Significance
Coassembly, in which peptides A and B selectively associate to form β-sheet structures, is an emerging approach to fabricate multicomponent biomaterials. As coassembly is rare in nature, designing synthetic peptide pairs must be informed by insights into molecular organization within existing systems. For charge-complementary peptide pairs, it is assumed that molecules organize into an alternating arrangement, yet there are no experimental data to support this assumption. Here, a combination of molecular dynamics simulations, biophysical measurements, electron microscopy, and solid-state NMR demonstrates that an established pair of charge-complementary peptides [CATCH(+) and CATCH(−)] coassemble into bilayers of AB-alternating β-sheets. However, significant mismatched AA and BB neighbors do occur. Off-pathway β-barrel oligomers were predicted by simulations, and concentration-dependent oligomer formation was observed experimentally.
Keywords: coassembly, peptides, fibril, β-barrel, coarse-grained simulation
Abstract
Peptide self-assembly, wherein molecule A associates with other A molecules to form fibrillar β-sheet structures, is common in nature and widely used to fabricate synthetic biomaterials. Selective coassembly of peptide pairs A and B with complementary partial charges is gaining interest due to its potential for expanding the form and function of biomaterials that can be realized. It has been hypothesized that charge-complementary peptides organize into alternating ABAB-type arrangements within assembled β-sheets, but no direct molecular-level evidence exists to support this interpretation. We report a computational and experimental approach to characterize molecular-level organization of the established peptide pair, CATCH. Discontinuous molecular dynamics simulations predict that CATCH(+) and CATCH(−) peptides coassemble but do not self-assemble. Two-layer β-sheet amyloid structures predominate, but off-pathway β-barrel oligomers are also predicted. At low concentration, transmission electron microscopy and dynamic light scattering identified nonfibrillar ∼20-nm oligomers, while at high concentrations elongated fibers predominated. Thioflavin T fluorimetry estimates rapid and near-stoichiometric coassembly of CATCH(+) and CATCH(−) at concentrations ≥100 μM. Natural abundance 13C NMR and isotope-edited Fourier transform infrared spectroscopy indicate that CATCH(+) and CATCH(−) coassemble into two-component nanofibers instead of self-sorting. However, 13C–13C dipolar recoupling solid-state NMR measurements also identify nonnegligible AA and BB interactions among a majority of AB pairs. Collectively, these results demonstrate that strictly alternating arrangements of β-strands predominate in coassembled CATCH structures, but deviations from perfect alternation occur. Off-pathway β-barrel oligomers are also suggested to occur in coassembled β-strand peptide systems.
Coassembly of peptides is a new research frontier in peptide and protein biophysics (1, 2). In general, peptides A and B are said to coassemble when they combine to form a stable supramolecular architecture. Many peptides that self-assemble into β-sheet nanofibers can also coassemble with an appropriate partner, as seen with charge-complementary and enantiomeric mixtures (3–5). Selective peptide coassembly describes the unique case wherein molecules A and B form nanofibers when combined in solution but independently remain unassembled in random coil configurations. Although charge-complementary peptides that selectively assemble into elongated α-helical coiled-coil fibers have been extensively characterized (6–8), only a few pairs of synthetic peptides that selectively coassemble into amyloid-like β-sheet nanofibers have been designed to date (9–12). No naturally occurring β-sheet–forming pairs are currently known.
Increasing interest in coassembling peptides is attributable to their potential for expanding the form and function of biomaterials that can be realized (13, 14). Nevertheless, we have little direct knowledge of the molecular-level interactions that cause two different peptides to coassemble into amyloid-like structures, in stark contrast to our deeper understanding of self-assembly. As a result, de novo design of selectively coassembling peptide pairs remains a challenge. In fact, most of the known pairs of peptides that selectively coassemble into β-sheets are simply charged variants of sequences that are known to self-assemble. Charge complementarity between the two peptides has led to the general working hypothesis that coassembled molecules adopt a strictly alternating -ABABAB- arrangement within each β-sheet. However, there is no direct evidence to support this hypothesis, and it is not possible to access this information experimentally via standard biophysical measurements.
Here we present a comprehensive investigation to test the hypothesis that charge-complementary peptides adopt a strictly alternating (AB)n arrangement when coassembled into amyloid-like β-sheet nanofibers. In particular, we use a combination of computer simulations, biophysical measurements, and solid-state NMR (ssNMR) spectroscopy to characterize the time evolution of aggregate size and structure, as well as the molecular-level organization of peptide strands coassembled into β-sheet–rich structures. The peptide pair is “CATCH(+)” (Ac-QQKFKFKFKQQ-Am) and “CATCH(−)” (Ac-EQEFEFEFEQE-Am), charge-complementary coassembling β-strand peptides whose overall charges (4+ and 6−) are determined by their different numbers of lysine (K) or glutamic acid (E) residues (9). Discontinuous molecular dynamics (DMD) simulations, a fast alternative to traditional molecular dynamics, are used with the coarse-grained peptide force field PRIME20 to simulate the coassembly dynamics of large (96-peptide) systems of CATCH(+) and CATCH(−) peptides starting from a random initial state. DMD/PRIME20 is among the most realistic of the protein coarse-grained models, does not build in any predetermined secondary structure or start from a preset ordered structure, provides a good representation of amyloid structure in comparison to experiment, and is fast enough to get to the fibrillar stage starting from the random-coil state (15–17). The DMD/PRIME20 combination allows us to explore in molecular detail the structure and rearrangement of the oligomers that form along the aggregation/fibrilization pathway at time scales up to 672 µs at millimolar concentrations (18). Informed by simulations, thioflavin T (ThT) fluorimetry, transmission electron microscopy (TEM), and dynamic light scattering (DLS) are used to probe CATCH peptide coassembly at different experimental conditions. Finally, Fourier transform infrared (FTIR) spectroscopy and ssNMR measurements are used to determine secondary structure and interstrand contacts between peptides within coassembled nanofibers. Together, these methods demonstrate that strictly alternating coassembly of CATCH(+) and CATCH(−) predominates, yet AA and BB neighbors do occur. In addition, DMD simulations suggest the formation of stable out-of-register off-pathway β-barrel oligomers. TEM and DLS also identified concentration-dependent formation of ∼20-nm nonfibrillar oligomers. Although β-barrel oligomer formation is now being explored as a common pathway in amyloid formation (19–22), here it is suggested to also result from charge-complementary β-strand peptide coassembly.
Results
CATCH(+) and CATCH(−) Exhibit Molecular-Level Coassembly.
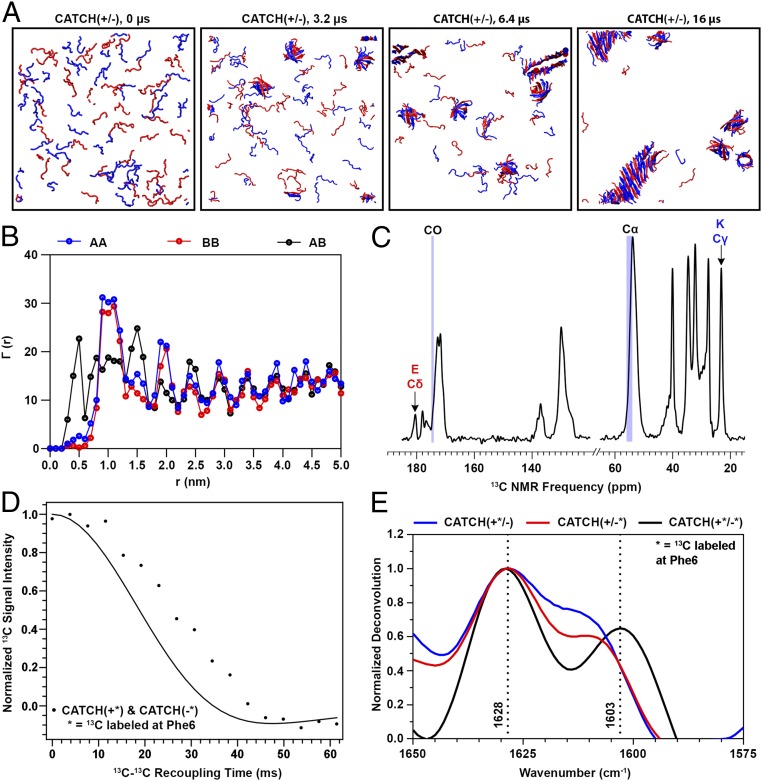
Data from simulations and experiments in Fig. 1 demonstrate molecular-level coassembly of CATCH(+) and CATCH(−) peptides into β-sheets. Here, we refer to the CATCH(+) peptide as “A” and the CATCH(−) peptide as “B” for simplicity. Ten DMD simulations were run at T* = 0.2 (equivalent to 342 K) for 16 µs, using the PRIME20 model. Each simulation started with a mixture of 48 A and 48 B molecules in random-coil configurations at a concentration of 20 mM. Representative snapshots of the simulations at 0, 3.2, 6.4, and 16 µs depict the coassembly process (Fig. 1A). Ordered aggregates containing both A and B emerge as early as 3.2 μs but they are β-barrel structures, not amyloid-like structures. As the simulation progresses, more and more peptides join the ordered aggregates. At 6.4 µs, an amyloid-like structure can be seen in addition to several β-barrel structures. The amyloid-like structure grows with time and by 8 µs differentiates itself significantly from the β-barrel structures. By 16 µs, nearly 90% of peptides have aggregated into two-component amyloid or β-barrel structures. The simulation kinetics for this run is described in SI Appendix and shown in SI Appendix, Fig. S1. The β-barrel structures do not convert to amyloid or serve as seeds for amyloid formation, suggesting that they are off-pathway for fibril formation. However, the simulation time scale of 16 µs is orders of magnitude smaller than the experimental time scale, so simulations cannot be used to predict whether or not the β-barrel is a kinetically trapped metastable state or transient in real-world settings.
Fig. 1.
Computational simulations and biophysical measurements of an equimolar CATCH(+) and CATCH(−) mixture show coassembly. (A) DMD/PRIME20 simulations of a mixture of 48 CATCH(+) and 48 CATCH(−) peptides at 20 mM concentration. Snapshots at 0, 3.2, 6.4, and 16 μs are presented. (B) , , and , defined in the text as computationally predicted average numbers of A or B central atoms as a function of distance r from central atoms within peptide A or B. (C) The 1H-13C CPMAS spectra of a CATCH(+/−) nanofiber sample. (D) PITHIRDS-CT decay curve of a CATCH nanofiber sample 13C-labeled on both CATCH(+) and CATCH(−) on the carbonyl C of Phe6. The solid black curve corresponds to the predicted signal decay in the PITHIRDS-CT experiment from a nuclear spin simulation of eight 13C atoms along an ideal coassembled antiparallel β-sheet. (E) FTIR spectra of equimolar mixtures of labeled CATCH(+) and CATCH(−) (black), labeled CATCH(+) and unlabeled CATCH(−) (blue), and unlabeled CATCH(+) and labeled CATCH(−) (red) at 10 mM in 1× phosphate-buffered saline. The dashed lines identify the wavenumbers corresponding to the location of the peaks in the black spectrum.
Quantitative analysis of the organization of A and B in the final configurations of the DMD simulations indicates that the majority of the peptides are arranged in an ordered alternating ABAB pattern. Fig. 1B shows the numbers of AB, AA, and BB neighbors, , , and , as a function of the distance r between the Phe6 carbonyl C atoms (central atoms) on the peptides in the final aggregated structure averaged over the 10 independent simulations. Choosing the central atom as a reference makes analysis of peptide organization insensitive to the relative directions of adjacent β-strands (parallel or antiparallel) within each β-sheet. The high-intensity peak for at r = 0.5 nm (the first-neighbor distance between peptides) demonstrates that the majority of A molecules are next to B molecules, and vice versa, in the final aggregated structures. Further, the peaks for and at r = 1.0 nm (the second-neighbor distance) demonstrate that the peptides are primarily organized into ABA or BAB configurations. The nonzero values for and when r < 1.0 nm indicate, however, that some of the A and B molecules do sit next to their own kind in the final structure, as will be discussed in a later section.
FTIR and ssNMR measurements demonstrate that structures rich in β-sheets are formed in equimolar mixtures of A and B at concentrations comparable to those used in simulations. We observed up-field shifts in 13C natural abundance ssNMR peak frequencies for the carbonyl carbon (CO) and α-carbon (Cα) of Gln, Lys, Glu, and Phe relative to reference peptides in random-coil configurations (Fig. 1C), which indicate that A and B aggregate into a β-sheet secondary structure (23, 24). A centrifuge pellet of coassembled A and B in which each peptide was labeled with 13C at its central atom (CO site of Phe6) yielded a strong 13C decay in PITHIRDS-CT NMR measurements (Fig. 1D), which indicates dipolar couplings between 13C-labeled sites (25). This decay maps to simulations of 8 13C spins spaced as predicted by an antiparallel β-sheet structure, with 13C–13C distances of either 0.49 nm or 0.65 nm. Likewise, the FTIR spectrum of an equimolar mixture of A and B has a strong maximum at 1,620 cm−1 (SI Appendix, Fig. S2), which is within the amide I region and indicative of peptides in a β-strand conformation.
FTIR and ssNMR measurements also indicate that A and B are coassembled. In a centrifuge pellet of coassembled A and B, we observed 13C natural abundance ssNMR signals that are uniquely attributable to the Lys and Glu residues which are exclusive to CATCH(+) or CATCH(−), respectively (Fig. 1C), indicating that both peptides are present in the sample (26). Likewise, in the deconvoluted FTIR spectrum of an equimolar mixture of 13C-labeled A and B, the amide I peak was split into maxima at 1,628 and 1,603 cm−1 (Fig. 1E), similar to a prior report of enantiomeric peptide mixtures with a single 13C label on each peptide (4). In contrast, the deconvoluted FTIR spectra of equimolar mixtures of labeled A and unlabeled B, as well as unlabeled A and labeled B, had maxima at 1,628 and 1,610 cm−1. The maximum at 1,603 cm−1 in mixtures of labeled A and B is attributed to vibrational coupling between 13C atoms in neighboring hydrogen-bonded β-strands, whereas the higher-frequency maximum at 1,610 cm−1 in mixtures of labeled and unlabeled peptides is likely due to the presence of 13C-labeled strands with interspersed 12C strands. The latter would only occur if unlabeled A is coassembled with labeled B, or vice versa, as has been reported recently for mixtures of 12C/13C-labeled amyloid-β (27). Collectively, these experimental results validate the computational observations that A and B coassemble into two-component β-sheet structures rather than self-sort.
CATCH(+) and CATCH(−) Peptides Resist Self-Assembly.
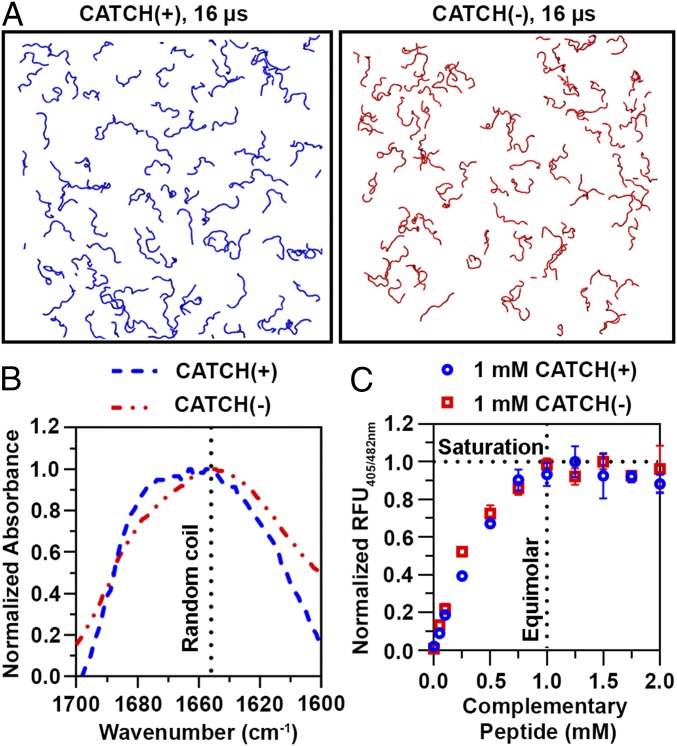
DMD/PRIME20 simulations of single-component systems of CATCH(+) or of CATCH(−) peptides show little evidence of aggregation. Representative snapshots depict DMD/PRIME20 simulations of 96 CATCH(+) and of 96 CATCH(−) peptides at 16 μs (Fig. 2A). The peptides are in random-coil configurations at this time point. CATCH(−) peptides remain dispersed throughout the entire simulation run, whereas some instances of unstable interactions are observed in snapshots of CATCH(+) peptide simulation runs (SI Appendix, Fig. S3). These computational observations are consistent with prior experimental data demonstrating that individual CATCH peptides resist self-assembly, albeit at much lower concentrations than those used in the models reported herein (9).
Fig. 2.
Complementary interactions are necessary for assembly. (A) Snapshots of DMD/PRIME20 simulations of a system containing 96 CATCH(+) peptides (blue) and a system containing 96 CATCH(−) peptides (red) at 16 μs. (B) FTIR spectra of 10 mM CATCH(+) and CATCH(−). (C) ThT fluorescence measurements at varying CATCH(+):CATCH(−) ratios indicating saturation at equimolar mixtures.
Biophysical measurements demonstrate that CATCH(+) and CATCH(−) do not self-associate in solution at concentrations comparable to those used in simulations. FTIR spectra of solutions containing only CATCH(+) or CATCH(−) have broad peaks at ∼1,645 cm−1 that are consistent with an unstructured and predominantly random-coil state (Fig. 2B). Likewise, solutions containing only CATCH(+) or CATCH(−) did not increase the fluorescence emission of ThT, a dye that emits increased fluorescence when bound to β-sheet structures (SI Appendix, Fig. S4A). In contrast, adding increasing amounts of CATCH(−) to a 1 mM CATCH(+) solution increased ThT fluorescence emission until the molar ratio of CATCH(+) to CATCH(−) approached unity (Fig. 2C). CATCH(+) present in a molar excess relative to CATCH(−) produced no further increase in ThT fluorescence, suggesting that coassembly of CATCH peptides proceeds until molar equivalency is reached, at which point excess peptide remains unassembled (Fig. 2C). Similar behavior was observed when the concentration of CATCH(−) was held constant and the concentration of CATCH(+) was varied. Collectively, computational models and experimental data demonstrate that CATCH(+) and CATCH(−) only assemble into β-sheets when the complementary peptide is present, and at a near-stoichiometric ratio.
Computational and Experimental Observation of Nonfibrillar Oligomer Formation.
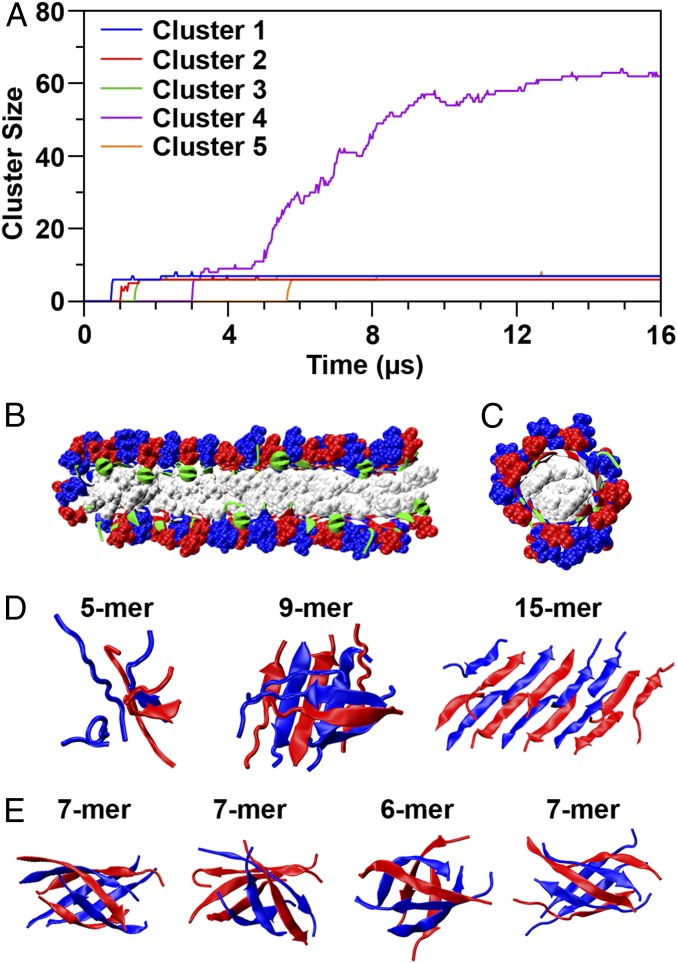
Unexpectedly, DMD/PRIME20 simulations predict that CATCH(+) and CATCH(−) coassemble into two types of stable aggregates: amyloid fibrils and β-barrel oligomers. The roles played by these two types of aggregates in the assembly process are best introduced by referring to Fig. 3A, which shows the growth profiles of the five aggregates that remain at the end of the simulation depicted in Fig. 1A. The biggest aggregate in the simulation contains >60 peptides and has an amyloid structure with two layers of β-sheets as shown in Fig. 3B. Peptides form hydrogen bonds with adjacent peptides in neighboring strands; the charged side chains point outward toward solvent and the hydrophobic side chains point inward between the sheets. The ABABAB pattern is apparent. The other four aggregates form β-barrels having six or seven peptides. These structures are quite stable; none breaks or joins the amyloid structures. A detailed analysis reveals that they are out-of-register β-strands rolled up into a cylinder, with hydrogen bonds between adjacent peptides, charged side chains pointing outward, and hydrophobic side chains pointing inward as shown in Fig. 3C. As will be discussed later, self-assembled versions of these types of structures have been observed experimentally in the literature and are hypothesized to be the toxic oligomers responsible for the cell damage in Alzheimer’s and related amyloid diseases. Fig. 3D shows snapshots of the structures formed along the pathway from a disordered 5-mer to a 20-mer β-sheet–rich double-layer amyloid. The latter eventually grows into the biggest cluster seen in that run and is likely the precursor of the amyloid observed in our biophysical experiments on mixtures of CATCH(+) and CATCH(−). Fig. 3E shows snapshots of the β-barrels. These contain both CATCH(+) and CATCH(−), are rich in β-strand, and remain stable and fixed at their initial size over the course of the simulation. As described in SI Appendix, the oligomers along the amyloid folding path are less structurally ordered than the β-barrels, allowing space for insertion of additional peptides. Also described is an interesting asymmetry in the ability of β-barrels containing even and odd numbers of strands to form mismatches (AA or BB neighbors), and hence to grow by adding strands.
Fig. 3.
Aggregation pathways and polymorphism for the coassembling system. (A) Number of peptide chains in cluster vs. simulation time for the five clusters. (B) Amyloid structure [blue: CATCH(+) side chain, red: CATCH(−) side chain, white: Phe side chain, green: backbone]. (C) Structure of one of the β-barrel oligomers represented as in B. (D) Snapshots of oligomers during the early stage of the amyloid formation leading to cluster 4 (5-mer: Nhb = 4.4, Nhp = 3.0; 9-mer: Nhb = 6.7, Nhp = 4.7; 15-mer: Nhb = 6.9, Nhp = 3.9). (E) Snapshots of four β-barrel oligomers (Left: Nhb = 7.3, Nhp = 5.6; Middle Left: Nhb = 7.7, Nhp = 5.6; Middle Right: Nhb = 7.5, Nhp = 5.4; Right: Nhb = 8.0, Nhp = 5.5).
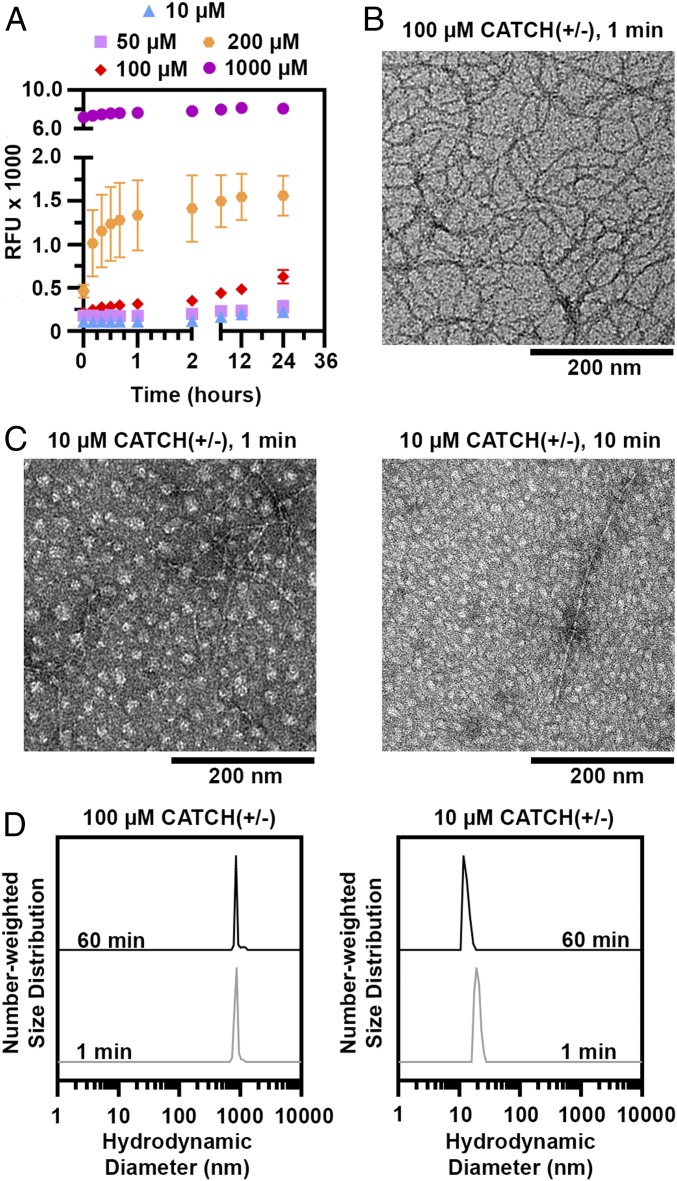
A combination of ThT fluorescence time-course evolution, TEM, and DLS suggests that CATCH(+) and CATCH(−) coassemble into elongated fibers and nonfibrillar oligomers in a concentration-dependent manner. In equimolar mixtures of CATCH(+/−) at 100 to 1,000 µM, ThT fluorescence increased rapidly to a plateau within a few hours with no apparent lag phase (Fig. 4A), whereas the signal from either peptide alone remained unchanged (SI Appendix, Fig. S4B), consistent with a prior report (9). No ThT fluorescence change was observed in equimolar mixtures of CATCH(+/−) at either 10 or 50 μM (Fig. 4A). These observations suggested that CATCH peptides may not coassemble into nanofibers below a critical concentration at or near 100 μM. Consistent with this, a previous report showed a transition from random-coil to β-sheet structures in equimolar mixtures of CATCH(+/−) over the range of 50 to 320 μM (9).
Fig. 4.
Concentration-dependent formation of nonfibrillar CATCH(+/−) oligomers. (A) ThT kinetic curves of CATCH(+/−) at 10 to 1,000 µM. (B) Transmission electron micrographs of 100 µM CATCH(+/−) at 1 min. (C) Transmission electron micrographs of 10 µM CATCH(+/−) at 1 min (Left) and 60 min (Right). (Scale bars, 200 nm.) (D) Number-weighted DLS of equimolar mixtures of CATCH(+/−) at 100 and 10 µM at 1 and 60 min.
TEM identified formation of two different types of structures in equimolar mixtures of CATCH(+/−). Elongated nanofibers were prevalent in samples of 100 μM CATCH(+/−) maintained for 1 min (Fig. 4B and SI Appendix, Fig. S5A), consistent with a prior report (9). In contrast, a combination of round, ∼20-nm particles, as well as elongated nanofibers, was observed in 50 and 10 μM CATCH(+/−) samples at 1 to 60 min (Fig. 4C and SI Appendix, Fig. S5 B–F). Particles with similar appearance have been reported in samples of amyloid-β and α-synuclein previously and are attributed to nonfibrillar oligomers (28–30). No oligomers or nanofibers were seen in samples of CATCH(+) or CATCH(−) alone (SI Appendix, Fig. S6), consistent with simulations and biophysical measurements demonstrating that these peptides resist self-assembly. In an effort to trap nonfibrillar oligomers in a metastable state that would allow biophysical characterization, we viewed coassembled CATCH(+/−) samples and controls at 4 °C with TEM as described in SI Appendix, Figs. S7–S9. Although lowering the temperature did slow the coassembly process, as exemplified by our observation of a combination of particles and nanofibers in 100 μM CATCH(+/−) samples, nonfibrillar CATCH oligomers did not persist under any conditions, likely because of their rapid coassembly kinetics and low coassembly concentration threshold. We also considered the possibility that TEM sample drying could account for variability in the observed structures and performed DLS analyses at various concentrations (Fig. 4D) as described in SI Appendix, Fig. S10. These observations suggested that the ∼20-nm oligomers observed in TEM images of 10 μM CATCH(+/−) samples (Fig. 4C) may be stable in aqueous conditions, whereas those in images of 50 μM mixtures may be of relatively low abundance. Further characterization of oligomer size, structure, and strand content, for example with size-exclusion chromatography or biophysical measurements, was precluded by the low concentration at which CATCH(+/−) forms stable oligomers (∼10 μM), as this falls below the detection limit of many analytical devices.
Taken together, the TEM and DLS measurements suggest concentration-dependent coassembly of CATCH(+) and CATCH(−) into nonfibrillar oligomers. However, it remains unclear if oligomer formation is a productive step along the pathway to elongated fiber assembly or if it is an off-pathway event. Further, it remains to be determined if the observed oligomers encompass the coassembled β-barrel structures predicted by simulation or some other nonfibrillar architecture.
CATCH Nanofibers Contain Detectable AA and BB Nearest Neighbors.
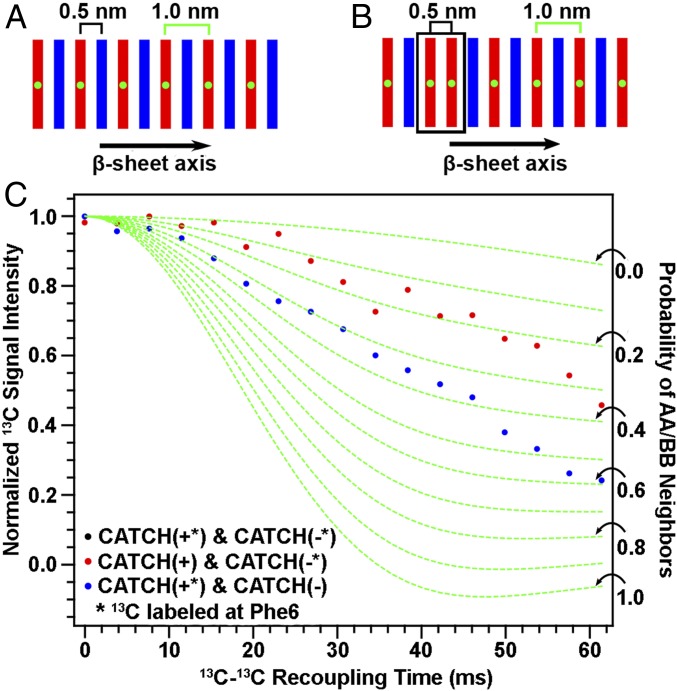
Simulations and experimental measurements identified a significant number of AA and BB neighbors within CATCH β-sheet assemblies; this behavior was unexpected based on the charge state of the peptides in neutral aqueous conditions. In the final configuration of the 10 DMD simulations, the percentage of AA [i.e., CATCH(+)] neighbors is 12.5 ± 6.9%, while that of BB [i.e., CATCH(−)] neighbors is 5.1 ± 4.3%. We probed for the presence of AA and BB neighbors using PITHIRDS-CT NMR experiments on nanofiber samples produced with 13C labeling of peptide A or peptide B, but not both. As illustrated in Fig. 5A, β-sheets with ideal alternation of A and B β-strands would correspond to a 1.0-nm 13C–13C nearest-neighbor distance if only the central atom of one peptide were 13C-labeled. Fig. 5B shows that the presence of some like-peptide nearest neighbors results in a fraction of 13C atoms having 0.5-nm nearest-neighbor distances. Centrifuge pellets of “isotopically diluted” equimolar mixtures of 13C-labeled A with unlabeled B (Fig. 5C, red data points) or 13C-labeled B with unlabeled A (blue data points) yielded reduced decays of 13C signal intensity in PITHIRDS-CT NMR measurements when compared to the decay observed for the “isotopically pure” mixture containing both labeled A and labeled B (Fig. 1D, black data points). Reduction of PITHIRDS-CT decay with this kind of isotopic dilution indicates that A and B are coassembled into the same β-sheets. As explained next, however, the degree of observed reduction in the decays with isotopic dilution is less than what would be anticipated with ideal AB alternation in the β-sheet structure.
Fig. 5.
Evaluating the propensity for CATCH(+) and CATCH(−) to self-associate. (A and B) Schematics of possible peptide organization within the nanofiber; green dots represent 13C labeling. (C) Isotopic dilution PITHIRDS-CT measurements of coassembled CATCH(+/−) nanofibers where only one peptide is 13C-labeled at a time. The asterisks in the plot legend indicate which peptide was isotopically labeled with 13C at the central atom. The dashed green curves correspond to simulations that account for the probabilities of like-neighbors for the 13C-labeled peptide as indicated by black arrows along the right vertical axis. Details describing these simulations can be found in SI Appendix.
To understand the effects of isotopic dilution in the presence of deviation from ideal AB alternation, we performed Monte Carlo simulations to generate the many possible arrangements of β-sheets that can form by coassembling two complementary peptides and combined this analysis with an analysis of the 13C–13C dipolar coupling during PITHIRDS-CT NMR experiments on the various arrangements. These simulations, described in detail in SI Appendix and illustrated in SI Appendix, Fig. S11, predict the relationship between the arrangements of A and B β-strands and the probabilities of having like-peptide (AA or BB) nearest neighbors within each β-sheet. The results of this analysis are the green dashed curves in Fig. 5C, which are PITHIRDS-CT decays predicted for the probabilities of like neighbors indicated by black arrows of the 13C-labeled peptide (AA or BB) for experiments in which only one peptide is 13C-labeled. The experimental data lie in between the predicted curves corresponding to the two extreme cases. The weakest predicted decay in Fig. 5C, corresponding to a 0 probability for AA or BB nearest neighbors, is the prediction for ideal alternation of A and B peptides within each β-sheet (Fig. 5A). The strongest predicted PITHIRDS-CT decay in Fig. 5C, corresponding to a 1.0 probability for AA/BB neighbors, is the prediction for the case in which the isotopically labeled peptide self-assembles into β-sheets that contain no unlabeled peptide. Intermediate predicted curves correspond to the illustration in Fig. 4B, in which some AA/BB neighbors occur in coassembled β-sheets. As illustrated in SI Appendix, Fig. S12 A and B, shapes of PITHIRDS-CT decays are sensitive to small changes in relative positions of 13C atoms within reasonable models of β-sheet structure, but this sensitivity is low for 13C–13C recoupling times below 30 ms. At short 13C–13C recoupling times, we expect decays to be more sensitive to nearest-neighbor 13C–13C interactions and less affected by longer-range interactions. SI Appendix, Fig. S12C shows that PITHIRDS-CT curves on isotopically diluted samples exhibit linear dependence on like-peptide nearest-neighbor probability (AA or BB) for 13C–13C recoupling times below 30 ms. Based on comparison of measured and simulated PITHIRDS-CT decays at recoupling times less than 30 ms, we estimate that the percentage of like-peptide nearest-neighbor pairs is between 9.4% and 32.8% in the coassembled nanofiber. Although there is significant sample-to-sample variation (SI Appendix, Fig. S13), like-peptide neighbors of CATCH(+) appear more likely than like-peptide neighbors of CATCH(−), consistent with the simulation predictions. This observation is also consistent with the smaller net charge of +4 for CATCH(+) in comparison to −6 for CATCH(−).
There are several possible explanations for the apparent discrepancy in the estimates of the numbers of AA and BB neighbors from PITHIRDS-CT experiments and simulations. First, the number of peptides assessed in simulations is orders of magnitude smaller than those present in samples analyzed using NMR; thus, the probability will change significantly if the number of like neighbors changes by only 1 or 2 in simulations (SI Appendix, Figs. S14 and S15). Second, the apparent discrepancies could be due to inaccuracies in the PRIME20 potential energy function. Third, variability in the PITHIRDS-CT measurements could result from errors introduced during sample preparation. Uncertainties associated with measuring peptide concentration could cause sample-to-sample variation in measured 13C signal decays as shown in SI Appendix, Fig. S13. Lyophilization of CATCH(+/−) samples for ssNMR analysis may induce residual unassembled peptide to self-associate into β-sheet structures upon dehydration as described in SI Appendix and demonstrated in SI Appendix, Fig. S16. Fourth, our efforts to correct for naturally abundant 13C, which accounts for ∼1% of the carbon in the sample, may be inadequate. This is especially complicating at longer recoupling times where natural abundance plays a larger role as signal decays. Nonetheless, despite reasonable differences in the probabilities estimated from simulations and experiments, these observations collectively demonstrate a nonnegligible frequency of like neighbors in CATCH β-sheet coassemblies.
Discussion
This work reports on the molecular-level coassembly of CATCH(+) and CATCH(−) peptides into two-component amyloid-like β-sheet nanofibers. When kept alone in aqueous solution, CATCH(+) and CATCH(−) largely resist self-assembly, although CATCH(+) demonstrates propensity for weak, transient association in DMD simulations, possibly due to its lower net charge compared to CATCH(−). When combined in aqueous solution, the two peptides coorganize into a bilayer of β-sheets, each having a predominantly alternating -ABABAB- type β-strand pattern. Bilayer formation is a result of hydrophobic collapse; the sheets have opposing hydrophilic and hydrophobic faces conferred by the alternating sequence of hydrophilic and hydrophobic amino acids in CATCH(+) and CATCH(−), and these hydrophobic faces are packed into the core of the bilayer. This general architecture is not surprising, as it is consistent with the structure hypothesized for the self-assembling zwitterionic Q11 peptide from which CATCH(+) and CATCH(−) were designed (31), as well as the closely related P11 variant (32). Taken together, these observations support the general hypothesis that electrostatic attraction and repulsion can encode molecular-level organization of β-strands within coassembled β-sheets.
The resultant assemblies are not, however, perfectly alternating. Rather, some CATCH(−):CATCH(−) and CATCH(+):CATCH(+) neighbors were found in DMD simulations and observed in ssNMR measurements. Not surprisingly, the tendency for CATCH(+):CATCH(+) mismatches appears greater than that for CATCH(−):CATCH(−) mismatches, presumably due to the lower net charge of the former. This tendency for like-charge mismatches in CATCH nanofibers may be due in part to charge shielding by counterions present in buffered aqueous solutions. Here, instances may exist in which favorable hydrophobic interactions between the Phe residues on a free peptide and those in the core of a CATCH bilayer can overcome weakened coulombic repulsion associated with peptide–ion complexation, thereby leading to like-charge peptide pairing within the growing amyloid. Similar results have been observed in ferricytochrome c fibrillization, where hydrophobic interactions can outweigh electrostatic repulsion in alkaline conditions even in the absence of counterions (33), as well as thermally induced assembly of the cationic MAX3 peptide (34). Thus, although electrostatic attraction and repulsion can encode peptide organization in general, our results suggest that achieving exquisite molecular-level precision in coassembled amyloid-like structures will require more sophisticated designs that incorporate other types of specific interactions between complementary β-strands. Such precision may ultimately be important in coassembling peptide systems including use as biomaterials for medical or biotechnological applications because like-charged neighbor “defects” could act as fracture points or facilitate fiber remodeling via strand swapping.
An unexpected computational observation of the coassembly process was the formation of off-pathway “β-barrels” consisting of both CATCH(+) and CATCH(−). Although the β-barrel motif is a well-known fold for single proteins, (e.g., membrane proteins, porins, and fluorescent proteins), β-barrels formed by six or seven small peptides have only recently come to the attention of the scientific community. This attention was initiated by the Eisenberg group’s 2012 X-ray crystallography-based finding that the atomic structure of “cylindrin,” an 11-residue fragment from the protein αB crystalline, is a β-barrel. They further suggested that β-barrel oligomers could serve as models for the toxic amyloid oligomers that are widely believed to be the cause of cell damage in amyloid diseases such as Alzheimer’s, prion disease, and type 2 diabetes (19). Since then more evidence has come to light that shows that fragments of prion protein, Aβ, and β2 microglobulin, as well as full-length Aβ, form β-barrel oligomers (20–22). Our computationally based observation of β-barrel oligomers for CATCH(+) and CATCH(−) mixtures suggests these structures occur during the coassembly of two different small peptides. In our view this lends more credence to the idea that β-barrels are a common oligomeric structure in the amyloid landscape.
It is difficult to differentiate amyloid and β-barrel structures experimentally because they assemble via similar intermolecular interactions that cannot be readily distinguished using FTIR, NMR, or circular dichroism. Concentration-dependent formation of nonfibrillar oligomers was observed by TEM and DLS in equimolar mixtures of CATCH(+) and CATCH(−). However, deeper characterization of these oligomers was hindered by the rapid coassembly kinetics with no appreciable lag time, the low concentration threshold for elongated fiber predominance, and the inability to identify experimental conditions that kinetically trap nonfibrillar oligomers in a metastable state. Future efforts will seek to close the remaining gap between computational models and experimental methods to determine if nonfibrillar CATCH(+/−) oligomers consist of the β-barrels predicted by simulations, as well as if the oligomers are on- or off-pathway.
A detailed analysis of the aggregated structures during DMD simulations revealed that the number of hydrogen bonds and hydrophobic associations may determine whether CATCH peptides coassemble into either amyloid-like or β-barrel structures. Our observations demonstrate that the on-pathway oligomers have fewer hydrogen bonds and hydrophobic associations than β-barrel structures, indicating that preamyloid oligomers are less ordered and less stable. Based on these observations, we speculate that the preamyloid oligomers must be “imperfect” so that they can accommodate insertion of more peptides and eventually grow into amyloids. In other words, in order for the amyloid to grow it needs to leave some hydrogen bond acceptors/donors or hydrophobic motifs free to entice other peptides to join the aggregates. This mechanism may be operative not only for coassembling peptides but also for those that self-associate.
Our experimental observations suggest that hydration is an important determinant of CATCH peptide self-association vs. coassembly propensity. CATCH(+) and CATCH(−) adopted random-coil configurations when kept alone in aqueous conditions, yet each peptide aggregated into β-sheet–rich structures when dehydrated (SI Appendix, Fig. S16). Consistent with this result, a previous report demonstrated that dehydration of amyloid-forming peptides can increase their fibrillization kinetics (35). Likewise, studies based on computational models have identified peptide dehydration as a key event in Aβ aggregation and self-assembly (36, 37). One plausible explanation for the role of water in preventing erroneous, “off-pathway” (AA) or (BB) self-association is rooted in observations that hydrophilic molecules that tightly bind water molecules experience a repulsive steric-hydration force that leads them to repel each other at small separations (<1 nm) (38). Here, we postulate that it is the combination of long-range electrostatic repulsion plus the energy needed to dehydrate two CATCH(+) or two CATCH(−) molecules as they approach each other that limits (AA) or (BB) interactions in aqueous conditions. When CATCH(+) and CATCH(−) are combined in solution, though, coulombic attraction between oppositely charged E and K residues coupled with hydrophobic collapse involving Phe residues is sufficient to overcome the dehydration energy barrier. Ultimately, coulombic interactions favor formation of β-sheets with an alternating (AB) strand arrangement over (AA) or (BB) pairings.
Conclusions.
Coassembly of charge-complementary peptide pairs into amyloid-like β-sheet nanofibers is an emerging area of biophysics that is gaining increasing interest as the basis for fabricating new nanomaterials for medical and biotechnological applications. It is often suggested that charge-complementary peptides precisely coassemble into β-sheets with an alternating -ABABAB- strand pattern based on intuition and inferences drawn from biophysical measurements, yet such molecular-level order has not previously been validated. Here, we demonstrate that a combination of computational modeling and biophysical measurement methodologies can close this gap. In particular, we observed that an alternating strand pattern does indeed predominate upon coassembly of CATCH(+) and CATCH(−), yet some CATCH(+):CATCH(+) and CATCH(−):CATCH(−) neighbors do occur. Thus, charge complementarity alone is insufficient to encode precise β-strand order in two-component amyloid-like nanofibers. Computational models predict that in some instances CATCH(+) and CATCH(−) can coassemble into oligomeric β-barrel structures that are not discernible with conventional biophysical techniques. TEM and DLS also demonstrate that CATCH(+/−) peptides at low concentrations (∼10 µM) form nonfibrillar oligomers ∼20 nm in diameter. Collectively, these examples demonstrate the power of our computational–experimental framework to provide previously inaccessible views of the process of peptide coassembly from initiation to equilibrium. Such insights are expected to yield advances in our understanding of the molecular-level interactions that drive peptide coassembly, which in turn will lead to guiding principles for a priori design of new peptide pairs demonstrating exquisite molecular-level organization. We envision that achieving fine control of coassembled β-strand structure will afford unprecedented opportunities to design new nanomaterials with precisely defined organization of integrated functional biomolecule components, such as cell-binding peptides, enzymes, or antigens. As a result, we may ultimately be able to realize supramolecular designs or patterns that are not possible with conventional self-assembling systems, thereby greatly expanding the range of functional nanomaterials available to medicine and biotechnology.
Materials and Methods
The materials and methods are described in detail in SI Appendix. DMD with the PRIME20 force field was used to simulate CATCH(+/−) mixtures and their pure component analogs as described in detail in SI Appendix. The 13C-labeled CATCH(+) and 13C-labeled CATCH(−) were synthesized using standard solid-phase methods, their mass was determined using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry, and their purity was characterized using high-performance liquid chromatography. CATCH(+/−) samples were prepared by mixing aqueous stock solutions of each peptide at different volume ratios to give the final working peptide concentration. CATCH(+/−) samples, as well as samples of CATCH(+) and CATCH(−) alone, were viewed using TEM and analyzed using FTIR, ThT fluorimetry, and DLS according to methods described in SI Appendix. Centrifuged and freeze-dried CATCH(+/−) nanofiber samples were packed into 3.2-mm ssNMR rotors. The 1H-13C cross-polarization magic-angle spinning (CPMAS) and 13C-13C PITHIRDS-CT measurements were performed on an 11.75-T Bruker Avance III spectrometer with a Bruker MAS probe as described in detail in SI Appendix. Simulation of NMR experiments was conducted using SpinEvolution software and simulated and experimental NMR data were analyzed using in-house software built in Wolfram Mathematica as detailed in SI Appendix.
Data Availability.
The data reported in this paper have been deposited in the Georgia Institute of Technology SMARTech Repository (https://smartech.gatech.edu/handle/1853/62416).
Supplementary Material
Acknowledgments
This research was supported by National Science Foundation Division of Chemical, Bioengineering, Environmental and Transport Systems Grant 1743432.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Georgia Institute of Technology SMARTech Repository (https://smartech.gatech.edu/handle/1853/62416).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1912810117/-/DCSupplemental.
References
- 1.Makam P., Gazit E., Minimalistic peptide supramolecular co-assembly: Expanding the conformational space for nanotechnology. Chem. Soc. Rev. 47, 3406–3420 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Raymond D. M., Nilsson B. L., Multicomponent peptide assemblies. Chem. Soc. Rev. 47, 3659–3720 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hu Y., et al. , Electrostatic-driven lamination and untwisting of β-sheet assemblies. ACS Nano 10, 880–888 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Swanekamp R. J., DiMaio J. T., Bowerman C. J., Nilsson B. L., Coassembly of enantiomeric amphipathic peptides into amyloid-inspired rippled β-sheet fibrils. J. Am. Chem. Soc. 134, 5556–5559 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li S., et al. , Design of asymmetric peptide bilayer membranes. J. Am. Chem. Soc. 138, 3579–3586 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Pandya M. J., et al. , Sticky-end assembly of a designed peptide fiber provides insight into protein fibrillogenesis. Biochemistry 39, 8728–8734 (2000). [DOI] [PubMed] [Google Scholar]
- 7.Papapostolou D., et al. , Engineering nanoscale order into a designed protein fiber. Proc. Natl. Acad. Sci. U.S.A. 104, 10853–10858 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sharp T. H., et al. , Cryo-transmission electron microscopy structure of a gigadalton peptide fiber of de novo design. Proc. Natl. Acad. Sci. U.S.A. 109, 13266–13271 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seroski D. T., et al. , Co-assembly tags based on charge complementarity (CATCH) for installing functional protein ligands into supramolecular biomaterials. Cell. Mol. Bioeng. 9, 335–350 (2016). [Google Scholar]
- 10.King P. J., et al. , A modular self-assembly approach to functionalised β-sheet peptide hydrogel biomaterials. Soft Matter 12, 1915–1923 (2016). [DOI] [PubMed] [Google Scholar]
- 11.Kyle S., Felton S. H., McPherson M. J., Aggeli A., Ingham E., Rational molecular design of complementary self-assembling peptide hydrogels. Adv. Healthc. Mater. 1, 640–645 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Candreva J., Chau E., Aoraha E., Nanda V., Kim J. R., Hetero-assembly of a dual β-amyloid variant peptide system. Chem. Commun. (Camb.) 54, 6380–6383 (2018). [DOI] [PubMed] [Google Scholar]
- 13.Hudalla G. A., et al. , Gradated assembly of multiple proteins into supramolecular nanomaterials. Nat. Mater. 13, 829–836 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu R., Hudalla G. A., Using self-assembling peptides to integrate biomolecules into functional supramolecular biomaterials. Molecules 24, E1450 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheon M., Chang I., Hall C. K., Spontaneous formation of twisted Aβ(16-22) fibrils in large-scale molecular-dynamics simulations. Biophys. J. 101, 2493–2501 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cheon M., Chang I., Hall C. K., Extending the PRIME model for protein aggregation to all 20 amino acids. Proteins 78, 2950–2960 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Y., et al. , Thermodynamic phase diagram of amyloid-β (16-22) peptide. Proc. Natl. Acad. Sci. U.S.A. 116, 2091–2096 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Y., Shao Q., Hall C. K., N-terminal prion protein peptides (PrP(120-144)) form parallel in-register β-sheets via multiple nucleation-dependent pathways. J. Biol. Chem. 291, 22093–22105 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laganowsky A., et al. , Atomic view of a toxic amyloid small oligomer. Science 335, 1228–1231 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun Y., Ge X., Xing Y., Wang B., Ding F., β-barrel oligomers as common intermediates of peptides self-assembling into cross-β aggregates. Sci. Rep. 8, 10353 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berhanu W. M., Hansmann U. H. E., The stability of cylindrin β-barrel amyloid oligomer models-a molecular dynamics study. Proteins 81, 1542–1555 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Do T. D., et al. , Amyloid β-protein C-terminal fragments: Formation of cylindrins and β-barrels. J. Am. Chem. Soc. 138, 549–557 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wishart D. S., Sykes B. D., The 13C chemical-shift index: A simple method for the identification of protein secondary structure using 13C chemical-shift data. J. Biomol. NMR 4, 171–180 (1994). [DOI] [PubMed] [Google Scholar]
- 24.Wishart D. S., Interpreting protein chemical shift data. Prog. Nucl. Magn. Reson. Spectrosc. 58, 62–87 (2011). [DOI] [PubMed] [Google Scholar]
- 25.Tycko R., Symmetry-based constant-time homonuclear dipolar recoupling in solid state NMR. J. Chem. Phys. 126, 064506 (2007). [DOI] [PubMed] [Google Scholar]
- 26.Ulrich E. L., et al. , BioMagResBank. Nucleic Acids Res. 36, D402–D408 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baronio C. M., Baldassarre M., Barth A., Insight into the internal structure of amyloid-β oligomers by isotope-edited Fourier transform infrared spectroscopy. Phys. Chem. Chem. Phys. 21, 8587–8597 (2019). [DOI] [PubMed] [Google Scholar]
- 28.Ahmed M., et al. , Structural conversion of neurotoxic amyloid-β(1-42) oligomers to fibrils. Nat. Struct. Mol. Biol. 17, 561–567 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen S. W., et al. , Structural characterization of toxic oligomers that are kinetically trapped during α-synuclein fibril formation. Proc. Natl. Acad. Sci. U.S.A. 112, E1994–E2003 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bieschke J., et al. , Small-molecule conversion of toxic oligomers to nontoxic β-sheet-rich amyloid fibrils. Nat. Chem. Biol. 8, 93–101 (2011). [DOI] [PubMed] [Google Scholar]
- 31.Collier J. H., Messersmith P. B., Enzymatic modification of self-assembled peptide structures with tissue transglutaminase. Bioconjug. Chem. 14, 748–755 (2003). [DOI] [PubMed] [Google Scholar]
- 32.Aggeli A., et al. , Hierarchical self-assembly of chiral rod-like molecules as a model for peptide beta-sheet tapes, ribbons, fibrils, and fibers. Proc. Natl. Acad. Sci. U.S.A. 98, 11857–11862 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ramakrishna D., Prasad M. D., Bhuyan A. K., Hydrophobic collapse overrides Coulombic repulsion in ferricytochrome c fibrillation under extremely alkaline condition. Arch. Biochem. Biophys. 528, 67–71 (2012). [DOI] [PubMed] [Google Scholar]
- 34.Pochan D. J., et al. , Thermally reversible hydrogels via intramolecular folding and consequent self-assembly of a de novo designed peptide. J. Am. Chem. Soc. 125, 11802–11803 (2003). [DOI] [PubMed] [Google Scholar]
- 35.Mukherjee S., Chowdhury P., Gai F., Effect of dehydration on the aggregation kinetics of two amyloid peptides. J. Phys. Chem. B 113, 531–535 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krone M. G., et al. , Role of water in mediating the assembly of Alzheimer amyloid-beta Abeta16-22 protofilaments. J. Am. Chem. Soc. 130, 11066–11072 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tarus B., Straub J. E., Thirumalai D., Probing the initial stage of aggregation of the Abeta(10-35)-protein: Assessing the propensity for peptide dimerization. J. Mol. Biol. 345, 1141–1156 (2005). [DOI] [PubMed] [Google Scholar]
- 38.Israelachvili J. N., Intermolecular and Surface Forces (Academic Press, Burlington, MA, ed. 3, 2011), p. 674. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data reported in this paper have been deposited in the Georgia Institute of Technology SMARTech Repository (https://smartech.gatech.edu/handle/1853/62416).