Abstract
ABCG2 belongs to the ABC transporter superfamily and functions as a poly‐specific efflux pump. As it can transport a broad spectrum of substrates out of cells, ABCG2 is thought to alter the pharmacokinetics of drugs applied to treat certain diseases. Especially, its potential to induce resistance to chemotherapy is currently the object of intense research. To foster understanding of mechanisms relevant for substrate recognition and selection of ABCG2 substrates and to finally develop selective therapeutic modulators (e.g. inhibitors) of ABCG2 transport activity, it is important to further explore the precise 3D structure of the transporter. While efforts to elucidate the three‐dimensional structure of ABCG2 using X‐ray crystal structure analysis have not been successful so far, high‐resolution cryo‐electron microscopy‐based investigations have revealed exciting new insights into the structure and function of the transporter. In this review, we will focus on these seminal publications to summarize the current understanding of tertiary and quaternary structure, homodimerization or oligomerization, and functions of the ABCG2 transporter protein.
Abbreviations
- ABC
ATP‐binding cassette
- ABCB1–7
ATP‐binding cassette transporter of subfamily B, members 1–7
- ABCC1
ATP‐binding cassette transporter of subfamily C, member 1
- ABCG2–8
ATP‐binding cassette transporter of subfamily G, members 2–8
- cryo‐EM
single particle cryogenic electron microscopy
- E1S
estrone‐3‐sulfate
- EL
extracellular loop
- NBD
nucleotide binding domain
- P‐gp
P‐glycoprotein (permeability glycoprotein)
- TM1–6
membrane spanning α‐helices 1–6
- TMD
transmembrane domain
1. INTRODUCTION
The multidrug efflux transporter https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=792&familyId=155&familyType=TRANSPORTER was discovered independently by three different research groups and initially named according to the respective research context either as breast cancer resistance protein (BCRP; L. A. Doyle et al., 1998), ABC transporter highly expressed in placenta (ABCP; Allikmets, Schriml, Hutchinson, Romano‐Spica, & Dean, 1998), or mitoxantrone resistance (MXR; Miyake et al., 1999). It is a member of the https://www.guidetopharmacology.org/GRAC/FamilyDisplayForward?familyId=136, which consists of at least 48 members divided into seven subfamilies (ABCA‐G; Dean, Hamon, & Chimini, 2001). The function of individual members of this protein family can range from receptors to ion channels to membrane transporters. Similar to other clinically relevant ABC transporters such as P‐glycoprotein (P‐gp, multidrug resistance protein 1, https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=768&familyId=152&familyType=TRANSPORTER; Y. H. Li, Wang, Li, & Yang, 2006) and multidrug resistance associated protein 1 (https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=779&familyId=153&familyType=TRANSPORTER; Johnson & Chen, 2018), ABCG2 functions as a rather non‐specific efflux pump which utilizes the energy of ATP hydrolysis to actively transport a variety of substrates across the membrane. A list of suggested ABCG2 substrates has been reviewed elsewhere (Mo & Zhang, 2012).
Physiologically, ABCG2 is highly expressed in placental syncytiotrophoblasts (Mao, 2008) and cerebral endothelium (Zhang et al., 2003) but can also be found at the apical membrane of polar epithelial cells in the intestine (Gutmann, Hruz, Zimmermann, Beglinger, & Drewe, 2005), liver (Maliepaard et al., 2001), and kidney (Woodward et al., 2009). Due to its expression pattern at entry and exit points of the body as well as in barriers (blood–brain and blood–placenta barrier), the protein is believed to function as a “bouncer” that protects critical tissue from entrance of unwanted substances (like toxins and xenobiotics). However, ABCG2's physiological role is also believed to potentially alter pharmacokinetics and distribution of drugs in the treatment of several diseases. In this regard, it has been suggested that drugs, such as https://www.guidetopharmacology.org/GRAC/FamilyIntroductionForward?familyId=6, that inhibit ABCG2 efflux function can induce drug–drug interactions via this route (Bajcetic et al., 2007; Ripperger, Krischer, Robaa, Sippl, & Benndorf, 2018; Schumacher & Benndorf, 2017; Weiss et al., 2010). Moreover, naturally occurring genetic polymorphisms, such as the Q141K ABCG2 variant (on transcript 421C>A), have been shown to affect the pharmacokinetics of drugs like https://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=6823 (Q. Zhou et al., 2005), https://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=2954 (Zhang et al., 2006), and 9‐aminocamptothecin (Zamboni et al., 2006). In addition, Q141K, which reduces the surface expression of ABCG2 (Deppe, Boger, Weiss, & Benndorf, 2010; Ripperger & Benndorf, 2016), is associated with https://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=4731 levels and may hence play a causal role in hyperuricaemia and gout (Furukawa et al., 2009; Kannangara et al., 2016; R. Li et al., 2015). Further information on ABCG2 polymorphisms and their effect on protein expression, function, and drug pharmacokinetics can be found elsewhere (Heyes, Kapoor, & Kerr, 2018).
However, ABCG2 has received most of its pharmacological attention as a potential candidate to cause multidrug resistance in cancer treatment with chemotherapy (L. Doyle & Ross, 2003; Robey, Polgar, Deeken, To, & Bates, 2007). Expression of ABCG2 was associated with poor prognosis in the treatment of acute myeloid leukaemia (Damiani et al., 2006) and diffuse large B‐cell lymphoma (Kim et al., 2009). In contrast, the role of ABCG2 in multidrug resistance of other cancer types is rather unclear, although these clinical observations are contradictory to the fact that many of the known ABCG2 substrates are actually chemotherapeutic agents (Polgar, Robey, & Bates, 2008). Thus, a more complete understanding of the structure of ABCG2 should help to answer many questions regarding the role of ABCG2 in cancer development and progression.
2. PRIMARY SEQUENCE AND STRUCTURE PREDICTION
After the first publication of the cDNA and protein sequence of ABCG2 in 1998 (L. A. Doyle et al., 1998), scientists were able to screen it for functional motifs and putative structural and functional domains. As there were no structural data present at that time, scientists had to develop and apply homology models to other known ABC transporters and structure predictions which gave rise to the first models of ABCG2 architecture. This basic knowledge on ABC transporter physiology was very helpful to infer the structure and functionality of ABCG2. In general, ABC transporters are built of two transmembrane domains (TMDs) with at least 12 membrane spanning α‐helices and two catalytic nucleotide binding domains (NBDs) facing the cytosol (Polgar et al., 2008). The TMD carries the substrate binding site and constitutes the translocation pathway for the substrate, whereas the NBD is responsible for ATP binding and ATP hydrolysis. In most plasma membrane‐associated ABC transporters, the order of these domains within the protein sequence is N‐TMD‐NBD‐TMD‐NBD‐C, as shown for P‐gp (ABCB1) and the multidrug resistance associated protein 1 (ABCC1). However, the sequence of ABCG2 only encompasses one NBD and one TMD, the latter consisting of only six membrane spanning α‐helices (TM1–6). Therefore, it is considered to be a so‐called half‐transporter that needs to dimerize to gain full functionality. Half‐transporters are also commonly located in intracellular compartments, such as the transporter associated with antigen presentation 1 (ABCB2) and the transporter associated with antigen presentation 2 (https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=770&familyId=152&familyType=TRANSPORTER), that are located in the endoplasmic reticulum, ABCB7 which is found primarily in mitochondria, and the whole https://www.guidetopharmacology.org/GRAC/FamilyDisplayForward?familyId=154 of ABC transporters is mostly restricted to peroxisomes (Dean et al., 2001; Higgins et al., 1986). In the https://www.guidetopharmacology.org/GRAC/FamilyDisplayForward?familyId=155, the order of the domains is reversed compared to other ABC transporters being N‐NBD‐TMD‐C on the primary protein sequence.
Within the NBD, there are several conserved motifs that are common to most ABC transporters. The ATP binding side is usually composed of an A‐loop, a Walker A motif (P‐loop), a Q‐loop, a Walker B motif, a histidine switch (H‐loop), a D‐loop, and a C‐signature motif (ABC signature, C‐loop; ter Beek, Guskov, & Slotboom, 2014). Walker A (P‐loop, consensus sequence: GxxGxGKS/T, with x representing any amino acid) is a glycine‐rich sequence that is able to bind the phosphates of ATP by hydrogen bonds. ATP binding is assisted by the C‐signature motif (ABC signature, C‐loop), which has a consensus sequence of L/VSGGQ/E and is a typical feature of the ABC transporter family (Manolaridis et al., 2018). The magnesium ion associated with the ATP and a water molecule for ATP hydrolysis are bound by charged amino acids within the Q‐loop (a variable sequence with a conserved glutamine) and Walker B (consensus sequence: hhhhDE, with h representing hydrophobic residues). According to new structural data, the A‐loop, which consists of an aromatic side chain residue to bind the nucleobase of ATP by ring stacking, is missing in ABCG2 (Manolaridis et al., 2018). Although this is also the case for ABCG5 and ABCG8, in silico homology models of ABCG2 predict the A‐loop to be present at residue F52 upstream of Walker A (Ferreira, Bonito, Cordeiro, Ferreira, & Dos Santos, 2017), a region that is not well resolved in current structural models (Jackson et al., 2018; Manolaridis et al., 2018). The D‐loop of ABCG2 differs from its published consensus sequence SALD (Horn, Jenewein, Sohn‐Bosser, Bremer, & Schmitt, 2005) having only leucine and aspartate in common. During the hydrolysis reaction, the bound water molecule is polarized with the help of Walker B, H‐loop, D‐loop, and a conserved glutamine within the Q‐loop for a nucleophilic attack on the γ‐phosphate of ATP (Locher, 2016; ter Beek et al., 2014). As a result, ADP and free phosphate are generated.
To fully bind an ATP molecule, a dimerization of the two NBDs is necessary, as not all of the structural elements within a single NBD can be arranged around the ATP molecule. Therefore, two ATP molecules glue two NBDs together using the missing structural elements from the opposing NBD in a complementary fashion (Walker A, Q‐loop, Walker B, and H‐loop from NBD1; C‐signature and D‐loop from NBD2). ATP binding‐induced conformational changes (two open NBDs to a dimer of closed NBDs) are thought to take place in order to provide the energy for substrate transport through the translocation pathway within the TMD (Manolaridis et al., 2018; McDevitt et al., 2008). To enable this, NBD and TMD have to be connected, a process possibly induced by two α‐helices (connecting helix and coupling helix) of the TMD (Manolaridis et al., 2018). Within the TMD, the substrate binding sites of ABC transporters are frequently located at the cytosolic side of the 12 membrane spanning α‐helices. There is much less conservation within the sequence of the TMD compared to the NBD. This is due to the fact that this domain had to adapt structural demands of substrate binding specificity among individual ABC transporter family members (Wilkens, 2015). Like P‐gp (ABCB1) and ABCC1, the substrate binding site of ABGG2 features even a polyspecificity to a variety of substrates (Wright, Muench, Goldman, & Baker, 2018). Hence, detailed structural information, especially regarding the TMD, is essential to identify potential new drugs as substrates for ABCG2‐related transport processes, and to design specific ABCG2 inhibitors.
One additional structural feature, unique to ABCG2 among other ABC transporters, is a much longer extracellular loop (EL3) between the TM5 and TM6 (Desuzinges‐Mandon et al., 2010). It bears three cysteine residues that form an intramolecular (C592–C608) and an intermolecular (C603–C603′ of opposing ABCG2 monomer) disulfide bond (Henriksen, Fog, Litman, & Gether, 2005). Although the intramolecular disulfide bond seems to be relevant for ABCG2 expression and function, the intermolecular disulfide bond is likely to be of minor importance in this respect. Within EL3, there is a functional N‐glycosylation site at N596 which most likely is modified by N‐acetylglucosamine (Nakagawa et al., 2009). A diagram of the domain structure and location of important residues is presented in Figure 1 and Table 1, respectively.
Figure 1.
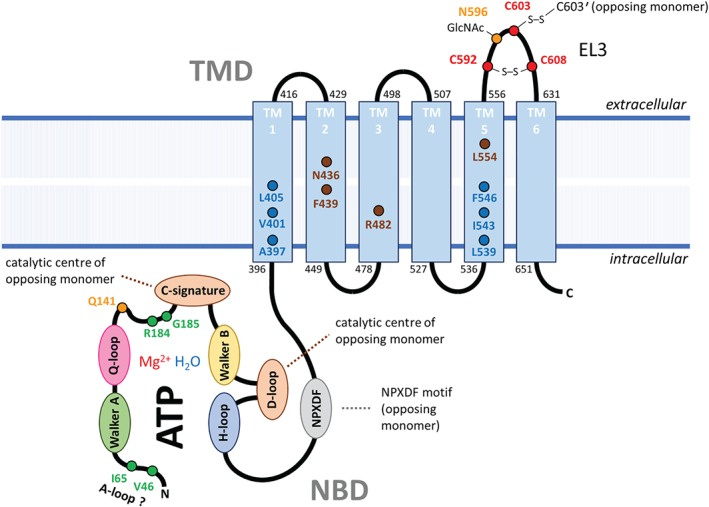
Schematic overview of the ABCG2 domain structure consisting of a nucleotide binding domain (NBD) and a transmembrane domain (TMD). Single membrane spanning α‐helices (TM1–6) were structured according to the information in published protein sequence (accession number: NP_001335914.1). The long extracellular loop 3 (EL3) is marked separately. Important sequence motifs or single residues are highlighted and further explained in Table 1. In ABCG2, the catalytic centre within the NBD is formed by the sequence motifs Walker A, Q‐loop, Walker B and H‐loop of one monomer, and the c‐signature and D‐loop from the other monomer. The A‐loop is either present upstream of Walker A or is missing and functionally replaced by the amino acids marked in green. Within the TMD, the membrane entrance site for hydrophobic substrates is highlighted by blue residues in TM1 and TM5. Residues involved in substrate binding or modification of substrate specificity are marked in brown. Cysteine bridge forming residues within EL3 are marked in red
Table 1.
Relevant residues/motifs within ABCG2 primary sequence
| Residuea | Remark | Citation | |
|---|---|---|---|
| Nucleotide binding domain (NBD) | V46, I65, G185, R184′ | Putative functional replacement for the A‐loop in ABCG2, van der Waals interactions with nucleobase of ATP, R184′ ring stacking | (Manolaridis et al., 2018) |
| F52 | Putative aromatic residue of the A‐loop according to in silico homology models of ABCG2 | (Ferreira et al., 2017) | |
| G80‐S87 | Walker A (P‐loop, consensus GxxGxGKS/T), GPTGGGKS in ABCG2, K86M mutation abolishes function and reduces surface expression | (Manolaridis et al., 2018; Ramakrishnan, Dani, & Ramasarma, 2002) | |
| Q126 | Conserved glutamine within the Q‐loop | (Manolaridis et al., 2018) | |
| Q141 | SNP Q141K, reduced ABCG2 surface expression, is associated with hyperuricaemia and gout, forms a H‐bond with N158 | (Furukawa et al., 2009; Jackson et al., 2018; Stiburkova, Pavelcova, Pavlikova, Jesina, & Pavelka, 2019) | |
| R184′ | Salt bridge with α‐phosphate of ATP | (Manolaridis et al., 2018) | |
| V186‐E200 | ABC signature (C‐signature, consensus L/VSGGQ/E), VSGGE in ABCG2 | (Manolaridis et al., 2018) | |
| I206‐E211 | Walker B (consensus hhhhDE), ILFLDE in ABCG2, E211Q mutation is catalytic inactive | (Y. Li, Huang, Zhang, Huang, & Li, 2013; Manolaridis et al., 2018; Taylor et al., 2017) | |
| D217 | Conserved aspartate within the D‐loop | (Jackson et al., 2018) | |
| H243 | Histidine switch (H‐loop) | (Manolaridis et al., 2018) | |
| N289‐F293 | NPXDF motif, NPADF in ABCG2, persistent contact interface of both NBDs | (Jackson et al., 2018) | |
| Transmembrane domain (TMD) | A397, V401, L405 | Residues from TM1 forming the membrane entrance for substrates between TM1 and TM5a | (Jackson et al., 2018) |
| N436 | Binding of ABCG2 substrate estrone‐3‐sulfate with H‐bonds | (Manolaridis et al., 2018) | |
| F439 | Binding of ABCG2 substrate estrone‐3‐sulfate with aromatic ring stacking | (Manolaridis et al., 2018) | |
| R482 | R482 mutations affect substrate specificity, H‐bond with S521 in TM4 | (Honjo et al., 2001; Taylor et al., 2017) | |
| L539, I543, F547 | Residues from TM5a forming the membrane entrance for substrates between TM1 and TM5a | (Jackson et al., 2018) | |
| L554, L554′ | Leucine plug involved in substrate binding | (Jackson et al., 2018; Manolaridis et al., 2018; Taylor et al., 2017) | |
| L555 | Structural relevant, L555A mutation leads to no functional protein | (Manolaridis et al., 2018) | |
| C592 | Intramolecular disulfide bond with C608 | (Henriksen et al., 2005; Taylor et al., 2017) | |
| N596 | Functional N‐glycosylation site binding GlcNAc | (Nakagawa et al., 2009; Taylor et al., 2017) | |
| C603 | Intermolecular disulfide bond with C603′ | (Henriksen et al., 2005; Taylor et al., 2017) | |
| C608 | Intramolecular disulfide bond with C592 | (Henriksen et al., 2005; Taylor et al., 2017) |
Apostrophe (') indicates residues from opposing monomer.
In recent years, structural high‐resolution X‐ray diffraction data of several ABC transporters have been published (bacterial Sav1866, Dawson & Locher, 2006, 2007; bacterial MsbA, A. Ward, Reyes, Yu, Roth, & Chang, 2007; P‐gp of mouse and Caenorhabditis elegans, Jin, Oldham, Zhang, & Chen, 2012; A. B. Ward et al., 2013; ABCG5/ABCG8 heterodimer, Lee et al., 2016). However, even more than 20 years after its discovery, a high‐resolution X‐ray diffraction structure of ABCG2 is still not available. As it is known that ABC transporters share strong structural homology with other ABC transporters, even without a strong homology in the overall protein sequence, scientists have tried to predict the 3D structure of ABCG2 using homology models. Thereby, in silico structural data of ABCG2 have been published using a homology model for its closest relative ABCG5/ABCG8 (Ferreira et al., 2017; Khunweeraphong, Stockner, & Kuchler, 2017; Laszlo, Sarkadi, & Hegedus, 2016). These structural predictions shared the problem that they might not be precise for the real 3D composition of ABCG2, underlying the need for high‐resolution structural data. As obtaining X‐ray diffraction structures of membrane proteins like ABCG2 is still challenging, recent structural analysis using electron microscopy might represent an appealing alternative for structural biologists interested in ABCG2.
3. FIRST STRUCTURAL DATA FROM ELECTRON MICROSCOPY
Electron microscopy in general lacks the high resolution obtained by X‐ray protein crystallography and is not able to provide 3D structures as such (Cheng, 2018). However, when capturing multiple 2D images of single protein complexes at different three‐dimensional orientations, a 3D structure of the protein can be reconstructed (De Rosier & Klug, 1968). Multiple images from different angles can be achieved by two ways. Either multiple images of a fixed 2D protein crystal are taken with different tilt angles (Henderson & Unwin, 1975), or multiple single protein complexes with an identical structure but a random orientation are imaged without the need of protein crystallization (Radermacher, Wagenknecht, Verschoor, & Frank, 1987). The latter technique is called single particle cryogenic electron microscopy (cryo‐EM), and new technological developments have recently enhanced its resolution to the near atomic level (Lyumkis, 2019) coming close to X‐ray protein crystallography. Therefore, this technique is an interesting alternative to obtain structural data, especially of proteins of which crystallization has not been successful so far.
One of the first studies trying to experimentally resolve the structure of ABCG2 by electron microscopy was conducted by McDevitt et al. (2006). In this study, a point‐mutated variant of ABCG2 (R482G) was investigated. The R482G mutation of ABCG2 broadens its substrate specificity, thereby enabling ABCG2 to transport substrates such as anthracyclines (Clark, Kerr, & Callaghan, 2006) but also dyes such as rhodamine 123 (Honjo et al., 2001). The substrate profile of the R482G variant shows a more pronounced overlap with the substrate profile of P‐gp, and thus, this mutant may represent an effector of persisting multidrug resistance following P‐gp inhibition. In the above‐mentioned study, R482G‐mutated ABCG2 was expressed in insect cells and isolated by a combination of detergent extraction and chromatography in conditions that were likely to preserve the native structure of the protein. ABCG2 was found to exist as a singular stable complex, which showed in non‐denaturation western blots a much higher MW than the ABCG2 monomer alone (430 kDa vs. 70 kDa), thereby indicating stable oligomerization of the protein. Single particle cryo‐EM of these protein complexes in combination with a homology model applied to the density map provided structural data at a resolution of ~18 Å. Although this resolution was not good enough to resolve single amino acids, this model was able to depict the tertiary and quaternary structure of the isolated ABCG2 complex, which seemed to be composed of a tetramer of ABCG2 dimers. The next approach to investigate ABCG2 protein structure by electron microscopy was undertaken by Rosenberg et al. (2015). ABCG2 was expressed in yeast cells (Pichia pastoris) and isolated in detergent solution. Without reconstitution of the protein complex in lipids, 2D crystals of ABCG2 were grown in the absence of nucleotides or substrates for ABCG2. Cryo‐EM using tilt angles ranging from 0° to 60° was applied on the 2D protein crystals to obtain a density map of ~20‐Å resolution. Again, the resolution was not good enough to model the protein structure but the boundaries of the protein complex and the orientation of its domains could be depicted. In contrast to the first cryo‐EM study (McDevitt et al., 2006), ABCG2 appeared to be only a homodimer of two ABCG2 molecules in a twofold symmetry. In a side view, the shape of the protein complex was constituted like a reversed V. A homology model using structural data from other ABC transporters was applied to the density map, and the best fit was achieved with the structure of ABCB1 in the so‐called inward‐facing conformation (A. B. Ward et al., 2013). In this conformation, the NBDs are spreading away from each other in an angle of about 60°. There was almost no fit with the so‐called outward‐facing conformation, where both NBDs remain tightly bound together, as had been shown for the structure of nucleotide bound Sav1866 (Dawson & Locher, 2006). In both studies (McDevitt and Rosenberg), the resolution of cryo‐EM was almost one order of magnitude below structural data obtained by X‐ray protein crystallography, thereby not allowing the primary sequence of ABCG2 to be modelled into the density map obtained by cryo‐EM.
4. RECENT HIGH‐RESOLUTION STRUCTURAL MODELS
The first high‐resolution 3D structure of ABCG2 was obtained by Taylor et al. (2017). The authors isolated human ABCG2 expressed in HEK cells and reconstituted the protein in membrane protein stabilizing nanodiscs (lipid composition:brain polar lipids and cholesterol, 4:1). Such nanodiscs, due to their bilayer of lipids, provide a more physiological environment. To further support structure formation and to aid single particle cryo‐EM imaging, the authors added the ABCG2‐specific monoclonal 5D3 Fab antibody fragment (S. Zhou et al., 2001) to the ABCG2 complexes. The 5D3 antibody is known to inhibit substrate transport and ATP hydrolysis of ABCG2 without having an effect on its substrate binding capabilities (Ozvegy‐Laczka et al., 2005). In native western blots, the 5D3–ABCG2 complex was running at a MW of 250 kDa and the ABCG2 complex alone at 144 kDa, which was smaller than the molecular size of the tetramer of ABCG2 dimers observed previously with detergent micelle preparation (McDevitt et al., 2006). These contradicting results are further discussed in Section 6. Single particle cryo‐EM of the 5D3–ABCG2 complex gave rise to a density map with an overall resolution of 3.8 Å which was sufficient to de novo build the structure of the protein except for the NBD. Due to higher flexibility and a lower amount of side views in single particle cryo‐EM, the resolution of this region was lower, and therefore, the authors used a homology model from available structural data of the ABCG5/ABCG8 complex (Lee et al., 2016) to structure the NBD.
The overall structure showed a twofold symmetric homodimer of two ABCG2 monomers with two 5D3 Fab bound to the long extracellular loop (EL3) of ABCG2 spreading away in an angle of 35° from the planar phospholipid membrane. Binding of at least one 5D3 Fab clamps EL3 together, thereby not allowing conformational changes to occur anymore. This represents a potential mechanistic explanation for the inhibitory effect of 5D3 Fab on ABCG2 activity. The structure of the clamped ABCG2 complex showed an inward‐facing conformation of an ABCG2 dimer with a slit‐like cavity at the cytosolic side (Cavity 1) formed by the TMD. Cavity 1 is accessible (open) from the cytosol and penetrates the plasma membrane by more than half of its distance. Due to its architecture and the composition of mostly hydrophobic amino acids, Cavity 1 is believed to constitute the binding site of ABCG2 substrates with flat, polycyclic, and hydrophobic characteristics. At the extracellular side, there is a second cavity (Cavity 2) which was formed by the TMD and the long extracellular loop between TM5 and TM6 (EL3). This much smaller cavity is not accessible (closed) from the extracellular space in the inward‐facing conformation of ABCG2 and cannot be found in the structure of ABCG2/ABCG5 dimers. Cavity 2 is mainly composed of less hydrophobic amino acids and thus believed to be the expel site for the (mostly) hydrophobic ABCG2 substrates. Cavity 1 and Cavity 2 are separated by a leucine plug composed of L554 from each monomer in the centre of the TMD, an amino acid residue believed to be involved in substrate binding as well. Within EL3, there are several known structural features, which have been mapped out now. The intramolecular disulfide bond C592–C608 (Henriksen et al., 2005) is likely to stabilize EL3, and the EL3 residing N‐glycosylation site at N596 (Nakagawa et al., 2009) bears the electron density fitting two molecules of N‐acetylglucosamine. Also, the intermolecular disulfide bond C603–C603′ (Henriksen et al., 2005) is clearly visible. The NBD was closely attached to TM1 in this model, indicating a structural interaction during ATP hydrolysis but, as this domain was not resolved in sufficient quality, the precise nature of this interaction could not be uncovered. In addition, the mode of substrate binding, ATP binding, and potential conformational changes during the substrate transport process could not be determined with this model.
To re‐address these issues, the same group published a follow‐up study (Jackson et al., 2018). In this study, the experimental design was similar to the one before but, this time, the nanodisc reconstitution was performed with ABCG2 bound to MZ29, a derivative of the selective ABCG2 inhibitor https://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=10007. By applying single particle cryo‐EM, the authors could resolve the structure of ABCG2 at an overall resolution of 3.6 Å and even enhance the resolution to 3.1 Å by adding structure stabilizing 5D3 Fab fragments during reconstitution of ABCG2. The resolution was good enough to even de novo build the 3D structure of the NBD, an attempt that had failed before (Taylor et al., 2017). The 3D model depicted an inward‐facing conformation of the ABCG2 dimer with two molecules of MZ29 bound inside the substrate binding Cavity 1. The inhibitory effect of MZ29 is therefore likely to be mediated by blocking the substrate binding site and substrate translocation pathway. Interestingly, five ordered cholesterols of the plasma membrane were bound to each monomer in an ABCG2 unique hydrophobic groove at the outside of the TMD potentially guiding the localization of ABCG2 to lipid rafts (Storch, Ehehalt, Haefeli, & Weiss, 2007). The non‐nucleotide bound NBDs had an open conformation and were facing away from each other leaving space for the entrance of substrates from the cytosol into Cavity 1 of the TMD. Q141 of the NBD interacted via a hydrogen bond with N158 of an α‐helix closely adjacent to TM1. This connection is believed to be mechanistically relevant as the polymorphism of Q141 has been related to dysfunction of ABCG2 in the kidney causing hyperuricaemia and gout (R. Li et al., 2015). TM1a might represent the so‐called connecting helix which was described in other ABC transporters before (Dawson & Locher, 2006; A. B. Ward et al., 2013) as being important for transmitting structural changes of the NBD to the TMD.
To further analyse the conformational changes during ATP‐dependent substrate transport, a third study with similar experimental design was published by Manolaridis et al. (2018). In this study, the authors expressed ABCG2 with the mutation E211Q which drastically reduced the catalytic activity for ATP hydrolysis within the NBD. This mutation was helpful to trap protein conformation in the respective state. Two structures were analysed by single particle cryo‐EM: a structure of mutated ABCG2 bound to the transporter substrate https://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=4749 (E1S) and one bound to https://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=1713 (resolutions of 3.6 and 3.1 Å, respectively, also see Figure 2). The structure of the substrate‐bound ABCG2 complex showed a dimer with an inward‐facing conformation, similar to that already described (Jackson et al., 2018; Taylor et al., 2017). One molecule of E1S was bound deep inside Cavity 1, almost halfway across the membrane. Due to the twofold symmetry of ABCG2, the substrate could bind in two possible orientations and was mainly coordinated by residues N436 and F439 side chains. Substrate binding boosted ATP hydrolysis by non‐mutated ABCG2 and mutations in ABCG2 that mimic substrate binding (V546F) were also able to strongly enhance ATPase activity. This indicated that substrate binding might support binding of ATP to the complex and that it might occur prior to ATP binding, as already shown for P‐gp (Martin et al., 2000). In contrast, the structure of catalytically silenced ABCG2 bound to ATP differed considerably from the substrate‐bound conformation. Binding of two ATP molecules, each to one NBD, caused a rotation of the domains by 35°, allowing a dimerization of the two elsewise separated NBDs. The large interface of the closed NBDs was likely to prevent exit of the substrate to the extracellular space again. Within the ATP‐binding site, the γ‐phosphate of ATP was bound to the residues Q211 (Walker B, conserved E in the wild‐type sequence), H243 (H‐loop), and Q126 (Q‐loop). In addition, there was a clear density representing a magnesium ion coordinated by Q211. The fact that the NBD is connected to the TMD via TM1 (connecting helix) and TM2 (coupling helix) had a marked effect on the structure of the TMD as well. TM1 and TM2 were rotated by 20° pushing the other TM helices closer together. As a consequence, Cavity 1 was collapsed completely, leaving no space for substrate binding. On the other hand, EL3 of each ABCG2 dimer was twisted away, thereby opening the smaller Cavity 2. In this outward‐facing conformation, the substrate might move peristaltically from the collapsed Cavity 1 to the now open Cavity 2 where it is repelled for substrate release. A scheme of the putative ABCG2 transport cycle according to the new cryo‐EM data is illustrated in Figure 3.
Figure 2.
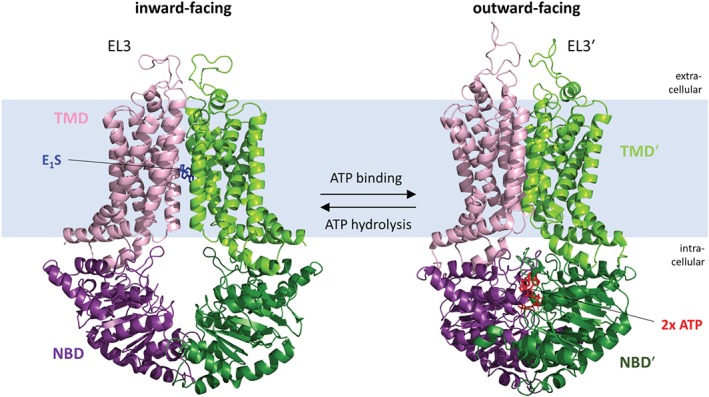
3D structure of substrate‐bound (left) and ATP‐bound (right) ABCG2 according to Manolaridis et al. (2018). ABCG2 is depicted as a dimer of two ABCG2 protein chains (purple, green) with their nucleotide binding domain (NBD) and transmembrane domain (TMD) displayed in a slightly different colour. Binding of one substrate molecule of estrone‐3‐sulfate (E1S) takes place in a cavity within the TMD. In this state, NBDs are facing away from each other (inward‐facing conformation). ABCG2 bound to ATP displays a markedly different conformation. Binding of two ATP molecules induces dimerization of NBDs forcing them to rotate by 35°. Rotation is transmitted to TMD by a link between NBD and transmembrane helix 1 (TM1) pushing all TM α‐helices closer together and thereby leaving no space for substrate binding. At the extracellular site, EL3 is twisted away opening a smaller cavity for substrate release (outward‐facing conformation)
Figure 3.
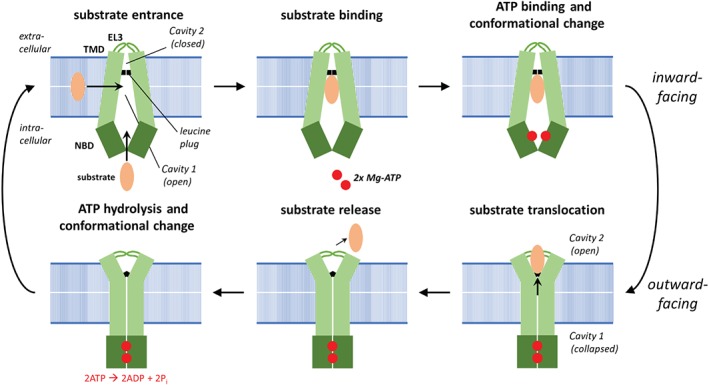
Schematic illustration of the ABCG2 transport cycle according to Manolaridis et al. (2018). In the apo state, the ABCG2 transporter displays an inward‐facing conformation which is comprised of two nucleotide binding domains (NBDs) facing away from each other and two transmembrane domains (TMDs) forming a large accessible cavity (open Cavity 1) and a small inaccessible cavity (closed Cavity 2) separated by a leucine plug. Substrates enter from the cytosol or through the membrane entrance site of ABCG2 and bind at hydrophobic residues of Cavity 1 close to the leucine plug. ATP binding induces a dimerization of both NBDs resulting in an overall conformational change (inward‐ to outward‐facing) of the ABCG2 complex. The collapse of substrate binding Cavity 1 and closing of the NBDs forces the substrate to move to Cavity 2 which is now open due to the bending away of the large extracellular loop 3 (EL3). Less hydrophobic amino acids in Cavity 2 help to expel substrates to the extracellular space. ATP hydrolysis within the NBD provides the energy to restore the apo state of the transporter (inward‐facing conformation). Please note that individual steps of the transport cycle might occur at the same time
Because it is not possible to capture short‐lived intermediate transitional states of the ABCG2 transport cycle with cryo‐EM techniques, the true nature of the ABCG2 transport cycle is very difficult to uncover. Currently, controversial issues are whether ATP binding or ATP hydrolysis drives the conformational changes that lead to the transport of the substrate across the membrane and whether ATP hydrolysis is necessary to restore the apo state after the transport process is completed or not. A different model of the substrate transport cycle based on the previous structural data from high‐resolution cryo‐EM (Jackson et al., 2018; Taylor et al., 2017) is discussed in a recent review (Kapoor, Horsey, Cox, & Kerr, 2018). In this model, substrates can bind at three different positions to ABCG2 (surface access site, binding site, and extrusion site). Substrate attachment occurs first at the surface access site of ABCG2. Binding of ATP to the NBDs causes a dimerization of the NBDs which leads to a transport of the attached substrate to the substrate binding site (corresponding to Cavity 1). ATP hydrolysis then induces conformational changes that transport the substrate to the substrate extrusion site (corresponding to Cavity 2) from where it is expelled. The apo state of the transporter is then restored by the dissociation of ADP. The idea of multiple different locations of the substrate in ABCG2 during the transport cycle is supported by in silico predictions of putative substrate binding sites along the substrate translocation pathway (Laszlo et al., 2016). This concept is further supported by findings that other residues more distant to the substrate binding site of the E1S bound cryo‐EM structure (Manolaridis et al., 2018) and outside Cavity 1 seem to be involved in substrate binding, as shown by R482 whose mutation leads to a change in ABCG2 substrate specificity (Clark et al., 2006; Honjo et al., 2001). As part of the substrate access site, R482 might be involved in defining the substrate selectivity of ABCG2. A similar mechanism of the ABCG2 transport cycle was postulated according to an in silico homology model of ABCG2 (Khunweeraphong et al., 2017). In this model, substrates are trapped in the central hydrophobic cavity (corresponding to Cavity 1) by the nucleotide free, inward‐facing apo state of the transporter. ATP binding and thereafter ATP hydrolysis lead to conformational changes of the NBD that are conveyed to the TMD via a transmission interface to an outward‐facing conformation. In this state, the central cavity is exposed to the extracellular space where the substrate is expelled. There is no Cavity 2 formed by EL3 in this model. Rather, a part of EL3 contributes as a so‐called re‐entry helix to remove a polar roof at the top of the central cavity and to open it to the outside. The transmission interface between NBD and TMD is formed by the intracellular loop 1 and the so‐called elbow helix (corresponding to connecting helix, TM1a). For this interaction, E451 of intracellular loop 1 seems to be crucial, as E451D mutants display increased ATP hydrolysis activity, while preventing substrate transport. This also indicates that ATP hydrolysis and substrate recognition are not necessarily coupled to each other and ATP hydrolysis and conformational changes might occur even in the absence of any bound substrate.
5. COMPARING PREVIOUSLY REPORTED STRUCTURAL FEATURES TO NEW HIGH‐RESOLUTION CRYO‐EM DATA
As mentioned earlier, before obtaining first high‐resolution structural data of ABCG2, scientist had to rely on structural predictions and first structural data from homology models to understand the structure of ABCG2. The first homology model (Laszlo et al., 2016) was based on structural data obtained from X‐ray diffraction of the ABCG5/G8 heterodimer (Lee et al., 2016). ABCG2 and ABCG5/G8 show a similar fold and, to some degree, even sequence homology (ABCG2 shares 26–27% sequence identity and ~45% sequence similarity to ABCG5/G8, Laszlo et al., 2016). Therefore, this model could describe quite well the overall structure of ABCG2 in both NBD and TMD. Even a few amino acid interactions could be correctly predicted before being proven true with cryo‐EM data. An example for this is the hydrogen bond of Q141‐R158 within the NBD close to the connecting helix of the TMD (Laszlo et al., 2016; Taylor et al., 2017). The link of this region of the NBD to the TMD is believed to be relevant for structure and function of ABCG2 as it might convey structural changes during the transport cycle. This assumption provides a potential structural explanation for ABCG2 misfolding and malfunction of the Q141K variant (Furukawa et al., 2009; R. Li et al., 2015).
However, there are also some major differences between the homology model and the recent structural models from cryo‐EM data. First of all, due to a backbone shift in TM2 and TM5a from opposing monomers, ABCG2 forms an almost membrane spanning, large inward‐facing Cavity 1 (Taylor et al., 2017), a feature that is not present in the ABCG5/G8 structure (Lee et al., 2016) and therefore could not be exploited for the corresponding ABCG2 homology models (Laszlo et al., 2016). As a consequence, the predicted substrate binding site was located closer to the cytosolic site. The residue R482 is located in this area (Clark et al., 2006; Honjo et al., 2001). Therefore, this residue was postulated to be directly involved in substrate binding. However, in high‐resolution structural data from cryo‐EM, R482 within TM3 forms a hydrogen bond with S521 of TM4, far away from the substrate binding site (Manolaridis et al., 2018; Taylor et al., 2017). This indicates that either allosteric effects or its postulated role as in the formation of a substrate access site (Kapoor et al., 2018) are likely to be the reason for R482 mutation phenotypes, which is further supported by findings that R482 is relevant for substrate transport, without being involved in direct substrate binding (Ejendal, Diop, Schweiger, & Hrycyna, 2006). Although the large shape of the central substrate binding cavity was missing in the first homology model (Laszlo et al., 2016), later homology models correctly identified this cavity (Ferreira et al., 2017; Laszlo et al., 2016) and showed a higher consistency with the cryo‐EM structure. In contrast, the smaller extracellular‐facing Cavity 2 is not present in all homology models. This is mostly due to the fact that EL3 differs considerably between ABCG2 and ABCG5/G8. In ABCG2, EL3 is involved in the formation of Cavity 2 (Taylor et al., 2017), whereas the shape of EL3 in ABCG5/G8 leaves no space to form this cavity (Lee et al., 2016). Nevertheless, in all homology models of ABCG2, cysteine bond‐forming residues (C592, C603, and C608) and the N‐glycosylation site at residue N596 are assigned to EL3. Probably the best match of the homology models with the high‐resolution cryo‐EM data is achieved within the NBD. The NBDs of ABCG2 and ABCG5/G8 share ~33% sequence identity, and, on the structural level, Walker A and ABC signature possess similar distances to each other (Jackson et al., 2018; Lee et al., 2016). The high similarity was also the reason why the first high‐resolution cryo‐EM structure used a NBD homology model based on structural data derived from ABCG5 to compensate for its low local resolution at that time (Taylor et al., 2017).
Apart from the homology models, there are a few previously postulated structural features of ABCG2 that are not supported by the high‐resolution cryo‐EM data. Within TM2 and TM5a of opposing monomers, two GXXXG motifs were thought to be involved in dimerization of ABCG2 (Polgar et al., 2004), whereas in the cryo‐EM structure, the respective residues are opposing each other (Taylor et al., 2017). Moreover, a C2 motif within the NBD was postulated to be involved in ATP binding and ATP hydrolysis (Macalou et al., 2016), whereas in the cryo‐EM structure, this motif is far from the catalytic centre at the bottom of the NBD (Taylor et al., 2017). The cysteine residues C284, C374, and C438 were believed to form intramolecular disulfide bonds important for ABCG2 structure (Liu, Yang, Qi, Peng, & Zhang, 2008) but, according to cryo‐EM data, these residues are located far apart from each other (Jackson et al., 2018). Also, it has been suggested that residues L555–L558 form a steroid‐binding element important for cholesterol sensing of ABCG2 within the plasma membrane (Telbisz, Hegedus, Varadi, Sarkadi, & Ozvegy‐Laczka, 2014). However, in the cryo‐EM structure, these residues face to the central core of ABCG2 and not outwards to the plasma membrane (Jackson et al., 2018).
6. EVIDENCE ON HIGHER ORDER ABCG2 OLIGOMERIZATION
The recent high‐resolution structural models of ABCG2 showed the functional protein to be only a dimer (Jackson et al., 2018; Manolaridis et al., 2018; Taylor et al., 2017). However, many other publications found ABCG2 to form higher order oligomers that are composed of a tetramer (Dezi et al., 2010; Xu, Liu, Yang, Bates, & Zhang, 2004), a tetramer of dimers (McDevitt et al., 2006) with a potential to even form dodecamers (Xu et al., 2004). One possible explanation for this discrepancy might be different preparation techniques used to isolate the ABCG2 protein. Most of the studies showing a higher order oligomerization used detergent (micelle) extraction procedures to purify ABCG2 from membranes which might have led to a concentration of ABCG2 and the formation of artificial oligomers. On the other hand, in high‐resolution cryo‐EM studies showing ABCG2 as a dimer, reconstitution of ABCG2 in small nanodiscs or the particle selection for cryo‐EM imaging might have missed possible oligomeric states of the protein. To answer the question whether oligomerization states of ABCG2 are present under physiological conditions, it is important to study the protein within the context of a living cell. One study trying to achieve that was performed by Wong, Briddon, Holliday, and Kerr (2016). In this study, the authors used HEK cells expressing GFP‐tagged ABCG2 at very low levels to be able to relate fluorescence intensity to single particles of ABCG2 complexes. Using fluorescence correlation spectroscopy, they imaged intensity fluctuation due to the diffusion mediated entrance and exit of single ABCG2 complexes within a confocal volume of a tiny part of the membrane. Photon counting histograms were collected for GFP‐tagged ABCG2 complexes and compared to statistical models considering the molecular brightness of a single GFP molecule. According to their findings, 69% of the ABCG2 complexes are likely to be formed of a tetrameric structure. This finding was supported by total internal reflection microscopy imaging and stepwise photobleaching of low GFP–ABCG2 expressing HEK cells that were fixed prior to the imaging experiments. In total internal reflection microscopy, the evanescent wave produced by the tilt angle illumination is able to limit fluorescence excitation to structures lying close to the glass bottom only. Thereby, it is possible to image single spots of GFP‐tagged ABCG2 that are at the very outer surface of the lower membrane. Stepwise photobleaching of these spots and fitting the results to dimer/tetramer models showed that 90% of the GFP–ABCG2 complexes were tetramers and only 10% dimers. However, all of the experiments performed by Wong and colleagues depend on fitting measured data into statistical models, a fact that makes interpretation of the results rather difficult. The question of how ABCG2 is organized within the membrane, either as a homodimer or higher order oligomers or even as a heterodimer as it was shown for the ABCG5/ABCG8 complex, remains unanswered and needs to be re‐addressed with more conclusive experimental designs.
7. CONCLUSIONS
Most recently, experimental attempts using the cryo‐EM technique were successful in obtaining the first high‐resolution structural information of the polyspecific efflux pump ABCG2. As previously assumed, these data indicate that ABCG2 forms a homodimer of two protein chains containing two major protein domains (NBD; TMD). ABCG2 can switch between two different conformations: in the nucleotide free state, ABCG2 faces inwards to the cytosol being accessible for substrates binding at the polyspecific substrate binding site within its TMD. In the ATP‐bound state, the dimerization of the NBDs induces conformational changes in the TMD which guide the substrate to the extracellular space. ATP binding and ATP hydrolysis are likely to mediate the conformational switch that runs the transport cycle helping to extrude a variety of substances out of the cells. Among those substances, many of them are drugs applied in cancer therapy which are additionally characterized by a narrow therapeutic index. It is therefore of great importance to use these new structural insights in ABCG2 structure to promote drug therapy safety management on the one hand and to develop clinically useful ABCG2 modulators on the other. Facing these challenges might help to further evolve our understanding of ABCG2 function in health and disease.
7.1. Nomenclature of targets and ligands
Key protein targets and ligands in this article are hyperlinked to corresponding entries in http://www.guidetopharmacology.org, the common portal for data from the IUPHAR/BPS Guide to PHARMACOLOGY (Harding et al., 2018), and are permanently archived in the Concise Guide to PHARMACOLOGY 2019/20 (Alexander et al., 2019).
CONFLICT OF INTEREST
The authors declare no conflicts of interest.
Eckenstaler R, Benndorf RA. 3D structure of the transporter ABCG2—What's new? Br J Pharmacol. 2020;177:1485–1496. 10.1111/bph.14991
REFERENCES
- Alexander, S. P. H. , Kelly, E. , Mathie, A. , Peters, J. A. , Veale, E. L. , Armstrong, J. F. , … Davies, J. A. (2019). The Concise Guide to PHARMACOLOGY 2019/20: Transporters. British Journal of Pharmacology, 176(Suppl 1), S397–s493. 10.1111/bph.14753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allikmets, R. , Schriml, L. M. , Hutchinson, A. , Romano‐Spica, V. , & Dean, M. (1998). A human placenta‐specific ATP‐binding cassette gene (ABCP) on chromosome 4q22 that is involved in multidrug resistance. Cancer Research, 58(23), 5337–5339. [PubMed] [Google Scholar]
- Bajcetic, M. , Benndorf, R. A. , Appel, D. , Schwedhelm, E. , Schulze, F. , Riekhof, D. , … Boger, R. H. (2007). Pharmacokinetics of oral doses of telmisartan and nisoldipine, given alone and in combination, in patients with essential hypertension. Journal of Clinical Pharmacology, 47(3), 295–304. 10.1177/0091270006297225 [DOI] [PubMed] [Google Scholar]
- ter Beek, J. , Guskov, A. , & Slotboom, D. J. (2014). Structural diversity of ABC transporters. The Journal of General Physiology, 143(4), 419–435. 10.1085/jgp.201411164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, Y. (2018). Single‐particle cryo‐EM‐How did it get here and where will it go. Science, 361(6405), 876–880. 10.1126/science.aat4346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark, R. , Kerr, I. D. , & Callaghan, R. (2006). Multiple drugbinding sites on the R482G isoform of the ABCG2 transporter. British Journal of Pharmacology, 149(5), 506–515. 10.1038/sj.bjp.0706904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damiani, D. , Tiribelli, M. , Calistri, E. , Geromin, A. , Chiarvesio, A. , Michelutti, A. , … Fanin, R. (2006). The prognostic value of P‐glycoprotein (ABCB) and breast cancer resistance protein (ABCG2) in adults with de novo acute myeloid leukemia with normal karyotype. Haematologica, 91(6), 825–828. [PubMed] [Google Scholar]
- Dawson, R. J. , & Locher, K. P. (2006). Structure of a bacterial multidrug ABC transporter. Nature, 443(7108), 180–185. 10.1038/nature05155 [DOI] [PubMed] [Google Scholar]
- Dawson, R. J. , & Locher, K. P. (2007). Structure of the multidrug ABC transporter Sav1866 from Staphylococcus aureus in complex with AMP‐PNP. FEBS Letters, 581(5), 935–938. 10.1016/j.febslet.2007.01.073 [DOI] [PubMed] [Google Scholar]
- De Rosier, D. J. , & Klug, A. (1968). Reconstruction of three dimensional structures from electron micrographs. Nature, 217(5124), 130–134. 10.1038/217130a0 [DOI] [PubMed] [Google Scholar]
- Dean, M. , Hamon, Y. , & Chimini, G. (2001). The human ATP‐binding cassette (ABC) transporter superfamily. Journal of Lipid Research, 42(7), 1007–1017. [PubMed] [Google Scholar]
- Deppe, S. , Boger, R. H. , Weiss, J. , & Benndorf, R. A. (2010). Telmisartan: A review of its pharmacodynamic and pharmacokinetic properties. Expert Opinion on Drug Metabolism & Toxicology, 6(7), 863–871. 10.1517/17425255.2010.494597 [DOI] [PubMed] [Google Scholar]
- Desuzinges‐Mandon, E. , Arnaud, O. , Martinez, L. , Huche, F. , Di Pietro, A. , & Falson, P. (2010). ABCG2 transports and transfers heme to albumin through its large extracellular loop. The Journal of Biological Chemistry, 285(43), 33123–33133. 10.1074/jbc.M110.139170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dezi, M. , Fribourg, P. F. , Di Cicco, A. , Arnaud, O. , Marco, S. , Falson, P. , … Levy, D. (2010). The multidrug resistance half‐transporter ABCG2 is purified as a tetramer upon selective extraction from membranes. Biochimica et Biophysica Acta, 1798(11), 2094–2101. 10.1016/j.bbamem.2010.07.034 [DOI] [PubMed] [Google Scholar]
- Doyle, L. , & Ross, D. D. (2003). Multidrug resistance mediated by the breast cancer resistance protein BCRP (ABCG2). Oncogene, 22(47), 7340–7358. 10.1038/sj.onc.1206938 [DOI] [PubMed] [Google Scholar]
- Doyle, L. A. , Yang, W. , Abruzzo, L. V. , Krogmann, T. , Gao, Y. , Rishi, A. K. , & Ross, D. D. (1998). A multidrug resistance transporter from human MCF‐7 breast cancer cells. Proceedings of the National Academy of Sciences of the United States of America, 95(26), 15665–15670. 10.1073/pnas.95.26.15665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ejendal, K. F. , Diop, N. K. , Schweiger, L. C. , & Hrycyna, C. A. (2006). The nature of amino acid 482 of human ABCG2 affects substrate transport and ATP hydrolysis but not substrate binding. Protein Science, 15(7), 1597–1607. 10.1110/ps.051998406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira, R. J. , Bonito, C. A. , Cordeiro, M. , Ferreira, M. U. , & Dos Santos, D. (2017). Structure‐function relationships in ABCG2: Insights from molecular dynamics simulations and molecular docking studies. Scientific Reports, 7(1), 1‐17, 15534. 10.1038/s41598-017-15452-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa, T. , Wakabayashi, K. , Tamura, A. , Nakagawa, H. , Morishima, Y. , Osawa, Y. , & Ishikawa, T. (2009). Major SNP (Q141K) variant of human ABC transporter ABCG2 undergoes lysosomal and proteasomal degradations. Pharmaceutical Research, 26(2), 469–479. 10.1007/s11095-008-9752-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutmann, H. , Hruz, P. , Zimmermann, C. , Beglinger, C. , & Drewe, J. (2005). Distribution of breast cancer resistance protein (BCRP/ABCG2) mRNA expression along the human GI tract. Biochemical Pharmacology, 70(5), 695–699. 10.1016/j.bcp.2005.05.031 [DOI] [PubMed] [Google Scholar]
- Harding, S. D. , Sharman, J. L. , Faccenda, E. , Southan, C. , Pawson, A. J. , Ireland, S. , … Davies, J. A. (2018). The IUPHAR/BPS Guide to PHARMACOLOGY in 2018: Updates and expansion to encompass the new guide to IMMUNOPHARMACOLOGY. Nucleic Acids Research, 46(D1), D1091–d1106. 10.1093/nar/gkx1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson, R. , & Unwin, P. N. (1975). Three‐dimensional model of purple membrane obtained by electron microscopy. Nature, 257(5521), 28–32. 10.1038/257028a0 [DOI] [PubMed] [Google Scholar]
- Henriksen, U. , Fog, J. U. , Litman, T. , & Gether, U. (2005). Identification of intra‐ and intermolecular disulfide bridges in the multidrug resistance transporter ABCG2. The Journal of Biological Chemistry, 280(44), 36926–36934. 10.1074/jbc.M502937200 [DOI] [PubMed] [Google Scholar]
- Heyes, N. , Kapoor, P. , & Kerr, I. D. (2018). Polymorphisms of the multidrug pump ABCG2: A systematic review of their effect on protein expression, function, and drug pharmacokinetics. Drug Metabolism and Disposition, 46(12), 1886–1899. 10.1124/dmd.118.083030 [DOI] [PubMed] [Google Scholar]
- Higgins, C. F. , Hiles, I. D. , Salmond, G. P. , Gill, D. R. , Downie, J. A. , Evans, I. J. , … Bell, A. W. (1986). A family of related ATP‐binding subunits coupled to many distinct biological processes in bacteria. Nature, 323(6087), 448–450. 10.1038/323448a0 [DOI] [PubMed] [Google Scholar]
- Honjo, Y. , Hrycyna, C. A. , Yan, Q. W. , Medina‐Perez, W. Y. , Robey, R. W. , van de Laar, A. , … Bates, S. E. (2001). Acquired mutations in the MXR/BCRP/ABCP gene alter substrate specificity in MXR/BCRP/ABCP‐overexpressing cells. Cancer Research, 61(18), 6635–6639. [PubMed] [Google Scholar]
- Horn, C. , Jenewein, S. , Sohn‐Bosser, L. , Bremer, E. , & Schmitt, L. (2005). Biochemical and structural analysis of the Bacillus subtilis ABC transporter OpuA and its isolated subunits. Journal of Molecular Microbiology and Biotechnology, 10(2–4), 76–91. 10.1159/000091556 [DOI] [PubMed] [Google Scholar]
- Jackson, S. M. , Manolaridis, I. , Kowal, J. , Zechner, M. , Taylor, N. M. I. , Bause, M. , … Locher, K. P. (2018). Structural basis of small‐molecule inhibition of human multidrug transporter ABCG2. Nature Structural & Molecular Biology, 25(4), 333–340. 10.1038/s41594-018-0049-1 [DOI] [PubMed] [Google Scholar]
- Jin, M. S. , Oldham, M. L. , Zhang, Q. , & Chen, J. (2012). Crystal structure of the multidrug transporter P‐glycoprotein from Caenorhabditis elegans . Nature, 490(7421), 566–569. 10.1038/nature11448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, Z. L. , & Chen, J. (2018). ATP binding enables substrate release from multidrug resistance protein 1. Cell, 172(1‐2), 81–89.e10. 10.1016/j.cell.2017.12.005 [DOI] [PubMed] [Google Scholar]
- Kannangara, D. R. , Phipps‐Green, A. J. , Dalbeth, N. , Stamp, L. K. , Williams, K. M. , Graham, G. G. , … Merriman, T. R. (2016). Hyperuricaemia: Contributions of urate transporter ABCG2 and the fractional renal clearance of urate. Annals of the Rheumatic Diseases, 75(7), 1363–1366. 10.1136/annrheumdis-2015-208111 [DOI] [PubMed] [Google Scholar]
- Kapoor, P. , Horsey, A. J. , Cox, M. H. , & Kerr, I. D. (2018). ABCG2: Does resolving its structure elucidate the mechanism? Biochemical Society Transactions, 46(6), 1485–1494. 10.1042/bst20180145 [DOI] [PubMed] [Google Scholar]
- Khunweeraphong, N. , Stockner, T. , & Kuchler, K. (2017). The structure of the human ABC transporter ABCG2 reveals a novel mechanism for drug extrusion. Scientific Reports, 7(1), 1‐15, 13767. 10.1038/s41598-017-11794-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J. E. , Singh, R. R. , Cho‐Vega, J. H. , Drakos, E. , Davuluri, Y. , Khokhar, F. A. , … Vega, F. (2009). Sonic hedgehog signaling proteins and ATP‐binding cassette G2 are aberrantly expressed in diffuse large B‐cell lymphoma. Modern Pathology, 22(10), 1312–1320. 10.1038/modpathol.2009.98 [DOI] [PubMed] [Google Scholar]
- Laszlo, L. , Sarkadi, B. , & Hegedus, T. (2016). Jump into a new fold—A homology based model for the ABCG2/BCRP multidrug transporter. PLoS ONE, 11(10), 1‐22, e0164426 10.1371/journal.pone.0164426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J. Y. , Kinch, L. N. , Borek, D. M. , Wang, J. , Wang, J. , Urbatsch, I. L. , … Rosenbaum, D. M. (2016). Crystal structure of the human sterol transporter ABCG5/ABCG8. Nature, 533(7604), 561–564. 10.1038/nature17666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, R. , Miao, L. , Qin, L. , Xiang, Y. , Zhang, X. , Peng, H. , … Yao, H. (2015). A meta‐analysis of the associations between the Q141K and Q126X ABCG2 gene variants and gout risk. International Journal of Clinical and Experimental Pathology, 8(9), 9812–9823. [PMC free article] [PubMed] [Google Scholar]
- Li, Y. , Huang, S. , Zhang, X. , Huang, T. , & Li, H. (2013). Cloning, expression, and functional analysis of molecular motor pilT and pilU genes of type IV pili in Acidithiobacillus ferrooxidans . Applied Microbiology and Biotechnology, 97(3), 1251–1257. 10.1007/s00253-012-4271-1 [DOI] [PubMed] [Google Scholar]
- Li, Y. H. , Wang, Y. H. , Li, Y. , & Yang, L. (2006). MDR1 gene polymorphisms and clinical relevance. Yi Chuan Xue Bao, 33(2), 93–104. 10.1016/s0379-4172(06)60027-9 [DOI] [PubMed] [Google Scholar]
- Liu, Y. , Yang, Y. , Qi, J. , Peng, H. , & Zhang, J. T. (2008). Effect of cysteine mutagenesis on the function and disulfide bond formation of human ABCG2. The Journal of Pharmacology and Experimental Therapeutics, 326(1), 33–40. 10.1124/jpet.108.138115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locher, K. P. (2016). Mechanistic diversity in ATP‐binding cassette (ABC) transporters. Nature Structural & Molecular Biology, 23(6), 487–493. 10.1038/nsmb.3216 [DOI] [PubMed] [Google Scholar]
- Lyumkis, D. (2019). Challenges and opportunities in cryo‐EM single‐particle analysis. The Journal of Biological Chemistry, 294(13), 5181–5197. 10.1074/jbc.REV118.005602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macalou, S. , Robey, R. W. , Jabor Gozzi, G. , Shukla, S. , Grosjean, I. , Hegedus, T. , … Di Pietro, A. (2016). The linker region of breast cancer resistance protein ABCG2 is critical for coupling of ATP‐dependent drug transport. Cellular and Molecular Life Sciences, 73(9), 1927–1937. 10.1007/s00018-015-2118-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maliepaard, M. , Scheffer, G. L. , Faneyte, I. F. , van Gastelen, M. A. , Pijnenborg, A. C. , Schinkel, A. H. , … Schellens, J. H. (2001). Subcellular localization and distribution of the breast cancer resistance protein transporter in normal human tissues. Cancer Research, 61(8), 3458–3464. [PubMed] [Google Scholar]
- Manolaridis, I. , Jackson, S. M. , Taylor, N. M. I. , Kowal, J. , Stahlberg, H. , & Locher, K. P. (2018). Cryo‐EM structures of a human ABCG2 mutant trapped in ATP‐bound and substrate‐bound states. Nature, 563(7731), 426–430. 10.1038/s41586-018-0680-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao, Q. (2008). BCRP/ABCG2 in the placenta: Expression, function and regulation. Pharmaceutical Research, 25(6), 1244–1255. 10.1007/s11095-008-9537-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, C. , Berridge, G. , Mistry, P. , Higgins, C. , Charlton, P. , & Callaghan, R. (2000). Drug binding sites on P‐glycoprotein are altered by ATP binding prior to nucleotide hydrolysis. Biochemistry, 39(39), 11901–11906. 10.1021/bi000559b [DOI] [PubMed] [Google Scholar]
- McDevitt, C. A. , Collins, R. F. , Conway, M. , Modok, S. , Storm, J. , Kerr, I. D. , … Callaghan, R. (2006). Purification and 3D structural analysis of oligomeric human multidrug transporter ABCG2. Structure, 14(11), 1623–1632. 10.1016/j.str.2006.08.014 [DOI] [PubMed] [Google Scholar]
- McDevitt, C. A. , Crowley, E. , Hobbs, G. , Starr, K. J. , Kerr, I. D. , & Callaghan, R. (2008). Is ATP binding responsible for initiating drug translocation by the multidrug transporter ABCG2? The FEBS Journal, 275(17), 4354–4362. 10.1111/j.1742-4658.2008.06578.x [DOI] [PubMed] [Google Scholar]
- Miyake, K. , Mickley, L. , Litman, T. , Zhan, Z. , Robey, R. , Cristensen, B. , … Bates, S. E. (1999). Molecular cloning of cDNAs which are highly overexpressed in mitoxantrone‐resistant cells: Demonstration of homology to ABC transport genes. Cancer Research, 59(1), 8–13. [PubMed] [Google Scholar]
- Mo, W. , & Zhang, J. T. (2012). Human ABCG2: Structure, function, and its role in multidrug resistance. International Journal of Biochemistry and Molecular Biology, 3(1), 1–27. [PMC free article] [PubMed] [Google Scholar]
- Nakagawa, H. , Wakabayashi‐Nakao, K. , Tamura, A. , Toyoda, Y. , Koshiba, S. , & Ishikawa, T. (2009). Disruption of N‐linked glycosylation enhances ubiquitin‐mediated proteasomal degradation of the human ATP‐binding cassette transporter ABCG2. The FEBS Journal, 276(24), 7237–7252. 10.1111/j.1742-4658.2009.07423.x [DOI] [PubMed] [Google Scholar]
- Ozvegy‐Laczka, C. , Varady, G. , Koblos, G. , Ujhelly, O. , Cervenak, J. , Schuetz, J. D. , … Sarkadi, B. (2005). Function‐dependent conformational changes of the ABCG2 multidrug transporter modify its interaction with a monoclonal antibody on the cell surface. The Journal of Biological Chemistry, 280(6), 4219–4227. 10.1074/jbc.M411338200 [DOI] [PubMed] [Google Scholar]
- Polgar, O. , Robey, R. W. , & Bates, S. E. (2008). ABCG2: Structure, function and role in drug response. Expert Opinion on Drug Metabolism & Toxicology, 4(1), 1–15. 10.1517/17425255.4.1.1 [DOI] [PubMed] [Google Scholar]
- Polgar, O. , Robey, R. W. , Morisaki, K. , Dean, M. , Michejda, C. , Sauna, Z. E. , … Bates, S. E. (2004). Mutational analysis of ABCG2: Role of the GXXXG motif. Biochemistry, 43(29), 9448–9456. 10.1021/bi0497953 [DOI] [PubMed] [Google Scholar]
- Radermacher, M. , Wagenknecht, T. , Verschoor, A. , & Frank, J. (1987). Three‐dimensional structure of the large ribosomal subunit from Escherichia coli . The EMBO Journal, 6(4), 1107–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishnan, C. , Dani, V. S. , & Ramasarma, T. (2002). A conformational analysis of Walker motif A [GXXXXGKT (S)] in nucleotide‐binding and other proteins. Protein Engineering, 15(10), 783–798. 10.1093/protein/15.10.783 [DOI] [PubMed] [Google Scholar]
- Ripperger, A. , & Benndorf, R. A. (2016). The C421A (Q141K) polymorphism enhances the 3′‐untranslated region (3′‐UTR)‐dependent regulation of ATP‐binding cassette transporter ABCG2. Biochemical Pharmacology, 104, 139–147. 10.1016/j.bcp.2016.02.011 [DOI] [PubMed] [Google Scholar]
- Ripperger, A. , Krischer, A. , Robaa, D. , Sippl, W. , & Benndorf, R. A. (2018). Pharmacogenetic aspects of the interaction of AT1 receptor antagonists with ATP‐binding cassette transporter ABCG2. Frontiers in Pharmacology, 9, 1‐13, 463. 10.3389/fphar.2018.00463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robey, R. W. , Polgar, O. , Deeken, J. , To, K. W. , & Bates, S. E. (2007). ABCG2: Determining its relevance in clinical drug resistance. Cancer Metastasis Reviews, 26(1), 39–57. 10.1007/s10555-007-9042-6 [DOI] [PubMed] [Google Scholar]
- Rosenberg, M. F. , Bikadi, Z. , Hazai, E. , Starborg, T. , Kelley, L. , Chayen, N. E. , … Mao, Q. (2015). Three‐dimensional structure of the human breast cancer resistance protein (BCRP/ABCG2) in an inward‐facing conformation. Acta Crystallographica. Section D, Biological Crystallography, 71(Pt 8), 1725–1735. 10.1107/s1399004715010676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher, T. , & Benndorf, R. A. (2017). ABC transport proteins in cardiovascular disease—A brief summary. Molecules, 22(4), 1‐19, 589. 10.3390/molecules22040589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiburkova, B. , Pavelcova, K. , Pavlikova, M. , Jesina, P. , & Pavelka, K. (2019). The impact of dysfunctional variants of ABCG2 on hyperuricemia and gout in pediatric‐onset patients. Arthritis Research & Therapy, 21(1), 77–10. 10.1186/s13075-019-1860-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storch, C. H. , Ehehalt, R. , Haefeli, W. E. , & Weiss, J. (2007). Localization of the human breast cancer resistance protein (BCRP/ABCG2) in lipid rafts/caveolae and modulation of its activity by cholesterol in vitro. The Journal of Pharmacology and Experimental Therapeutics, 323(1), 257–264. 10.1124/jpet.107.122994 [DOI] [PubMed] [Google Scholar]
- Taylor, N. M. I. , Manolaridis, I. , Jackson, S. M. , Kowal, J. , Stahlberg, H. , & Locher, K. P. (2017). Structure of the human multidrug transporter ABCG2. Nature, 546(7659), 504–509. 10.1038/nature22345 [DOI] [PubMed] [Google Scholar]
- Telbisz, A. , Hegedus, C. , Varadi, A. , Sarkadi, B. , & Ozvegy‐Laczka, C. (2014). Regulation of the function of the human ABCG2 multidrug transporter by cholesterol and bile acids: Effects of mutations in potential substrate and steroid binding sites. Drug Metabolism and Disposition, 42(4), 575–585. 10.1124/dmd.113.055731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward, A. , Reyes, C. L. , Yu, J. , Roth, C. B. , & Chang, G. (2007). Flexibility in the ABC transporter MsbA: Alternating access with a twist. Proceedings of the National Academy of Sciences of the United States of America, 104(48), 19005–19010. 10.1073/pnas.0709388104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward, A. B. , Szewczyk, P. , Grimard, V. , Lee, C. W. , Martinez, L. , Doshi, R. , … Chang, G. (2013). Structures of P‐glycoprotein reveal its conformational flexibility and an epitope on the nucleotide‐binding domain. Proceedings of the National Academy of Sciences of the United States of America, 110(33), 13386–13391. 10.1073/pnas.1309275110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss, J. , Sauer, A. , Divac, N. , Herzog, M. , Schwedhelm, E. , Boger, R. H. , … Benndorf, R. A. (2010). Interaction of angiotensin receptor type 1 blockers with ATP‐binding cassette transporters. Biopharmaceutics & Drug Disposition, 31(2–3), 150–161. 10.1002/bdd.699 [DOI] [PubMed] [Google Scholar]
- Wilkens, S. (2015). Structure and mechanism of ABC transporters. F1000Prime Rep, 7, 1‐9. 10.12703/p7-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong, K. , Briddon, S. J. , Holliday, N. D. , & Kerr, I. D. (2016). Plasma membrane dynamics and tetrameric organisation of ABCG2 transporters in mammalian cells revealed by single particle imaging techniques. Biochimica et Biophysica Acta, 1863(1), 19–29. 10.1016/j.bbamcr.2015.10.002 [DOI] [PubMed] [Google Scholar]
- Woodward, O. M. , Kottgen, A. , Coresh, J. , Boerwinkle, E. , Guggino, W. B. , & Kottgen, M. (2009). Identification of a urate transporter, ABCG2, with a common functional polymorphism causing gout. Proceedings of the National Academy of Sciences of the United States of America, 106(25), 10338–10342. 10.1073/pnas.0901249106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright, J. , Muench, S. P. , Goldman, A. , & Baker, A. (2018). Substrate polyspecificity and conformational relevance in ABC transporters: New insights from structural studies. Biochemical Society Transactions, 46(6), 1475–1484. 10.1042/bst20180146 [DOI] [PubMed] [Google Scholar]
- Xu, J. , Liu, Y. , Yang, Y. , Bates, S. , & Zhang, J. T. (2004). Characterization of oligomeric human half‐ABC transporter ATP‐binding cassette G2. The Journal of Biological Chemistry, 279(19), 19781–19789. 10.1074/jbc.M310785200 [DOI] [PubMed] [Google Scholar]
- Zamboni, W. C. , Ramanathan, R. K. , McLeod, H. L. , Mani, S. , Potter, D. M. , Strychor, S. , … Marsh, S. (2006). Disposition of 9‐nitrocamptothecin and its 9‐aminocamptothecin metabolite in relation to ABC transporter genotypes. Investigational New Drugs, 24(5), 393–401. 10.1007/s10637-006-6335-5 [DOI] [PubMed] [Google Scholar]
- Zhang, W. , Mojsilovic‐Petrovic, J. , Andrade, M. F. , Zhang, H. , Ball, M. , & Stanimirovic, D. B. (2003). The expression and functional characterization of ABCG2 in brain endothelial cells and vessels. The FASEB Journal, 17(14), 2085–2087. 10.1096/fj.02-1131fje [DOI] [PubMed] [Google Scholar]
- Zhang, W. , Yu, B. N. , He, Y. J. , Fan, L. , Li, Q. , Liu, Z. Q. , … Zhou, H. H. (2006). Role of BCRP 421C>A polymorphism on rosuvastatin pharmacokinetics in healthy Chinese males. Clinica Chimica Acta, 373(1–2), 99–103. 10.1016/j.cca.2006.05.010 [DOI] [PubMed] [Google Scholar]
- Zhou, Q. , Sparreboom, A. , Tan, E. H. , Cheung, Y. B. , Lee, A. , Poon, D. , … Chowbay, B. (2005). Pharmacogenetic profiling across the irinotecan pathway in Asian patients with cancer. British Journal of Clinical Pharmacology, 59(4), 415–424. 10.1111/j.1365-2125.2004.02330.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, S. , Schuetz, J. D. , Bunting, K. D. , Colapietro, A. M. , Sampath, J. , Morris, J. J. , … Sorrentino, B. P. (2001). The ABC transporter Bcrp1/ABCG2 is expressed in a wide variety of stem cells and is a molecular determinant of the side‐population phenotype. Nature Medicine, 7(9), 1028–1034. 10.1038/nm0901-1028 [DOI] [PubMed] [Google Scholar]


