age. The intact LTRs targeted by RdDM tend to be very large (5–10 kb and above) which could reflect secondary transposition events into existing elements29. However, the smaller intact elements of approximately 5 kb may be close homologs of functional transposons that have recently mobilized. According to this age-based hypothesis the NRPD1 targeted intact LTRs in distal chromatin would be older than those affected by CMT3a (Fig. 3d).
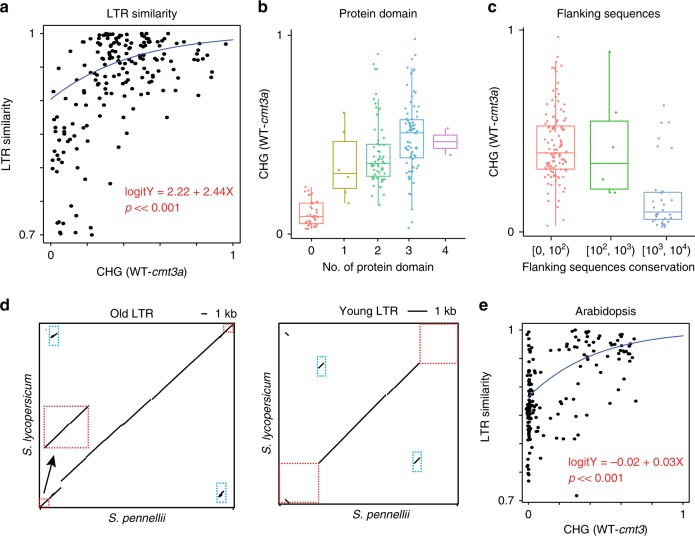
Preferential CHG methylation of younger LTRs by CMT3a
Three other features of the intact LTRs are consistent with this proposed age effect. First there was a positive correlation of the cmt3a hypoCHG methylation with 5′ and 3′LTR similarity that is a good indicator of LTR age30 (Fig. 4a). Second there is a correlation of cmt3a hypoCHG methylation with the presence of protein domains characteristic of transposition function (Fig. 4b). These protein domains are likely to be lost with transposon age. The third feature is in the flanking sequences of the intact LTRs that are more similar in Solanum pennelli and Solanum lycopersicum with the less cmt3a hypoCHG methylated elements (Fig. 4c, d). Conservation of flanking sequence is an indicator of older transposition events that occurred before the divergence of these two species.
Fig. 4. CMT3a-targeted intact LTRs are evolutionarily younger elements.
a Scatter plot of distal intact LTRs. X-axis represents the reduction of CHG methylation in cmt3a, y-axis represents the 5′ and 3′ LTR similarity. Regression-blue line. The regression model and p value are indicated in red. The p value is relative to the null hypothesis of there being a gradient of zero and so no causal relationship between the reduction of CHG methylation in cmt3a and the 5′ and 3′ LTR similarity. b Correlation of number of predicted protein domains of each intact LTRs (x-axis) and reduction of CHG methylation in cmt3a (y-axis). c Alignments results of intact LTR flanking sequences (2 kb upstream and 2 kb downstream) between S. lycopersicum and S. pennellii. X-axis represents three ranges of the alignment score by BLASTn. Y-axis represents the reduction of CHG methylation in cmt3a. d Dot plots of each one example of old and young element comparing the LTR sequence from S. lycopersicum and its homologous sequence in S. pennellii genome, plus 2 kb upstream and downstream regions. Blue boxes highlight the 5′ and 3′ LTRs. Red boxes highlight the flanking regions. e Scatter plot of Arabidopsis distal intact LTRs. X-axis represents the reduction of CHG methylation in cmt3, y-axis represents the 5′ and 3′ LTR similarity. Regression-blue line. The regression model and p value are indicated in red. The p value is relative to the null hypothesis of there being a gradient of zero and so no causal relationship between the reduction of CHG methylation in cmt3 and the 5′ and 3′ LTR similarity.
This pattern of CMT-dependent silencing of younger LTRs in distal chromatin is characteristic of the Rider family of LTR retrotransposons in tomato28 (Supplementary Fig. 12) and it features with Arabidopsis LTRs: intact LTRs with CMT-dependent CHG methylation in Arabidopsis are larger and also have more similar LTRs than other elements (Supplementary Figs. 13 and 14 and Fig. 4e). CMT3a-mediated CHG methylation plays silencing roles on these young intact LTRs because the mRNA levels of these LTRs were upregulated in cmt3a than in WT (Supplementary Fig. 15).
Discussion
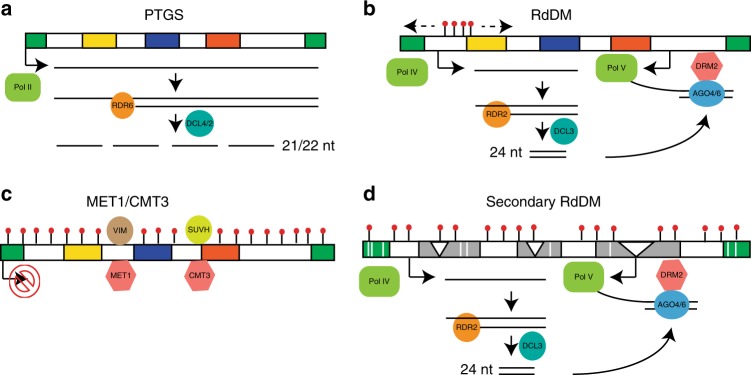
Post-transcriptional gene silencing and RdDM had been previously implicated in retrotransposon silencing during the phases of retrotransposition and copy number increase31 (Fig. 5a, b). The RdDM elements are then thought to progress to RNA-independent silencing in which the CMT and MET1 methyltransferases would be involved3,32 (Fig. 5c). We interpret the KYP/CMT3a-dependent silencing of intact LTR elements (Figs. 3 and 4) as corresponding to that RNA-independent stage.
Fig. 5. Model of LTR silencing.
a In early stages, LTR is transcribed by RNA Pol II. Post-transcriptional gene silencing (PTGS) is triggered with double-stranded RNA synthesis by RDR6 and afterwards 21-22nt sRNA generation by DCL4 and/or DCL2. b Along with increase of LTR copy number, 24nt sRNA is produced starting from certain region of LTR (gag for example) by RNA Pol IV, RDR2 and DCL3. These 24nt sRNAs are recruited into AGO4/6 which interact with DNA methyltransferase DRM2. By sequence pairing between 24nt sRNA and Pol V transcript, AGO-DRM2 complex function in cis to methylate LTR which is called RNA-directed DNA methylation (RdDM). RdDM then spreads to more regions of LTR. c After established by RdDM, DNA methylation is maintained by RNA-independent mechanism: CG methylation is maintained by VIM and MET1, CHG methylation is maintained by KYP/SUVHs and CMT3. LTR is rarely transcribed. d Accumulated mutations in 5′ and 3′ LTRs and coding sequences (represented by white bars) together with TE re-insertion (represented by white triangles) make it less possible for LTR to transpose. At this stage, LTR is transcribed again and generates 24 nt sRNA, which mediates secondary RdDM.
To account for our observations on H3K9me2, sRNA, transposon size and transposon age (Figs. 3 and 4) we propose that these elements would later undergo rearrangements and diminished transposon functionality. These rearranged elements would still be silenced by KYP/CMT3a (Fig. 5c) but, over time in the oldest elements, we envision that KYP/CMT3a-mediated CHG methylation would decrease and transcriptional silencing would be relaxed (Fig. 5d) so that RdDM would be re-established (Fig. 3).
According to this model there are separate phases of RdDM at two stages in the life and death cycle of retrotransposons. The first RdDM phase (Fig. 5b) would be short lived and observable only first few generations following invasion of a genome by a new transposon by introgression or horizontal gene transfer. This stage has been observed experimentally when the RNA-independent silencing of TEs has been released by mutation of MET1, DDM1, or CMT329,33,34. It is likely that there are very few intact elements in wild type genomes undergoing this primary RdDM because RdDM mutants of Arabidopsis do not exhibit transposon mobilization unless other stresses are applied that might release CMT- and MET1-mediated silencing35.
A possible function of the sRNAs from the older elements in the second phase of RdDM is similar to that of piRNAs in animal systems: they would be a reservoir of trans-acting silencing factors that could protect against re-invasion of the genome by TEs with similar sequence identity36. They could also act in trans within the genome to influence the gene expression profile and the elements could also act in cis to affect expression of adjacent genes. The younger elements, in contrast, with CMT-dependent silencing would only have an effect in cis on adjacent genes. Clearly the progression between different modes of LTR silencing will have a profound influence of the ways that these TEs are controlling elements of gene expression in the host genome.
Methods
Plasmid construction
The CRISPR/Cas9 constructs were made following the Golden Gate cloning strategy37. A pair of sgRNAs were designed to target KYP, CMT3a, or CMT3b and amplified using pICH86966:AtU6p:sgRNA_PDS construct as a template. PCR products containing each sgRNA, together with pICSL01009:AtU6p providing the Arabidopsis U6 promoter, were cloned into level 1 constructs pICH47751 and pICH47761 respectively using BsaI (Thermo Fisher) and T4 DNA ligase (NEB). Together with other level 1 constructs (pICH47732:NOSp:NPTII, pICH47742:35S:Cas9) and the linker pICH41766, level 1 sgRNA constructs were assembled into the level 2 vector pAGM4723 using BpiI (Thermo Fisher) and T4 DNA ligase (NEB).
Plant materials
All tomato plants used in this study are Solanum lycopersicum cv M82. A. tumefaciens strain AGL1 containing the CRISPR/Cas9 construct was used to transform tomato. In brief, tomato seeds were surfaced sterilized in 70% ethanol for 2 min followed by 2.2% sodium hypochlorite for 15 min and rinsed in sterilized water five times. The sterilized seeds were transferred to ½ strength Murashige-Skoog (MS) medium with vitamins, 0.8% agar, 1.5% sucrose. Cotyledons from 7-day-old plants were cut in two and submerged in a solution of Agrobacterium in MS liquid medium with 3% sucrose at OD600 = 0.5. The explants were then dried on filter paper and placed on a plate (1X MS medium, 0.6% agar, 0.5 mg/L 2,4-D) under dim light. After two days cultivation, the cotyledon segments were transferred to a selective regeneration medium (1X MS medium, 1X Nitsch vitamins, 0.1 g/L Myo inositol, 2% sucrose, 0.4% agar, 320 mg/L Timentin, 25 mg/L Cefotaxime, 2 mg/L Zeatin riboside, 100 mg/L Kanamycin). The later appearing shoots were transferred to a selective rooting medium (1X MS medium with vitamins, 2% sucrose, 2.25% gelrite, 320 mg/L Timentin, 50 mg/L Kanamycin). Regenerates with shoots and roots were then transferred into soil and genotyped. Primers for sgRNA amplification and mutant genotyping were listed in Supplementary Data 3.
Bisulfite-seq
Genomic DNA was extracted from 100 mg of leaf tissue using Dneasy Plant Mini Kit (Qiagen). Bisulfite-seq library preparation was performed with three biological replicates for each genotype. 1 µg of genomic DNA was sonicated using the E220 Covaris instrument (Covaris Inc., USA) with parameters of incident power = 140 W, duty factor = 10%, cycles/burst = 200, treat time = 120 s. After being purified on XP beads (Ampure, ratio 1.8×), fragmented DNA was end-repaired and A-tailed using T4 DNA polymerase and Klenow Fragment (NEB) and purified again using XP beads (ratio 1.8×). Methylated Illumina Y-shaped adapters for paired-end sequencing were ligated using Quick-Stick ligase (Bioline). Purified (ratio 1×) adapter-ligated DNA was bisulfite-converted using the EZ DNA Methylation-Gold Kit (Zymo research). DNA was barcoded using 15 cycles of PCR amplification with KAPA HiFi HotStart Uracil+Ready Mix (KAPA Biosystems) with forward universal primer and reverse index primers. Pooled libraries were sequenced to a depth of about 12X genome coverage on a NextSeq 500 150PE (Illumina).
Methylation analysis
Bisulfite-seq data generated in this study and Arabidopsis bisulfite-seq data generated by Stroud et al24 were used for further analysis. Tomato sequences from bisulfite-seq were trimmed and filtered by Trim Galore! with default parameters, and then mapped onto the tomato genome (Heinz SL3.0) using Bismark v0.15.038 with option -N 1. Reads were deduplicated with deduplicate_bismark and methylation calls were extracted using bismark_methylation_extractor with option -ignore_r2 2. Raw data of Stroud et al24 (WT and cmt3) were downloaded from NCBI and processed following the same analyzing pipeline as tomato sequences, with exception of mapping onto the Arabidopsis genome (TAIR10). Differentially Methylated Region (DMR) analysis were performed with segmentSeq v3.839. DMRs were called for each context separately, with cutoffs of width > 100 bp, FDR < 0.01 and likelihood > 0.99.
Average methylation profiles over genes and TEs were calculated from the cytosine reports with segmentSeq v3.839 using the function of averageProfiles39. Annotations of tomato coding genes and TEs are based on SOL ITAG3.2. Arabidopsis and tomato intact LTRs were predicted by LTRpred (https://github.com/HajkD/LTRpred)27,28.
Methylation results of each TE were calculated from the cytosine reports with segmentSeq v3.839 by averaging the methylation data of all cytosines of the same context within each TE.
sRNA-seq
Total RNA was extracted by following the standard protocol of TRIzol method. 10 µg total RNA was run on a 15% PAGE/7 M Urea gel (Bio-Rad) and the sRNA fraction (18–25 nt) was excised and eluted from the gel, which was subsequently cloned using the NEBNext multiplex small RNA library prep kit (NEB). Libraries were indexed during the PCR step with 12 cycles and gel size-selected and purified. Four biological replicates of libraries for each genotype were constructed. Pooled libraries were sequenced on a NextSeq 500 (Illumina). Sequencing reads were trimmed using Trim Galore! with default parameters and then mapped to the Heinz genome SL3.0 using Bowtie 1.1.140 with specified parameters of -m 1 and -v 0 for unique mapping and no mismatch allowed respectively. The output sam files were converted to bam files by Samtools41. The uniquely mapped bam files were used for RPKM analysis of sRNA.
RNA-seq
Three micrograms of total RNA was purified using Ribo-Zero rRNA Removal Kit (Illumina). 250 ng purified RNA was used for RNA-seq library constructions following the manufacturer’s protocol of ScriptSeq RNA-Seq Library Kit (Illumina). RNA-seq libraries of three biological replicates for each genotype were sequenced on Illumina NextSeq 500. Sequencing reads were trimmed using Trim Galore! with default parameters and then mapped to the Heinz genome SL3.0 using Tophat v2.1.042 with specified parameters of −N 0 and −r 330. The output sam files were converted to bam files by Samtools41 which were used for RPKM analysis.Differentially expressed genes were analyzed by DESeq243 with cutoff of p < 0.01.
ChIP-seq and distal/pericentric chromosome
ChIP-seq was performed following standard protocol44. Half gram of new leaves was ground in liquid nitrogen and in vitro cross-linked. The extracts were filtered through Miracloth and centrifuged for 20 min at 4000 rpm at 4 °C. The nuclei pellet was washed by extraction buffer (0.25 M sucrose, 10 mM Tris-HCl pH8.0, 10 mM MgCl2, 1 mM EDTA, 1% Triton X-100, 0.1 mM PMSF, 1 µM pepstatin, 1X protein inhibitor cocktails, 5 mM β-mercaptoethanol) and re-suspended in 0.6 ml of nuclear lysis buffer (50 mM Tris-HCl pH 8.0, 10 mM EDTA, 1% SDS, 0.1 mM PMSF, 1 µM pepstatin, 1X protein inhibitor cocktails) and sonicated using Bioruptor for 15 min (high, 30 s ON/30 s OFF). The lysate was centrifuged at 13,200 rpm for 10 min at 4 °C and the supernatant was kept as chromatin samples.
Antibodies of H3K9me2 (Abcam ab1220) and H3K9ac (Millipore 07-352) were used to perform ChIP on chromatin extracts. Libraries were constructed following manufacturer’s protocol of TruSeq Library Prep Kit (Illumina) and sequenced on Illumina NextSeq 500. Sequencing reads were trimmed using Trim Galore! with default parameters and then mapped to the Heinz genome SL3.0 using Bowtie240. The output sam files were converted to bam files by Samtools41 which were used for RPKM analysis.
Normalized H3K9me2 and H3K9ac across each 100 kb window were calculated based on the ChIP-seq mapping files. Ratio of H3K9me2/H3K9ac was used to define distal/pericentric chromatin: H3K9me2/H3K9ac < 0.6, distal; H3K9me2/H3K9ac > 1.2, pericentric. Arabidopsis H3K9me2 ChIP-seq data45 was downloaded and processed following the same analysing pipeline as tomato sequences, with exception of mapping onto the Arabidopsis genome (TAIR10). The normalized H3K9me2 in each 100 kb window was calculated as the following formula:
H3K9me2 normalized level = Number of reads in each 100 kb window / (Number of total mapped reads/Number of 100 kb window in Arabidopsis genome)
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Acknowledgements
The authors are grateful to Dr. Thomas J. Hardcastle for help with bioinformatics and Dr. Nik Cunniffe for help with data analysis. We thank Dr. Hajk-Georg Drost for assistance with LTRpred and communicating unpublished results. We thank Drs. Julie Ahringer and Alex Appert for help with the ChIP-seq experiments. We thank James Barlow, For Fan Chan, Shiyu He, Mel Steer, Shuoya Tang and Wing Hin Yip for technical assistance. We thank Drs. Ian Henderson, Sara Lopez-Gomollon, Claudia dos santos Martinho, Sebastian Müller and Nataliya E. Yelina for helpful comments on the manuscript. This work was supported by European Research Council Advanced Investigator grant ERC-2013-AdG 340642 (Transgressive Inheritance in plant Breeding and Evolution [TRIBE]), the Royal Society (RP170001), the Balzan Foundation and BBSRC grant BB/R018529/1. D.C.B. is the Royal Society Edward Penley Abraham Research Professor.
Source Data
Author contributions
Z.W. and D.C.B. designed the experiments. Z.W. performed the experiments. Z.W. and D.C.B. analyzed the data. Z.W. and D.C.B. wrote the paper.
Data availability
All raw sequencing data generated in this study have been deposited into in the NCBI Sequence Read Archive (SRA) under accession number PRJNA516166. The source data underlying Figs. 1a, 1c–e, 2a–b, 3a–d, 4a–c, 4e and Supplementary Figs 1c, 2e, 3a–c, 4a–c, 5d–e, 6, 7, 8, 9a–c, 10, 11a–d, 12, 13, 14, 15 are provided as Source Data file.
Competing interests
The authors declare no competing interests.
Footnotes
Peer review information Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. Peer reviewer reports are available.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information is available for this paper at 10.1038/s41467-020-14995-6.
References
- 1.Bird A. Perceptions of epigenetics. Nature. 2007;447:396–398. doi: 10.1038/nature05913. [DOI] [PubMed] [Google Scholar]
- 2.Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 2011;11:204–220. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim MY, Zilberman D. DNA methylation as a system of plant genomic immunity. Trends Plant Sci. 2014;19:320–326. doi: 10.1016/j.tplants.2014.01.014. [DOI] [PubMed] [Google Scholar]
- 4.Consortium Tomato Genome. The tomato genome sequence provides insights into fleshy fruit evolution. Nature. 2012;485:635–641. doi: 10.1038/nature11119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jouffroy O, Saha S, Mueller L, Quesneville H, Maumus F. Comprehensive repeatome annotation reveals strong potential impact of repetitive elements on tomato ripening. BMC Genomics. 2016;17:624. doi: 10.1186/s12864-016-2980-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kaul S, et al. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- 7.Saze H, Scheid OM, Paszkowski J. Maintenance of CpG methylation is essential for epigenetic inheritance during plant gametogenesis. Nat. Genet. 2003;34:65–69. doi: 10.1038/ng1138. [DOI] [PubMed] [Google Scholar]
- 8.Kankel MW, et al. Arabidopsis MET1 cytosine methyltransferase mutants. Genetics. 2003;163:1109–1122. doi: 10.1093/genetics/163.3.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reinders J, et al. Compromised stability of DNA methylation and transposon immobilization in mosaic Arabidopsis epigenomes. Genes Dev. 2009;23:939–950. doi: 10.1101/gad.524609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chan SWL, et al. RNAi, DRD1, and histone methylation actively target developmentally important Non-CG DNA methylation in Arabidopsis. PLoS Genet. 2006;2:e83. doi: 10.1371/journal.pgen.0020083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cokus SJ, et al. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature. 2008;452:215–219. doi: 10.1038/nature06745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lister R, et al. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell. 2008;133:523–536. doi: 10.1016/j.cell.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lindroth AM, et al. Requirement of CHROMOMETHYLASE3 for maintenance of CpXpG methylation. Science. 2001;292:2077–2080. doi: 10.1126/science.1059745. [DOI] [PubMed] [Google Scholar]
- 14.Jackson JP, Lindroth AM, Cao X, Jacobsen SE. Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature. 2002;416:556–560. doi: 10.1038/nature731. [DOI] [PubMed] [Google Scholar]
- 15.Gouil Q, Baulcombe DC. DNA methylation signatures of the plant chromomethyltransferases. PLoS Genet. 2016;12:e1006526. doi: 10.1371/journal.pgen.1006526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhong S, et al. Single-base resolution methylomes of tomato fruit development reveal epigenome modifications associated with ripening. Nat. Biotechnol. 2013;31:154–159. doi: 10.1038/nbt.2462. [DOI] [PubMed] [Google Scholar]
- 17.Corem S, et al. Redistribution of CHH methylation and small interfering RNAs across the genome of tomato ddm1 mutants. Plant Cell. 2018;30:1628–1644. doi: 10.1105/tpc.18.00167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang Y, et al. Critical function of DNA methyltransferase 1 in tomato development and regulation of the DNA methylome and transcriptome. J. Integr. Plant Biol. 2019;61:1224–1242. doi: 10.1111/jipb.12778. [DOI] [PubMed] [Google Scholar]
- 19.Du J, et al. Mechanism of DNA methylation-directed histone methylation by KRYPTONITE. Mol. Cell. 2014;55:495–504. doi: 10.1016/j.molcel.2014.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Du J, et al. Dual binding of chromomethylase domains to H3K9me2-containing nucleosomes directs DNA methylation in plants. Cell. 2012;151:167–180. doi: 10.1016/j.cell.2012.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cao D, et al. Genome-wide identification of cytosine-5 DNA methyltransferases and demethylases in Solanum lycopersicum. Gene. 2014;550:230–237. doi: 10.1016/j.gene.2014.08.034. [DOI] [PubMed] [Google Scholar]
- 22.Bewick AJ, et al. The evolution of CHROMOMETHYLASES and gene body DNA methylation in plants. Genome Biol. 2017;18:65. doi: 10.1186/s13059-017-1195-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aiese Cigliano R, et al. Genome-wide analysis of histone modifiers in tomato: gaining an insight into their developmental roles. BMC Genomics. 2013;14:57. doi: 10.1186/1471-2164-14-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stroud H, Greenberg MVC, Feng S, Bernatavichute YV, Jacobsen SE. Comprehensive analysis of silencing mutants reveals complex regulation of the Arabidopsis methylome. Cell. 2013;152:352–364. doi: 10.1016/j.cell.2012.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mehra M, Gangwar I, Shankar R. A deluge of complex repeats: the Solanum genome. PLoS ONE. 2015;10:e0133962. doi: 10.1371/journal.pone.0133962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zemach A, et al. The Arabidopsis nucleosome remodeler DDM1 allows DNA methyltransferases to access H1-containing heterochromatin. Cell. 2013;153:193–205. doi: 10.1016/j.cell.2013.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cho J, et al. Sensitive detection of pre-integration intermediates of long terminal repeat retrotransposons in crop plants. Nat. Plants. 2019;5:26–33. doi: 10.1038/s41477-018-0320-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Benoit M, et al. Environmental and epigenetic regulation of Rider retrotransposons in tomato. PLoS Genet. 2019;15:e1008370. doi: 10.1371/journal.pgen.1008370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lippman Z, et al. Role of transposable elements in heterochromatin and epigenetic control. Nature. 2004;430:471–476. doi: 10.1038/nature02651. [DOI] [PubMed] [Google Scholar]
- 30.Wicker T, Keller B. Genome-wide comparative analysis of copia retrotransposons in Triticeae, rice, and Arabidopsis reveals conserved ancient evolutionary lineages and distinct dynamics of individual copia families. Genome Res. 2007;17:1072–1081. doi: 10.1101/gr.6214107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marí-Ordóñez A, et al. Reconstructing de novo silencing of an active plant retrotransposon. Nat. Genet. 2013;45:1029–1039. doi: 10.1038/ng.2703. [DOI] [PubMed] [Google Scholar]
- 32.Bond DM, Baulcombe DC. Small RNAs and heritable epigenetic variation in plants. Trends Cell Biol. 2014;24:100–107. doi: 10.1016/j.tcb.2013.08.001. [DOI] [PubMed] [Google Scholar]
- 33.Mirouze M, et al. Selective epigenetic control of retrotransposition in Arabidopsis. Nature. 2009;461:427–430. doi: 10.1038/nature08328. [DOI] [PubMed] [Google Scholar]
- 34.Kato M, Miura A, Bender J, Jacobsen SE, Kakutani T. Role of CG and non-CG methylation in immobilization of transposons in Arabidopsis. Curr. Biol. 2003;13:421–426. doi: 10.1016/S0960-9822(03)00106-4. [DOI] [PubMed] [Google Scholar]
- 35.Ito H, et al. An siRNA pathway prevents transgenerational retrotransposition in plants subjected to stress. Nature. 2011;472:115–119. doi: 10.1038/nature09861. [DOI] [PubMed] [Google Scholar]
- 36.Czech B, et al. piRNA-guided genome defense: from biogenesis to silencing. Annu. Rev. Genet. 2018;52:131–157. doi: 10.1146/annurev-genet-120417-031441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weber E, Engler C, Gruetzner R, Werner S, Marillonnet S. A modular cloning system for standardized assembly of multigene constructs. PLoS ONE. 2011;6:e16765. doi: 10.1371/journal.pone.0016765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Krueger F, Andrews SR. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics. 2011;27:1571–1572. doi: 10.1093/bioinformatics/btr167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hardcastle TJ. Methods for discovering genomic loci exhibiting complex patterns of differential methylation. BMC Bioinform. 2017;18:416. doi: 10.1186/s12859-017-1836-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li H, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:1–21. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Choi K, et al. Nucleosomes and DNA methylation shape meiotic DSB frequency in Arabidopsis thaliana transposons and gene regulatory regions. Genome Res. 2018;28:1–15. doi: 10.1101/gr.225599.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stroud H, et al. Non-CG methylation patterns shape the epigenetic landscape in Arabidopsis. Nat. Struct. Mol. Biol. 2014;21:64–72. doi: 10.1038/nsmb.2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
All raw sequencing data generated in this study have been deposited into in the NCBI Sequence Read Archive (SRA) under accession number PRJNA516166. The source data underlying Figs. 1a, 1c–e, 2a–b, 3a–d, 4a–c, 4e and Supplementary Figs 1c, 2e, 3a–c, 4a–c, 5d–e, 6, 7, 8, 9a–c, 10, 11a–d, 12, 13, 14, 15 are provided as Source Data file.